Visualize and interpret droplet and pulse-seq topics
Peter Carbonetto
Last updated: 2020-09-23
Checks: 6 1
Knit directory: single-cell-topics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2.9000). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- diff-count-analysis-clusters-droplet
- diff-count-analysis-clusters-pulseseq
- diff-count-analysis-topics-droplet
- diff-count-analysis-topics-pulseseq
To ensure reproducibility of the results, delete the cache directory plots_tracheal_epithelium_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f0bdbda. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: data/droplet.RData
Ignored: data/pbmc_68k.RData
Ignored: data/pbmc_purified.RData
Ignored: data/pulseseq.RData
Ignored: output/droplet/fits-droplet.RData
Ignored: output/droplet/rds/
Ignored: output/pbmc-68k/fits-pbmc-68k.RData
Ignored: output/pbmc-68k/rds/
Ignored: output/pbmc-purified/fits-pbmc-purified.RData
Ignored: output/pbmc-purified/rds/
Ignored: output/pulseseq/fits-pulseseq.RData
Ignored: output/pulseseq/rds/
Untracked files:
Untracked: analysis/plots_tracheal_epithelium_cache/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/plots_tracheal_epithelium.Rmd) and HTML (docs/plots_tracheal_epithelium.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f0bdbda | Peter Carbonetto | 2020-09-23 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”, verbose = TRUE) |
| html | 361eded | Peter Carbonetto | 2020-09-23 | Added text to accompany ciliated cells volcano plots. |
| Rmd | 8198c07 | Peter Carbonetto | 2020-09-23 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”, verbose = TRUE) |
| html | 23f87bb | Peter Carbonetto | 2020-09-23 | Improved volcano plots for ciliated cell type in |
| Rmd | 516a32e | Peter Carbonetto | 2020-09-23 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”, verbose = TRUE) |
| html | 0ed5061 | Peter Carbonetto | 2020-09-22 | Added some rough, first-draft volcano plots to |
| Rmd | 6ccf998 | Peter Carbonetto | 2020-09-22 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”, verbose = TRUE) |
| Rmd | de36d31 | Peter Carbonetto | 2020-09-22 | Added volcano plots for ciliated cells to plots_tracheal_epithelium analysis. |
| Rmd | 082352a | Peter Carbonetto | 2020-09-22 | Added steps to plots_tracheal_epithelium analysis to compute differential expression statistics. |
| html | 06d0b30 | Peter Carbonetto | 2020-09-22 | I’m starting to revamp the plots_tracheal_epithelium analysis. |
| Rmd | 0877a1f | Peter Carbonetto | 2020-09-22 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| Rmd | 5af46f1 | Peter Carbonetto | 2020-09-20 | Working on Structure plot for droplet data. |
| Rmd | c072577 | Peter Carbonetto | 2020-09-19 | Re-created structure plot for pulseseq data. |
| Rmd | 942486b | Peter Carbonetto | 2020-09-18 | Fixing merge issue. |
| Rmd | 8cf758e | Peter Carbonetto | 2020-09-12 | Working on improvements to clustering of pulse-seq data. |
| Rmd | a128de5 | Peter Carbonetto | 2020-09-12 | Revamping the analysis of the pulseseq data in plots_tracheal_epithelium. |
| Rmd | 3ab7da1 | Peter Carbonetto | 2020-08-25 | A few minor edits. |
| html | f2e0b23 | Peter Carbonetto | 2020-08-25 | Fixed dimensions of PCA plots in plots_tracheal_epithelium analysis. |
| Rmd | 7b59815 | Peter Carbonetto | 2020-08-25 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 5589611 | Peter Carbonetto | 2020-08-25 | Added PCA plots and structure plots from pulseseq data. |
| Rmd | b731a4a | Peter Carbonetto | 2020-08-25 | workflowr::wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | c3c1b12 | Peter Carbonetto | 2020-08-25 | Build site. |
