Acute Tunicamycin mouse model
Last updated: 2021-02-27
Checks: 7 0
Knit directory: liver-disease-atlas/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201218) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 40203d5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/human-diehl-nafld_cache/
Ignored: analysis/human-hampe13-nash_cache/
Ignored: analysis/human-hampe14-misc_cache/
Ignored: analysis/human-hoang-nafld_cache/
Ignored: analysis/human-ramnath-fibrosis_cache/
Ignored: analysis/meta-chronic-vs-acute_cache/
Ignored: analysis/meta-mouse-vs-human_cache/
Ignored: analysis/mouse-acute-apap_cache/
Ignored: analysis/mouse-acute-bdl_cache/
Ignored: analysis/mouse-acute-ccl4_cache/
Ignored: analysis/mouse-acute-lps_cache/
Ignored: analysis/mouse-acute-ph_cache/
Ignored: analysis/mouse-chronic-ccl4_cache/
Ignored: analysis/plot-acute-apap_cache/
Ignored: analysis/plot-acute-bdl_cache/
Ignored: analysis/plot-acute-ccl4_cache/
Ignored: analysis/plot-acute-ph_cache/
Ignored: analysis/plot-chronic-ccl4_cache/
Ignored: analysis/plot-chronic-vs-acute_cache/
Ignored: analysis/plot-mouse-vs-human_cache/
Ignored: analysis/plot-precision-recall_cache/
Ignored: analysis/plot-study-overview_cache/
Ignored: analysis/plot-teufel-integration_cache/
Ignored: analysis/save-tables_cache/
Ignored: code/.DS_Store
Ignored: code/README.html
Ignored: code/meta-mouse-vs-human/.DS_Store
Ignored: data.zip
Ignored: data/.DS_Store
Ignored: data/annotation/
Ignored: data/human-diehl-nafld/
Ignored: data/human-hampe13-nash/
Ignored: data/human-hampe14-misc/
Ignored: data/human-hoang-nafld/
Ignored: data/human-ramnath-fibrosis/
Ignored: data/meta-chronic-vs-acute/
Ignored: data/meta-mouse-vs-human/
Ignored: data/mouse-acute-apap/
Ignored: data/mouse-acute-bdl/
Ignored: data/mouse-acute-ccl4/
Ignored: data/mouse-acute-lps/
Ignored: data/mouse-acute-ph/
Ignored: data/mouse-acute-tunicamycin/
Ignored: data/mouse-chronic-ccl4/
Ignored: external_software/.DS_Store
Ignored: external_software/README.html
Ignored: external_software/stem/.DS_Store
Ignored: figures/
Ignored: geo_submission/
Ignored: output/.DS_Store
Ignored: output/README.html
Ignored: output/human-diehl-nafld/
Ignored: output/human-hampe13-nash/
Ignored: output/human-hampe14-misc/
Ignored: output/human-hoang-nafld/
Ignored: output/human-ramnath-fibrosis/
Ignored: output/meta-chronic-vs-acute/
Ignored: output/meta-mouse-vs-human/
Ignored: output/mouse-acute-apap/
Ignored: output/mouse-acute-bdl/
Ignored: output/mouse-acute-ccl4/
Ignored: output/mouse-acute-lps/
Ignored: output/mouse-acute-ph/
Ignored: output/mouse-acute-tunicamycin/
Ignored: output/mouse-chronic-ccl4/
Ignored: renv/library/
Ignored: renv/staging/
Ignored: tables/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/mouse-acute-tunicamycin.Rmd) and HTML (docs/mouse-acute-tunicamycin.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 9c62197 | christianholland | 2021-01-07 | Build site. |
| html | 067c933 | christianholland | 2020-12-23 | Build site. |
| Rmd | d4f78fa | christianholland | 2020-12-23 | wflow_publish("analysis/*.Rmd", delete_cache = T) |
| html | 8a9b7bf | christianholland | 2020-12-20 | Build site. |
| html | e22a40b | christianholland | 2020-12-20 | Build site. |
| html | 413cd90 | christianholland | 2020-12-20 | Build site. |
| html | ae8a6f6 | christianholland | 2020-12-19 | Build site. |
| Rmd | 8ab21f2 | christianholland | 2020-12-19 | Added tunicamycin study |
Introduction
Here we analysis a mouse model of Tunicamycin induced acute liver damage.
