UMAP on 7 MOFA factors
## UMAP
plot.umap.data <- plot_dimred(mofa, method="UMAP", color_by = "condition",stroke = 0.001, dot_size =1, alpha = 0.2, return_data = T)
plot.umap.all <- ggplot(plot.umap.data, aes(x=x, y = y, fill = color_by))+
geom_point(size = 0.8, alpha = 0.6, shape = 21, stroke = 0) +
theme_half_open() +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg",)+
theme(legend.position="none")+
add.textsize +
labs(title = "UMAP on MOFA+ factors shows minute and \nhour time-scale cell-state transitions", x = "UMAP 1", y = "UMAP 2")
## UMAP legend
legend.umap <- get_legend( ggplot(plot.umap.data, aes(x=x, y = y, fill = color_by))+
geom_point(size = 2, alpha = 1, shape = 21, stroke = 0) +
theme_half_open() +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)",)+
add.textsize +
labs(title = "UMAP on MOFA+ factors shows minute and \nhour time-scale cell-state transitions", x = "UMAP 1", "y = UMAP 2"))
legend.umap <- as_ggplot(legend.umap)
plot_fig1_umap <- plot_grid(plot.umap.all,legend.umap, labels = c(""), label_size = 10, ncol = 2, rel_widths = c(1, 0.2))
ggsave(plot_fig1_umap, filename = "output/paper_figures/Fig1_UMAP.pdf", width = 68, height = 62, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(plot_fig1_umap, filename = "output/paper_figures/Fig1_UMAP.png", width = 68, height = 62, units = "mm", dpi = 300)
plot_fig1_umap
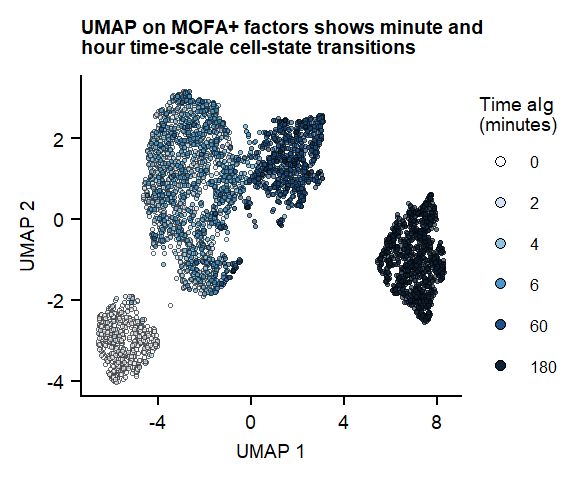 Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements
Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements
Suppl PCA
seu_combined_selectsamples <- RunPCA(seu_combined_selectsamples,assay = "SCT.RNA", features = genes.variable, verbose = FALSE, ndims.print = 0, reduction.name = "pca.RNA")
seu_combined_selectsamples <- RunPCA(seu_combined_selectsamples, assay = "PROT", features = proteins.all, verbose = FALSE, ndims.print = 0,reduction.name = "pca.PROT")
## PCA analysis
plot.PCA.RNA <- DimPlot(seu_combined_selectsamples, reduction = "pca.RNA", group.by = "condition", pt.size =0.2) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
labs(title = "RNA PCA separates cells \nat hour time-scale", x= "RNA PC 1", y = " RNA PC 2") +
add.textsize +
theme(legend.position = "none")
plot.PCA.PROT <- DimPlot(seu_combined_selectsamples, reduction = "pca.PROT", group.by = "condition", pt.size = 0.2) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
labs(title = "(phospho-)protein PCA separates cells\nat minutes time-scale", x= "Protein PC 1", y = "Protein PC 2") +
add.textsize +
# scale_x_reverse()+
theme(legend.position = "none")
legend <- get_legend(DimPlot(seu_combined_selectsamples, reduction = "pca.RNA", group.by = "condition", pt.size =0.1) +
scale_color_manual(values = colorgradient6_manual2, labels = c(labels), name = "Time aIg",)+
add.textsize)
legend <- as_ggplot(legend)
PCA.PROTPC1.data <- data.frame(rank = 1:80,
protein = names(sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1])),
