BCRABL_FunctionalKinaseAnalysis
Haider Inam
2022-11-02
Last updated: 2023-08-30
Checks: 6 1
Knit directory: duplex_sequencing_screen/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200402) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f9eea37. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Consensus_Data/.Rhistory
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered_sample5.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/variant_caller_outputs/variants_ann_sample6.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample8/
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample13/
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs.filt_1.fa.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14b/
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample1_combined/
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/duplex/
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample8/
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane2/
Ignored: data/Consensus_Data/Novogene_lane3/
Ignored: data/Consensus_Data/Novogene_lane4/
Ignored: data/Consensus_Data/Novogene_lane5/
Ignored: data/Consensus_Data/Novogene_lane6/
Ignored: data/Consensus_Data/Novogene_lane7/
Ignored: data/Consensus_Data/Ranomics_Pooled/
Ignored: data/Consensus_Data/archive/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/firstrun(lowsequencing)/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/firstrun(lowsequencing)/sscs/
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/.empty/
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/duplex/
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample11/
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/archive/
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/duplex/
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/low_depth/
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 2/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 3/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/slow_variantcaller_outputs_forcomparison/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/fastercaller/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/.DS_Store
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample7/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample8/
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/sscs_dcs_comparisons/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/NA/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_IL3_low_rep1vsrep2/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_Imat_Lowvsk562_Imat_Medium/
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Untracked files:
Untracked: analysis/ABL_sheared_libraries.Rmd
Untracked: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2ft.csv
Unstaged changes:
Modified: .DS_Store
Modified: analysis/ABL_cosmic_analysis.Rmd
Modified: analysis/ABL_kinasefunction_data_parser.Rmd
Deleted: analysis/ABL_unevenness_analysis.Rmd
Modified: analysis/BCRABL_FunctionalKinaseAnalysis.Rmd
Modified: analysis/index.Rmd
Modified: code/compare_screens.R
Modified: data/.DS_Store
Modified: data/Consensus_Data/.DS_Store
Modified: data/Consensus_Data/novogene_lane18/tlane18a_sample6/.DS_Store
Modified: output/.DS_Store
Modified: output/ABLEnrichmentScreens/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/.DS_Store
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/BCRABL_FunctionalKinaseAnalysis.Rmd) and HTML
(docs/BCRABL_FunctionalKinaseAnalysis.html) files. If
you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f9eea37 | haiderinam | 2023-08-30 | 2023 Updates |
| html | f9eea37 | haiderinam | 2023-08-30 | 2023 Updates |
| Rmd | 81a5af3 | haiderinam | 2023-02-25 | Added shortest codon finder function |
| Rmd | 7ac4a41 | haiderinam | 2023-01-20 | lane16,17 data added |
| Rmd | fa7baf1 | haiderinam | 2022-12-15 | Updates to the variant caller |
| Rmd | 077edd0 | haiderinam | 2022-12-09 | Added K562 Webpage |
| html | 2c1ccd3 | haiderinam | 2022-12-08 | Build site. |
| Rmd | 912824f | haiderinam | 2022-12-08 | wflow_publish("analysis/BCRABL_FunctionalKinaseAnalysis.Rmd") |
| Rmd | 9612cc9 | haiderinam | 2022-11-22 | Added analyses on mutant enrichment scores in the imatinib background |
| Rmd | accc9f5 | haiderinam | 2022-11-16 | Lane 15 additional sequencing |
| html | c1aa64b | haiderinam | 2022-11-12 | Build site. |
| Rmd | 5aea8c2 | haiderinam | 2022-11-12 | wflow_publish("analysis/BCRABL_FunctionalKinaseAnalysis.Rmd") |
| Rmd | b794806 | haiderinam | 2022-11-11 | Plotting imatinib enrichment scores distributions etc |
| html | efcc61d | haiderinam | 2022-11-04 | Build site. |
| Rmd | 261bf5f | haiderinam | 2022-11-04 | wflow_publish("analysis/BCRABL_FunctionalKinaseAnalysis.Rmd") |
| Rmd | 3a2f887 | haiderinam | 2022-11-04 | Added analyses of IL3 independence |
| Rmd | 6f54acf | haiderinam | 2022-11-04 | Added analyses of SSCS vs DCS error rates |
Plotting the correlation of allele frequencies of stuff that I see in two samples
# samplex=read.csv("data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
samplex=read.csv("data/Consensus_Data/Novogene_lane15/sample_6/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
# "Novogene_lane15/sample_6/sscs","Novogene_lane14/sample12/sscs"
samplex=samplex%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
samplex=samplex%>%mutate(maf=ct/depth)
samplex_simple=samplex%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf)
# sampley=read.csv("data/Consensus_Data/Novogene_lane15/sample_3/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
sampley=read.csv("data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
# sample7=read.csv("data/Consensus_Data/Novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
sampley=sampley%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
sampley=sampley%>%mutate(maf=ct/depth)
sampley_simple=sampley%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf)
samples_xy=merge(samplex_simple%>%filter(consequence_terms%in%"missense_variant"),sampley_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
plotly=ggplot(samples_xy%>%filter(protein_start>=242,protein_start<=492),aes(x=maf.x,y=maf.y))+
geom_point(color="black",shape=21,aes(fill=ct.x))+
scale_x_continuous(trans="log")+
scale_y_continuous(trans="log")+
geom_abline()+
theme_bw()
ggplotly(plotly)Warning in L$marker$color[idx] <- aes2plotly(data, params, "fill")[idx]: number
of items to replace is not a multiple of replacement length# cor(samples_xy$maf.x,samples_xy$maf.y)Background samples: Lane13 Sample 7, Lane 14 Sample 10, 11? D2 Post iL3 withdrawal: Lane 13 Sample 9,10 D4 Post iL3 withdrawal: Lane 13 Sample 11,12 D2 Post Imatinib treatment: Lane 14 Sample 12
SSCS
source("code/merge_samples.R")
source("code/depth_finder.R")
il3all=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
il3all=merge_samples(il3all,"Novogene_lane14/sample11/sscs")
il3all=merge_samples(il3all,"Novogene_lane15/sample_3/sscs")
il3all=merge_samples(il3all,"Novogene_lane13/Sample10/sscs")
il3all=merge_samples(il3all,"Novogene_lane13/Sample9/sscs")
il3all=merge_samples(il3all,"Novogene_lane15/sample_4/sscs")
il3all=merge_samples(il3all,"Novogene_lane15/sample_5/sscs")
il3all=merge_samples(il3all,"Novogene_lane15/sample_6/sscs")
il3all=merge_samples(il3all,"Novogene_lane15/sample_7/sscs")
# a=il3all%>%filter(protein_start%in%c(242:494),consequence_terms%in%"missense_variant")
# il3D0=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane15/sample_3/sscs")
# a=il3D0%>%filter(!protein_start%in%c(242:494),type%in%"mnv")
# a=il3D0%>%filter(protein_start%in%c(242:494),consequence_terms%in%"missense_variant")
il3D0=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
il3D0=merge_samples(il3D0,"Novogene_lane14/sample11/sscs")
il3D0=merge_samples(il3D0,"Novogene_lane15/sample_3/sscs")
il3D0=il3D0%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
il3D0=il3D0%>%mutate(maf=ct/depth)
il3D0=il3D0%>%mutate(totalcells=370,totalmutant=maf*totalcells)
il3D0_simple=il3D0%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
# a=il3D0%>%filter(protein_start%in%c(242:494),consequence_terms%in%"missense_variant")
# a=il3D0.D2.merge%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494))
il3D2=merge_samples("Novogene_lane13/Sample9/sscs","Novogene_lane13/Sample10/sscs")
il3D2=merge_samples(il3D2,"Novogene_lane15/Sample_4/sscs")
il3D2=il3D2%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
il3D2=il3D2%>%mutate(maf=ct/depth)
il3D2=il3D2%>%mutate(totalcells=1515,totalmutant=maf*totalcells)
il3D2_simple=il3D2%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
# imatD2=read.csv("data/Consensus_Data/Novogene_lane14/sample12/variant_caller_outputs/variants_unique_ann.csv",stringsAsFactors = F)
imatD2=merge_samples("Novogene_lane15/sample_6/sscs","Novogene_lane14/sample12/sscs")
# imatD2=merge_samples(imatD2,"Novogene_lane15/Sample4")
imatD2=imatD2%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
imatD2=imatD2%>%mutate(maf=ct/depth)
imatD2=imatD2%>%mutate(totalcells=1192,totalmutant=maf*totalcells)
imatD2_simple=imatD2%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
il3D4=read.csv("data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_unique_ann.csv")
# imatD2=merge_samples(imatD2,"Novogene_lane15/Sample4")
il3D4=il3D4%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
il3D4=il3D4%>%mutate(maf=ct/depth)
il3D4=il3D4%>%mutate(totalcells=17239,totalmutant=maf*totalcells)
il3D4_simple=il3D4%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
imatD4=read.csv("data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_unique_ann.csv")
# imatD2=merge_samples(imatD2,"Novogene_lane15/Sample4")
imatD4=imatD4%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
imatD4=imatD4%>%mutate(maf=ct/depth)
imatD4=imatD4%>%mutate(totalcells=10397,totalmutant=maf*totalcells)
imatD4_simple=imatD4%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
# D6
il3D6=read.csv("data/Consensus_Data/novogene_lane17/sample2/sscs/variant_caller_outputs/variants_unique_ann.csv")
il3D6=il3D6%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
il3D6=il3D6%>%mutate(maf=ct/depth)
il3D6=il3D6%>%mutate(totalcells=430000,totalmutant=maf*totalcells)
il3D6_simple=il3D6%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
# D6
imatD6=read.csv("data/Consensus_Data/novogene_lane17/sample3/sscs/variant_caller_outputs/variants_unique_ann.csv")
# imatD2=merge_samples(imatD2,"Novogene_lane15/Sample4")
imatD6=imatD6%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
imatD6=imatD6%>%mutate(maf=ct/depth)
imatD6=imatD6%>%mutate(totalcells=156000,totalmutant=maf*totalcells)
imatD6_simple=imatD6%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf,totalcells,totalmutant)
##########IL3 Day 2 vs IL3 D0############
il3D0.D2=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant")%>%mutate(totalmutant=totalmutant*107/370),il3D2_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
il3D0.D2$alt_aa=factor(il3D0.D2$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
il3D0.D2[il3D0.D2$ct.y%in%NA,"ct.y"]=.5
il3D0.D2=depth_finder(il3D0.D2,"depth.y")
il3D0.D2=depth_finder(il3D0.D2,"totalcells.y")
il3D0.D2=il3D0.D2%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
il3D0.D2[il3D0.D2$ct.x%in%NA,"ct.x"]=.5
il3D0.D2=il3D0.D2%>%mutate(score=log2(maf.y/maf.x))
# il3D0.D2[il3D0.D2$score%in%NA,"score"]=-6
il3D0.D2[il3D0.D2$totalmutant.y%in%NA,"totalmutant.y"]=0
il3D0.D2=il3D0.D2%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/48)
il3D0.D2[il3D0.D2$netgr_obs%in%NA,"netgr_obs"]=-.055
##########IL3 Day 4 vs IL3 D2############
il3D2.D4=merge(il3D2_simple%>%filter(consequence_terms%in%"missense_variant"),il3D4_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
il3D2.D4[il3D2.D4$ct.y%in%NA,"ct.y"]=.5
il3D2.D4=depth_finder(il3D2.D4,"depth.y")
il3D2.D4=depth_finder(il3D2.D4,"totalcells.y")
il3D2.D4=il3D2.D4%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
il3D2.D4[il3D2.D4$ct.x%in%NA,"ct.x"]=.5
il3D2.D4=il3D2.D4%>%mutate(score=log2(maf.y/maf.x))
# il3D2.D4[il3D2.D4$score%in%NA,"score"]=-6
il3D2.D4[il3D2.D4$totalmutant.y%in%NA,"totalmutant.y"]=0
il3D2.D4=il3D2.D4%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/48)
il3D2.D4[il3D2.D4$netgr_obs%in%NA,"netgr_obs"]=-.055
##########IL3 Day 4 vs Imat D0############
il3D0.D4=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant")%>%mutate(totalmutant=totalmutant*107/370),il3D4_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
# il3D0.D4[il3D0.D4$ct.y%in%NA,"ct.y"]=0
il3D0.D4[il3D0.D4$ct.y%in%NA,"ct.y"]=.5
il3D0.D4=depth_finder(il3D0.D4,"depth.y")
il3D0.D4=depth_finder(il3D0.D4,"totalcells.y")
il3D0.D4=il3D0.D4%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
il3D0.D4[il3D0.D4$ct.x%in%NA,"ct.x"]=.5
il3D0.D4=il3D0.D4%>%mutate(score=log2(maf.y/maf.x))
# il3D0.D4[il3D0.D4$score%in%NA,"score"]=-6
il3D0.D4[il3D0.D4$totalmutant.y%in%NA,"totalmutant.y"]=0
il3D0.D4=il3D0.D4%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/96)
il3D0.D4[il3D0.D4$netgr_obs%in%NA,"netgr_obs"]=-.055
##########Imatinib Day 2 vs Imat D0############
imatD0.D2=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant"),imatD2_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
# imatD0.D2[imatD0.D2$ct.y%in%NA,"ct.y"]=0
imatD0.D2[imatD0.D2$ct.y%in%NA,"ct.y"]=.5
imatD0.D2=depth_finder(imatD0.D2,"depth.y")
imatD0.D2=depth_finder(imatD0.D2,"totalcells.y")
imatD0.D2=imatD0.D2%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
imatD0.D2[imatD0.D2$ct.x%in%NA,"ct.x"]=.5
imatD0.D2=imatD0.D2%>%mutate(score=log2(maf.y/maf.x))