| Rmd | 2defe6d | Peter Carbonetto | 2020-08-25 | Added crosstab plot to plots_tracheal_epithelium analysis. |
| html | 97c13c2 | Peter Carbonetto | 2020-08-25 | Build site. |
| Rmd | e11855b | Peter Carbonetto | 2020-08-25 | Working on revised analysis of droplet and pulse-seq data sets. |
| html | e11855b | Peter Carbonetto | 2020-08-25 | Working on revised analysis of droplet and pulse-seq data sets. |
| Rmd | bf23ca0 | Peter Carbonetto | 2020-08-20 | Added manual labeling of purified PBMC data to plots_pbmc analysis. |
| Rmd | 077d3d5 | Peter Carbonetto | 2020-08-20 | Added k=9 and k=11 pulseseq fits to plots_tracheal_epithelium analysis. |
| html | 0ce9604 | Peter Carbonetto | 2020-08-20 | Re-built plots_tracheal_epithelium with fastTopics 0.3-162. |
| Rmd | 961570e | Peter Carbonetto | 2020-08-20 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | b17bfa4 | Peter Carbonetto | 2020-08-19 | Added pulseseq PCA plots to plots_tracheal_epithelium analysis. |
| Rmd | 76dc0c6 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| Rmd | c70612f | Peter Carbonetto | 2020-08-19 | Revised structure plot settings for abundant droplet samples in plots_tracheal_epithelium. |
| html | adda33f | Peter Carbonetto | 2020-08-19 | Fixed another structure plot in plots_tracheal_epithelium analysis. |
| Rmd | 29a9258 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 0a16b60 | Peter Carbonetto | 2020-08-19 | Fixed structure plot in plots_tracheal_epithelium analysis. |
| Rmd | 3a7bd74 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | f4bdf19 | Peter Carbonetto | 2020-08-19 | Added explanatory text and improved PC-based manual clustering of |
| Rmd | c7b77ee | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| Rmd | 70a4a60 | Peter Carbonetto | 2020-08-19 | Added note to plots_tracheal_epithelium.Rmd. |
| html | fb21b3b | Peter Carbonetto | 2020-08-19 | Added very initial Structure plots to plots_tracheal_epithelium analysis. |
| Rmd | d35cb03 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 368a74a | Peter Carbonetto | 2020-08-19 | Added some text to plots_tracheal_epithelium analysis. |
| Rmd | 223406b | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | aca46cc | Peter Carbonetto | 2020-08-19 | Added manual clustering of droplet samples based on PCs. |
| Rmd | 38f811b | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| Rmd | 343747e | Peter Carbonetto | 2020-08-19 | Small edit to figure dimensions. |
| html | 5a35bbd | Peter Carbonetto | 2020-08-19 | Added labeled PCA plot; adjusted plot dimensions in |
| Rmd | fb91075 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 8b9b528 | Peter Carbonetto | 2020-08-19 | Added more PCA plots to plots_tracheal_epithelium analysis. |
| Rmd | ee7cbf1 | Peter Carbonetto | 2020-08-19 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | c517ea2 | Peter Carbonetto | 2020-08-18 | Small fix to one of the PCA plots in plots_tracheal_epithelium. |
| Rmd | 8f5c210 | Peter Carbonetto | 2020-08-18 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 01afbd2 | Peter Carbonetto | 2020-08-18 | Added some PC plots to the plots_tracheal_epithelium analysis. |
| Rmd | f1c7d02 | Peter Carbonetto | 2020-08-18 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| html | 0a04fc1 | Peter Carbonetto | 2020-08-18 | Added abundance plots to plots_tracheal_epithelium analysis. |
| Rmd | f914f7e | Peter Carbonetto | 2020-08-18 | wflow_publish(“plots_tracheal_epithelium.Rmd”) |
| Rmd | 61917ad | Peter Carbonetto | 2020-08-18 | Working on new analysis, plots_tracheal_epithelium.Rmd. |
Add introductory text here.
