Last updated: 2020-04-28
Checks:
Knit directory: mr_mash_test/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200328) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 84ab89b . See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .sos/
Ignored: code/fit_mr_mash.66662433.err
Ignored: code/fit_mr_mash.66662433.out
Ignored: dsc/.sos/
Ignored: dsc/outfiles/
Ignored: output/dsc.html
Ignored: output/dsc/
Ignored: output/dsc_OLD.html
Ignored: output/dsc_OLD/
Untracked files:
Untracked: code/plot_test.R
Untracked: dsc/dsc.scripts.html
Unstaged changes:
Modified: dsc/midway2.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/results_accuracy.Rmd) and HTML (docs/results_accuracy.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
File
Version
Author
Date
Message
html
8ed3a7f
fmorgante
2020-04-28
Build site.
html
f2fa981
fmorgante
2020-04-28
Build site.
html
e2c9550
fmorgante
2020-04-16
Build site.
html
161eea7
fmorgante
2020-04-16
Build site.
html
dd40784
fmorgante
2020-04-16
Build site.
html
8f4456e
fmorgante
2020-04-16
Build site.
html
422658e
fmorgante
2020-04-16
Build site.
html
04f5485
fmorgante
2020-03-30
Build site.
html
cc50c05
fmorgante
2020-03-30
Build site.
Rmd
ec880d7
fmorgante
2020-03-30
Adjust date format
html
378e278
fmorgante
2020-03-30
Build site.
Rmd
e4b5d3f
fmorgante
2020-03-30
Add simulation 4
html
40cd0cf
fmorgante
2020-03-30
Build site.
html
b48d34d
fmorgante
2020-03-30
Build site.
Rmd
6ff19bc
fmorgante
2020-03-30
Fix typo in simulation 3
html
7a2afe7
fmorgante
2020-03-30
Build site.
Rmd
b3eb015
fmorgante
2020-03-30
Add simulation 3
html
f7e2f68
fmorgante
2020-03-30
Build site.
Rmd
6063335
fmorgante
2020-03-30
Fix typo in simulation 2
html
4f5c291
fmorgante
2020-03-30
Build site.
Rmd
f4eb72a
fmorgante
2020-03-30
Add simulation 2
html
8621d7d
fmorgante
2020-03-30
Build site.
Rmd
1cfb1f8
fmorgante
2020-03-30
Slight improvements
html
d5773c9
fmorgante
2020-03-30
Build site.
html
07a9b4d
fmorgante
2020-03-30
Build site.
Rmd
f416bb4
fmorgante
2020-03-30
Tweak prediction results
html
f2c716e
fmorgante
2020-03-30
Build site.
Rmd
f9a9283
fmorgante
2020-03-30
Tweak prediction results
html
8e7a5b5
fmorgante
2020-03-30
Build site.
Rmd
cf442a2
fmorgante
2020-03-30
Fix prediction results
html
4e9aad6
fmorgante
2020-03-30
Build site.
Rmd
1f94def
fmorgante
2020-03-30
Tweak prediction results
html
7911e81
fmorgante
2020-03-30
Build site.
Rmd
a6fd418
fmorgante
2020-03-30
Add prediction results
html
e1707b2
fmorgante
2020-03-30
Build site.
Rmd
2b58162
fmorgante
2020-03-30
Start adding prediction results
html
f2451af
fmorgante
2020-03-29
Build site.
html
2fe7214
fmorgante
2020-03-29
Build site.
Rmd
d43e6a6
fmorgante
2020-03-29
Set current date automatically
html
dbf0d8e
fmorgante
2020-03-29
Build site.
html
c71dfff
fmorgante
2020-03-29
Build site.
Rmd
bafd72f
fmorgante
2020-03-29
Modify titles and author
html
796c93e
fmorgante
2020-03-29
Build site.
Rmd
ed9843a
fmorgante
2020-03-29
Add additional pages
options(stringsAsFactors = FALSE)
accuracy <- function(Y, Yhat) {
bias <- rep(NA, ncol(Y))
r2 <- rep(NA, ncol(Y))
mse <- rep(NA, ncol(Y))
for(i in 1:ncol(Y)){
fit <- lm(Y[, i] ~ Yhat[, i])
bias[i] <- coef(fit)[2]
r2[i] <- summary(fit)$r.squared
mse[i] <- mean((Y[, i] - Yhat[, i])^2)
}
return(list(bias=bias, r2=r2, mse=mse))
}
Simulation 1 – Shared effects, independent variables
dat1 <- readRDS("output/fit_mr_mash_n600_p1000_p_caus50_r5_pve0.5_sigmaoffdiag1_sigmascale0.8_gammaoffdiag0_gammascale0.8_Voffdiag0.2_Vscale0_updatew0TRUE_updatew0TRUE_updatew0methodmixsqp_updateVTRUE.rds")
n1 <- dat1$params$n
p1 <- dat1$params$p
p_causal1 <- dat1$params$p_causal
r1 <- dat1$params$r
k1 <- length(dat1$fit$w0)
pve1 <- dat1$params$pve
prop_testset1 <- dat1$params$prop_testset
B1 <- dat1$inputs$B
V1 <- dat1$inputs$V
Sigma1 <- dat1$inputs$Sigma
Gamma1 <- dat1$inputs$Gamma
Ytrain1 <- dat1$Ytrain
Ytest1 <- dat1$Ytest
mu11 <- dat1$fit$mu1
fitted1 <- dat1$fit$fitted
Yhat_test1 <- dat1$Yhat_testThe results below are based on simulation with 600 samples, 1000 variables of which 50 were causal, 5 responses with a per-response proportion of variance explained (PVE) of 0.5. Variables, X, were drawn from MVN(0, Gamma), causal effects, B, were drawn from MVN(0, Sigma). The responses, Y, were drawn from MN(XB, I, V).
cat("Gamma (First 5 elements)")Gamma (First 5 elements)Gamma1[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.0 0.0 0.0 0.0
[2,] 0.0 0.8 0.0 0.0 0.0
[3,] 0.0 0.0 0.8 0.0 0.0
[4,] 0.0 0.0 0.0 0.8 0.0
[5,] 0.0 0.0 0.0 0.0 0.8cat("Sigma")SigmaSigma1 [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.8 0.8 0.8 0.8
[2,] 0.8 0.8 0.8 0.8 0.8
[3,] 0.8 0.8 0.8 0.8 0.8
[4,] 0.8 0.8 0.8 0.8 0.8
[5,] 0.8 0.8 0.8 0.8 0.8cat("V")VV1 [,1] [,2] [,3] [,4] [,5]
[1,] 25.55836 0.00000 0.00000 0.00000 0.00000
[2,] 0.00000 25.55836 0.00000 0.00000 0.00000
[3,] 0.00000 0.00000 25.55836 0.00000 0.00000
[4,] 0.00000 0.00000 0.00000 25.55836 0.00000
[5,] 0.00000 0.00000 0.00000 0.00000 25.55836mr.mash was fitted to the training data (80% of the data) updating V and updating the prior weights using mixSQP. Then, responses were predicted on the test data (20% of the data). The mixture prior consisted of 101 components.
In the plots below, each color/symbol defines a diffrent response.
