Test susieI using chr17 to 22
Last updated: 2020-09-03
Checks: 6 1
Knit directory: causal-TWAS/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191103) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 193a8df. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/workflow/.ipynb_checkpoints/
Ignored: data/
Unstaged changes:
Modified: analysis/simulation-multi-ukbchr17to22-gtex.adipose_susieprior.Rmd
Modified: analysis/simulation-susieI-ukbchr17to22-gtex.adipose.Rmd
Modified: code/run_test_susie.R
Modified: code/run_test_susieI.R
Modified: code/workflow/workflow-ashtest4.ipynb
Modified: code/workflow/workflow-susieI-20200813.ipynb
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/simulation-susieI-ukbchr17to22-gtex.adipose.Rmd) and HTML (docs/simulation-susieI-ukbchr17to22-gtex.adipose.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 193a8df | simingz | 2020-09-01 | increase gene pve |
| html | 193a8df | simingz | 2020-09-01 | increase gene pve |
| Rmd | 86681eb | simingz | 2020-08-28 | susieI all regions |
| html | 86681eb | simingz | 2020-08-28 | susieI all regions |
| Rmd | 41c8849 | simingz | 2020-08-20 | more parameter examples for susieI |
| html | 41c8849 | simingz | 2020-08-20 | more parameter examples for susieI |
| Rmd | 86ac96e | simingz | 2020-08-19 | more iterations |
| html | 86ac96e | simingz | 2020-08-19 | more iterations |
| Rmd | bd402eb | simingz | 2020-08-19 | susieI bug fix |
| html | bd402eb | simingz | 2020-08-19 | susieI bug fix |
| Rmd | f125882 | simingz | 2020-08-19 | susieI |
| html | f125882 | simingz | 2020-08-19 | susieI |
| Rmd | d299f60 | simingz | 2020-08-19 | susieI |
| html | d299f60 | simingz | 2020-08-19 | susieI |
| Rmd | 846fb96 | simingz | 2020-08-14 | susieI |
| html | 846fb96 | simingz | 2020-08-14 | susieI |
| Rmd | fd909a1 | simingz | 2020-08-14 | susieI |
