Results publication
Jens Daniel Müller
13 July, 2022
Last updated: 2022-07-13
Checks: 7 0
Knit directory: emlr_obs_analysis/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210412) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version defdcfe. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Ignored: output/other/
Ignored: output/publication/
Untracked files:
Untracked: figure/
Unstaged changes:
Modified: analysis/child/budget_analysis_plot_data.Rmd
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/results_publication.Rmd) and HTML (docs/results_publication.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | defdcfe | jens-daniel-mueller | 2022-07-13 | additional analyis |
| html | 17cd1d1 | jens-daniel-mueller | 2022-07-13 | Build site. |
| Rmd | 1bf1708 | jens-daniel-mueller | 2022-07-13 | rerun reoccupation |
| html | 26e9496 | jens-daniel-mueller | 2022-07-12 | Build site. |
| Rmd | 5d121f0 | jens-daniel-mueller | 2022-07-12 | applied dcant scaling and improved ensemble member analysis |
| html | 8fb595c | jens-daniel-mueller | 2022-07-12 | Build site. |
| Rmd | e2d97ef | jens-daniel-mueller | 2022-07-12 | excluded no adjustment from ensemble |
| html | 003b161 | jens-daniel-mueller | 2022-07-12 | Build site. |
| Rmd | 54a0d14 | jens-daniel-mueller | 2022-07-12 | added cruise based adjustment |
| html | b44c72a | jens-daniel-mueller | 2022-07-03 | Build site. |
| html | 37f56b3 | jens-daniel-mueller | 2022-07-01 | Build site. |
| Rmd | 8c4a9f8 | jens-daniel-mueller | 2022-07-01 | added basin separation analysis |
| html | 232909e | jens-daniel-mueller | 2022-07-01 | Build site. |
| Rmd | 7f85db3 | jens-daniel-mueller | 2022-07-01 | added basin separation analysis |
| html | df21d31 | jens-daniel-mueller | 2022-07-01 | Build site. |
| Rmd | 2bbfba0 | jens-daniel-mueller | 2022-07-01 | rebuild |
| html | 6be73e0 | jens-daniel-mueller | 2022-06-30 | Build site. |
| Rmd | 6e173bf | jens-daniel-mueller | 2022-06-30 | updated regional budget plots |
| html | 6e173bf | jens-daniel-mueller | 2022-06-30 | updated regional budget plots |
| html | 8ab4a87 | jens-daniel-mueller | 2022-06-29 | Build site. |
| Rmd | d8c7cb9 | jens-daniel-mueller | 2022-06-29 | ensemble with unadjusted data |
| html | 7629c78 | jens-daniel-mueller | 2022-06-29 | Build site. |
| Rmd | 4758911 | jens-daniel-mueller | 2022-06-29 | ensemble without unadjusted data |
| html | f6786c8 | jens-daniel-mueller | 2022-06-29 | Build site. |
| Rmd | 6f694a1 | jens-daniel-mueller | 2022-06-29 | included Cstar N in ensemble |
| html | f09080e | jens-daniel-mueller | 2022-06-28 | Build site. |
| Rmd | d457610 | jens-daniel-mueller | 2022-06-28 | save figures for publication |
| html | ee30748 | jens-daniel-mueller | 2022-06-28 | Build site. |
| Rmd | fe42644 | jens-daniel-mueller | 2022-06-28 | GCB emissions ratio included |
| html | 9393c07 | jens-daniel-mueller | 2022-06-28 | Build site. |
| Rmd | 2a3cf97 | jens-daniel-mueller | 2022-06-28 | included basin-hemisphere bias, and bias contributions |
| html | 0825298 | jens-daniel-mueller | 2022-06-28 | Build site. |
| Rmd | d35ddb1 | jens-daniel-mueller | 2022-06-28 | included GCB ocean sink data as boxplot |
| html | a13a7cf | jens-daniel-mueller | 2022-06-28 | Build site. |
| html | fb59a6f | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | 7ff568c | jens-daniel-mueller | 2022-06-27 | included GCB ocean sink data |
| html | a26a21d | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | 9425709 | jens-daniel-mueller | 2022-06-27 | scaled sabine 2004 to full area |
| html | 748aa43 | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | 82d5793 | jens-daniel-mueller | 2022-06-27 | ensemble scatter plot |
| html | 457e640 | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | 426c5bf | jens-daniel-mueller | 2022-06-27 | cleaned read-in section |
| html | 16dc3af | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | dd2063a | jens-daniel-mueller | 2022-06-27 | ensemble member analysis added |
| html | 87e9eb8 | jens-daniel-mueller | 2022-06-27 | Build site. |
| Rmd | 09a3348 | jens-daniel-mueller | 2022-06-27 | 1 as standard case and new ensemble members |
| html | b52b159 | jens-daniel-mueller | 2022-06-27 | Build site. |
| html | 09b0780 | jens-daniel-mueller | 2022-05-24 | Build site. |
| html | 25da2fb | jens-daniel-mueller | 2022-05-24 | Build site. |
| html | 1d73ec9 | jens-daniel-mueller | 2022-05-16 | Build site. |
| Rmd | e117b7b | jens-daniel-mueller | 2022-05-16 | rerun w/o data adjustments |
| html | 2ffbdda | jens-daniel-mueller | 2022-05-16 | Build site. |
| Rmd | 6fc8438 | jens-daniel-mueller | 2022-05-16 | plot individual zonal sections |
| html | c71227f | jens-daniel-mueller | 2022-05-16 | Build site. |
| Rmd | 55e9ac6 | jens-daniel-mueller | 2022-05-16 | plot individual zonal sections |
| html | 3c1100c | jens-daniel-mueller | 2022-05-16 | Build site. |
| Rmd | fdb111d | jens-daniel-mueller | 2022-05-16 | plot individual zonal sections |
| html | 2ca0109 | jens-daniel-mueller | 2022-05-02 | Build site. |
| Rmd | 63d51bd | jens-daniel-mueller | 2022-05-02 | rerun with adjusted data |
| html | dcf2eaf | jens-daniel-mueller | 2022-05-02 | Build site. |
| Rmd | a3a2cf9 | jens-daniel-mueller | 2022-05-02 | IO sections seperate |
| html | eff4fd7 | jens-daniel-mueller | 2022-05-02 | Build site. |
| Rmd | 39e920a | jens-daniel-mueller | 2022-05-02 | IO sections seperate |
| html | 08607eb | jens-daniel-mueller | 2022-05-02 | Build site. |
| Rmd | 0791c1b | jens-daniel-mueller | 2022-05-02 | modified plots |
| html | b018a9a | jens-daniel-mueller | 2022-04-29 | Build site. |
| Rmd | 02ede93 | jens-daniel-mueller | 2022-04-29 | standard case uncorrected data |
| html | e09320d | jens-daniel-mueller | 2022-04-12 | Build site. |
| Rmd | 0dec180 | jens-daniel-mueller | 2022-04-12 | 3 data adjustment procedures implemented |
| html | 8dca96a | jens-daniel-mueller | 2022-04-12 | Build site. |
| Rmd | e5e9288 | jens-daniel-mueller | 2022-04-12 | 3 data adjustment procedures implemented |
| html | 2f20ea6 | jens-daniel-mueller | 2022-04-11 | Build site. |
| html | 209c9b6 | jens-daniel-mueller | 2022-04-10 | Build site. |
| Rmd | 537aff7 | jens-daniel-mueller | 2022-04-10 | no data adjustment implemented |
| html | acad2e2 | jens-daniel-mueller | 2022-04-09 | Build site. |
| html | 3d81135 | jens-daniel-mueller | 2022-04-07 | Build site. |
| html | 0f5d372 | jens-daniel-mueller | 2022-04-04 | Build site. |
| Rmd | ac5121f | jens-daniel-mueller | 2022-04-04 | added zonal mean beta distribution new figure |
| html | 1b5a309 | jens-daniel-mueller | 2022-04-04 | Build site. |
| Rmd | 8b96a9d | jens-daniel-mueller | 2022-04-04 | added zonal mean beta distribution new figure |
| html | a74e341 | jens-daniel-mueller | 2022-04-04 | Build site. |
| Rmd | c0432be | jens-daniel-mueller | 2022-04-04 | added zonal mean beta distribution ensemble sd |
| html | b599680 | jens-daniel-mueller | 2022-04-04 | Build site. |
| Rmd | e406c39 | jens-daniel-mueller | 2022-04-04 | added zonal mean beta distribution |
| html | ca7e590 | jens-daniel-mueller | 2022-03-22 | Build site. |
| Rmd | 8cb4b78 | jens-daniel-mueller | 2022-03-22 | use 1800 as tref for sabine estimates |
| html | 5a6be34 | jens-daniel-mueller | 2022-03-22 | Build site. |
| Rmd | cee203c | jens-daniel-mueller | 2022-03-22 | rerun with NP 2021 talk correction |
| html | bd9e11d | jens-daniel-mueller | 2022-03-22 | Build site. |
| html | 2501978 | jens-daniel-mueller | 2022-03-21 | Build site. |
| Rmd | 9e12898 | jens-daniel-mueller | 2022-03-21 | use 1800 as tref for sabine estimates |
| html | c3a6238 | jens-daniel-mueller | 2022-03-08 | Build site. |
| Rmd | 775eb4f | jens-daniel-mueller | 2022-03-08 | moving eras analysis implemented |
| html | 094bfa0 | jens-daniel-mueller | 2022-02-18 | Build site. |
| Rmd | fa258cc | jens-daniel-mueller | 2022-02-18 | updated plots |
| html | ba2d62e | jens-daniel-mueller | 2022-02-17 | Build site. |
| Rmd | 5420131 | jens-daniel-mueller | 2022-02-17 | added Seaflux data |
| html | 192504c | jens-daniel-mueller | 2022-02-17 | Build site. |
| Rmd | 2394302 | jens-daniel-mueller | 2022-02-17 | added Seaflux data |
| html | 251c7cf | jens-daniel-mueller | 2022-02-17 | Build site. |
| Rmd | 9ea0d8c | jens-daniel-mueller | 2022-02-17 | adapted beta factor |
| html | 565224d | jens-daniel-mueller | 2022-02-17 | Build site. |
| Rmd | 33422e3 | jens-daniel-mueller | 2022-02-17 | scaled budgets to global coverage |
| html | 2116dd3 | jens-daniel-mueller | 2022-02-09 | Build site. |
| Rmd | e281bf3 | jens-daniel-mueller | 2022-02-09 | updated plots |
| html | 6fe70a1 | jens-daniel-mueller | 2022-02-05 | Build site. |
| Rmd | 8955c85 | jens-daniel-mueller | 2022-02-05 | cleaned plots |
| html | a6b33aa | jens-daniel-mueller | 2022-02-04 | Build site. |
| Rmd | 73efef8 | jens-daniel-mueller | 2022-02-04 | atm co2 time series plotted, uncertainties revised |
| html | 4b48475 | jens-daniel-mueller | 2022-02-04 | Build site. |
| Rmd | 6afd7b1 | jens-daniel-mueller | 2022-02-04 | calculated uncertainty of basin budget changes |
| html | fec5a1e | jens-daniel-mueller | 2022-02-04 | Build site. |
| Rmd | 325da3a | jens-daniel-mueller | 2022-02-04 | calculated uncertainty of basin budget changes |
| html | d2191ad | jens-daniel-mueller | 2022-02-04 | Build site. |
| Rmd | 79a54fe | jens-daniel-mueller | 2022-02-04 | new coaverage map |
| html | 4c5b079 | jens-daniel-mueller | 2022-02-03 | Build site. |
| Rmd | 66d89a9 | jens-daniel-mueller | 2022-02-03 | added surface flux model products |
| html | 4077397 | jens-daniel-mueller | 2022-02-03 | Build site. |
| Rmd | c0c3be1 | jens-daniel-mueller | 2022-02-03 | added surface flux products |
| html | 0d0f790 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 049346f | jens-daniel-mueller | 2022-02-02 | shifted year labels |
| html | 4673df5 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 1185397 | jens-daniel-mueller | 2022-02-02 | rearranged plots |
| html | 60727e6 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | db5308b | jens-daniel-mueller | 2022-02-02 | rearranged plots |
| html | c7b4984 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | a3d8469 | jens-daniel-mueller | 2022-02-02 | ensemble uncertainties in global buget |
| html | 7fb28a2 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | a332fe6 | jens-daniel-mueller | 2022-02-02 | ensemble uncertainties in time series |
| html | 49097e8 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | f98c33b | jens-daniel-mueller | 2022-02-02 | incl ensemble uncertainties in plot |
| html | fe11bfd | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | d3f7f05 | jens-daniel-mueller | 2022-02-02 | incl ensemble uncertainties |
| html | fa46251 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 33feacf | jens-daniel-mueller | 2022-02-02 | incl sabine budgets |
| html | 7655085 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 09dd555 | jens-daniel-mueller | 2022-02-02 | incl sabine budgets |
| html | 226d67d | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | ed056e2 | jens-daniel-mueller | 2022-02-02 | incl sabine budgets |
| html | ed903f7 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 9d11695 | jens-daniel-mueller | 2022-02-02 | scaled budget to atm pco2 increase |
| html | 32e9682 | jens-daniel-mueller | 2022-02-02 | Build site. |
| Rmd | 5cc5572 | jens-daniel-mueller | 2022-02-02 | included Sabine column inventory as reference |
| html | 913e42f | jens-daniel-mueller | 2022-02-01 | Build site. |
| Rmd | c4b2de9 | jens-daniel-mueller | 2022-02-01 | updated profile plots |
| html | 189de95 | jens-daniel-mueller | 2022-02-01 | Build site. |
| Rmd | b0c17bc | jens-daniel-mueller | 2022-02-01 | updated profile plots |
| html | ab001eb | jens-daniel-mueller | 2022-01-31 | Build site. |
| Rmd | ccf6723 | jens-daniel-mueller | 2022-01-31 | filled step plot for layer budgets |
| html | d2ae5fe | jens-daniel-mueller | 2022-01-31 | Build site. |
| Rmd | 278523c | jens-daniel-mueller | 2022-01-31 | step plot for layer budgets |
| html | b62308d | jens-daniel-mueller | 2022-01-31 | Build site. |
| Rmd | 714c5cc | jens-daniel-mueller | 2022-01-31 | step plot for layer budgets |
| html | ec7fe7e | jens-daniel-mueller | 2022-01-31 | Build site. |
| Rmd | 5c948ae | jens-daniel-mueller | 2022-01-31 | added time series vs atm pco2 |
| html | de557de | jens-daniel-mueller | 2022-01-28 | Build site. |
| html | 5f2aed0 | jens-daniel-mueller | 2022-01-27 | Build site. |
| Rmd | 54c9e26 | jens-daniel-mueller | 2022-01-27 | added layer budget profiles |
| html | eccd82b | jens-daniel-mueller | 2022-01-26 | Build site. |
| Rmd | c5577d3 | jens-daniel-mueller | 2022-01-26 | added meand sd to offset mean concentrations profiles |
| html | c6fe495 | jens-daniel-mueller | 2022-01-26 | Build site. |
| Rmd | e0e7974 | jens-daniel-mueller | 2022-01-26 | added offset mean concentrations profiles |
| html | 9753eb8 | jens-daniel-mueller | 2022-01-26 | Build site. |
| html | b1d7720 | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 0210ed5 | jens-daniel-mueller | 2022-01-21 | added mean concentrations profiles per 5 basins |
| html | d6b399a | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | da17a07 | jens-daniel-mueller | 2022-01-21 | added mean concentrations profiles |
| html | c499be8 | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | d941871 | jens-daniel-mueller | 2022-01-21 | run color map test |
| html | e572075 | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 99b6c92 | jens-daniel-mueller | 2022-01-21 | run color map test |
| html | 4fe7150 | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 0379e99 | jens-daniel-mueller | 2022-01-21 | script cleaning |
| html | 49b41cf | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 2c82651 | jens-daniel-mueller | 2022-01-21 | added map of scaled absolute change |
| html | c0807e8 | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 5dd3d7a | jens-daniel-mueller | 2022-01-21 | added map of scaled relative change |
| html | 22b421f | jens-daniel-mueller | 2022-01-21 | Build site. |
| Rmd | 2c3fa75 | jens-daniel-mueller | 2022-01-21 | cleaned alluvial plots |
| html | 1a35f1f | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | e58f510 | jens-daniel-mueller | 2022-01-20 | added relative changes to alluvial plots |
| html | b503ae1 | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | 2eb2567 | jens-daniel-mueller | 2022-01-20 | added relative changes to alluvial plots |
| html | cc31f4b | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | 416e107 | jens-daniel-mueller | 2022-01-20 | added delta dcant map |
| html | 11a800b | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | 81a40d5 | jens-daniel-mueller | 2022-01-20 | updated alluvial plots |
| html | 3087804 | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | 2ae5966 | jens-daniel-mueller | 2022-01-20 | updated alluvial plots |