Load the packages used in the analysis below, as well as additional functions that we will use to generate some of the plots.
library(Matrix)
library(dplyr)
library(fastTopics)
library(ggplot2)
library(ggrepel)
library(cowplot)
source("../code/plots.R")Load data and results
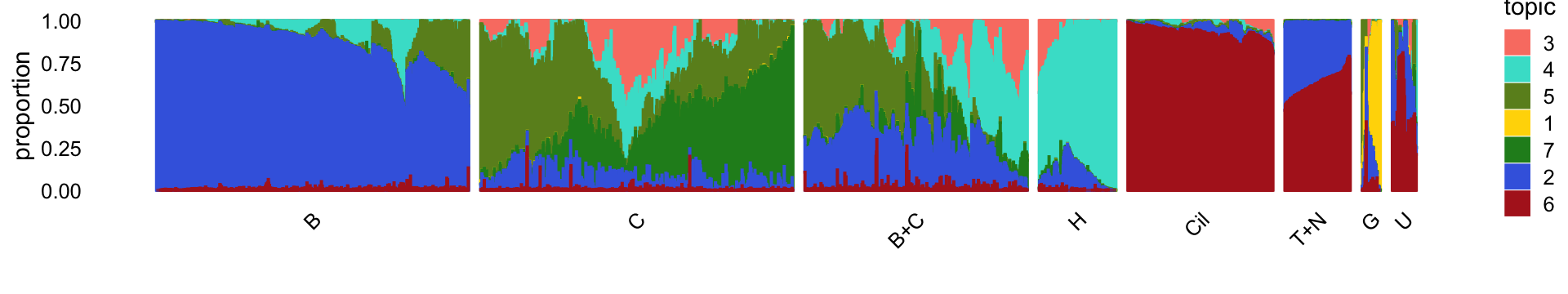
Load the smaller “droplet” data set, the \(k = 7\) Poisson NMF model fit for these data, and the 8 clusters identified in the clustering analysis.
load("../data/droplet.RData")
counts_droplet <- counts
samples_droplet <- readRDS("../output/droplet/clustering-droplet.rds")
fit_droplet <- readRDS("../output/droplet/rds/fit-droplet-scd-ex-k=7.rds")$fit
rm(samples,counts)For reference, we show here the Structure plot from the clustering analysis of the droplet data. This Structure plot summarizes the topic proportions in each of the 8 subsets.
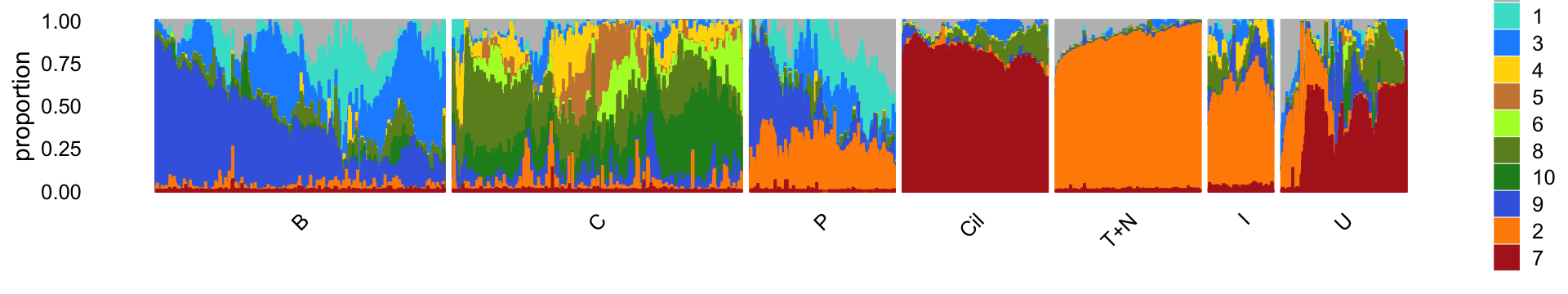
Next, load the larger “pulse-seq” data set, the \(k = 11\) Poisson NMF model fit for these data, and the 7 clusters identified in the clustering analysis.
load("../data/pulseseq.RData")
counts_pulseseq <- counts
samples_pulseseq <- readRDS("../output/pulseseq/clustering-pulseseq.rds")
fit_pulseseq <- readRDS("../output/pulseseq/rds/fit-pulseseq-scd-ex-k=11.rds")$fit
rm(samples,counts)For reference, we show here the Structure plot from the clustering analysis of the pulse-seq data. This Structure plot summarizes the topic proportions in each of the 7 subsets:
Differential expression analysis
Perform differential expression analysis using the topic model fitted to the droplet data.
