Supplementary figures
ERM
2023-08-10
Last updated: 2023-08-10
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b30e0e4. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/Arr_geneset.RDS
Ignored: data/BC_cell_lines.csv
Ignored: data/BurridgeDOXTOX.RDS
Ignored: data/CADGWASgene_table.csv
Ignored: data/CAD_geneset.RDS
Ignored: data/CALIMA_Data/
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/DRC48hoursdata.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/Ep24counts.txt
Ignored: data/Full_LD_rep.csv
Ignored: data/GOIsig.csv
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/GTEX_sig24.RDS
Ignored: data/GTEx_gene_list.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/HF_geneset.RDS
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Heatmap_mat.RDS
Ignored: data/Heatmap_sig.RDS
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/LD50_05via.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/NoRespDEG_final.csv
Ignored: data/RINsamplelist.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TMMnormed_x.RDS
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Var_test_list.RDS
Ignored: data/Var_test_list24.RDS
Ignored: data/Var_test_list24alt.RDS
Ignored: data/Var_test_list3.RDS
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allfinal3hour.RDS
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/allmymatrix.RDS
Ignored: data/annotation_data_frame.RDS
Ignored: data/averageviabilitytable.RDS
Ignored: data/avgLD50.RDS
Ignored: data/avg_LD50.RDS
Ignored: data/backGL.txt
Ignored: data/burr_genes.RDS
Ignored: data/calcium_data.RDS
Ignored: data/clamp_summary.RDS
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/counts24hours.RDS
Ignored: data/cpmcount.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/crispr_genes.csv
Ignored: data/ctnnt_results.txt
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/drug_noveh1.csv
Ignored: data/efit2.RDS
Ignored: data/efit2_final.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filcpm_counts.RDS
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/geneDoxonlyQTL.csv
Ignored: data/gene_corr_df.RDS
Ignored: data/gene_corr_frame.RDS
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/geneset_24.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/hsa_kegg_anno.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_DRC48.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/indv_noveh1.csv
Ignored: data/kegglistDEG.RDS
Ignored: data/kegglistDEG24.RDS
Ignored: data/kegglistDEG3.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/label_list.RDS
Ignored: data/ld50_table.csv
Ignored: data/mean_vardrug1.csv
Ignored: data/mean_varframe.csv
Ignored: data/mymatrix.RDS
Ignored: data/new_ld50avg.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/organized_drugframe.csv
Ignored: data/plan2plot.png
Ignored: data/plot_intv_list.RDS
Ignored: data/plot_list_DRC.RDS
Ignored: data/qval24hr.RDS
Ignored: data/qval3hr.RDS
Ignored: data/qvalueEPItemp.RDS
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/siglist_final.RDS
Ignored: data/siglist_old.RDS
Ignored: data/slope_table.csv
Ignored: data/supp10_24hlist.RDS
Ignored: data/supp10_3hlist.RDS
Ignored: data/supp_normLDH48.RDS
Ignored: data/supp_pca_all_anno.RDS
Ignored: data/table3a.omar
Ignored: data/testlist.txt
Ignored: data/toplistall.RDS
Ignored: data/trtonly_24h_genes.RDS
Ignored: data/trtonly_3h_genes.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Ignored: data/viability.RDS
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: Doxorubicin_vehicle_3_24.csv
Untracked: Doxtoplist.csv
Untracked: GWAS_list_of_interest.xlsx
Untracked: KEGGpathwaylist.R
Untracked: OmicNavigator_learn.R