| html | 6d566d5 | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | 4901b0f | jens-daniel-mueller | 2022-01-20 | updated alluvial plots |
| html | 44796b1 | jens-daniel-mueller | 2022-01-20 | Build site. |
| Rmd | cdbd92c | jens-daniel-mueller | 2022-01-20 | created alluvial plots |
| html | 48ec4c6 | jens-daniel-mueller | 2022-01-19 | Build site. |
| Rmd | 0fb2ae5 | jens-daniel-mueller | 2022-01-19 | printed column inv from AIP standard runs |
| html | f347cd7 | jens-daniel-mueller | 2022-01-18 | Build site. |
| Rmd | 86b711c | jens-daniel-mueller | 2022-01-18 | plot hemisphere budgets and publication results |
1 Libraries
2 Read files
2.1 Paths and Versions
version_id_pattern <- "103"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
print(Version_IDs)[1] "v_1103" "v_2103" "v_3103"version_id_pattern <- "1"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_x <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
version_id_pattern <- "o"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_o <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
version_id_pattern <- "d"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_d <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
version_id_pattern <- "g"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_g <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
version_id_pattern <- "n"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_n <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
rm(Version_IDs_1, Version_IDs_2, Version_IDs_3)
version_id_pattern <- "c"
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", version_id_pattern))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", version_id_pattern))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", version_id_pattern))
Version_IDs_c <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
rm(Version_IDs_1, Version_IDs_2, Version_IDs_3)
Version_IDs_ensemble <- c(Version_IDs_x, Version_IDs_o, Version_IDs_d, Version_IDs_g, Version_IDs_n, Version_IDs_c)
rm(Version_IDs_x, Version_IDs_o, Version_IDs_g, Version_IDs_n, Version_IDs_c)2.2 Parameters
for (i_Version_IDs in Version_IDs) {
# i_Version_IDs <- Version_IDs[1]
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
params_local <-
read_rds(paste(path_version_data,
"params_local.rds",
sep = ""))
params_local <- bind_cols(
Version_ID = i_Version_IDs,
tref1 = params_local$tref1,
tref2 = params_local$tref2,
MLR_basins = params_local$MLR_basins
)
tref <- read_csv(paste(path_version_data,
"tref.csv",
sep = ""))
params_local <- params_local %>%
mutate(
median_year_1 = sort(tref$median_year)[1],
median_year_2 = sort(tref$median_year)[2],
duration = median_year_2 - median_year_1,
period = paste(median_year_1, "-", median_year_2)
)
if (exists("params_local_all")) {
params_local_all <- bind_rows(params_local_all, params_local)
}
if (!exists("params_local_all")) {
params_local_all <- params_local
}
}
rm(params_local,
tref)
params_local_all <- params_local_all %>%
select(Version_ID, period, MLR_basins, tref1, tref2)for (i_Version_IDs in Version_IDs_ensemble) {
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
params_local <-
read_rds(paste(path_version_data,
"params_local.rds",
sep = ""))
params_local <- bind_cols(
Version_ID = i_Version_IDs,
tref1 = params_local$tref1,
tref2 = params_local$tref2,
MLR_basins = params_local$MLR_basins
)
tref <- read_csv(paste(path_version_data,
"tref.csv",
sep = ""))
params_local <- params_local %>%
mutate(
median_year_1 = sort(tref$median_year)[1],
median_year_2 = sort(tref$median_year)[2],
duration = median_year_2 - median_year_1,
period = paste(median_year_1, "-", median_year_2)
)
if (exists("params_local_all_ensemble")) {
params_local_all_ensemble <- bind_rows(params_local_all_ensemble, params_local)
}
if (!exists("params_local_all_ensemble")) {
params_local_all_ensemble <- params_local
}
}
rm(params_local,
tref)
params_local_all_ensemble <- params_local_all_ensemble %>%
select(Version_ID, period, MLR_basins, tref1, tref2)
params_local_all_ensemble <-
params_local_all_ensemble %>%
mutate(
Version_ID_group = str_sub(Version_ID, 4, 4),
Version_ID_group = case_when(
Version_ID_group == "1" ~ "standard",
Version_ID_group == "o" ~ "reoccupation filter",
Version_ID_group == "d" ~ "no data adjustment",
Version_ID_group == "g" ~ "no gap filling",
Version_ID_group == "n" ~ "C*(N)",
Version_ID_group == "c" ~ "cruise adjustment",
TRUE ~ Version_ID_group
)
)2.3 Budgets
for (i_Version_IDs in Version_IDs_ensemble) {
# i_Version_IDs <- Version_IDs[1]
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
dcant_budget_global <-
read_csv(paste(path_version_data,
"dcant_budget_global.csv",
sep = ""))
dcant_budget_global_mod_truth <-
read_csv(paste(
path_version_data,
"dcant_budget_global_mod_truth.csv",
sep = ""
))
dcant_budget_global <- bind_rows(dcant_budget_global,
dcant_budget_global_mod_truth)
dcant_budget_global <- dcant_budget_global %>%
mutate(Version_ID = i_Version_IDs)
if (exists("dcant_budget_global_all")) {
dcant_budget_global_all <-
bind_rows(dcant_budget_global_all, dcant_budget_global)
}
if (!exists("dcant_budget_global_all")) {
dcant_budget_global_all <- dcant_budget_global
}
}
rm(dcant_budget_global,
dcant_budget_global_mod_truth)for (i_Version_IDs in Version_IDs_ensemble) {
# i_Version_IDs <- Version_IDs[1]
print(i_Version_IDs)
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
dcant_budget_basin_MLR <-
read_csv(paste(path_version_data,
"dcant_budget_basin_MLR.csv",
sep = ""))
dcant_budget_basin_MLR_mod_truth <-
read_csv(paste(
path_version_data,
"dcant_budget_basin_MLR_mod_truth.csv",
sep = ""
))
dcant_budget_basin_MLR <- bind_rows(dcant_budget_basin_MLR,
dcant_budget_basin_MLR_mod_truth)
dcant_budget_basin_MLR <- dcant_budget_basin_MLR %>%
mutate(Version_ID = i_Version_IDs)
if (exists("dcant_budget_basin_MLR_all")) {
dcant_budget_basin_MLR_all <-
bind_rows(dcant_budget_basin_MLR_all, dcant_budget_basin_MLR)
}
if (!exists("dcant_budget_basin_MLR_all")) {
dcant_budget_basin_MLR_all <- dcant_budget_basin_MLR
}
}[1] "v_1101"
[1] "v_1102"
[1] "v_1103"
[1] "v_1104"
[1] "v_1105"
[1] "v_1106"
[1] "v_2101"
[1] "v_2102"
[1] "v_2103"
[1] "v_2104"
[1] "v_2105"
[1] "v_2106"
[1] "v_3101"
[1] "v_3102"
[1] "v_3103"
[1] "v_3104"
[1] "v_3105"
[1] "v_3106"
[1] "v_1o01"
[1] "v_1o02"
[1] "v_1o03"
[1] "v_1o04"
[1] "v_1o05"
[1] "v_1o06"
[1] "v_2o01"
[1] "v_2o02"
[1] "v_2o03"
[1] "v_2o04"
[1] "v_2o05"
[1] "v_2o06"
[1] "v_3o01"
[1] "v_3o02"
[1] "v_3o03"
[1] "v_3o04"
[1] "v_3o05"
[1] "v_3o06"
[1] "v_1d01"
[1] "v_1d02"
[1] "v_1d03"
[1] "v_1d04"
[1] "v_1d05"
[1] "v_1d06"
[1] "v_2d01"
[1] "v_2d02"
[1] "v_2d03"
[1] "v_2d04"
[1] "v_2d05"
[1] "v_2d06"
[1] "v_3d01"
[1] "v_3d02"
[1] "v_3d03"
[1] "v_3d04"
[1] "v_3d05"
[1] "v_3d06"
[1] "v_1g01"
[1] "v_1g02"
[1] "v_1g03"
[1] "v_1g04"
[1] "v_1g05"
[1] "v_1g06"
[1] "v_2g01"
[1] "v_2g02"
[1] "v_2g03"
[1] "v_2g04"
[1] "v_2g05"
[1] "v_2g06"
[1] "v_3g01"
[1] "v_3g02"
[1] "v_3g03"
[1] "v_3g04"
[1] "v_3g05"
[1] "v_3g06"
[1] "v_1n01"
[1] "v_1n02"
[1] "v_1n03"
[1] "v_1n04"
[1] "v_1n05"
[1] "v_1n06"
[1] "v_2n01"
[1] "v_2n02"
[1] "v_2n03"
[1] "v_2n04"
[1] "v_2n05"
[1] "v_2n06"
[1] "v_3n01"
[1] "v_3n02"
[1] "v_3n03"
[1] "v_3n04"
[1] "v_3n05"
[1] "v_3n06"
[1] "v_1c01"
[1] "v_1c02"
[1] "v_1c03"
[1] "v_1c04"
[1] "v_1c05"
[1] "v_1c06"
[1] "v_2c01"
[1] "v_2c02"
[1] "v_2c03"
[1] "v_2c04"
[1] "v_2c05"
[1] "v_2c06"
[1] "v_3c01"
[1] "v_3c02"
[1] "v_3c03"
[1] "v_3c04"
[1] "v_3c05"
[1] "v_3c06"rm(
dcant_budget_basin_MLR,
dcant_budget_basin_MLR_mod_truth
)dcant_budget_global_all <- dcant_budget_global_all %>%
filter(estimate == "dcant",
method == "total") %>%
select(-c(estimate, method)) %>%
rename(dcant = value)
dcant_budget_global_all <- dcant_budget_global_all %>%
filter(inv_depth == params_global$inventory_depth_standard)dcant_budget_basin_MLR_all <- dcant_budget_basin_MLR_all %>%
filter(estimate == "dcant",
method == "total") %>%
select(-c(estimate, method)) %>%
rename(dcant = value)
dcant_budget_basin_MLR_all <- dcant_budget_basin_MLR_all %>%
filter(inv_depth == params_global$inventory_depth_standard)2.4 Column inventories
for (i_Version_IDs in Version_IDs_ensemble) {
# i_Version_IDs <- Version_IDs[1]
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
dcant_inv <-
read_csv(paste(path_version_data,
"dcant_inv.csv",
sep = ""))
dcant_inv_mod_truth <-
read_csv(paste(path_version_data,
"dcant_inv_mod_truth.csv",
sep = "")) %>%
filter(method == "total") %>%
select(-method)
dcant_inv_bias <-
read_csv(paste(path_version_data,
"dcant_inv_bias.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
dcant_inv <- bind_rows(dcant_inv,
dcant_inv_mod_truth) %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_lat_grid <-
read_csv(paste(path_version_data,
"dcant_budget_lat_grid.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_lon_grid <-
read_csv(paste(path_version_data,
"dcant_budget_lon_grid.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
if (exists("dcant_inv_all")) {
dcant_inv_all <- bind_rows(dcant_inv_all, dcant_inv)
}
if (!exists("dcant_inv_all")) {
dcant_inv_all <- dcant_inv
}
if (exists("dcant_inv_bias_all")) {
dcant_inv_bias_all <- bind_rows(dcant_inv_bias_all, dcant_inv_bias)
}
if (!exists("dcant_inv_bias_all")) {
dcant_inv_bias_all <- dcant_inv_bias
}
if (exists("dcant_budget_lat_grid_all")) {
dcant_budget_lat_grid_all <- bind_rows(dcant_budget_lat_grid_all, dcant_budget_lat_grid)
}
if (!exists("dcant_budget_lat_grid_all")) {
dcant_budget_lat_grid_all <- dcant_budget_lat_grid
}
if (exists("dcant_budget_lon_grid_all")) {
dcant_budget_lon_grid_all <- bind_rows(dcant_budget_lon_grid_all, dcant_budget_lon_grid)
}
if (!exists("dcant_budget_lon_grid_all")) {
dcant_budget_lon_grid_all <- dcant_budget_lon_grid
}
}
rm(dcant_inv,
dcant_inv_bias,
dcant_inv_mod_truth,
dcant_budget_lat_grid,
dcant_budget_lon_grid)dcant_inv_all <- dcant_inv_all %>%
filter(inv_depth == params_global$inventory_depth_standard)
dcant_budget_lat_grid_all <- dcant_budget_lat_grid_all %>%
filter(inv_depth == params_global$inventory_depth_standard)
dcant_budget_lon_grid_all <- dcant_budget_lon_grid_all %>%
filter(inv_depth == params_global$inventory_depth_standard)dcant_budget_lat_grid_all <- dcant_budget_lat_grid_all %>%
pivot_wider(names_from = estimate,
values_from = value) %>%
filter(period != "1994 - 2014",
method == "total")
dcant_budget_lon_grid_all <- dcant_budget_lon_grid_all %>%
pivot_wider(names_from = estimate,
values_from = value) %>%
filter(period != "1994 - 2014",
method == "total")2.5 Sections / profiles
for (i_Version_IDs in Version_IDs_ensemble) {
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
dcant_zonal <-
read_csv(paste(path_version_data,
"dcant_zonal.csv",
sep = ""))
dcant_zonal_mod_truth <-
read_csv(paste(path_version_data,
"dcant_zonal_mod_truth.csv",
sep = ""))
dcant_zonal <- bind_rows(dcant_zonal,
dcant_zonal_mod_truth)
dcant_profile <-
read_csv(paste(path_version_data,
"dcant_profile.csv",
sep = ""))
dcant_profile_mod_truth <-
read_csv(paste(path_version_data,
"dcant_profile_mod_truth.csv",
sep = ""))
dcant_profile_basin_MLR <-
read_csv(paste(path_version_data,
"dcant_profile_basin_MLR.csv",
sep = ""))
dcant_profile <- bind_rows(dcant_profile,
dcant_profile_mod_truth)
dcant_budget_basin_AIP_layer <-
read_csv(paste(path_version_data,
"dcant_budget_basin_AIP_layer.csv",
sep = ""))
dcant_budget_basin_MLR_layer <-
read_csv(paste(path_version_data,
"dcant_budget_basin_MLR_layer.csv",
sep = ""))
dcant_zonal_bias <-
read_csv(paste(path_version_data,
"dcant_zonal_bias.csv",
sep = ""))
dcant_zonal <- dcant_zonal %>%
mutate(Version_ID = i_Version_IDs)
dcant_profile <- dcant_profile %>%
mutate(Version_ID = i_Version_IDs)
dcant_profile_basin_MLR <- dcant_profile_basin_MLR %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_basin_AIP_layer <- dcant_budget_basin_AIP_layer %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_basin_MLR_layer <- dcant_budget_basin_MLR_layer %>%
mutate(Version_ID = i_Version_IDs)
dcant_zonal_bias <- dcant_zonal_bias %>%
mutate(Version_ID = i_Version_IDs)
if (exists("dcant_zonal_all")) {
dcant_zonal_all <- bind_rows(dcant_zonal_all, dcant_zonal)
}
if (!exists("dcant_zonal_all")) {
dcant_zonal_all <- dcant_zonal
}
if (exists("dcant_profile_all")) {
dcant_profile_all <- bind_rows(dcant_profile_all, dcant_profile)
}
if (!exists("dcant_profile_all")) {
dcant_profile_all <- dcant_profile
}
if (exists("dcant_profile_basin_MLR_all")) {
dcant_profile_basin_MLR_all <- bind_rows(dcant_profile_basin_MLR_all, dcant_profile_basin_MLR)
}
if (!exists("dcant_profile_basin_MLR_all")) {
dcant_profile_basin_MLR_all <- dcant_profile_basin_MLR
}
if (exists("dcant_budget_basin_AIP_layer_all")) {
dcant_budget_basin_AIP_layer_all <-
bind_rows(dcant_budget_basin_AIP_layer_all,
dcant_budget_basin_AIP_layer)
}
if (!exists("dcant_budget_basin_AIP_layer_all")) {
dcant_budget_basin_AIP_layer_all <- dcant_budget_basin_AIP_layer
}
if (exists("dcant_budget_basin_MLR_layer_all")) {
dcant_budget_basin_MLR_layer_all <-
bind_rows(dcant_budget_basin_MLR_layer_all,
dcant_budget_basin_MLR_layer)
}
if (!exists("dcant_budget_basin_MLR_layer_all")) {
dcant_budget_basin_MLR_layer_all <- dcant_budget_basin_MLR_layer
}
if (exists("dcant_zonal_bias_all")) {
dcant_zonal_bias_all <- bind_rows(dcant_zonal_bias_all, dcant_zonal_bias)
}
if (!exists("dcant_zonal_bias_all")) {
dcant_zonal_bias_all <- dcant_zonal_bias
}
}
rm(dcant_zonal, dcant_zonal_bias, dcant_zonal_mod_truth,
dcant_budget_basin_AIP_layer, dcant_budget_basin_MLR_layer,
dcant_profile, dcant_profile_basin_MLR)2.6 Observations coverage
for (i_Version_IDs in Version_IDs) {
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
GLODAP_grid_era <-
read_csv(paste(
path_version_data,
"GLODAPv2.2020_clean_obs_grid_era.csv",
sep = ""
))
GLODAP_grid_era <- GLODAP_grid_era %>%
mutate(Version_ID = i_Version_IDs)
if (exists("GLODAP_grid_era_all")) {
GLODAP_grid_era_all <-
bind_rows(GLODAP_grid_era_all, GLODAP_grid_era)
}
if (!exists("GLODAP_grid_era_all")) {
GLODAP_grid_era_all <- GLODAP_grid_era
}
}
rm(GLODAP_grid_era)
GLODAP_grid_era_all <- full_join(GLODAP_grid_era_all,
params_local_all)2.7 Atm CO2
co2_atm <-
read_csv(paste(path_preprocessing,
"co2_atm.csv",
sep = ""))
co2_atm_reccap2 <-
read_csv(paste(path_preprocessing,
"co2_atm_reccap2.csv",
sep = ""))2.8 Sabine total Cant
tcant_inv <-
read_csv(paste(path_preprocessing,
"S04_tcant_inv.csv", sep = ""))
tcant_inv <- tcant_inv %>%
filter(inv_depth == 3000) %>%
rename(dcant = tcant,
dcant_pos = tcant_pos) %>%
mutate(tref1 = 1800,
tref2 = 1994)2.9 GCB carbon sink
Ocean_Sink <-
read_csv(paste(path_preprocessing,
"Ocean_Sink.csv",
sep = ""))
Global_Carbon_Budget <-
read_csv(paste(path_preprocessing,
"GCB_Global_Carbon_Budget.csv",
sep = ""))3 Define labels
dcant_pgc_label <- expression(Delta * C["ant"]~(PgC))
dcant_pgc_scaled_label <- expression(beta~(PgC~µatm^{-1}))
dcant_umol_label <- expression(Delta * C[ant]~(mu * mol ~ kg ^ {-1}))
ddcant_umol_label <- expression(Delta * Delta * C[ant]~(mu * mol ~ kg ^ {-1}))
dcant_layer_budget_label <-
expression(Delta ~ C[ant] ~ "budget per 500m depth layer (PgC)")