timing <- system.time(
diff_count_droplet <- diff_count_analysis(fit_droplet,counts_droplet))
cat(sprintf("Computation took %0.2f seconds.\n",timing["elapsed"]))
# Fitting 18388 x 7 = 128716 univariate Poisson models.
# Computing log-fold change statistics.
# Computation took 21.57 seconds.
Warning: The above code chunk cached its results, but it won’t be re-run if previous chunks it depends on are updated. If you need to use caching, it is highly recommended to also set knitr::opts_chunk$set(autodep = TRUE) at the top of the file (in a chunk that is not cached). Alternatively, you can customize the option dependson for each individual chunk that is cached. Using either autodep or dependson will remove this warning. See the knitr cache options for more details.
Perform a differential expression analysis using the topic model fitted to the pulse-seq data.
timing <- system.time(
diff_count_pulseseq <- diff_count_analysis(fit_pulseseq,counts_pulseseq))
cat(sprintf("Computation took %0.2f seconds.\n",timing["elapsed"]))
# Fitting 21621 x 11 = 237831 univariate Poisson models.
# Computing log-fold change statistics.
# Computation took 1068.47 seconds.
Warning: The above code chunk cached its results, but it won’t be re-run if previous chunks it depends on are updated. If you need to use caching, it is highly recommended to also set knitr::opts_chunk$set(autodep = TRUE) at the top of the file (in a chunk that is not cached). Alternatively, you can customize the option dependson for each individual chunk that is cached. Using either autodep or dependson will remove this warning. See the knitr cache options for more details.
Now, calculate differential expression statistics using the clusters identified in the droplet data.
fit_clusters_droplet <-
init_poisson_nmf_from_clustering(counts_droplet,samples_droplet$cluster)
timing <- system.time(
diff_count_clusters_droplet <- diff_count_analysis(fit_clusters_droplet,
counts_droplet))
# All topic proportions are either zero or one; using simpler single-topic calculations for model parameter estimates
cat(sprintf("Computation took %0.2f seconds.\n",timing["elapsed"]))
# Fitting 18388 x 8 = 147104 univariate Poisson models.
# Computing log-fold change statistics.
# Computation took 30.32 seconds.
Warning: The above code chunk cached its results, but it won’t be re-run if previous chunks it depends on are updated. If you need to use caching, it is highly recommended to also set knitr::opts_chunk$set(autodep = TRUE) at the top of the file (in a chunk that is not cached). Alternatively, you can customize the option dependson for each individual chunk that is cached. Using either autodep or dependson will remove this warning. See the knitr cache options for more details.
And do the same in the pulse-seq data.
fit_clusters_pulseseq <-
init_poisson_nmf_from_clustering(counts_pulseseq,samples_pulseseq$cluster)
timing <- system.time(
diff_count_clusters_pulseseq <- diff_count_analysis(fit_clusters_pulseseq,
counts_pulseseq))
# All topic proportions are either zero or one; using simpler single-topic calculations for model parameter estimates
cat(sprintf("Computation took %0.2f seconds.\n",timing["elapsed"]))
# Fitting 21621 x 7 = 151347 univariate Poisson models.
# Computing log-fold change statistics.
# Computation took 540.97 seconds.
Warning: The above code chunk cached its results, but it won’t be re-run if previous chunks it depends on are updated. If you need to use caching, it is highly recommended to also set knitr::opts_chunk$set(autodep = TRUE) at the top of the file (in a chunk that is not cached). Alternatively, you can customize the option dependson for each individual chunk that is cached. Using either autodep or dependson will remove this warning. See the knitr cache options for more details.
