QC and filtering
vanBuggenum
Last updated: 2021-10-07
Checks: 7 0
Knit directory: Multimodal-Plasmacell_manuscript/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211005) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8d9f708. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: code/Import_and_create_seuratObj.R
Untracked: code/load_packages.R
Untracked: code/plot_QC_function.R
Untracked: data/genelist.plots.diffmarkers.txt
Untracked: data/metadata.txt
Untracked: data/raw/
Untracked: output/paper_figures/
Untracked: output/seu.PROT_fix.rds
Untracked: output/seu.PROT_live.rds
Untracked: output/seu.RNA.rds
Untracked: output/seu.fix_norm.rds
Untracked: output/seu.live_norm.rds
Unstaged changes:
Deleted: analysis/about.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/QC.Rmd) and HTML (docs/QC.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8d9f708 | jessievb | 2021-10-07 | quality check page and remove header title |
Introduction
The single-cell multi-omics data contains single-cell transcriptomic and proteomic and phospho-proteomic data of in-vitro generated plasmacells. The code below shows Quality control plots, QC-filtering, normalization and scaling of the dataset.
Convert matrix to Seurat objects
Import all count matrixes (from data/raw folder), combine plates and create unfiltered Seurat objects.
source("code/Import_and_create_seuratObj.R")seu_RNA <- readRDS("output/seu.RNA.rds")
seu.PROT_live <- readRDS("output/seu.PROT_live.rds")
seu.PROT_fix <- readRDS("output/seu.PROT_fix.rds")QC plots
plot_RNA_nCount <- plot_QC_paper(seu_object = seu_RNA,
feature = "nCount_RNA",
ytext = "Total UMI counts per cell",
xtext = "Plate number",
paneltitle = "Fixed cells (1586 to 1589) show lower counts",
colorviolin = "dodgerblue2" ) +
geom_vline(xintercept =6.5, size = 0.3, color = "red") +
annotate(geom = "text", x = 6.6, y=20000, label = "Fixed cells", hjust = 0, size = 2.5) +
theme(axis.title.x = element_blank())
plot_RNA_ngenes <- plot_QC_paper(seu_object = seu_RNA,
feature = "nFeature_RNA",
ytext = "Total genes per cell",
xtext = "Plate number",
paneltitle = "Keep cells >300 genes",
colorviolin = "dodgerblue2" ) +
geom_hline(yintercept = 300, size = 0.3, color = "red") +
theme(axis.title.x = element_blank())
plot_percent.mt <- plot_QC_paper(seu_object = seu_RNA,
feature = "percent.mt",
ytext = "% Mitochondrial counts",
xtext = "Plate number",
paneltitle = "Keep cells < 5 % mitochondrial genecounts",
colorviolin = "dodgerblue2" ) +