dcant_CI_label <- expression(Delta * C[ant]~(mol ~ m ^ {-2}))
beta_CI_label <- expression(beta~(mol ~ m ^ {-2} ~ µatm ^ {-1}))4 Coverage maps
coverage_map <- map +
geom_tile(data = GLODAP_grid_era_all,
aes(lon, lat),
fill = "#377eb8",
col = "#377eb8") +
facet_wrap( ~ era, ncol = 2) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
coverage_map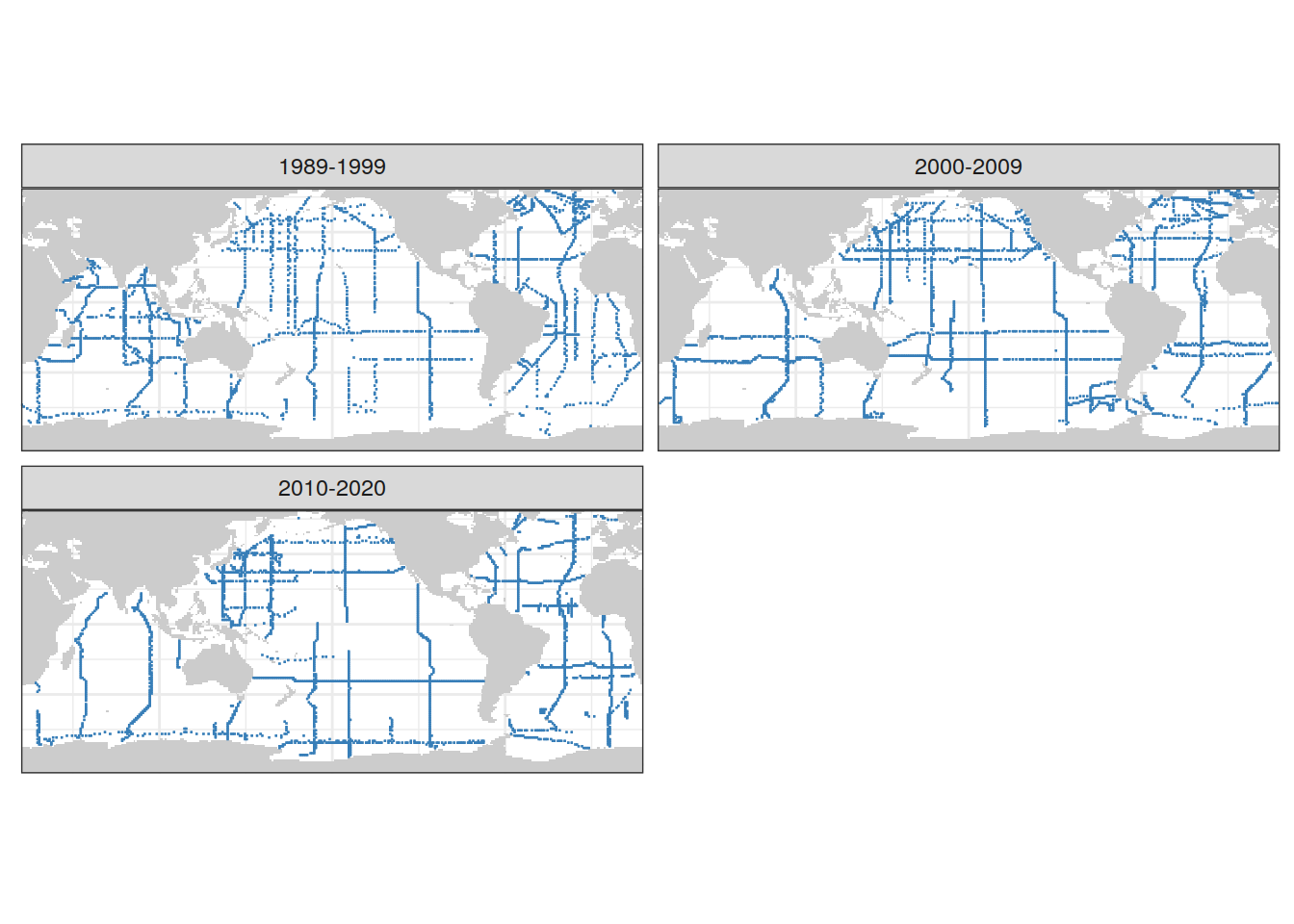
ggsave(plot = coverage_map,
path = here::here("output/publication"),
filename = "FigS_coverage_map_observations.png",
height = 4,
width = 8)5 Inventory maps
5.1 Absoulte values
5.1.1 dcant - absolute
dcant_inv_all <- dcant_inv_all %>%
filter(data_source %in% c("obs"),
period != "1994 - 2014")
dcant_inv_all <- left_join(dcant_inv_all,
co2_atm %>% rename(tref1 = year,
pCO21 = pCO2))
dcant_inv_all <- left_join(dcant_inv_all,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
tcant_inv <- tcant_inv %>%
mutate(pCO21 = 280)
tcant_inv <- left_join(tcant_inv,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
dcant_inv_all <- bind_rows(
dcant_inv_all,
tcant_inv %>% mutate(period = paste(tref1, tref2, sep = " - "))
)
dcant_inv_all <- dcant_inv_all %>%
mutate(delta_pCO2 = pCO22 - pCO21,
beta = dcant / delta_pCO2,
beta_pos = dcant_pos / delta_pCO2) %>%
select(-starts_with("pCO2"))
dcant_inv_all_ensemble <- dcant_inv_all
unique(dcant_inv_all$Version_ID) [1] "v_1101" "v_1102" "v_1103" "v_1104" "v_1105" "v_1106" "v_2101" "v_2102"
[9] "v_2103" "v_2104" "v_2105" "v_2106" "v_1o01" "v_1o02" "v_1o03" "v_1o04"
[17] "v_1o05" "v_1o06" "v_2o01" "v_2o02" "v_2o03" "v_2o04" "v_2o05" "v_2o06"
[25] "v_1d01" "v_1d02" "v_1d03" "v_1d04" "v_1d05" "v_1d06" "v_2d01" "v_2d02"
[33] "v_2d03" "v_2d04" "v_2d05" "v_2d06" "v_1g01" "v_1g02" "v_1g03" "v_1g04"
[41] "v_1g05" "v_1g06" "v_2g01" "v_2g02" "v_2g03" "v_2g04" "v_2g05" "v_2g06"
[49] "v_1n01" "v_1n02" "v_1n03" "v_1n04" "v_1n05" "v_1n06" "v_2n01" "v_2n02"
[57] "v_2n03" "v_2n04" "v_2n05" "v_2n06" "v_1c01" "v_1c02" "v_1c03" "v_1c04"
[65] "v_1c05" "v_1c06" "v_2c01" "v_2c02" "v_2c03" "v_2c04" "v_2c05" "v_2c06"
[73] NA dcant_inv_all <- dcant_inv_all %>%
filter(Version_ID %in% Version_IDs | is.na(Version_ID))p_CI_absolute <- dcant_inv_all %>%
filter(period != "1800 - 1994") %>%
p_map_cant_inv(var = "dcant",
title_text = NULL) +
facet_grid(period ~ .) +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank())
p_CI_absolute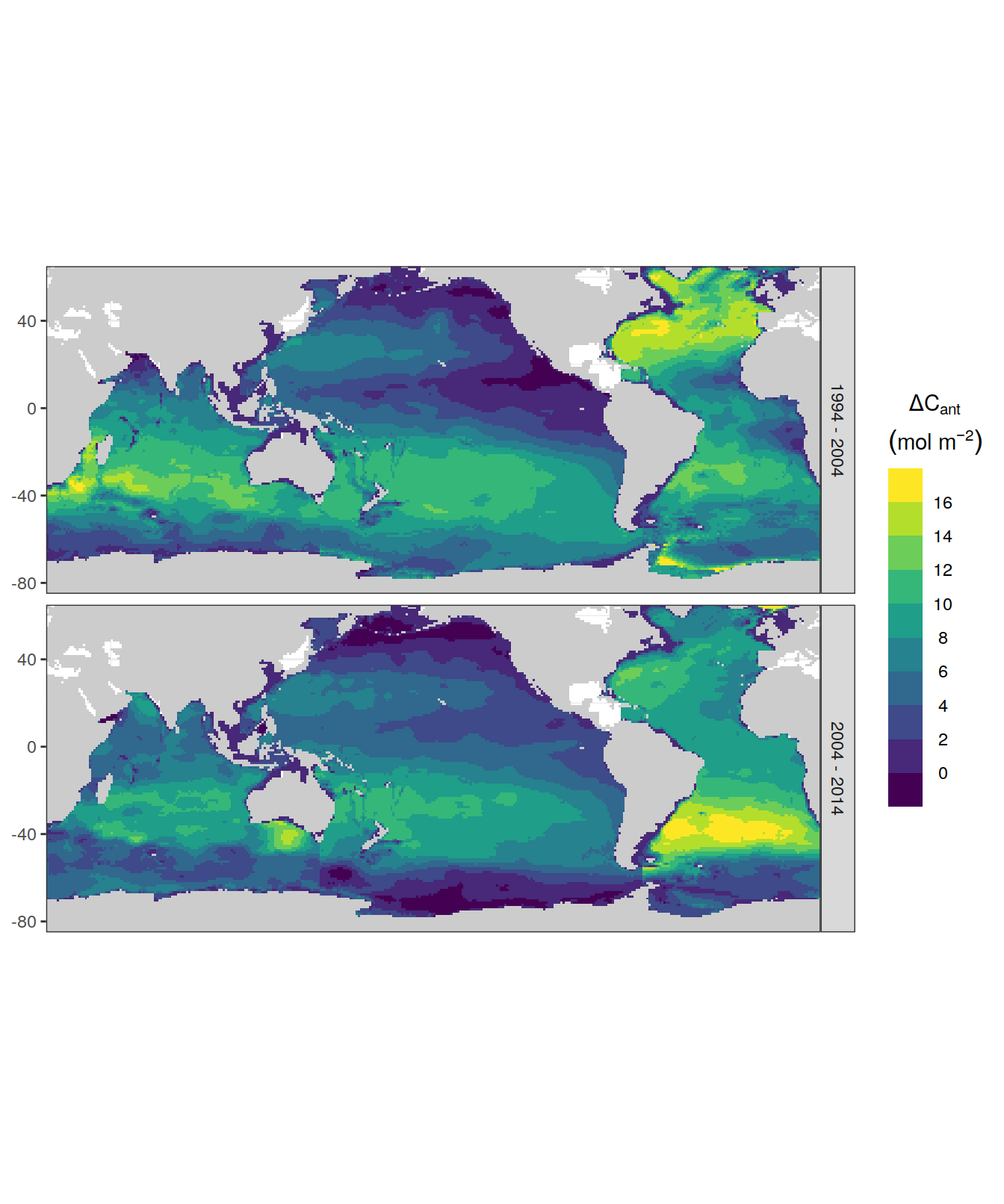
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 09b0780 | jens-daniel-mueller | 2022-05-24 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| 08607eb | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
dcant_inv_all %>%
p_map_cant_inv(var = "beta",
breaks = c(-Inf, seq(0, 1, 0.1), Inf),
title_text = NULL) +
facet_grid(period ~ .)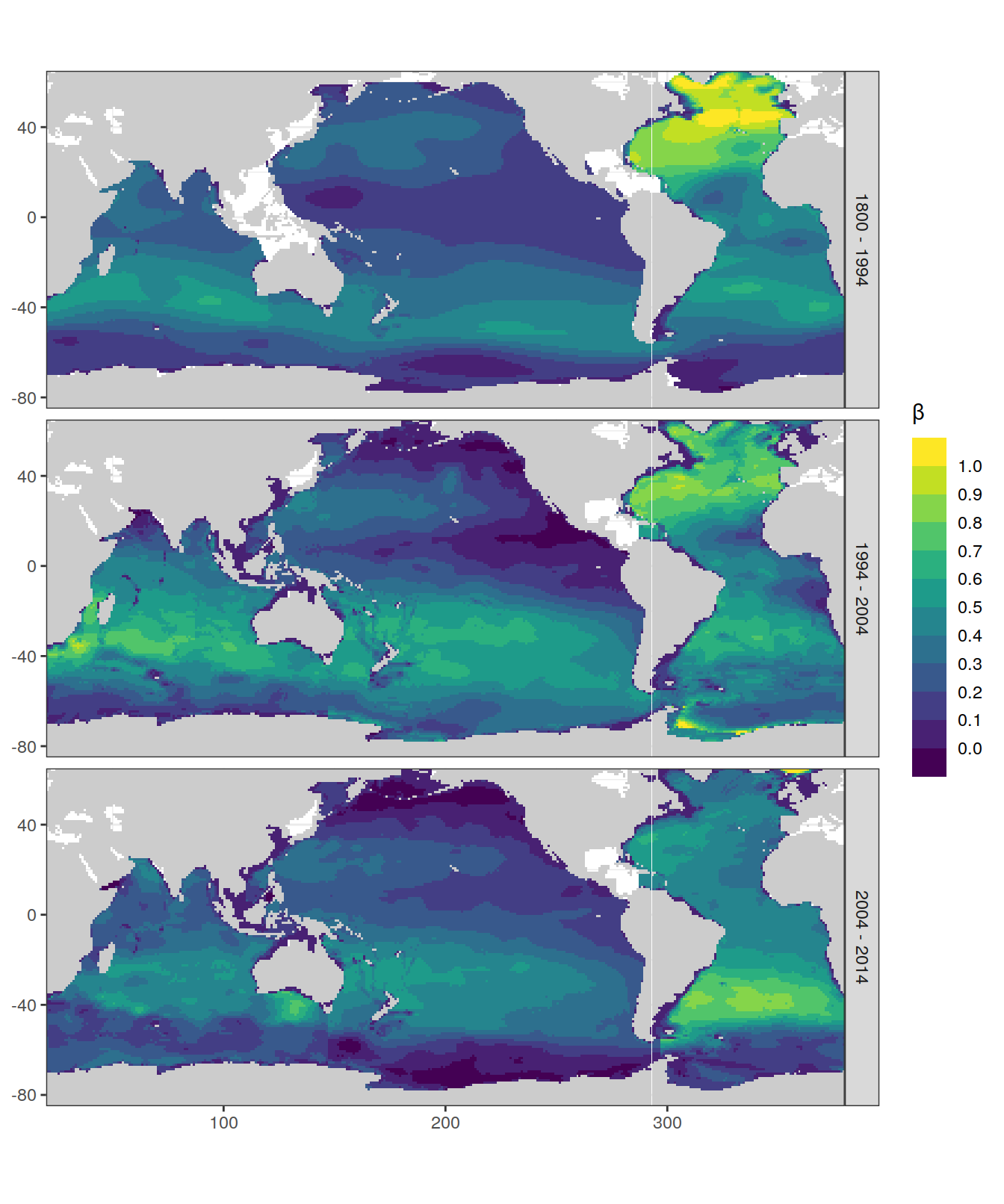
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 09b0780 | jens-daniel-mueller | 2022-05-24 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
ggsave(path = here::here("output/publication"),
filename = "FigS_column_inventory_map_beta.png",
height = 6,
width = 6)
dcant_inv_all %>%
filter(period != "1800 - 1994") %>%
p_map_cant_inv(var = "beta",
breaks = c(-Inf, seq(0, 1, 0.1), Inf),) +
facet_grid(period ~ .)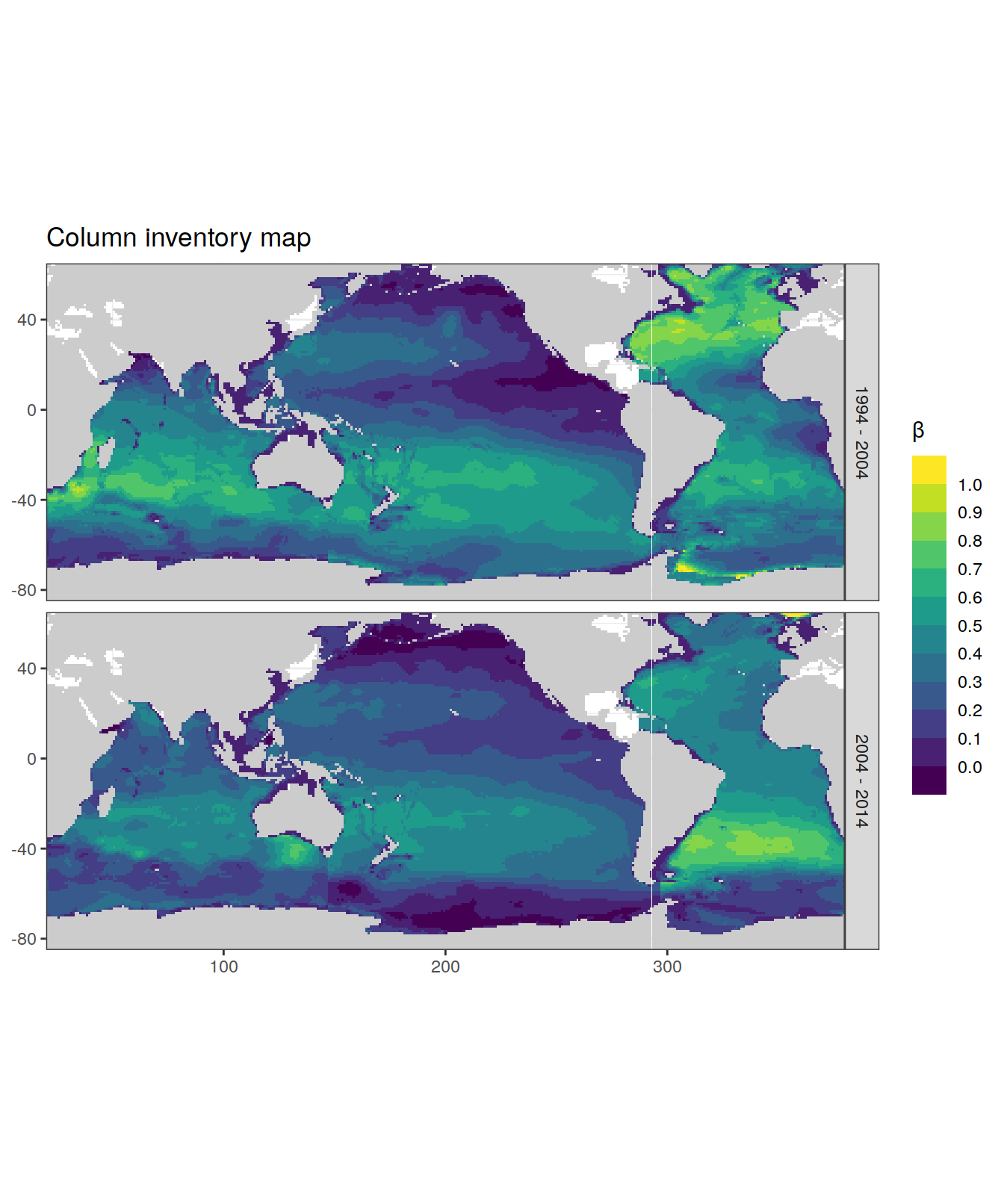
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 09b0780 | jens-daniel-mueller | 2022-05-24 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
5.1.2 dcant - absolute delta
p_CI_delta <-
dcant_inv_all %>%
filter(data_source %in% c("obs"),
period != "1994 - 2014") %>%
select(data_source, lon, lat, basin_AIP, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(dcant_offset = `2004 - 2014` - `1994 - 2004`) %>%
group_by(data_source) %>%
group_split() %>%
# head(1) %>%
map(
~ p_map_cant_inv(
df = .x,
var = "dcant_offset",
title_text = NULL,
subtitle_text = NULL,
col = "bias"
)
)
p_CI_absolute / p_CI_delta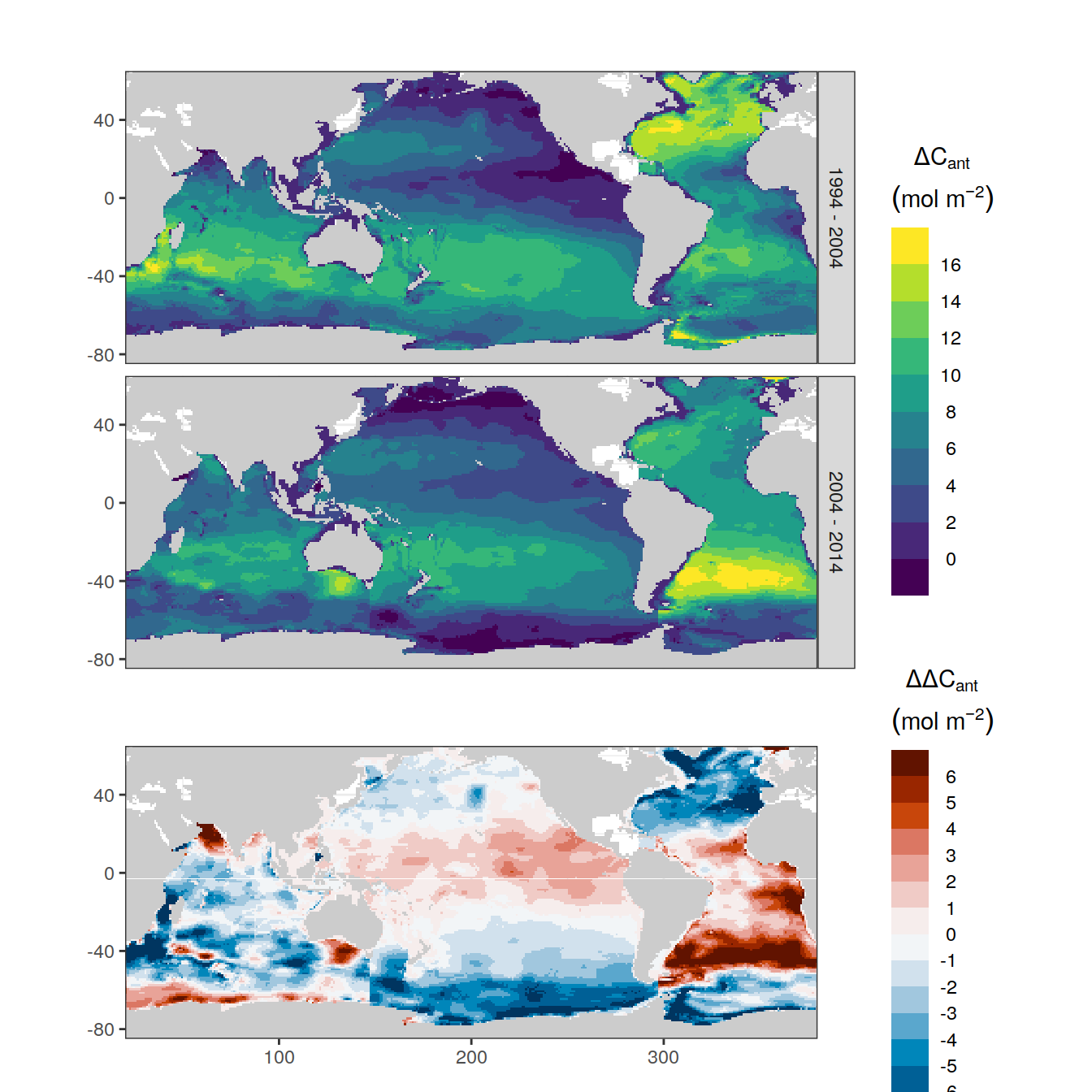
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
ggsave(path = here::here("output/publication"),
filename = "Fig_column_inventory_map_dcant.png",
height = 9,
width = 8)5.1.3 dcant - absolute delta scaled
dcant_budget_scaling <- dcant_budget_global_all %>%
filter(period != "1994 - 2014") %>%
select(data_source, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(dcant_scaling = `2004 - 2014` / `1994 - 2004`) %>%
select(data_source, dcant_scaling)
left_join(dcant_inv_all %>% filter(data_source == "obs"),
dcant_budget_scaling %>% filter(data_source == "obs")) %>%
filter(period != "1994 - 2014") %>%
select(data_source, lon, lat, basin_AIP, period, dcant, dcant_scaling) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(delta_dcant = `2004 - 2014` - `1994 - 2004`*dcant_scaling) %>%
group_by(data_source) %>%
group_split() %>%
# head(1) %>%
map(
~ p_map_cant_inv(df = .x,
var = "delta_dcant",
col = "bias") +
# facet_grid(period ~ .) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
)[[1]]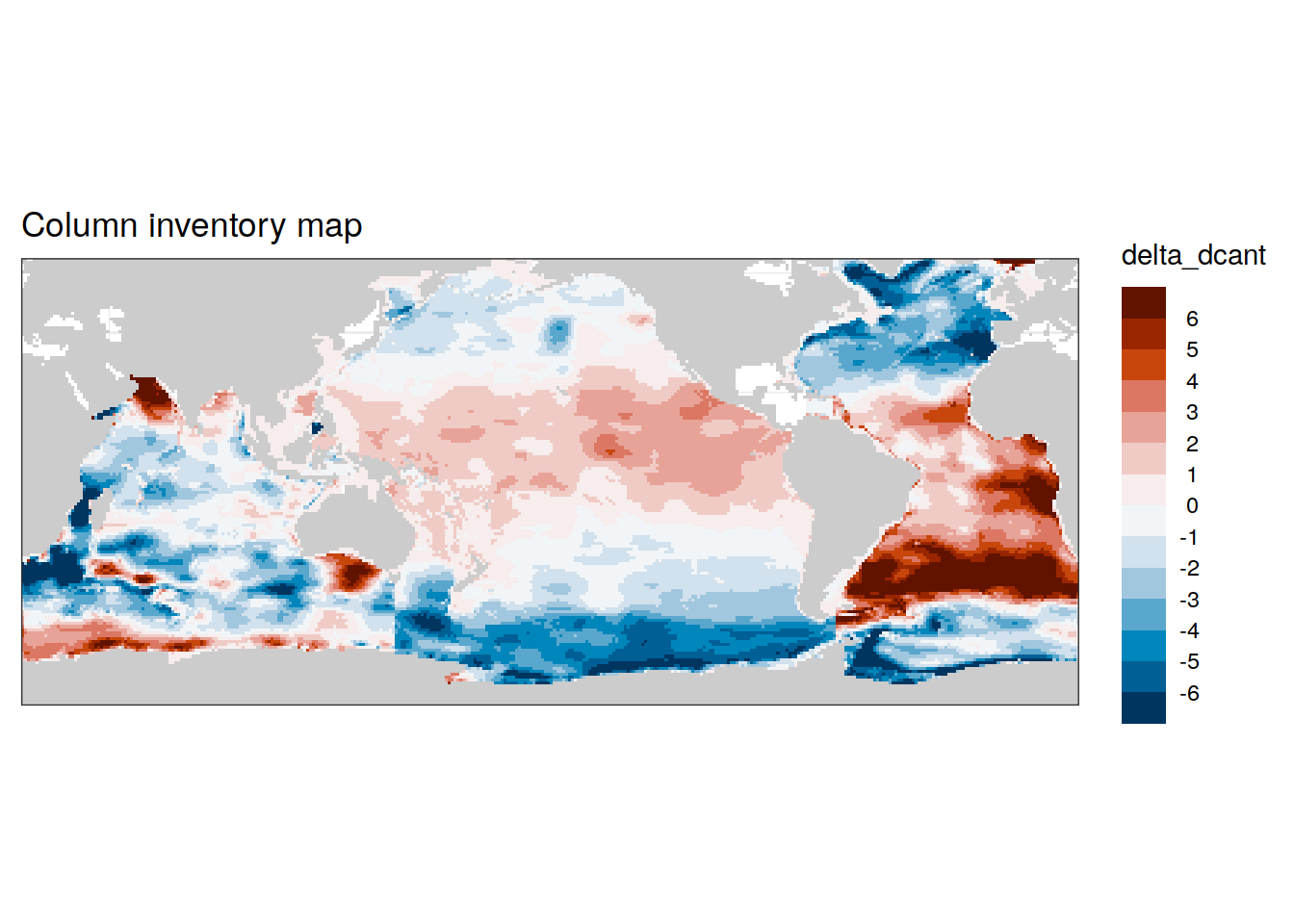
| Version | Author | Date |
|---|---|---|
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
dcant_inv_all_ensemble_mean <-
dcant_inv_all_ensemble %>%
filter(period != "1800 - 1994",
data_source == "obs") %>%
group_by(data_source, period, lon, lat) %>%
summarise(dcant_sd = sd(dcant)) %>%
ungroup()
dcant_inv_all_ensemble_mean %>%
p_map_cant_inv(var = "dcant_sd",
breaks = c(seq(0, 5, 0.5), Inf),
title_text = NULL) +
facet_grid(period ~ .) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())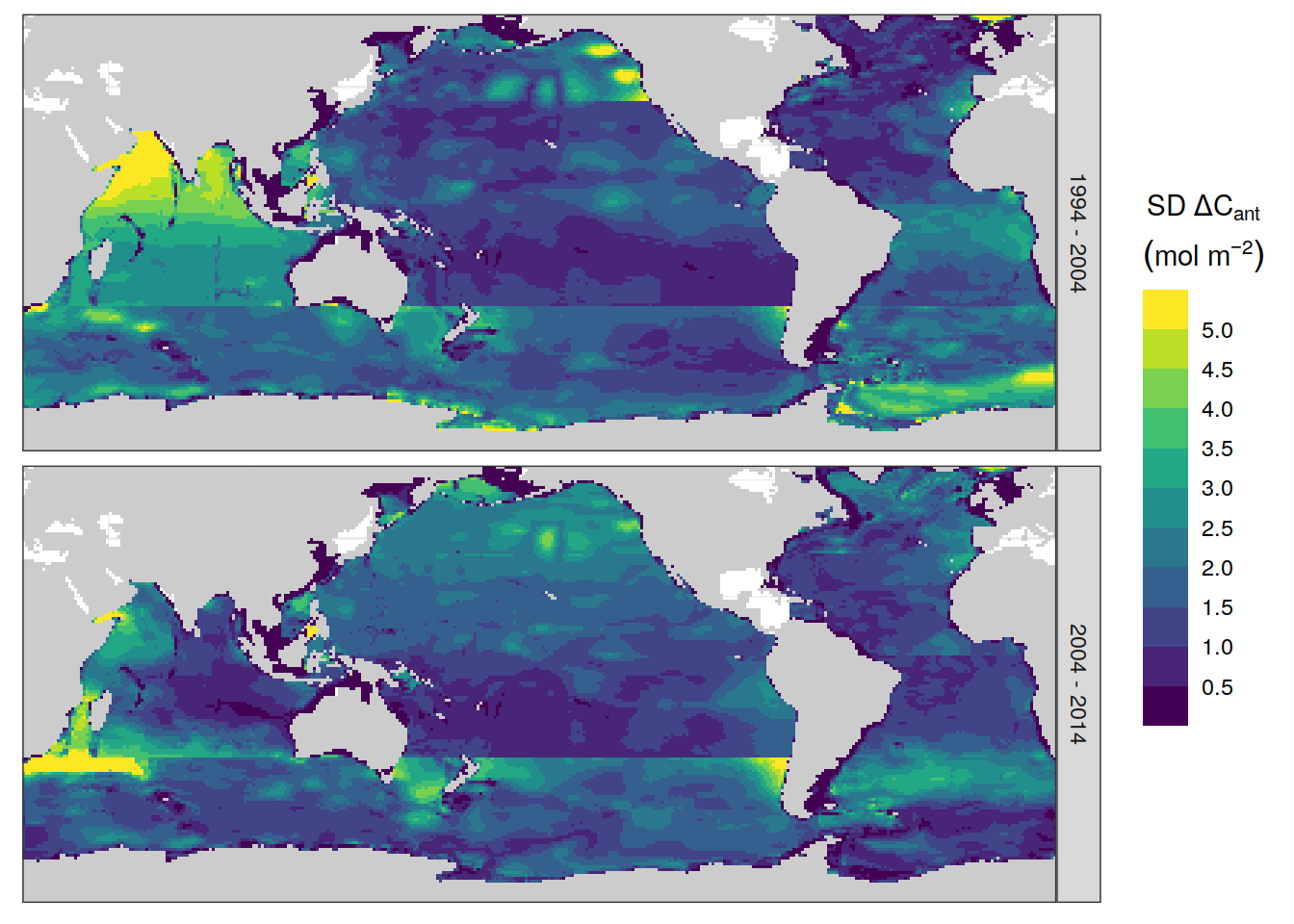
ggsave(path = here::here("output/publication"),
filename = "FigS_column_inventory_map_dcant_ensemble_standard_deviation.png",
height = 4,
width = 6)5.2 Zonal means
dcant_inv_all <- m_grid_horizontal_coarse(dcant_inv_all)
dcant_inv_all_lat_mean <- dcant_inv_all %>%
group_by(basin_AIP, period, lat_grid) %>%
summarise(beta = mean(beta, na.rm = TRUE),
dcant = mean(dcant, na.rm = TRUE)) %>%
ungroup()
dcant_inv_all_ensemble <- m_grid_horizontal_coarse(dcant_inv_all_ensemble)
dcant_inv_all_ensemble_lat_mean <- dcant_inv_all_ensemble %>%
group_by(basin_AIP, period, lat_grid, Version_ID) %>%
summarise(beta_mean = mean(beta, na.rm = TRUE),
dcant_mean = mean(dcant, na.rm = TRUE)) %>%
ungroup()
dcant_inv_all_ensemble_lat_mean_average <- dcant_inv_all_ensemble_lat_mean %>%
group_by(basin_AIP, period, lat_grid) %>%
summarise(beta_sd = sd(beta_mean, na.rm = TRUE),
beta_mean = mean(beta_mean, na.rm = TRUE),
dcant_sd = sd(dcant_mean, na.rm = TRUE),
dcant_mean = mean(dcant_mean, na.rm = TRUE)) %>%
ungroup()
dcant_inv_all_lat_mean <- full_join(dcant_inv_all_lat_mean,
dcant_inv_all_ensemble_lat_mean_average)5.2.1 dcant
dcant_inv_all_lat_mean %>%
filter(period != "1800 - 1994") %>%
ggplot(aes(lat_grid, dcant, fill = period)) +
geom_hline(yintercept = c(0,10)) +
geom_vline(xintercept = c(-10,10)) +
geom_vline(xintercept = c(-40,-30,30,40)) +
geom_ribbon(aes(ymin = dcant - dcant_sd,
ymax = dcant + dcant_sd),
alpha = 0.3) +
geom_path(aes(col = period)) +
geom_point(shape = 21) +
scale_color_brewer(palette = "Dark2", name = "Mean ± SD") +
scale_fill_brewer(palette = "Dark2", name = "Mean ± SD") +
coord_flip() +
facet_grid(. ~ basin_AIP) +
labs(x = "Latitude (°N)",
y = dcant_CI_label) +
scale_x_continuous(breaks = seq(-75,75, 15))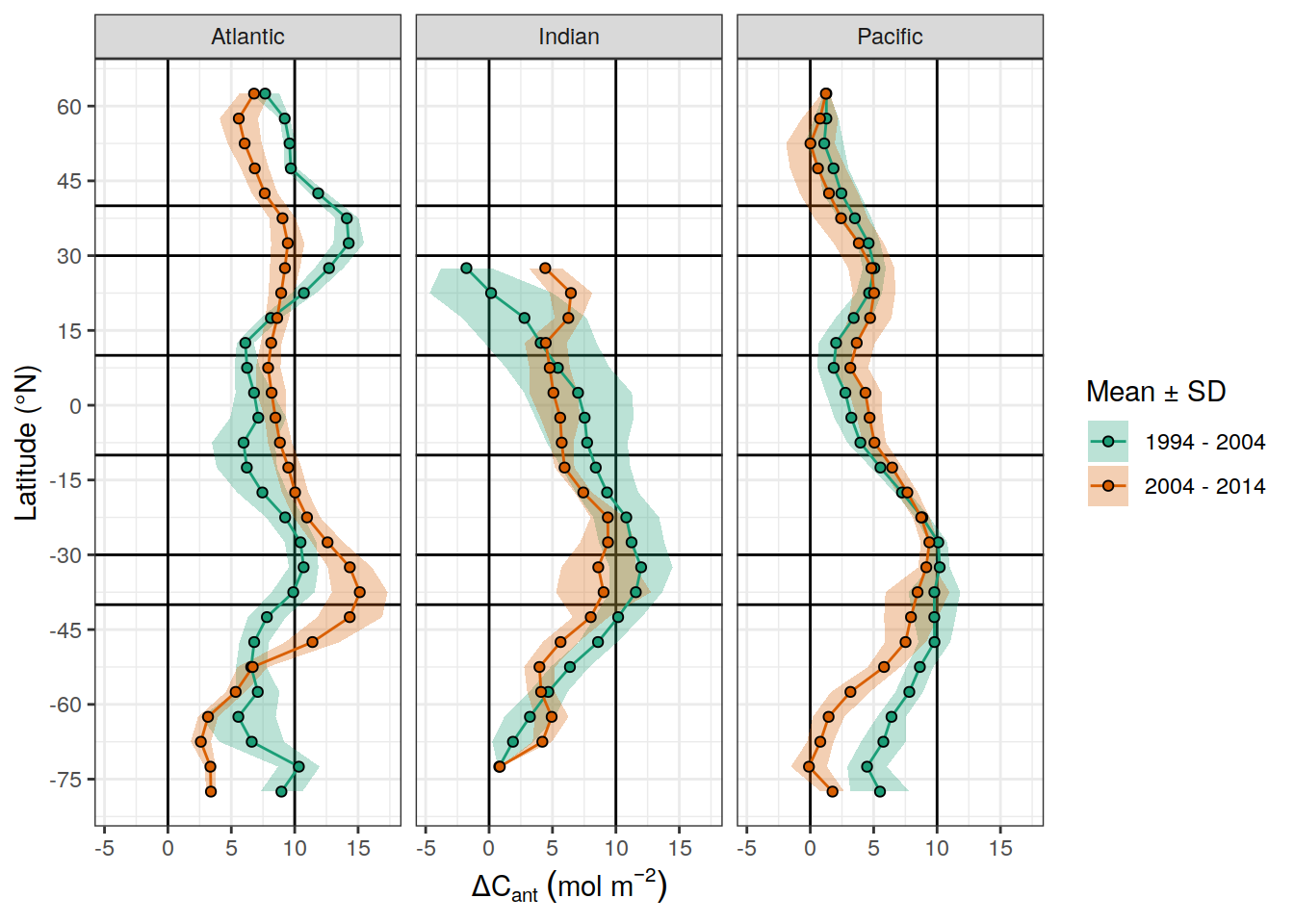
dcant_inv_all_lat_mean %>%
filter(period != "1800 - 1994") %>%
mutate(lat_grid = cut(lat_grid, c(-40,-30,-10,10,30,40))) %>%
group_by(lat_grid) %>%
summarise(dcant_mean = mean(dcant_mean, na_rm = TRUE)) %>%
ungroup()# A tibble: 6 × 2
lat_grid dcant_mean
<fct> <dbl>
1 (-40,-30] 10.9
2 (-30,-10] 8.72
3 (-10,10] 5.72
4 (10,30] 5.68
5 (30,40] 8.25
6 <NA> 5.51dcant_inv_all_lat_mean %>%
filter(period != "1800 - 1994") %>%
mutate(lat_grid = cut(lat_grid, c(-40,-30,-10,10,30,40))) %>%
group_by(lat_grid, basin_AIP) %>%
summarise(dcant_mean = mean(dcant_mean, na_rm = TRUE)) %>%
ungroup()# A tibble: 17 × 3
lat_grid basin_AIP dcant_mean
<fct> <chr> <dbl>
1 (-40,-30] Atlantic 11.7
2 (-40,-30] Indian 11.1
3 (-40,-30] Pacific 9.80
4 (-30,-10] Atlantic 8.68
5 (-30,-10] Indian 9.30
6 (-30,-10] Pacific 8.16
7 (-10,10] Atlantic 6.80
8 (-10,10] Indian 6.24
9 (-10,10] Pacific 4.11
10 (10,30] Atlantic 9.00
11 (10,30] Indian 3.06
12 (10,30] Pacific 4.97
13 (30,40] Atlantic 11.9
14 (30,40] Pacific 4.63
15 <NA> Atlantic 6.91
16 <NA> Indian 5.22
17 <NA> Pacific 4.28dcant_inv_all_ensemble_lat_mean %>%
ggplot(aes(lat_grid, dcant_mean, fill=period, group=Version_ID)) +
geom_hline(yintercept = 0) +
geom_path(aes(col=period)) +
scale_color_brewer(palette = "Dark2") +
scale_fill_brewer(palette = "Dark2") +
coord_flip() +
facet_grid(. ~ basin_AIP) +
labs(x = "Latitude (°N)",
y = dcant_CI_label)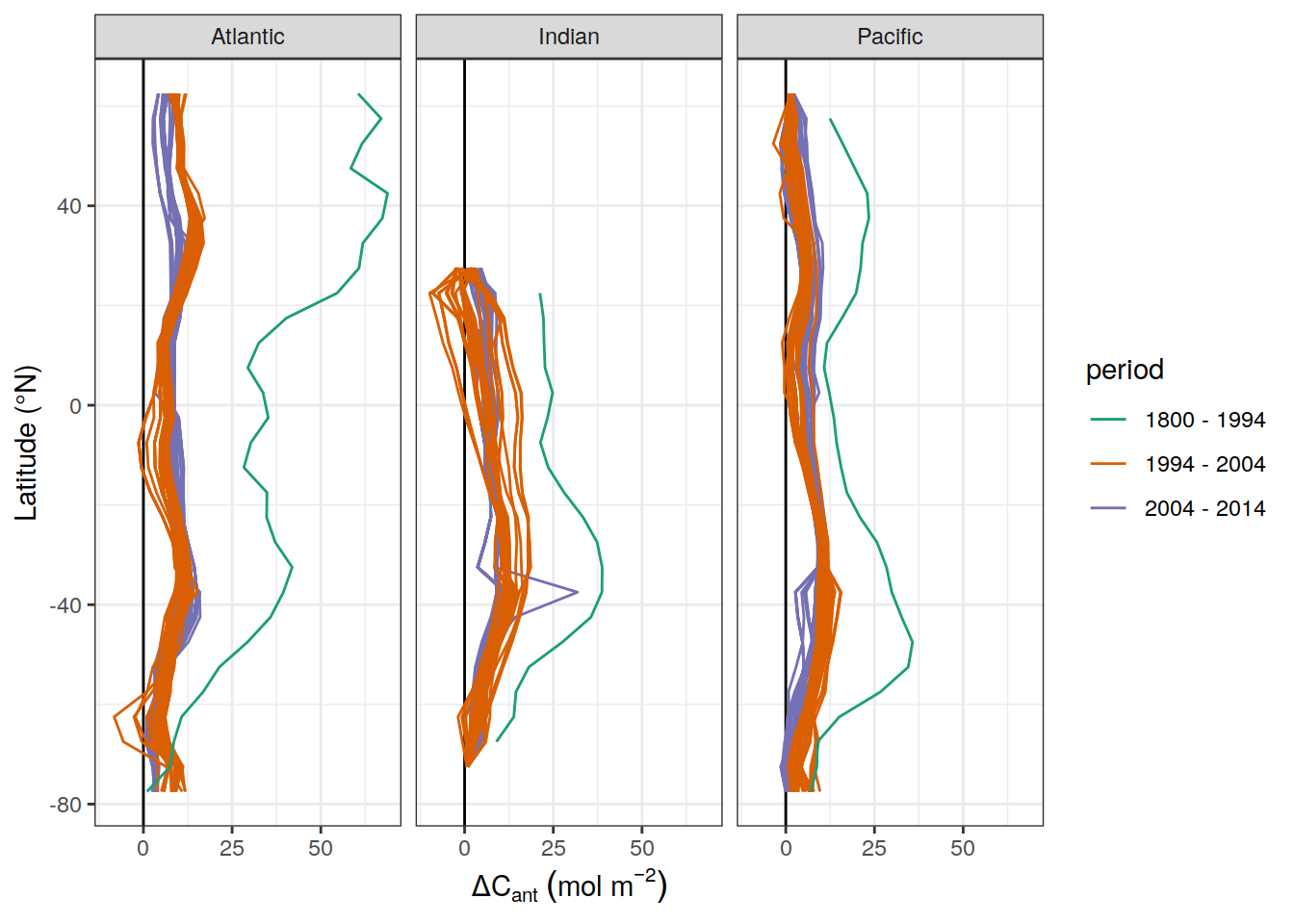
5.2.2 beta
dcant_inv_all_lat_mean %>%
ggplot(aes(lat_grid, beta, fill = period)) +
geom_hline(yintercept = 0) +
geom_ribbon(aes(ymin = beta - beta_sd,
ymax = beta + beta_sd),
alpha = 0.3) +
geom_path(aes(col = period)) +
geom_point(shape = 21) +
scale_color_brewer(palette = "Dark2", name = "Mean ± SD") +
scale_fill_brewer(palette = "Dark2", name = "Mean ± SD") +
coord_flip() +
facet_grid(. ~ basin_AIP) +
labs(x = "Latitude (°N)",
y = beta_CI_label) +
scale_x_continuous(breaks = seq(-75,75, 15))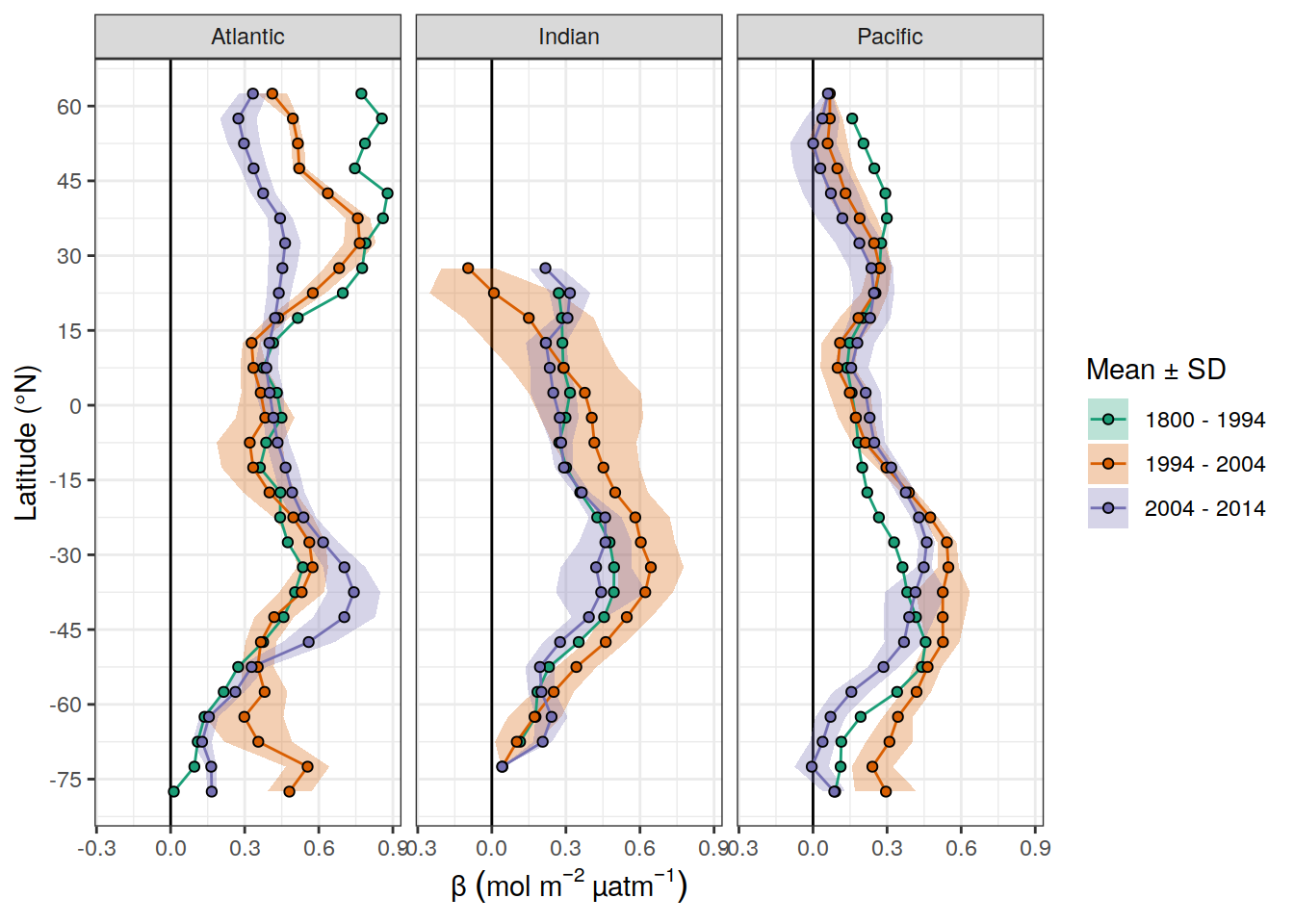
dcant_inv_all_ensemble_lat_mean %>%
ggplot(aes(lat_grid, beta_mean, fill=period, group=Version_ID)) +
geom_hline(yintercept = 0) +
geom_path(aes(col=period)) +
scale_color_brewer(palette = "Dark2") +
scale_fill_brewer(palette = "Dark2") +
coord_flip() +
facet_grid(. ~ basin_AIP) +
labs(x = "Latitude (°N)",
y = beta_CI_label)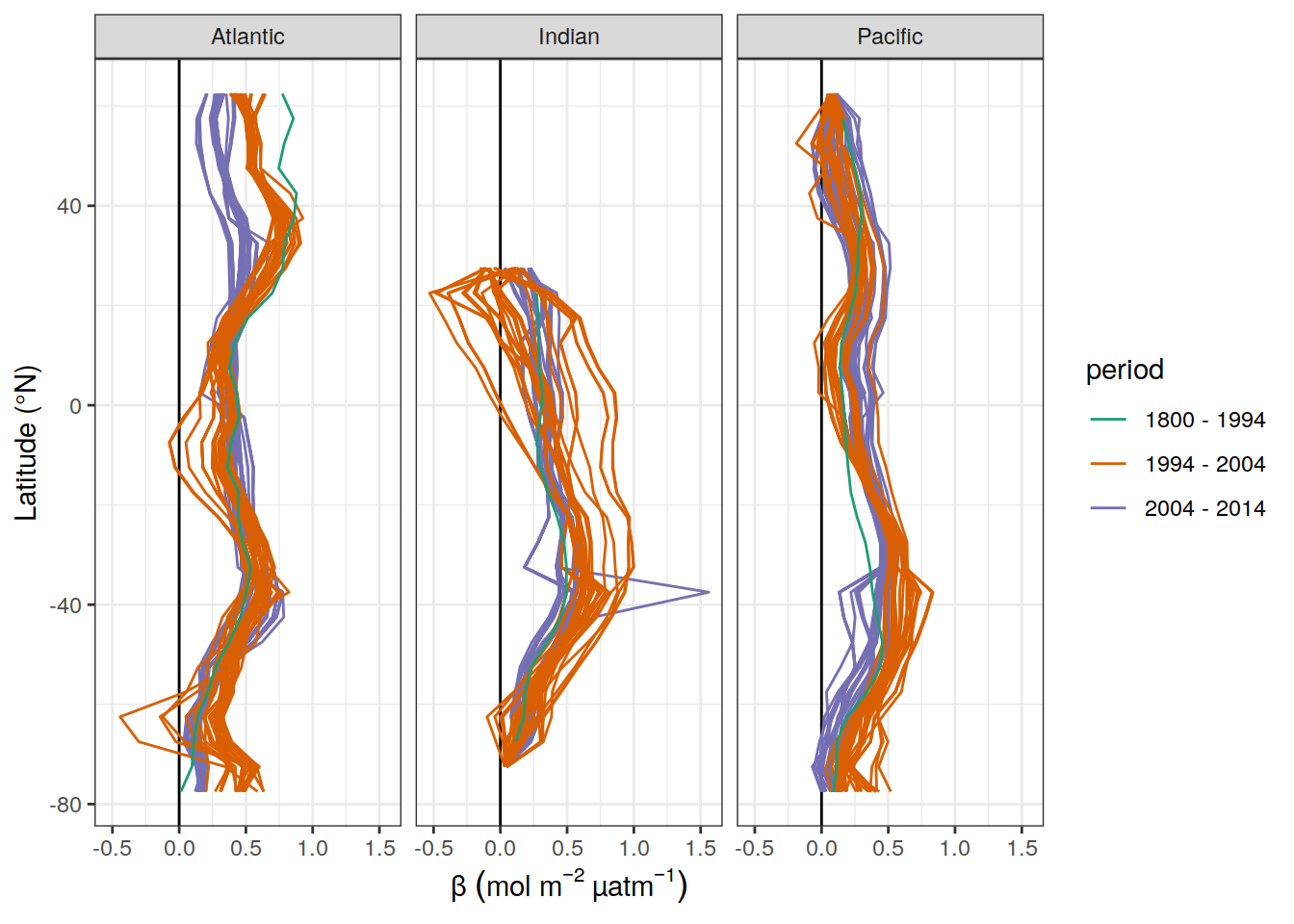
dcant_inv_all_lat_mean %>%
filter(basin_AIP == "Atlantic") %>%
ggplot(aes(lat_grid, beta, fill = period)) +
geom_hline(yintercept = 0) +
geom_ribbon(aes(ymin = beta - beta_sd,
ymax = beta + beta_sd),
alpha = 0.3) +
geom_path(aes(col = period)) +
geom_point(shape = 21) +
scale_color_brewer(palette = "Dark2", name = "Mean ± SD") +
scale_fill_brewer(palette = "Dark2", name = "Mean ± SD") +
coord_flip() +
facet_grid(. ~ basin_AIP) +
labs(x = "Latitude (°N)",
y = beta_CI_label) +
scale_x_continuous(breaks = seq(-75,75, 15))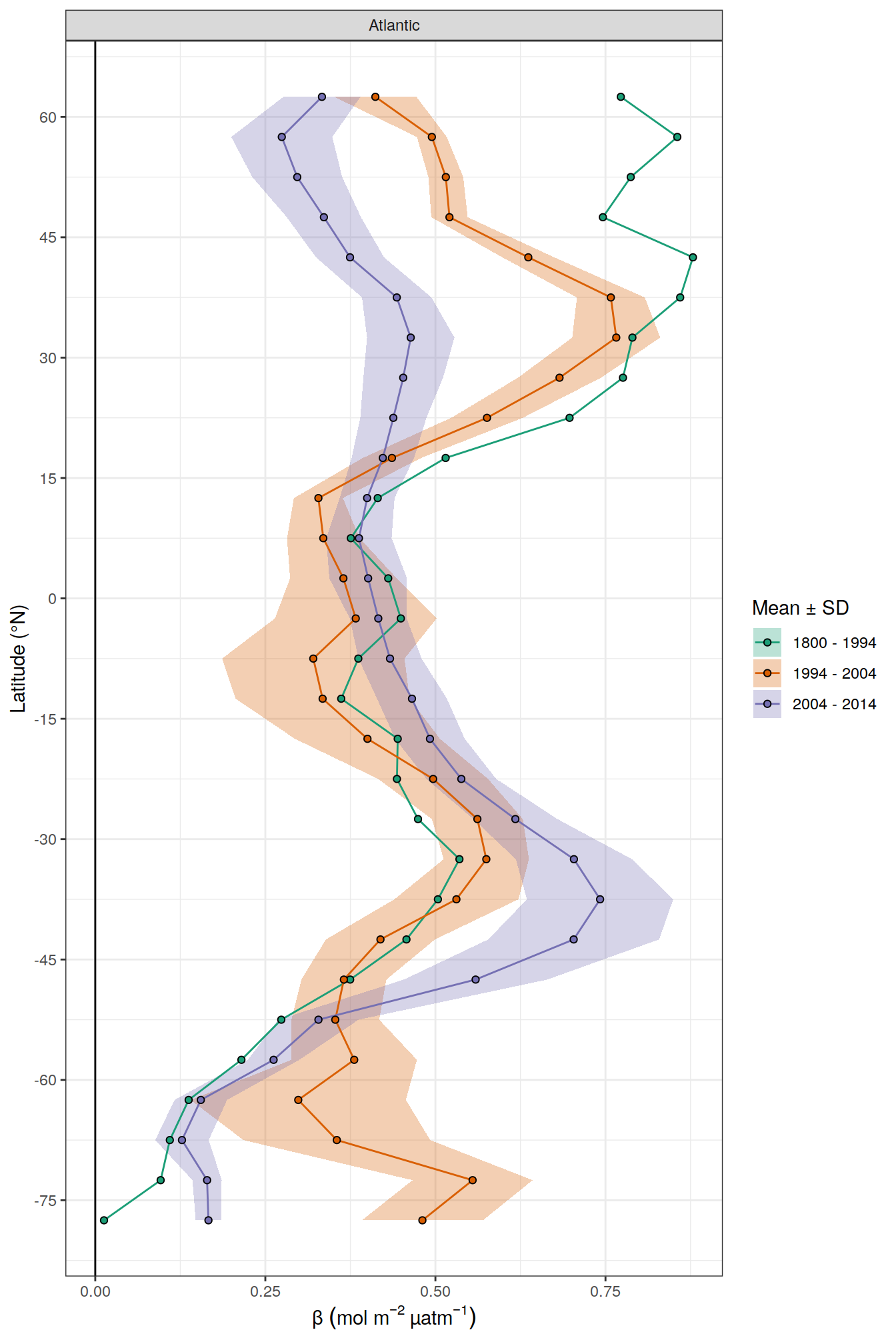
6 Zonal sections
6.1 dcant - absolut
p_dcant_zonal_absolute <-
dcant_zonal_all %>%
filter(
data_source == "obs",
period != "1994 - 2014",
depth <= params_global$inventory_depth_standard
) %>%
p_section_zonal_continous_depth(var = "dcant",
plot_slabs = "n",
title_text = NULL) +
geom_contour(aes(lat, depth, z = dcant),
breaks = 5,
col = "white") +
# geom_text_contour(
# aes(lat, depth, z = dcant),
# breaks = 5,
# stroke = 0.2,
# stroke.colour = "white",
# rotate = FALSE,
# label.placer = label_placer_n(n = 1)
# ) +
facet_grid(period ~ basin_AIP) +
theme(axis.text.x = element_blank(),
axis.title.x = element_blank())
p_dcant_zonal_absolute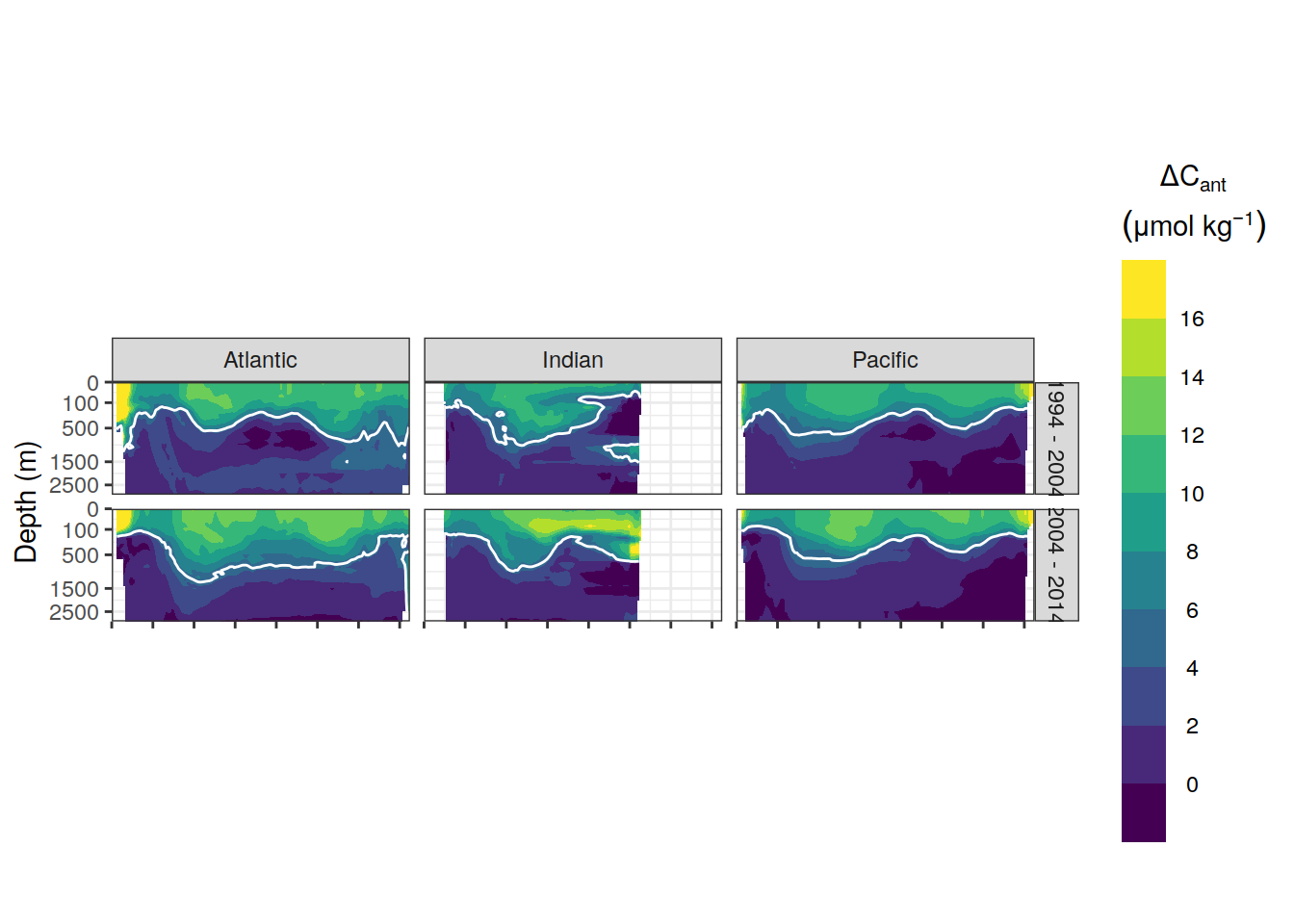
| Version | Author | Date |
|---|---|---|
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 09b0780 | jens-daniel-mueller | 2022-05-24 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| e572075 | jens-daniel-mueller | 2022-01-21 |
dcant_zonal_all %>%
filter(
data_source == "obs",
period != "1994 - 2014",
depth <= params_global$inventory_depth_standard
) %>%
group_split(basin_AIP) %>%
# head(1) %>%
map(
~ p_section_zonal_continous_depth(
df = .x,
var = "dcant",
breaks = c(-Inf, -1, 0, 1, seq(2,16,2), Inf),
plot_slabs = "n",
title_text = .x$basin_AIP,
subtitle_text = NULL
) +
facet_grid(period ~ .)
)6.2 dcant - absolute delta
dcant_zonal_all %>%
filter(data_source %in% c("mod", "obs"),
period != "1994 - 2014") %>%
select(data_source, lat, depth, basin_AIP, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(delta_dcant = `2004 - 2014` - `1994 - 2004`) %>%
group_by(data_source) %>%
group_split() %>%
# head(1) %>%
map(
~ p_section_zonal_continous_depth(
df = .x,
var = "delta_dcant",
plot_slabs = "n",
subtitle_text = unique(.x$data_source),
col = "bias"
) +
facet_grid(basin_AIP ~ .)
)[[1]]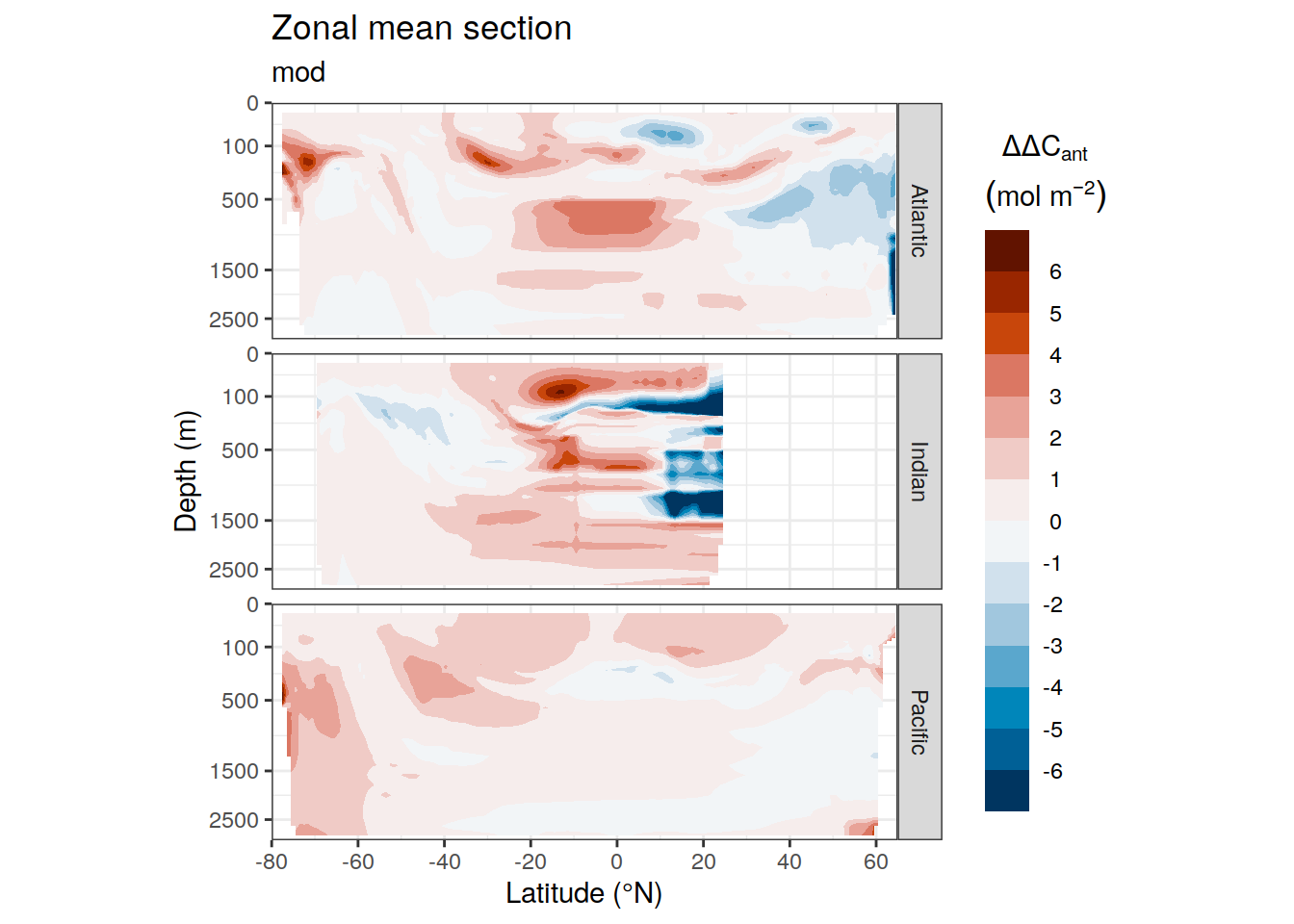
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| d6b399a | jens-daniel-mueller | 2022-01-21 |
[[2]]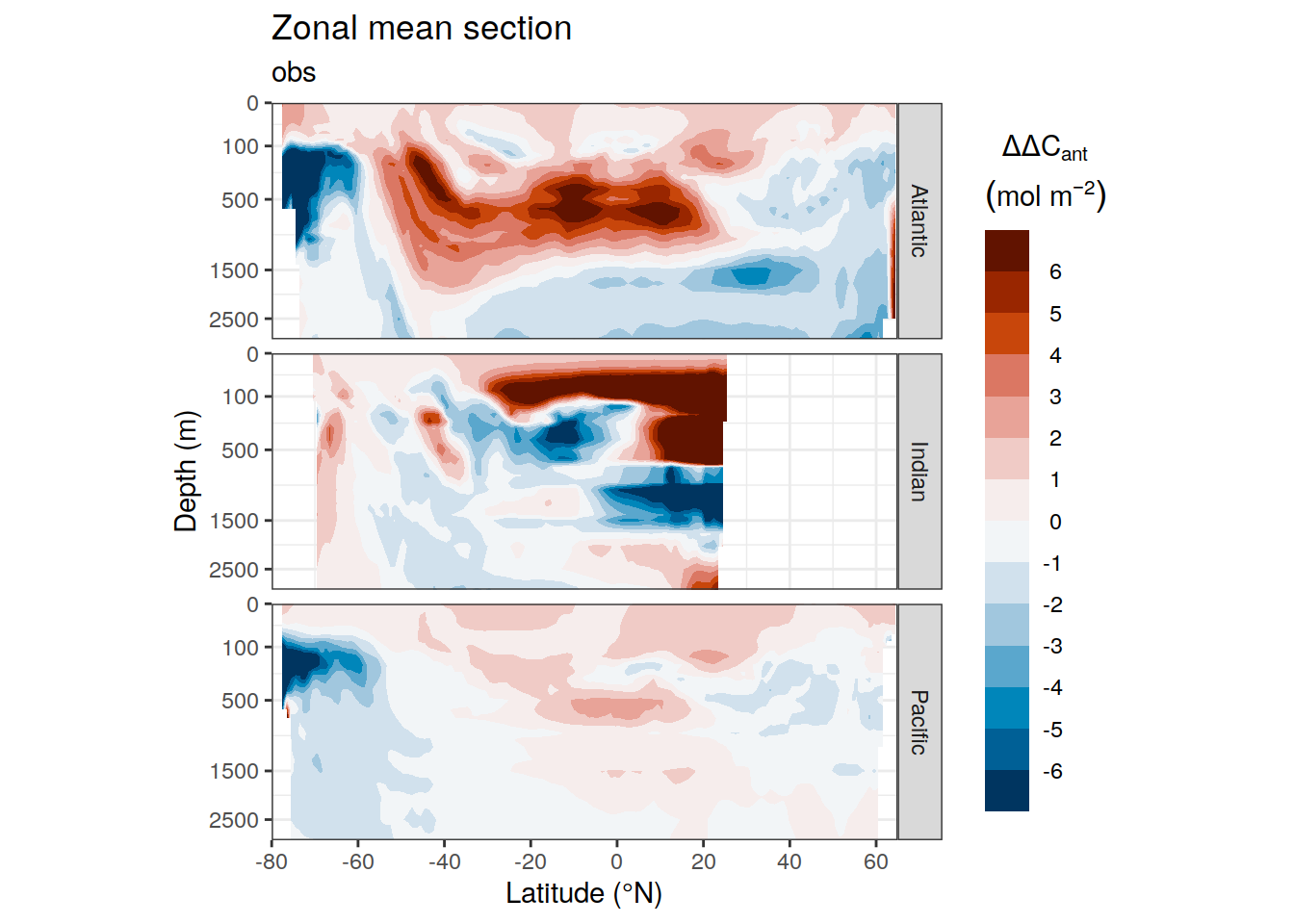
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| d6b399a | jens-daniel-mueller | 2022-01-21 |
p_dcant_zonal_delta <-
dcant_zonal_all %>%
filter(data_source %in% c("obs"),
period != "1994 - 2014") %>%
select(data_source, lat, depth, basin_AIP, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(delta_dcant = `2004 - 2014` - `1994 - 2004`) %>%
group_by(data_source) %>%
group_split() %>%
# head(1) %>%
map(
~ p_section_zonal_continous_depth(
df = .x,
var = "delta_dcant",
plot_slabs = "n",
subtitle_text = NULL,
title_text = NULL,
col = "bias"
) +
facet_grid(. ~ basin_AIP)
# theme(strip.text = element_blank(),
# strip.background = element_blank())
)
p_dcant_zonal_absolute / p_dcant_zonal_delta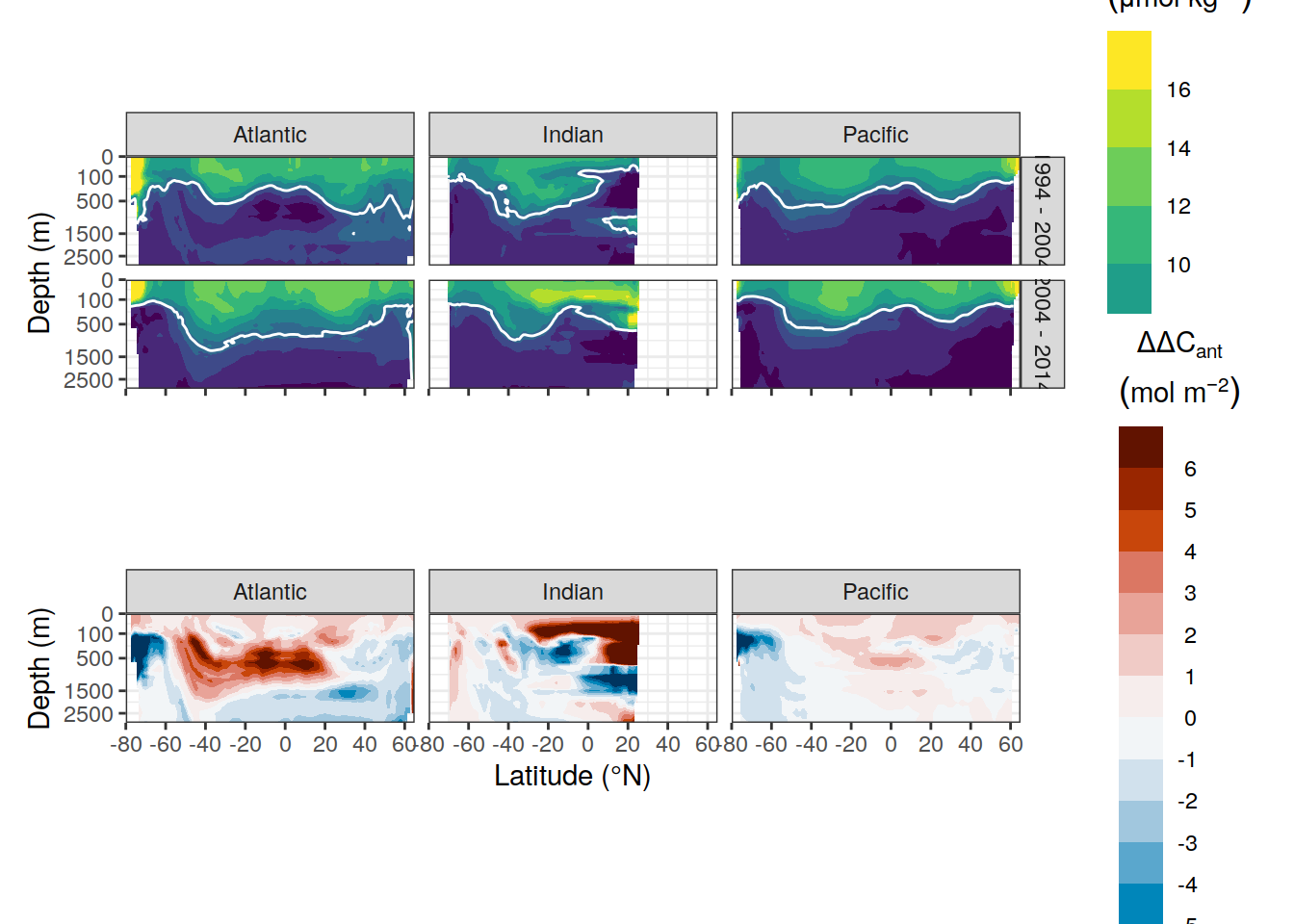
| Version | Author | Date |
|---|---|---|
| f09080e | jens-daniel-mueller | 2022-06-28 |
ggsave(path = here::here("output/publication"),
filename = "Fig_zonal_mean_sections_dcant.png",
height = 9,
width = 15)7 Concentration profiles
7.1 dcant - absolute
dcant_profile_global_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
depth <= params_global$inventory_depth_standard) %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = dcant - dcant_sd,
xmax = dcant + dcant_sd,
fill = period
),
alpha = 0.3
) +
geom_path(aes(col = period)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
# scale_y_reverse(breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(
palette = "Set1",
name = "mean \u00B1 sd",
direction = -1
) +
scale_fill_brewer(
palette = "Set1",
name = "mean \u00B1 sd",
direction = -1
) +
labs(
title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = dcant_umol_label
) +
facet_wrap( ~ basin, ncol = 3, dir = "v") +
theme(legend.position = c(0.8, 0.2))
)[[1]]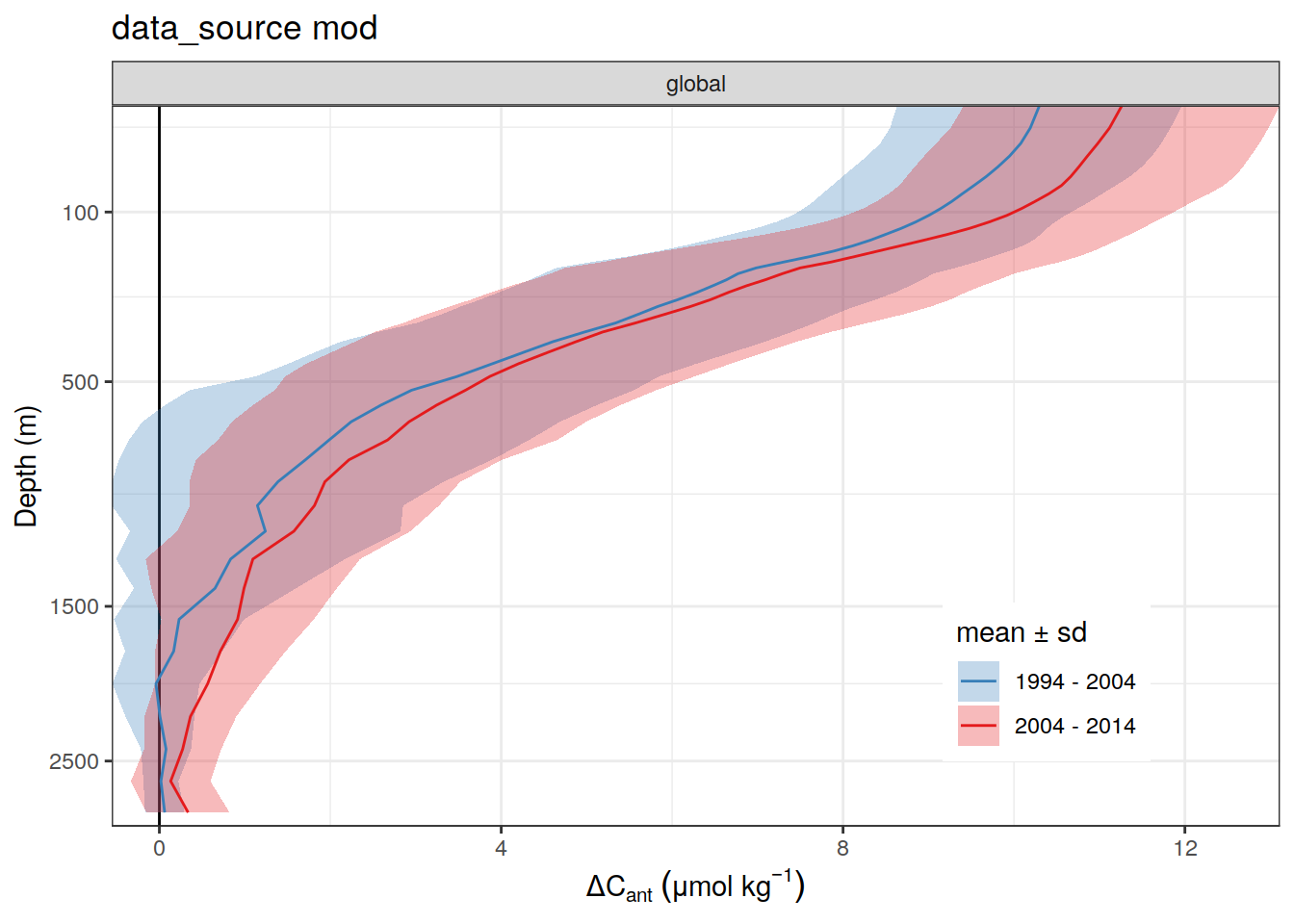
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
[[2]]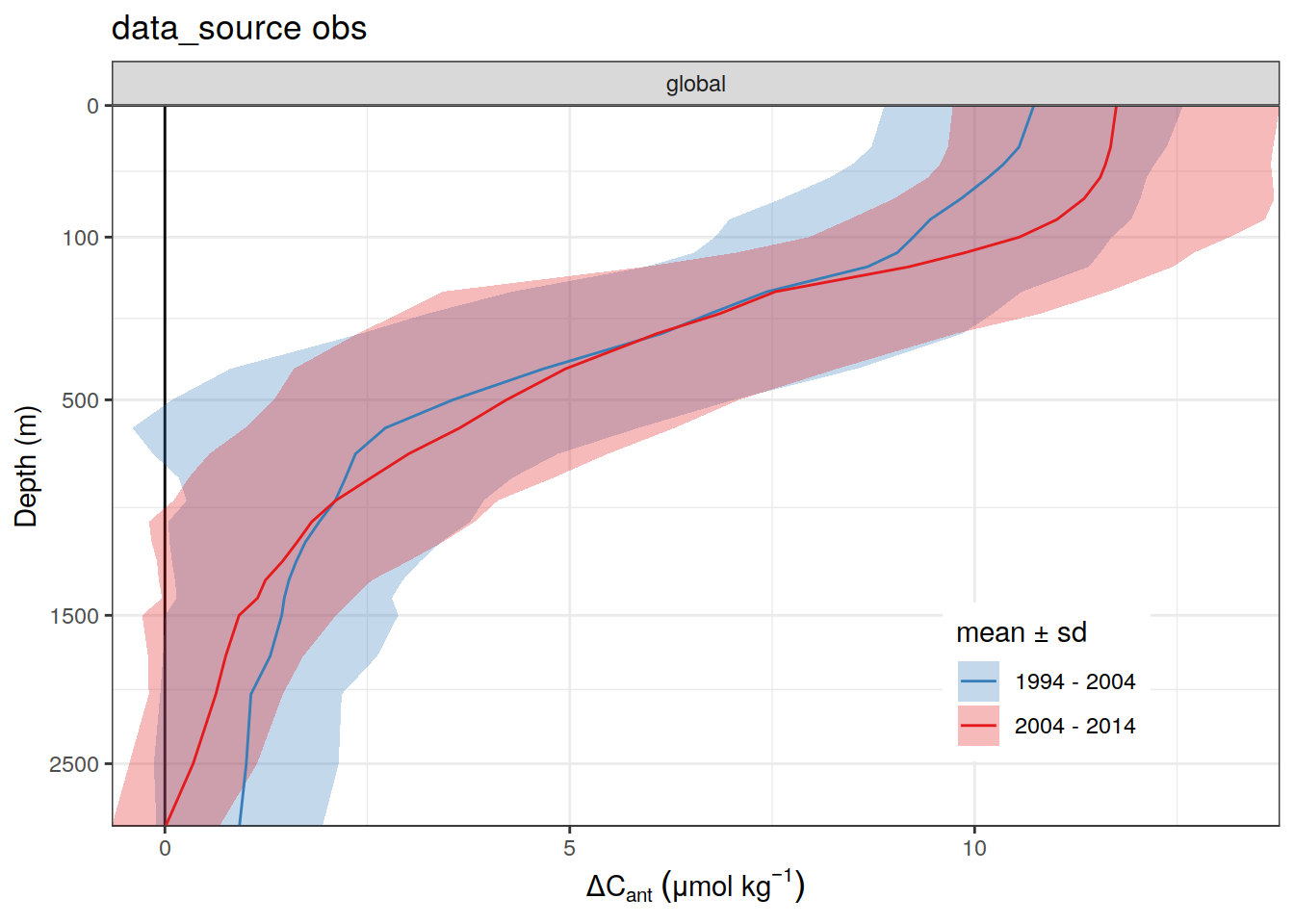
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
mean_surface_dcant <- dcant_profile_global_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
data_source == "obs") %>%
filter(depth <= 50) %>%
summarise(dcant = mean(dcant)) %>%
pull()
dcant_profile_global_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
data_source == "obs") %>%
filter(dcant >= mean_surface_dcant/2) %>%
group_by(period) %>%
summarise(max_depth = max(depth)) %>%
ungroup()# A tibble: 2 × 2
period max_depth
<chr> <dbl>
1 1994 - 2004 300
2 2004 - 2014 300dcant_profile_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014") %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = dcant - dcant_sd,
xmax = dcant + dcant_sd,
fill = period
),
alpha = 0.3
) +
geom_path(aes(col = period)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
scale_fill_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
labs(
title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = dcant_umol_label
) +
facet_grid(basin_AIP ~ .)
