KEGG
Ha Tran
22/08/2021
Last updated: 2022-11-05
Checks: 7 0
Knit directory: SRB_2022/1_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the
code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cc4bf5d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: .gitignore
Untracked: 0_data/
Untracked: 1_analysis/
Untracked: 3_output/
Untracked: LICENSE.md
Untracked: README.md
Untracked: SRB_2022.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Data Setup
# working with data
library(dplyr)
library(magrittr)
library(readr)
library(tibble)
library(reshape2)
library(tidyverse)
library(KEGGREST)
# Visualisation:
library(kableExtra)
library(ggplot2)
library(grid)
library(pander)
library(viridis)
library(cowplot)
library(pheatmap)
# Custom ggplot
library(ggplotify)
library(ggpubr)
library(ggbiplot)
library(ggrepel)
# Bioconductor packages:
library(edgeR)
library(limma)
library(Glimma)
library(clusterProfiler)
library(org.Mm.eg.db)
library(enrichplot)
library(pathview)
theme_set(theme_minimal())
pub <- readRDS(here::here("0_data/RDS_objects/pub.rds"))Import DGElist Data
DGElist object containing the raw feature count, sample metadata, and gene metadata, created in the Set Up stage.
# load DGElist previously created in the set up
dge <- readRDS(here::here("0_data/RDS_objects/dge.rds"))
fc <- readRDS(here::here("0_data/RDS_objects/fc.rds"))
lfc <- readRDS(here::here("0_data/RDS_objects/lfc.rds"))
lmTreat <- readRDS(here::here("0_data/RDS_objects/lmTreat.rds"))
lmTreat_sig <- readRDS(here::here("0_data/RDS_objects/lmTreat_sig.rds"))KEGG Analysis
KEGG enrichment analysis is performed with the significant DE genes that have absolute FC > 1.5 ( genes from Limma). Top 30 most significant KEGG are displayed. All enriched KEGG pathways are exported
# chosing the pathways of interest
kegg_id <- c("mmu04670", "mmu04640", "mmu04270", "mmu04151", "mmu04510", "mmu04060")
kegg_pathway <- KEGGREST::keggGet(kegg_id)enrichKEGG <- list()
enrichKEGG_all <- list()
enrichKEGG_sig <- list()
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
# find enriched KEGG pathways
enrichKEGG[[x]] <- clusterProfiler::enrichKEGG(
gene = lmTreat_sig[[x]]$entrezid,
keyType = "kegg",
organism = "mmu",
pvalueCutoff = 0.05,
pAdjustMethod = "none"
)
# save to csv
enrichKEGG_all[[x]] <- enrichKEGG[[x]]@result
# filter the significant and print top 30
enrichKEGG_sig[[x]] <- enrichKEGG_all[[x]] %>%
dplyr::filter(pvalue <= 0.05) %>%
separate(col = BgRatio, sep = "/", into = c("Total", "Universe")) %>%
dplyr::mutate(
logPval = -log(pvalue, 10),
GeneRatio = Count / as.numeric(Total)
) %>%
dplyr::select(c("Count", "GeneRatio", "pvalue", "logPval", "p.adjust", "qvalue", "Description", "geneID"))
# at the beginnning of a word (after 35 characters), add a newline. shorten the y axis for dot plot
enrichKEGG_sig[[x]]$Description <- sub(pattern = "(.{1,35})(?:$| )",
replacement = "\\1\n",
x = enrichKEGG_sig[[x]]$Description)
# remove the additional newline at the end of the string
enrichKEGG_sig[[x]]$Description <- sub(pattern = "\n$",
replacement = "",
x = enrichKEGG_sig[[x]]$Description)
}FC=1.05
p=1Top 20
enrichKEGG_sig[[p]][1:20, ] %>%
kable(caption = "Top 20 enriched KEGG pathways") %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Count | GeneRatio | pvalue | logPval | p.adjust | qvalue | Description | geneID | |
|---|---|---|---|---|---|---|---|---|
| mmu04713 | 24 | 0.2448980 | 0.00e+00 | 7.918458 | 0.00e+00 | 0.0000026 | Circadian entrainment | 16519/14802/12288/20190/14677/234889/14812/104111/14710/18795/54195/60596/104110/11513/66066/14702/11517/73086/18796/20192/58226/14697/14708/14696 |
| mmu04514 | 34 | 0.1868132 | 0.00e+00 | 7.668357 | 0.00e+00 | 0.0000026 | Cell adhesion molecules | 16401/241568/19266/192167/12558/16421/272381/15894/83964/54420/386750/19274/12562/225825/16408/69524/14961/245537/269116/15002/20339/18613/14969/18007/12741/14960/80883/241226/20737/12737/12504/13003/216856/67374 |
| mmu04724 | 24 | 0.2123894 | 2.00e-07 | 6.649484 | 2.00e-07 | 0.0000184 | Glutamatergic synapse | 20512/16519/14802/12288/14805/14677/14812/104111/210274/14710/18795/104110/11513/66066/14806/14702/18796/105727/13385/14697/14708/14696/12286/18806 |
| mmu04371 | 26 | 0.1897810 | 7.00e-07 | 6.137567 | 7.00e-07 | 0.0000449 | Apelin signaling pathway | 20190/20541/18127/14677/17260/104111/14710/104709/18795/110893/104110/11513/66066/23796/11607/14702/17897/18796/20192/23797/14219/14697/14708/30878/14696/16890 |
| mmu05414 | 20 | 0.2127660 | 2.20e-06 | 5.653207 | 2.20e-06 | 0.0001095 | Dilated cardiomyopathy | 12288/24052/16401/16000/20541/16421/12293/104111/109700/110893/104110/11513/16773/16404/17897/81904/24051/241226/192897/12298 |
| mmu04340 | 15 | 0.2586207 | 3.20e-06 | 5.489003 | 3.20e-06 | 0.0001190 | Hedgehog signaling pathway | 14632/16147/15245/68525/19207/59056/16576/12444/14634/14725/240888/109689/216869/19206/74239 |
| mmu04015 | 33 | 0.1542056 | 3.40e-06 | 5.470690 | 3.40e-06 | 0.0001190 | Rap1 signaling pathway | 15234/11600/16542/16000/14026/240168/14677/18645/78134/14812/18760/104111/21687/18795/19395/16590/104110/11513/16408/22341/50791/93737/17311/106952/70549/18796/23797/54519/16797/18596/16822/14062/20469 |
| mmu05032 | 19 | 0.2087912 | 5.30e-06 | 5.272311 | 5.30e-06 | 0.0001645 | Morphine addiction | 16519/14677/18574/104111/14710/104110/11513/14397/66066/18573/14702/109689/216869/14697/14708/14696/54393/18584/12286 |
| mmu04022 | 28 | 0.1618497 | 7.30e-06 | 5.134646 | 7.30e-06 | 0.0001863 | cGMP-PKG signaling pathway | 12288/11549/20541/18127/14677/13618/17260/234889/104111/104709/18795/110893/22068/54195/60596/104110/11513/11607/16531/13617/12062/73181/18796/242202/23797/381290/98660/16367 |
| mmu05412 | 17 | 0.2207792 | 7.60e-06 | 5.121322 | 7.60e-06 | 0.0001863 | Arrhythmogenic right ventricular cardiomyopathy | 12288/16842/24052/16401/20541/12558/16421/12293/109700/110893/16773/16404/81904/24051/241226/192897/12298 |
| mmu04512 | 18 | 0.2045455 | 1.27e-05 | 4.895147 | 1.27e-05 | 0.0002851 | ECM-receptor interaction | 16401/94216/21827/114249/12505/23928/16775/16421/14268/109700/12840/329872/16773/16404/75209/241226/192897/12830 |
| mmu04020 | 34 | 0.1416667 | 1.59e-05 | 4.798193 | 1.59e-05 | 0.0003267 | Calcium signaling pathway | 15234/12288/16542/20190/11549/20541/18127/231602/13618/18574/104111/12290/19220/18795/110893/104110/11513/22341/12494/11607/18573/13617/12062/15558/18796/65973/20192/58226/19218/381290/18596/12286/14062/52163 |
| mmu04924 | 16 | 0.2105263 | 2.63e-05 | 4.579335 | 2.63e-05 | 0.0004991 | Renin secretion | 12288/14677/234889/18574/13616/18795/54195/60596/11607/18573/16531/13617/11517/18796/229933/19217 |
| mmu04926 | 22 | 0.1705426 | 3.04e-05 | 4.516919 | 3.04e-05 | 0.0005351 | Relaxin signaling pathway | 94216/18127/14677/13618/17390/20418/104111/14710/18795/104110/11513/66066/22341/14702/109689/18796/216869/23797/14697/14708/14696/12830 |
| mmu04725 | 20 | 0.1785714 | 3.51e-05 | 4.454493 | 3.51e-05 | 0.0005766 | Cholinergic synapse | 16519/12288/14677/226922/104111/14710/104709/18795/104110/11513/66066/14702/18796/14360/23797/14697/14708/14696/60613/12286 |
| mmu04510 | 29 | 0.1442786 | 4.75e-05 | 4.323411 | 4.75e-05 | 0.0006931 | Focal adhesion | 15234/16401/16542/16000/94216/21827/23928/16775/12444/16421/14268/20418/18719/109700/12840/16773/22341/16404/64099/170736/68794/70549/14360/23797/18596/12389/241226/192897/12830 |
| mmu04360 | 27 | 0.1491713 | 4.78e-05 | 4.320237 | 4.78e-05 | 0.0006931 | Axon guidance | 13842/13643/20356/241568/13176/235611/20357/18186/20361/14677/20315/22068/238057/13837/218877/93737/73181/13841/18007/19206/14360/22253/53972/20358/80883/12162/20564 |
| mmu04014 | 32 | 0.1361702 | 6.22e-05 | 4.206315 | 6.22e-05 | 0.0008075 | Ras signaling pathway | 15234/11600/26565/16542/16000/23871/240168/20418/19414/14812/21687/14710/19395/14255/16590/66066/22341/16002/14702/17311/85031/23797/14697/14708/23872/240057/22637/14696/16797/18596/18806/320484 |
| mmu04728 | 22 | 0.1629630 | 6.23e-05 | 4.205596 | 6.23e-05 | 0.0008075 | Dopaminergic synapse | 16519/14802/12288/109731/13491/14677/20265/14812/17161/14710/18795/66066/14702/109689/18796/216869/23797/14697/14708/14696/12286/16572 |
| mmu04974 | 19 | 0.1759259 | 6.78e-05 | 4.169019 | 6.78e-05 | 0.0008345 | Protein digestion and absorption | 12814/140709/12818/94216/20541/58803/107581/234967/110893/12840/12817/17380/12832/12819/98660/20540/237759/12822/12830 |
Dot Plot
kegg_dot <- list()
upset=list()
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