Untracked: SigDoxtoplist.csv
Untracked: analysis/DRC_analysist.Rmd
Untracked: analysis/ciFIT.R
Untracked: analysis/enricher.Rmd
Untracked: analysis/export_to_excel.R
Untracked: analysis/untitled1.R
Untracked: code/DRC_plotfigure1.png
Untracked: code/constantcode.R
Untracked: code/cpm_boxplot.R
Untracked: code/extracting_ggplot_data.R
Untracked: code/fig1plot.png
Untracked: code/figurelegeddrc.png
Untracked: code/movingfilesto_ppl.R
Untracked: code/pearson_extract_func.R
Untracked: code/pearson_tox_extract.R
Untracked: code/plot1C.fun.R
Untracked: code/spearman_extract_func.R
Untracked: code/venndiagramcolor_control.R
Untracked: cormotif_probability_genelist.csv
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/DNRvenn.RDS
Untracked: output/DOXvenn.RDS
Untracked: output/EPIvenn.RDS
Untracked: output/Figures/
Untracked: output/MTXvenn.RDS
Untracked: output/Volcanoplot_10
Untracked: output/Volcanoplot_10.RDS
Untracked: output/allfinal_sup10.RDS
Untracked: output/gene_corr_fig9.RDS
Untracked: output/legend_b.RDS
Untracked: output/motif_ERrep.RDS
Untracked: output/motif_LRrep.RDS
Untracked: output/motif_NRrep.RDS
Untracked: output/motif_TI_rep.RDS
Untracked: output/output-old/
Untracked: output/rank24genes.csv
Untracked: output/rank3genes.csv
Untracked: output/supplementary_motif_list_GO.RDS
Untracked: output/toptablebydrug.RDS
Untracked: output/x_counts.RDS
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/Figure5.Rmd
Modified: analysis/Knowles2019.Rmd
Modified: analysis/variance_scrip.Rmd
Modified: output/TNNI_LDH_RNAnormlist.txt
Modified: output/daplot.RDS
Modified: output/dxplot.RDS
Modified: output/epplot.RDS
Modified: output/mtplot.RDS
Modified: output/plan2plot.png
Modified: output/toplistall.csv
Modified: output/trplot.RDS
Modified: output/veplot.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Supplementary_figures.Rmd)
and HTML (docs/Supplementary_figures.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b30e0e4 | reneeisnowhere | 2023-08-10 | adding fig3 |
| html | 021ae6d | reneeisnowhere | 2023-08-08 | Build site. |
| Rmd | 04026ce | reneeisnowhere | 2023-08-08 | adding svglite |
| Rmd | b0c1fb8 | reneeisnowhere | 2023-08-01 | updated code but not published yet |
| html | fa4c610 | reneeisnowhere | 2023-07-28 | Build site. |
| Rmd | b2ef59a | reneeisnowhere | 2023-07-28 | new figures update |
| html | b31f596 | reneeisnowhere | 2023-07-28 | Build site. |
| Rmd | fdb94fb | reneeisnowhere | 2023-07-28 | new figures update |
| Rmd | 62286c3 | reneeisnowhere | 2023-07-28 | Updateing figure code |
| html | 680af3a | reneeisnowhere | 2023-06-26 | Build site. |
| Rmd | d7b9ff1 | reneeisnowhere | 2023-06-26 | Adding supplementary figures |
| html | 537bc2e | reneeisnowhere | 2023-06-26 | Build site. |
| Rmd | 6e50959 | reneeisnowhere | 2023-06-26 | Adding supplementary figures |
| html | dace8ba | reneeisnowhere | 2023-06-23 | Build site. |
| Rmd | 2b109c3 | reneeisnowhere | 2023-06-23 | adding some supp graphs |
| Rmd | c1d667f | reneeisnowhere | 2023-06-23 | updating the codes at Friday start. |
library(tidyverse)
library(ggpubr)
library(rstatix)
library(zoo)
library(ggsignif)
library(RColorBrewer)
library(grid)
library(scales)
library(ComplexHeatmap)
library(gridExtra)
library(cowplot)
library(drc)
library(kableExtra)Error: package or namespace load failed for 'kableExtra':
.onLoad failed in loadNamespace() for 'kableExtra', details:
call: !is.null(rmarkdown::metadata$output) && rmarkdown::metadata$output %in%
error: 'length = 3' in coercion to 'logical(1)'library(broom)
library(ggVennDiagram)cpm_boxplot_time <-function(cpmcounts,timex, GOI,brewer_palette, fill_colors, ylab) {
