DLPFC
Last updated: 2024-07-14
Checks: 7 0
Knit directory: KODAMA-Analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240618) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0051854. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Untracked files:
Untracked: .h5seurat
Untracked: analysis/Single-cell.Rmd
Untracked: code/VisiumHD_CRC_download.sh
Untracked: data/Pathology.csv
Unstaged changes:
Deleted: _site.yml
Deleted: analysis/DLPFC.Rmd
Deleted: analysis/about.Rmd
Deleted: data/Moffitt_and_Bambah-Mukku_et_al_merfish_all_cells.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/DLPFC-4.Rmd) and HTML
(docs/DLPFC-4.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0051854 | Stefano Cacciatore | 2024-07-14 | Start my new project |
We will analyze all 4 slides
library("nnSVG")
library("scater")
library("scran")
library("scry")
library("SPARK")
library("harmony")
library("Seurat")
library("spatialLIBD")
library("KODAMA")
library("KODAMAextra")
sample_names=c("151507",
"151508",
"151509",
"151510",
"151669",
"151670",
"151671",
"151672",
"151673",
"151674",
"151675",
"151676")
subject_names= c("Br5292","Br5595", "Br8100")
#spe <- fetch_data(type = 'spe',destdir="../Temporary")
load("../DLFPC.RData")
metaData = SingleCellExperiment::colData(spe)
expr = SingleCellExperiment::counts(spe)
sample_names <- paste0("sample_", unique(colData(spe)$sample_id))
sample_names <- unique(colData(spe)$sample_id)
dim(spe)[1] 33538 47681identify mitochondrial genes
is_mito <- grepl("(^MT-)|(^mt-)", rowData(spe)$gene_name)
table(is_mito)is_mito
FALSE TRUE
33525 13 calculate per-spot QC metrics and select the QC threshoulds
spe <- addPerCellQC(spe, subsets = list(mito = is_mito))
# select QC thresholds
qc_lib_size <- colData(spe)$sum < 500
qc_detected <- colData(spe)$detected < 250
qc_mito <- colData(spe)$subsets_mito_percent > 30
qc_cell_count <- colData(spe)$cell_count > 12Spot to discard and filter low-quality
discard <- qc_lib_size | qc_detected | qc_mito | qc_cell_count
table(discard)discard
FALSE TRUE
46653 1028 colData(spe)$discard <- discard
spe <- spe[, !colData(spe)$discard]
dim(spe)[1] 33538 46653Horizontalization
# readjust xy
xy=spatialCoords(spe)
samples=unique(colData(spe)$sample_id)
for(j in 1:length(samples)){
sel=samples[j]==colData(spe)$sample_id
xy[sel,1]=spatialCoords(spe)[sel,1]+12000*(j-1)
}
spatialCoords(spe)=xyGene filtering
spe <- filter_genes(
spe,
filter_genes_ncounts = 2, #ncounts
filter_genes_pcspots = 0.5,
filter_mito = TRUE
)
dim(spe)[1] 6623 46653REmove the spots that have not been manually assigned
sel= !is.na(colData(spe)$layer_guess_reordered)
spe = spe[,sel]
dim(spe)[1] 6623 46318normalization
spe <- computeLibraryFactors(spe)
spe <- logNormCounts(spe)Identification of the gene spatially relevvant
gene_i=NULL
pvalue_i=NULL
pvalue_mat=matrix(nrow=nrow(spe),ncol=length(sample_names))
for(i in 1:length(sample_names)){
sel=colData(spe)$sample_id==sample_names[i]
spe_sub= spe[,sel]
sparkX <- sparkx(logcounts(spe_sub),spatialCoords(spe_sub),numCores=1,option="mixture")
gene_i=c(gene_i,rowData(spe)$gene_id)
pvalue_i=c(pvalue_i,sparkX$res_mtest$combinedPval)
pvalue_mat[,i]=sparkX$res_mtest$combinedPval
print(sample_names[i])
}## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 4170
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5
[1] "151507"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 4285
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151508"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 4708
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5
[1] "151509"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 4571
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151510"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3587
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151669"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3274
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151670"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 4013
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5
[1] "151671"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3772
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151672"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3568
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5
[1] "151673"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3576
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5
[1] "151674"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3468
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151675"
## ===== SPARK-X INPUT INFORMATION ====
## number of total samples: 3326
## number of total genes: 6623
## Running with single core, may take some time
## Testing With Projection Kernel
## Testing With Gaussian Kernel 1
## Testing With Gaussian Kernel 2
## Testing With Gaussian Kernel 3
## Testing With Gaussian Kernel 4
## Testing With Gaussian Kernel 5
## Testing With Cosine Kernel 1
## Testing With Cosine Kernel 2
## Testing With Cosine Kernel 3
## Testing With Cosine Kernel 4
## Testing With Cosine Kernel 5Warning in FUN(newX[, i], ...): There are p-values that are exactly 1![1] "151676"oo=order(pvalue_i)
top_genes=gene_i[oo]
n=ave(1:length(top_genes), top_genes, FUN = seq_along)
top_genes=top_genes[n==1]
oo=order(apply(pvalue_mat,1,function(x) median(-log(x))),decreasing = TRUE)
top=gene_i[oo]Selection of the patient Br5595
subject_names=="Br5595"[1] FALSE TRUE FALSE spe_sub <- spe[, colData(spe)$subject == subject_names]Warning in colData(spe)$subject == subject_names: longer object length is not a
multiple of shorter object length subjects=colData(spe_sub)$subject
dim(spe_sub)[1] 6623 15440 spe_sub <- runPCA(spe_sub, 50,subset_row = top[1:2000], scale=TRUE)
pca=reducedDim(spe_sub,type = "PCA")[,1:50]
labels=as.factor(colData(spe_sub)$layer_guess_reordered)
xy=as.matrix(spatialCoords(spe_sub))
samples=colData(spe_sub)$sample_id
data=t(logcounts(spe_sub)[top[1:2000],])
plot(pca, col=as.factor(colData(spe_sub)$sample_id))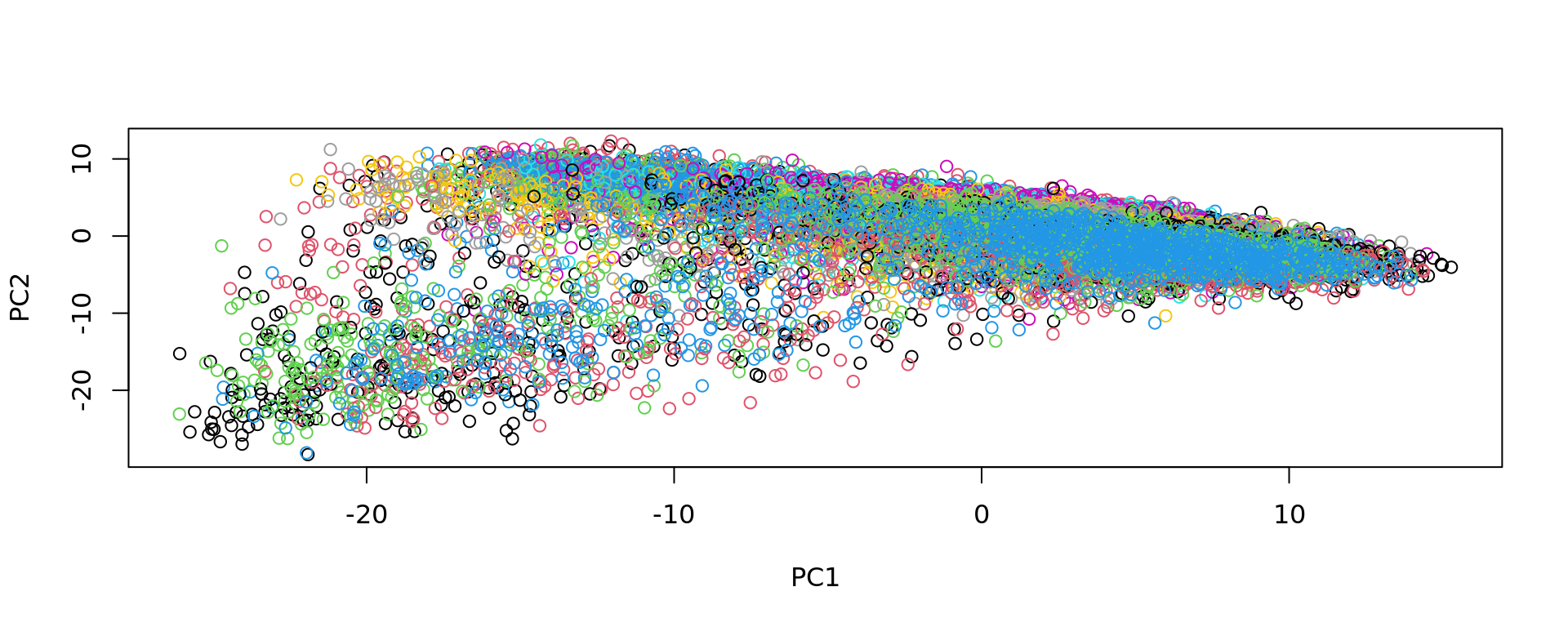
KODAMA
kk=KODAMA.matrix.parallel(pca,
spatial = xy,
FUN= "PLS" ,
landmarks = 100000,
splitting = 100,
f.par.pls = 50,
spatial.resolution = 0.4,
n.cores=4)socket cluster with 4 nodes on host 'localhost'
================================================================================[1] "Finished parallel computation"
[1] "Calculation of dissimilarity matrix..."
