GO Analysis
Ha Tran
22/08/2021
Last updated: 2023-01-20
Checks: 6 1
Knit directory: SRB_2022/1_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the
code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 6e0f643. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: .gitignore
Untracked: test plz delete me.Rmd
Untracked: test-plz-delete-me.html
Unstaged changes:
Modified: 0_data/RDS_objects/enrichGO_sig.rds
Modified: 1_analysis/_site.yml
Modified: 1_analysis/deAnalysis.Rmd
Modified: 1_analysis/go.Rmd
Modified: 1_analysis/index.Rmd
Modified: 1_analysis/kegg.Rmd
Modified: 1_analysis/reactome.Rmd
Modified: 1_analysis/setUp.Rmd
Modified: 2_plots/de/ma_1.05.png
Modified: 2_plots/de/ma_1.1.png
Modified: 2_plots/de/ma_1.5.png
Modified: 2_plots/go/bp_dot_1.05.svg
Modified: 2_plots/go/bp_dot_1.1.svg
Modified: 2_plots/go/bp_dot_1.5.svg
Modified: 2_plots/go/cc_dot_1.05.svg
Modified: 2_plots/go/cc_dot_1.1.svg
Modified: 2_plots/go/cc_dot_1.5.svg
Modified: 2_plots/go/mf_dot_1.05.svg
Modified: 2_plots/go/mf_dot_1.1.svg
Modified: 2_plots/go/mf_dot_1.5.svg
Deleted: 2_plots/ipa/funct_cat_alluvial.svg
Deleted: 2_plots/ipa/up_cat_alluvial.svg
Deleted: 2_plots/ipa/upstream_funct_alluvial.svg
Modified: 2_plots/kegg/kegg_dot_1.05.svg
Modified: 2_plots/kegg/kegg_dot_1.1.svg
Modified: 2_plots/kegg/kegg_dot_1.5.svg
Deleted: 2_plots/reactome/combine_GO.svg
Modified: 2_plots/reactome/react_dot_1.05.svg
Modified: 2_plots/reactome/react_dot_1.1.svg
Modified: 2_plots/reactome/react_dot_1.5.svg
Deleted: 2_plots/reactome/react_dot_INT vs CONT.svg
Deleted: 2_plots/reactome/react_dot_INT vs SVX_VAS.svg
Deleted: 2_plots/reactome/react_dot_SVX vs SVX_VAS.svg
Deleted: 2_plots/reactome/react_dot_VAS vs SVX_VAS.svg
Modified: 2_plots/reactome/upset_react_1.05.svg
Modified: 2_plots/reactome/upset_react_1.1.svg
Modified: 2_plots/reactome/upset_react_1.5.svg
Deleted: 2_plots/reactome/upset_react_INT vs CONT.svg
Deleted: 2_plots/reactome/upset_react_INT vs SVX_VAS.svg
Deleted: 2_plots/reactome/upset_react_SVX vs SVX_VAS.svg
Deleted: 2_plots/reactome/upset_react_VAS vs SVX_VAS.svg
Modified: 3_output/enrichKEGG_all.xlsx
Modified: 3_output/enrichKEGG_sig.xlsx
Modified: 3_output/lmTreat_all.xlsx
Modified: 3_output/lmTreat_fc1.5_voom2_all_fdr.xlsx
Modified: 3_output/lmTreat_sig.xlsx
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (1_analysis/go.Rmd) and HTML
(docs/go.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 691cf34 | Ha Manh Tran | 2023-01-20 | Build site. |
| Rmd | 7f6bab2 | Ha Manh Tran | 2023-01-20 | workflowr::wflow_publish(here::here("1_analysis/*.Rmd")) |
| Rmd | b6cf190 | tranmanhha135 | 2023-01-19 | quick commit |
| Rmd | 3119fad | tranmanhha135 | 2022-11-05 | build website |
| html | 3119fad | tranmanhha135 | 2022-11-05 | build website |
Data Setup
# working with data
library(dplyr)
library(magrittr)
library(readr)
library(tibble)
library(reshape2)
library(tidyverse)
# Visualisation:
library(kableExtra)
library(ggplot2)
library(grid)
# Custom ggplot
library(gridExtra)
library(ggbiplot)
library(ggrepel)
# Bioconductor packages:
library(edgeR)
library(limma)
library(Glimma)
library(clusterProfiler)
library(org.Mm.eg.db)
library(enrichplot)
theme_set(theme_minimal())
pub <- readRDS(here::here("0_data/RDS_objects/pub.rds"))Import DGElist Data
DGElist object containing the raw feature count, sample metadata, and gene metadata, created in the Set Up stage.
# load DGElist previously created in the set up
dge <- readRDS(here::here("0_data/RDS_objects/dge.rds"))
fc <- readRDS(here::here("0_data/RDS_objects/fc.rds"))
lfc <- readRDS(here::here("0_data/RDS_objects/lfc.rds"))
lmTreat <- readRDS(here::here("0_data/RDS_objects/lmTreat.rds"))
lmTreat_sig <- readRDS(here::here("0_data/RDS_objects/lmTreat_sig.rds"))GO Analysis
goSummaries is a package created by Dr Stephen Pederson
for filtering GO terms based on ontology level.
# circumvent rerunning of lengthy analysis.
enrichGO <- readRDS(here::here("0_data/RDS_objects/enrichGO.rds"))
enrichGO_sig <- readRDS(here::here("0_data/RDS_objects/enrichGO_sig.rds"))# download go summaries and set the minimum ontology level
goSummaries <- url("https://uofabioinformaticshub.github.io/summaries2GO/data/goSummaries.RDS") %>%
readRDS()
minPath <- 3
enrichGO=list()
enrichGO_sig <- list()
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
# find enriched GO terms
enrichGO[[x]] <- clusterProfiler::enrichGO(
gene = lmTreat_sig[[x]]$entrezid,
OrgDb = org.Mm.eg.db,
keyType = "ENTREZID",
ont = "ALL",
pAdjustMethod = "fdr",
pvalueCutoff = 0.05
)
# bind to goSummaries to elminate go terms with ontology levels 1 and 2.
enrichGO_sig[[x]] <- enrichGO[[x]] %>%
clusterProfiler::setReadable(OrgDb = org.Mm.eg.db, keyType = "auto")
enrichGO_sig[[x]] <- enrichGO_sig[[x]] %>%
as.data.frame() %>%
rownames_to_column("id") %>%
left_join(goSummaries) %>%
dplyr::filter(shortest_path >= minPath) %>%
column_to_rownames("id")
# adjust go results, separate compound column, add FDR column, adjust the GeneRatio column
enrichGO_sig[[x]] <- enrichGO_sig[[x]] %>%
separate(col = BgRatio, sep = "/", into = c("Total", "Universe")) %>%
dplyr::mutate(
logFDR = -log(p.adjust, 10),
GeneRatio = Count / as.numeric(Total)) %>%
dplyr::select(c("Description", "ontology", "GeneRatio", "pvalue", "p.adjust", "logFDR", "qvalue", "geneID", "Count"))
# at the beginnning of a word (after 35 characters), add a newline. shorten the y axis for dot plot
# enrichGO_sig[[x]]$Description <- sub(pattern = "(.{1,35})(?:$| )",
# replacement = "\\1\n",
# x = enrichGO_sig[[x]]$Description)
# # remove the additional newline at the end of the string
# enrichGO_sig[[x]]$Description <- sub(pattern = "\n$",
# replacement = "",
# x = enrichGO_sig[[x]]$Description)
}FC=1.05
# display the top 30 most sig
enrichGO_sig[[1]] %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(.,3)))) %>%
kable(caption = "Significantly enriched GO terms") %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Description | ontology | GeneRatio | pvalue | p.adjust | logFDR | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0030198 | extracellular matrix organization | BP | 0.21 | 2e-24 | 5.36e-21 | 20.3 | 3.47e-21 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Ddr2/Adamts20/Col4a6/Prdm5/Phldb2/Npnt/Spock2/Ets1/Col16a1/Tgfbi/Adamts16/Adamts19/Fn1/Mmp2/Foxf1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Loxl3/Adamts10/Smoc1/Emilin1/Tnfrsf11b/Tie1/Col5a2/Ndnf/Adamts7/Ltbp3/Mmp19/Col15a1/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Efemp2/Crtap/Olfml2a/Cav1/Ntng1/Col23a1/Col18a1/Itga8/Adamts17/Lum/Ccdc80/Fbln5/Col4a5/Eng | 66 |
| GO:0043062 | extracellular structure organization | BP | 0.209 | 2.41e-24 | 5.36e-21 | 20.3 | 3.47e-21 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Ddr2/Adamts20/Col4a6/Prdm5/Phldb2/Npnt/Spock2/Ets1/Col16a1/Tgfbi/Adamts16/Adamts19/Fn1/Mmp2/Foxf1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Loxl3/Adamts10/Smoc1/Emilin1/Tnfrsf11b/Tie1/Col5a2/Ndnf/Adamts7/Ltbp3/Mmp19/Col15a1/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Efemp2/Crtap/Olfml2a/Cav1/Ntng1/Col23a1/Col18a1/Itga8/Adamts17/Lum/Ccdc80/Fbln5/Col4a5/Eng | 66 |
| GO:0045229 | external encapsulating structure organization | BP | 0.208 | 2.91e-24 | 5.36e-21 | 20.3 | 3.47e-21 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Ddr2/Adamts20/Col4a6/Prdm5/Phldb2/Npnt/Spock2/Ets1/Col16a1/Tgfbi/Adamts16/Adamts19/Fn1/Mmp2/Foxf1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Loxl3/Adamts10/Smoc1/Emilin1/Tnfrsf11b/Tie1/Col5a2/Ndnf/Adamts7/Ltbp3/Mmp19/Col15a1/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Efemp2/Crtap/Olfml2a/Cav1/Ntng1/Col23a1/Col18a1/Itga8/Adamts17/Lum/Ccdc80/Fbln5/Col4a5/Eng | 66 |
| GO:0045785 | positive regulation of cell adhesion | BP | 0.164 | 8.25e-22 | 1.14e-18 | 17.9 | 7.39e-19 | Ihh/Efnb3/Vit/Col26a1/Angpt1/Mdk/Lef1/Itga4/Cd83/Kdr/Igf1/Npnt/Nrp1/Chst2/Cd44/Fermt2/Spock2/Ets1/Col16a1/Fn1/Foxf1/Icam1/Nid1/Tek/Cdh13/Cxcl12/Has2/Egflam/Pik3r6/Gli3/Podxl/Calca/Thy1/Plpp3/Tnfsf13b/Coro1a/Cd1d1/Ppm1f/Smoc1/Itgal/H2-Ab1/Il27ra/Emilin1/Vegfc/Sox12/Havcr2/H2-Ob/Sele/Igf2/Ndnf/Dysf/Nckap1l/Xcl1/Pcsk5/H2-Eb1/Stat5b/Net1/Abi3bp/Adam19/H2-Aa/Gimap5/Efemp2/St3gal4/Zap70/Gimap3/Apbb1ip/Ptn/Il12rb1/Cav1/Pld2/Il2rg/Hacd1/Spn/Enpp2/Bmp7/Il7r/Cd4/Rasal3/Ccdc80 | 79 |
| GO:0006816 | calcium ion transport | BP | 0.166 | 1.48e-20 | 1.64e-17 | 16.8 | 1.06e-17 | Nalcn/Cacna1c/Ryr1/Jph4/Spink1/Igf1/Adra1a/Slc8a1/Nos3/Drd4/Cachd1/Cacna2d1/Ednrb/Myo5a/Slc24a4/Icam1/Grin2b/Prkd1/Stc1/Cxcr3/Cxcl12/Slc24a3/Cacna1e/Ank2/Slc8a3/Calca/Trpc6/Thy1/Nalf1/Coro1a/Gja4/Aplnr/Cracr2a/Agtr1a/Ednra/Dysf/Bin1/Adcyap1r1/Htr2a/Xcl1/Pkd1/Tspan13/Cacng7/Stac2/Selenon/Arrb2/Asph/Pkd2/Fyn/Xcr1/Ryr3/Cd84/Rrad/Calcrl/Cacna1h/Slc25a23/Ptger3/Atp2b4/Ccn2/Gnb5/Gimap5/Gstm7/Atp1a2/Gimap3/Psen1/Sri/Pdgfrb/Cav1/Cacna1a/Usp2/Cacnb4/F2r/Cd4 | 73 |
| GO:0050808 | synapse organization | BP | 0.156 | 6.02e-20 | 5.55e-17 | 16.3 | 3.59e-17 | Gpc6/Gap43/Sdk2/Lrrc4c/Plxnb1/Ptprd/St8sia2/Nlgn1/Nrp1/Ptpro/Rims3/Pdzrn3/P2rx2/Cdh2/Wasf3/Lrrk2/Lrrc4b/Myo5a/Mef2c/Pfn2/Nptx1/Grin2b/Shank2/Pcdhgc3/Obsl1/Adgrl1/Ppfia2/Slc8a3/Slitrk3/Plxnd1/Dock10/Glrb/Abi2/Kif1a/Ppfia4/Palld/Epha3/Nlgn3/Dgkb/Nptxr/Apoe/Arhgef15/Nfasc/Magi2/Prrt1/Sparcl1/Caprin2/Pcdh17/Nfatc4/Epha7/Bhlhb9/Erc2/Lrfn2/Fyn/Dlg4/Cux2/Palm/Dlg5/Syngap1/Ngef/Arhgef9/Rab39b/Psen1/Ptn/Ntng1/Cacna1a/Pcdhgc4/Dact1/Iqsec3/Zmynd8/Cacnb4/F2r/Lrp4/Camk1/Nlgn2/Zfp365/Col4a5 | 77 |
| GO:0034329 | cell junction assembly | BP | 0.161 | 2.8e-19 | 2.21e-16 | 15.7 | 1.43e-16 | Gpc6/Gap43/Sdk2/Wnt11/Kdr/Plxnb1/Ptprd/St8sia2/Nlgn1/Epb41l3/Phldb2/Mpdz/Nrp1/Ptpro/Fermt2/Col16a1/Cdh2/Fn1/Lrrc4b/Cdh6/Mef2c/Nptx1/Jam3/Thsd1/Tek/Shank2/Obsl1/Adgrl1/Cldn8/Cdh10/Slitrk3/Myo9a/Plxnd1/Cdh5/Thy1/Abi2/Ppm1f/Epha3/Aplnr/Esam/Nlgn3/Sh3bp1/Limch1/Arhgap6/Nptxr/Nfasc/Cdh11/Magi2/Gjc1/Pecam1/Snai1/Pcdh17/Epha7/Heg1/Dlc1/Dchs1/Bhlhb9/Nphp1/Cldn5/Cux2/Dlg5/Arhgef9/Hipk1/Cav1/Cacna1a/Itgb4/Pdcd6ip/Enpp2/Cldn1/Lrp4/Nlgn2 | 71 |
| GO:0031589 | cell-substrate adhesion | BP | 0.166 | 1.09e-17 | 7.51e-15 | 14.1 | 4.87e-15 | Ptprz1/Vit/Col26a1/Angpt1/Mdk/Itga4/Kdr/Phldb2/Npnt/Nrp1/Lamc3/Fermt2/Spock2/Col16a1/Itgb7/Fn1/Foxf1/Nid1/Jam3/Thsd1/Tek/Cdh13/Has2/Egflam/Itga1/Col13a1/Frem1/Thy1/Postn/Coro1a/Ppm1f/Smoc1/Epha3/Itgal/Limch1/Arhgap6/Emilin1/Vegfc/Sned1/Hoxd3/Pecam1/Tmeff2/Ndnf/Itga7/Parvg/Pcsk5/Parvb/Dlc1/Epdr1/Net1/Abi3bp/Ccn2/Efemp2/Ptn/Cd96/Ntng1/Itga8/Itgb4/Hacd1/Enpp2/Nid2/Ccdc80 | 62 |
| GO:0048754 | branching morphogenesis of an epithelial tube | BP | 0.224 | 1.64e-17 | 1.01e-14 | 14 | 6.52e-15 | Ihh/Sfrp2/Sema5a/Wnt6/Ar/Angpt1/Hhip/Mdk/Lef1/Hoxd11/Kdr/Nog/Igf1/Tbx2/Npnt/Nrp1/Cd44/Hoxa11/Adamts16/Gpc3/Areg/Foxf1/Tek/Cxcl12/Tcf21/Gli3/Plxnd1/Gdf7/Mgp/Agtr1a/Tbx3/Rasip1/Tie1/Ednra/Pkd1/Nfatc4/Ptch1/Dchs1/Dlg5/Fat4/Bmp7/Myc/Eng | 43 |
| GO:0007224 | smoothened signaling pathway | BP | 0.245 | 1.86e-17 | 1.03e-14 | 14 | 6.67e-15 | Gli1/Ihh/Hhip/Evc2/Nog/Ptch2/Evc/Kif7/Arl6/Dzip1l/Gpc3/Foxf1/Cibar1/Serpine2/Bbs7/Armc9/Zfp423/Gli3/Ptpdc1/Ift57/Wnt9a/Rab34/Gpr161/Enpp1/Dync2h1/Hipk2/Cfap410/Ttc26/Arl3/Ptch1/Tmem231/Ttbk2/Rab23/Dzip1/Dlg5/Glis2/Hipk1/Gpc2/Ift81 | 39 |
| GO:0001655 | urogenital system development | BP | 0.16 | 6.83e-17 | 3.15e-14 | 13.5 | 2.04e-14 | Stra6/Wnt6/Ar/Angpt1/Wnt11/Pygo1/Hoxd11/Cyp7b1/Nog/Igf1/Npnt/Nrp1/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Lrrk2/Ednrb/Mmp2/Mef2c/Foxf1/Id3/Id4/Osr2/Nid1/Enpep/Tek/Gdf11/Adamts1/Tcf21/Gli3/Plxnd1/Lrp2/Tshz3/Prdm1/Cplane1/Enpp1/Cdkn1c/Agtr1a/Dync2h1/Ednra/Heyl/Pkd1/Pcsk5/Arl3/Smad9/Epha7/Ptch1/Dchs1/Osr1/Pkd2/Aqp11/Dlg5/Fat4/Pdgfrb/Glis2/Itga8/Bmp7/Lrp4/Vcan/Myc | 62 |
| GO:0001822 | kidney development | BP | 0.17 | 1.62e-16 | 6.88e-14 | 13.2 | 4.46e-14 | Stra6/Wnt6/Angpt1/Wnt11/Pygo1/Hoxd11/Nog/Npnt/Nrp1/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Lrrk2/Ednrb/Mef2c/Id3/Osr2/Nid1/Enpep/Tek/Gdf11/Adamts1/Tcf21/Gli3/Plxnd1/Lrp2/Tshz3/Prdm1/Cplane1/Enpp1/Cdkn1c/Agtr1a/Dync2h1/Ednra/Heyl/Pkd1/Pcsk5/Arl3/Smad9/Epha7/Ptch1/Dchs1/Osr1/Pkd2/Aqp11/Dlg5/Fat4/Pdgfrb/Glis2/Itga8/Bmp7/Lrp4/Vcan/Myc | 56 |
| GO:0072001 | renal system development | BP | 0.165 | 3e-16 | 1.11e-13 | 13 | 7.2e-14 | Stra6/Wnt6/Angpt1/Wnt11/Pygo1/Hoxd11/Nog/Npnt/Nrp1/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Lrrk2/Ednrb/Mef2c/Foxf1/Id3/Osr2/Nid1/Enpep/Tek/Gdf11/Adamts1/Tcf21/Gli3/Plxnd1/Lrp2/Tshz3/Prdm1/Cplane1/Enpp1/Cdkn1c/Agtr1a/Dync2h1/Ednra/Heyl/Pkd1/Pcsk5/Arl3/Smad9/Epha7/Ptch1/Dchs1/Osr1/Pkd2/Aqp11/Dlg5/Fat4/Pdgfrb/Glis2/Itga8/Bmp7/Lrp4/Vcan/Myc | 57 |
| GO:0034765 | regulation of ion transmembrane transport | BP | 0.141 | 7.76e-16 | 2.58e-13 | 12.6 | 1.67e-13 | Kcnb2/Nalcn/Kcnj3/Cacna1c/Hecw2/Kcna6/Kcnd2/Kcnd3/Nlgn1/Tcaf1/Slc8a1/Scn4b/Drd4/Fxyd5/Scn1a/Cacna2d1/Kcnq5/Myo5a/Mef2c/Prkd1/Cxcr3/Cacna1e/Ank2/Edn3/Plcb1/Trpc6/Thy1/Coro1a/Scn2a/Aplnr/Nlgn3/Cracr2a/Agtr1a/Prrt1/Kcnma1/Ednra/Kcnc4/Dysf/Bin1/Tspan13/Cacng7/Stac2/Kcnip3/Selenon/Asph/Osr1/Pkd2/Fhl1/Fyn/Rrad/Cacna1h/Dlg4/Kcng4/Ptger3/Atp2b4/Scn3a/Gnb5/Gstm7/Atp1a2/Psen1/Abcb1a/Sri/Cav1/Kcnq4/Cacna1a/Cacnb4/F2r/Kcnh1/Twist1/Nlgn2 | 70 |
| GO:0001667 | ameboidal-type cell migration | BP | 0.146 | 7.94e-16 | 2.58e-13 | 12.6 | 1.67e-13 | Cyp1b1/Sema5a/Angpt1/Wnt11/Itga4/Kdr/Igf1/Evl/Zeb2/Ddr2/Sema5b/Slc8a1/Nos3/Nrp1/Sema7a/Ets1/Cdh2/Itgb7/Fn1/Ednrb/Mef2c/Pfn2/Ptp4a3/Prkd1/Stc1/Tek/Cdh13/Cxcl12/Has2/Akap12/Vash1/Edn3/Ptprm/Calca/Plxnd1/Cdh5/Syde1/Plpp3/Ppm1f/Kit/Jcad/Sh3bp1/Vegfc/Prr5l/Apoe/Ednra/Sema3g/Daam2/Pecam1/Igf2/Kitl/Hdac9/Akt3/Atp2b4/Fstl1/Arsb/Adgra2/Sema6a/Mapre2/Cygb/Itgb4/Enpp2/Bmp7/Hand2/Twist1/Plekhg5 | 66 |
| GO:0048736 | appendage development | BP | 0.206 | 9.52e-16 | 2.77e-13 | 12.6 | 1.79e-13 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Fermt2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Cibar1/Osr2/Bbs7/Flvcr1/Gli3/Wnt9a/Smoc1/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Psen1/Itgb4/Iqce/Bmp7/Lrp4/Hand2/Twist1 | 42 |
| GO:0060173 | limb development | BP | 0.206 | 9.52e-16 | 2.77e-13 | 12.6 | 1.79e-13 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Fermt2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Cibar1/Osr2/Bbs7/Flvcr1/Gli3/Wnt9a/Smoc1/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Psen1/Itgb4/Iqce/Bmp7/Lrp4/Hand2/Twist1 | 42 |
| GO:0031345 | negative regulation of cell projection organization | BP | 0.2 | 1.31e-15 | 3.61e-13 | 12.4 | 2.34e-13 | Efnb3/Ptprz1/Map2/Sema5a/Evl/Sema5b/Nlgn1/Nrp1/Sema7a/Cspg4/Lrrk2/Ntm/Abi3/Dab1/Pfn2/Grin2b/Trpc6/Thy1/Epha3/Minar1/Nlgn3/Tsku/Cd38/Apoe/Sema3g/Dennd5a/Nfatc4/Rtn4rl1/Epha7/Neo1/Fyn/Fat3/Xylt1/Dpysl3/Syngap1/Cep97/Ngef/Psen1/Sema6a/Ulk2/Lrp4/Draxin/Zfp365 | 43 |
| GO:0060562 | epithelial tube morphogenesis | BP | 0.152 | 2.68e-15 | 7.05e-13 | 12.2 | 4.57e-13 | Ihh/Sfrp2/Sema5a/Wnt6/Ar/Angpt1/Hhip/Mdk/Lef1/Hoxd11/Kdr/Nog/Igf1/Zeb2/Tbx2/Npnt/Nrp1/Cd44/Hoxa11/Adamts16/Gpc3/Areg/Mef2c/Foxf1/Tek/Cxcl12/Tcf21/Gli3/Ift57/Plxnd1/Gdf7/Lrp2/Cecr2/Mgp/Aplnr/Agtr1a/Tbx3/Rasip1/Tie1/Ednra/Prickle1/Pkd1/Nfatc4/Epha7/Ipmk/Dlc1/Ptch1/Dchs1/Osr1/Pkd2/Rab23/Fzd2/Dlg5/Fat4/Psen1/Bmp7/Hand2/Myc/Twist1/Eng | 60 |
| GO:0035107 | appendage morphogenesis | BP | 0.218 | 2.93e-15 | 7.05e-13 | 12.2 | 4.57e-13 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Gpc3/Hoxd9/Hoxa10/Cibar1/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Psen1/Iqce/Bmp7/Lrp4/Hand2/Twist1 | 38 |
| GO:0035108 | limb morphogenesis | BP | 0.218 | 2.93e-15 | 7.05e-13 | 12.2 | 4.57e-13 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Gpc3/Hoxd9/Hoxa10/Cibar1/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Psen1/Iqce/Bmp7/Lrp4/Hand2/Twist1 | 38 |
| GO:0006874 | cellular calcium ion homeostasis | BP | 0.138 | 3.21e-15 | 7.39e-13 | 12.1 | 4.79e-13 | Kcnk3/Cacna1c/Kdr/Ryr1/Adra1a/Slc8a1/P2rx2/Pygm/Bok/Cacna2d1/Ednrb/Myo5a/Slc24a4/Inpp4b/Lpar4/Grin2b/Prkd1/Stc1/Cxcr3/Slc24a3/Cacna1e/Ptgfr/Ank2/Edn3/Slc8a3/Calca/Trpc6/Cdh5/Thy1/Pth1r/Nalf1/Coro1a/Grik2/Aplnr/Kctd17/Cd38/Agtr1a/Apoe/Ednra/Bdkrb2/Adcyap1r1/Htr2a/Xcl1/Pkd1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Slc25a23/Dlg4/Fzd2/Ptger3/Atp2b4/Gimap5/Gstm7/Atp1a2/Gimap3/Psen1/Sri/Cav1/Cacna1a/Cacnb4/F2r/Cd4/Kcnh1/Cd52/Ptger2 | 69 |
| GO:0061138 | morphogenesis of a branching epithelium | BP | 0.191 | 3.36e-15 | 7.42e-13 | 12.1 | 4.81e-13 | Ihh/Hgf/Sfrp2/Sema5a/Wnt6/Ar/Angpt1/Hhip/Mdk/Lef1/Hoxd11/Kdr/Nog/Igf1/Tbx2/Npnt/Nrp1/Cd44/Hoxa11/Adamts16/Gpc3/Areg/Foxf1/Tek/Cxcl12/Tcf21/Gli3/Plxnd1/Gdf7/Mgp/Agtr1a/Tbx3/Rasip1/Tie1/Ednra/Pkd1/Nfatc4/Ptch1/Dchs1/Dlg5/Fat4/Bmp7/Myc/Eng | 44 |
| GO:0007409 | axonogenesis | BP | 0.138 | 5.24e-15 | 1.11e-12 | 12 | 7.22e-13 | Gap43/Epha8/Prtg/Efnb3/Ptprz1/Map2/Sema5a/Itga4/Lrrc4c/Pla2g10/Dcc/Plxnb1/Nog/Evl/Zeb2/Sema5b/Rnf165/Nrp1/Ptpro/Sema7a/Cdh2/Fn1/Dab1/Mef2c/Nptx1/Arhgef25/Cxcl12/Edn3/Gli3/Slitrk3/Ptprm/Plxnd1/Gdf7/Lrp2/Thy1/Ndn/Lama2/Epha3/Matn2/Nlgn3/Tsku/Apoe/Nfasc/Cdh11/Ednra/Sema3g/Nin/Rtn4rl1/Epha7/Neo1/Chodl/Ptch1/Unc5c/Syngap1/Brsk1/Psen1/Sema6a/Ulk2/Nrn1/Ntng1/Cacna1a/Bmp7/Lrp4/Bcl11b/Kif5a/Slit3/Draxin/Numbl | 68 |
| GO:0051924 | regulation of calcium ion transport | BP | 0.171 | 9.91e-15 | 2.03e-12 | 11.7 | 1.31e-12 | Cacna1c/Jph4/Spink1/Igf1/Slc8a1/Nos3/Drd4/Cacna2d1/Myo5a/Icam1/Prkd1/Stc1/Cxcr3/Cxcl12/Ank2/Calca/Trpc6/Thy1/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Dysf/Bin1/Adcyap1r1/Tspan13/Stac2/Selenon/Arrb2/Asph/Pkd2/Fyn/Cd84/Rrad/Ptger3/Gnb5/Gimap5/Gstm7/Atp1a2/Gimap3/Sri/Pdgfrb/Cav1/Cacna1a/Usp2/Cacnb4/F2r/Cd4 | 49 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | BP | 0.192 | 2.77e-14 | 5.48e-12 | 11.3 | 3.55e-12 | Gpc6/Prtg/Pcdh18/Hmcn1/Sdk2/Mcam/Itga4/Lrrc4c/Ptprd/Nlgn1/Cdh2/Lrrc4b/Cdh6/Tenm3/Dab1/Cdh13/Pcdhgc3/Obsl1/Adgrl1/Cdh10/Slitrk3/Ptprm/Cdh5/Pcdh19/Pcdh12/Palld/Epha3/Itgal/Esam/Cdh11/Magi2/Sparcl1/Sele/Pecam1/Pcdh17/Dchs1/Fat3/Fat4/Pcdhb14/Ntng1/Cldn1 | 41 |
| GO:0050673 | epithelial cell proliferation | BP | 0.137 | 3.4e-14 | 6.48e-12 | 11.2 | 4.2e-12 | Gli1/Ihh/Hgf/Sfrp2/Sema5a/Ar/Mdk/Itga4/Cd109/Kdr/Agap2/Cyp7b1/Nog/Igf1/Tbx2/Gkn3/Fst/Ccnd2/Gpc3/Ednrb/Areg/Mef2c/Osr2/Prkd1/Cxcr3/Tek/Cdh13/Cxcl12/Has2/Vash1/Stxbp4/Ptprm/Calca/Ift57/Kit/Ppp1r16b/Aplnr/Jcad/Cdkn1c/Vegfc/Agtr1a/Apoe/Tie1/Ednra/Igf2/Dysf/Klf9/Twist2/Ptch1/Osr1/Aqp11/Akt3/Apln/Egfl7/Psen1/Ptn/Cav1/Igfbp4/Vash2/Cldn1/Wfdc1/Bcl11b/Myc/Twist1/Eng | 65 |
| GO:0060402 | calcium ion transport into cytosol | BP | 0.207 | 4.24e-14 | 7.73e-12 | 11.1 | 5.01e-12 | Cacna1c/Ryr1/Adra1a/Slc8a1/Cacna2d1/Myo5a/Slc24a4/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Calca/Thy1/Nalf1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Htr2a/Xcl1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Gimap5/Gstm7/Gimap3/Sri/Cav1/Cacna1a/F2r/Cd4 | 37 |
| GO:0031346 | positive regulation of cell projection organization | BP | 0.14 | 4.34e-14 | 7.73e-12 | 11.1 | 5.01e-12 | Ptprz1/Hgf/Sema5a/Acsl6/Fez1/Mdk/Dcc/Plxnb1/Ptprd/Zeb2/Nlgn1/Ccdc88a/Lrrc7/Nrp1/Sema7a/Fn1/Tenm3/Prkd1/Serpine2/Cxcl12/Tbc1d24/Obsl1/Adamts1/Gprc5b/Plxnd1/Serpini1/Abi2/Kit/Epha3/Kctd17/Rp1/Eps8l1/Apoe/Magi2/Crocc/Caprin2/Ndnf/Nin/P3h1/Chodl/Bhlhb9/Ttbk2/Fyn/Dzip1/Dlg4/Cux2/Palm/Dpysl3/Cdc42ep5/Arsb/Def8/Ptn/Gpc2/Cpeb1/Atmin/Zmynd8/Enpp2/Ror1/Bmp7/Setx/Camk1/Numbl | 62 |
| GO:0010977 | negative regulation of neuron projection development | BP | 0.216 | 5.11e-14 | 8.83e-12 | 11.1 | 5.72e-12 | Efnb3/Ptprz1/Map2/Sema5a/Sema5b/Nlgn1/Nrp1/Sema7a/Cspg4/Lrrk2/Ntm/Dab1/Thy1/Epha3/Minar1/Nlgn3/Tsku/Cd38/Apoe/Sema3g/Dennd5a/Rtn4rl1/Epha7/Neo1/Fat3/Xylt1/Dpysl3/Syngap1/Ngef/Psen1/Sema6a/Ulk2/Lrp4/Draxin/Zfp365 | 35 |
| GO:0070588 | calcium ion transmembrane transport | BP | 0.163 | 5.78e-14 | 9.62e-12 | 11 | 6.23e-12 | Nalcn/Cacna1c/Ryr1/Slc8a1/Drd4/Cacna2d1/Ednrb/Myo5a/Slc24a4/Grin2b/Prkd1/Cxcr3/Slc24a3/Cacna1e/Ank2/Slc8a3/Trpc6/Thy1/Nalf1/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Dysf/Bin1/Htr2a/Xcl1/Pkd1/Tspan13/Cacng7/Stac2/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Rrad/Slc25a23/Ptger3/Atp2b4/Gnb5/Gstm7/Atp1a2/Sri/Cacna1a/Cacnb4/F2r | 49 |
| GO:0010959 | regulation of metal ion transport | BP | 0.139 | 5.91e-14 | 9.62e-12 | 11 | 6.23e-12 | Cacna1c/Hecw2/Jph4/Spink1/Igf1/Slc8a1/Nos3/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Icam1/Prkd1/Serpine2/Stc1/Cxcr3/Cxcl12/Ank2/Edn3/Plcb1/Calca/Trpc6/Thy1/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Dysf/Bin1/Adcyap1r1/Htr2a/Tspan13/Stac2/Kcnip3/Selenon/Arrb2/Asph/Osr1/Pkd2/Fhl1/Fyn/Cd84/Rrad/Akt3/Ptger3/Atp2b4/Gnb5/Gimap5/Gstm7/Atp1a2/Gimap3/Sri/Pdgfrb/Cav1/Cacna1a/Usp2/Cacnb4/F2r/Cd4/Mllt6 | 62 |
| GO:0060401 | cytosolic calcium ion transport | BP | 0.194 | 7.68e-14 | 1.21e-11 | 10.9 | 7.86e-12 | Cacna1c/Ryr1/Adra1a/Slc8a1/Cacna2d1/Myo5a/Slc24a4/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Calca/Thy1/Nalf1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Htr2a/Xcl1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Slc25a23/Ccn2/Gimap5/Gstm7/Gimap3/Sri/Cav1/Cacna1a/F2r/Cd4 | 39 |
| GO:0051480 | regulation of cytosolic calcium ion concentration | BP | 0.144 | 1.56e-13 | 2.39e-11 | 10.6 | 1.55e-11 | Kcnk3/Cacna1c/Kdr/Ryr1/Adra1a/Slc8a1/P2rx2/Bok/Cacna2d1/Ednrb/Myo5a/Slc24a4/Lpar4/Grin2b/Prkd1/Cxcr3/Cacna1e/Ptgfr/Ank2/Slc8a3/Calca/Trpc6/Thy1/Pth1r/Nalf1/Coro1a/Aplnr/Cd38/Agtr1a/Ednra/Bdkrb2/Adcyap1r1/Htr2a/Xcl1/Pkd1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Dlg4/Fzd2/Ptger3/Atp2b4/Gimap5/Gstm7/Atp1a2/Gimap3/Sri/Cav1/Cacna1a/F2r/Cd4/Kcnh1/Cd52/Ptger2 | 57 |
| GO:0050678 | regulation of epithelial cell proliferation | BP | 0.141 | 3.64e-13 | 5.44e-11 | 10.3 | 3.53e-11 | Gli1/Ihh/Sfrp2/Sema5a/Ar/Mdk/Itga4/Cd109/Kdr/Agap2/Cyp7b1/Nog/Igf1/Gkn3/Ccnd2/Gpc3/Ednrb/Mef2c/Osr2/Prkd1/Cxcr3/Tek/Cdh13/Cxcl12/Has2/Vash1/Stxbp4/Ptprm/Ift57/Ppp1r16b/Aplnr/Jcad/Cdkn1c/Vegfc/Agtr1a/Apoe/Tie1/Ednra/Igf2/Dysf/Klf9/Twist2/Ptch1/Osr1/Aqp11/Akt3/Apln/Egfl7/Ptn/Cav1/Vash2/Cldn1/Wfdc1/Bcl11b/Myc/Twist1/Eng | 57 |
| GO:0007159 | leukocyte cell-cell adhesion | BP | 0.144 | 4.13e-13 | 5.96e-11 | 10.2 | 3.86e-11 | Ihh/Efnb3/Mdk/Lef1/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Itgb7/Icam1/Podxl2/Cxcl12/Has2/Pik3r6/Gli3/Calca/Thy1/Tnfsf13b/Loxl3/Msn/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Sox12/Havcr2/H2-Ob/Sele/Pecam1/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/Pde5a/H2-Aa/Dlg5/Gimap5/St3gal4/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Bmp7/Il7r/Cd4/Rasal3/Jam2/Lax1 | 55 |
| GO:0016358 | dendrite development | BP | 0.155 | 4.92e-13 | 6.81e-11 | 10.2 | 4.41e-11 | Ptprz1/Map2/Hecw2/Ptprd/Mark1/Nlgn1/Tet1/Mpdz/Nrp1/Lrrk2/Dab1/Mef2c/Tbc1d24/Obsl1/Ppfia2/D16Ertd472e/Trpc6/Dock10/Abi2/Kif1a/Matn2/Nlgn3/Apoe/Caprin2/Prex2/Nfatc4/Sarm1/Bhlhb9/Fyn/Dlg4/Cux2/Palm/Dlg5/Fat3/Syngap1/Ngef/Bbs1/Psen1/Ptn/Cacna1a/Dact1/Zmynd8/Bmp7/Lrp4/Camk1/Nlgn2/Zfp365/Numbl/Rap2a | 49 |
| GO:1903039 | positive regulation of leukocyte cell-cell adhesion | BP | 0.169 | 1.05e-12 | 1.42e-10 | 9.85 | 9.17e-11 | Ihh/Efnb3/Mdk/Lef1/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Has2/Pik3r6/Gli3/Thy1/Tnfsf13b/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Sox12/Havcr2/H2-Ob/Sele/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/H2-Aa/Gimap5/St3gal4/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Il7r/Cd4/Rasal3 | 42 |
| GO:0030326 | embryonic limb morphogenesis | BP | 0.217 | 1.28e-12 | 1.64e-10 | 9.78 | 1.06e-10 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Tbx2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Tbx3/Dync2h1/Hoxa9/Ptch1/Tmem231/Osr1/Ttbk2/Rab23/Psen1/Bmp7/Lrp4/Hand2/Twist1 | 31 |
| GO:0035113 | embryonic appendage morphogenesis | BP | 0.217 | 1.28e-12 | 1.64e-10 | 9.78 | 1.06e-10 | Ihh/Sfrp2/Cacna1c/Lef1/Hoxd10/Hoxd11/Nog/Aff3/Tbx2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Tbx3/Dync2h1/Hoxa9/Ptch1/Tmem231/Osr1/Ttbk2/Rab23/Psen1/Bmp7/Lrp4/Hand2/Twist1 | 31 |
| GO:0007204 | positive regulation of cytosolic calcium ion concentration | BP | 0.144 | 2.6e-12 | 3.21e-10 | 9.49 | 2.08e-10 | Cacna1c/Kdr/Ryr1/Adra1a/Slc8a1/Cacna2d1/Ednrb/Myo5a/Slc24a4/Lpar4/Grin2b/Prkd1/Cxcr3/Cacna1e/Ptgfr/Ank2/Slc8a3/Calca/Trpc6/Thy1/Pth1r/Nalf1/Coro1a/Aplnr/Cd38/Agtr1a/Ednra/Bdkrb2/Adcyap1r1/Htr2a/Xcl1/Pkd1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Dlg4/Fzd2/Ptger3/Gimap5/Gstm7/Gimap3/Sri/Cav1/Cacna1a/F2r/Cd4/Cd52/Ptger2 | 51 |
| GO:0007160 | cell-matrix adhesion | BP | 0.171 | 2.67e-12 | 3.21e-10 | 9.49 | 2.08e-10 | Itga4/Kdr/Phldb2/Npnt/Nrp1/Fermt2/Col16a1/Itgb7/Fn1/Nid1/Jam3/Thsd1/Tek/Cdh13/Itga1/Col13a1/Frem1/Thy1/Postn/Ppm1f/Epha3/Itgal/Limch1/Arhgap6/Emilin1/Vegfc/Sned1/Hoxd3/Pecam1/Itga7/Pcsk5/Dlc1/Epdr1/Ccn2/Efemp2/Cd96/Itga8/Itgb4/Enpp2/Nid2 | 40 |
| GO:0022409 | positive regulation of cell-cell adhesion | BP | 0.154 | 2.67e-12 | 3.21e-10 | 9.49 | 2.08e-10 | Ihh/Efnb3/Mdk/Lef1/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Has2/Pik3r6/Gli3/Podxl/Thy1/Plpp3/Tnfsf13b/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Sox12/Havcr2/H2-Ob/Sele/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/Adam19/H2-Aa/Gimap5/St3gal4/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Bmp7/Il7r/Cd4/Rasal3 | 46 |
| GO:0060485 | mesenchyme development | BP | 0.154 | 3.01e-12 | 3.53e-10 | 9.45 | 2.29e-10 | Sfrp2/Sema5a/Mdk/Lef1/Wnt11/Itga4/Nog/Zeb2/Mark1/Tbx2/Sema5b/Phldb2/Nos3/Nrp1/Fermt2/Sema7a/Cdh2/Fn1/Ednrb/Mef2c/Foxf1/Erg/Has2/Tcf21/Edn3/Loxl3/Epha3/Aplnr/Tbx3/Ednra/Spred3/Heyl/Sema3g/Snai1/Kitl/Dchs1/Osr1/Il17rd/Dlg5/Sema6a/Bmp7/Hey2/Hand2/Myc/Twist1/Eng | 46 |
| GO:0003018 | vascular process in circulatory system | BP | 0.17 | 3.07e-12 | 3.53e-10 | 9.45 | 2.29e-10 | Cacna1c/Angpt1/Kdr/Ddah1/Adra1a/Slc8a1/Nos3/Cbs/Npr3/Fermt2/Ednrb/Mmp2/Ptp4a3/Icam1/Wdr35/Abcg2/Akap12/Edn3/Itga1/Ptprm/Calca/Cdh5/Htr1b/Gucy1a1/Cd38/Agtr1a/Apoe/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Cldn5/Ptger3/Apln/Atp1a2/Abcb1a/Cav1/Abcg3/F2r | 40 |
| GO:0010811 | positive regulation of cell-substrate adhesion | BP | 0.214 | 4.02e-12 | 4.54e-10 | 9.34 | 2.94e-10 | Vit/Col26a1/Mdk/Kdr/Npnt/Nrp1/Fermt2/Spock2/Col16a1/Fn1/Foxf1/Nid1/Tek/Cdh13/Has2/Egflam/Thy1/Ppm1f/Smoc1/Emilin1/Vegfc/Ndnf/Pcsk5/Net1/Abi3bp/Efemp2/Ptn/Hacd1/Enpp2/Ccdc80 | 30 |
| GO:0097485 | neuron projection guidance | BP | 0.162 | 4.59e-12 | 5.07e-10 | 9.29 | 3.29e-10 | Gap43/Epha8/Prtg/Efnb3/Sema5a/Pla2g10/Dcc/Plxnb1/Nog/Evl/Sema5b/Rnf165/Nrp1/Ptpro/Sema7a/Mef2c/Dpysl4/Arhgef25/Cxcl12/Edn3/Gli3/Ptprm/Plxnd1/Gdf7/Lrp2/Lama2/Epha3/Matn2/Nfasc/Ednra/Sema3g/Rtn4rl1/Epha7/Neo1/Ptch1/Unc5c/Sema6a/Bmp7/Bcl11b/Kif5a/Slit3/Draxin | 42 |
| GO:0022407 | regulation of cell-cell adhesion | BP | 0.128 | 5.04e-12 | 5.46e-10 | 9.26 | 3.54e-10 | Ihh/Efnb3/Mdk/Lef1/Itga4/Cd83/Igf1/Abat/Chst2/Cd44/Ets1/Tenm3/Icam1/Serpine2/Cxcl12/Has2/Pik3r6/Gli3/Podxl/Thy1/Plpp3/Tnfsf13b/Loxl3/Coro1a/Cd1d1/Ppm1f/Ccm2l/Itgal/H2-Ab1/Il27ra/Btla/Magi2/Sox12/Havcr2/H2-Ob/Sele/Igf2/Nckap1l/Xcl1/Epha7/H2-Eb1/Stat5b/Pde5a/Adam19/Ptger3/H2-Aa/Dlg5/Gimap5/St3gal4/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Bmp7/Il7r/Cd4/Rasal3/Lax1 | 60 |
| GO:0043113 | receptor clustering | BP | 0.286 | 6.82e-12 | 7.25e-10 | 9.14 | 4.7e-10 | Itga4/Nlgn1/Lrrc7/Cdh2/Itgb7/Grin2b/Slitrk3/Thy1/Glrb/Dlg2/Grik2/Itgal/Apoe/Magi2/Tnfaip6/Dlg4/Syngap1/Arhgef9/Ptn/Cacna1a/Lrp4/Nlgn2 | 22 |
| GO:0032970 | regulation of actin filament-based process | BP | 0.133 | 9.66e-12 | 1.01e-09 | 9 | 6.53e-10 | Kank4/Sema5a/Cacna1c/Mdk/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Hcls1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Stc1/Jam3/Tek/Ank2/Abi2/Coro1a/Ppm1f/Epha3/Esam/Sh3bp1/Limch1/Arhgap6/Nox4/Arhgef15/Cnn2/Rhobtb3/Daam2/Pecam1/Tmeff2/Ppm1e/Scin/Bin1/Nckap1l/Carmil3/Dlc1/Myo1f/Arhgap28/Cacna1h/Fgr/Ptger3/Ccn2/Atp1a2/Cdc42ep5/Sri/Pdgfrb/Cav1/Plekhh2/Cyfip2/Plek | 55 |
| GO:0016055 | Wnt signaling pathway | BP | 0.129 | 1.01e-11 | 1.03e-09 | 8.99 | 6.68e-10 | Gli1/Ndp/Sfrp2/Sema5a/Wnt6/Mdk/Lef1/Apcdd1/Wnt11/Pygo1/Nog/Zeb2/Mark1/Nkd1/Ptpro/Cd44/Fermt2/Arl6/Tert/Cdh2/Gpc3/Lrrk2/Ednrb/Rbms3/Tmem237/Gprc5b/Gli3/Peg12/Wnt9a/Plpp3/Cpe/Znrf3/Dkk3/Enpp1/Tsku/Apoe/Ednra/Daam2/Prickle1/Caprin2/Pkd1/Nfatc4/Tle1/Pkd2/Mgat3/Fzd2/Lats2/Psen1/Adgra2/Cav1/Dact1/Nkd2/Grb10/Tle2/Ror1/Lrp4/Myc/Draxin | 58 |
| GO:0198738 | cell-cell signaling by wnt | BP | 0.128 | 1.21e-11 | 1.19e-09 | 8.92 | 7.71e-10 | Gli1/Ndp/Sfrp2/Sema5a/Wnt6/Mdk/Lef1/Apcdd1/Wnt11/Pygo1/Nog/Zeb2/Mark1/Nkd1/Ptpro/Cd44/Fermt2/Arl6/Tert/Cdh2/Gpc3/Lrrk2/Ednrb/Rbms3/Tmem237/Gprc5b/Gli3/Peg12/Wnt9a/Plpp3/Cpe/Znrf3/Dkk3/Enpp1/Tsku/Apoe/Ednra/Daam2/Prickle1/Caprin2/Pkd1/Nfatc4/Tle1/Pkd2/Mgat3/Fzd2/Lats2/Psen1/Adgra2/Cav1/Dact1/Nkd2/Grb10/Tle2/Ror1/Lrp4/Myc/Draxin | 58 |
| GO:0090287 | regulation of cellular response to growth factor stimulus | BP | 0.15 | 1.25e-11 | 1.21e-09 | 8.92 | 7.87e-10 | Sfrp2/Angpt1/Hhip/Cd109/Kdr/Nog/Zeb2/Rnf165/Npnt/Fst/Gpc3/Ptp4a3/Tek/Zfp423/Cdh5/Lrp2/Pbld2/Zeb1/Jcad/Cdkn1c/Tsku/Emilin1/Vegfc/Tcf4/Spred3/Hipk2/Kcp/Tnfaip6/Neo1/Nrros/Zfyve9/Il17rd/Atp2b4/Apln/Fstl1/Lats2/Adgra2/Sema6a/Cav1/Cyfip2/Itga8/Grb10/Tmprss6/Nrep/Eng | 45 |
| GO:0050890 | cognition | BP | 0.139 | 1.51e-11 | 1.44e-09 | 8.84 | 9.3e-10 | Stra6/Ptprz1/Cacna1c/Mdk/Cyp7b1/Jph4/Nog/Igf1/Aph1b/Aff2/Drd4/Tmod2/Ccnd2/Shc3/Mef2c/Chst10/B4galt2/Grin2b/Pde1b/Adcy3/Shank2/Cacna1e/Plcb1/Eif2ak4/Slc8a3/Abi2/Kit/Mme/Scn2a/Nlgn3/Apoe/Prrt1/Igf2/Htr2a/Nfatc4/Bhlhb9/Pde5a/Mgat3/Cux2/Ddhd2/St3gal4/Atp1a2/Syngap1/Brsk1/Psen1/Ptn/Itga8/Prkar1b/Lins1/Bche | 50 |
| GO:0007411 | axon guidance | BP | 0.159 | 1.59e-11 | 1.49e-09 | 8.83 | 9.68e-10 | Gap43/Epha8/Prtg/Efnb3/Sema5a/Pla2g10/Dcc/Plxnb1/Nog/Evl/Sema5b/Rnf165/Nrp1/Ptpro/Sema7a/Mef2c/Arhgef25/Cxcl12/Edn3/Gli3/Ptprm/Plxnd1/Gdf7/Lrp2/Lama2/Epha3/Matn2/Nfasc/Ednra/Sema3g/Rtn4rl1/Epha7/Neo1/Ptch1/Unc5c/Sema6a/Bmp7/Bcl11b/Kif5a/Slit3/Draxin | 41 |
| GO:0042391 | regulation of membrane potential | BP | 0.126 | 1.66e-11 | 1.53e-09 | 8.82 | 9.91e-10 | Kcnk3/Nalcn/Cacna1c/Grik1/Begain/Kcnd2/Kdr/Kcnd3/Nlgn1/Adra1a/Abat/Slc8a1/Celf4/Rims3/Scn4b/Stx1b/P2rx2/Drd4/Bok/Lrrk2/Scn1a/Cacna2d1/Slc24a4/Mef2c/Ano1/Grin2b/Tbc1d24/Ank2/Slc8a3/Glrb/Slc29a1/Scn2a/Gabra4/Grik2/Nlgn3/Kcnma1/Kcnc4/Bin1/Mpp2/Fhl1/Cacna1h/Dlg4/Cux2/Scn3a/Gimap5/Slc4a8/Gimap3/Psen1/Piezo2/Ptn/Cav1/Pid1/Cacna1a/Zmynd8/Cacnb4/Prkar1b/Kcnh1/Myc/Nlgn2 | 59 |
| GO:0099173 | postsynapse organization | BP | 0.174 | 1.96e-11 | 1.77e-09 | 8.75 | 1.15e-09 | Gap43/Ptprd/Nlgn1/Nrp1/Cdh2/Lrrk2/Lrrc4b/Nptx1/Grin2b/Shank2/Ppfia2/Slitrk3/Dock10/Glrb/Abi2/Kif1a/Nlgn3/Dgkb/Nptxr/Apoe/Magi2/Caprin2/Epha7/Bhlhb9/Lrfn2/Fyn/Dlg4/Cux2/Syngap1/Ngef/Arhgef9/Ptn/Zmynd8/Lrp4/Nlgn2/Zfp365 | 36 |
| GO:0048762 | mesenchymal cell differentiation | BP | 0.161 | 3.21e-11 | 2.84e-09 | 8.55 | 1.84e-09 | Sfrp2/Sema5a/Mdk/Lef1/Wnt11/Nog/Zeb2/Mark1/Sema5b/Phldb2/Nrp1/Fermt2/Sema7a/Cdh2/Fn1/Ednrb/Mef2c/Erg/Has2/Tcf21/Edn3/Loxl3/Epha3/Tbx3/Ednra/Spred3/Heyl/Sema3g/Snai1/Kitl/Osr1/Il17rd/Dlg5/Sema6a/Bmp7/Hey2/Hand2/Twist1/Eng | 39 |
| GO:1903037 | regulation of leukocyte cell-cell adhesion | BP | 0.14 | 3.24e-11 | 2.84e-09 | 8.55 | 1.84e-09 | Ihh/Efnb3/Mdk/Lef1/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Cxcl12/Has2/Pik3r6/Gli3/Thy1/Tnfsf13b/Loxl3/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Sox12/Havcr2/H2-Ob/Sele/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/Pde5a/H2-Aa/Dlg5/Gimap5/St3gal4/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Il7r/Cd4/Rasal3/Lax1 | 48 |
| GO:0050767 | regulation of neurogenesis | BP | 0.125 | 4.32e-11 | 3.68e-09 | 8.43 | 2.38e-09 | Prtg/Efnb3/Ptprz1/Map2/Sema5a/Mdk/Kdr/Plxnb1/Ptprd/Nog/Igf1/Zeb2/Sema5b/Nrp1/Sema7a/Tert/Fn1/Dab1/Id4/Serpine2/Cxcl12/Tbc1d24/Obsl1/Gli3/Trpc6/Plxnd1/Lrp2/Thy1/Kit/Mme/Vegfc/Apoe/Heyl/Sema3g/Daam2/Caprin2/Bin1/Nin/Zcchc24/Nfatc4/Epha7/Chodl/Bhlhb9/Cux2/Syngap1/Psen1/Sema6a/Ulk2/Ptn/Enpp2/Bmp7/Hey2/Lrp4/Myc/Draxin/Zfp365/Numbl | 57 |
| GO:0007264 | small GTPase mediated signal transduction | BP | 0.128 | 4.66e-11 | 3.91e-09 | 8.41 | 2.53e-09 | Igf1/Adra1a/Ccdc88a/Nrp1/Rasgrp3/Arl6/Cdh2/Garnl3/Dab1/Rasa3/Lpar4/Prkd1/Arhgef25/Cdh13/Arhgap25/Dock2/Rasgrp2/Sh2b2/Syde1/Dock10/Abi2/Sh3bp1/Eps8l1/Agtr1a/Apoe/Rasip1/Rhobtb3/Ednra/Pecam1/Adcyap1r1/Kitl/Arap3/Arrb1/Arl3/Heg1/Dlc1/Arhgap28/Net1/Arhgap31/Syngap1/Cdc42ep5/Arhgef9/Rab39b/Mapre2/Lat/Pdgfrb/Dennd4a/Sh2d3c/Iqsec3/Nradd/F2r/Sipa1/Rasal3/Rap2a/Plekhg5 | 55 |
| GO:0022604 | regulation of cell morphogenesis | BP | 0.139 | 6.47e-11 | 5.34e-09 | 8.27 | 3.46e-09 | Mdk/Kdr/Plxnb1/Ptprd/Epb41l3/Cpne5/Nrp1/Cd44/Fermt2/Fn1/Wasf3/Icam1/Tbc1d24/Has2/Obsl1/Myo9a/Plxnd1/Postn/Msn/Coro1a/Kit/Rhobtb3/Caprin2/Slc23a2/Itga7/Arap3/Cfap410/Parvb/Cacng7/Syne3/Dlc1/Bhlhb9/Net1/Fyn/Dlg4/Fgd1/Fgr/Cux2/Palm/Cdc42ep5/Plekho1/Arhgap15/Ptn/Ntng1/Enpp2/Fmnl3/Numbl | 47 |
| GO:0010810 | regulation of cell-substrate adhesion | BP | 0.163 | 7.3e-11 | 5.93e-09 | 8.23 | 3.84e-09 | Ptprz1/Vit/Col26a1/Mdk/Kdr/Phldb2/Npnt/Nrp1/Fermt2/Spock2/Col16a1/Fn1/Foxf1/Nid1/Tek/Cdh13/Has2/Egflam/Thy1/Postn/Ppm1f/Smoc1/Epha3/Limch1/Arhgap6/Emilin1/Vegfc/Ndnf/Pcsk5/Dlc1/Net1/Abi3bp/Efemp2/Ptn/Hacd1/Enpp2/Ccdc80 | 37 |
| GO:1902903 | regulation of supramolecular fiber organization | BP | 0.131 | 8.08e-11 | 6.48e-09 | 8.19 | 4.2e-09 | Kank4/Map2/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Hcls1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Cdh5/Abi2/Coro1a/Ppm1f/Esam/Rp1/Sh3bp1/Limch1/Arhgap6/Tubb4a/Emilin1/Apoe/Nox4/Arhgef15/Daam2/Pecam1/Tmeff2/Ppm1e/Scin/Dysf/Bin1/Nckap1l/Nin/Slain1/Carmil3/Dlc1/Arhgap28/Ttbk2/Ccn2/Efemp2/Cdc42ep5/Psen1/Cav1/Plekhh2/Cyfip2/Plek | 51 |
| GO:0048705 | skeletal system morphogenesis | BP | 0.153 | 8.79e-11 | 6.94e-09 | 8.16 | 4.5e-09 | Col11a1/Ihh/Mmp16/Sfrp2/Hhip/Hoxd10/Hoxd11/Nog/Thbs3/Cbs/Hoxa11/Hoxd9/Mmp2/Mef2c/Hoxd8/Osr2/Hoxd4/Stc1/Has2/Flvcr1/Gli3/Col13a1/Frem1/Wnt9a/Zeb1/Enpp1/Hoxd3/Ltbp3/Pkd1/Hoxa9/Twist2/Osr1/Rab23/Fgr/Ccn2/Psen1/Pdgfrb/Bmp7/Myc/Twist1 | 40 |
| GO:0003007 | heart morphogenesis | BP | 0.148 | 9.5e-11 | 7.4e-09 | 8.13 | 4.79e-09 | Col11a1/Ihh/Sfrp2/Angpt1/Wnt11/Ryr1/Nog/Tbx2/Slc8a1/Nos3/Nrp1/Mef2c/Foxf1/Erg/Tek/Has2/Adamts1/Ift57/Plxnd1/Lrp2/Cpe/Ccm2l/Aplnr/Tbx3/Ednra/Heyl/Snai1/Myl3/Heg1/Dlc1/Ptch1/Dchs1/Pkd2/Fzd2/Fat4/Psen1/Bmp7/Hey2/Hand2/Slit3/Twist1/Eng | 42 |
| GO:0045765 | regulation of angiogenesis | BP | 0.143 | 9.71e-11 | 7.46e-09 | 8.13 | 4.83e-09 | Cyp1b1/Hgf/Sfrp2/Sema5a/Hhip/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Cxcr3/Tek/Pik3r6/Adamts1/Vash1/Ptprm/Plxnd1/Cdh5/Ppp1r16b/Aplnr/Minar1/Jcad/Emilin1/Ecscr/Vegfc/Agtr1a/Tcf4/Tie1/Igf2/Hipk2/Cldn5/Akt3/Atp2b4/Adgra2/Sema6a/Ptn/Vash2/Enpp2/Stab1/Fbln5/Eng | 44 |
| GO:0003012 | muscle system process | BP | 0.126 | 1.28e-10 | 9.7e-09 | 8.01 | 6.28e-09 | Ar/Cacna1c/Sgcd/Col14a1/Ryr1/Igf1/Kcnd3/Tbx2/Npnt/Adra1a/Abat/Slc8a1/Nos3/Mybpc1/Scn4b/P2rx2/Tmod2/Scn1a/Cacna2d1/Ednrb/Mef2c/Stc1/Ank2/Edn3/Slc8a3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Tbx3/Kcnma1/Ednra/Bin1/Bdkrb2/Htr2a/Myl3/Cmya5/Stac2/Selenon/Pde5a/Ryr3/Pi16/Calcrl/Cacna1h/Ptger3/Atp2b4/Ccn2/Atp1a2/Sri/Cav1/F2r/Hey2/Hand2 | 54 |
| GO:0001505 | regulation of neurotransmitter levels | BP | 0.154 | 1.42e-10 | 1.06e-08 | 7.97 | 6.89e-09 | Syn2/Maob/Slc6a2/Nlgn1/Adra1a/Abat/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Pde1b/Maoa/Edn3/Ppfia2/Syde1/Htr1b/Naalad2/Slc29a1/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Ncs1/Erc2/Atp1a2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Kcnh1/Bche | 39 |
| GO:1901342 | regulation of vasculature development | BP | 0.141 | 1.49e-10 | 1.1e-08 | 7.96 | 7.11e-09 | Cyp1b1/Hgf/Sfrp2/Sema5a/Hhip/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Cxcr3/Tek/Pik3r6/Adamts1/Vash1/Ptprm/Plxnd1/Cdh5/Ppp1r16b/Aplnr/Minar1/Jcad/Emilin1/Ecscr/Vegfc/Agtr1a/Tcf4/Tie1/Igf2/Hipk2/Cldn5/Akt3/Atp2b4/Adgra2/Sema6a/Ptn/Vash2/Enpp2/Stab1/Fbln5/Eng | 44 |
| GO:0006936 | muscle contraction | BP | 0.14 | 2.04e-10 | 1.48e-08 | 7.83 | 9.61e-09 | Cacna1c/Sgcd/Ryr1/Kcnd3/Tbx2/Npnt/Adra1a/Abat/Slc8a1/Mybpc1/Scn4b/P2rx2/Tmod2/Scn1a/Cacna2d1/Ednrb/Stc1/Ank2/Edn3/Slc8a3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Tbx3/Kcnma1/Ednra/Bin1/Bdkrb2/Htr2a/Myl3/Stac2/Pde5a/Ryr3/Calcrl/Cacna1h/Ptger3/Atp2b4/Ccn2/Atp1a2/Sri/Cav1/F2r | 44 |
| GO:0060840 | artery development | BP | 0.216 | 2.15e-10 | 1.54e-08 | 7.81 | 9.98e-09 | Apob/Stra6/Mdk/Wnt11/Nog/Tbx2/Nrp1/Kif7/Foxf1/Fkbp10/Gli3/Plxnd1/Lrp2/Prdm1/Aplnr/Enpp1/Apoe/Ednra/Prickle1/Akt3/Efemp2/Pdgfrb/Hey2/Hand2/Eng | 25 |
| GO:0018212 | peptidyl-tyrosine modification | BP | 0.138 | 2.32e-10 | 1.64e-08 | 7.78 | 1.07e-08 | Ptprz1/Hgf/Sfrp2/Angpt1/Kdr/Spink1/Igf1/Ddr2/Pkdcc/Adra1a/Nrp1/Hcls1/Cd44/Cspg4/Areg/Icam1/Tpst1/Itk/Tek/Gprc5b/Thy1/Plpp3/Abi2/Kit/Epha3/Vegfc/Agtr1a/Nox4/Pecam1/Igf2/Kitl/Htr2a/Epha7/Arrb2/Fyn/Dlg4/Fgr/Zap70/Psen1/Pdgfrb/Il12rb1/Cav1/Enpp2/Cd4/Lrp4 | 45 |
| GO:1903169 | regulation of calcium ion transmembrane transport | BP | 0.178 | 2.58e-10 | 1.78e-08 | 7.75 | 1.15e-08 | Cacna1c/Slc8a1/Drd4/Cacna2d1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Dysf/Bin1/Tspan13/Stac2/Selenon/Asph/Pkd2/Fyn/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Cacna1a/Cacnb4/F2r | 31 |
| GO:0006939 | smooth muscle contraction | BP | 0.214 | 2.61e-10 | 1.78e-08 | 7.75 | 1.15e-08 | Cacna1c/Tbx2/Npnt/Adra1a/Abat/Slc8a1/P2rx2/Ednrb/Edn3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Tbx3/Kcnma1/Ednra/Bdkrb2/Htr2a/Calcrl/Ptger3/Atp2b4/Atp1a2/Cav1/F2r | 25 |
| GO:0097553 | calcium ion transmembrane import into cytosol | BP | 0.187 | 2.91e-10 | 1.96e-08 | 7.71 | 1.27e-08 | Cacna1c/Ryr1/Slc8a1/Myo5a/Slc24a4/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Thy1/Nalf1/Coro1a/Aplnr/Agtr1a/Ednra/Htr2a/Xcl1/Selenon/Asph/Pkd2/Fyn/Xcr1/Ryr3/Gstm7/Sri/Cacna1a/F2r | 29 |
| GO:0050679 | positive regulation of epithelial cell proliferation | BP | 0.161 | 3.17e-10 | 2.11e-08 | 7.67 | 1.37e-08 | Ihh/Sema5a/Ar/Mdk/Itga4/Kdr/Agap2/Cyp7b1/Nog/Igf1/Ccnd2/Osr2/Prkd1/Cdh13/Cxcl12/Has2/Stxbp4/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Igf2/Dysf/Twist2/Osr1/Akt3/Apln/Egfl7/Ptn/Cav1/Vash2/Cldn1/Myc/Twist1 | 35 |
| GO:0009100 | glycoprotein metabolic process | BP | 0.136 | 3.83e-10 | 2.52e-08 | 7.6 | 1.63e-08 | Col11a1/Ihh/St6gal2/Itm2a/Igf1/St8sia2/St8sia4/Gxylt2/Tet1/Man2a2/Uggt2/Spock2/Soat1/Chst10/Tmtc1/B3galnt1/Fut10/Egflam/Plcb1/Galnt16/St3gal2/Mamdc2/St6galnac4/Fbxo17/Eogt/Ggta1/Ndnf/Adamts7/St8sia6/Galntl6/Bgn/Poglut2/Dse/Mgat3/Pomt2/Aqp11/Large1/St6galnac6/B3gnt5/Xylt1/St3gal4/Psen1/Alg14/Galnt18/Dpy19l1 | 45 |
| GO:1904062 | regulation of cation transmembrane transport | BP | 0.129 | 4.05e-10 | 2.63e-08 | 7.58 | 1.7e-08 | Cacna1c/Hecw2/Nlgn1/Slc8a1/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Mef2c/Prkd1/Cxcr3/Ank2/Edn3/Plcb1/Thy1/Coro1a/Aplnr/Nlgn3/Cracr2a/Agtr1a/Prrt1/Ednra/Dysf/Bin1/Tspan13/Cacng7/Stac2/Kcnip3/Selenon/Asph/Osr1/Pkd2/Fhl1/Fyn/Rrad/Dlg4/Ptger3/Atp2b4/Gnb5/Gstm7/Atp1a2/Sri/Cav1/Cacna1a/Cacnb4/F2r/Twist1/Nlgn2 | 49 |
| GO:0045055 | regulated exocytosis | BP | 0.143 | 4.64e-10 | 2.98e-08 | 7.53 | 1.93e-08 | Syn2/Cacna1c/Nlgn1/Syt13/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Foxf1/Pfn2/Mical1/Ppfia2/Syde1/Htr1b/Coro1a/Kit/Cadps/Syt7/Prrt2/Nckap1l/Htr2a/Myo1f/Ncs1/Erc2/Cd84/Cacna1h/Fgr/Slc4a8/Stx2/Psen1/Rab27a/Lat/Clnk/Nrn1/Cacna1a/Pld2/Cacnb4/Kcnh1/Plek | 41 |
| GO:0008589 | regulation of smoothened signaling pathway | BP | 0.234 | 4.83e-10 | 3.07e-08 | 7.51 | 1.99e-08 | Gli1/Ihh/Hhip/Evc/Kif7/Arl6/Gpc3/Cibar1/Serpine2/Armc9/Gli3/Wnt9a/Rab34/Gpr161/Enpp1/Dync2h1/Ptch1/Ttbk2/Rab23/Dlg5/Glis2/Ift81 | 22 |
| GO:0003205 | cardiac chamber development | BP | 0.165 | 5.53e-10 | 3.47e-08 | 7.46 | 2.25e-08 | Col11a1/Stra6/Sfrp2/Wnt11/Nog/Tbx2/Nos3/Nrp1/Kif7/Mef2c/Foxf1/Tek/Adamts1/Ank2/Plxnd1/Lrp2/Cpe/Prdm1/Cplane1/Ccm2l/Aplnr/Tbx3/Ednra/Heyl/Myl3/Pcsk5/Heg1/Fzd2/Bmp7/Hey2/Hand2/Slit3/Eng | 33 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | BP | 0.136 | 5.63e-10 | 3.49e-08 | 7.46 | 2.26e-08 | Ptprz1/Hgf/Sfrp2/Angpt1/Kdr/Spink1/Igf1/Ddr2/Pkdcc/Adra1a/Nrp1/Hcls1/Cd44/Cspg4/Areg/Icam1/Itk/Tek/Gprc5b/Thy1/Plpp3/Abi2/Kit/Epha3/Vegfc/Agtr1a/Nox4/Pecam1/Igf2/Kitl/Htr2a/Epha7/Arrb2/Fyn/Dlg4/Fgr/Zap70/Psen1/Pdgfrb/Il12rb1/Cav1/Enpp2/Cd4/Lrp4 | 44 |
| GO:0060070 | canonical Wnt signaling pathway | BP | 0.138 | 8.12e-10 | 4.99e-08 | 7.3 | 3.23e-08 | Gli1/Sfrp2/Sema5a/Wnt6/Mdk/Lef1/Wnt11/Pygo1/Nog/Zeb2/Nkd1/Ptpro/Cdh2/Gpc3/Lrrk2/Ednrb/Rbms3/Gprc5b/Gli3/Peg12/Wnt9a/Plpp3/Znrf3/Dkk3/Apoe/Ednra/Daam2/Prickle1/Caprin2/Tle1/Mgat3/Fzd2/Lats2/Psen1/Adgra2/Cav1/Dact1/Nkd2/Tle2/Lrp4/Myc/Draxin | 42 |
| GO:0051145 | smooth muscle cell differentiation | BP | 0.25 | 8.46e-10 | 5.12e-08 | 7.29 | 3.31e-08 | Tbx2/Npnt/Ednrb/Mef2c/Foxf1/Zeb1/Tshz3/Kit/Cth/Aplnr/Tbx3/Ednra/Nfatc4/Rbpms2/Efemp2/Sgcb/Pdgfrb/Itga8/Hey2/Eng | 20 |
| GO:0072073 | kidney epithelium development | BP | 0.184 | 8.51e-10 | 5.12e-08 | 7.29 | 3.31e-08 | Wnt6/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Gdf11/Tcf21/Gli3/Tshz3/Agtr1a/Ednra/Heyl/Smad9/Epha7/Ptch1/Dchs1/Osr1/Aqp11/Fat4/Bmp7/Vcan/Myc | 28 |
| GO:0035296 | regulation of tube diameter | BP | 0.173 | 9.81e-10 | 5.69e-08 | 7.24 | 3.69e-08 | Cacna1c/Kdr/Adra1a/Slc8a1/Nos3/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Ptprm/Calca/Htr1b/Gucy1a1/Cd38/Agtr1a/Apoe/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1/F2r | 30 |
| GO:0097746 | blood vessel diameter maintenance | BP | 0.173 | 9.81e-10 | 5.69e-08 | 7.24 | 3.69e-08 | Cacna1c/Kdr/Adra1a/Slc8a1/Nos3/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Ptprm/Calca/Htr1b/Gucy1a1/Cd38/Agtr1a/Apoe/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1/F2r | 30 |
| GO:0050803 | regulation of synapse structure or activity | BP | 0.144 | 9.88e-10 | 5.69e-08 | 7.24 | 3.69e-08 | Gpc6/Ptprd/St8sia2/Nlgn1/Ptpro/Rims3/Cdh2/Lrrk2/Lrrc4b/Mef2c/Nptx1/Grin2b/Shank2/Adgrl1/Ppfia2/Slitrk3/Abi2/Kif1a/Nlgn3/Dgkb/Apoe/Arhgef15/Magi2/Sparcl1/Caprin2/Nfatc4/Epha7/Bhlhb9/Lrfn2/Fyn/Cux2/Dlg5/Syngap1/Ngef/Ptn/Zmynd8/Lrp4/Camk1/Nlgn2 | 39 |
| GO:0035150 | regulation of tube size | BP | 0.172 | 1.13e-09 | 6.45e-08 | 7.19 | 4.18e-08 | Cacna1c/Kdr/Adra1a/Slc8a1/Nos3/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Ptprm/Calca/Htr1b/Gucy1a1/Cd38/Agtr1a/Apoe/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1/F2r | 30 |
| GO:0048562 | embryonic organ morphogenesis | BP | 0.132 | 1.35e-09 | 7.59e-08 | 7.12 | 4.92e-08 | Gli1/Col11a1/Stra6/Ihh/Mmp16/Wnt11/Hoxd10/Hoxd11/Nog/Tbx2/Hoxa11/Hoxd9/Mef2c/Foxf1/Lrig3/Osr2/Hoxd4/Tshz1/Flvcr1/Tcf21/Gli3/Ift57/Wnt9a/Zeb1/Aplnr/Enpp1/Tbx3/Hoxd3/Ednra/Hipk2/Hoxa9/Twist2/Osr1/Pkd2/Fzd2/Psen1/Hipk1/Kcnq4/Itga8/Bmp7/Hand2/Myc/Twist1/Eng | 44 |
| GO:0050807 | regulation of synapse organization | BP | 0.144 | 1.44e-09 | 8.04e-08 | 7.09 | 5.21e-08 | Gpc6/Ptprd/St8sia2/Nlgn1/Ptpro/Rims3/Cdh2/Lrrk2/Lrrc4b/Mef2c/Nptx1/Grin2b/Shank2/Adgrl1/Ppfia2/Slitrk3/Abi2/Kif1a/Nlgn3/Dgkb/Apoe/Arhgef15/Magi2/Sparcl1/Caprin2/Nfatc4/Epha7/Bhlhb9/Lrfn2/Fyn/Cux2/Dlg5/Ngef/Ptn/Zmynd8/Lrp4/Camk1/Nlgn2 | 38 |
| GO:0048813 | dendrite morphogenesis | BP | 0.166 | 1.62e-09 | 8.98e-08 | 7.05 | 5.82e-08 | Ptprz1/Hecw2/Ptprd/Nlgn1/Tet1/Nrp1/Lrrk2/Tbc1d24/Obsl1/Ppfia2/Trpc6/Dock10/Abi2/Kif1a/Nlgn3/Caprin2/Prex2/Nfatc4/Sarm1/Bhlhb9/Fyn/Dlg4/Cux2/Ngef/Ptn/Cacna1a/Dact1/Lrp4/Zfp365/Numbl/Rap2a | 31 |
| GO:1901888 | regulation of cell junction assembly | BP | 0.155 | 1.79e-09 | 9.78e-08 | 7.01 | 6.34e-08 | Gpc6/Kdr/Ptprd/St8sia2/Nlgn1/Phldb2/Nrp1/Fermt2/Col16a1/Lrrc4b/Mef2c/Nptx1/Tek/Adgrl1/Slitrk3/Thy1/Ppm1f/Epha3/Aplnr/Nlgn3/Limch1/Arhgap6/Snai1/Epha7/Dlc1/Bhlhb9/Nphp1/Cldn5/Cux2/Dlg5/Cav1/Enpp2/Cldn1/Nlgn2 | 34 |
| GO:0072006 | nephron development | BP | 0.178 | 1.83e-09 | 9.91e-08 | 7 | 6.42e-08 | Wnt6/Angpt1/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Nid1/Enpep/Tek/Tcf21/Gli3/Agtr1a/Ednra/Heyl/Ptch1/Dchs1/Osr1/Aqp11/Fat4/Pdgfrb/Bmp7/Myc | 28 |
| GO:0007162 | negative regulation of cell adhesion | BP | 0.134 | 1.99e-09 | 1.06e-07 | 6.98 | 6.85e-08 | Ihh/Cyp1b1/Ptprz1/Sema5a/Angpt1/Mdk/Lef1/Plxnb1/Phldb2/Abat/Cd44/Mmp2/Dab1/Serpine2/Jam3/Cdh13/Cxcl12/Gli3/Podxl/Plxnd1/Postn/Loxl3/Ppm1f/Ccm2l/H2-Ab1/Btla/Arhgap6/Havcr2/Nckap1l/Dlc1/Myo1f/Pde5a/Ptger3/H2-Aa/Dlg5/Gimap5/Gimap3/Sema6a/Spn/Enpp2/Jam2/Lax1 | 42 |
| GO:0010631 | epithelial cell migration | BP | 0.134 | 1.99e-09 | 1.06e-07 | 6.98 | 6.85e-08 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Evl/Zeb2/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Ptp4a3/Prkd1/Stc1/Tek/Cdh13/Has2/Vash1/Ptprm/Calca/Plxnd1/Cdh5/Plpp3/Ppm1f/Kit/Jcad/Sh3bp1/Vegfc/Apoe/Daam2/Pecam1/Igf2/Hdac9/Akt3/Atp2b4/Fstl1/Arsb/Adgra2/Mapre2/Enpp2/Plekhg5 | 42 |
| GO:0014848 | urinary tract smooth muscle contraction | BP | 0.643 | 2.15e-09 | 1.13e-07 | 6.95 | 7.33e-08 | Cacna1c/Tbx2/P2rx2/Tshz3/Tbx3/Kcnma1/Htr2a/Ptger3/Atp2b4 | 9 |
| GO:0001649 | osteoblast differentiation | BP | 0.153 | 2.27e-09 | 1.18e-07 | 6.93 | 7.68e-08 | Gli1/Ihh/Sfrp2/Wnt11/Nog/Igf1/Ddr2/Npnt/Fermt2/Areg/Mef2c/Id3/Id4/Prkd1/Gli3/Glis1/Nell1/Pth1r/Gdf10/Smoc1/Enpp1/Vegfc/Igf2/Rorb/Tnfaip6/Twist2/Rassf2/Ptch1/Lrp3/Gabbr1/Bmp7/Hand2/Twist1/Zfp932 | 34 |
| GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.145 | 2.34e-09 | 1.21e-07 | 6.92 | 7.83e-08 | Sfrp2/Cd109/Kdr/Nog/Zeb2/Rnf165/Npnt/Fst/Gpc3/Gdf11/Zfp423/Cdh5/Gdf7/Lrp2/Pbld2/Zeb1/Gdf10/Cdkn1c/Tsku/Emilin1/Magi2/Spred3/Hipk2/Kcp/Tnfaip6/Neo1/Nrros/Zfyve9/Il17rd/Fstl1/Lats2/Cav1/Itga8/Tmprss6/Bmp7/Nrep/Eng | 37 |
| GO:0090132 | epithelium migration | BP | 0.133 | 2.41e-09 | 1.23e-07 | 6.91 | 8e-08 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Evl/Zeb2/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Ptp4a3/Prkd1/Stc1/Tek/Cdh13/Has2/Vash1/Ptprm/Calca/Plxnd1/Cdh5/Plpp3/Ppm1f/Kit/Jcad/Sh3bp1/Vegfc/Apoe/Daam2/Pecam1/Igf2/Hdac9/Akt3/Atp2b4/Fstl1/Arsb/Adgra2/Mapre2/Enpp2/Plekhg5 | 42 |
| GO:0099068 | postsynapse assembly | BP | 0.382 | 2.49e-09 | 1.26e-07 | 6.9 | 8.19e-08 | Gap43/Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Slitrk3/Nlgn3/Nptxr/Magi2/Arhgef9/Lrp4/Nlgn2 | 13 |
| GO:0010522 | regulation of calcium ion transport into cytosol | BP | 0.207 | 2.53e-09 | 1.27e-07 | 6.9 | 8.25e-08 | Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Calca/Thy1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Selenon/Asph/Pkd2/Fyn/Gimap5/Gstm7/Gimap3/Sri/Cav1/F2r/Cd4 | 23 |
| GO:0050870 | positive regulation of T cell activation | BP | 0.152 | 2.56e-09 | 1.27e-07 | 6.89 | 8.26e-08 | Ihh/Efnb3/Mdk/Lef1/Cd83/Igf1/Pik3r6/Gli3/Thy1/Tnfsf13b/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Sox12/Havcr2/H2-Ob/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Il7r/Cd4/Rasal3 | 34 |
| GO:0048880 | sensory system development | BP | 0.119 | 2.6e-09 | 1.28e-07 | 6.89 | 8.31e-08 | Ndp/Tmem132e/Stra6/Ihh/Cyp1b1/Sdk2/Cacna1c/Nhs/Kdr/Zeb2/Rcn1/Tub/Tbx2/Celf4/Nrp1/Lamc3/Arl6/Tenm3/Osr2/Obsl1/Bbs7/Sall2/Gdf11/Gli3/Ptprm/Thy1/Tgif2/Zeb1/Prdm1/Abi2/Smoc1/Rp1/Cdkn1c/Tsku/Arhgef15/Spred3/Col5a2/Pbx4/Lpcat1/Hipk2/Rorb/Twist2/Tmem231/Nphp1/Fat3/Pdgfrb/Ptn/Hipk1/Bmp7/Bcl11b/Twist1 | 51 |
| GO:0030336 | negative regulation of cell migration | BP | 0.137 | 2.62e-09 | 1.28e-07 | 6.89 | 8.31e-08 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Phldb2/Tcaf1/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Ptprm/Thy1/Gpr173/Robo4/Il27ra/Limch1/Emilin1/Apoe/Magi2/Cnn2/Tie1/Cd200/Tmeff2/Arap3/Tnfaip6/Dlc1/Cldn5/Atp2b4/Dlg5/Dpysl3/Egfl7/Ptn/Cygb/Zmynd8/Rap2a/Eng | 40 |
| GO:0048015 | phosphatidylinositol-mediated signaling | BP | 0.171 | 2.75e-09 | 1.33e-07 | 6.88 | 8.64e-08 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Car8/Hcls1/Npr3/Fn1/Serpine2/Tek/Plcb1/Ppp1r16b/Prr5l/Htr2a/Prex2/Plcb2/Ncs1/Fyn/Fgr/Pdgfrb/Pld2/Ror1/F2r/Kcnh1/Twist1/Irs1 | 29 |
| GO:0072132 | mesenchyme morphogenesis | BP | 0.296 | 2.78e-09 | 1.34e-07 | 6.87 | 8.67e-08 | Lef1/Wnt11/Nog/Tbx2/Nos3/Foxf1/Aplnr/Tbx3/Heyl/Snai1/Dchs1/Osr1/Bmp7/Myc/Twist1/Eng | 16 |
| GO:0043270 | positive regulation of ion transport | BP | 0.131 | 2.89e-09 | 1.36e-07 | 6.87 | 8.83e-08 | Cacna1c/Grik1/Pla2g10/Tcaf1/Abat/Scn4b/Drd4/Cacna2d1/Myo5a/Grin2b/Stc1/Cxcr3/Cxcl12/Ank2/Edn3/Plcb1/Trpc6/Thy1/Aplnr/Nlgn3/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Stac2/Arrb2/Asph/Pkd2/Fhl1/Akt3/Gimap5/Gstm7/Gimap3/Psen1/Abcb1a/Sri/Pdgfrb/Gabbr1/Cav1/Cacnb4/F2r/Cd4/Mllt6 | 43 |
| GO:0032956 | regulation of actin cytoskeleton organization | BP | 0.126 | 2.9e-09 | 1.36e-07 | 6.87 | 8.83e-08 | Kank4/Sema5a/Mdk/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Hcls1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Jam3/Tek/Abi2/Coro1a/Ppm1f/Epha3/Esam/Sh3bp1/Limch1/Arhgap6/Nox4/Arhgef15/Rhobtb3/Daam2/Pecam1/Tmeff2/Ppm1e/Scin/Bin1/Nckap1l/Carmil3/Dlc1/Myo1f/Arhgap28/Fgr/Ccn2/Cdc42ep5/Pdgfrb/Plekhh2/Cyfip2/Plek | 46 |
| GO:0060348 | bone development | BP | 0.143 | 2.91e-09 | 1.36e-07 | 6.87 | 8.83e-08 | Ihh/Mmp16/Sfrp2/Hoxd11/Kdr/Ryr1/Plxnb1/Igf1/Ddr2/Thbs3/Evc/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Has2/Lrrc17/Gli3/Col13a1/Frem1/Fli1/Kit/Enpp1/Adamts7/Ltbp3/Tal1/Bgn/Smad9/P3h1/Dchs1/Rab23/Xylt1/Fat4/Twist1/Papss2/Fbln5 | 37 |
| GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | BP | 0.205 | 3.04e-09 | 1.41e-07 | 6.85 | 9.16e-08 | Prtg/Pcdh18/Hmcn1/Sdk2/Cdh2/Cdh6/Tenm3/Cdh13/Pcdhgc3/Obsl1/Cdh10/Ptprm/Cdh5/Pcdh19/Pcdh12/Palld/Esam/Cdh11/Pecam1/Dchs1/Fat3/Fat4/Pcdhb14 | 23 |
| GO:0007015 | actin filament organization | BP | 0.116 | 3.57e-09 | 1.64e-07 | 6.78 | 1.07e-07 | Kank4/Hmcn1/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Hcls1/Fermt2/Aif1l/Tmod2/Wasf3/Myo5a/Pfn2/Icam1/Mical1/Arhgap25/Abi2/Catip/Coro1a/Ppm1f/Esam/Sh3bp1/Limch1/Arhgap6/Nox4/Arhgef15/Rhobtb3/Daam2/Pecam1/Hip1/Tmeff2/Ppm1e/Scin/Bin1/Nckap1l/Efs/Carmil3/Arrb1/Dlc1/Myo1f/Arhgap28/Wipf1/Ccn2/Dpysl3/Cdc42ep5/Elmo2/Plekhh2/Cyfip2/Shroom1/Plek/Pstpip1 | 53 |
| GO:0099504 | synaptic vesicle cycle | BP | 0.15 | 4.09e-09 | 1.87e-07 | 6.73 | 1.21e-07 | Syn2/Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Cdh2/Lrrk2/Pfn2/Tbc1d24/Ppfia2/Syde1/Htr1b/Cadps/Nlgn3/Magi2/Syt7/Prrt2/Bin1/Htr2a/Pcdh17/Ncs1/Erc2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Prkar1b/Kcnh1/Nlgn2 | 34 |
| GO:0001654 | eye development | BP | 0.119 | 4.16e-09 | 1.87e-07 | 6.73 | 1.21e-07 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Cacna1c/Nhs/Kdr/Zeb2/Rcn1/Tub/Tbx2/Celf4/Nrp1/Lamc3/Arl6/Tenm3/Osr2/Obsl1/Bbs7/Sall2/Gdf11/Gli3/Ptprm/Thy1/Tgif2/Zeb1/Prdm1/Abi2/Smoc1/Rp1/Cdkn1c/Tsku/Arhgef15/Spred3/Col5a2/Pbx4/Lpcat1/Hipk2/Rorb/Twist2/Tmem231/Nphp1/Fat3/Pdgfrb/Ptn/Hipk1/Bmp7/Bcl11b/Twist1 | 50 |
| GO:0048017 | inositol lipid-mediated signaling | BP | 0.168 | 4.16e-09 | 1.87e-07 | 6.73 | 1.21e-07 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Car8/Hcls1/Npr3/Fn1/Serpine2/Tek/Plcb1/Ppp1r16b/Prr5l/Htr2a/Prex2/Plcb2/Ncs1/Fyn/Fgr/Pdgfrb/Pld2/Ror1/F2r/Kcnh1/Twist1/Irs1 | 29 |
| GO:0019932 | second-messenger-mediated signaling | BP | 0.133 | 4.3e-09 | 1.92e-07 | 6.72 | 1.24e-07 | Sgcd/Kdr/Nr5a2/Spink1/Igf1/Slc8a1/Nos3/Cbs/Drd4/Gnai1/Lrrk2/Ednrb/Slc24a4/Gucy1a2/Cxcr3/Cdh13/Ptgfr/Adgrl1/Ank2/Edn3/Gucy1b1/Gucy1a1/Aplnr/Itgal/Agtr1a/Apoe/Ndnf/Adcyap1r1/Cap2/Cmya5/Nfatc4/Rasd2/Xcr1/Pde5a/Ptger3/Atp2b4/Zap70/Lat/Rcan3/Cd4/Tbc1d10c | 41 |
| GO:0042733 | embryonic digit morphogenesis | BP | 0.254 | 4.61e-09 | 2.04e-07 | 6.69 | 1.32e-07 | Ihh/Sfrp2/Hoxd11/Nog/Tbx2/Hoxa11/Osr2/Flvcr1/Gli3/Cplane1/Tbx3/Tmem231/Osr1/Ttbk2/Rab23/Lrp4/Hand2/Twist1 | 18 |
| GO:0007269 | neurotransmitter secretion | BP | 0.162 | 5.05e-09 | 2.2e-07 | 6.66 | 1.42e-07 | Syn2/Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Syde1/Htr1b/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Ncs1/Erc2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 30 |
| GO:0099643 | signal release from synapse | BP | 0.162 | 5.05e-09 | 2.2e-07 | 6.66 | 1.42e-07 | Syn2/Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Syde1/Htr1b/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Ncs1/Erc2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 30 |
| GO:0150063 | visual system development | BP | 0.118 | 5.27e-09 | 2.28e-07 | 6.64 | 1.47e-07 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Cacna1c/Nhs/Kdr/Zeb2/Rcn1/Tub/Tbx2/Celf4/Nrp1/Lamc3/Arl6/Tenm3/Osr2/Obsl1/Bbs7/Sall2/Gdf11/Gli3/Ptprm/Thy1/Tgif2/Zeb1/Prdm1/Abi2/Smoc1/Rp1/Cdkn1c/Tsku/Arhgef15/Spred3/Col5a2/Pbx4/Lpcat1/Hipk2/Rorb/Twist2/Tmem231/Nphp1/Fat3/Pdgfrb/Ptn/Hipk1/Bmp7/Bcl11b/Twist1 | 50 |
| GO:0050863 | regulation of T cell activation | BP | 0.126 | 5.31e-09 | 2.28e-07 | 6.64 | 1.47e-07 | Ihh/Efnb3/Mdk/Lef1/Cd83/Igf1/Cd44/Mapk8ip1/Pik3r6/Gli3/Thy1/Tnfsf13b/Loxl3/Zeb1/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Sox12/Havcr2/H2-Ob/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/Pde5a/H2-Aa/Dlg5/Gimap5/Zap70/Gimap3/Lat/Il12rb1/Cav1/Il2rg/Spn/Il7r/Cd4/Rasal3/Lax1 | 44 |
| GO:0007043 | cell-cell junction assembly | BP | 0.174 | 6.02e-09 | 2.56e-07 | 6.59 | 1.66e-07 | Wnt11/Epb41l3/Mpdz/Ptpro/Cdh2/Cdh6/Jam3/Cldn8/Cdh10/Cdh5/Abi2/Aplnr/Esam/Nfasc/Cdh11/Gjc1/Pecam1/Snai1/Heg1/Dchs1/Nphp1/Cldn5/Dlg5/Hipk1/Cav1/Pdcd6ip/Cldn1 | 27 |
| GO:0051928 | positive regulation of calcium ion transport | BP | 0.179 | 6.11e-09 | 2.58e-07 | 6.59 | 1.67e-07 | Cacna1c/Cacna2d1/Stc1/Cxcr3/Cxcl12/Ank2/Trpc6/Thy1/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Stac2/Arrb2/Asph/Pkd2/Gimap5/Gstm7/Gimap3/Sri/Pdgfrb/Cav1/Cacnb4/F2r/Cd4 | 26 |
| GO:0006836 | neurotransmitter transport | BP | 0.145 | 6.18e-09 | 2.59e-07 | 6.59 | 1.68e-07 | Syn2/Slc6a2/Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Syde1/Htr1b/Slc6a15/Slc29a1/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Ncs1/Sv2c/Erc2/Atp1a2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 35 |
| GO:1903522 | regulation of blood circulation | BP | 0.139 | 6.76e-09 | 2.81e-07 | 6.55 | 1.82e-07 | Cacna1c/Kcnd3/Adra1a/Slc8a1/Nos3/Scn4b/Cacna2d1/Ednrb/Mmp2/Icam1/Wdr35/Stc1/Hopx/Cacna1e/Ank2/Edn3/Calca/Cd38/Agtr1a/Gjc1/Ednra/Bin1/Htr2a/Myl3/Pde5a/Slc4a3/Cacna1h/Ptger3/Atp2b4/Ccn2/Apln/Atp1a2/Sri/Cav1/F2r/Hey2/Ptger2 | 37 |
| GO:0051271 | negative regulation of cellular component movement | BP | 0.131 | 6.89e-09 | 2.84e-07 | 6.55 | 1.84e-07 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Phldb2/Tcaf1/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Ptprm/Thy1/Gpr173/Robo4/Il27ra/Limch1/Emilin1/Apoe/Magi2/Cnn2/Tie1/Cd200/Tmeff2/Bin1/Arap3/Tnfaip6/Dlc1/Cldn5/Atp2b4/Dlg5/Dpysl3/Egfl7/Ptn/Cygb/Zmynd8/Rap2a/Eng | 41 |
| GO:0048844 | artery morphogenesis | BP | 0.235 | 7.02e-09 | 2.87e-07 | 6.54 | 1.86e-07 | Apob/Stra6/Mdk/Wnt11/Nog/Tbx2/Nrp1/Foxf1/Fkbp10/Lrp2/Prdm1/Apoe/Ednra/Akt3/Efemp2/Pdgfrb/Hey2/Hand2/Eng | 19 |
| GO:0035249 | synaptic transmission, glutamatergic | BP | 0.197 | 7.38e-09 | 2.98e-07 | 6.53 | 1.93e-07 | Grid1/Gria4/Grik1/Nlgn1/Aph1b/Cdh2/Lrrk2/Myo5a/Shc3/Mef2c/Grin2b/Serpine2/Shank2/Htr1b/Tshz3/Grik2/Nlgn3/Htr2a/Cacng7/Psen1/Cacna1a/Cacnb4/Nlgn2 | 23 |
| GO:0045446 | endothelial cell differentiation | BP | 0.197 | 7.38e-09 | 2.98e-07 | 6.53 | 1.93e-07 | Kdr/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Zeb1/Msn/Ppp1r16b/Robo4/Tie1/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Fstl1/Akap11/Col18a1/Cldn1/Hey2/S1pr3 | 23 |
| GO:0001936 | regulation of endothelial cell proliferation | BP | 0.168 | 7.63e-09 | 3.06e-07 | 6.51 | 1.98e-07 | Sema5a/Mdk/Itga4/Kdr/Igf1/Mef2c/Prkd1/Cxcr3/Tek/Cdh13/Cxcl12/Vash1/Ptprm/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Apoe/Tie1/Igf2/Dysf/Akt3/Apln/Egfl7/Cav1/Vash2/Eng | 28 |
| GO:0008360 | regulation of cell shape | BP | 0.172 | 8.02e-09 | 3.19e-07 | 6.5 | 2.07e-07 | Kdr/Plxnb1/Epb41l3/Fermt2/Fn1/Wasf3/Icam1/Plxnd1/Msn/Coro1a/Kit/Rhobtb3/Itga7/Arap3/Cfap410/Parvb/Syne3/Dlc1/Fyn/Fgd1/Fgr/Palm/Cdc42ep5/Plekho1/Arhgap15/Ptn/Fmnl3 | 27 |
| GO:0045123 | cellular extravasation | BP | 0.232 | 8.72e-09 | 3.44e-07 | 6.46 | 2.23e-07 | Mdk/Itga4/Chst2/Itgb7/Icam1/Podxl2/Jam3/Cxcl12/Itga1/Plcb1/Thy1/Itgal/Il27ra/Sele/Pecam1/Ptger3/St3gal4/Spn/Jam2 | 19 |
| GO:2000146 | negative regulation of cell motility | BP | 0.131 | 9.26e-09 | 3.63e-07 | 6.44 | 2.35e-07 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Phldb2/Tcaf1/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Ptprm/Thy1/Gpr173/Robo4/Il27ra/Limch1/Emilin1/Apoe/Magi2/Cnn2/Tie1/Cd200/Tmeff2/Arap3/Tnfaip6/Dlc1/Cldn5/Atp2b4/Dlg5/Dpysl3/Egfl7/Ptn/Cygb/Zmynd8/Rap2a/Eng | 40 |
| GO:0001885 | endothelial cell development | BP | 0.243 | 9.4e-09 | 3.66e-07 | 6.44 | 2.37e-07 | Ednrb/Icam1/Stc1/Plcb1/Cdh5/Msn/Ppp1r16b/Robo4/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Akap11/Col18a1/Cldn1/S1pr3 | 18 |
| GO:0099084 | postsynaptic specialization organization | BP | 0.318 | 9.74e-09 | 3.77e-07 | 6.42 | 2.44e-07 | Gap43/Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Shank2/Slitrk3/Nlgn3/Nptxr/Syngap1/Arhgef9/Zmynd8/Nlgn2 | 14 |
| GO:0051216 | cartilage development | BP | 0.15 | 1.08e-08 | 4.13e-07 | 6.38 | 2.68e-07 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Nog/Pkdcc/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Gli3/Pth1r/Wnt9a/Zeb1/Mgp/Enpp1/Hoxd3/Adamts7/Snai1/Ltbp3/Pkd1/Bgn/Smad9/Osr1/Ccn2/Bbs1/Bmp7/Hand2/Mboat2 | 32 |
| GO:0001823 | mesonephros development | BP | 0.2 | 1.12e-08 | 4.25e-07 | 6.37 | 2.76e-07 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Osr2/Gdf11/Tcf21/Gli3/Tshz3/Agtr1a/Smad9/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Vcan/Myc | 22 |
| GO:0060079 | excitatory postsynaptic potential | BP | 0.217 | 1.15e-08 | 4.33e-07 | 6.36 | 2.81e-07 | Grik1/Begain/Nlgn1/Celf4/Stx1b/Drd4/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Slc29a1/Grik2/Nlgn3/Mpp2/Dlg4/Cux2/Zmynd8/Prkar1b/Nlgn2 | 20 |
| GO:0030199 | collagen fibril organization | BP | 0.271 | 1.15e-08 | 4.33e-07 | 6.36 | 2.81e-07 | Col11a1/Cyp1b1/Sfrp2/Col14a1/Ddr2/Fkbp10/Col13a1/Loxl3/Emilin1/Col5a2/Adamts7/P3h1/Dpt/Efemp2/Crtap/Lum | 16 |
| GO:0033674 | positive regulation of kinase activity | BP | 0.115 | 1.23e-08 | 4.59e-07 | 6.34 | 2.97e-07 | Epha8/Angpt1/Kdr/Agap2/Igf1/Zeb2/Ddr2/Adra1a/Ccdc88a/Fermt2/Drd4/Ccnd2/Lrrk2/Areg/Dab1/Tek/Pik3r6/Gprc5b/Edn3/Calca/Flt3/Kit/Mmd/Epha3/Vegfc/Agtr1a/Nox4/Tie1/Kitl/Htr2a/Pkd1/Epha7/Rassf2/Arrb2/Pkd2/Pde5a/Dlg4/Fgr/Atp2b4/Acsl1/Psen1/Lat/Pdgfrb/Map4k2/C1qtnf9/Ror1/Fgd2/Cd4/Camk1/Irs1 | 50 |
| GO:0001569 | branching involved in blood vessel morphogenesis | BP | 0.311 | 1.35e-08 | 5.01e-07 | 6.3 | 3.25e-07 | Ihh/Sfrp2/Sema5a/Angpt1/Mdk/Lef1/Kdr/Nrp1/Tek/Cxcl12/Plxnd1/Ednra/Nfatc4/Eng | 14 |
| GO:0001935 | endothelial cell proliferation | BP | 0.158 | 1.54e-08 | 5.68e-07 | 6.25 | 3.68e-07 | Sema5a/Mdk/Itga4/Kdr/Igf1/Mef2c/Prkd1/Cxcr3/Tek/Cdh13/Cxcl12/Vash1/Ptprm/Calca/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Apoe/Tie1/Igf2/Dysf/Akt3/Apln/Egfl7/Cav1/Vash2/Eng | 29 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | BP | 0.204 | 1.67e-08 | 6.12e-07 | 6.21 | 3.97e-07 | Grik1/Begain/Nlgn1/Abat/Celf4/Stx1b/Drd4/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Slc29a1/Grik2/Nlgn3/Mpp2/Dlg4/Cux2/Zmynd8/Prkar1b/Nlgn2 | 21 |
| GO:0003206 | cardiac chamber morphogenesis | BP | 0.171 | 1.69e-08 | 6.15e-07 | 6.21 | 3.99e-07 | Col11a1/Sfrp2/Wnt11/Nog/Tbx2/Nos3/Nrp1/Mef2c/Foxf1/Tek/Adamts1/Lrp2/Cpe/Ccm2l/Aplnr/Tbx3/Ednra/Heyl/Myl3/Heg1/Fzd2/Bmp7/Hey2/Hand2/Slit3/Eng | 26 |
| GO:0006887 | exocytosis | BP | 0.116 | 1.82e-08 | 6.58e-07 | 6.18 | 4.26e-07 | Syn2/Cacna1c/Nlgn1/Syt13/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Myo5a/Foxf1/Pfn2/Mical1/Exoc3l/Ppfia2/Syde1/Htr1b/Coro1a/Kit/Cadps/Lin7a/Syt7/Prrt2/Nckap1l/Htr2a/Myo1f/Ncs1/Erc2/Cd84/Cacna1h/Fgr/Septin4/Slc4a8/Stx2/Psen1/Rab27a/Lat/Clnk/Nrn1/Cacna1a/Pld2/Nkd2/Pdcd6ip/Cacnb4/Kcnh1/Plek/Prss12 | 48 |
| GO:0110053 | regulation of actin filament organization | BP | 0.134 | 1.84e-08 | 6.58e-07 | 6.18 | 4.26e-07 | Kank4/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Hcls1/Fermt2/Tmod2/Wasf3/Pfn2/Icam1/Abi2/Coro1a/Ppm1f/Esam/Sh3bp1/Limch1/Arhgap6/Nox4/Arhgef15/Daam2/Pecam1/Tmeff2/Ppm1e/Scin/Bin1/Nckap1l/Carmil3/Dlc1/Arhgap28/Ccn2/Cdc42ep5/Plekhh2/Cyfip2/Plek | 37 |
| GO:0060976 | coronary vasculature development | BP | 0.234 | 1.84e-08 | 6.58e-07 | 6.18 | 4.26e-07 | Angpt1/Sgcd/Nrp1/Kif7/Gpc3/Plxnd1/Lrp2/Prdm1/Cplane1/Aplnr/Dync2h1/Prickle1/Pcsk5/Apln/Pdgfrb/Prokr1/Hey2/Hand2 | 18 |
| GO:0099003 | vesicle-mediated transport in synapse | BP | 0.138 | 2.19e-08 | 7.75e-07 | 6.11 | 5.02e-07 | Syn2/Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Cdh2/Lrrk2/Pfn2/Tbc1d24/Ppfia2/Syde1/Htr1b/Cadps/Nlgn3/Magi2/Syt7/Prrt2/Bin1/Htr2a/Pcdh17/Nsg1/Ncs1/Erc2/Slc4a8/Brsk1/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Prkar1b/Kcnh1/Nlgn2 | 35 |
| GO:0046631 | alpha-beta T cell activation | BP | 0.156 | 2.24e-08 | 7.88e-07 | 6.1 | 5.11e-07 | Ihh/Lef1/Cd83/Wdfy4/Cd44/Itk/Mapk8ip1/Gli3/Dock2/Loxl3/Prdm1/Cd1d1/H2-Ab1/Btla/Cracr2a/Nckap1l/Xcl1/Satb1/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg/Batf/Spn/Ly9/Bcl11b/Myc/Rasal3 | 29 |
| GO:1901890 | positive regulation of cell junction assembly | BP | 0.185 | 2.34e-08 | 8.18e-07 | 6.09 | 5.3e-07 | Kdr/Ptprd/St8sia2/Nlgn1/Nrp1/Fermt2/Col16a1/Lrrc4b/Tek/Adgrl1/Slitrk3/Thy1/Ppm1f/Nlgn3/Bhlhb9/Nphp1/Cldn5/Cux2/Dlg5/Cav1/Enpp2/Cldn1/Nlgn2 | 23 |
| GO:0007265 | Ras protein signal transduction | BP | 0.123 | 2.74e-08 | 9.42e-07 | 6.03 | 6.1e-07 | Igf1/Adra1a/Nrp1/Rasgrp3/Arl6/Cdh2/Rasa3/Lpar4/Prkd1/Arhgef25/Cdh13/Dock2/Rasgrp2/Sh2b2/Syde1/Abi2/Sh3bp1/Eps8l1/Agtr1a/Apoe/Rasip1/Ednra/Pecam1/Kitl/Arap3/Arrb1/Heg1/Dlc1/Net1/Syngap1/Cdc42ep5/Rab39b/Mapre2/Lat/Pdgfrb/Dennd4a/Iqsec3/Nradd/F2r/Rasal3/Rap2a/Plekhg5 | 42 |
| GO:0030111 | regulation of Wnt signaling pathway | BP | 0.126 | 3.01e-08 | 1.02e-06 | 5.99 | 6.61e-07 | Gli1/Sfrp2/Sema5a/Mdk/Lef1/Apcdd1/Wnt11/Nog/Zeb2/Nkd1/Ptpro/Tert/Cdh2/Gpc3/Lrrk2/Rbms3/Tmem237/Gprc5b/Gli3/Peg12/Plpp3/Znrf3/Dkk3/Tsku/Apoe/Daam2/Prickle1/Caprin2/Nfatc4/Tle1/Lats2/Psen1/Adgra2/Cav1/Dact1/Nkd2/Grb10/Tle2/Lrp4/Draxin | 40 |
| GO:0048706 | embryonic skeletal system development | BP | 0.171 | 3.1e-08 | 1.04e-06 | 5.98 | 6.75e-07 | Col11a1/Ihh/Mmp16/Hoxd10/Hoxd11/Nog/Hoxa11/Hoxd9/Mef2c/Osr2/Hoxd4/Flvcr1/Gli3/Wnt9a/Zeb1/Hoxd3/Ednra/Pcsk5/Hoxa9/Twist2/Osr1/Xylt1/Bmp7/Hand2/Twist1 | 25 |
| GO:0030178 | negative regulation of Wnt signaling pathway | BP | 0.162 | 3.11e-08 | 1.04e-06 | 5.98 | 6.75e-07 | Gli1/Sfrp2/Mdk/Apcdd1/Wnt11/Nog/Nkd1/Ptpro/Cdh2/Gpc3/Rbms3/Gli3/Znrf3/Dkk3/Tsku/Apoe/Prickle1/Nfatc4/Tle1/Lats2/Cav1/Dact1/Nkd2/Grb10/Tle2/Lrp4/Draxin | 27 |
| GO:0001657 | ureteric bud development | BP | 0.196 | 3.38e-08 | 1.11e-06 | 5.96 | 7.17e-07 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Tcf21/Gli3/Tshz3/Agtr1a/Smad9/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Vcan/Myc | 21 |
| GO:0072163 | mesonephric epithelium development | BP | 0.196 | 3.38e-08 | 1.11e-06 | 5.96 | 7.17e-07 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Tcf21/Gli3/Tshz3/Agtr1a/Smad9/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Vcan/Myc | 21 |
| GO:0072164 | mesonephric tubule development | BP | 0.196 | 3.38e-08 | 1.11e-06 | 5.96 | 7.17e-07 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Tcf21/Gli3/Tshz3/Agtr1a/Smad9/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Vcan/Myc | 21 |
| GO:0043087 | regulation of GTPase activity | BP | 0.122 | 3.53e-08 | 1.15e-06 | 5.94 | 7.43e-07 | Wnt11/Plxnb1/Rgs6/Fermt2/Rgs16/Rasgrp3/Garnl3/Lrrk2/Arhgap45/Rasa3/Icam1/Arhgap25/Rasgrp2/Myo9a/Plxnd1/Thy1/Dock10/Epha3/Sh3bp1/Arhgap6/Arhgef15/Rasip1/Pecam1/Bin1/Arap3/Xcl1/Arrb1/Arrb2/Arhgef26/Net1/Fgd1/Gnb5/Syngap1/Ngef/Mapre2/Arhgap15/Iqsec3/Gpsm1/F2r/Sipa1/Tbc1d10c/Rasal3 | 42 |
| GO:0007229 | integrin-mediated signaling pathway | BP | 0.204 | 3.59e-08 | 1.16e-06 | 5.94 | 7.51e-07 | Itga4/Nrp1/Fermt2/Sema7a/Itgb7/Fn1/Itga1/Thy1/Plpp3/Loxl3/Itgal/Itga7/Pcsk5/Fgr/Ccn2/Lat/Ptn/Itga8/Itgb4/Plek | 20 |
| GO:0072009 | nephron epithelium development | BP | 0.188 | 3.63e-08 | 1.17e-06 | 5.93 | 7.56e-07 | Wnt6/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Tcf21/Gli3/Agtr1a/Ednra/Heyl/Ptch1/Dchs1/Osr1/Aqp11/Fat4/Myc | 22 |
| GO:0009581 | detection of external stimulus | BP | 0.175 | 3.7e-08 | 1.18e-06 | 5.93 | 7.62e-07 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Foxf1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Htr2a/Pkd1/Arrb1/Arrb2/Pkd2/Fyn/Piezo2/Pkdrej/Cacnb4/Myc | 24 |
| GO:0016079 | synaptic vesicle exocytosis | BP | 0.175 | 3.7e-08 | 1.18e-06 | 5.93 | 7.62e-07 | Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Pfn2/Ppfia2/Syde1/Htr1b/Cadps/Syt7/Prrt2/Htr2a/Ncs1/Erc2/Slc4a8/Stx2/Psen1/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 24 |
| GO:0045667 | regulation of osteoblast differentiation | BP | 0.165 | 3.84e-08 | 1.21e-06 | 5.92 | 7.87e-07 | Gli1/Sfrp2/Nog/Igf1/Ddr2/Npnt/Fermt2/Areg/Mef2c/Id3/Prkd1/Gli3/Nell1/Gdf10/Smoc1/Vegfc/Rorb/Tnfaip6/Twist2/Rassf2/Ptch1/Lrp3/Bmp7/Hand2/Twist1/Zfp932 | 26 |
| GO:0003002 | regionalization | BP | 0.117 | 3.96e-08 | 1.24e-06 | 5.9 | 8.07e-07 | Gli1/Sfrp2/Ar/Hhip/Lef1/Wnt11/Hoxd10/Hoxd11/Nog/Zeb2/Nrp1/Hoxa11/Dzip1l/Gpc3/Hoxd9/Hoxa10/Mef2c/Foxf1/Hoxd8/Hoxd4/Tshz1/Gdf11/Gli3/Ift57/Lrp2/Lfng/Gpr161/Tbx3/Dync2h1/Hoxd3/Ednra/Heyl/Snai1/Hipk2/Pcsk5/Hoxa9/Ptch1/Osr1/Nr2f1/Rab23/Ets2/Psen1/Hipk1/Hey2/Lrp4 | 45 |
| GO:0070997 | neuron death | BP | 0.112 | 4.06e-08 | 1.27e-06 | 5.9 | 8.21e-07 | Ptprz1/Angpt1/Mdk/Dcc/Kdr/Agap2/Igf1/Ddr2/St8sia2/Nrp1/Tert/Bok/Lrrk2/Mef2c/Dpysl4/Grin2b/Pcdhgc3/Tbc1d24/Itga1/Coro1a/Scn2a/Grik2/Il27ra/Rilpl1/Enpp1/Apoe/Kcnma1/Cd200/Slc23a2/Ndnf/Hipk2/Arrb1/Nfatc4/Epha7/Neo1/Kcnip3/Sarm1/Arrb2/Bhlhb9/Fyn/Nlrp1b/Syngap1/Psen1/Meak7/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 49 |
| GO:0060078 | regulation of postsynaptic membrane potential | BP | 0.186 | 4.26e-08 | 1.32e-06 | 5.88 | 8.57e-07 | Grik1/Begain/Nlgn1/Abat/Celf4/Stx1b/Drd4/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Slc29a1/Gabra4/Grik2/Nlgn3/Mpp2/Dlg4/Cux2/Zmynd8/Prkar1b/Nlgn2 | 22 |
| GO:0001938 | positive regulation of endothelial cell proliferation | BP | 0.202 | 4.29e-08 | 1.33e-06 | 5.88 | 8.59e-07 | Sema5a/Mdk/Itga4/Kdr/Igf1/Prkd1/Cdh13/Cxcl12/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Igf2/Dysf/Akt3/Apln/Egfl7/Cav1/Vash2 | 20 |
| GO:1901215 | negative regulation of neuron death | BP | 0.134 | 4.38e-08 | 1.35e-06 | 5.87 | 8.72e-07 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Igf1/Nrp1/Tert/Bok/Lrrk2/Mef2c/Pcdhgc3/Tbc1d24/Coro1a/Grik2/Il27ra/Apoe/Cd200/Slc23a2/Ndnf/Hipk2/Arrb1/Nfatc4/Neo1/Arrb2/Bhlhb9/Fyn/Syngap1/Psen1/Meak7/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 35 |
| GO:0043010 | camera-type eye development | BP | 0.118 | 4.62e-08 | 1.4e-06 | 5.85 | 9.1e-07 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Cacna1c/Nhs/Kdr/Zeb2/Rcn1/Tub/Tbx2/Celf4/Nrp1/Lamc3/Arl6/Tenm3/Osr2/Obsl1/Gdf11/Gli3/Ptprm/Thy1/Tgif2/Zeb1/Abi2/Rp1/Cdkn1c/Tsku/Arhgef15/Spred3/Lpcat1/Hipk2/Rorb/Twist2/Tmem231/Nphp1/Fat3/Pdgfrb/Ptn/Hipk1/Bmp7/Bcl11b/Twist1 | 44 |
| GO:0030318 | melanocyte differentiation | BP | 0.379 | 4.72e-08 | 1.43e-06 | 5.85 | 9.25e-07 | Zeb2/Adamts20/Ednrb/Myo5a/Mef2c/Edn3/Gli3/Kit/Enpp1/Kitl/Rab27a | 11 |
| GO:0070374 | positive regulation of ERK1 and ERK2 cascade | BP | 0.141 | 5.05e-08 | 1.52e-06 | 5.82 | 9.82e-07 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Npnt/Adra1a/Nrp1/Cd44/Fermt2/Sema7a/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Apoe/Nox4/Havcr2/Ednra/Htr2a/Xcl1/Arrb1/Cavin3/Arrb2/Ccn2/Pdgfrb/Pde8a/F2r/Cd4/Hand2 | 32 |
| GO:0010463 | mesenchymal cell proliferation | BP | 0.263 | 5.13e-08 | 1.53e-06 | 5.81 | 9.94e-07 | Ihh/Wnt11/Kdr/Tbx2/Gpc3/Foxf1/Zeb1/Dchs1/Osr1/Fat4/Ptn/Bmp7/Hand2/Myc/Irs1 | 15 |
| GO:0045216 | cell-cell junction organization | BP | 0.144 | 5.35e-08 | 1.59e-06 | 5.8 | 1.03e-06 | Wnt11/Epb41l3/Mpdz/Ptpro/Fermt2/Cdh2/Cdh6/Jam3/Cldn8/Cdh10/Cdh5/Abi2/Aplnr/Esam/Nfasc/Cdh11/Gjc1/Pecam1/Snai1/Heg1/Dchs1/Svep1/Nphp1/Cldn5/Dlg5/Hipk1/Cav1/Pdcd6ip/Cldn1/F2r/Numbl | 31 |
| GO:0043542 | endothelial cell migration | BP | 0.14 | 5.6e-08 | 1.66e-06 | 5.78 | 1.07e-06 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Ptp4a3/Prkd1/Stc1/Tek/Cdh13/Vash1/Ptprm/Calca/Plxnd1/Cdh5/Plpp3/Jcad/Sh3bp1/Vegfc/Apoe/Pecam1/Igf2/Hdac9/Akt3/Atp2b4/Fstl1/Adgra2/Plekhg5 | 32 |
| GO:0048608 | reproductive structure development | BP | 0.107 | 5.93e-08 | 1.74e-06 | 5.76 | 1.13e-06 | Ndp/Tex15/Stra6/Sfrp2/Ar/Angpt1/Lef1/Itga4/Kdr/Cyp7b1/Nog/Igf1/Slc8a1/Nos3/Fst/Cd44/Hoxa11/Il11ra1/Hoxa10/Mmp2/Icam1/Id4/Serpine2/Adamts1/Tcf21/Vash1/Gdf7/Lrp2/Syde1/Lfng/Prdm1/Pcdh12/Kit/Cdkn1c/Tbx3/Rhobtb3/Igf2/Snai1/Kitl/Pkd1/Arrb1/Tnfaip6/Hoxa9/Arrb2/Stat5b/Osr1/Pkd2/Abcb1a/Ptn/Vash2/Bmp7/Hey2 | 52 |
| GO:0010563 | negative regulation of phosphorus metabolic process | BP | 0.109 | 6e-08 | 1.75e-06 | 5.76 | 1.13e-06 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Slc8a1/Ptpro/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Ppp1r16b/Enpp1/Cdkn1c/Emilin1/Ppef2/Prr5l/Apoe/Rasip1/Pecam1/Ppm1e/Nckap1l/Bdkrb2/Pkib/Lpcat1/Cmya5/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Ppp1r26/Smpd1/Psen1/Cav1/Pid1/Grb10/Bmp7/Prkar1b/Twist1/Plek/Lax1/Eng | 50 |
| GO:0045936 | negative regulation of phosphate metabolic process | BP | 0.109 | 6e-08 | 1.75e-06 | 5.76 | 1.13e-06 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Slc8a1/Ptpro/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Ppp1r16b/Enpp1/Cdkn1c/Emilin1/Ppef2/Prr5l/Apoe/Rasip1/Pecam1/Ppm1e/Nckap1l/Bdkrb2/Pkib/Lpcat1/Cmya5/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Ppp1r26/Smpd1/Psen1/Cav1/Pid1/Grb10/Bmp7/Prkar1b/Twist1/Plek/Lax1/Eng | 50 |
| GO:0003197 | endocardial cushion development | BP | 0.28 | 6.06e-08 | 1.75e-06 | 5.76 | 1.14e-06 | Nog/Tbx2/Nos3/Foxf1/Erg/Epha3/Aplnr/Tbx3/Heyl/Snai1/Dchs1/Bmp7/Twist1/Eng | 14 |
| GO:0060021 | roof of mouth development | BP | 0.198 | 6.1e-08 | 1.76e-06 | 5.76 | 1.14e-06 | Lef1/Wnt11/Pkdcc/Tbx2/Mef2c/Osr2/Tshz1/Bbs7/Gdf11/Tcf21/Gli3/Loxl3/Jag2/Cplane1/Tbx3/Snai1/Asph/Osr1/Fzd2/Hand2 | 20 |
| GO:0001933 | negative regulation of protein phosphorylation | BP | 0.119 | 6.24e-08 | 1.78e-06 | 5.75 | 1.15e-06 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Slc8a1/Ptpro/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Enpp1/Cdkn1c/Emilin1/Ppef2/Prr5l/Apoe/Rasip1/Pecam1/Ppm1e/Bdkrb2/Pkib/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Smpd1/Psen1/Cav1/Pid1/Bmp7/Prkar1b/Twist1/Lax1/Eng | 42 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.115 | 6.27e-08 | 1.78e-06 | 5.75 | 1.15e-06 | Sfrp2/Lef1/Cd109/Kdr/Nog/Zeb2/Rnf165/Npnt/Fst/Fermt2/Btbd11/Gpc3/Gdf11/Zfp423/Cdh5/Gdf7/Lrp2/Pbld2/Tgif2/Zeb1/Gdf10/Cdkn1c/Tsku/Emilin1/Magi2/Spred3/Ltbp3/Hipk2/Kcp/Tnfaip6/Smad9/Neo1/Nrros/Arrb2/Zfyve9/Cldn5/Il17rd/Fstl1/Lats2/Cav1/Itga8/Tmprss6/Bmp7/Nrep/Eng | 45 |
| GO:0010720 | positive regulation of cell development | BP | 0.116 | 6.31e-08 | 1.78e-06 | 5.75 | 1.15e-06 | Ptprz1/Sema5a/Mdk/Hoxd11/Kdr/Plxnb1/Ptprd/Nog/Igf1/Zeb2/Nrp1/Fermt2/Sema7a/Hoxa11/Fn1/Serpine2/Cxcl12/Tbc1d24/Has2/Obsl1/Gli3/Plxnd1/Cdh5/Lrp2/Kit/Mme/Vegfc/Apoe/Caprin2/Bin1/Nin/Chodl/Bhlhb9/Net1/Pde5a/Cldn5/Cux2/Abcb1a/Ptn/Akap11/Enpp2/Myc/Zfp365/Numbl | 44 |
| GO:0099175 | regulation of postsynapse organization | BP | 0.189 | 6.6e-08 | 1.85e-06 | 5.73 | 1.2e-06 | Ptprd/Nlgn1/Cdh2/Lrrk2/Lrrc4b/Nptx1/Grin2b/Ppfia2/Abi2/Kif1a/Nlgn3/Dgkb/Apoe/Caprin2/Epha7/Bhlhb9/Lrfn2/Fyn/Cux2/Ngef/Zmynd8 | 21 |
| GO:1901214 | regulation of neuron death | BP | 0.115 | 6.76e-08 | 1.89e-06 | 5.72 | 1.22e-06 | Ptprz1/Angpt1/Mdk/Dcc/Kdr/Agap2/Igf1/Ddr2/St8sia2/Nrp1/Tert/Bok/Lrrk2/Mef2c/Grin2b/Pcdhgc3/Tbc1d24/Itga1/Coro1a/Grik2/Il27ra/Rilpl1/Apoe/Kcnma1/Cd200/Slc23a2/Ndnf/Hipk2/Arrb1/Nfatc4/Epha7/Neo1/Kcnip3/Sarm1/Arrb2/Bhlhb9/Fyn/Syngap1/Psen1/Meak7/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 45 |
| GO:0050770 | regulation of axonogenesis | BP | 0.151 | 7.45e-08 | 2.07e-06 | 5.68 | 1.34e-06 | Efnb3/Map2/Sema5a/Lrrc4c/Plxnb1/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Dab1/Cxcl12/Plxnd1/Thy1/Apoe/Sema3g/Nin/Epha7/Chodl/Syngap1/Brsk1/Psen1/Sema6a/Ulk2/Cacna1a/Lrp4/Draxin | 28 |
| GO:0061458 | reproductive system development | BP | 0.107 | 7.74e-08 | 2.14e-06 | 5.67 | 1.39e-06 | Ndp/Tex15/Stra6/Sfrp2/Ar/Angpt1/Lef1/Itga4/Kdr/Cyp7b1/Nog/Igf1/Slc8a1/Nos3/Fst/Cd44/Hoxa11/Il11ra1/Hoxa10/Mmp2/Icam1/Id4/Serpine2/Adamts1/Tcf21/Vash1/Gdf7/Lrp2/Syde1/Lfng/Prdm1/Pcdh12/Kit/Cdkn1c/Tbx3/Rhobtb3/Igf2/Snai1/Kitl/Pkd1/Arrb1/Tnfaip6/Hoxa9/Arrb2/Stat5b/Osr1/Pkd2/Abcb1a/Ptn/Vash2/Bmp7/Hey2 | 52 |
| GO:0003158 | endothelium development | BP | 0.173 | 9.05e-08 | 2.49e-06 | 5.6 | 1.61e-06 | Kdr/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Zeb1/Msn/Ppp1r16b/Robo4/Tie1/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Fstl1/Akap11/Col18a1/Cldn1/Hey2/S1pr3 | 23 |
| GO:1990778 | protein localization to cell periphery | BP | 0.116 | 9.29e-08 | 2.54e-06 | 5.59 | 1.65e-06 | Gpc6/Kcnb2/Ar/Pkdcc/Tub/Epb41l3/Ccdc88a/Stx1b/Arl6/Gnai1/Efr3b/Cdh2/Gga2/Abi3/Myo5a/Nptx1/Ank2/Rab34/Dlg2/Ttc7b/Epha3/Lin7a/Rilpl1/Nptxr/Nfasc/Magi2/Arl3/Nsg1/Cacng7/Stac2/Kcnip3/Ptch1/Dchs1/Tspan5/Dlg4/Palm/Atp2b4/Bbs1/Pid1/Nkd2/Golph3l/Rabep1/Rap2a | 43 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | BP | 0.137 | 9.37e-08 | 2.55e-06 | 5.59 | 1.65e-06 | Igf1/Adra1a/Nrp1/Cdh2/Garnl3/Rasa3/Lpar4/Arhgef25/Arhgap25/Dock2/Sh2b2/Syde1/Sh3bp1/Eps8l1/Apoe/Rasip1/Adcyap1r1/Kitl/Arap3/Arrb1/Heg1/Dlc1/Arhgap28/Net1/Syngap1/Mapre2/Pdgfrb/Dennd4a/Iqsec3/F2r/Sipa1/Rasal3 | 32 |
| GO:0051966 | regulation of synaptic transmission, glutamatergic | BP | 0.212 | 9.44e-08 | 2.56e-06 | 5.59 | 1.66e-06 | Gria4/Grik1/Nlgn1/Cdh2/Lrrk2/Myo5a/Mef2c/Grin2b/Serpine2/Shank2/Htr1b/Tshz3/Grik2/Nlgn3/Htr2a/Cacng7/Psen1/Nlgn2 | 18 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | BP | 0.172 | 1.04e-07 | 2.8e-06 | 5.55 | 1.82e-06 | Gli1/Sfrp2/Mdk/Wnt11/Nog/Nkd1/Ptpro/Cdh2/Gpc3/Rbms3/Gli3/Znrf3/Dkk3/Apoe/Prickle1/Tle1/Lats2/Cav1/Dact1/Nkd2/Tle2/Lrp4/Draxin | 23 |
| GO:0042692 | muscle cell differentiation | BP | 0.11 | 1.05e-07 | 2.8e-06 | 5.55 | 1.82e-06 | AW551984/Sgcd/Col14a1/Ryr1/Igf1/Tbx2/Npnt/Adra1a/Slc8a1/P2rx2/Tmod2/Cdh2/Ednrb/Mef2c/Foxf1/Cxcl12/Hopx/Ank2/Zeb1/Tshz3/Kit/Cth/Aplnr/Nox4/Tbx3/Ednra/Igf2/Dysf/Bin1/Hdac9/Nfatc4/Flnc/Neo1/Selenon/Arrb2/Pi16/Rbpms2/Efemp2/Sgcb/Plekho1/Pdgfrb/Itga8/Hacd1/Cacnb4/Hey2/Hand2/Camk1/Eng | 48 |
| GO:0061448 | connective tissue development | BP | 0.125 | 1.06e-07 | 2.82e-06 | 5.55 | 1.83e-06 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Nog/Igf1/Pkdcc/Thbs3/Cbs/Hoxa11/Mef2c/Id4/Osr2/Stc1/Gli3/Gdf7/Pth1r/Wnt9a/Zeb1/Selenom/Mgp/Enpp1/Hoxd3/Adamts7/Snai1/Ltbp3/Pkd1/Bgn/Smad9/Osr1/Ccn2/Bbs1/Pdgfrb/Bmp7/Hand2/Mboat2 | 37 |
| GO:0051208 | sequestering of calcium ion | BP | 0.177 | 1.07e-07 | 2.84e-06 | 5.55 | 1.84e-06 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Htr2a/Xcl1/Selenon/Asph/Pkd2/Xcr1/Ryr3/Slc25a23/Gstm7/Psen1/Sri/F2r | 22 |
| GO:0043271 | negative regulation of ion transport | BP | 0.153 | 1.07e-07 | 2.84e-06 | 5.55 | 1.84e-06 | Maob/Hecw2/Spink1/Abat/Nos3/Drd4/Myo5a/Icam1/Serpine2/Stc1/Calca/Htr1b/Dysf/Bin1/Htr2a/Osr1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Gabbr1/Cav1/Usp2/Twist1 | 27 |
| GO:0051962 | positive regulation of nervous system development | BP | 0.117 | 1.08e-07 | 2.86e-06 | 5.54 | 1.85e-06 | Ptprz1/Sema5a/Mdk/Kdr/Plxnb1/Ptprd/Nog/Igf1/Zeb2/St8sia2/Nlgn1/Nrp1/Sema7a/Fn1/Lrrc4b/Serpine2/Cxcl12/Tbc1d24/Obsl1/Adgrl1/Gli3/Slitrk3/Plxnd1/Lrp2/Kit/Mme/Nlgn3/Vegfc/Apoe/Caprin2/Bin1/Nin/Chodl/Bhlhb9/Cux2/Dlg5/Ptn/Enpp2/Myc/Nlgn2/Zfp365/Numbl | 42 |
| GO:0022898 | regulation of transmembrane transporter activity | BP | 0.127 | 1.2e-07 | 3.14e-06 | 5.5 | 2.03e-06 | Hecw2/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Mef2c/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Dysf/Cacng7/Stac2/Selenon/Asph/Osr1/Pkd2/Fhl1/Rrad/Dlg4/Gnb5/Gstm7/Atp1a2/Abcb1a/Sri/Cav1/Cacna1a/Cacnb4/Twist1/Nlgn2 | 36 |
| GO:0009612 | response to mechanical stimulus | BP | 0.152 | 1.21e-07 | 3.15e-06 | 5.5 | 2.04e-06 | Col11a1/Stra6/Slc1a3/Mdk/Wnt11/Igf1/Slc8a1/Nos3/Scn1a/Mmp2/Grin2b/Serpine2/Cxcl12/Kit/Cnn2/Ednra/Htr2a/Pkd1/Pkd2/Fyn/Habp4/Atp1a2/Piezo2/Ptn/Pkdrej/Cav1/Myc | 27 |
| GO:0009101 | glycoprotein biosynthetic process | BP | 0.129 | 1.23e-07 | 3.2e-06 | 5.49 | 2.07e-06 | St6gal2/Itm2a/Igf1/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Soat1/Chst10/Tmtc1/B3galnt1/Fut10/Plcb1/Galnt16/St3gal2/St6galnac4/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Dse/Mgat3/Pomt2/Aqp11/Large1/St6galnac6/B3gnt5/Xylt1/St3gal4/Psen1/Alg14/Galnt18/Dpy19l1 | 35 |
| GO:0006814 | sodium ion transport | BP | 0.133 | 1.27e-07 | 3.28e-06 | 5.48 | 2.12e-06 | Nalcn/Hecw2/Slc6a2/Slc5a11/Slc13a5/Slc8a1/Nos3/Scn4b/Drd4/Fxyd5/Scn1a/Ednrb/Slc9a7/Slc24a4/Serpine2/Slc24a3/Plcb1/Slc8a3/Slc6a15/Maged2/Scn2a/Ednra/Slc9a5/Slc23a2/Osr1/Slc38a1/Pkd2/Akt3/Atp2b4/Scn3a/Atp1a2/Slc4a8/Mllt6 | 33 |
| GO:0051926 | negative regulation of calcium ion transport | BP | 0.232 | 1.28e-07 | 3.28e-06 | 5.48 | 2.13e-06 | Spink1/Nos3/Drd4/Icam1/Stc1/Calca/Dysf/Bin1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Usp2 | 16 |
| GO:0006486 | protein glycosylation | BP | 0.147 | 1.32e-07 | 3.36e-06 | 5.47 | 2.18e-06 | St6gal2/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Tmtc1/B3galnt1/Fut10/Galnt16/St3gal2/St6galnac4/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Mgat3/Pomt2/Aqp11/Large1/St6galnac6/B3gnt5/St3gal4/Psen1/Alg14/Galnt18/Dpy19l1 | 28 |
| GO:0043413 | macromolecule glycosylation | BP | 0.147 | 1.32e-07 | 3.36e-06 | 5.47 | 2.18e-06 | St6gal2/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Tmtc1/B3galnt1/Fut10/Galnt16/St3gal2/St6galnac4/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Mgat3/Pomt2/Aqp11/Large1/St6galnac6/B3gnt5/St3gal4/Psen1/Alg14/Galnt18/Dpy19l1 | 28 |
| GO:0043524 | negative regulation of neuron apoptotic process | BP | 0.147 | 1.32e-07 | 3.36e-06 | 5.47 | 2.18e-06 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Coro1a/Grik2/Il27ra/Apoe/Ndnf/Hipk2/Arrb1/Nfatc4/Arrb2/Bhlhb9/Fyn/Syngap1/Psen1/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 28 |
| GO:0070371 | ERK1 and ERK2 cascade | BP | 0.117 | 1.37e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Npnt/Adra1a/Nrp1/Cd44/Fermt2/Sema7a/Fn1/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Emilin1/Apoe/Nox4/Havcr2/Ednra/Spred3/Htr2a/Xcl1/Arrb1/Cavin3/Epha7/Arrb2/Ccn2/Sema6a/Pdgfrb/Pde8a/Cav1/Grb10/F2r/Cd4/Hand2/Tbc1d10c/Myc | 41 |
| GO:0032409 | regulation of transporter activity | BP | 0.124 | 1.37e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Hecw2/Slc5a11/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Mef2c/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Dysf/Cacng7/Stac2/Selenon/Asph/Osr1/Pkd2/Fhl1/Rrad/Dlg4/Gnb5/Gstm7/Atp1a2/Abcb1a/Sri/Cav1/Cacna1a/Cacnb4/Twist1/Nlgn2 | 37 |
| GO:0010976 | positive regulation of neuron projection development | BP | 0.138 | 1.37e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Ptprz1/Hgf/Acsl6/Fez1/Mdk/Dcc/Lrrc7/Tenm3/Prkd1/Serpine2/Adamts1/Gprc5b/Serpini1/Epha3/Apoe/Magi2/Caprin2/Ndnf/P3h1/Bhlhb9/Fyn/Cux2/Dpysl3/Arsb/Ptn/Gpc2/Cpeb1/Ror1/Bmp7/Setx/Camk1 | 31 |
| GO:0050866 | negative regulation of cell activation | BP | 0.138 | 1.37e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Ihh/Mdk/Abat/Cd44/Foxf1/Serpine2/Gli3/Flt3/Loxl3/Prdm1/H2-Ab1/Btla/Emilin1/Apoe/Havcr2/Cd200/Nckap1l/Pde5a/Cd84/Fgr/Ptger3/H2-Aa/Dlg5/Gimap5/Gimap3/Clnk/Cygb/Nr1h3/Spn/Tbc1d10c/Lax1 | 31 |
| GO:0009582 | detection of abiotic stimulus | BP | 0.169 | 1.38e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Htr2a/Pkd1/Arrb1/Arrb2/Pkd2/Fyn/Piezo2/Pkdrej/Cacnb4/Myc | 23 |
| GO:0014065 | phosphatidylinositol 3-kinase signaling | BP | 0.169 | 1.38e-07 | 3.41e-06 | 5.47 | 2.21e-06 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Serpine2/Tek/Ppp1r16b/Prr5l/Htr2a/Prex2/Fyn/Fgr/Pdgfrb/Pld2/Ror1/F2r/Twist1/Irs1 | 23 |
| GO:0032412 | regulation of ion transmembrane transporter activity | BP | 0.128 | 1.48e-07 | 3.63e-06 | 5.44 | 2.35e-06 | Hecw2/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Mef2c/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Dysf/Cacng7/Stac2/Selenon/Asph/Osr1/Pkd2/Fhl1/Rrad/Dlg4/Gnb5/Gstm7/Atp1a2/Abcb1a/Sri/Cav1/Cacna1a/Cacnb4/Nlgn2 | 35 |
| GO:0003015 | heart process | BP | 0.132 | 1.54e-07 | 3.76e-06 | 5.43 | 2.43e-06 | Cacna1c/Sgcd/Kcnd3/Adra1a/Slc8a1/Nos3/Scn4b/Scn1a/Cacna2d1/Ednrb/Stc1/Hopx/Cacna1e/Ank2/Edn3/Calca/Rps6ka2/Nox4/Gjc1/Ednra/Bin1/Myl3/Fyn/Pde5a/Slc4a3/Cacna1h/Atp2b4/Ccn2/Apln/Atp1a2/Sri/Cav1/Hey2 | 33 |
| GO:0050771 | negative regulation of axonogenesis | BP | 0.229 | 1.58e-07 | 3.85e-06 | 5.42 | 2.49e-06 | Efnb3/Map2/Sema5a/Sema5b/Nrp1/Sema7a/Dab1/Thy1/Sema3g/Epha7/Syngap1/Psen1/Sema6a/Ulk2/Lrp4/Draxin | 16 |
| GO:0002691 | regulation of cellular extravasation | BP | 0.283 | 1.59e-07 | 3.85e-06 | 5.42 | 2.49e-06 | Mdk/Itga4/Chst2/Icam1/Jam3/Cxcl12/Plcb1/Thy1/Il27ra/Sele/Pecam1/Ptger3/St3gal4 | 13 |
| GO:0035136 | forelimb morphogenesis | BP | 0.283 | 1.59e-07 | 3.85e-06 | 5.42 | 2.49e-06 | Cacna1c/Hoxd10/Hoxd11/Rnf165/Hoxa11/Hoxd9/Osr2/Wnt9a/Enpp1/Tbx3/Hoxa9/Osr1/Twist1 | 13 |
| GO:0071695 | anatomical structure maturation | BP | 0.123 | 1.62e-07 | 3.9e-06 | 5.41 | 2.53e-06 | Epha8/Ihh/Pla2g10/Kdr/Ryr1/Plxnb1/Spink1/Igf1/Thbs3/Cspg4/Lrrk2/Ednrb/Mmp2/Myo5a/Slc24a4/Gdf11/Flvcr1/Plcb1/Calca/Rps6ka2/Cdh5/Pth1r/Dlg2/Enpp1/Cdkn1c/Nfasc/Kcnma1/Ednra/Adamts7/Tal1/Lhx6/Dchs1/Septin4/Xylt1/Fat4/Psen1/Cacna1a | 37 |
| GO:0003151 | outflow tract morphogenesis | BP | 0.205 | 1.65e-07 | 3.95e-06 | 5.4 | 2.56e-06 | Sfrp2/Wnt11/Ryr1/Nog/Tbx2/Nrp1/Mef2c/Plxnd1/Lrp2/Tbx3/Ednra/Heyl/Fzd2/Bmp7/Hey2/Hand2/Twist1/Eng | 18 |
| GO:0050931 | pigment cell differentiation | BP | 0.308 | 1.69e-07 | 4.04e-06 | 5.39 | 2.61e-06 | Zeb2/Adamts20/Ednrb/Myo5a/Mef2c/Ap1m1/Edn3/Gli3/Kit/Enpp1/Kitl/Rab27a | 12 |
| GO:0070372 | regulation of ERK1 and ERK2 cascade | BP | 0.12 | 1.71e-07 | 4.05e-06 | 5.39 | 2.62e-06 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Npnt/Adra1a/Nrp1/Cd44/Fermt2/Sema7a/Fn1/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Emilin1/Apoe/Nox4/Havcr2/Ednra/Spred3/Htr2a/Xcl1/Arrb1/Cavin3/Epha7/Arrb2/Ccn2/Sema6a/Pdgfrb/Pde8a/Cav1/F2r/Cd4/Hand2/Tbc1d10c | 39 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | BP | 0.116 | 1.73e-07 | 4.08e-06 | 5.39 | 2.64e-06 | Sfrp2/Igf1/Zeb2/Slc8a1/Cbs/Fermt2/Drd4/Ccnd2/Lrrk2/Mapk8ip1/Pik3r6/Edn3/Slc8a3/Pkia/Thy1/Kit/Cdkn1c/Apoe/Nox4/Rasip1/Kitl/Htr2a/Pkib/Pkd1/Heg1/Pkd2/Pde5a/Atp2b4/Acsl1/Lats2/Smpd1/Psen1/Pdgfrb/Map4k2/Cav1/C1qtnf9/Fgd2/Bmp7/Prkar1b/Camk1/Lax1 | 41 |
| GO:0007416 | synapse assembly | BP | 0.145 | 1.85e-07 | 4.35e-06 | 5.36 | 2.82e-06 | Gpc6/Gap43/Sdk2/Plxnb1/Ptprd/St8sia2/Nlgn1/Cdh2/Lrrc4b/Mef2c/Nptx1/Shank2/Obsl1/Adgrl1/Slitrk3/Plxnd1/Nlgn3/Nptxr/Magi2/Pcdh17/Epha7/Bhlhb9/Cux2/Dlg5/Arhgef9/Cacna1a/Lrp4/Nlgn2 | 28 |
| GO:0061326 | renal tubule development | BP | 0.185 | 1.95e-07 | 4.54e-06 | 5.34 | 2.94e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Tcf21/Gli3/Agtr1a/Heyl/Ptch1/Dchs1/Osr1/Pkd2/Aqp11/Fat4/Myc | 20 |
| GO:0050805 | negative regulation of synaptic transmission | BP | 0.202 | 1.98e-07 | 4.57e-06 | 5.34 | 2.96e-06 | Grik1/Grid2ip/Celf4/Drd4/Gnai1/Lrrk2/Myo5a/Shank2/Htr1b/Grik2/Nlgn3/Sorcs2/Cd38/Prrt1/Htr2a/Pcdh17/Gabbr1/Bche | 18 |
| GO:0061333 | renal tubule morphogenesis | BP | 0.202 | 1.98e-07 | 4.57e-06 | 5.34 | 2.96e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Osr1/Pkd2/Fat4/Myc | 18 |
| GO:0045664 | regulation of neuron differentiation | BP | 0.133 | 2.06e-07 | 4.73e-06 | 5.32 | 3.07e-06 | Sfrp2/Fez1/Nlgn1/Rgs6/Lrrk2/Ednrb/Dab1/Mef2c/Id4/Cxcl12/Gdf11/Gprc5b/Gli3/Eif2ak4/Trpc6/Gdf7/Tgif2/Zeb1/Mmd/Tcf4/Hoxd3/Heyl/Zc4h2/Bin1/Nin/Ptger3/Cav1/Bend6/Bmp7/Hey2/Nrep/Bcl11b | 32 |
| GO:0031644 | regulation of nervous system process | BP | 0.144 | 2.06e-07 | 4.73e-06 | 5.32 | 3.07e-06 | Hgf/Begain/Igf1/Nlgn1/Abat/Nos3/Celf4/Stx1b/Drd4/Wasf3/Lrrk2/Ednrb/Nptx1/Grin2b/Tbc1d24/Slc8a3/Calca/Nlgn3/Nptxr/Kcnc4/Dlg4/Cux2/Tppp/Zmynd8/F2r/Prkar1b/Nlgn2/Jam2 | 28 |
| GO:0045766 | positive regulation of angiogenesis | BP | 0.148 | 2.15e-07 | 4.87e-06 | 5.31 | 3.16e-06 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Cxcr3/Tek/Pik3r6/Cdh5/Aplnr/Jcad/Emilin1/Vegfc/Agtr1a/Tie1/Igf2/Hipk2/Akt3/Vash2/Eng | 27 |
| GO:1904018 | positive regulation of vasculature development | BP | 0.148 | 2.15e-07 | 4.87e-06 | 5.31 | 3.16e-06 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Cxcr3/Tek/Pik3r6/Cdh5/Aplnr/Jcad/Emilin1/Vegfc/Agtr1a/Tie1/Igf2/Hipk2/Akt3/Vash2/Eng | 27 |
| GO:0001755 | neural crest cell migration | BP | 0.238 | 2.17e-07 | 4.87e-06 | 5.31 | 3.16e-06 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Fn1/Ednrb/Edn3/Ednra/Sema3g/Kitl/Sema6a/Bmp7/Hand2/Twist1 | 15 |
| GO:0010721 | negative regulation of cell development | BP | 0.141 | 2.17e-07 | 4.87e-06 | 5.31 | 3.16e-06 | Prtg/Efnb3/Map2/Sema5a/Nog/Igf1/Sema5b/Nrp1/Sema7a/Tert/Ednrb/Dab1/Id4/Trpc6/Thy1/Postn/Sema3g/Daam2/Nfatc4/Epha7/Syngap1/Psen1/Sema6a/Ulk2/Ptn/Bmp7/Lrp4/S1pr3/Draxin | 29 |
| GO:0071805 | potassium ion transmembrane transport | BP | 0.141 | 2.17e-07 | 4.87e-06 | 5.31 | 3.16e-06 | Kcnk3/Kcnb2/Nalcn/Slc1a3/Kcnj3/Kcna6/Kcnd2/Kcnd3/Slc12a8/Kcnq5/Slc9a7/Slc24a4/Slc24a3/Ank2/Edn3/Kcnma1/Slc9a5/Kcnc4/Bin1/Kcnip3/Slc12a4/Pkd2/Fhl1/Kcng4/Ptger3/Atp1a2/Cav1/Kcnq4/Kcnh1 | 29 |
| GO:0042476 | odontogenesis | BP | 0.171 | 2.21e-07 | 4.94e-06 | 5.31 | 3.2e-06 | Wnt6/Lef1/Apcdd1/Fst/Edaradd/Myo5a/Slc24a4/Id3/Osr2/Gli3/Jag2/Enpp1/Tnfrsf11b/Ednra/Osr1/Hacd1/Edar/Bmp7/Lrp4/Hand2/Bcl11b/Ank | 22 |
| GO:0042310 | vasoconstriction | BP | 0.183 | 2.28e-07 | 5.05e-06 | 5.3 | 3.27e-06 | Cacna1c/Adra1a/Slc8a1/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Htr1b/Cd38/Agtr1a/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1/F2r | 20 |
| GO:0048704 | embryonic skeletal system morphogenesis | BP | 0.183 | 2.28e-07 | 5.05e-06 | 5.3 | 3.27e-06 | Col11a1/Mmp16/Hoxd10/Hoxd11/Nog/Hoxa11/Hoxd9/Mef2c/Osr2/Hoxd4/Flvcr1/Gli3/Wnt9a/Zeb1/Hoxd3/Hoxa9/Twist2/Osr1/Bmp7/Twist1 | 20 |
| GO:0003231 | cardiac ventricle development | BP | 0.159 | 2.47e-07 | 5.47e-06 | 5.26 | 3.54e-06 | Col11a1/Stra6/Sfrp2/Wnt11/Nog/Nos3/Mef2c/Foxf1/Lrp2/Cpe/Prdm1/Cplane1/Ccm2l/Aplnr/Tbx3/Ednra/Heyl/Myl3/Heg1/Fzd2/Hey2/Hand2/Slit3/Eng | 24 |
| GO:0003279 | cardiac septum development | BP | 0.169 | 2.54e-07 | 5.56e-06 | 5.25 | 3.6e-06 | Stra6/Wnt11/Nog/Tbx2/Nos3/Nrp1/Kif7/Ank2/Plxnd1/Lrp2/Prdm1/Cplane1/Aplnr/Tbx3/Heyl/Pcsk5/Heg1/Fzd2/Bmp7/Hey2/Slit3/Eng | 22 |
| GO:0060996 | dendritic spine development | BP | 0.169 | 2.54e-07 | 5.56e-06 | 5.25 | 3.6e-06 | Nlgn1/Lrrk2/Mef2c/Ppfia2/D16Ertd472e/Dock10/Abi2/Kif1a/Nlgn3/Apoe/Caprin2/Bhlhb9/Dlg4/Cux2/Palm/Dlg5/Ngef/Psen1/Zmynd8/Camk1/Nlgn2/Zfp365 | 22 |
| GO:0060537 | muscle tissue development | BP | 0.103 | 2.59e-07 | 5.66e-06 | 5.25 | 3.67e-06 | Gli1/Col11a1/Stra6/Ihh/AW551984/Angpt1/Lef1/Sgcd/Hoxd10/Col14a1/Ryr1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/P2rx2/Hoxd9/Mef2c/Tcf21/Lrp2/Tshz3/Ccm2l/Nox4/Tbx3/Gjc1/Ednra/Heyl/Igf2/Dysf/Myl3/Hdac9/Heg1/Selenon/Arrb2/Osr1/Pi16/Efemp2/Sgcb/Hlf/Pdgfrb/Zfp689/Cav1/Itga8/Usp2/Bmp7/Hey2/Hand2/Myc/Twist1/Eng | 51 |
| GO:1903305 | regulation of regulated secretory pathway | BP | 0.146 | 2.69e-07 | 5.86e-06 | 5.23 | 3.8e-06 | Nlgn1/Syt13/Adra1a/Rims3/P2rx2/Drd4/Lrrk2/Foxf1/Pfn2/Mical1/Ppfia2/Htr1b/Cadps/Syt7/Nckap1l/Htr2a/Ncs1/Cd84/Cacna1h/Fgr/Slc4a8/Rab27a/Nrn1/Cacna1a/Pld2/Cacnb4/Kcnh1 | 27 |
| GO:0090596 | sensory organ morphogenesis | BP | 0.119 | 2.73e-07 | 5.91e-06 | 5.23 | 3.83e-06 | Col11a1/Stra6/Ihh/Sdk2/Kdr/Nog/Tbx2/Arl6/Tenm3/Lrig3/Osr2/Tshz1/Obsl1/Gdf11/Gli3/Ptprm/Thy1/Zeb1/Prdm1/Abi2/Rp1/Enpp1/Tsku/Tbx3/Ednra/Col5a2/Hipk2/Rorb/Osr1/Fzd2/Fat3/Ptn/Hipk1/Kcnq4/Itga8/Bmp7/Myc/Twist1 | 38 |
| GO:0050768 | negative regulation of neurogenesis | BP | 0.153 | 2.83e-07 | 6.1e-06 | 5.21 | 3.95e-06 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Nrp1/Sema7a/Tert/Dab1/Id4/Trpc6/Thy1/Sema3g/Daam2/Nfatc4/Epha7/Syngap1/Psen1/Sema6a/Ulk2/Ptn/Bmp7/Lrp4/Draxin | 25 |
| GO:0008016 | regulation of heart contraction | BP | 0.142 | 2.85e-07 | 6.13e-06 | 5.21 | 3.97e-06 | Cacna1c/Kcnd3/Adra1a/Slc8a1/Nos3/Scn4b/Cacna2d1/Ednrb/Stc1/Hopx/Cacna1e/Ank2/Edn3/Calca/Gjc1/Ednra/Bin1/Myl3/Pde5a/Slc4a3/Cacna1h/Atp2b4/Ccn2/Apln/Atp1a2/Sri/Cav1/Hey2 | 28 |
| GO:0072577 | endothelial cell apoptotic process | BP | 0.25 | 2.86e-07 | 6.13e-06 | 5.21 | 3.97e-06 | Sema5a/Angpt1/Itga4/Kdr/Tert/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Angptl4/Hipk1/Col18a1 | 14 |
| GO:1902414 | protein localization to cell junction | BP | 0.174 | 3.05e-07 | 6.51e-06 | 5.19 | 4.22e-06 | Gpc6/Nlgn1/Stx1b/Fermt2/Nptx1/Jam3/Slitrk3/Cdh5/Dlg2/Nlgn3/Nptxr/Magi2/Pecam1/Nsg1/Cacng7/Heg1/Dlg4/Dlg5/Abcb1a/Kif5a/Nlgn2 | 21 |
| GO:0008344 | adult locomotory behavior | BP | 0.18 | 3.1e-07 | 6.58e-06 | 5.18 | 4.26e-06 | Efnb3/Cacna1c/Hoxd10/Drd4/Ccnd2/Hoxd9/Scn1a/Dab1/Cxcl12/Shank2/Glrb/Enpp1/Kcnma1/Hipk2/Prex2/Arrb2/Atp1a2/Cacna1a/Cacnb4/Nlgn2 | 20 |
| GO:0099560 | synaptic membrane adhesion | BP | 0.324 | 3.13e-07 | 6.62e-06 | 5.18 | 4.29e-06 | Gpc6/Lrrc4c/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Epha3/Magi2/Sparcl1/Pcdh17/Ntng1 | 11 |
| GO:0042326 | negative regulation of phosphorylation | BP | 0.11 | 3.16e-07 | 6.67e-06 | 5.18 | 4.32e-06 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Slc8a1/Ptpro/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Enpp1/Cdkn1c/Emilin1/Ppef2/Prr5l/Apoe/Rasip1/Pecam1/Ppm1e/Bdkrb2/Pkib/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Smpd1/Psen1/Cav1/Pid1/Grb10/Bmp7/Prkar1b/Twist1/Lax1/Eng | 44 |
| GO:0051282 | regulation of sequestering of calcium ion | BP | 0.172 | 3.52e-07 | 7.39e-06 | 5.13 | 4.79e-06 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Htr2a/Xcl1/Selenon/Asph/Pkd2/Xcr1/Ryr3/Slc25a23/Gstm7/Sri/F2r | 21 |
| GO:0072080 | nephron tubule development | BP | 0.186 | 3.55e-07 | 7.43e-06 | 5.13 | 4.82e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Tcf21/Gli3/Agtr1a/Heyl/Ptch1/Dchs1/Osr1/Aqp11/Fat4/Myc | 19 |
| GO:0098698 | postsynaptic specialization assembly | BP | 0.357 | 3.78e-07 | 7.88e-06 | 5.1 | 5.11e-06 | Gap43/Ptprd/Nlgn1/Lrrc4b/Nptx1/Slitrk3/Nlgn3/Nptxr/Arhgef9/Nlgn2 | 10 |
| GO:0010524 | positive regulation of calcium ion transport into cytosol | BP | 0.227 | 4.16e-07 | 8.55e-06 | 5.07 | 5.54e-06 | Cxcr3/Thy1/Aplnr/Agtr1a/Ednra/Adcyap1r1/Asph/Pkd2/Gimap5/Gstm7/Gimap3/Sri/Cav1/F2r/Cd4 | 15 |
| GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | BP | 0.184 | 4.16e-07 | 8.55e-06 | 5.07 | 5.54e-06 | Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Serpine2/Tek/Ppp1r16b/Prr5l/Fyn/Fgr/Pdgfrb/Pld2/Ror1/F2r/Twist1 | 19 |
| GO:0021510 | spinal cord development | BP | 0.184 | 4.16e-07 | 8.55e-06 | 5.07 | 5.54e-06 | Vit/Hoxd10/Dcc/Nog/Dab1/Gdf11/Gli3/Gdf7/Loxl3/Enpp1/Phgdh/Sox12/Dync2h1/Daam2/Tal1/Ptch1/Rab23/Cacna1a/Draxin | 19 |
