T2D - Pancreas
sheng Qian
2021-2-6
Last updated: 2022-02-26
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5c37a5d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Amygdala_S.Rmd
Modified: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/BMI_Brain_Cerebellum_S.Rmd
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/BMI_Brain_Cortex_S.Rmd
Modified: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/BMI_Brain_Hippocampus_S.Rmd
Modified: analysis/BMI_Brain_Hypothalamus_S.Rmd
Modified: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Pancreas.Rmd) and HTML (docs/T2D_Pancreas.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5c37a5d | sq-96 | 2022-02-26 | update |
| html | 3fa3a64 | sq-96 | 2022-02-26 | Build site. |
| Rmd | 0e6a2f2 | sq-96 | 2022-02-26 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 7605#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
748 581 480 310 401 436 387 270 287 327 513 475 161 273 282 292 407 119 367 228
21 22
92 169 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 4452#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.5854Check convergence of parameters
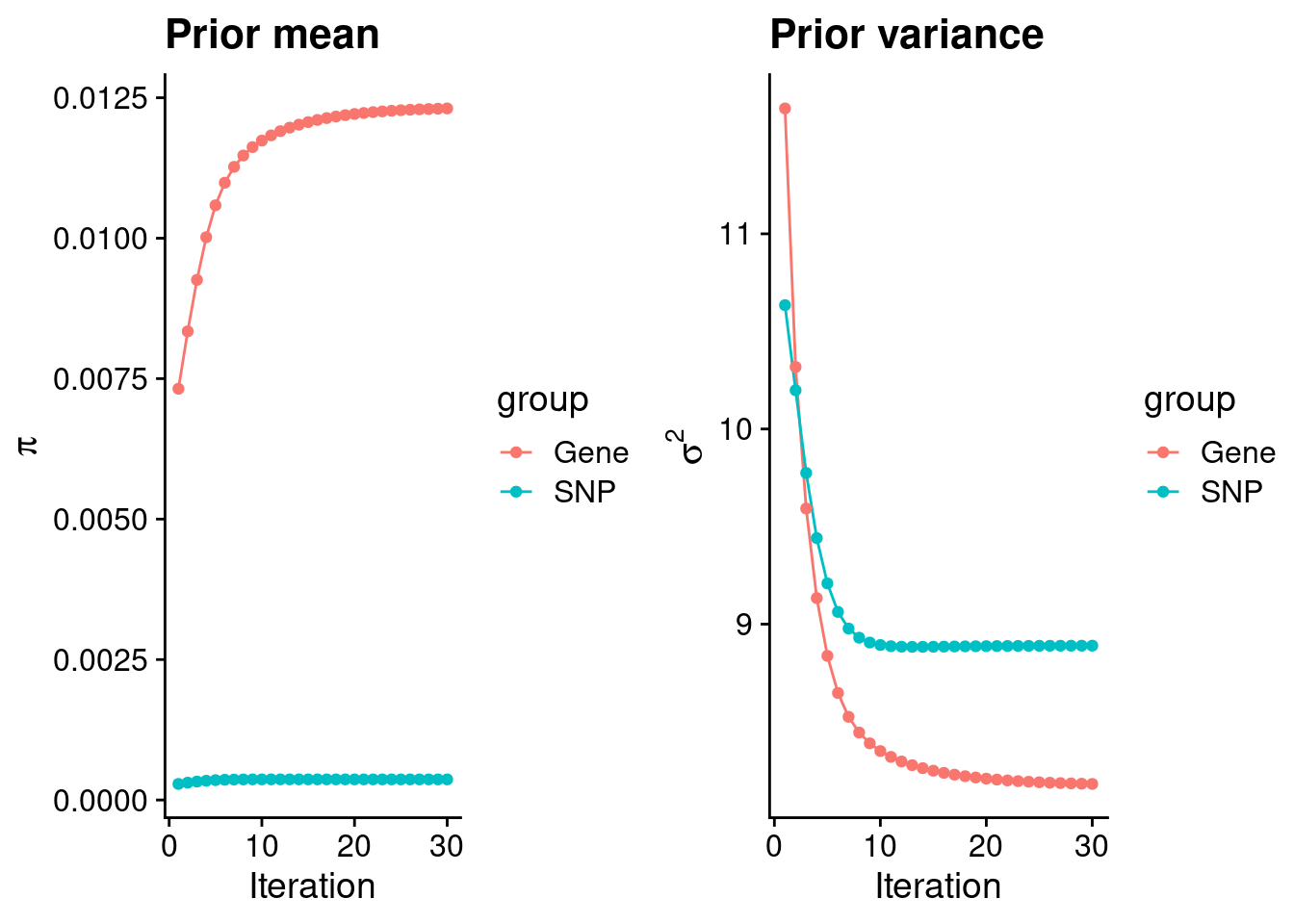
