eMLR - assumption testing
Jens Daniel Müller
16 March, 2021
Last updated: 2021-03-16
Checks: 7 0
Knit directory: emlr_obs_v_XXX/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a9930ea. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Unstaged changes:
Modified: code/Workflowr_project_managment.R
Modified: data/auxillary/params_local.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/eMLR_assumption_testing.Rmd) and HTML (docs/eMLR_assumption_testing.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | a1d52ff | jens-daniel-mueller | 2021-03-15 | Build site. |
| html | 0bade3b | jens-daniel-mueller | 2021-03-15 | Build site. |
| html | 27c1f4b | jens-daniel-mueller | 2021-03-14 | Build site. |
| html | af75ebf | jens-daniel-mueller | 2021-03-14 | Build site. |
| html | 5017709 | jens-daniel-mueller | 2021-03-11 | Build site. |
| html | 585b07f | jens-daniel-mueller | 2021-03-11 | Build site. |
| html | 6482ed7 | jens-daniel-mueller | 2021-03-11 | Build site. |
| html | 85a5ed2 | jens-daniel-mueller | 2021-03-10 | Build site. |
| html | 00688a1 | jens-daniel-mueller | 2021-03-05 | Build site. |
| html | 6c0bec6 | jens-daniel-mueller | 2021-03-05 | Build site. |
| html | 3c2ec33 | jens-daniel-mueller | 2021-03-05 | Build site. |
| html | af70b94 | jens-daniel-mueller | 2021-03-04 | Build site. |
| html | 86406d5 | jens-daniel-mueller | 2021-02-24 | Build site. |
| html | 3d3b4cc | jens-daniel-mueller | 2021-02-23 | Build site. |
| html | 7b672f7 | jens-daniel-mueller | 2021-01-11 | Build site. |
| html | 33ba23c | jens-daniel-mueller | 2021-01-07 | Build site. |
| html | 318609d | jens-daniel-mueller | 2020-12-23 | adapted more variable predictor selection |
| html | 9d0b2d0 | jens-daniel-mueller | 2020-12-23 | Build site. |
| html | 0aa2b50 | jens-daniel-mueller | 2020-12-23 | remove html before duplication |
| html | 39113c3 | jens-daniel-mueller | 2020-12-23 | Build site. |
| html | 2886da0 | jens-daniel-mueller | 2020-12-19 | Build site. |
| html | 02f0ee9 | jens-daniel-mueller | 2020-12-18 | cleaned up for copying template |
| html | 965dba3 | jens-daniel-mueller | 2020-12-18 | Build site. |
| html | 5d452fe | jens-daniel-mueller | 2020-12-18 | Build site. |
| Rmd | ca65bf5 | jens-daniel-mueller | 2020-12-18 | rebuild after final cleaning |
| html | 7bcb4eb | jens-daniel-mueller | 2020-12-18 | Build site. |
| html | d397028 | jens-daniel-mueller | 2020-12-18 | Build site. |
| html | 7131186 | jens-daniel-mueller | 2020-12-17 | Build site. |
| html | 22b07fb | jens-daniel-mueller | 2020-12-17 | Build site. |
| html | f3a708f | jens-daniel-mueller | 2020-12-17 | Build site. |
| html | e4ca289 | jens-daniel-mueller | 2020-12-16 | Build site. |
| html | 158fe26 | jens-daniel-mueller | 2020-12-15 | Build site. |
| html | 7a9a4cb | jens-daniel-mueller | 2020-12-15 | Build site. |
| html | 61b263c | jens-daniel-mueller | 2020-12-15 | Build site. |
| html | 4d612dd | jens-daniel-mueller | 2020-12-15 | Build site. |
| Rmd | e7e5ff1 | jens-daniel-mueller | 2020-12-15 | rebuild with eMLR target variable selection |
| html | 953caf3 | jens-daniel-mueller | 2020-12-15 | Build site. |
| html | 42daf5c | jens-daniel-mueller | 2020-12-14 | Build site. |
| Rmd | 923aa7f | jens-daniel-mueller | 2020-12-14 | rebuild with new path and auto folder creation |
| html | 984697e | jens-daniel-mueller | 2020-12-12 | Build site. |
| html | 3ebff89 | jens-daniel-mueller | 2020-12-12 | Build site. |
| html | 5d96d3c | jens-daniel-mueller | 2020-12-11 | Build site. |
| Rmd | 3d33a37 | jens-daniel-mueller | 2020-12-11 | selectable basinmask, try 5 |
| html | b01a367 | jens-daniel-mueller | 2020-12-09 | Build site. |
| html | 24a632f | jens-daniel-mueller | 2020-12-07 | Build site. |
| html | 92dca91 | jens-daniel-mueller | 2020-12-07 | Build site. |
| html | 6a8004b | jens-daniel-mueller | 2020-12-07 | Build site. |
| html | 70bf1a5 | jens-daniel-mueller | 2020-12-07 | Build site. |
| html | 7555355 | jens-daniel-mueller | 2020-12-07 | Build site. |
| html | 143d6fa | jens-daniel-mueller | 2020-12-07 | Build site. |
| Rmd | 33b1973 | jens-daniel-mueller | 2020-12-07 | run with WOCE flag 2 only |
| html | abc6818 | jens-daniel-mueller | 2020-12-03 | Build site. |
| Rmd | 992ba15 | jens-daniel-mueller | 2020-12-03 | rebuild with variable inventory depth |
| html | c8c2e7b | jens-daniel-mueller | 2020-12-03 | Build site. |
| Rmd | 83203db | jens-daniel-mueller | 2020-12-03 | calculate cant with variable inventory depth |
| html | 090e4d5 | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | 7c25f7a | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | ec8dc38 | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | c987de1 | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | f8358f8 | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | b03ddb8 | jens-daniel-mueller | 2020-12-02 | Build site. |
| Rmd | 9183e8f | jens-daniel-mueller | 2020-12-02 | revised assignment of era to eras |
| html | 22d0127 | jens-daniel-mueller | 2020-12-01 | Build site. |
| html | 0ff728b | jens-daniel-mueller | 2020-12-01 | Build site. |
| html | 91435ae | jens-daniel-mueller | 2020-12-01 | Build site. |
| Rmd | 17d09be | jens-daniel-mueller | 2020-12-01 | auto eras naming |
| html | cf19652 | jens-daniel-mueller | 2020-11-30 | Build site. |
| Rmd | 2842970 | jens-daniel-mueller | 2020-11-30 | cleaned for eMLR part only |
| html | 196be51 | jens-daniel-mueller | 2020-11-30 | Build site. |
| Rmd | 7a4b015 | jens-daniel-mueller | 2020-11-30 | first rebuild on ETH server |
| Rmd | bc61ce3 | Jens Müller | 2020-11-30 | Initial commit |
| html | bc61ce3 | Jens Müller | 2020-11-30 | Initial commit |
1 Required data
Required are:
- cleaned and prepared GLODAPv2.2020 file
GLODAP <-
read_csv(paste(path_version_data,
"GLODAPv2.2020_MLR_fitting_ready.csv",
sep = ""))2 Predictor correlation
The correlation between:
- pairs of seven potential predictor variables and
- C* and seven potential predictor variables
were investigated based on:
- property-property plots and
- calculated correlation coeffcients.
