Extreme pH Profiles
Pasqualina Vonlanthen & Jens Daniel Müller
28 January, 2022
Last updated: 2022-01-28
Checks: 7 0
Knit directory: bgc_argo_r_argodata/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5024768. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Ignored: output/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/extreme_pH.Rmd) and HTML (docs/extreme_pH.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5024768 | jens-daniel-mueller | 2022-01-28 | code review: basinmask and regression |
| html | 5635ef2 | pasqualina-vonlanthendinenna | 2022-01-27 | Build site. |
| Rmd | 23dc282 | pasqualina-vonlanthendinenna | 2022-01-27 | failed attempt at updating basinmask and regression |
| html | c44ff0f | pasqualina-vonlanthendinenna | 2022-01-25 | Build site. |
| Rmd | 3851824 | pasqualina-vonlanthendinenna | 2022-01-25 | added basin-mean profiles |
| html | 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 | Build site. |
| Rmd | 825a50a | pasqualina-vonlanthendinenna | 2022-01-25 | added seasonal and biome profiles |
| html | 3ae43e4 | pasqualina-vonlanthendinenna | 2022-01-24 | Build site. |
| Rmd | 3f8e824 | pasqualina-vonlanthendinenna | 2022-01-24 | updated 24/01 |
| html | 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | e72d7ca | pasqualina-vonlanthendinenna | 2022-01-21 | updated linear regression to monthly |
| html | 587755e | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | 7a9209b | pasqualina-vonlanthendinenna | 2022-01-21 | updated threshold calculation 2 |
| html | c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | 58b3b3b | pasqualina-vonlanthendinenna | 2022-01-21 | updated threshold calculation |
| html | ed3fef2 | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | 3d2f8fc | jens-daniel-mueller | 2022-01-07 | code review |
| html | 486c9c8 | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | e9ad067 | jens-daniel-mueller | 2022-01-07 | code review |
| html | 343689f | pasqualina-vonlanthendinenna | 2022-01-06 | Build site. |
| Rmd | f53cc2d | pasqualina-vonlanthendinenna | 2022-01-06 | updated profile page |
| html | b8a6482 | pasqualina-vonlanthendinenna | 2022-01-03 | Build site. |
| Rmd | 054f8a6 | pasqualina-vonlanthendinenna | 2022-01-03 | added Argo profiles |
Task
Compare depth profiles of normal pH and of extreme pH, as identified in the surface OceanSODA pH data product
theme_set(theme_bw())
HNL_colors <- c("H" = "#b2182b",
"N" = "#636363",
"L" = "#2166ac")Load data