)[[1]]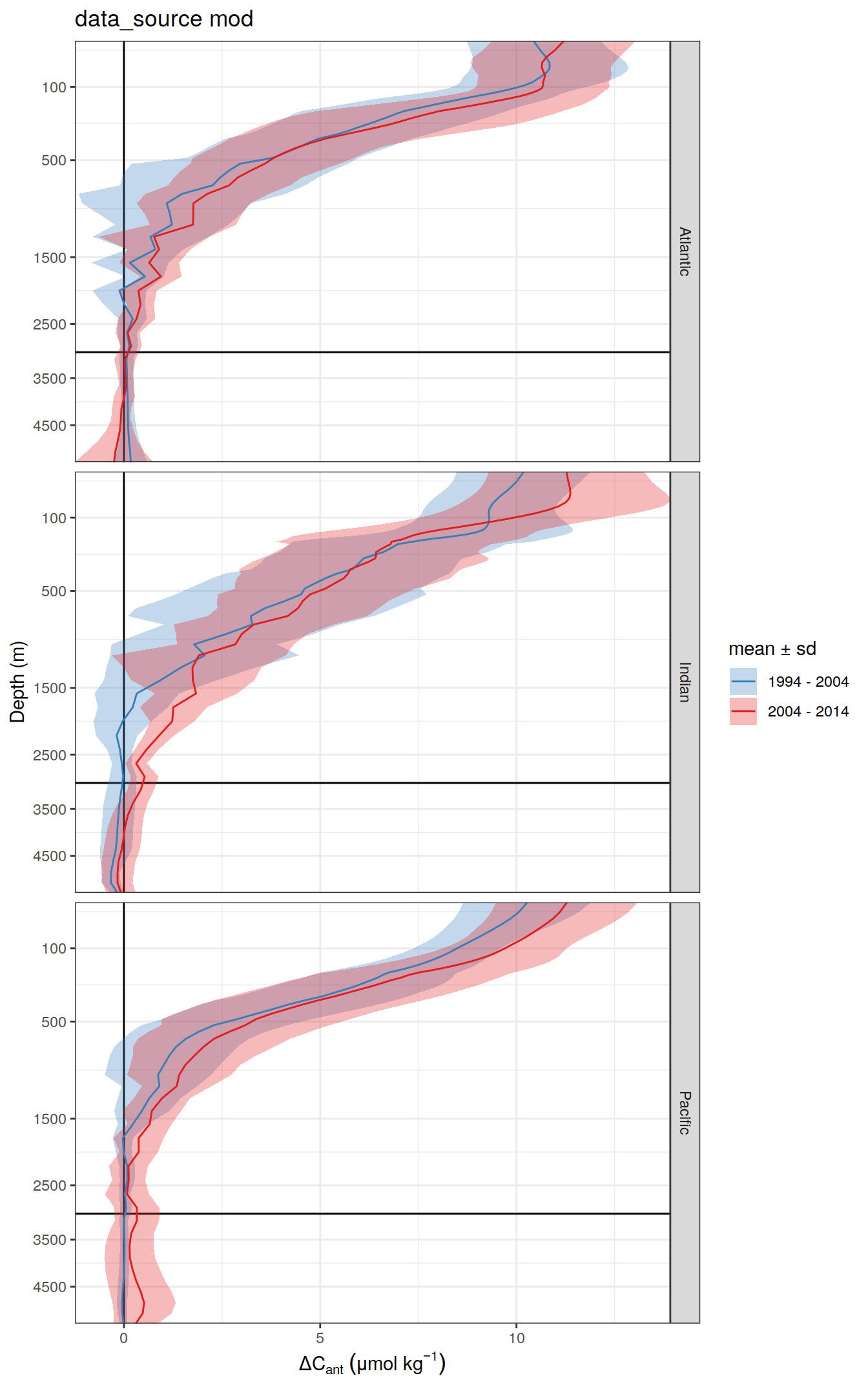
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
[[2]]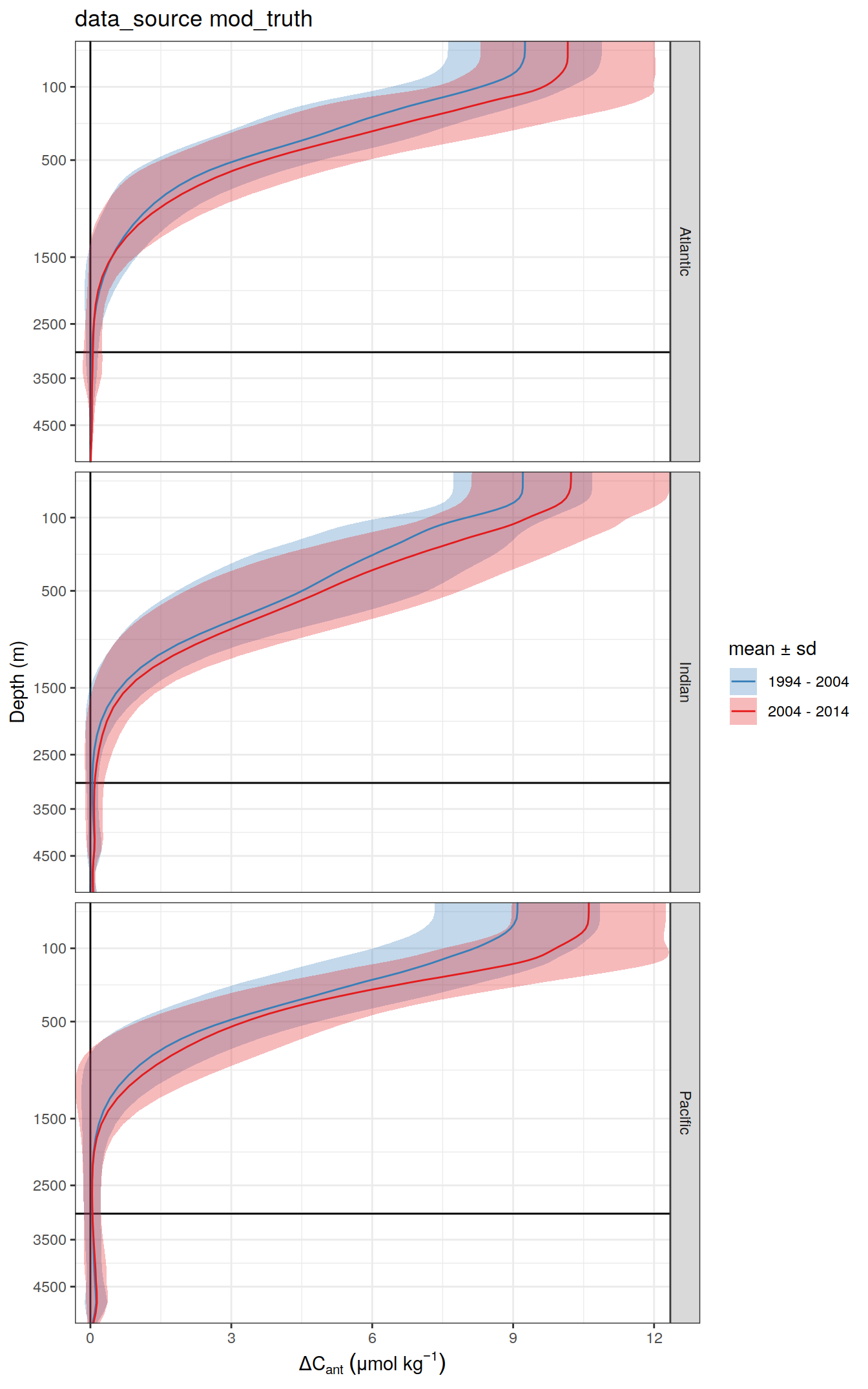
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
[[3]]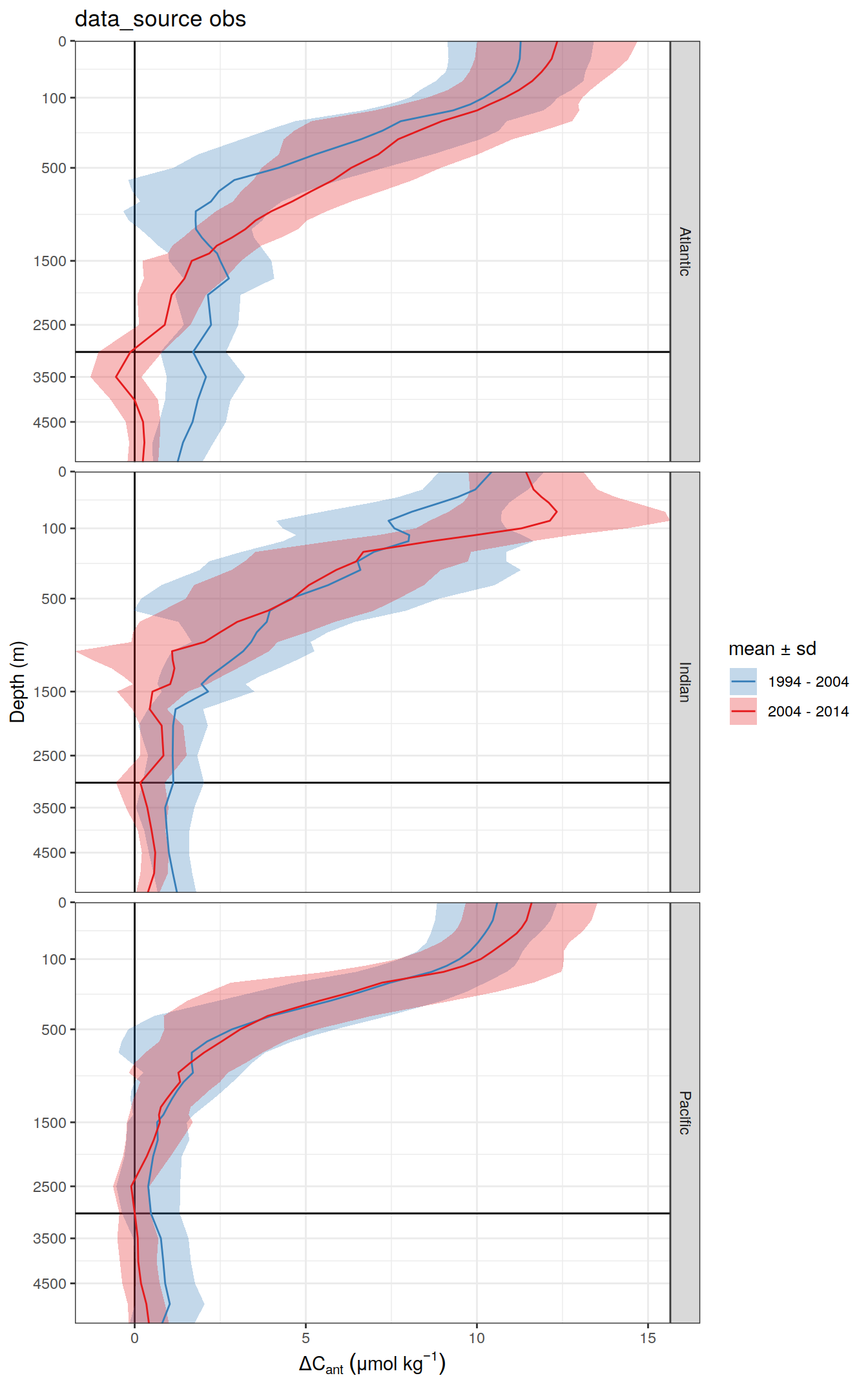
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
dcant_profile_basin_MLR_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
depth <= params_global$inventory_depth_standard) %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = dcant - dcant_sd,
xmax = dcant + dcant_sd,
fill = period
),
alpha = 0.3
) +
geom_path(aes(col = period)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
scale_fill_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
labs(
title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = dcant_umol_label
) +
facet_wrap(~ basin, ncol = 3, dir = "v") +
theme(legend.position = c(0.8,0.2))
)[[1]]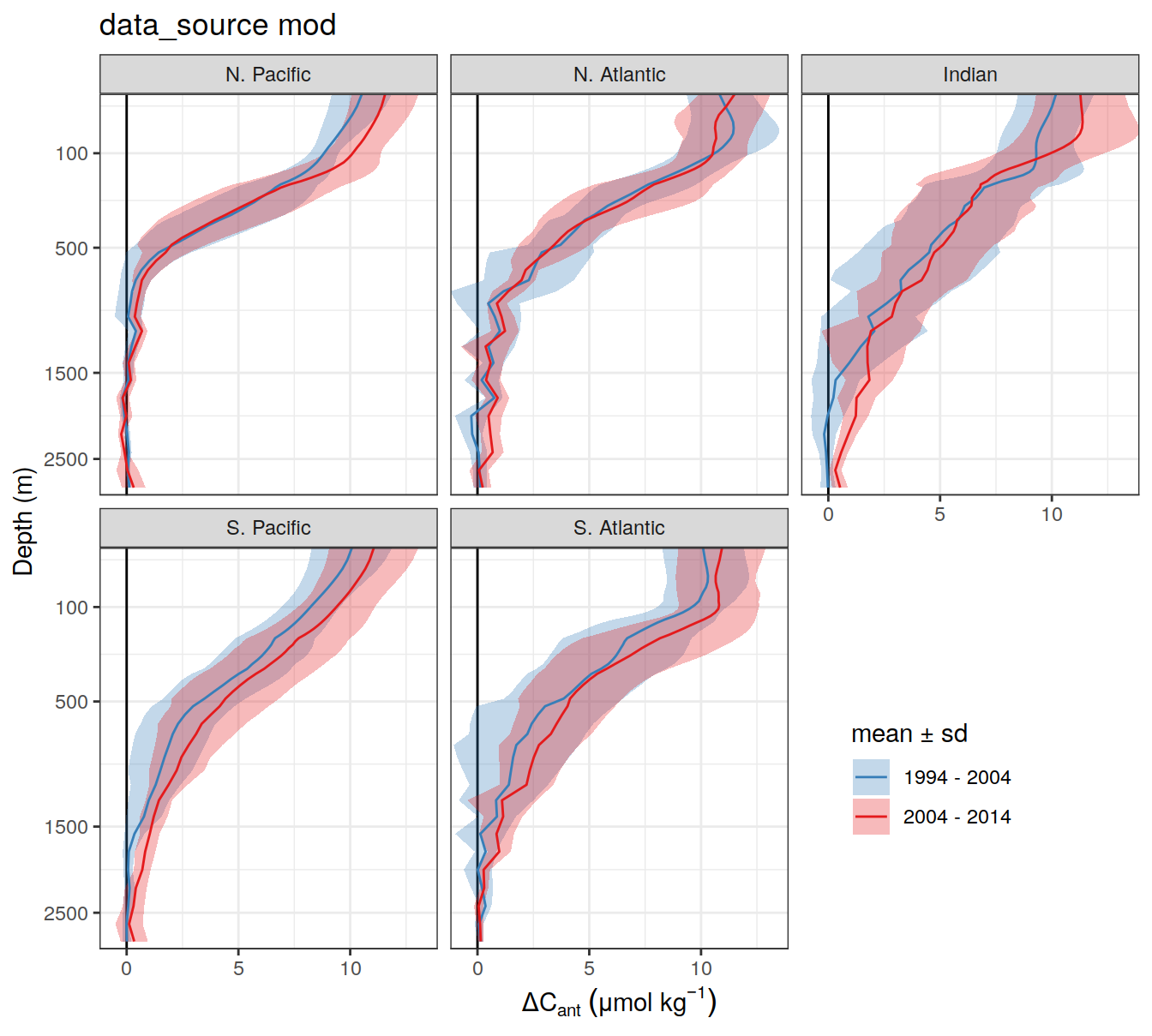
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 4673df5 | jens-daniel-mueller | 2022-02-02 |
| 60727e6 | jens-daniel-mueller | 2022-02-02 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
[[2]]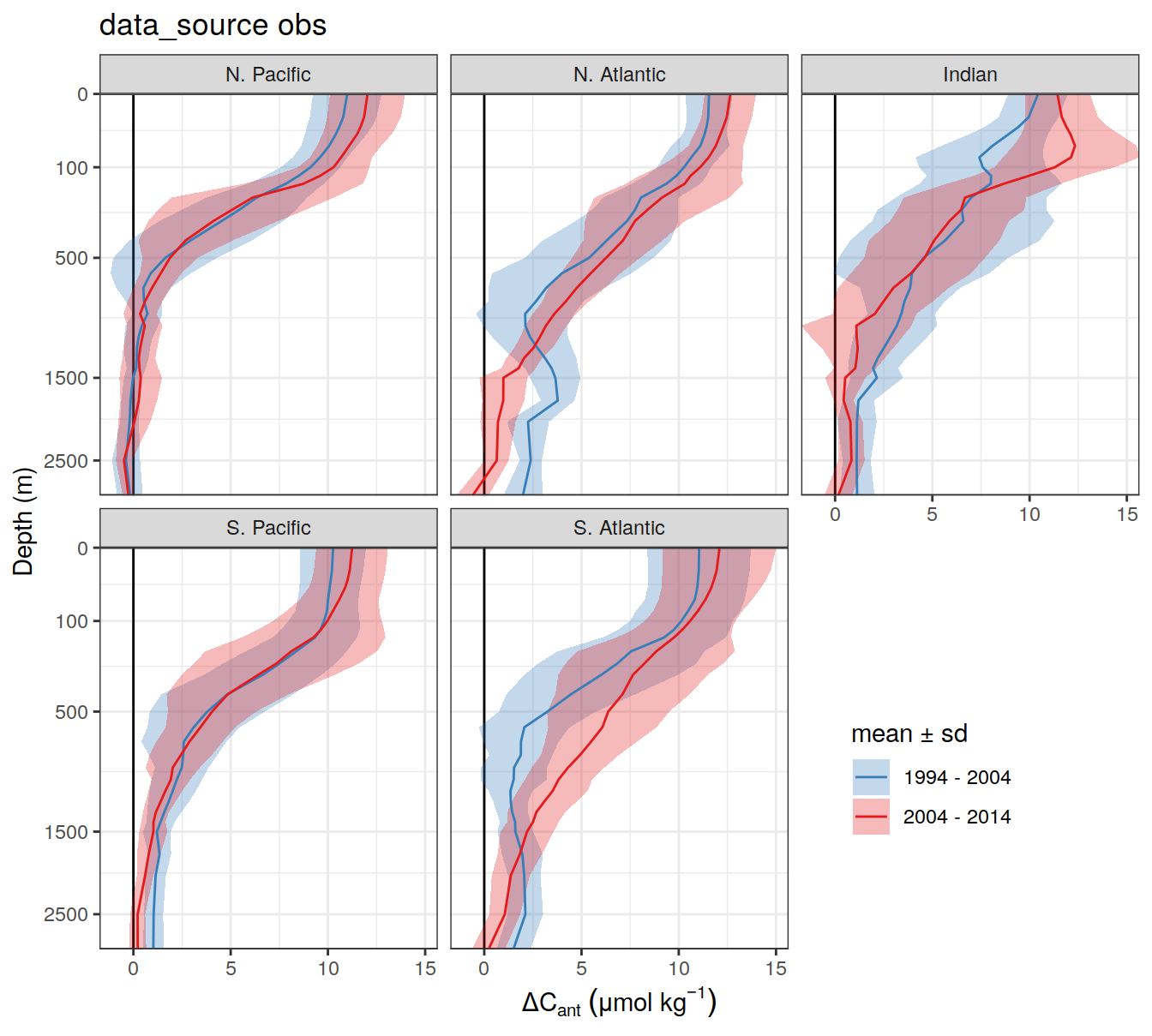
| Version | Author | Date |
|---|---|---|
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 4673df5 | jens-daniel-mueller | 2022-02-02 |
| 60727e6 | jens-daniel-mueller | 2022-02-02 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
dcant_profile_global_all_ensemble %>%
arrange(depth) %>%
filter(
period != "1994 - 2014",
depth <= params_global$inventory_depth_standard,
data_source == "obs"
) %>%
group_split(data_source) %>%
map(
~ ggplot() +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_path(
data = .x,
aes(
dcant,
depth,
col = period,
group = Version_ID,
linetype = "ensemble member",
size = "ensemble member"
), alpha = 0.5
) +
geom_path(
data = .x %>% filter(Version_ID %in% Version_IDs),
aes(
dcant,
depth,
col = period,
group = Version_ID,
linetype = "standard case",
size = "standard case"
)
) +
scale_size_manual(values = c(0.5, 2), name="x") +
scale_linetype_manual(values = c(2, 1), name="x") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(
palette = "Set1",
direction = -1
) +
scale_fill_brewer(
palette = "Set1",
direction = -1
) +
labs(
# title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = dcant_umol_label
) +
facet_wrap( ~ basin, ncol = 3, dir = "v") +
theme(legend.position = c(0.85, 0.3),
legend.title = element_blank())
)[[1]]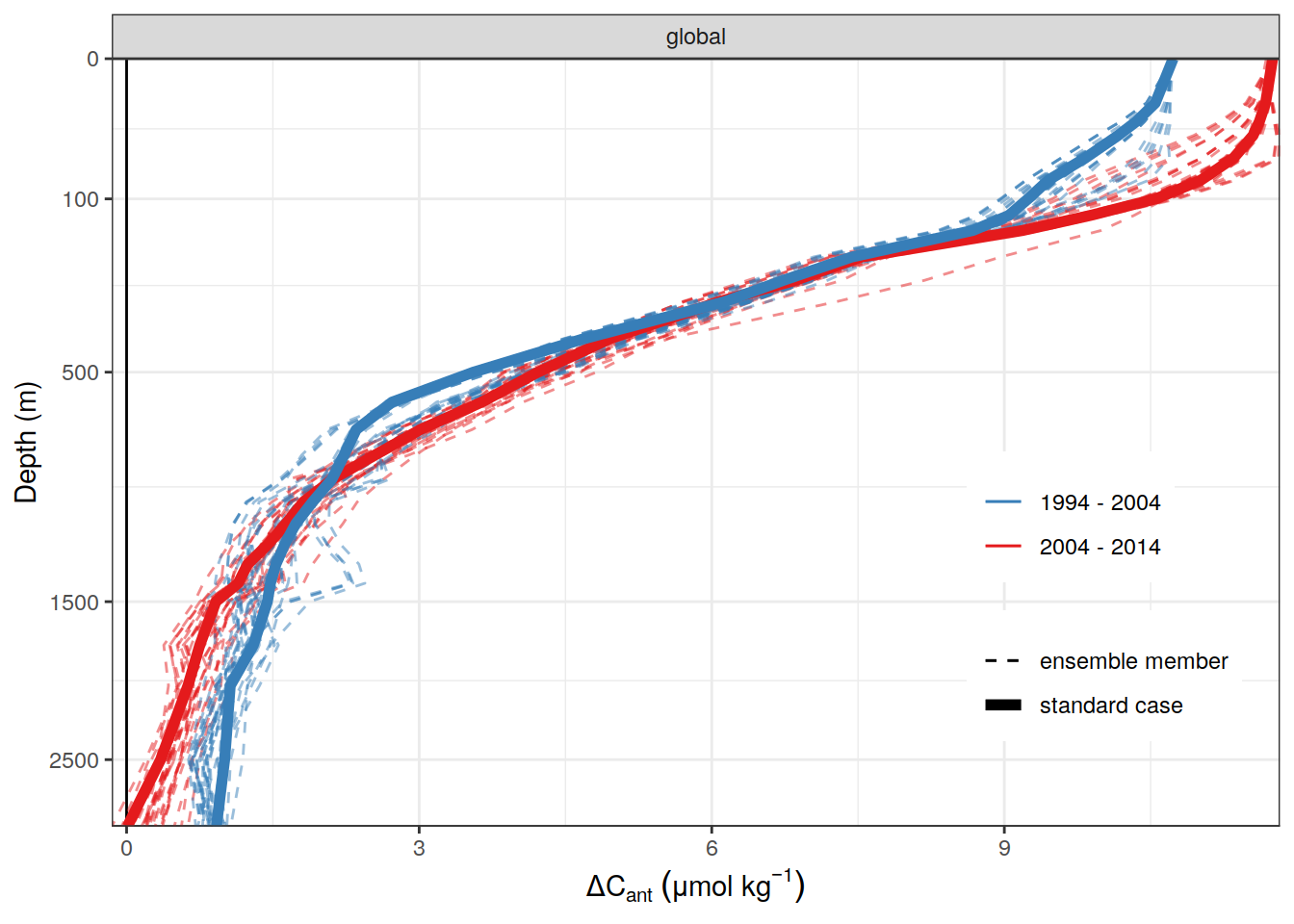
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
ggsave(path = here::here("output/publication"),
filename = "Fig_concentration_profiles_dcant.png",
height = 6,
width = 8)dcant_profile_basin_MLR_all_ensemble %>%
arrange(depth) %>%
filter(
period != "1994 - 2014",
depth <= params_global$inventory_depth_standard,
data_source == "obs"
) %>%
group_split(data_source) %>%
map(
~ ggplot() +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_path(
data = .x,
aes(
dcant,
depth,
col = period,
group = Version_ID,
linetype = "ensemble member",
size = "ensemble member"
), alpha = 0.5
) +
geom_path(
data = .x %>% filter(Version_ID %in% Version_IDs),
aes(
dcant,
depth,
col = period,
group = Version_ID,
linetype = "standard case",
size = "standard case"
)
) +
scale_size_manual(values = c(0.5, 2), name="x") +
scale_linetype_manual(values = c(2, 1), name="x") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(
palette = "Set1",
direction = -1
) +
scale_fill_brewer(
palette = "Set1",
direction = -1
) +
labs(
# title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = dcant_umol_label
) +
facet_wrap( ~ basin, ncol = 3, dir = "v") +
theme(legend.position = c(0.85, 0.3),
legend.title = element_blank())
)[[1]]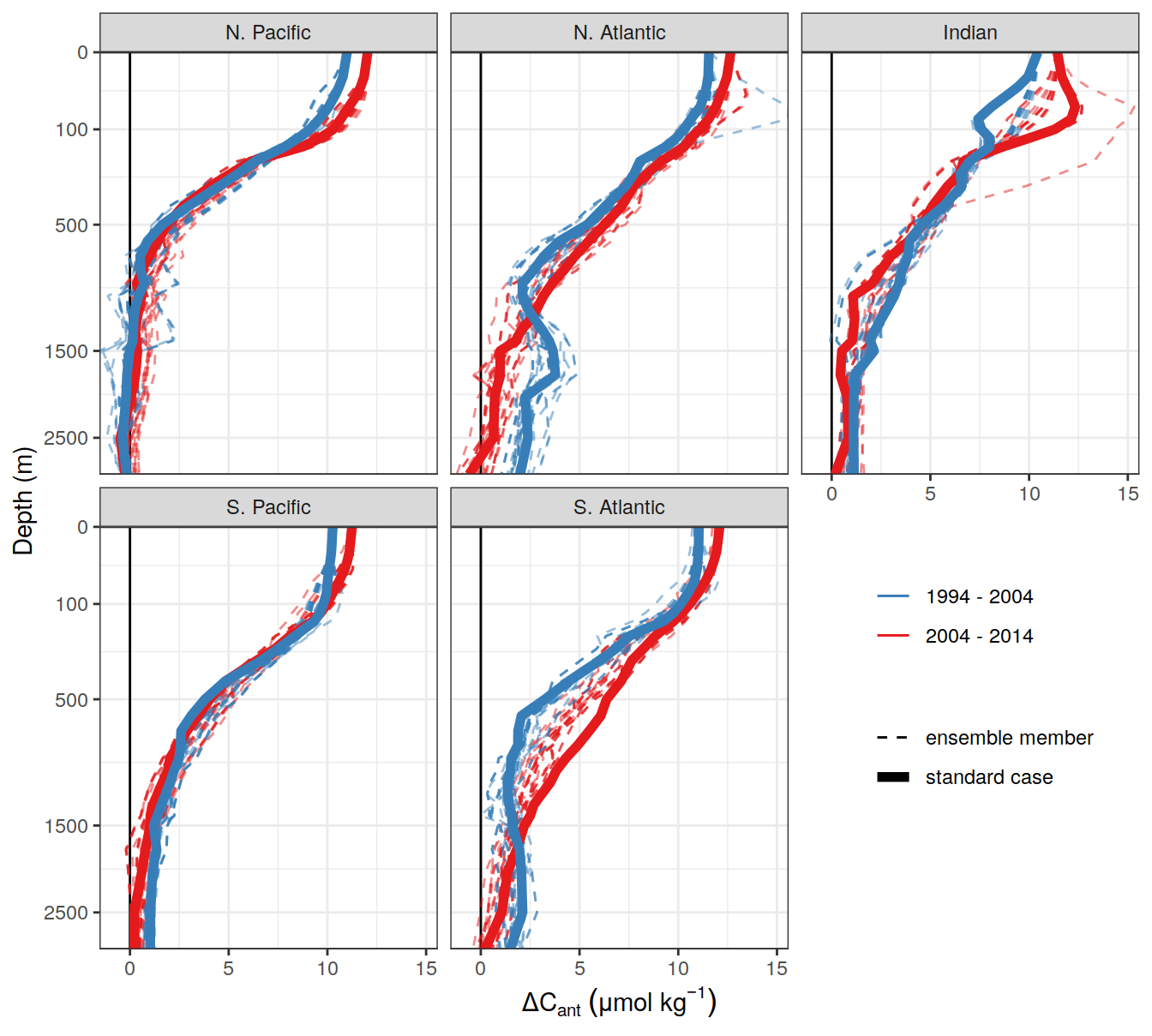
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| f09080e | jens-daniel-mueller | 2022-06-28 |
| 16dc3af | jens-daniel-mueller | 2022-06-27 |
ggsave(path = here::here("output/publication"),
filename = "Fig_concentration_profiles_dcant.png",
height = 6,
width = 8)
mean_surface_dcant <- dcant_profile_basin_MLR_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
data_source == "obs") %>%
filter(depth <= 50) %>%
group_by(basin) %>%
summarise(mean_surface_dcant = mean(dcant)) %>%
ungroup()
full_join(dcant_profile_basin_MLR_all,
mean_surface_dcant) %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
data_source == "obs") %>%
filter(dcant >= mean_surface_dcant/2) %>%
group_by(period, basin) %>%
summarise(max_depth = max(depth)) %>%
ungroup()# A tibble: 10 × 3
period basin max_depth
<chr> <fct> <dbl>
1 1994 - 2004 N. Pacific 200
2 1994 - 2004 S. Pacific 300
3 1994 - 2004 N. Atlantic 400
4 1994 - 2004 S. Atlantic 300
5 1994 - 2004 Indian 400
6 2004 - 2014 N. Pacific 200
7 2004 - 2014 S. Pacific 300
8 2004 - 2014 N. Atlantic 500
9 2004 - 2014 S. Atlantic 600
10 2004 - 2014 Indian 300dcant_profile_basin_MLR_all_ensemble %>%
arrange(depth) %>%
filter(
period != "1994 - 2014",
depth <= params_global$inventory_depth_standard,
data_source == "obs"
) %>%
group_split(data_source) %>%
map(
~ ggplot() +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_path(
data = .x,
aes(dcant,
depth,
col = MLR_basins,
group = Version_ID),
alpha = 0.5
) +
scale_size_manual(values = c(0.5, 2), name = "x") +
scale_linetype_manual(values = c(2, 1), name = "x") +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Dark2") +
labs(y = "Depth (m)",
x = dcant_umol_label) +
facet_grid(basin ~ period)
)[[1]]
dcant_profile_basin_MLR_all_ensemble %>%
arrange(depth) %>%
filter(
# period != "1994 - 2014",
depth <= params_global$inventory_depth_standard
# data_source == "obs"
) %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot() +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_path(
data = .x,
aes(
dcant,
depth,
col = Version_ID_group,
group = Version_ID
), alpha = 0.5
) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(
palette = "Dark2",
direction = -1
) +
scale_fill_brewer(
palette = "Set1",
direction = -1
) +
labs(
y = "Depth (m)",
x = dcant_umol_label,
title = unique(.x$data_source)
) +
facet_grid(basin ~ period)
)[[1]]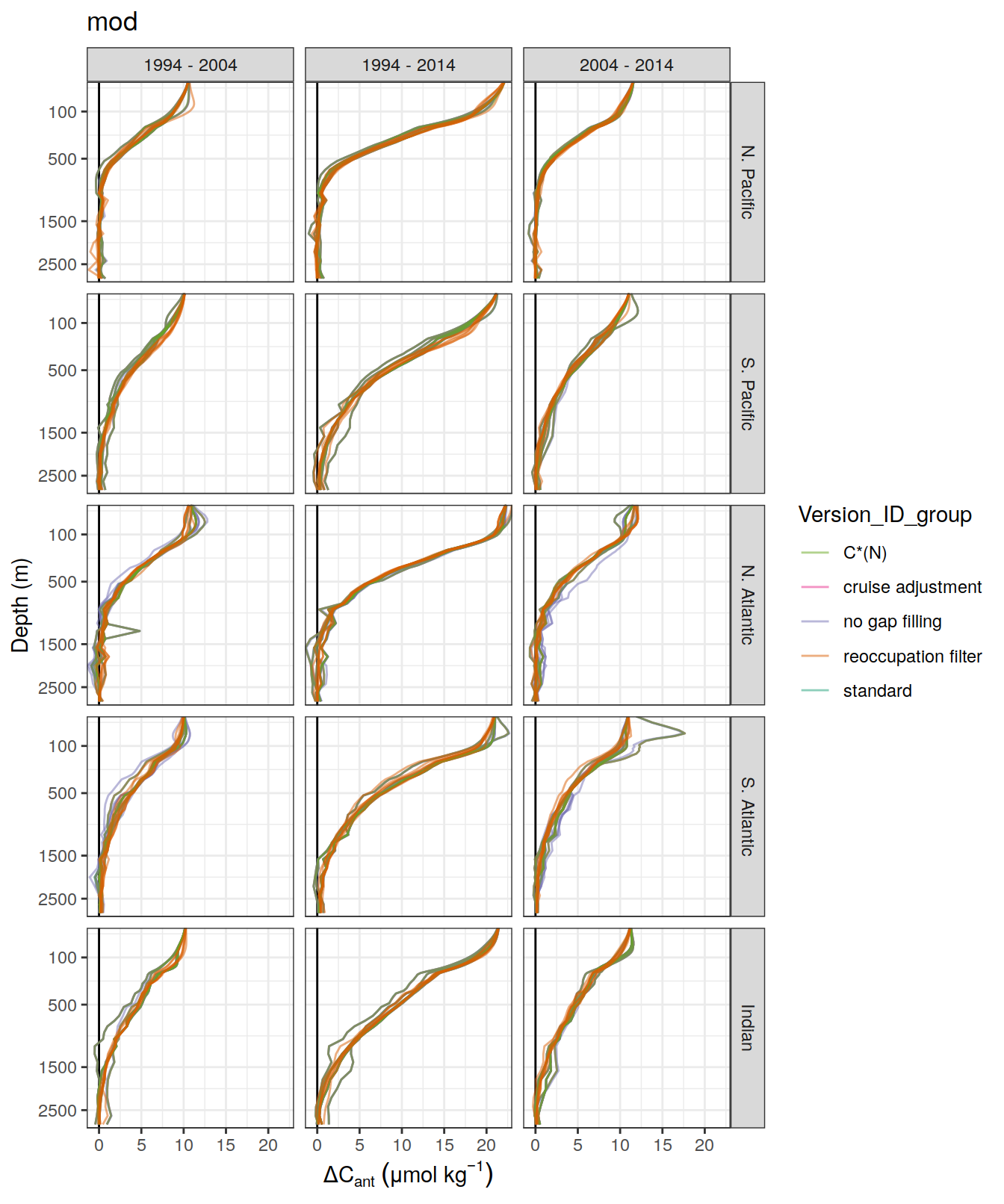
[[2]]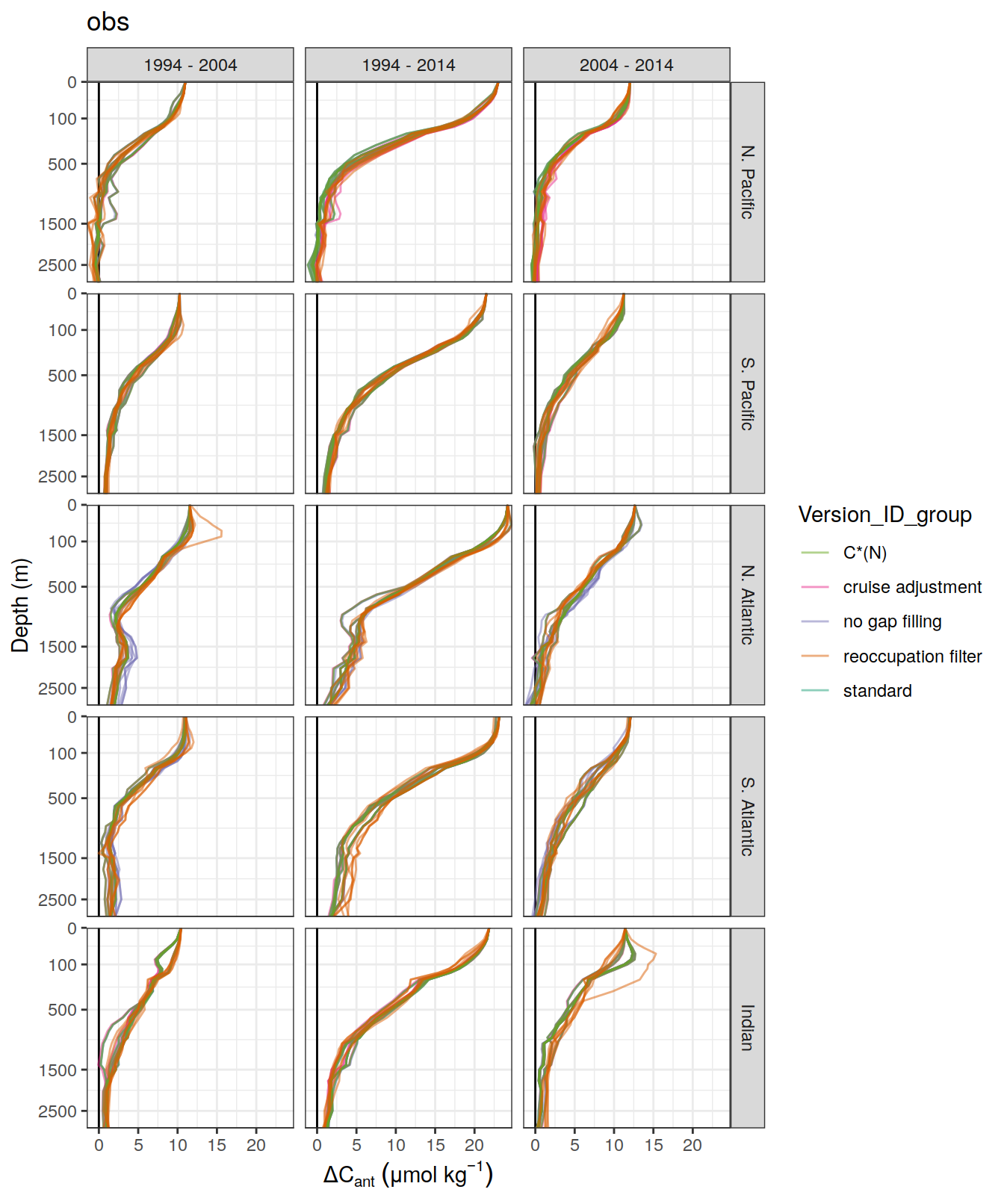
ggsave(path = here::here("output/publication"),
filename = "Fig_concentration_profiles_dcant_ensemble_member.png",
height = 6,
width = 8)dcant_profile_basin_MLR_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014",
depth <= params_global$inventory_depth_standard) %>%
group_split(data_source, basin) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = dcant - dcant_sd,
xmax = dcant + dcant_sd,
fill = period
),
alpha = 0.3
) +
geom_path(aes(col = period)) +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
scale_fill_brewer(palette = "Set1", name = "mean \u00B1 sd", direction = -1) +
labs(
title = paste("data_source", unique(.x$data_source),
"|", unique(.x$basin)),
y = "Depth (m)",
x = dcant_umol_label
) +
# facet_wrap(~ basin, ncol = 3, dir = "v") +
theme(legend.position = c(0.8,0.2))
)7.2 dcant - absolute delta
delta <- dcant_profile_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014") %>%
select(data_source, depth, basin_AIP, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(delta_dcant = `2004 - 2014` - `1994 - 2004`) %>%
select(-c(`2004 - 2014`, `1994 - 2004`))
delta_sd <- dcant_profile_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014") %>%
select(data_source, depth, basin_AIP, period, dcant_sd) %>%
pivot_wider(names_from = period,
values_from = dcant_sd) %>%
mutate(delta_dcant_sd = (`2004 - 2014` + `1994 - 2004`) / 2) %>%
select(-c(`2004 - 2014`, `1994 - 2004`))
dcant_profile_all_delta <- full_join(delta, delta_sd)
rm(delta, delta_sd)
dcant_profile_all_delta %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(delta_dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = delta_dcant - delta_dcant_sd,
xmax = delta_dcant + delta_dcant_sd
),
alpha = 0.3
) +
geom_path() +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Set1", name = "mean \u00B1 sd") +
scale_fill_brewer(palette = "Set1", name = "mean \u00B1 sd") +
labs(title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = ddcant_umol_label) +
facet_grid(basin_AIP ~ .))[[1]]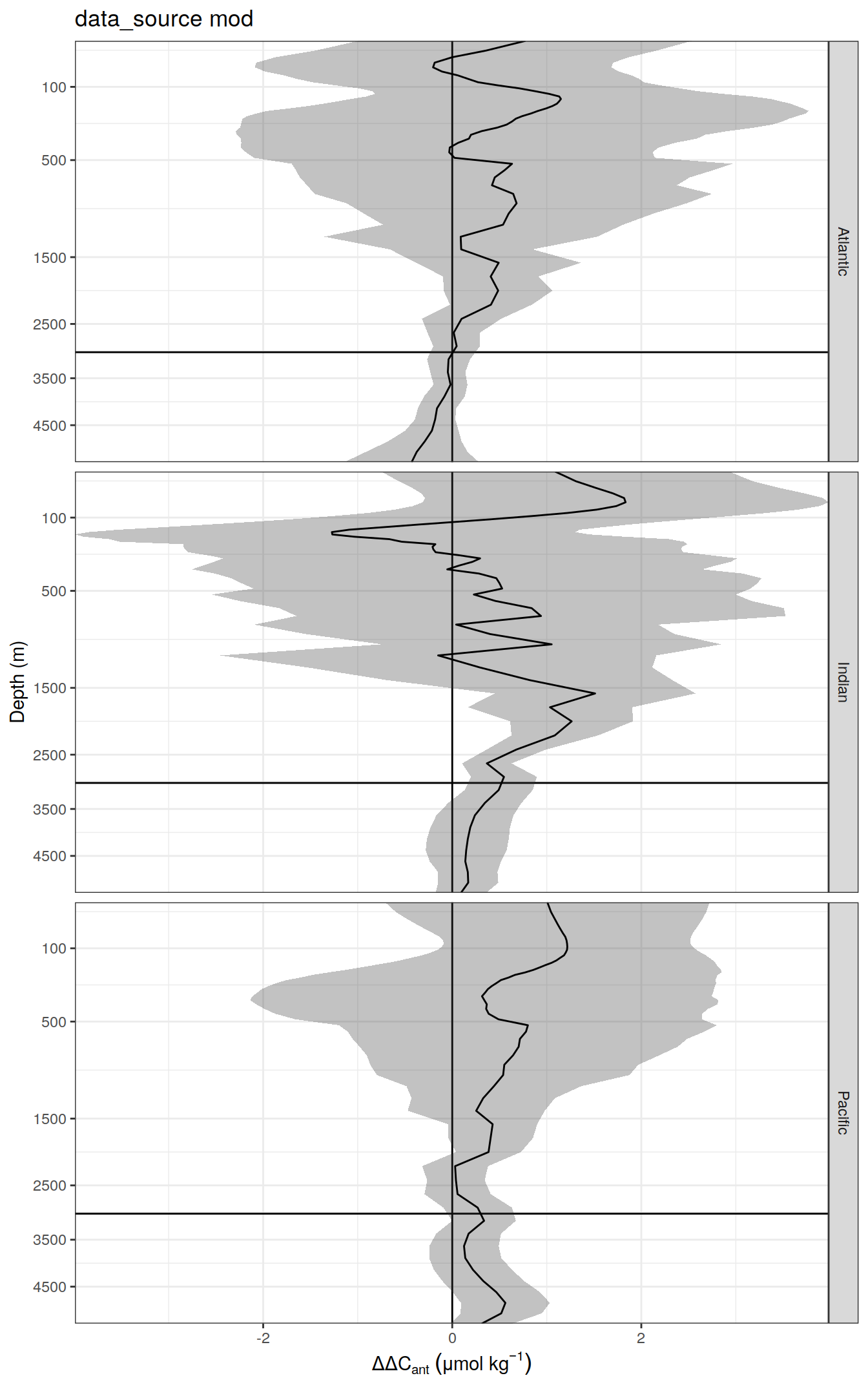
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| eccd82b | jens-daniel-mueller | 2022-01-26 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
[[2]]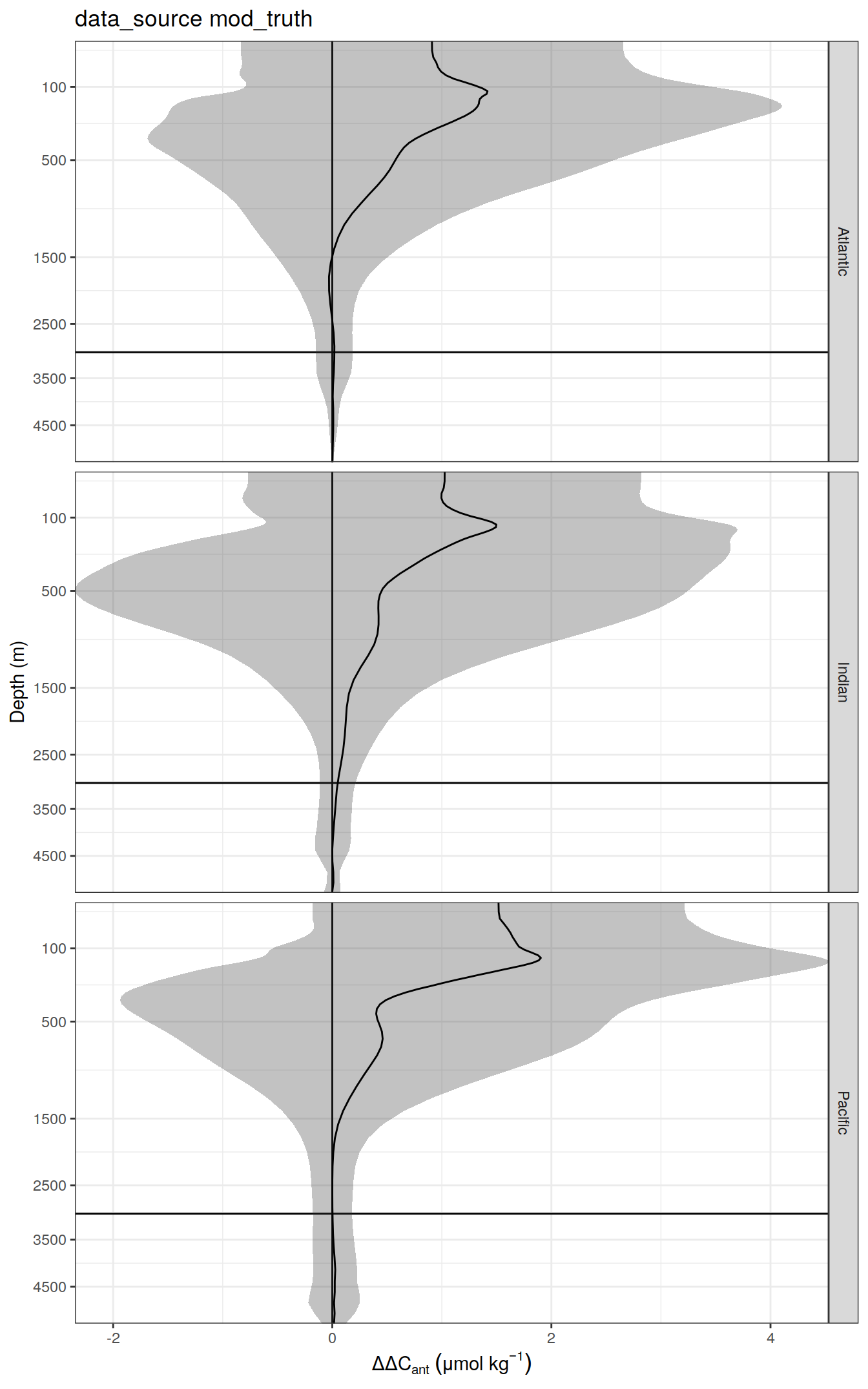
[[3]]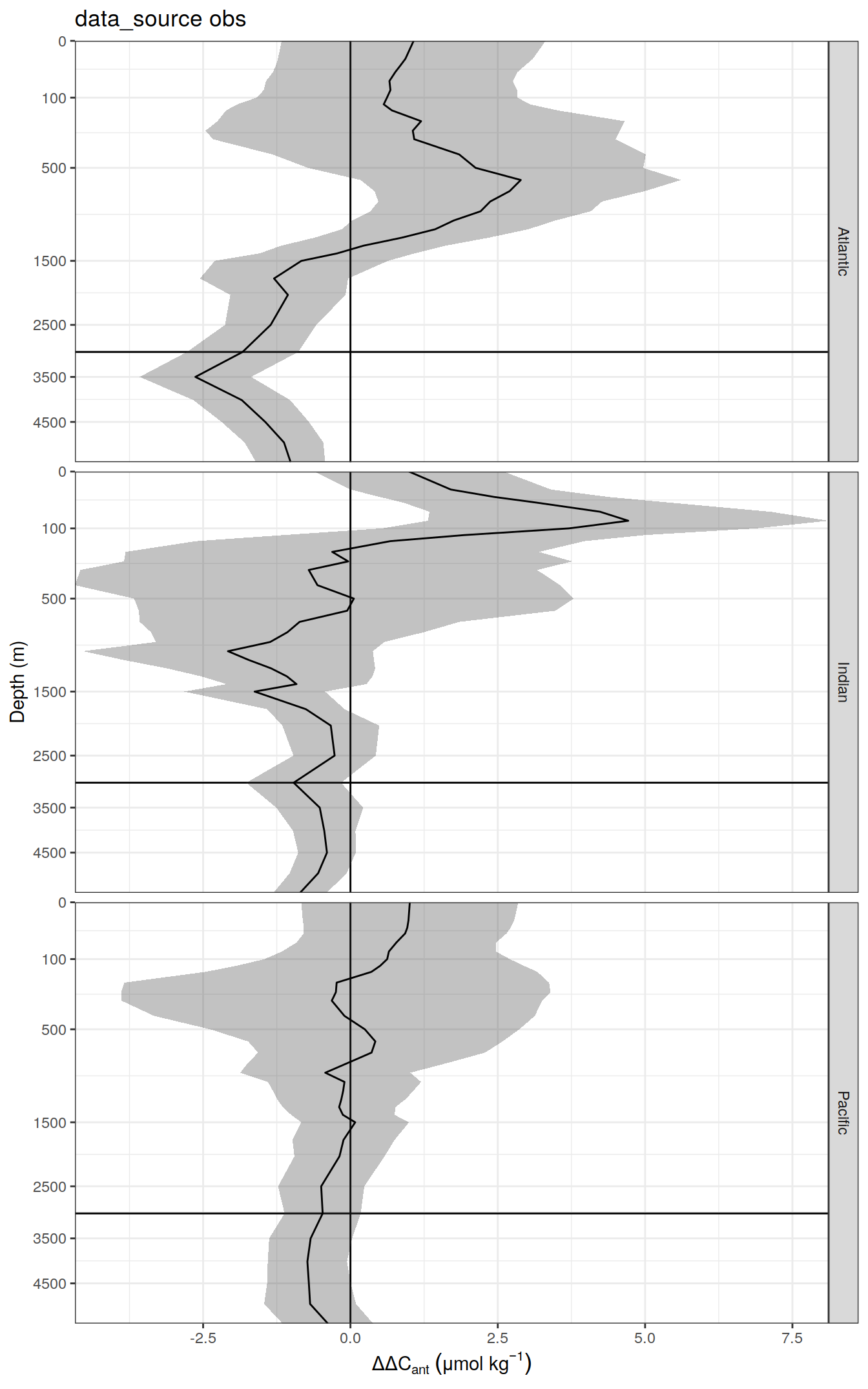
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| eccd82b | jens-daniel-mueller | 2022-01-26 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
delta <- dcant_profile_basin_MLR_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014") %>%
select(data_source, depth, basin, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(delta_dcant = `2004 - 2014` - `1994 - 2004`) %>%
select(-c(`2004 - 2014`, `1994 - 2004`))
delta_sd <- dcant_profile_basin_MLR_all %>%
arrange(depth) %>%
filter(period != "1994 - 2014") %>%
select(data_source, depth, basin, period, dcant_sd) %>%
pivot_wider(names_from = period,
values_from = dcant_sd) %>%
mutate(delta_dcant_sd = (`2004 - 2014` + `1994 - 2004`) / 2) %>%
select(-c(`2004 - 2014`, `1994 - 2004`))
dcant_profile_basin_MLR_all_delta <- full_join(delta, delta_sd)
rm(delta, delta_sd)
dcant_profile_basin_MLR_all_delta %>%
group_split(data_source) %>%
# head(3) %>%
map(
~ ggplot(data = .x,
aes(delta_dcant,
depth)) +
geom_hline(yintercept = params_global$inventory_depth_standard) +
geom_vline(xintercept = 0) +
geom_ribbon(
aes(
xmin = delta_dcant - delta_dcant_sd,
xmax = delta_dcant + delta_dcant_sd
),
alpha = 0.3
) +
geom_path() +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(0, 100, 500, seq(1500, 5000, 1000))) +
coord_cartesian(expand = 0) +
scale_color_brewer(palette = "Set1", name = "mean \u00B1 sd") +
scale_fill_brewer(palette = "Set1", name = "mean \u00B1 sd") +
labs(
title = paste("data_source", unique(.x$data_source)),
y = "Depth (m)",
x = ddcant_umol_label
) +
facet_grid(basin ~ .)
)[[1]]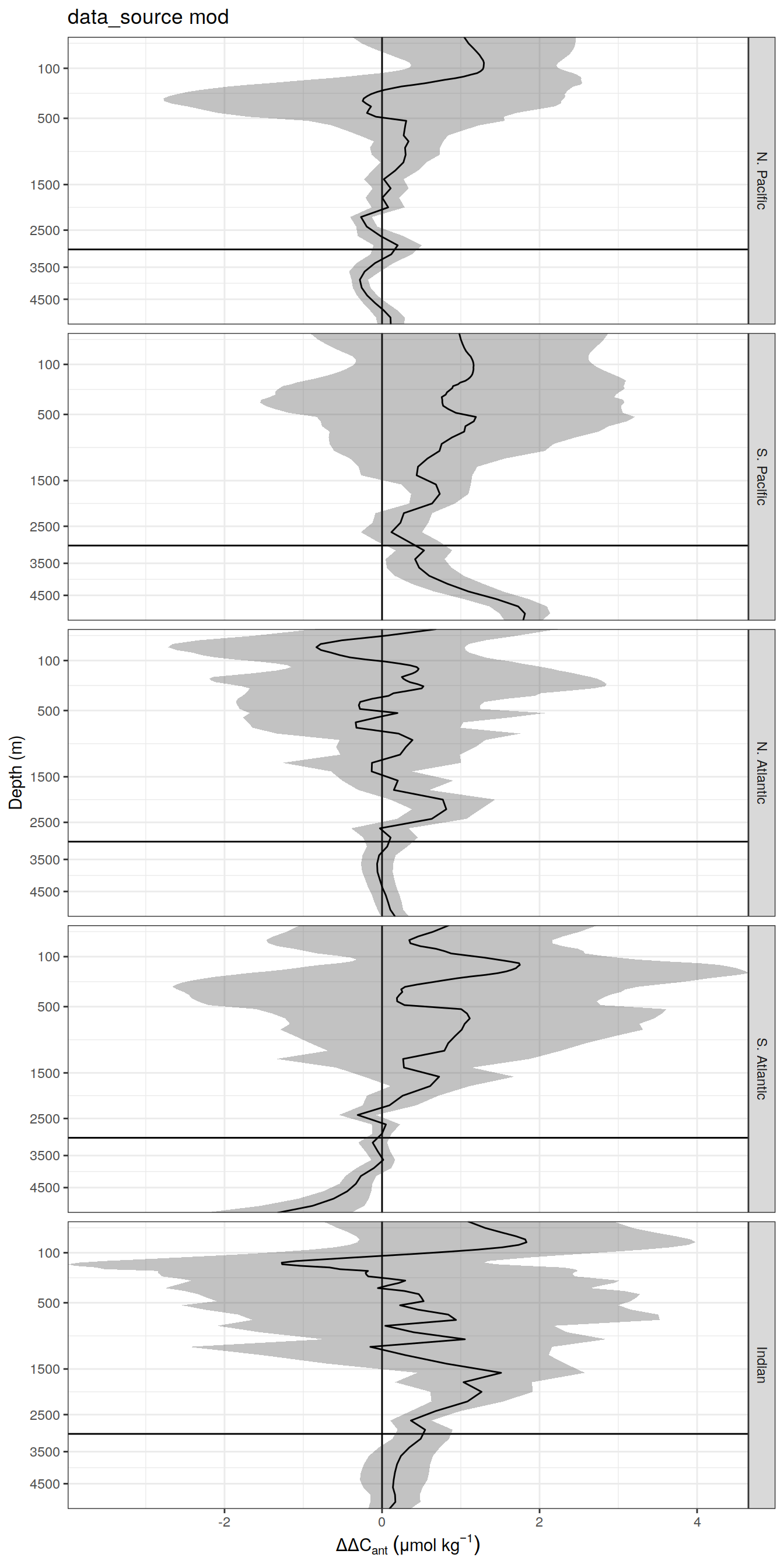
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| eccd82b | jens-daniel-mueller | 2022-01-26 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
[[2]]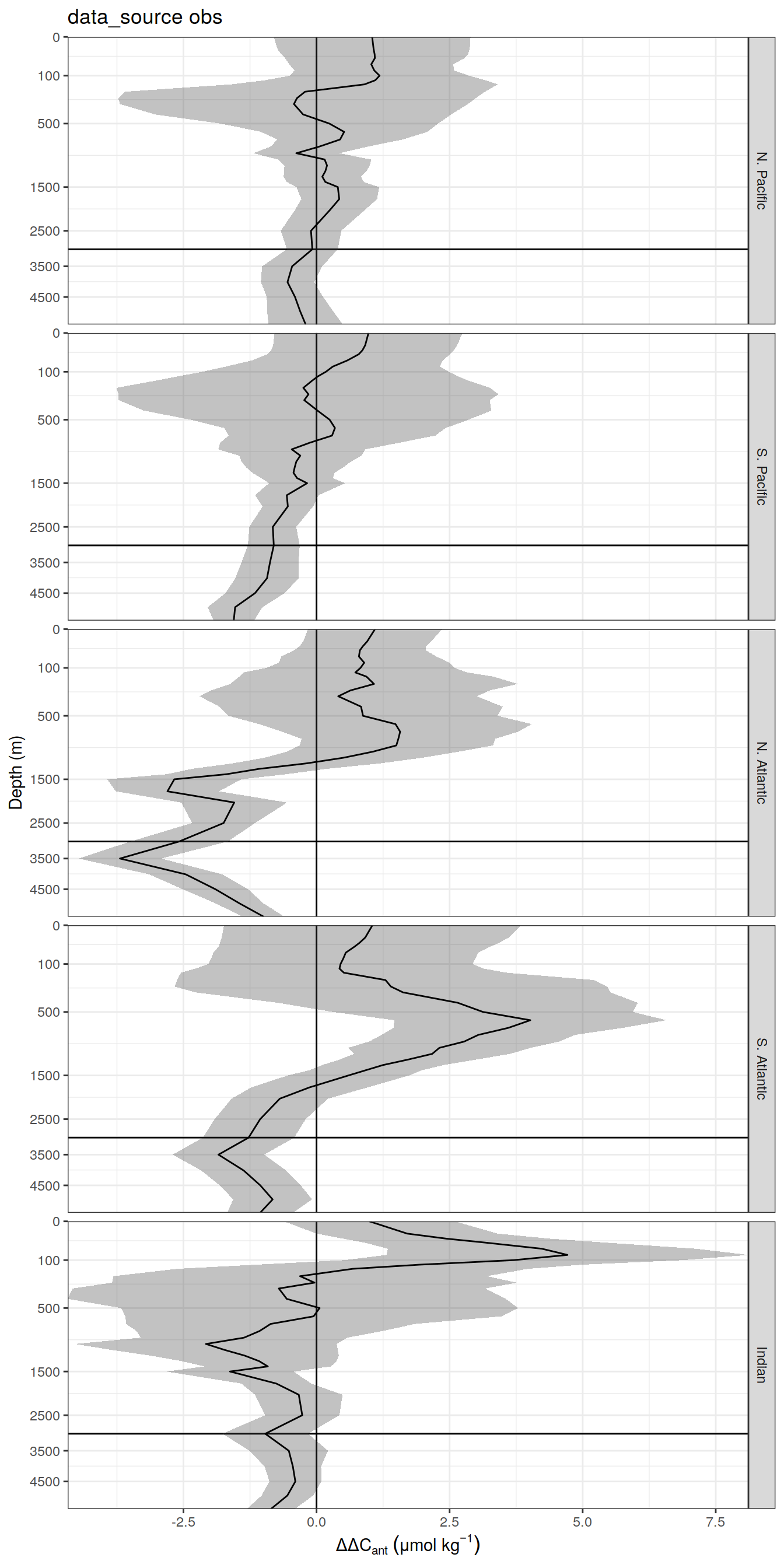
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| eccd82b | jens-daniel-mueller | 2022-01-26 |
| c6fe495 | jens-daniel-mueller | 2022-01-26 |
8 Budgets
8.1 Scatterplots
8.1.1 Global
bind_rows(dcant_budget_global_all_no_adj,
dcant_budget_global_all_ensemble) %>%
ggplot(aes(period, dcant, col = Version_ID_group)) +
geom_jitter() +
scale_color_brewer(palette = "Dark2") +
facet_grid(. ~ data_source)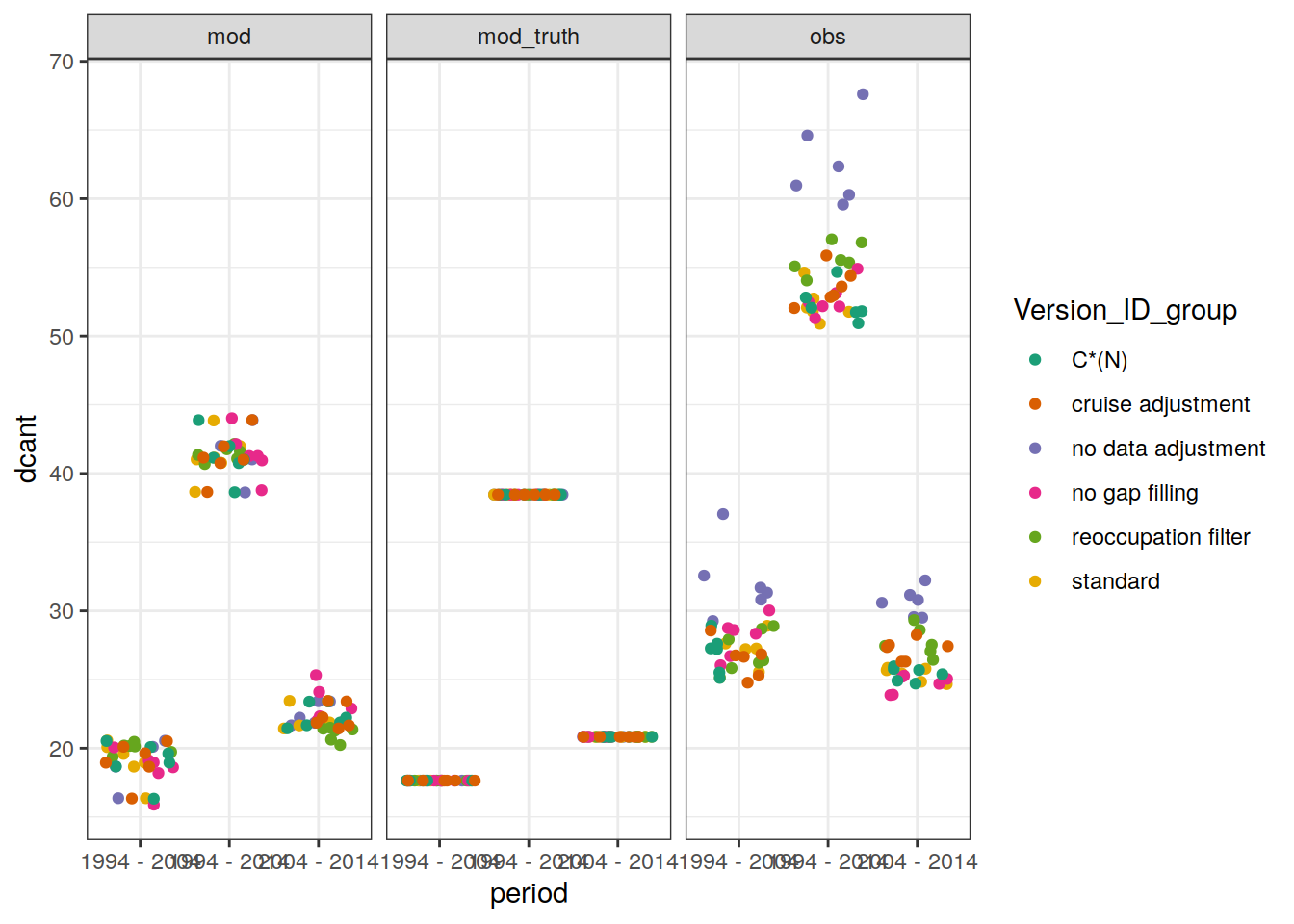
dcant_budget_global_all_ensemble %>%
mutate(ensemble_role = case_when(Version_ID %in% Version_IDs ~ "standard case",
TRUE ~ "others")) %>%
filter(data_source == "obs") %>%
ggplot(aes(period, dcant)) +
scale_fill_brewer(palette = "Dark2",name = "ensemble group") +
scale_color_manual(values = c( "black", "red", "grey"),
name = "ensemble member") +
geom_boxplot(
width = .1,
outlier.shape = NA,
fill = "grey",
alpha = 0.7,
position = position_nudge(x = 0.3)
) +
geom_point(data = dcant_budget_global_all_no_adj %>%
mutate(ensemble_role = "unadjusted") %>%
filter(data_source == "obs"),
aes(fill = Version_ID_group,
col = ensemble_role),
size = 2,
alpha = .7,
shape = 21,
position = position_dodge2(width = 0.3)
) +
geom_point(
aes(fill = Version_ID_group,
col = ensemble_role),
size = 2,
alpha = .5,
shape = 21,
position = position_dodge2(width = 0.3)
) +
# stat_summary(
# fun.data = "mean_sdl",
# fun.args = list(mult = 2),
# position = position_nudge(x = -0.3),
# col = "grey"
# ) +
# stat_summary(
# fun.data = "mean_sdl",
# fun.args = list(mult = 1),
# position = position_nudge(x = -0.3)
# ) +
scale_y_continuous(name = dcant_pgc_label, breaks = seq(0, 100, 2))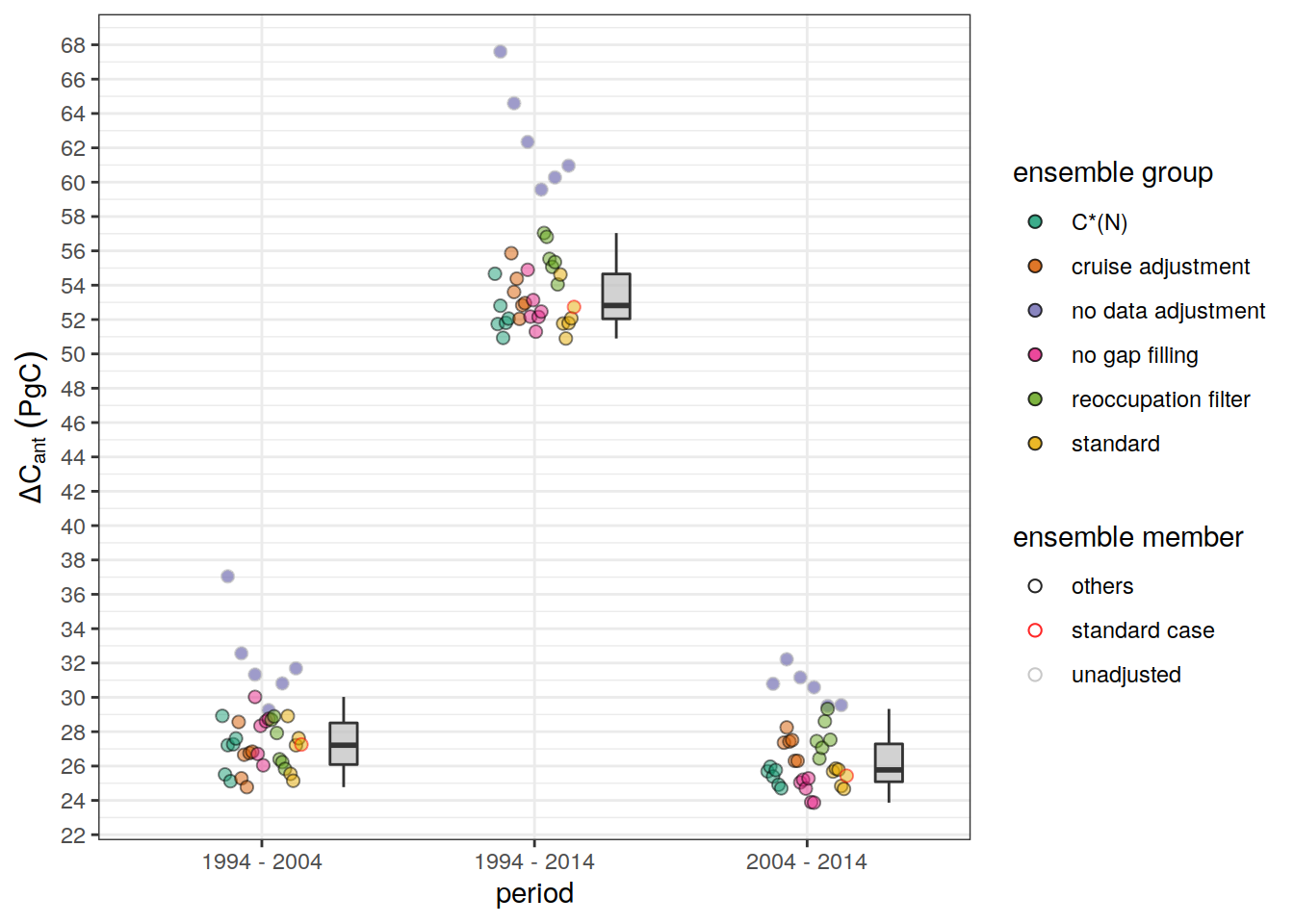
ggsave(path = here::here("output/publication"),
filename = "FigS_budget_dcant_global_ensemble_member.png",
height = 6,
width = 6)
dcant_budget_global_all_ensemble %>%
mutate(ensemble_role = if_else(Version_ID %in% Version_IDs,
"standard case",
"others")) %>%
filter(data_source == "obs") %>%
ggplot(aes(period, dcant)) +
scale_fill_brewer(palette = "Dark2") +
scale_color_manual(values = c("grey", "red"),
name = "ensemble member") +
geom_boxplot(
width = .1,
outlier.shape = NA,
