Home
Last updated: 2020-08-28
Checks: 1 1
Knit directory: jesslyn_ovca/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cd835db. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: GO_results/.DS_Store
Ignored: GO_results/GO_PCA/.DS_Store
Ignored: GO_results/GO_Vln/.DS_Store
Ignored: GO_results/GO_Vln/PDX/.DS_Store
Ignored: GO_results/GO_plots/.DS_Store
Ignored: GO_results/GO_plots/PDX/.DS_Store
Ignored: GO_results/GO_plots/PDX/F/
Ignored: GO_results/GO_plots/SS2/.DS_Store
Ignored: GO_results/Tables/.DS_Store
Ignored: GO_results/Tables/PDX/.DS_Store
Ignored: GO_results/Tables/PDX/F/
Ignored: GO_results/Tables/SS2/.DS_Store
Ignored: analysis/.DS_Store
Ignored: code/.DS_Store
Ignored: data/.DS_Store
Ignored: data/HTAPP/
Ignored: data/Izar_2020/
Ignored: data/gene_lists/.DS_Store
Ignored: data/gene_lists/GO_PDX/.DS_Store
Ignored: data/gene_lists/GO_SS2/.DS_Store
Ignored: data/gene_lists/extra/.DS_Store
Ignored: jesslyn_plots/
Ignored: mike_plots/
Ignored: old/.DS_Store
Ignored: old/edited/.DS_Store
Ignored: renv/.DS_Store
Ignored: renv/library/
Ignored: renv/python/
Ignored: renv/staging/
Ignored: vignettes/
Unstaged changes:
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/index.Rmd) and HTML (docs/index.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | b7634b5 | jgoh2 | 2020-08-23 | Build site. |
| html | 2c1accf | jgoh2 | 2020-08-19 | Build site. |
| Rmd | 729ebf4 | jgoh2 | 2020-08-19 | workflowr::wflow_publish(files = files) |
| html | 92568b8 | jgoh2 | 2020-08-17 | Build site. |
| Rmd | d60aeff | jgoh2 | 2020-08-17 | More DE Analysis of other hallmarks |
| html | 80345fe | jgoh2 | 2020-08-15 | Build site. |
| html | 789982d | jgoh2 | 2020-08-14 | Build site. |
| html | e05c328 | jgoh2 | 2020-08-12 | Build site. |
| html | 84edf85 | jgoh2 | 2020-08-12 | Build site. |
| html | a3ddf54 | jgoh2 | 2020-08-07 | Build site. |
| html | d801a7a | jgoh2 | 2020-08-04 | Build site. |
| html | 26f64a3 | jgoh2 | 2020-08-03 | Build site. |
| html | 2dc1bee | jgoh2 | 2020-07-31 | Build site. |
| Rmd | 284aad4 | jgoh2 | 2020-07-31 | workflowr::wflow_publish(files = files) |
| html | 5c5305b | jgoh2 | 2020-07-30 | Build site. |
| Rmd | c8bb9fc | jgoh2 | 2020-07-30 | PDX Exploratory + DE + Cell Cycle Analyses |
| html | 6be6c85 | jgoh2 | 2020-07-28 | Build site. |
| Rmd | cdd10f9 | jgoh2 | 2020-07-28 | SS2 DE Analysis |
| html | cdd10f9 | jgoh2 | 2020-07-28 | SS2 DE Analysis |
| html | 35c7947 | jgoh2 | 2020-07-27 | SS2 Analysis Part 1 and 2 |
| html | f1acd7b | jgoh2 | 2020-07-24 | Move PDX_choices.Rmd to old |
| Rmd | 2b6116d | Mike Cuoco | 2020-07-24 | added 03.1_Izar2020_PDX_DEAnalysis |
| html | 0058815 | Mike Cuoco | 2020-07-15 | Build site. |
| html | a526dc6 | Mike Cuoco | 2020-07-15 | add workflowr website |
| Rmd | 086db99 | Mike Cuoco | 2020-07-15 | add project description |
| Rmd | e407c2d | Mike Cuoco | 2020-07-14 | Start workflowr project. |
CHARACTERIZATION OF METABOLIC DYNAMICS OF OVARIAN PERSISTER CELLS FOLLOWING CARBOPLATIN TREATMENT
The purpose of this project is to identify gene expression signatures from High-grade Serous Ovarian Cancer single-cell RNA-seq data that are associated with a drug tolerant state that is characteristic of cancer persister cells.
