Project-1: STING-seq
<<<<<<< HEAD Last updated: 2022-08-04 ======= Last updated: 2022-07-19 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838
Checks: 7 0
Knit directory: rotation2/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220607) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
<<<<<<< HEAD Repository version: 3da8afa
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
<<<<<<< HEAD The results in this page were generated with repository version 3da8afa. ======= The results in this page were generated with repository version 3576593. >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
<<<<<<< HEAD
Unstaged changes:
Modified: .RData
Modified: .Rhistory
=======
Ignored files:
Ignored: data/
>>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/project_1.Rmd) and HTML
(docs/project_1.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | <<<<<<< HEAD 3da8afa | Hang Chen | 2022-08-04 | Build site. |
| html | 37d15c9 ======= 8f6816e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-19 | Build site. |
| html | <<<<<<< HEAD 8f6816e | chenh19 | 2022-07-19 | Build site. |
| html | 4a94b94 ======= 4a94b94 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-19 | Build site. |
| Rmd | <<<<<<< HEAD a18fc1f ======= a18fc1f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-19 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 870115f ======= 870115f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-19 | Build site. |
| Rmd | <<<<<<< HEAD 6307f5c ======= 6307f5c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-19 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 9241fe6 ======= 9241fe6 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-08 | Build site. |
| Rmd | <<<<<<< HEAD abd8a0c ======= abd8a0c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-08 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 0a7633b ======= 0a7633b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 06d53d1 ======= 06d53d1 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 63535ad ======= 63535ad >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD fe2a82b ======= fe2a82b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| Rmd | <<<<<<< HEAD 9898acd ======= 9898acd >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | update |
| html | <<<<<<< HEAD feb5923 ======= feb5923 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 9eae283 ======= 9eae283 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD c849244 ======= c849244 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 29eb161 ======= 29eb161 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 7ab5d71 ======= 7ab5d71 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 2baffc8 ======= 2baffc8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 625e7ca ======= 625e7ca >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD bb3e1aa ======= bb3e1aa >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 0e1d92d ======= 0e1d92d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 138b4fe ======= 138b4fe >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 24d6bcf ======= 24d6bcf >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 941e24b ======= 941e24b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 3a14766 ======= 3a14766 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD d425ac9 ======= d425ac9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD ca77db2 ======= ca77db2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | Build site. |
| Rmd | <<<<<<< HEAD 608cdf8 ======= 608cdf8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-07 | wflow_publish("./analysis/*.Rmd") |
| Rmd | <<<<<<< HEAD c639389 ======= c639389 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-07-06 | update |
| html | <<<<<<< HEAD e0cb7a2 ======= e0cb7a2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD ac33ff0 ======= ac33ff0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD bf05c98 ======= bf05c98 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD 1c8bb9a ======= 1c8bb9a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 7cfb685 ======= 7cfb685 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD 82fce8f ======= 82fce8f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 43dad9c ======= 43dad9c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD 20718e9 ======= 20718e9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 7aa3172 ======= 7aa3172 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD e0425d8 ======= e0425d8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 2254608 ======= 2254608 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | Build site. |
