Single-cell PBMC Example
karltayeb
2022-03-16
Last updated: 2022-03-29
Checks: 7 0
Knit directory: logistic-susie-gsea/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220105) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9a5aad9. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: library/
Ignored: renv/library/
Ignored: renv/staging/
Ignored: staging/
Untracked files:
Untracked: _targets.R
Untracked: _targets.html
Untracked: _targets.md
Untracked: _targets/
Untracked: _targets_r/
Untracked: analysis/fetal_reference_cellid_gsea.Rmd
Untracked: analysis/fixed_intercept.Rmd
Untracked: analysis/iDEA_examples.Rmd
Untracked: analysis/latent_gene_list.Rmd
Untracked: analysis/latent_logistic_susie.Rmd
Untracked: analysis/libra_setup.Rmd
Untracked: analysis/linear_method_failure_modes.Rmd
Untracked: analysis/linear_regression_failure_regime.Rmd
Untracked: analysis/logistic_susie_veb_boost_vs_vb.Rmd
Untracked: analysis/references.bib
Untracked: analysis/simulations.Rmd
Untracked: analysis/test.Rmd
Untracked: analysis/wenhe_baboon_example.Rmd
Untracked: build_site.R
Untracked: cache/
Untracked: code/latent_logistic_susie.R
Untracked: code/marginal_sumstat_gsea_collapsed.R
Untracked: data/adipose_2yr_topsnp.txt
Untracked: data/fetal_reference_cellid_gene_sets.RData
Untracked: data/pbmc-purified/
Untracked: docs.zip
Untracked: index.md
Untracked: latent_logistic_susie_cache/
Untracked: simulation_targets/
Untracked: single_cell_pbmc_cache/
Untracked: summary_stat_gsea_exploration_cache/
Unstaged changes:
Modified: _simulation_targets.R
Modified: _targets.Rmd
Modified: analysis/gseabenchmark_tcga.Rmd
Modified: code/fit_baselines.R
Modified: code/fit_logistic_susie.R
Modified: code/fit_mr_ash.R
Modified: code/fit_susie.R
Modified: code/load_gene_sets.R
Modified: code/marginal_sumstat_gsea.R
Modified: code/simulate_gene_lists.R
Modified: code/utils.R
Modified: target_components/factories.R
Modified: target_components/methods.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/single_cell_pbmc.Rmd) and HTML (docs/single_cell_pbmc.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9a5aad9 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | cdd6193 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 9cf1df0 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 0369ee2 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 9fcb3b4 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 2fa21ff | karltayeb | 2022-03-29 | Build site. |
| Rmd | 7ee315c | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 8314424 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 524d94b | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 61540e8 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 54209af | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 67bf8cf | karltayeb | 2022-03-29 | Build site. |
| Rmd | 170e480 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 90c6006 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 8159b83 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 9d7fd29 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 51647d4 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 3da7c5b | karltayeb | 2022-03-29 | Build site. |
| Rmd | fc1f3c1 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 8646723 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 6143c44 | karltayeb | 2022-03-29 | wflow_publish(“analysis/single_cell_pbmc.Rmd”) |
| html | 56f8130 | karltayeb | 2022-03-29 | Build site. |
| html | a2bdb56 | karltayeb | 2022-03-29 | Build site. |
| Rmd | 122deec | karltayeb | 2022-03-29 | wflow_publish(pages) |
Introduction
Our goals here are to run Logistic SuSiE on differential expression results from TCGA. We want to assess:
- If the resulting enrichment results look good/interpretable across multiple/concatenated gene sets
- Assess sensitivity to a range of p-value thresholds
- Evaluate the potential of the summary stat latent model
library(GSEABenchmarkeR)
library(EnrichmentBrowser)
library(tidyverse)
library(susieR)
library(DT)
library(kableExtra)
source('code/load_gene_sets.R')
source('code/utils.R')
source('code/logistic_susie_vb.R')
source('code/logistic_susie_veb_boost.R')
source('code/latent_logistic_susie.R')Setup
Load Gene Sets
loadGeneSetX uniformly formats gene sets and generates the \(X\) matrix We can source any gene set from WebGestaltR::listGeneSet()
gs_list <- WebGestaltR::listGeneSet()
gobp <- loadGeneSetX('geneontology_Biological_Process', min.size=50) # just huge number of gene sets
gobp_nr <- loadGeneSetX('geneontology_Biological_Process_noRedundant', min.size=1)
gomf <- loadGeneSetX('geneontology_Molecular_Function', min.size=1)
kegg <- loadGeneSetX('pathway_KEGG', min.size=1)
reactome <- loadGeneSetX('pathway_Reactome', min.size=1)
wikipathway_cancer <- loadGeneSetX('pathway_Wikipathway_cancer', min.size=1)
wikipathway <- loadGeneSetX('pathway_Wikipathway', min.size=1)
genesets <- list(
gobp=gobp,
gobp_nr=gobp_nr,
gomf=gomf,
kegg=kegg,
reactome=reactome,
wikipathway_cancer=wikipathway_cancer,
wikipathway=wikipathway
)load('data/pbmc-purified/deseq2-pbmc-purified.RData')
convert_labels <- function(y, from='SYMBOL', to='ENTREZID'){
hs <- org.Hs.eg.db::org.Hs.eg.db
gene_symbols <- names(y)
symbol2entrez <- AnnotationDbi::select(hs, keys=gene_symbols, columns=c(to, from), keytype = from)