path_argo <- '/nfs/kryo/work/updata/bgc_argo_r_argodata'
path_argo_preprocessed <- paste0(path_argo, "/preprocessed_bgc_data")
path_emlr_utilities <- "/nfs/kryo/work/jenmueller/emlr_cant/utilities/files/"# RECCAP2-ocean region mask
region_masks_all_2x2 <- read_rds(file = paste0(path_argo_preprocessed,
"/region_masks_all_2x2.rds"))
region_masks_all_2x2 <- region_masks_all_2x2 %>%
rename(biome = value) %>%
mutate(coast = as.character(coast))
# WOA 18 basin mask
basinmask <-
read_csv(
paste(path_emlr_utilities,
"basin_mask_WOA18.csv",
sep = ""),
col_types = cols("MLR_basins" = col_character())
)
basinmask <- basinmask %>%
filter(MLR_basins == unique(basinmask$MLR_basins)[1]) %>%
select(-c(MLR_basins, basin))
# OceanSODA
OceanSODA <- read_rds(file = paste0(path_argo_preprocessed, "/OceanSODA.rds"))
OceanSODA <- OceanSODA %>%
mutate(month = month(date))
# load in the full argo data
full_argo <- read_rds(file = paste0(path_argo_preprocessed, "/bgc_merge_pH_qc_1.rds"))
# change the date format for compatibility with OceanSODA pH data
full_argo <- full_argo %>%
mutate(year = year(date),
month = month(date)) %>%
mutate(date = ymd(format(date, "%Y-%m-15")))Regions
Biomes
region_masks_all_2x2 <- region_masks_all_2x2 %>%
filter(region == 'southern',
biome != 0) %>%
select(-region)Remove coastal data
basemap(limits = -32) +
geom_spatial_tile(
data = region_masks_all_2x2,
aes(x = lon,
y = lat,
fill = coast),
col = 'transparent'
) +
scale_fill_brewer(palette = "Dark2")
region_masks_all_2x2 <- region_masks_all_2x2 %>%
filter(coast == "0")Grid reduction
basemap(limits = -32) +
geom_spatial_tile(
data = region_masks_all_2x2,
aes(x = lon,
y = lat,
fill = biome),
col = 'transparent'
) +
scale_fill_brewer(palette = "Dark2")
region_masks_all_2x2 <- region_masks_all_2x2 %>%
count(lon, lat, biome) %>%
group_by(lon, lat) %>%
slice_max(n, with_ties = FALSE) %>%
ungroup()basemap(limits = -32) +
geom_spatial_tile(
data = region_masks_all_2x2,
aes(x = lon,
y = lat,
fill = biome),
col = 'transparent'
) +
scale_fill_brewer(palette = "Dark2")
Basins
basinmask <- basinmask %>%
filter(lat < -30)Grid reduction
basemap(limits = -32) +
geom_spatial_tile(
data = basinmask,
aes(x = lon,
y = lat,
fill = basin_AIP),
col = 'transparent'
) +
scale_fill_brewer(palette = "Dark2")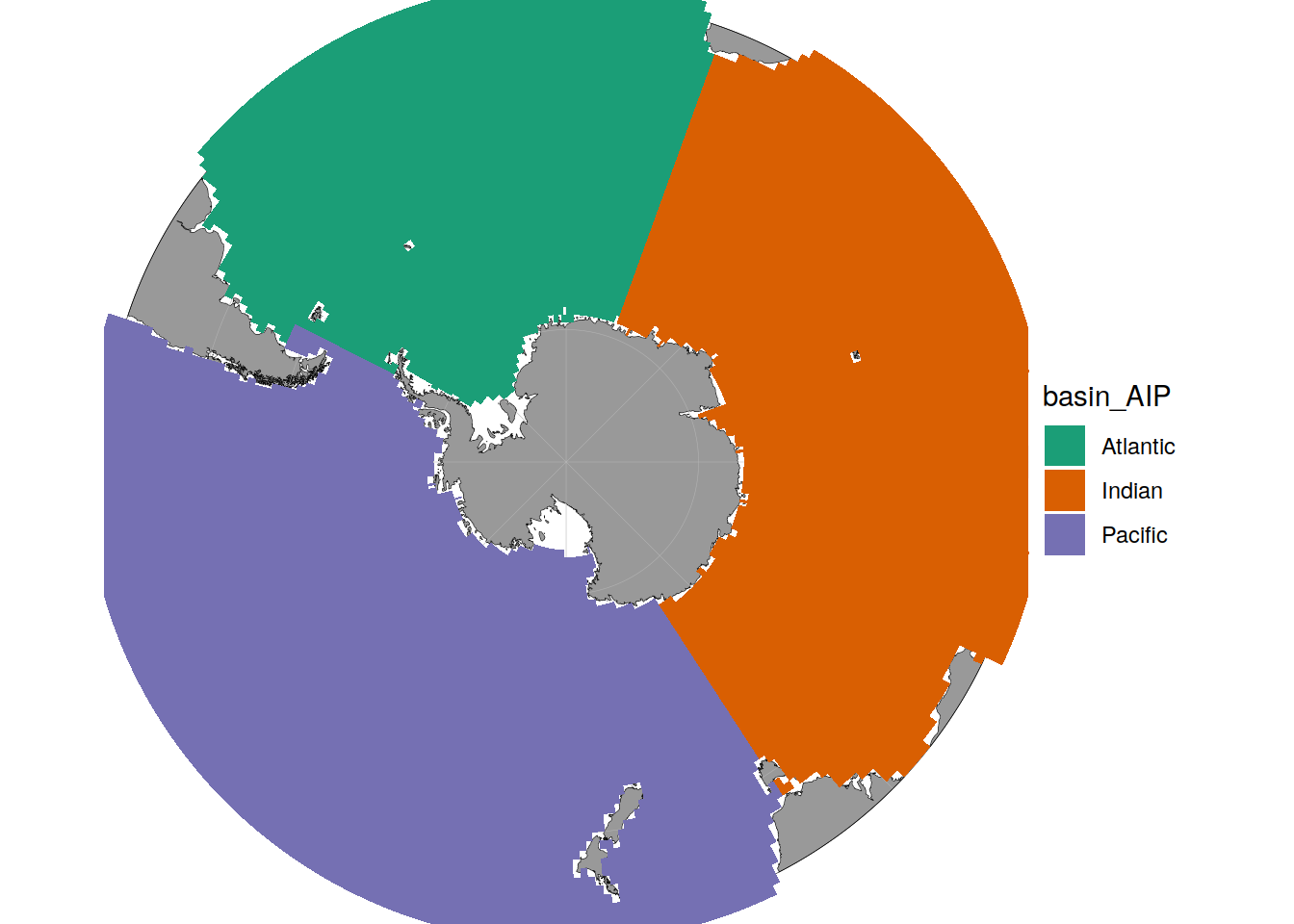
basinmask_2x2 <- basinmask %>%
mutate(
lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
lat = as.numeric(as.character(lat)),
lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