OVARIAN CANCER
Ovarian cancer (OVCA) is one of the deadliest cancers in women (Matulonis et al., 2016). OVCA is a disease with multiple origins and subtypes that share a common anatomical site upon presentation. 90% of the cases are of epithelial origin, the majority of which are High-grade serous tumors, which are known to have the worst prognosis. Despite not being listed as one of the top 10 leading sites of (new) cancer cases, OVCA is among the top 5 leading sites of estimated cancer deaths, with an estimate of 13,940 new deaths in 2020, taking up 5% of all cancer deaths according to the American Cancer Society. Unfortunately, HGSOC is typically detected at late stage due to the lack of symptoms, and the low specificity and sensitivity of the common biomarker CA 125, and in combination with the lack of druggable target molecules, makes the prognosis of HGSOC poor.
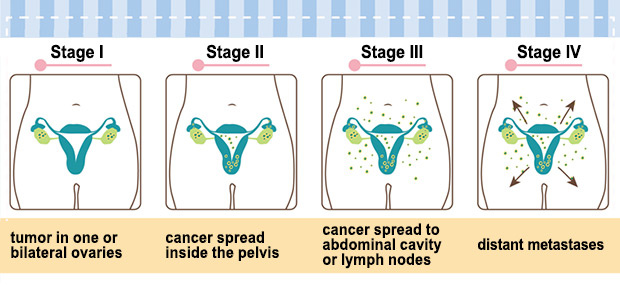
Ovarian Cancer Stages
TREATMENT Late stage OVCA detection is frequently associated with metastasis, and along with the lack of druggable target molecules in ovarian cancer, treatment of ovarian cancer is fairly difficult. Current treatment for ovarian cancer often includes surgery and chemotherapy. The goal of the surgery is to stage the cancer and to debulk the tumor (remove as much as possible). The treatment for ovarian cancer is different at different stages.
- Stage 1: limited to one or both ovaries, and can be found on the surface of the ovary.
- Stage 2: tumor has invaded one or both ovaries and extended to the pelvic region but not to the abdomen
- Stage 3: tumor has extended to the abdominal organs
- Stage 4: cancer has spread to the lungs, liver, or lymph nodes in the neck
The lack of druggable target molecules makes cytotoxic platinum-based chemotherapy, such as cisplatin or carboplatin, the standard chemotherapy drugs for ovarian cancer.
RESISTANCE Despite improved clinical response and survival in recent years, numerous patients succumb to the disease as a result of chemotherapy resistance. Although 70% of ovarian cancer patients initially respond to platinum-based chemotherapy, relapse occurs in 25% of patients with early stage ovarian cancer, and more than 80% in patients with advanced disease. The majority of these patients experience relapse within 2 years of initial treatment.
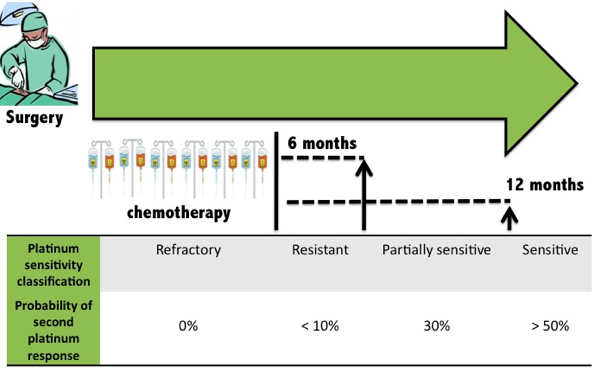
Tapia and Dias-Padilla 2013
Being able to understand the molecular mechanisms of chemotherapy resistance would therefore have important clinical relevance as researchers design new therapeutic strategies to overcome resistance and prevent cancer relapse.
NONGENETIC DRUG RESISTANCE
Drug resistance is associated with the heterogeneity within the tumor cell population, where a subpopulation of tumor cells acquires the ability to survive treatment. These cells, termed minimal residual disease cells (MRD) or drug-tolerant persister cells (DTPs) re-proliferate once the environment becomes appropriate to do so, leading to cancer relapse. Earlier studies focused on genetic mechanisms of drug resistance, where pro-survival genetic mutations can either be present in cells of the initial tumor cell population or be acquired during therapy. These mutations allow cancer cells to survive treatment while also passing down this capability permanently to their progeny.
However, non-genetic mechanisms have also been found to play an important role in drug resistance and cancer relapse, one of which was found in the PC9 cell line (epidermal growth factor receptor EFGR mutant non-small-cell lung carcinoma NSCLC), where a subpopulation of cells acquires a slow-cycling, transiently drug-tolerant cell state following treatment (EGFR tyrosine kinase inhibitor) (Sharma et al. 2010). Interestingly, the progeny of these drug-tolerant persisters (drug-tolerant expanded persisters) reacquire cancer drug sensitivity following 30+ passages in drug-free medium, which demonstrates a nongenetic mechanism of drug resistance in the original drug-tolerant persisters.