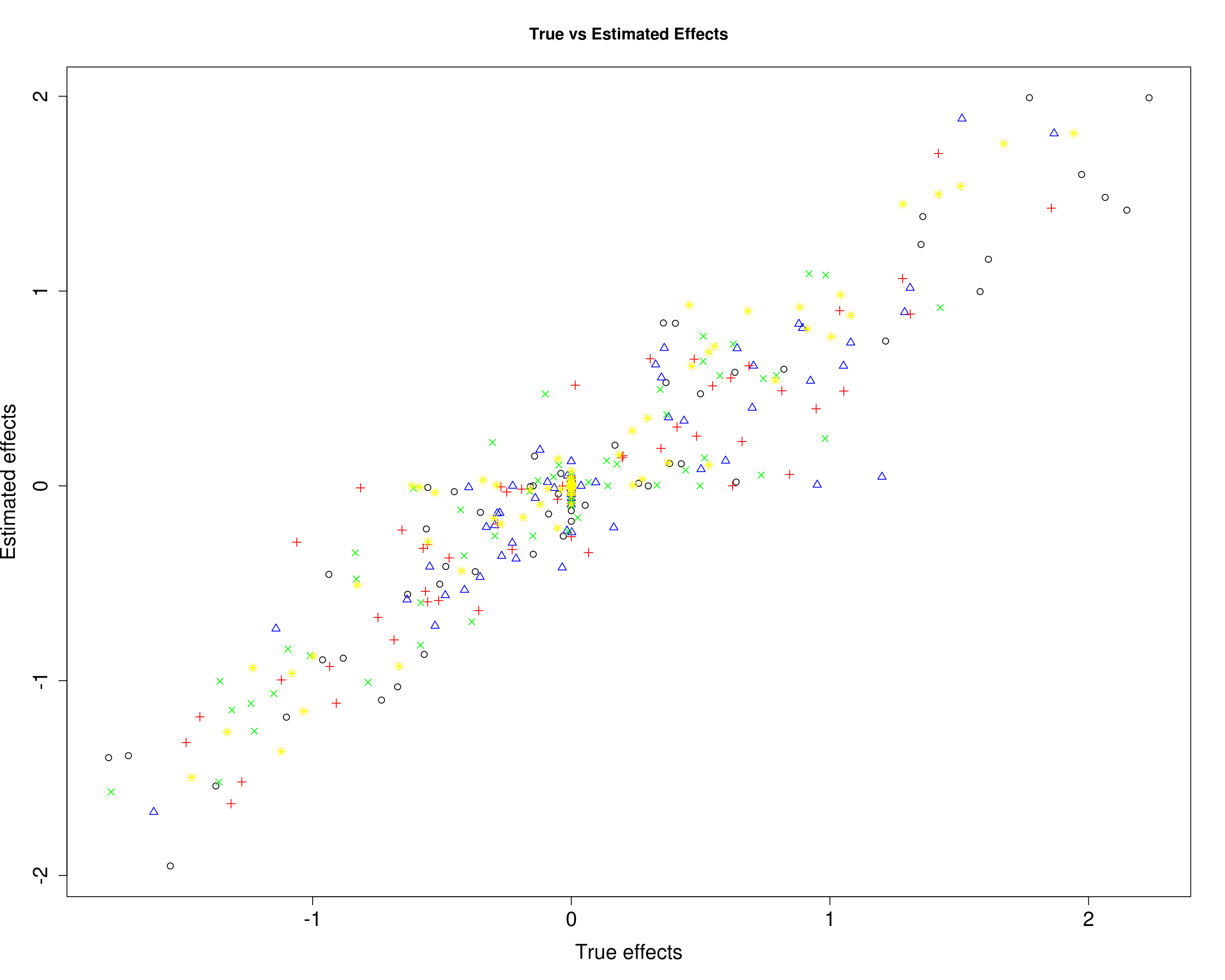
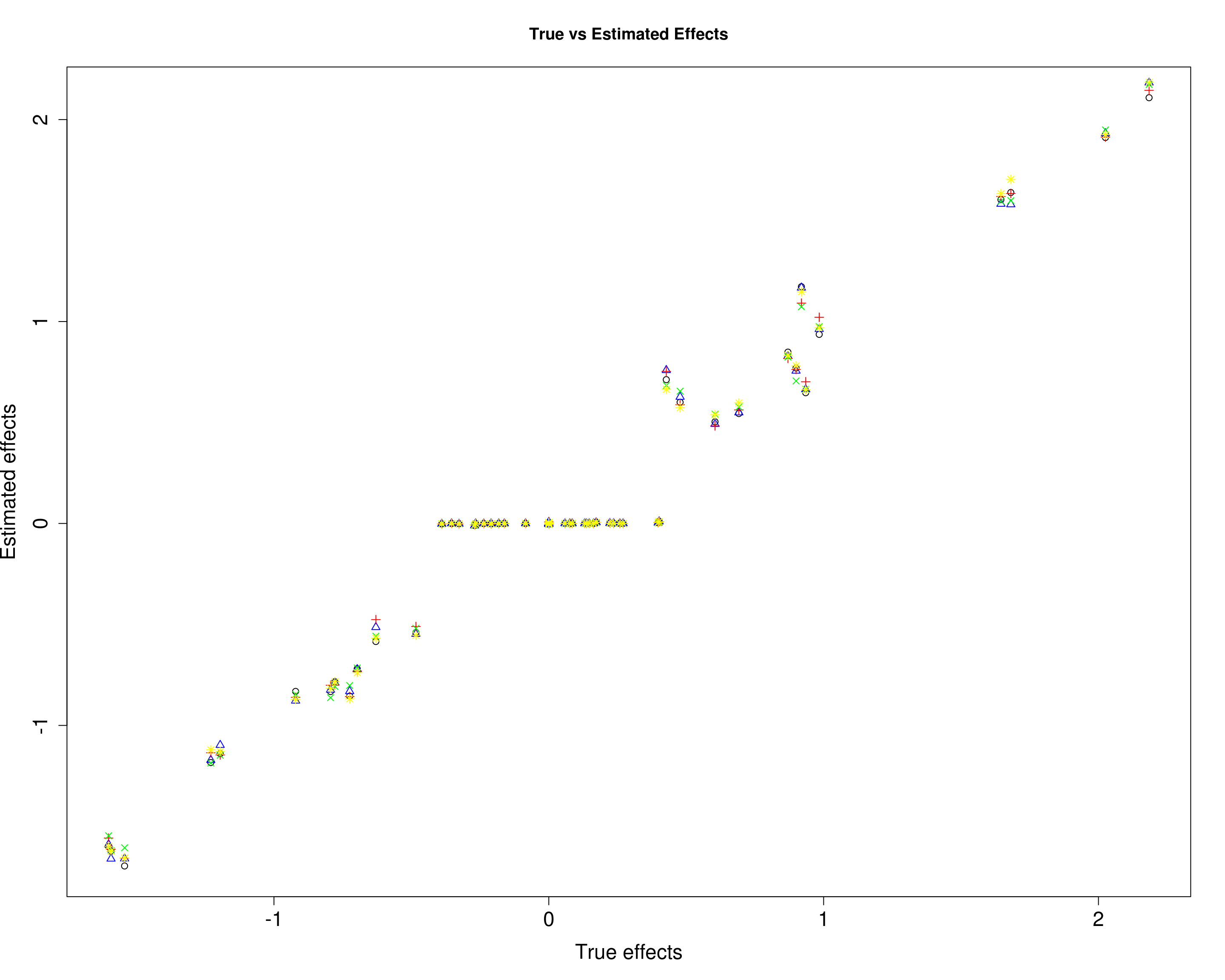
Here, we compare the estimated effects with the true effects.
