Report
Francisco Bischoff
on Jan 31, 2022
Last updated: 2022-02-02
Checks: 6 1
Knit directory: false.alarm/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201020) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8d2fda3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Renviron
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .devcontainer/exts/
Ignored: .devcontainer/git-hours.tgz
Ignored: .devcontainer/mathjax.tgz
Ignored: .history
Ignored: .httr-oauth
Ignored: R/RcppExports.R
Ignored: _classifier/.gittargets
Ignored: _classifier/meta/process
Ignored: _classifier/meta/progress
Ignored: _classifier/objects/
Ignored: _classifier/user/
Ignored: _targets/meta/process
Ignored: _targets/meta/progress
Ignored: _targets/objects/
Ignored: _targets/user/
Ignored: dev/
Ignored: inst/extdata/
Ignored: output/images/
Ignored: papers/aime2021/aime2021.md
Ignored: presentations/MEDCIDS21/MEDCIDS21-10min_files/
Ignored: presentations/MEDCIDS21/MEDCIDS21_files/
Ignored: presentations/Report/Midterm-Report.md
Ignored: presentations/Report/Midterm-Report_cache/
Ignored: presentations/Report/Midterm-Report_files/
Ignored: protocol/SecondReport_cache/
Ignored: protocol/SecondReport_files/
Ignored: renv/library/
Ignored: renv/python/
Ignored: renv/staging/
Ignored: src/RcppExports.cpp
Ignored: src/RcppExports.o
Ignored: src/contrast.o
Ignored: src/false.alarm.so
Ignored: src/fft.o
Ignored: src/mass.o
Ignored: src/math.o
Ignored: src/mpx.o
Ignored: src/scrimp.o
Ignored: src/stamp.o
Ignored: src/stomp.o
Ignored: src/windowfunc.o
Ignored: tmp/
Untracked files:
Untracked: protocol/figure/floss_eval.pdf
Untracked: protocol/figure/floss_eval.svg
Untracked: protocol/figure/kappa.svg
Unstaged changes:
Modified: analysis/report.Rmd
Modified: analysis/style.css
Modified: codemeta.json
Modified: protocol/SecondReport.Rmd
Modified: protocol/figure/regime_filter.pdf
Modified: renv.lock
Staged changes:
Modified: protocol/FirstReport.Rmd
Modified: protocol/Protocol.Rmd
Deleted: protocol/figure/floss_eval1.png
Deleted: protocol/figure/kappa.png
Deleted: protocol/figure/targets.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/report.Rmd) and HTML
(docs/report.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | c0d48a7 | Francisco Bischoff | 2022-01-18 | remote some temps |
| html | 867bcf2 | Francisco Bischoff | 2022-01-16 | workflowr |
| Rmd | 571ac34 | Francisco Bischoff | 2022-01-15 | premerge |
| Rmd | dc34ece | Francisco Bischoff | 2022-01-10 | k_shapelets |
| html | 95ae431 | Francisco Bischoff | 2022-01-05 | blogdog |
| html | 7278108 | Francisco Bischoff | 2021-12-21 | update dataset on zenodo |
| Rmd | 1ef8e75 | Francisco Bischoff | 2021-10-14 | freeze for presentation |
| html | ca1941e | GitHub Actions | 2021-10-12 | Build site. |
| Rmd | 6b03f43 | Francisco Bischoff | 2021-10-11 | Squashed commit of the following: |
| html | c19ec01 | Francisco Bischoff | 2021-08-17 | Build site. |
| html | a5ec160 | Francisco Bischoff | 2021-08-17 | Build site. |
| html | b51dba2 | GitHub Actions | 2021-08-17 | Build site. |
| Rmd | c88cbd5 | Francisco Bischoff | 2021-08-17 | targets workflowr |
| html | c88cbd5 | Francisco Bischoff | 2021-08-17 | targets workflowr |
| html | e7e5d48 | GitHub Actions | 2021-07-15 | Build site. |
| Rmd | 1473a05 | Francisco Bischoff | 2021-07-15 | report |
| html | 1473a05 | Francisco Bischoff | 2021-07-15 | report |
| Rmd | 7436fbe | Francisco Bischoff | 2021-07-11 | stage cpp code |
| html | 7436fbe | Francisco Bischoff | 2021-07-11 | stage cpp code |
| html | 52e7f0b | GitHub Actions | 2021-03-24 | Build site. |
| Rmd | 7c3cc31 | Francisco Bischoff | 2021-03-23 | Targets |
| html | 7c3cc31 | Francisco Bischoff | 2021-03-23 | Targets |
Objectives and the research question
While this research was inspired on the CinC/Physionet Challenge 2015, its purpose is not to beat the state of the art on that challenge, but to identify, on streaming data, abnormal hearth electric patterns, specifically those which are life-threatening, using low CPU and low memory requirements in order to be able to generalize the use of such information on lower-end devices, outside the ICU, as ward devices, home devices, and wearable devices.
The main questions is: can we accomplish this objective using a minimalist approach (low CPU, low memory) while maintaining robustness?
Principles
This research is being conducted using the Research Compendium principles1:
- Stick with the convention of your peers;
- Keep data, methods, and output separated;
- Specify your computational environment as clearly as you can.
Data management follows the FAIR principle (findable, accessible, interoperable, reusable)2. Concerning these principles, the dataset was converted from Matlab’s format to CSV format, allowing more interoperability. Additionally, all the project, including the dataset, is in conformity with the Codemeta Project3.
Materials and methods
Softwares
Pipeline management
All steps of the process are being managed using the R package
targets4 from data
extraction to the final report. An example of a pipeline visualization
created with targets is shown in Fig. 1. This package helps
to keep record of the random seeds (allowing reproducibility), changes
in some part of the code (or dependencies) and then running only the
branches that need to be updated, and several other features to keep a
reproducible workflow avoiding unnecessary repetitions.
Figure 1 - Example of pipeline visualization using targets.
From left to right we see ‘Stems’ (steps that do not create branches)
and ‘Patterns’ (that contains two or more branches) and the flow of the
information. The green color means that the step is up to date to the
current code and dependencies.
Reports management
The report is available on the main webpage5, allowing inspection of previous
versions managed by the R package workflowr6. This package complements the
targets package by taking care of the versioning of every
report. It is like a Log Book that keeps track of every important
milestone of the project, while summarize the computational environment
where it was run. Fig. 2 shows only a fraction of the generated website,
where we can see that this version passed the required checks (system is
up-to-date, no caches, session information was recorded, and others) and
we see a table of previous versions.