Although the actual mechanisms of non-genetic drug resistance remain to be fully understood, several studies suggest that chemotherapy resistance may be linked to cancer stem-like cell characteristics such as cancer quiescence or metabolic reprogramming.
CANCER STEM CELLS AND DRUG RESISTANT PERSISTER CELLS Chemotherapy resistant cells are found to have overlapping biological features with those of cancer stem cells, which includes plasticity, tumor-initiating capacity, and slow proliferation. Cancer stem cells are slow-proliferating cells at the top of the cancer hierarchy, and are involved in tumor-initiation as they give rise to intermediate progenitors and to differentiated progeny. However, the boundaries between stem and non-stem cells have been found to be relatively weak in cancer, and studies report phenomena of therapy-induced enrichment of cancer stem-like cells, where differentiated cells are found to transition to a stem-like cell state that confer resistance.For example, Sharma et al observed that the putative CSC marker CD133 is expressed in all DTPs (but only in approximately 2% of the parental PC9 population), while another CSC marker CD24 is also enriched in some settings.
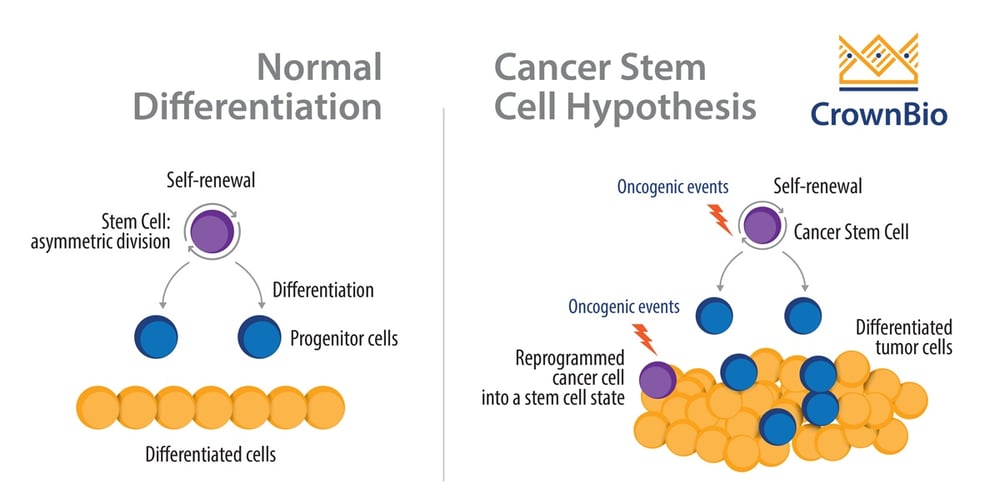
Cancer Stem Cells vs. Normal Stem Cells
So how does cancer stem-like characteristics confer drug resistance?
CANCER DORMANCY AND DRUG RESISTANCE One of the many ways that a stem-like cell state may confer resistance is quiescent. Drug resistance is commonly characterized by a suppression of proliferation.Cancer cells can escape from the mitotic cell cycle and stay arrested at the non or low cycling G0-G1 phase to wait for an appropriate environment. Cancer cells can therefore make use of dormancy to overcome hostile microenvironments and re-enter the cell cycle to proliferate once unfavourable conditions are ruled out.