plot(B1[, 1], mu11[, 1], xlab="True effects", ylab="Estimated effects", main="True vs Estimated Effects", pch=1, cex.lab=1.5, cex.axis=1.5)
points(B1[, 2], mu11[, 2], col="blue", pch=2)
points(B1[, 3], mu11[, 3], col="red", pch=3)
points(B1[, 4], mu11[, 4], col="green", pch=4)
points(B1[, 5], mu11[, 5], col="yellow", pch=8)
Past versions of “effects 1-1.png”
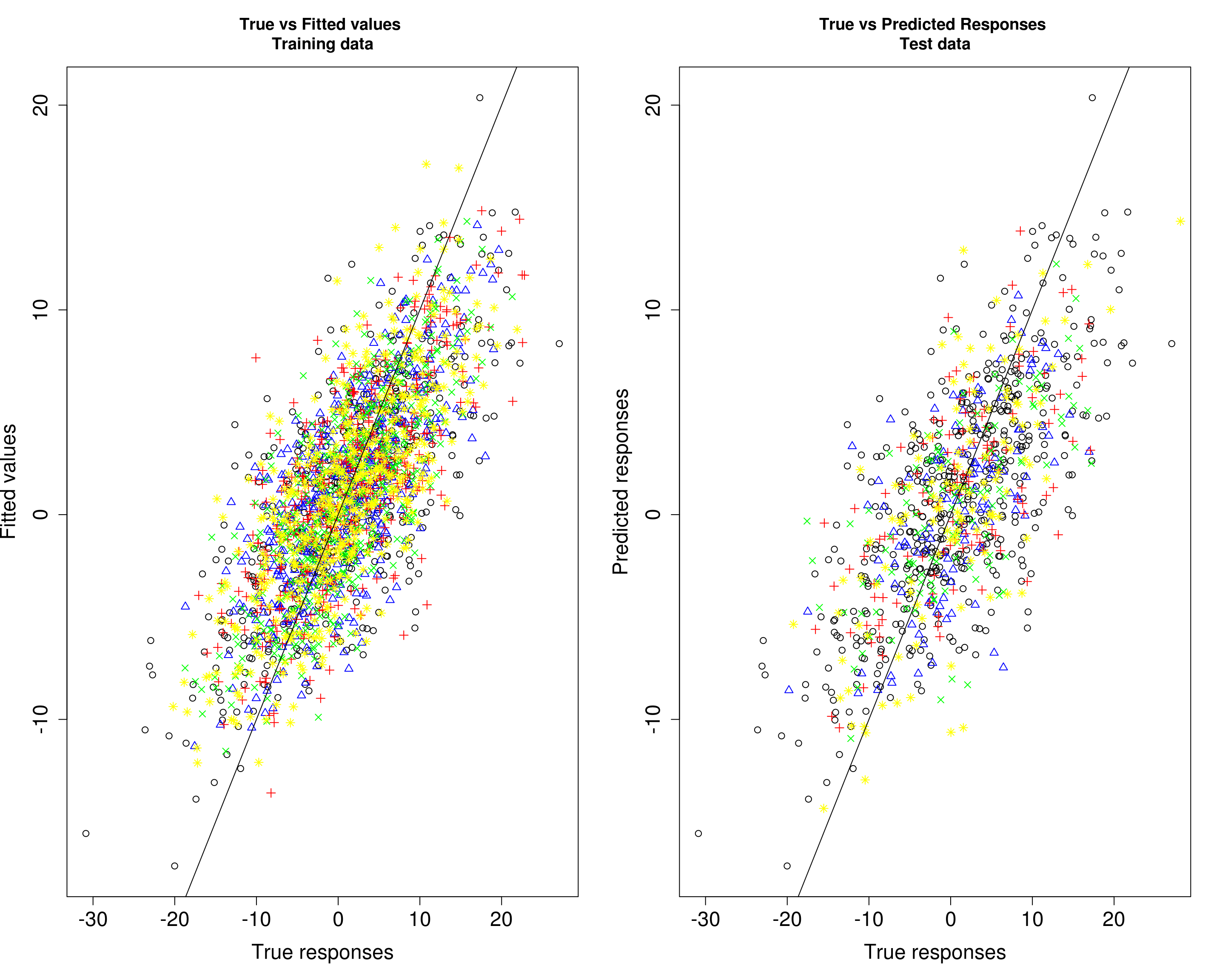
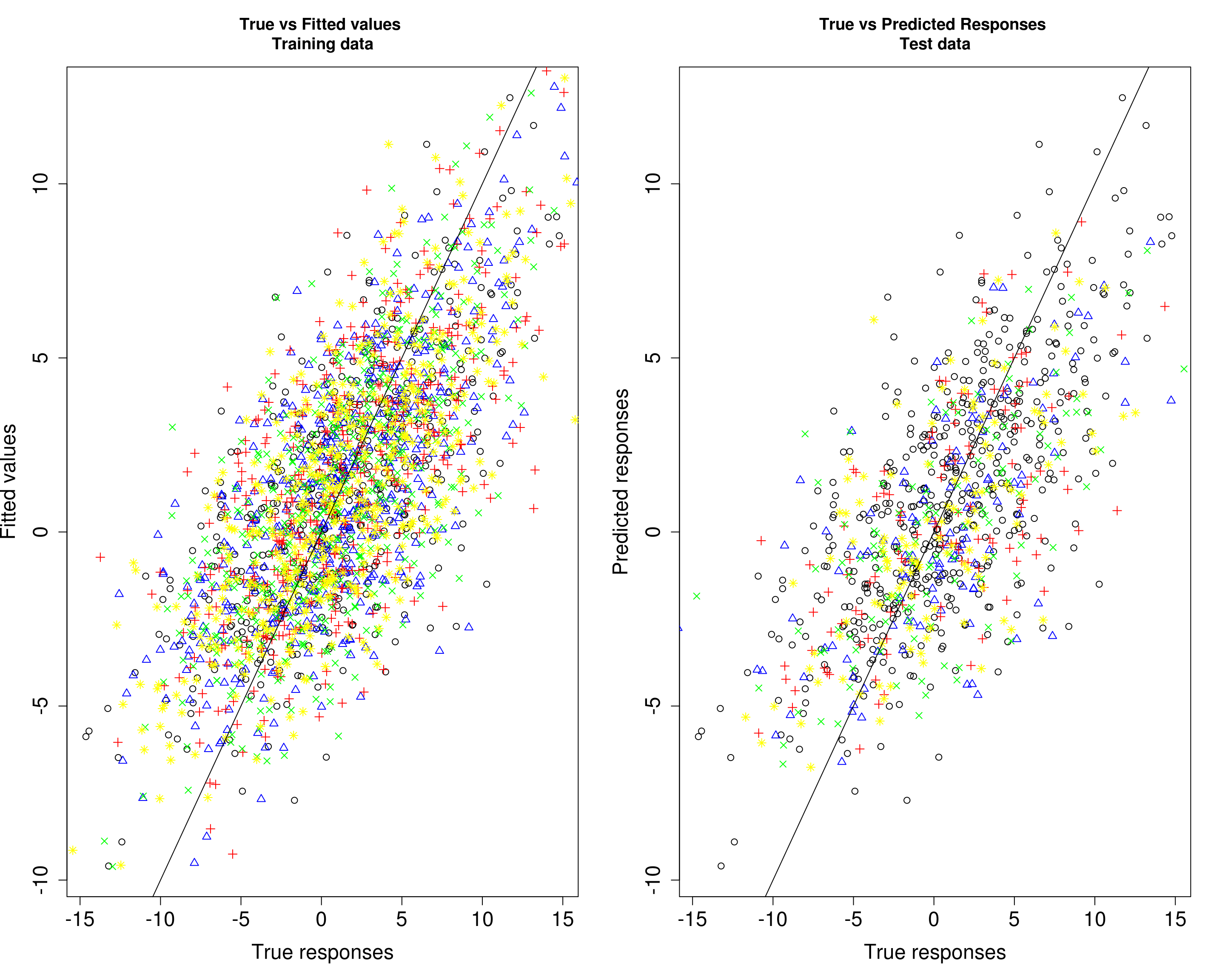
Then, we compare the predicted responses with the true responses in the training data (left panel) and test data (right panel).