##GOI needs to be ENTREZID
df <- cpmcounts
df_plot <- df %>%
dplyr::filter(rownames(.)==GOI) %>%
pivot_longer(everything(),
names_to = "treatment",
values_to = "counts") %>%
separate(treatment, c("drug","indv","time")) %>%
dplyr::filter(time == paste(timex)) %>%
mutate(time=factor(time, levels =c("3h", "24h"), labels=c("3 hours", "24 hours"))) %>%
mutate(indv=factor(indv, levels = c(1,2,3,4,5,6))) %>%
mutate(drug =case_match(drug, "Da"~"DNR",
"Do"~"DOX",
"Ep"~"EPI",
"Mi"~"MTX",
"Tr"~"TRZ",
"Ve"~"VEH", .default = drug)) %>%
mutate(drug=factor(drug, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH')))
plot <- ggplot2::ggplot(df_plot, aes(x=drug, y=counts))+
geom_boxplot(position="identity",aes(fill=drug))+
geom_point(aes(col=indv, size=1.5, alpha=0.5))+
guides(alpha= "none", size= "none")+
scale_color_brewer(palette = brewer_palette, guide = "none")+
scale_fill_manual(values=fill_colors)+
# try(facet_wrap("time", nrow=1, ncol=2))+
theme_bw()+
ylab(ylab)+
xlab("")+
theme(strip.background = element_rect(fill = "white",linetype=1, linewidth = 0.5),
plot.title = element_text(size=10,hjust = 0.5,face="bold"),
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(linewidth = 1.0),
axis.line = element_line(linewidth = 1.0),
axis.text.x = element_blank(),
strip.text.x = element_text(margin = margin(2,0,2,0, "pt"),face = "bold"))
print(plot)
}Fig. S1
ctnnt <- read.csv("data/ctnnt_results.txt", row.names = 1)
ctnnt %>%
mutate(Individual=fct_inorder(Individual)) %>%
ggplot(., aes(Individual,Percent , fill=Individual))+
geom_boxplot()+
geom_point()+
geom_hline(yintercept =70,linetype="dashed", alpha=0.75)+###adds a line indicating high positivity +
coord_cartesian(ylim = c(0,105))+ ##set those limits
theme_bw()+ ##white background
labs(title="Cardiomyocyte Purity")+ #subtitle = "from n>3 differentiations")+
geom_boxplot(color="black",alpha =0.2, fill=NA, fatten=0, show.legend = FALSE)+
scale_fill_brewer(palette = "Dark2",name="" )+
xlab(NULL)+
ylab("% cTNNT+ ")+
guides(fill = NULL)+
theme(plot.title = element_text(hjust = 0.5, size =20, face= "bold"),
axis.title.x=element_blank(),
axis.text.x=element_blank(),###removes all axis names and tick names etc.####
axis.ticks.x=element_blank(),
# legend.text=element_text(size=15),
axis.title.y=element_text(size=15),
axis.ticks.y=element_line(size =2),
axis.text.y=element_text(size=10, face = "bold"),
panel.grid.major = element_line(colour = 'grey'),
panel.border=element_rect(fill = NA, size = 3),
plot.subtitle=element_text(size=18, hjust=0.5, face="italic", color="black")) 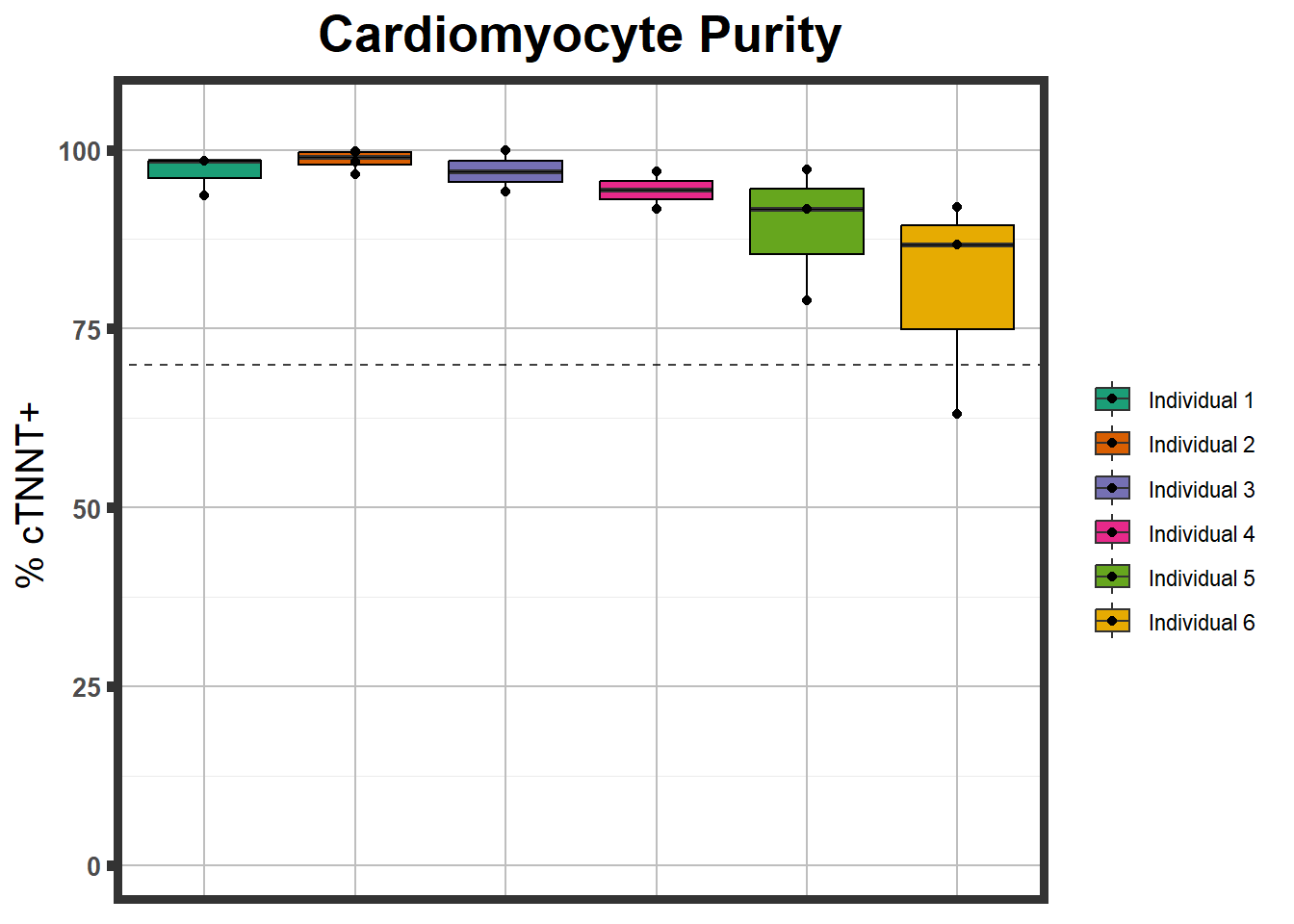
# ctnnt %>%
# summary() %>%
# kable(., caption= "Stats summary of cTNNT+ FACs readings") %>%
# kable_paper("striped", full_width = FALSE) #%>%
# kable_styling(full_width = FALSE,font_size = 18) #%>%
# # scroll_box(width = "60%", height = "400px")Fig. S2
drug_pal_fact <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
library(data.table)
conf_int <- readRDS("data/plot_intv_list.RDS")
DRC_list <- readRDS("data/plot_list_DRC.RDS")
pull_drc2 <- data.frame("ind1", "ind2","ind3","ind4","ind5","ind6")
doubl_plot <- data.frame("ind3a", "ind3b", "ind5a", "ind5b")
lvl_order <- c('1','2','3','4','5','6')
intervals <- rbindlist(conf_int,idcol="trt")
drug_list <- c("DNR","DOX","EPI","MTX", "TRZ", "VEH")
# GeomRibbon$handle_na <- function(data, params) { data }
# brewer.pal(n=6,"Dark2")
# [1] "#1B9E77" "#D95F02" "#7570B3" "#E7298A" "#66A61E" "#E6AB02"
# > display.brewer.pal(n=6,"Dark2")
for (each in 1:6){
newdata <- intervals %>%
separate("trt", into=c("sDrug",NA)) %>%
dplyr::filter(sDrug ==drug_list[each]) %>%
mutate(SampleID=indv) %>%