Ciliated cells
We begin with the cluster that captures ciliated cells in the droplet data. This cluster is one of the most distinctive in both the droplet and pulse-seq data sets.
cilitated_genes <- c("Ccdc113","Ccdc153","Cdhr3","Foxj1","Lztfl1","Mlf1")
p1 <- volcano_plot_with_highlighted_genes(diff_count_clusters_droplet,"Cil",
cilitated_genes,
label_above_quantile = 0.998)
print(p1)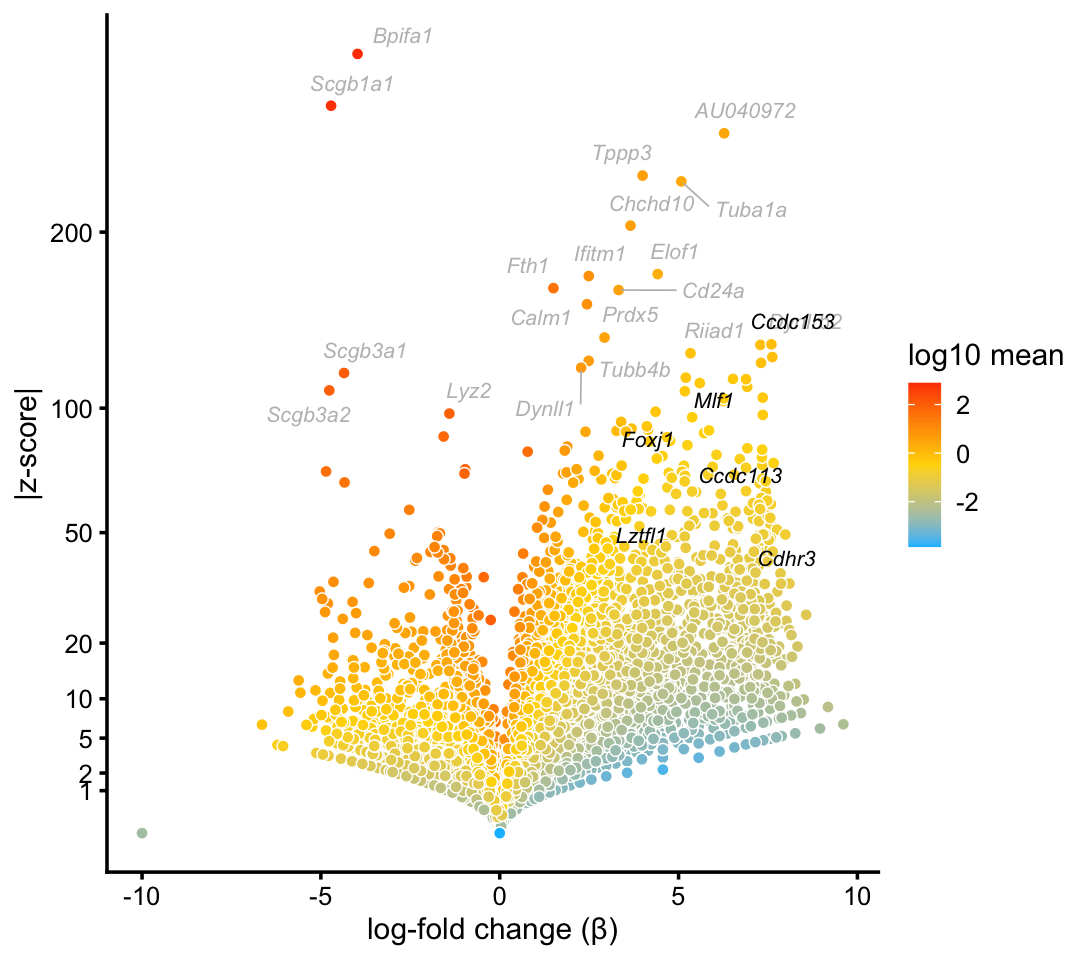
In this volcano plot, marker genes and transcription factors identified in Montoro et al (2018) are highlighted in black, and other top differentially expressed genes are shown with gray labels.