# dot plot, save
kegg_dot[[x]] <- ggplot(enrichKEGG_sig[[x]][1:15, ]) +
geom_point(aes(x = GeneRatio, y = reorder(Description, GeneRatio), colour = logPval, size = Count)) +
scale_color_gradient(low = "dodgerblue3", high = "firebrick3", limits = c(0, NA)) +
scale_size(range = c(.3,3)) +
ggtitle("KEGG Pathways") +
ylab(label = "") +
xlab(label = "Gene Ratio") +
labs(color = expression("-log"[10] * "Pvalue"), size = "Gene Counts")
ggsave(filename = paste0("kegg_dot_", fc[i], ".svg"), plot = kegg_dot[[x]], path = here::here("2_plots/kegg/"),
width = 88, height = 95, units = "mm")
upset[[x]] <- upsetplot(x = enrichKEGG[[x]], 10)
ggsave(filename = paste0("upset_kegg_", fc[i], ".svg"), plot = upset[[x]], path = here::here("2_plots/kegg/"))
}
kegg_dot[[p]]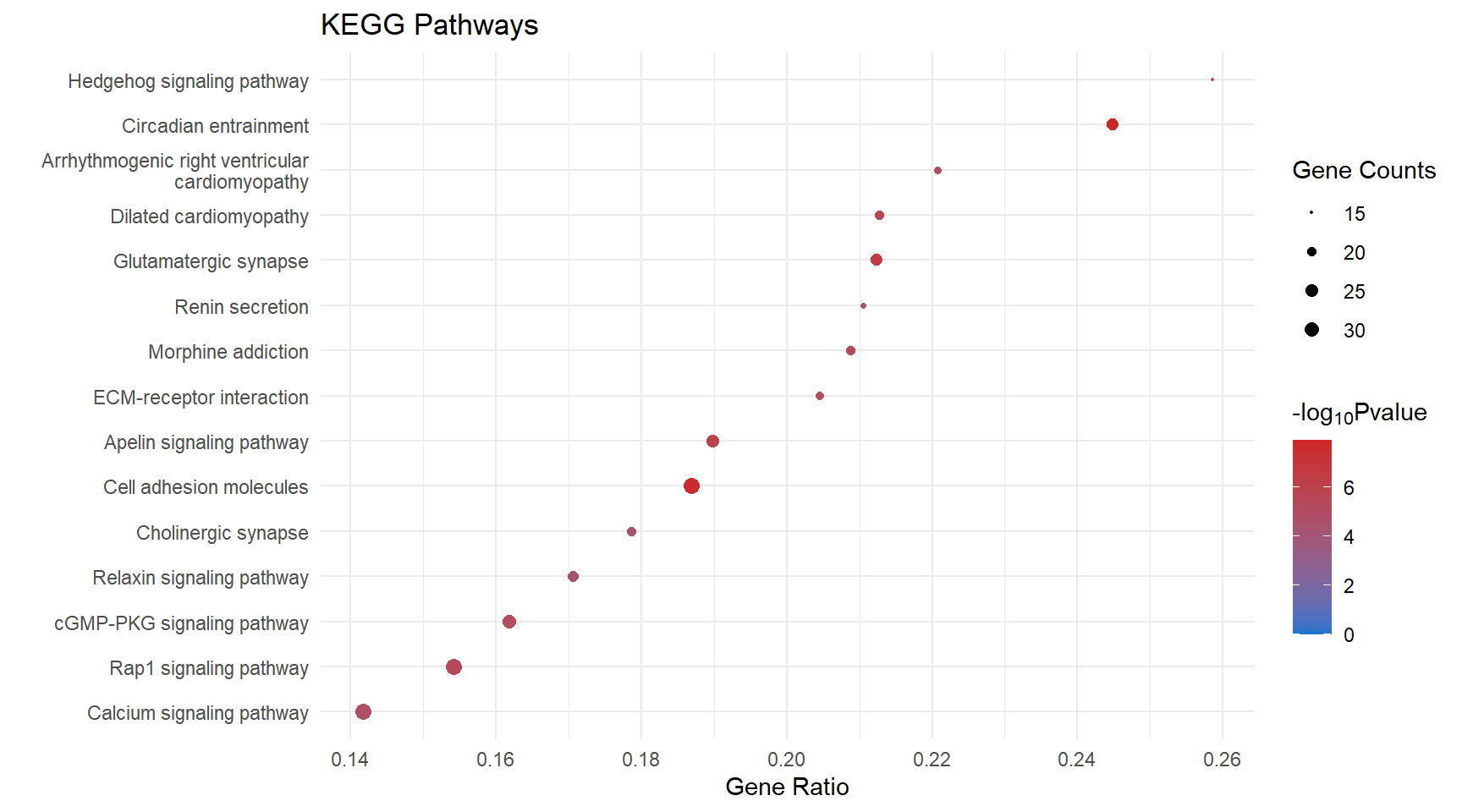
Upset Plot
upset[[p]]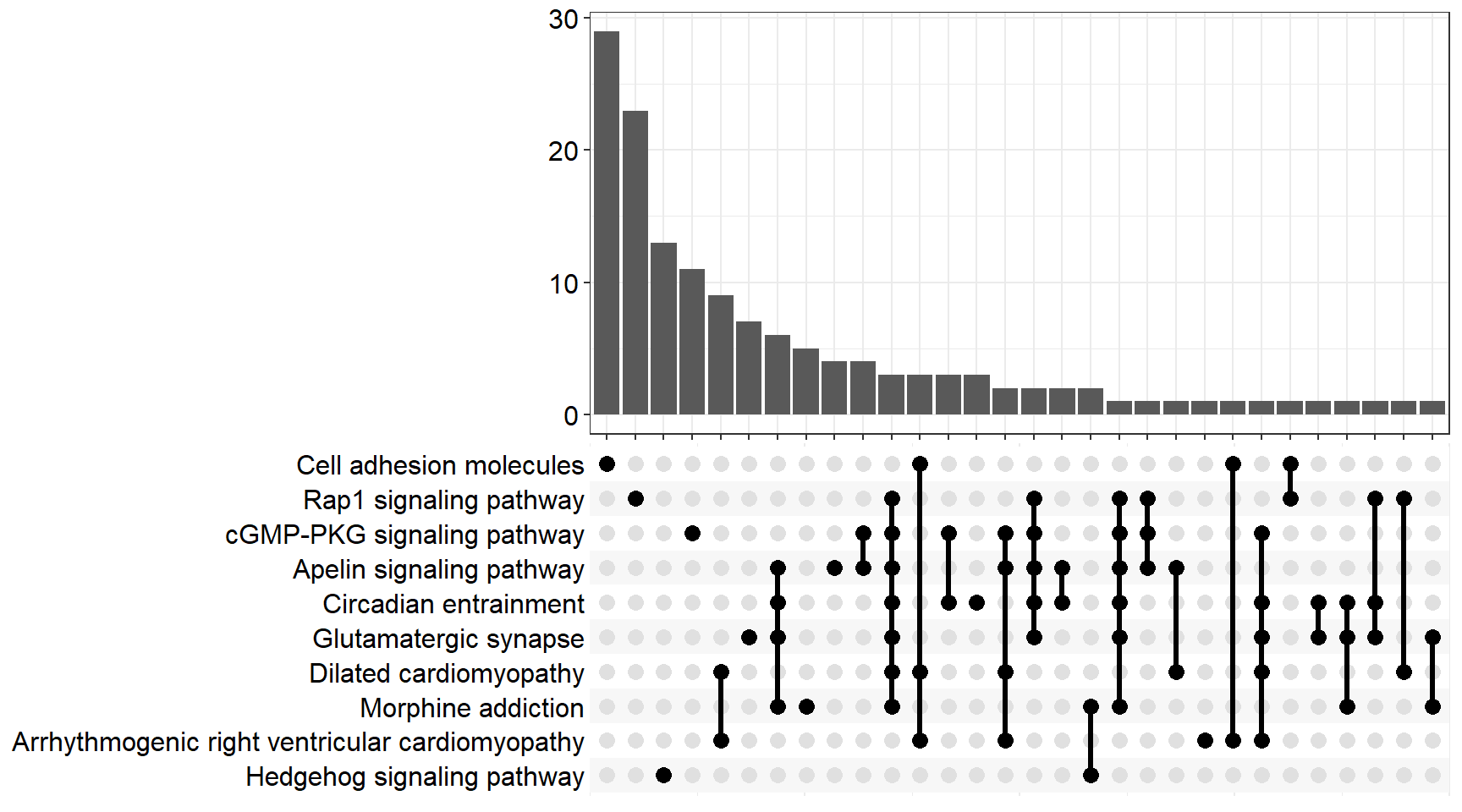
Leukocyte transendothelial migration
q=1Heatmap
# create df with normalised read counts with an additional entrezid column for binding
logCPM <- cpm(dge, prior.count = 3, log = TRUE)
logCPM <- logCPM[,1:7]
logCPM <- cbind(logCPM, dge$genes$entrezid)
rownames(logCPM) <- dge$genes$gene_name
colnames(logCPM) <- c("Control 1", "Control 2", "Control 4", "Intact 1", "Intact 2", "Intact 3", "Intact 4", "entrezid")
### full pathway method
# complete_pathway <- kegg_pathway[[1]]$GENE %>% as.data.frame()
# complete_pathway <- focal_adhesion[seq(1, nrow(focal_adhesion), 2),]
# match_complete_pathway <- logCPM[,"entrezid"] %in% complete_pathway
# my_palette <- colorRampPalette(c("dodgerblue3", "white", "firebrick3"))(n = 201)
my_palette <- viridis_pal(option = "viridis")(300)
# df for heatmap annotation of sample group
anno <- as.factor(dge$samples$group) %>% as.data.frame()
anno <- anno[1:7,] %>% as.data.frame()
colnames(anno) <- "Sample Groups"
anno$`Sample Groups` <- gsub("CONT", "Control", anno$`Sample Groups`)
anno$`Sample Groups` <- gsub("INT", "Intact", anno$`Sample Groups`)
rownames(anno) <- colnames(logCPM[, 1:7])
# setting colour of sample group annotation
# original sample colours
# anno_colours <- c("#66C2A5", "#FC8D62")
# new sample colours
anno_colours <- c("#f8766d", "#00bdc4")
names(anno_colours) <- c("Control", "Intact")matrix <- list()
display_matrix <- list()
kegg_heat=list()
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
for (j in 1:length(kegg_id)) {
y <- kegg_pathway[[j]]$PATHWAY_MAP
partial <- enrichKEGG_all[[x]][, c("ID", "geneID")]
partial <- partial[kegg_id[j], "geneID"] %>% as.data.frame()
partial <- separate_rows(partial, ., sep = "/")
colnames(partial) <- "entrezid"
# heatmap matrix
match <- logCPM[, "entrezid"] %in% partial$entrezid
matrix[[x]][[y]] <- logCPM[match, c("Control 1", "Control 2", "Control 4", "Intact 1", "Intact 2", "Intact 3", "Intact 4")] %>%
as.data.frame()
# changing the colname to numeric for some reason, cant remember
matrix[[x]][[y]][, c("Control 1", "Control 2", "Control 4", "Intact 1", "Intact 2", "Intact 3", "Intact 4")] <- as.numeric(as.character(unlist(matrix[[x]][[y]][, c("Control 1", "Control 2", "Control 4", "Intact 1", "Intact 2", "Intact 3", "Intact 4")])))
# display matrix
match2 <- lmTreat_sig[[x]][, "entrezid"] %in% partial$entrezid
display_matrix[[x]][[y]] <- lmTreat_sig[[x]][match2, c("gene_name", "logFC", "P.Value", "adj.P.Val", "description")] %>%
as.data.frame()
colnames(display_matrix[[x]][[y]]) <- c("Gene Name", "logFC", "P Value", "Adjusted P Value", "Description")
## Heatmap
kegg_heat[[x]][[y]] <- pheatmap(
mat = matrix[[x]][[y]],
### Publish
show_colnames = T,
main = paste0(y, "\n"),
legend = F,
annotation_legend = F,
fontsize = 8,
fontsize_col = 9,
fontsize_number = 7,
fontsize_row = 8,
treeheight_row = 25,
treeheight_col = 10,
cluster_cols = T,
clustering_distance_rows = "euclidean",
legend_breaks = c(seq(-3, 11, by = .5), 1.4),
legend_labels = c(seq(-3, 11, by = .5), "Z-Score"),
angle_col = 90,
cutree_cols = 2,
cutree_rows = 2,
color = my_palette,
scale = "row",
border_color = NA,
annotation_col = anno,
annotation_colors = list("Sample Groups" = anno_colours),
annotation_names_col = F,
annotation = T,
silent = T,
labels_row = as.expression(lapply(rownames(matrix[[x]][[y]]), function(a) bquote(italic(.(a)))))
) %>% as.ggplot()
# save
ggsave(filename = paste0("heat_", x, "_", y, ".svg"),
plot = kegg_heat[[x]][[y]],
path = here::here("2_plots/kegg/"),
width = 58,
height = 110,
units = "mm")}
}kegg_heat[[p]][[q]]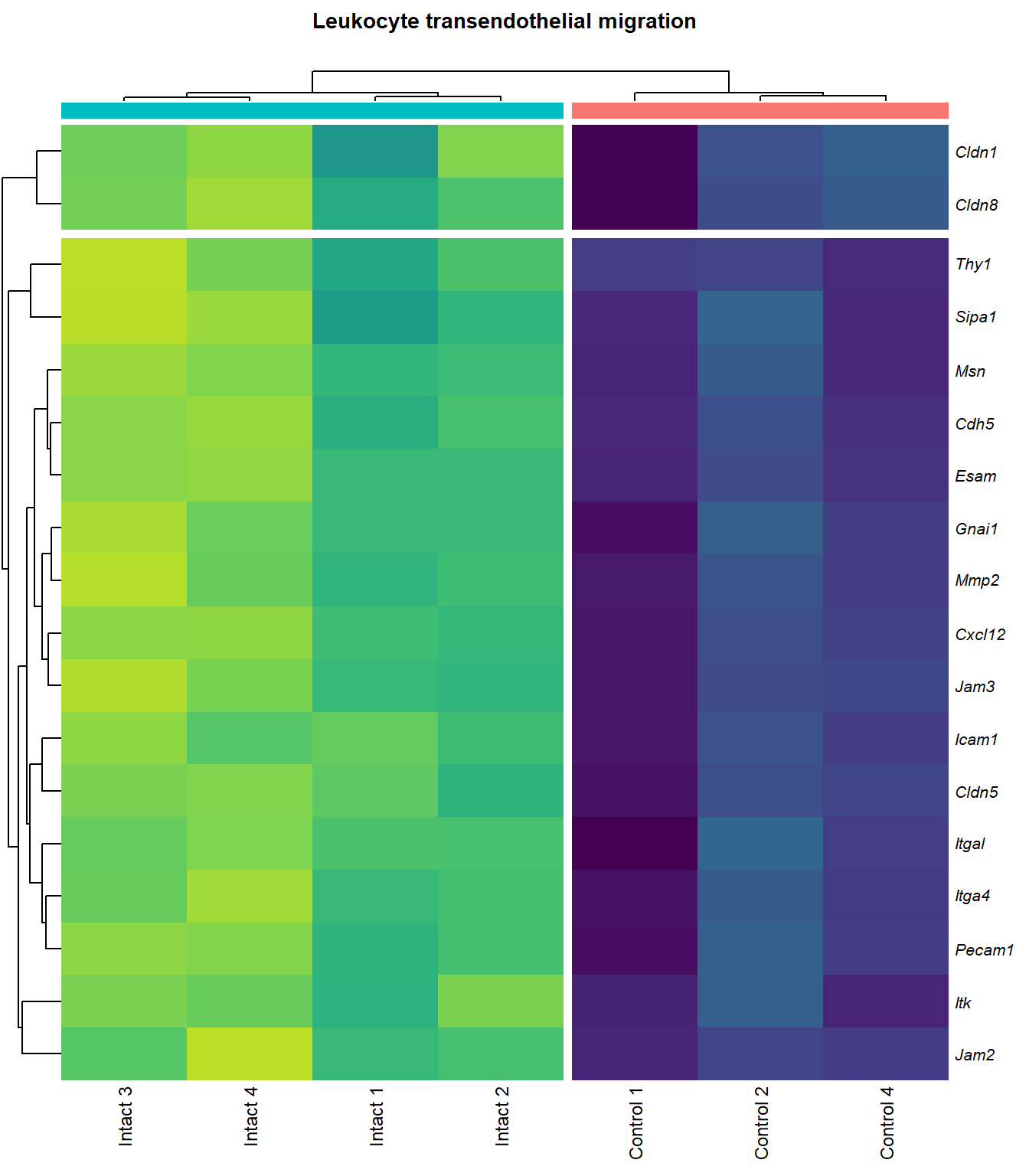
Table
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000155 | 0.0025120 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000057614 | Gnai1 | 2.405667 | 0.0001011 | 0.0055045 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 [Source:MGI Symbol;Acc:MGI:95771] |
| ENSMUSG00000031740 | Mmp2 | 2.270956 | 0.0001333 | 0.0062896 | matrix metallopeptidase 2 [Source:MGI Symbol;Acc:MGI:97009] |
| ENSMUSG00000037405 | Icam1 | 1.989282 | 0.0001592 | 0.0068723 | intercellular adhesion molecule 1 [Source:MGI Symbol;Acc:MGI:96392] |
| ENSMUSG00000020395 | Itk | 2.082706 | 0.0001896 | 0.0074705 | IL2 inducible T cell kinase [Source:MGI Symbol;Acc:MGI:96621] |
| ENSMUSG00000031990 | Jam3 | 1.932372 | 0.0001934 | 0.0075172 | junction adhesion molecule 3 [Source:MGI Symbol;Acc:MGI:1933825] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0002010 | 0.0076920 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| ENSMUSG00000050520 | Cldn8 | 1.948402 | 0.0002705 | 0.0093445 | claudin 8 [Source:MGI Symbol;Acc:MGI:1859286] |
| ENSMUSG00000031871 | Cdh5 | 2.498728 | 0.0004002 | 0.0116843 | cadherin 5 [Source:MGI Symbol;Acc:MGI:105057] |
| ENSMUSG00000032011 | Thy1 | 1.783613 | 0.0004132 | 0.0118214 | thymus cell antigen 1, theta [Source:MGI Symbol;Acc:MGI:98747] |
| ENSMUSG00000031207 | Msn | 1.375211 | 0.0004493 | 0.0121659 | moesin [Source:MGI Symbol;Acc:MGI:97167] |
| ENSMUSG00000030830 | Itgal | 1.816463 | 0.0006482 | 0.0150234 | integrin alpha L [Source:MGI Symbol;Acc:MGI:96606] |
| ENSMUSG00000001946 | Esam | 1.936624 | 0.0006493 | 0.0150234 | endothelial cell-specific adhesion molecule [Source:MGI Symbol;Acc:MGI:1916774] |
| ENSMUSG00000020717 | Pecam1 | 1.731782 | 0.0009000 | 0.0178528 | platelet/endothelial cell adhesion molecule 1 [Source:MGI Symbol;Acc:MGI:97537] |
| ENSMUSG00000041378 | Cldn5 | 1.919823 | 0.0018784 | 0.0281045 | claudin 5 [Source:MGI Symbol;Acc:MGI:1276112] |
| ENSMUSG00000022512 | Cldn1 | 1.838024 | 0.0037807 | 0.0435854 | claudin 1 [Source:MGI Symbol;Acc:MGI:1276109] |
| ENSMUSG00000056917 | Sipa1 | 1.252646 | 0.0042264 | 0.0465693 | signal-induced proliferation associated gene 1 [Source:MGI Symbol;Acc:MGI:107576] |
| ENSMUSG00000053062 | Jam2 | 3.164427 | 0.0046329 | 0.0493355 | junction adhesion molecule 2 [Source:MGI Symbol;Acc:MGI:1933820] |
Pathview
# adjusting the kegg id to suit the parameters of the pathview funtion
adj.keggID <- gsub("mmu", "", kegg_id)
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
# extract the logFC from the DE gene list
pathview_table <- dplyr::select(.data = lmTreat_sig[[x]], c("logFC")) %>% as.matrix()
# run pathview with Ensembl ID instead of entrezID
pathview <- pathview(
gene.data = pathview_table[, 1],
gene.idtype = "ENSEMBL",
pathway.id = adj.keggID,
species = "mmu",
out.suffix = "pv",
kegg.dir = here::here("2_plots/kegg/"),
kegg.native = T
)
# move the result file to the plot directory
file.rename(
from = paste0("mmu", adj.keggID, ".pv.png"),
to = here::here(paste0("2_plots/kegg/pv_", x, "_", kegg_id, ".png"))
)
}[1] "Note: 166 of 1629 unique input IDs unmapped."[1] "Note: 151 of 1447 unique input IDs unmapped."[1] "Note: 63 of 388 unique input IDs unmapped."# display the pathview graph. doesnt work when knitting!!!!
# knitr::include_graphics(here::here(paste0("2_plots/kegg/pv_", fc[p], "_", kegg_id[q], ".png")))Hematopoietic cell lineage
q=q+1Heatmap
kegg_heat[[p]][[q]]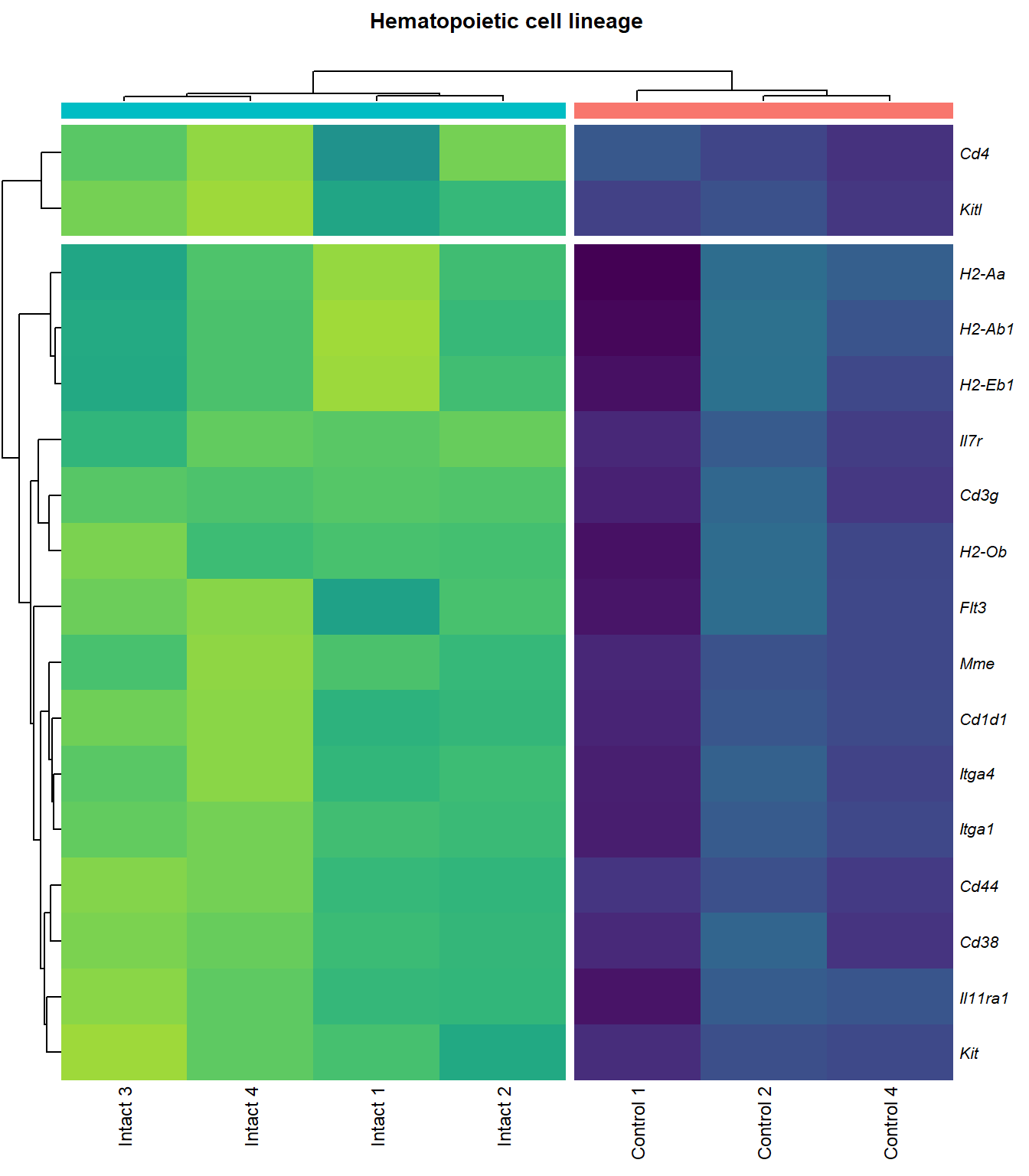
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000155 | 0.0025120 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000005087 | Cd44 | 2.271713 | 0.0000724 | 0.0046732 | CD44 antigen [Source:MGI Symbol;Acc:MGI:88338] |
| ENSMUSG00000073889 | Il11ra1 | 1.542982 | 0.0001081 | 0.0056640 | interleukin 11 receptor, alpha chain 1 [Source:MGI Symbol;Acc:MGI:107426] |
| ENSMUSG00000002033 | Cd3g | 3.645674 | 0.0002123 | 0.0079664 | CD3 antigen, gamma polypeptide [Source:MGI Symbol;Acc:MGI:88333] |