geom_hline(yintercept = 5, color = "red", size = 0.3) +
theme(axis.title.x = element_blank())
plot_percent.rb <- plot_QC_paper(seu_object = seu_RNA,
feature = "percent.rb",
ytext = "% Ribosomal counts",
xtext = "Plate number",
paneltitle = "comparable % ribosomal counts in all plates",
colorviolin = "dodgerblue2" ) +
theme(axis.title.x = element_blank())
plot_PROT_nCount.live <- plot_QC_paper(seu_object = seu.PROT_live,
feature = "nCount_PROT",
ytext = "Total UMI counts per cell",
xtext = "Plate number",
paneltitle = "Keep cells > 1500 & < 9000 PROT counts",
colorviolin = "deeppink3" ) +
geom_hline(yintercept = 1500, size = 0.3) +
geom_hline(yintercept = 9000, size = 0.3) +
theme(axis.title.x = element_blank())
plot_PROT_nCount.fix <- plot_QC_paper(seu_object = seu.PROT_fix,
feature = "nCount_PROT",
ytext = "Total UMI counts per cell",
xtext = "Plate number",
paneltitle = "Keep cells > 2500 & < 20000 PROT counts",
colorviolin = "deeppink3" ) +
geom_hline(yintercept = 2500, size = 0.3) +
geom_hline(yintercept = 20000, size = 0.3) +
theme(axis.title.x = element_blank())
plot_PROT_nproteins.live <- plot_QC_paper(seu_object = seu.PROT_live,
feature = "nFeature_PROT",
ytext = "Total proteins per cell",
xtext = "Plate number",
paneltitle = "Keep cells >40 proteins",
colorviolin = "deeppink3" ) +
geom_hline(yintercept = 40, size = 0.3, color = "red")+
theme(axis.title.x = element_blank())
plot_PROT_nproteins.fix <- plot_QC_paper(seu_object = seu.PROT_fix,
feature = "nFeature_PROT",
ytext = "Total proteins per cell",
xtext = "Plate number",
paneltitle = "Keep cells >65 proteins",
colorviolin = "deeppink3" ) +
geom_hline(yintercept = 65, size = 0.3, color = "red")+
theme(axis.title.x = element_blank())
plot.QC <- plot_grid(plot_RNA_nCount,
plot_RNA_ngenes,
plot_percent.mt,
plot_percent.rb,
plot_PROT_nCount.live,
plot_PROT_nCount.fix,
plot_PROT_nproteins.live,
plot_PROT_nproteins.fix,
labels = c('a', 'b', 'c','d' , 'e', 'f', 'g', 'h'), label_size = 10, ncol = 2)
ggsave(plot.QC, filename = "output/paper_figures/Suppl_QC_filters.pdf", width = 183, height = 200, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot.QC, filename = "output/paper_figures/Suppl_QC_filters.png", width = 183, height = 200, units = "mm", dpi = 300)plot.QC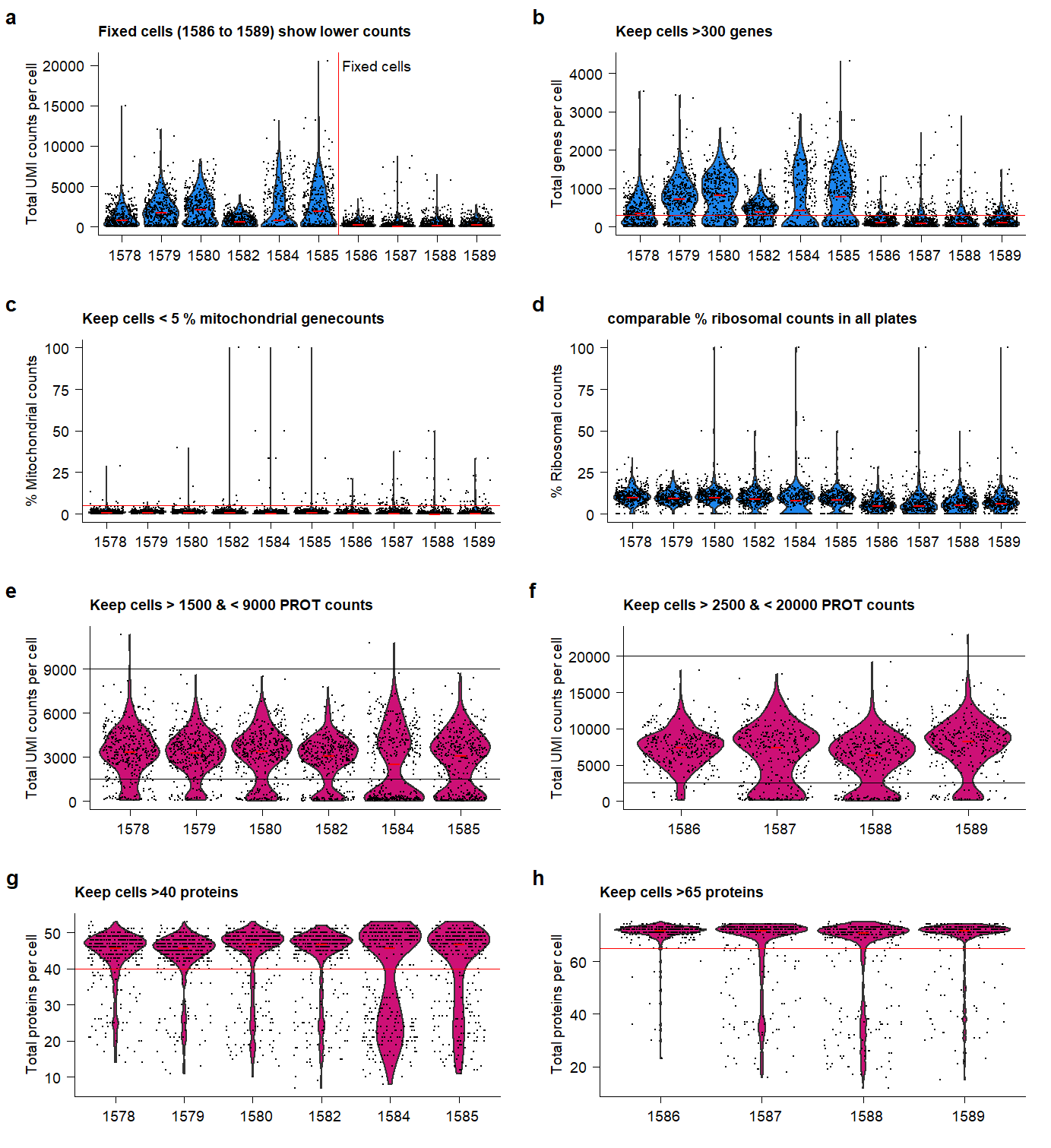 Supplementary Figure Thresholds for selection of high-quality samples and cells from the QuRIE-seq datasets.
Supplementary Figure Thresholds for selection of high-quality samples and cells from the QuRIE-seq datasets.