We obtain similar top differentially expressed genes in the pulse-seq data:
p2 <- volcano_plot_with_highlighted_genes(diff_count_clusters_pulseseq,"Cil",
cilitated_genes,
label_above_quantile = 0.998)
print(p2)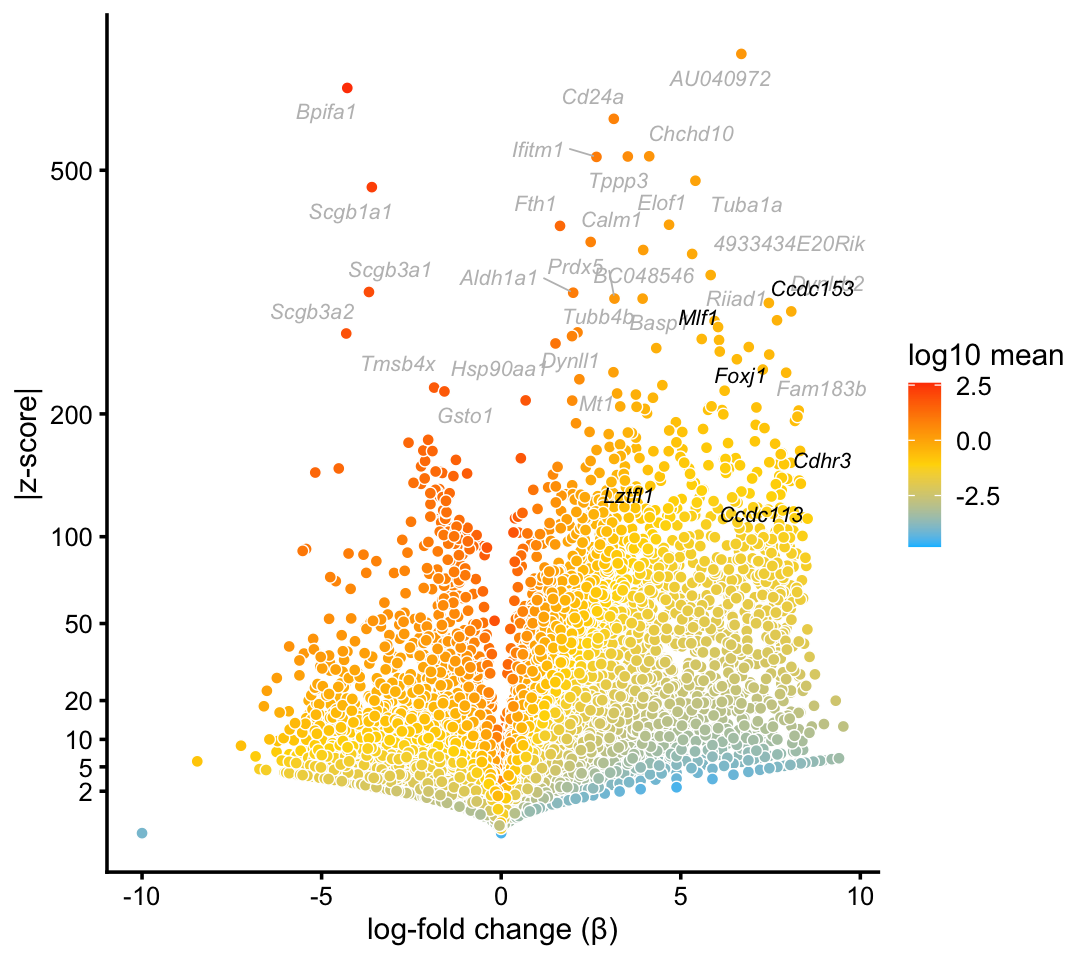
Ionocytes
In the pulse-seq data, we identify a distinctive cluster for the newly discovered ionocyte cell type. Gene expression in these cells is not fully captured by any single topic.