| ENSMUSG00000042284 | Itga1 | 1.938967 | 0.0003108 | 0.0102149 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000042817 | Flt3 | 2.075768 | 0.0004099 | 0.0118214 | FMS-like tyrosine kinase 3 [Source:MGI Symbol;Acc:MGI:95559] |
| ENSMUSG00000028076 | Cd1d1 | 2.344493 | 0.0005063 | 0.0130930 | CD1d1 antigen [Source:MGI Symbol;Acc:MGI:107674] |
| ENSMUSG00000005672 | Kit | 1.518478 | 0.0005311 | 0.0134106 | KIT proto-oncogene receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:96677] |
| ENSMUSG00000027820 | Mme | 3.737788 | 0.0005351 | 0.0134543 | membrane metallo endopeptidase [Source:MGI Symbol;Acc:MGI:97004] |
| ENSMUSG00000073421 | H2-Ab1 | 1.742382 | 0.0006499 | 0.0150234 | histocompatibility 2, class II antigen A, beta 1 [Source:MGI Symbol;Acc:MGI:103070] |
| ENSMUSG00000029084 | Cd38 | 1.711230 | 0.0007558 | 0.0163113 | CD38 antigen [Source:MGI Symbol;Acc:MGI:107474] |
| ENSMUSG00000041538 | H2-Ob | 1.716311 | 0.0008312 | 0.0170615 | histocompatibility 2, O region beta locus [Source:MGI Symbol;Acc:MGI:95925] |
| ENSMUSG00000019966 | Kitl | 1.310648 | 0.0010693 | 0.0200607 | kit ligand [Source:MGI Symbol;Acc:MGI:96974] |
| ENSMUSG00000060586 | H2-Eb1 | 1.690764 | 0.0013795 | 0.0231779 | histocompatibility 2, class II antigen E beta [Source:MGI Symbol;Acc:MGI:95901] |
| ENSMUSG00000036594 | H2-Aa | 1.672690 | 0.0021648 | 0.0308677 | histocompatibility 2, class II antigen A, alpha [Source:MGI Symbol;Acc:MGI:95895] |
| ENSMUSG00000003882 | Il7r | 2.257016 | 0.0040536 | 0.0454821 | interleukin 7 receptor [Source:MGI Symbol;Acc:MGI:96562] |
| ENSMUSG00000023274 | Cd4 | 2.413573 | 0.0040842 | 0.0457300 | CD4 antigen [Source:MGI Symbol;Acc:MGI:88335] |
Pathview
Vascular smooth muscle contraction
q=q+1Heatmap
kegg_heat[[p]][[q]]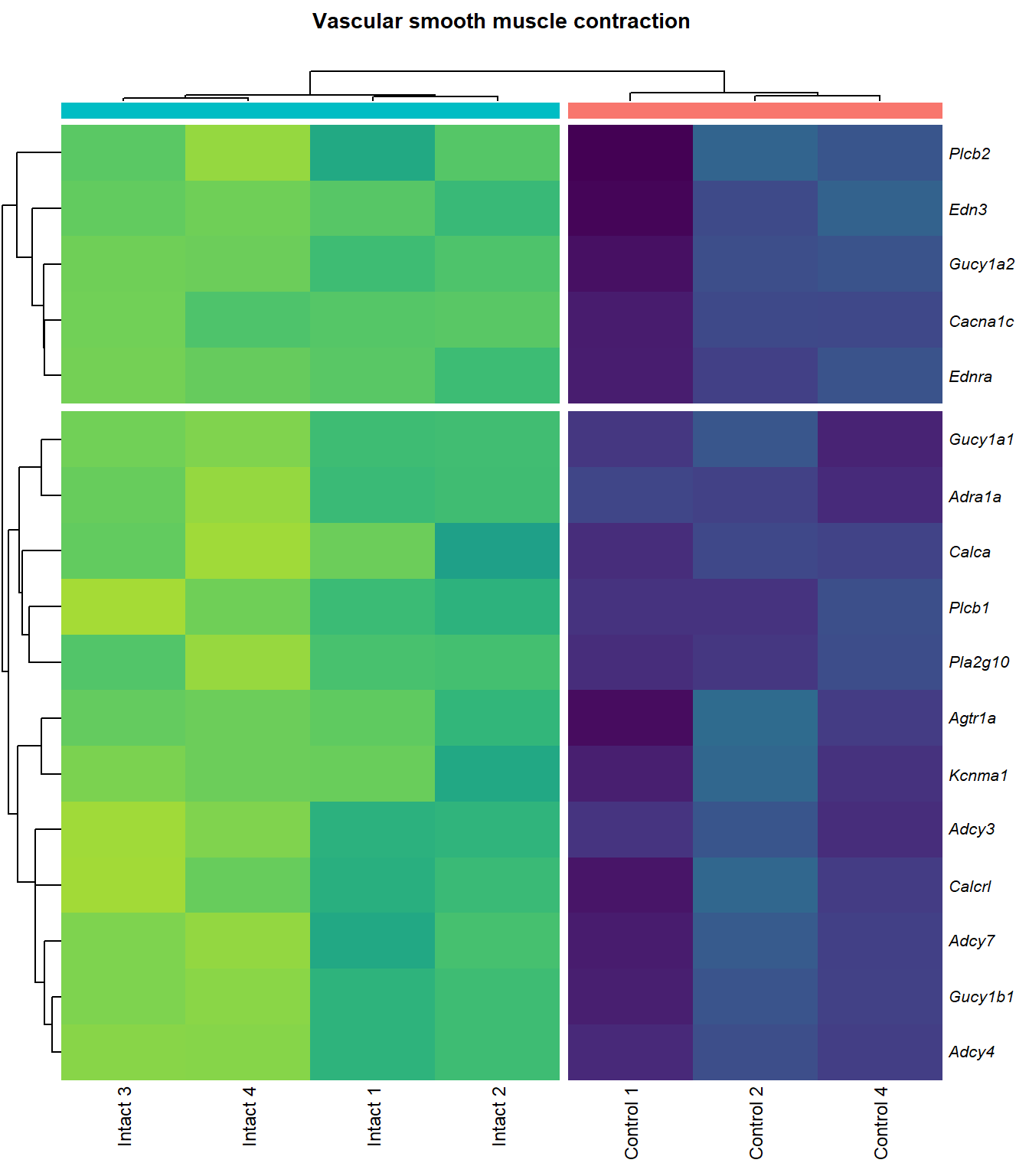
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000051331 | Cacna1c | 1.120969 | 0.0000094 | 0.0019883 | calcium channel, voltage-dependent, L type, alpha 1C subunit [Source:MGI Symbol;Acc:MGI:103013] |
| ENSMUSG00000022683 | Pla2g10 | 5.773220 | 0.0000180 | 0.0025584 | phospholipase A2, group X [Source:MGI Symbol;Acc:MGI:1347522] |
| ENSMUSG00000045875 | Adra1a | 4.131590 | 0.0000560 | 0.0042689 | adrenergic receptor, alpha 1a [Source:MGI Symbol;Acc:MGI:104773] |
| ENSMUSG00000041624 | Gucy1a2 | 2.199471 | 0.0001654 | 0.0070122 | guanylate cyclase 1, soluble, alpha 2 [Source:MGI Symbol;Acc:MGI:2660877] |
| ENSMUSG00000020654 | Adcy3 | 1.648297 | 0.0001903 | 0.0074705 | adenylate cyclase 3 [Source:MGI Symbol;Acc:MGI:99675] |
| ENSMUSG00000027524 | Edn3 | 3.849271 | 0.0003001 | 0.0100411 | endothelin 3 [Source:MGI Symbol;Acc:MGI:95285] |
| ENSMUSG00000051177 | Plcb1 | 1.960299 | 0.0003205 | 0.0104215 | phospholipase C, beta 1 [Source:MGI Symbol;Acc:MGI:97613] |
| ENSMUSG00000030669 | Calca | 5.762174 | 0.0003662 | 0.0111446 | calcitonin/calcitonin-related polypeptide, alpha [Source:MGI Symbol;Acc:MGI:2151253] |
| ENSMUSG00000028005 | Gucy1b1 | 1.861114 | 0.0004126 | 0.0118214 | guanylate cyclase 1, soluble, beta 1 [Source:MGI Symbol;Acc:MGI:1860604] |
| ENSMUSG00000033910 | Gucy1a1 | 1.985306 | 0.0005168 | 0.0132227 | guanylate cyclase 1, soluble, alpha 1 [Source:MGI Symbol;Acc:MGI:1926562] |
| ENSMUSG00000022220 | Adcy4 | 1.867108 | 0.0005490 | 0.0136242 | adenylate cyclase 4 [Source:MGI Symbol;Acc:MGI:99674] |
| ENSMUSG00000031659 | Adcy7 | 1.309423 | 0.0005572 | 0.0137479 | adenylate cyclase 7 [Source:MGI Symbol;Acc:MGI:102891] |
| ENSMUSG00000049115 | Agtr1a | 1.761758 | 0.0007646 | 0.0164599 | angiotensin II receptor, type 1a [Source:MGI Symbol;Acc:MGI:87964] |
| ENSMUSG00000063142 | Kcnma1 | 2.434426 | 0.0008434 | 0.0171847 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 [Source:MGI Symbol;Acc:MGI:99923] |
| ENSMUSG00000031616 | Ednra | 1.779614 | 0.0008698 | 0.0175204 | endothelin receptor type A [Source:MGI Symbol;Acc:MGI:105923] |
| ENSMUSG00000040061 | Plcb2 | 1.498256 | 0.0015357 | 0.0249268 | phospholipase C, beta 2 [Source:MGI Symbol;Acc:MGI:107465] |
| ENSMUSG00000059588 | Calcrl | 1.494208 | 0.0018856 | 0.0281484 | calcitonin receptor-like [Source:MGI Symbol;Acc:MGI:1926944] |
Pathview
PI3K-Akt signaling pathway
q=q+1Heatmap
kegg_heat[[p]][[q]]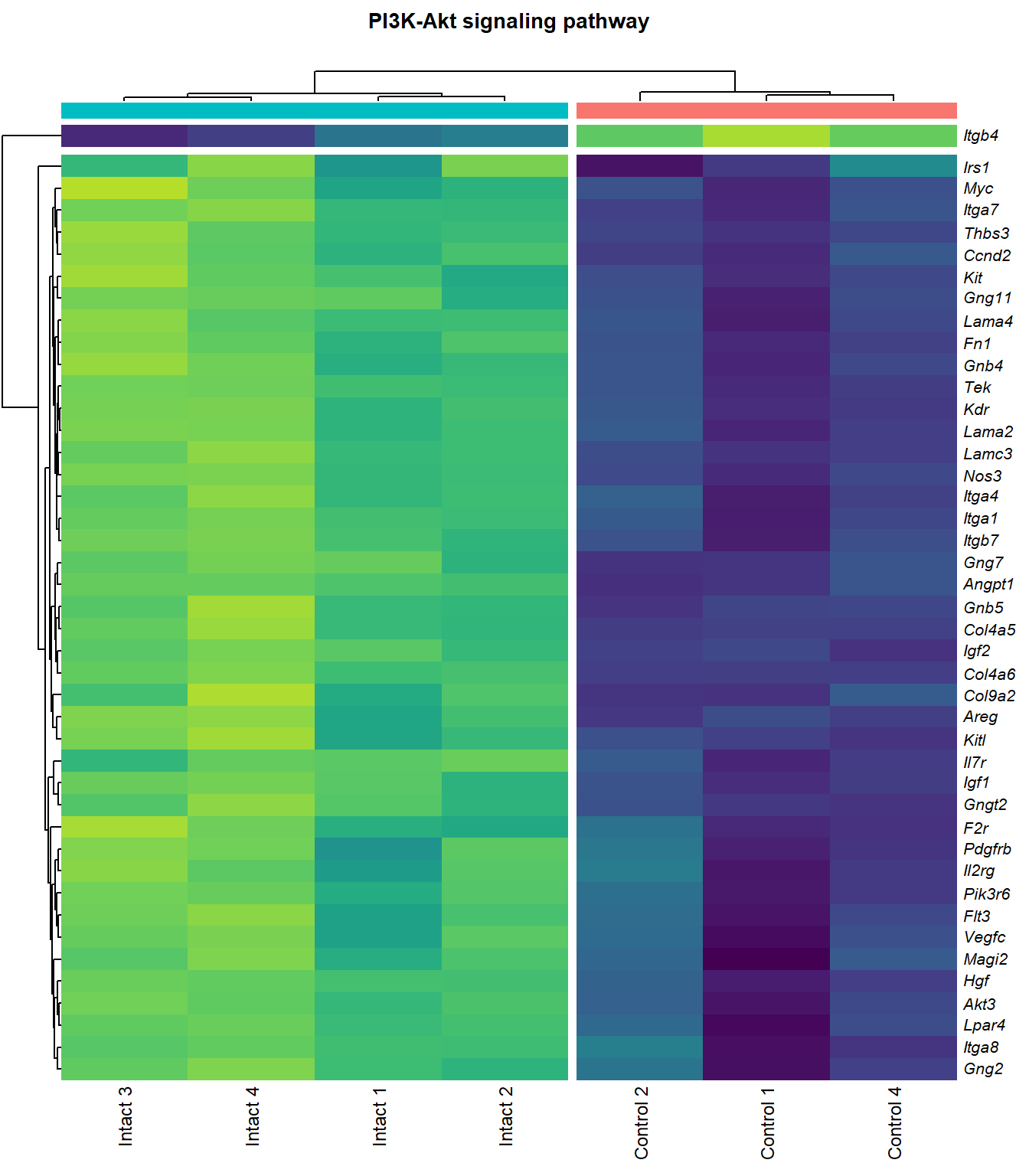
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.1253117 | 0.0000044 | 0.0015028 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000022309 | Angpt1 | 3.3885860 | 0.0000102 | 0.0021117 | angiopoietin 1 [Source:MGI Symbol;Acc:MGI:108448] |
| ENSMUSG00000027009 | Itga4 | 2.0741848 | 0.0000155 | 0.0025120 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.0456427 | 0.0000226 | 0.0028011 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000020053 | Igf1 | 2.7804446 | 0.0000309 | 0.0032608 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000031273 | Col4a6 | 3.4668103 | 0.0000409 | 0.0037259 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000028047 | Thbs3 | 1.6738088 | 0.0000461 | 0.0039924 | thrombospondin 3 [Source:MGI Symbol;Acc:MGI:98739] |
| ENSMUSG00000028978 | Nos3 | 2.1486818 | 0.0000581 | 0.0043512 | nitric oxide synthase 3, endothelial cell [Source:MGI Symbol;Acc:MGI:97362] |
| ENSMUSG00000026840 | Lamc3 | 3.8038416 | 0.0000783 | 0.0048313 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000019846 | Lama4 | 1.9402802 | 0.0000823 | 0.0049752 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
| ENSMUSG00000000184 | Ccnd2 | 1.7053911 | 0.0001079 | 0.0056640 | cyclin D2 [Source:MGI Symbol;Acc:MGI:88314] |
| ENSMUSG00000001281 | Itgb7 | 1.8062915 | 0.0001092 | 0.0056640 | integrin beta 7 [Source:MGI Symbol;Acc:MGI:96616] |
| ENSMUSG00000026193 | Fn1 | 1.9696894 | 0.0001143 | 0.0057552 | fibronectin 1 [Source:MGI Symbol;Acc:MGI:95566] |
| ENSMUSG00000029378 | Areg | 5.2465226 | 0.0001304 | 0.0062202 | amphiregulin [Source:MGI Symbol;Acc:MGI:88068] |
| ENSMUSG00000049929 | Lpar4 | 2.1460743 | 0.0001661 | 0.0070122 | lysophosphatidic acid receptor 4 [Source:MGI Symbol;Acc:MGI:1925384] |
| ENSMUSG00000006386 | Tek | 2.3390336 | 0.0001955 | 0.0075310 | TEK receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:98664] |
| ENSMUSG00000038811 | Gngt2 | 1.8196583 | 0.0002207 | 0.0081751 | guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 [Source:MGI Symbol;Acc:MGI:893584] |
| ENSMUSG00000046207 | Pik3r6 | 1.6924253 | 0.0002696 | 0.0093445 | phosphoinositide-3-kinase regulatory subunit 5 [Source:MGI Symbol;Acc:MGI:2144613] |
| ENSMUSG00000042284 | Itga1 | 1.9389666 | 0.0003108 | 0.0102149 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000028626 | Col9a2 | 4.1297594 | 0.0003600 | 0.0110798 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000042817 | Flt3 | 2.0757680 | 0.0004099 | 0.0118214 | FMS-like tyrosine kinase 3 [Source:MGI Symbol;Acc:MGI:95559] |
| ENSMUSG00000005672 | Kit | 1.5184784 | 0.0005311 | 0.0134106 | KIT proto-oncogene receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:96677] |
| ENSMUSG00000032766 | Gng11 | 2.0192291 | 0.0005737 | 0.0139360 | guanine nucleotide binding protein (G protein), gamma 11 [Source:MGI Symbol;Acc:MGI:1913316] |
| ENSMUSG00000019899 | Lama2 | 1.8339350 | 0.0005773 | 0.0140035 | laminin, alpha 2 [Source:MGI Symbol;Acc:MGI:99912] |
| ENSMUSG00000031520 | Vegfc | 2.1245474 | 0.0007514 | 0.0162972 | vascular endothelial growth factor C [Source:MGI Symbol;Acc:MGI:109124] |
| ENSMUSG00000040003 | Magi2 | 1.5230701 | 0.0007976 | 0.0166697 | membrane associated guanylate kinase, WW and PDZ domain containing 2 [Source:MGI Symbol;Acc:MGI:1354953] |
| ENSMUSG00000048583 | Igf2 | 2.4269395 | 0.0009328 | 0.0183389 | insulin-like growth factor 2 [Source:MGI Symbol;Acc:MGI:96434] |
| ENSMUSG00000043004 | Gng2 | 1.2390569 | 0.0009402 | 0.0184405 | guanine nucleotide binding protein (G protein), gamma 2 [Source:MGI Symbol;Acc:MGI:102705] |
| ENSMUSG00000025348 | Itga7 | 1.5797819 | 0.0009870 | 0.0189401 | integrin alpha 7 [Source:MGI Symbol;Acc:MGI:102700] |
| ENSMUSG00000019966 | Kitl | 1.3106480 | 0.0010693 | 0.0200607 | kit ligand [Source:MGI Symbol;Acc:MGI:96974] |
| ENSMUSG00000019699 | Akt3 | 1.2001093 | 0.0020912 | 0.0301880 | thymoma viral proto-oncogene 3 [Source:MGI Symbol;Acc:MGI:1345147] |
| ENSMUSG00000032192 | Gnb5 | 1.0340057 | 0.0021925 | 0.0310772 | guanine nucleotide binding protein (G protein), beta 5 [Source:MGI Symbol;Acc:MGI:101848] |
| ENSMUSG00000048240 | Gng7 | 2.3093807 | 0.0022679 | 0.0316071 | guanine nucleotide binding protein (G protein), gamma 7 [Source:MGI Symbol;Acc:MGI:95787] |