mutate(indv=substr(indv,4,4)) %>%
mutate(indv=factor(indv, levels=lvl_order)) %>%
dplyr::filter(SampleID %in% doubl_plot)
# newdata <- sub_intv %>%
# filter(indv %in% doubl_plot) %>%
# mutate(sub_ind=substr(indv,4,4)) %>%
# mutate(sub_ind=factor(sub_ind,levels=lvl_order))
drug_plot <- DRC_list[[each]]
f <-
drug_plot %>%
filter(SampleID %in% doubl_plot) %>%
ggplot(., aes(x=Conc, y= Percent, group=SampleID,linetype=SampleID, color= indv,alpha =0.6 )) +
guides(color="none", alpha = "none")+
stat_smooth(method = "drm",
method.args = list(fct = L.4(c(NA,NA,1,NA))),
se = FALSE)+
geom_ribbon(data = newdata,
aes(x = Conc, y = Prediction,
ymin = Lower,
ymax = Upper,
fill=indv),
alpha = 0.1,
color = "transparent")+
# ylim(-.2,1.5)+
coord_cartesian(ylim = c(-0.1, 1.5)) +
scale_linetype_manual(values = c("dotted","solid","dotted","solid"),
name="replicate",
labels=c("Rep 1", "Rep 2","Rep 1", "Rep 2"))+
scale_color_brewer(palette = "Dark2")+
scale_fill_brewer(palette = "Dark2")+
scale_x_log10() + # Change the x-axis scale to log 10 scale
theme_classic() +
xlab(NULL)+
ylab(NULL)+
# scale_y_continuous(oob=scales::rescale_max, limits = -.4, 1.5)+
ggtitle(drug_list[each])+
theme(plot.title = element_text(hjust = 0.5, size =15, face ="bold"),
axis.title=element_text(size=10),
axis.ticks=element_line(linewidth = 2),
axis.text=element_text(size=10, face = "bold", color="black"),
panel.grid.major = element_line(colour = 'lightgrey'),
panel.border=element_rect(fill = NA, linewidth = 2),
plot.background = element_rect(fill = "white", colour = NA))
print(f)
} 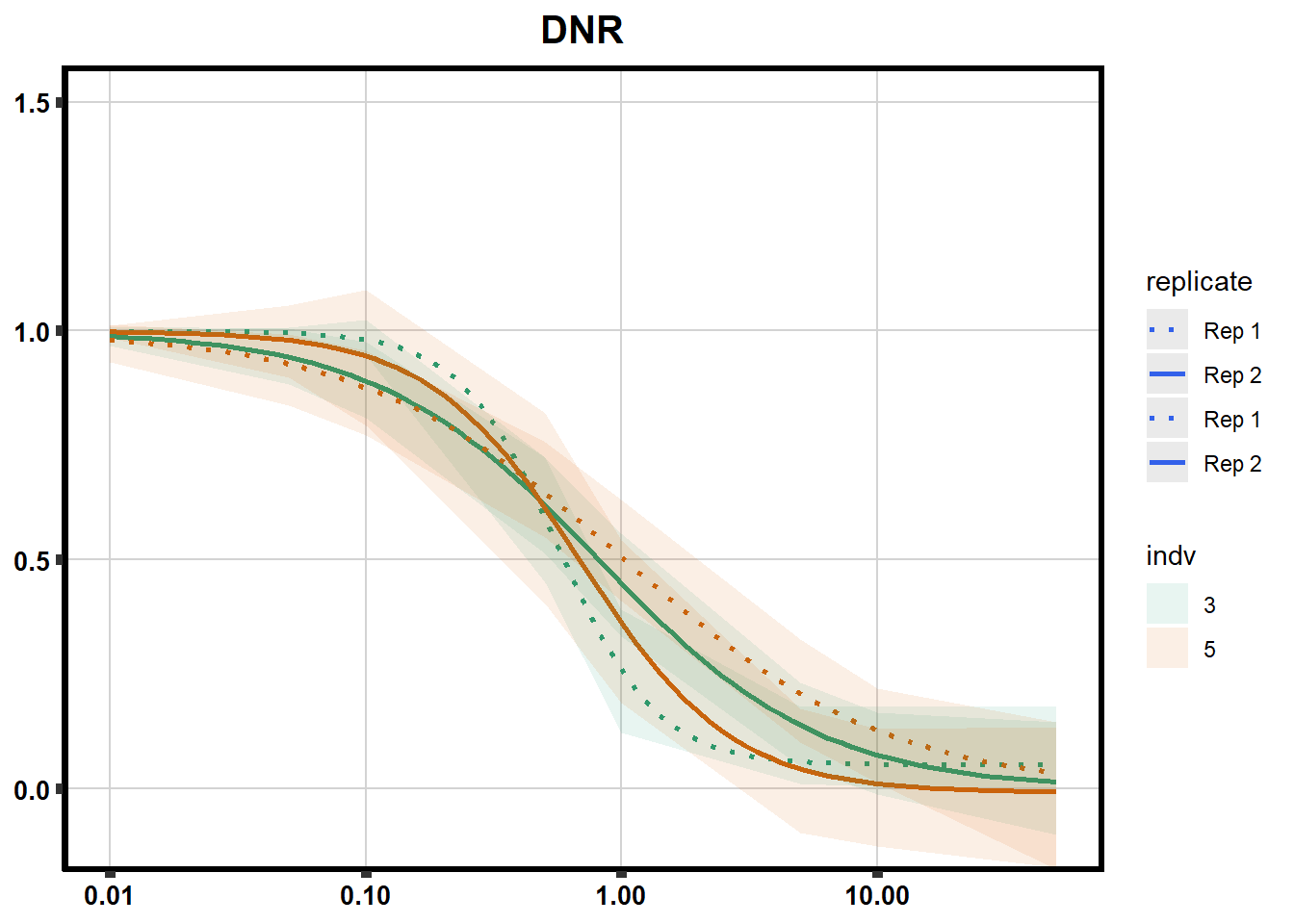
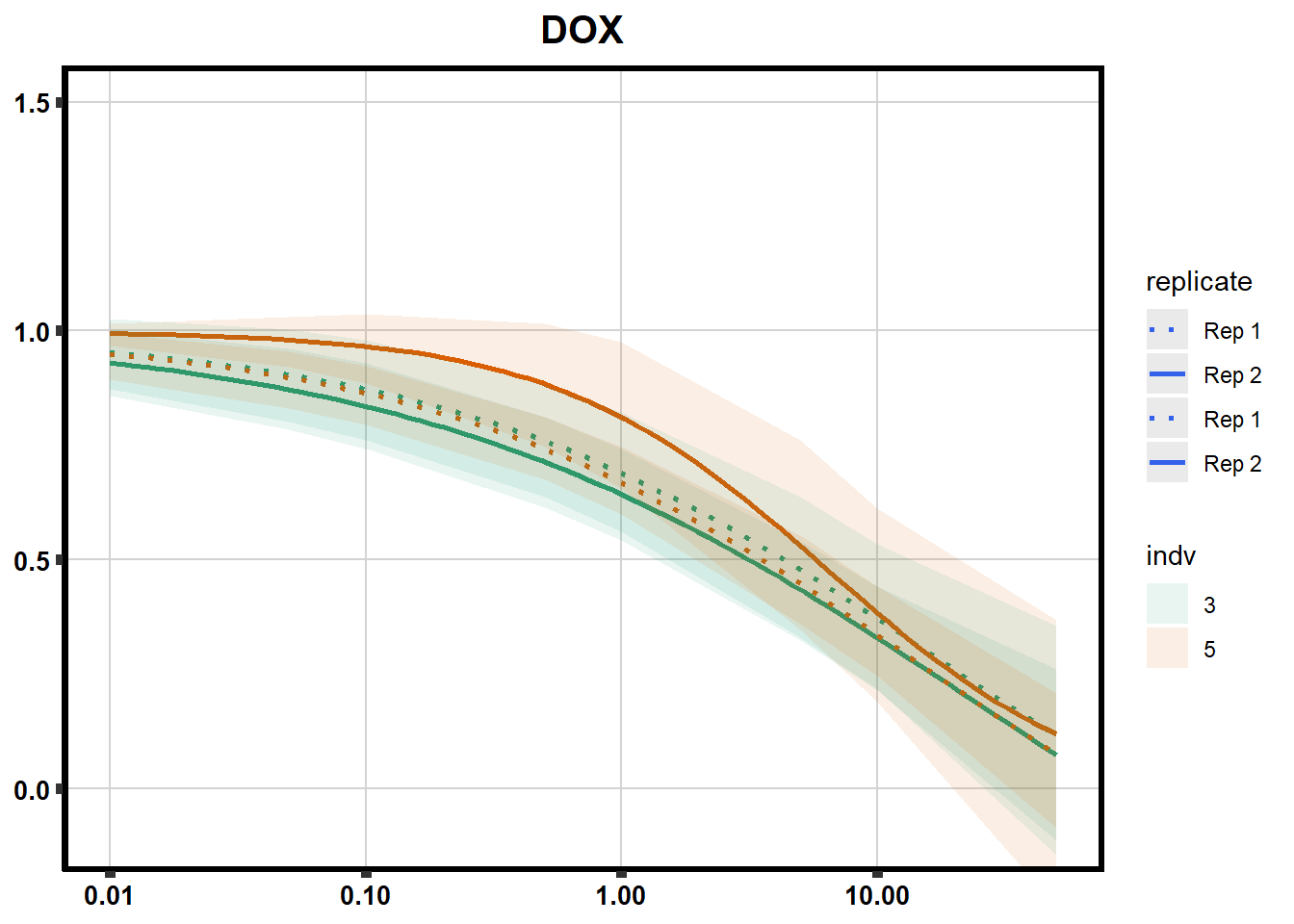
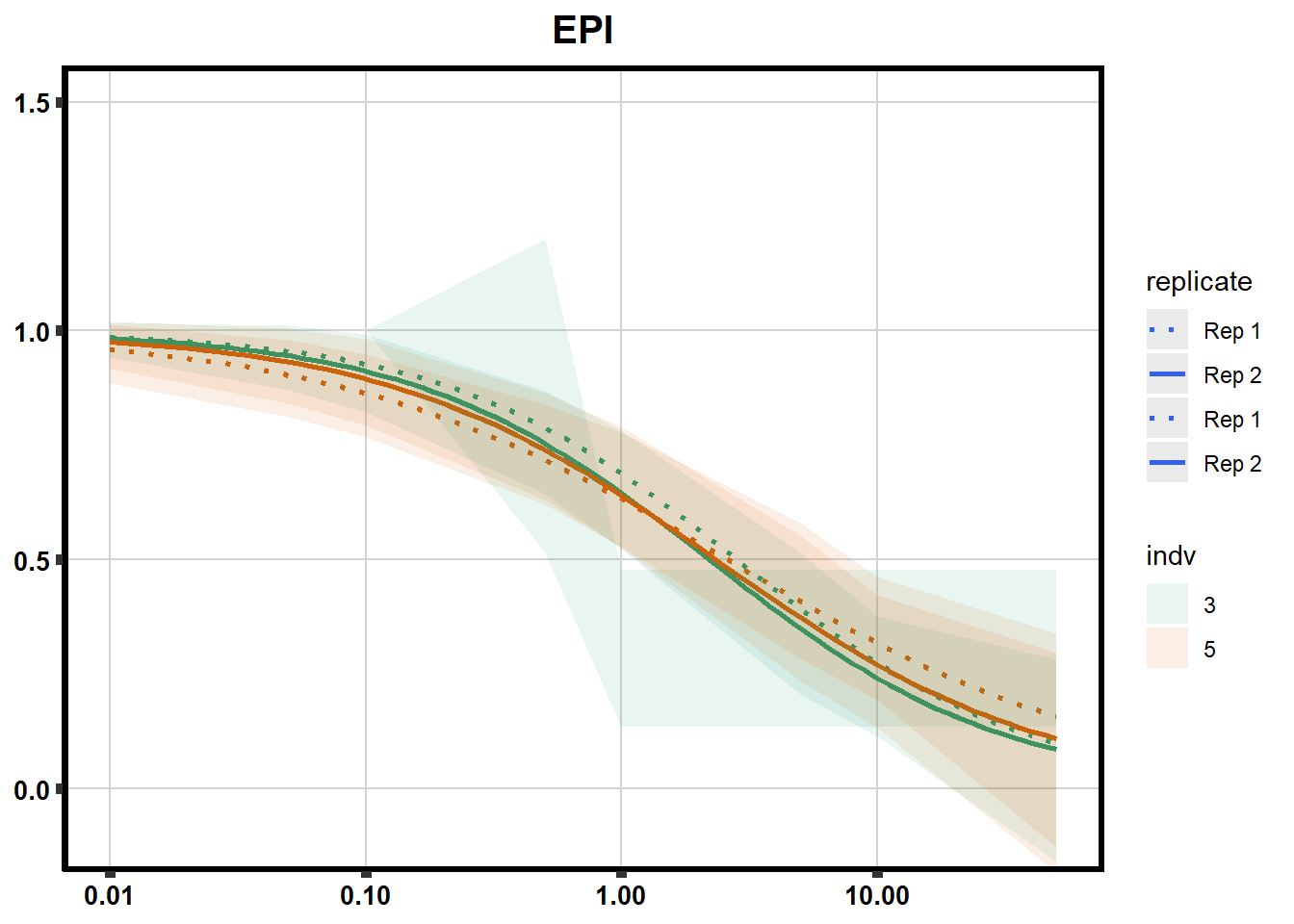
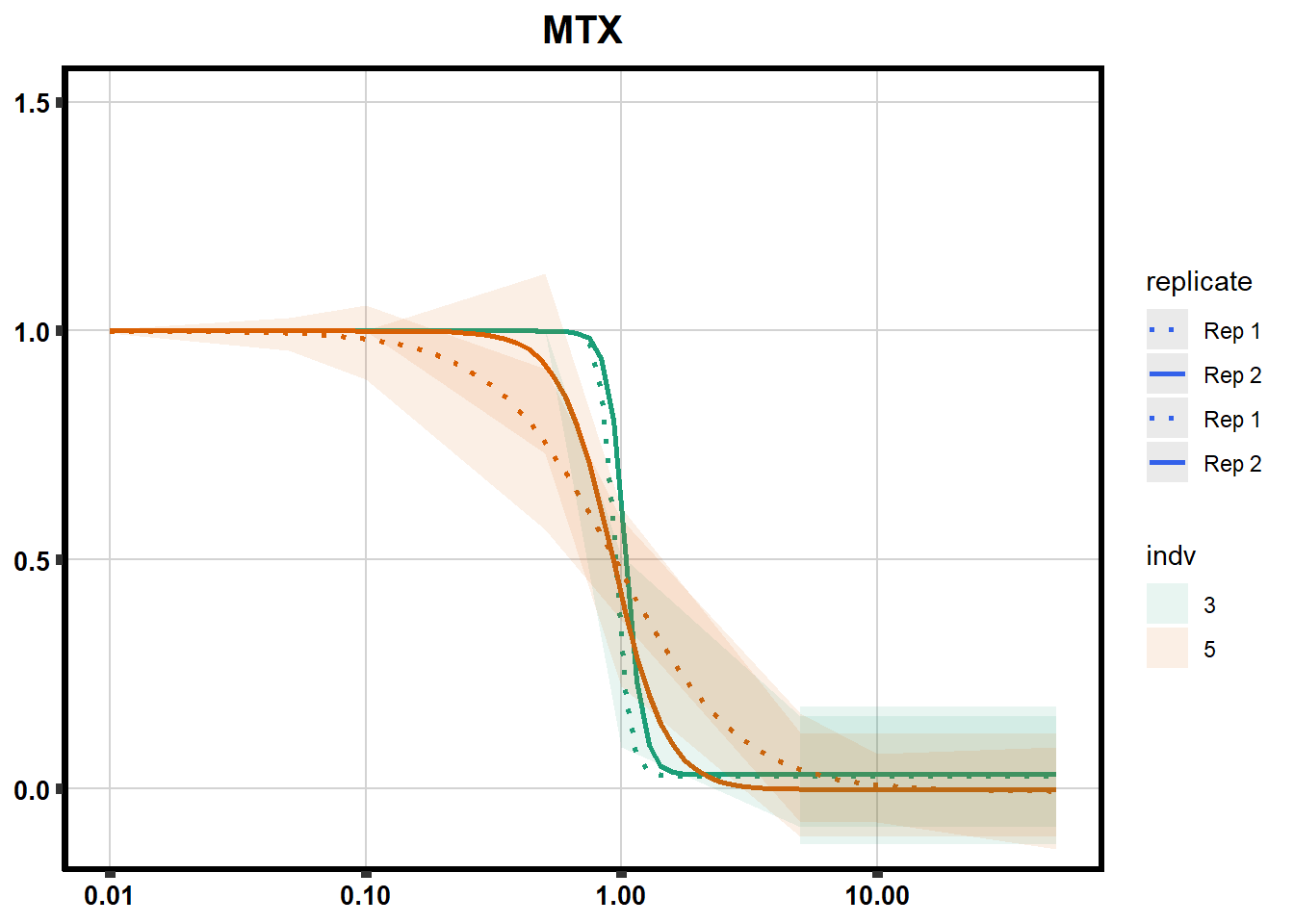
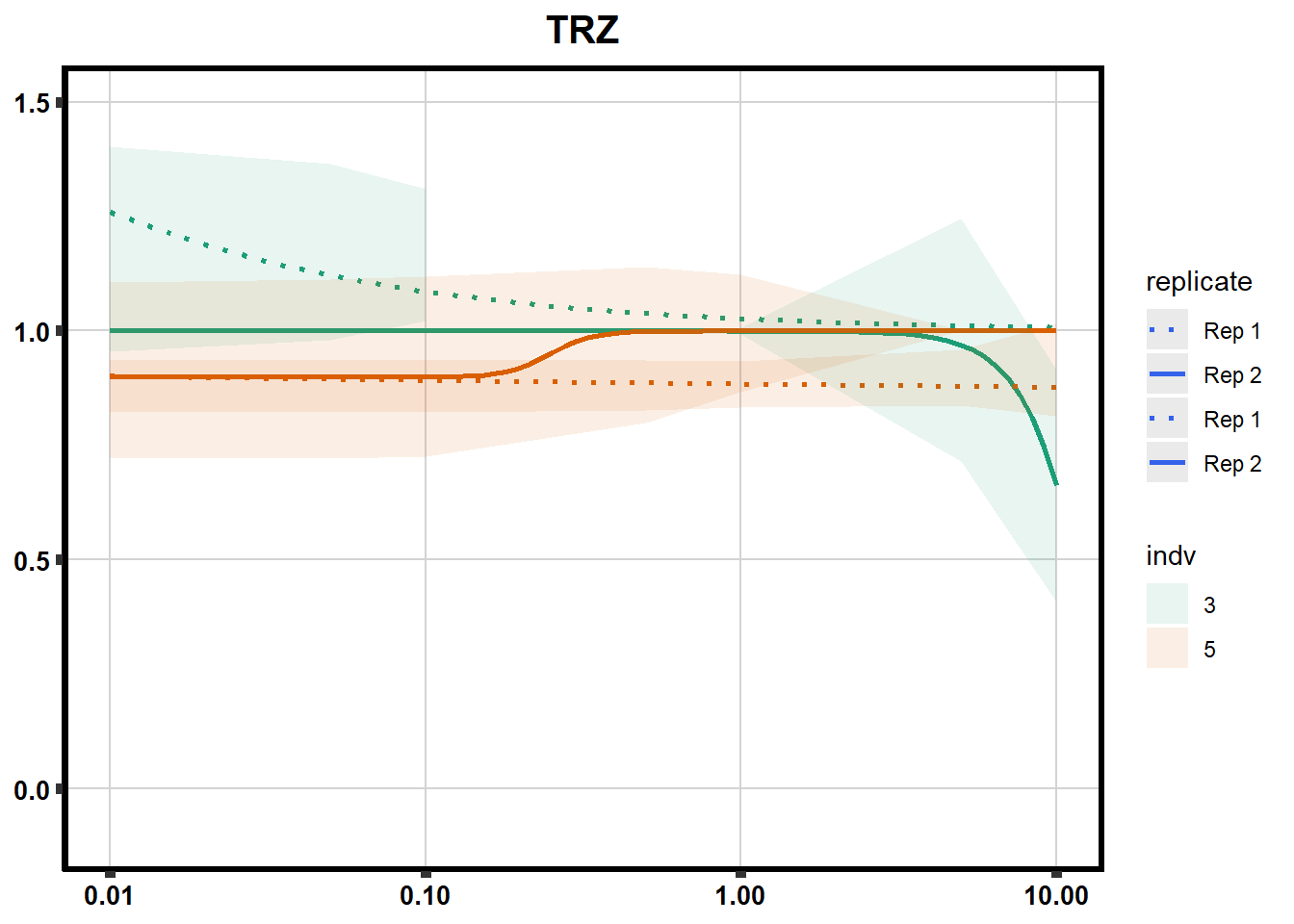
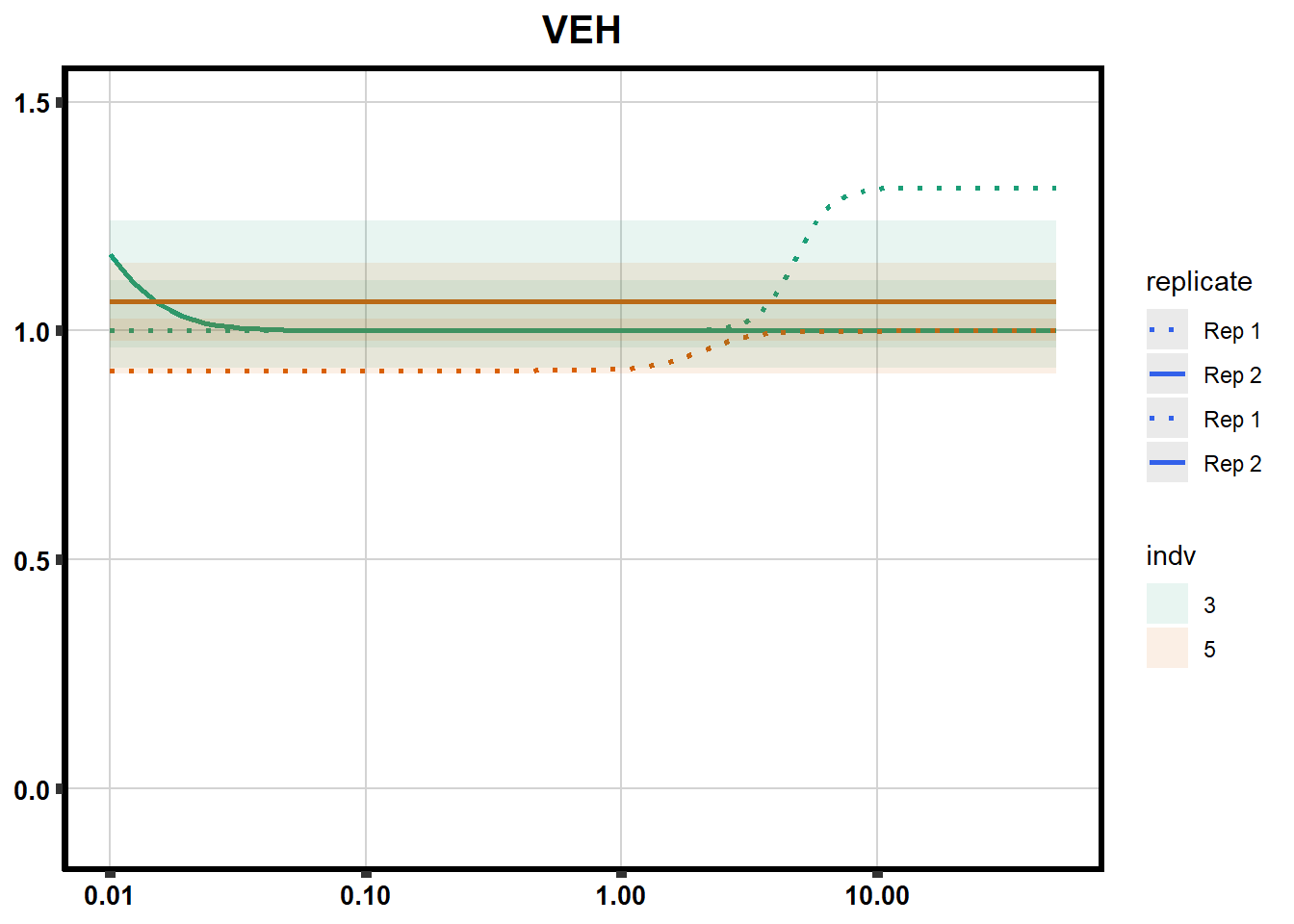
Fig. S3
viability <- readRDS("data/viability.RDS")
norm_LDH48 <- readRDS("data/supp_normLDH48.RDS")
viability %>%
left_join(., norm_LDH48,by = c("indv","Drug","Conc")) %>%
ggplot(., aes(x=per.live, y=ldh))+
geom_point(aes(col=indv))+
geom_smooth(method="lm")+
facet_wrap(~Drug)+
theme_bw()+
xlab("Average viability of cardiomyocytes/100") +
ylab("Average LDH") +
ggtitle("Cell stress and viability correlation")+
scale_color_brewer(palette = "Dark2",name = "Individual", label = c("1","2","3","4","5","6"))+
ggpubr::stat_cor(method="pearson",
aes(label = paste(..r.label.., ..p.label.., sep = "*`,`~")),
color = "red")+
theme(plot.title = element_text(size = rel(1), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"),
strip.background = element_rect(fill = "transparent")) 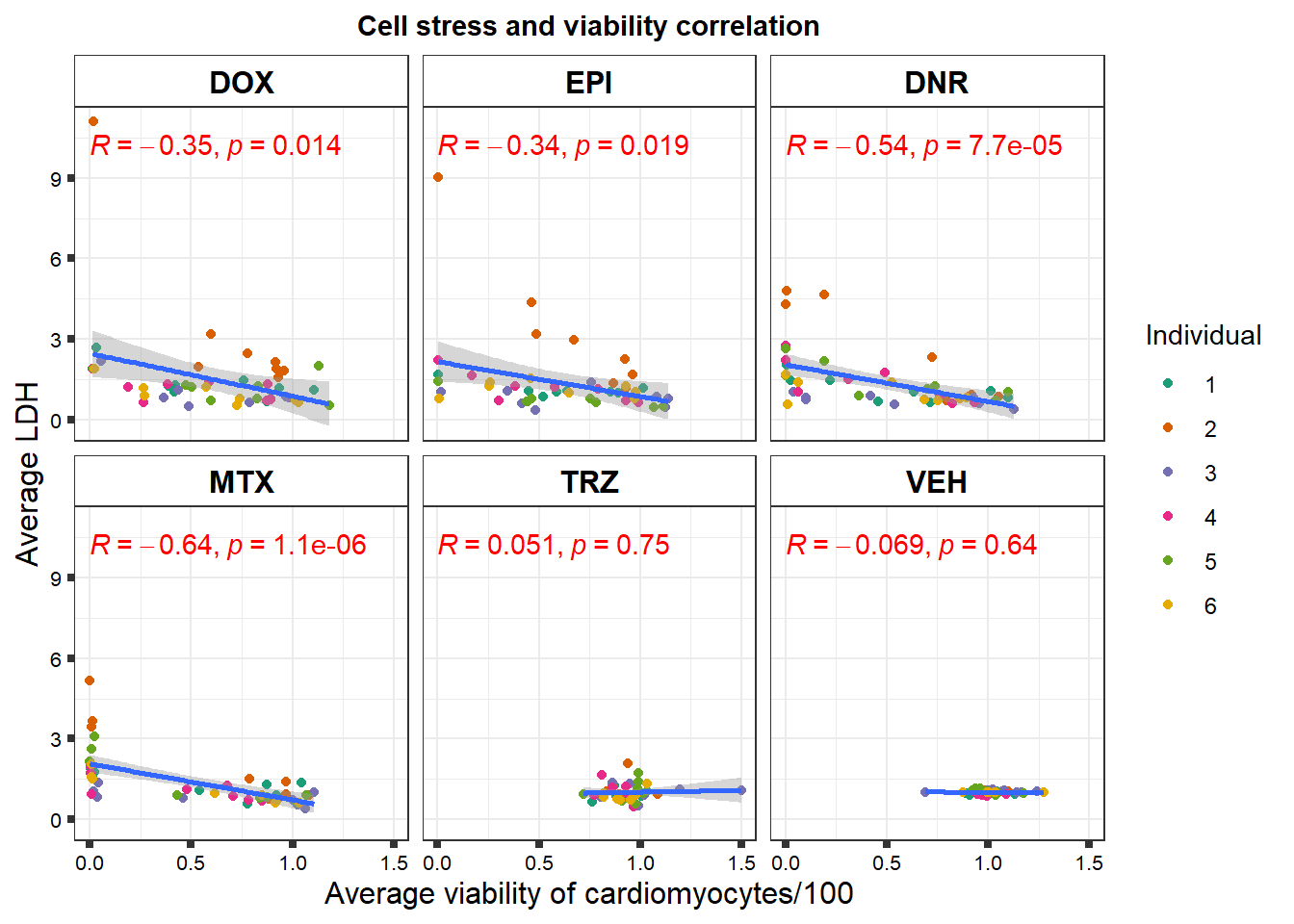
Fig. S4
viabilitytable <- readRDS("data/averageviabilitytable.RDS")
viabilitytable %>%
ungroup() %>%
mutate(indv=substr(SampleID,4,4)) %>%
mutate(indv=factor(indv, levels= c('1','2','3','4','5','6'))) %>%
dplyr::filter(Conc <5) %>%
mutate(Conc= factor(as.numeric(Conc))) %>%
group_by(indv,sDrug,Conc) %>%
dplyr::summarize(Viability=mean(Mean)) %>%
ggplot(., aes(x=sDrug, y= Viability*100 )) +
geom_boxplot(position="dodge",
aes(fill=sDrug))+
geom_point(aes(color=indv))+
guides(alpha = "none")+
ylim(0,150.5)+
scale_color_brewer(palette = "Dark2",
guide="legend",
name ="individual",
labels(c(1,2,3,4,5,6)))+
scale_fill_manual(values=drug_pal_fact, name ="treatment")+
theme_classic() +
xlab("")+
ylab("% Viability") +
facet_wrap(~Conc)+
theme(axis.title=element_text(size=10),
axis.ticks=element_line(size =2),
axis.text.y=element_text(size=9, face = "bold"),
axis.text.x=element_blank(),
panel.grid.major = element_line(colour = 'darkgrey'),
panel.border=element_rect(fill = NA, size = 2),
