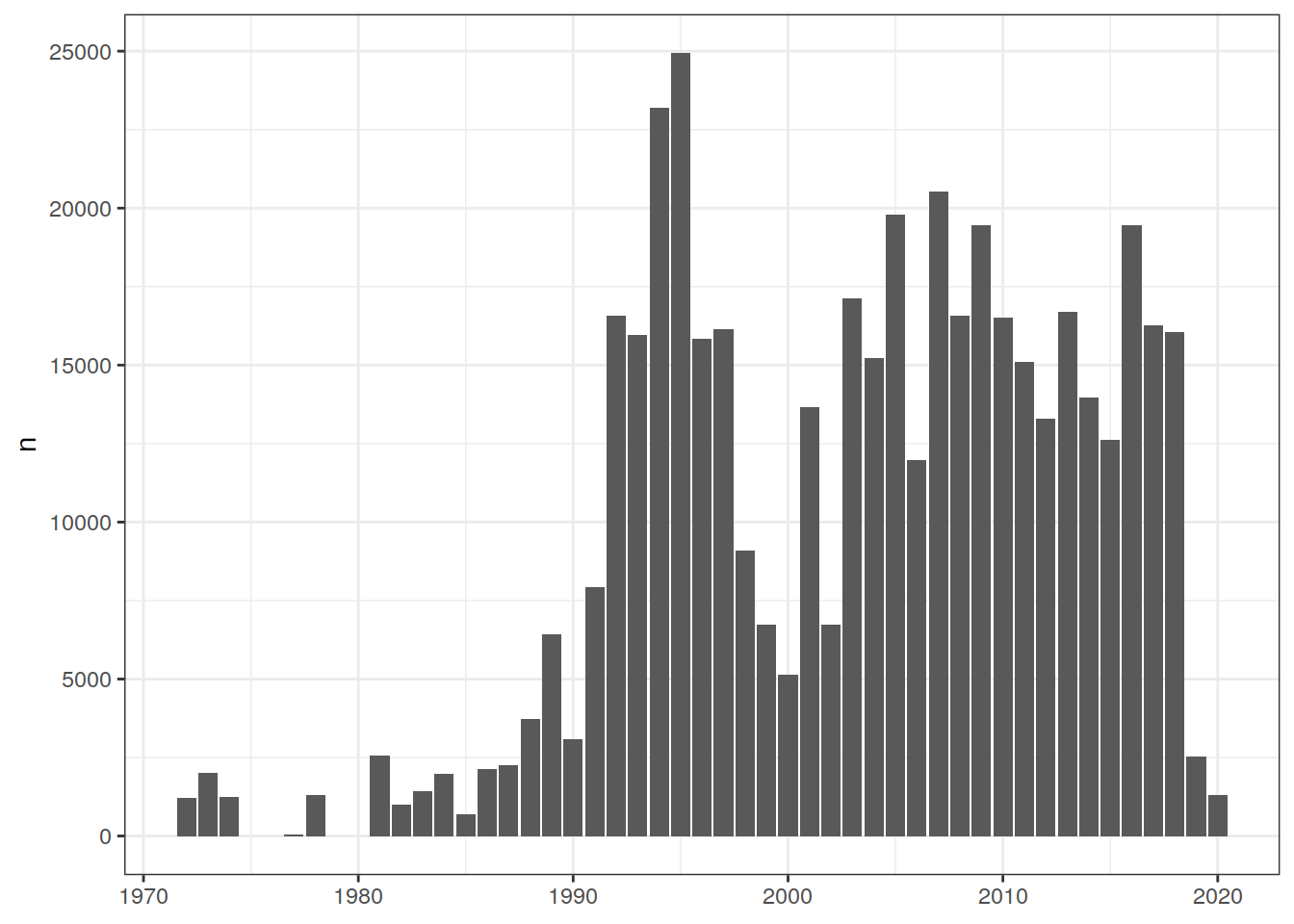
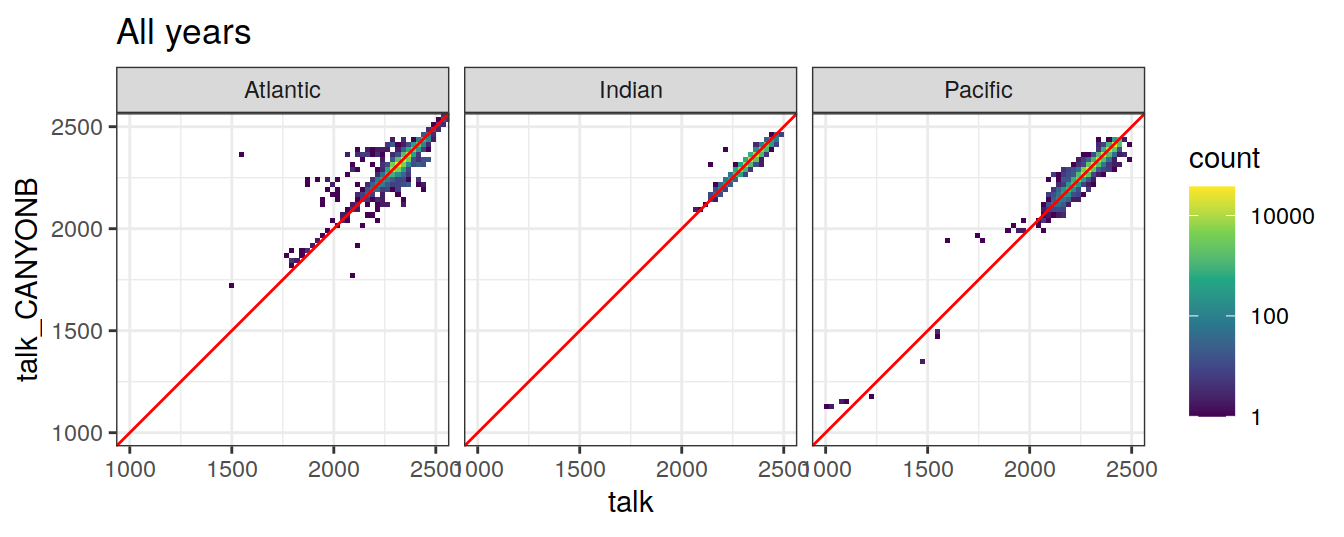
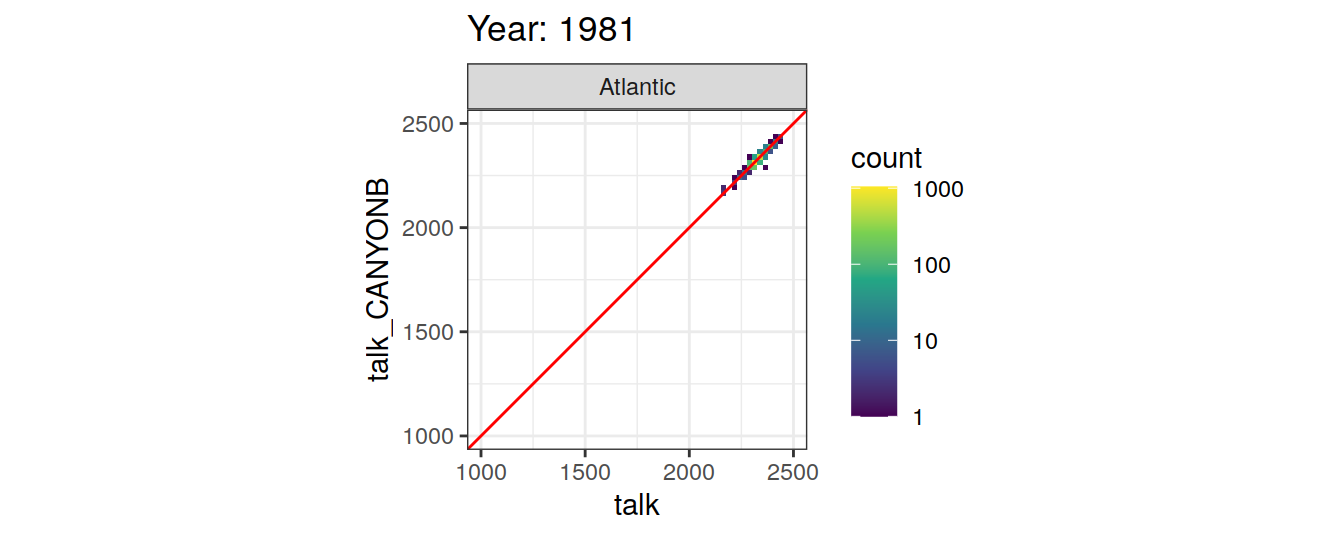
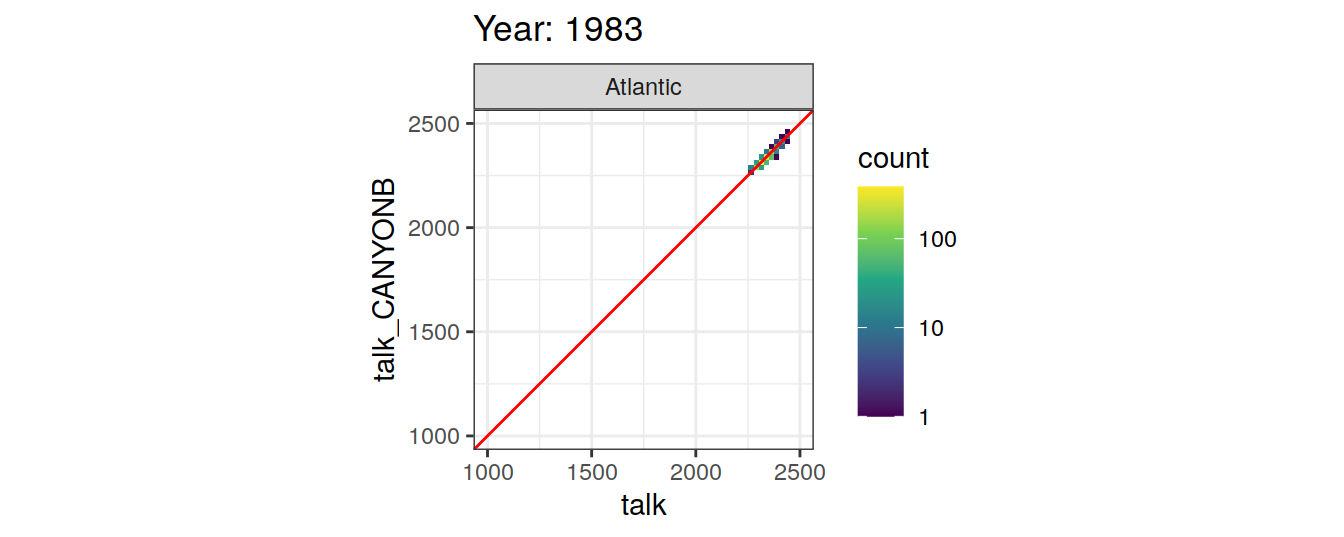
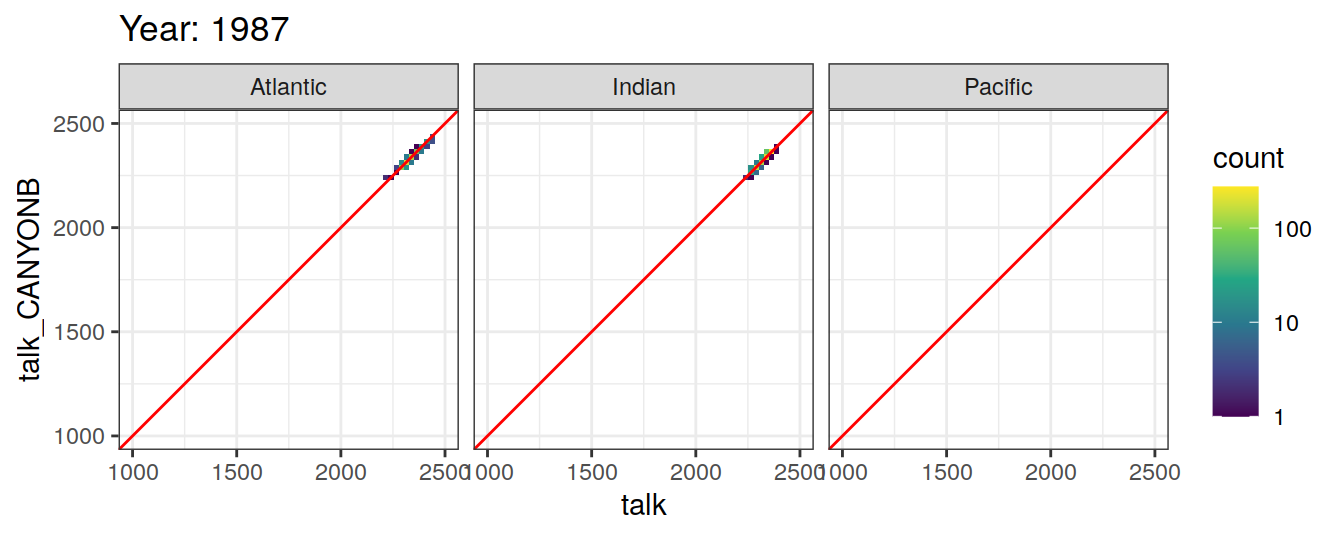
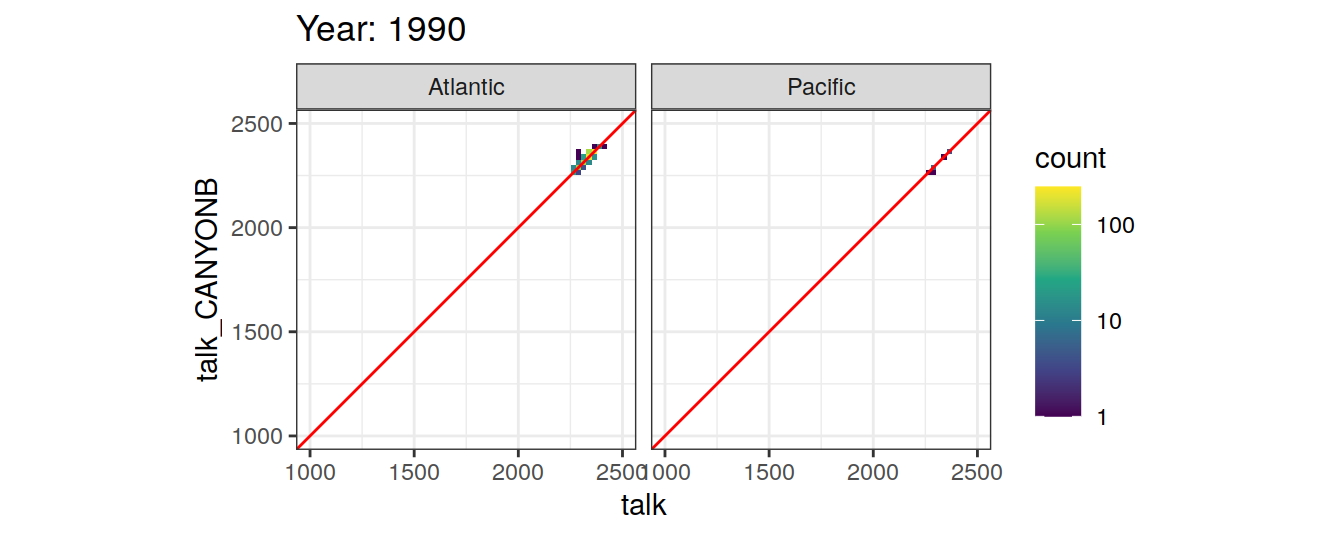
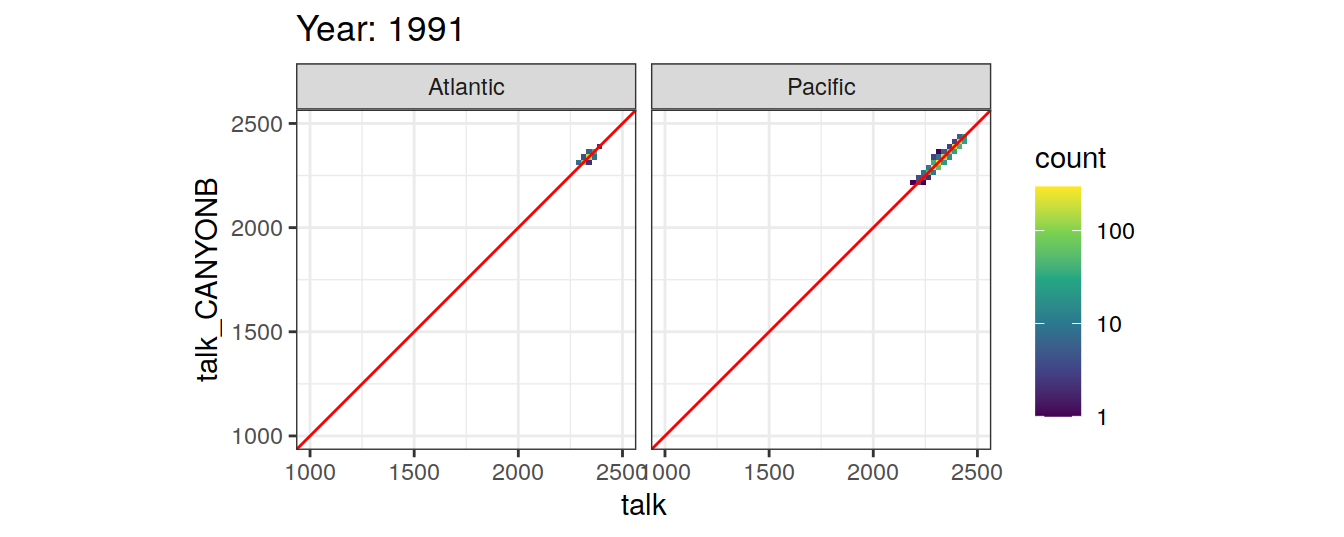
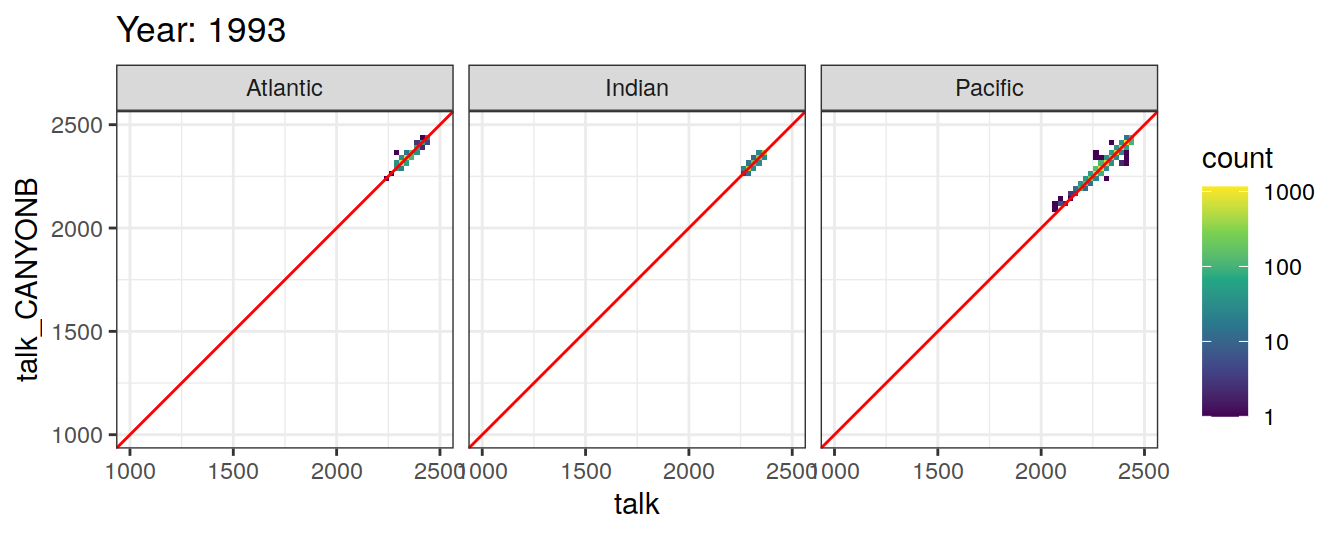
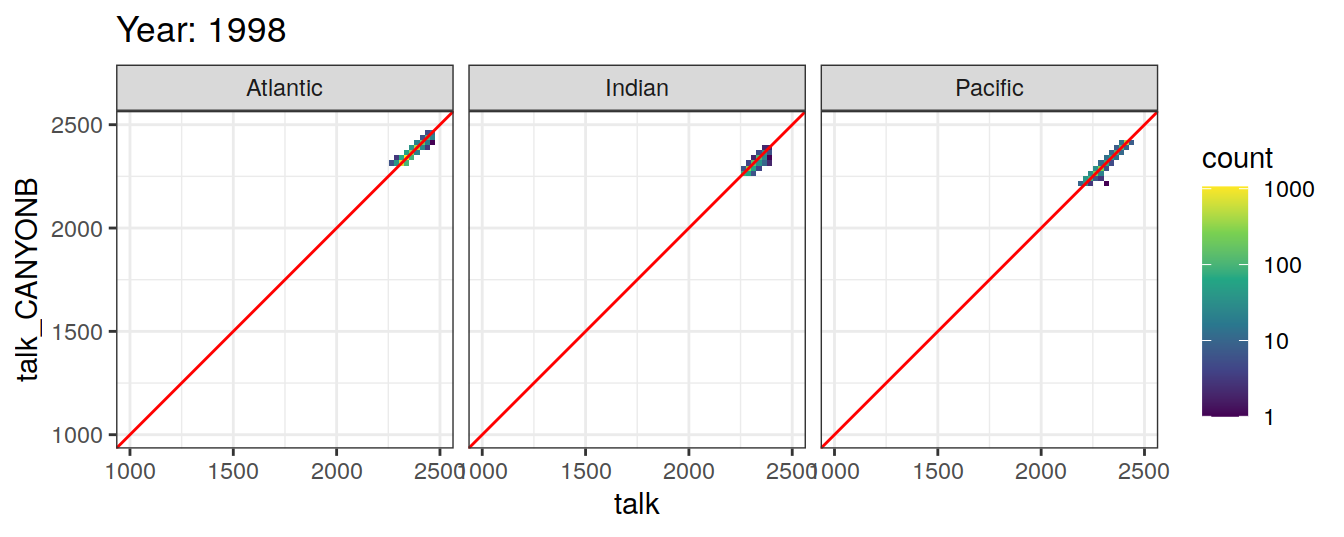
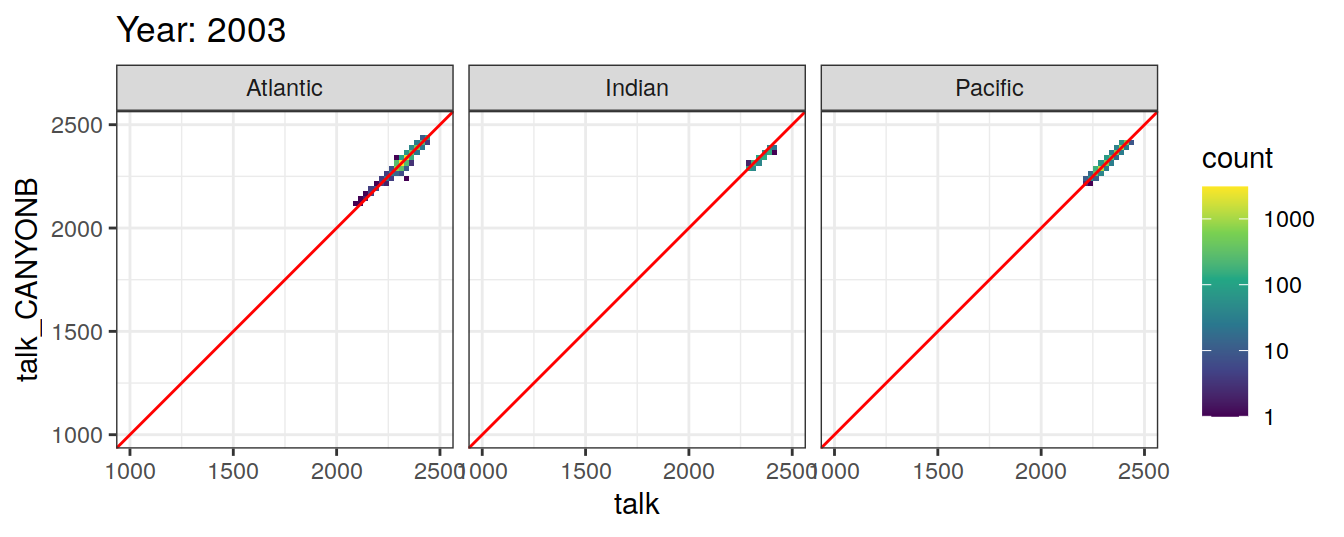
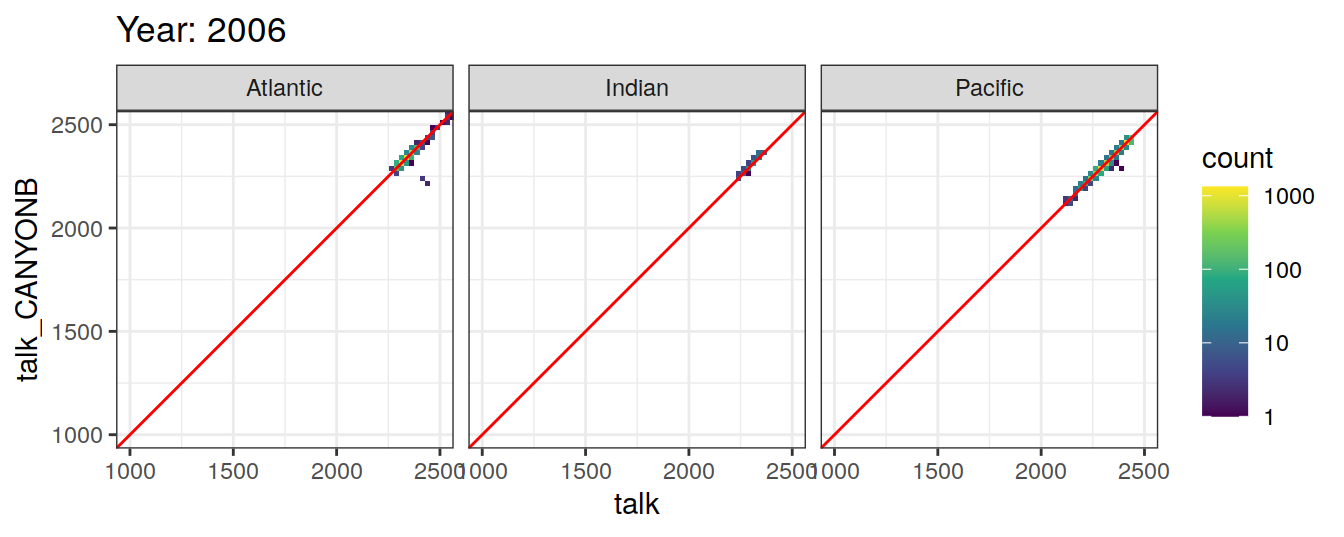
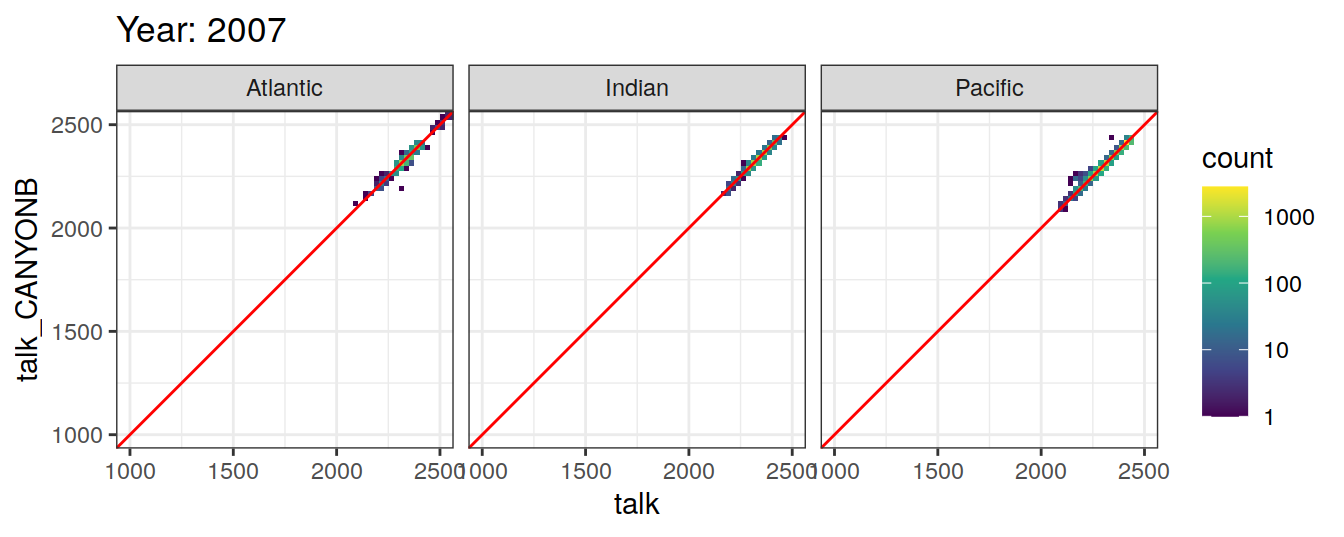
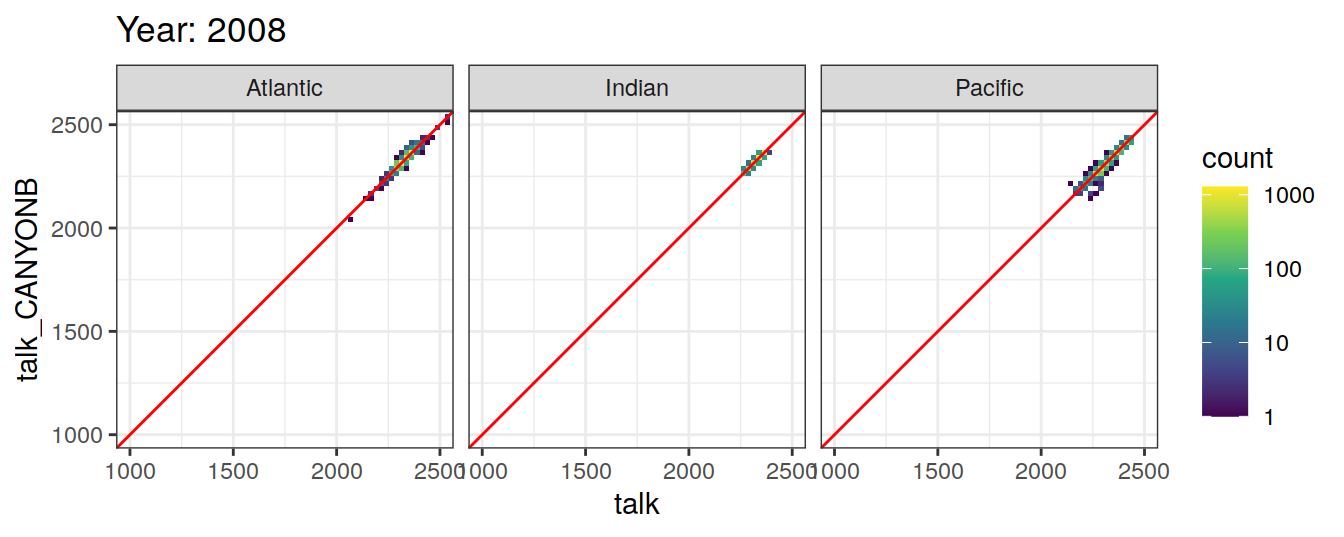
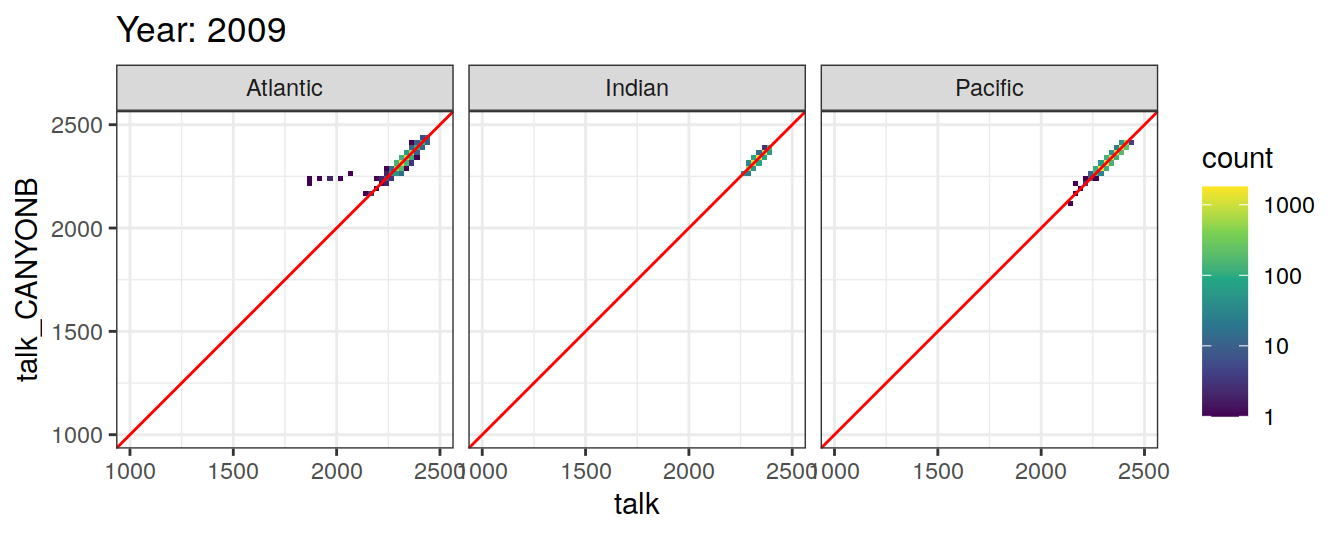
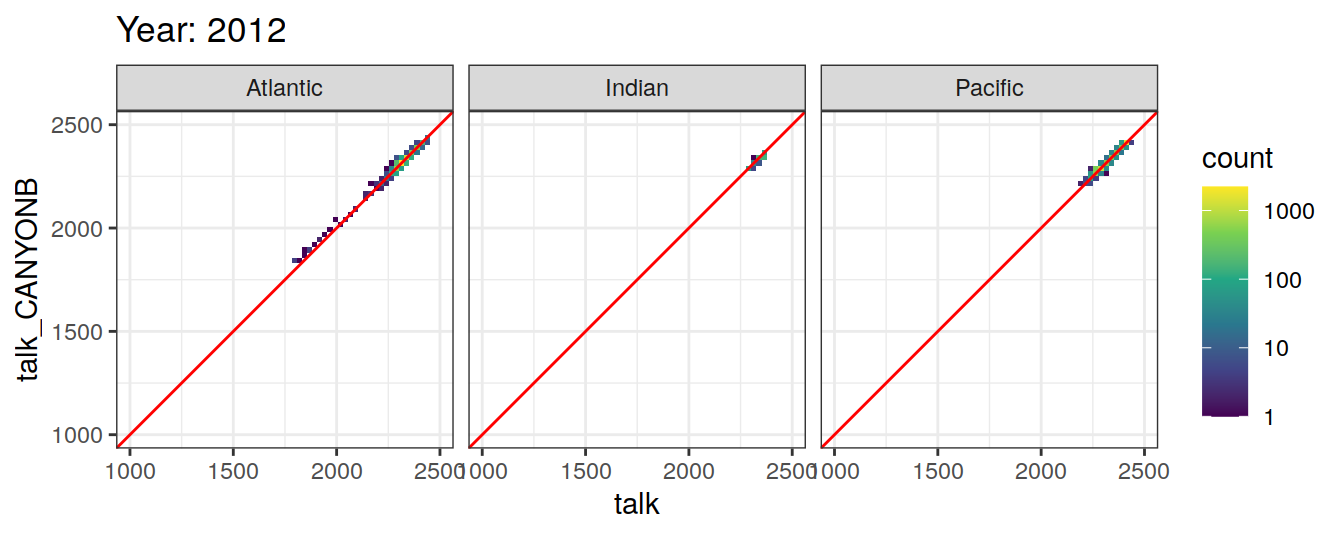
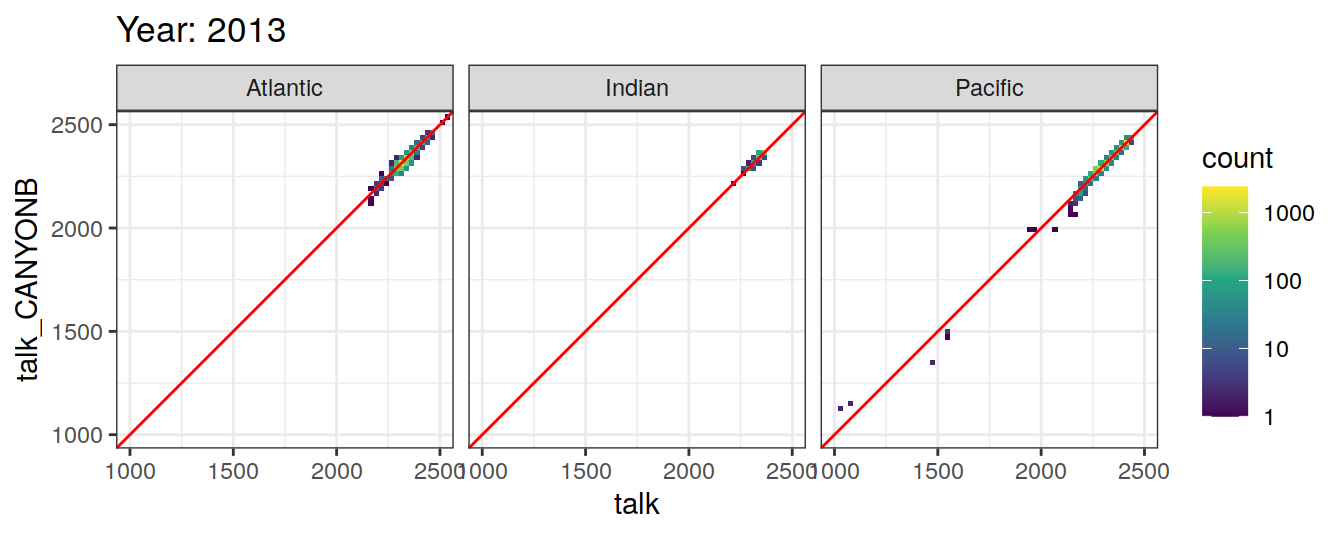
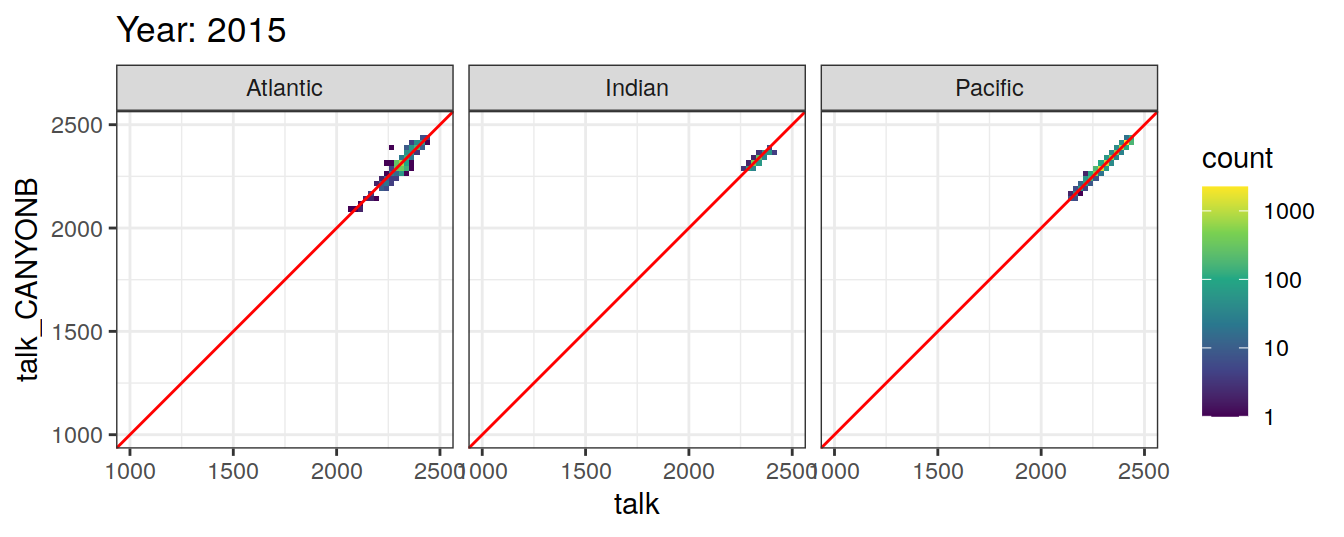
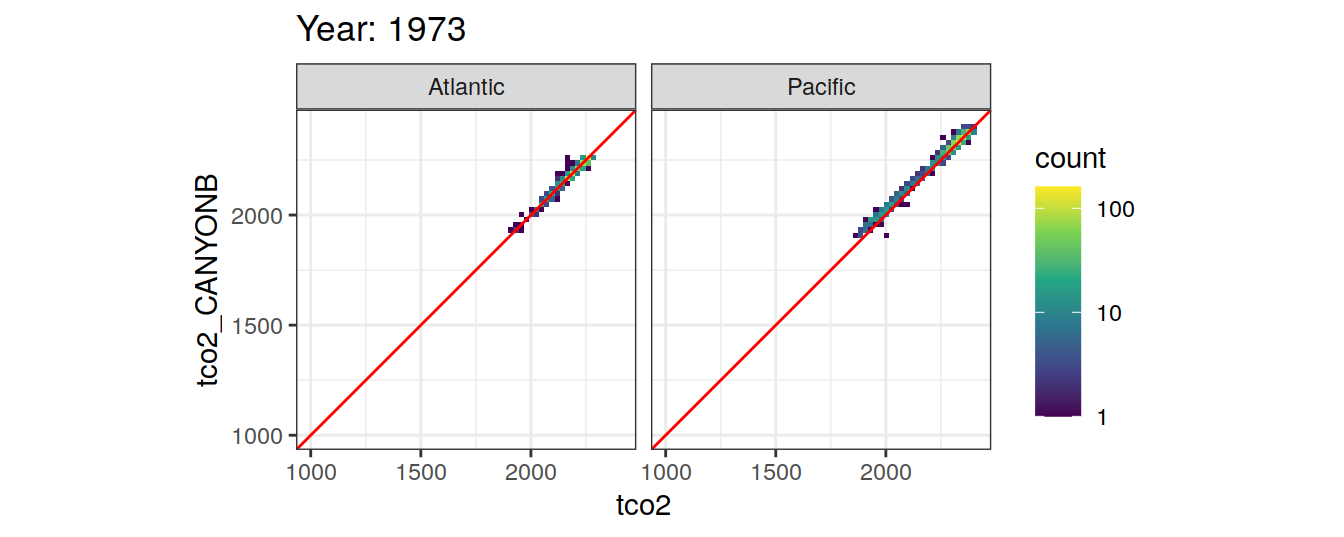
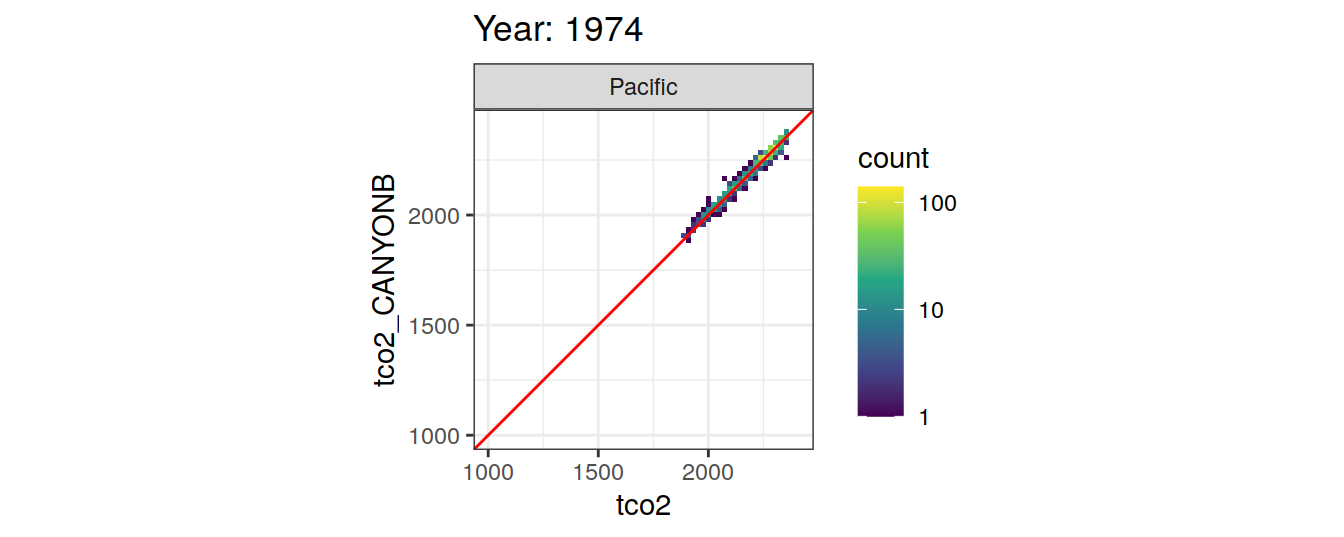
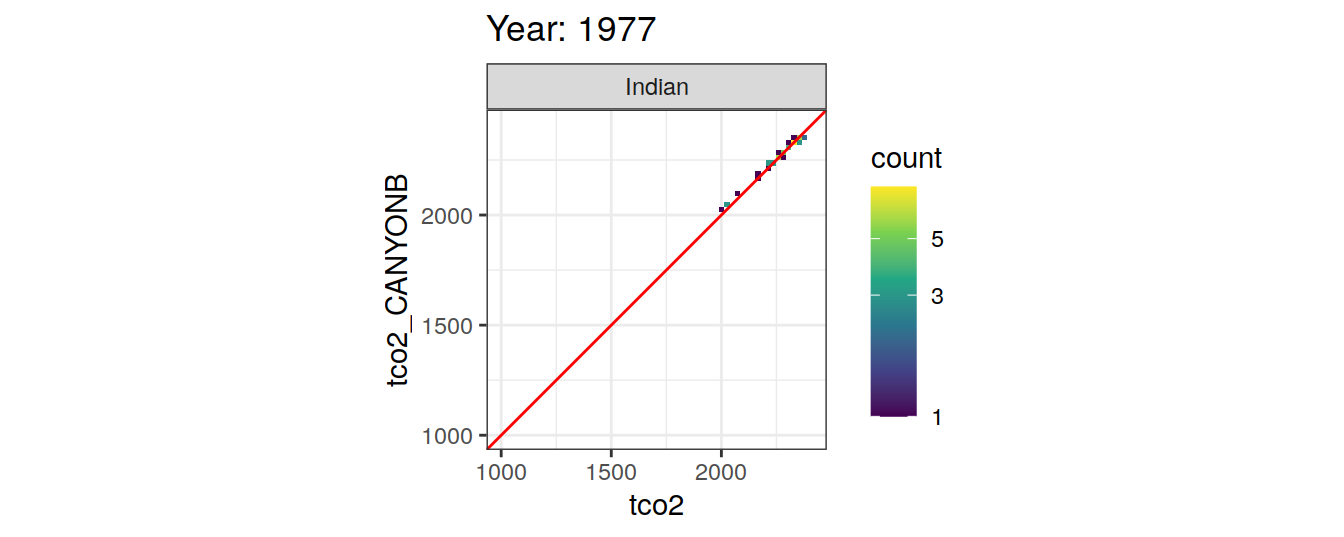
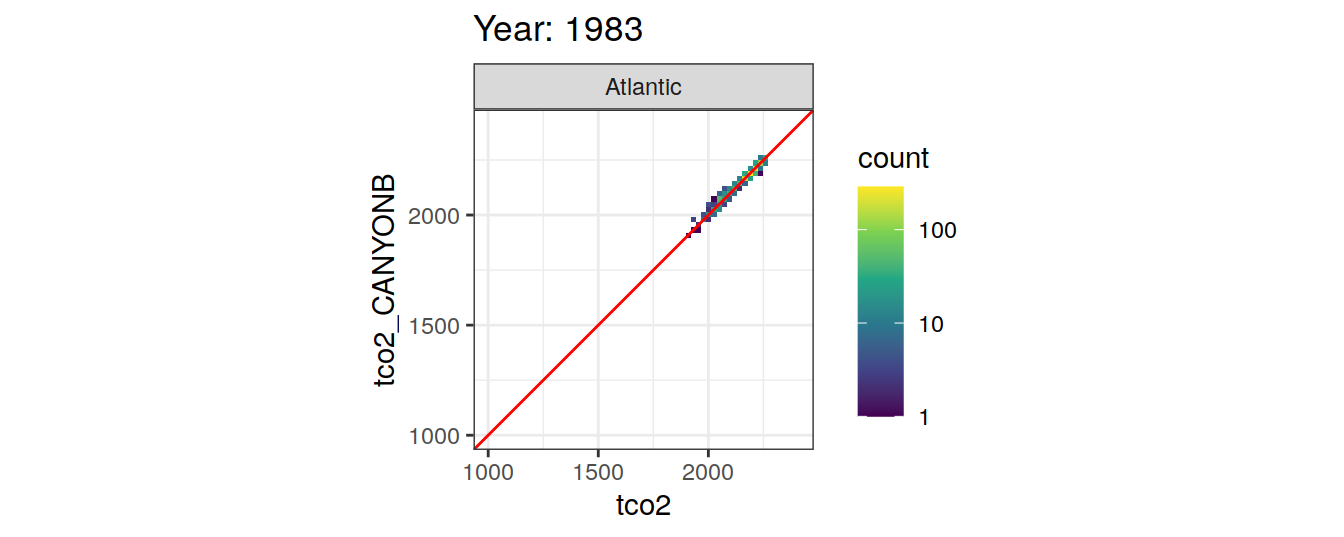
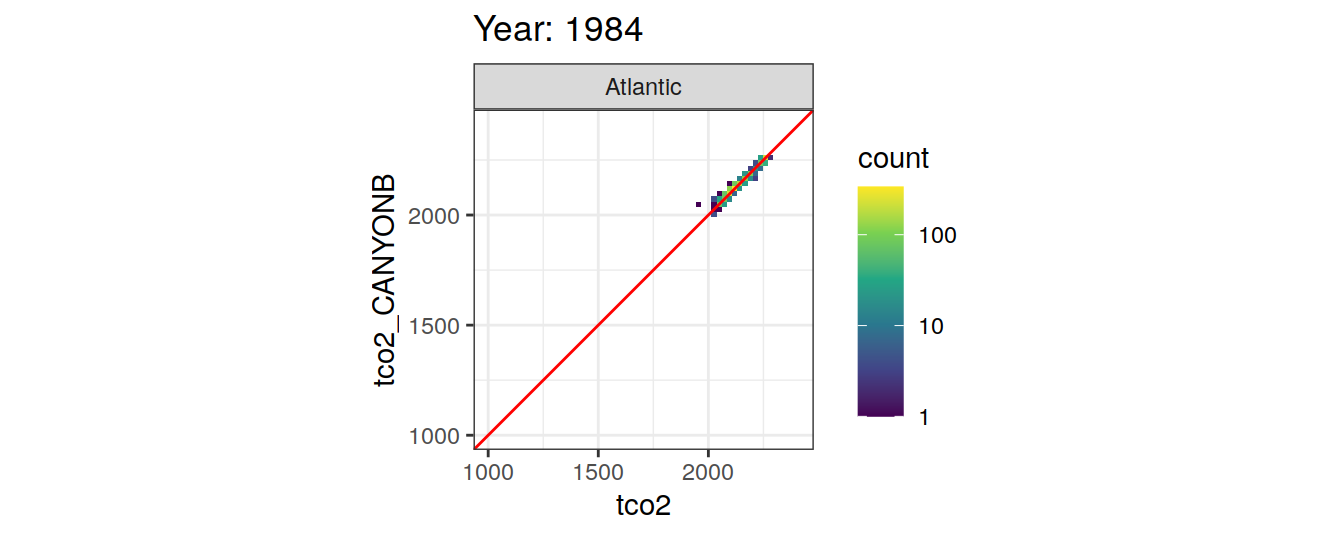
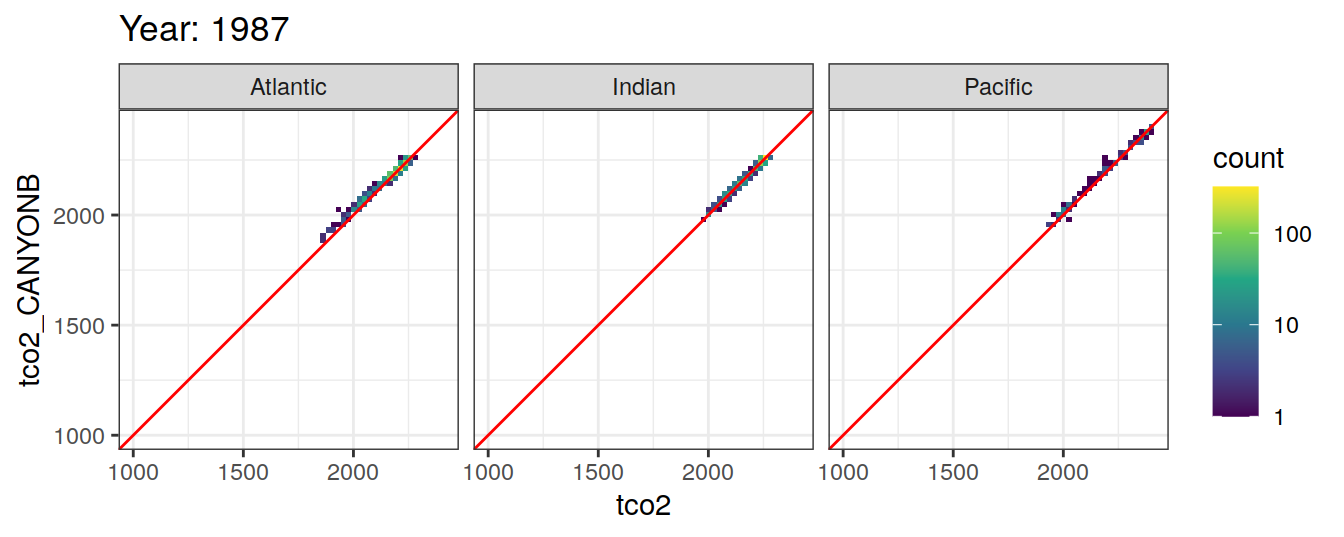
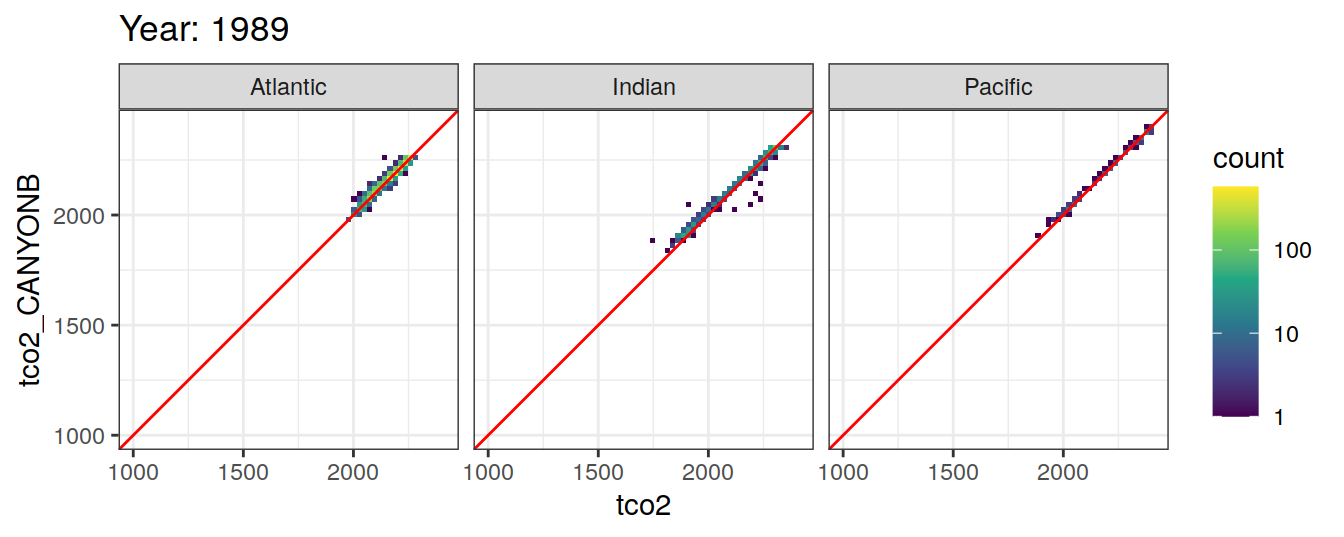
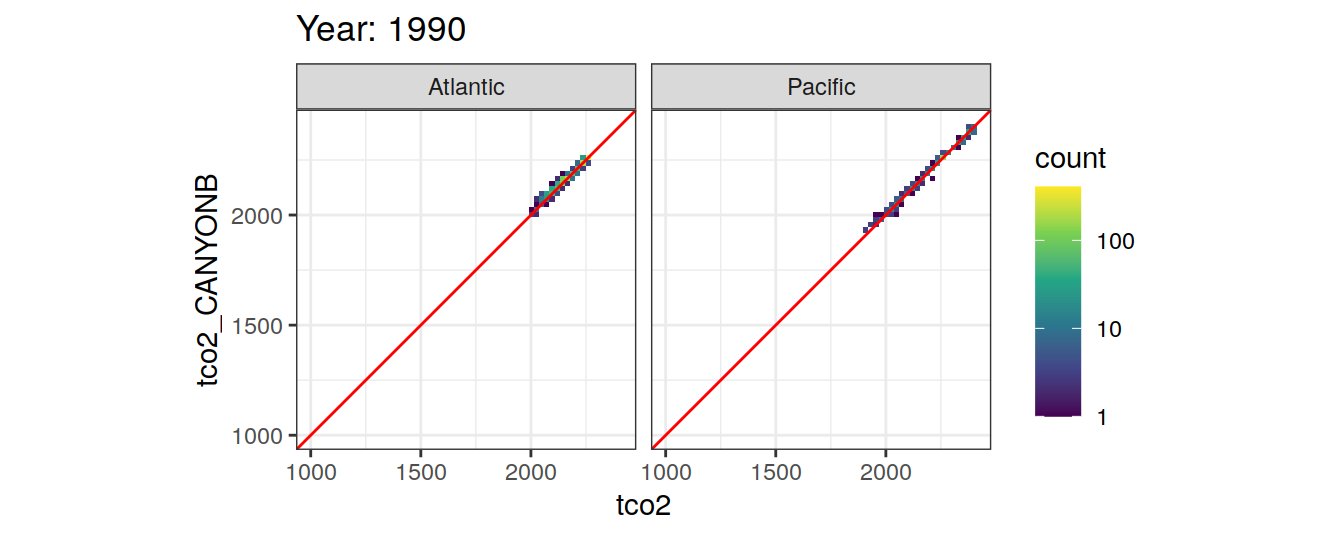
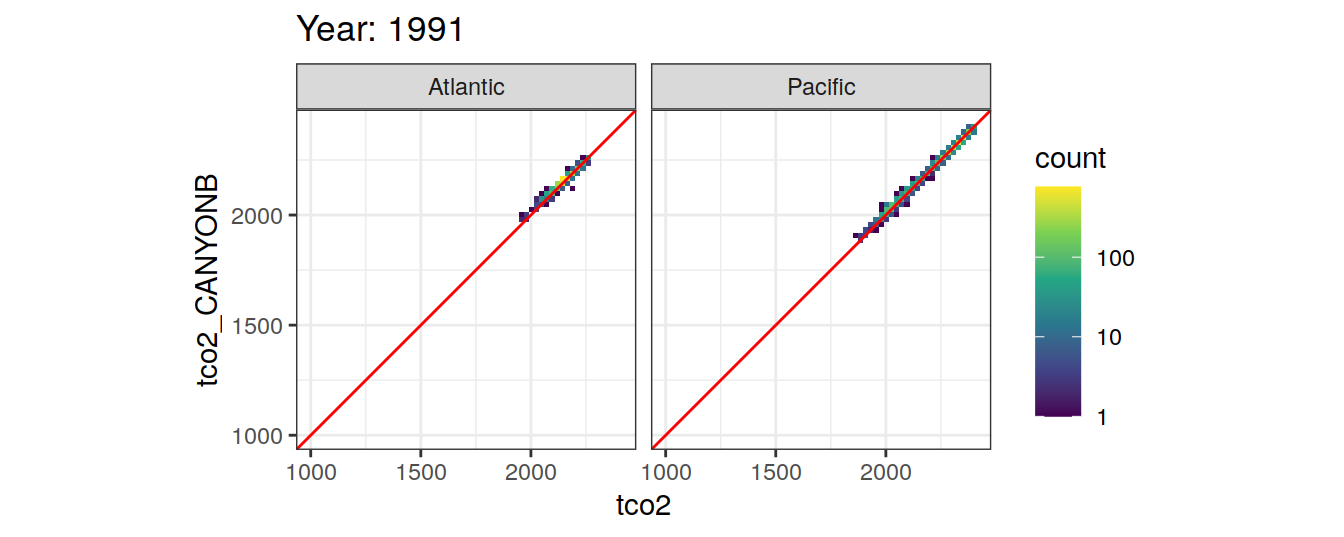
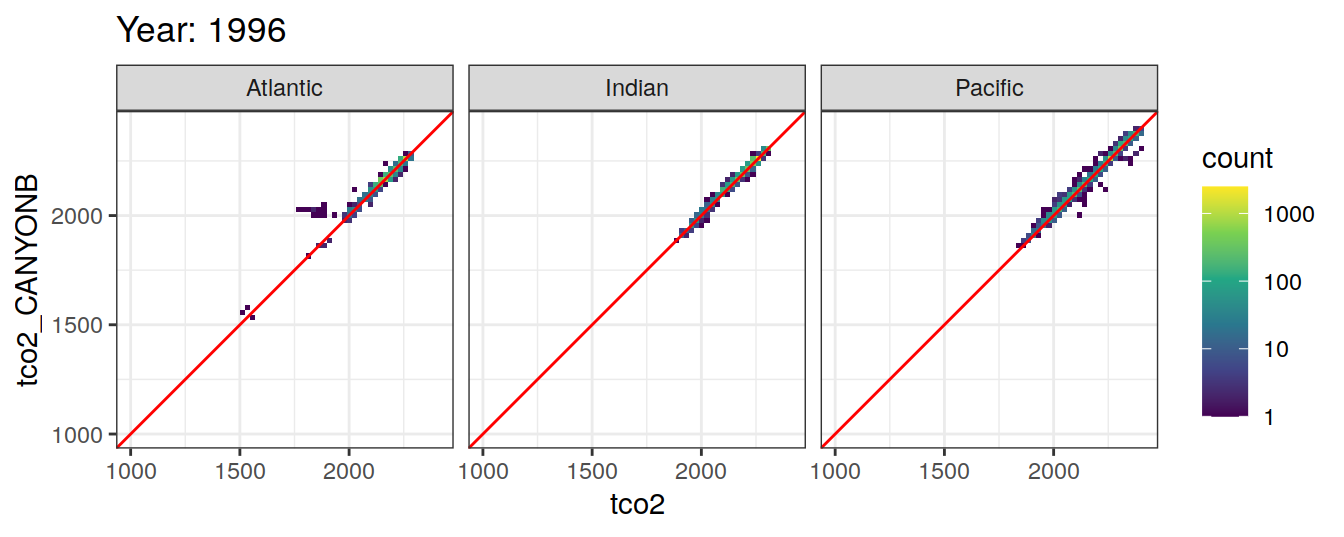
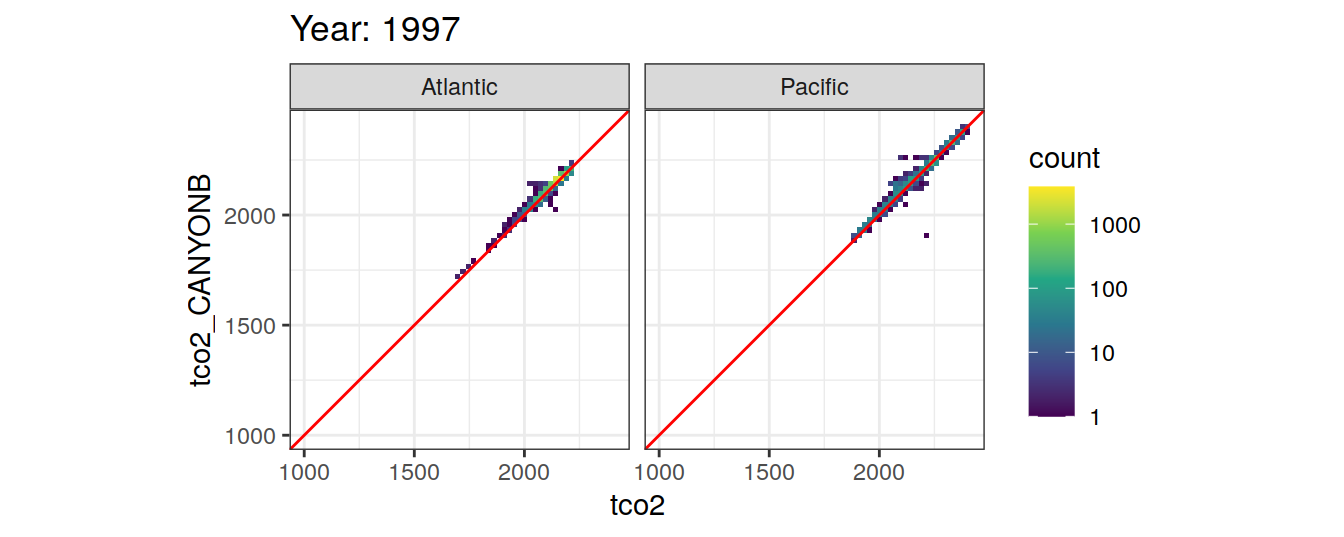
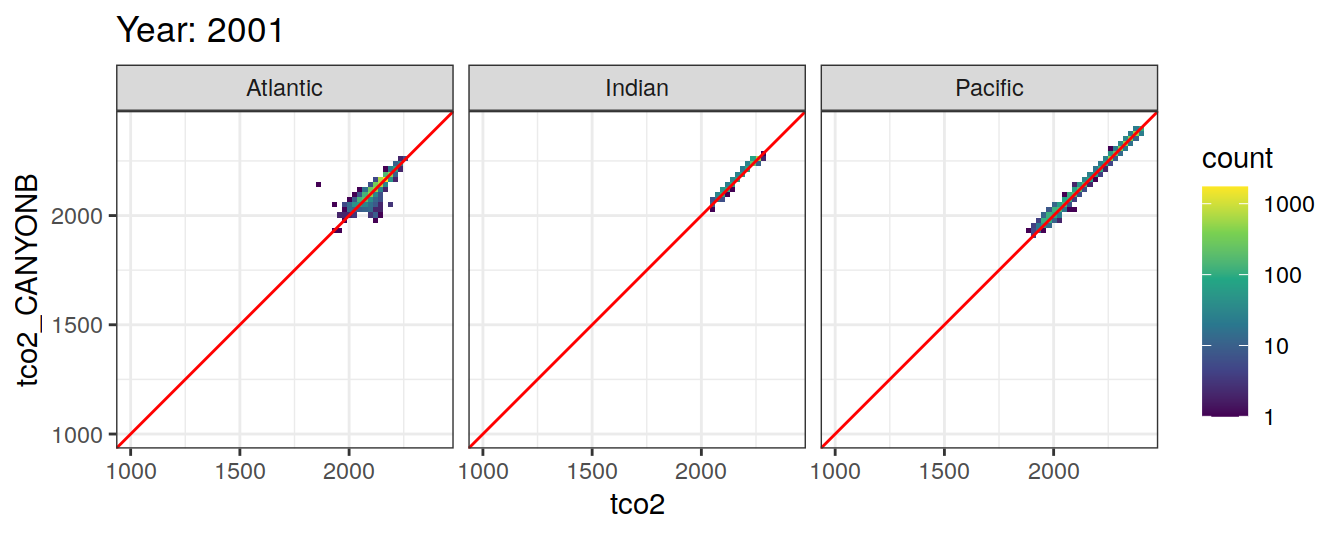
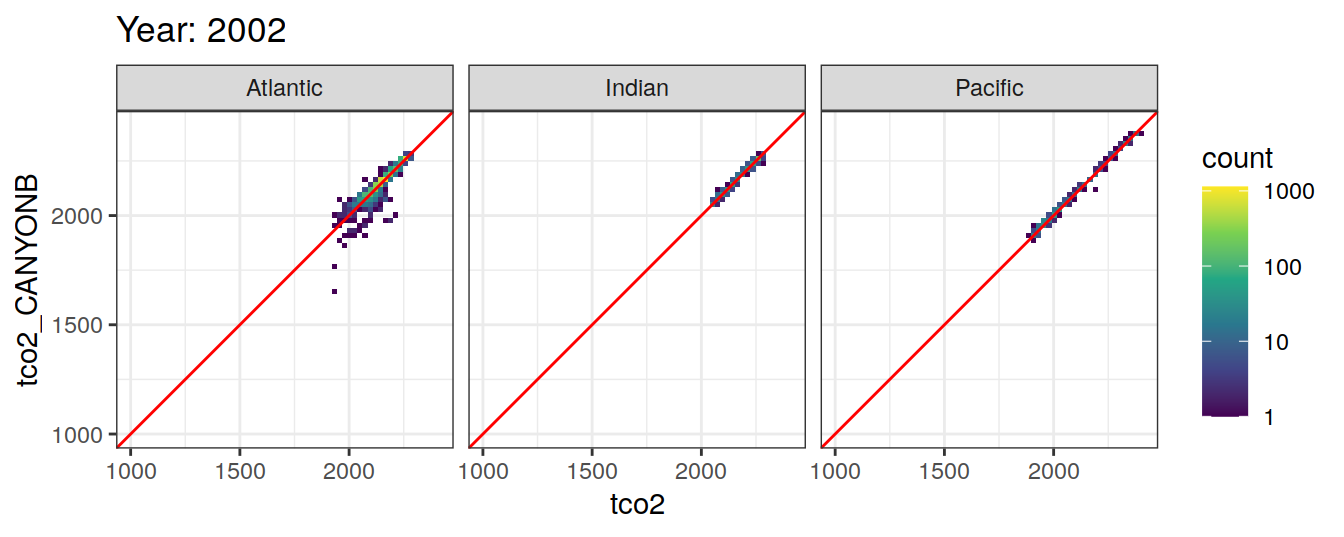
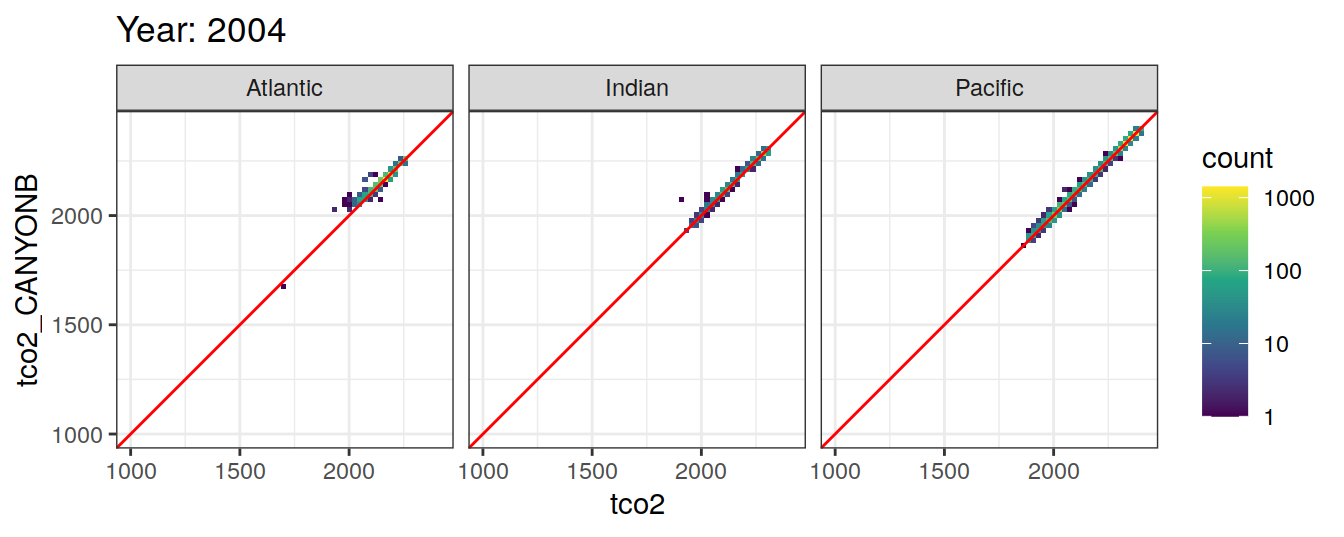
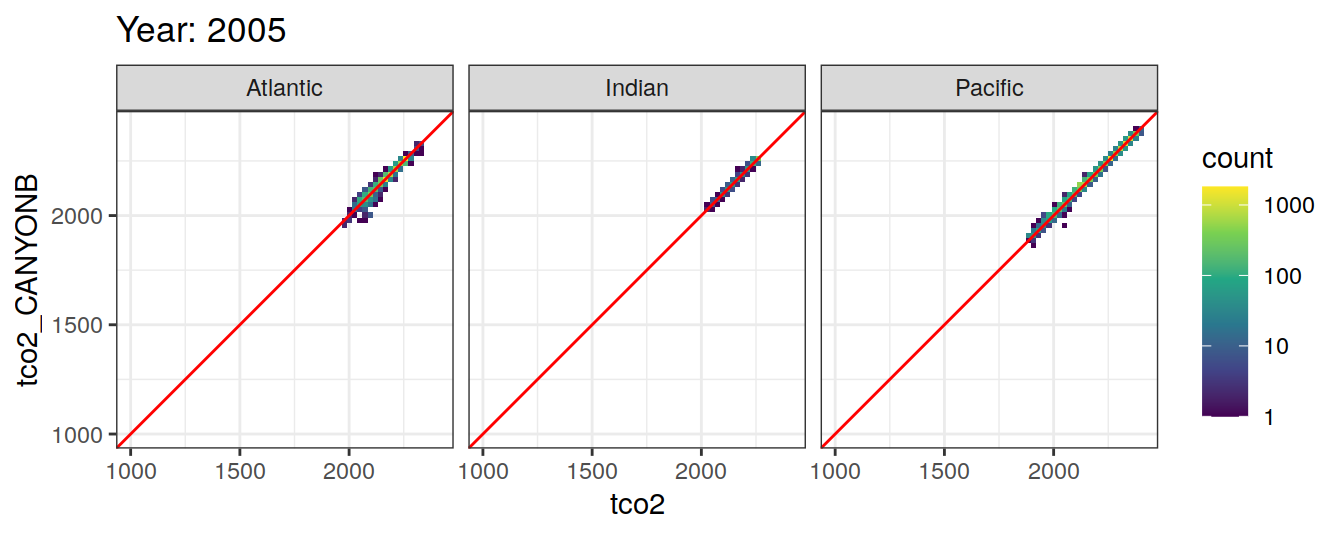
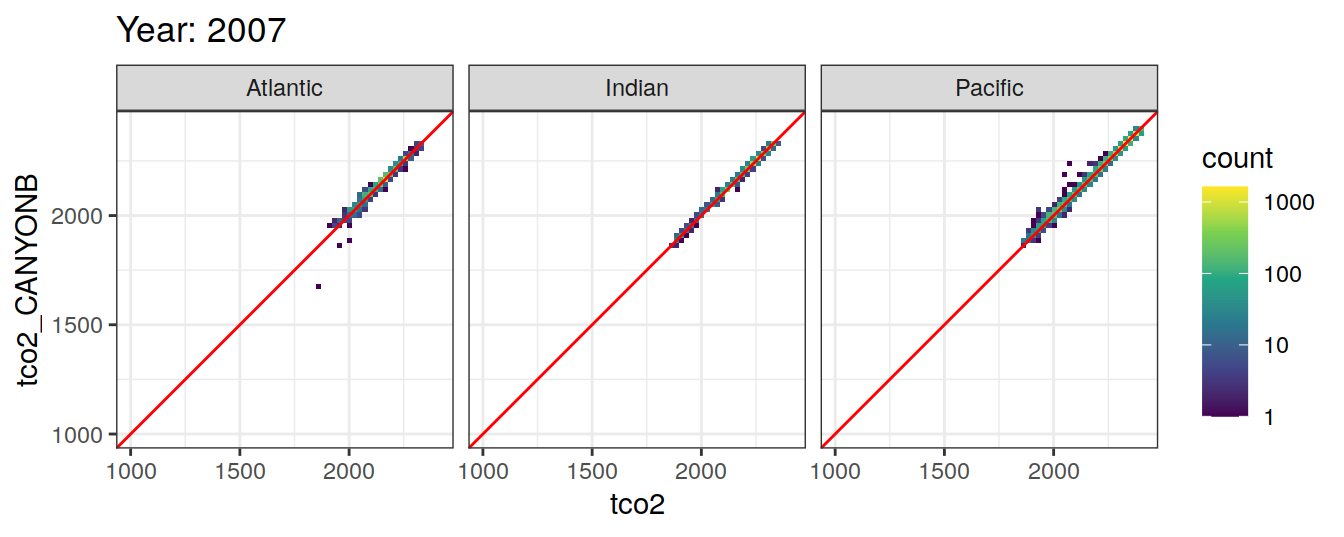
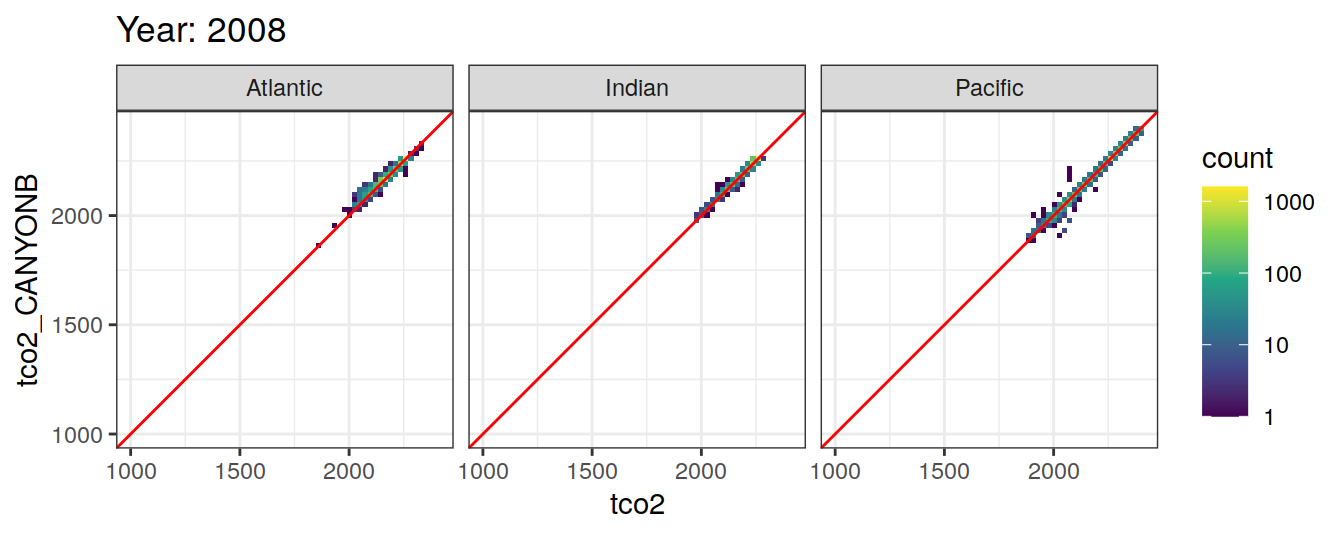
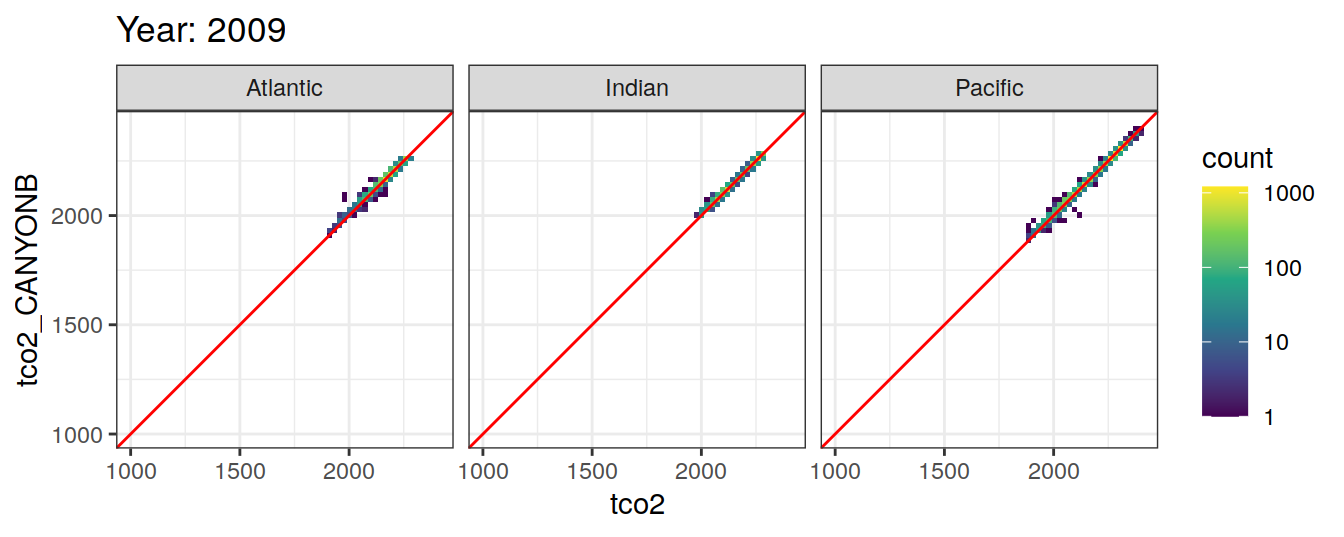
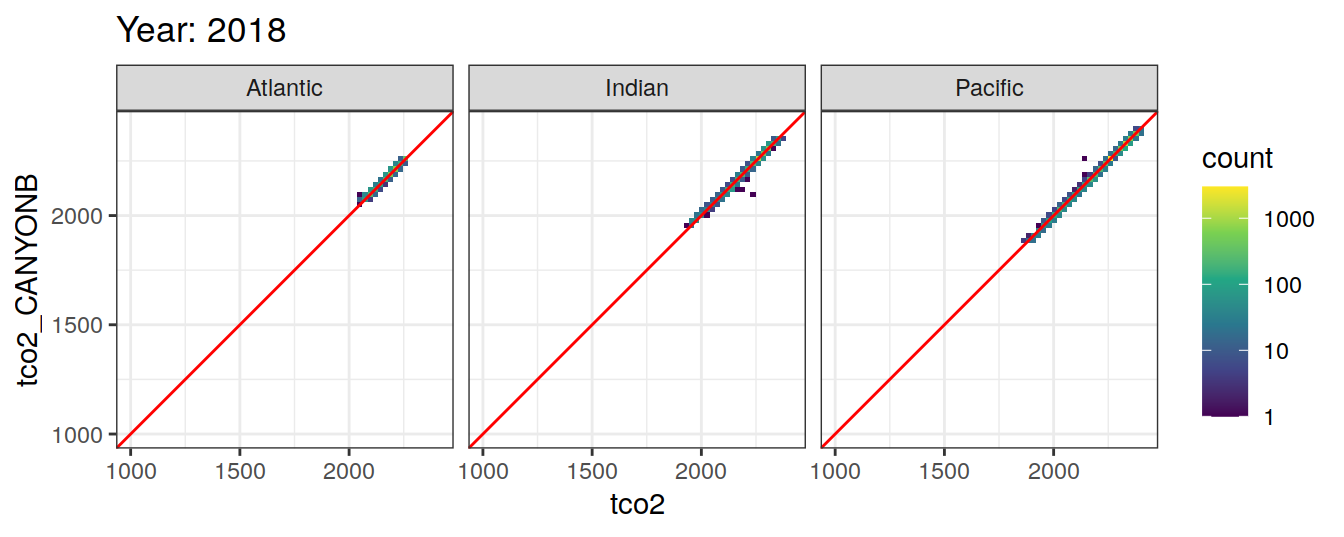
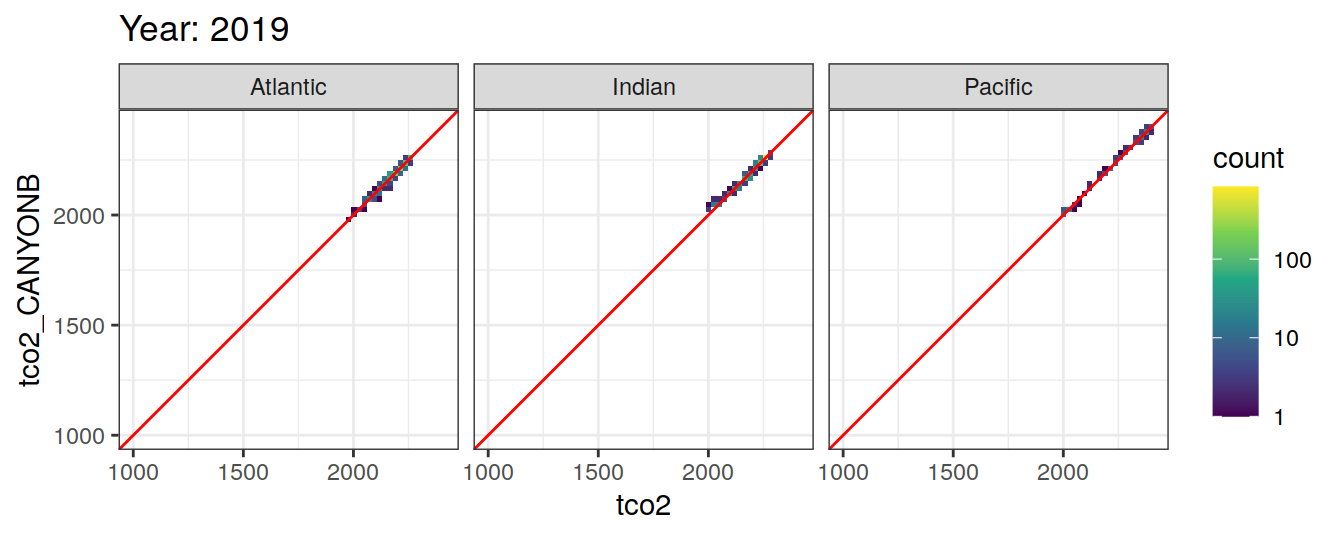
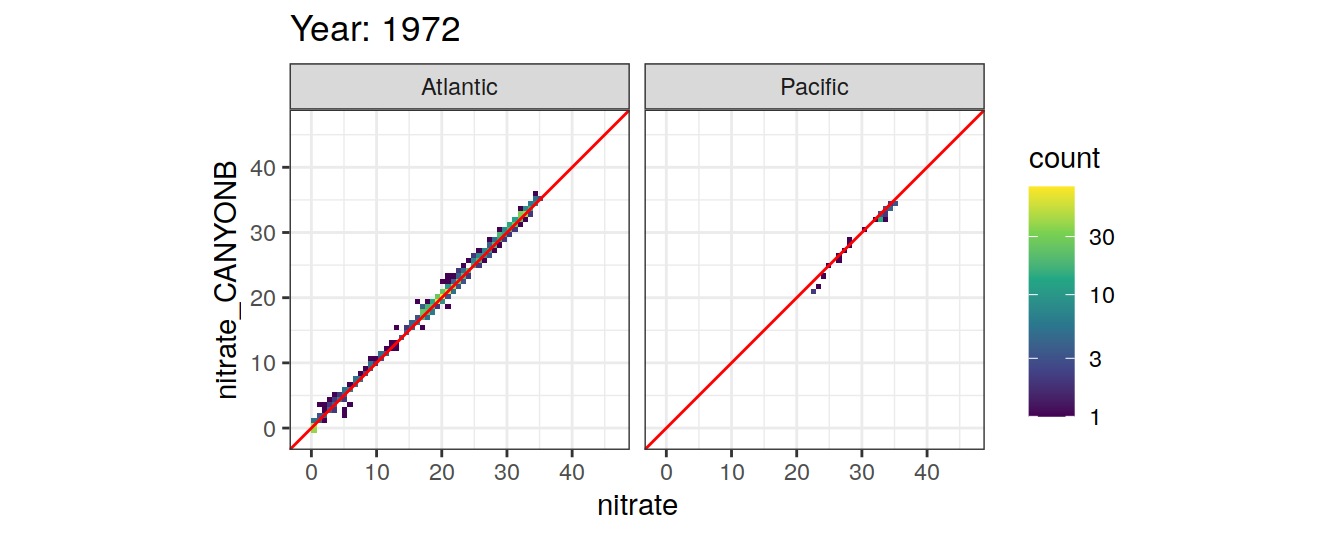
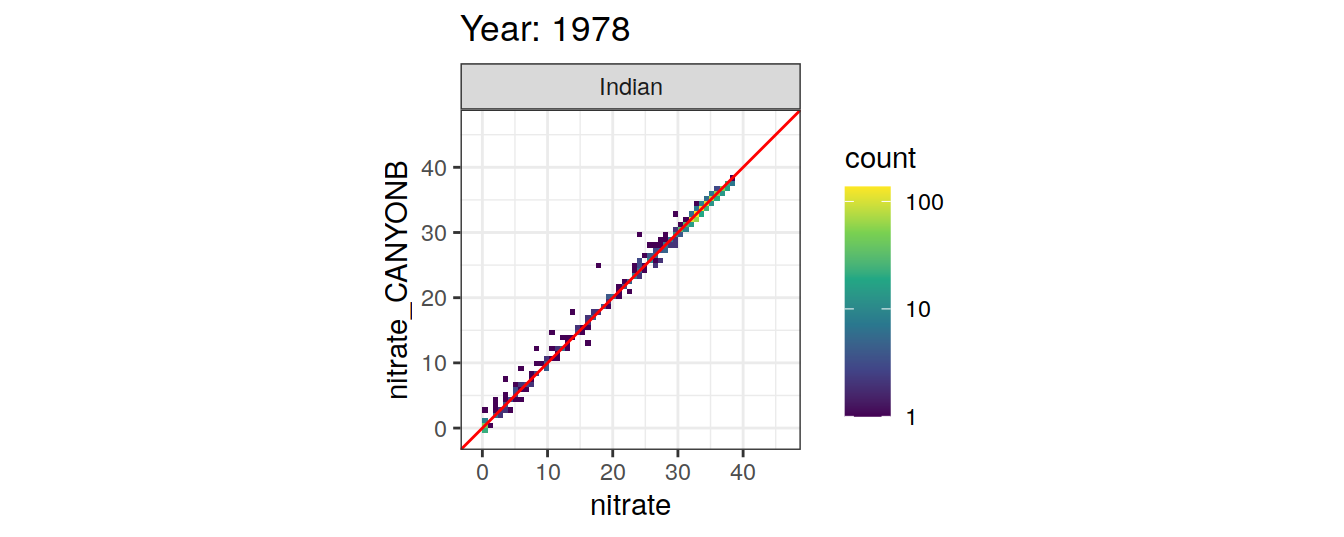
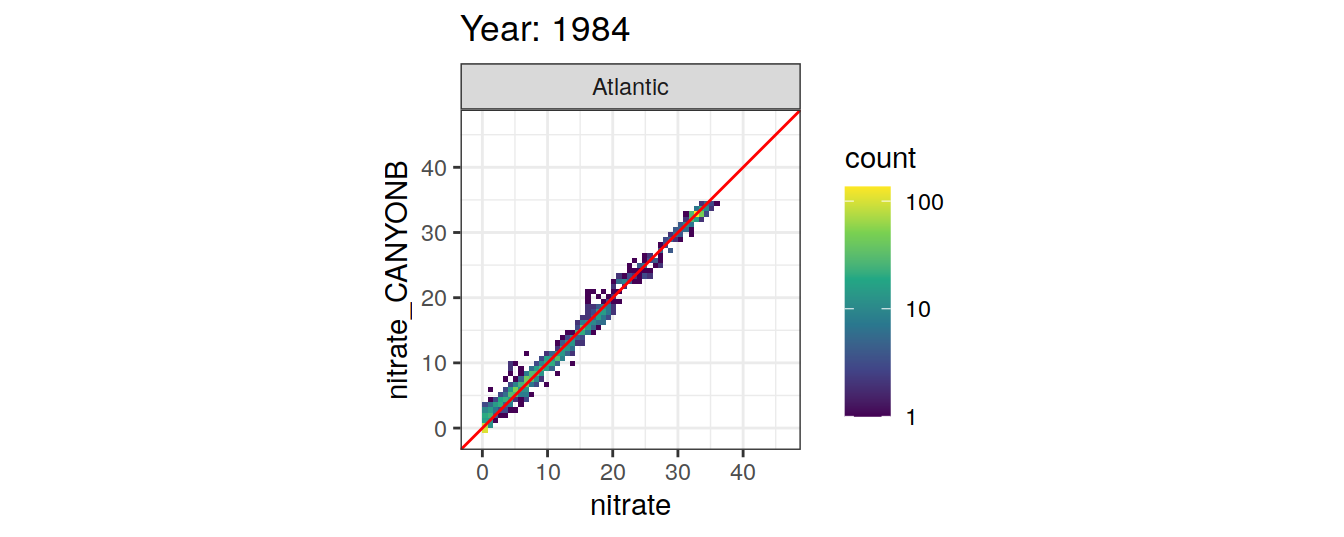
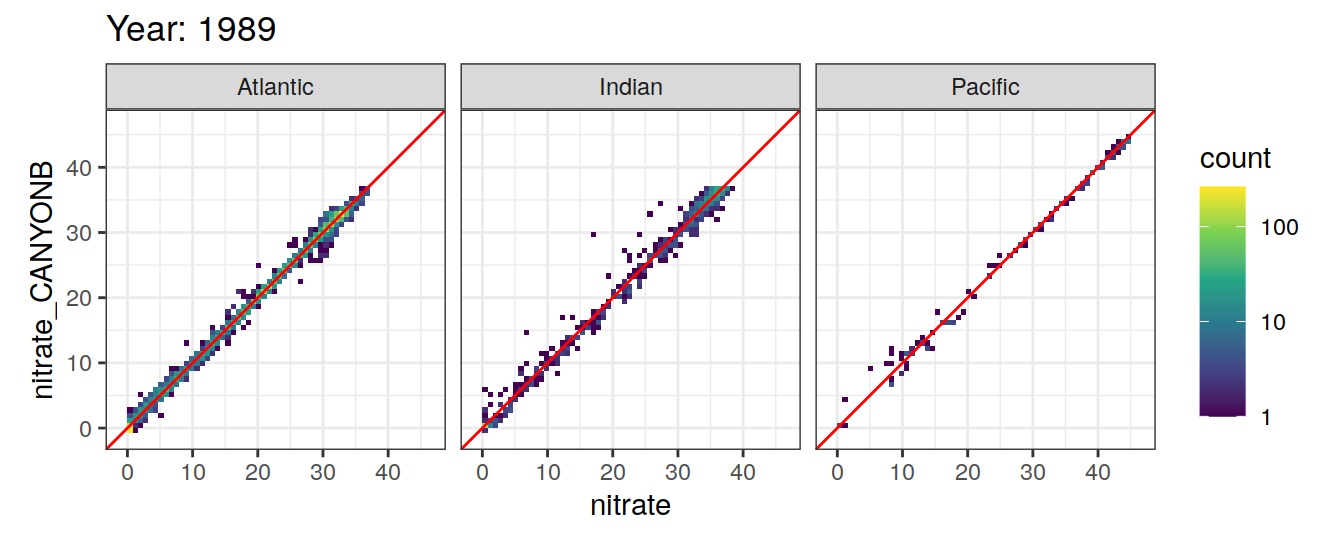
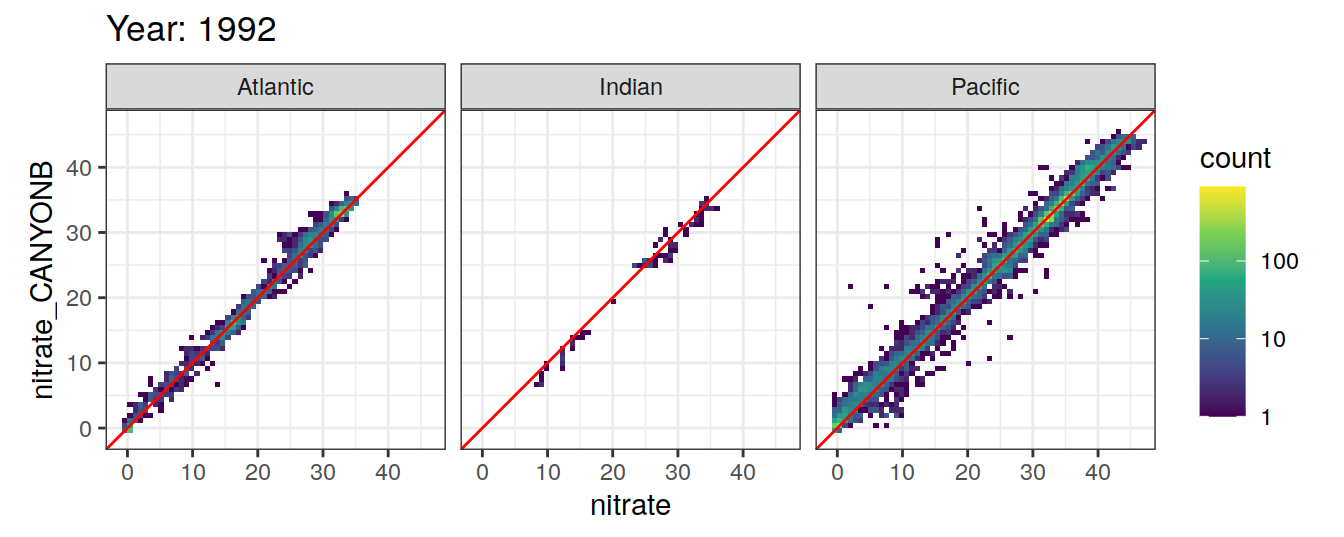
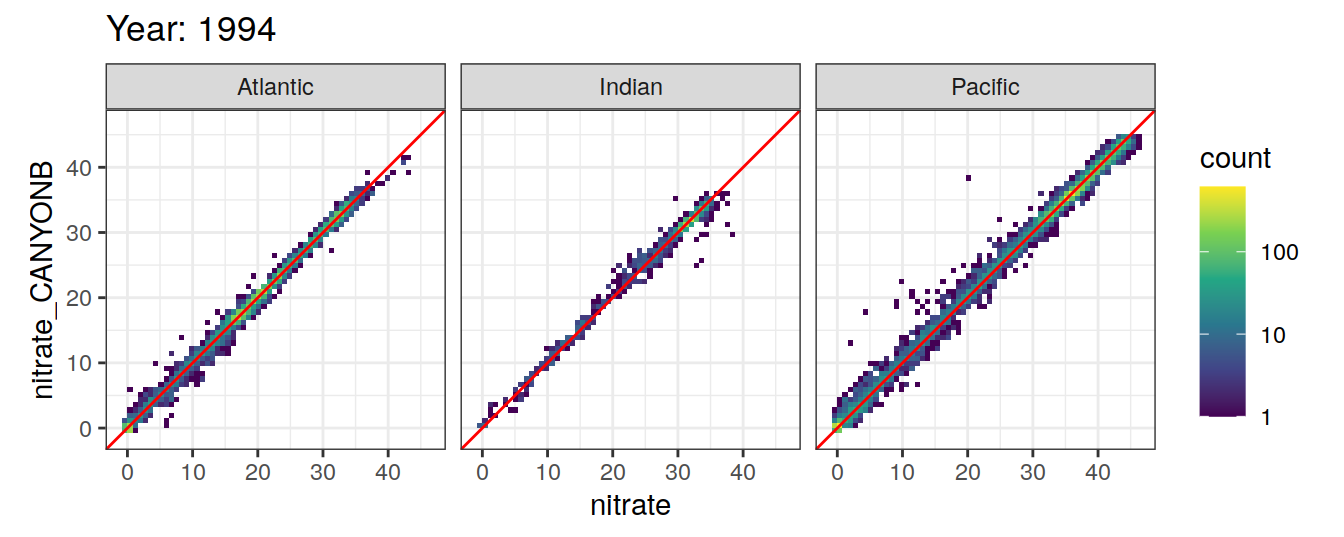
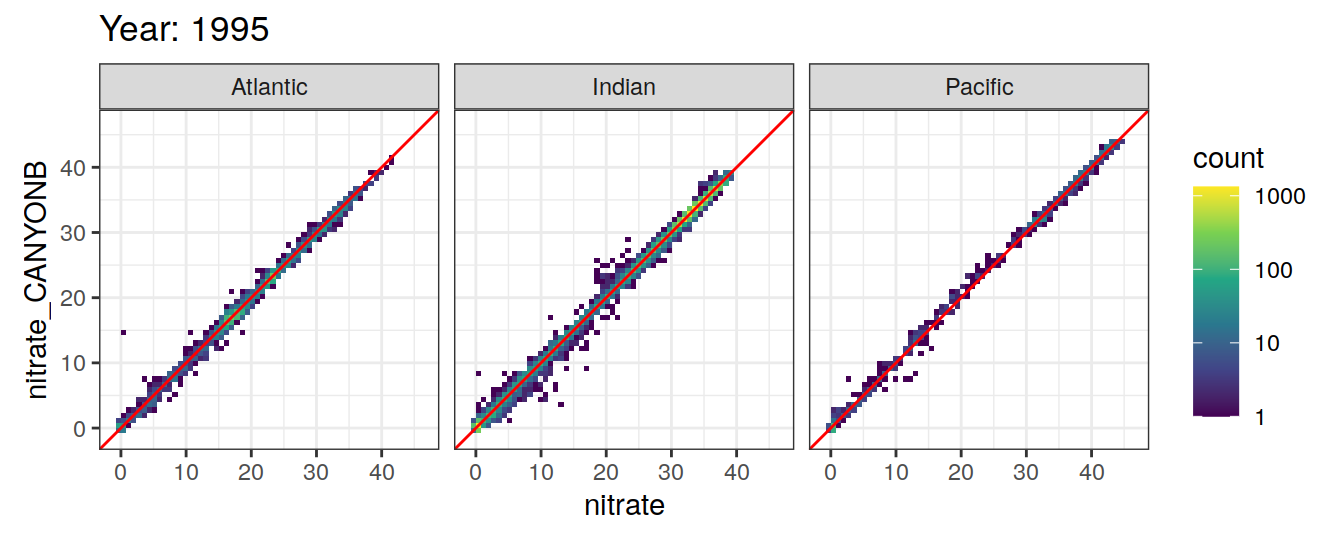
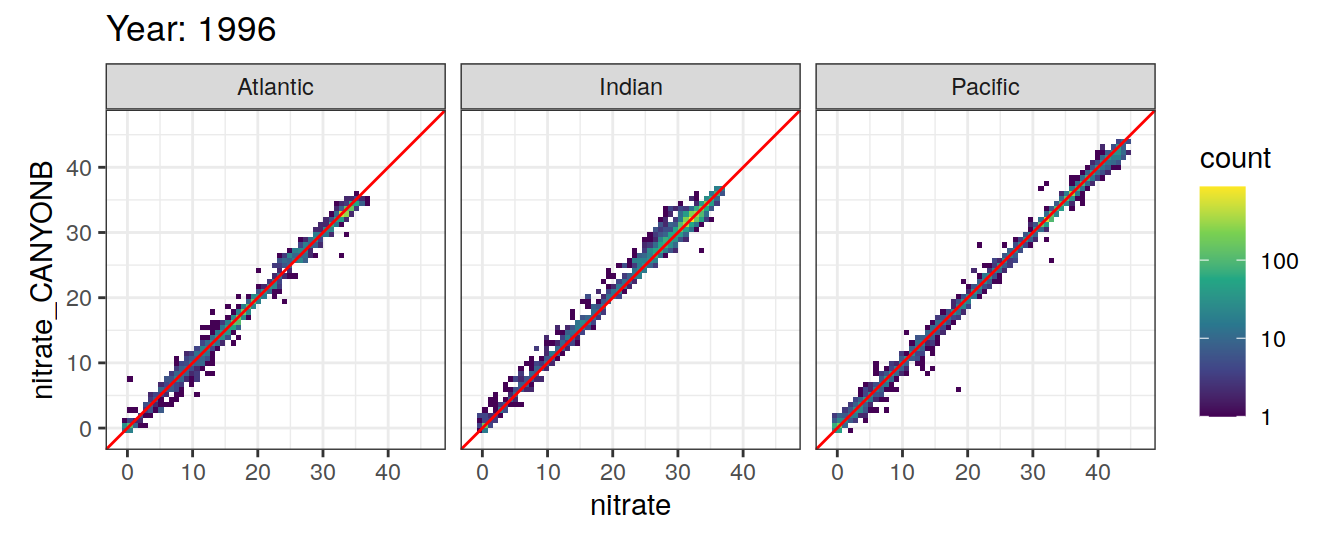
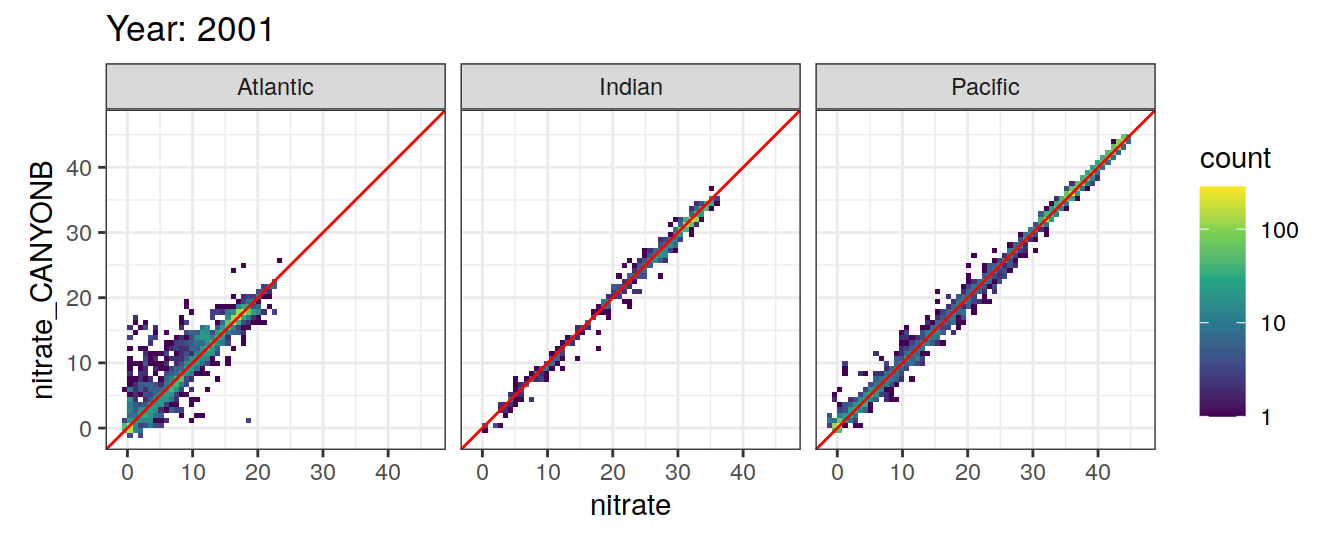
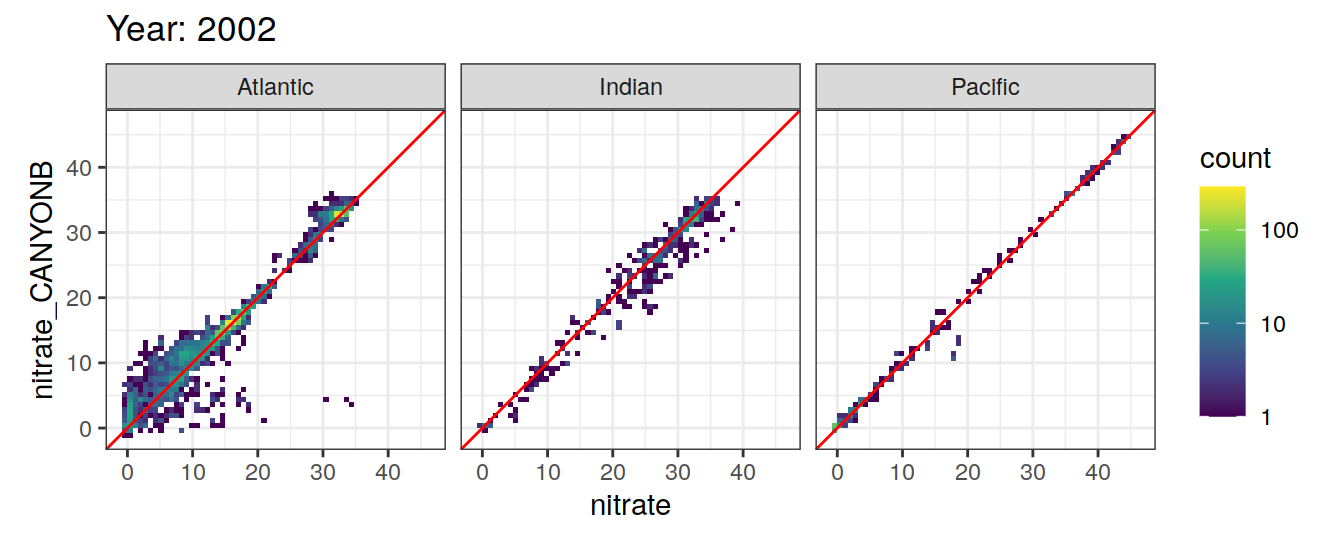
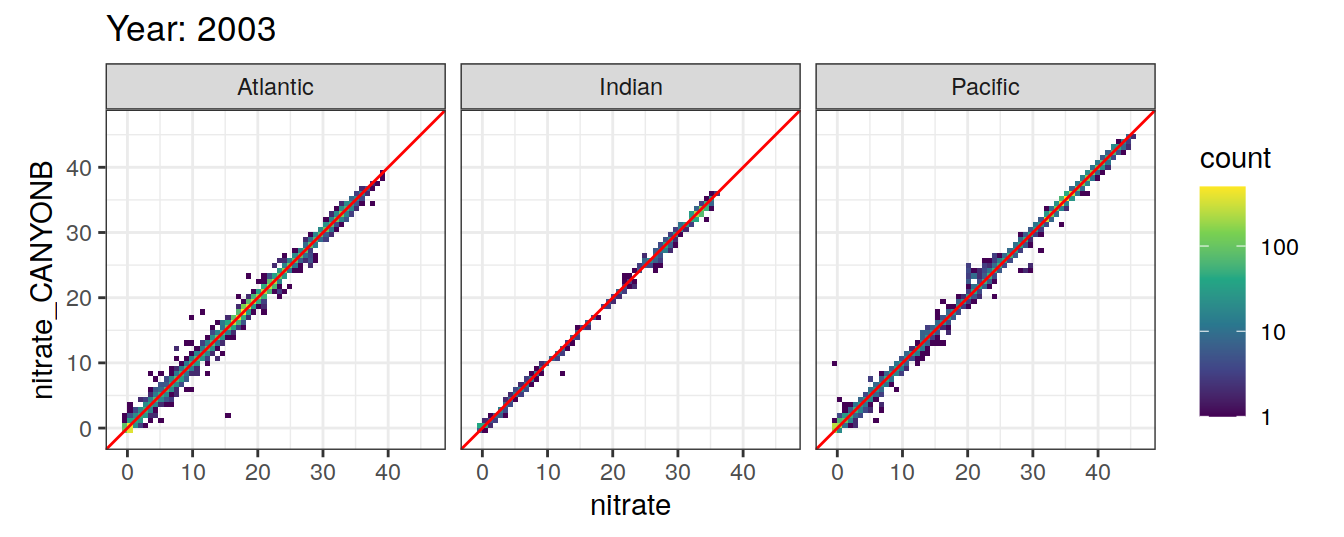
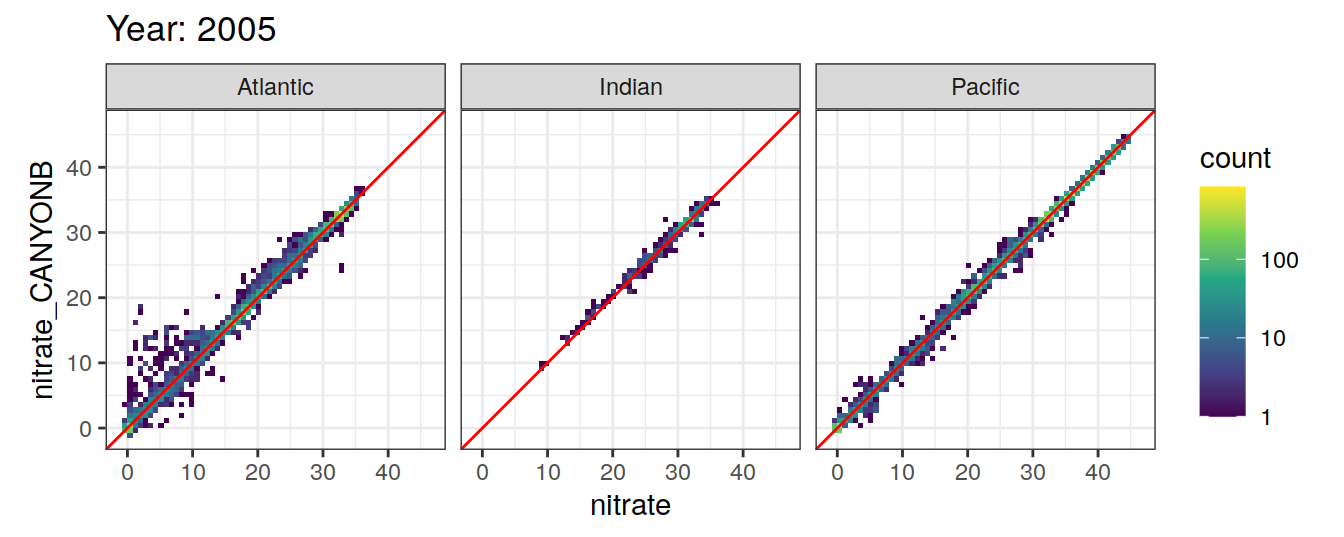
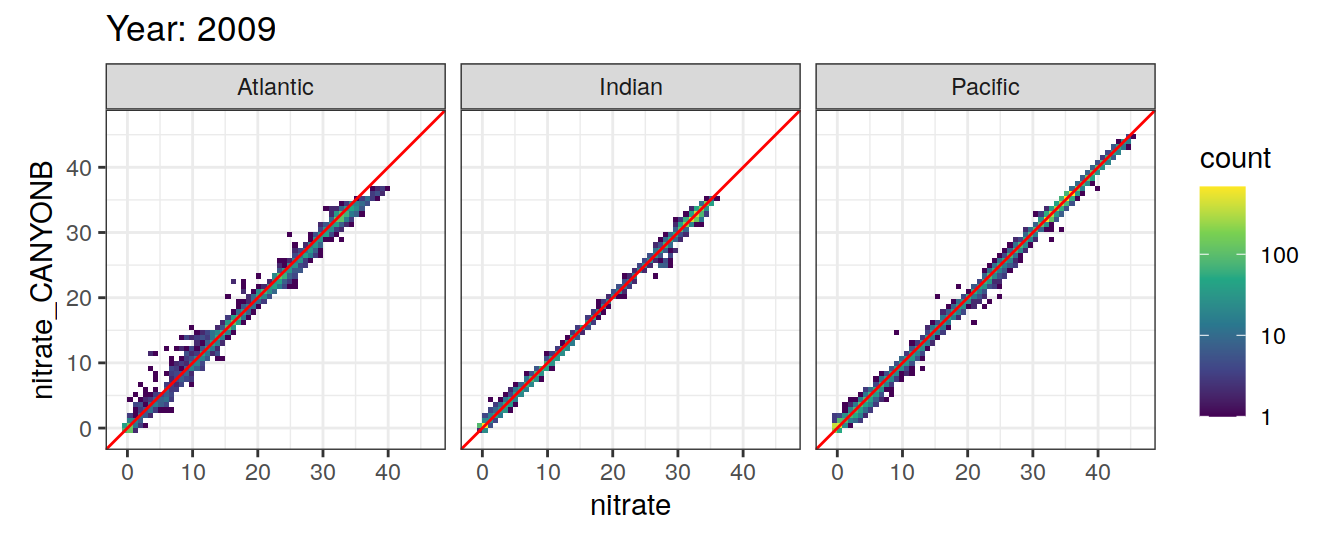
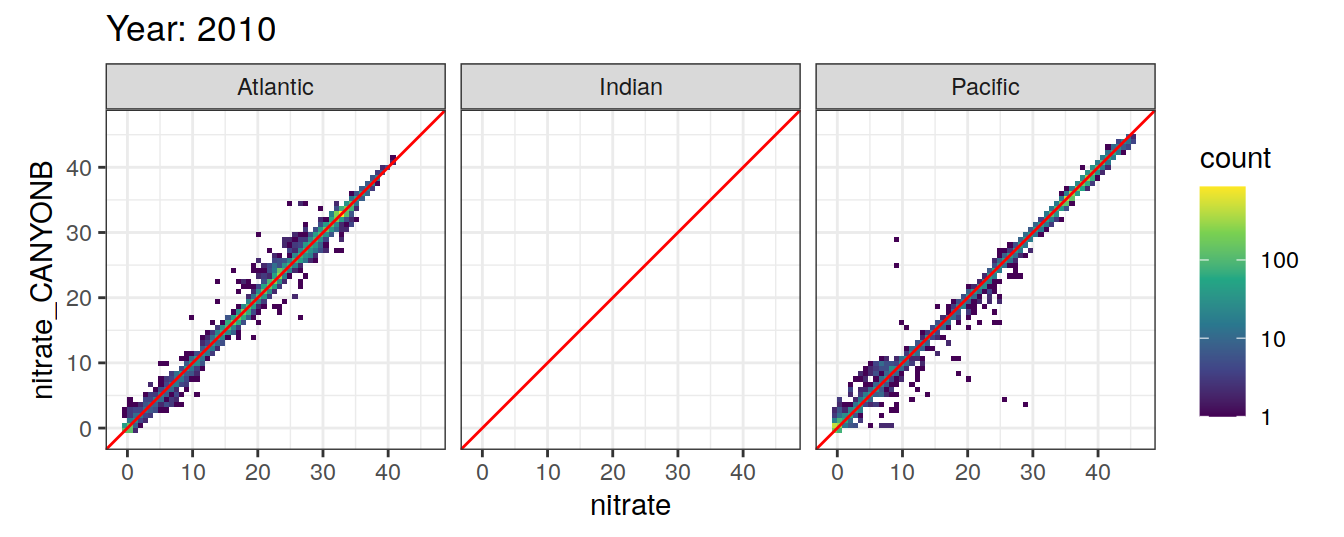
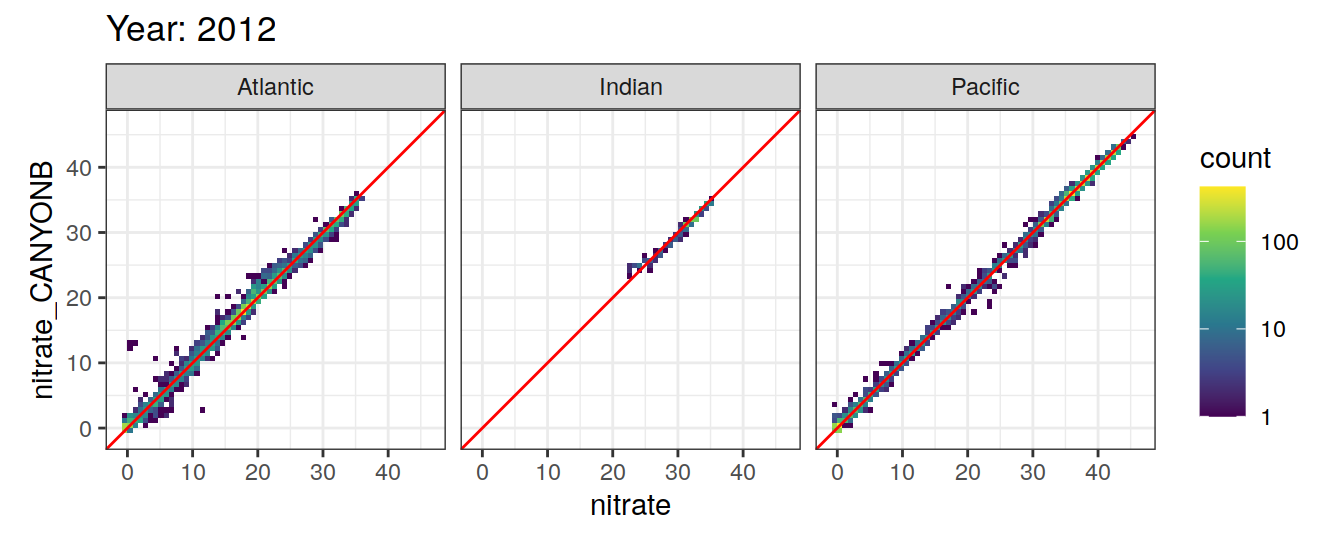
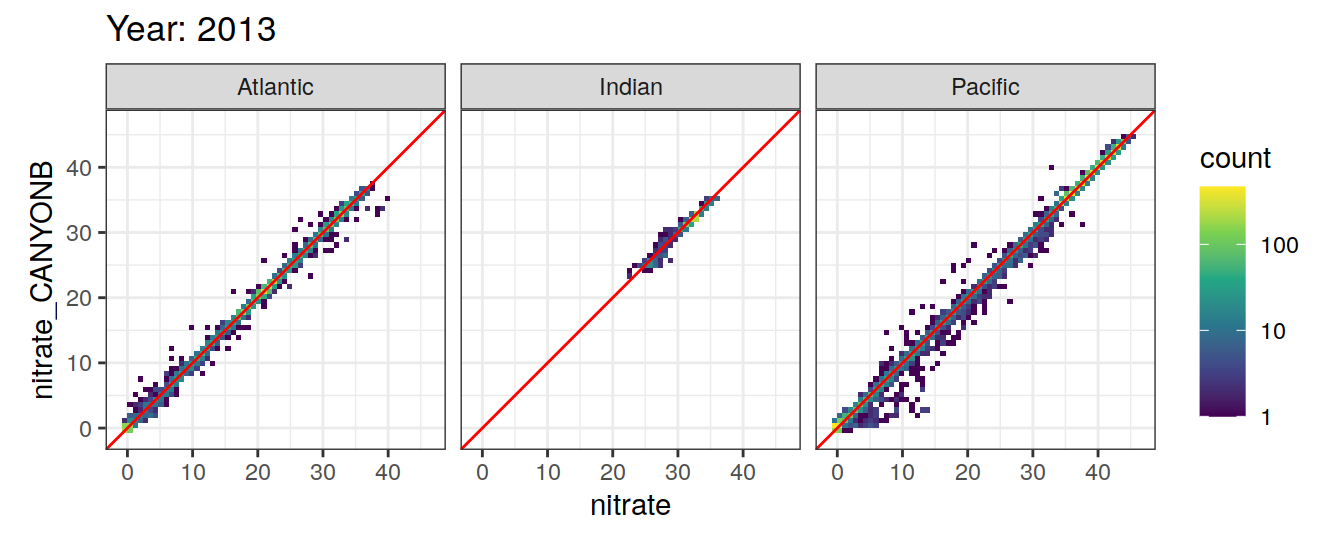
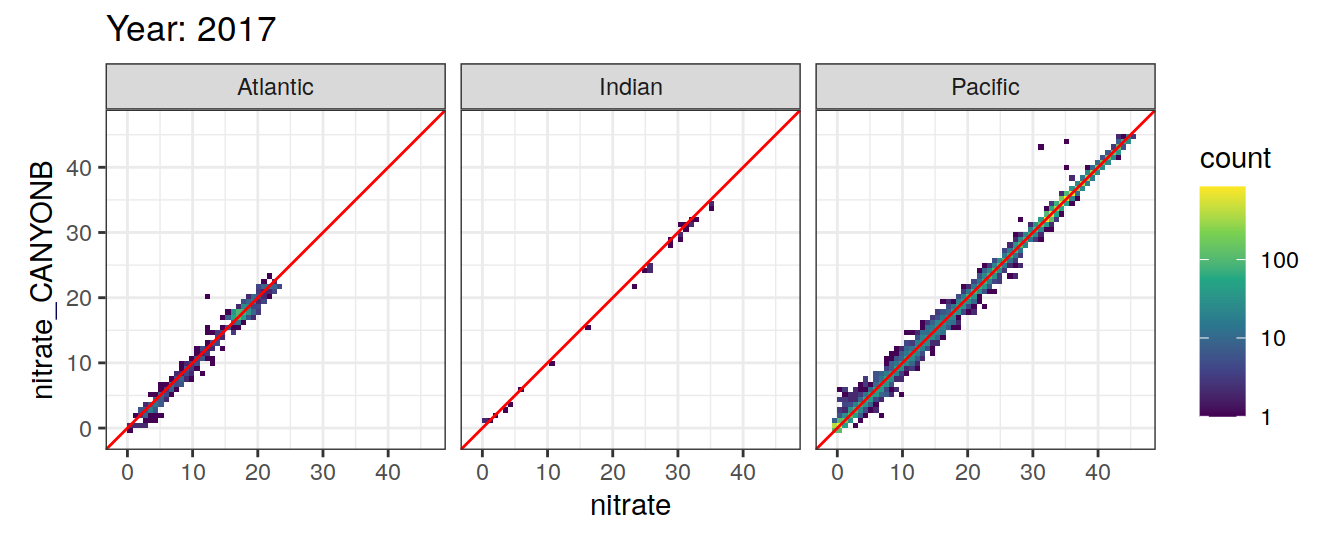
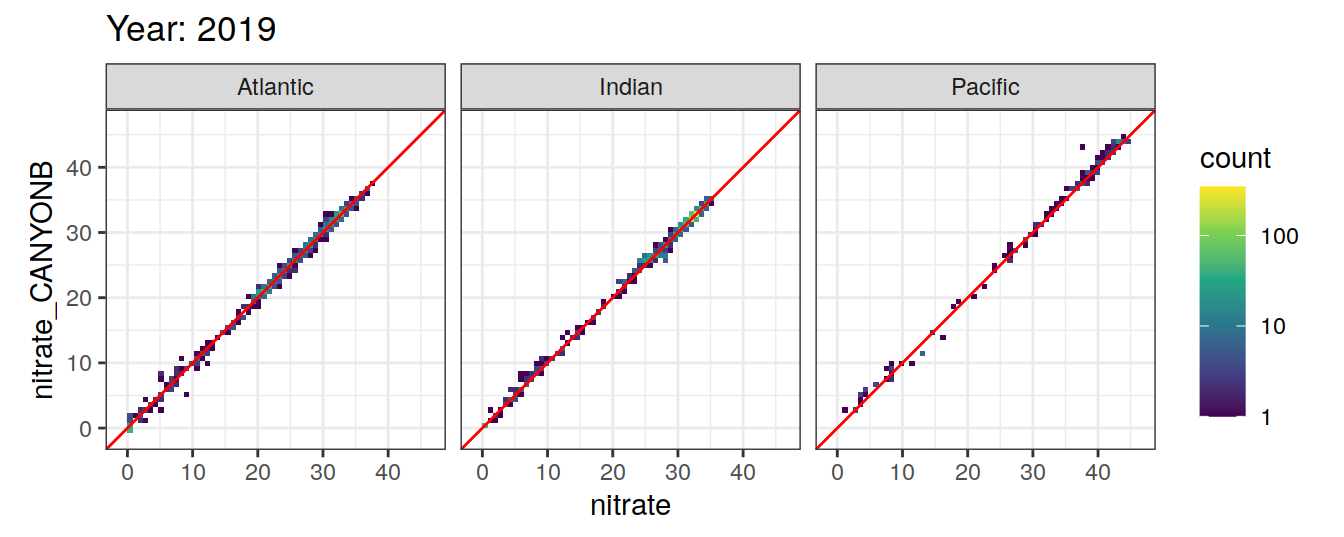
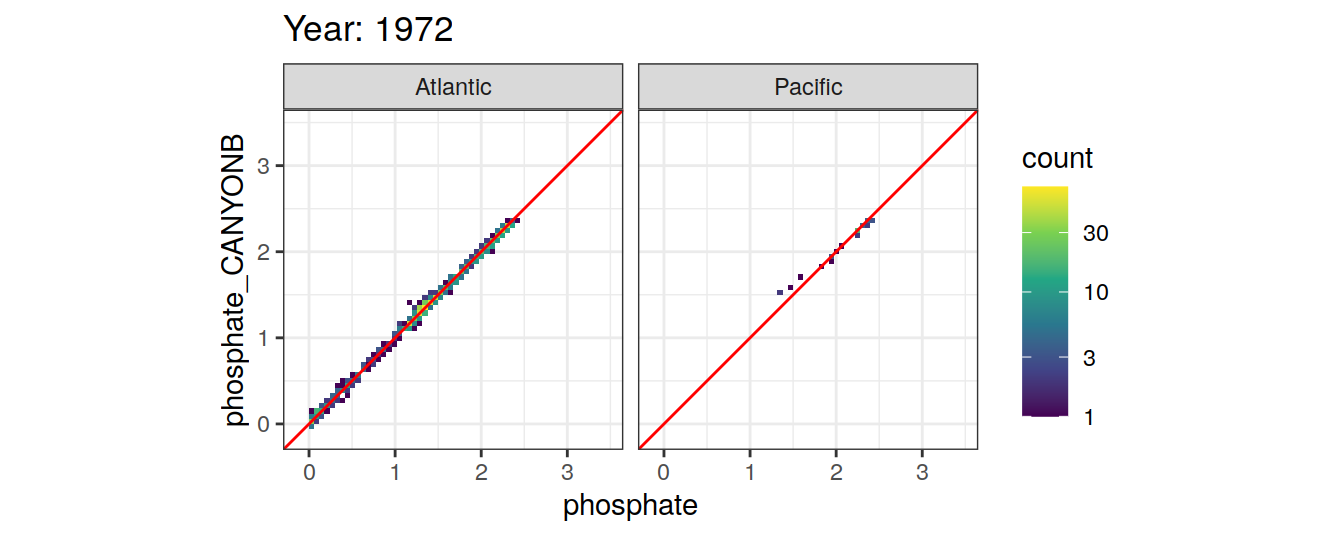
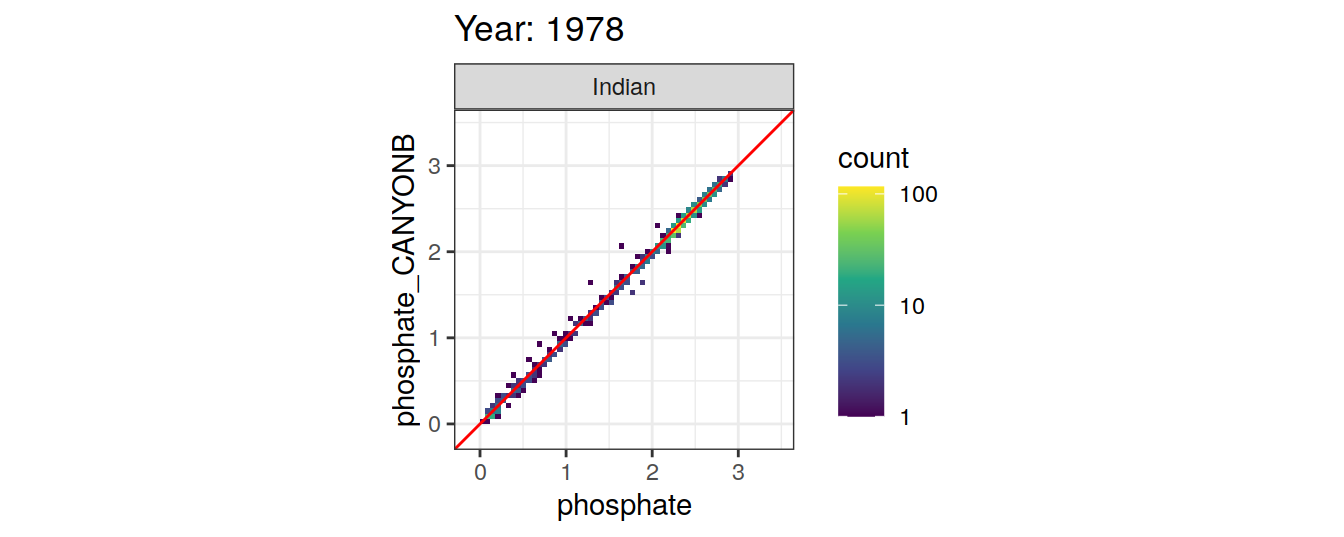
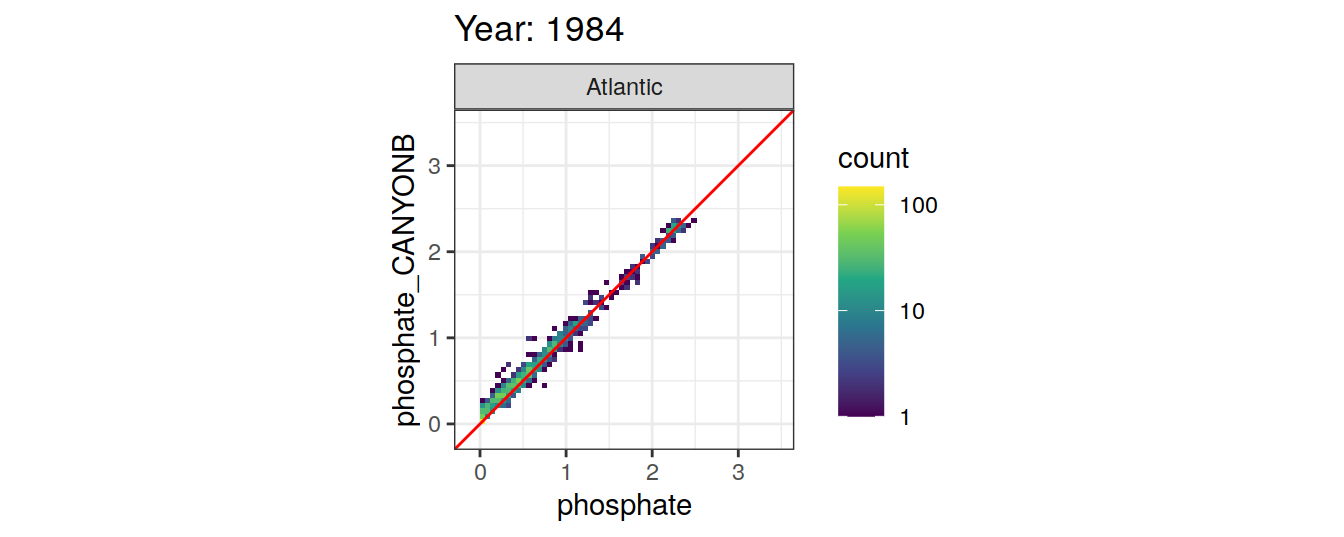
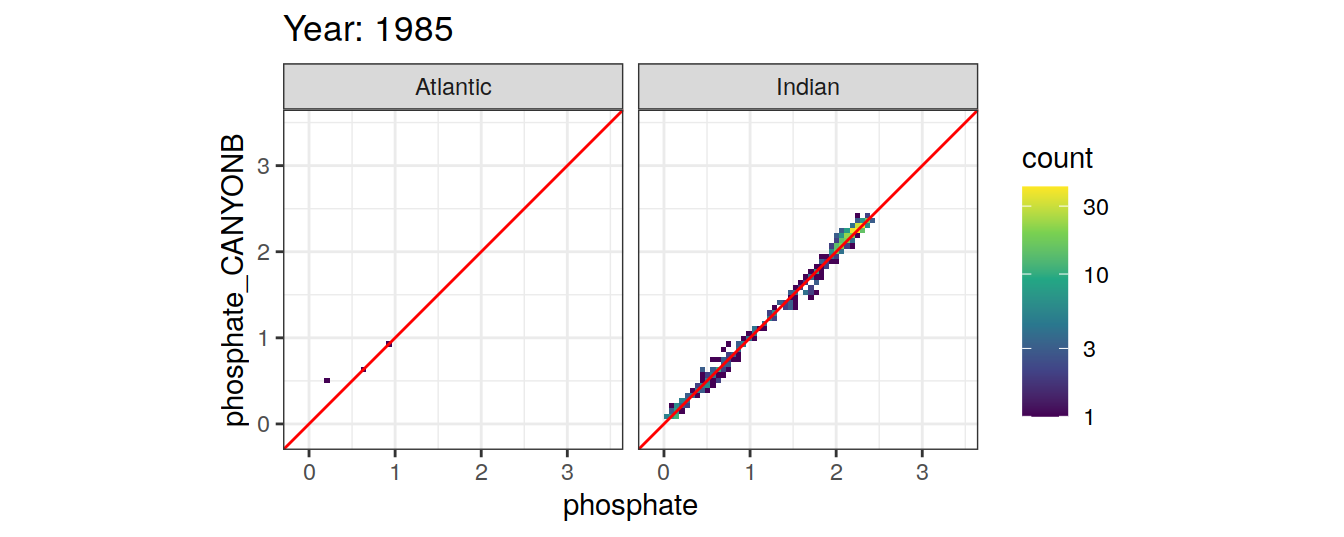
