Enichment-analysis-goseq
unawaz1996
2023-03-16
Last updated: 2023-05-11
Checks: 7 0
Knit directory: NMD-analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230314) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 6025d0f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Differential-transcript-usage.nb.html
Ignored: analysis/Enichment-analysis-fgsea.nb.html
Ignored: analysis/Enichment-analysis-goseq.nb.html
Untracked files:
Untracked: PCA.png
Untracked: PCA_plot.pdf
Untracked: PCA_transcript.png
Untracked: analysis/Differential-transcript-usage.Rmd
Untracked: analysis/UPF3B_KD.Rmd
Untracked: analysis/transcript-preprocessing.Rmd
Untracked: code/eisaR.R
Untracked: code/external_code/
Untracked: data/LTK_Sample Metafile_V3.txt
Untracked: data/Mus_musculus.GRCm39.105__nifs.tsv
Untracked: data/data.txt
Untracked: data/data2.txt
Untracked: data/fastqc/
Untracked: data/nif_output/
Untracked: data/samples.txt
Untracked: output/DEG-limma-results.Rda
Untracked: output/DEG-list.Rda
Untracked: output/DEG/
Untracked: output/EISA/
Untracked: output/ISAR/
Untracked: output/QC/
Untracked: output/Transcript/
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_complete.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_1_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_2_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_3_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_nt.fasta
Untracked: output/limma-matrices.Rda
Untracked: tmp/
Unstaged changes:
Modified: analysis/RNA-stability.Rmd
Modified: analysis/_site.yml
Modified: code/functions.R
Modified: code/libraries.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Enichment-analysis-goseq.Rmd) and HTML
(docs/Enichment-analysis-goseq.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 6025d0f | unawaz1996 | 2023-05-11 | wflow_publish(c("analysis/index.Rmd", "analysis/Differential-transcript-expression.Rmd", |
| html | 650317e | unawaz1996 | 2023-03-22 | Build site. |
| Rmd | f2ed11a | unawaz1996 | 2023-03-22 | wflow_publish(c("analysis/index.Rmd", "analysis/Enichment-analysis-fgsea.Rmd", |
| html | 1f4ee28 | unawaz1996 | 2023-03-22 | Build site. |
| Rmd | 917d8df | unawaz1996 | 2023-03-22 | wflow_publish(c("analysis/index.Rmd", "analysis/Enichment-analysis-fgsea.Rmd", |
| html | 0e23a3b | unawaz1996 | 2023-03-17 | Build site. |
| Rmd | 84be588 | unawaz1996 | 2023-03-17 | Enrichment analysis |
In this workbook, we are testing for enrichment within discrete sets of DE genes as defined in the DEG analysis.