Libraries and sources
These libraries and sources are used for this analysis.
library(mouse4302.db)
library(tidyverse)
library(tidylog)
library(here)
library(oligo)
library(annotate)
library(limma)
library(biobroom)
library(janitor)
library(AachenColorPalette)
library(cowplot)
library(lemon)
library(patchwork)
options("tidylog.display" = list(print))
source(here("code/utils-microarray.R"))
source(here("code/utils-utils.R"))
source(here("code/utils-plots.R"))Definition of global variables that are used throughout this analysis.
# i/o
data_path <- "data/mouse-acute-tunicamycin"
output_path <- "output/mouse-acute-tunicamycin"
# graphical parameters
# fontsize
fz <- 9Data processing
Load .CEL files and quality control
The array quality is controlled based on the relative log expression values (RLE) and the normalized unscaled standard errors (NUSE).
# load cel files and check quality
platforms <- readRDS(here("data/annotation/platforms.rds"))
raw_eset <- list.celfiles(here(data_path), listGzipped = T, full.names = T) %>%
read.celfiles() %>%
ma_qc()
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740952.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740953.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740954.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740955.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740956.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740957.CEL.gz
#> Reading in : /Users/cholland/Google Drive/Projects/liver-disease-atlas/data/mouse-acute-tunicamycin/GSM740958.CEL.gzNormalization and probe annotation
Probe intensities are normalized with the rma() function. Probes are annotated with MGI symbols.
eset <- rma(raw_eset)
#> Background correcting
#> Normalizing
#> Calculating Expression
# annotate microarray probes with mgi symbols
expr <- ma_annotate(eset, platforms)
colnames(expr) = str_remove(colnames(expr), ".CEL.gz")
# gene "BC001981" has a constant value across all samples and must be thus removed
constant_genes = which(apply(expr, 1, var) == 0)
expr = expr[-constant_genes,]
# save normalized expression
saveRDS(expr, here(output_path, "normalized_expression.rds"))Build meta data
Meta information are parsed from the sample names.