symbol2entrez <- symbol2entrez[!duplicated(symbol2entrez[[from]]),]
symbol2entrez <- symbol2entrez[!is.na(symbol2entrez[[to]]),]
symbol2entrez <- symbol2entrez[!is.na(symbol2entrez[[from]]),]
rownames(symbol2entrez) <- symbol2entrez[[from]]
ysub <- y[names(y) %in% symbol2entrez[[from]]]
names(ysub) <- symbol2entrez[names(ysub),][[to]]
return(ysub)
}
par(mfrow=c(1,1))
deseq$`CD19+ B` %>% .$padj %>% hist(main='CD19+B p-values')Loading required package: DESeq2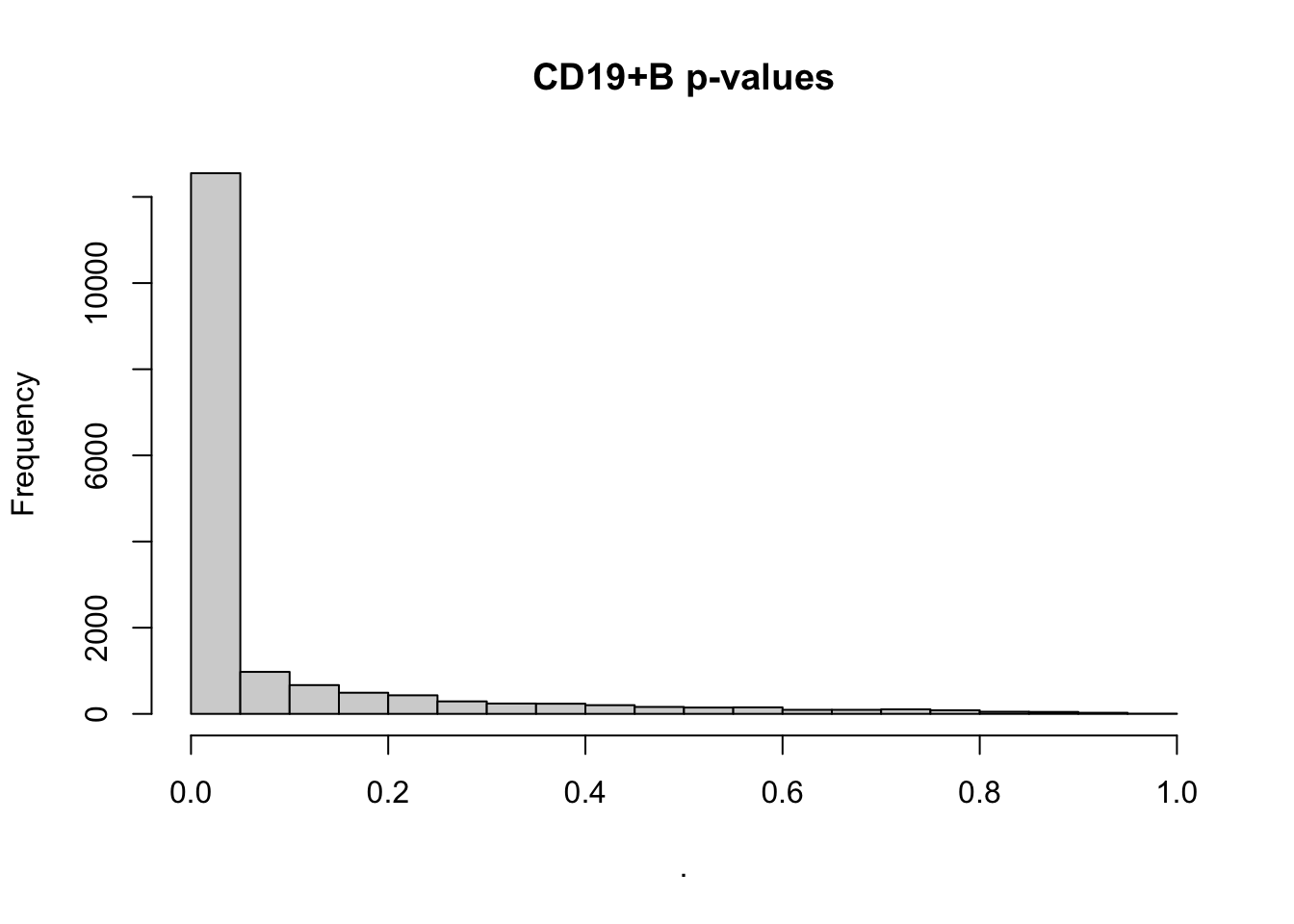
| Version | Author | Date |
|---|---|---|
| a2bdb56 | karltayeb | 2022-03-29 |
Fit logistic SuSiE
logistic_susie_driver = function(db, celltype, thresh){
gs <- genesets[[db]]
data <- deseq[[celltype]]
# set up binary y
y <- data %>%
as.data.frame %>%
rownames_to_column('gene') %>%
dplyr::select(gene, padj) %>%
filter(!is.na(padj)) %>%
mutate(y = as.integer(padj < thresh)) %>%
select(gene, y) %>%
tibble2namedlist %>%
convert_labels('ENSEMBL')
u <- process_input(gs$X, y) # subset to common genes
vb.fit <- logistic.susie( # fit model
u$X, u$y, L=10, init.intercept = 0, verbose=1, maxit=100)
# summarise results
set.summary <- vb.fit$pip %>%
as_tibble(rownames='geneSet') %>%
rename(pip=value) %>%
mutate(
top_component = apply(vb.fit$alpha, 2, which.max),
active_set = top_component %in% vb.fit$sets$cs_index,
top_component = paste0('L', top_component),
cs = purrr::map(top_component, ~tryCatch(
colnames(gs$X)[get(.x, vb.fit$sets$cs)], error = function(e) list())),
in_cs = geneSet %in% cs,
beta = colSums(vb.fit$mu * vb.fit$alpha),
geneListSize = sum(u$y),
geneSetSize = colSums(u$X),
overlap = (u$y %*% u$X)[1,],
nGenes = length(u$y),
propSetInList = overlap / geneSetSize,
oddsRatio = (overlap / (geneListSize - overlap)) / (
(geneSetSize - overlap) / (nGenes - geneSetSize + overlap)),
pValueHypergeometric = phyper(
overlap-1, geneListSize, nGenes, geneSetSize, lower.tail= FALSE),
db = db,
celltype = celltype,
thresh = thresh
) %>% left_join(gs$geneSet$geneSetDes)
return(list(fit = vb.fit, set.summary=set.summary))
}For each celltype, we fit logistic SuSiE using multiple gene set sources at various threshold of padj.
celltypes <- names(deseq)
pthresh <- c(0.1, 0.01, 0.001, 0.0001, 0.00001, 0.000001)
db_name <- names(genesets)
crossed <- cross3(db_name, celltypes, pthresh)
pbmc_res <- xfun::cache_rds({
res <- purrr::map(crossed, purrr::lift_dl(logistic_susie_driver))
for (i in 1:length(res)){ # save some space
res[[i]]$fit$dat <- NULL
}
res
}, file = 'logistic_susie_pbmc_genesets_pthresh.rds'
)
pbmc_res_set_summary <- dplyr::bind_rows(purrr::map(pbmc_res, ~ pluck(.x, 'set.summary')))Summary functions
Just a few functions to help streamline looking at output
pval_focussed_table = function(thresh=1e-3, filter_db=NULL, filter_celltype=NULL, top.n=50){
pbmc_res_set_summary %>%
filter(
case_when(
is.null(filter_db) ~ TRUE,
!is.null(filter_db) ~ db %in% filter_db
) &
thresh == thresh &
case_when(
is.null(filter_celltype) ~ TRUE,
!is.null(filter_celltype) ~ celltype %in% filter_celltype
)
) %>%
dplyr::arrange(celltype, db, pValueHypergeometric) %>%
group_by(celltype, db) %>% slice(1:top.n) %>%
select(celltype, db, geneSet, description, pip, top_component, oddsRatio, propSetInList, pValueHypergeometric) %>%
mutate_at(vars(celltype, db), factor) %>%
datatable(filter = 'top')
}
set_focussed_table = function(thresh=1e-3, filter_db=NULL, filter_celltype=NULL){
pbmc_res_set_summary %>%
filter(
case_when(
is.null(filter_db) ~ TRUE,
!is.null(filter_db) ~ db %in% filter_db
) &
thresh == 1e-3 &
in_cs & active_set &
case_when(
is.null(filter_celltype) ~ TRUE,
!is.null(filter_celltype) ~ celltype %in% filter_celltype
)
) %>%
dplyr::arrange(celltype, db, desc(pip)) %>%
select(celltype, db, geneSet, description, pip, top_component, oddsRatio, propSetInList, pValueHypergeometric) %>%
mutate_at(vars(celltype, geneSet, db), factor) %>%
datatable(filter = 'top')
}
#' takes a tibble
#' organize by database and component
#' report credible set, descriptions, pips, and hypergeometric pvalue
#' in one row, with cs ordered by pip
db_component_kable = function(tbl){
tbl %>%
filter(active_set, thresh==1e-4) %>%
group_by(db, celltype, top_component) %>%
arrange(db, celltype, top_component, desc(pip)) %>%
select(geneSet, description, pip, pValueHypergeometric) %>%
chop(c(geneSet, description, pip, pValueHypergeometric)) %>%
knitr::kable()
}
#' takes a tibble
#' organize by database and component
#' report credible set, descriptions, pips, and hypergeometric pvalue
#' in one row, with cs ordered by pip
db_component_kable = function(tbl){
tbl %>%
filter(active_set, thresh==1e-4) %>%
group_by(db, celltype, top_component) %>%
arrange(db, celltype, top_component, desc(pip)) %>%
mutate(pip = cell_spec(pip, color=ifelse(in_cs, 'green', 'red'))) %>%
select(geneSet, description, pip, pValueHypergeometric) %>%
#chop(c(geneSet, description, pip, pValueHypergeometric)) %>%
knitr::kable()
}Results/Explore enrichments
Our goal is to assess 1. The quality of the gene set enrichments we get from each celltype - do reported gene set enrichments seem celltype specific/celltype relevant? - how much “interesting” marginal enrichment do we fail to capture in the multivariate model - how sensitive are we to the choice of pvalue threshold
Results
Lets take a look at what enrichment we’re getting across cell-types.