| ENSMUSG00000027669 | Gnb4 | 1.1575446 | 0.0027290 | 0.0356047 | guanine nucleotide binding protein (G protein), beta 4 [Source:MGI Symbol;Acc:MGI:104581] |
| ENSMUSG00000024620 | Pdgfrb | 0.9572539 | 0.0029764 | 0.0376073 | platelet derived growth factor receptor, beta polypeptide [Source:MGI Symbol;Acc:MGI:97531] |
| ENSMUSG00000031304 | Il2rg | 1.8830596 | 0.0033557 | 0.0405276 | interleukin 2 receptor, gamma chain [Source:MGI Symbol;Acc:MGI:96551] |
| ENSMUSG00000026768 | Itga8 | 1.5873015 | 0.0033743 | 0.0407238 | integrin alpha 8 [Source:MGI Symbol;Acc:MGI:109442] |
| ENSMUSG00000020758 | Itgb4 | -1.4990466 | 0.0035398 | 0.0419565 | integrin beta 4 [Source:MGI Symbol;Acc:MGI:96613] |
| ENSMUSG00000048376 | F2r | 2.1925090 | 0.0040492 | 0.0454821 | coagulation factor II (thrombin) receptor [Source:MGI Symbol;Acc:MGI:101802] |
| ENSMUSG00000003882 | Il7r | 2.2570158 | 0.0040536 | 0.0454821 | interleukin 7 receptor [Source:MGI Symbol;Acc:MGI:96562] |
| ENSMUSG00000022346 | Myc | 1.8123908 | 0.0043062 | 0.0470865 | myelocytomatosis oncogene [Source:MGI Symbol;Acc:MGI:97250] |
| ENSMUSG00000031274 | Col4a5 | 1.6976014 | 0.0047060 | 0.0498134 | collagen, type IV, alpha 5 [Source:MGI Symbol;Acc:MGI:88456] |
| ENSMUSG00000055980 | Irs1 | 0.9503609 | 0.0047208 | 0.0499093 | insulin receptor substrate 1 [Source:MGI Symbol;Acc:MGI:99454] |
Pathview
Focal adhesion
q=q+1Heatmap
kegg_heat[[p]][[q]]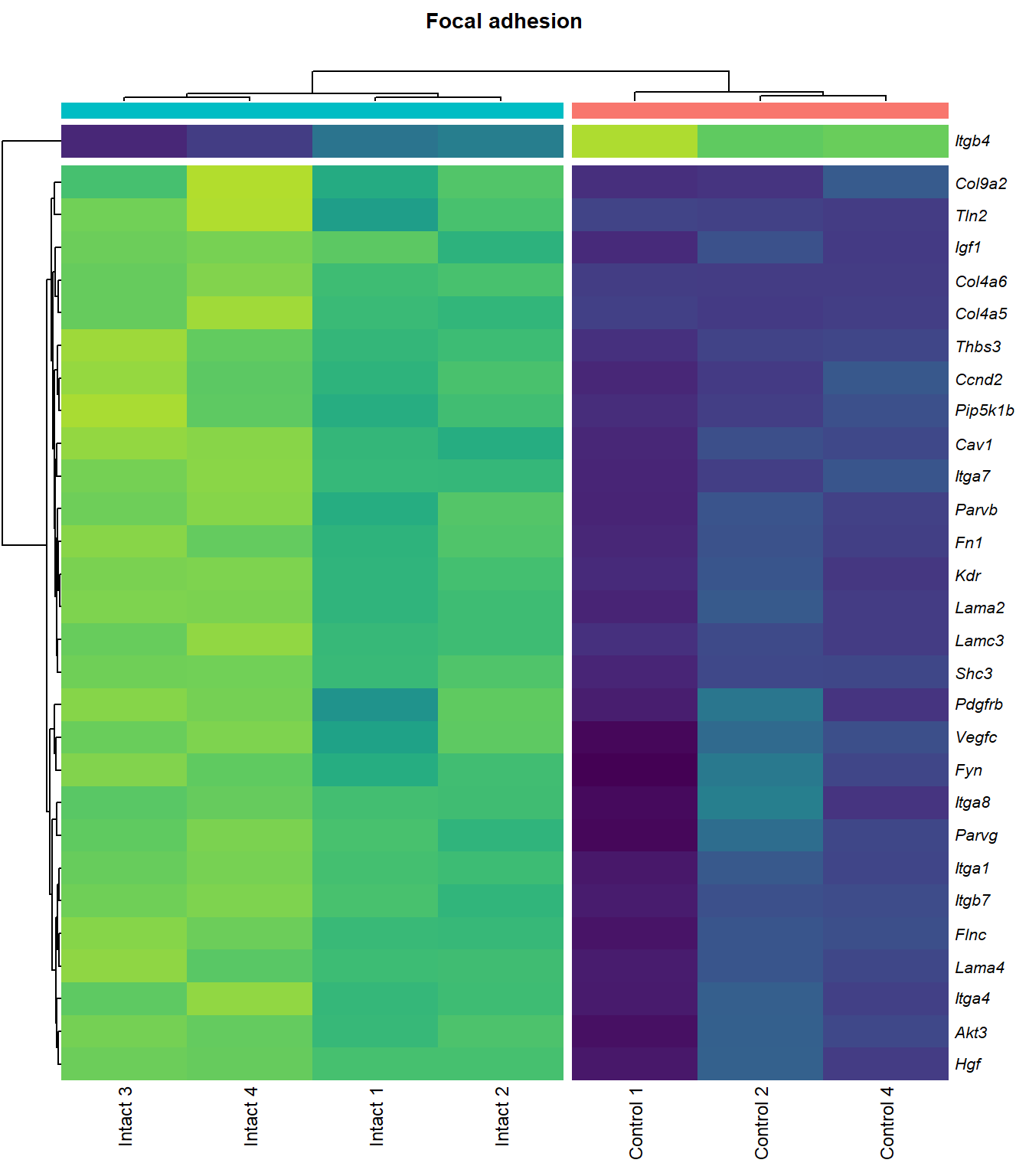
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.1253117 | 0.0000044 | 0.0015028 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000027009 | Itga4 | 2.0741848 | 0.0000155 | 0.0025120 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.0456427 | 0.0000226 | 0.0028011 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000020053 | Igf1 | 2.7804446 | 0.0000309 | 0.0032608 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000031273 | Col4a6 | 3.4668103 | 0.0000409 | 0.0037259 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000028047 | Thbs3 | 1.6738088 | 0.0000461 | 0.0039924 | thrombospondin 3 [Source:MGI Symbol;Acc:MGI:98739] |
| ENSMUSG00000026840 | Lamc3 | 3.8038416 | 0.0000783 | 0.0048313 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000019846 | Lama4 | 1.9402802 | 0.0000823 | 0.0049752 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
| ENSMUSG00000000184 | Ccnd2 | 1.7053911 | 0.0001079 | 0.0056640 | cyclin D2 [Source:MGI Symbol;Acc:MGI:88314] |
| ENSMUSG00000001281 | Itgb7 | 1.8062915 | 0.0001092 | 0.0056640 | integrin beta 7 [Source:MGI Symbol;Acc:MGI:96616] |
| ENSMUSG00000026193 | Fn1 | 1.9696894 | 0.0001143 | 0.0057552 | fibronectin 1 [Source:MGI Symbol;Acc:MGI:95566] |
| ENSMUSG00000021448 | Shc3 | 2.1490360 | 0.0001372 | 0.0063240 | src homology 2 domain-containing transforming protein C3 [Source:MGI Symbol;Acc:MGI:106179] |
| ENSMUSG00000024867 | Pip5k1b | 1.5003642 | 0.0001491 | 0.0066795 | phosphatidylinositol-4-phosphate 5-kinase, type 1 beta [Source:MGI Symbol;Acc:MGI:107930] |
| ENSMUSG00000042284 | Itga1 | 1.9389666 | 0.0003108 | 0.0102149 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000028626 | Col9a2 | 4.1297594 | 0.0003600 | 0.0110798 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000019899 | Lama2 | 1.8339350 | 0.0005773 | 0.0140035 | laminin, alpha 2 [Source:MGI Symbol;Acc:MGI:99912] |
| ENSMUSG00000031520 | Vegfc | 2.1245474 | 0.0007514 | 0.0162972 | vascular endothelial growth factor C [Source:MGI Symbol;Acc:MGI:109124] |
| ENSMUSG00000025348 | Itga7 | 1.5797819 | 0.0009870 | 0.0189401 | integrin alpha 7 [Source:MGI Symbol;Acc:MGI:102700] |
| ENSMUSG00000022439 | Parvg | 1.7706088 | 0.0010067 | 0.0191779 | parvin, gamma [Source:MGI Symbol;Acc:MGI:2158329] |
| ENSMUSG00000022438 | Parvb | 1.6425559 | 0.0012361 | 0.0215474 | parvin, beta [Source:MGI Symbol;Acc:MGI:2153063] |
| ENSMUSG00000068699 | Flnc | 1.1797740 | 0.0013967 | 0.0234216 | filamin C, gamma [Source:MGI Symbol;Acc:MGI:95557] |
| ENSMUSG00000052698 | Tln2 | 1.2448359 | 0.0014789 | 0.0243963 | talin 2 [Source:MGI Symbol;Acc:MGI:1917799] |
| ENSMUSG00000019843 | Fyn | 1.1786653 | 0.0017407 | 0.0266942 | Fyn proto-oncogene [Source:MGI Symbol;Acc:MGI:95602] |
| ENSMUSG00000019699 | Akt3 | 1.2001093 | 0.0020912 | 0.0301880 | thymoma viral proto-oncogene 3 [Source:MGI Symbol;Acc:MGI:1345147] |
| ENSMUSG00000024620 | Pdgfrb | 0.9572539 | 0.0029764 | 0.0376073 | platelet derived growth factor receptor, beta polypeptide [Source:MGI Symbol;Acc:MGI:97531] |
| ENSMUSG00000007655 | Cav1 | 1.6011191 | 0.0031814 | 0.0392979 | caveolin 1, caveolae protein [Source:MGI Symbol;Acc:MGI:102709] |
| ENSMUSG00000026768 | Itga8 | 1.5873015 | 0.0033743 | 0.0407238 | integrin alpha 8 [Source:MGI Symbol;Acc:MGI:109442] |
| ENSMUSG00000020758 | Itgb4 | -1.4990466 | 0.0035398 | 0.0419565 | integrin beta 4 [Source:MGI Symbol;Acc:MGI:96613] |
| ENSMUSG00000031274 | Col4a5 | 1.6976014 | 0.0047060 | 0.0498134 | collagen, type IV, alpha 5 [Source:MGI Symbol;Acc:MGI:88456] |
Pathview
Cytokine-cytokine receptor interaction
q=q+1Heatmap
kegg_heat[[p]][[q]]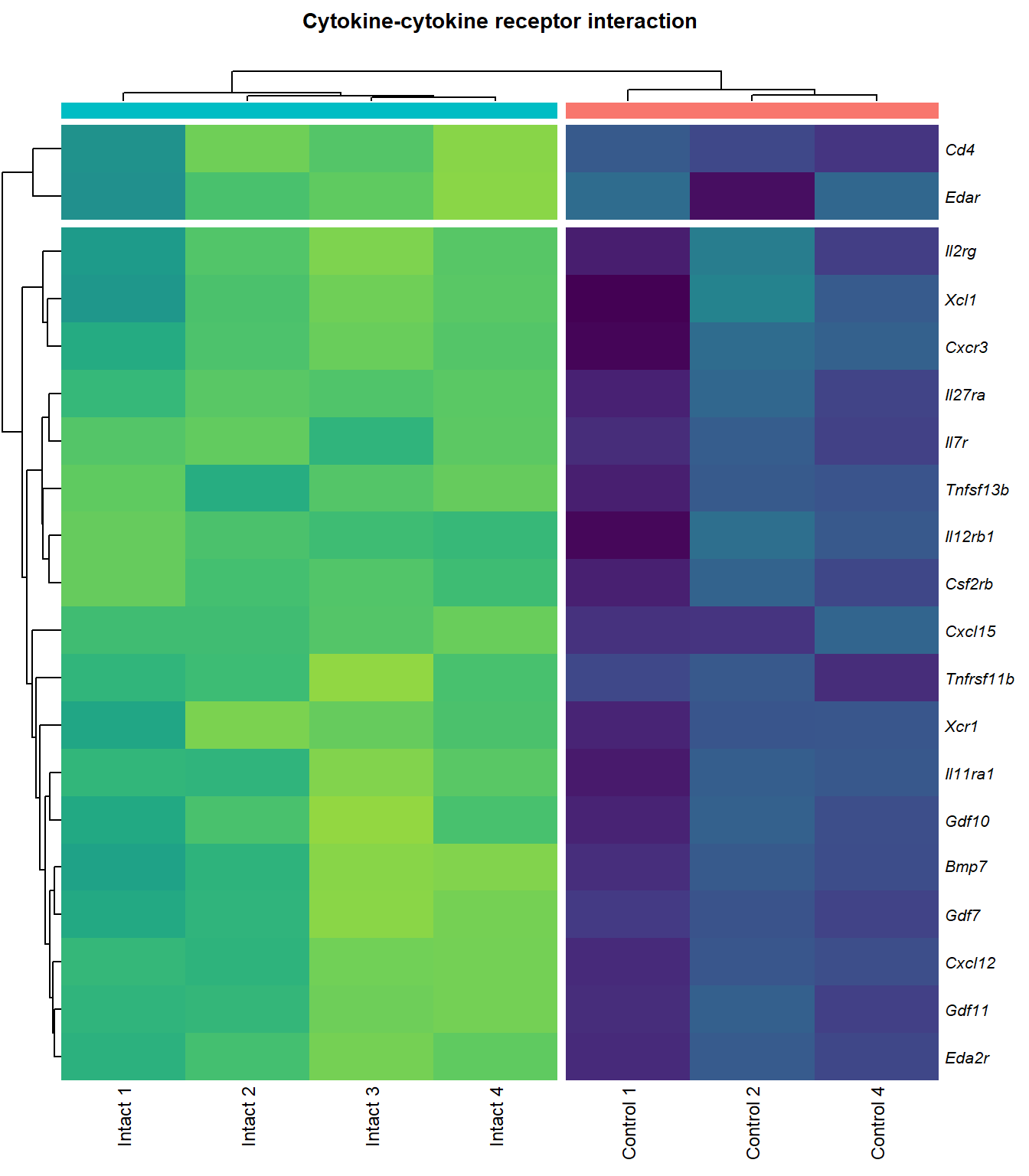
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000029375 | Cxcl15 | 6.916834 | 0.0000000 | 0.0001479 | chemokine (C-X-C motif) ligand 15 [Source:MGI Symbol;Acc:MGI:1339941] |
| ENSMUSG00000034457 | Eda2r | 3.524309 | 0.0000387 | 0.0036589 | ectodysplasin A2 receptor [Source:MGI Symbol;Acc:MGI:2442860] |
| ENSMUSG00000073889 | Il11ra1 | 1.542982 | 0.0001081 | 0.0056640 | interleukin 11 receptor, alpha chain 1 [Source:MGI Symbol;Acc:MGI:107426] |
| ENSMUSG00000050232 | Cxcr3 | 2.280257 | 0.0001954 | 0.0075310 | chemokine (C-X-C motif) receptor 3 [Source:MGI Symbol;Acc:MGI:1277207] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0002010 | 0.0076920 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| ENSMUSG00000025352 | Gdf11 | 2.251448 | 0.0002430 | 0.0087564 | growth differentiation factor 11 [Source:MGI Symbol;Acc:MGI:1338027] |
| ENSMUSG00000037660 | Gdf7 | 3.410398 | 0.0004003 | 0.0116843 | growth differentiation factor 7 [Source:MGI Symbol;Acc:MGI:95690] |
| ENSMUSG00000031497 | Tnfsf13b | 2.218370 | 0.0004343 | 0.0120034 | tumor necrosis factor (ligand) superfamily, member 13b [Source:MGI Symbol;Acc:MGI:1344376] |
| ENSMUSG00000021943 | Gdf10 | 2.741543 | 0.0005206 | 0.0132671 | growth differentiation factor 10 [Source:MGI Symbol;Acc:MGI:95684] |
| ENSMUSG00000005465 | Il27ra | 1.937581 | 0.0006697 | 0.0153371 | interleukin 27 receptor, alpha [Source:MGI Symbol;Acc:MGI:1355318] |
| ENSMUSG00000063727 | Tnfrsf11b | 2.131252 | 0.0007401 | 0.0161954 | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) [Source:MGI Symbol;Acc:MGI:109587] |
| ENSMUSG00000026573 | Xcl1 | 2.671247 | 0.0011054 | 0.0204395 | chemokine (C motif) ligand 1 [Source:MGI Symbol;Acc:MGI:104593] |
| ENSMUSG00000060509 | Xcr1 | 2.001440 | 0.0017559 | 0.0268808 | chemokine (C motif) receptor 1 [Source:MGI Symbol;Acc:MGI:1346338] |
| ENSMUSG00000071713 | Csf2rb | 2.031328 | 0.0017896 | 0.0271305 | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) [Source:MGI Symbol;Acc:MGI:1339759] |
| ENSMUSG00000000791 | Il12rb1 | 2.130604 | 0.0031159 | 0.0386619 | interleukin 12 receptor, beta 1 [Source:MGI Symbol;Acc:MGI:104579] |
| ENSMUSG00000031304 | Il2rg | 1.883060 | 0.0033557 | 0.0405276 | interleukin 2 receptor, gamma chain [Source:MGI Symbol;Acc:MGI:96551] |
| ENSMUSG00000003227 | Edar | 2.573653 | 0.0038496 | 0.0439969 | ectodysplasin-A receptor [Source:MGI Symbol;Acc:MGI:1343498] |
| ENSMUSG00000008999 | Bmp7 | 1.585592 | 0.0040494 | 0.0454821 | bone morphogenetic protein 7 [Source:MGI Symbol;Acc:MGI:103302] |
| ENSMUSG00000003882 | Il7r | 2.257016 | 0.0040536 | 0.0454821 | interleukin 7 receptor [Source:MGI Symbol;Acc:MGI:96562] |
| ENSMUSG00000023274 | Cd4 | 2.413573 | 0.0040842 | 0.0457300 | CD4 antigen [Source:MGI Symbol;Acc:MGI:88335] |
Pathview
FC=1.1
p=p+1Top 20
enrichKEGG_sig[[p]][1:20, ] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Count | GeneRatio | pvalue | logPval | p.adjust | qvalue | Description | geneID | |
|---|---|---|---|---|---|---|---|---|
| mmu04713 | 24 | 0.2448980 | 0.00e+00 | 9.071978 | 0.00e+00 | 0.0000002 | Circadian entrainment | 16519/14802/12288/20190/14677/234889/14812/104111/14710/18795/54195/60596/104110/66066/11513/11517/14702/73086/18796/20192/58226/14708/14697/14696 |
| mmu04371 | 26 | 0.1897810 | 1.00e-07 | 7.277696 | 1.00e-07 | 0.0000064 | Apelin signaling pathway | 20190/18127/20541/14677/17260/104111/14710/104709/110893/18795/104110/66066/23796/11513/11607/17897/14702/18796/20192/14219/30878/14708/23797/14697/14696/16890 |
