Read raw data
Jens Daniel Müller
11 February, 2021
Last updated: 2021-02-11
Checks: 7 0
Knit directory: BloomSail/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191021) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 40f9a07. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Ignored: output/Plots/Figures_publication/.tmp.drivedownload/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/read-in.Rmd) and HTML (docs/read-in.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 40f9a07 | jens-daniel-mueller | 2021-02-11 | cleaning |
| html | 8051798 | jens-daniel-mueller | 2021-02-10 | Build site. |
| Rmd | 9477a18 | jens-daniel-mueller | 2021-02-10 | rerun all with empty folders |
| html | ed2a1b2 | jens-daniel-mueller | 2021-02-08 | Build site. |
| Rmd | 833d0f6 | jens-daniel-mueller | 2021-02-08 | code and output cleaning |
| html | 7027b12 | jens-daniel-mueller | 2021-02-08 | Build site. |
| Rmd | 3d6eaa1 | jens-daniel-mueller | 2021-02-08 | code and output cleaning |
| html | c5fc34c | jens-daniel-mueller | 2021-01-22 | Build site. |
| html | 4277235 | jens-daniel-mueller | 2021-01-05 | Build site. |
| html | 9a3f42a | jens-daniel-mueller | 2020-10-24 | Build site. |
| html | 05248bf | jens-daniel-mueller | 2020-10-20 | Build site. |
| html | 1c4fe8e | jens-daniel-mueller | 2020-10-20 | table with time series in depth intervals added |
| html | a43fcac | jens-daniel-mueller | 2020-10-13 | Build site. |
| Rmd | a23fb5d | jens-daniel-mueller | 2020-10-13 | removed unnecessary output prints |
| html | 6896725 | jens-daniel-mueller | 2020-10-01 | Build site. |
| Rmd | f2f9226 | jens-daniel-mueller | 2020-10-01 | tower data converted to UTC |
| html | 9f66019 | jens-daniel-mueller | 2020-10-01 | Build site. |
| html | 27c5431 | jens-daniel-mueller | 2020-09-29 | Build site. |
| Rmd | 2e0f902 | jens-daniel-mueller | 2020-09-29 | all parameters separate, rebuild |
| html | 1d01685 | jens-daniel-mueller | 2020-09-28 | Build site. |
| Rmd | d28129f | jens-daniel-mueller | 2020-09-28 | republish after tau factor set to 1 and using final pCO2 data |
| html | 02a1609 | jens-daniel-mueller | 2020-09-25 | Build site. |
| Rmd | 99e69cf | jens-daniel-mueller | 2020-09-25 | activated read-in of th and ts data |
| Rmd | 616c27f | jens-daniel-mueller | 2020-09-25 | updated repo manually |
| html | 904f0f7 | jens-daniel-mueller | 2020-09-23 | Build site. |
| html | 66bf52a | jens-daniel-mueller | 2020-09-23 | Build site. |
| Rmd | 0c8eed6 | jens-daniel-mueller | 2020-09-23 | included postprocessed cleaned data |
| html | c919fb7 | jens-daniel-mueller | 2020-06-29 | Build site. |
| Rmd | 1461cb6 | jens-daniel-mueller | 2020-06-29 | Fig update for talk |
| html | 603af23 | jens-daniel-mueller | 2020-05-25 | Build site. |
| html | 3414c23 | jens-daniel-mueller | 2020-05-25 | Build site. |
| html | 9ccd9a3 | jens-daniel-mueller | 2020-05-25 | Build site. |
| Rmd | 9bedac5 | jens-daniel-mueller | 2020-05-25 | revised pp time series plot |
| html | dd3bd89 | jens-daniel-mueller | 2020-05-07 | Build site. |
| Rmd | ad98da2 | jens-daniel-mueller | 2020-05-07 | harmonized parameter labeling |
| html | 3832733 | jens-daniel-mueller | 2020-04-30 | Build site. |
| Rmd | 4f4ab08 | jens-daniel-mueller | 2020-04-30 | harmonized code until RT determination |
| html | 1b6480f | jens-daniel-mueller | 2020-04-30 | Build site. |
| Rmd | fe72316 | jens-daniel-mueller | 2020-04-30 | revised variable and object names, used temp-dependent tau only, rerun code |
| html | d9248a6 | jens-daniel-mueller | 2020-04-29 | Build site. |
| Rmd | 70bd3f0 | jens-daniel-mueller | 2020-04-29 | correct interpolation, new d pco2 plot |
| html | aa52c73 | jens-daniel-mueller | 2020-04-28 | Build site. |
| Rmd | 3044ec0 | jens-daniel-mueller | 2020-04-28 | completely revised |
| html | 57f7231 | jens-daniel-mueller | 2020-04-28 | Build site. |
| Rmd | 5ebd364 | jens-daniel-mueller | 2020-04-28 | revised |
| html | b5722a7 | jens-daniel-mueller | 2020-04-28 | Build site. |
| html | 472c2b4 | jens-daniel-mueller | 2020-04-21 | Build site. |
| html | f8fcf50 | jens-daniel-mueller | 2020-04-19 | created pub figures for time series |
| html | 87658c3 | jens-daniel-mueller | 2020-04-14 | Build site. |