CD19+ B
pbmc_res_set_summary %>%
filter(celltype == 'CD19+ B') %>%
filter(active_set, thresh==1e-4) %>%
db_component_kableAdding missing grouping variables: `db`, `celltype`, `top_component`| db | celltype | top_component | geneSet | description | pip | pValueHypergeometric |
|---|---|---|---|---|---|---|
| gobp | CD19+ B | L1 | GO:0002376 | immune system process | <span style=" color: green !important;" >0.999957465451611</span> | 0 |
| gobp | CD19+ B | L2 | GO:0045047 | protein targeting to ER | <span style=" color: green !important;" >0.962052077569581</span> | 0 |
| gobp | CD19+ B | L2 | GO:0006613 | cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0216561609975944</span> | 0 |
| gobp | CD19+ B | L2 | GO:0072599 | establishment of protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.0145368812974931</span> | 0 |
| gobp | CD19+ B | L2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.00410197009826074</span> | 0 |
| gobp | CD19+ B | L3 | GO:0042773 | ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.624496308507348</span> | 0 |
| gobp | CD19+ B | L3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.360022211508933</span> | 0 |
| gobp | CD19+ B | L3 | GO:0006119 | oxidative phosphorylation | <span style=" color: red !important;" >0.0175238678609325</span> | 0 |
| gobp | CD19+ B | L5 | GO:0001775 | cell activation | <span style=" color: green !important;" >0.978054175097094</span> | 0 |
| gobp | CD19+ B | L5 | GO:0045321 | leukocyte activation | <span style=" color: red !important;" >0.0218972720769344</span> | 0 |
| gobp | CD19+ B | L5 | GO:0002366 | leukocyte activation involved in immune response | <span style=" color: red !important;" >0.000595713062097269</span> | 0 |
| gobp | CD19+ B | L5 | GO:0002263 | cell activation involved in immune response | <span style=" color: red !important;" >0.000577006708652683</span> | 0 |
| gobp | CD19+ B | L6 | GO:0008380 | RNA splicing | <span style=" color: red !important;" >0.822736133183297</span> | 0 |
| gobp | CD19+ B | L6 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | <span style=" color: red !important;" >0.049644134105862</span> | 0 |
| gobp | CD19+ B | L6 | GO:0000398 | mRNA splicing, via spliceosome | <span style=" color: red !important;" >0.049644134105862</span> | 0 |
| gobp | CD19+ B | L6 | GO:0000375 | RNA splicing, via transesterification reactions | <span style=" color: red !important;" >0.0330051165719797</span> | 0 |
| gobp | CD19+ B | L6 | GO:0006397 | mRNA processing | <span style=" color: red !important;" >0.0172340532174977</span> | 0 |
| gobp | CD19+ B | L6 | GO:0050684 | regulation of mRNA processing | <span style=" color: red !important;" >0.0094172042437507</span> | 0 |
| gobp | CD19+ B | L6 | GO:0043484 | regulation of RNA splicing | <span style=" color: red !important;" >0.00722067130606185</span> | 0 |
| gobp | CD19+ B | L6 | GO:0000380 | alternative mRNA splicing, via spliceosome | <span style=" color: red !important;" >0.00293948333200866</span> | 0 |
| gobp | CD19+ B | L6 | GO:0048024 | regulation of mRNA splicing, via spliceosome | <span style=" color: red !important;" >0.00183491753300979</span> | 0 |
| gobp | CD19+ B | L6 | GO:0016071 | mRNA metabolic process | <span style=" color: red !important;" >0.00105163745213244</span> | 0 |
| gobp_nr | CD19+ B | L1 | GO:0002446 | neutrophil mediated immunity | <span style=" color: red !important;" >0.515144038268755</span> | 0 |
| gobp_nr | CD19+ B | L1 | GO:0036230 | granulocyte activation | <span style=" color: red !important;" >0.485735788920244</span> | 0 |
| gobp_nr | CD19+ B | L2 | GO:0070972 | protein localization to endoplasmic reticulum | <span style=" color: green !important;" >0.999958026971368</span> | 0 |
| gobp_nr | CD19+ B | L4 | GO:0009123 | nucleoside monophosphate metabolic process | <span style=" color: green !important;" >0.984497563614406</span> | 0 |
| gobp_nr | CD19+ B | L4 | GO:0009141 | nucleoside triphosphate metabolic process | <span style=" color: red !important;" >0.0164867485447392</span> | 0 |
| gobp_nr | CD19+ B | L5 | GO:0002764 | immune response-regulating signaling pathway | <span style=" color: green !important;" >0.996185347850837</span> | 0 |
| gomf | CD19+ B | L1 | GO:0003723 | RNA binding | <span style=" color: green !important;" >0.999743954825178</span> | 0 |
| gomf | CD19+ B | L2 | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | <span style=" color: green !important;" >0.995003782967434</span> | 0 |
| gomf | CD19+ B | L2 | GO:0003700 | DNA-binding transcription factor activity | <span style=" color: red !important;" >0.00505824116754672</span> | 0 |
| gomf | CD19+ B | L3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.487251391312216</span> | 0 |
| gomf | CD19+ B | L3 | GO:0001012 | RNA polymerase II regulatory region DNA binding | <span style=" color: red !important;" >0.431399316819835</span> | 0 |
| gomf | CD19+ B | L3 | GO:0000987 | proximal promoter sequence-specific DNA binding | <span style=" color: red !important;" >0.0365953412200314</span> | 0 |
| gomf | CD19+ B | L3 | GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | <span style=" color: red !important;" >0.0243679373454444</span> | 0 |
| gomf | CD19+ B | L3 | GO:0000976 | transcription regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.0132413813040726</span> | 0 |
| gomf | CD19+ B | L3 | GO:0003690 | double-stranded DNA binding | <span style=" color: red !important;" >0.00592789564076235</span> | 0 |
| gomf | CD19+ B | L3 | GO:1990837 | sequence-specific double-stranded DNA binding | <span style=" color: red !important;" >0.00136790369019235</span> | 0 |
| gomf | CD19+ B | L3 | GO:0001067 | regulatory region nucleic acid binding | <span style=" color: red !important;" >0.00046527212966796</span> | 0 |
| gomf | CD19+ B | L3 | GO:0044212 | transcription regulatory region DNA binding | <span style=" color: red !important;" >0.000355705894771408</span> | 0 |
| gomf | CD19+ B | L6 | GO:0003735 | structural constituent of ribosome | <span style=" color: green !important;" >0.995344082600683</span> | 0 |
| gomf | CD19+ B | L7 | GO:0003954 | NADH dehydrogenase activity | <span style=" color: red !important;" >0.325953617177259</span> | 0 |
| gomf | CD19+ B | L7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity | <span style=" color: red !important;" >0.218583180359325</span> | 0 |
| gomf | CD19+ B | L7 | GO:0050136 | NADH dehydrogenase (quinone) activity | <span style=" color: red !important;" >0.218583180359325</span> | 0 |
| gomf | CD19+ B | L7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | <span style=" color: red !important;" >0.177265521649769</span> | 0 |
| gomf | CD19+ B | L7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H | <span style=" color: red !important;" >0.0387976083993536</span> | 0 |
| kegg | CD19+ B | L1 | hsa00190 | Oxidative phosphorylation | <span style=" color: green !important;" >0.997990176564082</span> | 0 |
| kegg | CD19+ B | L1 | hsa05012 | Parkinson disease | <span style=" color: red !important;" >0.0125303037367086</span> | 0 |
| kegg | CD19+ B | L2 | hsa04640 | Hematopoietic cell lineage | <span style=" color: green !important;" >0.999795101338638</span> | 0 |
| kegg | CD19+ B | L3 | hsa03010 | Ribosome | <span style=" color: green !important;" >0.999999982887443</span> | 0 |
| reactome | CD19+ B | L1 | R-HSA-168256 | Immune System | <span style=" color: green !important;" >1</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-156842 | Eukaryotic Translation Elongation | <span style=" color: red !important;" >0.760651463839203</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-156902 | Peptide chain elongation | <span style=" color: red !important;" >0.12438973980464</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-192823 | Viral mRNA Translation | <span style=" color: red !important;" >0.0671329230529306</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-2408557 | Selenocysteine synthesis | <span style=" color: red !important;" >0.0260066810882063</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-72764 | Eukaryotic Translation Termination | <span style=" color: red !important;" >0.0172676775018812</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-72689 | Formation of a pool of free 40S subunits | <span style=" color: red !important;" >0.00330537630812389</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.00313197971436374</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | <span style=" color: red !important;" >0.00122676226108998</span> | 0 |
| reactome | CD19+ B | L2 | R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | <span style=" color: red !important;" >0.00100083013030172</span> | 0 |
| reactome | CD19+ B | L3 | R-HSA-74160 | Gene expression (Transcription) | <span style=" color: red !important;" >0.885679352792777</span> | 0 |