| mmu04724 | 23 | 0.2035398 | 1.00e-07 | 7.087147 | 1.00e-07 | 0.0000066 | Glutamatergic synapse | 16519/20512/14802/14805/12288/14677/14812/104111/210274/14710/18795/104110/66066/11513/14806/14702/18796/105727/13385/14708/14697/14696/12286 |
| mmu05414 | 20 | 0.2127660 | 3.00e-07 | 6.568740 | 3.00e-07 | 0.0000164 | Dilated cardiomyopathy | 24052/12288/16401/16000/20541/16421/12293/104111/110893/109700/104110/16773/11513/16404/17897/81904/24051/241226/192897/12298 |
| mmu04514 | 29 | 0.1593407 | 5.00e-07 | 6.328504 | 5.00e-07 | 0.0000228 | Cell adhesion molecules | 16401/241568/19266/192167/12558/272381/16421/15894/83964/54420/386750/12562/19274/225825/245537/69524/16408/14961/269116/20339/15002/18613/14969/12741/18007/14960/80883/241226/12737 |
| mmu05412 | 17 | 0.2207792 | 1.20e-06 | 5.909386 | 1.20e-06 | 0.0000499 | Arrhythmogenic right ventricular cardiomyopathy | 16842/24052/12288/16401/20541/12558/16421/12293/110893/109700/16773/16404/81904/24051/241226/192897/12298 |
| mmu04022 | 27 | 0.1560694 | 1.80e-06 | 5.751113 | 1.80e-06 | 0.0000616 | cGMP-PKG signaling pathway | 12288/11549/18127/20541/14677/13618/17260/234889/104111/104709/110893/18795/54195/22068/60596/104110/11513/11607/16531/13617/12062/18796/242202/73181/381290/23797/98660 |
| mmu05032 | 18 | 0.1978022 | 3.30e-06 | 5.486280 | 3.30e-06 | 0.0000949 | Morphine addiction | 16519/14677/18574/104111/14710/14397/104110/66066/11513/18573/14702/109689/216869/14708/14697/14696/54393/12286 |
| mmu04340 | 14 | 0.2413793 | 3.50e-06 | 5.454437 | 3.50e-06 | 0.0000949 | Hedgehog signaling pathway | 14632/16147/15245/68525/19207/59056/16576/12444/14634/14725/240888/109689/19206/216869 |
| mmu04725 | 20 | 0.1785714 | 4.90e-06 | 5.306682 | 4.90e-06 | 0.0001200 | Cholinergic synapse | 16519/12288/14677/226922/104111/14710/104709/18795/104110/66066/11513/14702/18796/14360/14708/23797/14697/14696/60613/12286 |
| mmu04512 | 17 | 0.1931818 | 8.50e-06 | 5.068903 | 8.50e-06 | 0.0001886 | ECM-receptor interaction | 16401/94216/114249/21827/23928/12505/16775/16421/14268/109700/12840/329872/16773/16404/75209/241226/192897 |
| mmu04926 | 21 | 0.1627907 | 1.29e-05 | 4.888815 | 1.29e-05 | 0.0002207 | Relaxin signaling pathway | 94216/18127/14677/13618/17390/20418/104111/14710/18795/104110/66066/11513/22341/14702/109689/18796/216869/14708/23797/14697/14696 |
| mmu04015 | 29 | 0.1355140 | 1.31e-05 | 4.881899 | 1.31e-05 | 0.0002207 | Rap1 signaling pathway | 15234/11600/16542/16000/14026/240168/14677/18645/14812/78134/18760/21687/104111/18795/19395/104110/16590/11513/16408/22341/50791/106952/93737/17311/18796/70549/23797/16797/54519 |
| mmu04062 | 27 | 0.1406250 | 1.32e-05 | 4.879785 | 1.32e-05 | 0.0002207 | Chemokine signaling pathway | 20309/14677/20418/16428/12766/20315/104111/14710/104709/18795/19395/94176/104110/66066/11513/16963/14702/109689/18796/23832/216869/14708/14191/20851/23797/14697/14696 |
| mmu05410 | 17 | 0.1868132 | 1.36e-05 | 4.865970 | 1.36e-05 | 0.0002207 | Hypertrophic cardiomyopathy | 24052/12288/16401/16000/20541/16421/12293/110893/109700/16773/16404/17897/81904/24051/241226/192897/12298 |
| mmu04020 | 31 | 0.1291667 | 1.76e-05 | 4.753641 | 1.76e-05 | 0.0002680 | Calcium signaling pathway | 15234/12288/20190/16542/11549/18127/20541/231602/13618/18574/104111/12290/19220/110893/18795/104110/11513/22341/18573/12494/11607/13617/12062/15558/18796/65973/20192/58226/19218/381290/12286 |
| mmu04970 | 16 | 0.1882353 | 2.20e-05 | 4.656709 | 2.20e-05 | 0.0002979 | Salivary secretion | 11549/234889/104111/18795/54195/60596/104110/11513/12494/17105/16531/18796/20192/17110/381290/98660 |
| mmu04924 | 15 | 0.1973684 | 2.20e-05 | 4.656604 | 2.20e-05 | 0.0002979 | Renin secretion | 12288/14677/234889/18574/13616/18795/54195/60596/18573/11607/16531/13617/11517/18796/229933 |
| mmu04728 | 21 | 0.1555556 | 2.63e-05 | 4.579580 | 2.63e-05 | 0.0003369 | Dopaminergic synapse | 16519/14802/109731/12288/13491/14677/20265/14812/17161/14710/18795/66066/14702/109689/18796/216869/14708/23797/14697/14696/12286 |
| mmu04510 | 27 | 0.1343284 | 3.05e-05 | 4.515260 | 3.05e-05 | 0.0003712 | Focal adhesion | 15234/16401/16542/16000/94216/21827/23928/16775/16421/12444/14268/20418/18719/109700/12840/16773/22341/64099/16404/170736/68794/70549/14360/23797/12389/241226/192897 |
Dot plot
kegg_dot[[p]]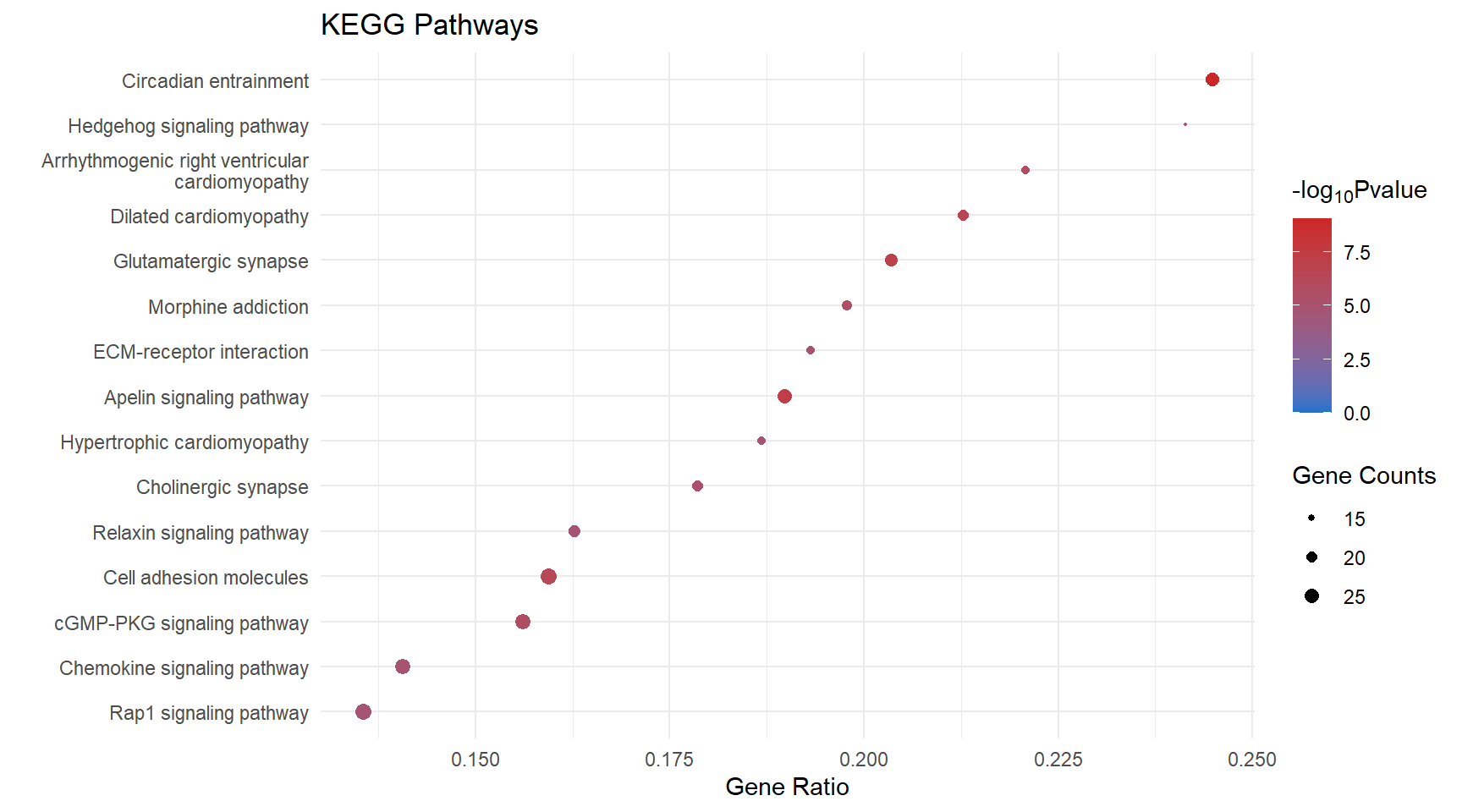
Upset Plot
upset[[p]]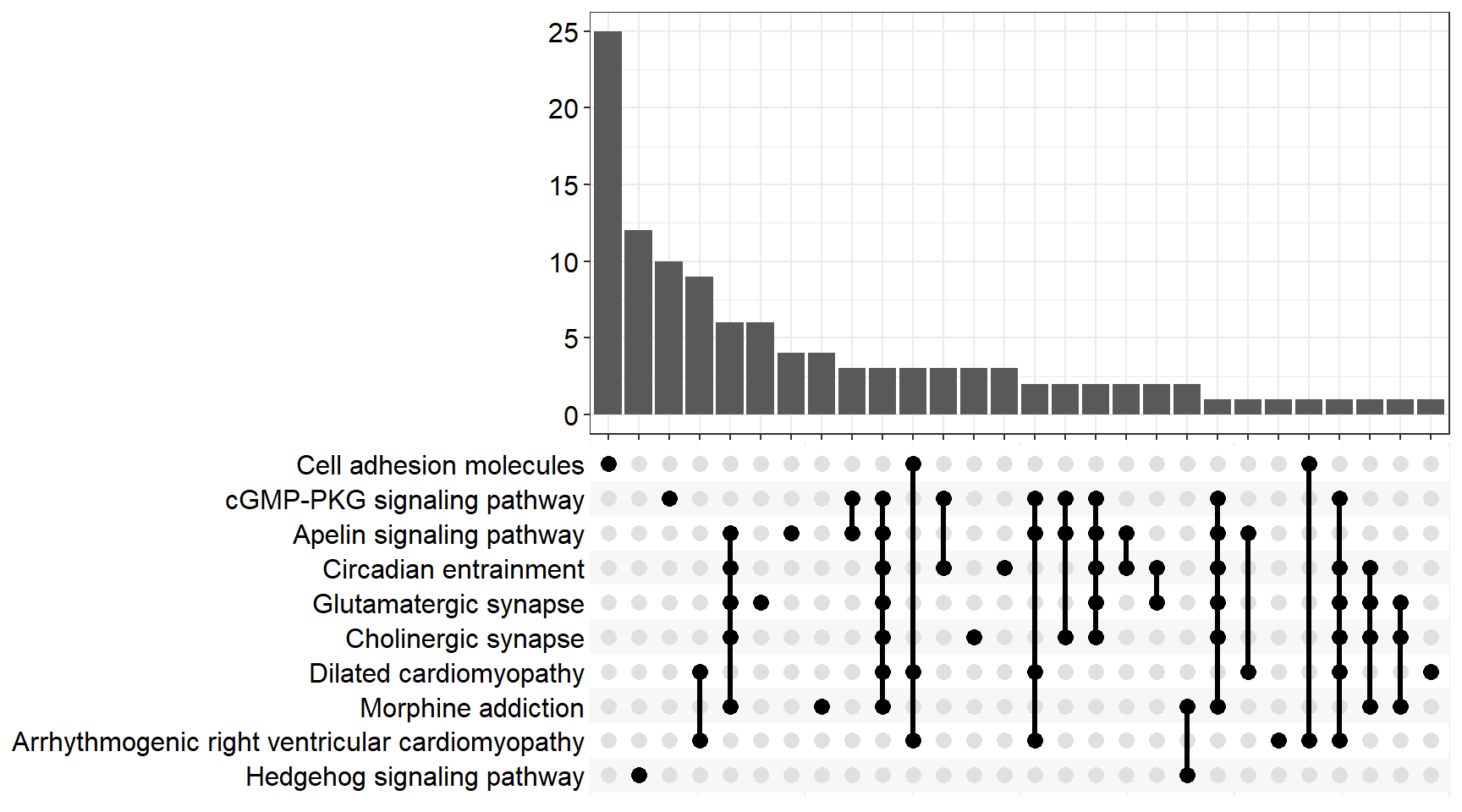
Leukocyte transendothelial migration
q=1Heatmap
kegg_heat[[p]][[q]]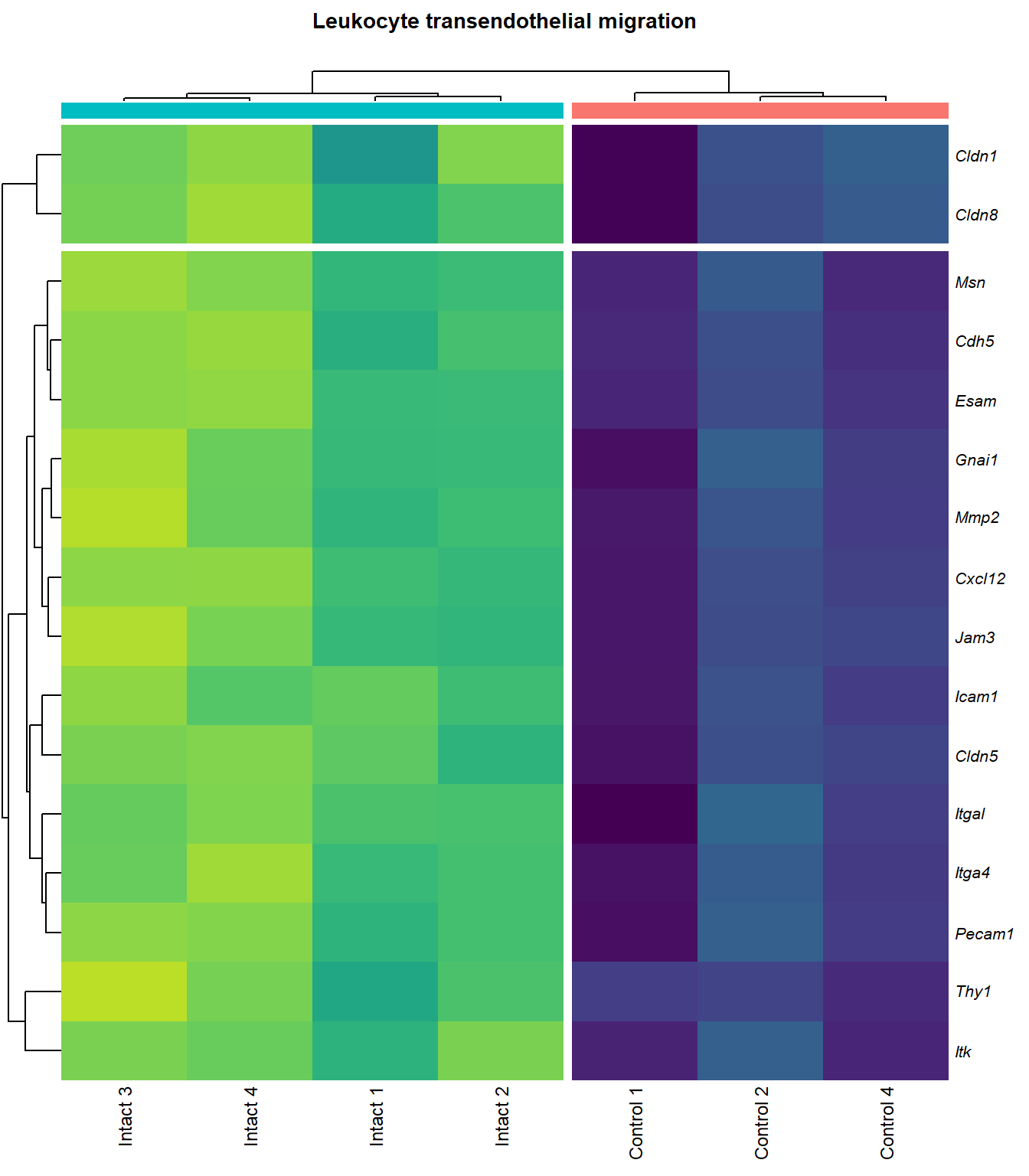
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000191 | 0.0028514 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000057614 | Gnai1 | 2.405667 | 0.0001147 | 0.0063953 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 [Source:MGI Symbol;Acc:MGI:95771] |
| ENSMUSG00000031740 | Mmp2 | 2.270956 | 0.0001526 | 0.0071629 | matrix metallopeptidase 2 [Source:MGI Symbol;Acc:MGI:97009] |
| ENSMUSG00000037405 | Icam1 | 1.989282 | 0.0001885 | 0.0082607 | intercellular adhesion molecule 1 [Source:MGI Symbol;Acc:MGI:96392] |
| ENSMUSG00000020395 | Itk | 2.082706 | 0.0002206 | 0.0087952 | IL2 inducible T cell kinase [Source:MGI Symbol;Acc:MGI:96621] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0002222 | 0.0088088 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| ENSMUSG00000031990 | Jam3 | 1.932372 | 0.0002299 | 0.0089179 | junction adhesion molecule 3 [Source:MGI Symbol;Acc:MGI:1933825] |
| ENSMUSG00000050520 | Cldn8 | 1.948402 | 0.0003180 | 0.0110875 | claudin 8 [Source:MGI Symbol;Acc:MGI:1859286] |
| ENSMUSG00000031871 | Cdh5 | 2.498728 | 0.0004399 | 0.0132230 | cadherin 5 [Source:MGI Symbol;Acc:MGI:105057] |
| ENSMUSG00000032011 | Thy1 | 1.783613 | 0.0004934 | 0.0139533 | thymus cell antigen 1, theta [Source:MGI Symbol;Acc:MGI:98747] |
| ENSMUSG00000031207 | Msn | 1.375211 | 0.0005939 | 0.0153674 | moesin [Source:MGI Symbol;Acc:MGI:97167] |
| ENSMUSG00000001946 | Esam | 1.936624 | 0.0007470 | 0.0176708 | endothelial cell-specific adhesion molecule [Source:MGI Symbol;Acc:MGI:1916774] |
| ENSMUSG00000030830 | Itgal | 1.816463 | 0.0007591 | 0.0178553 | integrin alpha L [Source:MGI Symbol;Acc:MGI:96606] |
| ENSMUSG00000020717 | Pecam1 | 1.731782 | 0.0010578 | 0.0211587 | platelet/endothelial cell adhesion molecule 1 [Source:MGI Symbol;Acc:MGI:97537] |
| ENSMUSG00000041378 | Cldn5 | 1.919823 | 0.0021023 | 0.0324715 | claudin 5 [Source:MGI Symbol;Acc:MGI:1276112] |
| ENSMUSG00000022512 | Cldn1 | 1.838024 | 0.0041828 | 0.0498233 | claudin 1 [Source:MGI Symbol;Acc:MGI:1276109] |
Pathview
Hematopoietic cell lineage
q=q+1Heatmap
kegg_heat[[p]][[q]]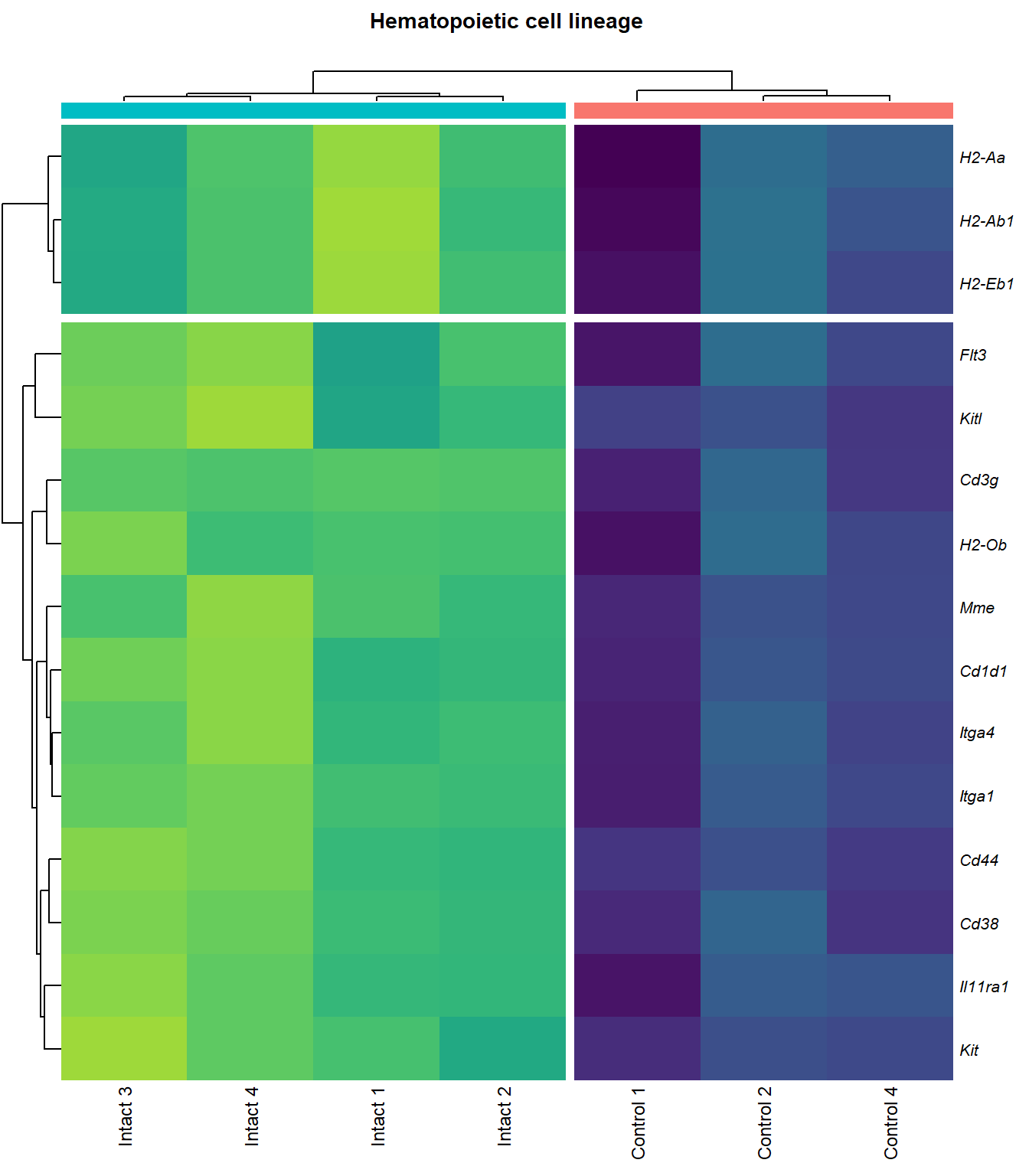
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000191 | 0.0028514 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000005087 | Cd44 | 2.271713 | 0.0000838 | 0.0053858 | CD44 antigen [Source:MGI Symbol;Acc:MGI:88338] |
| ENSMUSG00000073889 | Il11ra1 | 1.542982 | 0.0001432 | 0.0068940 | interleukin 11 receptor, alpha chain 1 [Source:MGI Symbol;Acc:MGI:107426] |
| ENSMUSG00000002033 | Cd3g | 3.645674 | 0.0002234 | 0.0088088 | CD3 antigen, gamma polypeptide [Source:MGI Symbol;Acc:MGI:88333] |
| ENSMUSG00000042284 | Itga1 | 1.938967 | 0.0003646 | 0.0117800 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000042817 | Flt3 | 2.075768 | 0.0004687 | 0.0137336 | FMS-like tyrosine kinase 3 [Source:MGI Symbol;Acc:MGI:95559] |
| ENSMUSG00000027820 | Mme | 3.737788 | 0.0005576 | 0.0148797 | membrane metallo endopeptidase [Source:MGI Symbol;Acc:MGI:97004] |
| ENSMUSG00000028076 | Cd1d1 | 2.344493 | 0.0005609 | 0.0148797 | CD1d1 antigen [Source:MGI Symbol;Acc:MGI:107674] |
| ENSMUSG00000005672 | Kit | 1.518478 | 0.0006666 | 0.0164221 | KIT proto-oncogene receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:96677] |
| ENSMUSG00000073421 | H2-Ab1 | 1.742382 | 0.0007707 | 0.0178553 | histocompatibility 2, class II antigen A, beta 1 [Source:MGI Symbol;Acc:MGI:103070] |
| ENSMUSG00000029084 | Cd38 | 1.711230 | 0.0008968 | 0.0193746 | CD38 antigen [Source:MGI Symbol;Acc:MGI:107474] |
| ENSMUSG00000041538 | H2-Ob | 1.716311 | 0.0009822 | 0.0203799 | histocompatibility 2, O region beta locus [Source:MGI Symbol;Acc:MGI:95925] |
| ENSMUSG00000019966 | Kitl | 1.310648 | 0.0013846 | 0.0249431 | kit ligand [Source:MGI Symbol;Acc:MGI:96974] |
| ENSMUSG00000060586 | H2-Eb1 | 1.690764 | 0.0016090 | 0.0274087 | histocompatibility 2, class II antigen E beta [Source:MGI Symbol;Acc:MGI:95901] |