# build meta data
meta = colnames(expr) %>%
enframe(name = NULL, value = "sample") %>%
mutate(group = c(rep("treated", 4), rep("control", 3))) %>%
mutate(group = factor(group, levels = c("control", "treated")))
#> mutate: new variable 'group' (character) with 2 unique values and 0% NA
#> mutate: converted 'group' from character to factor (0 new NA)
# save meta data
saveRDS(meta, here(output_path, "meta_data.rds"))Exploratory analysis
PCA of normalized data
PCA plot of normalized expression data contextualized based on the treatment. Only the top 1000 most variable genes are used as features.
expr <- readRDS(here(output_path, "normalized_expression.rds"))
meta <- readRDS(here(output_path, "meta_data.rds"))
pca_result <- do_pca(expr, meta, top_n_var_genes = 1000)
#> left_join: added one column (group)
#> > rows only in x 0
#> > rows only in y (0)
#> > matched rows 7
#> > ===
#> > rows total 7
saveRDS(pca_result, here(output_path, "pca_result.rds"))
plot_pca(pca_result, feature = "group") +
my_theme()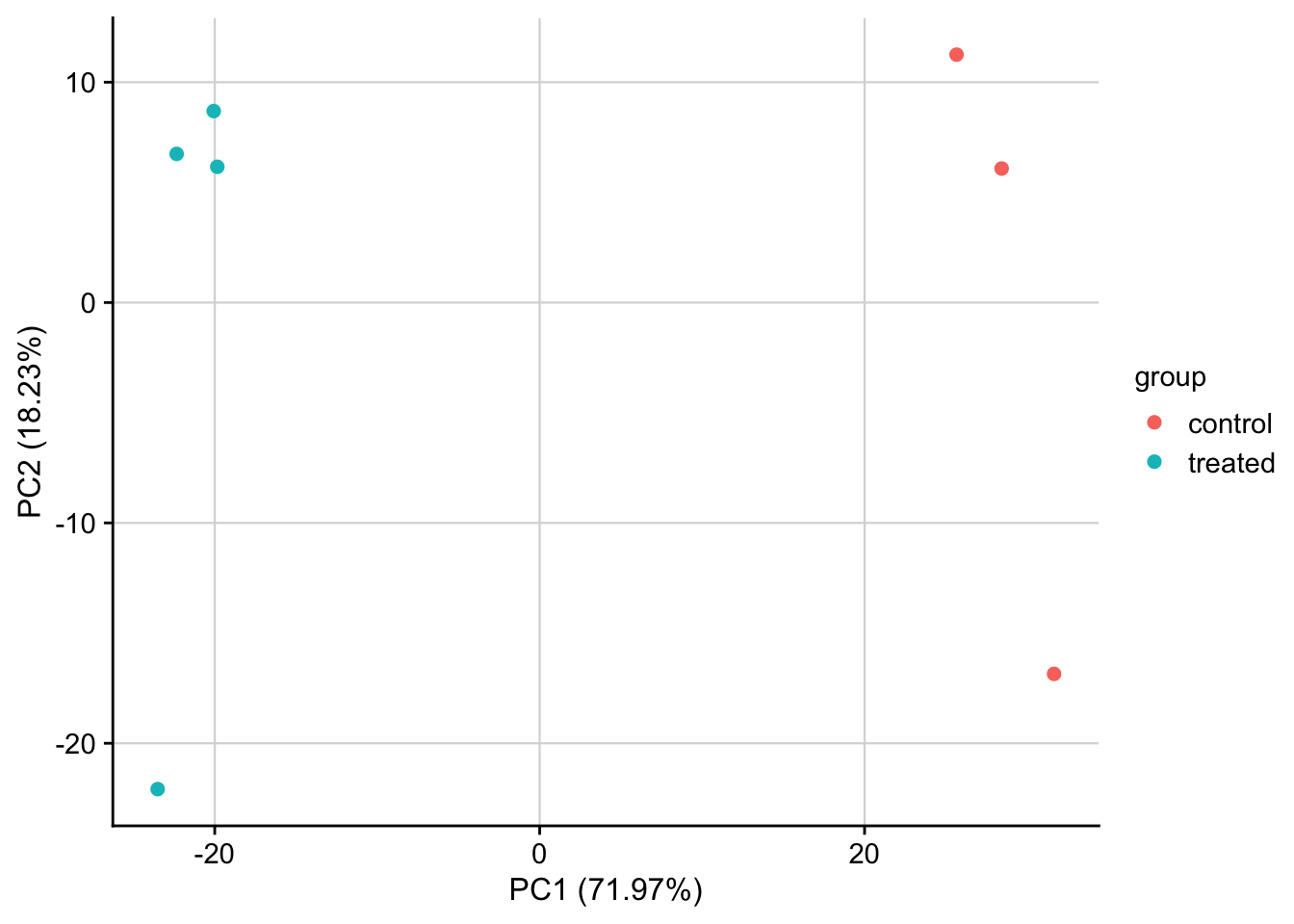
Differential gene expression analysis
Running limma
Differential gene expression analysis via limma with the aim to identify the effect of Tunicamycin intoxication.
# load expression and meta data
expr <- readRDS(here(output_path, "normalized_expression.rds"))
meta <- readRDS(here(output_path, "meta_data.rds"))
stopifnot(colnames(expr) == meta$sample)
# build design matrix
design = model.matrix(~0+group, data=meta)
rownames(design) = meta$sample
colnames(design) = levels(meta$group)
# define contrasts
contrasts = makeContrasts(
treat_vs_ctrl = treated - control,
levels = design
)
limma_result = run_limma(expr, design, contrasts) %>%
assign_deg()
#> select: renamed 3 variables (contrast, logFC, pval) and dropped one variable
#> group_by: one grouping variable (contrast)
#> mutate (grouped): new variable 'fdr' (double) with 13,527 unique values and 0% NA
#> ungroup: no grouping variables
#> mutate: new variable 'regulation' (character) with 3 unique values and 0% NA
#> mutate: converted 'regulation' from character to factor (0 new NA)
saveRDS(limma_result, here(output_path, "limma_result.rds"))Volcano plots
Volcano plots visualizing the effect of Tunicamycin on gene expression.