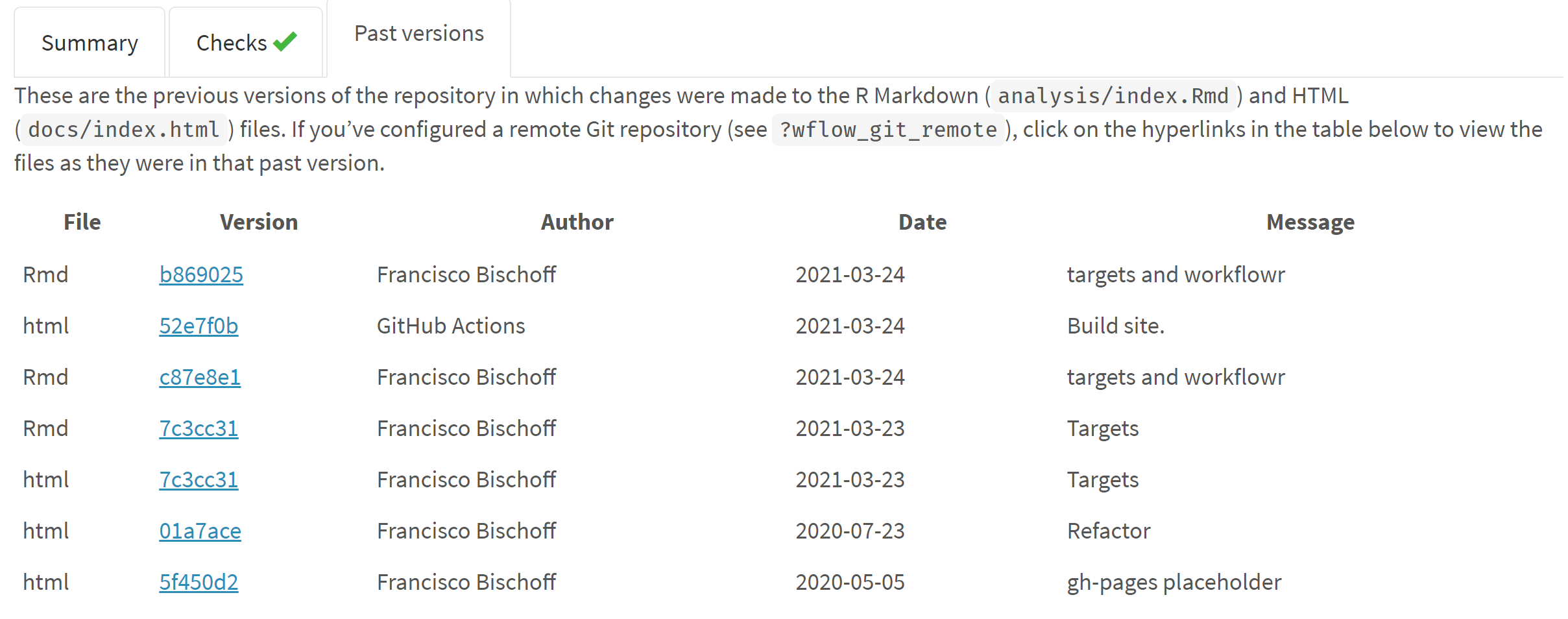
Figure 2 - Fraction of the website generated by workflowr.
On top we see that this version passed all checks, and in the middle we
see a table referring to the previous versions of the report.
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
Modeling and parameter tuning
The well known package used for data science in R is the
caret (short for Classification
And REgression
Training)7.
Nevertheless, the author of caret recognizes several
limitations of his (great) package, and is now in charge of the
development of the tidymodels8
collection. For sure, there are other available frameworks and
opinions9. Notwithstanding, this project
will follow the tidymodels road. Three significant
arguments 1) constantly improving and constantly being re-checked for
bugs; large community contribution; 2) allows to plug in a custom
modeling algorithm that, in this case, will be the one needed for
developing this work; 3) caret is not in active
development.
Continuous integration
Meanwhile, the project pipeline has been set up on GitHub, Inc.10 leveraging on Github Actions11 for the Continuous Integration lifecycle. The repository is available at10, and the resulting report is available at5. It is also public available the roadmap and tasks status of this thesis on Zenhub12.
Developed software
Matrix Profile
Matrix Profile (MP)13, is a state-of-the-art14,15 time series analysis technique that once computed, allows us to derive frameworks to all sorts of tasks, as motif discovery, anomaly detection, regime change detection and others13.
Before MP, time series analysis relied on what is called distance matrix (DM), a matrix that stores all the distances between two time series (or itself, in case of a Self-Join). This was very power consuming, and several methods of pruning and dimensionality reduction were researched16.
For brevity, let’s just understand that the MP and the companion Profile Index (PI) are two vectors that hold one floating point value and one integer value, respectively, regarding the original time series: (1) the similarity distance between that point on time (let’s call these points “indexes”) and its first nearest-neighbor (1-NN), (2) The index where this this 1-NN is located. The original paper has more detailed information13. It is computed using a rolling window but instead of creating a whole DM, only the minimum values and the index of these minimum are stored (in the MP and PI respectively). We can have an idea of the relationship of both on Fig. 3.
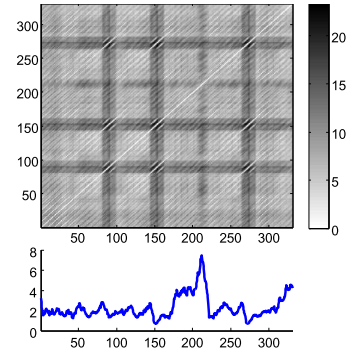
Figure 3 - A distance matrix (top), and a matrix profile (bottom). The matrix profile stores only the minimum values of the distance matrix.
This research has already yielded two R packages concerning the MP
algorithms from UCR17. The first package is called
tsmp, and a paper has also been published in the R
Journal18 (Journal Impact Factor™, 2020
of 3.984). The second package is called matrixprofiler and
enhances the first one, using low-level language to improve
computational speed. The author has also joined the Matrix Profile
Foundation as co-founder together with contributors from Python and Go
languages19,20.
This implementation in R is being used for computing the MP and MP-based algorithms of this thesis.
The data
The current dataset used is the CinC/Physionet Challenge 2015 public dataset, modified to include only the actual data and the header files in order to be read by the pipeline and is hosted by Zenodo21 under the same license as Physionet.
The dataset is composed of 750 patients with at least five minutes records. All signals have been resampled (using anti-alias filters) to 12 bit, 250 Hz and have had FIR band-pass (0.05 to 40Hz) and mains notch filters applied to remove noise. Pacemaker and other artifacts are still present on the ECG22. Furthermore, this dataset contains at least two ECG derivations and one or more variables like arterial blood pressure, photoplethysmograph readings, and respiration movements.
The events we seek to identify are the life-threatening arrhythmias as defined by Physionet in Table 1.
| Alarm | Definition |
|---|---|
| Asystole | No QRS for at least 4 seconds |
| Extreme Bradycardia | Heart rate lower than 40 bpm for 5 consecutive beats |
| Extreme Tachycardia | Heart rate higher than 140 bpm for 17 consecutive beats |
| Ventricular Tachycardia | 5 or more ventricular beats with heart rate higher than 100 bpm |
| Ventricular Flutter/Fibrillation | Fibrillatory, flutter, or oscillatory waveform for at least 4 seconds |
The fifth minute is precisely where the alarm has been triggered on the original recording set. To meet the ANSI/AAMI EC13 Cardiac Monitor Standards23, the onset of the event is within 10 seconds of the alarm (i.e., between 4:50 and 5:00 of the record). That doesn’t mean that there are no other arrhythmias before.
For comparison, on Table 2 we collected the score of the five best participants of the challenge24–28.
| Score | Authors |
|---|---|
| 81.39 | Filip Plesinger, Petr Klimes, Josef Halamek, Pavel Jurak |
| 79.44 | Vignesh Kalidas |
| 79.02 | Paula Couto, Ruben Ramalho, Rui Rodrigues |
| 76.11 | Sibylle Fallet, Sasan Yazdani, Jean-Marc Vesin |
| 75.55 | Christoph Hoog Antink, Steffen Leonhardt |
The equation used on this challenge to compute the score of the algorithms is in the Equation \(\eqref{score}\). This equation is the accuracy formula, with penalization of the false negatives. The reasoning pointed out by the authors22 is the clinical impact of existing a genuine life-threatening event that was considered unimportant. Accuracy is known to be misleading when there is a high class imbalance29.
\[
Score = \frac{TP+TN}{TP+TN+FP+5*FN} \tag{1} \label{score}
\]
Assuming that this is a finite dataset, the pathologic cases (1) \(\lim_{TP \to \infty}\) (whenever there is an event, it is positive) or (2) \(\lim_{TN \to \infty}\) (whenever there is an event, it is false), cannot happen. This dataset has 292 True alarms and 458 False alarms. Experimentally, this equation yields:
- 0.24 if all guesses are on False class
- 0.28 if random guesses
- 0.39 if all guesses are on True class
- 0.45 if no false positives plus random on True class
- 0.69 if no false negatives plus random on False class
This small experiment (knowing the data in advance) shows that “a single line of code and a few minutes of effort”30 algorithm could achieve at most a score of 0.39 in this challenge (the last two lines, the algorithm must to be very good on one class).