| GO:0060993 | kidney morphogenesis | BP | 0.184 | 4.16e-07 | 8.55e-06 | 5.07 | 5.54e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Lrrk2/Tcf21/Gli3/Tshz3/Agtr1a/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Myc | 19 |
| GO:0060828 | regulation of canonical Wnt signaling pathway | BP | 0.129 | 4.34e-07 | 8.83e-06 | 5.05 | 5.72e-06 | Gli1/Sfrp2/Sema5a/Mdk/Wnt11/Nog/Zeb2/Nkd1/Ptpro/Cdh2/Gpc3/Lrrk2/Rbms3/Gprc5b/Gli3/Peg12/Znrf3/Dkk3/Apoe/Daam2/Prickle1/Caprin2/Tle1/Lats2/Psen1/Adgra2/Cav1/Dact1/Nkd2/Tle2/Lrp4/Draxin | 32 |
| GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | BP | 0.213 | 4.34e-07 | 8.83e-06 | 5.05 | 5.72e-06 | Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Tek/Prr5l/Fyn/Fgr/Pdgfrb/Pld2/Ror1/F2r | 16 |
| GO:0072171 | mesonephric tubule morphogenesis | BP | 0.213 | 4.34e-07 | 8.83e-06 | 5.05 | 5.72e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Osr1/Fat4/Myc | 16 |
| GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.314 | 4.36e-07 | 8.84e-06 | 5.05 | 5.73e-06 | Pcdh18/Nlgn1/Cdh2/Cdh6/Cdh13/Pcdhgc3/Cdh10/Cdh5/Pcdh12/Cdh11/Dchs1 | 11 |
| GO:0051961 | negative regulation of nervous system development | BP | 0.15 | 4.52e-07 | 9.13e-06 | 5.04 | 5.91e-06 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Nrp1/Sema7a/Tert/Dab1/Id4/Trpc6/Thy1/Sema3g/Daam2/Nfatc4/Epha7/Syngap1/Psen1/Sema6a/Ulk2/Ptn/Bmp7/Lrp4/Draxin | 25 |
| GO:0045165 | cell fate commitment | BP | 0.122 | 4.55e-07 | 9.16e-06 | 5.04 | 5.93e-06 | Gap43/Ihh/Sfrp2/Wnt6/Ar/AW551984/Wnt11/Hoxd10/Kdr/Ptch2/Tbx2/Hoxa11/Mef2c/Gli3/Glis1/Gdf7/Wnt9a/Loxl3/Prdm1/Jag2/Prickle2/Tbx3/Sox12/Ednra/Tal1/Lhx6/Ptch1/Ets2/Lats2/Psen1/Batf/Spn/Ly9/Hey2/Bcl11b | 35 |
| GO:0060047 | heart contraction | BP | 0.13 | 4.86e-07 | 9.73e-06 | 5.01 | 6.31e-06 | Cacna1c/Sgcd/Kcnd3/Adra1a/Slc8a1/Nos3/Scn4b/Scn1a/Cacna2d1/Ednrb/Stc1/Hopx/Cacna1e/Ank2/Edn3/Calca/Rps6ka2/Gjc1/Ednra/Bin1/Myl3/Pde5a/Slc4a3/Cacna1h/Atp2b4/Ccn2/Apln/Atp1a2/Sri/Cav1/Hey2 | 31 |
| GO:0042098 | T cell proliferation | BP | 0.133 | 4.91e-07 | 9.81e-06 | 5.01 | 6.35e-06 | Ihh/Igf1/Cd44/Cxcl12/Mapk8ip1/Dock2/Tnfsf13b/Msn/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Havcr2/Igf2/Nckap1l/Xcl1/Satb1/Stat5b/Fyn/Pde5a/H2-Aa/Dlg5/Zap70/Il12rb1/Spn/Cd4/Myc/Rasal3 | 30 |
| GO:0060998 | regulation of dendritic spine development | BP | 0.2 | 5.07e-07 | 1.01e-05 | 5 | 6.51e-06 | Nlgn1/Mef2c/Ppfia2/D16Ertd472e/Kif1a/Nlgn3/Apoe/Caprin2/Bhlhb9/Cux2/Palm/Dlg5/Ngef/Psen1/Zmynd8/Camk1/Nlgn2 | 17 |
| GO:0001658 | branching involved in ureteric bud morphogenesis | BP | 0.224 | 5.12e-07 | 1.01e-05 | 5 | 6.55e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Fat4/Myc | 15 |
| GO:1903829 | positive regulation of protein localization | BP | 0.102 | 5.43e-07 | 1.06e-05 | 4.97 | 6.89e-06 | Igf1/Nlgn1/Tcaf1/Abat/Ccdc88a/Hcls1/Fermt2/Gnai1/Tert/Myo5a/Ano1/Ptp4a3/Prkd1/Tek/Gli3/Lrp2/Rab34/Msn/Epha3/Nlgn3/Vegfc/Cd38/Prr5l/Crocc/Pecam1/Tmem35a/Arrb1/Tnfaip6/Stac2/Abcg1/Asph/Frmd4a/Fyn/Wipf1/Mgat3/Dzip1/Dlg4/Atp2b4/Psen1/Sri/Ptn/Meak7/Glis2/Nkd2/Pfkm/Golph3l/Cacnb4/Lrp4/Camk1/Nlgn2 | 50 |
| GO:0014031 | mesenchymal cell development | BP | 0.189 | 5.51e-07 | 1.08e-05 | 4.97 | 6.98e-06 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Ednrb/Edn3/Ednra/Heyl/Sema3g/Kitl/Sema6a/Bmp7/Hey2/Hand2/Twist1 | 18 |
| GO:0003203 | endocardial cushion morphogenesis | BP | 0.306 | 6.01e-07 | 1.17e-05 | 4.93 | 7.58e-06 | Nog/Tbx2/Nos3/Aplnr/Tbx3/Heyl/Snai1/Dchs1/Bmp7/Twist1/Eng | 11 |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | BP | 0.124 | 6.51e-07 | 1.26e-05 | 4.9 | 8.15e-06 | Ptprz1/Hgf/Sfrp2/Angpt1/Spink1/Igf1/Adra1a/Nrp1/Hcls1/Cd44/Cspg4/Areg/Icam1/Gprc5b/Thy1/Plpp3/Kit/Vegfc/Nox4/Pecam1/Igf2/Kitl/Htr2a/Epha7/Arrb2/Fyn/Dlg4/Psen1/Pdgfrb/Cav1/Enpp2/Cd4/Lrp4 | 33 |
| GO:0120032 | regulation of plasma membrane bounded cell projection assembly | BP | 0.137 | 6.52e-07 | 1.26e-05 | 4.9 | 8.15e-06 | Gap43/Evl/Nlgn1/Ccdc88a/Nrp1/Adamts16/Abi3/Pfn2/Icam1/Podxl/Abi2/Kit/Kctd17/Rp1/Eps8l1/Crocc/Daam2/Ttbk2/Dzip1/Palm/Dpysl3/Dync2li1/Cdc42ep5/Cep97/Def8/Cav1/Atmin/Zmynd8 | 28 |
| GO:0051098 | regulation of binding | BP | 0.108 | 6.53e-07 | 1.26e-05 | 4.9 | 8.15e-06 | Map2/Angpt1/Lef1/Itga4/Nog/Igf1/Zfp462/Runx1t1/Tert/Lrrk2/Hopx/Plxnd1/Lfng/Ccm2l/Apoe/Caprin2/Zc4h2/Dysf/Pkd1/Hipk2/Arrb1/Nfatc4/Twist2/Arrb2/Arhgap28/Ttbk2/Cldn5/Dzip1/Habp4/Mfng/Zfp90/Psen1/Sri/Nes/Hipk1/Cav1/Dact1/Gpsm1/Hey2/Hand2/Camk1/Myc/Twist1 | 43 |
| GO:0098815 | modulation of excitatory postsynaptic potential | BP | 0.233 | 7.14e-07 | 1.37e-05 | 4.86 | 8.88e-06 | Nlgn1/Celf4/Stx1b/Drd4/Lrrk2/Grin2b/Tbc1d24/Slc8a3/Nlgn3/Dlg4/Cux2/Zmynd8/Prkar1b/Nlgn2 | 14 |
| GO:0072028 | nephron morphogenesis | BP | 0.195 | 7.18e-07 | 1.37e-05 | 4.86 | 8.89e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Osr1/Fat4/Bmp7/Myc | 17 |
| GO:0045860 | positive regulation of protein kinase activity | BP | 0.112 | 7.19e-07 | 1.37e-05 | 4.86 | 8.89e-06 | Angpt1/Agap2/Igf1/Zeb2/Ddr2/Adra1a/Ccdc88a/Fermt2/Drd4/Ccnd2/Lrrk2/Areg/Dab1/Pik3r6/Gprc5b/Edn3/Calca/Kit/Mmd/Vegfc/Agtr1a/Nox4/Kitl/Htr2a/Pkd1/Rassf2/Arrb2/Pkd2/Pde5a/Dlg4/Atp2b4/Acsl1/Psen1/Lat/Pdgfrb/Map4k2/C1qtnf9/Fgd2/Cd4/Camk1 | 40 |
| GO:0007266 | Rho protein signal transduction | BP | 0.159 | 7.33e-07 | 1.39e-05 | 4.86 | 9.03e-06 | Adra1a/Nrp1/Cdh2/Lpar4/Arhgef25/Cdh13/Eps8l1/Agtr1a/Apoe/Rasip1/Ednra/Pecam1/Arap3/Arrb1/Heg1/Dlc1/Net1/Cdc42ep5/Pdgfrb/Nradd/F2r/Plekhg5 | 22 |
| GO:0019233 | sensory perception of pain | BP | 0.154 | 7.43e-07 | 1.41e-05 | 4.85 | 9.12e-06 | Kcnd2/Scn1a/Ednrb/Ano1/Grin2b/Cxcl12/Cacna1e/Calca/Ndn/Mme/Dlg2/Ednra/Kcnc4/Htr2a/Kcnip3/Arrb2/Fyn/Cacna1h/Scn3a/Cacna1a/F2r/Prkar1b/Nlgn2 | 23 |
| GO:0048872 | homeostasis of number of cells | BP | 0.11 | 7.51e-07 | 1.42e-05 | 4.85 | 9.18e-06 | Tex15/Pla2g10/Col14a1/Gprasp2/Emcn/Nos3/Crispld1/P4htm/Hcls1/Cd44/Ets1/Cdh2/Mef2c/Jam3/Flvcr1/Plcb1/Myct1/Sh2b2/Flt3/Tnfsf13b/Dock10/Coro1a/Armcx1/Kit/Heph/Enpp1/Nckap1l/Zfp251/Kitl/Tal1/Rassf2/Stat5b/Akt3/Septin4/Gimap5/Fstl1/Gimap3/Lat/Glis2/F2r/Il7r | 41 |
| GO:1902905 | positive regulation of supramolecular fiber organization | BP | 0.142 | 7.55e-07 | 1.42e-05 | 4.85 | 9.19e-06 | Sema5a/Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Wasf3/Pfn2/Icam1/Abi2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Scin/Bin1/Nckap1l/Nin/Slain1/Ccn2/Efemp2/Cdc42ep5/Psen1/Cav1/Plek | 26 |
| GO:0061028 | establishment of endothelial barrier | BP | 0.25 | 7.59e-07 | 1.42e-05 | 4.85 | 9.19e-06 | Ednrb/Icam1/Plcb1/Cdh5/Msn/Ppp1r16b/Robo4/Ednra/Pecam1/Cldn5/Akap11/Cldn1/S1pr3 | 13 |
| GO:0006813 | potassium ion transport | BP | 0.128 | 7.67e-07 | 1.43e-05 | 4.85 | 9.26e-06 | Kcnk3/Kcnb2/Nalcn/Slc1a3/Kcnj3/Kcna6/Kcnd2/Kcnd3/Slc12a8/Nos3/Kcnq5/Slc9a7/Slc24a4/Slc24a3/Ank2/Edn3/Kcnma1/Slc9a5/Kcnc4/Bin1/Htr2a/Kcnip3/Slc12a4/Pkd2/Fhl1/Kcng4/Ptger3/Atp1a2/Cav1/Kcnq4/Kcnh1 | 31 |
| GO:0060560 | developmental growth involved in morphogenesis | BP | 0.121 | 7.93e-07 | 1.47e-05 | 4.83 | 9.54e-06 | Sfrp2/Map2/Sema5a/Wnt11/Itga4/Zeb2/St8sia2/Tbx2/Sema5b/Cpne5/Nrp1/Nkd1/Sema7a/Fn1/Areg/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Nin/Epha7/Cacng7/Sema6a/Ulk2/Nrn1/Cyfip2/Slit3/Draxin | 34 |
| GO:0030282 | bone mineralization | BP | 0.164 | 8.02e-07 | 1.48e-05 | 4.83 | 9.6e-06 | Wnt11/Igf1/Ddr2/Pkdcc/Slc8a1/Gpc3/Mef2c/Osr2/Slc24a3/Nell1/Pth1r/Mgp/Enpp1/Ltbp3/Osr1/Fgr/Ptn/Dnm3os/Bmp7/Ank/Twist1 | 21 |
| GO:0090257 | regulation of muscle system process | BP | 0.125 | 8.08e-07 | 1.49e-05 | 4.83 | 9.65e-06 | Cacna1c/Ryr1/Igf1/Npnt/Adra1a/Abat/Slc8a1/Nos3/Mef2c/Stc1/Ank2/Slc8a3/Calca/Gucy1a1/Kit/Kcnma1/Bin1/Myl3/Cmya5/Selenon/Pde5a/Pi16/Calcrl/Cacna1h/Ptger3/Atp2b4/Ccn2/Atp1a2/Sri/Cav1/F2r/Hand2 | 32 |
| GO:0007613 | memory | BP | 0.149 | 8.25e-07 | 1.52e-05 | 4.82 | 9.82e-06 | Mdk/Cyp7b1/Nog/Igf1/Aph1b/Drd4/Ccnd2/Chst10/B4galt2/Grin2b/Shank2/Plcb1/Eif2ak4/Slc8a3/Scn2a/Apoe/Igf2/Htr2a/Nfatc4/Pde5a/Cux2/Psen1/Ptn/Itga8 | 24 |
| GO:0001738 | morphogenesis of a polarized epithelium | BP | 0.193 | 8.5e-07 | 1.56e-05 | 4.81 | 1.01e-05 | Sfrp2/Wnt11/Nkd1/Gpc3/Foxf1/Myo9a/Msn/Znrf3/Cplane1/Sh3bp1/Dchs1/Nphp1/Fzd2/Dlg5/Brsk1/Fat4/Dact1 | 17 |
| GO:0008217 | regulation of blood pressure | BP | 0.132 | 8.76e-07 | 1.6e-05 | 4.8 | 1.03e-05 | Ar/Cacna1c/Kdr/Ddah1/Adra1a/Abat/Nos3/Ptpro/Npr3/P2rx2/Adamts16/Ednrb/Enpep/Edn3/Calca/Rps6ka2/Postn/Gucy1a1/Emilin1/Vegfc/Agtr1a/Ednra/Bdkrb2/Trhde/Apln/Atp1a2/Lvrn/F2r/Eng | 29 |
| GO:0060491 | regulation of cell projection assembly | BP | 0.135 | 8.78e-07 | 1.6e-05 | 4.8 | 1.03e-05 | Gap43/Evl/Nlgn1/Ccdc88a/Nrp1/Adamts16/Abi3/Pfn2/Icam1/Podxl/Abi2/Kit/Kctd17/Rp1/Eps8l1/Crocc/Daam2/Ttbk2/Dzip1/Palm/Dpysl3/Dync2li1/Cdc42ep5/Cep97/Def8/Cav1/Atmin/Zmynd8 | 28 |
| GO:0097178 | ruffle assembly | BP | 0.267 | 9.53e-07 | 1.73e-05 | 4.76 | 1.12e-05 | Evl/Nlgn1/Cspg4/Aif1l/Pfn2/Icam1/Sh3bp1/Eps8l1/Arhgef26/Def8/Pdgfrb/Cav1 | 12 |
| GO:2000351 | regulation of endothelial cell apoptotic process | BP | 0.245 | 9.62e-07 | 1.74e-05 | 4.76 | 1.13e-05 | Sema5a/Angpt1/Itga4/Kdr/Tert/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Angptl4/Col18a1 | 13 |
| GO:0051209 | release of sequestered calcium ion into cytosol | BP | 0.168 | 9.83e-07 | 1.77e-05 | 4.75 | 1.15e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Htr2a/Xcl1/Selenon/Asph/Pkd2/Xcr1/Ryr3/Gstm7/Sri/F2r | 20 |
| GO:0033627 | cell adhesion mediated by integrin | BP | 0.191 | 1e-06 | 1.8e-05 | 4.75 | 1.16e-05 | Epha8/Cyp1b1/Itga4/Npnt/Col16a1/Itgb7/Icam1/Jam3/Itga1/Podxl/Plpp3/Itgal/Emilin1/Itga7/Nckap1l/Itga8/Itgb4 | 17 |
| GO:1901019 | regulation of calcium ion transmembrane transporter activity | BP | 0.191 | 1e-06 | 1.8e-05 | 4.75 | 1.16e-05 | Drd4/Cacna2d1/Myo5a/Ank2/Cracr2a/Dysf/Stac2/Selenon/Asph/Pkd2/Rrad/Gnb5/Gstm7/Atp1a2/Sri/Cacna1a/Cacnb4 | 17 |
| GO:0106027 | neuron projection organization | BP | 0.174 | 1.03e-06 | 1.83e-05 | 4.74 | 1.19e-05 | Nlgn1/Lrrk2/Grin2b/Ppfia2/Dock10/Abi2/Kif1a/Nlgn3/Apoe/Caprin2/Bhlhb9/Fyn/Dlg4/Cux2/Abcd2/Ngef/Psen1/Zmynd8/Zfp365 | 19 |
| GO:0051492 | regulation of stress fiber assembly | BP | 0.182 | 1.04e-06 | 1.85e-05 | 4.73 | 1.2e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Dlc1/Arhgap28/Ccn2 | 18 |
| GO:0042063 | gliogenesis | BP | 0.111 | 1.06e-06 | 1.87e-05 | 4.73 | 1.21e-05 | Gap43/Ptprz1/Mdk/Lef1/Apcdd1/Nog/Igf1/Lamc3/Cspg4/Tert/Cdh2/Fn1/Wasf3/Areg/Dab1/Id4/Serpine2/Gli3/Slc8a3/Lrp2/Plpp3/Ndn/Matn2/Nlgn3/Enpp1/Phgdh/Daam2/Bin1/Zcchc24/Tal1/Nrros/Psen1/Ptn/Tppp/Itgb4/Enpp2/Vcan/Myc/Zfp365 | 39 |
| GO:0017157 | regulation of exocytosis | BP | 0.126 | 1.09e-06 | 1.93e-05 | 4.72 | 1.25e-05 | Nlgn1/Syt13/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Myo5a/Foxf1/Pfn2/Mical1/Ppfia2/Htr1b/Cadps/Syt7/Nckap1l/Htr2a/Ncs1/Cd84/Cacna1h/Fgr/Septin4/Slc4a8/Rab27a/Nrn1/Cacna1a/Pld2/Pdcd6ip/Cacnb4/Kcnh1 | 31 |
| GO:0050982 | detection of mechanical stimulus | BP | 0.226 | 1.09e-06 | 1.93e-05 | 4.72 | 1.25e-05 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12/Kit/Htr2a/Pkd1/Pkd2/Fyn/Piezo2/Pkdrej/Myc | 14 |
| GO:0051283 | negative regulation of sequestering of calcium ion | BP | 0.167 | 1.13e-06 | 1.98e-05 | 4.7 | 1.28e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Htr2a/Xcl1/Selenon/Asph/Pkd2/Xcr1/Ryr3/Gstm7/Sri/F2r | 20 |
| GO:0006937 | regulation of muscle contraction | BP | 0.146 | 1.16e-06 | 2.03e-05 | 4.69 | 1.31e-05 | Cacna1c/Ryr1/Npnt/Adra1a/Abat/Slc8a1/Stc1/Ank2/Slc8a3/Calca/Gucy1a1/Kit/Kcnma1/Bin1/Myl3/Pde5a/Calcrl/Cacna1h/Ptger3/Ccn2/Atp1a2/Sri/Cav1/F2r | 24 |
| GO:1904019 | epithelial cell apoptotic process | BP | 0.165 | 1.29e-06 | 2.24e-05 | 4.65 | 1.45e-05 | Sema5a/Angpt1/Mdk/Itga4/Kdr/Igf1/Tert/Bok/Icam1/Tek/Cdh5/Jag2/Cd248/Ecscr/Ednra/Ndnf/Arrb2/Angptl4/Hipk1/Col18a1 | 20 |
| GO:0007263 | nitric oxide mediated signal transduction | BP | 0.36 | 1.35e-06 | 2.35e-05 | 4.63 | 1.52e-05 | Spink1/Nos3/Cbs/Gucy1a2/Gucy1a1/Apoe/Ndnf/Pde5a/Atp2b4 | 9 |
| GO:0110020 | regulation of actomyosin structure organization | BP | 0.171 | 1.37e-06 | 2.37e-05 | 4.63 | 1.53e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Mef2c/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Dlc1/Arhgap28/Ccn2 | 19 |
| GO:0001656 | metanephros development | BP | 0.187 | 1.39e-06 | 2.39e-05 | 4.62 | 1.55e-05 | Hoxd11/Hoxa11/Gpc3/Id3/Osr2/Gdf11/Tcf21/Gli3/Tshz3/Ptch1/Osr1/Dlg5/Fat4/Pdgfrb/Itga8/Bmp7/Myc | 17 |
| GO:0031032 | actomyosin structure organization | BP | 0.131 | 1.42e-06 | 2.43e-05 | 4.61 | 1.58e-05 | Kank4/Wnt11/Evl/Frmd3/Epb41l3/Phldb2/Ccdc88a/Nrp1/Fermt2/Tmod2/Mef2c/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Cnn2/Tmeff2/Ppm1e/Arrb1/Flnc/Dlc1/Arhgap28/Cnn3/Ccn2/Pdgfrb/Pdcd6ip | 28 |
| GO:0046578 | regulation of Ras protein signal transduction | BP | 0.134 | 1.43e-06 | 2.43e-05 | 4.61 | 1.58e-05 | Igf1/Adra1a/Nrp1/Cdh2/Rasa3/Lpar4/Arhgef25/Dock2/Sh2b2/Syde1/Sh3bp1/Eps8l1/Apoe/Rasip1/Kitl/Arap3/Arrb1/Heg1/Dlc1/Net1/Syngap1/Mapre2/Pdgfrb/Dennd4a/Iqsec3/F2r/Rasal3 | 27 |
| GO:0070661 | leukocyte proliferation | BP | 0.109 | 1.45e-06 | 2.47e-05 | 4.61 | 1.6e-05 | Ihh/Lef1/Igf1/Cd44/Npr3/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Msn/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Kitl/Xcl1/Satb1/Stat5b/Fyn/Pde5a/Csf2rb/H2-Aa/Dlg5/Impdh1/Zap70/Il12rb1/Spn/Il7r/Cd4/Myc/Rasal3 | 40 |
| GO:0006790 | sulfur compound metabolic process | BP | 0.113 | 1.47e-06 | 2.49e-05 | 4.6 | 1.61e-05 | Acsl6/Angpt1/Hmgcs2/Acacb/Cbs/Chst2/Spock2/Ednrb/Mical1/Tpst1/Egflam/Gamt/Gstm6/Them5/Mamdc2/Cth/Chst1/Enpp1/Phgdh/Nox4/Ednra/Ndnf/Bgn/Acot7/Stat5b/Dse/Acsl1/Xylt1/Gstm7/Gstm1/Ggt7/Cacna1a/Sult1a1/Gstk1/Papss2/Suclg2/Acot6 | 37 |
| GO:0046928 | regulation of neurotransmitter secretion | BP | 0.164 | 1.47e-06 | 2.49e-05 | 4.6 | 1.61e-05 | Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Kcnc4/Htr2a/Ncs1/Slc4a8/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 20 |
| GO:0001837 | epithelial to mesenchymal transition | BP | 0.148 | 1.51e-06 | 2.53e-05 | 4.6 | 1.64e-05 | Sfrp2/Mdk/Lef1/Wnt11/Nog/Mark1/Phldb2/Fermt2/Erg/Has2/Loxl3/Epha3/Tbx3/Ednra/Spred3/Heyl/Snai1/Il17rd/Dlg5/Bmp7/Hey2/Twist1/Eng | 23 |
| GO:0048593 | camera-type eye morphogenesis | BP | 0.148 | 1.51e-06 | 2.53e-05 | 4.6 | 1.64e-05 | Stra6/Ihh/Sdk2/Kdr/Tbx2/Arl6/Tenm3/Obsl1/Gdf11/Gli3/Ptprm/Thy1/Zeb1/Abi2/Rp1/Tsku/Hipk2/Rorb/Fat3/Ptn/Hipk1/Bmp7/Twist1 | 23 |
| GO:0030217 | T cell differentiation | BP | 0.116 | 1.51e-06 | 2.53e-05 | 4.6 | 1.64e-05 | Ihh/Mdk/Lef1/Cd83/Cd44/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Lfng/Loxl3/Zeb1/Prdm1/Jag2/Cd1d1/Kit/Enpp1/Cracr2a/Sox12/Nckap1l/Satb1/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Batf/Spn/Ly9/Il7r/Cd4/Bcl11b | 35 |
| GO:0048588 | developmental cell growth | BP | 0.119 | 1.62e-06 | 2.71e-05 | 4.57 | 1.75e-05 | Map2/Sema5a/Itga4/Col14a1/Igf1/Zeb2/St8sia2/Sema5b/Adra1a/Cpne5/Nrp1/Sema7a/Fn1/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Nin/Epha7/Cacng7/Pi16/Sema6a/Ulk2/Nrn1/Cyfip2/Slit3/Draxin | 33 |
| GO:0002685 | regulation of leukocyte migration | BP | 0.128 | 1.67e-06 | 2.79e-05 | 4.55 | 1.81e-05 | Mdk/Itga4/Chst2/Icam1/Jam3/Cxcr3/Cxcl12/Edn3/Plcb1/Thy1/Msn/Il27ra/Emilin1/Vegfc/Sele/Ednra/Pecam1/Cd200/Dysf/Nckap1l/Kitl/Xcl1/Tnfaip6/Ptger3/St3gal4/Mpp1/Ptn/Spn/Jam2 | 29 |
| GO:0072078 | nephron tubule morphogenesis | BP | 0.193 | 1.83e-06 | 3.03e-05 | 4.52 | 1.96e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Osr1/Fat4/Myc | 16 |
| GO:0034764 | positive regulation of transmembrane transport | BP | 0.123 | 1.83e-06 | 3.03e-05 | 4.52 | 1.96e-05 | Cacna1c/Acsl6/Igf1/Tcaf1/Drd4/Tert/Gpc3/Cacna2d1/Myo5a/Cxcr3/Ank2/Edn3/Plcb1/Trpc6/Thy1/Aplnr/Nlgn3/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Acsl1/Gstm7/Psen1/Abcb1a/Sri/Cacnb4/F2r/Irs1 | 31 |
| GO:0001666 | response to hypoxia | BP | 0.13 | 1.88e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Kcnk3/Kcnd2/Kdr/Ryr1/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Grin2b/Tek/Slc8a3/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Pld2/Hif3a/Myc/Twist1/Eng | 28 |
| GO:0022612 | gland morphogenesis | BP | 0.146 | 1.89e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Gli1/Hgf/Ar/Mdk/Cyp7b1/Nog/Igf1/Tbx2/Nrp1/Cd44/Areg/Mmp2/Id4/Gli3/Plxnd1/Gdf7/Msn/Tbx3/Ptch1/Ptn/Cav1/Edar/Bmp7 | 23 |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | BP | 0.135 | 1.9e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Ptprz1/Hgf/Angpt1/Igf1/Adra1a/Nrp1/Hcls1/Cd44/Cspg4/Areg/Icam1/Gprc5b/Plpp3/Kit/Vegfc/Nox4/Pecam1/Igf2/Kitl/Htr2a/Arrb2/Fyn/Dlg4/Enpp2/Cd4/Lrp4 | 26 |
| GO:0043535 | regulation of blood vessel endothelial cell migration | BP | 0.183 | 1.9e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Prkd1/Vash1/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Akt3/Atp2b4 | 17 |
| GO:1904035 | regulation of epithelial cell apoptotic process | BP | 0.183 | 1.9e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Sema5a/Angpt1/Mdk/Itga4/Kdr/Igf1/Tert/Bok/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Arrb2/Angptl4/Col18a1 | 17 |
| GO:0031503 | protein-containing complex localization | BP | 0.132 | 1.91e-06 | 3.1e-05 | 4.51 | 2.01e-05 | Gpc6/Sgcd/Tub/Nlgn1/Lrrc7/Cep112/Stx1b/Drd4/Wdr35/Nptx1/Exoc3l/Ift57/Dlg2/Nptxr/Magi2/Dync2h1/Ttc26/Arl3/Nsg1/Cacng7/Dzip1/Dlg4/Dync2li1/Bbs1/Ift81/Kif5a/Zfp365 | 27 |
| GO:0010632 | regulation of epithelial cell migration | BP | 0.124 | 1.92e-06 | 3.12e-05 | 4.51 | 2.02e-05 | Sema5a/Angpt1/Kdr/Igf1/Evl/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Prkd1/Stc1/Tek/Has2/Vash1/Ptprm/Plpp3/Ppm1f/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Akt3/Atp2b4/Arsb/Adgra2/Mapre2/Enpp2 | 30 |
| GO:0060675 | ureteric bud morphogenesis | BP | 0.203 | 1.97e-06 | 3.17e-05 | 4.5 | 2.06e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Fat4/Myc | 15 |
| GO:1903510 | mucopolysaccharide metabolic process | BP | 0.203 | 1.97e-06 | 3.17e-05 | 4.5 | 2.06e-05 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Dse/Habp4/Xylt1 | 15 |
| GO:0051100 | negative regulation of binding | BP | 0.142 | 2e-06 | 3.21e-05 | 4.49 | 2.08e-05 | Map2/Lef1/Itga4/Nog/Zfp462/Lrrk2/Ccm2l/Dysf/Nfatc4/Twist2/Arrb2/Arhgap28/Ttbk2/Habp4/Zfp90/Psen1/Sri/Nes/Cav1/Dact1/Hey2/Hand2/Camk1/Myc | 24 |
| GO:0032835 | glomerulus development | BP | 0.215 | 2.01e-06 | 3.21e-05 | 4.49 | 2.08e-05 | Angpt1/Nog/Ptpro/Ednrb/Mef2c/Nid1/Enpep/Tek/Tcf21/Ednra/Heyl/Osr1/Pdgfrb/Bmp7 | 14 |
| GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | BP | 0.25 | 2.02e-06 | 3.23e-05 | 4.49 | 2.09e-05 | Wnt11/Lrrc7/Foxf1/Myo9a/Msn/Dlg2/Prickle2/Lin7a/Sh3bp1/Pard3b/Dlg5/Pdcd6ip | 12 |
| GO:2001257 | regulation of cation channel activity | BP | 0.138 | 2.07e-06 | 3.3e-05 | 4.48 | 2.14e-05 | Nlgn1/Drd4/Cacna2d1/Myo5a/Mef2c/Ank2/Nlgn3/Cracr2a/Prrt1/Ednra/Dysf/Cacng7/Stac2/Selenon/Asph/Pkd2/Rrad/Dlg4/Gnb5/Gstm7/Sri/Cav1/Cacna1a/Cacnb4/Nlgn2 | 25 |
| GO:0060973 | cell migration involved in heart development | BP | 0.4 | 2.09e-06 | 3.33e-05 | 4.48 | 2.16e-05 | Nrp1/Ednra/Dchs1/Pdgfrb/Bmp7/Hand2/Twist1/Eng | 8 |
| GO:0072203 | cell proliferation involved in metanephros development | BP | 0.6 | 2.17e-06 | 3.42e-05 | 4.47 | 2.22e-05 | Gpc3/Ptch1/Osr1/Pdgfrb/Bmp7/Myc | 6 |
| GO:2000807 | regulation of synaptic vesicle clustering | BP | 0.6 | 2.17e-06 | 3.42e-05 | 4.47 | 2.22e-05 | Nlgn1/Cdh2/Nlgn3/Magi2/Pcdh17/Nlgn2 | 6 |
| GO:0042060 | wound healing | BP | 0.107 | 2.17e-06 | 3.42e-05 | 4.47 | 2.22e-05 | Cd109/Kdr/Nog/Igf1/Evl/Ddr2/Phldb2/Tfpi/Abat/Cd44/Fermt2/Fn1/Fkbp10/Serpine2/Plpp3/Procr/Ccm2l/Tsku/Apoe/Syt7/Cnn2/Pecam1/Tmeff2/Ndnf/Dysf/Pros1/Abi3bp/Clec7a/Ptger3/St3gal4/Smpd1/Rab27a/Cav1/Fgfr1op2/Cldn1/F2r/Wfdc1/Plek/Papss2/Eng | 40 |
| GO:0043393 | regulation of protein binding | BP | 0.128 | 2.26e-06 | 3.55e-05 | 4.45 | 2.3e-05 | Map2/Angpt1/Itga4/Nog/Tert/Lrrk2/Hopx/Plxnd1/Lfng/Ccm2l/Apoe/Caprin2/Dysf/Pkd1/Hipk2/Arrb1/Nfatc4/Arrb2/Ttbk2/Cldn5/Dzip1/Mfng/Psen1/Nes/Cav1/Dact1/Camk1/Myc | 28 |
| GO:0031529 | ruffle organization | BP | 0.228 | 2.34e-06 | 3.66e-05 | 4.44 | 2.37e-05 | Evl/Nlgn1/Cspg4/Aif1l/Pfn2/Icam1/Sh3bp1/Eps8l1/Arhgef26/Def8/Pdgfrb/Cav1/Plek | 13 |
| GO:0032231 | regulation of actin filament bundle assembly | BP | 0.165 | 2.37e-06 | 3.71e-05 | 4.43 | 2.4e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Dlc1/Arhgap28/Ccn2/Plek | 19 |
| GO:0046651 | lymphocyte proliferation | BP | 0.11 | 2.44e-06 | 3.79e-05 | 4.42 | 2.46e-05 | Ihh/Lef1/Igf1/Cd44/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Msn/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Satb1/Stat5b/Fyn/Pde5a/H2-Aa/Dlg5/Impdh1/Zap70/Il12rb1/Spn/Il7r/Cd4/Myc/Rasal3 | 37 |
| GO:0072088 | nephron epithelium morphogenesis | BP | 0.188 | 2.54e-06 | 3.94e-05 | 4.4 | 2.56e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Tcf21/Gli3/Agtr1a/Ptch1/Dchs1/Osr1/Fat4/Myc | 16 |
| GO:0071526 | semaphorin-plexin signaling pathway | BP | 0.245 | 2.56e-06 | 3.96e-05 | 4.4 | 2.57e-05 | Sema5a/Plxnb1/Sema5b/Nrp1/Sema7a/Mef2c/Plxnd1/Sh3bp1/Ednra/Sema3g/Sema6a/Hand2 | 12 |
| GO:0030038 | contractile actin filament bundle assembly | BP | 0.164 | 2.71e-06 | 4.18e-05 | 4.38 | 2.71e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Arrb1/Dlc1/Arhgap28/Ccn2 | 19 |
| GO:0043149 | stress fiber assembly | BP | 0.164 | 2.71e-06 | 4.18e-05 | 4.38 | 2.71e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Arrb1/Dlc1/Arhgap28/Ccn2 | 19 |
| GO:0018209 | peptidyl-serine modification | BP | 0.107 | 2.77e-06 | 4.26e-05 | 4.37 | 2.76e-05 | Hgf/Sfrp2/Angpt1/Hcls1/Cd44/Spock2/Lrrk2/Pfn2/Prkd1/Tek/Egflam/Galnt16/Rps6ka2/Mamdc2/Ppm1f/Caprin2/Ndnf/Dclk2/Bdkrb2/Pkd1/Hipk2/Rps6ka5/Arrb1/Bgn/Poglut2/Rassf2/Arrb2/Ttbk2/Akt3/Atp2b4/Lats2/Brsk1/Ulk2/Naa11/Hipk1/Cav1/Sh2d3c/Mknk2/Camk1 | 39 |
| GO:0001764 | neuron migration | BP | 0.133 | 2.81e-06 | 4.3e-05 | 4.37 | 2.79e-05 | Ptprz1/Mdk/Dcc/Mark1/Nrp1/Dab1/Mef2c/Cxcl12/Tbc1d24/Cep85l/Ndn/Gpr173/Matn2/Ndnf/Lhx6/Lrp12/Neo1/Nr2f1/Fyn/Septin4/Fat3/Bbs1/Psen1/Sema6a/Ntng1/Twist1 | 26 |
| GO:0014821 | phasic smooth muscle contraction | BP | 0.333 | 2.85e-06 | 4.35e-05 | 4.36 | 2.82e-05 | Tbx2/P2rx2/Ednrb/Edn3/Tshz3/Kit/Tbx3/Kcnma1/Ptger3 | 9 |
| GO:0070509 | calcium ion import | BP | 0.177 | 3e-06 | 4.57e-05 | 4.34 | 2.96e-05 | Cacna1c/Spink1/Slc8a1/Slc24a4/Stc1/Cxcl12/Cacna1e/Slc8a3/Nalf1/Agtr1a/Dysf/Pkd2/Fyn/Cacna1h/Slc25a23/Pdgfrb/Cacna1a | 17 |
| GO:0050769 | positive regulation of neurogenesis | BP | 0.114 | 3.01e-06 | 4.57e-05 | 4.34 | 2.96e-05 | Ptprz1/Sema5a/Mdk/Kdr/Plxnb1/Ptprd/Nog/Igf1/Zeb2/Nrp1/Sema7a/Fn1/Serpine2/Cxcl12/Tbc1d24/Obsl1/Gli3/Plxnd1/Lrp2/Kit/Mme/Vegfc/Apoe/Caprin2/Bin1/Nin/Chodl/Bhlhb9/Cux2/Ptn/Enpp2/Myc/Zfp365/Numbl | 34 |
| GO:0035725 | sodium ion transmembrane transport | BP | 0.147 | 3.07e-06 | 4.65e-05 | 4.33 | 3.02e-05 | Nalcn/Hecw2/Slc6a2/Slc8a1/Scn4b/Drd4/Fxyd5/Scn1a/Slc9a7/Slc24a4/Slc24a3/Plcb1/Slc8a3/Slc6a15/Scn2a/Slc9a5/Osr1/Pkd2/Atp2b4/Scn3a/Atp1a2/Slc4a8 | 22 |
| GO:0032943 | mononuclear cell proliferation | BP | 0.109 | 3.22e-06 | 4.83e-05 | 4.32 | 3.13e-05 | Ihh/Lef1/Igf1/Cd44/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Msn/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Satb1/Stat5b/Fyn/Pde5a/H2-Aa/Dlg5/Impdh1/Zap70/Il12rb1/Spn/Il7r/Cd4/Myc/Rasal3 | 37 |
| GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.24 | 3.22e-06 | 4.83e-05 | 4.32 | 3.13e-05 | Hmcn1/Mcam/Itga4/Ptprd/Nlgn1/Cdh2/Tenm3/Adgrl1/Itgal/Sele/Dchs1/Fat4 | 12 |
| GO:0048592 | eye morphogenesis | BP | 0.134 | 3.41e-06 | 5.12e-05 | 4.29 | 3.32e-05 | Stra6/Ihh/Sdk2/Kdr/Tbx2/Arl6/Tenm3/Obsl1/Gdf11/Gli3/Ptprm/Thy1/Zeb1/Prdm1/Abi2/Rp1/Tsku/Col5a2/Hipk2/Rorb/Fat3/Ptn/Hipk1/Bmp7/Twist1 | 25 |
| GO:0001570 | vasculogenesis | BP | 0.175 | 3.48e-06 | 5.2e-05 | 4.28 | 3.37e-05 | Angpt1/Kdr/Nrp1/Foxf1/Tek/Has2/Aplnr/Enpp1/Rasip1/Gjc1/Tie1/Heg1/Egfl7/Pdgfrb/Cav1/Hey2/Eng | 17 |
| GO:0003170 | heart valve development | BP | 0.22 | 3.54e-06 | 5.27e-05 | 4.28 | 3.42e-05 | Stra6/Nos3/Mef2c/Erg/Prdm1/Aplnr/Emilin1/Tie1/Heyl/Dchs1/Hey2/Slit3/Twist1 | 13 |
| GO:0030048 | actin filament-based movement | BP | 0.155 | 3.59e-06 | 5.34e-05 | 4.27 | 3.46e-05 | Cacna1c/Sgcd/Evl/Kcnd3/Scn4b/Scn1a/Cacna2d1/Myo5a/Stc1/Ank2/Limch1/Bin1/Epdr1/Myo1f/Wipf1/Cacna1h/Ptger3/Atp1a2/Sri/Cav1 | 20 |
| GO:0060326 | cell chemotaxis | BP | 0.111 | 3.67e-06 | 5.44e-05 | 4.26 | 3.53e-05 | Cxcl15/Hgf/Sema5a/Mdk/Lef1/Kdr/Cyp7b1/Nrp1/Ptpro/Ednrb/Prkd1/Jam3/Cxcr3/Cxcl12/Edn3/Itga1/Calca/Coro1a/Kit/Vegfc/Agtr1a/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Arrb2/Xcr1/Mpp1/Pdgfrb/Ptn/Elmo2/Enpp2/Plekhg5/Eng | 35 |
| GO:0071772 | response to BMP | BP | 0.134 | 3.76e-06 | 5.55e-05 | 4.26 | 3.6e-05 | Sfrp2/Lef1/Kdr/Nog/Kcnd3/Rnf165/Fst/Vstm2a/Gpc3/Zfp423/Cdh5/Gdf7/Lrp2/Heyl/Adamts7/Hipk2/Kcp/Tnfaip6/Smad9/Neo1/Fstl1/Cav1/Tmprss6/Bmp7/Eng | 25 |
| GO:0071773 | cellular response to BMP stimulus | BP | 0.134 | 3.76e-06 | 5.55e-05 | 4.26 | 3.6e-05 | Sfrp2/Lef1/Kdr/Nog/Kcnd3/Rnf165/Fst/Vstm2a/Gpc3/Zfp423/Cdh5/Gdf7/Lrp2/Heyl/Adamts7/Hipk2/Kcp/Tnfaip6/Smad9/Neo1/Fstl1/Cav1/Tmprss6/Bmp7/Eng | 25 |
| GO:0060249 | anatomical structure homeostasis | BP | 0.11 | 3.83e-06 | 5.63e-05 | 4.25 | 3.65e-05 | Tex15/Ihh/Angpt1/Kdr/Col14a1/Tub/Nos3/P2rx2/Calca/Flt3/Pth1r/Pbld2/Coro1a/Ldb2/Rp1/Enpp1/Tnfrsf11b/Cd38/Nox4/Crocc/Bdkrb2/Ltbp3/Lpcat1/Rcn3/Iqcb1/Cldn5/Large1/Akt3/Ccn2/Def8/Bbs1/Abcb1a/Pdgfrb/Pfkm/F2r/Kcnh1 | 36 |
| GO:0003272 | endocardial cushion formation | BP | 0.321 | 4.02e-06 | 5.86e-05 | 4.23 | 3.8e-05 | Nog/Tbx2/Aplnr/Tbx3/Heyl/Snai1/Dchs1/Bmp7/Eng | 9 |
| GO:0043931 | ossification involved in bone maturation | BP | 0.321 | 4.02e-06 | 5.86e-05 | 4.23 | 3.8e-05 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:0061484 | hematopoietic stem cell homeostasis | BP | 0.321 | 4.02e-06 | 5.86e-05 | 4.23 | 3.8e-05 | Gprasp2/Emcn/Crispld1/Myct1/Armcx1/Zfp251/Septin4/Fstl1/Glis2 | 9 |
| GO:0030900 | forebrain development | BP | 0.103 | 4.18e-06 | 6.07e-05 | 4.22 | 3.94e-05 | Gli1/Sema5a/Mdk/Lef1/Nog/Zeb2/Nrp1/Sema7a/Cdh2/Lrrk2/Dab1/Id4/Fut10/Cxcl12/Dnah5/Gli3/Plcb1/D16Ertd472e/Gdf7/Lrp2/Kif1a/Tsku/Tbx3/Dync2h1/Ndnf/Dclk2/Nin/Lhx6/Rtn4rl1/Dlc1/Nr2f1/Ttbk2/Fyn/Ptger3/Atp1a2/Fat4/Bbs1/Psen1/Bcl11b/Draxin/Numbl | 41 |
| GO:0045880 | positive regulation of smoothened signaling pathway | BP | 0.256 | 4.22e-06 | 6.12e-05 | 4.21 | 3.97e-05 | Gli1/Ihh/Evc/Kif7/Gpc3/Cibar1/Armc9/Wnt9a/Rab34/Dync2h1/Dlg5 | 11 |
| GO:0150116 | regulation of cell-substrate junction organization | BP | 0.203 | 4.25e-06 | 6.15e-05 | 4.21 | 3.98e-05 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Mapre2/Enpp2 | 14 |
| GO:0014812 | muscle cell migration | BP | 0.158 | 4.55e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Myo5a/Mef2c/Has2/Adamts1/Postn/Enpp1/Nox4/Pcsk5/Net1/Plekho1/Pdgfrb/Grb10/Myc | 19 |
| GO:0072498 | embryonic skeletal joint development | BP | 0.438 | 4.55e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Ihh/Hoxd11/Nog/Hoxa11/Osr2/Osr1/Bmp7 | 7 |
| GO:0010749 | regulation of nitric oxide mediated signal transduction | BP | 0.545 | 4.58e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Spink1/Cbs/Gucy1a2/Gucy1a1/Pde5a/Atp2b4 | 6 |
| GO:0014832 | urinary bladder smooth muscle contraction | BP | 0.545 | 4.58e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Cacna1c/P2rx2/Kcnma1/Htr2a/Ptger3/Atp2b4 | 6 |
| GO:0097104 | postsynaptic membrane assembly | BP | 0.545 | 4.58e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Nlgn1/Cdh2/Nlgn3/Magi2/Lrp4/Nlgn2 | 6 |
| GO:0071560 | cellular response to transforming growth factor beta stimulus | BP | 0.124 | 4.58e-06 | 6.52e-05 | 4.19 | 4.23e-05 | Cd109/Zeb2/Npnt/Nos3/Fermt2/Mef2c/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Ltbp3/Xcl1/Hipk2/Smad9/Nrros/Arrb2/Zfyve9/Fyn/Cldn5/Il17rd/Lats2/Cav1/Itga8/Nrep/Eng | 28 |
| GO:0014033 | neural crest cell differentiation | BP | 0.172 | 4.64e-06 | 6.6e-05 | 4.18 | 4.28e-05 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Ednrb/Mef2c/Edn3/Ednra/Sema3g/Kitl/Sema6a/Bmp7/Hand2/Twist1 | 17 |
| GO:0014032 | neural crest cell development | BP | 0.18 | 4.76e-06 | 6.72e-05 | 4.17 | 4.36e-05 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Ednrb/Edn3/Ednra/Sema3g/Kitl/Sema6a/Bmp7/Hand2/Twist1 | 16 |
| GO:1904427 | positive regulation of calcium ion transmembrane transport | BP | 0.18 | 4.76e-06 | 6.72e-05 | 4.17 | 4.36e-05 | Cacna1c/Cacna2d1/Cxcr3/Ank2/Thy1/Aplnr/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Sri/Cacnb4/F2r | 16 |
| GO:0051402 | neuron apoptotic process | BP | 0.11 | 4.87e-06 | 6.87e-05 | 4.16 | 4.45e-05 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Itga1/Coro1a/Scn2a/Grik2/Il27ra/Enpp1/Apoe/Kcnma1/Ndnf/Hipk2/Arrb1/Nfatc4/Epha7/Kcnip3/Arrb2/Bhlhb9/Fyn/Nlrp1b/Syngap1/Psen1/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 35 |
| GO:0060349 | bone morphogenesis | BP | 0.164 | 5e-06 | 7.02e-05 | 4.15 | 4.55e-05 | Ihh/Mmp16/Sfrp2/Hoxd11/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Has2/Gli3/Col13a1/Frem1/Enpp1/Ltbp3/Rab23/Twist1 | 18 |
| GO:0061756 | leukocyte adhesion to vascular endothelial cell | BP | 0.231 | 5.01e-06 | 7.02e-05 | 4.15 | 4.55e-05 | Mdk/Itga4/Chst2/Ets1/Itgb7/Icam1/Podxl2/Cxcl12/Sele/St3gal4/Spn/Jam2 | 12 |
| GO:0035904 | aorta development | BP | 0.2 | 5.08e-06 | 7.1e-05 | 4.15 | 4.6e-05 | Tbx2/Kif7/Fkbp10/Plxnd1/Lrp2/Prdm1/Aplnr/Enpp1/Ednra/Prickle1/Efemp2/Pdgfrb/Hey2/Eng | 14 |
| GO:0035115 | embryonic forelimb morphogenesis | BP | 0.278 | 5.15e-06 | 7.18e-05 | 4.14 | 4.66e-05 | Cacna1c/Hoxd11/Hoxa11/Hoxd9/Osr2/Wnt9a/Tbx3/Hoxa9/Osr1/Twist1 | 10 |
| GO:1903706 | regulation of hemopoiesis | BP | 0.102 | 5.34e-06 | 7.42e-05 | 4.13 | 4.81e-05 | Ihh/Mdk/Lef1/Cd83/P4htm/Hcls1/Cd44/Ets1/Mef2c/Inpp4b/Pik3r6/Lrrc17/Gli3/Calca/Flt3/Loxl3/Zeb1/Prdm1/Cd1d1/Tnfrsf11b/Gpr171/Sox12/Zbtb46/Scin/Nckap1l/Kitl/Tal1/Tnfaip6/Hoxa9/Twist2/Rassf2/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Ptn/Il2rg/Il7r/Cd4/Myc | 41 |
| GO:0044782 | cilium organization | BP | 0.103 | 5.34e-06 | 7.42e-05 | 4.13 | 4.81e-05 | Tub/Ccdc88a/Arl6/Dzip1l/Adamts16/Cibar1/Wdr35/Tmem237/Bbs7/Armc9/Dnah5/Zfp423/Ptpdc1/Ift57/Catip/Cplane1/Kctd17/Rp1/Rilpl1/Dync2h1/Crocc/Cfap410/Ttc26/Arl3/Cep19/Tmem231/Pkd2/Ttbk2/Iqcb1/Dnah7b/Rab23/Dzip1/Dync2li1/Lrrc49/Dnah1/Cep97/Bbs1/Atmin/Ift81/Septin6 | 40 |
| GO:0032102 | negative regulation of response to external stimulus | BP | 0.102 | 5.67e-06 | 7.86e-05 | 4.1 | 5.09e-05 | Hgf/Sema5a/Acod1/Mdk/Pla2g10/Cd109/Igf1/Sema5b/Aph1b/Phldb2/Tfpi/Abat/Nrp1/Cd44/Sema7a/Ets1/Foxf1/Serpine2/Fndc4/Cdh5/Prdm1/Nlgn3/Apoe/Havcr2/Sema3g/Cd200/Igf2/Pros1/Tnfaip6/Rtn4rl1/Neo1/Mapkbp1/Arrb2/Calcrl/Ptger3/Xylt1/Sema6a/Cd96/Nr1h3/Spn/Wfdc1 | 41 |
| GO:1990138 | neuron projection extension | BP | 0.127 | 5.91e-06 | 8.17e-05 | 4.09 | 5.29e-05 | Map2/Sema5a/Itga4/St8sia2/Sema5b/Cpne5/Nrp1/Sema7a/Fn1/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Cacng7/Sema6a/Ulk2/Nrn1/Cyfip2/Slit3/Draxin | 26 |
| GO:0097061 | dendritic spine organization | BP | 0.168 | 6.14e-06 | 8.47e-05 | 4.07 | 5.49e-05 | Nlgn1/Lrrk2/Grin2b/Ppfia2/Dock10/Abi2/Kif1a/Nlgn3/Apoe/Caprin2/Bhlhb9/Fyn/Dlg4/Cux2/Ngef/Zmynd8/Zfp365 | 17 |
| GO:0050806 | positive regulation of synaptic transmission | BP | 0.107 | 6.18e-06 | 8.49e-05 | 4.07 | 5.5e-05 | Slc1a3/Gria4/Grik1/Nog/Nlgn1/Adra1a/Rims3/Stx1b/Grin2b/Serpine2/Shank2/Eif2ak4/Slc8a3/Syde1/Tshz3/Mme/Lama2/Grik2/Nlgn3/Enpp1/Nrgn/Apoe/Prrt1/Nsg1/Nfatc4/Cacng7/Cyp46a1/Arrb2/Ncs1/Mpp2/Dlg4/Slc4a8/Psen1/Ptn/Prkar1b/Nlgn2 | 36 |
| GO:0048732 | gland development | BP | 0.0966 | 6.2e-06 | 8.5e-05 | 4.07 | 5.51e-05 | Gli1/Stra6/Hgf/Ar/Mdk/Lef1/Agap2/Cyp7b1/Nog/Igf1/Tbx2/Nrp1/Cd44/Edaradd/Hoxd9/Areg/Mmp2/Foxf1/Id4/Serpine2/Tcf21/Gli3/Plxnd1/Gdf7/Msn/Cdkn1c/Tbx3/Hoxd3/Ednra/Igf2/Pkd1/Hoxa9/Ptch1/Stat5b/Pkd2/Asl/Psen1/Pdgfrb/Ptn/Cav1/Cacnb4/Edar/Bmp7/Hand2/Bcl11b/Irs1 | 46 |
| GO:0003208 | cardiac ventricle morphogenesis | BP | 0.185 | 6.41e-06 | 8.74e-05 | 4.06 | 5.67e-05 | Col11a1/Sfrp2/Nog/Mef2c/Foxf1/Lrp2/Cpe/Ccm2l/Ednra/Heyl/Myl3/Heg1/Hey2/Hand2/Eng | 15 |
| GO:0043279 | response to alkaloid | BP | 0.185 | 6.41e-06 | 8.74e-05 | 4.06 | 5.67e-05 | Slc1a3/Ryr1/Abat/Slc8a1/Drd4/Htr1b/Ednra/Htr2a/Selenon/Dlg4/Gstm7/Smpd1/Bche/Myc/Irs1 | 15 |
| GO:0071559 | response to transforming growth factor beta | BP | 0.122 | 6.42e-06 | 8.74e-05 | 4.06 | 5.67e-05 | Cd109/Zeb2/Npnt/Nos3/Fermt2/Mef2c/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Ltbp3/Xcl1/Hipk2/Smad9/Nrros/Arrb2/Zfyve9/Fyn/Cldn5/Il17rd/Lats2/Cav1/Itga8/Nrep/Eng | 28 |
| GO:0051495 | positive regulation of cytoskeleton organization | BP | 0.127 | 6.46e-06 | 8.78e-05 | 4.06 | 5.69e-05 | Sema5a/Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Wasf3/Pfn2/Icam1/Tek/Abi2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Scin/Bin1/Nckap1l/Nin/Slain1/Ccn2/Cdc42ep5/Nes/Cav1/Plek | 26 |
| GO:0034767 | positive regulation of ion transmembrane transport | BP | 0.13 | 6.64e-06 | 9e-05 | 4.05 | 5.83e-05 | Cacna1c/Tcaf1/Drd4/Cacna2d1/Myo5a/Cxcr3/Ank2/Edn3/Plcb1/Trpc6/Thy1/Aplnr/Nlgn3/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Psen1/Abcb1a/Sri/Cacnb4/F2r | 25 |
| GO:0009954 | proximal/distal pattern formation | BP | 0.27 | 6.75e-06 | 9.12e-05 | 4.04 | 5.91e-05 | Gli1/Hoxd10/Hoxd11/Hoxa11/Hoxd9/Hoxa10/Gli3/Hoxa9/Osr1/Lrp4 | 10 |
| GO:0043523 | regulation of neuron apoptotic process | BP | 0.113 | 7e-06 | 9.44e-05 | 4.02 | 6.12e-05 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Itga1/Coro1a/Grik2/Il27ra/Apoe/Kcnma1/Ndnf/Hipk2/Arrb1/Nfatc4/Epha7/Kcnip3/Arrb2/Bhlhb9/Fyn/Syngap1/Psen1/Nes/Cacna1a/Pcdhgc4/F2r/Draxin | 32 |
| GO:0150115 | cell-substrate junction organization | BP | 0.167 | 7.04e-06 | 9.48e-05 | 4.02 | 6.14e-05 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Fn1/Thsd1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Mapre2/Itgb4/Enpp2 | 17 |
| GO:0051090 | regulation of DNA-binding transcription factor activity | BP | 0.0986 | 7.44e-06 | 9.98e-05 | 4 | 6.47e-05 | Gli1/Ndp/Cyp1b1/4930447C04Rik/Acod1/Ar/Pla2g10/Ddr2/Eda2r/Hcls1/Id3/Icam1/Prkd1/Gli3/Eif2ak4/Plpp3/Ndn/Kit/Cth/Enpp1/Havcr2/Heyl/Cd200/Zc4h2/Pkd1/Hipk2/Rps6ka5/Arrb1/Foxs1/Arrb2/Ptch1/Pkd2/Cd84/Nwd1/Fzd2/Sri/Glis2/Cav1/Commd7/Ror1/Hand2/Twist1/Zfp932 | 43 |
| GO:0051279 | regulation of release of sequestered calcium ion into cytosol | BP | 0.183 | 7.5e-06 | 1e-04 | 4 | 6.5e-05 | Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Selenon/Asph/Pkd2/Gstm7/Sri/F2r | 15 |
| GO:0035088 | establishment or maintenance of apical/basal cell polarity | BP | 0.222 | 7.61e-06 | 0.000101 | 4 | 6.55e-05 | Wnt11/Lrrc7/Foxf1/Myo9a/Msn/Dlg2/Prickle2/Lin7a/Sh3bp1/Pard3b/Dlg5/Pdcd6ip | 12 |
| GO:0061245 | establishment or maintenance of bipolar cell polarity | BP | 0.222 | 7.61e-06 | 0.000101 | 4 | 6.55e-05 | Wnt11/Lrrc7/Foxf1/Myo9a/Msn/Dlg2/Prickle2/Lin7a/Sh3bp1/Pard3b/Dlg5/Pdcd6ip | 12 |
| GO:0070977 | bone maturation | BP | 0.3 | 7.63e-06 | 0.000101 | 4 | 6.55e-05 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:0140058 | neuron projection arborization | BP | 0.3 | 7.63e-06 | 0.000101 | 4 | 6.55e-05 | Nlgn1/Nrp1/Lrrk2/Myo9a/Lrp2/Dlg4/Ptn/Ntng1/Zfp365 | 9 |
| GO:1903131 | mononuclear cell differentiation | BP | 0.0949 | 7.88e-06 | 0.000104 | 3.98 | 6.75e-05 | Ihh/Mdk/Lef1/Cd83/Itm2a/Cd44/Il11ra1/Mef2c/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Lfng/Tnfsf13b/Loxl3/Dock10/Zeb1/Prdm1/Jag2/Cd1d1/Kit/H2-Ab1/Enpp1/Cracr2a/Sox12/Irf8/Zbtb46/Nckap1l/Satb1/Hdac9/Stat5b/H2-Aa/Gimap5/Mfng/Zap70/Gimap3/Psen1/Il2rg/Batf/Spn/Ly9/Il7r/Cd4/Bcl11b/Myc | 47 |
| GO:0033002 | muscle cell proliferation | BP | 0.116 | 8.33e-06 | 0.00011 | 3.96 | 7.12e-05 | Gli1/Angpt1/Vipr2/Nog/Igf1/Ddr2/Tbx2/Nos3/Npr3/Drd4/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Enpp1/Apoe/Rgs5/Ednra/Selenon/Stat5b/Irak4/Calcrl/Efemp2/Apln/Pdgfrb/Cav1/Hey2/Myc | 30 |
| GO:0061512 | protein localization to cilium | BP | 0.192 | 8.49e-06 | 0.000112 | 3.95 | 7.24e-05 | Tub/Ccdc88a/Arl6/Dzip1l/Wdr35/Zfp423/Cplane1/Dync2h1/Crocc/Ttc26/Arl3/Dzip1/Bbs1/Rabep1 | 14 |
| GO:0001894 | tissue homeostasis | BP | 0.112 | 8.71e-06 | 0.000114 | 3.94 | 7.41e-05 | Tex15/Ihh/Angpt1/Kdr/Col14a1/Tub/Nos3/Calca/Flt3/Pth1r/Pbld2/Coro1a/Ldb2/Rp1/Enpp1/Tnfrsf11b/Cd38/Nox4/Crocc/Bdkrb2/Ltbp3/Lpcat1/Rcn3/Iqcb1/Cldn5/Akt3/Ccn2/Def8/Bbs1/Abcb1a/Pdgfrb/F2r | 32 |
| GO:0019722 | calcium-mediated signaling | BP | 0.128 | 8.74e-06 | 0.000114 | 3.94 | 7.42e-05 | Sgcd/Kdr/Nr5a2/Igf1/Slc8a1/Drd4/Lrrk2/Ednrb/Slc24a4/Cxcr3/Cdh13/Ptgfr/Adgrl1/Ank2/Itgal/Agtr1a/Cmya5/Nfatc4/Xcr1/Atp2b4/Zap70/Lat/Rcan3/Cd4/Tbc1d10c | 25 |
| GO:0097106 | postsynaptic density organization | BP | 0.263 | 8.77e-06 | 0.000115 | 3.94 | 7.42e-05 | Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Shank2/Slitrk3/Syngap1/Zmynd8/Nlgn2 | 10 |
| GO:0007219 | Notch signaling pathway | BP | 0.13 | 8.95e-06 | 0.000117 | 3.93 | 7.56e-05 | Tbx2/Aph1b/Cdh6/Ptp4a3/Zfp423/Postn/Lfng/Jag2/Kit/Hoxd3/Heyl/Snai1/Arrb1/Dtx3/Tspan5/Egfl7/Mfng/Fat4/Psen1/Bend6/Maml3/Bmp7/Hey2/S1pr3 | 24 |
| GO:0014911 | positive regulation of smooth muscle cell migration | BP | 0.203 | 9.18e-06 | 0.000118 | 3.93 | 7.67e-05 | Cyp1b1/Mdk/Igf1/Ddr2/Tert/Myo5a/Has2/Adamts1/Postn/Nox4/Pcsk5/Pdgfrb/Myc | 13 |
| GO:0033344 | cholesterol efflux | BP | 0.203 | 9.18e-06 | 0.000118 | 3.93 | 7.67e-05 | Apob/Pla2g10/Soat1/Pltp/Tsku/Apoe/Abca5/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3/Abcg4 | 13 |
| GO:0051893 | regulation of focal adhesion assembly | BP | 0.203 | 9.18e-06 | 0.000118 | 3.93 | 7.67e-05 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Enpp2 | 13 |
| GO:0090109 | regulation of cell-substrate junction assembly | BP | 0.203 | 9.18e-06 | 0.000118 | 3.93 | 7.67e-05 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Enpp2 | 13 |
| GO:0030510 | regulation of BMP signaling pathway | BP | 0.163 | 9.21e-06 | 0.000118 | 3.93 | 7.67e-05 | Sfrp2/Kdr/Nog/Rnf165/Fst/Gpc3/Zfp423/Cdh5/Lrp2/Hipk2/Kcp/Tnfaip6/Neo1/Fstl1/Cav1/Tmprss6/Eng | 17 |
| GO:0070167 | regulation of biomineral tissue development | BP | 0.163 | 9.21e-06 | 0.000118 | 3.93 | 7.67e-05 | Wnt6/Ddr2/Pkdcc/Slc8a1/Nos3/Mef2c/Osr2/Nell1/Mgp/Enpp1/Ltbp3/Osr1/Ptn/Bmp7/Hey2/Ank/Twist1 | 17 |
| GO:0060271 | cilium assembly | BP | 0.104 | 9.28e-06 | 0.000119 | 3.92 | 7.71e-05 | Ccdc88a/Arl6/Dzip1l/Adamts16/Cibar1/Wdr35/Tmem237/Bbs7/Armc9/Dnah5/Zfp423/Ptpdc1/Ift57/Cplane1/Kctd17/Rp1/Rilpl1/Dync2h1/Crocc/Cfap410/Ttc26/Arl3/Cep19/Tmem231/Ttbk2/Iqcb1/Dnah7b/Rab23/Dzip1/Dync2li1/Lrrc49/Dnah1/Cep97/Bbs1/Atmin/Ift81/Septin6 | 37 |
| GO:0046634 | regulation of alpha-beta T cell activation | BP | 0.157 | 9.48e-06 | 0.000121 | 3.92 | 7.86e-05 | Ihh/Cd83/Cd44/Mapk8ip1/Gli3/Loxl3/Prdm1/Cd1d1/H2-Ab1/Btla/Nckap1l/Xcl1/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg/Rasal3 | 18 |
| GO:0009952 | anterior/posterior pattern specification | BP | 0.119 | 9.67e-06 | 0.000123 | 3.91 | 8e-05 | Sfrp2/Lef1/Hoxd10/Hoxd11/Nog/Zeb2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Foxf1/Hoxd8/Hoxd4/Tshz1/Gdf11/Gli3/Lfng/Tbx3/Hoxd3/Heyl/Hipk2/Pcsk5/Hoxa9/Osr1/Ets2/Psen1/Hipk1/Hey2 | 28 |
| GO:0030203 | glycosaminoglycan metabolic process | BP | 0.17 | 9.86e-06 | 0.000126 | 3.9 | 8.14e-05 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Dse/Habp4/Xylt1/Pdgfrb | 16 |
| GO:0051017 | actin filament bundle assembly | BP | 0.133 | 9.95e-06 | 0.000126 | 3.9 | 8.19e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Aif1l/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Arrb1/Dlc1/Arhgap28/Ccn2/Dpysl3/Shroom1/Plek | 23 |
| GO:2000300 | regulation of synaptic vesicle exocytosis | BP | 0.179 | 1.02e-05 | 0.000129 | 3.89 | 8.37e-05 | Nlgn1/Adra1a/Rims3/P2rx2/Drd4/Lrrk2/Pfn2/Ppfia2/Htr1b/Htr2a/Ncs1/Slc4a8/Nrn1/Cacnb4/Kcnh1 | 15 |
| GO:0002026 | regulation of the force of heart contraction | BP | 0.29 | 1.03e-05 | 0.00013 | 3.89 | 8.4e-05 | Adra1a/Slc8a1/Nos3/Myl3/Pde5a/Atp2b4/Apln/Atp1a2/Cav1 | 9 |
| GO:0060317 | cardiac epithelial to mesenchymal transition | BP | 0.29 | 1.03e-05 | 0.00013 | 3.89 | 8.4e-05 | Nog/Erg/Has2/Tbx3/Heyl/Snai1/Hey2/Twist1/Eng | 9 |
| GO:2000352 | negative regulation of endothelial cell apoptotic process | BP | 0.29 | 1.03e-05 | 0.00013 | 3.89 | 8.4e-05 | Sema5a/Angpt1/Kdr/Tert/Icam1/Tek/Cdh5/Ndnf/Angptl4 | 9 |
| GO:0008154 | actin polymerization or depolymerization | BP | 0.126 | 1.04e-05 | 0.000131 | 3.88 | 8.51e-05 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Mical1/Abi2/Catip/Coro1a/Esam/Sh3bp1/Daam2/Pecam1/Scin/Bin1/Nckap1l/Arhgap28/Wipf1/Cdc42ep5/Plekhh2/Cyfip2/Plek/Pstpip1 | 25 |
| GO:0007628 | adult walking behavior | BP | 0.234 | 1.07e-05 | 0.000134 | 3.87 | 8.65e-05 | Efnb3/Cacna1c/Scn1a/Dab1/Glrb/Enpp1/Kcnma1/Hipk2/Arrb2/Cacna1a/Cacnb4 | 11 |
| GO:0043616 | keratinocyte proliferation | BP | 0.234 | 1.07e-05 | 0.000134 | 3.87 | 8.65e-05 | Mdk/Cd109/Fst/Prkd1/Cdh13/Has2/Stxbp4/Klf9/Twist2/Ptch1/Bcl11b | 11 |
| GO:0050974 | detection of mechanical stimulus involved in sensory perception | BP | 0.234 | 1.07e-05 | 0.000134 | 3.87 | 8.65e-05 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12/Kit/Htr2a/Fyn/Piezo2/Myc | 11 |
| GO:0043534 | blood vessel endothelial cell migration | BP | 0.155 | 1.07e-05 | 0.000134 | 3.87 | 8.65e-05 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Prkd1/Vash1/Cdh5/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Akt3/Atp2b4 | 18 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | BP | 0.129 | 1.08e-05 | 0.000134 | 3.87 | 8.67e-05 | Cd109/Zeb2/Npnt/Fermt2/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Ltbp3/Hipk2/Smad9/Nrros/Arrb2/Zfyve9/Cldn5/Il17rd/Lats2/Cav1/Itga8/Nrep/Eng | 24 |
| GO:0010594 | regulation of endothelial cell migration | BP | 0.132 | 1.09e-05 | 0.000136 | 3.87 | 8.79e-05 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Prkd1/Stc1/Tek/Vash1/Ptprm/Plpp3/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Akt3/Atp2b4/Adgra2 | 23 |
| GO:0060999 | positive regulation of dendritic spine development | BP | 0.2 | 1.1e-05 | 0.000136 | 3.87 | 8.8e-05 | Nlgn1/D16Ertd472e/Nlgn3/Apoe/Caprin2/Bhlhb9/Cux2/Palm/Dlg5/Psen1/Zmynd8/Camk1/Nlgn2 | 13 |
| GO:0045879 | negative regulation of smoothened signaling pathway | BP | 0.256 | 1.13e-05 | 0.000139 | 3.86 | 9.02e-05 | Gli1/Hhip/Kif7/Gpc3/Serpine2/Gli3/Gpr161/Enpp1/Ptch1/Glis2 | 10 |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | BP | 0.214 | 1.13e-05 | 0.000139 | 3.86 | 9.03e-05 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Jcad/Vegfc/Igf2/Hdac9/Akt3 | 12 |
| GO:0048864 | stem cell development | BP | 0.168 | 1.13e-05 | 0.000139 | 3.86 | 9.03e-05 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Ednrb/Edn3/Ednra/Sema3g/Kitl/Sema6a/Bmp7/Hand2/Twist1 | 16 |
| GO:0051588 | regulation of neurotransmitter transport | BP | 0.144 | 1.14e-05 | 0.000139 | 3.86 | 9.03e-05 | Nlgn1/Adra1a/Rims3/Stx1b/P2rx2/Drd4/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Kcnc4/Htr2a/Ncs1/Slc4a8/Nrn1/Cacna1a/Cacnb4/Kcnh1 | 20 |
| GO:0055123 | digestive system development | BP | 0.144 | 1.14e-05 | 0.000139 | 3.86 | 9.03e-05 | Stra6/Ihh/Sfrp2/Clmp/Igf1/Pkdcc/Tbx2/Ednrb/Foxf1/Tcf21/Gli3/Prdm1/Kit/Cdkn1c/Igf2/Pcsk5/Dchs1/Fat4/Stx2/Dact1 | 20 |
| GO:0097091 | synaptic vesicle clustering | BP | 0.389 | 1.16e-05 | 0.000142 | 3.85 | 9.2e-05 | Syn2/Nlgn1/Cdh2/Nlgn3/Magi2/Pcdh17/Nlgn2 | 7 |
| GO:0042129 | regulation of T cell proliferation | BP | 0.128 | 1.18e-05 | 0.000144 | 3.84 | 9.31e-05 | Ihh/Igf1/Cd44/Mapk8ip1/Tnfsf13b/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Pde5a/H2-Aa/Dlg5/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 24 |
| GO:0014909 | smooth muscle cell migration | BP | 0.16 | 1.2e-05 | 0.000145 | 3.84 | 9.39e-05 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Myo5a/Mef2c/Has2/Adamts1/Postn/Enpp1/Nox4/Pcsk5/Pdgfrb/Grb10/Myc | 17 |
| GO:0048659 | smooth muscle cell proliferation | BP | 0.128 | 1.29e-05 | 0.000156 | 3.81 | 0.000101 | Vipr2/Igf1/Ddr2/Nos3/Npr3/Drd4/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Enpp1/Apoe/Rgs5/Ednra/Stat5b/Irak4/Calcrl/Efemp2/Apln/Pdgfrb/Cav1/Myc | 24 |
| GO:0007044 | cell-substrate junction assembly | BP | 0.167 | 1.3e-05 | 0.000157 | 3.81 | 0.000101 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Fn1/Thsd1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Itgb4/Enpp2 | 16 |
| GO:0061572 | actin filament bundle organization | BP | 0.131 | 1.32e-05 | 0.000159 | 3.8 | 0.000103 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Aif1l/Pfn2/Ppm1f/Limch1/Arhgap6/Nox4/Arhgef15/Tmeff2/Ppm1e/Arrb1/Dlc1/Arhgap28/Ccn2/Dpysl3/Shroom1/Plek | 23 |
| GO:0001941 | postsynaptic membrane organization | BP | 0.229 | 1.33e-05 | 0.000159 | 3.8 | 0.000103 | Nlgn1/Cdh2/Glrb/Nlgn3/Apoe/Magi2/Dlg4/Ptn/Zmynd8/Lrp4/Nlgn2 | 11 |
| GO:0003179 | heart valve morphogenesis | BP | 0.229 | 1.33e-05 | 0.000159 | 3.8 | 0.000103 | Stra6/Nos3/Mef2c/Erg/Emilin1/Tie1/Heyl/Dchs1/Hey2/Slit3/Twist1 | 11 |
| GO:0003382 | epithelial cell morphogenesis | BP | 0.229 | 1.33e-05 | 0.000159 | 3.8 | 0.000103 | Ihh/Ar/Stc1/Palld/Rilpl1/Pecam1/Col15a1/Heg1/Arhgef26/Col18a1/Bcl11b | 11 |
| GO:0046632 | alpha-beta T cell differentiation | BP | 0.147 | 1.33e-05 | 0.000159 | 3.8 | 0.000103 | Ihh/Lef1/Cd83/Itk/Gli3/Loxl3/Prdm1/Cd1d1/Cracr2a/Nckap1l/Satb1/Gimap5/Zap70/Gimap3/Il2rg/Batf/Spn/Ly9/Bcl11b | 19 |
| GO:0019229 | regulation of vasoconstriction | BP | 0.174 | 1.37e-05 | 0.000162 | 3.79 | 0.000105 | Cacna1c/Adra1a/Mmp2/Icam1/Wdr35/Edn3/Cd38/Agtr1a/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1/F2r | 15 |
| GO:0098703 | calcium ion import across plasma membrane | BP | 0.281 | 1.37e-05 | 0.000162 | 3.79 | 0.000105 | Cacna1c/Slc8a1/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Fyn/Cacna1a | 9 |
| GO:0061383 | trabecula morphogenesis | BP | 0.211 | 1.37e-05 | 0.000162 | 3.79 | 0.000105 | Plxnb1/Nog/Thbs3/Mmp2/Tek/Adamts1/Ccm2l/Enpp1/Heg1/Bmp7/Hey2/Eng | 12 |
| GO:0030098 | lymphocyte differentiation | BP | 0.0962 | 1.37e-05 | 0.000162 | 3.79 | 0.000105 | Ihh/Mdk/Lef1/Cd83/Itm2a/Cd44/Il11ra1/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Lfng/Tnfsf13b/Loxl3/Dock10/Zeb1/Prdm1/Jag2/Cd1d1/Kit/H2-Ab1/Enpp1/Cracr2a/Sox12/Irf8/Nckap1l/Satb1/Hdac9/Stat5b/H2-Aa/Gimap5/Mfng/Zap70/Gimap3/Il2rg/Batf/Spn/Ly9/Il7r/Cd4/Bcl11b | 43 |
| GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | BP | 0.119 | 1.43e-05 | 0.000168 | 3.77 | 0.000109 | Vipr2/Adra1a/Adgrl4/Npr3/Drd4/Gnai1/Adcy3/Cxcr3/Akap12/Ptgfr/Adgrl1/Calca/Htr1b/Pth1r/Adcy4/Adcy7/Gpr161/Ednra/Adcyap1r1/Calcrl/Gnaz/Ptger3/Palm/Atp2b4/Gabbr1/S1pr3/Ptger2 | 27 |
| GO:0048265 | response to pain | BP | 0.25 | 1.44e-05 | 0.000169 | 3.77 | 0.00011 | Grik1/Slc6a2/P2rx2/Ednrb/Grin2b/Cacna1e/Calca/Kcnip3/Vwa1/Cacna1a | 10 |
| GO:0002695 | negative regulation of leukocyte activation | BP | 0.124 | 1.48e-05 | 0.000173 | 3.76 | 0.000112 | Ihh/Mdk/Cd44/Foxf1/Gli3/Flt3/Loxl3/Prdm1/H2-Ab1/Btla/Havcr2/Cd200/Nckap1l/Pde5a/Cd84/Fgr/H2-Aa/Dlg5/Gimap5/Gimap3/Clnk/Nr1h3/Spn/Tbc1d10c/Lax1 | 25 |
| GO:0045666 | positive regulation of neuron differentiation | BP | 0.151 | 1.54e-05 | 0.00018 | 3.75 | 0.000116 | Fez1/Rgs6/Dab1/Mef2c/Cxcl12/Gprc5b/Trpc6/Gdf7/Tgif2/Zeb1/Mmd/Tcf4/Hoxd3/Heyl/Zc4h2/Nin/Bend6/Bmp7 | 18 |
| GO:0060272 | embryonic skeletal joint morphogenesis | BP | 0.462 | 1.56e-05 | 0.000182 | 3.74 | 0.000118 | Hoxd11/Nog/Hoxa11/Osr2/Osr1/Bmp7 | 6 |
| GO:0097090 | presynaptic membrane organization | BP | 0.462 | 1.56e-05 | 0.000182 | 3.74 | 0.000118 | Ptprd/Nlgn1/Nlgn3/Apoe/Lrp4/Nlgn2 | 6 |
| GO:0048167 | regulation of synaptic plasticity | BP | 0.101 | 1.61e-05 | 0.000187 | 3.73 | 0.000121 | Grik1/Kdr/Jph4/Nog/Grid2ip/Nlgn1/Rims3/Mef2c/Grin2b/Serpine2/Shank2/Eif2ak4/Slc8a3/Tshz3/Kit/Mme/Grik2/Nlgn3/Enpp1/Nrgn/Sorcs2/Cd38/Apoe/Prrt1/Syt7/Prrt2/Nsg1/Nfatc4/Cyp46a1/Mpp2/Erc2/Dlg4/Syngap1/Brsk1/Psen1/Ptn/Cpeb1/Prkar1b | 38 |
| GO:0045761 | regulation of adenylate cyclase activity | BP | 0.224 | 1.64e-05 | 0.00019 | 3.72 | 0.000123 | Cacna1c/Vipr2/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Palm/Gng7/Gabbr1 | 11 |
| GO:0051496 | positive regulation of stress fiber assembly | BP | 0.207 | 1.66e-05 | 0.000191 | 3.72 | 0.000124 | Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Ccn2 | 12 |
| GO:0032535 | regulation of cellular component size | BP | 0.0992 | 1.69e-05 | 0.000194 | 3.71 | 0.000126 | Kank4/Map2/Sema5a/Evl/Sema5b/Slc12a8/Nrp1/Hcls1/Sema7a/Tmod2/Fn1/Pfn2/Icam1/Cxcl12/Coro1a/Esam/Sh3bp1/Apoe/Kcnma1/Sema3g/Daam2/Pecam1/Scin/Bin1/Nckap1l/Epha7/Slc12a4/Arhgap28/Aqp11/Akt3/Cdc42ep5/Sema6a/Ulk2/Plekhh2/Cyfip2/C1qtnf9/Il7r/Plek/Draxin | 39 |
| GO:0051648 | vesicle localization | BP | 0.126 | 1.69e-05 | 0.000194 | 3.71 | 0.000126 | Syn2/Map2/Itga4/Nlgn1/Sec16b/Cdh2/Lrrk2/Myo5a/Ppfia2/Kif1a/Lin7a/Nlgn3/Magi2/5730455P16Rik/Pcdh17/Myo1f/Cep19/Arfgap3/Psen1/Rab27a/Map4k2/Pdcd6ip/Kif5a/Nlgn2 | 24 |
| GO:0030148 | sphingolipid biosynthetic process | BP | 0.163 | 1.7e-05 | 0.000195 | 3.71 | 0.000126 | St8sia2/St8sia4/Agk/Elovl4/Cerkl/Sptlc3/Cers4/St3gal2/St6galnac4/Degs2/St8sia6/Abca8b/St6galnac6/B3gnt5/Smpd1/Hacd1 | 16 |
| GO:0070252 | actin-mediated cell contraction | BP | 0.163 | 1.7e-05 | 0.000195 | 3.71 | 0.000126 | Cacna1c/Sgcd/Kcnd3/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Limch1/Bin1/Epdr1/Cacna1h/Ptger3/Atp1a2/Sri/Cav1 | 16 |
| GO:0043547 | positive regulation of GTPase activity | BP | 0.113 | 1.79e-05 | 0.000204 | 3.69 | 0.000132 | Wnt11/Plxnb1/Rgs6/Fermt2/Rgs16/Rasgrp3/Garnl3/Arhgap45/Icam1/Arhgap25/Rasgrp2/Myo9a/Thy1/Dock10/Sh3bp1/Arhgap6/Arhgef15/Bin1/Arap3/Xcl1/Arhgef26/Net1/Gnb5/Ngef/Mapre2/Iqsec3/F2r/Sipa1/Tbc1d10c | 29 |
| GO:0150117 | positive regulation of cell-substrate junction organization | BP | 0.273 | 1.8e-05 | 0.000205 | 3.69 | 0.000133 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Mapre2/Enpp2 | 9 |
| GO:1902656 | calcium ion import into cytosol | BP | 0.273 | 1.8e-05 | 0.000205 | 3.69 | 0.000133 | Cacna1c/Slc8a1/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Fyn/Cacna1a | 9 |
| GO:0036293 | response to decreased oxygen levels | BP | 0.115 | 1.81e-05 | 0.000205 | 3.69 | 0.000133 | Kcnk3/Kcnd2/Kdr/Ryr1/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Grin2b/Tek/Slc8a3/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Pld2/Hif3a/Myc/Twist1/Eng | 28 |
| GO:2000463 | positive regulation of excitatory postsynaptic potential | BP | 0.244 | 1.82e-05 | 0.000206 | 3.69 | 0.000134 | Nlgn1/Stx1b/Drd4/Grin2b/Tbc1d24/Nlgn3/Dlg4/Cux2/Prkar1b/Nlgn2 | 10 |
| GO:0032233 | positive regulation of actin filament bundle assembly | BP | 0.191 | 1.83e-05 | 0.000207 | 3.68 | 0.000134 | Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Ccn2/Plek | 13 |
| GO:0006665 | sphingolipid metabolic process | BP | 0.139 | 1.94e-05 | 0.000219 | 3.66 | 0.000142 | St8sia2/St8sia4/Agk/Elovl4/Cerkl/Sptlc3/Cers4/St3gal2/Plpp3/St6galnac4/Kit/Degs2/St8sia6/Abca8b/Plpp1/St6galnac6/B3gnt5/Smpd1/Hacd1/Enpp2 | 20 |
| GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | BP | 0.149 | 1.94e-05 | 0.000219 | 3.66 | 0.000142 | Cd109/Zeb2/Npnt/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Hipk2/Nrros/Zfyve9/Il17rd/Lats2/Cav1/Itga8/Nrep/Eng | 18 |
| GO:0016525 | negative regulation of angiogenesis | BP | 0.155 | 1.97e-05 | 0.000221 | 3.66 | 0.000143 | Hhip/Cxcr3/Tek/Adamts1/Vash1/Ptprm/Minar1/Emilin1/Ecscr/Tcf4/Tie1/Cldn5/Atp2b4/Sema6a/Ptn/Stab1/Fbln5 | 17 |
| GO:0034446 | substrate adhesion-dependent cell spreading | BP | 0.155 | 1.97e-05 | 0.000221 | 3.66 | 0.000143 | Mdk/Itga4/Nrp1/Lamc3/Fermt2/Itgb7/Fn1/Tek/Has2/Postn/Tmeff2/Parvg/Parvb/Net1/Ntng1/Itga8/Enpp2 | 17 |
| GO:0031279 | regulation of cyclase activity | BP | 0.203 | 1.99e-05 | 0.000223 | 3.65 | 0.000144 | Cacna1c/Vipr2/Nos3/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Palm/Gng7/Gabbr1 | 12 |
| GO:0034332 | adherens junction organization | BP | 0.22 | 2.01e-05 | 0.000224 | 3.65 | 0.000145 | Fermt2/Cdh6/Jam3/Cdh10/Cdh5/Abi2/Cdh11/Dchs1/Dlg5/Hipk1/Numbl | 11 |
| GO:0090659 | walking behavior | BP | 0.22 | 2.01e-05 | 0.000224 | 3.65 | 0.000145 | Efnb3/Cacna1c/Scn1a/Dab1/Glrb/Enpp1/Kcnma1/Hipk2/Arrb2/Cacna1a/Cacnb4 | 11 |
| GO:0060041 | retina development in camera-type eye | BP | 0.127 | 2.09e-05 | 0.000233 | 3.63 | 0.000151 | Ndp/Ihh/Cyp1b1/Sdk2/Tub/Celf4/Nrp1/Lamc3/Arl6/Obsl1/Ptprm/Thy1/Tgif2/Rp1/Arhgef15/Lpcat1/Hipk2/Rorb/Nphp1/Fat3/Pdgfrb/Ptn/Hipk1 | 23 |
| GO:0038084 | vascular endothelial growth factor signaling pathway | BP | 0.238 | 2.29e-05 | 0.000254 | 3.59 | 0.000165 | Kdr/Nrp1/Ptp4a3/Prkd1/Jcad/Vegfc/Tcf4/Adgra2/Sema6a/Pdgfrb | 10 |
| GO:0048799 | animal organ maturation | BP | 0.265 | 2.35e-05 | 0.00026 | 3.58 | 0.000169 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:0002027 | regulation of heart rate | BP | 0.152 | 2.51e-05 | 0.000276 | 3.56 | 0.000179 | Cacna1c/Kcnd3/Adra1a/Slc8a1/Scn4b/Cacna2d1/Ednrb/Cacna1e/Ank2/Edn3/Calca/Ednra/Bin1/Apln/Sri/Cav1/Hey2 | 17 |
| GO:2000181 | negative regulation of blood vessel morphogenesis | BP | 0.152 | 2.51e-05 | 0.000276 | 3.56 | 0.000179 | Hhip/Cxcr3/Tek/Adamts1/Vash1/Ptprm/Minar1/Emilin1/Ecscr/Tcf4/Tie1/Cldn5/Atp2b4/Sema6a/Ptn/Stab1/Fbln5 | 17 |
| GO:0086003 | cardiac muscle cell contraction | BP | 0.186 | 2.54e-05 | 0.000278 | 3.56 | 0.00018 | Cacna1c/Sgcd/Kcnd3/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Bin1/Cacna1h/Atp1a2/Sri/Cav1 | 13 |
| GO:0002031 | G protein-coupled receptor internalization | BP | 0.35 | 2.6e-05 | 0.000285 | 3.55 | 0.000185 | Calca/Htr1b/Arrb1/Arrb2/Ptger3/Apln/Pld2 | 7 |
| GO:0002064 | epithelial cell development | BP | 0.115 | 2.67e-05 | 0.000291 | 3.54 | 0.000189 | Ihh/Ar/Cdh2/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Msn/Prdm1/Abi2/Palld/Ppp1r16b/Robo4/Rilpl1/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Abcb1a/Akap11/Col18a1/Cldn1/Bcl11b/S1pr3 | 27 |
| GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | BP | 0.145 | 2.72e-05 | 0.000297 | 3.53 | 0.000192 | Cd109/Zeb2/Npnt/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Hipk2/Nrros/Zfyve9/Il17rd/Lats2/Cav1/Itga8/Nrep/Eng | 18 |
| GO:0035023 | regulation of Rho protein signal transduction | BP | 0.165 | 2.75e-05 | 0.000299 | 3.52 | 0.000194 | Adra1a/Nrp1/Cdh2/Lpar4/Arhgef25/Eps8l1/Apoe/Rasip1/Arap3/Arrb1/Heg1/Dlc1/Net1/Pdgfrb/F2r | 15 |
| GO:0048638 | regulation of developmental growth | BP | 0.097 | 2.81e-05 | 0.000305 | 3.52 | 0.000198 | Gli1/Map2/Sema5a/Ar/Cacna1c/Col14a1/H19/Nog/Igf1/Tbx2/Sema5b/Cpne5/Nrp1/Nkd1/Acacb/Sema7a/Fn1/Mef2c/Cxcl12/Hopx/Flvcr1/Gamt/Plcb1/Flt3/Apoe/Sema3g/Igf2/Slc23a2/Foxs1/Epha7/Cacng7/Ptch1/Stat5b/Pi16/Lats2/Sema6a/Ulk2/Hey2/Draxin | 39 |
| GO:1901343 | negative regulation of vasculature development | BP | 0.15 | 2.82e-05 | 0.000306 | 3.51 | 0.000198 | Hhip/Cxcr3/Tek/Adamts1/Vash1/Ptprm/Minar1/Emilin1/Ecscr/Tcf4/Tie1/Cldn5/Atp2b4/Sema6a/Ptn/Stab1/Fbln5 | 17 |
| GO:0010874 | regulation of cholesterol efflux | BP | 0.233 | 2.86e-05 | 0.000308 | 3.51 | 2e-04 | Pla2g10/Pltp/Apoe/Abca5/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3/Abcg4 | 10 |
| GO:0085029 | extracellular matrix assembly | BP | 0.233 | 2.86e-05 | 0.000308 | 3.51 | 2e-04 | Ihh/Phldb2/Fkbp10/Has2/Emilin1/Tie1/Ltbp3/Efemp2/Ntng1/Fbln5 | 10 |
| GO:0014706 | striated muscle tissue development | BP | 0.107 | 2.94e-05 | 0.000316 | 3.5 | 0.000205 | Gli1/Col11a1/AW551984/Angpt1/Lef1/Sgcd/Col14a1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/Mef2c/Lrp2/Ccm2l/Nox4/Tbx3/Gjc1/Ednra/Myl3/Hdac9/Heg1/Arrb2/Pi16/Sgcb/Pdgfrb/Bmp7/Hey2/Hand2/Twist1/Eng | 31 |
| GO:0061005 | cell differentiation involved in kidney development | BP | 0.212 | 2.98e-05 | 0.00032 | 3.49 | 0.000208 | Ptpro/Ednrb/Mef2c/Tcf21/Gli3/Tshz3/Ednra/Ptch1/Osr1/Fat4/Glis2 | 11 |
| GO:1904738 | vascular associated smooth muscle cell migration | BP | 0.257 | 3.03e-05 | 0.000325 | 3.49 | 0.000211 | Ddr2/Drd4/Tert/Myo5a/Mef2c/Adamts1/Enpp1/Pcsk5/Grb10 | 9 |
| GO:0021915 | neural tube development | BP | 0.121 | 3.07e-05 | 0.000329 | 3.48 | 0.000213 | Sfrp2/Nog/Zeb2/Dzip1l/Sall2/Gli3/Ift57/Gdf7/Lrp2/Cecr2/Gpr161/Phgdh/Prickle1/Ipmk/Dlc1/Ptch1/Dchs1/Ttbk2/Rab23/Fzd2/Psen1/Dact1/Bmp7/Twist1 | 24 |
| GO:0050905 | neuromuscular process | BP | 0.121 | 3.07e-05 | 0.000329 | 3.48 | 0.000213 | Ankfn1/Stra6/Slc1a3/Hoxd10/Jph4/Aph1b/Scn1a/Myo5a/Grin2b/Slc8a3/Tshz3/Glrb/Nlgn3/Npas3/Prrt2/Kcnma1/Ednra/Hipk2/Foxs1/Stac2/Dlg4/Cacna1a/Kcnh1/Nlgn2 | 24 |
| GO:0006643 | membrane lipid metabolic process | BP | 0.124 | 3.25e-05 | 0.000346 | 3.46 | 0.000224 | Cyp1b1/St8sia2/St8sia4/Agk/Elovl4/Cerkl/Sptlc3/Cers4/St3gal2/Plpp3/St6galnac4/Kit/Degs2/St8sia6/Sccpdh/Abca8b/Plpp1/St6galnac6/B3gnt5/St3gal4/Smpd1/Hacd1/Enpp2 | 23 |
| GO:0001736 | establishment of planar polarity | BP | 0.194 | 3.36e-05 | 0.000357 | 3.45 | 0.000231 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Cplane1/Dchs1/Nphp1/Fzd2/Brsk1/Fat4/Dact1 | 12 |
| GO:0051339 | regulation of lyase activity | BP | 0.194 | 3.36e-05 | 0.000357 | 3.45 | 0.000231 | Cacna1c/Vipr2/Nos3/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Palm/Gng7/Gabbr1 | 12 |
| GO:1903034 | regulation of response to wounding | BP | 0.126 | 3.39e-05 | 0.00036 | 3.44 | 0.000233 | Mdk/Cd109/Ddr2/Phldb2/Tfpi/Abat/Fermt2/Serpine2/Plpp3/Apoe/Pros1/Rtn4rl1/Neo1/Clec7a/Ptger3/Xylt1/St3gal4/Ptn/Cav1/Cldn1/F2r/Wfdc1 | 22 |
| GO:0002062 | chondrocyte differentiation | BP | 0.148 | 3.56e-05 | 0.000375 | 3.43 | 0.000243 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Pkdcc/Hoxa11/Mef2c/Osr2/Gli3/Pth1r/Wnt9a/Adamts7/Ltbp3/Osr1/Ccn2/Mboat2 | 17 |
| GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | BP | 0.148 | 3.56e-05 | 0.000375 | 3.43 | 0.000243 | Nlgn1/Ccdc88a/Nrp1/Abi2/Kit/Kctd17/Rp1/Eps8l1/Crocc/Ttbk2/Dzip1/Palm/Dpysl3/Cdc42ep5/Def8/Atmin/Zmynd8 | 17 |
| GO:1904036 | negative regulation of epithelial cell apoptotic process | BP | 0.208 | 3.6e-05 | 0.000379 | 3.42 | 0.000245 | Sema5a/Angpt1/Mdk/Kdr/Igf1/Tert/Icam1/Tek/Cdh5/Ndnf/Angptl4 | 11 |
| GO:0033622 | integrin activation | BP | 0.286 | 3.66e-05 | 0.000383 | 3.42 | 0.000248 | Fermt2/Col16a1/Fn1/Jam3/Cxcl12/Rasip1/Pcsk5/Plek | 8 |
| GO:0051894 | positive regulation of focal adhesion assembly | BP | 0.286 | 3.66e-05 | 0.000383 | 3.42 | 0.000248 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Enpp2 | 8 |
| GO:0061437 | renal system vasculature development | BP | 0.286 | 3.66e-05 | 0.000383 | 3.42 | 0.000248 | Angpt1/Nrp1/Tek/Tcf21/Ednra/Osr1/Pdgfrb/Bmp7 | 8 |
| GO:0061440 | kidney vasculature development | BP | 0.286 | 3.66e-05 | 0.000383 | 3.42 | 0.000248 | Angpt1/Nrp1/Tek/Tcf21/Ednra/Osr1/Pdgfrb/Bmp7 | 8 |
| GO:0046629 | gamma-delta T cell activation | BP | 0.333 | 3.74e-05 | 0.000391 | 3.41 | 0.000253 | Lef1/Itk/Jag2/Nckap1l/Stat5b/Gimap5/Gimap3 | 7 |
| GO:0003073 | regulation of systemic arterial blood pressure | BP | 0.142 | 3.77e-05 | 0.000393 | 3.41 | 0.000255 | Ar/Kdr/Ddah1/Adra1a/Nos3/Ptpro/P2rx2/Adamts16/Ednrb/Enpep/Edn3/Calca/Rps6ka2/Postn/Agtr1a/Apln/F2r/Eng | 18 |
| GO:0098657 | import into cell | BP | 0.112 | 3.87e-05 | 0.000402 | 3.4 | 0.000261 | Slc1a3/Kcnj3/Cacna1c/Acsl6/Itga4/Slc6a2/Kcnd3/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Slc7a2/Cacna1e/Slc8a3/Lrp2/Nalf1/Slc29a1/Agtr1a/Slc9a5/Slc12a4/Fyn/Acsl1/Atp1a2/Steap4/Psen1/Cacna1a/Lrrc8c | 27 |
| GO:0007164 | establishment of tissue polarity | BP | 0.19 | 3.97e-05 | 0.000412 | 3.39 | 0.000267 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Cplane1/Dchs1/Nphp1/Fzd2/Brsk1/Fat4/Dact1 | 12 |
| GO:0060997 | dendritic spine morphogenesis | BP | 0.178 | 4.04e-05 | 0.000418 | 3.38 | 0.000271 | Nlgn1/Lrrk2/Ppfia2/Dock10/Abi2/Kif1a/Nlgn3/Caprin2/Bhlhb9/Dlg4/Cux2/Ngef/Zfp365 | 13 |
| GO:1902105 | regulation of leukocyte differentiation | BP | 0.102 | 4.11e-05 | 0.000424 | 3.37 | 0.000275 | Ihh/Mdk/Lef1/Cd83/Hcls1/Cd44/Inpp4b/Pik3r6/Lrrc17/Gli3/Calca/Flt3/Loxl3/Zeb1/Prdm1/Cd1d1/Tnfrsf11b/Sox12/Zbtb46/Nckap1l/Kitl/Tal1/Tnfaip6/Rassf2/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r/Cd4/Myc | 33 |
| GO:0006820 | anion transport | BP | 0.0904 | 4.12e-05 | 0.000424 | 3.37 | 0.000275 | Slc1a3/Car4/Grik1/Pla2g10/Slc13a5/Slc2a3/Tcaf1/Slc12a8/Abat/Drd4/Ano1/Slc7a2/Grin2b/Abcg2/Stc1/Slc36a4/Ano2/Ttyh2/Lrp2/Htr1b/Slc6a15/Glrb/Gabra4/Enpp1/Slc23a2/Bdkrb2/Sfxn5/Slc12a4/Slc38a1/Nfkbie/Slco4c1/Slc4a3/Ptger3/Clca2/Slc4a8/Psen1/Abcb1a/Abcc5/Gabbr1/Abcg3/Cacna1a/Lrrc8c/Cacnb4/Ank/Myc | 45 |
| GO:0034763 | negative regulation of transmembrane transport | BP | 0.136 | 4.29e-05 | 0.000442 | 3.35 | 0.000286 | Hecw2/Drd4/Sh2b2/Enpp1/Dysf/Bin1/Osr1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Cav1/Pid1/Grb10/Myc/Twist1 | 19 |
| GO:0070663 | regulation of leukocyte proliferation | BP | 0.108 | 4.48e-05 | 0.000461 | 3.34 | 0.000298 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Kitl/Xcl1/Stat5b/Pde5a/Csf2rb/H2-Aa/Dlg5/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 29 |
| GO:0045582 | positive regulation of T cell differentiation | BP | 0.151 | 4.61e-05 | 0.000472 | 3.33 | 0.000306 | Ihh/Mdk/Lef1/Cd83/Pik3r6/Gli3/Cd1d1/Sox12/Nckap1l/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r | 16 |
| GO:0014910 | regulation of smooth muscle cell migration | BP | 0.158 | 4.63e-05 | 0.000472 | 3.33 | 0.000306 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Myo5a/Mef2c/Has2/Adamts1/Postn/Nox4/Pcsk5/Pdgfrb/Myc | 15 |
| GO:0019226 | transmission of nerve impulse | BP | 0.158 | 4.63e-05 | 0.000472 | 3.33 | 0.000306 | Kcnd2/P2rx2/Scn1a/Jam3/Cacna1e/Scn2a/Grik2/Nfasc/Kcnma1/Kcnc4/Cacng7/Cacna1h/Scn3a/Cacna1a/Cacnb4 | 15 |
| GO:0001952 | regulation of cell-matrix adhesion | BP | 0.14 | 4.66e-05 | 0.000475 | 3.32 | 0.000308 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Cdh13/Thy1/Postn/Ppm1f/Epha3/Limch1/Arhgap6/Vegfc/Pcsk5/Dlc1/Efemp2/Enpp2 | 18 |
| GO:0050901 | leukocyte tethering or rolling | BP | 0.243 | 4.89e-05 | 0.000496 | 3.3 | 0.000322 | Itga4/Chst2/Itgb7/Podxl2/Cxcl12/Sele/St3gal4/Spn/Jam2 | 9 |
| GO:0050920 | regulation of chemotaxis | BP | 0.113 | 4.92e-05 | 0.000498 | 3.3 | 0.000323 | Sema5a/Mdk/Kdr/Sema5b/Nrp1/Sema7a/Fn1/Prkd1/Jam3/Cxcr3/Cdh13/Cxcl12/Edn3/Ppm1f/Vegfc/Ednra/Sema3g/Dysf/Nckap1l/Xcl1/Tnfaip6/Mpp1/Adgra2/Sema6a/Pdgfrb/Ptn | 26 |
| GO:0098739 | import across plasma membrane | BP | 0.12 | 4.94e-05 | 0.000499 | 3.3 | 0.000324 | Slc1a3/Kcnj3/Cacna1c/Acsl6/Kcnd3/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Slc7a2/Cacna1e/Slc8a3/Lrp2/Nalf1/Agtr1a/Slc9a5/Slc12a4/Fyn/Acsl1/Atp1a2/Psen1/Cacna1a/Lrrc8c | 23 |
| GO:0030308 | negative regulation of cell growth | BP | 0.118 | 5e-05 | 0.000503 | 3.3 | 0.000326 | Tro/Sfrp2/Map2/Sema5a/Wnt11/Sema5b/Nrp1/Sema7a/Serpine2/Cth/Minar1/Enpp1/Sema3g/Caprin2/Epha7/P3h1/Fhl1/Pi16/Rrad/Tspyl2/Sema6a/Ulk2/Slit3/Draxin | 24 |
| GO:0030500 | regulation of bone mineralization | BP | 0.165 | 5.09e-05 | 0.000512 | 3.29 | 0.000332 | Ddr2/Pkdcc/Slc8a1/Mef2c/Osr2/Nell1/Mgp/Enpp1/Ltbp3/Osr1/Ptn/Bmp7/Ank/Twist1 | 14 |
| GO:0097105 | presynaptic membrane assembly | BP | 0.5 | 5.24e-05 | 0.000525 | 3.28 | 0.00034 | Ptprd/Nlgn1/Nlgn3/Lrp4/Nlgn2 | 5 |
| GO:0002366 | leukocyte activation involved in immune response | BP | 0.101 | 5.24e-05 | 0.000525 | 3.28 | 0.00034 | Mdk/Lef1/Itm2a/Foxf1/Icam1/Eif2ak4/Dock2/Lfng/Loxl3/Dock10/Coro1a/Kit/Itgal/Il27ra/Enpp1/Cracr2a/Havcr2/Irf8/Dysf/Nckap1l/Myo1f/Cd84/Fgr/Mfng/Psen1/Apbb1ip/Rab27a/Lat/Clnk/Pld2/Batf/Spn/Ly9 | 33 |
| GO:1903170 | negative regulation of calcium ion transmembrane transport | BP | 0.217 | 5.32e-05 | 0.000532 | 3.27 | 0.000345 | Drd4/Dysf/Bin1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri | 10 |
| GO:0042982 | amyloid precursor protein metabolic process | BP | 0.173 | 5.42e-05 | 0.00054 | 3.27 | 0.00035 | Itm2a/Igf1/Aph1b/Rtn1/Soat1/Slc2a13/Apoe/Bin1/Nsg1/Abcg1/Adam19/Psen1/Tmcc2 | 13 |
| GO:0097120 | receptor localization to synapse | BP | 0.173 | 5.42e-05 | 0.00054 | 3.27 | 0.00035 | Gpc6/Nlgn1/Lrrc7/Cep112/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4/Kif5a | 13 |
| GO:0001954 | positive regulation of cell-matrix adhesion | BP | 0.185 | 5.48e-05 | 0.000545 | 3.26 | 0.000353 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Cdh13/Thy1/Ppm1f/Vegfc/Pcsk5/Efemp2/Enpp2 | 12 |
| GO:0050670 | regulation of lymphocyte proliferation | BP | 0.11 | 5.54e-05 | 0.00055 | 3.26 | 0.000356 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Pde5a/H2-Aa/Dlg5/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 27 |
| GO:0048660 | regulation of smooth muscle cell proliferation | BP | 0.122 | 5.7e-05 | 0.000565 | 3.25 | 0.000366 | Vipr2/Igf1/Ddr2/Nos3/Npr3/Drd4/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Apoe/Rgs5/Stat5b/Irak4/Calcrl/Efemp2/Apln/Pdgfrb/Cav1/Myc | 22 |
| GO:0010001 | glial cell differentiation | BP | 0.108 | 5.73e-05 | 0.000566 | 3.25 | 0.000367 | Gap43/Ptprz1/Mdk/Lef1/Nog/Igf1/Lamc3/Cdh2/Wasf3/Dab1/Id4/Serpine2/Gli3/Slc8a3/Plpp3/Nlgn3/Enpp1/Phgdh/Daam2/Bin1/Tal1/Nrros/Psen1/Ptn/Tppp/Itgb4/Enpp2/Zfp365 | 28 |
| GO:0042102 | positive regulation of T cell proliferation | BP | 0.148 | 5.82e-05 | 0.000573 | 3.24 | 0.000372 | Igf1/Tnfsf13b/Coro1a/Cd1d1/Itgal/Il27ra/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 16 |
| GO:0048041 | focal adhesion assembly | BP | 0.163 | 5.82e-05 | 0.000573 | 3.24 | 0.000372 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Thsd1/Tek/Thy1/Ppm1f/Epha3/Limch1/Arhgap6/Dlc1/Enpp2 | 14 |
| GO:1901385 | regulation of voltage-gated calcium channel activity | BP | 0.237 | 6.14e-05 | 0.000604 | 3.22 | 0.000391 | Drd4/Cacna2d1/Dysf/Stac2/Rrad/Gnb5/Sri/Cacna1a/Cacnb4 | 9 |
| GO:0071709 | membrane assembly | BP | 0.196 | 6.15e-05 | 0.000604 | 3.22 | 0.000392 | Ptprd/Nlgn1/Cdh2/Ubxn2b/Nlgn3/Magi2/Stx2/Cav1/Tlcd2/Lrp4/Nlgn2 | 11 |
| GO:0010634 | positive regulation of epithelial cell migration | BP | 0.128 | 6.22e-05 | 0.000609 | 3.22 | 0.000395 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Tek/Has2/Plpp3/Ppm1f/Jcad/Vegfc/Igf2/Hdac9/Akt3/Adgra2/Mapre2/Enpp2 | 20 |
| GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | BP | 0.267 | 6.33e-05 | 0.000615 | 3.21 | 0.000398 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln/Pld2 | 8 |
| GO:0022401 | negative adaptation of signaling pathway | BP | 0.267 | 6.33e-05 | 0.000615 | 3.21 | 0.000398 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln/Pld2 | 8 |
| GO:0045932 | negative regulation of muscle contraction | BP | 0.267 | 6.33e-05 | 0.000615 | 3.21 | 0.000398 | Calca/Gucy1a1/Kcnma1/Bin1/Pde5a/Calcrl/Atp1a2/Sri | 8 |
| GO:0048596 | embryonic camera-type eye morphogenesis | BP | 0.267 | 6.33e-05 | 0.000615 | 3.21 | 0.000398 | Stra6/Ihh/Tbx2/Zeb1/Hipk2/Hipk1/Bmp7/Twist1 | 8 |
| GO:0048745 | smooth muscle tissue development | BP | 0.267 | 6.33e-05 | 0.000615 | 3.21 | 0.000398 | Stra6/Ihh/Tshz3/Osr1/Efemp2/Pdgfrb/Itga8/Eng | 8 |
| GO:0060048 | cardiac muscle contraction | BP | 0.136 | 6.34e-05 | 0.000615 | 3.21 | 0.000398 | Cacna1c/Sgcd/Kcnd3/Adra1a/Slc8a1/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Bin1/Myl3/Pde5a/Cacna1h/Ccn2/Atp1a2/Sri/Cav1 | 18 |
| GO:0070482 | response to oxygen levels | BP | 0.103 | 6.35e-05 | 0.000615 | 3.21 | 0.000398 | Kcnk3/Kcnd2/Kdr/Ryr1/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Gucy1a2/Grin2b/Tek/Slc8a3/Gucy1b1/Gucy1a1/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Pld2/Hif3a/Myc/Twist1/Eng | 31 |
| GO:0001574 | ganglioside biosynthetic process | BP | 0.375 | 6.44e-05 | 0.000622 | 3.21 | 0.000403 | St8sia2/St8sia4/St3gal2/St6galnac4/St8sia6/St6galnac6 | 6 |
| GO:0010595 | positive regulation of endothelial cell migration | BP | 0.147 | 6.52e-05 | 0.000628 | 3.2 | 0.000407 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Tek/Plpp3/Jcad/Vegfc/Igf2/Hdac9/Akt3/Adgra2 | 16 |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | BP | 0.147 | 6.52e-05 | 0.000628 | 3.2 | 0.000407 | Sfrp2/Slc8a1/Mapk8ip1/Slc8a3/Pkia/Cdkn1c/Apoe/Rasip1/Pkib/Heg1/Lats2/Smpd1/Cav1/Bmp7/Prkar1b/Lax1 | 16 |
| GO:0035418 | protein localization to synapse | BP | 0.161 | 6.63e-05 | 0.000637 | 3.2 | 0.000412 | Gpc6/Nlgn1/Stx1b/Nptx1/Slitrk3/Dlg2/Nlgn3/Nptxr/Magi2/Nsg1/Cacng7/Dlg4/Kif5a/Nlgn2 | 14 |
| GO:0060411 | cardiac septum morphogenesis | BP | 0.161 | 6.63e-05 | 0.000637 | 3.2 | 0.000412 | Wnt11/Nog/Tbx2/Nos3/Nrp1/Lrp2/Aplnr/Tbx3/Heyl/Fzd2/Bmp7/Hey2/Slit3/Eng | 14 |
| GO:0002263 | cell activation involved in immune response | BP | 0.0997 | 6.65e-05 | 0.000637 | 3.2 | 0.000413 | Mdk/Lef1/Itm2a/Foxf1/Icam1/Eif2ak4/Dock2/Lfng/Loxl3/Dock10/Coro1a/Kit/Itgal/Il27ra/Enpp1/Cracr2a/Havcr2/Irf8/Dysf/Nckap1l/Myo1f/Cd84/Fgr/Mfng/Psen1/Apbb1ip/Rab27a/Lat/Clnk/Pld2/Batf/Spn/Ly9 | 33 |
| GO:0022408 | negative regulation of cell-cell adhesion | BP | 0.115 | 6.82e-05 | 0.000652 | 3.19 | 0.000423 | Ihh/Mdk/Lef1/Abat/Cd44/Serpine2/Cxcl12/Gli3/Podxl/Loxl3/Ppm1f/Ccm2l/H2-Ab1/Btla/Havcr2/Nckap1l/Pde5a/Ptger3/H2-Aa/Dlg5/Gimap5/Gimap3/Spn/Lax1 | 24 |
| GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.14 | 6.87e-05 | 0.000656 | 3.18 | 0.000425 | Kdr/Zeb2/Rnf165/Npnt/Gpc3/Gdf11/Zfp423/Cdh5/Gdf7/Gdf10/Cdkn1c/Hipk2/Kcp/Neo1/Itga8/Bmp7/Eng | 17 |
| GO:0046467 | membrane lipid biosynthetic process | BP | 0.135 | 7.01e-05 | 0.000668 | 3.18 | 0.000433 | St8sia2/St8sia4/Agk/Elovl4/Cerkl/Sptlc3/Cers4/St3gal2/St6galnac4/Degs2/St8sia6/Sccpdh/Abca8b/St6galnac6/B3gnt5/St3gal4/Smpd1/Hacd1 | 18 |
| GO:0051250 | negative regulation of lymphocyte activation | BP | 0.124 | 7.13e-05 | 0.000678 | 3.17 | 0.00044 | Ihh/Mdk/Cd44/Gli3/Flt3/Loxl3/Prdm1/H2-Ab1/Btla/Havcr2/Nckap1l/Pde5a/Fgr/H2-Aa/Dlg5/Gimap5/Gimap3/Clnk/Spn/Tbc1d10c/Lax1 | 21 |
| GO:0046635 | positive regulation of alpha-beta T cell activation | BP | 0.169 | 7.19e-05 | 0.000683 | 3.17 | 0.000443 | Ihh/Cd83/Gli3/Cd1d1/H2-Ab1/Nckap1l/Xcl1/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg/Rasal3 | 13 |
| GO:0006022 | aminoglycan metabolic process | BP | 0.145 | 7.29e-05 | 0.00069 | 3.16 | 0.000447 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Dse/Habp4/Xylt1/Pdgfrb | 16 |
| GO:0006493 | protein O-linked glycosylation | BP | 0.193 | 7.3e-05 | 0.00069 | 3.16 | 0.000447 | Gxylt2/Tet1/Tmtc1/Galnt16/Eogt/St8sia6/Galntl6/Poglut2/Pomt2/Large1/Galnt18 | 11 |
| GO:0032964 | collagen biosynthetic process | BP | 0.193 | 7.3e-05 | 0.00069 | 3.16 | 0.000447 | Ihh/P3h3/Ddr2/Fn1/Emilin1/Rcn3/Arrb2/Ccn2/Pdgfrb/Cygb/F2r | 11 |
| GO:0032944 | regulation of mononuclear cell proliferation | BP | 0.108 | 7.32e-05 | 0.00069 | 3.16 | 0.000447 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Btla/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Pde5a/H2-Aa/Dlg5/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 27 |
| GO:1990874 | vascular associated smooth muscle cell proliferation | BP | 0.179 | 7.47e-05 | 0.000704 | 3.15 | 0.000456 | Igf1/Ddr2/Drd4/Tert/Mmp2/Mef2c/Adamts1/Htr1b/Enpp1/Rgs5/Efemp2/Apln | 12 |
| GO:0003281 | ventricular septum development | BP | 0.159 | 7.54e-05 | 0.000708 | 3.15 | 0.000459 | Stra6/Wnt11/Nog/Nos3/Lrp2/Prdm1/Cplane1/Aplnr/Tbx3/Heyl/Heg1/Fzd2/Hey2/Slit3 | 14 |
| GO:0006942 | regulation of striated muscle contraction | BP | 0.159 | 7.54e-05 | 0.000708 | 3.15 | 0.000459 | Cacna1c/Adra1a/Slc8a1/Stc1/Ank2/Slc8a3/Bin1/Myl3/Pde5a/Cacna1h/Ccn2/Atp1a2/Sri/Cav1 | 14 |
| GO:0045621 | positive regulation of lymphocyte differentiation | BP | 0.139 | 7.63e-05 | 0.000715 | 3.15 | 0.000463 | Ihh/Mdk/Lef1/Cd83/Pik3r6/Gli3/Prdm1/Cd1d1/Sox12/Nckap1l/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r | 17 |
| GO:0043583 | ear development | BP | 0.108 | 8.39e-05 | 0.000784 | 3.11 | 0.000508 | Col11a1/Stra6/Nog/Igf1/Tbx2/Lrig3/Osr2/Tshz1/Gli3/Zeb1/Jag2/Cecr2/Lin7a/Enpp1/Tsku/Tbx3/Kcnma1/Ednra/Dchs1/Osr1/Fzd2/Fat4/Kcnq4/Itga8/Ror1/Hey2/Myc | 27 |
| GO:0001508 | action potential | BP | 0.129 | 8.41e-05 | 0.000785 | 3.1 | 0.000509 | Cacna1c/Kcnd2/Kcnd3/Adra1a/Scn4b/P2rx2/Scn1a/Cacna2d1/Grin2b/Ank2/Scn2a/Grik2/Kcnma1/Kcnc4/Bin1/Cacna1h/Scn3a/Cav1/Cacnb4 | 19 |
| GO:0015849 | organic acid transport | BP | 0.0997 | 8.52e-05 | 0.000795 | 3.1 | 0.000515 | Slc1a3/Acsl6/Grik1/Pla2g10/Slc13a5/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Slc36a4/Rbp7/Lrp2/Htr1b/Slc6a15/Apoe/Slc23a2/Bdkrb2/Sfxn5/Slc38a1/Nfkbie/Abcd2/Acsl1/Slc7a7/Psen1/Abcb1a/Abcc5/Gabbr1/Cacna1a/Lrrc8c/Cacnb4/Myc | 32 |
| GO:0007517 | muscle organ development | BP | 0.0959 | 8.92e-05 | 0.00083 | 3.08 | 0.000538 | Col11a1/Stra6/Angpt1/Lef1/Hoxd10/Ryr1/Nog/P2rx2/Hoxd9/Mef2c/Id3/Tcf21/Lrp2/Ccm2l/Ednra/Heyl/Igf2/Dysf/Myl3/Hdac9/Heg1/Selenon/Fhl1/Large1/Fzd2/Efemp2/Sgcb/Hlf/Zfp689/Cav1/Usp2/Hey2/Myc/Twist1/Eng | 35 |
| GO:0035701 | hematopoietic stem cell migration | BP | 0.455 | 9.23e-05 | 0.000854 | 3.07 | 0.000554 | Jam3/Kit/Enpp1/Bcl11b/Jam2 | 5 |
| GO:0045588 | positive regulation of gamma-delta T cell differentiation | BP | 0.455 | 9.23e-05 | 0.000854 | 3.07 | 0.000554 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0046645 | positive regulation of gamma-delta T cell activation | BP | 0.455 | 9.23e-05 | 0.000854 | 3.07 | 0.000554 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0061351 | neural precursor cell proliferation | BP | 0.116 | 9.33e-05 | 0.000862 | 3.06 | 0.000559 | Gli1/Ptprz1/Acsl6/Hhip/Mdk/Lef1/Nog/Igf1/Zeb2/Cdh2/Lrrk2/Smarca1/Id4/Zfp423/Gli3/Lrp2/Kif1a/Eml1/Vegfc/Bbs1/Ptn/Nes/Numbl | 23 |
| GO:0008277 | regulation of G protein-coupled receptor signaling pathway | BP | 0.132 | 9.4e-05 | 0.000867 | 3.06 | 0.000562 | Tub/Tmod2/Slc24a4/Plcb1/Calca/Htr1b/Aplnr/Arrb1/Arrb2/Pde5a/Ptger3/Atp2b4/Apln/Pld2/Kctd12b/Gpsm1/Kctd12/Plek | 18 |
| GO:0009311 | oligosaccharide metabolic process | BP | 0.225 | 9.44e-05 | 0.00087 | 3.06 | 0.000564 | St6gal2/St8sia2/St8sia4/B3galnt1/St3gal2/St6galnac4/St8sia6/St6galnac6/St3gal4 | 9 |
| GO:0001886 | endothelial cell morphogenesis | BP | 0.353 | 9.55e-05 | 0.000877 | 3.06 | 0.000568 | Stc1/Pecam1/Col15a1/Heg1/Arhgef26/Col18a1 | 6 |
| GO:1905564 | positive regulation of vascular endothelial cell proliferation | BP | 0.353 | 9.55e-05 | 0.000877 | 3.06 | 0.000568 | Mdk/Itga4/Igf1/Igf2/Akt3/Apln | 6 |
| GO:0019915 | lipid storage | BP | 0.156 | 9.69e-05 | 0.000889 | 3.05 | 0.000576 | Apob/Pla2g10/Acacb/Vstm2a/Mest/Soat1/Enpp1/Apoe/Dysf/Abcg1/Stat5b/Cav1/Nr1h3/Cds1 | 14 |
| GO:0010842 | retina layer formation | BP | 0.292 | 9.82e-05 | 0.000898 | 3.05 | 0.000582 | Sdk2/Arl6/Obsl1/Ptprm/Hipk2/Fat3/Hipk1 | 7 |
| GO:0006941 | striated muscle contraction | BP | 0.121 | 9.99e-05 | 0.000913 | 3.04 | 0.000591 | Cacna1c/Sgcd/Kcnd3/Adra1a/Slc8a1/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Slc8a3/Bin1/Myl3/Stac2/Pde5a/Ryr3/Cacna1h/Ccn2/Atp1a2/Sri/Cav1 | 21 |
| GO:0048863 | stem cell differentiation | BP | 0.108 | 1e-04 | 0.000914 | 3.04 | 0.000592 | Sema5a/Zeb2/Sema5b/Nrp1/Sema7a/Cdh2/Fn1/Ednrb/Mef2c/Hoxd4/Edn3/Kit/Ednra/Sema3g/Prickle1/Kitl/Ltbp3/Tal1/Osr1/Pus7/Sema6a/Ptn/Batf/Bmp7/Hand2/Twist1 | 26 |
| GO:0006940 | regulation of smooth muscle contraction | BP | 0.174 | 0.000101 | 0.000914 | 3.04 | 0.000592 | Npnt/Adra1a/Abat/Calca/Gucy1a1/Kit/Kcnma1/Calcrl/Ptger3/Atp1a2/Cav1/F2r | 12 |
| GO:0045668 | negative regulation of osteoblast differentiation | BP | 0.174 | 0.000101 | 0.000914 | 3.04 | 0.000592 | Nog/Areg/Id3/Gdf10/Vegfc/Rorb/Tnfaip6/Twist2/Ptch1/Hand2/Twist1/Zfp932 | 12 |
| GO:0007612 | learning | BP | 0.118 | 0.000101 | 0.000917 | 3.04 | 0.000594 | Stra6/Cacna1c/Jph4/Nog/Drd4/Chst10/B4galt2/Grin2b/Pde1b/Adcy3/Shank2/Cacna1e/Eif2ak4/Slc8a3/Kit/Nlgn3/Ddhd2/Atp1a2/Syngap1/Brsk1/Ptn/Bche | 22 |
| GO:0030278 | regulation of ossification | BP | 0.131 | 0.000103 | 0.000937 | 3.03 | 0.000607 | Mdk/Ddr2/Pkdcc/Slc8a1/Mef2c/Osr2/Calca/Nell1/Mgp/Enpp1/Ltbp3/P3h1/Osr1/Ptn/Bmp7/Lrp4/Ank/Twist1 | 18 |
| GO:0010875 | positive regulation of cholesterol efflux | BP | 0.25 | 0.000104 | 0.000943 | 3.03 | 0.000611 | Pltp/Apoe/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3/Abcg4 | 8 |
| GO:0023058 | adaptation of signaling pathway | BP | 0.25 | 0.000104 | 0.000943 | 3.03 | 0.000611 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln/Pld2 | 8 |
| GO:0055001 | muscle cell development | BP | 0.11 | 0.000111 | 0.001 | 3 | 0.000651 | Sgcd/Col14a1/Ryr1/Igf1/Adra1a/Slc8a1/P2rx2/Tmod2/Mef2c/Ank2/Tbx3/Ednra/Dysf/Bin1/Hdac9/Nfatc4/Flnc/Selenon/Pi16/Efemp2/Sgcb/Pdgfrb/Cacnb4/Hey2/Eng | 25 |
| GO:0048565 | digestive tract development | BP | 0.135 | 0.000115 | 0.00103 | 2.99 | 0.000668 | Stra6/Ihh/Sfrp2/Clmp/Pkdcc/Tbx2/Ednrb/Foxf1/Tcf21/Gli3/Prdm1/Kit/Pcsk5/Dchs1/Fat4/Stx2/Dact1 | 17 |
| GO:0035886 | vascular associated smooth muscle cell differentiation | BP | 0.22 | 0.000116 | 0.00104 | 2.98 | 0.000673 | Kit/Cth/Aplnr/Ednra/Efemp2/Sgcb/Pdgfrb/Hey2/Eng | 9 |
| GO:0043368 | positive T cell selection | BP | 0.22 | 0.000116 | 0.00104 | 2.98 | 0.000673 | Cd3g/Dock2/Loxl3/Cd1d1/Zap70/Batf/Spn/Ly9/Bcl11b | 9 |
| GO:0086065 | cell communication involved in cardiac conduction | BP | 0.22 | 0.000116 | 0.00104 | 2.98 | 0.000673 | Cacna1c/Kcnd3/Slc8a1/Scn4b/Cacna2d1/Ank2/Gjc1/Sri/Cav1 | 9 |
| GO:0030509 | BMP signaling pathway | BP | 0.119 | 0.000118 | 0.00105 | 2.98 | 0.000682 | Sfrp2/Lef1/Kdr/Nog/Rnf165/Fst/Gpc3/Zfp423/Cdh5/Gdf7/Lrp2/Hipk2/Kcp/Tnfaip6/Smad9/Neo1/Fstl1/Cav1/Tmprss6/Bmp7/Eng | 21 |
| GO:0048738 | cardiac muscle tissue development | BP | 0.104 | 0.000118 | 0.00105 | 2.98 | 0.000682 | Gli1/Col11a1/AW551984/Angpt1/Sgcd/Col14a1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/Mef2c/Lrp2/Ccm2l/Nox4/Tbx3/Gjc1/Ednra/Myl3/Heg1/Arrb2/Pi16/Sgcb/Pdgfrb/Bmp7/Hey2/Hand2/Eng | 28 |
| GO:1902107 | positive regulation of leukocyte differentiation | BP | 0.116 | 0.000118 | 0.00105 | 2.98 | 0.000682 | Ihh/Mdk/Lef1/Cd83/Hcls1/Pik3r6/Gli3/Calca/Prdm1/Cd1d1/Sox12/Zbtb46/Nckap1l/Kitl/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r/Cd4 | 22 |
| GO:1903708 | positive regulation of hemopoiesis | BP | 0.116 | 0.000118 | 0.00105 | 2.98 | 0.000682 | Ihh/Mdk/Lef1/Cd83/Hcls1/Pik3r6/Gli3/Calca/Prdm1/Cd1d1/Sox12/Zbtb46/Nckap1l/Kitl/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r/Cd4 | 22 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | BP | 0.183 | 0.000119 | 0.00105 | 2.98 | 0.000683 | Kdr/Nrp1/Ptp4a3/Prkd1/Jcad/Vegfc/Tcf4/Atp2b4/Adgra2/Sema6a/Pdgfrb | 11 |
| GO:0097479 | synaptic vesicle localization | BP | 0.183 | 0.000119 | 0.00105 | 2.98 | 0.000683 | Syn2/Map2/Nlgn1/Cdh2/Lrrk2/Lin7a/Nlgn3/Magi2/Pcdh17/Kif5a/Nlgn2 | 11 |
| GO:0001558 | regulation of cell growth | BP | 0.0889 | 0.000125 | 0.00111 | 2.96 | 0.000718 | Tro/Sfrp2/Map2/Sema5a/Wnt11/Col14a1/Igf1/Ptch2/Sema5b/Epb41l3/Cpne5/Nrp1/Cd44/Sema7a/Fn1/Serpine2/Cxcl12/Klhl22/Kif26a/Cth/Minar1/Enpp1/Cd38/Apoe/Sema3g/Caprin2/Slc23a2/Epha7/Cacng7/P3h1/Fhl1/Pi16/Csf2rb/Rrad/Tspyl2/Sema6a/Ulk2/Igfbp4/Wfdc1/Slit3/Draxin | 41 |
| GO:0097529 | myeloid leukocyte migration | BP | 0.109 | 0.000128 | 0.00113 | 2.95 | 0.000734 | Cxcl15/Mdk/Ptpro/Ednrb/Jam3/Cxcl12/Edn3/Itga1/Plcb1/Calca/Kit/Enpp1/Emilin1/Vegfc/Ednra/Pecam1/Cd200/Dysf/Nckap1l/Xcl1/Tnfaip6/Stat5b/Irak4/Ptger3/Mpp1 | 25 |
| GO:0007202 | activation of phospholipase C activity | BP | 0.28 | 0.000131 | 0.00115 | 2.94 | 0.000746 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1 | 7 |
| GO:0101023 | vascular endothelial cell proliferation | BP | 0.28 | 0.000131 | 0.00115 | 2.94 | 0.000746 | Mdk/Itga4/Igf1/Mef2c/Igf2/Akt3/Apln | 7 |
| GO:1905562 | regulation of vascular endothelial cell proliferation | BP | 0.28 | 0.000131 | 0.00115 | 2.94 | 0.000746 | Mdk/Itga4/Igf1/Mef2c/Igf2/Akt3/Apln | 7 |
| GO:0060740 | prostate gland epithelium morphogenesis | BP | 0.242 | 0.000132 | 0.00116 | 2.94 | 0.00075 | Ar/Cyp7b1/Nog/Igf1/Cd44/Mmp2/Id4/Bmp7 | 8 |
| GO:0062149 | detection of stimulus involved in sensory perception of pain | BP | 0.242 | 0.000132 | 0.00116 | 2.94 | 0.00075 | Scn1a/Ano1/Grin2b/Cxcl12/Calca/Htr2a/Arrb2/Fyn | 8 |
| GO:1905314 | semi-lunar valve development | BP | 0.242 | 0.000132 | 0.00116 | 2.94 | 0.00075 | Stra6/Nos3/Emilin1/Tie1/Heyl/Hey2/Slit3/Twist1 | 8 |
| GO:0046849 | bone remodeling | BP | 0.144 | 0.000134 | 0.00117 | 2.93 | 0.000756 | Ihh/Mdk/Inpp4b/Calca/Htr1b/Pth1r/Enpp1/Tnfrsf11b/Cd38/Nox4/Syt7/Ltbp3/Rassf2/Def8/Ptn | 15 |
| GO:0055117 | regulation of cardiac muscle contraction | BP | 0.169 | 0.000134 | 0.00117 | 2.93 | 0.000756 | Cacna1c/Adra1a/Slc8a1/Stc1/Ank2/Bin1/Pde5a/Cacna1h/Ccn2/Atp1a2/Sri/Cav1 | 12 |
| GO:0008064 | regulation of actin polymerization or depolymerization | BP | 0.121 | 0.000136 | 0.00119 | 2.93 | 0.000769 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Sh3bp1/Daam2/Pecam1/Scin/Bin1/Nckap1l/Arhgap28/Cdc42ep5/Plekhh2/Cyfip2/Plek | 20 |
| GO:0070665 | positive regulation of leukocyte proliferation | BP | 0.121 | 0.000136 | 0.00119 | 2.93 | 0.000769 | Igf1/Mef2c/Tnfsf13b/Coro1a/Cd1d1/Itgal/Il27ra/Cd38/Havcr2/Igf2/Nckap1l/Kitl/Xcl1/Stat5b/Csf2rb/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 20 |
| GO:0042492 | gamma-delta T cell differentiation | BP | 0.333 | 0.000137 | 0.00119 | 2.92 | 0.000773 | Lef1/Jag2/Nckap1l/Stat5b/Gimap5/Gimap3 | 6 |
| GO:0099150 | regulation of postsynaptic specialization assembly | BP | 0.333 | 0.000137 | 0.00119 | 2.92 | 0.000773 | Gap43/Ptprd/Lrrc4b/Nptx1/Nptxr/Arhgef9 | 6 |
| GO:1902904 | negative regulation of supramolecular fiber organization | BP | 0.118 | 0.000138 | 0.0012 | 2.92 | 0.000775 | Kank4/Map2/Evl/Phldb2/Tmod2/Pfn2/Cdh5/Coro1a/Rp1/Arhgap6/Tubb4a/Emilin1/Apoe/Pecam1/Tmeff2/Scin/Dysf/Dlc1/Arhgap28/Ttbk2/Plekhh2 | 21 |
| GO:0098659 | inorganic cation import across plasma membrane | BP | 0.138 | 0.000139 | 0.0012 | 2.92 | 0.000775 | Kcnj3/Cacna1c/Kcnd3/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Slc9a5/Slc12a4/Fyn/Atp1a2/Cacna1a | 16 |
| GO:0099587 | inorganic ion import across plasma membrane | BP | 0.138 | 0.000139 | 0.0012 | 2.92 | 0.000775 | Kcnj3/Cacna1c/Kcnd3/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Slc9a5/Slc12a4/Fyn/Atp1a2/Cacna1a | 16 |
| GO:0042596 | fear response | BP | 0.18 | 0.000139 | 0.0012 | 2.92 | 0.000775 | Ankfn1/Mdk/Drd4/Mef2c/Grin2b/Cacna1e/Grik2/Apoe/Gng7/Atp1a2/Prkar1b | 11 |
| GO:0045807 | positive regulation of endocytosis | BP | 0.133 | 0.000139 | 0.0012 | 2.92 | 0.000778 | Angpt1/Nlgn1/Gpc3/Lrrk2/Lrp2/Apoe/Magi2/Sele/Hip1/Bin1/Nckap1l/Arrb1/Arrb2/Apln/Smpd1/Cav1/Pld2 | 17 |
| GO:0006066 | alcohol metabolic process | BP | 0.0957 | 0.000146 | 0.00126 | 2.9 | 0.000816 | Apob/Cyp1b1/Ttr/Cyp7b1/Hmgcs2/Ednrb/Soat1/Inpp4b/Sptlc3/Plcb1/Pth1r/Plpp3/Dkk3/Enpp1/Tsku/Apoe/Bpnt1/Kcnma1/Dysf/Degs2/Adcyap1r1/Abca5/Ipmk/Cyp46a1/Abcg1/Cacna1h/Sord/Plpp1/Smpd1/Lipe/Rdh1/Abcg4/Plek | 33 |
| GO:0072659 | protein localization to plasma membrane | BP | 0.0993 | 0.000149 | 0.00128 | 2.89 | 0.000827 | Kcnb2/Ar/Pkdcc/Epb41l3/Ccdc88a/Arl6/Efr3b/Cdh2/Gga2/Abi3/Myo5a/Ank2/Rab34/Ttc7b/Epha3/Rilpl1/Nfasc/Nsg1/Stac2/Kcnip3/Ptch1/Dchs1/Tspan5/Palm/Atp2b4/Bbs1/Pid1/Nkd2/Golph3l/Rap2a | 30 |
| GO:0007369 | gastrulation | BP | 0.117 | 0.00015 | 0.00128 | 2.89 | 0.000831 | Sfrp2/Lef1/Wnt11/Nog/Phldb2/Hoxa11/Fn1/Foxf1/Osr2/Plpp3/Tgif2/Aplnr/Col5a2/Snai1/Tal1/Osr1/Apln/Ets2/Dact1/Kif16b/Bmp7 | 21 |
| GO:0042135 | neurotransmitter catabolic process | BP | 0.417 | 0.000152 | 0.0013 | 2.89 | 0.000842 | Maob/Abat/Maoa/Naalad2/Bche | 5 |
| GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.132 | 0.000153 | 0.00131 | 2.88 | 0.00085 | Sfrp2/Cd109/Nog/Fst/Lrp2/Pbld2/Tsku/Emilin1/Magi2/Spred3/Hipk2/Tnfaip6/Nrros/Il17rd/Cav1/Tmprss6/Eng | 17 |
| GO:0042475 | odontogenesis of dentin-containing tooth | BP | 0.149 | 0.000156 | 0.00133 | 2.88 | 0.000863 | Wnt6/Lef1/Apcdd1/Fst/Edaradd/Slc24a4/Gli3/Jag2/Tnfrsf11b/Edar/Bmp7/Lrp4/Hand2/Bcl11b | 14 |
| GO:0051899 | membrane depolarization | BP | 0.149 | 0.000156 | 0.00133 | 2.88 | 0.000863 | Cacna1c/Grik1/Kdr/Scn4b/Bok/Lrrk2/Scn1a/Cacna2d1/Scn2a/Fhl1/Cacna1h/Scn3a/Cav1/Cacna1a | 14 |
| GO:0045580 | regulation of T cell differentiation | BP | 0.12 | 0.000161 | 0.00136 | 2.87 | 0.000884 | Ihh/Mdk/Lef1/Cd83/Cd44/Pik3r6/Gli3/Loxl3/Zeb1/Prdm1/Cd1d1/Sox12/Nckap1l/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r | 20 |
| GO:0006584 | catecholamine metabolic process | BP | 0.177 | 0.000161 | 0.00136 | 2.87 | 0.000884 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Sult1a1/Hand2 | 11 |
| GO:0009712 | catechol-containing compound metabolic process | BP | 0.177 | 0.000161 | 0.00136 | 2.87 | 0.000884 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Sult1a1/Hand2 | 11 |
| GO:0010171 | body morphogenesis | BP | 0.177 | 0.000161 | 0.00136 | 2.87 | 0.000884 | Stra6/Ihh/Lef1/Nog/Phldb2/Crispld1/Gpc3/Mmp2/Flvcr1/Twist2/Asph | 11 |
| GO:0031102 | neuron projection regeneration | BP | 0.177 | 0.000161 | 0.00136 | 2.87 | 0.000884 | Gap43/Hgf/Mmp2/Thy1/Matn2/Enpp1/Rtn4rl1/Neo1/Xylt1/Ptn/Nrep | 11 |
| GO:0050906 | detection of stimulus involved in sensory perception | BP | 0.117 | 0.000162 | 0.00137 | 2.86 | 0.000885 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Ppef2/Htr2a/Plcb2/Arrb2/Fyn/Piezo2/Cacnb4/Myc | 21 |
| GO:0010863 | positive regulation of phospholipase C activity | BP | 0.235 | 0.000165 | 0.00139 | 2.86 | 0.000898 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1/Pdgfrb | 8 |
| GO:0050654 | chondroitin sulfate proteoglycan metabolic process | BP | 0.235 | 0.000165 | 0.00139 | 2.86 | 0.000898 | Igf1/Spock2/Egflam/Mamdc2/Ndnf/Bgn/Dse/Xylt1 | 8 |
| GO:0060512 | prostate gland morphogenesis | BP | 0.235 | 0.000165 | 0.00139 | 2.86 | 0.000898 | Ar/Cyp7b1/Nog/Igf1/Cd44/Mmp2/Id4/Bmp7 | 8 |
| GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | BP | 0.235 | 0.000165 | 0.00139 | 2.86 | 0.000898 | Drd4/Dysf/Pkd2/Rrad/Gnb5/Gstm7/Atp1a2/Sri | 8 |
| GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.235 | 0.000165 | 0.00139 | 2.86 | 0.000898 | Mdk/Itga4/Chst2/Ets1/Icam1/Cxcl12/Sele/St3gal4 | 8 |
| GO:0021953 | central nervous system neuron differentiation | BP | 0.111 | 0.000169 | 0.00141 | 2.85 | 0.000915 | Map2/Hoxd10/Dcc/Zeb2/Nrp1/Id4/Gli3/D16Ertd472e/Gdf7/Tsku/Cdh11/Dync2h1/Ndnf/Dclk2/Nin/Tal1/Lhx6/Ptch1/Ptger3/Psen1/Cacna1a/Bcl11b/Draxin | 23 |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | BP | 0.111 | 0.000169 | 0.00141 | 2.85 | 0.000915 | Igf1/Zeb2/Fermt2/Drd4/Ccnd2/Lrrk2/Pik3r6/Edn3/Kit/Nox4/Kitl/Htr2a/Pkd1/Pkd2/Pde5a/Atp2b4/Acsl1/Psen1/Pdgfrb/Map4k2/C1qtnf9/Fgd2/Camk1 | 23 |
| GO:0042474 | middle ear morphogenesis | BP | 0.269 | 0.000171 | 0.00143 | 2.85 | 0.000925 | Nog/Osr2/Tshz1/Enpp1/Ednra/Osr1/Myc | 7 |
| GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.269 | 0.000171 | 0.00143 | 2.85 | 0.000925 | Mdk/Itga4/Chst2/Ets1/Icam1/Sele/St3gal4 | 7 |
| GO:0030832 | regulation of actin filament length | BP | 0.119 | 0.000174 | 0.00145 | 2.84 | 0.000938 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Sh3bp1/Daam2/Pecam1/Scin/Bin1/Nckap1l/Arhgap28/Cdc42ep5/Plekhh2/Cyfip2/Plek | 20 |
| GO:0055067 | monovalent inorganic cation homeostasis | BP | 0.119 | 0.000174 | 0.00145 | 2.84 | 0.000938 | Slc1a3/Car4/Slc12a8/Slc8a1/Lrrk2/Ednrb/Slc9a7/Dmxl2/Agtr1a/Lacc1/Kcnma1/Ednra/Slc9a5/Slc12a4/Slc4a3/Aqp11/Atp1a2/Slc4a8/Mllt6/Rab20 | 20 |
| GO:0010717 | regulation of epithelial to mesenchymal transition | BP | 0.147 | 0.000175 | 0.00145 | 2.84 | 0.000942 | Sfrp2/Mdk/Lef1/Nog/Mark1/Phldb2/Fermt2/Epha3/Spred3/Snai1/Il17rd/Bmp7/Twist1/Eng | 14 |
| GO:0031099 | regeneration | BP | 0.122 | 0.000186 | 0.00154 | 2.81 | 0.000998 | Gli1/Gap43/Hgf/Mdk/Igf1/Mmp2/Cxcl12/Hopx/Thy1/Matn2/Enpp1/Dysf/Rtn4rl1/Neo1/Selenon/Large1/Xylt1/Ptn/Nrep | 19 |
| GO:0044091 | membrane biogenesis | BP | 0.175 | 0.000187 | 0.00154 | 2.81 | 0.001 | Ptprd/Nlgn1/Cdh2/Ubxn2b/Nlgn3/Magi2/Stx2/Cav1/Tlcd2/Lrp4/Nlgn2 | 11 |
| GO:0072678 | T cell migration | BP | 0.175 | 0.000187 | 0.00154 | 2.81 | 0.001 | Itga4/Itgb7/Icam1/Cxcr3/Cxcl12/Msn/Itgal/Il27ra/Cd200/Xcl1/Spn | 11 |
| GO:0006525 | arginine metabolic process | BP | 0.316 | 0.000193 | 0.00158 | 2.8 | 0.00103 | Ddah1/Nos3/Fah/Atp2b4/Slc7a7/Asl | 6 |
| GO:0014051 | gamma-aminobutyric acid secretion | BP | 0.316 | 0.000193 | 0.00158 | 2.8 | 0.00103 | Grik1/Abat/Htr1b/Gabbr1/Cacna1a/Cacnb4 | 6 |
| GO:0035641 | locomotory exploration behavior | BP | 0.316 | 0.000193 | 0.00158 | 2.8 | 0.00103 | Lrrk2/Apoe/Dlg4/Atp1a2/Lsamp/Nlgn2 | 6 |
| GO:0032271 | regulation of protein polymerization | BP | 0.11 | 0.000195 | 0.0016 | 2.8 | 0.00104 | Kank4/Map2/Evl/Hcls1/Tmod2/Pfn2/Icam1/Cdh5/Coro1a/Esam/Tubb4a/Daam2/Pecam1/Scin/Bin1/Nckap1l/Nin/Slain1/Arhgap28/Cdc42ep5/Cav1/Cyfip2/Tppp | 23 |
| GO:0002687 | positive regulation of leukocyte migration | BP | 0.121 | 0.000202 | 0.00166 | 2.78 | 0.00107 | Mdk/Itga4/Icam1/Jam3/Cxcl12/Edn3/Thy1/Vegfc/Sele/Ednra/Pecam1/Dysf/Nckap1l/Kitl/Xcl1/Ptger3/Ptn/Spn/Jam2 | 19 |
| GO:0030856 | regulation of epithelial cell differentiation | BP | 0.121 | 0.000202 | 0.00166 | 2.78 | 0.00107 | Cd109/Macroh2a2/Zeb2/Ptch2/Plcb1/Cdh5/Zeb1/Cdkn1c/Tbx3/Spred3/Ptch1/Stat5b/Osr1/Cldn5/Fat4/Cav1/Akap11/Hey2/S1pr3 | 19 |
| GO:1904064 | positive regulation of cation transmembrane transport | BP | 0.118 | 0.000205 | 0.00167 | 2.78 | 0.00108 | Cacna1c/Drd4/Cacna2d1/Cxcr3/Ank2/Edn3/Plcb1/Thy1/Aplnr/Nlgn3/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Sri/Cacnb4/F2r | 20 |
| GO:0010837 | regulation of keratinocyte proliferation | BP | 0.229 | 0.000205 | 0.00168 | 2.78 | 0.00109 | Mdk/Cd109/Prkd1/Has2/Stxbp4/Klf9/Twist2/Bcl11b | 8 |
| GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | BP | 0.172 | 0.000216 | 0.00176 | 2.75 | 0.00114 | Igf1/Ddr2/Drd4/Tert/Mmp2/Mef2c/Adamts1/Htr1b/Rgs5/Efemp2/Apln | 11 |
| GO:0032330 | regulation of chondrocyte differentiation | BP | 0.185 | 0.00022 | 0.00179 | 2.75 | 0.00116 | Mdk/Hoxd11/Pkdcc/Hoxa11/Gli3/Wnt9a/Adamts7/Ltbp3/Ccn2/Mboat2 | 10 |
| GO:0030204 | chondroitin sulfate metabolic process | BP | 0.259 | 0.000222 | 0.0018 | 2.75 | 0.00116 | Spock2/Egflam/Mamdc2/Ndnf/Bgn/Dse/Xylt1 | 7 |
| GO:0072012 | glomerulus vasculature development | BP | 0.259 | 0.000222 | 0.0018 | 2.75 | 0.00116 | Angpt1/Tek/Tcf21/Ednra/Osr1/Pdgfrb/Bmp7 | 7 |
| GO:0052547 | regulation of peptidase activity | BP | 0.088 | 0.000222 | 0.0018 | 2.75 | 0.00116 | Hgf/Sfrp2/Cd109/Spink1/Igf1/Aph1b/Tfpi/Cd44/Bok/Gpc3/Wdr35/Grin2b/Mical1/Serpine2/Ift57/Wfdc3/Serpini1/Wnt9a/Ppm1f/Hip1/Bin1/Arrb1/Rcn3/Epha7/Dlc1/Arrb2/Asph/Fyn/Nlrp1b/Pi16/Ccn2/Simc1/Pi15/Mgmt/Cav1/Cyfip2/F2r/Wfdc1/Myc | 39 |
| GO:0061041 | regulation of wound healing | BP | 0.128 | 0.000223 | 0.0018 | 2.75 | 0.00116 | Cd109/Ddr2/Phldb2/Tfpi/Abat/Fermt2/Serpine2/Plpp3/Apoe/Pros1/Clec7a/Ptger3/St3gal4/Cav1/Cldn1/F2r/Wfdc1 | 17 |
| GO:2000027 | regulation of animal organ morphogenesis | BP | 0.128 | 0.000223 | 0.0018 | 2.75 | 0.00116 | Hgf/Sfrp2/Ar/Apcdd1/Wnt11/Hoxd11/Nog/Nkd1/Hoxa11/Gpc3/Znrf3/Tnfrsf11b/Agtr1a/Ednra/Fzd2/Dact1/Bmp7 | 17 |
| GO:0045669 | positive regulation of osteoblast differentiation | BP | 0.16 | 0.000229 | 0.00184 | 2.74 | 0.00119 | Sfrp2/Igf1/Ddr2/Npnt/Fermt2/Mef2c/Prkd1/Gli3/Nell1/Gdf10/Lrp3/Bmp7 | 12 |