================================================================================ print("KODAMA finished")[1] "KODAMA finished" config=umap.defaults
config$n_threads = 4
config$n_sgd_threads = "auto"
kk_UMAP=KODAMA.visualization(kk,method="UMAP",config=config)
plot(kk_UMAP,col=as.factor(labels))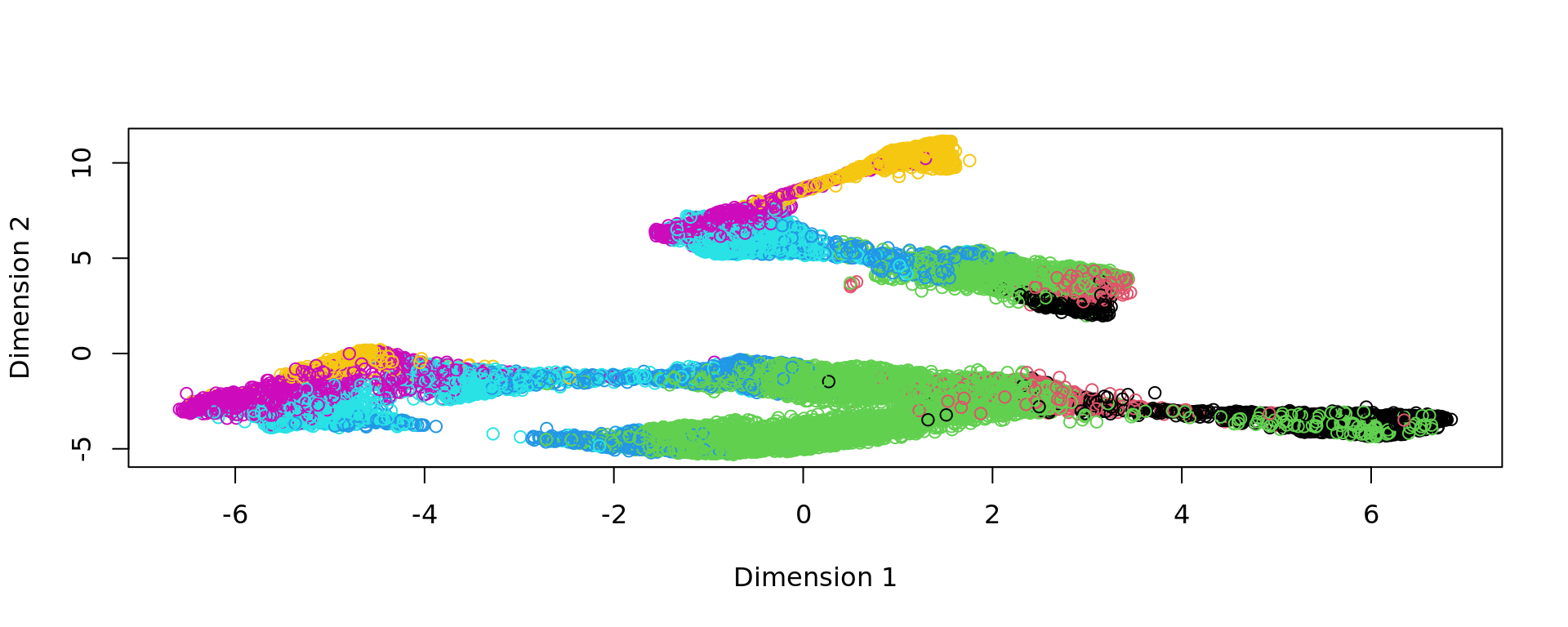
CLUSTER
library("mclust")
clu=kmeans(kk_UMAP,7,nstart = 100)$cluster
plot(kk_UMAP,col=labels,pch=20)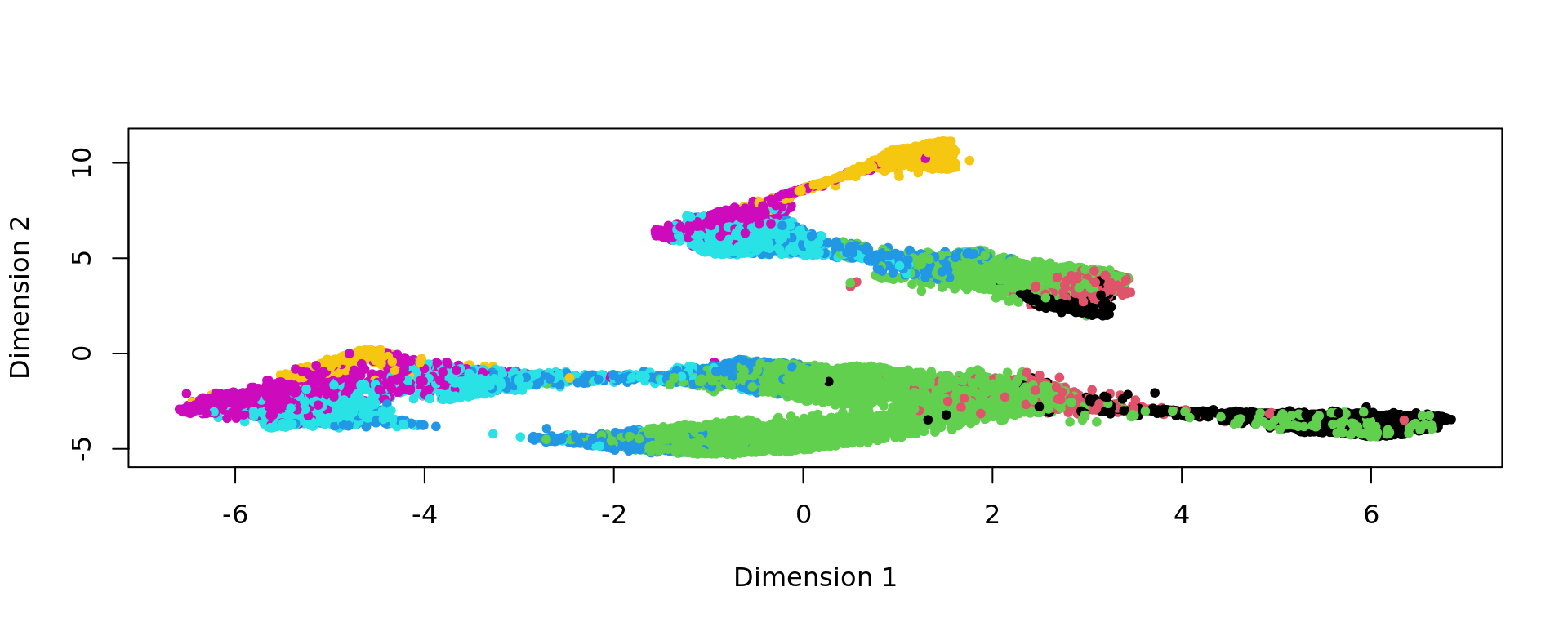
plot(kk_UMAP,col=clu,pch=20)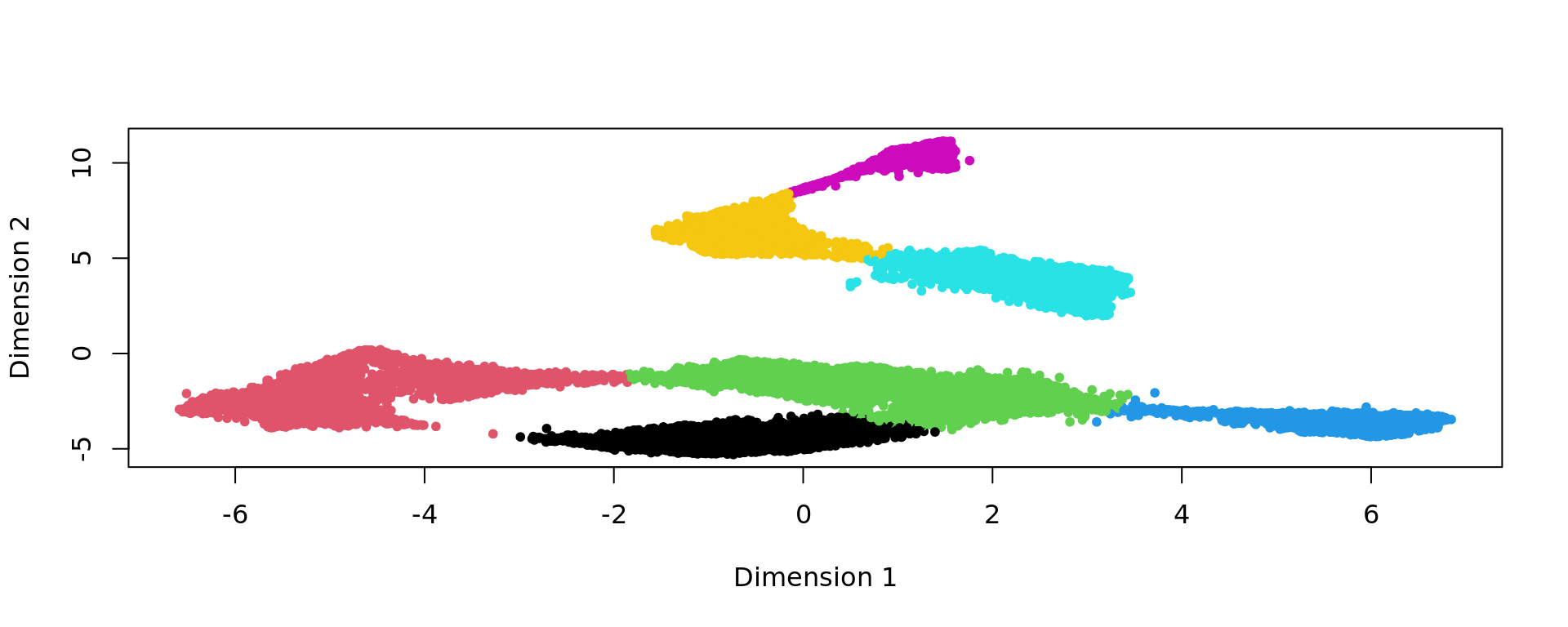
plot(xy,col=clu,pch=20)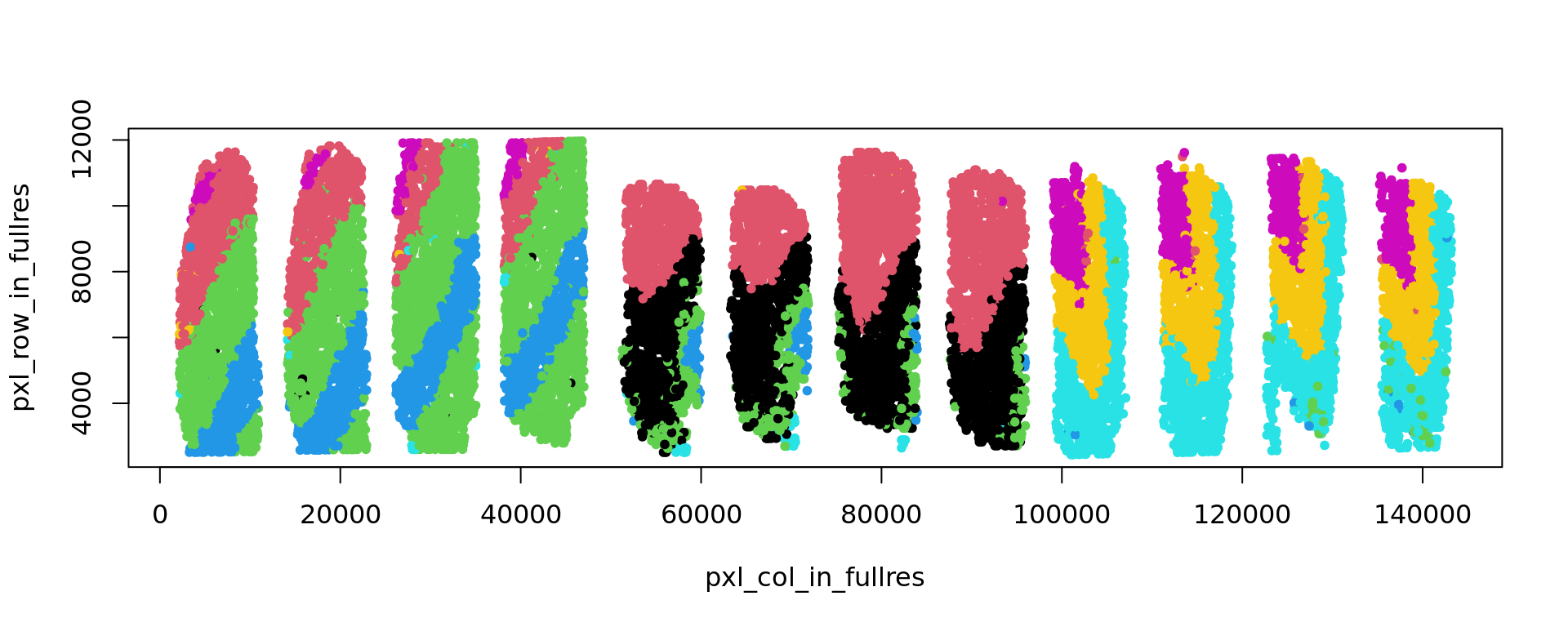
u=unique(samples)
for(i in 1:length(u)){
sel=samples==u[i]
print(adjustedRandIndex(labels[sel],clu[sel]))
}[1] 0.5205199
[1] 0.5609284
[1] 0.5564057
[1] 0.5484287
[1] 0.3519972
[1] 0.3489148
[1] 0.4441065
[1] 0.4489396
[1] 0.475645
[1] 0.4401037
[1] 0.4238303
[1] 0.4239171ref=refine_SVM(xy,clu,samples,cost=100)[1] "151507"
