DLPFC (Maynard et al., 2021)
Zheng Li
2022-02-24
Last updated: 2022-03-05
Checks: 7 0
Knit directory: BASS-analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(0) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b3e7dcf. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Unstaged changes:
Deleted: data/spatialLIBD.RData
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/DLPFC.Rmd) and HTML (docs/DLPFC.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b3e7dcf | zhengli09 | 2022-03-05 | Separate spatialLIBD data into three parts, each corresponding to an |
| html | 68275b2 | zhengli09 | 2022-03-05 | Build site. |
| Rmd | e9eebcb | zhengli09 | 2022-03-05 | Add DLPFC analysis |
Introduction
Here, we apply BASS to analyze the DLPFC (human dorsolateral prefrontal cortex) dataset from Maynard et al., 2021. DLPFC data contains expression values of 33,538 genes measured on two pairs of tissue sections from three independent neurotypical adult donors. Each pair consisted of two directly adjacent, 10 \(\mu m\) serial tissue sections with the second pair located 300 \(\mu m\) posterior to the first, resulting in a total of 12 tissue sections. The original data can be downloaded from here. We excluded spots that are not mapped to the tissue region in the histology image and retained a total of 33,538 genes measured on 4,226 (151507), 4,384 (151508), 4,789 (151509), 4,634 (151510), 3,661 (151669), 3,498 (151670), 4,110 (151671), 4,015 (151672), 3,639 (151673), 3,673 (151674), 3,592 (151675), and 3,460 (151676) spots along with their spatial locations for further analysis. The processed data can be download from the data directory. We focused our analysis only on spatial domain detection because the clustering of spatial spots no longer has the cell type interpretation. For single-sample analysis, we analyzed each of the 12 tissue sections separately. For multi-sample analysis, we jointly analyzed four tissue sections from each adult donor because they contain similar tissue structures.
library(BASS)
# cntm: a list of expression count matrices for 4 tissue sections
# (151673-151676) from the same individual
# infom: a list of manually annotated labels of seven laminar
# clusters by the original study for 4 tissue sections
# (151673-151676) from the same individual
# xym: a list of spatial coordinates for 4 tissue sections
# (151673-151676) from the same individual
load("data/spatialLIBD_p3.RData")
# hyper-parameters
# We set the number of cell types to a relatively large
# number (20) to capture the expression heterogeneity.
C <- 20
# number of spatial domains
R <- 7Single-sample analysis
Run BASS
smp <- "151673"
set.seed(0)
# Set up BASS object
BASS <- createBASSObject(cntm[smp], xym[smp], C = C, R = R,
beta_est_approach = "ACCUR_EST", init_method = "mclust",
burn_in = 10000, samples = 10000)Loading required package: MatrixExpression data coerced to a matrix
***************************************
Bayesian Analytics for Spatial Segmentation (BASS)
Authors: Zheng Li, Xiang Zhou
Affiliate: Department of Biostatistics, University of Michigan
INPUT INFO:
- Number of samples: 1
- Number of spots/cells: 3639
- Number of genes: 33538
- Potts interaction parameter estimation approach: ACCUR_EST
- Variance-covariance structure of gene expression features: EEE
To list all hyper-parameters, Type listAllHyper(BASS_object)
***************************************# Data pre-processing:
# 1.Library size normalization followed with a log2 transformation
# 2.Select top 3000 spatially expressed genes with SPARK-X
# 3.Dimension reduction with PCA
BASS <- BASS.preprocess(BASS, doLogNormalize = TRUE,
geneSelect = "sparkx", nSE = 3000,
doPCA = TRUE, scaleFeature = FALSE, nPC = 20)***** Log-normalize gene expression data *****
***** Select spatially expressed genes with sparkx *****
***** Exclude genes with 0 expression *****
***** Reduce data dimension with PCA *****# Run BASS algorithm
BASS <- BASS.run(BASS)# post-process posterior samples:
# 1.Adjust for label switching with the ECR-1 algorithm
# 2.Summarize the posterior samples to obtain the spatial domain labels
BASS <- BASS.postprocess(BASS)
......................................................................................
. Method Time (sec) Status .
......................................................................................
. ECR-ITERATIVE-1 178.602 Converged (3 iterations) .
......................................................................................
Relabelling all methods according to method ECR-ITERATIVE-1 ... done!