par(mfrow=c(1,2))
plot(Ytrain1[, 1], fitted1[, 1], xlab="True responses", ylab="Fitted values", main="True vs Fitted values \nTraining data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytrain1[, 2], fitted1[, 2], col="blue", pch=2)
points(Ytrain1[, 3], fitted1[, 3], col="red", pch=3)
points(Ytrain1[, 4], fitted1[, 4], col="green", pch=4)
points(Ytrain1[, 5], fitted1[, 5], col="yellow", pch=8)
abline(0, 1)
plot(Ytrain1[, 1], fitted1[, 1], xlab="True responses", ylab="Predicted responses", main="True vs Predicted Responses \nTest data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytest1[, 2], Yhat_test1[, 2], col="blue", pch=2)
points(Ytest1[, 3], Yhat_test1[, 3], col="red", pch=3)
points(Ytest1[, 4], Yhat_test1[, 4], col="green", pch=4)
points(Ytest1[, 5], Yhat_test1[, 5], col="yellow", pch=8)
abline(0, 1)
Past versions of “predict 1-1.png”
par(mfrow=c(1,1))
r2_train1 <- round(accuracy(Ytrain1, fitted1)$r2, 4)
r2_test1 <- round(accuracy(Ytest1, Yhat_test1)$r2, 4)
bias_train1 <- round(accuracy(Ytrain1, fitted1)$bias, 4)
bias_test1 <- round(accuracy(Ytest1, Yhat_test1)$bias, 4)
mse_train1 <- round(accuracy(Ytrain1, fitted1)$mse, 4)
mse_test1 <- round(accuracy(Ytest1, Yhat_test1)$mse, 4)
acc1 <- rbind(r2_train1, r2_test1, bias_train1, bias_test1, mse_train1, mse_test1)
colnames(acc1) <- paste0("Y", seq(1, r1))
part_metric1 <- c("Training data r2", "Test data r2", "Training data bias", "Test data bias" , "Training data MSE" , "Test data MSE")
res1 <- data.frame(part_metric1, acc1)
colnames(res1)[1] <- c("Partition_metric")
rownames(res1) <- NULL
print(res1) Partition_metric Y1 Y2 Y3 Y4 Y5
1 Training data r2 0.5338 0.5418 0.4904 0.5239 0.5620
2 Test data r2 0.4591 0.4476 0.4630 0.4523 0.4237
3 Training data bias 1.0606 1.0586 0.9935 1.0013 1.0971
4 Test data bias 0.9892 1.0660 1.0666 1.0905 0.9659
5 Training data MSE 24.6985 23.8540 25.3913 22.4603 23.9106
6 Test data MSE 24.5054 29.4890 27.1642 29.6734 26.7496
Simulation 2 – Independent effects, independent variables
dat2 <- readRDS("output/fit_mr_mash_n600_p1000_p_caus50_r5_pve0.5_sigmaoffdiag0_sigmascale0.8_gammaoffdiag0_gammascale0.8_Voffdiag0.2_Vscale0_updatew0TRUE_updatew0TRUE_updatew0methodmixsqp_updateVTRUE.rds")
n2 <- dat2$params$n
p2 <- dat2$params$p
p_causal2 <- dat2$params$p_causal
r2 <- dat2$params$r
k2 <- length(dat2$fit$w0)
pve2 <- dat2$params$pve
prop_testset2 <- dat2$params$prop_testset
B2 <- dat2$inputs$B
V2 <- dat2$inputs$V
Sigma2 <- dat2$inputs$Sigma
Gamma2 <- dat2$inputs$Gamma
Ytrain2 <- dat2$Ytrain
Ytest2 <- dat2$Ytest
mu12 <- dat2$fit$mu1
fitted2 <- dat2$fit$fitted
Yhat_test2 <- dat2$Yhat_testThe results below are based on simulation with 600 samples, 1000 variables of which 50 were causal, 5 responses with a per-response proportion of variance explained (PVE) of 0.5. Variables, X, were drawn from MVN(0, Gamma), causal effects, B, were drawn from MVN(0, Sigma). The responses, Y, were drawn from MN(XB, I, V).
cat("Gamma (First 5 elements)")Gamma (First 5 elements)Gamma2[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.0 0.0 0.0 0.0
[2,] 0.0 0.8 0.0 0.0 0.0
[3,] 0.0 0.0 0.8 0.0 0.0
[4,] 0.0 0.0 0.0 0.8 0.0
[5,] 0.0 0.0 0.0 0.0 0.8cat("Sigma")SigmaSigma2 [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.0 0.0 0.0 0.0
[2,] 0.0 0.8 0.0 0.0 0.0
[3,] 0.0 0.0 0.8 0.0 0.0
[4,] 0.0 0.0 0.0 0.8 0.0
[5,] 0.0 0.0 0.0 0.0 0.8cat("V")VV2 [,1] [,2] [,3] [,4] [,5]
[1,] 39.87305 0.00000 0.00000 0.00000 0.00000
[2,] 0.00000 24.41042 0.00000 0.00000 0.00000
[3,] 0.00000 0.00000 27.69452 0.00000 0.00000
[4,] 0.00000 0.00000 0.00000 25.53166 0.00000
[5,] 0.00000 0.00000 0.00000 0.00000 29.00472mr.mash was fitted to the training data (80% of the data) updating V and updating the prior weights using mixSQP. Then, responses were predicted on the test data (20% of the data). The mixture prior consisted of 101 components.
In the plots below, each color/symbol defines a diffrent response.
Here, we compare the estimated effects with the true effects.
plot(B2[, 1], mu12[, 1], xlab="True effects", ylab="Estimated effects", main="True vs Estimated Effects", pch=1, cex.lab=1.5, cex.axis=1.5)
points(B2[, 2], mu12[, 2], col="blue", pch=2)
points(B2[, 3], mu12[, 3], col="red", pch=3)
points(B2[, 4], mu12[, 4], col="green", pch=4)
points(B2[, 5], mu12[, 5], col="yellow", pch=8)
Past versions of “effects 2-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
4f5c291
fmorgante
2020-03-30
Then, we compare the predicted responses with the true responses in the training data (left panel) and test data (right panel).