lon = as.numeric(as.character(lon))
) # regrid into 2x2º grid
# assign basins from each pixel to to each 2 Lon x Lat pixel, based on the majority of basins in each 2x2 grid
basinmask_2x2 <- basinmask_2x2 %>%
count(lon, lat, basin_AIP) %>%
group_by(lon, lat) %>%
slice_max(n, with_ties = FALSE) %>%
ungroup() %>%
select(-n)
rm(basinmask)basemap(limits = -32) +
geom_spatial_tile(
data = basinmask_2x2 %>% filter(lat < -30),
aes(x = lon,
y = lat,
fill = basin_AIP),
col = 'transparent'
) +
scale_fill_brewer(palette = "Dark2")
OceanSODA
Grid reduction
# Note: While reducing lon x lat grid,
# we keep the original number of observations
OceanSODA_2x2 <- OceanSODA %>%
mutate(
lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
lat = as.numeric(as.character(lat)),
lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
lon = as.numeric(as.character(lon))) # regrid into 2x2º grid Apply region masks
# keep only Southern Ocean data
OceanSODA_2x2_SO <- inner_join(OceanSODA_2x2, region_masks_all_2x2)
# add in basin separations
OceanSODA_2x2_SO <- inner_join(OceanSODA_2x2_SO, basinmask_2x2)
# expected number of rows from -30 to -70º latitude, 360º longitude, for 12 months, 8 years:
# 40 lat x 360 lon x 12 months x 8 years = 1 382 400 rows
OceanSODA_2x2_SO <- OceanSODA_2x2_SO %>%
filter(!is.na(ph_total))OceanSODA pH anomalies
Grid level
Climatological thresholds
Fit lm models
# fit a linear regression of OceanSODA pH against time (temporal trend)
# in each lat/lon/month grid
OceanSODA_2x2_SO <- OceanSODA_2x2_SO %>%
mutate(year = year(date))
OceanSODA_regression <- OceanSODA_2x2_SO %>%
# filter(basin_AIP == "Indian",
# biome == "2",
# lon < 40) %>%
nest(data = -c(lon, lat, month)) %>%
mutate(fit = map(.x = data,
.f = ~ lm(ph_total ~ year, data = .x)),
tidied = map(.x = fit, .f = tidy),
glanced = map(.x = fit, .f = glance),
augmented = map(.x = fit, .f = augment))
OceanSODA_regression_tidied <- OceanSODA_regression %>%
select(-c(data, fit, augmented, glanced)) %>%
unnest(tidied)
OceanSODA_regression_tidied <- OceanSODA_regression_tidied %>%
select(lat:estimate) %>%
pivot_wider(names_from = term,
values_from = estimate) %>%
rename(intercept = `(Intercept)`,
slope = year)
OceanSODA_regression_augmented <- OceanSODA_regression %>%
select(-c(data, fit, tidied, glanced)) %>%
unnest(augmented)
OceanSODA_regression_glanced <- OceanSODA_regression %>%
select(-c(data, fit, tidied, augmented)) %>%
unnest(glanced)Slope maps
basemap(limits = -32) +
geom_spatial_tile(data = OceanSODA_regression_tidied,