Retrieve the 1 permutation arrays by typing:
[...]$permutations$"ECR-ITERATIVE-1"
Retrieve the 1 best clusterings: [...]$clusters
Retrieve the 1 CPU times: [...]$timings
Retrieve the 1 X 1 similarity matrix: [...]$similarity
Label switching finished. Total time: 185.9 seconds.
......................................................................................
. Method Time (sec) Status .
......................................................................................
. ECR-ITERATIVE-1 98.758 Converged (3 iterations) .
......................................................................................
Relabelling all methods according to method ECR-ITERATIVE-1 ... done!
Retrieve the 1 permutation arrays by typing:
[...]$permutations$"ECR-ITERATIVE-1"
Retrieve the 1 best clusterings: [...]$clusters
Retrieve the 1 CPU times: [...]$timings
Retrieve the 1 X 1 similarity matrix: [...]$similarity
Label switching finished. Total time: 105.8 seconds. zlabels <- BASS@res_postprocess$z_ls # spatial domain labelsVisualization
You can refer to visualization for some useful plotting functions or you can write your own code for plotting.
library(ggplot2)
source("code/viz.R")
# Spatial domains
zlabels_l <- factor(zlabels[[1]])
levels(zlabels_l) <- c("L4", "L3", "L5", "L6", "L2", "L1", "WM")
zlabels_l <- factor(zlabels_l, c("L1", "L2", "L3", "L4", "L5", "L6", "WM"))
plotClusters(xym[[smp]], labels = zlabels_l, title = "Spatial domains",
flip_xy = T, flip_y = T, ratio = 1.5) +
theme(legend.position = "bottom") +
scale_color_viridis_d(name = "Spatial domain")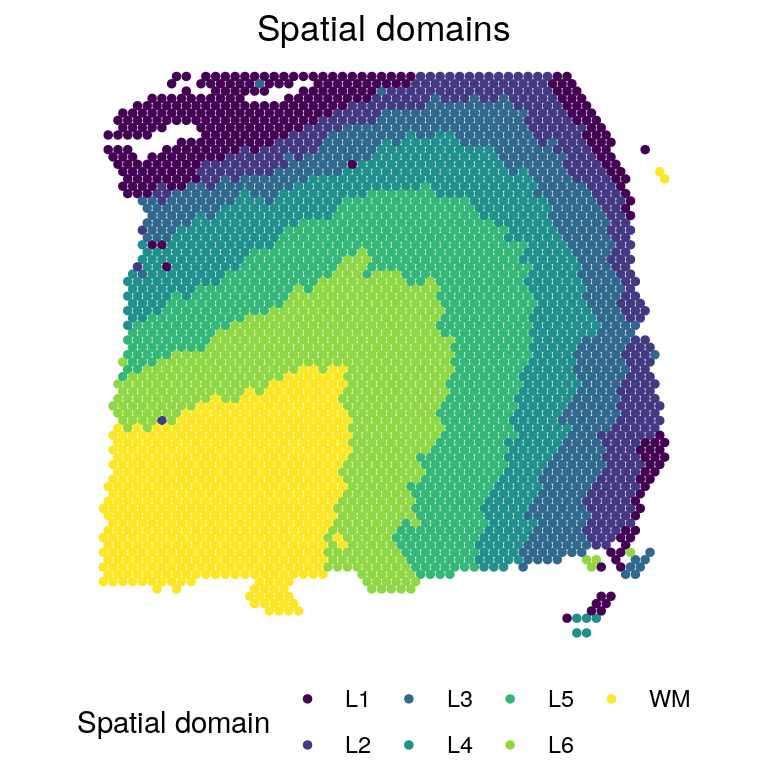
| Version | Author | Date |
|---|---|---|
| 68275b2 | zhengli09 | 2022-03-05 |
Multi-sample analysis
Run BASS
smps <- c("151673", "151674", "151675", "151676")
set.seed(0)
# Set up BASS object
BASS <- createBASSObject(cntm[smps], xym[smps], C = C, R = R,
beta_est_approach = "ACCUR_EST", init_method = "mclust",
burn_in = 10000, samples = 10000)Expression data coerced to a matrix
Expression data coerced to a matrix
Expression data coerced to a matrix
Expression data coerced to a matrix
***************************************
Bayesian Analytics for Spatial Segmentation (BASS)
Authors: Zheng Li, Xiang Zhou
Affiliate: Department of Biostatistics, University of Michigan
INPUT INFO:
- Number of samples: 4
- Number of spots/cells: 3639 3673 3592 3460
- Number of genes: 33538
- Potts interaction parameter estimation approach: ACCUR_EST
- Variance-covariance structure of gene expression features: EEE
To list all hyper-parameters, Type listAllHyper(BASS_object)