Test susieI for ukb chr 17 to chr 22 combined. SNPs are downsampled to 1/10, eQTLs defined by FUSION-TWAS (Adipose/GTEx) lasso effect size > 0 were kept, p= 86k, n = 20k.
susieI is an iterative version of susie, learning parameters (prior weights) iteratively using the top region from mr.ash for genes only.
library(mr.ash.alpha)
library(data.table)
suppressMessages({library(plotly)})
library(tidyr)
library(plyr)
library(stringr)
library(kableExtra)
source("analysis/summarize_twas_plots.R")simdatadir <- "~/causalTWAS/simulations/simulation_ashtest_20200721/"
outputdir <- "~/causalTWAS/simulations/simulation_susieI_20200813/"
susiedir <- "~/causalTWAS/simulations/simulation_susieI_20200813/"
mrashdir <- "~/causalTWAS/simulations/simulation_ashtest_20200721/"
tag <- '20200813-1-1'
Niter <- 30MR.ASH pip distribution
- Use mr.ash for gene only:
rpipf <- paste0(outputdir, tag, "-mr.ash.rPIP.txt")
a <- read.table(rpipf, header = T)
a.c <- a[a$nCausal > 0, ]
ax <- pretty(0:max(a$rPIP), n = 30)
par(mfrow=c(2,1))
h1 <- hist(a$rPIP, breaks = 100, xlab = "PIP", main = "PIP distribution-all", col = "grey"); grid()
h2 <- hist(a.c$rPIP, breaks = h1$breaks, xlab = "PIP", main = "PIP distribution-causal", col = "salmon");grid()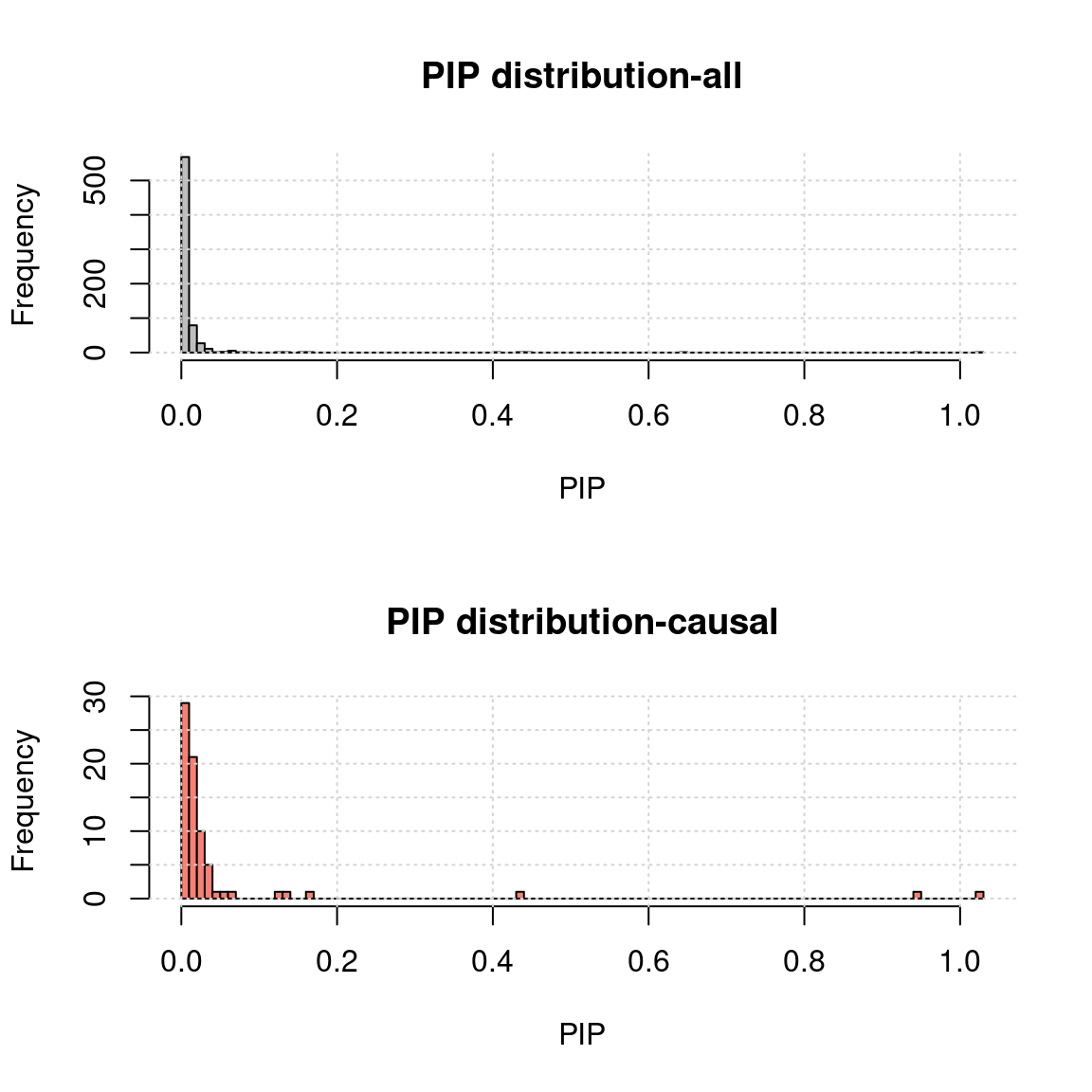
- MR.ash2s results for the same simulated data
rpipf <- "~/causalTWAS/simulations/simulation_ashtest_20200721/20200721-1-1-mr.ash2s.lassoes-es.rPIP.txt"
a <- read.table(rpipf, header = T)
a.c <- a[a$nCausal > 0 ,]
ax <- pretty(0:max(a$rPIP), n = 30) # Make a neat vector for the breakpoints
par(mfrow=c(2,1))
h1 <- hist(a$rPIP, breaks = 100, xlab = "PIP", main = "PIP distribution-all", col = "grey"); grid()
h2 <- hist(a.c$rPIP, breaks = h1$breaks, xlab = "PIP", main = "PIP distribution-causal", col = "salmon");grid()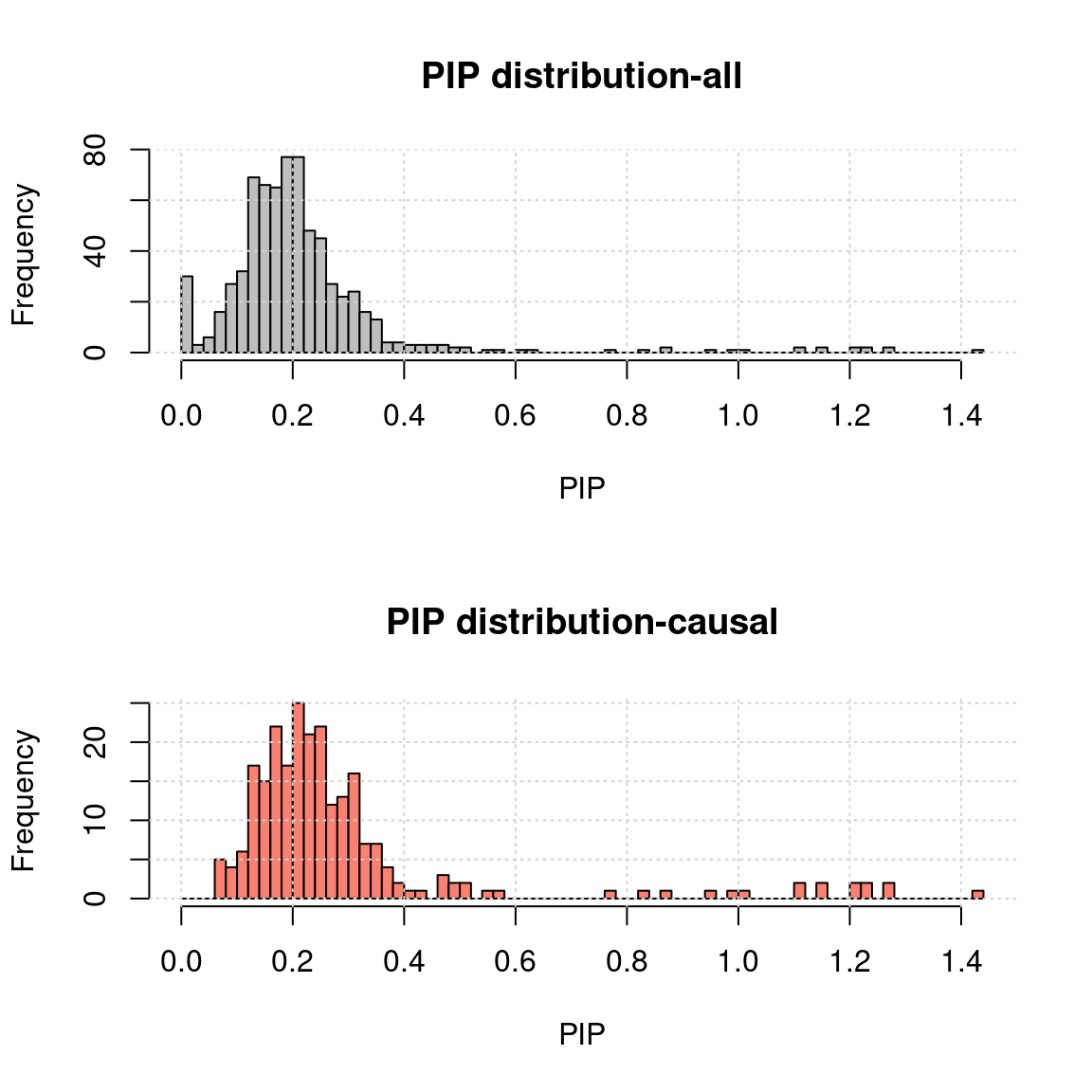
susieI genes - setting 1
Regions: We use regional PIP of both SNP and gene from mr.ash2s > 0.3, and also require at least one causal gene or SNP is present, also, at least one gene.