plot.title = element_text(hjust = 0.5, size =15, face = "bold"))
Fig. S5
library(limma)
library(edgeR)
library(cowplot)
filcpm_matrix <- readRDS("data/filcpm_counts.RDS")
x <- readRDS("data/filtermatrix_x.RDS")
x$samples %>%
mutate(drug=case_match(drug, "Daunorubicin"~"DNR",
"Doxorubicin"~"DOX",
"Epirubicin"~"EPI",
"Mitoxantrone"~"MTX",
"Trastuzumab"~"TRZ",
"Vehicle"~"VEH", .default = drug)) %>%
mutate(drug=factor(drug, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
mutate(time=factor(time, labels= c("3 hours","24 hours"))) %>%
ggplot(., aes(x = as.factor(time), y = RIN)) +
geom_boxplot(aes(fill=as.factor(time)))+
theme_bw()+
ylim(c(0,10))+
labs(x= "", fill ="Time in hours",y ="RNA Integrity Number")+
ggtitle("Boxplot of RIN by time and drug")+
facet_wrap(~drug)+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 0, hjust = 1, vjust = 0.2),
strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.background = element_rect(fill = "white"))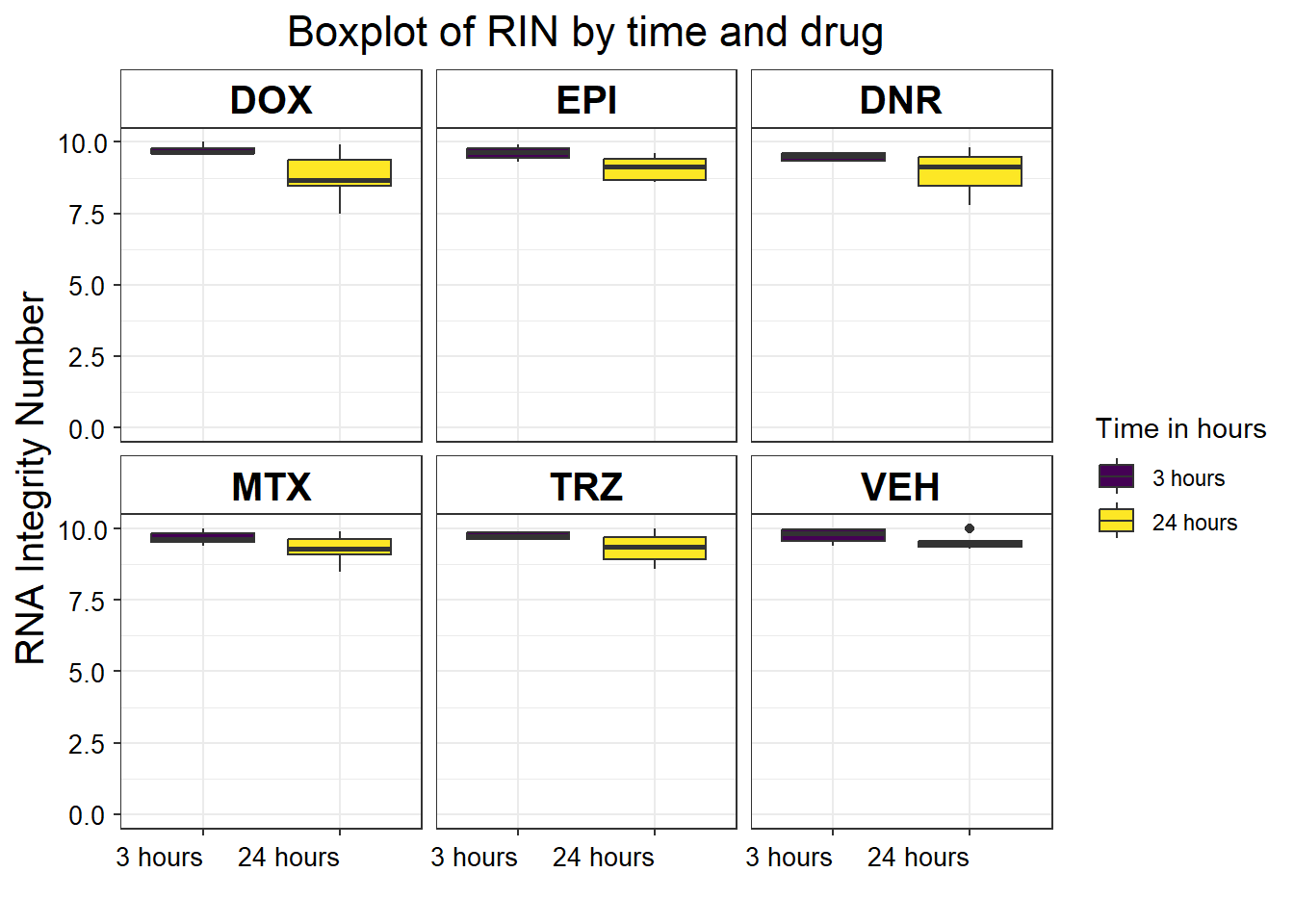
seq_info <-read.csv("output/sequencing_info.txt", row.names = 1)
seq_info %>%
filter(type=="Total_reads") %>%
mutate(drug=case_match(drug, "Daunorubicin"~"DNR",
"Doxorubicin"~"DOX",
"Epirubicin"~"EPI",
"Mitoxantrone"~"MTX",
"Trastuzumab"~"TRZ",
"Vehicle"~"VEH", .default = drug)) %>%
mutate(drug=factor(drug, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
ggplot(., aes (x =drug, y=Total.Sequences, fill = drug))+
geom_boxplot()+
scale_fill_manual(values=drug_pal_fact)+
ggtitle(expression("Total number of reads by treatment"))+
xlab(" ")+
ylab(expression("RNA -sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "white"),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))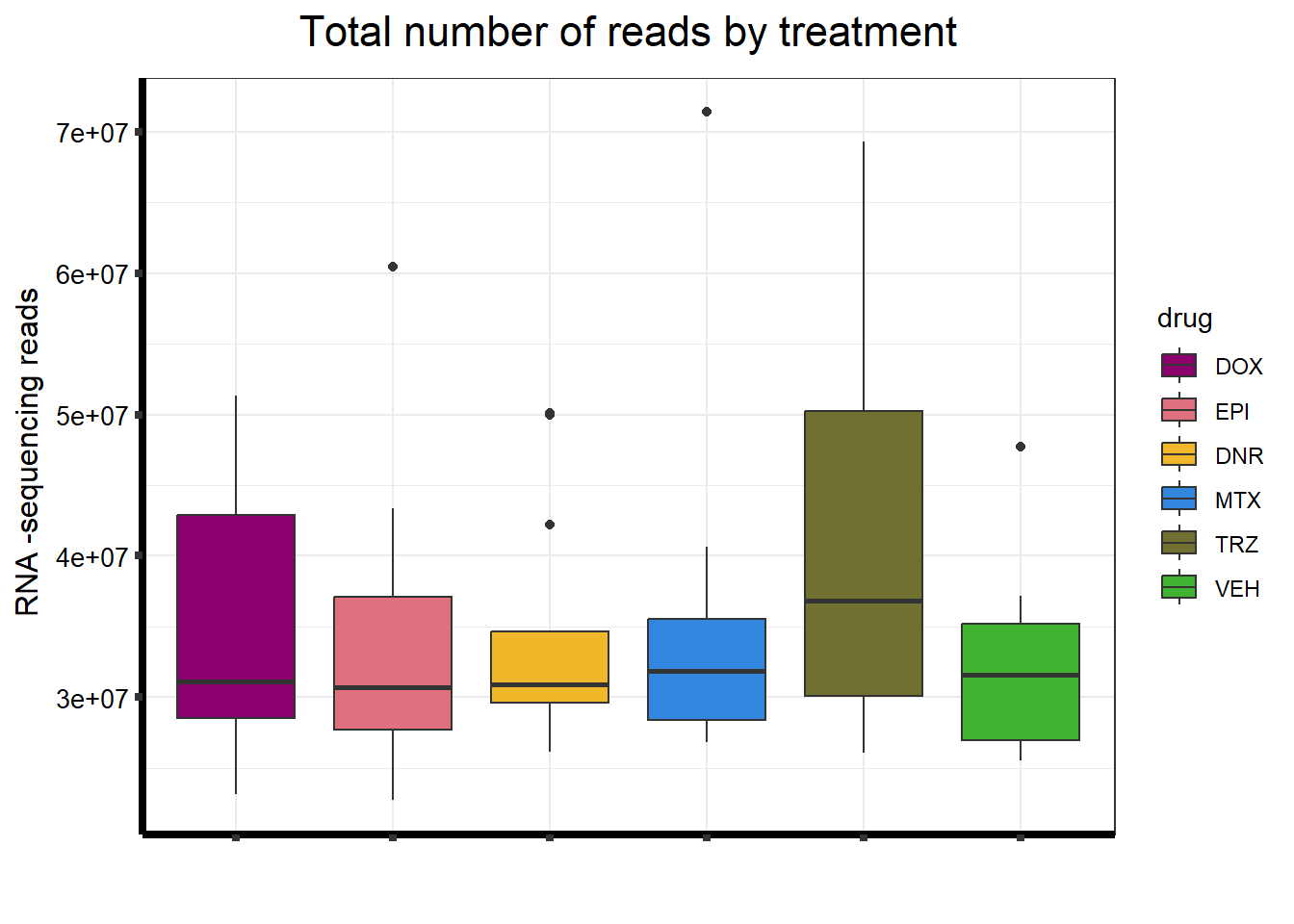
seq_info %>%
mutate(drug=case_match(drug, "Daunorubicin"~"DNR",
"Doxorubicin"~"DOX",
"Epirubicin"~"EPI",
"Mitoxantrone"~"MTX",
"Trastuzumab"~"TRZ",
"Vehicle"~"VEH", .default = drug)) %>%
mutate(drug=factor(drug, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
# separate(samplenames, into=c(NA,NA,NA,"samplenames")) %>%
# mutate(shortnames = paste("Sample",str_trim(samplenames))) %>%
filter(type=="Total_reads") %>%
mutate(sampleID=colnames(filcpm_matrix)) %>%
ggplot(., aes (x =sampleID, y=Total.Sequences, fill = drug, group_by=indv))+
geom_col()+
geom_hline(aes(yintercept=20000000))+
scale_fill_manual(values=drug_pal_fact)+
ggtitle(expression("Total number of reads by sample"))+
xlab("")+
ylab(expression("RNA -sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =6, color = "black", angle = 90, hjust = 1, vjust = 0.2),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))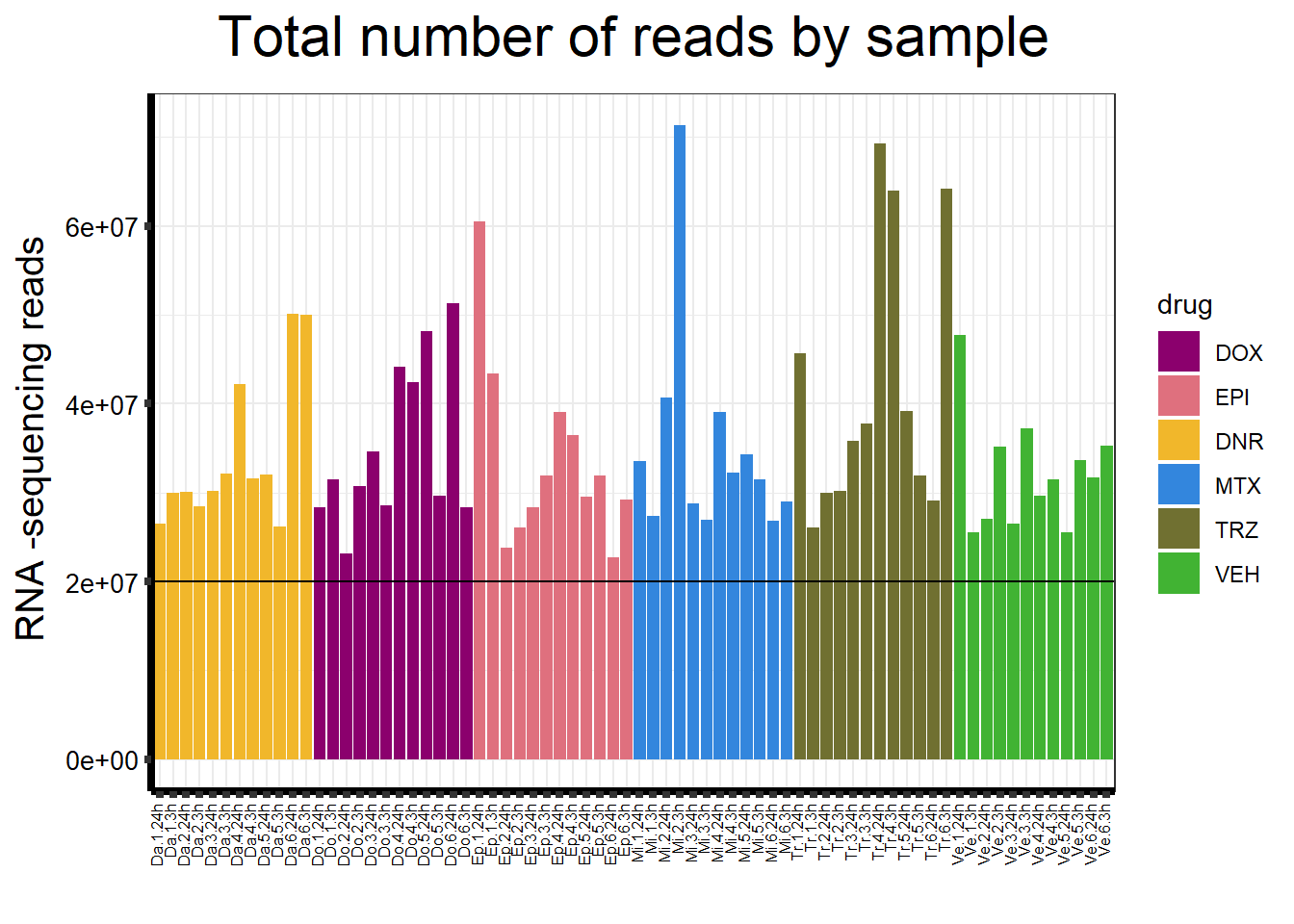 ### Fig. S6
### Fig. S6
filcpm_matrix <- readRDS("data/filcpm_counts.RDS")
mcor <- cor(filcpm_matrix)
filmat_groupmat_col <- data.frame(timeset = colnames(filcpm_matrix))
counts_corr_mat<- filmat_groupmat_col %>%
separate(timeset, into= c("drug","indv","time")) %>%
mutate(class = if_else(drug=="Da","AC", if_else(drug=="Do","AC", if_else(drug=="Ep","AC","nAC")))) %>%
mutate(TOP2i = if_else(drug=="Da","yes", if_else(drug=="Do","yes", if_else(drug=="Ep","yes",if_else(drug=="Mi","yes","no")))))
mat_colors <- list(
drug= c("#F1B72B","#8B006D","#DF707E","#3386DD","#707031","#41B333"),
indv=c("#1B9E77", "#D95F02" ,"#7570B3", "#E7298A" ,"#66A61E", "#E6AB02"),
time=c("pink", "chocolate4"),
class=c("yellow1","lightgreen"),
TOP2i =c("darkgreen","goldenrod"))
names(mat_colors$drug) <- unique(counts_corr_mat$drug)
names(mat_colors$indv) <- unique(counts_corr_mat$indv)
names(mat_colors$time) <- unique(counts_corr_mat$time)
names(mat_colors$class) <- unique(counts_corr_mat$class)
names(mat_colors$TOP2i) <- unique(counts_corr_mat$TOP2i)