In order to do this, we will be using goseq, an R
package that performs enrichment analysis whilst taking length bias into
account. It does so by calculating a Probability Weighting Function
(PWF), which gives the probability that a gene will be differentially
expressed based on length alone.
The PWD is calculated by fitting a monotonic spline to the binary data series of differential expression (1 = DE, 0= Not DE) as a function of gene length. The PWF is used to weight the chance of selecting each gene when forming a null distribution for GO category membership. The fact that the PWF is calculated directly from the dataset under consideration makes this approach robust, only correcting for the length bias present in the data.
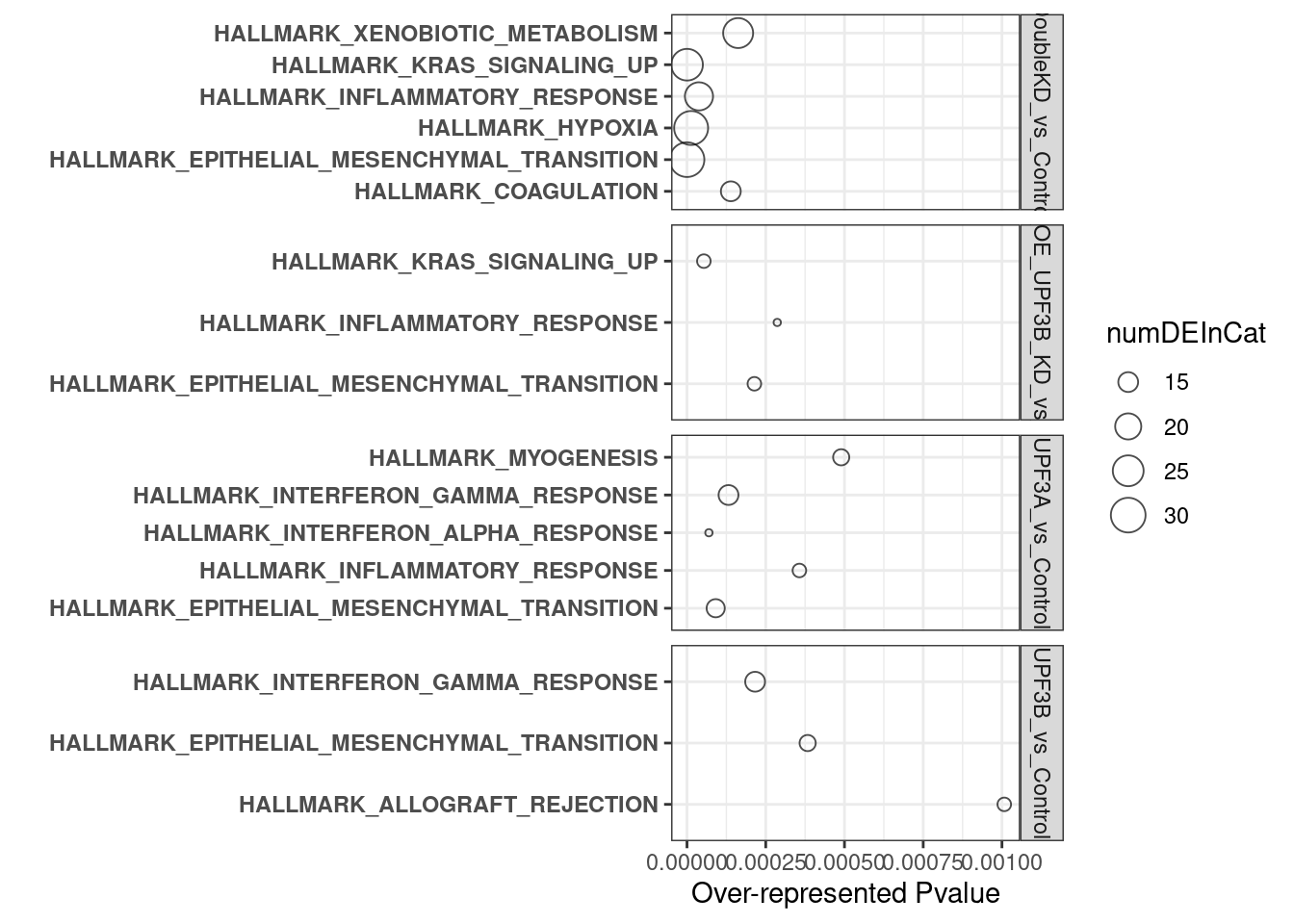
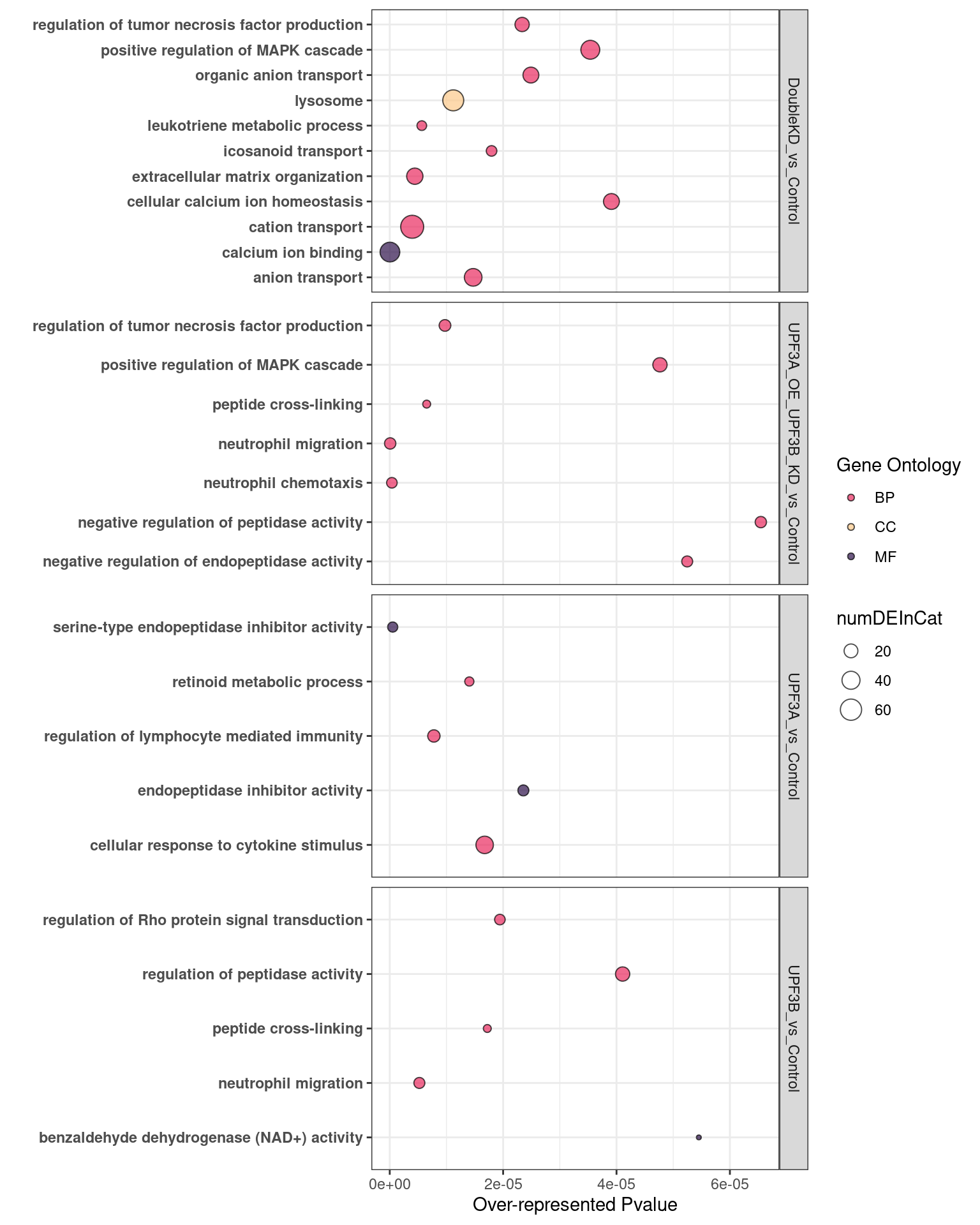
Results can be interpreted as following:
overrpresented p-value: Over representation in this analysis means that there are more DE genes in categort than expected. P-value relates to the probability of observing this number of DE genes in category by chancenumDEinCat: number of DE genes in categoryExpected: Expected number of genes in categoryadjP: Bonferroni adjustment of over represented p-valueFDR: FDR adjustment of over represented p-value