2.1 Correlation plots
For an overview, a random subset of data from all eras was plotted separately for both basins, with color indicating neutral density slabs (high density = dark-purple color).
for (i_basin in unique(GLODAP$basin)) {
# i_basin <- unique(GLODAP$basin)[1]
print(
GLODAP %>%
filter(basin == i_basin) %>%
sample_frac(0.05) %>%
ggpairs(columns = c(params_local$MLR_target,
params_local$MLR_predictors),
upper = "blank",
ggplot2::aes(col = gamma_slab, fill = gamma_slab, alpha = 0.01)) +
scale_fill_viridis_d(direction = -1) +
scale_color_viridis_d(direction = -1) +
labs(title = paste("Basin:", i_basin ,"| era: all | subsample size: 5 % of",
nrow(GLODAP %>% filter(basin == i_basin))))
)
}
| Version | Author | Date |
|---|---|---|
| a1d52ff | jens-daniel-mueller | 2021-03-15 |
| 0bade3b | jens-daniel-mueller | 2021-03-15 |
| 27c1f4b | jens-daniel-mueller | 2021-03-14 |
| af75ebf | jens-daniel-mueller | 2021-03-14 |
| 5017709 | jens-daniel-mueller | 2021-03-11 |
| 585b07f | jens-daniel-mueller | 2021-03-11 |
| 85a5ed2 | jens-daniel-mueller | 2021-03-10 |
| 6c0bec6 | jens-daniel-mueller | 2021-03-05 |
| af70b94 | jens-daniel-mueller | 2021-03-04 |
| 7b672f7 | jens-daniel-mueller | 2021-01-11 |
| 33ba23c | jens-daniel-mueller | 2021-01-07 |
| 318609d | jens-daniel-mueller | 2020-12-23 |
| 9d0b2d0 | jens-daniel-mueller | 2020-12-23 |
| 0aa2b50 | jens-daniel-mueller | 2020-12-23 |
| 2886da0 | jens-daniel-mueller | 2020-12-19 |
| 02f0ee9 | jens-daniel-mueller | 2020-12-18 |
| 7bcb4eb | jens-daniel-mueller | 2020-12-18 |
| 158fe26 | jens-daniel-mueller | 2020-12-15 |
| 7a9a4cb | jens-daniel-mueller | 2020-12-15 |
| 61b263c | jens-daniel-mueller | 2020-12-15 |
| 4d612dd | jens-daniel-mueller | 2020-12-15 |
| 984697e | jens-daniel-mueller | 2020-12-12 |
| 3ebff89 | jens-daniel-mueller | 2020-12-12 |
| 5d96d3c | jens-daniel-mueller | 2020-12-11 |

| Version | Author | Date |
|---|---|---|
| a1d52ff | jens-daniel-mueller | 2021-03-15 |
| 0bade3b | jens-daniel-mueller | 2021-03-15 |
| 27c1f4b | jens-daniel-mueller | 2021-03-14 |
| af75ebf | jens-daniel-mueller | 2021-03-14 |
| 5017709 | jens-daniel-mueller | 2021-03-11 |
| 585b07f | jens-daniel-mueller | 2021-03-11 |
| 85a5ed2 | jens-daniel-mueller | 2021-03-10 |
| 6c0bec6 | jens-daniel-mueller | 2021-03-05 |
| af70b94 | jens-daniel-mueller | 2021-03-04 |
| 7b672f7 | jens-daniel-mueller | 2021-01-11 |
| 33ba23c | jens-daniel-mueller | 2021-01-07 |
| 318609d | jens-daniel-mueller | 2020-12-23 |
| 9d0b2d0 | jens-daniel-mueller | 2020-12-23 |
| 0aa2b50 | jens-daniel-mueller | 2020-12-23 |
| 2886da0 | jens-daniel-mueller | 2020-12-19 |
| 02f0ee9 | jens-daniel-mueller | 2020-12-18 |
| 7bcb4eb | jens-daniel-mueller | 2020-12-18 |
| 158fe26 | jens-daniel-mueller | 2020-12-15 |
| 7a9a4cb | jens-daniel-mueller | 2020-12-15 |
| 61b263c | jens-daniel-mueller | 2020-12-15 |
| 4d612dd | jens-daniel-mueller | 2020-12-15 |
| 984697e | jens-daniel-mueller | 2020-12-12 |
| 3ebff89 | jens-daniel-mueller | 2020-12-12 |
| 5d96d3c | jens-daniel-mueller | 2020-12-11 |
Individual correlation plots for each basin, era and neutral density (gamma) slab are available at:
/nfs/kryo/work/jenmueller/emlr_cant/observations/v_XXX/figures/Observations_correlation/
if (params_local$plot_all_figures == "y") {
for (i_basin in unique(GLODAP$basin)) {
for (i_era in unique(GLODAP$era)) {
# i_basin <- unique(GLODAP$basin)[1]
# i_era <- unique(GLODAP$era)[1]
GLODAP_basin_era <- GLODAP %>%
filter(basin == i_basin,
era == i_era)
for (i_gamma_slab in unique(GLODAP_basin_era$gamma_slab)) {
# i_gamma_slab <- unique(GLODAP_basin_era$gamma_slab)[5]
GLODAP_highlight <- GLODAP_basin_era %>%
mutate(gamma_highlight = if_else(gamma_slab == i_gamma_slab,
"in", "out")) %>%
arrange(desc(gamma_highlight))
p <- GLODAP_highlight %>%
ggpairs(
columns = c(params_local$MLR_target,
params_local$MLR_predictors),
ggplot2::aes(
col = gamma_highlight,
fill = gamma_highlight,
alpha = 0.01
)
) +
scale_fill_manual(values = c("red", "grey")) +
scale_color_manual(values = c("red", "grey")) +
labs(
title = paste(
i_era,
"|",
i_basin,
"| Gamma slab",
i_gamma_slab,
"| # obs total",
nrow(GLODAP_basin_era),
"| # obs slab",
nrow(GLODAP_highlight %>%
filter(gamma_highlight == "in"))
)
)
png(
filename = paste(
path_version_figures,
"Observations_correlation/",
paste(
"Predictor_correlation",
i_era,
i_basin,
i_gamma_slab,
".png",
sep = "_"
),
sep = ""),
width = 12,
height = 12,
units = "in",
res = 300
)
print(p)
dev.off()
}
}
}
}2.2 Correlation assesment
2.2.1 Calculation of correlation coeffcients
Correlation coefficients were calculated individually within each slabs, era and basin.