***************************************# Data pre-processing:
# In addition to the log-normalization, feature selection, and dimension
# reduction with PCA, we also conduct a batch effect adjustment using the
# Harmony package.
BASS <- BASS.preprocess(BASS, doLogNormalize = TRUE,
geneSelect = "sparkx", nSE = 3000,
doPCA = TRUE, scaleFeature = FALSE, nPC = 20)***** Log-normalize gene expression data *****
***** Select spatially expressed genes with sparkx *****Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!
Warning in FUN(newX[, i], ...): There are p-values that are exactly 1!***** Exclude genes with 0 expression *****
***** Reduce data dimension with PCA *****
***** Correct batch effect with Harmony *****# Run BASS algorithm
BASS <- BASS.run(BASS)# post-process posterior samples:
BASS <- BASS.postprocess(BASS)
......................................................................................
. Method Time (sec) Status .
......................................................................................
. ECR-ITERATIVE-1 494.959 Converged (3 iterations) .
......................................................................................
Relabelling all methods according to method ECR-ITERATIVE-1 ... done!
Retrieve the 1 permutation arrays by typing:
[...]$permutations$"ECR-ITERATIVE-1"
Retrieve the 1 best clusterings: [...]$clusters
Retrieve the 1 CPU times: [...]$timings
Retrieve the 1 X 1 similarity matrix: [...]$similarity
Label switching finished. Total time: 524.9 seconds.
......................................................................................
. Method Time (sec) Status .
......................................................................................
. ECR-ITERATIVE-1 482.04 Converged (4 iterations) .
......................................................................................
Relabelling all methods according to method ECR-ITERATIVE-1 ... done!
Retrieve the 1 permutation arrays by typing:
[...]$permutations$"ECR-ITERATIVE-1"
Retrieve the 1 best clusterings: [...]$clusters
Retrieve the 1 CPU times: [...]$timings
Retrieve the 1 X 1 similarity matrix: [...]$similarity