ionocyte_genes <- c("Ascl3","Asgr1","Atp6v0d2","Atp6v1c2","Cftr","Foxi1",
"Moxd1","P2ry14","Stap1")
p3 <- volcano_plot_with_highlighted_genes(diff_count_clusters_pulseseq,"I",
ionocyte_genes,
label_above_quantile = 0.998)
print(p3)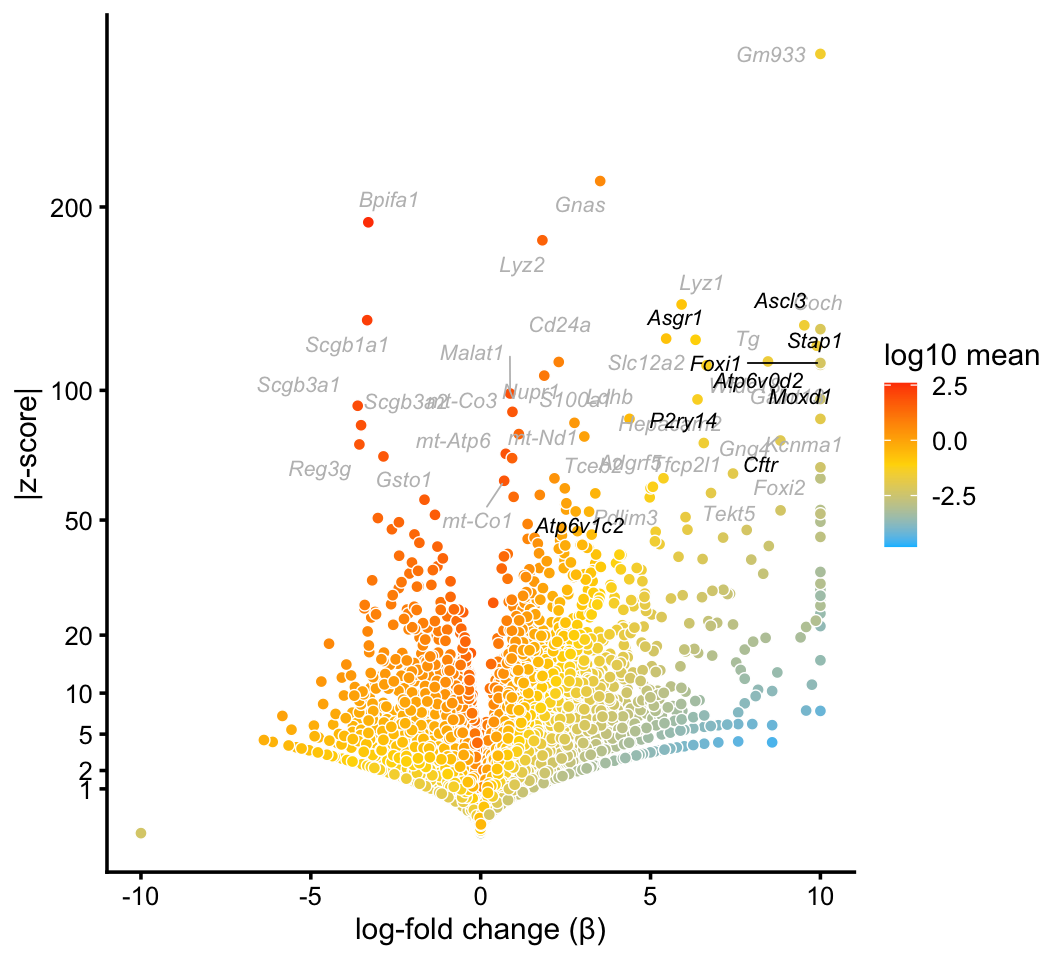
We do not identify a cluster for ionocytes in the droplet data, probably because only a handful of cells in this data set appear to be ionocytes, at least judging by expression of the Foxi1 ionocyte marker gene:
pca <- prcomp(poisson2multinom(fit_droplet)$L)$x
pdat <- data.frame(PC1 = pca[,1],
PC2 = pca[,2],
Foxi1 = counts_droplet[,"Foxi1"])
p4 <- ggplot(pdat,aes(x = PC1,y = PC2,fill = Foxi1)) +
geom_point(shape = 21,color = "white",size = 1.25) +
scale_fill_gradientn(colors = c("skyblue","gold","darkorange","magenta"),
na.value = "lightskyblue") +
theme_cowplot(font_size = 10) +
labs(fill = "expression level")
print(p4)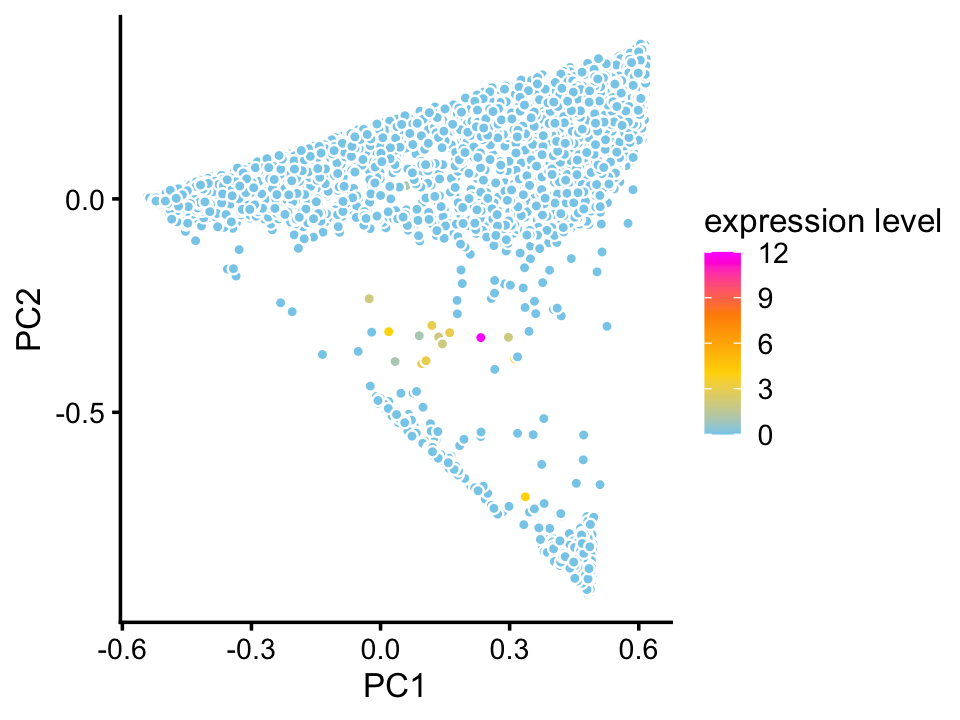
Goblet cells
Volcano plot for “G” cluster in droplet data:
genes_goblet <- "Gp2"
genes <- colnames(counts_droplet)
p5 <- volcano_plot(diff_count_clusters_droplet,k = "G",labels = genes) +
labs(title = "")
print(p5)
sessionInfo()