[1] "151508"
[1] "151509"
[1] "151510"
[1] "151669"
[1] "151670"
[1] "151671"
[1] "151672"
[1] "151673"
[1] "151674"
[1] "151675"
[1] "151676"u=unique(samples)
for(i in 1:length(u)){
sel=samples==u[i]
print(adjustedRandIndex(labels[sel],ref[sel]))
}[1] 0.5254138
[1] 0.5915947
[1] 0.5895009
[1] 0.584852
[1] 0.3494789
[1] 0.3576089
[1] 0.4547297
[1] 0.4837599
[1] 0.4871104
[1] 0.4617376
[1] 0.4491465
[1] 0.4400833plot(xy,col=ref,pch=20)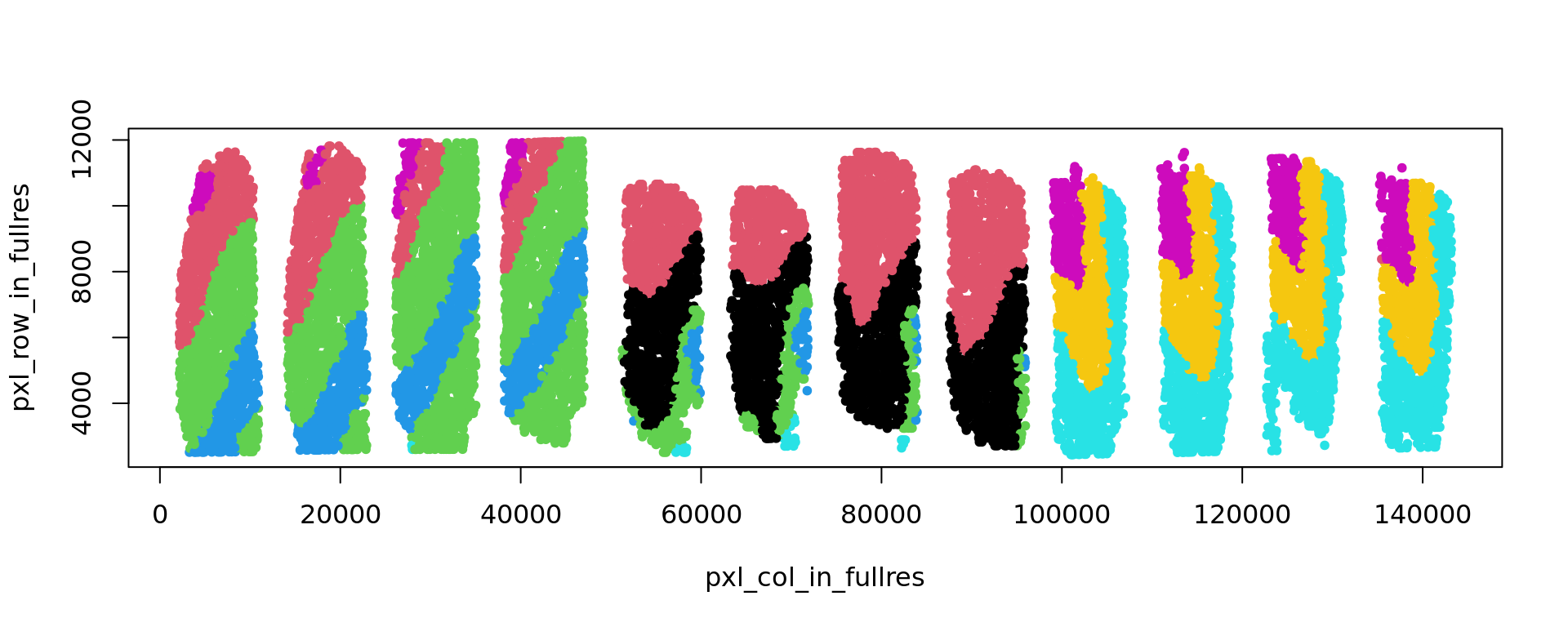
sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] mclust_6.1.1 KODAMAextra_1.0
[3] e1071_1.7-14 doParallel_1.0.17
[5] iterators_1.0.14 foreach_1.5.2
[7] KODAMA_3.1 umap_0.2.10.0
[9] Rtsne_0.17 minerva_1.5.10
[11] spatialLIBD_1.16.2 SpatialExperiment_1.14.0
[13] Seurat_5.1.0 SeuratObject_5.0.2
[15] sp_2.1-4 harmony_1.2.0
[17] Rcpp_1.0.12 SPARK_1.1.1
[19] scry_1.16.0 scran_1.32.0
[21] scater_1.32.0 ggplot2_3.5.1
[23] scuttle_1.14.0 SingleCellExperiment_1.26.0