| GO:0030323 | respiratory tube development | BP | 0.105 | 0.000233 | 0.00187 | 2.73 | 0.00121 | Gli1/Stra6/Hhip/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Gpc3/Foxf1/Hopx/Tcf21/Gli3/Loxl3/Tshz3/Mgp/Ltbp3/Pcsk5/Rcn3/Heg1/Selenon/Ccn2/Dlg5/Pdgfrb | 25 |
| GO:0060541 | respiratory system development | BP | 0.101 | 0.000234 | 0.00187 | 2.73 | 0.00121 | Gli1/Stra6/Hhip/Lef1/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Edaradd/Gpc3/Ano1/Foxf1/Hopx/Tcf21/Gli3/Loxl3/Tshz3/Mgp/Ltbp3/Rcn3/Heg1/Selenon/Ccn2/Dlg5/Pdgfrb | 27 |
| GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | BP | 0.385 | 0.000237 | 0.00189 | 2.72 | 0.00123 | Nlgn1/Tshz3/Nlgn3/Atp1a2/Nlgn2 | 5 |
| GO:0097084 | vascular associated smooth muscle cell development | BP | 0.385 | 0.000237 | 0.00189 | 2.72 | 0.00123 | Ednra/Efemp2/Sgcb/Hey2/Eng | 5 |
| GO:0032091 | negative regulation of protein binding | BP | 0.143 | 0.000245 | 0.00195 | 2.71 | 0.00126 | Map2/Itga4/Nog/Lrrk2/Ccm2l/Dysf/Nfatc4/Arrb2/Ttbk2/Nes/Cav1/Dact1/Camk1/Myc | 14 |
| GO:1904063 | negative regulation of cation transmembrane transport | BP | 0.143 | 0.000245 | 0.00195 | 2.71 | 0.00126 | Hecw2/Drd4/Dysf/Bin1/Osr1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Cav1/Twist1 | 14 |
| GO:0050918 | positive chemotaxis | BP | 0.2 | 0.000247 | 0.00196 | 2.71 | 0.00127 | Sema5a/Angpt1/Kdr/Nrp1/Cdh13/Cxcl12/Lrp2/Coro1a/Vegfc | 9 |
| GO:0071825 | protein-lipid complex subunit organization | BP | 0.2 | 0.000247 | 0.00196 | 2.71 | 0.00127 | Apob/Pla2g10/Soat1/Pltp/Apoe/Bin1/Abca5/Abcg1/Pcdhga3 | 9 |
| GO:0003229 | ventricular cardiac muscle tissue development | BP | 0.169 | 0.000249 | 0.00197 | 2.7 | 0.00128 | Col11a1/Col14a1/Nog/Lrp2/Ccm2l/Tbx3/Ednra/Myl3/Heg1/Hey2/Eng | 11 |
| GO:0048048 | embryonic eye morphogenesis | BP | 0.222 | 0.000253 | 0.002 | 2.7 | 0.0013 | Stra6/Ihh/Tbx2/Zeb1/Hipk2/Hipk1/Bmp7/Twist1 | 8 |
| GO:0050671 | positive regulation of lymphocyte proliferation | BP | 0.122 | 0.000254 | 0.00201 | 2.7 | 0.0013 | Igf1/Mef2c/Tnfsf13b/Coro1a/Cd1d1/Itgal/Il27ra/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 18 |
| GO:0001662 | behavioral fear response | BP | 0.182 | 0.000258 | 0.00203 | 2.69 | 0.00132 | Ankfn1/Mdk/Drd4/Mef2c/Grin2b/Cacna1e/Grik2/Apoe/Gng7/Atp1a2 | 10 |
| GO:0062012 | regulation of small molecule metabolic process | BP | 0.0916 | 0.000263 | 0.00207 | 2.68 | 0.00134 | Apob/Igf1/Nos3/Acacb/Pth1r/Kit/Dkk3/Enpp1/Apoe/Lacc1/Kcnma1/Igf2/Snai1/Adcyap1r1/Htr2a/Zbtb20/Abcg1/Atp2b4/Fam3c/Abcd2/Slc7a7/Psen1/Cav1/Nr1h3/Pid1/Cacna1a/Igfbp4/Grb10/Rdh1/Abcg4/Myc/Twist1/Plek/Irs1 | 34 |
| GO:0043116 | negative regulation of vascular permeability | BP | 0.3 | 0.000264 | 0.00208 | 2.68 | 0.00135 | Angpt1/Ddah1/Fermt2/Akap12/Apoe/Cldn5 | 6 |
| GO:0043405 | regulation of MAP kinase activity | BP | 0.11 | 0.000267 | 0.0021 | 2.68 | 0.00136 | Sfrp2/Zeb2/Cbs/Drd4/Lrrk2/Mapk8ip1/Pik3r6/Edn3/Kit/Apoe/Nox4/Kitl/Htr2a/Pde5a/Smpd1/Psen1/Pdgfrb/Map4k2/Cav1/Fgd2/Bmp7/Lax1 | 22 |
| GO:1905475 | regulation of protein localization to membrane | BP | 0.11 | 0.000267 | 0.0021 | 2.68 | 0.00136 | Gpc6/Ar/Pkdcc/Tcaf1/Cdh2/Gpc3/Abi3/Myo5a/Epha3/Magi2/Tnfaip6/Stac2/Fyn/Dlg4/Atp2b4/Ptn/Pid1/Gpc2/Nkd2/Zmynd8/Lrp4/Nlgn2 | 22 |
| GO:0003014 | renal system process | BP | 0.13 | 0.000276 | 0.00216 | 2.67 | 0.0014 | Cacna1c/Adra1a/Ptpro/Npr3/Ednrb/Abcg2/Stc1/Maged2/Agtr1a/Kcnma1/Ednra/Ptger3/Akap11/Abcg3/F2r/Mllt6 | 16 |
| GO:0045061 | thymic T cell selection | BP | 0.25 | 0.000284 | 0.00222 | 2.65 | 0.00144 | Cd3g/Gli3/Dock2/Jag2/Cd1d1/Zap70/Spn | 7 |
| GO:0060323 | head morphogenesis | BP | 0.196 | 0.000294 | 0.00229 | 2.64 | 0.00148 | Stra6/Ihh/Lef1/Nog/Crispld1/Mmp2/Flvcr1/Twist2/Asph | 9 |
| GO:1903115 | regulation of actin filament-based movement | BP | 0.196 | 0.000294 | 0.00229 | 2.64 | 0.00148 | Cacna1c/Stc1/Ank2/Bin1/Cacna1h/Ptger3/Atp1a2/Sri/Cav1 | 9 |
| GO:0006029 | proteoglycan metabolic process | BP | 0.156 | 0.000294 | 0.00229 | 2.64 | 0.00149 | Col11a1/Ihh/Igf1/Spock2/Chst10/Egflam/Mamdc2/Ndnf/Adamts7/Bgn/Dse/Xylt1 | 12 |
| GO:0007588 | excretion | BP | 0.179 | 0.000301 | 0.00233 | 2.63 | 0.00151 | Cacna1c/Mdk/Adra1a/Ednrb/Abcg2/Stc1/Kcnma1/Ptger3/Abcg3/Mllt6 | 10 |
| GO:0061001 | regulation of dendritic spine morphogenesis | BP | 0.179 | 0.000301 | 0.00233 | 2.63 | 0.00151 | Nlgn1/Lrrk2/Ppfia2/Abi2/Kif1a/Nlgn3/Caprin2/Bhlhb9/Cux2/Ngef | 10 |
| GO:0032946 | positive regulation of mononuclear cell proliferation | BP | 0.121 | 0.000301 | 0.00233 | 2.63 | 0.00151 | Igf1/Mef2c/Tnfsf13b/Coro1a/Cd1d1/Itgal/Il27ra/Cd38/Havcr2/Igf2/Nckap1l/Xcl1/Stat5b/Zap70/Il12rb1/Spn/Cd4/Rasal3 | 18 |
| GO:0046777 | protein autophosphorylation | BP | 0.103 | 0.000301 | 0.00233 | 2.63 | 0.00151 | Epha8/Kdr/Ddr2/Lrrk2/Prkd1/Itk/Tek/Eif2ak4/Stk33/Flt3/Thy1/Kit/Enpp1/Vegfc/Epha7/Rassf2/Fyn/Fgr/Zap70/Ulk2/Pdgfrb/Cav1/Mknk2/Rap2a/Eng | 25 |
| GO:0042552 | myelination | BP | 0.114 | 0.000302 | 0.00233 | 2.63 | 0.00151 | Ptprz1/Hgf/Igf1/Epb41l3/Mpdz/Cdh2/Wasf3/Myo5a/Id4/Jam3/Slc8a3/Enpp1/Nfasc/Fyn/Cldn5/Abcd2/Ptn/Tppp/Itgb4/Jam2 | 20 |
| GO:0032092 | positive regulation of protein binding | BP | 0.14 | 0.000303 | 0.00234 | 2.63 | 0.00152 | Tert/Lrrk2/Plxnd1/Lfng/Apoe/Caprin2/Pkd1/Hipk2/Arrb1/Cldn5/Mfng/Psen1/Cav1/Dact1 | 14 |
| GO:0048846 | axon extension involved in axon guidance | BP | 0.216 | 0.000309 | 0.00237 | 2.62 | 0.00154 | Sema5a/Sema5b/Nrp1/Sema7a/Cxcl12/Sema3g/Sema6a/Slit3 | 8 |
| GO:0061384 | heart trabecula morphogenesis | BP | 0.216 | 0.000309 | 0.00237 | 2.62 | 0.00154 | Nog/Tek/Adamts1/Ccm2l/Heg1/Bmp7/Hey2/Eng | 8 |
| GO:0090022 | regulation of neutrophil chemotaxis | BP | 0.216 | 0.000309 | 0.00237 | 2.62 | 0.00154 | Mdk/Jam3/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Mpp1 | 8 |
| GO:1902284 | neuron projection extension involved in neuron projection guidance | BP | 0.216 | 0.000309 | 0.00237 | 2.62 | 0.00154 | Sema5a/Sema5b/Nrp1/Sema7a/Cxcl12/Sema3g/Sema6a/Slit3 | 8 |
| GO:0030258 | lipid modification | BP | 0.106 | 0.000315 | 0.00241 | 2.62 | 0.00156 | Acacb/Soat1/Agk/Inpp4b/St3gal2/Plpp3/Bdh2/Dgkb/Agtr1a/Apoe/Ggta1/Echs1/Abcg1/Plpp1/Abcd2/St3gal4/Cygb/C1qtnf9/Twist1/Mboat2/Plppr2/Echdc2/Irs1 | 23 |
| GO:0050773 | regulation of dendrite development | BP | 0.124 | 0.000318 | 0.00243 | 2.61 | 0.00158 | Ptprz1/Hecw2/Ptprd/Mark1/Tbc1d24/Obsl1/Trpc6/Caprin2/Nfatc4/Sarm1/Bhlhb9/Cux2/Fat3/Ptn/Bmp7/Numbl/Rap2a | 17 |
| GO:0021537 | telencephalon development | BP | 0.101 | 0.000319 | 0.00243 | 2.61 | 0.00158 | Mdk/Lef1/Zeb2/Sema7a/Cdh2/Lrrk2/Dab1/Id4/Fut10/Cxcl12/Dnah5/Gli3/Plcb1/Tsku/Dclk2/Nin/Lhx6/Rtn4rl1/Nr2f1/Atp1a2/Fat4/Bbs1/Psen1/Bcl11b/Draxin/Numbl | 26 |
| GO:0045137 | development of primary sexual characteristics | BP | 0.102 | 0.00032 | 0.00244 | 2.61 | 0.00158 | Sfrp2/Ar/Angpt1/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Rhobtb3/Kitl/Arrb1/Tnfaip6/Hoxa9/Arrb2/Stat5b/Osr1/Abcb1a/Lhfpl2 | 25 |
| GO:0002040 | sprouting angiogenesis | BP | 0.128 | 0.000332 | 0.00253 | 2.6 | 0.00164 | Sema5a/Angpt1/Lef1/Kdr/Nrp1/Tek/Cdh13/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Hdac9/Akt3/Adgra2/Sema6a | 16 |
| GO:0045586 | regulation of gamma-delta T cell differentiation | BP | 0.357 | 0.000354 | 0.00268 | 2.57 | 0.00173 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0046643 | regulation of gamma-delta T cell activation | BP | 0.357 | 0.000354 | 0.00268 | 2.57 | 0.00173 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0051152 | positive regulation of smooth muscle cell differentiation | BP | 0.357 | 0.000354 | 0.00268 | 2.57 | 0.00173 | Tshz3/Kit/Cth/Efemp2/Eng | 5 |
| GO:0051967 | negative regulation of synaptic transmission, glutamatergic | BP | 0.357 | 0.000354 | 0.00268 | 2.57 | 0.00173 | Grik1/Myo5a/Htr1b/Grik2/Htr2a | 5 |
| GO:1905809 | negative regulation of synapse organization | BP | 0.357 | 0.000354 | 0.00268 | 2.57 | 0.00173 | Apoe/Arhgef15/Nfatc4/Epha7/Ptn | 5 |
| GO:0050921 | positive regulation of chemotaxis | BP | 0.119 | 0.000355 | 0.00268 | 2.57 | 0.00173 | Sema5a/Mdk/Kdr/Nrp1/Fn1/Prkd1/Cxcr3/Cdh13/Cxcl12/Edn3/Ppm1f/Vegfc/Ednra/Dysf/Nckap1l/Xcl1/Pdgfrb/Ptn | 18 |
| GO:0007338 | single fertilization | BP | 0.116 | 0.000355 | 0.00268 | 2.57 | 0.00174 | Ar/Pla2g10/Hoxd10/Hoxd11/Spink1/Hoxa11/Hoxd9/Hoxa10/Adcy3/Plcb1/Trpc6/Glrb/Cecr2/Hoxa9/Cacna1h/Ccdc136/Stx2/Pkdrej/Lhfpl2 | 19 |
| GO:1900027 | regulation of ruffle assembly | BP | 0.241 | 0.000359 | 0.0027 | 2.57 | 0.00175 | Evl/Nlgn1/Pfn2/Icam1/Eps8l1/Def8/Cav1 | 7 |
| GO:0002228 | natural killer cell mediated immunity | BP | 0.144 | 0.000361 | 0.00272 | 2.57 | 0.00176 | Pik3r6/Coro1a/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gzmb/Gimap3/Rab27a/Clnk/Cd96 | 13 |
| GO:0032414 | positive regulation of ion transmembrane transporter activity | BP | 0.127 | 0.000363 | 0.00273 | 2.56 | 0.00177 | Tcaf1/Drd4/Cacna2d1/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Cacnb4 | 16 |
| GO:0030099 | myeloid cell differentiation | BP | 0.0866 | 0.00037 | 0.00278 | 2.56 | 0.0018 | Lef1/Pla2g10/Cd109/P4htm/Hcls1/Ets1/Gpc3/Mef2c/Inpp4b/Flvcr1/Lrrc17/Plcb1/Calca/Fli1/Kit/Heph/Enpp1/Cdkn1c/Tnfrsf11b/Gpr171/Irf8/Zbtb46/Scin/Nckap1l/Kitl/Tal1/Tnfaip6/Hoxa9/Twist2/Rassf2/Nrros/Stat5b/Gimap5/Gimap3/Psen1/Batf/Cd4/Myc | 38 |
| GO:0061337 | cardiac conduction | BP | 0.162 | 0.000374 | 0.0028 | 2.55 | 0.00182 | Cacna1c/Kcnd3/Slc8a1/Scn4b/Cacna2d1/Ank2/Gjc1/Bin1/Slc4a3/Sri/Cav1 | 11 |
| GO:0086004 | regulation of cardiac muscle cell contraction | BP | 0.211 | 0.000376 | 0.0028 | 2.55 | 0.00182 | Cacna1c/Stc1/Ank2/Bin1/Cacna1h/Atp1a2/Sri/Cav1 | 8 |
| GO:1900274 | regulation of phospholipase C activity | BP | 0.211 | 0.000376 | 0.0028 | 2.55 | 0.00182 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1/Pdgfrb | 8 |
| GO:0008366 | axon ensheathment | BP | 0.112 | 0.000377 | 0.00281 | 2.55 | 0.00182 | Ptprz1/Hgf/Igf1/Epb41l3/Mpdz/Cdh2/Wasf3/Myo5a/Id4/Jam3/Slc8a3/Enpp1/Nfasc/Fyn/Cldn5/Abcd2/Ptn/Tppp/Itgb4/Jam2 | 20 |
| GO:0051258 | protein polymerization | BP | 0.0978 | 0.000396 | 0.00294 | 2.53 | 0.00191 | Kank4/Map2/Evl/Hcls1/Tmod2/Pfn2/Icam1/Cdh5/Catip/Coro1a/Esam/Rp1/Tubb4a/Daam2/Pecam1/Scin/Bin1/Nckap1l/Nin/Slain1/Arhgap28/Cdc42ep5/Cav1/Cyfip2/Tppp/Map7d3/Pstpip1 | 27 |
| GO:0051101 | regulation of DNA binding | BP | 0.126 | 0.000398 | 0.00295 | 2.53 | 0.00191 | Lef1/Igf1/Zfp462/Runx1t1/Zc4h2/Hipk2/Twist2/Habp4/Zfp90/Psen1/Sri/Hipk1/Hey2/Hand2/Myc/Twist1 | 16 |
| GO:0042311 | vasodilation | BP | 0.172 | 0.000404 | 0.00299 | 2.52 | 0.00194 | Kdr/Npr3/Ednrb/Itga1/Ptprm/Calca/Gucy1a1/Apoe/Kcnma1/Bdkrb2 | 10 |
| GO:0032965 | regulation of collagen biosynthetic process | BP | 0.188 | 0.00041 | 0.00303 | 2.52 | 0.00196 | Ihh/Ddr2/Fn1/Emilin1/Arrb2/Ccn2/Pdgfrb/Cygb/F2r | 9 |
| GO:0035137 | hindlimb morphogenesis | BP | 0.188 | 0.00041 | 0.00303 | 2.52 | 0.00196 | Hoxd10/Aff3/Gpc3/Hoxd9/Osr2/Tbx3/Ptch1/Osr1/Twist1 | 9 |
| GO:0042073 | intraciliary transport | BP | 0.188 | 0.00041 | 0.00303 | 2.52 | 0.00196 | Tub/Wdr35/Ift57/Dync2h1/Ttc26/Arl3/Dync2li1/Bbs1/Ift81 | 9 |
| GO:0046513 | ceramide biosynthetic process | BP | 0.159 | 0.000426 | 0.00314 | 2.5 | 0.00203 | St8sia2/St8sia4/Agk/Sptlc3/Cers4/St3gal2/St6galnac4/Degs2/St8sia6/St6galnac6/Smpd1 | 11 |
| GO:0051251 | positive regulation of lymphocyte activation | BP | 0.0858 | 0.000441 | 0.00325 | 2.49 | 0.0021 | Ihh/Efnb3/Mdk/Lef1/Cd83/Igf1/Mef2c/Pik3r6/Gli3/Thy1/Tnfsf13b/Prdm1/Coro1a/Cd1d1/Itgal/H2-Ab1/Il27ra/Cd38/Sox12/Havcr2/H2-Ob/Igf2/Nckap1l/Xcl1/H2-Eb1/Stat5b/Clec7a/H2-Aa/Gimap5/Zap70/Gimap3/Il12rb1/Cav1/Il2rg/Spn/Il7r/Cd4/Rasal3 | 38 |
| GO:0045724 | positive regulation of cilium assembly | BP | 0.233 | 0.000448 | 0.0033 | 2.48 | 0.00214 | Ccdc88a/Kctd17/Rp1/Crocc/Ttbk2/Dzip1/Atmin | 7 |
| GO:1903035 | negative regulation of response to wounding | BP | 0.141 | 0.000449 | 0.0033 | 2.48 | 0.00214 | Mdk/Cd109/Phldb2/Tfpi/Abat/Serpine2/Apoe/Pros1/Rtn4rl1/Neo1/Ptger3/Xylt1/Wfdc1 | 13 |
| GO:1904645 | response to amyloid-beta | BP | 0.205 | 0.000453 | 0.00332 | 2.48 | 0.00215 | Itga4/Igf1/Cacna2d1/Mmp2/Icam1/Fyn/Psen1/Cacna1a | 8 |
| GO:0030324 | lung development | BP | 0.102 | 0.000466 | 0.0034 | 2.47 | 0.0022 | Gli1/Stra6/Hhip/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Gpc3/Foxf1/Hopx/Tcf21/Gli3/Loxl3/Tshz3/Mgp/Ltbp3/Rcn3/Heg1/Selenon/Ccn2/Dlg5/Pdgfrb | 24 |
| GO:0002285 | lymphocyte activation involved in immune response | BP | 0.104 | 0.000466 | 0.0034 | 2.47 | 0.0022 | Mdk/Lef1/Itm2a/Icam1/Eif2ak4/Lfng/Loxl3/Dock10/Coro1a/Itgal/Il27ra/Enpp1/Cracr2a/Havcr2/Irf8/Nckap1l/Mfng/Psen1/Apbb1ip/Rab27a/Batf/Spn/Ly9 | 23 |
| GO:1903828 | negative regulation of protein localization | BP | 0.104 | 0.000466 | 0.0034 | 2.47 | 0.0022 | Angpt1/Pkdcc/Drd4/Lrrk2/Abi3/Pkia/Ppm1f/Ubxn2b/Apoe/Cd200/Dclk2/Nin/Neo1/Frmd4a/Ttbk2/Rab23/Ptger3/Lats2/Nr1h3/Pid1/F2r/Ankrd13a/Irs1 | 23 |
| GO:0003180 | aortic valve morphogenesis | BP | 0.273 | 0.000468 | 0.0034 | 2.47 | 0.0022 | Nos3/Emilin1/Tie1/Hey2/Slit3/Twist1 | 6 |
| GO:0060080 | inhibitory postsynaptic potential | BP | 0.273 | 0.000468 | 0.0034 | 2.47 | 0.0022 | Grik1/Abat/Drd4/Grik2/Nlgn3/Nlgn2 | 6 |
| GO:0072111 | cell proliferation involved in kidney development | BP | 0.273 | 0.000468 | 0.0034 | 2.47 | 0.0022 | Gpc3/Ptch1/Osr1/Pdgfrb/Bmp7/Myc | 6 |
| GO:0072578 | neurotransmitter-gated ion channel clustering | BP | 0.273 | 0.000468 | 0.0034 | 2.47 | 0.0022 | Nlgn1/Slitrk3/Glrb/Apoe/Dlg4/Nlgn2 | 6 |
| GO:0008202 | steroid metabolic process | BP | 0.0917 | 0.000473 | 0.00343 | 2.46 | 0.00222 | Apob/Cyp3a59/Cyp1b1/Cyp3a25/Cyp7b1/Nr5a2/Igf1/Hmgcs2/Ednrb/Soat1/Chst10/Kit/Dkk3/Enpp1/Tsku/Agtr1a/Apoe/Kcnma1/Igf2/Snai1/Ebpl/Abca5/Cyp46a1/Abcg1/Stat5b/Cacna1h/Smpd1/Lipe/Sult1a1/Rdh1/Abcg4 | 31 |
| GO:0015711 | organic anion transport | BP | 0.0907 | 0.000474 | 0.00343 | 2.46 | 0.00222 | Slc1a3/Car4/Grik1/Pla2g10/Slc13a5/Slc2a3/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Slc36a4/Lrp2/Htr1b/Slc6a15/Slc23a2/Bdkrb2/Sfxn5/Slc38a1/Nfkbie/Slco4c1/Slc4a3/Ptger3/Slc4a8/Psen1/Abcc5/Gabbr1/Abcg3/Cacna1a/Lrrc8c/Cacnb4/Myc | 32 |
| GO:0014013 | regulation of gliogenesis | BP | 0.124 | 0.000474 | 0.00343 | 2.46 | 0.00222 | Ptprz1/Mdk/Nog/Igf1/Tert/Dab1/Id4/Serpine2/Lrp2/Daam2/Bin1/Zcchc24/Ptn/Enpp2/Myc/Zfp365 | 16 |
| GO:0003407 | neural retina development | BP | 0.148 | 0.000475 | 0.00343 | 2.46 | 0.00222 | Ihh/Sdk2/Arl6/Obsl1/Ptprm/Thy1/Rp1/Hipk2/Rorb/Fat3/Ptn/Hipk1 | 12 |
| GO:0019228 | neuronal action potential | BP | 0.184 | 0.000481 | 0.00347 | 2.46 | 0.00225 | Kcnd2/P2rx2/Scn1a/Scn2a/Grik2/Kcnma1/Kcnc4/Cacna1h/Scn3a | 9 |
| GO:0046633 | alpha-beta T cell proliferation | BP | 0.184 | 0.000481 | 0.00347 | 2.46 | 0.00225 | Cd44/Mapk8ip1/Dock2/Btla/Xcl1/Zap70/Il12rb1/Myc/Rasal3 | 9 |
| GO:0048675 | axon extension | BP | 0.12 | 0.000486 | 0.0035 | 2.46 | 0.00227 | Map2/Sema5a/Sema5b/Nrp1/Sema7a/Fn1/Cxcl12/Edn3/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Sema6a/Ulk2/Slit3/Draxin | 17 |
| GO:0001892 | embryonic placenta development | BP | 0.128 | 0.000494 | 0.00355 | 2.45 | 0.0023 | Lef1/Itga4/Slc8a1/Vash1/Syde1/Prdm1/Pcdh12/Cdkn1c/Igf2/Snai1/Pkd1/Pkd2/Vash2/Bmp7/Hey2 | 15 |
| GO:0030100 | regulation of endocytosis | BP | 0.101 | 0.000495 | 0.00355 | 2.45 | 0.0023 | Angpt1/Nlgn1/Drd4/Gpc3/Lrrk2/Cdh13/Lrp2/Epha3/Apoe/Magi2/Syt7/Sele/Hip1/Bin1/Nckap1l/Arrb1/Arrb2/Dlg4/Apln/Smpd1/Cav1/Nr1h3/Pld2/Ankrd13a | 24 |
| GO:0018105 | peptidyl-serine phosphorylation | BP | 0.0914 | 0.000498 | 0.00357 | 2.45 | 0.00231 | Hgf/Sfrp2/Angpt1/Hcls1/Cd44/Lrrk2/Pfn2/Prkd1/Tek/Rps6ka2/Ppm1f/Caprin2/Dclk2/Bdkrb2/Pkd1/Hipk2/Rps6ka5/Arrb1/Rassf2/Arrb2/Ttbk2/Akt3/Atp2b4/Lats2/Brsk1/Ulk2/Hipk1/Cav1/Sh2d3c/Mknk2/Camk1 | 31 |
| GO:0048814 | regulation of dendrite morphogenesis | BP | 0.14 | 0.000499 | 0.00358 | 2.45 | 0.00232 | Ptprz1/Hecw2/Ptprd/Tbc1d24/Obsl1/Trpc6/Caprin2/Nfatc4/Sarm1/Bhlhb9/Cux2/Numbl/Rap2a | 13 |
| GO:0031000 | response to caffeine | BP | 0.333 | 0.00051 | 0.00364 | 2.44 | 0.00236 | Ryr1/Slc8a1/Selenon/Gstm7/Irs1 | 5 |
| GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | BP | 0.333 | 0.00051 | 0.00364 | 2.44 | 0.00236 | Sfrp2/Nkd1/Gpc3/Znrf3/Dact1 | 5 |
| GO:0071248 | cellular response to metal ion | BP | 0.112 | 0.000518 | 0.0037 | 2.43 | 0.0024 | Slc13a5/Nlgn1/Syt13/Cpne5/Fn1/Lrrk2/Mef2c/Nptx1/Rasgrp2/Adcy7/Dlg2/Syt7/Nfatc4/Asph/Pkd2/Slc25a23/Dlg4/Smpd1/Kcnh1 | 19 |
| GO:0060042 | retina morphogenesis in camera-type eye | BP | 0.146 | 0.000532 | 0.00379 | 2.42 | 0.00246 | Ihh/Sdk2/Arl6/Obsl1/Ptprm/Thy1/Rp1/Hipk2/Rorb/Fat3/Ptn/Hipk1 | 12 |
| GO:0050818 | regulation of coagulation | BP | 0.155 | 0.000548 | 0.0039 | 2.41 | 0.00253 | Tfpi/Abat/Serpine2/Procr/Apoe/Pros1/Ptger3/St3gal4/Psen1/Cav1/F2r | 11 |
| GO:0001945 | lymph vessel development | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Kdr/Vash1/Vegfc/Tie1/Pkd1/Heg1/Svep1 | 7 |
| GO:0050951 | sensory perception of temperature stimulus | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Ano1/Cxcl12/Calca/Grik2/Enpp1/Htr2a/Arrb2 | 7 |
| GO:0086010 | membrane depolarization during action potential | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Cacna1c/Scn4b/Scn1a/Cacna2d1/Scn2a/Cacna1h/Scn3a | 7 |
| GO:0099633 | protein localization to postsynaptic specialization membrane | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Gpc6/Nptx1/Dlg2/Nptxr/Magi2/Cacng7/Dlg4 | 7 |
| GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Gpc6/Nptx1/Dlg2/Nptxr/Magi2/Cacng7/Dlg4 | 7 |
| GO:1904752 | regulation of vascular associated smooth muscle cell migration | BP | 0.226 | 0.000555 | 0.00392 | 2.41 | 0.00254 | Ddr2/Drd4/Tert/Myo5a/Mef2c/Adamts1/Pcsk5 | 7 |
| GO:1903053 | regulation of extracellular matrix organization | BP | 0.18 | 0.000562 | 0.00396 | 2.4 | 0.00257 | Sema5a/Ddr2/Prdm5/Phldb2/Ets1/Has2/Emilin1/Tie1/Efemp2 | 9 |
| GO:0050728 | negative regulation of inflammatory response | BP | 0.118 | 0.000571 | 0.00402 | 2.4 | 0.00261 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Ets1/Foxf1/Fndc4/Cdh5/Apoe/Cd200/Tnfaip6/Calcrl/Nr1h3/Spn/Wfdc1 | 17 |
| GO:0008406 | gonad development | BP | 0.1 | 0.000593 | 0.00417 | 2.38 | 0.0027 | Sfrp2/Ar/Angpt1/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Rhobtb3/Kitl/Arrb1/Tnfaip6/Hoxa9/Arrb2/Stat5b/Osr1/Abcb1a | 24 |
| GO:2000116 | regulation of cysteine-type endopeptidase activity | BP | 0.1 | 0.000593 | 0.00417 | 2.38 | 0.0027 | Hgf/Sfrp2/Igf1/Cd44/Bok/Wdr35/Grin2b/Mical1/Ift57/Wnt9a/Ppm1f/Hip1/Arrb1/Epha7/Dlc1/Arrb2/Asph/Fyn/Nlrp1b/Ccn2/Mgmt/Cyfip2/F2r/Myc | 24 |
| GO:0009065 | glutamine family amino acid catabolic process | BP | 0.261 | 0.000608 | 0.00425 | 2.37 | 0.00276 | Ddah1/Nos3/Asrgl1/Fah/Atp2b4/Prodh | 6 |
| GO:0015812 | gamma-aminobutyric acid transport | BP | 0.261 | 0.000608 | 0.00425 | 2.37 | 0.00276 | Grik1/Abat/Htr1b/Gabbr1/Cacna1a/Cacnb4 | 6 |
| GO:0050961 | detection of temperature stimulus involved in sensory perception | BP | 0.261 | 0.000608 | 0.00425 | 2.37 | 0.00276 | Ano1/Cxcl12/Calca/Grik2/Htr2a/Arrb2 | 6 |
| GO:0034766 | negative regulation of ion transmembrane transport | BP | 0.131 | 0.000612 | 0.00427 | 2.37 | 0.00277 | Hecw2/Drd4/Dysf/Bin1/Osr1/Pkd2/Rrad/Ptger3/Gnb5/Gstm7/Atp1a2/Sri/Cav1/Twist1 | 14 |
| GO:0007368 | determination of left/right symmetry | BP | 0.121 | 0.000613 | 0.00428 | 2.37 | 0.00277 | Ihh/Tbx2/Mef2c/Foxf1/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1/Psen1/Hand2/Eng | 16 |
| GO:0033077 | T cell differentiation in thymus | BP | 0.137 | 0.000615 | 0.00428 | 2.37 | 0.00277 | Ihh/Cd3g/Gli3/Dock2/Zeb1/Jag2/Cd1d1/Stat5b/Zap70/Il2rg/Spn/Il7r/Bcl11b | 13 |
| GO:1901879 | regulation of protein depolymerization | BP | 0.137 | 0.000615 | 0.00428 | 2.37 | 0.00277 | Map2/Sema5a/Evl/Tmod2/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2/Plek | 13 |
| GO:0061035 | regulation of cartilage development | BP | 0.153 | 0.000619 | 0.0043 | 2.37 | 0.00279 | Mdk/Hoxd11/Nog/Pkdcc/Hoxa11/Gli3/Wnt9a/Adamts7/Ltbp3/Ccn2/Mboat2 | 11 |
| GO:0016331 | morphogenesis of embryonic epithelium | BP | 0.108 | 0.000621 | 0.00431 | 2.37 | 0.00279 | Sfrp2/Wnt6/Ar/Nog/Zeb2/Ift57/Gdf7/Lrp2/Jag2/Cecr2/Vegfc/Prickle1/Ipmk/Dlc1/Ptch1/Osr1/Rab23/Fzd2/Bmp7/Twist1 | 20 |
| GO:0035051 | cardiocyte differentiation | BP | 0.108 | 0.000621 | 0.00431 | 2.37 | 0.00279 | AW551984/Sgcd/Col14a1/Igf1/Adra1a/Slc8a1/Mef2c/Nox4/Tbx3/Ednra/Prickle1/Arrb2/Pi16/Abi3bp/Sgcb/Pdgfrb/Bmp7/Hey2/Hand2/Twist1 | 20 |
| GO:0030513 | positive regulation of BMP signaling pathway | BP | 0.195 | 0.000646 | 0.00447 | 2.35 | 0.0029 | Kdr/Rnf165/Gpc3/Zfp423/Cdh5/Kcp/Neo1/Eng | 8 |
| GO:0051968 | positive regulation of synaptic transmission, glutamatergic | BP | 0.195 | 0.000646 | 0.00447 | 2.35 | 0.0029 | Gria4/Nlgn1/Grin2b/Shank2/Tshz3/Nlgn3/Cacng7/Nlgn2 | 8 |
| GO:0008306 | associative learning | BP | 0.125 | 0.000648 | 0.00448 | 2.35 | 0.0029 | Cacna1c/Nog/Drd4/B4galt2/Grin2b/Pde1b/Adcy3/Shank2/Cacna1e/Kit/Nlgn3/Ddhd2/Atp1a2/Syngap1/Brsk1 | 15 |
| GO:0010518 | positive regulation of phospholipase activity | BP | 0.176 | 0.000654 | 0.00451 | 2.35 | 0.00292 | Adra1a/Itk/Arhgap6/Agtr1a/Sele/Adcyap1r1/Htr2a/Dlc1/Pdgfrb | 9 |
| GO:0002260 | lymphocyte homeostasis | BP | 0.143 | 0.000665 | 0.00457 | 2.34 | 0.00296 | Cd44/Mef2c/Sh2b2/Tnfsf13b/Dock10/Coro1a/Enpp1/Nckap1l/Stat5b/Gimap5/Gimap3/Lat | 12 |
| GO:0006809 | nitric oxide biosynthetic process | BP | 0.143 | 0.000665 | 0.00457 | 2.34 | 0.00296 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0050848 | regulation of calcium-mediated signaling | BP | 0.143 | 0.000665 | 0.00457 | 2.34 | 0.00296 | Kdr/Igf1/Drd4/Lrrk2/Slc24a4/Cdh13/Itgal/Cmya5/Atp2b4/Zap70/Cd4/Tbc1d10c | 12 |
| GO:0060191 | regulation of lipase activity | BP | 0.143 | 0.000665 | 0.00457 | 2.34 | 0.00296 | Adra1a/Itk/Arhgap6/Agtr1a/Sele/Adcyap1r1/Htr2a/Hdac9/Dlc1/Angptl4/Pdgfrb/Nr1h3 | 12 |
| GO:0050868 | negative regulation of T cell activation | BP | 0.12 | 0.000666 | 0.00457 | 2.34 | 0.00296 | Ihh/Mdk/Cd44/Gli3/Loxl3/H2-Ab1/Btla/Havcr2/Nckap1l/Pde5a/H2-Aa/Dlg5/Gimap5/Gimap3/Spn/Lax1 | 16 |
| GO:0002573 | myeloid leukocyte differentiation | BP | 0.0992 | 0.000668 | 0.00457 | 2.34 | 0.00296 | Lef1/Cd109/Hcls1/Gpc3/Mef2c/Inpp4b/Lrrc17/Plcb1/Calca/Kit/Enpp1/Tnfrsf11b/Zbtb46/Kitl/Tal1/Tnfaip6/Rassf2/Nrros/Gimap5/Gimap3/Psen1/Batf/Cd4/Myc | 24 |
| GO:0051150 | regulation of smooth muscle cell differentiation | BP | 0.219 | 0.000682 | 0.00466 | 2.33 | 0.00302 | Zeb1/Tshz3/Kit/Cth/Rbpms2/Efemp2/Eng | 7 |
| GO:0060977 | coronary vasculature morphogenesis | BP | 0.219 | 0.000682 | 0.00466 | 2.33 | 0.00302 | Angpt1/Sgcd/Nrp1/Lrp2/Pdgfrb/Hey2/Hand2 | 7 |
| GO:0050680 | negative regulation of epithelial cell proliferation | BP | 0.11 | 0.000692 | 0.00472 | 2.33 | 0.00306 | Sfrp2/Ar/Cd109/Gkn3/Gpc3/Mef2c/Cxcr3/Vash1/Ptprm/Ift57/Cdkn1c/Apoe/Klf9/Ptch1/Aqp11/Ptn/Cav1/Wfdc1/Eng | 19 |
| GO:0043954 | cellular component maintenance | BP | 0.151 | 0.000698 | 0.00474 | 2.32 | 0.00307 | Epb41l3/Fermt2/Grin2b/Shank2/Apoe/Erc2/Fyn/Syngap1/Zmynd8/Cldn1/F2r | 11 |
| GO:0046637 | regulation of alpha-beta T cell differentiation | BP | 0.151 | 0.000698 | 0.00474 | 2.32 | 0.00307 | Ihh/Cd83/Gli3/Loxl3/Prdm1/Cd1d1/Nckap1l/Gimap5/Zap70/Gimap3/Il2rg | 11 |
| GO:0048678 | response to axon injury | BP | 0.151 | 0.000698 | 0.00474 | 2.32 | 0.00307 | Gap43/Mmp2/Matn2/Enpp1/Rtn4rl1/Neo1/Sarm1/Xylt1/Dpysl3/Ptn/Nrep | 11 |
| GO:0001947 | heart looping | BP | 0.161 | 0.000699 | 0.00474 | 2.32 | 0.00307 | Ihh/Tbx2/Mef2c/Ift57/Aplnr/Tbx3/Pkd2/Psen1/Hand2/Eng | 10 |
| GO:0051058 | negative regulation of small GTPase mediated signal transduction | BP | 0.161 | 0.000699 | 0.00474 | 2.32 | 0.00307 | Adra1a/Rasa3/Arhgap25/Sh3bp1/Rasip1/Arap3/Heg1/Dlc1/Syngap1/Rasal3 | 10 |
| GO:0051963 | regulation of synapse assembly | BP | 0.124 | 0.000707 | 0.00479 | 2.32 | 0.0031 | Gpc6/Ptprd/St8sia2/Nlgn1/Lrrc4b/Mef2c/Nptx1/Adgrl1/Slitrk3/Nlgn3/Epha7/Bhlhb9/Cux2/Dlg5/Nlgn2 | 15 |
| GO:0072521 | purine-containing compound metabolic process | BP | 0.0867 | 0.000708 | 0.00479 | 2.32 | 0.0031 | Ttr/Nme4/Acsl6/Igf1/Hmgcs2/Nos3/Acacb/Lrrk2/Adcy3/Gamt/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Lacc1/Htr2a/Acot7/Zbtb20/Pde5a/Rab23/Acsl1/Impdh1/Dpyd/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 34 |
| GO:0006865 | amino acid transport | BP | 0.112 | 0.000713 | 0.0048 | 2.32 | 0.00311 | Slc1a3/Grik1/Abat/Slc7a2/Grin2b/Slc36a4/Htr1b/Slc6a15/Sfxn5/Slc38a1/Nfkbie/Slc7a7/Psen1/Gabbr1/Cacna1a/Lrrc8c/Cacnb4/Myc | 18 |
| GO:0044065 | regulation of respiratory system process | BP | 0.312 | 0.000713 | 0.0048 | 2.32 | 0.00311 | Nlgn1/Tshz3/Nlgn3/Atp1a2/Nlgn2 | 5 |
| GO:0060046 | regulation of acrosome reaction | BP | 0.312 | 0.000713 | 0.0048 | 2.32 | 0.00311 | Pla2g10/Spink1/Plcb1/Cacna1h/Pkdrej | 5 |
| GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | BP | 0.312 | 0.000713 | 0.0048 | 2.32 | 0.00311 | Plxnb1/Nrp1/Mef2c/Plxnd1/Ednra | 5 |
| GO:0045619 | regulation of lymphocyte differentiation | BP | 0.104 | 0.000722 | 0.00485 | 2.31 | 0.00314 | Ihh/Mdk/Lef1/Cd83/Cd44/Pik3r6/Gli3/Flt3/Loxl3/Zeb1/Prdm1/Cd1d1/Sox12/Nckap1l/Stat5b/H2-Aa/Gimap5/Zap70/Gimap3/Il2rg/Il7r | 21 |
| GO:0032411 | positive regulation of transporter activity | BP | 0.119 | 0.000723 | 0.00485 | 2.31 | 0.00314 | Tcaf1/Drd4/Cacna2d1/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Cacnb4 | 16 |
| GO:1903038 | negative regulation of leukocyte cell-cell adhesion | BP | 0.116 | 0.000724 | 0.00485 | 2.31 | 0.00314 | Ihh/Mdk/Cd44/Cxcl12/Gli3/Loxl3/H2-Ab1/Btla/Havcr2/Nckap1l/Pde5a/H2-Aa/Dlg5/Gimap5/Gimap3/Spn/Lax1 | 17 |
| GO:0007548 | sex differentiation | BP | 0.0915 | 0.000738 | 0.00494 | 2.31 | 0.0032 | Tex15/Stra6/Sfrp2/Ar/Angpt1/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Tbx3/Rhobtb3/Kitl/Arrb1/Tnfaip6/Hoxa9/Smad9/Arrb2/Stat5b/Osr1/Abcb1a/Lhfpl2 | 29 |
| GO:0034103 | regulation of tissue remodeling | BP | 0.141 | 0.000741 | 0.00495 | 2.31 | 0.00321 | Mdk/Ddr2/Inpp4b/Erg/Calca/Tnfrsf11b/Cd38/Syt7/Ltbp3/Abi3bp/Def8/Hand2 | 12 |
| GO:0002028 | regulation of sodium ion transport | BP | 0.134 | 0.000751 | 0.00501 | 2.3 | 0.00325 | Hecw2/Slc8a1/Nos3/Scn4b/Drd4/Fxyd5/Serpine2/Plcb1/Osr1/Akt3/Atp2b4/Atp1a2/Mllt6 | 13 |
| GO:0002718 | regulation of cytokine production involved in immune response | BP | 0.134 | 0.000751 | 0.00501 | 2.3 | 0.00325 | Angpt1/Sema7a/Tek/Gprc5b/Tril/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Clnk/Cd96 | 13 |
| GO:0032715 | negative regulation of interleukin-6 production | BP | 0.173 | 0.000757 | 0.00504 | 2.3 | 0.00326 | Hgf/Flt3/Il27ra/Havcr2/Cd200/Nckap1l/Arrb1/Arrb2/Cd84 | 9 |
| GO:1902622 | regulation of neutrophil migration | BP | 0.173 | 0.000757 | 0.00504 | 2.3 | 0.00326 | Mdk/Jam3/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Ptger3/Mpp1 | 9 |
| GO:0014046 | dopamine secretion | BP | 0.19 | 0.000765 | 0.00507 | 2.3 | 0.00328 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 8 |
| GO:0014059 | regulation of dopamine secretion | BP | 0.19 | 0.000765 | 0.00507 | 2.3 | 0.00328 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 8 |
| GO:0031076 | embryonic camera-type eye development | BP | 0.19 | 0.000765 | 0.00507 | 2.3 | 0.00328 | Stra6/Ihh/Tbx2/Zeb1/Hipk2/Hipk1/Bmp7/Twist1 | 8 |
| GO:0070507 | regulation of microtubule cytoskeleton organization | BP | 0.112 | 0.000767 | 0.00508 | 2.29 | 0.00329 | Map2/Phldb2/Mpdz/Gnai1/Cdh5/Map9/Epha3/Rp1/Tubb4a/Dysf/Pkd1/Nin/Slain1/Ttbk2/Cep97/Mapre2/Cav1/Tppp | 18 |
| GO:0002693 | positive regulation of cellular extravasation | BP | 0.25 | 0.000777 | 0.00513 | 2.29 | 0.00332 | Mdk/Icam1/Jam3/Thy1/Pecam1/Ptger3 | 6 |
| GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | BP | 0.25 | 0.000777 | 0.00513 | 2.29 | 0.00332 | Ptp4a3/Jcad/Tcf4/Atp2b4/Adgra2/Sema6a | 6 |
| GO:0009064 | glutamine family amino acid metabolic process | BP | 0.159 | 0.000796 | 0.00523 | 2.28 | 0.00339 | Ddah1/Nos3/Asrgl1/Fah/Amdhd1/Phgdh/Atp2b4/Slc7a7/Asl/Prodh | 10 |
| GO:0035306 | positive regulation of dephosphorylation | BP | 0.159 | 0.000796 | 0.00523 | 2.28 | 0.00339 | Npnt/Mef2c/Itga1/Cdh5/Ppp1r16b/Magi2/Dlc1/Smpd1/Pdgfrb/Plek | 10 |
| GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | BP | 0.159 | 0.000796 | 0.00523 | 2.28 | 0.00339 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Atp2b4/Apln/Pld2/Plek | 10 |
| GO:0044282 | small molecule catabolic process | BP | 0.0898 | 0.000813 | 0.00534 | 2.27 | 0.00346 | Ddah1/Npl/Abat/Nos3/Acacb/Cbs/Asrgl1/Inpp4b/Fah/Amdhd1/Bdh2/Hdc/Apoe/Bpnt1/Echs1/Acot7/Cyp46a1/Sord/Atp2b4/Abcd2/Dpyd/Dhdh/Lipe/Sardh/Prodh/Pfkm/Twist1/Abhd3/Echdc2/Irs1 | 30 |
| GO:0006469 | negative regulation of protein kinase activity | BP | 0.103 | 0.000821 | 0.00538 | 2.27 | 0.00349 | Sfrp2/Slc8a1/Ptpro/Mapk8ip1/Slc8a3/Pkia/Thy1/Ppm1f/Cdkn1c/Apoe/Rasip1/Ppm1e/Pkib/Heg1/Lats2/Smpd1/Psen1/Cav1/Bmp7/Prkar1b/Lax1 | 21 |
| GO:0060415 | muscle tissue morphogenesis | BP | 0.14 | 0.000824 | 0.0054 | 2.27 | 0.0035 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Fzd2/Efemp2/Hey2/Eng | 12 |
| GO:0042632 | cholesterol homeostasis | BP | 0.133 | 0.000829 | 0.0054 | 2.27 | 0.0035 | Apob/Pla2g10/Cyp7b1/Nr5a2/Soat1/Tsku/Apoe/Hdac9/Abca5/Abcg1/Cav1/Nr1h3/Abcg4 | 13 |
| GO:0009187 | cyclic nucleotide metabolic process | BP | 0.212 | 0.00083 | 0.0054 | 2.27 | 0.0035 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7/Pde5a/Cacnb4 | 7 |
| GO:0048566 | embryonic digestive tract development | BP | 0.212 | 0.00083 | 0.0054 | 2.27 | 0.0035 | Stra6/Ihh/Pkdcc/Foxf1/Tcf21/Gli3/Pcsk5 | 7 |
| GO:0048841 | regulation of axon extension involved in axon guidance | BP | 0.212 | 0.00083 | 0.0054 | 2.27 | 0.0035 | Sema5a/Sema5b/Nrp1/Sema7a/Cxcl12/Sema3g/Sema6a | 7 |
| GO:1904646 | cellular response to amyloid-beta | BP | 0.212 | 0.00083 | 0.0054 | 2.27 | 0.0035 | Itga4/Igf1/Cacna2d1/Icam1/Fyn/Psen1/Cacna1a | 7 |
| GO:0051494 | negative regulation of cytoskeleton organization | BP | 0.108 | 0.000854 | 0.00555 | 2.26 | 0.0036 | Kank4/Map2/Evl/Phldb2/Tmod2/Pfn2/Cdh5/Coro1a/Rp1/Arhgap6/Tubb4a/Pecam1/Tmeff2/Scin/Dysf/Dlc1/Arhgap28/Ttbk2/Plekhh2 | 19 |
| GO:0043392 | negative regulation of DNA binding | BP | 0.17 | 0.000874 | 0.00565 | 2.25 | 0.00366 | Lef1/Zfp462/Twist2/Habp4/Zfp90/Psen1/Sri/Hey2/Hand2 | 9 |
| GO:0045907 | positive regulation of vasoconstriction | BP | 0.17 | 0.000874 | 0.00565 | 2.25 | 0.00366 | Adra1a/Icam1/Wdr35/Cd38/Htr2a/Pde5a/Ptger3/Cav1/F2r | 9 |
| GO:0046638 | positive regulation of alpha-beta T cell differentiation | BP | 0.17 | 0.000874 | 0.00565 | 2.25 | 0.00366 | Ihh/Cd83/Gli3/Cd1d1/Nckap1l/Gimap5/Zap70/Gimap3/Il2rg | 9 |
| GO:0060412 | ventricular septum morphogenesis | BP | 0.17 | 0.000874 | 0.00565 | 2.25 | 0.00366 | Wnt11/Nog/Nos3/Aplnr/Tbx3/Heyl/Fzd2/Hey2/Slit3 | 9 |
| GO:0070169 | positive regulation of biomineral tissue development | BP | 0.17 | 0.000874 | 0.00565 | 2.25 | 0.00366 | Wnt6/Pkdcc/Slc8a1/Mef2c/Osr2/Nell1/Osr1/Ptn/Bmp7 | 9 |
| GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Ednra/Bmp7/Hand2/Twist1 | 4 |
| GO:0006527 | arginine catabolic process | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Ddah1/Nos3/Fah/Atp2b4 | 4 |
| GO:0060385 | axonogenesis involved in innervation | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Itga4/Nrp1/Nptx1/Ednra | 4 |
| GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Slc8a1/Gjc1/Sri/Cav1 | 4 |
| GO:0097119 | postsynaptic density protein 95 clustering | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Nlgn1/Cdh2/Zmynd8/Nlgn2 | 4 |
| GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.4 | 0.000884 | 0.00567 | 2.25 | 0.00368 | Ppp1r16b/Aplnr/Jcad/Agtr1a | 4 |
| GO:0062197 | cellular response to chemical stress | BP | 0.0893 | 0.000894 | 0.00573 | 2.24 | 0.00372 | Cyp1b1/Hgf/Ddr2/Slc8a1/Nos3/Lrrk2/Mmp2/Prkd1/Tbc1d24/Trpc6/Scn2a/Aifm2/Prr5l/Ednra/Dysf/Bdkrb2/Selenon/Pkd2/Net1/Fyn/Mgat3/Slc25a23/Stx2/Pdgfrb/Meak7/Pde8a/Cav1/Lrrc8c/Setx/Fbln5 | 30 |
| GO:0030501 | positive regulation of bone mineralization | BP | 0.186 | 9e-04 | 0.00575 | 2.24 | 0.00373 | Pkdcc/Slc8a1/Mef2c/Osr2/Nell1/Osr1/Ptn/Bmp7 | 8 |
| GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | BP | 0.186 | 9e-04 | 0.00575 | 2.24 | 0.00373 | Cxcr3/Thy1/Aplnr/Ednra/Pkd2/Gstm7/Sri/F2r | 8 |
| GO:0086002 | cardiac muscle cell action potential involved in contraction | BP | 0.186 | 9e-04 | 0.00575 | 2.24 | 0.00373 | Cacna1c/Kcnd3/Scn4b/Scn1a/Cacna2d1/Ank2/Bin1/Cav1 | 8 |
| GO:0008347 | glial cell migration | BP | 0.156 | 0.000903 | 0.00575 | 2.24 | 0.00373 | Ptprz1/Apcdd1/Cspg4/Fn1/Dab1/Gli3/Ndn/Matn2/Enpp1/Vcan | 10 |
| GO:0050435 | amyloid-beta metabolic process | BP | 0.156 | 0.000903 | 0.00575 | 2.24 | 0.00373 | Igf1/Aph1b/Rtn1/Slc2a13/Mme/Apoe/Bin1/Abcg1/Mgat3/Psen1 | 10 |
| GO:0060193 | positive regulation of lipase activity | BP | 0.156 | 0.000903 | 0.00575 | 2.24 | 0.00373 | Adra1a/Itk/Arhgap6/Agtr1a/Sele/Adcyap1r1/Htr2a/Dlc1/Pdgfrb/Nr1h3 | 10 |
| GO:0006672 | ceramide metabolic process | BP | 0.131 | 0.000913 | 0.00579 | 2.24 | 0.00375 | St8sia2/St8sia4/Agk/Sptlc3/Cers4/St3gal2/Plpp3/St6galnac4/Degs2/St8sia6/Plpp1/St6galnac6/Smpd1 | 13 |
| GO:0055092 | sterol homeostasis | BP | 0.131 | 0.000913 | 0.00579 | 2.24 | 0.00375 | Apob/Pla2g10/Cyp7b1/Nr5a2/Soat1/Tsku/Apoe/Hdac9/Abca5/Abcg1/Cav1/Nr1h3/Abcg4 | 13 |
| GO:0002286 | T cell activation involved in immune response | BP | 0.121 | 0.000914 | 0.00579 | 2.24 | 0.00375 | Mdk/Lef1/Icam1/Eif2ak4/Loxl3/Itgal/Cracr2a/Havcr2/Nckap1l/Psen1/Apbb1ip/Rab27a/Batf/Spn/Ly9 | 15 |
| GO:0032410 | negative regulation of transporter activity | BP | 0.138 | 0.000915 | 0.00579 | 2.24 | 0.00375 | Hecw2/Drd4/Dysf/Osr1/Pkd2/Rrad/Gnb5/Gstm7/Atp1a2/Sri/Cav1/Twist1 | 12 |
| GO:0031623 | receptor internalization | BP | 0.117 | 0.000921 | 0.00582 | 2.23 | 0.00377 | Angpt1/Drd4/Calca/Htr1b/Magi2/Sele/Arrb1/Cacng7/Arrb2/Calcrl/Dlg4/Ptger3/Apln/Cav1/Pld2/Ankrd13a | 16 |
| GO:0046395 | carboxylic acid catabolic process | BP | 0.1 | 0.000925 | 0.00584 | 2.23 | 0.00379 | Ddah1/Npl/Abat/Nos3/Acacb/Cbs/Asrgl1/Fah/Amdhd1/Bdh2/Hdc/Echs1/Acot7/Atp2b4/Abcd2/Lipe/Sardh/Prodh/Twist1/Abhd3/Echdc2/Irs1 | 22 |
| GO:0001845 | phagolysosome assembly | BP | 0.294 | 0.00097 | 0.00611 | 2.21 | 0.00396 | Rab34/Srpx/Coro1a/Syt7/Rab20 | 5 |
| GO:0097094 | craniofacial suture morphogenesis | BP | 0.294 | 0.00097 | 0.00611 | 2.21 | 0.00396 | Mmp16/Gli3/Frem1/Rab23/Twist1 | 5 |
| GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | BP | 0.294 | 0.00097 | 0.00611 | 2.21 | 0.00396 | Plxnb1/Nrp1/Mef2c/Plxnd1/Ednra | 5 |
| GO:0007620 | copulation | BP | 0.24 | 0.000982 | 0.00616 | 2.21 | 0.00399 | Ar/Abat/Drd4/Ednrb/Serpine2/Ednra | 6 |
| GO:0035988 | chondrocyte proliferation | BP | 0.24 | 0.000982 | 0.00616 | 2.21 | 0.00399 | Ihh/Mmp16/Lef1/Ddr2/Stc1/Ccn2 | 6 |
| GO:2000311 | regulation of AMPA receptor activity | BP | 0.24 | 0.000982 | 0.00616 | 2.21 | 0.00399 | Nlgn1/Mef2c/Nlgn3/Prrt1/Cacng7/Nlgn2 | 6 |
| GO:0046847 | filopodium assembly | BP | 0.145 | 0.000985 | 0.00617 | 2.21 | 0.004 | Gap43/Nlgn1/Nrp1/Sh3bp1/Daam2/Fgd1/Palm/Dpysl3/Itgb4/Zmynd8/Fgd2 | 11 |
| GO:0099054 | presynapse assembly | BP | 0.167 | 0.001 | 0.00628 | 2.2 | 0.00407 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn3/Pcdh17/Lrp4/Nlgn2 | 9 |
| GO:0016054 | organic acid catabolic process | BP | 0.0995 | 0.00104 | 0.00651 | 2.19 | 0.00422 | Ddah1/Npl/Abat/Nos3/Acacb/Cbs/Asrgl1/Fah/Amdhd1/Bdh2/Hdc/Echs1/Acot7/Atp2b4/Abcd2/Lipe/Sardh/Prodh/Twist1/Abhd3/Echdc2/Irs1 | 22 |
| GO:0010464 | regulation of mesenchymal cell proliferation | BP | 0.182 | 0.00105 | 0.00657 | 2.18 | 0.00426 | Ihh/Wnt11/Kdr/Foxf1/Zeb1/Ptn/Myc/Irs1 | 8 |
| GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | BP | 0.182 | 0.00105 | 0.00657 | 2.18 | 0.00426 | Cacna2d1/Ank2/Cracr2a/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 8 |
| GO:0046660 | female sex differentiation | BP | 0.112 | 0.00106 | 0.00657 | 2.18 | 0.00426 | Stra6/Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lrp2/Lfng/Kit/Tbx3/Kitl/Arrb1/Tnfaip6/Arrb2/Stat5b/Lhfpl2 | 17 |
| GO:0051099 | positive regulation of binding | BP | 0.104 | 0.00106 | 0.00657 | 2.18 | 0.00426 | Igf1/Tert/Lrrk2/Plxnd1/Lfng/Apoe/Caprin2/Pkd1/Hipk2/Arrb1/Cldn5/Mfng/Psen1/Hipk1/Cav1/Dact1/Gpsm1/Hand2/Myc/Twist1 | 20 |
| GO:0010876 | lipid localization | BP | 0.0824 | 0.00109 | 0.00674 | 2.17 | 0.00437 | Apob/Stra6/Nme4/Acsl6/Pla2g10/Acacb/Drd4/Vstm2a/Mest/Soat1/Abcg2/Rbp7/Osbp2/Pltp/Selenom/Enpp1/Agtr1a/Apoe/Syt7/Dysf/Bdkrb2/Abca5/Abcg1/Stat5b/Abca8b/Abcd2/Acsl1/Atp10a/Abcb1a/Osbpl1a/Cav1/Nr1h3/Xkr5/Atp8b4/Prelid3a/Abcg4/Cds1 | 37 |
| GO:0009410 | response to xenobiotic stimulus | BP | 0.0892 | 0.00109 | 0.00676 | 2.17 | 0.00438 | Kcnk3/Cyp1b1/Slc1a3/Sfrp2/Kdr/Adra1a/Abat/Mmp2/Mef2c/Shank2/Ptprm/Htr1b/Fmo1/Ppm1f/Cd38/Igf2/Ppm1e/Nckap1l/Htr2a/Cyp46a1/Acsl1/Gstm7/Dpyd/Gstm1/Abcb1a/Sult1a1/Bche/Myc/Rap2a | 29 |
| GO:0033135 | regulation of peptidyl-serine phosphorylation | BP | 0.108 | 0.0011 | 0.00679 | 2.17 | 0.0044 | Hgf/Sfrp2/Angpt1/Hcls1/Cd44/Pfn2/Prkd1/Tek/Ppm1f/Caprin2/Bdkrb2/Arrb1/Rassf2/Arrb2/Atp2b4/Cav1/Sh2d3c/Camk1 | 18 |
| GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | BP | 0.129 | 0.0011 | 0.00683 | 2.17 | 0.00442 | Adra1a/Npr3/Ednrb/Ano1/Lpar4/Calca/Pth1r/Agtr1a/Ednra/Htr2a/Plcb2/Ptger3/F2r | 13 |
| GO:0045637 | regulation of myeloid cell differentiation | BP | 0.0991 | 0.00111 | 0.00683 | 2.17 | 0.00442 | Lef1/P4htm/Hcls1/Ets1/Mef2c/Inpp4b/Lrrc17/Calca/Tnfrsf11b/Gpr171/Zbtb46/Scin/Nckap1l/Kitl/Tal1/Tnfaip6/Hoxa9/Twist2/Rassf2/Stat5b/Cd4/Myc | 22 |
| GO:0048469 | cell maturation | BP | 0.0991 | 0.00111 | 0.00683 | 2.17 | 0.00442 | Epha8/Ihh/Pla2g10/Kdr/Spink1/Cspg4/Lrrk2/Ednrb/Gdf11/Flvcr1/Plcb1/Calca/Rps6ka2/Pth1r/Dlg2/Cdkn1c/Nfasc/Kcnma1/Ednra/Tal1/Lhx6/Septin4 | 22 |
| GO:0030010 | establishment of cell polarity | BP | 0.111 | 0.00114 | 0.00701 | 2.15 | 0.00454 | Ankfn1/Map2/Fez1/Foxf1/Jam3/Dock2/Myo9a/Cdh5/Msn/Frmd4b/Ubxn2b/Prickle2/Sh3bp1/Pkd1/Pard3b/Frmd4a/Brsk1 | 17 |
| GO:0010712 | regulation of collagen metabolic process | BP | 0.164 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Ihh/Ddr2/Fn1/Emilin1/Arrb2/Ccn2/Pdgfrb/Cygb/F2r | 9 |
| GO:0055010 | ventricular cardiac muscle tissue morphogenesis | BP | 0.164 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Col11a1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Hey2/Eng | 9 |
| GO:0001818 | negative regulation of cytokine production | BP | 0.0912 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Hgf/Acod1/Angpt1/Lef1/Wnt11/Pla2g10/Cd83/Igf1/Fn1/Flt3/Adcy7/Il27ra/Tsku/Havcr2/Cd200/Nckap1l/Arrb1/Hdac9/Twist2/Mapkbp1/Arrb2/Cd84/Gimap5/Abcd2/Gimap3/Cd96/Twist1 | 27 |
| GO:0031532 | actin cytoskeleton reorganization | BP | 0.123 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Hmcn1/Mdk/Nrp1/Thsd7b/Tek/Syde1/Kit/Esam/Parvg/Efs/Carmil3/Parvb/Plek/Rap2a | 14 |
| GO:0002715 | regulation of natural killer cell mediated immunity | BP | 0.152 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Pik3r6/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gimap3/Clnk/Cd96 | 10 |
| GO:0002720 | positive regulation of cytokine production involved in immune response | BP | 0.152 | 0.00115 | 0.00707 | 2.15 | 0.00458 | Sema7a/Tek/Gprc5b/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Clnk | 10 |
| GO:0001573 | ganglioside metabolic process | BP | 0.231 | 0.00122 | 0.00749 | 2.13 | 0.00485 | St8sia2/St8sia4/St3gal2/St6galnac4/St8sia6/St6galnac6 | 6 |
| GO:2000050 | regulation of non-canonical Wnt signaling pathway | BP | 0.231 | 0.00122 | 0.00749 | 2.13 | 0.00485 | Sfrp2/Nkd1/Gpc3/Znrf3/Daam2/Dact1 | 6 |
| GO:0032413 | negative regulation of ion transmembrane transporter activity | BP | 0.141 | 0.00123 | 0.00749 | 2.13 | 0.00485 | Hecw2/Drd4/Dysf/Osr1/Pkd2/Rrad/Gnb5/Gstm7/Atp1a2/Sri/Cav1 | 11 |
| GO:0046209 | nitric oxide metabolic process | BP | 0.133 | 0.00124 | 0.00756 | 2.12 | 0.0049 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0009855 | determination of bilateral symmetry | BP | 0.113 | 0.00125 | 0.00764 | 2.12 | 0.00495 | Ihh/Tbx2/Mef2c/Foxf1/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1/Psen1/Hand2/Eng | 16 |
| GO:0051146 | striated muscle cell differentiation | BP | 0.0884 | 0.00125 | 0.00764 | 2.12 | 0.00495 | AW551984/Sgcd/Col14a1/Ryr1/Igf1/Adra1a/Slc8a1/P2rx2/Tmod2/Cdh2/Mef2c/Cxcl12/Hopx/Nox4/Tbx3/Igf2/Dysf/Hdac9/Flnc/Neo1/Selenon/Arrb2/Pi16/Sgcb/Plekho1/Pdgfrb/Hacd1/Hey2/Hand2 | 29 |
| GO:0048640 | negative regulation of developmental growth | BP | 0.117 | 0.00127 | 0.00771 | 2.11 | 0.00499 | Map2/Sema5a/H19/Nog/Sema5b/Nrp1/Nkd1/Sema7a/Sema3g/Epha7/Ptch1/Pi16/Sema6a/Ulk2/Draxin | 15 |
| GO:0007614 | short-term memory | BP | 0.278 | 0.00129 | 0.00783 | 2.11 | 0.00507 | Mdk/Aph1b/Drd4/Pde5a/Cux2 | 5 |
| GO:0070486 | leukocyte aggregation | BP | 0.278 | 0.00129 | 0.00783 | 2.11 | 0.00507 | Cd44/Has2/Msn/Bmp7/Jam2 | 5 |
| GO:0060350 | endochondral bone morphogenesis | BP | 0.149 | 0.0013 | 0.00787 | 2.1 | 0.0051 | Ihh/Mmp16/Hoxd11/Thbs3/Cbs/Hoxa11/Mef2c/Stc1/Col13a1/Enpp1 | 10 |
| GO:1903539 | protein localization to postsynaptic membrane | BP | 0.161 | 0.00131 | 0.00795 | 2.1 | 0.00515 | Gpc6/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 9 |
| GO:0001704 | formation of primary germ layer | BP | 0.126 | 0.00132 | 0.008 | 2.1 | 0.00518 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Fn1/Foxf1/Col5a2/Snai1/Tal1/Ets2/Kif16b/Bmp7 | 13 |
| GO:0021761 | limbic system development | BP | 0.126 | 0.00132 | 0.008 | 2.1 | 0.00518 | Mdk/Lef1/Zeb2/Nrp1/Dab1/Id4/Gli3/Tsku/Tbx3/Ndnf/Dclk2/Atp1a2/Bbs1 | 13 |
| GO:0002399 | MHC class II protein complex assembly | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0002503 | peptide antigen assembly with MHC class II protein complex | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0035336 | long-chain fatty-acyl-CoA metabolic process | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | Acsl6/Them5/Acot7/Acsl1 | 4 |
| GO:0036302 | atrioventricular canal development | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | Tbx2/Has2/Tbx3/Eng | 4 |
| GO:0061299 | retina vasculature morphogenesis in camera-type eye | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | Ndp/Cyp1b1/Nrp1/Arhgef15 | 4 |
| GO:0071415 | cellular response to purine-containing compound | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | Ryr1/Slc8a1/Selenon/Gstm7 | 4 |
| GO:0072075 | metanephric mesenchyme development | BP | 0.364 | 0.00134 | 0.00801 | 2.1 | 0.00519 | Tcf21/Osr1/Bmp7/Myc | 4 |
| GO:0030041 | actin filament polymerization | BP | 0.107 | 0.00135 | 0.00806 | 2.09 | 0.00522 | Kank4/Evl/Hcls1/Tmod2/Pfn2/Icam1/Catip/Coro1a/Esam/Daam2/Pecam1/Scin/Bin1/Nckap1l/Arhgap28/Cdc42ep5/Cyfip2/Pstpip1 | 18 |
| GO:0009799 | specification of symmetry | BP | 0.113 | 0.00135 | 0.00808 | 2.09 | 0.00524 | Ihh/Tbx2/Mef2c/Foxf1/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1/Psen1/Hand2/Eng | 16 |
| GO:0071277 | cellular response to calcium ion | BP | 0.139 | 0.00136 | 0.00814 | 2.09 | 0.00528 | Nlgn1/Syt13/Cpne5/Mef2c/Rasgrp2/Syt7/Asph/Pkd2/Slc25a23/Smpd1/Kcnh1 | 11 |
| GO:0043297 | apical junction assembly | BP | 0.132 | 0.00137 | 0.00814 | 2.09 | 0.00528 | Wnt11/Cldn8/Cdh5/Abi2/Esam/Pecam1/Snai1/Nphp1/Cldn5/Dlg5/Pdcd6ip/Cldn1 | 12 |
| GO:0035710 | CD4-positive, alpha-beta T cell activation | BP | 0.121 | 0.00137 | 0.00814 | 2.09 | 0.00528 | Lef1/Cd83/Cd44/Loxl3/Cracr2a/Nckap1l/Xcl1/Satb1/Gimap5/Gimap3/Il2rg/Batf/Spn/Ly9 | 14 |
| GO:0015850 | organic hydroxy compound transport | BP | 0.0941 | 0.00138 | 0.00822 | 2.09 | 0.00533 | Apob/Stra6/Maob/Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Slc2a13/Htr1b/Selenom/Cadps/Agtr1a/Apoe/Syt7/Htr2a/Abca5/Abcg1/Abca8b/Ptger3/Gabbr1/Nr1h3/Abcg4/Myc | 24 |
| GO:0001838 | embryonic epithelial tube formation | BP | 0.109 | 0.00141 | 0.00837 | 2.08 | 0.00543 | Sfrp2/Wnt6/Nog/Zeb2/Ift57/Gdf7/Lrp2/Cecr2/Prickle1/Ipmk/Dlc1/Ptch1/Osr1/Rab23/Fzd2/Bmp7/Twist1 | 17 |
| GO:2001258 | negative regulation of cation channel activity | BP | 0.174 | 0.00143 | 0.00848 | 2.07 | 0.00549 | Drd4/Dysf/Pkd2/Rrad/Gnb5/Gstm7/Sri/Cav1 | 8 |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | BP | 0.194 | 0.00143 | 0.00849 | 2.07 | 0.0055 | Grik1/Kdr/Grin2b/Kit/Grik2/Dlg4/Syngap1 | 7 |
| GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | BP | 0.125 | 0.00145 | 0.00859 | 2.07 | 0.00557 | Mdk/Ptprd/Nrp1/Fermt2/Tbc1d24/Has2/Obsl1/Caprin2/Bhlhb9/Net1/Cux2/Enpp2/Numbl | 13 |
| GO:0061371 | determination of heart left/right asymmetry | BP | 0.147 | 0.00146 | 0.00864 | 2.06 | 0.0056 | Ihh/Tbx2/Mef2c/Ift57/Aplnr/Tbx3/Pkd2/Psen1/Hand2/Eng | 10 |
| GO:0033673 | negative regulation of kinase activity | BP | 0.0969 | 0.00147 | 0.00873 | 2.06 | 0.00565 | Sfrp2/Ddr2/Slc8a1/Ptpro/Mapk8ip1/Slc8a3/Pkia/Thy1/Ppm1f/Cdkn1c/Apoe/Rasip1/Ppm1e/Pkib/Heg1/Lats2/Smpd1/Psen1/Cav1/Bmp7/Prkar1b/Lax1 | 22 |
| GO:0062237 | protein localization to postsynapse | BP | 0.158 | 0.00149 | 0.00881 | 2.05 | 0.00571 | Gpc6/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 9 |
| GO:0086001 | cardiac muscle cell action potential | BP | 0.158 | 0.00149 | 0.00881 | 2.05 | 0.00571 | Cacna1c/Kcnd2/Kcnd3/Scn4b/Scn1a/Cacna2d1/Ank2/Bin1/Cav1 | 9 |
| GO:1900271 | regulation of long-term synaptic potentiation | BP | 0.158 | 0.00149 | 0.00881 | 2.05 | 0.00571 | Eif2ak4/Mme/Nlgn3/Nrgn/Apoe/Nsg1/Cyp46a1/Ptn/Prkar1b | 9 |
| GO:0035050 | embryonic heart tube development | BP | 0.13 | 0.0015 | 0.00882 | 2.05 | 0.00572 | Ihh/Tbx2/Slc8a1/Mef2c/Ift57/Aplnr/Tbx3/Ednra/Pkd2/Psen1/Hand2/Eng | 12 |
| GO:0043367 | CD4-positive, alpha-beta T cell differentiation | BP | 0.13 | 0.0015 | 0.00882 | 2.05 | 0.00572 | Lef1/Cd83/Loxl3/Cracr2a/Nckap1l/Satb1/Gimap5/Gimap3/Il2rg/Batf/Spn/Ly9 | 12 |
| GO:2001057 | reactive nitrogen species metabolic process | BP | 0.13 | 0.0015 | 0.00882 | 2.05 | 0.00572 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0003176 | aortic valve development | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Nos3/Emilin1/Tie1/Hey2/Slit3/Twist1 | 6 |
| GO:0014829 | vascular associated smooth muscle contraction | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Slc8a1/Ednrb/Edn3/Cd38/Ednra/Htr2a | 6 |
| GO:0016048 | detection of temperature stimulus | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Ano1/Cxcl12/Calca/Grik2/Htr2a/Arrb2 | 6 |
| GO:0034368 | protein-lipid complex remodeling | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:0034369 | plasma lipoprotein particle remodeling | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:0048843 | negative regulation of axon extension involved in axon guidance | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Sema5a/Sema5b/Nrp1/Sema7a/Sema3g/Sema6a | 6 |
| GO:0060004 | reflex | BP | 0.222 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Npnt/Adra1a/Glrb/Kcnma1/Satb1/Cacna1a | 6 |
| GO:0051965 | positive regulation of synapse assembly | BP | 0.138 | 0.00151 | 0.00882 | 2.05 | 0.00572 | Ptprd/St8sia2/Nlgn1/Lrrc4b/Adgrl1/Slitrk3/Nlgn3/Bhlhb9/Cux2/Dlg5/Nlgn2 | 11 |
| GO:1903532 | positive regulation of secretion by cell | BP | 0.0854 | 0.00152 | 0.00884 | 2.05 | 0.00573 | Pla2g10/Igf1/Nlgn1/Abat/Stx1b/Ano1/Grin2b/Cxcl12/Edn3/Rab34/Cadps/Vegfc/Cd38/Syt7/Arrb1/Abcg1/Frmd4a/Ncs1/Cacna1h/Fgr/Apln/Slc4a8/Rab27a/Sri/Gabbr1/Pld2/Pfkm/Golph3l/Pdcd6ip/Nlgn2/Irs1 | 31 |
| GO:0008361 | regulation of cell size | BP | 0.0981 | 0.0016 | 0.00931 | 2.03 | 0.00603 | Map2/Sema5a/Sema5b/Slc12a8/Nrp1/Sema7a/Fn1/Cxcl12/Apoe/Kcnma1/Sema3g/Epha7/Slc12a4/Aqp11/Akt3/Sema6a/Ulk2/C1qtnf9/Il7r/Plek/Draxin | 21 |
| GO:0007009 | plasma membrane organization | BP | 0.108 | 0.00162 | 0.0094 | 2.03 | 0.00609 | Ar/Epb41l3/Ank2/Syt7/Tie1/Tmeff2/Dysf/Bin1/Cavin2/Abcd2/Atp10a/Fat4/Stx2/Smpd1/Cav1/Tlcd2/Xkr5 | 17 |
| GO:0003143 | embryonic heart tube morphogenesis | BP | 0.145 | 0.00163 | 0.00947 | 2.02 | 0.00614 | Ihh/Tbx2/Mef2c/Ift57/Aplnr/Tbx3/Pkd2/Psen1/Hand2/Eng | 10 |
| GO:0099601 | regulation of neurotransmitter receptor activity | BP | 0.145 | 0.00163 | 0.00947 | 2.02 | 0.00614 | Begain/Nlgn1/Mef2c/Nptx1/Nlgn3/Nptxr/Prrt1/Cacng7/Dlg4/Nlgn2 | 10 |
| GO:0051650 | establishment of vesicle localization | BP | 0.105 | 0.00164 | 0.00951 | 2.02 | 0.00616 | Map2/Itga4/Nlgn1/Sec16b/Lrrk2/Myo5a/Ppfia2/Kif1a/Lin7a/5730455P16Rik/Myo1f/Cep19/Arfgap3/Psen1/Rab27a/Map4k2/Pdcd6ip/Kif5a | 18 |
| GO:0003016 | respiratory system process | BP | 0.17 | 0.00165 | 0.00953 | 2.02 | 0.00617 | Nlgn1/Tshz3/Ndn/Jag2/Nlgn3/Selenon/Atp1a2/Nlgn2 | 8 |
| GO:0043114 | regulation of vascular permeability | BP | 0.17 | 0.00165 | 0.00953 | 2.02 | 0.00617 | Angpt1/Ddah1/Fermt2/Ptp4a3/Akap12/Cdh5/Apoe/Cldn5 | 8 |
| GO:0003214 | cardiac left ventricle morphogenesis | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Sfrp2/Foxf1/Cpe/Ednra/Hey2 | 5 |
| GO:0030011 | maintenance of cell polarity | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Wnt11/Lin7a/Nckap1l/Dlg5/Pdcd6ip | 5 |
| GO:0033623 | regulation of integrin activation | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Fermt2/Jam3/Rasip1/Pcsk5/Plek | 5 |
| GO:0045821 | positive regulation of glycolytic process | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Igf1/Htr2a/Zbtb20/Psen1/Myc | 5 |
| GO:1901550 | regulation of endothelial cell development | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Plcb1/Cdh5/Cldn5/Akap11/S1pr3 | 5 |
| GO:1903140 | regulation of establishment of endothelial barrier | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Plcb1/Cdh5/Cldn5/Akap11/S1pr3 | 5 |
| GO:1905874 | regulation of postsynaptic density organization | BP | 0.263 | 0.00168 | 0.00966 | 2.02 | 0.00626 | Ptprd/Cdh2/Lrrc4b/Nptx1/Zmynd8 | 5 |
| GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | BP | 0.189 | 0.00169 | 0.00968 | 2.01 | 0.00627 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Fzd2/Dact1 | 7 |
| GO:0086091 | regulation of heart rate by cardiac conduction | BP | 0.189 | 0.00169 | 0.00968 | 2.01 | 0.00627 | Cacna1c/Kcnd3/Scn4b/Cacna2d1/Ank2/Bin1/Cav1 | 7 |
| GO:0015872 | dopamine transport | BP | 0.155 | 0.00169 | 0.00968 | 2.01 | 0.00627 | Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 9 |
| GO:1903531 | negative regulation of secretion by cell | BP | 0.104 | 0.00175 | 0.01 | 2 | 0.00648 | Maob/Abat/Drd4/Myo5a/Foxf1/Edn3/Htr1b/Apoe/Cd200/Nckap1l/Neo1/Frmd4a/Cd84/Ptger3/Gabbr1/Nr1h3/F2r/Irs1 | 18 |
| GO:0050727 | regulation of inflammatory response | BP | 0.0855 | 0.00177 | 0.0101 | 2 | 0.00654 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Sema7a/Ets1/Lrrk2/Ednrb/Foxf1/Slc7a2/Fndc4/Gprc5b/Cdh5/Bcl6b/Apoe/Lacc1/Ednra/Cd200/Xcl1/Tnfaip6/Stat5b/Pde5a/Nlrp1b/Calcrl/Ptger3/Nr1h3/Spn/Wfdc1 | 30 |
| GO:0030833 | regulation of actin filament polymerization | BP | 0.11 | 0.00181 | 0.0103 | 1.99 | 0.00668 | Kank4/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Daam2/Pecam1/Scin/Bin1/Nckap1l/Arhgap28/Cdc42ep5/Cyfip2 | 16 |
| GO:0048644 | muscle organ morphogenesis | BP | 0.128 | 0.00181 | 0.0103 | 1.99 | 0.00668 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Fzd2/Efemp2/Hey2/Eng | 12 |
| GO:0120193 | tight junction organization | BP | 0.128 | 0.00181 | 0.0103 | 1.99 | 0.00668 | Wnt11/Mpdz/Cldn8/Cdh5/Esam/Pecam1/Snai1/Svep1/Nphp1/Cldn5/Pdcd6ip/Cldn1 | 12 |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | BP | 0.143 | 0.00183 | 0.0104 | 1.98 | 0.00671 | Vipr2/Nos3/Npr3/Drd4/Mef2c/Apoe/Rgs5/Efemp2/Apln/Cav1 | 10 |
| GO:1902017 | regulation of cilium assembly | BP | 0.143 | 0.00183 | 0.0104 | 1.98 | 0.00671 | Ccdc88a/Adamts16/Kctd17/Rp1/Crocc/Ttbk2/Dzip1/Dync2li1/Cep97/Atmin | 10 |
| GO:0003171 | atrioventricular valve development | BP | 0.214 | 0.00185 | 0.0105 | 1.98 | 0.00678 | Aplnr/Heyl/Dchs1/Hey2/Slit3/Twist1 | 6 |
| GO:0018126 | protein hydroxylation | BP | 0.214 | 0.00185 | 0.0105 | 1.98 | 0.00678 | P3h3/P4htm/Fkbp10/P3h1/Asph/Crtap | 6 |
| GO:0014015 | positive regulation of gliogenesis | BP | 0.134 | 0.00185 | 0.0105 | 1.98 | 0.0068 | Ptprz1/Mdk/Nog/Igf1/Serpine2/Lrp2/Bin1/Ptn/Enpp2/Myc/Zfp365 | 11 |
| GO:2000177 | regulation of neural precursor cell proliferation | BP | 0.117 | 0.00189 | 0.0107 | 1.97 | 0.00692 | Gli1/Ptprz1/Mdk/Nog/Igf1/Cdh2/Lrrk2/Smarca1/Zfp423/Gli3/Lrp2/Vegfc/Ptn/Nes | 14 |
| GO:0030517 | negative regulation of axon extension | BP | 0.167 | 0.00189 | 0.0107 | 1.97 | 0.00692 | Map2/Sema5a/Sema5b/Nrp1/Sema7a/Sema3g/Sema6a/Draxin | 8 |
| GO:0034205 | amyloid-beta formation | BP | 0.167 | 0.00189 | 0.0107 | 1.97 | 0.00692 | Igf1/Aph1b/Rtn1/Slc2a13/Apoe/Bin1/Abcg1/Psen1 | 8 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | BP | 0.167 | 0.00189 | 0.0107 | 1.97 | 0.00692 | Angpt1/Kdr/Nrp1/Prkd1/Tek/Emilin1/Vegfc/Grb10 | 8 |
| GO:0045778 | positive regulation of ossification | BP | 0.153 | 0.00192 | 0.0107 | 1.97 | 0.00696 | Pkdcc/Slc8a1/Mef2c/Osr2/Calca/Nell1/Osr1/Ptn/Bmp7 | 9 |
| GO:1903350 | response to dopamine | BP | 0.153 | 0.00192 | 0.0107 | 1.97 | 0.00696 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Gnb5 | 9 |
| GO:1903351 | cellular response to dopamine | BP | 0.153 | 0.00192 | 0.0107 | 1.97 | 0.00696 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Gnb5 | 9 |
| GO:0034333 | adherens junction assembly | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Jam3/Abi2/Dlg5/Hipk1 | 4 |
| GO:0048251 | elastic fiber assembly | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Emilin1/Ltbp3/Efemp2/Fbln5 | 4 |
| GO:0060073 | micturition | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Cacna1c/Adra1a/Kcnma1/Ptger3 | 4 |
| GO:0060536 | cartilage morphogenesis | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Mef2c/Stc1/Snai1/Hand2 | 4 |
| GO:0070444 | oligodendrocyte progenitor proliferation | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0070445 | regulation of oligodendrocyte progenitor proliferation | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0099519 | dense core granule cytoskeletal transport | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Map2/Ppfia2/Kif1a/Kif5a | 4 |
| GO:1903911 | positive regulation of receptor clustering | BP | 0.333 | 0.00193 | 0.0107 | 1.97 | 0.00696 | Tnfaip6/Dlg4/Ptn/Lrp4 | 4 |
| GO:0006163 | purine nucleotide metabolic process | BP | 0.084 | 0.00196 | 0.0109 | 1.96 | 0.00707 | Nme4/Acsl6/Igf1/Hmgcs2/Nos3/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Lacc1/Htr2a/Acot7/Zbtb20/Pde5a/Rab23/Acsl1/Impdh1/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 31 |
| GO:0030866 | cortical actin cytoskeleton organization | BP | 0.184 | 0.00199 | 0.011 | 1.96 | 0.00716 | Epb41l3/Nckap1l/Cavin3/Akap11/Pdcd6ip/Fmnl3/Plek | 7 |
| GO:0098868 | bone growth | BP | 0.184 | 0.00199 | 0.011 | 1.96 | 0.00716 | Kdr/Ddr2/Thbs3/Evc/Stc1/Enpp1/Fbln5 | 7 |
| GO:0001841 | neural tube formation | BP | 0.112 | 0.00201 | 0.0111 | 1.95 | 0.00722 | Sfrp2/Nog/Zeb2/Ift57/Gdf7/Lrp2/Cecr2/Prickle1/Ipmk/Dlc1/Ptch1/Rab23/Fzd2/Bmp7/Twist1 | 15 |
| GO:0048332 | mesoderm morphogenesis | BP | 0.141 | 0.00203 | 0.0113 | 1.95 | 0.0073 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Tbx3/Snai1/Tal1/Bmp7 | 10 |
| GO:0050432 | catecholamine secretion | BP | 0.141 | 0.00203 | 0.0113 | 1.95 | 0.0073 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 10 |
| GO:0072676 | lymphocyte migration | BP | 0.12 | 0.00204 | 0.0113 | 1.95 | 0.00733 | Itga4/Cyp7b1/Itgb7/Icam1/Cxcr3/Cxcl12/Msn/Itgal/Il27ra/Cd200/Xcl1/Spn/Jam2 | 13 |
| GO:0046545 | development of primary female sexual characteristics | BP | 0.116 | 0.00205 | 0.0113 | 1.95 | 0.00733 | Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lfng/Kit/Kitl/Arrb1/Tnfaip6/Arrb2/Stat5b/Lhfpl2 | 14 |
| GO:0016485 | protein processing | BP | 0.0899 | 0.00208 | 0.0115 | 1.94 | 0.00745 | Cpxm2/Ihh/Mmp16/Aph1b/Lrrk2/Serpine2/Gli3/Rps6ka2/Cpe/Mme/Ldlrad3/C1rl/Dync2h1/Pcsk5/C1ra/Ptch1/Asph/Ece2/Adam19/Arxes2/Psen1/Cyfip2/Nkd2/Myc/Prss12 | 25 |
| GO:0055088 | lipid homeostasis | BP | 0.102 | 0.00212 | 0.0117 | 1.93 | 0.00758 | Apob/Pla2g10/Cyp7b1/Nr5a2/Soat1/Tsku/Apoe/Hdac9/Abca5/Rcn3/Zbtb20/Abcg1/Angptl4/Abhd8/Cav1/Nr1h3/Tlcd2/Abcg4 | 18 |
| GO:0032252 | secretory granule localization | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Map2/Myo5a/Ppfia2/Kif1a/Kif5a | 5 |
| GO:0045019 | negative regulation of nitric oxide biosynthetic process | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Wdr35/Atp2b4/Gimap5/Gimap3/Cav1 | 5 |
| GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Ano1/Cxcl12/Calca/Htr2a/Arrb2 | 5 |
| GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Scn1a/Grin2b/Cxcl12/Htr2a/Fyn | 5 |
| GO:1903236 | regulation of leukocyte tethering or rolling | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Itga4/Chst2/Cxcl12/Sele/St3gal4 | 5 |
| GO:1904406 | negative regulation of nitric oxide metabolic process | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Wdr35/Atp2b4/Gimap5/Gimap3/Cav1 | 5 |
| GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | BP | 0.25 | 0.00216 | 0.0118 | 1.93 | 0.00765 | Ddr2/Tert/Myo5a/Adamts1/Pcsk5 | 5 |
| GO:0042987 | amyloid precursor protein catabolic process | BP | 0.15 | 0.00216 | 0.0118 | 1.93 | 0.00766 | Igf1/Aph1b/Rtn1/Slc2a13/Apoe/Bin1/Abcg1/Adam19/Psen1 | 9 |
| GO:0045444 | fat cell differentiation | BP | 0.0909 | 0.00219 | 0.012 | 1.92 | 0.00777 | Sfrp2/Igf1/Fermt2/Lama4/Runx1t1/Arl6/Vstm2a/Id4/Bbs7/Plcb1/Glis1/Sh2b2/Gdf10/Enpp1/Dysf/Htr2a/Lrp3/Arxes2/Steap4/Bbs1/Dact1/Lrrc8c/Bmp7/Cds1 | 24 |
| GO:0010769 | regulation of cell morphogenesis involved in differentiation | BP | 0.115 | 0.00221 | 0.0121 | 1.92 | 0.00783 | Mdk/Ptprd/Nrp1/Fermt2/Tbc1d24/Has2/Obsl1/Postn/Caprin2/Bhlhb9/Net1/Cux2/Enpp2/Numbl | 14 |
| GO:0034367 | protein-containing complex remodeling | BP | 0.207 | 0.00223 | 0.0122 | 1.91 | 0.00789 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:1904861 | excitatory synapse assembly | BP | 0.207 | 0.00223 | 0.0122 | 1.91 | 0.00789 | Ptprd/Nlgn1/Lrrc4b/Nptx1/Slitrk3/Nlgn2 | 6 |
| GO:0048639 | positive regulation of developmental growth | BP | 0.0955 | 0.00224 | 0.0122 | 1.91 | 0.00792 | Gli1/Sema5a/Igf1/Tbx2/Cpne5/Nrp1/Acacb/Sema7a/Fn1/Mef2c/Cxcl12/Hopx/Plcb1/Flt3/Apoe/Igf2/Slc23a2/Foxs1/Cacng7/Stat5b/Hey2 | 21 |
| GO:0072089 | stem cell proliferation | BP | 0.131 | 0.00225 | 0.0123 | 1.91 | 0.00795 | Sfrp2/Fermt2/Tert/Znrf3/Vegfc/Tbx3/Kitl/Ltbp3/Septin4/Abcb1a/Nes | 11 |
| GO:0007405 | neuroblast proliferation | BP | 0.139 | 0.00226 | 0.0123 | 1.91 | 0.00795 | Acsl6/Hhip/Lef1/Lrrk2/Id4/Gli3/Eml1/Vegfc/Ptn/Numbl | 10 |
| GO:0055008 | cardiac muscle tissue morphogenesis | BP | 0.139 | 0.00226 | 0.0123 | 1.91 | 0.00795 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Hey2/Eng | 10 |
| GO:0071868 | cellular response to monoamine stimulus | BP | 0.139 | 0.00226 | 0.0123 | 1.91 | 0.00795 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Atp2b4/Gnb5 | 10 |
| GO:0071870 | cellular response to catecholamine stimulus | BP | 0.139 | 0.00226 | 0.0123 | 1.91 | 0.00795 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Atp2b4/Gnb5 | 10 |
| GO:0031281 | positive regulation of cyclase activity | BP | 0.179 | 0.00232 | 0.0125 | 1.9 | 0.00812 | Cacna1c/Vipr2/Nos3/Adcy3/Calca/Adcy4/Adcy7 | 7 |
| GO:0045981 | positive regulation of nucleotide metabolic process | BP | 0.179 | 0.00232 | 0.0125 | 1.9 | 0.00812 | Igf1/Nos3/Htr2a/Zbtb20/Psen1/Pid1/Myc | 7 |
| GO:0051180 | vitamin transport | BP | 0.179 | 0.00232 | 0.0125 | 1.9 | 0.00812 | Stra6/Slc2a3/Abcg2/Lrp2/Slc23a2/Abcc5/Abcg3 | 7 |
| GO:0051481 | negative regulation of cytosolic calcium ion concentration | BP | 0.179 | 0.00232 | 0.0125 | 1.9 | 0.00812 | Kcnk3/Slc8a1/Calca/Pkd2/Gstm7/Atp1a2/Sri | 7 |
| GO:1900544 | positive regulation of purine nucleotide metabolic process | BP | 0.179 | 0.00232 | 0.0125 | 1.9 | 0.00812 | Igf1/Nos3/Htr2a/Zbtb20/Psen1/Pid1/Myc | 7 |
| GO:0001890 | placenta development | BP | 0.101 | 0.0024 | 0.0129 | 1.89 | 0.00838 | Ndp/Lef1/Itga4/Slc8a1/Il11ra1/Vash1/Syde1/Prdm1/Pcdh12/Cdkn1c/Igf2/Snai1/Pkd1/Pkd2/Ptn/Vash2/Bmp7/Hey2 | 18 |
| GO:0010656 | negative regulation of muscle cell apoptotic process | BP | 0.148 | 0.00243 | 0.013 | 1.88 | 0.00845 | Sfrp2/Gria4/Mdk/Igf1/Agtr1a/Arrb2/Cav1/Hey2/Hand2 | 9 |
| GO:0060324 | face development | BP | 0.148 | 0.00243 | 0.013 | 1.88 | 0.00845 | Stra6/Lef1/Itga4/Nog/Crispld1/Mmp2/Ednra/Twist2/Asph | 9 |
| GO:0099172 | presynapse organization | BP | 0.148 | 0.00243 | 0.013 | 1.88 | 0.00845 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn3/Pcdh17/Lrp4/Nlgn2 | 9 |
| GO:1902930 | regulation of alcohol biosynthetic process | BP | 0.16 | 0.00248 | 0.0133 | 1.88 | 0.00861 | Apob/Pth1r/Dkk3/Apoe/Adcyap1r1/Abcg1/Abcg4/Plek | 8 |
| GO:0046530 | photoreceptor cell differentiation | BP | 0.129 | 0.00248 | 0.0133 | 1.88 | 0.00861 | Ihh/Sdk2/Thy1/Prdm1/Rp1/Rorb/Ttc26/Arl3/Nphp1/Bbs1/Ptn | 11 |
| GO:0008542 | visual learning | BP | 0.137 | 0.00251 | 0.0134 | 1.87 | 0.00871 | Cacna1c/Nog/B4galt2/Pde1b/Cacna1e/Kit/Nlgn3/Ddhd2/Atp1a2/Syngap1 | 10 |
| GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | BP | 0.106 | 0.00255 | 0.0137 | 1.86 | 0.00885 | Adra1a/Adgrl4/Adcy3/Cxcr3/Ptgfr/Adgrl1/Calca/Pth1r/Adcy4/Adcy7/Gpr161/Calcrl/Ptger3/Atp2b4/S1pr3/Ptger2 | 16 |
| GO:0043491 | protein kinase B signaling | BP | 0.0962 | 0.00259 | 0.0138 | 1.86 | 0.00896 | Sema5a/Angpt1/Kdr/Igf1/Hcls1/Fermt2/Inpp4b/Tek/Cxcl12/Lrp2/Nox4/Magi2/Igf2/Cavin3/Rcn3/Arrb2/Rasd2/Setx/Eng/Irs1 | 20 |
| GO:0019693 | ribose phosphate metabolic process | BP | 0.0824 | 0.00261 | 0.0139 | 1.86 | 0.00901 | Nme4/Acsl6/Igf1/Hmgcs2/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Htr2a/Acot7/Zbtb20/Pde5a/Rab23/Acsl1/Impdh1/Dpyd/Psen1/Pid1/Pfkm/Cacnb4/Pygl/Myc/Papss2/Suclg2/Acot6 | 31 |
| GO:0030004 | cellular monovalent inorganic cation homeostasis | BP | 0.117 | 0.00261 | 0.0139 | 1.86 | 0.00901 | Slc1a3/Slc8a1/Lrrk2/Slc9a7/Dmxl2/Lacc1/Kcnma1/Slc9a5/Slc4a3/Aqp11/Atp1a2/Slc4a8/Rab20 | 13 |
| GO:1902106 | negative regulation of leukocyte differentiation | BP | 0.117 | 0.00261 | 0.0139 | 1.86 | 0.00901 | Ihh/Mdk/Cd44/Inpp4b/Lrrc17/Gli3/Calca/Flt3/Loxl3/Tnfrsf11b/Zbtb46/Tnfaip6/Myc | 13 |
| GO:0016042 | lipid catabolic process | BP | 0.0851 | 0.00262 | 0.0139 | 1.86 | 0.00903 | Apob/Cyp1b1/Aspg/Pla2g10/Acacb/Plcb1/Bdh2/Apoe/Pla1a/Echs1/Acot7/Cyp46a1/Plcb2/Ddhd2/Gimap5/Abcd2/Gimap3/Smpd1/Lipe/Pld2/Enpp2/Pafah2/Plbd1/Twist1/Abhd3/Acap1/Echdc2/Irs1 | 28 |
| GO:0009150 | purine ribonucleotide metabolic process | BP | 0.0841 | 0.00266 | 0.014 | 1.85 | 0.00909 | Nme4/Acsl6/Igf1/Hmgcs2/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Htr2a/Acot7/Zbtb20/Pde5a/Rab23/Acsl1/Impdh1/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 29 |
| GO:0002274 | myeloid leukocyte activation | BP | 0.0896 | 0.00267 | 0.014 | 1.85 | 0.00909 | Pla2g10/Fn1/Lrrk2/Foxf1/Slc7a2/Dock2/Cd1d1/Kit/Cd226/Havcr2/Cd200/Dysf/Myo1f/Cd84/Fgr/Gimap5/Gimap3/Psen1/Lat/Clnk/Nr1h3/Pld2/Batf/Lcp2 | 24 |
| GO:1905039 | carboxylic acid transmembrane transport | BP | 0.109 | 0.00268 | 0.014 | 1.85 | 0.00909 | Slc1a3/Acsl6/Slc7a2/Slc36a4/Lrp2/Slc23a2/Slc38a1/Abcd2/Acsl1/Slc7a7/Psen1/Abcb1a/Abcc5/Lrrc8c/Myc | 15 |
| GO:0010838 | positive regulation of keratinocyte proliferation | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Mdk/Has2/Stxbp4/Twist2 | 4 |
| GO:0014052 | regulation of gamma-aminobutyric acid secretion | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Grik1/Abat/Htr1b/Gabbr1 | 4 |
| GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Angpt1/Kdr/Tek/Grb10 | 4 |
| GO:0033625 | positive regulation of integrin activation | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Fermt2/Rasip1/Pcsk5/Plek | 4 |
| GO:0034374 | low-density lipoprotein particle remodeling | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Apob/Pla2g10/Apoe/Abcg1 | 4 |
| GO:0034375 | high-density lipoprotein particle remodeling | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Pltp/Apoe/Abca5/Abcg1 | 4 |
| GO:0034392 | negative regulation of smooth muscle cell apoptotic process | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Gria4/Igf1/Agtr1a/Arrb2 | 4 |
| GO:0086067 | AV node cell to bundle of His cell communication | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Cacna1c/Scn4b/Ank2/Gjc1 | 4 |
| GO:1901841 | regulation of high voltage-gated calcium channel activity | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Cacna2d1/Dysf/Rrad/Sri | 4 |
| GO:1902950 | regulation of dendritic spine maintenance | BP | 0.308 | 0.00268 | 0.014 | 1.85 | 0.00909 | Grin2b/Apoe/Fyn/Zmynd8 | 4 |
| GO:0006688 | glycosphingolipid biosynthetic process | BP | 0.2 | 0.00268 | 0.014 | 1.85 | 0.00909 | St8sia2/St8sia4/St3gal2/St6galnac4/St8sia6/St6galnac6 | 6 |
| GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | BP | 0.2 | 0.00268 | 0.014 | 1.85 | 0.00909 | Angpt1/Kdr/Tek/Emilin1/Vegfc/Grb10 | 6 |
| GO:0060343 | trabecula formation | BP | 0.2 | 0.00268 | 0.014 | 1.85 | 0.00909 | Thbs3/Mmp2/Tek/Adamts1/Enpp1/Hey2 | 6 |
| GO:0071312 | cellular response to alkaloid | BP | 0.2 | 0.00268 | 0.014 | 1.85 | 0.00909 | Slc1a3/Ryr1/Slc8a1/Htr1b/Selenon/Gstm7 | 6 |
| GO:0001782 | B cell homeostasis | BP | 0.175 | 0.0027 | 0.0141 | 1.85 | 0.00914 | Cd44/Mef2c/Sh2b2/Tnfsf13b/Dock10/Enpp1/Nckap1l | 7 |
| GO:0051349 | positive regulation of lyase activity | BP | 0.175 | 0.0027 | 0.0141 | 1.85 | 0.00914 | Cacna1c/Vipr2/Nos3/Adcy3/Calca/Adcy4/Adcy7 | 7 |
| GO:0060325 | face morphogenesis | BP | 0.175 | 0.0027 | 0.0141 | 1.85 | 0.00914 | Stra6/Lef1/Nog/Crispld1/Mmp2/Twist2/Asph | 7 |
| GO:0010952 | positive regulation of peptidase activity | BP | 0.1 | 0.00271 | 0.0141 | 1.85 | 0.00915 | Sfrp2/Aph1b/Bok/Wdr35/Grin2b/Ift57/Ppm1f/Hip1/Arrb1/Rcn3/Dlc1/Asph/Fyn/Nlrp1b/Ccn2/Cyfip2/F2r/Myc | 18 |
| GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | BP | 0.238 | 0.00272 | 0.0141 | 1.85 | 0.00915 | Adra1a/P2rx2/Calca/Rps6ka2/Agtr1a | 5 |
| GO:0007190 | activation of adenylate cyclase activity | BP | 0.238 | 0.00272 | 0.0141 | 1.85 | 0.00915 | Vipr2/Adcy3/Calca/Adcy4/Adcy7 | 5 |
| GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | BP | 0.238 | 0.00272 | 0.0141 | 1.85 | 0.00915 | Kdr/Nrp1/Jcad/Hdac9/Akt3 | 5 |
| GO:0097107 | postsynaptic density assembly | BP | 0.238 | 0.00272 | 0.0141 | 1.85 | 0.00915 | Ptprd/Lrrc4b/Nptx1/Slitrk3/Nlgn2 | 5 |
| GO:0051261 | protein depolymerization | BP | 0.112 | 0.00278 | 0.0144 | 1.84 | 0.00933 | Map2/Sema5a/Evl/Tmod2/Mical1/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2/Plek | 14 |
| GO:0071867 | response to monoamine | BP | 0.135 | 0.00278 | 0.0144 | 1.84 | 0.00933 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Atp2b4/Gnb5 | 10 |
| GO:0071869 | response to catecholamine | BP | 0.135 | 0.00278 | 0.0144 | 1.84 | 0.00933 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Atp2b4/Gnb5 | 10 |
| GO:0008038 | neuron recognition | BP | 0.157 | 0.00282 | 0.0145 | 1.84 | 0.00943 | Gap43/Prtg/Efnb3/Ptprz1/Sema5a/Nrp1/Ndn/Epha3 | 8 |
| GO:0042088 | T-helper 1 type immune response | BP | 0.157 | 0.00282 | 0.0145 | 1.84 | 0.00943 | Lef1/H2-Ab1/Il27ra/Cracr2a/Havcr2/Xcl1/Il12rb1/Spn | 8 |
| GO:0030177 | positive regulation of Wnt signaling pathway | BP | 0.108 | 0.00287 | 0.0148 | 1.83 | 0.0096 | Sfrp2/Sema5a/Lef1/Zeb2/Nkd1/Tert/Gpc3/Lrrk2/Gprc5b/Peg12/Daam2/Caprin2/Adgra2/Cav1/Dact1 | 15 |
| GO:1903825 | organic acid transmembrane transport | BP | 0.108 | 0.00287 | 0.0148 | 1.83 | 0.0096 | Slc1a3/Acsl6/Slc7a2/Slc36a4/Lrp2/Slc23a2/Slc38a1/Abcd2/Acsl1/Slc7a7/Psen1/Abcb1a/Abcc5/Lrrc8c/Myc | 15 |
| GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | BP | 0.0952 | 0.00289 | 0.0149 | 1.83 | 0.00965 | Hgf/Sfrp2/Igf1/Cd44/Bok/Wdr35/Mical1/Ift57/Wnt9a/Ppm1f/Hip1/Arrb1/Epha7/Dlc1/Arrb2/Nlrp1b/Ccn2/Mgmt/F2r/Myc | 20 |
| GO:0016311 | dephosphorylation | BP | 0.0836 | 0.0029 | 0.0149 | 1.83 | 0.00966 | Ptprz1/Ptprd/Npnt/Ptpro/Mef2c/Ptp4a3/Inpp4b/Itga1/Ptpdc1/Ptprm/Cdh5/Plpp3/Lhpp/Ppm1f/Ppp1r16b/Bpnt1/Magi2/Ppm1e/Nckap1l/Cmya5/Dlc1/Plpp1/Ppp1r26/Smpd1/Pdgfrb/Vcan/Plek/Ptpn7/Plppr2 | 29 |
| GO:0072175 | epithelial tube formation | BP | 0.102 | 0.00292 | 0.015 | 1.82 | 0.00971 | Sfrp2/Wnt6/Nog/Zeb2/Ift57/Gdf7/Lrp2/Cecr2/Prickle1/Ipmk/Dlc1/Ptch1/Osr1/Rab23/Fzd2/Bmp7/Twist1 | 17 |
| GO:0042267 | natural killer cell mediated cytotoxicity | BP | 0.126 | 0.00298 | 0.0153 | 1.81 | 0.00993 | Pik3r6/Coro1a/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gzmb/Gimap3/Rab27a | 11 |
| GO:0010517 | regulation of phospholipase activity | BP | 0.143 | 0.00304 | 0.0156 | 1.81 | 0.0101 | Adra1a/Itk/Arhgap6/Agtr1a/Sele/Adcyap1r1/Htr2a/Dlc1/Pdgfrb | 9 |
| GO:0030865 | cortical cytoskeleton organization | BP | 0.143 | 0.00304 | 0.0156 | 1.81 | 0.0101 | Nlgn1/Epb41l3/Rhobtb3/Nckap1l/Cavin3/Akap11/Pdcd6ip/Fmnl3/Plek | 9 |
| GO:0009259 | ribonucleotide metabolic process | BP | 0.0824 | 0.00304 | 0.0156 | 1.81 | 0.0101 | Nme4/Acsl6/Igf1/Hmgcs2/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Htr2a/Acot7/Zbtb20/Pde5a/Rab23/Acsl1/Impdh1/Dpyd/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 30 |
| GO:0035148 | tube formation | BP | 0.0989 | 0.00306 | 0.0156 | 1.81 | 0.0101 | Sfrp2/Wnt6/Nog/Zeb2/Ift57/Gdf7/Lrp2/Cecr2/Prickle1/Ipmk/Dlc1/Ptch1/Osr1/Rab23/Fzd2/Edar/Bmp7/Twist1 | 18 |
| GO:1903707 | negative regulation of hemopoiesis | BP | 0.115 | 0.00306 | 0.0156 | 1.81 | 0.0101 | Ihh/Mdk/Cd44/Inpp4b/Lrrc17/Gli3/Calca/Flt3/Loxl3/Tnfrsf11b/Zbtb46/Tnfaip6/Myc | 13 |
| GO:0006885 | regulation of pH | BP | 0.12 | 0.00307 | 0.0157 | 1.81 | 0.0101 | Car4/Lrrk2/Ednrb/Slc9a7/Dmxl2/Agtr1a/Lacc1/Slc9a5/Slc4a3/Aqp11/Slc4a8/Rab20 | 12 |
| GO:0048645 | animal organ formation | BP | 0.133 | 0.00307 | 0.0157 | 1.8 | 0.0102 | Ar/Wnt11/Hoxd11/Nog/Hoxa11/Mef2c/Gli3/Lrp2/Bmp7/Hand2 | 10 |
| GO:0051149 | positive regulation of muscle cell differentiation | BP | 0.133 | 0.00307 | 0.0157 | 1.8 | 0.0102 | Igf1/Mef2c/Hopx/Tshz3/Kit/Cth/Arrb2/Efemp2/Camk1/Eng | 10 |
| GO:0062013 | positive regulation of small molecule metabolic process | BP | 0.101 | 0.0031 | 0.0158 | 1.8 | 0.0102 | Igf1/Nos3/Pth1r/Igf2/Adcyap1r1/Htr2a/Zbtb20/Abcg1/Abcd2/Psen1/Nr1h3/Pid1/Rdh1/Abcg4/Myc/Twist1/Irs1 | 17 |
| GO:0031348 | negative regulation of defense response | BP | 0.0913 | 0.00311 | 0.0158 | 1.8 | 0.0103 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Ets1/Foxf1/Fndc4/Cdh5/Apoe/Havcr2/Cd200/Igf2/Tnfaip6/Mapkbp1/Arrb2/Calcrl/Cd96/Nr1h3/Spn/Wfdc1 | 22 |
| GO:0003230 | cardiac atrium development | BP | 0.171 | 0.00312 | 0.0159 | 1.8 | 0.0103 | Wnt11/Nog/Ank2/Ccm2l/Heg1/Hey2/Eng | 7 |
| GO:0071604 | transforming growth factor beta production | BP | 0.171 | 0.00312 | 0.0159 | 1.8 | 0.0103 | Wnt11/Fn1/Tsku/Cd200/Ltbp3/Nrros/Lum | 7 |
| GO:0043254 | regulation of protein-containing complex assembly | BP | 0.0793 | 0.00316 | 0.016 | 1.79 | 0.0104 | Kank4/Map2/Evl/Hcls1/Stx1b/Fermt2/Tmod2/Pfn2/Icam1/Jam3/Cdh5/Msn/Coro1a/Esam/Tubb4a/Apoe/Rasip1/Prrt2/Daam2/Pecam1/Scin/Bin1/Nckap1l/Nin/Slain1/Pcsk5/Tal1/Arhgap28/Cdc42ep5/Cav1/Cyfip2/Dact1/Tppp/Plek | 34 |
| GO:0016050 | vesicle organization | BP | 0.0871 | 0.00318 | 0.016 | 1.79 | 0.0104 | Zeb2/Syt13/Sec16b/Stx1b/Ap1m1/Serpine2/Rab34/Srpx/Coro1a/Cadps/Syt7/Prrt2/Dysf/Abcg1/Fhip1b/Aqp11/Dlg4/Ccdc136/Arfgap3/Stx2/Rab27a/Nkd2/Golph3l/Pdcd6ip/Rab20 | 25 |
| GO:0050798 | activated T cell proliferation | BP | 0.154 | 0.00319 | 0.016 | 1.79 | 0.0104 | Igf1/Itgal/Il27ra/Igf2/Satb1/Stat5b/Fyn/Il12rb1 | 8 |
| GO:0090279 | regulation of calcium ion import | BP | 0.154 | 0.00319 | 0.016 | 1.79 | 0.0104 | Spink1/Stc1/Cxcl12/Agtr1a/Dysf/Pkd2/Fyn/Pdgfrb | 8 |
| GO:0002313 | mature B cell differentiation involved in immune response | BP | 0.194 | 0.00319 | 0.016 | 1.79 | 0.0104 | Itm2a/Lfng/Dock10/Enpp1/Irf8/Mfng | 6 |
| GO:0021952 | central nervous system projection neuron axonogenesis | BP | 0.194 | 0.00319 | 0.016 | 1.79 | 0.0104 | Dcc/Zeb2/Tsku/Cdh11/Nin/Draxin | 6 |
| GO:0032967 | positive regulation of collagen biosynthetic process | BP | 0.194 | 0.00319 | 0.016 | 1.79 | 0.0104 | Ihh/Ddr2/Arrb2/Ccn2/Pdgfrb/F2r | 6 |
| GO:0045762 | positive regulation of adenylate cyclase activity | BP | 0.194 | 0.00319 | 0.016 | 1.79 | 0.0104 | Cacna1c/Vipr2/Adcy3/Calca/Adcy4/Adcy7 | 6 |
| GO:2000648 | positive regulation of stem cell proliferation | BP | 0.194 | 0.00319 | 0.016 | 1.79 | 0.0104 | Fermt2/Tert/Vegfc/Tbx3/Kitl/Ltbp3 | 6 |
| GO:0071674 | mononuclear cell migration | BP | 0.0984 | 0.00324 | 0.0163 | 1.79 | 0.0106 | Mdk/Itga4/Cyp7b1/Ptpro/Itgb7/Icam1/Cxcr3/Cxcl12/Plcb1/Calca/Msn/Itgal/Il27ra/Pecam1/Cd200/Xcl1/Spn/Jam2 | 18 |
| GO:0051937 | catecholamine transport | BP | 0.125 | 0.00326 | 0.0164 | 1.79 | 0.0106 | Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 11 |
| GO:0120192 | tight junction assembly | BP | 0.125 | 0.00326 | 0.0164 | 1.79 | 0.0106 | Wnt11/Mpdz/Cldn8/Cdh5/Esam/Pecam1/Snai1/Nphp1/Cldn5/Pdcd6ip/Cldn1 | 11 |
| GO:0051224 | negative regulation of protein transport | BP | 0.106 | 0.0033 | 0.0165 | 1.78 | 0.0107 | Angpt1/Pkdcc/Drd4/Lrrk2/Pkia/Ppm1f/Apoe/Cd200/Neo1/Frmd4a/Rab23/Ptger3/Nr1h3/F2r/Irs1 | 15 |
| GO:0019216 | regulation of lipid metabolic process | BP | 0.0812 | 0.00331 | 0.0166 | 1.78 | 0.0107 | Apob/Epha8/Igf1/Acacb/Tek/Kit/Dkk3/Agtr1a/Apoe/Kcnma1/Igf2/Snai1/Htr2a/Lpcat1/Zbtb20/Abcg1/Stat5b/Fgr/Angptl4/Gimap5/Abcd2/Gimap3/Pdgfrb/Cav1/Nr1h3/Rdh1/Bcl11b/Abcg4/Twist1/Asxl3/Irs1 | 31 |
| GO:0043576 | regulation of respiratory gaseous exchange | BP | 0.227 | 0.00338 | 0.0169 | 1.77 | 0.0109 | Nlgn1/Tshz3/Nlgn3/Atp1a2/Nlgn2 | 5 |
| GO:0046325 | negative regulation of glucose import | BP | 0.227 | 0.00338 | 0.0169 | 1.77 | 0.0109 | Sh2b2/Enpp1/Pid1/Grb10/Myc | 5 |
| GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | BP | 0.227 | 0.00338 | 0.0169 | 1.77 | 0.0109 | Rims3/Stx1b/Cadps/Syt7/Cacna1a | 5 |
| GO:0071398 | cellular response to fatty acid | BP | 0.227 | 0.00338 | 0.0169 | 1.77 | 0.0109 | Id3/Plcb1/Nfatc4/Pid1/Irs1 | 5 |
| GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | BP | 0.227 | 0.00338 | 0.0169 | 1.77 | 0.0109 | Ptp4a3/Jcad/Tcf4/Adgra2/Sema6a | 5 |
| GO:1902115 | regulation of organelle assembly | BP | 0.0939 | 0.0034 | 0.0169 | 1.77 | 0.011 | Gap43/Fez1/Ptprd/Ccdc88a/Adamts16/Lrrk2/Lrrc4b/Nptx1/Msn/Kctd17/Rp1/Nptxr/Crocc/Ttbk2/Dzip1/Dync2li1/Cep97/Arhgef9/Atmin/Pdcd6ip | 20 |
| GO:0006644 | phospholipid metabolic process | BP | 0.0817 | 0.00343 | 0.0171 | 1.77 | 0.0111 | Acsl6/Aspg/Pla2g10/Hmgcs2/Efr3b/Pip5k1b/Inpp4b/Plcb1/Them5/Plpp3/Ttc7b/Dgkb/Bpnt1/Htr2a/Lpcat1/Plcb2/Abca8b/Plpp1/Abhd8/Smpd1/Pdgfrb/Nr1h3/Pld2/Enpp2/Plbd1/Plek/Mboat2/Cds1/Abhd3/Plppr2 | 30 |
| GO:0030595 | leukocyte chemotaxis | BP | 0.0921 | 0.00343 | 0.0171 | 1.77 | 0.0111 | Cxcl15/Mdk/Cyp7b1/Ptpro/Ednrb/Jam3/Cxcr3/Cxcl12/Edn3/Itga1/Calca/Coro1a/Kit/Vegfc/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Mpp1/Ptn | 21 |
| GO:0046942 | carboxylic acid transport | BP | 0.0865 | 0.00348 | 0.0173 | 1.76 | 0.0112 | Slc1a3/Grik1/Pla2g10/Slc13a5/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Slc36a4/Lrp2/Htr1b/Slc6a15/Slc23a2/Bdkrb2/Sfxn5/Slc38a1/Nfkbie/Psen1/Abcc5/Gabbr1/Cacna1a/Lrrc8c/Cacnb4/Myc | 25 |
| GO:0007565 | female pregnancy | BP | 0.1 | 0.0035 | 0.0174 | 1.76 | 0.0113 | Acod1/Ar/Cbs/Il11ra1/Mmp2/Abcg2/Calca/Syde1/Prdm1/Maged2/Havcr2/Klf9/Pcsk5/Stat5b/Stx2/Ptn/Itgb4 | 17 |
| GO:0010950 | positive regulation of endopeptidase activity | BP | 0.1 | 0.0035 | 0.0174 | 1.76 | 0.0113 | Sfrp2/Aph1b/Bok/Wdr35/Grin2b/Ift57/Ppm1f/Hip1/Arrb1/Dlc1/Asph/Fyn/Nlrp1b/Ccn2/Cyfip2/F2r/Myc | 17 |
| GO:0002367 | cytokine production involved in immune response | BP | 0.113 | 0.00356 | 0.0176 | 1.76 | 0.0114 | Angpt1/Sema7a/Tek/Gprc5b/Tril/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Clnk/Cd96 | 13 |
| GO:0045638 | negative regulation of myeloid cell differentiation | BP | 0.124 | 0.00357 | 0.0176 | 1.76 | 0.0114 | Inpp4b/Lrrc17/Calca/Tnfrsf11b/Gpr171/Zbtb46/Tnfaip6/Hoxa9/Twist2/Stat5b/Myc | 11 |
| GO:0010092 | specification of animal organ identity | BP | 0.167 | 0.0036 | 0.0176 | 1.76 | 0.0114 | Ar/Wnt11/Hoxd11/Hoxa11/Mef2c/Gli3/Lrp2 | 7 |
| GO:0090175 | regulation of establishment of planar polarity | BP | 0.167 | 0.0036 | 0.0176 | 1.76 | 0.0114 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Fzd2/Dact1 | 7 |
| GO:0099174 | regulation of presynapse organization | BP | 0.167 | 0.0036 | 0.0176 | 1.76 | 0.0114 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn3/Nlgn2 | 7 |
| GO:0030850 | prostate gland development | BP | 0.151 | 0.0036 | 0.0176 | 1.76 | 0.0114 | Ar/Cyp7b1/Nog/Igf1/Cd44/Mmp2/Id4/Bmp7 | 8 |
| GO:0003207 | cardiac chamber formation | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Tbx2/Mef2c/Ednra/Hand2 | 4 |
| GO:0009312 | oligosaccharide biosynthetic process | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | B3galnt1/St3gal2/St6galnac6/St3gal4 | 4 |
| GO:0030432 | peristalsis | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Tbx2/P2rx2/Tshz3/Tbx3 | 4 |
| GO:0032253 | dense core granule localization | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Map2/Ppfia2/Kif1a/Kif5a | 4 |
| GO:0032341 | aldosterone metabolic process | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Ednrb/Dkk3/Kcnma1/Cacna1h | 4 |
| GO:0045060 | negative thymic T cell selection | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Gli3/Dock2/Zap70/Spn | 4 |
| GO:0046058 | cAMP metabolic process | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Adcy3/Adcy4/Adcy7/Cacnb4 | 4 |
| GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Ednra/Bmp7/Hand2/Twist1 | 4 |
| GO:0098840 | protein transport along microtubule | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Myo5a/Kif1a/Dlg2/Kif5a | 4 |
| GO:0099118 | microtubule-based protein transport | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Myo5a/Kif1a/Dlg2/Kif5a | 4 |
| GO:1901201 | regulation of extracellular matrix assembly | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Phldb2/Has2/Emilin1/Tie1 | 4 |
| GO:1901950 | dense core granule transport | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Map2/Ppfia2/Kif1a/Kif5a | 4 |
| GO:1903238 | positive regulation of leukocyte tethering or rolling | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Itga4/Chst2/Sele/St3gal4 | 4 |
| GO:1903441 | protein localization to ciliary membrane | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Tub/Arl6/Arl3/Rabep1 | 4 |
| GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | BP | 0.286 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Kit/Cth/Efemp2/Eng | 4 |
| GO:0001708 | cell fate specification | BP | 0.118 | 0.00361 | 0.0176 | 1.76 | 0.0114 | Ihh/Sfrp2/Ar/Wnt11/Hoxd10/Ptch2/Tbx2/Hoxa11/Gli3/Tbx3/Ptch1/Psen1 | 12 |
| GO:0045834 | positive regulation of lipid metabolic process | BP | 0.0973 | 0.00364 | 0.0177 | 1.75 | 0.0115 | Epha8/Igf1/Tek/Agtr1a/Apoe/Igf2/Htr2a/Zbtb20/Abcg1/Fgr/Angptl4/Abcd2/Pdgfrb/Nr1h3/Rdh1/Abcg4/Twist1/Irs1 | 18 |
| GO:0051047 | positive regulation of secretion | BP | 0.0791 | 0.00368 | 0.0179 | 1.75 | 0.0116 | Grik1/Pla2g10/Igf1/Nlgn1/Abat/Stx1b/Ednrb/Ano1/Grin2b/Cxcl12/Edn3/Rab34/Cadps/Vegfc/Cd38/Syt7/Arrb1/Abcg1/Frmd4a/Ncs1/Cacna1h/Fgr/Apln/Slc4a8/Rab27a/Sri/Gabbr1/Pld2/Pfkm/Golph3l/Pdcd6ip/Nlgn2/Irs1 | 33 |
| GO:0034219 | carbohydrate transmembrane transport | BP | 0.109 | 0.0037 | 0.018 | 1.75 | 0.0116 | Igf1/Slc2a3/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Slc23a2/Abcb1a/Pid1/Grb10/Myc/Irs1 | 14 |
| GO:0042471 | ear morphogenesis | BP | 0.105 | 0.00377 | 0.0182 | 1.74 | 0.0118 | Col11a1/Nog/Tbx2/Lrig3/Osr2/Tshz1/Zeb1/Enpp1/Tbx3/Ednra/Osr1/Fzd2/Kcnq4/Itga8/Myc | 15 |
| GO:0019751 | polyol metabolic process | BP | 0.112 | 0.00384 | 0.0186 | 1.73 | 0.012 | Inpp4b/Sptlc3/Plcb1/Pth1r/Plpp3/Bpnt1/Dysf/Degs2/Adcyap1r1/Ipmk/Sord/Plpp1/Plek | 13 |
| GO:0051952 | regulation of amine transport | BP | 0.112 | 0.00384 | 0.0186 | 1.73 | 0.012 | Grik1/Syt13/Abat/Myo5a/Grin2b/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Psen1/Gabbr1/Cacna1a | 13 |
| GO:0008593 | regulation of Notch signaling pathway | BP | 0.122 | 0.00389 | 0.0188 | 1.73 | 0.0122 | Postn/Lfng/Jag2/Kit/Arrb1/Tspan5/Egfl7/Mfng/Bend6/Bmp7/Hey2 | 11 |
| GO:0010827 | regulation of glucose transmembrane transport | BP | 0.122 | 0.00389 | 0.0188 | 1.73 | 0.0122 | Igf1/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Pid1/Grb10/Myc/Irs1 | 11 |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | BP | 0.108 | 0.00397 | 0.0191 | 1.72 | 0.0124 | Sfrp2/Angpt1/Hcls1/Cd44/Pfn2/Prkd1/Tek/Caprin2/Arrb1/Arrb2/Atp2b4/Cav1/Sh2d3c/Camk1 | 14 |
| GO:0043406 | positive regulation of MAP kinase activity | BP | 0.108 | 0.00397 | 0.0191 | 1.72 | 0.0124 | Zeb2/Drd4/Lrrk2/Pik3r6/Edn3/Kit/Nox4/Kitl/Htr2a/Pde5a/Psen1/Pdgfrb/Map4k2/Fgd2 | 14 |
| GO:1990266 | neutrophil migration | BP | 0.108 | 0.00397 | 0.0191 | 1.72 | 0.0124 | Cxcl15/Mdk/Jam3/Edn3/Itga1/Ednra/Pecam1/Dysf/Nckap1l/Xcl1/Tnfaip6/Irak4/Ptger3/Mpp1 | 14 |
| GO:0031103 | axon regeneration | BP | 0.148 | 0.00405 | 0.0195 | 1.71 | 0.0126 | Gap43/Mmp2/Enpp1/Rtn4rl1/Neo1/Xylt1/Ptn/Nrep | 8 |
| GO:0032892 | positive regulation of organic acid transport | BP | 0.148 | 0.00405 | 0.0195 | 1.71 | 0.0126 | Acsl6/Grik1/Pla2g10/Abat/Grin2b/Acsl1/Psen1/Gabbr1 | 8 |
| GO:0034331 | cell junction maintenance | BP | 0.163 | 0.00412 | 0.0197 | 1.7 | 0.0128 | Epb41l3/Fermt2/Shank2/Erc2/Syngap1/Cldn1/F2r | 7 |
| GO:0042220 | response to cocaine | BP | 0.163 | 0.00412 | 0.0197 | 1.7 | 0.0128 | Slc1a3/Abat/Drd4/Htr1b/Htr2a/Dlg4/Smpd1 | 7 |
| GO:0048873 | homeostasis of number of cells within a tissue | BP | 0.163 | 0.00412 | 0.0197 | 1.7 | 0.0128 | Tex15/Col14a1/Nos3/Flt3/Coro1a/Akt3/F2r | 7 |
| GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.217 | 0.00415 | 0.0199 | 1.7 | 0.0129 | Sema5a/Ppp1r16b/Aplnr/Jcad/Agtr1a | 5 |
| GO:0098656 | anion transmembrane transport | BP | 0.0941 | 0.00416 | 0.0199 | 1.7 | 0.0129 | Slc1a3/Slc13a5/Tcaf1/Slc12a8/Ano1/Slc7a2/Ano2/Lrp2/Glrb/Gabra4/Slc23a2/Slc12a4/Slc4a8/Psen1/Abcb1a/Abcc5/Lrrc8c/Ank/Myc | 19 |
| GO:0048286 | lung alveolus development | BP | 0.136 | 0.00418 | 0.02 | 1.7 | 0.013 | Stra6/Kdr/Igf1/Pkdcc/Foxf1/Hopx/Tcf21/Ltbp3/Selenon | 9 |
| GO:0090288 | negative regulation of cellular response to growth factor stimulus | BP | 0.115 | 0.00424 | 0.0202 | 1.69 | 0.0131 | Sfrp2/Nog/Lrp2/Emilin1/Hipk2/Tnfaip6/Atp2b4/Apln/Adgra2/Sema6a/Cav1/Tmprss6 | 12 |
| GO:0010639 | negative regulation of organelle organization | BP | 0.0804 | 0.00433 | 0.0206 | 1.69 | 0.0134 | Kank4/Hgf/Map2/Fez1/Igf1/Evl/Phldb2/Tmod2/Bok/Lrrk2/Pfn2/Klhl22/Rps6ka2/Cdh5/Coro1a/Rp1/Arhgap6/Tubb4a/Prrt2/Pecam1/Tmeff2/Scin/Dysf/Dlc1/Arrb2/Arhgap28/Ttbk2/Cep97/Plekhh2/Bmp7 | 30 |
| GO:0051259 | protein complex oligomerization | BP | 0.0887 | 0.00438 | 0.0209 | 1.68 | 0.0135 | Kcnb2/Kcna6/Kcnd2/Evl/Kcnd3/Acacb/Bok/Grin2b/Ikzf4/Cth/Kcnc4/Pkd1/Mpp2/Pkd2/Nlrp1b/Aqp11/Kcng4/Steap4/Lrrc8c/Pdcd6ip/Cldn1/Kctd12 | 22 |
| GO:0050999 | regulation of nitric-oxide synthase activity | BP | 0.182 | 0.00442 | 0.021 | 1.68 | 0.0136 | Npr3/Tert/Apoe/Atp2b4/Cav1/Eng | 6 |
| GO:0008585 | female gonad development | BP | 0.11 | 0.00445 | 0.0211 | 1.68 | 0.0137 | Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lfng/Kit/Kitl/Arrb1/Tnfaip6/Arrb2/Stat5b | 13 |
| GO:0051897 | positive regulation of protein kinase B signaling | BP | 0.11 | 0.00445 | 0.0211 | 1.68 | 0.0137 | Sema5a/Angpt1/Igf1/Hcls1/Fermt2/Tek/Cxcl12/Lrp2/Nox4/Igf2/Arrb2/Rasd2/Eng | 13 |
| GO:0071692 | protein localization to extracellular region | BP | 0.0788 | 0.00445 | 0.0211 | 1.68 | 0.0137 | Cacna1c/Igf1/Abat/Drd4/Ednrb/Myo5a/Ano1/Cacna1e/Stxbp4/Rab34/Vegfc/Cd38/Agtr1a/Apoe/Syt7/Cd200/Arrb1/Rcn3/Neo1/Nrros/P3h1/Abcg1/Frmd4a/Ptger3/Sri/Nr1h3/Pfkm/Golph3l/F2r/Nlgn2/Plek/Irs1 | 32 |
| GO:0051057 | positive regulation of small GTPase mediated signal transduction | BP | 0.127 | 0.0045 | 0.0213 | 1.67 | 0.0138 | Igf1/Lpar4/Dock2/Adcyap1r1/Kitl/Arrb1/Net1/Mapre2/Pdgfrb/F2r | 10 |
| GO:0052548 | regulation of endopeptidase activity | BP | 0.081 | 0.0045 | 0.0213 | 1.67 | 0.0138 | Hgf/Sfrp2/Spink1/Igf1/Aph1b/Cd44/Bok/Wdr35/Grin2b/Mical1/Serpine2/Ift57/Serpini1/Wnt9a/Ppm1f/Hip1/Bin1/Arrb1/Epha7/Dlc1/Arrb2/Asph/Fyn/Nlrp1b/Ccn2/Mgmt/Cyfip2/F2r/Myc | 29 |
| GO:0010038 | response to metal ion | BP | 0.0847 | 0.00453 | 0.0214 | 1.67 | 0.0139 | Slc13a5/Nlgn1/Syt13/Abat/Cpne5/Fn1/Lrrk2/Mef2c/Nptx1/Rasgrp2/Adcy7/Dlg2/Enpp1/Syt7/Kcnma1/Nfatc4/Kcnip3/Asph/Pkd2/Slc25a23/Sord/Dlg4/Smpd1/Cav1/Kcnh1 | 25 |
| GO:0046394 | carboxylic acid biosynthetic process | BP | 0.0847 | 0.00453 | 0.0214 | 1.67 | 0.0139 | Slc1a3/Gulo/Pla2g10/Fads2/Cyp7b1/Fads1/Abat/Acacb/Cbs/Myo5a/Elovl4/Gamt/Cth/Phgdh/Fads3/Acot7/Atp2b4/Abcd2/Gstm7/Asl/Nr1h3/Hacd1/Rdh1/Olah/Abhd3 | 25 |
| GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | BP | 0.145 | 0.00455 | 0.0214 | 1.67 | 0.0139 | Adra1a/Nos3/Ednrb/Enpep/Edn3/Rps6ka2/Agtr1a/F2r | 8 |
| GO:0034113 | heterotypic cell-cell adhesion | BP | 0.145 | 0.00455 | 0.0214 | 1.67 | 0.0139 | Itga4/Cd44/Itgb7/Jam3/Thy1/Cd200/Itga7/Bmp7 | 8 |
| GO:0046580 | negative regulation of Ras protein signal transduction | BP | 0.145 | 0.00455 | 0.0214 | 1.67 | 0.0139 | Adra1a/Rasa3/Rasip1/Arap3/Heg1/Dlc1/Syngap1/Rasal3 | 8 |
| GO:0048546 | digestive tract morphogenesis | BP | 0.145 | 0.00455 | 0.0214 | 1.67 | 0.0139 | Stra6/Ihh/Sfrp2/Foxf1/Tcf21/Gli3/Stx2/Dact1 | 8 |
| GO:1902476 | chloride transmembrane transport | BP | 0.145 | 0.00455 | 0.0214 | 1.67 | 0.0139 | Slc1a3/Slc12a8/Ano1/Ano2/Glrb/Gabra4/Slc12a4/Slc4a8 | 8 |
| GO:0043244 | regulation of protein-containing complex disassembly | BP | 0.106 | 0.00456 | 0.0214 | 1.67 | 0.0139 | Map2/Sema5a/Evl/Tmod2/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2/Setx/Plek | 14 |
| GO:0097581 | lamellipodium organization | BP | 0.12 | 0.00461 | 0.0217 | 1.66 | 0.014 | Ccdc88a/Ptpro/Cd44/Wasf3/Abi3/Cdh13/Abi2/Kit/Parvb/Plekho1/Enpp2 | 11 |
| GO:0001707 | mesoderm formation | BP | 0.134 | 0.00463 | 0.0217 | 1.66 | 0.0141 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Snai1/Tal1/Bmp7 | 9 |
| GO:0030193 | regulation of blood coagulation | BP | 0.134 | 0.00463 | 0.0217 | 1.66 | 0.0141 | Tfpi/Abat/Serpine2/Apoe/Pros1/Ptger3/St3gal4/Cav1/F2r | 9 |
| GO:0090398 | cellular senescence | BP | 0.134 | 0.00463 | 0.0217 | 1.66 | 0.0141 | Tbx2/Tert/Abi3/Vash1/Tbx3/Smc5/Akt3/Cav1/Twist1 | 9 |
| GO:0046640 | regulation of alpha-beta T cell proliferation | BP | 0.159 | 0.0047 | 0.022 | 1.66 | 0.0142 | Cd44/Mapk8ip1/Btla/Xcl1/Zap70/Il12rb1/Rasal3 | 7 |
| GO:0097028 | dendritic cell differentiation | BP | 0.159 | 0.0047 | 0.022 | 1.66 | 0.0142 | Flt3/Irf8/Zbtb46/Gimap5/Gimap3/Psen1/Batf | 7 |
| GO:0016053 | organic acid biosynthetic process | BP | 0.0845 | 0.00473 | 0.022 | 1.66 | 0.0142 | Slc1a3/Gulo/Pla2g10/Fads2/Cyp7b1/Fads1/Abat/Acacb/Cbs/Myo5a/Elovl4/Gamt/Cth/Phgdh/Fads3/Acot7/Atp2b4/Abcd2/Gstm7/Asl/Nr1h3/Hacd1/Rdh1/Olah/Abhd3 | 25 |
| GO:0002396 | MHC protein complex assembly | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0002501 | peptide antigen assembly with MHC protein complex | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0003188 | heart valve formation | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Erg/Dchs1/Hey2/Twist1 | 4 |
| GO:0035721 | intraciliary retrograde transport | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Wdr35/Dync2h1/Dync2li1/Bbs1 | 4 |
| GO:0042693 | muscle cell fate commitment | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | AW551984/Tbx2/Mef2c/Tbx3 | 4 |
| GO:0043383 | negative T cell selection | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Gli3/Dock2/Zap70/Spn | 4 |
| GO:0072074 | kidney mesenchyme development | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Tcf21/Osr1/Bmp7/Myc | 4 |
| GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Efnb3/Gucy1b1/Gucy1a1/F2r | 4 |
| GO:1901386 | negative regulation of voltage-gated calcium channel activity | BP | 0.267 | 0.00474 | 0.022 | 1.66 | 0.0142 | Drd4/Dysf/Rrad/Gnb5 | 4 |
| GO:0014020 | primary neural tube formation | BP | 0.109 | 0.00478 | 0.0221 | 1.65 | 0.0143 | Sfrp2/Nog/Zeb2/Ift57/Lrp2/Cecr2/Prickle1/Dlc1/Ptch1/Rab23/Fzd2/Bmp7/Twist1 | 13 |
| GO:0048709 | oligodendrocyte differentiation | BP | 0.109 | 0.00478 | 0.0221 | 1.65 | 0.0143 | Ptprz1/Mdk/Wasf3/Id4/Gli3/Slc8a3/Nlgn3/Enpp1/Daam2/Ptn/Tppp/Enpp2/Zfp365 | 13 |
| GO:0010469 | regulation of signaling receptor activity | BP | 0.102 | 0.00488 | 0.0226 | 1.65 | 0.0146 | Begain/Nog/Nlgn1/Areg/Slc24a4/Mef2c/Nptx1/Cnrip1/Nlgn3/Nptxr/Prrt1/Cacng7/Dlg4/Psen1/Nlgn2 | 15 |
| GO:0032103 | positive regulation of response to external stimulus | BP | 0.0764 | 0.00493 | 0.0228 | 1.64 | 0.0148 | Sema5a/Acod1/Mdk/Kdr/Nrp1/Ets1/Fn1/Lrrk2/Mmp2/Prkd1/Cxcr3/Cdh13/Cxcl12/Gprc5b/Edn3/Rab34/Ppm1f/Cd226/Emilin1/Vegfc/Havcr2/Ednra/Dysf/Nckap1l/Xcl1/Masp1/Stat5b/Pde5a/Nlrp1b/Ptger3/Gimap5/Gimap3/Pdgfrb/Ptn/Clnk | 35 |
| GO:0009953 | dorsal/ventral pattern formation | BP | 0.113 | 0.00494 | 0.0228 | 1.64 | 0.0148 | Gli1/Hhip/Hoxd11/Nog/Hoxa11/Gli3/Gpr161/Dync2h1/Ptch1/Rab23/Psen1/Lrp4 | 12 |
| GO:0015844 | monoamine transport | BP | 0.113 | 0.00494 | 0.0228 | 1.64 | 0.0148 | Maob/Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 12 |
| GO:0006898 | receptor-mediated endocytosis | BP | 0.0865 | 0.00496 | 0.0229 | 1.64 | 0.0148 | Angpt1/Itga4/Clec9a/Drd4/Calca/Lrp2/Htr1b/Ldlrad3/Nlgn3/Apoe/Magi2/Sele/Hip1/Arrb1/Cacng7/Arrb2/Calcrl/Dlg4/Ptger3/Apln/Cav1/Pld2/Ankrd13a | 23 |
| GO:0051896 | regulation of protein kinase B signaling | BP | 0.0966 | 0.00498 | 0.0229 | 1.64 | 0.0149 | Sema5a/Angpt1/Igf1/Hcls1/Fermt2/Inpp4b/Tek/Cxcl12/Lrp2/Nox4/Magi2/Igf2/Cavin3/Rcn3/Arrb2/Rasd2/Eng | 17 |
| GO:0044272 | sulfur compound biosynthetic process | BP | 0.118 | 0.00501 | 0.023 | 1.64 | 0.0149 | Acsl6/Angpt1/Acacb/Cbs/Cth/Chst1/Dse/Acsl1/Xylt1/Ggt7/Papss2 | 11 |
| GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | BP | 0.118 | 0.00501 | 0.023 | 1.64 | 0.0149 | Gpc6/Stx1b/Drd4/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Cpt1c/Dlg4 | 11 |
| GO:0003181 | atrioventricular valve morphogenesis | BP | 0.208 | 0.00504 | 0.0231 | 1.64 | 0.015 | Heyl/Dchs1/Hey2/Slit3/Twist1 | 5 |
| GO:0009190 | cyclic nucleotide biosynthetic process | BP | 0.208 | 0.00504 | 0.0231 | 1.64 | 0.015 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0030903 | notochord development | BP | 0.208 | 0.00504 | 0.0231 | 1.64 | 0.015 | Gli1/Wnt11/Nog/Id3/Col13a1 | 5 |
| GO:0036303 | lymph vessel morphogenesis | BP | 0.208 | 0.00504 | 0.0231 | 1.64 | 0.015 | Vash1/Vegfc/Tie1/Pkd1/Svep1 | 5 |