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0123097 0.0003667 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
8.181 8.890 #report sample size
print(sample_size)[1] 62892#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 7605 5017190#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01218 0.26008 #compare sum(PIP*mu2/sample_size) with above PVE calculation
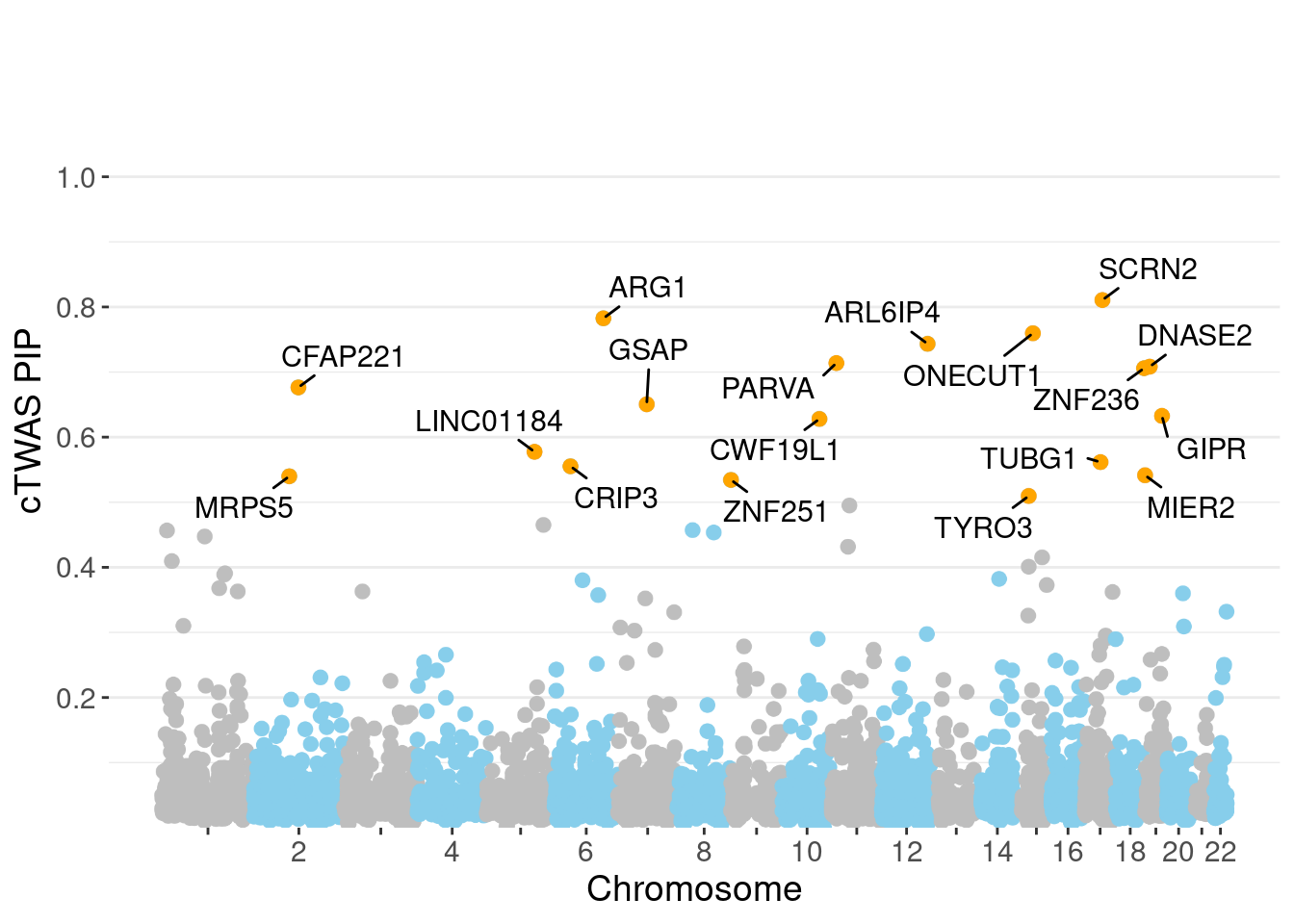
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.06992 1.44417Genes with highest PIPs
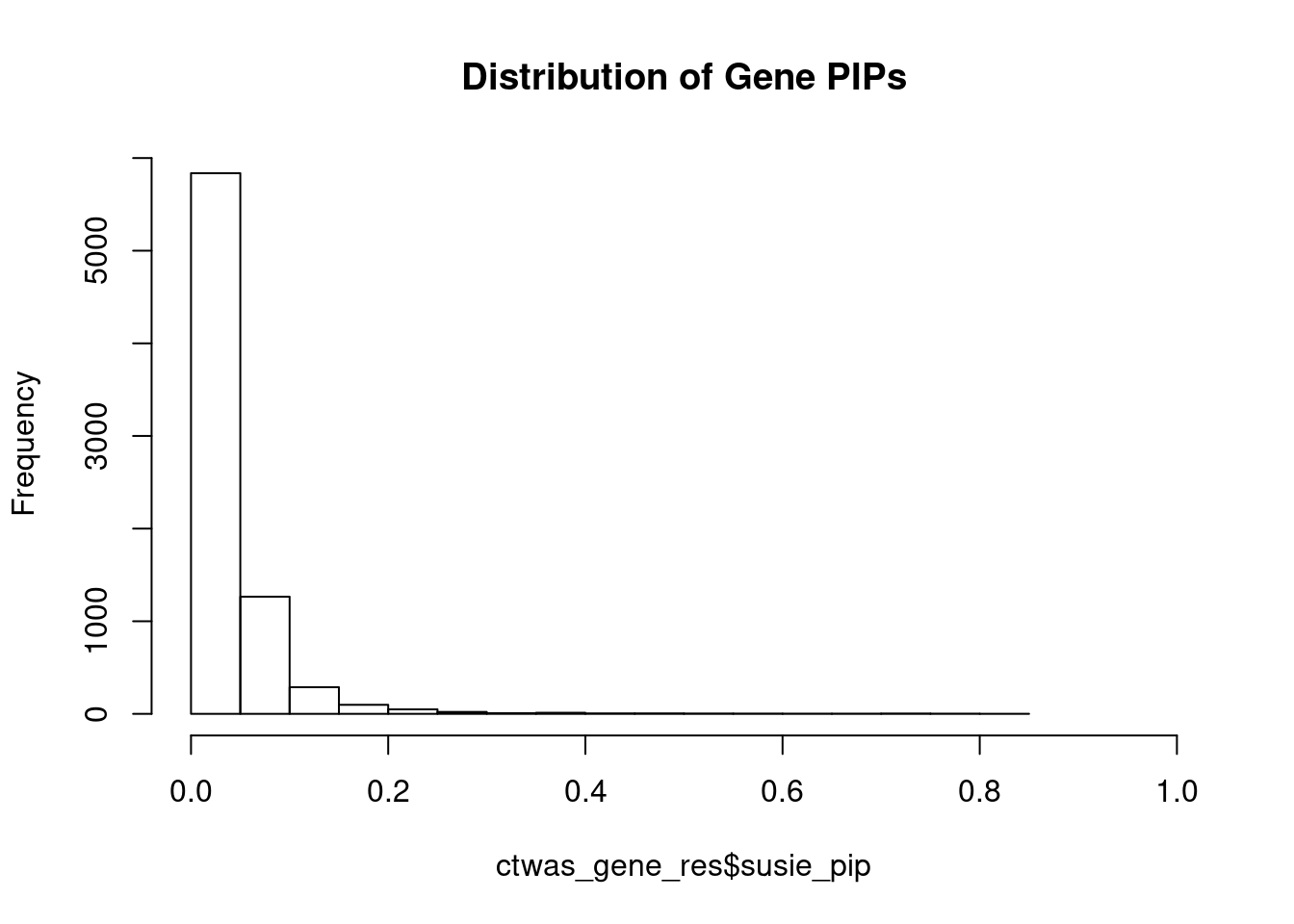
genename region_tag susie_pip mu2 PVE z num_eqtl
5708 SCRN2 17_28 0.8107 23.10 0.0002977 -4.970 2
3452 ARG1 6_87 0.7825 28.71 0.0003572 -5.418 2
8741 ONECUT1 15_22 0.7596 27.19 0.0003284 5.079 1
10062 ARL6IP4 12_75 0.7433 36.01 0.0004257 5.620 1
11158 PARVA 11_9 0.7138 21.89 0.0002484 -3.862 2
2227 DNASE2 19_10 0.7083 19.09 0.0002150 -3.744 1
4461 ZNF236 18_45 0.7059 20.70 0.0002323 -4.378 1
7604 CFAP221 2_69 0.6764 20.29 0.0002183 -4.050 2
10527 GSAP 7_49 0.6505 24.56 0.0002541 -4.185 1
183 GIPR 19_32 0.6326 36.32 0.0003653 -6.632 1
1451 CWF19L1 10_64 0.6280 33.14 0.0003309 -5.803 2
12493 LINC01184 5_78 0.5774 20.30 0.0001864 3.793 1