| Rmd | 5c96a65 | jens-daniel-mueller | 2020-04-14 | temperature penetration depth |
| html | 624835e | jens-daniel-mueller | 2020-04-02 | Build site. |
| Rmd | a7ac65d | jens-daniel-mueller | 2020-04-02 | BloomSail data 1-5m and sd in time series plots |
| html | 26cf733 | jens-daniel-mueller | 2020-04-02 | Build site. |
| Rmd | 57b77af | jens-daniel-mueller | 2020-04-02 | corrected Finnmaid lat borders and plotted fm track in map |
| html | a6c4c22 | jens-daniel-mueller | 2020-03-30 | Build site. |
| html | 80c78b3 | jens-daniel-mueller | 2020-03-30 | Build site. |
| html | 5f8ca30 | jens-daniel-mueller | 2020-03-20 | Build site. |
| Rmd | 1ebd01a | jens-daniel-mueller | 2020-03-20 | reorganitzed filenames and navbar |
library(tidyverse)
library(data.table)
library(lubridate)
library(DataExplorer)
library(leaflet)
library(readxl)
library(gsubfn)1 Scope of this script
For each data source:
- read raw data files
- harmonize column names
- clean obvious erroneous data
- merged into one file
- write merged file
2 Sea-Bird SBE 16 sensor data (ts)
CTD sensor data including recordings from the analog output of auxiliary pH, O2, Chla and pCO2 sensors were recorded with a measurement frequency of 15 sec. (In addition, pCO2 data were also internally recorded on the Contros HydroC instrument with higher temporal resolution and will later be used for further analysis after merging with CTD data.)
2.1 Read regular profiles and transects
files <-
list.files(path = "data/input/TinaV/Sensor/Profiles_Transects/", pattern = "[.]cnv$")
for (file in files) {
start_date <-
data.table(read.delim(
here::here("data/input/TinaV/Sensor/Profiles_Transects/", file),
sep = "#",
nrows = 160
))[[78, 1]]
start_date <- substr(start_date, 15, 34)
start_date <- mdy_hms(start_date, tz = "UTC")
temp <-
read.delim(
here::here("data/input/TinaV/Sensor/Profiles_Transects/", file),
sep = "",
skip = 160,
header = FALSE
)
temp <- data.table(temp[, c(2, 3, 4, 5, 6, 7, 9, 11, 13)])
names(temp) <-
c("date_time",
"dep",
"tem",
"sal",
"V_pH",
"pH",
"Chl",
"O2",
"pCO2_analog")
temp$start_date <- start_date
temp$date_time <- temp$date_time + temp$start_date
temp$ID <- substr(file, 1, 6)
temp$type <- substr(file, 8, 8)
temp$station <- substr(file, 8, 10)
temp$cast <- "up"
temp[date_time < mean(temp[dep == max(temp$dep)]$date_time)]$cast <-
"down"
if (exists("dataset")) {
dataset <- rbind(dataset, temp)
}
if (!exists("dataset")) {
dataset <- temp
}
rm(start_date)
rm(temp)
}
ts <- dataset
rm(dataset, file, files)2.2 Read profiles and transects around Östergarnsholm
files <-
list.files(path = "data/input/TinaV/Sensor/Ostergarnsholm/", pattern = "[.]cnv$")
for (file in files) {
start_date <-
data.table(read.delim(
here::here("data/input/TinaV/Sensor/Ostergarnsholm/", file),
sep = "#",
nrows = 160
))[[78, 1]]
start_date <- substr(start_date, 15, 34)
start_date <- mdy_hms(start_date, tz = "UTC")
temp <-
read.delim(
here::here("data/input/TinaV/Sensor/Ostergarnsholm/", file),
sep = "",
skip = 160,
header = FALSE
)
temp <- data.table(temp[, c(2, 3, 4, 5, 6, 7, 9, 11, 13)])
names(temp) <-
c("date_time",
"dep",
"tem",
"sal",
"V_pH",
"pH",
"Chl",
"O2",
"pCO2_analog")
temp$start_date <- start_date
temp$date_time <- temp$date_time + temp$start_date
temp$ID <- substr(file, 1, 6)
temp$type <- substr(file, 8, 8)
temp$station <- substr(file, 11, 12)
temp$cast <- "up"
temp[date_time < mean(temp[dep == max(temp$dep)]$date_time)]$cast <-
"down"
if (exists("dataset")) {
dataset <- rbind(dataset, temp)
}
if (!exists("dataset")) {
dataset <- temp
}
rm(start_date)
rm(temp)
}
ts_OGB <- dataset
rm(dataset, file, files)
ts_OGB <- ts_OGB %>%
mutate(
type = if_else(station == "bo", "P", "T"),
station = if_else(station == "bo", "P14", station),
station = if_else(station == "in", "T14", station),
station = if_else(station == "ou", "T15", station)
)ts <- bind_rows(ts, ts_OGB) %>%
arrange(date_time)
rm(ts_OGB)2.3 EDA raw data
source("code/eda.R")
eda(ts, "ts-raw")
rm(eda)The output of an automated Exploratory Data Analysis (EDA) performed on the raw data with the package DataExplorer can be accessed here:
2.4 Clean data set
Sensor recordings were cleaned from obviously erroneous readings, by setting suspicious values to NA.