# imatD0.D2[imatD0.D2$score%in%NA,"score"]=-6
imatD0.D2[imatD0.D2$totalmutant.y%in%NA,"totalmutant.y"]=0
imatD0.D2=imatD0.D2%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/48)
imatD0.D2[imatD0.D2$netgr_obs%in%NA,"netgr_obs"]=-.055
##########Imatinib Day 4 vs Imat D2############
imatD2.D4=merge(imatD2_simple%>%filter(consequence_terms%in%"missense_variant"),imatD4_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
# imatD2.D4[imatD2.D4$ct.y%in%NA,"ct.y"]=0
imatD2.D4[imatD2.D4$ct.y%in%NA,"ct.y"]=.5
imatD2.D4=depth_finder(imatD2.D4,"depth.y")
imatD2.D4=depth_finder(imatD2.D4,"totalcells.y")
imatD2.D4=imatD2.D4%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
imatD2.D4[imatD2.D4$ct.x%in%NA,"ct.x"]=.5
imatD2.D4=imatD2.D4%>%mutate(score=log2(maf.y/maf.x))
# imatD2.D4[imatD2.D4$score%in%NA,"score"]=-6
imatD2.D4[imatD2.D4$totalmutant.y%in%NA,"totalmutant.y"]=0
imatD2.D4=imatD2.D4%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/48)
imatD2.D4[imatD2.D4$netgr_obs%in%NA,"netgr_obs"]=-.055
##########Imatinib Day 4 vs Imat D0############
imatD0.D4=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant"),imatD4_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
# imatD0.D4[imatD0.D4$ct.y%in%NA,"ct.y"]=0
imatD0.D4[imatD0.D4$ct.y%in%NA,"ct.y"]=.5
imatD0.D4=depth_finder(imatD0.D4,"depth.y")
imatD0.D4=depth_finder(imatD0.D4,"totalcells.y")
imatD0.D4=imatD0.D4%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
imatD0.D4[imatD0.D4$ct.x%in%NA,"ct.x"]=.5
imatD0.D4=imatD0.D4%>%mutate(score=log2(maf.y/maf.x))
# imatD0.D4[imatD0.D4$score%in%NA,"score"]=-6
imatD0.D4[imatD0.D4$totalmutant.y%in%NA,"totalmutant.y"]=0
imatD0.D4=imatD0.D4%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/96)
imatD0.D4[imatD0.D4$netgr_obs%in%NA,"netgr_obs"]=-.055
##########IL3 Day 6 vs Imat D0############
il3D0.D6=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant"),il3D4_simple%>%
filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
# il3D0.D6[il3D0.D6$ct.y%in%NA,"ct.y"]=0
il3D0.D6[il3D0.D6$ct.y%in%NA,"ct.y"]=.5
il3D0.D6=depth_finder(il3D0.D6,"depth.y")
il3D0.D6=depth_finder(il3D0.D6,"totalcells.y")
il3D0.D6=il3D0.D6%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
il3D0.D6[il3D0.D6$ct.x%in%NA,"ct.x"]=.5
il3D0.D6=il3D0.D6%>%mutate(score=log2(maf.y/maf.x))
# il3D0.D6[il3D0.D6$score%in%NA,"score"]=-6
il3D0.D6[il3D0.D6$totalmutant.y%in%NA,"totalmutant.y"]=0
il3D0.D6=il3D0.D6%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/96)
il3D0.D6[il3D0.D6$netgr_obs%in%NA,"netgr_obs"]=-.055
##########Imatinib Day 6 vs Imat D0############
imatD0.D6=merge(il3D0_simple%>%filter(consequence_terms%in%"missense_variant"),imatD6_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all.x = T)
imatD0.D6[imatD0.D6$ct.y%in%NA,"ct.y"]=.5
imatD0.D6=depth_finder(imatD0.D6,"depth.y")
imatD0.D6=depth_finder(imatD0.D6,"totalcells.y")
imatD0.D6=imatD0.D6%>%mutate(maf.y=ct.y/depth.y,
totalmutant.y=maf.y*totalcells.y)
imatD0.D6[imatD0.D6$ct.x%in%NA,"ct.x"]=.5
imatD0.D6=imatD0.D6%>%mutate(score=log2(maf.y/maf.x))
imatD0.D6[imatD0.D6$totalmutant.y%in%NA,"totalmutant.y"]=0
imatD0.D6=imatD0.D6%>%mutate(netgr_obs=log(totalmutant.y/totalmutant.x)/96)
imatD0.D6[imatD0.D6$netgr_obs%in%NA,"netgr_obs"]=-.055
######Plotting imat D0 D2 Heatmap####
ggplot(imatD0.D2%>%filter(nchar(as.character(alt_aa))%in%1,protein_start>=242,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="darkblue",midpoint=-1,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(250,253,255,276,299,315,317,351,355,359,396,459,486))+
ylab("Mutant Amino Acid")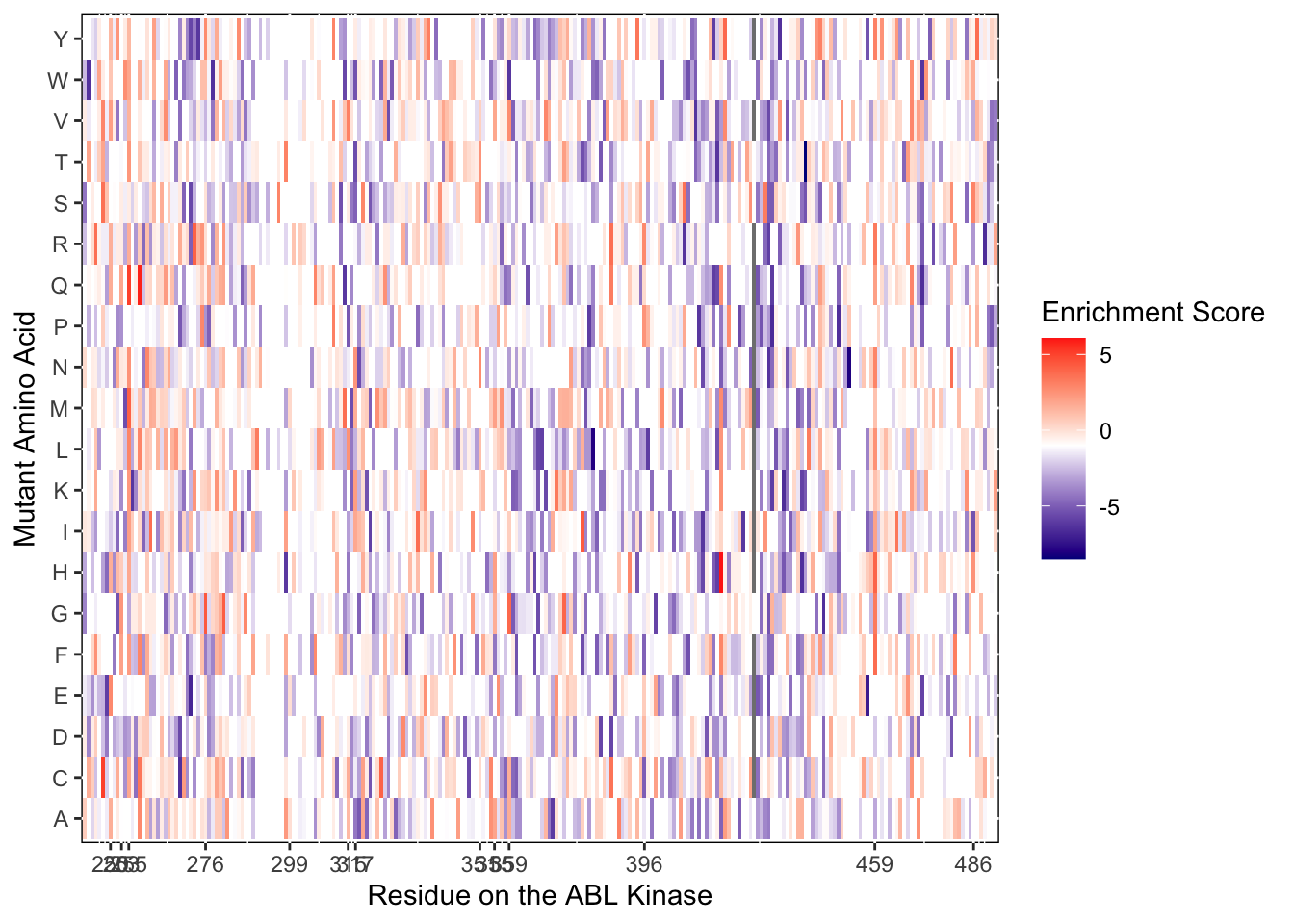
# ggsave("BCRABL_imatinib_D2.pdf",height=6,width=24,units="in",useDingbats=F)
######Plotting resistant imat D0 D2 Heatmap####
ggplot(imatD0.D2%>%filter(nchar(as.character(alt_aa))%in%1,protein_start%in%c(250,253,255,276,299,315,317,351,355,359,396,459,486)),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="darkblue",midpoint=-1,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks =c(250,253,255,276,299,315,317,351,355,359,396,459,486))+
ylab("Mutant Amino Acid")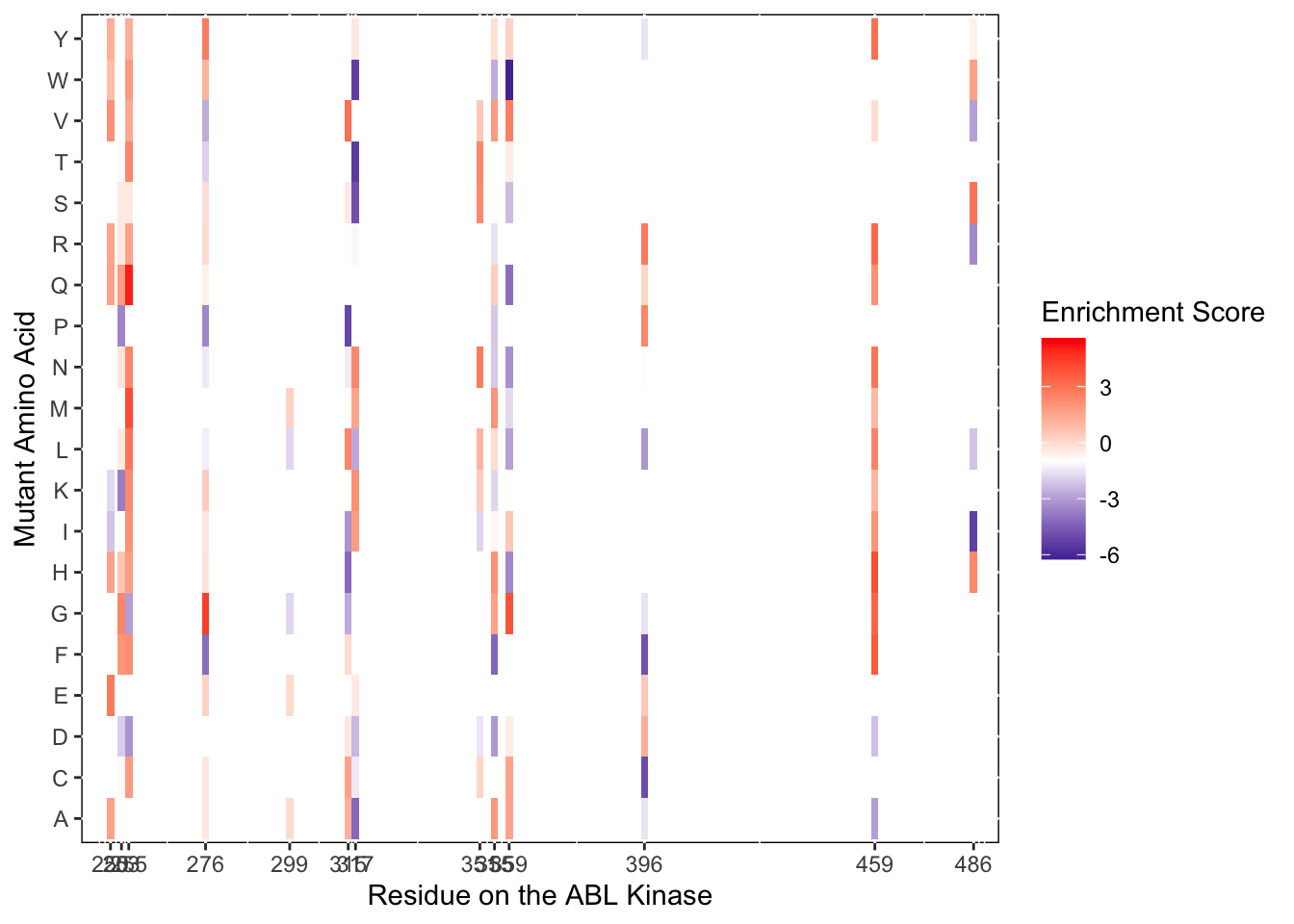
# ggsave("BCRABL_imatinib_D2_resistantresidues.pdf",height=6,width=24,units="in",useDingbats=F)
######Plotting iL3 D0 D2 Heatmap####
il3D0.D2$alt_aa=factor(il3D0.D2$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
ggplot(il3D0.D2%>%filter(nchar(as.character(alt_aa))%in%1,protein_start>=242,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="darkblue",midpoint=-1,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")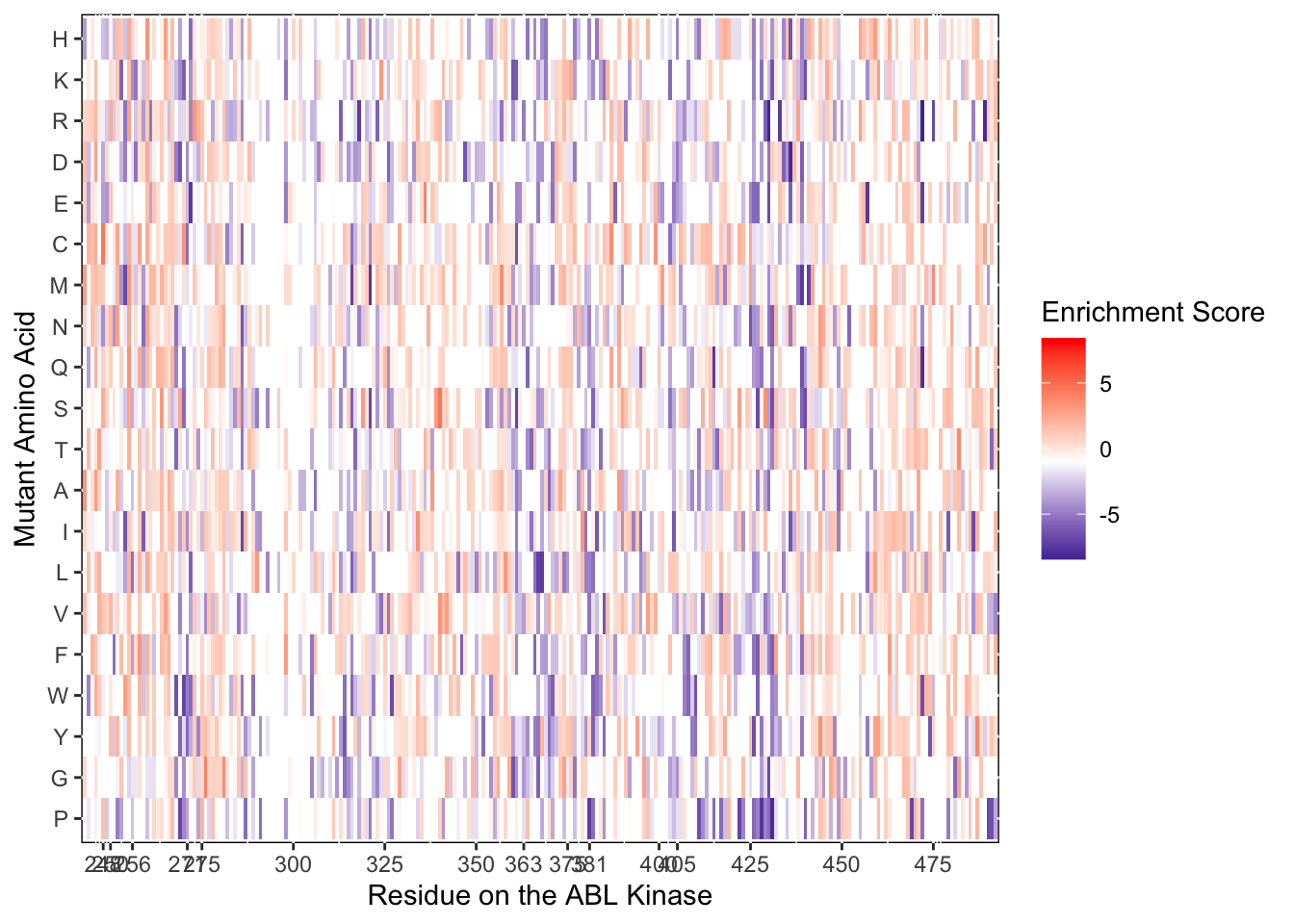
# a=il3D0.D2%>%filter(netgr_obs%in%"-Inf",consequence_terms%in%"missense_variant",protein_start%in%c(242:494))
# ggsave("BCRABL_iL3Independence_D2.pdf",height=6,width=24,units="in",useDingbats=F)
# write.csv(il3D0.D2,"BCRABL_Il3Independence_D2.csv")
#####Focusing on conserved residues#####
ggplot(il3D0.D4%>%filter(nchar(as.character(alt_aa))%in%1,protein_start%in%c(271,363,381:383)),aes(x=protein_start,y=alt_aa,fill=netgr_obs))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")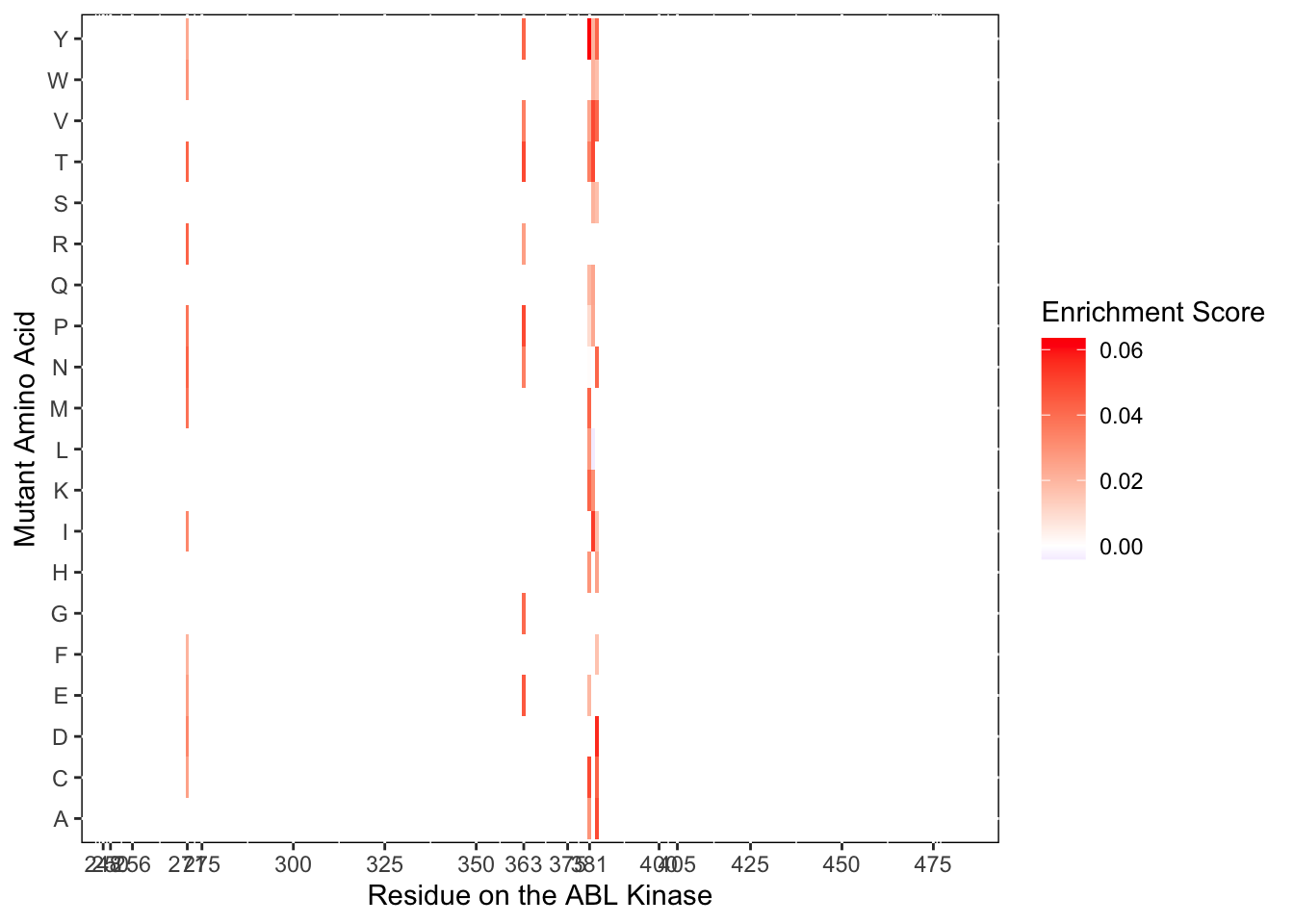
# ggsave("BCRABL_iL3Independence_D2_essential.pdf",height=6,width=24,units="in",useDingbats=F)
#####Graying out unseen residues#####
###D0 D2 IL3
df_grid = expand.grid(protein_start = c(242:493),alt_aa = unique(il3D0.D2$alt_aa))
il3D0.D2.merge=merge(df_grid,il3D0.D2,by=c("protein_start","alt_aa"),all=T)
il3D0.D2.merge=il3D0.D2.merge%>%filter(nchar(as.character(alt_aa))%in%1,protein_start>=242,protein_start<=494)
###Trying to add in the score as WT for residues that are wt
reference_seq=read.table("data/Refs/ABL/abl_cds_translation.txt")
il3D0.D2.merge=il3D0.D2.merge%>%
rowwise()%>%
# group_by(protein_start)%>%
mutate(ref_aa=case_when(ref_aa%in%NA~substr(reference_seq,protein_start,protein_start),
T~ref_aa))%>%
mutate(wt=case_when(ref_aa==alt_aa~T,
T~F))
##########Plotting gray and yellow heatmap
il3D0.D2.merge$alt_aa=factor(il3D0.D2.merge$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
il3D0.D2.merge.filtered=il3D0.D2.merge%>%filter(!protein_start%in%c(290:305))
a=il3D0.D2.merge.filtered%>%filter(wt%in%T)
ggplot(il3D0.D2.merge.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(il3D0.D2.merge.filtered,!is.na(score)),aes(fill=score))+
scale_fill_gradient2(low ="darkblue",midpoint=-1,mid="white", high ="red",name="Enrichment Score")+
geom_tile(data=subset(il3D0.D2.merge.filtered,is.na(score)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(il3D0.D2.merge.filtered,is.na(score)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")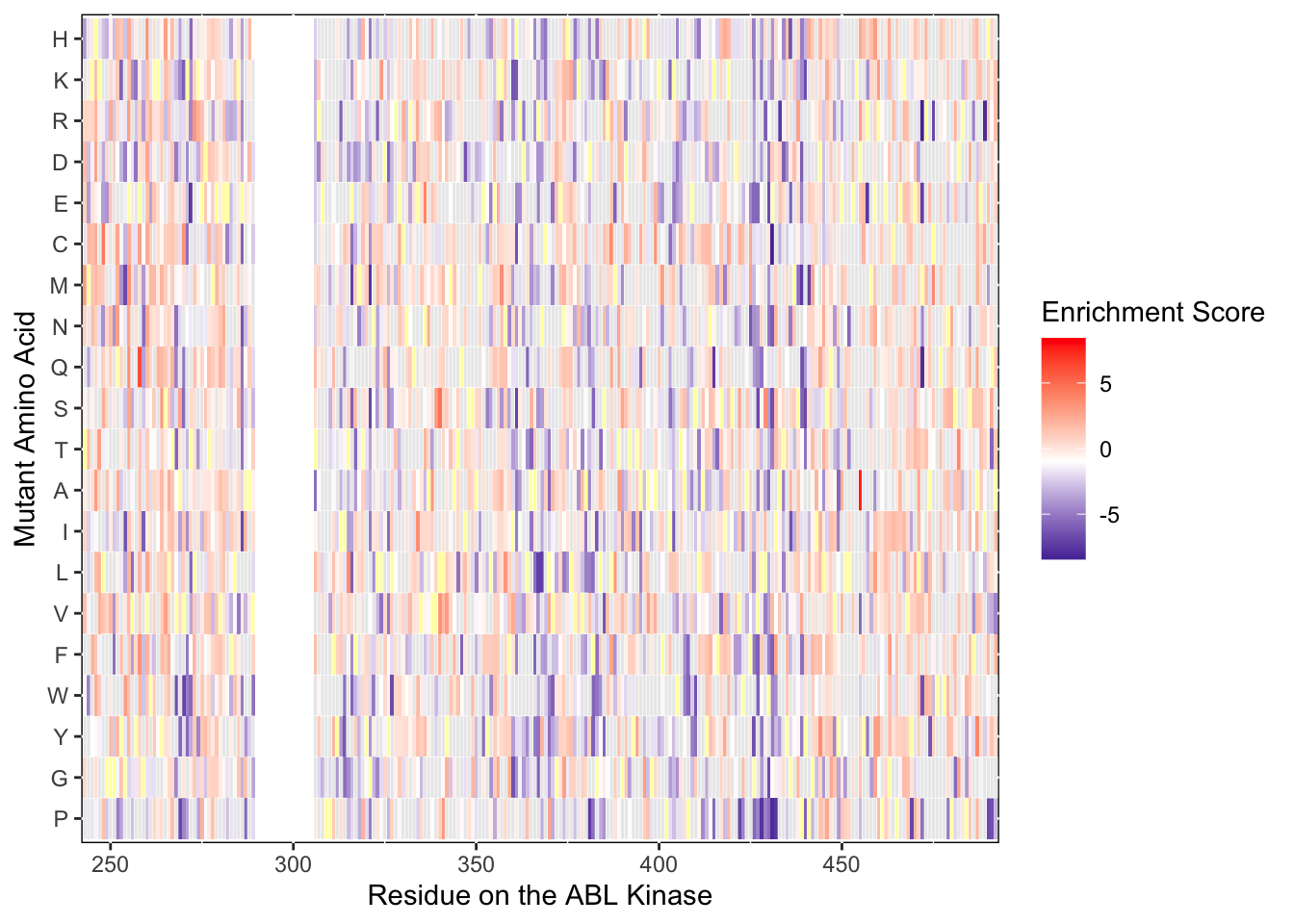
# ggsave("BCRABL_il3_D2_gray.pdf",height=6,width=24,units="in",useDingbats=F)
###D0 D4 IL3
#####Graying out unseen residues#####
df_grid = expand.grid(protein_start = c(242:493),alt_aa = unique(il3D0.D4$alt_aa))
il3D0.D4.merge=merge(df_grid,il3D0.D4,by=c("protein_start","alt_aa"),all=T)
il3D0.D4.merge=il3D0.D4.merge%>%filter(nchar(as.character(alt_aa))%in%1,protein_start>=242,protein_start<=494)
###Trying to add in the score as WT for residues that are wt
reference_seq=read.table("data/Refs/ABL/abl_cds_translation.txt")
il3D0.D4.merge=il3D0.D4.merge%>%
rowwise()%>%
# group_by(protein_start)%>%
mutate(ref_aa=case_when(ref_aa%in%NA~substr(reference_seq,protein_start,protein_start),
T~ref_aa))%>%
mutate(wt=case_when(ref_aa==alt_aa~T,
T~F))
##########Plotting gray and yellow heatmap
il3D0.D4.merge$alt_aa=factor(il3D0.D4.merge$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
il3D0.D4.merge.filtered=il3D0.D4.merge%>%filter(!protein_start%in%c(290:305))
# a=il3D0.D4.merge.filtered%>%filter(wt%in%T)
ggplot(il3D0.D4.merge.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(il3D0.D4.merge.filtered,!is.na(score)),aes(fill=netgr_obs))+
scale_fill_gradient2(low ="darkblue",midpoint=.04,mid="white", high ="red",name="Net growth rate")+
geom_tile(data=subset(il3D0.D4.merge.filtered,is.na(score)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(il3D0.D4.merge.filtered,is.na(score)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")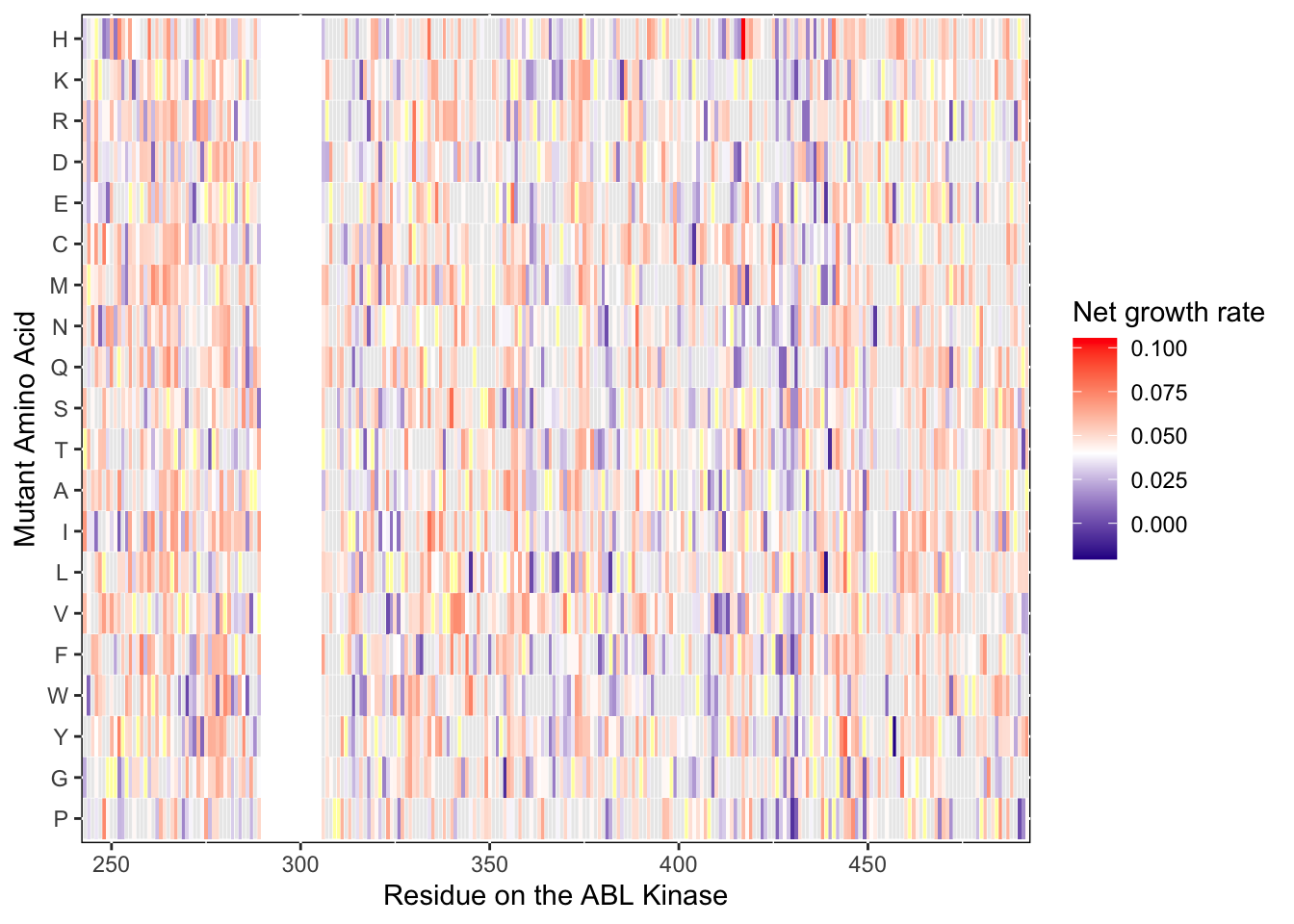
ggplot(il3D0.D4.merge.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(il3D0.D4.merge.filtered,!is.na(score)),aes(fill=score))+
scale_fill_gradient2(low ="darkblue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
geom_tile(data=subset(il3D0.D4.merge.filtered,is.na(score)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(il3D0.D4.merge.filtered,is.na(score)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")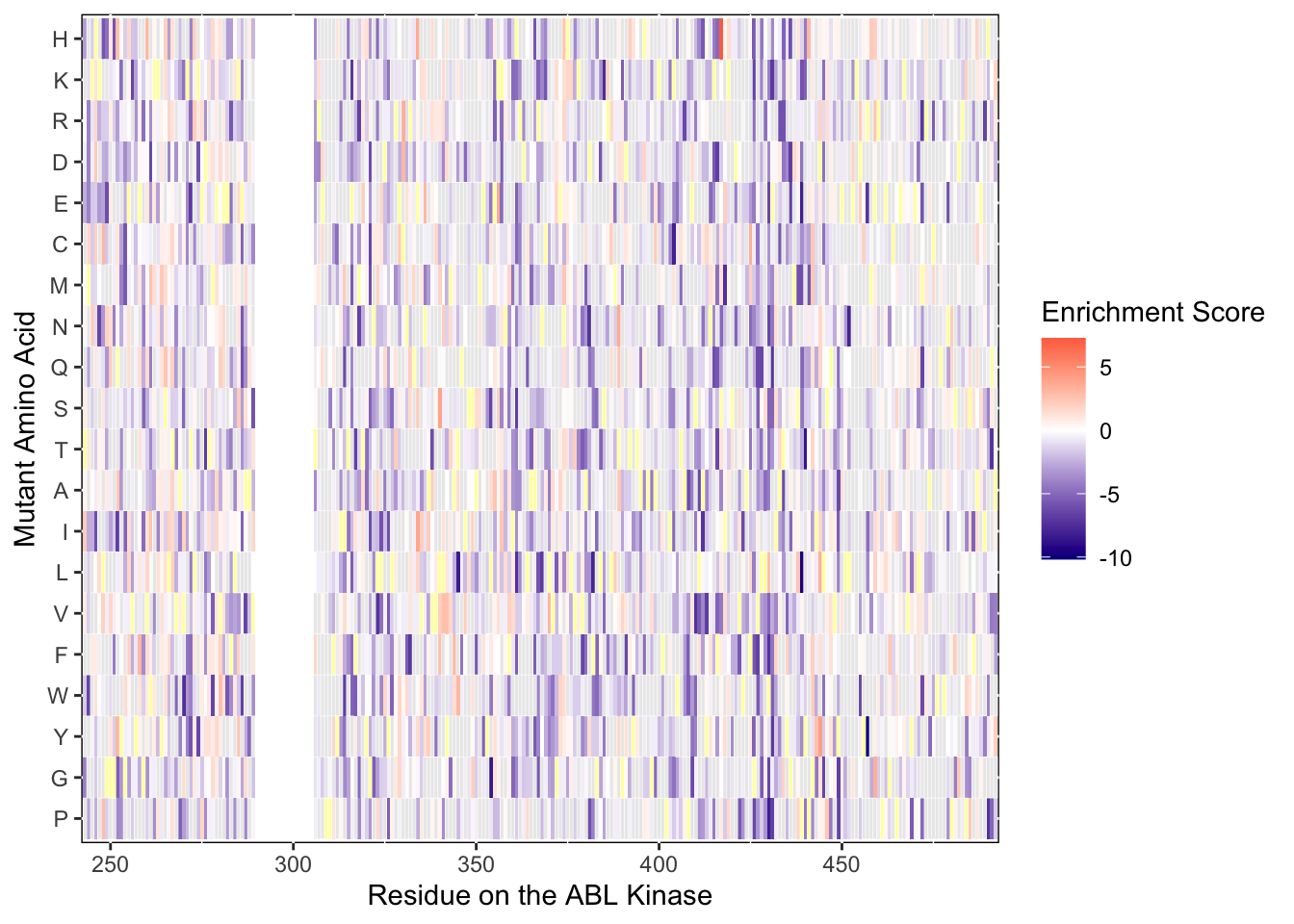
# ggsave("BCRABL_il3_D4_scores_updated.pdf",height=6,width=24,units="in",useDingbats=F)
###D0 D4 Imat
#####Graying out unseen residues#####
df_grid = expand.grid(protein_start = c(242:493),alt_aa = unique(imatD0.D4$alt_aa))
imatD0.D4.merge=merge(df_grid,imatD0.D4,by=c("protein_start","alt_aa"),all=T)
imatD0.D4.merge=imatD0.D4.merge%>%filter(nchar(as.character(alt_aa))%in%1,protein_start>=242,protein_start<=494)
###Trying to add in the score as WT for residues that are wt
reference_seq=read.table("data/Refs/ABL/abl_cds_translation.txt")
imatD0.D4.merge=imatD0.D4.merge%>%
rowwise()%>%
# group_by(protein_start)%>%
mutate(ref_aa=case_when(ref_aa%in%NA~substr(reference_seq,protein_start,protein_start),
T~ref_aa))%>%
mutate(wt=case_when(ref_aa==alt_aa~T,
T~F))
##########Plotting gray and yellow heatmap
imatD0.D4.merge$alt_aa=factor(imatD0.D4.merge$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
imatD0.D4.merge.filtered=imatD0.D4.merge%>%filter(!protein_start%in%c(290:305))
# a=il3D0.D4.merge.filtered%>%filter(wt%in%T)
ggplot(imatD0.D4.merge.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(imatD0.D4.merge.filtered,!is.na(score)),aes(fill=netgr_obs))+
scale_fill_gradient2(low ="darkblue",midpoint=.03,mid="white", high ="red",name="Net growth rate")+
geom_tile(data=subset(imatD0.D4.merge.filtered,is.na(score)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(imatD0.D4.merge.filtered,is.na(score)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")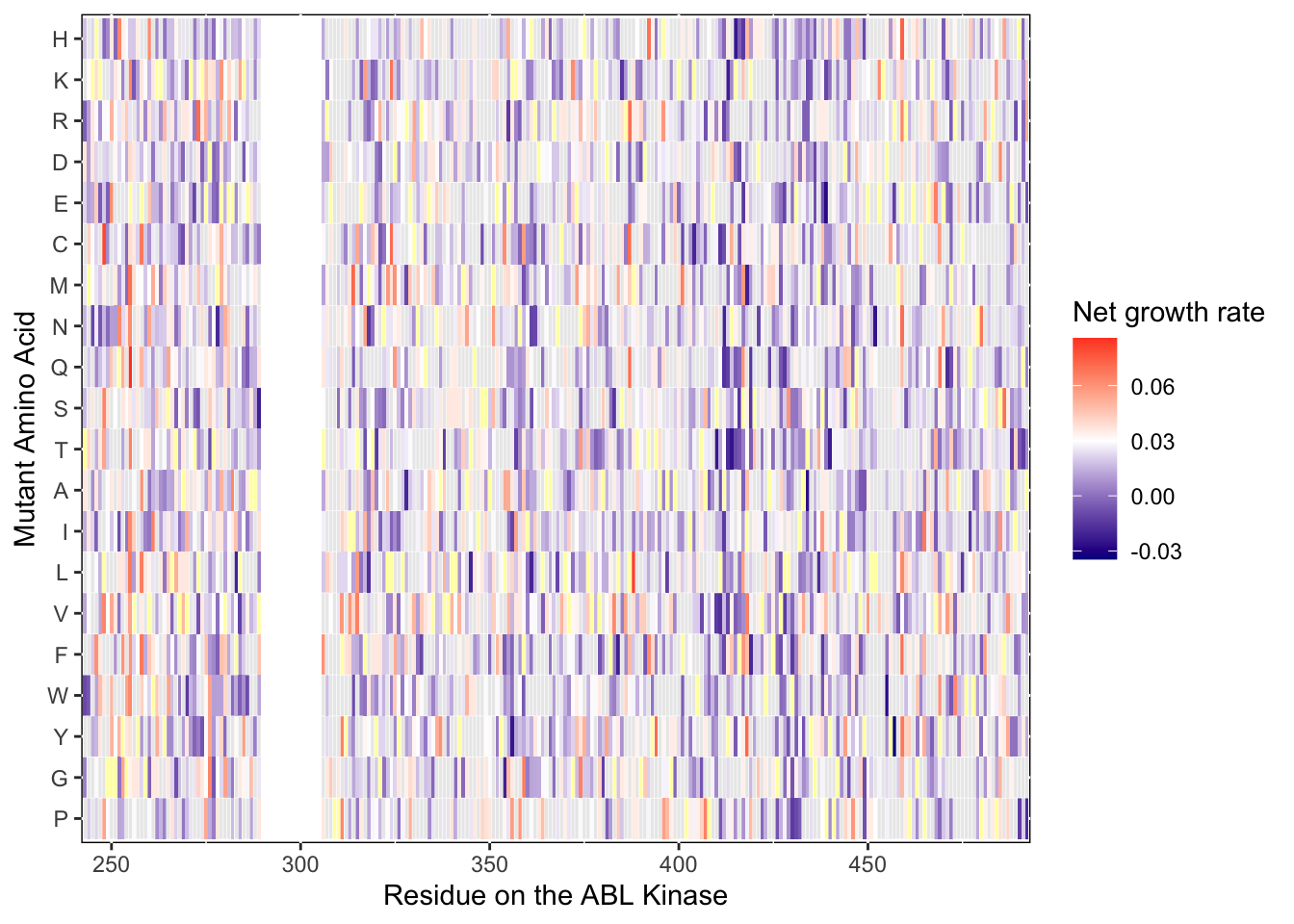
# ggsave("BCRABL_imat_D4_gray_updated.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(imatD0.D4.merge.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(imatD0.D4.merge.filtered,!is.na(score)),aes(fill=score))+
scale_fill_gradient2(low ="darkblue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
geom_tile(data=subset(imatD0.D4.merge.filtered,is.na(score)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(imatD0.D4.merge.filtered,is.na(score)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")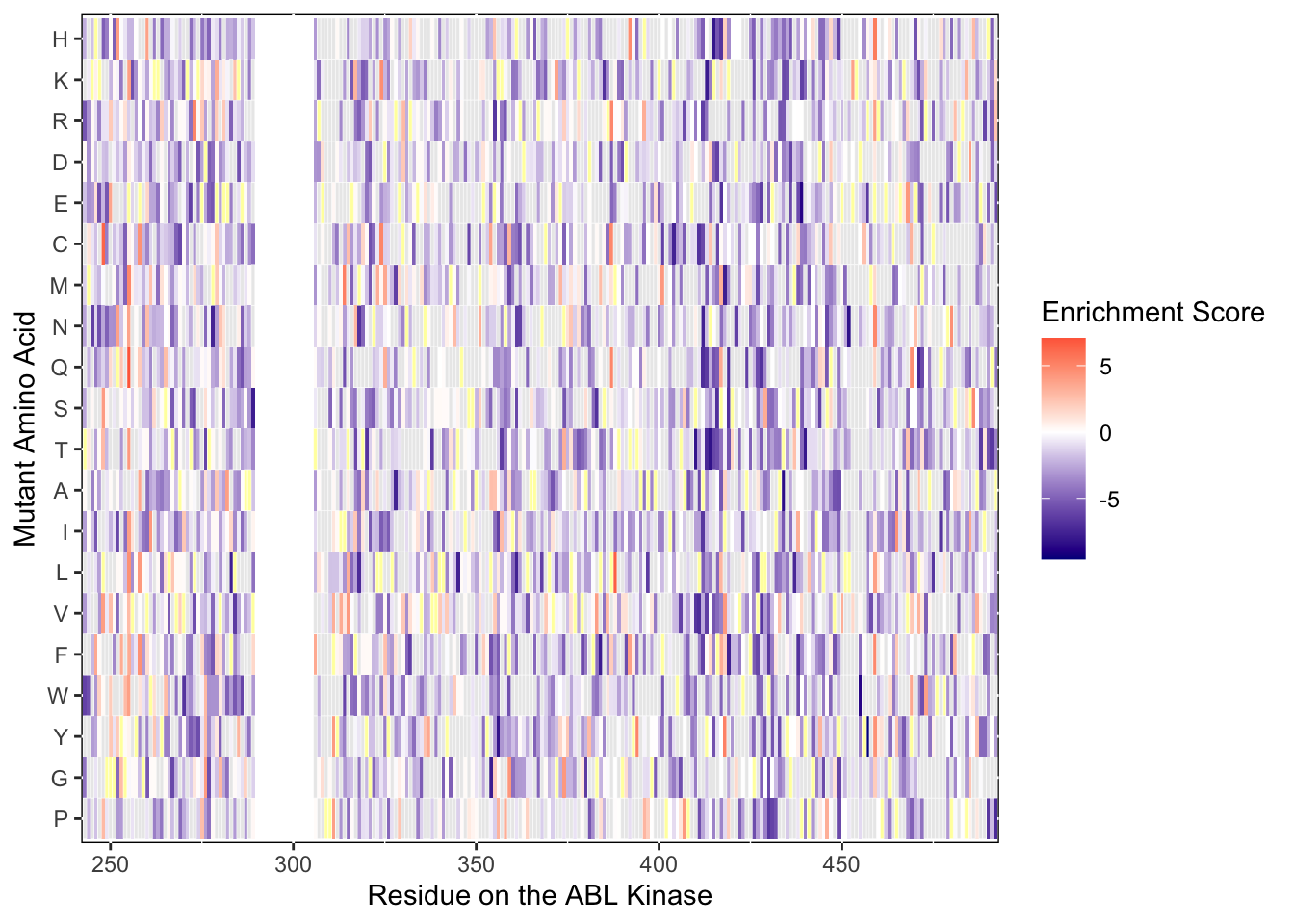
# ggsave("BCRABL_imat_D4_scores.pdf",height=6,width=24,units="in",useDingbats=F)
b=imatD0.D4%>%filter(consequence_terms%in%"missense_variant",protein_start>=242,protein_start<=494)Plotting enrichment scores
####Plotting Imatinib D4 D2 scores vs Imatinib D2 D0 scores
#First merging IL3 D0 D2 to IL3 D2 D4
scores_all=merge(il3D0.D2%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),il3D2.D4%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),suffixes = c(".il3d0.d2",".il3d2.d4"),all = T)
#Second, adding Imat D0 D2 to dataframe
scores_all=merge(scores_all,imatD0.D2%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.imatd0.d2=score,netgr_obs.imatd0.d2=netgr_obs)
#Third, adding Imat D2 D4 to dataframe
scores_all=merge(scores_all,imatD2.D4%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.imatd2.d4=score,netgr_obs.imatd2.d4=netgr_obs)
#Fourth, adding IL3 D4 D0 to dataframe
scores_all=merge(scores_all,il3D0.D4%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.il3d0.d4=score,netgr_obs.il3d0.d4=netgr_obs)
#Fifth, adding Imatinib D4 D0 to dataframe
scores_all=merge(scores_all,imatD0.D4%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.imatd0.d4=score,netgr_obs.imatd0.d4=netgr_obs)
#Sixth, adding IL3 D6 D0 to dataframe
scores_all=merge(scores_all,il3D0.D6%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.il3d0.d6=score,netgr_obs.il3d0.d6=netgr_obs)
#Seventh, adding Imatinib D4 D0 to dataframe
scores_all=merge(scores_all,imatD0.D6%>%dplyr::select(-ct.x,-depth.x,-maf.x,-totalmutant.x,-ct.y,-depth.y,-maf.y,-totalmutant.y,-totalcells.x,-totalcells.y),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"),all = T)
scores_all=scores_all%>%rename(score.imatd0.d6=score,netgr_obs.imatd0.d6=netgr_obs)
scores_all=scores_all%>%mutate(resmuts=case_when(protein_start%in%253&alt_aa%in%"H"~T,
protein_start%in%255&alt_aa%in%"V"~T,
protein_start%in%486&alt_aa%in%"S"~T,
protein_start%in%396&alt_aa%in%"P"~T,
protein_start%in%255&alt_aa%in%"K"~T,
protein_start%in%315&alt_aa%in%"I"~T,
protein_start%in%252&alt_aa%in%"H"~T,
protein_start%in%253&alt_aa%in%"F"~T,
protein_start%in%250&alt_aa%in%"E"~T,
protein_start%in%359&alt_aa%in%"C"~T,
protein_start%in%351&alt_aa%in%"T"~T,
protein_start%in%355&alt_aa%in%"G"~T,
protein_start%in%317&alt_aa%in%"L"~T,
protein_start%in%359&alt_aa%in%"I"~T,
protein_start%in%355&alt_aa%in%"A"~T,
protein_start%in%459&alt_aa%in%"K"~T,
protein_start%in%276&alt_aa%in%"G"~T,
protein_start%in%299&alt_aa%in%"L"~T,
T~F))
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.il3d2.d4%in%-6),aes(x=score.il3d0.d2,y=score.il3d2.d4))+geom_point()+geom_abline()Warning: Removed 1351 rows containing missing values (geom_point).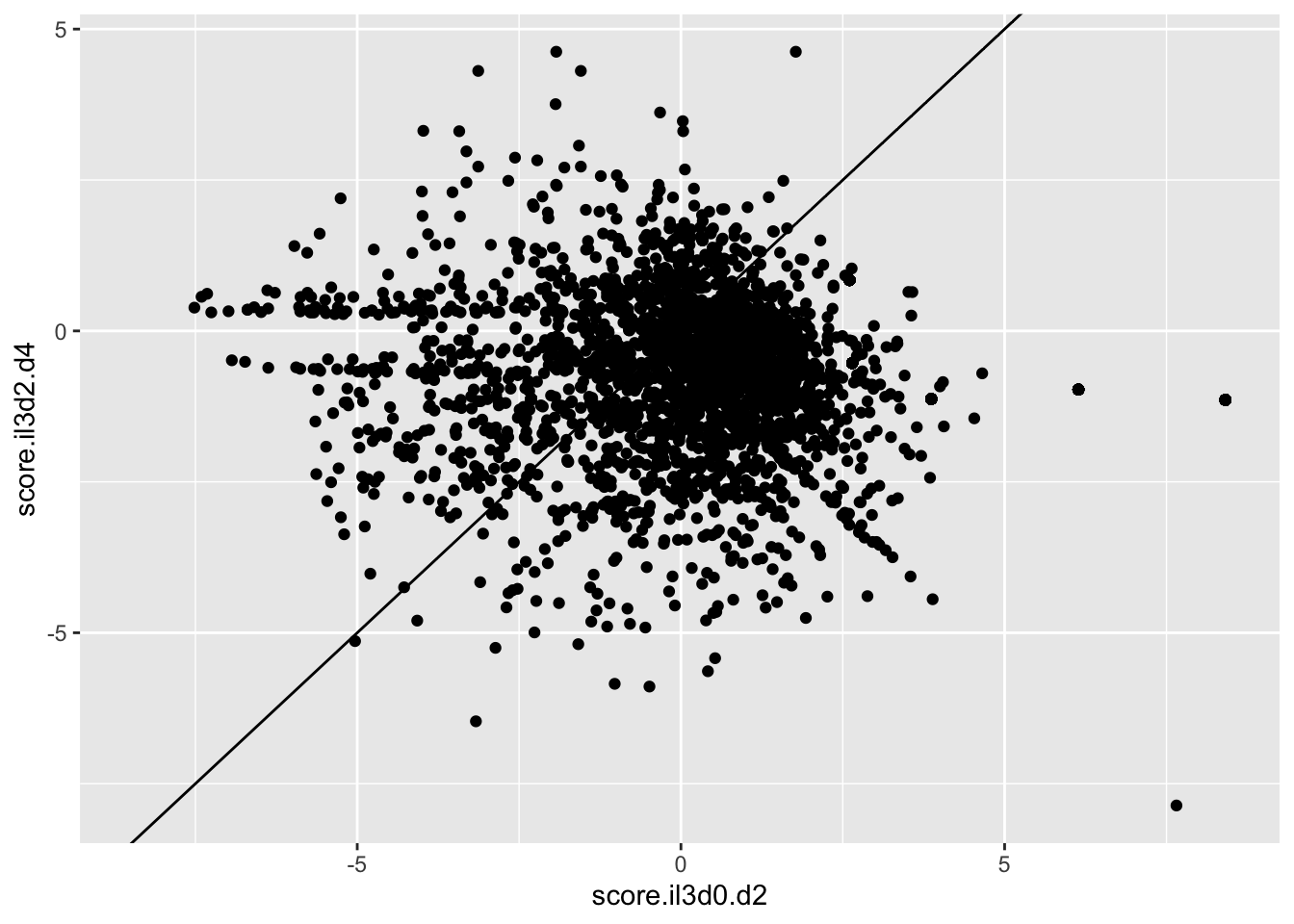
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.il3d2.d4%in%-6),aes(x=score.il3d0.d2,y=score.il3d2.d4))+geom_point()+geom_abline()Warning: Removed 1351 rows containing missing values (geom_point).