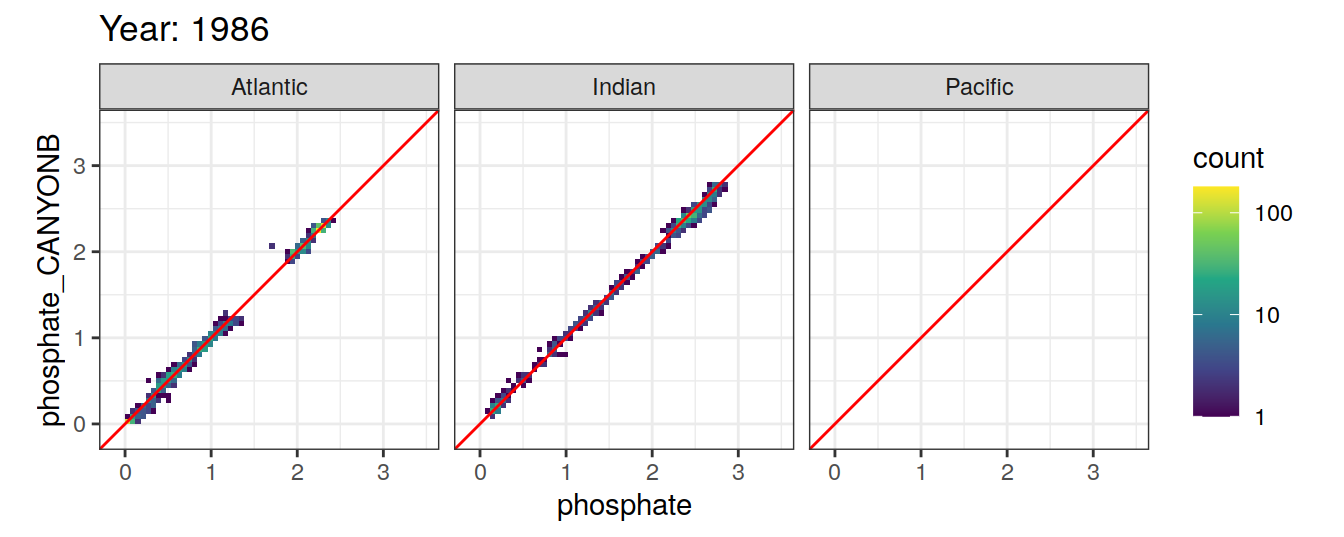
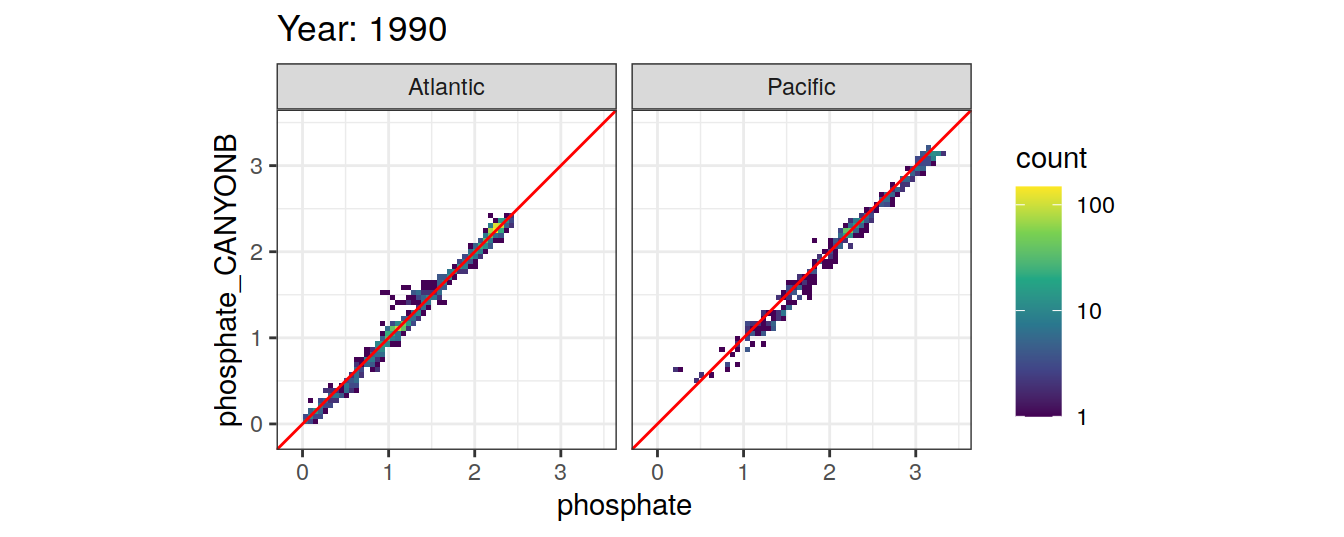
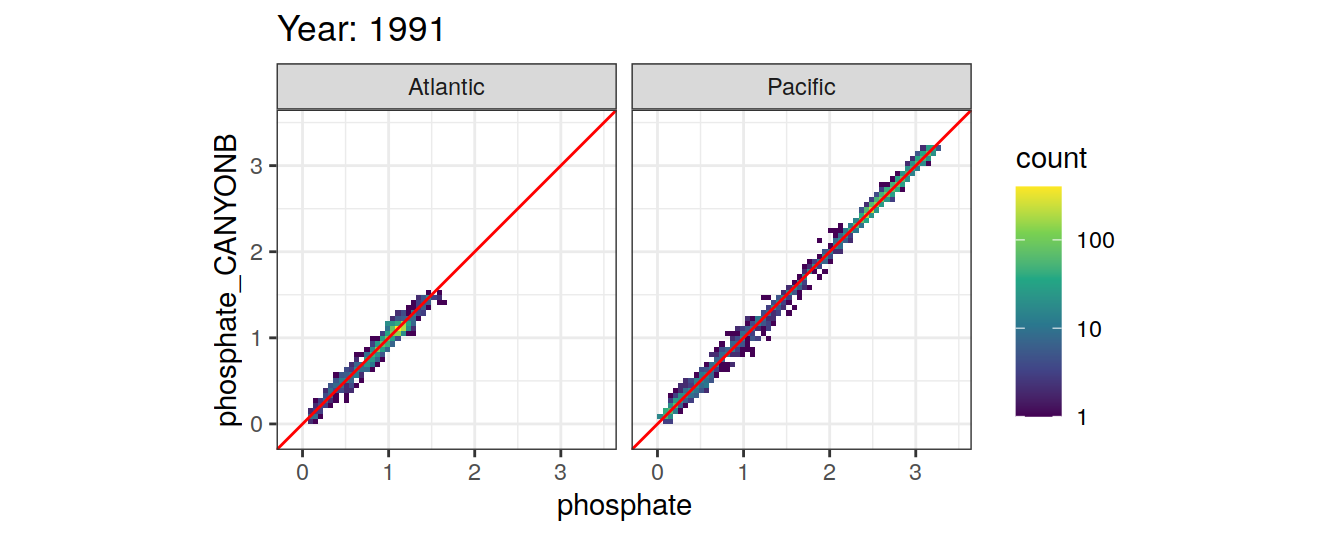
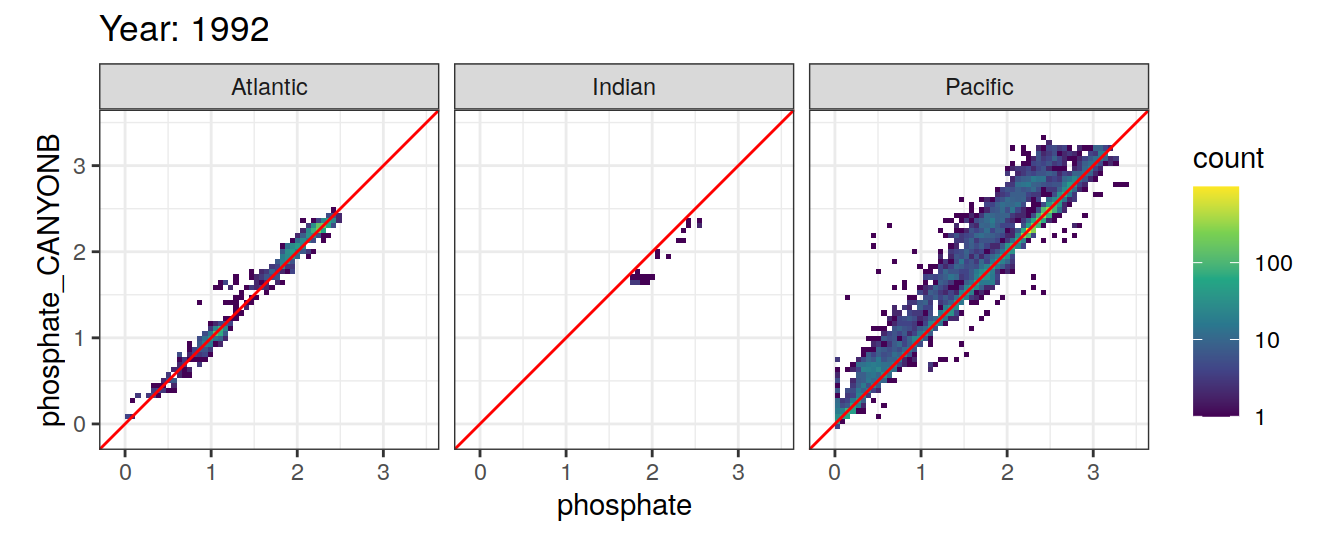
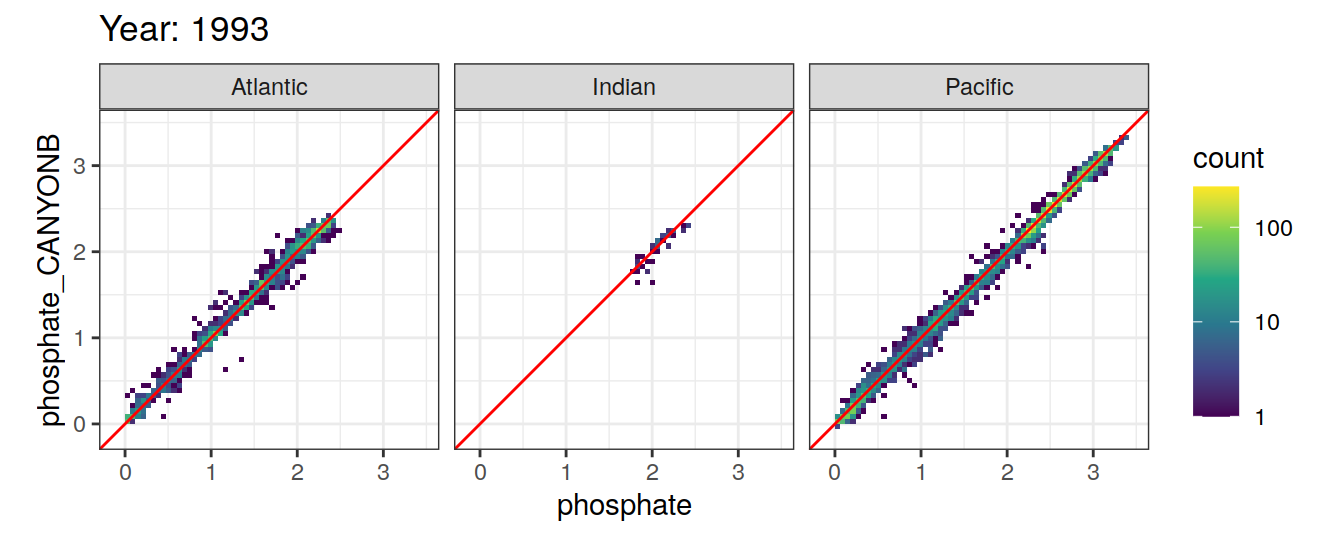
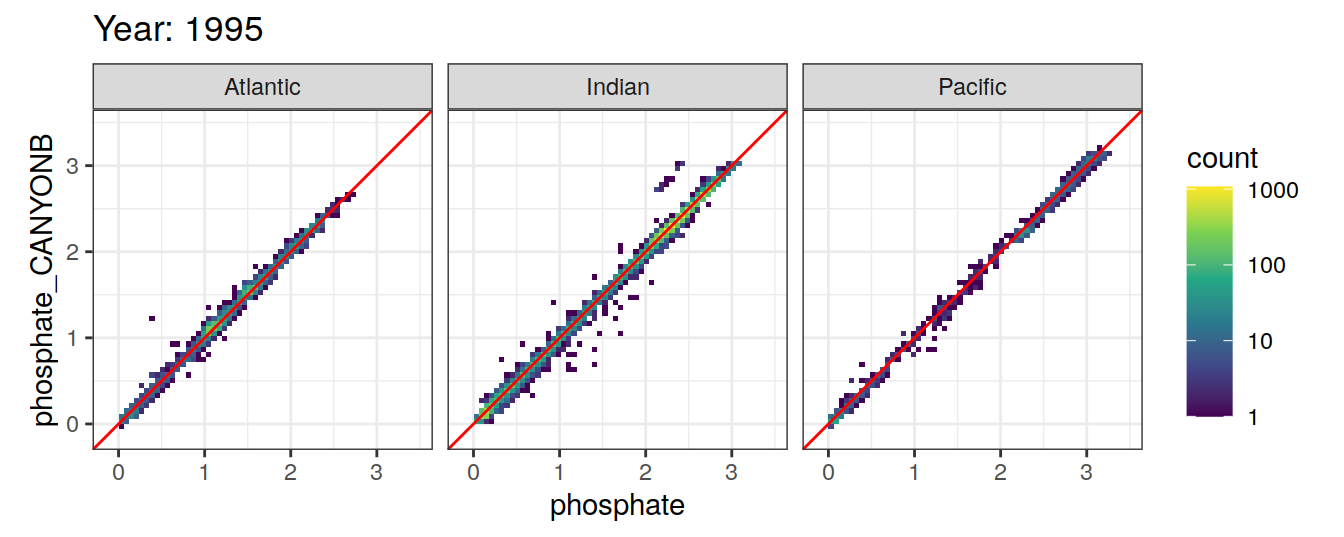
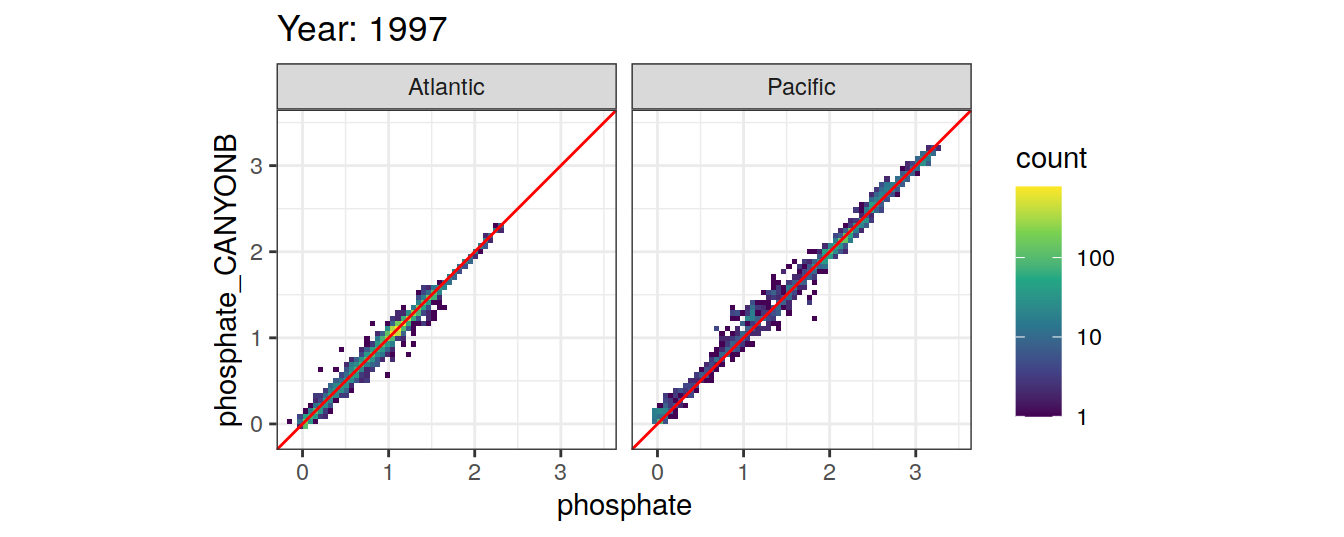
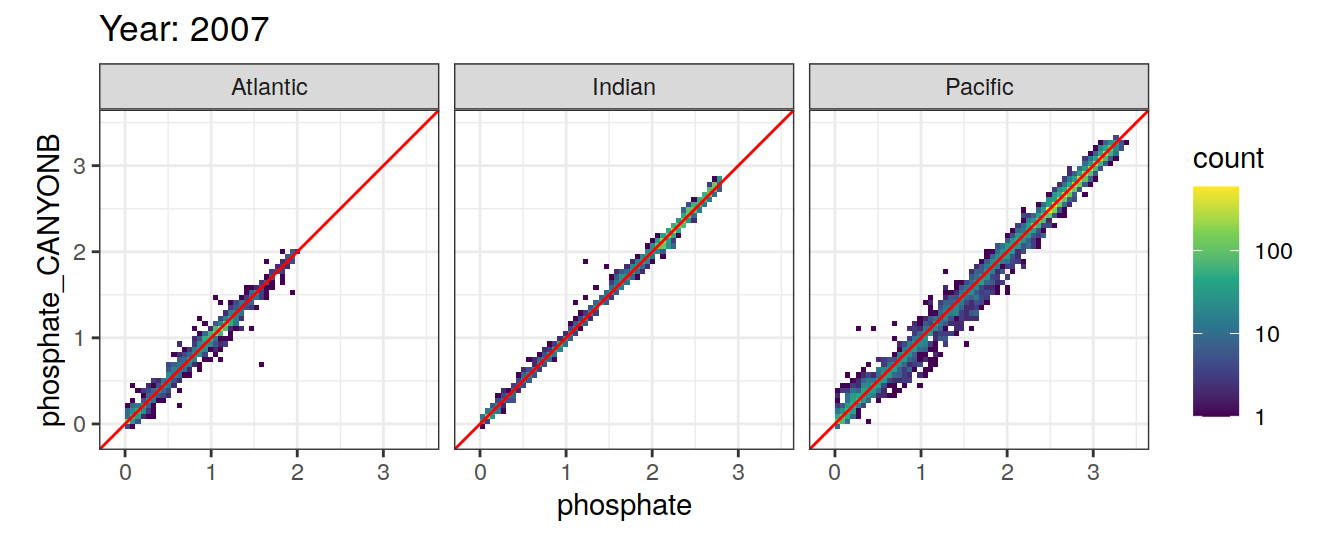
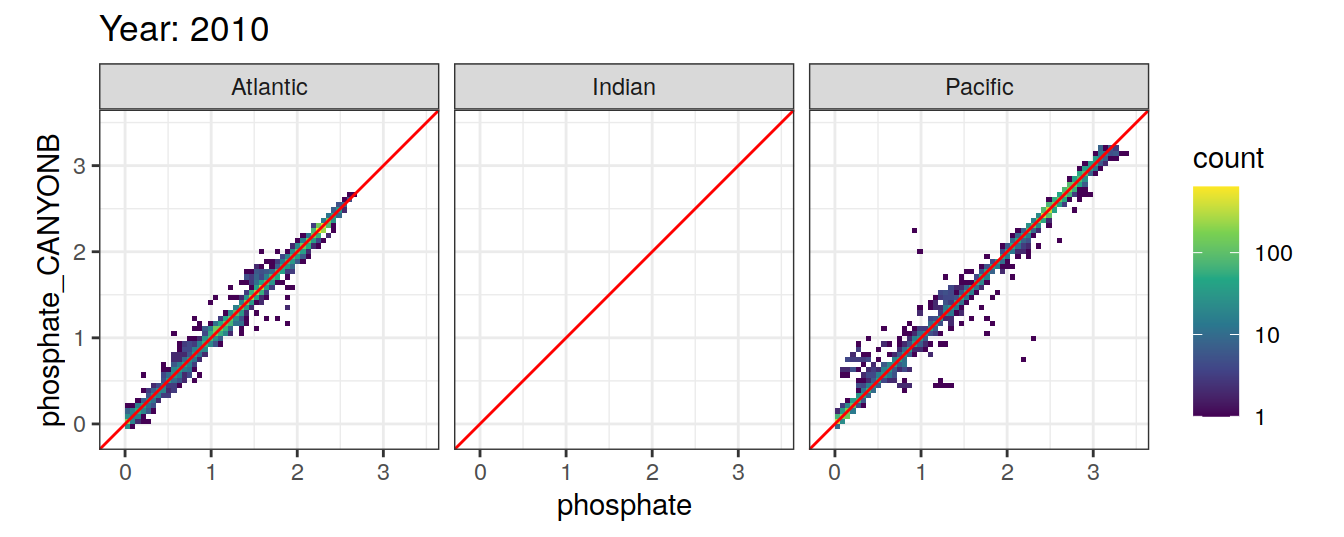
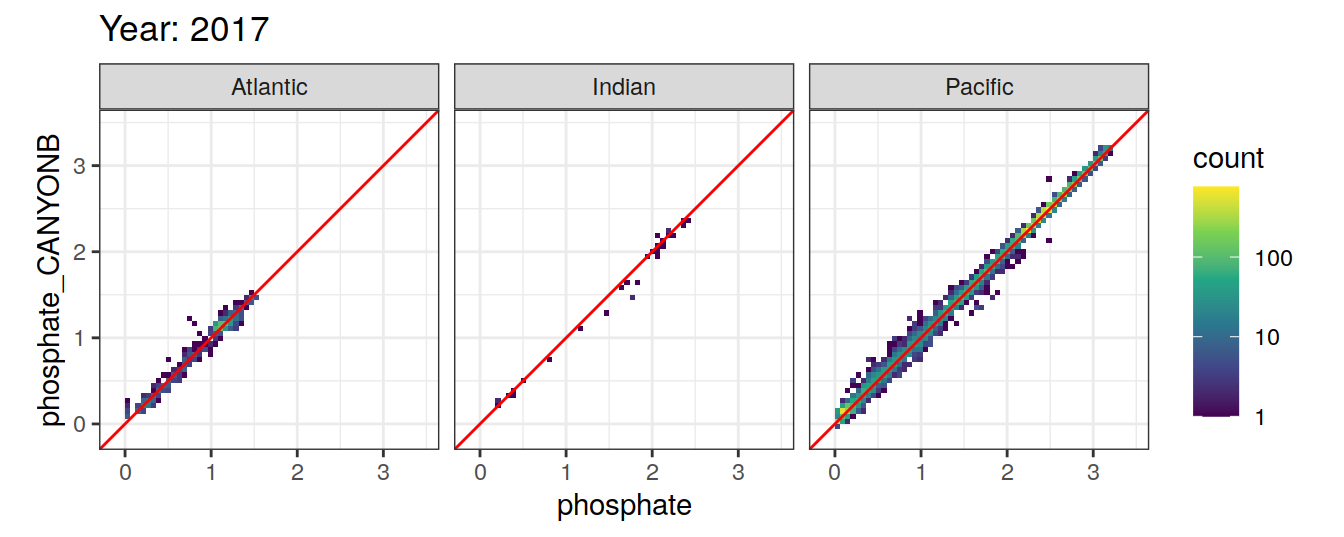
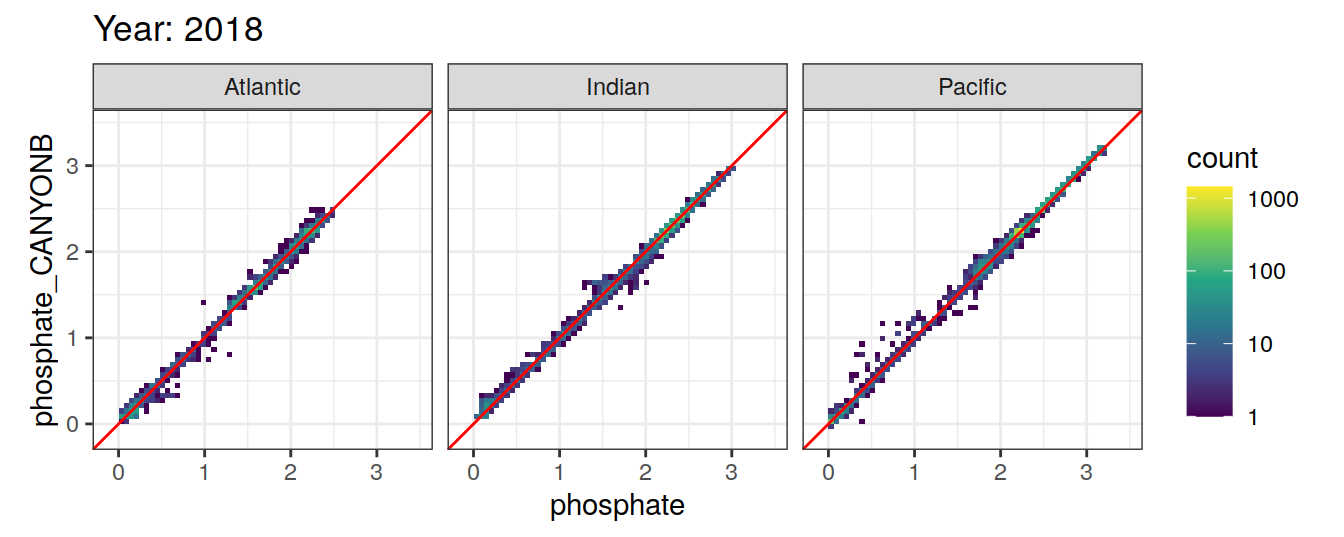
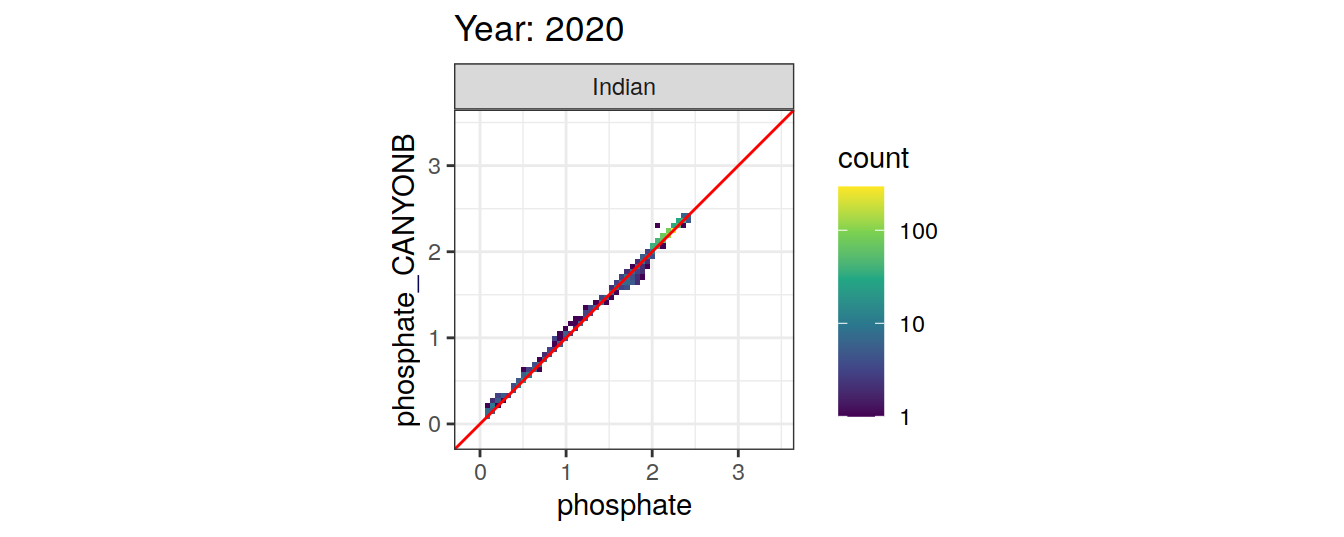
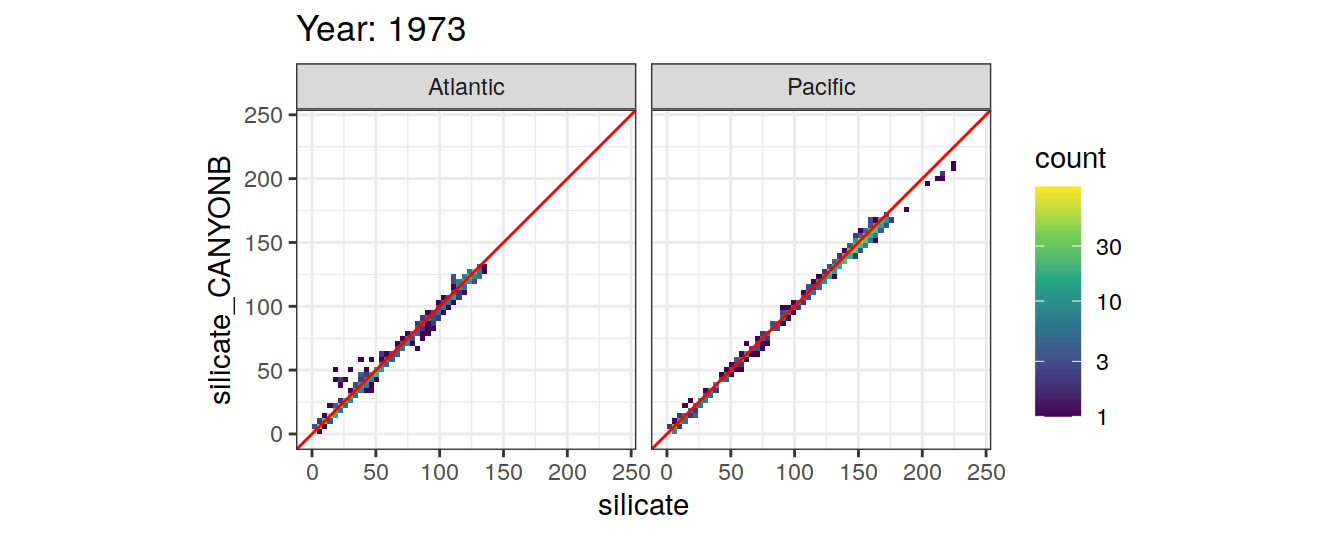
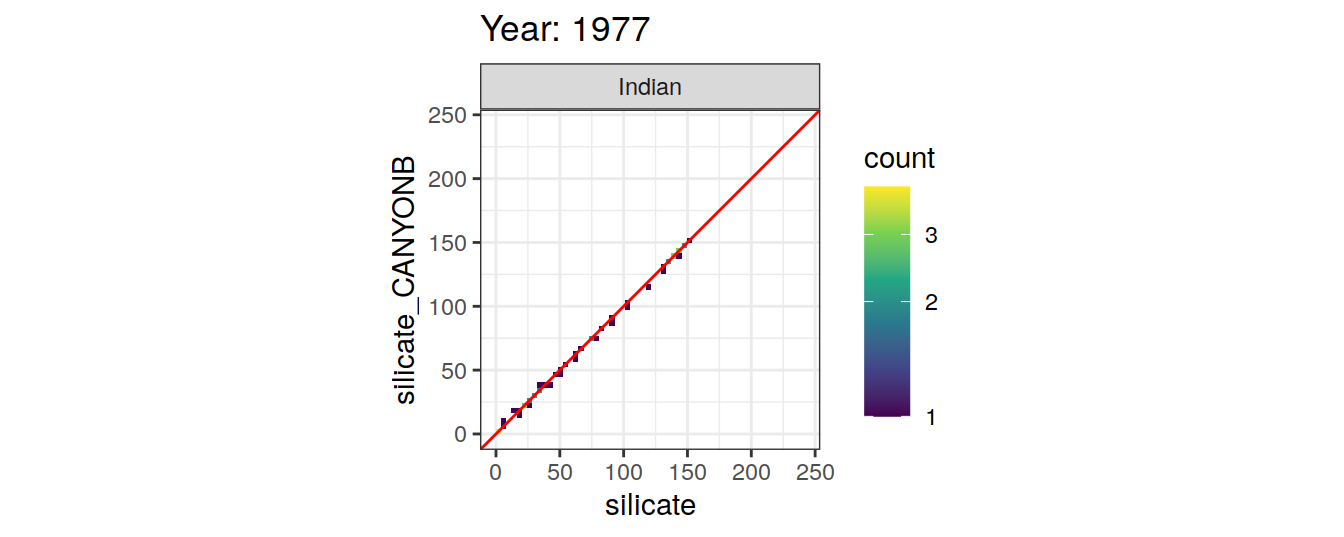
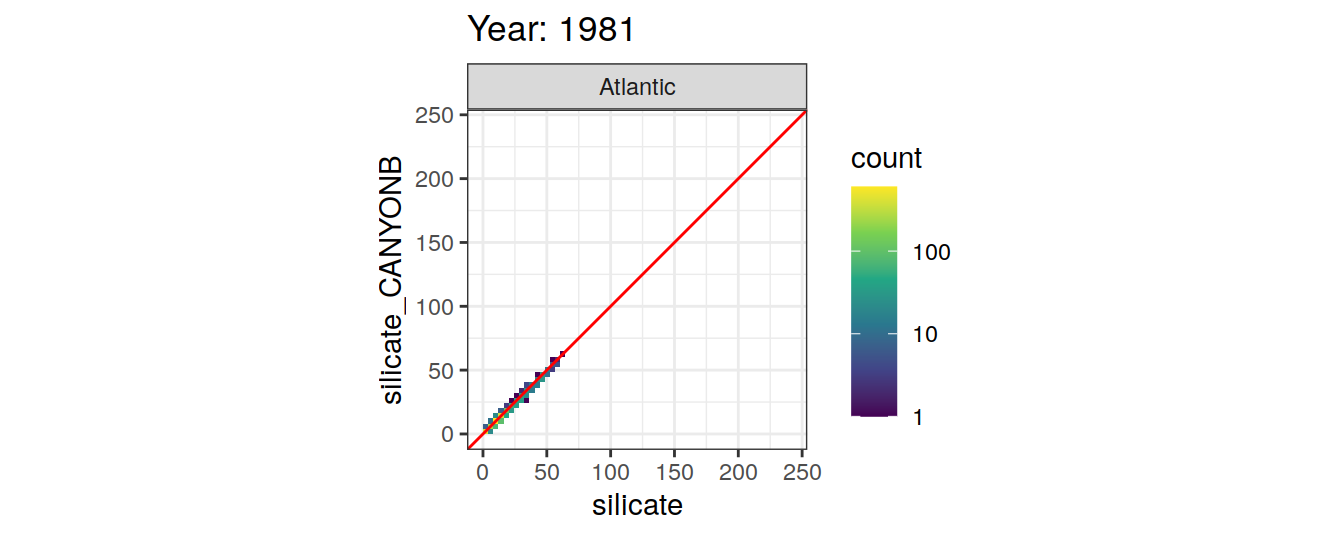
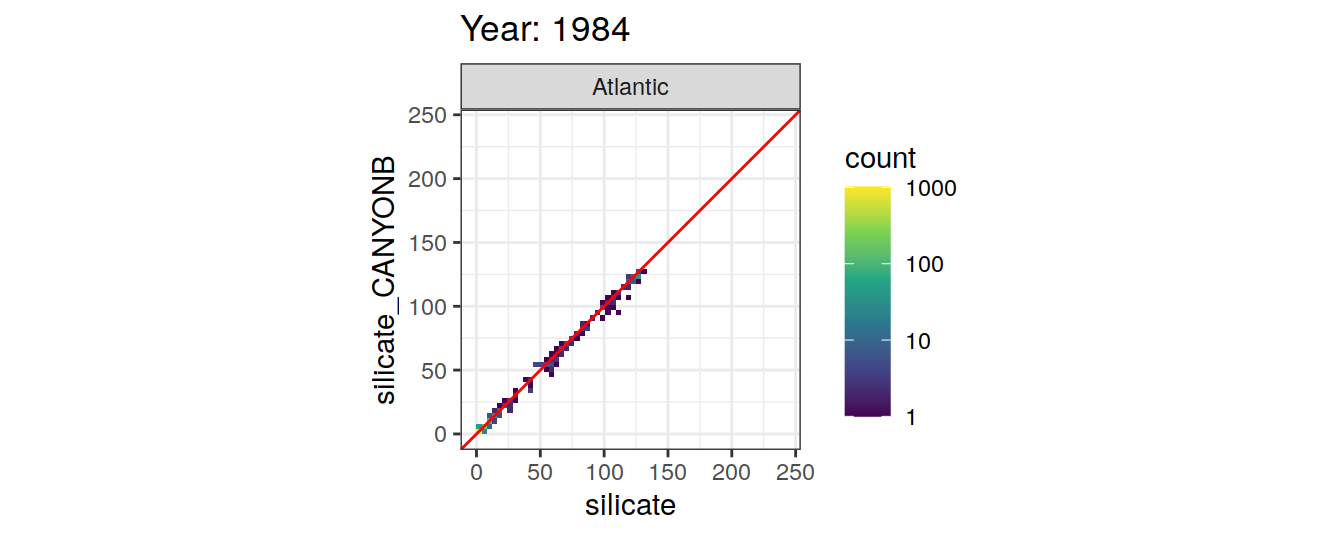
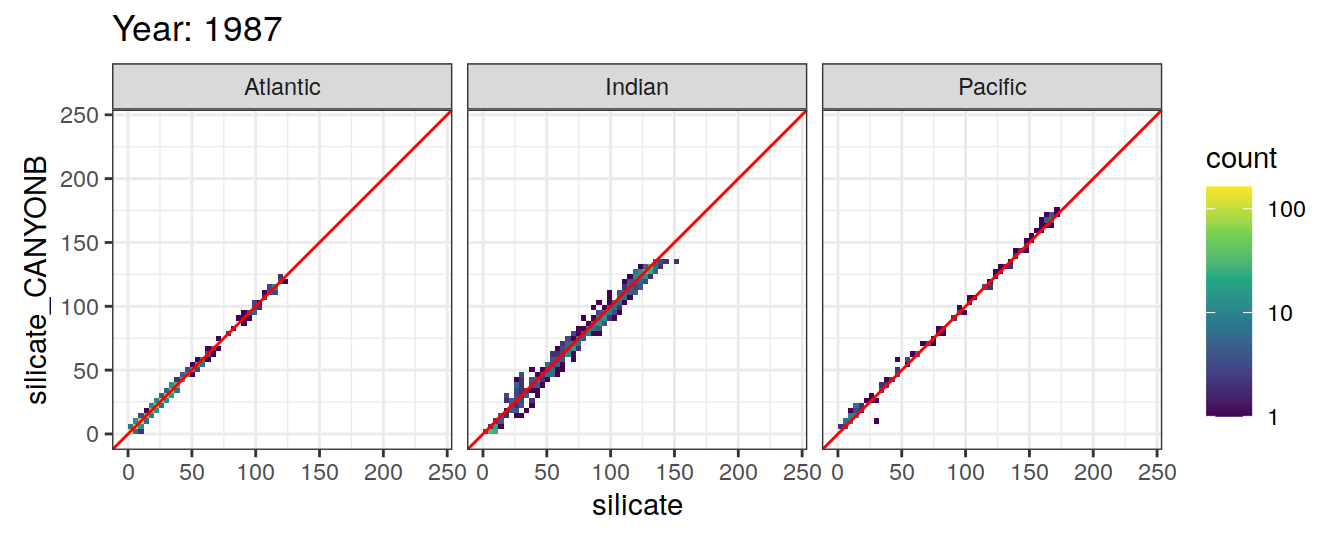
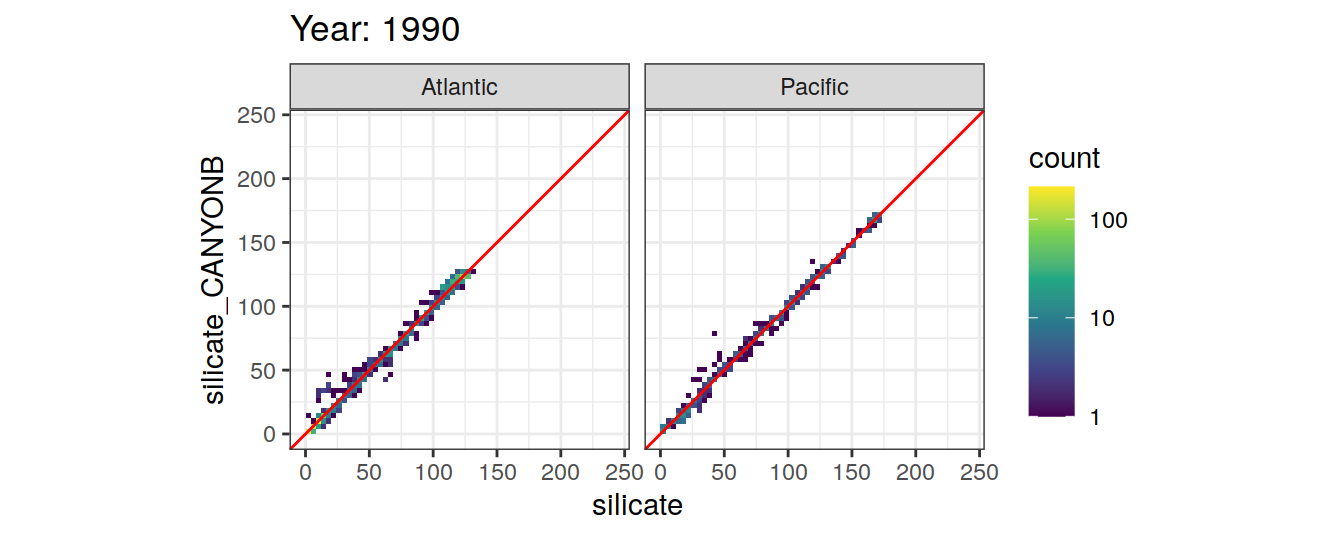
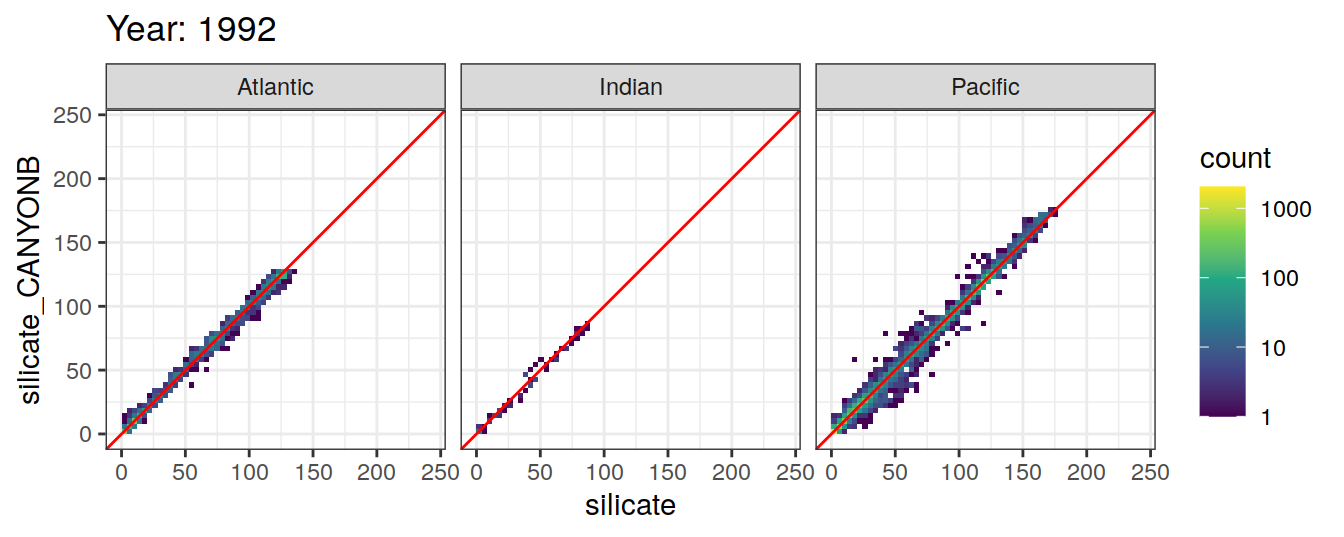
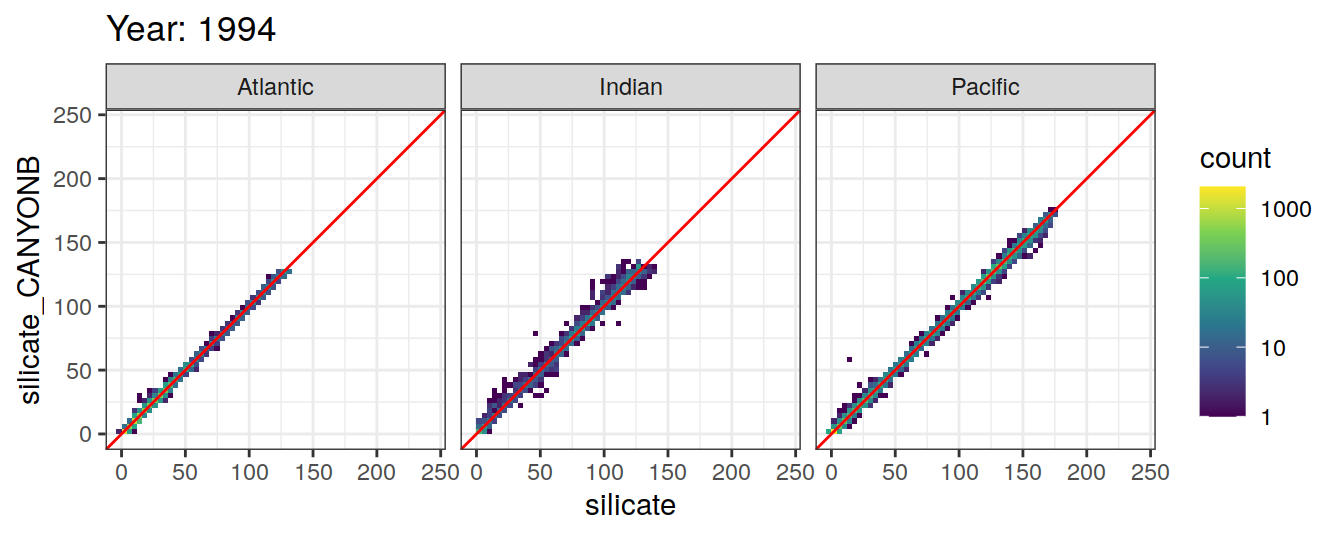
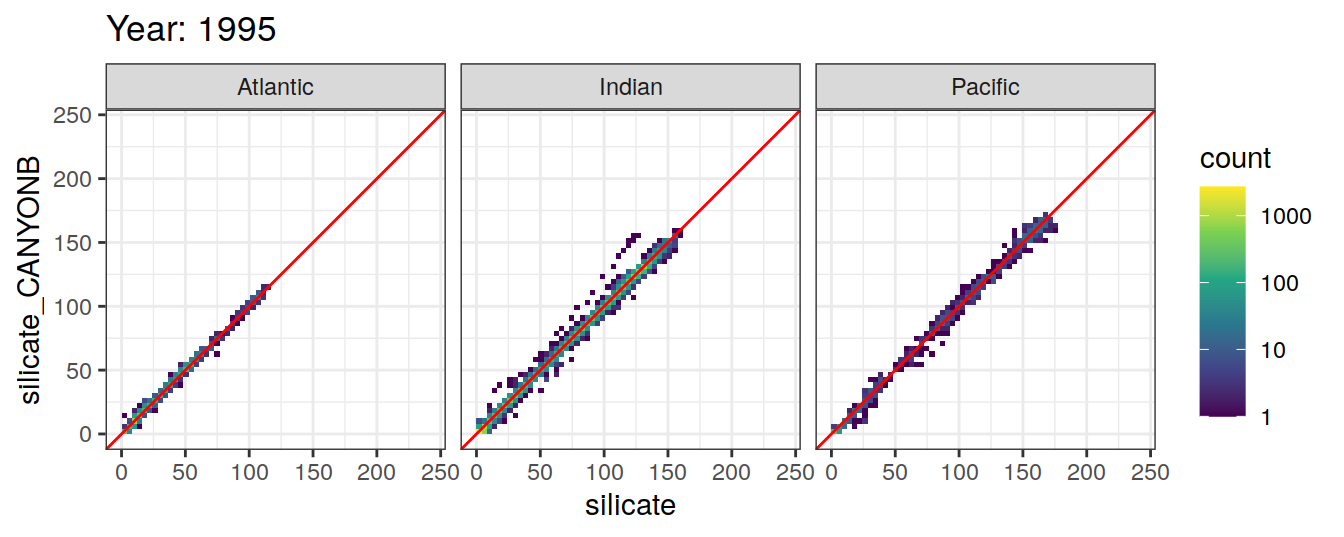
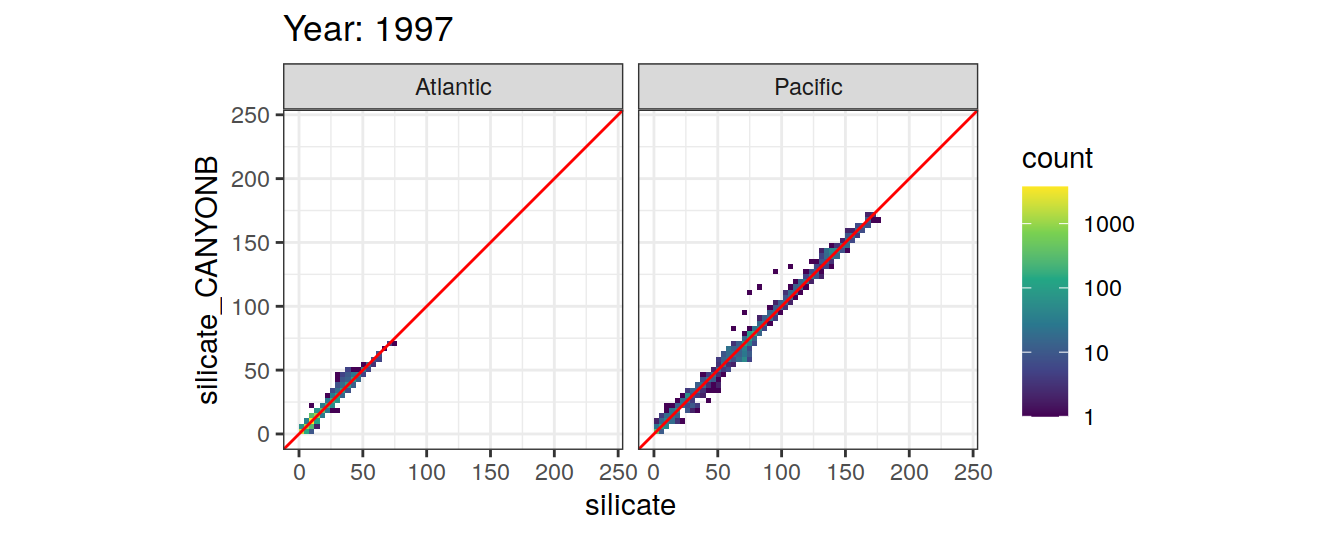
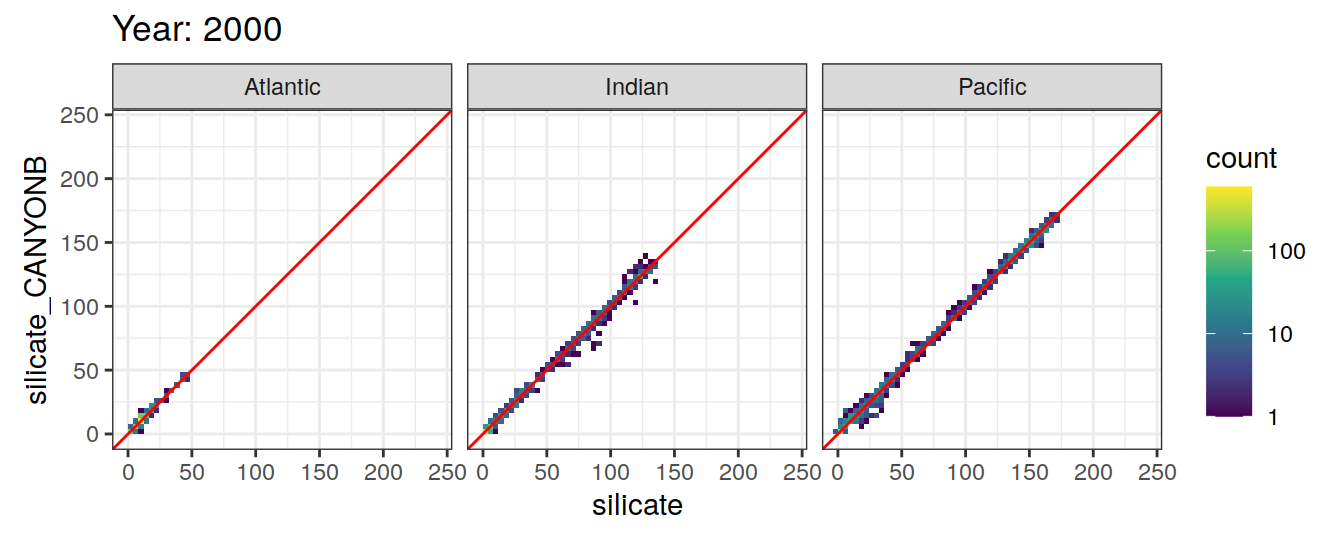
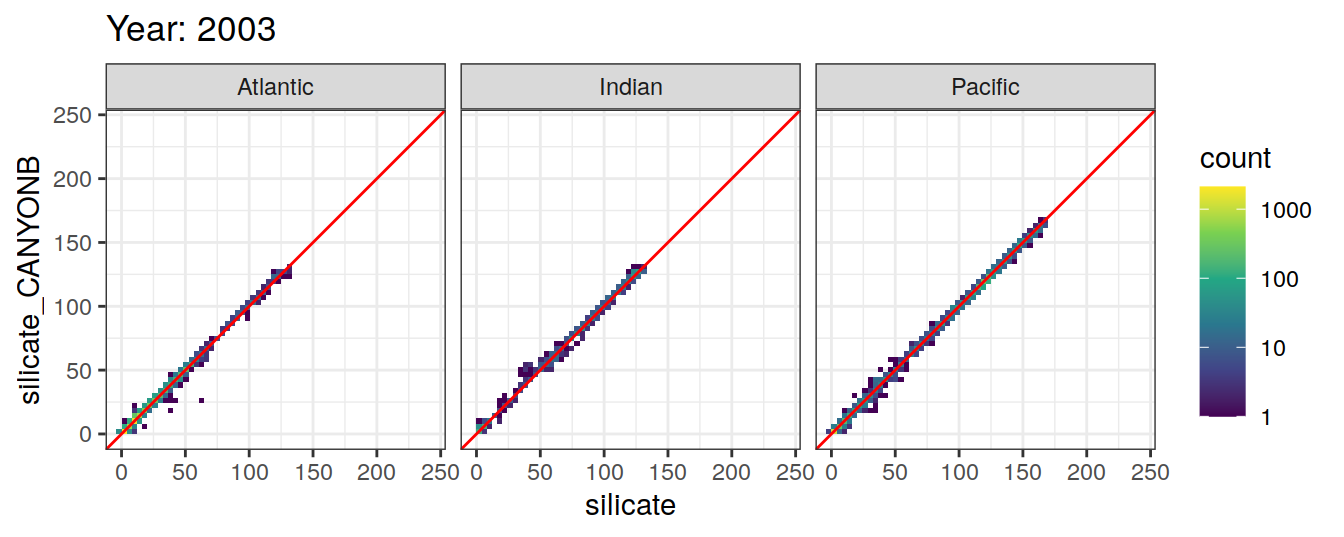
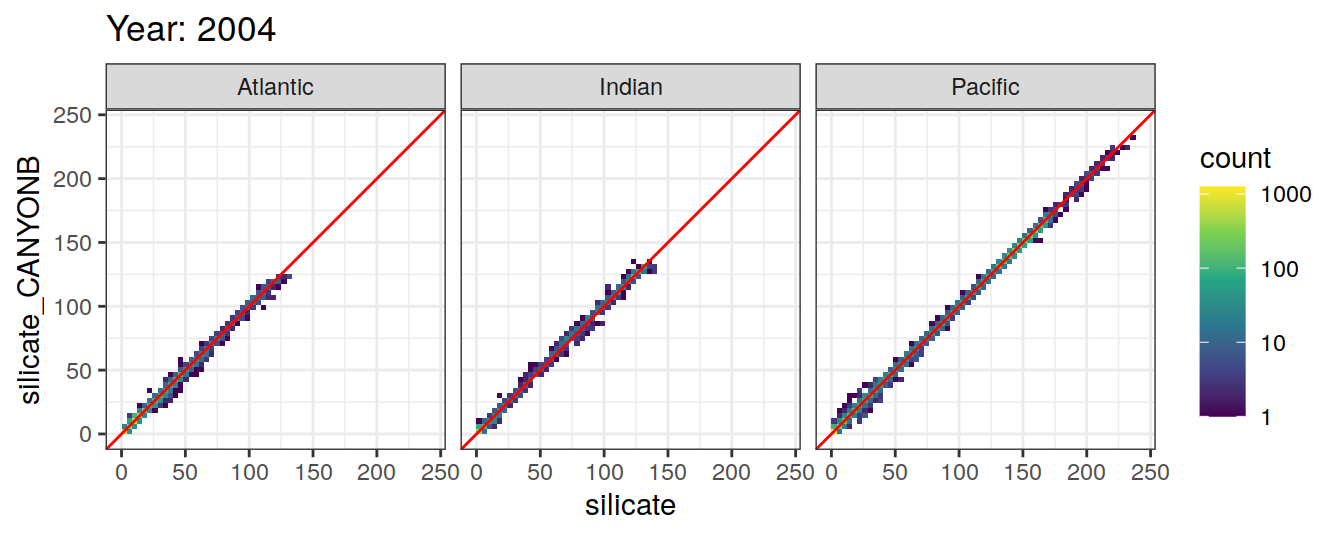
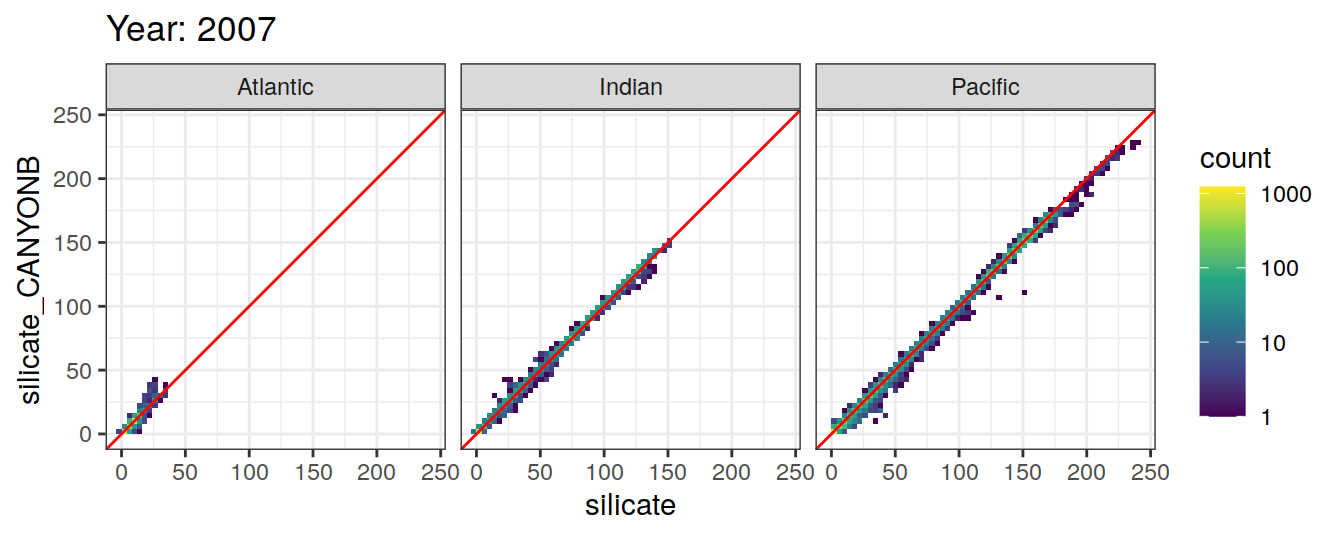
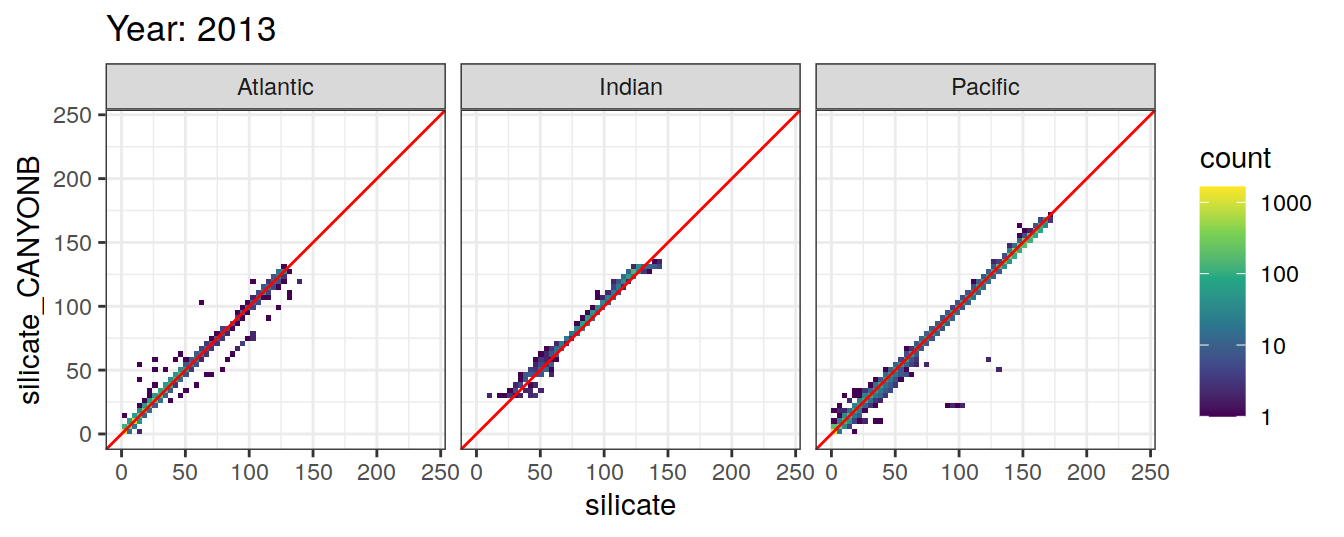
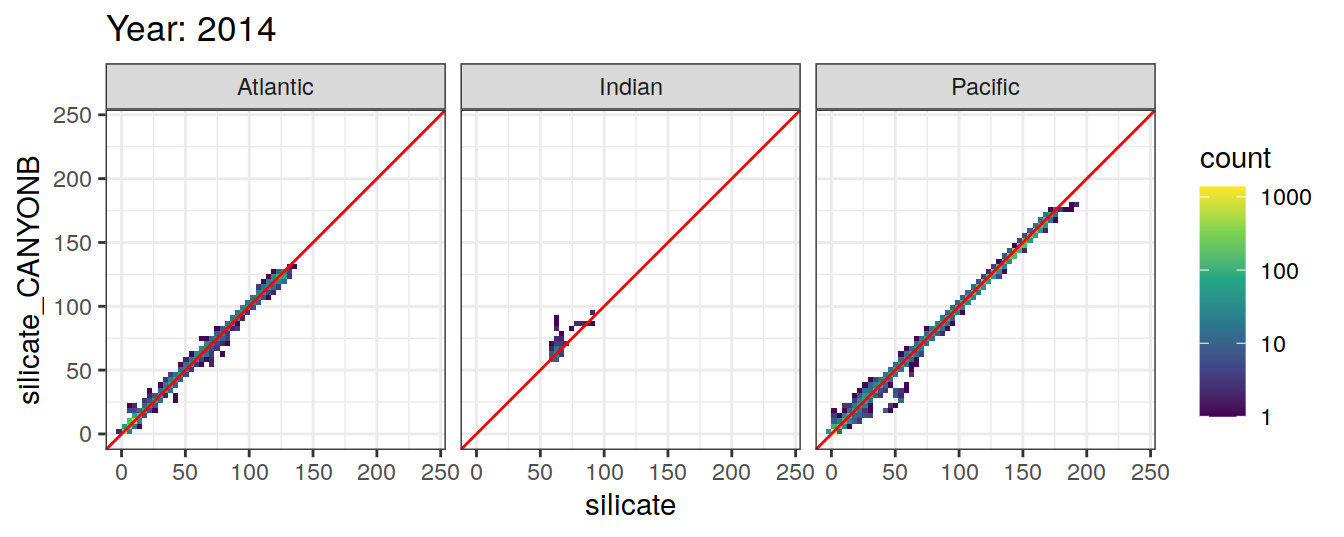
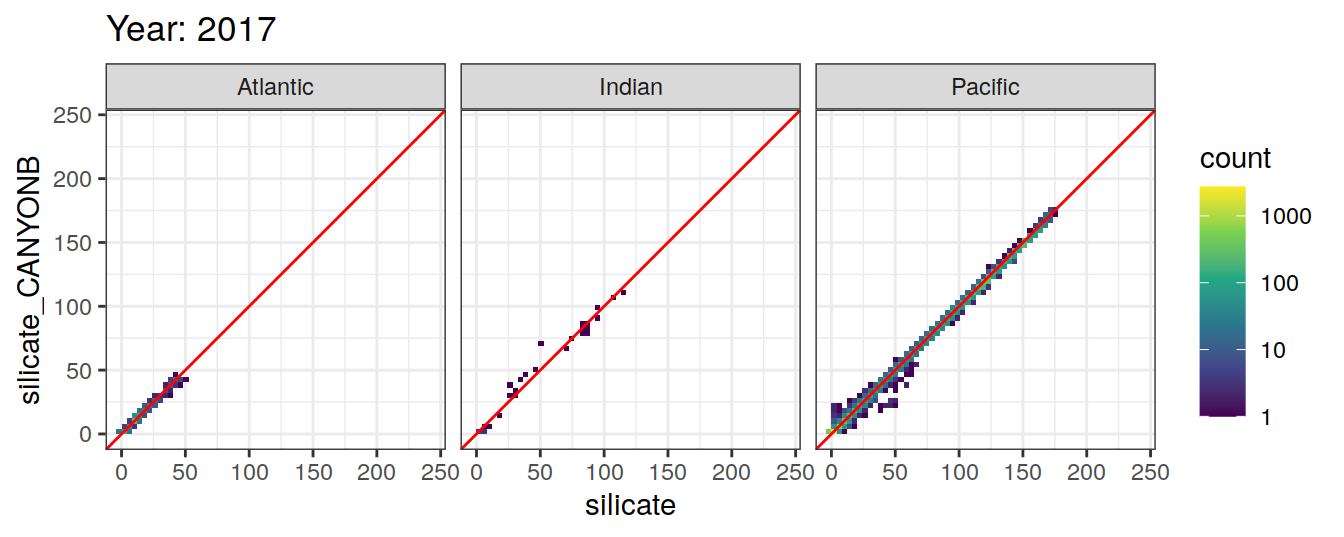
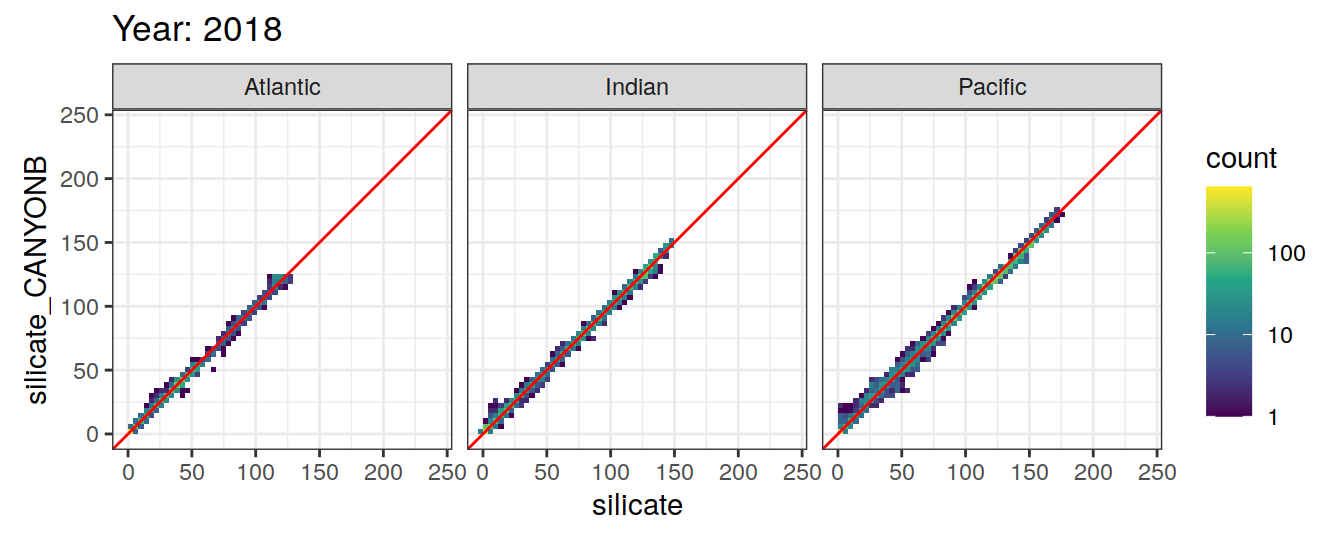
Comparison to GLODAP
source("/net/kryo/work/uptools/co2_calculation/CANYON-B/CANYONB.R")
GLODAP_Can_B <- GLODAP %>%
mutate(lon = if_else(lon > 180, lon - 360, lon)) %>%
arrange(year) %>%
select(row_number, year, date, lat, lon, depth, basin_AIP,
temp, sal, oxygen,
talk, tco2, nitrate, phosphate, silicate)
# filter rows with essential variables for Canyon-B
GLODAP_Can_B <- GLODAP_Can_B %>%
filter(across(c(lat, lon, depth,
temp, sal, oxygen), ~ !is.na(.x)))
GLODAP_Can_B <- GLODAP_Can_B %>%
mutate(as_tibble(
CANYONB(
date = paste0(as.character(date), " 12:00"),
lat = lat,
lon = lon,
pres = depth,
temp = temp,
psal = sal,
doxy = oxygen,
param = c("AT", "CT", "NO3", "PO4", "SiOH4")
)
))
GLODAP_Can_B <- GLODAP_Can_B %>%
select(-ends_with(c("_ci", "_cim", "_cin", "_cii")))
GLODAP_Can_B <- GLODAP_Can_B %>%
rename(
"talk_CANYONB" = "AT",
"tco2_CANYONB" = "CT",
"nitrate_CANYONB" = "NO3",
"phosphate_CANYONB" = "PO4",
"silicate_CANYONB" = "SiOH4"
)
variables <- c("talk", "tco2", "nitrate", "phosphate", "silicate")
for (i_variable in variables) {
# i_variable <- variables[1]
# calculate equal axis limits and binwidth
axis_lims <- GLODAP_Can_B %>%