# running the commented code for plotting
# before and after the cleaning steps
# allows to visualize the removal of errorneous readings
ts <- data.table(ts)
# Profiling data
# temperature
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(tem, dep, col=station, linetype = cast))+
# geom_line()+
# scale_y_reverse()+
# geom_vline(xintercept = c(10, 20))+
# facet_wrap(~ID)
ts[ID == "180723" & station == "P07" & dep < 2 & cast == "up"]$tem <- NA
# salinity
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(sal, dep, col=station, linetype = cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
ts[sal < 6]$sal <- NA
# pH
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(pH, dep, col=station, linetype=cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
#
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(V_pH, dep, col=station, linetype=cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
ts[pH < 7.5]$V_pH <- NA
ts[pH < 7.5]$pH <- NA
ts[ID == "180709" & station == "P03" & dep < 5 & cast == "down"]$pH <- NA
ts[ID == "180709" & station == "P05" & dep < 10 & cast == "down"]$pH <- NA
ts[ID == "180718" & station == "P10" & dep < 3 & cast == "down"]$pH <- NA
ts[ID == "180815" & station == "P03" & dep < 2 & cast == "down"]$pH <- NA
ts[ID == "180820" & station == "P11" & dep < 15 & cast == "down"]$pH <- NA
ts[ID == "180709" & station == "P03" & dep < 5 & cast == "down"]$V_pH <- NA
ts[ID == "180709" & station == "P05" & dep < 10 & cast == "down"]$V_pH <- NA
ts[ID == "180718" & station == "P10" & dep < 3 & cast == "down"]$V_pH <- NA
ts[ID == "180815" & station == "P03" & dep < 2 & cast == "down"]$V_pH <- NA
ts[ID == "180820" & station == "P11" & dep < 15 & cast == "down"]$V_pH <- NA
# pCO2
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(pCO2, dep, col=station, linetype = cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
ts[ID == "180616"]$pCO2_analog <- NA
# O2
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(O2, dep, col=station, linetype = cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
# Chlorophyll
# ts %>%
# filter(type == "P") %>%
# ggplot(aes(Chl, dep, col=station, linetype = cast))+
# geom_path()+
# scale_y_reverse()+
# facet_wrap(~ID)
ts[Chl > 100]$Chl <- NA