| GO:0052652 | cyclic purine nucleotide metabolic process | BP | 0.208 | 0.00504 | 0.0231 | 1.64 | 0.015 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0050433 | regulation of catecholamine secretion | BP | 0.132 | 0.00511 | 0.0234 | 1.63 | 0.0152 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Gabbr1 | 9 |
| GO:0007498 | mesoderm development | BP | 0.108 | 0.00513 | 0.0234 | 1.63 | 0.0152 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Tbx3/Snai1/Tal1/Osr1/Pus7/Ets2/Bmp7 | 13 |
| GO:0002092 | positive regulation of receptor internalization | BP | 0.176 | 0.00516 | 0.0235 | 1.63 | 0.0152 | Angpt1/Magi2/Sele/Arrb1/Arrb2/Apln | 6 |
| GO:0010714 | positive regulation of collagen metabolic process | BP | 0.176 | 0.00516 | 0.0235 | 1.63 | 0.0152 | Ihh/Ddr2/Arrb2/Ccn2/Pdgfrb/F2r | 6 |
| GO:0051497 | negative regulation of stress fiber assembly | BP | 0.176 | 0.00516 | 0.0235 | 1.63 | 0.0152 | Kank4/Phldb2/Arhgap6/Tmeff2/Dlc1/Arhgap28 | 6 |
| GO:0035303 | regulation of dephosphorylation | BP | 0.104 | 0.00521 | 0.0237 | 1.63 | 0.0154 | Npnt/Mef2c/Itga1/Cdh5/Ppp1r16b/Magi2/Nckap1l/Cmya5/Dlc1/Ppp1r26/Smpd1/Pdgfrb/Vcan/Plek | 14 |
| GO:0071214 | cellular response to abiotic stimulus | BP | 0.0818 | 0.00524 | 0.0238 | 1.62 | 0.0154 | Sfrp2/Wnt11/Nos3/Ccnd2/Mmp2/Slc24a4/Eif2ak4/Mme/Scn2a/Rp1/Cnn2/Ednra/Dysf/Bdkrb2/Nfatc4/Pkd2/Net1/Slc25a23/Habp4/Palm/Gnb5/Atp1a2/Smpd1/Piezo2/Cav1/Lrrc8c/Sipa1 | 27 |
| GO:0104004 | cellular response to environmental stimulus | BP | 0.0818 | 0.00524 | 0.0238 | 1.62 | 0.0154 | Sfrp2/Wnt11/Nos3/Ccnd2/Mmp2/Slc24a4/Eif2ak4/Mme/Scn2a/Rp1/Cnn2/Ednra/Dysf/Bdkrb2/Nfatc4/Pkd2/Net1/Slc25a23/Habp4/Palm/Gnb5/Atp1a2/Smpd1/Piezo2/Cav1/Lrrc8c/Sipa1 | 27 |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | BP | 0.112 | 0.00533 | 0.0242 | 1.62 | 0.0157 | Igf1/Ddr2/Tert/Mmp2/Cdh13/Adamts1/Htr1b/Stat5b/Irak4/Calcrl/Pdgfrb/Myc | 12 |
| GO:0071456 | cellular response to hypoxia | BP | 0.112 | 0.00533 | 0.0242 | 1.62 | 0.0157 | Kcnk3/Kcnd2/Ddah1/Cbs/Tert/Slc8a3/Scn2a/Ndnf/Cpeb1/Hif3a/Myc/Twist1 | 12 |
| GO:0030225 | macrophage differentiation | BP | 0.156 | 0.00534 | 0.0242 | 1.62 | 0.0157 | Hcls1/Plcb1/Calca/Enpp1/Zbtb46/Nrros/Cd4 | 7 |
| GO:0021543 | pallium development | BP | 0.0982 | 0.00542 | 0.0245 | 1.61 | 0.0159 | Mdk/Lef1/Zeb2/Cdh2/Dab1/Id4/Fut10/Gli3/Plcb1/Tsku/Dclk2/Lhx6/Nr2f1/Fat4/Bbs1/Psen1 | 16 |
| GO:0021954 | central nervous system neuron development | BP | 0.117 | 0.00543 | 0.0246 | 1.61 | 0.0159 | Map2/Dcc/Zeb2/Nrp1/Tsku/Cdh11/Ndnf/Dclk2/Nin/Lhx6/Draxin | 11 |
| GO:0015749 | monosaccharide transmembrane transport | BP | 0.107 | 0.0055 | 0.0248 | 1.6 | 0.0161 | Igf1/Slc2a3/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Slc23a2/Pid1/Grb10/Myc/Irs1 | 13 |
| GO:0015837 | amine transport | BP | 0.107 | 0.0055 | 0.0248 | 1.6 | 0.0161 | Grik1/Syt13/Abat/Myo5a/Grin2b/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Psen1/Gabbr1/Cacna1a | 13 |
| GO:0044106 | cellular amine metabolic process | BP | 0.107 | 0.0055 | 0.0248 | 1.6 | 0.0161 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Atp2b4/Slc7a7/Sult1a1/Hand2 | 13 |
| GO:0055007 | cardiac muscle cell differentiation | BP | 0.101 | 0.00552 | 0.0249 | 1.6 | 0.0161 | AW551984/Sgcd/Col14a1/Igf1/Adra1a/Slc8a1/Mef2c/Nox4/Tbx3/Arrb2/Pi16/Sgcb/Pdgfrb/Hey2/Hand2 | 15 |
| GO:2001056 | positive regulation of cysteine-type endopeptidase activity | BP | 0.101 | 0.00552 | 0.0249 | 1.6 | 0.0161 | Bok/Wdr35/Grin2b/Ift57/Ppm1f/Hip1/Arrb1/Dlc1/Asph/Fyn/Nlrp1b/Ccn2/Cyfip2/F2r/Myc | 15 |
| GO:0030902 | hindbrain development | BP | 0.0955 | 0.00557 | 0.0251 | 1.6 | 0.0163 | Gli1/Mdk/Lef1/Nog/Igf1/Cbs/Dab1/B4galt2/Serpine2/Zfp423/Cplane1/Smad9/Dlc1/Ttbk2/Cacna1a/Bmp7/Zfp365 | 17 |
| GO:0031646 | positive regulation of nervous system process | BP | 0.14 | 0.00567 | 0.0255 | 1.59 | 0.0165 | Hgf/Igf1/Abat/Wasf3/Nlgn3/Tppp/Prkar1b/Nlgn2 | 8 |
| GO:0035567 | non-canonical Wnt signaling pathway | BP | 0.14 | 0.00567 | 0.0255 | 1.59 | 0.0165 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Daam2/Fzd2/Dact1 | 8 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | BP | 0.111 | 0.00574 | 0.0258 | 1.59 | 0.0167 | Sfrp2/Sema5a/Zeb2/Gpc3/Lrrk2/Gprc5b/Peg12/Daam2/Caprin2/Adgra2/Cav1/Dact1 | 12 |
| GO:0045913 | positive regulation of carbohydrate metabolic process | BP | 0.122 | 0.00587 | 0.0263 | 1.58 | 0.017 | Igf1/Has2/Pth1r/Igf2/Adcyap1r1/Htr2a/Zbtb20/Psen1/Myc/Irs1 | 10 |
| GO:0070830 | bicellular tight junction assembly | BP | 0.122 | 0.00587 | 0.0263 | 1.58 | 0.017 | Wnt11/Cldn8/Cdh5/Esam/Pecam1/Snai1/Nphp1/Cldn5/Pdcd6ip/Cldn1 | 10 |
| GO:0009063 | cellular amino acid catabolic process | BP | 0.116 | 0.00588 | 0.0263 | 1.58 | 0.017 | Ddah1/Abat/Nos3/Cbs/Asrgl1/Fah/Amdhd1/Hdc/Atp2b4/Sardh/Prodh | 11 |
| GO:0045685 | regulation of glial cell differentiation | BP | 0.116 | 0.00588 | 0.0263 | 1.58 | 0.017 | Ptprz1/Mdk/Nog/Dab1/Id4/Serpine2/Daam2/Bin1/Ptn/Enpp2/Zfp365 | 11 |
| GO:0061053 | somite development | BP | 0.116 | 0.00588 | 0.0263 | 1.58 | 0.017 | Ihh/Sfrp2/Lef1/Wnt11/Nog/Zeb2/Foxf1/Lfng/Loxl3/Ptch1/Psen1 | 11 |
| GO:0009117 | nucleotide metabolic process | BP | 0.0761 | 0.00593 | 0.0265 | 1.58 | 0.0171 | Nme4/Acsl6/Igf1/Hmgcs2/Nos3/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Lacc1/Htr2a/Acot7/Zbtb20/Sarm1/Pde5a/Rab23/Nt5c3b/Acsl1/Impdh1/Dpyd/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 34 |
| GO:0035116 | embryonic hindlimb morphogenesis | BP | 0.171 | 0.00597 | 0.0266 | 1.58 | 0.0172 | Aff3/Gpc3/Osr2/Tbx3/Osr1/Twist1 | 6 |
| GO:0042908 | xenobiotic transport | BP | 0.171 | 0.00597 | 0.0266 | 1.58 | 0.0172 | Abcg2/Abca8b/Abcb1a/Abcc5/Abcg3/Cldn1 | 6 |
| GO:0045987 | positive regulation of smooth muscle contraction | BP | 0.171 | 0.00597 | 0.0266 | 1.58 | 0.0172 | Npnt/Adra1a/Abat/Kit/Ptger3/F2r | 6 |
| GO:0090162 | establishment of epithelial cell polarity | BP | 0.171 | 0.00597 | 0.0266 | 1.58 | 0.0172 | Foxf1/Myo9a/Msn/Frmd4b/Sh3bp1/Frmd4a | 6 |
| GO:0001990 | regulation of systemic arterial blood pressure by hormone | BP | 0.152 | 0.00604 | 0.0267 | 1.57 | 0.0173 | Nos3/Ednrb/Enpep/Edn3/Rps6ka2/Agtr1a/F2r | 7 |
| GO:0031952 | regulation of protein autophosphorylation | BP | 0.152 | 0.00604 | 0.0267 | 1.57 | 0.0173 | Enpp1/Vegfc/Epha7/Rassf2/Cav1/Rap2a/Eng | 7 |
| GO:0042417 | dopamine metabolic process | BP | 0.152 | 0.00604 | 0.0267 | 1.57 | 0.0173 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Agtr1a | 7 |
| GO:0007413 | axonal fasciculation | BP | 0.2 | 0.00605 | 0.0267 | 1.57 | 0.0173 | Ptprz1/Sema5a/Nrp1/Ndn/Epha3 | 5 |
| GO:0010996 | response to auditory stimulus | BP | 0.2 | 0.00605 | 0.0267 | 1.57 | 0.0173 | Stra6/Slc1a3/Mdk/Atp1a2/Ptn | 5 |
| GO:0035024 | negative regulation of Rho protein signal transduction | BP | 0.2 | 0.00605 | 0.0267 | 1.57 | 0.0173 | Adra1a/Rasip1/Arap3/Heg1/Dlc1 | 5 |
| GO:0036010 | protein localization to endosome | BP | 0.2 | 0.00605 | 0.0267 | 1.57 | 0.0173 | Nrp1/Msn/Mgat3/Akap11/Ankrd13a | 5 |
| GO:0106030 | neuron projection fasciculation | BP | 0.2 | 0.00605 | 0.0267 | 1.57 | 0.0173 | Ptprz1/Sema5a/Nrp1/Ndn/Epha3 | 5 |
| GO:0008212 | mineralocorticoid metabolic process | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Ednrb/Dkk3/Kcnma1/Cacna1h | 4 |
| GO:0031650 | regulation of heat generation | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Abat/Ednrb/Ptger3/Apln | 4 |
| GO:0034433 | steroid esterification | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Soat1/Agtr1a/Apoe/Abcg1 | 4 |
| GO:0034434 | sterol esterification | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Soat1/Agtr1a/Apoe/Abcg1 | 4 |
| GO:0034435 | cholesterol esterification | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Soat1/Agtr1a/Apoe/Abcg1 | 4 |
| GO:0038065 | collagen-activated signaling pathway | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Ddr2/Col4a6/Enpp1/Col4a5 | 4 |
| GO:0043084 | penile erection | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Ar/Drd4/Ednrb/Ednra | 4 |
| GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Cd44/Snai1/Pcbp4/Twist1 | 4 |
| GO:2000479 | regulation of cAMP-dependent protein kinase activity | BP | 0.25 | 0.00608 | 0.0267 | 1.57 | 0.0173 | Pkia/Pkib/Atp2b4/Prkar1b | 4 |
| GO:0051346 | negative regulation of hydrolase activity | BP | 0.0792 | 0.00609 | 0.0267 | 1.57 | 0.0173 | Hgf/Sfrp2/Cd109/Spink1/Igf1/Tfpi/Nos3/Cd44/Gpc3/Lrrk2/Mical1/Serpine2/Wfdc3/Serpini1/Wnt9a/Pecam1/Bin1/Nckap1l/Cmya5/Arrb1/Hdac9/Arrb2/Pi16/Angptl4/Simc1/Ppp1r26/Pi15/Gpsm1/Wfdc1 | 29 |
| GO:0009306 | protein secretion | BP | 0.0777 | 0.0062 | 0.0271 | 1.57 | 0.0176 | Cacna1c/Igf1/Abat/Drd4/Ednrb/Myo5a/Ano1/Cacna1e/Stxbp4/Rab34/Vegfc/Cd38/Agtr1a/Apoe/Syt7/Cd200/Arrb1/Rcn3/Neo1/P3h1/Abcg1/Frmd4a/Ptger3/Sri/Nr1h3/Pfkm/Golph3l/F2r/Nlgn2/Plek/Irs1 | 31 |
| GO:0046324 | regulation of glucose import | BP | 0.129 | 0.0062 | 0.0271 | 1.57 | 0.0176 | Igf1/Tert/Gpc3/Sh2b2/Enpp1/Pid1/Grb10/Myc/Irs1 | 9 |
| GO:0050885 | neuromuscular process controlling balance | BP | 0.129 | 0.0062 | 0.0271 | 1.57 | 0.0176 | Ankfn1/Slc1a3/Jph4/Myo5a/Kcnma1/Foxs1/Dlg4/Cacna1a/Nlgn2 | 9 |
| GO:0007254 | JNK cascade | BP | 0.0944 | 0.00622 | 0.0272 | 1.57 | 0.0176 | Sfrp2/Eda2r/Lrrk2/Mapk8ip1/Plcb1/Grik2/Cracr2a/Hipk2/Mapkbp1/Rassf2/Irak4/Ccn2/Cdc42ep5/Map4k2/Dact1/Edar/Rap2a | 17 |
| GO:0051147 | regulation of muscle cell differentiation | BP | 0.0993 | 0.00624 | 0.0272 | 1.56 | 0.0177 | Igf1/Mef2c/Hopx/Zeb1/Tshz3/Kit/Cth/Igf2/Hdac9/Arrb2/Pi16/Rbpms2/Efemp2/Camk1/Eng | 15 |
| GO:1900180 | regulation of protein localization to nucleus | BP | 0.0993 | 0.00624 | 0.0272 | 1.56 | 0.0177 | Angpt1/Hcls1/Fermt2/Tert/Prkd1/Tek/Gli3/Pkia/Dclk2/Fyn/Rab23/Lats2/Psen1/Glis2/Cacnb4 | 15 |
| GO:0042542 | response to hydrogen peroxide | BP | 0.106 | 0.00631 | 0.0275 | 1.56 | 0.0178 | Cyp1b1/Hgf/Ddr2/Lrrk2/Mmp2/Trpc6/Ednra/Net1/Fyn/Pdgfrb/Pde8a/Pld2/Setx | 13 |
| GO:0043030 | regulation of macrophage activation | BP | 0.138 | 0.00631 | 0.0275 | 1.56 | 0.0178 | Pla2g10/Lrrk2/Slc7a2/Cd1d1/Havcr2/Cd200/Cd84/Nr1h3 | 8 |
| GO:1902808 | positive regulation of cell cycle G1/S phase transition | BP | 0.138 | 0.00631 | 0.0275 | 1.56 | 0.0178 | Gli1/Ddr2/Tbx2/Tert/Ccnd2/Adamts1/Plcb1/Stxbp4 | 8 |
| GO:0042633 | hair cycle | BP | 0.102 | 0.00632 | 0.0275 | 1.56 | 0.0178 | Apcdd1/Pla2g10/Cd109/Ptch2/Fst/Tert/Edaradd/Myo5a/Ldb2/Tsku/Snai1/Psen1/Edar/Lrp4 | 14 |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | BP | 0.115 | 0.00636 | 0.0276 | 1.56 | 0.0179 | Hhip/Nog/Tbx2/Cd44/Enpp1/Ndnf/Ccn2/Apln/Fat4/Kif16b/Setx | 11 |
| GO:0035592 | establishment of protein localization to extracellular region | BP | 0.0775 | 0.00642 | 0.0279 | 1.55 | 0.0181 | Cacna1c/Igf1/Abat/Drd4/Ednrb/Myo5a/Ano1/Cacna1e/Stxbp4/Rab34/Vegfc/Cd38/Agtr1a/Apoe/Syt7/Cd200/Arrb1/Rcn3/Neo1/P3h1/Abcg1/Frmd4a/Ptger3/Sri/Nr1h3/Pfkm/Golph3l/F2r/Nlgn2/Plek/Irs1 | 31 |
| GO:0050708 | regulation of protein secretion | BP | 0.0822 | 0.0066 | 0.0286 | 1.54 | 0.0186 | Igf1/Abat/Drd4/Ano1/Cacna1e/Stxbp4/Rab34/Vegfc/Cd38/Apoe/Syt7/Cd200/Arrb1/Neo1/P3h1/Abcg1/Frmd4a/Ptger3/Sri/Nr1h3/Pfkm/Golph3l/F2r/Nlgn2/Irs1 | 25 |
| GO:0046620 | regulation of organ growth | BP | 0.105 | 0.00675 | 0.0293 | 1.53 | 0.019 | Gli1/Cacna1c/Col14a1/Nog/Igf1/Tbx2/Acacb/Mef2c/Flvcr1/Igf2/Pi16/Lats2/Hey2 | 13 |
| GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | BP | 0.127 | 0.0068 | 0.0294 | 1.53 | 0.0191 | Npr3/Drd4/Akap12/Htr1b/Ednra/Gnaz/Palm/Gabbr1/S1pr3 | 9 |
| GO:0017156 | calcium-ion regulated exocytosis | BP | 0.127 | 0.0068 | 0.0294 | 1.53 | 0.0191 | Syn2/Cacna1c/Syt13/Rims3/Stx1b/Cadps/Syt7/Cacna1h/Cacna1a | 9 |
| GO:0035307 | positive regulation of protein dephosphorylation | BP | 0.149 | 0.00681 | 0.0294 | 1.53 | 0.0191 | Itga1/Cdh5/Ppp1r16b/Magi2/Dlc1/Smpd1/Pdgfrb | 7 |
| GO:1903524 | positive regulation of blood circulation | BP | 0.149 | 0.00681 | 0.0294 | 1.53 | 0.0191 | Adra1a/Edn3/Ptger3/Ccn2/Apln/Hey2/Ptger2 | 7 |
| GO:0010922 | positive regulation of phosphatase activity | BP | 0.167 | 0.00688 | 0.0297 | 1.53 | 0.0192 | Npnt/Mef2c/Itga1/Magi2/Pdgfrb/Plek | 6 |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | BP | 0.167 | 0.00688 | 0.0297 | 1.53 | 0.0192 | Ar/Igf1/Plcb1/Igfbp4/Grb10/Irs1 | 6 |
| GO:0009416 | response to light stimulus | BP | 0.081 | 0.00689 | 0.0297 | 1.53 | 0.0193 | Slc1a3/Sema5a/Cacna1c/Nog/Sema5b/Mmp2/Slc24a4/B4galt2/Pde1b/Cacna1e/Eif2ak4/Fbxl21/Kit/Mme/Rp1/Nlgn3/Arrb1/Nfatc4/Gnb5/Ddhd2/Atp1a2/Syngap1/Brsk1/Smpd1/Usp2/Cacnb4 | 26 |
| GO:0051260 | protein homooligomerization | BP | 0.0934 | 0.00693 | 0.0299 | 1.52 | 0.0193 | Kcnb2/Kcna6/Kcnd2/Evl/Kcnd3/Acacb/Ikzf4/Cth/Kcnc4/Mpp2/Pkd2/Nlrp1b/Aqp11/Kcng4/Steap4/Pdcd6ip/Kctd12 | 17 |
| GO:2001259 | positive regulation of cation channel activity | BP | 0.119 | 0.00695 | 0.0299 | 1.52 | 0.0194 | Cacna2d1/Ank2/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 10 |
| GO:0007528 | neuromuscular junction development | BP | 0.136 | 0.007 | 0.0301 | 1.52 | 0.0195 | Pdzrn3/P2rx2/Lrrk2/Ptn/Cacnb4/F2r/Lrp4/Col4a5 | 8 |
| GO:0071622 | regulation of granulocyte chemotaxis | BP | 0.136 | 0.007 | 0.0301 | 1.52 | 0.0195 | Mdk/Jam3/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Mpp1 | 8 |
| GO:1903793 | positive regulation of anion transport | BP | 0.136 | 0.007 | 0.0301 | 1.52 | 0.0195 | Grik1/Pla2g10/Tcaf1/Abat/Grin2b/Psen1/Abcb1a/Gabbr1 | 8 |
| GO:0034599 | cellular response to oxidative stress | BP | 0.0839 | 0.00704 | 0.0302 | 1.52 | 0.0196 | Cyp1b1/Hgf/Ddr2/Slc8a1/Nos3/Lrrk2/Mmp2/Prkd1/Tbc1d24/Trpc6/Aifm2/Prr5l/Ednra/Selenon/Net1/Fyn/Mgat3/Stx2/Pdgfrb/Meak7/Pde8a/Setx/Fbln5 | 23 |
| GO:0030859 | polarized epithelial cell differentiation | BP | 0.192 | 0.0072 | 0.0308 | 1.51 | 0.02 | Foxf1/Myo9a/Msn/Sh3bp1/Dlg5 | 5 |
| GO:0043171 | peptide catabolic process | BP | 0.192 | 0.0072 | 0.0308 | 1.51 | 0.02 | Enpep/Cpe/Trhde/Ggt7/Lvrn | 5 |
| GO:0060065 | uterus development | BP | 0.192 | 0.0072 | 0.0308 | 1.51 | 0.02 | Stra6/Hoxa11/Hoxa10/Cdkn1c/Hoxa9 | 5 |
| GO:1902473 | regulation of protein localization to synapse | BP | 0.192 | 0.0072 | 0.0308 | 1.51 | 0.02 | Gpc6/Nlgn1/Nlgn3/Magi2/Nlgn2 | 5 |
| GO:0007259 | receptor signaling pathway via JAK-STAT | BP | 0.0909 | 0.00733 | 0.0313 | 1.5 | 0.0203 | Stra6/Cyp1b1/Agap2/Ptprd/Igf1/Hcls1/Dab1/Kit/Agtr1a/Pecam1/Pkd1/Stat5b/Pkd2/Fyn/Csf2rb/Il12rb1/Cav1/Il7r | 18 |
| GO:0030705 | cytoskeleton-dependent intracellular transport | BP | 0.0909 | 0.00733 | 0.0313 | 1.5 | 0.0203 | Map2/Fez1/Tub/Ccdc88a/Myo5a/Wdr35/Ppfia2/Ift57/Kif1a/Dlg2/Dync2h1/Ttc26/Arl3/Myo1f/Dync2li1/Bbs1/Ift81/Kif5a | 18 |
| GO:0071774 | response to fibroblast growth factor | BP | 0.112 | 0.00741 | 0.0316 | 1.5 | 0.0205 | Hhip/Nog/Tbx2/Cd44/Enpp1/Ndnf/Ccn2/Apln/Fat4/Kif16b/Setx | 11 |
| GO:0045428 | regulation of nitric oxide biosynthetic process | BP | 0.125 | 0.00745 | 0.0317 | 1.5 | 0.0205 | Igf1/Ddah1/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 9 |
| GO:0046579 | positive regulation of Ras protein signal transduction | BP | 0.125 | 0.00745 | 0.0317 | 1.5 | 0.0205 | Igf1/Lpar4/Dock2/Kitl/Arrb1/Net1/Mapre2/Pdgfrb/F2r | 9 |
| GO:0048168 | regulation of neuronal synaptic plasticity | BP | 0.125 | 0.00745 | 0.0317 | 1.5 | 0.0205 | Grik1/Kdr/Nog/Grin2b/Kit/Grik2/Dlg4/Syngap1/Cpeb1 | 9 |
| GO:2000179 | positive regulation of neural precursor cell proliferation | BP | 0.125 | 0.00745 | 0.0317 | 1.5 | 0.0205 | Gli1/Mdk/Nog/Igf1/Zfp423/Gli3/Lrp2/Vegfc/Nes | 9 |
| GO:0001843 | neural tube closure | BP | 0.107 | 0.00764 | 0.0321 | 1.49 | 0.0208 | Sfrp2/Nog/Zeb2/Ift57/Lrp2/Cecr2/Prickle1/Dlc1/Ptch1/Rab23/Fzd2/Twist1 | 12 |
| GO:1905477 | positive regulation of protein localization to membrane | BP | 0.107 | 0.00764 | 0.0321 | 1.49 | 0.0208 | Tcaf1/Myo5a/Epha3/Tnfaip6/Stac2/Fyn/Dlg4/Atp2b4/Ptn/Nkd2/Lrp4/Nlgn2 | 12 |
| GO:0002295 | T-helper cell lineage commitment | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Loxl3/Batf/Spn/Ly9 | 4 |
| GO:0003184 | pulmonary valve morphogenesis | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Stra6/Nos3/Heyl/Hey2 | 4 |
| GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Nog/Heyl/Snai1/Eng | 4 |
| GO:0003222 | ventricular trabecula myocardium morphogenesis | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Ccm2l/Heg1/Hey2/Eng | 4 |
| GO:0006491 | N-glycan processing | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | St8sia2/St8sia4/Man2a2/St8sia6 | 4 |
| GO:0035337 | fatty-acyl-CoA metabolic process | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Acsl6/Them5/Acot7/Acsl1 | 4 |
| GO:0042756 | drinking behavior | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Slc24a4/Htr1b/Agtr1a/Apln | 4 |
| GO:0045059 | positive thymic T cell selection | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Cd3g/Dock2/Cd1d1/Zap70 | 4 |
| GO:0051132 | NK T cell activation | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Cd1d1/Il12rb1/Myc/Rasal3 | 4 |
| GO:0061000 | negative regulation of dendritic spine development | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Nlgn1/Nlgn3/Apoe/Ngef | 4 |
| GO:0061308 | cardiac neural crest cell development involved in heart development | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Ednra/Bmp7/Hand2/Twist1 | 4 |
| GO:0070571 | negative regulation of neuron projection regeneration | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Thy1/Rtn4rl1/Neo1/Xylt1 | 4 |
| GO:0090136 | epithelial cell-cell adhesion | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Ihh/Cyp1b1/Kit/Svep1 | 4 |
| GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.235 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Ppp1r16b/Aplnr/Jcad/Agtr1a | 4 |
| GO:0010718 | positive regulation of epithelial to mesenchymal transition | BP | 0.146 | 0.00765 | 0.0321 | 1.49 | 0.0208 | Mdk/Lef1/Fermt2/Snai1/Bmp7/Twist1/Eng | 7 |
| GO:0006753 | nucleoside phosphate metabolic process | BP | 0.0747 | 0.00769 | 0.0322 | 1.49 | 0.0209 | Nme4/Acsl6/Igf1/Hmgcs2/Nos3/Acacb/Lrrk2/Adcy3/Them5/Gucy1b1/Gucy1a1/Adcy4/Dhtkd1/Adcy7/Enpp1/Lacc1/Htr2a/Acot7/Zbtb20/Sarm1/Pde5a/Rab23/Nt5c3b/Acsl1/Impdh1/Dpyd/Psen1/Pid1/Pfkm/Cacnb4/Myc/Papss2/Suclg2/Acot6 | 34 |
| GO:0009308 | amine metabolic process | BP | 0.103 | 0.0077 | 0.0322 | 1.49 | 0.0209 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Atp2b4/Slc7a7/Sult1a1/Hand2 | 13 |
| GO:0018958 | phenol-containing compound metabolic process | BP | 0.103 | 0.0077 | 0.0322 | 1.49 | 0.0209 | Maob/Zeb2/Abat/Drd4/Myo5a/Pde1b/Maoa/Fah/Hdc/Agtr1a/Ednra/Sult1a1/Hand2 | 13 |
| GO:1901605 | alpha-amino acid metabolic process | BP | 0.0924 | 0.0077 | 0.0322 | 1.49 | 0.0209 | Aspg/Ddah1/Nos3/Cbs/Asrgl1/Gamt/Kyat1/Fah/Amdhd1/Cth/Phgdh/Nox4/Atp2b4/Slc7a7/Asl/Sardh/Prodh | 17 |
| GO:0016051 | carbohydrate biosynthetic process | BP | 0.0905 | 0.00771 | 0.0322 | 1.49 | 0.0209 | Gulo/Igf1/Chst10/B3galnt1/Has2/St3gal2/Pth1r/Enpp1/Igf2/St8sia6/Adcyap1r1/Sord/St6galnac6/Fam3c/St3gal4/Grb10/Plek/Irs1 | 18 |
| GO:0021515 | cell differentiation in spinal cord | BP | 0.133 | 0.00775 | 0.0323 | 1.49 | 0.021 | Hoxd10/Gli3/Gdf7/Dync2h1/Tal1/Ptch1/Cacna1a/Draxin | 8 |
| GO:0030042 | actin filament depolymerization | BP | 0.133 | 0.00775 | 0.0323 | 1.49 | 0.021 | Sema5a/Evl/Tmod2/Mical1/Sh3bp1/Scin/Plekhh2/Plek | 8 |
| GO:0046717 | acid secretion | BP | 0.133 | 0.00775 | 0.0323 | 1.49 | 0.021 | Grik1/Abat/Htr1b/Ptger3/Gabbr1/Cacna1a/Cacnb4/Myc | 8 |
| GO:0001964 | startle response | BP | 0.162 | 0.00789 | 0.0328 | 1.48 | 0.0213 | Aph1b/Grin2b/Glrb/Nlgn3/Npas3/Kcnh1 | 6 |
| GO:0035909 | aorta morphogenesis | BP | 0.162 | 0.00789 | 0.0328 | 1.48 | 0.0213 | Tbx2/Fkbp10/Efemp2/Pdgfrb/Hey2/Eng | 6 |
| GO:0045746 | negative regulation of Notch signaling pathway | BP | 0.162 | 0.00789 | 0.0328 | 1.48 | 0.0213 | Lfng/Arrb1/Egfl7/Bend6/Bmp7/Hey2 | 6 |
| GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | BP | 0.162 | 0.00789 | 0.0328 | 1.48 | 0.0213 | Igf1/Ddr2/Tert/Mmp2/Adamts1/Htr1b | 6 |
| GO:0045862 | positive regulation of proteolysis | BP | 0.0784 | 0.00793 | 0.033 | 1.48 | 0.0214 | Sfrp2/Aph1b/Bok/Lrrk2/Wdr35/Grin2b/Bbs7/Ift57/Ppm1f/Rnft2/Ecscr/Apoe/Hip1/Prickle1/Arrb1/Rcn3/Dlc1/Asph/Fyn/Nlrp1b/Ccn2/Psen1/Cav1/Cyfip2/Nkd2/F2r/Rnf144a/Myc | 28 |
| GO:0019935 | cyclic-nucleotide-mediated signaling | BP | 0.111 | 0.00799 | 0.0332 | 1.48 | 0.0215 | Gnai1/Ednrb/Gucy1a2/Gucy1b1/Gucy1a1/Aplnr/Apoe/Adcyap1r1/Cap2/Rasd2/Ptger3 | 11 |
| GO:0043299 | leukocyte degranulation | BP | 0.111 | 0.00799 | 0.0332 | 1.48 | 0.0215 | Foxf1/Coro1a/Kit/Nckap1l/Myo1f/Cd84/Fgr/Rab27a/Lat/Clnk/Pld2 | 11 |
| GO:0002244 | hematopoietic progenitor cell differentiation | BP | 0.0941 | 0.00805 | 0.0334 | 1.48 | 0.0216 | Ptprz1/Pygo1/Kdr/Fst/Jam3/Thsd1/Slc8a3/Flt3/Kit/Eml1/Dhtkd1/Kcp/Tal1/Pus7/Psen1/Batf | 16 |
| GO:2001234 | negative regulation of apoptotic signaling pathway | BP | 0.0854 | 0.00809 | 0.0336 | 1.47 | 0.0218 | Hgf/Sfrp2/Ar/Agap2/Nog/Igf1/Nrp1/Cd44/Tert/Bok/Lrrk2/Icam1/Cxcl12/Mapk8ip1/Tnfrsf22/Cth/Snai1/Bdkrb2/Arrb2/Fyn/Psen1 | 21 |
| GO:0001541 | ovarian follicle development | BP | 0.123 | 0.00815 | 0.0337 | 1.47 | 0.0218 | Angpt1/Kdr/Mmp2/Lfng/Kit/Kitl/Arrb1/Tnfaip6/Arrb2 | 9 |
| GO:0061045 | negative regulation of wound healing | BP | 0.123 | 0.00815 | 0.0337 | 1.47 | 0.0218 | Cd109/Phldb2/Tfpi/Abat/Serpine2/Apoe/Pros1/Ptger3/Wfdc1 | 9 |
| GO:0010921 | regulation of phosphatase activity | BP | 0.116 | 0.00819 | 0.0338 | 1.47 | 0.0219 | Npnt/Mef2c/Itga1/Magi2/Nckap1l/Cmya5/Ppp1r26/Pdgfrb/Vcan/Plek | 10 |
| GO:0060606 | tube closure | BP | 0.106 | 0.00819 | 0.0338 | 1.47 | 0.0219 | Sfrp2/Nog/Zeb2/Ift57/Lrp2/Cecr2/Prickle1/Dlc1/Ptch1/Rab23/Fzd2/Twist1 | 12 |
| GO:0010829 | negative regulation of glucose transmembrane transport | BP | 0.185 | 0.0085 | 0.035 | 1.46 | 0.0227 | Sh2b2/Enpp1/Pid1/Grb10/Myc | 5 |
| GO:0042104 | positive regulation of activated T cell proliferation | BP | 0.185 | 0.0085 | 0.035 | 1.46 | 0.0227 | Igf1/Il27ra/Igf2/Stat5b/Il12rb1 | 5 |
| GO:0062009 | secondary palate development | BP | 0.185 | 0.0085 | 0.035 | 1.46 | 0.0227 | Lef1/Wnt11/Tshz1/Jag2/Fzd2 | 5 |
| GO:0090382 | phagosome maturation | BP | 0.185 | 0.0085 | 0.035 | 1.46 | 0.0227 | Rab34/Srpx/Coro1a/Syt7/Rab20 | 5 |
| GO:1902806 | regulation of cell cycle G1/S phase transition | BP | 0.0936 | 0.0085 | 0.035 | 1.46 | 0.0227 | Gli1/Myo16/Ddr2/Tbx2/Tert/Ccnd2/Adamts1/Plcb1/Stxbp4/Pkd1/Pkd2/Fhl1/Atp2b4/Dact1/Hacd1/Cacnb4 | 16 |
| GO:0031641 | regulation of myelination | BP | 0.131 | 0.00855 | 0.0351 | 1.45 | 0.0228 | Ptprz1/Hgf/Igf1/Cdh2/Wasf3/Ptn/Tppp/Jam2 | 8 |
| GO:0042551 | neuron maturation | BP | 0.131 | 0.00855 | 0.0351 | 1.45 | 0.0228 | Epha8/Cspg4/Lrrk2/Ednrb/Dlg2/Cdkn1c/Nfasc/Ednra | 8 |
| GO:0140115 | export across plasma membrane | BP | 0.131 | 0.00855 | 0.0351 | 1.45 | 0.0228 | Kcnd3/Slc8a1/Slc24a4/Abcg2/Slc8a3/Atp1a2/Abcb1a/Abcc5 | 8 |
| GO:0051348 | negative regulation of transferase activity | BP | 0.0837 | 0.00859 | 0.0352 | 1.45 | 0.0228 | Sfrp2/Ddr2/Slc8a1/Ptpro/Mapk8ip1/Slc8a3/Pkia/Thy1/Ppm1f/Cdkn1c/Apoe/Rasip1/Ppm1e/Pkib/Heg1/Lats2/Smpd1/Psen1/Cav1/Bmp7/Prkar1b/Lax1 | 22 |
| GO:1902882 | regulation of response to oxidative stress | BP | 0.11 | 0.0086 | 0.0353 | 1.45 | 0.0228 | Hgf/Ddr2/Lrrk2/Tbc1d24/Aifm2/Selenon/Fyn/Ggt7/Meak7/Pde8a/Fbln5 | 11 |
| GO:0006979 | response to oxidative stress | BP | 0.0758 | 0.00872 | 0.0357 | 1.45 | 0.0232 | Cyp1b1/Hgf/Ddr2/Slc8a1/Nos3/Lrrk2/Mmp2/Prkd1/Tbc1d24/Trpc6/Msrb3/Aifm2/Cd38/Prr5l/Apoe/Ednra/Selenon/Net1/Fyn/Mgat3/Gpx7/Ggt7/Stx2/Psen1/Pdgfrb/Meak7/Cygb/Pde8a/Pld2/Setx/Fbln5 | 31 |
| GO:0061387 | regulation of extent of cell growth | BP | 0.102 | 0.00874 | 0.0358 | 1.45 | 0.0232 | Map2/Sema5a/Sema5b/Nrp1/Sema7a/Fn1/Cxcl12/Apoe/Sema3g/Epha7/Sema6a/Ulk2/Draxin | 13 |
| GO:0034394 | protein localization to cell surface | BP | 0.122 | 0.00889 | 0.0364 | 1.44 | 0.0236 | Angpt1/Wnt11/Gga2/Jam3/Ank2/Prrt1/Tmem35a/Nlgn2/Fbln5 | 9 |
| GO:0051604 | protein maturation | BP | 0.0785 | 0.00892 | 0.0365 | 1.44 | 0.0236 | Cpxm2/Ihh/Mmp16/Aph1b/Lrrk2/Serpine2/Gli3/Rps6ka2/Cpe/Mme/Ldlrad3/C1rl/Dync2h1/Pcsk5/C1ra/Ptch1/Asph/Tspan5/Ece2/Adam19/Arxes2/Psen1/Naa11/Cyfip2/Nkd2/Myc/Prss12 | 27 |
| GO:0051048 | negative regulation of secretion | BP | 0.0891 | 0.00893 | 0.0365 | 1.44 | 0.0236 | Maob/Abat/Drd4/Myo5a/Foxf1/Edn3/Htr1b/Apoe/Cd200/Nckap1l/Neo1/Frmd4a/Cd84/Ptger3/Gabbr1/Nr1h3/F2r/Irs1 | 18 |
| GO:0046661 | male sex differentiation | BP | 0.093 | 0.00896 | 0.0366 | 1.44 | 0.0237 | Tex15/Sfrp2/Ar/Kdr/Hoxa11/Hoxa10/Icam1/Tcf21/Lrp2/Tbx3/Rhobtb3/Hoxa9/Smad9/Stat5b/Abcb1a/Lhfpl2 | 16 |
| GO:0007212 | dopamine receptor signaling pathway | BP | 0.158 | 0.00899 | 0.0366 | 1.44 | 0.0237 | Nsg2/Drd4/Lrrk2/Nsg1/Palm/Gnb5 | 6 |
| GO:0045589 | regulation of regulatory T cell differentiation | BP | 0.158 | 0.00899 | 0.0366 | 1.44 | 0.0237 | Mdk/Cd44/Sox12/Gimap5/Gimap3/Il2rg | 6 |
| GO:0060674 | placenta blood vessel development | BP | 0.158 | 0.00899 | 0.0366 | 1.44 | 0.0237 | Vash1/Syde1/Pkd1/Pkd2/Vash2/Hey2 | 6 |
| GO:1902003 | regulation of amyloid-beta formation | BP | 0.158 | 0.00899 | 0.0366 | 1.44 | 0.0237 | Igf1/Rtn1/Slc2a13/Apoe/Bin1/Abcg1 | 6 |
| GO:1905606 | regulation of presynapse assembly | BP | 0.158 | 0.00899 | 0.0366 | 1.44 | 0.0237 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn2 | 6 |
| GO:0034614 | cellular response to reactive oxygen species | BP | 0.0979 | 0.00912 | 0.037 | 1.43 | 0.024 | Cyp1b1/Hgf/Ddr2/Slc8a1/Nos3/Lrrk2/Mmp2/Trpc6/Ednra/Net1/Pdgfrb/Pde8a/Setx/Fbln5 | 14 |
| GO:0051656 | establishment of organelle localization | BP | 0.0749 | 0.00913 | 0.037 | 1.43 | 0.024 | Ankfn1/Map2/Fez1/Itga4/Mark1/Nlgn1/Sec16b/Lrrk2/Myo5a/Foxf1/Ppfia2/Kif1a/Kit/Ubxn2b/Lin7a/Crocc/5730455P16Rik/Syne3/Pard3b/Myo1f/Cep19/Cd84/Fgr/Arfgap3/Psen1/Rab27a/Lat/Map4k2/Clnk/Pld2/Pdcd6ip/Kif5a | 32 |
| GO:0050772 | positive regulation of axonogenesis | BP | 0.109 | 0.00924 | 0.0375 | 1.43 | 0.0243 | Sema5a/Plxnb1/Zeb2/Nrp1/Sema7a/Fn1/Cxcl12/Plxnd1/Apoe/Nin/Chodl | 11 |
| GO:0060538 | skeletal muscle organ development | BP | 0.0872 | 0.00924 | 0.0375 | 1.43 | 0.0243 | Stra6/Hoxd10/Ryr1/P2rx2/Hoxd9/Mef2c/Tcf21/Heyl/Igf2/Dysf/Hdac9/Selenon/Large1/Hlf/Zfp689/Cav1/Usp2/Myc/Twist1 | 19 |
| GO:0008643 | carbohydrate transport | BP | 0.0949 | 0.00935 | 0.0378 | 1.42 | 0.0245 | Slc5a11/Igf1/Slc2a3/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Slc23a2/Abcb1a/Pid1/Grb10/Myc/Irs1 | 15 |
| GO:1901653 | cellular response to peptide | BP | 0.0799 | 0.00939 | 0.038 | 1.42 | 0.0246 | Itga4/Igf1/Cacna2d1/Myo5a/Ano1/Icam1/Plcb1/Stxbp4/Sh2b2/Enpp1/Agtr1a/Ednra/Igf2/Hdac9/Stat5b/Fyn/Akt3/Psen1/Cav1/Pid1/C1qtnf9/Cacna1a/Cpeb1/Grb10/Irs1 | 25 |
| GO:0021872 | forebrain generation of neurons | BP | 0.129 | 0.00941 | 0.038 | 1.42 | 0.0246 | Lef1/Nrp1/Gli3/Ndnf/Dclk2/Lhx6/Ptger3/Bcl11b | 8 |
| GO:0046850 | regulation of bone remodeling | BP | 0.129 | 0.00941 | 0.038 | 1.42 | 0.0246 | Mdk/Inpp4b/Calca/Tnfrsf11b/Cd38/Syt7/Ltbp3/Def8 | 8 |
| GO:0060711 | labyrinthine layer development | BP | 0.129 | 0.00941 | 0.038 | 1.42 | 0.0246 | Lef1/Itga4/Vash1/Syde1/Pcdh12/Vash2/Bmp7/Hey2 | 8 |
| GO:0010470 | regulation of gastrulation | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Phldb2/Osr2/Tgif2/Osr1 | 4 |
| GO:0010644 | cell communication by electrical coupling | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Slc8a1/Gjc1/Sri/Cav1 | 4 |
| GO:0015701 | bicarbonate transport | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Car4/Slc4a3/Ptger3/Slc4a8 | 4 |
| GO:0032230 | positive regulation of synaptic transmission, GABAergic | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Grik1/Nlgn1/Adra1a/Nlgn2 | 4 |
| GO:0033631 | cell-cell adhesion mediated by integrin | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Itga4/Npnt/Podxl/Plpp3 | 4 |
| GO:0035633 | maintenance of blood-brain barrier | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Angpt1/Bdkrb2/Cldn5/Abcb1a | 4 |
| GO:0061307 | cardiac neural crest cell differentiation involved in heart development | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Ednra/Bmp7/Hand2/Twist1 | 4 |
| GO:0061430 | bone trabecula morphogenesis | BP | 0.222 | 0.00947 | 0.038 | 1.42 | 0.0246 | Plxnb1/Thbs3/Mmp2/Enpp1 | 4 |
| GO:1901617 | organic hydroxy compound biosynthetic process | BP | 0.0855 | 0.00947 | 0.038 | 1.42 | 0.0246 | Apob/Cyp7b1/Zeb2/Hmgcs2/Myo5a/Sptlc3/Pth1r/Pltp/Hdc/Dkk3/Agtr1a/Apoe/Snai1/Adcyap1r1/Ipmk/Abcg1/Cacna1h/Hand2/Abcg4/Plek | 20 |
| GO:0050819 | negative regulation of coagulation | BP | 0.14 | 0.00956 | 0.0383 | 1.42 | 0.0248 | Tfpi/Abat/Serpine2/Procr/Apoe/Pros1/Ptger3 | 7 |
| GO:0021675 | nerve development | BP | 0.114 | 0.00958 | 0.0384 | 1.42 | 0.0249 | Slc1a3/Itga4/Rnf165/Nrp1/Slc24a4/Nptx1/Serpine2/Gli3/Hoxd3/Ednra | 10 |
| GO:0048839 | inner ear development | BP | 0.0868 | 0.00968 | 0.0387 | 1.41 | 0.0251 | Col11a1/Igf1/Tbx2/Lrig3/Gli3/Zeb1/Jag2/Cecr2/Lin7a/Tsku/Tbx3/Kcnma1/Dchs1/Fzd2/Fat4/Kcnq4/Itga8/Ror1/Hey2 | 19 |
| GO:0042130 | negative regulation of T cell proliferation | BP | 0.12 | 0.00968 | 0.0387 | 1.41 | 0.0251 | Ihh/Cd44/H2-Ab1/Btla/Havcr2/Pde5a/H2-Aa/Dlg5/Spn | 9 |
| GO:0010657 | muscle cell apoptotic process | BP | 0.108 | 0.00992 | 0.0396 | 1.4 | 0.0256 | Sfrp2/Gria4/Mdk/Igf1/Rps6ka2/Agtr1a/Arrb1/Arrb2/Cav1/Hey2/Hand2 | 11 |
| GO:0034390 | smooth muscle cell apoptotic process | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Gria4/Igf1/Agtr1a/Arrb1/Arrb2 | 5 |
| GO:0034391 | regulation of smooth muscle cell apoptotic process | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Gria4/Igf1/Agtr1a/Arrb1/Arrb2 | 5 |
| GO:0045822 | negative regulation of heart contraction | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Adra1a/Bin1/Pde5a/Atp1a2/Sri | 5 |
| GO:0061298 | retina vasculature development in camera-type eye | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Ndp/Cyp1b1/Nrp1/Arhgef15/Pdgfrb | 5 |
| GO:1903055 | positive regulation of extracellular matrix organization | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Sema5a/Ddr2/Phldb2/Emilin1/Efemp2 | 5 |
| GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | BP | 0.179 | 0.00994 | 0.0396 | 1.4 | 0.0256 | Drd4/Mef2c/Rgs5/Efemp2/Apln | 5 |
| GO:0001773 | myeloid dendritic cell activation | BP | 0.154 | 0.0102 | 0.0403 | 1.39 | 0.0261 | Dock2/Havcr2/Gimap5/Gimap3/Psen1/Batf | 6 |
| GO:0002335 | mature B cell differentiation | BP | 0.154 | 0.0102 | 0.0403 | 1.39 | 0.0261 | Itm2a/Lfng/Dock10/Enpp1/Irf8/Mfng | 6 |
| GO:0032885 | regulation of polysaccharide biosynthetic process | BP | 0.154 | 0.0102 | 0.0403 | 1.39 | 0.0261 | Igf1/Has2/Enpp1/Igf2/Grb10/Irs1 | 6 |
| GO:0033238 | regulation of cellular amine metabolic process | BP | 0.154 | 0.0102 | 0.0403 | 1.39 | 0.0261 | Maob/Abat/Drd4/Pde1b/Atp2b4/Slc7a7 | 6 |
| GO:0070570 | regulation of neuron projection regeneration | BP | 0.154 | 0.0102 | 0.0403 | 1.39 | 0.0261 | Hgf/Thy1/Rtn4rl1/Neo1/Xylt1/Ptn | 6 |
| GO:0002315 | marginal zone B cell differentiation | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Lfng/Dock10/Mfng | 3 |
| GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Gimap5/Gimap3/Il2rg | 3 |
| GO:0002692 | negative regulation of cellular extravasation | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Cxcl12/Plcb1/Il27ra | 3 |
| GO:0021957 | corticospinal tract morphogenesis | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Zeb2/Cdh11/Nin | 3 |
| GO:0035864 | response to potassium ion | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Nptx1/Dlg2/Dlg4 | 3 |
| GO:0035865 | cellular response to potassium ion | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Nptx1/Dlg2/Dlg4 | 3 |
| GO:0043201 | response to leucine | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Klhl22/Rragd/Ubr1 | 3 |
| GO:0045199 | maintenance of epithelial cell apical/basal polarity | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Wnt11/Lin7a/Pdcd6ip | 3 |
| GO:0051001 | negative regulation of nitric-oxide synthase activity | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Atp2b4/Cav1/Eng | 3 |
| GO:0060513 | prostatic bud formation | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Ar/Nog/Bmp7 | 3 |
| GO:0060982 | coronary artery morphogenesis | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Nrp1/Lrp2/Hand2 | 3 |
| GO:0070294 | renal sodium ion absorption | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Ednrb/Maged2/Ednra | 3 |
| GO:0071233 | cellular response to leucine | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Klhl22/Rragd/Ubr1 | 3 |
| GO:0090269 | fibroblast growth factor production | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Wnt11/Ccm2l/Heg1 | 3 |
| GO:0090270 | regulation of fibroblast growth factor production | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Wnt11/Ccm2l/Heg1 | 3 |
| GO:0099640 | axo-dendritic protein transport | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Myo5a/Dlg2/Kif5a | 3 |
| GO:1900119 | positive regulation of execution phase of apoptosis | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Bok/Cxcr3/Dlc1 | 3 |
| GO:1902746 | regulation of lens fiber cell differentiation | BP | 0.3 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Zeb2/Cdkn1c/Spred3 | 3 |
| GO:0097696 | receptor signaling pathway via STAT | BP | 0.0878 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Stra6/Cyp1b1/Agap2/Ptprd/Igf1/Hcls1/Dab1/Kit/Agtr1a/Pecam1/Pkd1/Stat5b/Pkd2/Fyn/Csf2rb/Il12rb1/Cav1/Il7r | 18 |
| GO:0046486 | glycerolipid metabolic process | BP | 0.0761 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Apob/Acsl6/Pla2g10/Efr3b/Pip5k1b/Inpp4b/Plcb1/Them5/Ttc7b/Dgkb/Apoe/Bpnt1/Htr2a/Lpcat1/Plcb2/Plpp1/Ddhd2/Abhd8/Acsl1/Pdgfrb/Cav1/Nr1h3/Lipe/Pld2/Enpp2/Plek/Mboat2/Cds1/Abhd3 | 29 |
| GO:0042269 | regulation of natural killer cell mediated cytotoxicity | BP | 0.127 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Pik3r6/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gimap3 | 8 |
| GO:0048260 | positive regulation of receptor-mediated endocytosis | BP | 0.127 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Angpt1/Magi2/Sele/Hip1/Arrb1/Arrb2/Apln/Pld2 | 8 |
| GO:2000401 | regulation of lymphocyte migration | BP | 0.127 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Itga4/Cxcl12/Msn/Il27ra/Cd200/Xcl1/Spn/Jam2 | 8 |
| GO:0046323 | glucose import | BP | 0.112 | 0.0103 | 0.0403 | 1.39 | 0.0261 | Igf1/Slc2a3/Tert/Gpc3/Sh2b2/Enpp1/Pid1/Grb10/Myc/Irs1 | 10 |
| GO:0009314 | response to radiation | BP | 0.0742 | 0.0104 | 0.0404 | 1.39 | 0.0261 | Slc1a3/Sfrp2/Sema5a/Cacna1c/Nog/Sema5b/Ccnd2/Mmp2/Slc24a4/B4galt2/Pde1b/Cacna1e/Eif2ak4/Lrp2/Fbxl21/Kit/Mme/Rp1/Nlgn3/Arrb1/Nfatc4/Net1/Gnb5/Ddhd2/Atp1a2/Syngap1/Brsk1/Smpd1/Usp2/Ints3/Cacnb4/Myc | 32 |
| GO:0046822 | regulation of nucleocytoplasmic transport | BP | 0.0992 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Angpt1/Hcls1/Inpp4b/Prkd1/Tek/Gli3/Pkia/Peg12/Dnajc27/Wipf1/Rab23/Psen1/Camk1 | 13 |
| GO:0051592 | response to calcium ion | BP | 0.0992 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Nlgn1/Syt13/Cpne5/Mef2c/Rasgrp2/Syt7/Kcnma1/Asph/Pkd2/Slc25a23/Smpd1/Cav1/Kcnh1 | 13 |
| GO:0002704 | negative regulation of leukocyte mediated immunity | BP | 0.118 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Foxf1/Havcr2/Igf2/Nckap1l/Arrb2/Cd84/Cd96/Spn/Il7r | 9 |
| GO:0021766 | hippocampus development | BP | 0.118 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Mdk/Lef1/Zeb2/Dab1/Id4/Gli3/Tsku/Dclk2/Bbs1 | 9 |
| GO:0080164 | regulation of nitric oxide metabolic process | BP | 0.118 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Igf1/Ddah1/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 9 |
| GO:1903036 | positive regulation of response to wounding | BP | 0.118 | 0.0105 | 0.0408 | 1.39 | 0.0264 | Mdk/Ddr2/Fermt2/Plpp3/Clec7a/St3gal4/Ptn/Cldn1/F2r | 9 |
| GO:0032886 | regulation of microtubule-based process | BP | 0.0821 | 0.0106 | 0.0409 | 1.39 | 0.0265 | Map2/Fez1/Phldb2/Mpdz/Gnai1/Cdh5/Map9/Epha3/Rp1/Tubb4a/Dysf/Pkd1/Nin/Slain1/Klhl42/Ttbk2/Cep97/Bbs1/Mapre2/Cav1/Tppp/Pdcd6ip | 22 |
| GO:0006687 | glycosphingolipid metabolic process | BP | 0.137 | 0.0106 | 0.0411 | 1.39 | 0.0266 | St8sia2/St8sia4/St3gal2/St6galnac4/Kit/St8sia6/St6galnac6 | 7 |
| GO:0045933 | positive regulation of muscle contraction | BP | 0.137 | 0.0106 | 0.0411 | 1.39 | 0.0266 | Npnt/Adra1a/Abat/Kit/Ptger3/Ccn2/F2r | 7 |
| GO:0030593 | neutrophil chemotaxis | BP | 0.107 | 0.0106 | 0.0411 | 1.39 | 0.0266 | Cxcl15/Mdk/Jam3/Edn3/Itga1/Ednra/Dysf/Nckap1l/Xcl1/Tnfaip6/Mpp1 | 11 |
| GO:0019218 | regulation of steroid metabolic process | BP | 0.103 | 0.0107 | 0.0412 | 1.39 | 0.0267 | Apob/Igf1/Kit/Dkk3/Agtr1a/Apoe/Kcnma1/Igf2/Snai1/Abcg1/Stat5b/Abcg4 | 12 |
| GO:1904659 | glucose transmembrane transport | BP | 0.103 | 0.0107 | 0.0412 | 1.39 | 0.0267 | Igf1/Slc2a3/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Pid1/Grb10/Myc/Irs1 | 12 |
| GO:0008645 | hexose transmembrane transport | BP | 0.102 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Igf1/Slc2a3/Tert/Gpc3/Stxbp4/Sh2b2/Enpp1/Ednra/Pid1/Grb10/Myc/Irs1 | 12 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | BP | 0.106 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Fam83a/Ccdc88a/Areg/Cdh13/Itga1/Hip1/Rassf2/Afap1l2/Mvb12b/Psen1/Kif16b | 11 |
| GO:0002090 | regulation of receptor internalization | BP | 0.117 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Angpt1/Drd4/Magi2/Sele/Arrb1/Arrb2/Dlg4/Apln/Ankrd13a | 9 |
| GO:0005977 | glycogen metabolic process | BP | 0.117 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Igf1/Pygm/Pcdh12/Enpp1/Igf2/Grb10/Pfkm/Pygl/Irs1 | 9 |
| GO:0006073 | cellular glucan metabolic process | BP | 0.117 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Igf1/Pygm/Pcdh12/Enpp1/Igf2/Grb10/Pfkm/Pygl/Irs1 | 9 |
| GO:0098661 | inorganic anion transmembrane transport | BP | 0.117 | 0.0114 | 0.0436 | 1.36 | 0.0283 | Slc1a3/Slc12a8/Ano1/Ano2/Glrb/Gabra4/Slc12a4/Slc4a8/Ank | 9 |
| GO:0032232 | negative regulation of actin filament bundle assembly | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Kank4/Phldb2/Arhgap6/Tmeff2/Dlc1/Arhgap28 | 6 |
| GO:0050850 | positive regulation of calcium-mediated signaling | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Kdr/Igf1/Cdh13/Itgal/Zap70/Cd4 | 6 |
| GO:0060219 | camera-type eye photoreceptor cell differentiation | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Ihh/Sdk2/Thy1/Rp1/Rorb/Ptn | 6 |
| GO:0060351 | cartilage development involved in endochondral bone morphogenesis | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Ihh/Hoxd11/Thbs3/Cbs/Hoxa11/Stc1 | 6 |
| GO:1902624 | positive regulation of neutrophil migration | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Mdk/Ednra/Dysf/Nckap1l/Xcl1/Ptger3 | 6 |
| GO:1903580 | positive regulation of ATP metabolic process | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Igf1/Htr2a/Zbtb20/Psen1/Pid1/Myc | 6 |
| GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | BP | 0.15 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Cd83/Nckap1l/Xcl1/Gimap5/Gimap3/Il2rg | 6 |
| GO:0002827 | positive regulation of T-helper 1 type immune response | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | H2-Ab1/Il27ra/Xcl1/Il12rb1 | 4 |
| GO:0003228 | atrial cardiac muscle tissue development | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Nog/Tbx3/Ednra/Eng | 4 |
| GO:0010523 | negative regulation of calcium ion transport into cytosol | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Calca/Pkd2/Gstm7/Sri | 4 |
| GO:0010544 | negative regulation of platelet activation | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Abat/Serpine2/Apoe/Ptger3 | 4 |
| GO:0022038 | corpus callosum development | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Zeb2/Tsku/Nin/Rtn4rl1 | 4 |
| GO:0032570 | response to progesterone | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Acod1/Cav1/Nr1h3/Ptger2 | 4 |
| GO:0033604 | negative regulation of catecholamine secretion | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Abat/Myo5a/Ptger3/Gabbr1 | 4 |
| GO:0034111 | negative regulation of homotypic cell-cell adhesion | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Abat/Serpine2/Ccm2l/Ptger3 | 4 |
| GO:0035461 | vitamin transmembrane transport | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Stra6/Lrp2/Slc23a2/Abcc5 | 4 |
| GO:0045198 | establishment of epithelial cell apical/basal polarity | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Foxf1/Myo9a/Msn/Sh3bp1 | 4 |
| GO:0045579 | positive regulation of B cell differentiation | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Prdm1/Nckap1l/Stat5b/Il2rg | 4 |
| GO:0071514 | genomic imprinting | BP | 0.211 | 0.0115 | 0.0436 | 1.36 | 0.0283 | H19/Tet1/Ndn/Cdkn1c | 4 |
| GO:0033198 | response to ATP | BP | 0.172 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Ryr1/Slc8a1/P2rx2/Enpp1/Asph | 5 |
| GO:0042133 | neurotransmitter metabolic process | BP | 0.172 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Maob/Abat/Maoa/Naalad2/Bche | 5 |
| GO:0060571 | morphogenesis of an epithelial fold | BP | 0.172 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Ar/Nog/Gdf7/Cecr2/Bmp7 | 5 |
| GO:0061311 | cell surface receptor signaling pathway involved in heart development | BP | 0.172 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Wnt11/Nog/Snai1/Hey2/Hand2 | 5 |
| GO:0090023 | positive regulation of neutrophil chemotaxis | BP | 0.172 | 0.0115 | 0.0436 | 1.36 | 0.0283 | Mdk/Ednra/Dysf/Nckap1l/Xcl1 | 5 |
| GO:0060291 | long-term synaptic potentiation | BP | 0.0824 | 0.0119 | 0.0448 | 1.35 | 0.029 | Nog/Nlgn1/Grin2b/Serpine2/Shank2/Eif2ak4/Slc8a3/Tshz3/Mme/Nlgn3/Enpp1/Nrgn/Apoe/Prrt1/Nsg1/Nfatc4/Cyp46a1/Mpp2/Psen1/Ptn/Prkar1b | 21 |
| GO:0044242 | cellular lipid catabolic process | BP | 0.0848 | 0.0121 | 0.0457 | 1.34 | 0.0296 | Apob/Cyp1b1/Pla2g10/Acacb/Plcb1/Bdh2/Echs1/Acot7/Ddhd2/Abcd2/Smpd1/Lipe/Pld2/Enpp2/Plbd1/Twist1/Abhd3/Echdc2/Irs1 | 19 |
| GO:0050764 | regulation of phagocytosis | BP | 0.101 | 0.0121 | 0.0457 | 1.34 | 0.0296 | Tub/Dock2/Syt7/Cnn2/Dysf/Pros1/Nckap1l/Masp1/Fgr/Clec7a/Rab27a/Il2rg | 12 |
| GO:0044042 | glucan metabolic process | BP | 0.115 | 0.0124 | 0.0466 | 1.33 | 0.0302 | Igf1/Pygm/Pcdh12/Enpp1/Igf2/Grb10/Pfkm/Pygl/Irs1 | 9 |
| GO:0045453 | bone resorption | BP | 0.115 | 0.0124 | 0.0466 | 1.33 | 0.0302 | Ihh/Calca/Pth1r/Enpp1/Tnfrsf11b/Cd38/Nox4/Ltbp3/Def8 | 9 |
| GO:0009247 | glycolipid biosynthetic process | BP | 0.123 | 0.0124 | 0.0466 | 1.33 | 0.0302 | St8sia2/St8sia4/St3gal2/St6galnac4/St8sia6/Sccpdh/St6galnac6/St3gal4 | 8 |
| GO:0071260 | cellular response to mechanical stimulus | BP | 0.123 | 0.0124 | 0.0466 | 1.33 | 0.0302 | Wnt11/Nos3/Cnn2/Ednra/Habp4/Atp1a2/Piezo2/Cav1 | 8 |
| GO:0051222 | positive regulation of protein transport | BP | 0.0779 | 0.0126 | 0.0473 | 1.32 | 0.0307 | Igf1/Tcaf1/Abat/Hcls1/Ano1/Prkd1/Tek/Gli3/Lrp2/Rab34/Vegfc/Cd38/Prr5l/Arrb1/Abcg1/Asph/Frmd4a/Fyn/Wipf1/Psen1/Sri/Pfkm/Golph3l/Camk1/Nlgn2 | 25 |
| GO:0050672 | negative regulation of lymphocyte proliferation | BP | 0.109 | 0.0129 | 0.0483 | 1.32 | 0.0313 | Ihh/Cd44/Prdm1/H2-Ab1/Btla/Havcr2/Pde5a/H2-Aa/Dlg5/Spn | 10 |
| GO:1900182 | positive regulation of protein localization to nucleus | BP | 0.109 | 0.0129 | 0.0483 | 1.32 | 0.0313 | Hcls1/Fermt2/Tert/Prkd1/Tek/Gli3/Fyn/Psen1/Glis2/Cacnb4 | 10 |
| GO:0035850 | epithelial cell differentiation involved in kidney development | BP | 0.146 | 0.013 | 0.0486 | 1.31 | 0.0315 | Ptpro/Ednrb/Mef2c/Ednra/Osr1/Fat4 | 6 |
| GO:2000772 | regulation of cellular senescence | BP | 0.146 | 0.013 | 0.0486 | 1.31 | 0.0315 | Tbx2/Tert/Abi3/Vash1/Akt3/Twist1 | 6 |
| GO:0072091 | regulation of stem cell proliferation | BP | 0.132 | 0.013 | 0.0487 | 1.31 | 0.0316 | Fermt2/Tert/Vegfc/Tbx3/Kitl/Ltbp3/Septin4 | 7 |
| GO:2000677 | regulation of transcription regulatory region DNA binding | BP | 0.132 | 0.013 | 0.0487 | 1.31 | 0.0316 | Igf1/Zc4h2/Psen1/Sri/Hey2/Hand2/Twist1 | 7 |
| GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Havcr2/Igf2/Nckap1l/Arrb2/Il7r | 5 |
| GO:0001963 | synaptic transmission, dopaminergic | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Slc6a2/Aph1b/Drd4/Arrb2/Rasd2 | 5 |
| GO:0005979 | regulation of glycogen biosynthetic process | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Igf1/Enpp1/Igf2/Grb10/Irs1 | 5 |
| GO:0010962 | regulation of glucan biosynthetic process | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Igf1/Enpp1/Igf2/Grb10/Irs1 | 5 |
| GO:0051957 | positive regulation of amino acid transport | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Grik1/Abat/Grin2b/Psen1/Gabbr1 | 5 |
| GO:1903523 | negative regulation of blood circulation | BP | 0.167 | 0.0133 | 0.0495 | 1.3 | 0.0321 | Adra1a/Bin1/Pde5a/Atp1a2/Sri | 5 |
| GO:0045776 | negative regulation of blood pressure | BP | 0.121 | 0.0135 | 0.0499 | 1.3 | 0.0323 | Kdr/Adra1a/Abat/Nos3/Calca/Vegfc/Bdkrb2/Apln | 8 |
| GO:0060688 | regulation of morphogenesis of a branching structure | BP | 0.121 | 0.0135 | 0.0499 | 1.3 | 0.0323 | Hgf/Ar/Mdk/Nog/Lrrk2/Cpe/Agtr1a/Bmp7 | 8 |
| GO:0090311 | regulation of protein deacetylation | BP | 0.121 | 0.0135 | 0.0499 | 1.3 | 0.0323 | Lrrk2/Prkd1/Tcf21/Ndn/Aplnr/Dysf/Tada2a/Tppp | 8 |
| GO:1903307 | positive regulation of regulated secretory pathway | BP | 0.121 | 0.0135 | 0.0499 | 1.3 | 0.0323 | Nlgn1/Cadps/Syt7/Cacna1h/Fgr/Slc4a8/Rab27a/Pld2 | 8 |
| GO:0030879 | mammary gland development | BP | 0.0909 | 0.0136 | 0.0499 | 1.3 | 0.0323 | Ar/Lef1/Agap2/Igf1/Tbx2/Hoxd9/Areg/Foxf1/Gli3/Tbx3/Hoxa9/Ptch1/Stat5b/Cav1/Irs1 | 15 |
| GO:0002318 | myeloid progenitor cell differentiation | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Jam3/Flt3/Kit | 3 |
| GO:0003139 | secondary heart field specification | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Wnt11/Mef2c/Lrp2 | 3 |
| GO:0006171 | cAMP biosynthetic process | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Adcy3/Adcy4/Adcy7 | 3 |
| GO:0007158 | neuron cell-cell adhesion | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Nlgn1/Nlgn3/Nlgn2 | 3 |
| GO:0009415 | response to water | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Pkd2/Atp2b4/Sipa1 | 3 |
| GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Gli1/Igf1/Zfp423 | 3 |
| GO:0030202 | heparin metabolic process | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Angpt1/Ednrb/Ednra | 3 |
| GO:0032815 | negative regulation of natural killer cell activation | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Havcr2/Fgr/Clnk | 3 |
| GO:0034454 | microtubule anchoring at centrosome | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Ninl/Nin/Cep19 | 3 |
| GO:0035090 | maintenance of apical/basal cell polarity | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Wnt11/Lin7a/Pdcd6ip | 3 |
| GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Ar/Igf1/Igfbp4 | 3 |
| GO:0051135 | positive regulation of NK T cell activation | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Cd1d1/Il12rb1/Rasal3 | 3 |
| GO:0060405 | regulation of penile erection | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Ar/Drd4/Ednrb | 3 |
| GO:0060600 | dichotomous subdivision of an epithelial terminal unit | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Nrp1/Areg/Plxnd1 | 3 |
| GO:0071372 | cellular response to follicle-stimulating hormone stimulus | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Epha8/Epha3/Ednra | 3 |
| GO:0072102 | glomerulus morphogenesis | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Mef2c/Tcf21/Pdgfrb | 3 |
| GO:0072178 | nephric duct morphogenesis | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Gpc3/Epha7/Osr1 | 3 |
| GO:0086016 | AV node cell action potential | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Cacna1c/Scn4b/Ank2 | 3 |
| GO:0086027 | AV node cell to bundle of His cell signaling | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Cacna1c/Scn4b/Ank2 | 3 |
| GO:0090184 | positive regulation of kidney development | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Nog/Ednra/Myc | 3 |
| GO:0099545 | trans-synaptic signaling by trans-synaptic complex | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Efnb3/Ptprd/Nlgn1 | 3 |
| GO:1902946 | protein localization to early endosome | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Nrp1/Msn/Mgat3 | 3 |
| GO:1903564 | regulation of protein localization to cilium | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Ccdc88a/Crocc/Dzip1 | 3 |
| GO:2000344 | positive regulation of acrosome reaction | BP | 0.273 | 0.0137 | 0.0499 | 1.3 | 0.0323 | Pla2g10/Plcb1/Cacna1h | 3 |
| GO:0097060 | synaptic membrane | CC | 0.192 | 1.58e-24 | 8.5e-22 | 21.1 | 5.62e-22 | Efnb3/Ptprz1/Kcnj3/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Slc6a2/Kcnd2/Dcc/Ptprd/Kcnd3/Grid2ip/Nlgn1/Slc8a1/Nrp1/Ptpro/Rims3/Stx1b/P2rx2/Cdh2/Lrrc4b/Grin2b/Shank2/Cacna1e/Adgrl1/Ank2/Slitrk3/Col13a1/Syde1/Htr1b/Cpe/Glrb/Scn2a/Gabra4/Dlg2/Grik2/Lin7a/Nlgn3/Dgkb/Sorcs2/Prrt1/Syt7/Prrt2/Kcnma1/Hip1/Zc4h2/Arrb1/Septin3/Pcdh17/Nsg1/Epha7/Cacng7/Arrb2/Mpp2/Erc2/Lrfn2/Cacna1h/Magee1/Dlg4/Palm/Slc4a8/Stx2/Psen1/Gabbr1/Ntng1/Sspn/Itga8/Iqsec3/F2r/Lrp4/Kcnh1/Kctd12/Nlgn2 | 75 |
| GO:0062023 | collagen-containing extracellular matrix | CC | 0.179 | 1.89e-21 | 5.08e-19 | 18.3 | 3.36e-19 | Ndp/Gpc6/Col11a1/Ptprz1/Hmcn1/Vit/Angpt1/Emid1/Col14a1/Colec12/Igf1/Col4a6/Thbs3/Npnt/Lamc3/Lama4/Spock2/Cspg4/Col16a1/Tgfbi/Gpc3/Fn1/Angptl2/Nid1/Serpine2/Egflam/Adamts1/Col9a2/Col13a1/Frem1/Mamdc2/Postn/Loxl3/Adamts10/Smoc1/Mgp/Lama2/Matn2/Emilin1/Sparcl1/Tmeff2/Igf2/Col5a2/Ltbp3/Bgn/Col15a1/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Adam19/Angptl4/Efemp2/Egfl7/Plxdc2/Ptn/Ntng1/Col23a1/Col18a1/Gpc2/Adamts17/Itgb4/Spn/Lum/Nid2/Bmp7/Vcan/Ccdc80/Fbln5/Col4a5 | 71 |
| GO:0045211 | postsynaptic membrane | CC | 0.194 | 6.73e-18 | 1.21e-15 | 14.9 | 7.98e-16 | Ptprz1/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Grid2ip/Nlgn1/Slc8a1/Nrp1/Ptpro/Cdh2/Lrrc4b/Grin2b/Shank2/Cacna1e/Ank2/Slitrk3/Col13a1/Glrb/Gabra4/Dlg2/Grik2/Lin7a/Nlgn3/Dgkb/Sorcs2/Prrt1/Prrt2/Kcnma1/Hip1/Zc4h2/Arrb1/Pcdh17/Nsg1/Epha7/Cacng7/Arrb2/Mpp2/Lrfn2/Cacna1h/Magee1/Dlg4/Gabbr1/Sspn/Iqsec3/F2r/Kcnh1/Kctd12/Nlgn2 | 53 |
| GO:0098978 | glutamatergic synapse | CC | 0.161 | 4.36e-14 | 5.87e-12 | 11.2 | 3.88e-12 | Efnb3/Syn2/Grid1/Gria4/Lrrc4c/Begain/Kcnd2/Ptprd/Nlgn1/Nrp1/Ptpro/Drd4/Wasf3/Lrrk2/Lrrc4b/Myo5a/Pfn2/Nptx1/Grin2b/Plcb1/Ppfia2/Abi2/Coro1a/Gucy1a1/Scn2a/Dlg2/Cadps/Grik2/Dgkb/Nptxr/Apoe/Cdh11/Syt7/Prrt2/Sparcl1/Pcdh17/Nsg1/Epha7/Cacng7/Mpp2/Erc2/Cpt1c/Cacna1h/Dlg4/Syngap1/Slc4a8/Ngef/Nrn1/Ntng1/Cacna1a/Cacnb4/Prkar1b | 52 |
| GO:0098984 | neuron to neuron synapse | CC | 0.152 | 8.6e-14 | 9.25e-12 | 11 | 6.12e-12 | Gap43/Efnb3/Syn2/Map2/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Epb41l3/Slc8a1/Lrrc7/Ptpro/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Rps6kc1/Adgrl1/Ank2/Slc8a3/Slitrk3/Dlg2/Grik2/Nrgn/Sorcs2/Prrt1/Cap2/Arrb1/Cacng7/Pdzd2/Arrb2/Ptch1/Mpp2/Erc2/Lrfn2/Fyn/Cnn3/Dlg4/Palm/Dlg5/Syngap1/Slc4a8/Arhgef9/Cpeb1/Itga8/Iqsec3/Prkar1b/Lrp4 | 55 |
| GO:0099572 | postsynaptic specialization | CC | 0.153 | 1.09e-13 | 9.73e-12 | 11 | 6.44e-12 | Gap43/Syn2/Map2/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Epb41l3/Slc8a1/Lrrc7/Ptpro/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Rps6kc1/Adgrl1/Ank2/Slc8a3/Slitrk3/Dlg2/Grik2/Nlgn3/Nrgn/Sorcs2/Prrt1/Cap2/Arrb1/Cacng7/Pdzd2/Arrb2/Ptch1/Mpp2/Erc2/Lrfn2/Fyn/Cnn3/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Cpeb1/Itga8/Iqsec3/Lrp4/Nlgn2 | 54 |
| GO:0032279 | asymmetric synapse | CC | 0.153 | 3.12e-13 | 1.86e-11 | 10.7 | 1.23e-11 | Gap43/Syn2/Map2/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Epb41l3/Slc8a1/Lrrc7/Ptpro/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Rps6kc1/Adgrl1/Ank2/Slc8a3/Slitrk3/Dlg2/Grik2/Nrgn/Sorcs2/Prrt1/Cap2/Arrb1/Cacng7/Pdzd2/Arrb2/Ptch1/Mpp2/Erc2/Lrfn2/Fyn/Cnn3/Dlg4/Palm/Dlg5/Syngap1/Slc4a8/Arhgef9/Cpeb1/Itga8/Iqsec3/Lrp4 | 52 |
| GO:0014069 | postsynaptic density | CC | 0.155 | 3.92e-13 | 2.11e-11 | 10.7 | 1.39e-11 | Gap43/Syn2/Map2/Grid1/Gria4/Cacna1c/Grik1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Epb41l3/Slc8a1/Lrrc7/Ptpro/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Rps6kc1/Adgrl1/Ank2/Slc8a3/Slitrk3/Dlg2/Grik2/Nrgn/Sorcs2/Prrt1/Cap2/Arrb1/Cacng7/Pdzd2/Arrb2/Ptch1/Mpp2/Erc2/Lrfn2/Fyn/Cnn3/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Cpeb1/Itga8/Iqsec3/Lrp4 | 51 |
| GO:0042383 | sarcolemma | CC | 0.206 | 9.35e-13 | 4.32e-11 | 10.4 | 2.86e-11 | Kcnj3/Car4/Cacna1c/Sgcd/Kcnd2/Ryr1/Adra1a/Slc8a1/Nos3/Cdh2/Scn1a/Cacna2d1/Ank2/Slc8a3/Msn/Scn2a/Lama2/Ednra/Itga7/Dysf/Bin1/Bgn/Cacng7/Flnc/Ryr3/Cacna1h/Atp2b4/Sgcb/Atp1a2/Psen1/Sri/Cav1/Sspn/Pld2 | 34 |
| GO:0098857 | membrane microdomain | CC | 0.142 | 9.65e-13 | 4.32e-11 | 10.4 | 2.86e-11 | Cacna1c/Angpt1/Kcnd2/Dcc/Kdr/Kcnd3/Tfpi/Nos3/Rgs16/Gnai1/Cdh2/Lrrk2/Ednrb/Ano1/Icam1/Abcg2/Adcy3/Tek/Cdh13/Has2/Tnfrsf22/Gprc5b/Itga1/Lrp2/Thy1/Plpp3/Cpe/Mme/Kcnma1/Sele/Ednra/Pecam1/Dysf/Adcyap1r1/Cavin3/Rtn4rl1/Dlc1/Ptch1/Fyn/Cacna1h/Cavin2/Plpp1/Atp1a2/Zap70/Stx2/Psen1/Gabbr1/Cav1/Abcg3/Lipe/Pld2/Lcp2/F2r/Cd4/Lrp4/Irs1 | 56 |
| GO:0042734 | presynaptic membrane | CC | 0.227 | 1.42e-12 | 5.87e-11 | 10.2 | 3.88e-11 | Efnb3/Kcnj3/Cacna1c/Grik1/Slc6a2/Ptprd/Rims3/Stx1b/P2rx2/Cdh2/Grin2b/Cacna1e/Adgrl1/Htr1b/Scn2a/Grik2/Syt7/Prrt2/Kcnma1/Hip1/Septin3/Pcdh17/Erc2/Cacna1h/Slc4a8/Stx2/Gabbr1/Ntng1/Kcnh1/Kctd12 | 30 |
| GO:0045121 | membrane raft | CC | 0.14 | 2.88e-12 | 1.03e-10 | 9.99 | 6.84e-11 | Cacna1c/Angpt1/Kcnd2/Dcc/Kdr/Kcnd3/Tfpi/Nos3/Rgs16/Gnai1/Cdh2/Lrrk2/Ednrb/Ano1/Icam1/Abcg2/Adcy3/Tek/Cdh13/Has2/Tnfrsf22/Gprc5b/Itga1/Lrp2/Thy1/Plpp3/Cpe/Mme/Kcnma1/Sele/Ednra/Pecam1/Adcyap1r1/Cavin3/Rtn4rl1/Dlc1/Ptch1/Fyn/Cacna1h/Cavin2/Plpp1/Atp1a2/Zap70/Stx2/Psen1/Gabbr1/Cav1/Abcg3/Lipe/Pld2/Lcp2/F2r/Cd4/Lrp4/Irs1 | 55 |
| GO:0005604 | basement membrane | CC | 0.227 | 1.93e-11 | 6.1e-10 | 9.21 | 4.03e-10 | Hmcn1/Col4a6/Npnt/Lamc3/Lama4/Tgfbi/Fn1/Nid1/Egflam/Adamts1/Frem1/Smoc1/Lama2/Matn2/Tmeff2/Col15a1/P3h1/Vwa1/Efemp2/Ptn/Ntng1/Col18a1/Itgb4/Spn/Nid2/Ccdc80/Col4a5 | 27 |
| GO:0034703 | cation channel complex | CC | 0.165 | 9.29e-11 | 2.78e-09 | 8.56 | 1.84e-09 | Kcnb2/Kcnj3/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Grin2b/Cacna1e/Unc80/Trpc6/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Pkd2/Ryr3/Cpt1c/Cacna1h/Dlg4/Kcng4/Scn3a/Nrn1/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 37 |
| GO:0034702 | ion channel complex | CC | 0.146 | 1.66e-10 | 4.69e-09 | 8.33 | 3.1e-09 | Kcnb2/Kcnj3/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Ano1/Grin2b/Cacna1e/Ano2/Unc80/Ttyh2/Trpc6/Glrb/Scn2a/Gabra4/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Pkd2/Ryr3/Cpt1c/Cacna1h/Dlg4/Kcng4/Scn3a/Nrn1/Kcnq4/Cacna1a/Lrrc8c/Cacnb4/Kcnh1 | 43 |
| GO:0099240 | intrinsic component of synaptic membrane | CC | 0.211 | 2.4e-10 | 6.47e-09 | 8.19 | 4.28e-09 | Efnb3/Kcnj3/Grid1/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Nlgn1/Nrp1/Ptpro/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Htr1b/Scn2a/Nlgn3/Prrt2/Pcdh17/Mpp2/Lrfn2/Cacna1h/Ntng1/Nlgn2 | 26 |
| GO:0099699 | integral component of synaptic membrane | CC | 0.212 | 1.06e-09 | 2.58e-08 | 7.59 | 1.71e-08 | Efnb3/Kcnj3/Grid1/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Nlgn1/Nrp1/Ptpro/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Htr1b/Scn2a/Nlgn3/Prrt2/Pcdh17/Lrfn2/Cacna1h/Nlgn2 | 24 |
| GO:0005901 | caveola | CC | 0.229 | 1.13e-09 | 2.65e-08 | 7.58 | 1.75e-08 | Cacna1c/Kcnd2/Kcnd3/Tfpi/Nos3/Lrrk2/Cdh13/Kcnma1/Sele/Adcyap1r1/Cavin3/Dlc1/Ptch1/Cacna1h/Cavin2/Plpp1/Atp1a2/Cav1/Lipe/Pld2/F2r/Irs1 | 22 |
| GO:0044853 | plasma membrane raft | CC | 0.191 | 2.37e-09 | 5.31e-08 | 7.27 | 3.51e-08 | Cacna1c/Kcnd2/Kcnd3/Tfpi/Nos3/Cdh2/Lrrk2/Cdh13/Has2/Kcnma1/Sele/Adcyap1r1/Cavin3/Dlc1/Ptch1/Cacna1h/Cavin2/Plpp1/Atp1a2/Cav1/Lipe/Pld2/Lcp2/F2r/Lrp4/Irs1 | 26 |
| GO:0005938 | cell cortex | CC | 0.134 | 5.57e-09 | 1.2e-07 | 6.92 | 7.93e-08 | Hmcn1/Ryr1/Phldb2/Hcls1/Rims3/Drd4/Gnai1/Cdh2/Prkd1/Exoc3l/Gypc/Coro1a/Sh3bp1/Rhobtb3/Sele/Pard6g/Septin3/Dlc1/Pard3b/BC034090/Myo1f/Asph/Pkd2/Erc2/Cldn5/Dlg4/Septin4/Ccn2/Arhgef9/Psen1/Mpp1/Cav1/Plekhh2/Shroom1/Usp2/Itgb4/Septin6/Cd302/Gpsm1/Cytip/Pstpip1 | 41 |
| GO:0031253 | cell projection membrane | CC | 0.131 | 7.45e-09 | 1.54e-07 | 6.81 | 1.02e-07 | Gap43/Ptprz1/Map2/Car4/Evc2/Evc/Epb41l3/Cd44/Aif1l/Abcg2/Adcy3/Enpep/Shank2/Bbs7/Podxl/Lrp2/Thy1/Pth1r/Msn/Gabra4/Jcad/Eps8l1/Kcnc4/Dlc1/Tmem231/Mpp2/Pkd2/Fgr/Ptger3/Palm/Bbs1/Plekho1/Abcb1a/Gabbr1/Abcg3/Pld2/Itga8/Nradd/Iqce/Kcnh1/Tbc1d10c/Plek | 42 |
| GO:0030027 | lamellipodium | CC | 0.165 | 1.03e-08 | 1.97e-07 | 6.7 | 1.31e-07 | Ptprz1/Evl/Ccdc88a/Cd44/Cdh2/Wasf3/Abi3/Podxl/Ptprm/Plxnd1/Abi2/Coro1a/Palld/Sh3bp1/Dysf/Kitl/Arap3/Carmil3/Parvb/Pkd2/Fgd1/Unc5c/Arhgap31/Dpysl3/Stx2/Plekhh2/Pld2/Nradd/Plekhg5 | 29 |
| GO:0043197 | dendritic spine | CC | 0.157 | 1.13e-08 | 2.1e-07 | 6.68 | 1.39e-07 | Ptprz1/Slc1a3/Gria4/Kcnd2/Kcnd3/Grid2ip/Nlgn1/Slc8a1/Lrrc7/Ptpro/P2rx2/Drd4/Grin2b/Shank2/Ppfia2/Slc8a3/Dock10/Abi2/Nrgn/Sla/Arrb1/Arrb2/Mpp2/Cnn3/Dlg4/Palm/Atp1a2/Sri/Gabbr1/Itga8/Zmynd8 | 31 |
| GO:0044309 | neuron spine | CC | 0.153 | 2.06e-08 | 3.69e-07 | 6.43 | 2.44e-07 | Ptprz1/Slc1a3/Gria4/Kcnd2/Kcnd3/Grid2ip/Nlgn1/Slc8a1/Lrrc7/Ptpro/P2rx2/Drd4/Grin2b/Shank2/Ppfia2/Slc8a3/Dock10/Abi2/Nrgn/Sla/Arrb1/Arrb2/Mpp2/Cnn3/Dlg4/Palm/Atp1a2/Sri/Gabbr1/Itga8/Zmynd8 | 31 |
| GO:0099634 | postsynaptic specialization membrane | CC | 0.224 | 2.35e-08 | 4.08e-07 | 6.39 | 2.7e-07 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Dlg2/Nlgn3/Prrt1/Cacng7/Mpp2/Lrfn2/Dlg4/Nlgn2 | 19 |
| GO:0098936 | intrinsic component of postsynaptic membrane | CC | 0.213 | 5.2e-08 | 8.55e-07 | 6.07 | 5.65e-07 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Nrp1/Ptpro/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Nlgn3/Pcdh17/Mpp2/Lrfn2/Cacna1h/Nlgn2 | 19 |
| GO:0016323 | basolateral plasma membrane | CC | 0.138 | 5.52e-08 | 8.73e-07 | 6.06 | 5.77e-07 | Slc1a3/Slc13a5/Slc8a1/Lrrc7/Cd44/Cdh2/Tek/Cldn8/Ank2/Pth1r/Plpp3/Msn/Slc29a1/Dlg2/Heph/Lin7a/Cd38/Agtr1a/Slc23a2/Pkd1/Arrb1/Arrb2/Slc38a1/Abca8b/Slco4c1/Palm/Atp2b4/Stx2/Abcc5/Cav1/Nkd2/Cldn1/S100g/Slc43a3 | 34 |
| GO:1902495 | transmembrane transporter complex | CC | 0.118 | 7.51e-08 | 1.15e-06 | 5.94 | 7.63e-07 | Kcnb2/Kcnj3/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Ano1/Grin2b/Cacna1e/Ano2/Unc80/Ttyh2/Trpc6/Glrb/Scn2a/Gabra4/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Pkd2/Ryr3/Cpt1c/Cacna1h/Dlg4/Kcng4/Scn3a/Atp1a2/Nrn1/Kcnq4/Cacna1a/Lrrc8c/Cacnb4/Kcnh1 | 44 |
| GO:0009925 | basal plasma membrane | CC | 0.13 | 1.01e-07 | 1.51e-06 | 5.82 | 9.98e-07 | Slc1a3/Slc13a5/Slc8a1/Lrrc7/Cd44/Cdh2/Tek/Cldn8/Ank2/Pth1r/Plpp3/Msn/Slc29a1/Dlg2/Heph/Lin7a/Cd38/Agtr1a/Slc23a2/Pkd1/Arrb1/Arrb2/Slc38a1/Pkd2/Abca8b/Slco4c1/Palm/Atp2b4/Stx2/Abcc5/Cav1/Itgb4/Nkd2/Cldn1/S100g/Slc43a3 | 36 |
| GO:0099055 | integral component of postsynaptic membrane | CC | 0.212 | 1.32e-07 | 1.84e-06 | 5.73 | 1.22e-06 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Nrp1/Ptpro/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Nlgn3/Pcdh17/Lrfn2/Cacna1h/Nlgn2 | 18 |
| GO:0030133 | transport vesicle | CC | 0.123 | 1.34e-07 | 1.84e-06 | 5.73 | 1.22e-06 | Syn2/Car4/Igf1/Aph1b/Syt13/Sec16b/Stx1b/Lrrk2/Myo5a/Grin2b/Kif1a/Mme/Dlg2/Lin7a/Dmxl2/Nrgn/Lyz2/Prrt1/Syt7/Prrt2/Kcnc4/Bin1/Synrg/Bgn/Sv2c/Dlg4/Septin4/Lyz1/Dpysl3/Slc4a8/Brsk1/Stx2/Psen1/Rab27a/Gabbr1/Sspn/Nkd2/Septin6/Sipa1 | 39 |
| GO:0019897 | extrinsic component of plasma membrane | CC | 0.151 | 2.13e-07 | 2.79e-06 | 5.55 | 1.85e-06 | Ryr1/Fermt2/Gnai1/Cdh2/Cdh6/Serpine2/Cdh13/Gngt2/Cdh10/Cdh5/Gng11/Sla/Apoe/Cdh11/Hip1/Gng2/Stac2/Dchs1/Fyn/Gnaz/Dlg4/Fgr/Gnb5/Gng7/Zap70/Gnb4/Styk1 | 27 |
| GO:0098948 | intrinsic component of postsynaptic specialization membrane | CC | 0.238 | 2.89e-07 | 3.71e-06 | 5.43 | 2.45e-06 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Nlgn3/Mpp2/Lrfn2/Nlgn2 | 15 |
| GO:0005614 | interstitial matrix | CC | 0.471 | 5.47e-07 | 6.84e-06 | 5.17 | 4.52e-06 | Vit/Col14a1/Igf1/Egflam/Mamdc2/Vwa1/Abi3bp/Ccdc80 | 8 |
| GO:0098982 | GABA-ergic synapse | CC | 0.217 | 1.02e-06 | 1.24e-05 | 4.91 | 8.23e-06 | Kcnd2/Kcnd3/Ptpro/Plcb1/Slitrk3/Glrb/Gucy1a1/Epha3/Syt7/Pcdh17/Erc2/Arhgef9/Gabbr1/Iqsec3/Nlgn2 | 15 |
| GO:0099060 | integral component of postsynaptic specialization membrane | CC | 0.23 | 1.16e-06 | 1.39e-05 | 4.86 | 9.16e-06 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Nlgn3/Lrfn2/Nlgn2 | 14 |
| GO:0031594 | neuromuscular junction | CC | 0.187 | 1.89e-06 | 2.16e-05 | 4.67 | 1.43e-05 | Pdzrn3/Stx1b/Lama4/Serpine2/Tbc1d24/Slc8a3/Postn/Dlg2/Lama2/Kcnc4/Itga7/Dlg4/Psen1/Ptn/F2r/Lrp4/Col4a5 | 17 |
| GO:0043679 | axon terminus | CC | 0.137 | 2.36e-06 | 2.59e-05 | 4.59 | 1.71e-05 | Gria4/Grik1/Kcna6/Slc8a1/P2rx2/Drd4/Lrrk2/Pfn2/Grin2b/Mical1/Slc8a3/Calca/Grik2/Syt7/Prrt2/Kcnma1/Kcnc4/Bin1/Kcnip3/Erc2/Septin4/Slc4a8/Sri/Septin6/Ror1/Prss12 | 26 |
| GO:0030426 | growth cone | CC | 0.133 | 2.64e-06 | 2.84e-05 | 4.55 | 1.88e-05 | Ptprz1/Map2/Fez1/Itga4/Dcc/Nrp1/Tmod2/Lrrk2/Shank2/Mapk8ip1/Adgrl1/Myo9a/Slc2a13/Lrp2/Thy1/Tshz3/Nin/Neo1/Ptch1/Erc2/Pcdhgb1/Unc5c/Dpysl3/Psen1/Cpeb1/Kif5a/Setx | 27 |
| GO:0016324 | apical plasma membrane | CC | 0.108 | 2.69e-06 | 2.84e-05 | 4.55 | 1.88e-05 | Car4/Ddr2/Mpdz/Ptpro/Cd44/P2rx2/Cdh2/Fn1/Slc24a4/Ano1/Abcg2/Enpep/Stc1/Tek/Shank2/Podxl/Lrp2/Thy1/Pth1r/Msn/Slc29a1/Nox4/Kcnma1/Slc23a2/Pard6g/Pard3b/BC034090/Slc38a1/Plpp1/Psen1/Abcb1a/Rab27a/Pdgfrb/Abcc5/Cav1/Shroom1/Pfkm/Cldn1/S100g/Atp6v1b2 | 40 |
| GO:0098839 | postsynaptic density membrane | CC | 0.217 | 5.52e-06 | 5.5e-05 | 4.26 | 3.64e-05 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Dlg2/Prrt1/Cacng7/Mpp2/Lrfn2/Dlg4 | 13 |
| GO:0001726 | ruffle | CC | 0.144 | 6.13e-06 | 6e-05 | 4.22 | 3.97e-05 | Ptprz1/Map2/Cspg4/Aif1l/Myo5a/Podxl/Sh2b2/Rab34/Frmd4b/Jcad/Eps8l1/Pecam1/Arap3/Tln2/Dlc1/Fgd1/Fgr/Plekho1/Pdgfrb/Fgd2/Plek/Atp6v1b2 | 22 |
| GO:0098802 | plasma membrane signaling receptor complex | CC | 0.132 | 7.41e-06 | 7.12e-05 | 4.15 | 4.71e-05 | Gria4/Grik1/Itga4/Itgb7/Grin2b/Cd3g/Itga1/Grik2/Itgal/Il27ra/Emilin1/Itga7/Cacng7/Csf2rb/Cpt1c/Calcrl/Dlg4/Zap70/Apbb1ip/Il12rb1/Gabbr1/Nrn1/Itga8/Itgb4/Irs1 | 25 |
| GO:0070382 | exocytic vesicle | CC | 0.123 | 8.26e-06 | 7.8e-05 | 4.11 | 5.15e-05 | Syn2/Igf1/Syt13/Stx1b/Lrrk2/Myo5a/Grin2b/Kif1a/Mme/Dlg2/Dmxl2/Prrt1/Syt7/Prrt2/Kcnc4/Bin1/Sv2c/Dlg4/Septin4/Dpysl3/Slc4a8/Brsk1/Stx2/Psen1/Rab27a/Gabbr1/Nkd2/Septin6 | 28 |
| GO:0030055 | cell-substrate junction | CC | 0.132 | 1.08e-05 | 1e-04 | 4 | 6.61e-05 | Hmcn1/Evl/Phldb2/Nrp1/Fermt2/Aif1l/Itgb7/Thsd1/Itga1/Tnfsf13b/Msn/Palld/Sla/Nox4/Itga7/Parvg/Efs/Parvb/Tln2/Dlc1/Mapre2/Cav1/Itgb4/Tle2 | 24 |
| GO:0098562 | cytoplasmic side of membrane | CC | 0.128 | 1.17e-05 | 0.000107 | 3.97 | 7.07e-05 | Ryr1/Fermt2/Gnai1/Lrrk2/Rasa3/Gngt2/Msn/Kit/Gng11/Sla/Hip1/Gng2/Stac2/Pkd2/Fyn/Gnaz/Dlg4/Fgr/Gnb5/Gng7/Zap70/Gnb4/Styk1/Ptpn7/Rasal3 | 25 |
| GO:0005912 | adherens junction | CC | 0.141 | 2.17e-05 | 0.000195 | 3.71 | 0.000129 | Hmcn1/Fermt2/Cdh2/Cdh6/Cdh10/Ptprm/Cdh5/Msn/Abi2/Frmd4b/Esam/Jcad/Lin7a/Sh3bp1/Cdh11/Pard3b/Dchs1/Frmd4a/Dlg5/Shroom1 | 20 |
| GO:0034705 | potassium channel complex | CC | 0.169 | 2.72e-05 | 0.00024 | 3.62 | 0.000159 | Kcnb2/Kcnj3/Gria4/Grik1/Kcna6/Kcnd2/Kcnd3/Kcnq5/Grik2/Kcnma1/Kcnc4/Kcnip3/Kcng4/Kcnq4/Kcnh1 | 15 |
| GO:0098889 | intrinsic component of presynaptic membrane | CC | 0.216 | 3.02e-05 | 0.000258 | 3.59 | 0.000171 | Efnb3/Kcnj3/Ptprd/Cdh2/Cacna1e/Htr1b/Scn2a/Prrt2/Pcdh17/Cacna1h/Ntng1 | 11 |
| GO:0005925 | focal adhesion | CC | 0.13 | 3.02e-05 | 0.000258 | 3.59 | 0.000171 | Evl/Phldb2/Nrp1/Fermt2/Aif1l/Itgb7/Thsd1/Itga1/Tnfsf13b/Msn/Palld/Sla/Nox4/Parvg/Efs/Parvb/Tln2/Dlc1/Mapre2/Cav1/Itgb4/Tle2 | 22 |
| GO:0034706 | sodium channel complex | CC | 0.296 | 3.21e-05 | 0.00027 | 3.57 | 0.000179 | Gria4/Grik1/Scn4b/Scn1a/Scn2a/Grik2/Cacna1h/Scn3a | 8 |
| GO:0099056 | integral component of presynaptic membrane | CC | 0.227 | 4.29e-05 | 0.000355 | 3.45 | 0.000235 | Efnb3/Kcnj3/Ptprd/Cdh2/Cacna1e/Htr1b/Scn2a/Prrt2/Pcdh17/Cacna1h | 10 |
| GO:0005923 | bicellular tight junction | CC | 0.138 | 4.75e-05 | 0.000387 | 3.41 | 0.000256 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Frmd4b/Esam/Lin7a/Sh3bp1/Magi2/Pard6g/Adcyap1r1/Pard3b/Frmd4a/Nphp1/Cldn5/Pdcd6ip/Cldn1/Jam2 | 19 |
| GO:0098685 | Schaffer collateral - CA1 synapse | CC | 0.169 | 4.96e-05 | 0.000398 | 3.4 | 0.000263 | Syn2/Lrrc4c/Dcc/Ptprd/Pfn2/Akap12/Dgkb/Cdh11/Mpp2/Lrfn2/Fyn/Gabbr1/Ntng1/Prkar1b | 14 |
| GO:0043296 | apical junction complex | CC | 0.129 | 5.3e-05 | 0.000416 | 3.38 | 0.000275 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Frmd4b/Esam/Lin7a/Sh3bp1/Nfasc/Magi2/Pard6g/Adcyap1r1/Pard3b/Frmd4a/Nphp1/Cldn5/Shroom1/Pdcd6ip/Cldn1/Jam2 | 21 |
| GO:0070160 | tight junction | CC | 0.132 | 5.33e-05 | 0.000416 | 3.38 | 0.000275 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Frmd4b/Esam/Lin7a/Sh3bp1/Nfasc/Magi2/Pard6g/Adcyap1r1/Pard3b/Frmd4a/Nphp1/Cldn5/Pdcd6ip/Cldn1/Jam2 | 20 |
| GO:0009898 | cytoplasmic side of plasma membrane | CC | 0.124 | 6.16e-05 | 0.000466 | 3.33 | 0.000308 | Ryr1/Fermt2/Gnai1/Rasa3/Gngt2/Msn/Kit/Gng11/Sla/Hip1/Gng2/Stac2/Fyn/Gnaz/Dlg4/Fgr/Gnb5/Gng7/Zap70/Gnb4/Styk1/Ptpn7 | 22 |
| GO:0036064 | ciliary basal body | CC | 0.124 | 6.16e-05 | 0.000466 | 3.33 | 0.000308 | Evc/Ccdc88a/Kif7/Dzip1l/Cibar1/Wdr35/Bbs7/Armc9/Ift57/Rilpl1/Cfap410/Ttc26/Arl3/Cep19/Pkd2/Ttbk2/Dzip1/Dlg5/Dync2li1/Bbs1/Ift81/Irs1 | 22 |
| GO:0098858 | actin-based cell projection | CC | 0.114 | 6.62e-05 | 0.000488 | 3.31 | 0.000323 | Gap43/Ptprz1/Map2/Angpt1/Nlgn1/Lrrc7/Cd44/Lrrk2/Myo5a/Jam3/Tek/Podxl/Msn/Abi2/Coro1a/Nfasc/Lyz2/Kitl/Myo1f/Unc5c/Lyz1/Palm/Spn/Cd302/Tbc1d10c/Atp6v1b2 | 26 |
| GO:0043195 | terminal bouton | CC | 0.156 | 0.000123 | 0.000848 | 3.07 | 0.000561 | Gria4/Grik1/P2rx2/Drd4/Lrrk2/Pfn2/Grin2b/Mical1/Calca/Grik2/Kcnma1/Erc2/Slc4a8/Prss12 | 14 |
| GO:0099568 | cytoplasmic region | CC | 0.111 | 0.000137 | 0.000925 | 3.03 | 0.000612 | Gli1/Map2/Phldb2/Rims3/Arl6/Gnai1/Dzip1l/Lrrk2/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Grik2/Rp1/Tubb4a/Dync2h1/Arl3/Pkd2/Erc2/Dnah7b/Dlg4/Dync2li1/Dnah1/Bbs1 | 25 |
| GO:0099146 | intrinsic component of postsynaptic density membrane | CC | 0.22 | 0.000138 | 0.000925 | 3.03 | 0.000612 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Mpp2/Lrfn2 | 9 |
| GO:0008305 | integrin complex | CC | 0.235 | 0.000193 | 0.00128 | 2.89 | 0.000849 | Itga4/Itgb7/Itga1/Itgal/Emilin1/Itga7/Itga8/Itgb4 | 8 |
| GO:0008021 | synaptic vesicle | CC | 0.112 | 0.000203 | 0.00133 | 2.88 | 0.00088 | Syn2/Stx1b/Lrrk2/Myo5a/Grin2b/Kif1a/Mme/Dlg2/Dmxl2/Prrt1/Syt7/Prrt2/Kcnc4/Bin1/Sv2c/Dlg4/Septin4/Slc4a8/Brsk1/Stx2/Psen1/Gabbr1/Septin6 | 23 |
| GO:0098644 | complex of collagen trimers | CC | 0.316 | 0.000218 | 0.00141 | 2.85 | 0.000934 | Col11a1/Col4a6/Col16a1/Col5a2/Lum/Col4a5 | 6 |
| GO:0005790 | smooth endoplasmic reticulum | CC | 0.229 | 0.00024 | 0.00152 | 2.82 | 0.001 | Map2/Ryr1/Jph4/Rtn1/Myo5a/Ryr3/Psen1/Sri | 8 |
| GO:0005834 | heterotrimeric G-protein complex | CC | 0.229 | 0.00024 | 0.00152 | 2.82 | 0.001 | Gnai1/Gngt2/Gng11/Gng2/Gnaz/Gnb5/Gng7/Gnb4 | 8 |
| GO:0008076 | voltage-gated potassium channel complex | CC | 0.16 | 0.000281 | 0.0017 | 2.77 | 0.00112 | Kcnb2/Kcnj3/Kcna6/Kcnd2/Kcnd3/Kcnq5/Kcnma1/Kcnc4/Kcnip3/Kcng4/Kcnq4/Kcnh1 | 12 |
| GO:0098791 | Golgi apparatus subcompartment | CC | 0.0977 | 0.000291 | 0.00174 | 2.76 | 0.00115 | B4galnt4/Car4/Nsg2/Chst2/Tgfbi/Bok/Gga2/Lrrk2/Slc9a7/Ap1m1/Prkd1/Postn/Plpp3/Rab34/Mme/Chst1/Cracr2a/Ggta1/Lyz2/Dennd5a/5730455P16Rik/Pcsk5/Ms4a7/Nsg1/Ece2/Lyz1/Xylt1/Vps53/Golph3l/Atp8b4 | 30 |
| GO:0030175 | filopodium | CC | 0.141 | 0.000343 | 0.00203 | 2.69 | 0.00134 | Gap43/Ptprz1/Map2/Nlgn1/Lrrc7/Myo5a/Podxl/Msn/Abi2/Kitl/Unc5c/Palm/Cd302/Tbc1d10c | 14 |
| GO:1905360 | GTPase complex | CC | 0.216 | 0.00036 | 0.00208 | 2.68 | 0.00138 | Gnai1/Gngt2/Gng11/Gng2/Gnaz/Gnb5/Gng7/Gnb4 | 8 |
| GO:0034704 | calcium channel complex | CC | 0.164 | 0.000397 | 0.00227 | 2.64 | 0.0015 | Cacna1c/Ryr1/Cachd1/Cacna2d1/Cacna1e/Pkd1/Cacng7/Ryr3/Cacna1h/Cacna1a/Cacnb4 | 11 |
| GO:0099061 | integral component of postsynaptic density membrane | CC | 0.205 | 0.000526 | 0.00298 | 2.53 | 0.00197 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Lrfn2 | 8 |
| GO:0097730 | non-motile cilium | CC | 0.114 | 0.000588 | 0.00329 | 2.48 | 0.00218 | D630045J12Rik/Myo5a/Slc24a4/Tmem237/Shank2/Bbs7/Cerkl/Ano2/Ift57/Rp1/Cfap410/Arl3/Pkd2/Nphp1/Iqcb1/Gnb5/C130074G19Rik/Rab27a/Glis2 | 19 |
| GO:0097733 | photoreceptor cell cilium | CC | 0.128 | 0.000624 | 0.00346 | 2.46 | 0.00229 | D630045J12Rik/Myo5a/Slc24a4/Tmem237/Shank2/Bbs7/Cerkl/Ift57/Rp1/Cfap410/Arl3/Nphp1/Iqcb1/Gnb5/Rab27a | 15 |
| GO:0016342 | catenin complex | CC | 0.226 | 0.000636 | 0.00349 | 2.46 | 0.00231 | Cdh2/Cdh6/Cdh13/Cdh10/Cdh5/Cdh11/Dchs1 | 7 |
| GO:0097731 | 9+0 non-motile cilium | CC | 0.123 | 0.000659 | 0.00358 | 2.45 | 0.00237 | D630045J12Rik/Myo5a/Slc24a4/Tmem237/Shank2/Bbs7/Cerkl/Ift57/Rp1/Cfap410/Arl3/Nphp1/Iqcb1/Gnb5/C130074G19Rik/Rab27a | 16 |
| GO:0043292 | contractile fiber | CC | 0.101 | 0.00068 | 0.00366 | 2.44 | 0.00242 | Cacna1c/Ryr1/Npnt/Adra1a/Slc8a1/Pygm/Tmod2/Scn1a/Mmp2/Mef2c/Grin2b/Arhgef25/Ank2/Palld/Pecam1/Bin1/Myl3/Cmya5/Ryr3/Habp4/Atp2b4/Simc1/Psen1/Sri | 24 |
| GO:0032838 | plasma membrane bounded cell projection cytoplasm | CC | 0.109 | 0.000767 | 0.00408 | 2.39 | 0.0027 | Gli1/Map2/Arl6/Dzip1l/Lrrk2/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Grik2/Rp1/Tubb4a/Dync2h1/Arl3/Dnah7b/Dlg4/Dync2li1/Dnah1/Bbs1 | 20 |
| GO:0016528 | sarcoplasm | CC | 0.141 | 0.000902 | 0.00476 | 2.32 | 0.00315 | Car4/Sgcd/Ryr1/Jph4/Pygm/Cacna2d1/Cmya5/Flnc/Asph/Ryr3/Gstm7/Sri | 12 |
| GO:0030863 | cortical cytoskeleton | CC | 0.123 | 0.000969 | 0.00506 | 2.3 | 0.00335 | Hcls1/Rims3/Cdh2/Gypc/Coro1a/Sele/Dlc1/Myo1f/Erc2/Cldn5/Dlg4/Mpp1/Plekhh2/Shroom1/Pstpip1 | 15 |
| GO:0005874 | microtubule | CC | 0.0845 | 0.00104 | 0.00534 | 2.27 | 0.00353 | Map2/Fez1/Clmp/Capn6/Slc8a1/Kif7/Cep170/Klhl22/Dnah5/Slc8a3/Kif26a/Kif1a/Reep1/Eml1/Map9/Tubb4a/Katnal1/Apoe/Dync2h1/Ninl/Kif6/Dysf/Nin/Slain1/Arl3/Sarm1/Stard9/Dync2li1/Lrrc49/Tcp11l1/Dnah1/Mapre2/Shroom1/Tppp/Kif16b/Rmdn2/Kif5a | 37 |
| GO:0005891 | voltage-gated calcium channel complex | CC | 0.186 | 0.00104 | 0.00534 | 2.27 | 0.00353 | Cacna1c/Cachd1/Cacna2d1/Cacna1e/Cacng7/Cacna1h/Cacna1a/Cacnb4 | 8 |
| GO:0001518 | voltage-gated sodium channel complex | CC | 0.294 | 0.00107 | 0.00545 | 2.26 | 0.00361 | Scn4b/Scn1a/Scn2a/Cacna1h/Scn3a | 5 |
| GO:0016327 | apicolateral plasma membrane | CC | 0.24 | 0.0011 | 0.00555 | 2.26 | 0.00367 | Mpdz/Cdh2/Cldn8/Prickle2/Cldn5/Palm | 6 |
| GO:0005802 | trans-Golgi network | CC | 0.0976 | 0.00114 | 0.0057 | 2.24 | 0.00377 | Car4/Nsg2/Chst2/Tgfbi/Bok/Gga2/Lrrk2/Slc9a7/Ap1m1/Prkd1/Postn/Plpp3/Mme/Chst1/Cracr2a/Dennd5a/5730455P16Rik/Pcsk5/Ms4a7/Nsg1/Ece2/Vps53/Golph3l/Atp8b4 | 24 |
| GO:0008328 | ionotropic glutamate receptor complex | CC | 0.182 | 0.00122 | 0.00602 | 2.22 | 0.00398 | Gria4/Grik1/Grin2b/Grik2/Cacng7/Cpt1c/Dlg4/Nrn1 | 8 |
| GO:0043083 | synaptic cleft | CC | 0.278 | 0.00143 | 0.00692 | 2.16 | 0.00458 | Lama4/Grin2b/Lama2/Apoe/Prss12 | 5 |
| GO:0005583 | fibrillar collagen trimer | CC | 0.364 | 0.00145 | 0.00692 | 2.16 | 0.00458 | Col11a1/Col16a1/Col5a2/Lum | 4 |
| GO:0098643 | banded collagen fibril | CC | 0.364 | 0.00145 | 0.00692 | 2.16 | 0.00458 | Col11a1/Col16a1/Col5a2/Lum | 4 |
| GO:0030667 | secretory granule membrane | CC | 0.133 | 0.0015 | 0.00708 | 2.15 | 0.00468 | Car4/Slc2a3/Cpe/Msn/Kif1a/Cd38/Pla1a/Ccdc136/Rab27a/Sri/Cav1/Tmem184a | 12 |
| GO:0097014 | ciliary plasm | CC | 0.113 | 0.00158 | 0.0074 | 2.13 | 0.00489 | Gli1/Arl6/Dzip1l/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Rp1/Tubb4a/Dync2h1/Arl3/Dnah7b/Dync2li1/Dnah1/Bbs1 | 16 |
| GO:0098878 | neurotransmitter receptor complex | CC | 0.174 | 0.00165 | 0.00759 | 2.12 | 0.00502 | Gria4/Grik1/Grin2b/Grik2/Cacng7/Cpt1c/Dlg4/Nrn1 | 8 |
| GO:0030016 | myofibril | CC | 0.0982 | 0.00165 | 0.00759 | 2.12 | 0.00502 | Cacna1c/Ryr1/Adra1a/Slc8a1/Pygm/Tmod2/Scn1a/Mmp2/Mef2c/Grin2b/Arhgef25/Ank2/Palld/Bin1/Myl3/Cmya5/Ryr3/Habp4/Atp2b4/Simc1/Psen1/Sri | 22 |
| GO:0060170 | ciliary membrane | CC | 0.158 | 0.00175 | 0.00796 | 2.1 | 0.00526 | Evc2/Evc/Adcy3/Shank2/Bbs7/Tmem231/Pkd2/Bbs1/Iqce | 9 |
| GO:0032391 | photoreceptor connecting cilium | CC | 0.17 | 0.0019 | 0.00859 | 2.07 | 0.00568 | D630045J12Rik/Tmem237/Ift57/Rp1/Cfap410/Arl3/Nphp1/Iqcb1 | 8 |
| GO:0005902 | microvillus | CC | 0.124 | 0.00193 | 0.00866 | 2.06 | 0.00572 | Angpt1/Cd44/Lrrk2/Jam3/Tek/Podxl/Msn/Nfasc/Lyz2/Myo1f/Lyz1/Spn/Atp6v1b2 | 13 |
| GO:0048787 | presynaptic active zone membrane | CC | 0.214 | 0.00207 | 0.00913 | 2.04 | 0.00604 | Stx1b/Cdh2/Kcnma1/Cacna1h/Stx2/Ntng1 | 6 |
| GO:0042613 | MHC class II protein complex | CC | 0.333 | 0.0021 | 0.00917 | 2.04 | 0.00606 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0005884 | actin filament | CC | 0.113 | 0.00232 | 0.0101 | 2 | 0.00665 | Hcls1/Aif1l/Myo5a/Jam3/Tek/Sh2b2/Abi2/Coro1a/Palld/Dlc1/Myo1f/Fyn/Wipf1/Dpysl3/Pstpip1 | 15 |
| GO:0032589 | neuron projection membrane | CC | 0.139 | 0.00267 | 0.0115 | 1.94 | 0.00758 | Epb41l3/Thy1/Gabra4/Kcnc4/Mpp2/Palm/Gabbr1/Itga8/Nradd/Kcnh1 | 10 |
| GO:0031985 | Golgi cisterna | CC | 0.148 | 0.00282 | 0.012 | 1.92 | 0.00791 | B4galnt4/Nsg2/Rab34/Ggta1/Lyz2/Pcsk5/Lyz1/Xylt1/Golph3l | 9 |
| GO:0000137 | Golgi cis cisterna | CC | 0.308 | 0.00291 | 0.0122 | 1.91 | 0.00809 | Nsg2/Lyz2/Lyz1/Xylt1 | 4 |
| GO:0019867 | outer membrane | CC | 0.0957 | 0.00353 | 0.0145 | 1.84 | 0.00962 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx1/Armcx6/Aifm2/Snn/Agtr1a/Syne3/Sarm1/Cpt1c/Armcx2/Acsl1/Gimap3/Asl/Psen1/Qtrt2 | 20 |
| GO:0031968 | organelle outer membrane | CC | 0.0957 | 0.00353 | 0.0145 | 1.84 | 0.00962 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx1/Armcx6/Aifm2/Snn/Agtr1a/Syne3/Sarm1/Cpt1c/Armcx2/Acsl1/Gimap3/Asl/Psen1/Qtrt2 | 20 |
| GO:0031045 | dense core granule | CC | 0.194 | 0.00357 | 0.0145 | 1.84 | 0.00962 | Cpe/Kif1a/Cadps/Dmxl2/Syt7/Ncs1 | 6 |
| GO:0016529 | sarcoplasmic reticulum | CC | 0.132 | 0.00398 | 0.0161 | 1.79 | 0.0106 | Car4/Sgcd/Ryr1/Pygm/Cacna2d1/Cmya5/Asph/Ryr3/Gstm7/Sri | 10 |
| GO:0044291 | cell-cell contact zone | CC | 0.122 | 0.00461 | 0.0185 | 1.73 | 0.0122 | Slc8a1/Scn4b/Cdh2/Scn1a/Jam3/Ank2/Scn2a/Pecam1/Tln2/Atp1a2/Jam2 | 11 |
| GO:0044295 | axonal growth cone | CC | 0.159 | 0.00531 | 0.021 | 1.68 | 0.0139 | Dcc/Mapk8ip1/Myo9a/Lrp2/Nin/Neo1/Ptch1 | 7 |
| GO:0031526 | brush border membrane | CC | 0.125 | 0.00576 | 0.0224 | 1.65 | 0.0148 | Car4/Abcg2/Enpep/Shank2/Lrp2/Pth1r/Ptger3/Abcb1a/Abcg3/Pld2 | 10 |
| GO:0005769 | early endosome | CC | 0.0826 | 0.00622 | 0.0241 | 1.62 | 0.0159 | Nsg2/Kdr/St8sia2/Nrp1/Bok/Myo5a/Rps6kc1/Rab34/Coro1a/Cd1d1/Mme/Epha3/H2-Ab1/Sorcs2/Apoe/Havcr2/Dysf/Nsg1/Cacng7/Zfyve9/Mvb12b/Psen1/Kif16b/Fgd2/F2r/Kcnh1/Tmem184a | 27 |
| GO:0005788 | endoplasmic reticulum lumen | CC | 0.119 | 0.0081 | 0.0302 | 1.52 | 0.02 | Wnt6/Selenom/Eogt/Lyz2/Pcsk5/Poglut2/Tor4a/Lyz1/Cd4/Bche | 10 |
| GO:0031225 | anchored component of membrane | CC | 0.0939 | 0.00814 | 0.0302 | 1.52 | 0.02 | Gpc6/Cntn3/Car4/Cd109/Sema7a/Gpc3/Ntm/Cdh13/Thy1/Mmp19/Rtn4rl1/Mpp2/Nrn1/Ntng1/Gpc2/Lsamp/Cd52 | 17 |
| GO:0031527 | filopodium membrane | CC | 0.235 | 0.00828 | 0.0305 | 1.52 | 0.0202 | Gap43/Msn/Palm/Tbc1d10c | 4 |
| GO:0046658 | anchored component of plasma membrane | CC | 0.125 | 0.00858 | 0.0314 | 1.5 | 0.0208 | Gpc6/Car4/Gpc3/Thy1/Rtn4rl1/Mpp2/Nrn1/Ntng1/Gpc2 | 9 |
| GO:0032587 | ruffle membrane | CC | 0.123 | 0.00937 | 0.0341 | 1.47 | 0.0225 | Ptprz1/Map2/Aif1l/Jcad/Eps8l1/Dlc1/Fgr/Plekho1/Plek | 9 |
| GO:0005814 | centriole | CC | 0.102 | 0.00981 | 0.0354 | 1.45 | 0.0234 | Ccdc88a/Dzip1l/Cibar1/Cep170/Cntln/Armc9/Crocc/Nin/Cep19/Ttbk2/Dzip1/Stard9/Cep97 | 13 |
| GO:0032809 | neuronal cell body membrane | CC | 0.158 | 0.01 | 0.0358 | 1.45 | 0.0237 | Kcnb2/Slc6a2/Kcnd2/Thy1/Kcnc4/Slc4a8 | 6 |
| GO:0098690 | glycinergic synapse | CC | 0.3 | 0.011 | 0.0376 | 1.42 | 0.0249 | Glrb/Iqsec3/Nlgn2 | 3 |
| GO:0099569 | presynaptic cytoskeleton | CC | 0.3 | 0.011 | 0.0376 | 1.42 | 0.0249 | Rims3/Septin3/Erc2 | 3 |
| GO:1990454 | L-type voltage-gated calcium channel complex | CC | 0.3 | 0.011 | 0.0376 | 1.42 | 0.0249 | Cacna1c/Cacna2d1/Cacng7 | 3 |
| GO:0005741 | mitochondrial outer membrane | CC | 0.0904 | 0.0116 | 0.0396 | 1.4 | 0.0262 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx1/Armcx6/Aifm2/Snn/Sarm1/Cpt1c/Armcx2/Acsl1/Gimap3/Asl/Qtrt2 | 17 |
| GO:0005903 | brush border | CC | 0.0966 | 0.0123 | 0.0416 | 1.38 | 0.0275 | Car4/Dab1/Abcg2/Enpep/Shank2/Lrp2/Pth1r/Mme/Scart1/Scin/Ptger3/Abcb1a/Abcg3/Pld2 | 14 |
| GO:0032281 | AMPA glutamate receptor complex | CC | 0.172 | 0.0126 | 0.042 | 1.38 | 0.0278 | Gria4/Cacng7/Cpt1c/Dlg4/Nrn1 | 5 |
| GO:0097440 | apical dendrite | CC | 0.172 | 0.0126 | 0.042 | 1.38 | 0.0278 | Map2/Grin2b/Osbp2/Enpp1/Kif5a | 5 |
| GO:0005871 | kinesin complex | CC | 0.135 | 0.0132 | 0.0437 | 1.36 | 0.0289 | Kif7/Kif26a/Kif1a/Kif6/Stard9/Kif16b/Kif5a | 7 |
| GO:0001931 | uropod | CC | 0.273 | 0.0145 | 0.0474 | 1.32 | 0.0314 | Pip5k1b/Msn/Spn | 3 |
| GO:0016010 | dystrophin-associated glycoprotein complex | CC | 0.2 | 0.015 | 0.0486 | 1.31 | 0.0321 | Sgcd/Sntb1/Sgcb/Sspn | 4 |
| GO:0050839 | cell adhesion molecule binding | MF | 0.195 | 2.13e-20 | 2.03e-17 | 16.7 | 1.56e-17 | Prtg/Ptprz1/Itga4/Lrrc4c/Kdr/Ptprd/Igf1/Nlgn1/Npnt/Nos3/Ptpro/Fermt2/Sema7a/Col16a1/Tgfbi/Fxyd5/Cdh2/Itgb7/Fn1/Cdh6/Tenm3/Icam1/Grin2b/Jam3/Cdh13/Cxcl12/Adgrl1/Cdh10/Itga1/Ptprm/Cdh5/Thy1/Postn/Plpp3/Cpe/Msn/Cd1d1/Cd226/Itgal/Esam/Nlgn3/Emilin1/Nfasc/Cdh11/Cd200/Igf2/Itga7/Neo1/Tln2/Dchs1/Ccn2/Psen1/Ptn/Ntng1/Itga8/Itgb4/S1pr3/Nlgn2/Jam2/Fbln5 | 60 |
| GO:0046873 | metal ion transmembrane transporter activity | MF | 0.138 | 1.05e-12 | 3.33e-10 | 9.48 | 2.56e-10 | Kcnk3/Kcnb2/Nalcn/Slc1a3/Kcnj3/Cacna1c/Grik1/Kcna6/Slc6a2/Kcnd2/Slc5a11/Slc13a5/Ryr1/Kcnd3/Slc12a8/Slc8a1/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Slc9a7/Slc24a4/Rasa3/Grin2b/Slc24a3/Cacna1e/Slc8a3/Trpc6/Htr1b/Nalf1/Slc6a15/Scn2a/Grik2/Kcnma1/Slc9a5/Slc23a2/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Slc12a4/Slc38a1/Pkd2/Ryr3/Cacna1h/Kcng4/Atp2b4/Scn3a/Atp1a2/Slc4a8/Psen1/Pkdrej/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 58 |
| GO:0030695 | GTPase regulator activity | MF | 0.135 | 3.73e-12 | 7.1e-10 | 9.15 | 5.46e-10 | Wnt11/Agap2/Ccdc88a/Nrp1/Rgs6/Rgs16/Rasgrp3/Garnl3/Lrrk2/Arhgap45/Rasa3/Arhgef25/Tbc1d24/Arhgap25/Plcb1/Dock2/Rasgrp2/Myo9a/Denn2b/Syde1/Thy1/Dock10/Sh3bp1/Arhgap6/Eps8l1/Arhgef40/Rgs5/Arhgef15/Dennd5a/Nckap1l/Arap3/Arrb1/Prex2/Arhgap10/Dlc1/Arhgap28/Net1/Fgd1/Gnb5/Arhgap31/Syngap1/Arfgap3/Ngef/Arhgef9/Fgd5/Arhgap15/Dennd4a/Sh2d3c/Iqsec3/Gpsm1/Fgd2/Sipa1/Tbc1d10c/Rasal3/Rabep1/Acap1/Plekhg5 | 57 |
| GO:0060589 | nucleoside-triphosphatase regulator activity | MF | 0.135 | 3.73e-12 | 7.1e-10 | 9.15 | 5.46e-10 | Wnt11/Agap2/Ccdc88a/Nrp1/Rgs6/Rgs16/Rasgrp3/Garnl3/Lrrk2/Arhgap45/Rasa3/Arhgef25/Tbc1d24/Arhgap25/Plcb1/Dock2/Rasgrp2/Myo9a/Denn2b/Syde1/Thy1/Dock10/Sh3bp1/Arhgap6/Eps8l1/Arhgef40/Rgs5/Arhgef15/Dennd5a/Nckap1l/Arap3/Arrb1/Prex2/Arhgap10/Dlc1/Arhgap28/Net1/Fgd1/Gnb5/Arhgap31/Syngap1/Arfgap3/Ngef/Arhgef9/Fgd5/Arhgap15/Dennd4a/Sh2d3c/Iqsec3/Gpsm1/Fgd2/Sipa1/Tbc1d10c/Rasal3/Rabep1/Acap1/Plekhg5 | 57 |
| GO:0005539 | glycosaminoglycan binding | MF | 0.167 | 1.53e-11 | 2.43e-09 | 8.61 | 1.87e-09 | Apob/Col11a1/Vit/Sema5a/Mdk/Thbs3/Nrp1/Cd44/Spock2/Fn1/Serpine2/Egflam/Adamts1/Col13a1/Mamdc2/Postn/Nell1/Smoc1/Apoe/Layn/Ndnf/Tnfaip6/Bgn/Rtn4rl1/Hapln3/Ptch1/Abi3bp/Habp4/Ccn2/Efemp2/Dpysl3/Fstl1/Ptn/Stab1/Bmp7/Vcan/Slit3/Ccdc80/Eng | 39 |
| GO:0005261 | cation channel activity | MF | 0.144 | 1.89e-11 | 2.57e-09 | 8.59 | 1.97e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Cachd1/Scn1a/Cacna2d1/Kcnq5/Slc24a4/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Unc80/Trpc6/Htr1b/Nalf1/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Pkd2/Ryr3/Cacna1h/Kcng4/Scn3a/Psen1/Piezo2/Pkdrej/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 48 |
| GO:0005216 | ion channel activity | MF | 0.128 | 5.11e-11 | 6.08e-09 | 8.22 | 4.68e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Cachd1/Scn1a/Cacna2d1/Kcnq5/Slc24a4/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Ttyh2/Trpc6/Htr1b/Nalf1/Glrb/Scn2a/Gabra4/Grik2/Kcnma1/Gjc1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Pkd2/Ryr3/Cacna1h/Kcng4/Scn3a/Clca2/Psen1/Piezo2/Pkdrej/Kcnq4/Cacna1a/Lrrc8c/Cacnb4/Kcnh1 | 56 |
| GO:0022836 | gated channel activity | MF | 0.142 | 7.49e-11 | 7.85e-09 | 8.1 | 6.04e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Cachd1/Scn1a/Cacna2d1/Kcnq5/Ano1/Rasa3/Grin2b/Cacna1e/Ano2/Ttyh2/Htr1b/Nalf1/Glrb/Scn2a/Gabra4/Grik2/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Pkd2/Ryr3/Cacna1h/Kcng4/Scn3a/Clca2/Piezo2/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 46 |
| GO:0005178 | integrin binding | MF | 0.194 | 8.67e-11 | 7.85e-09 | 8.1 | 6.04e-09 | Ptprz1/Itga4/Kdr/Igf1/Npnt/Fermt2/Sema7a/Col16a1/Tgfbi/Itgb7/Fn1/Icam1/Jam3/Cxcl12/Itga1/Thy1/Plpp3/Cd226/Itgal/Emilin1/Igf2/Itga7/Tln2/Ccn2/Ptn/Itga8/Itgb4/S1pr3/Jam2/Fbln5 | 30 |
| GO:0005096 | GTPase activator activity | MF | 0.161 | 9.08e-11 | 7.85e-09 | 8.1 | 6.04e-09 | Wnt11/Agap2/Nrp1/Rgs6/Rgs16/Rasgrp3/Garnl3/Lrrk2/Arhgap45/Rasa3/Tbc1d24/Arhgap25/Plcb1/Dock2/Myo9a/Syde1/Thy1/Sh3bp1/Arhgap6/Rgs5/Arhgef15/Nckap1l/Arap3/Arrb1/Prex2/Arhgap10/Dlc1/Arhgap28/Gnb5/Arhgap31/Syngap1/Arfgap3/Arhgap15/Sipa1/Tbc1d10c/Rasal3/Rabep1/Acap1 | 38 |
| GO:0015267 | channel activity | MF | 0.12 | 2.56e-10 | 1.88e-08 | 7.73 | 1.44e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Cachd1/Scn1a/Cacna2d1/Kcnq5/Slc24a4/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Ttyh2/Trpc6/Htr1b/Nalf1/Glrb/Scn2a/Gja4/Gabra4/Grik2/Kcnma1/Gjc1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Pkd2/Ryr3/Cacna1h/Aqp11/Kcng4/Scn3a/Clca2/Psen1/Piezo2/Pkdrej/Kcnq4/Cacna1a/Lrrc8c/Cacnb4/Kcnh1 | 58 |
| GO:0022803 | passive transmembrane transporter activity | MF | 0.12 | 2.56e-10 | 1.88e-08 | 7.73 | 1.44e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Cacna1c/Grik1/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Cachd1/Scn1a/Cacna2d1/Kcnq5/Slc24a4/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Ttyh2/Trpc6/Htr1b/Nalf1/Glrb/Scn2a/Gja4/Gabra4/Grik2/Kcnma1/Gjc1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Pkd2/Ryr3/Cacna1h/Aqp11/Kcng4/Scn3a/Clca2/Psen1/Piezo2/Pkdrej/Kcnq4/Cacna1a/Lrrc8c/Cacnb4/Kcnh1 | 58 |
| GO:0005244 | voltage-gated ion channel activity | MF | 0.167 | 1.12e-09 | 7.12e-08 | 7.15 | 5.48e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Cacna1c/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Ano1/Grin2b/Cacna1e/Htr1b/Scn2a/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Pkd2/Cacna1h/Kcng4/Scn3a/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 32 |
| GO:0005516 | calmodulin binding | MF | 0.167 | 1.12e-09 | 7.12e-08 | 7.15 | 5.48e-08 | Gap43/Map2/Cacna1c/Ryr1/Slc8a1/Nos3/Kcnq5/Myo5a/Slc24a4/Pde1b/Adcy3/Akap12/Plcb1/Slc8a3/Sntb1/Cth/Nrgn/Pde1a/Syt7/Cnn2/Myo1f/Cnn3/Ryr3/Rrad/Iqcb1/Atp2b4/Scn3a/Kcnq4/Cacna1a/Mknk2/Kcnh1/Camk1 | 32 |
| GO:0022832 | voltage-gated channel activity | MF | 0.166 | 1.28e-09 | 7.64e-08 | 7.12 | 5.88e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Cacna1c/Kcna6/Kcnd2/Ryr1/Kcnd3/Scn4b/Cachd1/Scn1a/Cacna2d1/Kcnq5/Ano1/Grin2b/Cacna1e/Htr1b/Scn2a/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Pkd2/Cacna1h/Kcng4/Scn3a/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 32 |
| GO:0003779 | actin binding | MF | 0.117 | 5.49e-09 | 3.07e-07 | 6.51 | 2.37e-07 | Lsp1/Myo16/Map2/Slc6a2/Evl/Epb41l3/Nos3/Ccdc88a/Hcls1/Fermt2/Aif1l/Fxyd5/Tmod2/Wasf3/Lrrk2/Myo5a/Pfn2/Mical1/Vash1/Myo9a/Trpc6/Msn/Coro1a/Sntb1/Palld/Limch1/Eps8l1/Cnn2/Kcnma1/Crocc/Daam2/Hip1/Scin/Parvg/Bin1/Myl3/Cap2/Parvb/Rai14/Flnc/Syne3/Tln2/Myo1f/Cnn3/Wipf1/Pknox2/Plekhh2/Actr3b/Shroom1/Vash2/Fmnl3/Pstpip1 | 52 |
| GO:0015085 | calcium ion transmembrane transporter activity | MF | 0.187 | 6.94e-09 | 3.67e-07 | 6.44 | 2.83e-07 | Cacna1c/Ryr1/Slc8a1/Cachd1/Cacna2d1/Slc24a4/Rasa3/Grin2b/Slc24a3/Cacna1e/Slc8a3/Trpc6/Htr1b/Nalf1/Pkd1/Cacng7/Ncs1/Pkd2/Ryr3/Cacna1h/Atp2b4/Psen1/Pkdrej/Cacna1a/Cacnb4 | 25 |
| GO:0022843 | voltage-gated cation channel activity | MF | 0.176 | 2.35e-08 | 1.18e-06 | 5.93 | 9.05e-07 | Kcnk3/Kcnb2/Kcnj3/Cacna1c/Kcna6/Kcnd2/Ryr1/Kcnd3/Cachd1/Cacna2d1/Kcnq5/Grin2b/Cacna1e/Htr1b/Kcnma1/Kcnc4/Cacng7/Ncs1/Pkd2/Cacna1h/Kcng4/Kcnq4/Cacna1a/Cacnb4/Kcnh1 | 25 |
| GO:0005262 | calcium channel activity | MF | 0.19 | 4.03e-08 | 1.92e-06 | 5.72 | 1.48e-06 | Cacna1c/Ryr1/Cachd1/Cacna2d1/Slc24a4/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpc6/Htr1b/Nalf1/Pkd1/Cacng7/Ncs1/Pkd2/Ryr3/Cacna1h/Psen1/Pkdrej/Cacna1a/Cacnb4 | 22 |
| GO:0015081 | sodium ion transmembrane transporter activity | MF | 0.166 | 1.5e-07 | 6.78e-06 | 5.17 | 5.22e-06 | Nalcn/Slc1a3/Grik1/Slc6a2/Slc5a11/Slc13a5/Slc8a1/Scn4b/Scn1a/Slc9a7/Slc24a4/Slc24a3/Slc8a3/Slc6a15/Scn2a/Grik2/Slc9a5/Slc23a2/Slc38a1/Pkd2/Cacna1h/Scn3a/Atp1a2/Slc4a8 | 24 |
| GO:0016757 | glycosyltransferase activity | MF | 0.127 | 3.69e-07 | 1.53e-05 | 4.82 | 1.18e-05 | B4galnt4/St6gal2/St8sia2/St8sia4/Gxylt2/Uggt2/Pygm/Tmtc1/B4galt2/B3galnt1/Fut10/Has2/Galnt16/St3gal2/Lfng/St6galnac4/Eogt/Ggta1/Lacc1/St8sia6/Galntl6/Poglut2/Mgat3/Pomt2/Large1/St6galnac6/B3gnt5/Xylt1/St3gal4/Mfng/Qtrt2/Pygl/Galnt18/Dpy19l1 | 34 |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | MF | 0.117 | 4.02e-07 | 1.59e-05 | 4.8 | 1.23e-05 | Lef1/Zeb2/Tbx2/Prdm5/Zfp37/Hoxd9/Osr2/Klf8/Zfp239/Tcf21/Glis1/Bcl6b/Zeb1/Prdm1/Zfp13/Zfp248/Tbx3/Heyl/Irf8/Zbtb46/Snai1/Satb1/Peg3/Zfp382/Klf12/Foxs1/Nfatc4/Zbtb20/Kcnip3/Zbtb8a/Nr2f1/Zfp512b/Cux2/Ets2/Zfp566/Zfp90/Zfp763/Hey2/Myc | 39 |
| GO:0001217 | DNA-binding transcription repressor activity | MF | 0.116 | 5.06e-07 | 1.93e-05 | 4.72 | 1.48e-05 | Lef1/Zeb2/Tbx2/Prdm5/Zfp37/Hoxd9/Osr2/Klf8/Zfp239/Tcf21/Glis1/Bcl6b/Zeb1/Prdm1/Zfp13/Zfp248/Tbx3/Heyl/Irf8/Zbtb46/Snai1/Satb1/Peg3/Zfp382/Klf12/Foxs1/Nfatc4/Zbtb20/Kcnip3/Zbtb8a/Nr2f1/Zfp512b/Cux2/Ets2/Zfp566/Zfp90/Zfp763/Hey2/Myc | 39 |
| GO:0035591 | signaling adaptor activity | MF | 0.214 | 1.13e-06 | 4.13e-05 | 4.38 | 3.18e-05 | Lrrk2/Shank2/Mapk8ip1/Akap12/Sh2b2/Abi2/Magi2/Sarm1/Afap1l2/Gnb5/Gnb4/Lat/Sh2d3c/Grb10/Irs1 | 15 |
| GO:0008237 | metallopeptidase activity | MF | 0.141 | 1.24e-06 | 4.2e-05 | 4.38 | 3.23e-05 | Cpxm2/Mmp16/Adamts20/Adamts16/Adamts19/Mmp2/Enpep/Adam33/Adamts1/Vash1/Naalad2/Cpe/Adamts10/Mme/Adamts7/Adam11/Agbl3/Mmp19/Trhde/Ece2/Adam19/Clca2/Pappa/Adamts17/Vash2/Lvrn | 26 |
| GO:0030020 | extracellular matrix structural constituent conferring tensile strength | MF | 0.289 | 1.28e-06 | 4.2e-05 | 4.38 | 3.23e-05 | Col11a1/Col14a1/Col4a6/Col16a1/Col9a2/Col13a1/Col5a2/Col15a1/Col23a1/Col18a1/Col4a5 | 11 |
| GO:0030159 | signaling receptor complex adaptor activity | MF | 0.289 | 1.28e-06 | 4.2e-05 | 4.38 | 3.23e-05 | Lrrk2/Shank2/Akap12/Sh2b2/Magi2/Gnb5/Gnb4/Lat/Sh2d3c/Grb10/Irs1 | 11 |
| GO:0019838 | growth factor binding | MF | 0.15 | 1.55e-06 | 4.9e-05 | 4.31 | 3.77e-05 | Epha8/Ptprz1/Cd109/Kdr/Nrp1/Il11ra1/Tek/Lrp2/Flt3/Epha3/Tsku/Ltbp3/Epha7/Nrros/Ccn2/Pdgfrb/Ptn/Il2rg/Igfbp4/Itgb4/Nkd2/Nradd/Eng | 23 |
| GO:0005245 | voltage-gated calcium channel activity | MF | 0.255 | 1.86e-06 | 5.67e-05 | 4.25 | 4.37e-05 | Cacna1c/Ryr1/Cachd1/Cacna2d1/Cacna1e/Htr1b/Cacng7/Ncs1/Pkd2/Cacna1h/Cacna1a/Cacnb4 | 12 |
| GO:0017124 | SH3 domain binding | MF | 0.158 | 1.94e-06 | 5.67e-05 | 4.25 | 4.37e-05 | Evl/Hcls1/Drd4/Abi3/Mical1/Shank2/Lrp2/Abi2/Sh3bp1/Arhgap6/Afap1l1/Prrt2/Efs/Wipf1/Afap1l2/Adam19/Arhgap31/Dpysl3/Syngap1/Elmo2/Pdcd6ip | 21 |
| GO:0005080 | protein kinase C binding | MF | 0.219 | 1.97e-06 | 5.67e-05 | 4.25 | 4.37e-05 | Fez1/Ccdc88a/Nell1/Adcy4/Pard6g/Hdac9/Cavin3/BC034090/Cavin2/Akt3/Pld2/Dact1/Plek/Irs1 | 14 |
| GO:0015079 | potassium ion transmembrane transporter activity | MF | 0.146 | 2.43e-06 | 6.81e-05 | 4.17 | 5.24e-05 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcna6/Kcnd2/Kcnd3/Slc12a8/Kcnq5/Slc9a7/Slc24a4/Slc24a3/Grik2/Kcnma1/Slc9a5/Kcnc4/Kcnip3/Slc12a4/Pkd2/Kcng4/Atp1a2/Kcnq4/Kcnh1 | 23 |
| GO:0008201 | heparin binding | MF | 0.141 | 2.89e-06 | 7.85e-05 | 4.1 | 6.04e-05 | Apob/Col11a1/Mdk/Thbs3/Nrp1/Fn1/Serpine2/Adamts1/Col13a1/Postn/Nell1/Smoc1/Apoe/Ndnf/Rtn4rl1/Ptch1/Abi3bp/Ccn2/Efemp2/Fstl1/Ptn/Bmp7/Slit3/Ccdc80 | 24 |
| GO:0008373 | sialyltransferase activity | MF | 0.381 | 3.63e-06 | 9.6e-05 | 4.02 | 7.39e-05 | St6gal2/St8sia2/St8sia4/St3gal2/St6galnac4/St8sia6/St6galnac6/St3gal4 | 8 |
| GO:0004713 | protein tyrosine kinase activity | MF | 0.156 | 3.99e-06 | 0.000103 | 3.99 | 7.9e-05 | Epha8/Kdr/Ddr2/Pkdcc/Nrp1/Itk/Tek/Flt3/Kit/Epha3/Tie1/Hipk2/Epha7/Fyn/Fgr/Zap70/Styk1/Pdgfrb/Hipk1/Ror1 | 20 |
| GO:0045296 | cadherin binding | MF | 0.203 | 5.06e-06 | 0.000127 | 3.9 | 9.75e-05 | Kdr/Nos3/Ptpro/Fxyd5/Cdh2/Cdh6/Cdh13/Cdh10/Ptprm/Cdh5/Cdh11/Neo1/Dchs1/Psen1 | 14 |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | MF | 0.207 | 1.93e-05 | 0.00047 | 3.33 | 0.000362 | Epha8/Kdr/Ddr2/Nrp1/Tek/Flt3/Kit/Epha3/Tie1/Epha7/Pdgfrb/Ror1 | 12 |
| GO:0044325 | transmembrane transporter binding | MF | 0.136 | 3.26e-05 | 0.000775 | 3.11 | 0.000597 | Kcnb2/Cacna1c/Kcnd3/Tcaf1/Slc8a1/Rims3/Scn4b/Lrrk2/Ank2/Cdh5/Pkd1/Arrb1/Kcnip3/Mpp2/Pkd2/Fhl1/Fyn/Kcng4/Cav1/Kcnh1 | 20 |
| GO:0001664 | G protein-coupled receptor binding | MF | 0.103 | 4.59e-05 | 0.00106 | 2.97 | 0.000819 | Cxcl15/Ndp/Wnt6/Wnt11/Ptch2/Gprasp2/Tub/Gnai1/Ednrb/Grin2b/Cxcl12/Cnrip1/Edn3/Calca/Wnt9a/Znrf3/Reep1/Agtr1a/Magi2/Bdkrb2/Xcl1/Arrb1/Arrb2/Ptch1/Fyn/Gnaz/Dlg4/Palm/Apln/Bbs1/Nes/Itgb4/Pdcd6ip | 33 |
| GO:0004222 | metalloendopeptidase activity | MF | 0.151 | 5.52e-05 | 0.00125 | 2.9 | 0.000961 | Mmp16/Adamts20/Adamts16/Adamts19/Mmp2/Adam33/Adamts1/Adamts10/Mme/Adamts7/Adam11/Mmp19/Ece2/Adam19/Pappa/Adamts17 | 16 |
| GO:0098879 | structural constituent of postsynaptic specialization | MF | 0.5 | 5.64e-05 | 0.00125 | 2.9 | 0.000961 | Shank2/Dlg2/Magi2/Mpp2/Dlg4 | 5 |
| GO:0008509 | anion transmembrane transporter activity | MF | 0.102 | 7e-05 | 0.00151 | 2.82 | 0.00117 | Slc1a3/Grik1/Slc6a2/Slc13a5/Slc2a3/Slc12a8/Slc9a7/Slc24a4/Ano1/Slc7a2/Abcg2/Slc36a4/Slc24a3/Ano2/Ttyh2/Glrb/Slc29a1/Gabra4/Slc9a5/Slc23a2/Sfxn5/Slc12a4/Slc38a1/Slco4c1/Slc4a3/Slc25a23/Clca2/Slc4a8/Abcc5/Abcg3/Lrrc8c/Ank | 32 |