Nevertheless, this equation will only be useful to allow us to compare the results of this thesis with other algorithms.
Work structure
Project start
The project started with a literature survey on the databases Scopus, PubMed, Web of Science, and Google Scholar with the following query (the syntax was adapted for each database):
TITLE-ABS-KEY ( algorithm OR ‘point of care’ OR ‘signal processing’ OR
‘computer assisted’ OR ‘support vector machine’ OR ‘decision support
system’ OR ’neural network’ OR ‘automatic interpretation’ OR
‘machine learning’) AND TITLE-ABS-KEY ( electrocardiography OR
cardiography OR ‘electrocardiographic tracing’ OR ecg OR
electrocardiogram OR cardiogram ) AND TITLE-ABS-KEY ( ‘Intensive care
unit’ OR ‘cardiologic care unit’ OR ‘intensive care center’ OR
‘cardiologic care center’ )
The inclusion and exclusion criteria were defined as in Table 3.
| Inclusion criteria | Exclusion criteria |
|---|---|
| ECG automatic interpretation | Manual interpretation |
| ECG anomaly detection | Publication older than ten years |
| ECG context change detection | Do not attempt to identify life-threatening arrhythmias, namely asystole, extreme bradycardia, extreme tachycardia, ventricular tachycardia, and ventricular flutter/fibrillation |
| Online Stream ECG analysis | No performance measurements reported |
| Specific diagnosis (like a flutter, hyperkalemia, etc.) |
The survey is being conducted with peer review, all articles on full-text phase were obtained and assessed for the extraction phase, with exception of 5 articles that were not available. The survey is currently staled on the Data Extraction phase due to external factors.
Fig. 4 shows the flow diagram of the resulting screening using PRISMA format.
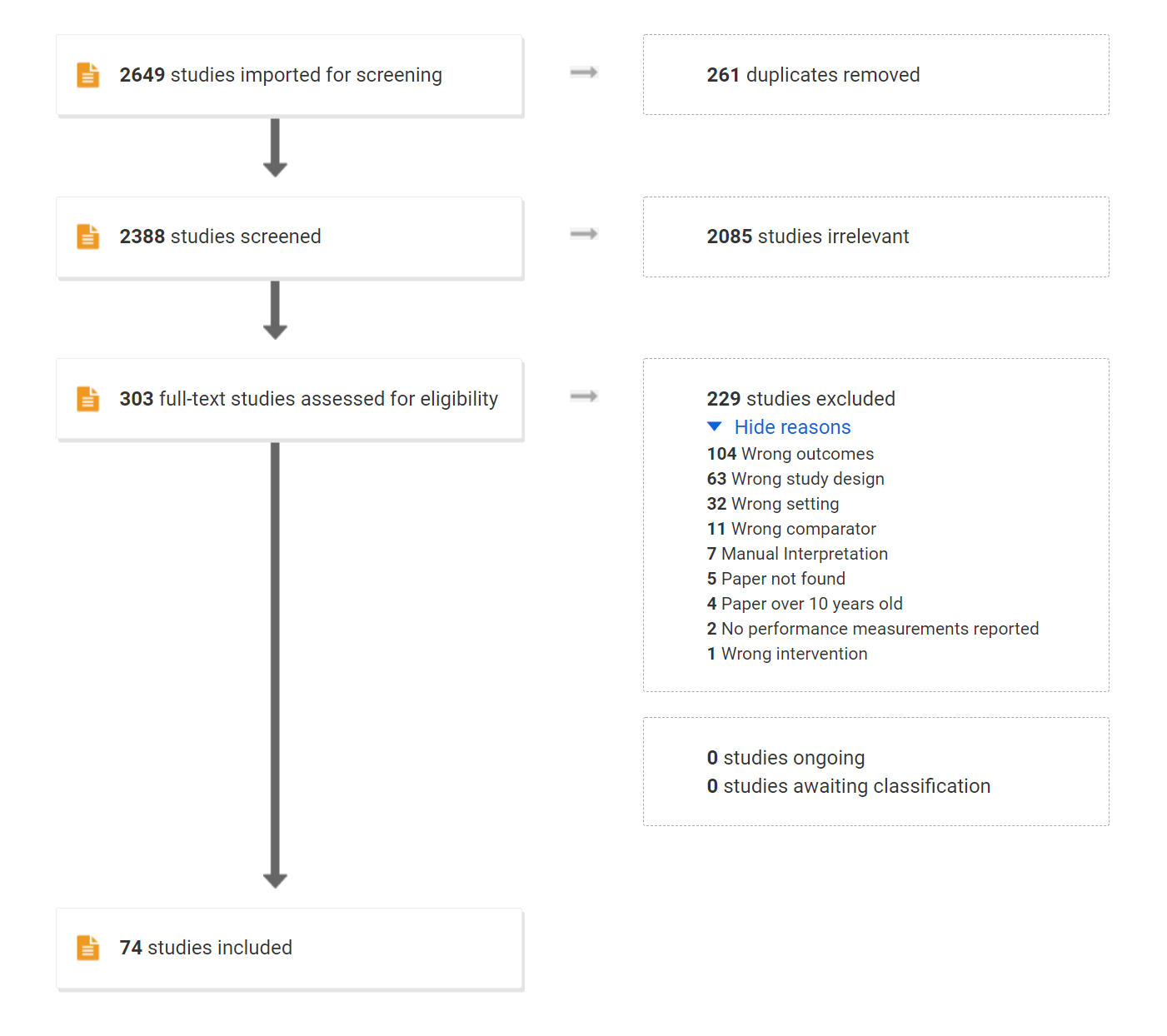
Figure 2 - Flowchart of the literature survey.
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
The peer review is being conducted by the author of this thesis together with another coleague, Dr. Andrew Van Benschoten from the Matrix Profile Foundation19.
Table. 4 shows the Inter-rater Reliability (IRR) of the screening phases, using Cohen’s \(\kappa\) statistic. The bottom line shows the estimated accuracy after corrected for possible confounders31.
|
Title-Abstract (2388 articles) |
Full-Review (303 articles) |
|||||
|---|---|---|---|---|---|---|
| Reviewer #2 | Reviewer #2 | |||||
| Include | Exclude | Include | Exclude | |||
| Reviewer #1 | Include | 185 | 381 | 63 | 58 | |
| Exclude | 129 | 1693 | 13 | 169 | ||
| Cohen’s omnibus \(\kappa\) | 0.30 | 0.48 | ||||
| Maximum possible \(\kappa\) | 0.66 | 0.67 | ||||
| Std Err for \(\kappa\) | 0.02 | 0.05 | ||||
| Observed Agreement | 79% | 77% | ||||
| Random Agreement | 69% | 55% | ||||
| Agreement corrected with KappaAcc | 82% | 85% | ||||
The purpose of using Cohen’s \(\kappa\) in such review is to allow us to gauge the agreement of both reviewers on the task of selecting the articles according to the goal of the survey. The most naive way to verify this would be simply to measure the overall agreement (the number of articles included and excluded by both, divided by the total number of articles). Nevertheless, this would not take into account the agreement we could expect purely by chance.
However, the \(\kappa\) statistic must be assessed carefully. This topic is beyond the scope of this work therefore it will be explained briefly.
While it is widely used, the \(\kappa\) statistic is also well criticized. The direct interpretation of its value depends on several assumptions that are often violated. (1) It is assumed that both reviewers have the same level of experience; (2) The “codes” (include, exclude) are identified with same accuracy; (3) The “codes” prevalence are the same; (4) There is no reviewer bias towards one of the choices32,33.