| ENSMUSG00000036594 | H2-Aa | 1.672690 | 0.0024922 | 0.0360270 | histocompatibility 2, class II antigen A, alpha [Source:MGI Symbol;Acc:MGI:95895] |
Pathview
Vascular smooth muscle contraction
q=q+1Heatmap
kegg_heat[[p]][[q]]
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000051331 | Cacna1c | 1.120969 | 0.0000172 | 0.0028514 | calcium channel, voltage-dependent, L type, alpha 1C subunit [Source:MGI Symbol;Acc:MGI:103013] |
| ENSMUSG00000022683 | Pla2g10 | 5.773220 | 0.0000185 | 0.0028514 | phospholipase A2, group X [Source:MGI Symbol;Acc:MGI:1347522] |
| ENSMUSG00000045875 | Adra1a | 4.131590 | 0.0000588 | 0.0047985 | adrenergic receptor, alpha 1a [Source:MGI Symbol;Acc:MGI:104773] |
| ENSMUSG00000041624 | Gucy1a2 | 2.199471 | 0.0001902 | 0.0082833 | guanylate cyclase 1, soluble, alpha 2 [Source:MGI Symbol;Acc:MGI:2660877] |
| ENSMUSG00000020654 | Adcy3 | 1.648297 | 0.0002399 | 0.0092013 | adenylate cyclase 3 [Source:MGI Symbol;Acc:MGI:99675] |
| ENSMUSG00000027524 | Edn3 | 3.849271 | 0.0003134 | 0.0110143 | endothelin 3 [Source:MGI Symbol;Acc:MGI:95285] |
| ENSMUSG00000030669 | Calca | 5.762174 | 0.0003731 | 0.0119508 | calcitonin/calcitonin-related polypeptide, alpha [Source:MGI Symbol;Acc:MGI:2151253] |
| ENSMUSG00000051177 | Plcb1 | 1.960299 | 0.0003744 | 0.0119561 | phospholipase C, beta 1 [Source:MGI Symbol;Acc:MGI:97613] |
| ENSMUSG00000028005 | Gucy1b1 | 1.861114 | 0.0004861 | 0.0139237 | guanylate cyclase 1, soluble, beta 1 [Source:MGI Symbol;Acc:MGI:1860604] |
| ENSMUSG00000033910 | Gucy1a1 | 1.985306 | 0.0005943 | 0.0153674 | guanylate cyclase 1, soluble, alpha 1 [Source:MGI Symbol;Acc:MGI:1926562] |
| ENSMUSG00000022220 | Adcy4 | 1.867108 | 0.0006410 | 0.0161149 | adenylate cyclase 4 [Source:MGI Symbol;Acc:MGI:99674] |
| ENSMUSG00000031659 | Adcy7 | 1.309423 | 0.0007469 | 0.0176708 | adenylate cyclase 7 [Source:MGI Symbol;Acc:MGI:102891] |
| ENSMUSG00000049115 | Agtr1a | 1.761758 | 0.0008988 | 0.0193746 | angiotensin II receptor, type 1a [Source:MGI Symbol;Acc:MGI:87964] |
| ENSMUSG00000063142 | Kcnma1 | 2.434426 | 0.0009192 | 0.0195760 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 [Source:MGI Symbol;Acc:MGI:99923] |
| ENSMUSG00000031616 | Ednra | 1.779614 | 0.0010153 | 0.0206446 | endothelin receptor type A [Source:MGI Symbol;Acc:MGI:105923] |
| ENSMUSG00000040061 | Plcb2 | 1.498256 | 0.0018527 | 0.0299595 | phospholipase C, beta 2 [Source:MGI Symbol;Acc:MGI:107465] |
| ENSMUSG00000059588 | Calcrl | 1.494208 | 0.0022567 | 0.0340616 | calcitonin receptor-like [Source:MGI Symbol;Acc:MGI:1926944] |
Pathview
PI3K-Akt signaling pathway
q=q+1Heatmap
kegg_heat[[p]][[q]]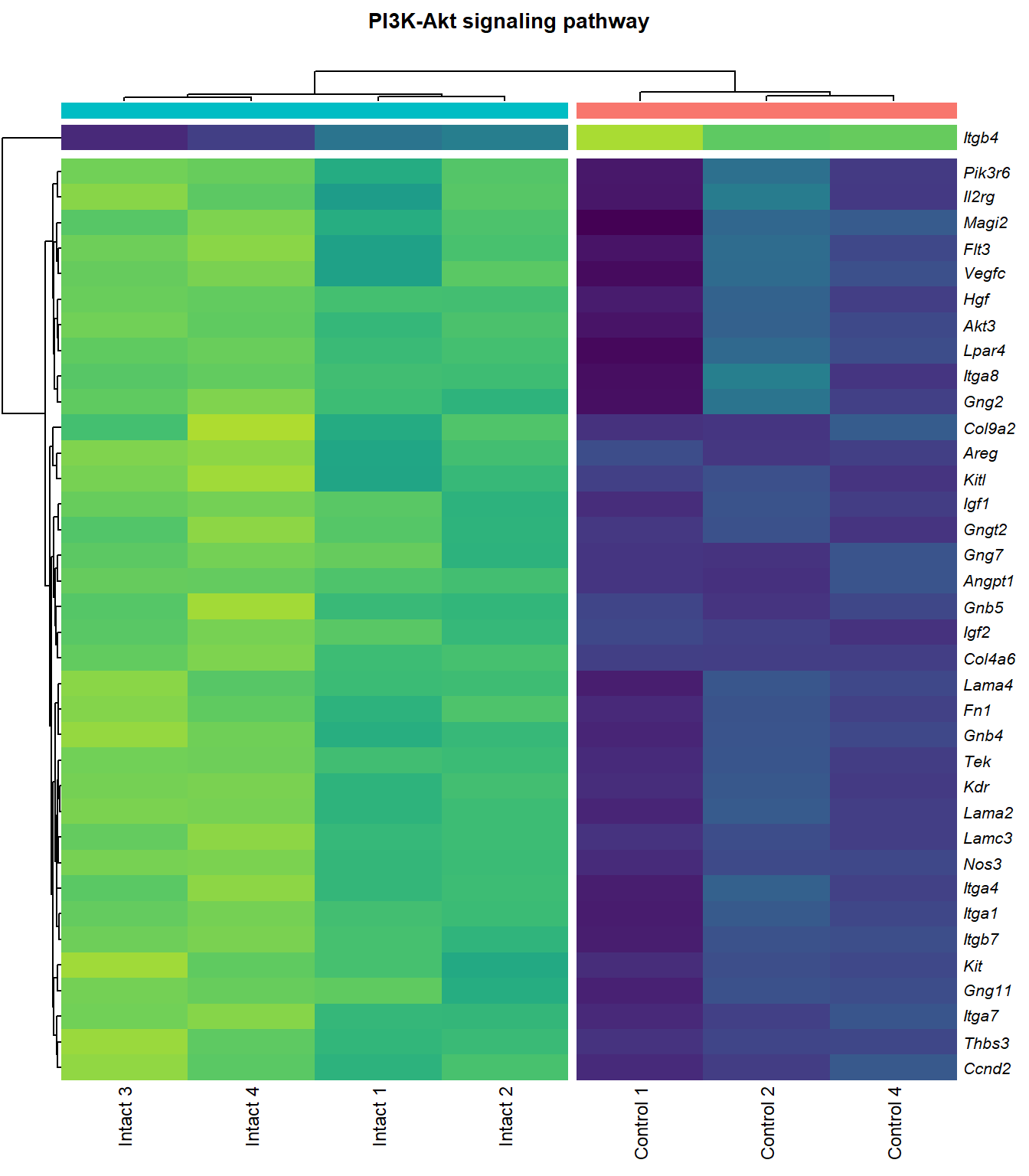
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.125312 | 0.0000047 | 0.0015780 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000022309 | Angpt1 | 3.388586 | 0.0000111 | 0.0023074 | angiopoietin 1 [Source:MGI Symbol;Acc:MGI:108448] |
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000191 | 0.0028514 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.045643 | 0.0000277 | 0.0033785 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000020053 | Igf1 | 2.780445 | 0.0000345 | 0.0036412 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000031273 | Col4a6 | 3.466810 | 0.0000439 | 0.0041268 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000028047 | Thbs3 | 1.673809 | 0.0000604 | 0.0048021 | thrombospondin 3 [Source:MGI Symbol;Acc:MGI:98739] |
| ENSMUSG00000028978 | Nos3 | 2.148682 | 0.0000687 | 0.0051480 | nitric oxide synthase 3, endothelial cell [Source:MGI Symbol;Acc:MGI:97362] |
| ENSMUSG00000026840 | Lamc3 | 3.803842 | 0.0000826 | 0.0053543 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000019846 | Lama4 | 1.940280 | 0.0000999 | 0.0059120 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
| ENSMUSG00000029378 | Areg | 5.246523 | 0.0001339 | 0.0067067 | amphiregulin [Source:MGI Symbol;Acc:MGI:88068] |
| ENSMUSG00000001281 | Itgb7 | 1.806291 | 0.0001350 | 0.0067067 | integrin beta 7 [Source:MGI Symbol;Acc:MGI:96616] |
| ENSMUSG00000000184 | Ccnd2 | 1.705391 | 0.0001366 | 0.0067339 | cyclin D2 [Source:MGI Symbol;Acc:MGI:88314] |
| ENSMUSG00000026193 | Fn1 | 1.969689 | 0.0001369 | 0.0067339 | fibronectin 1 [Source:MGI Symbol;Acc:MGI:95566] |
| ENSMUSG00000049929 | Lpar4 | 2.146074 | 0.0001922 | 0.0082958 | lysophosphatidic acid receptor 4 [Source:MGI Symbol;Acc:MGI:1925384] |
| ENSMUSG00000006386 | Tek | 2.339034 | 0.0002206 | 0.0087952 | TEK receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:98664] |
| ENSMUSG00000038811 | Gngt2 | 1.819658 | 0.0002668 | 0.0098590 | guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 [Source:MGI Symbol;Acc:MGI:893584] |
| ENSMUSG00000046207 | Pik3r6 | 1.692425 | 0.0003324 | 0.0113962 | phosphoinositide-3-kinase regulatory subunit 5 [Source:MGI Symbol;Acc:MGI:2144613] |
| ENSMUSG00000042284 | Itga1 | 1.938967 | 0.0003646 | 0.0117800 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000028626 | Col9a2 | 4.129759 | 0.0003733 | 0.0119508 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000042817 | Flt3 | 2.075768 | 0.0004687 | 0.0137336 | FMS-like tyrosine kinase 3 [Source:MGI Symbol;Acc:MGI:95559] |
| ENSMUSG00000032766 | Gng11 | 2.019229 | 0.0006551 | 0.0163107 | guanine nucleotide binding protein (G protein), gamma 11 [Source:MGI Symbol;Acc:MGI:1913316] |
| ENSMUSG00000005672 | Kit | 1.518478 | 0.0006666 | 0.0164221 | KIT proto-oncogene receptor tyrosine kinase [Source:MGI Symbol;Acc:MGI:96677] |
| ENSMUSG00000019899 | Lama2 | 1.833935 | 0.0006765 | 0.0165713 | laminin, alpha 2 [Source:MGI Symbol;Acc:MGI:99912] |
| ENSMUSG00000031520 | Vegfc | 2.124547 | 0.0008424 | 0.0189346 | vascular endothelial growth factor C [Source:MGI Symbol;Acc:MGI:109124] |
| ENSMUSG00000040003 | Magi2 | 1.523070 | 0.0009834 | 0.0203802 | membrane associated guanylate kinase, WW and PDZ domain containing 2 [Source:MGI Symbol;Acc:MGI:1354953] |
| ENSMUSG00000048583 | Igf2 | 2.426940 | 0.0010153 | 0.0206446 | insulin-like growth factor 2 [Source:MGI Symbol;Acc:MGI:96434] |
| ENSMUSG00000025348 | Itga7 | 1.579782 | 0.0011908 | 0.0226598 | integrin alpha 7 [Source:MGI Symbol;Acc:MGI:102700] |
| ENSMUSG00000043004 | Gng2 | 1.239057 | 0.0012599 | 0.0235328 | guanine nucleotide binding protein (G protein), gamma 2 [Source:MGI Symbol;Acc:MGI:102705] |
| ENSMUSG00000019966 | Kitl | 1.310648 | 0.0013846 | 0.0249431 | kit ligand [Source:MGI Symbol;Acc:MGI:96974] |
| ENSMUSG00000048240 | Gng7 | 2.309381 | 0.0024454 | 0.0355698 | guanine nucleotide binding protein (G protein), gamma 7 [Source:MGI Symbol;Acc:MGI:95787] |
| ENSMUSG00000019699 | Akt3 | 1.200109 | 0.0027128 | 0.0380460 | thymoma viral proto-oncogene 3 [Source:MGI Symbol;Acc:MGI:1345147] |
| ENSMUSG00000032192 | Gnb5 | 1.034006 | 0.0030610 | 0.0413459 | guanine nucleotide binding protein (G protein), beta 5 [Source:MGI Symbol;Acc:MGI:101848] |
| ENSMUSG00000027669 | Gnb4 | 1.157545 | 0.0035385 | 0.0449227 | guanine nucleotide binding protein (G protein), beta 4 [Source:MGI Symbol;Acc:MGI:104581] |
| ENSMUSG00000031304 | Il2rg | 1.883060 | 0.0037085 | 0.0462470 | interleukin 2 receptor, gamma chain [Source:MGI Symbol;Acc:MGI:96551] |
| ENSMUSG00000026768 | Itga8 | 1.587301 | 0.0038730 | 0.0475073 | integrin alpha 8 [Source:MGI Symbol;Acc:MGI:109442] |
| ENSMUSG00000020758 | Itgb4 | -1.499047 | 0.0041179 | 0.0494897 | integrin beta 4 [Source:MGI Symbol;Acc:MGI:96613] |
Pathview
Focal adhesion
q=q+1Heatmap
kegg_heat[[p]][[q]]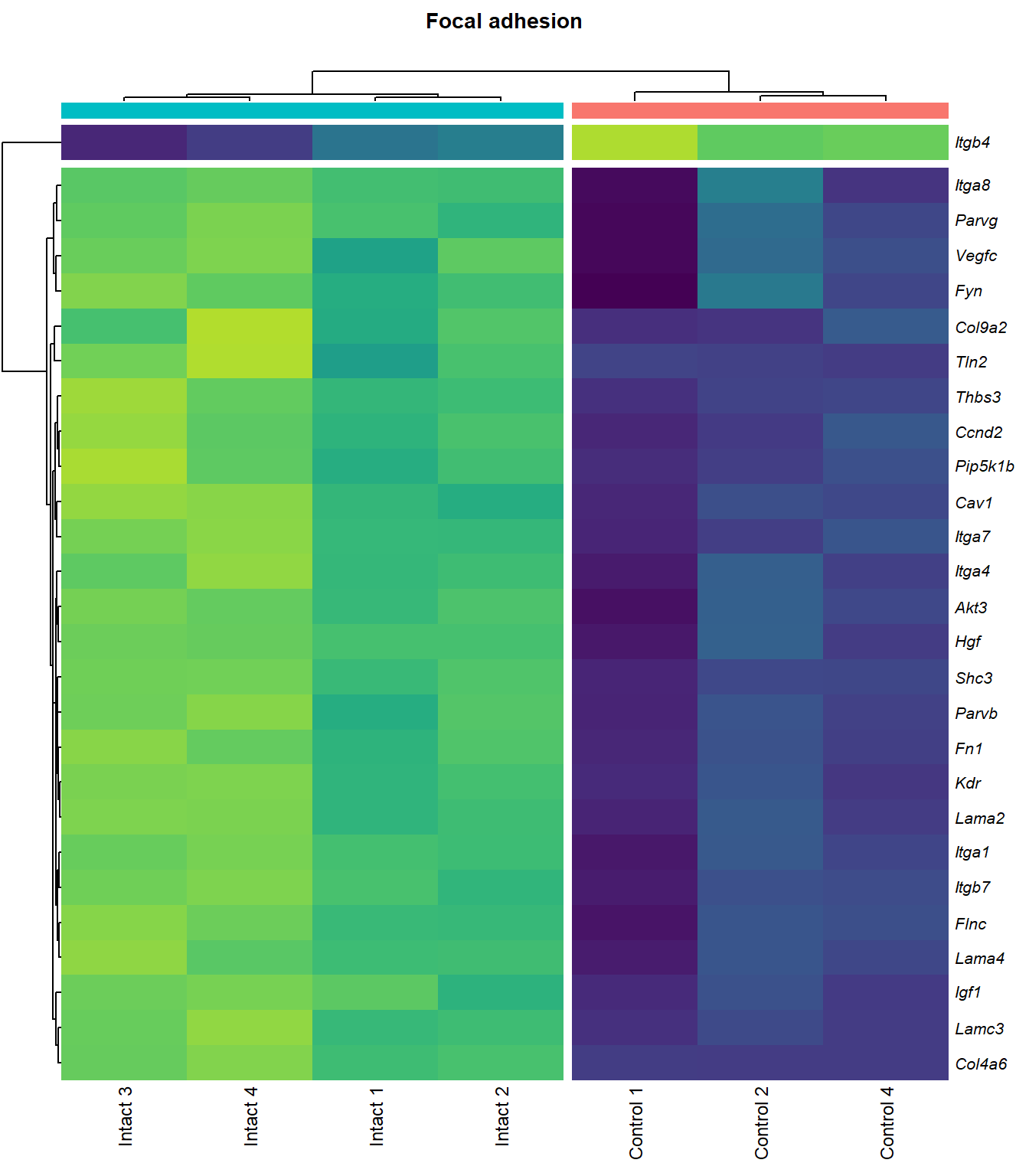
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.125312 | 0.0000047 | 0.0015780 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0000191 | 0.0028514 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.045643 | 0.0000277 | 0.0033785 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000020053 | Igf1 | 2.780445 | 0.0000345 | 0.0036412 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000031273 | Col4a6 | 3.466810 | 0.0000439 | 0.0041268 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000028047 | Thbs3 | 1.673809 | 0.0000604 | 0.0048021 | thrombospondin 3 [Source:MGI Symbol;Acc:MGI:98739] |
| ENSMUSG00000026840 | Lamc3 | 3.803842 | 0.0000826 | 0.0053543 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000019846 | Lama4 | 1.940280 | 0.0000999 | 0.0059120 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