Susie run parameters:
L=1,null_weight = 0, didn’t updatenull_weightin iterations. We initialize with prior for genes and SNPs as uniform.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.00853819 0.006018864
Iteration 2 0.01049847 0.005956966
Iteration 3 0.01202068 0.005908900
Iteration 4 0.01318421 0.005872160
Iteration 5 0.01405232 0.005844749
Iteration 6 0.01469379 0.005824494
Iteration 7 0.01516465 0.005809626
Iteration 8 0.01550865 0.005798763
Iteration 9 0.01575915 0.005790854
Iteration 10 0.01594113 0.005785108
Iteration 11 0.01607310 0.005780940
Iteration 12 0.01616870 0.005777922
Iteration 13 0.01623788 0.005775737
Iteration 14 0.01628792 0.005774157
Iteration 15 0.01632410 0.005773015
Iteration 16 0.01635024 0.005772189
Iteration 17 0.01636913 0.005771593
Iteration 18 0.01638277 0.005771162
Iteration 19 0.01639263 0.005770851
Iteration 20 0.01639975 0.005770626
Iteration 21 0.01640489 0.005770464
Iteration 22 0.01640860 0.005770346
Iteration 23 0.01641129 0.005770262
Iteration 24 0.01641322 0.005770201
Iteration 25 0.01641462 0.005770157
Iteration 26 0.01641563 0.005770125
Iteration 27 0.01641636 0.005770102
Iteration 28 0.01641688 0.005770085
Iteration 29 0.01641726 0.005770073
Iteration 30 0.01641754 0.005770064more examples
Each row represents one simulation run. Runs labeled 1-x have the same simulation parameters, as you can tell from the true parameters. (and so are runs labled 2-x and 3-x)
show_param <- function(mrashfs, susieIfs, susieIfs2){
param <- do.call(rbind, lapply(mr.ashfs, function(x) t(read.table(x))[2:1,]))
pars <- cbind(param[seq(1,nrow(param), 2), 2], param[seq(1,nrow(param), 2), 4], param[seq(1,nrow(param), 2), 1], param[seq(2,nrow(param), 2), 1], param[seq(1,nrow(param), 2), 3], param[seq(2,nrow(param), 2), 3])
colnames(pars) <- c("PVE.gene_truth", "PVE.SNP_truth", paste(rep(c("pi1.gene_", "pi1.SNP_"), each = 2), c("truth", "mr.ash2"), sep = ""))
param.s <- do.call(rbind, lapply(susieIfs, function(x) {load(x); c(tail(prior.gene_rec, 1), tail(prior.SNP_rec,1))}))
param.s.truth <- do.call(rbind, lapply(susieIfs2, function(x) {
a <- read.table(x, header = T);
c(nrow(a[a$ifcausal == 1 & a$type == "gene", , drop = F])/ nrow(a[a$type == "gene", , drop = F]),
nrow(a[a$ifcausal == 1 & a$type == "SNP", , drop = F])/ nrow(a[a$type == "SNP", , drop = F]))
}))
pars.s <- cbind(param.s.truth, param.s)[, c(1,3,2,4)]
colnames(pars.s) <- paste(rep(c("pi1.gene_", "pi1.SNP_"), each = 2), c("susietruth", "susieI"), sep = "")
df <- cbind(tags, format(pars, digits = 4), format(pars.s, digits =4))
rownames(df) <- NULL
df %>%
kable("html", escape = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(c(1:5, 11:15), background = "#FEF3B9") %>%
scroll_box(width = "100%", height = "600px", fixed_thead = T)
}
tags <- paste(rep(1:3, each = 5), 1:5, sep = "-")
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".1.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 6.356e-02 | 1.642e-02 | 6.288e-03 | 5.770e-03 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 8.989e-02 | 2.728e-02 | 6.885e-03 | 5.521e-03 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 9.610e-02 | 5.144e-02 | 5.420e-03 | 5.042e-03 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 9.900e-02 | 4.260e-02 | 5.384e-03 | 5.350e-03 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 1.042e-01 | 7.617e-02 | 5.410e-03 | 4.203e-03 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 1.429e-01 | 2.165e-08 | 6.279e-03 | 6.279e-03 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 1.367e-01 | 3.291e-02 | 9.089e-03 | 5.263e-03 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 9.302e-02 | 7.277e-02 | 1.237e-02 | 5.775e-03 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 1.379e-01 | 4.098e-02 | 9.026e-03 | 5.805e-03 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 1.881e-01 | 3.701e-02 | 8.109e-03 | 6.363e-03 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 7.826e-02 | 6.582e-02 | 5.666e-03 | 4.227e-03 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 8.589e-02 | 5.083e-02 | 8.970e-03 | 4.532e-03 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 6.667e-02 | 4.392e-02 | 6.864e-03 | 5.631e-03 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 8.333e-02 | 1.055e-01 | 6.423e-03 | 3.100e-03 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 4.950e-02 | 3.959e-02 | 8.435e-03 | 5.624e-03 |
susieI genes - setting 2
Regions: All regions.