for (i_basin in unique(GLODAP$basin)) {
for (i_era in unique(GLODAP$era)) {
# i_basin <- unique(GLODAP$basin)[1]
# i_era <- unique(GLODAP$era)[1]
GLODAP_basin_era <- GLODAP %>%
filter(basin == i_basin,
era == i_era) %>%
select(basin,
era,
gamma_slab,
params_local$MLR_target,
params_local$MLR_predictors)
for (i_gamma_slab in unique(GLODAP_basin_era$gamma_slab)) {
# i_gamma_slab <- unique(GLODAP_basin_era$gamma_slab)[5]
print(i_gamma_slab)
GLODAP_basin_era_slab <- GLODAP_basin_era %>%
filter(gamma_slab == i_gamma_slab)
# calculate correlation table
cor_target_predictor_temp <- GLODAP_basin_era_slab %>%
select(-c(basin, era, gamma_slab)) %>%
correlate() %>%
focus(params_local$MLR_target) %>%
mutate(basin = i_basin,
era = i_era,
gamma_slab = i_gamma_slab)
if (exists("cor_target_predictor")) {
cor_target_predictor <-
bind_rows(cor_target_predictor, cor_target_predictor_temp)
}
if (!exists("cor_target_predictor")) {
cor_target_predictor <- cor_target_predictor_temp
}
cor_predictors_temp <- GLODAP_basin_era_slab %>%
select(-c(basin, era, gamma_slab)) %>%
correlate() %>%
shave %>%
stretch() %>%
filter(!is.na(r),
x != params_local$MLR_target,
y != params_local$MLR_target) %>%
mutate(pair = paste(x, y, sep = " + ")) %>%
select(-c(x, y)) %>%
mutate(basin = i_basin,
era = i_era,
gamma_slab = i_gamma_slab)
if (exists("cor_predictors")) {
cor_predictors <- bind_rows(cor_predictors, cor_predictors_temp)
}
if (!exists("cor_predictors")) {
cor_predictors <- cor_predictors_temp
}
}
}
}
rm(cor_predictors_temp, cor_target_predictor_temp,
i_gamma_slab, i_era, i_basin,
GLODAP_basin_era, GLODAP_basin_era_slab)2.2.2 Predictor pairs
Below, the range of correlations coefficients for each predictor pair is plotted per basin (facet) and density slab (color). Note that the range indicates the min and max values of in total 3 calculated coefficients (one per era).
# calculate min, max, mean across all eras
cor_predictors_stats <- cor_predictors %>%
group_by(pair, basin, gamma_slab) %>%
summarise(mean_r = mean(r),
min_r = min(r),
max_r = max(r)) %>%
ungroup()
# plot figure
cor_predictors_stats %>%
mutate(pair = reorder(pair, mean_r)) %>%
ggplot() +
geom_vline(xintercept = c(-0.9, 0.9), col = "red") +
geom_vline(xintercept = 0) +
geom_linerange(
aes(y = pair, xmin = min_r, xmax = max_r, col = gamma_slab),
position = position_dodge(width = 0.6)) +
facet_wrap(~basin) +
scale_color_viridis_d(direction = -1) +
labs(x = "correlation coefficient", y = "") +
theme(legend.position = "top")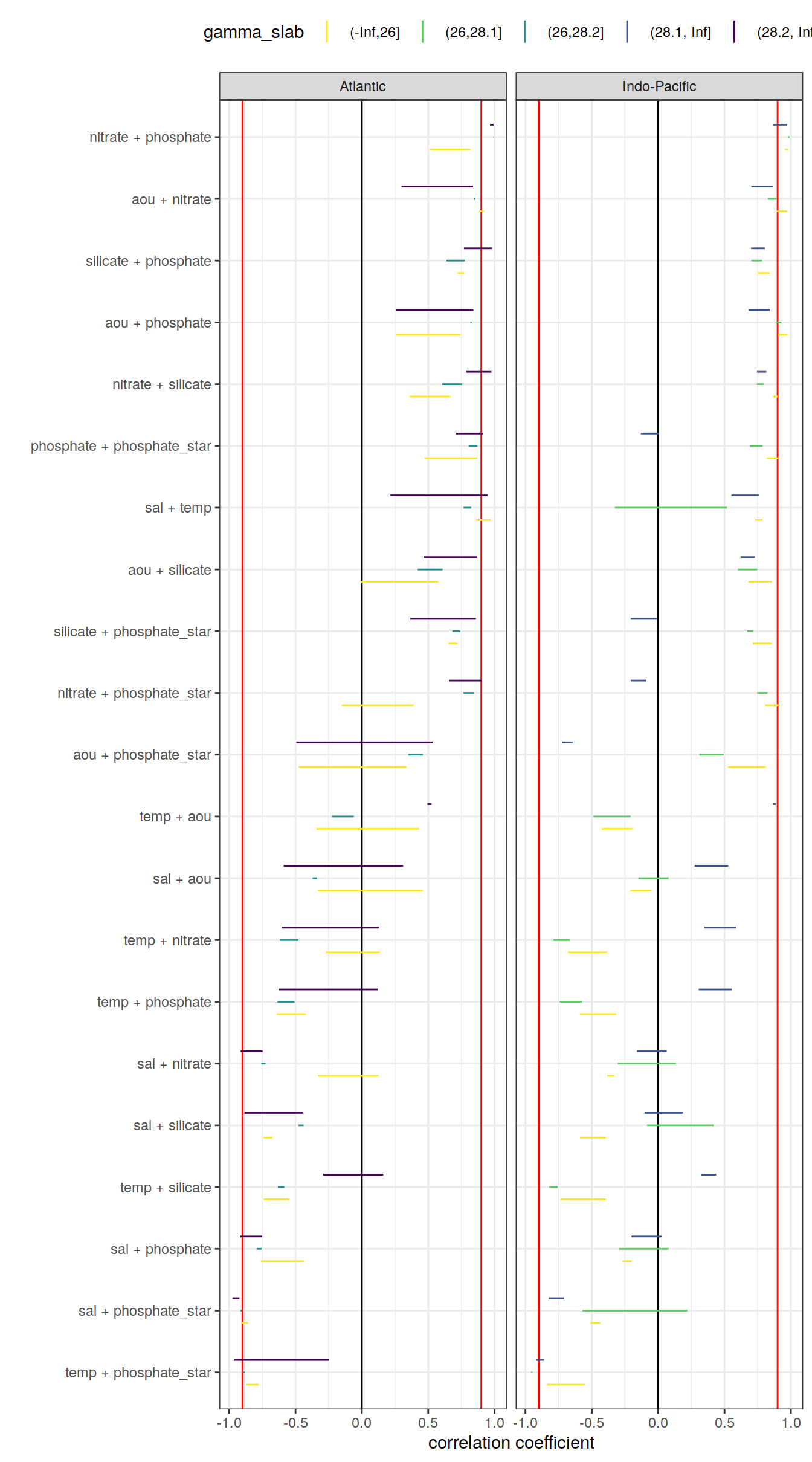
| Version | Author | Date |
|---|---|---|
| a1d52ff | jens-daniel-mueller | 2021-03-15 |
| 0bade3b | jens-daniel-mueller | 2021-03-15 |
| 27c1f4b | jens-daniel-mueller | 2021-03-14 |
| af75ebf | jens-daniel-mueller | 2021-03-14 |
| 5017709 | jens-daniel-mueller | 2021-03-11 |
| 585b07f | jens-daniel-mueller | 2021-03-11 |
| 85a5ed2 | jens-daniel-mueller | 2021-03-10 |
| 6c0bec6 | jens-daniel-mueller | 2021-03-05 |
| 7b672f7 | jens-daniel-mueller | 2021-01-11 |
| 33ba23c | jens-daniel-mueller | 2021-01-07 |
| 318609d | jens-daniel-mueller | 2020-12-23 |
| 9d0b2d0 | jens-daniel-mueller | 2020-12-23 |
| 0aa2b50 | jens-daniel-mueller | 2020-12-23 |
| 2886da0 | jens-daniel-mueller | 2020-12-19 |
| 02f0ee9 | jens-daniel-mueller | 2020-12-18 |
| 158fe26 | jens-daniel-mueller | 2020-12-15 |
| 984697e | jens-daniel-mueller | 2020-12-12 |
| 3ebff89 | jens-daniel-mueller | 2020-12-12 |
| 5d96d3c | jens-daniel-mueller | 2020-12-11 |
| 24a632f | jens-daniel-mueller | 2020-12-07 |
| 6a8004b | jens-daniel-mueller | 2020-12-07 |
| 70bf1a5 | jens-daniel-mueller | 2020-12-07 |
| 7555355 | jens-daniel-mueller | 2020-12-07 |
| 143d6fa | jens-daniel-mueller | 2020-12-07 |
| 0ff728b | jens-daniel-mueller | 2020-12-01 |