| reactome | CD19+ B | L3 | R-HSA-212436 | Generic Transcription Pathway | <span style=" color: red !important;" >0.0726625717379175</span> | 0 |
| reactome | CD19+ B | L3 | R-HSA-73857 | RNA Polymerase II Transcription | <span style=" color: red !important;" >0.041834760675025</span> | 0 |
| reactome | CD19+ B | L4 | R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | <span style=" color: red !important;" >0.845323000060671</span> | 0 |
| reactome | CD19+ B | L4 | R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | <span style=" color: red !important;" >0.14938336590149</span> | 0 |
| reactome | CD19+ B | L4 | R-HSA-611105 | Respiratory electron transport | <span style=" color: red !important;" >0.00596574446721121</span> | 0 |
| reactome | CD19+ B | L5 | R-HSA-983168 | Antigen processing: Ubiquitination & Proteasome degradation | <span style=" color: green !important;" >0.997049696139591</span> | 0 |
| reactome | CD19+ B | L5 | R-HSA-983169 | Class I MHC mediated antigen processing & presentation | <span style=" color: red !important;" >0.00311879186973374</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-180585 | Vif-mediated degradation of APOBEC3G | <span style=" color: red !important;" >0.749039310761527</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | <span style=" color: red !important;" >0.0319137208638701</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-211733 | Regulation of activated PAK-2p34 by proteasome mediated degradation | <span style=" color: red !important;" >0.0256472437189241</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | <span style=" color: red !important;" >0.0242873384108474</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69610 | p53-Independent DNA Damage Response | <span style=" color: red !important;" >0.0242873384108474</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69613 | p53-Independent G1/S DNA damage checkpoint | <span style=" color: red !important;" >0.0242873384108474</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69541 | Stabilization of p53 | <span style=" color: red !important;" >0.0225422403485689</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69229 | Ubiquitin-dependent degradation of Cyclin D1 | <span style=" color: red !important;" >0.0120133659466743</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-75815 | Ubiquitin-dependent degradation of Cyclin D | <span style=" color: red !important;" >0.0120133659466743</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-180534 | Vpu mediated degradation of CD4 | <span style=" color: red !important;" >0.0119846232558787</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-169911 | Regulation of Apoptosis | <span style=" color: red !important;" >0.0108046763000044</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | <span style=" color: red !important;" >0.0101911528157629</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-9604323 | Negative regulation of NOTCH4 signaling | <span style=" color: red !important;" >0.0071579855747933</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-8854050 | FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | <span style=" color: red !important;" >0.00703778872278604</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-1236978 | Cross-presentation of soluble exogenous antigens (endosomes) | <span style=" color: red !important;" >0.00406554835881989</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-174154 | APC/C:Cdc20 mediated degradation of Securin | <span style=" color: red !important;" >0.00385891217123191</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-1169091 | Activation of NF-kappaB in B cells | <span style=" color: red !important;" >0.00376792382128477</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-174084 | Autodegradation of Cdh1 by Cdh1:APC/C | <span style=" color: red !important;" >0.00361414870965138</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-1234174 | Regulation of Hypoxia-inducible Factor (HIF) by oxygen | <span style=" color: red !important;" >0.00305797077052683</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-2262749 | Cellular response to hypoxia | <span style=" color: red !important;" >0.00305797077052683</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-983705 | Signaling by the B Cell Receptor (BCR) | <span style=" color: red !important;" >0.00286836400043933</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-174113 | SCF-beta-TrCP mediated degradation of Emi1 | <span style=" color: red !important;" >0.0024109403813769</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-450408 | AUF1 (hnRNP D0) binds and destabilizes mRNA | <span style=" color: red !important;" >0.00171267983373247</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69563 | p53-Dependent G1 DNA Damage Response | <span style=" color: red !important;" >0.00150097375541414</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69580 | p53-Dependent G1/S DNA damage checkpoint | <span style=" color: red !important;" >0.00150097375541414</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | <span style=" color: red !important;" >0.00133190881117529</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-351202 | Metabolism of polyamines | <span style=" color: red !important;" >0.00131731594365259</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-68867 | Assembly of the pre-replicative complex | <span style=" color: red !important;" >0.00121050465862294</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-176409 | APC/C:Cdc20 mediated degradation of mitotic proteins | <span style=" color: red !important;" >0.00116496309289926</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-350562 | Regulation of ornithine decarboxylase (ODC) | <span style=" color: red !important;" >0.00102467992167676</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-69615 | G1/S DNA Damage Checkpoints | <span style=" color: red !important;" >0.000957829552408529</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-5362768 | Hh mutants that don’t undergo autocatalytic processing are degraded by ERAD | <span style=" color: red !important;" >0.00095140800156257</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | <span style=" color: red !important;" >0.000862233445453997</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | <span style=" color: red !important;" >0.000720719571974726</span> | 0 |
| reactome | CD19+ B | L6 | R-HSA-5610780 | Degradation of GLI1 by the proteasome | <span style=" color: red !important;" >0.000661022828454261</span> | 0 |
| reactome | CD19+ B | L7 | R-HSA-72163 | mRNA Splicing - Major Pathway | <span style=" color: red !important;" >0.545788750456562</span> | 0 |
| reactome | CD19+ B | L7 | R-HSA-72172 | mRNA Splicing | <span style=" color: red !important;" >0.443954636390887</span> | 0 |
| reactome | CD19+ B | L7 | R-HSA-109688 | Cleavage of Growing Transcript in the Termination Region | <span style=" color: red !important;" >0.00214822879004417</span> | 0 |
| reactome | CD19+ B | L7 | R-HSA-73856 | RNA Polymerase II Transcription Termination | <span style=" color: red !important;" >0.00214822879004417</span> | 0 |
| reactome | CD19+ B | L7 | R-HSA-72203 | Processing of Capped Intron-Containing Pre-mRNA | <span style=" color: red !important;" >0.00064614111580874</span> | 0 |
| reactome | CD19+ B | L8 | R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | <span style=" color: red !important;" >0.706384848748338</span> | 0 |
| reactome | CD19+ B | L8 | R-HSA-114608 | Platelet degranulation | <span style=" color: red !important;" >0.231432202567839</span> | 0 |
| reactome | CD19+ B | L8 | R-HSA-76002 | Platelet activation, signaling and aggregation | <span style=" color: red !important;" >0.0252623002452426</span> | 0 |
| reactome | CD19+ B | L8 | R-HSA-109582 | Hemostasis | <span style=" color: red !important;" >0.000407534393526032</span> | 0 |
| wikipathway | CD19+ B | L1 | WP477 | Cytoplasmic Ribosomal Proteins | <span style=" color: green !important;" >0.999999999999907</span> | 0 |
| wikipathway | CD19+ B | L2 | WP111 | Electron Transport Chain (OXPHOS system in mitochondria) | <span style=" color: green !important;" >0.999918859916203</span> | 0 |
| wikipathway_cancer | CD19+ B | L1 | WP619 | Type II interferon signaling (IFNG) | <span style=" color: green !important;" >0.999863579914409</span> | 0 |
CD56+ NK
pbmc_res_set_summary %>%
filter(celltype == 'CD56+ NK') %>%
filter(active_set, thresh==1e-4) %>%
db_component_kableAdding missing grouping variables: `db`, `celltype`, `top_component`| db | celltype | top_component | geneSet | description | pip | pValueHypergeometric |
|---|---|---|---|---|---|---|