| ENSMUSG00000001281 | Itgb7 | 1.806291 | 0.0001350 | 0.0067067 | integrin beta 7 [Source:MGI Symbol;Acc:MGI:96616] |
| ENSMUSG00000000184 | Ccnd2 | 1.705391 | 0.0001366 | 0.0067339 | cyclin D2 [Source:MGI Symbol;Acc:MGI:88314] |
| ENSMUSG00000026193 | Fn1 | 1.969689 | 0.0001369 | 0.0067339 | fibronectin 1 [Source:MGI Symbol;Acc:MGI:95566] |
| ENSMUSG00000021448 | Shc3 | 2.149036 | 0.0001593 | 0.0073164 | src homology 2 domain-containing transforming protein C3 [Source:MGI Symbol;Acc:MGI:106179] |
| ENSMUSG00000024867 | Pip5k1b | 1.500364 | 0.0001979 | 0.0083928 | phosphatidylinositol-4-phosphate 5-kinase, type 1 beta [Source:MGI Symbol;Acc:MGI:107930] |
| ENSMUSG00000042284 | Itga1 | 1.938967 | 0.0003646 | 0.0117800 | integrin alpha 1 [Source:MGI Symbol;Acc:MGI:96599] |
| ENSMUSG00000028626 | Col9a2 | 4.129759 | 0.0003733 | 0.0119508 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000019899 | Lama2 | 1.833935 | 0.0006765 | 0.0165713 | laminin, alpha 2 [Source:MGI Symbol;Acc:MGI:99912] |
| ENSMUSG00000031520 | Vegfc | 2.124547 | 0.0008424 | 0.0189346 | vascular endothelial growth factor C [Source:MGI Symbol;Acc:MGI:109124] |
| ENSMUSG00000022439 | Parvg | 1.770609 | 0.0011712 | 0.0225219 | parvin, gamma [Source:MGI Symbol;Acc:MGI:2158329] |
| ENSMUSG00000025348 | Itga7 | 1.579782 | 0.0011908 | 0.0226598 | integrin alpha 7 [Source:MGI Symbol;Acc:MGI:102700] |
| ENSMUSG00000022438 | Parvb | 1.642556 | 0.0014600 | 0.0259169 | parvin, beta [Source:MGI Symbol;Acc:MGI:2153063] |
| ENSMUSG00000068699 | Flnc | 1.179774 | 0.0018739 | 0.0300212 | filamin C, gamma [Source:MGI Symbol;Acc:MGI:95557] |
| ENSMUSG00000052698 | Tln2 | 1.244836 | 0.0019261 | 0.0305158 | talin 2 [Source:MGI Symbol;Acc:MGI:1917799] |
| ENSMUSG00000019843 | Fyn | 1.178665 | 0.0023041 | 0.0344884 | Fyn proto-oncogene [Source:MGI Symbol;Acc:MGI:95602] |
| ENSMUSG00000019699 | Akt3 | 1.200109 | 0.0027128 | 0.0380460 | thymoma viral proto-oncogene 3 [Source:MGI Symbol;Acc:MGI:1345147] |
| ENSMUSG00000007655 | Cav1 | 1.601119 | 0.0036517 | 0.0457046 | caveolin 1, caveolae protein [Source:MGI Symbol;Acc:MGI:102709] |
| ENSMUSG00000026768 | Itga8 | 1.587301 | 0.0038730 | 0.0475073 | integrin alpha 8 [Source:MGI Symbol;Acc:MGI:109442] |
| ENSMUSG00000020758 | Itgb4 | -1.499047 | 0.0041179 | 0.0494897 | integrin beta 4 [Source:MGI Symbol;Acc:MGI:96613] |
Pathview
Cytokine-cytokine receptor interaction
q=q+1Heatmap
kegg_heat[[p]][[q]]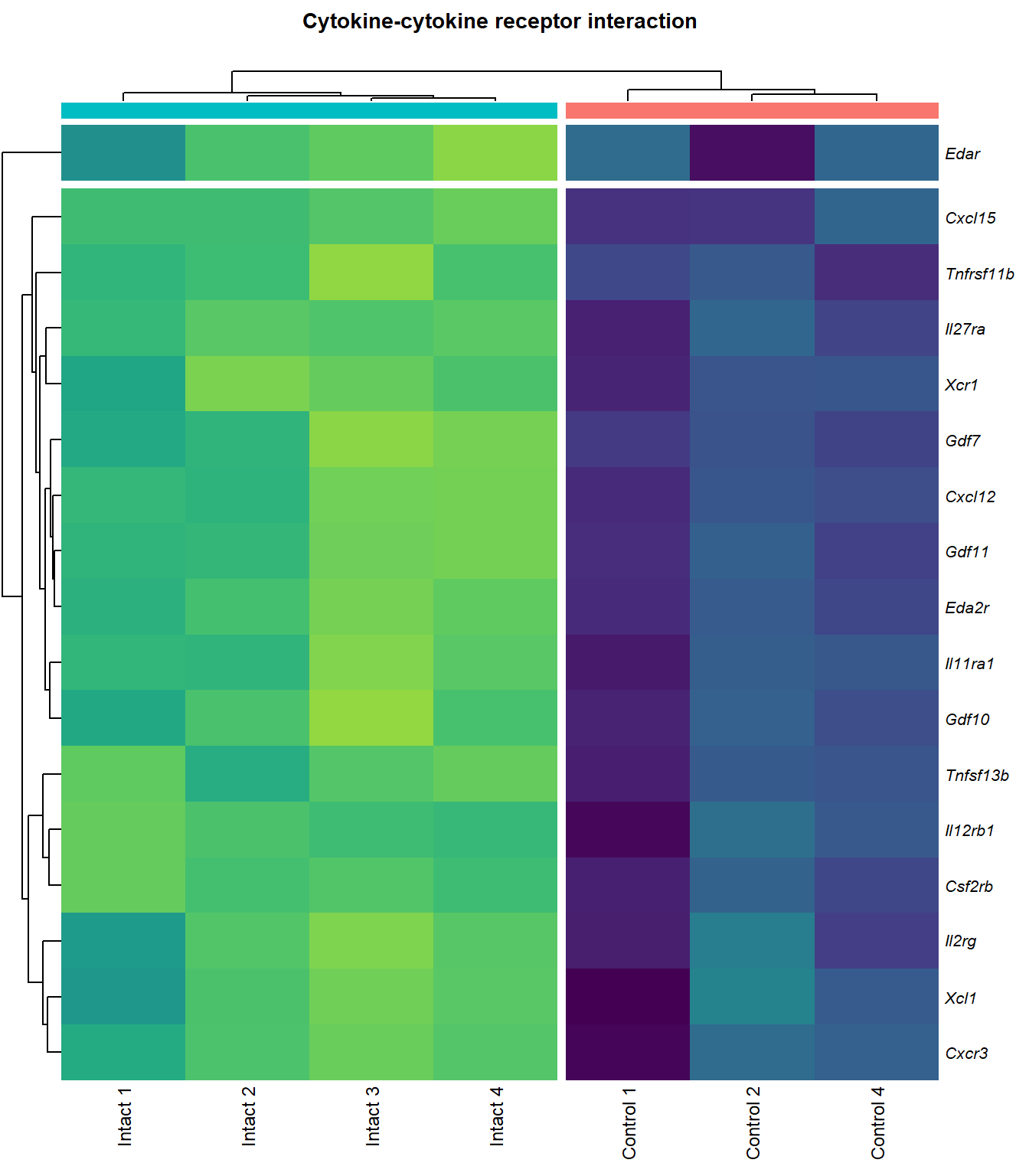
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000029375 | Cxcl15 | 6.916834 | 0.0000000 | 0.0001526 | chemokine (C-X-C motif) ligand 15 [Source:MGI Symbol;Acc:MGI:1339941] |
| ENSMUSG00000034457 | Eda2r | 3.524309 | 0.0000414 | 0.0040532 | ectodysplasin A2 receptor [Source:MGI Symbol;Acc:MGI:2442860] |
| ENSMUSG00000073889 | Il11ra1 | 1.542982 | 0.0001432 | 0.0068940 | interleukin 11 receptor, alpha chain 1 [Source:MGI Symbol;Acc:MGI:107426] |
| ENSMUSG00000050232 | Cxcr3 | 2.280257 | 0.0002218 | 0.0088088 | chemokine (C-X-C motif) receptor 3 [Source:MGI Symbol;Acc:MGI:1277207] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0002222 | 0.0088088 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| ENSMUSG00000025352 | Gdf11 | 2.251448 | 0.0002756 | 0.0100961 | growth differentiation factor 11 [Source:MGI Symbol;Acc:MGI:1338027] |
| ENSMUSG00000037660 | Gdf7 | 3.410398 | 0.0004216 | 0.0129205 | growth differentiation factor 7 [Source:MGI Symbol;Acc:MGI:95690] |
| ENSMUSG00000031497 | Tnfsf13b | 2.218370 | 0.0004882 | 0.0139237 | tumor necrosis factor (ligand) superfamily, member 13b [Source:MGI Symbol;Acc:MGI:1344376] |
| ENSMUSG00000021943 | Gdf10 | 2.741543 | 0.0005614 | 0.0148797 | growth differentiation factor 10 [Source:MGI Symbol;Acc:MGI:95684] |
| ENSMUSG00000005465 | Il27ra | 1.937581 | 0.0007697 | 0.0178553 | interleukin 27 receptor, alpha [Source:MGI Symbol;Acc:MGI:1355318] |
| ENSMUSG00000063727 | Tnfrsf11b | 2.131252 | 0.0008294 | 0.0187463 | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) [Source:MGI Symbol;Acc:MGI:109587] |
| ENSMUSG00000026573 | Xcl1 | 2.671247 | 0.0011830 | 0.0225865 | chemokine (C motif) ligand 1 [Source:MGI Symbol;Acc:MGI:104593] |
| ENSMUSG00000060509 | Xcr1 | 2.001440 | 0.0019520 | 0.0308129 | chemokine (C motif) receptor 1 [Source:MGI Symbol;Acc:MGI:1346338] |
| ENSMUSG00000071713 | Csf2rb | 2.031328 | 0.0019826 | 0.0310486 | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) [Source:MGI Symbol;Acc:MGI:1339759] |
| ENSMUSG00000000791 | Il12rb1 | 2.130604 | 0.0033775 | 0.0436691 | interleukin 12 receptor, beta 1 [Source:MGI Symbol;Acc:MGI:104579] |
| ENSMUSG00000031304 | Il2rg | 1.883060 | 0.0037085 | 0.0462470 | interleukin 2 receptor, gamma chain [Source:MGI Symbol;Acc:MGI:96551] |
| ENSMUSG00000003227 | Edar | 2.573653 | 0.0040571 | 0.0490666 | ectodysplasin-A receptor [Source:MGI Symbol;Acc:MGI:1343498] |
Pathview
FC=1.5
p=p+1Top 20
enrichKEGG_sig[[p]][1:20, ] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Count | GeneRatio | pvalue | logPval | p.adjust | qvalue | Description | geneID | |
|---|---|---|---|---|---|---|---|---|
| mmu04340 | 8 | 0.1379310 | 0.0000028 | 5.557687 | 0.0000028 | 0.0006063 | Hedgehog signaling pathway | 14632/16147/15245/19207/68525/59056/14725/16576 |
| mmu05217 | 7 | 0.1111111 | 0.0000504 | 4.297777 | 0.0000504 | 0.0045810 | Basal cell carcinoma | 14632/22420/15245/22411/16842/19207/16576 |
| mmu04512 | 8 | 0.0909091 | 0.0000628 | 4.202261 | 0.0000628 | 0.0045810 | ECM-receptor interaction | 94216/23928/16401/114249/12505/12840/329872/16775 |
| mmu04360 | 11 | 0.0607735 | 0.0001136 | 3.944594 | 0.0001136 | 0.0062185 | Axon guidance | 13842/13643/20356/13176/241568/20357/235611/14677/20361/20315/238057 |
| mmu05205 | 10 | 0.0487805 | 0.0013117 | 2.882157 | 0.0013117 | 0.0519047 | Proteoglycans in cancer | 16147/15234/22420/22411/15430/16000/16542/14734/12505/17390 |
| mmu04974 | 7 | 0.0648148 | 0.0014224 | 2.846982 | 0.0014224 | 0.0519047 | Protein digestion and absorption | 12814/140709/12818/94216/58803/12840/110893 |
| mmu04020 | 10 | 0.0416667 | 0.0041455 | 2.382427 | 0.0041455 | 0.1296624 | Calcium signaling pathway | 15234/20190/11549/16542/13618/18127/231602/19220/12290/110893 |
| mmu04728 | 7 | 0.0518519 | 0.0049990 | 2.301116 | 0.0049990 | 0.1368149 | Dopaminergic synapse | 16519/109731/14802/13491/20265/14677/14812 |
| mmu05412 | 5 | 0.0649351 | 0.0068058 | 2.167120 | 0.0068058 | 0.1520052 | Arrhythmogenic right ventricular cardiomyopathy | 16842/24052/16401/12558/110893 |
| mmu04080 | 13 | 0.0336788 | 0.0069425 | 2.158481 | 0.0069425 | 0.1520052 | Neuroactive ligand-receptor interaction | 14803/14805/22355/14802/11549/13491/13618/12310/13616/231602/19220/14812/14397 |
| mmu04724 | 6 | 0.0530973 | 0.0082096 | 2.085675 | 0.0082096 | 0.1634073 | Glutamatergic synapse | 16519/20512/14805/14802/14677/14812 |
| mmu04510 | 8 | 0.0398010 | 0.0129835 | 1.886608 | 0.0129835 | 0.2368921 | Focal adhesion | 15234/94216/16000/23928/16401/16542/12840/16775 |
| mmu04713 | 5 | 0.0510204 | 0.0180879 | 1.742612 | 0.0180879 | 0.2856467 | Circadian entrainment | 16519/14802/20190/14677/14812 |
| mmu04015 | 8 | 0.0373832 | 0.0183037 | 1.737462 | 0.0183037 | 0.2856467 | Rap1 signaling pathway | 15234/11600/16000/16542/14677/14026/14812/240168 |
| mmu04916 | 5 | 0.0500000 | 0.0195695 | 1.708419 | 0.0195695 | 0.2856467 | Melanogenesis | 22420/22411/16842/13618/14677 |
| mmu05033 | 3 | 0.0750000 | 0.0238642 | 1.622254 | 0.0238642 | 0.3078051 | Nicotine addiction | 14802/14812/14397 |
| mmu04151 | 11 | 0.0306407 | 0.0238993 | 1.621615 | 0.0238993 | 0.3078051 | PI3K-Akt signaling pathway | 15234/11600/94216/16000/23928/11839/16401/16542/18127/12840/16775 |
| mmu00910 | 2 | 0.1176471 | 0.0279885 | 1.553020 | 0.0279885 | 0.3404451 | Nitrogen metabolism | 12351/12319 |
| mmu04390 | 6 | 0.0382166 | 0.0354622 | 1.450235 | 0.0354622 | 0.4086500 | Hippo signaling pathway | 22420/22411/16842/11839/93960/238057 |
| mmu05030 | 3 | 0.0625000 | 0.0382166 | 1.417748 | 0.0382166 | 0.4183712 | Cocaine addiction | 109731/14677/14812 |
Dot plot
kegg_dot[[p]]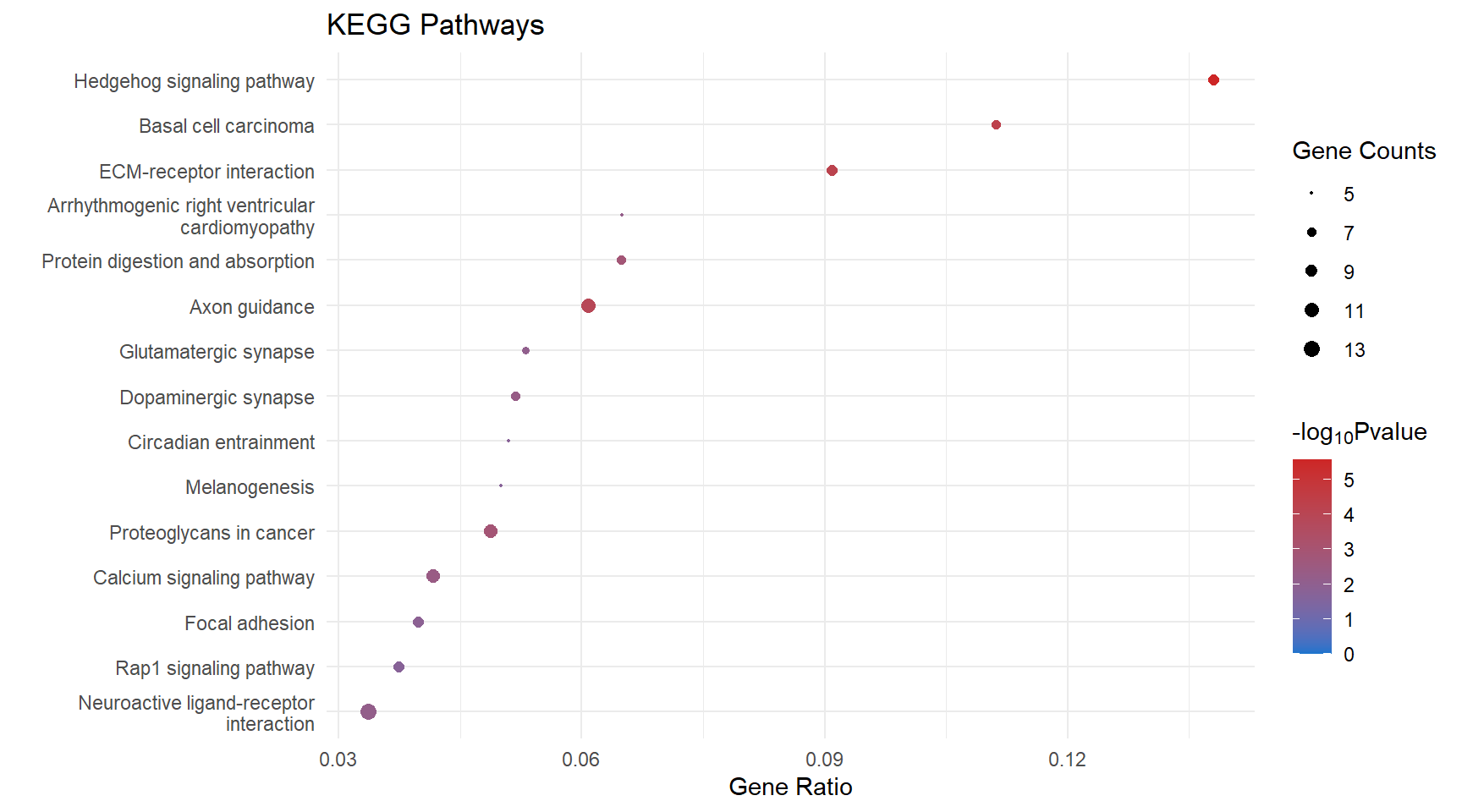
Upset Plot
upset[[p]]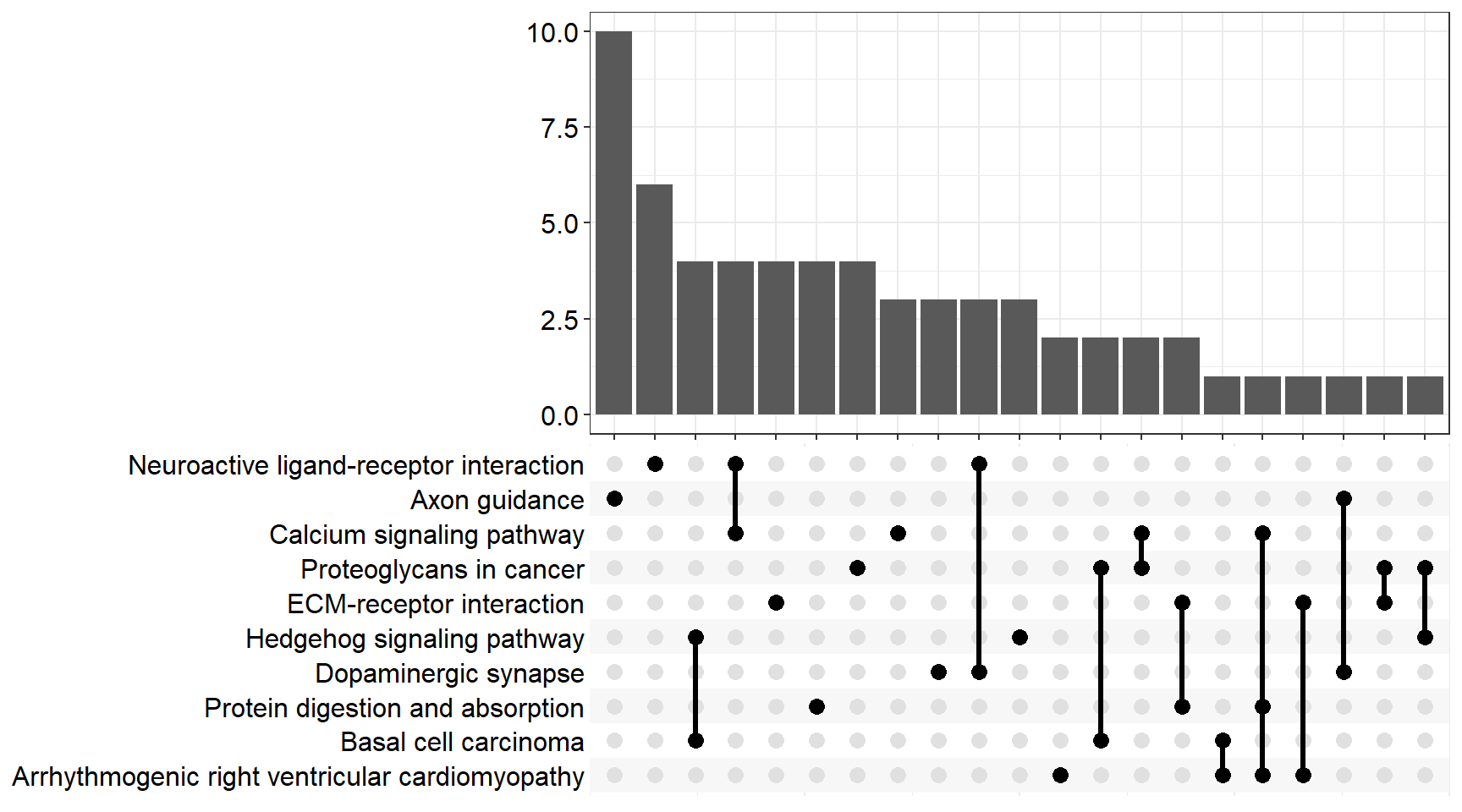
Leukocyte transendothelial migration
q=1Heatmap
kegg_heat[[p]][[q]]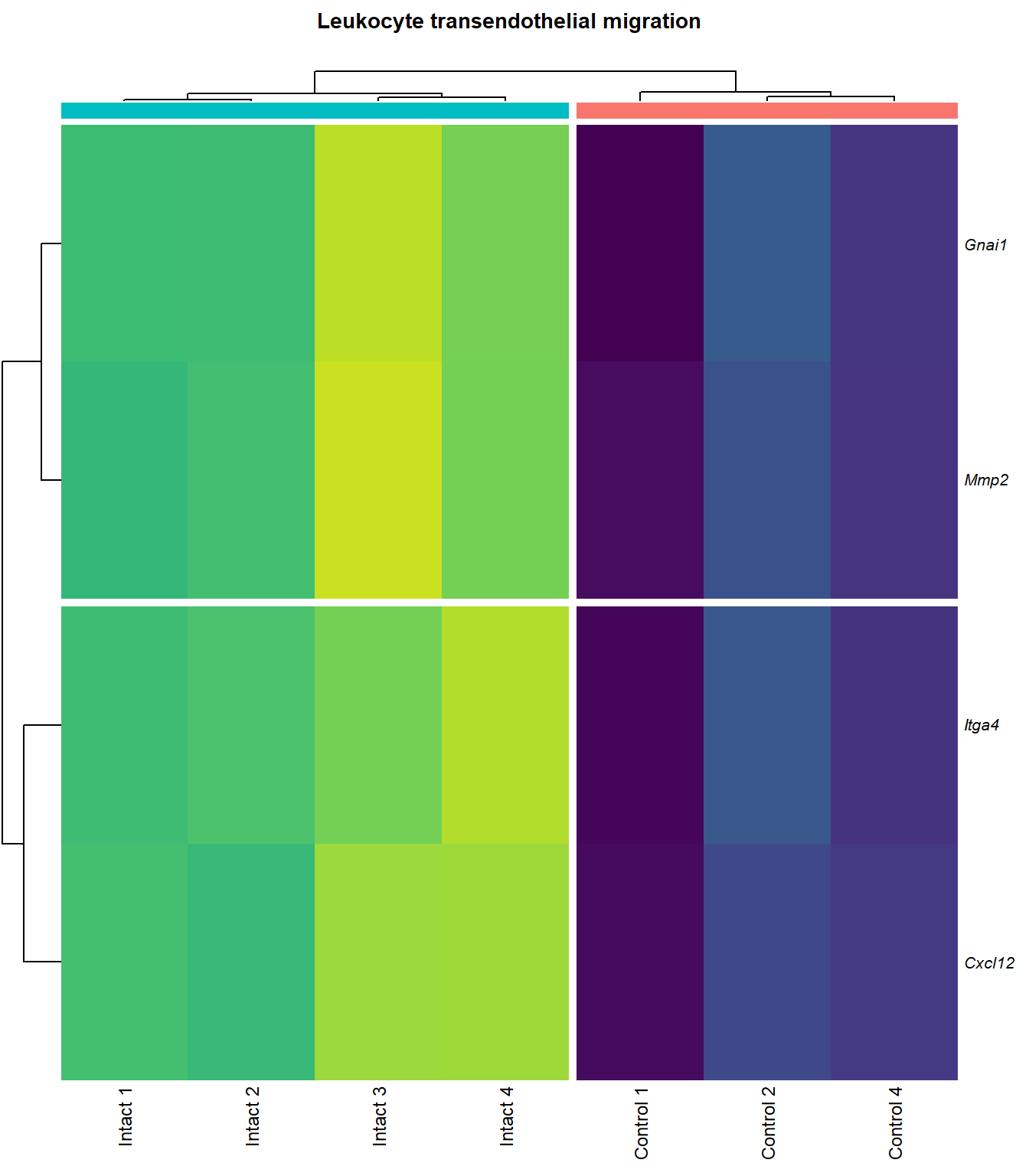
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0002323 | 0.0204195 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000057614 | Gnai1 | 2.405667 | 0.0006615 | 0.0368704 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 [Source:MGI Symbol;Acc:MGI:95771] |