| 91435ae | jens-daniel-mueller | 2020-12-01 |
| 196be51 | jens-daniel-mueller | 2020-11-30 |
| bc61ce3 | Jens Müller | 2020-11-30 |
# print table
kable(cor_predictors_stats) %>%
add_header_above() %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| pair | basin | gamma_slab | mean_r | min_r | max_r |
|---|---|---|---|---|---|
| aou + nitrate | Atlantic | (-Inf,26] | 0.9023751 | 0.8823703 | 0.9188740 |
| aou + nitrate | Atlantic | (26,28.2] | 0.8497682 | 0.8460103 | 0.8559783 |
| aou + nitrate | Atlantic | (28.2, Inf] | 0.6543331 | 0.2987857 | 0.8382487 |
| aou + nitrate | Indo-Pacific | (-Inf,26] | 0.9279977 | 0.8900168 | 0.9727810 |
| aou + nitrate | Indo-Pacific | (26,28.1] | 0.8698723 | 0.8270094 | 0.8918242 |
| aou + nitrate | Indo-Pacific | (28.1, Inf] | 0.7726827 | 0.7020998 | 0.8661750 |
| aou + phosphate | Atlantic | (-Inf,26] | 0.5422936 | 0.2598012 | 0.7434863 |
| aou + phosphate | Atlantic | (26,28.2] | 0.8226072 | 0.8185323 | 0.8257789 |
| aou + phosphate | Atlantic | (28.2, Inf] | 0.6402840 | 0.2585115 | 0.8400544 |
| aou + phosphate | Indo-Pacific | (-Inf,26] | 0.9421463 | 0.9043755 | 0.9734355 |
| aou + phosphate | Indo-Pacific | (26,28.1] | 0.9131789 | 0.8887854 | 0.9291946 |
| aou + phosphate | Indo-Pacific | (28.1, Inf] | 0.7634814 | 0.6804230 | 0.8401635 |
| aou + phosphate_star | Atlantic | (-Inf,26] | -0.1733598 | -0.4766688 | 0.3362197 |
| aou + phosphate_star | Atlantic | (26,28.2] | 0.4116665 | 0.3496995 | 0.4604067 |
| aou + phosphate_star | Atlantic | (28.2, Inf] | 0.1881227 | -0.4929523 | 0.5332518 |
| aou + phosphate_star | Indo-Pacific | (-Inf,26] | 0.6907002 | 0.5252778 | 0.8117232 |
| aou + phosphate_star | Indo-Pacific | (26,28.1] | 0.3973642 | 0.3099802 | 0.4946232 |
| aou + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.6877056 | -0.7235766 | -0.6447285 |
| aou + silicate | Atlantic | (-Inf,26] | 0.2575781 | -0.0090087 | 0.5754309 |
| aou + silicate | Atlantic | (26,28.2] | 0.5046701 | 0.4213469 | 0.6080963 |
| aou + silicate | Atlantic | (28.2, Inf] | 0.7231502 | 0.4655243 | 0.8667673 |
| aou + silicate | Indo-Pacific | (-Inf,26] | 0.7612281 | 0.6806252 | 0.8539364 |
| aou + silicate | Indo-Pacific | (26,28.1] | 0.6506044 | 0.6013762 | 0.7472330 |
| aou + silicate | Indo-Pacific | (28.1, Inf] | 0.6632363 | 0.6264779 | 0.7284222 |
| nitrate + phosphate | Atlantic | (-Inf,26] | 0.6939950 | 0.5117436 | 0.8173702 |
| nitrate + phosphate | Atlantic | (26,28.2] | 0.9924594 | 0.9914240 | 0.9939900 |
| nitrate + phosphate | Atlantic | (28.2, Inf] | 0.9825189 | 0.9655311 | 0.9931844 |
| nitrate + phosphate | Indo-Pacific | (-Inf,26] | 0.9692465 | 0.9543975 | 0.9774665 |
| nitrate + phosphate | Indo-Pacific | (26,28.1] | 0.9848393 | 0.9773303 | 0.9903013 |
| nitrate + phosphate | Indo-Pacific | (28.1, Inf] | 0.9302885 | 0.8663697 | 0.9714872 |
| nitrate + phosphate_star | Atlantic | (-Inf,26] | 0.0541938 | -0.1506424 | 0.3908953 |
| nitrate + phosphate_star | Atlantic | (26,28.2] | 0.8108042 | 0.7643849 | 0.8441759 |
| nitrate + phosphate_star | Atlantic | (28.2, Inf] | 0.8161710 | 0.6579978 | 0.9018502 |
| nitrate + phosphate_star | Indo-Pacific | (-Inf,26] | 0.8680068 | 0.8048000 | 0.9153999 |
| nitrate + phosphate_star | Indo-Pacific | (26,28.1] | 0.7772318 | 0.7452512 | 0.8216409 |
| nitrate + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.1396453 | -0.2058096 | -0.0882964 |
| nitrate + silicate | Atlantic | (-Inf,26] | 0.4833689 | 0.3594654 | 0.6667275 |
| nitrate + silicate | Atlantic | (26,28.2] | 0.6723928 | 0.6054068 | 0.7547735 |
| nitrate + silicate | Atlantic | (28.2, Inf] | 0.9109951 | 0.7869083 | 0.9761971 |
| nitrate + silicate | Indo-Pacific | (-Inf,26] | 0.8861818 | 0.8635500 | 0.8992106 |
| nitrate + silicate | Indo-Pacific | (26,28.1] | 0.7614373 | 0.7440481 | 0.7932007 |