Susie run parameters:
L=1. initialize withnull_weight = 0and updatenull_weightbased on last iteration results. We initialize with prior for genes and SNPs as uniform.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".config1.susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.0042939376 3.927826e-03
Iteration 2 0.0022949947 1.834679e-03
Iteration 3 0.0010887830 6.261872e-04
Iteration 4 0.0006670077 1.887757e-04
Iteration 5 0.0006069632 8.284829e-05
Iteration 6 0.0006029904 5.534014e-05
Iteration 7 0.0006020495 4.568706e-05
Iteration 8 0.0006012293 4.161309e-05
Iteration 9 0.0006007693 3.976660e-05
Iteration 10 0.0006004784 3.890228e-05
Iteration 11 0.0006003633 3.849407e-05
Iteration 12 0.0006003019 3.829982e-05
Iteration 13 0.0006002719 3.820714e-05
Iteration 14 0.0006002574 3.816285e-05
Iteration 15 0.0006002504 3.814168e-05
Iteration 16 0.0006002471 3.813156e-05
Iteration 17 0.0006002455 3.812671e-05
Iteration 18 0.0006002447 3.812440e-05
Iteration 19 0.0006002444 3.812329e-05
Iteration 20 0.0006002442 3.812276e-05
Iteration 21 0.0006002441 3.812251e-05
Iteration 22 0.0006002441 3.812238e-05
Iteration 23 0.0006002441 3.812233e-05
Iteration 24 0.0006002441 3.812230e-05
Iteration 25 0.0006002441 3.812228e-05
Iteration 26 0.0006002441 3.812228e-05
Iteration 27 0.0006002441 3.812228e-05
Iteration 28 0.0006002440 3.812227e-05
Iteration 29 0.0006002440 3.812227e-05
Iteration 30 0.0006002440 3.812227e-05More examples
tags <- paste(rep(1:4, each = 5), 1:5, sep = "-")
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".config1.susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".config1.susieI.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 5.021e-02 | 6.002e-04 | 2.498e-03 | 3.812e-05 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 5.021e-02 | 1.873e-03 | 2.498e-03 | 1.567e-04 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 5.021e-02 | 5.378e-15 | 2.498e-03 | 4.347e-05 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 5.021e-02 | 2.501e-04 | 2.498e-03 | 2.254e-04 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 5.021e-02 | 0.000e+00 | 2.498e-03 | 4.876e-05 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 9.982e-02 | 0.000e+00 | 2.498e-03 | 4.081e-05 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 9.982e-02 | 1.145e-03 | 2.498e-03 | 1.518e-04 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 9.982e-02 | 3.852e-04 | 2.498e-03 | 6.283e-05 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 9.982e-02 | 8.148e-07 | 2.498e-03 | 3.860e-05 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 9.982e-02 | 5.420e-03 | 2.498e-03 | 3.004e-03 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 1.996e-02 | 2.628e-03 | 2.498e-03 | 1.246e-04 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 1.996e-02 | 3.672e-03 | 2.498e-03 | 1.447e-04 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 1.996e-02 | 4.068e-03 | 2.498e-03 | 2.391e-04 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 1.996e-02 | 7.020e-03 | 2.498e-03 | 1.548e-04 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 1.996e-02 | 5.143e-04 | 2.498e-03 | 8.153e-05 |
| 4-1 | 0.057488 | 0.048139 | 0.019964 | 0.029905 | 0.002498 | 0.002066 | 1.996e-02 | 9.918e-03 | 2.498e-03 | 1.638e-04 |
| 4-2 | 0.059249 | 0.052328 | 0.019964 | 0.013600 | 0.002498 | 0.001880 | 1.996e-02 | 1.046e-02 | 2.498e-03 | 1.876e-04 |
| 4-3 | 0.030658 | 0.062869 | 0.019964 | 0.027449 | 0.002498 | 0.001603 | 1.996e-02 | 1.116e-02 | 2.498e-03 | 2.920e-04 |
| 4-4 | 0.082776 | 0.051363 | 0.019964 | 0.018648 | 0.002498 | 0.001731 | 1.996e-02 | 1.556e-02 | 2.498e-03 | 3.080e-04 |
| 4-5 | 0.037037 | 0.053187 | 0.019964 | 0.009693 | 0.002498 | 0.002194 | 1.996e-02 | 8.222e-03 | 2.498e-03 | 2.546e-04 |
susieI genes - setting 3
Regions: All regions.