aes(x = lon,
y = lat,
fill = slope),
col = 'transparent') +
scale_fill_scico(palette = "vik", midpoint = 0) +
facet_wrap( ~ month, ncol = 2)
Thresholds
OceanSODA_regression_augmented_stats <- OceanSODA_regression_augmented %>%
group_by(lat, lon, month) %>%
summarise(residual_sd = sd(.resid)) %>%
ungroup()
compare <- full_join(OceanSODA_regression_augmented_stats,
OceanSODA_regression_glanced)
compare %>%
ggplot(aes(residual_sd -sigma)) +
geom_histogram()
OceanSODA_2x2_SO_extreme_grid <-
full_join(OceanSODA_regression_augmented %>%
select(lat:year, .resid),
OceanSODA_regression_glanced %>%
select(lat:month, sigma))
# # calculate H and L pH thresholds for climatological monthly pH
# OceanSODA_2x2_SO_extreme_grid <- OceanSODA_2x2_SO_extreme_grid %>%
# mutate(ph_L = .fitted - 2*(.sigma),
# ph_H = .fitted + 2*(.sigma))# calculate climatological average OceanSODA pH
# and the 95th percentile of the monthly OceanSODA pH
#
# OceanSODA_2x2_SO_clim_grid <- OceanSODA_2x2_SO %>%
# group_by(lon, lat, month) %>%
# summarise(
# ph_N = mean(ph_total, na.rm = TRUE),
# ph_H = quantile(ph_total, 0.95, na.rm = TRUE),
# ph_L = quantile(ph_total, 0.05, na.rm = TRUE)
# ) %>%
# ungroup()
#
# OceanSODA_2x2_SO_extreme_grid <- inner_join(OceanSODA_2x2_SO, OceanSODA_2x2_SO_clim_grid)Anomaly identification
Calculate OceanSODA pH anomalies: L for abnormally low, H for abnormally high, N for normal pH
# when the in-situ OceanSODA pH is lower than the 5th percentile (predicted - 2*residual.st.dev), assign 'L' for low extreme
# when the in-situ OceanSODA pH exceeds the 95th percentile (predicted + 2*residual.st.dev), assign 'H' for high extreme
# when the in-situ OceanSODA pH is within 95% of the range, then assign 'N' for normal pH
OceanSODA_2x2_SO_extreme_grid <- OceanSODA_2x2_SO_extreme_grid %>%
mutate(
ph_extreme = case_when(
.resid < -sigma*2 ~ 'L',
.resid > sigma*2 ~ 'H',
TRUE ~ 'N'
)
)
# table(is.na(OceanSODA_2x2_SO_extreme_grid))
OceanSODA_2x2_SO_extreme_grid <- OceanSODA_2x2_SO_extreme_grid %>%
mutate(ph_extreme = fct_relevel(ph_extreme, "H", "N", "L"))
OceanSODA_2x2_SO_extreme_grid <-
full_join(OceanSODA_2x2_SO_extreme_grid,
OceanSODA_regression_tidied)