| nitrate + silicate | Indo-Pacific | (28.1, Inf] | 0.7854278 | 0.7442723 | 0.8147302 |
| phosphate + phosphate_star | Atlantic | (-Inf,26] | 0.6851429 | 0.4715637 | 0.8695592 |
| phosphate + phosphate_star | Atlantic | (26,28.2] | 0.8434871 | 0.8039118 | 0.8708875 |
| phosphate + phosphate_star | Atlantic | (28.2, Inf] | 0.8432769 | 0.7107125 | 0.9143822 |
| phosphate + phosphate_star | Indo-Pacific | (-Inf,26] | 0.8765585 | 0.8191963 | 0.9092677 |
| phosphate + phosphate_star | Indo-Pacific | (26,28.1] | 0.7265507 | 0.6908297 | 0.7867655 |
| phosphate + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.0673758 | -0.1306307 | 0.0077083 |
| sal + aou | Atlantic | (-Inf,26] | 0.1369277 | -0.3310034 | 0.4592283 |
| sal + aou | Atlantic | (26,28.2] | -0.3518209 | -0.3708266 | -0.3376168 |
| sal + aou | Atlantic | (28.2, Inf] | -0.2722572 | -0.5887224 | 0.3108904 |
| sal + aou | Indo-Pacific | (-Inf,26] | -0.1427131 | -0.2119195 | -0.0525508 |
| sal + aou | Indo-Pacific | (26,28.1] | -0.0104237 | -0.1487352 | 0.0791408 |
| sal + aou | Indo-Pacific | (28.1, Inf] | 0.4381001 | 0.2748165 | 0.5285879 |
| sal + nitrate | Atlantic | (-Inf,26] | -0.0473435 | -0.3308324 | 0.1247558 |
| sal + nitrate | Atlantic | (26,28.2] | -0.7366711 | -0.7578511 | -0.7256783 |
| sal + nitrate | Atlantic | (28.2, Inf] | -0.8520824 | -0.9129323 | -0.7478006 |
| sal + nitrate | Indo-Pacific | (-Inf,26] | -0.3628061 | -0.3843728 | -0.3316537 |
| sal + nitrate | Indo-Pacific | (26,28.1] | -0.1531361 | -0.3037177 | 0.1348102 |
| sal + nitrate | Indo-Pacific | (28.1, Inf] | -0.0464716 | -0.1597012 | 0.0640355 |
| sal + phosphate | Atlantic | (-Inf,26] | -0.5657649 | -0.7613570 | -0.4334744 |
| sal + phosphate | Atlantic | (26,28.2] | -0.7696367 | -0.7902254 | -0.7540245 |
| sal + phosphate | Atlantic | (28.2, Inf] | -0.8561618 | -0.9146446 | -0.7512426 |
| sal + phosphate | Indo-Pacific | (-Inf,26] | -0.2331966 | -0.2688773 | -0.2002269 |
| sal + phosphate | Indo-Pacific | (26,28.1] | -0.1483684 | -0.2954408 | 0.0791816 |
| sal + phosphate | Indo-Pacific | (28.1, Inf] | -0.0608389 | -0.2007789 | 0.0297573 |
| sal + phosphate_star | Atlantic | (-Inf,26] | -0.8881511 | -0.9084437 | -0.8595142 |
| sal + phosphate_star | Atlantic | (26,28.2] | -0.9093510 | -0.9139892 | -0.9037945 |
| sal + phosphate_star | Atlantic | (28.2, Inf] | -0.9559723 | -0.9752144 | -0.9223373 |
| sal + phosphate_star | Indo-Pacific | (-Inf,26] | -0.4825926 | -0.5100133 | -0.4363244 |
| sal + phosphate_star | Indo-Pacific | (26,28.1] | -0.2359722 | -0.5698878 | 0.2191250 |
| sal + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.7756090 | -0.8264981 | -0.7071927 |
| sal + silicate | Atlantic | (-Inf,26] | -0.7094091 | -0.7405506 | -0.6732885 |
| sal + silicate | Atlantic | (26,28.2] | -0.4615601 | -0.4773367 | -0.4394293 |
| sal + silicate | Atlantic | (28.2, Inf] | -0.7202054 | -0.8846868 | -0.4463975 |
| sal + silicate | Indo-Pacific | (-Inf,26] | -0.5099271 | -0.5881732 | -0.3952421 |
| sal + silicate | Indo-Pacific | (26,28.1] | 0.1317132 | -0.0832250 | 0.4172679 |
| sal + silicate | Indo-Pacific | (28.1, Inf] | 0.0636811 | -0.1011285 | 0.1893801 |
| sal + temp | Atlantic | (-Inf,26] | 0.9282488 | 0.8591490 | 0.9718026 |
| sal + temp | Atlantic | (26,28.2] | 0.7919537 | 0.7661651 | 0.8236916 |
| sal + temp | Atlantic | (28.2, Inf] | 0.4643470 | 0.2151571 | 0.9466866 |
| sal + temp | Indo-Pacific | (-Inf,26] | 0.7511059 | 0.7322291 | 0.7860495 |
| sal + temp | Indo-Pacific | (26,28.1] | 0.1394241 | -0.3257970 | 0.5179756 |
| sal + temp | Indo-Pacific | (28.1, Inf] | 0.6640090 | 0.5516618 | 0.7574075 |
| silicate + phosphate | Atlantic | (-Inf,26] | 0.7463328 | 0.7218927 | 0.7719039 |
| silicate + phosphate | Atlantic | (26,28.2] | 0.6997652 | 0.6370500 | 0.7751127 |