In addition, the number of “codes” affects the relation between the value of \(\kappa\) and the actual agreement between the reviewers. For example, given equiprobable “codes” and reviewers who are 85% accurate, the value of \(\kappa\) are 0.49, 0.60, 0.66, and 0.69 when number of codes is 2, 3, 5, and 10, respectively33,34.
In order to take these limitations in account, the agreement between reviewers was calculated using the KappaAcc31 from Professor Emeritus Roger Bakeman, Georgia State University, which computes the estimated accuracy of simulated reviewers.
RAW data
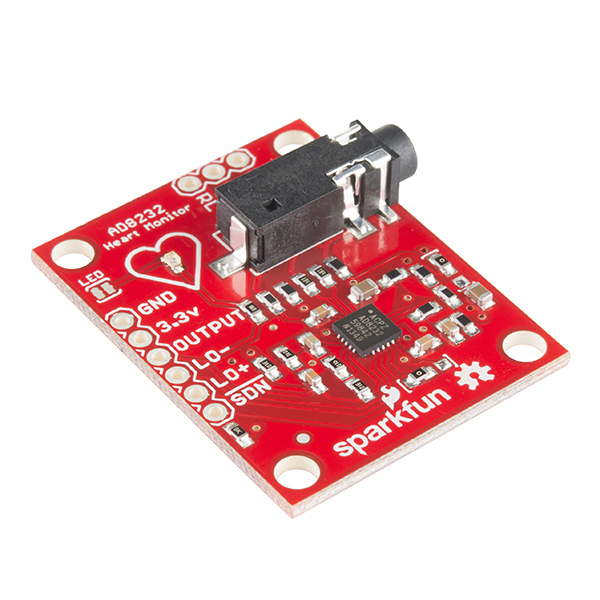
In order to better understand the data acquisition, it has been acquired a Single Lead Heart Rate Monitor breakout from Sparkfun™35 using the AD823236 microchip from Analog Devices Inc., compatible with Arduino®37, for an in-house experiment (Fig. 5).
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
The output gives us a RAW signal, as shown in Fig. 6.
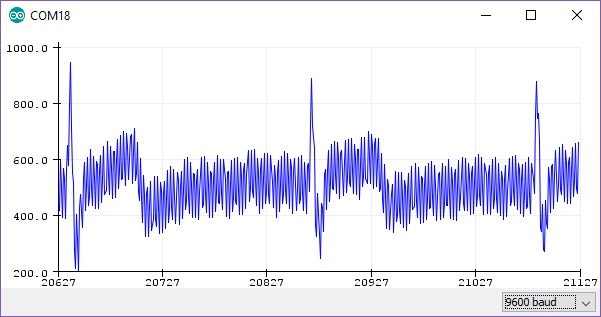
Figure 6 - RAW output from Arduino at ~300hz
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
After applying the same settings as the Physionet database (collecting the data at 500hz, resample to 250hz, pass-filter, and notch filter), the signal is much better, as shown in Fig. 7.
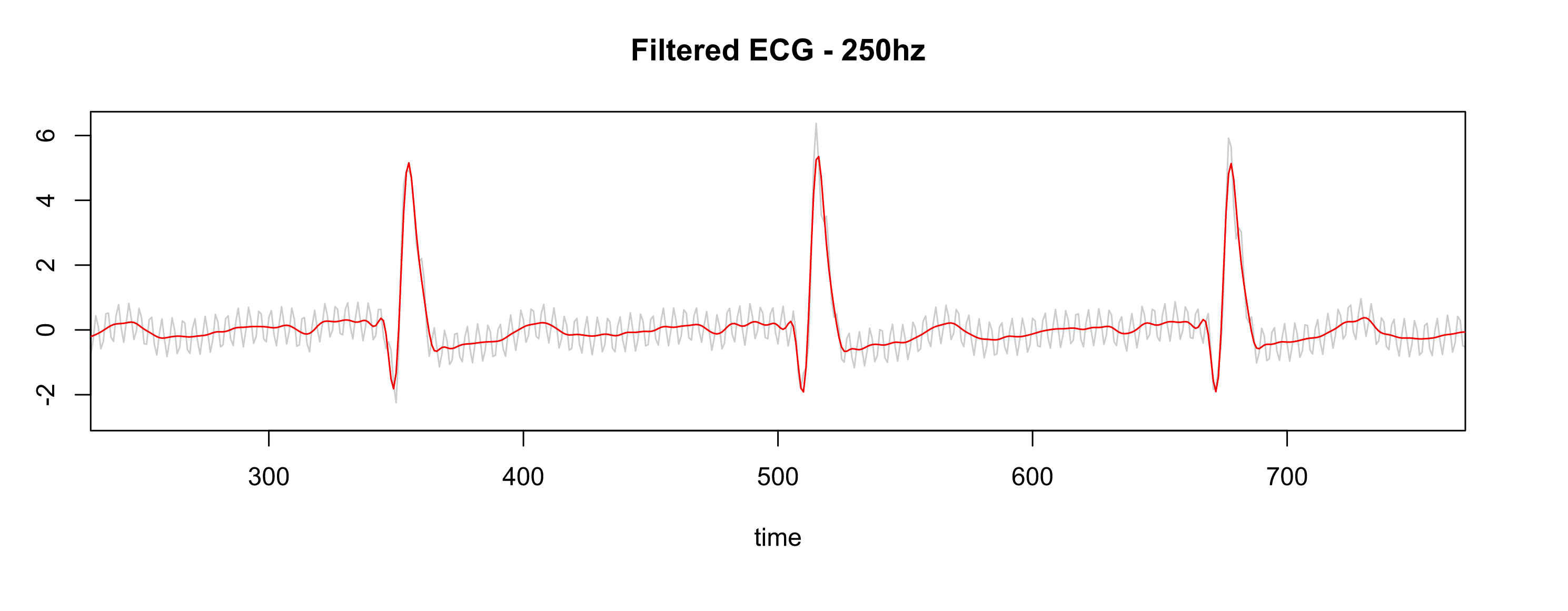
Figure 7 - Gray is RAW, Red is filtered
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
Preparing the data
Usually, data obtained by sensors needs to be “cleaned” for proper evaluation. That is different from the initial filtering process where the purpose is to enhance the signal. Here we are dealing with artifacts, disconnected cables, wandering baselines and others.
Several SQIs (Signal Quality Indexes) are used in the literature38, some trivial measures as kurtosis, skewness, median local noise level, other more complex as pcaSQI (the ratio of the sum of the five largest eigenvalues associated with the principal components over the sum of all eigenvalues obtained by principal component analysis applied to the time aligned ECG segments in the window). By experimentation (yet to be validated), a simple formula gives us the “complexity” of the signal and correlates well with the noisy data is shown in Equation \(\eqref{complex}\).
\[
\sqrt{\sum_{i=1}^w((x_{i+1}-x_i)^2)}, \quad \text{where}\; w \; \text{is
the window size} \tag{2} \label{complex}
\]
The Fig. 8 shows some SQIs and their relation with the data.