- Based on the indicated cut-offs, high-quality cells are filtered for further analysis:
## Filter fixed protein dataset
seu.PROT.fix.subset <- subset(seu.PROT_fix, subset = nCount_PROT >= 2500 & nCount_PROT < 20000)
## Filter live-cell protein dataset
seu.PROT.live.subset <- subset(seu.PROT_live, subset = nCount_PROT >= 1500 & nCount_PROT <= 9000)
## RNA quality of fixed dataset is too low (very low gene numbers and counts). Therefore continue only with live-cell dataset.
seu.RNA_live <- subset(seu_RNA, idents = c(1586:1589), invert = TRUE)
seu.RNA_fix <- subset(seu_RNA, idents = c(1586:1589))
## Filter RNA live dataset
seu.RNA_live.subset <- subset(seu.RNA_live, subset = percent.mt <=5 & nFeature_RNA >= 300)
seu.RNA_fix.subset <- subset(seu.RNA_fix) #, subset = percent.mt <= 5 & nFeature_RNA >= 300 # Nofilter because RNA not taken along.- Filter low-detected genes
Keep genes that are >1% cells detected.
## Additional filter features (genes) detected in 1% of cells
seu.RNA_live.subset <- CreateSeuratObject(seu.RNA_live.subset[["RNA"]]@counts, min.cells = round(ncol(seu.RNA_live.subset)/100)) ## keep features detected in 1% of cells
seu.RNA_fix.subset <- CreateSeuratObject(seu.RNA_fix.subset[["RNA"]]@counts, min.cells = round(ncol(seu.RNA_fix.subset)/100)) ## keep features detected in min 1% cells- Merge Seurat objects from Protein & RNA modalities
## Merge Seurat objects live dataset
intersect <- colnames(seu.RNA_live.subset)[colnames(seu.RNA_live.subset) %in% colnames(seu.PROT.live.subset)]
intersect <- colnames(seu.PROT.live.subset)[colnames(seu.PROT.live.subset) %in% intersect]
seu.RNA_combined.live <- subset(seu.RNA_live.subset, cells = intersect )
Prot.live.intersect <- seu.PROT.live.subset@assays$PROT@counts[,colnames(seu.PROT.live.subset) %in% intersect]
seu.RNA_combined.live[["PROT"]] <- CreateAssayObject(counts = Prot.live.intersect)
seu.RNA_combined.liveAn object of class Seurat
10211 features across 1433 samples within 2 assays
Active assay: RNA (10158 features, 0 variable features)
1 other assay present: PROT## fix dataset
intersect <- colnames(seu.RNA_fix.subset)[colnames(seu.RNA_fix.subset) %in% colnames(seu.PROT.fix.subset)]
intersect <- colnames(seu.PROT.fix.subset)[colnames(seu.PROT.fix.subset) %in% intersect]
seu.RNA_combined.fix <- subset(seu.RNA_fix.subset, cells = intersect )