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.imatd2.d4%in%-6),aes(x=score.imatd0.d2,y=score.imatd0.d4))+geom_point()+geom_abline()Warning: Removed 719 rows containing missing values (geom_point).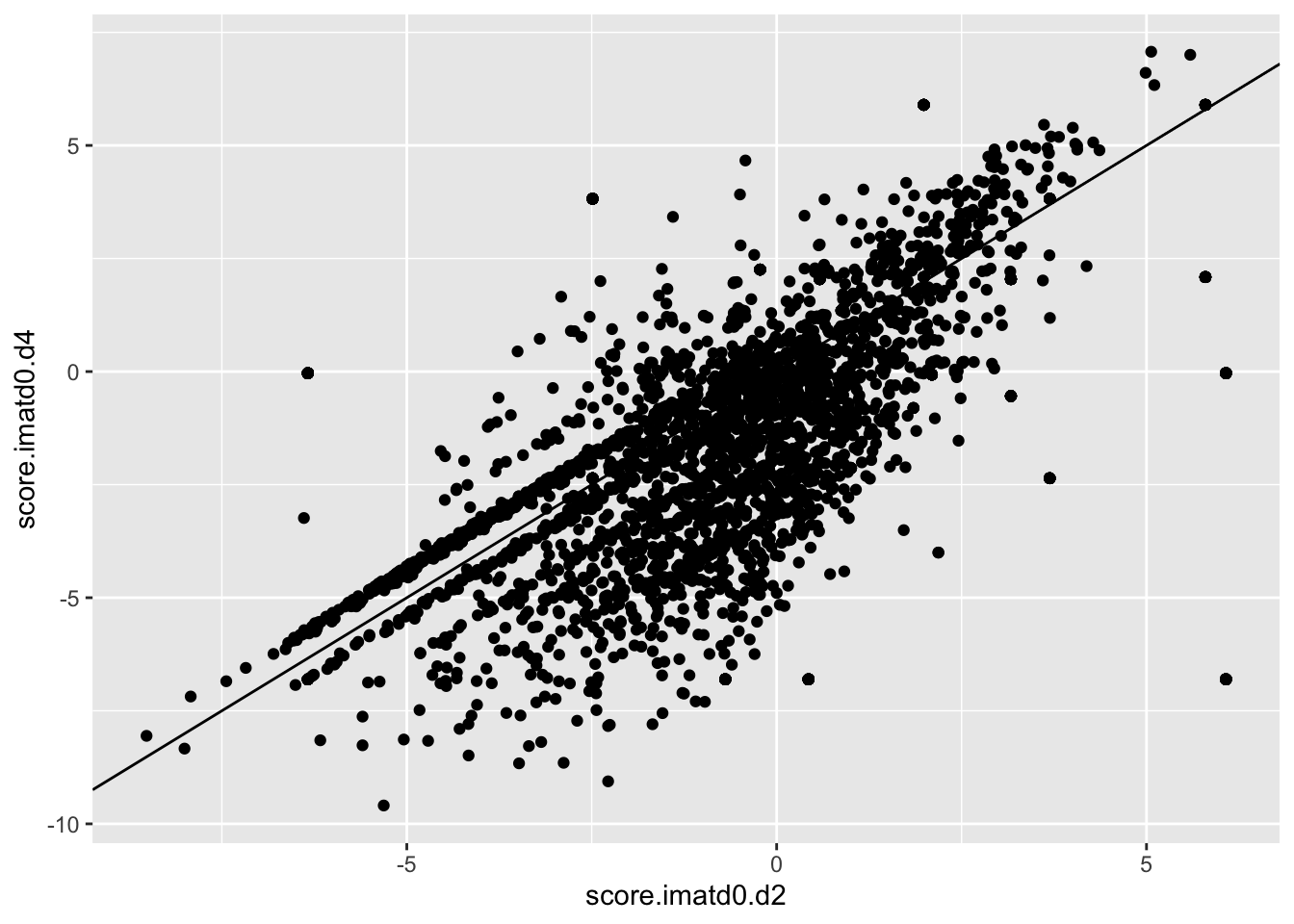
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.imatd2.d4%in%-6),aes(x=netgr_obs.imatd0.d2,y=netgr_obs.imatd0.d4))+geom_point()+geom_abline()Warning: Removed 719 rows containing missing values (geom_point).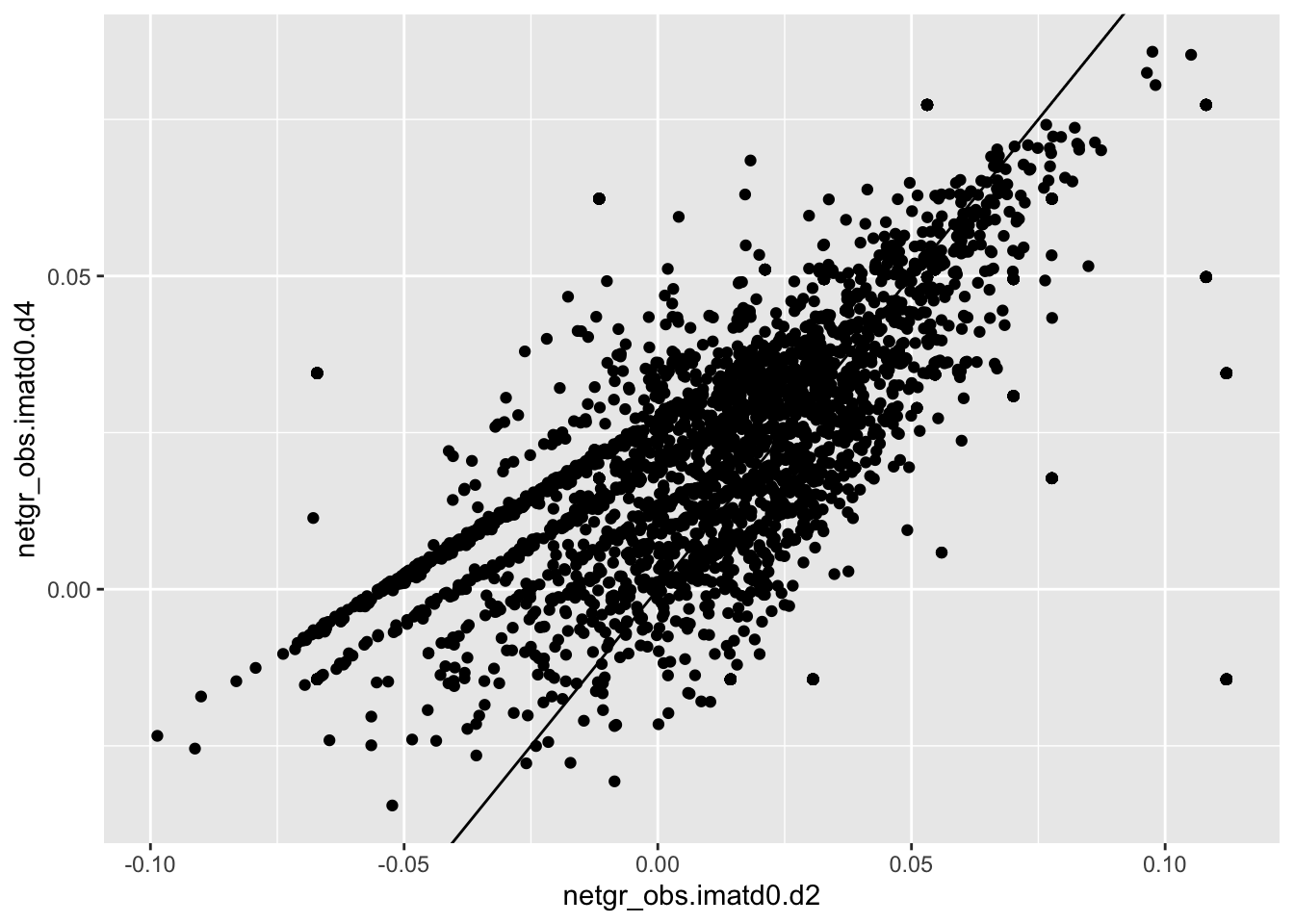
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.imatd2.d4%in%-6),aes(x=score.il3d0.d2,y=score.il3d0.d4))+geom_point()+geom_abline()Warning: Removed 703 rows containing missing values (geom_point).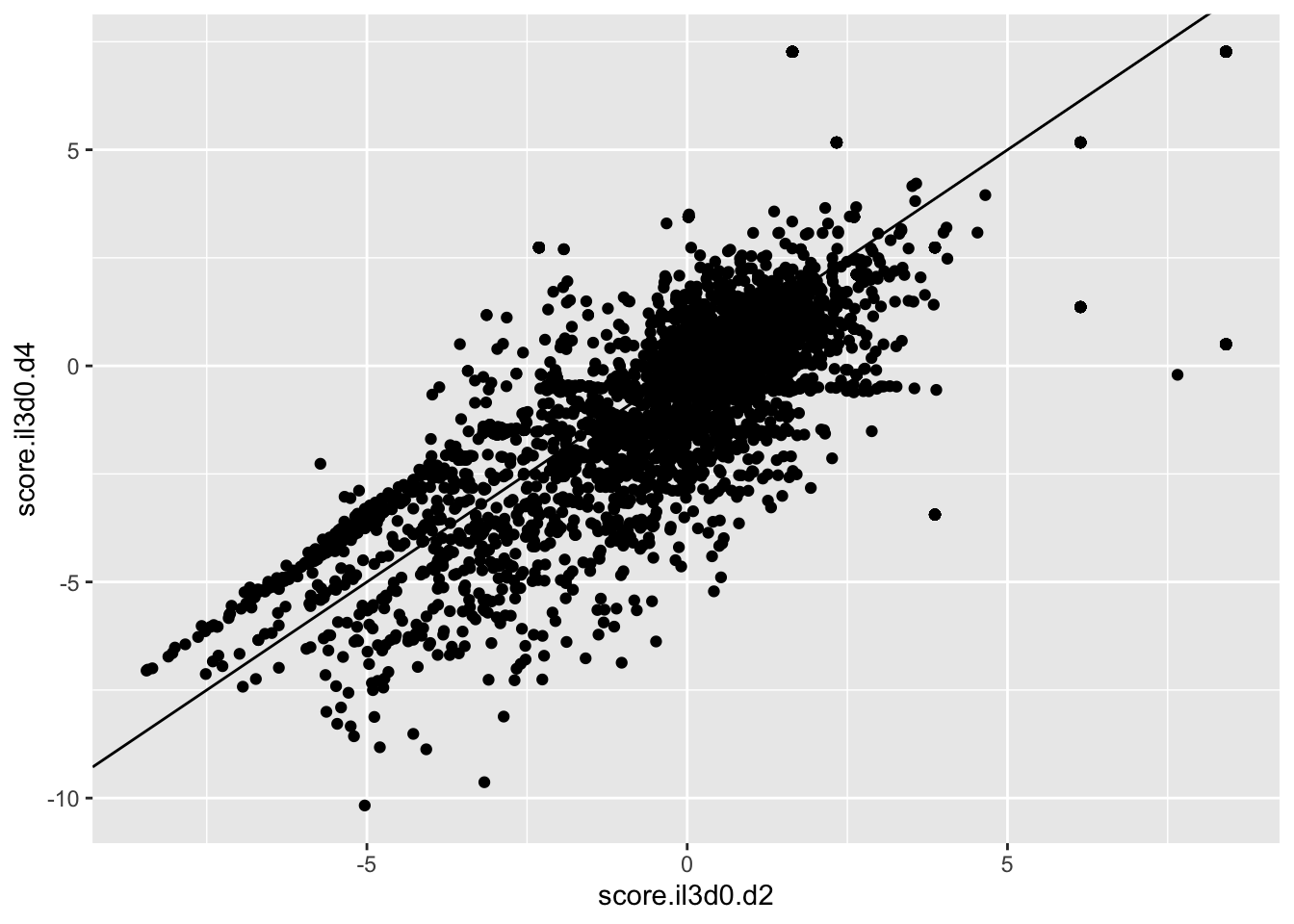
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.imatd2.d4%in%-6),aes(x=score.imatd0.d2,y=score.imatd2.d4))+geom_point()+geom_abline()Warning: Removed 1915 rows containing missing values (geom_point).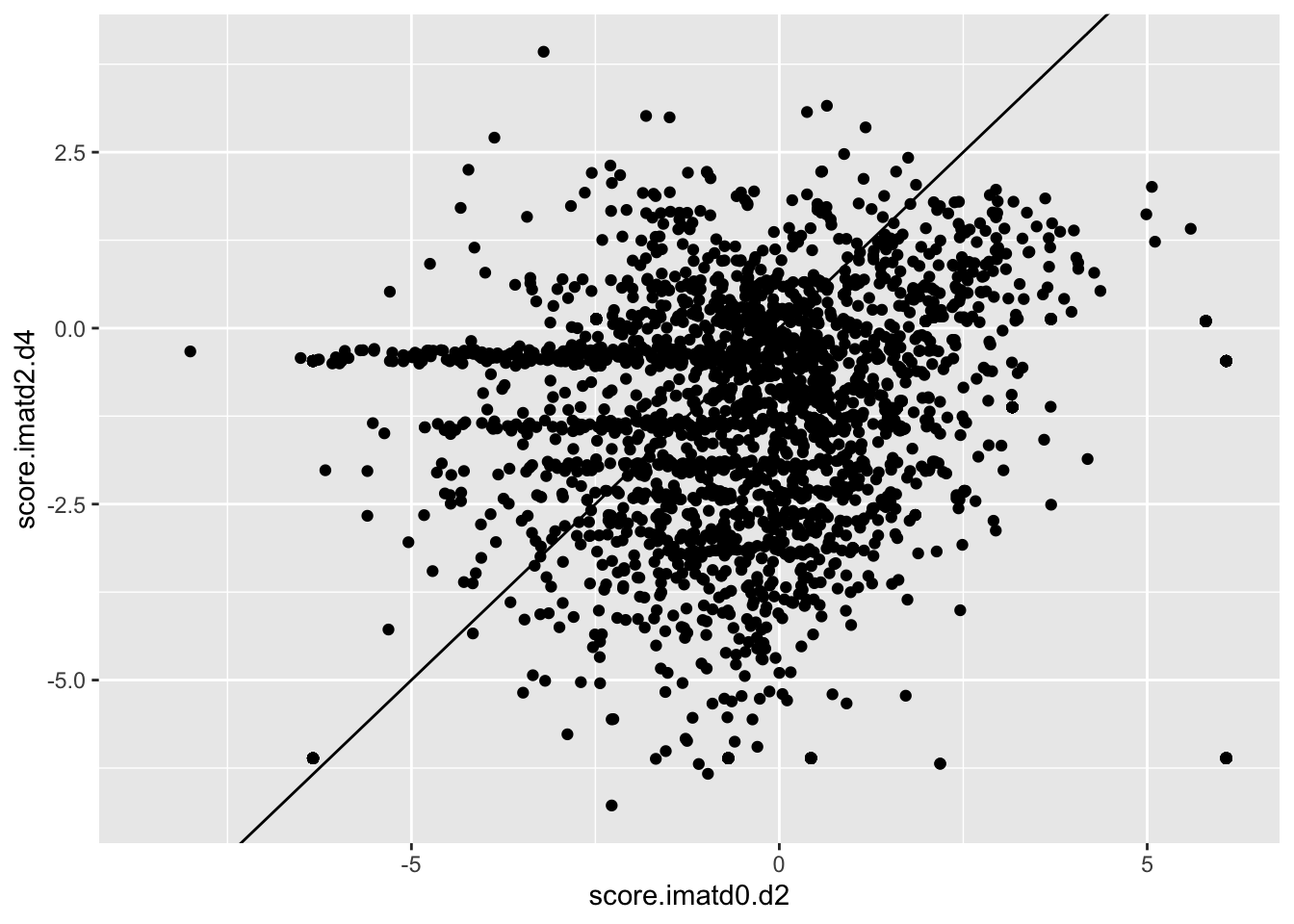
ggplot(scores_all%>%filter(consequence_terms%in%"missense_variant",protein_start%in%c(242:494),!score.imatd2.d4%in%-6),aes(x=score.il3d0.d2,y=score.il3d2.d4))+geom_point()+geom_abline()Warning: Removed 1351 rows containing missing values (geom_point).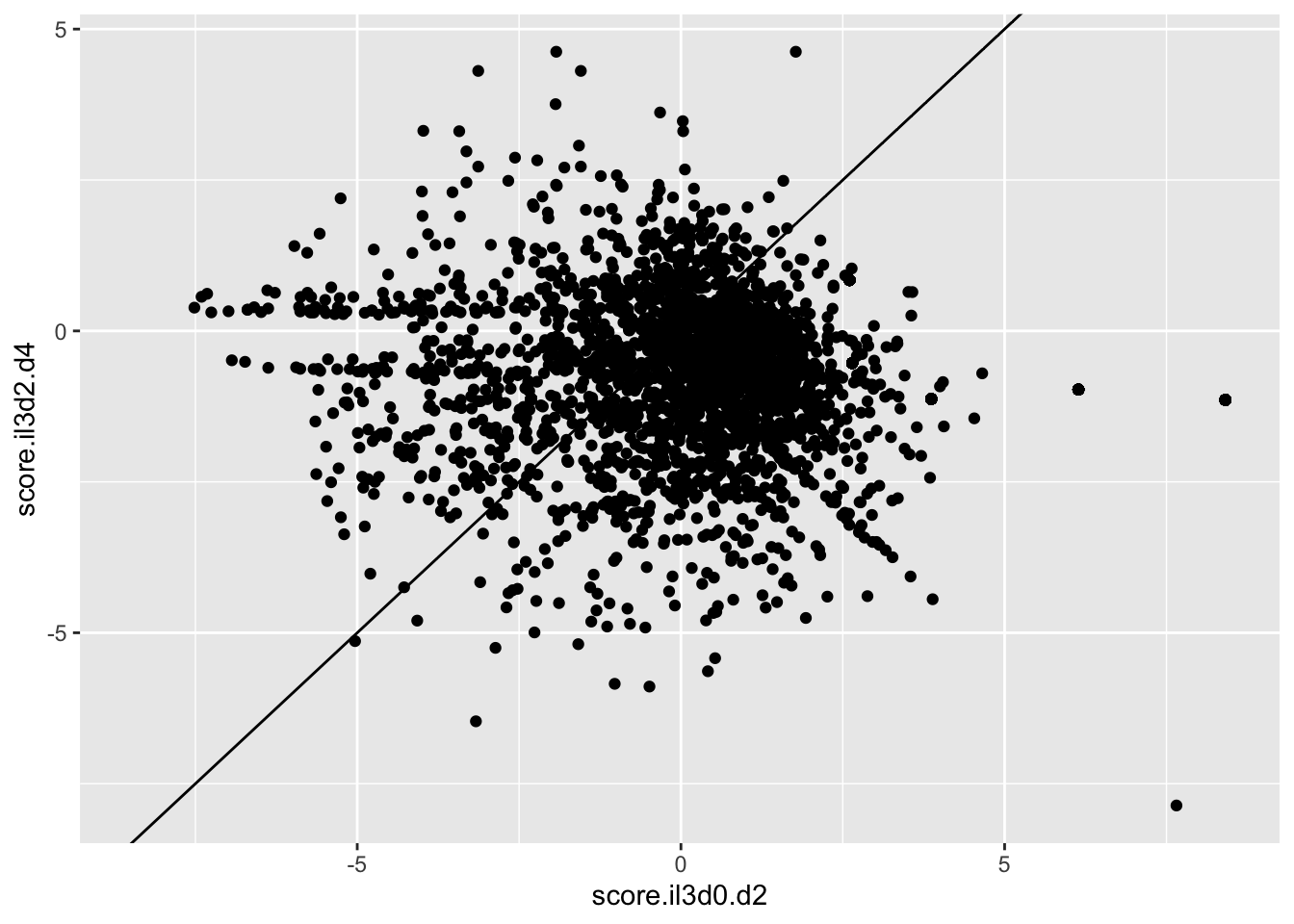
ggplot(scores_all%>%filter(!score.imatd2.d4%in%-6),aes(x=score.imatd2.d4))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1991 rows containing non-finite values (stat_bin).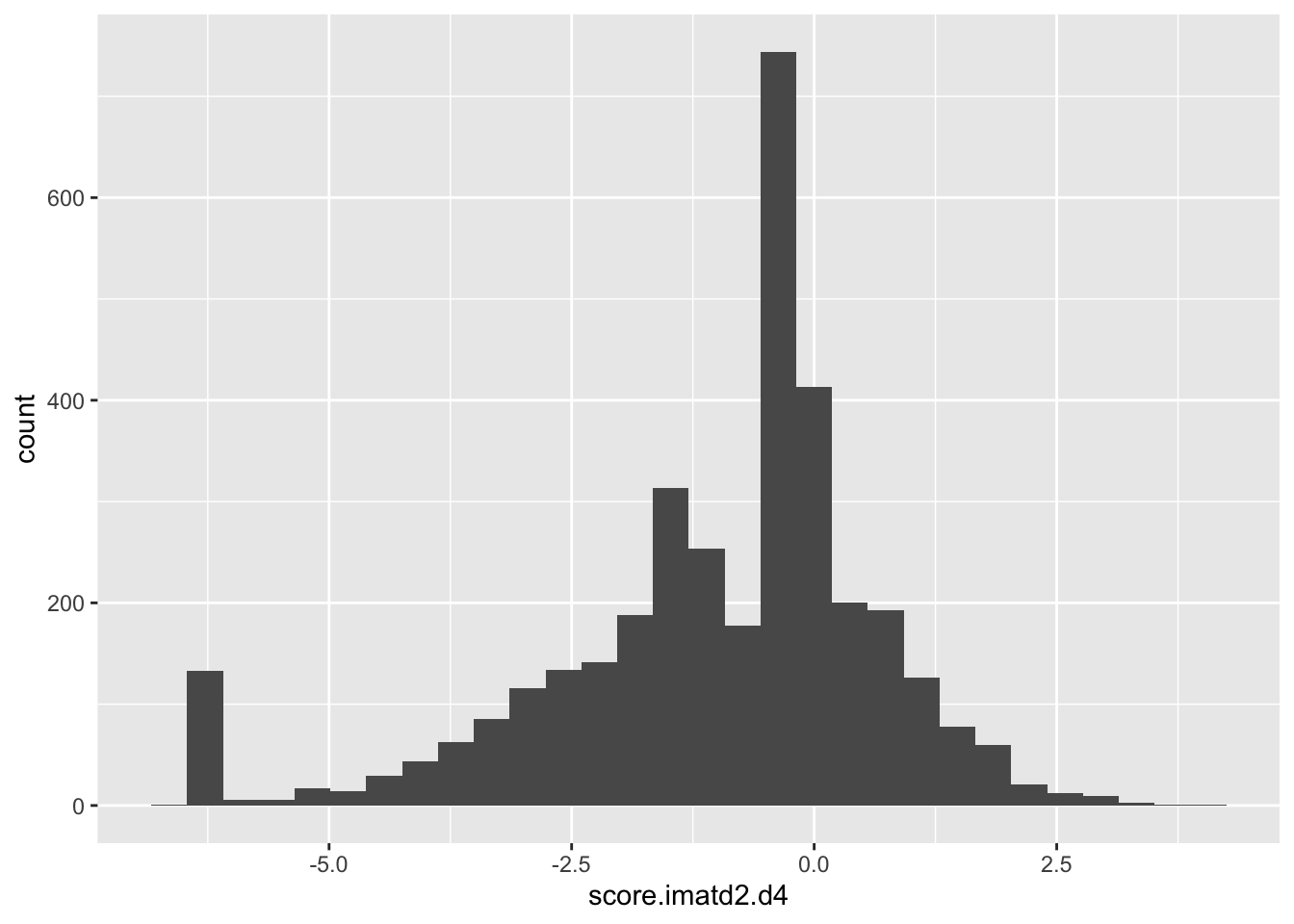
ggplot(scores_all%>%filter(!score.imatd0.d4%in%-6),aes(x=score.imatd0.d4))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1417 rows containing non-finite values (stat_bin).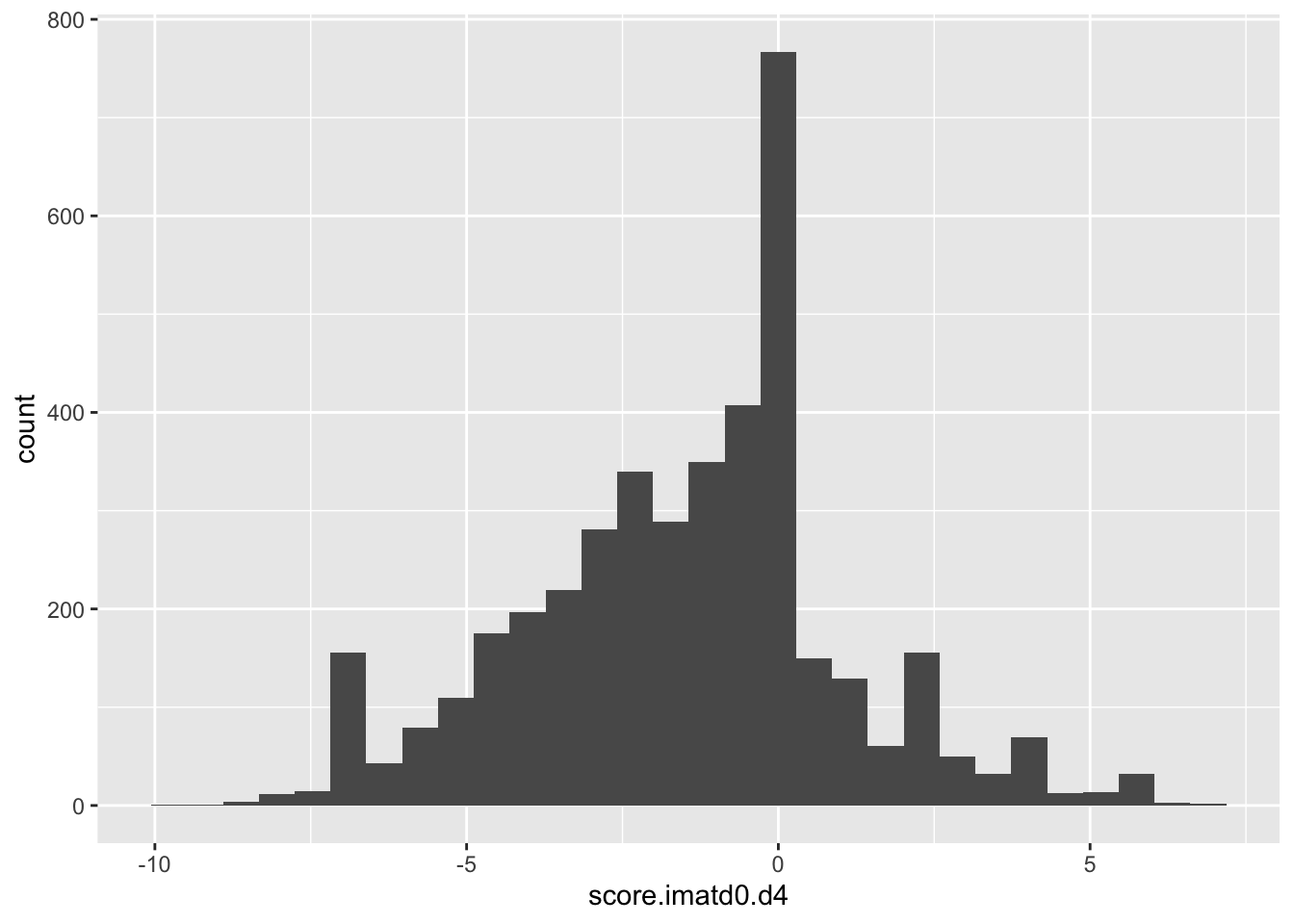
ggplot(scores_all%>%filter(!score.imatd0.d2%in%-6),aes(x=score.imatd0.d2))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1433 rows containing non-finite values (stat_bin).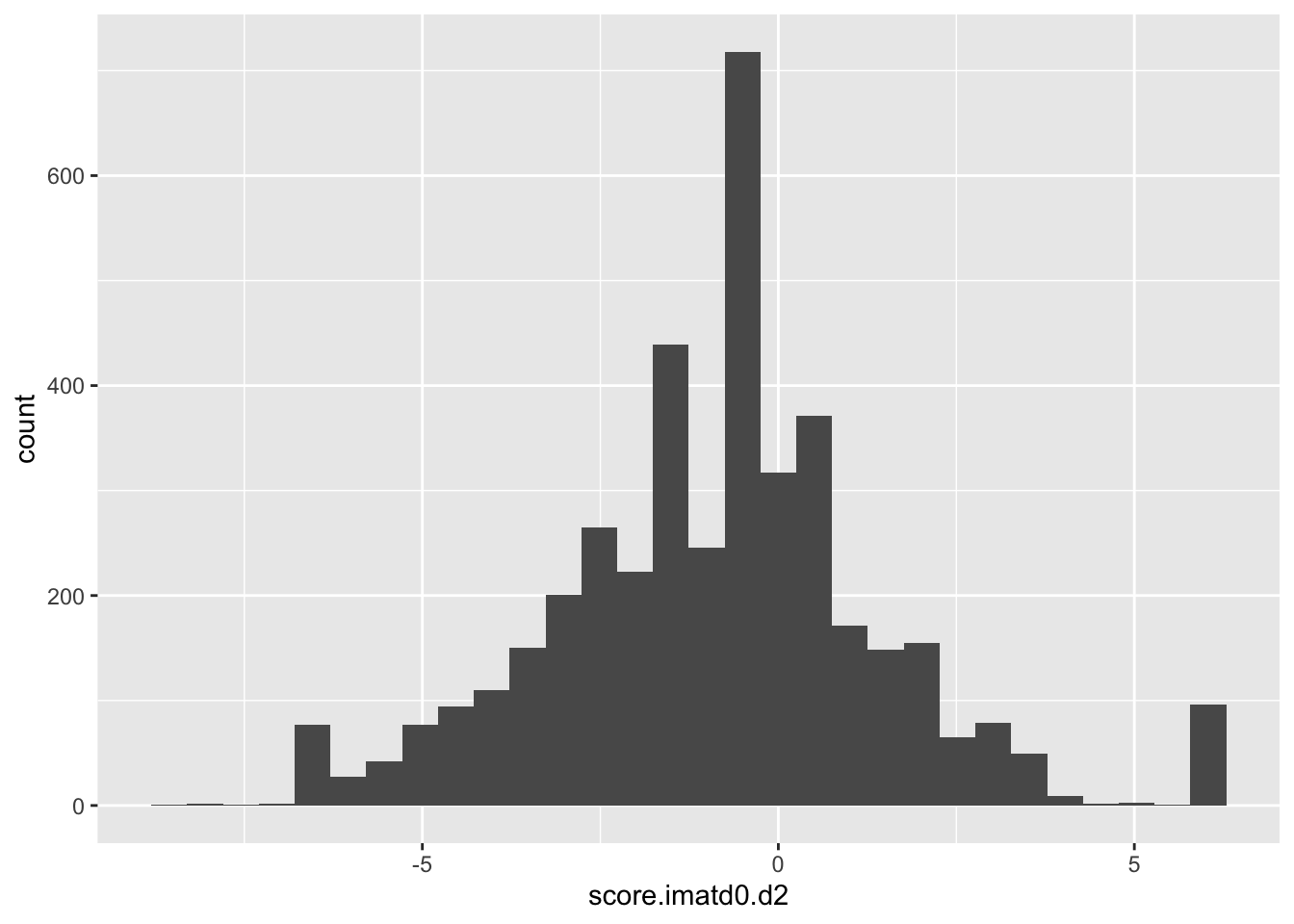
ggplot(scores_all%>%filter(!score.il3d0.d4%in%-6),aes(x=score.il3d0.d4))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1417 rows containing non-finite values (stat_bin).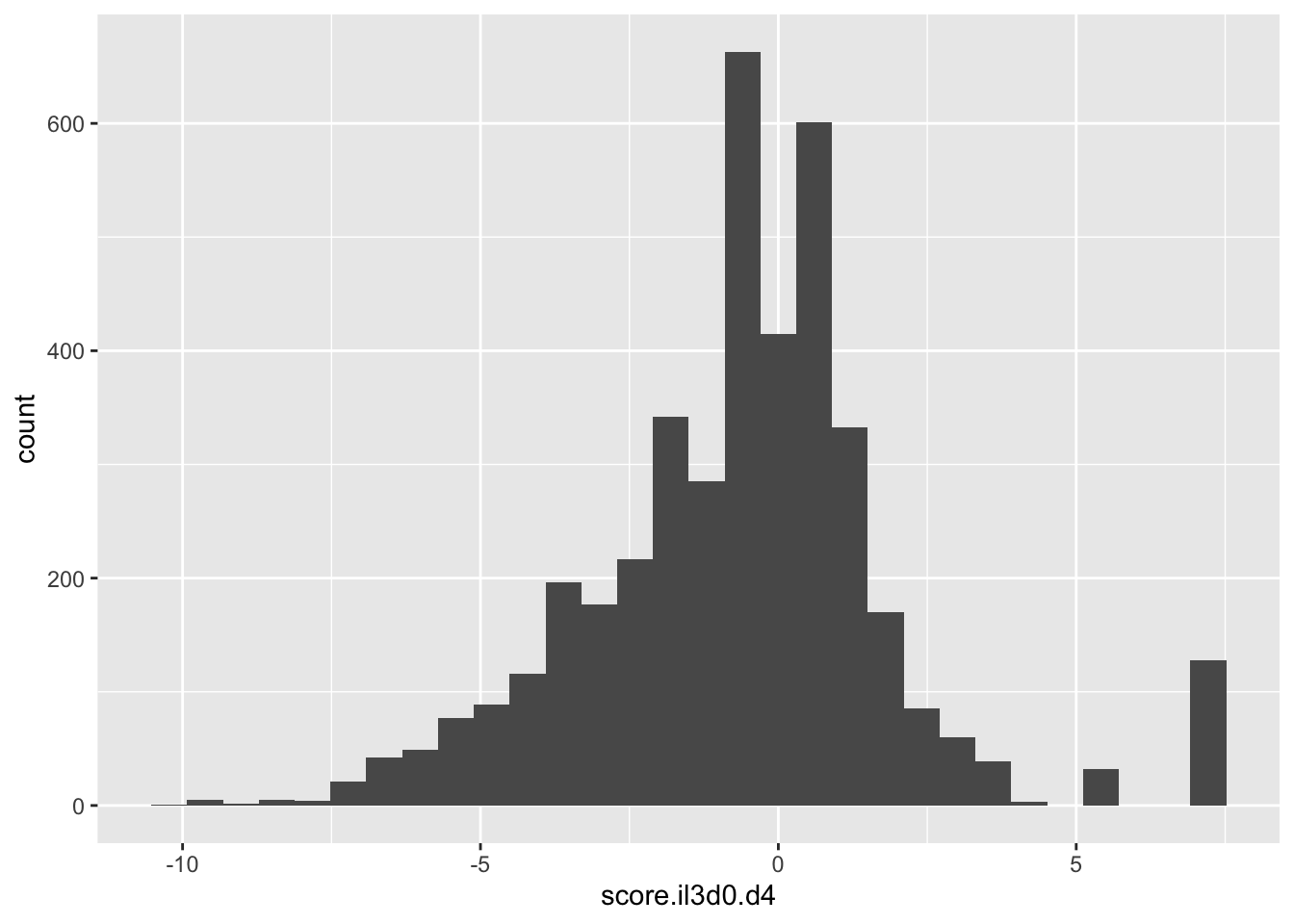
ggplot(scores_all%>%filter(!score.il3d2.d4%in%-6),aes(x=score.il3d2.d4))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 928 rows containing non-finite values (stat_bin).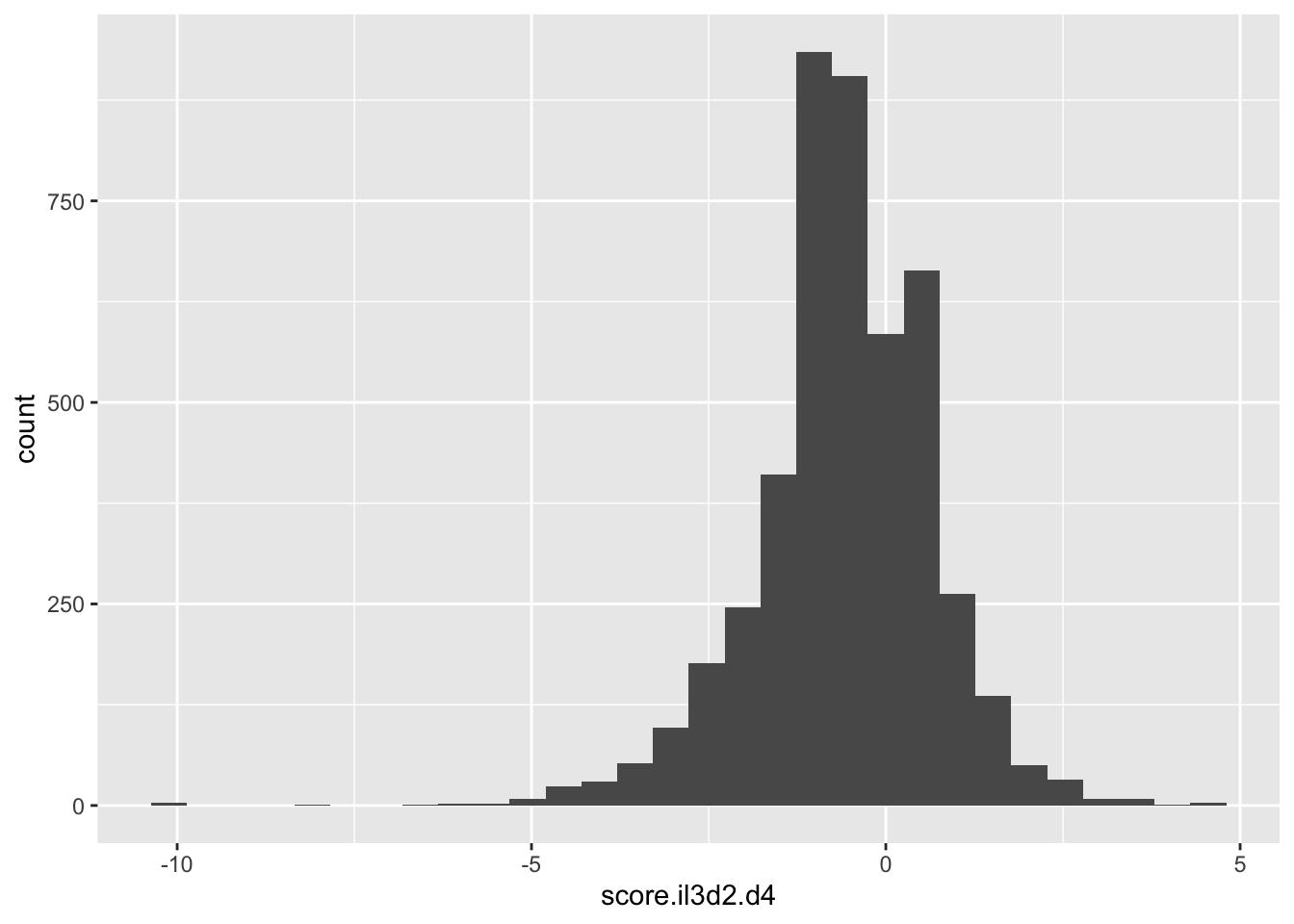
ggplot(scores_all%>%filter(!score.il3d0.d2%in%-6),aes(x=score.il3d0.d2))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 1417 rows containing non-finite values (stat_bin).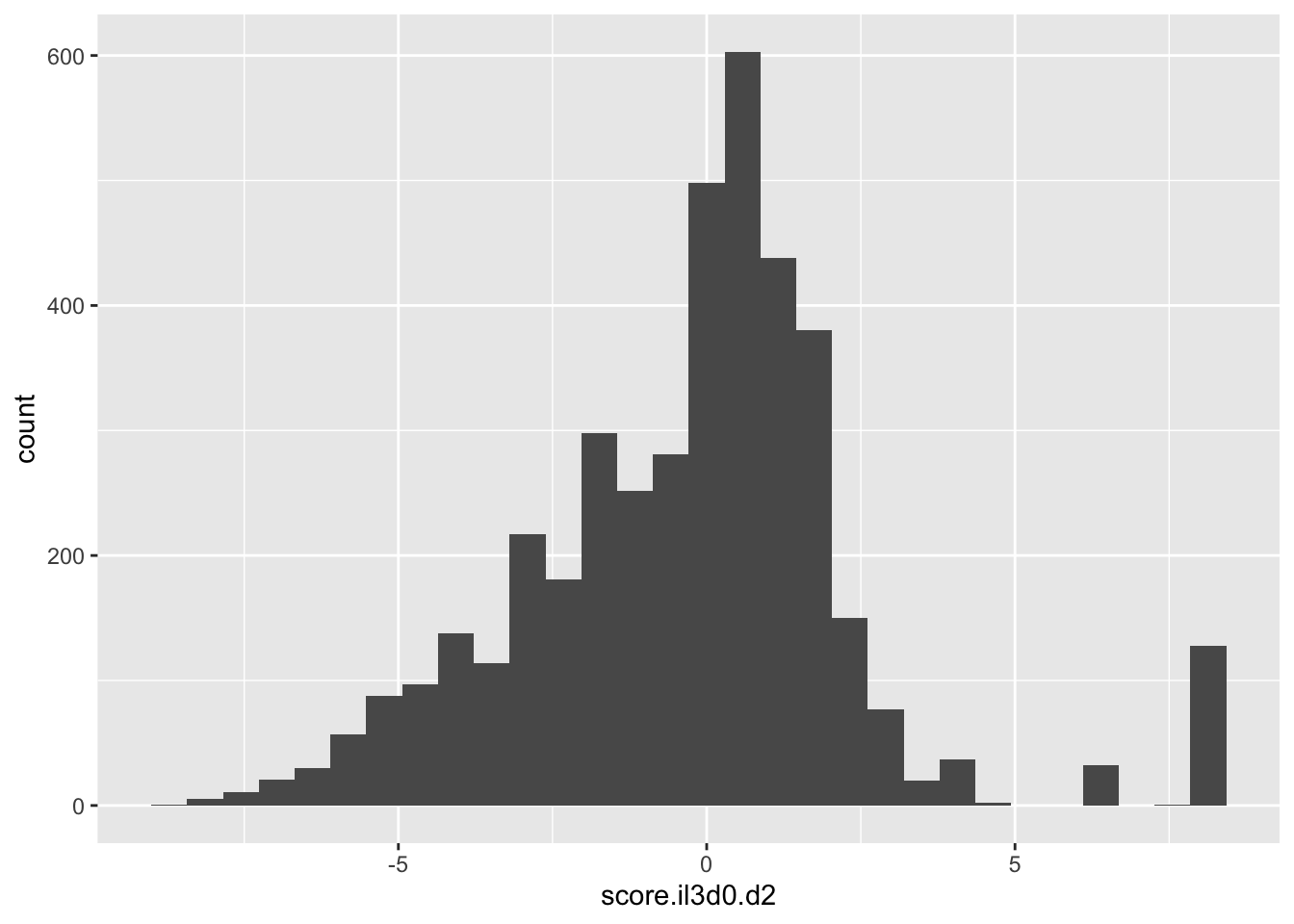
ggplot(il3D0_simple,aes(x=maf))+geom_density()+scale_x_continuous(trans = "log10")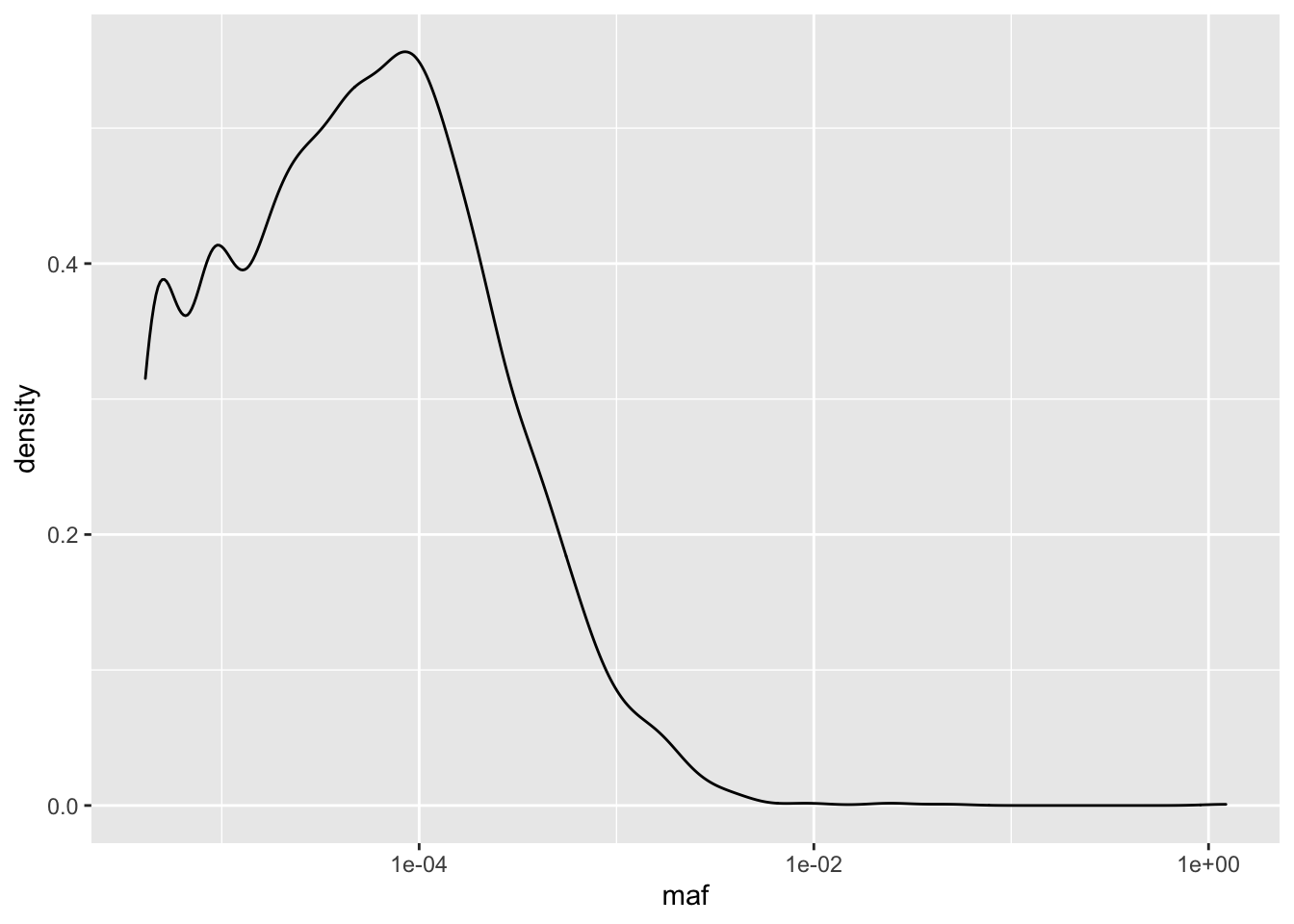
ggplot(il3D2_simple,aes(x=maf))+geom_density()+scale_x_continuous(trans = "log10")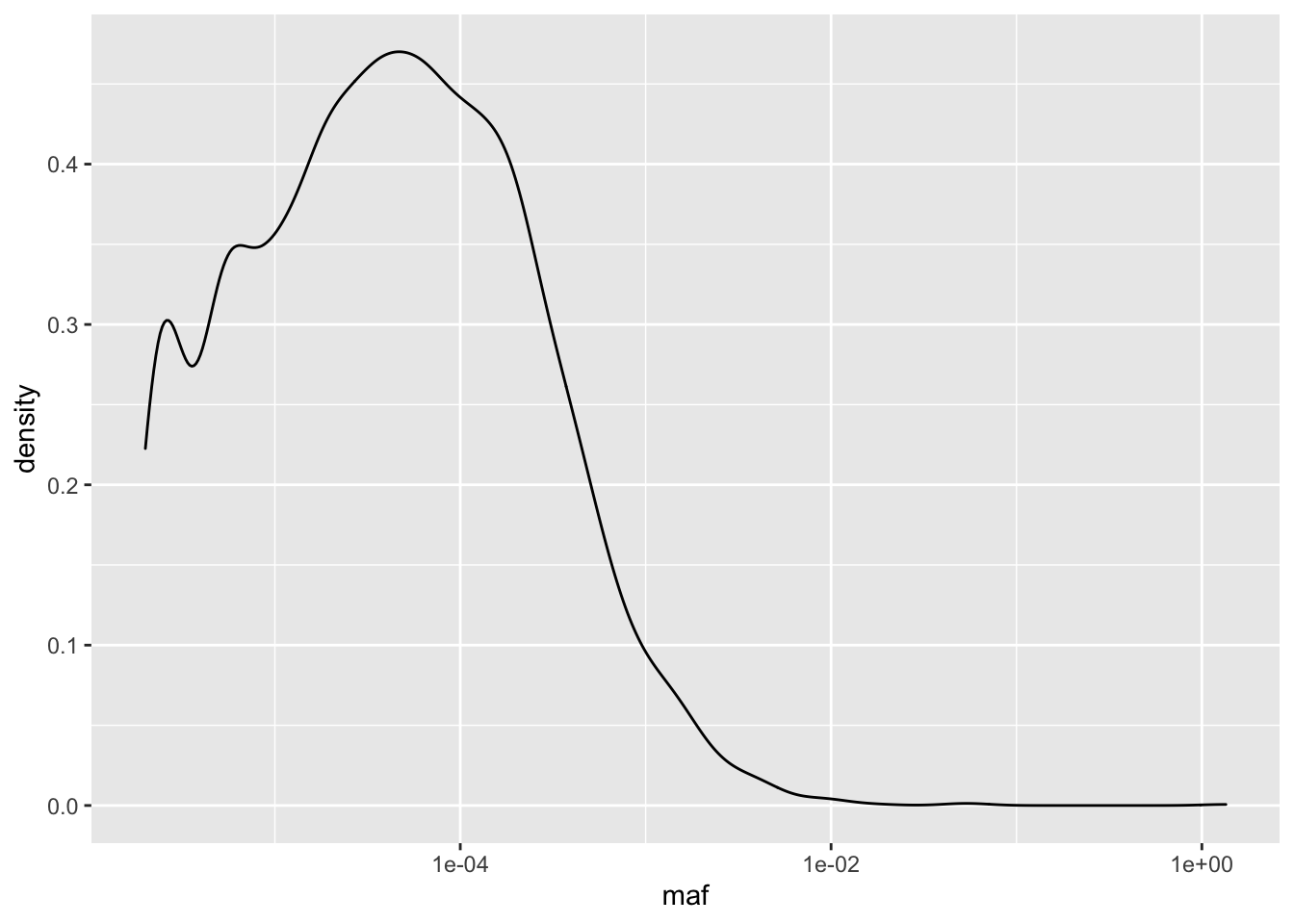