weight.PC1 = sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1]),
highlights = c(names(sort(seu_combined_selectsamples@reductions$pca.PROT@feature.loadings[,1]))[1:4],rep("",76))
)
plot.PCA.PROTweightsPC1 <- ggplot(PCA.PROTPC1.data, aes(x=weight.PC1, y = rank, label = highlights)) +
geom_point(size=0.1) +
labs(title = "Top 4 protein loadings \n", x= "Weight Protein PC1") +
geom_point(color = ifelse(PCA.PROTPC1.data$highlights == "", "grey50", "red")) +
geom_text_repel(size = 2, segment.size = 0.25)+
theme_half_open()+
add.textsize +
theme(axis.title.y=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
) +
scale_x_reverse()+
scale_y_reverse()
PCA.RNAPC1.data <- data.frame(rank = 1:2371,
RNA = names(sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1])),
weight.PC1 = sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1]),
highlights = c(rep("",2366),names(sort(seu_combined_selectsamples@reductions$pca.RNA@feature.loadings[,1]))[2367:2371])
)
## Loadings RNA
plot.PCA.RNAweightsPC1 <- ggplot(PCA.RNAPC1.data, aes(x=weight.PC1, y = rank, label = highlights)) +
geom_point(size=0.1) +
labs(title = "Top 4 RNA loadings \n", x= "Weight RNA PC1") +
geom_point(color = ifelse(PCA.RNAPC1.data$highlights == "", "grey50", "red")) +
geom_text_repel(size = 2, segment.size = 0.25)+
theme_half_open()+
add.textsize +
theme(axis.title.y=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank()
)