fill = "grey",
alpha = 0.7,
position = position_nudge(x = 0.3)
) +
geom_point(
aes(group = Version_ID_group,
fill = MLR_basins,
col = ensemble_role),
size = 1.3,
alpha = .7,
shape = 21,
position = position_dodge2(width = 0.3)
) +
stat_summary(
fun.data = "mean_sdl",
fun.args = list(mult = 2),
position = position_nudge(x = -0.3),
col = "grey"
) +
stat_summary(
fun.data = "mean_sdl",
fun.args = list(mult = 1),
position = position_nudge(x = -0.3)
) +
scale_y_continuous(name = dcant_pgc_label, breaks = seq(0, 100, 2))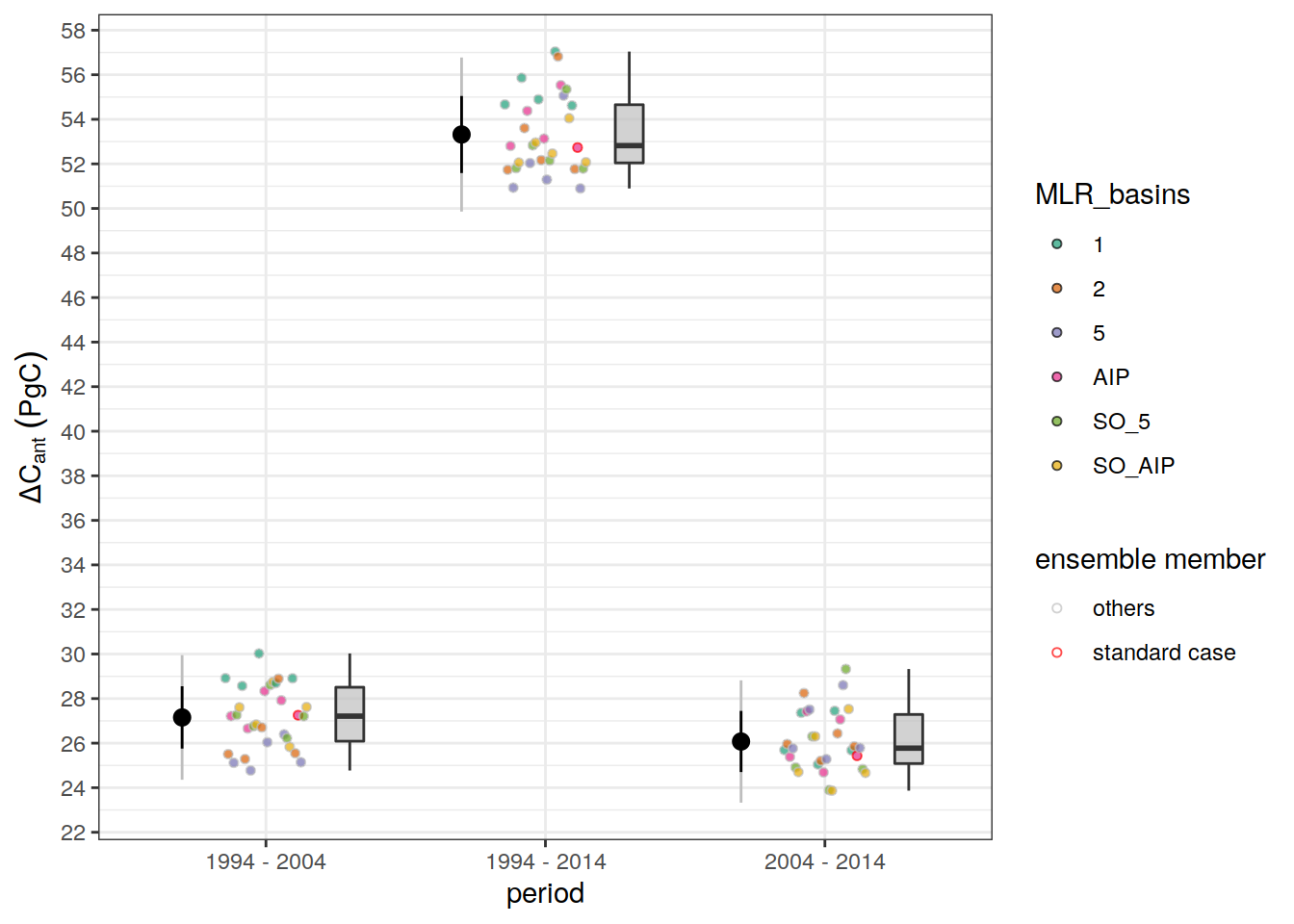
# dcant_budget_global_all_ensemble_summary <- dcant_budget_global_all_ensemble %>%
# group_by(data_source, period) %>%
# summarise(dcant_mean = mean(dcant),
# dcant_sd = sd(dcant)*2) %>%
# ungroup()8.1.2 basin hemisphere
dcant_budget_basin_MLR_all_ensemble %>%
ggplot(aes(period, dcant, col = Version_ID_group)) +
geom_jitter() +
scale_color_brewer(palette = "Dark2") +
facet_grid(basin ~ data_source)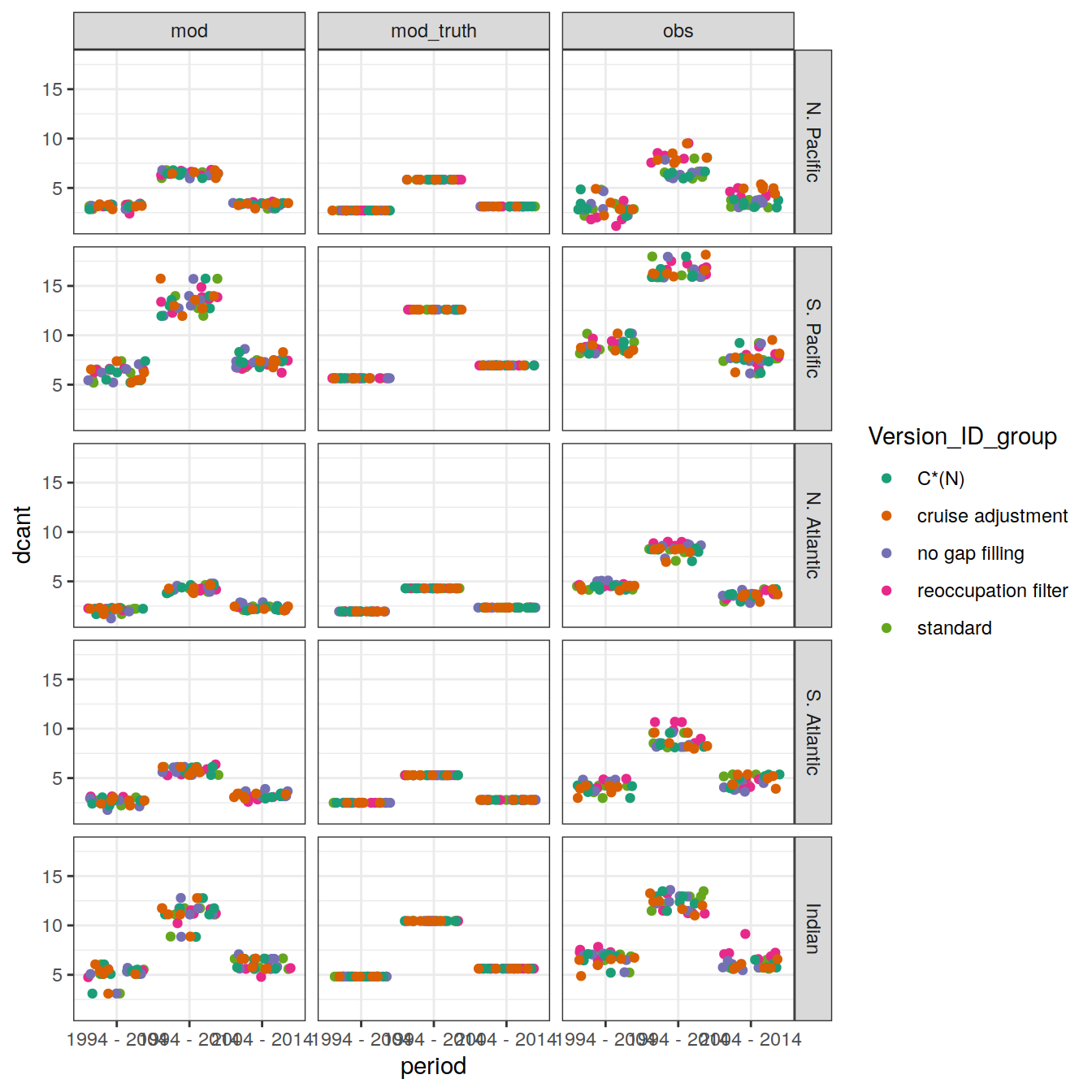
dcant_budget_basin_MLR_all_ensemble %>%
mutate(ensemble_role = if_else(Version_ID %in% Version_IDs,
"standard case",
"others")) %>%
filter(data_source == "obs") %>%
ggplot(aes(period, dcant)) +
scale_fill_brewer(palette = "Dark2", name = "ensemble group") +
scale_color_manual(values = c("grey", "black", "red"),
name = "ensemble member") +
geom_boxplot(
width = .1,
outlier.shape = NA,
fill = "grey",
alpha = 0.7,
position = position_nudge(x = 0.3)
) +
geom_point(
data = dcant_budget_basin_MLR_all_no_adj %>%
mutate(ensemble_role = "no adjustment") %>%
filter(data_source == "obs"),
aes(fill = Version_ID_group,
col = ensemble_role),
size = 1.3,
alpha = .7,
shape = 21,
position = position_dodge2(width = 0.3)
) +
geom_point(
aes(fill = Version_ID_group,
col = ensemble_role),
size = 1.3,
alpha = .7,
shape = 21,
position = position_dodge2(width = 0.3)
) +
# stat_summary(
# fun.data = "mean_sdl",
# fun.args = list(mult = 2),
# position = position_nudge(x = -0.3),
# col = "grey"
# ) +
# stat_summary(
# fun.data = "mean_sdl",
# fun.args = list(mult = 1),
# position = position_nudge(x = -0.3)
# ) +
scale_y_continuous(name = dcant_pgc_label, breaks = seq(0, 100, 2)) +
facet_grid(basin ~ ., scales = "free_y")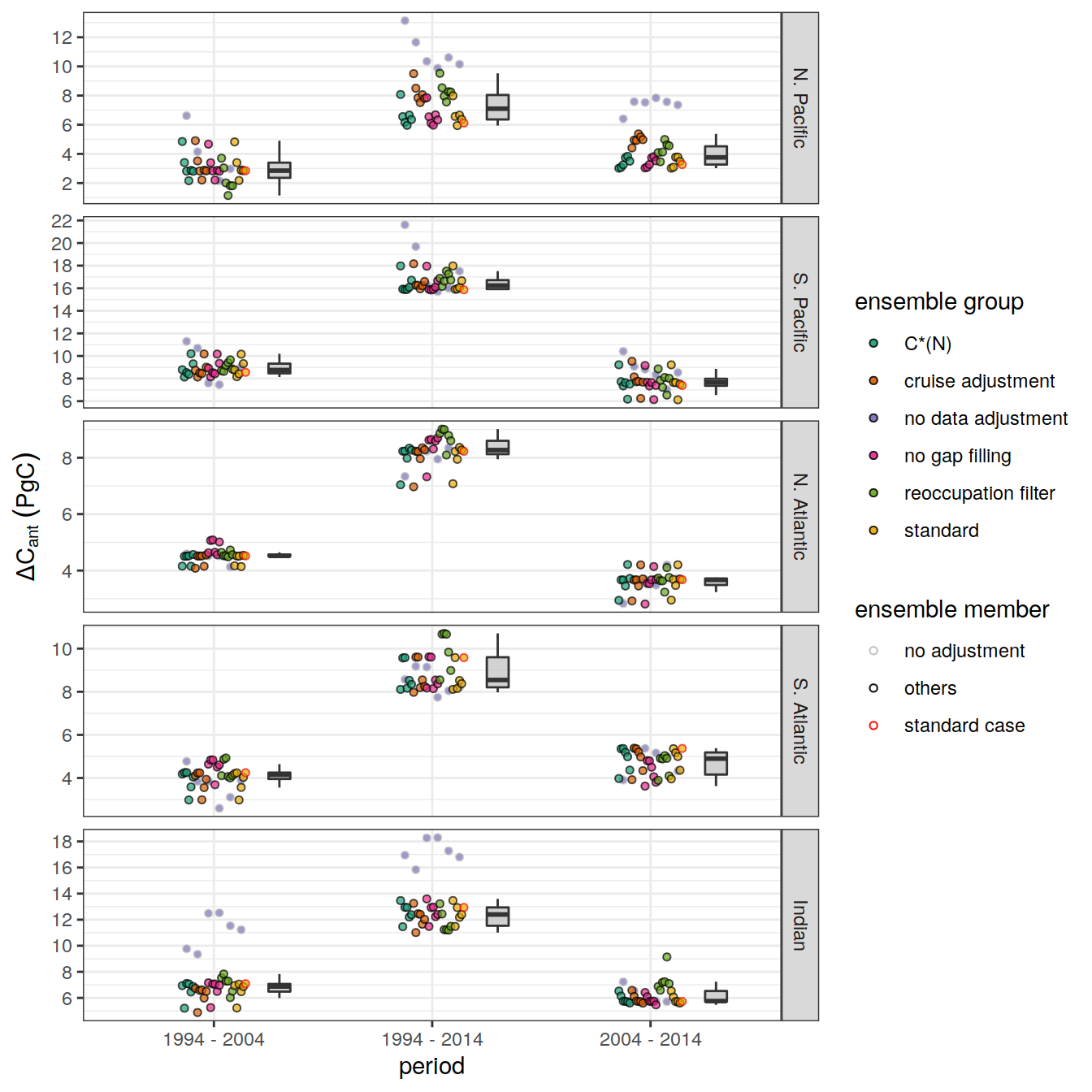
ggsave(path = here::here("output/publication"),
filename = "FigS_budget_dcant_basin_hemisphere_ensemble_member.png",
height = 6,
width = 6)
dcant_budget_basin_MLR_all_ensemble %>%
mutate(ensemble_role = if_else(Version_ID %in% Version_IDs,
"standard case",
"others")) %>%
filter(data_source == "obs") %>%
ggplot(aes(period, dcant)) +
scale_fill_brewer(palette = "Dark2") +
scale_color_manual(values = c("grey", "red"),
name = "ensemble member") +
geom_boxplot(
width = .1,
outlier.shape = NA,
fill = "grey",
alpha = 0.7,
position = position_nudge(x = 0.3)
) +
geom_point(
aes(fill = MLR_basins,
col = ensemble_role),
size = 1.3,
alpha = .7,
shape = 21,
position = position_dodge2(width = 0.3)
) +
stat_summary(
fun.data = "mean_sdl",
fun.args = list(mult = 2),
position = position_nudge(x = -0.3),
col = "grey"
) +
stat_summary(
fun.data = "mean_sdl",
fun.args = list(mult = 1),
position = position_nudge(x = -0.3)
) +
scale_y_continuous(name = dcant_pgc_label, breaks = seq(0, 100, 2)) +
facet_grid(basin ~ ., scales = "free_y")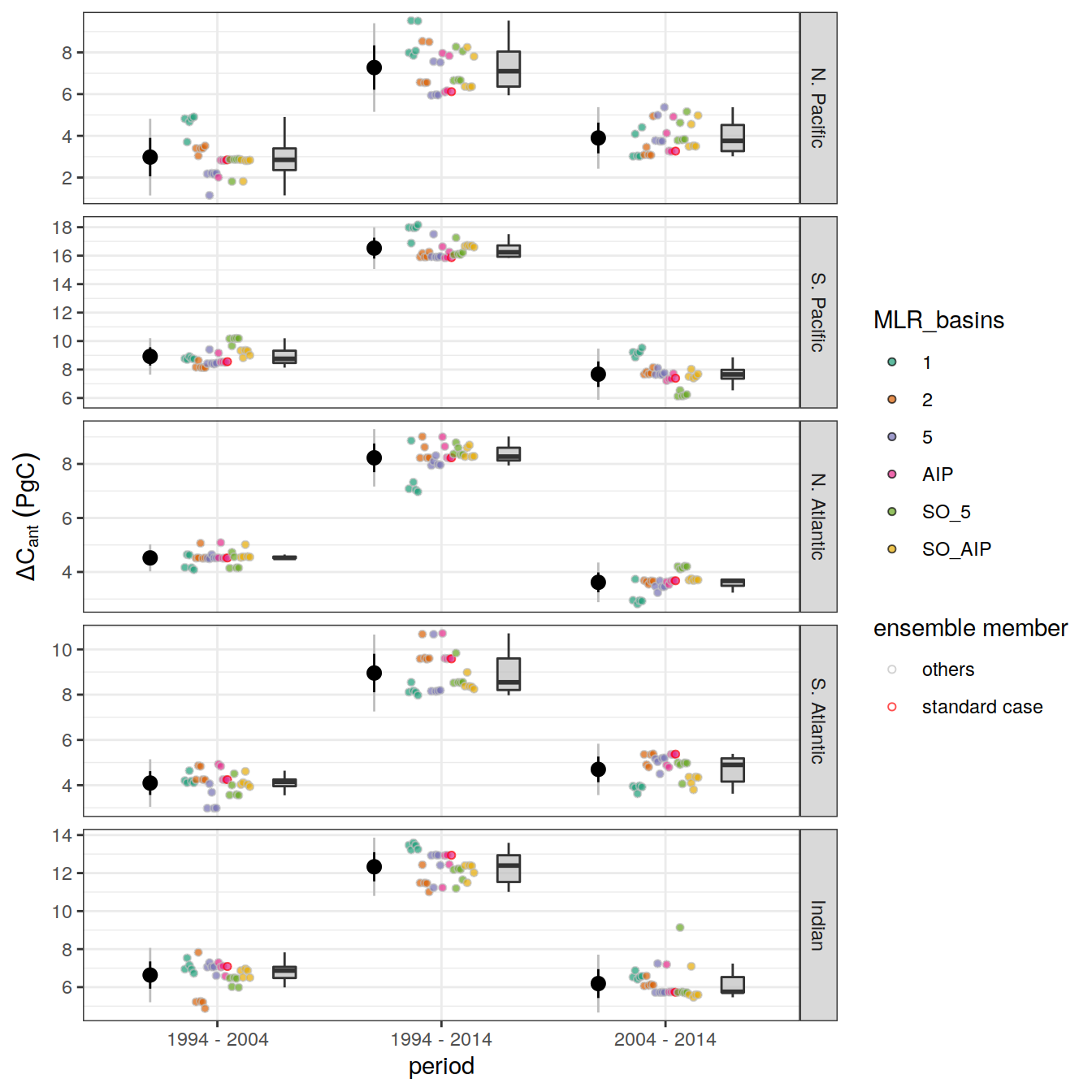
dcant_budget_basin_MLR_all_ensemble_summary <- dcant_budget_basin_MLR_all_ensemble %>%
group_by(data_source, basin, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant)*2) %>%
ungroup()
dcant_budget_global_all_ensemble_summary <-
dcant_budget_basin_MLR_all_ensemble %>%
filter(period != "1994 - 2014") %>%
group_by(data_source, Version_ID, tref1, tref2, period) %>%
summarise(dcant = sum(dcant)) %>%
ungroup() %>%
group_by(data_source, tref1, tref2, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant)*2) %>%
ungroup() %>%
filter(data_source == "obs")8.2 Flow chart
dcant_budget_basin_MLR_all <- left_join(dcant_budget_basin_MLR_all,
co2_atm %>% rename(tref1 = year,
pCO21 = pCO2))
dcant_budget_basin_MLR_all <- left_join(dcant_budget_basin_MLR_all,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
dcant_budget_basin_MLR_all_plot <- dcant_budget_basin_MLR_all %>%
filter(period != "1994 - 2014",
data_source == "obs")tcant_inv <-
inner_join(tcant_inv, basinmask)
dcant_inv_all <-
inner_join(dcant_inv_all, basinmask)
area_scaling <-
bind_rows(
tcant_inv %>% distinct(lat, lon, basin) %>%
mutate(source = "SO4"),
dcant_inv_all %>% distinct(lat, lon, basin) %>%
mutate(source = "M22")
)
area_scaling %>%
ggplot(aes(lon, lat, fill = basin)) +
geom_tile() +
facet_wrap( ~ source) +
coord_quickmap()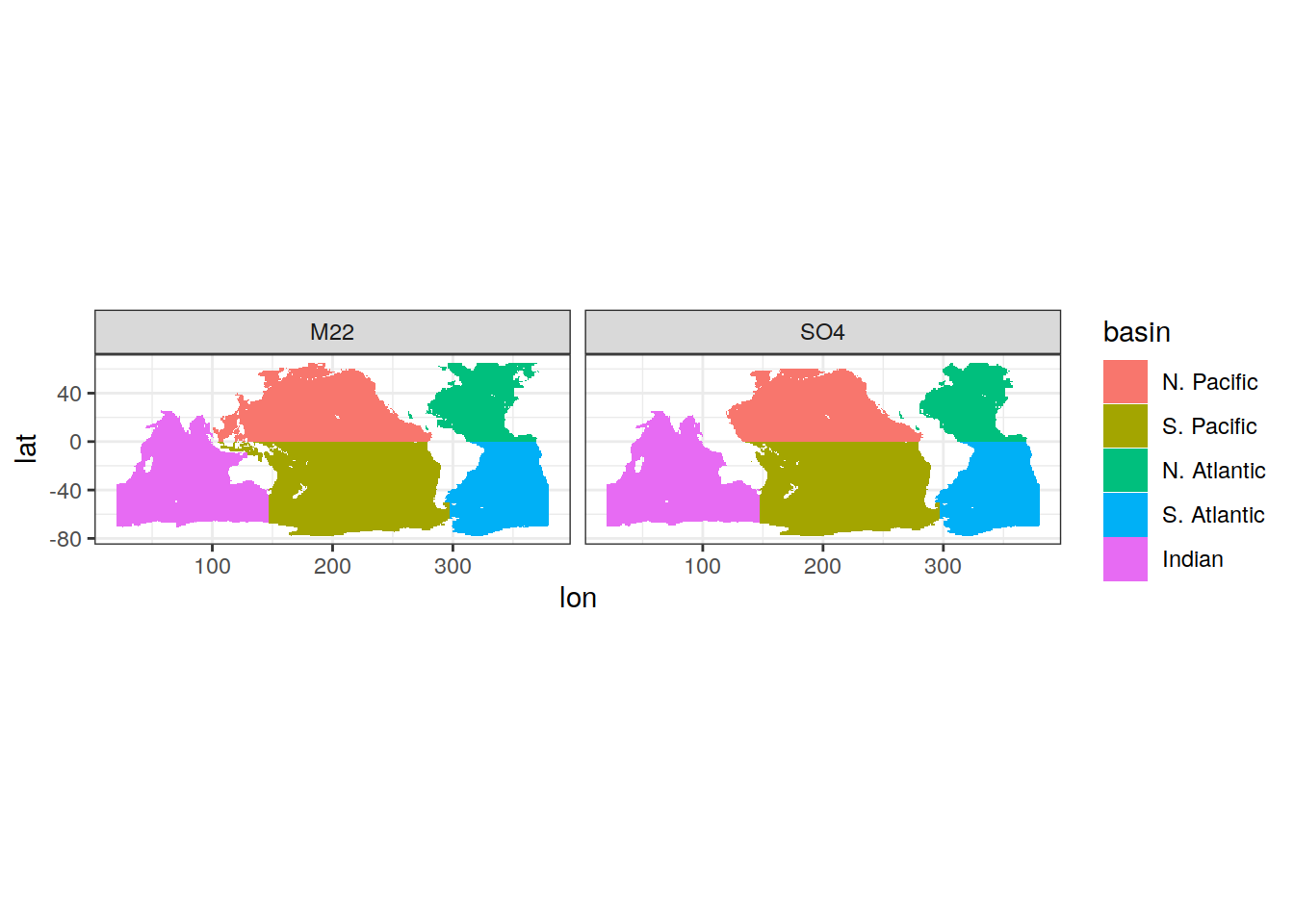
| Version | Author | Date |
|---|---|---|
| a26a21d | jens-daniel-mueller | 2022-06-27 |
area_scaling <- area_scaling %>%
mutate(area = earth_surf(lat, lon)) %>%
group_by(basin, source) %>%
summarise(area_total = sum(area)) %>%
ungroup() %>%
pivot_wider(values_from = area_total,
names_from = source) %>%
mutate(scaling_factor = M22 / SO4)
tcant_budget_basin_MLR <-
tcant_inv %>%
mutate(method = "layer",
data_source = "obs") %>%
group_by(basin) %>%
nest() %>%
mutate(budget = map(.x = data, ~m_dcant_budget(.x))) %>%
select(-data) %>%
unnest(budget)
tcant_budget_basin_MLR <-
full_join(tcant_budget_basin_MLR,
area_scaling %>% select(basin, scaling_factor)) %>%
mutate(value = value * scaling_factor) %>%
select(-scaling_factor)
tcant_budget_basin_MLR <- tcant_budget_basin_MLR %>%
mutate(pCO21 = 281,
tref1 = 1800,
tref2 = 1994)
tcant_budget_basin_MLR <- left_join(tcant_budget_basin_MLR,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
tcant_budget_basin_MLR <- tcant_budget_basin_MLR %>%
mutate(period = paste(tref1, tref2, sep = " - ")) %>%
filter(estimate == "dcant") %>%
select(-estimate) %>%
rename(dcant = value)
dcant_budget_basin_MLR_all_plot <- bind_rows(
dcant_budget_basin_MLR_all_plot,
tcant_budget_basin_MLR)
dcant_budget_basin_MLR_all_plot <-
left_join(dcant_budget_basin_MLR_all_plot,
dcant_budget_basin_MLR_all_ensemble_summary)8.2.1 Absolute
g1 <- dcant_budget_basin_MLR_all_plot %>%
filter(period != "1800 - 1994") %>%
ggplot(aes(
y = dcant,
x = period,
alluvium = basin,
fill = basin,
stratum = basin
)) +
stat_alluvium(decreasing = FALSE) +
stat_stratum(decreasing = FALSE) +
stat_stratum(geom = "text",
decreasing = FALSE,
aes(label = paste(
round(after_stat(max-min),1)
# 100*round(after_stat(prop), 2), "%"
))) +
ggrepel::geom_label_repel(
data = dcant_budget_basin_MLR_all_plot %>% filter(period == "2004 - 2014"),
stat = "stratum",
size = 4,
nudge_x = .5,
point.padding = 3,
aes(fill = basin, label = basin),
decreasing = FALSE
)+
scale_fill_brewer(palette = "Paired", guide = "none") +
scale_y_continuous(limits = c(0, 32), expand = c(0, 0)) +
labs(y = dcant_pgc_label) +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
axis.title.x = element_blank()) +
theme_classic()
newdat <- tibble(layer_data(g1))
change <-
newdat %>%
select(x, alluvium, count) %>%
pivot_wider(names_from = x,
values_from = count) %>%
mutate(dcant_change = round(100*(`2` - `1`) / `1`)) %>%
select(alluvium, dcant_change)
coord <- newdat %>%
filter(x == 2) %>%
select(x, y, alluvium)
new_layer <- full_join(
change,
coord
)
new_layer <- new_layer %>%
mutate(dcant_change = as.character(dcant_change),
dcant_change = if_else(str_detect(dcant_change, "-"),
dcant_change,
paste0("+", dcant_change)),
dcant_change = paste(dcant_change, "%"))
g1 +
geom_text(data = new_layer,
aes(
x = x - 0.3,
y = y,
label = dcant_change
),
inherit.aes = FALSE)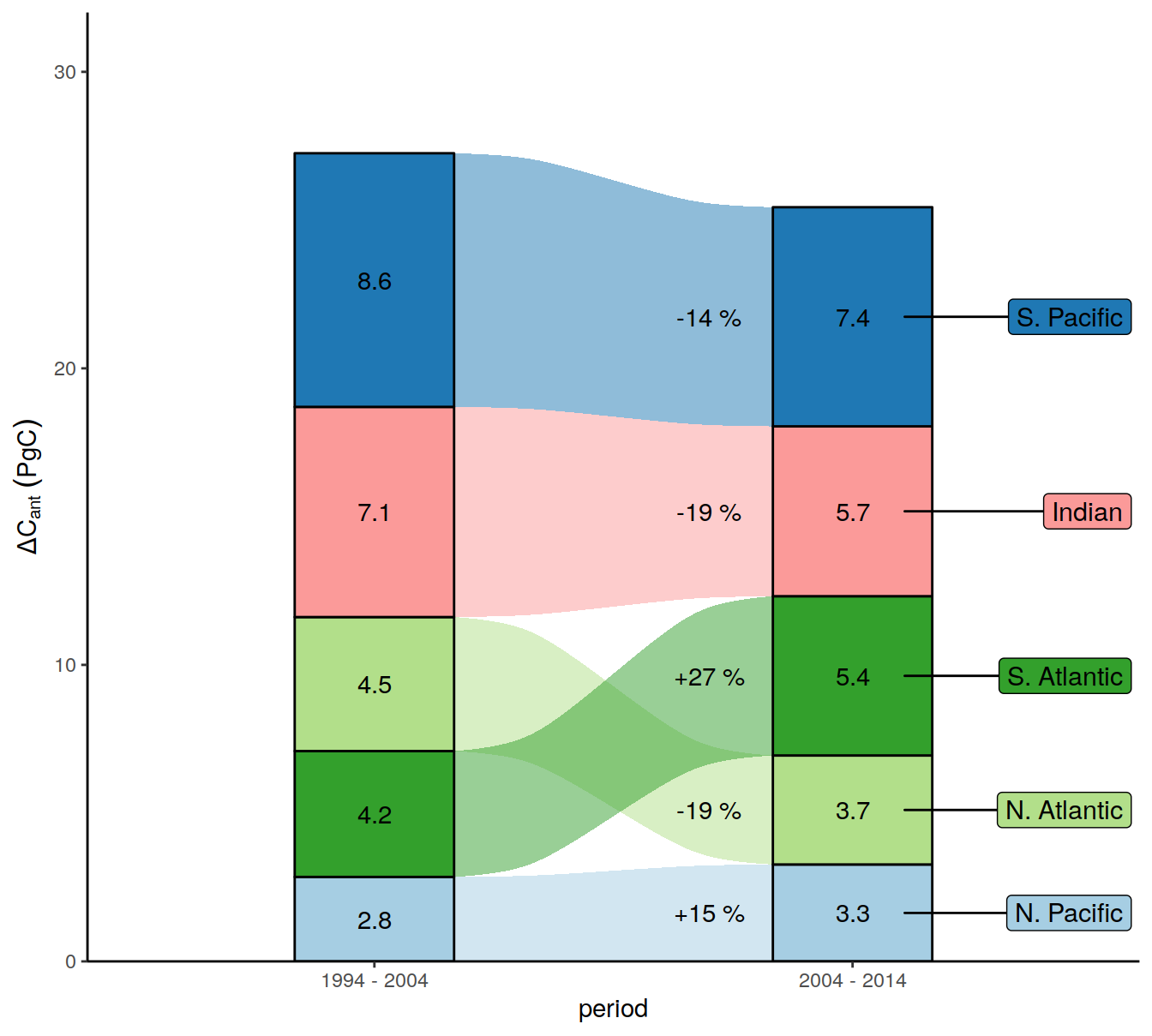
| Version | Author | Date |
|---|---|---|
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| a26a21d | jens-daniel-mueller | 2022-06-27 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
g2 <- dcant_budget_basin_MLR_all_plot %>%
filter(period != "1800 - 1994") %>%
ggplot(aes(
y = dcant,
x = period,
alluvium = basin,
fill = basin,
stratum = basin,
label = basin
)) +
geom_alluvium() +
geom_stratum() +
stat_stratum(geom = "text",
aes(label = paste(
round(after_stat(count),1)
# 100*round(after_stat(prop), 2), "%"
))) +
ggrepel::geom_label_repel(
data = dcant_budget_basin_MLR_all_plot %>% filter(period == "2004 - 2014"),
stat = "stratum",
size = 4,
nudge_x = .5,
point.padding = 3,
aes(fill = basin)
)+
scale_fill_brewer(palette = "Paired", guide = "none") +
scale_color_brewer(palette = "Paired", guide = "none") +
scale_y_continuous(limits = c(0, 33), expand = c(0, 0)) +
guides(y = "none") +
labs(title = dcant_pgc_label) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
plot.title = element_text(hjust = 0.5))
newdat <- tibble(layer_data(g2))
change_basin <-
newdat %>%
select(x, alluvium, count) %>%
pivot_wider(names_from = x,
values_from = count) %>%
mutate(dcant_change = round(100*(`2` - `1`) / `1`)) %>%
select(alluvium, dcant_change)
coord_basin <- newdat %>%
filter(x == 2) %>%
select(x, y, alluvium)
new_layer_basin <- full_join(
change_basin,
coord_basin
)
new_layer_basin <- new_layer_basin %>%
mutate(dcant_change = as.character(dcant_change),
dcant_change = if_else(str_detect(dcant_change, "-"),
dcant_change,
paste0("+", dcant_change)),
dcant_change = paste(dcant_change, "%"))
new_layer_total <-
newdat %>%
select(x, alluvium, count) %>%
group_by(x) %>%
summarise(dcant_change = sum(count)) %>%
ungroup()
new_layer_total <- new_layer_total %>%
mutate(y = dcant_change,
dcant_change = as.character(round(dcant_change,1)),
dcant_change = paste("global:",dcant_change))
g2 +
geom_text(data = new_layer_basin,
aes(
x = x - 0.3,
y = y,
label = dcant_change
),
inherit.aes = FALSE) +
geom_label(data = new_layer_total,
aes(
x = x,
y = y + 1,
label = dcant_change
),
inherit.aes = FALSE)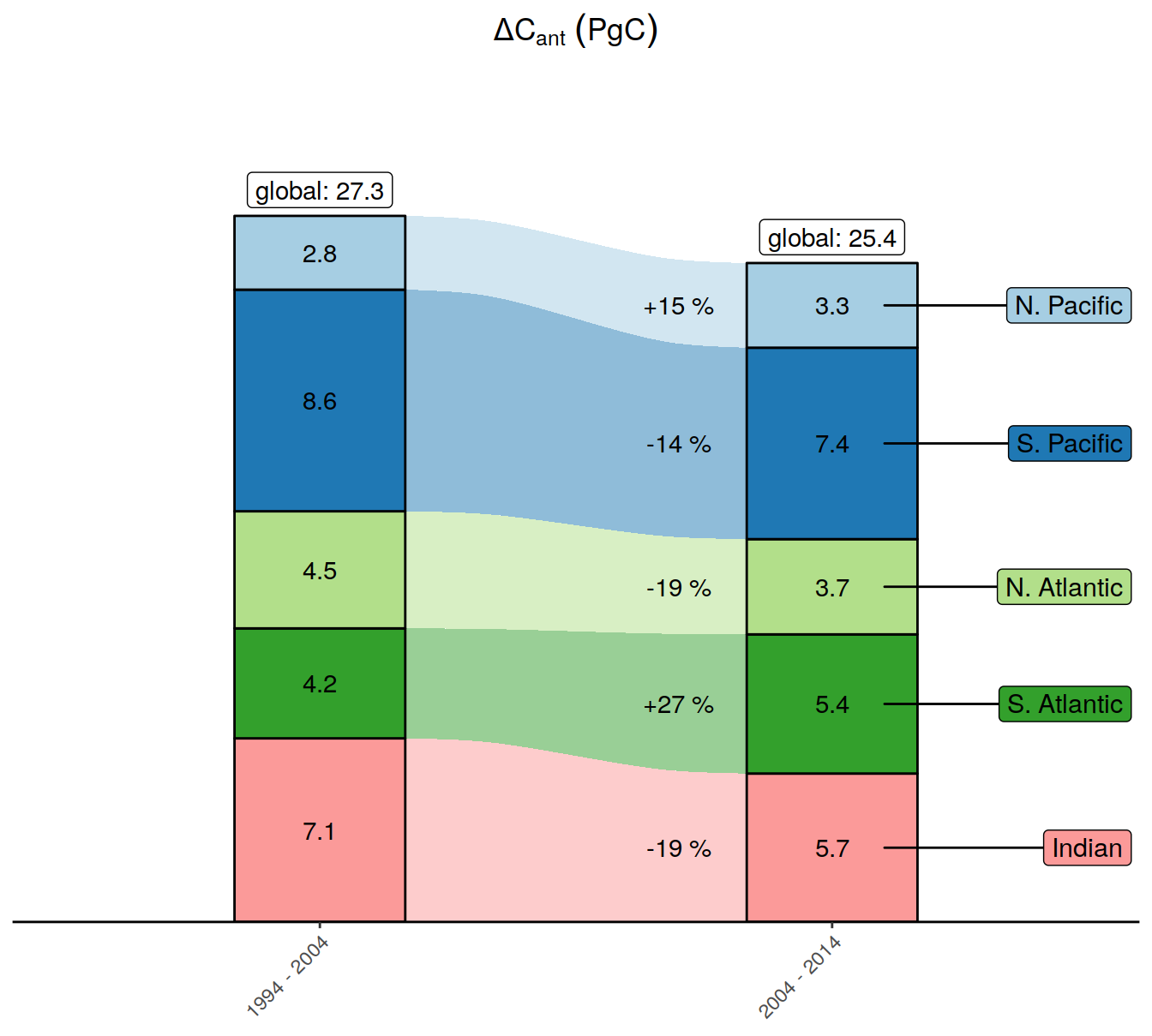
| Version | Author | Date |
|---|---|---|
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| a26a21d | jens-daniel-mueller | 2022-06-27 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| ec7fe7e | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| b1d7720 | jens-daniel-mueller | 2022-01-21 |
| 4fe7150 | jens-daniel-mueller | 2022-01-21 |
8.2.2 Scaled
dcant_budget_basin_MLR_all_plot <- left_join(dcant_budget_basin_MLR_all_plot,
co2_atm %>% rename(tref1 = year,
pCO21 = pCO2))
dcant_budget_basin_MLR_all_plot <- left_join(dcant_budget_basin_MLR_all_plot,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
dcant_budget_basin_MLR_all_plot <- dcant_budget_basin_MLR_all_plot %>%
mutate(delta_pCO2 = pCO22 - pCO21,
beta = dcant / delta_pCO2,
dcant_sd_scaled = dcant_sd / delta_pCO2) %>%
select(-starts_with("pCO2"))
dcant_budget_global_all_ensemble_summary <- left_join(dcant_budget_global_all_ensemble_summary,
co2_atm %>% rename(tref1 = year,
pCO21 = pCO2))
dcant_budget_global_all_ensemble_summary <- left_join(dcant_budget_global_all_ensemble_summary,
co2_atm %>% rename(tref2 = year,
pCO22 = pCO2))
dcant_budget_global_all_ensemble_summary <- dcant_budget_global_all_ensemble_summary %>%
mutate(delta_pCO2 = pCO22 - pCO21,
beta = dcant_mean / delta_pCO2,
dcant_sd_scaled = dcant_sd / delta_pCO2) %>%
select(-starts_with("pCO2"))g2 <- dcant_budget_basin_MLR_all_plot %>%
filter(period != "1800 - 1994") %>%
ggplot(aes(
y = beta,
x = period,
alluvium = basin,
fill = basin,
stratum = basin,
label = basin
)) +
geom_alluvium() +
geom_stratum() +
stat_stratum(geom = "text",
aes(label = paste(
round(after_stat(count),2)
# 100*round(after_stat(prop), 2), "%"
))) +
ggrepel::geom_label_repel(
data = dcant_budget_basin_MLR_all_plot %>% filter(period == "2004 - 2014"),
stat = "stratum",
size = 4,
nudge_x = .5,
point.padding = 3,
aes(fill = basin)
)+
scale_fill_brewer(palette = "Paired", guide = "none") +
scale_color_brewer(palette = "Paired", guide = "none") +
# scale_y_continuous(limits = c(0, 33), expand = c(0, 0)) +
guides(y = "none") +
labs(title = dcant_pgc_scaled_label) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
plot.title = element_text(hjust = 0.5))
newdat <- tibble(layer_data(g2))
change_basin <-
newdat %>%
select(x, alluvium, count) %>%
pivot_wider(names_from = x,
values_from = count) %>%
mutate(dcant_change = round(100*(`2` - `1`) / `1`)) %>%
select(alluvium, dcant_change)
coord_basin <- newdat %>%
filter(x == 2) %>%
select(x, y, alluvium)
new_layer_basin <- full_join(
change_basin,
coord_basin
)
new_layer_basin <- new_layer_basin %>%
mutate(dcant_change = as.character(dcant_change),