| gobp | CD56+ NK | L1 | GO:0002376 | immune system process | <span style=" color: green !important;" >0.999998415304339</span> | 0 |
| gobp | CD56+ NK | L2 | GO:0045047 | protein targeting to ER | <span style=" color: red !important;" >0.682786144147153</span> | 0 |
| gobp | CD56+ NK | L2 | GO:0072599 | establishment of protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.209785127668247</span> | 0 |
| gobp | CD56+ NK | L2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0712305252918061</span> | 0 |
| gobp | CD56+ NK | L2 | GO:0006613 | cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0382267562392382</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0006119 | oxidative phosphorylation | <span style=" color: green !important;" >0.981575408284152</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0009123 | nucleoside monophosphate metabolic process | <span style=" color: red !important;" >0.00751725679613058</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0042773 | ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00504616694105076</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00356583700729807</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0009161 | ribonucleoside monophosphate metabolic process | <span style=" color: red !important;" >0.00314627190684258</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0009126 | purine nucleoside monophosphate metabolic process | <span style=" color: red !important;" >0.00184701119525377</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0009167 | purine ribonucleoside monophosphate metabolic process | <span style=" color: red !important;" >0.00184701119525377</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0022900 | electron transport chain | <span style=" color: red !important;" >0.0017166414433879</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0022904 | respiratory electron transport chain | <span style=" color: red !important;" >0.00153917641145329</span> | 0 |
| gobp | CD56+ NK | L4 | GO:0046034 | ATP metabolic process | <span style=" color: red !important;" >0.000947236633707993</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0008380 | RNA splicing | <span style=" color: red !important;" >0.590744524371383</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0006397 | mRNA processing | <span style=" color: red !important;" >0.222524709416129</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0000375 | RNA splicing, via transesterification reactions | <span style=" color: red !important;" >0.0582561122282698</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | <span style=" color: red !important;" >0.0544142179241016</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0000398 | mRNA splicing, via spliceosome | <span style=" color: red !important;" >0.0544142179241016</span> | 0 |
| gobp | CD56+ NK | L5 | GO:0016071 | mRNA metabolic process | <span style=" color: red !important;" >0.0145223502033037</span> | 0 |
| gobp_nr | CD56+ NK | L1 | GO:0036230 | granulocyte activation | <span style=" color: red !important;" >0.665507403243785</span> | 0 |
| gobp_nr | CD56+ NK | L1 | GO:0002446 | neutrophil mediated immunity | <span style=" color: red !important;" >0.335442878271956</span> | 0 |
| gobp_nr | CD56+ NK | L2 | GO:0006413 | translational initiation | <span style=" color: green !important;" >0.99548773368734</span> | 0 |
| gobp_nr | CD56+ NK | L3 | GO:0042110 | T cell activation | <span style=" color: green !important;" >0.985660130026994</span> | 0 |
| gobp_nr | CD56+ NK | L3 | GO:0007159 | leukocyte cell-cell adhesion | <span style=" color: red !important;" >0.00817191764319258</span> | 0 |
| gobp_nr | CD56+ NK | L4 | GO:0009123 | nucleoside monophosphate metabolic process | <span style=" color: green !important;" >0.981592710152146</span> | 0 |
| gobp_nr | CD56+ NK | L4 | GO:0009141 | nucleoside triphosphate metabolic process | <span style=" color: red !important;" >0.0159047739538065</span> | 0 |
| gobp_nr | CD56+ NK | L4 | GO:0009259 | ribonucleotide metabolic process | <span style=" color: red !important;" >0.00137244907750789</span> | 0 |
| gobp_nr | CD56+ NK | L5 | GO:0042113 | B cell activation | <span style=" color: green !important;" >0.99727245196416</span> | 0 |
| gomf | CD56+ NK | L1 | GO:0003735 | structural constituent of ribosome | <span style=" color: green !important;" >0.999999999999999</span> | 0 |
| gomf | CD56+ NK | L2 | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | <span style=" color: green !important;" >0.996770584349283</span> | 0 |
| gomf | CD56+ NK | L2 | GO:0003700 | DNA-binding transcription factor activity | <span style=" color: red !important;" >0.00335654880277059</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.52466380080945</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0000976 | transcription regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.281690588134962</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0001012 | RNA polymerase II regulatory region DNA binding | <span style=" color: red !important;" >0.176078508119199</span> | 0 |
| gomf | CD56+ NK | L3 | GO:1990837 | sequence-specific double-stranded DNA binding | <span style=" color: red !important;" >0.0110809288112284</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0003690 | double-stranded DNA binding | <span style=" color: red !important;" >0.00671717319054521</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0043565 | sequence-specific DNA binding | <span style=" color: red !important;" >0.000626085792263753</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0000987 | proximal promoter sequence-specific DNA binding | <span style=" color: red !important;" >0.00036790679417642</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0001067 | regulatory region nucleic acid binding | <span style=" color: red !important;" >0.000251707443836069</span> | 0 |
| gomf | CD56+ NK | L3 | GO:0044212 | transcription regulatory region DNA binding | <span style=" color: red !important;" >0.000226812998128212</span> | 0 |
| kegg | CD56+ NK | L1 | hsa03010 | Ribosome | <span style=" color: green !important;" >0.99999999999974</span> | 0 |
| kegg | CD56+ NK | L2 | hsa05012 | Parkinson disease | <span style=" color: green !important;" >0.999997842304999</span> | 0 |
| kegg | CD56+ NK | L3 | hsa04640 | Hematopoietic cell lineage | <span style=" color: green !important;" >0.997854126506646</span> | 0 |
| reactome | CD56+ NK | L1 | R-HSA-168256 | Immune System | <span style=" color: green !important;" >0.999999999999966</span> | 0 |
| reactome | CD56+ NK | L2 | R-HSA-156842 | Eukaryotic Translation Elongation | <span style=" color: red !important;" >0.864742536856063</span> | 0 |
| reactome | CD56+ NK | L2 | R-HSA-156902 | Peptide chain elongation | <span style=" color: red !important;" >0.106897566460362</span> | 0 |
| reactome | CD56+ NK | L2 | R-HSA-192823 | Viral mRNA Translation | <span style=" color: red !important;" >0.0271582604054716</span> | 0 |
| reactome | CD56+ NK | L2 | R-HSA-72764 | Eukaryotic Translation Termination | <span style=" color: red !important;" >0.00241317499950555</span> | 0 |
| reactome | CD56+ NK | L2 | R-HSA-72689 | Formation of a pool of free 40S subunits | <span style=" color: red !important;" >0.00141870838965241</span> | 0 |
| reactome | CD56+ NK | L3 | R-HSA-212436 | Generic Transcription Pathway | <span style=" color: red !important;" >0.68127739585357</span> | 0 |
| reactome | CD56+ NK | L3 | R-HSA-74160 | Gene expression (Transcription) | <span style=" color: red !important;" >0.296792949917508</span> | 0 |
| reactome | CD56+ NK | L3 | R-HSA-73857 | RNA Polymerase II Transcription | <span style=" color: red !important;" >0.0222263519097332</span> | 0 |
| reactome | CD56+ NK | L4 | R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | <span style=" color: green !important;" >0.993324533392242</span> | 0 |
| reactome | CD56+ NK | L4 | R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | <span style=" color: red !important;" >0.00603401657316116</span> | 0 |
| reactome | CD56+ NK | L4 | R-HSA-611105 | Respiratory electron transport | <span style=" color: red !important;" >0.00157556665476488</span> | 0 |
| reactome | CD56+ NK | L5 | R-HSA-8878171 | Transcriptional regulation by RUNX1 | <span style=" color: green !important;" >0.97128180665404</span> | 0 |
| reactome | CD56+ NK | L5 | R-HSA-157118 | Signaling by NOTCH | <span style=" color: red !important;" >0.0439386134612366</span> | 0 |
| reactome | CD56+ NK | L5 | R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | <span style=" color: red !important;" >0.0199801192168844</span> | 0 |
| reactome | CD56+ NK | L6 | R-HSA-983168 | Antigen processing: Ubiquitination & Proteasome degradation | <span style=" color: red !important;" >0.880824832409136</span> | 0 |