ComplexHeatmap::pheatmap(mcor,
# column_title=(paste0("RNA-seq log"[2]~"cpm correlation")),
annotation_col = counts_corr_mat,
annotation_colors = mat_colors,
fontsize=10,
fontsize_row = 8,
angle_col="90",
treeheight_row=25,
fontsize_col = 8,
treeheight_col = 20)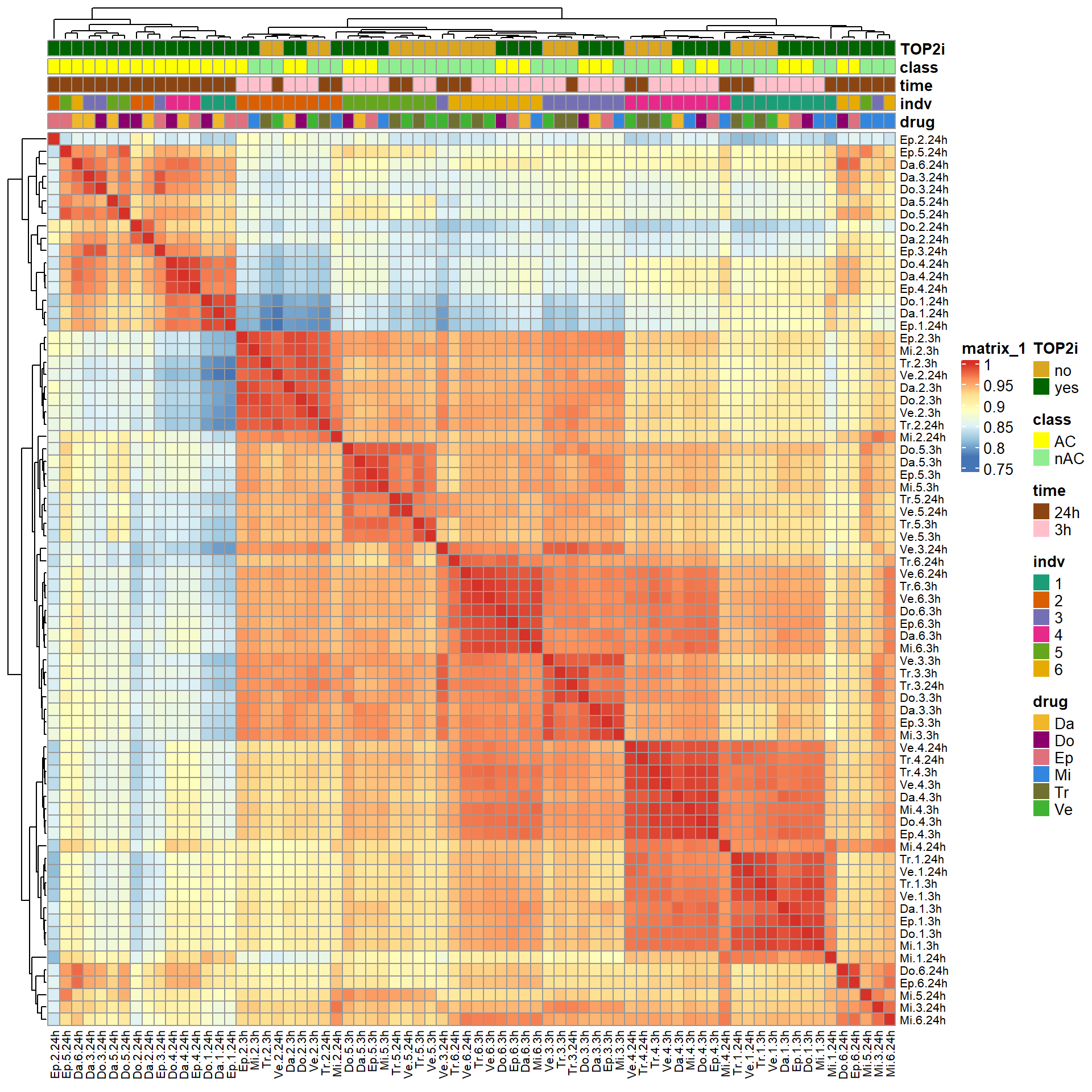
Fig. S7
pca_all_anno <- readRDS("data/supp_pca_all_anno.RDS")
pca_all_anno <- pca_all_anno %>%
mutate(drug = case_match(drug, "Daunorubicin"~"DNR","Doxorubicin"~"DOX", "Epirubicin"~"EPI","Mitoxantrone"~"MTX","Trastuzumab"~"TRX","Vehicle"~"VEH", .default = drug))
facs <- c("indv", "drug", "time")
names(facs) <- c("Individual", "Treatment", "Time")
get_regr_pval <- function(mod) {
# Returns the p-value for the Fstatistic of a linear model
# mod: class lm
stopifnot(class(mod) == "lm")
fstat <- summary(mod)$fstatistic
pval <- 1 - pf(fstat[1], fstat[2], fstat[3])
return(pval)
}
plot_versus_pc <- function(df, pc_num, fac) {
# df: data.frame
# pc_num: numeric, specific PC for plotting
# fac: column name of df for plotting against PC
pc_char <- paste0("PC", pc_num)
# Calculate F-statistic p-value for linear model
pval <- get_regr_pval(lm(df[, pc_char] ~ df[, fac]))
if (is.numeric(df[, f])) {
ggplot(df, aes_string(x = f, y = pc_char)) + geom_point() +
geom_smooth(method = "lm") + labs(title = sprintf("p-val: %.2f", pval))
} else {
ggplot(df, aes_string(x = f, y = pc_char)) + geom_boxplot() +
labs(title = sprintf("p-val: %.2f", pval))
}
}
for (f in facs) {
# Plot f versus PC1 and PC2
f_v_pc1 <- arrangeGrob(plot_versus_pc(pca_all_anno, 1, f)+theme_bw())
f_v_pc2 <- arrangeGrob(plot_versus_pc(pca_all_anno, 2, f)+theme_bw())
grid.arrange(f_v_pc1, f_v_pc2, ncol = 2, top = names(facs)[which(facs == f)])
}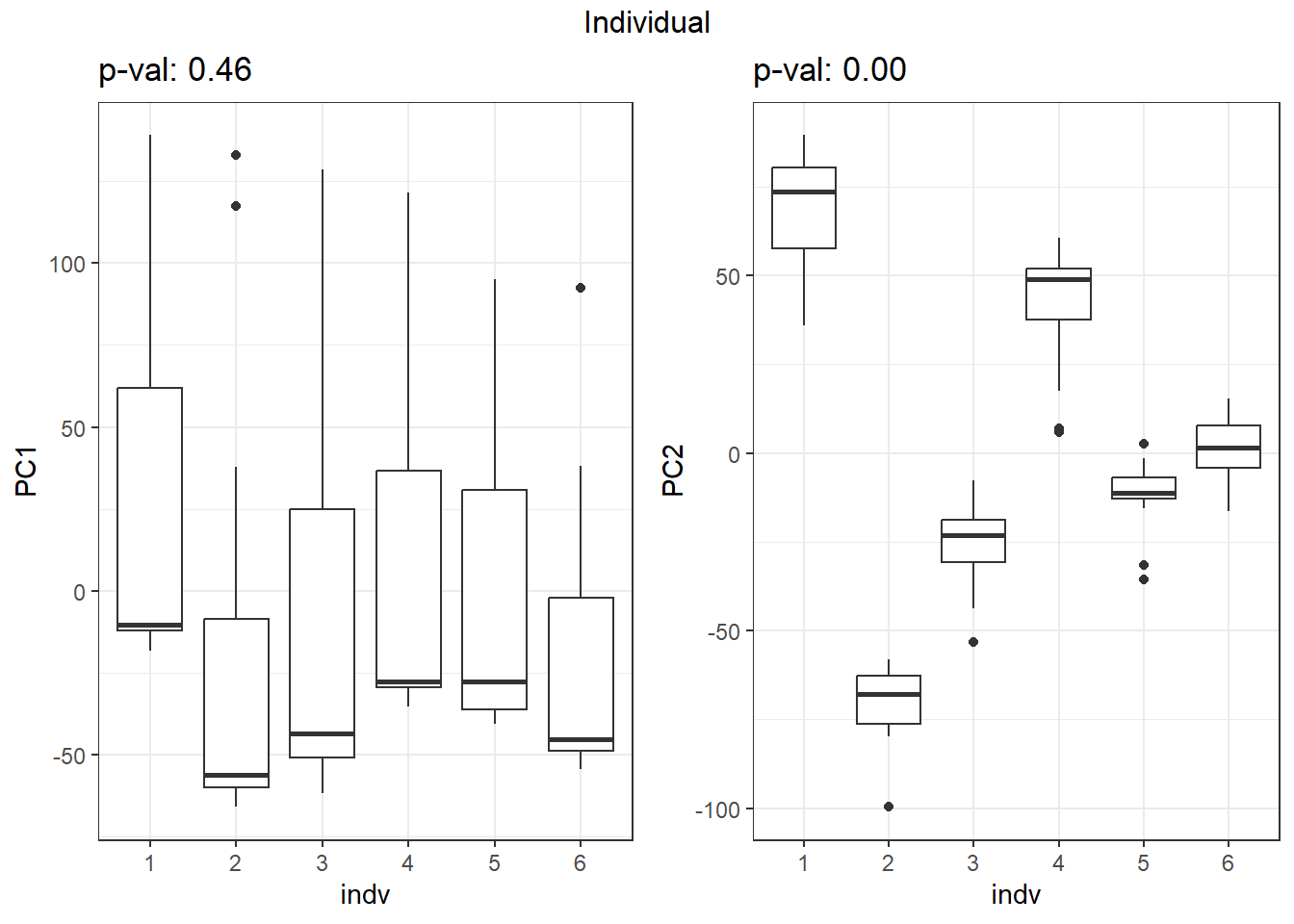
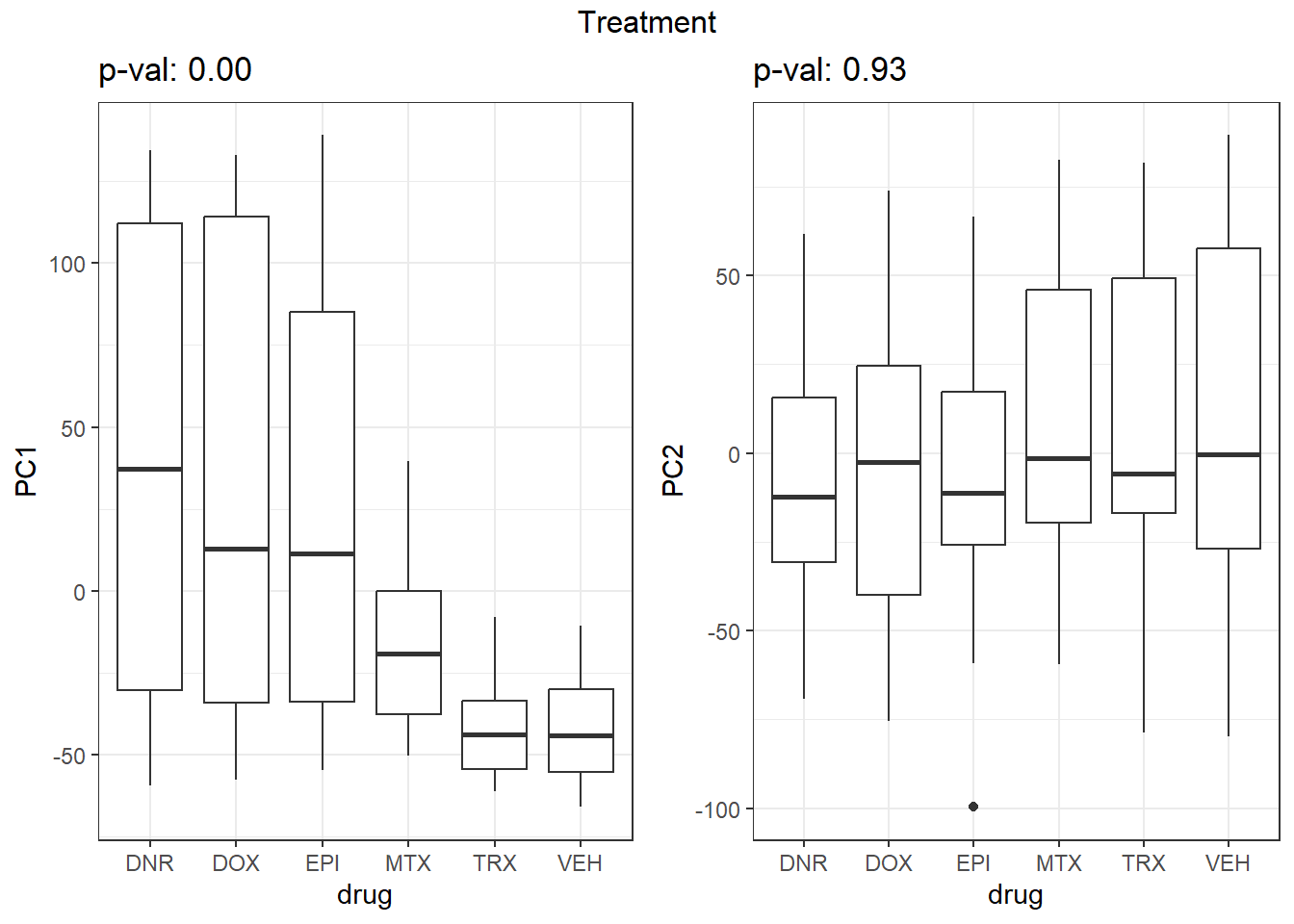
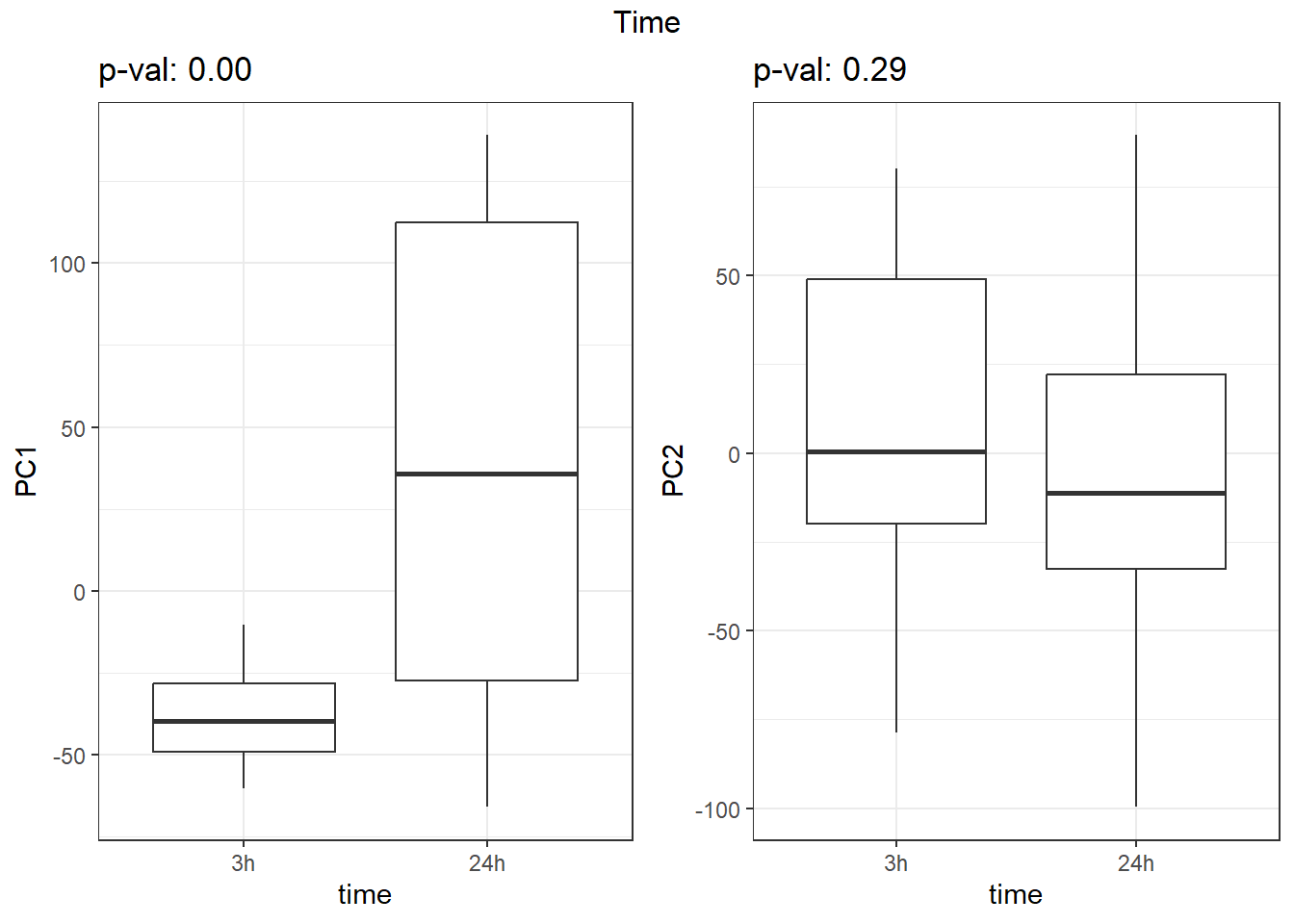
Fig. S8
Volcanoplots <- readRDS("output/Volcanoplot_10.RDS")
Volcanoplots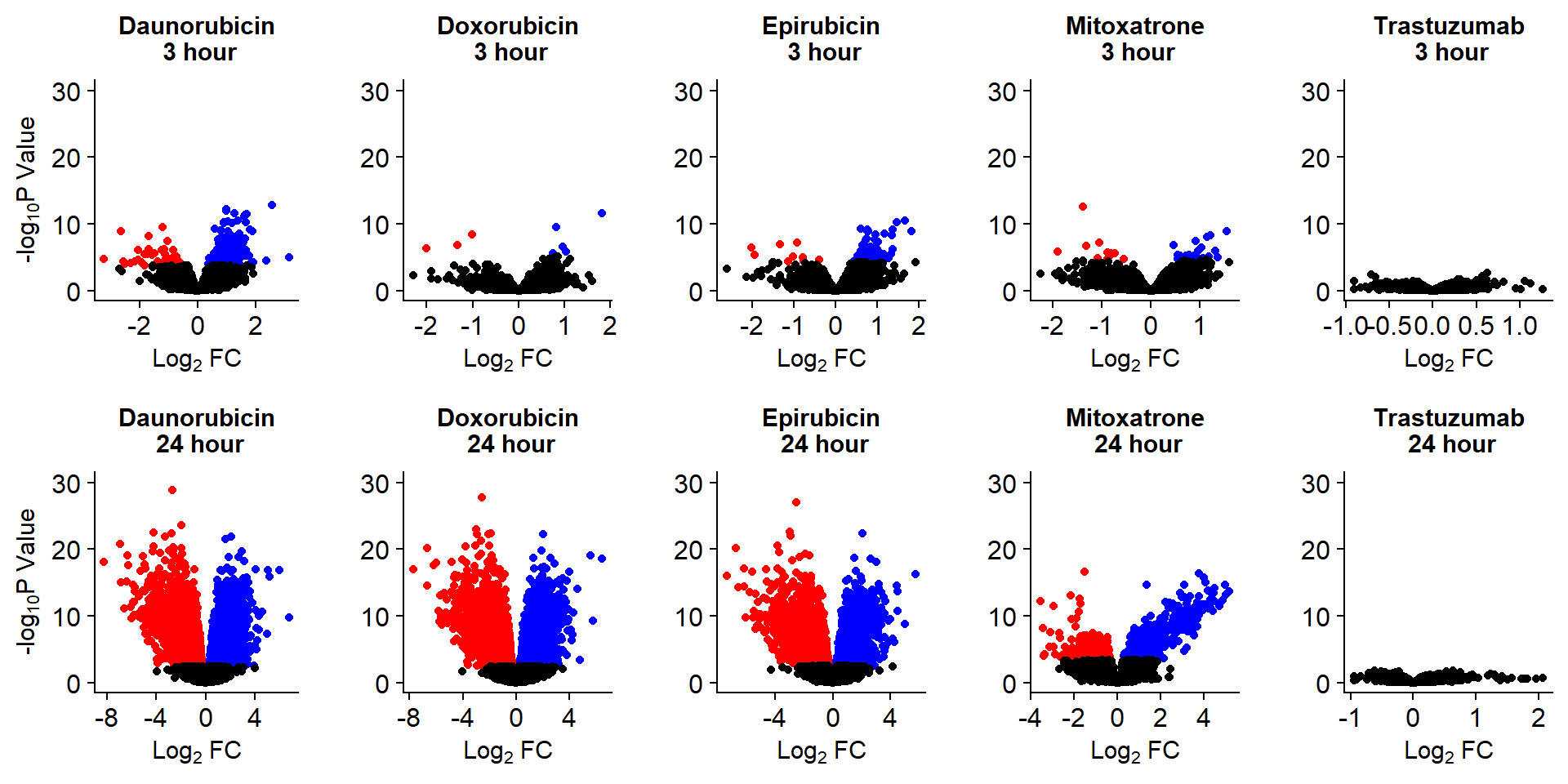
Fig. S9
toplistall <- readRDS("data/toplistall.RDS")
toplistall %>%
group_by(time, id) %>%
mutate(id=factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ','VEH'))) %>%
mutate(time= factor(time,
levels=c("3_hours","24_hours"),
labels=c("3 hours","24 hours"))) %>%
ggplot(., aes(x=id, y=logFC))+
geom_boxplot(aes(fill=id))+
ggpubr::fill_palette(palette =drug_pal_fact)+
guides(fill=guide_legend(title = "Treatment"))+
# facet_wrap(sigcount~time)+
theme_bw()+
xlab("")+
ylab(expression("Log"[2]*" fold change"))+
theme_bw()+
facet_wrap(~time)+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "transparent"),
axis.text.x = element_blank(),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))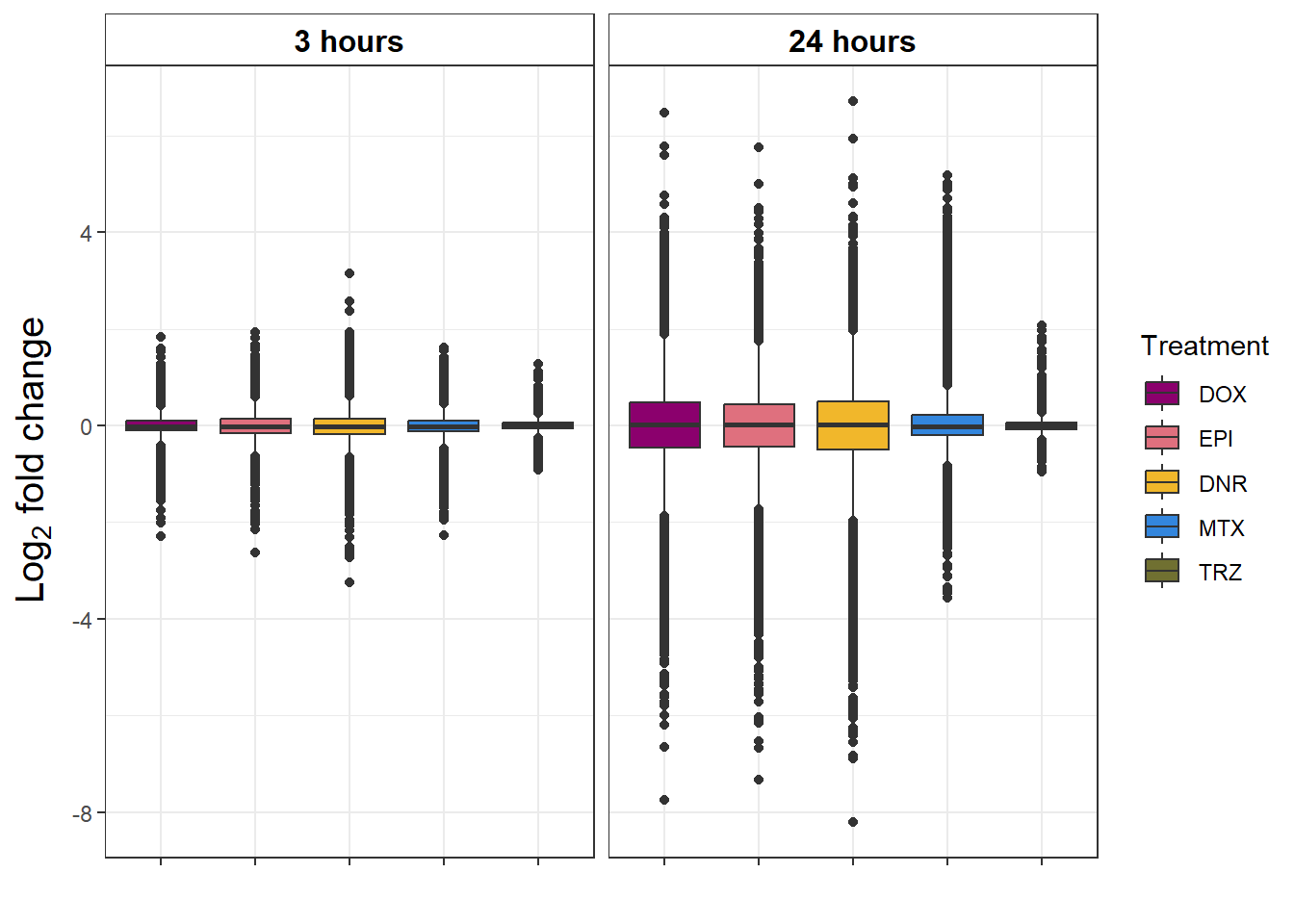
# drug_palNoVeh <- c("#8B006D" ,"#DF707E", "#F1B72B" ,"#3386DD", "#707031")
ggsave("output/Figures/Percent_DEG-1.eps",width = 6, height =4, units = "in")
toplistall %>%
mutate(id=factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ','VEH'))) %>%
mutate(time= factor(time,
levels=c("3_hours","24_hours"),
labels=c("3 hours","24 hours"))) %>%
group_by(time, id) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05,'sig','notsig'))%>%
count(sigcount) %>%
pivot_wider(id_cols = c(time,id), names_from=sigcount, values_from=n) %>%
mutate(prop = sig/(sig+notsig)*100) %>%
mutate(prop=if_else(is.na(prop),0,prop)) %>%
ggplot(., aes(x=id, y= prop))+
geom_col(aes(fill=id))+
geom_text(aes(label = sprintf("%.2f",prop)),
position=position_dodge(0.9),vjust=-.2 )+
scale_fill_manual(values =drug_pal_fact)+
guides(fill=guide_legend(title = "Treatment"))+
facet_wrap(~time)+#labeller = (time = facettimelabel) )+
theme_bw()+
xlab("")+