Label switching finished. Total time: 510.3 seconds. zlabels <- BASS@res_postprocess$z_ls # spatial domain labels
names(zlabels) <- smpsVisualization
library(cowplot)
# Spatial domains
zTypes <- c("L3", "L2", "L6", "L4", "WM", "L1", "L5")
zlabels <- lapply(zlabels, function(zlabels.l){
zlabels.l <- factor(zlabels.l)
levels(zlabels.l) <- zTypes
zlabels.l <- factor(zlabels.l, levels = c(
"L1", "L2", "L3", "L4", "L5", "L6", "WM"))
})
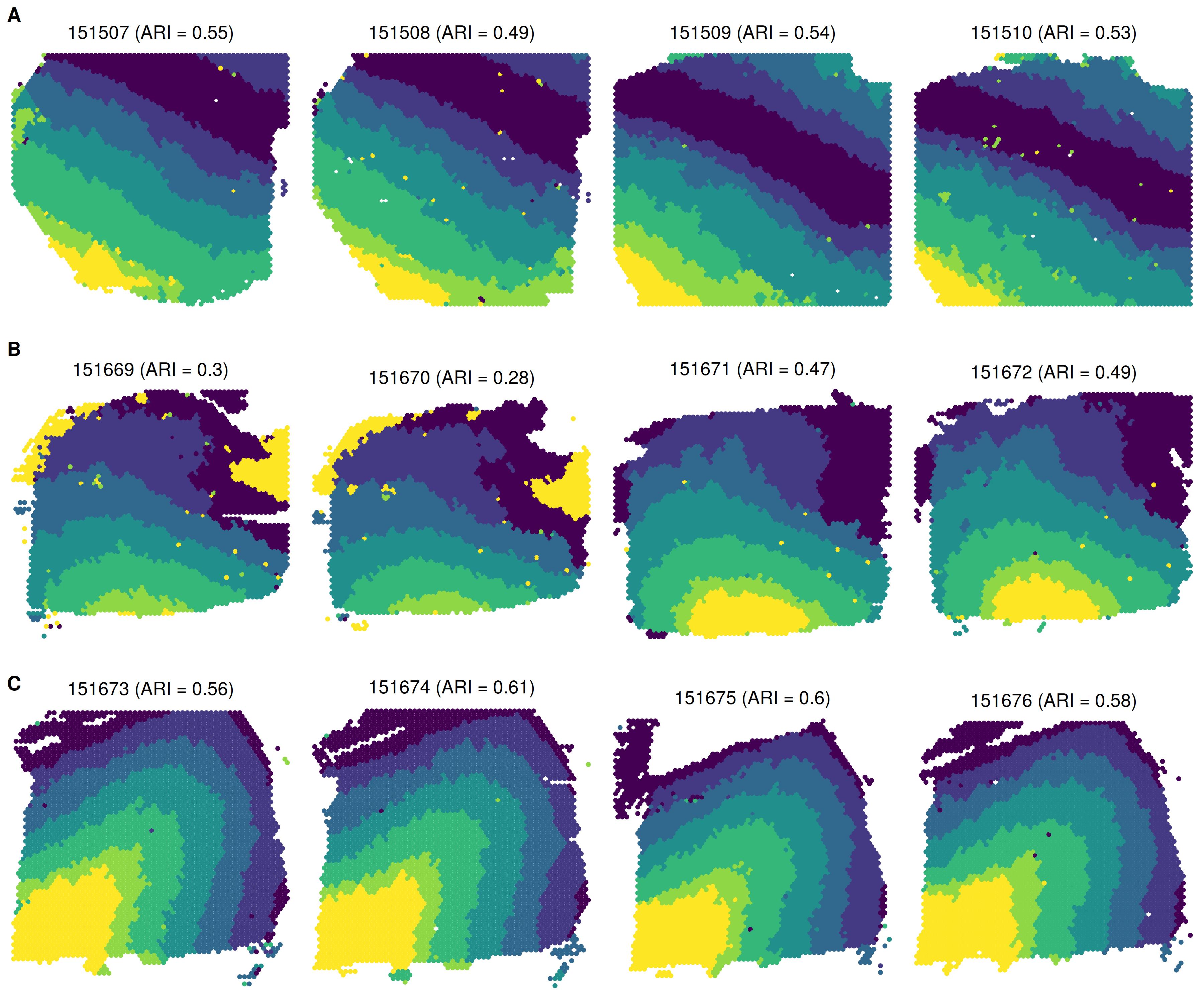
plist <- lapply(smps, function(smp){
ARI <- round(adjustedRandIndex(zlabels[[smp]],
infom[[smp]]$layer_guess_reordered_short),
digits = 2)
plotClusters(xym[[smp]], zlabels[[smp]],
title = paste0(smp, " (ARI =", ARI, ") "),
flip_xy = T, flip_y = T, ratio = 1.5) +
scale_color_viridis_d(name = "Spatial domain")
})plot_grid(plotlist = plist, ncol = 4)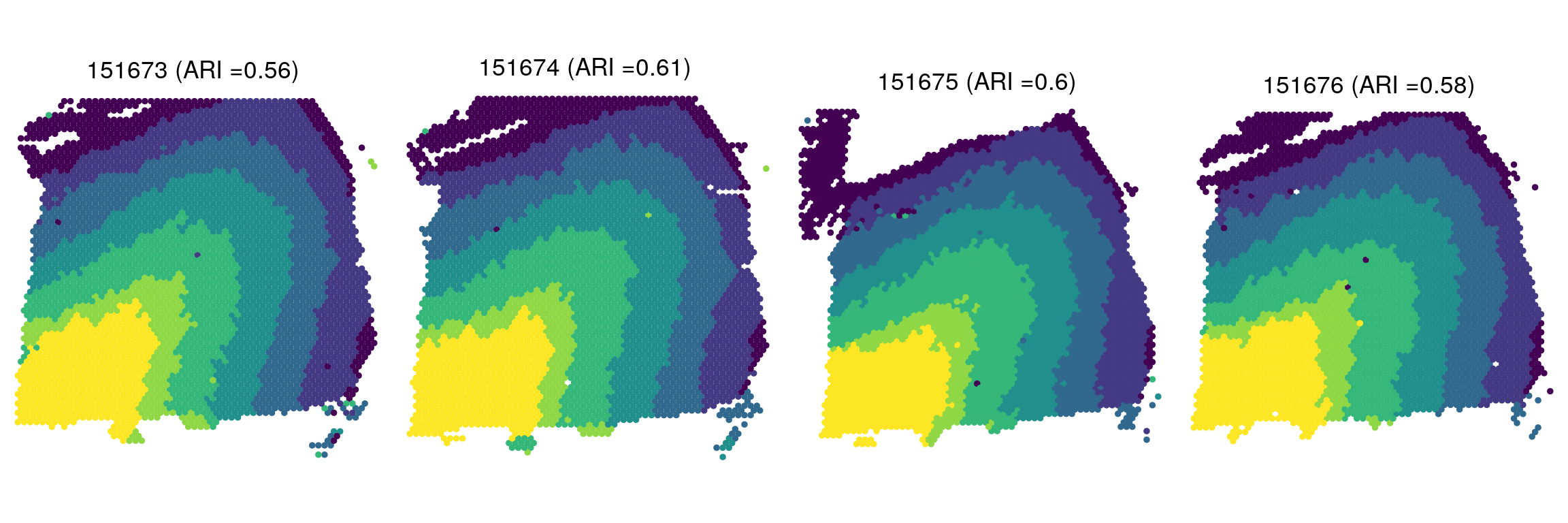
| Version | Author | Date |
|---|---|---|
| 68275b2 | zhengli09 | 2022-03-05 |
Summary of spatial domain detection across all 12 tissue sections
Manually annotated spatial domain labels