| reactome | CD56+ NK | L6 | R-HSA-983169 | Class I MHC mediated antigen processing & presentation | <span style=" color: red !important;" >0.118994963852104</span> | 0 |
| reactome | CD56+ NK | L6 | R-HSA-8951664 | Neddylation | <span style=" color: red !important;" >0.000972083746535013</span> | 0 |
| wikipathway | CD56+ NK | L1 | WP477 | Cytoplasmic Ribosomal Proteins | <span style=" color: green !important;" >1</span> | 0 |
| wikipathway | CD56+ NK | L2 | WP111 | Electron Transport Chain (OXPHOS system in mitochondria) | <span style=" color: green !important;" >0.990862019433003</span> | 0 |
| wikipathway | CD56+ NK | L2 | WP623 | Oxidative phosphorylation | <span style=" color: red !important;" >0.0189766446579098</span> | 0 |
T cell
pbmc_res_set_summary %>%
filter(celltype == 'T cell') %>%
filter(active_set, thresh==1e-4) %>%
db_component_kableAdding missing grouping variables: `db`, `celltype`, `top_component`| db | celltype | top_component | geneSet | description | pip | pValueHypergeometric |
|---|---|---|---|---|---|---|
| gobp | T cell | L1 | GO:0001775 | cell activation | <span style=" color: green !important;" >0.998243490581058</span> | 0 |
| gobp | T cell | L1 | GO:0045321 | leukocyte activation | <span style=" color: red !important;" >0.00286420092825412</span> | 0 |
| gobp | T cell | L2 | GO:0006119 | oxidative phosphorylation | <span style=" color: green !important;" >0.998867320720395</span> | 0 |
| gobp | T cell | L2 | GO:0042773 | ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.0023066924918852</span> | 0 |
| gobp | T cell | L2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00209073341052279</span> | 0 |
| gobp | T cell | L3 | GO:0010941 | regulation of cell death | <span style=" color: red !important;" >0.404035437148868</span> | 0 |
| gobp | T cell | L3 | GO:0008219 | cell death | <span style=" color: red !important;" >0.367067203128673</span> | 0 |
| gobp | T cell | L3 | GO:0043067 | regulation of programmed cell death | <span style=" color: red !important;" >0.0598125813344138</span> | 0 |
| gobp | T cell | L3 | GO:0042981 | regulation of apoptotic process | <span style=" color: red !important;" >0.0595901782002641</span> | 0 |
| gobp | T cell | L3 | GO:0012501 | programmed cell death | <span style=" color: red !important;" >0.0428480095786762</span> | 0 |
| gobp | T cell | L3 | GO:0006915 | apoptotic process | <span style=" color: red !important;" >0.0385478112991525</span> | 0 |
| gobp | T cell | L3 | GO:0097190 | apoptotic signaling pathway | <span style=" color: red !important;" >0.0206934598276991</span> | 0 |
| gobp | T cell | L3 | GO:0048518 | positive regulation of biological process | <span style=" color: red !important;" >0.00970233532368381</span> | 0 |
| gobp | T cell | L3 | GO:0010942 | positive regulation of cell death | <span style=" color: red !important;" >0.00624600959039168</span> | 0 |
| gobp | T cell | L3 | GO:0048522 | positive regulation of cellular process | <span style=" color: red !important;" >0.00603763209058406</span> | 0 |
| gobp | T cell | L3 | GO:0048519 | negative regulation of biological process | <span style=" color: red !important;" >0.00546887738370505</span> | 0 |
| gobp | T cell | L3 | GO:0043068 | positive regulation of programmed cell death | <span style=" color: red !important;" >0.00209014350358461</span> | 0 |
| gobp | T cell | L3 | GO:0043065 | positive regulation of apoptotic process | <span style=" color: red !important;" >0.00193399448533771</span> | 0 |
| gobp | T cell | L4 | GO:0045047 | protein targeting to ER | <span style=" color: red !important;" >0.422517754846463</span> | 0 |
| gobp | T cell | L4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.370059387377491</span> | 0 |
| gobp | T cell | L4 | GO:0072599 | establishment of protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.138094584905281</span> | 0 |
| gobp | T cell | L4 | GO:0070972 | protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.0438561363685445</span> | 0 |
| gobp | T cell | L4 | GO:0006613 | cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0317425197962778</span> | 0 |
| gobp | T cell | L5 | GO:0002376 | immune system process | <span style=" color: green !important;" >0.998468900817696</span> | 0 |
| gobp | T cell | L5 | GO:0006955 | immune response | <span style=" color: red !important;" >0.00288124340072227</span> | 0 |
| gobp_nr | T cell | L1 | GO:0036230 | granulocyte activation | <span style=" color: red !important;" >0.732429735587743</span> | 0 |
| gobp_nr | T cell | L1 | GO:0002446 | neutrophil mediated immunity | <span style=" color: red !important;" >0.270336034815327</span> | 0 |
| gobp_nr | T cell | L2 | GO:0042110 | T cell activation | <span style=" color: green !important;" >0.999990596278583</span> | 0 |
| gobp_nr | T cell | L3 | GO:0070972 | protein localization to endoplasmic reticulum | <span style=" color: green !important;" >0.999855861356505</span> | 0 |
| gomf | T cell | L1 | GO:0005515 | protein binding | <span style=" color: green !important;" >0.999999999999996</span> | 0 |
| kegg | T cell | L1 | hsa05010 | Alzheimer disease | <span style=" color: green !important;" >0.999999276670234</span> | 0 |
| reactome | T cell | L1 | R-HSA-6798695 | Neutrophil degranulation | <span style=" color: green !important;" >0.999999999999371</span> | 0 |
| reactome | T cell | L2 | R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | <span style=" color: red !important;" >0.586005479354723</span> | 0 |
| reactome | T cell | L2 | R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | <span style=" color: red !important;" >0.400596683089486</span> | 0 |
| reactome | T cell | L2 | R-HSA-72689 | Formation of a pool of free 40S subunits | <span style=" color: red !important;" >0.00918003962007052</span> | 0 |
| reactome | T cell | L2 | R-HSA-72613 | Eukaryotic Translation Initiation | <span style=" color: red !important;" >0.00466852361335934</span> | 0 |
| reactome | T cell | L2 | R-HSA-72737 | Cap-dependent Translation Initiation | <span style=" color: red !important;" >0.00466852361335934</span> | 0 |
| reactome | T cell | L2 | R-HSA-192823 | Viral mRNA Translation | <span style=" color: red !important;" >0.00349644548656469</span> | 0 |
| reactome | T cell | L2 | R-HSA-156902 | Peptide chain elongation | <span style=" color: red !important;" >0.00317407099445022</span> | 0 |
CD14+ Monocyte
pbmc_res_set_summary %>%
filter(celltype == 'CD14+ Monocyte') %>%
filter(active_set, thresh==1e-4) %>%
db_component_kableAdding missing grouping variables: `db`, `celltype`, `top_component`| db | celltype | top_component | geneSet | description | pip | pValueHypergeometric |
|---|---|---|---|---|---|---|
| gobp | CD14+ Monocyte | L1 | GO:0045321 | leukocyte activation | <span style=" color: red !important;" >0.913955996656784</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L1 | GO:0001775 | cell activation | <span style=" color: red !important;" >0.0861038961576571</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L2 | GO:0045047 | protein targeting to ER | <span style=" color: red !important;" >0.861539198812174</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0574691369768895</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L2 | GO:0006613 | cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.0477713854410204</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L2 | GO:0072599 | establishment of protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.0336165786420544</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L2 | GO:0070972 | protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.000401591156238168</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L3 | GO:0006119 | oxidative phosphorylation | <span style=" color: green !important;" >0.999048492769061</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L3 | GO:0042773 | ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00061526966849168</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.000437951615106402</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0016192 | vesicle-mediated transport | <span style=" color: green !important;" >0.985143389925145</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0002376 | immune system process | <span style=" color: red !important;" >0.0210152324386211</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0006955 | immune response | <span style=" color: red !important;" >0.00598414884177589</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0045055 | regulated exocytosis | <span style=" color: red !important;" >0.00105846105456608</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0006810 | transport | <span style=" color: red !important;" >0.000928956786439117</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0042119 | neutrophil activation | <span