dcant_change = if_else(str_detect(dcant_change, "-"),
dcant_change,
paste0("+", dcant_change)),
dcant_change = paste(dcant_change, "%"))
new_layer_total <-
newdat %>%
select(x, alluvium, count) %>%
group_by(x) %>%
summarise(dcant_change = sum(count)) %>%
ungroup()
new_layer_total <- new_layer_total %>%
mutate(y = dcant_change,
dcant_change = as.character(round(dcant_change,2)),
dcant_change = paste("global:",dcant_change))
g2 +
geom_text(data = new_layer_basin,
aes(
x = x - 0.3,
y = y,
label = dcant_change
),
inherit.aes = FALSE) +
geom_label(data = new_layer_total,
aes(
x = x,
y = y + 0.05,
label = dcant_change
),
inherit.aes = FALSE)
| Version | Author | Date |
|---|---|---|
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| a26a21d | jens-daniel-mueller | 2022-06-27 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| ed903f7 | jens-daniel-mueller | 2022-02-02 |
g2 <- dcant_budget_basin_MLR_all_plot %>%
ggplot(aes(
y = beta,
x = period,
alluvium = basin,
fill = basin,
stratum = basin,
label = basin
)) +
geom_alluvium() +
geom_stratum() +
stat_stratum(geom = "text",
aes(label = paste(
round(after_stat(count),2)
# 100*round(after_stat(prop), 2), "%"
))) +
ggrepel::geom_label_repel(
data = dcant_budget_basin_MLR_all_plot %>% filter(period == "2004 - 2014"),
stat = "stratum",
size = 4,
nudge_x = .5,
point.padding = 3,
aes(fill = basin)
)+
scale_fill_brewer(palette = "Paired", guide = "none") +
scale_color_brewer(palette = "Paired", guide = "none") +
scale_y_continuous(limits = c(0, 1.7), expand = c(0, 0)) +
guides(y = "none") +
labs(title = dcant_pgc_scaled_label) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
plot.title = element_text(hjust = 0.5))
newdat <- tibble(layer_data(g2))
change_basin <-
newdat %>%
select(x, alluvium, count) %>%
group_by(alluvium) %>%
arrange(x) %>%
mutate(dcant_change = round(100*(count - lag(count)) / lag(count))) %>%
ungroup()
coord_basin <- newdat %>%
select(x, y, alluvium)
new_layer_basin <- left_join(
change_basin %>% drop_na(),
coord_basin
)
new_layer_basin <- new_layer_basin %>%
mutate(dcant_change = as.character(dcant_change),
dcant_change = if_else(str_detect(dcant_change, "-"),
dcant_change,
paste0("+", dcant_change)),
dcant_change = paste(dcant_change, "%"))
new_layer_total <-
newdat %>%
select(x, alluvium, count) %>%
group_by(x) %>%
summarise(dcant_change = sum(count)) %>%
ungroup()
new_layer_total <- new_layer_total %>%
mutate(y = dcant_change,
dcant_change = as.character(round(dcant_change,2)),
dcant_change = paste("global:",dcant_change))
g2 +
geom_text(data = new_layer_basin,
aes(
x = x - 0.3,
y = y,
label = dcant_change
),
inherit.aes = FALSE) +
geom_label(data = new_layer_total,
aes(
x = x,
y = y + 0.1,
label = dcant_change
),
inherit.aes = FALSE)
| Version | Author | Date |
|---|---|---|
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| a26a21d | jens-daniel-mueller | 2022-06-27 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| fa46251 | jens-daniel-mueller | 2022-02-02 |
| 7655085 | jens-daniel-mueller | 2022-02-02 |
| 226d67d | jens-daniel-mueller | 2022-02-02 |
g2 <- dcant_budget_basin_MLR_all_plot %>%
ggplot(aes(
y = beta,
x = period,
alluvium = basin,
fill = basin,
stratum = basin,
label = basin
)) +
geom_alluvium() +
geom_stratum() +
ggrepel::geom_label_repel(
data = dcant_budget_basin_MLR_all_plot %>% filter(period == "2004 - 2014"),
stat = "stratum",
size = 4,
nudge_x = .5,
point.padding = 3,
aes(fill = basin)
)+
scale_fill_brewer(palette = "Paired", guide = "none") +
scale_color_brewer(palette = "Paired", guide = "none") +
scale_y_continuous(limits = c(0, 1.6), expand = c(0, 0)) +
guides(y = "none") +
labs(title = dcant_pgc_scaled_label) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
plot.title = element_text(hjust = 0.5))
newdat <- tibble(layer_data(g2))
# construct budget labels
budget_basin <-
newdat %>%
select(x, y, alluvium, count)
budget_layer_basin <- budget_basin %>%
mutate(dcant = as.character(round(count,2)))
# construct uncertainty labels
uncertainty_basin <-
dcant_budget_basin_MLR_all_plot %>%
mutate(x = fct_recode(period,
"2" = "1994 - 2004",
"3" = "2004 - 2014")) %>%
select(x, alluvium = basin, dcant_sd_scaled) %>%
drop_na() %>%
arrange(x, alluvium)
coord_basin <- newdat %>%
select(x, y, alluvium) %>%
filter(x != 1)
uncertainty_layer_basin <- bind_cols(
coord_basin,
uncertainty_basin %>% select(dcant_sd_scaled),
)
# construct change labels
change_basin <-
newdat %>%
select(x, alluvium, count) %>%
group_by(alluvium) %>%
arrange(x) %>%
mutate(dcant_change_abs = count - lag(count),
dcant_change = round(100*(dcant_change_abs) / lag(count))) %>%
ungroup()
coord_basin <- newdat %>%
select(x, y, alluvium)
new_layer_basin <- left_join(
change_basin %>% drop_na(),
coord_basin
)
new_layer_basin <- new_layer_basin %>%
mutate(dcant_change = as.character(dcant_change),
dcant_change = if_else(str_detect(dcant_change, "-"),
dcant_change,
paste0("+", dcant_change)),
dcant_change = paste0(dcant_change, "%"))
# construct global budget labels
new_layer_total <-
newdat %>%
select(x, alluvium, count) %>%
group_by(x) %>%
summarise(dcant_change = sum(count)) %>%
ungroup()
dcant_sd_scaled <- round(c(
0.24,
dcant_budget_global_all_ensemble_summary %>%
pull(dcant_sd_scaled)
), 2)
new_layer_total <- bind_cols(new_layer_total, dcant_sd_scaled = dcant_sd_scaled)
new_layer_total <- new_layer_total %>%
mutate(y = dcant_change,
dcant_sd_scaled_rel = round(100 * dcant_sd_scaled/dcant_change),
dcant_change = as.character(round(dcant_change,2)),
dcant_sd_scaled = round(dcant_sd_scaled,1),
dcant_change = paste0(dcant_change, "\n(±", dcant_sd_scaled, ")"))
# basin change + uncertainty
budget_layer_basin <- full_join(budget_layer_basin,
uncertainty_layer_basin)
budget_layer_basin <-
budget_layer_basin %>%
mutate(dcant_sd_scaled_rel = 100*dcant_sd_scaled/count,
dcant_sd_scaled_rel = as.character(round(dcant_sd_scaled_rel)),
dcant_sd_scaled_numeric = dcant_sd_scaled,
dcant_sd_scaled = round(dcant_sd_scaled, 2),
dcant_sd_scaled = paste0("(±", dcant_sd_scaled, ")"),
dcant_label = if_else(dcant_sd_scaled != "(±NA)",
paste0(dcant, "\n", dcant_sd_scaled),
dcant))
# uncertainty changes
uncertainty_changes <- budget_layer_basin %>%
select(x, alluvium, dcant_sd_scaled = dcant_sd_scaled_numeric) %>%
group_by(alluvium) %>%
mutate(dcant_change_uncert = sqrt(dcant_sd_scaled^2 + lag(dcant_sd_scaled)^2)) %>%
ungroup()
new_layer_basin <- left_join(new_layer_basin, uncertainty_changes) %>%
mutate(dcant_change_uncert_rel = 100*dcant_change_uncert/abs((count + lag(count))/2),
dcant_change_label = if_else(!is.na(dcant_change_uncert),
paste0(dcant_change, "\n(±", round(dcant_change_uncert_rel), "%)"),
dcant_change))
g2 +
geom_text(data = budget_layer_basin,
aes(
x = x,
y = y,
label = dcant_label
),
size = 3,
inherit.aes = FALSE) +
geom_text(data = new_layer_basin,
aes(
x = x - 0.3,
y = y,
label = dcant_change_label
),
size = 4,
inherit.aes = FALSE)
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 6be73e0 | jens-daniel-mueller | 2022-06-30 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| a26a21d | jens-daniel-mueller | 2022-06-27 |
| 748aa43 | jens-daniel-mueller | 2022-06-27 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| b599680 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| a6b33aa | jens-daniel-mueller | 2022-02-04 |
| 4b48475 | jens-daniel-mueller | 2022-02-04 |
| fec5a1e | jens-daniel-mueller | 2022-02-04 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| c7b4984 | jens-daniel-mueller | 2022-02-02 |
| 7fb28a2 | jens-daniel-mueller | 2022-02-02 |
| 49097e8 | jens-daniel-mueller | 2022-02-02 |
| fe11bfd | jens-daniel-mueller | 2022-02-02 |
# geom_label(data = new_layer_total,
# aes(
# x = x,
# y = y + 0.13,
# label = dcant_change
# ),
# size = 4,
# inherit.aes = FALSE)
ggsave(path = here::here("output/publication"),
filename = "Fig_budget_beta_basin_hemisphere.png",
height = 6,
width = 8)8.3 Depth layer
8.3.1 Global
dcant_budget_change <- dcant_budget_global_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
select(-c(tref1, tref2, Version_ID)) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(sign = if_else(`2004 - 2014` - `1994 - 2004` > 0,
"increase",
"decrease"))
dcant_budget_layer <- dcant_budget_global_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3400,
data_source == "obs") %>%
rename(dcant = value) %>%
group_by(basin, period, data_source) %>%
mutate(dcant = if_else(is.na(lead(dcant)), 888, dcant),
dcant = na_if(dcant, 888)) %>%
fill(dcant) %>%
ungroup()
dcant_budget_layer %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_rect(
data = dcant_budget_change,
aes(
xmin = inv_depth - 250,
xmax = inv_depth + 250,
ymin = `1994 - 2004`,
ymax = `2004 - 2014`,
fill = sign
),
alpha = 0.4
) +
geom_step(aes(inv_depth - 250, dcant, col = period), direction = "vh",
size = 1) +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500)) +
scale_color_brewer(palette = "Set1", direction = -1, name = "period") +
scale_fill_brewer(palette = "Set1", direction = -1, name = "") +
facet_wrap(~ basin, ncol = 3, dir = "v") +
labs(y = dcant_layer_budget_label,
x = "Depth (m)") +
theme(legend.position = c(0.8,0.2))
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
dcant_budget_change <- dcant_budget_global_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
arrange(inv_depth) %>%
group_by(data_source, basin, period) %>%
mutate(dcant_cum = cumsum(dcant)) %>%
ungroup() %>%
select(-c(tref2, Version_ID, dcant)) %>%
pivot_wider(names_from = period,
values_from = dcant_cum) %>%
mutate(sign = if_else(`2004 - 2014` - `1994 - 2004` > 0,
"increase",
"decrease"))
dcant_budget_layer <- dcant_budget_global_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3400,
data_source == "obs") %>%
rename(dcant = value) %>%
arrange(inv_depth) %>%
group_by(data_source, basin, period) %>%
mutate(dcant_cum = cumsum(dcant)) %>%
ungroup() %>%
group_by(basin, period, data_source) %>%
mutate(dcant_cum = if_else(is.na(lead(dcant_cum)), 888, dcant_cum),
dcant_cum = na_if(dcant_cum, 888)) %>%
fill(dcant_cum) %>%
ungroup()
dcant_budget_layer %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_rect(
data = dcant_budget_change,
aes(
xmin = inv_depth - 250,
xmax = inv_depth + 250,
ymin = `1994 - 2004`,
ymax = `2004 - 2014`,
fill = sign
),
alpha = 0.3
) +
geom_step(aes(inv_depth - 250, dcant_cum, col = period), direction = "vh",
size = 1) +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500)) +
scale_color_brewer(palette = "Set1",
direction = -1,
name = "Decade") +
scale_fill_brewer(palette = "Set1",
direction = -1,
name = "Decadal change") +
facet_wrap( ~ basin, ncol = 3, dir = "v") +
labs(y = dcant_layer_budget_label,
x = "Depth (m)",
title = "Cumulative layer inventory change") +
theme(legend.position = c(0.8,0.2))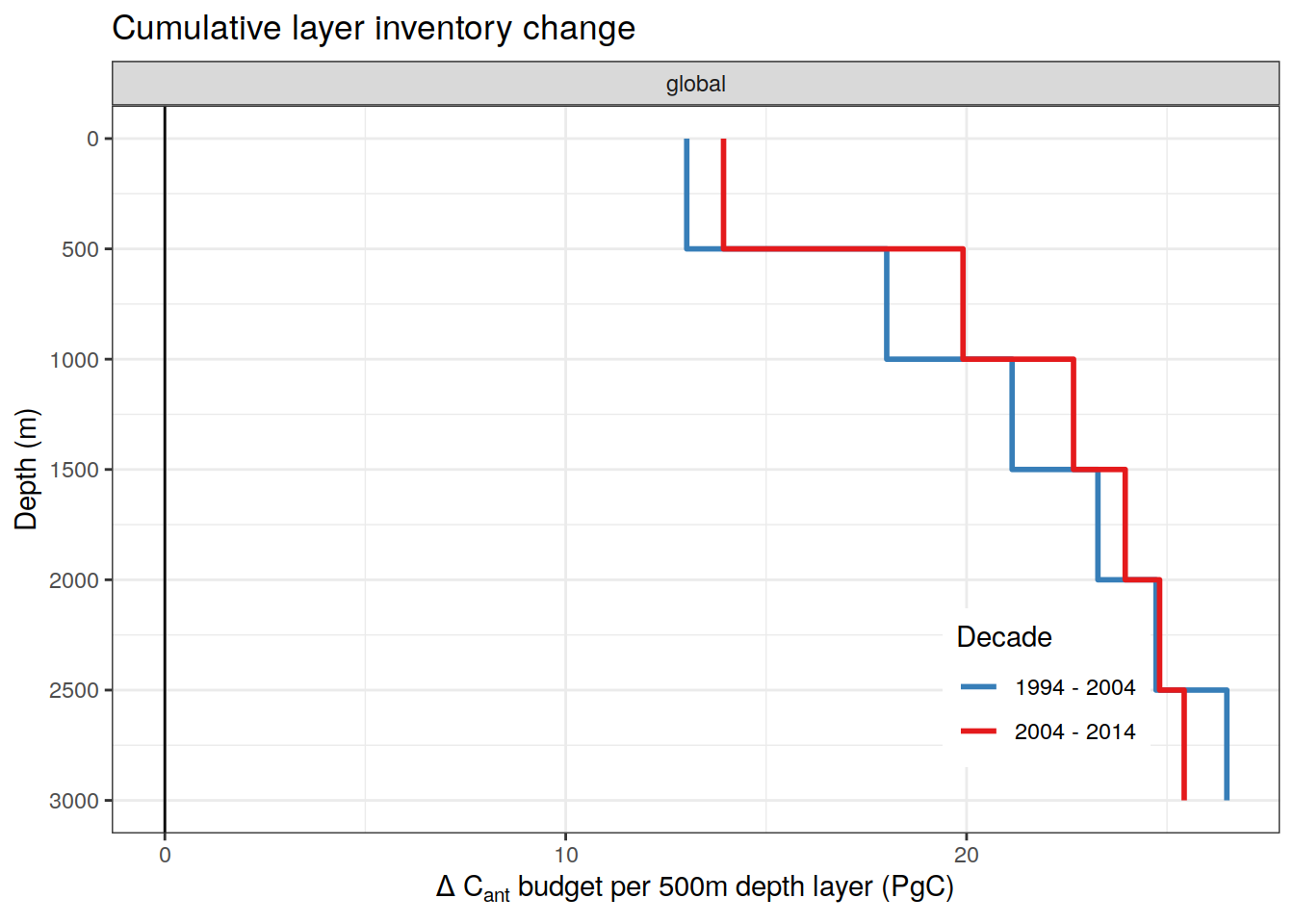
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
8.3.2 Basin AIP
dcant_budget_basin_AIP_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
ggplot(aes(inv_depth-250, dcant, col=period)) +
geom_hline(yintercept = 0) +
geom_step(direction = "vh") +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500)) +
scale_color_brewer(palette = "Set1", direction = -1) +
facet_grid(basin_AIP ~ .) +
labs(y = dcant_layer_budget_label,
x = "Depth (m)")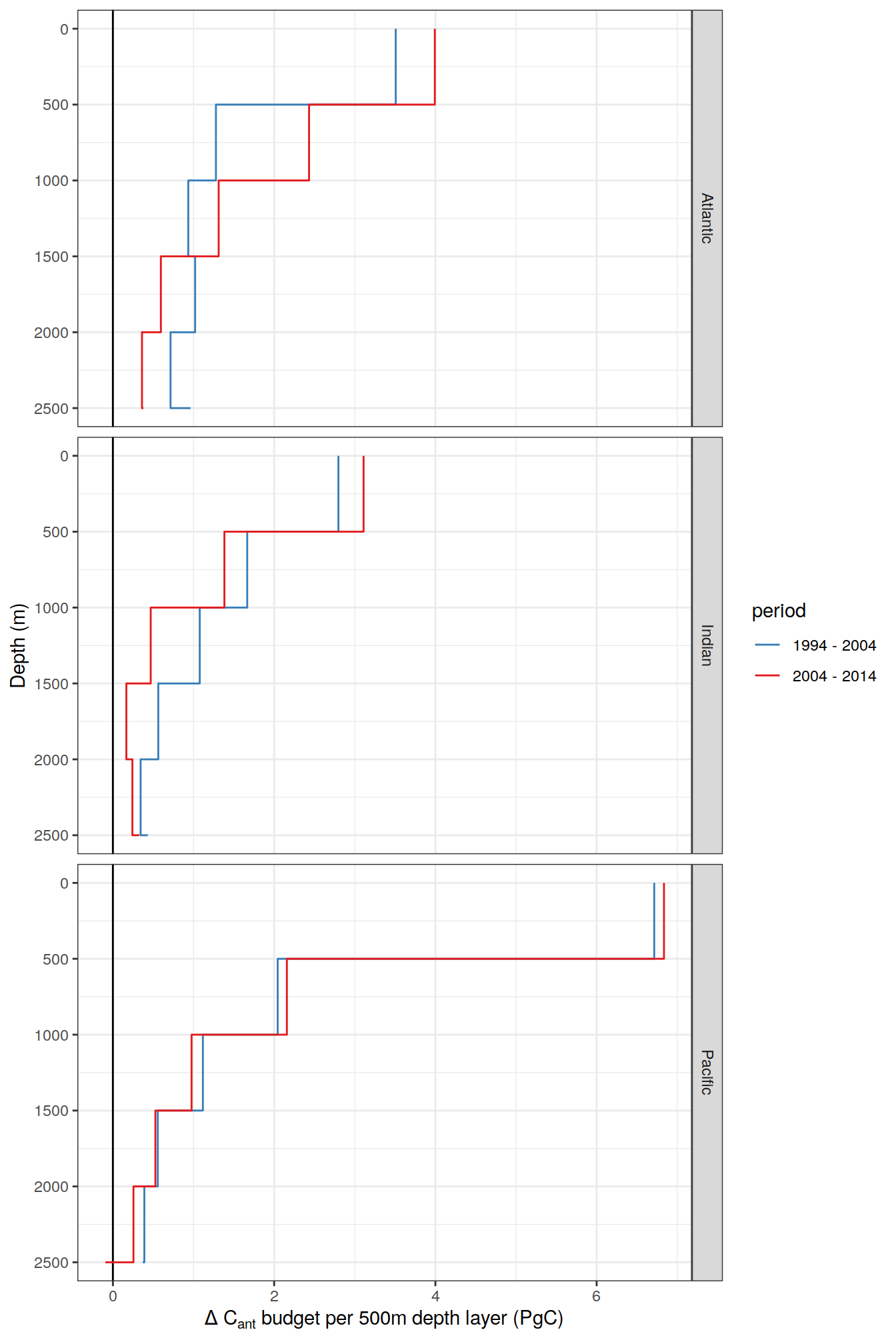
| Version | Author | Date |
|---|---|---|
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
8.3.3 5 basins
dcant_budget_change <- dcant_budget_basin_MLR_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
select(-c(tref1, tref2, Version_ID)) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
mutate(sign = if_else(`2004 - 2014` - `1994 - 2004` > 0,
"increase",
"decrease"))
dcant_budget_layer <- dcant_budget_basin_MLR_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3400,
data_source == "obs") %>%
rename(dcant = value) %>%
group_by(basin, period, data_source) %>%
mutate(dcant = if_else(is.na(lead(dcant)), 888, dcant),
dcant = na_if(dcant, 888)) %>%
fill(dcant) %>%
ungroup()
dcant_budget_layer %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_rect(
data = dcant_budget_change,
aes(
xmin = inv_depth - 250,
xmax = inv_depth + 250,
ymin = `1994 - 2004`,
ymax = `2004 - 2014`,
fill = sign
),
alpha = 0.4
) +
geom_step(aes(inv_depth - 250, dcant, col = period), direction = "vh",
size = 1) +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500)) +