## ridgeplots
PC1.data <- data.frame(sample = rownames(seu_combined_selectsamples@reductions$pca.PROT@cell.embeddings),
PC1_PROT = seu_combined_selectsamples@reductions$pca.PROT@cell.embeddings[,1],
PC1_RNA = seu_combined_selectsamples@reductions$pca.RNA@cell.embeddings[,1]) %>%
left_join(meta.allcells)
plot_ridge_PC1Prot <- ggplot(PC1.data, aes(x = PC1_PROT, y = condition, fill = condition)) +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)")+
geom_density_ridges2() +
scale_y_discrete(labels = labels, name = "Time aIg (minutes)")+
scale_x_continuous(name = "PC1 Proteins") +
theme_half_open() +
add.textsize+
theme(legend.position = "none")
plot_ridge_PC1RNA <- ggplot(PC1.data, aes(x = PC1_RNA, y = condition, fill = condition)) +
scale_fill_manual(values = colorgradient6_manual, labels = c(labels), name = "Time aIg \n(minutes)")+
geom_density_ridges2() +
scale_y_discrete(labels = labels, name = "Time aIg (minutes)")+
scale_x_continuous(name = "PC1 RNA") +
theme_half_open() +
add.textsize+
theme(legend.position = "none")
Fig1.pca <- plot_grid(plot.PCA.PROT,plot.PCA.RNA,legend, plot_ridge_PC1Prot, plot_ridge_PC1RNA,NULL, plot.PCA.PROTweightsPC1, plot.PCA.RNAweightsPC1, labels = c(panellabels[1:2], "","","","",panellabels[3:4]), label_size = 10, ncol = 3, rel_widths = c(0.9,0.9,0.2,0.9,0.9), rel_heights = c(1,0.8,1.3))
Picking joint bandwidth of 0.37
Picking joint bandwidth of 0.368