# pivot_wider two columns (slope and intercept), values_from = estimate, names_from = terms, names.repair = 'unique'
# gives a slope and intercept column
# rename date...26 = slope and date...2 = date
# OceanSODA_2x2_SO_extreme_grid <- OceanSODA_2x2_SO_extreme_grid %>%
# pivot_wider(names_from = term,
# values_from = estimate,
# names_repair = 'unique') %>%
# rename(date = date...2,
# regression_slope = date...24,
# regression_intercept = `(Intercept)`)
# fill in NAs in the slope and intercept columns (values from above for regression_slope and values from below for regression_intercept) (creates duplicate rows) and remove duplicate rows
# OceanSODA_2x2_SO_extreme_grid <- OceanSODA_2x2_SO_extreme_grid %>%
# group_by(lon, lat, date, year, month) %>%
# fill(regression_slope, .direction = 'up') %>%
# fill(regression_intercept, .direction = 'down') %>%
# distinct()OceanSODA_2x2_SO_extreme_grid %>%
group_split(lon, lat, month) %>%
head(6) %>%
map(~ ggplot(data = .x) +
geom_point(aes(x = year,
y = ph_total,
col = ph_extreme)) +
geom_abline(data = .x, aes(slope = slope,
intercept = intercept)) +
geom_abline(data = .x, aes(slope = slope,
intercept = intercept + 2*sigma),
linetype = 2) +
geom_abline(data = .x, aes(slope = slope,
intercept = intercept - 2*sigma),
linetype = 2) +
labs(title = paste(fititle = paste(
"lon:", unique(.x$lon),
"| lat:", unique(.x$lat),
"| month:", unique(.x$month)
))) +
scale_color_manual(values = HNL_colors))[[1]]
[[2]]
[[3]]
[[4]]
[[5]]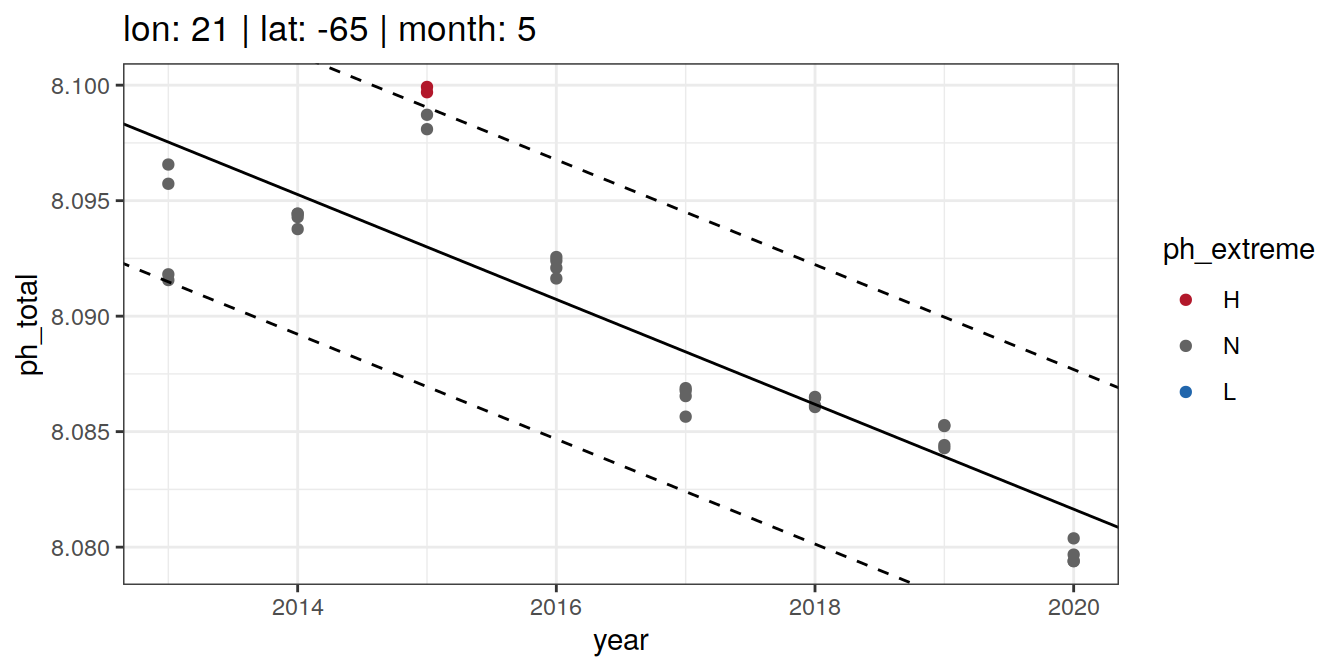
[[6]]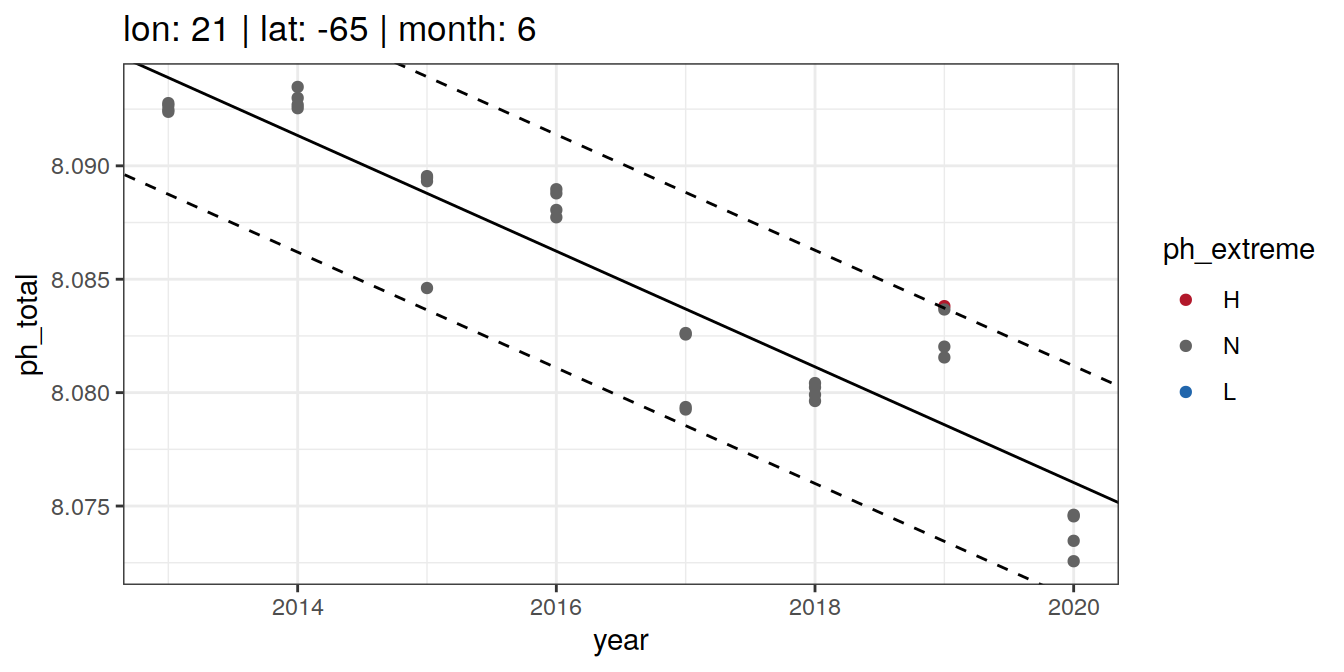
Anomaly maps
Location of OceanSODA pH extremes
OceanSODA_2x2_SO_extreme_grid %>%
group_split(year) %>%
head(1) %>%
map(
~ basemap(limits = -32, data = .x)+
geom_spatial_tile(data = .x,
aes(x = lon,
y = lat,
fill = ph_extreme),
linejoin = 'mitre',
col = 'transparent',
detail = 60
) +
scale_fill_manual(values = HNL_colors) +
facet_wrap(~month, ncol = 2)+
labs(title = paste("Year:", unique(.x$year)),
fill = 'pH')
)[[1]]
Anomaly time series
# calculate a regional mean pH for each biome, basin, and ph extreme (H/L/N) and plot a timeseries
OceanSODA_2x2_SO_extreme_grid <- left_join(
OceanSODA_2x2_SO_extreme_grid,
basinmask_2x2
)
OceanSODA_2x2_SO_extreme_grid <- left_join(
OceanSODA_2x2_SO_extreme_grid,
region_masks_all_2x2
)
OceanSODA_2x2_SO_extreme_grid %>%
group_by(year, biome, basin_AIP, ph_extreme) %>%
summarise(ph_regional = mean(ph_total, na.rm = TRUE)) %>%
ungroup() %>%
ggplot(aes(x = year, y = ph_regional, col = ph_extreme))+
geom_point(size = 0.3)+
geom_line()+
scale_color_manual(values = HNL_colors) +
facet_grid(basin_AIP~biome)+
theme(legend.position = 'bottom')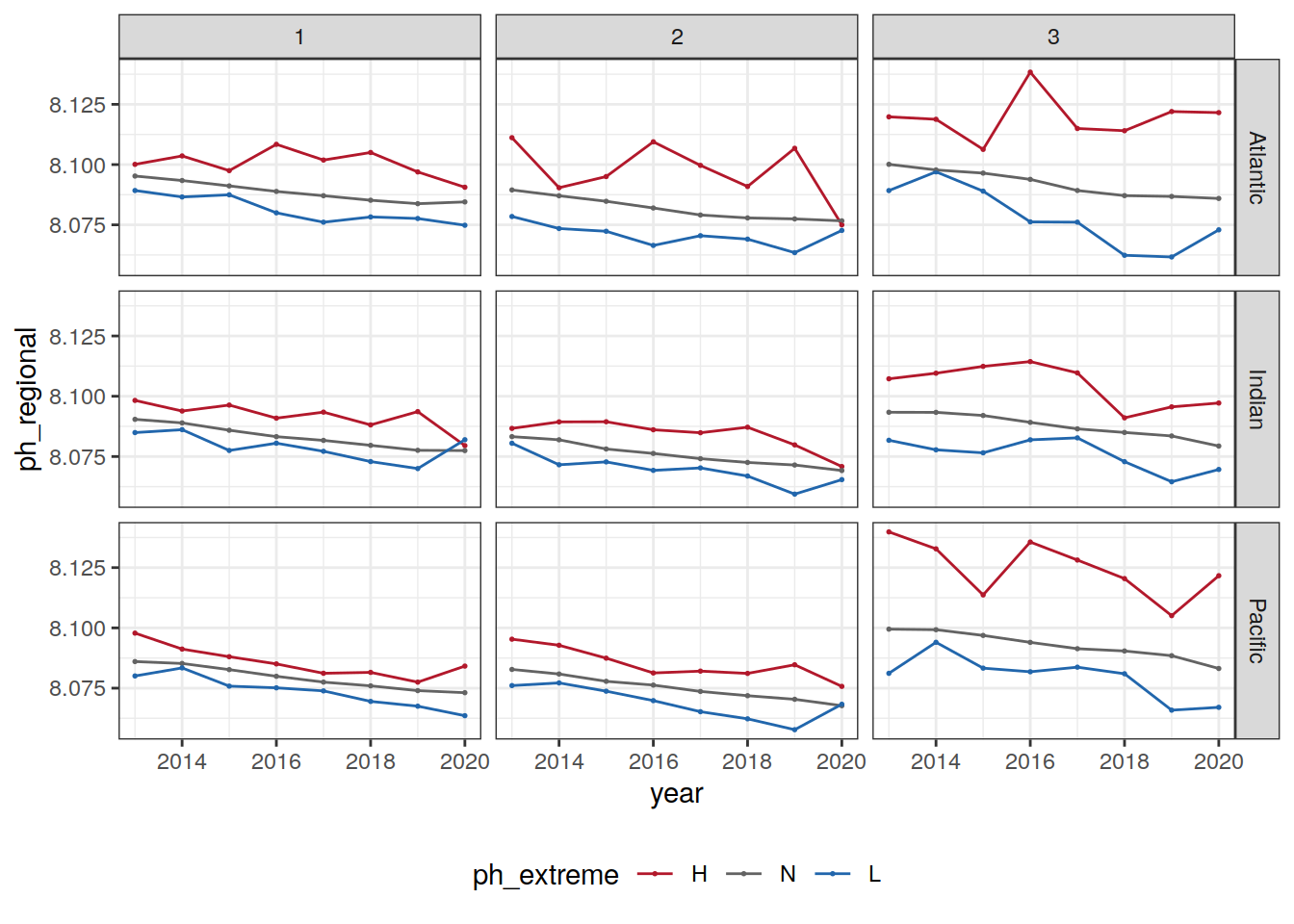
Anomaly histogram
OceanSODA_2x2_SO_extreme_grid %>%
ggplot(aes(ph_total, col = ph_extreme)) +
geom_density() +
scale_color_manual(values = HNL_colors) +