ggplot(imatD2_simple,aes(x=maf))+geom_density()+scale_x_continuous(trans = "log10")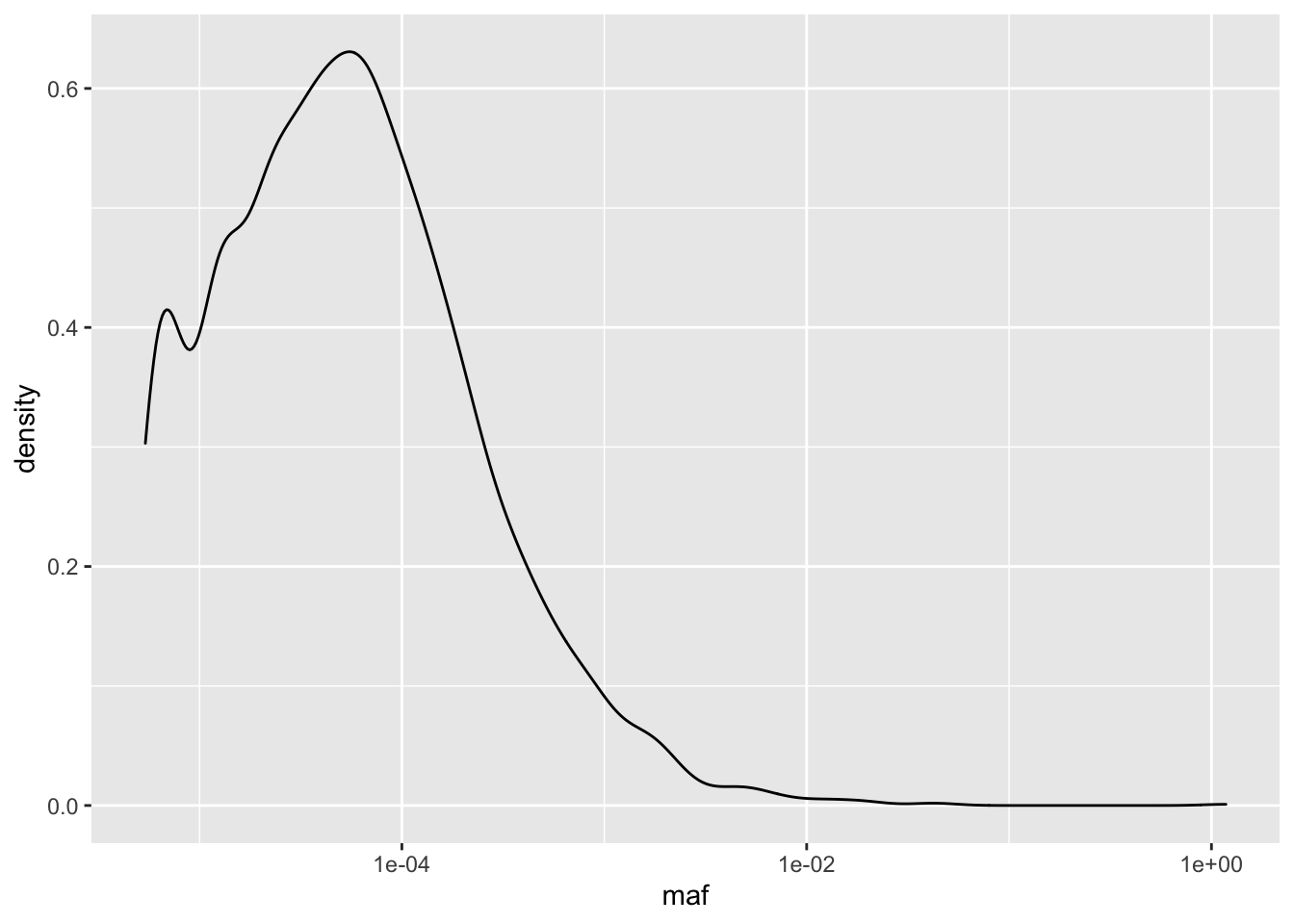
ggplot(imatD4_simple,aes(x=maf))+geom_density()+scale_x_continuous(trans = "log10")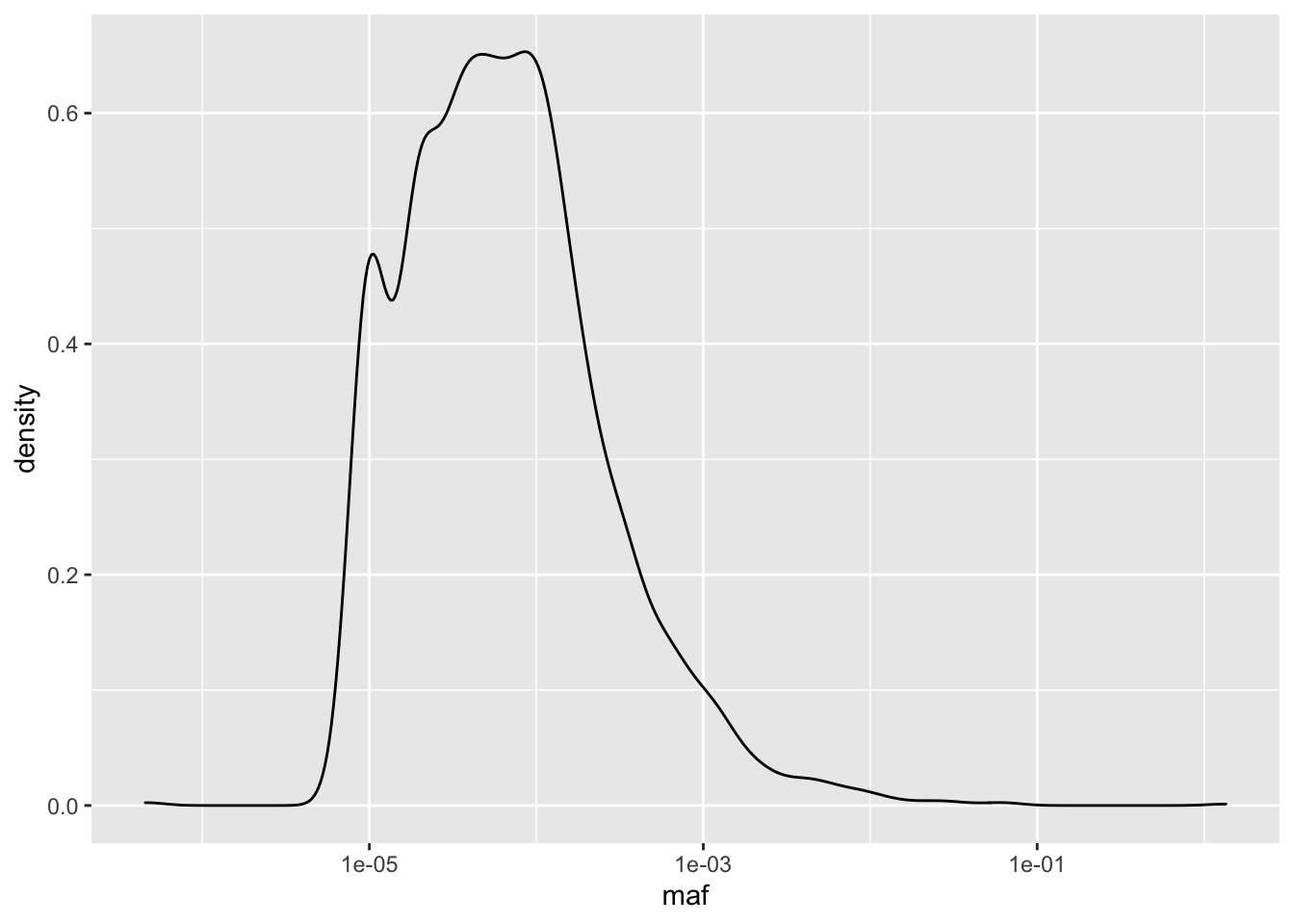
a=imatD0.D4%>%filter(protein_start%in%c(253,252,459,276))Plotting specific resistant mutants
resmuts=scores_all%>%filter(resmuts%in%T)%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
ggplot(resmuts%>%filter(!protein_start%in%317),aes(x=score.imatd0.d2,y=score.imatd2.d4,label=species))+geom_text()+geom_abline()Warning: Removed 5 rows containing missing values (geom_text).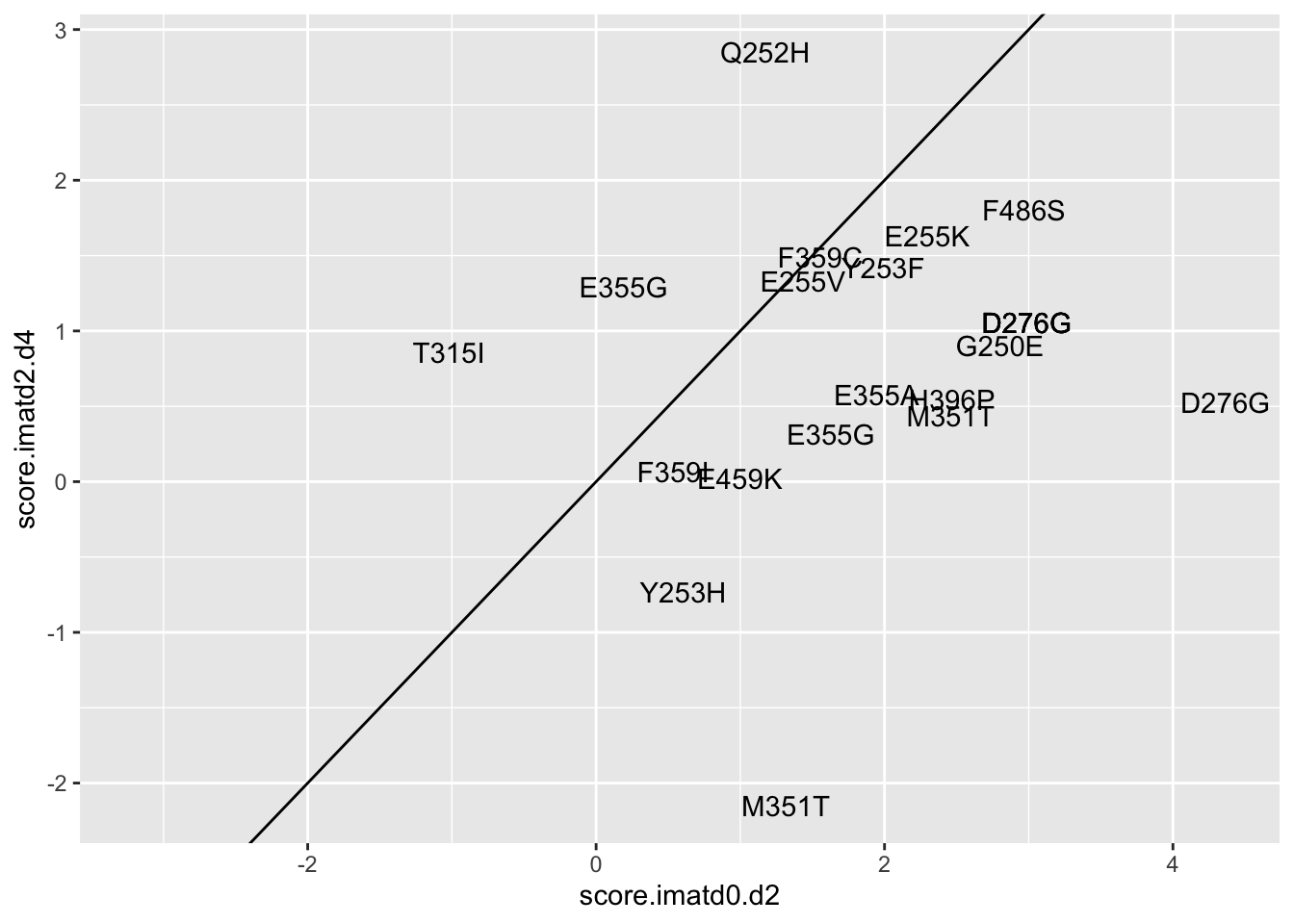
# ggplot(resmuts%>%filter(!protein_start%in%317),aes(x=netgr_obs.imatd0.d2,y=netgr_obs.imatd2.d4,label=species))+geom_text()+geom_abline()
ic50data_all_sum=read.csv("output/ic50data_all_confidence_intervals_raw_data.csv",row.names = 1)
resmuts_merged=merge(resmuts,ic50data_all_sum,by="species")
# ggplot(resmuts_merged,aes(x=netgr_pred_model,y=netgr_obs.imatd0.d6,label=species))+geom_text()
#Plotting Imatinib D0 vs D2
ggplot(resmuts_merged,aes(x=netgr_pred_model,y=score.imatd0.d2,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).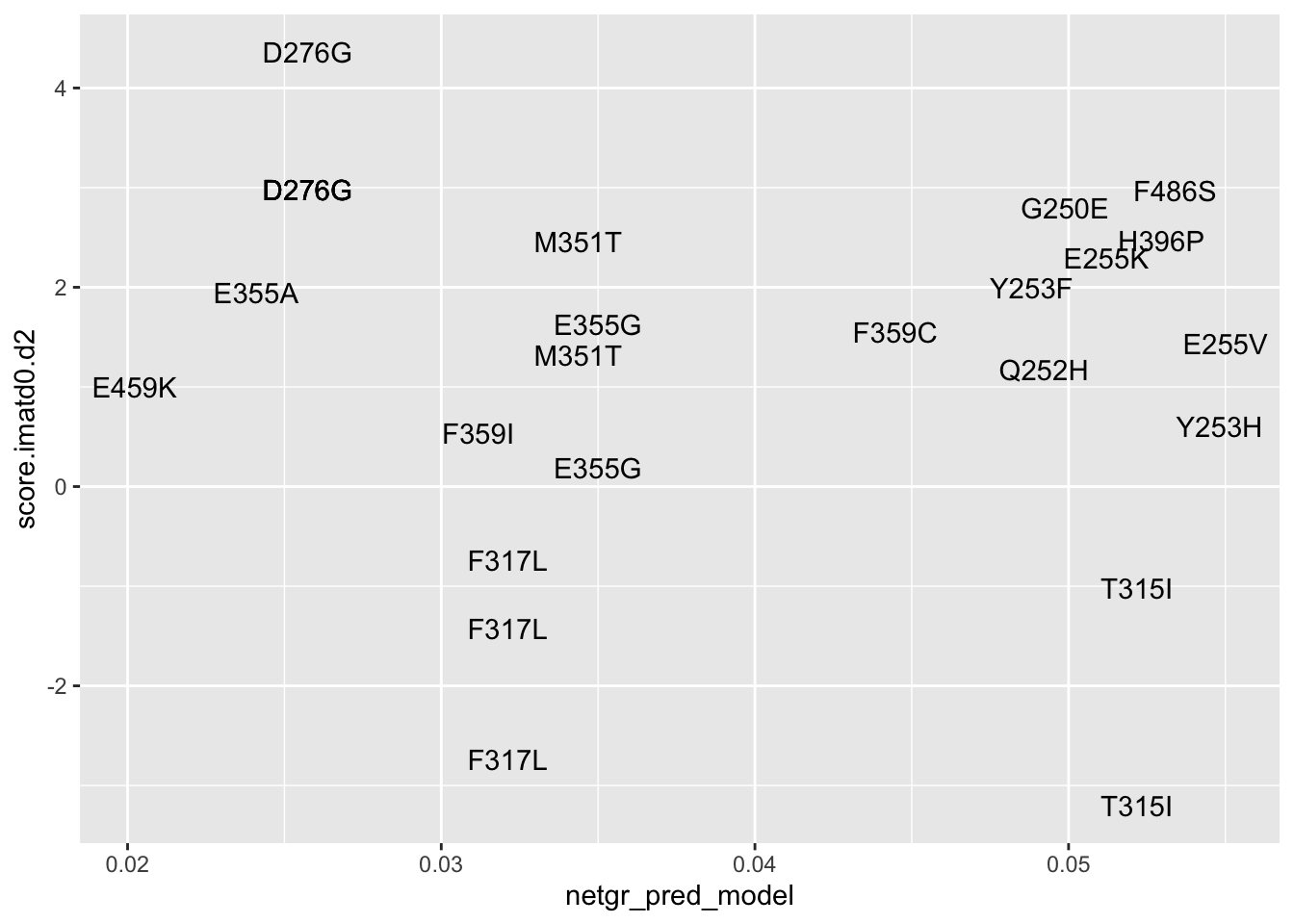
ggplot(resmuts_merged,aes(x=netgr_pred_model,y=netgr_obs.imatd0.d2,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).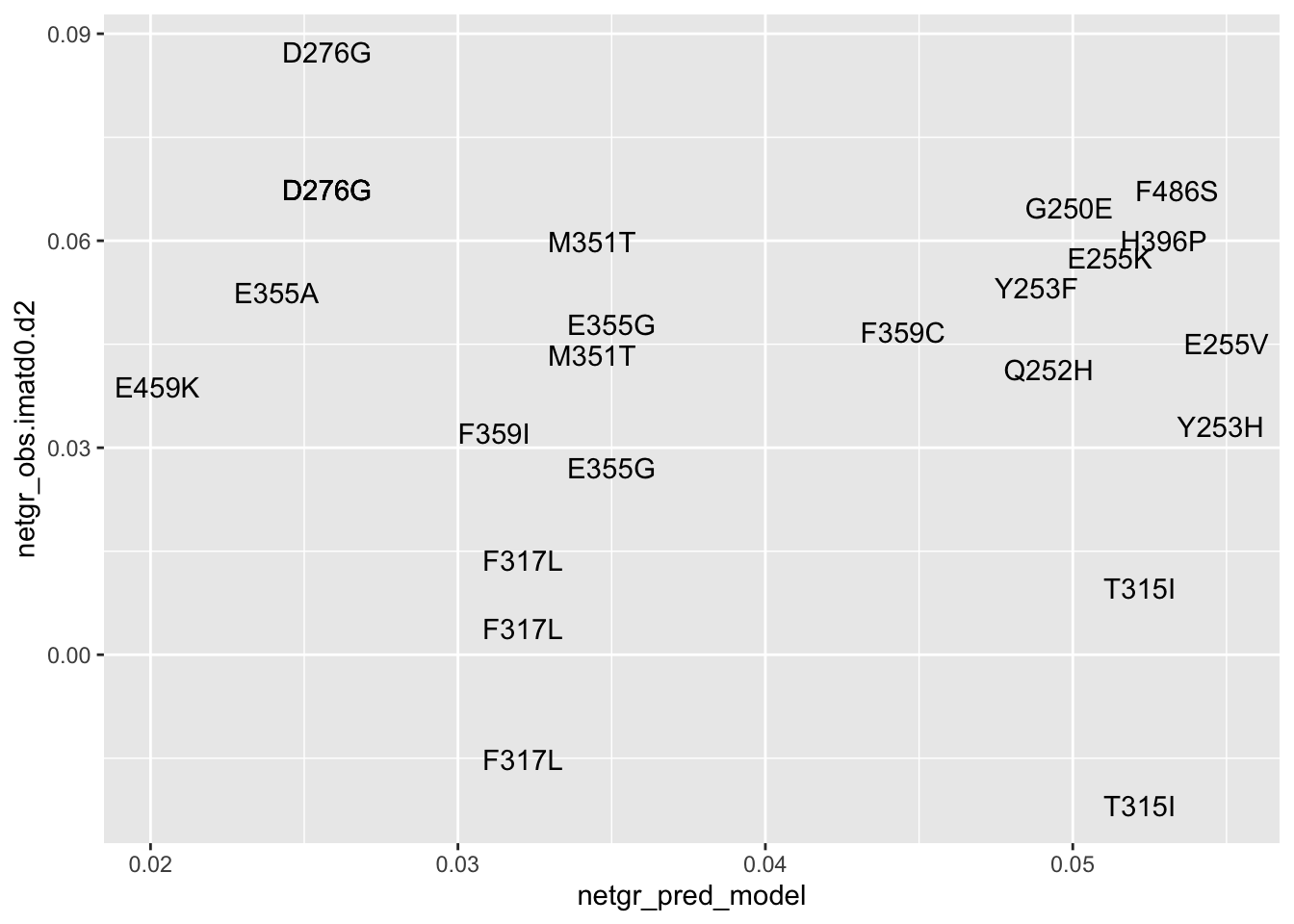
#Plotting Imatinib D2 vs D4
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd2.d4,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).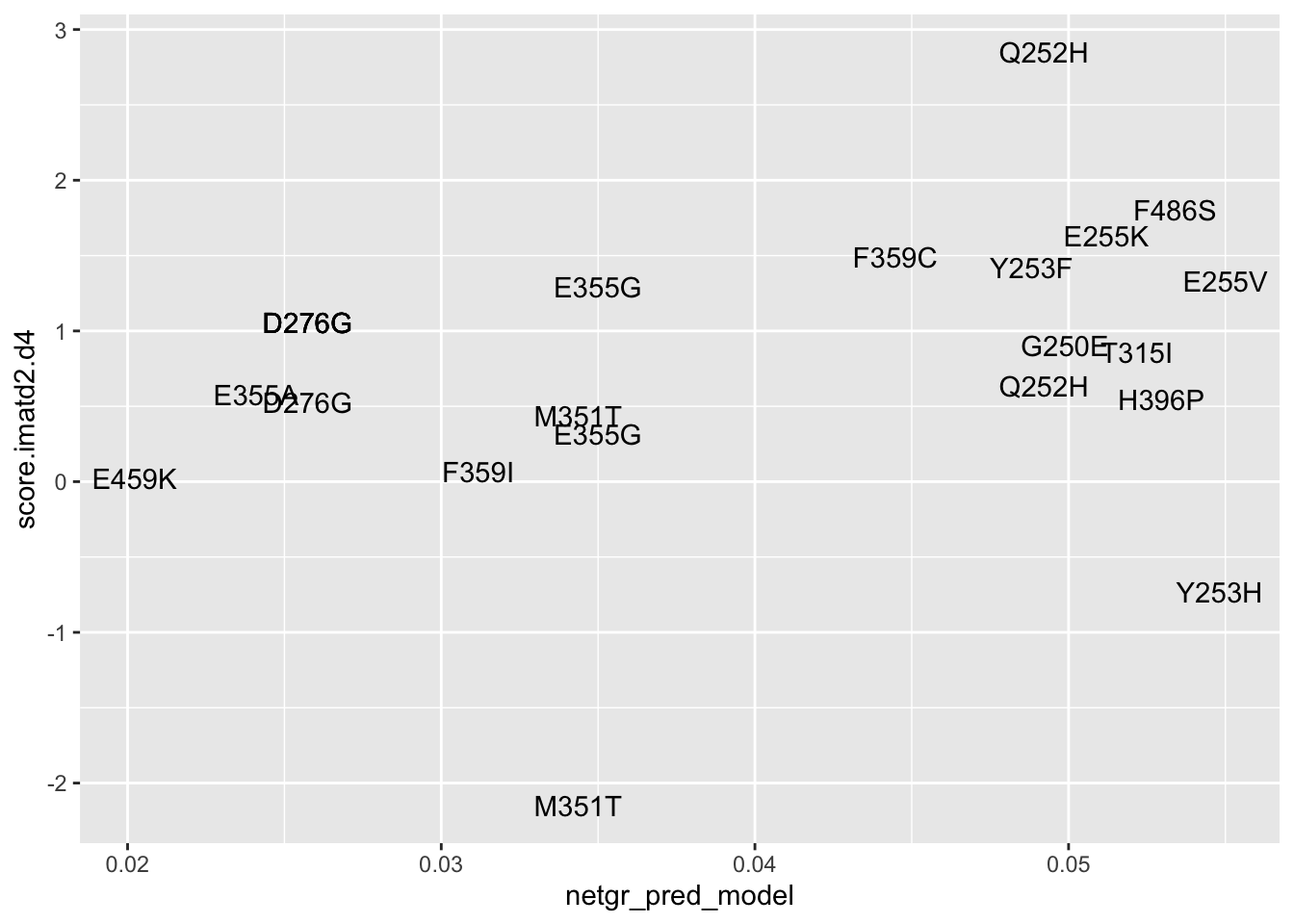
#Plotting Imatinib D0 vs D4
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd0.d4,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).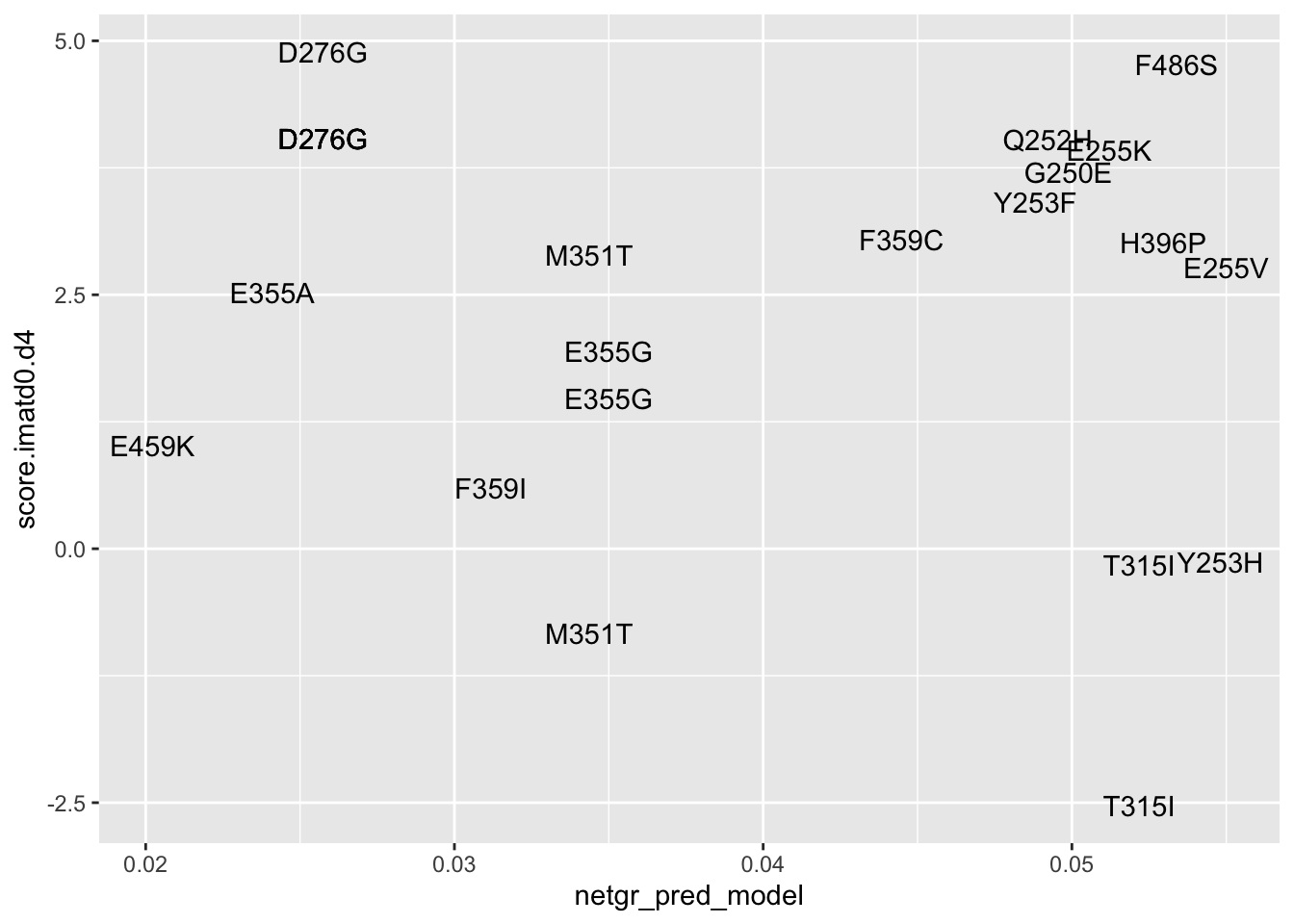
#Plotting Imatinib D0 vs D2 Adjusted
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd0.d2-score.il3d0.d2,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).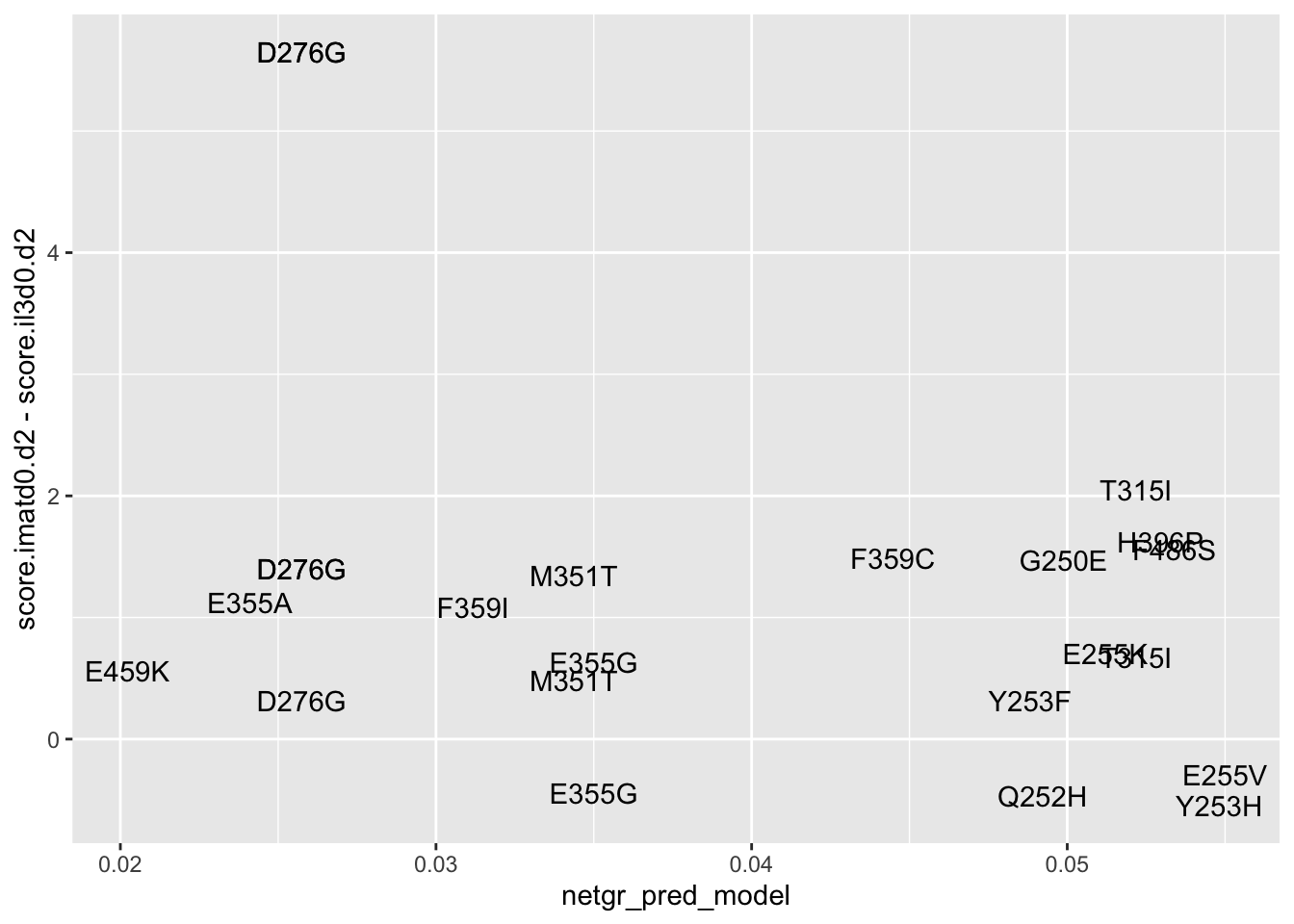
#Plotting Imatinib D2 vs D4 Adjusted
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd2.d4-score.il3d2.d4,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).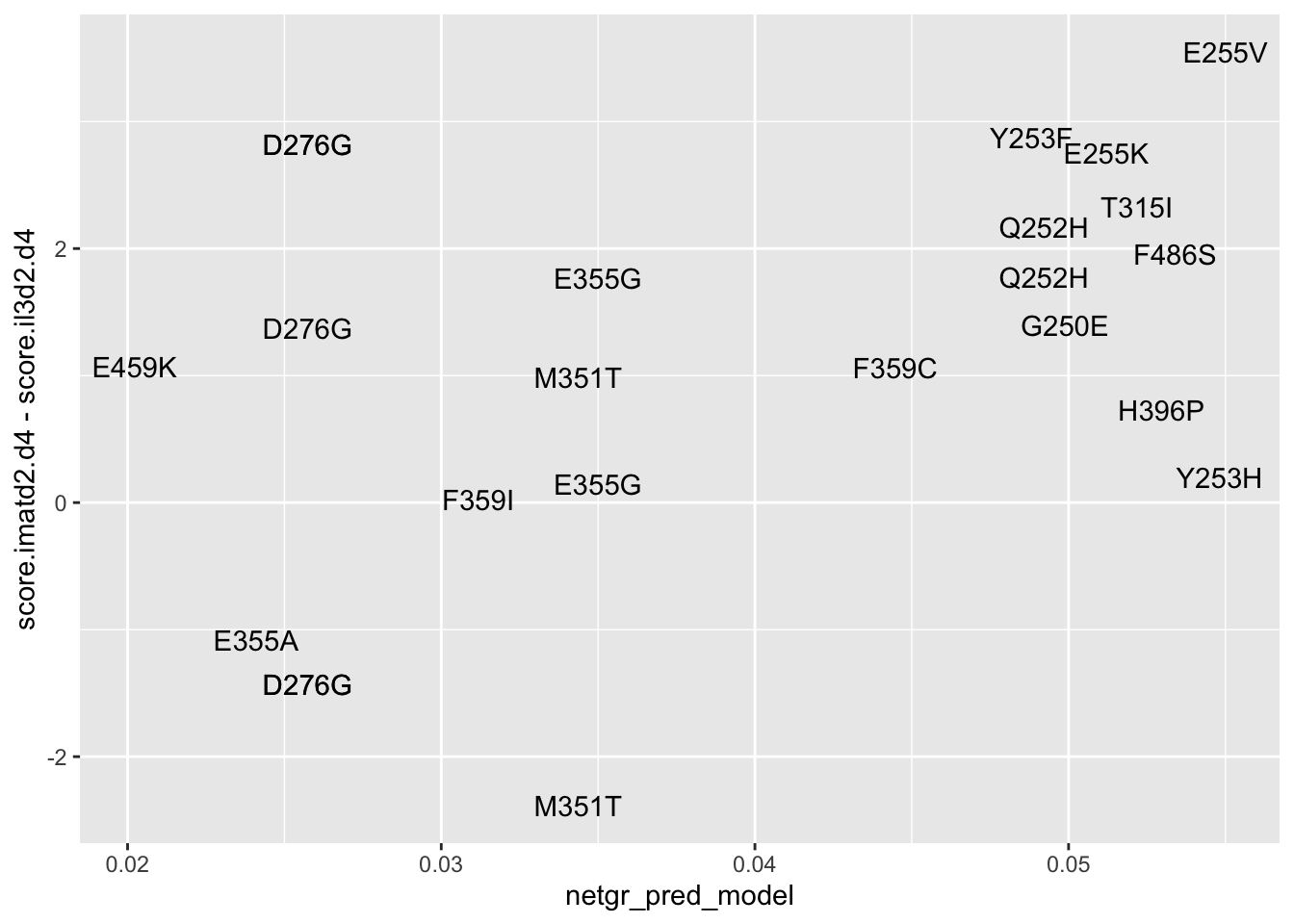
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=-netgr_obs.imatd2.d4+netgr_obs.il3d2.d4+.055,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).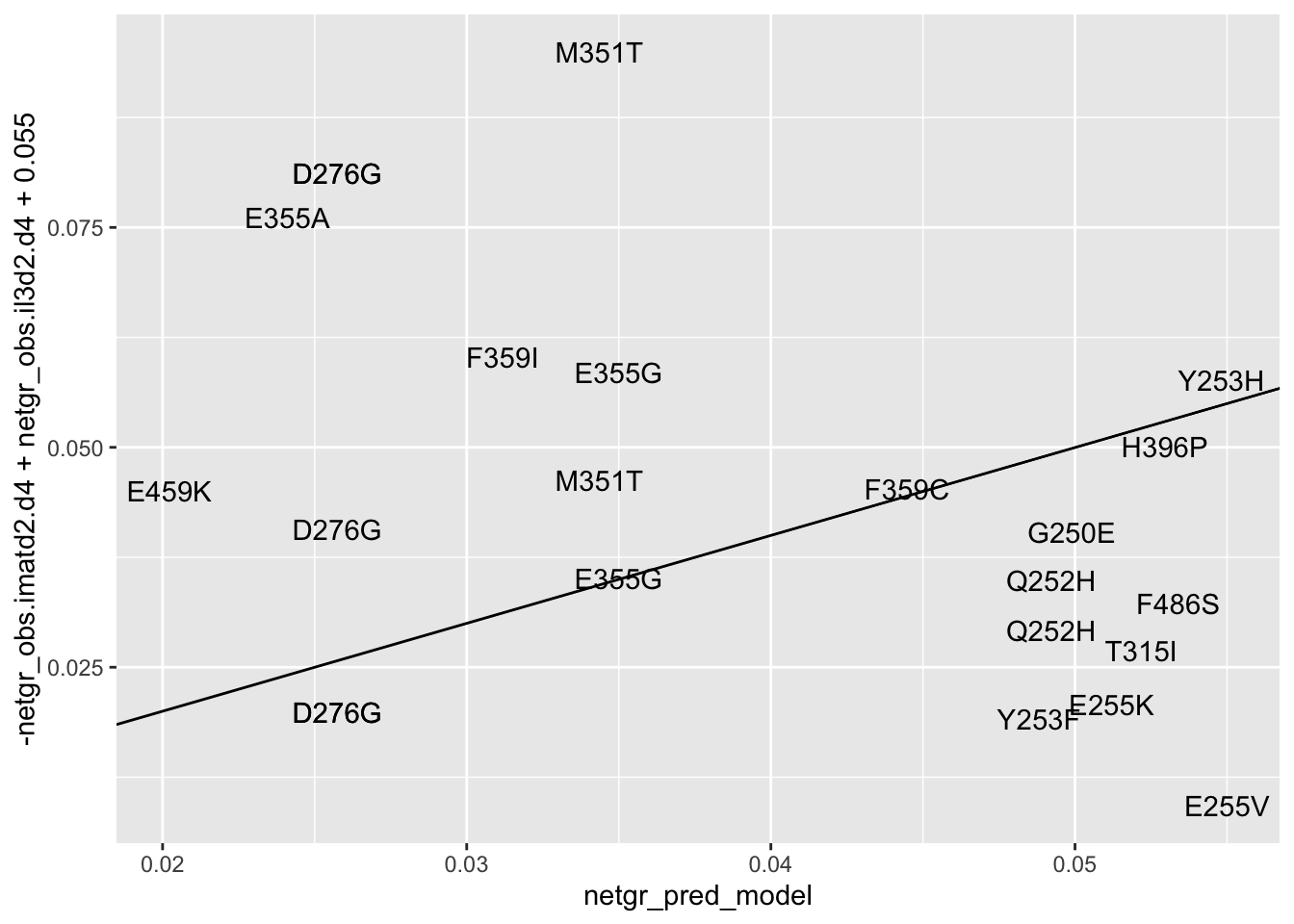
#Plotting Imatinib D0 vs D4 Adjusted
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd0.d4-score.il3d0.d4,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).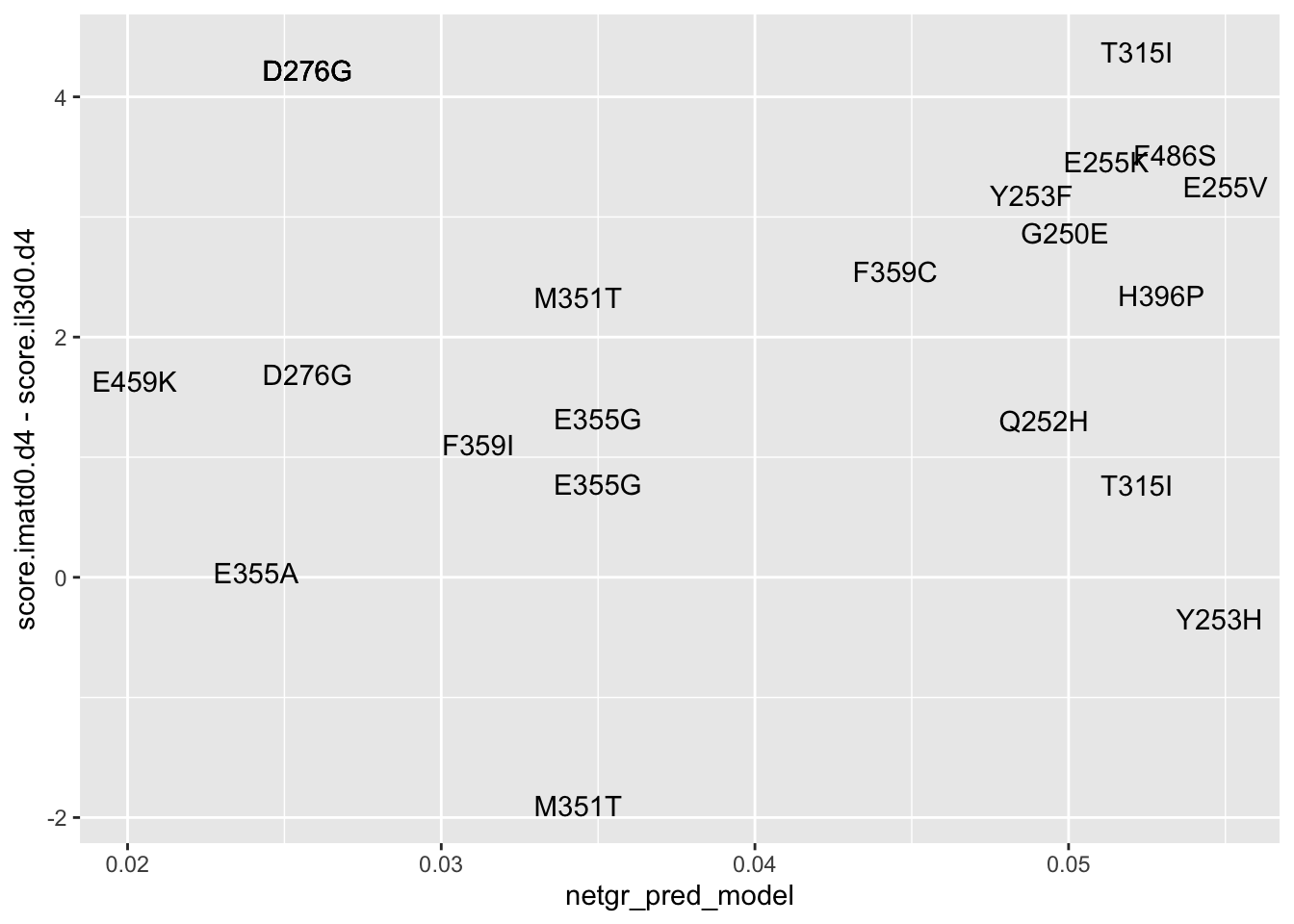
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=netgr_obs.imatd0.d4-netgr_obs.il3d0.d4,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).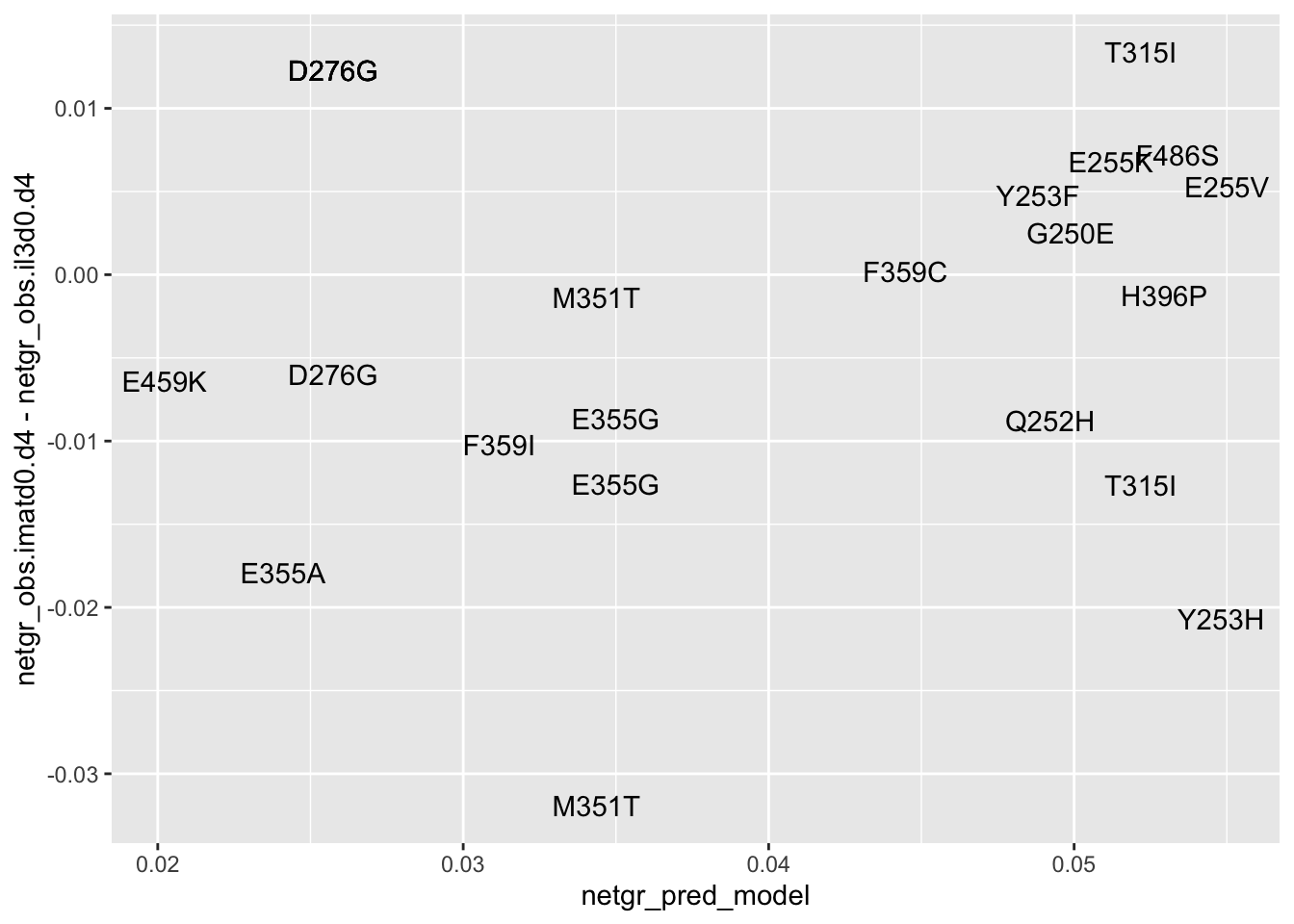
#Plotting Imatinib D0 vs D6 Adjusted