Databases used for testing
Data was sourced using the misigdbr package.
Hallmark gene sets
Mappings were required from gene to pathway, and Ensembl identifies were used to map from gene to pathway. A total of 4,391 Ensembl IDs were mapped to pathways from the Hallmark set.
C2 gene set
The same mapping process was applied to datasets from c2 signatures. For this analysis, only Wikipathways, KEGG and Reactome gene sets were retrieved, and a total of 11,774 Ensembl IDs were mapped to c2 genesets.
Gene Ontology Gene set
For the analysis of gene-sets from GO database, gene-sets were restricted to those with 3 or more steps back to the ontology root terms.
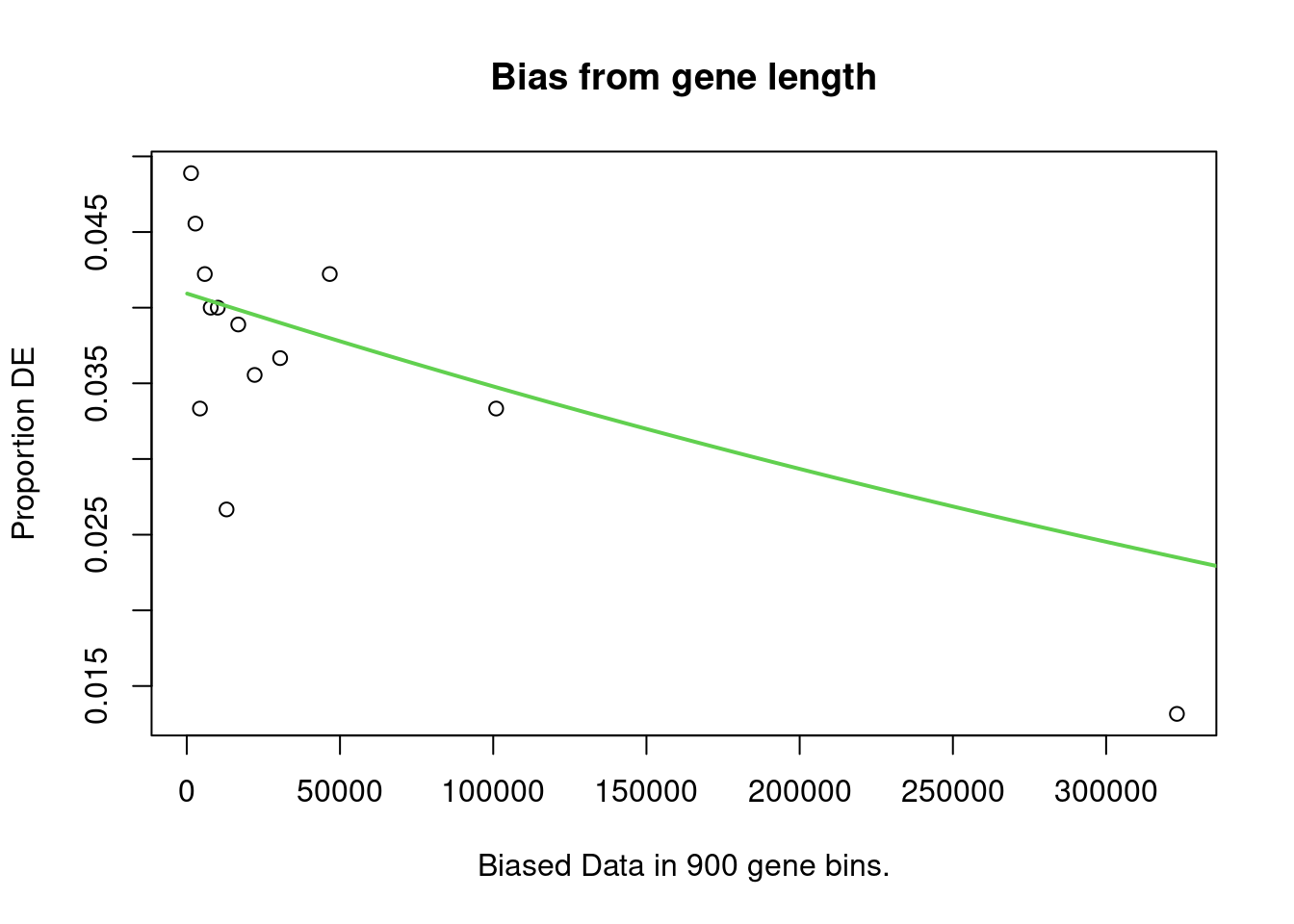
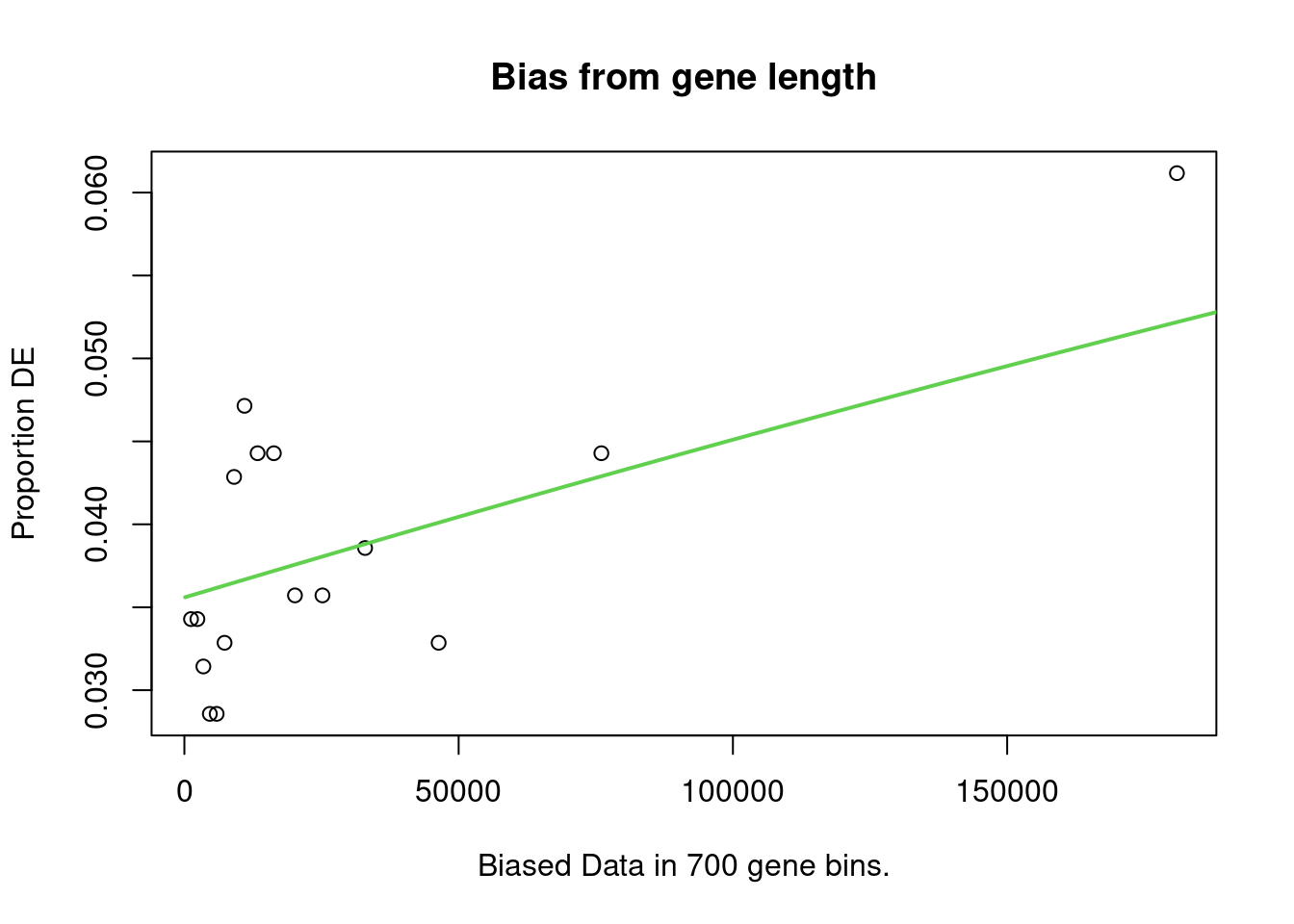
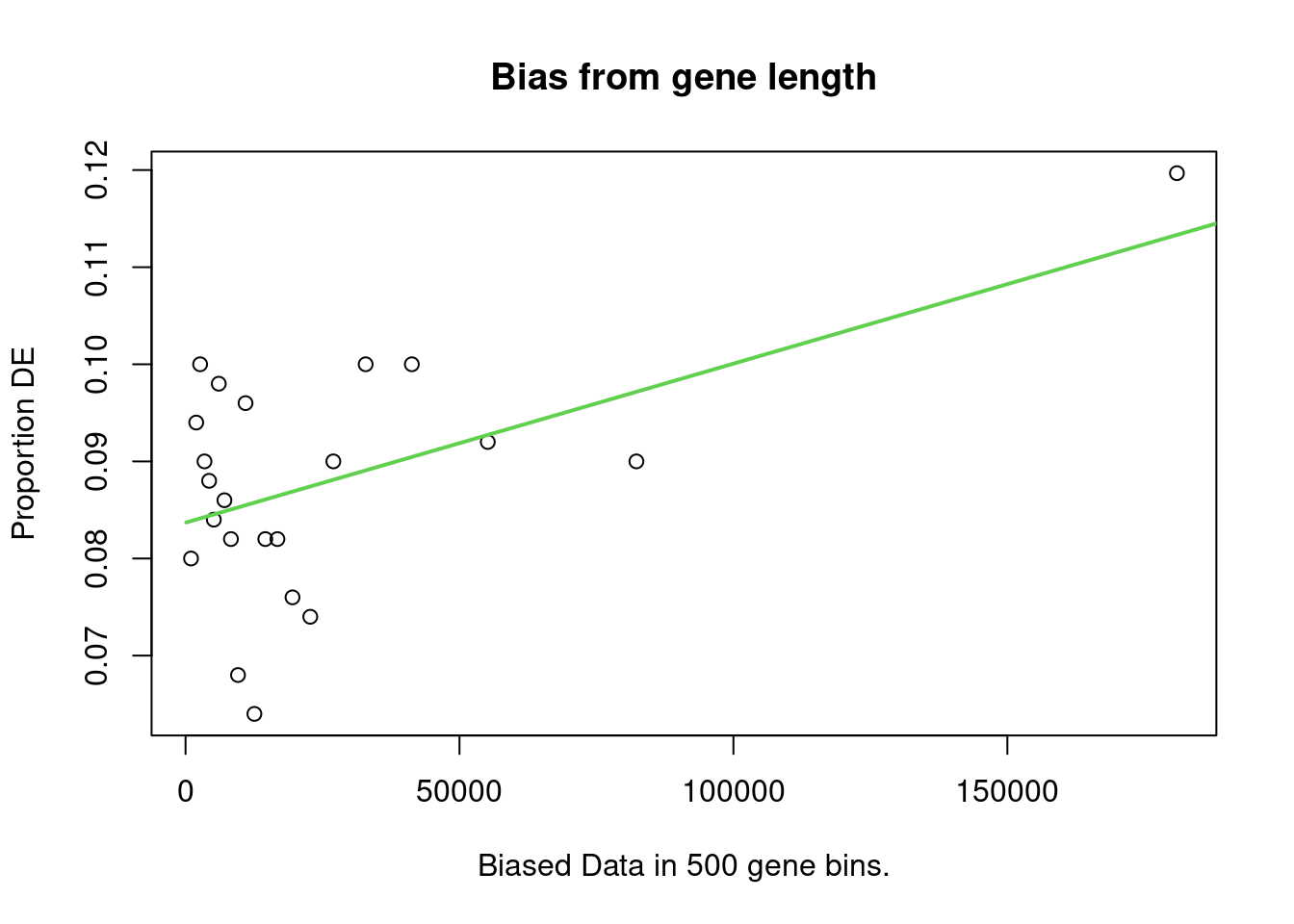
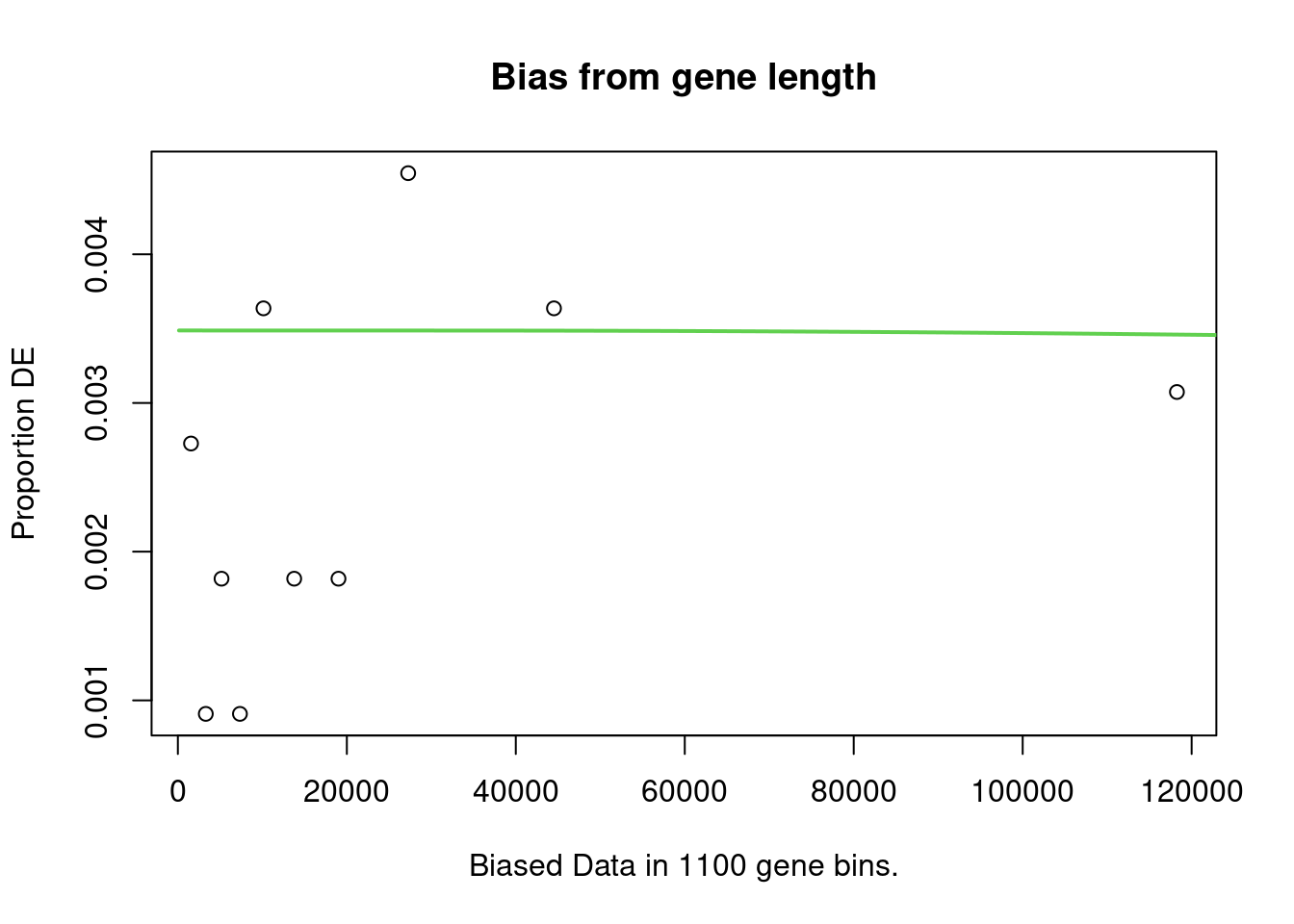
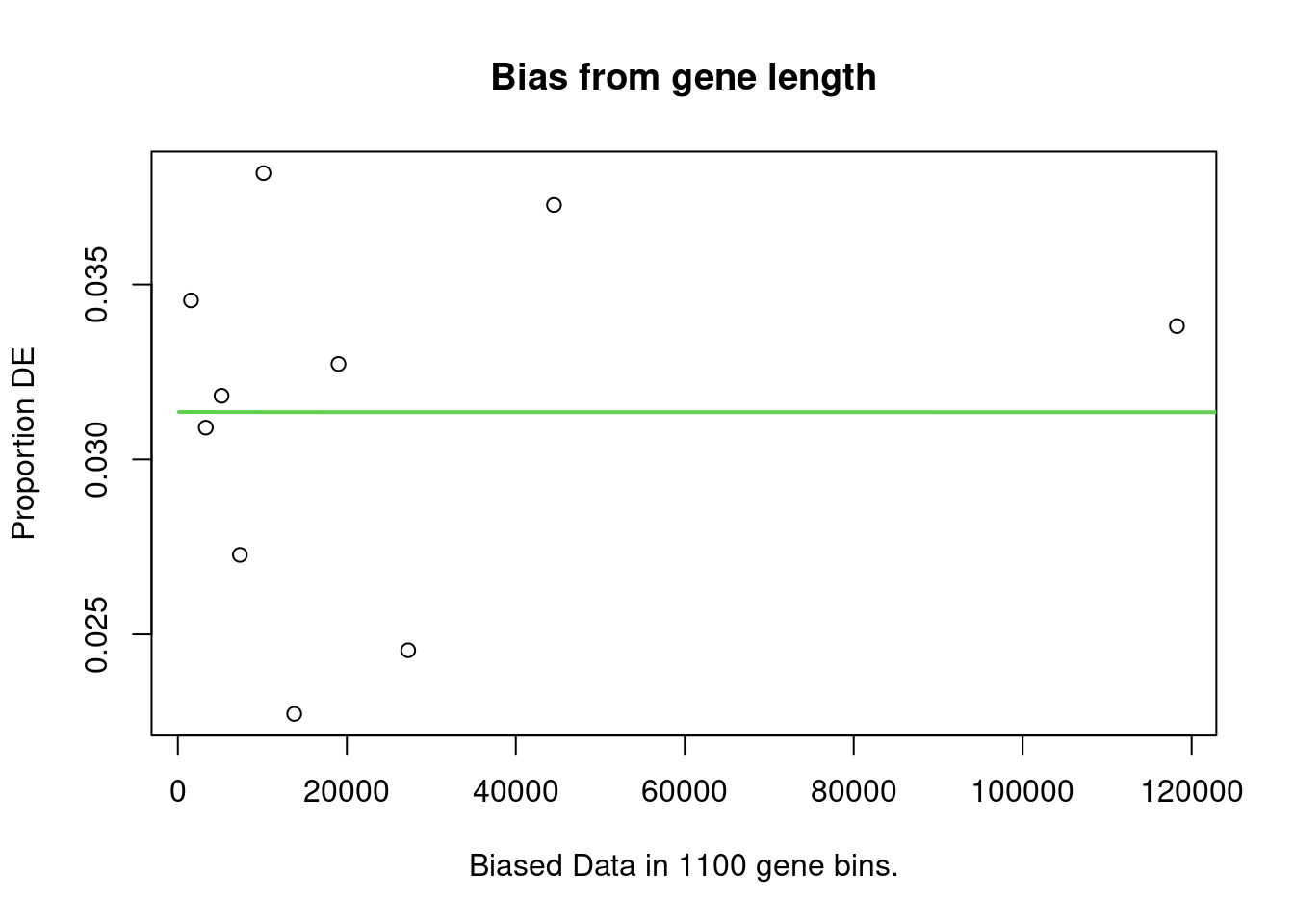
Enrichment in the DE Gene Set
The first step of analysis using goseq, regardless of the gene-set, is estimation of the probability weight function (PWF) which quantifies the probability of a gene being considered as DE based on a single covariate.
GO terms
For gene ontology analysis, we will be using the GO summaries method.