| Rmd | <<<<<<< HEAD 754150e ======= 754150e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-28 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 968a4b6 ======= 968a4b6 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | Build site. |
| html | <<<<<<< HEAD 229f924 ======= 229f924 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | Build site. |
| Rmd | <<<<<<< HEAD c7847bd ======= c7847bd >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 9750134 ======= 9750134 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | Build site. |
| Rmd | <<<<<<< HEAD 0064ff3 ======= 0064ff3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 4b38bf6 ======= 4b38bf6 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | Build site. |
| Rmd | <<<<<<< HEAD 4eb11c9 ======= 4eb11c9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 8a7980d ======= 8a7980d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-23 | Build site. |
| html | <<<<<<< HEAD 3e1478e ======= 3e1478e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | Build site. |
| html | <<<<<<< HEAD 5258612 ======= 5258612 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | Build site. |
| Rmd | <<<<<<< HEAD ecfd58d ======= ecfd58d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 2382868 ======= 2382868 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | Build site. |
| Rmd | <<<<<<< HEAD 694470f ======= 694470f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 1144127 ======= 1144127 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-22 | Build site. |
| html | <<<<<<< HEAD da2c0fe ======= da2c0fe >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| html | <<<<<<< HEAD 8aa5960 ======= 8aa5960 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 3dea9b1 ======= 3dea9b1 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 6783fa3 ======= 6783fa3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 6699b8f ======= 6699b8f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD d9be701 ======= d9be701 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD f93179d ======= f93179d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 82e6e50 ======= 82e6e50 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD f753baa ======= f753baa >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD d376ad0 ======= d376ad0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 4a9db6a ======= 4a9db6a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 325a212 ======= 325a212 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD d54cc77 ======= d54cc77 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD e14c55c ======= e14c55c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 2ab041a ======= 2ab041a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD f971bbd ======= f971bbd >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| html | <<<<<<< HEAD dca882f ======= dca882f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| html | <<<<<<< HEAD 3a80eaf ======= 3a80eaf >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD b6fb1d2 ======= b6fb1d2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 33829a5 ======= 33829a5 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD a949aec ======= a949aec >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 5b446cf ======= 5b446cf >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD c4ad45d ======= c4ad45d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 28a06ee ======= 28a06ee >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 53f2292 ======= 53f2292 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD d5b4ff0 ======= d5b4ff0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD e982ac7 ======= e982ac7 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 16400a7 ======= 16400a7 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD d6aa5b2 ======= d6aa5b2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD a324166 ======= a324166 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD ab8cbc3 ======= ab8cbc3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 27a4d51 ======= 27a4d51 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 624e791 ======= 624e791 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD f765024 ======= f765024 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| html | <<<<<<< HEAD bc55dbb ======= bc55dbb >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 8b66c3b ======= 8b66c3b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | update |
| html | <<<<<<< HEAD b3b7ed6 ======= b3b7ed6 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 405116a ======= 405116a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | update |