facet_grid(basin_AIP ~ biome) +
coord_cartesian(xlim = c(8, 8.2)) +
labs(x = 'value',
y = 'density',
col = 'pH anomaly') +
theme(legend.position = 'bottom')
Threshold histogram
OceanSODA_2x2_SO_extreme_grid %>%
mutate(ph_extreme = as.double(ph_extreme)) %>%
pivot_longer(starts_with("ph_"),
names_to = "level",
values_to = "value",
names_prefix = "ph_") %>%
distinct() %>%
ggplot(aes(value, col = level)) +
geom_density() +
scale_color_manual(values = HNL_colors, name = "threshold") +
coord_cartesian(xlim = c(8, 8.2)) +
lims(y = c(0, 230))+
theme(legend.position = 'bottom')
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 3ae43e4 | pasqualina-vonlanthendinenna | 2022-01-24 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| 587755e | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| ed3fef2 | jens-daniel-mueller | 2022-01-07 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
Argo
Grid reduction
# Note: While reducing lon x lat grid,
# we keep the original number of observations
full_argo_2x2 <- full_argo %>%
mutate(
lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
lat = as.numeric(as.character(lat)),
lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
lon = as.numeric(as.character(lon))) # re-grid to 2x2Apply region masks
# keep only Southern Ocean argo data
full_argo_2x2_SO <- inner_join(full_argo_2x2, region_masks_all_2x2)
# add in basin separations
full_argo_2x2_SO <- inner_join(full_argo_2x2_SO, basinmask_2x2)
# # remove duplicate rows (keep only distinct rows)
# full_argo_2x2_SO <- full_argo_2x2_SO %>%
# distinct()Join OceanSODA
# rename OceanSODA columns
OceanSODA_2x2_SO_extreme <- OceanSODA_2x2_SO_extreme_grid %>%
rename(OceanSODA_ph = ph_total)
# combine the argo profile data to the surface extreme data
profile_extreme <- inner_join(full_argo_2x2_SO, OceanSODA_2x2_SO_extreme)Plot profiles
Argo profiles plotted according to the surface OceanSODA pH
L profiles correspond to a surface acidification event (low pH), as recorded in OceanSODA
H profiles correspond to an event of high surface pH, as recorded in OceanSODA
N profiles correspond to normal surface OceanSODA pH
pH
profile_extreme %>%
group_split(biome, basin_AIP, year) %>%
head(1) %>%
map(
~ ggplot(
data = .x,
aes(
x = ph_in_situ_total_adjusted,
y = depth,
group = ph_extreme,
col = ph_extreme
)
) +
geom_point(pch = 19, size = 0.3) +
scale_y_reverse() +
scale_color_manual(values = HNL_colors) +
facet_wrap(~ month, ncol = 6) +
labs(
x = 'Argo pH (total scale)',