| GO:0019199 | transmembrane receptor protein kinase activity | MF | 0.171 | 7.29e-05 | 0.00154 | 2.81 | 0.00119 | Epha8/Kdr/Ddr2/Nrp1/Tek/Flt3/Kit/Epha3/Tie1/Epha7/Pdgfrb/Ror1/Eng | 13 |
| GO:0005540 | hyaluronic acid binding | MF | 0.292 | 0.000108 | 0.00219 | 2.66 | 0.00168 | Cd44/Layn/Tnfaip6/Hapln3/Habp4/Stab1/Vcan | 7 |
| GO:0005248 | voltage-gated sodium channel activity | MF | 0.28 | 0.000144 | 0.00279 | 2.55 | 0.00215 | Nalcn/Scn4b/Scn1a/Scn2a/Pkd2/Cacna1h/Scn3a | 7 |
| GO:0016849 | phosphorus-oxygen lyase activity | MF | 0.28 | 0.000144 | 0.00279 | 2.55 | 0.00215 | Gucy1a2/Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7/Cd38 | 7 |
| GO:0004016 | adenylate cyclase activity | MF | 0.417 | 0.000163 | 0.00311 | 2.51 | 0.00239 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0005525 | GTP binding | MF | 0.0947 | 0.000188 | 0.00351 | 2.45 | 0.0027 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Arl6/Gnai1/Lrrk2/Gucy1b1/Gimap8/Dnajc27/Rab34/Rragd/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Rhobtb3/Nin/Arl3/Septin3/Rasd2/Rrad/Gnaz/Rab23/Septin4/Gimap5/Gimap3/Rab39b/Rab27a/Septin6/Suclg2/Rap2a/Rab20 | 34 |
| GO:0003774 | cytoskeletal motor activity | MF | 0.142 | 0.000196 | 0.00357 | 2.45 | 0.00275 | Myo16/Kif7/Myo5a/Dnah5/Myo9a/Kif1a/Dync2h1/Kif6/Myl3/Myo1f/Dnah7b/Stard9/Dnah1/Kif16b/Kif5a | 15 |
| GO:0016758 | hexosyltransferase activity | MF | 0.114 | 0.000199 | 0.00357 | 2.45 | 0.00275 | B4galnt4/Uggt2/Pygm/Tmtc1/B4galt2/B3galnt1/Fut10/Has2/Galnt16/Lfng/Eogt/Ggta1/Galntl6/Poglut2/Mgat3/Pomt2/Large1/B3gnt5/Mfng/Pygl/Galnt18/Dpy19l1 | 22 |
| GO:0005272 | sodium channel activity | MF | 0.205 | 0.000231 | 0.00389 | 2.41 | 0.00299 | Nalcn/Grik1/Scn4b/Scn1a/Scn2a/Grik2/Pkd2/Cacna1h/Scn3a | 9 |
| GO:0023026 | MHC class II protein complex binding | MF | 0.385 | 0.000255 | 0.00404 | 2.39 | 0.00311 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa/Cd4 | 5 |
| GO:0099511 | voltage-gated calcium channel activity involved in regulation of cytosolic calcium levels | MF | 0.385 | 0.000255 | 0.00404 | 2.39 | 0.00311 | Cacna1e/Htr1b/Ncs1/Cacna1a/Cacnb4 | 5 |
| GO:0099626 | voltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels | MF | 0.385 | 0.000255 | 0.00404 | 2.39 | 0.00311 | Cacna1e/Htr1b/Ncs1/Cacna1a/Cacnb4 | 5 |
| GO:0005267 | potassium channel activity | MF | 0.132 | 0.00027 | 0.00422 | 2.38 | 0.00324 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcna6/Kcnd2/Kcnd3/Kcnq5/Grik2/Kcnma1/Kcnc4/Kcnip3/Pkd2/Kcng4/Kcnq4/Kcnh1 | 16 |
| GO:0019001 | guanyl nucleotide binding | MF | 0.0916 | 0.000287 | 0.00433 | 2.36 | 0.00333 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Arl6/Gnai1/Lrrk2/Gucy1b1/Gimap8/Dnajc27/Rab34/Rragd/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Rhobtb3/Nin/Arl3/Septin3/Rasd2/Pde5a/Rrad/Gnaz/Rab23/Septin4/Gimap5/Gimap3/Rab39b/Rab27a/Septin6/Suclg2/Rap2a/Rab20 | 35 |
| GO:0032561 | guanyl ribonucleotide binding | MF | 0.0916 | 0.000287 | 0.00433 | 2.36 | 0.00333 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Arl6/Gnai1/Lrrk2/Gucy1b1/Gimap8/Dnajc27/Rab34/Rragd/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Rhobtb3/Nin/Arl3/Septin3/Rasd2/Pde5a/Rrad/Gnaz/Rab23/Septin4/Gimap5/Gimap3/Rab39b/Rab27a/Septin6/Suclg2/Rap2a/Rab20 | 35 |
| GO:0140359 | ABC-type transporter activity | MF | 0.25 | 0.000311 | 0.00463 | 2.33 | 0.00356 | Abcg2/Abcg1/Abca8b/Abcb1a/Abcc5/Abcg3/Abcg4 | 7 |
| GO:0005249 | voltage-gated potassium channel activity | MF | 0.148 | 0.000334 | 0.00481 | 2.32 | 0.0037 | Kcnk3/Kcnb2/Kcnj3/Kcna6/Kcnd2/Kcnd3/Kcnq5/Kcnma1/Kcnc4/Pkd2/Kcng4/Kcnq4/Kcnh1 | 13 |
| GO:0008194 | UDP-glycosyltransferase activity | MF | 0.121 | 0.000361 | 0.00512 | 2.29 | 0.00394 | B4galnt4/Gxylt2/Uggt2/B4galt2/B3galnt1/Has2/Galnt16/Lfng/Eogt/Ggta1/Galntl6/Poglut2/Mgat3/Large1/B3gnt5/Xylt1/Mfng/Galnt18 | 18 |
| GO:0005251 | delayed rectifier potassium channel activity | MF | 0.233 | 0.000491 | 0.00678 | 2.17 | 0.00522 | Kcnb2/Kcna6/Kcnq5/Kcnc4/Kcng4/Kcnq4/Kcnh1 | 7 |
| GO:0030215 | semaphorin receptor binding | MF | 0.273 | 0.000508 | 0.0069 | 2.16 | 0.00531 | Sema5a/Sema5b/Sema7a/Sh3bp1/Sema3g/Sema6a | 6 |
| GO:0042043 | neurexin family protein binding | MF | 0.333 | 0.000547 | 0.00734 | 2.13 | 0.00565 | Nlgn1/Cpe/Nlgn3/Dlg4/Nlgn2 | 5 |
| GO:0005543 | phospholipid binding | MF | 0.085 | 0.000601 | 0.00794 | 2.1 | 0.00611 | Apob/Gap43/Nme4/Pla2g10/Nr5a2/Mark1/Syt13/Ccdc88a/Cpne5/Fermt2/P2rx2/Dab1/Pfn2/Cibar1/Lpar4/Rps6kc1/Plcb1/Thy1/Pltp/Mme/Cadps/Nrgn/Apoe/Syt7/Hip1/Scin/Dysf/Bin1/Arap3/Epdr1/Pard3b/Plcb2/Zfyve9/Cavin2/Pld2/Kif16b/Golph3l/Fgd2/Kcnh1 | 39 |
| GO:0003924 | GTPase activity | MF | 0.0952 | 0.000609 | 0.00794 | 2.1 | 0.00611 | Rragb/Agap2/Rasl12/Rab36/Arl6/Gnai1/Lrrk2/Dnajc27/Rab34/Rragd/Cracr2a/Tubb4a/Rhobtb3/Gng2/Arl3/Septin3/Rasd2/Rrad/Gnaz/Rab23/Septin4/Gnb4/Rab39b/Rab27a/Tppp/Septin6/Rap2a/Rab20 | 28 |
| GO:0008235 | metalloexopeptidase activity | MF | 0.164 | 0.000689 | 0.00887 | 2.05 | 0.00682 | Cpxm2/Mmp16/Enpep/Vash1/Naalad2/Cpe/Agbl3/Trhde/Vash2/Lvrn | 10 |
| GO:0015631 | tubulin binding | MF | 0.0881 | 0.000702 | 0.00891 | 2.05 | 0.00686 | Gli1/Map2/Fez1/Capn6/Ccdc88a/Kif7/Lrrk2/Kif26a/Ndn/Kif1a/Reep1/Eml1/Map9/Rp1/Katnal1/Kif6/Dysf/Dclk2/Nin/Arl3/Tbcel/Fyn/Cnn3/Unc5c/Stard9/Brsk1/Mapre2/Tppp/Vash2/Ift81/Kif16b/Map7d3/Rmdn2/Kif5a | 34 |
| GO:0043548 | phosphatidylinositol 3-kinase binding | MF | 0.195 | 0.000714 | 0.00894 | 2.05 | 0.00688 | Fam83a/Dab1/Flt3/Coro1a/Rasd2/Fyn/Pdgfrb/Irs1 | 8 |
| GO:0015248 | sterol transporter activity | MF | 0.219 | 0.000746 | 0.00922 | 2.04 | 0.0071 | Apob/Osbp2/Pltp/Apoe/Abcg1/Osbpl1a/Abcg4 | 7 |
| GO:0015491 | cation:cation antiporter activity | MF | 0.25 | 0.000842 | 0.0101 | 1.99 | 0.00781 | Slc8a1/Slc9a7/Slc24a4/Slc24a3/Slc8a3/Slc9a5 | 6 |
| GO:0031681 | G-protein beta-subunit binding | MF | 0.25 | 0.000842 | 0.0101 | 1.99 | 0.00781 | Gngt2/Gng11/Gng2/Rasd2/Gng7/F2r | 6 |
| GO:0015368 | calcium:cation antiporter activity | MF | 0.4 | 0.000936 | 0.0109 | 1.96 | 0.0084 | Slc8a1/Slc24a4/Slc24a3/Slc8a3 | 4 |
| GO:0035374 | chondroitin sulfate binding | MF | 0.4 | 0.000936 | 0.0109 | 1.96 | 0.0084 | Mdk/Rtn4rl1/Dpysl3/Ptn | 4 |
| GO:0050660 | flavin adenine dinucleotide binding | MF | 0.14 | 0.00094 | 0.0109 | 1.96 | 0.0084 | Gulo/Maob/Nos3/Mical1/Maoa/Fmo1/Aifm2/Dpyd/Steap4/Sardh/Prodh/Aox4 | 12 |
| GO:0019842 | vitamin binding | MF | 0.118 | 0.000998 | 0.0114 | 1.94 | 0.00881 | Stra6/P3h3/Abat/P4htm/Cbs/Pygm/Rbp7/Sptlc3/Kyat1/Hdc/Dhtkd1/Cth/P3h1/Sardh/S100g/Pygl | 16 |
| GO:0003777 | microtubule motor activity | MF | 0.156 | 0.00101 | 0.0115 | 1.94 | 0.00885 | Kif7/Dnah5/Kif1a/Dync2h1/Kif6/Dnah7b/Stard9/Dnah1/Kif16b/Kif5a | 10 |
| GO:0008559 | ABC-type xenobiotic transporter activity | MF | 0.294 | 0.00104 | 0.0116 | 1.93 | 0.00896 | Abcg2/Abca8b/Abcb1a/Abcc5/Abcg3 | 5 |
| GO:0022804 | active transmembrane transporter activity | MF | 0.0859 | 0.00109 | 0.0118 | 1.93 | 0.00912 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Abcg2/Slc36a4/Slc24a3/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Atp6v0e2/Abca5/Abcg1/Slc12a4/Slc38a1/Abca8b/Slco4c1/Slc4a3/Atp2b4/Cox4i2/Abcd2/Atp1a2/Slc4a8/Abcb1a/Abcc5/Abcg3/Ank/Abcg4 | 34 |
| GO:0098632 | cell-cell adhesion mediator activity | MF | 0.206 | 0.0011 | 0.0118 | 1.93 | 0.00912 | Prtg/Lrrc4c/Jam3/Esam/Nfasc/Cd200/Ntng1 | 7 |
| GO:0004181 | metallocarboxypeptidase activity | MF | 0.231 | 0.00132 | 0.0142 | 1.85 | 0.0109 | Cpxm2/Vash1/Naalad2/Cpe/Agbl3/Vash2 | 6 |
| GO:0004970 | ionotropic glutamate receptor activity | MF | 0.278 | 0.00138 | 0.0146 | 1.84 | 0.0112 | Grid1/Gria4/Grik1/Grin2b/Grik2 | 5 |
| GO:0008331 | high voltage-gated calcium channel activity | MF | 0.364 | 0.00141 | 0.0146 | 1.83 | 0.0113 | Cacna1c/Cacna1e/Cacna1a/Cacnb4 | 4 |
| GO:0042285 | xylosyltransferase activity | MF | 0.364 | 0.00141 | 0.0146 | 1.83 | 0.0113 | Gxylt2/Poglut2/Large1/Xylt1 | 4 |
| GO:0030674 | protein-macromolecule adaptor activity | MF | 0.089 | 0.00144 | 0.0147 | 1.83 | 0.0113 | Aph1b/Epb41l3/Stx1b/Lrrk2/Ap1m1/Klhdc2/Shank2/Mapk8ip1/Akap12/Cntln/Ank2/Sh2b2/Abi2/Spsb4/Magi2/Hip1/Arrb1/Syne3/Sarm1/Frmd4a/Afap1l2/Gnb5/Stx2/Gnb4/Lat/Cav1/Sh2d3c/Grb10/Irs1 | 29 |
| GO:0022853 | active ion transmembrane transporter activity | MF | 0.0966 | 0.00148 | 0.0149 | 1.83 | 0.0115 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Slc24a3/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Atp6v0e2/Slc12a4/Slc38a1/Slco4c1/Slc4a3/Atp2b4/Cox4i2/Atp1a2/Slc4a8 | 23 |
| GO:0042169 | SH2 domain binding | MF | 0.174 | 0.00157 | 0.0156 | 1.81 | 0.012 | Ccdc88a/Dab1/Sh2b2/Kit/Dlc1/Afap1l2/Lax1/Irs1 | 8 |
| GO:0070888 | E-box binding | MF | 0.174 | 0.00157 | 0.0156 | 1.81 | 0.012 | Tcf21/Zeb1/Tcf4/Snai1/Tal1/Hand2/Myc/Twist1 | 8 |
| GO:0019955 | cytokine binding | MF | 0.112 | 0.0017 | 0.0167 | 1.78 | 0.0128 | Itga4/Cd109/Nog/Cd44/Il11ra1/Cxcr3/Kit/Il27ra/Tsku/Ltbp3/Nrros/Xcr1/Il12rb1/Il2rg/Cd4/Eng | 16 |
| GO:0008017 | microtubule binding | MF | 0.0923 | 0.0018 | 0.0175 | 1.76 | 0.0135 | Gli1/Map2/Capn6/Ccdc88a/Kif7/Kif26a/Kif1a/Reep1/Eml1/Map9/Rp1/Katnal1/Kif6/Dysf/Dclk2/Nin/Arl3/Cnn3/Stard9/Mapre2/Vash2/Kif16b/Map7d3/Rmdn2/Kif5a | 25 |
| GO:0005158 | insulin receptor binding | MF | 0.214 | 0.00199 | 0.0192 | 1.72 | 0.0148 | Igf1/Ccdc88a/Enpp1/Igf2/Grb10/Irs1 | 6 |
| GO:0031402 | sodium ion binding | MF | 0.333 | 0.00204 | 0.0194 | 1.71 | 0.0149 | Scn1a/Scn2a/Scn3a/Atp1a2 | 4 |
| GO:0030165 | PDZ domain binding | MF | 0.117 | 0.00218 | 0.0205 | 1.69 | 0.0158 | Mpp3/Nlgn1/Nkd1/Lrp2/Sntb1/Dlg2/Grik2/Lin7a/Mpp2/Erc2/Dlg4/Fzd2/Atp2b4/Psen1 | 14 |
| GO:0099186 | structural constituent of postsynapse | MF | 0.25 | 0.0023 | 0.0215 | 1.67 | 0.0166 | Shank2/Dlg2/Magi2/Mpp2/Dlg4 | 5 |
| GO:0015298 | solute:cation antiporter activity | MF | 0.207 | 0.00241 | 0.022 | 1.66 | 0.0169 | Slc8a1/Slc9a7/Slc24a4/Slc24a3/Slc8a3/Slc9a5 | 6 |
| GO:0045499 | chemorepellent activity | MF | 0.207 | 0.00241 | 0.022 | 1.66 | 0.0169 | Sema5a/Sema5b/Sema7a/Sema3g/Epha7/Sema6a | 6 |
| GO:0008083 | growth factor activity | MF | 0.108 | 0.00242 | 0.022 | 1.66 | 0.0169 | Hgf/Mdk/Igf1/Areg/Cxcl12/Gdf11/Gdf7/Jag2/Gdf10/Vegfc/Igf2/Kitl/Ccn2/Ptn/Bmp7/Rabep1 | 16 |
| GO:0008238 | exopeptidase activity | MF | 0.125 | 0.00246 | 0.0221 | 1.66 | 0.017 | Cpxm2/Mmp16/Enpep/Vash1/Naalad2/Cpe/Mme/Agbl3/Trhde/Mindy1/Vash2/Lvrn | 12 |
| GO:0015276 | ligand-gated ion channel activity | MF | 0.111 | 0.0025 | 0.0222 | 1.65 | 0.0171 | Kcnj3/Grid1/Gria4/Grik1/Ryr1/P2rx2/Rasa3/Grin2b/Glrb/Gabra4/Grik2/Kcnma1/Pkd2/Ryr3/Clca2 | 15 |
| GO:0004896 | cytokine receptor activity | MF | 0.124 | 0.00269 | 0.0237 | 1.63 | 0.0182 | Cd44/Il11ra1/Cxcr3/Flt3/Il27ra/Xcr1/Csf2rb/Il17rd/Il12rb1/Il2rg/Il7r/Cd4 | 12 |
| GO:0051219 | phosphoprotein binding | MF | 0.118 | 0.00275 | 0.024 | 1.62 | 0.0185 | Cd44/Shc3/Dpysl4/She/Arrb1/Pkd2/Fgr/Dpysl3/Zap70/Bbs1/Sh2d3c/Grb10/Irs1 | 13 |
| GO:0042578 | phosphoric ester hydrolase activity | MF | 0.084 | 0.00285 | 0.0247 | 1.61 | 0.019 | Ptprz1/Pald1/Ptprd/Ptpro/Lrrk2/Ptp4a3/Inpp4b/Pde1b/Plcb1/Ptpdc1/Ptprm/Plpp3/Lhpp/Ppm1f/Enpp1/Ppef2/Pde1a/Hddc3/Bpnt1/Ppm1e/Plcb2/Pde5a/Plpp1/Nt5c3b/Smpd1/Pde8a/Pld2/Enpp2/Ptpn7/Plppr2 | 30 |
| GO:0022834 | ligand-gated channel activity | MF | 0.109 | 0.00288 | 0.0247 | 1.61 | 0.019 | Kcnj3/Grid1/Gria4/Grik1/Ryr1/P2rx2/Rasa3/Grin2b/Glrb/Gabra4/Grik2/Kcnma1/Pkd2/Ryr3/Clca2 | 15 |
| GO:0016836 | hydro-lyase activity | MF | 0.143 | 0.00336 | 0.0283 | 1.55 | 0.0218 | Cyp1b1/Car4/Car8/Cbs/L3hypdh/Echs1/Naxd/Hacd1/Echdc2 | 9 |
| GO:0004180 | carboxypeptidase activity | MF | 0.171 | 0.0034 | 0.0284 | 1.55 | 0.0219 | Cpxm2/Vash1/Naalad2/Cpe/Agbl3/Mindy1/Vash2 | 7 |
| GO:0030170 | pyridoxal phosphate binding | MF | 0.154 | 0.0035 | 0.029 | 1.54 | 0.0223 | Abat/Cbs/Pygm/Sptlc3/Kyat1/Hdc/Cth/Pygl | 8 |
| GO:0032794 | GTPase activating protein binding | MF | 0.227 | 0.00361 | 0.0296 | 1.53 | 0.0228 | Plxnb1/Gnai1/Arl3/Gnb5/Fmnl3 | 5 |
| GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | MF | 0.286 | 0.00381 | 0.0305 | 1.52 | 0.0235 | Fads2/Fads1/Fads3/Degs2 | 4 |
| GO:0030021 | extracellular matrix structural constituent conferring compression resistance | MF | 0.286 | 0.00381 | 0.0305 | 1.52 | 0.0235 | Spock2/Bgn/Lum/Vcan | 4 |
| GO:0047555 | 3’,5’-cyclic-GMP phosphodiesterase activity | MF | 0.286 | 0.00381 | 0.0305 | 1.52 | 0.0235 | Pde1b/Pde1a/Pde5a/Pde8a | 4 |
| GO:0070279 | vitamin B6 binding | MF | 0.151 | 0.00395 | 0.0313 | 1.5 | 0.0241 | Abat/Cbs/Pygm/Sptlc3/Kyat1/Hdc/Cth/Pygl | 8 |
| GO:0042910 | xenobiotic transmembrane transporter activity | MF | 0.188 | 0.00406 | 0.0318 | 1.5 | 0.0245 | Abcg2/Abca8b/Abcb1a/Abcc5/Abcg3/Slc43a3 | 6 |
| GO:0042626 | ATPase-coupled transmembrane transporter activity | MF | 0.118 | 0.00408 | 0.0318 | 1.5 | 0.0245 | Abcg2/Atp6v0e2/Abca5/Abcg1/Abca8b/Atp2b4/Abcd2/Atp1a2/Abcb1a/Abcc5/Abcg3/Abcg4 | 12 |
| GO:0030276 | clathrin binding | MF | 0.138 | 0.00417 | 0.0322 | 1.49 | 0.0248 | Nsg2/Syt13/Lrrk2/Ap1m1/Trpc6/Syt7/Hip1/Arrb1/Nsg1 | 9 |
| GO:0023023 | MHC protein complex binding | MF | 0.217 | 0.00443 | 0.0335 | 1.48 | 0.0258 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa/Cd4 | 5 |
| GO:0050431 | transforming growth factor beta binding | MF | 0.217 | 0.00443 | 0.0335 | 1.48 | 0.0258 | Cd109/Tsku/Ltbp3/Nrros/Eng | 5 |
| GO:0051020 | GTPase binding | MF | 0.0841 | 0.00443 | 0.0335 | 1.48 | 0.0258 | Rragb/Rims3/Rasgrp3/Gga2/Lrrk2/Myo5a/Mical1/Ranbp17/Dock2/Dock10/Rab34/Abi2/Rragd/Dmxl2/Rasip1/Rhobtb3/Daam2/Dennd5a/Bin1/Adcyap1r1/Net1/Fgd1/Cdc42ep5/Cav1/Cyfip2/Kif16b/Fmnl3 | 27 |
| GO:0051018 | protein kinase A binding | MF | 0.145 | 0.00498 | 0.0361 | 1.44 | 0.0278 | Wasf3/Lrrk2/Akap12/Pkia/Akap17b/Akap11/Dact1/Prkar1b | 8 |
| GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential | MF | 0.145 | 0.00498 | 0.0361 | 1.44 | 0.0278 | Grid1/Gria4/Grik1/Grin2b/Glrb/Gabra4/Grik2/Gabbr1 | 8 |
| GO:0005229 | intracellular calcium activated chloride channel activity | MF | 0.267 | 0.005 | 0.0361 | 1.44 | 0.0278 | Ano1/Ano2/Ttyh2/Clca2 | 4 |
| GO:0015278 | calcium-release channel activity | MF | 0.267 | 0.005 | 0.0361 | 1.44 | 0.0278 | Ryr1/Rasa3/Pkd2/Ryr3 | 4 |
| GO:0022821 | potassium ion antiporter activity | MF | 0.267 | 0.005 | 0.0361 | 1.44 | 0.0278 | Slc9a7/Slc24a4/Slc24a3/Slc9a5 | 4 |
| GO:0061778 | intracellular chloride channel activity | MF | 0.267 | 0.005 | 0.0361 | 1.44 | 0.0278 | Ano1/Ano2/Ttyh2/Clca2 | 4 |
| GO:0008528 | G protein-coupled peptide receptor activity | MF | 0.103 | 0.00525 | 0.0376 | 1.42 | 0.0289 | Vipr2/Npr3/Ednrb/Cxcr3/Pth1r/Aplnr/Agtr1a/Ednra/Ogfrl1/Bdkrb2/Adcyap1r1/Xcr1/Calcrl/Prokr1/F2r | 15 |
| GO:0031690 | adrenergic receptor binding | MF | 0.208 | 0.00537 | 0.0376 | 1.42 | 0.0289 | Magi2/Bdkrb2/Arrb1/Arrb2/Dlg4 | 5 |
| GO:0099604 | ligand-gated calcium channel activity | MF | 0.208 | 0.00537 | 0.0376 | 1.42 | 0.0289 | Ryr1/Rasa3/Grin2b/Pkd2/Ryr3 | 5 |
| GO:0098960 | postsynaptic neurotransmitter receptor activity | MF | 0.13 | 0.00621 | 0.0432 | 1.36 | 0.0332 | Grid1/Gria4/Grik1/Drd4/Grin2b/Glrb/Gabra4/Grik2/Gabbr1 | 9 |
| GO:0005159 | insulin-like growth factor receptor binding | MF | 0.25 | 0.00641 | 0.0436 | 1.36 | 0.0335 | Igf1/Igf2/Arrb1/Irs1 | 4 |
| GO:0005172 | vascular endothelial growth factor receptor binding | MF | 0.25 | 0.00641 | 0.0436 | 1.36 | 0.0335 | Angpt1/Ccdc88a/Cdh5/Vegfc | 4 |
| GO:0015562 | efflux transmembrane transporter activity | MF | 0.25 | 0.00641 | 0.0436 | 1.36 | 0.0335 | Abcg2/Abcb1a/Abcc5/Abcg3 | 4 |
| GO:0050780 | dopamine receptor binding | MF | 0.2 | 0.00645 | 0.0436 | 1.36 | 0.0335 | Grin2b/Agtr1a/Arrb2/Dlg4/Palm | 5 |
| GO:0015294 | solute:cation symporter activity | MF | 0.116 | 0.00657 | 0.0437 | 1.36 | 0.0337 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc2a13/Slc6a15/Slc23a2/Slc12a4/Slc38a1/Slc4a8 | 11 |
| GO:0048306 | calcium-dependent protein binding | MF | 0.116 | 0.00657 | 0.0437 | 1.36 | 0.0337 | Syn2/Myo5a/Slc24a4/Mgp/Masp1/Kcnip3/S100pbp/Atp2b4/Stx2/Pdcd6ip/S100g | 11 |
| GO:0016597 | amino acid binding | MF | 0.129 | 0.00683 | 0.0451 | 1.35 | 0.0347 | Slc1a3/Grik1/Ddah1/Nos3/Grin2b/Hdc/Glrb/Ubr1/Prodh | 9 |
| GO:0031267 | small GTPase binding | MF | 0.0842 | 0.00691 | 0.0453 | 1.34 | 0.0348 | Rims3/Rasgrp3/Gga2/Lrrk2/Myo5a/Mical1/Ranbp17/Dock2/Dock10/Rab34/Abi2/Dmxl2/Rasip1/Rhobtb3/Daam2/Dennd5a/Adcyap1r1/Net1/Fgd1/Cdc42ep5/Cav1/Cyfip2/Kif16b/Fmnl3 | 24 |
| GO:0008013 | beta-catenin binding | MF | 0.115 | 0.0071 | 0.0457 | 1.34 | 0.0352 | Ar/Lef1/Nos3/Cdh2/Grin2b/Gli3/Cdh5/Tcf4/Dlg5/Psen1/Dact1 | 11 |
| GO:0001784 | phosphotyrosine residue binding | MF | 0.149 | 0.00739 | 0.0472 | 1.33 | 0.0363 | Shc3/She/Fgr/Zap70/Sh2d3c/Grb10/Irs1 | 7 |
| GO:0008066 | glutamate receptor activity | MF | 0.192 | 0.00767 | 0.0486 | 1.31 | 0.0374 | Grid1/Gria4/Grik1/Grin2b/Grik2 | 5 |
| GO:0097110 | scaffold protein binding | MF | 0.119 | 0.00771 | 0.0486 | 1.31 | 0.0374 | Nlgn1/Nos3/Grin2b/Nlgn3/Fyn/Cacna1h/Dlg4/Atp2b4/Lrp4/Kif5a | 10 |
| GO:0015291 | secondary active transmembrane transporter activity | MF | 0.0868 | 0.00797 | 0.0499 | 1.3 | 0.0384 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc9a7/Slc24a4/Slc36a4/Slc24a3/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Slc12a4/Slc38a1/Slco4c1/Slc4a3/Slc4a8/Ank | 21 |
| GO:0001653 | peptide receptor activity | MF | 0.098 | 0.00803 | 0.0499 | 1.3 | 0.0384 | Vipr2/Npr3/Ednrb/Cxcr3/Pth1r/Aplnr/Agtr1a/Ednra/Ogfrl1/Bdkrb2/Adcyap1r1/Xcr1/Calcrl/Prokr1/F2r | 15 |
| GO:0008574 | plus-end-directed microtubule motor activity | MF | 0.235 | 0.00807 | 0.0499 | 1.3 | 0.0384 | Kif1a/Stard9/Kif16b/Kif5a | 4 |
Visualisation
bp_dot=list()
mf_dot=list()
cc_dot=list()
upset=list()
for (i in 1:length(fc)) {
x <- fc[i] %>% as.character()
# extract the enriched GO terms from each ontology
bp <- enrichGO_sig[[x]] %>% dplyr::filter(ontology == "BP")
mf <- enrichGO_sig[[x]] %>% dplyr::filter(ontology == "MF")
cc <- enrichGO_sig[[x]] %>% dplyr::filter(ontology == "CC")
# bp dot plot, save
bp_dot[[x]] <- ggplot(bp[1:15, ]) +
geom_point(aes(x = GeneRatio, y = reorder(Description, GeneRatio), colour = logFDR, size = Count)) +
scale_color_gradient(low = "dodgerblue3", high = "firebrick3", limits = c(0, NA)) +
scale_size(range = c(.5,3)) +
ggtitle("Biological Process") +
ylab(label = "") +
xlab(label = "Gene Ratio") +
labs(color = expression("-log"[10] * "FDR"), size = "Gene Counts")
ggsave(filename = paste0("bp_dot_", fc[i], ".svg"), plot = bp_dot[[x]] + pub, path = here::here("2_plots/go/"),
width = 200, height = 120, units = "mm")
# mf dot plot, save
mf_dot[[x]] <- ggplot(mf[1:15, ]) +
geom_point(aes(x = GeneRatio, y = reorder(Description, GeneRatio), colour = logFDR, size = Count)) +
scale_color_gradient(low = "dodgerblue3", high = "firebrick3", limits = c(0, NA)) +
scale_size(range = c(.5,3)) +
ggtitle("Molecular Function") +
ylab(label = "") +
xlab(label = "Gene Ratio") +
labs(color = expression("-log"[10] * "FDR"), size = "Gene Counts")
ggsave(filename = paste0("mf_dot_", fc[i], ".svg"), plot = mf_dot[[x]] + pub, path = here::here("2_plots/go/"),
width = 200, height = 120, units = "mm")
# cc dot plot, save
cc_dot[[x]] <- ggplot(cc[1:15, ]) +
geom_point(aes(x = GeneRatio, y = reorder(Description, GeneRatio), colour = logFDR, size = Count)) +
scale_color_gradient(low = "dodgerblue3", high = "firebrick3", limits = c(0, NA)) +
scale_size(range = c(.5,3)) +
ggtitle("Cellular Components") +
ylab(label = "") +
xlab(label = "Gene Ratio") +
labs(color = expression("-log"[10] * "FDR"), size = "Gene Counts")
ggsave(filename = paste0("cc_dot_", fc[i], ".svg"), plot = cc_dot[[x]] + pub, path = here::here("2_plots/go/"),
width = 200, height = 120, units = "mm")
upset[[x]] <- upsetplot(x = enrichGO[[x]], 10)
ggsave(filename = paste0("upset_", fc[i], ".svg"), plot = upset[[x]], path = here::here("2_plots/go/"), width = 250, height = 166, units = "mm")
}Biological Process
bp_dot[[1]] 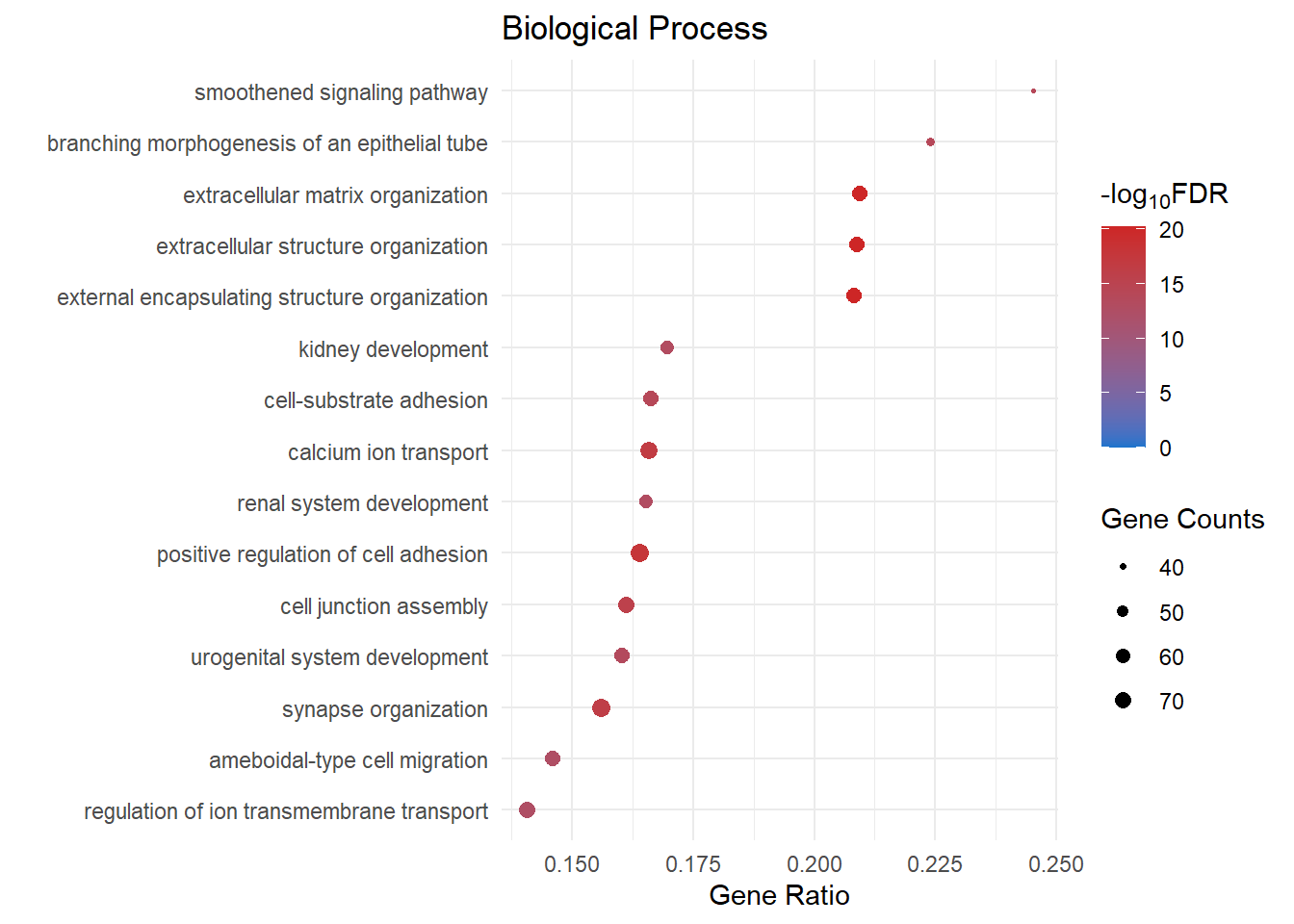
Molecular Function
mf_dot[[1]]
Cellular Components
cc_dot[[1]]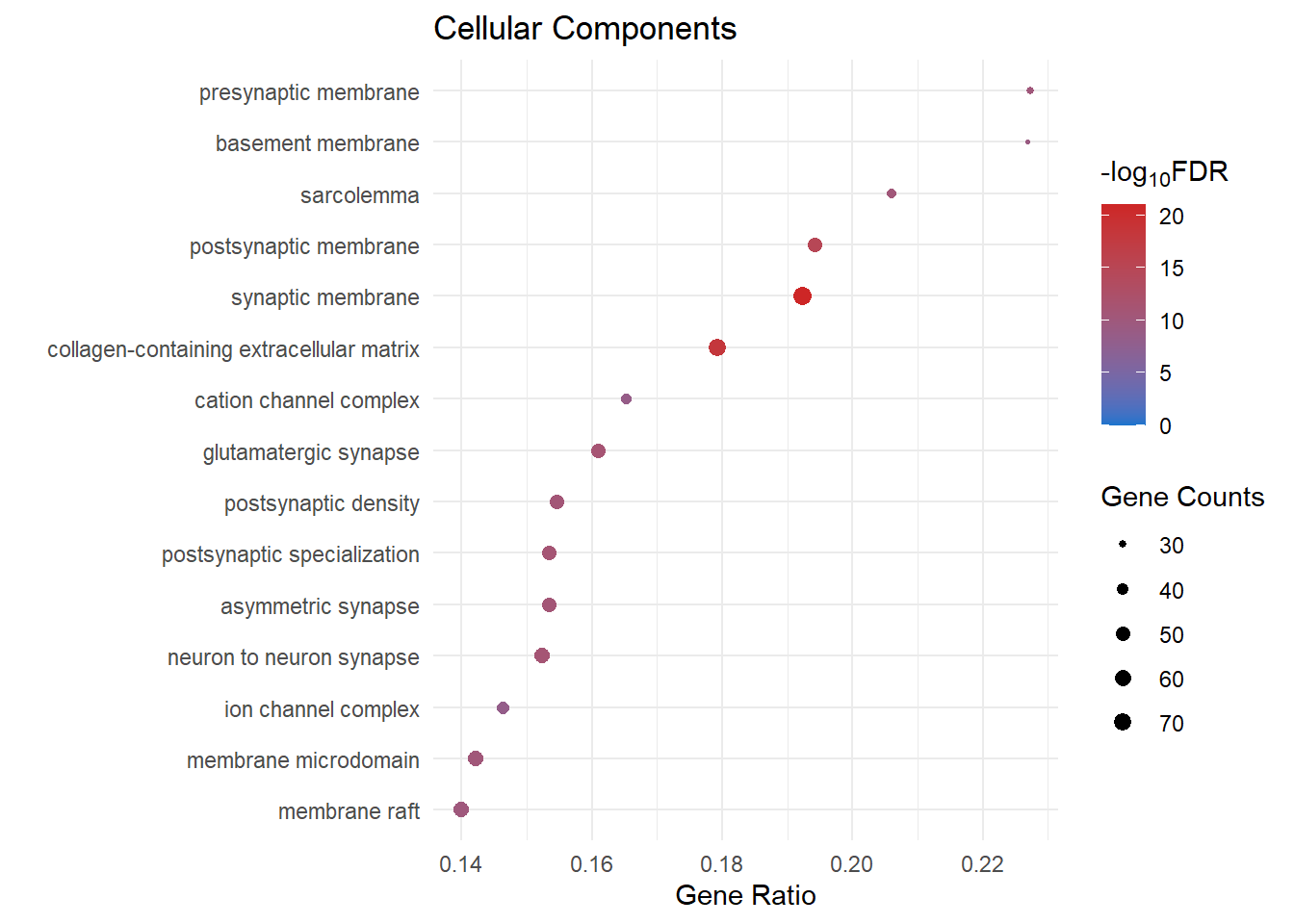
Upset
upset[[1]]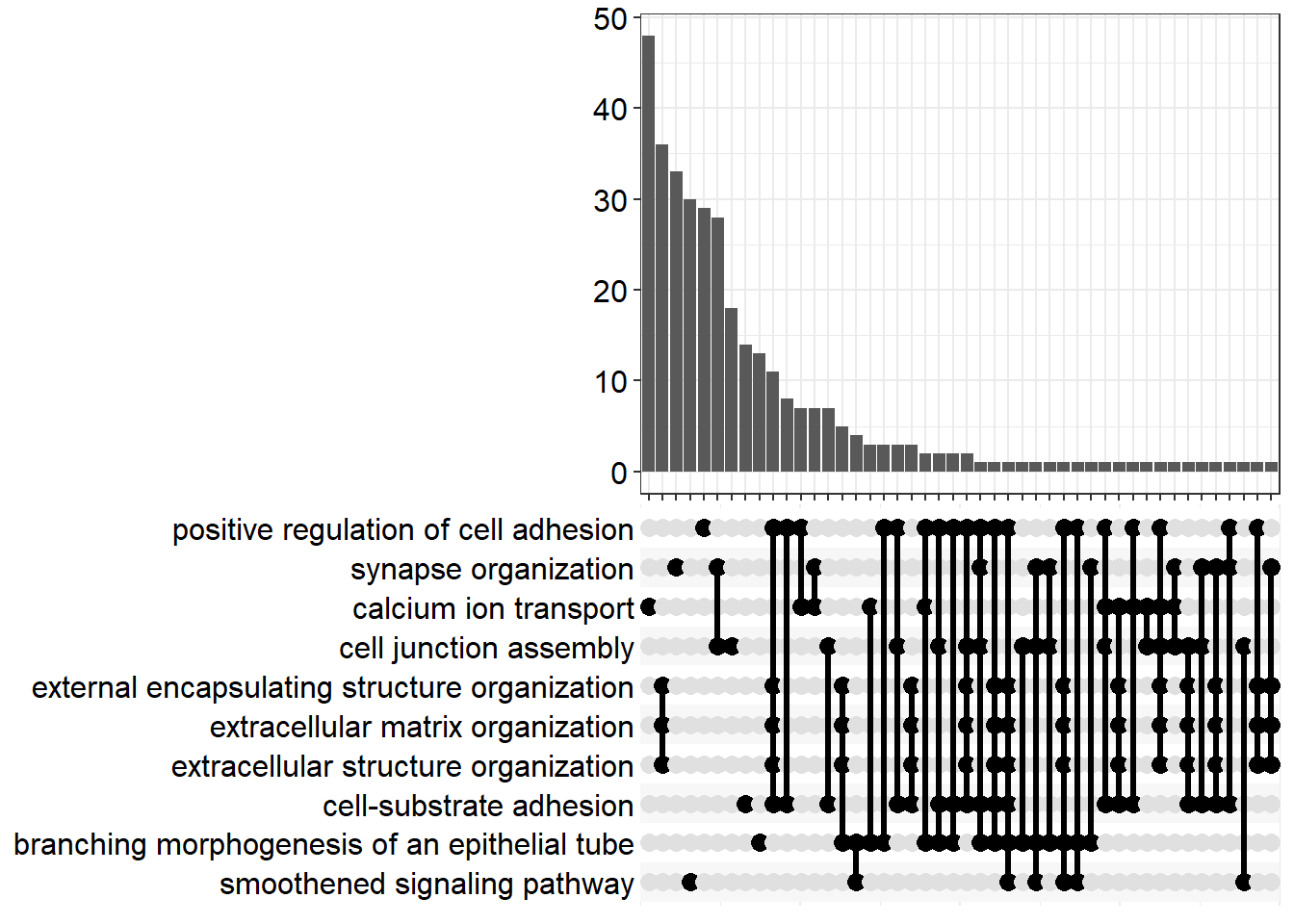
FC=1.1
# display the top 30 most sig
enrichGO_sig[[2]] %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(.,3)))) %>%
kable(caption = "Significantly enriched GO terms") %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Description | ontology | GeneRatio | pvalue | p.adjust | logFDR | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0030198 | extracellular matrix organization | BP | 0.194 | 1.46e-23 | 3.71e-20 | 19.4 | 2.52e-20 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Adamts20/Col4a6/Ddr2/Prdm5/Npnt/Phldb2/Spock2/Adamts16/Tgfbi/Ets1/Adamts19/Fn1/Mmp2/Foxf1/Col16a1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Smoc1/Loxl3/Adamts10/Tnfrsf11b/Emilin1/Tie1/Ndnf/Adamts7/Col5a2/Col15a1/Ltbp3/Mmp19/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Crtap/Efemp2/Olfml2a/Col23a1/Ntng1/Adamts17/Cav1/Col18a1/Itga8 | 61 |
| GO:0043062 | extracellular structure organization | BP | 0.193 | 1.74e-23 | 3.71e-20 | 19.4 | 2.52e-20 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Adamts20/Col4a6/Ddr2/Prdm5/Npnt/Phldb2/Spock2/Adamts16/Tgfbi/Ets1/Adamts19/Fn1/Mmp2/Foxf1/Col16a1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Smoc1/Loxl3/Adamts10/Tnfrsf11b/Emilin1/Tie1/Ndnf/Adamts7/Col5a2/Col15a1/Ltbp3/Mmp19/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Crtap/Efemp2/Olfml2a/Col23a1/Ntng1/Adamts17/Cav1/Col18a1/Itga8 | 61 |
| GO:0045229 | external encapsulating structure organization | BP | 0.192 | 2.07e-23 | 3.71e-20 | 19.4 | 2.52e-20 | Col11a1/Ihh/Cyp1b1/Mmp16/Hmcn1/Sfrp2/Vit/Sema5a/Scara3/Col14a1/Adamts20/Col4a6/Ddr2/Prdm5/Npnt/Phldb2/Spock2/Adamts16/Tgfbi/Ets1/Adamts19/Fn1/Mmp2/Foxf1/Col16a1/Fkbp10/Nid1/Olfml2b/Has2/Egflam/Adamts1/Col9a2/Col13a1/Postn/Smoc1/Loxl3/Adamts10/Tnfrsf11b/Emilin1/Tie1/Ndnf/Adamts7/Col5a2/Col15a1/Ltbp3/Mmp19/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Ccn2/Crtap/Efemp2/Olfml2a/Col23a1/Ntng1/Adamts17/Cav1/Col18a1/Itga8 | 61 |
| GO:0006816 | calcium ion transport | BP | 0.155 | 2.15e-20 | 2.89e-17 | 16.5 | 1.97e-17 | Nalcn/Cacna1c/Ryr1/Spink1/Jph4/Igf1/Adra1a/Nos3/Slc8a1/Drd4/Ednrb/Slc24a4/Cachd1/Cacna2d1/Myo5a/Icam1/Grin2b/Stc1/Prkd1/Cxcr3/Cxcl12/Slc24a3/Cacna1e/Ank2/Slc8a3/Calca/Nalf1/Thy1/Trpc6/Gja4/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Xcl1/Htr2a/Tspan13/Bin1/Stac2/Pkd1/Cacng7/Dysf/Asph/Ryr3/Selenon/Xcr1/Rrad/Cd84/Cacna1h/Arrb2/Ptger3/Calcrl/Ccn2/Fyn/Pkd2/Slc25a23/Gstm7/Atp2b4/Gimap5/Atp1a2/Gnb5/Gimap3/Cav1/Usp2/Cacna1a/Cacnb4 | 68 |
| GO:0045785 | positive regulation of cell adhesion | BP | 0.145 | 1.99e-19 | 2.14e-16 | 15.7 | 1.46e-16 | Ihh/Efnb3/Vit/Col26a1/Angpt1/Lef1/Mdk/Itga4/Cd83/Kdr/Igf1/Npnt/Chst2/Cd44/Spock2/Nrp1/Fermt2/Ets1/Fn1/Foxf1/Col16a1/Icam1/Nid1/Tek/Cxcl12/Has2/Cdh13/Egflam/Pik3r6/Gli3/Calca/Podxl/Tnfsf13b/Thy1/Plpp3/Cd1d1/Smoc1/Coro1a/Ppm1f/Itgal/Il27ra/H2-Ab1/Vegfc/Sele/Havcr2/Sox12/H2-Ob/Emilin1/Igf2/Ndnf/Xcl1/Pcsk5/Nckap1l/H2-Eb1/Dysf/Abi3bp/Net1/H2-Aa/Adam19/Stat5b/Gimap5/Zap70/Gimap3/Efemp2/St3gal4/Apbb1ip/Ptn/Il12rb1/Cav1/Il2rg | 70 |
| GO:0034329 | cell junction assembly | BP | 0.15 | 4.44e-19 | 3.98e-16 | 15.4 | 2.71e-16 | Gpc6/Gap43/Sdk2/Wnt11/Kdr/Plxnb1/Ptprd/St8sia2/Nlgn1/Epb41l3/Ptpro/Phldb2/Mpdz/Nrp1/Fermt2/Cdh2/Lrrc4b/Cdh6/Fn1/Col16a1/Nptx1/Mef2c/Tek/Thsd1/Jam3/Shank2/Obsl1/Cldn8/Cdh10/Adgrl1/Slitrk3/Cdh5/Thy1/Plxnd1/Abi2/Epha3/Ppm1f/Nlgn3/Aplnr/Esam/Myo9a/Nptxr/Sh3bp1/Nfasc/Limch1/Arhgap6/Cdh11/Magi2/Gjc1/Pecam1/Snai1/Epha7/Heg1/Pcdh17/Dchs1/Dlc1/Cldn5/Bhlhb9/Cux2/Nphp1/Dlg5/Arhgef9/Cav1/Cacna1a/Itgb4/Cldn1 | 66 |
| GO:0031589 | cell-substrate adhesion | BP | 0.155 | 1.05e-17 | 8.06e-15 | 14.1 | 5.48e-15 | Ptprz1/Vit/Col26a1/Angpt1/Mdk/Itga4/Kdr/Npnt/Phldb2/Lamc3/Spock2/Nrp1/Fermt2/Itgb7/Fn1/Foxf1/Col16a1/Nid1/Tek/Thsd1/Has2/Jam3/Cdh13/Egflam/Itga1/Frem1/Col13a1/Postn/Thy1/Smoc1/Coro1a/Epha3/Ppm1f/Itgal/Limch1/Vegfc/Arhgap6/Hoxd3/Sned1/Tmeff2/Emilin1/Ndnf/Pecam1/Parvg/Itga7/Pcsk5/Parvb/Epdr1/Dlc1/Abi3bp/Net1/Ccn2/Efemp2/Ptn/Ntng1/Cd96/Itga8/Itgb4 | 58 |
| GO:0050808 | synapse organization | BP | 0.136 | 4.39e-17 | 2.47e-14 | 13.6 | 1.68e-14 | Gpc6/Gap43/Sdk2/Lrrc4c/Plxnb1/Ptprd/St8sia2/Nlgn1/Ptpro/Rims3/Pdzrn3/P2rx2/Nrp1/Cdh2/Lrrc4b/Wasf3/Lrrk2/Nptx1/Mef2c/Myo5a/Pfn2/Grin2b/Shank2/Pcdhgc3/Obsl1/Adgrl1/Ppfia2/Slc8a3/Slitrk3/Plxnd1/Dock10/Glrb/Kif1a/Abi2/Ppfia4/Epha3/Nlgn3/Dgkb/Nptxr/Nfasc/Arhgef15/Apoe/Prrt1/Palld/Sparcl1/Magi2/Caprin2/Epha7/Erc2/Pcdh17/Lrfn2/Nfatc4/Fyn/Bhlhb9/Dlg4/Cux2/Palm/Dlg5/Syngap1/Rab39b/Ngef/Arhgef9/Ptn/Ntng1/Dact1/Cacna1a/Cacnb4 | 67 |
| GO:0048754 | branching morphogenesis of an epithelial tube | BP | 0.208 | 4.59e-17 | 2.47e-14 | 13.6 | 1.68e-14 | Ihh/Sfrp2/Sema5a/Wnt6/Angpt1/Hhip/Ar/Lef1/Mdk/Hoxd11/Kdr/Nog/Igf1/Tbx2/Npnt/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Areg/Foxf1/Tek/Cxcl12/Gli3/Tcf21/Gdf7/Plxnd1/Mgp/Agtr1a/Rasip1/Tbx3/Tie1/Ednra/Pkd1/Ptch1/Dchs1/Nfatc4/Fat4/Dlg5 | 40 |
| GO:0007224 | smoothened signaling pathway | BP | 0.226 | 1.03e-16 | 5.02e-14 | 13.3 | 3.42e-14 | Gli1/Ihh/Hhip/Evc2/Nog/Ptch2/Evc/Kif7/Dzip1l/Gpc3/Arl6/Foxf1/Serpine2/Cibar1/Bbs7/Zfp423/Armc9/Gli3/Ift57/Ptpdc1/Rab34/Wnt9a/Gpr161/Enpp1/Dync2h1/Hipk2/Arl3/Cfap410/Ptch1/Tmem231/Ttc26/Ttbk2/Dzip1/Rab23/Dlg5/Gpc2 | 36 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | BP | 0.192 | 4.21e-16 | 1.89e-13 | 12.7 | 1.28e-13 | Gpc6/Prtg/Hmcn1/Pcdh18/Sdk2/Mcam/Itga4/Lrrc4c/Ptprd/Nlgn1/Cdh2/Lrrc4b/Cdh6/Tenm3/Dab1/Cdh13/Pcdhgc3/Obsl1/Cdh10/Adgrl1/Slitrk3/Cdh5/Pcdh19/Ptprm/Pcdh12/Epha3/Esam/Itgal/Palld/Sele/Sparcl1/Cdh11/Magi2/Pecam1/Pcdh17/Dchs1/Fat3/Fat4/Ntng1/Pcdhb14/Cldn1 | 41 |
| GO:0001655 | urogenital system development | BP | 0.145 | 1e-15 | 4.15e-13 | 12.4 | 2.83e-13 | Stra6/Wnt6/Angpt1/Ar/Wnt11/Pygo1/Hoxd11/Nog/Cyp7b1/Igf1/Npnt/Ptpro/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Ednrb/Lrrk2/Mmp2/Foxf1/Mef2c/Id3/Osr2/Nid1/Id4/Tek/Enpep/Gdf11/Adamts1/Gli3/Tcf21/Lrp2/Plxnd1/Tshz3/Prdm1/Cdkn1c/Cplane1/Enpp1/Agtr1a/Heyl/Dync2h1/Ednra/Pcsk5/Smad9/Pkd1/Epha7/Arl3/Ptch1/Osr1/Dchs1/Aqp11/Pkd2/Fat4/Dlg5/Itga8 | 56 |
| GO:0001667 | ameboidal-type cell migration | BP | 0.135 | 1.45e-15 | 5.56e-13 | 12.3 | 3.78e-13 | Cyp1b1/Sema5a/Angpt1/Wnt11/Itga4/Kdr/Igf1/Evl/Sema5b/Ddr2/Zeb2/Nos3/Slc8a1/Sema7a/Nrp1/Ets1/Cdh2/Ednrb/Itgb7/Fn1/Mef2c/Pfn2/Ptp4a3/Stc1/Prkd1/Tek/Cxcl12/Has2/Cdh13/Akap12/Edn3/Vash1/Calca/Cdh5/Plxnd1/Syde1/Plpp3/Ptprm/Kit/Ppm1f/Jcad/Sh3bp1/Prr5l/Vegfc/Apoe/Ednra/Igf2/Daam2/Pecam1/Sema3g/Kitl/Hdac9/Atp2b4/Akt3/Fstl1/Sema6a/Arsb/Adgra2/Mapre2/Cygb/Itgb4 | 61 |
| GO:0034765 | regulation of ion transmembrane transport | BP | 0.129 | 3.11e-15 | 1.04e-12 | 12 | 7.1e-13 | Kcnb2/Nalcn/Kcnj3/Hecw2/Kcna6/Cacna1c/Kcnd2/Kcnd3/Nlgn1/Tcaf1/Slc8a1/Scn4b/Drd4/Scn1a/Fxyd5/Kcnq5/Cacna2d1/Mef2c/Myo5a/Prkd1/Cxcr3/Cacna1e/Edn3/Ank2/Plcb1/Thy1/Trpc6/Scn2a/Coro1a/Nlgn3/Aplnr/Cracr2a/Prrt1/Agtr1a/Kcnma1/Kcnc4/Ednra/Tspan13/Bin1/Stac2/Cacng7/Dysf/Kcnip3/Fhl1/Asph/Selenon/Osr1/Rrad/Cacna1h/Kcng4/Ptger3/Fyn/Pkd2/Scn3a/Dlg4/Gstm7/Atp2b4/Atp1a2/Gnb5/Abcb1a/Kcnq4/Cav1/Cacna1a/Cacnb4 | 64 |
| GO:0001822 | kidney development | BP | 0.152 | 5.49e-15 | 1.7e-12 | 11.8 | 1.16e-12 | Stra6/Wnt6/Angpt1/Wnt11/Pygo1/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Ednrb/Lrrk2/Mef2c/Id3/Osr2/Nid1/Tek/Enpep/Gdf11/Adamts1/Gli3/Tcf21/Lrp2/Plxnd1/Tshz3/Prdm1/Cdkn1c/Cplane1/Enpp1/Agtr1a/Heyl/Dync2h1/Ednra/Pcsk5/Smad9/Pkd1/Epha7/Arl3/Ptch1/Osr1/Dchs1/Aqp11/Pkd2/Fat4/Dlg5/Itga8 | 50 |
| GO:0061138 | morphogenesis of a branching epithelium | BP | 0.178 | 5.69e-15 | 1.7e-12 | 11.8 | 1.16e-12 | Ihh/Hgf/Sfrp2/Sema5a/Wnt6/Angpt1/Hhip/Ar/Lef1/Mdk/Hoxd11/Kdr/Nog/Igf1/Tbx2/Npnt/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Areg/Foxf1/Tek/Cxcl12/Gli3/Tcf21/Gdf7/Plxnd1/Mgp/Agtr1a/Rasip1/Tbx3/Tie1/Ednra/Pkd1/Ptch1/Dchs1/Nfatc4/Fat4/Dlg5 | 41 |
| GO:0072001 | renal system development | BP | 0.148 | 8.09e-15 | 2.29e-12 | 11.6 | 1.56e-12 | Stra6/Wnt6/Angpt1/Wnt11/Pygo1/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Ednrb/Lrrk2/Foxf1/Mef2c/Id3/Osr2/Nid1/Tek/Enpep/Gdf11/Adamts1/Gli3/Tcf21/Lrp2/Plxnd1/Tshz3/Prdm1/Cdkn1c/Cplane1/Enpp1/Agtr1a/Heyl/Dync2h1/Ednra/Pcsk5/Smad9/Pkd1/Epha7/Arl3/Ptch1/Osr1/Dchs1/Aqp11/Pkd2/Fat4/Dlg5/Itga8 | 51 |
| GO:0070588 | calcium ion transmembrane transport | BP | 0.157 | 9.96e-15 | 2.68e-12 | 11.6 | 1.82e-12 | Nalcn/Cacna1c/Ryr1/Slc8a1/Drd4/Ednrb/Slc24a4/Cacna2d1/Myo5a/Grin2b/Prkd1/Cxcr3/Slc24a3/Cacna1e/Ank2/Slc8a3/Nalf1/Thy1/Trpc6/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Xcl1/Htr2a/Tspan13/Bin1/Stac2/Pkd1/Cacng7/Dysf/Asph/Ryr3/Selenon/Xcr1/Rrad/Ptger3/Fyn/Pkd2/Slc25a23/Gstm7/Atp2b4/Atp1a2/Gnb5/Cacna1a/Cacnb4 | 47 |
| GO:0051924 | regulation of calcium ion transport | BP | 0.157 | 3.58e-14 | 8.92e-12 | 11 | 6.07e-12 | Cacna1c/Spink1/Jph4/Igf1/Nos3/Slc8a1/Drd4/Cacna2d1/Myo5a/Icam1/Stc1/Prkd1/Cxcr3/Cxcl12/Ank2/Calca/Thy1/Trpc6/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Tspan13/Bin1/Stac2/Dysf/Asph/Selenon/Rrad/Cd84/Arrb2/Ptger3/Fyn/Pkd2/Gstm7/Gimap5/Atp1a2/Gnb5/Gimap3/Cav1/Usp2/Cacna1a/Cacnb4 | 45 |
| GO:0060562 | epithelial tube morphogenesis | BP | 0.137 | 3.65e-14 | 8.92e-12 | 11 | 6.07e-12 | Ihh/Sfrp2/Sema5a/Wnt6/Angpt1/Hhip/Ar/Lef1/Mdk/Hoxd11/Kdr/Nog/Igf1/Tbx2/Zeb2/Npnt/Cd44/Hoxa11/Nrp1/Adamts16/Gpc3/Areg/Foxf1/Mef2c/Tek/Cxcl12/Gli3/Tcf21/Lrp2/Gdf7/Ift57/Plxnd1/Cecr2/Mgp/Aplnr/Agtr1a/Rasip1/Tbx3/Tie1/Ednra/Prickle1/Pkd1/Epha7/Ptch1/Ipmk/Osr1/Dchs1/Dlc1/Nfatc4/Pkd2/Fzd2/Rab23/Fat4/Dlg5 | 54 |
| GO:0006874 | cellular calcium ion homeostasis | BP | 0.124 | 4.41e-14 | 1.03e-11 | 11 | 7.02e-12 | Kcnk3/Cacna1c/Ryr1/Kdr/Adra1a/Slc8a1/P2rx2/Pygm/Bok/Ednrb/Slc24a4/Cacna2d1/Myo5a/Inpp4b/Grin2b/Lpar4/Stc1/Prkd1/Cxcr3/Slc24a3/Cacna1e/Ptgfr/Edn3/Ank2/Slc8a3/Calca/Cdh5/Pth1r/Nalf1/Thy1/Trpc6/Coro1a/Aplnr/Kctd17/Grik2/Apoe/Cd38/Agtr1a/Ednra/Adcyap1r1/Bdkrb2/Xcl1/Htr2a/Pkd1/Asph/Ryr3/Selenon/Xcr1/Ptger3/Fyn/Pkd2/Slc25a23/Fzd2/Dlg4/Gstm7/Atp2b4/Gimap5/Atp1a2/Gimap3/Cav1/Cacna1a/Cacnb4 | 62 |
| GO:0031345 | negative regulation of cell projection organization | BP | 0.177 | 7.42e-14 | 1.66e-11 | 10.8 | 1.13e-11 | Ptprz1/Efnb3/Map2/Sema5a/Evl/Sema5b/Nlgn1/Sema7a/Nrp1/Cspg4/Ntm/Dab1/Lrrk2/Abi3/Pfn2/Grin2b/Thy1/Trpc6/Epha3/Minar1/Nlgn3/Tsku/Apoe/Cd38/Sema3g/Dennd5a/Rtn4rl1/Epha7/Nfatc4/Neo1/Fyn/Fat3/Xylt1/Dpysl3/Syngap1/Sema6a/Ngef/Cep97 | 38 |
| GO:0010959 | regulation of metal ion transport | BP | 0.128 | 1.31e-13 | 2.83e-11 | 10.5 | 1.92e-11 | Hecw2/Cacna1c/Spink1/Jph4/Igf1/Nos3/Slc8a1/Scn4b/Drd4/Fxyd5/Cacna2d1/Myo5a/Icam1/Stc1/Serpine2/Prkd1/Cxcr3/Cxcl12/Edn3/Ank2/Calca/Plcb1/Thy1/Trpc6/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Htr2a/Tspan13/Bin1/Stac2/Dysf/Kcnip3/Fhl1/Asph/Selenon/Osr1/Rrad/Cd84/Arrb2/Ptger3/Fyn/Pkd2/Gstm7/Atp2b4/Gimap5/Akt3/Atp1a2/Gnb5/Gimap3/Cav1/Usp2/Cacna1a/Cacnb4 | 57 |
| GO:0060402 | calcium ion transport into cytosol | BP | 0.19 | 1.76e-13 | 3.65e-11 | 10.4 | 2.48e-11 | Cacna1c/Ryr1/Adra1a/Slc8a1/Slc24a4/Cacna2d1/Myo5a/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Calca/Nalf1/Thy1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Fyn/Pkd2/Gstm7/Gimap5/Gimap3/Cav1/Cacna1a | 34 |
| GO:0050673 | epithelial cell proliferation | BP | 0.124 | 1.86e-13 | 3.71e-11 | 10.4 | 2.52e-11 | Gli1/Ihh/Hgf/Sfrp2/Sema5a/Ar/Mdk/Itga4/Cd109/Kdr/Agap2/Nog/Cyp7b1/Igf1/Tbx2/Gkn3/Fst/Gpc3/Ednrb/Areg/Ccnd2/Mef2c/Osr2/Prkd1/Tek/Cxcr3/Cxcl12/Has2/Cdh13/Vash1/Calca/Ift57/Stxbp4/Ptprm/Kit/Ppp1r16b/Aplnr/Cdkn1c/Jcad/Vegfc/Apoe/Agtr1a/Tie1/Ednra/Igf2/Klf9/Twist2/Dysf/Ptch1/Osr1/Aqp11/Apln/Akt3/Egfl7/Ptn/Cav1/Vash2/Igfbp4/Cldn1 | 59 |
| GO:0060401 | cytosolic calcium ion transport | BP | 0.179 | 2.22e-13 | 4.26e-11 | 10.4 | 2.9e-11 | Cacna1c/Ryr1/Adra1a/Slc8a1/Slc24a4/Cacna2d1/Myo5a/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Calca/Nalf1/Thy1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Ccn2/Fyn/Pkd2/Slc25a23/Gstm7/Gimap5/Gimap3/Cav1/Cacna1a | 36 |
| GO:0003018 | vascular process in circulatory system | BP | 0.166 | 2.85e-13 | 5.29e-11 | 10.3 | 3.6e-11 | Angpt1/Cacna1c/Kdr/Ddah1/Adra1a/Nos3/Slc8a1/Cbs/Npr3/Fermt2/Ednrb/Mmp2/Icam1/Wdr35/Ptp4a3/Abcg2/Akap12/Edn3/Itga1/Calca/Cdh5/Htr1b/Ptprm/Gucy1a1/Apoe/Cd38/Agtr1a/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Cldn5/Ptger3/Apln/Atp1a2/Abcb1a/Abcg3/Cav1 | 39 |
| GO:0048736 | appendage development | BP | 0.176 | 3.52e-13 | 6.11e-11 | 10.2 | 4.16e-11 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Fermt2/Gpc3/Hoxd9/Hoxa10/Osr2/Cibar1/Bbs7/Flvcr1/Gli3/Wnt9a/Smoc1/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Itgb4 | 36 |
| GO:0060173 | limb development | BP | 0.176 | 3.52e-13 | 6.11e-11 | 10.2 | 4.16e-11 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Fermt2/Gpc3/Hoxd9/Hoxa10/Osr2/Cibar1/Bbs7/Flvcr1/Gli3/Wnt9a/Smoc1/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23/Itgb4 | 36 |
| GO:0007409 | axonogenesis | BP | 0.12 | 8.43e-13 | 1.42e-10 | 9.85 | 9.64e-11 | Gap43/Epha8/Prtg/Ptprz1/Efnb3/Map2/Sema5a/Pla2g10/Itga4/Lrrc4c/Dcc/Nog/Plxnb1/Evl/Sema5b/Rnf165/Zeb2/Ptpro/Sema7a/Nrp1/Cdh2/Fn1/Dab1/Nptx1/Mef2c/Cxcl12/Arhgef25/Edn3/Gli3/Lrp2/Slitrk3/Gdf7/Thy1/Plxnd1/Ptprm/Ndn/Lama2/Epha3/Nlgn3/Matn2/Tsku/Nfasc/Apoe/Cdh11/Ednra/Sema3g/Rtn4rl1/Nin/Epha7/Chodl/Ptch1/Neo1/Unc5c/Brsk1/Syngap1/Sema6a/Ntng1/Nrn1/Cacna1a | 59 |
| GO:0007160 | cell-matrix adhesion | BP | 0.162 | 1.15e-12 | 1.87e-10 | 9.73 | 1.27e-10 | Itga4/Kdr/Npnt/Phldb2/Nrp1/Fermt2/Itgb7/Fn1/Col16a1/Nid1/Tek/Thsd1/Jam3/Cdh13/Itga1/Frem1/Col13a1/Postn/Thy1/Epha3/Ppm1f/Itgal/Limch1/Vegfc/Arhgap6/Hoxd3/Sned1/Emilin1/Pecam1/Itga7/Pcsk5/Epdr1/Dlc1/Ccn2/Efemp2/Cd96/Itga8/Itgb4 | 38 |
| GO:0050678 | regulation of epithelial cell proliferation | BP | 0.128 | 1.31e-12 | 2.07e-10 | 9.68 | 1.41e-10 | Gli1/Ihh/Sfrp2/Sema5a/Ar/Mdk/Itga4/Cd109/Kdr/Agap2/Nog/Cyp7b1/Igf1/Gkn3/Gpc3/Ednrb/Ccnd2/Mef2c/Osr2/Prkd1/Tek/Cxcr3/Cxcl12/Has2/Cdh13/Vash1/Ift57/Stxbp4/Ptprm/Ppp1r16b/Aplnr/Cdkn1c/Jcad/Vegfc/Apoe/Agtr1a/Tie1/Ednra/Igf2/Klf9/Twist2/Dysf/Ptch1/Osr1/Aqp11/Apln/Akt3/Egfl7/Ptn/Cav1/Vash2/Cldn1 | 52 |
| GO:0051480 | regulation of cytosolic calcium ion concentration | BP | 0.128 | 2.11e-12 | 3.24e-10 | 9.49 | 2.2e-10 | Kcnk3/Cacna1c/Ryr1/Kdr/Adra1a/Slc8a1/P2rx2/Bok/Ednrb/Slc24a4/Cacna2d1/Myo5a/Grin2b/Lpar4/Prkd1/Cxcr3/Cacna1e/Ptgfr/Ank2/Slc8a3/Calca/Pth1r/Nalf1/Thy1/Trpc6/Coro1a/Aplnr/Cd38/Agtr1a/Ednra/Adcyap1r1/Bdkrb2/Xcl1/Htr2a/Pkd1/Asph/Ryr3/Selenon/Xcr1/Ptger3/Fyn/Pkd2/Fzd2/Dlg4/Gstm7/Atp2b4/Gimap5/Atp1a2/Gimap3/Cav1/Cacna1a | 51 |
| GO:0035107 | appendage morphogenesis | BP | 0.184 | 2.26e-12 | 3.28e-10 | 9.48 | 2.23e-10 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Cibar1/Flvcr1/Gli3/Wnt9a/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23 | 32 |
| GO:0035108 | limb morphogenesis | BP | 0.184 | 2.26e-12 | 3.28e-10 | 9.48 | 2.23e-10 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Pkdcc/Tbx2/Rnf165/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Cibar1/Flvcr1/Gli3/Wnt9a/Cplane1/Enpp1/Tbx3/Dync2h1/Pcsk5/Hoxa9/Ptch1/Tmem231/Asph/Osr1/Ttbk2/Rab23 | 32 |
| GO:0032970 | regulation of actin filament-based process | BP | 0.126 | 3.07e-12 | 4.34e-10 | 9.36 | 2.95e-10 | Kank4/Sema5a/Mdk/Wnt11/Cacna1c/Evl/Phldb2/Ccdc88a/Hcls1/Nrp1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Stc1/Tek/Jam3/Ank2/Coro1a/Abi2/Epha3/Ppm1f/Esam/Sh3bp1/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Daam2/Pecam1/Ppm1e/Scin/Cnn2/Nckap1l/Carmil3/Rhobtb3/Bin1/Myo1f/Arhgap28/Dlc1/Cacna1h/Ptger3/Ccn2/Fgr/Atp1a2/Cdc42ep5/Cav1/Cyfip2/Plekhh2 | 52 |
| GO:1903039 | positive regulation of leukocyte cell-cell adhesion | BP | 0.153 | 7.12e-12 | 9.58e-10 | 9.02 | 6.52e-10 | Ihh/Efnb3/Lef1/Mdk/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Has2/Pik3r6/Gli3/Tnfsf13b/Thy1/Cd1d1/Coro1a/Itgal/Il27ra/H2-Ab1/Sele/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/St3gal4/Il12rb1/Cav1/Il2rg | 38 |
| GO:0010977 | negative regulation of neuron projection development | BP | 0.185 | 9.09e-12 | 1.17e-09 | 8.93 | 7.93e-10 | Ptprz1/Efnb3/Map2/Sema5a/Sema5b/Nlgn1/Sema7a/Nrp1/Cspg4/Ntm/Dab1/Lrrk2/Thy1/Epha3/Minar1/Nlgn3/Tsku/Apoe/Cd38/Sema3g/Dennd5a/Rtn4rl1/Epha7/Neo1/Fat3/Xylt1/Dpysl3/Syngap1/Sema6a/Ngef | 30 |
| GO:1902903 | regulation of supramolecular fiber organization | BP | 0.126 | 1.07e-11 | 1.33e-09 | 8.87 | 9.08e-10 | Kank4/Map2/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Hcls1/Nrp1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Cdh5/Coro1a/Abi2/Ppm1f/Rp1/Esam/Sh3bp1/Limch1/Tubb4a/Nox4/Arhgef15/Apoe/Arhgap6/Tmeff2/Emilin1/Daam2/Pecam1/Ppm1e/Scin/Nckap1l/Slain1/Carmil3/Nin/Bin1/Dysf/Arhgap28/Ttbk2/Dlc1/Ccn2/Efemp2/Cdc42ep5/Cav1/Cyfip2/Plekhh2 | 49 |
| GO:0090287 | regulation of cellular response to growth factor stimulus | BP | 0.14 | 1.17e-11 | 1.43e-09 | 8.85 | 9.7e-10 | Sfrp2/Angpt1/Hhip/Cd109/Kdr/Nog/Rnf165/Zeb2/Npnt/Fst/Gpc3/Ptp4a3/Tek/Zfp423/Lrp2/Cdh5/Pbld2/Zeb1/Cdkn1c/Jcad/Tsku/Vegfc/Emilin1/Tcf4/Spred3/Kcp/Tnfaip6/Hipk2/Nrros/Zfyve9/Neo1/Il17rd/Apln/Atp2b4/Fstl1/Sema6a/Lats2/Adgra2/Cav1/Cyfip2/Itga8/Tmprss6 | 42 |
| GO:0031346 | positive regulation of cell projection organization | BP | 0.12 | 1.25e-11 | 1.49e-09 | 8.83 | 1.01e-09 | Ptprz1/Hgf/Sema5a/Acsl6/Fez1/Mdk/Dcc/Plxnb1/Ptprd/Nlgn1/Zeb2/Lrrc7/Ccdc88a/Sema7a/Nrp1/Fn1/Tenm3/Serpine2/Prkd1/Cxcl12/Tbc1d24/Obsl1/Gprc5b/Adamts1/Serpini1/Plxnd1/Abi2/Kit/Epha3/Rp1/Kctd17/Apoe/Magi2/Eps8l1/Caprin2/Ndnf/Crocc/Nin/Chodl/P3h1/Ttbk2/Dzip1/Fyn/Bhlhb9/Dlg4/Cux2/Palm/Dpysl3/Cdc42ep5/Arsb/Ptn/Gpc2/Cpeb1 | 53 |
| GO:0007204 | positive regulation of cytosolic calcium ion concentration | BP | 0.13 | 1.71e-11 | 1.96e-09 | 8.71 | 1.33e-09 | Cacna1c/Ryr1/Kdr/Adra1a/Slc8a1/Ednrb/Slc24a4/Cacna2d1/Myo5a/Grin2b/Lpar4/Prkd1/Cxcr3/Cacna1e/Ptgfr/Ank2/Slc8a3/Calca/Pth1r/Nalf1/Thy1/Trpc6/Coro1a/Aplnr/Cd38/Agtr1a/Ednra/Adcyap1r1/Bdkrb2/Xcl1/Htr2a/Pkd1/Asph/Ryr3/Selenon/Xcr1/Ptger3/Fyn/Pkd2/Fzd2/Dlg4/Gstm7/Gimap5/Gimap3/Cav1/Cacna1a | 46 |
| GO:0007159 | leukocyte cell-cell adhesion | BP | 0.125 | 2.27e-11 | 2.55e-09 | 8.59 | 1.73e-09 | Ihh/Efnb3/Lef1/Mdk/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Itgb7/Icam1/Podxl2/Cxcl12/Has2/Pik3r6/Gli3/Calca/Tnfsf13b/Thy1/Cd1d1/Msn/Loxl3/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Sele/Havcr2/Sox12/H2-Ob/Igf2/Pecam1/Xcl1/Nckap1l/H2-Eb1/Pde5a/H2-Aa/Stat5b/Gimap5/Zap70/Dlg5/Gimap3/St3gal4/Il12rb1/Cav1/Il2rg | 48 |
| GO:0050890 | cognition | BP | 0.128 | 2.78e-11 | 3.05e-09 | 8.52 | 2.07e-09 | Stra6/Ptprz1/Mdk/Cacna1c/Nog/Cyp7b1/Jph4/Igf1/Aph1b/Aff2/Drd4/Tmod2/Ccnd2/Chst10/Shc3/Mef2c/Grin2b/B4galt2/Pde1b/Adcy3/Shank2/Cacna1e/Slc8a3/Plcb1/Eif2ak4/Mme/Scn2a/Abi2/Kit/Nlgn3/Apoe/Prrt1/Igf2/Htr2a/Pde5a/Nfatc4/Mgat3/Bhlhb9/Cux2/Atp1a2/Brsk1/Syngap1/St3gal4/Ptn/Ddhd2/Itga8 | 46 |
| GO:0022407 | regulation of cell-cell adhesion | BP | 0.115 | 3.22e-11 | 3.47e-09 | 8.46 | 2.36e-09 | Ihh/Efnb3/Lef1/Mdk/Itga4/Cd83/Igf1/Abat/Chst2/Cd44/Ets1/Tenm3/Icam1/Serpine2/Cxcl12/Has2/Pik3r6/Gli3/Podxl/Tnfsf13b/Thy1/Plpp3/Cd1d1/Loxl3/Coro1a/Ccm2l/Ppm1f/Btla/Itgal/Il27ra/H2-Ab1/Sele/Havcr2/Sox12/H2-Ob/Magi2/Igf2/Xcl1/Nckap1l/Epha7/H2-Eb1/Pde5a/Ptger3/H2-Aa/Adam19/Stat5b/Gimap5/Zap70/Dlg5/Gimap3/St3gal4/Il12rb1/Cav1/Il2rg | 54 |
| GO:0022409 | positive regulation of cell-cell adhesion | BP | 0.138 | 3.57e-11 | 3.76e-09 | 8.42 | 2.56e-09 | Ihh/Efnb3/Lef1/Mdk/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Has2/Pik3r6/Gli3/Podxl/Tnfsf13b/Thy1/Plpp3/Cd1d1/Coro1a/Itgal/Il27ra/H2-Ab1/Sele/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/H2-Aa/Adam19/Stat5b/Gimap5/Zap70/Gimap3/St3gal4/Il12rb1/Cav1/Il2rg | 41 |
| GO:0010811 | positive regulation of cell-substrate adhesion | BP | 0.193 | 3.72e-11 | 3.85e-09 | 8.41 | 2.62e-09 | Vit/Col26a1/Mdk/Kdr/Npnt/Spock2/Nrp1/Fermt2/Fn1/Foxf1/Col16a1/Nid1/Tek/Has2/Cdh13/Egflam/Thy1/Smoc1/Ppm1f/Vegfc/Emilin1/Ndnf/Pcsk5/Abi3bp/Net1/Efemp2/Ptn | 27 |
| GO:0022604 | regulation of cell morphogenesis | BP | 0.13 | 4.43e-11 | 4.5e-09 | 8.35 | 3.06e-09 | Mdk/Kdr/Plxnb1/Ptprd/Epb41l3/Cpne5/Cd44/Nrp1/Fermt2/Wasf3/Fn1/Icam1/Has2/Tbc1d24/Obsl1/Postn/Plxnd1/Msn/Coro1a/Kit/Myo9a/Caprin2/Itga7/Arap3/Slc23a2/Rhobtb3/Parvb/Cfap410/Cacng7/Syne3/Dlc1/Net1/Fyn/Bhlhb9/Fgd1/Dlg4/Cux2/Fgr/Palm/Cdc42ep5/Ptn/Arhgap15/Ntng1/Plekho1 | 44 |
| GO:0043113 | receptor clustering | BP | 0.26 | 4.53e-11 | 4.52e-09 | 8.35 | 3.07e-09 | Itga4/Nlgn1/Lrrc7/Cdh2/Itgb7/Grin2b/Slitrk3/Thy1/Glrb/Dlg2/Itgal/Grik2/Apoe/Magi2/Tnfaip6/Dlg4/Syngap1/Arhgef9/Ptn/Cacna1a | 20 |
| GO:0006936 | muscle contraction | BP | 0.134 | 5.14e-11 | 5.02e-09 | 8.3 | 3.42e-09 | Sgcd/Cacna1c/Ryr1/Kcnd3/Tbx2/Adra1a/Npnt/Abat/Slc8a1/Scn4b/Mybpc1/P2rx2/Tmod2/Scn1a/Ednrb/Cacna2d1/Stc1/Edn3/Ank2/Slc8a3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Kcnma1/Tbx3/Ednra/Bdkrb2/Htr2a/Myl3/Bin1/Stac2/Ryr3/Pde5a/Cacna1h/Ptger3/Calcrl/Ccn2/Atp2b4/Atp1a2/Cav1 | 42 |
| GO:0042391 | regulation of membrane potential | BP | 0.113 | 1.07e-10 | 1.02e-08 | 7.99 | 6.92e-09 | Kcnk3/Nalcn/Grik1/Cacna1c/Begain/Kcnd2/Kdr/Kcnd3/Nlgn1/Adra1a/Abat/Celf4/Rims3/Slc8a1/Scn4b/Drd4/Stx1b/P2rx2/Scn1a/Bok/Slc24a4/Cacna2d1/Lrrk2/Ano1/Mef2c/Grin2b/Tbc1d24/Ank2/Slc8a3/Glrb/Gabra4/Slc29a1/Scn2a/Nlgn3/Grik2/Kcnma1/Kcnc4/Bin1/Fhl1/Mpp2/Cacna1h/Scn3a/Dlg4/Cux2/Gimap5/Gimap3/Ptn/Piezo2/Cav1/Slc4a8/Cacna1a/Pid1/Cacnb4 | 53 |
| GO:0097485 | neuron projection guidance | BP | 0.143 | 1.08e-10 | 1.02e-08 | 7.99 | 6.92e-09 | Gap43/Epha8/Prtg/Efnb3/Sema5a/Pla2g10/Dcc/Nog/Plxnb1/Evl/Sema5b/Rnf165/Ptpro/Sema7a/Nrp1/Dpysl4/Mef2c/Cxcl12/Arhgef25/Edn3/Gli3/Lrp2/Gdf7/Plxnd1/Ptprm/Lama2/Epha3/Matn2/Nfasc/Ednra/Sema3g/Rtn4rl1/Epha7/Ptch1/Neo1/Unc5c/Sema6a | 37 |
| GO:0006939 | smooth muscle contraction | BP | 0.205 | 1.18e-10 | 1.09e-08 | 7.96 | 7.44e-09 | Cacna1c/Tbx2/Adra1a/Npnt/Abat/Slc8a1/P2rx2/Ednrb/Edn3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Kcnma1/Tbx3/Ednra/Bdkrb2/Htr2a/Ptger3/Calcrl/Atp2b4/Atp1a2/Cav1 | 24 |
| GO:0003012 | muscle system process | BP | 0.116 | 1.32e-10 | 1.2e-08 | 7.92 | 8.17e-09 | Ar/Sgcd/Cacna1c/Col14a1/Ryr1/Igf1/Kcnd3/Tbx2/Adra1a/Npnt/Abat/Nos3/Slc8a1/Scn4b/Mybpc1/P2rx2/Tmod2/Scn1a/Ednrb/Cacna2d1/Mef2c/Stc1/Edn3/Ank2/Slc8a3/Calca/Tshz3/Gucy1a1/Kit/Cd38/Kcnma1/Tbx3/Ednra/Bdkrb2/Htr2a/Myl3/Cmya5/Bin1/Stac2/Ryr3/Selenon/Pde5a/Cacna1h/Ptger3/Calcrl/Ccn2/Pi16/Atp2b4/Atp1a2/Cav1 | 50 |
| GO:0060485 | mesenchyme development | BP | 0.134 | 1.47e-10 | 1.32e-08 | 7.88 | 8.95e-09 | Sfrp2/Sema5a/Lef1/Mdk/Wnt11/Itga4/Nog/Tbx2/Sema5b/Mark1/Zeb2/Nos3/Phldb2/Sema7a/Nrp1/Fermt2/Cdh2/Ednrb/Fn1/Foxf1/Mef2c/Erg/Has2/Edn3/Tcf21/Loxl3/Epha3/Aplnr/Tbx3/Heyl/Ednra/Snai1/Sema3g/Spred3/Kitl/Osr1/Dchs1/Il17rd/Dlg5/Sema6a | 40 |
| GO:0010810 | regulation of cell-substrate adhesion | BP | 0.15 | 1.69e-10 | 1.49e-08 | 7.83 | 1.01e-08 | Ptprz1/Vit/Col26a1/Mdk/Kdr/Npnt/Phldb2/Spock2/Nrp1/Fermt2/Fn1/Foxf1/Col16a1/Nid1/Tek/Has2/Cdh13/Egflam/Postn/Thy1/Smoc1/Epha3/Ppm1f/Limch1/Vegfc/Arhgap6/Emilin1/Ndnf/Pcsk5/Dlc1/Abi3bp/Net1/Efemp2/Ptn | 34 |
| GO:0050679 | positive regulation of epithelial cell proliferation | BP | 0.152 | 2.08e-10 | 1.77e-08 | 7.75 | 1.21e-08 | Ihh/Sema5a/Ar/Mdk/Itga4/Kdr/Agap2/Nog/Cyp7b1/Igf1/Ccnd2/Osr2/Prkd1/Cxcl12/Has2/Cdh13/Stxbp4/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Igf2/Twist2/Dysf/Osr1/Apln/Akt3/Egfl7/Ptn/Cav1/Vash2/Cldn1 | 33 |
| GO:0016055 | Wnt signaling pathway | BP | 0.113 | 2.34e-10 | 1.93e-08 | 7.71 | 1.31e-08 | Gli1/Ndp/Sfrp2/Sema5a/Wnt6/Lef1/Mdk/Apcdd1/Wnt11/Pygo1/Nog/Mark1/Zeb2/Ptpro/Nkd1/Cd44/Fermt2/Tert/Cdh2/Gpc3/Arl6/Ednrb/Lrrk2/Tmem237/Rbms3/Gprc5b/Gli3/Peg12/Cpe/Plpp3/Wnt9a/Dkk3/Znrf3/Tsku/Apoe/Enpp1/Caprin2/Ednra/Daam2/Prickle1/Pkd1/Tle1/Nfatc4/Mgat3/Pkd2/Fzd2/Lats2/Adgra2/Cav1/Dact1/Nkd2 | 51 |
| GO:0001505 | regulation of neurotransmitter levels | BP | 0.142 | 2.4e-10 | 1.93e-08 | 7.71 | 1.31e-08 | Syn2/Maob/Slc6a2/Nlgn1/Adra1a/Abat/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Pde1b/Maoa/Edn3/Ppfia2/Htr1b/Naalad2/Syde1/Cadps/Slc29a1/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Erc2/Ncs1/Atp1a2/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 36 |
| GO:0035296 | regulation of tube diameter | BP | 0.168 | 2.45e-10 | 1.93e-08 | 7.71 | 1.31e-08 | Cacna1c/Kdr/Adra1a/Nos3/Slc8a1/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Calca/Htr1b/Ptprm/Gucy1a1/Apoe/Cd38/Agtr1a/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1 | 29 |
| GO:0097746 | blood vessel diameter maintenance | BP | 0.168 | 2.45e-10 | 1.93e-08 | 7.71 | 1.31e-08 | Cacna1c/Kdr/Adra1a/Nos3/Slc8a1/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Calca/Htr1b/Ptprm/Gucy1a1/Apoe/Cd38/Agtr1a/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1 | 29 |
| GO:1903037 | regulation of leukocyte cell-cell adhesion | BP | 0.125 | 2.47e-10 | 1.93e-08 | 7.71 | 1.31e-08 | Ihh/Efnb3/Lef1/Mdk/Itga4/Cd83/Igf1/Chst2/Cd44/Ets1/Icam1/Cxcl12/Has2/Pik3r6/Gli3/Tnfsf13b/Thy1/Cd1d1/Loxl3/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Sele/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/Pde5a/H2-Aa/Stat5b/Gimap5/Zap70/Dlg5/Gimap3/St3gal4/Il12rb1/Cav1/Il2rg | 43 |
| GO:0099173 | postsynapse organization | BP | 0.155 | 2.53e-10 | 1.95e-08 | 7.71 | 1.32e-08 | Gap43/Ptprd/Nlgn1/Nrp1/Cdh2/Lrrc4b/Lrrk2/Nptx1/Grin2b/Shank2/Ppfia2/Slitrk3/Dock10/Glrb/Kif1a/Abi2/Nlgn3/Dgkb/Nptxr/Apoe/Magi2/Caprin2/Epha7/Lrfn2/Fyn/Bhlhb9/Dlg4/Cux2/Syngap1/Ngef/Arhgef9/Ptn | 32 |
| GO:0198738 | cell-cell signaling by wnt | BP | 0.113 | 2.74e-10 | 2.05e-08 | 7.69 | 1.39e-08 | Gli1/Ndp/Sfrp2/Sema5a/Wnt6/Lef1/Mdk/Apcdd1/Wnt11/Pygo1/Nog/Mark1/Zeb2/Ptpro/Nkd1/Cd44/Fermt2/Tert/Cdh2/Gpc3/Arl6/Ednrb/Lrrk2/Tmem237/Rbms3/Gprc5b/Gli3/Peg12/Cpe/Plpp3/Wnt9a/Dkk3/Znrf3/Tsku/Apoe/Enpp1/Caprin2/Ednra/Daam2/Prickle1/Pkd1/Tle1/Nfatc4/Mgat3/Pkd2/Fzd2/Lats2/Adgra2/Cav1/Dact1/Nkd2 | 51 |
| GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | BP | 0.205 | 2.79e-10 | 2.05e-08 | 7.69 | 1.39e-08 | Prtg/Hmcn1/Pcdh18/Sdk2/Cdh2/Cdh6/Tenm3/Cdh13/Pcdhgc3/Obsl1/Cdh10/Cdh5/Pcdh19/Ptprm/Pcdh12/Esam/Palld/Cdh11/Pecam1/Dchs1/Fat3/Fat4/Pcdhb14 | 23 |
| GO:0035150 | regulation of tube size | BP | 0.167 | 2.82e-10 | 2.05e-08 | 7.69 | 1.39e-08 | Cacna1c/Kdr/Adra1a/Nos3/Slc8a1/Cbs/Npr3/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Itga1/Calca/Htr1b/Ptprm/Gucy1a1/Apoe/Cd38/Agtr1a/Kcnma1/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1 | 29 |
| GO:1903169 | regulation of calcium ion transmembrane transport | BP | 0.167 | 2.82e-10 | 2.05e-08 | 7.69 | 1.39e-08 | Cacna1c/Slc8a1/Drd4/Cacna2d1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Cracr2a/Agtr1a/Ednra/Tspan13/Bin1/Stac2/Dysf/Asph/Selenon/Rrad/Ptger3/Fyn/Pkd2/Gstm7/Atp1a2/Gnb5/Cacna1a/Cacnb4 | 29 |
| GO:0045765 | regulation of angiogenesis | BP | 0.13 | 3.28e-10 | 2.32e-08 | 7.63 | 1.58e-08 | Cyp1b1/Hgf/Sfrp2/Sema5a/Hhip/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Tek/Cxcr3/Pik3r6/Adamts1/Vash1/Cdh5/Plxnd1/Ptprm/Minar1/Ppp1r16b/Aplnr/Jcad/Vegfc/Ecscr/Agtr1a/Emilin1/Tie1/Igf2/Tcf4/Hipk2/Cldn5/Atp2b4/Akt3/Sema6a/Ptn/Adgra2/Vash2 | 40 |
| GO:0030326 | embryonic limb morphogenesis | BP | 0.182 | 3.32e-10 | 2.32e-08 | 7.63 | 1.58e-08 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Tbx2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Tbx3/Dync2h1/Hoxa9/Ptch1/Tmem231/Osr1/Ttbk2/Rab23 | 26 |
| GO:0035113 | embryonic appendage morphogenesis | BP | 0.182 | 3.32e-10 | 2.32e-08 | 7.63 | 1.58e-08 | Ihh/Sfrp2/Lef1/Cacna1c/Hoxd10/Hoxd11/Nog/Aff3/Tbx2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Osr2/Flvcr1/Gli3/Wnt9a/Cplane1/Tbx3/Dync2h1/Hoxa9/Ptch1/Tmem231/Osr1/Ttbk2/Rab23 | 26 |
| GO:0007411 | axon guidance | BP | 0.14 | 3.74e-10 | 2.58e-08 | 7.59 | 1.75e-08 | Gap43/Epha8/Prtg/Efnb3/Sema5a/Pla2g10/Dcc/Nog/Plxnb1/Evl/Sema5b/Rnf165/Ptpro/Sema7a/Nrp1/Mef2c/Cxcl12/Arhgef25/Edn3/Gli3/Lrp2/Gdf7/Plxnd1/Ptprm/Lama2/Epha3/Matn2/Nfasc/Ednra/Sema3g/Rtn4rl1/Epha7/Ptch1/Neo1/Unc5c/Sema6a | 36 |
| GO:0097553 | calcium ion transmembrane import into cytosol | BP | 0.174 | 4.16e-10 | 2.83e-08 | 7.55 | 1.93e-08 | Cacna1c/Ryr1/Slc8a1/Slc24a4/Myo5a/Grin2b/Prkd1/Cxcr3/Cacna1e/Ank2/Slc8a3/Nalf1/Thy1/Coro1a/Aplnr/Agtr1a/Ednra/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Fyn/Pkd2/Gstm7/Cacna1a | 27 |
| GO:1901342 | regulation of vasculature development | BP | 0.129 | 4.84e-10 | 3.25e-08 | 7.49 | 2.21e-08 | Cyp1b1/Hgf/Sfrp2/Sema5a/Hhip/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Tek/Cxcr3/Pik3r6/Adamts1/Vash1/Cdh5/Plxnd1/Ptprm/Minar1/Ppp1r16b/Aplnr/Jcad/Vegfc/Ecscr/Agtr1a/Emilin1/Tie1/Igf2/Tcf4/Hipk2/Cldn5/Atp2b4/Akt3/Sema6a/Ptn/Adgra2/Vash2 | 40 |
| GO:0010631 | epithelial cell migration | BP | 0.128 | 5.86e-10 | 3.87e-08 | 7.41 | 2.63e-08 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Evl/Zeb2/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Ptp4a3/Stc1/Prkd1/Tek/Has2/Cdh13/Vash1/Calca/Cdh5/Plxnd1/Plpp3/Ptprm/Kit/Ppm1f/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Daam2/Pecam1/Hdac9/Atp2b4/Akt3/Fstl1/Arsb/Adgra2/Mapre2 | 40 |
| GO:0032956 | regulation of actin cytoskeleton organization | BP | 0.12 | 5.9e-10 | 3.87e-08 | 7.41 | 2.63e-08 | Kank4/Sema5a/Mdk/Wnt11/Evl/Phldb2/Ccdc88a/Hcls1/Nrp1/Fermt2/Tmod2/Wasf3/Mef2c/Pfn2/Icam1/Tek/Jam3/Coro1a/Abi2/Epha3/Ppm1f/Esam/Sh3bp1/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Daam2/Pecam1/Ppm1e/Scin/Nckap1l/Carmil3/Rhobtb3/Bin1/Myo1f/Arhgap28/Dlc1/Ccn2/Fgr/Cdc42ep5/Cyfip2/Plekhh2 | 44 |
| GO:1904062 | regulation of cation transmembrane transport | BP | 0.118 | 6.68e-10 | 4.33e-08 | 7.36 | 2.95e-08 | Hecw2/Cacna1c/Nlgn1/Slc8a1/Scn4b/Drd4/Fxyd5/Cacna2d1/Mef2c/Myo5a/Prkd1/Cxcr3/Edn3/Ank2/Plcb1/Thy1/Coro1a/Nlgn3/Aplnr/Cracr2a/Prrt1/Agtr1a/Ednra/Tspan13/Bin1/Stac2/Cacng7/Dysf/Kcnip3/Fhl1/Asph/Selenon/Osr1/Rrad/Ptger3/Fyn/Pkd2/Dlg4/Gstm7/Atp2b4/Atp1a2/Gnb5/Cav1/Cacna1a/Cacnb4 | 45 |
| GO:0090132 | epithelium migration | BP | 0.127 | 7.09e-10 | 4.54e-08 | 7.34 | 3.09e-08 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Evl/Zeb2/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Ptp4a3/Stc1/Prkd1/Tek/Has2/Cdh13/Vash1/Calca/Cdh5/Plxnd1/Plpp3/Ptprm/Kit/Ppm1f/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Daam2/Pecam1/Hdac9/Atp2b4/Akt3/Fstl1/Arsb/Adgra2/Mapre2 | 40 |
| GO:0014848 | urinary tract smooth muscle contraction | BP | 0.643 | 7.17e-10 | 4.54e-08 | 7.34 | 3.09e-08 | Cacna1c/Tbx2/P2rx2/Tshz3/Kcnma1/Tbx3/Htr2a/Ptger3/Atp2b4 | 9 |
| GO:0016358 | dendrite development | BP | 0.126 | 8.56e-10 | 5.35e-08 | 7.27 | 3.64e-08 | Ptprz1/Map2/Hecw2/Ptprd/Nlgn1/Mark1/Tet1/Mpdz/Nrp1/Dab1/Lrrk2/Mef2c/Tbc1d24/Obsl1/Ppfia2/D16Ertd472e/Dock10/Trpc6/Kif1a/Abi2/Nlgn3/Matn2/Apoe/Caprin2/Prex2/Sarm1/Nfatc4/Fyn/Bhlhb9/Dlg4/Cux2/Fat3/Palm/Dlg5/Syngap1/Ngef/Ptn/Bbs1/Dact1/Cacna1a | 40 |
| GO:0048762 | mesenchymal cell differentiation | BP | 0.14 | 9.52e-10 | 5.89e-08 | 7.23 | 4.01e-08 | Sfrp2/Sema5a/Lef1/Mdk/Wnt11/Nog/Sema5b/Mark1/Zeb2/Phldb2/Sema7a/Nrp1/Fermt2/Cdh2/Ednrb/Fn1/Mef2c/Erg/Has2/Edn3/Tcf21/Loxl3/Epha3/Tbx3/Heyl/Ednra/Snai1/Sema3g/Spred3/Kitl/Osr1/Il17rd/Dlg5/Sema6a | 34 |
| GO:0009100 | glycoprotein metabolic process | BP | 0.123 | 1.01e-09 | 6.19e-08 | 7.21 | 4.21e-08 | Col11a1/Ihh/St6gal2/Itm2a/Igf1/St8sia2/St8sia4/Gxylt2/Tet1/Man2a2/Spock2/Uggt2/Chst10/Soat1/Tmtc1/B3galnt1/Fut10/Egflam/Plcb1/Galnt16/Mamdc2/St6galnac4/Fbxo17/St3gal2/Eogt/Ggta1/Ndnf/Adamts7/St8sia6/Galntl6/Bgn/Poglut2/Aqp11/Mgat3/B3gnt5/Xylt1/Dse/St6galnac6/St3gal4/Large1/Alg14 | 41 |
| GO:0007015 | actin filament organization | BP | 0.109 | 1.18e-09 | 7.15e-08 | 7.15 | 4.87e-08 | Kank4/Hmcn1/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Hcls1/Nrp1/Fermt2/Aif1l/Tmod2/Wasf3/Myo5a/Pfn2/Icam1/Mical1/Arhgap25/Catip/Coro1a/Abi2/Ppm1f/Esam/Sh3bp1/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Daam2/Pecam1/Ppm1e/Scin/Nckap1l/Carmil3/Hip1/Efs/Rhobtb3/Bin1/Arrb1/Myo1f/Arhgap28/Dlc1/Ccn2/Wipf1/Dpysl3/Cdc42ep5/Cyfip2/Plekhh2/Shroom1 | 50 |
| GO:1901888 | regulation of cell junction assembly | BP | 0.145 | 1.22e-09 | 7.32e-08 | 7.14 | 4.98e-08 | Gpc6/Kdr/Ptprd/St8sia2/Nlgn1/Phldb2/Nrp1/Fermt2/Lrrc4b/Col16a1/Nptx1/Mef2c/Tek/Adgrl1/Slitrk3/Thy1/Epha3/Ppm1f/Nlgn3/Aplnr/Limch1/Arhgap6/Snai1/Epha7/Dlc1/Cldn5/Bhlhb9/Cux2/Nphp1/Dlg5/Cav1/Cldn1 | 32 |
| GO:0048705 | skeletal system morphogenesis | BP | 0.134 | 1.92e-09 | 1.13e-07 | 6.95 | 7.71e-08 | Col11a1/Ihh/Mmp16/Sfrp2/Hhip/Hoxd10/Hoxd11/Nog/Thbs3/Cbs/Hoxa11/Hoxd9/Mmp2/Mef2c/Osr2/Hoxd8/Stc1/Hoxd4/Has2/Flvcr1/Gli3/Frem1/Col13a1/Wnt9a/Zeb1/Enpp1/Hoxd3/Hoxa9/Ltbp3/Twist2/Pkd1/Osr1/Ccn2/Fgr/Rab23 | 35 |
| GO:0045055 | regulated exocytosis | BP | 0.129 | 2.01e-09 | 1.18e-07 | 6.93 | 8.01e-08 | Syn2/Cacna1c/Nlgn1/Syt13/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Foxf1/Pfn2/Mical1/Ppfia2/Htr1b/Syde1/Cadps/Coro1a/Kit/Syt7/Prrt2/Htr2a/Nckap1l/Erc2/Ncs1/Myo1f/Cd84/Cacna1h/Fgr/Lat/Rab27a/Clnk/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 37 |
| GO:0008589 | regulation of smoothened signaling pathway | BP | 0.213 | 2.09e-09 | 1.21e-07 | 6.92 | 8.24e-08 | Gli1/Ihh/Hhip/Evc/Kif7/Gpc3/Arl6/Serpine2/Cibar1/Armc9/Gli3/Rab34/Wnt9a/Gpr161/Enpp1/Dync2h1/Ptch1/Ttbk2/Rab23/Dlg5 | 20 |
| GO:0018212 | peptidyl-tyrosine modification | BP | 0.122 | 2.14e-09 | 1.22e-07 | 6.91 | 8.32e-08 | Ptprz1/Hgf/Sfrp2/Angpt1/Kdr/Spink1/Igf1/Pkdcc/Ddr2/Adra1a/Cd44/Hcls1/Nrp1/Cspg4/Areg/Icam1/Itk/Tek/Tpst1/Gprc5b/Thy1/Plpp3/Abi2/Kit/Epha3/Vegfc/Nox4/Agtr1a/Igf2/Pecam1/Htr2a/Kitl/Epha7/Arrb2/Fyn/Dlg4/Fgr/Zap70/Il12rb1/Cav1 | 40 |
| GO:0001936 | regulation of endothelial cell proliferation | BP | 0.162 | 2.3e-09 | 1.3e-07 | 6.89 | 8.84e-08 | Sema5a/Mdk/Itga4/Kdr/Igf1/Mef2c/Prkd1/Tek/Cxcr3/Cxcl12/Cdh13/Vash1/Ptprm/Ppp1r16b/Aplnr/Jcad/Vegfc/Apoe/Agtr1a/Tie1/Igf2/Dysf/Apln/Akt3/Egfl7/Cav1/Vash2 | 27 |
| GO:0110053 | regulation of actin filament organization | BP | 0.13 | 2.42e-09 | 1.34e-07 | 6.87 | 9.12e-08 | Kank4/Sema5a/Wnt11/Evl/Phldb2/Ccdc88a/Hcls1/Nrp1/Fermt2/Tmod2/Wasf3/Pfn2/Icam1/Coro1a/Abi2/Ppm1f/Esam/Sh3bp1/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Daam2/Pecam1/Ppm1e/Scin/Nckap1l/Carmil3/Bin1/Arhgap28/Dlc1/Ccn2/Cdc42ep5/Cyfip2/Plekhh2 | 36 |
| GO:0008360 | regulation of cell shape | BP | 0.166 | 2.68e-09 | 1.47e-07 | 6.83 | 1e-07 | Kdr/Plxnb1/Epb41l3/Fermt2/Wasf3/Fn1/Icam1/Plxnd1/Msn/Coro1a/Kit/Itga7/Arap3/Rhobtb3/Parvb/Cfap410/Syne3/Dlc1/Fyn/Fgd1/Fgr/Palm/Cdc42ep5/Ptn/Arhgap15/Plekho1 | 26 |
| GO:0030336 | negative regulation of cell migration | BP | 0.127 | 3.25e-09 | 1.77e-07 | 6.75 | 1.2e-07 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Tcaf1/Phldb2/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Thy1/Ptprm/Gpr173/Il27ra/Robo4/Limch1/Apoe/Tmeff2/Magi2/Emilin1/Tie1/Cd200/Cnn2/Arap3/Tnfaip6/Dlc1/Cldn5/Atp2b4/Egfl7/Dpysl3/Dlg5/Ptn/Cygb | 37 |
| GO:0006836 | neurotransmitter transport | BP | 0.136 | 3.61e-09 | 1.94e-07 | 6.71 | 1.32e-07 | Syn2/Slc6a2/Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Slc6a15/Syde1/Cadps/Slc29a1/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Sv2c/Erc2/Ncs1/Atp1a2/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 33 |
| GO:0001935 | endothelial cell proliferation | BP | 0.153 | 4.17e-09 | 2.21e-07 | 6.66 | 1.5e-07 | Sema5a/Mdk/Itga4/Kdr/Igf1/Mef2c/Prkd1/Tek/Cxcr3/Cxcl12/Cdh13/Vash1/Calca/Ptprm/Ppp1r16b/Aplnr/Jcad/Vegfc/Apoe/Agtr1a/Tie1/Igf2/Dysf/Apln/Akt3/Egfl7/Cav1/Vash2 | 28 |
| GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.133 | 4.18e-09 | 2.21e-07 | 6.66 | 1.5e-07 | Sfrp2/Cd109/Kdr/Nog/Rnf165/Zeb2/Npnt/Fst/Gpc3/Gdf11/Zfp423/Lrp2/Gdf7/Cdh5/Pbld2/Zeb1/Gdf10/Cdkn1c/Tsku/Magi2/Emilin1/Spred3/Kcp/Tnfaip6/Hipk2/Nrros/Zfyve9/Neo1/Il17rd/Fstl1/Lats2/Cav1/Itga8/Tmprss6 | 34 |
| GO:0019932 | second-messenger-mediated signaling | BP | 0.123 | 4.63e-09 | 2.42e-07 | 6.62 | 1.64e-07 | Sgcd/Kdr/Spink1/Nr5a2/Igf1/Nos3/Slc8a1/Cbs/Drd4/Gnai1/Ednrb/Slc24a4/Lrrk2/Gucy1a2/Cxcr3/Cdh13/Ptgfr/Edn3/Ank2/Adgrl1/Gucy1b1/Gucy1a1/Aplnr/Itgal/Apoe/Agtr1a/Ndnf/Adcyap1r1/Cap2/Cmya5/Rasd2/Xcr1/Pde5a/Nfatc4/Ptger3/Atp2b4/Zap70/Lat | 38 |
| GO:0060348 | bone development | BP | 0.132 | 5.12e-09 | 2.65e-07 | 6.58 | 1.8e-07 | Ihh/Mmp16/Sfrp2/Hoxd11/Ryr1/Kdr/Plxnb1/Igf1/Ddr2/Evc/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Has2/Lrrc17/Gli3/Frem1/Col13a1/Fli1/Kit/Enpp1/Adamts7/Tal1/Smad9/Ltbp3/Bgn/P3h1/Dchs1/Xylt1/Rab23/Fat4 | 34 |
| GO:0050803 | regulation of synapse structure or activity | BP | 0.129 | 5.21e-09 | 2.66e-07 | 6.57 | 1.81e-07 | Gpc6/Ptprd/St8sia2/Nlgn1/Ptpro/Rims3/Cdh2/Lrrc4b/Lrrk2/Nptx1/Mef2c/Grin2b/Shank2/Adgrl1/Ppfia2/Slitrk3/Kif1a/Abi2/Nlgn3/Dgkb/Arhgef15/Apoe/Sparcl1/Magi2/Caprin2/Epha7/Lrfn2/Nfatc4/Fyn/Bhlhb9/Cux2/Dlg5/Syngap1/Ngef/Ptn | 35 |
| GO:0007269 | neurotransmitter secretion | BP | 0.151 | 5.34e-09 | 2.66e-07 | 6.57 | 1.81e-07 | Syn2/Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Syde1/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Erc2/Ncs1/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 28 |
| GO:0099643 | signal release from synapse | BP | 0.151 | 5.34e-09 | 2.66e-07 | 6.57 | 1.81e-07 | Syn2/Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Syde1/Cadps/Lin7a/Syt7/Prrt2/Kcnc4/Htr2a/Erc2/Ncs1/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 28 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | BP | 0.12 | 5.35e-09 | 2.66e-07 | 6.57 | 1.81e-07 | Ptprz1/Hgf/Sfrp2/Angpt1/Kdr/Spink1/Igf1/Pkdcc/Ddr2/Adra1a/Cd44/Hcls1/Nrp1/Cspg4/Areg/Icam1/Itk/Tek/Gprc5b/Thy1/Plpp3/Abi2/Kit/Epha3/Vegfc/Nox4/Agtr1a/Igf2/Pecam1/Htr2a/Kitl/Epha7/Arrb2/Fyn/Dlg4/Fgr/Zap70/Il12rb1/Cav1 | 39 |
| GO:0001938 | positive regulation of endothelial cell proliferation | BP | 0.202 | 5.43e-09 | 2.68e-07 | 6.57 | 1.82e-07 | Sema5a/Mdk/Itga4/Kdr/Igf1/Prkd1/Cxcl12/Cdh13/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Igf2/Dysf/Apln/Akt3/Egfl7/Cav1/Vash2 | 20 |
| GO:0072073 | kidney epithelium development | BP | 0.164 | 6.22e-09 | 3.02e-07 | 6.52 | 2.05e-07 | Wnt6/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Gdf11/Gli3/Tcf21/Tshz3/Agtr1a/Heyl/Ednra/Smad9/Epha7/Ptch1/Osr1/Dchs1/Aqp11/Fat4 | 25 |
| GO:0007162 | negative regulation of cell adhesion | BP | 0.121 | 6.61e-09 | 3.18e-07 | 6.5 | 2.16e-07 | Ihh/Cyp1b1/Ptprz1/Sema5a/Angpt1/Lef1/Mdk/Plxnb1/Abat/Phldb2/Cd44/Dab1/Mmp2/Serpine2/Cxcl12/Jam3/Cdh13/Gli3/Podxl/Postn/Plxnd1/Loxl3/Ccm2l/Ppm1f/Btla/H2-Ab1/Arhgap6/Havcr2/Nckap1l/Myo1f/Pde5a/Dlc1/Ptger3/H2-Aa/Gimap5/Dlg5/Gimap3/Sema6a | 38 |
| GO:0051271 | negative regulation of cellular component movement | BP | 0.121 | 7.22e-09 | 3.44e-07 | 6.46 | 2.34e-07 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Tcaf1/Phldb2/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Thy1/Ptprm/Gpr173/Il27ra/Robo4/Limch1/Apoe/Tmeff2/Magi2/Emilin1/Tie1/Cd200/Cnn2/Arap3/Tnfaip6/Bin1/Dlc1/Cldn5/Atp2b4/Egfl7/Dpysl3/Dlg5/Ptn/Cygb | 38 |
| GO:0048608 | reproductive structure development | BP | 0.103 | 7.53e-09 | 3.55e-07 | 6.45 | 2.42e-07 | Ndp/Tex15/Stra6/Sfrp2/Angpt1/Ar/Lef1/Itga4/Kdr/Nog/Cyp7b1/Igf1/Nos3/Fst/Slc8a1/Cd44/Hoxa11/Hoxa10/Il11ra1/Mmp2/Icam1/Id4/Serpine2/Adamts1/Vash1/Tcf21/Lrp2/Gdf7/Syde1/Prdm1/Lfng/Pcdh12/Kit/Cdkn1c/Tbx3/Igf2/Snai1/Hoxa9/Rhobtb3/Kitl/Tnfaip6/Arrb1/Pkd1/Osr1/Arrb2/Pkd2/Stat5b/Abcb1a/Ptn/Vash2 | 50 |
| GO:0050807 | regulation of synapse organization | BP | 0.129 | 8.38e-09 | 3.92e-07 | 6.41 | 2.67e-07 | Gpc6/Ptprd/St8sia2/Nlgn1/Ptpro/Rims3/Cdh2/Lrrc4b/Lrrk2/Nptx1/Mef2c/Grin2b/Shank2/Adgrl1/Ppfia2/Slitrk3/Kif1a/Abi2/Nlgn3/Dgkb/Arhgef15/Apoe/Sparcl1/Magi2/Caprin2/Epha7/Lrfn2/Nfatc4/Fyn/Bhlhb9/Cux2/Dlg5/Ngef/Ptn | 34 |
| GO:0048880 | sensory system development | BP | 0.107 | 8.85e-09 | 4.11e-07 | 6.39 | 2.79e-07 | Ndp/Tmem132e/Stra6/Ihh/Cyp1b1/Sdk2/Nhs/Cacna1c/Kdr/Tbx2/Tub/Rcn1/Zeb2/Celf4/Lamc3/Nrp1/Arl6/Tenm3/Osr2/Gdf11/Bbs7/Obsl1/Sall2/Gli3/Thy1/Zeb1/Prdm1/Tgif2/Smoc1/Ptprm/Abi2/Rp1/Cdkn1c/Tsku/Arhgef15/Pbx4/Spred3/Rorb/Lpcat1/Col5a2/Twist2/Hipk2/Tmem231/Fat3/Nphp1/Ptn | 46 |
| GO:0007043 | cell-cell junction assembly | BP | 0.161 | 9.37e-09 | 4.31e-07 | 6.37 | 2.93e-07 | Wnt11/Epb41l3/Ptpro/Mpdz/Cdh2/Cdh6/Jam3/Cldn8/Cdh10/Cdh5/Abi2/Aplnr/Esam/Nfasc/Cdh11/Gjc1/Pecam1/Snai1/Heg1/Dchs1/Cldn5/Nphp1/Dlg5/Cav1/Cldn1 | 25 |
| GO:0060070 | canonical Wnt signaling pathway | BP | 0.122 | 9.74e-09 | 4.44e-07 | 6.35 | 3.02e-07 | Gli1/Sfrp2/Sema5a/Wnt6/Lef1/Mdk/Wnt11/Pygo1/Nog/Zeb2/Ptpro/Nkd1/Cdh2/Gpc3/Ednrb/Lrrk2/Rbms3/Gprc5b/Gli3/Peg12/Plpp3/Wnt9a/Dkk3/Znrf3/Apoe/Caprin2/Ednra/Daam2/Prickle1/Tle1/Mgat3/Fzd2/Lats2/Adgra2/Cav1/Dact1/Nkd2 | 37 |
| GO:0061458 | reproductive system development | BP | 0.102 | 9.86e-09 | 4.46e-07 | 6.35 | 3.03e-07 | Ndp/Tex15/Stra6/Sfrp2/Angpt1/Ar/Lef1/Itga4/Kdr/Nog/Cyp7b1/Igf1/Nos3/Fst/Slc8a1/Cd44/Hoxa11/Hoxa10/Il11ra1/Mmp2/Icam1/Id4/Serpine2/Adamts1/Vash1/Tcf21/Lrp2/Gdf7/Syde1/Prdm1/Lfng/Pcdh12/Kit/Cdkn1c/Tbx3/Igf2/Snai1/Hoxa9/Rhobtb3/Kitl/Tnfaip6/Arrb1/Pkd1/Osr1/Arrb2/Pkd2/Stat5b/Abcb1a/Ptn/Vash2 | 50 |
| GO:2000146 | negative regulation of cell motility | BP | 0.121 | 1.06e-08 | 4.77e-07 | 6.32 | 3.24e-07 | Cyp1b1/Sfrp2/Wnt11/Nog/Evl/Tcaf1/Phldb2/Drd4/Mef2c/Pfn2/Stc1/Cxcl12/Vash1/Plcb1/Thy1/Ptprm/Gpr173/Il27ra/Robo4/Limch1/Apoe/Tmeff2/Magi2/Emilin1/Tie1/Cd200/Cnn2/Arap3/Tnfaip6/Dlc1/Cldn5/Atp2b4/Egfl7/Dpysl3/Dlg5/Ptn/Cygb | 37 |
| GO:0043542 | endothelial cell migration | BP | 0.136 | 1.14e-08 | 5.08e-07 | 6.29 | 3.46e-07 | Cyp1b1/Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Ptp4a3/Stc1/Prkd1/Tek/Cdh13/Vash1/Calca/Cdh5/Plxnd1/Plpp3/Ptprm/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Pecam1/Hdac9/Atp2b4/Akt3/Fstl1/Adgra2 | 31 |
| GO:0072006 | nephron development | BP | 0.159 | 1.22e-08 | 5.36e-07 | 6.27 | 3.64e-07 | Wnt6/Angpt1/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Nid1/Tek/Enpep/Gli3/Tcf21/Agtr1a/Heyl/Ednra/Ptch1/Osr1/Dchs1/Aqp11/Fat4 | 25 |
| GO:0030318 | melanocyte differentiation | BP | 0.379 | 1.32e-08 | 5.67e-07 | 6.25 | 3.86e-07 | Adamts20/Zeb2/Ednrb/Mef2c/Myo5a/Edn3/Gli3/Kit/Enpp1/Kitl/Rab27a | 11 |
| GO:0001654 | eye development | BP | 0.107 | 1.51e-08 | 6.47e-07 | 6.19 | 4.4e-07 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Nhs/Cacna1c/Kdr/Tbx2/Tub/Rcn1/Zeb2/Celf4/Lamc3/Nrp1/Arl6/Tenm3/Osr2/Gdf11/Bbs7/Obsl1/Sall2/Gli3/Thy1/Zeb1/Prdm1/Tgif2/Smoc1/Ptprm/Abi2/Rp1/Cdkn1c/Tsku/Arhgef15/Pbx4/Spred3/Rorb/Lpcat1/Col5a2/Twist2/Hipk2/Tmem231/Fat3/Nphp1/Ptn | 45 |
| GO:0030199 | collagen fibril organization | BP | 0.254 | 1.65e-08 | 7e-07 | 6.15 | 4.76e-07 | Col11a1/Cyp1b1/Sfrp2/Col14a1/Ddr2/Fkbp10/Col13a1/Loxl3/Emilin1/Adamts7/Col5a2/P3h1/Dpt/Crtap/Efemp2 | 15 |
| GO:0009581 | detection of external stimulus | BP | 0.168 | 1.67e-08 | 7.02e-07 | 6.15 | 4.78e-07 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Foxf1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Htr2a/Arrb1/Pkd1/Arrb2/Fyn/Pkd2/Pkdrej/Piezo2/Cacnb4 | 23 |
| GO:0003007 | heart morphogenesis | BP | 0.123 | 1.76e-08 | 7.32e-07 | 6.14 | 4.98e-07 | Col11a1/Ihh/Sfrp2/Angpt1/Wnt11/Ryr1/Nog/Tbx2/Nos3/Slc8a1/Nrp1/Foxf1/Mef2c/Erg/Tek/Has2/Adamts1/Lrp2/Ift57/Cpe/Plxnd1/Ccm2l/Aplnr/Tbx3/Heyl/Ednra/Snai1/Myl3/Heg1/Ptch1/Dchs1/Dlc1/Pkd2/Fzd2/Fat4 | 35 |
| GO:0060840 | artery development | BP | 0.181 | 1.77e-08 | 7.32e-07 | 6.14 | 4.98e-07 | Apob/Stra6/Mdk/Wnt11/Nog/Tbx2/Nrp1/Kif7/Foxf1/Fkbp10/Gli3/Lrp2/Plxnd1/Prdm1/Aplnr/Apoe/Enpp1/Ednra/Prickle1/Akt3/Efemp2 | 21 |
| GO:0150063 | visual system development | BP | 0.106 | 1.88e-08 | 7.71e-07 | 6.11 | 5.24e-07 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Nhs/Cacna1c/Kdr/Tbx2/Tub/Rcn1/Zeb2/Celf4/Lamc3/Nrp1/Arl6/Tenm3/Osr2/Gdf11/Bbs7/Obsl1/Sall2/Gli3/Thy1/Zeb1/Prdm1/Tgif2/Smoc1/Ptprm/Abi2/Rp1/Cdkn1c/Tsku/Arhgef15/Pbx4/Spred3/Rorb/Lpcat1/Col5a2/Twist2/Hipk2/Tmem231/Fat3/Nphp1/Ptn | 45 |
| GO:0035249 | synaptic transmission, glutamatergic | BP | 0.179 | 2.07e-08 | 8.38e-07 | 6.08 | 5.7e-07 | Grid1/Gria4/Grik1/Nlgn1/Aph1b/Cdh2/Lrrk2/Shc3/Mef2c/Myo5a/Grin2b/Serpine2/Shank2/Htr1b/Tshz3/Nlgn3/Grik2/Htr2a/Cacng7/Cacna1a/Cacnb4 | 21 |
| GO:0072009 | nephron epithelium development | BP | 0.179 | 2.07e-08 | 8.38e-07 | 6.08 | 5.7e-07 | Wnt6/Hoxd11/Nog/Npnt/Ptpro/Cd44/Hoxa11/Adamts16/Gpc3/Ednrb/Mef2c/Gli3/Tcf21/Agtr1a/Heyl/Ednra/Ptch1/Osr1/Dchs1/Aqp11/Fat4 | 21 |
| GO:0050767 | regulation of neurogenesis | BP | 0.103 | 2.12e-08 | 8.51e-07 | 6.07 | 5.79e-07 | Prtg/Ptprz1/Efnb3/Map2/Sema5a/Mdk/Kdr/Nog/Plxnb1/Ptprd/Igf1/Sema5b/Zeb2/Sema7a/Nrp1/Tert/Fn1/Dab1/Id4/Serpine2/Cxcl12/Tbc1d24/Obsl1/Gli3/Lrp2/Thy1/Plxnd1/Trpc6/Mme/Kit/Vegfc/Apoe/Heyl/Caprin2/Daam2/Sema3g/Nin/Bin1/Zcchc24/Epha7/Chodl/Nfatc4/Bhlhb9/Cux2/Syngap1/Sema6a/Ptn | 47 |
| GO:0071695 | anatomical structure maturation | BP | 0.12 | 2.22e-08 | 8.87e-07 | 6.05 | 6.03e-07 | Epha8/Ihh/Pla2g10/Ryr1/Kdr/Spink1/Plxnb1/Igf1/Thbs3/Cspg4/Ednrb/Slc24a4/Lrrk2/Mmp2/Myo5a/Gdf11/Flvcr1/Calca/Plcb1/Rps6ka2/Cdh5/Pth1r/Dlg2/Cdkn1c/Nfasc/Enpp1/Kcnma1/Ednra/Adamts7/Lhx6/Tal1/Dchs1/Septin4/Xylt1/Fat4/Cacna1a | 36 |
| GO:0001649 | osteoblast differentiation | BP | 0.135 | 2.26e-08 | 8.93e-07 | 6.05 | 6.08e-07 | Gli1/Ihh/Sfrp2/Wnt11/Nog/Igf1/Ddr2/Npnt/Fermt2/Areg/Mef2c/Id3/Id4/Prkd1/Gli3/Glis1/Nell1/Pth1r/Gdf10/Smoc1/Vegfc/Enpp1/Igf2/Rorb/Tnfaip6/Twist2/Rassf2/Ptch1/Lrp3/Gabbr1 | 30 |
| GO:0006814 | sodium ion transport | BP | 0.129 | 2.39e-08 | 9.38e-07 | 6.03 | 6.38e-07 | Nalcn/Hecw2/Slc6a2/Slc5a11/Slc13a5/Nos3/Slc8a1/Scn4b/Drd4/Scn1a/Fxyd5/Ednrb/Slc24a4/Slc9a7/Serpine2/Slc24a3/Slc8a3/Plcb1/Slc6a15/Scn2a/Maged2/Slc9a5/Ednra/Slc23a2/Osr1/Slc38a1/Pkd2/Scn3a/Atp2b4/Akt3/Atp1a2/Slc4a8 | 32 |
| GO:0050870 | positive regulation of T cell activation | BP | 0.135 | 2.51e-08 | 9.77e-07 | 6.01 | 6.64e-07 | Ihh/Efnb3/Lef1/Mdk/Cd83/Igf1/Pik3r6/Gli3/Tnfsf13b/Thy1/Cd1d1/Coro1a/Itgal/Il27ra/H2-Ab1/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il12rb1/Cav1/Il2rg | 30 |
| GO:0007229 | integrin-mediated signaling pathway | BP | 0.194 | 2.64e-08 | 1.02e-06 | 5.99 | 6.95e-07 | Itga4/Sema7a/Nrp1/Fermt2/Itgb7/Fn1/Itga1/Thy1/Plpp3/Loxl3/Itgal/Itga7/Pcsk5/Ccn2/Fgr/Lat/Ptn/Itga8/Itgb4 | 19 |
| GO:0048813 | dendrite morphogenesis | BP | 0.144 | 2.77e-08 | 1.06e-06 | 5.97 | 7.24e-07 | Ptprz1/Hecw2/Ptprd/Nlgn1/Tet1/Nrp1/Lrrk2/Tbc1d24/Obsl1/Ppfia2/Dock10/Trpc6/Kif1a/Abi2/Nlgn3/Caprin2/Prex2/Sarm1/Nfatc4/Fyn/Bhlhb9/Dlg4/Cux2/Ngef/Ptn/Dact1/Cacna1a | 27 |
| GO:0001569 | branching involved in blood vessel morphogenesis | BP | 0.289 | 2.8e-08 | 1.07e-06 | 5.97 | 7.27e-07 | Ihh/Sfrp2/Sema5a/Angpt1/Lef1/Mdk/Kdr/Nrp1/Tek/Cxcl12/Plxnd1/Ednra/Nfatc4 | 13 |
| GO:0003205 | cardiac chamber development | BP | 0.14 | 3.05e-08 | 1.15e-06 | 5.94 | 7.85e-07 | Col11a1/Stra6/Sfrp2/Wnt11/Nog/Tbx2/Nos3/Nrp1/Kif7/Foxf1/Mef2c/Tek/Ank2/Adamts1/Lrp2/Cpe/Plxnd1/Prdm1/Ccm2l/Aplnr/Cplane1/Tbx3/Heyl/Ednra/Myl3/Pcsk5/Heg1/Fzd2 | 28 |
| GO:0007264 | small GTPase mediated signal transduction | BP | 0.104 | 3.07e-08 | 1.15e-06 | 5.94 | 7.85e-07 | Igf1/Adra1a/Ccdc88a/Rasgrp3/Nrp1/Cdh2/Arl6/Garnl3/Dab1/Rasa3/Lpar4/Prkd1/Arhgef25/Cdh13/Rasgrp2/Arhgap25/Dock2/Sh2b2/Syde1/Dock10/Abi2/Sh3bp1/Apoe/Agtr1a/Rasip1/Eps8l1/Ednra/Pecam1/Adcyap1r1/Arap3/Rhobtb3/Kitl/Arrb1/Arl3/Heg1/Arhgap28/Dlc1/Net1/Arhgap31/Lat/Syngap1/Rab39b/Cdc42ep5/Arhgef9/Mapre2 | 45 |
| GO:0051216 | cartilage development | BP | 0.136 | 3.26e-08 | 1.22e-06 | 5.91 | 8.29e-07 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Nog/Pkdcc/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Gli3/Pth1r/Wnt9a/Zeb1/Mgp/Enpp1/Hoxd3/Snai1/Adamts7/Smad9/Ltbp3/Bgn/Pkd1/Osr1/Ccn2/Bbs1 | 29 |
| GO:0051145 | smooth muscle cell differentiation | BP | 0.212 | 3.41e-08 | 1.26e-06 | 5.9 | 8.6e-07 | Tbx2/Npnt/Ednrb/Foxf1/Mef2c/Tshz3/Zeb1/Kit/Cth/Aplnr/Tbx3/Ednra/Nfatc4/Rbpms2/Efemp2/Sgcb/Itga8 | 17 |
| GO:0002691 | regulation of cellular extravasation | BP | 0.283 | 3.75e-08 | 1.38e-06 | 5.86 | 9.38e-07 | Mdk/Itga4/Chst2/Icam1/Cxcl12/Jam3/Plcb1/Thy1/Il27ra/Sele/Pecam1/Ptger3/St3gal4 | 13 |
| GO:0099504 | synaptic vesicle cycle | BP | 0.132 | 3.77e-08 | 1.38e-06 | 5.86 | 9.38e-07 | Syn2/Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Cdh2/Lrrk2/Pfn2/Tbc1d24/Ppfia2/Htr1b/Syde1/Cadps/Nlgn3/Syt7/Prrt2/Magi2/Htr2a/Bin1/Erc2/Ncs1/Pcdh17/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 30 |
| GO:1903522 | regulation of blood circulation | BP | 0.124 | 3.81e-08 | 1.38e-06 | 5.86 | 9.41e-07 | Cacna1c/Kcnd3/Adra1a/Nos3/Slc8a1/Scn4b/Ednrb/Cacna2d1/Mmp2/Icam1/Wdr35/Stc1/Hopx/Cacna1e/Edn3/Ank2/Calca/Cd38/Agtr1a/Gjc1/Ednra/Htr2a/Myl3/Bin1/Pde5a/Cacna1h/Ptger3/Slc4a3/Ccn2/Apln/Atp2b4/Atp1a2/Cav1 | 33 |
| GO:0050863 | regulation of T cell activation | BP | 0.112 | 3.86e-08 | 1.39e-06 | 5.86 | 9.47e-07 | Ihh/Efnb3/Lef1/Mdk/Cd83/Igf1/Cd44/Mapk8ip1/Pik3r6/Gli3/Tnfsf13b/Thy1/Zeb1/Prdm1/Cd1d1/Loxl3/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/Pde5a/H2-Aa/Stat5b/Gimap5/Zap70/Dlg5/Gimap3/Lat/Il12rb1/Cav1/Il2rg | 39 |
| GO:0009612 | response to mechanical stimulus | BP | 0.146 | 3.92e-08 | 1.41e-06 | 5.85 | 9.56e-07 | Col11a1/Stra6/Slc1a3/Mdk/Wnt11/Igf1/Nos3/Slc8a1/Scn1a/Mmp2/Grin2b/Serpine2/Cxcl12/Kit/Ednra/Htr2a/Cnn2/Pkd1/Fyn/Pkd2/Atp1a2/Habp4/Ptn/Pkdrej/Piezo2/Cav1 | 26 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.107 | 3.95e-08 | 1.41e-06 | 5.85 | 9.58e-07 | Sfrp2/Lef1/Cd109/Kdr/Nog/Rnf165/Zeb2/Npnt/Fst/Fermt2/Gpc3/Btbd11/Gdf11/Zfp423/Lrp2/Gdf7/Cdh5/Pbld2/Zeb1/Gdf10/Tgif2/Cdkn1c/Tsku/Magi2/Emilin1/Spred3/Kcp/Smad9/Ltbp3/Tnfaip6/Hipk2/Nrros/Zfyve9/Cldn5/Arrb2/Neo1/Il17rd/Fstl1/Lats2/Cav1/Itga8/Tmprss6 | 42 |
| GO:0010522 | regulation of calcium ion transport into cytosol | BP | 0.18 | 4.18e-08 | 1.47e-06 | 5.83 | 1e-06 | Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Calca/Thy1/Coro1a/Aplnr/Agtr1a/Ednra/Adcyap1r1/Asph/Selenon/Fyn/Pkd2/Gstm7/Gimap5/Gimap3/Cav1 | 20 |
| GO:0099175 | regulation of postsynapse organization | BP | 0.18 | 4.18e-08 | 1.47e-06 | 5.83 | 1e-06 | Ptprd/Nlgn1/Cdh2/Lrrc4b/Lrrk2/Nptx1/Grin2b/Ppfia2/Kif1a/Abi2/Nlgn3/Dgkb/Apoe/Caprin2/Epha7/Lrfn2/Fyn/Bhlhb9/Cux2/Ngef | 20 |
| GO:0050931 | pigment cell differentiation | BP | 0.308 | 4.37e-08 | 1.53e-06 | 5.82 | 1.04e-06 | Adamts20/Zeb2/Ednrb/Mef2c/Myo5a/Ap1m1/Edn3/Gli3/Kit/Enpp1/Kitl/Rab27a | 12 |
| GO:0060021 | roof of mouth development | BP | 0.188 | 4.4e-08 | 1.53e-06 | 5.82 | 1.04e-06 | Lef1/Wnt11/Pkdcc/Tbx2/Mef2c/Osr2/Tshz1/Gdf11/Bbs7/Gli3/Tcf21/Jag2/Loxl3/Cplane1/Tbx3/Snai1/Asph/Osr1/Fzd2 | 19 |
| GO:0045123 | cellular extravasation | BP | 0.207 | 5.02e-08 | 1.73e-06 | 5.76 | 1.18e-06 | Mdk/Itga4/Chst2/Itgb7/Icam1/Podxl2/Cxcl12/Jam3/Itga1/Plcb1/Thy1/Itgal/Il27ra/Sele/Pecam1/Ptger3/St3gal4 | 17 |
| GO:0071805 | potassium ion transmembrane transport | BP | 0.136 | 5.8e-08 | 1.99e-06 | 5.7 | 1.35e-06 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Kcna6/Kcnd2/Kcnd3/Slc12a8/Slc24a4/Kcnq5/Slc9a7/Slc24a3/Edn3/Ank2/Kcnma1/Slc9a5/Kcnc4/Bin1/Kcnip3/Fhl1/Kcng4/Ptger3/Pkd2/Slc12a4/Atp1a2/Kcnq4/Cav1 | 28 |
| GO:1901890 | positive regulation of cell junction assembly | BP | 0.169 | 5.93e-08 | 2.02e-06 | 5.69 | 1.37e-06 | Kdr/Ptprd/St8sia2/Nlgn1/Nrp1/Fermt2/Lrrc4b/Col16a1/Tek/Adgrl1/Slitrk3/Thy1/Ppm1f/Nlgn3/Cldn5/Bhlhb9/Cux2/Nphp1/Dlg5/Cav1/Cldn1 | 21 |
| GO:0001885 | endothelial cell development | BP | 0.216 | 6.63e-08 | 2.24e-06 | 5.65 | 1.53e-06 | Ednrb/Icam1/Stc1/Plcb1/Cdh5/Msn/Ppp1r16b/Robo4/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Col18a1/Cldn1 | 16 |
| GO:0009582 | detection of abiotic stimulus | BP | 0.162 | 6.78e-08 | 2.28e-06 | 5.64 | 1.55e-06 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Htr2a/Arrb1/Pkd1/Arrb2/Fyn/Pkd2/Pkdrej/Piezo2/Cacnb4 | 22 |
| GO:0045766 | positive regulation of angiogenesis | BP | 0.142 | 6.94e-08 | 2.3e-06 | 5.64 | 1.56e-06 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Tek/Cxcr3/Pik3r6/Cdh5/Aplnr/Jcad/Vegfc/Agtr1a/Emilin1/Tie1/Igf2/Hipk2/Akt3/Vash2 | 26 |
| GO:1904018 | positive regulation of vasculature development | BP | 0.142 | 6.94e-08 | 2.3e-06 | 5.64 | 1.56e-06 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Kdr/Ddah1/Nos3/Nrp1/Ets1/Tert/Prkd1/Tek/Cxcr3/Pik3r6/Cdh5/Aplnr/Jcad/Vegfc/Agtr1a/Emilin1/Tie1/Igf2/Hipk2/Akt3/Vash2 | 26 |
| GO:1990778 | protein localization to cell periphery | BP | 0.108 | 6.95e-08 | 2.3e-06 | 5.64 | 1.56e-06 | Gpc6/Kcnb2/Ar/Pkdcc/Tub/Epb41l3/Ccdc88a/Stx1b/Gnai1/Cdh2/Efr3b/Arl6/Abi3/Nptx1/Myo5a/Gga2/Ank2/Rab34/Dlg2/Epha3/Lin7a/Ttc7b/Nptxr/Nfasc/Rilpl1/Magi2/Nsg1/Stac2/Arl3/Cacng7/Ptch1/Kcnip3/Dchs1/Tspan5/Dlg4/Atp2b4/Palm/Bbs1/Pid1/Nkd2 | 40 |
| GO:0003002 | regionalization | BP | 0.106 | 7.26e-08 | 2.38e-06 | 5.62 | 1.62e-06 | Gli1/Sfrp2/Hhip/Ar/Lef1/Wnt11/Hoxd10/Hoxd11/Nog/Zeb2/Hoxa11/Nrp1/Dzip1l/Gpc3/Hoxd9/Hoxa10/Foxf1/Mef2c/Hoxd8/Hoxd4/Tshz1/Gdf11/Gli3/Lrp2/Ift57/Lfng/Gpr161/Hoxd3/Tbx3/Heyl/Dync2h1/Ednra/Snai1/Pcsk5/Hoxa9/Hipk2/Ptch1/Nr2f1/Osr1/Ets2/Rab23 | 41 |
| GO:0016079 | synaptic vesicle exocytosis | BP | 0.161 | 7.75e-08 | 2.52e-06 | 5.6 | 1.71e-06 | Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Pfn2/Ppfia2/Htr1b/Syde1/Cadps/Syt7/Prrt2/Htr2a/Erc2/Ncs1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 22 |
| GO:0032412 | regulation of ion transmembrane transporter activity | BP | 0.12 | 7.78e-08 | 2.52e-06 | 5.6 | 1.71e-06 | Hecw2/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Mef2c/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Stac2/Cacng7/Dysf/Fhl1/Asph/Selenon/Osr1/Rrad/Pkd2/Dlg4/Gstm7/Atp1a2/Gnb5/Abcb1a/Cav1/Cacna1a/Cacnb4 | 33 |
| GO:0043270 | positive regulation of ion transport | BP | 0.112 | 7.82e-08 | 2.52e-06 | 5.6 | 1.71e-06 | Grik1/Cacna1c/Pla2g10/Abat/Tcaf1/Scn4b/Drd4/Cacna2d1/Myo5a/Grin2b/Stc1/Cxcr3/Cxcl12/Edn3/Ank2/Plcb1/Thy1/Trpc6/Nlgn3/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Stac2/Fhl1/Asph/Arrb2/Pkd2/Gstm7/Gimap5/Akt3/Gimap3/Abcb1a/Cav1/Gabbr1/Cacnb4 | 37 |
| GO:0099068 | postsynapse assembly | BP | 0.324 | 8.95e-08 | 2.85e-06 | 5.55 | 1.94e-06 | Gap43/Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Slitrk3/Nlgn3/Nptxr/Magi2/Arhgef9 | 11 |
| GO:0099560 | synaptic membrane adhesion | BP | 0.324 | 8.95e-08 | 2.85e-06 | 5.55 | 1.94e-06 | Gpc6/Lrrc4c/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Epha3/Sparcl1/Magi2/Pcdh17/Ntng1 | 11 |
| GO:0003015 | heart process | BP | 0.124 | 9.86e-08 | 3.12e-06 | 5.51 | 2.12e-06 | Sgcd/Cacna1c/Kcnd3/Adra1a/Nos3/Slc8a1/Scn4b/Scn1a/Ednrb/Cacna2d1/Stc1/Hopx/Cacna1e/Edn3/Ank2/Calca/Rps6ka2/Nox4/Gjc1/Ednra/Myl3/Bin1/Pde5a/Cacna1h/Slc4a3/Ccn2/Fyn/Apln/Atp2b4/Atp1a2/Cav1 | 31 |
| GO:0045446 | endothelial cell differentiation | BP | 0.171 | 1.04e-07 | 3.27e-06 | 5.49 | 2.22e-06 | Kdr/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Zeb1/Msn/Ppp1r16b/Robo4/Tie1/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Fstl1/Col18a1/Cldn1 | 20 |
| GO:0048562 | embryonic organ morphogenesis | BP | 0.111 | 1.07e-07 | 3.33e-06 | 5.48 | 2.27e-06 | Gli1/Col11a1/Stra6/Ihh/Mmp16/Wnt11/Hoxd10/Hoxd11/Nog/Tbx2/Hoxa11/Hoxd9/Foxf1/Mef2c/Osr2/Hoxd4/Tshz1/Lrig3/Flvcr1/Gli3/Tcf21/Ift57/Wnt9a/Zeb1/Aplnr/Enpp1/Hoxd3/Tbx3/Ednra/Hoxa9/Twist2/Hipk2/Osr1/Pkd2/Fzd2/Kcnq4/Itga8 | 37 |
| GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.314 | 1.26e-07 | 3.91e-06 | 5.41 | 2.66e-06 | Pcdh18/Nlgn1/Cdh2/Cdh6/Cdh13/Pcdhgc3/Cdh10/Cdh5/Pcdh12/Cdh11/Dchs1 | 11 |
| GO:0070997 | neuron death | BP | 0.1 | 1.32e-07 | 4.09e-06 | 5.39 | 2.78e-06 | Ptprz1/Angpt1/Mdk/Dcc/Kdr/Agap2/Igf1/St8sia2/Ddr2/Nrp1/Tert/Bok/Lrrk2/Dpysl4/Mef2c/Grin2b/Pcdhgc3/Tbc1d24/Itga1/Scn2a/Coro1a/Il27ra/Grik2/Apoe/Rilpl1/Enpp1/Kcnma1/Cd200/Ndnf/Slc23a2/Arrb1/Hipk2/Epha7/Sarm1/Kcnip3/Nlrp1b/Nfatc4/Arrb2/Neo1/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 44 |
| GO:0061326 | renal tubule development | BP | 0.176 | 1.34e-07 | 4.11e-06 | 5.39 | 2.8e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Gli3/Tcf21/Agtr1a/Heyl/Ptch1/Osr1/Dchs1/Aqp11/Pkd2/Fat4 | 19 |
| GO:0043271 | negative regulation of ion transport | BP | 0.141 | 1.38e-07 | 4.21e-06 | 5.38 | 2.86e-06 | Maob/Hecw2/Spink1/Abat/Nos3/Drd4/Myo5a/Icam1/Stc1/Serpine2/Calca/Htr1b/Htr2a/Bin1/Dysf/Osr1/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1/Gabbr1/Usp2 | 25 |
| GO:0099003 | vesicle-mediated transport in synapse | BP | 0.122 | 1.42e-07 | 4.28e-06 | 5.37 | 2.91e-06 | Syn2/Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Cdh2/Lrrk2/Pfn2/Tbc1d24/Ppfia2/Htr1b/Syde1/Cadps/Nlgn3/Syt7/Prrt2/Magi2/Htr2a/Nsg1/Bin1/Erc2/Ncs1/Pcdh17/Brsk1/Nrn1/Stx2/Slc4a8/Cacna1a/Cacnb4 | 31 |
| GO:0001738 | morphogenesis of a polarized epithelium | BP | 0.193 | 1.49e-07 | 4.49e-06 | 5.35 | 3.06e-06 | Sfrp2/Wnt11/Nkd1/Gpc3/Foxf1/Msn/Myo9a/Znrf3/Cplane1/Sh3bp1/Dchs1/Fzd2/Nphp1/Fat4/Dlg5/Brsk1/Dact1 | 17 |
| GO:0042310 | vasoconstriction | BP | 0.174 | 1.56e-07 | 4.65e-06 | 5.33 | 3.16e-06 | Cacna1c/Adra1a/Slc8a1/Ednrb/Mmp2/Icam1/Wdr35/Edn3/Htr1b/Cd38/Agtr1a/Ednra/Bdkrb2/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1 | 19 |
| GO:0051926 | negative regulation of calcium ion transport | BP | 0.217 | 1.58e-07 | 4.66e-06 | 5.33 | 3.17e-06 | Spink1/Nos3/Drd4/Icam1/Stc1/Calca/Bin1/Dysf/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5/Usp2 | 15 |
| GO:0051492 | regulation of stress fiber assembly | BP | 0.182 | 1.7e-07 | 5.01e-06 | 5.3 | 3.41e-06 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arhgap28/Dlc1/Ccn2 | 18 |
| GO:0006813 | potassium ion transport | BP | 0.123 | 1.74e-07 | 5.09e-06 | 5.29 | 3.46e-06 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Kcna6/Kcnd2/Kcnd3/Slc12a8/Nos3/Slc24a4/Kcnq5/Slc9a7/Slc24a3/Edn3/Ank2/Kcnma1/Slc9a5/Kcnc4/Htr2a/Bin1/Kcnip3/Fhl1/Kcng4/Ptger3/Pkd2/Slc12a4/Atp1a2/Kcnq4/Cav1 | 30 |
| GO:0033627 | cell adhesion mediated by integrin | BP | 0.191 | 1.77e-07 | 5.11e-06 | 5.29 | 3.47e-06 | Epha8/Cyp1b1/Itga4/Npnt/Itgb7/Col16a1/Icam1/Jam3/Itga1/Podxl/Plpp3/Itgal/Emilin1/Itga7/Nckap1l/Itga8/Itgb4 | 17 |
| GO:0050805 | negative regulation of synaptic transmission | BP | 0.191 | 1.77e-07 | 5.11e-06 | 5.29 | 3.47e-06 | Grik1/Grid2ip/Celf4/Drd4/Gnai1/Lrrk2/Myo5a/Shank2/Htr1b/Nlgn3/Sorcs2/Grik2/Prrt1/Cd38/Htr2a/Pcdh17/Gabbr1 | 17 |
| GO:0061333 | renal tubule morphogenesis | BP | 0.191 | 1.77e-07 | 5.11e-06 | 5.29 | 3.47e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Gli3/Tcf21/Agtr1a/Ptch1/Osr1/Dchs1/Pkd2/Fat4 | 17 |
| GO:0001823 | mesonephros development | BP | 0.173 | 1.82e-07 | 5.17e-06 | 5.29 | 3.52e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Osr2/Gdf11/Gli3/Tcf21/Tshz3/Agtr1a/Smad9/Ptch1/Osr1/Dchs1/Fat4 | 19 |
| GO:0022898 | regulation of transmembrane transporter activity | BP | 0.116 | 1.82e-07 | 5.17e-06 | 5.29 | 3.52e-06 | Hecw2/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Mef2c/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Stac2/Cacng7/Dysf/Fhl1/Asph/Selenon/Osr1/Rrad/Pkd2/Dlg4/Gstm7/Atp1a2/Gnb5/Abcb1a/Cav1/Cacna1a/Cacnb4 | 33 |
| GO:0032409 | regulation of transporter activity | BP | 0.114 | 1.85e-07 | 5.2e-06 | 5.28 | 3.54e-06 | Hecw2/Slc5a11/Nlgn1/Tcaf1/Scn4b/Drd4/Fxyd5/Cacna2d1/Mef2c/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Prrt1/Ednra/Stac2/Cacng7/Dysf/Fhl1/Asph/Selenon/Osr1/Rrad/Pkd2/Dlg4/Gstm7/Atp1a2/Gnb5/Abcb1a/Cav1/Cacna1a/Cacnb4 | 34 |
| GO:0099084 | postsynaptic specialization organization | BP | 0.273 | 1.94e-07 | 5.41e-06 | 5.27 | 3.68e-06 | Gap43/Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Shank2/Slitrk3/Nlgn3/Nptxr/Syngap1/Arhgef9 | 12 |
| GO:0006887 | exocytosis | BP | 0.101 | 1.94e-07 | 5.41e-06 | 5.27 | 3.68e-06 | Syn2/Cacna1c/Nlgn1/Syt13/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Foxf1/Myo5a/Pfn2/Mical1/Ppfia2/Exoc3l/Htr1b/Syde1/Cadps/Coro1a/Kit/Lin7a/Syt7/Prrt2/Htr2a/Nckap1l/Erc2/Ncs1/Myo1f/Cd84/Cacna1h/Septin4/Fgr/Lat/Rab27a/Clnk/Nrn1/Stx2/Slc4a8/Cacna1a/Nkd2/Cacnb4 | 42 |
| GO:0007416 | synapse assembly | BP | 0.135 | 2.04e-07 | 5.66e-06 | 5.25 | 3.85e-06 | Gpc6/Gap43/Sdk2/Plxnb1/Ptprd/St8sia2/Nlgn1/Cdh2/Lrrc4b/Nptx1/Mef2c/Shank2/Obsl1/Adgrl1/Slitrk3/Plxnd1/Nlgn3/Nptxr/Magi2/Epha7/Pcdh17/Bhlhb9/Cux2/Dlg5/Arhgef9/Cacna1a | 26 |
| GO:0008344 | adult locomotory behavior | BP | 0.171 | 2.11e-07 | 5.75e-06 | 5.24 | 3.91e-06 | Efnb3/Cacna1c/Hoxd10/Drd4/Scn1a/Hoxd9/Ccnd2/Dab1/Cxcl12/Shank2/Glrb/Enpp1/Kcnma1/Prex2/Hipk2/Arrb2/Atp1a2/Cacna1a/Cacnb4 | 19 |
| GO:0110020 | regulation of actomyosin structure organization | BP | 0.171 | 2.11e-07 | 5.75e-06 | 5.24 | 3.91e-06 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Mef2c/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arhgap28/Dlc1/Ccn2 | 19 |
| GO:0051928 | positive regulation of calcium ion transport | BP | 0.152 | 2.17e-07 | 5.89e-06 | 5.23 | 4.01e-06 | Cacna1c/Cacna2d1/Stc1/Cxcr3/Cxcl12/Ank2/Thy1/Trpc6/Aplnr/Cracr2a/Agtr1a/Ednra/Adcyap1r1/Stac2/Asph/Arrb2/Pkd2/Gstm7/Gimap5/Gimap3/Cav1/Cacnb4 | 22 |
| GO:0042733 | embryonic digit morphogenesis | BP | 0.211 | 2.35e-07 | 6.33e-06 | 5.2 | 4.3e-06 | Ihh/Sfrp2/Hoxd11/Nog/Tbx2/Hoxa11/Osr2/Flvcr1/Gli3/Cplane1/Tbx3/Tmem231/Osr1/Ttbk2/Rab23 | 15 |
| GO:2000351 | regulation of endothelial cell apoptotic process | BP | 0.245 | 2.35e-07 | 6.33e-06 | 5.2 | 4.3e-06 | Sema5a/Angpt1/Itga4/Kdr/Tert/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Angptl4/Col18a1 | 13 |
| GO:0048706 | embryonic skeletal system development | BP | 0.151 | 2.45e-07 | 6.57e-06 | 5.18 | 4.47e-06 | Col11a1/Ihh/Mmp16/Hoxd10/Hoxd11/Nog/Hoxa11/Hoxd9/Mef2c/Osr2/Hoxd4/Flvcr1/Gli3/Wnt9a/Zeb1/Hoxd3/Ednra/Pcsk5/Hoxa9/Twist2/Osr1/Xylt1 | 22 |
| GO:0043010 | camera-type eye development | BP | 0.104 | 2.6e-07 | 6.91e-06 | 5.16 | 4.7e-06 | Ndp/Stra6/Ihh/Cyp1b1/Sdk2/Nhs/Cacna1c/Kdr/Tbx2/Tub/Rcn1/Zeb2/Celf4/Lamc3/Nrp1/Arl6/Tenm3/Osr2/Gdf11/Obsl1/Gli3/Thy1/Zeb1/Tgif2/Ptprm/Abi2/Rp1/Cdkn1c/Tsku/Arhgef15/Spred3/Rorb/Lpcat1/Twist2/Hipk2/Tmem231/Fat3/Nphp1/Ptn | 39 |
| GO:0072080 | nephron tubule development | BP | 0.176 | 2.72e-07 | 7.22e-06 | 5.14 | 4.91e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Mef2c/Gli3/Tcf21/Agtr1a/Heyl/Ptch1/Osr1/Dchs1/Aqp11/Fat4 | 18 |
| GO:0060079 | excitatory postsynaptic potential | BP | 0.185 | 2.93e-07 | 7.7e-06 | 5.11 | 5.24e-06 | Grik1/Begain/Nlgn1/Celf4/Drd4/Stx1b/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Slc29a1/Nlgn3/Grik2/Mpp2/Dlg4/Cux2 | 17 |
| GO:0030111 | regulation of Wnt signaling pathway | BP | 0.11 | 2.93e-07 | 7.7e-06 | 5.11 | 5.24e-06 | Gli1/Sfrp2/Sema5a/Lef1/Mdk/Apcdd1/Wnt11/Nog/Zeb2/Ptpro/Nkd1/Tert/Cdh2/Gpc3/Lrrk2/Tmem237/Rbms3/Gprc5b/Gli3/Peg12/Plpp3/Dkk3/Znrf3/Tsku/Apoe/Caprin2/Daam2/Prickle1/Tle1/Nfatc4/Lats2/Adgra2/Cav1/Dact1/Nkd2 | 35 |
| GO:0008016 | regulation of heart contraction | BP | 0.132 | 3.07e-07 | 8.03e-06 | 5.1 | 5.46e-06 | Cacna1c/Kcnd3/Adra1a/Nos3/Slc8a1/Scn4b/Ednrb/Cacna2d1/Stc1/Hopx/Cacna1e/Edn3/Ank2/Calca/Gjc1/Ednra/Myl3/Bin1/Pde5a/Cacna1h/Slc4a3/Ccn2/Apln/Atp2b4/Atp1a2/Cav1 | 26 |
| GO:0021510 | spinal cord development | BP | 0.175 | 3.17e-07 | 8.2e-06 | 5.09 | 5.58e-06 | Vit/Hoxd10/Dcc/Nog/Dab1/Gdf11/Gli3/Gdf7/Loxl3/Phgdh/Enpp1/Sox12/Dync2h1/Daam2/Tal1/Ptch1/Rab23/Cacna1a | 18 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | BP | 0.175 | 3.17e-07 | 8.2e-06 | 5.09 | 5.58e-06 | Grik1/Begain/Nlgn1/Abat/Celf4/Drd4/Stx1b/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Slc29a1/Nlgn3/Grik2/Mpp2/Dlg4/Cux2 | 18 |
| GO:1901214 | regulation of neuron death | BP | 0.102 | 3.24e-07 | 8.33e-06 | 5.08 | 5.67e-06 | Ptprz1/Angpt1/Mdk/Dcc/Kdr/Agap2/Igf1/St8sia2/Ddr2/Nrp1/Tert/Bok/Lrrk2/Mef2c/Grin2b/Pcdhgc3/Tbc1d24/Itga1/Coro1a/Il27ra/Grik2/Apoe/Rilpl1/Kcnma1/Cd200/Ndnf/Slc23a2/Arrb1/Hipk2/Epha7/Sarm1/Kcnip3/Nfatc4/Arrb2/Neo1/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 40 |
| GO:1903305 | regulation of regulated secretory pathway | BP | 0.135 | 3.28e-07 | 8.4e-06 | 5.08 | 5.71e-06 | Nlgn1/Syt13/Adra1a/Rims3/Drd4/P2rx2/Lrrk2/Foxf1/Pfn2/Mical1/Ppfia2/Htr1b/Cadps/Syt7/Htr2a/Nckap1l/Ncs1/Cd84/Cacna1h/Fgr/Rab27a/Nrn1/Slc4a8/Cacna1a/Cacnb4 | 25 |
| GO:0035136 | forelimb morphogenesis | BP | 0.261 | 3.3e-07 | 8.43e-06 | 5.07 | 5.73e-06 | Cacna1c/Hoxd10/Hoxd11/Rnf165/Hoxa11/Hoxd9/Osr2/Wnt9a/Enpp1/Tbx3/Hoxa9/Osr1 | 12 |
| GO:0010720 | positive regulation of cell development | BP | 0.103 | 3.41e-07 | 8.65e-06 | 5.06 | 5.89e-06 | Ptprz1/Sema5a/Mdk/Hoxd11/Kdr/Nog/Plxnb1/Ptprd/Igf1/Zeb2/Sema7a/Hoxa11/Nrp1/Fermt2/Fn1/Serpine2/Cxcl12/Has2/Tbc1d24/Obsl1/Gli3/Lrp2/Cdh5/Plxnd1/Mme/Kit/Vegfc/Apoe/Caprin2/Nin/Bin1/Chodl/Pde5a/Cldn5/Net1/Bhlhb9/Cux2/Abcb1a/Ptn | 39 |
| GO:0043535 | regulation of blood vessel endothelial cell migration | BP | 0.183 | 3.44e-07 | 8.65e-06 | 5.06 | 5.89e-06 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Prkd1/Vash1/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Atp2b4/Akt3 | 17 |
| GO:1904035 | regulation of epithelial cell apoptotic process | BP | 0.183 | 3.44e-07 | 8.65e-06 | 5.06 | 5.89e-06 | Sema5a/Angpt1/Mdk/Itga4/Kdr/Igf1/Tert/Bok/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Arrb2/Angptl4/Col18a1 | 17 |
| GO:0007613 | memory | BP | 0.143 | 3.52e-07 | 8.82e-06 | 5.05 | 6e-06 | Mdk/Nog/Cyp7b1/Igf1/Aph1b/Drd4/Ccnd2/Chst10/Grin2b/B4galt2/Shank2/Slc8a3/Plcb1/Eif2ak4/Scn2a/Apoe/Igf2/Htr2a/Pde5a/Nfatc4/Cux2/Ptn/Itga8 | 23 |
| GO:0060047 | heart contraction | BP | 0.122 | 3.67e-07 | 9.13e-06 | 5.04 | 6.21e-06 | Sgcd/Cacna1c/Kcnd3/Adra1a/Nos3/Slc8a1/Scn4b/Scn1a/Ednrb/Cacna2d1/Stc1/Hopx/Cacna1e/Edn3/Ank2/Calca/Rps6ka2/Gjc1/Ednra/Myl3/Bin1/Pde5a/Cacna1h/Slc4a3/Ccn2/Apln/Atp2b4/Atp1a2/Cav1 | 29 |
| GO:0050866 | negative regulation of cell activation | BP | 0.124 | 3.76e-07 | 9.31e-06 | 5.03 | 6.34e-06 | Ihh/Mdk/Abat/Cd44/Foxf1/Serpine2/Gli3/Flt3/Prdm1/Loxl3/Btla/H2-Ab1/Apoe/Havcr2/Emilin1/Cd200/Nckap1l/Pde5a/Cd84/Ptger3/Fgr/H2-Aa/Gimap5/Dlg5/Gimap3/Clnk/Cygb/Nr1h3 | 28 |
| GO:0035725 | sodium ion transmembrane transport | BP | 0.147 | 3.96e-07 | 9.78e-06 | 5.01 | 6.66e-06 | Nalcn/Hecw2/Slc6a2/Slc8a1/Scn4b/Drd4/Scn1a/Fxyd5/Slc24a4/Slc9a7/Slc24a3/Slc8a3/Plcb1/Slc6a15/Scn2a/Slc9a5/Osr1/Pkd2/Scn3a/Atp2b4/Atp1a2/Slc4a8 | 22 |
| GO:1903510 | mucopolysaccharide metabolic process | BP | 0.203 | 4.16e-07 | 1.02e-05 | 4.99 | 6.95e-06 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Xylt1/Dse/Habp4 | 15 |
| GO:0030038 | contractile actin filament bundle assembly | BP | 0.164 | 4.29e-07 | 1.04e-05 | 4.98 | 7.1e-06 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arrb1/Arhgap28/Dlc1/Ccn2 | 19 |
| GO:0043149 | stress fiber assembly | BP | 0.164 | 4.29e-07 | 1.04e-05 | 4.98 | 7.1e-06 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arrb1/Arhgap28/Dlc1/Ccn2 | 19 |
| GO:0061448 | connective tissue development | BP | 0.112 | 4.38e-07 | 1.06e-05 | 4.97 | 7.23e-06 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Nog/Igf1/Pkdcc/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Id4/Gli3/Gdf7/Pth1r/Wnt9a/Zeb1/Mgp/Selenom/Enpp1/Hoxd3/Snai1/Adamts7/Smad9/Ltbp3/Bgn/Pkd1/Osr1/Ccn2/Bbs1 | 33 |
| GO:0072577 | endothelial cell apoptotic process | BP | 0.232 | 4.69e-07 | 1.13e-05 | 4.95 | 7.7e-06 | Sema5a/Angpt1/Itga4/Kdr/Tert/Icam1/Tek/Cdh5/Cd248/Ecscr/Ndnf/Angptl4/Col18a1 | 13 |
| GO:0010632 | regulation of epithelial cell migration | BP | 0.12 | 4.78e-07 | 1.15e-05 | 4.94 | 7.79e-06 | Sema5a/Angpt1/Kdr/Igf1/Evl/Nos3/Nrp1/Ets1/Mef2c/Pfn2/Stc1/Prkd1/Tek/Has2/Vash1/Plpp3/Ptprm/Ppm1f/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Atp2b4/Akt3/Arsb/Adgra2/Mapre2 | 29 |
| GO:0007263 | nitric oxide mediated signal transduction | BP | 0.36 | 4.79e-07 | 1.15e-05 | 4.94 | 7.79e-06 | Spink1/Nos3/Cbs/Gucy1a2/Gucy1a1/Apoe/Ndnf/Pde5a/Atp2b4 | 9 |
| GO:0072171 | mesonephric tubule morphogenesis | BP | 0.2 | 4.99e-07 | 1.18e-05 | 4.93 | 8.03e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Osr1/Dchs1/Fat4 | 15 |
| GO:0051966 | regulation of synaptic transmission, glutamatergic | BP | 0.188 | 5e-07 | 1.18e-05 | 4.93 | 8.03e-06 | Gria4/Grik1/Nlgn1/Cdh2/Lrrk2/Mef2c/Myo5a/Grin2b/Serpine2/Shank2/Htr1b/Tshz3/Nlgn3/Grik2/Htr2a/Cacng7 | 16 |
| GO:0045216 | cell-cell junction organization | BP | 0.125 | 5.52e-07 | 1.3e-05 | 4.89 | 8.82e-06 | Wnt11/Epb41l3/Ptpro/Mpdz/Fermt2/Cdh2/Cdh6/Jam3/Cldn8/Cdh10/Cdh5/Abi2/Aplnr/Esam/Nfasc/Cdh11/Gjc1/Pecam1/Snai1/Heg1/Svep1/Dchs1/Cldn5/Nphp1/Dlg5/Cav1/Cldn1 | 27 |
| GO:0060078 | regulation of postsynaptic membrane potential | BP | 0.161 | 5.63e-07 | 1.32e-05 | 4.88 | 8.96e-06 | Grik1/Begain/Nlgn1/Abat/Celf4/Drd4/Stx1b/Lrrk2/Mef2c/Grin2b/Tbc1d24/Slc8a3/Gabra4/Slc29a1/Nlgn3/Grik2/Mpp2/Dlg4/Cux2 | 19 |
| GO:0001657 | ureteric bud development | BP | 0.168 | 5.71e-07 | 1.32e-05 | 4.88 | 8.96e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Gli3/Tcf21/Tshz3/Agtr1a/Smad9/Ptch1/Osr1/Dchs1/Fat4 | 18 |
| GO:0072163 | mesonephric epithelium development | BP | 0.168 | 5.71e-07 | 1.32e-05 | 4.88 | 8.96e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Gli3/Tcf21/Tshz3/Agtr1a/Smad9/Ptch1/Osr1/Dchs1/Fat4 | 18 |
| GO:0072164 | mesonephric tubule development | BP | 0.168 | 5.71e-07 | 1.32e-05 | 4.88 | 8.96e-06 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gdf11/Gli3/Tcf21/Tshz3/Agtr1a/Smad9/Ptch1/Osr1/Dchs1/Fat4 | 18 |
| GO:0048872 | homeostasis of number of cells | BP | 0.102 | 6.17e-07 | 1.42e-05 | 4.85 | 9.64e-06 | Tex15/Pla2g10/Col14a1/Gprasp2/Emcn/Crispld1/Nos3/P4htm/Cd44/Hcls1/Ets1/Cdh2/Mef2c/Jam3/Flvcr1/Plcb1/Myct1/Sh2b2/Flt3/Tnfsf13b/Dock10/Coro1a/Kit/Armcx1/Heph/Enpp1/Nckap1l/Tal1/Zfp251/Kitl/Rassf2/Septin4/Stat5b/Gimap5/Akt3/Gimap3/Lat/Fstl1 | 38 |
| GO:0009101 | glycoprotein biosynthetic process | BP | 0.114 | 6.48e-07 | 1.48e-05 | 4.83 | 1.01e-05 | St6gal2/Itm2a/Igf1/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Chst10/Soat1/Tmtc1/B3galnt1/Fut10/Plcb1/Galnt16/St6galnac4/St3gal2/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Aqp11/Mgat3/B3gnt5/Xylt1/Dse/St6galnac6/St3gal4/Large1/Alg14 | 31 |
| GO:0001658 | branching involved in ureteric bud morphogenesis | BP | 0.209 | 6.81e-07 | 1.55e-05 | 4.81 | 1.05e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Dchs1/Fat4 | 14 |
| GO:0030178 | negative regulation of Wnt signaling pathway | BP | 0.138 | 6.83e-07 | 1.55e-05 | 4.81 | 1.05e-05 | Gli1/Sfrp2/Mdk/Apcdd1/Wnt11/Nog/Ptpro/Nkd1/Cdh2/Gpc3/Rbms3/Gli3/Dkk3/Znrf3/Tsku/Apoe/Prickle1/Tle1/Nfatc4/Lats2/Cav1/Dact1/Nkd2 | 23 |
| GO:0051962 | positive regulation of nervous system development | BP | 0.103 | 6.95e-07 | 1.57e-05 | 4.8 | 1.07e-05 | Ptprz1/Sema5a/Mdk/Kdr/Nog/Plxnb1/Ptprd/Igf1/St8sia2/Nlgn1/Zeb2/Sema7a/Nrp1/Lrrc4b/Fn1/Serpine2/Cxcl12/Tbc1d24/Obsl1/Adgrl1/Gli3/Lrp2/Slitrk3/Plxnd1/Mme/Kit/Nlgn3/Vegfc/Apoe/Caprin2/Nin/Bin1/Chodl/Bhlhb9/Cux2/Dlg5/Ptn | 37 |
| GO:0060976 | coronary vasculature development | BP | 0.195 | 7.14e-07 | 1.61e-05 | 4.79 | 1.09e-05 | Angpt1/Sgcd/Nrp1/Kif7/Gpc3/Lrp2/Plxnd1/Prdm1/Aplnr/Cplane1/Dync2h1/Prickle1/Pcsk5/Apln/Prokr1 | 15 |
| GO:0043087 | regulation of GTPase activity | BP | 0.104 | 7.29e-07 | 1.63e-05 | 4.79 | 1.11e-05 | Wnt11/Plxnb1/Rgs6/Rgs16/Rasgrp3/Fermt2/Garnl3/Lrrk2/Rasa3/Arhgap45/Icam1/Rasgrp2/Arhgap25/Thy1/Plxnd1/Dock10/Epha3/Myo9a/Sh3bp1/Arhgef15/Arhgap6/Rasip1/Pecam1/Xcl1/Arap3/Bin1/Arrb1/Arhgef26/Net1/Arrb2/Fgd1/Gnb5/Syngap1/Ngef/Arhgap15/Mapre2 | 36 |