ylab("Percentage of expressed genes")+
theme_bw()+
ggtitle("Percent DEGs (adj. P value <0.05)")+
scale_y_continuous(expand=expansion(c(0.02,.2)))+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "transparent"),
axis.text.x = element_text(size = 8, color = "white", angle = 0),
axis.text.y = element_text(size = 8, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))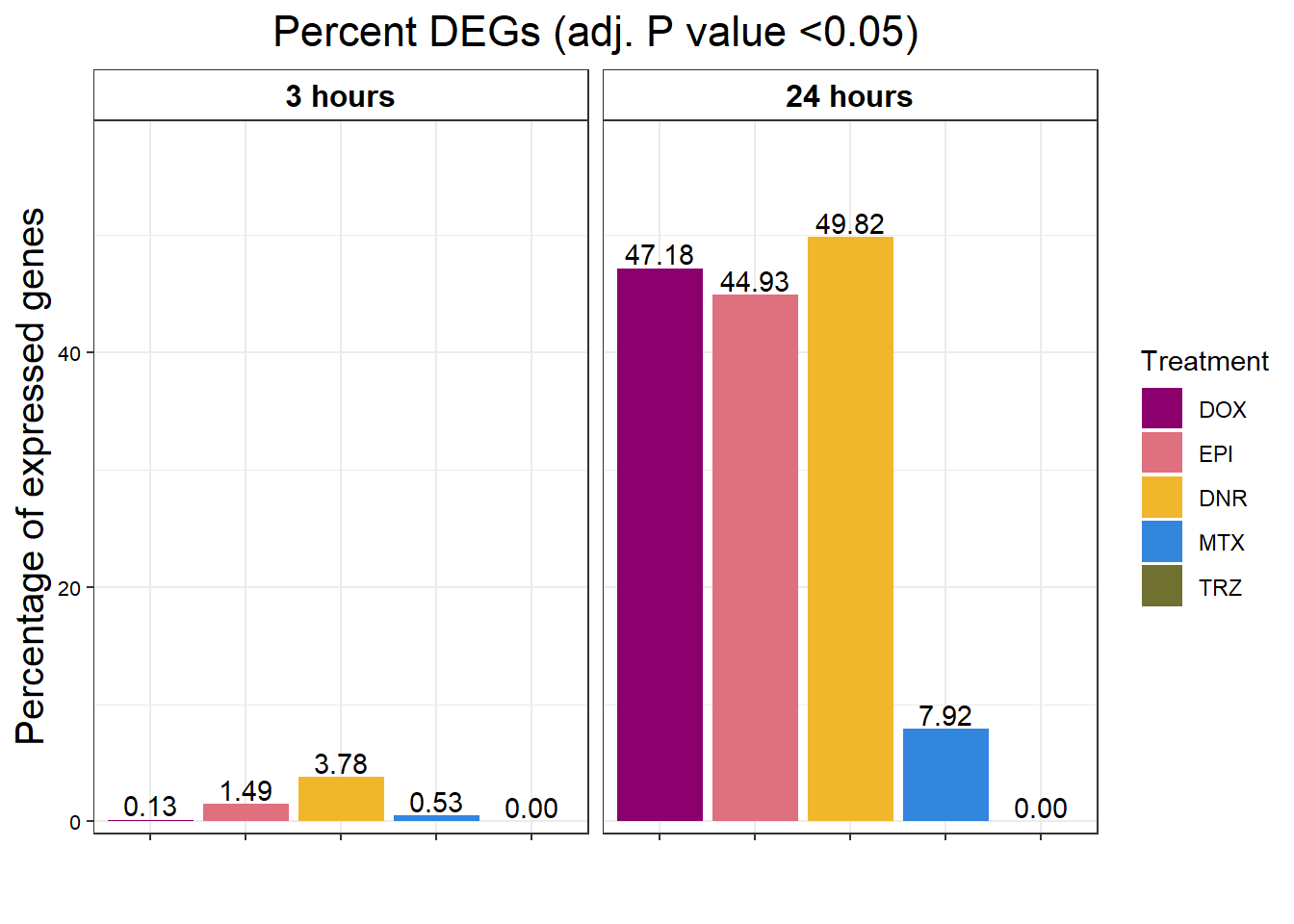
ggsave("output/Figures/Percent_DEG-2.eps",width = 6, height =4, units = "in")Fig. S10
Fig S10 Drug Specific Pathways
gostres3Dnrdeg_sp <- readRDS("data/DEG-GO/gostres3Dnrdeg_sp.RDS")
Dnr3_sp_DEGtable <- gostres3Dnrdeg_sp$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
Dnr3_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels =scales::label_wrap(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('DNR 3 hour specific(stringent)\n gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))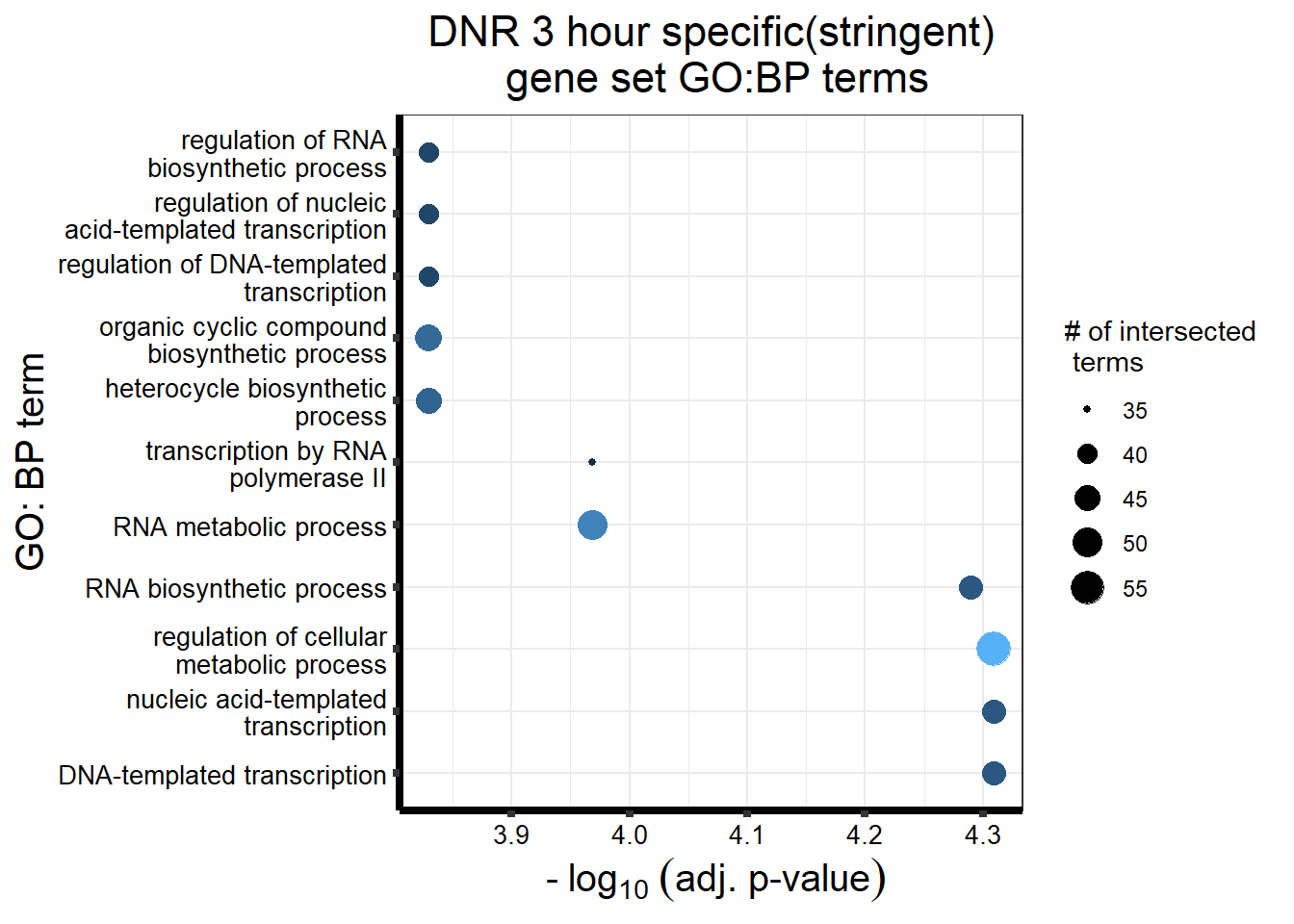
gostres3Mtxdeg_sp <- readRDS("data/DEG-GO/gostres3Mtxdeg_sp.RDS")
Mtx3_sp_DEGtable <- gostres3Mtxdeg_sp$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
Mtx3_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=5,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30))+
geom_vline(xintercept = (-log10(0.05)))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('MTX 3 hour specific(stringent)\n gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))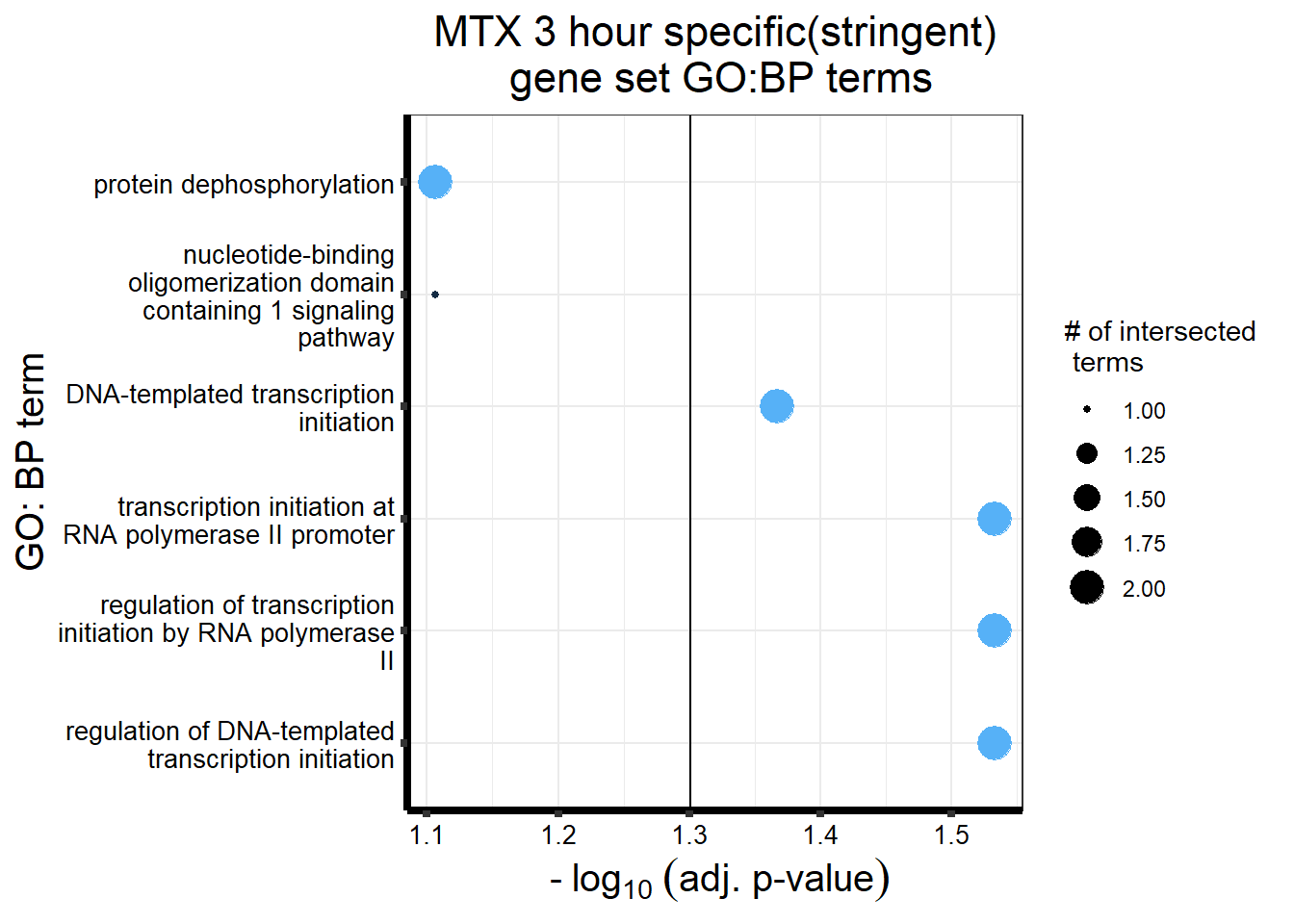
DX_sp_DEGgostres <- readRDS("data/DEG-GO/gostresDOXdeg_sp.RDS")
MT_sp_DEGgostres <- readRDS("data/DEG-GO/gostresMTXdeg_sp.RDS")
DX_sp_DEGtable <- DX_sp_DEGgostres$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
MT_sp_DEGtable <- MT_sp_DEGgostres$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
DX_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::wrap_format(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('DOX specific 24 hour gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))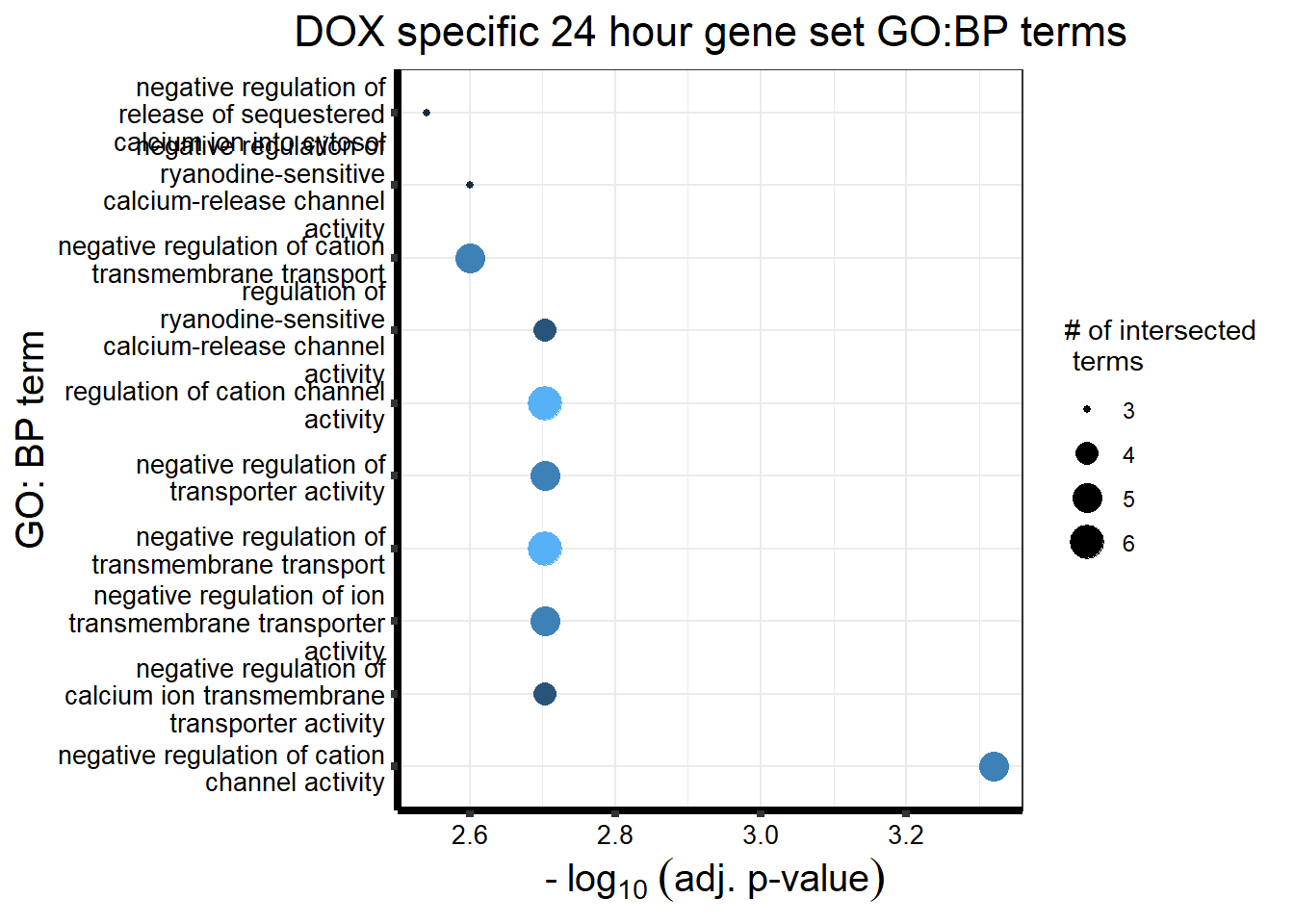
MT_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::wrap_format(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('MTX specific 24 hour gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))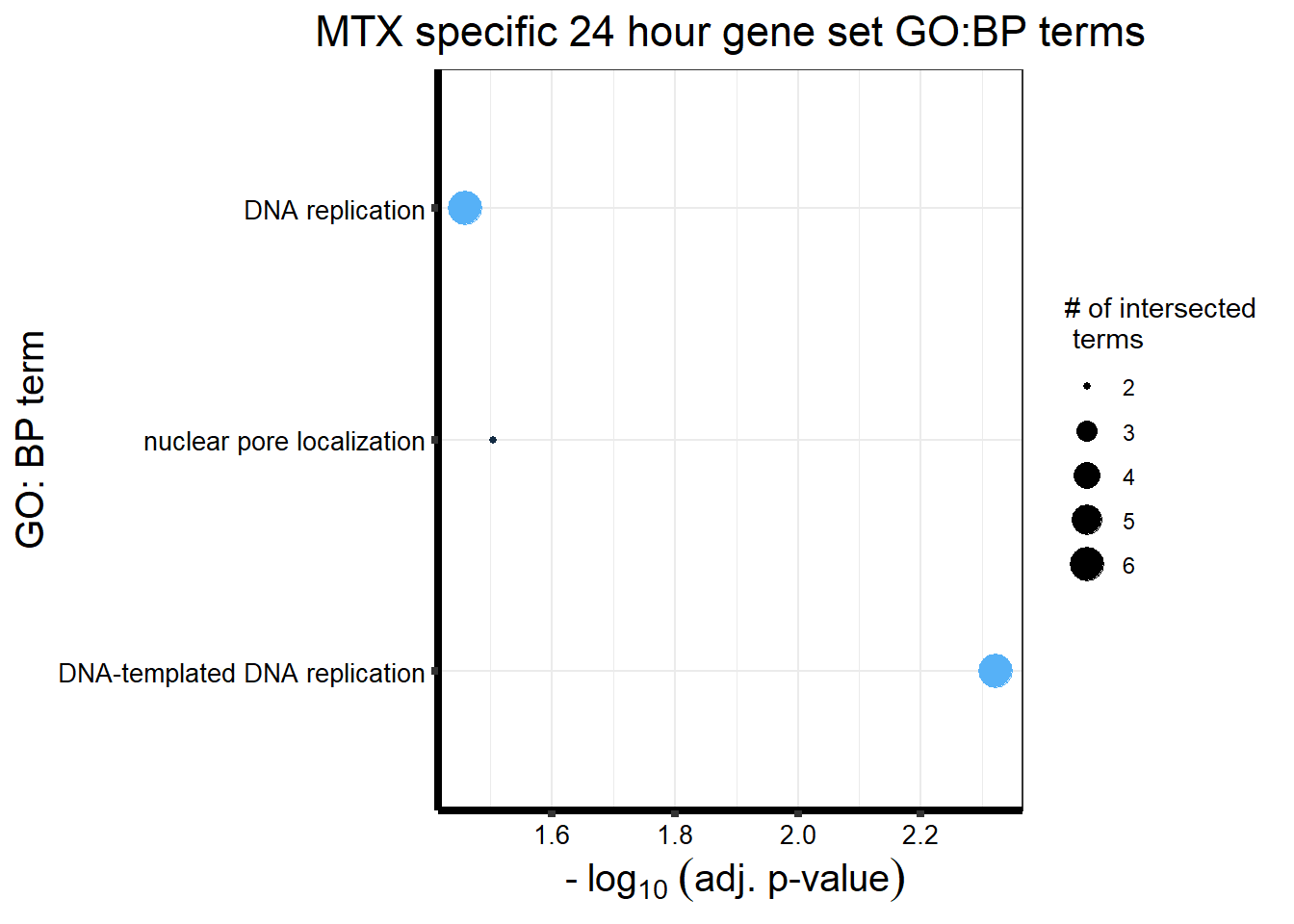
Fig. S12
DNRvenn<- readRDS ("output/DNRvenn.RDS")
DOXvenn<- readRDS ("output/DOXvenn.RDS")
EPIvenn<- readRDS ("output/EPIvenn.RDS")
MTXvenn<- readRDS ("output/MTXvenn.RDS")
plot_grid(DNRvenn,DOXvenn,EPIvenn,MTXvenn,nrow=2, ncol = 2)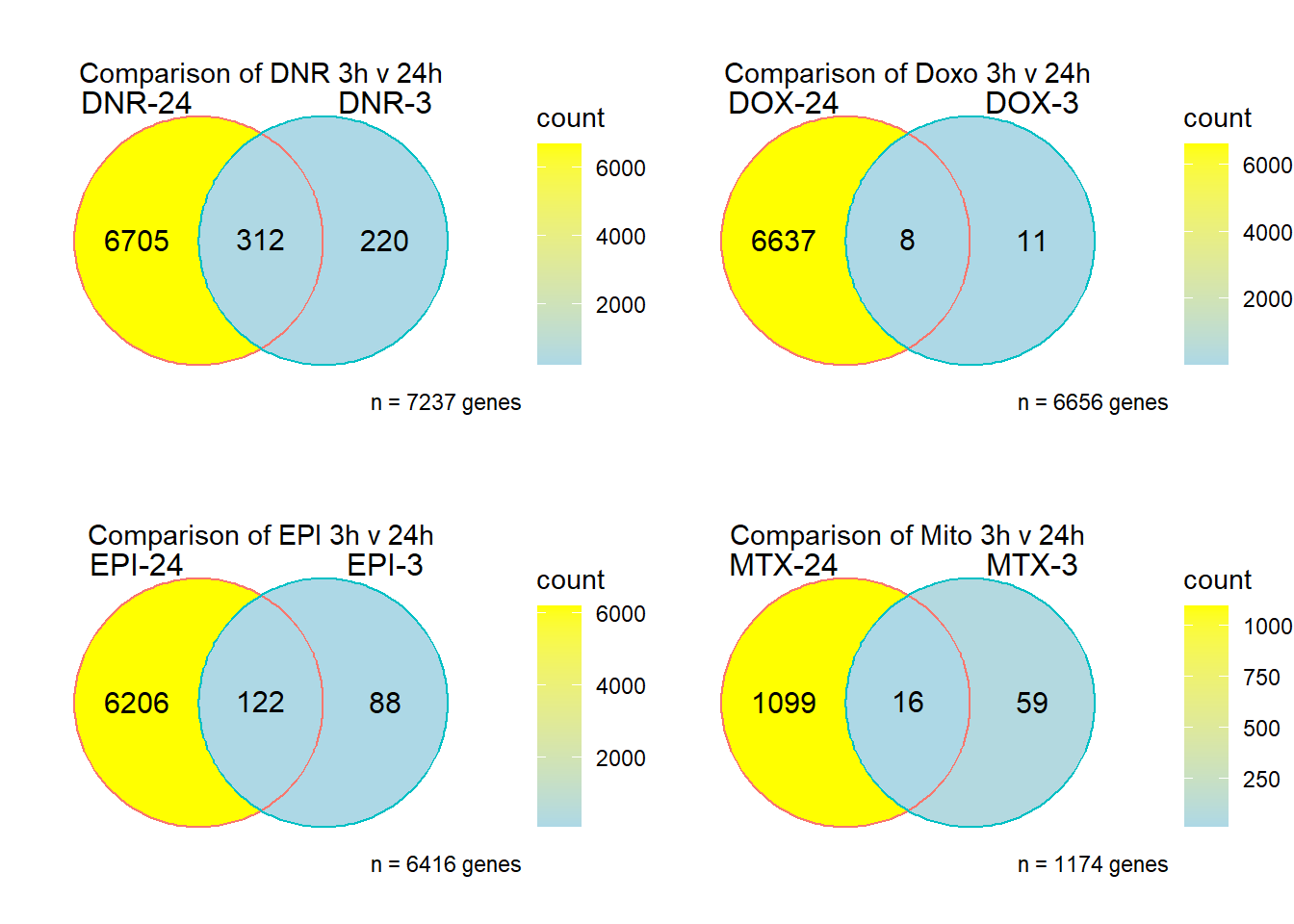
Fig. S12
motif_NRrep <- readRDS("output/motif_NRrep.RDS")
motif_ERrep <- readRDS("output/motif_ERrep.RDS")
motif_TIrep <- readRDS("output/motif_TI_rep.RDS")
motif_LRrep <- readRDS("output/motif_LRrep.RDS")
motif_NRrep <- motif_NRrep +
scale_y_continuous(labels = scales::number_format(accuracy = 0.1))+
theme(legend.position = "NULL", axis.title.y =element_text(size =12))
motif_ERrep <- motif_ERrep+
scale_y_continuous(labels = scales::number_format(accuracy = 0.1))+
theme(legend.position = "NULL", axis.title.y =element_text(size =12))
motif_TIrep <- motif_TIrep+
scale_y_continuous(labels = scales::number_format(accuracy = 0.1))+
theme(legend.position = "NULL", axis.title.y =element_text(size =12))
motif_LRrep <- motif_LRrep+
scale_y_continuous(labels = scales::number_format(accuracy = 0.1))+
theme(legend.position = "NULL", axis.title.y =element_text(size =12))
plot_grid(motif_ERrep,motif_TIrep,motif_LRrep,motif_NRrep,nrow = 4,ncol = 1)
<environment: R_GlobalEnv>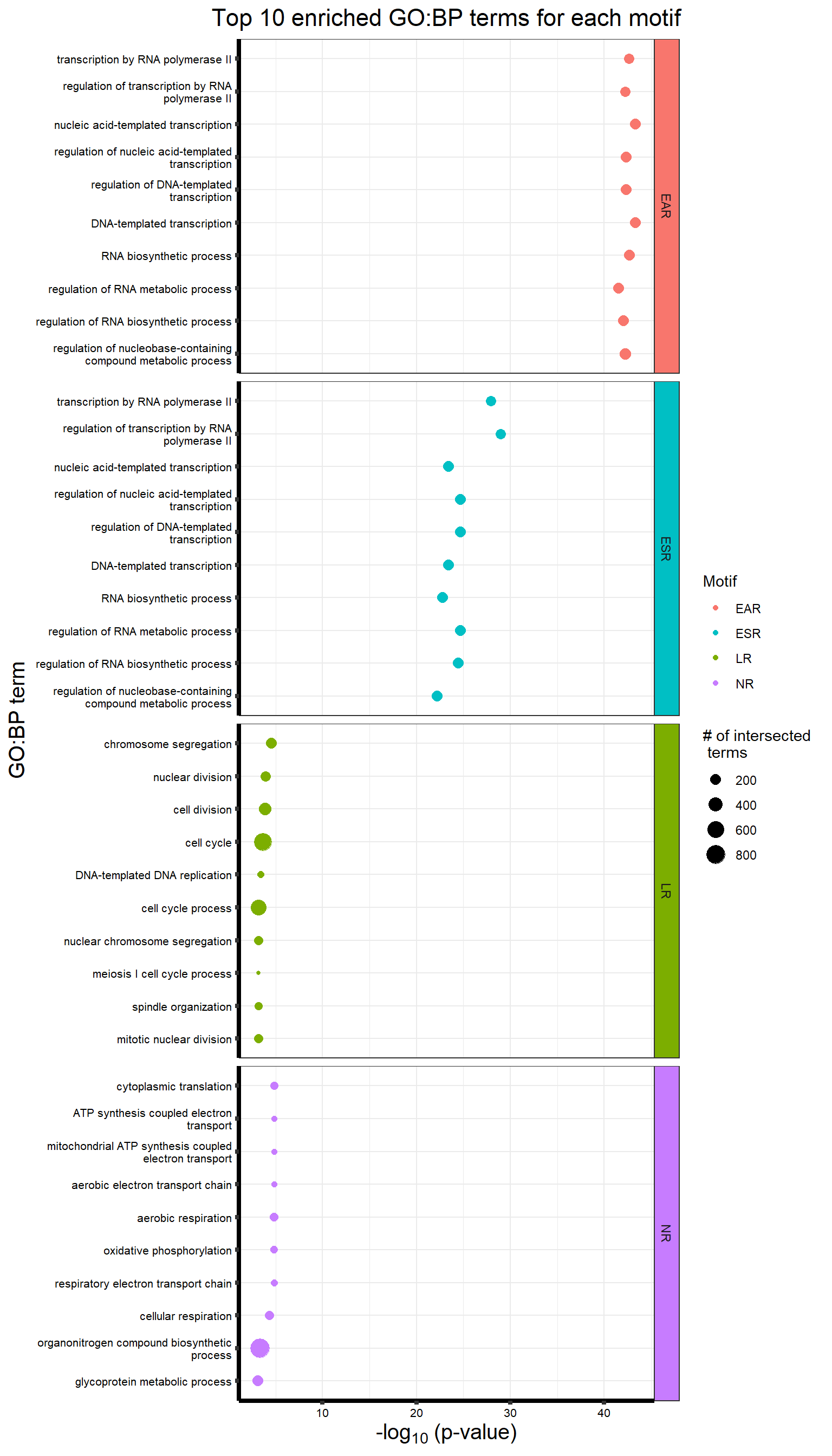
Fig. S15
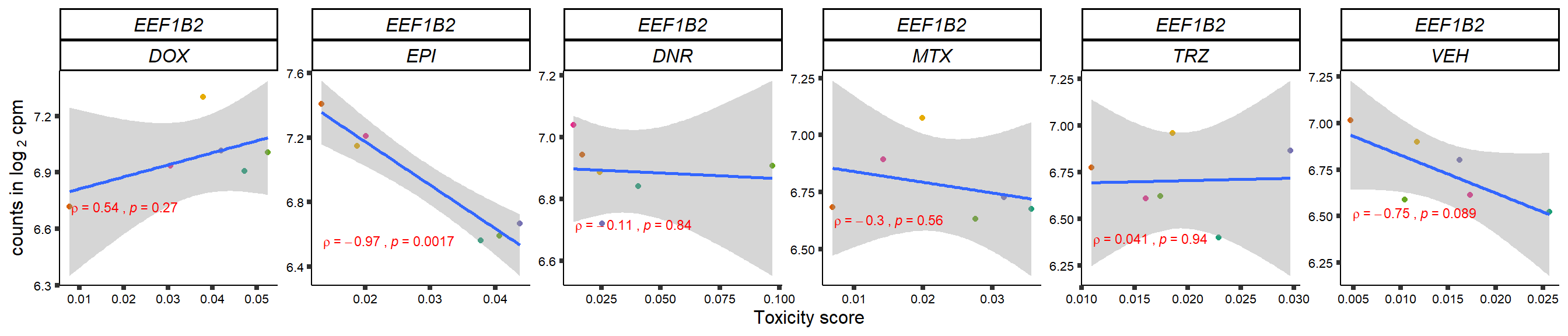
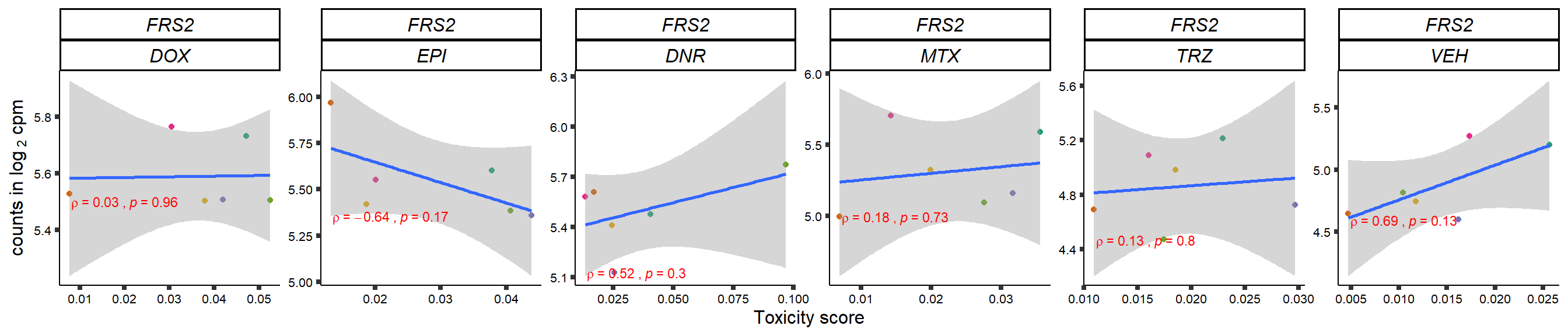
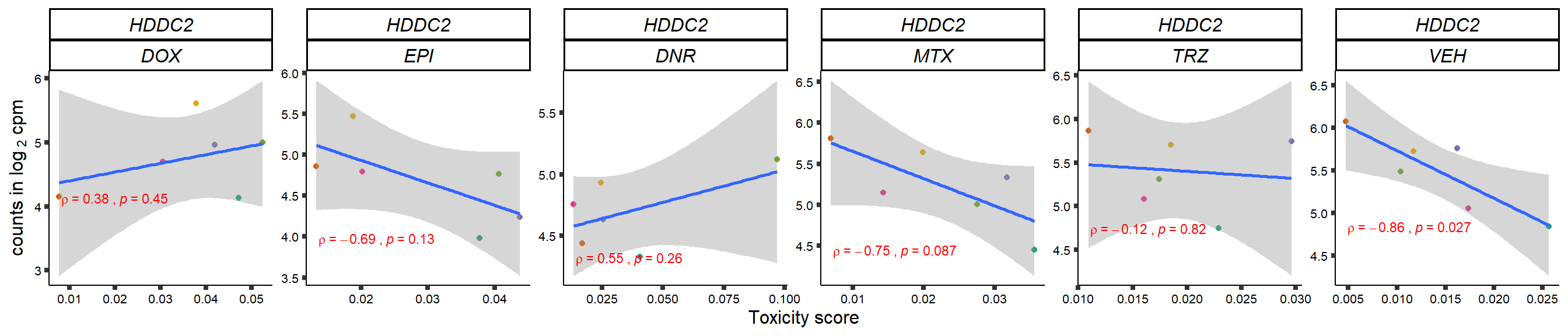