y = 'depth (m)',
title = paste(
unique(.x$basin_AIP),
"|",
unique(.x$year),
"| biome:",
unique(.x$biome)
),
col = 'OceanSODA pH \nanomaly'
)
)[[1]]
Plot monthly profiles
# calculate mean profiles in each basin and biome, for each month between 2014 and 2021
# cut depth levels at 10, 20, .... etc m
# add seasons
# Dec, Jan, Feb <- summer
# Mar, Apr, May <- autumn
# Jun, Jul, Aug <- winter
# Sep, Oct, Nov <- spring
profile_extreme_monthly <- profile_extreme %>%
mutate(
depth = Hmisc::cut2(
depth,
cuts = c(10, 20, 30, 50, 70, 100, 300, 500, 800, 1000, 1500, 2000, 2500),
m = 5,
levels.mean = TRUE
),
depth = as.numeric(as.character(depth))
) %>%
mutate(
season = case_when(
between(month, 3, 5) ~ 'autumn',
between(month, 6, 8) ~ 'winter',
between(month, 9, 11) ~ 'spring',
month == 12 | 1 | 2 ~ 'summer'
),
.after = date
) %>%
group_by(season, biome, basin_AIP, ph_extreme, depth) %>%
summarise(
ph_mean = mean(ph_in_situ_total_adjusted, na.rm = TRUE),
temp_mean = mean(temp_adjusted, na.rm = TRUE)
) %>%
ungroup()pH
By season
profile_extreme_monthly %>%
arrange(depth) %>%
group_split(season) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(
x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme
)) +
geom_path() +
scale_color_manual(values = HNL_colors) +
labs(title = paste("season:", unique(.x$season)),
col = 'OceanSODA\npH\nanomaly') +
scale_y_reverse() +
facet_grid(basin_AIP ~ biome)
)[[1]]
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
[[2]]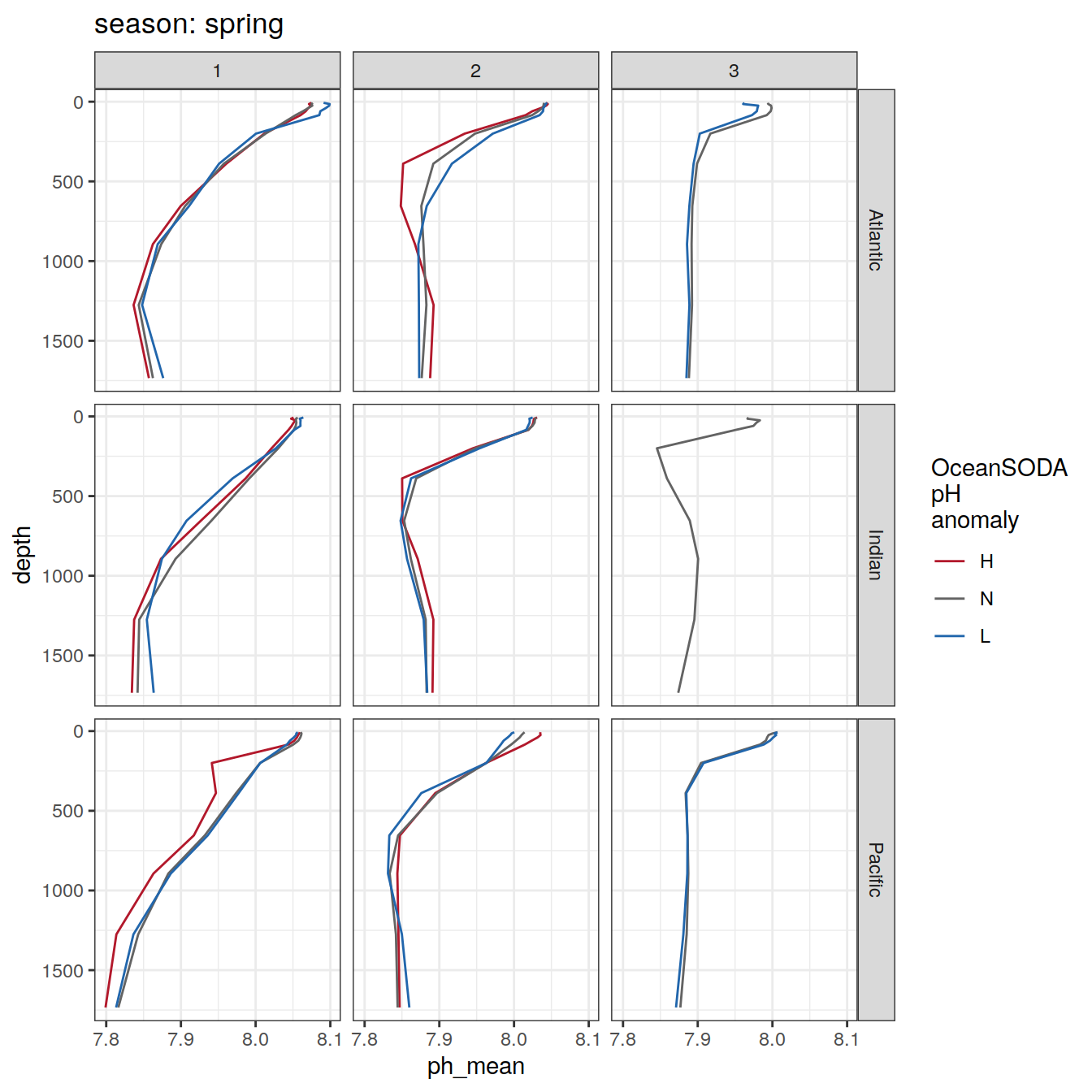
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
[[3]]
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
[[4]]
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
Averaged over biomes
profile_extreme_biome <- profile_extreme_monthly %>%
group_by(season, biome, ph_extreme, depth) %>%
summarise(ph_biome = mean(ph_mean, na.rm = TRUE)) %>%
ungroup()
profile_extreme_biome %>%
ggplot(aes(
x = ph_biome,
y = depth,
group = ph_extreme,
col = ph_extreme
)) +
geom_path() +
scale_color_manual(values = HNL_colors) +
labs(col = 'OceanSODA\npH\nanomaly') +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500))) +
facet_grid(season ~ biome)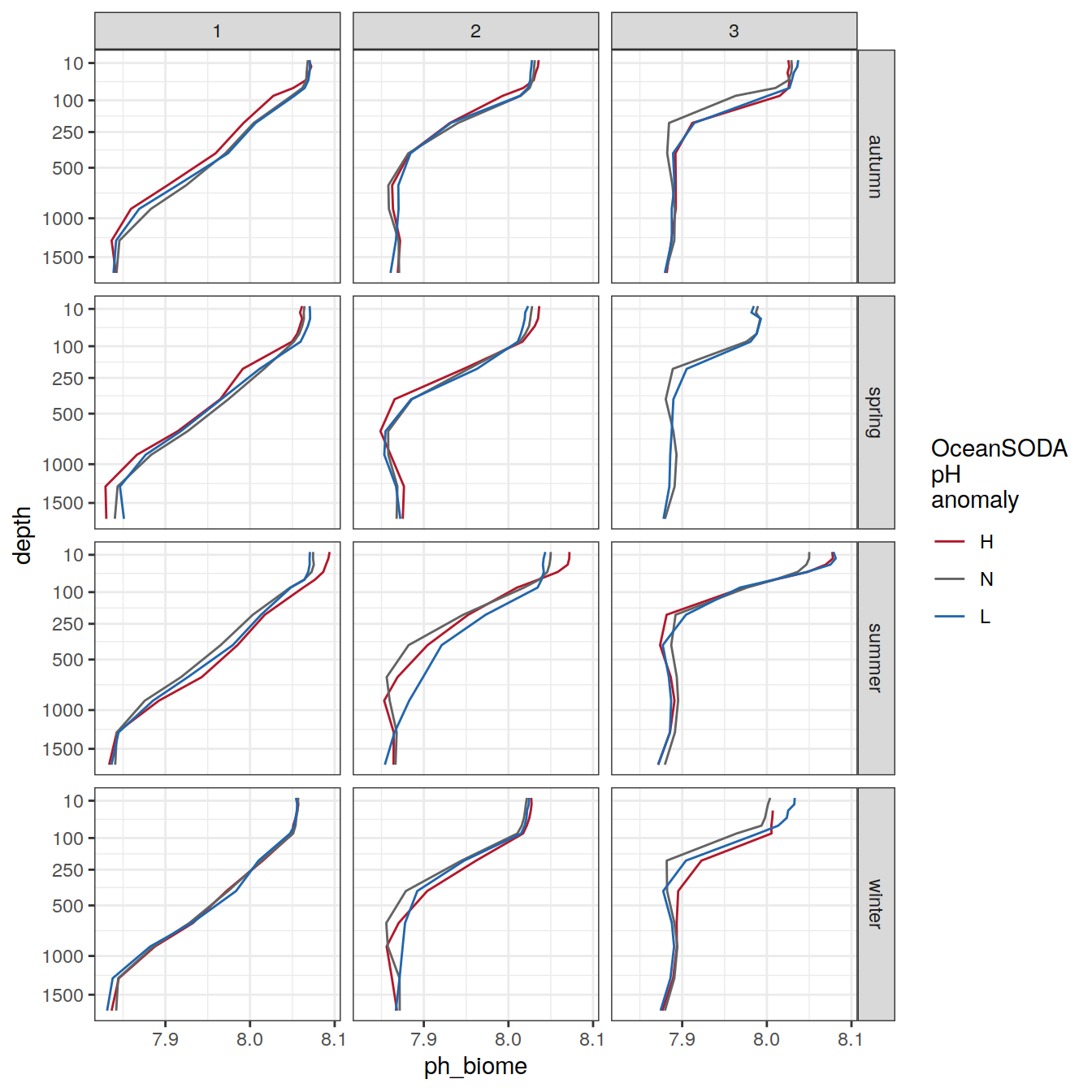
| Version | Author | Date |
|---|---|---|
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
Averaged over ocean basins
profile_extreme_basin <- profile_extreme_monthly %>%
group_by(season, basin_AIP, ph_extreme, depth) %>%
summarise(ph_basin = mean(ph_mean, na.rm = TRUE)) %>%
ungroup()