[25] SummarizedExperiment_1.34.0 Biobase_2.64.0
[27] GenomicRanges_1.56.1 GenomeInfoDb_1.40.1
[29] IRanges_2.38.1 S4Vectors_0.42.1
[31] BiocGenerics_0.50.0 MatrixGenerics_1.16.0
[33] matrixStats_1.3.0 nnSVG_1.8.0
[35] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] goftest_1.2-3 DT_0.33
[3] Biostrings_2.72.1 vctrs_0.6.5
[5] spatstat.random_3.2-3 digest_0.6.36
[7] png_0.1-8 proxy_0.4-27
[9] git2r_0.33.0 ggrepel_0.9.5
[11] deldir_2.0-4 parallelly_1.37.1
[13] magick_2.8.3 MASS_7.3-61
[15] reshape2_1.4.4 httpuv_1.6.15
[17] withr_3.0.0 xfun_0.45
[19] survival_3.7-0 memoise_2.0.1
[21] benchmarkme_1.0.8 ggbeeswarm_0.7.2
[23] zoo_1.8-12 pbapply_1.7-2
[25] rematch2_2.1.2 KEGGREST_1.44.1
[27] promises_1.3.0 httr_1.4.7
[29] restfulr_0.0.15 globals_0.16.3
[31] fitdistrplus_1.1-11 ps_1.7.7
[33] rstudioapi_0.16.0 UCSC.utils_1.0.0
[35] miniUI_0.1.1.1 generics_0.1.3
[37] processx_3.8.4 curl_5.2.1