| html | <<<<<<< HEAD 405d57e ======= 405d57e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD bbbaab0 ======= bbbaab0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD c10a1a8 ======= c10a1a8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD a8f7999 ======= a8f7999 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 2443e38 ======= 2443e38 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 8e0c8ad ======= 8e0c8ad >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 5d192d2 ======= 5d192d2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 356550c ======= 356550c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 21e9501 ======= 21e9501 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | Build site. |
| Rmd | <<<<<<< HEAD 4a8f7ee ======= 4a8f7ee >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD b002776 ======= b002776 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-20 | Build site. |
| Rmd | <<<<<<< HEAD 6d78198 ======= 6d78198 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-20 | wflow_publish("./analysis/*.Rmd") |
| Rmd | <<<<<<< HEAD 0e1817a ======= 0e1817a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-19 | update |
| html | <<<<<<< HEAD 6211c60 ======= 6211c60 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | Build site. |
| Rmd | <<<<<<< HEAD 46e8cc3 ======= 46e8cc3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 0da18e2 ======= 0da18e2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | Build site. |
| Rmd | <<<<<<< HEAD d999122 ======= d999122 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD 0aff555 ======= 0aff555 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | Build site. |
| Rmd | <<<<<<< HEAD eda2d56 ======= eda2d56 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD a4e1e73 ======= a4e1e73 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | Build site. |
| Rmd | <<<<<<< HEAD 7229c17 ======= 7229c17 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-15 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD f0e98f9 ======= f0e98f9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD e0aa022 ======= e0aa022 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD eafa16b ======= eafa16b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 69b29f1 ======= 69b29f1 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD dfd60ce ======= dfd60ce >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 49f1922 ======= 49f1922 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("./analysis/*.Rmd") |
| html | <<<<<<< HEAD fd7271e ======= fd7271e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD 1b4d12e ======= 1b4d12e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD a6c402d ======= a6c402d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD cedad99 ======= cedad99 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 6b3f021 ======= 6b3f021 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD e3b9788 ======= e3b9788 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD aed5eed ======= aed5eed >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD c552123 ======= c552123 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 45bb6ed ======= 45bb6ed >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 81fdf42 ======= 81fdf42 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD c23765a ======= c23765a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD b5fb71c ======= b5fb71c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 10f6641 ======= 10f6641 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 152325b ======= 152325b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 1739dd9 ======= 1739dd9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 543ef4c ======= 543ef4c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| html | <<<<<<< HEAD 9b6cb27 ======= 9b6cb27 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD d8908c0 ======= d8908c0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 8674c8a ======= 8674c8a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 2972ce6 ======= 2972ce6 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD ada4068 ======= ada4068 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 7c5402e ======= 7c5402e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 0d08121 ======= 0d08121 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 6226526 ======= 6226526 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD dd2046d ======= dd2046d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 71f5d04 ======= 71f5d04 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 2c4cab1 ======= 2c4cab1 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 