4519 TUBG1 17_25 0.5616 23.49 0.0002098 5.268 2
6236 CRIP3 6_33 0.5552 21.66 0.0001912 4.545 2
2221 MIER2 19_1 0.5414 23.96 0.0002063 3.684 1
6019 MRPS5 2_57 0.5399 21.72 0.0001865 -3.737 1
11243 ZNF251 8_94 0.5343 24.42 0.0002075 -4.886 1
1411 TYRO3 15_15 0.5096 23.09 0.0001871 4.866 1
3830 KBTBD4 11_29 0.4950 25.44 0.0002003 -5.098 1
12664 LINC01933 5_89 0.4651 20.74 0.0001534 3.780 1Genes with largest effect sizes
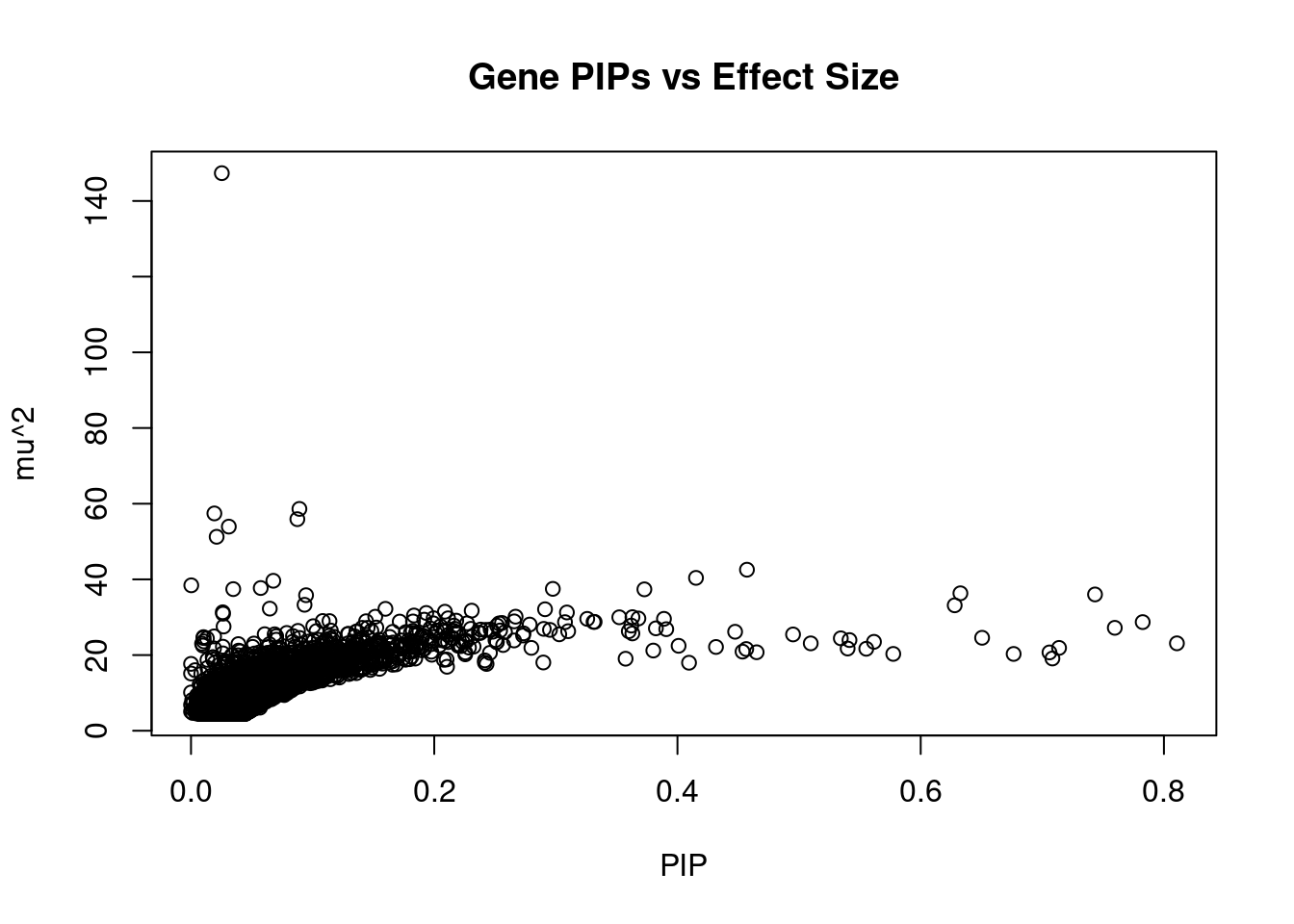
genename region_tag susie_pip mu2 PVE z num_eqtl
6795 JAZF1 7_23 0.0253350 147.34 5.935e-05 -13.082 2
474 BCAR1 16_40 0.0891404 58.62 8.308e-05 -7.720 1
10584 ARAP1 11_41 0.0192136 57.43 1.754e-05 7.886 1
6345 CDKN2B 9_16 0.0874433 55.91 7.773e-05 -8.192 1
13639 LINC01126 2_27 0.0310930 53.94 2.667e-05 -8.377 1
10742 NCR3LG1 11_12 0.0210812 51.25 1.718e-05 -7.370 1
7991 NKX6-3 8_36 0.4571379 42.55 3.093e-04 -6.781 2
9232 PEAK1 15_36 0.4152426 40.39 2.667e-04 6.885 1
7192 ATP5G1 17_28 0.0676819 39.63 4.264e-05 6.400 1
11440 RBM20 10_70 0.0002241 38.43 1.369e-07 -3.000 1
7194 SNF8 17_28 0.0573206 37.71 3.437e-05 6.300 1
1291 P2RX7 12_74 0.2975092 37.49 1.773e-04 5.484 2