Essentially, this analysis involves creating a graph for each ontology
term, and removing the node all, as this redundant. For
each GO term, we get the ontology it belongs to, the shortest path back
to the root note, the longest path to the root node and whether it GO
term is a terminal node.
UPF3B KD vs Controls
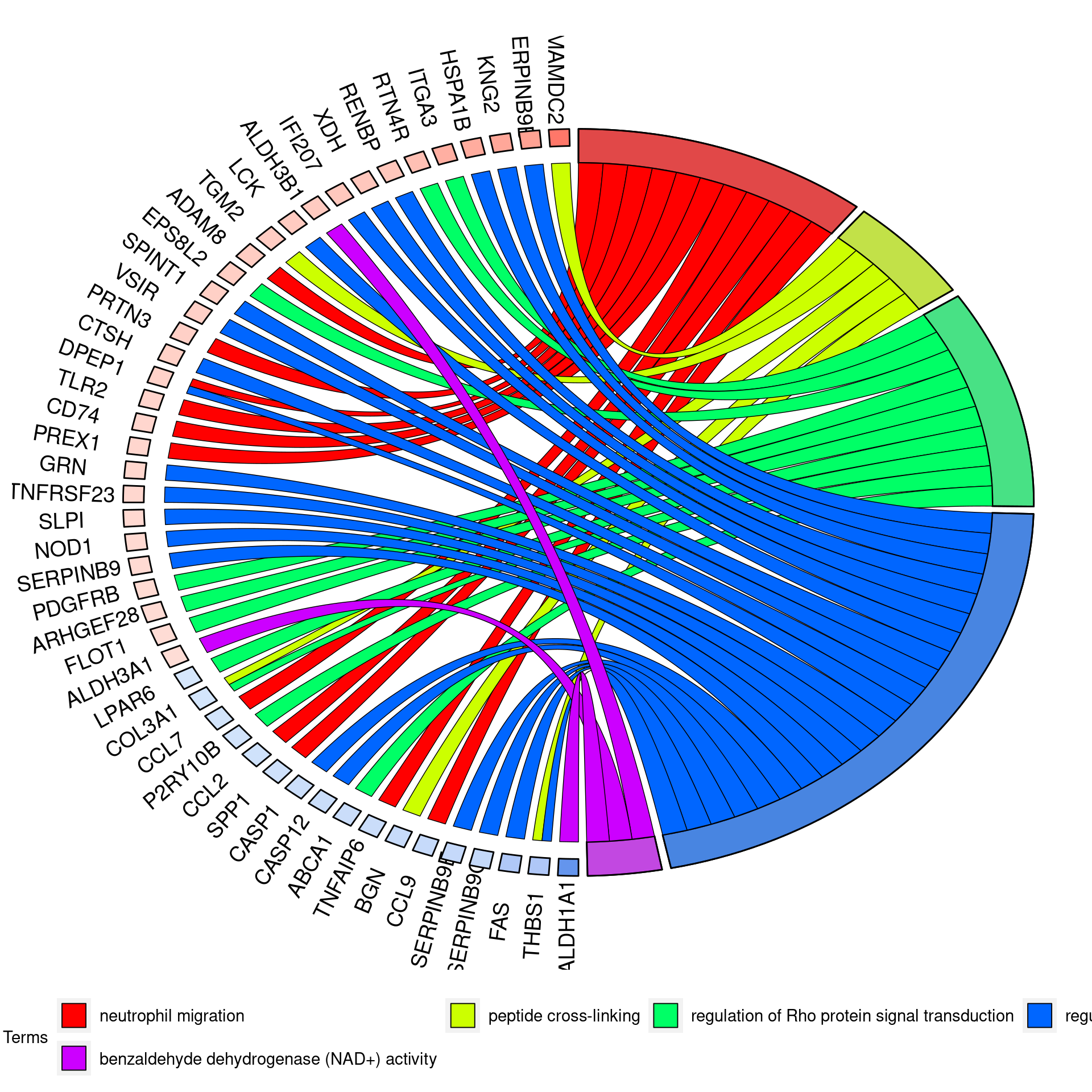
UPF3A KD vs Controls
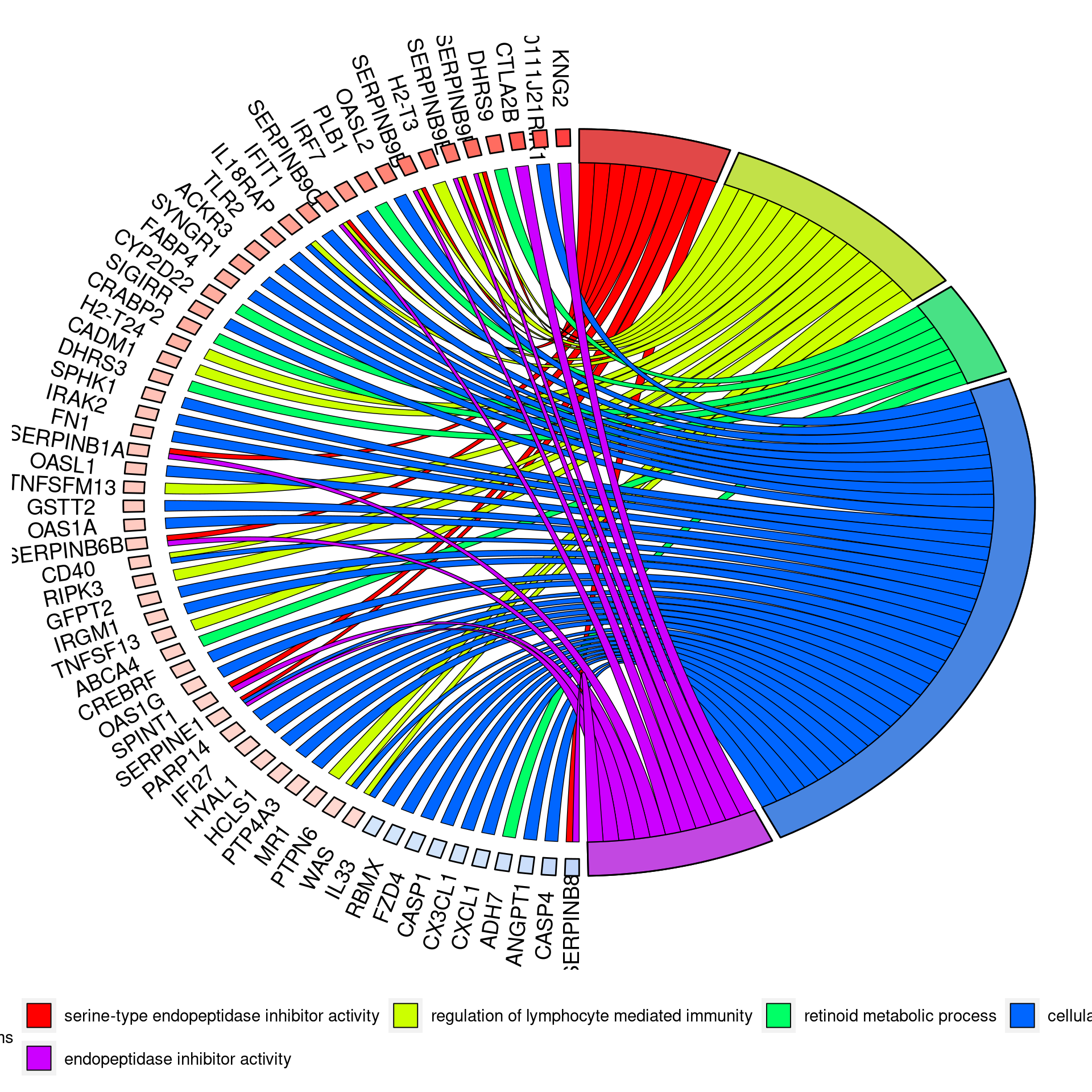
Double KD
UPF3A OE
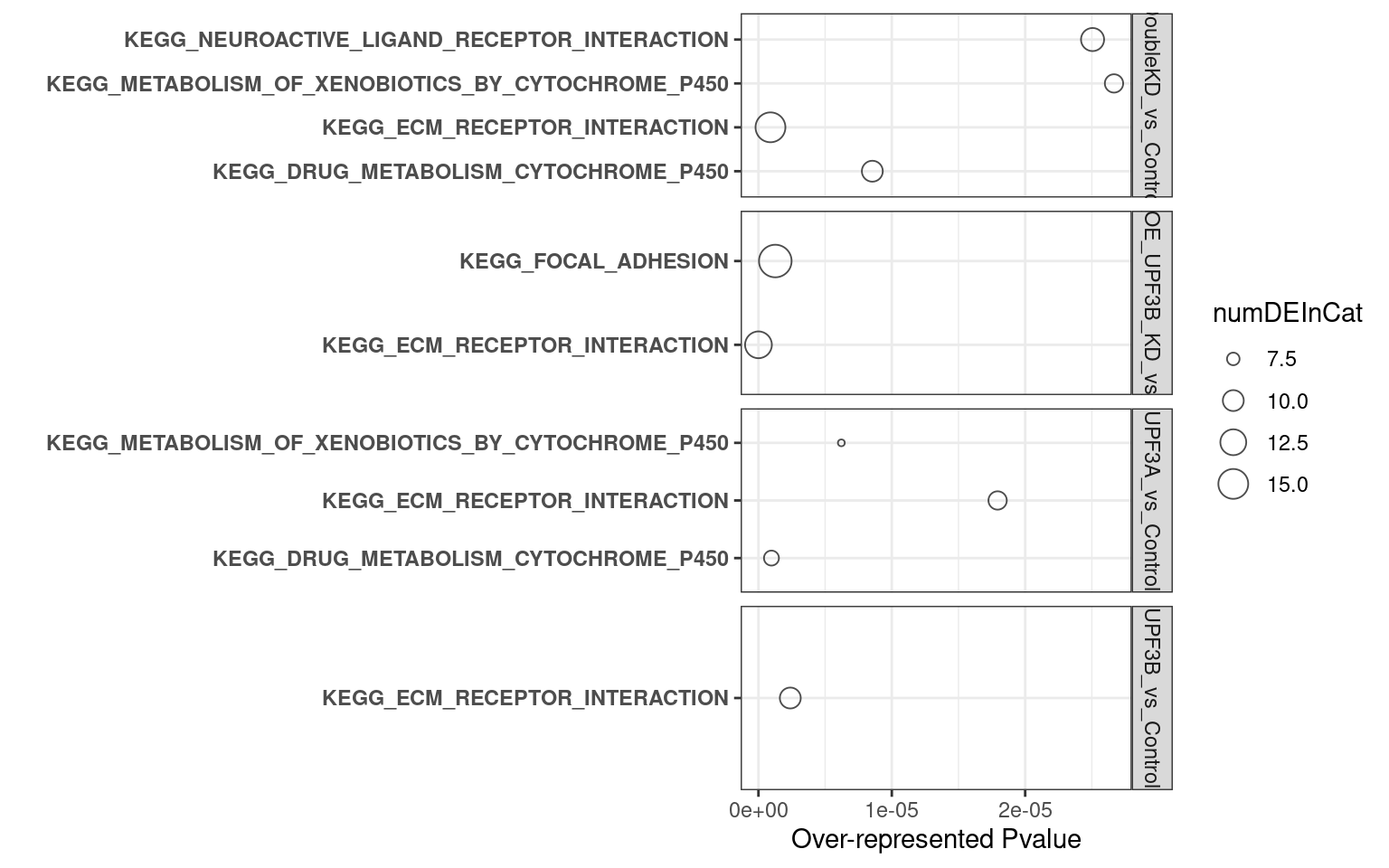
2. C2 database
- UPF3B KD vs Controls
- UPF3A KD vs Controls