6e73c04 ======= 6e73c04 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD d46eaab ======= d46eaab >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD be56d9d ======= be56d9d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 02a0c26 ======= 02a0c26 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD e2c15c0 ======= e2c15c0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD abad46e ======= abad46e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 68948e3 ======= 68948e3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 741027b ======= 741027b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD 065d7e9 ======= 065d7e9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD bb15812 ======= bb15812 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD b6e0993 ======= b6e0993 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 93a27ae ======= 93a27ae >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD b4a7331 ======= b4a7331 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 27121a9 ======= 27121a9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | Build site. |
| Rmd | <<<<<<< HEAD fce1ffd ======= fce1ffd >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 44517c1 ======= 44517c1 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD cc6a40a ======= cc6a40a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD da08f11 ======= da08f11 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 05ccc35 ======= 05ccc35 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 572f6ba ======= 572f6ba >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD b8870d3 ======= b8870d3 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 719925e ======= 719925e >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD e7541fa ======= e7541fa >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 9d9615d ======= 9d9615d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 04feaa7 ======= 04feaa7 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD bbd8978 ======= bbd8978 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 0ec2bfa ======= 0ec2bfa >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD e5a5b52 ======= e5a5b52 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD c43ae1f ======= c43ae1f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 4d8bd72 ======= 4d8bd72 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 3373521 ======= 3373521 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD af21ea8 ======= af21ea8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 6e56d75 ======= 6e56d75 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD f653f7b ======= f653f7b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 2723e7f ======= 2723e7f >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD d69c892 ======= d69c892 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 34d877d ======= 34d877d >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD e72400b ======= e72400b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD c411223 ======= c411223 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 1daccd2 ======= 1daccd2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 63f46d2 ======= 63f46d2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 26adb45 ======= 26adb45 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD a6022a8 ======= a6022a8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 1215832 ======= 1215832 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 9abc4b8 ======= 9abc4b8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 7efcfe0 ======= 7efcfe0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD f18d385 ======= f18d385 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD a7c1ce0 ======= a7c1ce0 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD e991f56 ======= e991f56 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 3c9b1d9 ======= 3c9b1d9 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD ae1553a ======= ae1553a >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 34e0d02 ======= 34e0d02 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD e69aa83 ======= e69aa83 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 9be31af ======= 9be31af >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD ead84c2 ======= ead84c2 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | <<<<<<< HEAD 0f41de8 ======= 0f41de8 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 31ad035 ======= 31ad035 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD bdf3b44 ======= bdf3b44 >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| html | <<<<<<< HEAD 8d0890c ======= 8d0890c >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | Build site. |