| silicate + phosphate | Atlantic | (28.2, Inf] | 0.9058355 | 0.7689431 | 0.9797417 |
| silicate + phosphate | Indo-Pacific | (-Inf,26] | 0.8054182 | 0.7494571 | 0.8389260 |
| silicate + phosphate | Indo-Pacific | (26,28.1] | 0.7347214 | 0.7007987 | 0.7829188 |
| silicate + phosphate | Indo-Pacific | (28.1, Inf] | 0.7655142 | 0.6999056 | 0.8049697 |
| silicate + phosphate_star | Atlantic | (-Inf,26] | 0.6900571 | 0.6533103 | 0.7219868 |
| silicate + phosphate_star | Atlantic | (26,28.2] | 0.7129083 | 0.6823614 | 0.7403741 |
| silicate + phosphate_star | Atlantic | (28.2, Inf] | 0.6936300 | 0.3652654 | 0.8594449 |
| silicate + phosphate_star | Indo-Pacific | (-Inf,26] | 0.7739023 | 0.7126891 | 0.8563502 |
| silicate + phosphate_star | Indo-Pacific | (26,28.1] | 0.6896547 | 0.6708858 | 0.7159326 |
| silicate + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.1321441 | -0.2062023 | -0.0096971 |
| temp + aou | Atlantic | (-Inf,26] | 0.1097169 | -0.3425090 | 0.4329165 |
| temp + aou | Atlantic | (26,28.2] | -0.1541990 | -0.2251707 | -0.0603008 |
| temp + aou | Atlantic | (28.2, Inf] | 0.5099566 | 0.4938223 | 0.5246358 |
| temp + aou | Indo-Pacific | (-Inf,26] | -0.3033151 | -0.4239825 | -0.1918473 |
| temp + aou | Indo-Pacific | (26,28.1] | -0.3393591 | -0.4878334 | -0.2078194 |
| temp + aou | Indo-Pacific | (28.1, Inf] | 0.8742121 | 0.8635041 | 0.8872273 |
| temp + nitrate | Atlantic | (-Inf,26] | -0.0354229 | -0.2709976 | 0.1353095 |
| temp + nitrate | Atlantic | (26,28.2] | -0.5572719 | -0.6178836 | -0.4772867 |
| temp + nitrate | Atlantic | (28.2, Inf] | -0.1232512 | -0.6051405 | 0.1284445 |
| temp + nitrate | Indo-Pacific | (-Inf,26] | -0.5317602 | -0.6794288 | -0.3861544 |
| temp + nitrate | Indo-Pacific | (26,28.1] | -0.7066781 | -0.7885778 | -0.6645480 |
| temp + nitrate | Indo-Pacific | (28.1, Inf] | 0.4439504 | 0.3483953 | 0.5869496 |
| temp + phosphate | Atlantic | (-Inf,26] | -0.5062464 | -0.6413374 | -0.4215209 |
| temp + phosphate | Atlantic | (26,28.2] | -0.5831156 | -0.6362513 | -0.5078295 |
| temp + phosphate | Atlantic | (28.2, Inf] | -0.1393499 | -0.6278047 | 0.1197551 |
| temp + phosphate | Indo-Pacific | (-Inf,26] | -0.4439968 | -0.5911527 | -0.3145635 |
| temp + phosphate | Indo-Pacific | (26,28.1] | -0.6359600 | -0.7418304 | -0.5739118 |
| temp + phosphate | Indo-Pacific | (28.1, Inf] | 0.4307803 | 0.3053523 | 0.5540156 |
| temp + phosphate_star | Atlantic | (-Inf,26] | -0.8198183 | -0.8713230 | -0.7758658 |
| temp + phosphate_star | Atlantic | (26,28.2] | -0.8914803 | -0.8987917 | -0.8823009 |
| temp + phosphate_star | Atlantic | (28.2, Inf] | -0.4893200 | -0.9602729 | -0.2471928 |
| temp + phosphate_star | Indo-Pacific | (-Inf,26] | -0.7272788 | -0.8389880 | -0.5535823 |
| temp + phosphate_star | Indo-Pacific | (26,28.1] | -0.9510068 | -0.9573038 | -0.9474141 |
| temp + phosphate_star | Indo-Pacific | (28.1, Inf] | -0.8918001 | -0.9174238 | -0.8626825 |
| temp + silicate | Atlantic | (-Inf,26] | -0.6378258 | -0.7393488 | -0.5464948 |
| temp + silicate | Atlantic | (26,28.2] | -0.6058317 | -0.6321507 | -0.5843090 |
| temp + silicate | Atlantic | (28.2, Inf] | 0.0061799 | -0.2917156 | 0.1611065 |
| temp + silicate | Indo-Pacific | (-Inf,26] | -0.5738389 | -0.7373553 | -0.3945553 |
| temp + silicate | Indo-Pacific | (26,28.1] | -0.7875580 | -0.8205969 | -0.7572852 |
| temp + silicate | Indo-Pacific | (28.1, Inf] | 0.3797466 | 0.3232790 | 0.4363778 |
rm(cor_predictors, cor_predictors_stats)2.2.3 C* vs individual predictors
Below, the range of correlations coefficients for C* with each predictor is plotted per basin (facet) and density slab (color). Note that the range indicates the min and max values of in total 3 calculated coefficients (one per era).