# Surface transect data
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, dep, col=station))+
# geom_point()+
# scale_y_reverse()+
# facet_wrap(~ID, scales = "free_x")
#
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, tem, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")
#
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, sal, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")
#
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, pCO2, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")
#
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, pH, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")
#
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, Chl, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")
ts[type == "T" & Chl > 10]$Chl <- NA
# ts %>%
# filter(type == "T") %>%
# ggplot(aes(date, O2, col=station))+
# geom_point()+
# facet_wrap(~ID, scales = "free_x")2.5 Write summary file
Relevant columns were selected and renamed, only observations from regular stations (P01-P13) and transects (T01-T13) were selected and summarized data were written to file.
ts <- ts %>%
select(date_time,
ID,
type,
station,
dep,
sal,
tem,
pCO2_analog)
ts %>%
write_csv(here::here("data/intermediate/_summarized_data_files", "ts.csv"))2.6 EDA clean data
source("code/eda.R")
eda(ts, "ts_clean")
rm(eda)The output of an automated Exploratory Data Analysis (EDA) performed on the cleaned data with the package DataExplorer can be accessed here:
2.7 Overview plots
ts %>%
arrange(date_time) %>%
filter(type == "P",!(station %in% c("PX1", "PX2"))) %>%
ggplot(aes(tem, dep, col = ymd(ID), group = ID)) +
geom_path() +
scale_y_reverse() +
scale_color_viridis_c(trans = "date", name = "") +
facet_wrap( ~ station)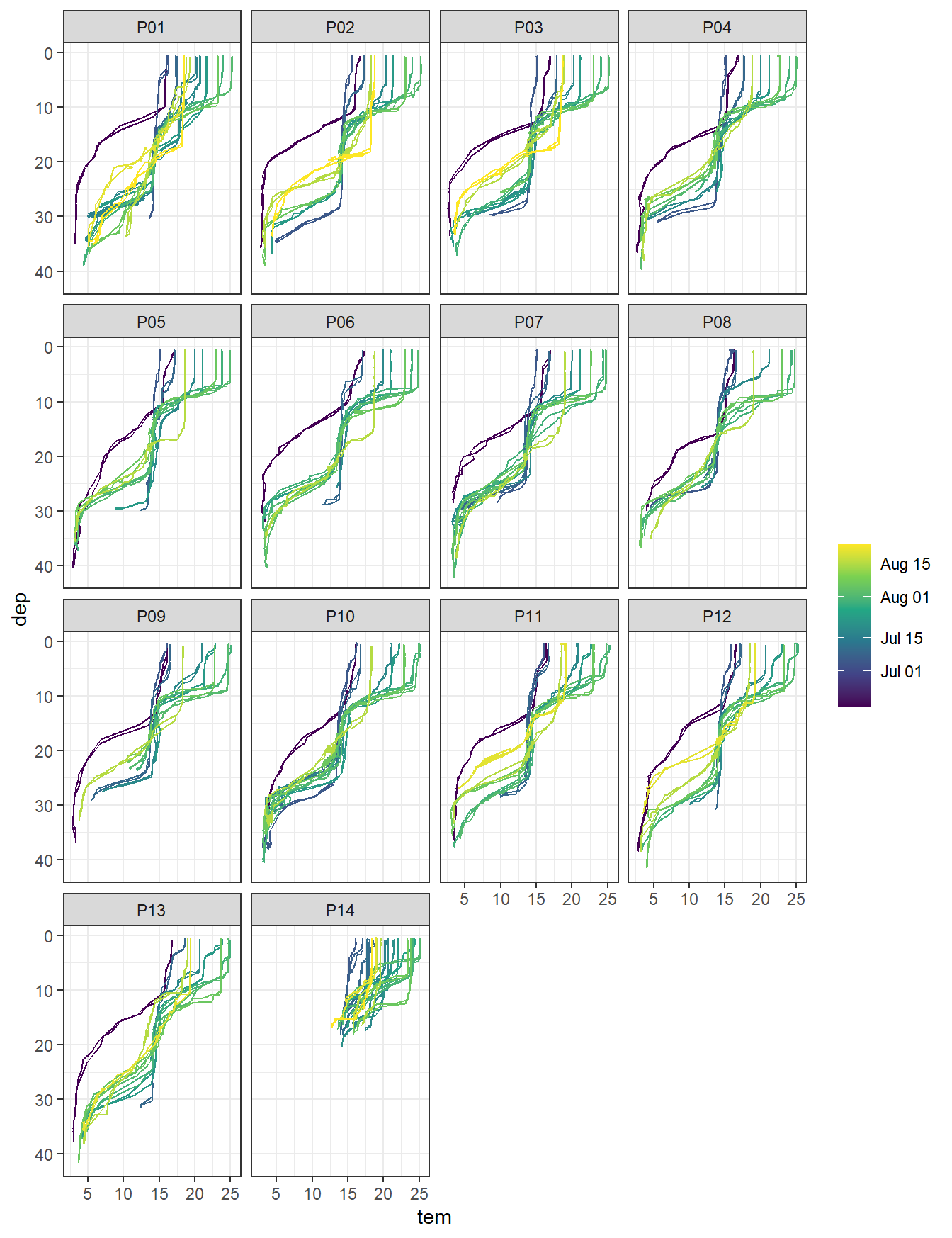
Temperature profiles by stations. Color refers to the starting date of each cruise.
ts %>%
arrange(date_time) %>%
filter(type == "P",!(station %in% c("PX1", "PX2"))) %>%
ggplot(aes(pCO2_analog, dep, col = ymd(ID), group = ID)) +
geom_path() +
scale_y_reverse() +
scale_color_viridis_c(trans = "date", name = "") +
facet_wrap( ~ station)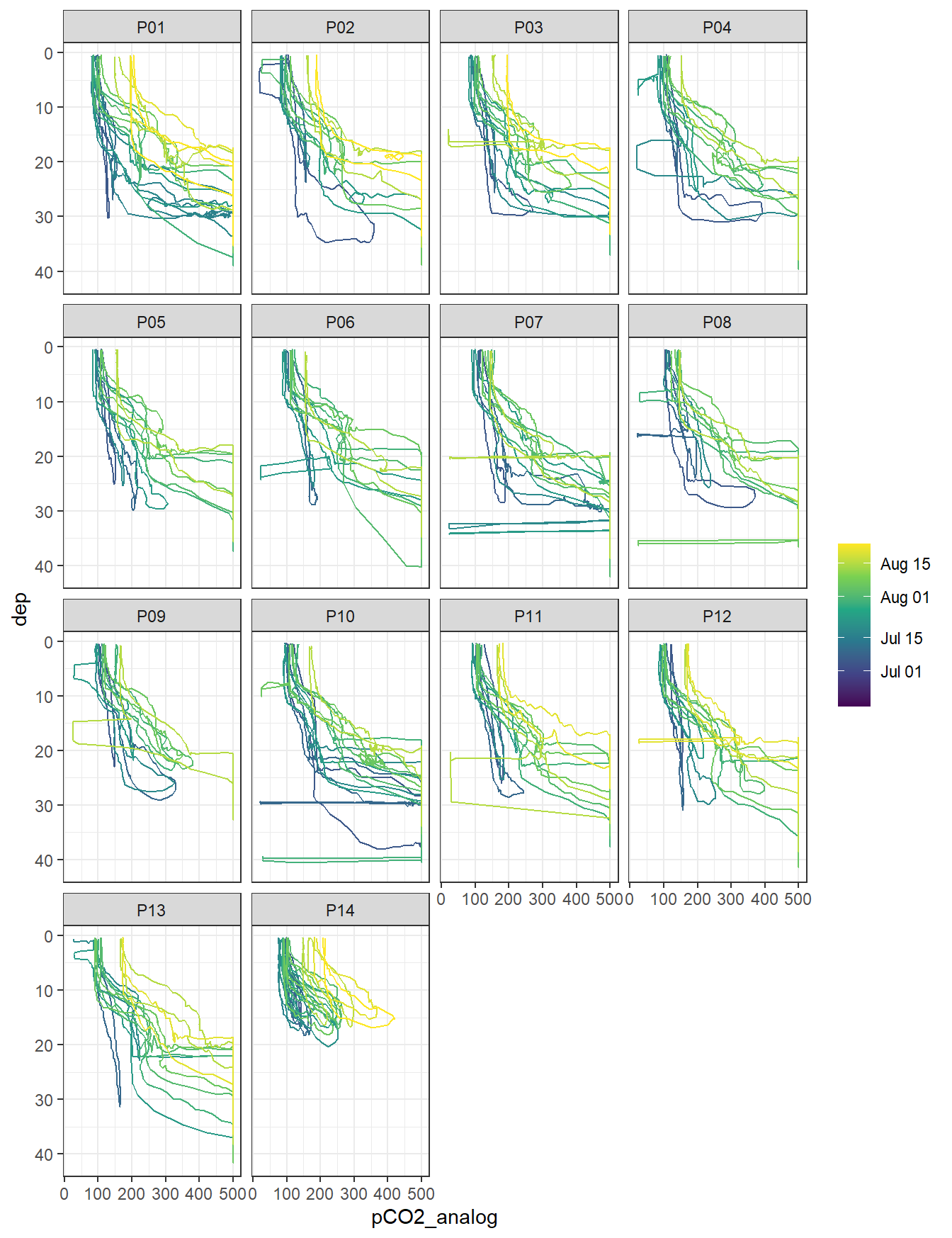
pCO2 (analog signal) profiles by stations. Color refers to the starting date of each cruise.