| Rmd | <<<<<<< HEAD 26f455b ======= 26f455b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | update |
| html | <<<<<<< HEAD 26f455b ======= 26f455b >>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838 | chenh19 | 2022-06-13 | update |
1. Understand RNA-seq
a. Read about RNA-seq analysis
Yalamanchili et al. 2017: RNA-seq analysis pipeline
Some key points:
Protocol-1 (differential expression of genes):
- demuxed raw reads (FastQC)
- trimming reads (awk)
- aligning reads (TopHat2)
- counting reads (HTSeq; may filter out genes with low counts before next step)
- detect DE using counted reads (DEseq2)
- more QC (PCA/correlation heatmap)
Protocol-2 (differential usage of isoforms):
- Protocol-1
- counting isoforms (Kallisto, also check cell ranger)
- detect DU using counted isoforms (Sleuth)
- more QC (aslo PCA/correlation heatmap)
Protocol-3 (crypic splicing):
- Protocol-1
- detect differential junstions (CrypSplice)
b. Read more about RNA-seq analysis
Luecken
et al. 2019: RNA-seq analysis pipeline
Supplementary
code
c. Read the Morris paper
Some key points:
Some key ideas:
- STING-seq: Systematic Targeting and Inbition of Noncoding GWAS loci with scRNA-seq
- prioritizes candidate cis-regulatory elements (cCREs, 1kb<distance to TSS<1Mb) using fine-mapped GWAS
- selected 88 variants (in 56 loci) with enhancer activity
- dual CRISPR inhibition: dCas9 as the GPS, MeCP2 and KRAB as the repressors
- confirming dual CRISPRi efficacy: gRNAs target TSS of MRPS23, CTSB, FSCN1
- CRIPSRi on the 88 variants: two gRNAs for each variant, both within 200bp of the variant
- ECCITE-seq: captures gRNAs and epitopes
Some data processing steps and results:
- QC: remove cells with low total reads or excessive mitochondrial reads, gRNA assignment UMI>5 (9,343 cells after QC)
- Kallisto: counting read more on the official website
- Seurat: QC and
reference mapping?read more on the official website - SCEPTRE: gRNA_to_gene-expression pairwise test
- non-targeting gRNA-gene pairs: not significant (negative ctrl)
- TSS-targeting gRNA-gene pairs: expression significantly decreased (positive ctrl)
- 37 of the 88 variants were significant
- Trans-regulatory elements:
I'll come back later
Note:
- Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq)
- Expanded CRISPR-compatible CITE-seq (ECCITE-Seq)
- cDNA, HTO (Hashtag oligos), GDO (gRNAs)
- ECCITE-seq:

2. Prelim QC for raw STING-seq data
a. Download all data
Code: download.sh
b. Perform FastQC on all fastq files
Code: fastqc.sh
SRR14141135:
SRR14141136:
SRR14141137:
SRR14141138:
SRR14141139:
SRR14141140:
SRR14141141:
SRR14141142:
SRR14141143:
SRR14141144:
SRR14141145:
SRR14141146:
A brief summary:
- length:
26bpor57bp(trimmed?) - depth:
30-35x - overall quality: good (within
~40 bp)
d. Kallisto | bustools pipeline
Code: pip3-kb.sh
Code: anaconda_kallisto.sh
3. Analyze QC’ed STING-seq data
a. Install packages
Code: seurat.sh
b. Data overview
Code: overview.R
Note: about sparse
matrix
The [Expression] matrix has:
- 35,606 rows/genes/targets
- 686,612 columns/barcodes/cells
- 24,447,506,872 values in total
- 82,507,471 values that are non-zero
- 50,421,358 values that are 1
- 32,086,113 values that are bigger than 1
- 3,370,699 values that are bigger than 10
- 259,734 values that are bigger than 100
- 2,515 values that are bigger than 1,000
- 0 values that are bigger than 10,000
- 0 values that are bigger than 100,000
The [gRNA] matrix has:
- 210 rows/genes/targets
- 137,347 columns/barcodes/cells
- 28,842,870 values in total
- 2,506,474 values that are non-zero
- 1,510,919 values that are 1
- 995,555 values that are bigger than 1
- 121,554 values that are bigger than 10
- 41,071 values that are bigger than 100
- 2,232 values that are bigger than 1,000
- 20 values that are bigger than 10,000
- 0 values that are bigger than 100,000
The [Hashtag] matrix has:
- 4 rows/genes/targets
- 410,228 columns/barcodes/cells
- 1,640,912 values in total
- 739,820 values that are non-zero
- 409,830 values that are 1
- 329,990 values that are bigger than 1
- 218,280 values that are bigger than 10
- 8,155 values that are bigger than 100
- 282 values that are bigger than 1,000
- 46 values that are bigger than 10,000
- 0 values that are bigger than 100,000
c. Calculate means and non-zeros
Code: Expression_barcode_stats.R
Code: Expression_target_stats.R
Code: gRNA_barcode_stats.R
Code: gRNA_target_stats.R
Code: Hashtag_barcode_stats.R
Code: Hashtag_target_stats.R
d. Expression (cDNA) dataset
i) Expression barcodes
Code: Expression_barcode_dist_plot.R
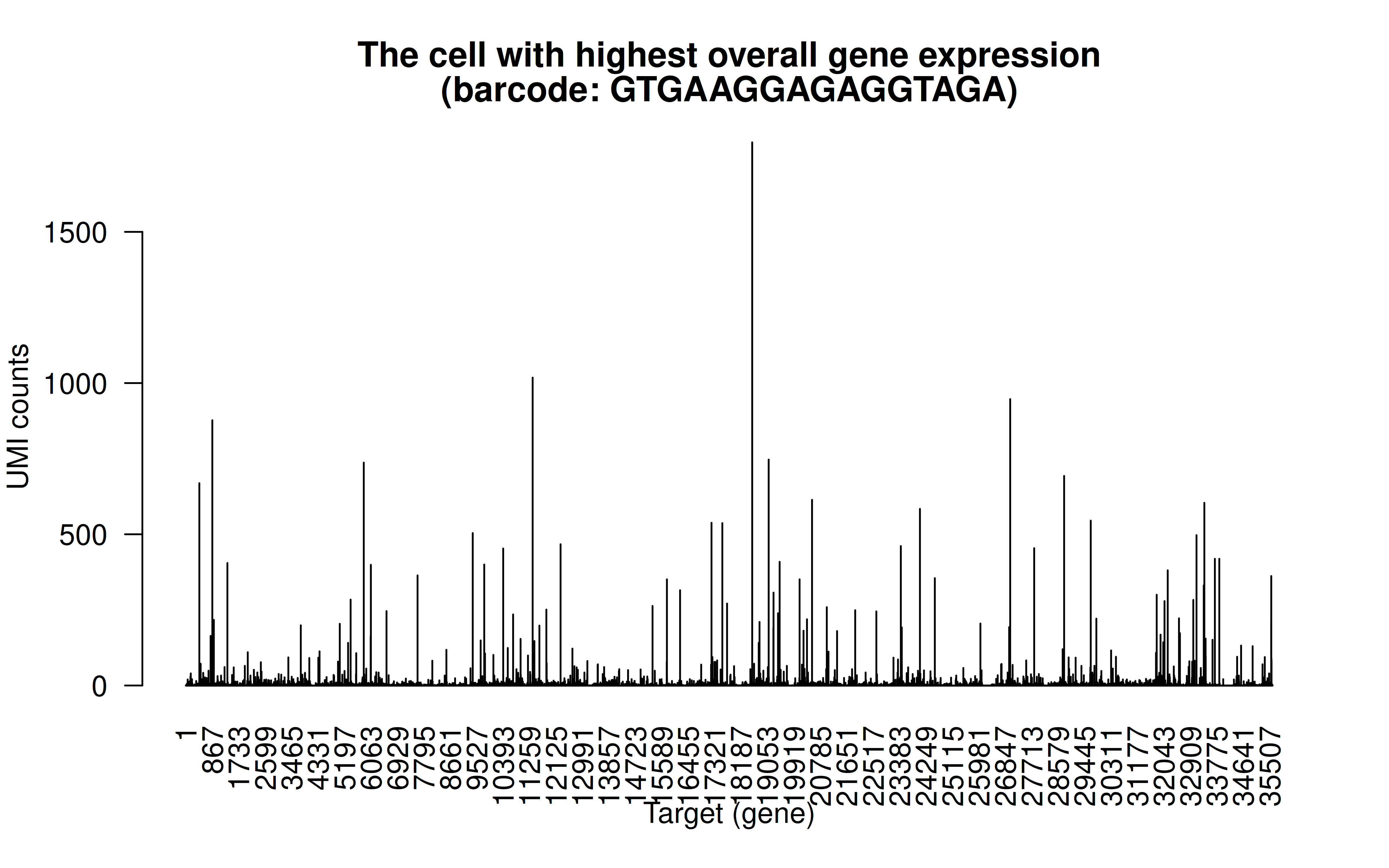