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=score.imatd0.d6-score.il3d0.d6,label=species))+geom_text()Warning: Removed 2 rows containing missing values (geom_text).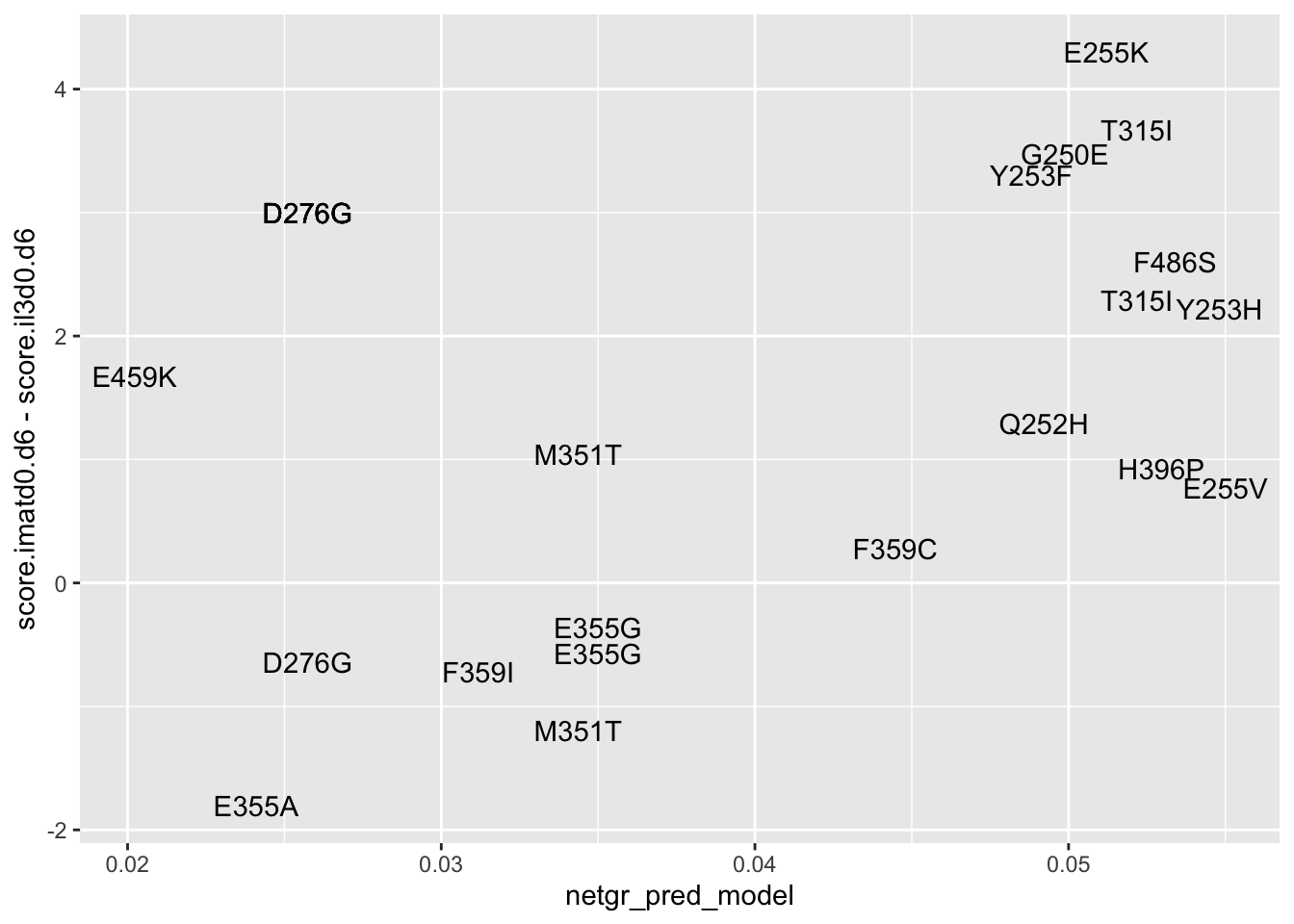
ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=netgr_pred_model,y=netgr_obs.imatd0.d6-netgr_obs.il3d0.d6,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).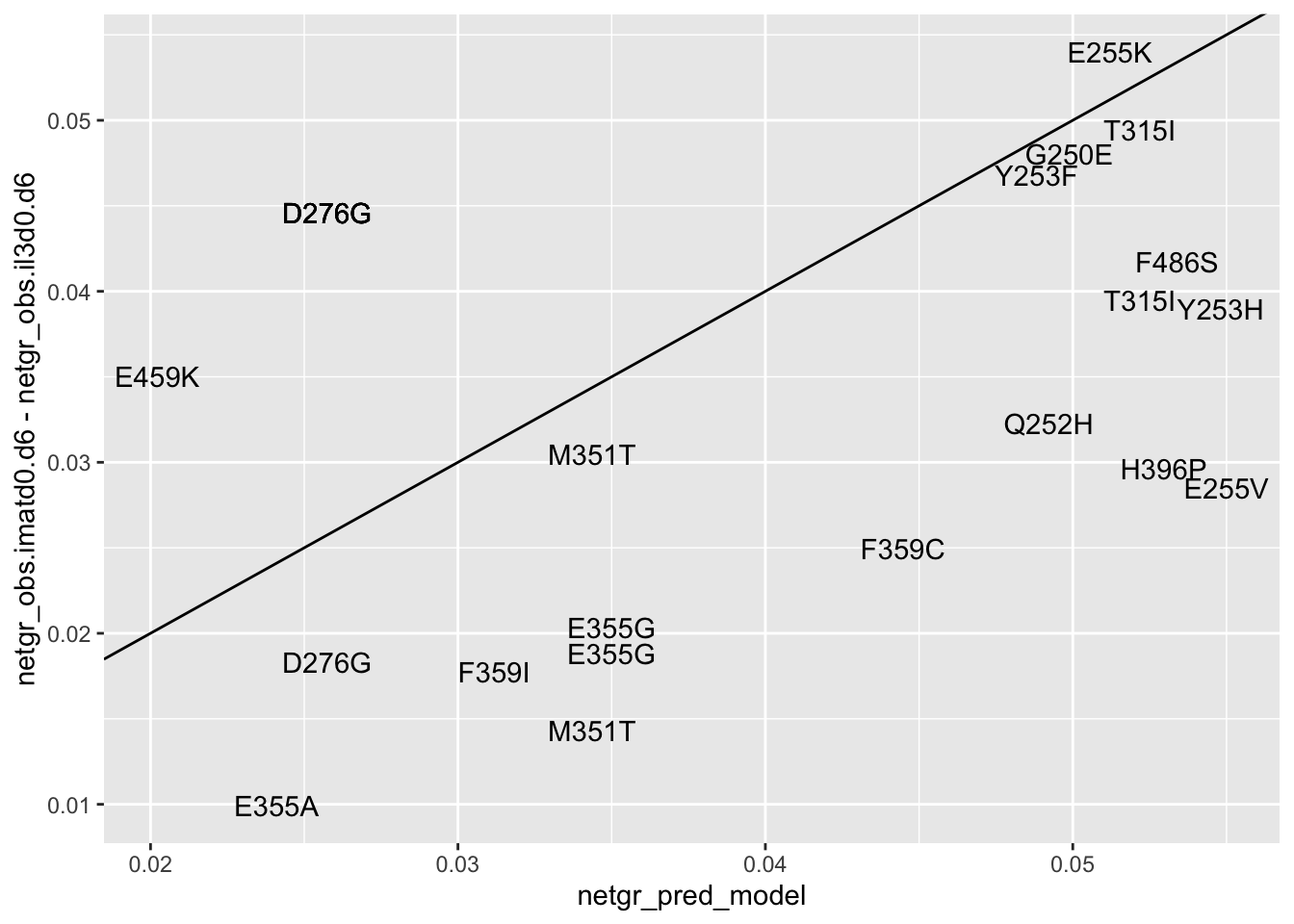
# Also plot IL3 D4 D2 scores vs iL3 D2 D0 scores
# Also update imatininb scores by considering without drug (fitness in the absence of iL3). Do these scores match up better with the IC50 predictions?
# Also, when looking at the distribution of enrichment scores, color the multiple nucleotide variantsLooking at how well the observed and predicted growth rates match up
ic50data_all_sum2=read.csv("output/ic50data_all_conc.csv",row.names = 1)
ic50data_all_sum2=ic50data_all_sum2%>%filter(conc%in%.7)%>%rename(species=mutant)%>%dplyr::select(species,drug_effect)
resmuts_merged=merge(resmuts,ic50data_all_sum2,by="species")
#Looking at D2 D4
ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=.055-drug_effect,y=.055-(-netgr_obs.imatd2.d4+netgr_obs.il3d2.d4),label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).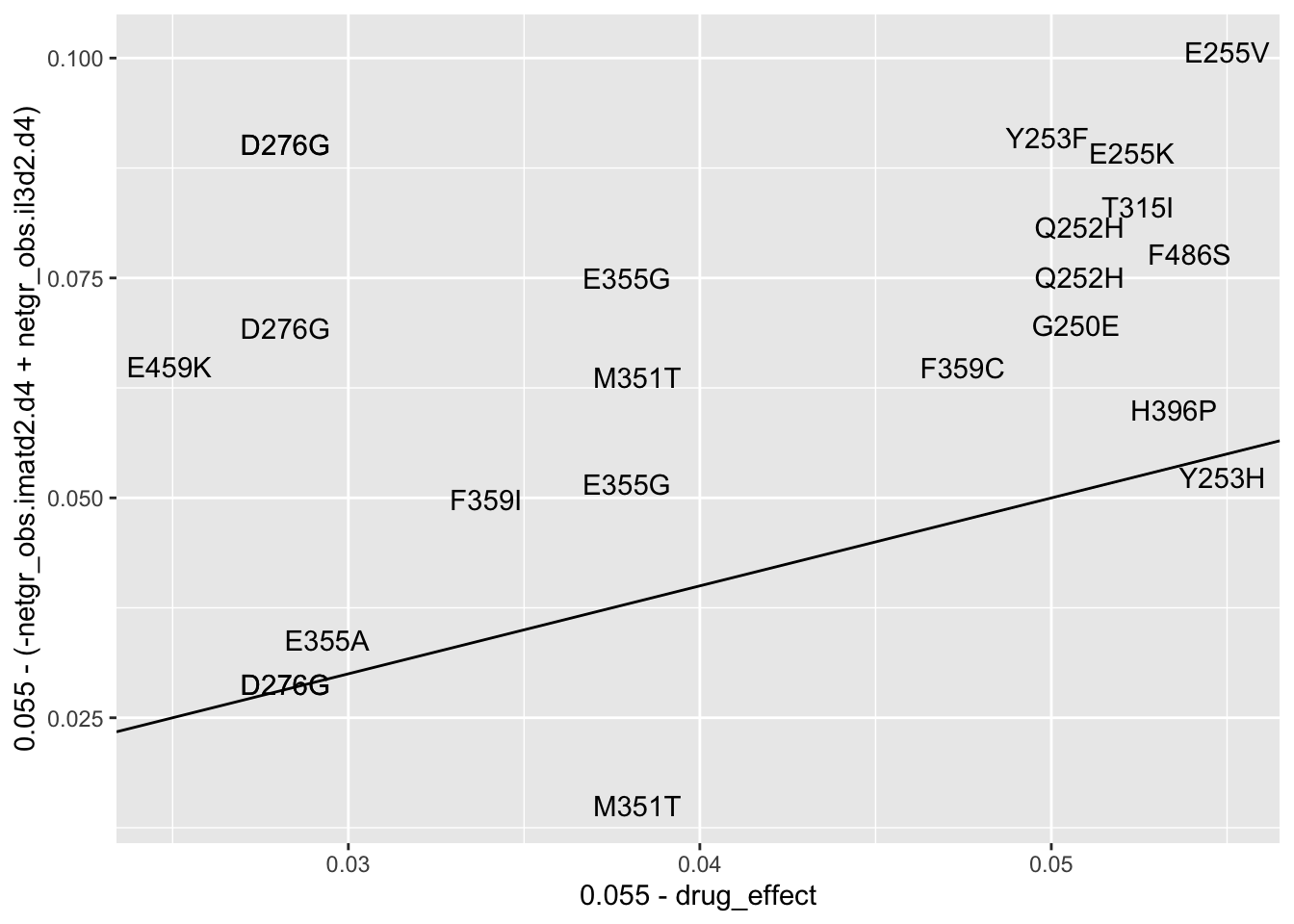
ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=.055-drug_effect,y=netgr_obs.imatd2.d4,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).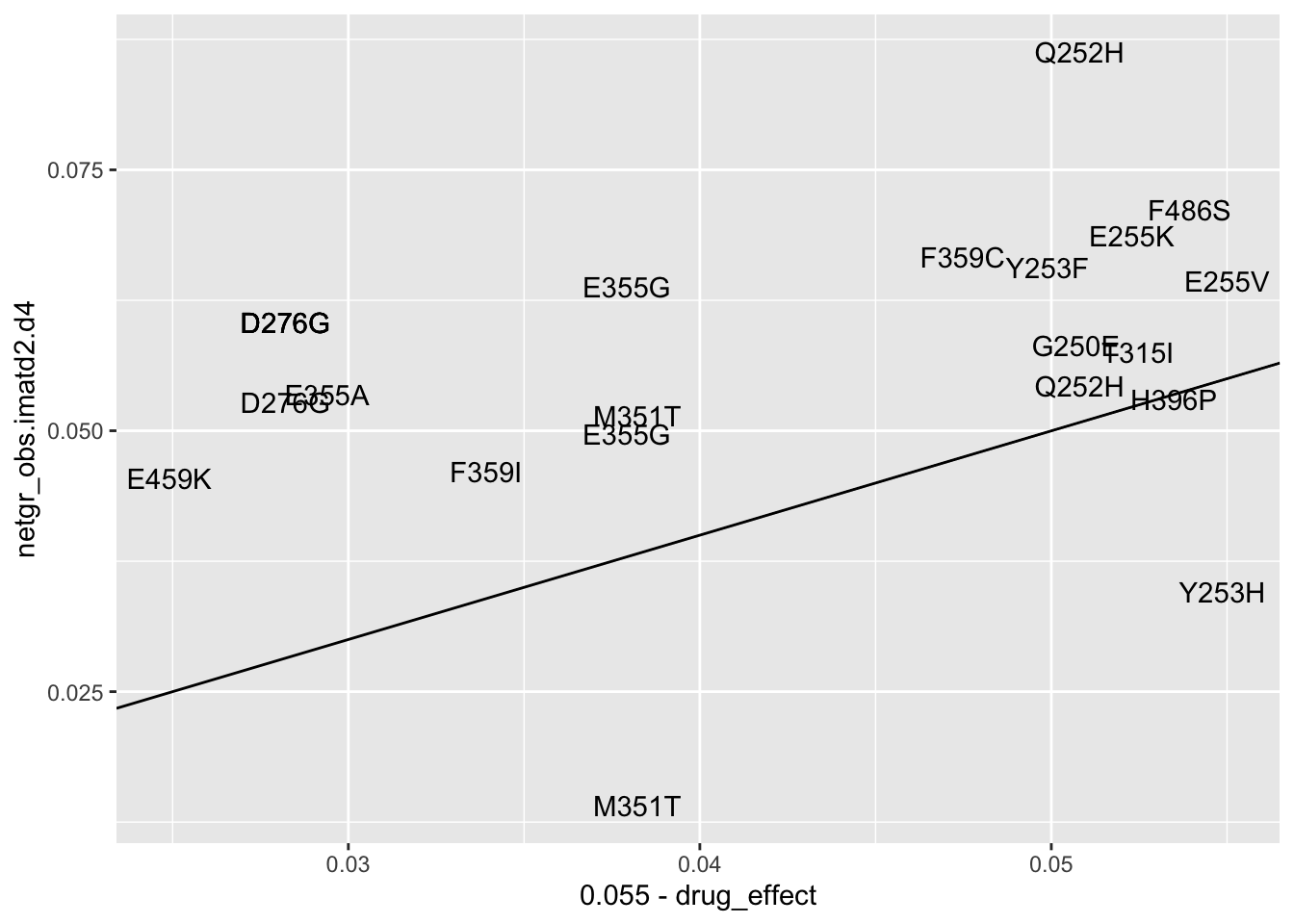
# ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=netgr_obs.il3d0.d2-drug_effect,y=netgr_obs.imatd0.d2,label=species))+geom_text()+geom_abline()
#Looking at D0 D4
ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=.055-drug_effect,y=.055-(-netgr_obs.imatd0.d4+netgr_obs.il3d0.d4),label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).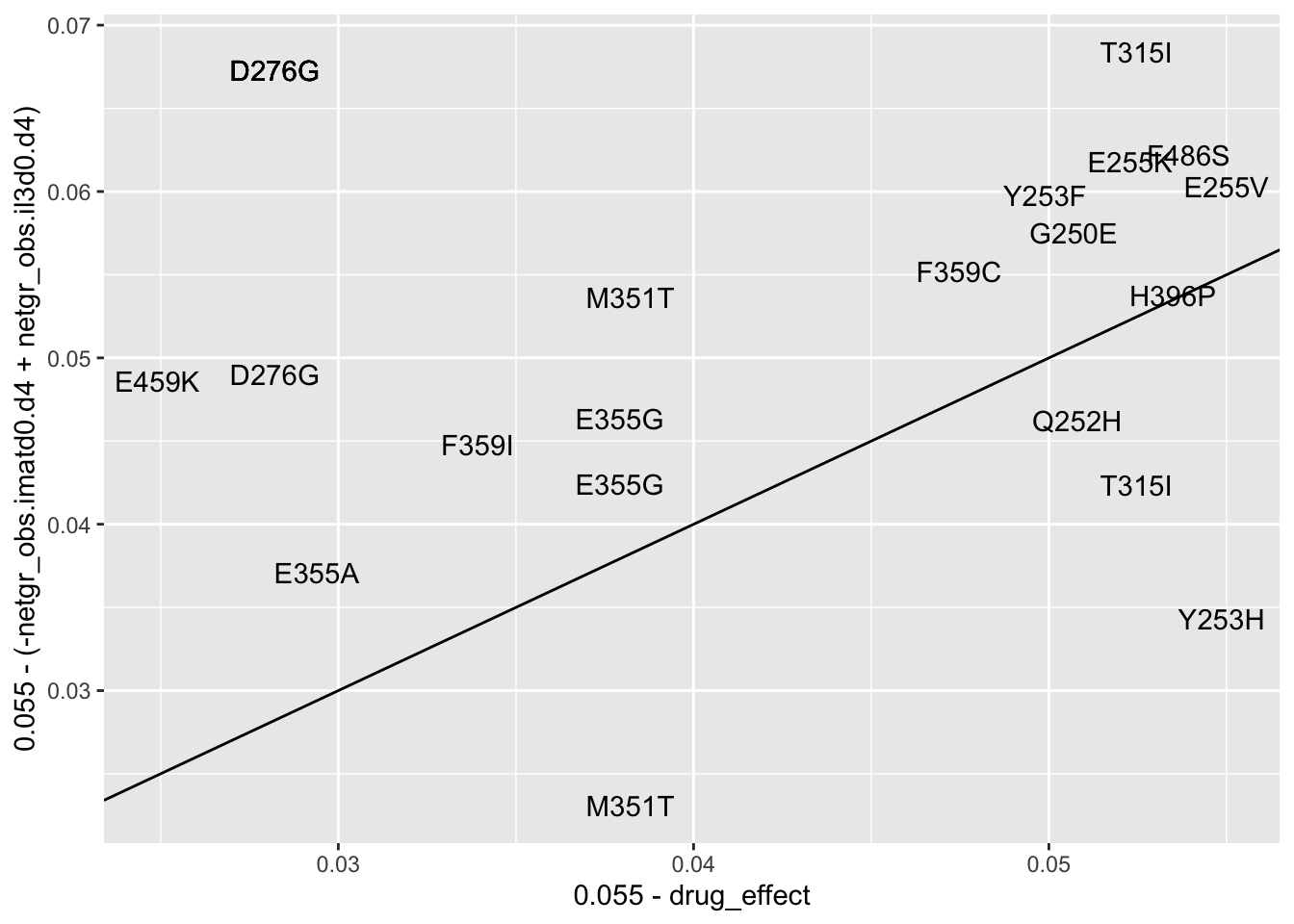
ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=.055-drug_effect,y=netgr_obs.imatd0.d4,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).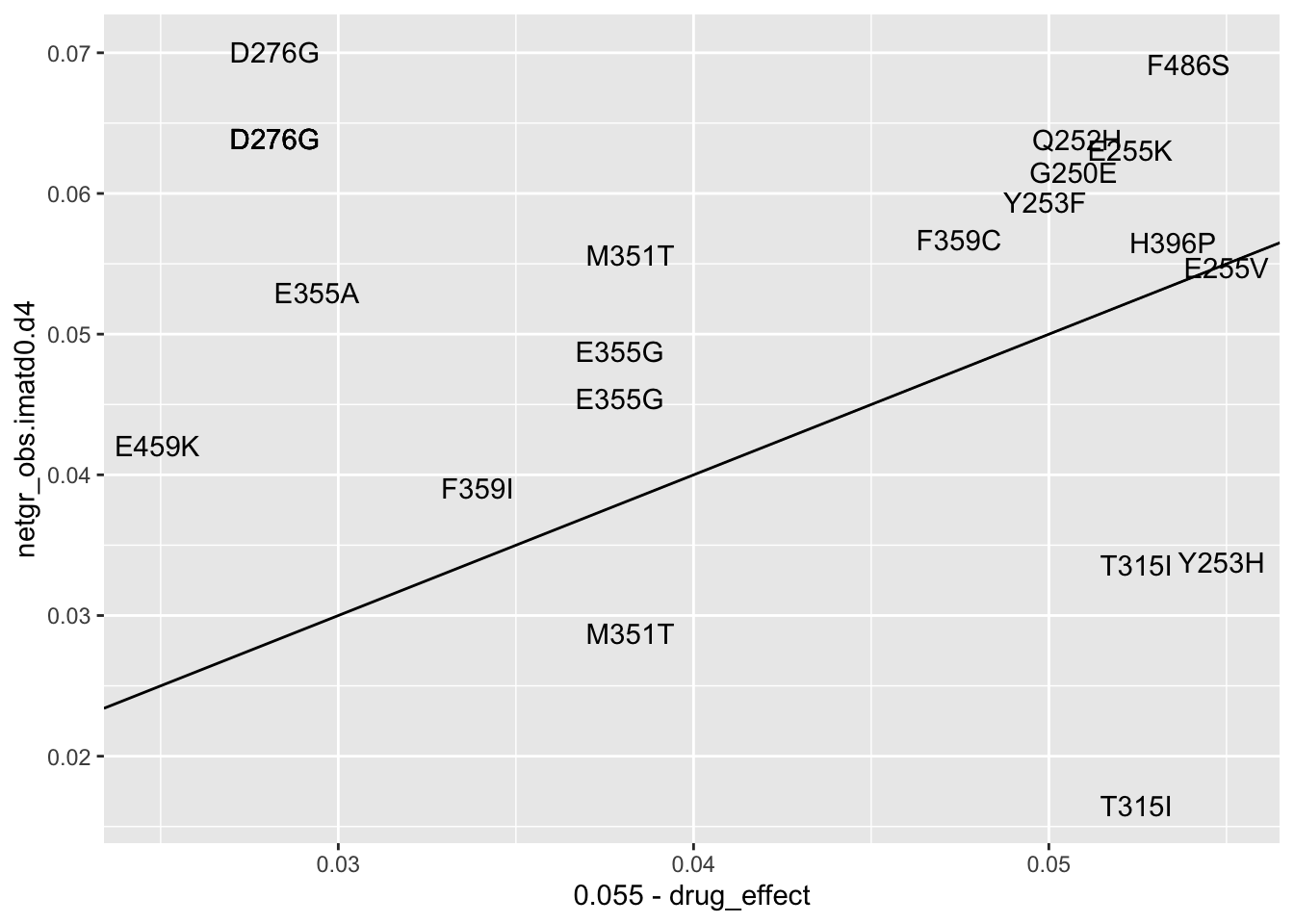
# resmuts_merged[netgr_obs.imat]=resmuts_merged%>%mutate(drugfit=)
ggplot(resmuts_merged%>%filter(!protein_start%in%317,!species%in%"V299L"),aes(x=netgr_obs.il3d0.d4-drug_effect,y=netgr_obs.imatd0.d4,label=species))+geom_text()+geom_abline()Warning: Removed 2 rows containing missing values (geom_text).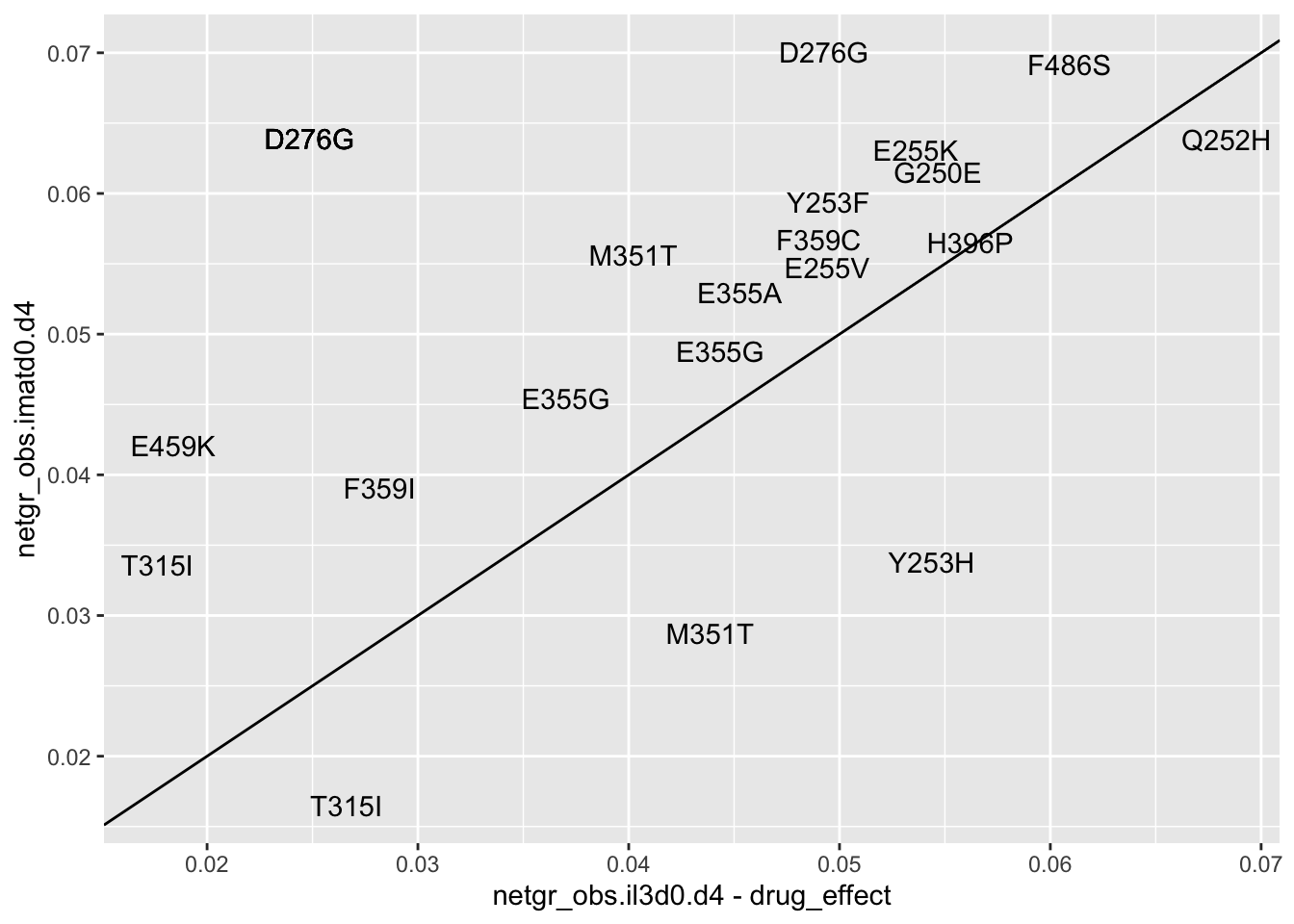
# ggplot(resmuts_merged%>%filter(!protein_start%in%317),aes(x=.025-drug_effect,y=netgr_obs.imatd2.d4,label=species))+geom_text()+geom_abline()
ggplot(resmuts,aes(x=species,y=netgr_obs.il3d0.d2))+geom_col()Warning: Removed 3 rows containing missing values (position_stack).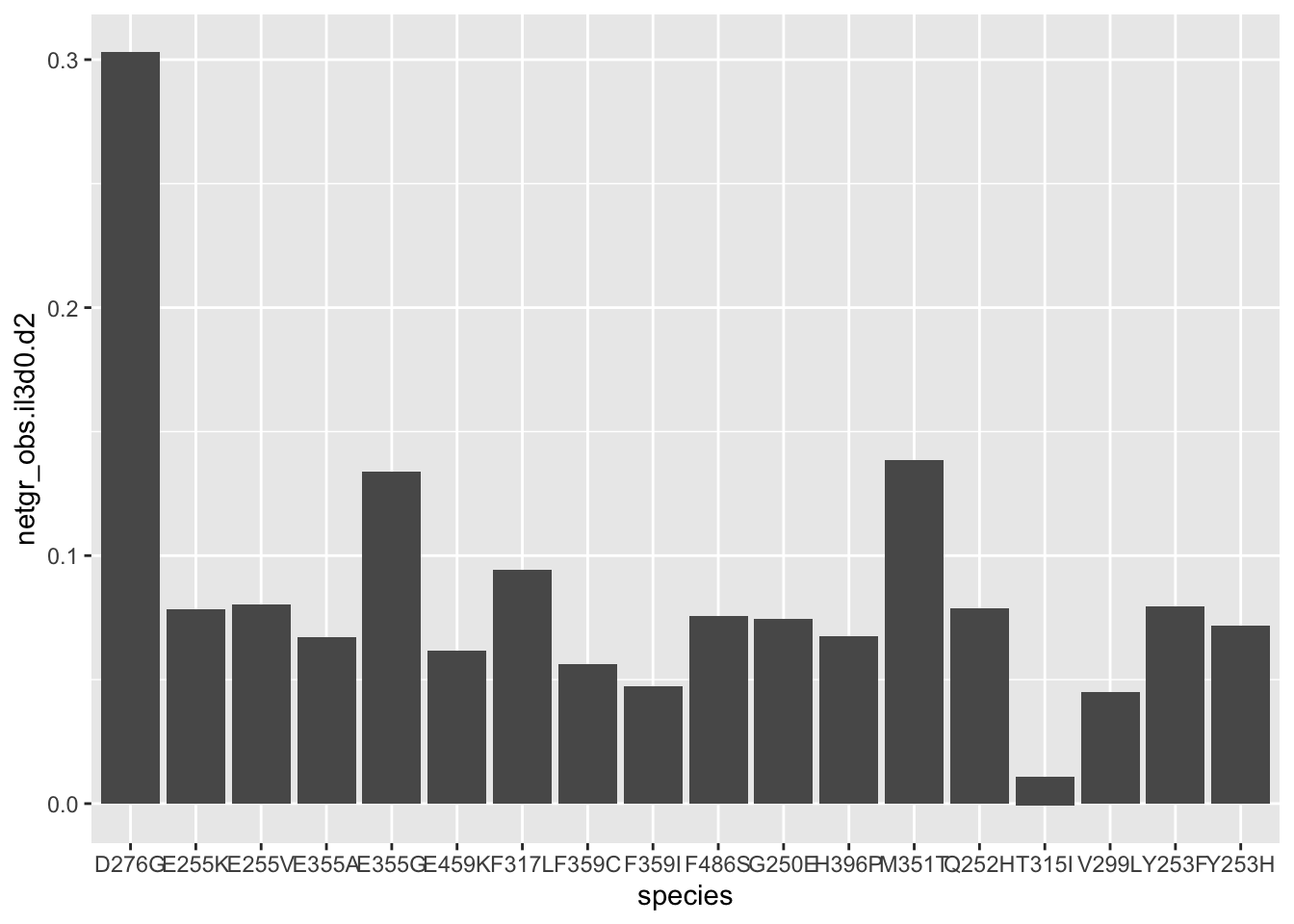
ggplot(resmuts,aes(x=species,y=netgr_obs.il3d2.d4))+geom_col()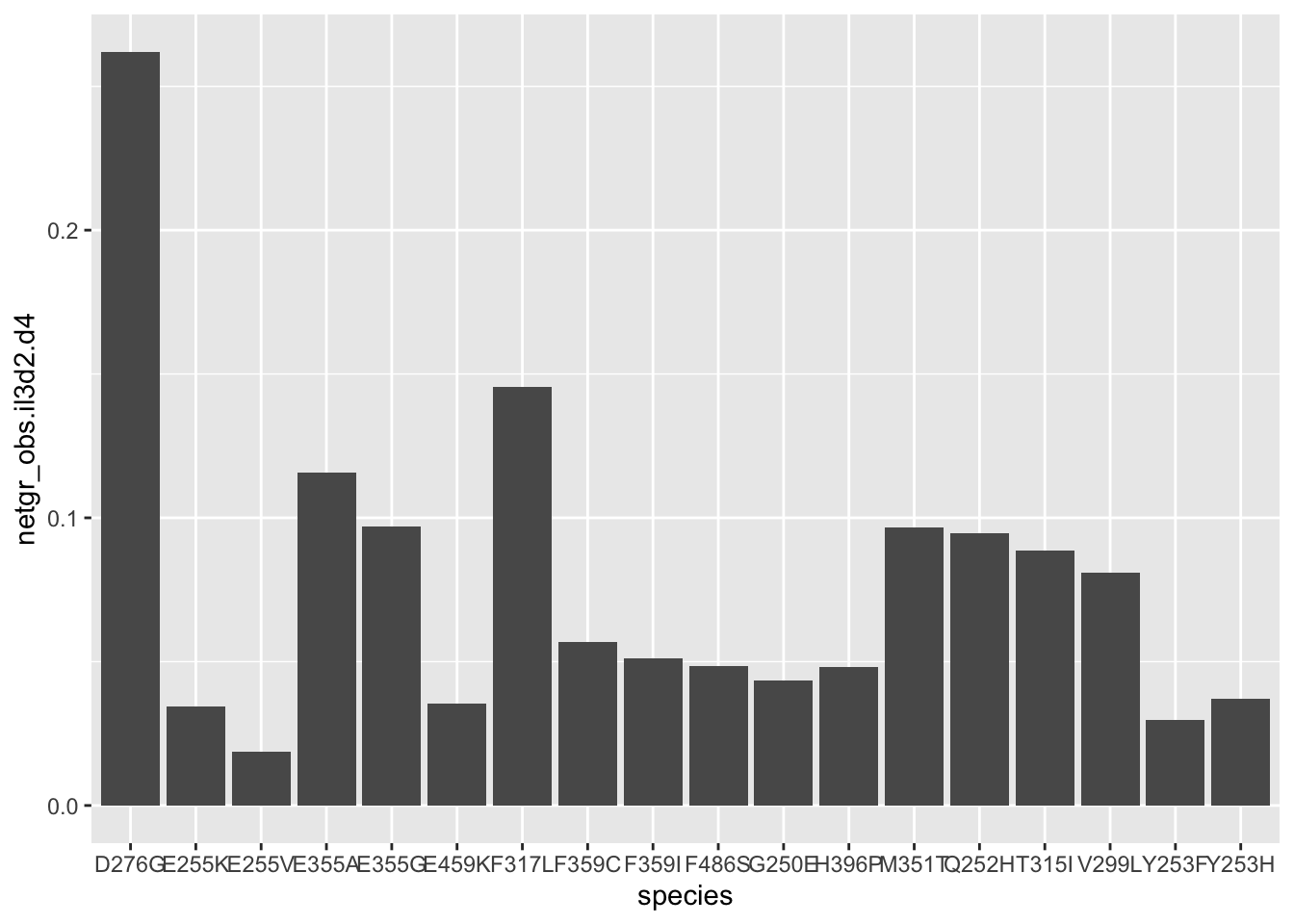