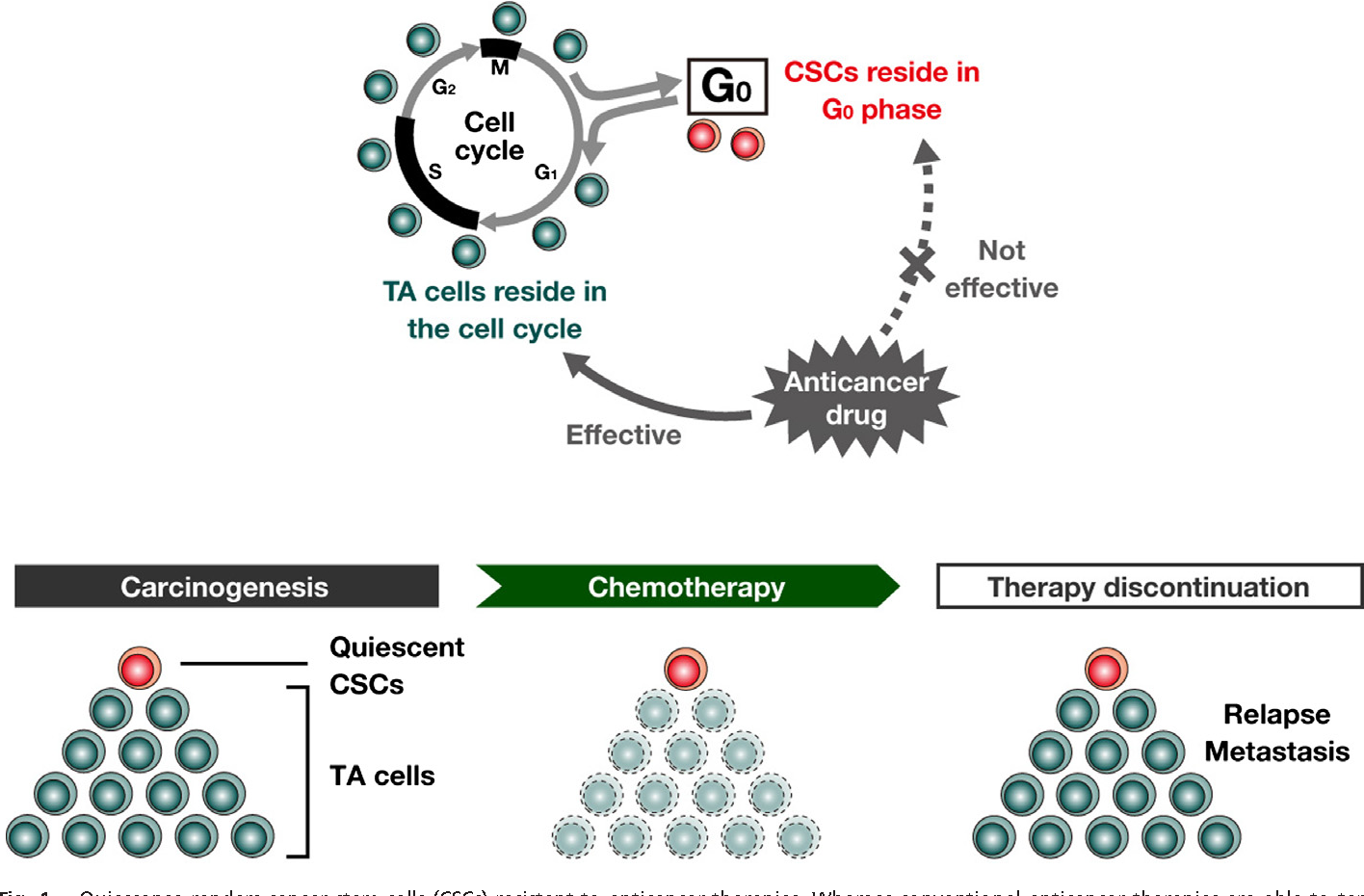
Takeishi and Nakayama, 2016
METABOLIC REPROGRAMMING AND DRUG RESISTANCE Another hypothesized way that a stem-like cell state may confer resistance is plasticity, specifically metabolic plasticity. Several studies report that drug resistant persister cells are transcriptionally and/or metabolically different from their drug-sensitive proliferating counterparts.
A recent study reports a metabolic shift from aerobic glycolysis to oxidative phosphorylation (OXPHOS) in cisplatin-resistant (SKOV3/DPP) ovarian cancer cells (Xu et al., 2018). This is a crucial finding because cancer cells are known to favor glycolysis over OXPHOS even under aerobic conditions (the Warburg effect).

Thomas and Yu, 2017
Other studies report a correlation between the activation of the unfolded protein response (UPR) and tumor progression (Madden et al., 2019; Sisinni et al., 2019). UPR is an ER stress response mechanism that can help to restore ER homeostasis or induce apoptosis under overloaded ER stress. ER stress can be induced by adverse conditions such as the accumulation of ROS, nutrient deprivation, or mutations that lead to misfolded proteins in the ER, and UPR helps relieve this stress by fixing misfolded and unfolded proteins through three main pathways.
- PERK (Protein Kinase R-like Endoplasmic Reticulum Kinase):
- Global inhibition of protein translation by phosphorylating and inactivating the eukaryotic translation initiation factor 2 (eIF2)
- IRE1a (Inositol-requiring enzyme 1)
- Dimeric kinase and endoribonuclease splices the mRNA of the X-box binding protein-1 (XBP1), and the changed ORF is translated into XBP1s, which serves as a transcription factor for genes involved in folding, glycosylation, and ER associated degradation.