Susie run parameters:
L=2. initialize withnull_weight = 0and updatenull_weightbased on last iteration results. We initialize with prior for genes and SNPs as uniform.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".config2.susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.0066993297 6.584306e-03
Iteration 2 0.0056787027 5.455210e-03
Iteration 3 0.0050337726 4.612196e-03
Iteration 4 0.0043467028 3.754690e-03
Iteration 5 0.0036153401 2.851934e-03
Iteration 6 0.0026731079 1.875408e-03
Iteration 7 0.0016861836 9.690431e-04
Iteration 8 0.0009732381 3.791106e-04
Iteration 9 0.0006853260 1.445436e-04
Iteration 10 0.0006245655 7.569032e-05
Iteration 11 0.0006136886 5.453603e-05
Iteration 12 0.0006106502 4.650609e-05
Iteration 13 0.0006090173 4.302479e-05
Iteration 14 0.0006082797 4.140850e-05
Iteration 15 0.0006079140 4.063401e-05
Iteration 16 0.0006076843 4.025587e-05
Iteration 17 0.0006075553 4.006912e-05
Iteration 18 0.0006075079 3.997749e-05
Iteration 19 0.0006074857 3.993250e-05
Iteration 20 0.0006074749 3.991039e-05
Iteration 21 0.0006074696 3.989953e-05
Iteration 22 0.0006074670 3.989419e-05
Iteration 23 0.0006074657 3.989157e-05
Iteration 24 0.0006074650 3.989028e-05
Iteration 25 0.0006074647 3.988965e-05
Iteration 26 0.0006074646 3.988933e-05
Iteration 27 0.0006074645 3.988918e-05
Iteration 28 0.0006074645 3.988911e-05
Iteration 29 0.0006074644 3.988907e-05
Iteration 30 0.0006074644 3.988905e-05More examples
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".config2.susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".config2.susieI.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 5.021e-02 | 6.075e-04 | 2.498e-03 | 3.989e-05 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 5.021e-02 | 2.523e-02 | 2.498e-03 | 5.641e-03 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 5.021e-02 | 2.436e-02 | 2.498e-03 | 8.315e-03 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 5.021e-02 | 7.602e-04 | 2.498e-03 | 2.410e-04 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 5.021e-02 | 3.049e-17 | 2.498e-03 | 5.010e-05 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 9.982e-02 | 2.384e-02 | 2.498e-03 | 8.353e-03 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 9.982e-02 | 1.165e-03 | 2.498e-03 | 1.863e-04 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 9.982e-02 | 4.951e-04 | 2.498e-03 | 6.351e-05 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 9.982e-02 | 1.763e-05 | 2.498e-03 | 4.017e-05 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 9.982e-02 | 1.354e-02 | 2.498e-03 | 1.159e-02 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 1.996e-02 | 1.216e-02 | 2.498e-03 | 9.847e-03 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 1.996e-02 | 4.399e-03 | 2.498e-03 | 1.622e-04 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 1.996e-02 | 3.834e-02 | 2.498e-03 | 7.942e-03 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 1.996e-02 | 7.789e-03 | 2.498e-03 | 1.657e-04 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 1.996e-02 | 2.044e-02 | 2.498e-03 | 6.743e-03 |
| 4-1 | 0.057488 | 0.048139 | 0.019964 | 0.029905 | 0.002498 | 0.002066 | 1.996e-02 | 1.226e-02 | 2.498e-03 | 2.320e-04 |
| 4-2 | 0.059249 | 0.052328 | 0.019964 | 0.013600 | 0.002498 | 0.001880 | 1.996e-02 | 1.313e-02 | 2.498e-03 | 2.217e-04 |
| 4-3 | 0.030658 | 0.062869 | 0.019964 | 0.027449 | 0.002498 | 0.001603 | 1.996e-02 | 1.420e-02 | 2.498e-03 | 3.708e-04 |
| 4-4 | 0.082776 | 0.051363 | 0.019964 | 0.018648 | 0.002498 | 0.001731 | 1.996e-02 | 4.826e-02 | 2.498e-03 | 6.057e-03 |
| 4-5 | 0.037037 | 0.053187 | 0.019964 | 0.009693 | 0.002498 | 0.002194 | 1.996e-02 | 4.448e-02 | 2.498e-03 | 3.679e-03 |
susieI genes - setting 4
Regions: All regions.
Susie run parameters:
L=1. Did not updatenull_weightin each iteration. We initialize with prior for genes and SNPs as uniform.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".config3.susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.004293938 0.003927826
Iteration 2 0.004628671 0.003921336
Iteration 3 0.004887227 0.003916324
Iteration 4 0.005108334 0.003900309
Iteration 5 0.005320663 0.003896192
Iteration 6 0.005511267 0.003892497
Iteration 7 0.005681650 0.003889194
Iteration 8 0.005833397 0.003886252
Iteration 9 0.005942095 0.003872416
Iteration 10 0.006053236 0.003870262
Iteration 11 0.006151198 0.003868363
Iteration 12 0.006300427 0.003877198
Iteration 13 0.006416455 0.003874948
Iteration 14 0.006518892 0.003872962
Iteration 15 0.006609166 0.003871212
Iteration 16 0.006645557 0.003858779
Iteration 17 0.006770023 0.003868094
Iteration 18 0.006863567 0.003866280
Iteration 19 0.006946413 0.003864674
Iteration 20 0.007019537 0.003863257
Iteration 21 0.007123598 0.003872967
Iteration 22 0.007197194 0.003871541
Iteration 23 0.007261948 0.003870285
Iteration 24 0.007318829 0.003869182
Iteration 25 0.007368722 0.003868215
Iteration 26 0.007412434 0.003867368
Iteration 27 0.007450689 0.003866626
Iteration 28 0.007484139 0.003865978
Iteration 29 0.007513365 0.003865411
Iteration 30 0.007538883 0.003864916More examples