# R version 3.6.2 (2019-12-12)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS Catalina 10.15.6
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] cowplot_1.0.0 ggrepel_0.9.0 ggplot2_3.3.0 fastTopics_0.3-175
# [5] dplyr_0.8.3 Matrix_1.2-18
#
# loaded via a namespace (and not attached):
# [1] Rcpp_1.0.5 lattice_0.20-38 tidyr_1.0.0
# [4] prettyunits_1.1.1 assertthat_0.2.1 zeallot_0.1.0
# [7] rprojroot_1.3-2 digest_0.6.23 R6_2.4.1
# [10] backports_1.1.5 MatrixModels_0.4-1 evaluate_0.14
# [13] coda_0.19-3 httr_1.4.1 pillar_1.4.3
# [16] rlang_0.4.5 progress_1.2.2 lazyeval_0.2.2
# [19] data.table_1.12.8 irlba_2.3.3 SparseM_1.78
# [22] whisker_0.4 rmarkdown_2.3 labeling_0.3
# [25] Rtsne_0.15 stringr_1.4.0 htmlwidgets_1.5.1
# [28] munsell_0.5.0 compiler_3.6.2 httpuv_1.5.2
# [31] xfun_0.11 pkgconfig_2.0.3 mcmc_0.9-6
# [34] htmltools_0.4.0 tidyselect_0.2.5 tibble_2.1.3
# [37] workflowr_1.6.2.9000 quadprog_1.5-8 viridisLite_0.3.0
# [40] crayon_1.3.4 withr_2.1.2 later_1.0.0
# [43] MASS_7.3-51.4 grid_3.6.2 jsonlite_1.6
# [46] gtable_0.3.0 lifecycle_0.1.0 git2r_0.26.1
# [49] magrittr_1.5 scales_1.1.0 RcppParallel_4.4.2
# [52] stringi_1.4.3 farver_2.0.1 fs_1.3.1
# [55] promises_1.1.0 vctrs_0.2.1 tools_3.6.2
# [58] glue_1.3.1 purrr_0.3.3 hms_0.5.2
# [61] yaml_2.2.0 colorspace_1.4-1 plotly_4.9.2
# [64] knitr_1.26 quantreg_5.54 MCMCpack_1.4-5