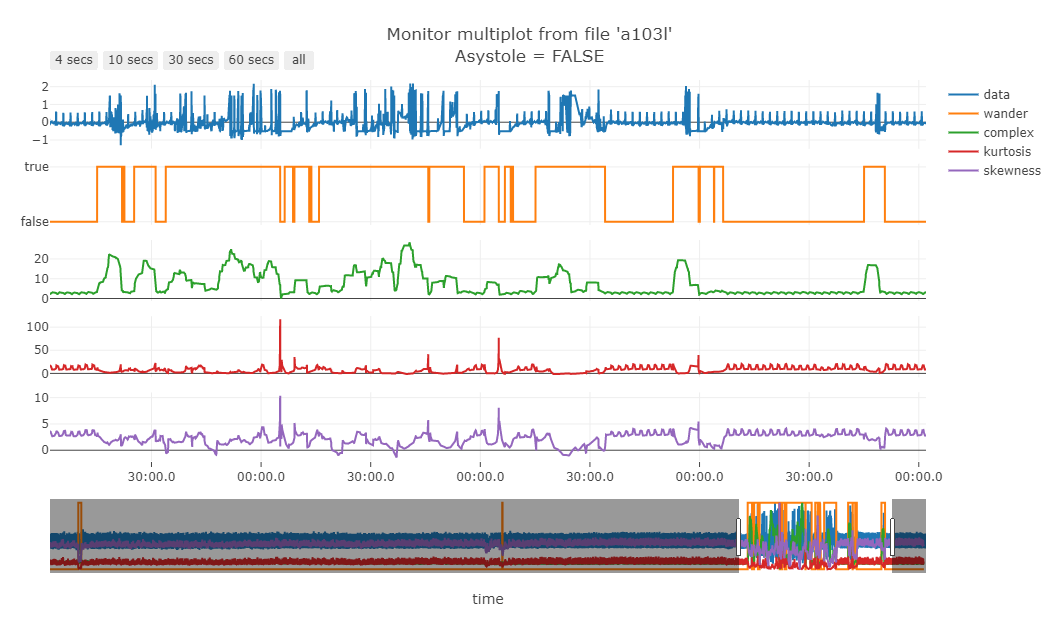
Figure 8 - Green line is the “complexity” of the signal
| Version | Author | Date |
|---|---|---|
| 1473a05 | Francisco Bischoff | 2021-07-15 |
Fig. 9 shows that noisy data (probably patient muscle movements) are marked with a blue point and thus are ignored by the algorithm.
Figure 9 - Noisy data marked by the “complexity” filter
Although this step of “cleaning” the data is often used, this step will also be tested if it is really necessary and the performance with and without “cleaning” will be reported.
Detecting regime changes
The regime change approach will be using the Arc Counts concept, used on the FLUSS (Fast Low-cost Unipotent Semantic Segmentation) algorithm, as explained by Gharghabi, et al.,39.
The FLUSS (and FLOSS, the on-line version) algorithm is built on top of the Matrix Profile (MP)13, described on section 1. Recalling that the MP and the companion Profile Index (PI) are two vectors holding information about the 1-NN. One can imagine several “arcs” starting from one “index” to another. This algorithm is based on the assumption that between two regimes, the most similar shape (its nearest neighbor) is located on “the same side”, so the number of “arcs” decreases when there is a change on the regime, and increases again. As show on Fig. 10. This drop on the Arc Counts is a signal that a change on the shape of the signal has happened.
Figure 10 - FLUSS algorithm, using arc counts.
The choice of the FLOSS algorithm (on-line version of FLUSS) is founded on the following arguments:
- Domain Agnosticism: the algorithm makes no assumptions about the data as opposed to most available algorithms to date.
- Streaming: the algorithm can provide real-time information.
- Real-World Data Suitability: the objective is not to explain all the data. Therefore, areas marked as “don’t know” areas are acceptable.
- FLOSS is not: a change point detection algorithm40. The interest here is changes in the shapes of a sequence of measurements.
Other algorithms we can cite are based on Hidden Markov Models (HMM) that require at least two parameters to be set by domain experts: cardinality and dimensionality reduction. The most attractive alternative could be the Autoplait41, which is also domain agnostic and parameter-free. It segments the time series using Minimum Description Length (MDL) and recursively tests if the region is best modeled by one or two HMM. However, Autoplait is designed for batch operation, not streaming, and also requires discrete data. FLOSS was demonstrated to be superior in several datasets in its original paper. In addition, FLOSS is robust to several changes in data like downsampling, bit depth reduction, baseline wandering, noise, smoothing, and even deleting 3% of the data and filling with simple interpolation. Finally, the most important, the algorithm is light and suitable for low-power devices.
In the MP domain, it is worth also mentioning other possible algorithm: the Time Series Snippets42, based on MPdist43. The latter measures the distance between two sequences considering how many similar sub-sequences they share, no matter the order of matching. It proved to be a useful measure (not a metric) for meaningfully clustering similar sequences. Time Series Snippets exploits MPdist properties to summarize a dataset extracting the \(k\) sequences that represent most of the data. The final result seems to be an alternative for detecting regime changes, but it is not. The purpose of this algorithm is to find which pattern(s) explains most of the dataset. Also, it is not suitable for streaming data. Lastly, MPdist is quite expensive compared to the trivial Euclidean distance.
The regime change detection will be evaluated following the criterias explained on section 1.
Classification of the new regime
The next step towards the objective of this work is to verify if the new regime detected by the previous step is indeed a life-threatening pattern that we should trigger the alarm.
First let’s dismiss some apparent solutions: (1) Clustering. It is well understood that we cannot cluster time series subsequences meaningfully with any distance measure, or with any algorithm44. The main argument is that in a meaningfull algorithm, the output depends on the input, and this has been proven to not happen in time series subsequence clustering44. (2) Anomaly detection. In this work we are not looking for surprises, but for patterns that are known to be life-threatening. (3) Forecasting. We may be tempted to make predictions, but clearly this is not the idea here.
The method of choice is classification. The simplest algorithm could
be a TRUE/FALSE binary classification.
Nevertheless, the five life-threatening patterns have well defined
characteristics that may seem more plausible to classify the new regime
using some kind of ensamble of binary classifiers or a “six-class”
classifier (being the sixth class the FALSE class).
Since the model doesn’t know which life-threatening pattern will be
present in the regime (or if it will be a FALSE case), the
model will need to check for all five TRUE cases and if
none of these cases are identified, it will classify the regime as
FALSE.
In order to avoid exceeding processor capacity, an initial set of
shapelets45 can be sufficient to build the
TRUE/FALSE classifier. And to build such set
of shapelets, leveraging on the MP, we will use the Contrast
Profile46.
The Contrast Profile (CP) looks for patterns that are at the same time very similar to its neighbors in class A while is very different from the nearest neighbor from class B. In other words, this means that such pattern represents well class A and may be taken as a “signature” of that class.
In this case we need to compute two MP, one self-join MP using the positive class \(MP^{(++)}\) (the class that has the signature we want to find) and one AB-join MP using the positive and negative classes \(MP^{(+-)}\). Then we subtract the first \(MP^{(++)}\) from the last \(MP^{(+-)}\), resulting in the \(CP\). The high values on \(CP\) are the locations for the signature candidates we look for (the author of CP calls these segments Plato’s).
Due to the nature of this approach, the MP’s (containing values in Euclidean Distance) are truncated for values above \(\sqrt{2w}\), where \(w\) is the window size. This because values above this threshold are negatively correlated in the Pearson Correlation space. Finally, we normalize the values by \(\sqrt{2w}\). The formula \(\eqref{contrast}\) synthesizes this computation.
\[
CP_w = \frac{MP_{w}^{(+-)} - MP_{w}^{(++)}}{\sqrt{2w}} \quad
\text{where}\; w \; \text{is the window size} \tag{3} \label{contrast}
\]
For a more complete understanding of the process, Fig. 11 shows a practical example from the original article46.
Figure 11 - Top to bottom: two weakly-labeled snippets of a larger time series. T(-) contains only normal beats. T(+) also contains PVC (premature ventricular contractions). Next, two Matrix Profiles with window size 91; AB-join is in red and self-join in blue. Bottom, the Contrast Profile showing the highest location.
After extracting candidates for each class signature, a classification algorithm will be fitted and evaluated using the criterias explained on section 1.