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".config3.susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".config3.susieI.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 0.050212 | 0.007539 | 0.002498 | 0.003865 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 0.050212 | 0.016912 | 0.002498 | 0.004516 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 0.050212 | 0.020157 | 0.002498 | 0.005145 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 0.050212 | 0.019758 | 0.002498 | 0.004074 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 0.050212 | 0.031572 | 0.002498 | 0.003880 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 0.099819 | 0.018729 | 0.002498 | 0.005137 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 0.099819 | 0.027621 | 0.002498 | 0.003722 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 0.099819 | 0.034116 | 0.002498 | 0.003092 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 0.099819 | 0.028365 | 0.002498 | 0.003555 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 0.099819 | 0.007000 | 0.002498 | 0.006890 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 0.019964 | 0.011169 | 0.002498 | 0.005870 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 0.019964 | 0.028055 | 0.002498 | 0.003948 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 0.019964 | 0.027042 | 0.002498 | 0.005164 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 0.019964 | 0.028733 | 0.002498 | 0.003583 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 0.019964 | 0.017204 | 0.002498 | 0.004792 |
| 4-1 | 0.057488 | 0.048139 | 0.019964 | 0.029905 | 0.002498 | 0.002066 | 0.019964 | 0.037781 | 0.002498 | 0.004850 |
| 4-2 | 0.059249 | 0.052328 | 0.019964 | 0.013600 | 0.002498 | 0.001880 | 0.019964 | 0.037458 | 0.002498 | 0.003860 |
| 4-3 | 0.030658 | 0.062869 | 0.019964 | 0.027449 | 0.002498 | 0.001603 | 0.019964 | 0.039563 | 0.002498 | 0.004569 |
| 4-4 | 0.082776 | 0.051363 | 0.019964 | 0.018648 | 0.002498 | 0.001731 | 0.019964 | 0.035083 | 0.002498 | 0.005583 |
| 4-5 | 0.037037 | 0.053187 | 0.019964 | 0.009693 | 0.002498 | 0.002194 | 0.019964 | 0.034297 | 0.002498 | 0.005504 |
susieI genes - setting 5
Regions: All regions.
Susie run parameters:
L=5. Updatenull_weightin each iteration. We initialize prior for gene as0.05and prior for SNPs as2.5E-3, we initializenull_weightas 0.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".config4.susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.128448763 0.01158279
Iteration 2 0.094168925 0.01149096
Iteration 3 0.076063912 0.01178310
Iteration 4 0.064524110 0.01212570
Iteration 5 0.055119850 0.01229631
Iteration 6 0.048102245 0.01233622
Iteration 7 0.042120368 0.01232301
Iteration 8 0.037456936 0.01240276
Iteration 9 0.033310491 0.01238190
Iteration 10 0.030195811 0.01238656
Iteration 11 0.027329944 0.01236479
Iteration 12 0.024712911 0.01228908
Iteration 13 0.022706543 0.01232025
Iteration 14 0.021054701 0.01237728
Iteration 15 0.019502100 0.01238979
Iteration 16 0.017963015 0.01238032
Iteration 17 0.016656702 0.01239216
Iteration 18 0.015540301 0.01240985
Iteration 19 0.014511437 0.01238932
Iteration 20 0.013736011 0.01245430
Iteration 21 0.013082786 0.01246666
Iteration 22 0.012476337 0.01245043
Iteration 23 0.011952295 0.01244994
Iteration 24 0.011490005 0.01245502
Iteration 25 0.011078000 0.01246174
Iteration 26 0.010708610 0.01246864
Iteration 27 0.010376228 0.01247518
Iteration 28 0.010076332 0.01248121
Iteration 29 0.009805134 0.01248671
Iteration 30 0.009559407 0.01249170More examples
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".config4.susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".config4.susieI.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 0.050212 | 0.009559 | 0.002498 | 0.012492 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 0.050212 | 0.023603 | 0.002498 | 0.015701 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 0.050212 | 0.029077 | 0.002498 | 0.018980 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 0.050212 | 0.028906 | 0.002498 | 0.014768 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 0.050212 | 0.050957 | 0.002498 | 0.012467 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 0.099819 | 0.027189 | 0.002498 | 0.018974 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 0.099819 | 0.024997 | 0.002498 | 0.012936 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 0.099819 | 0.052765 | 0.002498 | 0.010592 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 0.099819 | 0.056156 | 0.002498 | 0.011524 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 0.099819 | 0.023854 | 0.002498 | 0.024732 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 0.019964 | 0.017983 | 0.002498 | 0.021351 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 0.019964 | 0.036252 | 0.002498 | 0.014132 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 0.019964 | 0.056365 | 0.002498 | 0.017616 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 0.019964 | 0.044908 | 0.002498 | 0.011689 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 0.019964 | 0.025246 | 0.002498 | 0.016104 |
| 4-1 | 0.057488 | 0.048139 | 0.019964 | 0.029905 | 0.002498 | 0.002066 | 0.019964 | 0.052581 | 0.002498 | 0.017496 |
| 4-2 | 0.059249 | 0.052328 | 0.019964 | 0.013600 | 0.002498 | 0.001880 | 0.019964 | 0.055914 | 0.002498 | 0.013129 |
| 4-3 | 0.030658 | 0.062869 | 0.019964 | 0.027449 | 0.002498 | 0.001603 | 0.019964 | 0.077002 | 0.002498 | 0.015812 |
| 4-4 | 0.082776 | 0.051363 | 0.019964 | 0.018648 | 0.002498 | 0.001731 | 0.019964 | 0.061251 | 0.002498 | 0.021152 |
| 4-5 | 0.037037 | 0.053187 | 0.019964 | 0.009693 | 0.002498 | 0.002194 | 0.019964 | 0.054815 | 0.002498 | 0.019761 |
susieI genes - setting 6
Regions: All regions.