par(mfrow=c(1,2))
plot(Ytrain2[, 1], fitted2[, 1], xlab="True responses", ylab="Fitted values", main="True vs Fitted values \nTraining data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytrain2[, 2], fitted2[, 2], col="blue", pch=2)
points(Ytrain2[, 3], fitted2[, 3], col="red", pch=3)
points(Ytrain2[, 4], fitted2[, 4], col="green", pch=4)
points(Ytrain2[, 5], fitted2[, 5], col="yellow", pch=8)
abline(0, 1)
plot(Ytrain2[, 1], fitted2[, 1], xlab="True responses", ylab="Predicted responses", main="True vs Predicted Responses \nTest data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytest2[, 2], Yhat_test2[, 2], col="blue", pch=2)
points(Ytest2[, 3], Yhat_test2[, 3], col="red", pch=3)
points(Ytest2[, 4], Yhat_test2[, 4], col="green", pch=4)
points(Ytest2[, 5], Yhat_test2[, 5], col="yellow", pch=8)
abline(0, 1)
Past versions of “predict 2-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
4f5c291
fmorgante
2020-03-30
par(mfrow=c(1,1))
r2_train2 <- round(accuracy(Ytrain2, fitted2)$r2, 4)
r2_test2 <- round(accuracy(Ytest2, Yhat_test2)$r2, 4)
bias_train2 <- round(accuracy(Ytrain2, fitted2)$bias, 4)
bias_test2 <- round(accuracy(Ytest2, Yhat_test2)$bias, 4)
mse_train2 <- round(accuracy(Ytrain2, fitted2)$mse, 4)
mse_test2 <- round(accuracy(Ytest2, Yhat_test2)$mse, 4)
acc2 <- rbind(r2_train2, r2_test2, bias_train2, bias_test2, mse_train2, mse_test2)
colnames(acc2) <- paste0("Y", seq(1, r2))
part_metric2 <- c("Training data r2", "Test data r2", "Training data bias", "Test data bias" , "Training data MSE" , "Test data MSE")
res2 <- data.frame(part_metric2, acc2)
colnames(res2)[1] <- c("Partition_metric")
rownames(res2) <- NULL
print(res2) Partition_metric Y1 Y2 Y3 Y4 Y5
1 Training data r2 0.5373 0.5341 0.5146 0.5716 0.5570
2 Test data r2 0.4600 0.3854 0.4395 0.4473 0.4948
3 Training data bias 1.1737 1.1069 1.1264 1.1019 1.1270
4 Test data bias 0.9847 0.9370 1.1124 1.1859 0.9449
5 Training data MSE 38.1690 22.6568 25.8972 21.0610 26.7705
6 Test data MSE 41.8959 28.1329 34.3800 34.3338 30.6942
Simulation 3 – Shared effects, correlated variables
dat3 <- readRDS("output/fit_mr_mash_n600_p1000_p_caus50_r5_pve0.5_sigmaoffdiag1_sigmascale0.8_gammaoffdiag0.5_gammascale0.8_Voffdiag0.2_Vscale0_updatew0TRUE_updatew0TRUE_updatew0methodmixsqp_updateVTRUE.rds")
n3 <- dat3$params$n
p3 <- dat3$params$p
p_causal3 <- dat3$params$p_causal
r3 <- dat3$params$r