Summary of the methodology
In order to summarize the steps taken on this thesis to accomplish the main objective, Figs. 12, 13 and 14 show the overview of the processes involved.
First let us introduce the concept of Nested Resampling47. It is known that when increasing model complexity, overfitting on the training set becomes more likely to happen48. This is an issue that this work has to countermeasure as there are many steps that requires parameter tuning, even for algorithms that are almost parameter-free like the MP.
The rule that must be followed is simple: do not evaluate a model on the same resampling split used to perform its own parameter tuning. Using simple cross-validation, the information about the test set “leaks” into the evaluation, which leads to overfitting/overtuning, and gives us an optimistic biased estimative of the performance. Bernd Bischl, 201247 describes more deeply these factors, and also gives us a countermeasure for that: (1) from preprocessing the data to model selection use the training set; (2) the test set should be touched once, on the evaluation step; (3) repeat. This guarantees that a “new” separated data is only used after the model is trained/tuned.
Fig. 15 shows us this principle. The steps (1) and (2) described above are part of the Outer resampling, which in each loop splits the data in two sets: the training set and the test set. The training set is then used in the Inner resampling where, for example, the usual cross-validation may be used (creating an Analysis set and an Assessment set, to avoid conflict of terminology), and the best model/parameters is selected. Then, this best model is evaluated against the unseen test set that was created for this resampling.
The resulting (aggregated) performance of all outer samples gives us a more honest estimative of the expected performance on new data.
Figure 15 - Nested resampling. The full dataset is resampled several times (outer resampling), so each branch has its own Test set (yellow). On each branch, the Training set is used as if it were a full dataset, being resampled again (inner resampling); here the Assessment set (blue) is used to test the learning model and tune parameters. The best model then, is finally evaluated on its own Test set.
After the understanding of the Nested Resampling47, the following flowcharts can be better interpreted. Fig. 12 starts with the “Full Dataset” that contains all time series from the dataset described on section 1. Each time series represents one file from the database, and represents one patient.
The regime change detection will use subsampling (bootstrapping can lead to substantial bias toward more complex models) in the Outer resampling and cross-validation in the Inner resampling. How the evaluation will be performed and why the use of cross-validation will be explained on section 1.
Figure 12 - Pipeline for regime change detection. The full dataset (containing several patients) is divided on a Training set and a Test set. The Training set is then resampled in an Analysis set and an Assessment set. The former is used for training/parameter tuning and the latter for assessing the result. The best parameters are then used for evaluation on the Test set. This may be repeated several times.
Fig. 13 shows the processes for training the classification model.
First, the last ten seconds of each time series will be identified (the
even occurs in this segment). Then the dataset will be grouped by class
(type of event) and TRUE/FALSE (alarm), so the
Outer/Inner resampling will produce a Training/Analysis set and
Test/Assessment set with similar frequency of the full dataset.
The next step will be to extract shapelet candidates using the Contrast Profile and train the classifier.
This pipeline will use subsampling (for the same reason above) in the Outer resampling and cross-validation in the Inner resampling. How the evaluation will be performed and why the use of cross-validation will be explained on section 1.
Figure 13 - Pipeline for alarm classification. The full dataset (containing several patients) is grouped by class and by TRUE/FALSE alarm. This grouping allows resampling to keep a similar frequency of classes and TRUE/FALSE of the full dataset. Then the full dataset is divided on a Training set and a Test set. The Training set is then resampled in an Analysis set and an Assessment set. The former is used for extracting shapelets, training the model and parameter tuning; the latter for assessing the performance of the model. Finally, the best model is evaluated on the Test set. This may be repeated several times.
Finally, Fig. 14 shows how the final model will be used on the field. In a streaming scenario, the data will be collected and processed in real-time to maintain an up to date Matrix Profile. The FLOSS algorithm will be looking for a regime change. When a regime change is detected, a sample of this new regime will be presented to the trained classifier that will evaluate if this new regime is a life-threatening condition or not.
Pipeline of the final process. The streaming data, coming from one patient, is processed to create its Matrix Profile. Then, the FLOSS algorithm is computed for detecting a regime change. When a new regime is detected, a sample of this new regime is analysed by the model and a decision is made. If the new regime is life-threatening, the alarm will be fired.
Evaluation of the algorithms
The subsampling method used on both algorithms, regime change and classification, will be the Cross Validation, as the learning task will be in batches.
Other options dismissed47:
Leave-One-Out Cross Validation: has better properties for regression than for classification. It has a high variance as an estimator of the mean loss. It also is asymptotically inconsistent and tends to select too complex models. It is demonstrates empirically that 10-fold CV is often superior.
Bootstrapping: while it has low variance, it may be optimistic biased on more complex models. Also, its resampling method with replacement can leak information into the assessment set.
Subsampling: is like bootstrapping, but without replacement. The only argument for not choosing it, is that with Cross Validation we make sure all the data is used for analysis and assessment.
Regime change
A detailed discussion about the evaluation process of segmentation algorithms is made by the FLUSS/FLOSS author39. Previous researches have used precision/recall or derived measures for performance. The main issue is how to assume that the algorithm was correct? If the ground truth says the change occurred at location 10,000, and the algorithm detects a change at location 10,001, is this a miss?
As pointed out by the author, several independent researchers have suggested a temporal tolerance, that solves one issue, but also has a hard time on penalize any tiny miss beyond this tolerance.
The second issue is a over-penalization of an algorithm in which most of the detections are good, but just one (or a few) is poor.
The author proposes the solution depicted in Fig. 16. It gives 0 as the best score and 1 as the worst. The function sums the distances between the ground truth locations and the locations suggested by the algorithm. The sum is then divided by the length of the time series to normalize the range to [0, 1].
The goal is minimizing this score.
Figure 16 - Regime change evaluation. The top line illustrates the ground truth, and the bottom line the locations reported by the algorithm. Note that multiple proposed locations can be mapped to a single ground truth point.
Classification
As described on section 1, the model for classification will use a
set of shapelets to identify if we have a TRUE
(life-threatening) regime or a FALSE (non life-threatening)
regime.
Although the implementation of the final process will be using streaming data, the classification algorithm will work in batches, because it will not be applied on every single data point, but on samples that are extracted when a regime change is detected. During the training phase, the data is also analyzed in batches.
One important factor we must consider is that, on real world, the
majority of regime changes will be FALSE (i.e., not
life-threatening). Thus, a performance measure that is robust to class
imbalance is needed if we want to be able to assess the model after it
was trained, on the field.
It is well known that the Accuracy measure is not reliable for unbalanced data29,49 as it returns optimistic results for a classifier on the majority class. A description of common measures used on classification is available29,50. Here we will focus on three candidate measures that can be used: F-score (well discussed on50), Matthew’s Correlation Coefficient (MCC)51 and \(\kappa_m\) statistic52.
The F-score (let’s abbreviate to F1 as this is the more
common setting), is widely used on information retrieval, where
the classes are usually classified as “relevant” and “irrelevant”, and
combines the recall (also known as sensitivity) and the
precision (the positive predicted value). Recall
assess how well the algorithm retrieves relevant examples among the
(usually few) relevant items in dataset, while precision assess
the proportion of indeed relevant items are contained in the retrieved
examples. It ranges from [0, 1]. It ignores completely the irrelevant
items that were not retrieved (usually this set contain lots of items).