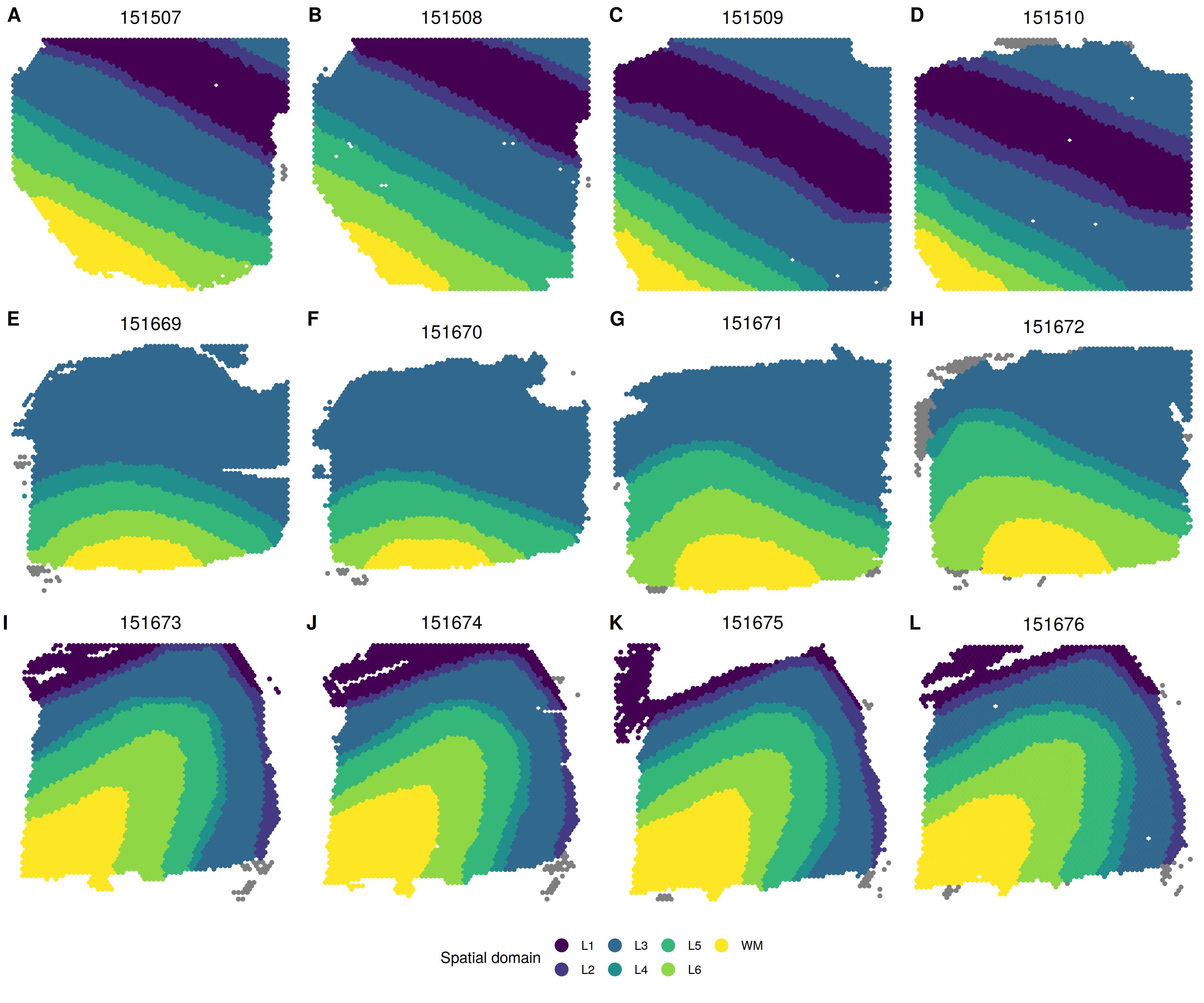
Single-sample analysis by BASS
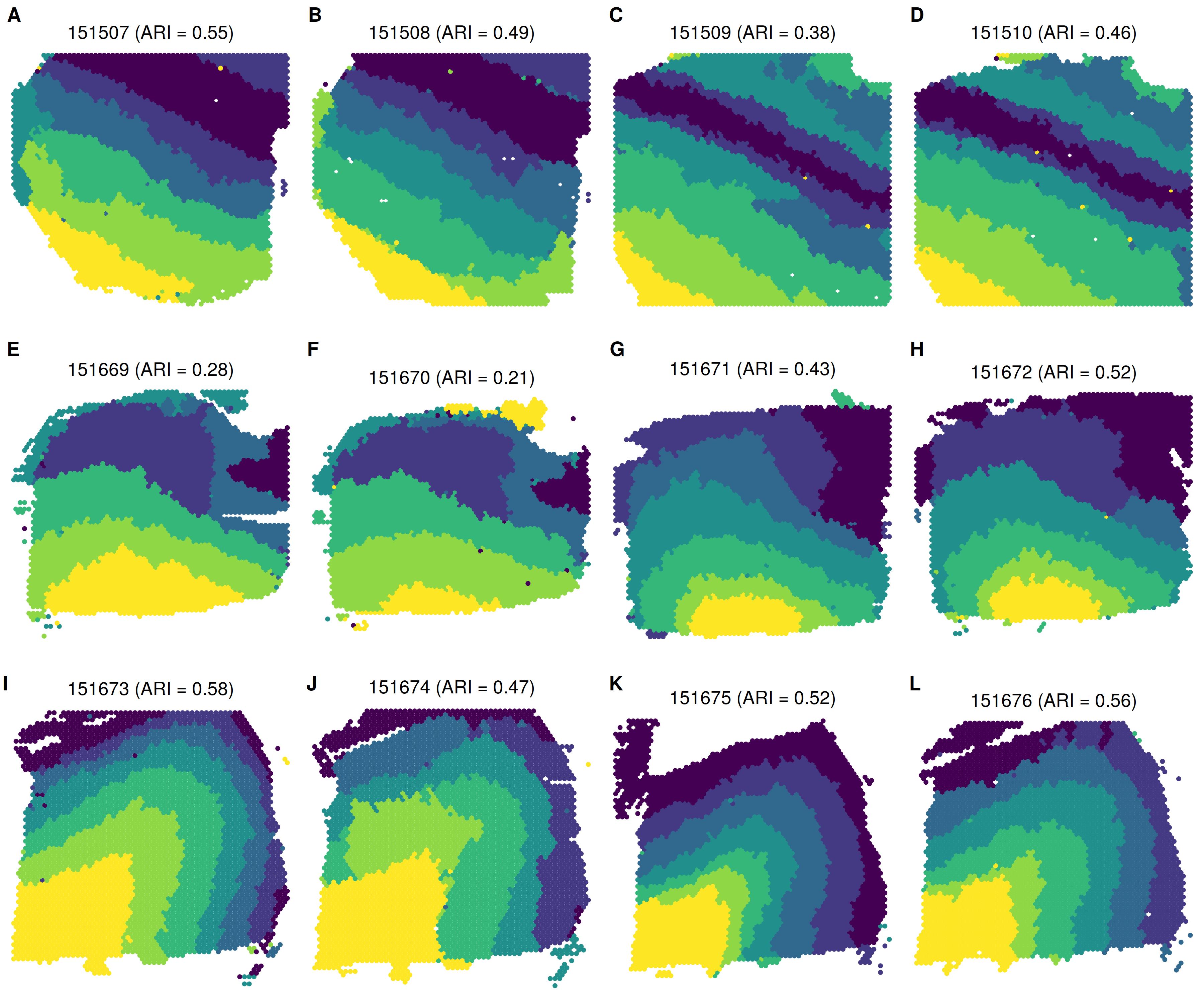
Multi-sample analysis by BASS
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 ggplot2_3.3.5 Matrix_1.3-4 BASS_1.0
[5] mclust_5.4.9 GIGrvg_0.5 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] bitops_1.0-7 matrixStats_0.61.0
[3] fs_1.5.2 doParallel_1.0.15
[5] httr_1.4.2 rprojroot_2.0.2
[7] GenomeInfoDb_1.24.2 tools_4.1.2
[9] bslib_0.3.1 utf8_1.2.2
[11] R6_2.5.1 irlba_2.3.3
[13] matlab_1.0.2 vipor_0.4.5
[15] DBI_1.1.1 BiocGenerics_0.38.0
[17] colorspace_2.0-3 withr_2.4.3
[19] label.switching_1.8 tidyselect_1.1.2
[21] gridExtra_2.3 processx_3.5.2
[23] compiler_4.1.2 git2r_0.28.0
[25] cli_3.2.0 Biobase_2.48.0
[27] SPARK_1.1.1 BiocNeighbors_1.6.0
[29] DelayedArray_0.18.0 labeling_0.4.2
[31] sass_0.4.0 scales_1.1.1
[33] callr_3.7.0 stringr_1.4.0
[35] digest_0.6.29 rmarkdown_2.12.1
[37] harmony_0.1.0 XVector_0.32.0
[39] scater_1.16.2 pkgconfig_2.0.3
[41] htmltools_0.5.2 sparseMatrixStats_1.4.2
[43] MatrixGenerics_1.4.3 highr_0.9