Prot.fix.intersect <- seu.PROT.fix.subset@assays$PROT@counts[,colnames(seu.PROT.fix.subset) %in% intersect]
seu.RNA_combined.fix[["PROT"]] <- CreateAssayObject(counts = Prot.fix.intersect)
seu.RNA_combined.fixAn object of class Seurat
5095 features across 1038 samples within 2 assays
Active assay: RNA (5019 features, 0 variable features)
1 other assay present: PROT- Antibody quality (Non-detected proteins) filter
scale_x_reordered <- function(..., sep = "___") {
reg <- paste0(sep, ".+$")
ggplot2::scale_x_discrete(labels = function(x) gsub(reg, "", x), ...)
}
reorder_within <- function(x, by, within, fun = mean, sep = "___", ...) {
new_x <- paste(x, within, sep = sep)
stats::reorder(new_x, by, FUN = fun)
}PROT_tbl_subset.fix <- as.data.frame(seu.PROT.fix.subset@assays$PROT@counts) %>%
mutate(protein = rownames(seu.PROT.fix.subset)) %>%
dplyr::select(protein, everything()) %>%
gather("cell", "count", 2:c(ncol(seu.PROT.fix.subset)+1)) %>%
mutate(sample = gsub('.{5}$', '', cell) )ggplot(PROT_tbl_subset.fix, aes(reorder_within(protein, count, sample), count)) +
geom_boxplot(outlier.colour="black", outlier.shape=16,
outlier.size=0.5, notch=FALSE) +
scale_x_reordered() +
facet_wrap(.~ sample, ncol=4,scales = "free_y") +
theme_few()+
theme(axis.text.x=element_text(angle=90,hjust=1,vjust=.5,colour='gray50')) +
scale_y_log10() +
add.textsize +
coord_flip() +
labs(y="count (log10)", x = "Antibody (ordered per plate)")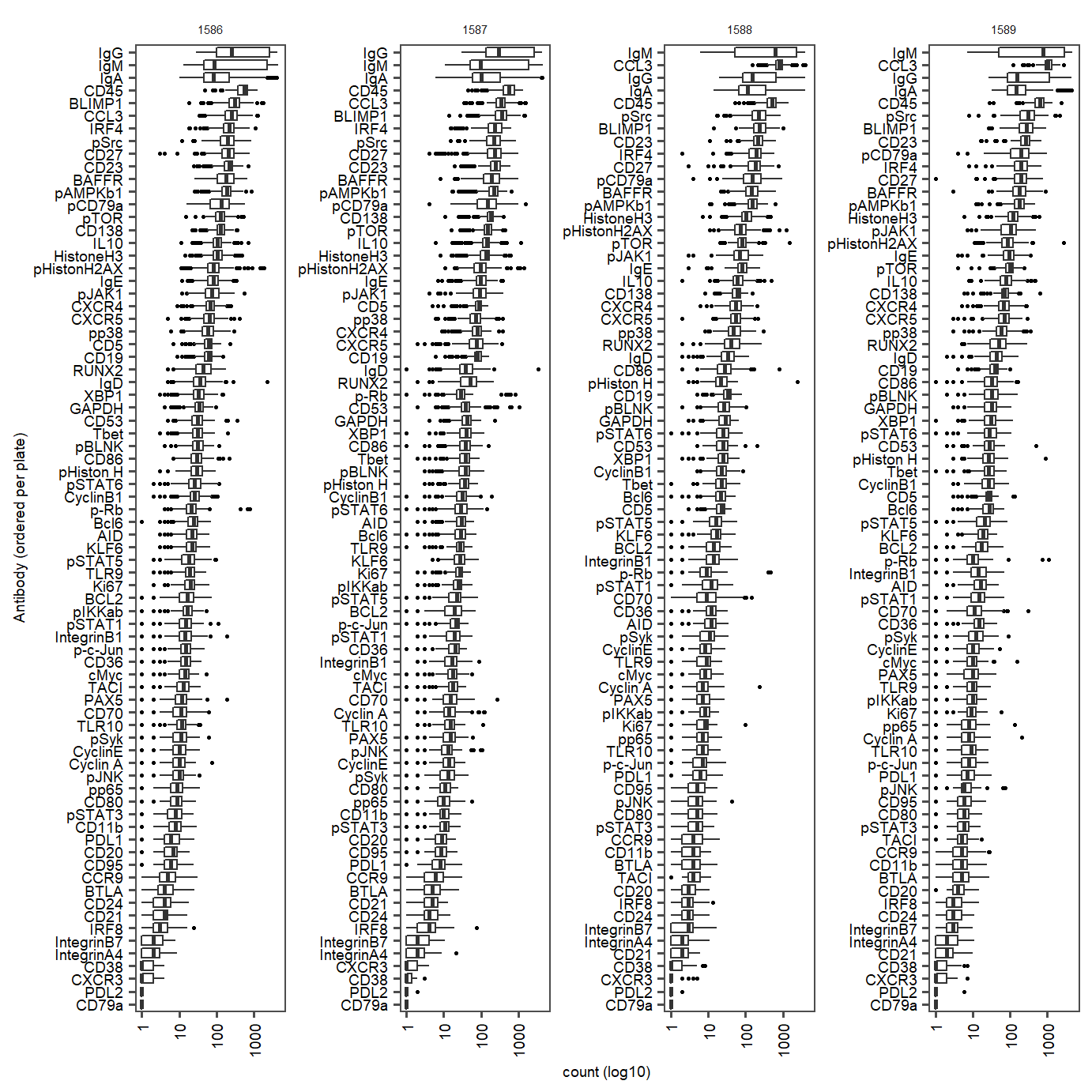 Supplementary Figure Fixed dataset antibody counts per plate (ordered from high to low per plate (1586-1589))
Supplementary Figure Fixed dataset antibody counts per plate (ordered from high to low per plate (1586-1589))
prot.median.fix <- aggregate(PROT_tbl_subset.fix[, 3], list(protein =PROT_tbl_subset.fix$protein), mean)
prot.fix.toremove <- prot.median.fix$protein[prot.median.fix$x <=0.2]
filtered.prot.counts <- seu.PROT.fix.subset[["PROT"]]@counts[!c(rownames(seu.PROT.fix.subset[["PROT"]]@counts) %chin% prot.fix.toremove),]
seu.PROT.fix.subset <- CreateSeuratObject(filtered.prot.counts, assay = "PROT")PROT_tbl_subset.live <- as.data.frame(seu.RNA_combined.live@assays$PROT@counts) %>%
mutate(protein = rownames(seu.RNA_combined.live[["PROT"]])) %>%
dplyr::select(protein, everything()) %>%
gather("cell", "count", 2:c(ncol(seu.RNA_combined.live[["PROT"]])+1)) %>%