k3 <- length(dat3$fit$w0)
pve3 <- dat3$params$pve
prop_testset3 <- dat3$params$prop_testset
B3 <- dat3$inputs$B
V3 <- dat3$inputs$V
Sigma3 <- dat3$inputs$Sigma
Gamma3 <- dat3$inputs$Gamma
Ytrain3 <- dat3$Ytrain
Ytest3 <- dat3$Ytest
mu13 <- dat3$fit$mu1
fitted3 <- dat3$fit$fitted
Yhat_test3 <- dat3$Yhat_testThe results below are based on simulation with 600 samples, 1000 variables of which 50 were causal, 5 responses with a per-response proportion of variance explained (PVE) of 0.5. Variables, X, were drawn from MVN(0, Gamma), causal effects, B, were drawn from MVN(0, Sigma). The responses, Y, were drawn from MN(XB, I, V).
cat("Gamma (First 5 elements)")Gamma (First 5 elements)Gamma3[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.4 0.4 0.4 0.4
[2,] 0.4 0.8 0.4 0.4 0.4
[3,] 0.4 0.4 0.8 0.4 0.4
[4,] 0.4 0.4 0.4 0.8 0.4
[5,] 0.4 0.4 0.4 0.4 0.8cat("Sigma")SigmaSigma3 [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.8 0.8 0.8 0.8
[2,] 0.8 0.8 0.8 0.8 0.8
[3,] 0.8 0.8 0.8 0.8 0.8
[4,] 0.8 0.8 0.8 0.8 0.8
[5,] 0.8 0.8 0.8 0.8 0.8cat("V")VV3 [,1] [,2] [,3] [,4] [,5]
[1,] 13.98626 0.00000 0.00000 0.00000 0.00000
[2,] 0.00000 13.98625 0.00000 0.00000 0.00000
[3,] 0.00000 0.00000 13.98625 0.00000 0.00000
[4,] 0.00000 0.00000 0.00000 13.98625 0.00000
[5,] 0.00000 0.00000 0.00000 0.00000 13.98625mr.mash was fitted to the training data (80% of the data) updating V and updating the prior weights using mixSQP. Then, responses were predicted on the test data (20% of the data). The mixture prior consisted of 101 components.
In the plots below, each color/symbol defines a diffrent response.
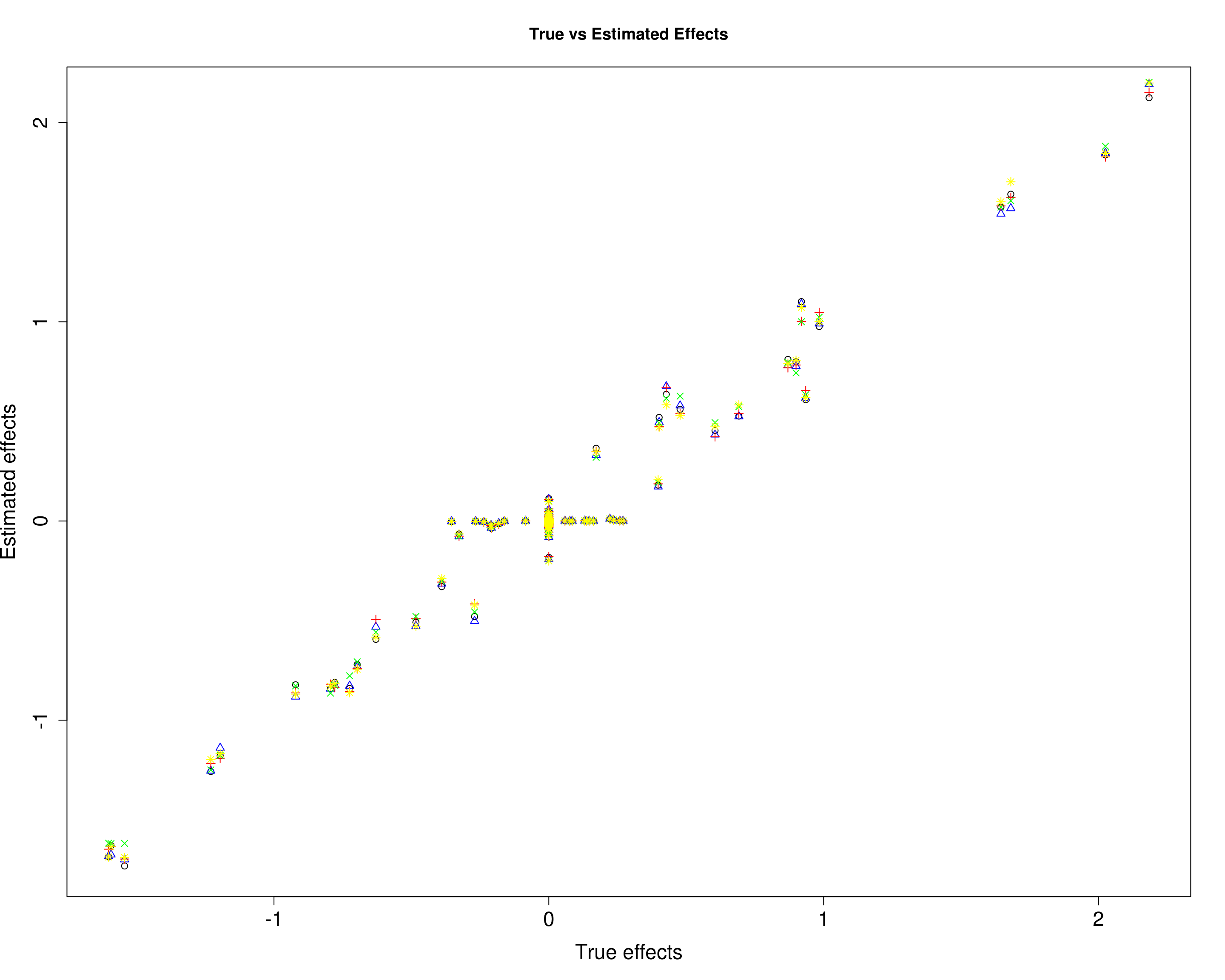
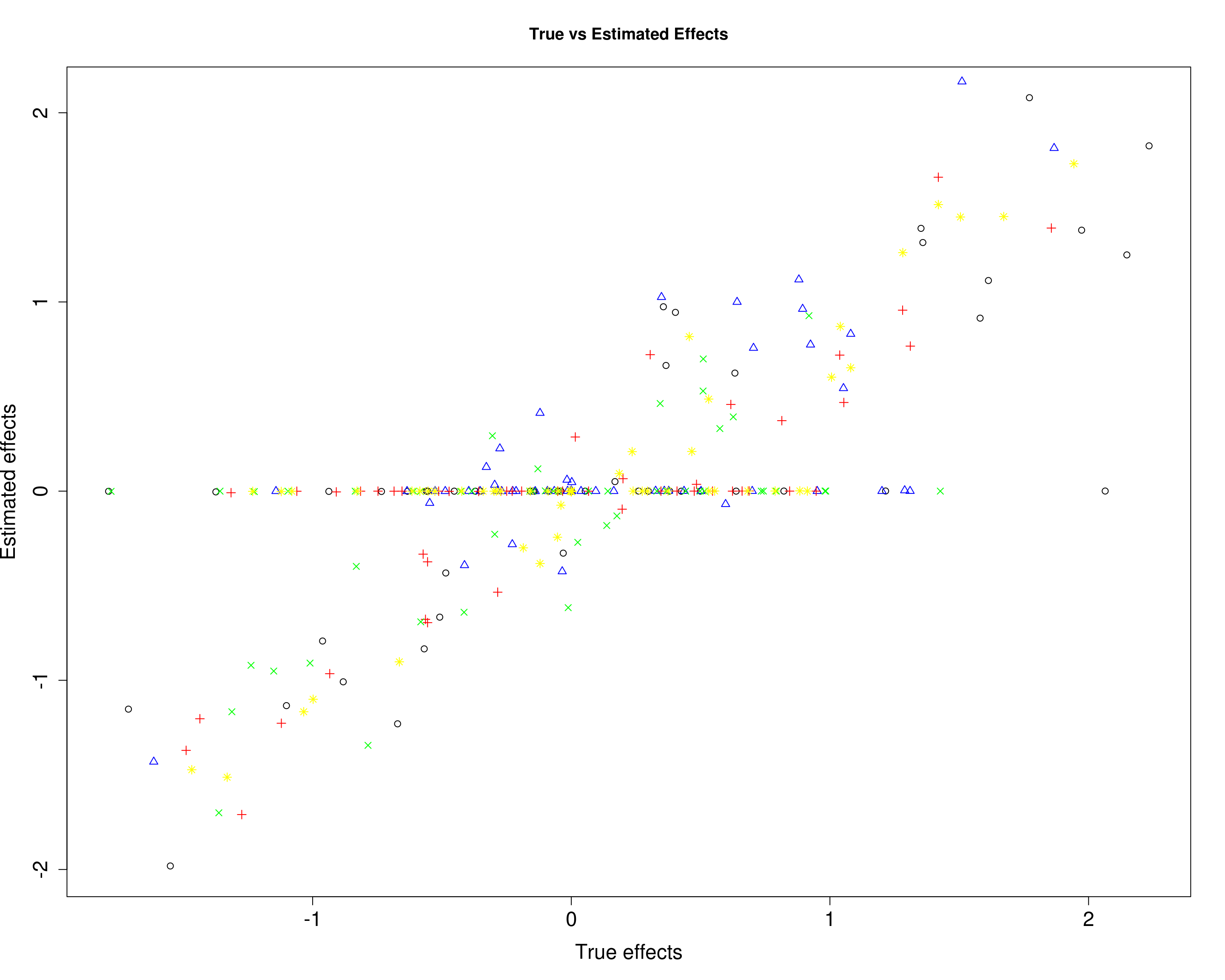
Here, we compare the estimated effects with the true effects.
plot(B3[, 1], mu13[, 1], xlab="True effects", ylab="Estimated effects", main="True vs Estimated Effects", pch=1, cex.lab=1.5, cex.axis=1.5)
points(B3[, 2], mu13[, 2], col="blue", pch=2)
points(B3[, 3], mu13[, 3], col="red", pch=3)
points(B3[, 4], mu13[, 4], col="green", pch=4)
points(B3[, 5], mu13[, 5], col="yellow", pch=8)
Past versions of “effects 3-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
b48d34d
fmorgante
2020-03-30
7a2afe7
fmorgante
2020-03-30
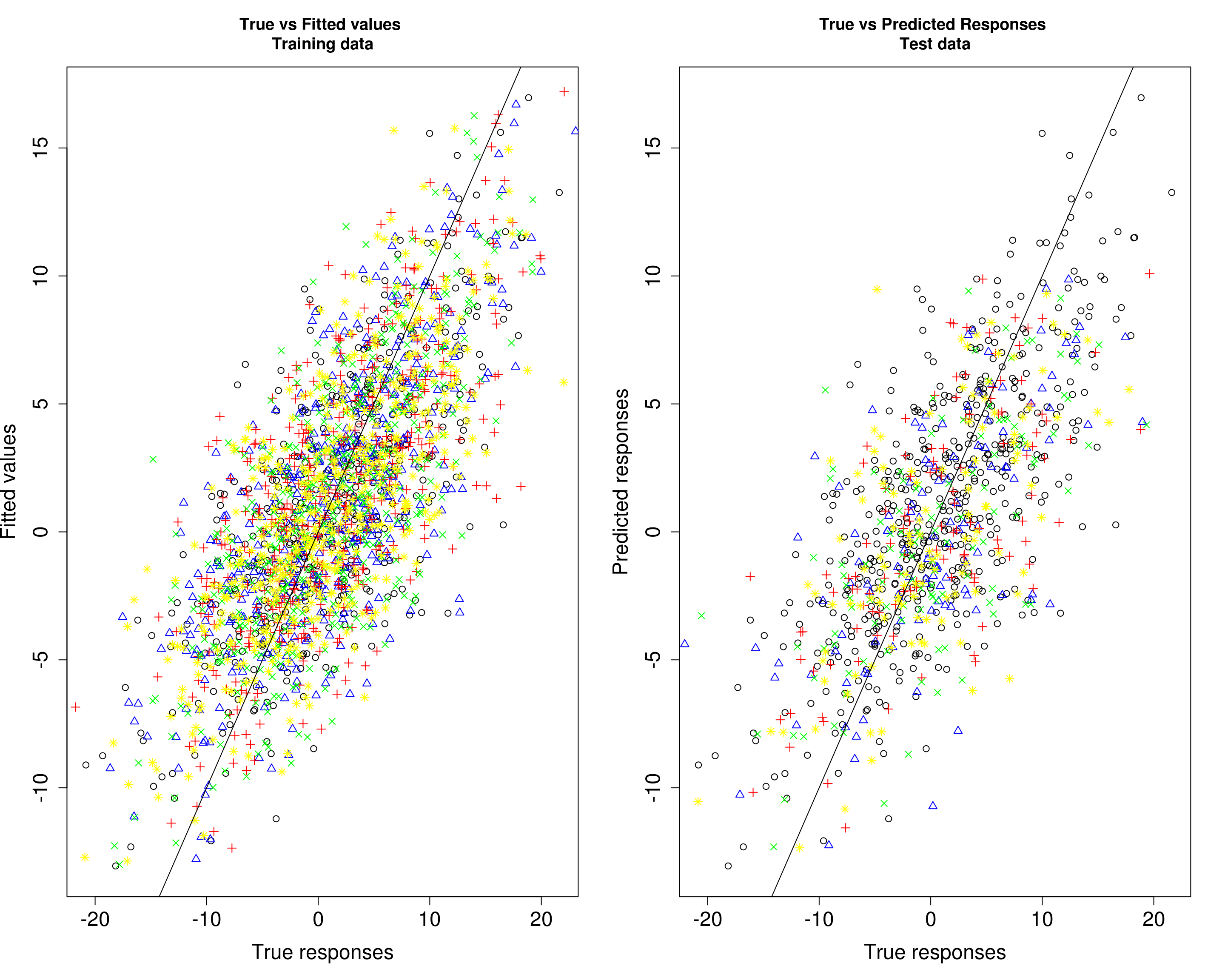
Then, we compare the predicted responses with the true responses in the training data (left panel) and test data (right panel).