[39] fields_16.2 zlibbioc_1.50.0
[41] ScaledMatrix_1.12.0 polyclip_1.10-6
[43] doSNOW_1.0.20 GenomeInfoDbData_1.2.12
[45] ExperimentHub_2.12.0 SparseArray_1.4.8
[47] golem_0.4.1 xtable_1.8-4
[49] stringr_1.5.1 pracma_2.4.4
[51] evaluate_0.24.0 S4Arrays_1.4.1
[53] BiocFileCache_2.12.0 irlba_2.3.5.1
[55] colorspace_2.1-0 filelock_1.0.3
[57] ROCR_1.0-11 reticulate_1.38.0
[59] spatstat.data_3.1-2 shinyWidgets_0.8.6
[61] magrittr_2.0.3 lmtest_0.9-40
[63] later_1.3.2 viridis_0.6.5
[65] lattice_0.22-6 spatstat.geom_3.2-9
[67] future.apply_1.11.2 getPass_0.2-4
[69] scattermore_1.2 XML_3.99-0.17
[71] cowplot_1.1.3 RcppAnnoy_0.0.22
[73] class_7.3-22 pillar_1.9.0
[75] nlme_3.1-165 compiler_4.4.0
[77] beachmat_2.20.0 RSpectra_0.16-1
[79] stringi_1.8.4 tensor_1.5
[81] GenomicAlignments_1.40.0 plyr_1.8.9
[83] crayon_1.5.3 abind_1.4-5
[85] BiocIO_1.14.0 locfit_1.5-9.10
[87] bit_4.0.5 dplyr_1.1.4
[89] whisker_0.4.1 codetools_0.2-20