Susie run parameters:
L=2. Updatenull_weightin each iteration. We initialize prior for gene as0.05and prior for SNPs as2.5E-3, we initializenull_weightas 0.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".config5.susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.0697892636 5.440846e-03
Iteration 2 0.0398270510 3.419298e-03
Iteration 3 0.0252924568 2.149136e-03
Iteration 4 0.0142952300 1.180874e-03
Iteration 5 0.0060263016 5.228431e-04
Iteration 6 0.0018044565 2.045111e-04
Iteration 7 0.0007474008 9.506614e-05
Iteration 8 0.0006249124 6.107471e-05
Iteration 9 0.0006122135 4.911233e-05
Iteration 10 0.0006094090 4.413298e-05
Iteration 11 0.0006083671 4.188944e-05
Iteration 12 0.0006078704 4.082639e-05
Iteration 13 0.0006075778 4.031127e-05
Iteration 14 0.0006074180 4.005856e-05
Iteration 15 0.0006073554 3.993494e-05
Iteration 16 0.0006073258 3.987438e-05
Iteration 17 0.0006073114 3.984470e-05
Iteration 18 0.0006073043 3.983013e-05
Iteration 19 0.0006073008 3.982299e-05
Iteration 20 0.0006072991 3.981949e-05
Iteration 21 0.0006072982 3.981777e-05
Iteration 22 0.0006072978 3.981692e-05
Iteration 23 0.0006072976 3.981651e-05
Iteration 24 0.0006072975 3.981630e-05
Iteration 25 0.0006072975 3.981620e-05
Iteration 26 0.0006072975 3.981616e-05
Iteration 27 0.0006072975 3.981613e-05
Iteration 28 0.0006072974 3.981612e-05
Iteration 29 0.0006072974 3.981611e-05
Iteration 30 0.0006072974 3.981611e-05More examples
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt")
susieIfs <- paste0(outputdir, "20200813-", tags, ".config5.susieIres.Rd")
susieIfs2 <- paste0(outputdir, "20200813-", tags, ".config5.susieI.txt")
show_param(mrashfs, susieIfs, susieIfs2)| tags | PVE.gene_truth | PVE.SNP_truth | pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.006683 | 0.051070 | 0.050212 | 0.003163 | 0.002498 | 0.001744 | 5.021e-02 | 6.073e-04 | 2.498e-03 | 3.982e-05 |
| 1-2 | 0.009196 | 0.043706 | 0.050212 | 0.002314 | 0.002498 | 0.001938 | 5.021e-02 | 2.011e-03 | 2.498e-03 | 1.654e-04 |
| 1-3 | 0.011473 | 0.047521 | 0.050212 | 0.023767 | 0.002498 | 0.002285 | 5.021e-02 | 6.954e-09 | 2.498e-03 | 4.569e-05 |
| 1-4 | 0.011460 | 0.054859 | 0.050212 | 0.015942 | 0.002498 | 0.002540 | 5.021e-02 | 4.962e-04 | 2.498e-03 | 2.415e-04 |
| 1-5 | 0.011086 | 0.047848 | 0.050212 | 0.058862 | 0.002498 | 0.001532 | 5.021e-02 | 0.000e+00 | 2.498e-03 | 5.009e-05 |
| 2-1 | 0.009457 | 0.044345 | 0.099819 | 0.010598 | 0.002498 | 0.002237 | 9.982e-02 | 1.054e-17 | 2.498e-03 | 4.287e-05 |
| 2-2 | 0.011067 | 0.056852 | 0.099819 | 0.022076 | 0.002498 | 0.002191 | 9.982e-02 | 1.163e-03 | 2.498e-03 | 1.850e-04 |
| 2-3 | 0.010324 | 0.045895 | 0.099819 | 0.035246 | 0.002498 | 0.001585 | 9.982e-02 | 4.815e-04 | 2.498e-03 | 6.338e-05 |
| 2-4 | 0.011733 | 0.047672 | 0.099819 | 0.019076 | 0.002498 | 0.001622 | 9.982e-02 | 1.392e-05 | 2.498e-03 | 4.019e-05 |
| 2-5 | 0.008494 | 0.047920 | 0.099819 | 0.011194 | 0.002498 | 0.002138 | 9.982e-02 | 1.538e-02 | 2.498e-03 | 9.494e-03 |
| 3-1 | 0.011578 | 0.048473 | 0.019964 | 0.013220 | 0.002498 | 0.003176 | 1.996e-02 | 1.315e-02 | 2.498e-03 | 6.146e-03 |
| 3-2 | 0.011964 | 0.052832 | 0.019964 | 0.018732 | 0.002498 | 0.002654 | 1.996e-02 | 4.334e-03 | 2.498e-03 | 1.609e-04 |
| 3-3 | 0.006031 | 0.061836 | 0.019964 | 0.009304 | 0.002498 | 0.001802 | 1.996e-02 | 4.663e-03 | 2.498e-03 | 2.917e-04 |
| 3-4 | 0.017024 | 0.052816 | 0.019964 | 0.025294 | 0.002498 | 0.002402 | 1.996e-02 | 7.648e-03 | 2.498e-03 | 1.641e-04 |
| 3-5 | 0.007322 | 0.052574 | 0.019964 | 0.012802 | 0.002498 | 0.002439 | 1.996e-02 | 6.495e-04 | 2.498e-03 | 8.696e-05 |
| 4-1 | 0.057488 | 0.048139 | 0.019964 | 0.029905 | 0.002498 | 0.002066 | 1.996e-02 | 1.225e-02 | 2.498e-03 | 2.319e-04 |
| 4-2 | 0.059249 | 0.052328 | 0.019964 | 0.013600 | 0.002498 | 0.001880 | 1.996e-02 | 1.313e-02 | 2.498e-03 | 2.217e-04 |
| 4-3 | 0.030658 | 0.062869 | 0.019964 | 0.027449 | 0.002498 | 0.001603 | 1.996e-02 | 1.420e-02 | 2.498e-03 | 3.708e-04 |
| 4-4 | 0.082776 | 0.051363 | 0.019964 | 0.018648 | 0.002498 | 0.001731 | 1.996e-02 | 4.835e-02 | 2.498e-03 | 6.025e-03 |
| 4-5 | 0.037037 | 0.053187 | 0.019964 | 0.009693 | 0.002498 | 0.002194 | 1.996e-02 | 4.195e-02 | 2.498e-03 | 2.883e-03 |
susieI shuffle genes
one example
Shuffle genes to different regions. Regions: We use regional PIP of both SNP and gene from mr.ash2s > 0.3 , and also require at least one causal gene or SNP is present, also, at least one gene.