3 HydroC CO2 data (th)
3.1 Read data
Originally, HydroC pCO2 data were provided by KM Contros after applying a drift correction to the raw data, which was based on pre- and post-deployment calibration results. Those preliminary data are read-in here. However, some data recorded during testing and configuration of the sensor were later on removed, and the post-processing was repeated based on a cleaned data set. This revised post-processed file is use in the merging script.
# Read Contros corrected data file, based on all recordings
th <-
read_csv2(here::here("data/input/TinaV/Sensor/HydroC-pCO2/corrected_Contros",
"parameter&pCO2s(method 43).txt"),
col_names = c("date_time", "Zero", "Flush", "p_NDIR",
"p_in", "T_control", "T_gas", "%rH_gas",
"Signal_raw", "Signal_ref", "T_sensor",
"pCO2_corr", "Runtime", "nr.ave")) %>%
mutate(date_time = dmy_hms(date_time),
Flush = as.factor(as.character(Flush)),
Zero = as.factor(as.character(Zero)))3.2 Deployment identification and subsetting
Individual deployments (periods of observations with less than 30 sec between recordings) were identified and relevant deployment periods were selected. This procedure removes some data recorded during sensor testing and set-up.
# identify individual deployments
th <- th %>%
arrange(date_time) %>%
mutate(deployment = cumsum(c(TRUE, diff(date_time) >= 30)))
# write pre-cleaning file for later comparison
th %>%
select(date_time, pCO2_corr, deployment) %>%
write_csv(here::here(
"data/intermediate/_summarized_data_files",
"th_pre_cleaning.csv"
))
# filter relevant deployments
th <- th %>%
filter(deployment %in% c(2, 6, 9, 14, 17, 21, 23, 27, 31, 33, 34, 35, 37))3.3 Removal of duplicated time stamps
A low number of the recorded HydroC data revealed the exact same time stamp. This was corrected either by filling a corresponding gap before or after the duplicate, or by removing one of the duplicated rows if no such gap existed.
# add counter for date_time observations
th <- th %>%
add_count(date_time)
# find triplicated time stamp and select only first observation, and merge
th_no_triple <- th %>%
filter(n <= 2)
th_triple_clean <- th %>%
filter(n > 2) %>%
slice(1)
th <- full_join(th_no_triple, th_triple_clean)
rm(list = setdiff(ls(), c("th", "parameters")))
# find duplicated time stamps and shift first by one second backward, and merge
# th %>%
# distinct(date_time)
th <- th %>%
select(-n) %>%
add_count(date_time)
# unique(th$n)
th_no_duplicated <- th %>%
filter(n == 1)
th_duplicated <- th %>%
filter(n == 2)
th_duplicated_first <- th_duplicated %>%
group_by(date_time) %>%
slice(1) %>%
ungroup() %>%
mutate(date_time = date_time - 1)
th_duplicated_second <- th_duplicated %>%
group_by(date_time) %>%
slice(2) %>%
ungroup()
th_duplicated_clean <-
full_join(th_duplicated_first, th_duplicated_second) %>%
arrange(date_time)
th <- full_join(th_no_duplicated, th_duplicated_clean)
# th %>%
# distinct(date_time)
rm(list = setdiff(ls(), c("th", "parameters")))
# find duplicated time stamps and shift first by two seconds forward, and merge
# th %>%
# distinct(date_time)
th <- th %>%
select(-n) %>%
add_count(date_time)
# unique(th$n)
th_no_duplicated <- th %>%
filter(n == 1)
th_duplicated <- th %>%
filter(n == 2)
th_duplicated_first <- th_duplicated %>%
group_by(date_time) %>%
slice(1) %>%
ungroup() %>%
mutate(date_time = date_time + 2)
th_duplicated_second <- th_duplicated %>%
group_by(date_time) %>%
slice(2) %>%
ungroup()
th_duplicated_clean <-
full_join(th_duplicated_first, th_duplicated_second) %>%
arrange(date_time)
th <- full_join(th_no_duplicated, th_duplicated_clean)
# th %>%
# distinct(date_time)
rm(list = setdiff(ls(), c("th", "parameters")))
# remaining duplicates are observations where other observations with a +/- 1 sec timestamp exist
# for those cases, only the first duplicated observation is selected (similar to triplicate treatment)
# th %>%
# distinct(date_time)
th <- th %>%
select(-n) %>%
add_count(date_time)
# unique(th$n)
th_still_no_duplicated <- th %>%
filter(n == 1)
th_still_duplicated_first <- th %>%
filter(n == 2) %>%
group_by(date_time) %>%
slice(1)
th <- full_join(th_still_no_duplicated, th_still_duplicated_first)
# th %>%
# distinct(date_time)
rm(list = setdiff(ls(), c("th", "parameters")))
th <- th %>%
select(-n)3.4 Flush and Zeroing identification
Flush and zeroing periods of the sensor are identified and assigned with unique IDs.