ggplot(resmuts,aes(x=species,y=netgr_obs.imatd0.d2))+geom_col()Warning: Removed 3 rows containing missing values (position_stack).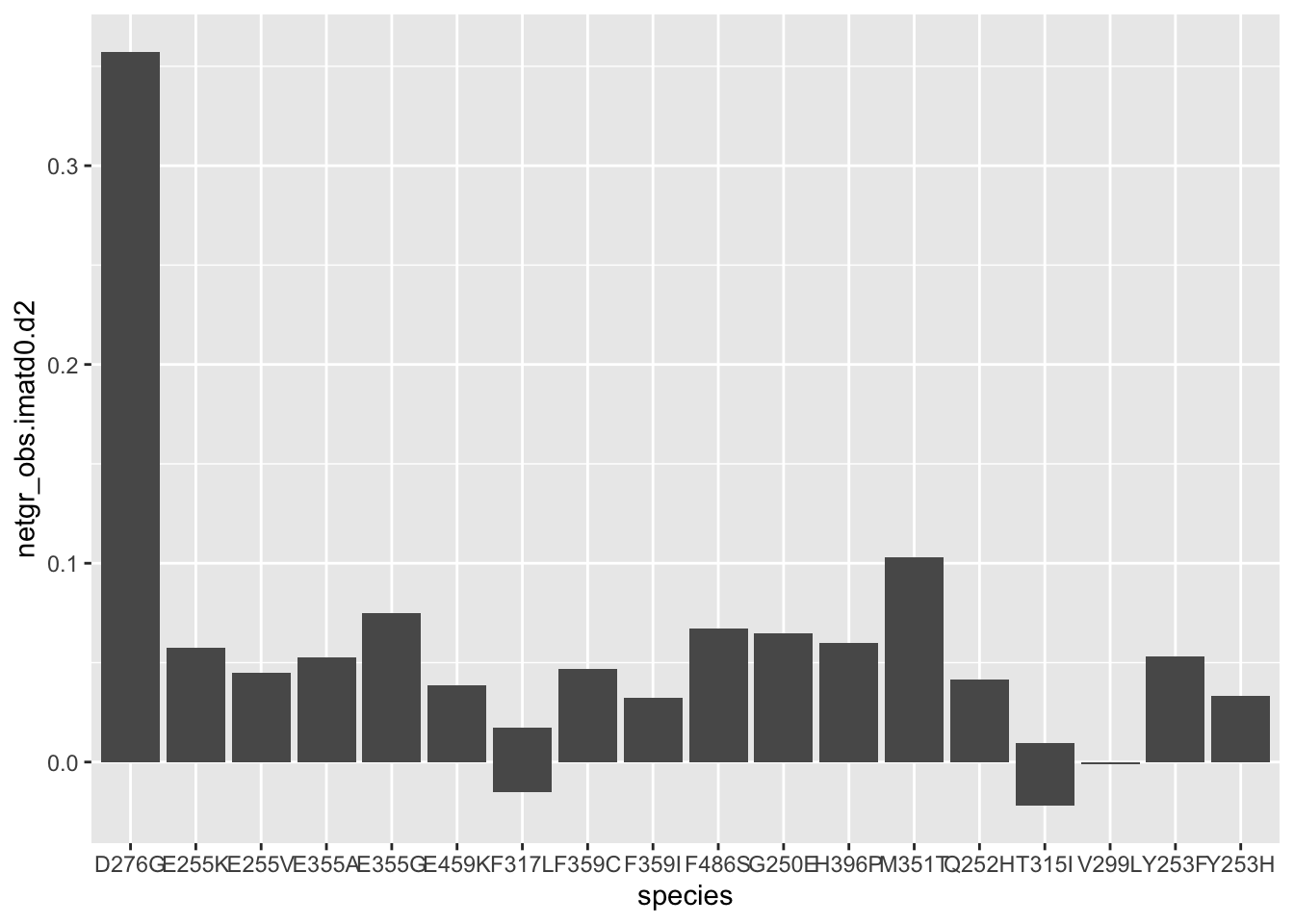
ggplot(resmuts%>%filter(!(protein_start%in%252&alt%in%"T")),aes(x=species,y=netgr_obs.imatd2.d4))+geom_col()Warning: Removed 6 rows containing missing values (position_stack).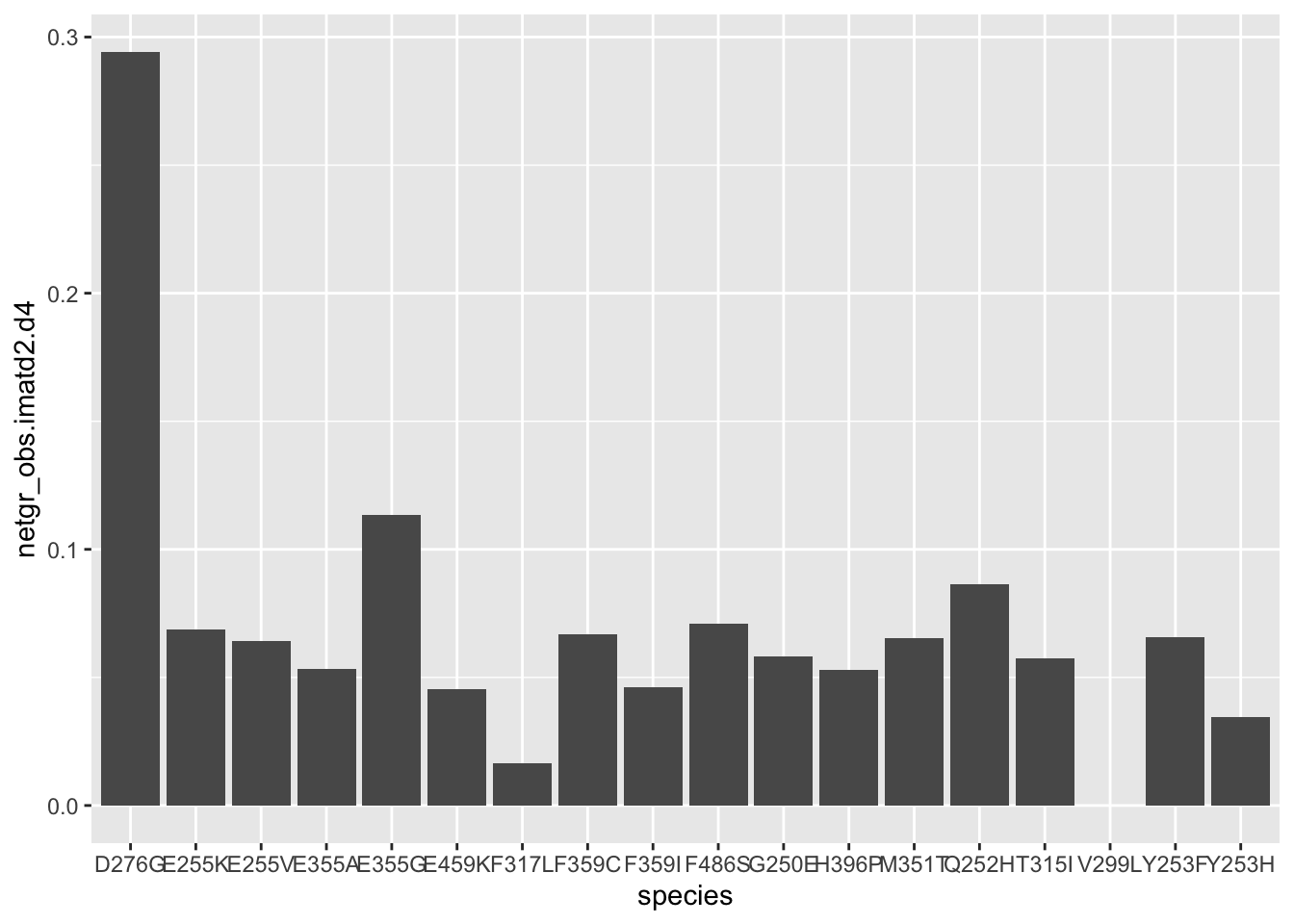
ggplot(resmuts,aes(x=species,y=score.il3d0.d2))+geom_col()Warning: Removed 3 rows containing missing values (position_stack).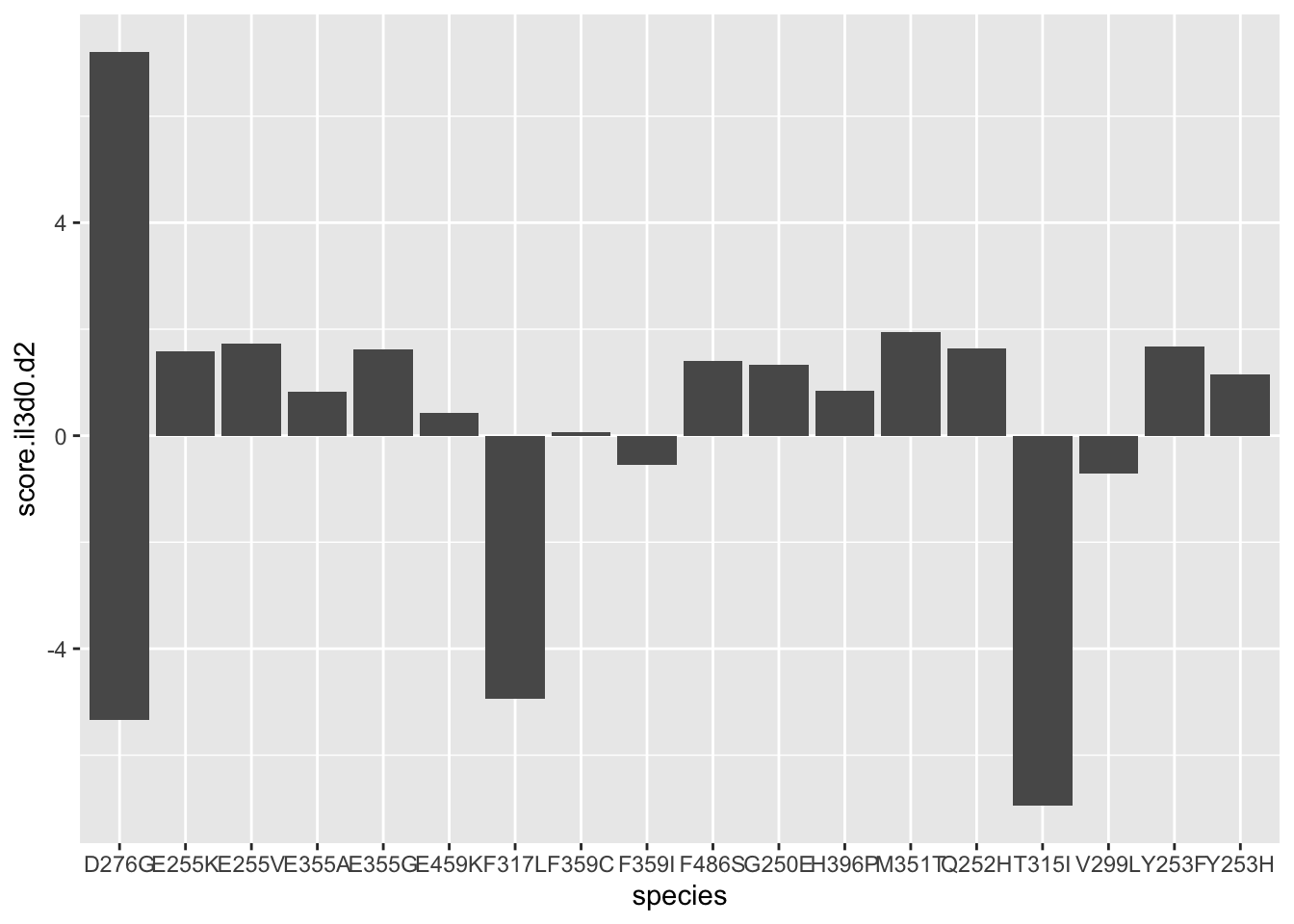
ggplot(resmuts,aes(x=species,y=score.il3d2.d4))+geom_col()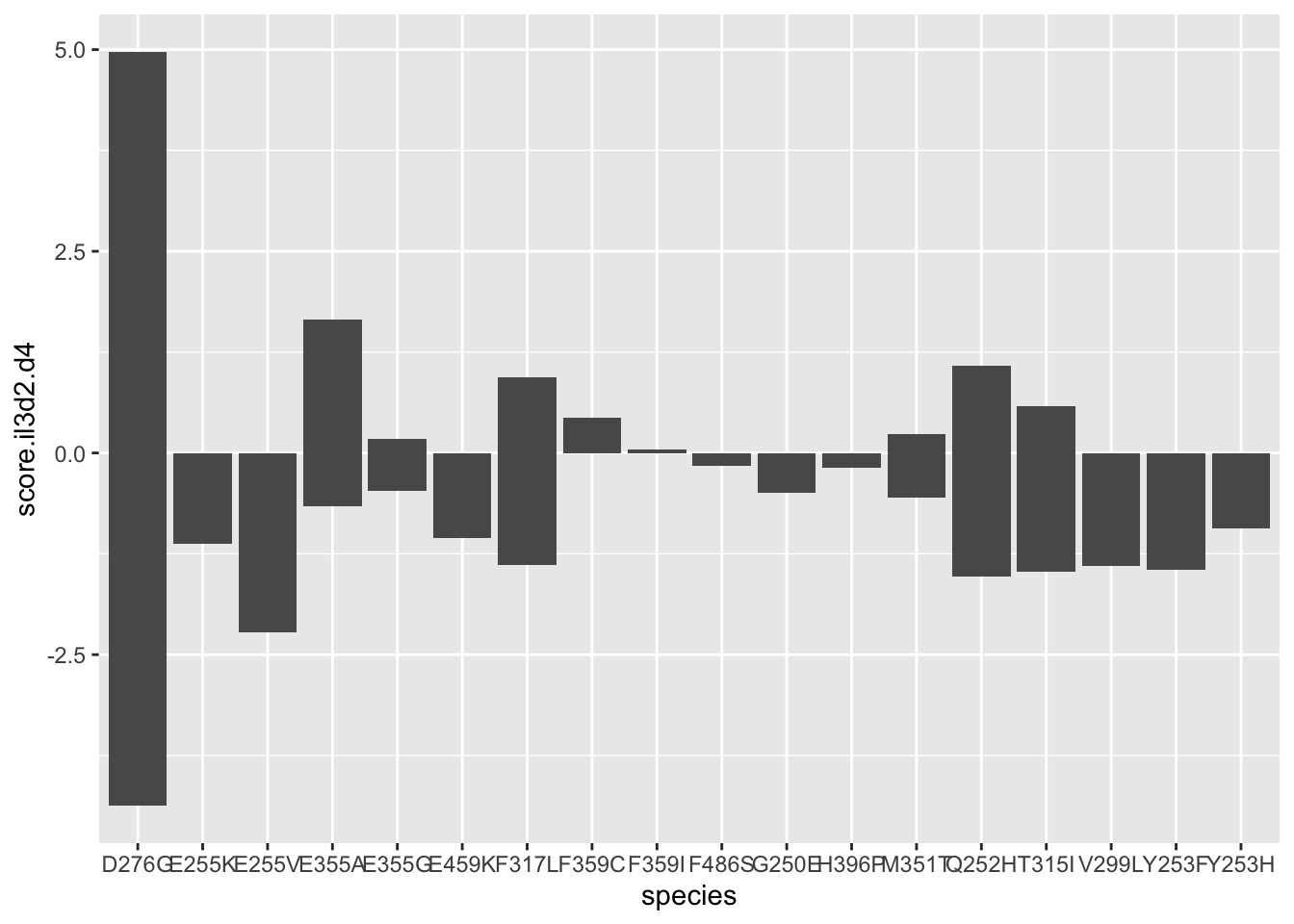
ggplot(resmuts,aes(x=species,y=score.imatd0.d2))+geom_col()Warning: Removed 3 rows containing missing values (position_stack).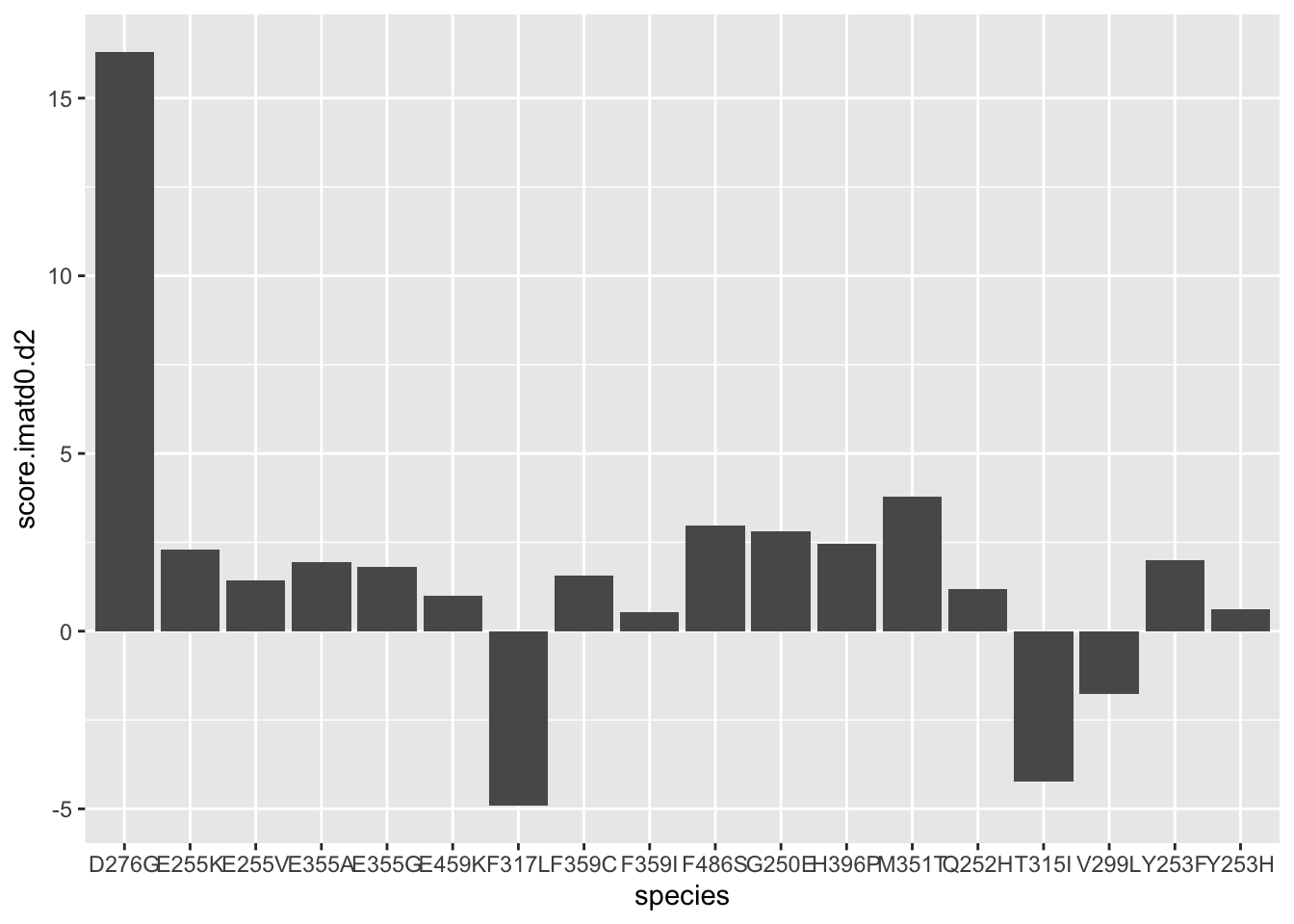
ggplot(resmuts%>%filter(!protein_start%in%317),aes(x=species,y=score.imatd2.d4))+geom_col()Warning: Removed 4 rows containing missing values (position_stack).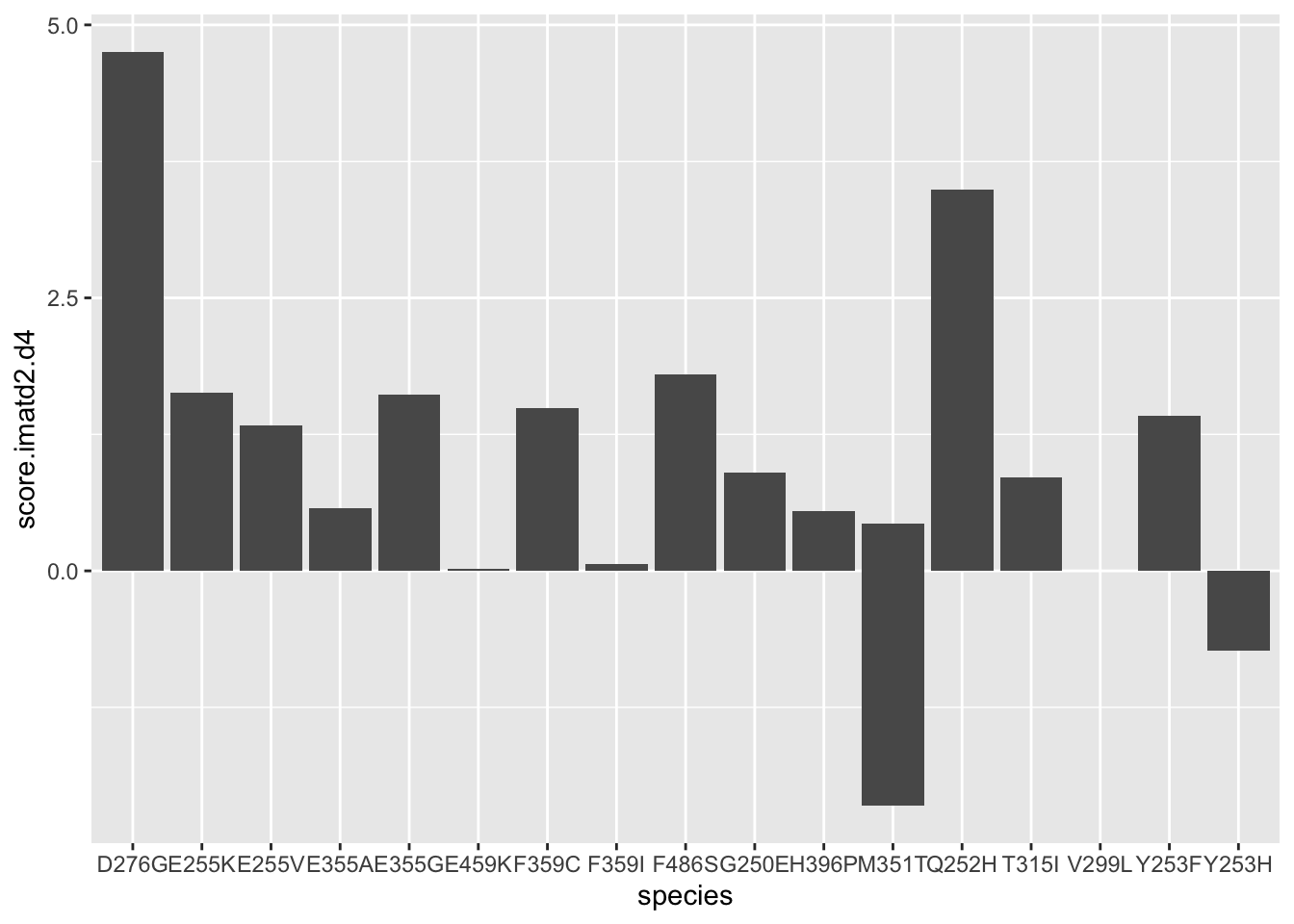
a=resmuts_merged%>%filter(!netgr_obs.imatd0.d4%in%c(NA,"-Inf"),!netgr_obs.il3d0.d4%in%c(NA,"-Inf"))
a=a%>%mutate(netgr_pred_mean=.055-drug_effect,netgr_obs=.055-(netgr_obs.il3d0.d4-netgr_obs.imatd0.d4))
a=a%>%mutate(netgr_pred_mean=.055-drug_effect,netgr_obs=netgr_obs.imatd0.d4)
a=a%>%mutate(netgr_pred_mean=netgr_obs.il3d0.d4-drug_effect,netgr_obs=netgr_obs.imatd0.d4)
cor(a$netgr_pred_mean,a$netgr_obs)[1] 0.5207143How many unique mutants are there per residue in the IL3D0 library
calls_sum=il3D0%>%filter(consequence_terms%in%"missense_variant")%>%group_by(protein_start)%>%summarize(unique_mutants=n(),count=sum(ct))
calls_sum=calls_sum%>%mutate(region=case_when(protein_start<250&protein_start>=242~1,
protein_start<258&protein_start>=250~2,
protein_start<266&protein_start>=258~3,
protein_start<274&protein_start>=266~4,
protein_start<282&protein_start>=274~5,
protein_start<290&protein_start>=282~6,
protein_start<298&protein_start>=290~7,
protein_start<306&protein_start>=298~8,
protein_start<314&protein_start>=306~9,
protein_start<322&protein_start>=314~10,
protein_start<330&protein_start>=322~11,
protein_start<338&protein_start>=330~12,
protein_start<346&protein_start>=338~13,
protein_start<354&protein_start>=346~14,
protein_start<362&protein_start>=354~15,
protein_start<370&protein_start>=362~16,
protein_start<378&protein_start>=370~17,
protein_start<386&protein_start>=378~18,
protein_start<394&protein_start>=386~19,
protein_start<402&protein_start>=394~20,
protein_start<410&protein_start>=402~21,
protein_start<418&protein_start>=410~22,
protein_start<426&protein_start>=418~23,
protein_start<434&protein_start>=426~24,
protein_start<442&protein_start>=434~25,
protein_start<450&protein_start>=442~26,
protein_start<458&protein_start>=450~27,
protein_start<466&protein_start>=458~28,
protein_start<474&protein_start>=466~29,