Susie run parameters:
L=1,null_weight = 0.
Prior change for each iteration:
tag <- '20200813-1-1-shuffleg'
load(paste0(outputdir, tag, ".susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:nrow(df))
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.01786469 0.005724369
Iteration 2 0.02870735 0.005382000
Iteration 3 0.03553571 0.005166386
Iteration 4 0.03936470 0.005045482
Iteration 5 0.04142174 0.004980528
Iteration 6 0.04250774 0.004946237
Iteration 7 0.04307662 0.004928274
Iteration 8 0.04337351 0.004918899
Iteration 9 0.04352816 0.004914016
Iteration 10 0.04360865 0.004911474
Iteration 11 0.04365052 0.004910152
Iteration 12 0.04367229 0.004909465
Iteration 13 0.04368361 0.004909107
Iteration 14 0.04368950 0.004908921
Iteration 15 0.04369256 0.004908825
Iteration 16 0.04369415 0.004908775
Iteration 17 0.04369498 0.004908748
Iteration 18 0.04369541 0.004908735
Iteration 19 0.04369563 0.004908728
Iteration 20 0.04369575 0.004908724
Iteration 21 0.04369581 0.004908722
Iteration 22 0.04369584 0.004908721
Iteration 23 0.04369585 0.004908721
Iteration 24 0.04369586 0.004908720
Iteration 25 0.04369587 0.004908720
Iteration 26 0.04369587 0.004908720
Iteration 27 0.04369587 0.004908720
Iteration 28 0.04369587 0.004908720
Iteration 29 0.04369587 0.004908720
Iteration 30 0.04369587 0.004908720
Iteration 31 0.04369587 0.004908720
Iteration 32 0.04369587 0.004908720
Iteration 33 0.04369587 0.004908720
Iteration 34 0.04369587 0.004908720
Iteration 35 0.04369587 0.004908720
Iteration 36 0.04369587 0.004908720
Iteration 37 0.04369587 0.004908720
Iteration 38 0.04369587 0.004908720
Iteration 39 0.04369587 0.004908720
Iteration 40 0.04369587 0.004908720
Iteration 41 0.04369587 0.004908720
Iteration 42 0.04369587 0.004908720
Iteration 43 0.04369587 0.004908720
Iteration 44 0.04369587 0.004908720
Iteration 45 0.04369587 0.004908720
Iteration 46 0.04369587 0.004908720
Iteration 47 0.04369587 0.004908720
Iteration 48 0.04369587 0.004908720
Iteration 49 0.04369587 0.004908720
Iteration 50 0.04369587 0.004908720
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.2.1 stringr_1.4.0 plyr_1.8.6
[4] tidyr_0.8.3 plotly_4.9.2.9000 ggplot2_3.3.1
[7] data.table_1.12.7 mr.ash.alpha_0.1-34
loaded via a namespace (and not attached):
[1] Rcpp_1.0.4.6 compiler_3.5.1 pillar_1.4.4
[4] later_0.7.5 git2r_0.26.1 workflowr_1.6.2
[7] tools_3.5.1 digest_0.6.25 viridisLite_0.3.0
[10] jsonlite_1.6.1 evaluate_0.12 tibble_3.0.1
[13] lifecycle_0.2.0 gtable_0.2.0 lattice_0.20-38
[16] pkgconfig_2.0.2 rlang_0.4.6 Matrix_1.2-15
[19] rstudioapi_0.11 yaml_2.2.0 xml2_1.2.0
[22] httr_1.4.1 withr_2.1.2 dplyr_1.0.0
[25] knitr_1.20 htmlwidgets_1.3 generics_0.0.2
[28] fs_1.3.1 vctrs_0.3.1 webshot_0.5.1
[31] tidyselect_1.1.0 rprojroot_1.3-2 grid_3.5.1
[34] glue_1.4.1 R6_2.3.0 rmarkdown_1.10
[37] purrr_0.3.4 magrittr_1.5 whisker_0.3-2
[40] backports_1.1.2 scales_1.0.0 promises_1.0.1
[43] htmltools_0.3.6 ellipsis_0.3.1 rvest_0.3.2
[46] colorspace_1.3-2 httpuv_1.4.5 stringi_1.3.1
[49] lazyeval_0.2.1 munsell_0.5.0 crayon_1.3.4