# Zeroing ID labeling
th <- th %>%
arrange(date_time) %>%
group_by(Zero) %>%
mutate(Zero_counter = as.factor(cumsum(c(
TRUE, diff(date_time) >= 30
)))) %>%
ungroup()
# Flush: Identification
th <- th %>%
mutate(Flush = 0) %>%
group_by(Zero, Zero_counter) %>%
mutate(
start = min(date_time),
duration = date_time - start,
Flush = if_else(Zero == 0 &
duration < parameters$HC_flush_duration, "1", "0")
) %>%
ungroup()
# Flush: Identify equilibration and internal gas mixing periods
th <- th %>%
mutate(mixing = if_else(
duration < parameters$HC_mixing_duration,
"mixing",
"equilibration"
))3.5 Deployment plots
pdf(file=here::here("output/Plots/read_in",
"th_deployments.pdf"), onefile = TRUE, width = 7, height = 4)
for (i in unique(th$deployment)) {
#i <- unique(th$deployment)[3]
sub <- th %>%
filter(deployment == i)
start_date <- min(sub$date_time)
print(
sub %>%
ggplot(aes(date_time, pCO2_corr, col=Zero_counter))+
geom_line()+
labs(title = paste("Deployment: ",i, "| Start time: ", start_date))
)
}
dev.off()
rm(sub, start_date, i)A pdf with pCO2 timeseries plots of all individual deployments can be found here:
source("code/eda.R")
eda(th, "th")
rm(eda)The output of an automated Exploratory Data Analysis (EDA) performed with the package DataExplorer can be accessed here:
3.6 Write summary file
Summarized clean pCO2 data were written to file.
th %>%
select(date_time,
Zero,
Flush,
pCO2_corr,
deployment,
Zero_counter,
duration,
mixing) %>%
write_csv(here::here("data/intermediate/_summarized_data_files",
"th.csv"))
rm(th)4 Bottle data CO2 (tb)
Discrete samples were collected with a Niskin bottle and analyzed for CT and AT at IOW’s CO2 lab.
4.1 Read data
# Read CO2 system bottle data
tb <-
read_csv(
here::here(
"data/input/TinaV/Bottle/Tracegases",
"BloomSail_bottle_CO2_all.csv"
),
col_types = list("c", "c", "n", "n", "n", "n", "n")
)
# select and rename relevant columns
tb <- tb %>%
select(
ID = transect.ID,
station = label,
dep = Dep,
sal = Sal,
CT,
AT
)4.2 Write summary file
tb %>% write_csv(here::here("data/intermediate/_summarized_data_files",
"tb.csv"))
rm(tb)5 Bottle data plankton (tp)
Discrete samples were collected with a Niskin bottle and analysed for phytoplankton composition and biomass at IOW’s phytoplankton lab (Norbert Wasmund).
5.1 Read data
tp <- read_csv(
here::here(
"data/input/TinaV/Bottle/Phytoplankton",
"181205_BloomSail_Plankton_counts.csv"
)
)
# delete colomns that contain counts, not calculated biomass
tp <- tp[, -seq(4, 21, 1)]
# assign new column names
# for species:
# nr = size class,
# HV = Heterocyst per Volume,
# Hl = Heterocyst per length,
# t = total
names(tp) <-
c(
"date",
"station",
"dep",
"Aphanizomenon.1",
"Aphanizomenon.2",
"Aphanizomenon.3",
"Aphanizomenon.t",
"Aphanizomenon.HV",
"Aphanizomenon.Hl",
"Dolichospermum.1",
"Dolichospermum.2",
"Dolichospermum.3",
"Dolichospermum.4",
"Dolichospermum.t",
"Dolichospermum.HV",
"Dolichospermum.Hl",
"Nodularia.1",
"Nodularia.2",
"Nodularia.3",
"Nodularia.t",
"Nodularia.HV",
"Nodularia.Hl",
"Nodulariadead.1",
"Nodulariadead.2",
"Nodulariadead.3",
"Nodulariadead.t",
"total.t"
)
# change format of data table and separate into 2 columns for species and class
tp <-
gather(tp, para, value, Aphanizomenon.1:total.t, factor_key = TRUE)
tp <- separate(tp, col = para, into = c("Species", "class"))
# change class of columns
tp <- tp %>%
mutate(ID = date,
date = ymd(date))5.2 Write summary file
tp %>% write_csv(here::here("data/intermediate/_summarized_data_files",
"tp.csv"))
rm(tp)6 GPS track (tt)
GPS track data were recorded with a Samsung Galaxy tablet.