Comment: The cell with highest overall detected gene expression
Comment: The cell with lowest overall detected gene expression
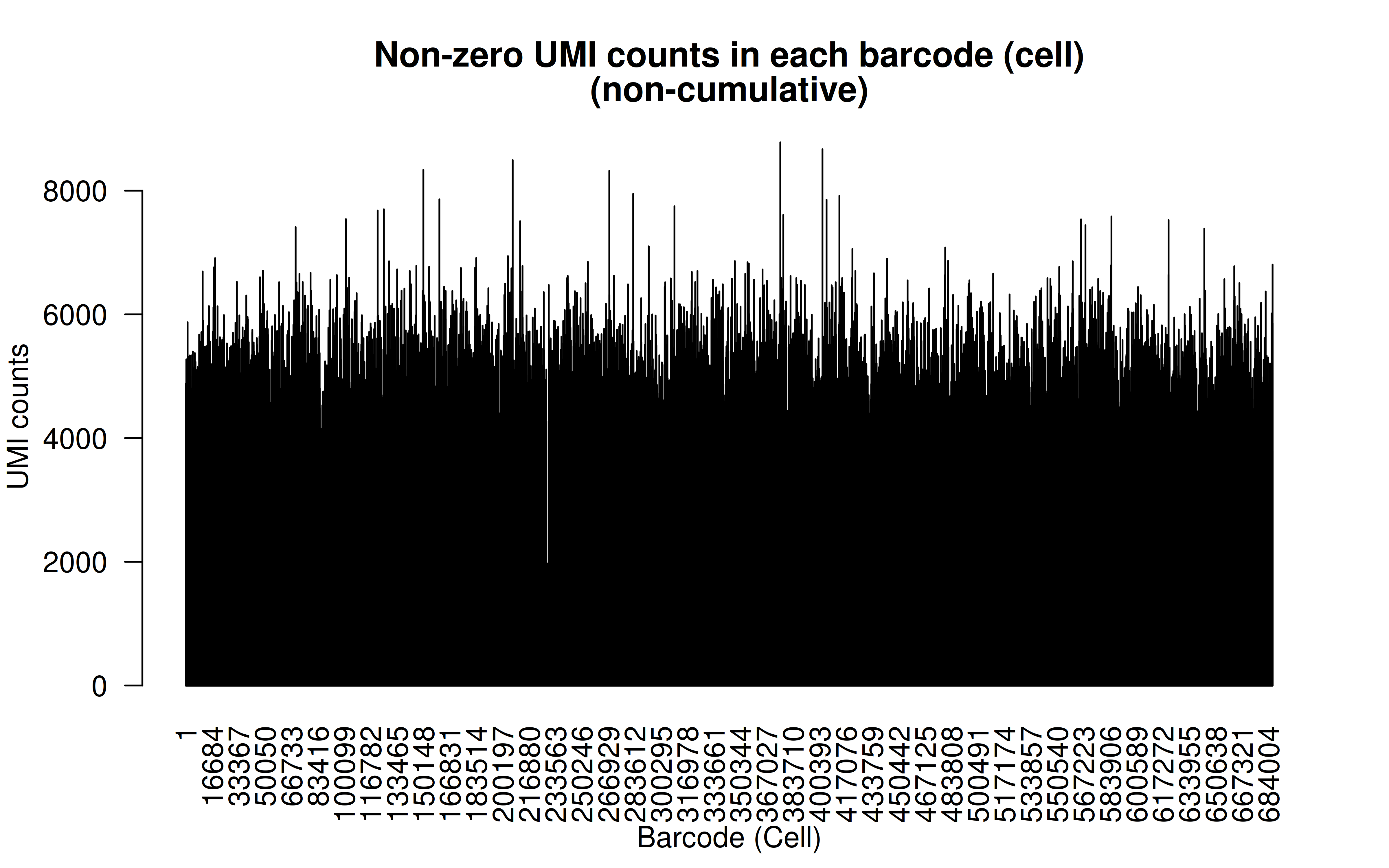
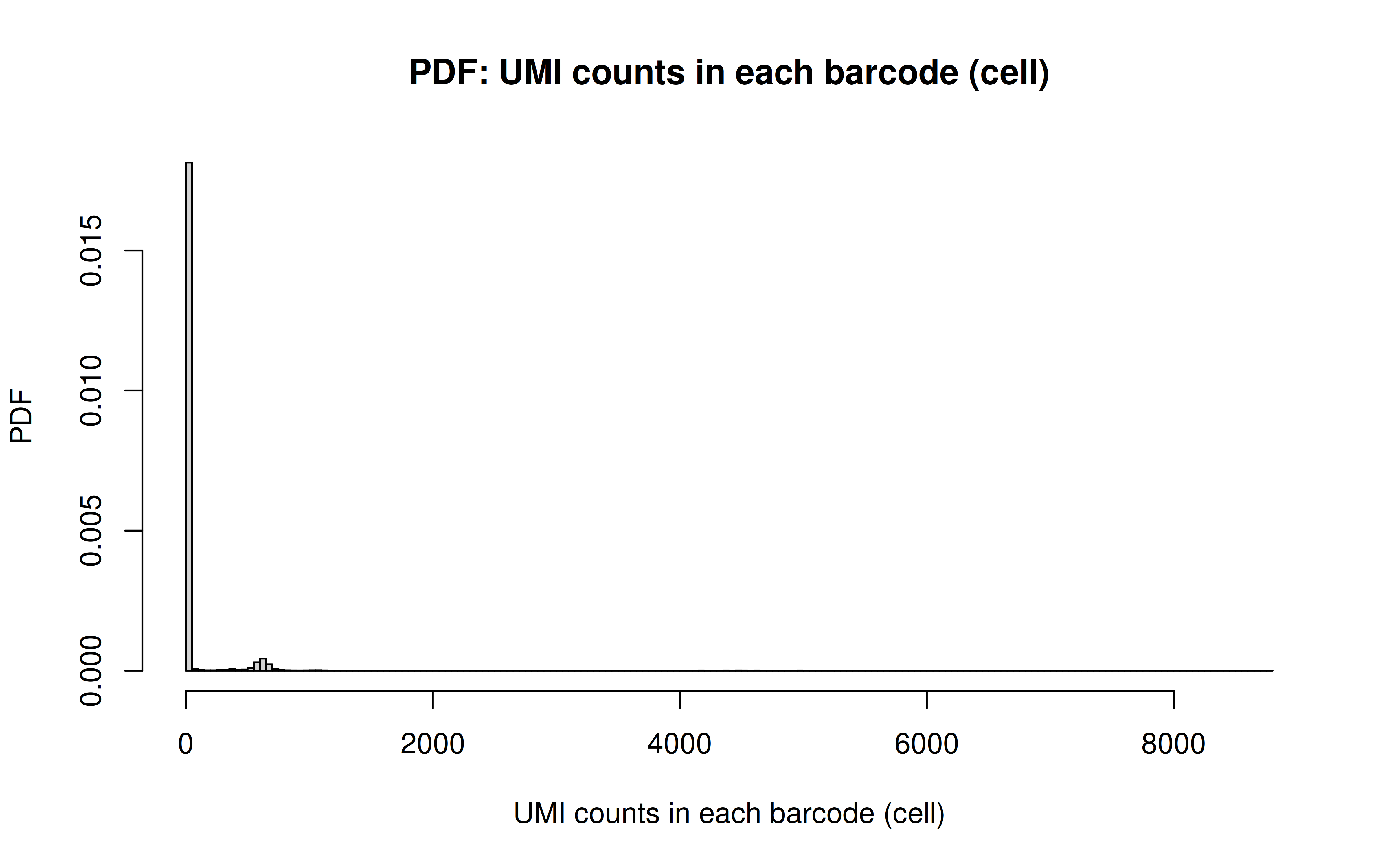
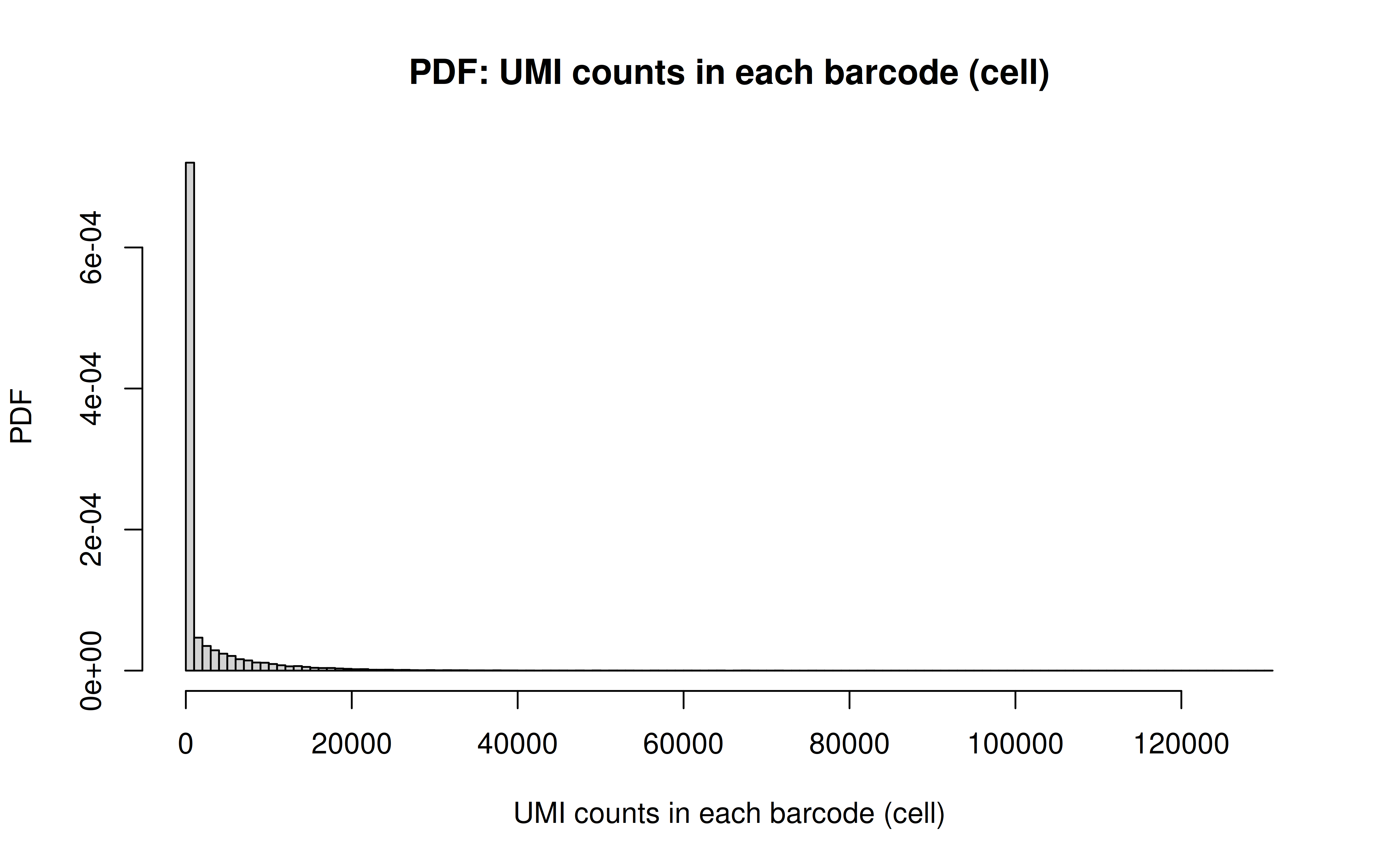
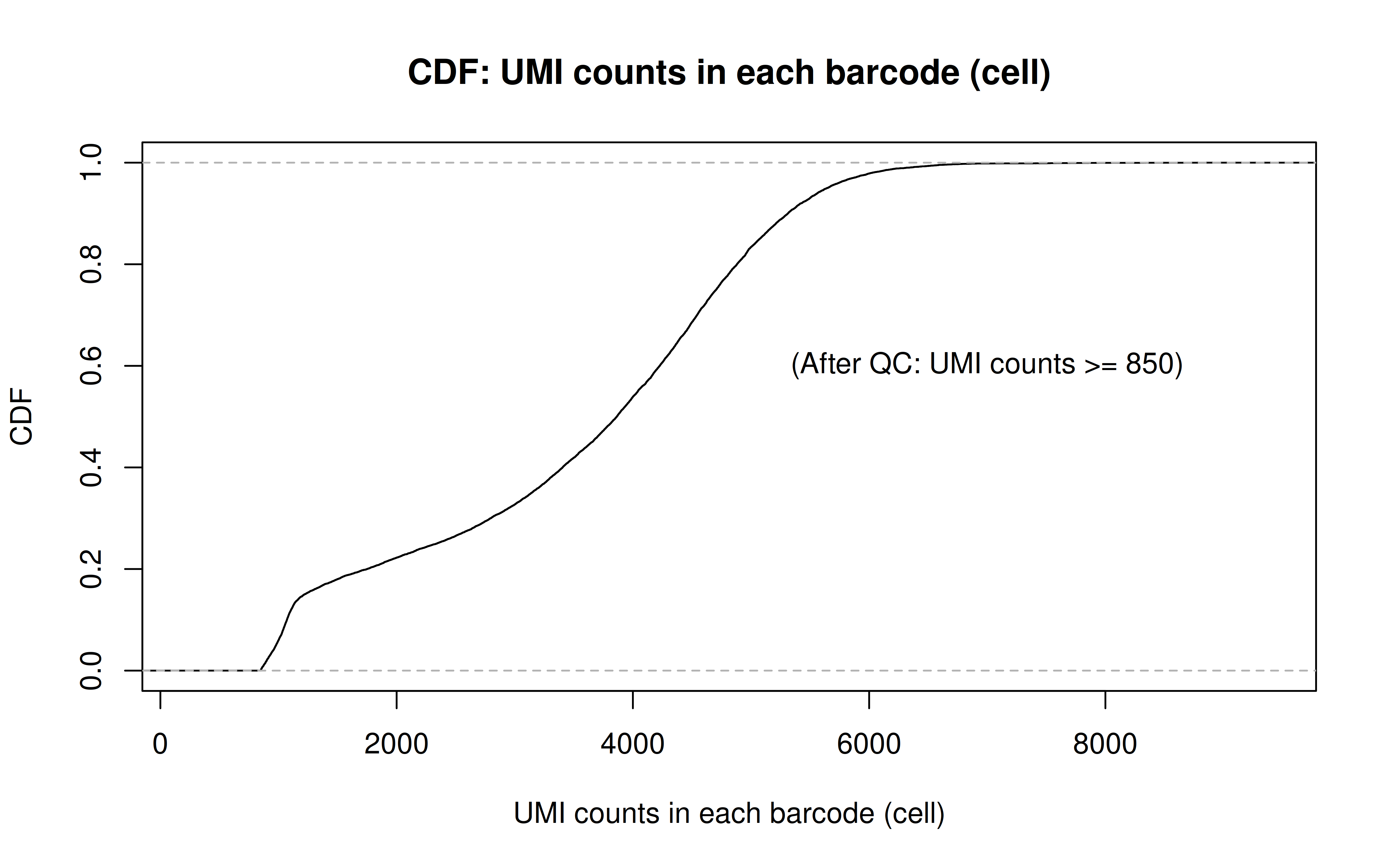
Comment: As Xuanyao said, this kind bar plot is too dense and can’t really see the overall distribution, the CDF plot below is more clear
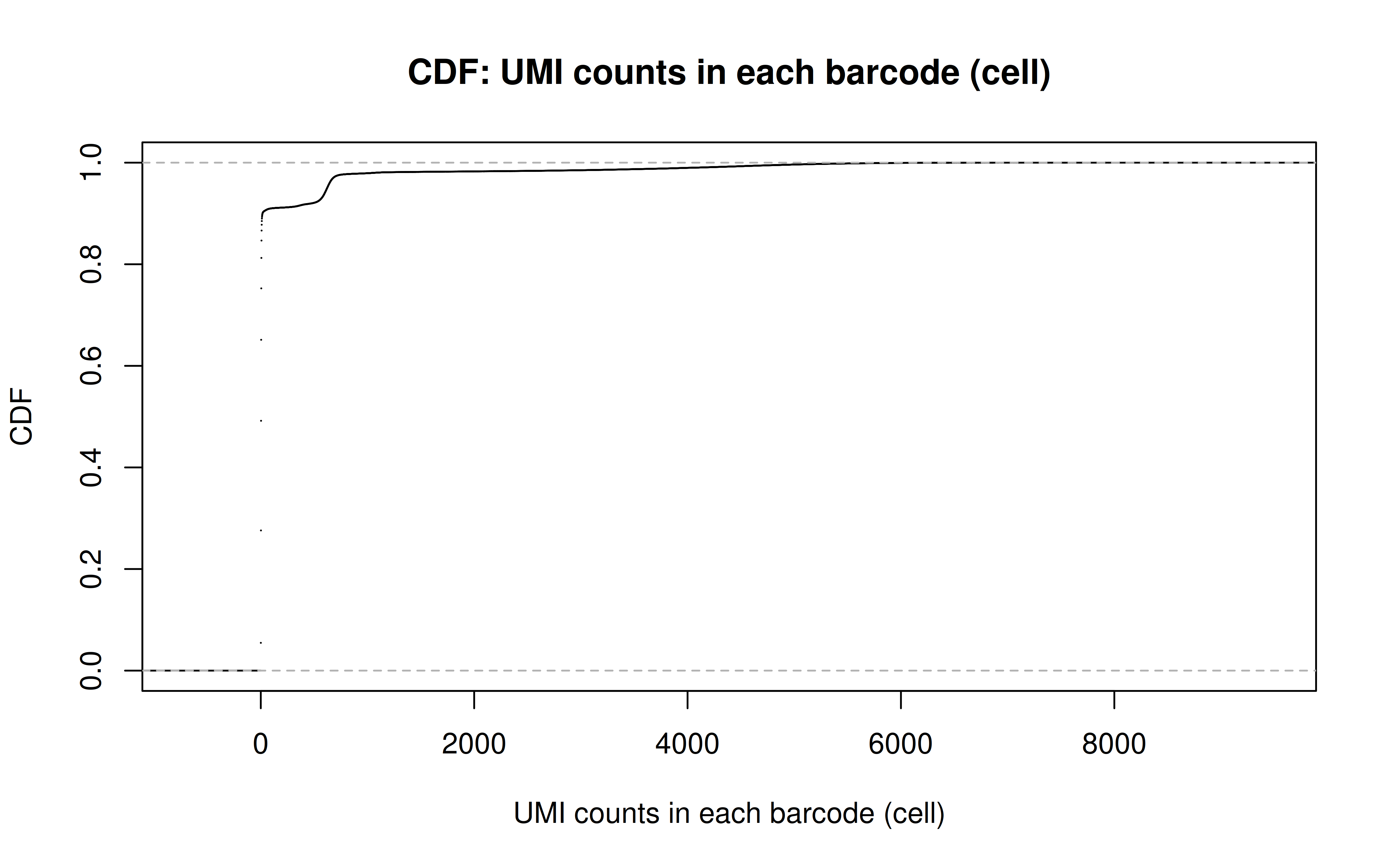
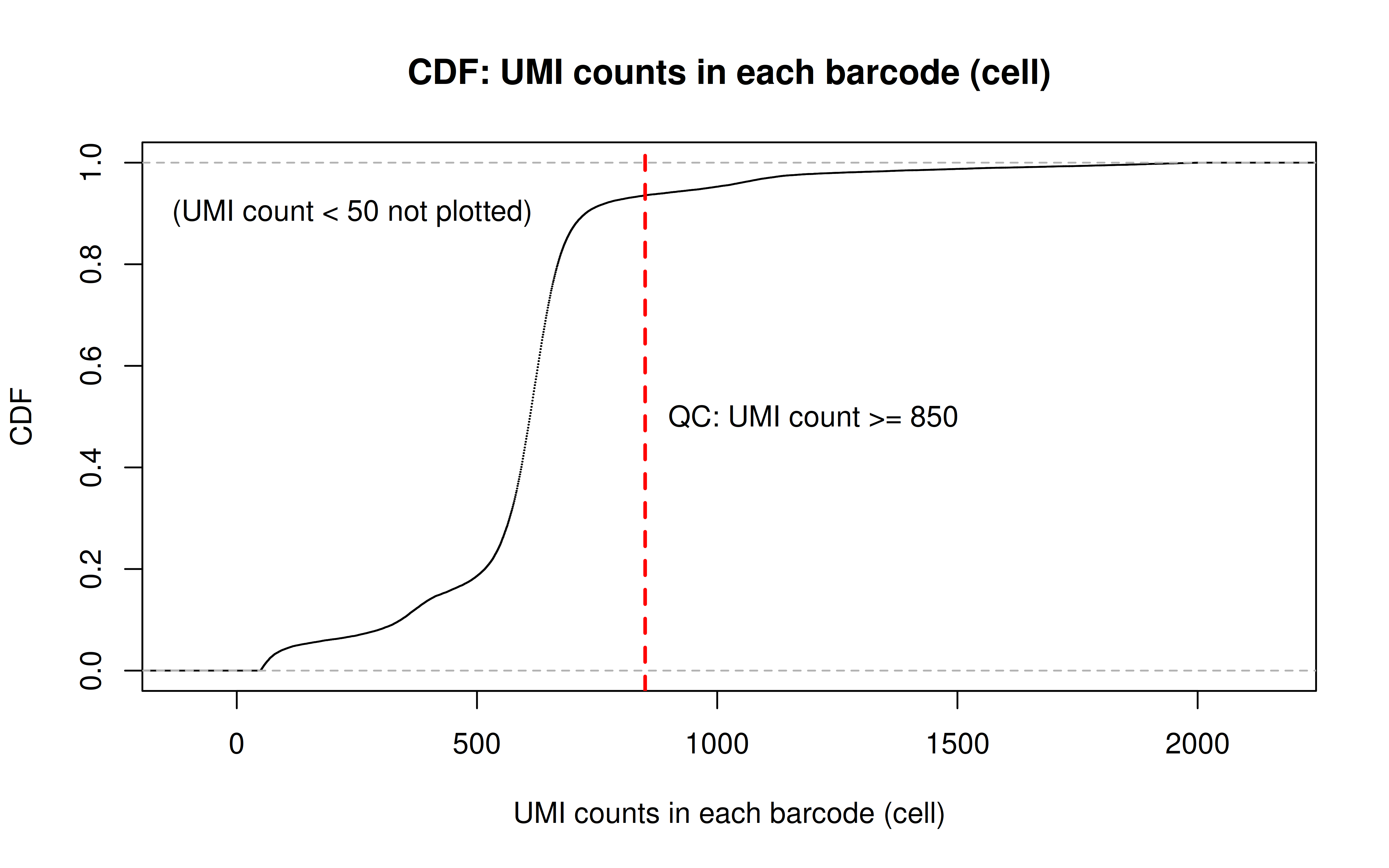
Comment: From this CDF figure I kind of know why there were only ~9000 cells used after Qc’ing with UMI>=850 filter. Less than 10% cells have UMI>=850. But still, why exactly 850 is still a question for me to explore
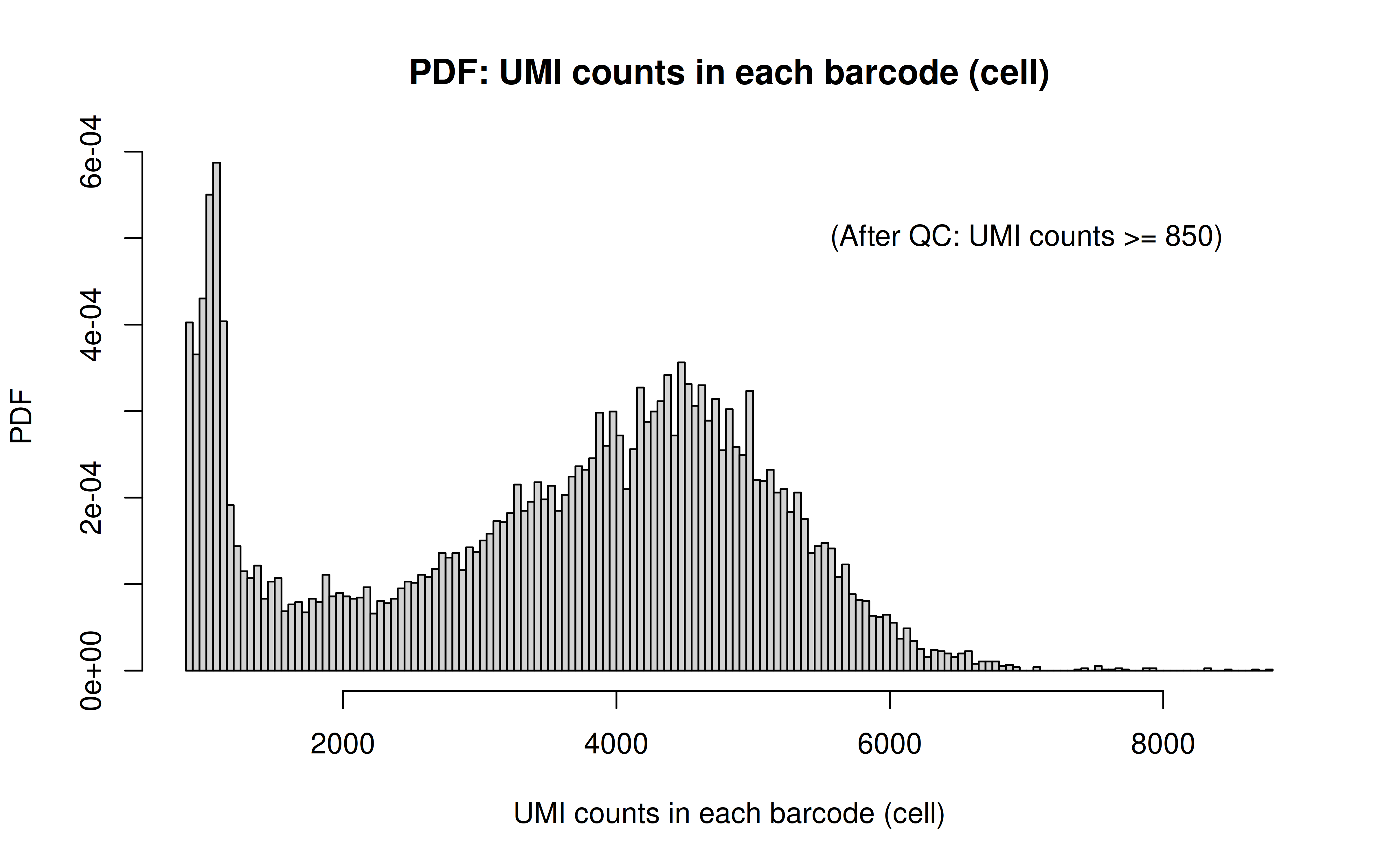
Comment: Many zeros (consistent with the observation that the matrix was very sparse); UMI>850 is invisible in this plot. As Xuanyao said, I should exclude the outliers or add y-axis break
ii) Expression targets
Code: Expression_target_dist_plot.R
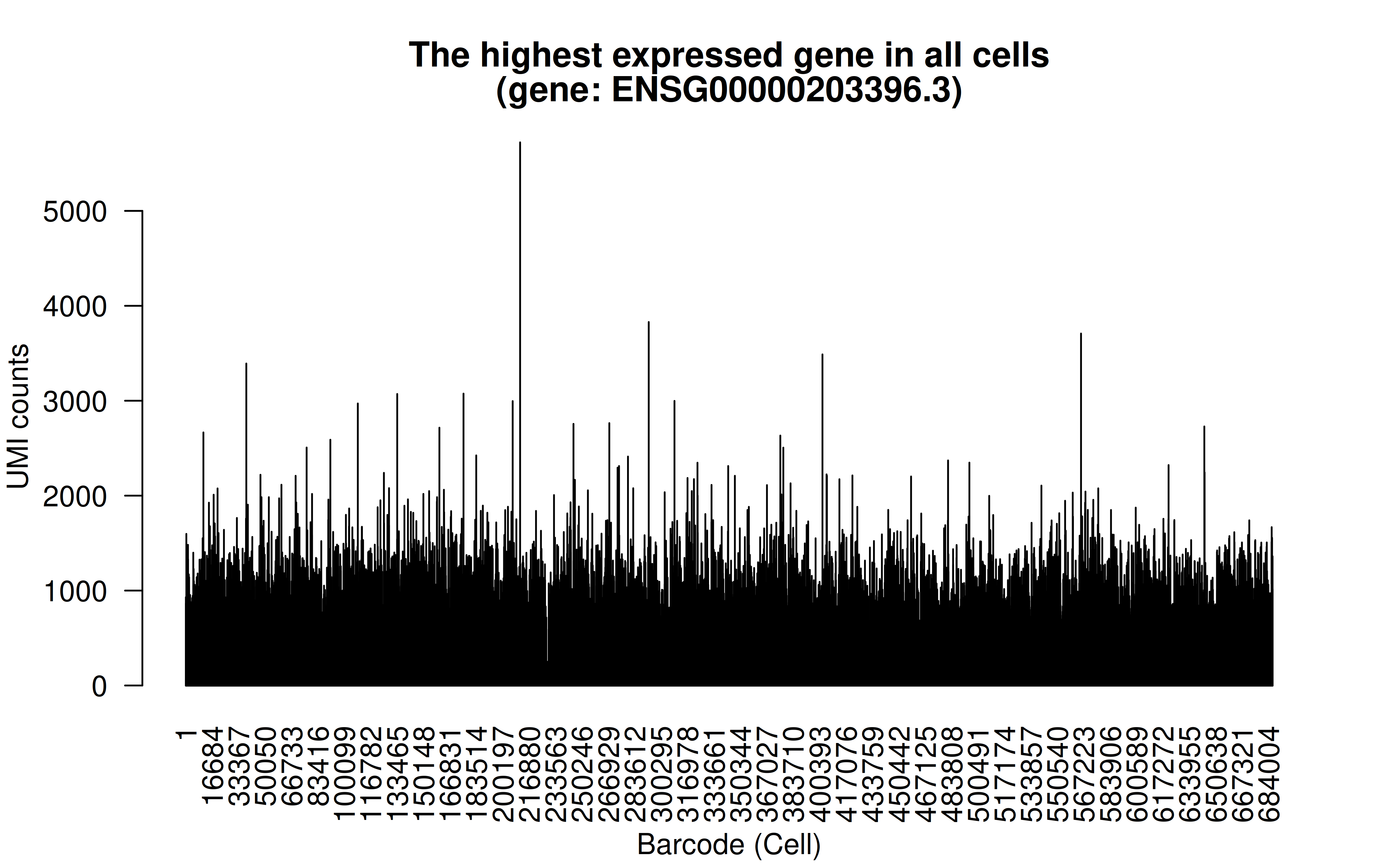
Comment: The highest (mean) expressed gene is WDR45-like (WDR45L) pseudogene (high UMI counts in all cells)
Comment: The lowest (mean) expressed gene is RP4-669L17.1 pseudogene (zero UMI counts in all cells)
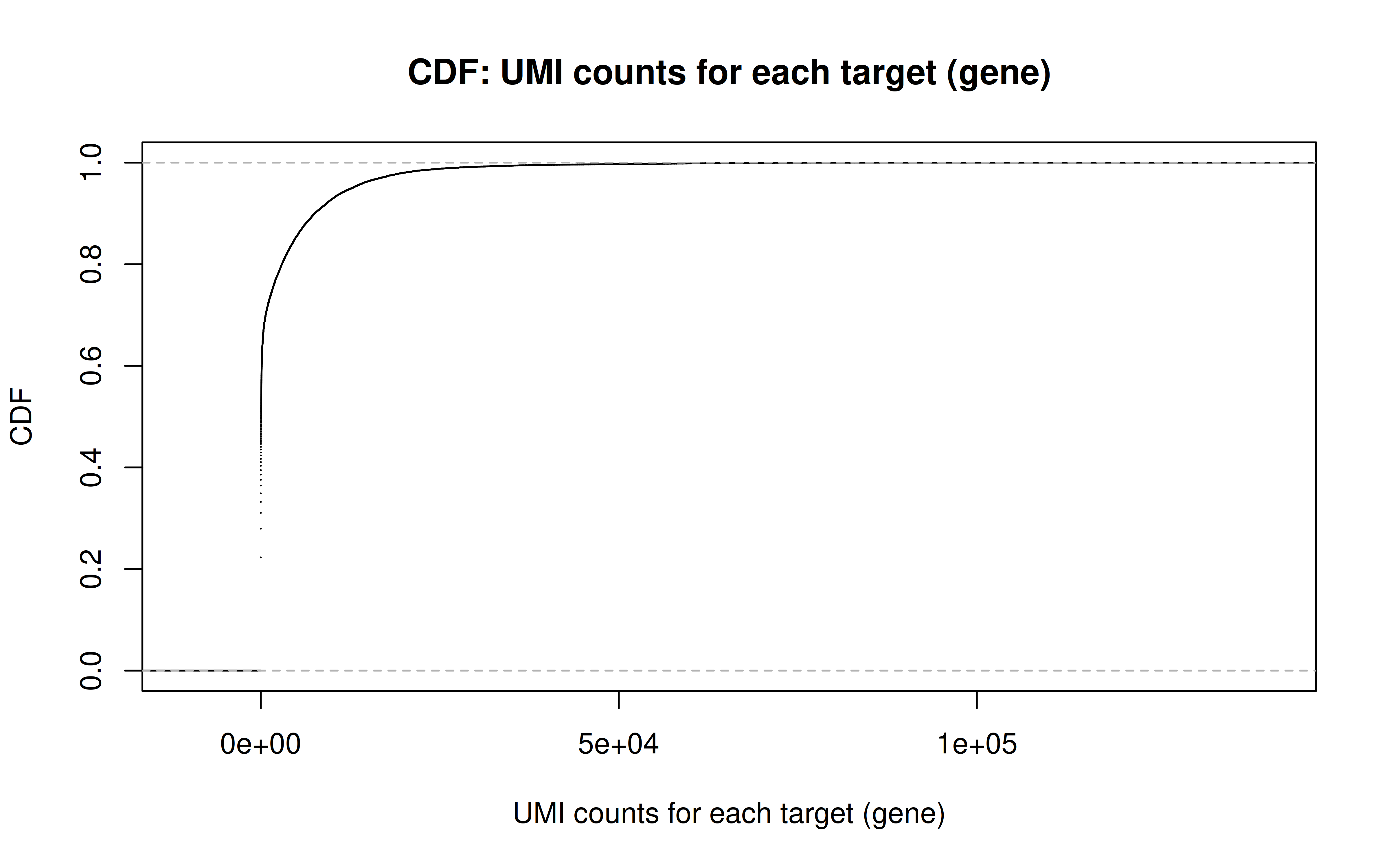
Comment: Non-zero UMI counts for all genes (~35k, including mito genes; 686,612 cells intotal)

Comment: CDF plot: ~80% genes have < ~5000 UMI counts in all cells (not all genes captured in each cell, but I guess still a lot)
Comment: PDF plot: same conclusion as above
e. gRNA dataset
i) gRNA barcodes
Code: gRNA_barcode_dist_plot.R
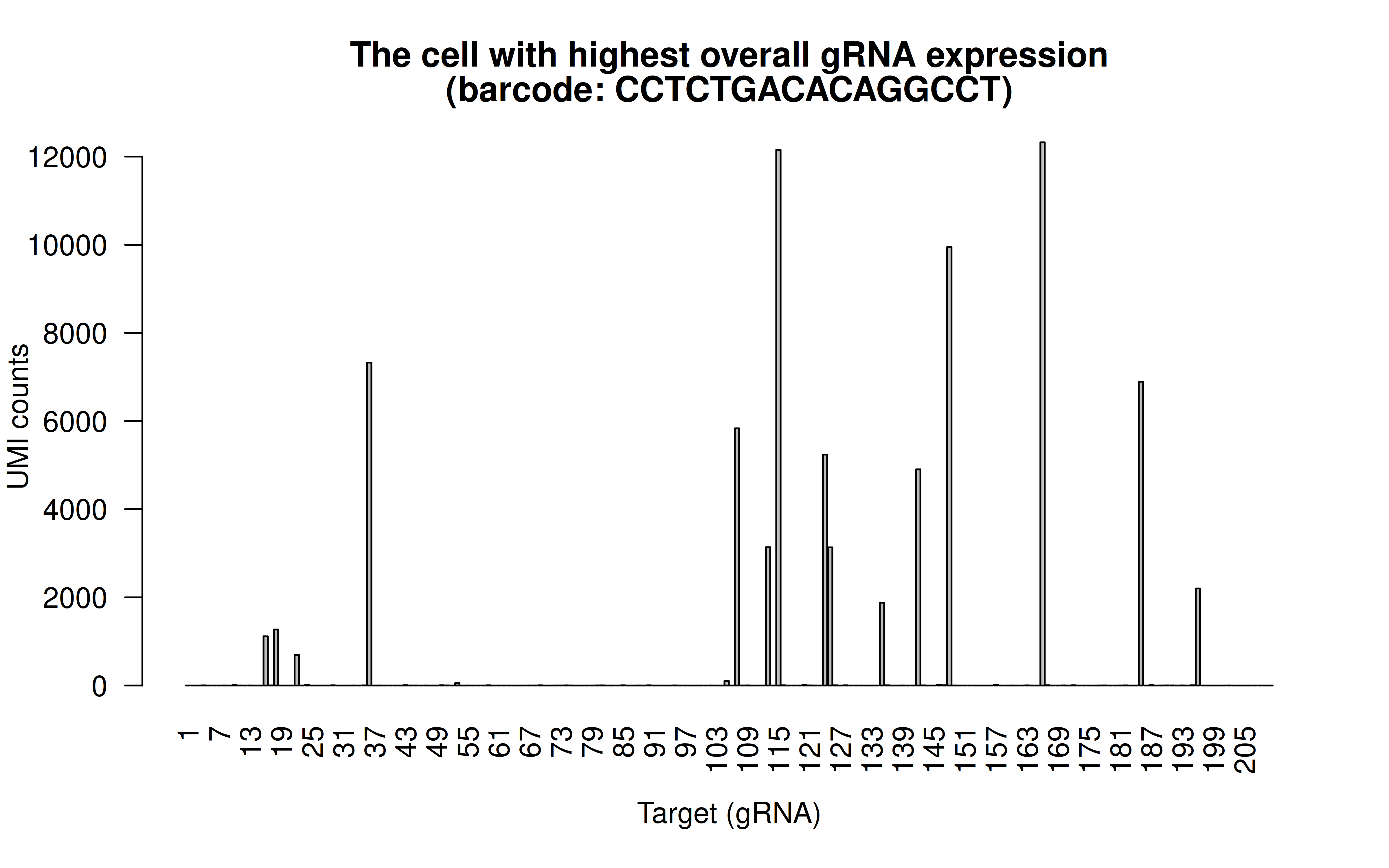
Comment: The cell with highest overall (mean) gRNAs, and it has 15 highly expressed gRNAs
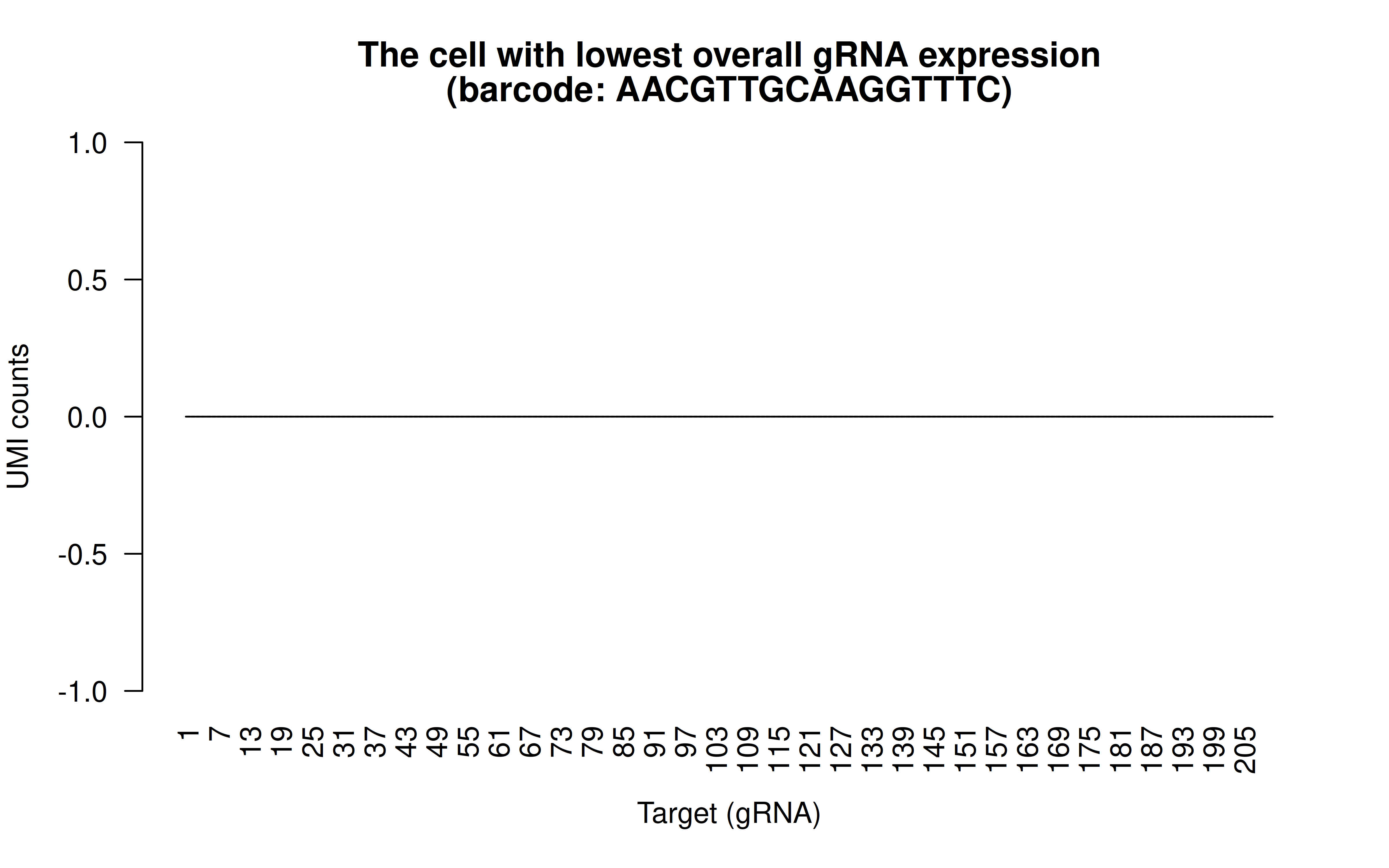
Comment: The cell with lowest overall (mean) gRNAs (transfection/transduction failed in this cell)
Comment: Non-zero UMI counts in all cells (I’d say the transfection/transduction relatively even across all cells)
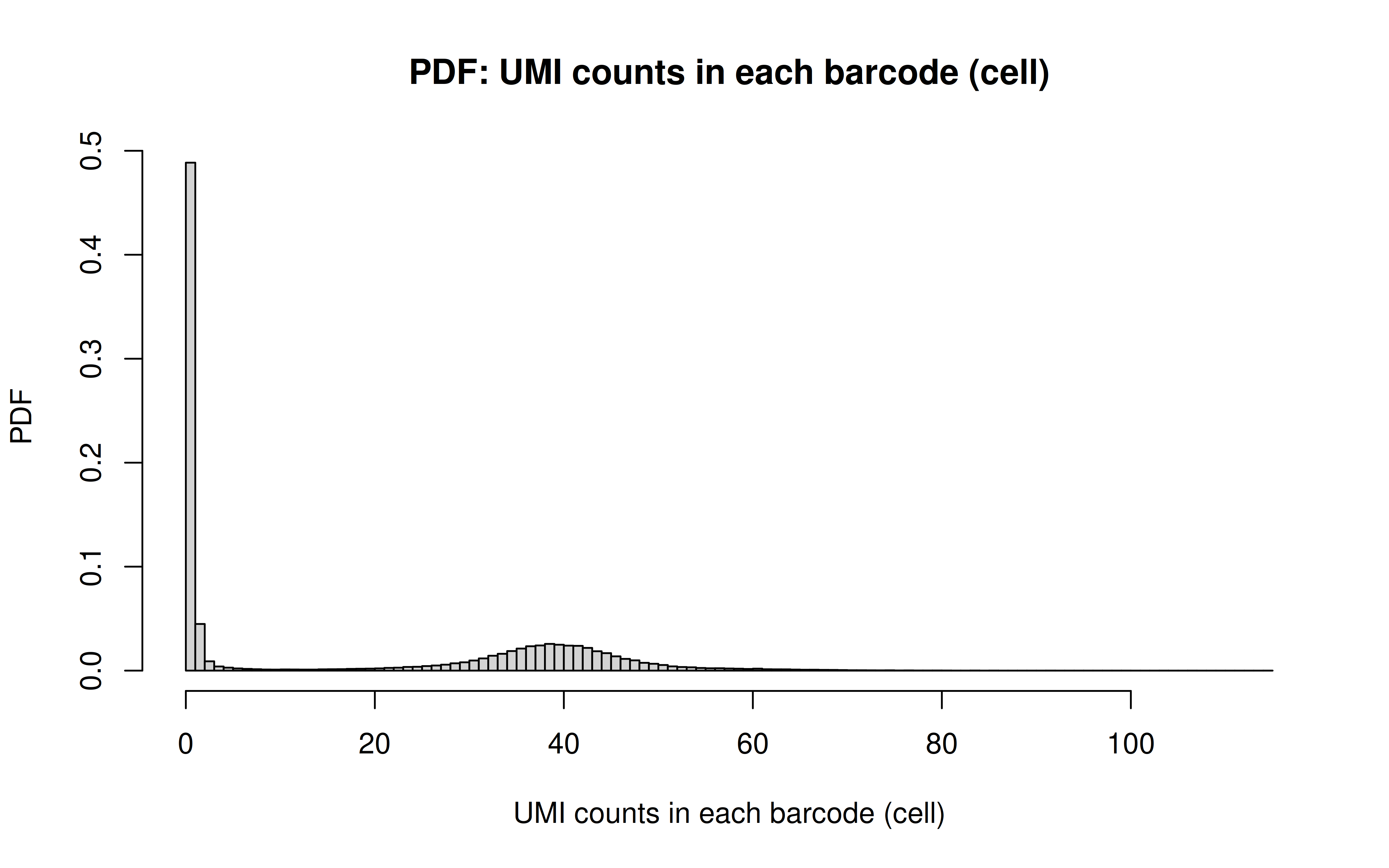
Comment: CDF plot: ~80% cells have < ~40 UMI counts for each gRNA (note: the authors mentioned MOI ~ 10)
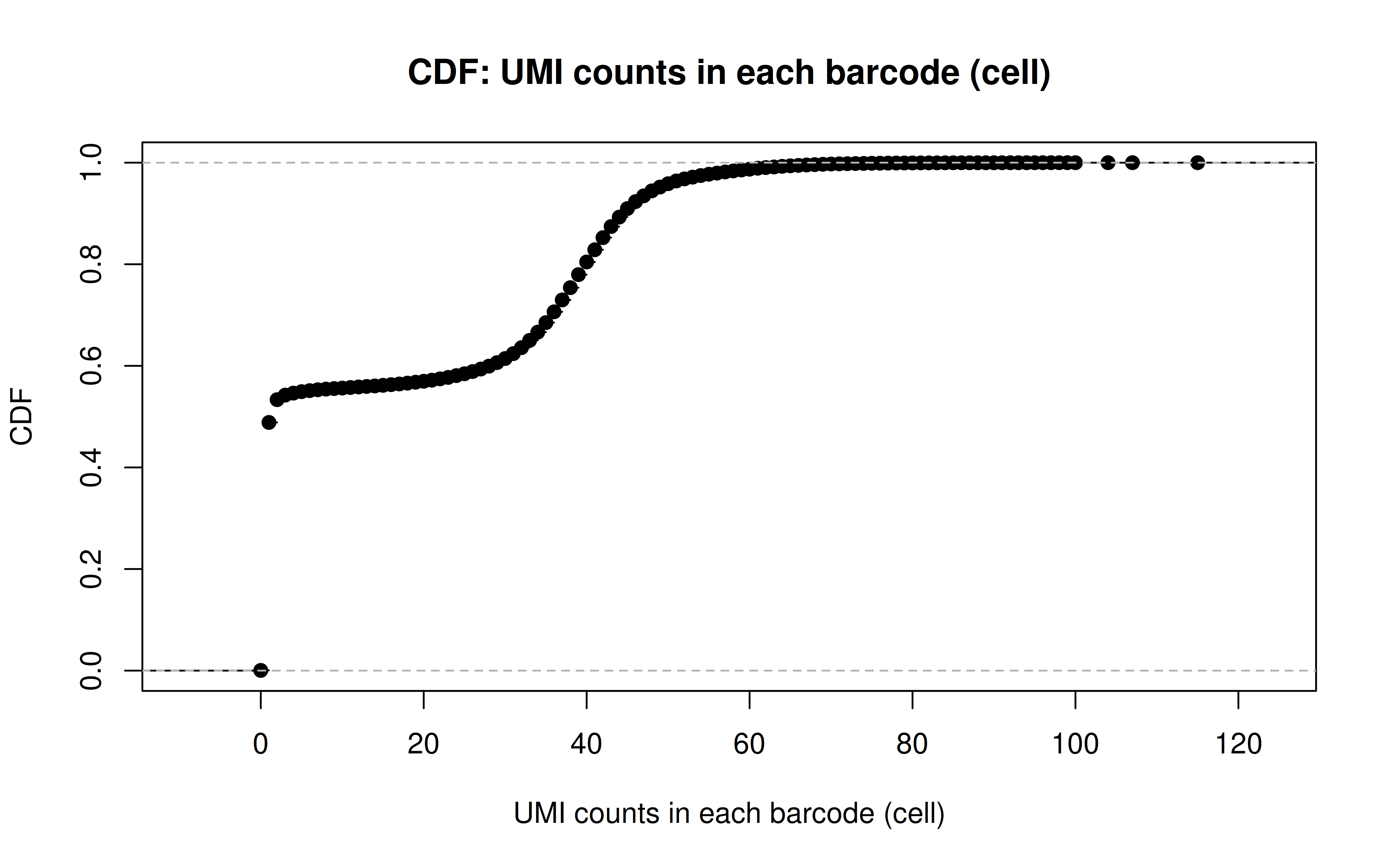
Comment: PDF plot: same conclusion as above
ii) gRNA targets
Code: gRNA_target_dist_plot.R
Comment: The highest (mean) gRNA in all cells (gRNA targeting PPIA-2, which is a control)
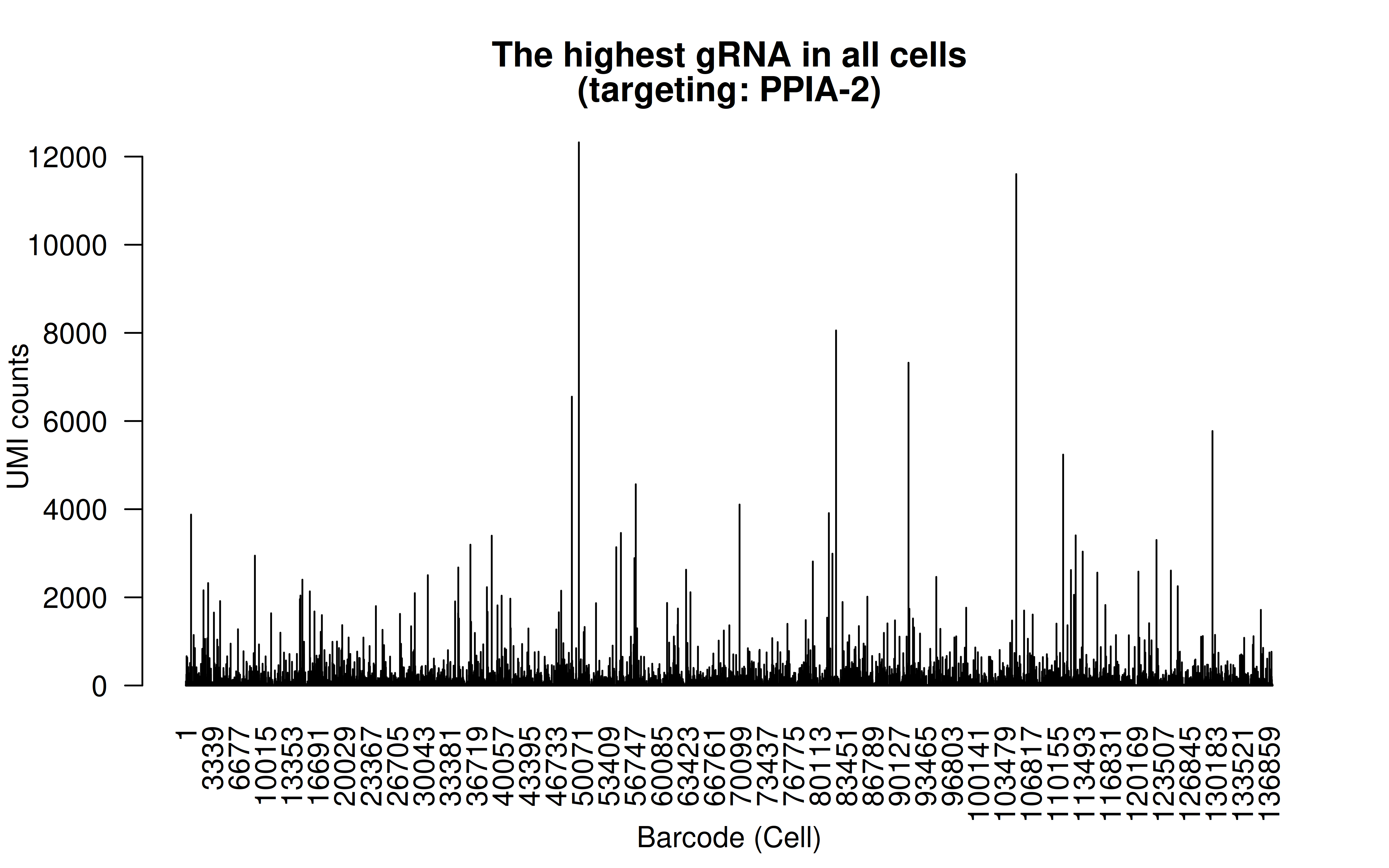
Comment: The lowest (mean) gRNA in all cells (likely it’s a low score gRNA site but the authors didn’t have better choices)
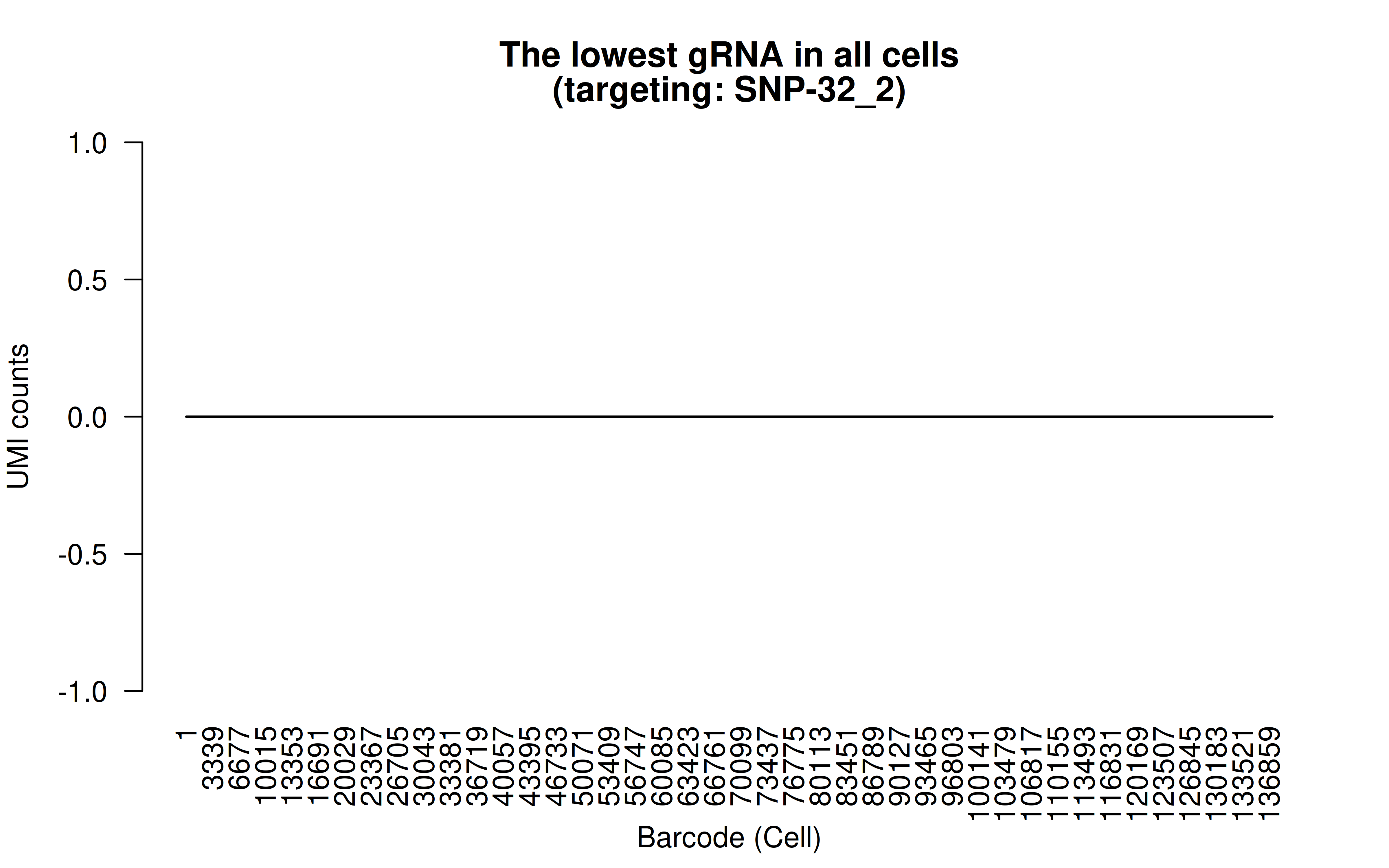
Comment: Non-zero UMI counst for all gRNAs (137,347 cells in total; I’d say the transfection/transduction efficiency varies among gRNAs. The authors designed all the gRNAs within 200bp of the targeted variants,there must be limitations in terms of gRNA options)
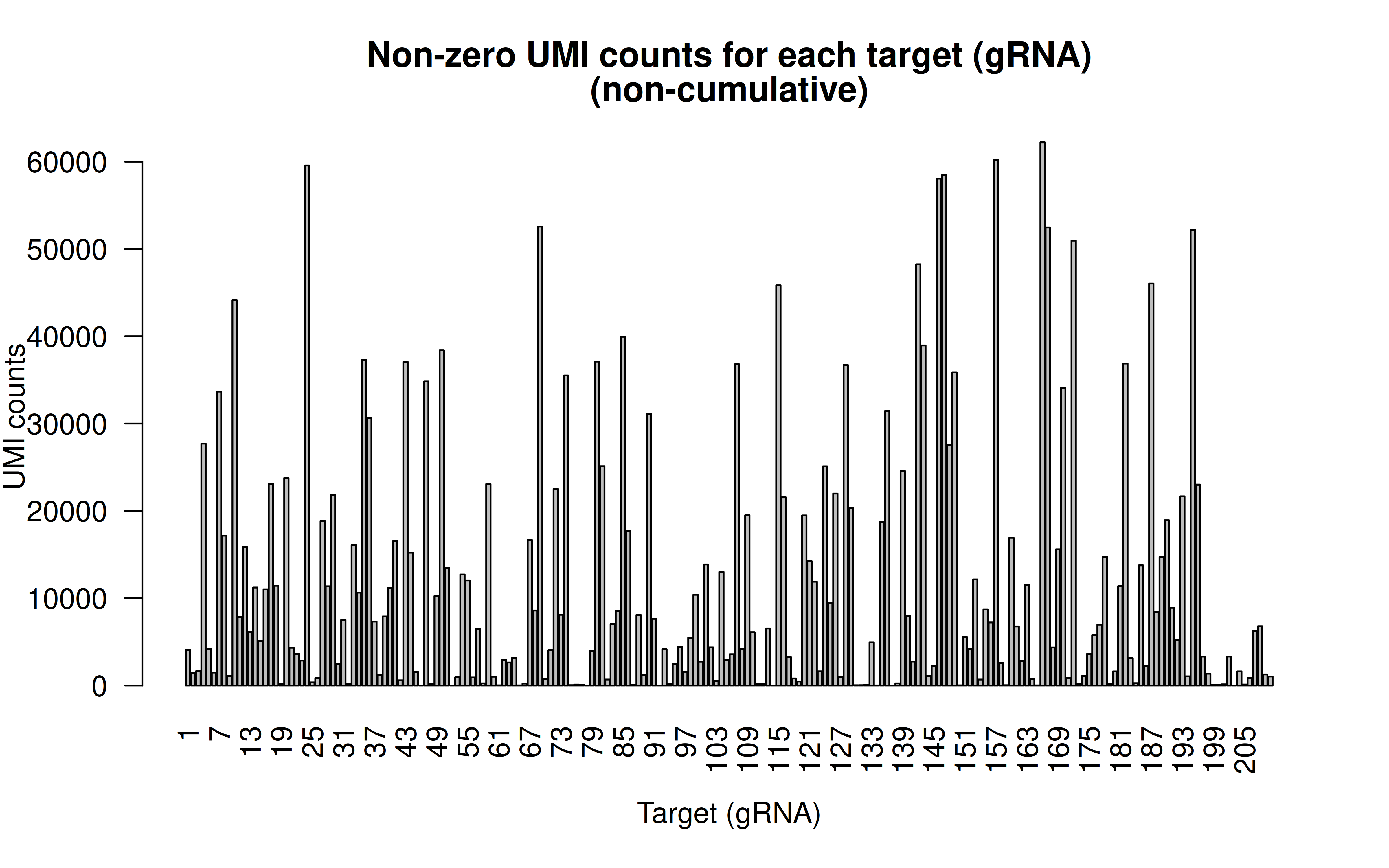
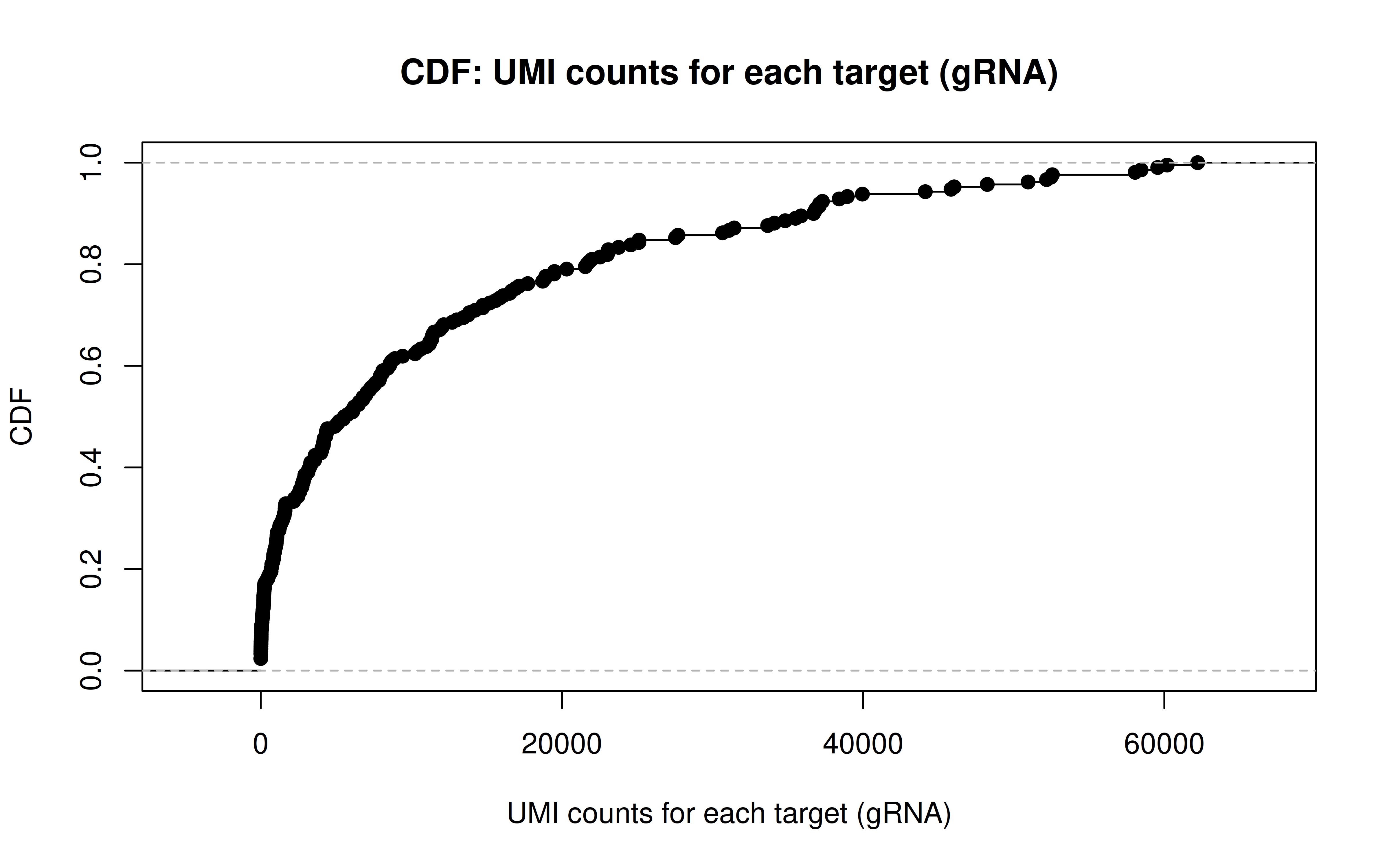
Comment: CDF plot: ~80% gRNAs have < ~20,000 UMI counts in all cells (137,347 cells in total, ~15% transfection/transduction success rate, acceptable)
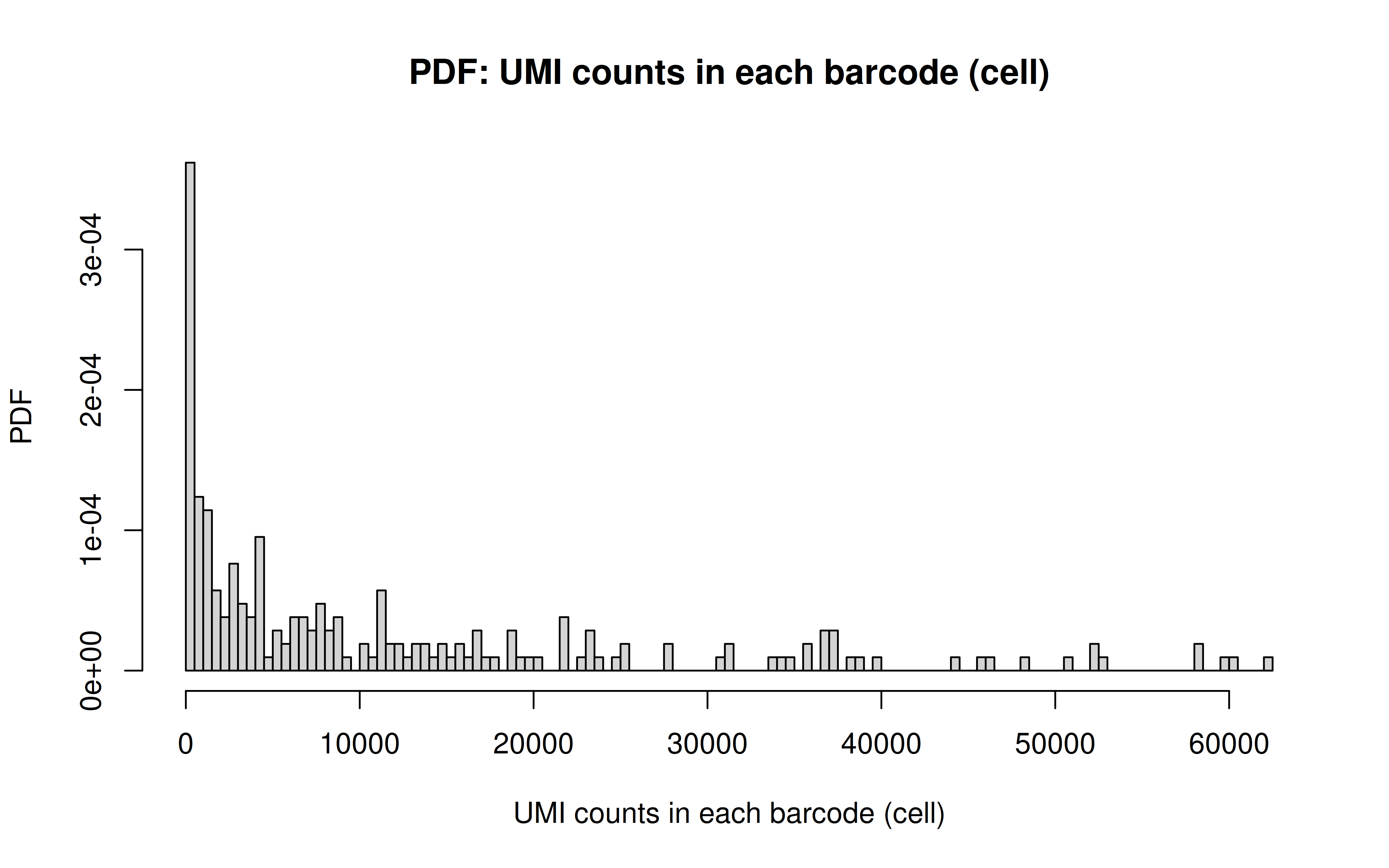
Comment: PDF plot: same conclusion as above
f. Hashtag dataset
i) Hashtag barcodes
Code: Hashtag_barcode_dist_plot.R
Comment: The cell with highest (mean) Hashtags (note: the authors used only 4 Hashtags, I might check which antibodies they are when performing association)

Comment: The cell with lowest (mean) Hashtags (not tagged by any of the antibodies)

Comment: This figure is not an error. All cells have 1/2/3/4 UMI counts, and because many of them have 4, it looks like a block when it’s such dense

Comment: CDF plot: ~80% cells have < ~2 UMI counts for each Hashtag (It make sense to me because the authors are likely trying to label different cell types)

Comment: PDF plot: same conclusion as above

i) Hashtag targets
Code: Hashtag_target_dist_plot.R
Comment: The highest (mean) Hashtag (HTO23) in all cells (I would guess this is the relatively more common cell type, also, there were some non-specific antibody binding)

Comment: The lowest (mean) Hashtag (HTO25) in all cells (I would guess this is the relatively less common cell type, also, it dosen’t seem to overlap with HTO25, which is a good thing)

Comment: Non-zero UMI counts for the 4 Hashtags (I’d say the 4 cell types are relatively even)

Comment: CDF plot: ~80% Hashtags have < ~200,000 UMI counts in all cells (410,228 cells in total, I thinking the antibody binding efficiency is pretty good)

Comment: PDF plot: same conclusion as above

g. QC filtering
- After preliminary filtering, the authors got
14,775 cellswith3,875 median genes per cell.