| GO:0033674 | positive regulation of kinase activity | BP | 0.0966 | 7.31e-07 | 1.63e-05 | 4.79 | 1.11e-05 | Epha8/Angpt1/Kdr/Agap2/Igf1/Ddr2/Zeb2/Adra1a/Ccdc88a/Drd4/Fermt2/Areg/Ccnd2/Dab1/Lrrk2/Tek/Edn3/Gprc5b/Pik3r6/Calca/Flt3/Kit/Epha3/Mmd/Vegfc/Nox4/Agtr1a/Tie1/Htr2a/Kitl/Pkd1/Epha7/Rassf2/Pde5a/Arrb2/Pkd2/Dlg4/Fgr/Atp2b4/Lat/Acsl1/C1qtnf9 | 42 |
| GO:0048704 | embryonic skeletal system morphogenesis | BP | 0.165 | 7.57e-07 | 1.68e-05 | 4.77 | 1.15e-05 | Col11a1/Mmp16/Hoxd10/Hoxd11/Nog/Hoxa11/Hoxd9/Mef2c/Osr2/Hoxd4/Flvcr1/Gli3/Wnt9a/Zeb1/Hoxd3/Hoxa9/Twist2/Osr1 | 18 |
| GO:0008217 | regulation of blood pressure | BP | 0.123 | 7.95e-07 | 1.76e-05 | 4.75 | 1.2e-05 | Ar/Cacna1c/Kdr/Ddah1/Adra1a/Abat/Nos3/Ptpro/Npr3/P2rx2/Adamts16/Ednrb/Enpep/Edn3/Calca/Rps6ka2/Postn/Gucy1a1/Vegfc/Agtr1a/Emilin1/Ednra/Bdkrb2/Trhde/Apln/Atp1a2/Lvrn | 27 |
| GO:1901215 | negative regulation of neuron death | BP | 0.115 | 8.16e-07 | 1.8e-05 | 4.75 | 1.22e-05 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Igf1/Nrp1/Tert/Bok/Lrrk2/Mef2c/Pcdhgc3/Tbc1d24/Coro1a/Il27ra/Grik2/Apoe/Cd200/Ndnf/Slc23a2/Arrb1/Hipk2/Nfatc4/Arrb2/Neo1/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 30 |
| GO:1902414 | protein localization to cell junction | BP | 0.157 | 8.37e-07 | 1.83e-05 | 4.74 | 1.25e-05 | Gpc6/Nlgn1/Stx1b/Fermt2/Nptx1/Jam3/Slitrk3/Cdh5/Dlg2/Nlgn3/Nptxr/Magi2/Pecam1/Nsg1/Cacng7/Heg1/Dlg4/Dlg5/Abcb1a | 19 |
| GO:1904019 | epithelial cell apoptotic process | BP | 0.157 | 8.37e-07 | 1.83e-05 | 4.74 | 1.25e-05 | Sema5a/Angpt1/Mdk/Itga4/Kdr/Igf1/Tert/Bok/Icam1/Tek/Cdh5/Jag2/Cd248/Ecscr/Ednra/Ndnf/Arrb2/Angptl4/Col18a1 | 19 |
| GO:0042692 | muscle cell differentiation | BP | 0.0959 | 8.76e-07 | 1.9e-05 | 4.72 | 1.29e-05 | AW551984/Sgcd/Col14a1/Ryr1/Igf1/Tbx2/Adra1a/Npnt/Slc8a1/P2rx2/Tmod2/Cdh2/Ednrb/Foxf1/Mef2c/Cxcl12/Hopx/Ank2/Tshz3/Zeb1/Kit/Cth/Aplnr/Nox4/Tbx3/Ednra/Igf2/Bin1/Hdac9/Dysf/Flnc/Selenon/Nfatc4/Arrb2/Neo1/Pi16/Rbpms2/Efemp2/Sgcb/Itga8/Plekho1/Cacnb4 | 42 |
| GO:0022612 | gland morphogenesis | BP | 0.14 | 8.79e-07 | 1.9e-05 | 4.72 | 1.29e-05 | Gli1/Hgf/Ar/Mdk/Nog/Cyp7b1/Igf1/Tbx2/Cd44/Nrp1/Areg/Mmp2/Id4/Gli3/Gdf7/Plxnd1/Msn/Tbx3/Ptch1/Ptn/Cav1/Edar | 22 |
| GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.24 | 8.81e-07 | 1.9e-05 | 4.72 | 1.29e-05 | Hmcn1/Mcam/Itga4/Ptprd/Nlgn1/Cdh2/Tenm3/Adgrl1/Itgal/Sele/Dchs1/Fat4 | 12 |
| GO:0003158 | endothelium development | BP | 0.15 | 8.81e-07 | 1.9e-05 | 4.72 | 1.29e-05 | Kdr/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Zeb1/Msn/Ppp1r16b/Robo4/Tie1/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Fstl1/Col18a1/Cldn1 | 20 |
| GO:0090596 | sensory organ morphogenesis | BP | 0.107 | 9.07e-07 | 1.94e-05 | 4.71 | 1.32e-05 | Col11a1/Stra6/Ihh/Sdk2/Kdr/Nog/Tbx2/Arl6/Tenm3/Osr2/Tshz1/Lrig3/Gdf11/Obsl1/Gli3/Thy1/Zeb1/Prdm1/Ptprm/Abi2/Rp1/Tsku/Enpp1/Tbx3/Ednra/Rorb/Col5a2/Hipk2/Osr1/Fzd2/Fat3/Ptn/Kcnq4/Itga8 | 34 |
| GO:0046928 | regulation of neurotransmitter secretion | BP | 0.156 | 9.52e-07 | 2.03e-05 | 4.69 | 1.38e-05 | Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Kcnc4/Htr2a/Ncs1/Nrn1/Slc4a8/Cacna1a/Cacnb4 | 19 |
| GO:0051282 | regulation of sequestering of calcium ion | BP | 0.156 | 9.52e-07 | 2.03e-05 | 4.69 | 1.38e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Pkd2/Slc25a23/Gstm7 | 19 |
| GO:1901019 | regulation of calcium ion transmembrane transporter activity | BP | 0.18 | 9.57e-07 | 2.03e-05 | 4.69 | 1.38e-05 | Drd4/Cacna2d1/Myo5a/Ank2/Cracr2a/Stac2/Dysf/Asph/Selenon/Rrad/Pkd2/Gstm7/Atp1a2/Gnb5/Cacna1a/Cacnb4 | 16 |
| GO:1902905 | positive regulation of supramolecular fiber organization | BP | 0.131 | 9.72e-07 | 2.05e-05 | 4.69 | 1.39e-05 | Sema5a/Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Wasf3/Pfn2/Icam1/Abi2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Scin/Nckap1l/Slain1/Nin/Bin1/Ccn2/Efemp2/Cdc42ep5/Cav1 | 24 |
| GO:0045667 | regulation of osteoblast differentiation | BP | 0.139 | 9.81e-07 | 2.06e-05 | 4.69 | 1.4e-05 | Gli1/Sfrp2/Nog/Igf1/Ddr2/Npnt/Fermt2/Areg/Mef2c/Id3/Prkd1/Gli3/Nell1/Gdf10/Smoc1/Vegfc/Rorb/Tnfaip6/Twist2/Rassf2/Ptch1/Lrp3 | 22 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | BP | 0.149 | 9.95e-07 | 2.08e-05 | 4.68 | 1.42e-05 | Gli1/Sfrp2/Mdk/Wnt11/Nog/Ptpro/Nkd1/Cdh2/Gpc3/Rbms3/Gli3/Dkk3/Znrf3/Apoe/Prickle1/Tle1/Lats2/Cav1/Dact1/Nkd2 | 20 |
| GO:0014821 | phasic smooth muscle contraction | BP | 0.333 | 1.02e-06 | 2.12e-05 | 4.67 | 1.44e-05 | Tbx2/P2rx2/Ednrb/Edn3/Tshz3/Kit/Kcnma1/Tbx3/Ptger3 | 9 |
| GO:0001933 | negative regulation of protein phosphorylation | BP | 0.102 | 1.17e-06 | 2.44e-05 | 4.61 | 1.66e-05 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ptpro/Slc8a1/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Cdkn1c/Prr5l/Apoe/Enpp1/Ppef2/Rasip1/Emilin1/Pecam1/Ppm1e/Bdkrb2/Pkib/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Smpd1/Cav1/Pid1 | 36 |
| GO:0050770 | regulation of axonogenesis | BP | 0.13 | 1.18e-06 | 2.45e-05 | 4.61 | 1.67e-05 | Efnb3/Map2/Sema5a/Lrrc4c/Plxnb1/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Fn1/Dab1/Cxcl12/Thy1/Plxnd1/Apoe/Sema3g/Nin/Epha7/Chodl/Brsk1/Syngap1/Sema6a/Cacna1a | 24 |
| GO:0051208 | sequestering of calcium ion | BP | 0.153 | 1.23e-06 | 2.53e-05 | 4.6 | 1.72e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Pkd2/Slc25a23/Gstm7 | 19 |
| GO:0010976 | positive regulation of neuron projection development | BP | 0.12 | 1.24e-06 | 2.54e-05 | 4.6 | 1.73e-05 | Ptprz1/Hgf/Acsl6/Fez1/Mdk/Dcc/Lrrc7/Tenm3/Serpine2/Prkd1/Gprc5b/Adamts1/Serpini1/Epha3/Apoe/Magi2/Caprin2/Ndnf/P3h1/Fyn/Bhlhb9/Cux2/Dpysl3/Arsb/Ptn/Gpc2/Cpeb1 | 27 |
| GO:0045880 | positive regulation of smoothened signaling pathway | BP | 0.256 | 1.27e-06 | 2.59e-05 | 4.59 | 1.76e-05 | Gli1/Ihh/Evc/Kif7/Gpc3/Cibar1/Armc9/Rab34/Wnt9a/Dync2h1/Dlg5 | 11 |
| GO:0060560 | developmental growth involved in morphogenesis | BP | 0.11 | 1.31e-06 | 2.66e-05 | 4.57 | 1.81e-05 | Sfrp2/Map2/Sema5a/Wnt11/Itga4/St8sia2/Tbx2/Sema5b/Zeb2/Cpne5/Nkd1/Sema7a/Nrp1/Areg/Fn1/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Nin/Epha7/Cacng7/Sema6a/Nrn1/Cyfip2 | 31 |
| GO:0031032 | actomyosin structure organization | BP | 0.122 | 1.4e-06 | 2.84e-05 | 4.55 | 1.93e-05 | Kank4/Wnt11/Frmd3/Evl/Epb41l3/Phldb2/Ccdc88a/Nrp1/Fermt2/Tmod2/Mef2c/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Cnn2/Arrb1/Flnc/Cnn3/Arhgap28/Dlc1/Ccn2 | 26 |
| GO:0048844 | artery morphogenesis | BP | 0.185 | 1.41e-06 | 2.84e-05 | 4.55 | 1.93e-05 | Apob/Stra6/Mdk/Wnt11/Nog/Tbx2/Nrp1/Foxf1/Fkbp10/Lrp2/Prdm1/Apoe/Ednra/Akt3/Efemp2 | 15 |
| GO:0010594 | regulation of endothelial cell migration | BP | 0.132 | 1.41e-06 | 2.85e-05 | 4.55 | 1.94e-05 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Stc1/Prkd1/Tek/Vash1/Plpp3/Ptprm/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Atp2b4/Akt3/Adgra2 | 23 |
| GO:0043931 | ossification involved in bone maturation | BP | 0.321 | 1.44e-06 | 2.89e-05 | 4.54 | 1.96e-05 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:0098698 | postsynaptic specialization assembly | BP | 0.321 | 1.44e-06 | 2.89e-05 | 4.54 | 1.96e-05 | Gap43/Ptprd/Nlgn1/Lrrc4b/Nptx1/Slitrk3/Nlgn3/Nptxr/Arhgef9 | 9 |
| GO:0002685 | regulation of leukocyte migration | BP | 0.119 | 1.47e-06 | 2.92e-05 | 4.53 | 1.99e-05 | Mdk/Itga4/Chst2/Icam1/Cxcr3/Cxcl12/Jam3/Edn3/Plcb1/Thy1/Msn/Il27ra/Vegfc/Sele/Emilin1/Cd200/Ednra/Pecam1/Xcl1/Nckap1l/Kitl/Tnfaip6/Dysf/Ptger3/St3gal4/Mpp1/Ptn | 27 |
| GO:0070374 | positive regulation of ERK1 and ERK2 cascade | BP | 0.119 | 1.47e-06 | 2.92e-05 | 4.53 | 1.99e-05 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Adra1a/Npnt/Cd44/Sema7a/Nrp1/Fermt2/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Nox4/Apoe/Havcr2/Ednra/Xcl1/Htr2a/Cavin3/Arrb1/Arrb2/Ccn2 | 27 |
| GO:0045664 | regulation of neuron differentiation | BP | 0.116 | 1.52e-06 | 3e-05 | 4.52 | 2.04e-05 | Sfrp2/Fez1/Nlgn1/Rgs6/Ednrb/Dab1/Lrrk2/Mef2c/Id4/Cxcl12/Gdf11/Gprc5b/Gli3/Gdf7/Eif2ak4/Zeb1/Trpc6/Tgif2/Mmd/Hoxd3/Heyl/Zc4h2/Tcf4/Nin/Bin1/Ptger3/Bend6/Cav1 | 28 |
| GO:0060993 | kidney morphogenesis | BP | 0.165 | 1.54e-06 | 3.03e-05 | 4.52 | 2.06e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Lrrk2/Gli3/Tcf21/Tshz3/Agtr1a/Ptch1/Osr1/Dchs1/Fat4 | 17 |
| GO:0050982 | detection of mechanical stimulus | BP | 0.21 | 1.63e-06 | 3.19e-05 | 4.5 | 2.17e-05 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12/Kit/Htr2a/Pkd1/Fyn/Pkd2/Pkdrej/Piezo2 | 13 |
| GO:0090257 | regulation of muscle system process | BP | 0.113 | 1.66e-06 | 3.24e-05 | 4.49 | 2.21e-05 | Cacna1c/Ryr1/Igf1/Adra1a/Npnt/Abat/Nos3/Slc8a1/Mef2c/Stc1/Ank2/Slc8a3/Calca/Gucy1a1/Kit/Kcnma1/Myl3/Cmya5/Bin1/Selenon/Pde5a/Cacna1h/Ptger3/Calcrl/Ccn2/Pi16/Atp2b4/Atp1a2/Cav1 | 29 |
| GO:0032231 | regulation of actin filament bundle assembly | BP | 0.157 | 1.7e-06 | 3.29e-05 | 4.48 | 2.24e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arhgap28/Dlc1/Ccn2 | 18 |
| GO:0032102 | negative regulation of response to external stimulus | BP | 0.0968 | 1.7e-06 | 3.29e-05 | 4.48 | 2.24e-05 | Hgf/Acod1/Sema5a/Mdk/Pla2g10/Cd109/Igf1/Sema5b/Tfpi/Abat/Aph1b/Phldb2/Cd44/Sema7a/Nrp1/Ets1/Foxf1/Serpine2/Fndc4/Cdh5/Prdm1/Nlgn3/Apoe/Havcr2/Cd200/Igf2/Sema3g/Pros1/Rtn4rl1/Tnfaip6/Mapkbp1/Arrb2/Neo1/Ptger3/Calcrl/Xylt1/Sema6a/Cd96/Nr1h3 | 39 |
| GO:0055123 | digestive system development | BP | 0.144 | 1.79e-06 | 3.46e-05 | 4.46 | 2.35e-05 | Stra6/Ihh/Sfrp2/Igf1/Clmp/Pkdcc/Tbx2/Ednrb/Foxf1/Gli3/Tcf21/Prdm1/Kit/Cdkn1c/Igf2/Pcsk5/Dchs1/Fat4/Dact1/Stx2 | 20 |
| GO:0006937 | regulation of muscle contraction | BP | 0.134 | 1.85e-06 | 3.56e-05 | 4.45 | 2.42e-05 | Cacna1c/Ryr1/Adra1a/Npnt/Abat/Slc8a1/Stc1/Ank2/Slc8a3/Calca/Gucy1a1/Kit/Kcnma1/Myl3/Bin1/Pde5a/Cacna1h/Ptger3/Calcrl/Ccn2/Atp1a2/Cav1 | 22 |
| GO:0006486 | protein glycosylation | BP | 0.126 | 1.92e-06 | 3.64e-05 | 4.44 | 2.48e-05 | St6gal2/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Tmtc1/B3galnt1/Fut10/Galnt16/St6galnac4/St3gal2/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Aqp11/Mgat3/B3gnt5/St6galnac6/St3gal4/Large1/Alg14 | 24 |
| GO:0043413 | macromolecule glycosylation | BP | 0.126 | 1.92e-06 | 3.64e-05 | 4.44 | 2.48e-05 | St6gal2/St8sia2/St8sia4/Gxylt2/Tet1/Uggt2/Tmtc1/B3galnt1/Fut10/Galnt16/St6galnac4/St3gal2/Eogt/Ggta1/St8sia6/Galntl6/Poglut2/Aqp11/Mgat3/B3gnt5/St6galnac6/St3gal4/Large1/Alg14 | 24 |
| GO:0043524 | negative regulation of neuron apoptotic process | BP | 0.126 | 1.92e-06 | 3.64e-05 | 4.44 | 2.48e-05 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Coro1a/Il27ra/Grik2/Apoe/Ndnf/Arrb1/Hipk2/Nfatc4/Arrb2/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 24 |
| GO:0043534 | blood vessel endothelial cell migration | BP | 0.155 | 1.93e-06 | 3.65e-05 | 4.44 | 2.48e-05 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Mef2c/Prkd1/Vash1/Cdh5/Jcad/Sh3bp1/Vegfc/Apoe/Igf2/Hdac9/Atp2b4/Akt3 | 18 |
| GO:0072078 | nephron tubule morphogenesis | BP | 0.181 | 1.94e-06 | 3.66e-05 | 4.44 | 2.49e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Osr1/Dchs1/Fat4 | 15 |
| GO:0003206 | cardiac chamber morphogenesis | BP | 0.138 | 1.95e-06 | 3.67e-05 | 4.43 | 2.5e-05 | Col11a1/Sfrp2/Wnt11/Nog/Tbx2/Nos3/Nrp1/Foxf1/Mef2c/Tek/Adamts1/Lrp2/Cpe/Ccm2l/Aplnr/Tbx3/Heyl/Ednra/Myl3/Heg1/Fzd2 | 21 |
| GO:0072132 | mesenchyme morphogenesis | BP | 0.222 | 2.13e-06 | 3.99e-05 | 4.4 | 2.71e-05 | Lef1/Wnt11/Nog/Tbx2/Nos3/Foxf1/Aplnr/Tbx3/Heyl/Snai1/Osr1/Dchs1 | 12 |
| GO:0010749 | regulation of nitric oxide mediated signal transduction | BP | 0.545 | 2.22e-06 | 4.13e-05 | 4.38 | 2.81e-05 | Spink1/Cbs/Gucy1a2/Gucy1a1/Pde5a/Atp2b4 | 6 |
| GO:0014832 | urinary bladder smooth muscle contraction | BP | 0.545 | 2.22e-06 | 4.13e-05 | 4.38 | 2.81e-05 | Cacna1c/P2rx2/Kcnma1/Htr2a/Ptger3/Atp2b4 | 6 |
| GO:0051495 | positive regulation of cytoskeleton organization | BP | 0.122 | 2.26e-06 | 4.16e-05 | 4.38 | 2.83e-05 | Sema5a/Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Wasf3/Pfn2/Icam1/Tek/Abi2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Scin/Nckap1l/Slain1/Nin/Bin1/Ccn2/Cdc42ep5/Nes/Cav1 | 25 |
| GO:0120032 | regulation of plasma membrane bounded cell projection assembly | BP | 0.122 | 2.26e-06 | 4.16e-05 | 4.38 | 2.83e-05 | Gap43/Evl/Nlgn1/Ccdc88a/Nrp1/Adamts16/Abi3/Pfn2/Icam1/Podxl/Abi2/Kit/Rp1/Kctd17/Eps8l1/Daam2/Crocc/Ttbk2/Dzip1/Palm/Dync2li1/Dpysl3/Cdc42ep5/Cep97/Cav1 | 25 |
| GO:0030048 | actin filament-based movement | BP | 0.147 | 2.26e-06 | 4.16e-05 | 4.38 | 2.83e-05 | Sgcd/Cacna1c/Kcnd3/Evl/Scn4b/Scn1a/Cacna2d1/Myo5a/Stc1/Ank2/Limch1/Bin1/Epdr1/Myo1f/Cacna1h/Ptger3/Wipf1/Atp1a2/Cav1 | 19 |
| GO:0060675 | ureteric bud morphogenesis | BP | 0.189 | 2.41e-06 | 4.42e-05 | 4.35 | 3e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Dchs1/Fat4 | 14 |
| GO:0017157 | regulation of exocytosis | BP | 0.113 | 2.46e-06 | 4.51e-05 | 4.35 | 3.07e-05 | Nlgn1/Syt13/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Foxf1/Myo5a/Pfn2/Mical1/Ppfia2/Htr1b/Cadps/Syt7/Htr2a/Nckap1l/Ncs1/Cd84/Cacna1h/Septin4/Fgr/Rab27a/Nrn1/Slc4a8/Cacna1a/Cacnb4 | 28 |
| GO:0010563 | negative regulation of phosphorus metabolic process | BP | 0.0919 | 2.62e-06 | 4.75e-05 | 4.32 | 3.23e-05 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Ptpro/Slc8a1/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Ppp1r16b/Cdkn1c/Prr5l/Apoe/Enpp1/Ppef2/Rasip1/Emilin1/Pecam1/Ppm1e/Bdkrb2/Nckap1l/Pkib/Cmya5/Lpcat1/Arrb1/Heg1/Rassf2/Arrb2/Ppp1r26/Lats2/Smpd1/Cav1/Pid1 | 42 |
| GO:0045936 | negative regulation of phosphate metabolic process | BP | 0.0919 | 2.62e-06 | 4.75e-05 | 4.32 | 3.23e-05 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Ptpro/Slc8a1/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Ppp1r16b/Cdkn1c/Prr5l/Apoe/Enpp1/Ppef2/Rasip1/Emilin1/Pecam1/Ppm1e/Bdkrb2/Nckap1l/Pkib/Cmya5/Lpcat1/Arrb1/Heg1/Rassf2/Arrb2/Ppp1r26/Lats2/Smpd1/Cav1/Pid1 | 42 |
| GO:0072088 | nephron epithelium morphogenesis | BP | 0.176 | 2.65e-06 | 4.8e-05 | 4.32 | 3.26e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Osr1/Dchs1/Fat4 | 15 |
| GO:0070509 | calcium ion import | BP | 0.167 | 2.72e-06 | 4.91e-05 | 4.31 | 3.34e-05 | Cacna1c/Spink1/Slc8a1/Slc24a4/Stc1/Cxcl12/Cacna1e/Slc8a3/Nalf1/Agtr1a/Dysf/Cacna1h/Fyn/Pkd2/Slc25a23/Cacna1a | 16 |
| GO:0070977 | bone maturation | BP | 0.3 | 2.77e-06 | 4.98e-05 | 4.3 | 3.39e-05 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:2001257 | regulation of cation channel activity | BP | 0.127 | 2.81e-06 | 5.01e-05 | 4.3 | 3.41e-05 | Nlgn1/Drd4/Cacna2d1/Mef2c/Myo5a/Ank2/Nlgn3/Cracr2a/Prrt1/Ednra/Stac2/Cacng7/Dysf/Asph/Selenon/Rrad/Pkd2/Dlg4/Gstm7/Gnb5/Cav1/Cacna1a/Cacnb4 | 23 |
| GO:0051209 | release of sequestered calcium ion into cytosol | BP | 0.151 | 2.81e-06 | 5.01e-05 | 4.3 | 3.41e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Pkd2/Gstm7 | 18 |
| GO:0048588 | developmental cell growth | BP | 0.108 | 2.81e-06 | 5.01e-05 | 4.3 | 3.41e-05 | Map2/Sema5a/Itga4/Col14a1/Igf1/St8sia2/Sema5b/Zeb2/Adra1a/Cpne5/Sema7a/Nrp1/Fn1/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Nin/Epha7/Cacng7/Pi16/Sema6a/Nrn1/Cyfip2 | 30 |
| GO:0060828 | regulation of canonical Wnt signaling pathway | BP | 0.112 | 2.89e-06 | 5.13e-05 | 4.29 | 3.49e-05 | Gli1/Sfrp2/Sema5a/Mdk/Wnt11/Nog/Zeb2/Ptpro/Nkd1/Cdh2/Gpc3/Lrrk2/Rbms3/Gprc5b/Gli3/Peg12/Dkk3/Znrf3/Apoe/Caprin2/Daam2/Prickle1/Tle1/Lats2/Adgra2/Cav1/Dact1/Nkd2 | 28 |
| GO:0060491 | regulation of cell projection assembly | BP | 0.12 | 2.94e-06 | 5.21e-05 | 4.28 | 3.54e-05 | Gap43/Evl/Nlgn1/Ccdc88a/Nrp1/Adamts16/Abi3/Pfn2/Icam1/Podxl/Abi2/Kit/Rp1/Kctd17/Eps8l1/Daam2/Crocc/Ttbk2/Dzip1/Palm/Dync2li1/Dpysl3/Cdc42ep5/Cep97/Cav1 | 25 |
| GO:0051283 | negative regulation of sequestering of calcium ion | BP | 0.15 | 3.18e-06 | 5.6e-05 | 4.25 | 3.81e-05 | Ryr1/Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Xcl1/Htr2a/Asph/Ryr3/Selenon/Xcr1/Pkd2/Gstm7 | 18 |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | BP | 0.214 | 3.2e-06 | 5.63e-05 | 4.25 | 3.83e-05 | Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Jcad/Vegfc/Igf2/Hdac9/Akt3 | 12 |
| GO:0060537 | muscle tissue development | BP | 0.0892 | 3.28e-06 | 5.73e-05 | 4.24 | 3.9e-05 | Gli1/Col11a1/Stra6/Ihh/AW551984/Angpt1/Lef1/Sgcd/Hoxd10/Col14a1/Ryr1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/P2rx2/Hoxd9/Mef2c/Tcf21/Lrp2/Tshz3/Ccm2l/Nox4/Tbx3/Heyl/Gjc1/Ednra/Igf2/Myl3/Hdac9/Heg1/Dysf/Selenon/Osr1/Arrb2/Pi16/Hlf/Efemp2/Sgcb/Cav1/Zfp689/Usp2/Itga8 | 44 |
| GO:0007628 | adult walking behavior | BP | 0.234 | 3.28e-06 | 5.73e-05 | 4.24 | 3.9e-05 | Efnb3/Cacna1c/Scn1a/Dab1/Glrb/Enpp1/Kcnma1/Hipk2/Arrb2/Cacna1a/Cacnb4 | 11 |
| GO:0001764 | neuron migration | BP | 0.122 | 3.33e-06 | 5.79e-05 | 4.24 | 3.94e-05 | Ptprz1/Mdk/Dcc/Mark1/Nrp1/Dab1/Mef2c/Cxcl12/Tbc1d24/Cep85l/Ndn/Gpr173/Matn2/Ndnf/Lhx6/Lrp12/Nr2f1/Neo1/Fyn/Septin4/Fat3/Sema6a/Ntng1/Bbs1 | 24 |
| GO:0048015 | phosphatidylinositol-mediated signaling | BP | 0.129 | 3.39e-06 | 5.88e-05 | 4.23 | 4e-05 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Car8/Npr3/Hcls1/Fn1/Serpine2/Tek/Plcb1/Ppp1r16b/Prr5l/Htr2a/Prex2/Ncs1/Plcb2/Fyn/Fgr | 22 |
| GO:0072028 | nephron morphogenesis | BP | 0.172 | 3.58e-06 | 6.19e-05 | 4.21 | 4.21e-05 | Wnt6/Hoxd11/Nog/Npnt/Cd44/Hoxa11/Adamts16/Gpc3/Gli3/Tcf21/Agtr1a/Ptch1/Osr1/Dchs1/Fat4 | 15 |
| GO:0002026 | regulation of the force of heart contraction | BP | 0.29 | 3.76e-06 | 6.46e-05 | 4.19 | 4.39e-05 | Adra1a/Nos3/Slc8a1/Myl3/Pde5a/Apln/Atp2b4/Atp1a2/Cav1 | 9 |
| GO:2000352 | negative regulation of endothelial cell apoptotic process | BP | 0.29 | 3.76e-06 | 6.46e-05 | 4.19 | 4.39e-05 | Sema5a/Angpt1/Kdr/Tert/Icam1/Tek/Cdh5/Ndnf/Angptl4 | 9 |
| GO:0060349 | bone morphogenesis | BP | 0.155 | 3.91e-06 | 6.69e-05 | 4.17 | 4.55e-05 | Ihh/Mmp16/Sfrp2/Hoxd11/Thbs3/Cbs/Hoxa11/Mef2c/Osr2/Stc1/Has2/Gli3/Frem1/Col13a1/Enpp1/Ltbp3/Rab23 | 17 |
| GO:1903706 | regulation of hemopoiesis | BP | 0.0945 | 4.01e-06 | 6.86e-05 | 4.16 | 4.66e-05 | Ihh/Lef1/Mdk/Cd83/P4htm/Cd44/Hcls1/Ets1/Mef2c/Inpp4b/Pik3r6/Lrrc17/Gli3/Calca/Flt3/Zeb1/Prdm1/Cd1d1/Loxl3/Tnfrsf11b/Gpr171/Sox12/Scin/Zbtb46/Nckap1l/Tal1/Hoxa9/Kitl/Tnfaip6/Twist2/Rassf2/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Ptn/Il2rg | 38 |
| GO:0070372 | regulation of ERK1 and ERK2 cascade | BP | 0.101 | 4.03e-06 | 6.86e-05 | 4.16 | 4.67e-05 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Adra1a/Npnt/Cd44/Sema7a/Nrp1/Fermt2/Fn1/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Nox4/Apoe/Havcr2/Emilin1/Ednra/Spred3/Xcl1/Htr2a/Cavin3/Arrb1/Epha7/Arrb2/Ccn2/Sema6a/Cav1 | 33 |
| GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | BP | 0.229 | 4.1e-06 | 6.95e-05 | 4.16 | 4.73e-05 | Wnt11/Lrrc7/Foxf1/Dlg2/Msn/Lin7a/Prickle2/Myo9a/Sh3bp1/Pard3b/Dlg5 | 11 |
| GO:0042098 | T cell proliferation | BP | 0.115 | 4.25e-06 | 7.16e-05 | 4.15 | 4.87e-05 | Ihh/Igf1/Cd44/Cxcl12/Mapk8ip1/Dock2/Tnfsf13b/Cd1d1/Msn/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Havcr2/Igf2/Xcl1/Nckap1l/Satb1/Pde5a/Fyn/H2-Aa/Stat5b/Zap70/Dlg5/Il12rb1 | 26 |
| GO:0071560 | cellular response to transforming growth factor beta stimulus | BP | 0.115 | 4.25e-06 | 7.16e-05 | 4.15 | 4.87e-05 | Cd109/Zeb2/Npnt/Nos3/Fermt2/Mef2c/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Xcl1/Smad9/Ltbp3/Hipk2/Nrros/Zfyve9/Cldn5/Arrb2/Fyn/Il17rd/Lats2/Cav1/Itga8 | 26 |
| GO:0007265 | Ras protein signal transduction | BP | 0.0994 | 4.3e-06 | 7.24e-05 | 4.14 | 4.92e-05 | Igf1/Adra1a/Rasgrp3/Nrp1/Cdh2/Arl6/Rasa3/Lpar4/Prkd1/Arhgef25/Cdh13/Rasgrp2/Dock2/Sh2b2/Syde1/Abi2/Sh3bp1/Apoe/Agtr1a/Rasip1/Eps8l1/Ednra/Pecam1/Arap3/Kitl/Arrb1/Heg1/Dlc1/Net1/Lat/Syngap1/Rab39b/Cdc42ep5/Mapre2 | 34 |
| GO:0044782 | cilium organization | BP | 0.0954 | 4.38e-06 | 7.33e-05 | 4.13 | 4.99e-05 | Tub/Ccdc88a/Adamts16/Dzip1l/Arl6/Wdr35/Cibar1/Tmem237/Bbs7/Dnah5/Zfp423/Armc9/Ift57/Ptpdc1/Catip/Rp1/Kctd17/Cplane1/Rilpl1/Dync2h1/Crocc/Arl3/Cfap410/Tmem231/Ttc26/Ttbk2/Dzip1/Cep19/Pkd2/Dnah7b/Iqcb1/Rab23/Dnah1/Dync2li1/Lrrc49/Cep97/Bbs1 | 37 |
| GO:0046631 | alpha-beta T cell activation | BP | 0.124 | 4.48e-06 | 7.48e-05 | 4.13 | 5.09e-05 | Ihh/Lef1/Cd83/Wdfy4/Cd44/Itk/Mapk8ip1/Gli3/Dock2/Prdm1/Cd1d1/Loxl3/Btla/H2-Ab1/Cracr2a/Xcl1/Nckap1l/Satb1/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg | 23 |
| GO:0048017 | inositol lipid-mediated signaling | BP | 0.127 | 4.53e-06 | 7.52e-05 | 4.12 | 5.12e-05 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Car8/Npr3/Hcls1/Fn1/Serpine2/Tek/Plcb1/Ppp1r16b/Prr5l/Htr2a/Prex2/Ncs1/Plcb2/Fyn/Fgr | 22 |
| GO:0051017 | actin filament bundle assembly | BP | 0.127 | 4.53e-06 | 7.52e-05 | 4.12 | 5.12e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Aif1l/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arrb1/Arhgap28/Dlc1/Ccn2/Dpysl3/Shroom1 | 22 |
| GO:0006790 | sulfur compound metabolic process | BP | 0.101 | 4.59e-06 | 7.6e-05 | 4.12 | 5.17e-05 | Acsl6/Angpt1/Hmgcs2/Chst2/Cbs/Acacb/Spock2/Ednrb/Mical1/Tpst1/Egflam/Gamt/Gstm6/Them5/Mamdc2/Chst1/Cth/Phgdh/Nox4/Enpp1/Ednra/Ndnf/Acot7/Bgn/Gstm7/Gstm1/Stat5b/Xylt1/Dse/Ggt7/Acsl1/Cacna1a/Sult1a1 | 33 |
| GO:0051496 | positive regulation of stress fiber assembly | BP | 0.207 | 4.73e-06 | 7.81e-05 | 4.11 | 5.31e-05 | Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Ccn2 | 12 |
| GO:0048265 | response to pain | BP | 0.25 | 4.84e-06 | 7.96e-05 | 4.1 | 5.42e-05 | Grik1/Slc6a2/P2rx2/Ednrb/Grin2b/Cacna1e/Calca/Kcnip3/Vwa1/Cacna1a | 10 |
| GO:0071772 | response to BMP | BP | 0.123 | 4.9e-06 | 8.02e-05 | 4.1 | 5.45e-05 | Sfrp2/Lef1/Kdr/Nog/Kcnd3/Rnf165/Fst/Gpc3/Vstm2a/Zfp423/Lrp2/Gdf7/Cdh5/Heyl/Adamts7/Kcp/Smad9/Tnfaip6/Hipk2/Neo1/Fstl1/Cav1/Tmprss6 | 23 |
| GO:0071773 | cellular response to BMP stimulus | BP | 0.123 | 4.9e-06 | 8.02e-05 | 4.1 | 5.45e-05 | Sfrp2/Lef1/Kdr/Nog/Kcnd3/Rnf165/Fst/Gpc3/Vstm2a/Zfp423/Lrp2/Gdf7/Cdh5/Heyl/Adamts7/Kcp/Smad9/Tnfaip6/Hipk2/Neo1/Fstl1/Cav1/Tmprss6 | 23 |
| GO:0098703 | calcium ion import across plasma membrane | BP | 0.281 | 5.03e-06 | 8.2e-05 | 4.09 | 5.58e-05 | Cacna1c/Slc8a1/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Fyn/Cacna1a | 9 |
| GO:0045761 | regulation of adenylate cyclase activity | BP | 0.224 | 5.08e-06 | 8.23e-05 | 4.08 | 5.6e-05 | Cacna1c/Vipr2/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Gng7/Palm/Gabbr1 | 11 |
| GO:0071526 | semaphorin-plexin signaling pathway | BP | 0.224 | 5.08e-06 | 8.23e-05 | 4.08 | 5.6e-05 | Sema5a/Plxnb1/Sema5b/Sema7a/Nrp1/Mef2c/Plxnd1/Sh3bp1/Ednra/Sema3g/Sema6a | 11 |
| GO:0019233 | sensory perception of pain | BP | 0.134 | 5.32e-06 | 8.59e-05 | 4.07 | 5.85e-05 | Kcnd2/Scn1a/Ednrb/Ano1/Grin2b/Cxcl12/Cacna1e/Calca/Mme/Dlg2/Ndn/Kcnc4/Ednra/Htr2a/Kcnip3/Cacna1h/Arrb2/Fyn/Scn3a/Cacna1a | 20 |
| GO:0031279 | regulation of cyclase activity | BP | 0.203 | 5.71e-06 | 9.17e-05 | 4.04 | 6.24e-05 | Cacna1c/Vipr2/Nos3/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Gng7/Palm/Gabbr1 | 12 |
| GO:0150116 | regulation of cell-substrate junction organization | BP | 0.188 | 5.75e-06 | 9.21e-05 | 4.04 | 6.26e-05 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1/Mapre2 | 13 |
| GO:0071559 | response to transforming growth factor beta | BP | 0.113 | 5.86e-06 | 9.35e-05 | 4.03 | 6.36e-05 | Cd109/Zeb2/Npnt/Nos3/Fermt2/Mef2c/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Xcl1/Smad9/Ltbp3/Hipk2/Nrros/Zfyve9/Cldn5/Arrb2/Fyn/Il17rd/Lats2/Cav1/Itga8 | 26 |
| GO:0061572 | actin filament bundle organization | BP | 0.125 | 6.01e-06 | 9.56e-05 | 4.02 | 6.5e-05 | Kank4/Wnt11/Evl/Phldb2/Ccdc88a/Nrp1/Fermt2/Aif1l/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Arhgap6/Tmeff2/Ppm1e/Arrb1/Arhgap28/Dlc1/Ccn2/Dpysl3/Shroom1 | 22 |
| GO:0048167 | regulation of synaptic plasticity | BP | 0.0952 | 6.04e-06 | 9.58e-05 | 4.02 | 6.52e-05 | Grik1/Kdr/Nog/Jph4/Grid2ip/Nlgn1/Rims3/Mef2c/Grin2b/Serpine2/Shank2/Slc8a3/Eif2ak4/Tshz3/Mme/Kit/Nlgn3/Nrgn/Sorcs2/Grik2/Apoe/Prrt1/Cd38/Enpp1/Syt7/Prrt2/Nsg1/Cyp46a1/Erc2/Mpp2/Nfatc4/Dlg4/Brsk1/Syngap1/Ptn/Cpeb1 | 36 |
| GO:0150115 | cell-substrate junction organization | BP | 0.157 | 6.13e-06 | 9.7e-05 | 4.01 | 6.6e-05 | Kdr/Phldb2/Nrp1/Fermt2/Fn1/Col16a1/Tek/Thsd1/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1/Mapre2/Itgb4 | 16 |
| GO:0003197 | endocardial cushion development | BP | 0.22 | 6.26e-06 | 9.85e-05 | 4.01 | 6.7e-05 | Nog/Tbx2/Nos3/Foxf1/Erg/Epha3/Aplnr/Tbx3/Heyl/Snai1/Dchs1 | 11 |
| GO:0090659 | walking behavior | BP | 0.22 | 6.26e-06 | 9.85e-05 | 4.01 | 6.7e-05 | Efnb3/Cacna1c/Scn1a/Dab1/Glrb/Enpp1/Kcnma1/Hipk2/Arrb2/Cacna1a/Cacnb4 | 11 |
| GO:0003231 | cardiac ventricle development | BP | 0.132 | 6.53e-06 | 0.000102 | 3.99 | 6.97e-05 | Col11a1/Stra6/Sfrp2/Wnt11/Nog/Nos3/Foxf1/Mef2c/Lrp2/Cpe/Prdm1/Ccm2l/Aplnr/Cplane1/Tbx3/Heyl/Ednra/Myl3/Heg1/Fzd2 | 20 |
| GO:1902656 | calcium ion import into cytosol | BP | 0.273 | 6.65e-06 | 0.000104 | 3.98 | 7.08e-05 | Cacna1c/Slc8a1/Slc24a4/Cacna1e/Slc8a3/Nalf1/Agtr1a/Fyn/Cacna1a | 9 |
| GO:0031503 | protein-containing complex localization | BP | 0.118 | 6.68e-06 | 0.000104 | 3.98 | 7.09e-05 | Gpc6/Sgcd/Tub/Nlgn1/Lrrc7/Cep112/Drd4/Stx1b/Nptx1/Wdr35/Exoc3l/Ift57/Dlg2/Nptxr/Magi2/Dync2h1/Nsg1/Arl3/Cacng7/Ttc26/Dzip1/Dlg4/Dync2li1/Bbs1 | 24 |
| GO:0043393 | regulation of protein binding | BP | 0.115 | 6.82e-06 | 0.000106 | 3.97 | 7.22e-05 | Map2/Angpt1/Itga4/Nog/Tert/Lrrk2/Hopx/Plxnd1/Lfng/Ccm2l/Apoe/Caprin2/Arrb1/Hipk2/Pkd1/Dysf/Ttbk2/Nfatc4/Cldn5/Arrb2/Dzip1/Mfng/Nes/Cav1/Dact1 | 25 |
| GO:0051588 | regulation of neurotransmitter transport | BP | 0.137 | 6.94e-06 | 0.000108 | 3.97 | 7.32e-05 | Nlgn1/Adra1a/Rims3/Drd4/Stx1b/P2rx2/Lrrk2/Mef2c/Pfn2/Edn3/Ppfia2/Htr1b/Kcnc4/Htr2a/Ncs1/Nrn1/Slc4a8/Cacna1a/Cacnb4 | 19 |
| GO:0042063 | gliogenesis | BP | 0.0971 | 7.1e-06 | 0.00011 | 3.96 | 7.47e-05 | Gap43/Ptprz1/Lef1/Mdk/Apcdd1/Nog/Igf1/Lamc3/Cspg4/Tert/Cdh2/Wasf3/Areg/Fn1/Dab1/Id4/Serpine2/Gli3/Slc8a3/Lrp2/Plpp3/Ndn/Nlgn3/Matn2/Phgdh/Enpp1/Daam2/Tal1/Bin1/Zcchc24/Nrros/Ptn/Tppp/Itgb4 | 34 |
| GO:0050806 | positive regulation of synaptic transmission | BP | 0.0985 | 7.19e-06 | 0.000111 | 3.96 | 7.54e-05 | Slc1a3/Gria4/Grik1/Nog/Nlgn1/Adra1a/Rims3/Stx1b/Grin2b/Serpine2/Shank2/Slc8a3/Eif2ak4/Tshz3/Syde1/Mme/Lama2/Nlgn3/Nrgn/Grik2/Apoe/Prrt1/Enpp1/Nsg1/Cacng7/Cyp46a1/Ncs1/Mpp2/Nfatc4/Arrb2/Dlg4/Ptn/Slc4a8 | 33 |
| GO:0046634 | regulation of alpha-beta T cell activation | BP | 0.148 | 7.23e-06 | 0.000111 | 3.95 | 7.56e-05 | Ihh/Cd83/Cd44/Mapk8ip1/Gli3/Prdm1/Cd1d1/Loxl3/Btla/H2-Ab1/Xcl1/Nckap1l/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg | 17 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | BP | 0.112 | 7.41e-06 | 0.000114 | 3.94 | 7.72e-05 | Igf1/Adra1a/Nrp1/Cdh2/Garnl3/Rasa3/Lpar4/Arhgef25/Arhgap25/Dock2/Sh2b2/Syde1/Sh3bp1/Apoe/Rasip1/Eps8l1/Adcyap1r1/Arap3/Kitl/Arrb1/Heg1/Arhgap28/Dlc1/Net1/Syngap1/Mapre2 | 26 |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | BP | 0.12 | 7.64e-06 | 0.000117 | 3.93 | 7.94e-05 | Ptprz1/Hgf/Angpt1/Igf1/Adra1a/Cd44/Hcls1/Nrp1/Cspg4/Areg/Icam1/Gprc5b/Plpp3/Kit/Vegfc/Nox4/Igf2/Pecam1/Htr2a/Kitl/Arrb2/Fyn/Dlg4 | 23 |
| GO:0030900 | forebrain development | BP | 0.093 | 7.81e-06 | 0.000119 | 3.92 | 8.1e-05 | Gli1/Sema5a/Lef1/Mdk/Nog/Zeb2/Sema7a/Nrp1/Cdh2/Dab1/Lrrk2/Id4/Fut10/Cxcl12/Dnah5/Gli3/Plcb1/Lrp2/Gdf7/D16Ertd472e/Kif1a/Tsku/Tbx3/Dync2h1/Ndnf/Dclk2/Lhx6/Rtn4rl1/Nin/Nr2f1/Ttbk2/Dlc1/Ptger3/Fyn/Atp1a2/Fat4/Bbs1 | 37 |
| GO:0030510 | regulation of BMP signaling pathway | BP | 0.154 | 7.92e-06 | 0.00012 | 3.92 | 8.19e-05 | Sfrp2/Kdr/Nog/Rnf165/Fst/Gpc3/Zfp423/Lrp2/Cdh5/Kcp/Tnfaip6/Hipk2/Neo1/Fstl1/Cav1/Tmprss6 | 16 |
| GO:0030282 | bone mineralization | BP | 0.141 | 8.05e-06 | 0.000122 | 3.91 | 8.3e-05 | Wnt11/Igf1/Pkdcc/Ddr2/Slc8a1/Gpc3/Mef2c/Osr2/Slc24a3/Nell1/Pth1r/Mgp/Enpp1/Ltbp3/Osr1/Fgr/Ptn/Dnm3os | 18 |
| GO:0070661 | leukocyte proliferation | BP | 0.0951 | 8.33e-06 | 0.000126 | 3.9 | 8.56e-05 | Ihh/Lef1/Igf1/Cd44/Npr3/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Prdm1/Cd1d1/Msn/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Satb1/Kitl/Csf2rb/Pde5a/Fyn/H2-Aa/Stat5b/Zap70/Impdh1/Dlg5/Il12rb1 | 35 |
| GO:0048799 | animal organ maturation | BP | 0.265 | 8.71e-06 | 0.000131 | 3.88 | 8.93e-05 | Ryr1/Plxnb1/Igf1/Thbs3/Enpp1/Adamts7/Dchs1/Xylt1/Fat4 | 9 |
| GO:0042326 | negative regulation of phosphorylation | BP | 0.0925 | 8.75e-06 | 0.000131 | 3.88 | 8.94e-05 | Hgf/Sfrp2/Angpt1/Cd109/Spink1/Nog/Igf1/Ddr2/Ptpro/Slc8a1/Lrrk2/Mical1/Mapk8ip1/Slc8a3/Pkia/Thy1/Plpp3/Ppm1f/Cdkn1c/Prr5l/Apoe/Enpp1/Ppef2/Rasip1/Emilin1/Pecam1/Ppm1e/Bdkrb2/Pkib/Arrb1/Heg1/Rassf2/Arrb2/Lats2/Smpd1/Cav1/Pid1 | 37 |
| GO:0031644 | regulation of nervous system process | BP | 0.119 | 9.08e-06 | 0.000136 | 3.87 | 9.25e-05 | Hgf/Begain/Igf1/Nlgn1/Abat/Celf4/Nos3/Drd4/Stx1b/Ednrb/Wasf3/Lrrk2/Nptx1/Grin2b/Tbc1d24/Slc8a3/Calca/Nlgn3/Nptxr/Kcnc4/Dlg4/Cux2/Tppp | 23 |
| GO:0061028 | establishment of endothelial barrier | BP | 0.212 | 9.37e-06 | 0.00014 | 3.85 | 9.53e-05 | Ednrb/Icam1/Plcb1/Cdh5/Msn/Ppp1r16b/Robo4/Ednra/Pecam1/Cldn5/Cldn1 | 11 |
| GO:0030203 | glycosaminoglycan metabolic process | BP | 0.16 | 9.58e-06 | 0.000143 | 3.85 | 9.71e-05 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Xylt1/Dse/Habp4 | 15 |
| GO:0060271 | cilium assembly | BP | 0.0958 | 9.61e-06 | 0.000143 | 3.85 | 9.72e-05 | Ccdc88a/Adamts16/Dzip1l/Arl6/Wdr35/Cibar1/Tmem237/Bbs7/Dnah5/Zfp423/Armc9/Ift57/Ptpdc1/Rp1/Kctd17/Cplane1/Rilpl1/Dync2h1/Crocc/Arl3/Cfap410/Tmem231/Ttc26/Ttbk2/Dzip1/Cep19/Dnah7b/Iqcb1/Rab23/Dnah1/Dync2li1/Lrrc49/Cep97/Bbs1 | 34 |
| GO:0048593 | camera-type eye morphogenesis | BP | 0.129 | 9.73e-06 | 0.000144 | 3.84 | 9.81e-05 | Stra6/Ihh/Sdk2/Kdr/Tbx2/Arl6/Tenm3/Gdf11/Obsl1/Gli3/Thy1/Zeb1/Ptprm/Abi2/Rp1/Tsku/Rorb/Hipk2/Fat3/Ptn | 20 |
| GO:0001736 | establishment of planar polarity | BP | 0.194 | 9.8e-06 | 0.000144 | 3.84 | 9.83e-05 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Cplane1/Dchs1/Fzd2/Nphp1/Fat4/Brsk1/Dact1 | 12 |
| GO:0051339 | regulation of lyase activity | BP | 0.194 | 9.8e-06 | 0.000144 | 3.84 | 9.83e-05 | Cacna1c/Vipr2/Nos3/Npr3/Adcy3/Calca/Adcy4/Adcy7/Cap2/Gng7/Palm/Gabbr1 | 12 |
| GO:0003279 | cardiac septum development | BP | 0.138 | 1e-05 | 0.000147 | 3.83 | 1e-04 | Stra6/Wnt11/Nog/Tbx2/Nos3/Nrp1/Kif7/Ank2/Lrp2/Plxnd1/Prdm1/Aplnr/Cplane1/Tbx3/Heyl/Pcsk5/Heg1/Fzd2 | 18 |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | BP | 0.105 | 1.02e-05 | 0.00015 | 3.82 | 0.000102 | Ptprz1/Hgf/Sfrp2/Angpt1/Spink1/Igf1/Adra1a/Cd44/Hcls1/Nrp1/Cspg4/Areg/Icam1/Gprc5b/Thy1/Plpp3/Kit/Vegfc/Nox4/Igf2/Pecam1/Htr2a/Kitl/Epha7/Arrb2/Fyn/Dlg4/Cav1 | 28 |
| GO:0019226 | transmission of nerve impulse | BP | 0.158 | 1.09e-05 | 0.00016 | 3.8 | 0.000109 | Kcnd2/P2rx2/Scn1a/Jam3/Cacna1e/Scn2a/Nfasc/Grik2/Kcnma1/Kcnc4/Cacng7/Cacna1h/Scn3a/Cacna1a/Cacnb4 | 15 |
| GO:0061512 | protein localization to cilium | BP | 0.178 | 1.1e-05 | 0.00016 | 3.8 | 0.000109 | Tub/Ccdc88a/Dzip1l/Arl6/Wdr35/Zfp423/Cplane1/Dync2h1/Crocc/Arl3/Ttc26/Dzip1/Bbs1 | 13 |
| GO:2000300 | regulation of synaptic vesicle exocytosis | BP | 0.167 | 1.13e-05 | 0.000164 | 3.79 | 0.000112 | Nlgn1/Adra1a/Rims3/Drd4/P2rx2/Lrrk2/Pfn2/Ppfia2/Htr1b/Htr2a/Ncs1/Nrn1/Slc4a8/Cacnb4 | 14 |
| GO:1904036 | negative regulation of epithelial cell apoptotic process | BP | 0.208 | 1.14e-05 | 0.000165 | 3.78 | 0.000112 | Sema5a/Angpt1/Mdk/Kdr/Igf1/Tert/Icam1/Tek/Cdh5/Ndnf/Angptl4 | 11 |
| GO:0034764 | positive regulation of transmembrane transport | BP | 0.107 | 1.14e-05 | 0.000165 | 3.78 | 0.000112 | Acsl6/Cacna1c/Igf1/Tcaf1/Drd4/Tert/Gpc3/Cacna2d1/Myo5a/Cxcr3/Edn3/Ank2/Plcb1/Thy1/Trpc6/Nlgn3/Aplnr/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Acsl1/Cacnb4 | 27 |
| GO:0045860 | positive regulation of protein kinase activity | BP | 0.095 | 1.15e-05 | 0.000166 | 3.78 | 0.000113 | Angpt1/Agap2/Igf1/Ddr2/Zeb2/Adra1a/Ccdc88a/Drd4/Fermt2/Areg/Ccnd2/Dab1/Lrrk2/Edn3/Gprc5b/Pik3r6/Calca/Kit/Mmd/Vegfc/Nox4/Agtr1a/Htr2a/Kitl/Pkd1/Rassf2/Pde5a/Arrb2/Pkd2/Dlg4/Atp2b4/Lat/Acsl1/C1qtnf9 | 34 |
| GO:0045666 | positive regulation of neuron differentiation | BP | 0.143 | 1.15e-05 | 0.000166 | 3.78 | 0.000113 | Fez1/Rgs6/Dab1/Mef2c/Cxcl12/Gprc5b/Gdf7/Zeb1/Trpc6/Tgif2/Mmd/Hoxd3/Heyl/Zc4h2/Tcf4/Nin/Bend6 | 17 |
| GO:0001755 | neural crest cell migration | BP | 0.19 | 1.16e-05 | 0.000167 | 3.78 | 0.000113 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Ednrb/Fn1/Edn3/Ednra/Sema3g/Kitl/Sema6a | 12 |
| GO:0007164 | establishment of tissue polarity | BP | 0.19 | 1.16e-05 | 0.000167 | 3.78 | 0.000113 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Cplane1/Dchs1/Fzd2/Nphp1/Fat4/Brsk1/Dact1 | 12 |
| GO:0050769 | positive regulation of neurogenesis | BP | 0.101 | 1.21e-05 | 0.000173 | 3.76 | 0.000117 | Ptprz1/Sema5a/Mdk/Kdr/Nog/Plxnb1/Ptprd/Igf1/Zeb2/Sema7a/Nrp1/Fn1/Serpine2/Cxcl12/Tbc1d24/Obsl1/Gli3/Lrp2/Plxnd1/Mme/Kit/Vegfc/Apoe/Caprin2/Nin/Bin1/Chodl/Bhlhb9/Cux2/Ptn | 30 |
| GO:0007044 | cell-substrate junction assembly | BP | 0.156 | 1.25e-05 | 0.000177 | 3.75 | 0.000121 | Kdr/Phldb2/Nrp1/Fermt2/Fn1/Col16a1/Tek/Thsd1/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1/Itgb4 | 15 |
| GO:0008154 | actin polymerization or depolymerization | BP | 0.116 | 1.27e-05 | 0.00018 | 3.74 | 0.000123 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Mical1/Catip/Coro1a/Abi2/Esam/Sh3bp1/Daam2/Pecam1/Scin/Nckap1l/Bin1/Arhgap28/Wipf1/Cdc42ep5/Cyfip2/Plekhh2 | 23 |
| GO:0035088 | establishment or maintenance of apical/basal cell polarity | BP | 0.204 | 1.37e-05 | 0.000193 | 3.71 | 0.000131 | Wnt11/Lrrc7/Foxf1/Dlg2/Msn/Lin7a/Prickle2/Myo9a/Sh3bp1/Pard3b/Dlg5 | 11 |
| GO:0061245 | establishment or maintenance of bipolar cell polarity | BP | 0.204 | 1.37e-05 | 0.000193 | 3.71 | 0.000131 | Wnt11/Lrrc7/Foxf1/Dlg2/Msn/Lin7a/Prickle2/Myo9a/Sh3bp1/Pard3b/Dlg5 | 11 |
| GO:0033344 | cholesterol efflux | BP | 0.188 | 1.38e-05 | 0.000193 | 3.71 | 0.000131 | Apob/Pla2g10/Soat1/Pltp/Tsku/Apoe/Abca5/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3 | 12 |
| GO:0051893 | regulation of focal adhesion assembly | BP | 0.188 | 1.38e-05 | 0.000193 | 3.71 | 0.000131 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1 | 12 |
| GO:0090109 | regulation of cell-substrate junction assembly | BP | 0.188 | 1.38e-05 | 0.000193 | 3.71 | 0.000131 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1 | 12 |
| GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | BP | 0.11 | 1.38e-05 | 0.000193 | 3.71 | 0.000131 | Vipr2/Adra1a/Adgrl4/Drd4/Npr3/Gnai1/Cxcr3/Adcy3/Akap12/Ptgfr/Adgrl1/Calca/Htr1b/Pth1r/Adcy4/Gpr161/Adcy7/Ednra/Adcyap1r1/Gnaz/Ptger3/Calcrl/Atp2b4/Palm/Gabbr1 | 25 |
| GO:0035115 | embryonic forelimb morphogenesis | BP | 0.25 | 1.45e-05 | 0.000202 | 3.7 | 0.000137 | Cacna1c/Hoxd11/Hoxa11/Hoxd9/Osr2/Wnt9a/Tbx3/Hoxa9/Osr1 | 9 |
| GO:0010595 | positive regulation of endothelial cell migration | BP | 0.147 | 1.46e-05 | 0.000202 | 3.7 | 0.000137 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Tek/Plpp3/Jcad/Vegfc/Igf2/Hdac9/Akt3/Adgra2 | 16 |
| GO:0106027 | neuron projection organization | BP | 0.147 | 1.46e-05 | 0.000202 | 3.7 | 0.000137 | Nlgn1/Lrrk2/Grin2b/Ppfia2/Dock10/Kif1a/Abi2/Nlgn3/Apoe/Caprin2/Fyn/Bhlhb9/Dlg4/Cux2/Abcd2/Ngef | 16 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | BP | 0.118 | 1.46e-05 | 0.000202 | 3.7 | 0.000137 | Cd109/Zeb2/Npnt/Fermt2/Cdh5/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Smad9/Ltbp3/Hipk2/Nrros/Zfyve9/Cldn5/Arrb2/Il17rd/Lats2/Cav1/Itga8 | 22 |
| GO:0048592 | eye morphogenesis | BP | 0.118 | 1.46e-05 | 0.000202 | 3.7 | 0.000137 | Stra6/Ihh/Sdk2/Kdr/Tbx2/Arl6/Tenm3/Gdf11/Obsl1/Gli3/Thy1/Zeb1/Prdm1/Ptprm/Abi2/Rp1/Tsku/Rorb/Col5a2/Hipk2/Fat3/Ptn | 22 |
| GO:0001894 | tissue homeostasis | BP | 0.101 | 1.47e-05 | 0.000202 | 3.7 | 0.000137 | Tex15/Ihh/Angpt1/Col14a1/Kdr/Tub/Nos3/Calca/Flt3/Pth1r/Pbld2/Coro1a/Ldb2/Rp1/Tnfrsf11b/Nox4/Cd38/Enpp1/Bdkrb2/Crocc/Lpcat1/Ltbp3/Rcn3/Cldn5/Ccn2/Iqcb1/Akt3/Abcb1a/Bbs1 | 29 |
| GO:0019229 | regulation of vasoconstriction | BP | 0.163 | 1.49e-05 | 0.000205 | 3.69 | 0.000139 | Cacna1c/Adra1a/Mmp2/Icam1/Wdr35/Edn3/Cd38/Agtr1a/Htr2a/Pde5a/Ptger3/Apln/Atp1a2/Cav1 | 14 |
| GO:0061484 | hematopoietic stem cell homeostasis | BP | 0.286 | 1.5e-05 | 0.000205 | 3.69 | 0.000139 | Gprasp2/Emcn/Crispld1/Myct1/Armcx1/Zfp251/Septin4/Fstl1 | 8 |
| GO:0097178 | ruffle assembly | BP | 0.222 | 1.5e-05 | 0.000205 | 3.69 | 0.00014 | Evl/Nlgn1/Cspg4/Aif1l/Pfn2/Icam1/Sh3bp1/Eps8l1/Arhgef26/Cav1 | 10 |
| GO:0045165 | cell fate commitment | BP | 0.101 | 1.57e-05 | 0.000213 | 3.67 | 0.000145 | Gap43/Ihh/Sfrp2/Wnt6/AW551984/Ar/Wnt11/Hoxd10/Kdr/Ptch2/Tbx2/Hoxa11/Mef2c/Gli3/Glis1/Gdf7/Wnt9a/Prdm1/Jag2/Loxl3/Prickle2/Sox12/Tbx3/Ednra/Lhx6/Tal1/Ptch1/Ets2/Lats2 | 29 |
| GO:0001508 | action potential | BP | 0.129 | 1.57e-05 | 0.000213 | 3.67 | 0.000145 | Cacna1c/Kcnd2/Kcnd3/Adra1a/Scn4b/P2rx2/Scn1a/Cacna2d1/Grin2b/Ank2/Scn2a/Grik2/Kcnma1/Kcnc4/Bin1/Cacna1h/Scn3a/Cav1/Cacnb4 | 19 |
| GO:0030148 | sphingolipid biosynthetic process | BP | 0.153 | 1.61e-05 | 0.000217 | 3.66 | 0.000148 | St8sia2/St8sia4/Elovl4/Agk/Cerkl/Sptlc3/Cers4/St6galnac4/St3gal2/St8sia6/Degs2/Abca8b/B3gnt5/St6galnac6/Smpd1 | 15 |
| GO:0070252 | actin-mediated cell contraction | BP | 0.153 | 1.61e-05 | 0.000217 | 3.66 | 0.000148 | Sgcd/Cacna1c/Kcnd3/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Limch1/Bin1/Epdr1/Cacna1h/Ptger3/Atp1a2/Cav1 | 15 |
| GO:0032835 | glomerulus development | BP | 0.185 | 1.63e-05 | 0.000219 | 3.66 | 0.000149 | Angpt1/Nog/Ptpro/Ednrb/Mef2c/Nid1/Tek/Enpep/Tcf21/Heyl/Ednra/Osr1 | 12 |
| GO:0018209 | peptidyl-serine modification | BP | 0.0934 | 1.63e-05 | 0.000219 | 3.66 | 0.000149 | Hgf/Sfrp2/Angpt1/Cd44/Spock2/Hcls1/Lrrk2/Pfn2/Prkd1/Tek/Egflam/Galnt16/Rps6ka2/Mamdc2/Ppm1f/Caprin2/Ndnf/Dclk2/Bdkrb2/Rps6ka5/Bgn/Arrb1/Hipk2/Pkd1/Rassf2/Ttbk2/Poglut2/Arrb2/Atp2b4/Akt3/Brsk1/Naa11/Lats2/Cav1 | 34 |
| GO:0034446 | substrate adhesion-dependent cell spreading | BP | 0.145 | 1.64e-05 | 0.00022 | 3.66 | 0.00015 | Mdk/Itga4/Lamc3/Nrp1/Fermt2/Itgb7/Fn1/Tek/Has2/Postn/Tmeff2/Parvg/Parvb/Net1/Ntng1/Itga8 | 16 |
| GO:0051402 | neuron apoptotic process | BP | 0.0975 | 1.65e-05 | 0.000221 | 3.66 | 0.00015 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Itga1/Scn2a/Coro1a/Il27ra/Grik2/Apoe/Enpp1/Kcnma1/Ndnf/Arrb1/Hipk2/Epha7/Kcnip3/Nlrp1b/Nfatc4/Arrb2/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 31 |
| GO:0046629 | gamma-delta T cell activation | BP | 0.333 | 1.68e-05 | 0.000224 | 3.65 | 0.000152 | Lef1/Itk/Jag2/Nckap1l/Stat5b/Gimap5/Gimap3 | 7 |
| GO:0070371 | ERK1 and ERK2 cascade | BP | 0.0946 | 1.68e-05 | 0.000224 | 3.65 | 0.000152 | Angpt1/Dcc/Kdr/Igf1/Ddr2/Adra1a/Npnt/Cd44/Sema7a/Nrp1/Fermt2/Fn1/Icam1/Tek/Cxcl12/Akap12/Denn2b/Dnajc27/Nox4/Apoe/Havcr2/Emilin1/Ednra/Spred3/Xcl1/Htr2a/Cavin3/Arrb1/Epha7/Arrb2/Ccn2/Sema6a/Cav1 | 33 |
| GO:0048659 | smooth muscle cell proliferation | BP | 0.117 | 1.73e-05 | 0.000229 | 3.64 | 0.000156 | Vipr2/Igf1/Ddr2/Nos3/Drd4/Npr3/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Rgs5/Apoe/Enpp1/Ednra/Irak4/Calcrl/Apln/Stat5b/Efemp2/Cav1 | 22 |
| GO:0050920 | regulation of chemotaxis | BP | 0.109 | 1.73e-05 | 0.000229 | 3.64 | 0.000156 | Sema5a/Mdk/Kdr/Sema5b/Sema7a/Nrp1/Fn1/Prkd1/Cxcr3/Cxcl12/Jam3/Cdh13/Edn3/Ppm1f/Vegfc/Ednra/Sema3g/Xcl1/Nckap1l/Tnfaip6/Dysf/Sema6a/Mpp1/Ptn/Adgra2 | 25 |
| GO:0097529 | myeloid leukocyte migration | BP | 0.109 | 1.73e-05 | 0.000229 | 3.64 | 0.000156 | Cxcl15/Mdk/Ptpro/Ednrb/Cxcl12/Jam3/Edn3/Itga1/Calca/Plcb1/Kit/Vegfc/Enpp1/Emilin1/Cd200/Ednra/Pecam1/Xcl1/Nckap1l/Tnfaip6/Dysf/Irak4/Ptger3/Stat5b/Mpp1 | 25 |
| GO:0033002 | muscle cell proliferation | BP | 0.104 | 1.74e-05 | 0.000229 | 3.64 | 0.000156 | Gli1/Angpt1/Vipr2/Nog/Igf1/Tbx2/Ddr2/Nos3/Drd4/Npr3/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Rgs5/Apoe/Enpp1/Ednra/Selenon/Irak4/Calcrl/Apln/Stat5b/Efemp2/Cav1 | 27 |
| GO:0001666 | response to hypoxia | BP | 0.111 | 1.76e-05 | 0.000232 | 3.64 | 0.000157 | Kcnk3/Kcnd2/Ryr1/Kdr/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Grin2b/Tek/Slc8a3/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Hif3a | 24 |
| GO:0046651 | lymphocyte proliferation | BP | 0.0955 | 1.83e-05 | 0.00024 | 3.62 | 0.000163 | Ihh/Lef1/Igf1/Cd44/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Prdm1/Cd1d1/Msn/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Satb1/Pde5a/Fyn/H2-Aa/Stat5b/Zap70/Impdh1/Dlg5/Il12rb1 | 32 |
| GO:0009954 | proximal/distal pattern formation | BP | 0.243 | 1.84e-05 | 0.000241 | 3.62 | 0.000164 | Gli1/Hoxd10/Hoxd11/Hoxa11/Hoxd9/Hoxa10/Gli3/Hoxa9/Osr1 | 9 |
| GO:0010524 | positive regulation of calcium ion transport into cytosol | BP | 0.182 | 1.91e-05 | 0.000249 | 3.6 | 0.00017 | Cxcr3/Thy1/Aplnr/Agtr1a/Ednra/Adcyap1r1/Asph/Pkd2/Gstm7/Gimap5/Gimap3/Cav1 | 12 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | BP | 0.0938 | 2e-05 | 0.000261 | 3.58 | 0.000177 | Sfrp2/Igf1/Zeb2/Slc8a1/Cbs/Drd4/Fermt2/Ccnd2/Lrrk2/Mapk8ip1/Edn3/Pik3r6/Slc8a3/Pkia/Thy1/Kit/Cdkn1c/Nox4/Apoe/Rasip1/Htr2a/Pkib/Kitl/Pkd1/Heg1/Pde5a/Pkd2/Atp2b4/Lats2/Smpd1/Acsl1/Cav1/C1qtnf9 | 33 |
| GO:0050768 | negative regulation of neurogenesis | BP | 0.123 | 2.06e-05 | 0.000268 | 3.57 | 0.000182 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Sema7a/Nrp1/Tert/Dab1/Id4/Thy1/Trpc6/Daam2/Sema3g/Epha7/Nfatc4/Syngap1/Sema6a/Ptn | 20 |
| GO:1990138 | neuron projection extension | BP | 0.113 | 2.07e-05 | 0.000268 | 3.57 | 0.000182 | Map2/Sema5a/Itga4/St8sia2/Sema5b/Cpne5/Sema7a/Nrp1/Fn1/Cxcl12/Edn3/Eif2ak4/Postn/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Slc23a2/Cacng7/Sema6a/Nrn1/Cyfip2 | 23 |
| GO:1904427 | positive regulation of calcium ion transmembrane transport | BP | 0.157 | 2.23e-05 | 0.000287 | 3.54 | 0.000195 | Cacna1c/Cacna2d1/Cxcr3/Ank2/Thy1/Aplnr/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 14 |
| GO:1990874 | vascular associated smooth muscle cell proliferation | BP | 0.179 | 2.24e-05 | 0.000288 | 3.54 | 0.000196 | Igf1/Ddr2/Drd4/Tert/Mmp2/Mef2c/Adamts1/Htr1b/Rgs5/Enpp1/Apln/Efemp2 | 12 |
| GO:0043616 | keratinocyte proliferation | BP | 0.213 | 2.26e-05 | 0.000289 | 3.54 | 0.000197 | Mdk/Cd109/Fst/Prkd1/Has2/Cdh13/Stxbp4/Klf9/Twist2/Ptch1 | 10 |
| GO:0050974 | detection of mechanical stimulus involved in sensory perception | BP | 0.213 | 2.26e-05 | 0.000289 | 3.54 | 0.000197 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12/Kit/Htr2a/Fyn/Piezo2 | 10 |
| GO:0007266 | Rho protein signal transduction | BP | 0.13 | 2.3e-05 | 0.000294 | 3.53 | 2e-04 | Adra1a/Nrp1/Cdh2/Lpar4/Arhgef25/Cdh13/Apoe/Agtr1a/Rasip1/Eps8l1/Ednra/Pecam1/Arap3/Arrb1/Heg1/Dlc1/Net1/Cdc42ep5 | 18 |
| GO:0048638 | regulation of developmental growth | BP | 0.0896 | 2.32e-05 | 0.000294 | 3.53 | 2e-04 | Gli1/Map2/Sema5a/Ar/Cacna1c/Col14a1/H19/Nog/Igf1/Tbx2/Sema5b/Cpne5/Nkd1/Acacb/Sema7a/Nrp1/Fn1/Mef2c/Cxcl12/Hopx/Flvcr1/Gamt/Plcb1/Flt3/Apoe/Igf2/Sema3g/Slc23a2/Foxs1/Epha7/Cacng7/Ptch1/Pi16/Stat5b/Sema6a/Lats2 | 36 |
| GO:0032943 | mononuclear cell proliferation | BP | 0.0944 | 2.32e-05 | 0.000294 | 3.53 | 2e-04 | Ihh/Lef1/Igf1/Cd44/Mef2c/Cxcl12/Mapk8ip1/Dock2/Flt3/Tnfsf13b/Prdm1/Cd1d1/Msn/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Satb1/Pde5a/Fyn/H2-Aa/Stat5b/Zap70/Impdh1/Dlg5/Il12rb1 | 32 |
| GO:0097061 | dendritic spine organization | BP | 0.149 | 2.32e-05 | 0.000294 | 3.53 | 2e-04 | Nlgn1/Lrrk2/Grin2b/Ppfia2/Dock10/Kif1a/Abi2/Nlgn3/Apoe/Caprin2/Fyn/Bhlhb9/Dlg4/Cux2/Ngef | 15 |
| GO:0010463 | mesenchymal cell proliferation | BP | 0.193 | 2.36e-05 | 0.000297 | 3.53 | 0.000202 | Ihh/Wnt11/Kdr/Tbx2/Gpc3/Foxf1/Zeb1/Osr1/Dchs1/Fat4/Ptn | 11 |
| GO:1902904 | negative regulation of supramolecular fiber organization | BP | 0.118 | 2.36e-05 | 0.000297 | 3.53 | 0.000202 | Kank4/Map2/Evl/Phldb2/Tmod2/Pfn2/Cdh5/Coro1a/Rp1/Tubb4a/Apoe/Arhgap6/Tmeff2/Emilin1/Pecam1/Scin/Dysf/Arhgap28/Ttbk2/Dlc1/Plekhh2 | 21 |
| GO:0010721 | negative regulation of cell development | BP | 0.112 | 2.42e-05 | 0.000304 | 3.52 | 0.000207 | Prtg/Efnb3/Map2/Sema5a/Nog/Igf1/Sema5b/Sema7a/Nrp1/Tert/Ednrb/Dab1/Id4/Postn/Thy1/Trpc6/Daam2/Sema3g/Epha7/Nfatc4/Syngap1/Sema6a/Ptn | 23 |
| GO:0048565 | digestive tract development | BP | 0.135 | 2.47e-05 | 0.00031 | 3.51 | 0.000211 | Stra6/Ihh/Sfrp2/Clmp/Pkdcc/Tbx2/Ednrb/Foxf1/Gli3/Tcf21/Prdm1/Kit/Pcsk5/Dchs1/Fat4/Dact1/Stx2 | 17 |
| GO:0009952 | anterior/posterior pattern specification | BP | 0.106 | 2.5e-05 | 0.000313 | 3.51 | 0.000213 | Sfrp2/Lef1/Hoxd10/Hoxd11/Nog/Zeb2/Hoxa11/Gpc3/Hoxd9/Hoxa10/Foxf1/Hoxd8/Hoxd4/Tshz1/Gdf11/Gli3/Lfng/Hoxd3/Tbx3/Heyl/Pcsk5/Hoxa9/Hipk2/Osr1/Ets2 | 25 |
| GO:0032233 | positive regulation of actin filament bundle assembly | BP | 0.176 | 2.61e-05 | 0.000325 | 3.49 | 0.000221 | Wnt11/Evl/Ccdc88a/Nrp1/Fermt2/Pfn2/Ppm1f/Limch1/Nox4/Arhgef15/Ppm1e/Ccn2 | 12 |
| GO:0034767 | positive regulation of ion transmembrane transport | BP | 0.114 | 2.61e-05 | 0.000325 | 3.49 | 0.000221 | Cacna1c/Tcaf1/Drd4/Cacna2d1/Myo5a/Cxcr3/Edn3/Ank2/Plcb1/Thy1/Trpc6/Nlgn3/Aplnr/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Cacnb4 | 22 |
| GO:0140058 | neuron projection arborization | BP | 0.267 | 2.61e-05 | 0.000325 | 3.49 | 0.000221 | Nlgn1/Nrp1/Lrrk2/Lrp2/Myo9a/Dlg4/Ptn/Ntng1 | 8 |
| GO:0003382 | epithelial cell morphogenesis | BP | 0.208 | 2.74e-05 | 0.00034 | 3.47 | 0.000231 | Ihh/Ar/Stc1/Rilpl1/Palld/Pecam1/Col15a1/Heg1/Arhgef26/Col18a1 | 10 |
| GO:0042060 | wound healing | BP | 0.0909 | 2.85e-05 | 0.000352 | 3.45 | 0.000239 | Cd109/Kdr/Nog/Igf1/Evl/Ddr2/Tfpi/Abat/Phldb2/Cd44/Fermt2/Fn1/Fkbp10/Serpine2/Plpp3/Procr/Ccm2l/Tsku/Apoe/Syt7/Tmeff2/Ndnf/Pecam1/Pros1/Cnn2/Dysf/Abi3bp/Ptger3/Clec7a/St3gal4/Rab27a/Smpd1/Cav1/Cldn1 | 34 |
| GO:2000807 | regulation of synaptic vesicle clustering | BP | 0.5 | 2.88e-05 | 0.000355 | 3.45 | 0.000241 | Nlgn1/Cdh2/Nlgn3/Magi2/Pcdh17 | 5 |
| GO:0001656 | metanephros development | BP | 0.154 | 2.88e-05 | 0.000355 | 3.45 | 0.000241 | Hoxd11/Hoxa11/Gpc3/Id3/Osr2/Gdf11/Gli3/Tcf21/Tshz3/Ptch1/Osr1/Fat4/Dlg5/Itga8 | 14 |
| GO:0002062 | chondrocyte differentiation | BP | 0.139 | 2.89e-05 | 0.000355 | 3.45 | 0.000241 | Col11a1/Ihh/Sfrp2/Mdk/Hoxd11/Pkdcc/Hoxa11/Mef2c/Osr2/Gli3/Pth1r/Wnt9a/Adamts7/Ltbp3/Osr1/Ccn2 | 16 |
| GO:0045879 | negative regulation of smoothened signaling pathway | BP | 0.231 | 2.91e-05 | 0.000356 | 3.45 | 0.000242 | Gli1/Hhip/Kif7/Gpc3/Serpine2/Gli3/Gpr161/Enpp1/Ptch1 | 9 |
| GO:0051961 | negative regulation of nervous system development | BP | 0.12 | 2.94e-05 | 0.00036 | 3.44 | 0.000245 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Sema7a/Nrp1/Tert/Dab1/Id4/Thy1/Trpc6/Daam2/Sema3g/Epha7/Nfatc4/Syngap1/Sema6a/Ptn | 20 |
| GO:0060249 | anatomical structure homeostasis | BP | 0.0945 | 3.02e-05 | 0.000368 | 3.43 | 0.00025 | Tex15/Ihh/Angpt1/Col14a1/Kdr/Tub/Nos3/P2rx2/Calca/Flt3/Pth1r/Pbld2/Coro1a/Ldb2/Rp1/Tnfrsf11b/Nox4/Cd38/Enpp1/Bdkrb2/Crocc/Lpcat1/Ltbp3/Rcn3/Cldn5/Ccn2/Iqcb1/Akt3/Abcb1a/Bbs1/Large1 | 31 |
| GO:0060041 | retina development in camera-type eye | BP | 0.116 | 3.03e-05 | 0.000369 | 3.43 | 0.000251 | Ndp/Ihh/Cyp1b1/Sdk2/Tub/Celf4/Lamc3/Nrp1/Arl6/Obsl1/Thy1/Tgif2/Ptprm/Rp1/Arhgef15/Rorb/Lpcat1/Hipk2/Fat3/Nphp1/Ptn | 21 |
| GO:0032271 | regulation of protein polymerization | BP | 0.11 | 3.04e-05 | 0.00037 | 3.43 | 0.000251 | Kank4/Map2/Evl/Hcls1/Tmod2/Pfn2/Icam1/Cdh5/Coro1a/Esam/Tubb4a/Daam2/Pecam1/Scin/Nckap1l/Slain1/Nin/Bin1/Arhgap28/Cdc42ep5/Cav1/Cyfip2/Tppp | 23 |
| GO:0043523 | regulation of neuron apoptotic process | BP | 0.0989 | 3.19e-05 | 0.000386 | 3.41 | 0.000262 | Ptprz1/Angpt1/Mdk/Kdr/Agap2/Nrp1/Tert/Bok/Mef2c/Pcdhgc3/Itga1/Coro1a/Il27ra/Grik2/Apoe/Kcnma1/Ndnf/Arrb1/Hipk2/Epha7/Kcnip3/Nfatc4/Arrb2/Fyn/Bhlhb9/Syngap1/Nes/Cacna1a | 28 |
| GO:0001574 | ganglioside biosynthetic process | BP | 0.375 | 3.2e-05 | 0.000386 | 3.41 | 0.000262 | St8sia2/St8sia4/St6galnac4/St3gal2/St8sia6/St6galnac6 | 6 |
| GO:0072498 | embryonic skeletal joint development | BP | 0.375 | 3.2e-05 | 0.000386 | 3.41 | 0.000262 | Ihh/Hoxd11/Nog/Hoxa11/Osr2/Osr1 | 6 |
| GO:0098659 | inorganic cation import across plasma membrane | BP | 0.138 | 3.22e-05 | 0.000386 | 3.41 | 0.000263 | Kcnj3/Cacna1c/Kcnd3/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Cacna1e/Slc8a3/Nalf1/Agtr1a/Slc9a5/Fyn/Slc12a4/Atp1a2/Cacna1a | 16 |
| GO:0099587 | inorganic ion import across plasma membrane | BP | 0.138 | 3.22e-05 | 0.000386 | 3.41 | 0.000263 | Kcnj3/Cacna1c/Kcnd3/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Cacna1e/Slc8a3/Nalf1/Agtr1a/Slc9a5/Fyn/Slc12a4/Atp1a2/Cacna1a | 16 |
| GO:0060326 | cell chemotaxis | BP | 0.0955 | 3.29e-05 | 0.000393 | 3.41 | 0.000267 | Cxcl15/Hgf/Sema5a/Lef1/Mdk/Kdr/Cyp7b1/Ptpro/Nrp1/Ednrb/Prkd1/Cxcr3/Cxcl12/Jam3/Edn3/Itga1/Calca/Coro1a/Kit/Vegfc/Agtr1a/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Xcr1/Arrb2/Mpp1/Ptn | 30 |
| GO:0019722 | calcium-mediated signaling | BP | 0.112 | 3.32e-05 | 0.000396 | 3.4 | 0.000269 | Sgcd/Kdr/Nr5a2/Igf1/Slc8a1/Drd4/Ednrb/Slc24a4/Lrrk2/Cxcr3/Cdh13/Ptgfr/Ank2/Adgrl1/Itgal/Agtr1a/Cmya5/Xcr1/Nfatc4/Atp2b4/Zap70/Lat | 22 |
| GO:0001837 | epithelial to mesenchymal transition | BP | 0.123 | 3.33e-05 | 0.000397 | 3.4 | 0.00027 | Sfrp2/Lef1/Mdk/Wnt11/Nog/Mark1/Phldb2/Fermt2/Erg/Has2/Loxl3/Epha3/Tbx3/Heyl/Ednra/Snai1/Spred3/Il17rd/Dlg5 | 19 |
| GO:1903829 | positive regulation of protein localization | BP | 0.0835 | 3.34e-05 | 0.000397 | 3.4 | 0.00027 | Igf1/Nlgn1/Abat/Tcaf1/Ccdc88a/Hcls1/Fermt2/Gnai1/Tert/Ano1/Myo5a/Ptp4a3/Prkd1/Tek/Gli3/Lrp2/Rab34/Msn/Epha3/Nlgn3/Prr5l/Vegfc/Cd38/Pecam1/Tmem35a/Crocc/Tnfaip6/Stac2/Arrb1/Asph/Abcg1/Dzip1/Frmd4a/Mgat3/Fyn/Wipf1/Dlg4/Atp2b4/Ptn/Nkd2/Cacnb4 | 41 |
| GO:0032535 | regulation of cellular component size | BP | 0.0891 | 3.35e-05 | 0.000397 | 3.4 | 0.00027 | Kank4/Map2/Sema5a/Evl/Sema5b/Slc12a8/Hcls1/Sema7a/Nrp1/Tmod2/Fn1/Pfn2/Icam1/Cxcl12/Coro1a/Esam/Sh3bp1/Apoe/Kcnma1/Daam2/Pecam1/Scin/Sema3g/Nckap1l/Bin1/Epha7/Arhgap28/Aqp11/Slc12a4/Akt3/Sema6a/Cdc42ep5/C1qtnf9/Cyfip2/Plekhh2 | 35 |
| GO:0001952 | regulation of cell-matrix adhesion | BP | 0.132 | 3.36e-05 | 0.000398 | 3.4 | 0.000271 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Cdh13/Postn/Thy1/Epha3/Ppm1f/Limch1/Vegfc/Arhgap6/Pcsk5/Dlc1/Efemp2 | 17 |
| GO:0050771 | negative regulation of axonogenesis | BP | 0.171 | 3.53e-05 | 0.000416 | 3.38 | 0.000283 | Efnb3/Map2/Sema5a/Sema5b/Sema7a/Nrp1/Dab1/Thy1/Sema3g/Epha7/Syngap1/Sema6a | 12 |
| GO:0086003 | cardiac muscle cell contraction | BP | 0.171 | 3.53e-05 | 0.000416 | 3.38 | 0.000283 | Sgcd/Cacna1c/Kcnd3/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Bin1/Cacna1h/Atp1a2/Cav1 | 12 |
| GO:0098657 | import into cell | BP | 0.104 | 3.56e-05 | 0.000418 | 3.38 | 0.000284 | Kcnj3/Slc1a3/Acsl6/Cacna1c/Itga4/Slc6a2/Kcnd3/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Slc7a2/Cacna1e/Slc8a3/Lrp2/Nalf1/Slc29a1/Agtr1a/Slc9a5/Fyn/Slc12a4/Atp1a2/Steap4/Acsl1/Cacna1a | 25 |
| GO:0048732 | gland development | BP | 0.084 | 3.6e-05 | 0.000422 | 3.37 | 0.000287 | Gli1/Stra6/Hgf/Ar/Lef1/Mdk/Agap2/Nog/Cyp7b1/Igf1/Tbx2/Cd44/Nrp1/Edaradd/Hoxd9/Areg/Mmp2/Foxf1/Id4/Serpine2/Gli3/Tcf21/Gdf7/Plxnd1/Msn/Cdkn1c/Hoxd3/Tbx3/Ednra/Igf2/Hoxa9/Pkd1/Ptch1/Pkd2/Stat5b/Asl/Ptn/Cav1/Edar/Cacnb4 | 40 |
| GO:0009311 | oligosaccharide metabolic process | BP | 0.225 | 3.61e-05 | 0.000422 | 3.37 | 0.000287 | St6gal2/St8sia2/St8sia4/B3galnt1/St6galnac4/St3gal2/St8sia6/St6galnac6/St3gal4 | 9 |
| GO:0010634 | positive regulation of epithelial cell migration | BP | 0.122 | 3.65e-05 | 0.000426 | 3.37 | 0.00029 | Sema5a/Angpt1/Kdr/Igf1/Nos3/Nrp1/Ets1/Prkd1/Tek/Has2/Plpp3/Ppm1f/Jcad/Vegfc/Igf2/Hdac9/Akt3/Adgra2/Mapre2 | 19 |
| GO:0060996 | dendritic spine development | BP | 0.131 | 3.72e-05 | 0.000433 | 3.36 | 0.000295 | Nlgn1/Lrrk2/Mef2c/Ppfia2/D16Ertd472e/Dock10/Kif1a/Abi2/Nlgn3/Apoe/Caprin2/Bhlhb9/Dlg4/Cux2/Palm/Dlg5/Ngef | 17 |
| GO:0050905 | neuromuscular process | BP | 0.111 | 3.88e-05 | 0.00045 | 3.35 | 0.000306 | Ankfn1/Stra6/Slc1a3/Hoxd10/Jph4/Aph1b/Scn1a/Myo5a/Grin2b/Slc8a3/Tshz3/Glrb/Nlgn3/Npas3/Kcnma1/Prrt2/Ednra/Foxs1/Stac2/Hipk2/Dlg4/Cacna1a | 22 |
| GO:0098815 | modulation of excitatory postsynaptic potential | BP | 0.183 | 3.9e-05 | 0.000452 | 3.34 | 0.000307 | Nlgn1/Celf4/Drd4/Stx1b/Lrrk2/Grin2b/Tbc1d24/Slc8a3/Nlgn3/Dlg4/Cux2 | 11 |
| GO:0051279 | regulation of release of sequestered calcium ion into cytosol | BP | 0.159 | 3.98e-05 | 0.000461 | 3.34 | 0.000314 | Slc8a1/Myo5a/Prkd1/Cxcr3/Ank2/Thy1/Coro1a/Aplnr/Ednra/Asph/Selenon/Pkd2/Gstm7 | 13 |
| GO:0030217 | T cell differentiation | BP | 0.096 | 4.03e-05 | 0.000465 | 3.33 | 0.000316 | Ihh/Lef1/Mdk/Cd83/Cd44/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Zeb1/Prdm1/Cd1d1/Jag2/Lfng/Loxl3/Kit/Cracr2a/Enpp1/Sox12/Nckap1l/Satb1/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 29 |
| GO:0006665 | sphingolipid metabolic process | BP | 0.125 | 4.1e-05 | 0.000472 | 3.33 | 0.000321 | St8sia2/St8sia4/Elovl4/Agk/Cerkl/Sptlc3/Cers4/Plpp3/St6galnac4/St3gal2/Kit/St8sia6/Degs2/Abca8b/B3gnt5/St6galnac6/Plpp1/Smpd1 | 18 |
| GO:0045582 | positive regulation of T cell differentiation | BP | 0.142 | 4.16e-05 | 0.000478 | 3.32 | 0.000325 | Ihh/Lef1/Mdk/Cd83/Pik3r6/Gli3/Cd1d1/Sox12/Nckap1l/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 15 |
| GO:0051899 | membrane depolarization | BP | 0.149 | 4.18e-05 | 0.000479 | 3.32 | 0.000326 | Grik1/Cacna1c/Kdr/Scn4b/Scn1a/Bok/Cacna2d1/Lrrk2/Scn2a/Fhl1/Cacna1h/Scn3a/Cav1/Cacna1a | 14 |
| GO:0061351 | neural precursor cell proliferation | BP | 0.111 | 4.19e-05 | 0.000479 | 3.32 | 0.000326 | Gli1/Ptprz1/Acsl6/Hhip/Lef1/Mdk/Nog/Igf1/Zeb2/Cdh2/Smarca1/Lrrk2/Id4/Zfp423/Gli3/Lrp2/Kif1a/Eml1/Vegfc/Nes/Ptn/Bbs1 | 22 |
| GO:0060048 | cardiac muscle contraction | BP | 0.129 | 4.53e-05 | 0.000517 | 3.29 | 0.000352 | Sgcd/Cacna1c/Kcnd3/Adra1a/Slc8a1/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Myl3/Bin1/Pde5a/Cacna1h/Ccn2/Atp1a2/Cav1 | 17 |
| GO:0006643 | membrane lipid metabolic process | BP | 0.113 | 4.55e-05 | 0.000519 | 3.28 | 0.000353 | Cyp1b1/St8sia2/St8sia4/Elovl4/Agk/Cerkl/Sptlc3/Cers4/Plpp3/St6galnac4/St3gal2/Kit/St8sia6/Degs2/Sccpdh/Abca8b/B3gnt5/St6galnac6/Plpp1/St3gal4/Smpd1 | 21 |
| GO:0006820 | anion transport | BP | 0.0823 | 4.6e-05 | 0.000523 | 3.28 | 0.000356 | Slc1a3/Car4/Grik1/Pla2g10/Slc13a5/Slc2a3/Slc12a8/Abat/Tcaf1/Drd4/Ano1/Slc7a2/Grin2b/Stc1/Abcg2/Slc36a4/Ano2/Lrp2/Htr1b/Ttyh2/Slc6a15/Glrb/Gabra4/Enpp1/Bdkrb2/Slc23a2/Sfxn5/Slco4c1/Ptger3/Nfkbie/Slc4a3/Slc38a1/Slc12a4/Clca2/Abcb1a/Abcc5/Abcg3/Gabbr1/Slc4a8/Cacna1a/Cacnb4 | 41 |
| GO:0014031 | mesenchymal cell development | BP | 0.147 | 4.71e-05 | 0.000534 | 3.27 | 0.000364 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Ednrb/Fn1/Edn3/Heyl/Ednra/Sema3g/Kitl/Sema6a | 14 |
| GO:0001886 | endothelial cell morphogenesis | BP | 0.353 | 4.77e-05 | 0.00054 | 3.27 | 0.000367 | Stc1/Pecam1/Col15a1/Heg1/Arhgef26/Col18a1 | 6 |
| GO:1905564 | positive regulation of vascular endothelial cell proliferation | BP | 0.353 | 4.77e-05 | 0.00054 | 3.27 | 0.000367 | Mdk/Itga4/Igf1/Igf2/Apln/Akt3 | 6 |
| GO:0046578 | regulation of Ras protein signal transduction | BP | 0.109 | 4.88e-05 | 0.00055 | 3.26 | 0.000374 | Igf1/Adra1a/Nrp1/Cdh2/Rasa3/Lpar4/Arhgef25/Dock2/Sh2b2/Syde1/Sh3bp1/Apoe/Rasip1/Eps8l1/Arap3/Kitl/Arrb1/Heg1/Dlc1/Net1/Syngap1/Mapre2 | 22 |
| GO:0014812 | muscle cell migration | BP | 0.133 | 4.91e-05 | 0.000552 | 3.26 | 0.000376 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Mef2c/Myo5a/Has2/Adamts1/Postn/Nox4/Enpp1/Pcsk5/Net1/Plekho1 | 16 |
| GO:0007612 | learning | BP | 0.112 | 4.93e-05 | 0.000552 | 3.26 | 0.000376 | Stra6/Cacna1c/Nog/Jph4/Drd4/Chst10/Grin2b/B4galt2/Pde1b/Adcy3/Shank2/Cacna1e/Slc8a3/Eif2ak4/Kit/Nlgn3/Atp1a2/Brsk1/Syngap1/Ptn/Ddhd2 | 21 |
| GO:0042129 | regulation of T cell proliferation | BP | 0.112 | 4.93e-05 | 0.000552 | 3.26 | 0.000376 | Ihh/Igf1/Cd44/Mapk8ip1/Tnfsf13b/Cd1d1/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Havcr2/Igf2/Xcl1/Nckap1l/Pde5a/H2-Aa/Stat5b/Zap70/Dlg5/Il12rb1 | 21 |
| GO:0046467 | membrane lipid biosynthetic process | BP | 0.128 | 4.99e-05 | 0.000558 | 3.25 | 0.00038 | St8sia2/St8sia4/Elovl4/Agk/Cerkl/Sptlc3/Cers4/St6galnac4/St3gal2/St8sia6/Degs2/Sccpdh/Abca8b/B3gnt5/St6galnac6/St3gal4/Smpd1 | 17 |
| GO:0045588 | positive regulation of gamma-delta T cell differentiation | BP | 0.455 | 5.09e-05 | 0.000568 | 3.25 | 0.000386 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0046645 | positive regulation of gamma-delta T cell activation | BP | 0.455 | 5.09e-05 | 0.000568 | 3.25 | 0.000386 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0002695 | negative regulation of leukocyte activation | BP | 0.109 | 5.26e-05 | 0.000584 | 3.23 | 0.000397 | Ihh/Mdk/Cd44/Foxf1/Gli3/Flt3/Prdm1/Loxl3/Btla/H2-Ab1/Havcr2/Cd200/Nckap1l/Pde5a/Cd84/Fgr/H2-Aa/Gimap5/Dlg5/Gimap3/Clnk/Nr1h3 | 22 |
| GO:0006941 | striated muscle contraction | BP | 0.115 | 5.31e-05 | 0.000588 | 3.23 | 4e-04 | Sgcd/Cacna1c/Kcnd3/Adra1a/Slc8a1/Scn4b/Scn1a/Cacna2d1/Stc1/Ank2/Slc8a3/Myl3/Bin1/Stac2/Ryr3/Pde5a/Cacna1h/Ccn2/Atp1a2/Cav1 | 20 |
| GO:1903034 | regulation of response to wounding | BP | 0.115 | 5.31e-05 | 0.000588 | 3.23 | 4e-04 | Mdk/Cd109/Ddr2/Tfpi/Abat/Phldb2/Fermt2/Serpine2/Plpp3/Apoe/Pros1/Rtn4rl1/Neo1/Ptger3/Clec7a/Xylt1/St3gal4/Ptn/Cav1/Cldn1 | 20 |
| GO:0010171 | body morphogenesis | BP | 0.177 | 5.35e-05 | 0.000591 | 3.23 | 0.000402 | Stra6/Ihh/Lef1/Nog/Crispld1/Phldb2/Gpc3/Mmp2/Flvcr1/Twist2/Asph | 11 |
| GO:0060997 | dendritic spine morphogenesis | BP | 0.164 | 5.43e-05 | 0.000598 | 3.22 | 0.000407 | Nlgn1/Lrrk2/Ppfia2/Dock10/Kif1a/Abi2/Nlgn3/Caprin2/Bhlhb9/Dlg4/Cux2/Ngef | 12 |
| GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | BP | 0.132 | 5.44e-05 | 0.000598 | 3.22 | 0.000407 | Cd109/Zeb2/Npnt/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Hipk2/Nrros/Zfyve9/Il17rd/Lats2/Cav1/Itga8 | 16 |
| GO:0038084 | vascular endothelial growth factor signaling pathway | BP | 0.214 | 5.45e-05 | 0.000599 | 3.22 | 0.000407 | Kdr/Nrp1/Ptp4a3/Prkd1/Jcad/Vegfc/Tcf4/Sema6a/Adgra2 | 9 |
| GO:0062149 | detection of stimulus involved in sensory perception of pain | BP | 0.242 | 5.53e-05 | 0.000605 | 3.22 | 0.000412 | Scn1a/Ano1/Grin2b/Cxcl12/Calca/Htr2a/Arrb2/Fyn | 8 |
| GO:0150117 | positive regulation of cell-substrate junction organization | BP | 0.242 | 5.53e-05 | 0.000605 | 3.22 | 0.000412 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f/Mapre2 | 8 |
| GO:1902105 | regulation of leukocyte differentiation | BP | 0.0929 | 5.55e-05 | 0.000606 | 3.22 | 0.000412 | Ihh/Lef1/Mdk/Cd83/Cd44/Hcls1/Inpp4b/Pik3r6/Lrrc17/Gli3/Calca/Flt3/Zeb1/Prdm1/Cd1d1/Loxl3/Tnfrsf11b/Sox12/Zbtb46/Nckap1l/Tal1/Kitl/Tnfaip6/Rassf2/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 30 |
| GO:0061005 | cell differentiation involved in kidney development | BP | 0.192 | 5.69e-05 | 0.000618 | 3.21 | 0.000421 | Ptpro/Ednrb/Mef2c/Gli3/Tcf21/Tshz3/Ednra/Ptch1/Osr1/Fat4 | 10 |
| GO:0061756 | leukocyte adhesion to vascular endothelial cell | BP | 0.192 | 5.69e-05 | 0.000618 | 3.21 | 0.000421 | Mdk/Itga4/Chst2/Ets1/Itgb7/Icam1/Podxl2/Cxcl12/Sele/St3gal4 | 10 |
| GO:0060998 | regulation of dendritic spine development | BP | 0.153 | 5.87e-05 | 0.000636 | 3.2 | 0.000433 | Nlgn1/Mef2c/Ppfia2/D16Ertd472e/Kif1a/Nlgn3/Apoe/Caprin2/Bhlhb9/Cux2/Palm/Dlg5/Ngef | 13 |
| GO:0001570 | vasculogenesis | BP | 0.144 | 5.96e-05 | 0.000644 | 3.19 | 0.000438 | Angpt1/Kdr/Nrp1/Foxf1/Tek/Has2/Aplnr/Enpp1/Rasip1/Tie1/Gjc1/Heg1/Egfl7/Cav1 | 14 |
| GO:0007202 | activation of phospholipase C activity | BP | 0.28 | 5.98e-05 | 0.000644 | 3.19 | 0.000438 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1 | 7 |
| GO:0101023 | vascular endothelial cell proliferation | BP | 0.28 | 5.98e-05 | 0.000644 | 3.19 | 0.000438 | Mdk/Itga4/Igf1/Mef2c/Igf2/Apln/Akt3 | 7 |
| GO:1905562 | regulation of vascular endothelial cell proliferation | BP | 0.28 | 5.98e-05 | 0.000644 | 3.19 | 0.000438 | Mdk/Itga4/Igf1/Mef2c/Igf2/Apln/Akt3 | 7 |
| GO:0045621 | positive regulation of lymphocyte differentiation | BP | 0.131 | 6.02e-05 | 0.000646 | 3.19 | 0.00044 | Ihh/Lef1/Mdk/Cd83/Pik3r6/Gli3/Prdm1/Cd1d1/Sox12/Nckap1l/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 16 |
| GO:0006022 | aminoglycan metabolic process | BP | 0.136 | 6.44e-05 | 0.000687 | 3.16 | 0.000468 | Angpt1/Cd44/Spock2/Ednrb/Has2/Egflam/Mamdc2/Chst1/Ednra/Ndnf/Tnfaip6/Bgn/Xylt1/Dse/Habp4 | 15 |
| GO:0016525 | negative regulation of angiogenesis | BP | 0.136 | 6.44e-05 | 0.000687 | 3.16 | 0.000468 | Hhip/Tek/Cxcr3/Adamts1/Vash1/Ptprm/Minar1/Ecscr/Emilin1/Tie1/Tcf4/Cldn5/Atp2b4/Sema6a/Ptn | 15 |
| GO:0014065 | phosphatidylinositol 3-kinase signaling | BP | 0.125 | 6.63e-05 | 0.000704 | 3.15 | 0.000479 | Myo16/Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Serpine2/Tek/Ppp1r16b/Prr5l/Htr2a/Prex2/Fyn/Fgr | 17 |
| GO:0010874 | regulation of cholesterol efflux | BP | 0.209 | 6.64e-05 | 0.000704 | 3.15 | 0.000479 | Pla2g10/Pltp/Apoe/Abca5/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3 | 9 |
| GO:0085029 | extracellular matrix assembly | BP | 0.209 | 6.64e-05 | 0.000704 | 3.15 | 0.000479 | Ihh/Phldb2/Fkbp10/Has2/Emilin1/Tie1/Ltbp3/Efemp2/Ntng1 | 9 |
| GO:0048041 | focal adhesion assembly | BP | 0.151 | 6.65e-05 | 0.000704 | 3.15 | 0.000479 | Kdr/Phldb2/Nrp1/Fermt2/Col16a1/Tek/Thsd1/Thy1/Epha3/Ppm1f/Limch1/Arhgap6/Dlc1 | 13 |
| GO:0098739 | import across plasma membrane | BP | 0.11 | 6.71e-05 | 0.00071 | 3.15 | 0.000483 | Kcnj3/Slc1a3/Acsl6/Cacna1c/Kcnd3/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Slc7a2/Cacna1e/Slc8a3/Lrp2/Nalf1/Agtr1a/Slc9a5/Fyn/Slc12a4/Atp1a2/Acsl1/Cacna1a | 21 |
| GO:0042492 | gamma-delta T cell differentiation | BP | 0.333 | 6.9e-05 | 0.000724 | 3.14 | 0.000493 | Lef1/Jag2/Nckap1l/Stat5b/Gimap5/Gimap3 | 6 |
| GO:0097091 | synaptic vesicle clustering | BP | 0.333 | 6.9e-05 | 0.000724 | 3.14 | 0.000493 | Syn2/Nlgn1/Cdh2/Nlgn3/Magi2/Pcdh17 | 6 |
| GO:0099150 | regulation of postsynaptic specialization assembly | BP | 0.333 | 6.9e-05 | 0.000724 | 3.14 | 0.000493 | Gap43/Ptprd/Lrrc4b/Nptx1/Nptxr/Arhgef9 | 6 |
| GO:0002064 | epithelial cell development | BP | 0.102 | 6.92e-05 | 0.000724 | 3.14 | 0.000493 | Ihh/Ar/Cdh2/Ednrb/Icam1/Stc1/Plcb1/Cdh5/Prdm1/Msn/Abi2/Ppp1r16b/Robo4/Rilpl1/Palld/Ednra/Pecam1/Col15a1/Heg1/Arhgef26/Cldn5/Abcb1a/Col18a1/Cldn1 | 24 |
| GO:0050654 | chondroitin sulfate proteoglycan metabolic process | BP | 0.235 | 6.97e-05 | 0.000727 | 3.14 | 0.000494 | Igf1/Spock2/Egflam/Mamdc2/Ndnf/Bgn/Xylt1/Dse | 8 |
| GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.235 | 6.97e-05 | 0.000727 | 3.14 | 0.000494 | Mdk/Itga4/Chst2/Ets1/Icam1/Cxcl12/Sele/St3gal4 | 8 |
| GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | BP | 0.16 | 7.14e-05 | 0.00074 | 3.13 | 0.000503 | Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Tek/Prr5l/Fyn/Fgr | 12 |
| GO:0042982 | amyloid precursor protein metabolic process | BP | 0.16 | 7.14e-05 | 0.00074 | 3.13 | 0.000503 | Itm2a/Igf1/Aph1b/Rtn1/Soat1/Slc2a13/Apoe/Nsg1/Bin1/Abcg1/Adam19/Tmcc2 | 12 |
| GO:0097120 | receptor localization to synapse | BP | 0.16 | 7.14e-05 | 0.00074 | 3.13 | 0.000503 | Gpc6/Nlgn1/Lrrc7/Cep112/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 12 |
| GO:0014911 | positive regulation of smooth muscle cell migration | BP | 0.172 | 7.24e-05 | 0.000748 | 3.13 | 0.000509 | Cyp1b1/Mdk/Igf1/Ddr2/Tert/Myo5a/Has2/Adamts1/Postn/Nox4/Pcsk5 | 11 |
| GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | BP | 0.172 | 7.24e-05 | 0.000748 | 3.13 | 0.000509 | Igf1/Ddr2/Drd4/Tert/Mmp2/Mef2c/Adamts1/Htr1b/Rgs5/Apln/Efemp2 | 11 |
| GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | BP | 0.129 | 7.34e-05 | 0.000756 | 3.12 | 0.000514 | Cd109/Zeb2/Npnt/Pbld2/Zeb1/Cdkn1c/Tsku/Emilin1/Spred3/Hipk2/Nrros/Zfyve9/Il17rd/Lats2/Cav1/Itga8 | 16 |
| GO:0014033 | neural crest cell differentiation | BP | 0.141 | 7.48e-05 | 0.000769 | 3.11 | 0.000523 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Ednrb/Fn1/Mef2c/Edn3/Ednra/Sema3g/Kitl/Sema6a | 14 |
| GO:0060541 | respiratory system development | BP | 0.0974 | 7.84e-05 | 0.000804 | 3.09 | 0.000547 | Gli1/Stra6/Hhip/Lef1/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Edaradd/Gpc3/Ano1/Foxf1/Hopx/Gli3/Tcf21/Tshz3/Loxl3/Mgp/Ltbp3/Heg1/Rcn3/Selenon/Ccn2/Dlg5 | 26 |
| GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.269 | 7.89e-05 | 0.000807 | 3.09 | 0.000549 | Mdk/Itga4/Chst2/Ets1/Icam1/Sele/St3gal4 | 7 |
| GO:0008064 | regulation of actin polymerization or depolymerization | BP | 0.115 | 7.89e-05 | 0.000807 | 3.09 | 0.000549 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Sh3bp1/Daam2/Pecam1/Scin/Nckap1l/Bin1/Arhgap28/Cdc42ep5/Cyfip2/Plekhh2 | 19 |
| GO:0007369 | gastrulation | BP | 0.112 | 7.92e-05 | 0.000808 | 3.09 | 0.00055 | Sfrp2/Lef1/Wnt11/Nog/Phldb2/Hoxa11/Fn1/Foxf1/Osr2/Plpp3/Tgif2/Aplnr/Snai1/Tal1/Col5a2/Osr1/Apln/Ets2/Dact1/Kif16b | 20 |
| GO:0002027 | regulation of heart rate | BP | 0.134 | 7.94e-05 | 0.000808 | 3.09 | 0.00055 | Cacna1c/Kcnd3/Adra1a/Slc8a1/Scn4b/Ednrb/Cacna2d1/Cacna1e/Edn3/Ank2/Calca/Ednra/Bin1/Apln/Cav1 | 15 |
| GO:2000181 | negative regulation of blood vessel morphogenesis | BP | 0.134 | 7.94e-05 | 0.000808 | 3.09 | 0.00055 | Hhip/Tek/Cxcr3/Adamts1/Vash1/Ptprm/Minar1/Ecscr/Emilin1/Tie1/Tcf4/Cldn5/Atp2b4/Sema6a/Ptn | 15 |
| GO:0002040 | sprouting angiogenesis | BP | 0.128 | 8.09e-05 | 0.000821 | 3.09 | 0.000558 | Sema5a/Angpt1/Lef1/Kdr/Nrp1/Tek/Cdh13/Ppp1r16b/Aplnr/Jcad/Vegfc/Agtr1a/Hdac9/Akt3/Sema6a/Adgra2 | 16 |
| GO:0022408 | negative regulation of cell-cell adhesion | BP | 0.106 | 8.15e-05 | 0.000826 | 3.08 | 0.000562 | Ihh/Lef1/Mdk/Abat/Cd44/Serpine2/Cxcl12/Gli3/Podxl/Loxl3/Ccm2l/Ppm1f/Btla/H2-Ab1/Havcr2/Nckap1l/Pde5a/Ptger3/H2-Aa/Gimap5/Dlg5/Gimap3 | 22 |
| GO:0001954 | positive regulation of cell-matrix adhesion | BP | 0.169 | 8.39e-05 | 0.000848 | 3.07 | 0.000577 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Cdh13/Thy1/Ppm1f/Vegfc/Pcsk5/Efemp2 | 11 |
| GO:0003151 | outflow tract morphogenesis | BP | 0.148 | 8.48e-05 | 0.000854 | 3.07 | 0.000581 | Sfrp2/Wnt11/Ryr1/Nog/Tbx2/Nrp1/Mef2c/Lrp2/Plxnd1/Tbx3/Heyl/Ednra/Fzd2 | 13 |
| GO:0006942 | regulation of striated muscle contraction | BP | 0.148 | 8.48e-05 | 0.000854 | 3.07 | 0.000581 | Cacna1c/Adra1a/Slc8a1/Stc1/Ank2/Slc8a3/Myl3/Bin1/Pde5a/Cacna1h/Ccn2/Atp1a2/Cav1 | 13 |
| GO:0048660 | regulation of smooth muscle cell proliferation | BP | 0.111 | 8.56e-05 | 0.000859 | 3.07 | 0.000584 | Vipr2/Igf1/Ddr2/Nos3/Drd4/Npr3/Tert/Mmp2/Mef2c/Cdh13/Adamts1/Htr1b/Rgs5/Apoe/Irak4/Calcrl/Apln/Stat5b/Efemp2/Cav1 | 20 |
| GO:0050906 | detection of stimulus involved in sensory perception | BP | 0.111 | 8.56e-05 | 0.000859 | 3.07 | 0.000584 | Col11a1/Sema5a/Igf1/Sema5b/Scn1a/Slc24a4/Ano1/Grin2b/Serpine2/Cxcl12/Calca/Kit/Grik2/Ppef2/Htr2a/Plcb2/Arrb2/Fyn/Piezo2/Cacnb4 | 20 |
| GO:1904738 | vascular associated smooth muscle cell migration | BP | 0.229 | 8.7e-05 | 0.000872 | 3.06 | 0.000593 | Ddr2/Drd4/Tert/Mef2c/Myo5a/Adamts1/Enpp1/Pcsk5 | 8 |
| GO:1901343 | negative regulation of vasculature development | BP | 0.133 | 8.8e-05 | 0.00088 | 3.06 | 0.000599 | Hhip/Tek/Cxcr3/Adamts1/Vash1/Ptprm/Minar1/Ecscr/Emilin1/Tie1/Tcf4/Cldn5/Atp2b4/Sema6a/Ptn | 15 |
| GO:0070663 | regulation of leukocyte proliferation | BP | 0.0967 | 8.86e-05 | 0.000884 | 3.05 | 0.000602 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Cd1d1/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Kitl/Csf2rb/Pde5a/H2-Aa/Stat5b/Zap70/Dlg5/Il12rb1 | 26 |
| GO:0032414 | positive regulation of ion transmembrane transporter activity | BP | 0.127 | 8.9e-05 | 0.000887 | 3.05 | 0.000603 | Tcaf1/Drd4/Cacna2d1/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Cacnb4 | 16 |
| GO:0030323 | respiratory tube development | BP | 0.1 | 9.02e-05 | 0.000897 | 3.05 | 0.00061 | Gli1/Stra6/Hhip/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Gpc3/Foxf1/Hopx/Gli3/Tcf21/Tshz3/Loxl3/Mgp/Pcsk5/Ltbp3/Heg1/Rcn3/Selenon/Ccn2/Dlg5 | 24 |
| GO:0045580 | regulation of T cell differentiation | BP | 0.114 | 9.28e-05 | 0.000918 | 3.04 | 0.000625 | Ihh/Lef1/Mdk/Cd83/Cd44/Pik3r6/Gli3/Zeb1/Prdm1/Cd1d1/Loxl3/Sox12/Nckap1l/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 19 |
| GO:0006029 | proteoglycan metabolic process | BP | 0.156 | 9.29e-05 | 0.000918 | 3.04 | 0.000625 | Col11a1/Ihh/Igf1/Spock2/Chst10/Egflam/Mamdc2/Ndnf/Adamts7/Bgn/Xylt1/Dse | 12 |
| GO:0046635 | positive regulation of alpha-beta T cell activation | BP | 0.156 | 9.29e-05 | 0.000918 | 3.04 | 0.000625 | Ihh/Cd83/Gli3/Cd1d1/H2-Ab1/Xcl1/Nckap1l/Gimap5/Zap70/Gimap3/Il12rb1/Il2rg | 12 |
| GO:0051098 | regulation of binding | BP | 0.0856 | 9.32e-05 | 0.00092 | 3.04 | 0.000626 | Map2/Angpt1/Lef1/Itga4/Nog/Igf1/Zfp462/Runx1t1/Tert/Lrrk2/Hopx/Plxnd1/Lfng/Ccm2l/Apoe/Caprin2/Zc4h2/Twist2/Arrb1/Hipk2/Pkd1/Dysf/Arhgap28/Ttbk2/Nfatc4/Cldn5/Arrb2/Dzip1/Mfng/Habp4/Nes/Cav1/Dact1/Zfp90 | 34 |
| GO:0001662 | behavioral fear response | BP | 0.182 | 9.37e-05 | 0.000923 | 3.03 | 0.000628 | Ankfn1/Mdk/Drd4/Mef2c/Grin2b/Cacna1e/Grik2/Apoe/Gng7/Atp1a2 | 10 |
| GO:0030098 | lymphocyte differentiation | BP | 0.0828 | 9.47e-05 | 0.000932 | 3.03 | 0.000634 | Ihh/Lef1/Mdk/Cd83/Itm2a/Cd44/Il11ra1/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Tnfsf13b/Dock10/Zeb1/Prdm1/Cd1d1/Jag2/Lfng/Loxl3/Kit/H2-Ab1/Cracr2a/Enpp1/Sox12/Irf8/Nckap1l/Satb1/Hdac9/H2-Aa/Stat5b/Gimap5/Zap70/Mfng/Gimap3/Il2rg | 37 |
| GO:0014032 | neural crest cell development | BP | 0.146 | 9.55e-05 | 0.000937 | 3.03 | 0.000638 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Ednrb/Fn1/Edn3/Ednra/Sema3g/Kitl/Sema6a | 13 |
| GO:0008406 | gonad development | BP | 0.1 | 9.63e-05 | 0.000942 | 3.03 | 0.000641 | Sfrp2/Angpt1/Ar/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Hoxa9/Rhobtb3/Kitl/Tnfaip6/Arrb1/Osr1/Arrb2/Stat5b/Abcb1a | 24 |
| GO:0050918 | positive chemotaxis | BP | 0.2 | 9.67e-05 | 0.000942 | 3.03 | 0.000641 | Sema5a/Angpt1/Kdr/Nrp1/Cxcl12/Cdh13/Lrp2/Coro1a/Vegfc | 9 |
| GO:0071825 | protein-lipid complex subunit organization | BP | 0.2 | 9.67e-05 | 0.000942 | 3.03 | 0.000641 | Apob/Pla2g10/Soat1/Pltp/Apoe/Bin1/Abca5/Abcg1/Pcdhga3 | 9 |
| GO:0006525 | arginine metabolic process | BP | 0.316 | 9.73e-05 | 0.000945 | 3.02 | 0.000643 | Ddah1/Nos3/Fah/Atp2b4/Asl/Slc7a7 | 6 |
| GO:0014051 | gamma-aminobutyric acid secretion | BP | 0.316 | 9.73e-05 | 0.000945 | 3.02 | 0.000643 | Grik1/Abat/Htr1b/Gabbr1/Cacna1a/Cacnb4 | 6 |
| GO:0003073 | regulation of systemic arterial blood pressure | BP | 0.126 | 9.79e-05 | 0.000949 | 3.02 | 0.000646 | Ar/Kdr/Ddah1/Adra1a/Nos3/Ptpro/P2rx2/Adamts16/Ednrb/Enpep/Edn3/Calca/Rps6ka2/Postn/Agtr1a/Apln | 16 |
| GO:0072659 | protein localization to plasma membrane | BP | 0.0927 | 9.98e-05 | 0.000966 | 3.02 | 0.000657 | Kcnb2/Ar/Pkdcc/Epb41l3/Ccdc88a/Cdh2/Efr3b/Arl6/Abi3/Myo5a/Gga2/Ank2/Rab34/Epha3/Ttc7b/Nfasc/Rilpl1/Nsg1/Stac2/Ptch1/Kcnip3/Dchs1/Tspan5/Atp2b4/Palm/Bbs1/Pid1/Nkd2 | 28 |
| GO:0030832 | regulation of actin filament length | BP | 0.113 | 0.000101 | 0.000971 | 3.01 | 0.000661 | Kank4/Sema5a/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Sh3bp1/Daam2/Pecam1/Scin/Nckap1l/Bin1/Arhgap28/Cdc42ep5/Cyfip2/Plekhh2 | 19 |
| GO:0043547 | positive regulation of GTPase activity | BP | 0.0977 | 0.000102 | 0.000982 | 3.01 | 0.000668 | Wnt11/Plxnb1/Rgs6/Rgs16/Rasgrp3/Fermt2/Garnl3/Arhgap45/Icam1/Rasgrp2/Arhgap25/Thy1/Dock10/Myo9a/Sh3bp1/Arhgef15/Arhgap6/Xcl1/Arap3/Bin1/Arhgef26/Net1/Gnb5/Ngef/Mapre2 | 25 |
| GO:0030204 | chondroitin sulfate metabolic process | BP | 0.259 | 0.000103 | 0.000985 | 3.01 | 0.00067 | Spock2/Egflam/Mamdc2/Ndnf/Bgn/Xylt1/Dse | 7 |
| GO:0002228 | natural killer cell mediated immunity | BP | 0.144 | 0.000107 | 0.00103 | 2.99 | 0.000699 | Pik3r6/Coro1a/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gzmb/Gimap5/Gimap3/Rab27a/Cd96/Clnk | 13 |
| GO:0019915 | lipid storage | BP | 0.144 | 0.000107 | 0.00103 | 2.99 | 0.000699 | Apob/Pla2g10/Acacb/Vstm2a/Mest/Soat1/Apoe/Enpp1/Dysf/Abcg1/Stat5b/Cav1/Nr1h3 | 13 |
| GO:0045807 | positive regulation of endocytosis | BP | 0.125 | 0.000108 | 0.00103 | 2.99 | 0.000699 | Angpt1/Nlgn1/Gpc3/Lrrk2/Lrp2/Apoe/Sele/Magi2/Nckap1l/Hip1/Bin1/Arrb1/Arrb2/Apln/Smpd1/Cav1 | 16 |
| GO:0003203 | endocardial cushion morphogenesis | BP | 0.222 | 0.000108 | 0.00103 | 2.99 | 0.000699 | Nog/Tbx2/Nos3/Aplnr/Tbx3/Heyl/Snai1/Dchs1 | 8 |
| GO:0061001 | regulation of dendritic spine morphogenesis | BP | 0.179 | 0.00011 | 0.00104 | 2.98 | 0.00071 | Nlgn1/Lrrk2/Ppfia2/Kif1a/Abi2/Nlgn3/Caprin2/Bhlhb9/Cux2/Ngef | 10 |
| GO:0021915 | neural tube development | BP | 0.106 | 0.000113 | 0.00107 | 2.97 | 0.000727 | Sfrp2/Nog/Zeb2/Dzip1l/Sall2/Gli3/Lrp2/Gdf7/Ift57/Cecr2/Gpr161/Phgdh/Prickle1/Ptch1/Ipmk/Ttbk2/Dchs1/Dlc1/Fzd2/Rab23/Dact1 | 21 |
| GO:0060323 | head morphogenesis | BP | 0.196 | 0.000116 | 0.00109 | 2.96 | 0.000743 | Stra6/Ihh/Lef1/Nog/Crispld1/Mmp2/Flvcr1/Twist2/Asph | 9 |
| GO:1903170 | negative regulation of calcium ion transmembrane transport | BP | 0.196 | 0.000116 | 0.00109 | 2.96 | 0.000743 | Drd4/Bin1/Dysf/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5 | 9 |
| GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | BP | 0.136 | 0.000116 | 0.00109 | 2.96 | 0.000743 | Hgf/Angpt1/Kdr/Agap2/Plxnb1/Igf1/Hcls1/Fn1/Serpine2/Tek/Ppp1r16b/Prr5l/Fyn/Fgr | 14 |
| GO:0036293 | response to decreased oxygen levels | BP | 0.0988 | 0.000117 | 0.0011 | 2.96 | 0.000748 | Kcnk3/Kcnd2/Ryr1/Kdr/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Grin2b/Tek/Slc8a3/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Hif3a | 24 |
| GO:0031099 | regeneration | BP | 0.115 | 0.000117 | 0.0011 | 2.96 | 0.00075 | Gli1/Gap43/Hgf/Mdk/Igf1/Mmp2/Cxcl12/Hopx/Thy1/Matn2/Enpp1/Rtn4rl1/Dysf/Selenon/Neo1/Xylt1/Ptn/Large1 | 18 |
| GO:0042476 | odontogenesis | BP | 0.124 | 0.000118 | 0.00111 | 2.96 | 0.000753 | Wnt6/Lef1/Apcdd1/Fst/Edaradd/Slc24a4/Myo5a/Id3/Osr2/Gli3/Jag2/Tnfrsf11b/Enpp1/Ednra/Osr1/Edar | 16 |
| GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.124 | 0.000118 | 0.00111 | 2.96 | 0.000753 | Sfrp2/Cd109/Nog/Fst/Lrp2/Pbld2/Tsku/Magi2/Emilin1/Spred3/Tnfaip6/Hipk2/Nrros/Il17rd/Cav1/Tmprss6 | 16 |
| GO:0015849 | organic acid transport | BP | 0.0903 | 0.000119 | 0.00112 | 2.95 | 0.000759 | Slc1a3/Acsl6/Grik1/Pla2g10/Slc13a5/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Rbp7/Slc36a4/Lrp2/Htr1b/Slc6a15/Apoe/Bdkrb2/Slc23a2/Sfxn5/Nfkbie/Slc38a1/Abcd2/Abcb1a/Slc7a7/Acsl1/Abcc5/Gabbr1/Cacna1a/Cacnb4 | 29 |
| GO:0035023 | regulation of Rho protein signal transduction | BP | 0.143 | 0.00012 | 0.00112 | 2.95 | 0.000765 | Adra1a/Nrp1/Cdh2/Lpar4/Arhgef25/Apoe/Rasip1/Eps8l1/Arap3/Arrb1/Heg1/Dlc1/Net1 | 13 |
| GO:0010001 | glial cell differentiation | BP | 0.0965 | 0.000123 | 0.00114 | 2.94 | 0.000777 | Gap43/Ptprz1/Lef1/Mdk/Nog/Igf1/Lamc3/Cdh2/Wasf3/Dab1/Id4/Serpine2/Gli3/Slc8a3/Plpp3/Nlgn3/Phgdh/Enpp1/Daam2/Tal1/Bin1/Nrros/Ptn/Tppp/Itgb4 | 25 |
| GO:0051090 | regulation of DNA-binding transcription factor activity | BP | 0.0826 | 0.000123 | 0.00114 | 2.94 | 0.000778 | Gli1/Ndp/Cyp1b1/4930447C04Rik/Acod1/Ar/Pla2g10/Eda2r/Ddr2/Hcls1/Id3/Icam1/Prkd1/Gli3/Eif2ak4/Plpp3/Ndn/Kit/Cth/Enpp1/Havcr2/Heyl/Cd200/Zc4h2/Rps6ka5/Foxs1/Arrb1/Hipk2/Pkd1/Ptch1/Nwd1/Cd84/Arrb2/Pkd2/Fzd2/Cav1 | 36 |
| GO:0045137 | development of primary sexual characteristics | BP | 0.0984 | 0.000124 | 0.00116 | 2.94 | 0.000786 | Sfrp2/Angpt1/Ar/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Hoxa9/Rhobtb3/Kitl/Tnfaip6/Arrb1/Osr1/Arrb2/Stat5b/Abcb1a | 24 |
| GO:0031529 | ruffle organization | BP | 0.175 | 0.000128 | 0.00119 | 2.93 | 0.000807 | Evl/Nlgn1/Cspg4/Aif1l/Pfn2/Icam1/Sh3bp1/Eps8l1/Arhgef26/Cav1 | 10 |
| GO:0046849 | bone remodeling | BP | 0.135 | 0.000129 | 0.00119 | 2.92 | 0.000809 | Ihh/Mdk/Inpp4b/Calca/Htr1b/Pth1r/Tnfrsf11b/Nox4/Cd38/Enpp1/Syt7/Ltbp3/Rassf2/Ptn | 14 |
| GO:0003272 | endocardial cushion formation | BP | 0.25 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Nog/Tbx2/Aplnr/Tbx3/Heyl/Snai1/Dchs1 | 7 |
| GO:0033622 | integrin activation | BP | 0.25 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Fermt2/Fn1/Col16a1/Cxcl12/Jam3/Rasip1/Pcsk5 | 7 |
| GO:0051894 | positive regulation of focal adhesion assembly | BP | 0.25 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Kdr/Nrp1/Fermt2/Col16a1/Tek/Thy1/Ppm1f | 7 |
| GO:0060272 | embryonic skeletal joint morphogenesis | BP | 0.385 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Hoxd11/Nog/Hoxa11/Osr2/Osr1 | 5 |
| GO:0090022 | regulation of neutrophil chemotaxis | BP | 0.216 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Mdk/Jam3/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Mpp1 | 8 |
| GO:0050670 | regulation of lymphocyte proliferation | BP | 0.098 | 0.000132 | 0.00121 | 2.92 | 0.000822 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Cd1d1/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Pde5a/H2-Aa/Stat5b/Zap70/Dlg5/Il12rb1 | 24 |
| GO:0002031 | G protein-coupled receptor internalization | BP | 0.3 | 0.000134 | 0.00122 | 2.91 | 0.000828 | Calca/Htr1b/Arrb1/Arrb2/Ptger3/Apln | 6 |
| GO:0043116 | negative regulation of vascular permeability | BP | 0.3 | 0.000134 | 0.00122 | 2.91 | 0.000828 | Angpt1/Ddah1/Fermt2/Akap12/Apoe/Cldn5 | 6 |
| GO:0051258 | protein polymerization | BP | 0.0942 | 0.000135 | 0.00122 | 2.91 | 0.000831 | Kank4/Map2/Evl/Hcls1/Tmod2/Pfn2/Icam1/Cdh5/Catip/Coro1a/Rp1/Esam/Tubb4a/Daam2/Pecam1/Scin/Nckap1l/Slain1/Nin/Bin1/Arhgap28/Cdc42ep5/Cav1/Cyfip2/Tppp/Map7d3 | 26 |
| GO:0006940 | regulation of smooth muscle contraction | BP | 0.159 | 0.000146 | 0.00132 | 2.88 | 9e-04 | Adra1a/Npnt/Abat/Calca/Gucy1a1/Kit/Kcnma1/Ptger3/Calcrl/Atp1a2/Cav1 | 11 |
| GO:0046513 | ceramide biosynthetic process | BP | 0.159 | 0.000146 | 0.00132 | 2.88 | 9e-04 | St8sia2/St8sia4/Agk/Sptlc3/Cers4/St6galnac4/St3gal2/St8sia6/Degs2/St6galnac6/Smpd1 | 11 |
| GO:0042311 | vasodilation | BP | 0.172 | 0.000149 | 0.00134 | 2.87 | 0.000915 | Kdr/Npr3/Ednrb/Itga1/Calca/Ptprm/Gucy1a1/Apoe/Kcnma1/Bdkrb2 | 10 |
| GO:0003208 | cardiac ventricle morphogenesis | BP | 0.148 | 0.000153 | 0.00137 | 2.86 | 0.000935 | Col11a1/Sfrp2/Nog/Foxf1/Mef2c/Lrp2/Cpe/Ccm2l/Heyl/Ednra/Myl3/Heg1 | 12 |
| GO:0043279 | response to alkaloid | BP | 0.148 | 0.000153 | 0.00137 | 2.86 | 0.000935 | Slc1a3/Ryr1/Abat/Slc8a1/Drd4/Htr1b/Ednra/Htr2a/Selenon/Dlg4/Gstm7/Smpd1 | 12 |
| GO:0014909 | smooth muscle cell migration | BP | 0.132 | 0.000158 | 0.00142 | 2.85 | 0.000964 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Mef2c/Myo5a/Has2/Adamts1/Postn/Nox4/Enpp1/Pcsk5 | 14 |
| GO:0097106 | postsynaptic density organization | BP | 0.211 | 0.000161 | 0.00144 | 2.84 | 0.00098 | Ptprd/Nlgn1/Cdh2/Lrrc4b/Nptx1/Shank2/Slitrk3/Syngap1 | 8 |
| GO:1901385 | regulation of voltage-gated calcium channel activity | BP | 0.211 | 0.000161 | 0.00144 | 2.84 | 0.00098 | Drd4/Cacna2d1/Stac2/Dysf/Rrad/Gnb5/Cacna1a/Cacnb4 | 8 |
| GO:0035904 | aorta development | BP | 0.157 | 0.000167 | 0.00149 | 2.83 | 0.00101 | Tbx2/Kif7/Fkbp10/Lrp2/Plxnd1/Prdm1/Aplnr/Enpp1/Ednra/Prickle1/Efemp2 | 11 |
| GO:1902107 | positive regulation of leukocyte differentiation | BP | 0.106 | 0.000167 | 0.00149 | 2.83 | 0.00101 | Ihh/Lef1/Mdk/Cd83/Hcls1/Pik3r6/Gli3/Calca/Prdm1/Cd1d1/Sox12/Zbtb46/Nckap1l/Kitl/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 20 |
| GO:1903708 | positive regulation of hemopoiesis | BP | 0.106 | 0.000167 | 0.00149 | 2.83 | 0.00101 | Ihh/Lef1/Mdk/Cd83/Hcls1/Pik3r6/Gli3/Calca/Prdm1/Cd1d1/Sox12/Zbtb46/Nckap1l/Kitl/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 20 |
| GO:0052547 | regulation of peptidase activity | BP | 0.0813 | 0.000168 | 0.00149 | 2.83 | 0.00101 | Hgf/Sfrp2/Cd109/Spink1/Igf1/Tfpi/Aph1b/Cd44/Gpc3/Bok/Grin2b/Wdr35/Serpine2/Mical1/Wfdc3/Ift57/Serpini1/Wnt9a/Ppm1f/Hip1/Bin1/Arrb1/Epha7/Rcn3/Asph/Dlc1/Nlrp1b/Arrb2/Ccn2/Fyn/Pi16/Pi15/Mgmt/Simc1/Cav1/Cyfip2 | 36 |
| GO:2000027 | regulation of animal organ morphogenesis | BP | 0.12 | 0.000169 | 0.0015 | 2.82 | 0.00102 | Hgf/Sfrp2/Ar/Apcdd1/Wnt11/Hoxd11/Nog/Nkd1/Hoxa11/Gpc3/Znrf3/Tnfrsf11b/Agtr1a/Ednra/Fzd2/Dact1 | 16 |
| GO:0032944 | regulation of mononuclear cell proliferation | BP | 0.0964 | 0.00017 | 0.0015 | 2.82 | 0.00102 | Ihh/Igf1/Cd44/Mef2c/Mapk8ip1/Tnfsf13b/Prdm1/Cd1d1/Coro1a/Btla/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Pde5a/H2-Aa/Stat5b/Zap70/Dlg5/Il12rb1 | 24 |
| GO:0030308 | negative regulation of cell growth | BP | 0.103 | 0.000171 | 0.00151 | 2.82 | 0.00103 | Tro/Sfrp2/Map2/Sema5a/Wnt11/Sema5b/Sema7a/Nrp1/Serpine2/Cth/Minar1/Enpp1/Caprin2/Sema3g/Epha7/Fhl1/P3h1/Rrad/Pi16/Sema6a/Tspyl2 | 21 |
| GO:0003170 | heart valve development | BP | 0.169 | 0.000173 | 0.00152 | 2.82 | 0.00103 | Stra6/Nos3/Mef2c/Erg/Prdm1/Aplnr/Emilin1/Tie1/Heyl/Dchs1 | 10 |
| GO:0042552 | myelination | BP | 0.109 | 0.000173 | 0.00152 | 2.82 | 0.00103 | Ptprz1/Hgf/Igf1/Epb41l3/Mpdz/Cdh2/Wasf3/Myo5a/Id4/Jam3/Slc8a3/Nfasc/Enpp1/Cldn5/Fyn/Abcd2/Ptn/Tppp/Itgb4 | 19 |
| GO:0008306 | associative learning | BP | 0.125 | 0.000175 | 0.00153 | 2.82 | 0.00104 | Cacna1c/Nog/Drd4/Grin2b/B4galt2/Pde1b/Adcy3/Shank2/Cacna1e/Kit/Nlgn3/Atp1a2/Brsk1/Syngap1/Ddhd2 | 15 |
| GO:1903131 | mononuclear cell differentiation | BP | 0.0788 | 0.000175 | 0.00153 | 2.82 | 0.00104 | Ihh/Lef1/Mdk/Cd83/Itm2a/Cd44/Il11ra1/Mef2c/Itk/Cd3g/Pik3r6/Gli3/Dock2/Flt3/Tnfsf13b/Dock10/Zeb1/Prdm1/Cd1d1/Jag2/Lfng/Loxl3/Kit/H2-Ab1/Cracr2a/Enpp1/Sox12/Zbtb46/Irf8/Nckap1l/Satb1/Hdac9/H2-Aa/Stat5b/Gimap5/Zap70/Mfng/Gimap3/Il2rg | 39 |
| GO:0070507 | regulation of microtubule cytoskeleton organization | BP | 0.112 | 0.000175 | 0.00153 | 2.81 | 0.00104 | Map2/Phldb2/Mpdz/Gnai1/Cdh5/Epha3/Rp1/Map9/Tubb4a/Slain1/Nin/Pkd1/Dysf/Ttbk2/Cep97/Mapre2/Cav1/Tppp | 18 |
| GO:0001558 | regulation of cell growth | BP | 0.0803 | 0.000176 | 0.00154 | 2.81 | 0.00105 | Tro/Sfrp2/Map2/Sema5a/Wnt11/Col14a1/Igf1/Ptch2/Sema5b/Epb41l3/Cpne5/Cd44/Sema7a/Nrp1/Fn1/Serpine2/Cxcl12/Klhl22/Kif26a/Cth/Minar1/Apoe/Cd38/Enpp1/Caprin2/Sema3g/Slc23a2/Epha7/Cacng7/Fhl1/P3h1/Csf2rb/Rrad/Pi16/Sema6a/Tspyl2/Igfbp4 | 37 |
| GO:0006066 | alcohol metabolic process | BP | 0.087 | 0.00018 | 0.00157 | 2.8 | 0.00107 | Apob/Cyp1b1/Ttr/Cyp7b1/Hmgcs2/Ednrb/Soat1/Inpp4b/Sptlc3/Plcb1/Pth1r/Plpp3/Dkk3/Tsku/Apoe/Enpp1/Kcnma1/Bpnt1/Adcyap1r1/Degs2/Abca5/Cyp46a1/Dysf/Abcg1/Ipmk/Cacna1h/Sord/Plpp1/Smpd1/Lipe | 30 |
| GO:0032411 | positive regulation of transporter activity | BP | 0.119 | 0.000185 | 0.00161 | 2.79 | 0.00109 | Tcaf1/Drd4/Cacna2d1/Myo5a/Ank2/Plcb1/Trpc6/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Abcb1a/Cacnb4 | 16 |
| GO:0030509 | BMP signaling pathway | BP | 0.108 | 0.000186 | 0.00161 | 2.79 | 0.0011 | Sfrp2/Lef1/Kdr/Nog/Rnf165/Fst/Gpc3/Zfp423/Lrp2/Gdf7/Cdh5/Kcp/Smad9/Tnfaip6/Hipk2/Neo1/Fstl1/Cav1/Tmprss6 | 19 |
| GO:0051494 | negative regulation of cytoskeleton organization | BP | 0.108 | 0.000186 | 0.00161 | 2.79 | 0.0011 | Kank4/Map2/Evl/Phldb2/Tmod2/Pfn2/Cdh5/Coro1a/Rp1/Tubb4a/Arhgap6/Tmeff2/Pecam1/Scin/Dysf/Arhgap28/Ttbk2/Dlc1/Plekhh2 | 19 |
| GO:0014910 | regulation of smooth muscle cell migration | BP | 0.137 | 0.000187 | 0.00162 | 2.79 | 0.0011 | Cyp1b1/Mdk/Igf1/Ddr2/Drd4/Tert/Mef2c/Myo5a/Has2/Adamts1/Postn/Nox4/Pcsk5 | 13 |
| GO:0048864 | stem cell development | BP | 0.137 | 0.000187 | 0.00162 | 2.79 | 0.0011 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Ednrb/Fn1/Edn3/Ednra/Sema3g/Kitl/Sema6a | 13 |
| GO:0055117 | regulation of cardiac muscle contraction | BP | 0.155 | 0.00019 | 0.00164 | 2.78 | 0.00112 | Cacna1c/Adra1a/Slc8a1/Stc1/Ank2/Bin1/Pde5a/Cacna1h/Ccn2/Atp1a2/Cav1 | 11 |
| GO:0043583 | ear development | BP | 0.0956 | 0.000191 | 0.00165 | 2.78 | 0.00112 | Col11a1/Stra6/Nog/Igf1/Tbx2/Osr2/Tshz1/Lrig3/Gli3/Zeb1/Jag2/Cecr2/Lin7a/Tsku/Enpp1/Kcnma1/Tbx3/Ednra/Osr1/Dchs1/Fzd2/Fat4/Kcnq4/Itga8 | 24 |
| GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.124 | 0.000192 | 0.00165 | 2.78 | 0.00112 | Kdr/Rnf165/Zeb2/Npnt/Gpc3/Gdf11/Zfp423/Gdf7/Cdh5/Gdf10/Cdkn1c/Kcp/Hipk2/Neo1/Itga8 | 15 |
| GO:0019228 | neuronal action potential | BP | 0.184 | 0.000192 | 0.00165 | 2.78 | 0.00113 | Kcnd2/P2rx2/Scn1a/Scn2a/Grik2/Kcnma1/Kcnc4/Cacna1h/Scn3a | 9 |
| GO:0030324 | lung development | BP | 0.0975 | 0.000195 | 0.00168 | 2.78 | 0.00114 | Gli1/Stra6/Hhip/Kdr/Nog/Igf1/Pkdcc/Tbx2/Nos3/Gpc3/Foxf1/Hopx/Gli3/Tcf21/Tshz3/Loxl3/Mgp/Ltbp3/Heg1/Rcn3/Selenon/Ccn2/Dlg5 | 23 |
| GO:0045586 | regulation of gamma-delta T cell differentiation | BP | 0.357 | 0.000198 | 0.00169 | 2.77 | 0.00115 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0046643 | regulation of gamma-delta T cell activation | BP | 0.357 | 0.000198 | 0.00169 | 2.77 | 0.00115 | Lef1/Nckap1l/Stat5b/Gimap5/Gimap3 | 5 |
| GO:0051967 | negative regulation of synaptic transmission, glutamatergic | BP | 0.357 | 0.000198 | 0.00169 | 2.77 | 0.00115 | Grik1/Myo5a/Htr1b/Grik2/Htr2a | 5 |
| GO:1905809 | negative regulation of synapse organization | BP | 0.357 | 0.000198 | 0.00169 | 2.77 | 0.00115 | Arhgef15/Apoe/Epha7/Nfatc4/Ptn | 5 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | BP | 0.167 | 0.000199 | 0.0017 | 2.77 | 0.00115 | Kdr/Nrp1/Ptp4a3/Prkd1/Jcad/Vegfc/Tcf4/Atp2b4/Sema6a/Adgra2 | 10 |
| GO:0048469 | cell maturation | BP | 0.0991 | 0.00021 | 0.00178 | 2.75 | 0.00121 | Epha8/Ihh/Pla2g10/Kdr/Spink1/Cspg4/Ednrb/Lrrk2/Gdf11/Flvcr1/Calca/Plcb1/Rps6ka2/Pth1r/Dlg2/Cdkn1c/Nfasc/Kcnma1/Ednra/Lhx6/Tal1/Septin4 | 22 |
| GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | BP | 0.233 | 0.00021 | 0.00178 | 2.75 | 0.00121 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln | 7 |
| GO:0022401 | negative adaptation of signaling pathway | BP | 0.233 | 0.00021 | 0.00178 | 2.75 | 0.00121 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln | 7 |
| GO:0045932 | negative regulation of muscle contraction | BP | 0.233 | 0.00021 | 0.00178 | 2.75 | 0.00121 | Calca/Gucy1a1/Kcnma1/Bin1/Pde5a/Calcrl/Atp1a2 | 7 |
| GO:0008366 | axon ensheathment | BP | 0.107 | 0.000215 | 0.00181 | 2.74 | 0.00123 | Ptprz1/Hgf/Igf1/Epb41l3/Mpdz/Cdh2/Wasf3/Myo5a/Id4/Jam3/Slc8a3/Nfasc/Enpp1/Cldn5/Fyn/Abcd2/Ptn/Tppp/Itgb4 | 19 |
| GO:0007517 | muscle organ development | BP | 0.0849 | 0.000216 | 0.00181 | 2.74 | 0.00123 | Col11a1/Stra6/Angpt1/Lef1/Hoxd10/Ryr1/Nog/P2rx2/Hoxd9/Mef2c/Id3/Tcf21/Lrp2/Ccm2l/Heyl/Ednra/Igf2/Myl3/Hdac9/Heg1/Dysf/Fhl1/Selenon/Fzd2/Hlf/Efemp2/Sgcb/Large1/Cav1/Zfp689/Usp2 | 31 |
| GO:0002260 | lymphocyte homeostasis | BP | 0.143 | 0.000217 | 0.00182 | 2.74 | 0.00124 | Cd44/Mef2c/Sh2b2/Tnfsf13b/Dock10/Coro1a/Enpp1/Nckap1l/Stat5b/Gimap5/Gimap3/Lat | 12 |
| GO:0006809 | nitric oxide biosynthetic process | BP | 0.143 | 0.000217 | 0.00182 | 2.74 | 0.00124 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0007338 | single fertilization | BP | 0.11 | 0.000221 | 0.00185 | 2.73 | 0.00126 | Ar/Pla2g10/Hoxd10/Hoxd11/Spink1/Hoxa11/Hoxd9/Hoxa10/Adcy3/Plcb1/Trpc6/Glrb/Cecr2/Hoxa9/Cacna1h/Ccdc136/Pkdrej/Stx2 | 18 |
| GO:0007548 | sex differentiation | BP | 0.0883 | 0.000225 | 0.00188 | 2.73 | 0.00128 | Tex15/Stra6/Sfrp2/Angpt1/Ar/Kdr/Nos3/Fst/Hoxa11/Hoxa10/Mmp2/Icam1/Adamts1/Tcf21/Lrp2/Lfng/Kit/Tbx3/Hoxa9/Rhobtb3/Kitl/Smad9/Tnfaip6/Arrb1/Osr1/Arrb2/Stat5b/Abcb1a | 28 |
| GO:0034332 | adherens junction organization | BP | 0.18 | 0.000226 | 0.00188 | 2.73 | 0.00128 | Fermt2/Cdh6/Jam3/Cdh10/Cdh5/Abi2/Cdh11/Dchs1/Dlg5 | 9 |
| GO:1903053 | regulation of extracellular matrix organization | BP | 0.18 | 0.000226 | 0.00188 | 2.73 | 0.00128 | Sema5a/Ddr2/Prdm5/Phldb2/Ets1/Has2/Emilin1/Tie1/Efemp2 | 9 |
| GO:0042596 | fear response | BP | 0.164 | 0.000229 | 0.00191 | 2.72 | 0.0013 | Ankfn1/Mdk/Drd4/Mef2c/Grin2b/Cacna1e/Grik2/Apoe/Gng7/Atp1a2 | 10 |
| GO:0002718 | regulation of cytokine production involved in immune response | BP | 0.134 | 0.000232 | 0.00192 | 2.72 | 0.00131 | Angpt1/Sema7a/Tek/Gprc5b/Tril/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Cd96/Clnk | 13 |
| GO:0070482 | response to oxygen levels | BP | 0.0894 | 0.000238 | 0.00197 | 2.7 | 0.00134 | Kcnk3/Kcnd2/Ryr1/Kdr/Ddah1/Abat/Slc8a1/Cbs/P2rx2/Pygm/Tert/Mmp2/Gucy1a2/Grin2b/Tek/Slc8a3/Gucy1b1/Gucy1a1/Scn2a/Vegfc/Cd38/Kcnma1/Ednra/Ndnf/Cav1/Cpeb1/Hif3a | 27 |
| GO:0050921 | positive regulation of chemotaxis | BP | 0.113 | 0.000241 | 0.002 | 2.7 | 0.00136 | Sema5a/Mdk/Kdr/Nrp1/Fn1/Prkd1/Cxcr3/Cxcl12/Cdh13/Edn3/Ppm1f/Vegfc/Ednra/Xcl1/Nckap1l/Dysf/Ptn | 17 |
| GO:0046637 | regulation of alpha-beta T cell differentiation | BP | 0.151 | 0.000245 | 0.00202 | 2.69 | 0.00138 | Ihh/Cd83/Gli3/Prdm1/Cd1d1/Loxl3/Nckap1l/Gimap5/Zap70/Gimap3/Il2rg | 11 |
| GO:0001945 | lymph vessel development | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Kdr/Vash1/Vegfc/Tie1/Pkd1/Heg1/Svep1 | 7 |
| GO:0050951 | sensory perception of temperature stimulus | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Ano1/Cxcl12/Calca/Grik2/Enpp1/Htr2a/Arrb2 | 7 |
| GO:0086010 | membrane depolarization during action potential | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Cacna1c/Scn4b/Scn1a/Cacna2d1/Scn2a/Cacna1h/Scn3a | 7 |
| GO:0099633 | protein localization to postsynaptic specialization membrane | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Gpc6/Nptx1/Dlg2/Nptxr/Magi2/Cacng7/Dlg4 | 7 |
| GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Gpc6/Nptx1/Dlg2/Nptxr/Magi2/Cacng7/Dlg4 | 7 |
| GO:1904752 | regulation of vascular associated smooth muscle cell migration | BP | 0.226 | 0.000262 | 0.00214 | 2.67 | 0.00146 | Ddr2/Drd4/Tert/Mef2c/Myo5a/Adamts1/Pcsk5 | 7 |
| GO:0006584 | catecholamine metabolic process | BP | 0.161 | 0.000263 | 0.00215 | 2.67 | 0.00146 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Sult1a1 | 10 |
| GO:0009712 | catechol-containing compound metabolic process | BP | 0.161 | 0.000263 | 0.00215 | 2.67 | 0.00146 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Sult1a1 | 10 |
| GO:0031102 | neuron projection regeneration | BP | 0.161 | 0.000263 | 0.00215 | 2.67 | 0.00146 | Gap43/Hgf/Mmp2/Thy1/Matn2/Enpp1/Rtn4rl1/Neo1/Xylt1/Ptn | 10 |
| GO:0086065 | cell communication involved in cardiac conduction | BP | 0.195 | 0.000282 | 0.00229 | 2.64 | 0.00156 | Cacna1c/Kcnd3/Slc8a1/Scn4b/Cacna2d1/Ank2/Gjc1/Cav1 | 8 |
| GO:2000463 | positive regulation of excitatory postsynaptic potential | BP | 0.195 | 0.000282 | 0.00229 | 2.64 | 0.00156 | Nlgn1/Drd4/Stx1b/Grin2b/Tbc1d24/Nlgn3/Dlg4/Cux2 | 8 |
| GO:0030010 | establishment of cell polarity | BP | 0.111 | 0.000282 | 0.00229 | 2.64 | 0.00156 | Ankfn1/Map2/Fez1/Foxf1/Jam3/Dock2/Cdh5/Msn/Prickle2/Myo9a/Sh3bp1/Ubxn2b/Frmd4b/Pkd1/Pard3b/Frmd4a/Brsk1 | 17 |
| GO:0006672 | ceramide metabolic process | BP | 0.131 | 0.000284 | 0.0023 | 2.64 | 0.00157 | St8sia2/St8sia4/Agk/Sptlc3/Cers4/Plpp3/St6galnac4/St3gal2/St8sia6/Degs2/St6galnac6/Plpp1/Smpd1 | 13 |
| GO:0008202 | steroid metabolic process | BP | 0.0858 | 0.000287 | 0.00232 | 2.63 | 0.00158 | Apob/Cyp3a59/Cyp1b1/Cyp3a25/Nr5a2/Cyp7b1/Igf1/Hmgcs2/Ednrb/Chst10/Soat1/Dkk3/Kit/Tsku/Apoe/Enpp1/Agtr1a/Kcnma1/Igf2/Snai1/Ebpl/Abca5/Cyp46a1/Abcg1/Cacna1h/Stat5b/Smpd1/Lipe/Sult1a1 | 29 |
| GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | BP | 0.333 | 0.000287 | 0.00232 | 2.63 | 0.00158 | Sfrp2/Nkd1/Gpc3/Znrf3/Dact1 | 5 |
| GO:0055067 | monovalent inorganic cation homeostasis | BP | 0.107 | 0.000298 | 0.0024 | 2.62 | 0.00164 | Slc1a3/Car4/Slc12a8/Slc8a1/Ednrb/Lrrk2/Slc9a7/Dmxl2/Agtr1a/Kcnma1/Slc9a5/Ednra/Lacc1/Aqp11/Slc4a3/Slc12a4/Atp1a2/Slc4a8 | 18 |
| GO:0072678 | T cell migration | BP | 0.159 | 0.000301 | 0.00243 | 2.62 | 0.00165 | Itga4/Itgb7/Icam1/Cxcr3/Cxcl12/Msn/Itgal/Il27ra/Cd200/Xcl1 | 10 |
| GO:0035418 | protein localization to synapse | BP | 0.138 | 0.000304 | 0.00244 | 2.61 | 0.00166 | Gpc6/Nlgn1/Stx1b/Nptx1/Slitrk3/Dlg2/Nlgn3/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 12 |
| GO:0055001 | muscle cell development | BP | 0.0965 | 0.000306 | 0.00246 | 2.61 | 0.00167 | Sgcd/Col14a1/Ryr1/Igf1/Adra1a/Slc8a1/P2rx2/Tmod2/Mef2c/Ank2/Tbx3/Ednra/Bin1/Hdac9/Dysf/Flnc/Selenon/Nfatc4/Pi16/Efemp2/Sgcb/Cacnb4 | 22 |
| GO:0032715 | negative regulation of interleukin-6 production | BP | 0.173 | 0.000308 | 0.00246 | 2.61 | 0.00168 | Hgf/Flt3/Il27ra/Havcr2/Cd200/Nckap1l/Arrb1/Cd84/Arrb2 | 9 |
| GO:1902622 | regulation of neutrophil migration | BP | 0.173 | 0.000308 | 0.00246 | 2.61 | 0.00168 | Mdk/Jam3/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Ptger3/Mpp1 | 9 |
| GO:0045669 | positive regulation of osteoblast differentiation | BP | 0.147 | 0.000312 | 0.00249 | 2.6 | 0.0017 | Sfrp2/Igf1/Ddr2/Npnt/Fermt2/Mef2c/Prkd1/Gli3/Nell1/Gdf10/Lrp3 | 11 |
| GO:0015812 | gamma-aminobutyric acid transport | BP | 0.261 | 0.000313 | 0.00249 | 2.6 | 0.0017 | Grik1/Abat/Htr1b/Gabbr1/Cacna1a/Cacnb4 | 6 |
| GO:0050961 | detection of temperature stimulus involved in sensory perception | BP | 0.261 | 0.000313 | 0.00249 | 2.6 | 0.0017 | Ano1/Cxcl12/Calca/Grik2/Htr2a/Arrb2 | 6 |
| GO:0032092 | positive regulation of protein binding | BP | 0.13 | 0.000314 | 0.0025 | 2.6 | 0.0017 | Tert/Lrrk2/Plxnd1/Lfng/Apoe/Caprin2/Arrb1/Hipk2/Pkd1/Cldn5/Mfng/Cav1/Dact1 | 13 |
| GO:0051100 | negative regulation of binding | BP | 0.107 | 0.000321 | 0.00254 | 2.59 | 0.00173 | Map2/Lef1/Itga4/Nog/Zfp462/Lrrk2/Ccm2l/Twist2/Dysf/Arhgap28/Ttbk2/Nfatc4/Arrb2/Habp4/Nes/Cav1/Dact1/Zfp90 | 18 |
| GO:0071248 | cellular response to metal ion | BP | 0.107 | 0.000321 | 0.00254 | 2.59 | 0.00173 | Slc13a5/Nlgn1/Syt13/Cpne5/Fn1/Lrrk2/Nptx1/Mef2c/Rasgrp2/Dlg2/Adcy7/Syt7/Asph/Nfatc4/Pkd2/Slc25a23/Dlg4/Smpd1 | 18 |
| GO:0010875 | positive regulation of cholesterol efflux | BP | 0.219 | 0.000323 | 0.00256 | 2.59 | 0.00174 | Pltp/Apoe/Ptch1/Abcg1/Abca8b/Cav1/Nr1h3 | 7 |
| GO:0023058 | adaptation of signaling pathway | BP | 0.219 | 0.000323 | 0.00256 | 2.59 | 0.00174 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln | 7 |
| GO:0007219 | Notch signaling pathway | BP | 0.103 | 0.000329 | 0.0026 | 2.58 | 0.00177 | Tbx2/Aph1b/Cdh6/Ptp4a3/Zfp423/Postn/Jag2/Lfng/Kit/Hoxd3/Heyl/Snai1/Arrb1/Dtx3/Tspan5/Fat4/Egfl7/Mfng/Bend6 | 19 |
| GO:0014046 | dopamine secretion | BP | 0.19 | 0.000335 | 0.00264 | 2.58 | 0.0018 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 8 |
| GO:0014059 | regulation of dopamine secretion | BP | 0.19 | 0.000335 | 0.00264 | 2.58 | 0.0018 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 8 |
| GO:0003281 | ventricular septum development | BP | 0.136 | 0.000338 | 0.00266 | 2.57 | 0.00181 | Stra6/Wnt11/Nog/Nos3/Lrp2/Prdm1/Aplnr/Cplane1/Tbx3/Heyl/Heg1/Fzd2 | 12 |
| GO:0051250 | negative regulation of lymphocyte activation | BP | 0.106 | 0.000345 | 0.0027 | 2.57 | 0.00184 | Ihh/Mdk/Cd44/Gli3/Flt3/Prdm1/Loxl3/Btla/H2-Ab1/Havcr2/Nckap1l/Pde5a/Fgr/H2-Aa/Gimap5/Dlg5/Gimap3/Clnk | 18 |
| GO:1904064 | positive regulation of cation transmembrane transport | BP | 0.106 | 0.000345 | 0.0027 | 2.57 | 0.00184 | Cacna1c/Drd4/Cacna2d1/Cxcr3/Edn3/Ank2/Plcb1/Thy1/Nlgn3/Aplnr/Cracr2a/Agtr1a/Ednra/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 18 |
| GO:0046638 | positive regulation of alpha-beta T cell differentiation | BP | 0.17 | 0.000357 | 0.00279 | 2.55 | 0.0019 | Ihh/Cd83/Gli3/Cd1d1/Nckap1l/Gimap5/Zap70/Gimap3/Il2rg | 9 |
| GO:0002366 | leukocyte activation involved in immune response | BP | 0.0856 | 0.000373 | 0.00292 | 2.54 | 0.00198 | Lef1/Mdk/Itm2a/Foxf1/Icam1/Dock2/Eif2ak4/Dock10/Lfng/Loxl3/Coro1a/Kit/Itgal/Il27ra/Cracr2a/Enpp1/Havcr2/Irf8/Nckap1l/Dysf/Myo1f/Cd84/Fgr/Mfng/Lat/Rab27a/Apbb1ip/Clnk | 28 |
| GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | BP | 0.122 | 0.000374 | 0.00292 | 2.54 | 0.00198 | Nlgn1/Ccdc88a/Nrp1/Abi2/Kit/Rp1/Kctd17/Eps8l1/Crocc/Ttbk2/Dzip1/Palm/Dpysl3/Cdc42ep5 | 14 |
| GO:0045619 | regulation of lymphocyte differentiation | BP | 0.0995 | 0.000379 | 0.00295 | 2.53 | 0.00201 | Ihh/Lef1/Mdk/Cd83/Cd44/Pik3r6/Gli3/Flt3/Zeb1/Prdm1/Cd1d1/Loxl3/Sox12/Nckap1l/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il2rg | 20 |
| GO:0002687 | positive regulation of leukocyte migration | BP | 0.108 | 0.000383 | 0.00298 | 2.53 | 0.00203 | Mdk/Itga4/Icam1/Cxcl12/Jam3/Edn3/Thy1/Vegfc/Sele/Ednra/Pecam1/Xcl1/Nckap1l/Kitl/Dysf/Ptger3/Ptn | 17 |
| GO:0046632 | alpha-beta T cell differentiation | BP | 0.116 | 0.000388 | 0.00301 | 2.52 | 0.00205 | Ihh/Lef1/Cd83/Itk/Gli3/Prdm1/Cd1d1/Loxl3/Cracr2a/Nckap1l/Satb1/Gimap5/Zap70/Gimap3/Il2rg | 15 |
| GO:0009187 | cyclic nucleotide metabolic process | BP | 0.212 | 0.000395 | 0.00305 | 2.52 | 0.00207 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7/Pde5a/Cacnb4 | 7 |
| GO:0048566 | embryonic digestive tract development | BP | 0.212 | 0.000395 | 0.00305 | 2.52 | 0.00207 | Stra6/Ihh/Pkdcc/Foxf1/Gli3/Tcf21/Pcsk5 | 7 |
| GO:0048841 | regulation of axon extension involved in axon guidance | BP | 0.212 | 0.000395 | 0.00305 | 2.52 | 0.00207 | Sema5a/Sema5b/Sema7a/Nrp1/Cxcl12/Sema3g/Sema6a | 7 |
| GO:0060740 | prostate gland epithelium morphogenesis | BP | 0.212 | 0.000395 | 0.00305 | 2.52 | 0.00207 | Ar/Nog/Cyp7b1/Igf1/Cd44/Mmp2/Id4 | 7 |
| GO:0086002 | cardiac muscle cell action potential involved in contraction | BP | 0.186 | 0.000397 | 0.00306 | 2.51 | 0.00208 | Cacna1c/Kcnd3/Scn4b/Scn1a/Cacna2d1/Ank2/Bin1/Cav1 | 8 |
| GO:0002693 | positive regulation of cellular extravasation | BP | 0.25 | 0.000402 | 0.00309 | 2.51 | 0.0021 | Mdk/Icam1/Jam3/Thy1/Pecam1/Ptger3 | 6 |
| GO:0010842 | retina layer formation | BP | 0.25 | 0.000402 | 0.00309 | 2.51 | 0.0021 | Sdk2/Arl6/Obsl1/Ptprm/Hipk2/Fat3 | 6 |
| GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | BP | 0.25 | 0.000402 | 0.00309 | 2.51 | 0.0021 | Ptp4a3/Jcad/Tcf4/Atp2b4/Sema6a/Adgra2 | 6 |
| GO:0060046 | regulation of acrosome reaction | BP | 0.312 | 0.000403 | 0.00309 | 2.51 | 0.0021 | Pla2g10/Spink1/Plcb1/Cacna1h/Pkdrej | 5 |
| GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | BP | 0.312 | 0.000403 | 0.00309 | 2.51 | 0.0021 | Plxnb1/Nrp1/Mef2c/Plxnd1/Ednra | 5 |
| GO:0001818 | negative regulation of cytokine production | BP | 0.0878 | 0.000404 | 0.00309 | 2.51 | 0.0021 | Hgf/Acod1/Angpt1/Lef1/Wnt11/Pla2g10/Cd83/Igf1/Fn1/Flt3/Adcy7/Il27ra/Tsku/Havcr2/Cd200/Nckap1l/Hdac9/Twist2/Arrb1/Mapkbp1/Cd84/Arrb2/Abcd2/Gimap5/Gimap3/Cd96 | 26 |
| GO:0032330 | regulation of chondrocyte differentiation | BP | 0.167 | 0.000412 | 0.00314 | 2.5 | 0.00214 | Mdk/Hoxd11/Pkdcc/Hoxa11/Gli3/Wnt9a/Adamts7/Ltbp3/Ccn2 | 9 |
| GO:0007009 | plasma membrane organization | BP | 0.108 | 0.000413 | 0.00315 | 2.5 | 0.00214 | Ar/Epb41l3/Ank2/Syt7/Tmeff2/Tie1/Bin1/Dysf/Cavin2/Abcd2/Fat4/Smpd1/Atp10a/Cav1/Xkr5/Stx2/Tlcd2 | 17 |
| GO:0046209 | nitric oxide metabolic process | BP | 0.133 | 0.000418 | 0.00318 | 2.5 | 0.00216 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0021761 | limbic system development | BP | 0.126 | 0.000421 | 0.0032 | 2.5 | 0.00218 | Lef1/Mdk/Zeb2/Nrp1/Dab1/Id4/Gli3/Tsku/Tbx3/Ndnf/Dclk2/Atp1a2/Bbs1 | 13 |
| GO:0002715 | regulation of natural killer cell mediated immunity | BP | 0.152 | 0.000443 | 0.00336 | 2.47 | 0.00229 | Pik3r6/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gimap3/Cd96/Clnk | 10 |
| GO:0002720 | positive regulation of cytokine production involved in immune response | BP | 0.152 | 0.000443 | 0.00336 | 2.47 | 0.00229 | Sema7a/Tek/Gprc5b/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Clnk | 10 |
| GO:0002263 | cell activation involved in immune response | BP | 0.0846 | 0.000453 | 0.00343 | 2.46 | 0.00233 | Lef1/Mdk/Itm2a/Foxf1/Icam1/Dock2/Eif2ak4/Dock10/Lfng/Loxl3/Coro1a/Kit/Itgal/Il27ra/Cracr2a/Enpp1/Havcr2/Irf8/Nckap1l/Dysf/Myo1f/Cd84/Fgr/Mfng/Lat/Rab27a/Apbb1ip/Clnk | 28 |
| GO:0070167 | regulation of biomineral tissue development | BP | 0.125 | 0.000463 | 0.0035 | 2.46 | 0.00238 | Wnt6/Pkdcc/Ddr2/Nos3/Slc8a1/Mef2c/Osr2/Nell1/Mgp/Enpp1/Ltbp3/Osr1/Ptn | 13 |
| GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | BP | 0.182 | 0.000467 | 0.00352 | 2.45 | 0.0024 | Cacna2d1/Ank2/Cracr2a/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 8 |
| GO:0010863 | positive regulation of phospholipase C activity | BP | 0.206 | 0.000479 | 0.0036 | 2.44 | 0.00245 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1 | 7 |
| GO:0060512 | prostate gland morphogenesis | BP | 0.206 | 0.000479 | 0.0036 | 2.44 | 0.00245 | Ar/Nog/Cyp7b1/Igf1/Cd44/Mmp2/Id4 | 7 |
| GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | BP | 0.206 | 0.000479 | 0.0036 | 2.44 | 0.00245 | Drd4/Dysf/Rrad/Pkd2/Gstm7/Atp1a2/Gnb5 | 7 |
| GO:0030833 | regulation of actin filament polymerization | BP | 0.11 | 0.000492 | 0.00369 | 2.43 | 0.00251 | Kank4/Evl/Hcls1/Tmod2/Pfn2/Icam1/Coro1a/Esam/Daam2/Pecam1/Scin/Nckap1l/Bin1/Arhgap28/Cdc42ep5/Cyfip2 | 16 |
| GO:0060350 | endochondral bone morphogenesis | BP | 0.149 | 0.000502 | 0.00376 | 2.43 | 0.00256 | Ihh/Mmp16/Hoxd11/Thbs3/Cbs/Hoxa11/Mef2c/Stc1/Col13a1/Enpp1 | 10 |
| GO:0007620 | copulation | BP | 0.24 | 0.00051 | 0.00381 | 2.42 | 0.00259 | Ar/Abat/Drd4/Ednrb/Serpine2/Ednra | 6 |
| GO:0035988 | chondrocyte proliferation | BP | 0.24 | 0.00051 | 0.00381 | 2.42 | 0.00259 | Ihh/Mmp16/Lef1/Ddr2/Stc1/Ccn2 | 6 |
| GO:1903035 | negative regulation of response to wounding | BP | 0.13 | 0.000512 | 0.00382 | 2.42 | 0.0026 | Mdk/Cd109/Tfpi/Abat/Phldb2/Serpine2/Apoe/Pros1/Rtn4rl1/Neo1/Ptger3/Xylt1 | 12 |
| GO:2001057 | reactive nitrogen species metabolic process | BP | 0.13 | 0.000512 | 0.00382 | 2.42 | 0.0026 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 12 |
| GO:0030100 | regulation of endocytosis | BP | 0.0928 | 0.000521 | 0.00388 | 2.41 | 0.00264 | Angpt1/Nlgn1/Drd4/Gpc3/Lrrk2/Cdh13/Lrp2/Epha3/Apoe/Sele/Syt7/Magi2/Nckap1l/Hip1/Bin1/Arrb1/Arrb2/Apln/Dlg4/Smpd1/Cav1/Nr1h3 | 22 |
| GO:0051648 | vesicle localization | BP | 0.0995 | 0.000526 | 0.00391 | 2.41 | 0.00266 | Syn2/Map2/Itga4/Nlgn1/Sec16b/Cdh2/Lrrk2/Myo5a/Ppfia2/Kif1a/Nlgn3/Lin7a/Magi2/5730455P16Rik/Pcdh17/Myo1f/Cep19/Rab27a/Arfgap3 | 19 |
| GO:0061041 | regulation of wound healing | BP | 0.113 | 0.000538 | 0.00399 | 2.4 | 0.00271 | Cd109/Ddr2/Tfpi/Abat/Phldb2/Fermt2/Serpine2/Plpp3/Apoe/Pros1/Ptger3/Clec7a/St3gal4/Cav1/Cldn1 | 15 |
| GO:0007588 | excretion | BP | 0.161 | 0.000544 | 0.00402 | 2.4 | 0.00274 | Mdk/Cacna1c/Adra1a/Ednrb/Stc1/Abcg2/Kcnma1/Ptger3/Abcg3 | 9 |
| GO:0071709 | membrane assembly | BP | 0.161 | 0.000544 | 0.00402 | 2.4 | 0.00274 | Ptprd/Nlgn1/Cdh2/Nlgn3/Ubxn2b/Magi2/Cav1/Stx2/Tlcd2 | 9 |
| GO:1903539 | protein localization to postsynaptic membrane | BP | 0.161 | 0.000544 | 0.00402 | 2.4 | 0.00274 | Gpc6/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 9 |
| GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | BP | 0.294 | 0.000551 | 0.00406 | 2.39 | 0.00276 | Plxnb1/Nrp1/Mef2c/Plxnd1/Ednra | 5 |
| GO:0006527 | arginine catabolic process | BP | 0.4 | 0.000552 | 0.00406 | 2.39 | 0.00276 | Ddah1/Nos3/Fah/Atp2b4 | 4 |
| GO:0060385 | axonogenesis involved in innervation | BP | 0.4 | 0.000552 | 0.00406 | 2.39 | 0.00276 | Itga4/Nrp1/Nptx1/Ednra | 4 |
| GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.4 | 0.000552 | 0.00406 | 2.39 | 0.00276 | Ppp1r16b/Aplnr/Jcad/Agtr1a | 4 |
| GO:0021953 | central nervous system neuron differentiation | BP | 0.0966 | 0.000553 | 0.00406 | 2.39 | 0.00276 | Map2/Hoxd10/Dcc/Zeb2/Nrp1/Id4/Gli3/Gdf7/D16Ertd472e/Tsku/Cdh11/Dync2h1/Ndnf/Dclk2/Lhx6/Tal1/Nin/Ptch1/Ptger3/Cacna1a | 20 |
| GO:0061337 | cardiac conduction | BP | 0.147 | 0.000566 | 0.00414 | 2.38 | 0.00282 | Cacna1c/Kcnd3/Slc8a1/Scn4b/Cacna2d1/Ank2/Gjc1/Bin1/Slc4a3/Cav1 | 10 |
| GO:0010837 | regulation of keratinocyte proliferation | BP | 0.2 | 0.000577 | 0.00422 | 2.37 | 0.00287 | Mdk/Cd109/Prkd1/Has2/Stxbp4/Klf9/Twist2 | 7 |
| GO:2000177 | regulation of neural precursor cell proliferation | BP | 0.117 | 0.000578 | 0.00422 | 2.37 | 0.00287 | Gli1/Ptprz1/Mdk/Nog/Igf1/Cdh2/Smarca1/Lrrk2/Zfp423/Gli3/Lrp2/Vegfc/Nes/Ptn | 14 |
| GO:0015711 | organic anion transport | BP | 0.0822 | 0.000582 | 0.00424 | 2.37 | 0.00289 | Slc1a3/Car4/Grik1/Pla2g10/Slc13a5/Slc2a3/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Slc36a4/Lrp2/Htr1b/Slc6a15/Bdkrb2/Slc23a2/Sfxn5/Slco4c1/Ptger3/Nfkbie/Slc4a3/Slc38a1/Abcc5/Abcg3/Gabbr1/Slc4a8/Cacna1a/Cacnb4 | 29 |
| GO:0030099 | myeloid cell differentiation | BP | 0.0774 | 0.000598 | 0.00436 | 2.36 | 0.00296 | Lef1/Pla2g10/Cd109/P4htm/Hcls1/Ets1/Gpc3/Mef2c/Inpp4b/Flvcr1/Lrrc17/Calca/Plcb1/Fli1/Kit/Heph/Cdkn1c/Tnfrsf11b/Enpp1/Gpr171/Scin/Zbtb46/Irf8/Nckap1l/Tal1/Hoxa9/Kitl/Tnfaip6/Twist2/Rassf2/Nrros/Stat5b/Gimap5/Gimap3 | 34 |
| GO:0003407 | neural retina development | BP | 0.136 | 0.000613 | 0.00445 | 2.35 | 0.00303 | Ihh/Sdk2/Arl6/Obsl1/Thy1/Ptprm/Rp1/Rorb/Hipk2/Fat3/Ptn | 11 |
| GO:0048863 | stem cell differentiation | BP | 0.0917 | 0.000618 | 0.00448 | 2.35 | 0.00305 | Sema5a/Sema5b/Zeb2/Sema7a/Nrp1/Cdh2/Ednrb/Fn1/Mef2c/Hoxd4/Edn3/Kit/Ednra/Prickle1/Sema3g/Tal1/Kitl/Ltbp3/Osr1/Pus7/Sema6a/Ptn | 22 |
| GO:2000116 | regulation of cysteine-type endopeptidase activity | BP | 0.0917 | 0.000618 | 0.00448 | 2.35 | 0.00305 | Hgf/Sfrp2/Igf1/Cd44/Bok/Grin2b/Wdr35/Mical1/Ift57/Wnt9a/Ppm1f/Hip1/Arrb1/Epha7/Asph/Dlc1/Nlrp1b/Arrb2/Ccn2/Fyn/Mgmt/Cyfip2 | 22 |
| GO:0006493 | protein O-linked glycosylation | BP | 0.158 | 0.000622 | 0.00448 | 2.35 | 0.00305 | Gxylt2/Tet1/Tmtc1/Galnt16/Eogt/St8sia6/Galntl6/Poglut2/Large1 | 9 |
| GO:0032964 | collagen biosynthetic process | BP | 0.158 | 0.000622 | 0.00448 | 2.35 | 0.00305 | Ihh/P3h3/Ddr2/Fn1/Emilin1/Rcn3/Arrb2/Ccn2/Cygb | 9 |
| GO:0061383 | trabecula morphogenesis | BP | 0.158 | 0.000622 | 0.00448 | 2.35 | 0.00305 | Nog/Plxnb1/Thbs3/Mmp2/Tek/Adamts1/Ccm2l/Enpp1/Heg1 | 9 |
| GO:0062237 | protein localization to postsynapse | BP | 0.158 | 0.000622 | 0.00448 | 2.35 | 0.00305 | Gpc6/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Dlg4 | 9 |
| GO:0086001 | cardiac muscle cell action potential | BP | 0.158 | 0.000622 | 0.00448 | 2.35 | 0.00305 | Cacna1c/Kcnd2/Kcnd3/Scn4b/Scn1a/Cacna2d1/Ank2/Bin1/Cav1 | 9 |
| GO:0051963 | regulation of synapse assembly | BP | 0.116 | 0.000629 | 0.00452 | 2.34 | 0.00308 | Gpc6/Ptprd/St8sia2/Nlgn1/Lrrc4b/Nptx1/Mef2c/Adgrl1/Slitrk3/Nlgn3/Epha7/Bhlhb9/Cux2/Dlg5 | 14 |
| GO:1903115 | regulation of actin filament-based movement | BP | 0.174 | 0.000638 | 0.00458 | 2.34 | 0.00311 | Cacna1c/Stc1/Ank2/Bin1/Cacna1h/Ptger3/Atp1a2/Cav1 | 8 |
| GO:0001573 | ganglioside metabolic process | BP | 0.231 | 0.00064 | 0.00458 | 2.34 | 0.00311 | St8sia2/St8sia4/St6galnac4/St3gal2/St8sia6/St6galnac6 | 6 |
| GO:0042474 | middle ear morphogenesis | BP | 0.231 | 0.00064 | 0.00458 | 2.34 | 0.00311 | Nog/Osr2/Tshz1/Enpp1/Ednra/Osr1 | 6 |
| GO:2000050 | regulation of non-canonical Wnt signaling pathway | BP | 0.231 | 0.00064 | 0.00458 | 2.34 | 0.00311 | Sfrp2/Nkd1/Gpc3/Znrf3/Daam2/Dact1 | 6 |
| GO:0042102 | positive regulation of T cell proliferation | BP | 0.12 | 0.000666 | 0.00476 | 2.32 | 0.00324 | Igf1/Tnfsf13b/Cd1d1/Coro1a/Itgal/Il27ra/Havcr2/Igf2/Xcl1/Nckap1l/Stat5b/Zap70/Il12rb1 | 13 |
| GO:0014706 | striated muscle tissue development | BP | 0.0862 | 0.000679 | 0.00484 | 2.31 | 0.0033 | Gli1/Col11a1/AW551984/Angpt1/Lef1/Sgcd/Col14a1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/Mef2c/Lrp2/Ccm2l/Nox4/Tbx3/Gjc1/Ednra/Myl3/Hdac9/Heg1/Arrb2/Pi16/Sgcb | 25 |
| GO:0070665 | positive regulation of leukocyte proliferation | BP | 0.103 | 0.00068 | 0.00484 | 2.31 | 0.0033 | Igf1/Mef2c/Tnfsf13b/Cd1d1/Coro1a/Itgal/Il27ra/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Kitl/Csf2rb/Stat5b/Zap70/Il12rb1 | 17 |
| GO:0060042 | retina morphogenesis in camera-type eye | BP | 0.134 | 0.000681 | 0.00484 | 2.31 | 0.0033 | Ihh/Sdk2/Arl6/Obsl1/Thy1/Ptprm/Rp1/Rorb/Hipk2/Fat3/Ptn | 11 |
| GO:1901879 | regulation of protein depolymerization | BP | 0.126 | 0.000687 | 0.00488 | 2.31 | 0.00332 | Map2/Sema5a/Evl/Tmod2/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2 | 12 |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | BP | 0.194 | 0.000691 | 0.0049 | 2.31 | 0.00334 | Grik1/Kdr/Grin2b/Kit/Grik2/Dlg4/Syngap1 | 7 |
| GO:0051251 | positive regulation of lymphocyte activation | BP | 0.0767 | 0.000701 | 0.00497 | 2.3 | 0.00338 | Ihh/Efnb3/Lef1/Mdk/Cd83/Igf1/Mef2c/Pik3r6/Gli3/Tnfsf13b/Thy1/Prdm1/Cd1d1/Coro1a/Itgal/Il27ra/H2-Ab1/Cd38/Havcr2/Sox12/H2-Ob/Igf2/Xcl1/Nckap1l/H2-Eb1/Clec7a/H2-Aa/Stat5b/Gimap5/Zap70/Gimap3/Il12rb1/Cav1/Il2rg | 34 |
| GO:0015872 | dopamine transport | BP | 0.155 | 0.000709 | 0.00502 | 2.3 | 0.00342 | Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Gabbr1 | 9 |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | BP | 0.143 | 0.000715 | 0.00506 | 2.3 | 0.00344 | Vipr2/Nos3/Drd4/Npr3/Mef2c/Rgs5/Apoe/Apln/Efemp2/Cav1 | 10 |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | BP | 0.119 | 0.000728 | 0.00514 | 2.29 | 0.0035 | Sfrp2/Slc8a1/Mapk8ip1/Slc8a3/Pkia/Cdkn1c/Apoe/Rasip1/Pkib/Heg1/Lats2/Smpd1/Cav1 | 13 |
| GO:0007614 | short-term memory | BP | 0.278 | 0.000737 | 0.0052 | 2.28 | 0.00353 | Mdk/Aph1b/Drd4/Pde5a/Cux2 | 5 |
| GO:0043114 | regulation of vascular permeability | BP | 0.17 | 0.000741 | 0.00522 | 2.28 | 0.00355 | Angpt1/Ddah1/Fermt2/Ptp4a3/Akap12/Cdh5/Apoe/Cldn5 | 8 |
| GO:0030595 | leukocyte chemotaxis | BP | 0.0921 | 0.000764 | 0.00537 | 2.27 | 0.00365 | Cxcl15/Mdk/Cyp7b1/Ptpro/Ednrb/Cxcr3/Cxcl12/Jam3/Edn3/Itga1/Calca/Coro1a/Kit/Vegfc/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Mpp1/Ptn | 21 |
| GO:0046660 | female sex differentiation | BP | 0.105 | 0.000766 | 0.00538 | 2.27 | 0.00366 | Stra6/Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lrp2/Lfng/Kit/Tbx3/Kitl/Tnfaip6/Arrb1/Arrb2/Stat5b | 16 |
| GO:0014829 | vascular associated smooth muscle contraction | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Slc8a1/Ednrb/Edn3/Cd38/Ednra/Htr2a | 6 |
| GO:0016048 | detection of temperature stimulus | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Ano1/Cxcl12/Calca/Grik2/Htr2a/Arrb2 | 6 |
| GO:0034368 | protein-lipid complex remodeling | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:0034369 | plasma lipoprotein particle remodeling | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:0048843 | negative regulation of axon extension involved in axon guidance | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Sema5a/Sema5b/Sema7a/Nrp1/Sema3g/Sema6a | 6 |
| GO:0060004 | reflex | BP | 0.222 | 0.000793 | 0.00553 | 2.26 | 0.00376 | Adra1a/Npnt/Glrb/Kcnma1/Satb1/Cacna1a | 6 |
| GO:0050432 | catecholamine secretion | BP | 0.141 | 0.000802 | 0.00558 | 2.25 | 0.00379 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 10 |
| GO:0043254 | regulation of protein-containing complex assembly | BP | 0.0769 | 0.000802 | 0.00558 | 2.25 | 0.00379 | Kank4/Map2/Evl/Hcls1/Stx1b/Fermt2/Tmod2/Pfn2/Icam1/Jam3/Cdh5/Msn/Coro1a/Esam/Tubb4a/Apoe/Prrt2/Rasip1/Daam2/Pecam1/Scin/Pcsk5/Nckap1l/Slain1/Tal1/Nin/Bin1/Arhgap28/Cdc42ep5/Cav1/Dact1/Cyfip2/Tppp | 33 |
| GO:1903350 | response to dopamine | BP | 0.153 | 0.000806 | 0.00559 | 2.25 | 0.0038 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Gnb5 | 9 |
| GO:1903351 | cellular response to dopamine | BP | 0.153 | 0.000806 | 0.00559 | 2.25 | 0.0038 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Palm/Gnb5 | 9 |
| GO:0048846 | axon extension involved in axon guidance | BP | 0.189 | 0.000821 | 0.00565 | 2.25 | 0.00384 | Sema5a/Sema5b/Sema7a/Nrp1/Cxcl12/Sema3g/Sema6a | 7 |
| GO:0050901 | leukocyte tethering or rolling | BP | 0.189 | 0.000821 | 0.00565 | 2.25 | 0.00384 | Itga4/Chst2/Itgb7/Podxl2/Cxcl12/Sele/St3gal4 | 7 |
| GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | BP | 0.189 | 0.000821 | 0.00565 | 2.25 | 0.00384 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Fzd2/Dact1 | 7 |
| GO:0086091 | regulation of heart rate by cardiac conduction | BP | 0.189 | 0.000821 | 0.00565 | 2.25 | 0.00384 | Cacna1c/Kcnd3/Scn4b/Cacna2d1/Ank2/Bin1/Cav1 | 7 |
| GO:1902284 | neuron projection extension involved in neuron projection guidance | BP | 0.189 | 0.000821 | 0.00565 | 2.25 | 0.00384 | Sema5a/Sema5b/Sema7a/Nrp1/Cxcl12/Sema3g/Sema6a | 7 |
| GO:0002028 | regulation of sodium ion transport | BP | 0.124 | 0.000829 | 0.0057 | 2.24 | 0.00387 | Hecw2/Nos3/Slc8a1/Scn4b/Drd4/Fxyd5/Serpine2/Plcb1/Osr1/Atp2b4/Akt3/Atp1a2 | 12 |
| GO:0060191 | regulation of lipase activity | BP | 0.131 | 0.000836 | 0.0057 | 2.24 | 0.00388 | Adra1a/Itk/Arhgap6/Sele/Agtr1a/Adcyap1r1/Htr2a/Hdac9/Dlc1/Angptl4/Nr1h3 | 11 |
| GO:0072089 | stem cell proliferation | BP | 0.131 | 0.000836 | 0.0057 | 2.24 | 0.00388 | Sfrp2/Fermt2/Tert/Znrf3/Vegfc/Tbx3/Kitl/Ltbp3/Septin4/Abcb1a/Nes | 11 |
| GO:0002399 | MHC class II protein complex assembly | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0002503 | peptide antigen assembly with MHC class II protein complex | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0035336 | long-chain fatty-acyl-CoA metabolic process | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | Acsl6/Them5/Acot7/Acsl1 | 4 |
| GO:0061299 | retina vasculature morphogenesis in camera-type eye | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | Ndp/Cyp1b1/Nrp1/Arhgef15 | 4 |
| GO:0071415 | cellular response to purine-containing compound | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | Ryr1/Slc8a1/Selenon/Gstm7 | 4 |
| GO:0097104 | postsynaptic membrane assembly | BP | 0.364 | 0.000839 | 0.0057 | 2.24 | 0.00388 | Nlgn1/Cdh2/Nlgn3/Magi2 | 4 |
| GO:0030177 | positive regulation of Wnt signaling pathway | BP | 0.108 | 0.000854 | 0.00579 | 2.24 | 0.00394 | Sfrp2/Sema5a/Lef1/Zeb2/Nkd1/Tert/Gpc3/Lrrk2/Gprc5b/Peg12/Caprin2/Daam2/Adgra2/Cav1/Dact1 | 15 |
| GO:0001941 | postsynaptic membrane organization | BP | 0.167 | 0.000856 | 0.00579 | 2.24 | 0.00394 | Nlgn1/Cdh2/Glrb/Nlgn3/Apoe/Magi2/Dlg4/Ptn | 8 |
| GO:0003179 | heart valve morphogenesis | BP | 0.167 | 0.000856 | 0.00579 | 2.24 | 0.00394 | Stra6/Nos3/Mef2c/Erg/Emilin1/Tie1/Heyl/Dchs1 | 8 |
| GO:0035137 | hindlimb morphogenesis | BP | 0.167 | 0.000856 | 0.00579 | 2.24 | 0.00394 | Hoxd10/Aff3/Gpc3/Hoxd9/Osr2/Tbx3/Ptch1/Osr1 | 8 |
| GO:0042073 | intraciliary transport | BP | 0.167 | 0.000856 | 0.00579 | 2.24 | 0.00394 | Tub/Wdr35/Ift57/Dync2h1/Arl3/Ttc26/Dync2li1/Bbs1 | 8 |
| GO:0010876 | lipid localization | BP | 0.0757 | 0.000885 | 0.00597 | 2.22 | 0.00406 | Apob/Stra6/Nme4/Acsl6/Pla2g10/Acacb/Drd4/Vstm2a/Mest/Soat1/Abcg2/Rbp7/Osbp2/Pltp/Selenom/Apoe/Enpp1/Agtr1a/Syt7/Bdkrb2/Abca5/Dysf/Abcg1/Abca8b/Abcd2/Stat5b/Abcb1a/Atp10a/Acsl1/Osbpl1a/Cav1/Xkr5/Nr1h3/Prelid3a | 34 |
| GO:0030041 | actin filament polymerization | BP | 0.101 | 0.000891 | 0.00601 | 2.22 | 0.00409 | Kank4/Evl/Hcls1/Tmod2/Pfn2/Icam1/Catip/Coro1a/Esam/Daam2/Pecam1/Scin/Nckap1l/Bin1/Arhgap28/Cdc42ep5/Cyfip2 | 17 |
| GO:0061035 | regulation of cartilage development | BP | 0.139 | 0.000896 | 0.00602 | 2.22 | 0.00409 | Mdk/Hoxd11/Nog/Pkdcc/Hoxa11/Gli3/Wnt9a/Adamts7/Ltbp3/Ccn2 | 10 |
| GO:0071868 | cellular response to monoamine stimulus | BP | 0.139 | 0.000896 | 0.00602 | 2.22 | 0.00409 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Atp2b4/Palm/Gnb5 | 10 |
| GO:0071870 | cellular response to catecholamine stimulus | BP | 0.139 | 0.000896 | 0.00602 | 2.22 | 0.00409 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Atp2b4/Palm/Gnb5 | 10 |
| GO:0032091 | negative regulation of protein binding | BP | 0.122 | 0.000909 | 0.00608 | 2.22 | 0.00414 | Map2/Itga4/Nog/Lrrk2/Ccm2l/Dysf/Ttbk2/Nfatc4/Arrb2/Nes/Cav1/Dact1 | 12 |
| GO:0042632 | cholesterol homeostasis | BP | 0.122 | 0.000909 | 0.00608 | 2.22 | 0.00414 | Apob/Pla2g10/Nr5a2/Cyp7b1/Soat1/Tsku/Apoe/Hdac9/Abca5/Abcg1/Cav1/Nr1h3 | 12 |
| GO:1904063 | negative regulation of cation transmembrane transport | BP | 0.122 | 0.000909 | 0.00608 | 2.22 | 0.00414 | Hecw2/Drd4/Bin1/Dysf/Osr1/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1 | 12 |
| GO:0097479 | synaptic vesicle localization | BP | 0.15 | 0.000913 | 0.0061 | 2.21 | 0.00415 | Syn2/Map2/Nlgn1/Cdh2/Lrrk2/Nlgn3/Lin7a/Magi2/Pcdh17 | 9 |
| GO:0034763 | negative regulation of transmembrane transport | BP | 0.107 | 0.00092 | 0.00614 | 2.21 | 0.00418 | Hecw2/Drd4/Sh2b2/Enpp1/Bin1/Dysf/Osr1/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1/Pid1 | 15 |
| GO:1905475 | regulation of protein localization to membrane | BP | 0.095 | 0.000921 | 0.00614 | 2.21 | 0.00418 | Gpc6/Ar/Pkdcc/Tcaf1/Cdh2/Gpc3/Abi3/Myo5a/Epha3/Magi2/Tnfaip6/Stac2/Fyn/Dlg4/Atp2b4/Ptn/Gpc2/Pid1/Nkd2 | 19 |
| GO:0030500 | regulation of bone mineralization | BP | 0.129 | 0.000924 | 0.00615 | 2.21 | 0.00418 | Pkdcc/Ddr2/Slc8a1/Mef2c/Osr2/Nell1/Mgp/Enpp1/Ltbp3/Osr1/Ptn | 11 |
| GO:0046530 | photoreceptor cell differentiation | BP | 0.129 | 0.000924 | 0.00615 | 2.21 | 0.00418 | Ihh/Sdk2/Thy1/Prdm1/Rp1/Rorb/Arl3/Ttc26/Nphp1/Ptn/Bbs1 | 11 |
| GO:0016331 | morphogenesis of embryonic epithelium | BP | 0.0973 | 0.000943 | 0.00626 | 2.2 | 0.00426 | Sfrp2/Wnt6/Ar/Nog/Zeb2/Lrp2/Gdf7/Ift57/Jag2/Cecr2/Vegfc/Prickle1/Ptch1/Ipmk/Osr1/Dlc1/Fzd2/Rab23 | 18 |
| GO:0007565 | female pregnancy | BP | 0.1 | 0.000952 | 0.00632 | 2.2 | 0.0043 | Acod1/Ar/Cbs/Il11ra1/Mmp2/Abcg2/Calca/Syde1/Prdm1/Maged2/Havcr2/Pcsk5/Klf9/Stat5b/Ptn/Stx2/Itgb4 | 17 |
| GO:0086004 | regulation of cardiac muscle cell contraction | BP | 0.184 | 0.00097 | 0.00641 | 2.19 | 0.00436 | Cacna1c/Stc1/Ank2/Bin1/Cacna1h/Atp1a2/Cav1 | 7 |
| GO:1900274 | regulation of phospholipase C activity | BP | 0.184 | 0.00097 | 0.00641 | 2.19 | 0.00436 | Adra1a/Itk/Arhgap6/Sele/Adcyap1r1/Htr2a/Dlc1 | 7 |
| GO:0018126 | protein hydroxylation | BP | 0.214 | 0.000974 | 0.00641 | 2.19 | 0.00436 | P3h3/P4htm/Fkbp10/Asph/P3h1/Crtap | 6 |
| GO:0045061 | thymic T cell selection | BP | 0.214 | 0.000974 | 0.00641 | 2.19 | 0.00436 | Cd3g/Gli3/Dock2/Cd1d1/Jag2/Zap70 | 6 |
| GO:0061437 | renal system vasculature development | BP | 0.214 | 0.000974 | 0.00641 | 2.19 | 0.00436 | Angpt1/Nrp1/Tek/Tcf21/Ednra/Osr1 | 6 |
| GO:0061440 | kidney vasculature development | BP | 0.214 | 0.000974 | 0.00641 | 2.19 | 0.00436 | Angpt1/Nrp1/Tek/Tcf21/Ednra/Osr1 | 6 |
| GO:0055092 | sterol homeostasis | BP | 0.121 | 0.000995 | 0.00654 | 2.18 | 0.00445 | Apob/Pla2g10/Nr5a2/Cyp7b1/Soat1/Tsku/Apoe/Hdac9/Abca5/Abcg1/Cav1/Nr1h3 | 12 |
| GO:0008542 | visual learning | BP | 0.137 | 0.000999 | 0.00656 | 2.18 | 0.00446 | Cacna1c/Nog/B4galt2/Pde1b/Cacna1e/Kit/Nlgn3/Atp1a2/Syngap1/Ddhd2 | 10 |
| GO:0048678 | response to axon injury | BP | 0.137 | 0.000999 | 0.00656 | 2.18 | 0.00446 | Gap43/Mmp2/Matn2/Enpp1/Rtn4rl1/Sarm1/Neo1/Xylt1/Dpysl3/Ptn | 10 |
| GO:0060324 | face development | BP | 0.148 | 0.00103 | 0.00676 | 2.17 | 0.0046 | Stra6/Lef1/Itga4/Nog/Crispld1/Mmp2/Ednra/Twist2/Asph | 9 |
| GO:0030856 | regulation of epithelial cell differentiation | BP | 0.102 | 0.00108 | 0.0071 | 2.15 | 0.00483 | Cd109/Macroh2a2/Ptch2/Zeb2/Plcb1/Cdh5/Zeb1/Cdkn1c/Tbx3/Spred3/Ptch1/Osr1/Cldn5/Stat5b/Fat4/Cav1 | 16 |
| GO:0071867 | response to monoamine | BP | 0.135 | 0.00111 | 0.00726 | 2.14 | 0.00494 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Atp2b4/Palm/Gnb5 | 10 |
| GO:0071869 | response to catecholamine | BP | 0.135 | 0.00111 | 0.00726 | 2.14 | 0.00494 | Nsg2/Drd4/Lrrk2/Htr1b/Htr2a/Nsg1/Gnaz/Atp2b4/Palm/Gnb5 | 10 |
| GO:0050727 | regulation of inflammatory response | BP | 0.0798 | 0.00112 | 0.00732 | 2.14 | 0.00498 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Sema7a/Ets1/Ednrb/Lrrk2/Foxf1/Slc7a2/Fndc4/Gprc5b/Cdh5/Bcl6b/Apoe/Cd200/Ednra/Lacc1/Xcl1/Tnfaip6/Pde5a/Nlrp1b/Ptger3/Calcrl/Stat5b/Nr1h3 | 28 |
| GO:0042267 | natural killer cell mediated cytotoxicity | BP | 0.126 | 0.00112 | 0.00732 | 2.14 | 0.00498 | Pik3r6/Coro1a/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gzmb/Gimap5/Gimap3/Rab27a | 11 |
| GO:0031281 | positive regulation of cyclase activity | BP | 0.179 | 0.00114 | 0.00739 | 2.13 | 0.00502 | Cacna1c/Vipr2/Nos3/Adcy3/Calca/Adcy4/Adcy7 | 7 |
| GO:0051180 | vitamin transport | BP | 0.179 | 0.00114 | 0.00739 | 2.13 | 0.00502 | Stra6/Slc2a3/Abcg2/Lrp2/Slc23a2/Abcc5/Abcg3 | 7 |
| GO:1904645 | response to amyloid-beta | BP | 0.179 | 0.00114 | 0.00739 | 2.13 | 0.00502 | Itga4/Igf1/Cacna2d1/Mmp2/Icam1/Fyn/Cacna1a | 7 |
| GO:0050680 | negative regulation of epithelial cell proliferation | BP | 0.0983 | 0.00116 | 0.00748 | 2.13 | 0.00509 | Sfrp2/Ar/Cd109/Gkn3/Gpc3/Mef2c/Cxcr3/Vash1/Ift57/Ptprm/Cdkn1c/Apoe/Klf9/Ptch1/Aqp11/Ptn/Cav1 | 17 |
| GO:0051058 | negative regulation of small GTPase mediated signal transduction | BP | 0.145 | 0.00116 | 0.00751 | 2.12 | 0.00511 | Adra1a/Rasa3/Arhgap25/Sh3bp1/Rasip1/Arap3/Heg1/Dlc1/Syngap1 | 9 |
| GO:0048639 | positive regulation of developmental growth | BP | 0.0909 | 0.00118 | 0.00763 | 2.12 | 0.00519 | Gli1/Sema5a/Igf1/Tbx2/Cpne5/Acacb/Sema7a/Nrp1/Fn1/Mef2c/Cxcl12/Hopx/Plcb1/Flt3/Apoe/Igf2/Slc23a2/Foxs1/Cacng7/Stat5b | 20 |
| GO:0034367 | protein-containing complex remodeling | BP | 0.207 | 0.00118 | 0.00763 | 2.12 | 0.00519 | Apob/Pla2g10/Pltp/Apoe/Abca5/Abcg1 | 6 |
| GO:1900027 | regulation of ruffle assembly | BP | 0.207 | 0.00118 | 0.00763 | 2.12 | 0.00519 | Evl/Nlgn1/Pfn2/Icam1/Eps8l1/Cav1 | 6 |
| GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | BP | 0.119 | 0.00119 | 0.00764 | 2.12 | 0.0052 | Adra1a/Npr3/Ednrb/Ano1/Lpar4/Calca/Pth1r/Agtr1a/Ednra/Htr2a/Plcb2/Ptger3 | 12 |
| GO:0002367 | cytokine production involved in immune response | BP | 0.113 | 0.0012 | 0.00774 | 2.11 | 0.00527 | Angpt1/Sema7a/Tek/Gprc5b/Tril/Kit/Cd226/Lacc1/Xcl1/Gimap5/Gimap3/Cd96/Clnk | 13 |
| GO:0042135 | neurotransmitter catabolic process | BP | 0.333 | 0.00122 | 0.00778 | 2.11 | 0.00529 | Maob/Abat/Maoa/Naalad2 | 4 |
| GO:0060073 | micturition | BP | 0.333 | 0.00122 | 0.00778 | 2.11 | 0.00529 | Cacna1c/Adra1a/Kcnma1/Ptger3 | 4 |
| GO:0070444 | oligodendrocyte progenitor proliferation | BP | 0.333 | 0.00122 | 0.00778 | 2.11 | 0.00529 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0070445 | regulation of oligodendrocyte progenitor proliferation | BP | 0.333 | 0.00122 | 0.00778 | 2.11 | 0.00529 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0050728 | negative regulation of inflammatory response | BP | 0.104 | 0.00123 | 0.00784 | 2.11 | 0.00534 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Ets1/Foxf1/Fndc4/Cdh5/Apoe/Cd200/Tnfaip6/Calcrl/Nr1h3 | 15 |
| GO:0051937 | catecholamine transport | BP | 0.125 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 11 |
| GO:0032103 | positive regulation of response to external stimulus | BP | 0.0742 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Acod1/Sema5a/Mdk/Kdr/Nrp1/Ets1/Fn1/Lrrk2/Mmp2/Prkd1/Cxcr3/Cxcl12/Cdh13/Edn3/Gprc5b/Rab34/Cd226/Ppm1f/Vegfc/Havcr2/Emilin1/Ednra/Xcl1/Masp1/Nckap1l/Dysf/Pde5a/Nlrp1b/Ptger3/Stat5b/Gimap5/Gimap3/Ptn/Clnk | 34 |
| GO:0045019 | negative regulation of nitric oxide biosynthetic process | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Wdr35/Atp2b4/Gimap5/Gimap3/Cav1 | 5 |
| GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Ano1/Cxcl12/Calca/Htr2a/Arrb2 | 5 |
| GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Scn1a/Grin2b/Cxcl12/Htr2a/Fyn | 5 |
| GO:1903236 | regulation of leukocyte tethering or rolling | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Itga4/Chst2/Cxcl12/Sele/St3gal4 | 5 |
| GO:1904406 | negative regulation of nitric oxide metabolic process | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Wdr35/Atp2b4/Gimap5/Gimap3/Cav1 | 5 |
| GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | BP | 0.25 | 0.00124 | 0.00786 | 2.1 | 0.00535 | Ddr2/Tert/Myo5a/Adamts1/Pcsk5 | 5 |
| GO:0048738 | cardiac muscle tissue development | BP | 0.0852 | 0.00128 | 0.00807 | 2.09 | 0.00549 | Gli1/Col11a1/AW551984/Angpt1/Sgcd/Col14a1/Nog/Igf1/Tbx2/Adra1a/Slc8a1/Mef2c/Lrp2/Ccm2l/Nox4/Tbx3/Gjc1/Ednra/Myl3/Heg1/Arrb2/Pi16/Sgcb | 23 |
| GO:1990266 | neutrophil migration | BP | 0.108 | 0.00128 | 0.00807 | 2.09 | 0.00549 | Cxcl15/Mdk/Jam3/Edn3/Itga1/Ednra/Pecam1/Xcl1/Nckap1l/Tnfaip6/Dysf/Irak4/Ptger3/Mpp1 | 14 |
| GO:0008038 | neuron recognition | BP | 0.157 | 0.00129 | 0.00814 | 2.09 | 0.00553 | Gap43/Prtg/Ptprz1/Efnb3/Sema5a/Nrp1/Ndn/Epha3 | 8 |
| GO:0010518 | positive regulation of phospholipase activity | BP | 0.157 | 0.00129 | 0.00814 | 2.09 | 0.00553 | Adra1a/Itk/Arhgap6/Sele/Agtr1a/Adcyap1r1/Htr2a/Dlc1 | 8 |
| GO:0009064 | glutamine family amino acid metabolic process | BP | 0.143 | 0.0013 | 0.0082 | 2.09 | 0.00558 | Ddah1/Nos3/Asrgl1/Fah/Amdhd1/Phgdh/Atp2b4/Asl/Slc7a7 | 9 |
| GO:0044091 | membrane biogenesis | BP | 0.143 | 0.0013 | 0.0082 | 2.09 | 0.00558 | Ptprd/Nlgn1/Cdh2/Nlgn3/Ubxn2b/Magi2/Cav1/Stx2/Tlcd2 | 9 |
| GO:0045637 | regulation of myeloid cell differentiation | BP | 0.0901 | 0.00132 | 0.00827 | 2.08 | 0.00563 | Lef1/P4htm/Hcls1/Ets1/Mef2c/Inpp4b/Lrrc17/Calca/Tnfrsf11b/Gpr171/Scin/Zbtb46/Nckap1l/Tal1/Hoxa9/Kitl/Tnfaip6/Twist2/Rassf2/Stat5b | 20 |
| GO:1903828 | negative regulation of protein localization | BP | 0.0901 | 0.00132 | 0.00827 | 2.08 | 0.00563 | Angpt1/Pkdcc/Drd4/Lrrk2/Abi3/Pkia/Ppm1f/Apoe/Ubxn2b/Cd200/Dclk2/Nin/Ttbk2/Neo1/Ptger3/Frmd4a/Rab23/Lats2/Nr1h3/Pid1 | 20 |
| GO:0001782 | B cell homeostasis | BP | 0.175 | 0.00133 | 0.00832 | 2.08 | 0.00566 | Cd44/Mef2c/Sh2b2/Tnfsf13b/Dock10/Enpp1/Nckap1l | 7 |
| GO:0051349 | positive regulation of lyase activity | BP | 0.175 | 0.00133 | 0.00832 | 2.08 | 0.00566 | Cacna1c/Vipr2/Nos3/Adcy3/Calca/Adcy4/Adcy7 | 7 |
| GO:0060325 | face morphogenesis | BP | 0.175 | 0.00133 | 0.00832 | 2.08 | 0.00566 | Stra6/Lef1/Nog/Crispld1/Mmp2/Twist2/Asph | 7 |
| GO:0009416 | response to light stimulus | BP | 0.081 | 0.00133 | 0.00833 | 2.08 | 0.00566 | Slc1a3/Sema5a/Cacna1c/Nog/Sema5b/Slc24a4/Mmp2/B4galt2/Pde1b/Cacna1e/Eif2ak4/Fbxl21/Mme/Kit/Rp1/Nlgn3/Arrb1/Nfatc4/Atp1a2/Gnb5/Brsk1/Syngap1/Smpd1/Ddhd2/Usp2/Cacnb4 | 26 |
| GO:0015850 | organic hydroxy compound transport | BP | 0.0863 | 0.00137 | 0.00853 | 2.07 | 0.00581 | Apob/Stra6/Maob/Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Slc2a13/Htr1b/Cadps/Selenom/Apoe/Agtr1a/Syt7/Htr2a/Abca5/Abcg1/Abca8b/Ptger3/Gabbr1/Nr1h3 | 22 |
| GO:0010639 | negative regulation of organelle organization | BP | 0.0777 | 0.00137 | 0.00854 | 2.07 | 0.00581 | Kank4/Hgf/Map2/Fez1/Igf1/Evl/Phldb2/Tmod2/Bok/Lrrk2/Pfn2/Klhl22/Rps6ka2/Cdh5/Coro1a/Rp1/Tubb4a/Arhgap6/Prrt2/Tmeff2/Pecam1/Scin/Dysf/Arhgap28/Ttbk2/Dlc1/Arrb2/Cep97/Plekhh2 | 29 |
| GO:0055088 | lipid homeostasis | BP | 0.0966 | 0.0014 | 0.00868 | 2.06 | 0.0059 | Apob/Pla2g10/Nr5a2/Cyp7b1/Soat1/Tsku/Apoe/Hdac9/Abca5/Rcn3/Zbtb20/Abcg1/Angptl4/Abhd8/Cav1/Tlcd2/Nr1h3 | 17 |
| GO:0001704 | formation of primary germ layer | BP | 0.117 | 0.00141 | 0.00876 | 2.06 | 0.00596 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Fn1/Foxf1/Snai1/Tal1/Col5a2/Ets2/Kif16b | 12 |
| GO:0001892 | embryonic placenta development | BP | 0.111 | 0.00141 | 0.00876 | 2.06 | 0.00596 | Lef1/Itga4/Slc8a1/Vash1/Syde1/Prdm1/Pcdh12/Cdkn1c/Igf2/Snai1/Pkd1/Pkd2/Vash2 | 13 |
| GO:0006688 | glycosphingolipid biosynthetic process | BP | 0.2 | 0.00143 | 0.00882 | 2.05 | 0.006 | St8sia2/St8sia4/St6galnac4/St3gal2/St8sia6/St6galnac6 | 6 |
| GO:0045724 | positive regulation of cilium assembly | BP | 0.2 | 0.00143 | 0.00882 | 2.05 | 0.006 | Ccdc88a/Rp1/Kctd17/Crocc/Ttbk2/Dzip1 | 6 |
| GO:0048745 | smooth muscle tissue development | BP | 0.2 | 0.00143 | 0.00882 | 2.05 | 0.006 | Stra6/Ihh/Tshz3/Osr1/Efemp2/Itga8 | 6 |
| GO:0071312 | cellular response to alkaloid | BP | 0.2 | 0.00143 | 0.00882 | 2.05 | 0.006 | Slc1a3/Ryr1/Slc8a1/Htr1b/Selenon/Gstm7 | 6 |
| GO:0008347 | glial cell migration | BP | 0.141 | 0.00146 | 0.009 | 2.05 | 0.00612 | Ptprz1/Apcdd1/Cspg4/Fn1/Dab1/Gli3/Ndn/Matn2/Enpp1 | 9 |
| GO:0050435 | amyloid-beta metabolic process | BP | 0.141 | 0.00146 | 0.009 | 2.05 | 0.00612 | Igf1/Aph1b/Rtn1/Slc2a13/Mme/Apoe/Bin1/Abcg1/Mgat3 | 9 |
| GO:0060193 | positive regulation of lipase activity | BP | 0.141 | 0.00146 | 0.009 | 2.05 | 0.00612 | Adra1a/Itk/Arhgap6/Sele/Agtr1a/Adcyap1r1/Htr2a/Dlc1/Nr1h3 | 9 |
| GO:0050798 | activated T cell proliferation | BP | 0.154 | 0.00147 | 0.00904 | 2.04 | 0.00615 | Igf1/Itgal/Il27ra/Igf2/Satb1/Fyn/Stat5b/Il12rb1 | 8 |
| GO:0050671 | positive regulation of lymphocyte proliferation | BP | 0.102 | 0.00151 | 0.00927 | 2.03 | 0.00631 | Igf1/Mef2c/Tnfsf13b/Cd1d1/Coro1a/Itgal/Il27ra/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Stat5b/Zap70/Il12rb1 | 15 |
| GO:1903038 | negative regulation of leukocyte cell-cell adhesion | BP | 0.102 | 0.00151 | 0.00927 | 2.03 | 0.00631 | Ihh/Mdk/Cd44/Cxcl12/Gli3/Loxl3/Btla/H2-Ab1/Havcr2/Nckap1l/Pde5a/H2-Aa/Gimap5/Dlg5/Gimap3 | 15 |
| GO:0008585 | female gonad development | BP | 0.11 | 0.00153 | 0.00934 | 2.03 | 0.00636 | Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lfng/Kit/Kitl/Tnfaip6/Arrb1/Arrb2/Stat5b | 13 |
| GO:0090288 | negative regulation of cellular response to growth factor stimulus | BP | 0.115 | 0.00153 | 0.00938 | 2.03 | 0.00638 | Sfrp2/Nog/Lrp2/Emilin1/Tnfaip6/Hipk2/Apln/Atp2b4/Sema6a/Adgra2/Cav1/Tmprss6 | 12 |
| GO:0030513 | positive regulation of BMP signaling pathway | BP | 0.171 | 0.00155 | 0.00943 | 2.03 | 0.00641 | Kdr/Rnf165/Gpc3/Zfp423/Cdh5/Kcp/Neo1 | 7 |
| GO:0051968 | positive regulation of synaptic transmission, glutamatergic | BP | 0.171 | 0.00155 | 0.00943 | 2.03 | 0.00641 | Gria4/Nlgn1/Grin2b/Shank2/Tshz3/Nlgn3/Cacng7 | 7 |
| GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | BP | 0.238 | 0.00157 | 0.00956 | 2.02 | 0.0065 | Adra1a/P2rx2/Calca/Rps6ka2/Agtr1a | 5 |
| GO:0007190 | activation of adenylate cyclase activity | BP | 0.238 | 0.00157 | 0.00956 | 2.02 | 0.0065 | Vipr2/Adcy3/Calca/Adcy4/Adcy7 | 5 |
| GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | BP | 0.238 | 0.00157 | 0.00956 | 2.02 | 0.0065 | Kdr/Nrp1/Jcad/Hdac9/Akt3 | 5 |
| GO:0009410 | response to xenobiotic stimulus | BP | 0.08 | 0.00159 | 0.00963 | 2.02 | 0.00655 | Kcnk3/Cyp1b1/Slc1a3/Sfrp2/Kdr/Adra1a/Abat/Mmp2/Mef2c/Shank2/Htr1b/Ptprm/Fmo1/Ppm1f/Cd38/Igf2/Ppm1e/Htr2a/Nckap1l/Cyp46a1/Gstm7/Gstm1/Dpyd/Abcb1a/Acsl1/Sult1a1 | 26 |
| GO:0021537 | telencephalon development | BP | 0.0853 | 0.00159 | 0.00963 | 2.02 | 0.00655 | Lef1/Mdk/Zeb2/Sema7a/Cdh2/Dab1/Lrrk2/Id4/Fut10/Cxcl12/Dnah5/Gli3/Plcb1/Tsku/Dclk2/Lhx6/Rtn4rl1/Nin/Nr2f1/Atp1a2/Fat4/Bbs1 | 22 |
| GO:0050868 | negative regulation of T cell activation | BP | 0.105 | 0.00159 | 0.00964 | 2.02 | 0.00656 | Ihh/Mdk/Cd44/Gli3/Loxl3/Btla/H2-Ab1/Havcr2/Nckap1l/Pde5a/H2-Aa/Gimap5/Dlg5/Gimap3 | 14 |
| GO:0043297 | apical junction assembly | BP | 0.121 | 0.00163 | 0.00985 | 2.01 | 0.0067 | Wnt11/Cldn8/Cdh5/Abi2/Esam/Pecam1/Snai1/Cldn5/Nphp1/Dlg5/Cldn1 | 11 |
| GO:0003229 | ventricular cardiac muscle tissue development | BP | 0.138 | 0.00163 | 0.00986 | 2.01 | 0.00671 | Col11a1/Col14a1/Nog/Lrp2/Ccm2l/Tbx3/Ednra/Myl3/Heg1 | 9 |
| GO:0060999 | positive regulation of dendritic spine development | BP | 0.138 | 0.00163 | 0.00986 | 2.01 | 0.00671 | Nlgn1/D16Ertd472e/Nlgn3/Apoe/Caprin2/Bhlhb9/Cux2/Palm/Dlg5 | 9 |
| GO:0045907 | positive regulation of vasoconstriction | BP | 0.151 | 0.00167 | 0.01 | 2 | 0.00683 | Adra1a/Icam1/Wdr35/Cd38/Htr2a/Pde5a/Ptger3/Cav1 | 8 |
| GO:0070169 | positive regulation of biomineral tissue development | BP | 0.151 | 0.00167 | 0.01 | 2 | 0.00683 | Wnt6/Pkdcc/Slc8a1/Mef2c/Osr2/Nell1/Osr1/Ptn | 8 |
| GO:0032413 | negative regulation of ion transmembrane transporter activity | BP | 0.128 | 0.00167 | 0.0101 | 2 | 0.00684 | Hecw2/Drd4/Dysf/Osr1/Rrad/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1 | 10 |
| GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Nlgn1/Tshz3/Nlgn3/Atp1a2 | 4 |
| GO:0010838 | positive regulation of keratinocyte proliferation | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Mdk/Has2/Stxbp4/Twist2 | 4 |
| GO:0014052 | regulation of gamma-aminobutyric acid secretion | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Grik1/Abat/Htr1b/Gabbr1 | 4 |
| GO:0034374 | low-density lipoprotein particle remodeling | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Apob/Pla2g10/Apoe/Abcg1 | 4 |
| GO:0034375 | high-density lipoprotein particle remodeling | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Pltp/Apoe/Abca5/Abcg1 | 4 |
| GO:0034392 | negative regulation of smooth muscle cell apoptotic process | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Gria4/Igf1/Agtr1a/Arrb2 | 4 |
| GO:0086067 | AV node cell to bundle of His cell communication | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Cacna1c/Scn4b/Ank2/Gjc1 | 4 |
| GO:0097090 | presynaptic membrane organization | BP | 0.308 | 0.0017 | 0.0101 | 2 | 0.00688 | Ptprd/Nlgn1/Nlgn3/Apoe | 4 |
| GO:0002313 | mature B cell differentiation involved in immune response | BP | 0.194 | 0.00171 | 0.0101 | 1.99 | 0.00688 | Itm2a/Dock10/Lfng/Enpp1/Irf8/Mfng | 6 |
| GO:0045762 | positive regulation of adenylate cyclase activity | BP | 0.194 | 0.00171 | 0.0101 | 1.99 | 0.00688 | Cacna1c/Vipr2/Adcy3/Calca/Adcy4/Adcy7 | 6 |
| GO:0060317 | cardiac epithelial to mesenchymal transition | BP | 0.194 | 0.00171 | 0.0101 | 1.99 | 0.00688 | Nog/Erg/Has2/Tbx3/Heyl/Snai1 | 6 |
| GO:2000648 | positive regulation of stem cell proliferation | BP | 0.194 | 0.00171 | 0.0101 | 1.99 | 0.00688 | Fermt2/Tert/Vegfc/Tbx3/Kitl/Ltbp3 | 6 |
| GO:0032946 | positive regulation of mononuclear cell proliferation | BP | 0.101 | 0.00173 | 0.0102 | 1.99 | 0.00696 | Igf1/Mef2c/Tnfsf13b/Cd1d1/Coro1a/Itgal/Il27ra/Cd38/Havcr2/Igf2/Xcl1/Nckap1l/Stat5b/Zap70/Il12rb1 | 15 |
| GO:0010092 | specification of animal organ identity | BP | 0.167 | 0.00179 | 0.0106 | 1.98 | 0.00719 | Ar/Wnt11/Hoxd11/Hoxa11/Mef2c/Gli3/Lrp2 | 7 |
| GO:0090175 | regulation of establishment of planar polarity | BP | 0.167 | 0.00179 | 0.0106 | 1.98 | 0.00719 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Fzd2/Dact1 | 7 |
| GO:0015844 | monoamine transport | BP | 0.113 | 0.00181 | 0.0107 | 1.97 | 0.00726 | Maob/Slc6a2/Syt13/Abat/Myo5a/Cxcl12/Htr1b/Cadps/Syt7/Htr2a/Ptger3/Gabbr1 | 12 |
| GO:0048286 | lung alveolus development | BP | 0.136 | 0.00182 | 0.0107 | 1.97 | 0.0073 | Stra6/Kdr/Igf1/Pkdcc/Foxf1/Hopx/Tcf21/Ltbp3/Selenon | 9 |
| GO:0071277 | cellular response to calcium ion | BP | 0.127 | 0.00184 | 0.0109 | 1.96 | 0.00738 | Nlgn1/Syt13/Cpne5/Mef2c/Rasgrp2/Syt7/Asph/Pkd2/Slc25a23/Smpd1 | 10 |
| GO:0010038 | response to metal ion | BP | 0.0814 | 0.00189 | 0.0111 | 1.96 | 0.00754 | Slc13a5/Nlgn1/Syt13/Abat/Cpne5/Fn1/Lrrk2/Nptx1/Mef2c/Rasgrp2/Dlg2/Adcy7/Enpp1/Syt7/Kcnma1/Kcnip3/Asph/Nfatc4/Pkd2/Sord/Slc25a23/Dlg4/Smpd1/Cav1 | 24 |
| GO:0046545 | development of primary female sexual characteristics | BP | 0.107 | 0.00192 | 0.0112 | 1.95 | 0.00765 | Angpt1/Kdr/Nos3/Fst/Mmp2/Adamts1/Lfng/Kit/Kitl/Tnfaip6/Arrb1/Arrb2/Stat5b | 13 |
| GO:0033135 | regulation of peptidyl-serine phosphorylation | BP | 0.0964 | 0.00194 | 0.0114 | 1.94 | 0.00773 | Hgf/Sfrp2/Angpt1/Cd44/Hcls1/Pfn2/Prkd1/Tek/Ppm1f/Caprin2/Bdkrb2/Arrb1/Rassf2/Arrb2/Atp2b4/Cav1 | 16 |
| GO:0048814 | regulation of dendrite morphogenesis | BP | 0.118 | 0.00194 | 0.0114 | 1.94 | 0.00773 | Ptprz1/Hecw2/Ptprd/Tbc1d24/Obsl1/Trpc6/Caprin2/Sarm1/Nfatc4/Bhlhb9/Cux2 | 11 |
| GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | BP | 0.118 | 0.00194 | 0.0114 | 1.94 | 0.00773 | Gpc6/Drd4/Stx1b/Nptx1/Dlg2/Nptxr/Magi2/Nsg1/Cacng7/Cpt1c/Dlg4 | 11 |
| GO:0034766 | negative regulation of ion transmembrane transport | BP | 0.112 | 0.00196 | 0.0114 | 1.94 | 0.00778 | Hecw2/Drd4/Bin1/Dysf/Osr1/Rrad/Ptger3/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1 | 12 |
| GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | BP | 0.227 | 0.00197 | 0.0114 | 1.94 | 0.00778 | Rims3/Stx1b/Cadps/Syt7/Cacna1a | 5 |
| GO:0060080 | inhibitory postsynaptic potential | BP | 0.227 | 0.00197 | 0.0114 | 1.94 | 0.00778 | Grik1/Abat/Drd4/Nlgn3/Grik2 | 5 |
| GO:0072578 | neurotransmitter-gated ion channel clustering | BP | 0.227 | 0.00197 | 0.0114 | 1.94 | 0.00778 | Nlgn1/Slitrk3/Glrb/Apoe/Dlg4 | 5 |
| GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | BP | 0.227 | 0.00197 | 0.0114 | 1.94 | 0.00778 | Ptp4a3/Jcad/Tcf4/Sema6a/Adgra2 | 5 |
| GO:0051150 | regulation of smooth muscle cell differentiation | BP | 0.188 | 0.00203 | 0.0118 | 1.93 | 0.00801 | Tshz3/Zeb1/Kit/Cth/Rbpms2/Efemp2 | 6 |
| GO:0051965 | positive regulation of synapse assembly | BP | 0.125 | 0.00203 | 0.0118 | 1.93 | 0.00801 | Ptprd/St8sia2/Nlgn1/Lrrc4b/Adgrl1/Slitrk3/Nlgn3/Bhlhb9/Cux2/Dlg5 | 10 |
| GO:0030501 | positive regulation of bone mineralization | BP | 0.163 | 0.00206 | 0.0119 | 1.92 | 0.00812 | Pkdcc/Slc8a1/Mef2c/Osr2/Nell1/Osr1/Ptn | 7 |
| GO:0042220 | response to cocaine | BP | 0.163 | 0.00206 | 0.0119 | 1.92 | 0.00812 | Slc1a3/Abat/Drd4/Htr1b/Htr2a/Dlg4/Smpd1 | 7 |
| GO:0030278 | regulation of ossification | BP | 0.102 | 0.00211 | 0.0122 | 1.91 | 0.00828 | Mdk/Pkdcc/Ddr2/Slc8a1/Mef2c/Osr2/Calca/Nell1/Mgp/Enpp1/Ltbp3/P3h1/Osr1/Ptn | 14 |
| GO:0031623 | receptor internalization | BP | 0.102 | 0.00211 | 0.0122 | 1.91 | 0.00828 | Angpt1/Drd4/Calca/Htr1b/Sele/Magi2/Arrb1/Cacng7/Arrb2/Ptger3/Calcrl/Apln/Dlg4/Cav1 | 14 |
| GO:0050773 | regulation of dendrite development | BP | 0.102 | 0.00211 | 0.0122 | 1.91 | 0.00828 | Ptprz1/Hecw2/Ptprd/Mark1/Tbc1d24/Obsl1/Trpc6/Caprin2/Sarm1/Nfatc4/Bhlhb9/Cux2/Fat3/Ptn | 14 |
| GO:0120193 | tight junction organization | BP | 0.117 | 0.00212 | 0.0122 | 1.91 | 0.00831 | Wnt11/Mpdz/Cldn8/Cdh5/Esam/Pecam1/Snai1/Svep1/Cldn5/Nphp1/Cldn1 | 11 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | BP | 0.111 | 0.00212 | 0.0122 | 1.91 | 0.00831 | Sfrp2/Sema5a/Zeb2/Gpc3/Lrrk2/Gprc5b/Peg12/Caprin2/Daam2/Adgra2/Cav1/Dact1 | 12 |
| GO:0048546 | digestive tract morphogenesis | BP | 0.145 | 0.00213 | 0.0122 | 1.91 | 0.00832 | Stra6/Ihh/Sfrp2/Foxf1/Gli3/Tcf21/Dact1/Stx2 | 8 |
| GO:1902476 | chloride transmembrane transport | BP | 0.145 | 0.00213 | 0.0122 | 1.91 | 0.00832 | Slc1a3/Slc12a8/Ano1/Ano2/Glrb/Gabra4/Slc12a4/Slc4a8 | 8 |
| GO:0003014 | renal system process | BP | 0.106 | 0.00222 | 0.0127 | 1.89 | 0.00867 | Cacna1c/Adra1a/Ptpro/Npr3/Ednrb/Stc1/Abcg2/Maged2/Agtr1a/Kcnma1/Ednra/Ptger3/Abcg3 | 13 |
| GO:0050433 | regulation of catecholamine secretion | BP | 0.132 | 0.00225 | 0.0129 | 1.89 | 0.00878 | Syt13/Abat/Myo5a/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Gabbr1 | 9 |
| GO:0009312 | oligosaccharide biosynthetic process | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | B3galnt1/St3gal2/St6galnac6/St3gal4 | 4 |
| GO:0030432 | peristalsis | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Tbx2/P2rx2/Tshz3/Tbx3 | 4 |
| GO:0032341 | aldosterone metabolic process | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Ednrb/Dkk3/Kcnma1/Cacna1h | 4 |
| GO:0046058 | cAMP metabolic process | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Adcy3/Adcy4/Adcy7/Cacnb4 | 4 |
| GO:0051152 | positive regulation of smooth muscle cell differentiation | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Tshz3/Kit/Cth/Efemp2 | 4 |
| GO:1901201 | regulation of extracellular matrix assembly | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Phldb2/Has2/Emilin1/Tie1 | 4 |
| GO:1903238 | positive regulation of leukocyte tethering or rolling | BP | 0.286 | 0.0023 | 0.0131 | 1.88 | 0.0089 | Itga4/Chst2/Sele/St3gal4 | 4 |
| GO:0010717 | regulation of epithelial to mesenchymal transition | BP | 0.116 | 0.00231 | 0.0131 | 1.88 | 0.00892 | Sfrp2/Lef1/Mdk/Nog/Mark1/Phldb2/Fermt2/Epha3/Snai1/Spred3/Il17rd | 11 |
| GO:1904646 | cellular response to amyloid-beta | BP | 0.182 | 0.00239 | 0.0136 | 1.87 | 0.00923 | Itga4/Igf1/Cacna2d1/Icam1/Fyn/Cacna1a | 6 |
| GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.217 | 0.00243 | 0.0137 | 1.86 | 0.00935 | Sema5a/Ppp1r16b/Aplnr/Jcad/Agtr1a | 5 |
| GO:0009065 | glutamine family amino acid catabolic process | BP | 0.217 | 0.00243 | 0.0137 | 1.86 | 0.00935 | Ddah1/Nos3/Asrgl1/Fah/Atp2b4 | 5 |
| GO:0045668 | negative regulation of osteoblast differentiation | BP | 0.13 | 0.00249 | 0.0141 | 1.85 | 0.00957 | Nog/Areg/Id3/Gdf10/Vegfc/Rorb/Tnfaip6/Twist2/Ptch1 | 9 |
| GO:0099601 | regulation of neurotransmitter receptor activity | BP | 0.13 | 0.00249 | 0.0141 | 1.85 | 0.00957 | Begain/Nlgn1/Nptx1/Mef2c/Nlgn3/Nptxr/Prrt1/Cacng7/Dlg4 | 9 |
| GO:0051261 | protein depolymerization | BP | 0.104 | 0.00256 | 0.0144 | 1.84 | 0.00982 | Map2/Sema5a/Evl/Tmod2/Mical1/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2 | 13 |
| GO:0030004 | cellular monovalent inorganic cation homeostasis | BP | 0.108 | 0.00267 | 0.015 | 1.82 | 0.0102 | Slc1a3/Slc8a1/Lrrk2/Slc9a7/Dmxl2/Kcnma1/Slc9a5/Lacc1/Aqp11/Slc4a3/Atp1a2/Slc4a8 | 12 |
| GO:1902106 | negative regulation of leukocyte differentiation | BP | 0.108 | 0.00267 | 0.015 | 1.82 | 0.0102 | Ihh/Mdk/Cd44/Inpp4b/Lrrc17/Gli3/Calca/Flt3/Loxl3/Tnfrsf11b/Zbtb46/Tnfaip6 | 12 |
| GO:0035567 | non-canonical Wnt signaling pathway | BP | 0.14 | 0.00268 | 0.015 | 1.82 | 0.0102 | Sfrp2/Wnt11/Nkd1/Gpc3/Znrf3/Daam2/Fzd2/Dact1 | 8 |
| GO:1900271 | regulation of long-term synaptic potentiation | BP | 0.14 | 0.00268 | 0.015 | 1.82 | 0.0102 | Eif2ak4/Mme/Nlgn3/Nrgn/Apoe/Nsg1/Cyp46a1/Ptn | 8 |
| GO:0001838 | embryonic epithelial tube formation | BP | 0.0962 | 0.00271 | 0.0152 | 1.82 | 0.0103 | Sfrp2/Wnt6/Nog/Zeb2/Lrp2/Gdf7/Ift57/Cecr2/Prickle1/Ptch1/Ipmk/Osr1/Dlc1/Fzd2/Rab23 | 15 |
| GO:1902017 | regulation of cilium assembly | BP | 0.129 | 0.00275 | 0.0154 | 1.81 | 0.0105 | Ccdc88a/Adamts16/Rp1/Kctd17/Crocc/Ttbk2/Dzip1/Dync2li1/Cep97 | 9 |
| GO:0002092 | positive regulation of receptor internalization | BP | 0.176 | 0.0028 | 0.0156 | 1.81 | 0.0106 | Angpt1/Sele/Magi2/Arrb1/Arrb2/Apln | 6 |
| GO:0051497 | negative regulation of stress fiber assembly | BP | 0.176 | 0.0028 | 0.0156 | 1.81 | 0.0106 | Kank4/Phldb2/Arhgap6/Tmeff2/Arhgap28/Dlc1 | 6 |
| GO:0018105 | peptidyl-serine phosphorylation | BP | 0.0767 | 0.00284 | 0.0158 | 1.8 | 0.0108 | Hgf/Sfrp2/Angpt1/Cd44/Hcls1/Lrrk2/Pfn2/Prkd1/Tek/Rps6ka2/Ppm1f/Caprin2/Dclk2/Bdkrb2/Rps6ka5/Arrb1/Hipk2/Pkd1/Rassf2/Ttbk2/Arrb2/Atp2b4/Akt3/Brsk1/Lats2/Cav1 | 26 |
| GO:0050848 | regulation of calcium-mediated signaling | BP | 0.119 | 0.00292 | 0.0163 | 1.79 | 0.0111 | Kdr/Igf1/Drd4/Slc24a4/Lrrk2/Cdh13/Itgal/Cmya5/Atp2b4/Zap70 | 10 |
| GO:2001259 | positive regulation of cation channel activity | BP | 0.119 | 0.00292 | 0.0163 | 1.79 | 0.0111 | Cacna2d1/Ank2/Nlgn3/Cracr2a/Ednra/Stac2/Asph/Pkd2/Gstm7/Cacnb4 | 10 |
| GO:0048675 | axon extension | BP | 0.0986 | 0.00294 | 0.0163 | 1.79 | 0.0111 | Map2/Sema5a/Sema5b/Sema7a/Nrp1/Fn1/Cxcl12/Edn3/Ndn/Nlgn3/Apoe/Ednra/Sema3g/Sema6a | 14 |
| GO:1903531 | negative regulation of secretion by cell | BP | 0.0925 | 0.00295 | 0.0164 | 1.79 | 0.0111 | Maob/Abat/Drd4/Foxf1/Myo5a/Edn3/Htr1b/Apoe/Cd200/Nckap1l/Cd84/Neo1/Ptger3/Frmd4a/Gabbr1/Nr1h3 | 16 |
| GO:0009190 | cyclic nucleotide biosynthetic process | BP | 0.208 | 0.00296 | 0.0164 | 1.79 | 0.0111 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0030903 | notochord development | BP | 0.208 | 0.00296 | 0.0164 | 1.79 | 0.0111 | Gli1/Wnt11/Nog/Id3/Col13a1 | 5 |
| GO:0036303 | lymph vessel morphogenesis | BP | 0.208 | 0.00296 | 0.0164 | 1.79 | 0.0111 | Vash1/Vegfc/Tie1/Pkd1/Svep1 | 5 |
| GO:0052652 | cyclic purine nucleotide metabolic process | BP | 0.208 | 0.00296 | 0.0164 | 1.79 | 0.0111 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0043030 | regulation of macrophage activation | BP | 0.138 | 0.00299 | 0.0165 | 1.78 | 0.0112 | Pla2g10/Lrrk2/Slc7a2/Cd1d1/Havcr2/Cd200/Cd84/Nr1h3 | 8 |
| GO:1902808 | positive regulation of cell cycle G1/S phase transition | BP | 0.138 | 0.00299 | 0.0165 | 1.78 | 0.0112 | Gli1/Tbx2/Ddr2/Tert/Ccnd2/Adamts1/Plcb1/Stxbp4 | 8 |
| GO:0052548 | regulation of endopeptidase activity | BP | 0.0754 | 0.00301 | 0.0166 | 1.78 | 0.0113 | Hgf/Sfrp2/Spink1/Igf1/Aph1b/Cd44/Bok/Grin2b/Wdr35/Serpine2/Mical1/Ift57/Serpini1/Wnt9a/Ppm1f/Hip1/Bin1/Arrb1/Epha7/Asph/Dlc1/Nlrp1b/Arrb2/Ccn2/Fyn/Mgmt/Cyfip2 | 27 |
| GO:0002396 | MHC protein complex assembly | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0002501 | peptide antigen assembly with MHC protein complex | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0031000 | response to caffeine | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | Ryr1/Slc8a1/Selenon/Gstm7 | 4 |
| GO:0035721 | intraciliary retrograde transport | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | Wdr35/Dync2h1/Dync2li1/Bbs1 | 4 |
| GO:0042693 | muscle cell fate commitment | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | AW551984/Tbx2/Mef2c/Tbx3 | 4 |
| GO:1901386 | negative regulation of voltage-gated calcium channel activity | BP | 0.267 | 0.00303 | 0.0166 | 1.78 | 0.0113 | Drd4/Dysf/Rrad/Gnb5 | 4 |
| GO:0017156 | calcium-ion regulated exocytosis | BP | 0.127 | 0.00304 | 0.0166 | 1.78 | 0.0113 | Syn2/Cacna1c/Syt13/Rims3/Stx1b/Cadps/Syt7/Cacna1h/Cacna1a | 9 |
| GO:0048332 | mesoderm morphogenesis | BP | 0.127 | 0.00304 | 0.0166 | 1.78 | 0.0113 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Tbx3/Snai1/Tal1 | 9 |
| GO:0050818 | regulation of coagulation | BP | 0.127 | 0.00304 | 0.0166 | 1.78 | 0.0113 | Tfpi/Abat/Serpine2/Procr/Apoe/Pros1/Ptger3/St3gal4/Cav1 | 9 |
| GO:0042417 | dopamine metabolic process | BP | 0.152 | 0.00307 | 0.0167 | 1.78 | 0.0113 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Agtr1a | 7 |
| GO:2001258 | negative regulation of cation channel activity | BP | 0.152 | 0.00307 | 0.0167 | 1.78 | 0.0113 | Drd4/Dysf/Rrad/Pkd2/Gstm7/Gnb5/Cav1 | 7 |
| GO:0002285 | lymphocyte activation involved in immune response | BP | 0.0856 | 0.00307 | 0.0167 | 1.78 | 0.0113 | Lef1/Mdk/Itm2a/Icam1/Eif2ak4/Dock10/Lfng/Loxl3/Coro1a/Itgal/Il27ra/Cracr2a/Enpp1/Havcr2/Irf8/Nckap1l/Mfng/Rab27a/Apbb1ip | 19 |
| GO:1903707 | negative regulation of hemopoiesis | BP | 0.106 | 0.0031 | 0.0169 | 1.77 | 0.0115 | Ihh/Mdk/Cd44/Inpp4b/Lrrc17/Gli3/Calca/Flt3/Loxl3/Tnfrsf11b/Zbtb46/Tnfaip6 | 12 |
| GO:0042471 | ear morphogenesis | BP | 0.0979 | 0.00313 | 0.017 | 1.77 | 0.0116 | Col11a1/Nog/Tbx2/Osr2/Tshz1/Lrig3/Zeb1/Enpp1/Tbx3/Ednra/Osr1/Fzd2/Kcnq4/Itga8 | 14 |
| GO:0048640 | negative regulation of developmental growth | BP | 0.102 | 0.00315 | 0.0171 | 1.77 | 0.0116 | Map2/Sema5a/H19/Nog/Sema5b/Nkd1/Sema7a/Nrp1/Sema3g/Epha7/Ptch1/Pi16/Sema6a | 13 |
| GO:0034103 | regulation of tissue remodeling | BP | 0.118 | 0.00319 | 0.0173 | 1.76 | 0.0118 | Mdk/Ddr2/Inpp4b/Erg/Calca/Tnfrsf11b/Cd38/Syt7/Ltbp3/Abi3bp | 10 |
| GO:0019935 | cyclic-nucleotide-mediated signaling | BP | 0.111 | 0.00319 | 0.0173 | 1.76 | 0.0118 | Gnai1/Ednrb/Gucy1a2/Gucy1b1/Gucy1a1/Aplnr/Apoe/Adcyap1r1/Cap2/Rasd2/Ptger3 | 11 |
| GO:0042908 | xenobiotic transport | BP | 0.171 | 0.00326 | 0.0176 | 1.75 | 0.012 | Abcg2/Abca8b/Abcb1a/Abcc5/Abcg3/Cldn1 | 6 |
| GO:0090162 | establishment of epithelial cell polarity | BP | 0.171 | 0.00326 | 0.0176 | 1.75 | 0.012 | Foxf1/Msn/Myo9a/Sh3bp1/Frmd4b/Frmd4a | 6 |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | BP | 0.087 | 0.00328 | 0.0177 | 1.75 | 0.0121 | Igf1/Zeb2/Drd4/Fermt2/Ccnd2/Lrrk2/Edn3/Pik3r6/Kit/Nox4/Htr2a/Kitl/Pkd1/Pde5a/Pkd2/Atp2b4/Acsl1/C1qtnf9 | 18 |
| GO:0031532 | actin cytoskeleton reorganization | BP | 0.105 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Hmcn1/Mdk/Nrp1/Thsd7b/Tek/Syde1/Kit/Esam/Parvg/Carmil3/Efs/Parvb | 12 |
| GO:0045778 | positive regulation of ossification | BP | 0.136 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Pkdcc/Slc8a1/Mef2c/Osr2/Calca/Nell1/Osr1/Ptn | 8 |
| GO:0071622 | regulation of granulocyte chemotaxis | BP | 0.136 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Mdk/Jam3/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Mpp1 | 8 |
| GO:0007405 | neuroblast proliferation | BP | 0.125 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Acsl6/Hhip/Lef1/Lrrk2/Id4/Gli3/Eml1/Vegfc/Ptn | 9 |
| GO:0045428 | regulation of nitric oxide biosynthetic process | BP | 0.125 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Igf1/Ddah1/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 9 |
| GO:0048168 | regulation of neuronal synaptic plasticity | BP | 0.125 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Grik1/Kdr/Nog/Grin2b/Kit/Grik2/Dlg4/Syngap1/Cpeb1 | 9 |
| GO:2000179 | positive regulation of neural precursor cell proliferation | BP | 0.125 | 0.00334 | 0.0179 | 1.75 | 0.0122 | Gli1/Mdk/Nog/Igf1/Zfp423/Gli3/Lrp2/Vegfc/Nes | 9 |
| GO:0014013 | regulation of gliogenesis | BP | 0.101 | 0.00338 | 0.0181 | 1.74 | 0.0123 | Ptprz1/Mdk/Nog/Igf1/Tert/Dab1/Id4/Serpine2/Lrp2/Daam2/Bin1/Zcchc24/Ptn | 13 |
| GO:0031348 | negative regulation of defense response | BP | 0.083 | 0.00345 | 0.0184 | 1.73 | 0.0125 | Hgf/Acod1/Mdk/Pla2g10/Igf1/Cd44/Ets1/Foxf1/Fndc4/Cdh5/Apoe/Havcr2/Cd200/Igf2/Tnfaip6/Mapkbp1/Arrb2/Calcrl/Cd96/Nr1h3 | 20 |
| GO:0006865 | amino acid transport | BP | 0.0938 | 0.00345 | 0.0184 | 1.73 | 0.0125 | Slc1a3/Grik1/Abat/Slc7a2/Grin2b/Slc36a4/Htr1b/Slc6a15/Sfxn5/Nfkbie/Slc38a1/Slc7a7/Gabbr1/Cacna1a/Cacnb4 | 15 |
| GO:0006885 | regulation of pH | BP | 0.11 | 0.00345 | 0.0184 | 1.73 | 0.0125 | Car4/Ednrb/Lrrk2/Slc9a7/Dmxl2/Agtr1a/Slc9a5/Lacc1/Aqp11/Slc4a3/Slc4a8 | 11 |
| GO:0003016 | respiratory system process | BP | 0.149 | 0.00347 | 0.0185 | 1.73 | 0.0126 | Nlgn1/Tshz3/Jag2/Ndn/Nlgn3/Selenon/Atp1a2 | 7 |
| GO:0060415 | muscle tissue morphogenesis | BP | 0.116 | 0.00347 | 0.0185 | 1.73 | 0.0126 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Fzd2/Efemp2 | 10 |
| GO:0051896 | regulation of protein kinase B signaling | BP | 0.0909 | 0.0035 | 0.0186 | 1.73 | 0.0127 | Sema5a/Angpt1/Igf1/Hcls1/Fermt2/Inpp4b/Tek/Cxcl12/Lrp2/Nox4/Magi2/Igf2/Cavin3/Rcn3/Rasd2/Arrb2 | 16 |
| GO:0007413 | axonal fasciculation | BP | 0.2 | 0.00357 | 0.0189 | 1.72 | 0.0129 | Ptprz1/Sema5a/Nrp1/Ndn/Epha3 | 5 |
| GO:0010996 | response to auditory stimulus | BP | 0.2 | 0.00357 | 0.0189 | 1.72 | 0.0129 | Stra6/Slc1a3/Mdk/Atp1a2/Ptn | 5 |
| GO:0035024 | negative regulation of Rho protein signal transduction | BP | 0.2 | 0.00357 | 0.0189 | 1.72 | 0.0129 | Adra1a/Rasip1/Arap3/Heg1/Dlc1 | 5 |
| GO:0106030 | neuron projection fasciculation | BP | 0.2 | 0.00357 | 0.0189 | 1.72 | 0.0129 | Ptprz1/Sema5a/Nrp1/Ndn/Epha3 | 5 |
| GO:2000311 | regulation of AMPA receptor activity | BP | 0.2 | 0.00357 | 0.0189 | 1.72 | 0.0129 | Nlgn1/Mef2c/Nlgn3/Prrt1/Cacng7 | 5 |
| GO:0002573 | myeloid leukocyte differentiation | BP | 0.0826 | 0.00361 | 0.0191 | 1.72 | 0.013 | Lef1/Cd109/Hcls1/Gpc3/Mef2c/Inpp4b/Lrrc17/Calca/Plcb1/Kit/Tnfrsf11b/Enpp1/Zbtb46/Tal1/Kitl/Tnfaip6/Rassf2/Nrros/Gimap5/Gimap3 | 20 |
| GO:0051146 | striated muscle cell differentiation | BP | 0.0762 | 0.00366 | 0.0193 | 1.71 | 0.0132 | AW551984/Sgcd/Col14a1/Ryr1/Igf1/Adra1a/Slc8a1/P2rx2/Tmod2/Cdh2/Mef2c/Cxcl12/Hopx/Nox4/Tbx3/Igf2/Hdac9/Dysf/Flnc/Selenon/Arrb2/Neo1/Pi16/Sgcb/Plekho1 | 25 |
| GO:0001541 | ovarian follicle development | BP | 0.123 | 0.00367 | 0.0194 | 1.71 | 0.0132 | Angpt1/Kdr/Mmp2/Lfng/Kit/Kitl/Tnfaip6/Arrb1/Arrb2 | 9 |
| GO:0043954 | cellular component maintenance | BP | 0.123 | 0.00367 | 0.0194 | 1.71 | 0.0132 | Epb41l3/Fermt2/Grin2b/Shank2/Apoe/Erc2/Fyn/Syngap1/Cldn1 | 9 |
| GO:0042987 | amyloid precursor protein catabolic process | BP | 0.133 | 0.00371 | 0.0195 | 1.71 | 0.0133 | Igf1/Aph1b/Rtn1/Slc2a13/Apoe/Bin1/Abcg1/Adam19 | 8 |
| GO:0050772 | positive regulation of axonogenesis | BP | 0.109 | 0.00373 | 0.0196 | 1.71 | 0.0134 | Sema5a/Plxnb1/Zeb2/Sema7a/Nrp1/Fn1/Cxcl12/Plxnd1/Apoe/Nin/Chodl | 11 |
| GO:0032410 | negative regulation of transporter activity | BP | 0.115 | 0.00378 | 0.0198 | 1.7 | 0.0135 | Hecw2/Drd4/Dysf/Osr1/Rrad/Pkd2/Gstm7/Atp1a2/Gnb5/Cav1 | 10 |
| GO:0060411 | cardiac septum morphogenesis | BP | 0.115 | 0.00378 | 0.0198 | 1.7 | 0.0135 | Wnt11/Nog/Tbx2/Nos3/Nrp1/Lrp2/Aplnr/Tbx3/Heyl/Fzd2 | 10 |
| GO:0046777 | protein autophosphorylation | BP | 0.0823 | 0.00379 | 0.0199 | 1.7 | 0.0135 | Epha8/Kdr/Ddr2/Lrrk2/Prkd1/Itk/Tek/Stk33/Eif2ak4/Flt3/Thy1/Kit/Vegfc/Enpp1/Epha7/Rassf2/Fyn/Fgr/Zap70/Cav1 | 20 |
| GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | BP | 0.0857 | 0.00382 | 0.02 | 1.7 | 0.0136 | Hgf/Sfrp2/Igf1/Cd44/Bok/Wdr35/Mical1/Ift57/Wnt9a/Ppm1f/Hip1/Arrb1/Epha7/Dlc1/Nlrp1b/Arrb2/Ccn2/Mgmt | 18 |
| GO:0019751 | polyol metabolic process | BP | 0.103 | 0.00385 | 0.0201 | 1.7 | 0.0137 | Inpp4b/Sptlc3/Plcb1/Pth1r/Plpp3/Bpnt1/Adcyap1r1/Degs2/Dysf/Ipmk/Sord/Plpp1 | 12 |
| GO:0051952 | regulation of amine transport | BP | 0.103 | 0.00385 | 0.0201 | 1.7 | 0.0137 | Grik1/Syt13/Abat/Myo5a/Grin2b/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Gabbr1/Cacna1a | 12 |
| GO:0008212 | mineralocorticoid metabolic process | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Ednrb/Dkk3/Kcnma1/Cacna1h | 4 |
| GO:0031650 | regulation of heat generation | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Abat/Ednrb/Ptger3/Apln | 4 |
| GO:0034433 | steroid esterification | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Soat1/Apoe/Agtr1a/Abcg1 | 4 |
| GO:0034434 | sterol esterification | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Soat1/Apoe/Agtr1a/Abcg1 | 4 |
| GO:0034435 | cholesterol esterification | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Soat1/Apoe/Agtr1a/Abcg1 | 4 |
| GO:0043084 | penile erection | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Ar/Drd4/Ednrb/Ednra | 4 |
| GO:0044065 | regulation of respiratory system process | BP | 0.25 | 0.0039 | 0.0203 | 1.69 | 0.0138 | Nlgn1/Tshz3/Nlgn3/Atp1a2 | 4 |
| GO:0001890 | placenta development | BP | 0.0899 | 0.00391 | 0.0203 | 1.69 | 0.0138 | Ndp/Lef1/Itga4/Slc8a1/Il11ra1/Vash1/Syde1/Prdm1/Pcdh12/Cdkn1c/Igf2/Snai1/Pkd1/Pkd2/Ptn/Vash2 | 16 |
| GO:0030517 | negative regulation of axon extension | BP | 0.146 | 0.00392 | 0.0203 | 1.69 | 0.0138 | Map2/Sema5a/Sema5b/Sema7a/Nrp1/Sema3g/Sema6a | 7 |
| GO:0032965 | regulation of collagen biosynthetic process | BP | 0.146 | 0.00392 | 0.0203 | 1.69 | 0.0138 | Ihh/Ddr2/Fn1/Emilin1/Arrb2/Ccn2/Cygb | 7 |
| GO:0034205 | amyloid-beta formation | BP | 0.146 | 0.00392 | 0.0203 | 1.69 | 0.0138 | Igf1/Aph1b/Rtn1/Slc2a13/Apoe/Bin1/Abcg1 | 7 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | BP | 0.146 | 0.00392 | 0.0203 | 1.69 | 0.0138 | Angpt1/Kdr/Nrp1/Prkd1/Tek/Vegfc/Emilin1 | 7 |
| GO:0071214 | cellular response to abiotic stimulus | BP | 0.0758 | 0.00396 | 0.0204 | 1.69 | 0.0139 | Sfrp2/Wnt11/Nos3/Ccnd2/Slc24a4/Mmp2/Eif2ak4/Mme/Scn2a/Rp1/Ednra/Bdkrb2/Cnn2/Dysf/Nfatc4/Net1/Pkd2/Slc25a23/Palm/Atp1a2/Gnb5/Habp4/Smpd1/Piezo2/Cav1 | 25 |
| GO:0104004 | cellular response to environmental stimulus | BP | 0.0758 | 0.00396 | 0.0204 | 1.69 | 0.0139 | Sfrp2/Wnt11/Nos3/Ccnd2/Slc24a4/Mmp2/Eif2ak4/Mme/Scn2a/Rp1/Ednra/Bdkrb2/Cnn2/Dysf/Nfatc4/Net1/Pkd2/Slc25a23/Palm/Atp1a2/Gnb5/Habp4/Smpd1/Piezo2/Cav1 | 25 |
| GO:0016485 | protein processing | BP | 0.0791 | 0.00397 | 0.0205 | 1.69 | 0.0139 | Cpxm2/Ihh/Mmp16/Aph1b/Lrrk2/Serpine2/Gli3/Rps6ka2/Cpe/Mme/C1rl/Ldlrad3/Dync2h1/Pcsk5/C1ra/Ptch1/Asph/Ece2/Adam19/Arxes2/Cyfip2/Nkd2 | 22 |
| GO:0001708 | cell fate specification | BP | 0.108 | 0.00403 | 0.0207 | 1.68 | 0.0141 | Ihh/Sfrp2/Ar/Wnt11/Hoxd10/Ptch2/Tbx2/Hoxa11/Gli3/Tbx3/Ptch1 | 11 |
| GO:0051346 | negative regulation of hydrolase activity | BP | 0.0738 | 0.00405 | 0.0208 | 1.68 | 0.0142 | Hgf/Sfrp2/Cd109/Spink1/Igf1/Tfpi/Nos3/Cd44/Gpc3/Lrrk2/Serpine2/Mical1/Wfdc3/Serpini1/Wnt9a/Pecam1/Nckap1l/Cmya5/Bin1/Hdac9/Arrb1/Arrb2/Pi16/Angptl4/Pi15/Ppp1r26/Simc1 | 27 |
| GO:0021675 | nerve development | BP | 0.114 | 0.00411 | 0.021 | 1.68 | 0.0143 | Slc1a3/Itga4/Rnf165/Nrp1/Slc24a4/Nptx1/Serpine2/Gli3/Hoxd3/Ednra | 10 |
| GO:0120192 | tight junction assembly | BP | 0.114 | 0.00411 | 0.021 | 1.68 | 0.0143 | Wnt11/Mpdz/Cldn8/Cdh5/Esam/Pecam1/Snai1/Cldn5/Nphp1/Cldn1 | 10 |
| GO:0021543 | pallium development | BP | 0.092 | 0.00411 | 0.021 | 1.68 | 0.0143 | Lef1/Mdk/Zeb2/Cdh2/Dab1/Id4/Fut10/Gli3/Plcb1/Tsku/Dclk2/Lhx6/Nr2f1/Fat4/Bbs1 | 15 |
| GO:0007368 | determination of left/right symmetry | BP | 0.0985 | 0.00411 | 0.021 | 1.68 | 0.0143 | Ihh/Tbx2/Foxf1/Mef2c/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1 | 13 |
| GO:0042551 | neuron maturation | BP | 0.131 | 0.00412 | 0.021 | 1.68 | 0.0143 | Epha8/Cspg4/Ednrb/Lrrk2/Dlg2/Cdkn1c/Nfasc/Ednra | 8 |
| GO:0140115 | export across plasma membrane | BP | 0.131 | 0.00412 | 0.021 | 1.68 | 0.0143 | Kcnd3/Slc8a1/Slc24a4/Abcg2/Slc8a3/Atp1a2/Abcb1a/Abcc5 | 8 |
| GO:0030859 | polarized epithelial cell differentiation | BP | 0.192 | 0.00427 | 0.0218 | 1.66 | 0.0148 | Foxf1/Msn/Myo9a/Sh3bp1/Dlg5 | 5 |
| GO:0043171 | peptide catabolic process | BP | 0.192 | 0.00427 | 0.0218 | 1.66 | 0.0148 | Enpep/Cpe/Trhde/Ggt7/Lvrn | 5 |
| GO:0060065 | uterus development | BP | 0.192 | 0.00427 | 0.0218 | 1.66 | 0.0148 | Stra6/Hoxa11/Hoxa10/Cdkn1c/Hoxa9 | 5 |
| GO:0030593 | neutrophil chemotaxis | BP | 0.107 | 0.00434 | 0.0221 | 1.66 | 0.015 | Cxcl15/Mdk/Jam3/Edn3/Itga1/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Mpp1 | 11 |
| GO:2001234 | negative regulation of apoptotic signaling pathway | BP | 0.0813 | 0.00434 | 0.0221 | 1.66 | 0.015 | Hgf/Sfrp2/Ar/Agap2/Nog/Igf1/Cd44/Nrp1/Tert/Bok/Lrrk2/Icam1/Cxcl12/Mapk8ip1/Tnfrsf22/Cth/Snai1/Bdkrb2/Arrb2/Fyn | 20 |
| GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | BP | 0.162 | 0.00434 | 0.0221 | 1.66 | 0.015 | Igf1/Ddr2/Tert/Mmp2/Adamts1/Htr1b | 6 |
| GO:0010952 | positive regulation of peptidase activity | BP | 0.0889 | 0.00436 | 0.0221 | 1.65 | 0.0151 | Sfrp2/Aph1b/Bok/Grin2b/Wdr35/Ift57/Ppm1f/Hip1/Arrb1/Rcn3/Asph/Dlc1/Nlrp1b/Ccn2/Fyn/Cyfip2 | 16 |
| GO:0046633 | alpha-beta T cell proliferation | BP | 0.143 | 0.00441 | 0.0224 | 1.65 | 0.0152 | Cd44/Mapk8ip1/Dock2/Btla/Xcl1/Zap70/Il12rb1 | 7 |
| GO:0051897 | positive regulation of protein kinase B signaling | BP | 0.102 | 0.00442 | 0.0224 | 1.65 | 0.0152 | Sema5a/Angpt1/Igf1/Hcls1/Fermt2/Tek/Cxcl12/Lrp2/Nox4/Igf2/Rasd2/Arrb2 | 12 |
| GO:1902115 | regulation of organelle assembly | BP | 0.0845 | 0.00443 | 0.0225 | 1.65 | 0.0153 | Gap43/Fez1/Ptprd/Ccdc88a/Adamts16/Lrrc4b/Lrrk2/Nptx1/Msn/Rp1/Kctd17/Nptxr/Crocc/Ttbk2/Dzip1/Dync2li1/Arhgef9/Cep97 | 18 |
| GO:0045638 | negative regulation of myeloid cell differentiation | BP | 0.112 | 0.00445 | 0.0225 | 1.65 | 0.0153 | Inpp4b/Lrrc17/Calca/Tnfrsf11b/Gpr171/Zbtb46/Hoxa9/Tnfaip6/Twist2/Stat5b | 10 |
| GO:0045444 | fat cell differentiation | BP | 0.0795 | 0.00454 | 0.0229 | 1.64 | 0.0156 | Sfrp2/Igf1/Lama4/Fermt2/Runx1t1/Vstm2a/Arl6/Id4/Bbs7/Glis1/Plcb1/Sh2b2/Gdf10/Enpp1/Htr2a/Dysf/Lrp3/Arxes2/Steap4/Bbs1/Dact1 | 21 |
| GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | BP | 0.106 | 0.00467 | 0.0236 | 1.63 | 0.016 | Mdk/Ptprd/Nrp1/Fermt2/Has2/Tbc1d24/Obsl1/Caprin2/Net1/Bhlhb9/Cux2 | 11 |
| GO:0001841 | neural tube formation | BP | 0.097 | 0.00467 | 0.0236 | 1.63 | 0.016 | Sfrp2/Nog/Zeb2/Lrp2/Gdf7/Ift57/Cecr2/Prickle1/Ptch1/Ipmk/Dlc1/Fzd2/Rab23 | 13 |
| GO:0050764 | regulation of phagocytosis | BP | 0.101 | 0.00473 | 0.0238 | 1.62 | 0.0162 | Tub/Dock2/Syt7/Pros1/Masp1/Cnn2/Nckap1l/Dysf/Clec7a/Fgr/Rab27a/Il2rg | 12 |
| GO:0007259 | receptor signaling pathway via JAK-STAT | BP | 0.0859 | 0.00477 | 0.024 | 1.62 | 0.0163 | Stra6/Cyp1b1/Agap2/Ptprd/Igf1/Hcls1/Dab1/Kit/Agtr1a/Pecam1/Pkd1/Csf2rb/Fyn/Pkd2/Stat5b/Il12rb1/Cav1 | 17 |
| GO:0021766 | hippocampus development | BP | 0.118 | 0.00482 | 0.0242 | 1.62 | 0.0164 | Lef1/Mdk/Zeb2/Dab1/Id4/Gli3/Tsku/Dclk2/Bbs1 | 9 |
| GO:0046847 | filopodium assembly | BP | 0.118 | 0.00482 | 0.0242 | 1.62 | 0.0164 | Gap43/Nlgn1/Nrp1/Sh3bp1/Daam2/Fgd1/Palm/Dpysl3/Itgb4 | 9 |
| GO:0080164 | regulation of nitric oxide metabolic process | BP | 0.118 | 0.00482 | 0.0242 | 1.62 | 0.0164 | Igf1/Ddah1/Icam1/Wdr35/Pkd2/Atp2b4/Gimap5/Gimap3/Cav1 | 9 |
| GO:0035148 | tube formation | BP | 0.0879 | 0.00485 | 0.0243 | 1.61 | 0.0165 | Sfrp2/Wnt6/Nog/Zeb2/Lrp2/Gdf7/Ift57/Cecr2/Prickle1/Ptch1/Ipmk/Osr1/Dlc1/Fzd2/Rab23/Edar | 16 |
| GO:0001845 | phagolysosome assembly | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Rab34/Srpx/Coro1a/Syt7 | 4 |
| GO:0006491 | N-glycan processing | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | St8sia2/St8sia4/Man2a2/St8sia6 | 4 |
| GO:0035337 | fatty-acyl-CoA metabolic process | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Acsl6/Them5/Acot7/Acsl1 | 4 |
| GO:0042756 | drinking behavior | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Slc24a4/Htr1b/Agtr1a/Apln | 4 |
| GO:0045059 | positive thymic T cell selection | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Cd3g/Dock2/Cd1d1/Zap70 | 4 |
| GO:0061000 | negative regulation of dendritic spine development | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Nlgn1/Nlgn3/Apoe/Ngef | 4 |
| GO:0070571 | negative regulation of neuron projection regeneration | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Thy1/Rtn4rl1/Neo1/Xylt1 | 4 |
| GO:0090136 | epithelial cell-cell adhesion | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Ihh/Cyp1b1/Kit/Svep1 | 4 |
| GO:0097094 | craniofacial suture morphogenesis | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Mmp16/Gli3/Frem1/Rab23 | 4 |
| GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | BP | 0.235 | 0.00493 | 0.0245 | 1.61 | 0.0167 | Ppp1r16b/Aplnr/Jcad/Agtr1a | 4 |
| GO:0050819 | negative regulation of coagulation | BP | 0.14 | 0.00494 | 0.0245 | 1.61 | 0.0167 | Tfpi/Abat/Serpine2/Procr/Apoe/Pros1/Ptger3 | 7 |
| GO:0006898 | receptor-mediated endocytosis | BP | 0.0789 | 0.00494 | 0.0245 | 1.61 | 0.0167 | Angpt1/Itga4/Drd4/Clec9a/Calca/Lrp2/Htr1b/Nlgn3/Apoe/Sele/Ldlrad3/Magi2/Hip1/Arrb1/Cacng7/Arrb2/Ptger3/Calcrl/Apln/Dlg4/Cav1 | 21 |
| GO:0007212 | dopamine receptor signaling pathway | BP | 0.158 | 0.00498 | 0.0246 | 1.61 | 0.0167 | Nsg2/Drd4/Lrrk2/Nsg1/Palm/Gnb5 | 6 |
| GO:0045589 | regulation of regulatory T cell differentiation | BP | 0.158 | 0.00498 | 0.0246 | 1.61 | 0.0167 | Mdk/Cd44/Sox12/Gimap5/Gimap3/Il2rg | 6 |
| GO:0098868 | bone growth | BP | 0.158 | 0.00498 | 0.0246 | 1.61 | 0.0167 | Kdr/Ddr2/Evc/Thbs3/Stc1/Enpp1 | 6 |
| GO:1902003 | regulation of amyloid-beta formation | BP | 0.158 | 0.00498 | 0.0246 | 1.61 | 0.0167 | Igf1/Rtn1/Slc2a13/Apoe/Bin1/Abcg1 | 6 |
| GO:0010517 | regulation of phospholipase activity | BP | 0.127 | 0.00503 | 0.0247 | 1.61 | 0.0168 | Adra1a/Itk/Arhgap6/Sele/Agtr1a/Adcyap1r1/Htr2a/Dlc1 | 8 |
| GO:0035306 | positive regulation of dephosphorylation | BP | 0.127 | 0.00503 | 0.0247 | 1.61 | 0.0168 | Npnt/Mef2c/Itga1/Cdh5/Ppp1r16b/Magi2/Dlc1/Smpd1 | 8 |
| GO:0042269 | regulation of natural killer cell mediated cytotoxicity | BP | 0.127 | 0.00503 | 0.0247 | 1.61 | 0.0168 | Pik3r6/Cd226/Havcr2/Igf2/Arrb2/Stat5b/Gimap5/Gimap3 | 8 |
| GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | BP | 0.127 | 0.00503 | 0.0247 | 1.61 | 0.0168 | Calca/Htr1b/Aplnr/Arrb1/Arrb2/Ptger3/Apln/Atp2b4 | 8 |
| GO:0007498 | mesoderm development | BP | 0.1 | 0.00506 | 0.0248 | 1.61 | 0.0169 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Tbx3/Snai1/Tal1/Osr1/Pus7/Ets2 | 12 |
| GO:0042104 | positive regulation of activated T cell proliferation | BP | 0.185 | 0.00506 | 0.0248 | 1.61 | 0.0169 | Igf1/Il27ra/Igf2/Stat5b/Il12rb1 | 5 |
| GO:0062009 | secondary palate development | BP | 0.185 | 0.00506 | 0.0248 | 1.61 | 0.0169 | Lef1/Wnt11/Tshz1/Jag2/Fzd2 | 5 |
| GO:0072012 | glomerulus vasculature development | BP | 0.185 | 0.00506 | 0.0248 | 1.61 | 0.0169 | Angpt1/Tek/Tcf21/Ednra/Osr1 | 5 |
| GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | BP | 0.0927 | 0.00511 | 0.025 | 1.6 | 0.017 | Adra1a/Adgrl4/Cxcr3/Adcy3/Ptgfr/Adgrl1/Calca/Pth1r/Adcy4/Gpr161/Adcy7/Ptger3/Calcrl/Atp2b4 | 14 |
| GO:0071674 | mononuclear cell migration | BP | 0.0874 | 0.00511 | 0.025 | 1.6 | 0.017 | Mdk/Itga4/Cyp7b1/Ptpro/Itgb7/Icam1/Cxcr3/Cxcl12/Calca/Plcb1/Msn/Itgal/Il27ra/Cd200/Pecam1/Xcl1 | 16 |
| GO:0030258 | lipid modification | BP | 0.0833 | 0.00512 | 0.0251 | 1.6 | 0.017 | Acacb/Soat1/Inpp4b/Agk/Bdh2/Plpp3/St3gal2/Dgkb/Ggta1/Apoe/Agtr1a/Abcg1/Echs1/Abcd2/Plpp1/St3gal4/Cygb/C1qtnf9 | 18 |
| GO:0072175 | epithelial tube formation | BP | 0.0898 | 0.00514 | 0.0251 | 1.6 | 0.0171 | Sfrp2/Wnt6/Nog/Zeb2/Lrp2/Gdf7/Ift57/Cecr2/Prickle1/Ptch1/Ipmk/Osr1/Dlc1/Fzd2/Rab23 | 15 |
| GO:0008277 | regulation of G protein-coupled receptor signaling pathway | BP | 0.0956 | 0.00529 | 0.0258 | 1.59 | 0.0176 | Tub/Tmod2/Slc24a4/Calca/Plcb1/Htr1b/Aplnr/Arrb1/Pde5a/Arrb2/Ptger3/Apln/Atp2b4 | 13 |
| GO:0032886 | regulation of microtubule-based process | BP | 0.0784 | 0.00538 | 0.0262 | 1.58 | 0.0178 | Map2/Fez1/Phldb2/Mpdz/Gnai1/Cdh5/Epha3/Rp1/Map9/Tubb4a/Slain1/Nin/Pkd1/Dysf/Ttbk2/Klhl42/Cep97/Bbs1/Mapre2/Cav1/Tppp | 21 |
| GO:0072521 | purine-containing compound metabolic process | BP | 0.0714 | 0.00538 | 0.0262 | 1.58 | 0.0178 | Ttr/Nme4/Acsl6/Igf1/Hmgcs2/Nos3/Acacb/Lrrk2/Adcy3/Gamt/Them5/Gucy1b1/Gucy1a1/Dhtkd1/Adcy4/Adcy7/Enpp1/Lacc1/Htr2a/Acot7/Zbtb20/Pde5a/Dpyd/Rab23/Impdh1/Acsl1/Pid1/Cacnb4 | 28 |
| GO:0015837 | amine transport | BP | 0.0992 | 0.0054 | 0.0263 | 1.58 | 0.0179 | Grik1/Syt13/Abat/Myo5a/Grin2b/Cxcl12/Htr1b/Syt7/Htr2a/Ptger3/Gabbr1/Cacna1a | 12 |
| GO:0044106 | cellular amine metabolic process | BP | 0.0992 | 0.0054 | 0.0263 | 1.58 | 0.0179 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Atp2b4/Slc7a7/Sult1a1 | 12 |
| GO:0006687 | glycosphingolipid metabolic process | BP | 0.137 | 0.00552 | 0.0268 | 1.57 | 0.0182 | St8sia2/St8sia4/St6galnac4/St3gal2/Kit/St8sia6/St6galnac6 | 7 |
| GO:0042088 | T-helper 1 type immune response | BP | 0.137 | 0.00552 | 0.0268 | 1.57 | 0.0182 | Lef1/Il27ra/H2-Ab1/Cracr2a/Havcr2/Xcl1/Il12rb1 | 7 |
| GO:0097581 | lamellipodium organization | BP | 0.109 | 0.00564 | 0.0274 | 1.56 | 0.0186 | Ptpro/Ccdc88a/Cd44/Wasf3/Abi3/Cdh13/Abi2/Kit/Parvb/Plekho1 | 10 |
| GO:0002335 | mature B cell differentiation | BP | 0.154 | 0.00567 | 0.0274 | 1.56 | 0.0187 | Itm2a/Dock10/Lfng/Enpp1/Irf8/Mfng | 6 |
| GO:0033238 | regulation of cellular amine metabolic process | BP | 0.154 | 0.00567 | 0.0274 | 1.56 | 0.0187 | Maob/Abat/Drd4/Pde1b/Atp2b4/Slc7a7 | 6 |
| GO:0051481 | negative regulation of cytosolic calcium ion concentration | BP | 0.154 | 0.00567 | 0.0274 | 1.56 | 0.0187 | Kcnk3/Slc8a1/Calca/Pkd2/Gstm7/Atp1a2 | 6 |
| GO:0070570 | regulation of neuron projection regeneration | BP | 0.154 | 0.00567 | 0.0274 | 1.56 | 0.0187 | Hgf/Thy1/Rtn4rl1/Neo1/Xylt1/Ptn | 6 |
| GO:0010769 | regulation of cell morphogenesis involved in differentiation | BP | 0.0984 | 0.00577 | 0.0278 | 1.56 | 0.0189 | Mdk/Ptprd/Nrp1/Fermt2/Has2/Tbc1d24/Obsl1/Postn/Caprin2/Net1/Bhlhb9/Cux2 | 12 |
| GO:0006821 | chloride transport | BP | 0.103 | 0.00579 | 0.0279 | 1.55 | 0.019 | Slc1a3/Slc12a8/Ano1/Ano2/Ttyh2/Glrb/Gabra4/Slc12a4/Clca2/Abcb1a/Slc4a8 | 11 |
| GO:0009314 | response to radiation | BP | 0.0696 | 0.00584 | 0.0281 | 1.55 | 0.0191 | Slc1a3/Sfrp2/Sema5a/Cacna1c/Nog/Sema5b/Ccnd2/Slc24a4/Mmp2/B4galt2/Pde1b/Cacna1e/Lrp2/Eif2ak4/Fbxl21/Mme/Kit/Rp1/Nlgn3/Arrb1/Nfatc4/Net1/Atp1a2/Gnb5/Brsk1/Syngap1/Smpd1/Ddhd2/Usp2/Cacnb4 | 30 |
| GO:0034390 | smooth muscle cell apoptotic process | BP | 0.179 | 0.00595 | 0.0285 | 1.54 | 0.0194 | Gria4/Igf1/Agtr1a/Arrb1/Arrb2 | 5 |
| GO:0034391 | regulation of smooth muscle cell apoptotic process | BP | 0.179 | 0.00595 | 0.0285 | 1.54 | 0.0194 | Gria4/Igf1/Agtr1a/Arrb1/Arrb2 | 5 |
| GO:1903055 | positive regulation of extracellular matrix organization | BP | 0.179 | 0.00595 | 0.0285 | 1.54 | 0.0194 | Sema5a/Ddr2/Phldb2/Emilin1/Efemp2 | 5 |
| GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | BP | 0.179 | 0.00595 | 0.0285 | 1.54 | 0.0194 | Drd4/Mef2c/Rgs5/Apln/Efemp2 | 5 |
| GO:0010950 | positive regulation of endopeptidase activity | BP | 0.0882 | 0.00605 | 0.029 | 1.54 | 0.0197 | Sfrp2/Aph1b/Bok/Grin2b/Wdr35/Ift57/Ppm1f/Hip1/Arrb1/Asph/Dlc1/Nlrp1b/Ccn2/Fyn/Cyfip2 | 15 |
| GO:0009247 | glycolipid biosynthetic process | BP | 0.123 | 0.00608 | 0.0291 | 1.54 | 0.0198 | St8sia2/St8sia4/St6galnac4/St3gal2/St8sia6/Sccpdh/St6galnac6/St3gal4 | 8 |
| GO:0071260 | cellular response to mechanical stimulus | BP | 0.123 | 0.00608 | 0.0291 | 1.54 | 0.0198 | Wnt11/Nos3/Ednra/Cnn2/Atp1a2/Habp4/Piezo2/Cav1 | 8 |
| GO:0044272 | sulfur compound biosynthetic process | BP | 0.108 | 0.00608 | 0.0291 | 1.54 | 0.0198 | Acsl6/Angpt1/Cbs/Acacb/Chst1/Cth/Xylt1/Dse/Ggt7/Acsl1 | 10 |
| GO:0006469 | negative regulation of protein kinase activity | BP | 0.0837 | 0.0061 | 0.0292 | 1.54 | 0.0198 | Sfrp2/Ptpro/Slc8a1/Mapk8ip1/Slc8a3/Pkia/Thy1/Ppm1f/Cdkn1c/Apoe/Rasip1/Ppm1e/Pkib/Heg1/Lats2/Smpd1/Cav1 | 17 |
| GO:0010470 | regulation of gastrulation | BP | 0.222 | 0.00613 | 0.0292 | 1.54 | 0.0198 | Phldb2/Osr2/Tgif2/Osr1 | 4 |
| GO:0015701 | bicarbonate transport | BP | 0.222 | 0.00613 | 0.0292 | 1.54 | 0.0198 | Car4/Ptger3/Slc4a3/Slc4a8 | 4 |
| GO:0033631 | cell-cell adhesion mediated by integrin | BP | 0.222 | 0.00613 | 0.0292 | 1.54 | 0.0198 | Itga4/Npnt/Podxl/Plpp3 | 4 |
| GO:0035633 | maintenance of blood-brain barrier | BP | 0.222 | 0.00613 | 0.0292 | 1.54 | 0.0198 | Angpt1/Bdkrb2/Cldn5/Abcb1a | 4 |
| GO:0061430 | bone trabecula morphogenesis | BP | 0.222 | 0.00613 | 0.0292 | 1.54 | 0.0198 | Plxnb1/Thbs3/Mmp2/Enpp1 | 4 |
| GO:0090279 | regulation of calcium ion import | BP | 0.135 | 0.00615 | 0.0292 | 1.53 | 0.0199 | Spink1/Stc1/Cxcl12/Agtr1a/Dysf/Fyn/Pkd2 | 7 |
| GO:0072676 | lymphocyte migration | BP | 0.102 | 0.0062 | 0.0295 | 1.53 | 0.02 | Itga4/Cyp7b1/Itgb7/Icam1/Cxcr3/Cxcl12/Msn/Itgal/Il27ra/Cd200/Xcl1 | 11 |
| GO:0046942 | carboxylic acid transport | BP | 0.0761 | 0.00623 | 0.0296 | 1.53 | 0.0201 | Slc1a3/Grik1/Pla2g10/Slc13a5/Abat/Drd4/Slc7a2/Grin2b/Abcg2/Slc36a4/Lrp2/Htr1b/Slc6a15/Bdkrb2/Slc23a2/Sfxn5/Nfkbie/Slc38a1/Abcc5/Gabbr1/Cacna1a/Cacnb4 | 22 |
| GO:1902806 | regulation of cell cycle G1/S phase transition | BP | 0.0877 | 0.00638 | 0.0302 | 1.52 | 0.0206 | Gli1/Myo16/Tbx2/Ddr2/Tert/Ccnd2/Adamts1/Plcb1/Stxbp4/Pkd1/Fhl1/Pkd2/Atp2b4/Dact1/Cacnb4 | 15 |
| GO:0032232 | negative regulation of actin filament bundle assembly | BP | 0.15 | 0.00644 | 0.0304 | 1.52 | 0.0207 | Kank4/Phldb2/Arhgap6/Tmeff2/Arhgap28/Dlc1 | 6 |
| GO:0060219 | camera-type eye photoreceptor cell differentiation | BP | 0.15 | 0.00644 | 0.0304 | 1.52 | 0.0207 | Ihh/Sdk2/Thy1/Rp1/Rorb/Ptn | 6 |
| GO:0060351 | cartilage development involved in endochondral bone morphogenesis | BP | 0.15 | 0.00644 | 0.0304 | 1.52 | 0.0207 | Ihh/Hoxd11/Thbs3/Cbs/Hoxa11/Stc1 | 6 |
| GO:1902624 | positive regulation of neutrophil migration | BP | 0.15 | 0.00644 | 0.0304 | 1.52 | 0.0207 | Mdk/Ednra/Xcl1/Nckap1l/Dysf/Ptger3 | 6 |
| GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | BP | 0.15 | 0.00644 | 0.0304 | 1.52 | 0.0207 | Cd83/Xcl1/Nckap1l/Gimap5/Gimap3/Il2rg | 6 |
| GO:0046620 | regulation of organ growth | BP | 0.0968 | 0.00655 | 0.0308 | 1.51 | 0.021 | Gli1/Cacna1c/Col14a1/Nog/Igf1/Tbx2/Acacb/Mef2c/Flvcr1/Igf2/Pi16/Lats2 | 12 |
| GO:0021954 | central nervous system neuron development | BP | 0.106 | 0.00656 | 0.0308 | 1.51 | 0.021 | Map2/Dcc/Zeb2/Nrp1/Tsku/Cdh11/Ndnf/Dclk2/Lhx6/Nin | 10 |
| GO:0032273 | positive regulation of protein polymerization | BP | 0.106 | 0.00656 | 0.0308 | 1.51 | 0.021 | Evl/Pfn2/Icam1/Nckap1l/Slain1/Nin/Bin1/Cdc42ep5/Cav1/Tppp | 10 |
| GO:0042475 | odontogenesis of dentin-containing tooth | BP | 0.106 | 0.00656 | 0.0308 | 1.51 | 0.021 | Wnt6/Lef1/Apcdd1/Fst/Edaradd/Slc24a4/Gli3/Jag2/Tnfrsf11b/Edar | 10 |
| GO:0048644 | muscle organ morphogenesis | BP | 0.106 | 0.00656 | 0.0308 | 1.51 | 0.021 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1/Fzd2/Efemp2 | 10 |
| GO:0045776 | negative regulation of blood pressure | BP | 0.121 | 0.00667 | 0.0313 | 1.5 | 0.0213 | Kdr/Adra1a/Abat/Nos3/Calca/Vegfc/Bdkrb2/Apln | 8 |
| GO:0090311 | regulation of protein deacetylation | BP | 0.121 | 0.00667 | 0.0313 | 1.5 | 0.0213 | Lrrk2/Prkd1/Tcf21/Ndn/Aplnr/Dysf/Tada2a/Tppp | 8 |
| GO:0097696 | receptor signaling pathway via STAT | BP | 0.0829 | 0.00671 | 0.0315 | 1.5 | 0.0214 | Stra6/Cyp1b1/Agap2/Ptprd/Igf1/Hcls1/Dab1/Kit/Agtr1a/Pecam1/Pkd1/Csf2rb/Fyn/Pkd2/Stat5b/Il12rb1/Cav1 | 17 |
| GO:0046661 | male sex differentiation | BP | 0.0872 | 0.00672 | 0.0315 | 1.5 | 0.0214 | Tex15/Sfrp2/Ar/Kdr/Hoxa11/Hoxa10/Icam1/Tcf21/Lrp2/Tbx3/Hoxa9/Rhobtb3/Smad9/Stat5b/Abcb1a | 15 |
| GO:0030850 | prostate gland development | BP | 0.132 | 0.00684 | 0.0319 | 1.5 | 0.0217 | Ar/Nog/Cyp7b1/Igf1/Cd44/Mmp2/Id4 | 7 |
| GO:0060412 | ventricular septum morphogenesis | BP | 0.132 | 0.00684 | 0.0319 | 1.5 | 0.0217 | Wnt11/Nog/Nos3/Aplnr/Tbx3/Heyl/Fzd2 | 7 |
| GO:0072091 | regulation of stem cell proliferation | BP | 0.132 | 0.00684 | 0.0319 | 1.5 | 0.0217 | Fermt2/Tert/Vegfc/Tbx3/Kitl/Ltbp3/Septin4 | 7 |
| GO:0033198 | response to ATP | BP | 0.172 | 0.00694 | 0.0323 | 1.49 | 0.022 | Ryr1/Slc8a1/P2rx2/Enpp1/Asph | 5 |
| GO:0090023 | positive regulation of neutrophil chemotaxis | BP | 0.172 | 0.00694 | 0.0323 | 1.49 | 0.022 | Mdk/Ednra/Xcl1/Nckap1l/Dysf | 5 |
| GO:1904861 | excitatory synapse assembly | BP | 0.172 | 0.00694 | 0.0323 | 1.49 | 0.022 | Ptprd/Nlgn1/Lrrc4b/Nptx1/Slitrk3 | 5 |
| GO:0033077 | T cell differentiation in thymus | BP | 0.105 | 0.00706 | 0.0328 | 1.48 | 0.0223 | Ihh/Cd3g/Gli3/Dock2/Zeb1/Cd1d1/Jag2/Stat5b/Zap70/Il2rg | 10 |
| GO:0061053 | somite development | BP | 0.105 | 0.00706 | 0.0328 | 1.48 | 0.0223 | Ihh/Sfrp2/Lef1/Wnt11/Nog/Zeb2/Foxf1/Lfng/Loxl3/Ptch1 | 10 |
| GO:0009855 | determination of bilateral symmetry | BP | 0.0922 | 0.00714 | 0.0331 | 1.48 | 0.0225 | Ihh/Tbx2/Foxf1/Mef2c/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1 | 13 |
| GO:0051224 | negative regulation of protein transport | BP | 0.0922 | 0.00714 | 0.0331 | 1.48 | 0.0225 | Angpt1/Pkdcc/Drd4/Lrrk2/Pkia/Ppm1f/Apoe/Cd200/Neo1/Ptger3/Frmd4a/Rab23/Nr1h3 | 13 |
| GO:0035850 | epithelial cell differentiation involved in kidney development | BP | 0.146 | 0.00728 | 0.0333 | 1.48 | 0.0226 | Ptpro/Ednrb/Mef2c/Ednra/Osr1/Fat4 | 6 |
| GO:0035886 | vascular associated smooth muscle cell differentiation | BP | 0.146 | 0.00728 | 0.0333 | 1.48 | 0.0226 | Kit/Cth/Aplnr/Ednra/Efemp2/Sgcb | 6 |
| GO:0071604 | transforming growth factor beta production | BP | 0.146 | 0.00728 | 0.0333 | 1.48 | 0.0226 | Wnt11/Fn1/Tsku/Cd200/Ltbp3/Nrros | 6 |
| GO:0001707 | mesoderm formation | BP | 0.119 | 0.0073 | 0.0333 | 1.48 | 0.0226 | Sfrp2/Lef1/Wnt11/Nog/Hoxa11/Foxf1/Snai1/Tal1 | 8 |
| GO:0030193 | regulation of blood coagulation | BP | 0.119 | 0.0073 | 0.0333 | 1.48 | 0.0226 | Tfpi/Abat/Serpine2/Apoe/Pros1/Ptger3/St3gal4/Cav1 | 8 |
| GO:0042461 | photoreceptor cell development | BP | 0.119 | 0.0073 | 0.0333 | 1.48 | 0.0226 | Thy1/Prdm1/Rp1/Rorb/Arl3/Ttc26/Nphp1/Bbs1 | 8 |
| GO:0090398 | cellular senescence | BP | 0.119 | 0.0073 | 0.0333 | 1.48 | 0.0226 | Tbx2/Tert/Abi3/Vash1/Tbx3/Akt3/Smc5/Cav1 | 8 |
| GO:1902305 | regulation of sodium ion transmembrane transport | BP | 0.119 | 0.0073 | 0.0333 | 1.48 | 0.0226 | Hecw2/Scn4b/Drd4/Fxyd5/Plcb1/Osr1/Atp2b4/Atp1a2 | 8 |
| GO:0002315 | marginal zone B cell differentiation | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Dock10/Lfng/Mfng | 3 |
| GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Gimap5/Gimap3/Il2rg | 3 |
| GO:0002692 | negative regulation of cellular extravasation | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Cxcl12/Plcb1/Il27ra | 3 |
| GO:0021957 | corticospinal tract morphogenesis | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Zeb2/Cdh11/Nin | 3 |
| GO:0035864 | response to potassium ion | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Nptx1/Dlg2/Dlg4 | 3 |
| GO:0035865 | cellular response to potassium ion | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Nptx1/Dlg2/Dlg4 | 3 |
| GO:0043201 | response to leucine | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Klhl22/Rragd/Ubr1 | 3 |
| GO:0070294 | renal sodium ion absorption | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Ednrb/Maged2/Ednra | 3 |
| GO:0071233 | cellular response to leucine | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Klhl22/Rragd/Ubr1 | 3 |
| GO:0072203 | cell proliferation involved in metanephros development | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Gpc3/Ptch1/Osr1 | 3 |
| GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Slc8a1/Gjc1/Cav1 | 3 |
| GO:0090269 | fibroblast growth factor production | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Wnt11/Ccm2l/Heg1 | 3 |
| GO:0090270 | regulation of fibroblast growth factor production | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Wnt11/Ccm2l/Heg1 | 3 |
| GO:0097105 | presynaptic membrane assembly | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Ptprd/Nlgn1/Nlgn3 | 3 |
| GO:1900119 | positive regulation of execution phase of apoptosis | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Bok/Cxcr3/Dlc1 | 3 |
| GO:1902746 | regulation of lens fiber cell differentiation | BP | 0.3 | 0.00731 | 0.0333 | 1.48 | 0.0226 | Zeb2/Cdkn1c/Spred3 | 3 |
| GO:0002688 | regulation of leukocyte chemotaxis | BP | 0.0952 | 0.00742 | 0.0336 | 1.47 | 0.0229 | Mdk/Cxcl12/Jam3/Edn3/Vegfc/Ednra/Xcl1/Nckap1l/Tnfaip6/Dysf/Mpp1/Ptn | 12 |
| GO:0009308 | amine metabolic process | BP | 0.0952 | 0.00742 | 0.0336 | 1.47 | 0.0229 | Maob/Abat/Drd4/Myo5a/Pde1b/Maoa/Hdc/Agtr1a/Ednra/Atp2b4/Slc7a7/Sult1a1 | 12 |
| GO:0018958 | phenol-containing compound metabolic process | BP | 0.0952 | 0.00742 | 0.0336 | 1.47 | 0.0229 | Maob/Zeb2/Abat/Drd4/Myo5a/Pde1b/Maoa/Fah/Hdc/Agtr1a/Ednra/Sult1a1 | 12 |
| GO:0002827 | positive regulation of T-helper 1 type immune response | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Il27ra/H2-Ab1/Xcl1/Il12rb1 | 4 |
| GO:0003214 | cardiac left ventricle morphogenesis | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Sfrp2/Foxf1/Cpe/Ednra | 4 |
| GO:0010544 | negative regulation of platelet activation | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Abat/Serpine2/Apoe/Ptger3 | 4 |
| GO:0022038 | corpus callosum development | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Zeb2/Tsku/Rtn4rl1/Nin | 4 |
| GO:0030011 | maintenance of cell polarity | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Wnt11/Lin7a/Nckap1l/Dlg5 | 4 |
| GO:0033604 | negative regulation of catecholamine secretion | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Abat/Myo5a/Ptger3/Gabbr1 | 4 |
| GO:0033623 | regulation of integrin activation | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Fermt2/Jam3/Rasip1/Pcsk5 | 4 |
| GO:0034111 | negative regulation of homotypic cell-cell adhesion | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Abat/Serpine2/Ccm2l/Ptger3 | 4 |
| GO:0035461 | vitamin transmembrane transport | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Stra6/Lrp2/Slc23a2/Abcc5 | 4 |
| GO:0035641 | locomotory exploration behavior | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Lrrk2/Apoe/Dlg4/Atp1a2 | 4 |
| GO:0045198 | establishment of epithelial cell apical/basal polarity | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Foxf1/Msn/Myo9a/Sh3bp1 | 4 |
| GO:0045579 | positive regulation of B cell differentiation | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Prdm1/Nckap1l/Stat5b/Il2rg | 4 |
| GO:0071514 | genomic imprinting | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | H19/Tet1/Ndn/Cdkn1c | 4 |
| GO:1905874 | regulation of postsynaptic density organization | BP | 0.211 | 0.00751 | 0.0336 | 1.47 | 0.0229 | Ptprd/Cdh2/Lrrc4b/Nptx1 | 4 |
| GO:0009799 | specification of symmetry | BP | 0.0915 | 0.00756 | 0.0338 | 1.47 | 0.023 | Ihh/Tbx2/Foxf1/Mef2c/Dnah5/Ift57/Aplnr/Tbx3/Dync2h1/Daam2/Pcsk5/Pkd2/Dync2li1 | 13 |
| GO:0010883 | regulation of lipid storage | BP | 0.13 | 0.00757 | 0.0338 | 1.47 | 0.023 | Apob/Pla2g10/Acacb/Vstm2a/Mest/Abcg1/Nr1h3 | 7 |
| GO:0031103 | axon regeneration | BP | 0.13 | 0.00757 | 0.0338 | 1.47 | 0.023 | Gap43/Mmp2/Enpp1/Rtn4rl1/Neo1/Xylt1/Ptn | 7 |
| GO:0032892 | positive regulation of organic acid transport | BP | 0.13 | 0.00757 | 0.0338 | 1.47 | 0.023 | Acsl6/Grik1/Pla2g10/Abat/Grin2b/Acsl1/Gabbr1 | 7 |
| GO:0099054 | presynapse assembly | BP | 0.13 | 0.00757 | 0.0338 | 1.47 | 0.023 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn3/Pcdh17 | 7 |
| GO:0031110 | regulation of microtubule polymerization or depolymerization | BP | 0.104 | 0.00759 | 0.0338 | 1.47 | 0.023 | Map2/Cdh5/Rp1/Tubb4a/Slain1/Nin/Dysf/Ttbk2/Mapre2/Cav1 | 10 |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | BP | 0.104 | 0.00759 | 0.0338 | 1.47 | 0.023 | Hhip/Nog/Tbx2/Cd44/Enpp1/Ndnf/Ccn2/Apln/Fat4/Kif16b | 10 |
| GO:0043491 | protein kinase B signaling | BP | 0.0817 | 0.00772 | 0.0344 | 1.46 | 0.0234 | Sema5a/Angpt1/Kdr/Igf1/Hcls1/Fermt2/Inpp4b/Tek/Cxcl12/Lrp2/Nox4/Magi2/Igf2/Cavin3/Rcn3/Rasd2/Arrb2 | 17 |
| GO:0030183 | B cell differentiation | BP | 0.0857 | 0.00784 | 0.0349 | 1.46 | 0.0237 | Itm2a/Flt3/Tnfsf13b/Dock10/Prdm1/Lfng/Kit/H2-Ab1/Enpp1/Irf8/Nckap1l/Hdac9/Stat5b/Mfng/Il2rg | 15 |
| GO:0070830 | bicellular tight junction assembly | BP | 0.11 | 0.00792 | 0.0352 | 1.45 | 0.0239 | Wnt11/Cldn8/Cdh5/Esam/Pecam1/Snai1/Cldn5/Nphp1/Cldn1 | 9 |
| GO:0097530 | granulocyte migration | BP | 0.0881 | 0.00797 | 0.0354 | 1.45 | 0.0241 | Cxcl15/Mdk/Jam3/Edn3/Itga1/Ednra/Pecam1/Xcl1/Nckap1l/Tnfaip6/Dysf/Irak4/Ptger3/Mpp1 | 14 |
| GO:0001963 | synaptic transmission, dopaminergic | BP | 0.167 | 0.00804 | 0.0355 | 1.45 | 0.0242 | Slc6a2/Aph1b/Drd4/Rasd2/Arrb2 | 5 |
| GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | BP | 0.167 | 0.00804 | 0.0355 | 1.45 | 0.0242 | Angpt1/Kdr/Tek/Vegfc/Emilin1 | 5 |
| GO:0048596 | embryonic camera-type eye morphogenesis | BP | 0.167 | 0.00804 | 0.0355 | 1.45 | 0.0242 | Stra6/Ihh/Tbx2/Zeb1/Hipk2 | 5 |
| GO:0060343 | trabecula formation | BP | 0.167 | 0.00804 | 0.0355 | 1.45 | 0.0242 | Thbs3/Mmp2/Tek/Adamts1/Enpp1 | 5 |
| GO:0001843 | neural tube closure | BP | 0.0982 | 0.00811 | 0.0358 | 1.45 | 0.0244 | Sfrp2/Nog/Zeb2/Lrp2/Ift57/Cecr2/Prickle1/Ptch1/Dlc1/Fzd2/Rab23 | 11 |