par(mfrow=c(1,2))
plot(Ytrain3[, 1], fitted3[, 1], xlab="True responses", ylab="Fitted values", main="True vs Fitted values \nTraining data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytrain3[, 2], fitted3[, 2], col="blue", pch=2)
points(Ytrain3[, 3], fitted3[, 3], col="red", pch=3)
points(Ytrain3[, 4], fitted3[, 4], col="green", pch=4)
points(Ytrain3[, 5], fitted3[, 5], col="yellow", pch=8)
abline(0, 1)
plot(Ytrain3[, 1], fitted3[, 1], xlab="True responses", ylab="Predicted responses", main="True vs Predicted Responses \nTest data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytest3[, 2], Yhat_test3[, 2], col="blue", pch=2)
points(Ytest3[, 3], Yhat_test3[, 3], col="red", pch=3)
points(Ytest3[, 4], Yhat_test3[, 4], col="green", pch=4)
points(Ytest3[, 5], Yhat_test3[, 5], col="yellow", pch=8)
abline(0, 1)
Past versions of “predict 3-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
7a2afe7
fmorgante
2020-03-30
par(mfrow=c(1,1))
r2_train3 <- round(accuracy(Ytrain3, fitted3)$r2, 4)
r2_test3 <- round(accuracy(Ytest3, Yhat_test3)$r2, 4)
bias_train3 <- round(accuracy(Ytrain3, fitted3)$bias, 4)
bias_test3 <- round(accuracy(Ytest3, Yhat_test3)$bias, 4)
mse_train3 <- round(accuracy(Ytrain3, fitted3)$mse, 4)
mse_test3 <- round(accuracy(Ytest3, Yhat_test3)$mse, 4)
acc3 <- rbind(r2_train3, r2_test3, bias_train3, bias_test3, mse_train3, mse_test3)
colnames(acc3) <- paste0("Y", seq(1, r3))
part_metric3 <- c("Training data r2", "Test data r2", "Training data bias", "Test data bias" , "Training data MSE" , "Test data MSE")
res3 <- data.frame(part_metric3, acc3)
colnames(res3)[1] <- c("Partition_metric")
rownames(res3) <- NULL
print(res3) Partition_metric Y1 Y2 Y3 Y4 Y5
1 Training data r2 0.4892 0.5037 0.4710 0.4907 0.5390
2 Test data r2 0.4148 0.4341 0.4220 0.4639 0.4251
3 Training data bias 1.0358 1.0376 0.9876 0.9817 1.0887
4 Test data bias 1.0015 1.0979 1.0552 1.1736 1.0079
5 Training data MSE 14.4128 14.0238 14.5096 12.8917 13.6560
6 Test data MSE 14.0750 15.9244 15.3882 15.9627 14.0499
Simulation 4 – Independent effects, correlated variables
dat4 <- readRDS("output/fit_mr_mash_n600_p1000_p_caus50_r5_pve0.5_sigmaoffdiag0_sigmascale0.8_gammaoffdiag0.5_gammascale0.8_Voffdiag0.2_Vscale0_updatew0TRUE_updatew0TRUE_updatew0methodmixsqp_updateVTRUE.rds")
n4 <- dat4$params$n
p4 <- dat4$params$p
p_causal4 <- dat4$params$p_causal
r4 <- dat4$params$r
k4 <- length(dat4$fit$w0)
pve4 <- dat4$params$pve
prop_testset4 <- dat4$params$prop_testset
B4 <- dat4$inputs$B
V4 <- dat4$inputs$V
Sigma4 <- dat4$inputs$Sigma
Gamma4 <- dat4$inputs$Gamma
Ytrain4 <- dat4$Ytrain
Ytest4 <- dat4$Ytest
mu14 <- dat4$fit$mu1
fitted4 <- dat4$fit$fitted
Yhat_test4 <- dat4$Yhat_testThe results below are based on simulation with 600 samples, 1000 variables of which 50 were causal, 5 responses with a per-response proportion of variance explained (PVE) of 0.5. Variables, X, were drawn from MVN(0, Gamma), causal effects, B, were drawn from MVN(0, Sigma). The responses, Y, were drawn from MN(XB, I, V).
cat("Gamma (First 5 elements)")Gamma (First 5 elements)Gamma4[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.4 0.4 0.4 0.4
[2,] 0.4 0.8 0.4 0.4 0.4
[3,] 0.4 0.4 0.8 0.4 0.4
[4,] 0.4 0.4 0.4 0.8 0.4
[5,] 0.4 0.4 0.4 0.4 0.8cat("Sigma")SigmaSigma4 [,1] [,2] [,3] [,4] [,5]
[1,] 0.8 0.0 0.0 0.0 0.0
[2,] 0.0 0.8 0.0 0.0 0.0
[3,] 0.0 0.0 0.8 0.0 0.0
[4,] 0.0 0.0 0.0 0.8 0.0
[5,] 0.0 0.0 0.0 0.0 0.8cat("V")VV4 [,1] [,2] [,3] [,4] [,5]
[1,] 31.75545 0.00000 0.00000 0.00000 0.00000
[2,] 0.00000 31.47091 0.00000 0.00000 0.00000
[3,] 0.00000 0.00000 14.55202 0.00000 0.00000
[4,] 0.00000 0.00000 0.00000 42.12604 0.00000
[5,] 0.00000 0.00000 0.00000 0.00000 15.37456mr.mash was fitted to the training data (80% of the data) updating V and updating the prior weights using mixSQP. Then, responses were predicted on the test data (20% of the data). The mixture prior consisted of 101 components.
In the plots below, each color/symbol defines a diffrent response.