In classification tasks, its main weakness is not evaluate the True
Negatives, and if the proportion of a random classifier gets towards the
TRUE class (increasing the False Positives significantly),
this score actually gets better, thus not suitable to our case. The
F1 score is defined on equation \(\eqref{score}\).
\[ F_1 score = \frac{2 \cdot TP}{2 \cdot TP + FP + FN} = 2 \cdot \frac{precision \cdot recall}{precision + recall} \tag{4} \label{fscore} \]
The MCC is a good alternative to the F1 when we do care about the True negatives (both were considered to “provide more realistic estimates of real-world model performance”53). It is a method to compute the Pearson product-moment correlation coefficient54 between the actual and predicted values. It ranges from [-1, 1]. The MCC is the only binary classification rate that only gives a high score if the binary classifier was able to correctly classify the majority of the positive and negative instances50. One may argue that Cohen’s \(\kappa\) has the same behavior, but there are two main differences (1) MCC is undefined in the case of a majority voter while Cohen’s \(\kappa\) doesn’t discriminates this case from the random classifier (\(\kappa\) is zero for both cases) (2) It is proven that in an special case when the classifier is increasing the False Negatives, Cohen’s \(\kappa\) doesn’t get worse as spected, MCC doesn’t have this issue54. MCC is defined on equation \(\eqref{mccval}\).
\[ MCC = \frac{TP \cdot TN - FP \cdot FN}{\sqrt{(TP + FP) \cdot (TP + FN) \cdot (TN + FP) \cdot (TN + FN)}} \tag{5} \label{mccval} \]
The \(\kappa_m\) statistic52 is a measure that takes in account not the random classifier but the majority voter (a classifier that only votes on the larger class). It was introduced by Bifet et al.52 for being used in online settings, where the class balance may change over time. It is defined on equation \(\eqref{kappam}\), where \(p_0\) is the observed accuracy and \(p_m\) is the accuracy of the majority voter. The score ranges from (\(-\infty\), 1], theoretically, but in practice you see negative numbers if the classifier is performing worse than the majority voter and positive numbers if performing better than the majority number, until the maximum of 1, when the classifier is optimal.
\[ \kappa_m = \frac{p_0 - p_m}{1 - p_m} \tag{6} \label{kappam} \]
In the inner resampling (model training/tuning), the classification will be binary, and in our case we know that the data is slightly unbalanced (60% false alarms). For this step, the metric for model selection will be the MCC. Nevertheless, during the optimization process, the algorithm will seek to minimize the False Negative Rate (\(FNR = \frac{FN}{TP+FN}\)), and between ties, the smaller FNR wins.
In the outer resampling, the MCC and \(\kappa_m\) of all winning models will aggregated and reported using the median and interquartile range.
For different classifiers, we will use the Wilcoxon’s signed-rank test for comparing their performances, as this method is known to have low Type I and Type II errors in this kind of comparison52.
Full model (streaming setting)
For the final assessment, the best and the average model of the previous pipelines will be assembled and tested using the whole original dataset.
The algorithm will be tested in each of the five life-threatening event split individually, in order to evaluate its strengths and weakness.
For more transparency, the whole confusion matrix will be reported, as well as the MCC, \(\kappa_m\), and the FLOSS evaluation.
Current results
Regime change detection
The current status on regime change detection pipeline is the implementation of the resampling strategies and evaluation in order to start the parameter tuning. An example of the current implementation is shown on Fig. 17.
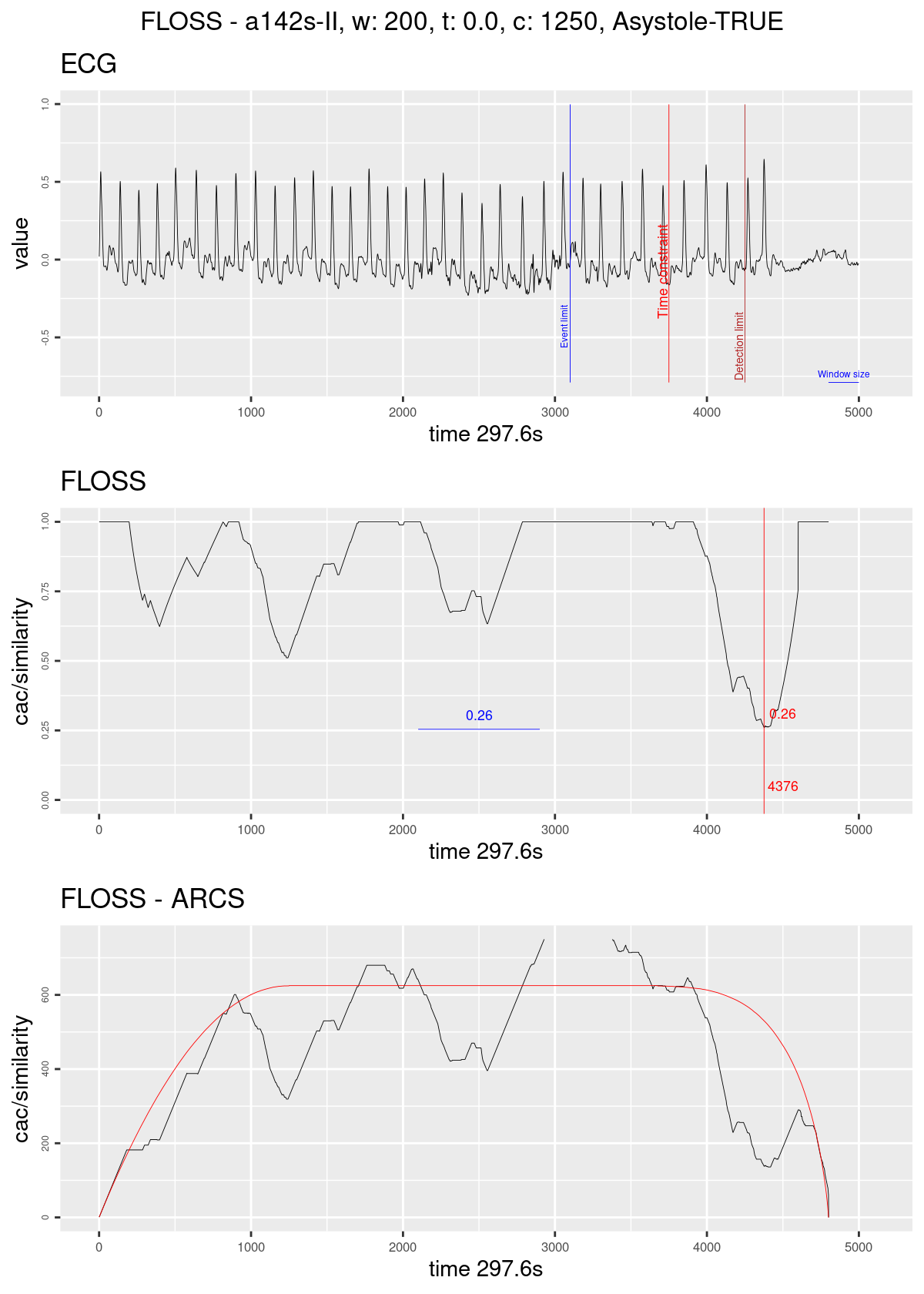
Figure 17 - Regime change detection example. The graph on top shows the ECG streaming; the blue line marks the ten seconds before the original alarm was fired; the red line marks the time constraint of 1250; the dark red line marks the limit for taking a decision in this case of Asystole the blue horizontal line represents the size of the sliding window. The graph on the middle shows the Arc counts as seen by the algorithm (with the corrected distribution); the red line marks the current minimum value and its index; the blue horizontal line shows the minimum value seen until then. The graph on the bottom shows the computed Arc counts (raw) and the red line is the theoretical distribution used for correction.
Classification
The current status on the classification pipeline is the implementation of the shapelets extraction using the Contrast Profile.
An example of candidates for ventricular tachycardia is presented on Fig. 18.
Figure 18 - Shapelet candidates for Ventricular Tachycardia.
Feasibility trial
A side-project called “false.alarm.io” has been derived from this work (an unfortunate mix of “false.alarm” and “PlatformIO”55, the IDE chosen to interface the panoply of embedded systems we can experiment with). The current results of this side-project are very enlightening and show that the final algorithm can indeed be used in small hardware. Further data will be available in the future.