[91] BiocSingular_1.20.0 openssl_2.2.0
[93] bslib_0.7.0 paletteer_1.6.0
[95] plotly_4.10.4 mime_0.12
[97] splines_4.4.0 fastDummies_1.7.3
[99] dbplyr_2.5.0 sparseMatrixStats_1.16.0
[101] attempt_0.3.1 knitr_1.48
[103] blob_1.2.4 utf8_1.2.4
[105] BiocVersion_3.19.1 fs_1.6.4
[107] listenv_0.9.1 DelayedMatrixStats_1.26.0
[109] rdist_0.0.5 tibble_3.2.1
[111] Matrix_1.7-0 callr_3.7.6
[113] statmod_1.5.0 pkgconfig_2.0.3
[115] tools_4.4.0 BRISC_1.0.5
[117] cachem_1.1.0 RSQLite_2.3.7
[119] viridisLite_0.4.2 DBI_1.2.3
[121] fastmap_1.2.0 rmarkdown_2.27
[123] scales_1.3.0 grid_4.4.0
[125] ica_1.0-3 Rsamtools_2.20.0
[127] AnnotationHub_3.12.0 sass_0.4.9
[129] patchwork_1.2.0 BiocManager_1.30.23
[131] dotCall64_1.1-1 RANN_2.6.1
[133] snow_0.4-4 yaml_2.3.9
[135] rtracklayer_1.64.0 cli_3.6.3
[137] purrr_1.0.2 leiden_0.4.3.1
[139] lifecycle_1.0.4 askpass_1.2.0
[141] uwot_0.2.2 bluster_1.14.0
[143] sessioninfo_1.2.2 BiocParallel_1.38.0