Here, we compare the estimated effects with the true effects.
plot(B4[, 1], mu14[, 1], xlab="True effects", ylab="Estimated effects", main="True vs Estimated Effects", pch=1, cex.lab=1.5, cex.axis=1.5)
points(B4[, 2], mu14[, 2], col="blue", pch=2)
points(B4[, 3], mu14[, 3], col="red", pch=3)
points(B4[, 4], mu14[, 4], col="green", pch=4)
points(B4[, 5], mu14[, 5], col="yellow", pch=8)
Past versions of “effects 4-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
378e278
fmorgante
2020-03-30
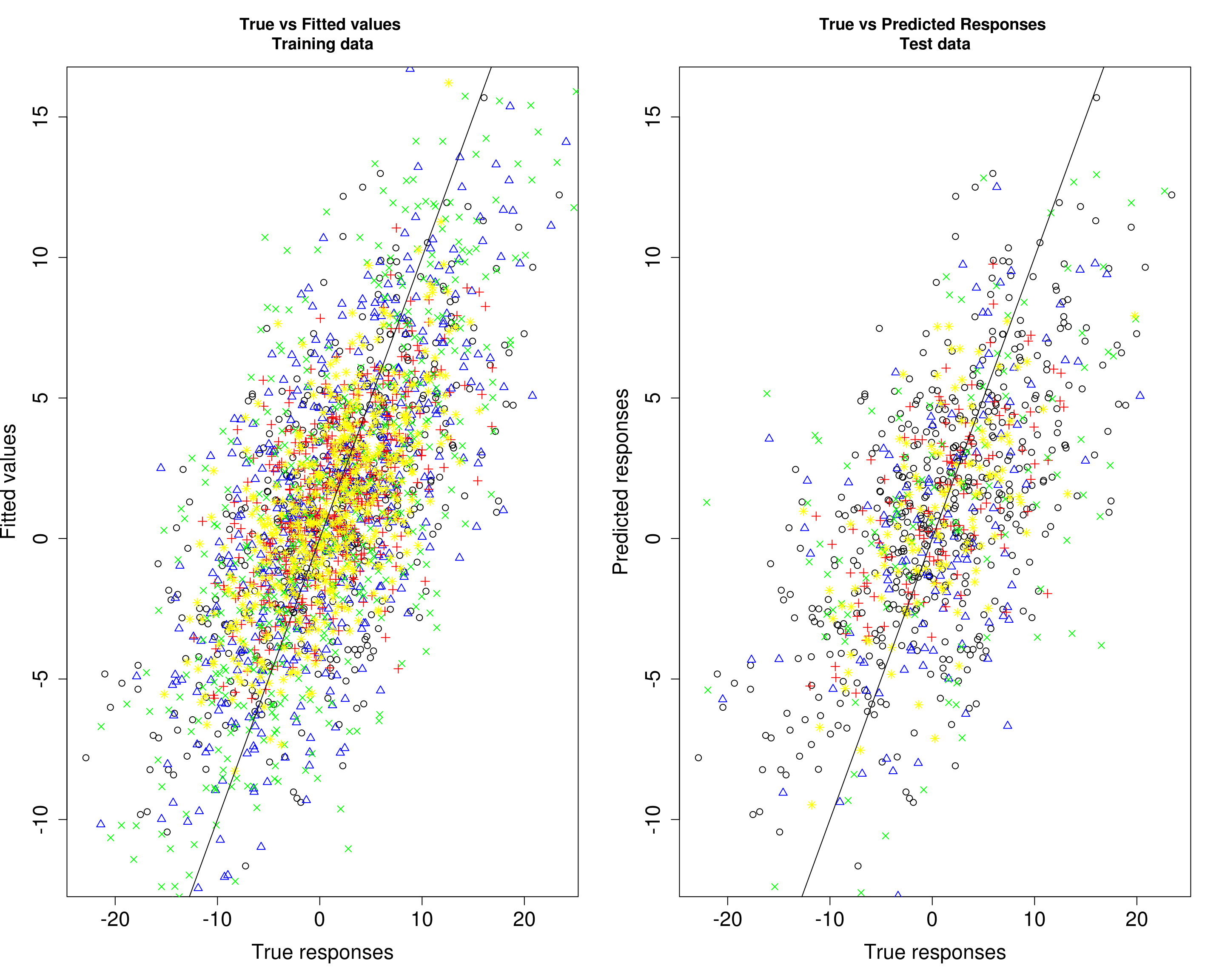
Then, we compare the predicted responses with the true responses in the training data (left panel) and test data (right panel).
par(mfrow=c(1,2))
plot(Ytrain4[, 1], fitted4[, 1], xlab="True responses", ylab="Fitted values", main="True vs Fitted values \nTraining data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytrain4[, 2], fitted4[, 2], col="blue", pch=2)
points(Ytrain4[, 3], fitted4[, 3], col="red", pch=3)
points(Ytrain4[, 4], fitted4[, 4], col="green", pch=4)
points(Ytrain4[, 5], fitted4[, 5], col="yellow", pch=8)
abline(0, 1)
plot(Ytrain4[, 1], fitted4[, 1], xlab="True responses", ylab="Predicted responses", main="True vs Predicted Responses \nTest data", pch=1, cex.lab=1.5, cex.axis=1.5)
points(Ytest4[, 2], Yhat_test4[, 2], col="blue", pch=2)
points(Ytest4[, 3], Yhat_test4[, 3], col="red", pch=3)
points(Ytest4[, 4], Yhat_test4[, 4], col="green", pch=4)
points(Ytest4[, 5], Yhat_test4[, 5], col="yellow", pch=8)
abline(0, 1)
Past versions of “predict 4-1.png”
Version
Author
Date
422658e
fmorgante
2020-04-16
378e278
fmorgante
2020-03-30
par(mfrow=c(1,1))
r2_train4 <- round(accuracy(Ytrain4, fitted4)$r2, 4)
r2_test4 <- round(accuracy(Ytest4, Yhat_test4)$r2, 4)
bias_train4 <- round(accuracy(Ytrain4, fitted4)$bias, 4)
bias_test4 <- round(accuracy(Ytest4, Yhat_test4)$bias, 4)
mse_train4 <- round(accuracy(Ytrain4, fitted4)$mse, 4)
mse_test4 <- round(accuracy(Ytest4, Yhat_test4)$mse, 4)
acc4 <- rbind(r2_train4, r2_test4, bias_train4, bias_test4, mse_train4, mse_test4)
colnames(acc4) <- paste0("Y", seq(1, r4))
part_metric4 <- c("Training data r2", "Test data r2", "Training data bias", "Test data bias" , "Training data MSE" , "Test data MSE")
res4 <- data.frame(part_metric4, acc4)
colnames(res4)[1] <- c("Partition_metric")
rownames(res4) <- NULL
print(res4) Partition_metric Y1 Y2 Y3 Y4 Y5
1 Training data r2 0.4107 0.4811 0.3574 0.5330 0.4272
2 Test data r2 0.3990 0.2959 0.3766 0.3298 0.3542
3 Training data bias 1.1022 1.0512 1.0676 1.0356 1.0803
4 Test data bias 1.0478 0.7843 1.1576 0.9455 0.9463
5 Training data MSE 36.2095 32.5375 17.8351 39.5427 18.2162
6 Test data MSE 36.5641 38.1858 20.4419 62.0936 19.4614
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] workflowr_1.6.1 Rcpp_1.0.4.6 digest_0.6.25 later_0.7.5
[5] rprojroot_1.3-2 R6_2.4.1 backports_1.1.5 git2r_0.26.1
[9] magrittr_1.5 evaluate_0.12 stringi_1.4.3 fs_1.3.1
[13] promises_1.0.1 whisker_0.3-2 rmarkdown_1.10 tools_3.5.1
[17] stringr_1.4.0 glue_1.4.0 httpuv_1.4.5 yaml_2.2.1
[21] compiler_3.5.1 htmltools_0.3.6 knitr_1.20