scale_color_brewer(palette = "Set1", direction = -1, name = "period") +
scale_fill_brewer(palette = "Set1", direction = -1, name = "") +
facet_wrap(~ basin, ncol = 3, dir = "v") +
labs(y = dcant_layer_budget_label,
x = "Depth (m)") +
theme(legend.position = c(0.8,0.2))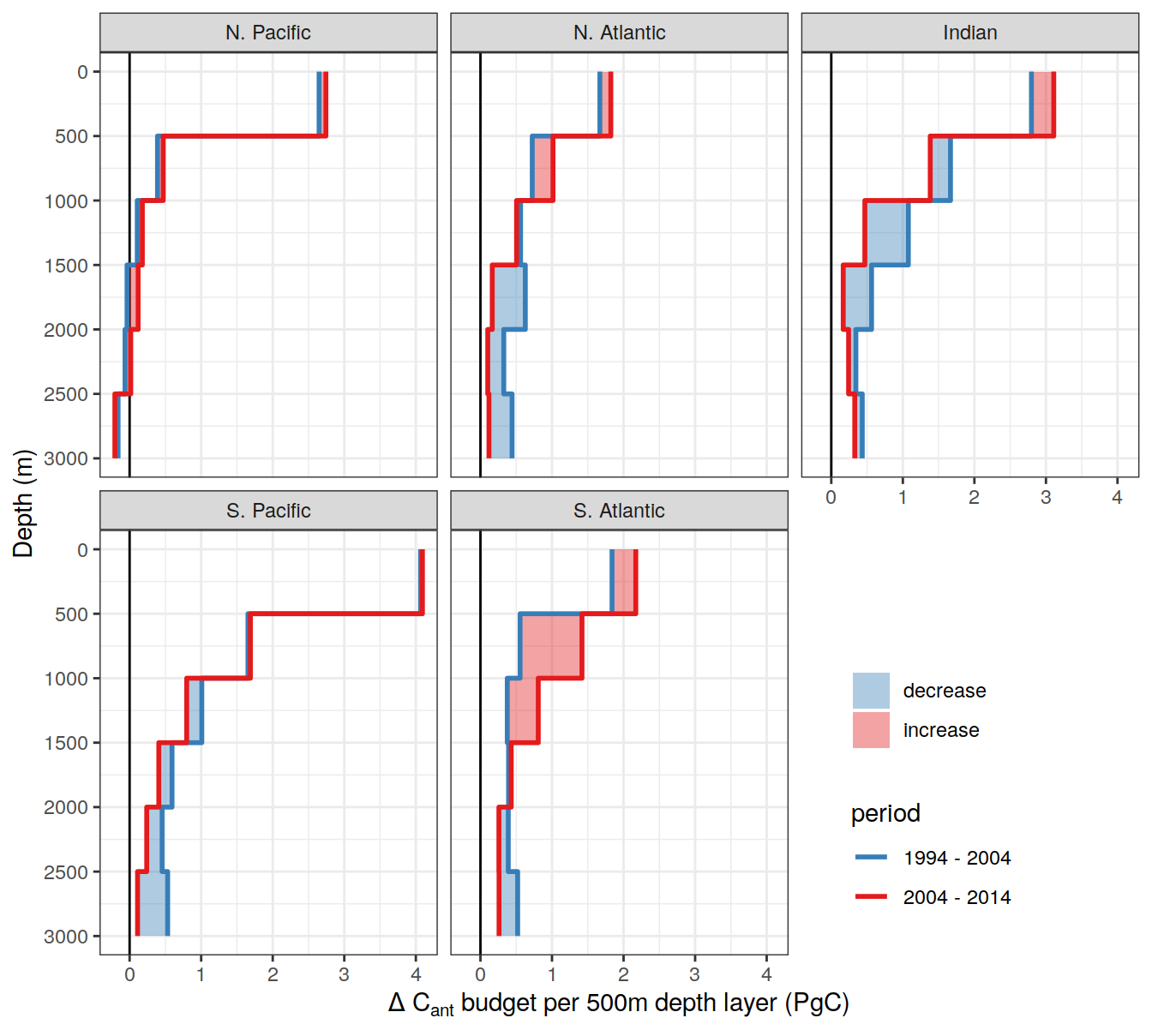
| Version | Author | Date |
|---|---|---|
| 26e9496 | jens-daniel-mueller | 2022-07-12 |
ggsave(path = here::here("output/publication"),
filename = "FigS_budget_dcant_basin_hemisphere_depth_layer.png",
height = 6,
width = 8)dcant_budget_range <- dcant_budget_basin_MLR_layer_all_ensemble %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
select(-c(tref1, tref2, Version_ID)) %>%
group_by(basin, inv_depth, period) %>%
summarise(dcant_sd = sd(dcant)*2) %>%
ungroup()
dcant_budget_layer <-
full_join(dcant_budget_range,
dcant_budget_layer)
dcant_budget_layer %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_rect(
aes(
xmin = inv_depth - 250,
xmax = inv_depth + 250,
ymin = dcant - dcant_sd,
ymax = dcant + dcant_sd,
fill = period
),
alpha = 0.4
) +
geom_step(aes(inv_depth - 250, dcant, col = period), direction = "vh",
size = 1) +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500), limits = c(3000,0)) +
scale_color_brewer(palette = "Set1", direction = -1, name = "period") +
scale_fill_brewer(palette = "Set1", direction = -1, name = "") +
facet_wrap(~ basin, ncol = 3, dir = "v") +
labs(y = dcant_layer_budget_label,
x = "Depth (m)") +
theme(legend.position = c(0.8,0.2))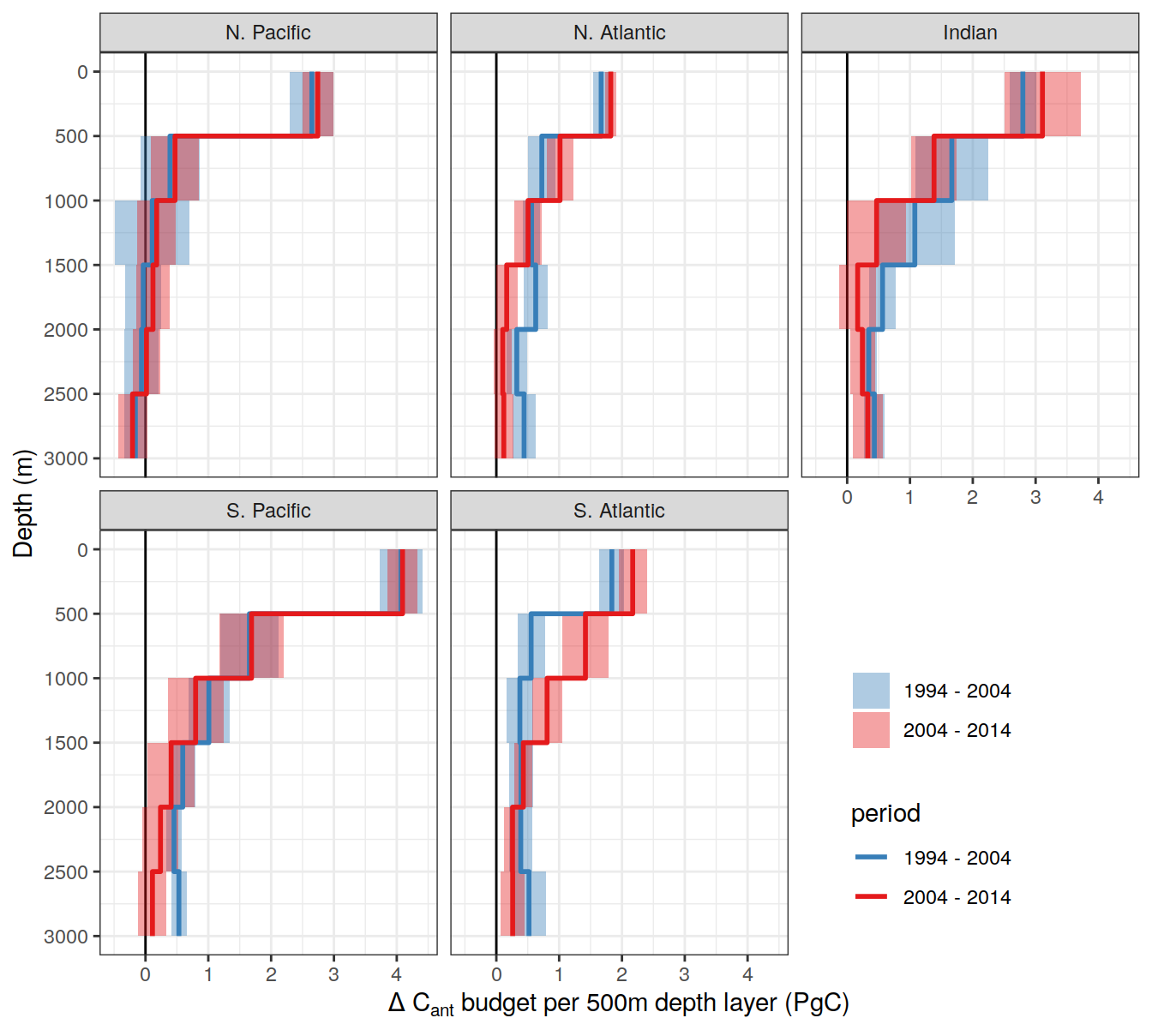
dcant_budget_change <- dcant_budget_basin_MLR_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3000,
data_source == "obs") %>%
rename(dcant = value) %>%
arrange(inv_depth) %>%
group_by(data_source, basin, period) %>%
mutate(dcant_cum = cumsum(dcant)) %>%
ungroup() %>%
select(-c(tref2, Version_ID, dcant)) %>%
pivot_wider(names_from = period,
values_from = dcant_cum) %>%
mutate(sign = if_else(`2004 - 2014` - `1994 - 2004` > 0,
"increase",
"decrease"))
dcant_budget_layer <- dcant_budget_basin_MLR_layer_all %>%
filter(estimate == "dcant",
period != "1994 - 2014",
inv_depth <= 3400,
data_source == "obs") %>%
rename(dcant = value) %>%
arrange(inv_depth) %>%
group_by(data_source, basin, period) %>%
mutate(dcant_cum = cumsum(dcant)) %>%
ungroup() %>%
group_by(basin, period, data_source) %>%
mutate(dcant_cum = if_else(is.na(lead(dcant_cum)), 888, dcant_cum),
dcant_cum = na_if(dcant_cum, 888)) %>%
fill(dcant_cum) %>%
ungroup()
dcant_budget_layer %>%
ggplot() +
geom_hline(yintercept = 0) +
geom_rect(
data = dcant_budget_change,
aes(
xmin = inv_depth - 250,
xmax = inv_depth + 250,
ymin = `1994 - 2004`,
ymax = `2004 - 2014`,
fill = sign
),
alpha = 0.3
) +
geom_step(aes(inv_depth - 250, dcant_cum, col = period), direction = "vh",
size = 1) +
coord_flip() +
scale_x_reverse(breaks = seq(0, 3000, 500)) +
scale_color_brewer(palette = "Set1",
direction = -1,
name = "Decade") +
scale_fill_brewer(palette = "Set1",
direction = -1,
name = "Decadal change") +
facet_wrap( ~ basin, ncol = 3, dir = "v") +
labs(y = dcant_layer_budget_label,
x = "Depth (m)",
title = "Cumulative layer inventory change") +
theme(legend.position = c(0.8,0.2))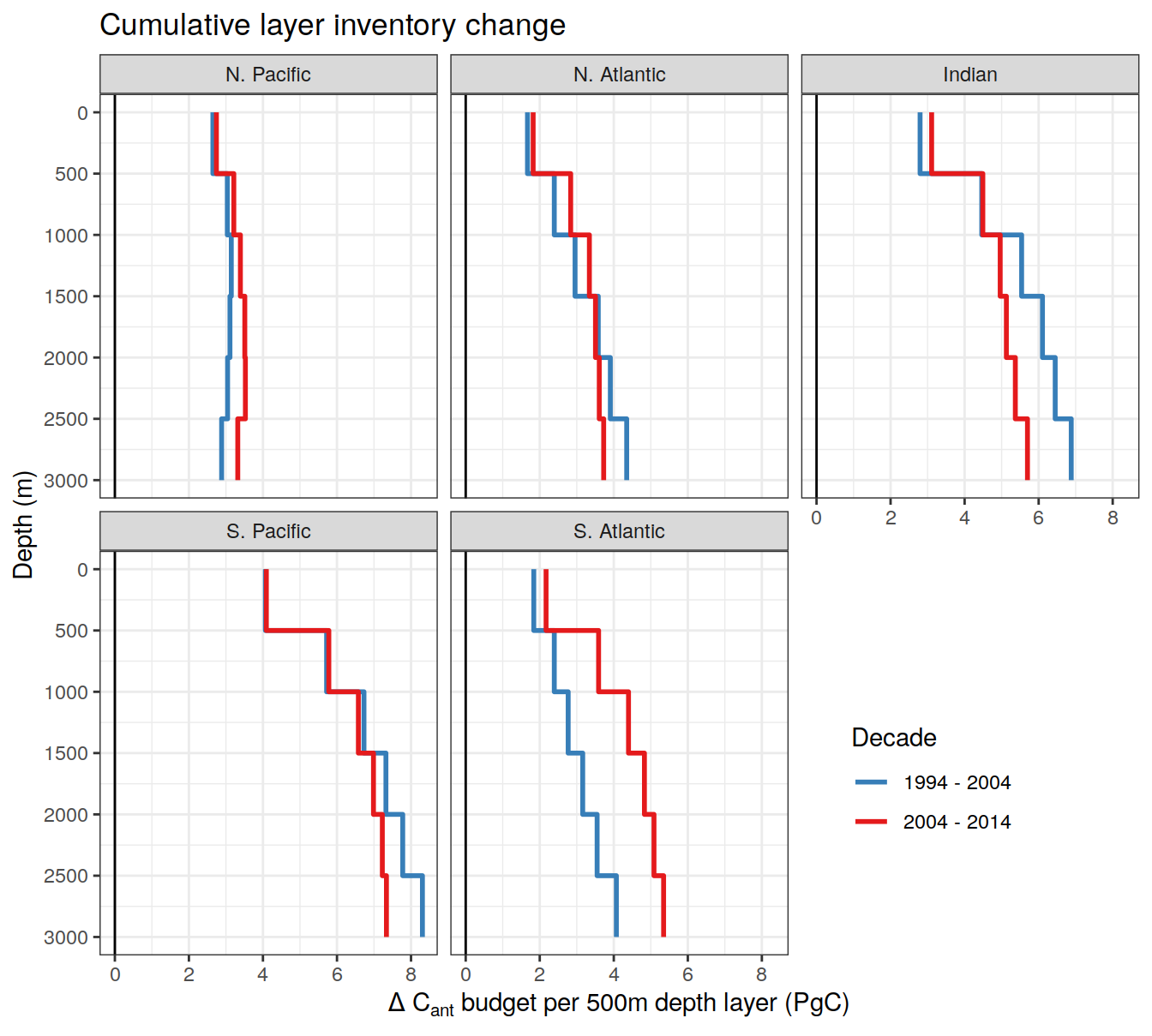
| Version | Author | Date |
|---|---|---|
| 26e9496 | jens-daniel-mueller | 2022-07-12 |
| 37f56b3 | jens-daniel-mueller | 2022-07-01 |
| df21d31 | jens-daniel-mueller | 2022-07-01 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 32e9682 | jens-daniel-mueller | 2022-02-02 |
| 913e42f | jens-daniel-mueller | 2022-02-01 |
| 189de95 | jens-daniel-mueller | 2022-02-01 |
| ab001eb | jens-daniel-mueller | 2022-01-31 |
| b62308d | jens-daniel-mueller | 2022-01-31 |
| 5f2aed0 | jens-daniel-mueller | 2022-01-27 |
9 Time series
9.1 Scale globale
- Accumulation beneath 3000m: +2%
- unmapped regions south of 65N: +1.5%
- Nordic Seas: +1%
- Arctic Ocean: +2%
dcant_scaling <- 1.02*1.015*1.01*1.029.2 eMLR only
dcant_budget_global_all_ensemble %>%
filter(data_source == "obs",
period != "1994 - 2014") %>%
ggplot(aes(tref2, dcant, group = interaction(MLR_basins, Version_ID_group),
col=Version_ID_group)) +
geom_path() +
geom_point()
dcant_budget_global_all_ensemble %>%
filter(data_source == "obs") %>%
select(MLR_basins, Version_ID_group, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
ggplot(aes(`1994 - 2004`, `2004 - 2014`,
col=Version_ID_group)) +
coord_equal() +
geom_point()
dcant_budget_global_all_ensemble %>%
filter(data_source == "obs") %>%
select(MLR_basins, Version_ID_group, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
ggplot(aes(`1994 - 2004`, `1994 - 2014`,
col=Version_ID_group)) +
coord_equal() +
geom_point()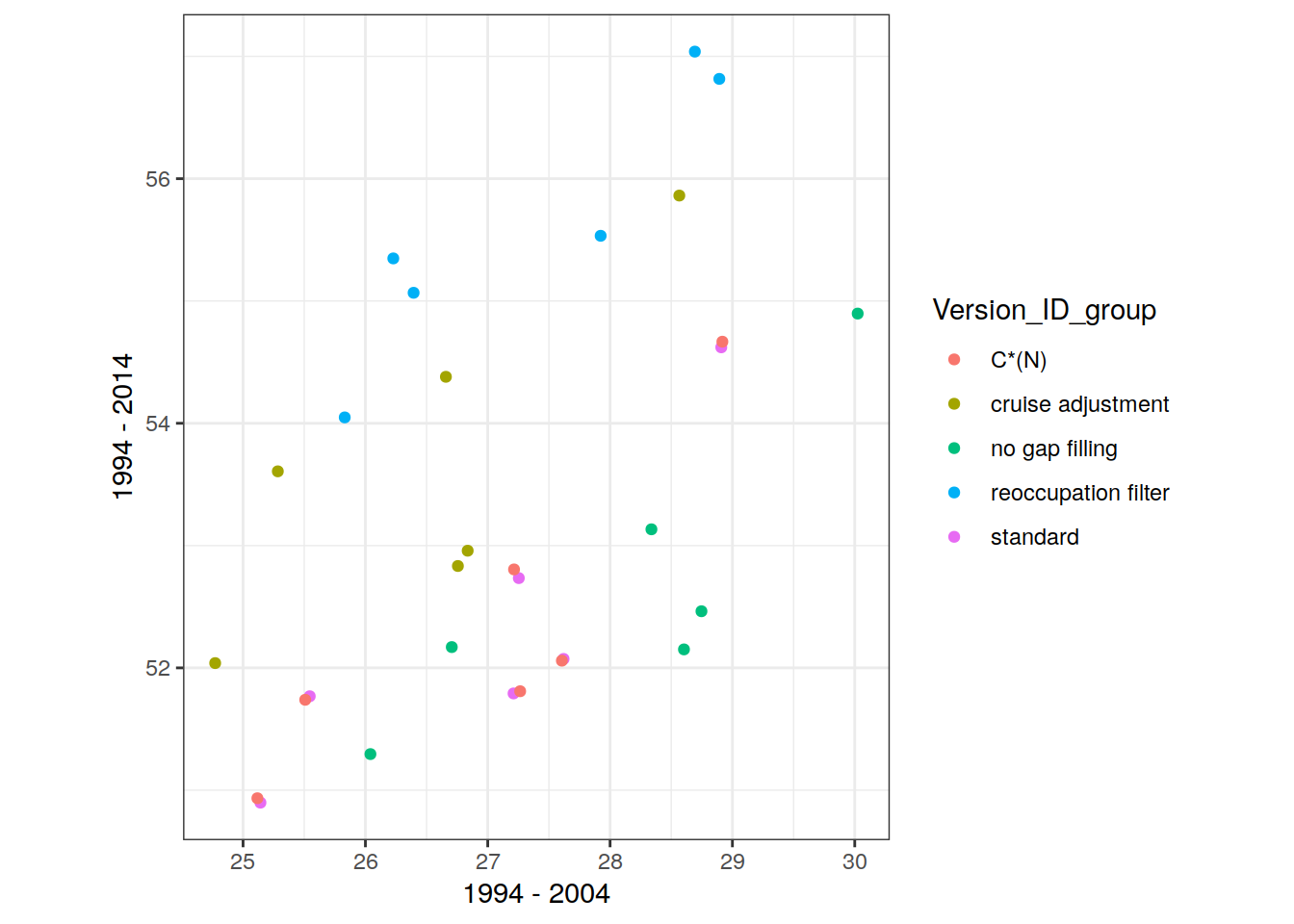
dcant_budget_global_all_ensemble %>%
filter(data_source == "obs") %>%
select(MLR_basins, Version_ID_group, period, dcant) %>%
pivot_wider(names_from = period,
values_from = dcant) %>%
ggplot(aes(`1994 - 2004` + `2004 - 2014`, `1994 - 2014`,
col=Version_ID_group)) +
geom_abline(intercept = 0, slope = 1) +
coord_equal() +
scale_x_continuous(breaks = seq(0,100,1)) +
scale_y_continuous(breaks = seq(0,100,1)) +
geom_point()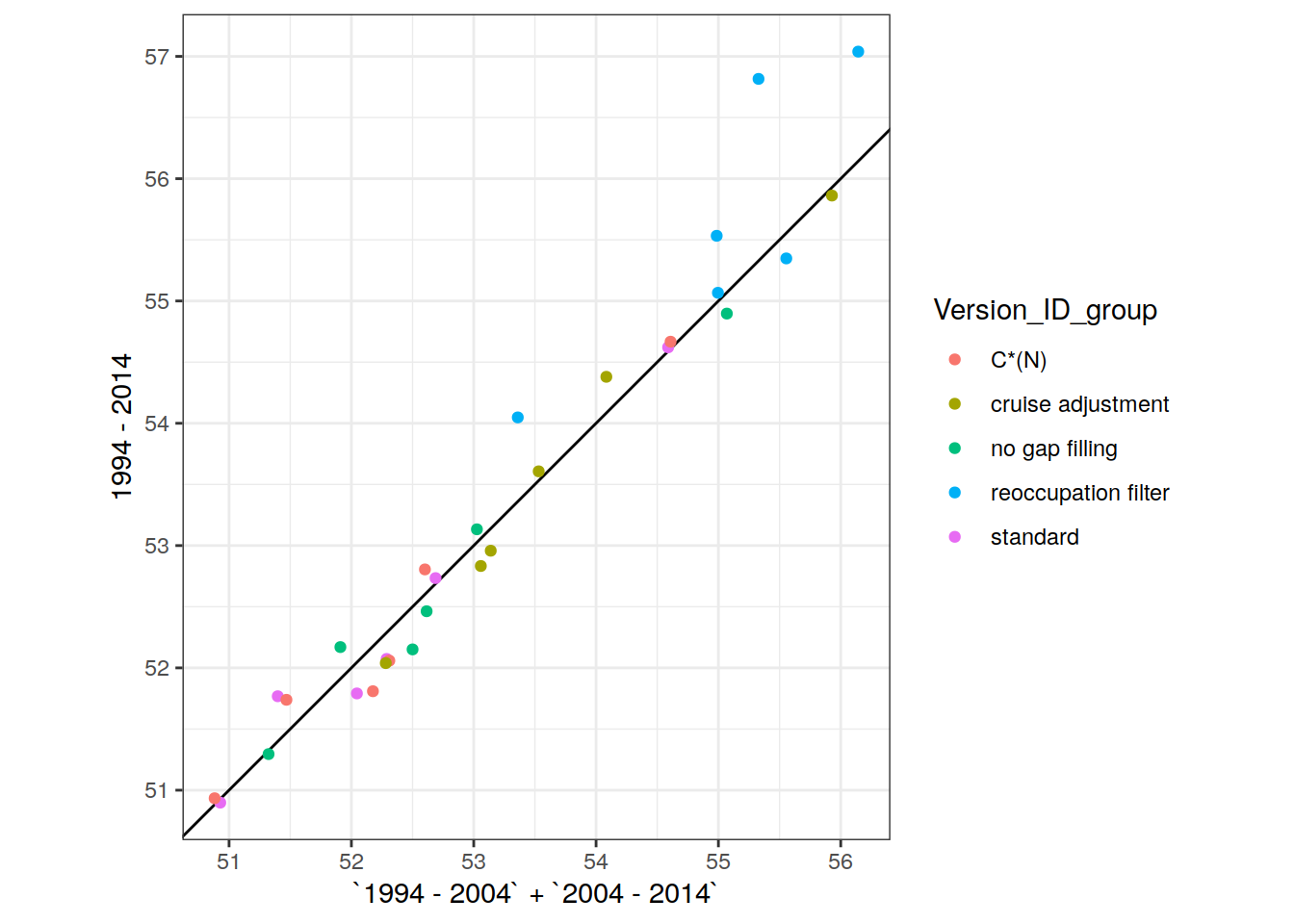
dcant_budget_global_ts <- dcant_budget_global_all %>%
filter(data_source == "obs",
period != "1994 - 2014") %>%
select(year = tref2, dcant_mean = dcant)
dcant_budget_global_ts <- dcant_budget_global_ts %>%
mutate(dcant_mean = dcant_mean * dcant_scaling)
dcant_budget_global_all_ensemble_summary <-
dcant_budget_basin_MLR_all_ensemble %>%
filter(period != "2004 - 2014") %>%
mutate(dcant = dcant * dcant_scaling) %>%
group_by(data_source, Version_ID, tref1, tref2, period) %>%
summarise(dcant = sum(dcant)) %>%
ungroup() %>%
group_by(data_source, tref1, tref2, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant)*2) %>%
ungroup() %>%
filter(data_source == "obs")
dcant_budget_global_ts <- left_join(dcant_budget_global_ts,
dcant_budget_global_all_ensemble_summary %>%
select(year = tref2, dcant_sd))
tcant_S04 <- bind_cols(year = 1994, dcant_mean = 118, dcant_sd = 19)
tcant_ts <- full_join(dcant_budget_global_ts, tcant_S04)
tcant_ts <- left_join(tcant_ts, co2_atm)
co2_atm_pi <- bind_cols(pCO2 = 280, dcant_mean = 0, year = 1800, dcant_sd = 0)
tcant_ts <- full_join(tcant_ts, co2_atm_pi)
tcant_ts <- tcant_ts %>%
arrange(year) %>%
mutate(tcant = cumsum(dcant_mean),
tcant_sd = cumsum(dcant_sd))
beta <- 1.57
co2_atm_pi_value <- 283
tCant_col <- "#56B4E9"
p_tcant_year <- tcant_ts %>%
ggplot(aes(year, tcant)) +
geom_line(data = co2_atm_reccap2 %>% filter(year > 1790),
aes(year, (pCO2 - co2_atm_pi_value) * beta),
col = tCant_col) +
geom_linerange(aes(
ymin = tcant - dcant_sd,
ymax = tcant + dcant_sd,
col = "interval\nuncertainty"
)) +
geom_point(fill = "white", shape = 21) +
geom_text(aes(label = year), nudge_x = -13, nudge_y = 6) +
scale_y_continuous(name = expression(Total ~ oceanic ~ C[ant] ~ (PgC)),
sec.axis = sec_axis( ~ (. / beta + co2_atm_pi_value),
name = expression(Atm. ~ pCO[2] ~ (µatm)))) +
scale_x_continuous(name = "Year", breaks = seq(1800, 2000, 50)) +
scale_color_manual(values = "black") +
theme(
axis.title.y.right = element_text(color = tCant_col),
axis.text.y.right = element_text(color = tCant_col),
axis.ticks.y.right = element_line(color = tCant_col),
legend.background = element_rect(fill = "transparent"),
legend.position = c(0.85, 0.2),
legend.title = element_blank()
)
p_tcant_year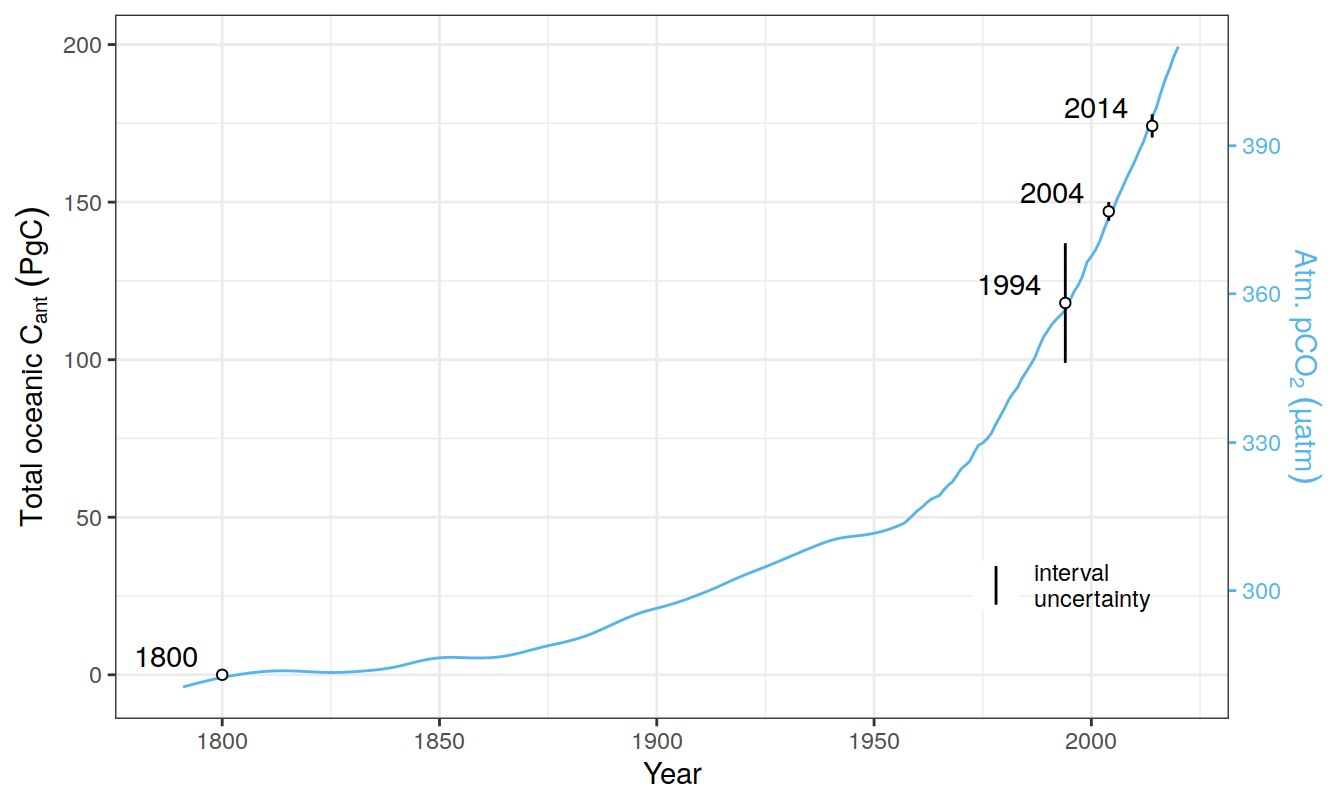
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| 5a6be34 | jens-daniel-mueller | 2022-03-22 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 251c7cf | jens-daniel-mueller | 2022-02-17 |
| 565224d | jens-daniel-mueller | 2022-02-17 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| 6fe70a1 | jens-daniel-mueller | 2022-02-05 |
| a6b33aa | jens-daniel-mueller | 2022-02-04 |
tCant_col <- "transparent"
p_tcant_year_trans <- tcant_ts %>%
ggplot(aes(year, tcant)) +
geom_line(data = co2_atm_reccap2 %>% filter(year > 1790),
aes(year, (pCO2 - co2_atm_pi_value) * beta),
col = tCant_col) +
geom_linerange(aes(
ymin = tcant - dcant_sd,
ymax = tcant + dcant_sd,
col = "interval\nuncertainty"
)) +
geom_point(fill = "white", shape = 21) +
geom_text(aes(label = year), nudge_x = -13, nudge_y = 6) +
scale_y_continuous(name = expression(Total ~ oceanic ~ C[ant] ~ (PgC)),
sec.axis = sec_axis( ~ (. / beta + co2_atm_pi_value),
name = expression(Atm. ~ pCO[2] ~ (µatm)))) +
scale_x_continuous(name = "Year", breaks = seq(1800, 2000, 50)) +
scale_color_manual(values = "black") +
theme(
axis.title.y.right = element_text(color = tCant_col),
axis.text.y.right = element_text(color = tCant_col),
axis.ticks.y.right = element_line(color = tCant_col),
legend.background = element_rect(fill = "transparent"),
legend.position = c(0.85, 0.2),
legend.title = element_blank()
)
p_tcant_year_trans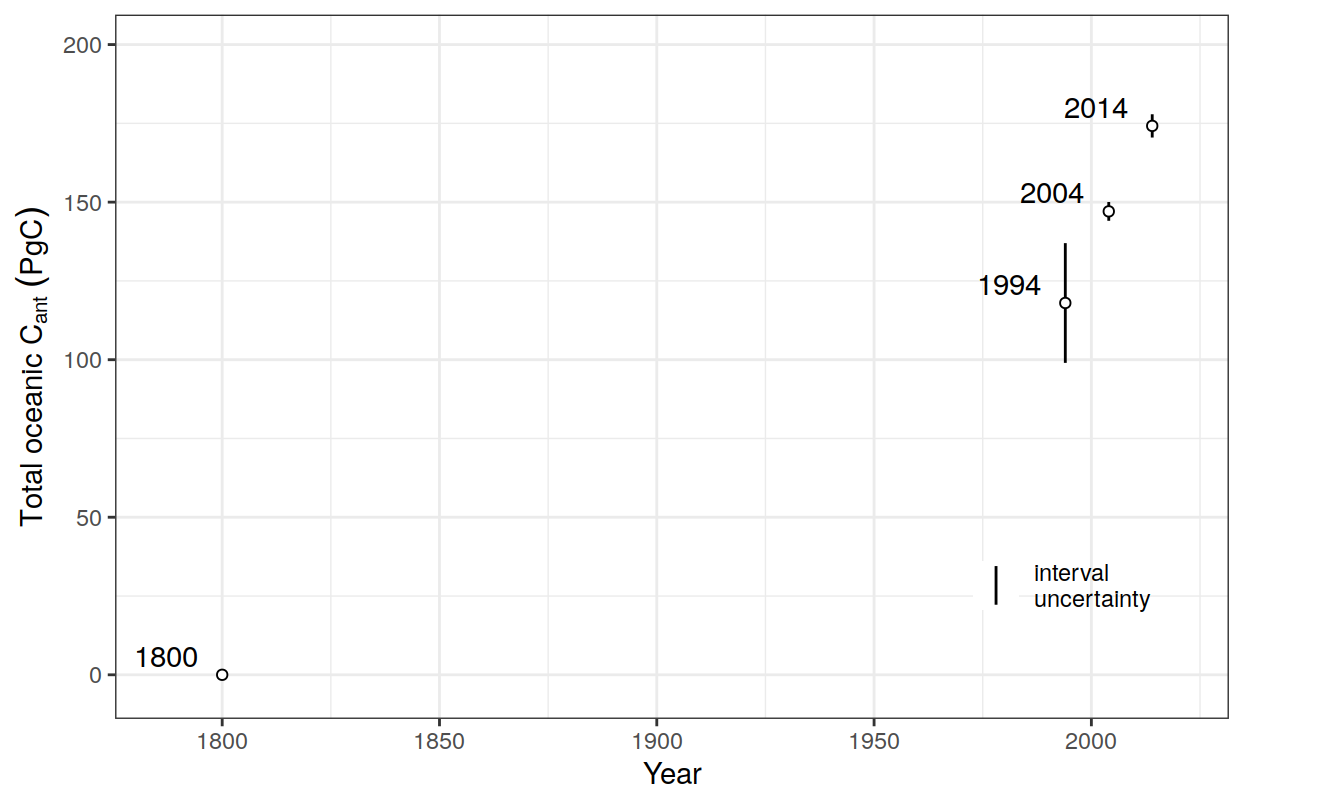
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| 5a6be34 | jens-daniel-mueller | 2022-03-22 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 251c7cf | jens-daniel-mueller | 2022-02-17 |
| 565224d | jens-daniel-mueller | 2022-02-17 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
dcant_uncert_2004 <- tcant_ts %>%
filter(year %in% c(1994, 2004)) %>%
mutate(dcant_sd = if_else(year == 1994, 0, dcant_sd),
period = "1994 - 2004")
dcant_uncert_2014 <- tcant_ts %>%
filter(year %in% c(2004, 2014)) %>%
mutate(dcant_sd = if_else(year == 2004, 0, dcant_sd),
period = "2004 - 2014")
dcant_uncert_ind <- bind_rows(
dcant_uncert_2004,
dcant_uncert_2014
)
dcant_uncert_2014_combined <-
tcant_ts %>%
filter(year %in% c(1994, 2004, 2014)) %>%
mutate(dcant_sd = if_else(year == 1994, 0, dcant_sd),
dcant_sd_combined = sqrt(dcant_sd^2 + lag(dcant_sd, default = 0)^2),
period = "1994 - 2014")
p_tcant_atm_co2 <- tcant_ts %>%
ggplot(aes(pCO2, tcant)) +
geom_ribbon(aes(ymin = tcant - tcant_sd, ymax = tcant + tcant_sd, fill = "1800 - 2014"),
alpha = 0.5) +
geom_ribbon(data = dcant_uncert_2014_combined,
aes(ymin = tcant - dcant_sd, ymax = tcant + dcant_sd, fill = "1994 - 2014"),
alpha = 0.5) +
# geom_linerange(aes(ymin = tcant - dcant_sd, ymax = tcant + dcant_sd, col = "interval\nuncertainty")) +
geom_line() +
geom_point(shape = 21, fill = "white") +
scale_x_continuous(breaks = seq(280, 400, 30)
# sec.axis = dup_axis(labels = c(1800, 1940, 1980, 2000, 2015),
# name = "Year")
) +
scale_fill_manual(values = c("grey60", "grey30"), name = "Cumulative\nuncertainty") +
scale_color_manual(values = "black", name = "") +
geom_text(aes(label = year), nudge_x = -6, nudge_y = 6) +
labs(x = expression(Atmospheric~pCO[2]~(µatm)),
y = expression(Total~oceanic~C[ant]~(PgC))) +
theme(legend.background = element_rect(fill = "transparent"),
legend.position=c(0.85, 0.2))
p_tcant_year / p_tcant_atm_co2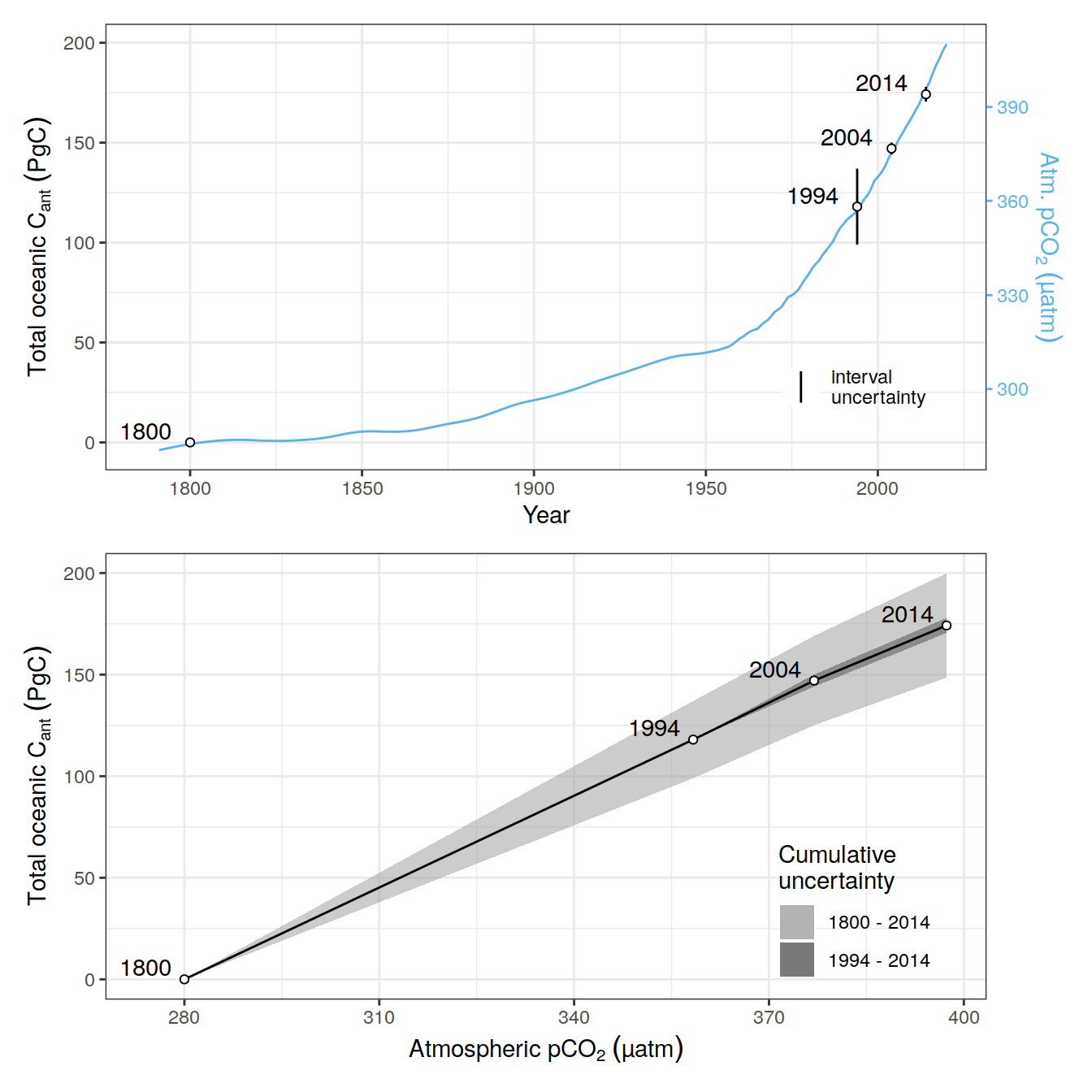
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| 5a6be34 | jens-daniel-mueller | 2022-03-22 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 251c7cf | jens-daniel-mueller | 2022-02-17 |
| 565224d | jens-daniel-mueller | 2022-02-17 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
| 6fe70a1 | jens-daniel-mueller | 2022-02-05 |
| a6b33aa | jens-daniel-mueller | 2022-02-04 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 0d0f790 | jens-daniel-mueller | 2022-02-02 |
| 60727e6 | jens-daniel-mueller | 2022-02-02 |
| c7b4984 | jens-daniel-mueller | 2022-02-02 |
| 7fb28a2 | jens-daniel-mueller | 2022-02-02 |
| 7655085 | jens-daniel-mueller | 2022-02-02 |
| ec7fe7e | jens-daniel-mueller | 2022-01-31 |
ggsave(path = here::here("output/publication"),
filename = "FigS_budget_dcant_global_vs_atm_pCO2.png",
height = 6,
width = 7)
p_tcant_year_trans / p_tcant_atm_co2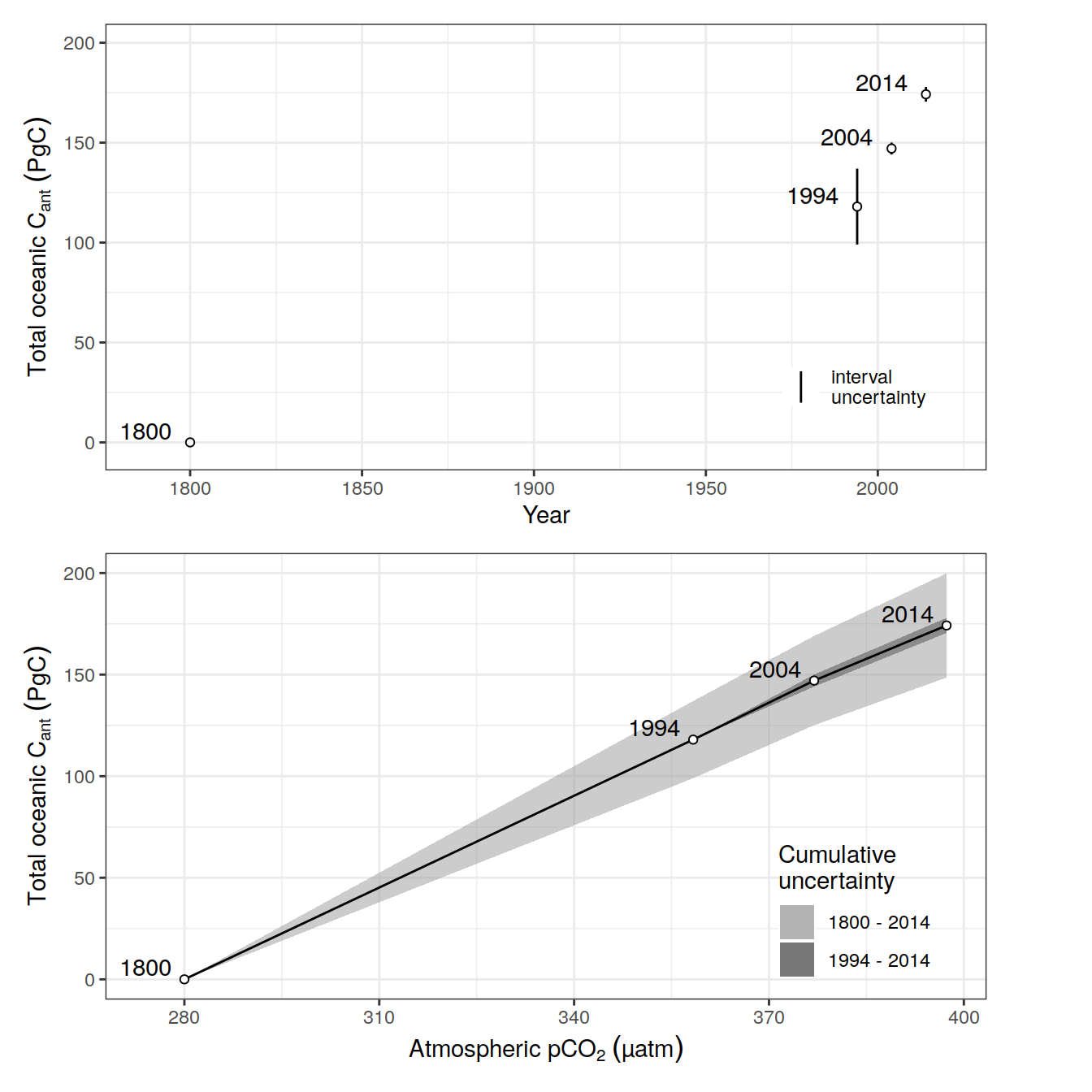
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
| 87e9eb8 | jens-daniel-mueller | 2022-06-27 |
| b52b159 | jens-daniel-mueller | 2022-06-27 |
| 1d73ec9 | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| 5a6be34 | jens-daniel-mueller | 2022-03-22 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| 251c7cf | jens-daniel-mueller | 2022-02-17 |
| 565224d | jens-daniel-mueller | 2022-02-17 |
| 2116dd3 | jens-daniel-mueller | 2022-02-09 |
# ggsave(path = here::here("output/publication"),
# filename = "Fig_global_dcant_budget_vs_atm_pCO2_trans.png",
# height = 6,
# width = 7)tcant_ts %>%
mutate(beta = dcant_mean / (pCO2-lag(pCO2)))# A tibble: 4 × 7