| ENSMUSG00000031740 | Mmp2 | 2.270956 | 0.0009560 | 0.0456079 | matrix metallopeptidase 2 [Source:MGI Symbol;Acc:MGI:97009] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0009762 | 0.0460594 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
Pathview
Hematopoietic cell lineage
q=q+1Heatmap
kegg_heat[[p]][[q]]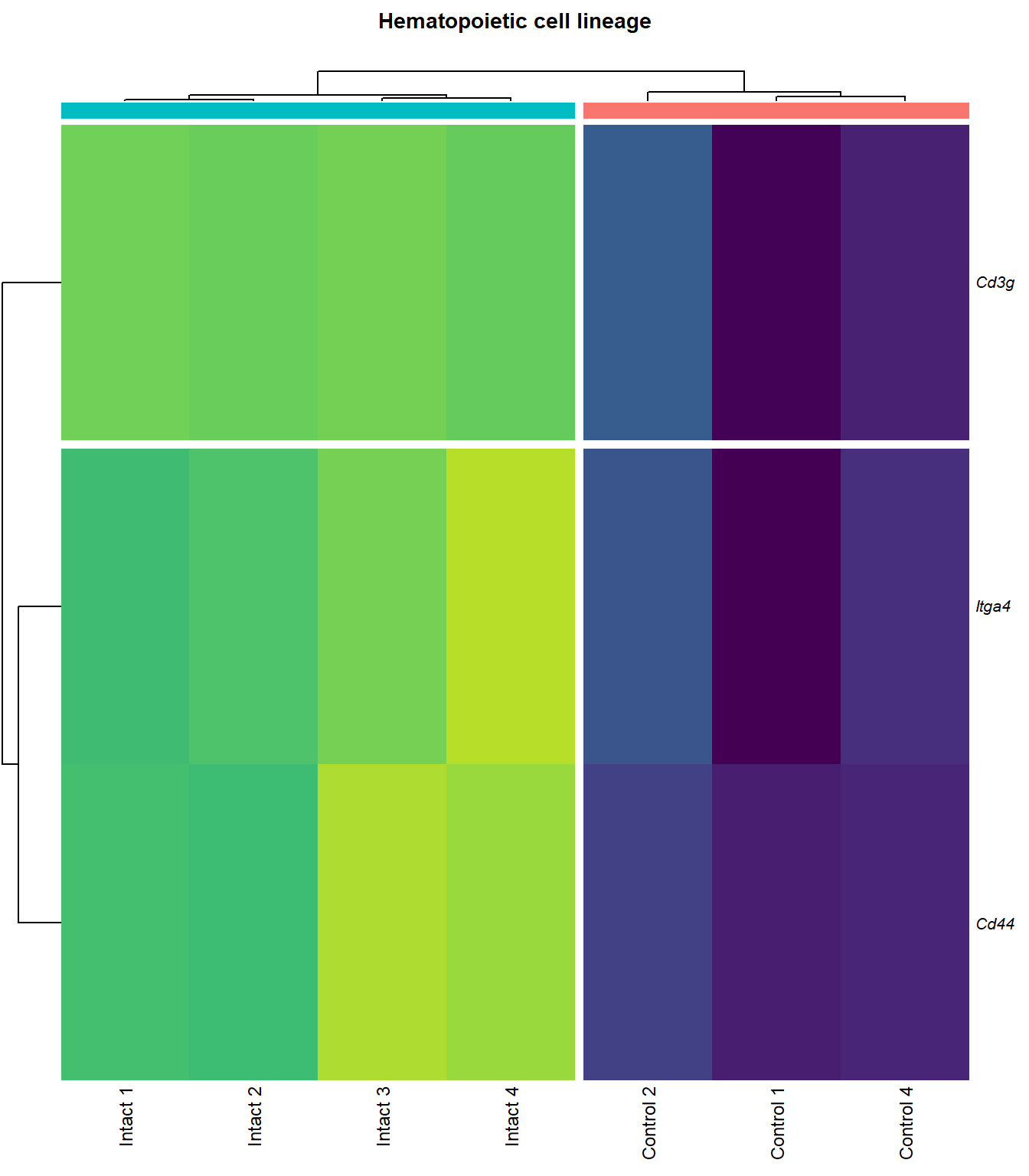
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0002323 | 0.0204195 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000002033 | Cd3g | 3.645674 | 0.0005422 | 0.0332987 | CD3 antigen, gamma polypeptide [Source:MGI Symbol;Acc:MGI:88333] |
| ENSMUSG00000005087 | Cd44 | 2.271713 | 0.0005912 | 0.0346913 | CD44 antigen [Source:MGI Symbol;Acc:MGI:88338] |
Pathview
Vascular smooth muscle contraction
q=q+1Heatmap
kegg_heat[[p]][[q]]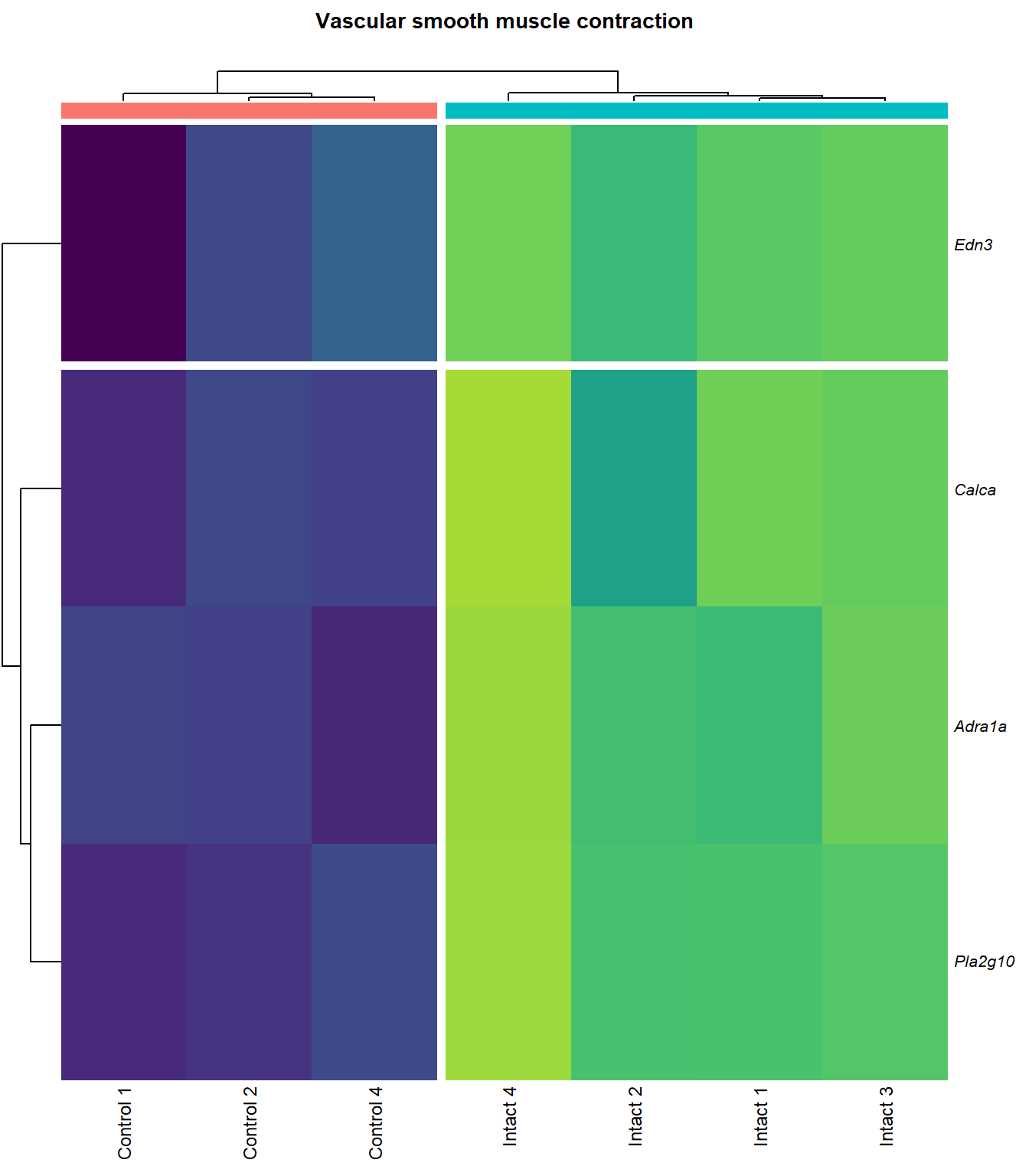
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000022683 | Pla2g10 | 5.773220 | 0.0000322 | 0.0076044 | phospholipase A2, group X [Source:MGI Symbol;Acc:MGI:1347522] |
| ENSMUSG00000045875 | Adra1a | 4.131590 | 0.0001383 | 0.0158943 | adrenergic receptor, alpha 1a [Source:MGI Symbol;Acc:MGI:104773] |
| ENSMUSG00000030669 | Calca | 5.762174 | 0.0005474 | 0.0333098 | calcitonin/calcitonin-related polypeptide, alpha [Source:MGI Symbol;Acc:MGI:2151253] |
| ENSMUSG00000027524 | Edn3 | 3.849271 | 0.0006811 | 0.0374735 | endothelin 3 [Source:MGI Symbol;Acc:MGI:95285] |
Pathview
PI3K-Akt signaling pathway
q=q+1Heatmap
kegg_heat[[p]][[q]]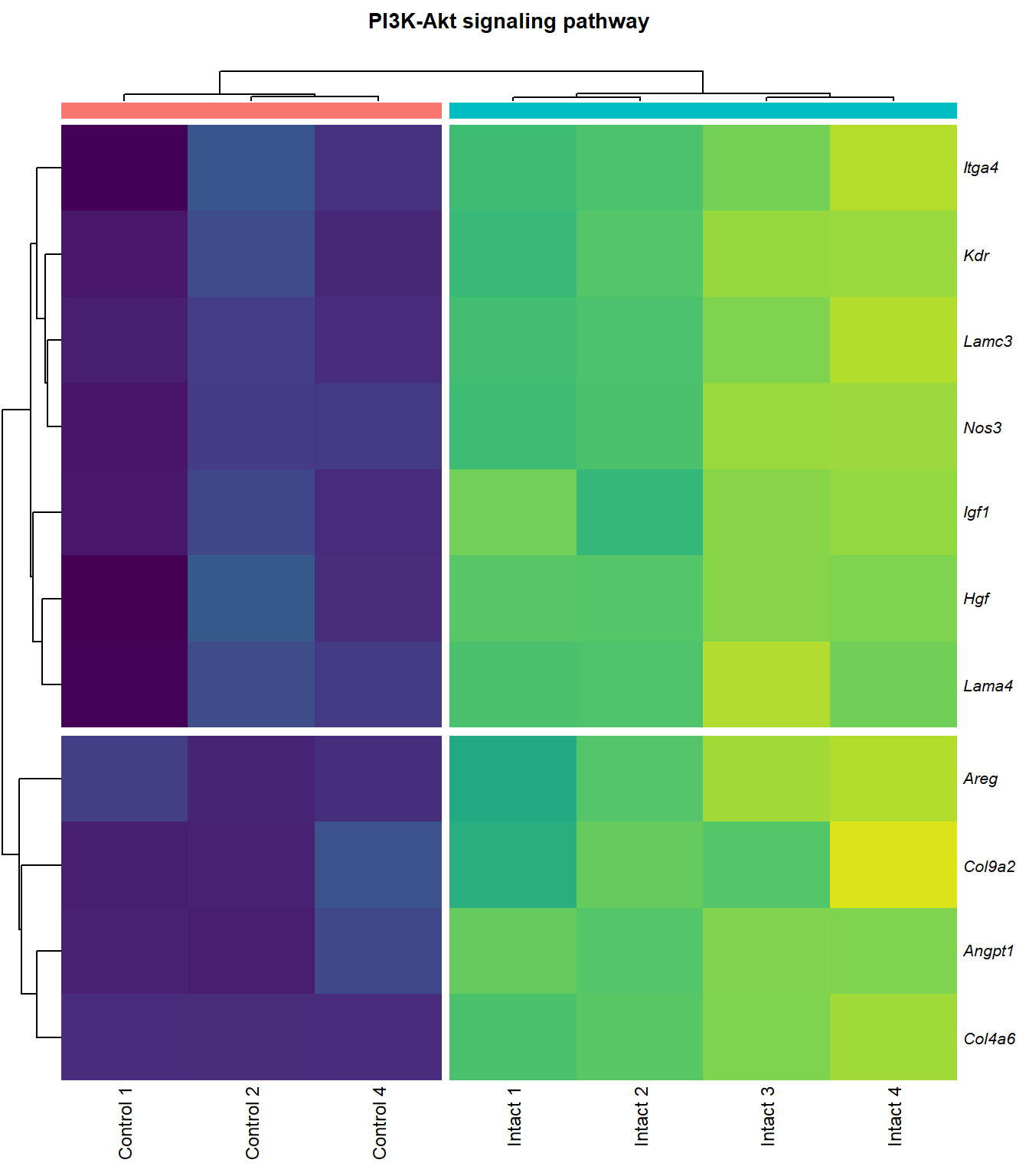
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.125312 | 0.0000135 | 0.0045661 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000022309 | Angpt1 | 3.388586 | 0.0000423 | 0.0085432 | angiopoietin 1 [Source:MGI Symbol;Acc:MGI:108448] |
| ENSMUSG00000031273 | Col4a6 | 3.466810 | 0.0001389 | 0.0158943 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000020053 | Igf1 | 2.780445 | 0.0001735 | 0.0173062 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000026840 | Lamc3 | 3.803842 | 0.0002111 | 0.0190302 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000029378 | Areg | 5.246523 | 0.0002264 | 0.0200946 | amphiregulin [Source:MGI Symbol;Acc:MGI:88068] |
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0002323 | 0.0204195 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.045643 | 0.0003273 | 0.0254005 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000028978 | Nos3 | 2.148682 | 0.0005850 | 0.0346913 | nitric oxide synthase 3, endothelial cell [Source:MGI Symbol;Acc:MGI:97362] |
| ENSMUSG00000028626 | Col9a2 | 4.129759 | 0.0007333 | 0.0389401 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000019846 | Lama4 | 1.940280 | 0.0010516 | 0.0475346 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
Pathview
Focal adhesion
q=q+1Heatmap
kegg_heat[[p]][[q]]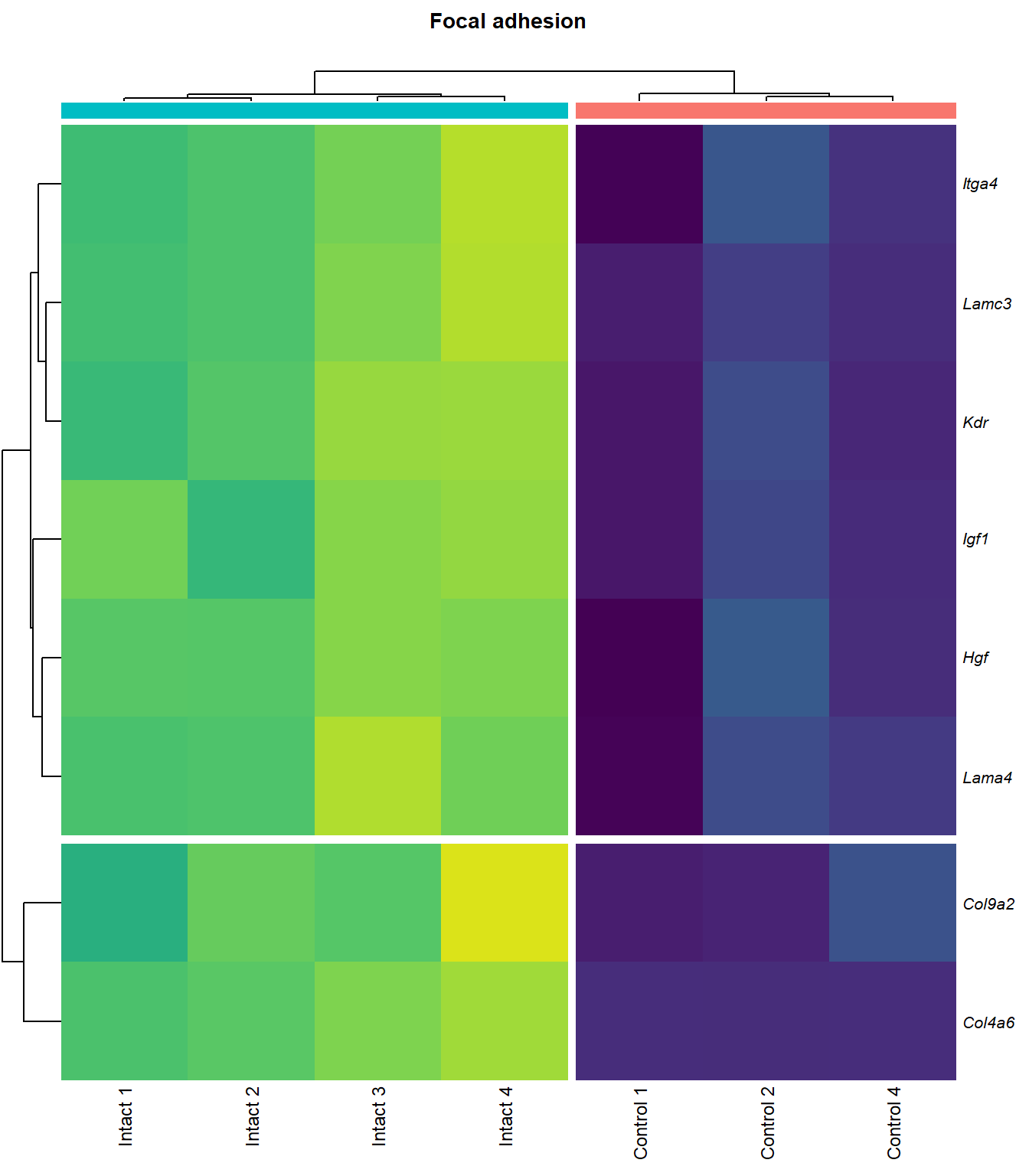
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000028864 | Hgf | 4.125312 | 0.0000135 | 0.0045661 | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| ENSMUSG00000031273 | Col4a6 | 3.466810 | 0.0001389 | 0.0158943 | collagen, type IV, alpha 6 [Source:MGI Symbol;Acc:MGI:2152695] |
| ENSMUSG00000020053 | Igf1 | 2.780445 | 0.0001735 | 0.0173062 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| ENSMUSG00000026840 | Lamc3 | 3.803842 | 0.0002111 | 0.0190302 | laminin gamma 3 [Source:MGI Symbol;Acc:MGI:1344394] |
| ENSMUSG00000027009 | Itga4 | 2.074185 | 0.0002323 | 0.0204195 | integrin alpha 4 [Source:MGI Symbol;Acc:MGI:96603] |
| ENSMUSG00000062960 | Kdr | 2.045643 | 0.0003273 | 0.0254005 | kinase insert domain protein receptor [Source:MGI Symbol;Acc:MGI:96683] |
| ENSMUSG00000028626 | Col9a2 | 4.129759 | 0.0007333 | 0.0389401 | collagen, type IX, alpha 2 [Source:MGI Symbol;Acc:MGI:88466] |
| ENSMUSG00000019846 | Lama4 | 1.940280 | 0.0010516 | 0.0475346 | laminin, alpha 4 [Source:MGI Symbol;Acc:MGI:109321] |
Pathview
Cytokine-cytokine receptor interaction
q=q+1Heatmap
kegg_heat[[p]][[q]]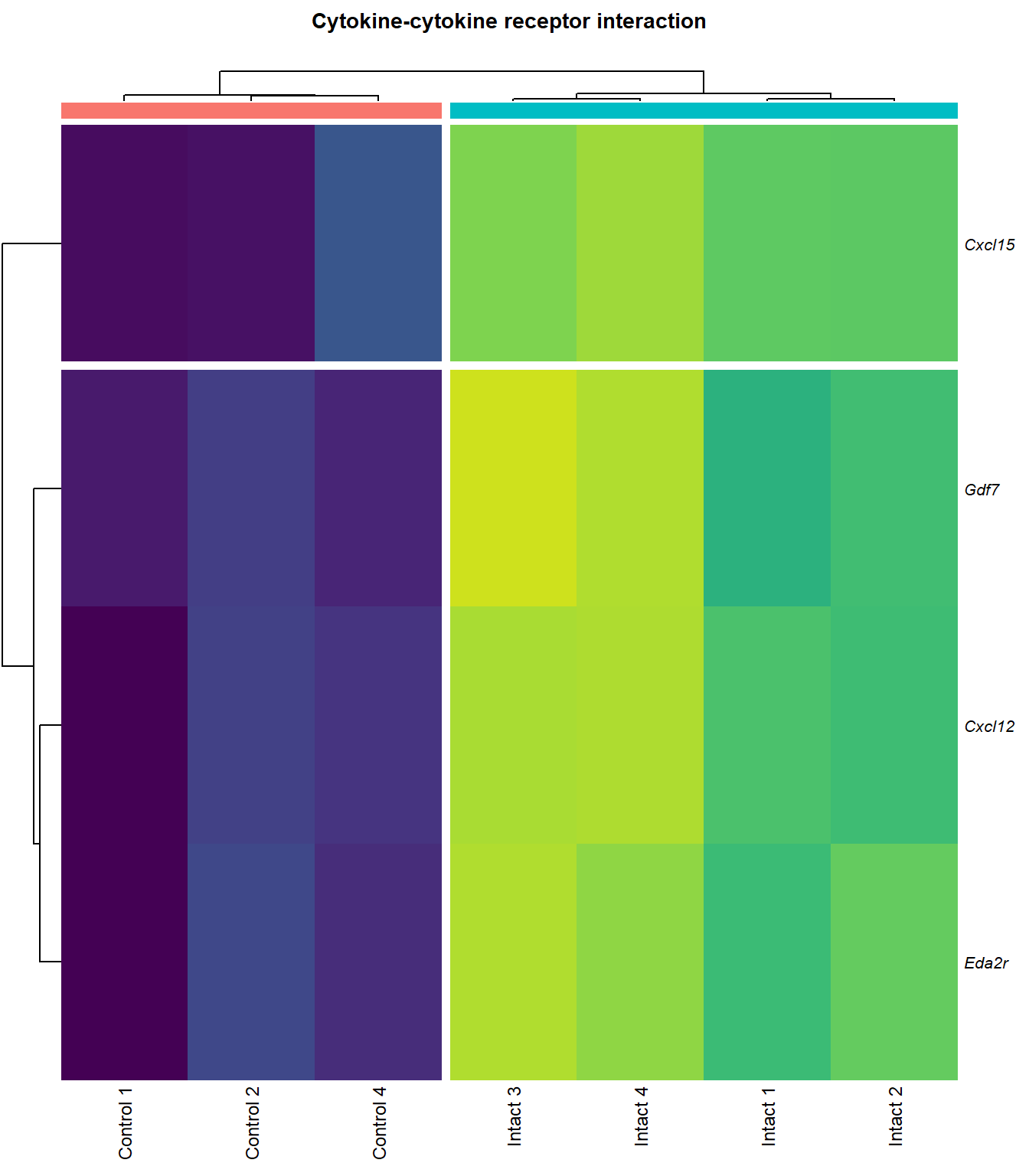
Tables
display_matrix[[p]][[q]] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Gene Name | logFC | P Value | Adjusted P Value | Description | |
|---|---|---|---|---|---|
| ENSMUSG00000029375 | Cxcl15 | 6.916834 | 0.0000000 | 0.0002795 | chemokine (C-X-C motif) ligand 15 [Source:MGI Symbol;Acc:MGI:1339941] |
| ENSMUSG00000034457 | Eda2r | 3.524309 | 0.0001283 | 0.0153427 | ectodysplasin A2 receptor [Source:MGI Symbol;Acc:MGI:2442860] |
| ENSMUSG00000061353 | Cxcl12 | 2.574792 | 0.0009762 | 0.0460594 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| ENSMUSG00000037660 | Gdf7 | 3.410398 | 0.0010346 | 0.0472634 | growth differentiation factor 7 [Source:MGI Symbol;Acc:MGI:95690] |
Pathview
Export Data
# save to csv
writexl::write_xlsx(x = enrichKEGG_all, here::here("3_output/enrichKEGG_all.xlsx"))
writexl::write_xlsx(x = enrichKEGG_sig, here::here("3_output/enrichKEGG_sig.xlsx"))
sessionInfo()R version 4.2.1 (2022-06-23 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_Australia.utf8 LC_CTYPE=English_Australia.utf8
[3] LC_MONETARY=English_Australia.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Australia.utf8
attached base packages:
[1] stats4 grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] pathview_1.36.1 enrichplot_1.16.2 org.Mm.eg.db_3.15.0
[4] AnnotationDbi_1.58.0 IRanges_2.30.1 S4Vectors_0.34.0
[7] Biobase_2.56.0 BiocGenerics_0.42.0 clusterProfiler_4.4.4
[10] Glimma_2.6.0 edgeR_3.38.4 limma_3.52.4
[13] ggrepel_0.9.1 ggbiplot_0.55 scales_1.2.1
[16] plyr_1.8.7 ggpubr_0.4.0 ggplotify_0.1.0
[19] pheatmap_1.0.12 cowplot_1.1.1 viridis_0.6.2
[22] viridisLite_0.4.1 pander_0.6.5 kableExtra_1.3.4
[25] KEGGREST_1.36.3 forcats_0.5.2 stringr_1.4.1
[28] purrr_0.3.5 tidyr_1.2.1 ggplot2_3.3.6
[31] tidyverse_1.3.2 reshape2_1.4.4 tibble_3.1.8
[34] readr_2.1.3 magrittr_2.0.3 dplyr_1.0.10
loaded via a namespace (and not attached):
[1] utf8_1.2.2 tidyselect_1.2.0
[3] RSQLite_2.2.18 htmlwidgets_1.5.4
[5] BiocParallel_1.30.3 scatterpie_0.1.8
[7] munsell_0.5.0 ragg_1.2.3
[9] codetools_0.2-18 withr_2.5.0
[11] colorspace_2.0-3 GOSemSim_2.22.0
[13] highr_0.9 knitr_1.40
[15] rstudioapi_0.14 ggsignif_0.6.3
[17] DOSE_3.22.1 labeling_0.4.2
[19] MatrixGenerics_1.8.1 KEGGgraph_1.56.0
[21] git2r_0.30.1 GenomeInfoDbData_1.2.8
[23] polyclip_1.10-0 bit64_4.0.5
[25] farver_2.1.1 rprojroot_2.0.3
[27] downloader_0.4 treeio_1.20.2
[29] vctrs_0.4.2 generics_0.1.3
[31] xfun_0.33 R6_2.5.1
[33] GenomeInfoDb_1.32.4 graphlayouts_0.8.2
[35] locfit_1.5-9.6 bitops_1.0-7
[37] cachem_1.0.6 fgsea_1.22.0
[39] gridGraphics_0.5-1 DelayedArray_0.22.0
[41] assertthat_0.2.1 promises_1.2.0.1
[43] ggraph_2.1.0 googlesheets4_1.0.1
[45] gtable_0.3.1 tidygraph_1.2.2
[47] workflowr_1.7.0 rlang_1.0.6
[49] genefilter_1.78.0 systemfonts_1.0.4
[51] splines_4.2.1 lazyeval_0.2.2
[53] rstatix_0.7.0 gargle_1.2.1
[55] broom_1.0.1 yaml_2.3.5
[57] abind_1.4-5 modelr_0.1.9
[59] backports_1.4.1 httpuv_1.6.6
[61] qvalue_2.28.0 tools_4.2.1
[63] ellipsis_0.3.2 jquerylib_0.1.4
[65] RColorBrewer_1.1-3 Rcpp_1.0.9
[67] zlibbioc_1.42.0 RCurl_1.98-1.9
[69] SummarizedExperiment_1.26.1 haven_2.5.1
[71] here_1.0.1 fs_1.5.2
[73] data.table_1.14.2 DO.db_2.9
[75] reprex_2.0.2 googledrive_2.0.0
[77] matrixStats_0.62.0 hms_1.1.2
[79] patchwork_1.1.2 evaluate_0.17
[81] xtable_1.8-4 XML_3.99-0.11
[83] readxl_1.4.1 gridExtra_2.3
[85] ggupset_0.3.0 compiler_4.2.1
[87] writexl_1.4.0 shadowtext_0.1.2
[89] crayon_1.5.2 htmltools_0.5.3
[91] ggfun_0.0.7 later_1.3.0
[93] tzdb_0.3.0 geneplotter_1.74.0
[95] aplot_0.1.8 lubridate_1.8.0
[97] DBI_1.1.3 tweenr_2.0.2
[99] dbplyr_2.2.1 MASS_7.3-57
[101] Matrix_1.5-1 car_3.1-0
[103] cli_3.4.1 parallel_4.2.1
[105] igraph_1.3.5 GenomicRanges_1.48.0
[107] pkgconfig_2.0.3 xml2_1.3.3
[109] ggtree_3.4.4 svglite_2.1.0
[111] annotate_1.74.0 bslib_0.4.0
[113] webshot_0.5.4 XVector_0.36.0
[115] rvest_1.0.3 yulab.utils_0.0.5
[117] digest_0.6.29 graph_1.74.0
[119] Biostrings_2.64.1 rmarkdown_2.17
[121] cellranger_1.1.0 fastmatch_1.1-3
[123] tidytree_0.4.1 curl_4.3.3
[125] nlme_3.1-157 lifecycle_1.0.3
[127] jsonlite_1.8.2 carData_3.0-5
[129] fansi_1.0.3 pillar_1.8.1
[131] lattice_0.20-45 fastmap_1.1.0
[133] httr_1.4.4 survival_3.3-1
[135] GO.db_3.15.0 glue_1.6.2
[137] png_0.1-7 Rgraphviz_2.40.0
[139] bit_4.0.4 ggforce_0.4.1
[141] stringi_1.7.8 sass_0.4.2
[143] blob_1.2.3 textshaping_0.3.6
[145] org.Hs.eg.db_3.15.0 DESeq2_1.36.0
[147] memoise_2.0.1 ape_5.6-2