- Double KDs vs Controls
- UPF3A OE vs Controls
- UPF3A OE UPF3B KD vs Controls
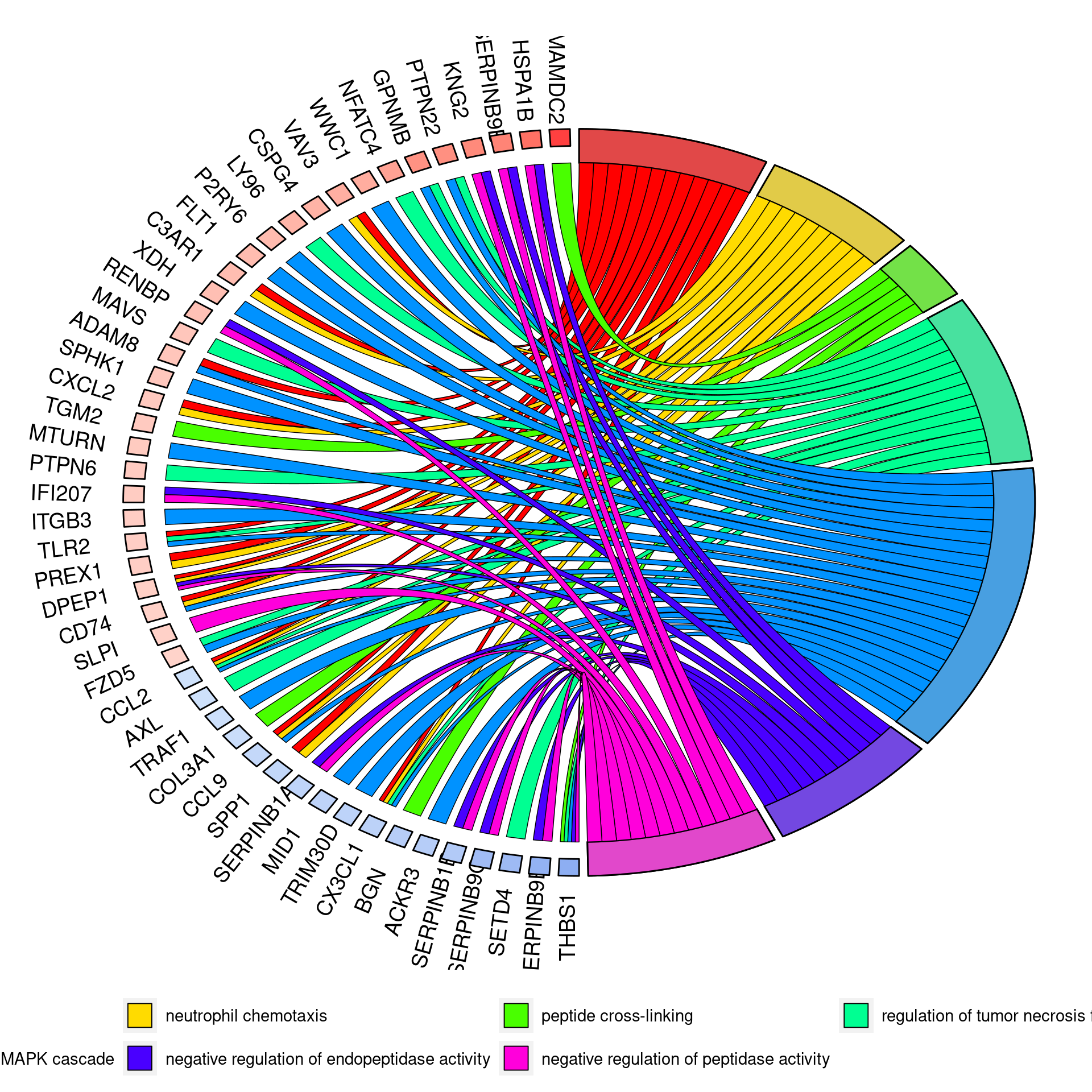
Results summary
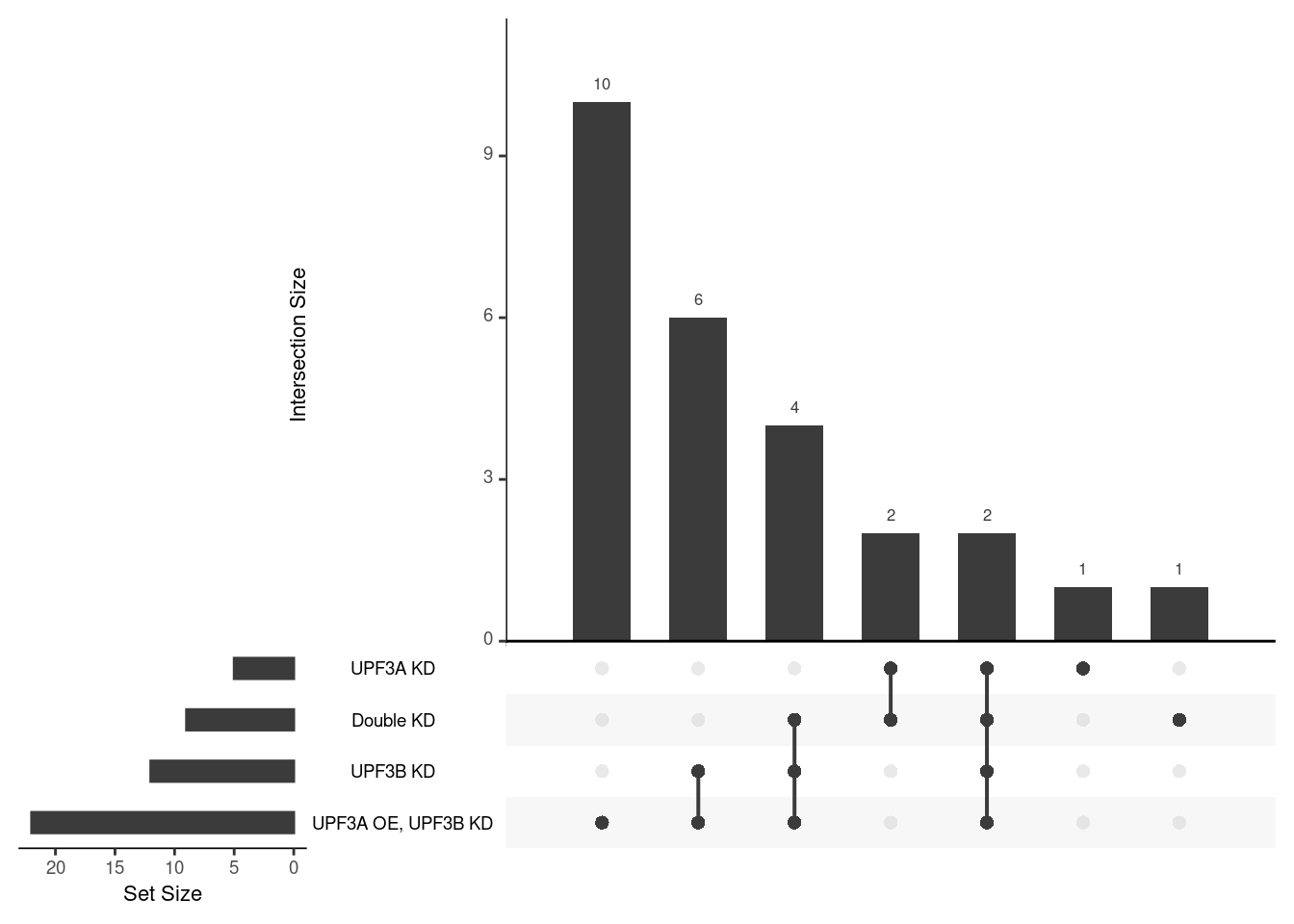
Overlap of terms between enrichment analyses of different groups
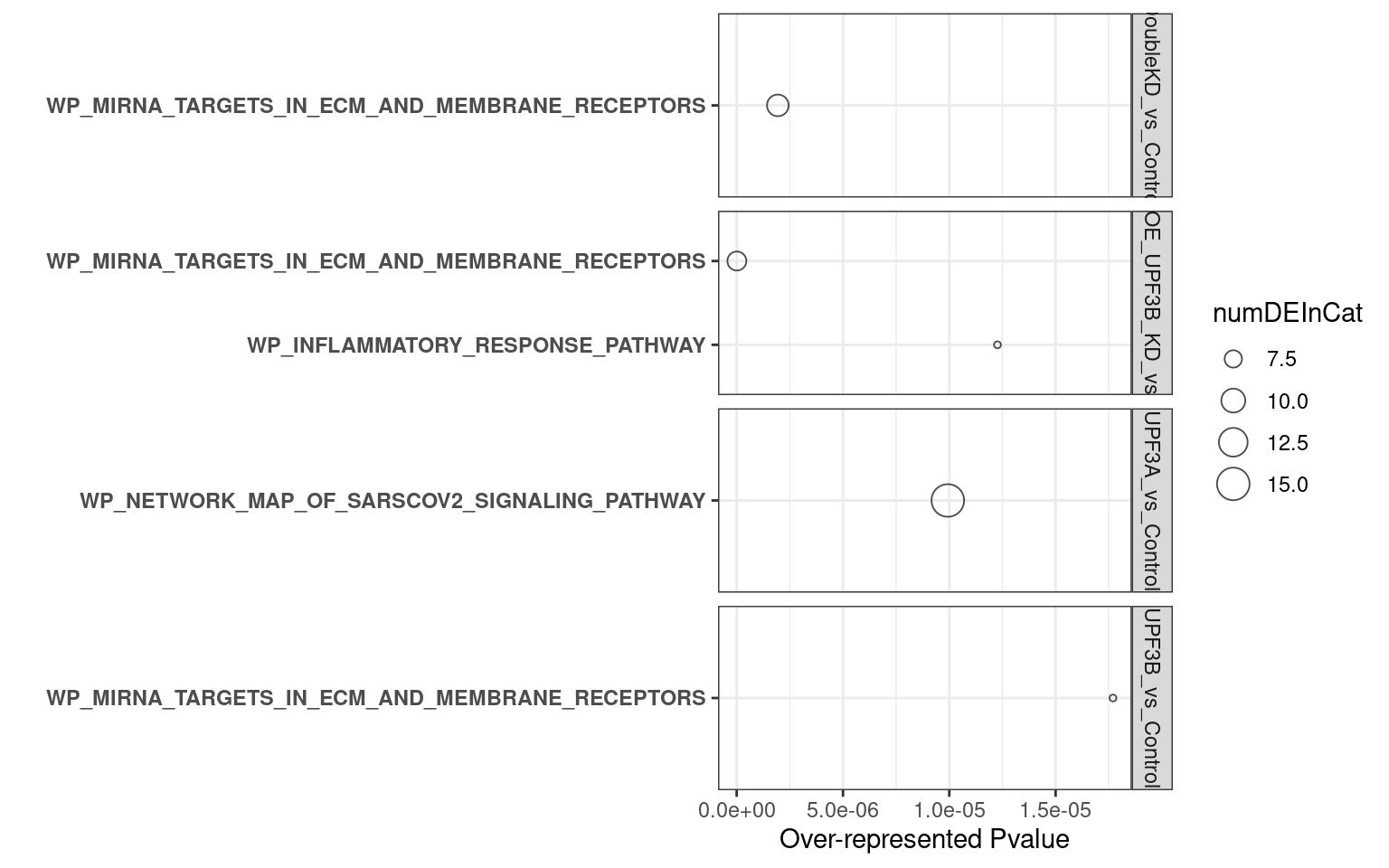