style=" color: red !important;" >0.000810401236960523</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0006887 | exocytosis | <span style=" color: red !important;" >0.000757076172462012</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L4 | GO:0051234 | establishment of localization | <span style=" color: red !important;" >0.000747810324681297</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0006915 | apoptotic process | <span style=" color: red !important;" >0.272017554202987</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0012501 | programmed cell death | <span style=" color: red !important;" >0.194857583380731</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0043067 | regulation of programmed cell death | <span style=" color: red !important;" >0.194088173818995</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0042981 | regulation of apoptotic process | <span style=" color: red !important;" >0.158326414667851</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0010941 | regulation of cell death | <span style=" color: red !important;" >0.10083762777805</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0008219 | cell death | <span style=" color: red !important;" >0.0513299924130467</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0097190 | apoptotic signaling pathway | <span style=" color: red !important;" >0.00254918838420504</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0006950 | response to stress | <span style=" color: red !important;" >0.000930732251314792</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0010942 | positive regulation of cell death | <span style=" color: red !important;" >0.000657647931395888</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0050790 | regulation of catalytic activity | <span style=" color: red !important;" >0.000573508659057254</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0048518 | positive regulation of biological process | <span style=" color: red !important;" >0.000545987494292133</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0010033 | response to organic substance | <span style=" color: red !important;" >0.000481190817748667</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0070887 | cellular response to chemical stimulus | <span style=" color: red !important;" >0.000450649515349011</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:2001233 | regulation of apoptotic signaling pathway | <span style=" color: red !important;" >0.000349195404672376</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0043065 | positive regulation of apoptotic process | <span style=" color: red !important;" >0.00032984108689027</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0043068 | positive regulation of programmed cell death | <span style=" color: red !important;" >0.000290053025053139</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0043069 | negative regulation of programmed cell death | <span style=" color: red !important;" >0.000181772401672053</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0043066 | negative regulation of apoptotic process | <span style=" color: red !important;" >0.000147644072101105</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0051716 | cellular response to stimulus | <span style=" color: red !important;" >0.000136512713189374</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0048522 | positive regulation of cellular process | <span style=" color: red !important;" >0.000135936601124764</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0060548 | negative regulation of cell death | <span style=" color: red !important;" >0.000122829004029934</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0050896 | response to stimulus | <span style=" color: red !important;" >0.00011255017314471</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0048523 | negative regulation of cellular process | <span style=" color: red !important;" >7.49396073772557e-05</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L5 | GO:0048519 | negative regulation of biological process | <span style=" color: red !important;" >6.92053062711917e-05</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L6 | GO:0006518 | peptide metabolic process | <span style=" color: red !important;" >0.790995030090949</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L6 | GO:0006412 | translation | <span style=" color: red !important;" >0.119614037058977</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L6 | GO:0043043 | peptide biosynthetic process | <span style=" color: red !important;" >0.0828579448699818</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L6 | GO:0043603 | cellular amide metabolic process | <span style=" color: red !important;" >0.00254329546224519</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L6 | GO:0043604 | amide biosynthetic process | <span style=" color: red !important;" >0.00124595731038435</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0008380 | RNA splicing | <span style=" color: red !important;" >0.422448316361525</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0000375 | RNA splicing, via transesterification reactions | <span style=" color: red !important;" >0.19679459335928</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | <span style=" color: red !important;" >0.178759471245491</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0000398 | mRNA splicing, via spliceosome | <span style=" color: red !important;" >0.178759471245491</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0006397 | mRNA processing | <span style=" color: red !important;" >0.0212448579074473</span> | 0.0000000 |
| gobp | CD14+ Monocyte | L7 | GO:0016071 | mRNA metabolic process | <span style=" color: red !important;" >0.00160999577015686</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L1 | GO:0036230 | granulocyte activation | <span style=" color: green !important;" >0.975928025123381</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L1 | GO:0002446 | neutrophil mediated immunity | <span style=" color: red !important;" >0.0246951417690048</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L2 | GO:0006413 | translational initiation | <span style=" color: green !important;" >0.999994018876987</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L3 | GO:0009123 | nucleoside monophosphate metabolic process | <span style=" color: green !important;" >0.988352798616283</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L3 | GO:0009141 | nucleoside triphosphate metabolic process | <span style=" color: red !important;" >0.0125932371081144</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L4 | GO:0002521 | leukocyte differentiation | <span style=" color: red !important;" >0.619222288628166</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L4 | GO:0042110 | T cell activation | <span style=" color: red !important;" >0.358464783041196</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L4 | GO:0002694 | regulation of leukocyte activation | <span style=" color: red !important;" >0.00793501697430843</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L4 | GO:1903706 | regulation of hemopoiesis | <span style=" color: red !important;" >0.00445659052005509</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L5 | GO:0008380 | RNA splicing | <span style=" color: red !important;" >0.876773132747017</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L5 | GO:0006397 | mRNA processing | <span style=" color: red !important;" >0.0896324054247939</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L5 | GO:1903311 | regulation of mRNA metabolic process | <span style=" color: red !important;" >0.00430389549836563</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L6 | GO:0090150 | establishment of protein localization to membrane | <span style=" color: red !important;" >0.756201036295744</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L6 | GO:0070972 | protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.243465201424837</span> | 0.0000000 |