mutate(sample = gsub('.{5}$', '', cell) )ggplot(PROT_tbl_subset.live, aes(reorder_within(protein, count, sample), count)) +
geom_boxplot(outlier.colour="black", outlier.shape=16,
outlier.size=0.5, notch=FALSE) +
scale_x_reordered() +
facet_wrap(.~ sample, ncol=4,scales = "free_y") +
theme_few()+
theme(axis.text.x=element_text(angle=90,hjust=1,vjust=.5,colour='gray50')) +
scale_y_log10() +
add.textsize +
coord_flip() +
labs(y="count (log10)", x = "Antibody (ordered per plate)")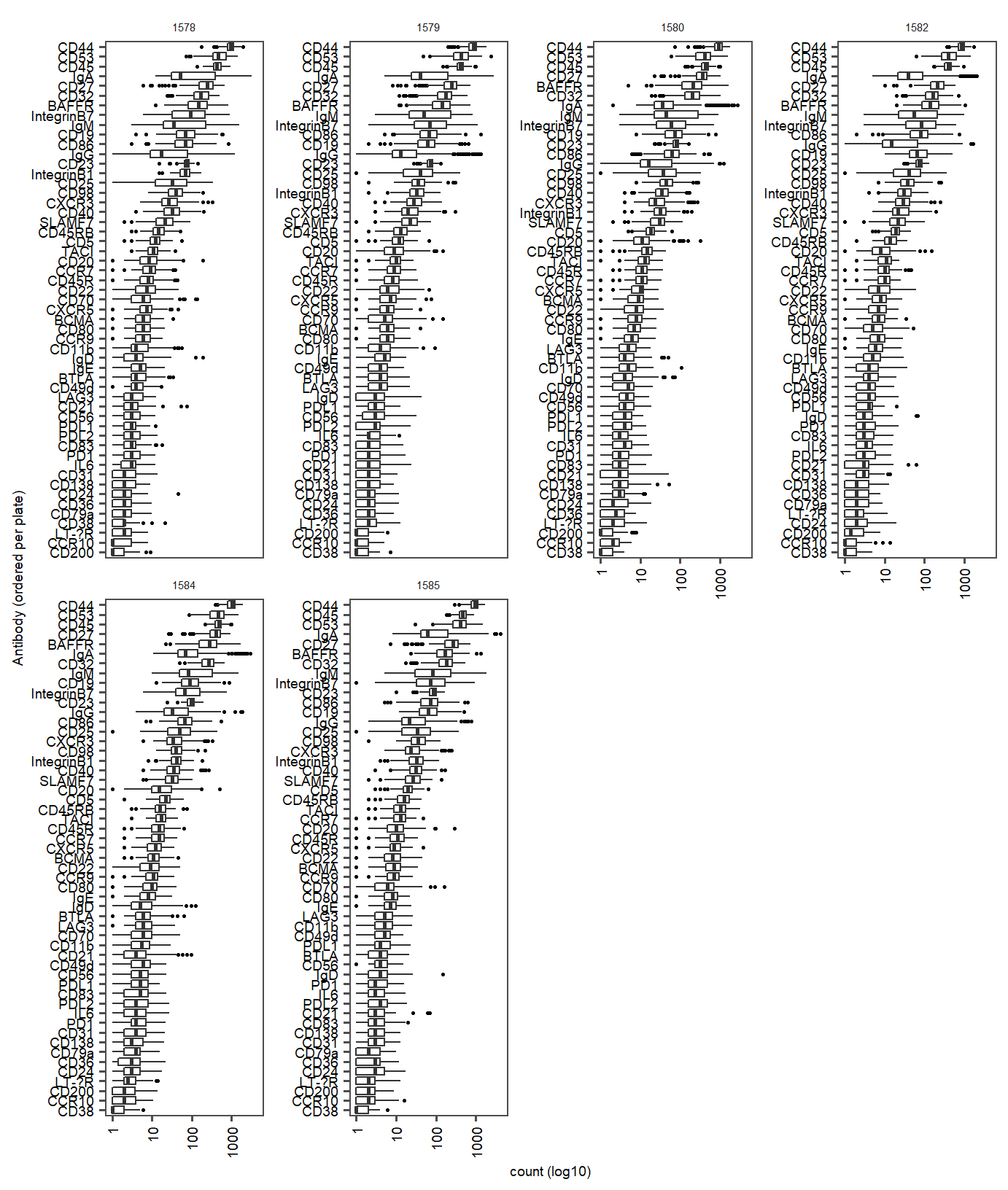 Supplementary Figure Live dataset antibody counts per plate (ordered from high to low per plate (1586-1589))
Supplementary Figure Live dataset antibody counts per plate (ordered from high to low per plate (1586-1589))
prot.median.live <- aggregate(PROT_tbl_subset.live[, 3], list(protein =PROT_tbl_subset.live$protein), mean)
prot.live.toremove <- prot.median.live$protein[prot.median.live$x <1]
filtered.prot.counts.live <- seu.RNA_combined.live[["PROT"]]@counts[!c(rownames(seu.RNA_combined.live[["PROT"]]@counts) %chin% prot.live.toremove),]
seu.RNA_combined.live[["PROT"]] <- CreateAssayObject(counts = filtered.prot.counts.live)- Add metadata to object:
## metadata import
metadata <- read_delim("data/metadata.txt", "\t", escape_double = FALSE, trim_ws = TRUE)
metadata$sample <- as.factor(metadata$sample)
## add metadata to fix dataset
meta.fix <- data.frame(seu.RNA_combined.fix@meta.data) %>%
mutate(sample = orig.ident ) %>%
left_join(metadata) %>%
mutate(group = sample)
meta.fix<-as.data.frame(meta.fix)
rownames(meta.fix) <- rownames(data.frame(seu.RNA_combined.fix@meta.data) )
seu.RNA_combined.fix <- AddMetaData(object = seu.RNA_combined.fix, metadata = meta.fix)
#meta.fix <- data.frame(seu.RNA_combined.fix@meta.data) %>%
# mutate(sample = rownames(seu.RNA_combined.fix@meta.data))
## add metadata to live dataset
meta.live <- data.frame(seu.RNA_combined.live@meta.data) %>%
mutate(sample = orig.ident ) %>%
left_join(metadata) %>%
mutate(group = sample)
meta.live<-as.data.frame(meta.live)
rownames(meta.live) <- rownames(data.frame(seu.RNA_combined.live@meta.data) )
seu.RNA_combined.live <- AddMetaData(object = seu.RNA_combined.live, metadata = meta.live)
#meta.live <- data.frame(seu.RNA_combined.live@meta.data) %>%