ggsave(filename = "output/paper_figures/Suppl_PCA_aIg.pdf", width = 143, height = 170, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(filename = "output/paper_figures/Suppl_PCA_aIg.png", width = 143, height = 170, units = "mm", dpi = 300)
Fig1.pca
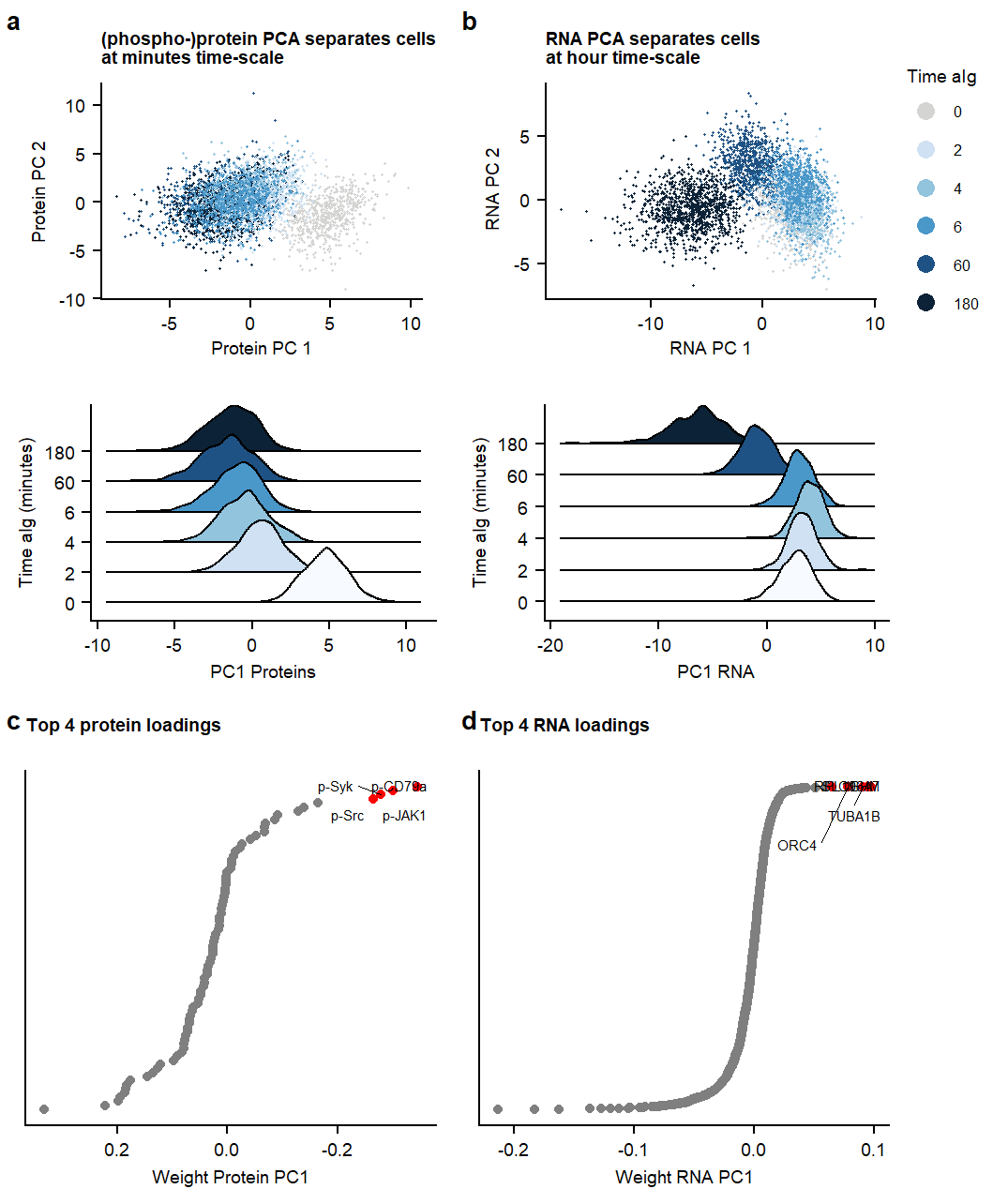 Supplemental Figure. PCA analysis on RNA ór Protein dataset
Supplemental Figure. PCA analysis on RNA ór Protein dataset
Suppl MOFA model properties
Fig.1.suppl.mofa.row1 <- plot_grid(plot.variance.perfactor.all, plot.variance.total,NULL, plot.heatmap.pval.covariates, labels = c(panellabels[1:3]), label_size = 10, ncol = 4, rel_widths = c(1.35, 0.27,0.2,0.38))
Fig.1.suppl.mofa.row2 <- plot_grid(plot.violin.factorall,legend.umap, labels = panellabels[4], label_size = 10, ncol = 2, rel_widths = c(1,0.1))
Suppl_mofa <- plot_grid(Fig.1.suppl.mofa.row1, Fig.1.suppl.mofa.row2, plot.rank.PROT.2.4to7, plot.rank.RNA.2.4to7, labels = c("","", panellabels[5:6]),label_size = 10, ncol = 1, rel_heights = c(1.45,1,1.1,1.1))
ggsave(Suppl_mofa, filename = "output/paper_figures/Suppl_MOFAaIg.pdf", width = 183, height = 220, units = "mm", dpi = 300, useDingbats = FALSE)
ggsave(Suppl_mofa, filename = "output/paper_figures/Suppl_MOFAaIg.png", width = 183, height = 220, units = "mm", dpi = 300)
Suppl_mofa
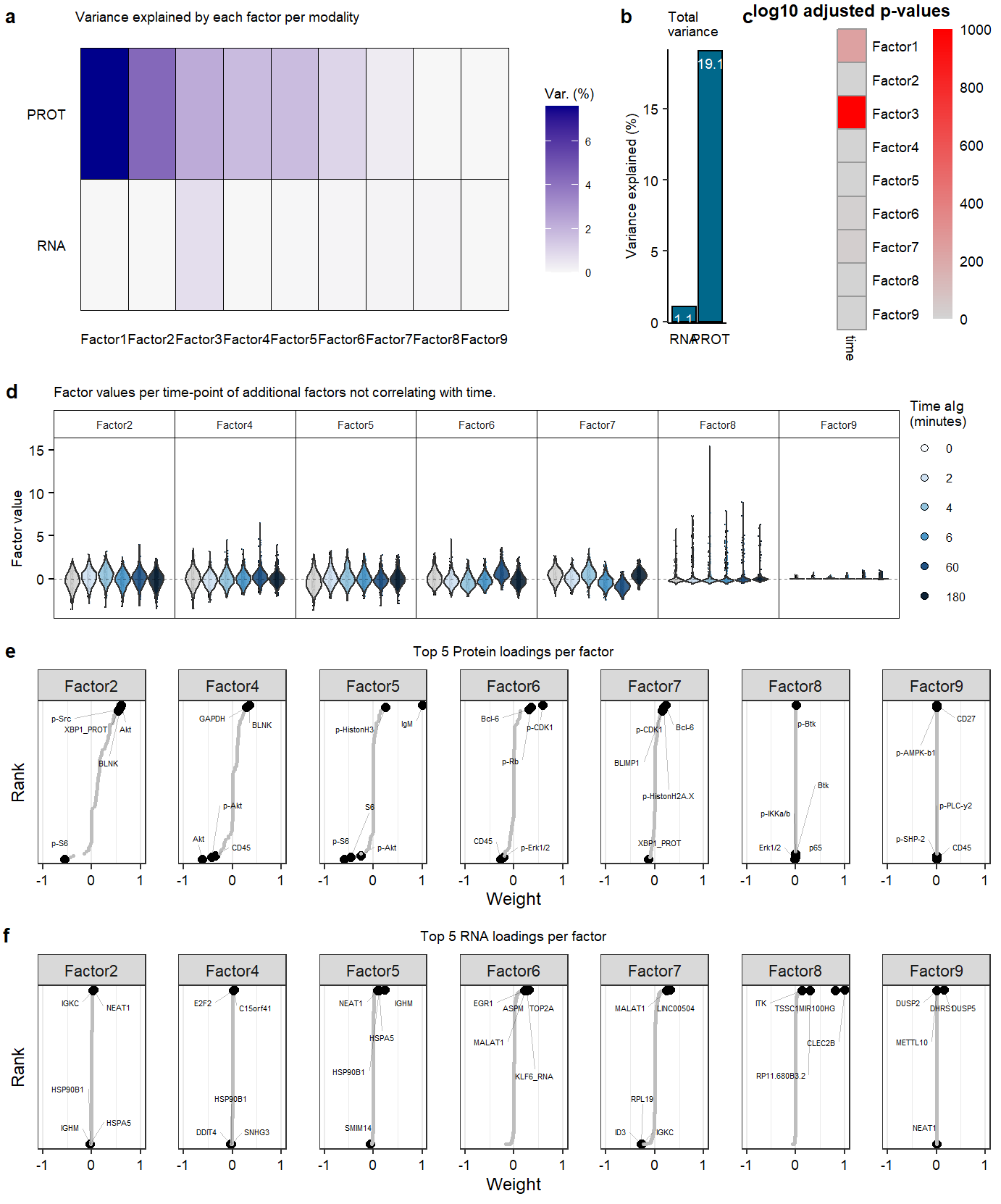 Supplemental Figure. MOFA model additional information
Supplemental Figure. MOFA model additional information
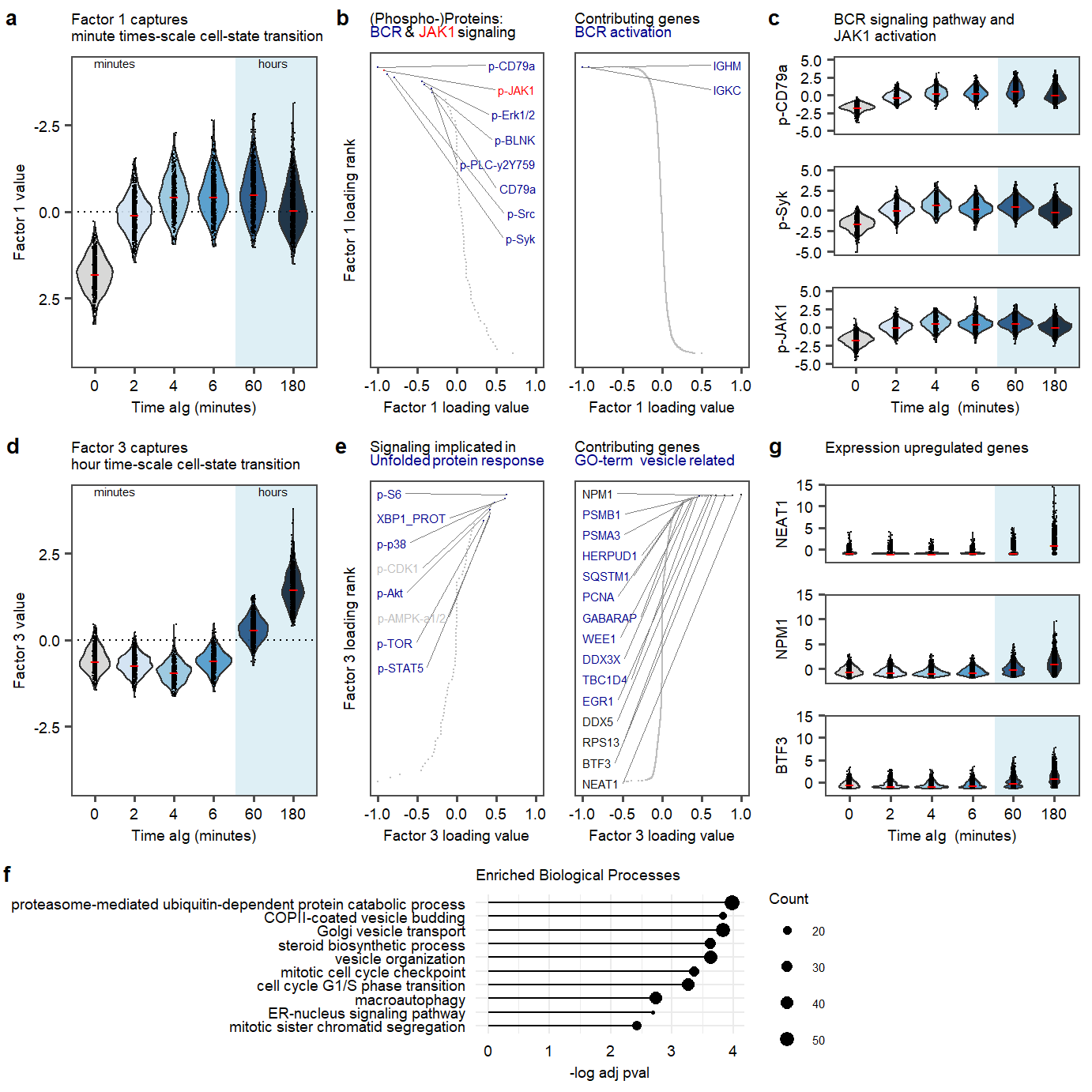 Figure 2. Factor 1 and 3 exploration.