cor_target_predictor <- cor_target_predictor %>%
rename(predictor = term)
# calculate min, max, mean across all eras
cor_target_predictor_stats <- cor_target_predictor %>%
select(-era) %>%
group_by(predictor, basin, gamma_slab) %>%
summarise_all(list(mean_r = mean, min_r = min, max_r = max)) %>%
ungroup()
# plot figure
cor_target_predictor_stats %>%
mutate(predictor = reorder(predictor, mean_r)) %>%
ggplot() +
geom_vline(xintercept = c(-0.9, 0.9), col = "red") +
geom_vline(xintercept = 0) +
geom_linerange(
aes(y = predictor, xmin = min_r, xmax = max_r, col = gamma_slab),
position = position_dodge(width = 0.6)) +
facet_wrap(~basin) +
scale_color_viridis_d(direction = -1) +
labs(x = "correlation coefficient", y = "C* correlation with...") +
theme(legend.position = "top")
| Version | Author | Date |
|---|---|---|
| a1d52ff | jens-daniel-mueller | 2021-03-15 |
| 0bade3b | jens-daniel-mueller | 2021-03-15 |
| 27c1f4b | jens-daniel-mueller | 2021-03-14 |
| af75ebf | jens-daniel-mueller | 2021-03-14 |
| 5017709 | jens-daniel-mueller | 2021-03-11 |
| 585b07f | jens-daniel-mueller | 2021-03-11 |
| 85a5ed2 | jens-daniel-mueller | 2021-03-10 |
| 6c0bec6 | jens-daniel-mueller | 2021-03-05 |
| 3c2ec33 | jens-daniel-mueller | 2021-03-05 |
| af70b94 | jens-daniel-mueller | 2021-03-04 |
| 7b672f7 | jens-daniel-mueller | 2021-01-11 |
| 33ba23c | jens-daniel-mueller | 2021-01-07 |
| 318609d | jens-daniel-mueller | 2020-12-23 |
| 9d0b2d0 | jens-daniel-mueller | 2020-12-23 |
| 0aa2b50 | jens-daniel-mueller | 2020-12-23 |
| 2886da0 | jens-daniel-mueller | 2020-12-19 |
| 02f0ee9 | jens-daniel-mueller | 2020-12-18 |
| 7bcb4eb | jens-daniel-mueller | 2020-12-18 |
| 158fe26 | jens-daniel-mueller | 2020-12-15 |
| 7a9a4cb | jens-daniel-mueller | 2020-12-15 |
| 61b263c | jens-daniel-mueller | 2020-12-15 |
| 984697e | jens-daniel-mueller | 2020-12-12 |
| 3ebff89 | jens-daniel-mueller | 2020-12-12 |
| 5d96d3c | jens-daniel-mueller | 2020-12-11 |
| 24a632f | jens-daniel-mueller | 2020-12-07 |
| 6a8004b | jens-daniel-mueller | 2020-12-07 |
| 70bf1a5 | jens-daniel-mueller | 2020-12-07 |
| 7555355 | jens-daniel-mueller | 2020-12-07 |
| 143d6fa | jens-daniel-mueller | 2020-12-07 |
| 090e4d5 | jens-daniel-mueller | 2020-12-02 |
| 0ff728b | jens-daniel-mueller | 2020-12-01 |
| 91435ae | jens-daniel-mueller | 2020-12-01 |
| 196be51 | jens-daniel-mueller | 2020-11-30 |
| bc61ce3 | Jens Müller | 2020-11-30 |
# print table
kable(cor_target_predictor_stats) %>%
add_header_above() %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| predictor | basin | gamma_slab | mean_r | min_r | max_r |
|---|---|---|---|---|---|
| aou | Atlantic | (-Inf,26] | 0.5954940 | 0.5403772 | 0.6259285 |
| aou | Atlantic | (26,28.2] | -0.6860104 | -0.7091660 | -0.6602374 |
| aou | Atlantic | (28.2, Inf] | -0.3578273 | -0.6996866 | 0.3171406 |
| aou | Indo-Pacific | (-Inf,26] | -0.2093870 | -0.4901407 | 0.0231798 |
| aou | Indo-Pacific | (26,28.1] | -0.1599882 | -0.2193908 | -0.0870938 |
| aou | Indo-Pacific | (28.1, Inf] | 0.3029332 | 0.1423855 | 0.5714053 |
| nitrate | Atlantic | (-Inf,26] | 0.3387740 | 0.3021479 | 0.4038121 |
| nitrate | Atlantic | (26,28.2] | -0.8658015 | -0.8988189 | -0.8227791 |
| nitrate | Atlantic | (28.2, Inf] | -0.8290772 | -0.9587749 | -0.5966614 |
| nitrate | Indo-Pacific | (-Inf,26] | -0.2364714 | -0.5229643 | -0.0065778 |
| nitrate | Indo-Pacific | (26,28.1] | -0.3732911 | -0.4458130 | -0.2499881 |
| nitrate | Indo-Pacific | (28.1, Inf] | 0.0945425 | -0.0526306 | 0.3856189 |
| phosphate | Atlantic | (-Inf,26] | -0.0542341 | -0.4118203 | 0.1927518 |
| phosphate | Atlantic | (26,28.2] | -0.8959814 | -0.9211525 | -0.8615260 |
| phosphate | Atlantic | (28.2, Inf] | -0.8683216 | -0.9683209 | -0.6881572 |
| phosphate | Indo-Pacific | (-Inf,26] | -0.2345097 | -0.5303663 | -0.0046817 |