# mutate(sample = rownames(seu.RNA_combined.live@meta.data))
seu.RNA_combined.live[["percent.mt"]] <- PercentageFeatureSet(seu.RNA_combined.live, pattern = "^MT")
seu.RNA_combined.fix[["percent.mt"]] <- PercentageFeatureSet(seu.RNA_combined.fix, pattern = "^MT")Normalize and Scale
## fix data normalize RNA
DefaultAssay(seu.RNA_combined.fix) <- 'RNA'
seu.RNA_combined.fix <- SCTransform(seu.RNA_combined.fix, assay = "RNA", new.assay.name = "SCT", vars.to.regress = c("nCount_RNA", "percent.mt", "plate"), return.only.var.genes = FALSE, verbose = FALSE)
# Add some metadata to normalized data (ncounts & percent mt)
seu.RNA_combined.fix <- AddMetaData(seu.RNA_combined.fix, as.data.frame(seu.RNA_combined.fix@assays$SCT@counts) %>% summarise_all(funs(sum)) %>% unlist(), col.name = "nCount_RNA_SCT")
seu.RNA_combined.fix <- PercentageFeatureSet(seu.RNA_combined.fix, pattern = "^MT\\.|^MTRN", col.name = "percent.mt.aftersct", assay = "SCT")
## Fixed dataset normalize protein
DefaultAssay(seu.RNA_combined.fix) <- 'PROT'
VariableFeatures(seu.RNA_combined.fix) <- rownames(seu.RNA_combined.fix[["PROT"]])
seu.RNA_combined.fix <- NormalizeData(seu.RNA_combined.fix, normalization.method = 'CLR', margin = 2, assay = "PROT") %>%
ScaleData(vars.to.regress = c("nCount_PROT", "plate"))
## live data normalize RNA
DefaultAssay(seu.RNA_combined.live) <- 'RNA'
seu.RNA_combined.live <- SCTransform(seu.RNA_combined.live, assay = "RNA", new.assay.name = "SCT", vars.to.regress = c("nCount_RNA", "percent.mt", "plate"), return.only.var.genes = FALSE, verbose = FALSE)
# Add some metadata to normalized data (ncounts & percent mt)
seu.RNA_combined.live <- AddMetaData(seu.RNA_combined.live, as.data.frame(seu.RNA_combined.live@assays$SCT@counts) %>% summarise_all(funs(sum)) %>% unlist(), col.name = "nCount_RNA_SCT")
seu.RNA_combined.live <- PercentageFeatureSet(seu.RNA_combined.live, pattern = "^MT\\.|^MTRN", col.name = "percent.mt.aftersct", assay = "SCT")
## live normalize & scale protein data
DefaultAssay(seu.RNA_combined.live) <- 'PROT'
VariableFeatures(seu.RNA_combined.live) <- rownames(seu.RNA_combined.live[["PROT"]])
seu.RNA_combined.live <- NormalizeData(seu.RNA_combined.live, normalization.method = 'CLR', margin = 2, assay = "PROT") %>%
ScaleData(vars.to.regress = c("nCount_PROT", "plate")) Filtered dataset properties
Overview of the number of cells and data properties of all plates.