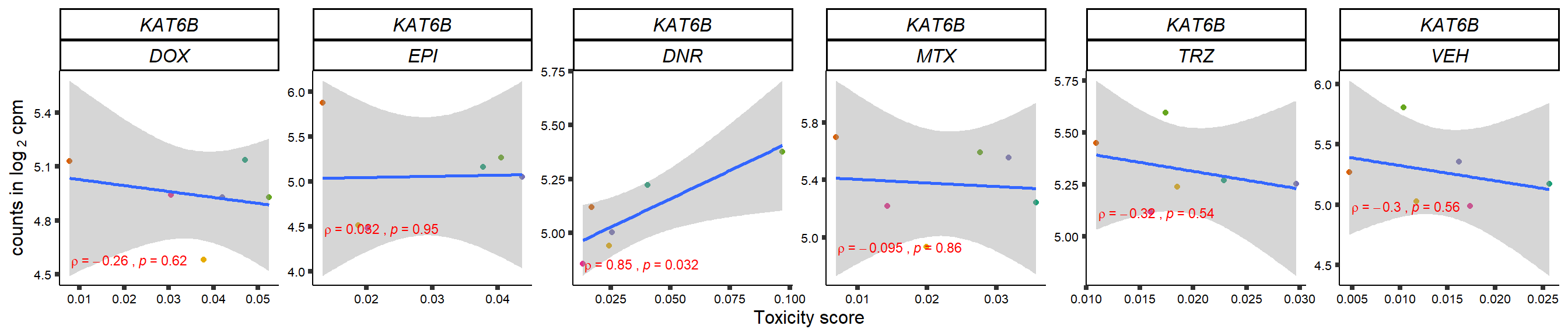
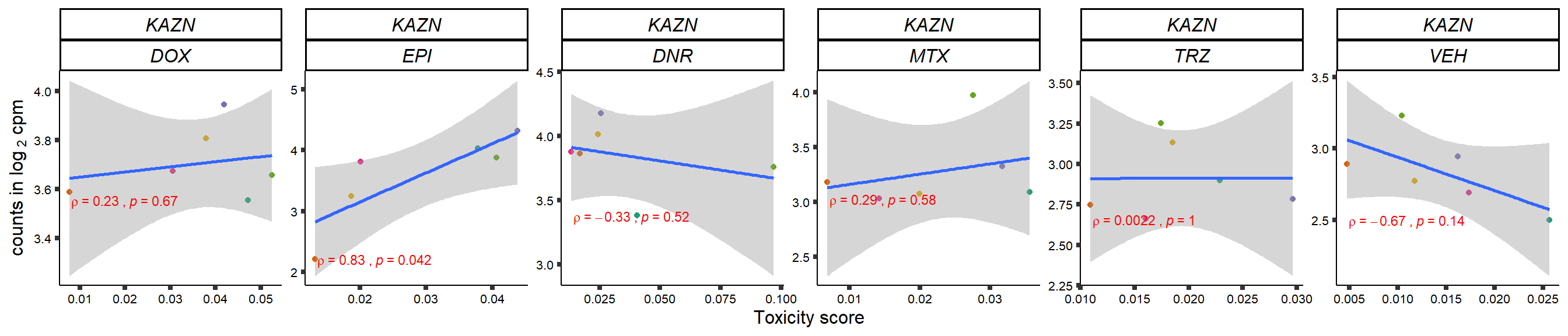
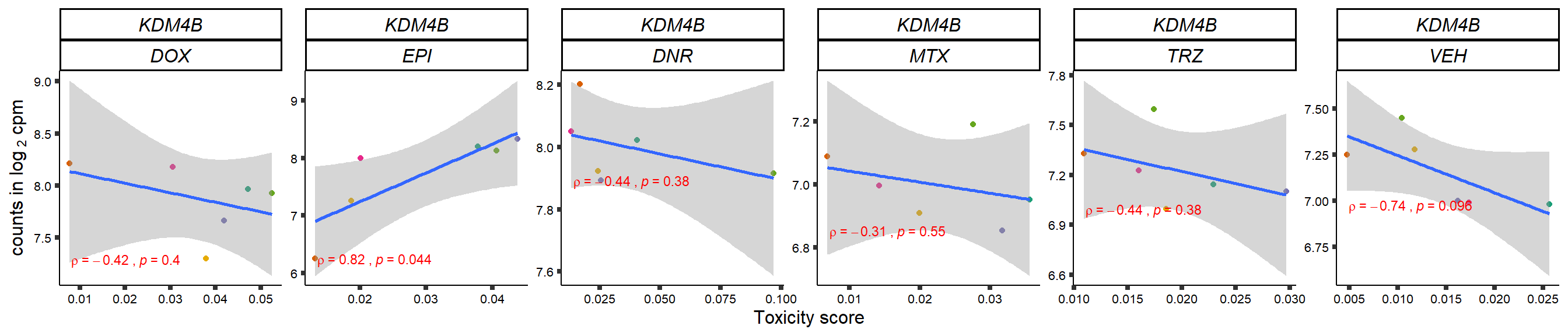
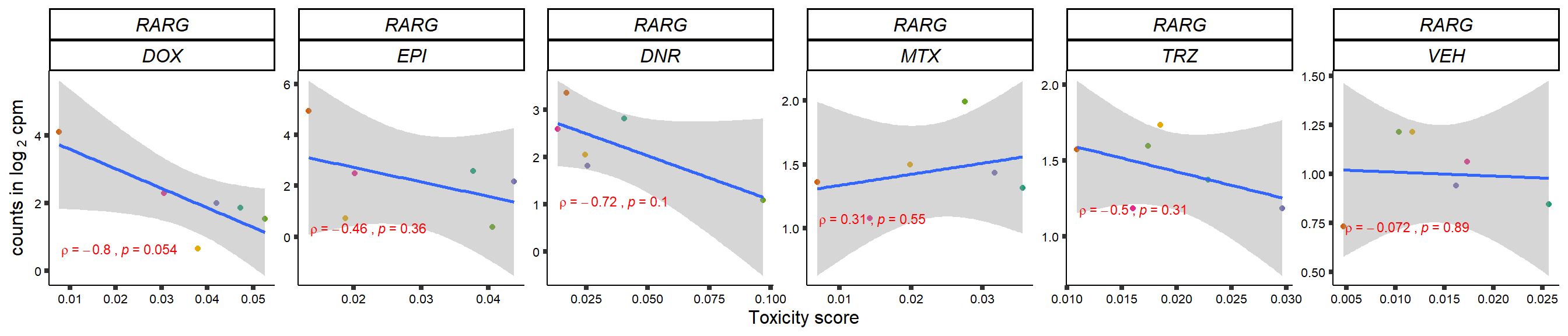
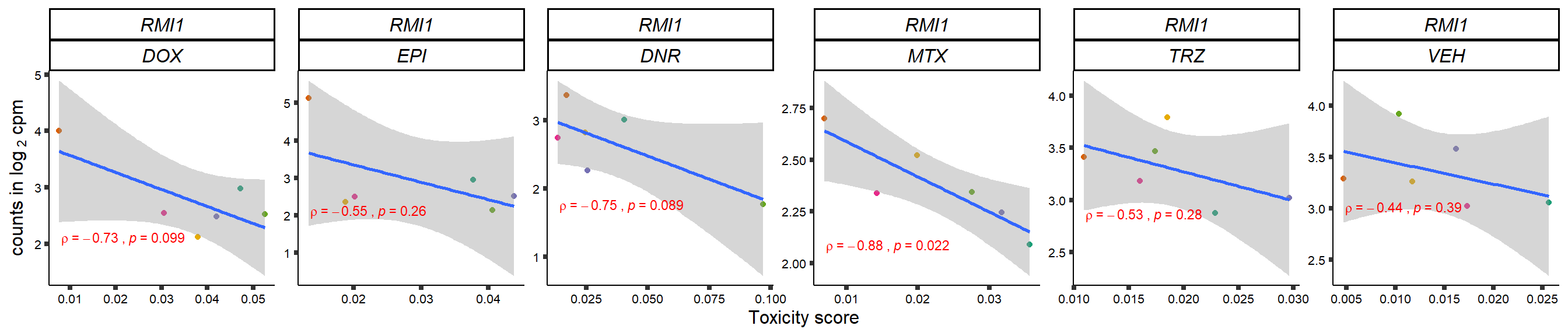
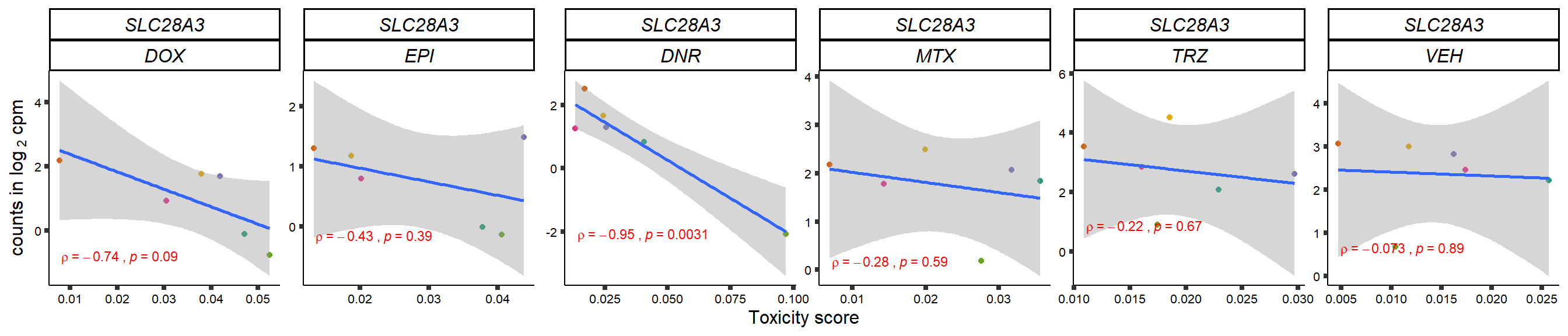
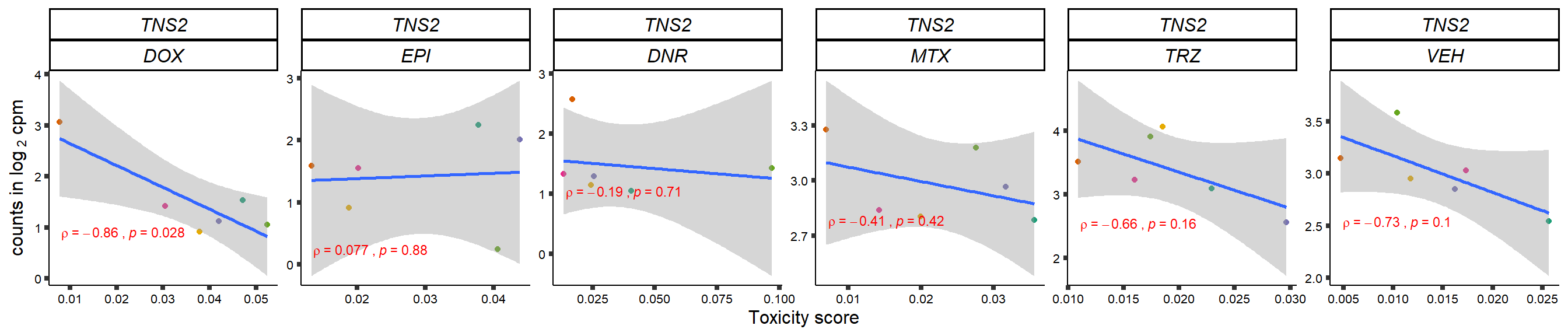
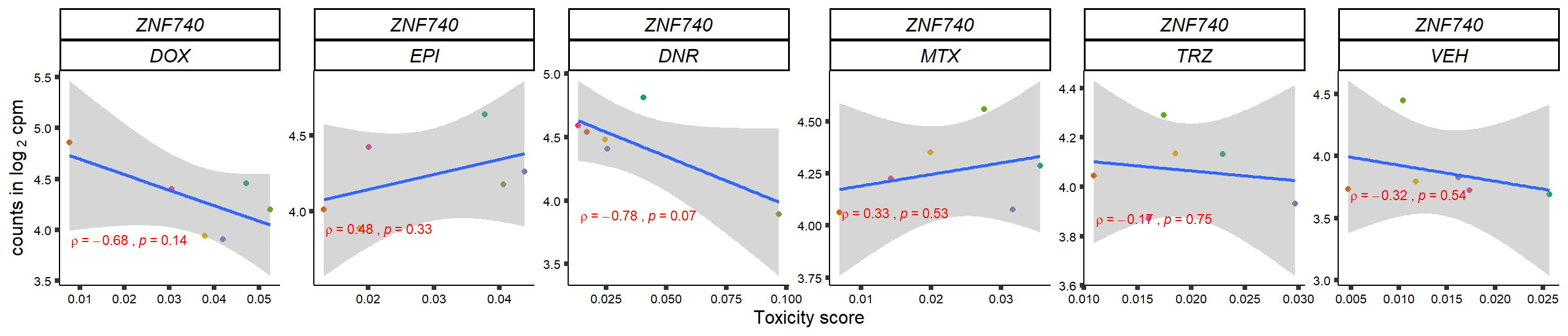
gene_corr_frame <- readRDS("data/gene_corr_frame.RDS")
GOI_genelist <- read.csv("output/GOI_genelist.txt",row.names = 1)
for (gene in GOI_genelist$entrezgene_id){
gene_plot <- gene_corr_frame %>%
dplyr::filter(entrezgene_id == gene) %>%
ggplot(., aes(x=tox, y=counts))+
geom_point(aes(col=indv))+
geom_smooth(method="lm")+
# scale_y_continuous(labels = scales::number_format(accuracy = 0.01))+
facet_wrap(hgnc_symbol~Drug, scales="free", nrow = 1)+
theme_classic()+
xlab("Toxicity score") +
ylab(expression("counts in log "[2]*" cpm")) +
# ggtitle(expression(paste("Correlation between counts and toxicity by drug")))+
scale_color_brewer(palette = "Dark2")+#,name = "Individual", label = c("1","2","3","4","5","6"))+
guides(color="none")+
ggpubr::stat_cor(method="pearson",
cor.coef.name="rho",
aes(label = paste(..r.label.., ..p.label.., sep = "~`,`~")),
color = "red",
label.x.npc = 0.01,
label.y.npc=0.01,
size = 3)+
theme(plot.title = element_text(hjust = 0.5,face = "bold"),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle =0),
strip.text.x = element_text(size = 12, color = "black", face = "italic"))
plot(gene_plot)
}
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] edgeR_3.42.4 limma_3.56.2 data.table_1.14.8
[4] ggVennDiagram_1.2.2 broom_1.0.5 drc_3.0-1
[7] MASS_7.3-60 cowplot_1.1.1 gridExtra_2.3
[10] ComplexHeatmap_2.16.0 scales_1.2.1 RColorBrewer_1.1-3
[13] ggsignif_0.6.4 zoo_1.8-12 rstatix_0.7.2
[16] ggpubr_0.6.0 lubridate_1.9.2 forcats_1.0.0
[19] stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1
[22] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[25] ggplot2_3.4.2 tidyverse_2.0.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rstudioapi_0.15.0 jsonlite_1.8.7 shape_1.4.6
[4] magrittr_2.0.3 TH.data_1.1-2 magick_2.7.4
[7] farver_2.1.1 rmarkdown_2.23 GlobalOptions_0.1.2
[10] fs_1.6.3 ragg_1.2.5 vctrs_0.6.3
[13] webshot_0.5.5 htmltools_0.5.5 plotrix_3.8-2
[16] sass_0.4.7 KernSmooth_2.23-22 bslib_0.5.0
[19] sandwich_3.0-2 cachem_1.0.8 whisker_0.4.1
[22] lifecycle_1.0.3 iterators_1.0.14 pkgconfig_2.0.3
[25] Matrix_1.6-0 R6_2.5.1 fastmap_1.1.1
[28] clue_0.3-64 digest_0.6.33 colorspace_2.1-0
[31] S4Vectors_0.38.1 ps_1.7.5 rprojroot_2.0.3
[34] textshaping_0.3.6 labeling_0.4.2 fansi_1.0.4
[37] timechange_0.2.0 httr_1.4.6 abind_1.4-5
[40] mgcv_1.9-0 compiler_4.3.1 proxy_0.4-27
[43] withr_2.5.0 doParallel_1.0.17 backports_1.4.1
[46] carData_3.0-5 DBI_1.1.3 highr_0.10
[49] rjson_0.2.21 classInt_0.4-9 gtools_3.9.4
[52] units_0.8-2 tools_4.3.1 httpuv_1.6.11
[55] glue_1.6.2 callr_3.7.3 nlme_3.1-162
[58] promises_1.2.0.1 sf_1.0-14 getPass_0.2-2
[61] cluster_2.1.4 generics_0.1.3 gtable_0.3.3
[64] tzdb_0.4.0 class_7.3-22 hms_1.1.3
[67] xml2_1.3.5 car_3.1-2 utf8_1.2.3
[70] BiocGenerics_0.46.0 foreach_1.5.2 pillar_1.9.0
[73] later_1.3.1 circlize_0.4.15 splines_4.3.1
[76] lattice_0.21-8 survival_3.5-5 tidyselect_1.2.0
[79] locfit_1.5-9.8 knitr_1.43 git2r_0.32.0
[82] IRanges_2.34.1 svglite_2.1.1 stats4_4.3.1
[85] xfun_0.39 matrixStats_1.0.0 stringi_1.7.12
[88] yaml_2.3.7 evaluate_0.21 codetools_0.2-19
[91] RVenn_1.1.0 cli_3.6.1 systemfonts_1.0.4
[94] munsell_0.5.0 processx_3.8.2 jquerylib_0.1.4
[97] Rcpp_1.0.11 png_0.1-8 parallel_4.3.1
[100] viridisLite_0.4.2 mvtnorm_1.2-2 e1071_1.7-13
[103] crayon_1.5.2 GetoptLong_1.0.5 rlang_1.1.1
[106] rvest_1.0.3 multcomp_1.4-25