| gobp_nr | CD14+ Monocyte | L6 | GO:0006605 | protein targeting | <span style=" color: red !important;" >0.00495025186440967</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L1 | GO:0003723 | RNA binding | <span style=" color: green !important;" >0.999999822304653</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L2 | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | <span style=" color: green !important;" >0.999827482454419</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L2 | GO:0003700 | DNA-binding transcription factor activity | <span style=" color: red !important;" >0.000227989524673178</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0001012 | RNA polymerase II regulatory region DNA binding | <span style=" color: red !important;" >0.660320565777245</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.292436321580928</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0000987 | proximal promoter sequence-specific DNA binding | <span style=" color: red !important;" >0.036062059217844</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | <span style=" color: red !important;" >0.00772449074681092</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0000976 | transcription regulatory region sequence-specific DNA binding | <span style=" color: red !important;" >0.00308292567214929</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:1990837 | sequence-specific double-stranded DNA binding | <span style=" color: red !important;" >0.00070252840459839</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0043565 | sequence-specific DNA binding | <span style=" color: red !important;" >0.000386897461082891</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L3 | GO:0044212 | transcription regulatory region DNA binding | <span style=" color: red !important;" >0.00010172217576454</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L4 | GO:0003735 | structural constituent of ribosome | <span style=" color: green !important;" >0.999999999996356</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | <span style=" color: red !important;" >0.827083558750439</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity | <span style=" color: red !important;" >0.0778621792476983</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L5 | GO:0050136 | NADH dehydrogenase (quinone) activity | <span style=" color: red !important;" >0.0778621792476983</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L5 | GO:0003954 | NADH dehydrogenase activity | <span style=" color: red !important;" >0.0178702201177219</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H | <span style=" color: red !important;" >0.00034694713121397</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L6 | GO:0045296 | cadherin binding | <span style=" color: red !important;" >0.789080093597068</span> | 0.0000000 |
| gomf | CD14+ Monocyte | L6 | GO:0050839 | cell adhesion molecule binding | <span style=" color: red !important;" >0.188160373616359</span> | 0.0000000 |
| kegg | CD14+ Monocyte | L1 | hsa03010 | Ribosome | <span style=" color: green !important;" >0.999999999999889</span> | 0.0000000 |
| kegg | CD14+ Monocyte | L2 | hsa05012 | Parkinson disease | <span style=" color: green !important;" >0.999902118704163</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L1 | R-HSA-6798695 | Neutrophil degranulation | <span style=" color: green !important;" >0.999995355573276</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L2 | R-HSA-72766 | Translation | <span style=" color: green !important;" >0.99999999999981</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L3 | R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | <span style=" color: green !important;" >0.99961105913657</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L3 | R-HSA-611105 | Respiratory electron transport | <span style=" color: red !important;" >0.000839226913661562</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L5 | R-HSA-72172 | mRNA Splicing | <span style=" color: red !important;" >0.674955580706303</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L5 | R-HSA-72163 | mRNA Splicing - Major Pathway | <span style=" color: red !important;" >0.306411442261019</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L5 | R-HSA-72203 | Processing of Capped Intron-Containing Pre-mRNA | <span style=" color: red !important;" >0.0182645263922769</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L6 | R-HSA-198933 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | <span style=" color: green !important;" >0.997487749726619</span> | 0.0000000 |
| reactome | CD14+ Monocyte | L7 | R-HSA-379726 | Mitochondrial tRNA aminoacylation | <span style=" color: green !important;" >0.99729860552702</span> | 0.6931782 |
| wikipathway | CD14+ Monocyte | L1 | WP477 | Cytoplasmic Ribosomal Proteins | <span style=" color: green !important;" >0.999999999999998</span> | 0.0000000 |
| wikipathway | CD14+ Monocyte | L2 | WP111 | Electron Transport Chain (OXPHOS system in mitochondria) | <span style=" color: green !important;" >0.999831154578974</span> | 0.0000000 |
CD34+
pbmc_res_set_summary %>%
filter(celltype == 'CD34+') %>%
filter(active_set, thresh==1e-4) %>%
db_component_kableAdding missing grouping variables: `db`, `celltype`, `top_component`| db | celltype | top_component | geneSet | description | pip | pValueHypergeometric |
|---|---|---|---|---|---|---|
| gobp | CD34+ | L1 | GO:0001775 | cell activation | <span style=" color: green !important;" >0.987053979028693</span> | 0 |
| gobp | CD34+ | L1 | GO:0045321 | leukocyte activation | <span style=" color: red !important;" >0.0137772991951518</span> | 0 |
| gobp | CD34+ | L2 | GO:0006613 | cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.813781011088962</span> | 0 |
| gobp | CD34+ | L2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | <span style=" color: red !important;" >0.153626340011531</span> | 0 |
| gobp | CD34+ | L2 | GO:0045047 | protein targeting to ER | <span style=" color: red !important;" >0.0290629664314263</span> | 0 |
| gobp | CD34+ | L2 | GO:0072599 | establishment of protein localization to endoplasmic reticulum | <span style=" color: red !important;" >0.00852780905068462</span> | 0 |
| gobp | CD34+ | L3 | GO:0006119 | oxidative phosphorylation | <span style=" color: green !important;" >0.998941297041509</span> | 0 |
| gobp | CD34+ | L3 | GO:0042773 | ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00219768696027844</span> | 0 |
| gobp | CD34+ | L3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | <span style=" color: red !important;" >0.00204003835356481</span> | 0 |
| gobp | CD34+ | L4 | GO:0044419 | interspecies interaction between organisms | <span style=" color: red !important;" >0.420351724582621</span> | 0 |
| gobp | CD34+ | L4 | GO:0016032 | viral process | <span style=" color: red !important;" >0.383314261542707</span> | 0 |
| gobp | CD34+ | L4 | GO:0044403 | symbiont process | <span style=" color: red !important;" >0.195249491605503</span> | 0 |
| gobp_nr | CD34+ | L1 | GO:0002446 | neutrophil mediated immunity | <span style=" color: red !important;" >0.607843923572364</span> | 0 |
| gobp_nr | CD34+ | L1 | GO:0036230 | granulocyte activation | <span style=" color: red !important;" >0.394659620593896</span> | 0 |
| gobp_nr | CD34+ | L2 | GO:0006413 | translational initiation | <span style=" color: green !important;" >0.999660829824325</span> | 0 |
| gomf | CD34+ | L1 | GO:0003735 | structural constituent of ribosome | <span style=" color: green !important;" >0.999999999995723</span> | 0 |
| gomf | CD34+ | L2 | GO:0005515 | protein binding | <span style=" color: green !important;" >0.999999999999647</span> | 0 |
| kegg | CD34+ | L1 | hsa00190 | Oxidative phosphorylation | <span style=" color: green !important;" >0.992175886824888</span> | 0 |
| kegg | CD34+ | L1 | hsa05012 | Parkinson disease | <span style=" color: red !important;" >0.0333607954124152</span> | 0 |
| kegg | CD34+ | L2 | hsa03010 | Ribosome | <span style=" color: green !important;" >0.999999994840815</span> | 0 |
| reactome | CD34+ | L1 | R-HSA-72764 | Eukaryotic Translation Termination | <span style=" color: green !important;" >0.99305623315484</span> | 0 |
| reactome | CD34+ | L1 | R-HSA-156902 | Peptide chain elongation | <span style=" color: red !important;" >0.00637721696595694</span> | 0 |
| reactome | CD34+ | L1 | R-HSA-156842 | Eukaryotic Translation Elongation | <span style=" color: red !important;" >0.00406036803807552</span> | 0 |
| reactome | CD34+ | L2 | R-HSA-6798695 | Neutrophil degranulation | <span style=" color: green !important;" >0.986271609979922</span> | 0 |
| reactome | CD34+ | L2 | R-HSA-168249 | Innate Immune System | <span style=" color: red !important;" >0.0166937283411153</span> | 0 |
| reactome | CD34+ | L3 | R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | <span style=" color: green !important;" >0.997732525415435</span> | 0 |
| reactome | CD34+ | L3 | R-HSA-611105 | Respiratory electron transport | <span style=" color: red !important;" >0.00375852255366282</span> | 0 |
| reactome | CD34+ | L4 | R-HSA-198933 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | <span style=" color: green !important;" >0.999950260516898</span> | 0 |
| wikipathway | CD34+ | L1 | WP477 | Cytoplasmic Ribosomal Proteins | <span style=" color: green !important;" >1</span> | 0 |
| wikipathway | CD34+ | L2 | WP111 | Electron Transport Chain (OXPHOS system in mitochondria) | <span style=" color: green !important;" >0.999997114937164</span> | 0 |
knitr::knit_exit()