Live-cells RNA & surface protein dataset
seu.RNA_combined.liveAn object of class Seurat
20366 features across 1433 samples within 3 assays
Active assay: PROT (50 features, 50 variable features)
2 other assays present: RNA, SCTTable Overview of per plate properties after filtering.
kable(seu.RNA_combined.live@meta.data %>%
group_by(donor,plate) %>%
summarise(`Number of cells` = round(n(),0),
`Median counts RNA` = round(median(nCount_RNA),0),
`Median Number genes` = round(median(nFeature_RNA),0),
`Median Mitochondrial counts (Median %)` = round(median(percent.mt),2),
`Median counts PROT` = round(median(nCount_PROT),0),
`Number proteins` = round(median(nFeature_PROT),0)
)) %>%
kable_styling(bootstrap_options = c("striped", "hover"))| donor | plate | Number of cells | Median counts RNA | Median Number genes | Median Mitochondrial counts (Median %) | Median counts PROT | Number proteins |
|---|---|---|---|---|---|---|---|
| D32 | P_1578 | 216 | 1624 | 556 | 1.14 | 3842 | 46 |
| D32 | P_1579 | 293 | 2333 | 861 | 1.21 | 3601 | 45 |
| D33 | P_1580 | 274 | 2888 | 1040 | 1.18 | 3908 | 47 |
| D33 | P_1584 | 184 | 3688 | 1220 | 1.21 | 4383 | 48 |
| D40 | P_1582 | 231 | 1150 | 524 | 1.11 | 3575 | 47 |
| D40 | P_1585 | 235 | 3706 | 1133 | 1.24 | 3831 | 47 |
Table Overview of per donor properties after filtering.
kable(seu.RNA_combined.live@meta.data %>%
group_by(donor) %>%
summarise(`Number of cells` = round(n(),0),
`Median counts RNA` = round(median(nCount_RNA),0),
`Median Number genes` = round(median(nFeature_RNA),0),
`Median Mitochondrial counts (Median %)` = round(median(percent.mt),2),
`Median counts PROT` = round(median(nCount_PROT),0),
`Number proteins` = round(median(nFeature_PROT),0)
)) %>%
kable_styling(bootstrap_options = c("striped", "hover"))| donor | Number of cells | Median counts RNA | Median Number genes | Median Mitochondrial counts (Median %) | Median counts PROT | Number proteins |
|---|---|---|---|---|---|---|
| D32 | 509 | 2008 | 732 | 1.18 | 3693 | 46 |
| D33 | 458 | 3168 | 1122 | 1.18 | 4098 | 47 |
| D40 | 466 | 1817 | 731 | 1.17 | 3694 | 47 |
Fixed cells intracellular proteins dataset
seu.RNA_combined.fixAn object of class Seurat
10114 features across 1038 samples within 3 assays
Active assay: PROT (76 features, 76 variable features)
2 other assays present: RNA, SCTTable Overview of per plate properties after filtering.
kable(seu.RNA_combined.fix@meta.data %>%
group_by(donor,plate) %>%
summarise(`Number of cells` = round(n(),0),
`Median counts RNA` = round(median(nCount_RNA),0),
`Median Number genes` = round(median(nFeature_RNA),0),
`Median Mitochondrial counts (Median %)` = round(median(percent.mt),2),
`Median counts PROT` = round(median(nCount_PROT),0),
`Number proteins` = round(median(nFeature_PROT),0)
)) %>%
kable_styling(bootstrap_options = c("striped", "hover"))| donor | plate | Number of cells | Median counts RNA | Median Number genes | Median Mitochondrial counts (Median %) | Median counts PROT | Number proteins |
|---|---|---|---|---|---|---|---|
| D33 | P_1586 | 290 | 280 | 106 | 0.52 | 7664 | 72 |
| D33 | P_1587 | 232 | 266 | 120 | 0.99 | 8492 | 72 |
| D40 | P_1588 | 254 | 322 | 140 | 0.57 | 7250 | 72 |
| D40 | P_1589 | 262 | 272 | 116 | 0.81 | 8704 | 72 |
Table Overview of per donor properties after filtering.
kable(seu.RNA_combined.fix@meta.data %>%
group_by(donor) %>%
summarise(`Number of cells` = round(n(),0),
`Median counts RNA` = round(median(nCount_RNA),0),
`Median Number genes` = round(median(nFeature_RNA),0),
`Median Mitochondrial counts (Median %)` = round(median(percent.mt),2),
`Median counts PROT` = round(median(nCount_PROT),0),
`Number proteins` = round(median(nFeature_PROT),0)
)) %>%
kable_styling(bootstrap_options = c("striped", "hover"))| donor | Number of cells | Median counts RNA | Median Number genes | Median Mitochondrial counts (Median %) | Median counts PROT | Number proteins |
|---|---|---|---|---|---|---|
| D33 | 522 | 272 | 112 | 0.73 | 7924 | 72 |
| D40 | 516 | 292 | 126 | 0.64 | 7942 | 72 |
Save dataset
Seurat object with filtered cells and normalized counts is stored in “output/seu.fix_norm.rds” (intracellular protein modality) and “output/seu.live_norm.rds”(RNA and surface protein modalities).