6.1 Read data
files <-
list.files(path = "data/input/TinaV/Track/GPS_Logger_Track/", pattern = "[.]txt$")
for (file in files) {
# if the merged dataset does exist, append to it
if (exists("dataset")) {
temp <-
data.table(read.delim(
here::here("data/input/TinaV/Track/GPS_Logger_Track", file),
sep = ","
)[, c(2, 3, 4)])
names(temp) <- c("date_time", "lat", "lon")
temp$date_time <- ymd_hms(temp$date, tz = "UTC")
dataset <- rbind(dataset, temp)
rm(temp)
}
# if the merged dataset doesn't exist, create it
if (!exists("dataset")) {
dataset <-
data.table(read.delim(
here::here("data/input/TinaV/Track/GPS_Logger_Track", file),
sep = ","
)[, c(2, 3, 4)])
names(dataset) <- c("date_time", "lat", "lon")
dataset$date_time <- ymd_hms(dataset$date_time, tz = "UTC")
}
}
tt <- dataset
rm(dataset, file, files)6.2 Write summary file
tt %>%
write_csv(here::here("data/intermediate/_summarized_data_files",
"tt.csv"))
rm(tt)7 Atmospheric observations
Atmospheric data were recorded at the ICOS station on Östergarnsholm.
7.1 Read data
og <-
read_delim(
here::here(
"data/input/Ostergarnsholm/Tower",
"Oes_Jens_atm_water_June_to_August_2018.csv"
),
delim = ";"
)
og <- og %>%
mutate(date_time = ymd_hms(paste(
paste(year, month, day, sep = "/"),
paste(hour, min, sec, sep = ":")
))) %>%
select(
"date_time",
"CO2 12m [ppm]",
"w_c [ppm m/s]",
"WS 12m [m/s]",
"WD 12m [degrees]",
"T 12m [degrees C]",
"RIS [W/m^2]"
)
# conversion from GMT+1 to UTC
og <- og %>%
mutate(date_time = date_time - 60 ^ 2)
og <- og %>%
select(date_time, pCO2_atm = "CO2 12m [ppm]", wind = "WS 12m [m/s]")7.2 Write summary file
og %>%
write_csv(here::here("data/intermediate/_summarized_data_files",
"og.csv"))
rm(og)8 SOOP Finnmaid
Here, we read in pCO2 and SST data recorded on SOOP Finnmaid in June-August 2018.
8.1 Read data
# LI-COR data
files <-
list.files(path = "data/input/Finnmaid_2018", pattern = "[.]xls$")
for (file in files) {
temp <- read_excel(here::here("data/input/Finnmaid_2018", file))
temp <- temp[c(1, 2, 3, 12, 7, 4, 15, 8, 5, 17)]
names(temp) <-
c("date_time",
"lon",
"lat",
"pCO2",
"sal",
"tem",
"cO2",
"patm",
"Teq",
"xCO2")
temp <- temp[-c(1), ]
temp$date_time <-
as.POSIXct(as.numeric(temp$date_time) * 60 * 60 * 24,
origin = "1899-12-30",
tz = "GMT")
temp$lon <- as.numeric(as.character(temp$lon))
temp$lat <- as.numeric(as.character(temp$lat))
temp$pCO2 <- as.numeric(as.character(temp$pCO2))
temp$sal <- as.numeric(as.character(temp$sal))
temp$tem <- as.numeric(as.character(temp$tem))
temp$cO2 <- as.numeric(as.character(temp$cO2))
temp$patm <- as.numeric(as.character(temp$patm))
temp$Teq <- as.numeric(as.character(temp$Teq))
temp$xCO2 <- as.numeric(as.character(temp$xCO2))
temp <- data.table(temp)
temp$route <-
strapplyc(as.character(file), ".*(.).xls*", simplify = TRUE)
temp$ID <- substr(as.character(file), 3, 10)
if (exists("dataset")) {
dataset <- rbind(dataset, temp)
} else{
dataset <- temp
}
}
rm(temp, files, file)
dataset <- dataset[pCO2 != 0]
# Los Gatos Research (LGR) data
# Please note that the LGR data were corrected manually before
# The correction procedure is outlined in the Appendix of the ms
files <-
list.files(path = "data/input/Finnmaid_2018/LGR", pattern = "[.]xls$")
for (file in files) {
temp <- read_excel(here::here("data/input/Finnmaid_2018/LGR", file))
temp <- temp[c(2, 3, 4, 8, 6, 5, 14, 7, 15, 9)]
names(temp) <-
c("date_time",
"lon",
"lat",
"pCO2",
"sal",
"tem",
"cO2",
"patm",
"Teq",
"xCO2")
temp <- temp[-c(1), ]
temp$date_time <- dmy_hms(temp$date_time)
temp <- data.table(temp)
temp$route <- substr(as.character(file), 12, 12)