| phosphate | Indo-Pacific | (26,28.1] | -0.3817305 | -0.4428987 | -0.2661418 |
| phosphate | Indo-Pacific | (28.1, Inf] | -0.0477819 | -0.2523948 | 0.2859410 |
| phosphate_star | Atlantic | (-Inf,26] | -0.4716389 | -0.7976096 | -0.2398918 |
| phosphate_star | Atlantic | (26,28.2] | -0.7456574 | -0.8020098 | -0.6679922 |
| phosphate_star | Atlantic | (28.2, Inf] | -0.9058320 | -0.9545451 | -0.8324191 |
| phosphate_star | Indo-Pacific | (-Inf,26] | -0.1073048 | -0.3944798 | 0.0764190 |
| phosphate_star | Indo-Pacific | (26,28.1] | -0.4517015 | -0.5978074 | -0.3226919 |
| phosphate_star | Indo-Pacific | (28.1, Inf] | -0.4908459 | -0.6174865 | -0.3838022 |
| sal | Atlantic | (-Inf,26] | 0.3229008 | 0.1394940 | 0.6429462 |
| sal | Atlantic | (26,28.2] | 0.7040261 | 0.6630134 | 0.7634276 |
| sal | Atlantic | (28.2, Inf] | 0.8332341 | 0.6700284 | 0.9224981 |
| sal | Indo-Pacific | (-Inf,26] | -0.0209090 | -0.0847747 | 0.0646838 |
| sal | Indo-Pacific | (26,28.1] | 0.3205295 | 0.1217238 | 0.4785595 |
| sal | Indo-Pacific | (28.1, Inf] | 0.2686642 | 0.2486360 | 0.2959155 |
| silicate | Atlantic | (-Inf,26] | -0.1481448 | -0.5178173 | 0.0956953 |
| silicate | Atlantic | (26,28.2] | -0.5830373 | -0.6663606 | -0.4932436 |
| silicate | Atlantic | (28.2, Inf] | -0.7224781 | -0.9318405 | -0.3368431 |
| silicate | Indo-Pacific | (-Inf,26] | -0.2413512 | -0.5023137 | -0.0276715 |
| silicate | Indo-Pacific | (26,28.1] | 0.1362193 | 0.0401413 | 0.2577697 |
| silicate | Indo-Pacific | (28.1, Inf] | -0.0063520 | -0.1043232 | 0.1746101 |
| temp | Atlantic | (-Inf,26] | 0.1430978 | -0.1455071 | 0.5281247 |
| temp | Atlantic | (26,28.2] | 0.4026251 | 0.2703236 | 0.4907233 |
| temp | Atlantic | (28.2, Inf] | 0.2448353 | -0.0490847 | 0.7371290 |
| temp | Indo-Pacific | (-Inf,26] | -0.3252096 | -0.3443171 | -0.2969229 |
| temp | Indo-Pacific | (26,28.1] | 0.1988360 | 0.0525078 | 0.3755751 |
| temp | Indo-Pacific | (28.1, Inf] | 0.3411732 | 0.1560601 | 0.6003943 |
rm(cor_target_predictor, cor_target_predictor_stats)
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.2
Matrix products: default
BLAS: /usr/local/R-4.0.3/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.0.3/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] corrr_0.4.3 broom_0.7.2 kableExtra_1.3.1 knitr_1.30
[5] olsrr_0.5.3 GGally_2.0.0 lubridate_1.7.9 metR_0.9.0
[9] scico_1.2.0 patchwork_1.1.1 collapse_1.5.0 forcats_0.5.0
[13] stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4 readr_1.4.0
[17] tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.2 tidyverse_1.3.0
[21] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.0 webshot_0.5.2 RColorBrewer_1.1-2
[4] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3
[7] backports_1.1.10 R6_2.5.0 nortest_1.0-4
[10] DBI_1.1.0 colorspace_1.4-1 withr_2.3.0
[13] gridExtra_2.3 tidyselect_1.1.0 curl_4.3
[16] compiler_4.0.3 git2r_0.27.1 cli_2.1.0
[19] rvest_0.3.6 xml2_1.3.2 labeling_0.4.2
[22] scales_1.1.1 checkmate_2.0.0 goftest_1.2-2
[25] digest_0.6.27 foreign_0.8-80 rmarkdown_2.5
[28] rio_0.5.16 pkgconfig_2.0.3 htmltools_0.5.0
[31] highr_0.8 dbplyr_1.4.4 rlang_0.4.9
[34] readxl_1.3.1 rstudioapi_0.13 farver_2.0.3
[37] generics_0.0.2 jsonlite_1.7.1 zip_2.1.1
[40] car_3.0-10 magrittr_1.5 Matrix_1.2-18
[43] Rcpp_1.0.5 munsell_0.5.0 fansi_0.4.1
[46] abind_1.4-5 lifecycle_0.2.0 stringi_1.5.3
[49] whisker_0.4 yaml_2.2.1 carData_3.0-4
[52] plyr_1.8.6 grid_4.0.3 blob_1.2.1
[55] parallel_4.0.3 promises_1.1.1 crayon_1.3.4
[58] lattice_0.20-41 haven_2.3.1 hms_0.5.3
[61] pillar_1.4.7 reprex_0.3.0 glue_1.4.2
[64] evaluate_0.14 RcppArmadillo_0.10.1.2.0 data.table_1.13.2
[67] modelr_0.1.8 vctrs_0.3.5 httpuv_1.5.4
[70] cellranger_1.1.0 gtable_0.3.0 reshape_0.8.8
[73] assertthat_0.2.1 xfun_0.18 openxlsx_4.2.3
[76] RcppEigen_0.3.3.7.0 later_1.1.0.1 viridisLite_0.3.0
[79] ellipsis_0.3.1 here_0.1