Figure 2. Factor 1 and 3 exploration. Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements
Figure 1. UMAP of 7 MOFA factors, integrating phospho-protein and RNA measurements Supplemental Figure. PCA analysis on RNA ór Protein dataset
Supplemental Figure. PCA analysis on RNA ór Protein dataset Supplemental Figure. MOFA model additional information
Supplemental Figure. MOFA model additional information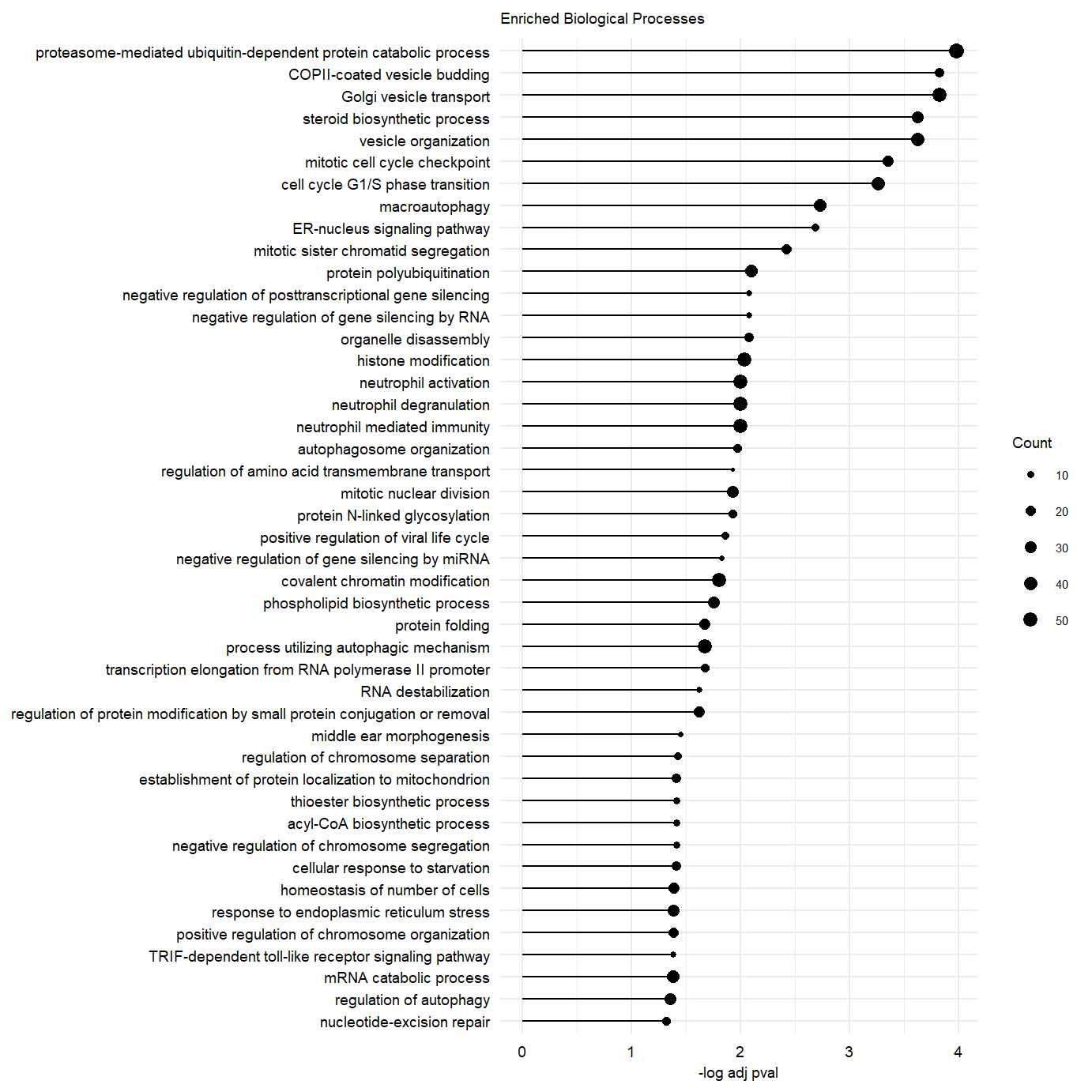 Supplemental Figure. Top 50 enriched gene-sets in postive loadings factor 3.
Supplemental Figure. Top 50 enriched gene-sets in postive loadings factor 3.