10224 BMP8A 1_24 0.0347387 37.42 2.067e-05 6.296 1
7087 AP3S2 15_41 0.3727948 37.36 2.215e-04 6.356 1
183 GIPR 19_32 0.6326493 36.32 3.653e-04 -6.632 1
10062 ARL6IP4 12_75 0.7433361 36.01 4.257e-04 5.620 1
10674 HAPLN4 19_15 0.0946101 35.81 5.387e-05 5.347 1
11533 MICB 6_25 0.0933549 33.28 4.940e-05 5.546 2
1451 CWF19L1 10_64 0.6279579 33.14 3.309e-04 -5.803 2
8540 TAP1 6_27 0.0647764 32.27 3.323e-05 -5.575 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
10062 ARL6IP4 12_75 0.7433 36.01 0.0004257 5.620 1
183 GIPR 19_32 0.6326 36.32 0.0003653 -6.632 1
3452 ARG1 6_87 0.7825 28.71 0.0003572 -5.418 2
1451 CWF19L1 10_64 0.6280 33.14 0.0003309 -5.803 2
8741 ONECUT1 15_22 0.7596 27.19 0.0003284 5.079 1
7991 NKX6-3 8_36 0.4571 42.55 0.0003093 -6.781 2
5708 SCRN2 17_28 0.8107 23.10 0.0002977 -4.970 2
9232 PEAK1 15_36 0.4152 40.39 0.0002667 6.885 1
10527 GSAP 7_49 0.6505 24.56 0.0002541 -4.185 1
11158 PARVA 11_9 0.7138 21.89 0.0002484 -3.862 2
4461 ZNF236 18_45 0.7059 20.70 0.0002323 -4.378 1
7087 AP3S2 15_41 0.3728 37.36 0.0002215 6.356 1
7604 CFAP221 2_69 0.6764 20.29 0.0002183 -4.050 2
2227 DNASE2 19_10 0.7083 19.09 0.0002150 -3.744 1
4519 TUBG1 17_25 0.5616 23.49 0.0002098 5.268 2