df <- readRDS(here(output_path, "limma_result.rds"))
df %>%
plot_volcano() +
my_theme(grid = "y", fsize = fz)
#> rename: renamed one variable (p)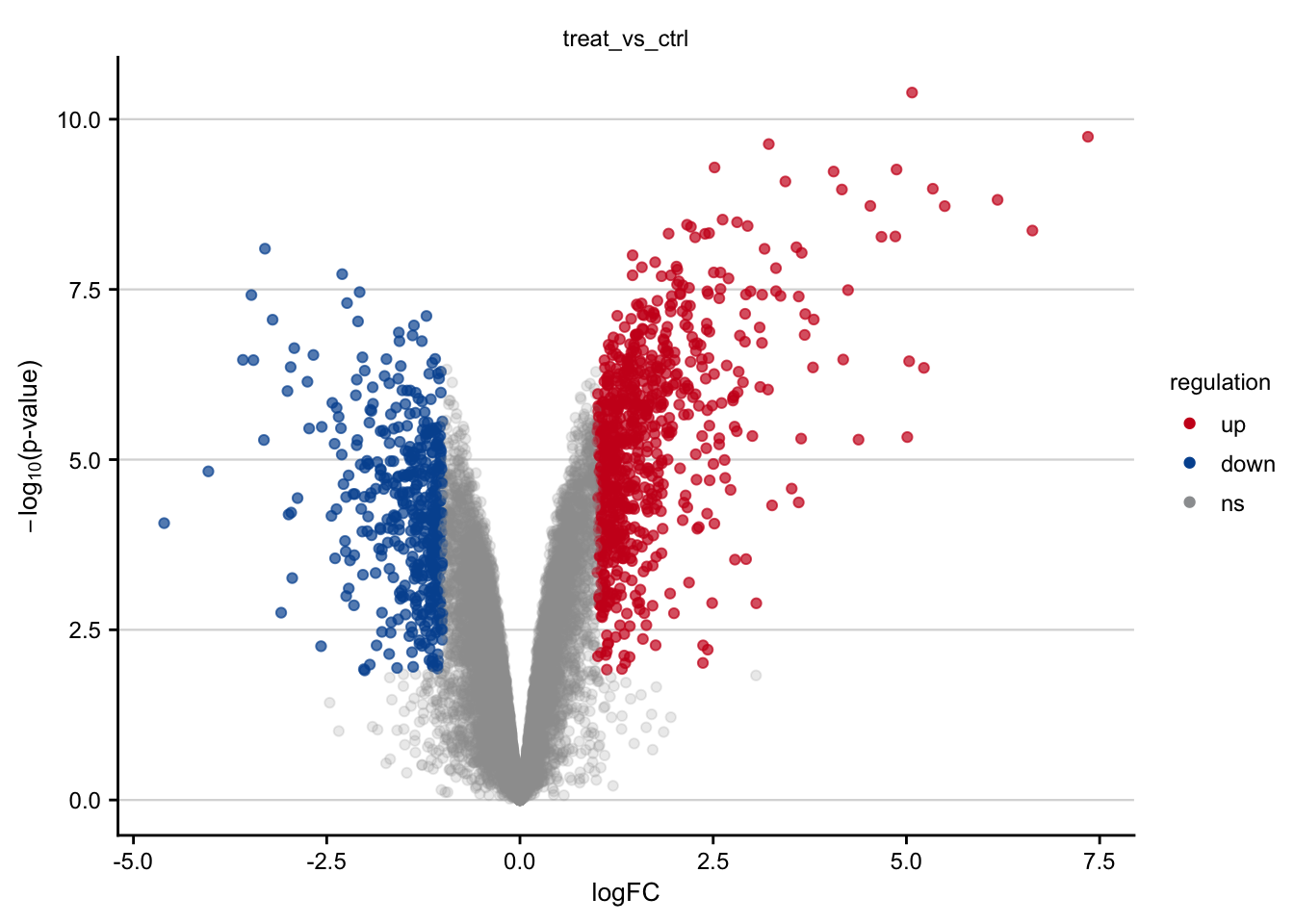
Time spend to execute this analysis: 00:38 minutes.