protein_start<482&protein_start>=474~30,
protein_start<490&protein_start>=482~31,
protein_start<498&protein_start>=490~32,
T~0))
getPalette = colorRampPalette(brewer.pal(33, "Set2"))Warning in brewer.pal(33, "Set2"): n too large, allowed maximum for palette Set2 is 8
Returning the palette you asked for with that many colorsplotly=ggplot(calls_sum%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=unique_mutants))+geom_col(color="black",aes(fill=factor(region)))+cleanup+scale_fill_manual(values=getPalette(33))
ggplotly(plotly)getPalette = colorRampPalette(brewer.pal(33, "Set2"))Warning in brewer.pal(33, "Set2"): n too large, allowed maximum for palette Set2 is 8
Returning the palette you asked for with that many colorsplotly=ggplot(calls_sum%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=count))+geom_col(color="black",aes(fill=factor(region)))+cleanup+scale_fill_manual(values=getPalette(33))
ggplotly(plotly)il3D0=il3D0%>%mutate(region=case_when(protein_start<250&protein_start>=242~1,
protein_start<258&protein_start>=250~2,
protein_start<266&protein_start>=258~3,
protein_start<274&protein_start>=266~4,
protein_start<282&protein_start>=274~5,
protein_start<290&protein_start>=282~6,
protein_start<298&protein_start>=290~7,
protein_start<306&protein_start>=298~8,
protein_start<314&protein_start>=306~9,
protein_start<322&protein_start>=314~10,
protein_start<330&protein_start>=322~11,
protein_start<338&protein_start>=330~12,
protein_start<346&protein_start>=338~13,
protein_start<354&protein_start>=346~14,
protein_start<362&protein_start>=354~15,
protein_start<370&protein_start>=362~16,
protein_start<378&protein_start>=370~17,
protein_start<386&protein_start>=378~18,
protein_start<394&protein_start>=386~19,
protein_start<402&protein_start>=394~20,
protein_start<410&protein_start>=402~21,
protein_start<418&protein_start>=410~22,
protein_start<426&protein_start>=418~23,
protein_start<434&protein_start>=426~24,
protein_start<442&protein_start>=434~25,
protein_start<450&protein_start>=442~26,
protein_start<458&protein_start>=450~27,
protein_start<466&protein_start>=458~28,
protein_start<474&protein_start>=466~29,
protein_start<482&protein_start>=474~30,
protein_start<490&protein_start>=482~31,
protein_start<498&protein_start>=490~32,
T~0))
getPalette = colorRampPalette(brewer.pal(33, "Set2"))Warning in brewer.pal(33, "Set2"): n too large, allowed maximum for palette Set2 is 8
Returning the palette you asked for with that many colorsplotly=ggplot(il3D0%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=ct))+geom_col(color="black",aes(fill=factor(region)))+cleanup+scale_fill_manual(values=getPalette(33))
ggplotly(plotly)#Next: plot iL3 D2 D4 scores #Correlate Il3 D2 D4 scores with il3 D0 D2 scores
Are any of the enriched mutants in the cosmic somatic database?