11243 ZNF251 8_94 0.5343 24.42 0.0002075 -4.886 1
2221 MIER2 19_1 0.5414 23.96 0.0002063 3.684 1
3830 KBTBD4 11_29 0.4950 25.44 0.0002003 -5.098 1
6236 CRIP3 6_33 0.5552 21.66 0.0001912 4.545 2
1411 TYRO3 15_15 0.5096 23.09 0.0001871 4.866 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
6795 JAZF1 7_23 0.02534 147.34 5.935e-05 -13.082 2
13639 LINC01126 2_27 0.03109 53.94 2.667e-05 -8.377 1
6345 CDKN2B 9_16 0.08744 55.91 7.773e-05 -8.192 1
10584 ARAP1 11_41 0.01921 57.43 1.754e-05 7.886 1
474 BCAR1 16_40 0.08914 58.62 8.308e-05 -7.720 1
10742 NCR3LG1 11_12 0.02108 51.25 1.718e-05 -7.370 1
9232 PEAK1 15_36 0.41524 40.39 2.667e-04 6.885 1
7991 NKX6-3 8_36 0.45714 42.55 3.093e-04 -6.781 2
183 GIPR 19_32 0.63265 36.32 3.653e-04 -6.632 1
7192 ATP5G1 17_28 0.06768 39.63 4.264e-05 6.400 1
7087 AP3S2 15_41 0.37279 37.36 2.215e-04 6.356 1
7194 SNF8 17_28 0.05732 37.71 3.437e-05 6.300 1
10224 BMP8A 1_24 0.03474 37.42 2.067e-05 6.296 1
1451 CWF19L1 10_64 0.62796 33.14 3.309e-04 -5.803 2
10062 ARL6IP4 12_75 0.74334 36.01 4.257e-04 5.620 1
3128 NRBP1 2_16 0.02609 31.31 1.298e-05 -5.595 1
8540 TAP1 6_27 0.06478 32.27 3.323e-05 -5.575 1