## Save Seurat objects
saveRDS(seu.RNA_combined.fix, file = "output/seu.fix_norm.rds")
saveRDS(seu.RNA_combined.live, file = "output/seu.live_norm.rds")
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_Netherlands.1252 LC_CTYPE=English_Netherlands.1252
[3] LC_MONETARY=English_Netherlands.1252 LC_NUMERIC=C
[5] LC_TIME=English_Netherlands.1252
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] ggupset_0.3.0 RColorBrewer_1.1-2 clusterProfiler_3.18.1
[4] enrichplot_1.10.2 UCell_1.0.0 data.table_1.13.6
[7] scales_1.1.1 cowplot_1.1.1 ggthemes_4.2.4
[10] kableExtra_1.3.1 knitr_1.31 org.Hs.eg.db_3.12.0
[13] AnnotationDbi_1.52.0 IRanges_2.24.1 S4Vectors_0.28.1
[16] Biobase_2.50.0 BiocGenerics_0.36.0 forcats_0.5.1
[19] stringr_1.4.0 dplyr_1.0.3 purrr_0.3.4
[22] readr_1.4.0 tidyr_1.1.2 tibble_3.0.5
[25] ggplot2_3.3.3 tidyverse_1.3.0 Matrix_1.2-18
[28] SeuratObject_4.0.0 Seurat_4.0.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] reticulate_1.18 tidyselect_1.1.0 RSQLite_2.2.3
[4] htmlwidgets_1.5.3 grid_4.0.3 BiocParallel_1.24.1
[7] Rtsne_0.15 scatterpie_0.1.5 munsell_0.5.0
[10] codetools_0.2-16 ica_1.0-2 future_1.21.0
[13] miniUI_0.1.1.1 withr_2.4.1 colorspace_2.0-0
[16] GOSemSim_2.16.1 highr_0.8 rstudioapi_0.13
[19] ROCR_1.0-11 tensor_1.5 DOSE_3.16.0
[22] listenv_0.8.0 labeling_0.4.2 git2r_0.28.0
[25] polyclip_1.10-0 bit64_4.0.5 farver_2.0.3
[28] downloader_0.4 rprojroot_2.0.2 parallelly_1.23.0
[31] vctrs_0.3.6 generics_0.1.0 xfun_0.23
[34] R6_2.5.0 graphlayouts_0.7.1 spatstat.utils_2.1-0
[37] cachem_1.0.1 fgsea_1.16.0 assertthat_0.2.1
[40] promises_1.1.1 ggraph_2.0.5 gtable_0.3.0
[43] globals_0.14.0 goftest_1.2-2 tidygraph_1.2.0
[46] rlang_0.4.10 splines_4.0.3 lazyeval_0.2.2
[49] broom_0.7.3 BiocManager_1.30.10 yaml_2.2.1
[52] reshape2_1.4.4 abind_1.4-5 modelr_0.1.8
[55] backports_1.2.1 httpuv_1.5.5 qvalue_2.22.0
[58] tools_4.0.3 ellipsis_0.3.1 ggridges_0.5.3
[61] Rcpp_1.0.6 plyr_1.8.6 ps_1.5.0
[64] rpart_4.1-15 deldir_0.2-10 pbapply_1.4-3
[67] viridis_0.5.1 zoo_1.8-8 haven_2.3.1
[70] ggrepel_0.9.1 cluster_2.1.0 fs_1.5.0
[73] magrittr_2.0.1 scattermore_0.7 DO.db_2.9
[76] lmtest_0.9-38 reprex_1.0.0 RANN_2.6.1
[79] whisker_0.4 fitdistrplus_1.1-3 matrixStats_0.57.0
[82] hms_1.0.0 patchwork_1.1.1 mime_0.9
[85] evaluate_0.14 xtable_1.8-4 readxl_1.3.1
[88] gridExtra_2.3 compiler_4.0.3 shadowtext_0.0.7
[91] KernSmooth_2.23-17 crayon_1.3.4 htmltools_0.5.1.1
[94] mgcv_1.8-33 later_1.1.0.1 lubridate_1.7.9.2
[97] DBI_1.1.1 tweenr_1.0.1 dbplyr_2.0.0
[100] MASS_7.3-53 cli_2.2.0 igraph_1.2.6
[103] pkgconfig_2.0.3 rvcheck_0.1.8 plotly_4.9.3
[106] xml2_1.3.2 webshot_0.5.2 rvest_0.3.6
[109] digest_0.6.27 sctransform_0.3.2 RcppAnnoy_0.0.18
[112] spatstat.data_2.1-0 rmarkdown_2.6 cellranger_1.1.0
[115] leiden_0.3.7 fastmatch_1.1-0 uwot_0.1.10
[118] shiny_1.6.0 lifecycle_0.2.0 nlme_3.1-149
[121] jsonlite_1.7.2 viridisLite_0.3.0 fansi_0.4.2
[124] pillar_1.4.7 lattice_0.20-41 fastmap_1.1.0
[127] httr_1.4.2 survival_3.2-7 GO.db_3.12.1
[130] glue_1.4.2 spatstat_1.64-1 png_0.1-7
[133] bit_4.0.4 ggforce_0.3.2 stringi_1.5.3
[136] blob_1.2.1 memoise_2.0.0 irlba_2.3.3
[139] future.apply_1.7.0