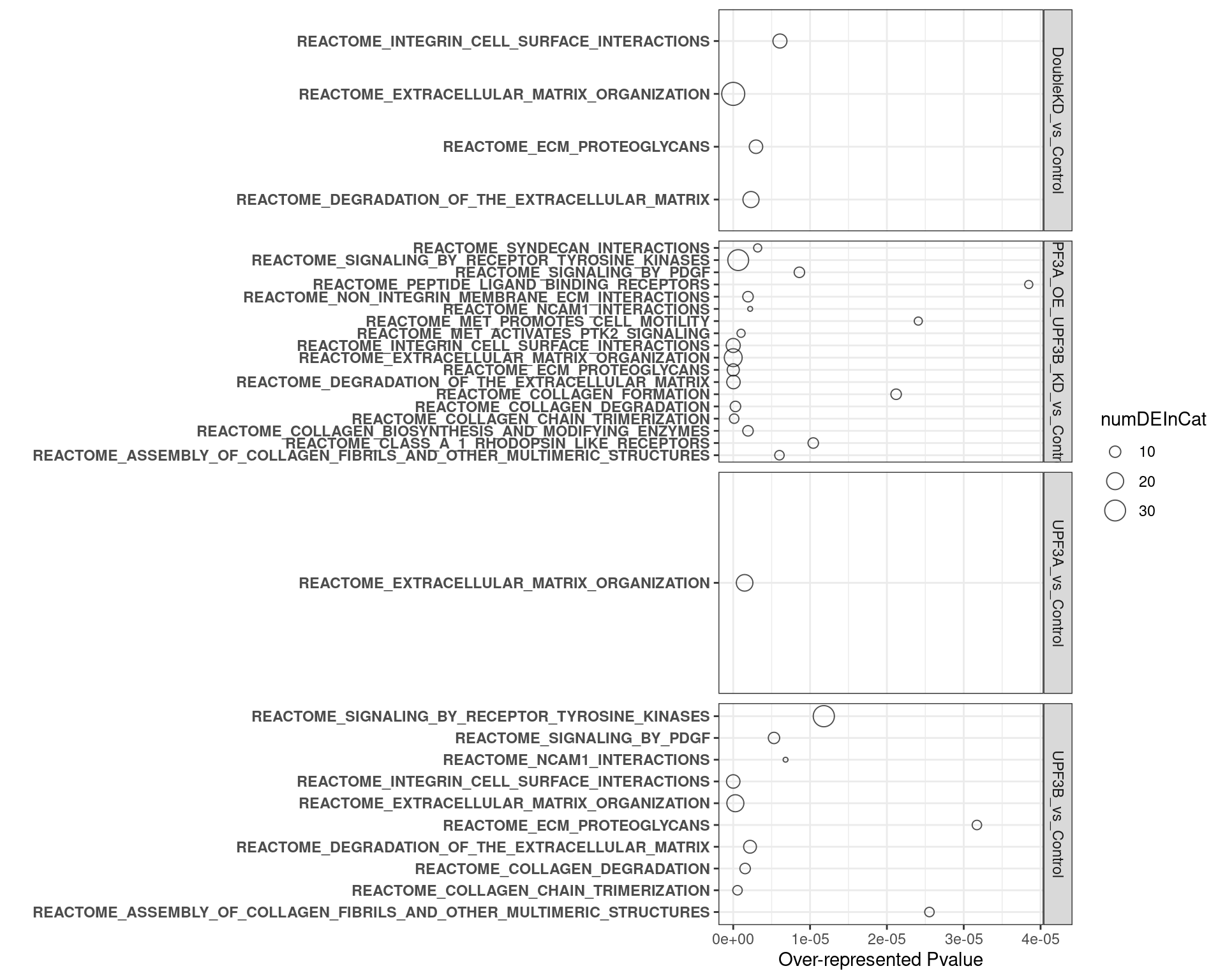
Hallmark datasets
- UPF3B KD vs Controls
- UPF3A KD vs Controls
- Double KD vs Controls
- UPF3A OE vs Controls
- UPF3A OE vs Controls
Results summary
- Overlap of Hallmark gene sets
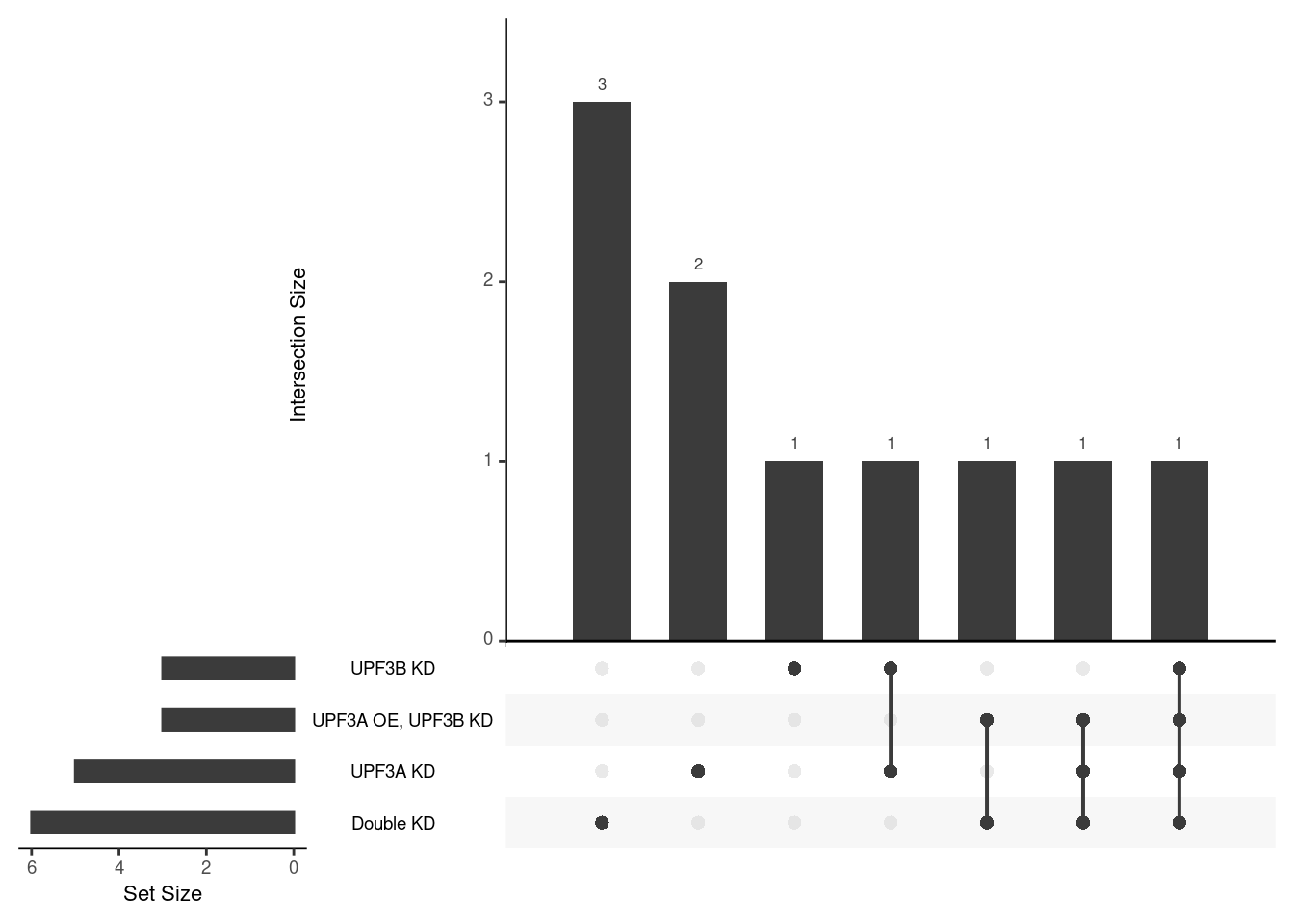
- Summary plots