drop_na() %>%
summarise(max_value = max(c(max(
!!sym(i_variable)
),
max(!!sym(
paste0(i_variable, "_CANYONB")
)))),
min_value = min(c(min(
!!sym(i_variable)
),
min(!!sym(
paste0(i_variable, "_CANYONB")
)))))
binwidth_value <- (axis_lims$max_value - axis_lims$min_value) / 60
axis_lims <- c(axis_lims$min_value, axis_lims$max_value)
print(
ggplot(GLODAP_Can_B, aes(
x = !!sym(i_variable),
y = !!sym(paste0(i_variable, "_CANYONB"))
)) +
geom_bin2d(binwidth = binwidth_value) +
scale_fill_viridis_c(trans = "log10") +
geom_abline(slope = 1, col = 'red') +
coord_equal(xlim = axis_lims,
ylim = axis_lims) +
facet_wrap( ~ basin_AIP) +
labs(title = "All years")
)
for (i_year in unique(GLODAP_Can_B$year)) {
# i_year <- 2017
print(
ggplot(
GLODAP_Can_B %>% filter(year == i_year),
aes(x = !!sym(i_variable),
y = !!sym(paste0(
i_variable, "_CANYONB"
)))
) +
geom_bin2d(binwidth = binwidth_value) +
scale_fill_viridis_c(trans = "log10") +
geom_abline(slope = 1, col = 'red') +
coord_equal(xlim = axis_lims,
ylim = axis_lims) +
facet_wrap( ~ basin_AIP) +
labs(title = paste("Year:", i_year))
)
}
}
Past versions of CANYON-B-1.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-2.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-3.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-4.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-5.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-6.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-7.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-8.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-9.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-10.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-11.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-12.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-13.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-14.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-15.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-16.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-17.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-18.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-19.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-20.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-21.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-22.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-23.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-24.