11533 MICB 6_25 0.09335 33.28 4.940e-05 5.546 2
3134 PPM1G 2_16 0.02637 30.93 1.297e-05 -5.544 1
1291 P2RX7 12_74 0.29751 37.49 1.773e-04 5.484 2Comparing z scores and PIPs
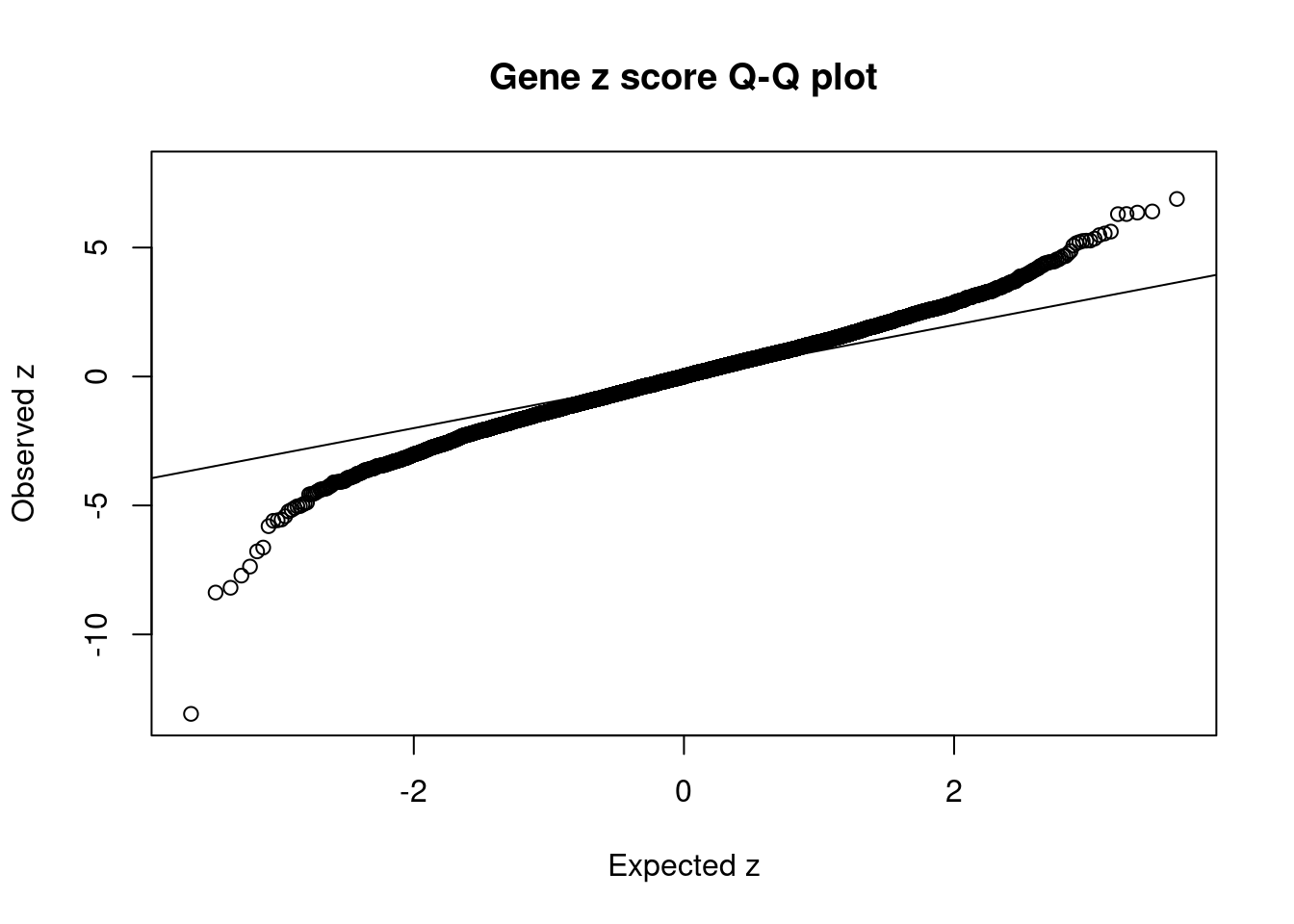
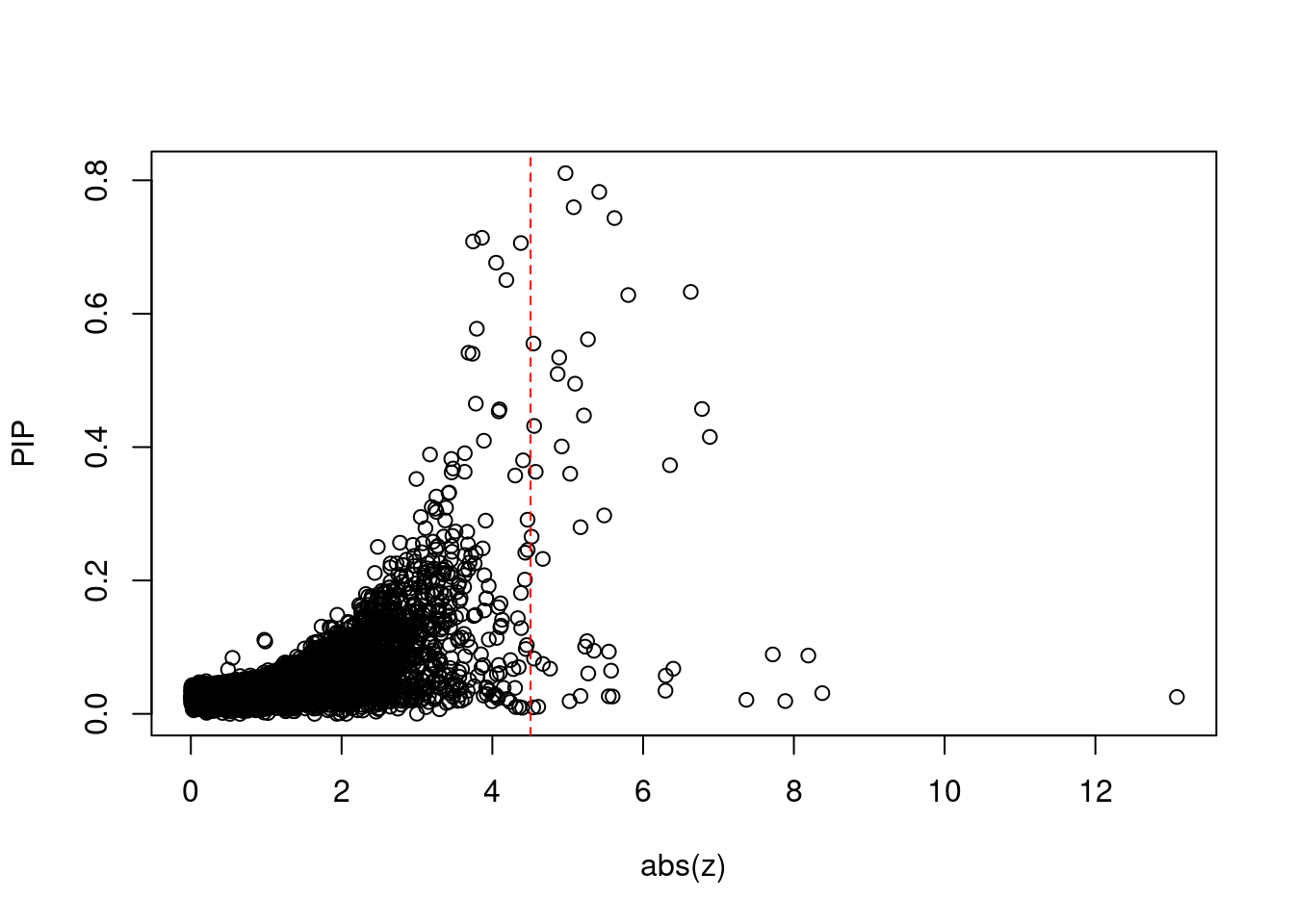
[1] 0.00618 genename region_tag susie_pip mu2 PVE z num_eqtl
6795 JAZF1 7_23 0.02534 147.34 5.935e-05 -13.082 2
13639 LINC01126 2_27 0.03109 53.94 2.667e-05 -8.377 1
6345 CDKN2B 9_16 0.08744 55.91 7.773e-05 -8.192 1
10584 ARAP1 11_41 0.01921 57.43 1.754e-05 7.886 1
474 BCAR1 16_40 0.08914 58.62 8.308e-05 -7.720 1
10742 NCR3LG1 11_12 0.02108 51.25 1.718e-05 -7.370 1
9232 PEAK1 15_36 0.41524 40.39 2.667e-04 6.885 1
7991 NKX6-3 8_36 0.45714 42.55 3.093e-04 -6.781 2
183 GIPR 19_32 0.63265 36.32 3.653e-04 -6.632 1
7192 ATP5G1 17_28 0.06768 39.63 4.264e-05 6.400 1
7087 AP3S2 15_41 0.37279 37.36 2.215e-04 6.356 1
7194 SNF8 17_28 0.05732 37.71 3.437e-05 6.300 1
10224 BMP8A 1_24 0.03474 37.42 2.067e-05 6.296 1
1451 CWF19L1 10_64 0.62796 33.14 3.309e-04 -5.803 2
10062 ARL6IP4 12_75 0.74334 36.01 4.257e-04 5.620 1
3128 NRBP1 2_16 0.02609 31.31 1.298e-05 -5.595 1
8540 TAP1 6_27 0.06478 32.27 3.323e-05 -5.575 1