[45] fastmap_1.1.0 rlang_1.0.1
[47] rstudioapi_0.13 DelayedMatrixStats_1.14.3
[49] farver_2.1.0 jquerylib_0.1.4
[51] generics_0.1.2 combinat_0.0-8
[53] jsonlite_1.8.0 BiocParallel_1.22.0
[55] dplyr_1.0.8 RCurl_1.98-1.5
[57] magrittr_2.0.2 BiocSingular_1.4.0
[59] GenomeInfoDbData_1.2.6 Rcpp_1.0.8
[61] ggbeeswarm_0.6.0 munsell_0.5.0
[63] S4Vectors_0.30.2 fansi_1.0.2
[65] viridis_0.5.1 lifecycle_1.0.1
[67] stringi_1.7.6 whisker_0.4
[69] yaml_2.3.5 CompQuadForm_1.4.3
[71] SummarizedExperiment_1.22.0 zlibbioc_1.34.0
[73] grid_4.1.2 blob_1.2.1
[75] parallel_4.1.2 promises_1.1.1
[77] crayon_1.5.0 lattice_0.20-45
[79] knitr_1.37 ps_1.6.0
[81] pillar_1.7.0 GenomicRanges_1.44.0
[83] lpSolve_5.6.15 codetools_0.2-18
[85] stats4_4.1.2 glue_1.6.2
[87] evaluate_0.15 getPass_0.2-2
[89] foreach_1.5.0 vctrs_0.3.8
[91] httpuv_1.5.4 tidyr_1.1.1
[93] gtable_0.3.0 purrr_0.3.4
[95] assertthat_0.2.1 xfun_0.29
[97] rsvd_1.0.3 pracma_2.2.9
[99] later_1.1.0.1 viridisLite_0.4.0
[101] SingleCellExperiment_1.14.1 tibble_3.1.6
[103] iterators_1.0.13 beeswarm_0.4.0
[105] IRanges_2.26.0 ellipsis_0.3.2