A brief mentioning, linking back to the objectives of this work, an initial trial was done using an ESP32 MCU (Fig. 19) in order to be sure if such small device can handle the task.
Figure 19 - ESP32 MCU
| Version | Author | Date |
|---|---|---|
| 571ac34 | Francisco Bischoff | 2022-01-15 |
Current results show that such device has enough computation power to handle the task in real-time using just one of its two microprocessors. The main limitation seen in advance is the on-chip SRAM that must be well managed.
Scientific contributions
Matrix Profile
Since the first paper presenting this new concept13, lots of investigations were made to speed up its computation. It is notable how all computations are not dependent on the rolling window size as previous works not using Matrix Profile. Aside from this, we can see that the first STAMP13 algorithm has the time complexity of \(O(n^2log{n})\) while STOMP56 \(O(n^2)\) (a significant improvement), but STOMP lacks the “any-time” property. Later SCRIMP57 solves this problem keeping the same time complexity of \(O(n^2)\). Here we are in the “exact” algorithms domain and we will not extend the scope for conciseness.
The main issue with the algorithms above is the dependency on a fast Fourier transform (FFT) library. FFT has been extensively optimized and architecture/CPU bounded to exploit the most of speed. Also, padding data to some power of 2 happens to increase the efficiency of the algorithm. We can argue that time complexity doesn’t mean “faster” when we can exploit low-level instructions. In our case, using FFT in a low-power device is overkilling. For example, a quick search over the internet gives us a hint that computing FFT on a 4096 data in an ESP32 takes about 21ms (~47 computations in 1 second). This means ~79 seconds for computing all FFT’s (~3797) required for STAMP using a window of 300. Currently, we can compute a full matrix of 5k data in about 9 seconds in an ESP32 MCU (Fig. 19), and keep updating it as fast as 1 min of data (at 250hz) in just 6 seconds.
Recent works using exact algorithms are using an unpublished algorithm called MPX, which computes the Matrix Profile using cross-correlation methods ending up faster and is easily portable.
On computing the Matrix Profile: the contribution of this work on this area is adding the Online capability to MPX, which means we can update the Matrix Profile as new data comes in.
On extending the Matrix Profile: the contribution of this work on this area is the use of an unexplored constraint that we could apply on building the Matrix Profile we are calling Similarity Threshold (ST). The original work outputs the similarity values in Euclidean Distance (ED) values, while MPX naturally outputs the values in Pearson’s correlation coefficients (CC). Both ED and CC are interchangeable using the equation \(\eqref{edcc}\). However, we may argue that it is easier to compare values that do not depend on the window size during an exploratory phase. MPX happens to naturally return values in CC, saving a few more computation time. The ST is an interesting factor that we can use, especially when detecting pattern changes during time. The FLOSS algorithm relies on counting references between indexes in the time series. ST can help remove “noise” from these references since only similar patterns above a certain threshold are referenced, and changes have more impact on these counts. The best ST threshold is still to be determined.
\[
CC = 1 - \frac{ED}{(2 \times WindowSize)} \tag{7} \label{edcc}
\]
Regime change detection
In the original paper, in chapter 3.5, the authors of FLOSS wisely introduce the temporal constraint, which improves the sensitivity of regime change detection on situations where a regime may alternate in short periods of time.
Nevertheless, the authors declare the correction curve typically used on the algorithm as “simply a uniform distribution”, but this is not an accurate statement. the Arc Counts of newly incoming data are truncated by the same amount of temporal constraint. This prevents completely the detection of a regime change in the last 10 seconds as this thesis requires.
The main contribution of this work on this area is overcoming this issue by computing the theoretical distribution using the temporal constraint parameters beforehand. as shown in Fig. 20. That gives us enough data to evaluate a regime change accurately utilizing a minimum of \(2 \times WindowSize\) datapoints.
Figure 20 - 1D-IAC distributions for earlier temporal constraint (on Matrix Profile)
Scientific outcomes
This research has already yielded two R packages concerning the MP
algorithms from UCR17. The first package is called
tsmp, and a paper has also been published in the R
Journal18 (Journal Impact Factor™, 2020
of 3.984). The second package is called matrixprofiler and
enhances the first one, using low-level language to improve
computational speed. The author has also joined the Matrix Profile
Foundation as co-founder together with contributors from Python and Go
languages19,20. The benchmarks of the R
implementation are available online58.
Additionally to the above publication and the publication of the ongoing literature survey it is planned to be published two articles about this thesis subject. The first regarding the application of the FLOSS algorithm on real-time ECG showing its potential on using it on low-power devices. The second regarding the use of combined shapelets for relevant ECG patterns identification.
Expected results and outcomes
At the end, this thesis will provide a framework for identify life-threatening conditions using biological streaming data on devices with low CPU and low memory specifications. We expect to achieve a high quality model on identifying these pathological conditions, maintaining its robustness in presence of noise and artifacts seen on real-world applications.
R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] stringr_1.4.0 ggplot2_3.3.5 gridExtra_2.3
[4] kableExtra_1.3.4 tibble_3.1.6 visNetwork_2.1.0
[7] glue_1.6.1 gittargets_0.0.1.9000 tarchetypes_0.4.1
[10] targets_0.10.0 workflowr_1.7.0 here_1.0.1
loaded via a namespace (and not attached):
[1] fs_1.5.2 bit64_4.0.5 webshot_0.5.2
[4] httr_1.4.2 rprojroot_2.0.2 tools_4.1.2
[7] backports_1.4.1 bslib_0.3.1 utf8_1.2.2
[10] R6_2.5.1 colorspace_2.0-2 withr_2.4.3
[13] tidyselect_1.1.1 processx_3.5.2 bit_4.0.4
[16] compiler_4.1.2 git2r_0.29.0 cli_3.1.1
[19] rvest_1.0.2 xml2_1.3.3 labeling_0.4.2
[22] sass_0.4.0 scales_1.1.1 checkmate_2.0.0
[25] readr_2.1.2 callr_3.7.0 askpass_1.1
[28] systemfonts_1.0.3 digest_0.6.29 rmarkdown_2.11.15
[31] svglite_2.0.0.9000 pkgconfig_2.0.3 htmltools_0.5.2
[34] fastmap_1.1.0 highr_0.9 htmlwidgets_1.5.4
[37] rlang_1.0.0.9000 rstudioapi_0.13 farver_2.1.0
[40] jquerylib_0.1.4 generics_0.1.2 jsonlite_1.7.3
[43] vroom_1.5.7 dplyr_1.0.7 magrittr_2.0.2
[46] credentials_1.3.2 Rcpp_1.0.8 munsell_0.5.0
[49] fansi_1.0.2 lifecycle_1.0.1 stringi_1.7.6
[52] whisker_0.4 yaml_2.2.2 debugme_1.1.0
[55] grid_4.1.2 parallel_4.1.2 promises_1.2.0.1
[58] crayon_1.4.2 hms_1.1.1 sys_3.4
[61] knitr_1.37 ps_1.6.0 languageserver_0.3.12
[64] pillar_1.7.0 igraph_1.2.11 uuid_1.0-3
[67] base64url_1.4 codetools_0.2-18 evaluate_0.14
[70] getPass_0.2-2 data.table_1.14.2 renv_0.15.2
[73] RcppParallel_5.1.5 vctrs_0.3.8 tzdb_0.2.0
[76] httpuv_1.6.5 gtable_0.3.0 openssl_1.4.6
[79] purrr_0.3.4 xfun_0.29 later_1.3.0
[82] viridisLite_0.4.0 gert_1.5.0 tsmp_0.4.14
[85] ellipsis_0.3.2