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-25.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-26.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-27.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-28.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-29.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-30.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-31.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-32.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-33.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-34.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-35.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-36.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-37.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-38.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-39.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-40.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-41.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-42.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-43.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-44.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-45.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-46.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-47.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-48.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-49.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-50.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-51.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-52.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-53.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-54.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-55.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-56.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-57.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-58.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-59.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-60.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-61.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-62.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-63.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-64.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-65.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-66.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-67.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-68.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-69.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-70.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-71.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-72.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-73.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-74.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-75.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-76.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-77.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-78.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-79.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-80.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-81.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-82.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-83.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-84.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-85.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-86.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-87.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-88.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-89.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-90.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-91.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-92.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-93.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-94.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-95.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-96.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-97.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-98.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-99.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-100.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-101.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-102.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-103.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-104.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-105.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-106.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-107.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-108.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-109.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-110.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-111.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-112.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-113.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-114.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-115.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-116.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-117.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-118.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-119.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-120.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-121.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-122.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-123.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-124.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-125.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-126.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-127.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-128.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-129.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-130.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-131.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-132.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-133.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-134.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-135.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-136.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-137.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-138.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-139.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-140.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-141.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-142.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-143.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-144.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-145.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-146.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-147.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-148.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-149.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-150.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-151.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-152.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-153.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-154.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-155.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-156.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-157.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-158.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-159.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-160.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-161.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-162.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-163.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-164.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-165.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-166.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-167.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-168.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-169.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-170.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-171.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-172.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-173.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-174.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-175.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-176.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-177.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-178.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-179.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-180.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-181.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-182.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-183.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-184.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-185.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-186.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-187.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-188.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-189.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-190.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-191.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-192.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-193.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-194.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-195.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-196.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-197.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-198.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-199.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-200.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-201.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-202.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-203.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-204.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-205.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-206.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-207.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-208.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-209.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-210.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-211.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-212.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-213.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-214.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-215.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-216.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-217.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-218.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-219.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-220.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-221.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-222.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-223.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-224.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-225.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-226.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-227.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-228.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-229.png
Version
Author
Date
98599d8
jens-daniel-mueller
2021-06-27
9d8353f
jens-daniel-mueller
2021-05-31
Past versions of CANYON-B-230.png
Version
Author
Date
9d8353f
jens-daniel-mueller
2021-05-31