# rm(list=ls())
cosmic_data=read.table("data/Cosmic_ABL/ABL_Cosmic_Gene_mutations.tsv",sep="\t",header = T,stringsAsFactors = )
cosmic_data=cosmic_data%>%mutate(AA.Mutation=gsub("p.","",AA.Mutation))
cosmic_data=cosmic_data[!grepl("ins|del",cosmic_data$CDS.Mutation),]
cosmic_data=cosmic_data[grepl("Missense",cosmic_data$Type),]
cosmic_data=cosmic_data%>%filter(!Type%in%"Substitution - coding silent")
cosmic_data=cosmic_data%>%filter(Position<=500,Position>=64,Count>=2)
# write.csv(cosmic_data,"cosmic_abl.csv")
cosmic_simple=cosmic_data%>%dplyr::select(subs_name=AA.Mutation,cosmic_count=Count)
cosmic_simple=cosmic_simple%>%group_by(subs_name)%>%summarize(cosmic_count=sum(cosmic_count))
cosmic_simple$cosmic_present=Tsource("code/merge_samples.R")
sample10=read.csv(file = "data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
sample10=sample10%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
sample10=sample10%>%mutate(maf=ct/depth)
sample10_simple=sample10%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf)
sample10=il3D0
sample12=read.csv(file = "data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/variants_unique_ann.csv",header=T,stringsAsFactors = F)
sample12=sample12%>%
rowwise()%>%
mutate(ref_aa=strsplit(amino_acids,"/")[[1]][1],
alt_aa=strsplit(amino_acids,"/")[[1]][2])
sample12=sample12%>%mutate(maf=ct/depth)
sample12=imatD4
sample12_simple=sample12%>%dplyr::select(alt_start_pos,protein_start,ref,alt,ref_aa,alt_aa,consequence_terms,ct,depth,maf)
samples_10.12=merge(sample10_simple%>%filter(consequence_terms%in%"missense_variant"),sample12_simple%>%filter(consequence_terms%in%"missense_variant"),by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms"))
samples_10.12=samples_10.12%>%mutate(score=log2(maf.y/maf.x),score2=log2(ct.y/ct.x))
samples_10.12_simple=samples_10.12%>%dplyr::select(ref_aa,protein_start,alt_aa,consequence_terms,ct.x,score)
ggplot(samples_10.12,aes(x=score))+geom_density()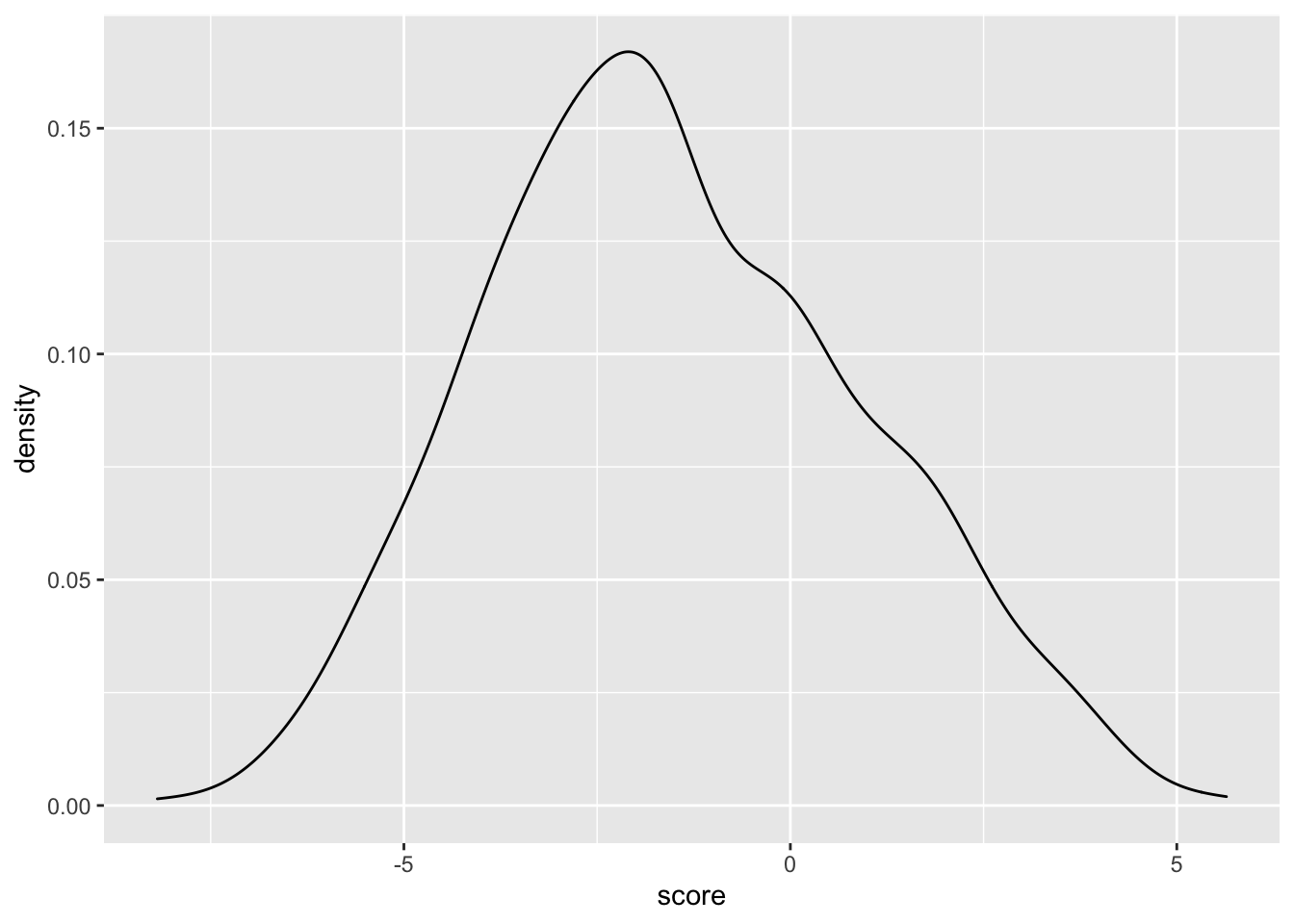
# ggplot(samples_14.16,aes(x=score))+geom_histogram(bins=100)
samples_10.12=samples_10.12%>%mutate(resmuts=case_when(protein_start%in%253&alt_aa%in%"H"~T,
protein_start%in%255&alt_aa%in%"V"~T,
protein_start%in%486&alt_aa%in%"S"~T,
protein_start%in%396&alt_aa%in%"P"~T,
protein_start%in%255&alt_aa%in%"K"~T,
protein_start%in%315&alt_aa%in%"I"~T,
protein_start%in%252&alt_aa%in%"H"~T,
protein_start%in%253&alt_aa%in%"F"~T,
protein_start%in%250&alt_aa%in%"E"~T,
protein_start%in%359&alt_aa%in%"C"~T,
protein_start%in%351&alt_aa%in%"T"~T,
protein_start%in%355&alt_aa%in%"G"~T,
protein_start%in%317&alt_aa%in%"L"~T,
protein_start%in%359&alt_aa%in%"I"~T,
protein_start%in%355&alt_aa%in%"A"~T,
protein_start%in%459&alt_aa%in%"K"~T,
protein_start%in%276&alt_aa%in%"G"~T,
protein_start%in%299&alt_aa%in%"L"~T,
T~F))
samples_10.12=samples_10.12%>%mutate(resresids=case_when(protein_start%in%253~T,
protein_start%in%255~T,
protein_start%in%486~T,
protein_start%in%396~T,
protein_start%in%255~T,
protein_start%in%315~T,
protein_start%in%252~T,
protein_start%in%253~T,
protein_start%in%250~T,
protein_start%in%359~T,
protein_start%in%351~T,
protein_start%in%355~T,
protein_start%in%317~T,
protein_start%in%359~T,
protein_start%in%355~T,
protein_start%in%459~T,
protein_start%in%276~T,
protein_start%in%299~T,
T~F))
highscore=samples_10.12%>%filter(!ct.x%in%1,protein_start>=242,protein_start<=494)
highscore=highscore%>%mutate(subs_name=paste(ref_aa,protein_start,alt_aa,sep = ""))
ggplot(highscore,aes(x=reorder(subs_name,-score),y=score,fill=resmuts))+geom_col()+theme(axis.text.x=element_text(angle=90, hjust=1))+scale_y_continuous(name="Enrithcment Score")+scale_x_discrete(name="Mutant")+guides(fill = guide_legend(title = "Clinically\n Observed \n Resmut"))+scale_fill_manual(values=c("gray90","red"))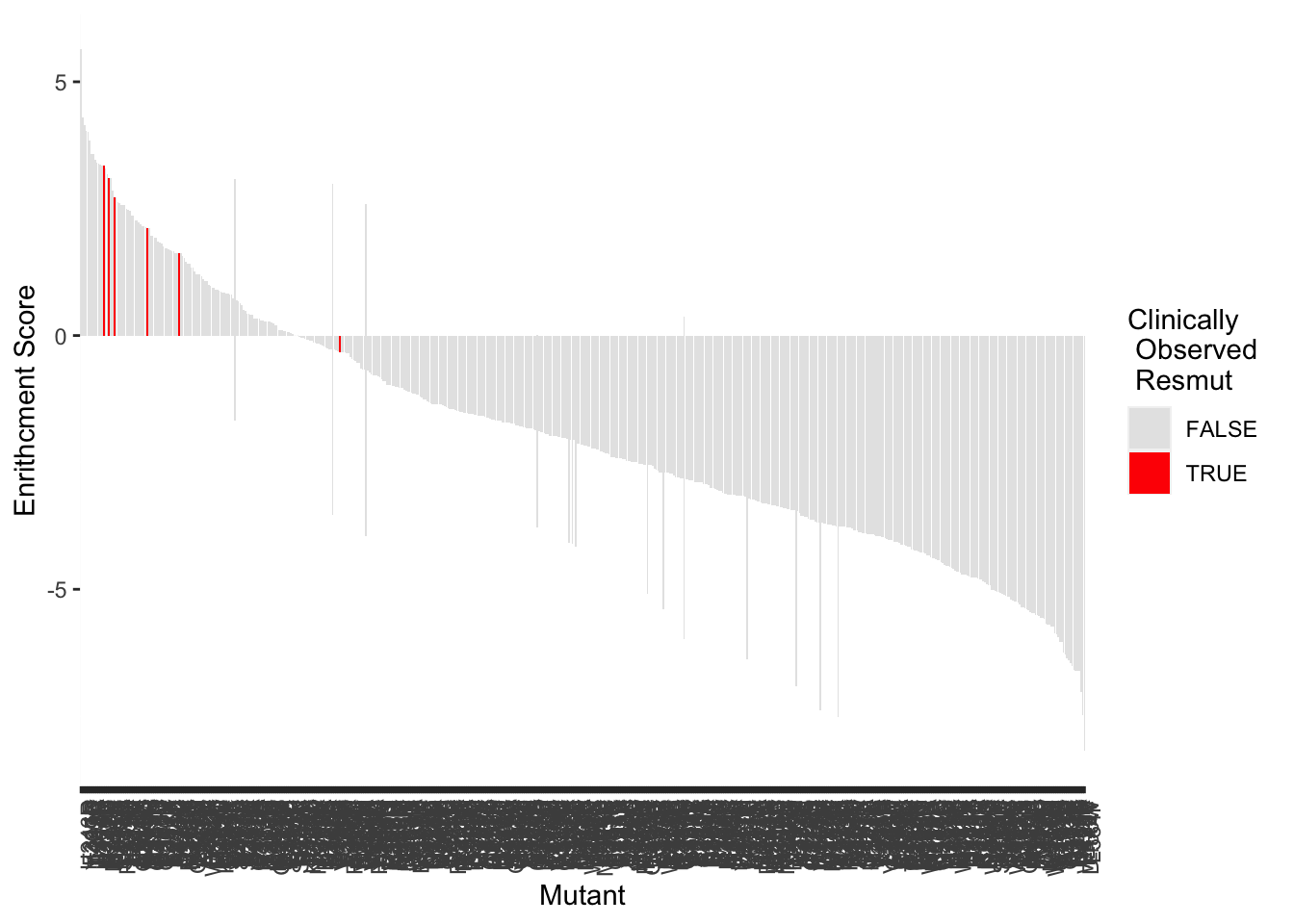
highscore2=highscore%>%group_by(alt_aa,protein_start,subs_name,resmuts,resresids)%>%summarize(score=mean(score))`summarise()` has grouped output by 'alt_aa', 'protein_start', 'subs_name', 'resmuts'. You can override using the `.groups` argument.ggplot(highscore2,aes(x=reorder(subs_name,-score),y=score,fill=resmuts))+geom_col()+theme(axis.text.x=element_blank(),axis.ticks.x=element_blank())+scale_y_continuous(name="Enrithcment Score")+scale_x_discrete(name="Mutant")+guides(fill = guide_legend(title = "Clinically\n Observed \n Resistant Mutant"))+scale_fill_manual(values=c("gray90","red"))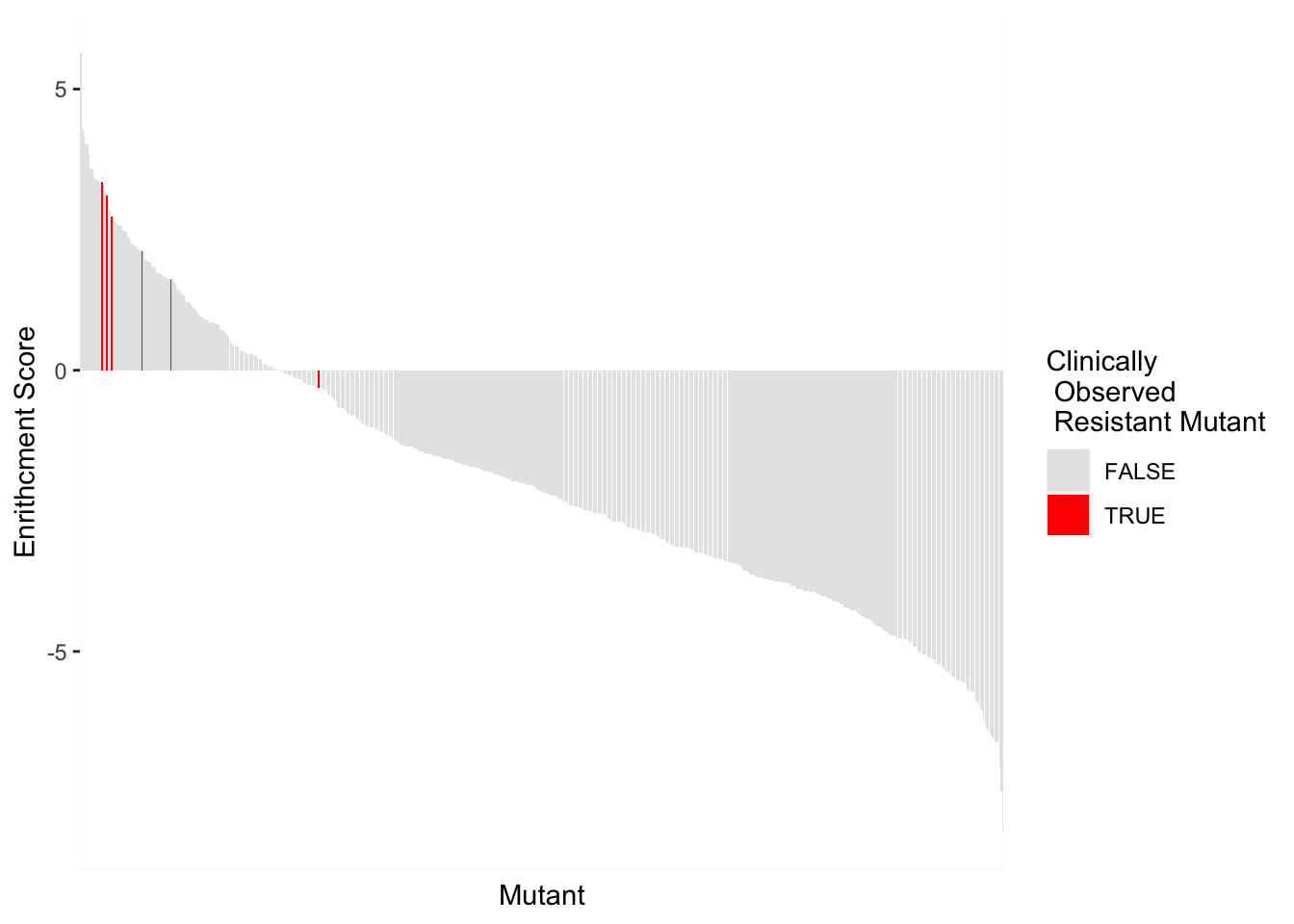
# ggsave("BCRABL_Imatinib_Scores_Resmuts.pdf",height=4,width=8,units="in",useDingbats=F)
ggplot(highscore2,aes(x=reorder(subs_name,-score),y=score,fill=resresids))+geom_col()+theme(axis.text.x=element_blank(),axis.ticks.x=element_blank())+scale_y_continuous(name="Enrithcment Score")+scale_x_discrete(name="Mutant")+guides(fill = guide_legend(title = "Clinically\n Observed \n Resistant Residue"))+scale_fill_manual(values=c("gray90","red"))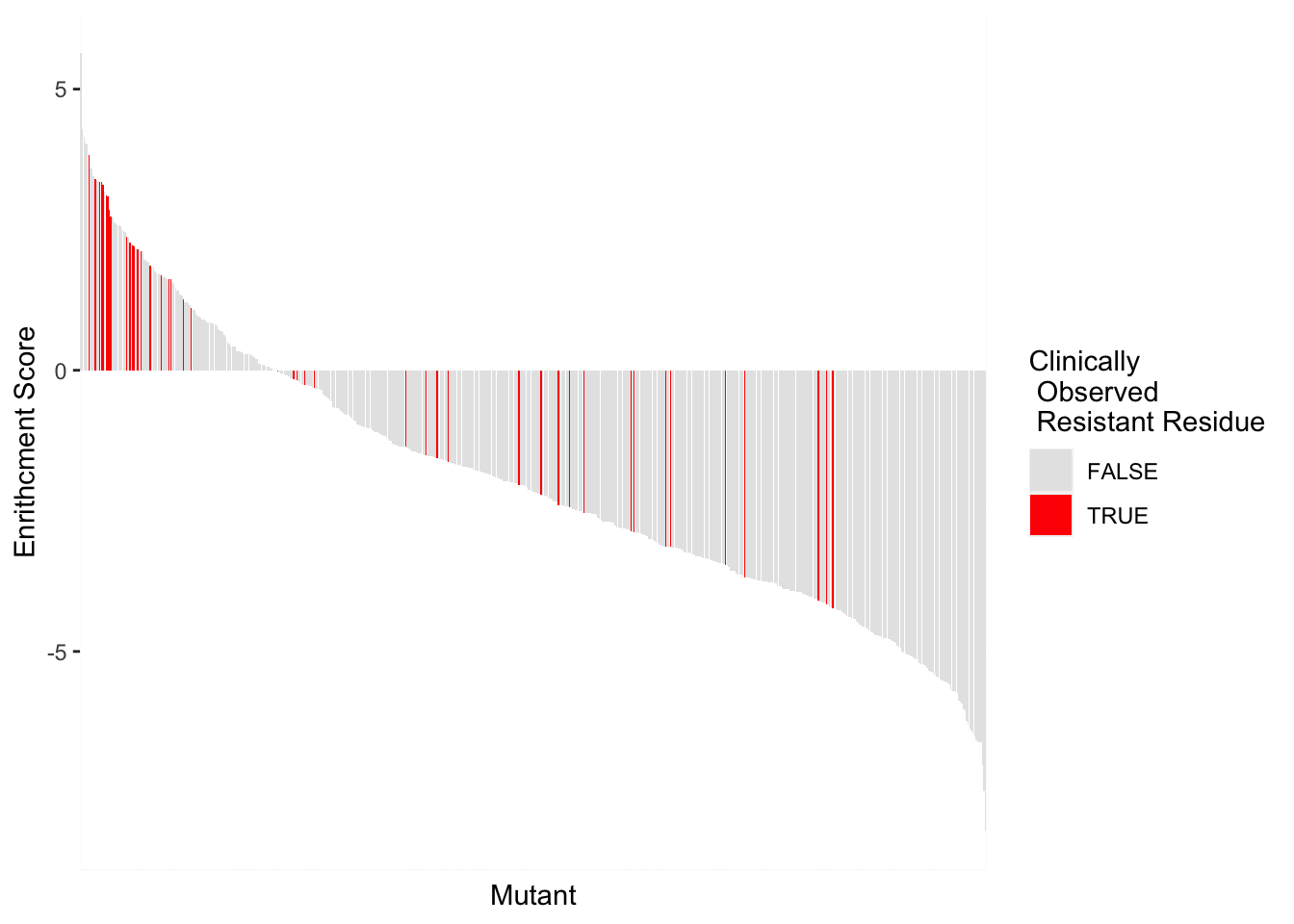
# ggsave("BCRABL_Imatinib_Scores_Resresids.pdf",height=4,width=8,units="in",useDingbats=F)
###Merging cosmic data and highscore
# highscore$cosmic_present=F
highscore_cosmic=merge(highscore,cosmic_simple,by="subs_name",all.x = T)
highscore_cosmic[highscore_cosmic$cosmic_present%in%NA,"cosmic_present"]=F
highscore_cosmic[highscore_cosmic$cosmic_count%in%NA,"cosmic_count"]=0
plotly=ggplot(highscore_cosmic,aes(x=reorder(subs_name,-score),y=score,fill=cosmic_present))+geom_col()+theme(axis.text.x=element_text(angle=90, hjust=1))+scale_y_continuous(name="Enrichment Score")+scale_x_discrete(name="Mutant")+guides(fill = guide_legend(title = "Cosmic\n Observed"))
ggplotly(plotly)a=cosmic_data%>%filter(AA.Mutation%in%"L248V")
a=highscore%>%filter(subs_name%in%"M388L")
plotly=ggplot(highscore_cosmic%>%filter(cosmic_present%in%T),aes(x=reorder(subs_name,-score),y=score,fill=cosmic_count))+geom_col()+theme_bw()
ggplotly(plotly)# a=highscore_cosmic%>%filter(cosmic_present%in%T)
# a=highscore2%>%filter(resmuts%in%T)
sessionInfo()R version 4.0.0 (2020-04-24)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] RColorBrewer_1.1-2 doParallel_1.0.15 iterators_1.0.12 foreach_1.5.0
[5] tictoc_1.0 plotly_4.9.2.1 ggplot2_3.3.3 dplyr_1.0.6
[9] stringr_1.4.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.31 bslib_0.3.1 purrr_0.3.4
[5] colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.5.2
[9] viridisLite_0.3.0 yaml_2.2.1 utf8_1.1.4 rlang_0.4.11
[13] jquerylib_0.1.4 later_1.0.0 pillar_1.6.1 glue_1.4.1
[17] withr_2.4.2 DBI_1.1.0 lifecycle_1.0.0 munsell_0.5.0
[21] gtable_0.3.0 workflowr_1.6.2 htmlwidgets_1.5.1 codetools_0.2-16
[25] evaluate_0.14 labeling_0.3 knitr_1.28 fastmap_1.1.0
[29] crosstalk_1.1.0.1 httpuv_1.5.2 fansi_0.4.1 Rcpp_1.0.4.6
[33] promises_1.1.0 backports_1.1.7 scales_1.1.1 jsonlite_1.7.2
[37] farver_2.0.3 fs_1.4.1 digest_0.6.25 stringi_1.7.5
[41] rprojroot_1.3-2 grid_4.0.0 tools_4.0.0 magrittr_2.0.1
[45] sass_0.4.1 lazyeval_0.2.2 tibble_3.1.2 crayon_1.4.1
[49] whisker_0.4 tidyr_1.1.3 pkgconfig_2.0.3 ellipsis_0.3.2
[53] data.table_1.12.8 assertthat_0.2.1 rmarkdown_2.14 httr_1.4.2
[57] R6_2.4.1 git2r_0.27.1 compiler_4.0.0