Previous question:
- why
percent-mito < 20%? - why
UMI > 850? - why
no UMI upper limit?
My understanding:
- Previously, 5% was usually the default threshold. But a recent paper did systematic evaluation and proposed a default cutoff of 10% for human cells. Also, according to Luecken et al. 2019, we can use a relatively loose QC cutoff at the beginning. Since the percent-mito was just in the first QC filtering step and the author further filtered by HTO and GDO, I think 20% makes sense.
- Again, according to Luecken et al. 2019, the dying/dead cells would be a small peak with low UMI counts. By the zoomed-in plot below, we can see 850 is a reasonable cutoff to remove the entire peak.
- Doublets was not filtered out by UMI, but by HTO demuxing, therefore the authors didn’t set an upper limit.
Notes:
- In the original cDNA feature txt file, there are only Ensembl gene IDs. To calculate percent-mito, I tried to convert the IDs to symbols using both this web tool and biomaRT package in R.
- If I directly use the gene symbols from these two methods and the
same filters by Nikita, I get exactly the same
results as Nikita (
14,813 cellsretained). - However, after taking a closer look at the converted gene lists,
there are still many “Mitochondrially Encoded” genes
starting with “
MT” rather than “MT-”, so I wrote a script to convert all these genes. - Then I performed filtering again and got
14,675 cellswith3,917 median genes per cell. - I don’t think we’ll know exactly how the authors filtered the cells unless they release their code.
- After QC filtering, there were
9,391 cellsretained (9,343 cellsby the authors in comparison, - I did cross comparison. There are
508 cellsfrom authors’ list not in my list.
Code: QC_filter.R
Code: UMI_plot.R
Before UMI count filtering:
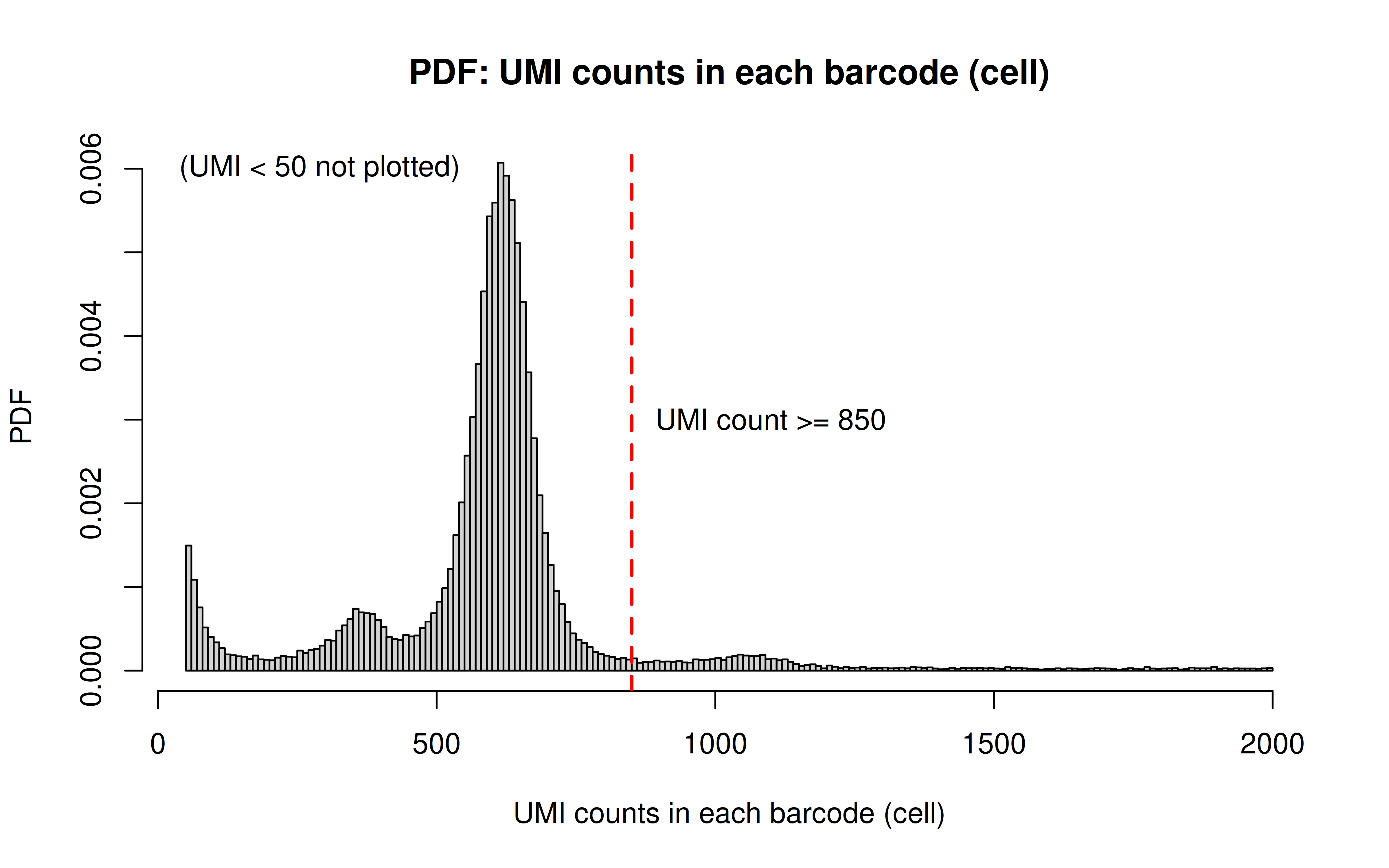
After UMI count filtering:
Before percent-mito filtering (generated by Seurat):
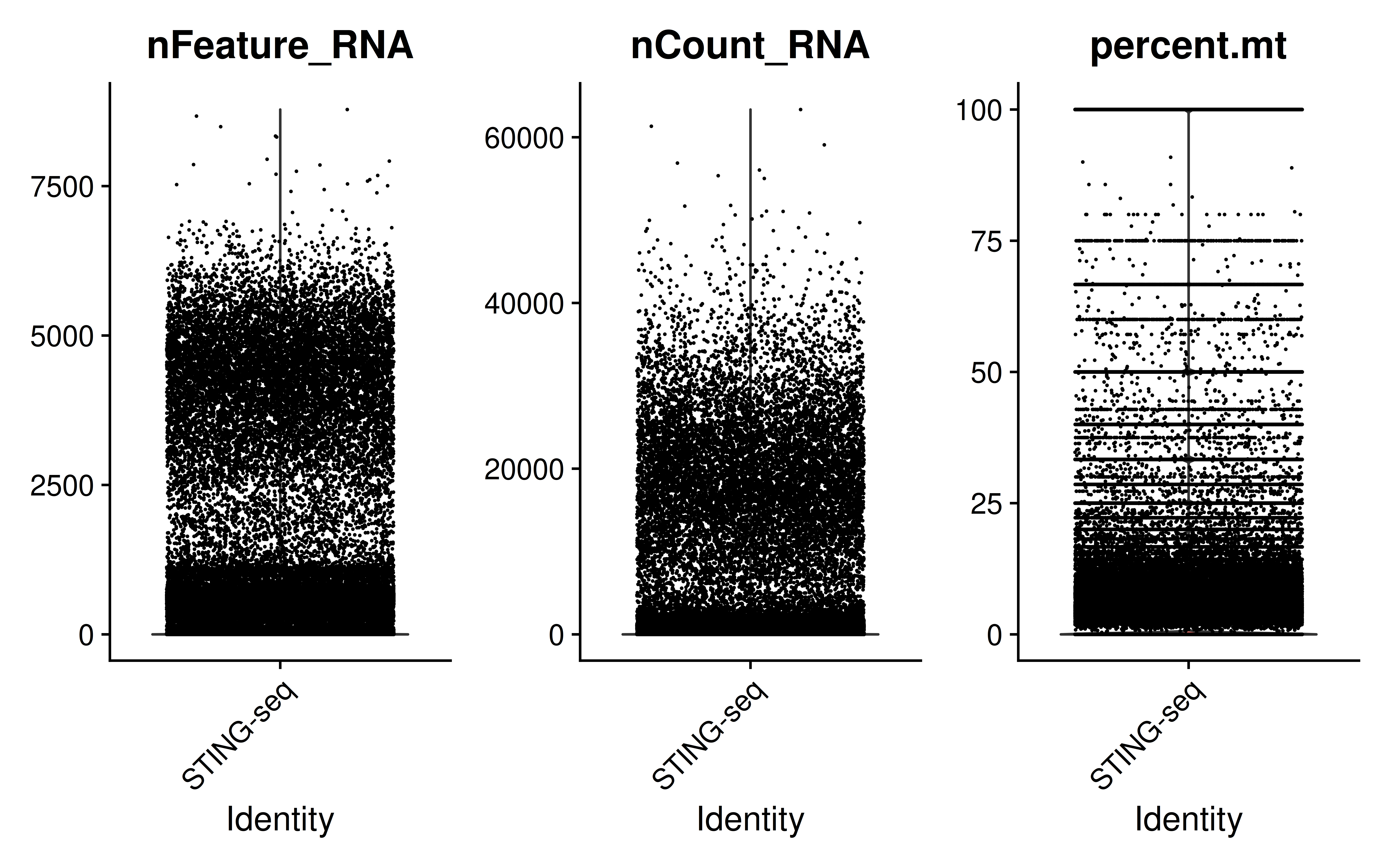
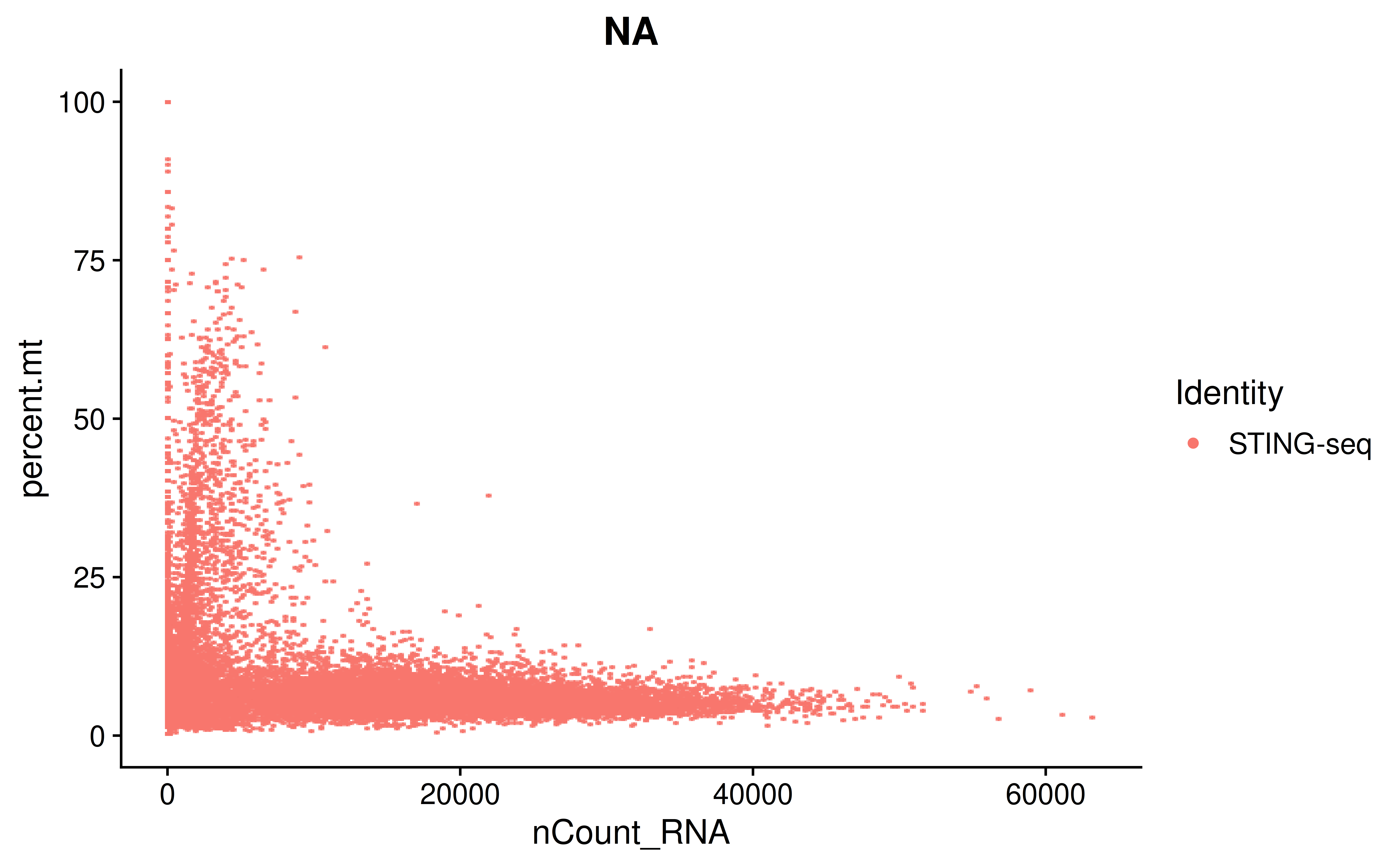
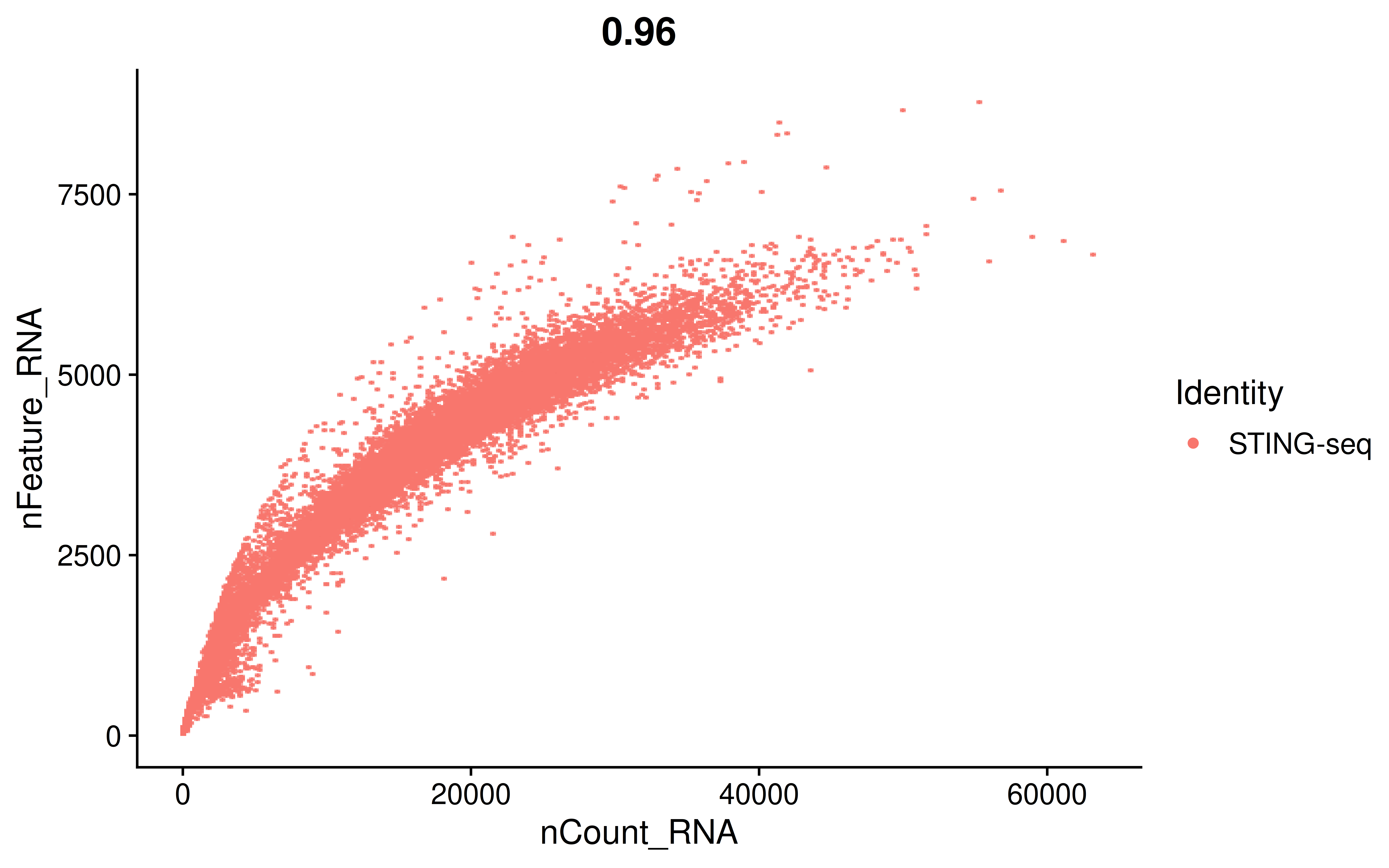
After percent-mito filtering (generated by Seurat):

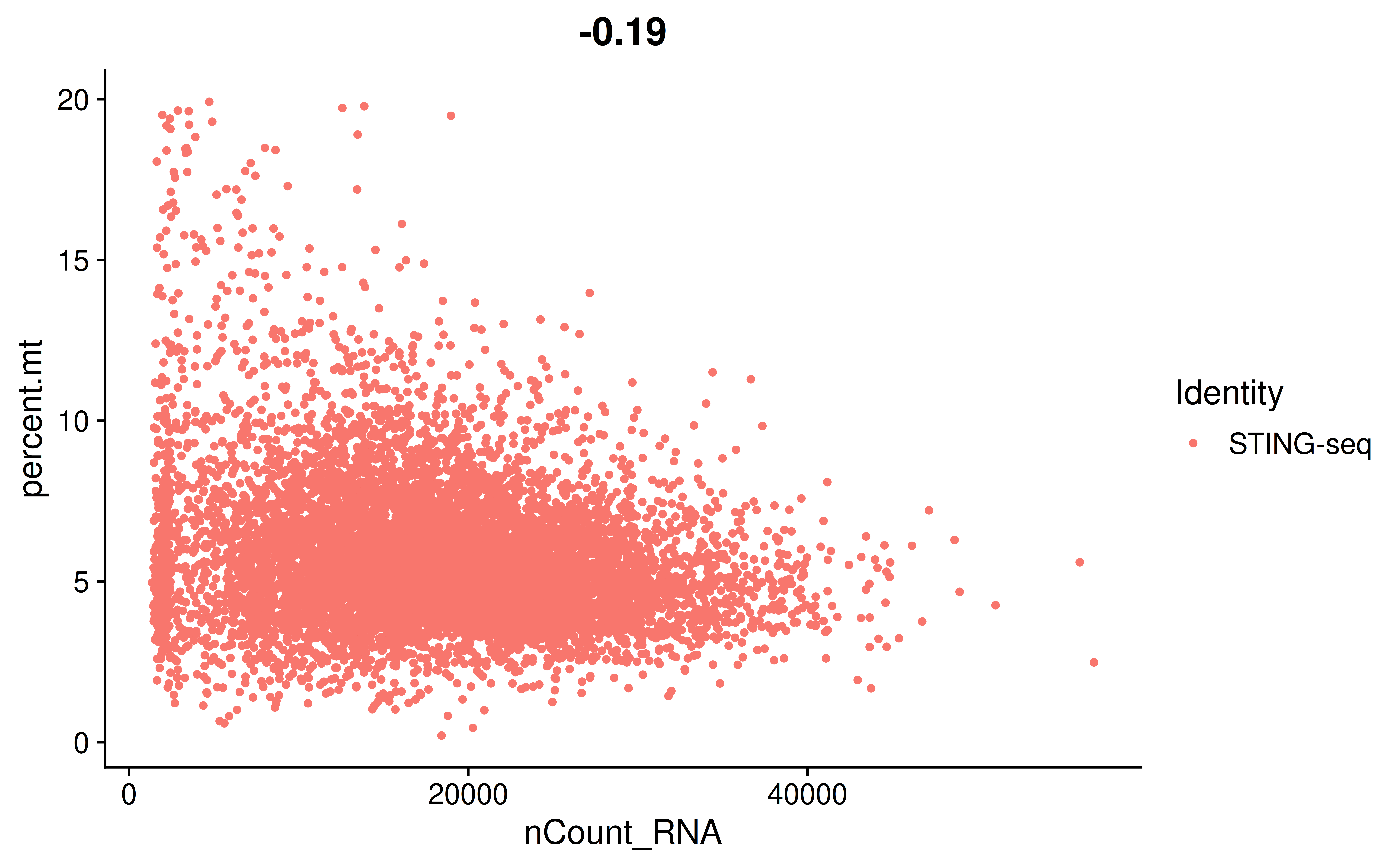
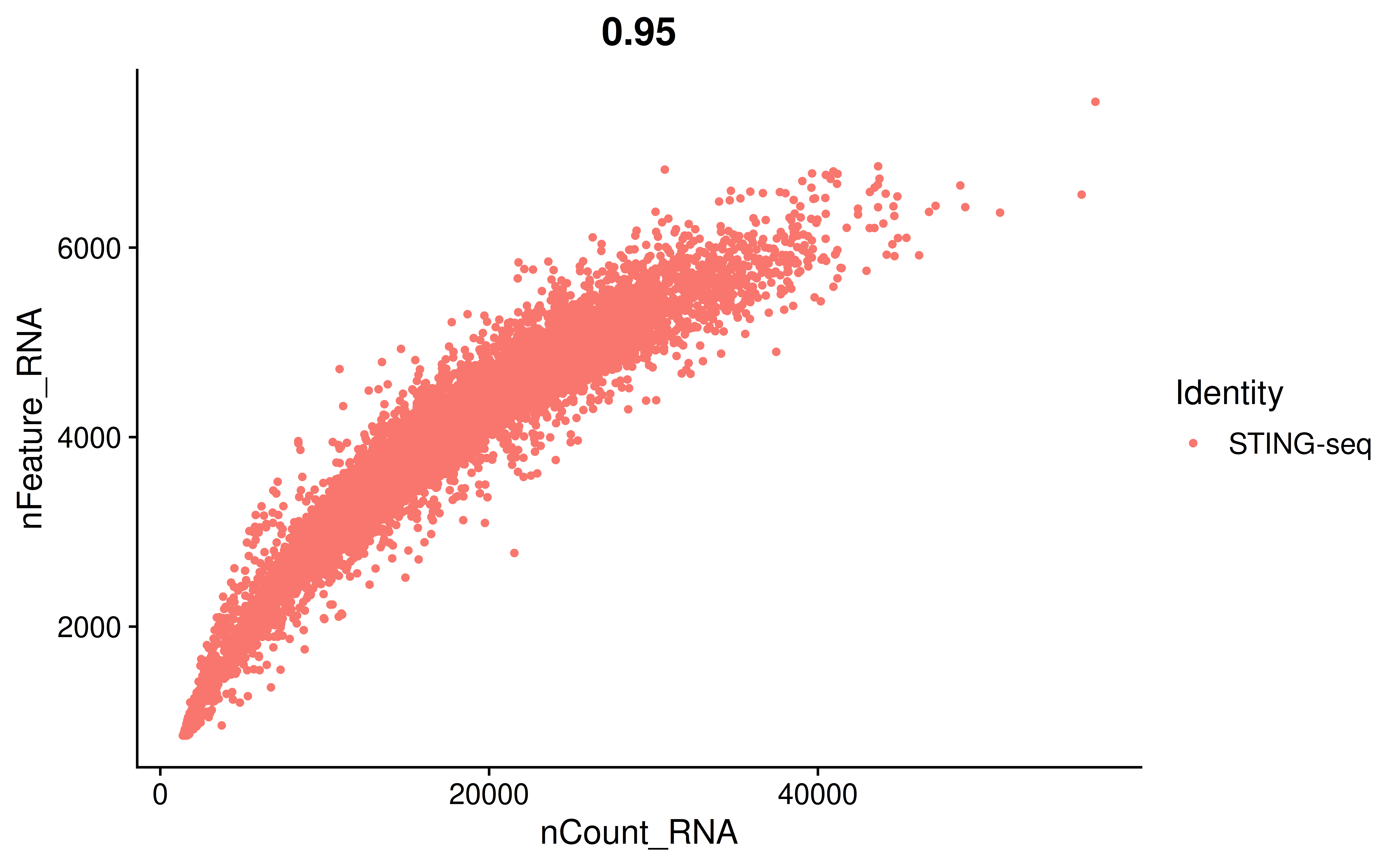
Barcodes comparison:
Code: QC_compare.R
QC_by_author.txt
QC_by_hang.txt
Comparison result:
[1] "There are 508 cells filtered out in comparison to authors' list."g2. PDF y axis issue
Previous question:
- Is the small values in the Y-axis of the previous PDF plot wrong?
My understanding: probably NOT wrong.Point 1: similar results by a package for epdf
Plotted with base R:
hist(barcode_dist, freq=F, breaks=150, main="PDF: UMI counts in each barcode (cell)", xlab="UMI counts in each barcode (cell)", ylab="PDF")