[145] gtable_0.3.5 rjson_0.2.21
[147] ggridges_0.5.6 progressr_0.14.0
[149] limma_3.60.3 jsonlite_1.8.8
[151] edgeR_4.2.0 RcppHNSW_0.6.0
[153] bitops_1.0-7 benchmarkmeData_1.0.4
[155] bit64_4.0.5 spatstat.utils_3.0-5
[157] BiocNeighbors_1.22.0 matlab_1.0.4.1
[159] jquerylib_0.1.4 highr_0.11
[161] metapod_1.12.0 config_0.3.2
[163] dqrng_0.4.1 lazyeval_0.2.2
[165] shiny_1.8.1.1 htmltools_0.5.8.1
[167] sctransform_0.4.1 rappdirs_0.3.3
[169] glue_1.7.0 spam_2.10-0
[171] XVector_0.44.0 RCurl_1.98-1.14
[173] rprojroot_2.0.4 gridExtra_2.3
[175] igraph_2.0.3 R6_2.5.1
[177] tidyr_1.3.1 CompQuadForm_1.4.3
[179] cluster_2.1.6 DelayedArray_0.30.1
[181] tidyselect_1.2.1 vipor_0.4.7
[183] maps_3.4.2 AnnotationDbi_1.66.0
[185] future_1.33.2 rsvd_1.0.5
[187] munsell_0.5.1 KernSmooth_2.23-24
[189] data.table_1.15.4 htmlwidgets_1.6.4
[191] RColorBrewer_1.1-3 rlang_1.1.4
[193] spatstat.sparse_3.1-0 spatstat.explore_3.2-7
[195] fansi_1.0.6 beeswarm_0.4.0