11533 MICB 6_25 0.09335 33.28 4.940e-05 5.546 2
3134 PPM1G 2_16 0.02637 30.93 1.297e-05 -5.544 1
1291 P2RX7 12_74 0.29751 37.49 1.773e-04 5.484 2GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 18Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio
50 CORTICAL DYSPLASIA, COMPLEX, WITH OTHER BRAIN MALFORMATIONS 4 0.03041 1/10
53 SPINOCEREBELLAR ATAXIA, AUTOSOMAL RECESSIVE 17 0.03041 1/10
32 Hyperargininemia 0.04052 1/10
11 Hair Diseases 0.04557 1/10
1 Amino Acid Metabolism, Inborn Errors 0.06042 1/10
2 Asbestosis 0.06042 1/10
4 Body Weight 0.06042 1/10
10 Glomerulonephritis, Membranoproliferative 0.06042 1/10
13 Immune System Diseases 0.06042 1/10
15 Leishmaniasis 0.06042 1/10
BgRatio
50 1/9703
53 1/9703
32 2/9703
11 3/9703
1 13/9703
2 14/9703
4 15/9703
10 7/9703
13 7/9703
15 9/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 72#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 27#significance threshold for TWAS
print(sig_thresh)[1] 4.507#number of ctwas genes
length(ctwas_genes)[1] 18#number of TWAS genes
length(twas_genes)[1] 47#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
6019 MRPS5 2_57 0.5399 21.72 0.0001865 -3.737 1
12493 LINC01184 5_78 0.5774 20.30 0.0001864 3.793 1
10527 GSAP 7_49 0.6505 24.56 0.0002541 -4.185 1
11158 PARVA 11_9 0.7138 21.89 0.0002484 -3.862 2
4461 ZNF236 18_45 0.7059 20.70 0.0002323 -4.378 1
2221 MIER2 19_1 0.5414 23.96 0.0002063 3.684 1
2227 DNASE2 19_10 0.7083 19.09 0.0002150 -3.744 1
7604 CFAP221 2_69 0.6764 20.29 0.0002183 -4.050 2#sensitivity / recall
print(sensitivity) ctwas TWAS
0.01389 0.01389 #specificity
print(specificity) ctwas TWAS
0.9978 0.9939 #precision / PPV
print(precision) ctwas TWAS
0.05556 0.02128
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.17
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.2 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 rvest_1.0.2 cli_3.1.0 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_1.0.1 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.2 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.8
[49] munsell_0.5.0 fansi_1.0.2 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.5.0 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.6.2 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.2 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.14 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2