sessionInfo()
#> R version 4.0.2 (2020-06-22)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Mojave 10.14.5
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] parallel stats4 stats graphics grDevices datasets utils
#> [8] methods base
#>
#> other attached packages:
#> [1] pd.mouse430.2_3.12.0 DBI_1.1.0 RSQLite_2.2.1
#> [4] patchwork_1.1.1 lemon_0.4.5 cowplot_1.1.0
#> [7] AachenColorPalette_1.1.2 janitor_2.0.1 biobroom_1.20.0
#> [10] broom_0.7.3 limma_3.44.3 annotate_1.66.0
#> [13] XML_3.99-0.5 oligo_1.52.1 Biostrings_2.56.0
#> [16] XVector_0.28.0 oligoClasses_1.50.4 here_1.0.1
#> [19] tidylog_1.0.2 forcats_0.5.0 stringr_1.4.0
#> [22] dplyr_1.0.2 purrr_0.3.4 readr_1.4.0
#> [25] tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.2
#> [28] tidyverse_1.3.0 mouse4302.db_3.2.3 org.Mm.eg.db_3.11.4
#> [31] AnnotationDbi_1.50.3 IRanges_2.22.2 S4Vectors_0.26.1
#> [34] Biobase_2.48.0 BiocGenerics_0.34.0 workflowr_1.6.2
#>
#> loaded via a namespace (and not attached):
#> [1] colorspace_2.0-0 ellipsis_0.3.1
#> [3] rprojroot_2.0.2 snakecase_0.11.0
#> [5] GenomicRanges_1.40.0 fs_1.5.0
#> [7] rstudioapi_0.13 farver_2.0.3
#> [9] affyio_1.58.0 bit64_4.0.5
#> [11] fansi_0.4.1 lubridate_1.7.9.2
#> [13] xml2_1.3.2 codetools_0.2-16
#> [15] splines_4.0.2 knitr_1.30
#> [17] jsonlite_1.7.2 dbplyr_2.0.0
#> [19] BiocManager_1.30.10 compiler_4.0.2
#> [21] httr_1.4.2 backports_1.2.1
#> [23] assertthat_0.2.1 Matrix_1.2-18
#> [25] cli_2.2.0 later_1.1.0.1
#> [27] htmltools_0.5.0 tools_4.0.2
#> [29] gtable_0.3.0 glue_1.4.2
#> [31] GenomeInfoDbData_1.2.3 affxparser_1.60.0
#> [33] Rcpp_1.0.5 cellranger_1.1.0
#> [35] vctrs_0.3.6 preprocessCore_1.50.0
#> [37] iterators_1.0.13 xfun_0.19
#> [39] rvest_0.3.6 lifecycle_0.2.0
#> [41] renv_0.12.3 zlibbioc_1.34.0
#> [43] scales_1.1.1 clisymbols_1.2.0
#> [45] hms_0.5.3 promises_1.1.1
#> [47] SummarizedExperiment_1.18.2 yaml_2.2.1
#> [49] gridExtra_2.3 memoise_1.1.0
#> [51] stringi_1.5.3 foreach_1.5.1
#> [53] GenomeInfoDb_1.24.2 rlang_0.4.9
#> [55] pkgconfig_2.0.3 bitops_1.0-6
#> [57] matrixStats_0.57.0 evaluate_0.14
#> [59] lattice_0.20-41 labeling_0.4.2
#> [61] bit_4.0.4 tidyselect_1.1.0
#> [63] plyr_1.8.6 magrittr_2.0.1
#> [65] R6_2.5.0 generics_0.1.0
#> [67] DelayedArray_0.14.1 pillar_1.4.7
#> [69] haven_2.3.1 whisker_0.4
#> [71] withr_2.3.0 RCurl_1.98-1.2
#> [73] modelr_0.1.8 crayon_1.3.4
#> [75] rmarkdown_2.6 grid_4.0.2
#> [77] readxl_1.3.1 blob_1.2.1
#> [79] git2r_0.27.1 reprex_0.3.0
#> [81] digest_0.6.27 xtable_1.8-4
#> [83] ff_4.0.4 httpuv_1.5.4
#> [85] munsell_0.5.0