year dcant_mean dcant_sd pCO2 tcant tcant_sd beta
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1800 0 0 280 0 0 NA
2 1994 118 19 358. 118 19 1.51
3 2004 29.1 2.98 377. 147. 22.0 1.56
4 2014 27.1 3.69 397. 174. 25.7 1.339.3 incl fluxes
Ocean_Sink_cum_1994 <-
Ocean_Sink %>%
filter(year >= 1994, year <= 2014) %>%
mutate(fgco2_glob = if_else(year == 1994, 0, GtC_yr),
fgco2_glob = fgco2_glob) %>%
arrange(year) %>%
group_by(product, type) %>%
mutate(fgco2_glob_cum = cumsum(fgco2_glob)) %>%
ungroup()
Ocean_Sink_cum_1994 <-
left_join(Ocean_Sink_cum_1994,
co2_atm)
Ocean_Sink_cum_1994 %>%
ggplot(aes(pCO2, fgco2_glob_cum, col = product)) +
geom_line() +
geom_point(shape = 21, fill = "white") +
theme(legend.position = "bottom") +
facet_grid(.~ type)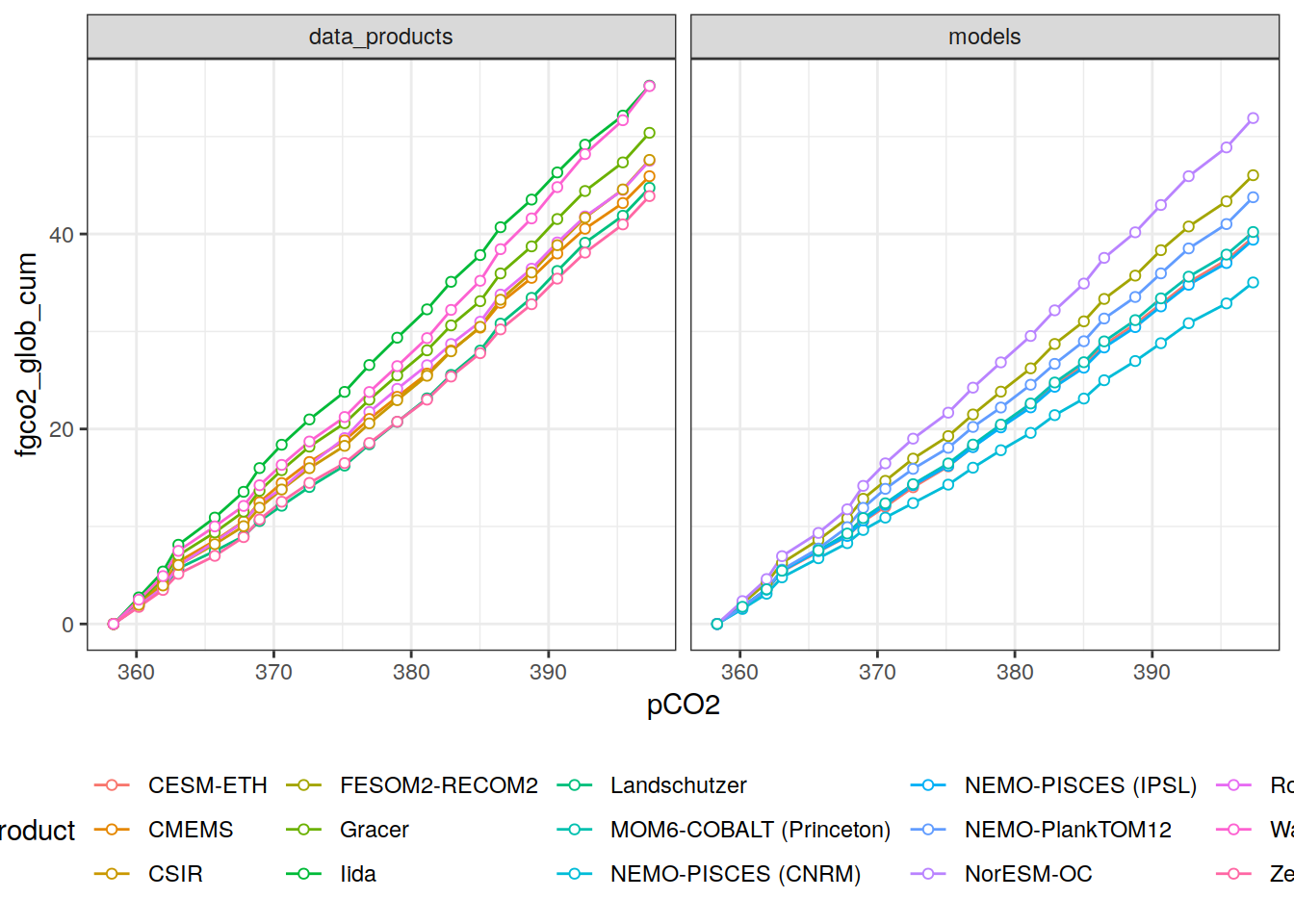
Ocean_Sink_cum_1994 <- Ocean_Sink_cum_1994 %>%
mutate(fgco2_glob_cum_total = fgco2_glob_cum + 118) %>%
select(year, pCO2, product, fgco2_glob_cum, fgco2_glob_cum_total, type)
Ocean_Sink_cum_1994 <- Ocean_Sink_cum_1994 %>%
mutate(type = case_when(
type == "data_products" ~ "Observation-based\nflux products",
TRUE ~ "Hindcast models"
))tcant_ts_zoom <- tcant_ts %>%
filter(year != 1800) %>%
mutate(dcant_sd = if_else(year == 1994,0,dcant_sd))
tcant_min = tcant_ts_zoom %>% pull(tcant) %>% min()
tcant_max =
tcant_ts_zoom %>% pull(tcant) %>% max() +
tcant_ts_zoom %>% pull(dcant_sd) %>% max()
p_tcant_ts_zoom <-
tcant_ts_zoom %>%
ggplot(aes(pCO2, tcant)) +
geom_ribbon(
data = dcant_uncert_2014_combined,
aes(
ymin = tcant - dcant_sd,
ymax = tcant + dcant_sd,
fill = "1994 - 2014"
),
alpha = 0.5
) +
geom_line(data = Ocean_Sink_cum_1994,
aes(pCO2, fgco2_glob_cum_total, group = product, col = type)) +
geom_line(aes(col = "eMLR(C*)")) +
geom_point(aes(col = "eMLR(C*)"),
shape = 21, fill = "white") +
geom_text(aes(label = year), nudge_x = -3, nudge_y = 3) +
scale_color_manual(values = c("black", "#E69F00", "#56B4E9", "pink"),
name = "Data source") +
scale_fill_manual(values = c("grey60", "grey30"), name = "Cumulative\nuncertainty") +
# scale_x_continuous(limits = c(NA, 409)) +
scale_y_continuous(limits = c(tcant_min, tcant_max)) +
labs(x = expression(Atmospheric ~ pCO[2] ~ (µatm)),
y = expression(Total ~ oceanic ~ C[ant] ~ (PgC))) +
guides(colour = guide_legend(order = 2),
fill = guide_legend(order = 1)) +
theme_classic() +
theme(
legend.background = element_rect(fill = "transparent"),
legend.position = c(0.8, 0.2),
legend.box.just = "bottom",
legend.box = "horizontal"
)
p_tcant_ts_zoom
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 9393c07 | jens-daniel-mueller | 2022-06-28 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
| fb59a6f | jens-daniel-mueller | 2022-06-27 |
| 3c1100c | jens-daniel-mueller | 2022-05-16 |
| 2ca0109 | jens-daniel-mueller | 2022-05-02 |
| b018a9a | jens-daniel-mueller | 2022-04-29 |
| e09320d | jens-daniel-mueller | 2022-04-12 |
| 8dca96a | jens-daniel-mueller | 2022-04-12 |
| 209c9b6 | jens-daniel-mueller | 2022-04-10 |
| acad2e2 | jens-daniel-mueller | 2022-04-09 |
| 3d81135 | jens-daniel-mueller | 2022-04-07 |
| a74e341 | jens-daniel-mueller | 2022-04-04 |
| bd9e11d | jens-daniel-mueller | 2022-03-22 |
| 2501978 | jens-daniel-mueller | 2022-03-21 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 094bfa0 | jens-daniel-mueller | 2022-02-18 |
| ba2d62e | jens-daniel-mueller | 2022-02-17 |
| 192504c | jens-daniel-mueller | 2022-02-17 |
| d2191ad | jens-daniel-mueller | 2022-02-04 |
| 4c5b079 | jens-daniel-mueller | 2022-02-03 |
| 4077397 | jens-daniel-mueller | 2022-02-03 |
dcant_type_box <-
bind_rows(
dcant_budget_global_all_ensemble %>%
filter(data_source == "obs",
period != "2004 - 2014") %>%
select(period, dcant) %>%
mutate(type = "eMLR(C*)",
dcant = dcant * dcant_scaling + 118),
Ocean_Sink_cum_1994 %>%
filter(year %in% c(2004, 2014)) %>%
mutate(period = if_else(year == 2004,
"1994 - 2004",
"1994 - 2014")) %>%
select(period, dcant = fgco2_glob_cum_total, type)
)
p_tcant_box <-
dcant_type_box %>%
filter(period == "1994 - 2014") %>%
ggplot(aes(period, dcant, col = type)) +
geom_boxplot(position = position_dodge(width = 1)) +
geom_jitter(position = position_dodge(width = 1),
shape = 21) +
scale_color_manual(values = c("black", "#E69F00", "#56B4E9", "pink"),
name = "Data source",
guide = "none") +
scale_y_continuous(limits = c(tcant_min, tcant_max))+
theme_classic() +
theme(
axis.line.y.left = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
legend.background = element_rect(fill = "transparent"),
legend.position = c(0.73, 0.25),
legend.box.just = "bottom",
legend.box = "horizontal",
axis.line.y =
)
p_tcant_ts_zoom + p_tcant_box +
plot_layout(ncol = 2, widths = c(7,1))
| Version | Author | Date |
|---|---|---|
| 17cd1d1 | jens-daniel-mueller | 2022-07-13 |
| 26e9496 | jens-daniel-mueller | 2022-07-12 |
| 8fb595c | jens-daniel-mueller | 2022-07-12 |
| 003b161 | jens-daniel-mueller | 2022-07-12 |
| 8ab4a87 | jens-daniel-mueller | 2022-06-29 |
| 7629c78 | jens-daniel-mueller | 2022-06-29 |
| f6786c8 | jens-daniel-mueller | 2022-06-29 |
| 9393c07 | jens-daniel-mueller | 2022-06-28 |
| 0825298 | jens-daniel-mueller | 2022-06-28 |
ggsave(path = here::here("output/publication"),
filename = "Fig_budget_dcant_global_vs_GCB_ocean_sink.png",
height = 6,
width = 9)
dcant_type_box %>%
filter(period == "1994 - 2014") %>%
mutate(dcant = dcant - 118) %>%
group_by(type) %>%
summarise(
dcant_sd = sd(dcant),
dcant_mean = mean(dcant),
dcant_median = median(dcant)
) %>%
ungroup() %>%
mutate(dcant_mean_rel = dcant_mean / first(dcant_mean))# A tibble: 3 × 5
type dcant_sd dcant_mean dcant_median dcant_mean_rel
<chr> <dbl> <dbl> <dbl> <dbl>
1 "eMLR(C*)" 1.84 56.9 56.3 1
2 "Hindcast models" 5.49 42.3 40.2 0.744
3 "Observation-based\nflux prod… 4.41 48.8 47.6 0.858dcant_type_box %>%
filter(period == "1994 - 2014",
type != "eMLR(C*)") %>%
mutate(dcant = dcant - 118) %>%
summarise(
dcant_sd = sd(dcant),
dcant_mean = mean(dcant),
dcant_median = median(dcant)
)# A tibble: 1 × 3
dcant_sd dcant_mean dcant_median
<dbl> <dbl> <dbl>
1 5.83 45.8 45.910 Emission removal
GCB_Cant_emissions <- Global_Carbon_Budget %>%
mutate(year = cut(year, c(1994, 2004, 2014), c("2004", "2014")),
year = as.numeric(as.character(year))) %>%
drop_na() %>%
filter(estimate %in% c(
"fossil emissions excluding carbonation",
"land-use change emissions"
)) %>%
group_by(year) %>%
summarise(cant_emissions = sum(GtC_yr)) %>%
ungroup()
left_join(GCB_Cant_emissions,
tcant_ts_zoom) %>%
mutate(
dcant_per_emission = 100 * dcant_mean / cant_emissions,
dcant_per_emission_sd = 100 * dcant_sd / cant_emissions
) %>%
kable() %>%
kable_styling() %>%
scroll_box(height = "300px")| year | cant_emissions | dcant_mean | dcant_sd | pCO2 | tcant | tcant_sd | dcant_per_emission | dcant_per_emission_sd |
|---|---|---|---|---|---|---|---|---|
| 2004 | 82.66365 | 29.06819 | 2.983080 | 376.95 | 147.0682 | 21.98308 | 35.16442 | 3.608696 |
| 2014 | 101.81399 | 27.12704 | 3.689235 | 397.34 | 174.1952 | 25.67231 | 26.64373 | 3.623505 |
10.0.0.1 Map test color palettes
breaks <- c(-Inf, seq(0, 16, 2), Inf)
legend_title <- expression(atop(Delta * C["ant"],
(mol ~ m ^ {
-2
})))
breaks_n <- length(breaks) - 1
dcant_inv_all_color_test <- dcant_inv_all %>%
filter(data_source %in% c("obs"),
period == "2004 - 2014") %>%
mutate(dcant_int = cut(dcant,
breaks,
right = FALSE))
scico_continous_palettes <- c(
"acton",
"bamako",
"batlow",
"bilbao",
"buda",
"davos",
"devon",
"grayC",
"hawaii",
"imola",
"lajolla",
"lapaz",
"nuuk",
"oslo",
"tokyo",
"turku"
)
p_test_scico <- function(df, i_palette) {
p_reg <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_scico_d(
palette = i_palette,
drop = FALSE,
name = legend_title,
guide = "none"
) +
labs(title = i_palette) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_rev <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_scico_d(
palette = i_palette,
drop = FALSE,
name = legend_title,
direction = -1,
guide = "none"
) +
labs(title = paste(i_palette, "rev")) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_comb <- p_reg / p_rev
return(p_comb)
}
all_plots_scico <- scico_continous_palettes %>%
# head(1) %>%
map(~p_test_scico(df = dcant_inv_all_color_test,
i_palette = .x))
viridis_continous_palettes <- c(
"viridis",
"magma",
"inferno",
"plasma"
)
p_test_viridis <- function(df, i_palette) {
p_reg <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_viridis_d(
option = i_palette,
drop = FALSE,
name = legend_title,
guide = "none"
) +
labs(title = i_palette) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_rev <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_viridis_d(
option = i_palette,
drop = FALSE,
name = legend_title,
direction = -1,
guide = "none"
) +
labs(title = paste(i_palette, "rev")) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_comb <- p_reg / p_rev
return(p_comb)
}
all_plots_viridis <- viridis_continous_palettes %>%
# head(1) %>%
map(~p_test_viridis(df = dcant_inv_all_color_test,
i_palette = .x))
library(colorspace)
colorspace_continous_palettes <- c(
# "Purple - Blue",
# "Red - Purple",
# "Blue - Yellow",
# "Heat",
# "Heat 2",
"Rocket"
# "Mako",
# "Dark",
# "Mint",
# "Mint",
# "BluGrn",
# "Teal",
# "Sunset",
# "SunsetDark",
# "ag_Sunset",
# "YlOrRd",
# "YlGnBu",
# "RdPu"
)
p_test_colorspace <- function(df, i_palette) {
i_palette <<- i_palette
p_reg <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_discrete_sequential(
palette = i_palette,
drop = FALSE,
name = legend_title,
guide = "none"
) +
labs(title = i_palette) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_rev <- map +
geom_raster(data = df,
aes(lon, lat, fill = dcant_int)) +
scale_fill_discrete_sequential(
palette = i_palette,
drop = FALSE,
name = legend_title,
rev = FALSE,
guide = "none"
) +
labs(title = paste(i_palette, "rev")) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
p_comb <- p_reg / p_rev
return(p_comb)
rm(i_palette)
}
all_plots_colorspace <- colorspace_continous_palettes %>%
# head(2) %>%
map(~p_test_colorspace(df = dcant_inv_all_color_test,
i_palette = .x))
all_plots_colorspace
all_plots <- c(all_plots_colorspace, all_plots_viridis, all_plots_scico)
pdf(here::here("output/publication/test_color_scales.pdf"),
width = 4, height = 4)
all_plots
dev.off()
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.3
Matrix products: default
BLAS: /usr/local/R-4.1.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.1.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.3.4 ggdist_3.1.1 ggalluvial_0.12.3 colorspace_2.0-2
[5] marelac_2.1.10 shape_1.4.6 ggforce_0.3.3 metR_0.11.0
[9] scico_1.3.0 patchwork_1.1.1 collapse_1.7.0 forcats_0.5.1
[13] stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[17] tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1
[21] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ellipsis_0.3.2 rprojroot_2.0.2 htmlTable_2.4.0
[4] base64enc_0.1-3 fs_1.5.2 rstudioapi_0.13
[7] farver_2.1.0 ggrepel_0.9.1 bit64_4.0.5
[10] fansi_1.0.2 lubridate_1.8.0 xml2_1.3.3
[13] splines_4.1.2 knitr_1.37 polyclip_1.10-0
[16] Formula_1.2-4 jsonlite_1.7.3 gsw_1.0-6
[19] broom_0.7.11 cluster_2.1.2 dbplyr_2.1.1
[22] png_0.1-7 compiler_4.1.2 httr_1.4.2
[25] backports_1.4.1 assertthat_0.2.1 Matrix_1.4-0
[28] fastmap_1.1.0 cli_3.1.1 later_1.3.0
[31] tweenr_1.0.2 htmltools_0.5.2 tools_4.1.2
[34] gtable_0.3.0 glue_1.6.0 Rcpp_1.0.8
[37] cellranger_1.1.0 jquerylib_0.1.4 vctrs_0.3.8
[40] svglite_2.0.0 xfun_0.29 ps_1.6.0
[43] rvest_1.0.2 lifecycle_1.0.1 oce_1.5-0
[46] getPass_0.2-2 MASS_7.3-55 scales_1.1.1
[49] vroom_1.5.7 hms_1.1.1 promises_1.2.0.1
[52] parallel_4.1.2 RColorBrewer_1.1-2 yaml_2.2.1
[55] gridExtra_2.3 sass_0.4.0 rpart_4.1-15
[58] latticeExtra_0.6-29 stringi_1.7.6 highr_0.9
[61] checkmate_2.0.0 rlang_1.0.2 pkgconfig_2.0.3
[64] systemfonts_1.0.3 distributional_0.3.0 evaluate_0.14
[67] lattice_0.20-45 SolveSAPHE_2.1.0 htmlwidgets_1.5.4
[70] labeling_0.4.2 bit_4.0.4 processx_3.5.2
[73] tidyselect_1.1.1 here_1.0.1 seacarb_3.3.0
[76] magrittr_2.0.1 R6_2.5.1 generics_0.1.1
[79] Hmisc_4.6-0 DBI_1.1.2 foreign_0.8-82
[82] pillar_1.6.4 haven_2.4.3 whisker_0.4
[85] withr_2.4.3 nnet_7.3-17 survival_3.2-13
[88] modelr_0.1.8 crayon_1.4.2 utf8_1.2.2
[91] tzdb_0.2.0 rmarkdown_2.11 jpeg_0.1-9
[94] grid_4.1.2 readxl_1.3.1 isoband_0.2.5
[97] data.table_1.14.2 callr_3.7.0 git2r_0.29.0
[100] reprex_2.0.1 digest_0.6.29 webshot_0.5.2
[103] httpuv_1.6.5 munsell_0.5.0 viridisLite_0.4.0
[106] bslib_0.3.1