- ATF6 (Activating Transcription Factor 6)
- The transmembrane ATF6 protein gets transported to the Golgi, where proteases cleaves of off the transmembrane domain and allows cATF6 to enter into the nucleus and act as a transcription factor of UPR target genes

So 2018
In addition to being a survival mechanism in normal cells, the dysregulation of ER stress response is reported to be one of the hallmarks of cancer, contributing to tumor progression and survival. For instance, studies report that UPR can ensure the survival of cancer cells and induce drug resistance through dormancy, angiogenesis, and the upregulation of antioxidant responses.
OUR PURPOSE
Despite these findings, the actual mechanisms behind drug resistance remain to be elucidated, and many of the hypotheses are contradicted by other evidence as well.
- For instance, although the expression of the putative CSC marker CD133 in all DTPs from the Sharma et al paper suggests a link between stemness and resistance, they also observed that CSC-specific markers are lost during the transition of DTPs to DTEPs despite the ability of both cell populations to be equally drug insensitive.
- Another study contradicts the hypothesis on the link between OXPHOS metabolism and drug resistance, where very different metabolic patterns are found in drug resistant cells (Morandi & Indraccolo, 2017). In fact, a recent study in 2019 showed that cancer cells are able to shift between OXPHOS and glycolysis in response to the inhibition of either process (Elgendy et al. 2019).
Together, these findings reveal the complex mechanisms that confer drug resistance in cancer cells, which are in all likelihood different in different cancer types, tumor microenvironment, and the drug used. The eradication of DTPs is the first step to overcoming resistance and preventing cancer relapse. It is therefore important for us to identify and characterize the underlying biological mechanisms that make them different from their drug sensitive counterparts and allow them to survive and proliferate. We aim to confirm the results found in previous papers and identify epigenetic differences in specific cell cycle and metabolic gene-sets by analyzing the gene expression profile of high grade serous ovarian cancer cells across different treatment time-points through Gene ontology (GO) and Gene-set enrichment analysis (GSEA) as well as differential expression (DE) analysis of single genes. We are interested in discovering whether the links between cancer stem cell-like quiescent, metabolic plasticity specifically in OXPHOS and UPR expression, and drug resistance reported in other studies are also found in our ovarian cancer data.
OUR DATA
Our single-cell Ovarian cancer data is kindly provided by Izar and Tirosh et al., from their paper A single-cell landscape of high-grade serous ovarian cancer.
For their study, they collected scRNA-seq data from three HGSOC cohorts, which includes a set of 22 ascites samples from 11 patients and three patient ascites-derived xenograft models. * 8 ascites samples out of the 22 samples were sequenced using droplet-based 10X scRNAseq, while the remaining 14 ascites samples were sequenced using plate-based single cell RNAseq (SS2). * Considering that only 1 patient (Patient 5) in the 10X cohort had samples from more than 1 treatment condition, and that 99% of the malignant cells from that patient is from the on-treatment condition, with only 8 cells from the treatment-naive condition(8 TN vs. 4885 1CCT cells), we focus our analysis on the SS2 and PDX cohorts, which have more equally distributed cells across all treatment statuses.
OUR FOCUS * The SS2 data from Izar2020 consists of cells from 14 ascites samples from 6 individuals: - Selected for malignant EPCAM+CD24+ cells by fluorophore-activated cell sorting into 96-well plates - We are only interested in the malignant ascites population, specifically samples from patient 8 and 9 because they are the only patients that have samples from different treatment statuses. * The PDX data from Izar2020 consists of only Malignant cells from three HGSOC PDX models derived from patients with different treatment histories were selected for implantation: - DF20 (BRCA WT treatment-naive, clinically platinum sensitive) - DF101 (BRCA1 mutant, 2 lines of prior therapy, clinically platinum resistant) - DF68 (BRCA1 mutant, 6 lines of prior therapy, clinically platinum resistant) * After tumors were established, animals were divided into two groups per model: - Vehicle (treated with DMSO) - Carboplatin (treated with IP carboplatin) * Carboplatin-treated mice for minimal residual disease (MRD) group were harvested for scRNA-seq * The remaining carboplatin-treated mice were harvested at endpoint (vehicle)
OUR ANALYSIS AND RESULTS
- A summary of the data from Izar 2020 et al.
- PDX Data Analysis
- SS2 Data Analysis
Decision Making Process
Credits
Developed by Jesslyn Goh and Mike Cuoco