temp$ID <- substr(as.character(file), 3, 10)
if (exists("dataset.LGR")) {
dataset.LGR <- rbind(dataset.LGR, temp)
} else{
dataset.LGR <- temp
}
}
rm(temp, files, file)# This code can be used to convert O2 units
# but is not applied, because O2 data are not used in this study
source(here::here("code", "O2stoO2c.R"))
dataset.LGR <- dataset.LGR %>%
filter() %>%
mutate(cO2 = O2stoO2c(
O2sat = cO2,
T = tem,
S = sal,
P = 3 / 10,
p_atm = 1013.5
))
rm(O2stoO2c, pH2Osat, sca_T, Scorr, TCorr, R, Vm)dataset$sensor <- "LICOR"
dataset.LGR$sensor <- "LosGatos"
fm <- bind_rows(dataset, dataset.LGR)
rm(dataset, dataset.LGR)8.2 Write summary file
fm %>%
write_csv(here::here("data/intermediate/_summarized_data_files",
"fm.csv"))
rm(fm)9 Interactive map
fm <-
read_csv(here::here("data/intermediate/_summarized_data_files",
"fm.csv"))
fm_sub <- fm %>%
arrange(date_time) %>%
slice(which(row_number() %% 20 == 1))
tt <-
read_csv(here::here("data/intermediate/_summarized_data_files", "tt.csv"))
tt_sub <- tt %>%
slice(which(row_number() %% 20 == 1))
rm(tt, fm)
leaflet() %>%
setView(lng = 20, lat = 57.3, zoom = 8) %>%
addLayersControl(
baseGroups = c("Ocean Basemap",
"Satellite"),
overlayGroups = c("BloomSail", "Finnmaid"),
options = layersControlOptions(collapsed = FALSE),
position = 'topright'
) %>%
addProviderTiles("Esri.WorldImagery", group = "Satellite") %>%
addProviderTiles(providers$Esri.OceanBasemap, group = "Ocean Basemap") %>%
addScaleBar(position = 'topright') %>%
addMeasure(
primaryLengthUnit = "kilometers",
secondaryLengthUnit = 'miles',
primaryAreaUnit = "sqmeters",
secondaryAreaUnit = "acres",
position = 'topleft'
) %>%
addCircles(data = fm_sub,
~ lon,
~ lat,
color = "white",
group = "Finnmaid") %>%
addPolylines(data = tt_sub,
~ lon,
~ lat,
color = "red",
group = "BloomSail")Overview map of SOOP Finnmaid and SV Tina V tracks. Please note that only a subet of the recorded track data is plotted.
rm(fm_sub, tt_sub)
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18363)
Matrix products: default
locale:
[1] LC_COLLATE=English_Germany.1252 LC_CTYPE=English_Germany.1252
[3] LC_MONETARY=English_Germany.1252 LC_NUMERIC=C
[5] LC_TIME=English_Germany.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] gsubfn_0.7 proto_1.0.0 readxl_1.3.1 leaflet_2.0.3
[5] DataExplorer_0.8.2 lubridate_1.7.9.2 data.table_1.13.6 forcats_0.5.0
[9] stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4 readr_1.4.0
[13] tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.3 tidyverse_1.3.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.2 jsonlite_1.7.2 viridisLite_0.3.0
[4] here_1.0.1 modelr_0.1.8 assertthat_0.2.1
[7] highr_0.8 cellranger_1.1.0 yaml_2.2.1
[10] pillar_1.4.7 backports_1.2.1 glue_1.4.2
[13] digest_0.6.27 promises_1.1.1 rvest_0.3.6
[16] leaflet.providers_1.9.0 colorspace_2.0-0 htmltools_0.5.0
[19] httpuv_1.5.4 pkgconfig_2.0.3 broom_0.7.3
[22] haven_2.3.1 scales_1.1.1 whisker_0.4
[25] later_1.1.0.1 git2r_0.27.1 generics_0.1.0
[28] farver_2.0.3 ellipsis_0.3.1 withr_2.3.0
[31] cli_2.2.0 magrittr_2.0.1 crayon_1.3.4
[34] evaluate_0.14 ps_1.5.0 fs_1.5.0
[37] fansi_0.4.1 xml2_1.3.2 tools_4.0.3
[40] hms_0.5.3 lifecycle_0.2.0 munsell_0.5.0
[43] reprex_0.3.0 networkD3_0.4 compiler_4.0.3
[46] rlang_0.4.10 grid_4.0.3 rstudioapi_0.13
[49] htmlwidgets_1.5.3 crosstalk_1.1.0.1 igraph_1.2.6
[52] tcltk_4.0.3 labeling_0.4.2 rmarkdown_2.6
[55] gtable_0.3.0 DBI_1.1.0 R6_2.5.0
[58] gridExtra_2.3 knitr_1.30 rprojroot_2.0.2
[61] stringi_1.5.3 parallel_4.0.3 Rcpp_1.0.5
[64] vctrs_0.3.6 dbplyr_2.0.0 tidyselect_1.1.0
[67] xfun_0.19