Plotted with EnvStats package:
EnvStats::epdfPlot(barcode_dist, epdf.col = "red").png)
Point 2: area of the plot is ~1 by eye
Didn’t bother to do calculus, just very roughly calculated
1.5e-04 x 7000 = 1.05
h. zero-inflated plot & regression
Ref: Kim et al. 2020
Code: zero-flated.R
.png)
.png)
regression summary
summary(glm(zero_prop ~ target_mean, family = poisson, data = whichmodel_10)) # poissonCall:
glm(formula = zero_prop ~ target_mean, family = poisson, data = whichmodel_10)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.13432 -0.00250 0.01255 0.01294 1.08289
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.012969 0.005915 -2.193 0.0283 *
target_mean -0.672805 0.019706 -34.141 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 2256.629 on 35374 degrees of freedom
Residual deviance: 88.113 on 35373 degrees of freedom
AIC: Inf
Number of Fisher Scoring iterations: 5summary(glm.nb(zero_prop ~ target_mean, data = whichmodel_10)) # negative binomialCall:
glm.nb(formula = zero_prop ~ target_mean, data = whichmodel_10,
init.theta = 18539.38634, link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.86791 -0.31620 0.01294 0.06140 1.32706
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.012969 0.005915 -2.193 0.0283 *
target_mean -0.672803 0.019707 -34.141 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for Negative Binomial(18539.39) family taken to be 1)
Null deviance: 6991.6 on 35374 degrees of freedom
Residual deviance: 4823.2 on 35373 degrees of freedom
AIC: 66516
Number of Fisher Scoring iterations: 1
Theta: 18539
Std. Err.: 7723
Warning while fitting theta: iteration limit reached
2 x log-likelihood: -66509.53 Note: negative binomial regression doesn’t seem to allow zeros.
sessionInfo()R version 4.2.1 (2022-06-23)
Platform: x86_64-pc-linux-gnu (64-bit)
<<<<<<< HEAD
Running under: Ubuntu 22.04.1 LTS
=======
Running under: Ubuntu 22.04 LTS
>>>>>>> 37d15c9e90ebd440e24cbf0e45836e069685a838
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.7.0
loaded via a namespace (and not attached):
<<<<<<< HEAD
[1] Rcpp_1.0.9 compiler_4.2.1 pillar_1.8.0 bslib_0.4.0
[5] later_1.3.0 git2r_0.30.1 jquerylib_0.1.4 tools_4.2.1
[9] getPass_0.2-2 digest_0.6.29 jsonlite_1.8.0 evaluate_0.15
[13] tibble_3.1.8 lifecycle_1.0.1 pkgconfig_2.0.3 rlang_1.0.4
[17] cli_3.3.0 rstudioapi_0.13 yaml_2.3.5 xfun_0.31
[21] fastmap_1.1.0 httr_1.4.3 stringr_1.4.0 knitr_1.39
[25] fs_1.5.2 vctrs_0.4.1 sass_0.4.2 rprojroot_2.0.3
[29] glue_1.6.2 R6_2.5.1 processx_3.7.0 fansi_1.0.3
[33] rmarkdown_2.14 callr_3.7.1 magrittr_2.0.3 whisker_0.4
[37] ps_1.7.1 promises_1.2.0.1 htmltools_0.5.3 httpuv_1.6.5
[41] utf8_1.2.2 stringi_1.7.8 cachem_1.0.6