profile_extreme_basin %>%
ggplot(aes(x = ph_basin,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_path()+
scale_color_manual(values = HNL_colors)+
labs(col = 'OceanSODA\npH\nanomaly')+
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500))) +
facet_grid(season~basin_AIP)
| Version | Author | Date |
|---|---|---|
| c44ff0f | pasqualina-vonlanthendinenna | 2022-01-25 |
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.3
Matrix products: default
BLAS: /usr/local/R-4.1.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.1.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggforce_0.3.3 metR_0.11.0 scico_1.3.0 ggOceanMaps_1.2.6
[5] ggspatial_1.1.5 broom_0.7.11 lubridate_1.8.0 forcats_0.5.1
[9] stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[13] tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1
[17] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.0-2 ellipsis_0.3.2 class_7.3-20
[4] rgdal_1.5-28 rprojroot_2.0.2 htmlTable_2.4.0
[7] base64enc_0.1-3 fs_1.5.2 rstudioapi_0.13
[10] proxy_0.4-26 farver_2.1.0 bit64_4.0.5
[13] fansi_1.0.2 xml2_1.3.3 codetools_0.2-18
[16] splines_4.1.2 knitr_1.37 polyclip_1.10-0
[19] Formula_1.2-4 jsonlite_1.7.3 cluster_2.1.2
[22] dbplyr_2.1.1 png_0.1-7 rgeos_0.5-9
[25] ggOceanMapsData_1.0.1 compiler_4.1.2 httr_1.4.2
[28] backports_1.4.1 assertthat_0.2.1 Matrix_1.4-0
[31] fastmap_1.1.0 cli_3.1.1 later_1.3.0
[34] tweenr_1.0.2 htmltools_0.5.2 tools_4.1.2
[37] gtable_0.3.0 glue_1.6.0 Rcpp_1.0.8
[40] cellranger_1.1.0 jquerylib_0.1.4 raster_3.5-11
[43] vctrs_0.3.8 xfun_0.29 ps_1.6.0
[46] rvest_1.0.2 lifecycle_1.0.1 terra_1.5-12
[49] getPass_0.2-2 MASS_7.3-55 scales_1.1.1
[52] vroom_1.5.7 hms_1.1.1 promises_1.2.0.1
[55] parallel_4.1.2 RColorBrewer_1.1-2 yaml_2.2.1
[58] gridExtra_2.3 sass_0.4.0 rpart_4.1-15
[61] latticeExtra_0.6-29 stringi_1.7.6 highr_0.9
[64] e1071_1.7-9 checkmate_2.0.0 rlang_0.4.12
[67] pkgconfig_2.0.3 evaluate_0.14 lattice_0.20-45
[70] sf_1.0-5 htmlwidgets_1.5.4 labeling_0.4.2
[73] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1
[76] magrittr_2.0.1 R6_2.5.1 generics_0.1.1
[79] Hmisc_4.6-0 DBI_1.1.2 foreign_0.8-82
[82] pillar_1.6.4 haven_2.4.3 whisker_0.4
[85] withr_2.4.3 units_0.7-2 nnet_7.3-17
[88] survival_3.2-13 sp_1.4-6 modelr_0.1.8
[91] crayon_1.4.2 KernSmooth_2.23-20 utf8_1.2.2
[94] tzdb_0.2.0 rmarkdown_2.11 jpeg_0.1-9
[97] grid_4.1.2 readxl_1.3.1 data.table_1.14.2
[100] callr_3.7.0 git2r_0.29.0 reprex_2.0.1
[103] digest_0.6.29 classInt_0.4-3 httpuv_1.6.5
[106] munsell_0.5.0 bslib_0.3.1