| GO:0018149 | peptide cross-linking | BP | 0.143 | 0.00819 | 0.0361 | 1.44 | 0.0246 | Spock2/Fn1/Egflam/Mamdc2/Ndnf/Bgn | 6 |
| GO:0045066 | regulatory T cell differentiation | BP | 0.143 | 0.00819 | 0.0361 | 1.44 | 0.0246 | Mdk/Cd44/Sox12/Gimap5/Gimap3/Il2rg | 6 |
| GO:0099174 | regulation of presynapse organization | BP | 0.143 | 0.00819 | 0.0361 | 1.44 | 0.0246 | Gpc6/Ptprd/Nlgn1/Lrrc4b/Slitrk3/Nlgn3 | 6 |
| GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Adra1a/Nos3/Ednrb/Enpep/Edn3/Rps6ka2/Agtr1a | 7 |
| GO:0010712 | regulation of collagen metabolic process | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Ihh/Ddr2/Fn1/Emilin1/Arrb2/Ccn2/Cygb | 7 |
| GO:0034113 | heterotypic cell-cell adhesion | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Itga4/Cd44/Itgb7/Jam3/Thy1/Cd200/Itga7 | 7 |
| GO:0046580 | negative regulation of Ras protein signal transduction | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Adra1a/Rasa3/Rasip1/Arap3/Heg1/Dlc1/Syngap1 | 7 |
| GO:0055010 | ventricular cardiac muscle tissue morphogenesis | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Col11a1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1 | 7 |
| GO:2001222 | regulation of neuron migration | BP | 0.127 | 0.00837 | 0.0367 | 1.44 | 0.025 | Ptprz1/Mdk/Tbc1d24/Gpr173/Neo1/Sema6a/Ntng1 | 7 |
| GO:0031109 | microtubule polymerization or depolymerization | BP | 0.0938 | 0.00838 | 0.0367 | 1.44 | 0.025 | Map2/Cdh5/Rp1/Tubb4a/Slain1/Nin/Dysf/Ttbk2/Mapre2/Cav1/Tppp/Map7d3 | 12 |
| GO:0033673 | negative regulation of kinase activity | BP | 0.0793 | 0.00844 | 0.0369 | 1.43 | 0.0251 | Sfrp2/Ddr2/Ptpro/Slc8a1/Mapk8ip1/Slc8a3/Pkia/Thy1/Ppm1f/Cdkn1c/Apoe/Rasip1/Ppm1e/Pkib/Heg1/Lats2/Smpd1/Cav1 | 18 |
| GO:0060606 | tube closure | BP | 0.0973 | 0.00865 | 0.0378 | 1.42 | 0.0257 | Sfrp2/Nog/Zeb2/Lrp2/Ift57/Cecr2/Prickle1/Ptch1/Dlc1/Fzd2/Rab23 | 11 |
| GO:0071774 | response to fibroblast growth factor | BP | 0.102 | 0.00874 | 0.0381 | 1.42 | 0.026 | Hhip/Nog/Tbx2/Cd44/Enpp1/Ndnf/Ccn2/Apln/Fat4/Kif16b | 10 |
| GO:0007018 | microtubule-based movement | BP | 0.0681 | 0.0089 | 0.0388 | 1.41 | 0.0264 | Apob/Map2/Fez1/Tub/Kif7/Myo5a/Wdr35/Adcy3/Cacna1e/Dnah5/Ift57/Pltp/Kif1a/Dlg2/Kif6/Dync2h1/Arl3/Ttc26/Dzip1/Sord/Septin4/Dnah7b/Atp2b4/Stard9/Dnah1/Dync2li1/Nsun7/Bbs1/Kif16b | 29 |
| GO:0062012 | regulation of small molecule metabolic process | BP | 0.0701 | 0.00901 | 0.0393 | 1.41 | 0.0267 | Apob/Igf1/Nos3/Acacb/Pth1r/Dkk3/Kit/Apoe/Enpp1/Kcnma1/Igf2/Snai1/Lacc1/Adcyap1r1/Htr2a/Zbtb20/Abcg1/Abcd2/Atp2b4/Fam3c/Slc7a7/Cav1/Nr1h3/Cacna1a/Pid1/Igfbp4 | 26 |
| GO:0010766 | negative regulation of sodium ion transport | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | Hecw2/Serpine2/Osr1/Atp1a2 | 4 |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0032252 | secretory granule localization | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | Map2/Myo5a/Ppfia2/Kif1a | 4 |
| GO:0045986 | negative regulation of smooth muscle contraction | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | Calca/Gucy1a1/Kcnma1/Calcrl | 4 |
| GO:0048557 | embryonic digestive tract morphogenesis | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | Ihh/Foxf1/Gli3/Tcf21 | 4 |
| GO:1902430 | negative regulation of amyloid-beta formation | BP | 0.2 | 0.00908 | 0.0394 | 1.4 | 0.0268 | Igf1/Rtn1/Apoe/Bin1 | 4 |
| GO:0030902 | hindbrain development | BP | 0.0843 | 0.0091 | 0.0395 | 1.4 | 0.0269 | Gli1/Lef1/Mdk/Nog/Igf1/Cbs/Dab1/B4galt2/Serpine2/Zfp423/Cplane1/Smad9/Ttbk2/Dlc1/Cacna1a | 15 |
| GO:0034331 | cell junction maintenance | BP | 0.14 | 0.00919 | 0.0397 | 1.4 | 0.027 | Epb41l3/Fermt2/Shank2/Erc2/Syngap1/Cldn1 | 6 |
| GO:0048873 | homeostasis of number of cells within a tissue | BP | 0.14 | 0.00919 | 0.0397 | 1.4 | 0.027 | Tex15/Col14a1/Nos3/Flt3/Coro1a/Akt3 | 6 |
| GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | BP | 0.14 | 0.00919 | 0.0397 | 1.4 | 0.027 | Cxcr3/Thy1/Aplnr/Ednra/Pkd2/Gstm7 | 6 |
| GO:0001974 | blood vessel remodeling | BP | 0.125 | 0.00922 | 0.0397 | 1.4 | 0.027 | Itga4/Igf1/Nos3/Cbs/Mef2c/Erg/Ednra | 7 |
| GO:2000649 | regulation of sodium ion transmembrane transporter activity | BP | 0.125 | 0.00922 | 0.0397 | 1.4 | 0.027 | Hecw2/Scn4b/Drd4/Fxyd5/Plcb1/Osr1/Atp1a2 | 7 |
| GO:0032272 | negative regulation of protein polymerization | BP | 0.107 | 0.00923 | 0.0397 | 1.4 | 0.027 | Kank4/Map2/Evl/Tmod2/Pfn2/Cdh5/Tubb4a/Pecam1/Scin | 9 |
| GO:0009743 | response to carbohydrate | BP | 0.0802 | 0.00925 | 0.0397 | 1.4 | 0.027 | Apob/Igf1/P2rx2/Ano1/Icam1/Cacna1e/Plcb1/Rps6ka2/Stxbp4/Rab34/Nox4/Enpp1/Arrb1/Sarm1/Zbtb20/Abcg1/Calcrl | 17 |
| GO:0010453 | regulation of cell fate commitment | BP | 0.161 | 0.00926 | 0.0397 | 1.4 | 0.027 | Sfrp2/Ar/Wnt11/Glis1/Loxl3 | 5 |
| GO:0021952 | central nervous system projection neuron axonogenesis | BP | 0.161 | 0.00926 | 0.0397 | 1.4 | 0.027 | Dcc/Zeb2/Tsku/Cdh11/Nin | 5 |
| GO:0033137 | negative regulation of peptidyl-serine phosphorylation | BP | 0.161 | 0.00926 | 0.0397 | 1.4 | 0.027 | Hgf/Ppm1f/Bdkrb2/Rassf2/Cav1 | 5 |
| GO:0061036 | positive regulation of cartilage development | BP | 0.161 | 0.00926 | 0.0397 | 1.4 | 0.027 | Mdk/Hoxd11/Pkdcc/Hoxa11/Gli3 | 5 |
| GO:0071624 | positive regulation of granulocyte chemotaxis | BP | 0.161 | 0.00926 | 0.0397 | 1.4 | 0.027 | Mdk/Ednra/Xcl1/Nckap1l/Dysf | 5 |
| GO:0043299 | leukocyte degranulation | BP | 0.101 | 0.00936 | 0.0401 | 1.4 | 0.0273 | Foxf1/Coro1a/Kit/Nckap1l/Myo1f/Cd84/Fgr/Lat/Rab27a/Clnk | 10 |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | BP | 0.0923 | 0.00943 | 0.0404 | 1.39 | 0.0275 | Sfrp2/Angpt1/Cd44/Hcls1/Pfn2/Prkd1/Tek/Caprin2/Arrb1/Arrb2/Atp2b4/Cav1 | 12 |
| GO:0062197 | cellular response to chemical stress | BP | 0.0714 | 0.00945 | 0.0404 | 1.39 | 0.0275 | Cyp1b1/Hgf/Ddr2/Nos3/Slc8a1/Lrrk2/Mmp2/Prkd1/Tbc1d24/Trpc6/Scn2a/Prr5l/Ednra/Aifm2/Bdkrb2/Dysf/Selenon/Net1/Mgat3/Fyn/Pkd2/Slc25a23/Cav1/Stx2 | 24 |
| GO:0042246 | tissue regeneration | BP | 0.114 | 0.00946 | 0.0404 | 1.39 | 0.0275 | Gap43/Mdk/Igf1/Hopx/Dysf/Selenon/Ptn/Large1 | 8 |
| GO:0050885 | neuromuscular process controlling balance | BP | 0.114 | 0.00946 | 0.0404 | 1.39 | 0.0275 | Ankfn1/Slc1a3/Jph4/Myo5a/Kcnma1/Foxs1/Dlg4/Cacna1a | 8 |
| GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | BP | 0.114 | 0.00946 | 0.0404 | 1.39 | 0.0275 | Cd83/Cd44/Loxl3/Xcl1/Nckap1l/Gimap5/Gimap3/Il2rg | 8 |
| GO:0002318 | myeloid progenitor cell differentiation | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Jam3/Flt3/Kit | 3 |
| GO:0003139 | secondary heart field specification | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Wnt11/Mef2c/Lrp2 | 3 |
| GO:0006171 | cAMP biosynthetic process | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Adcy3/Adcy4/Adcy7 | 3 |
| GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Gli1/Igf1/Zfp423 | 3 |
| GO:0030202 | heparin metabolic process | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Angpt1/Ednrb/Ednra | 3 |
| GO:0032815 | negative regulation of natural killer cell activation | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Havcr2/Fgr/Clnk | 3 |
| GO:0034454 | microtubule anchoring at centrosome | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Ninl/Nin/Cep19 | 3 |
| GO:0035701 | hematopoietic stem cell migration | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Jam3/Kit/Enpp1 | 3 |
| GO:0036302 | atrioventricular canal development | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Tbx2/Has2/Tbx3 | 3 |
| GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Ar/Igf1/Igfbp4 | 3 |
| GO:0060405 | regulation of penile erection | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Ar/Drd4/Ednrb | 3 |
| GO:0060600 | dichotomous subdivision of an epithelial terminal unit | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Nrp1/Areg/Plxnd1 | 3 |
| GO:0071372 | cellular response to follicle-stimulating hormone stimulus | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Epha8/Epha3/Ednra | 3 |
| GO:0072178 | nephric duct morphogenesis | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Gpc3/Epha7/Osr1 | 3 |
| GO:0086016 | AV node cell action potential | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Cacna1c/Scn4b/Ank2 | 3 |
| GO:0086027 | AV node cell to bundle of His cell signaling | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Cacna1c/Scn4b/Ank2 | 3 |
| GO:0099545 | trans-synaptic signaling by trans-synaptic complex | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Efnb3/Ptprd/Nlgn1 | 3 |
| GO:1902946 | protein localization to early endosome | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Nrp1/Msn/Mgat3 | 3 |
| GO:1903564 | regulation of protein localization to cilium | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Ccdc88a/Crocc/Dzip1 | 3 |
| GO:2000344 | positive regulation of acrosome reaction | BP | 0.273 | 0.00974 | 0.0409 | 1.39 | 0.0278 | Pla2g10/Plcb1/Cacna1h | 3 |
| GO:0051259 | protein complex oligomerization | BP | 0.0766 | 0.00985 | 0.0413 | 1.38 | 0.0281 | Kcnb2/Kcna6/Kcnd2/Kcnd3/Evl/Acacb/Bok/Grin2b/Ikzf4/Cth/Kcnc4/Pkd1/Mpp2/Nlrp1b/Aqp11/Kcng4/Pkd2/Steap4/Cldn1 | 19 |
| GO:0010469 | regulation of signaling receptor activity | BP | 0.0884 | 0.00998 | 0.0418 | 1.38 | 0.0285 | Begain/Nog/Nlgn1/Areg/Slc24a4/Nptx1/Mef2c/Cnrip1/Nlgn3/Nptxr/Prrt1/Cacng7/Dlg4 | 13 |
| GO:0051592 | response to calcium ion | BP | 0.0916 | 0.00999 | 0.0419 | 1.38 | 0.0285 | Nlgn1/Syt13/Cpne5/Mef2c/Rasgrp2/Syt7/Kcnma1/Asph/Pkd2/Slc25a23/Smpd1/Cav1 | 12 |
| GO:0007254 | JNK cascade | BP | 0.0833 | 0.01 | 0.042 | 1.38 | 0.0286 | Sfrp2/Eda2r/Lrrk2/Mapk8ip1/Plcb1/Cracr2a/Grik2/Hipk2/Rassf2/Mapkbp1/Irak4/Ccn2/Cdc42ep5/Dact1/Edar | 15 |
| GO:0008361 | regulation of cell size | BP | 0.0794 | 0.0101 | 0.0423 | 1.37 | 0.0288 | Map2/Sema5a/Sema5b/Slc12a8/Sema7a/Nrp1/Fn1/Cxcl12/Apoe/Kcnma1/Sema3g/Epha7/Aqp11/Slc12a4/Akt3/Sema6a/C1qtnf9 | 17 |
| GO:0030514 | negative regulation of BMP signaling pathway | BP | 0.123 | 0.0101 | 0.0424 | 1.37 | 0.0288 | Sfrp2/Nog/Lrp2/Tnfaip6/Hipk2/Cav1/Tmprss6 | 7 |
| GO:0008207 | C21-steroid hormone metabolic process | BP | 0.136 | 0.0103 | 0.0428 | 1.37 | 0.0291 | Ednrb/Dkk3/Kcnma1/Cyp46a1/Cacna1h/Stat5b | 6 |
| GO:0010464 | regulation of mesenchymal cell proliferation | BP | 0.136 | 0.0103 | 0.0428 | 1.37 | 0.0291 | Ihh/Wnt11/Kdr/Foxf1/Zeb1/Ptn | 6 |
| GO:0046640 | regulation of alpha-beta T cell proliferation | BP | 0.136 | 0.0103 | 0.0428 | 1.37 | 0.0291 | Cd44/Mapk8ip1/Btla/Xcl1/Zap70/Il12rb1 | 6 |
| GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | BP | 0.113 | 0.0103 | 0.0428 | 1.37 | 0.0291 | Drd4/Npr3/Akap12/Htr1b/Ednra/Gnaz/Palm/Gabbr1 | 8 |
| GO:0035710 | CD4-positive, alpha-beta T cell activation | BP | 0.0948 | 0.0104 | 0.0435 | 1.36 | 0.0296 | Lef1/Cd83/Cd44/Loxl3/Cracr2a/Xcl1/Nckap1l/Satb1/Gimap5/Gimap3/Il2rg | 11 |
| GO:0043244 | regulation of protein-containing complex disassembly | BP | 0.0909 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Map2/Sema5a/Evl/Tmod2/Rp1/Sh3bp1/Scin/Dysf/Asph/Ttbk2/Nes/Plekhh2 | 12 |
| GO:0030705 | cytoskeleton-dependent intracellular transport | BP | 0.0808 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Map2/Fez1/Tub/Ccdc88a/Myo5a/Wdr35/Ppfia2/Ift57/Kif1a/Dlg2/Dync2h1/Arl3/Ttc26/Myo1f/Dync2li1/Bbs1 | 16 |
| GO:0002825 | regulation of T-helper 1 type immune response | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Il27ra/H2-Ab1/Havcr2/Xcl1/Il12rb1 | 5 |
| GO:0003416 | endochondral bone growth | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Kdr/Ddr2/Evc/Thbs3/Stc1 | 5 |
| GO:0010259 | multicellular organism aging | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Rnf165/Adra1a/Enpp1/Agtr1a/Ednra | 5 |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Zeb2/Npnt/Cdkn1c/Hipk2/Itga8 | 5 |
| GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Cd83/Nckap1l/Gimap5/Gimap3/Il2rg | 5 |
| GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | BP | 0.156 | 0.0106 | 0.0439 | 1.36 | 0.0298 | Zeb2/Npnt/Cdkn1c/Hipk2/Itga8 | 5 |
| GO:0046785 | microtubule polymerization | BP | 0.105 | 0.0107 | 0.0442 | 1.35 | 0.0301 | Map2/Cdh5/Rp1/Tubb4a/Slain1/Nin/Cav1/Tppp/Map7d3 | 9 |
| GO:0002274 | myeloid leukocyte activation | BP | 0.0746 | 0.0108 | 0.0446 | 1.35 | 0.0304 | Pla2g10/Fn1/Lrrk2/Foxf1/Slc7a2/Dock2/Cd1d1/Kit/Cd226/Havcr2/Cd200/Dysf/Myo1f/Cd84/Fgr/Gimap5/Gimap3/Lat/Clnk/Nr1h3 | 20 |
| GO:0001946 | lymphangiogenesis | BP | 0.19 | 0.0108 | 0.0446 | 1.35 | 0.0304 | Vash1/Vegfc/Tie1/Pkd1 | 4 |
| GO:0032332 | positive regulation of chondrocyte differentiation | BP | 0.19 | 0.0108 | 0.0446 | 1.35 | 0.0304 | Hoxd11/Pkdcc/Hoxa11/Gli3 | 4 |
| GO:0097107 | postsynaptic density assembly | BP | 0.19 | 0.0108 | 0.0446 | 1.35 | 0.0304 | Ptprd/Lrrc4b/Nptx1/Slitrk3 | 4 |
| GO:2000353 | positive regulation of endothelial cell apoptotic process | BP | 0.19 | 0.0108 | 0.0446 | 1.35 | 0.0304 | Itga4/Cd248/Ecscr/Col18a1 | 4 |
| GO:0030879 | mammary gland development | BP | 0.0848 | 0.0109 | 0.0447 | 1.35 | 0.0304 | Ar/Lef1/Agap2/Igf1/Tbx2/Hoxd9/Areg/Foxf1/Gli3/Tbx3/Hoxa9/Ptch1/Stat5b/Cav1 | 14 |
| GO:0051260 | protein homooligomerization | BP | 0.0824 | 0.011 | 0.0454 | 1.34 | 0.0309 | Kcnb2/Kcna6/Kcnd2/Kcnd3/Evl/Acacb/Ikzf4/Cth/Kcnc4/Mpp2/Nlrp1b/Aqp11/Kcng4/Pkd2/Steap4 | 15 |
| GO:0019218 | regulation of steroid metabolic process | BP | 0.094 | 0.0111 | 0.0455 | 1.34 | 0.031 | Apob/Igf1/Dkk3/Kit/Apoe/Agtr1a/Kcnma1/Igf2/Snai1/Abcg1/Stat5b | 11 |
| GO:2001056 | positive regulation of cysteine-type endopeptidase activity | BP | 0.0872 | 0.0111 | 0.0456 | 1.34 | 0.031 | Bok/Grin2b/Wdr35/Ift57/Ppm1f/Hip1/Arrb1/Asph/Dlc1/Nlrp1b/Ccn2/Fyn/Cyfip2 | 13 |
| GO:0010633 | negative regulation of epithelial cell migration | BP | 0.111 | 0.0111 | 0.0456 | 1.34 | 0.031 | Evl/Mef2c/Pfn2/Stc1/Vash1/Ptprm/Apoe/Atp2b4 | 8 |
| GO:0055008 | cardiac muscle tissue morphogenesis | BP | 0.111 | 0.0111 | 0.0456 | 1.34 | 0.031 | Col11a1/Angpt1/Nog/Lrp2/Ccm2l/Ednra/Myl3/Heg1 | 8 |
| GO:0016050 | vesicle organization | BP | 0.0732 | 0.0113 | 0.0463 | 1.33 | 0.0315 | Syt13/Zeb2/Stx1b/Sec16b/Serpine2/Ap1m1/Rab34/Cadps/Srpx/Coro1a/Syt7/Prrt2/Dysf/Abcg1/Aqp11/Dlg4/Ccdc136/Rab27a/Arfgap3/Stx2/Nkd2 | 21 |
| GO:0006040 | amino sugar metabolic process | BP | 0.133 | 0.0114 | 0.0466 | 1.33 | 0.0317 | St6gal2/Npl/Chst2/Chst1/Mgat3/Uap1l1 | 6 |
| GO:0030225 | macrophage differentiation | BP | 0.133 | 0.0114 | 0.0466 | 1.33 | 0.0317 | Hcls1/Calca/Plcb1/Enpp1/Zbtb46/Nrros | 6 |
| GO:0086009 | membrane repolarization | BP | 0.133 | 0.0114 | 0.0466 | 1.33 | 0.0317 | Kcnd3/Scn4b/Slc24a4/Cacna2d1/Ank2/Cav1 | 6 |
| GO:1902991 | regulation of amyloid precursor protein catabolic process | BP | 0.133 | 0.0114 | 0.0466 | 1.33 | 0.0317 | Igf1/Rtn1/Slc2a13/Apoe/Bin1/Abcg1 | 6 |
| GO:0045995 | regulation of embryonic development | BP | 0.103 | 0.0115 | 0.0469 | 1.33 | 0.0319 | Sfrp2/Phldb2/Lama4/Osr2/Plcb1/Tgif2/Lama2/Osr1/Tada2a | 9 |
| GO:0043405 | regulation of MAP kinase activity | BP | 0.08 | 0.0116 | 0.0472 | 1.33 | 0.0321 | Sfrp2/Zeb2/Cbs/Drd4/Lrrk2/Mapk8ip1/Edn3/Pik3r6/Kit/Nox4/Apoe/Htr2a/Kitl/Pde5a/Smpd1/Cav1 | 16 |
| GO:0060538 | skeletal muscle organ development | BP | 0.078 | 0.012 | 0.0488 | 1.31 | 0.0332 | Stra6/Hoxd10/Ryr1/P2rx2/Hoxd9/Mef2c/Tcf21/Heyl/Igf2/Hdac9/Dysf/Selenon/Hlf/Large1/Cav1/Zfp689/Usp2 | 17 |
| GO:0061045 | negative regulation of wound healing | BP | 0.11 | 0.0121 | 0.049 | 1.31 | 0.0333 | Cd109/Tfpi/Abat/Phldb2/Serpine2/Apoe/Pros1/Ptger3 | 8 |
| GO:0050999 | regulation of nitric-oxide synthase activity | BP | 0.152 | 0.0121 | 0.049 | 1.31 | 0.0333 | Npr3/Tert/Apoe/Atp2b4/Cav1 | 5 |
| GO:1905314 | semi-lunar valve development | BP | 0.152 | 0.0121 | 0.049 | 1.31 | 0.0333 | Stra6/Nos3/Emilin1/Tie1/Heyl | 5 |
| GO:1901605 | alpha-amino acid metabolic process | BP | 0.0815 | 0.0121 | 0.0491 | 1.31 | 0.0334 | Aspg/Ddah1/Nos3/Cbs/Asrgl1/Gamt/Kyat1/Fah/Amdhd1/Cth/Phgdh/Nox4/Atp2b4/Asl/Slc7a7 | 15 |
| GO:1903793 | positive regulation of anion transport | BP | 0.119 | 0.0122 | 0.0493 | 1.31 | 0.0335 | Grik1/Pla2g10/Abat/Tcaf1/Grin2b/Abcb1a/Gabbr1 | 7 |
| GO:0051147 | regulation of muscle cell differentiation | BP | 0.0861 | 0.0123 | 0.0499 | 1.3 | 0.0339 | Igf1/Mef2c/Hopx/Tshz3/Zeb1/Kit/Cth/Igf2/Hdac9/Arrb2/Pi16/Rbpms2/Efemp2 | 13 |
| GO:1900180 | regulation of protein localization to nucleus | BP | 0.0861 | 0.0123 | 0.0499 | 1.3 | 0.0339 | Angpt1/Hcls1/Fermt2/Tert/Prkd1/Tek/Gli3/Pkia/Dclk2/Fyn/Rab23/Lats2/Cacnb4 | 13 |
| GO:0051604 | protein maturation | BP | 0.0698 | 0.0124 | 0.05 | 1.3 | 0.034 | Cpxm2/Ihh/Mmp16/Aph1b/Lrrk2/Serpine2/Gli3/Rps6ka2/Cpe/Mme/C1rl/Ldlrad3/Dync2h1/Pcsk5/C1ra/Ptch1/Asph/Tspan5/Ece2/Adam19/Arxes2/Naa11/Cyfip2/Nkd2 | 24 |
| GO:0097060 | synaptic membrane | CC | 0.174 | 7.95e-23 | 4.13e-20 | 19.4 | 2.76e-20 | Ptprz1/Efnb3/Kcnj3/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Slc6a2/Kcnd2/Dcc/Ptprd/Kcnd3/Grid2ip/Nlgn1/Ptpro/Rims3/Slc8a1/Stx1b/P2rx2/Nrp1/Cdh2/Lrrc4b/Grin2b/Shank2/Cacna1e/Ank2/Adgrl1/Slitrk3/Col13a1/Htr1b/Cpe/Syde1/Glrb/Gabra4/Dlg2/Scn2a/Nlgn3/Lin7a/Dgkb/Sorcs2/Grik2/Prrt1/Syt7/Kcnma1/Prrt2/Zc4h2/Hip1/Septin3/Nsg1/Arrb1/Epha7/Cacng7/Erc2/Pcdh17/Lrfn2/Mpp2/Cacna1h/Arrb2/Dlg4/Magee1/Palm/Ntng1/Stx2/Gabbr1/Slc4a8/Itga8/Sspn | 68 |
| GO:0062023 | collagen-containing extracellular matrix | CC | 0.159 | 3.96e-19 | 1.03e-16 | 16 | 6.88e-17 | Ndp/Gpc6/Col11a1/Ptprz1/Hmcn1/Vit/Angpt1/Emid1/Col14a1/Colec12/Igf1/Col4a6/Npnt/Thbs3/Lamc3/Spock2/Lama4/Cspg4/Tgfbi/Gpc3/Fn1/Col16a1/Nid1/Serpine2/Egflam/Angptl2/Adamts1/Col9a2/Frem1/Col13a1/Mamdc2/Postn/Smoc1/Loxl3/Mgp/Adamts10/Lama2/Matn2/Sparcl1/Tmeff2/Emilin1/Igf2/Col5a2/Col15a1/Ltbp3/Bgn/Adamtsl1/P3h1/Vwa1/Abi3bp/Dpt/Angptl4/Adam19/Egfl7/Efemp2/Plxdc2/Ptn/Col23a1/Ntng1/Gpc2/Adamts17/Col18a1/Itgb4 | 63 |
| GO:0045211 | postsynaptic membrane | CC | 0.176 | 1.21e-16 | 2.1e-14 | 13.7 | 1.4e-14 | Ptprz1/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Kcnd2/Dcc/Kcnd3/Grid2ip/Nlgn1/Ptpro/Slc8a1/Nrp1/Cdh2/Lrrc4b/Grin2b/Shank2/Cacna1e/Ank2/Slitrk3/Col13a1/Glrb/Gabra4/Dlg2/Nlgn3/Lin7a/Dgkb/Sorcs2/Grik2/Prrt1/Kcnma1/Prrt2/Zc4h2/Hip1/Nsg1/Arrb1/Epha7/Cacng7/Pcdh17/Lrfn2/Mpp2/Cacna1h/Arrb2/Dlg4/Magee1/Gabbr1/Sspn | 48 |
| GO:0098978 | glutamatergic synapse | CC | 0.158 | 1.35e-15 | 1.75e-13 | 12.8 | 1.17e-13 | Efnb3/Syn2/Grid1/Gria4/Lrrc4c/Begain/Kcnd2/Ptprd/Nlgn1/Ptpro/Drd4/Nrp1/Lrrc4b/Wasf3/Lrrk2/Nptx1/Myo5a/Pfn2/Grin2b/Ppfia2/Plcb1/Cadps/Dlg2/Gucy1a1/Scn2a/Coro1a/Abi2/Dgkb/Nptxr/Grik2/Apoe/Syt7/Prrt2/Sparcl1/Cdh11/Nsg1/Epha7/Cacng7/Erc2/Pcdh17/Mpp2/Cacna1h/Cpt1c/Dlg4/Syngap1/Ngef/Ntng1/Nrn1/Slc4a8/Cacna1a/Cacnb4 | 51 |
| GO:0098984 | neuron to neuron synapse | CC | 0.144 | 3.28e-14 | 3.41e-12 | 11.5 | 2.28e-12 | Gap43/Efnb3/Syn2/Map2/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Epb41l3/Lrrc7/Ptpro/Slc8a1/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Ank2/Adgrl1/Slc8a3/Slitrk3/Rps6kc1/Dlg2/Nrgn/Sorcs2/Grik2/Prrt1/Cap2/Arrb1/Pdzd2/Cacng7/Ptch1/Erc2/Lrfn2/Cnn3/Mpp2/Arrb2/Fyn/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Slc4a8/Itga8/Cpeb1 | 52 |
| GO:0032279 | asymmetric synapse | CC | 0.147 | 4.02e-14 | 3.41e-12 | 11.5 | 2.28e-12 | Gap43/Syn2/Map2/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Kcnd2/Dcc/Kcnd3/Epb41l3/Lrrc7/Ptpro/Slc8a1/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Ank2/Adgrl1/Slc8a3/Slitrk3/Rps6kc1/Dlg2/Nrgn/Sorcs2/Grik2/Prrt1/Cap2/Arrb1/Pdzd2/Cacng7/Ptch1/Erc2/Lrfn2/Cnn3/Mpp2/Arrb2/Fyn/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Slc4a8/Itga8/Cpeb1 | 50 |
| GO:0099572 | postsynaptic specialization | CC | 0.145 | 4.59e-14 | 3.41e-12 | 11.5 | 2.28e-12 | Gap43/Syn2/Map2/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Epb41l3/Lrrc7/Ptpro/Slc8a1/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Ank2/Adgrl1/Slc8a3/Slitrk3/Rps6kc1/Dlg2/Nlgn3/Nrgn/Sorcs2/Grik2/Prrt1/Cap2/Arrb1/Pdzd2/Cacng7/Ptch1/Erc2/Lrfn2/Cnn3/Mpp2/Arrb2/Fyn/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Itga8/Cpeb1 | 51 |
| GO:0014069 | postsynaptic density | CC | 0.148 | 5.57e-14 | 3.62e-12 | 11.4 | 2.42e-12 | Gap43/Syn2/Map2/Grid1/Gria4/Grik1/Cacna1c/Lrrc4c/Kcnd2/Dcc/Kcnd3/Epb41l3/Lrrc7/Ptpro/Slc8a1/P2rx2/Cdh2/Lrrc4b/Dab1/Grin2b/Shank2/Ank2/Adgrl1/Slc8a3/Slitrk3/Rps6kc1/Dlg2/Nrgn/Sorcs2/Grik2/Prrt1/Cap2/Arrb1/Pdzd2/Cacng7/Ptch1/Erc2/Lrfn2/Cnn3/Mpp2/Arrb2/Fyn/Dlg4/Palm/Dlg5/Syngap1/Arhgef9/Itga8/Cpeb1 | 49 |
| GO:0042734 | presynaptic membrane | CC | 0.212 | 2.51e-12 | 1.31e-10 | 9.88 | 8.73e-11 | Efnb3/Kcnj3/Grik1/Cacna1c/Slc6a2/Ptprd/Rims3/Stx1b/P2rx2/Cdh2/Grin2b/Cacna1e/Adgrl1/Htr1b/Scn2a/Grik2/Syt7/Kcnma1/Prrt2/Hip1/Septin3/Erc2/Pcdh17/Cacna1h/Ntng1/Stx2/Gabbr1/Slc4a8 | 28 |
| GO:0042383 | sarcolemma | CC | 0.188 | 5.06e-12 | 2.39e-10 | 9.62 | 1.6e-10 | Kcnj3/Car4/Sgcd/Cacna1c/Kcnd2/Ryr1/Adra1a/Nos3/Slc8a1/Cdh2/Scn1a/Cacna2d1/Ank2/Slc8a3/Msn/Scn2a/Lama2/Ednra/Itga7/Bgn/Bin1/Cacng7/Dysf/Ryr3/Flnc/Cacna1h/Atp2b4/Atp1a2/Sgcb/Cav1/Sspn | 31 |
| GO:0034703 | cation channel complex | CC | 0.161 | 1.2e-11 | 5.2e-10 | 9.28 | 3.47e-10 | Kcnb2/Kcnj3/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/Scn1a/Kcnq5/Cachd1/Cacna2d1/Grin2b/Cacna1e/Unc80/Trpc6/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ryr3/Cacna1h/Cpt1c/Kcng4/Pkd2/Scn3a/Dlg4/Nrn1/Kcnq4/Cacna1a/Cacnb4 | 36 |
| GO:0098857 | membrane microdomain | CC | 0.124 | 4.24e-11 | 1.47e-09 | 8.83 | 9.81e-10 | Angpt1/Cacna1c/Kcnd2/Dcc/Kdr/Kcnd3/Tfpi/Nos3/Rgs16/Gnai1/Cdh2/Ednrb/Lrrk2/Ano1/Icam1/Abcg2/Tek/Has2/Adcy3/Cdh13/Tnfrsf22/Gprc5b/Itga1/Lrp2/Cpe/Thy1/Plpp3/Mme/Sele/Kcnma1/Ednra/Pecam1/Adcyap1r1/Rtn4rl1/Cavin3/Dysf/Ptch1/Dlc1/Cacna1h/Cavin2/Fyn/Atp1a2/Zap70/Plpp1/Abcg3/Cav1/Stx2/Gabbr1/Lipe | 49 |
| GO:0034702 | ion channel complex | CC | 0.139 | 4.78e-11 | 1.55e-09 | 8.81 | 1.04e-09 | Kcnb2/Kcnj3/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/Scn1a/Kcnq5/Cachd1/Cacna2d1/Ano1/Grin2b/Cacna1e/Ano2/Unc80/Ttyh2/Trpc6/Glrb/Gabra4/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ryr3/Cacna1h/Cpt1c/Kcng4/Pkd2/Scn3a/Dlg4/Nrn1/Kcnq4/Cacna1a/Cacnb4 | 41 |
| GO:0099240 | intrinsic component of synaptic membrane | CC | 0.203 | 9.85e-11 | 3.01e-09 | 8.52 | 2.01e-09 | Efnb3/Kcnj3/Grid1/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Nlgn1/Ptpro/Nrp1/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Htr1b/Scn2a/Nlgn3/Prrt2/Pcdh17/Lrfn2/Mpp2/Cacna1h/Ntng1 | 25 |
| GO:0045121 | membrane raft | CC | 0.122 | 1.25e-10 | 3.63e-09 | 8.44 | 2.42e-09 | Angpt1/Cacna1c/Kcnd2/Dcc/Kdr/Kcnd3/Tfpi/Nos3/Rgs16/Gnai1/Cdh2/Ednrb/Lrrk2/Ano1/Icam1/Abcg2/Tek/Has2/Adcy3/Cdh13/Tnfrsf22/Gprc5b/Itga1/Lrp2/Cpe/Thy1/Plpp3/Mme/Sele/Kcnma1/Ednra/Pecam1/Adcyap1r1/Rtn4rl1/Cavin3/Ptch1/Dlc1/Cacna1h/Cavin2/Fyn/Atp1a2/Zap70/Plpp1/Abcg3/Cav1/Stx2/Gabbr1/Lipe | 48 |
| GO:0099699 | integral component of synaptic membrane | CC | 0.204 | 5.32e-10 | 1.46e-08 | 7.84 | 9.73e-09 | Efnb3/Kcnj3/Grid1/Lrrc4c/Kcnd2/Dcc/Ptprd/Kcnd3/Nlgn1/Ptpro/Nrp1/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Htr1b/Scn2a/Nlgn3/Prrt2/Pcdh17/Lrfn2/Cacna1h | 23 |
| GO:0005604 | basement membrane | CC | 0.193 | 1.55e-09 | 3.67e-08 | 7.44 | 2.45e-08 | Hmcn1/Col4a6/Npnt/Lamc3/Lama4/Tgfbi/Fn1/Nid1/Egflam/Adamts1/Frem1/Smoc1/Lama2/Matn2/Tmeff2/Col15a1/P3h1/Vwa1/Efemp2/Ptn/Ntng1/Col18a1/Itgb4 | 23 |
| GO:0016323 | basolateral plasma membrane | CC | 0.134 | 9.7e-09 | 2.19e-07 | 6.66 | 1.47e-07 | Slc1a3/Slc13a5/Lrrc7/Slc8a1/Cd44/Cdh2/Tek/Cldn8/Ank2/Pth1r/Plpp3/Dlg2/Msn/Slc29a1/Heph/Lin7a/Cd38/Agtr1a/Slc23a2/Arrb1/Pkd1/Slco4c1/Abca8b/Arrb2/Slc38a1/Atp2b4/Palm/Abcc5/Cav1/Stx2/Nkd2/S100g/Cldn1 | 33 |
| GO:0043197 | dendritic spine | CC | 0.146 | 1.04e-08 | 2.25e-07 | 6.65 | 1.5e-07 | Ptprz1/Slc1a3/Gria4/Kcnd2/Kcnd3/Grid2ip/Nlgn1/Lrrc7/Ptpro/Slc8a1/Drd4/P2rx2/Grin2b/Shank2/Ppfia2/Slc8a3/Dock10/Abi2/Nrgn/Sla/Arrb1/Cnn3/Mpp2/Arrb2/Dlg4/Palm/Atp1a2/Gabbr1/Itga8 | 29 |
| GO:0009925 | basal plasma membrane | CC | 0.127 | 1.52e-08 | 3.04e-07 | 6.52 | 2.03e-07 | Slc1a3/Slc13a5/Lrrc7/Slc8a1/Cd44/Cdh2/Tek/Cldn8/Ank2/Pth1r/Plpp3/Dlg2/Msn/Slc29a1/Heph/Lin7a/Cd38/Agtr1a/Slc23a2/Arrb1/Pkd1/Slco4c1/Abca8b/Arrb2/Slc38a1/Pkd2/Atp2b4/Palm/Abcc5/Cav1/Stx2/Nkd2/S100g/Itgb4/Cldn1 | 35 |
| GO:0019897 | extrinsic component of plasma membrane | CC | 0.151 | 1.75e-08 | 3.29e-07 | 6.48 | 2.2e-07 | Ryr1/Fermt2/Gnai1/Cdh2/Cdh6/Serpine2/Cdh13/Gngt2/Cdh10/Cdh5/Gng11/Sla/Apoe/Cdh11/Gng2/Hip1/Stac2/Dchs1/Gnaz/Fyn/Dlg4/Gng7/Fgr/Zap70/Gnb5/Styk1/Gnb4 | 27 |
| GO:0044309 | neuron spine | CC | 0.143 | 1.84e-08 | 3.29e-07 | 6.48 | 2.2e-07 | Ptprz1/Slc1a3/Gria4/Kcnd2/Kcnd3/Grid2ip/Nlgn1/Lrrc7/Ptpro/Slc8a1/Drd4/P2rx2/Grin2b/Shank2/Ppfia2/Slc8a3/Dock10/Abi2/Nrgn/Sla/Arrb1/Cnn3/Mpp2/Arrb2/Dlg4/Palm/Atp1a2/Gabbr1/Itga8 | 29 |
| GO:1902495 | transmembrane transporter complex | CC | 0.113 | 1.84e-08 | 3.29e-07 | 6.48 | 2.2e-07 | Kcnb2/Kcnj3/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/Scn1a/Kcnq5/Cachd1/Cacna2d1/Ano1/Grin2b/Cacna1e/Ano2/Unc80/Ttyh2/Trpc6/Glrb/Gabra4/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ryr3/Cacna1h/Cpt1c/Kcng4/Pkd2/Scn3a/Dlg4/Atp1a2/Nrn1/Kcnq4/Cacna1a/Cacnb4 | 42 |
| GO:0099634 | postsynaptic specialization membrane | CC | 0.212 | 2.06e-08 | 3.57e-07 | 6.45 | 2.38e-07 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Dlg2/Nlgn3/Prrt1/Cacng7/Lrfn2/Mpp2/Dlg4 | 18 |
| GO:0005901 | caveola | CC | 0.198 | 2.7e-08 | 4.53e-07 | 6.34 | 3.03e-07 | Cacna1c/Kcnd2/Kcnd3/Tfpi/Nos3/Lrrk2/Cdh13/Sele/Kcnma1/Adcyap1r1/Cavin3/Ptch1/Dlc1/Cacna1h/Cavin2/Atp1a2/Plpp1/Cav1/Lipe | 19 |
| GO:0098936 | intrinsic component of postsynaptic membrane | CC | 0.202 | 4.39e-08 | 6.92e-07 | 6.16 | 4.62e-07 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Nrp1/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Nlgn3/Pcdh17/Lrfn2/Mpp2/Cacna1h | 18 |
| GO:0030027 | lamellipodium | CC | 0.148 | 4.95e-08 | 7.57e-07 | 6.12 | 5.06e-07 | Ptprz1/Evl/Ccdc88a/Cd44/Cdh2/Wasf3/Abi3/Podxl/Plxnd1/Ptprm/Coro1a/Abi2/Sh3bp1/Palld/Carmil3/Arap3/Kitl/Parvb/Dysf/Pkd2/Fgd1/Unc5c/Dpysl3/Arhgap31/Stx2/Plekhh2 | 26 |
| GO:0099055 | integral component of postsynaptic membrane | CC | 0.2 | 1.23e-07 | 1.78e-06 | 5.75 | 1.19e-06 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Nrp1/Cdh2/Lrrc4b/Grin2b/Cacna1e/Slitrk3/Nlgn3/Pcdh17/Lrfn2/Cacna1h | 17 |
| GO:0030133 | transport vesicle | CC | 0.114 | 1.5e-07 | 2.05e-06 | 5.69 | 1.37e-06 | Syn2/Car4/Igf1/Syt13/Aph1b/Stx1b/Sec16b/Lrrk2/Myo5a/Grin2b/Mme/Kif1a/Dlg2/Lin7a/Nrgn/Dmxl2/Prrt1/Syt7/Lyz2/Prrt2/Kcnc4/Bgn/Bin1/Sv2c/Synrg/Lyz1/Septin4/Dlg4/Dpysl3/Brsk1/Rab27a/Stx2/Gabbr1/Slc4a8/Nkd2/Sspn | 36 |
| GO:0005938 | cell cortex | CC | 0.115 | 1.86e-07 | 2.49e-06 | 5.6 | 1.66e-06 | Hmcn1/Ryr1/Rims3/Phldb2/Drd4/Hcls1/Gnai1/Cdh2/Prkd1/Gypc/Exoc3l/Coro1a/Sh3bp1/Sele/Pard6g/Septin3/Rhobtb3/Erc2/BC034090/Pard3b/Asph/Myo1f/Dlc1/Cldn5/Ccn2/Pkd2/Septin4/Dlg4/Mpp1/Arhgef9/Cav1/Usp2/Plekhh2/Itgb4/Shroom1 | 35 |
| GO:0031253 | cell projection membrane | CC | 0.112 | 2.05e-07 | 2.67e-06 | 5.57 | 1.78e-06 | Gap43/Ptprz1/Car4/Map2/Evc2/Evc/Epb41l3/Cd44/Aif1l/Abcg2/Adcy3/Shank2/Enpep/Bbs7/Podxl/Lrp2/Pth1r/Thy1/Gabra4/Msn/Jcad/Kcnc4/Eps8l1/Tmem231/Mpp2/Dlc1/Ptger3/Pkd2/Fgr/Palm/Abcb1a/Bbs1/Abcg3/Gabbr1/Itga8/Plekho1 | 36 |
| GO:0098948 | intrinsic component of postsynaptic specialization membrane | CC | 0.222 | 4.05e-07 | 5.14e-06 | 5.29 | 3.43e-06 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Nlgn3/Lrfn2/Mpp2 | 14 |
| GO:0044853 | plasma membrane raft | CC | 0.154 | 4.42e-07 | 5.47e-06 | 5.26 | 3.66e-06 | Cacna1c/Kcnd2/Kcnd3/Tfpi/Nos3/Cdh2/Lrrk2/Has2/Cdh13/Sele/Kcnma1/Adcyap1r1/Cavin3/Ptch1/Dlc1/Cacna1h/Cavin2/Atp1a2/Plpp1/Cav1/Lipe | 21 |
| GO:0098839 | postsynaptic density membrane | CC | 0.217 | 1.43e-06 | 1.65e-05 | 4.78 | 1.1e-05 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Dlg2/Prrt1/Cacng7/Lrfn2/Mpp2/Dlg4 | 13 |
| GO:0099060 | integral component of postsynaptic specialization membrane | CC | 0.213 | 1.74e-06 | 1.97e-05 | 4.71 | 1.31e-05 | Grid1/Lrrc4c/Kcnd2/Dcc/Kcnd3/Nlgn1/Ptpro/Cdh2/Lrrc4b/Grin2b/Slitrk3/Nlgn3/Lrfn2 | 13 |
| GO:0098802 | plasma membrane signaling receptor complex | CC | 0.126 | 2.87e-06 | 3.18e-05 | 4.5 | 2.12e-05 | Gria4/Grik1/Itga4/Itgb7/Grin2b/Cd3g/Itga1/Itgal/Il27ra/Grik2/Emilin1/Itga7/Cacng7/Csf2rb/Cpt1c/Calcrl/Dlg4/Zap70/Apbb1ip/Il12rb1/Nrn1/Gabbr1/Itga8/Itgb4 | 24 |
| GO:0005912 | adherens junction | CC | 0.141 | 3.58e-06 | 3.88e-05 | 4.41 | 2.59e-05 | Hmcn1/Fermt2/Cdh2/Cdh6/Cdh10/Cdh5/Ptprm/Msn/Abi2/Lin7a/Esam/Jcad/Sh3bp1/Cdh11/Frmd4b/Pard3b/Dchs1/Frmd4a/Dlg5/Shroom1 | 20 |
| GO:0005614 | interstitial matrix | CC | 0.412 | 3.82e-06 | 4.05e-05 | 4.39 | 2.7e-05 | Vit/Col14a1/Igf1/Egflam/Mamdc2/Vwa1/Abi3bp | 7 |
| GO:0030055 | cell-substrate junction | CC | 0.126 | 4.56e-06 | 4.74e-05 | 4.32 | 3.17e-05 | Hmcn1/Evl/Phldb2/Nrp1/Fermt2/Aif1l/Itgb7/Thsd1/Itga1/Tnfsf13b/Msn/Nox4/Sla/Palld/Parvg/Itga7/Efs/Parvb/Tln2/Dlc1/Mapre2/Cav1/Itgb4 | 23 |
| GO:0016324 | apical plasma membrane | CC | 0.0976 | 5.91e-06 | 5.91e-05 | 4.23 | 3.95e-05 | Car4/Ddr2/Ptpro/Cd44/Mpdz/P2rx2/Cdh2/Fn1/Slc24a4/Ano1/Stc1/Abcg2/Tek/Shank2/Enpep/Podxl/Lrp2/Pth1r/Thy1/Msn/Slc29a1/Nox4/Kcnma1/Pard6g/Slc23a2/BC034090/Pard3b/Slc38a1/Plpp1/Abcb1a/Rab27a/Abcc5/Cav1/S100g/Cldn1/Shroom1 | 36 |
| GO:0098982 | GABA-ergic synapse | CC | 0.188 | 7.42e-06 | 7.28e-05 | 4.14 | 4.86e-05 | Kcnd2/Kcnd3/Ptpro/Plcb1/Slitrk3/Glrb/Gucy1a1/Epha3/Syt7/Erc2/Pcdh17/Arhgef9/Gabbr1 | 13 |
| GO:0070382 | exocytic vesicle | CC | 0.114 | 7.58e-06 | 7.3e-05 | 4.14 | 4.87e-05 | Syn2/Igf1/Syt13/Stx1b/Lrrk2/Myo5a/Grin2b/Mme/Kif1a/Dlg2/Dmxl2/Prrt1/Syt7/Prrt2/Kcnc4/Bin1/Sv2c/Septin4/Dlg4/Dpysl3/Brsk1/Rab27a/Stx2/Gabbr1/Slc4a8/Nkd2 | 26 |
| GO:0030426 | growth cone | CC | 0.118 | 9.09e-06 | 8.59e-05 | 4.07 | 5.74e-05 | Ptprz1/Map2/Fez1/Itga4/Dcc/Nrp1/Tmod2/Lrrk2/Shank2/Mapk8ip1/Adgrl1/Lrp2/Slc2a13/Thy1/Tshz3/Myo9a/Nin/Ptch1/Erc2/Neo1/Pcdhgb1/Unc5c/Dpysl3/Cpeb1 | 24 |
| GO:0098889 | intrinsic component of presynaptic membrane | CC | 0.216 | 9.6e-06 | 8.91e-05 | 4.05 | 5.95e-05 | Efnb3/Kcnj3/Ptprd/Cdh2/Cacna1e/Htr1b/Scn2a/Prrt2/Pcdh17/Cacna1h/Ntng1 | 11 |
| GO:0034706 | sodium channel complex | CC | 0.296 | 1.32e-05 | 0.00012 | 3.92 | 8.04e-05 | Gria4/Grik1/Scn4b/Scn1a/Scn2a/Grik2/Cacna1h/Scn3a | 8 |
| GO:0098562 | cytoplasmic side of membrane | CC | 0.118 | 1.44e-05 | 0.000129 | 3.89 | 8.63e-05 | Ryr1/Fermt2/Gnai1/Lrrk2/Rasa3/Gngt2/Msn/Gng11/Kit/Sla/Gng2/Hip1/Stac2/Gnaz/Fyn/Pkd2/Dlg4/Gng7/Fgr/Zap70/Gnb5/Styk1/Gnb4 | 23 |
| GO:0099056 | integral component of presynaptic membrane | CC | 0.227 | 1.49e-05 | 0.000131 | 3.88 | 8.77e-05 | Efnb3/Kcnj3/Ptprd/Cdh2/Cacna1e/Htr1b/Scn2a/Prrt2/Pcdh17/Cacna1h | 10 |
| GO:0005925 | focal adhesion | CC | 0.124 | 1.51e-05 | 0.000131 | 3.88 | 8.77e-05 | Evl/Phldb2/Nrp1/Fermt2/Aif1l/Itgb7/Thsd1/Itga1/Tnfsf13b/Msn/Nox4/Sla/Palld/Parvg/Efs/Parvb/Tln2/Dlc1/Mapre2/Cav1/Itgb4 | 21 |
| GO:0099568 | cytoplasmic region | CC | 0.111 | 1.9e-05 | 0.000158 | 3.8 | 0.000105 | Gli1/Map2/Rims3/Phldb2/Gnai1/Dzip1l/Arl6/Lrrk2/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Rp1/Grik2/Tubb4a/Dync2h1/Arl3/Erc2/Pkd2/Dlg4/Dnah7b/Dnah1/Dync2li1/Bbs1 | 25 |
| GO:0034705 | potassium channel complex | CC | 0.157 | 2.89e-05 | 0.000234 | 3.63 | 0.000156 | Kcnb2/Kcnj3/Gria4/Grik1/Kcna6/Kcnd2/Kcnd3/Kcnq5/Grik2/Kcnma1/Kcnc4/Kcnip3/Kcng4/Kcnq4 | 14 |
| GO:0043679 | axon terminus | CC | 0.116 | 2.92e-05 | 0.000234 | 3.63 | 0.000156 | Gria4/Grik1/Kcna6/Slc8a1/Drd4/P2rx2/Lrrk2/Pfn2/Grin2b/Mical1/Slc8a3/Calca/Grik2/Syt7/Kcnma1/Prrt2/Kcnc4/Bin1/Kcnip3/Erc2/Septin4/Slc4a8 | 22 |
| GO:0009898 | cytoplasmic side of plasma membrane | CC | 0.119 | 3.06e-05 | 0.000241 | 3.62 | 0.000161 | Ryr1/Fermt2/Gnai1/Rasa3/Gngt2/Msn/Gng11/Kit/Sla/Gng2/Hip1/Stac2/Gnaz/Fyn/Dlg4/Gng7/Fgr/Zap70/Gnb5/Styk1/Gnb4 | 21 |
| GO:0099146 | intrinsic component of postsynaptic density membrane | CC | 0.22 | 5.35e-05 | 0.000415 | 3.38 | 0.000277 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Lrfn2/Mpp2 | 9 |
| GO:0098685 | Schaffer collateral - CA1 synapse | CC | 0.157 | 5.78e-05 | 0.000442 | 3.35 | 0.000295 | Syn2/Lrrc4c/Dcc/Ptprd/Pfn2/Akap12/Dgkb/Cdh11/Lrfn2/Mpp2/Fyn/Ntng1/Gabbr1 | 13 |
| GO:0008305 | integrin complex | CC | 0.235 | 8.22e-05 | 0.000602 | 3.22 | 0.000402 | Itga4/Itgb7/Itga1/Itgal/Emilin1/Itga7/Itga8/Itgb4 | 8 |
| GO:0043296 | apical junction complex | CC | 0.117 | 9.12e-05 | 0.000658 | 3.18 | 0.00044 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Lin7a/Esam/Sh3bp1/Nfasc/Magi2/Adcyap1r1/Frmd4b/Pard6g/Pard3b/Cldn5/Frmd4a/Nphp1/Cldn1/Shroom1 | 19 |
| GO:0036064 | ciliary basal body | CC | 0.113 | 9.32e-05 | 0.000664 | 3.18 | 0.000444 | Evc/Ccdc88a/Kif7/Dzip1l/Wdr35/Cibar1/Bbs7/Armc9/Ift57/Rilpl1/Arl3/Cfap410/Ttc26/Ttbk2/Dzip1/Cep19/Pkd2/Dync2li1/Dlg5/Bbs1 | 20 |
| GO:0005834 | heterotrimeric G-protein complex | CC | 0.229 | 0.000103 | 0.000718 | 3.14 | 0.000479 | Gnai1/Gngt2/Gng11/Gng2/Gnaz/Gng7/Gnb5/Gnb4 | 8 |
| GO:0070160 | tight junction | CC | 0.119 | 0.000103 | 0.000718 | 3.14 | 0.000479 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Lin7a/Esam/Sh3bp1/Nfasc/Magi2/Adcyap1r1/Frmd4b/Pard6g/Pard3b/Cldn5/Frmd4a/Nphp1/Cldn1 | 18 |
| GO:0005923 | bicellular tight junction | CC | 0.123 | 0.000106 | 0.000727 | 3.14 | 0.000485 | Clmp/Mpdz/Jam3/Cldn8/Cdh5/Lin7a/Esam/Sh3bp1/Magi2/Adcyap1r1/Frmd4b/Pard6g/Pard3b/Cldn5/Frmd4a/Nphp1/Cldn1 | 17 |
| GO:0001726 | ruffle | CC | 0.118 | 0.000123 | 0.000818 | 3.09 | 0.000546 | Ptprz1/Map2/Cspg4/Aif1l/Myo5a/Podxl/Sh2b2/Rab34/Jcad/Eps8l1/Pecam1/Frmd4b/Arap3/Tln2/Dlc1/Fgd1/Fgr/Plekho1 | 18 |
| GO:0043195 | terminal bouton | CC | 0.144 | 0.000136 | 0.000892 | 3.05 | 0.000596 | Gria4/Grik1/Drd4/P2rx2/Lrrk2/Pfn2/Grin2b/Mical1/Calca/Grik2/Kcnma1/Erc2/Slc4a8 | 13 |
| GO:0034704 | calcium channel complex | CC | 0.164 | 0.000137 | 0.000892 | 3.05 | 0.000596 | Cacna1c/Ryr1/Cachd1/Cacna2d1/Cacna1e/Pkd1/Cacng7/Ryr3/Cacna1h/Cacna1a/Cacnb4 | 11 |
| GO:0031594 | neuromuscular junction | CC | 0.143 | 0.000152 | 0.000964 | 3.02 | 0.000644 | Pdzrn3/Stx1b/Lama4/Serpine2/Tbc1d24/Slc8a3/Postn/Dlg2/Lama2/Kcnc4/Itga7/Dlg4/Ptn | 13 |
| GO:1905360 | GTPase complex | CC | 0.216 | 0.000156 | 0.000976 | 3.01 | 0.000652 | Gnai1/Gngt2/Gng11/Gng2/Gnaz/Gng7/Gnb5/Gnb4 | 8 |
| GO:0032838 | plasma membrane bounded cell projection cytoplasm | CC | 0.109 | 0.000159 | 0.000984 | 3.01 | 0.000657 | Gli1/Map2/Dzip1l/Arl6/Lrrk2/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Rp1/Grik2/Tubb4a/Dync2h1/Arl3/Dlg4/Dnah7b/Dnah1/Dync2li1/Bbs1 | 20 |
| GO:0097731 | 9+0 non-motile cilium | CC | 0.123 | 0.000169 | 0.00103 | 2.99 | 0.000685 | D630045J12Rik/Slc24a4/Myo5a/Tmem237/Shank2/Bbs7/Cerkl/Ift57/Rp1/Arl3/Cfap410/Iqcb1/Nphp1/Gnb5/Rab27a/C130074G19Rik | 16 |
| GO:0097733 | photoreceptor cell cilium | CC | 0.128 | 0.00017 | 0.00103 | 2.99 | 0.000685 | D630045J12Rik/Slc24a4/Myo5a/Tmem237/Shank2/Bbs7/Cerkl/Ift57/Rp1/Arl3/Cfap410/Iqcb1/Nphp1/Gnb5/Rab27a | 15 |
| GO:0099061 | integral component of postsynaptic density membrane | CC | 0.205 | 0.00023 | 0.00136 | 2.87 | 0.000907 | Grid1/Lrrc4c/Dcc/Ptpro/Lrrc4b/Grin2b/Slitrk3/Lrfn2 | 8 |
| GO:0008021 | synaptic vesicle | CC | 0.102 | 0.000252 | 0.00147 | 2.83 | 0.000985 | Syn2/Stx1b/Lrrk2/Myo5a/Grin2b/Mme/Kif1a/Dlg2/Dmxl2/Prrt1/Syt7/Prrt2/Kcnc4/Bin1/Sv2c/Septin4/Dlg4/Brsk1/Stx2/Gabbr1/Slc4a8 | 21 |
| GO:0016342 | catenin complex | CC | 0.226 | 0.000302 | 0.00175 | 2.76 | 0.00117 | Cdh2/Cdh6/Cdh13/Cdh10/Cdh5/Cdh11/Dchs1 | 7 |
| GO:0097730 | non-motile cilium | CC | 0.108 | 0.000367 | 0.00208 | 2.68 | 0.00139 | D630045J12Rik/Slc24a4/Myo5a/Tmem237/Shank2/Bbs7/Cerkl/Ano2/Ift57/Rp1/Arl3/Cfap410/Pkd2/Iqcb1/Nphp1/Gnb5/Rab27a/C130074G19Rik | 18 |
| GO:0008076 | voltage-gated potassium channel complex | CC | 0.147 | 0.000381 | 0.00213 | 2.67 | 0.00142 | Kcnb2/Kcnj3/Kcna6/Kcnd2/Kcnd3/Kcnq5/Kcnma1/Kcnc4/Kcnip3/Kcng4/Kcnq4 | 11 |
| GO:0097014 | ciliary plasm | CC | 0.113 | 0.00043 | 0.00238 | 2.62 | 0.00159 | Gli1/Dzip1l/Arl6/Wdr35/Bbs7/Dnah5/Gli3/Ift57/Rp1/Tubb4a/Dync2h1/Arl3/Dnah7b/Dnah1/Dync2li1/Bbs1 | 16 |
| GO:0005874 | microtubule | CC | 0.0799 | 0.000434 | 0.00238 | 2.62 | 0.00159 | Map2/Fez1/Capn6/Clmp/Slc8a1/Kif7/Cep170/Dnah5/Klhl22/Slc8a3/Kif26a/Kif1a/Reep1/Eml1/Map9/Tubb4a/Apoe/Kif6/Dync2h1/Katnal1/Slain1/Ninl/Nin/Arl3/Sarm1/Dysf/Stard9/Dnah1/Dync2li1/Lrrc49/Tcp11l1/Mapre2/Tppp/Kif16b/Shroom1 | 35 |
| GO:0098791 | Golgi apparatus subcompartment | CC | 0.0879 | 0.000446 | 0.0024 | 2.62 | 0.0016 | B4galnt4/Car4/Nsg2/Chst2/Tgfbi/Bok/Lrrk2/Slc9a7/Gga2/Prkd1/Ap1m1/Postn/Rab34/Plpp3/Mme/Chst1/Cracr2a/Ggta1/Lyz2/Dennd5a/Pcsk5/Ms4a7/Nsg1/5730455P16Rik/Lyz1/Ece2/Xylt1 | 27 |
| GO:0098858 | actin-based cell projection | CC | 0.0961 | 0.000448 | 0.0024 | 2.62 | 0.0016 | Gap43/Ptprz1/Map2/Angpt1/Nlgn1/Lrrc7/Cd44/Lrrk2/Myo5a/Tek/Jam3/Podxl/Msn/Coro1a/Abi2/Nfasc/Lyz2/Kitl/Myo1f/Lyz1/Unc5c/Palm | 22 |
| GO:0005891 | voltage-gated calcium channel complex | CC | 0.186 | 0.000464 | 0.00246 | 2.61 | 0.00165 | Cacna1c/Cachd1/Cacna2d1/Cacna1e/Cacng7/Cacna1h/Cacna1a/Cacnb4 | 8 |
| GO:0008328 | ionotropic glutamate receptor complex | CC | 0.182 | 0.000546 | 0.00284 | 2.55 | 0.0019 | Gria4/Grik1/Grin2b/Grik2/Cacng7/Cpt1c/Dlg4/Nrn1 | 8 |
| GO:0016327 | apicolateral plasma membrane | CC | 0.24 | 0.000578 | 0.00298 | 2.53 | 0.00199 | Mpdz/Cdh2/Cldn8/Prickle2/Cldn5/Palm | 6 |
| GO:0001518 | voltage-gated sodium channel complex | CC | 0.294 | 0.000614 | 0.00313 | 2.5 | 0.00209 | Scn4b/Scn1a/Scn2a/Cacna1h/Scn3a | 5 |
| GO:0043292 | contractile fiber | CC | 0.0928 | 0.000712 | 0.00359 | 2.44 | 0.0024 | Cacna1c/Ryr1/Adra1a/Npnt/Slc8a1/Pygm/Tmod2/Scn1a/Mmp2/Mef2c/Grin2b/Arhgef25/Ank2/Palld/Pecam1/Myl3/Cmya5/Bin1/Ryr3/Atp2b4/Habp4/Simc1 | 22 |
| GO:0098878 | neurotransmitter receptor complex | CC | 0.174 | 0.000745 | 0.00369 | 2.43 | 0.00246 | Gria4/Grik1/Grin2b/Grik2/Cacng7/Cpt1c/Dlg4/Nrn1 | 8 |
| GO:0030863 | cortical cytoskeleton | CC | 0.115 | 0.000859 | 0.0042 | 2.38 | 0.0028 | Rims3/Hcls1/Cdh2/Gypc/Coro1a/Sele/Erc2/Myo1f/Dlc1/Cldn5/Dlg4/Mpp1/Plekhh2/Shroom1 | 14 |
| GO:0032391 | photoreceptor connecting cilium | CC | 0.17 | 0.000864 | 0.0042 | 2.38 | 0.0028 | D630045J12Rik/Tmem237/Ift57/Rp1/Arl3/Cfap410/Iqcb1/Nphp1 | 8 |
| GO:0048787 | presynaptic active zone membrane | CC | 0.214 | 0.0011 | 0.00525 | 2.28 | 0.00351 | Stx1b/Cdh2/Kcnma1/Cacna1h/Ntng1/Stx2 | 6 |
| GO:0016528 | sarcoplasm | CC | 0.129 | 0.00112 | 0.00529 | 2.28 | 0.00353 | Car4/Sgcd/Ryr1/Jph4/Pygm/Cacna2d1/Cmya5/Asph/Ryr3/Flnc/Gstm7 | 11 |
| GO:0030175 | filopodium | CC | 0.121 | 0.00122 | 0.00571 | 2.24 | 0.00381 | Gap43/Ptprz1/Map2/Nlgn1/Lrrc7/Myo5a/Podxl/Msn/Abi2/Kitl/Unc5c/Palm | 12 |
| GO:0042613 | MHC class II protein complex | CC | 0.333 | 0.00133 | 0.00616 | 2.21 | 0.00412 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0000137 | Golgi cis cisterna | CC | 0.308 | 0.00185 | 0.00852 | 2.07 | 0.00569 | Nsg2/Lyz2/Lyz1/Xylt1 | 4 |
| GO:0031045 | dense core granule | CC | 0.194 | 0.00193 | 0.00876 | 2.06 | 0.00585 | Cpe/Kif1a/Cadps/Dmxl2/Syt7/Ncs1 | 6 |
| GO:0030016 | myofibril | CC | 0.0893 | 0.00194 | 0.00876 | 2.06 | 0.00585 | Cacna1c/Ryr1/Adra1a/Slc8a1/Pygm/Tmod2/Scn1a/Mmp2/Mef2c/Grin2b/Arhgef25/Ank2/Palld/Myl3/Cmya5/Bin1/Ryr3/Atp2b4/Habp4/Simc1 | 20 |
| GO:0005884 | actin filament | CC | 0.105 | 0.00198 | 0.00888 | 2.05 | 0.00593 | Hcls1/Aif1l/Myo5a/Tek/Jam3/Sh2b2/Coro1a/Abi2/Palld/Myo1f/Dlc1/Fyn/Wipf1/Dpysl3 | 14 |
| GO:0005802 | trans-Golgi network | CC | 0.0854 | 0.00261 | 0.0116 | 1.93 | 0.00776 | Car4/Nsg2/Chst2/Tgfbi/Bok/Lrrk2/Slc9a7/Gga2/Prkd1/Ap1m1/Postn/Plpp3/Mme/Chst1/Cracr2a/Dennd5a/Pcsk5/Ms4a7/Nsg1/5730455P16Rik/Ece2 | 21 |
| GO:0044295 | axonal growth cone | CC | 0.159 | 0.0027 | 0.0118 | 1.93 | 0.00787 | Dcc/Mapk8ip1/Lrp2/Myo9a/Nin/Ptch1/Neo1 | 7 |
| GO:0060170 | ciliary membrane | CC | 0.14 | 0.0031 | 0.0133 | 1.88 | 0.00889 | Evc2/Evc/Adcy3/Shank2/Bbs7/Tmem231/Pkd2/Bbs1 | 8 |
| GO:0005814 | centriole | CC | 0.102 | 0.0036 | 0.0153 | 1.81 | 0.0102 | Ccdc88a/Dzip1l/Cibar1/Cep170/Cntln/Armc9/Crocc/Nin/Ttbk2/Dzip1/Cep19/Stard9/Cep97 | 13 |
| GO:0005790 | smooth endoplasmic reticulum | CC | 0.171 | 0.00366 | 0.0155 | 1.81 | 0.0103 | Map2/Ryr1/Jph4/Rtn1/Myo5a/Ryr3 | 6 |
| GO:0046658 | anchored component of plasma membrane | CC | 0.125 | 0.00391 | 0.0164 | 1.79 | 0.0109 | Gpc6/Car4/Gpc3/Thy1/Rtn4rl1/Mpp2/Ntng1/Gpc2/Nrn1 | 9 |
| GO:0019867 | outer membrane | CC | 0.0861 | 0.00464 | 0.019 | 1.72 | 0.0127 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx6/Armcx1/Agtr1a/Snn/Aifm2/Sarm1/Syne3/Cpt1c/Armcx2/Asl/Gimap3/Acsl1 | 18 |
| GO:0031968 | organelle outer membrane | CC | 0.0861 | 0.00464 | 0.019 | 1.72 | 0.0127 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx6/Armcx1/Agtr1a/Snn/Aifm2/Sarm1/Syne3/Cpt1c/Armcx2/Asl/Gimap3/Acsl1 | 18 |
| GO:0031985 | Golgi cisterna | CC | 0.131 | 0.00474 | 0.0191 | 1.72 | 0.0128 | B4galnt4/Nsg2/Rab34/Ggta1/Lyz2/Pcsk5/Lyz1/Xylt1 | 8 |
| GO:0032809 | neuronal cell body membrane | CC | 0.158 | 0.00558 | 0.0222 | 1.65 | 0.0148 | Kcnb2/Slc6a2/Kcnd2/Thy1/Kcnc4/Slc4a8 | 6 |
| GO:0016529 | sarcoplasmic reticulum | CC | 0.118 | 0.00561 | 0.0222 | 1.65 | 0.0148 | Car4/Sgcd/Ryr1/Pygm/Cacna2d1/Cmya5/Asph/Ryr3/Gstm7 | 9 |
| GO:0030667 | secretory granule membrane | CC | 0.111 | 0.00568 | 0.0222 | 1.65 | 0.0148 | Car4/Slc2a3/Cpe/Kif1a/Msn/Cd38/Pla1a/Ccdc136/Rab27a/Cav1 | 10 |
| GO:0044291 | cell-cell contact zone | CC | 0.111 | 0.00568 | 0.0222 | 1.65 | 0.0148 | Slc8a1/Scn4b/Cdh2/Scn1a/Jam3/Ank2/Scn2a/Pecam1/Tln2/Atp1a2 | 10 |
| GO:0005902 | microvillus | CC | 0.105 | 0.00597 | 0.0232 | 1.63 | 0.0155 | Angpt1/Cd44/Lrrk2/Tek/Jam3/Podxl/Msn/Nfasc/Lyz2/Myo1f/Lyz1 | 11 |
| GO:0043083 | synaptic cleft | CC | 0.222 | 0.00666 | 0.0257 | 1.59 | 0.0171 | Lama4/Grin2b/Lama2/Apoe | 4 |
| GO:0032281 | AMPA glutamate receptor complex | CC | 0.172 | 0.00765 | 0.029 | 1.54 | 0.0194 | Gria4/Cacng7/Cpt1c/Dlg4/Nrn1 | 5 |
| GO:0099569 | presynaptic cytoskeleton | CC | 0.3 | 0.00781 | 0.029 | 1.54 | 0.0194 | Rims3/Septin3/Erc2 | 3 |
| GO:1990454 | L-type voltage-gated calcium channel complex | CC | 0.3 | 0.00781 | 0.029 | 1.54 | 0.0194 | Cacna1c/Cacna2d1/Cacng7 | 3 |
| GO:0031526 | brush border membrane | CC | 0.112 | 0.00783 | 0.029 | 1.54 | 0.0194 | Car4/Abcg2/Shank2/Enpep/Lrp2/Pth1r/Ptger3/Abcb1a/Abcg3 | 9 |
| GO:0098644 | complex of collagen trimers | CC | 0.211 | 0.00816 | 0.0293 | 1.53 | 0.0196 | Col11a1/Col4a6/Col16a1/Col5a2 | 4 |
| GO:0005741 | mitochondrial outer membrane | CC | 0.0851 | 0.00817 | 0.0293 | 1.53 | 0.0196 | Acsl6/Maob/Bok/Lrrk2/Maoa/Slc8a3/Armcx6/Armcx1/Snn/Aifm2/Sarm1/Cpt1c/Armcx2/Asl/Gimap3/Acsl1 | 16 |
| GO:0014704 | intercalated disc | CC | 0.119 | 0.00837 | 0.0298 | 1.53 | 0.0199 | Slc8a1/Scn4b/Cdh2/Scn1a/Ank2/Scn2a/Tln2/Atp1a2 | 8 |
| GO:0016010 | dystrophin-associated glycoprotein complex | CC | 0.2 | 0.00985 | 0.0348 | 1.46 | 0.0233 | Sgcd/Sntb1/Sgcb/Sspn | 4 |
| GO:0005583 | fibrillar collagen trimer | CC | 0.273 | 0.0104 | 0.0363 | 1.44 | 0.0242 | Col11a1/Col16a1/Col5a2 | 3 |
| GO:0098643 | banded collagen fibril | CC | 0.273 | 0.0104 | 0.0363 | 1.44 | 0.0242 | Col11a1/Col16a1/Col5a2 | 3 |
| GO:0005903 | brush border | CC | 0.0897 | 0.0108 | 0.037 | 1.43 | 0.0247 | Car4/Dab1/Abcg2/Shank2/Enpep/Lrp2/Pth1r/Mme/Scart1/Scin/Ptger3/Abcb1a/Abcg3 | 13 |
| GO:0032589 | neuron projection membrane | CC | 0.111 | 0.0127 | 0.0429 | 1.37 | 0.0287 | Epb41l3/Thy1/Gabra4/Kcnc4/Mpp2/Palm/Gabbr1/Itga8 | 8 |
| GO:0031225 | anchored component of membrane | CC | 0.0829 | 0.0128 | 0.0431 | 1.37 | 0.0288 | Gpc6/Cntn3/Car4/Cd109/Sema7a/Gpc3/Ntm/Cdh13/Thy1/Rtn4rl1/Mmp19/Mpp2/Ntng1/Gpc2/Nrn1 | 15 |
| GO:0098945 | intrinsic component of presynaptic active zone membrane | CC | 0.25 | 0.0134 | 0.0444 | 1.35 | 0.0297 | Cdh2/Cacna1h/Ntng1 | 3 |
| GO:0032587 | ruffle membrane | CC | 0.11 | 0.0137 | 0.0452 | 1.34 | 0.0302 | Ptprz1/Map2/Aif1l/Jcad/Eps8l1/Dlc1/Fgr/Plekho1 | 8 |
| GO:0050839 | cell adhesion molecule binding | MF | 0.179 | 2.25e-19 | 2.05e-16 | 15.7 | 1.59e-16 | Prtg/Ptprz1/Itga4/Lrrc4c/Kdr/Ptprd/Igf1/Nlgn1/Npnt/Nos3/Ptpro/Sema7a/Fermt2/Tgfbi/Cdh2/Fxyd5/Cdh6/Itgb7/Fn1/Tenm3/Col16a1/Icam1/Grin2b/Cxcl12/Jam3/Cdh13/Cdh10/Adgrl1/Itga1/Cdh5/Postn/Cpe/Thy1/Plpp3/Cd1d1/Ptprm/Msn/Cd226/Nlgn3/Esam/Itgal/Nfasc/Cdh11/Emilin1/Cd200/Igf2/Itga7/Tln2/Dchs1/Neo1/Ccn2/Ptn/Ntng1/Itga8/Itgb4 | 55 |
| GO:0046873 | metal ion transmembrane transporter activity | MF | 0.133 | 8.27e-14 | 3.77e-11 | 10.4 | 2.93e-11 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Grik1/Kcna6/Cacna1c/Slc6a2/Kcnd2/Slc5a11/Slc13a5/Ryr1/Kcnd3/Slc12a8/Slc8a1/Scn4b/Scn1a/Slc24a4/Kcnq5/Cachd1/Cacna2d1/Slc9a7/Rasa3/Grin2b/Slc24a3/Cacna1e/Slc8a3/Htr1b/Nalf1/Slc6a15/Trpc6/Scn2a/Grik2/Kcnma1/Slc9a5/Kcnc4/Slc23a2/Pkd1/Cacng7/Kcnip3/Ncs1/Ryr3/Cacna1h/Kcng4/Slc38a1/Pkd2/Slc12a4/Scn3a/Atp2b4/Atp1a2/Pkdrej/Kcnq4/Slc4a8/Cacna1a/Cacnb4 | 56 |
| GO:0005261 | cation channel activity | MF | 0.138 | 3.66e-12 | 1.06e-09 | 8.97 | 8.26e-10 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Scn1a/Slc24a4/Kcnq5/Cachd1/Cacna2d1/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Unc80/Htr1b/Nalf1/Trpc6/Scn2a/Grik2/Kcnma1/Kcnc4/Pkd1/Cacng7/Kcnip3/Ncs1/Ryr3/Cacna1h/Kcng4/Pkd2/Scn3a/Pkdrej/Piezo2/Kcnq4/Cacna1a/Cacnb4 | 46 |
| GO:0022836 | gated channel activity | MF | 0.139 | 4.66e-12 | 1.06e-09 | 8.97 | 8.26e-10 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Scn1a/Kcnq5/Cachd1/Cacna2d1/Ano1/Rasa3/Grin2b/Cacna1e/Ano2/Htr1b/Ttyh2/Nalf1/Glrb/Gabra4/Scn2a/Grik2/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Ryr3/Cacna1h/Kcng4/Pkd2/Scn3a/Clca2/Piezo2/Kcnq4/Cacna1a/Cacnb4 | 45 |
| GO:0005216 | ion channel activity | MF | 0.121 | 1.57e-11 | 2.86e-09 | 8.54 | 2.22e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Scn1a/Slc24a4/Kcnq5/Cachd1/Cacna2d1/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Htr1b/Ttyh2/Nalf1/Trpc6/Glrb/Gabra4/Scn2a/Grik2/Kcnma1/Kcnc4/Gjc1/Pkd1/Cacng7/Kcnip3/Ncs1/Ryr3/Cacna1h/Kcng4/Pkd2/Scn3a/Clca2/Pkdrej/Piezo2/Kcnq4/Cacna1a/Cacnb4 | 53 |
| GO:0015267 | channel activity | MF | 0.114 | 6.35e-11 | 7.24e-09 | 8.14 | 5.62e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Scn1a/Slc24a4/Kcnq5/Cachd1/Cacna2d1/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Htr1b/Ttyh2/Nalf1/Trpc6/Glrb/Gabra4/Scn2a/Gja4/Grik2/Kcnma1/Kcnc4/Gjc1/Pkd1/Cacng7/Kcnip3/Ncs1/Ryr3/Aqp11/Cacna1h/Kcng4/Pkd2/Scn3a/Clca2/Pkdrej/Piezo2/Kcnq4/Cacna1a/Cacnb4 | 55 |
| GO:0022803 | passive transmembrane transporter activity | MF | 0.114 | 6.35e-11 | 7.24e-09 | 8.14 | 5.62e-09 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Gria4/Grik1/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/P2rx2/Scn1a/Slc24a4/Kcnq5/Cachd1/Cacna2d1/Ano1/Rasa3/Grin2b/Slc24a3/Cacna1e/Trpm3/Ano2/Unc80/Htr1b/Ttyh2/Nalf1/Trpc6/Glrb/Gabra4/Scn2a/Gja4/Grik2/Kcnma1/Kcnc4/Gjc1/Pkd1/Cacng7/Kcnip3/Ncs1/Ryr3/Aqp11/Cacna1h/Kcng4/Pkd2/Scn3a/Clca2/Pkdrej/Piezo2/Kcnq4/Cacna1a/Cacnb4 | 55 |
| GO:0005244 | voltage-gated ion channel activity | MF | 0.161 | 2.44e-10 | 2.48e-08 | 7.61 | 1.92e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/Scn1a/Kcnq5/Cachd1/Cacna2d1/Ano1/Grin2b/Cacna1e/Htr1b/Scn2a/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Cacna1h/Kcng4/Pkd2/Scn3a/Kcnq4/Cacna1a/Cacnb4 | 31 |
| GO:0022832 | voltage-gated channel activity | MF | 0.161 | 2.79e-10 | 2.55e-08 | 7.59 | 1.98e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Scn4b/Scn1a/Kcnq5/Cachd1/Cacna2d1/Ano1/Grin2b/Cacna1e/Htr1b/Scn2a/Kcnma1/Kcnc4/Cacng7/Kcnip3/Ncs1/Cacna1h/Kcng4/Pkd2/Scn3a/Kcnq4/Cacna1a/Cacnb4 | 31 |
| GO:0005178 | integrin binding | MF | 0.174 | 6.15e-10 | 5.1e-08 | 7.29 | 3.96e-08 | Ptprz1/Itga4/Kdr/Igf1/Npnt/Sema7a/Fermt2/Tgfbi/Itgb7/Fn1/Col16a1/Icam1/Cxcl12/Jam3/Itga1/Thy1/Plpp3/Cd226/Itgal/Emilin1/Igf2/Itga7/Tln2/Ccn2/Ptn/Itga8/Itgb4 | 27 |
| GO:0003779 | actin binding | MF | 0.113 | 7.42e-10 | 5.64e-08 | 7.25 | 4.38e-08 | Lsp1/Myo16/Map2/Slc6a2/Evl/Epb41l3/Nos3/Ccdc88a/Hcls1/Fermt2/Aif1l/Tmod2/Fxyd5/Wasf3/Lrrk2/Myo5a/Pfn2/Mical1/Vash1/Trpc6/Msn/Coro1a/Sntb1/Myo9a/Limch1/Palld/Kcnma1/Eps8l1/Daam2/Scin/Parvg/Myl3/Cap2/Crocc/Cnn2/Hip1/Parvb/Bin1/Syne3/Rai14/Myo1f/Flnc/Cnn3/Tln2/Wipf1/Pknox2/Actr3b/Plekhh2/Vash2/Shroom1 | 50 |
| GO:0005539 | glycosaminoglycan binding | MF | 0.142 | 2.12e-09 | 1.49e-07 | 6.83 | 1.16e-07 | Apob/Col11a1/Vit/Sema5a/Mdk/Thbs3/Cd44/Spock2/Nrp1/Fn1/Serpine2/Egflam/Adamts1/Nell1/Col13a1/Mamdc2/Postn/Smoc1/Apoe/Layn/Ndnf/Rtn4rl1/Tnfaip6/Bgn/Ptch1/Hapln3/Abi3bp/Ccn2/Dpysl3/Fstl1/Efemp2/Habp4/Ptn | 33 |
| GO:0005096 | GTPase activator activity | MF | 0.14 | 2.95e-09 | 1.86e-07 | 6.73 | 1.45e-07 | Wnt11/Agap2/Rgs6/Rgs16/Rasgrp3/Nrp1/Garnl3/Lrrk2/Rasa3/Arhgap45/Tbc1d24/Plcb1/Arhgap25/Dock2/Thy1/Syde1/Myo9a/Sh3bp1/Rgs5/Arhgef15/Arhgap6/Nckap1l/Arap3/Prex2/Arrb1/Arhgap10/Arhgap28/Dlc1/Gnb5/Arhgap31/Syngap1/Arhgap15/Arfgap3 | 33 |
| GO:0015085 | calcium ion transmembrane transporter activity | MF | 0.179 | 3.07e-09 | 1.86e-07 | 6.73 | 1.45e-07 | Cacna1c/Ryr1/Slc8a1/Slc24a4/Cachd1/Cacna2d1/Rasa3/Grin2b/Slc24a3/Cacna1e/Slc8a3/Htr1b/Nalf1/Trpc6/Pkd1/Cacng7/Ncs1/Ryr3/Cacna1h/Pkd2/Atp2b4/Pkdrej/Cacna1a/Cacnb4 | 24 |
| GO:0005516 | calmodulin binding | MF | 0.151 | 4.48e-09 | 2.55e-07 | 6.59 | 1.98e-07 | Gap43/Map2/Cacna1c/Ryr1/Nos3/Slc8a1/Slc24a4/Kcnq5/Myo5a/Pde1b/Adcy3/Akap12/Slc8a3/Plcb1/Cth/Sntb1/Nrgn/Pde1a/Syt7/Cnn2/Myo1f/Ryr3/Cnn3/Rrad/Scn3a/Iqcb1/Atp2b4/Kcnq4/Cacna1a | 29 |
| GO:0030695 | GTPase regulator activity | MF | 0.109 | 9.82e-09 | 4.85e-07 | 6.31 | 3.77e-07 | Wnt11/Agap2/Ccdc88a/Rgs6/Rgs16/Rasgrp3/Nrp1/Garnl3/Lrrk2/Rasa3/Arhgap45/Arhgef25/Tbc1d24/Plcb1/Rasgrp2/Arhgap25/Dock2/Thy1/Syde1/Dock10/Denn2b/Myo9a/Sh3bp1/Rgs5/Arhgef15/Arhgap6/Eps8l1/Arhgef40/Dennd5a/Nckap1l/Arap3/Prex2/Arrb1/Arhgap10/Arhgap28/Dlc1/Net1/Fgd1/Gnb5/Arhgap31/Syngap1/Ngef/Fgd5/Arhgef9/Arhgap15/Arfgap3 | 46 |
| GO:0060589 | nucleoside-triphosphatase regulator activity | MF | 0.109 | 9.82e-09 | 4.85e-07 | 6.31 | 3.77e-07 | Wnt11/Agap2/Ccdc88a/Rgs6/Rgs16/Rasgrp3/Nrp1/Garnl3/Lrrk2/Rasa3/Arhgap45/Arhgef25/Tbc1d24/Plcb1/Rasgrp2/Arhgap25/Dock2/Thy1/Syde1/Dock10/Denn2b/Myo9a/Sh3bp1/Rgs5/Arhgef15/Arhgap6/Eps8l1/Arhgef40/Dennd5a/Nckap1l/Arap3/Prex2/Arrb1/Arhgap10/Arhgap28/Dlc1/Net1/Fgd1/Gnb5/Arhgap31/Syngap1/Ngef/Fgd5/Arhgef9/Arhgap15/Arfgap3 | 46 |
| GO:0022843 | voltage-gated cation channel activity | MF | 0.169 | 1.01e-08 | 4.85e-07 | 6.31 | 3.77e-07 | Kcnk3/Kcnb2/Kcnj3/Kcna6/Cacna1c/Kcnd2/Ryr1/Kcnd3/Kcnq5/Cachd1/Cacna2d1/Grin2b/Cacna1e/Htr1b/Kcnma1/Kcnc4/Cacng7/Ncs1/Cacna1h/Kcng4/Pkd2/Kcnq4/Cacna1a/Cacnb4 | 24 |
| GO:0015081 | sodium ion transmembrane transporter activity | MF | 0.166 | 1.54e-08 | 7.03e-07 | 6.15 | 5.46e-07 | Nalcn/Slc1a3/Grik1/Slc6a2/Slc5a11/Slc13a5/Slc8a1/Scn4b/Scn1a/Slc24a4/Slc9a7/Slc24a3/Slc8a3/Slc6a15/Scn2a/Grik2/Slc9a5/Slc23a2/Cacna1h/Slc38a1/Pkd2/Scn3a/Atp1a2/Slc4a8 | 24 |
| GO:0005262 | calcium channel activity | MF | 0.181 | 2.41e-08 | 1.05e-06 | 5.98 | 8.12e-07 | Cacna1c/Ryr1/Slc24a4/Cachd1/Cacna2d1/Rasa3/Grin2b/Slc24a3/Cacna1e/Htr1b/Nalf1/Trpc6/Pkd1/Cacng7/Ncs1/Ryr3/Cacna1h/Pkd2/Pkdrej/Cacna1a/Cacnb4 | 21 |
| GO:0008237 | metallopeptidase activity | MF | 0.141 | 1.23e-07 | 5.1e-06 | 5.29 | 3.96e-06 | Cpxm2/Mmp16/Adamts20/Adamts16/Adamts19/Mmp2/Enpep/Adam33/Adamts1/Vash1/Cpe/Naalad2/Mme/Adamts10/Adamts7/Adam11/Trhde/Agbl3/Mmp19/Ece2/Adam19/Pappa/Clca2/Adamts17/Lvrn/Vash2 | 26 |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | MF | 0.108 | 4.41e-07 | 1.67e-05 | 4.78 | 1.3e-05 | Lef1/Tbx2/Zeb2/Prdm5/Zfp37/Hoxd9/Osr2/Klf8/Zfp239/Glis1/Tcf21/Bcl6b/Zeb1/Prdm1/Zfp248/Zfp13/Tbx3/Heyl/Snai1/Zbtb46/Irf8/Satb1/Klf12/Peg3/Foxs1/Zfp382/Zbtb20/Kcnip3/Nr2f1/Nfatc4/Cux2/Ets2/Zbtb8a/Zfp566/Zfp512b/Zfp90 | 36 |
| GO:0005245 | voltage-gated calcium channel activity | MF | 0.255 | 5.17e-07 | 1.88e-05 | 4.72 | 1.46e-05 | Cacna1c/Ryr1/Cachd1/Cacna2d1/Cacna1e/Htr1b/Cacng7/Ncs1/Cacna1h/Pkd2/Cacna1a/Cacnb4 | 12 |
| GO:0001217 | DNA-binding transcription repressor activity | MF | 0.107 | 5.48e-07 | 1.92e-05 | 4.72 | 1.49e-05 | Lef1/Tbx2/Zeb2/Prdm5/Zfp37/Hoxd9/Osr2/Klf8/Zfp239/Glis1/Tcf21/Bcl6b/Zeb1/Prdm1/Zfp248/Zfp13/Tbx3/Heyl/Snai1/Zbtb46/Irf8/Satb1/Klf12/Peg3/Foxs1/Zfp382/Zbtb20/Kcnip3/Nr2f1/Nfatc4/Cux2/Ets2/Zbtb8a/Zfp566/Zfp512b/Zfp90 | 36 |
| GO:0015079 | potassium ion transmembrane transporter activity | MF | 0.14 | 1.18e-06 | 3.99e-05 | 4.4 | 3.1e-05 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcna6/Kcnd2/Kcnd3/Slc12a8/Slc24a4/Kcnq5/Slc9a7/Slc24a3/Grik2/Kcnma1/Slc9a5/Kcnc4/Kcnip3/Kcng4/Pkd2/Slc12a4/Atp1a2/Kcnq4 | 22 |
| GO:0008373 | sialyltransferase activity | MF | 0.381 | 1.46e-06 | 4.77e-05 | 4.32 | 3.7e-05 | St6gal2/St8sia2/St8sia4/St6galnac4/St3gal2/St8sia6/St6galnac6/St3gal4 | 8 |
| GO:0030020 | extracellular matrix structural constituent conferring tensile strength | MF | 0.263 | 3.42e-06 | 0.000108 | 3.97 | 8.37e-05 | Col11a1/Col14a1/Col4a6/Col16a1/Col9a2/Col13a1/Col5a2/Col15a1/Col23a1/Col18a1 | 10 |
| GO:0017124 | SH3 domain binding | MF | 0.143 | 4.65e-06 | 0.000141 | 3.85 | 0.00011 | Evl/Drd4/Hcls1/Abi3/Mical1/Shank2/Lrp2/Abi2/Sh3bp1/Afap1l1/Arhgap6/Prrt2/Efs/Wipf1/Afap1l2/Adam19/Dpysl3/Arhgap31/Syngap1 | 19 |
| GO:0016757 | glycosyltransferase activity | MF | 0.108 | 5.87e-06 | 0.000173 | 3.76 | 0.000134 | B4galnt4/St6gal2/St8sia2/St8sia4/Gxylt2/Uggt2/Pygm/Tmtc1/B3galnt1/B4galt2/Fut10/Has2/Galnt16/St6galnac4/St3gal2/Lfng/Eogt/Ggta1/Lacc1/St8sia6/Galntl6/Poglut2/Mgat3/B3gnt5/Xylt1/Mfng/St6galnac6/St3gal4/Large1 | 29 |
| GO:0045296 | cadherin binding | MF | 0.188 | 6.98e-06 | 0.000199 | 3.7 | 0.000155 | Kdr/Nos3/Ptpro/Cdh2/Fxyd5/Cdh6/Cdh13/Cdh10/Cdh5/Ptprm/Cdh11/Dchs1/Neo1 | 13 |
| GO:0001664 | G protein-coupled receptor binding | MF | 0.1 | 1.04e-05 | 0.000278 | 3.56 | 0.000216 | Cxcl15/Ndp/Wnt6/Wnt11/Ptch2/Gprasp2/Tub/Gnai1/Ednrb/Grin2b/Cxcl12/Edn3/Cnrip1/Calca/Wnt9a/Reep1/Znrf3/Agtr1a/Magi2/Bdkrb2/Xcl1/Arrb1/Ptch1/Gnaz/Arrb2/Fyn/Apln/Dlg4/Palm/Nes/Bbs1/Itgb4 | 32 |
| GO:0019838 | growth factor binding | MF | 0.131 | 1.04e-05 | 0.000278 | 3.56 | 0.000216 | Epha8/Ptprz1/Cd109/Kdr/Nrp1/Il11ra1/Tek/Lrp2/Flt3/Epha3/Tsku/Ltbp3/Epha7/Nrros/Ccn2/Ptn/Il2rg/Nkd2/Igfbp4/Itgb4 | 20 |
| GO:0004222 | metalloendopeptidase activity | MF | 0.151 | 1.27e-05 | 0.000331 | 3.48 | 0.000257 | Mmp16/Adamts20/Adamts16/Adamts19/Mmp2/Adam33/Adamts1/Mme/Adamts10/Adamts7/Adam11/Mmp19/Ece2/Adam19/Pappa/Adamts17 | 16 |
| GO:0008201 | heparin binding | MF | 0.124 | 1.52e-05 | 0.000386 | 3.41 | 3e-04 | Apob/Col11a1/Mdk/Thbs3/Nrp1/Fn1/Serpine2/Adamts1/Nell1/Col13a1/Postn/Smoc1/Apoe/Ndnf/Rtn4rl1/Ptch1/Abi3bp/Ccn2/Fstl1/Efemp2/Ptn | 21 |
| GO:0044325 | transmembrane transporter binding | MF | 0.129 | 2.01e-05 | 0.000496 | 3.3 | 0.000385 | Kcnb2/Cacna1c/Kcnd3/Tcaf1/Rims3/Slc8a1/Scn4b/Lrrk2/Ank2/Cdh5/Arrb1/Pkd1/Kcnip3/Fhl1/Mpp2/Kcng4/Fyn/Pkd2/Cav1 | 19 |
| GO:0098879 | structural constituent of postsynaptic specialization | MF | 0.5 | 3.14e-05 | 0.000754 | 3.12 | 0.000586 | Shank2/Dlg2/Magi2/Mpp2/Dlg4 | 5 |
| GO:0004713 | protein tyrosine kinase activity | MF | 0.133 | 3.81e-05 | 0.000868 | 3.06 | 0.000674 | Epha8/Kdr/Pkdcc/Ddr2/Nrp1/Itk/Tek/Flt3/Kit/Epha3/Tie1/Hipk2/Epha7/Fyn/Fgr/Zap70/Styk1 | 17 |
| GO:0035591 | signaling adaptor activity | MF | 0.171 | 4.21e-05 | 0.000934 | 3.03 | 0.000725 | Lrrk2/Shank2/Akap12/Mapk8ip1/Sh2b2/Abi2/Magi2/Sarm1/Afap1l2/Gnb5/Lat/Gnb4 | 12 |
| GO:0008509 | anion transmembrane transporter activity | MF | 0.0958 | 4.3e-05 | 0.000934 | 3.03 | 0.000725 | Slc1a3/Grik1/Slc6a2/Slc13a5/Slc2a3/Slc12a8/Slc24a4/Slc9a7/Ano1/Slc7a2/Abcg2/Slc24a3/Slc36a4/Ano2/Ttyh2/Glrb/Gabra4/Slc29a1/Slc9a5/Slc23a2/Sfxn5/Slco4c1/Slc4a3/Slc38a1/Slc12a4/Slc25a23/Clca2/Abcc5/Abcg3/Slc4a8 | 30 |
| GO:0005248 | voltage-gated sodium channel activity | MF | 0.28 | 6.7e-05 | 0.00139 | 2.86 | 0.00108 | Nalcn/Scn4b/Scn1a/Scn2a/Cacna1h/Pkd2/Scn3a | 7 |
| GO:0016849 | phosphorus-oxygen lyase activity | MF | 0.28 | 6.7e-05 | 0.00139 | 2.86 | 0.00108 | Gucy1a2/Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7/Cd38 | 7 |
| GO:0005080 | protein kinase C binding | MF | 0.172 | 8.5e-05 | 0.00172 | 2.76 | 0.00134 | Fez1/Ccdc88a/Nell1/Adcy4/Pard6g/Hdac9/Cavin3/BC034090/Cavin2/Akt3/Dact1 | 11 |
| GO:0004016 | adenylate cyclase activity | MF | 0.417 | 9.18e-05 | 0.00175 | 2.76 | 0.00136 | Adcy3/Gucy1b1/Gucy1a1/Adcy4/Adcy7 | 5 |
| GO:0005272 | sodium channel activity | MF | 0.205 | 9.22e-05 | 0.00175 | 2.76 | 0.00136 | Nalcn/Grik1/Scn4b/Scn1a/Scn2a/Grik2/Cacna1h/Pkd2/Scn3a | 9 |
| GO:0099511 | voltage-gated calcium channel activity involved in regulation of cytosolic calcium levels | MF | 0.385 | 0.000144 | 0.00262 | 2.58 | 0.00204 | Cacna1e/Htr1b/Ncs1/Cacna1a/Cacnb4 | 5 |
| GO:0099626 | voltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels | MF | 0.385 | 0.000144 | 0.00262 | 2.58 | 0.00204 | Cacna1e/Htr1b/Ncs1/Cacna1a/Cacnb4 | 5 |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | MF | 0.172 | 0.000172 | 0.00308 | 2.51 | 0.0024 | Epha8/Kdr/Ddr2/Nrp1/Tek/Flt3/Kit/Epha3/Tie1/Epha7 | 10 |
| GO:0030159 | signaling receptor complex adaptor activity | MF | 0.211 | 0.000183 | 0.0032 | 2.49 | 0.00249 | Lrrk2/Shank2/Akap12/Sh2b2/Magi2/Gnb5/Lat/Gnb4 | 8 |
| GO:0003774 | cytoskeletal motor activity | MF | 0.132 | 0.00019 | 0.00328 | 2.48 | 0.00254 | Myo16/Kif7/Myo5a/Dnah5/Kif1a/Myo9a/Kif6/Dync2h1/Myl3/Myo1f/Dnah7b/Stard9/Dnah1/Kif16b | 14 |
| GO:0005267 | potassium channel activity | MF | 0.124 | 0.000233 | 0.0039 | 2.41 | 0.00303 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcna6/Kcnd2/Kcnd3/Kcnq5/Grik2/Kcnma1/Kcnc4/Kcnip3/Kcng4/Pkd2/Kcnq4 | 15 |
| GO:0008194 | UDP-glycosyltransferase activity | MF | 0.114 | 0.000254 | 0.00413 | 2.38 | 0.00321 | B4galnt4/Gxylt2/Uggt2/B3galnt1/B4galt2/Has2/Galnt16/Lfng/Eogt/Ggta1/Galntl6/Poglut2/Mgat3/B3gnt5/Xylt1/Mfng/Large1 | 17 |
| GO:0030215 | semaphorin receptor binding | MF | 0.273 | 0.000264 | 0.00417 | 2.38 | 0.00324 | Sema5a/Sema5b/Sema7a/Sh3bp1/Sema3g/Sema6a | 6 |
| GO:0008235 | metalloexopeptidase activity | MF | 0.164 | 0.000265 | 0.00417 | 2.38 | 0.00324 | Cpxm2/Mmp16/Enpep/Vash1/Cpe/Naalad2/Trhde/Agbl3/Lvrn/Vash2 | 10 |
| GO:0022853 | active ion transmembrane transporter activity | MF | 0.0966 | 0.000285 | 0.00441 | 2.36 | 0.00342 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Slc24a3/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Atp6v0e2/Slco4c1/Slc4a3/Slc38a1/Slc12a4/Cox4i2/Atp2b4/Atp1a2/Slc4a8 | 23 |
| GO:0005249 | voltage-gated potassium channel activity | MF | 0.136 | 0.000398 | 0.00605 | 2.22 | 0.0047 | Kcnk3/Kcnb2/Kcnj3/Kcna6/Kcnd2/Kcnd3/Kcnq5/Kcnma1/Kcnc4/Kcng4/Pkd2/Kcnq4 | 12 |
| GO:0015491 | cation:cation antiporter activity | MF | 0.25 | 0.000443 | 0.00662 | 2.18 | 0.00514 | Slc8a1/Slc24a4/Slc9a7/Slc24a3/Slc8a3/Slc9a5 | 6 |
| GO:0005525 | GTP binding | MF | 0.0836 | 0.000478 | 0.00702 | 2.15 | 0.00546 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Gnai1/Arl6/Lrrk2/Gucy1b1/Gimap8/Rab34/Rragd/Dnajc27/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Septin3/Rhobtb3/Nin/Arl3/Rasd2/Gnaz/Rrad/Septin4/Gimap5/Rab23/Gimap3/Rab39b/Rab27a | 30 |
| GO:0098632 | cell-cell adhesion mediator activity | MF | 0.206 | 0.000534 | 0.00773 | 2.11 | 0.006 | Prtg/Lrrc4c/Jam3/Esam/Nfasc/Cd200/Ntng1 | 7 |
| GO:0022804 | active transmembrane transporter activity | MF | 0.0808 | 0.000563 | 0.0079 | 2.1 | 0.00614 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Abcg2/Slc24a3/Slc36a4/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Atp6v0e2/Abca5/Slco4c1/Abcg1/Abca8b/Slc4a3/Slc38a1/Slc12a4/Cox4i2/Abcd2/Atp2b4/Atp1a2/Abcb1a/Abcc5/Abcg3/Slc4a8 | 32 |
| GO:0015368 | calcium:cation antiporter activity | MF | 0.4 | 0.000591 | 0.00803 | 2.1 | 0.00624 | Slc8a1/Slc24a4/Slc24a3/Slc8a3 | 4 |
| GO:0035374 | chondroitin sulfate binding | MF | 0.4 | 0.000591 | 0.00803 | 2.1 | 0.00624 | Mdk/Rtn4rl1/Dpysl3/Ptn | 4 |
| GO:0008559 | ABC-type xenobiotic transporter activity | MF | 0.294 | 0.000599 | 0.00803 | 2.1 | 0.00624 | Abcg2/Abca8b/Abcb1a/Abcc5/Abcg3 | 5 |
| GO:0019001 | guanyl nucleotide binding | MF | 0.0812 | 0.000635 | 0.00827 | 2.08 | 0.00643 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Gnai1/Arl6/Lrrk2/Gucy1b1/Gimap8/Rab34/Rragd/Dnajc27/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Septin3/Rhobtb3/Nin/Arl3/Rasd2/Pde5a/Gnaz/Rrad/Septin4/Gimap5/Rab23/Gimap3/Rab39b/Rab27a | 31 |
| GO:0032561 | guanyl ribonucleotide binding | MF | 0.0812 | 0.000635 | 0.00827 | 2.08 | 0.00643 | Rragb/Agap2/Rasl12/Rab36/Gimap4/Gnai1/Arl6/Lrrk2/Gucy1b1/Gimap8/Rab34/Rragd/Dnajc27/Gucy1a1/Gimap6/Cracr2a/Tubb4a/Septin3/Rhobtb3/Nin/Arl3/Rasd2/Pde5a/Gnaz/Rrad/Septin4/Gimap5/Rab23/Gimap3/Rab39b/Rab27a | 31 |
| GO:0004181 | metallocarboxypeptidase activity | MF | 0.231 | 0.000704 | 0.00904 | 2.04 | 0.00702 | Cpxm2/Vash1/Cpe/Naalad2/Agbl3/Vash2 | 6 |
| GO:0015631 | tubulin binding | MF | 0.0803 | 0.000754 | 0.00945 | 2.02 | 0.00734 | Gli1/Map2/Fez1/Capn6/Ccdc88a/Kif7/Lrrk2/Kif26a/Kif1a/Ndn/Reep1/Eml1/Rp1/Map9/Kif6/Katnal1/Dclk2/Nin/Arl3/Dysf/Cnn3/Fyn/Unc5c/Stard9/Tbcel/Brsk1/Mapre2/Vash2/Tppp/Kif16b/Map7d3 | 31 |
| GO:0015276 | ligand-gated ion channel activity | MF | 0.111 | 0.000756 | 0.00945 | 2.02 | 0.00734 | Kcnj3/Grid1/Gria4/Grik1/Ryr1/P2rx2/Rasa3/Grin2b/Glrb/Gabra4/Grik2/Kcnma1/Ryr3/Pkd2/Clca2 | 15 |
| GO:0004970 | ionotropic glutamate receptor activity | MF | 0.278 | 8e-04 | 0.00986 | 2.01 | 0.00766 | Grid1/Gria4/Grik1/Grin2b/Grik2 | 5 |
| GO:0022834 | ligand-gated channel activity | MF | 0.109 | 0.000881 | 0.0105 | 1.98 | 0.00812 | Kcnj3/Grid1/Gria4/Grik1/Ryr1/P2rx2/Rasa3/Grin2b/Glrb/Gabra4/Grik2/Kcnma1/Ryr3/Pkd2/Clca2 | 15 |
| GO:0008331 | high voltage-gated calcium channel activity | MF | 0.364 | 0.000897 | 0.0105 | 1.98 | 0.00812 | Cacna1c/Cacna1e/Cacna1a/Cacnb4 | 4 |
| GO:0042285 | xylosyltransferase activity | MF | 0.364 | 0.000897 | 0.0105 | 1.98 | 0.00812 | Gxylt2/Poglut2/Xylt1/Large1 | 4 |
| GO:0005543 | phospholipid binding | MF | 0.0763 | 0.000906 | 0.0105 | 1.98 | 0.00812 | Apob/Gap43/Nme4/Pla2g10/Nr5a2/Mark1/Syt13/Cpne5/Ccdc88a/P2rx2/Fermt2/Dab1/Pfn2/Lpar4/Cibar1/Plcb1/Rps6kc1/Thy1/Pltp/Mme/Cadps/Nrgn/Apoe/Syt7/Scin/Hip1/Arap3/Bin1/Dysf/Epdr1/Pard3b/Plcb2/Zfyve9/Cavin2/Kif16b | 35 |
| GO:0003924 | GTPase activity | MF | 0.085 | 0.00106 | 0.012 | 1.92 | 0.00935 | Rragb/Agap2/Rasl12/Rab36/Gnai1/Arl6/Lrrk2/Rab34/Rragd/Dnajc27/Cracr2a/Tubb4a/Gng2/Septin3/Rhobtb3/Arl3/Rasd2/Gnaz/Rrad/Septin4/Rab23/Rab39b/Rab27a/Gnb4/Tppp | 25 |
| GO:0140359 | ABC-type transporter activity | MF | 0.214 | 0.00107 | 0.012 | 1.92 | 0.00935 | Abcg2/Abcg1/Abca8b/Abcb1a/Abcc5/Abcg3 | 6 |
| GO:0015298 | solute:cation antiporter activity | MF | 0.207 | 0.0013 | 0.014 | 1.86 | 0.0108 | Slc8a1/Slc24a4/Slc9a7/Slc24a3/Slc8a3/Slc9a5 | 6 |
| GO:0045499 | chemorepellent activity | MF | 0.207 | 0.0013 | 0.014 | 1.86 | 0.0108 | Sema5a/Sema5b/Sema7a/Sema3g/Epha7/Sema6a | 6 |
| GO:0031402 | sodium ion binding | MF | 0.333 | 0.0013 | 0.014 | 1.86 | 0.0108 | Scn1a/Scn2a/Scn3a/Atp1a2 | 4 |
| GO:0099186 | structural constituent of postsynapse | MF | 0.25 | 0.00135 | 0.0143 | 1.85 | 0.0111 | Shank2/Dlg2/Magi2/Mpp2/Dlg4 | 5 |
| GO:0005251 | delayed rectifier potassium channel activity | MF | 0.2 | 0.00157 | 0.0162 | 1.79 | 0.0126 | Kcnb2/Kcna6/Kcnq5/Kcnc4/Kcng4/Kcnq4 | 6 |
| GO:0019199 | transmembrane receptor protein kinase activity | MF | 0.132 | 0.00157 | 0.0162 | 1.79 | 0.0126 | Epha8/Kdr/Ddr2/Nrp1/Tek/Flt3/Kit/Epha3/Tie1/Epha7 | 10 |
| GO:0003777 | microtubule motor activity | MF | 0.141 | 0.00166 | 0.017 | 1.77 | 0.0132 | Kif7/Dnah5/Kif1a/Kif6/Dync2h1/Dnah7b/Stard9/Dnah1/Kif16b | 9 |
| GO:0008017 | microtubule binding | MF | 0.0849 | 0.00169 | 0.0171 | 1.77 | 0.0133 | Gli1/Map2/Capn6/Ccdc88a/Kif7/Kif26a/Kif1a/Reep1/Eml1/Rp1/Map9/Kif6/Katnal1/Dclk2/Nin/Arl3/Dysf/Cnn3/Stard9/Mapre2/Vash2/Kif16b/Map7d3 | 23 |
| GO:0051020 | GTPase binding | MF | 0.081 | 0.00171 | 0.0172 | 1.77 | 0.0133 | Rragb/Rims3/Rasgrp3/Lrrk2/Myo5a/Gga2/Mical1/Ranbp17/Dock2/Rab34/Dock10/Rragd/Abi2/Dmxl2/Rasip1/Daam2/Dennd5a/Adcyap1r1/Rhobtb3/Bin1/Net1/Fgd1/Cdc42ep5/Cav1/Cyfip2/Kif16b | 26 |
| GO:0023026 | MHC class II protein complex binding | MF | 0.308 | 0.00181 | 0.018 | 1.75 | 0.014 | H2-Ab1/H2-Ob/H2-Eb1/H2-Aa | 4 |
| GO:0030276 | clathrin binding | MF | 0.138 | 0.00185 | 0.018 | 1.74 | 0.014 | Nsg2/Syt13/Lrrk2/Ap1m1/Trpc6/Syt7/Hip1/Nsg1/Arrb1 | 9 |
| GO:0016758 | hexosyltransferase activity | MF | 0.0933 | 0.00186 | 0.018 | 1.74 | 0.014 | B4galnt4/Uggt2/Pygm/Tmtc1/B3galnt1/B4galt2/Fut10/Has2/Galnt16/Lfng/Eogt/Ggta1/Galntl6/Poglut2/Mgat3/B3gnt5/Mfng/Large1 | 18 |
| GO:0030165 | PDZ domain binding | MF | 0.108 | 0.00208 | 0.02 | 1.7 | 0.0155 | Mpp3/Nlgn1/Nkd1/Lrp2/Dlg2/Sntb1/Lin7a/Grik2/Erc2/Mpp2/Fzd2/Dlg4/Atp2b4 | 13 |
| GO:0030674 | protein-macromolecule adaptor activity | MF | 0.0798 | 0.00212 | 0.0202 | 1.7 | 0.0157 | Epb41l3/Aph1b/Stx1b/Lrrk2/Ap1m1/Shank2/Akap12/Mapk8ip1/Klhdc2/Cntln/Ank2/Sh2b2/Abi2/Spsb4/Magi2/Hip1/Arrb1/Sarm1/Syne3/Frmd4a/Afap1l2/Gnb5/Lat/Gnb4/Cav1/Stx2 | 26 |
| GO:0015248 | sterol transporter activity | MF | 0.188 | 0.00222 | 0.0209 | 1.68 | 0.0162 | Apob/Osbp2/Pltp/Apoe/Abcg1/Osbpl1a | 6 |
| GO:0019842 | vitamin binding | MF | 0.103 | 0.00232 | 0.0216 | 1.67 | 0.0168 | Stra6/P3h3/Abat/P4htm/Cbs/Pygm/Rbp7/Sptlc3/Kyat1/Hdc/Dhtkd1/Cth/P3h1/S100g | 14 |
| GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential | MF | 0.145 | 0.00238 | 0.0219 | 1.66 | 0.017 | Grid1/Gria4/Grik1/Grin2b/Glrb/Gabra4/Grik2/Gabbr1 | 8 |
| GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | MF | 0.286 | 0.00245 | 0.0224 | 1.65 | 0.0174 | Fads2/Fads1/Fads3/Degs2 | 4 |
| GO:0015294 | solute:cation symporter activity | MF | 0.116 | 0.00265 | 0.0239 | 1.62 | 0.0186 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc2a13/Slc6a15/Slc23a2/Slc38a1/Slc12a4/Slc4a8 | 11 |
| GO:0098960 | postsynaptic neurotransmitter receptor activity | MF | 0.13 | 0.00281 | 0.0251 | 1.6 | 0.0195 | Grid1/Gria4/Grik1/Drd4/Grin2b/Glrb/Gabra4/Grik2/Gabbr1 | 9 |
| GO:0008514 | organic anion transmembrane transporter activity | MF | 0.0919 | 0.00286 | 0.0252 | 1.6 | 0.0196 | Slc1a3/Grik1/Slc13a5/Slc2a3/Slc7a2/Abcg2/Slc36a4/Slc29a1/Slc23a2/Sfxn5/Slco4c1/Slc4a3/Slc38a1/Slc25a23/Abcc5/Abcg3/Slc4a8 | 17 |
| GO:0008238 | exopeptidase activity | MF | 0.115 | 0.00288 | 0.0252 | 1.6 | 0.0196 | Cpxm2/Mmp16/Enpep/Vash1/Cpe/Naalad2/Mme/Trhde/Agbl3/Lvrn/Vash2 | 11 |
| GO:0005540 | hyaluronic acid binding | MF | 0.208 | 0.0032 | 0.0259 | 1.59 | 0.0201 | Cd44/Layn/Tnfaip6/Hapln3/Habp4 | 5 |
| GO:0031681 | G-protein beta-subunit binding | MF | 0.208 | 0.0032 | 0.0259 | 1.59 | 0.0201 | Gngt2/Gng11/Gng2/Rasd2/Gng7 | 5 |
| GO:0031690 | adrenergic receptor binding | MF | 0.208 | 0.0032 | 0.0259 | 1.59 | 0.0201 | Magi2/Bdkrb2/Arrb1/Arrb2/Dlg4 | 5 |
| GO:0099604 | ligand-gated calcium channel activity | MF | 0.208 | 0.0032 | 0.0259 | 1.59 | 0.0201 | Ryr1/Rasa3/Grin2b/Ryr3/Pkd2 | 5 |
| GO:0031267 | small GTPase binding | MF | 0.0807 | 0.0032 | 0.0259 | 1.59 | 0.0201 | Rims3/Rasgrp3/Lrrk2/Myo5a/Gga2/Mical1/Ranbp17/Dock2/Rab34/Dock10/Abi2/Dmxl2/Rasip1/Daam2/Dennd5a/Adcyap1r1/Rhobtb3/Net1/Fgd1/Cdc42ep5/Cav1/Cyfip2/Kif16b | 23 |
| GO:0005229 | intracellular calcium activated chloride channel activity | MF | 0.267 | 0.00323 | 0.0259 | 1.59 | 0.0201 | Ano1/Ano2/Ttyh2/Clca2 | 4 |
| GO:0015278 | calcium-release channel activity | MF | 0.267 | 0.00323 | 0.0259 | 1.59 | 0.0201 | Ryr1/Rasa3/Ryr3/Pkd2 | 4 |
| GO:0022821 | potassium ion antiporter activity | MF | 0.267 | 0.00323 | 0.0259 | 1.59 | 0.0201 | Slc24a4/Slc9a7/Slc24a3/Slc9a5 | 4 |
| GO:0042043 | neurexin family protein binding | MF | 0.267 | 0.00323 | 0.0259 | 1.59 | 0.0201 | Nlgn1/Cpe/Nlgn3/Dlg4 | 4 |
| GO:0061778 | intracellular chloride channel activity | MF | 0.267 | 0.00323 | 0.0259 | 1.59 | 0.0201 | Ano1/Ano2/Ttyh2/Clca2 | 4 |
| GO:0015293 | symporter activity | MF | 0.0979 | 0.00368 | 0.0289 | 1.54 | 0.0225 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc24a4/Slc24a3/Slc36a4/Slc2a13/Slc6a15/Slc23a2/Slc38a1/Slc12a4/Slc4a8 | 14 |
| GO:0019955 | cytokine binding | MF | 0.0979 | 0.00368 | 0.0289 | 1.54 | 0.0225 | Itga4/Cd109/Nog/Cd44/Il11ra1/Cxcr3/Kit/Il27ra/Tsku/Ltbp3/Nrros/Xcr1/Il12rb1/Il2rg | 14 |
| GO:0050780 | dopamine receptor binding | MF | 0.2 | 0.00386 | 0.0301 | 1.52 | 0.0234 | Grin2b/Agtr1a/Arrb2/Dlg4/Palm | 5 |
| GO:0050660 | flavin adenine dinucleotide binding | MF | 0.116 | 0.00395 | 0.0305 | 1.52 | 0.0237 | Gulo/Maob/Nos3/Mical1/Maoa/Fmo1/Aifm2/Dpyd/Steap4/Aox4 | 10 |
| GO:0005172 | vascular endothelial growth factor receptor binding | MF | 0.25 | 0.00416 | 0.0316 | 1.5 | 0.0246 | Angpt1/Ccdc88a/Cdh5/Vegfc | 4 |
| GO:0015562 | efflux transmembrane transporter activity | MF | 0.25 | 0.00416 | 0.0316 | 1.5 | 0.0246 | Abcg2/Abcb1a/Abcc5/Abcg3 | 4 |
| GO:0015291 | secondary active transmembrane transporter activity | MF | 0.0826 | 0.00441 | 0.0331 | 1.48 | 0.0257 | Slc1a3/Slc6a2/Slc5a11/Slc13a5/Slc12a8/Slc8a1/Slc24a4/Slc9a7/Slc24a3/Slc36a4/Slc8a3/Slc2a13/Slc6a15/Slc9a5/Slc23a2/Slco4c1/Slc4a3/Slc38a1/Slc12a4/Slc4a8 | 20 |
| GO:0008528 | G protein-coupled peptide receptor activity | MF | 0.0959 | 0.00443 | 0.0331 | 1.48 | 0.0257 | Vipr2/Npr3/Ednrb/Cxcr3/Pth1r/Aplnr/Agtr1a/Ednra/Adcyap1r1/Bdkrb2/Ogfrl1/Xcr1/Calcrl/Prokr1 | 14 |
| GO:0042626 | ATPase-coupled transmembrane transporter activity | MF | 0.108 | 0.0046 | 0.0339 | 1.47 | 0.0263 | Abcg2/Atp6v0e2/Abca5/Abcg1/Abca8b/Abcd2/Atp2b4/Atp1a2/Abcb1a/Abcc5/Abcg3 | 11 |
| GO:0008066 | glutamate receptor activity | MF | 0.192 | 0.00461 | 0.0339 | 1.47 | 0.0263 | Grid1/Gria4/Grik1/Grin2b/Grik2 | 5 |
| GO:0008083 | growth factor activity | MF | 0.0946 | 0.005 | 0.0365 | 1.44 | 0.0283 | Hgf/Mdk/Igf1/Areg/Cxcl12/Gdf11/Gdf7/Jag2/Gdf10/Vegfc/Igf2/Kitl/Ccn2/Ptn | 14 |
| GO:0000149 | SNARE binding | MF | 0.101 | 0.00544 | 0.0389 | 1.41 | 0.0302 | Syt13/Stx1b/Lrrk2/Myo5a/Exoc3l/Stxbp4/Syt7/Prrt2/Txlnb/Cav1/Stx2/Cacna1a | 12 |
| GO:0003785 | actin monomer binding | MF | 0.185 | 0.00546 | 0.0389 | 1.41 | 0.0302 | Nos3/Pfn2/Coro1a/Myl3/Pknox2 | 5 |
| GO:0090482 | vitamin transmembrane transporter activity | MF | 0.185 | 0.00546 | 0.0389 | 1.41 | 0.0302 | Stra6/Slc2a3/Abcg2/Slc23a2/Abcg3 | 5 |
| GO:0005496 | steroid binding | MF | 0.0984 | 0.00662 | 0.0464 | 1.33 | 0.0361 | Cyp3a59/Ar/Cyp3a25/Igf1/Soat1/Osbp2/Ptch1/Gstm1/Atp1a2/Osbpl1a/Sidt1/S100g | 12 |
| GO:0001653 | peptide receptor activity | MF | 0.0915 | 0.00668 | 0.0464 | 1.33 | 0.0361 | Vipr2/Npr3/Ednrb/Cxcr3/Pth1r/Aplnr/Agtr1a/Ednra/Adcyap1r1/Bdkrb2/Ogfrl1/Xcr1/Calcrl/Prokr1 | 14 |
| GO:0030170 | pyridoxal phosphate binding | MF | 0.135 | 0.00677 | 0.0464 | 1.33 | 0.0361 | Abat/Cbs/Pygm/Sptlc3/Kyat1/Hdc/Cth | 7 |
| GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential | MF | 0.135 | 0.00677 | 0.0464 | 1.33 | 0.0361 | Grid1/Gria4/Grik1/Grin2b/Glrb/Gabra4/Grik2 | 7 |
Visualisation
Biological Process
bp_dot[[2]]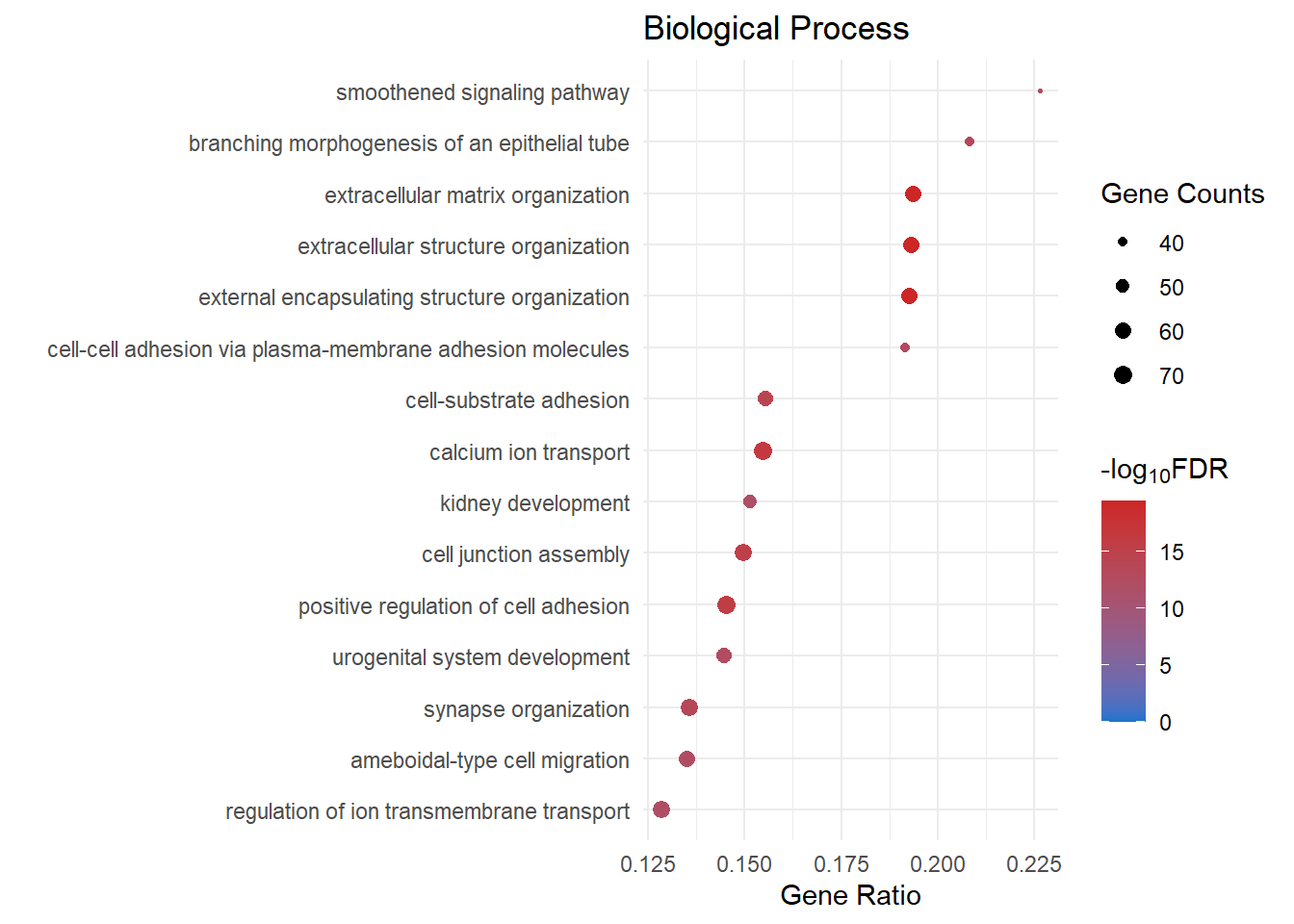
Molecular Function
mf_dot[[2]]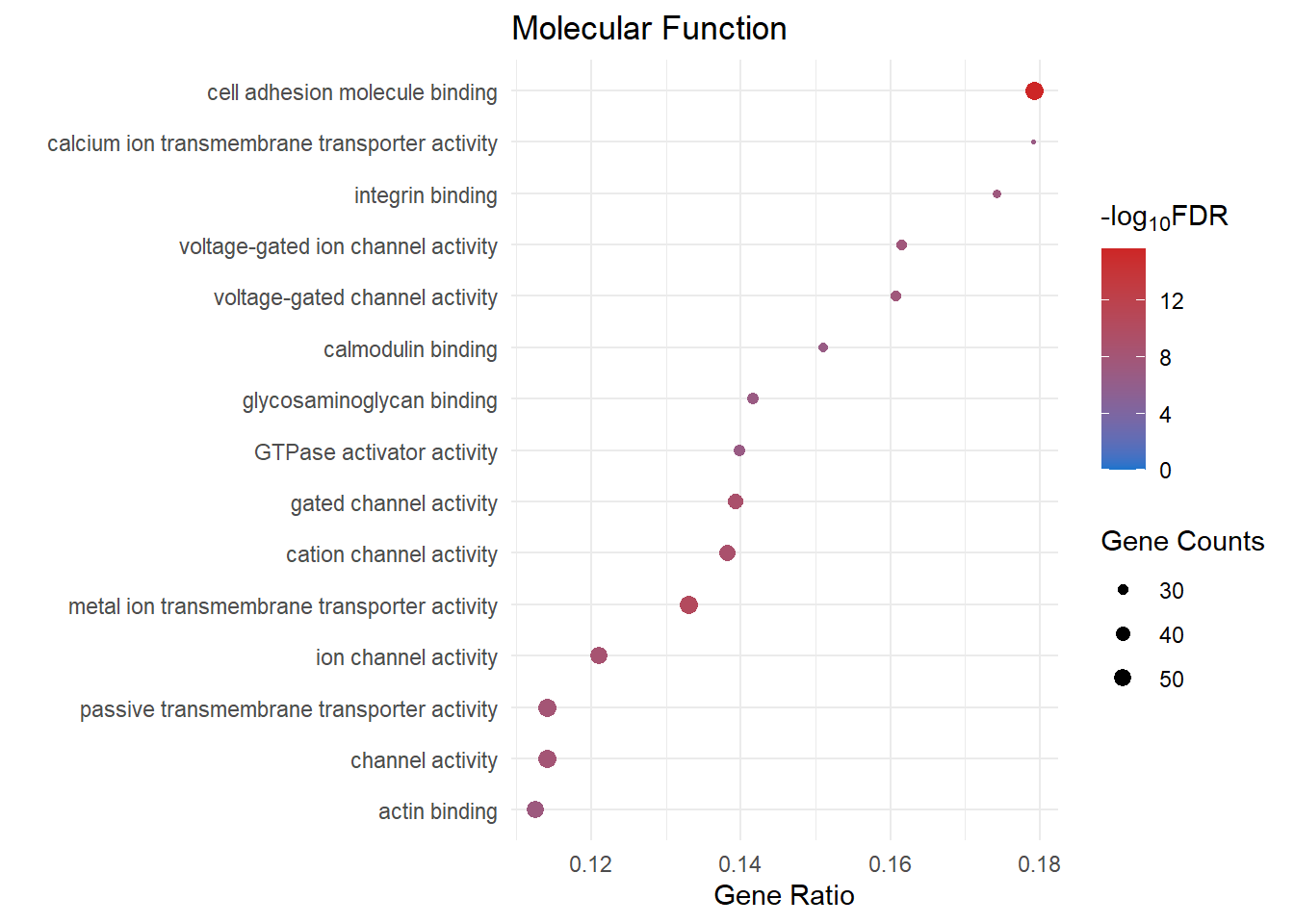
Cellular Components
cc_dot[[2]]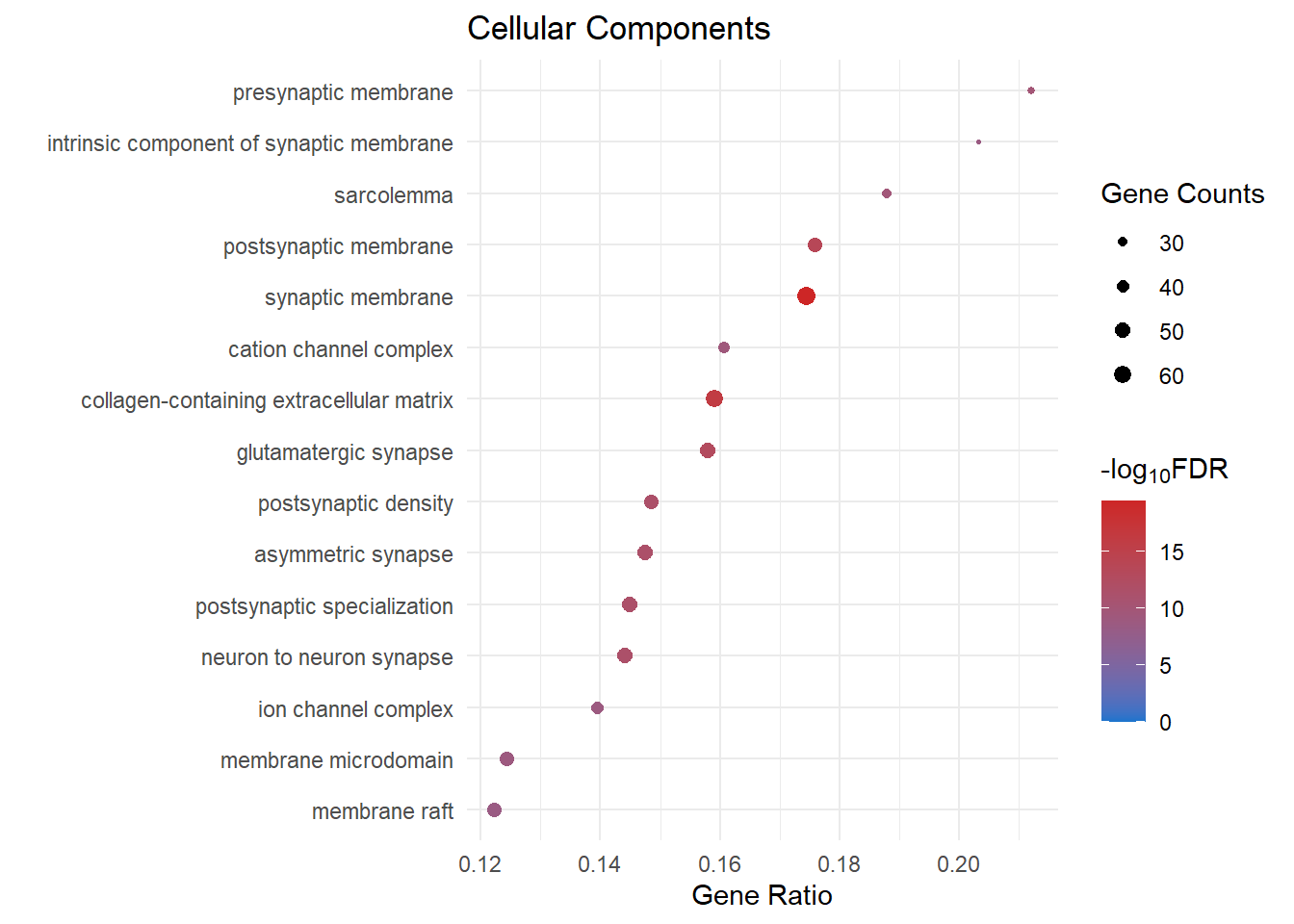
Upset
upset[[2]]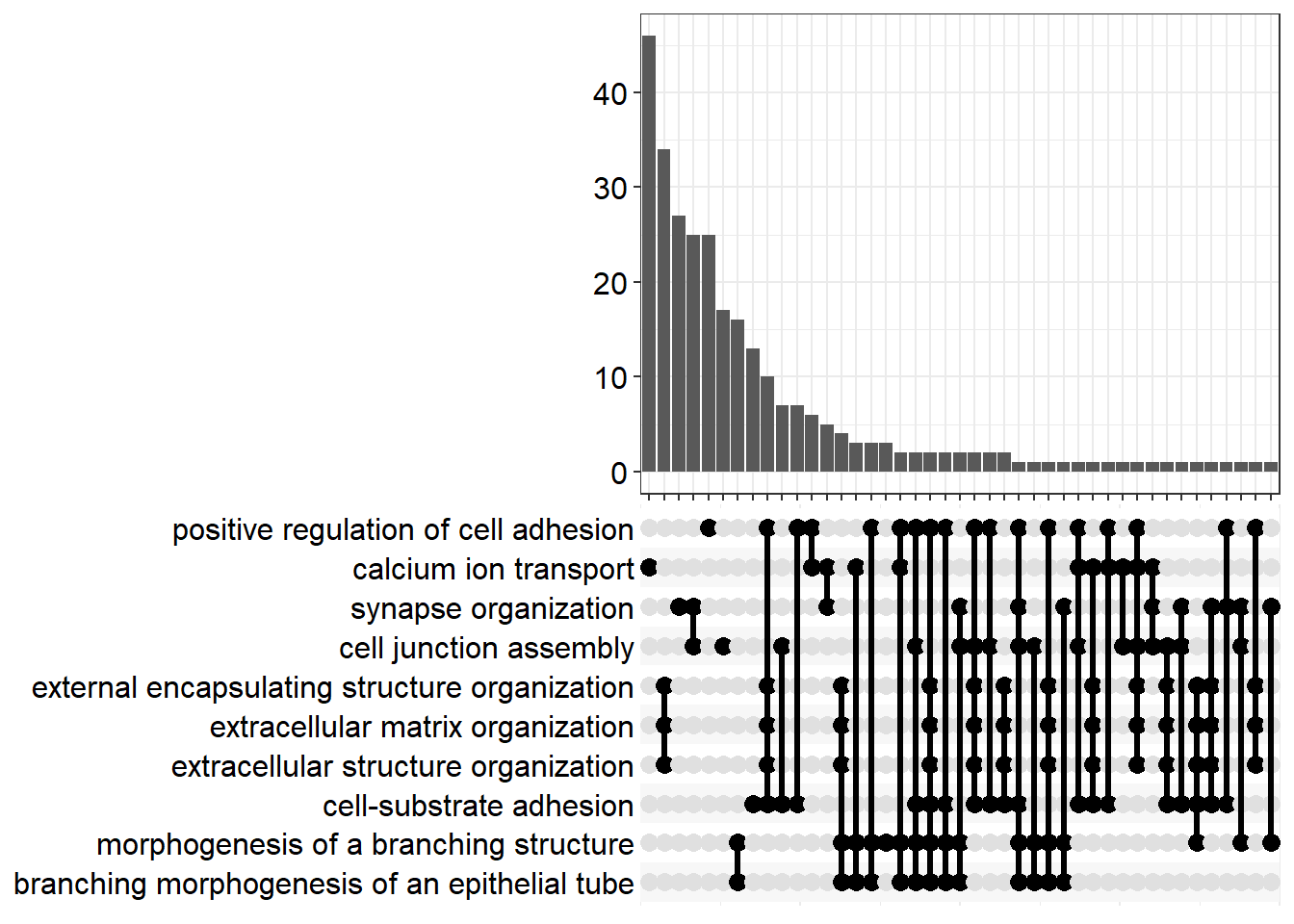
FC=1.5
# display the top 30 most sig
enrichGO_sig[[3]] %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(.,3)))) %>%
kable(caption = "Significantly enriched GO terms") %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
scroll_box(height = "600px")| Description | ontology | GeneRatio | pvalue | p.adjust | logFDR | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0048754 | branching morphogenesis of an epithelial tube | BP | 0.115 | 1.62e-16 | 5.72e-13 | 12.2 | 4.09e-13 | Ihh/Wnt6/Sfrp2/Hhip/Lef1/Angpt1/Sema5a/Mdk/Nog/Tbx2/Hoxd11/Adamts16/Igf1/Areg/Npnt/Kdr/Gpc3/Hoxa11/Foxf1/Cd44/Cxcl12/Gdf7 | 22 |
| GO:0061138 | morphogenesis of a branching epithelium | BP | 0.1 | 6.73e-16 | 1.19e-12 | 11.9 | 8.49e-13 | Ihh/Hgf/Wnt6/Sfrp2/Hhip/Lef1/Angpt1/Sema5a/Mdk/Nog/Tbx2/Hoxd11/Adamts16/Igf1/Areg/Npnt/Kdr/Gpc3/Hoxa11/Foxf1/Cd44/Cxcl12/Gdf7 | 23 |
| GO:0030198 | extracellular matrix organization | BP | 0.0762 | 6.91e-14 | 4.67e-11 | 10.3 | 3.34e-11 | Cyp1b1/Ihh/Col11a1/Hmcn1/Vit/Mmp16/Sfrp2/Sema5a/Adamts20/Col14a1/Col4a6/Adamts16/Scara3/Npnt/Spock2/Prdm5/Tgfbi/Foxf1/Has2/Col9a2/Olfml2b/Adamts19/Mmp2/Nid1 | 24 |
| GO:0043062 | extracellular structure organization | BP | 0.0759 | 7.41e-14 | 4.67e-11 | 10.3 | 3.34e-11 | Cyp1b1/Ihh/Col11a1/Hmcn1/Vit/Mmp16/Sfrp2/Sema5a/Adamts20/Col14a1/Col4a6/Adamts16/Scara3/Npnt/Spock2/Prdm5/Tgfbi/Foxf1/Has2/Col9a2/Olfml2b/Adamts19/Mmp2/Nid1 | 24 |
| GO:0045229 | external encapsulating structure organization | BP | 0.0757 | 7.94e-14 | 4.67e-11 | 10.3 | 3.34e-11 | Cyp1b1/Ihh/Col11a1/Hmcn1/Vit/Mmp16/Sfrp2/Sema5a/Adamts20/Col14a1/Col4a6/Adamts16/Scara3/Npnt/Spock2/Prdm5/Tgfbi/Foxf1/Has2/Col9a2/Olfml2b/Adamts19/Mmp2/Nid1 | 24 |
| GO:0097485 | neuron projection guidance | BP | 0.0734 | 5.71e-11 | 2.52e-08 | 7.6 | 1.8e-08 | Gap43/Epha8/Prtg/Efnb3/Pla2g10/Sema5a/Dcc/Nog/Sema5b/Ptpro/Rnf165/Plxnb1/Dpysl4/Edn3/Lrp2/Evl/Sema7a/Cxcl12/Gdf7 | 19 |
| GO:0060562 | epithelial tube morphogenesis | BP | 0.0582 | 5.71e-11 | 2.52e-08 | 7.6 | 1.8e-08 | Ihh/Wnt6/Sfrp2/Hhip/Lef1/Angpt1/Sema5a/Mdk/Nog/Tbx2/Hoxd11/Adamts16/Igf1/Areg/Npnt/Kdr/Gpc3/Hoxa11/Foxf1/Cd44/Lrp2/Cxcl12/Gdf7 | 23 |
| GO:0007409 | axonogenesis | BP | 0.0507 | 1.54e-10 | 6.03e-08 | 7.22 | 4.31e-08 | Gap43/Epha8/Ptprz1/Prtg/Efnb3/Map2/Pla2g10/Sema5a/Dcc/Nog/Lrrc4c/Sema5b/Ptpro/Rnf165/Itga4/Plxnb1/Nptx1/Cdh2/Dab1/Edn3/Lrp2/Evl/Sema7a/Cxcl12/Gdf7 | 25 |
| GO:0007411 | axon guidance | BP | 0.0698 | 4.17e-10 | 1.47e-07 | 6.83 | 1.05e-07 | Gap43/Epha8/Prtg/Efnb3/Pla2g10/Sema5a/Dcc/Nog/Sema5b/Ptpro/Rnf165/Plxnb1/Edn3/Lrp2/Evl/Sema7a/Cxcl12/Gdf7 | 18 |
| GO:0035107 | appendage morphogenesis | BP | 0.0862 | 6.71e-10 | 1.97e-07 | 6.71 | 1.41e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Pkdcc/Hoxd10/Tbx2/Hoxd11/Rnf165/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 15 |
| GO:0035108 | limb morphogenesis | BP | 0.0862 | 6.71e-10 | 1.97e-07 | 6.71 | 1.41e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Pkdcc/Hoxd10/Tbx2/Hoxd11/Rnf165/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 15 |
| GO:0001655 | urogenital system development | BP | 0.0543 | 1.42e-09 | 3.68e-07 | 6.43 | 2.63e-07 | Stra6/Wnt6/Wnt11/Angpt1/Nog/Hoxd11/Adamts16/Igf1/Ptpro/Cyp7b1/Npnt/Pygo1/Ednrb/Gpc3/Hoxa11/Foxf1/Cd44/Osr2/Lrp2/Mmp2/Nid1 | 21 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | BP | 0.0748 | 1.46e-09 | 3.68e-07 | 6.43 | 2.63e-07 | Gpc6/Prtg/Hmcn1/Sdk2/Pcdh18/Lrrc4c/Mcam/Nlgn1/Ptprd/Itga4/Lrrc4b/Cdh6/Cdh2/Dab1/Tenm3/Cdh10 | 16 |
| GO:0050673 | epithelial cell proliferation | BP | 0.0483 | 2.15e-09 | 5.07e-07 | 6.3 | 3.62e-07 | Gli1/Ihh/Hgf/Sfrp2/Sema5a/Cd109/Mdk/Nog/Tbx2/Gkn3/Igf1/Fst/Areg/Itga4/Cyp7b1/Kdr/Ednrb/Gpc3/Calca/Osr2/Has2/Cxcl12/Agap2 | 23 |
| GO:0030326 | embryonic limb morphogenesis | BP | 0.0909 | 4.91e-09 | 1.02e-06 | 5.99 | 7.29e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Hoxd10/Tbx2/Hoxd11/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 13 |
| GO:0035113 | embryonic appendage morphogenesis | BP | 0.0909 | 4.91e-09 | 1.02e-06 | 5.99 | 7.29e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Hoxd10/Tbx2/Hoxd11/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 13 |
| GO:0048736 | appendage development | BP | 0.0735 | 6.01e-09 | 1.12e-06 | 5.95 | 7.99e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Pkdcc/Hoxd10/Tbx2/Hoxd11/Rnf165/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 15 |
| GO:0060173 | limb development | BP | 0.0735 | 6.01e-09 | 1.12e-06 | 5.95 | 7.99e-07 | Ihh/Sfrp2/Lef1/Aff3/Nog/Pkdcc/Hoxd10/Tbx2/Hoxd11/Rnf165/Gpc3/Hoxa11/Hoxa10/Osr2/Hoxd9 | 15 |
| GO:0042391 | regulation of membrane potential | BP | 0.0469 | 8.32e-09 | 1.47e-06 | 5.83 | 1.05e-06 | Kcnk3/Nalcn/Grik1/Kcnd2/Begain/Kcnd3/Rims3/Adra1a/Drd4/Scn4b/Nlgn1/Slc24a4/Scn1a/Abat/Kdr/Celf4/P2rx2/Grin2b/Ano1/Bok/Slc8a3/Gabra4 | 22 |
| GO:0001569 | branching involved in blood vessel morphogenesis | BP | 0.178 | 2.32e-08 | 3.81e-06 | 5.42 | 2.73e-06 | Ihh/Sfrp2/Lef1/Angpt1/Sema5a/Mdk/Kdr/Cxcl12 | 8 |
| GO:0031644 | regulation of nervous system process | BP | 0.0722 | 2.46e-08 | 3.81e-06 | 5.42 | 2.73e-06 | Hgf/Begain/Drd4/Igf1/Nlgn1/Nptx1/Abat/Ednrb/Celf4/Calca/Nos3/Grin2b/Wasf3/Slc8a3 | 14 |
| GO:0048705 | skeletal system morphogenesis | BP | 0.0613 | 2.48e-08 | 3.81e-06 | 5.42 | 2.73e-06 | Ihh/Col11a1/Mmp16/Sfrp2/Hhip/Nog/Hoxd10/Hoxd11/Hoxa11/Stc1/Cbs/Osr2/Has2/Hoxd9/Frem1/Mmp2 | 16 |
| GO:0009581 | detection of external stimulus | BP | 0.0876 | 2.93e-08 | 4.31e-06 | 5.37 | 3.08e-06 | Col11a1/Sema5a/Sema5b/Igf1/Slc24a4/Scn1a/Calca/Foxf1/Grin2b/Serpine2/Ano1/Cxcl12 | 12 |
| GO:0072001 | renal system development | BP | 0.0522 | 3.97e-08 | 5.38e-06 | 5.27 | 3.85e-06 | Stra6/Wnt6/Wnt11/Angpt1/Nog/Hoxd11/Adamts16/Ptpro/Npnt/Pygo1/Ednrb/Gpc3/Hoxa11/Foxf1/Cd44/Osr2/Lrp2/Nid1 | 18 |
| GO:0003012 | muscle system process | BP | 0.0465 | 4.63e-08 | 6.06e-06 | 5.22 | 4.33e-06 | Kcnd3/Ryr1/Col14a1/Sgcd/Adra1a/Tbx2/Scn4b/Igf1/Scn1a/Npnt/Mybpc1/Abat/Ednrb/Tmod2/Stc1/Calca/Nos3/Edn3/P2rx2/Slc8a3 | 20 |
| GO:0006936 | muscle contraction | BP | 0.0541 | 5.58e-08 | 7.03e-06 | 5.15 | 5.03e-06 | Kcnd3/Ryr1/Sgcd/Adra1a/Tbx2/Scn4b/Scn1a/Npnt/Mybpc1/Abat/Ednrb/Tmod2/Stc1/Calca/Edn3/P2rx2/Slc8a3 | 17 |
| GO:0001822 | kidney development | BP | 0.0515 | 1.14e-07 | 1.34e-05 | 4.87 | 9.59e-06 | Stra6/Wnt6/Wnt11/Angpt1/Nog/Hoxd11/Adamts16/Ptpro/Npnt/Pygo1/Ednrb/Gpc3/Hoxa11/Cd44/Osr2/Lrp2/Nid1 | 17 |
| GO:0072006 | nephron development | BP | 0.0764 | 1.33e-07 | 1.5e-05 | 4.82 | 1.07e-05 | Wnt6/Angpt1/Nog/Hoxd11/Adamts16/Ptpro/Npnt/Ednrb/Gpc3/Hoxa11/Cd44/Nid1 | 12 |
| GO:0060348 | bone development | BP | 0.0581 | 1.36e-07 | 1.5e-05 | 4.82 | 1.07e-05 | Ihh/Mmp16/Sfrp2/Ryr1/Hoxd11/Igf1/Plxnb1/Kdr/Evc/Hoxa11/Stc1/Cbs/Osr2/Has2/Frem1 | 15 |
| GO:0007224 | smoothened signaling pathway | BP | 0.0755 | 1.53e-07 | 1.63e-05 | 4.79 | 1.17e-05 | Gli1/Ihh/Hhip/Ptch2/Nog/Evc2/Evc/Gpc3/Foxf1/Serpine2/Dzip1l/Kif7 | 12 |
| GO:0060485 | mesenchyme development | BP | 0.0535 | 1.62e-07 | 1.68e-05 | 4.78 | 1.2e-05 | Sfrp2/Wnt11/Lef1/Sema5a/Mdk/Nog/Sema5b/Tbx2/Itga4/Ednrb/Cdh2/Foxf1/Nos3/Edn3/Has2/Sema7a | 16 |
| GO:0048265 | response to pain | BP | 0.175 | 2.01e-07 | 2.02e-05 | 4.69 | 1.45e-05 | Grik1/Slc6a2/Ednrb/Calca/P2rx2/Cacna1e/Grin2b | 7 |
| GO:0060541 | respiratory system development | BP | 0.0562 | 2.12e-07 | 2.08e-05 | 4.68 | 1.49e-05 | Gli1/Stra6/Hhip/Lef1/Nog/Pkdcc/Tbx2/Igf1/Kdr/Gpc3/Edaradd/Foxf1/Nos3/Hopx/Ano1 | 15 |
| GO:0009582 | detection of abiotic stimulus | BP | 0.0809 | 2.51e-07 | 2.33e-05 | 4.63 | 1.67e-05 | Col11a1/Sema5a/Sema5b/Igf1/Slc24a4/Scn1a/Calca/Grin2b/Serpine2/Ano1/Cxcl12 | 11 |
| GO:0045785 | positive regulation of cell adhesion | BP | 0.0415 | 2.9e-07 | 2.63e-05 | 4.58 | 1.88e-05 | Ihh/Efnb3/Vit/Col26a1/Lef1/Angpt1/Cd83/Mdk/Igf1/Itga4/Npnt/Kdr/Spock2/Chst2/Calca/Foxf1/Cd44/Has2/Cxcl12/Nid1 | 20 |
| GO:0060349 | bone morphogenesis | BP | 0.0909 | 2.98e-07 | 2.63e-05 | 4.58 | 1.88e-05 | Ihh/Mmp16/Sfrp2/Hoxd11/Hoxa11/Stc1/Cbs/Osr2/Has2/Frem1 | 10 |
| GO:0048608 | reproductive structure development | BP | 0.0413 | 3.1e-07 | 2.67e-05 | 4.57 | 1.91e-05 | Ndp/Tex15/Stra6/Sfrp2/Lef1/Angpt1/Nog/Igf1/Fst/Itga4/Cyp7b1/Kdr/Hoxa11/Hoxa10/Nos3/Cd44/Lrp2/Serpine2/Mmp2/Gdf7 | 20 |
| GO:0061458 | reproductive system development | BP | 0.041 | 3.53e-07 | 2.96e-05 | 4.53 | 2.12e-05 | Ndp/Tex15/Stra6/Sfrp2/Lef1/Angpt1/Nog/Igf1/Fst/Itga4/Cyp7b1/Kdr/Hoxa11/Hoxa10/Nos3/Cd44/Lrp2/Serpine2/Mmp2/Gdf7 | 20 |
| GO:0071805 | potassium ion transmembrane transport | BP | 0.0631 | 3.7e-07 | 3.04e-05 | 4.52 | 2.17e-05 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Kcnd2/Kcna6/Kcnd3/Slc24a4/Slc12a8/Edn3/Kcnq5/Slc9a7 | 13 |
| GO:0006813 | potassium ion transport | BP | 0.0576 | 3.99e-07 | 3.2e-05 | 4.49 | 2.29e-05 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Kcnd2/Kcna6/Kcnd3/Slc24a4/Slc12a8/Nos3/Edn3/Kcnq5/Slc9a7 | 14 |
| GO:0050678 | regulation of epithelial cell proliferation | BP | 0.0444 | 4.29e-07 | 3.36e-05 | 4.47 | 2.4e-05 | Gli1/Ihh/Sfrp2/Sema5a/Cd109/Mdk/Nog/Gkn3/Igf1/Itga4/Cyp7b1/Kdr/Ednrb/Gpc3/Osr2/Has2/Cxcl12/Agap2 | 18 |
| GO:0006814 | sodium ion transport | BP | 0.0565 | 5.1e-07 | 3.83e-05 | 4.42 | 2.74e-05 | Nalcn/Slc5a11/Slc13a5/Slc6a2/Drd4/Scn4b/Slc24a4/Scn1a/Ednrb/Hecw2/Nos3/Slc9a7/Serpine2/Slc8a3 | 14 |
| GO:0009612 | response to mechanical stimulus | BP | 0.0674 | 5.17e-07 | 3.83e-05 | 4.42 | 2.74e-05 | Stra6/Col11a1/Slc1a3/Wnt11/Mdk/Igf1/Scn1a/Nos3/Grin2b/Serpine2/Mmp2/Cxcl12 | 12 |
| GO:0072498 | embryonic skeletal joint development | BP | 0.312 | 5.27e-07 | 3.83e-05 | 4.42 | 2.74e-05 | Ihh/Nog/Hoxd11/Hoxa11/Osr2 | 5 |
| GO:0072009 | nephron epithelium development | BP | 0.0855 | 5.32e-07 | 3.83e-05 | 4.42 | 2.74e-05 | Wnt6/Nog/Hoxd11/Adamts16/Ptpro/Npnt/Ednrb/Gpc3/Hoxa11/Cd44 | 10 |
| GO:0001658 | branching involved in ureteric bud morphogenesis | BP | 0.119 | 5.73e-07 | 4.04e-05 | 4.39 | 2.89e-05 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0001666 | response to hypoxia | BP | 0.0602 | 6.35e-07 | 4.4e-05 | 4.36 | 3.14e-05 | Kcnk3/Kcnd2/Ryr1/Ddah1/Abat/Kdr/Pygm/Cbs/P2rx2/Grin2b/Tert/Slc8a3/Mmp2 | 13 |
| GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.14 | 9.82e-07 | 6.66e-05 | 4.18 | 4.76e-05 | Hmcn1/Mcam/Nlgn1/Ptprd/Itga4/Cdh2/Tenm3 | 7 |
| GO:0022612 | gland morphogenesis | BP | 0.0701 | 1.06e-06 | 7.05e-05 | 4.15 | 5.04e-05 | Gli1/Hgf/Mdk/Nog/Tbx2/Igf1/Areg/Cyp7b1/Cd44/Mmp2/Gdf7 | 11 |
| GO:0003073 | regulation of systemic arterial blood pressure | BP | 0.0787 | 1.14e-06 | 7.42e-05 | 4.13 | 5.3e-05 | Adra1a/Adamts16/Ptpro/Ddah1/Kdr/Ednrb/Calca/Nos3/Edn3/P2rx2 | 10 |
| GO:0060675 | ureteric bud morphogenesis | BP | 0.108 | 1.24e-06 | 7.96e-05 | 4.1 | 5.7e-05 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0042063 | gliogenesis | BP | 0.0457 | 1.31e-06 | 8.27e-05 | 4.08 | 5.91e-05 | Gap43/Ptprz1/Lef1/Apcdd1/Mdk/Nog/Igf1/Lamc3/Areg/Cdh2/Dab1/Lrp2/Tert/Serpine2/Wasf3/Slc8a3 | 16 |
| GO:0072171 | mesonephric tubule morphogenesis | BP | 0.107 | 1.38e-06 | 8.52e-05 | 4.07 | 6.1e-05 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0099068 | postsynapse assembly | BP | 0.176 | 1.46e-06 | 8.87e-05 | 4.05 | 6.34e-05 | Gap43/Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2 | 6 |
| GO:0050808 | synapse organization | BP | 0.0385 | 1.78e-06 | 0.000106 | 3.97 | 7.61e-05 | Gap43/Gpc6/Sdk2/St8sia2/Lrrc4c/Rims3/Ptpro/Nlgn1/Ptprd/Plxnb1/Pdzrn3/Nptx1/Lrrc4b/Cdh2/P2rx2/Ppfia2/Grin2b/Wasf3/Slc8a3 | 19 |
| GO:0016055 | Wnt signaling pathway | BP | 0.0399 | 1.99e-06 | 0.000117 | 3.93 | 8.38e-05 | Ndp/Gli1/Wnt6/Sfrp2/Wnt11/Lef1/Sema5a/Apcdd1/Mdk/Nog/Ptpro/Pygo1/Ednrb/Gpc3/Cdh2/Nkd1/Cd44/Tert | 18 |
| GO:0001667 | ameboidal-type cell migration | BP | 0.0398 | 2.06e-06 | 0.000118 | 3.93 | 8.41e-05 | Cyp1b1/Wnt11/Angpt1/Sema5a/Sema5b/Igf1/Itga4/Kdr/Ednrb/Cdh2/Stc1/Calca/Nos3/Edn3/Has2/Evl/Sema7a/Cxcl12 | 18 |
| GO:0098657 | import into cell | BP | 0.0542 | 2.07e-06 | 0.000118 | 3.93 | 8.41e-05 | Kcnj3/Slc1a3/Slc6a2/Acsl6/Kcnd3/Slc24a4/Itga4/Slc12a8/Lrp2/Cacna1e/Slc9a7/Slc8a3/Slc7a2 | 13 |
| GO:0198738 | cell-cell signaling by wnt | BP | 0.0397 | 2.12e-06 | 0.000119 | 3.93 | 8.49e-05 | Ndp/Gli1/Wnt6/Sfrp2/Wnt11/Lef1/Sema5a/Apcdd1/Mdk/Nog/Ptpro/Pygo1/Ednrb/Gpc3/Cdh2/Nkd1/Cd44/Tert | 18 |
| GO:0060560 | developmental growth involved in morphogenesis | BP | 0.0498 | 2.24e-06 | 0.000122 | 3.91 | 8.76e-05 | Sfrp2/Map2/Wnt11/Sema5a/St8sia2/Sema5b/Tbx2/Cpne5/Areg/Itga4/Nkd1/Edn3/Sema7a/Cxcl12 | 14 |
| GO:0050767 | regulation of neurogenesis | BP | 0.0396 | 2.26e-06 | 0.000122 | 3.91 | 8.76e-05 | Ptprz1/Prtg/Efnb3/Map2/Sema5a/Mdk/Nog/Sema5b/Igf1/Ptprd/Plxnb1/Kdr/Dab1/Lrp2/Sema7a/Tert/Serpine2/Cxcl12 | 18 |
| GO:0036293 | response to decreased oxygen levels | BP | 0.0535 | 2.37e-06 | 0.000127 | 3.9 | 9.07e-05 | Kcnk3/Kcnd2/Ryr1/Ddah1/Abat/Kdr/Pygm/Cbs/P2rx2/Grin2b/Tert/Slc8a3/Mmp2 | 13 |
| GO:0009954 | proximal/distal pattern formation | BP | 0.162 | 2.45e-06 | 0.000129 | 3.89 | 9.22e-05 | Gli1/Hoxd10/Hoxd11/Hoxa11/Hoxa10/Hoxd9 | 6 |
| GO:0048844 | artery morphogenesis | BP | 0.0988 | 2.48e-06 | 0.000129 | 3.89 | 9.22e-05 | Apob/Stra6/Wnt11/Mdk/Nog/Tbx2/Foxf1/Lrp2 | 8 |
| GO:0001823 | mesonephros development | BP | 0.0818 | 2.85e-06 | 0.000146 | 3.84 | 0.000104 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44/Osr2 | 9 |
| GO:0072078 | nephron tubule morphogenesis | BP | 0.0964 | 2.99e-06 | 0.000151 | 3.82 | 0.000108 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0072088 | nephron epithelium morphogenesis | BP | 0.0941 | 3.58e-06 | 0.000178 | 3.75 | 0.000127 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0050906 | detection of stimulus involved in sensory perception | BP | 0.0611 | 4.02e-06 | 0.000197 | 3.71 | 0.000141 | Col11a1/Sema5a/Sema5b/Igf1/Slc24a4/Scn1a/Calca/Grin2b/Serpine2/Ano1/Cxcl12 | 11 |
| GO:0050679 | positive regulation of epithelial cell proliferation | BP | 0.0553 | 4.13e-06 | 2e-04 | 3.7 | 0.000143 | Ihh/Sema5a/Mdk/Nog/Igf1/Itga4/Cyp7b1/Kdr/Osr2/Has2/Cxcl12/Agap2 | 12 |
| GO:0048732 | gland development | BP | 0.0378 | 4.22e-06 | 0.000201 | 3.7 | 0.000144 | Gli1/Stra6/Hgf/Lef1/Mdk/Nog/Tbx2/Igf1/Areg/Cyp7b1/Edaradd/Foxf1/Cd44/Hoxd9/Serpine2/Mmp2/Gdf7/Agap2 | 18 |
| GO:0072028 | nephron morphogenesis | BP | 0.092 | 4.27e-06 | 0.000201 | 3.7 | 0.000144 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0010171 | body morphogenesis | BP | 0.113 | 4.33e-06 | 0.000201 | 3.7 | 0.000144 | Ihh/Stra6/Lef1/Nog/Crispld1/Gpc3/Mmp2 | 7 |
| GO:0060840 | artery development | BP | 0.0776 | 4.43e-06 | 0.000203 | 3.69 | 0.000145 | Apob/Stra6/Wnt11/Mdk/Nog/Tbx2/Foxf1/Lrp2/Kif7 | 9 |
| GO:0008217 | regulation of blood pressure | BP | 0.0545 | 4.76e-06 | 0.000215 | 3.67 | 0.000154 | Adra1a/Adamts16/Ptpro/Ddah1/Abat/Kdr/Ednrb/Calca/Nos3/Edn3/P2rx2/Npr3 | 12 |
| GO:0061333 | renal tubule morphogenesis | BP | 0.0899 | 5.07e-06 | 0.000225 | 3.65 | 0.000161 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0060078 | regulation of postsynaptic membrane potential | BP | 0.0763 | 5.1e-06 | 0.000225 | 3.65 | 0.000161 | Grik1/Begain/Drd4/Nlgn1/Abat/Celf4/Grin2b/Slc8a3/Gabra4 | 9 |
| GO:0060070 | canonical Wnt signaling pathway | BP | 0.0461 | 5.56e-06 | 0.00024 | 3.62 | 0.000172 | Gli1/Wnt6/Sfrp2/Wnt11/Lef1/Sema5a/Mdk/Nog/Ptpro/Pygo1/Ednrb/Gpc3/Cdh2/Nkd1 | 14 |
| GO:0072073 | kidney epithelium development | BP | 0.0658 | 5.76e-06 | 0.00024 | 3.62 | 0.000172 | Wnt6/Nog/Hoxd11/Adamts16/Ptpro/Npnt/Ednrb/Gpc3/Hoxa11/Cd44 | 10 |
| GO:0071772 | response to BMP | BP | 0.0588 | 5.79e-06 | 0.00024 | 3.62 | 0.000172 | Sfrp2/Lef1/Kcnd3/Nog/Fst/Rnf165/Kdr/Gpc3/Lrp2/Vstm2a/Gdf7 | 11 |
| GO:0071773 | cellular response to BMP stimulus | BP | 0.0588 | 5.79e-06 | 0.00024 | 3.62 | 0.000172 | Sfrp2/Lef1/Kcnd3/Nog/Fst/Rnf165/Kdr/Gpc3/Lrp2/Vstm2a/Gdf7 | 11 |
| GO:0070444 | oligodendrocyte progenitor proliferation | BP | 0.333 | 5.85e-06 | 0.00024 | 3.62 | 0.000172 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0070445 | regulation of oligodendrocyte progenitor proliferation | BP | 0.333 | 5.85e-06 | 0.00024 | 3.62 | 0.000172 | Ptprz1/Nog/Cdh2/Lrp2 | 4 |
| GO:0060537 | muscle tissue development | BP | 0.0365 | 6.83e-06 | 0.000277 | 3.56 | 0.000198 | Gli1/Ihh/Stra6/Col11a1/Lef1/AW551984/Angpt1/Ryr1/Col14a1/Sgcd/Nog/Hoxd10/Adra1a/Tbx2/Igf1/P2rx2/Lrp2/Hoxd9 | 18 |
| GO:0099084 | postsynaptic specialization organization | BP | 0.136 | 7e-06 | 0.000278 | 3.56 | 0.000199 | Gap43/Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2 | 6 |
| GO:1904035 | regulation of epithelial cell apoptotic process | BP | 0.086 | 7.04e-06 | 0.000278 | 3.56 | 0.000199 | Angpt1/Sema5a/Mdk/Igf1/Itga4/Kdr/Tert/Bok | 8 |
| GO:0098739 | import across plasma membrane | BP | 0.0576 | 7.09e-06 | 0.000278 | 3.56 | 0.000199 | Kcnj3/Slc1a3/Acsl6/Kcnd3/Slc24a4/Slc12a8/Lrp2/Cacna1e/Slc9a7/Slc8a3/Slc7a2 | 11 |
| GO:0007162 | negative regulation of cell adhesion | BP | 0.0447 | 7.75e-06 | 3e-04 | 3.52 | 0.000215 | Cyp1b1/Ihh/Ptprz1/Lef1/Angpt1/Sema5a/Mdk/Plxnb1/Abat/Dab1/Cd44/Serpine2/Mmp2/Cxcl12 | 14 |
| GO:0051962 | positive regulation of nervous system development | BP | 0.0418 | 8.33e-06 | 0.000318 | 3.5 | 0.000227 | Ptprz1/Sema5a/St8sia2/Mdk/Nog/Igf1/Nlgn1/Ptprd/Plxnb1/Kdr/Lrrc4b/Lrp2/Sema7a/Serpine2/Cxcl12 | 15 |
| GO:0060272 | embryonic skeletal joint morphogenesis | BP | 0.308 | 8.38e-06 | 0.000318 | 3.5 | 0.000227 | Nog/Hoxd11/Hoxa11/Osr2 | 4 |
| GO:0035136 | forelimb morphogenesis | BP | 0.13 | 9.12e-06 | 0.000339 | 3.47 | 0.000242 | Hoxd10/Hoxd11/Rnf165/Hoxa11/Osr2/Hoxd9 | 6 |
| GO:0060323 | head morphogenesis | BP | 0.13 | 9.12e-06 | 0.000339 | 3.47 | 0.000242 | Ihh/Stra6/Lef1/Nog/Crispld1/Mmp2 | 6 |
| GO:0030324 | lung development | BP | 0.0508 | 9.71e-06 | 0.000357 | 3.45 | 0.000255 | Gli1/Stra6/Hhip/Nog/Pkdcc/Tbx2/Igf1/Kdr/Gpc3/Foxf1/Nos3/Hopx | 12 |
| GO:0050974 | detection of mechanical stimulus involved in sensory perception | BP | 0.128 | 1.04e-05 | 0.000376 | 3.42 | 0.000269 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12 | 6 |
| GO:0061351 | neural precursor cell proliferation | BP | 0.0553 | 1.05e-05 | 0.000376 | 3.42 | 0.000269 | Gli1/Ptprz1/Hhip/Lef1/Acsl6/Mdk/Nog/Igf1/Cdh2/Lrp2/Smarca1 | 11 |
| GO:0060047 | heart contraction | BP | 0.0504 | 1.06e-05 | 0.000377 | 3.42 | 0.000269 | Kcnd3/Sgcd/Adra1a/Scn4b/Scn1a/Ednrb/Stc1/Calca/Nos3/Hopx/Edn3/Cacna1e | 12 |
| GO:0098698 | postsynaptic specialization assembly | BP | 0.179 | 1.07e-05 | 0.000377 | 3.42 | 0.00027 | Gap43/Nlgn1/Ptprd/Nptx1/Lrrc4b | 5 |
| GO:0042733 | embryonic digit morphogenesis | BP | 0.0986 | 1.08e-05 | 0.000377 | 3.42 | 0.00027 | Ihh/Sfrp2/Nog/Tbx2/Hoxd11/Hoxa11/Osr2 | 7 |
| GO:0030323 | respiratory tube development | BP | 0.0502 | 1.1e-05 | 0.000381 | 3.42 | 0.000273 | Gli1/Stra6/Hhip/Nog/Pkdcc/Tbx2/Igf1/Kdr/Gpc3/Foxf1/Nos3/Hopx | 12 |
| GO:0048762 | mesenchymal cell differentiation | BP | 0.0496 | 1.25e-05 | 0.000424 | 3.37 | 0.000303 | Sfrp2/Wnt11/Lef1/Sema5a/Mdk/Nog/Sema5b/Ednrb/Cdh2/Edn3/Has2/Sema7a | 12 |
| GO:0030178 | negative regulation of Wnt signaling pathway | BP | 0.0599 | 1.32e-05 | 0.000443 | 3.35 | 0.000317 | Gli1/Sfrp2/Wnt11/Apcdd1/Mdk/Nog/Ptpro/Gpc3/Cdh2/Nkd1 | 10 |
| GO:2000027 | regulation of animal organ morphogenesis | BP | 0.0677 | 1.35e-05 | 0.000449 | 3.35 | 0.000321 | Hgf/Sfrp2/Wnt11/Apcdd1/Nog/Hoxd11/Gpc3/Hoxa11/Nkd1 | 9 |
| GO:0072080 | nephron tubule development | BP | 0.0784 | 1.4e-05 | 0.00046 | 3.34 | 0.000329 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | BP | 0.0672 | 1.43e-05 | 0.000468 | 3.33 | 0.000335 | Gli1/Sfrp2/Wnt11/Mdk/Nog/Ptpro/Gpc3/Cdh2/Nkd1 | 9 |
| GO:0010721 | negative regulation of cell development | BP | 0.0534 | 1.45e-05 | 0.000468 | 3.33 | 0.000335 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Igf1/Ednrb/Dab1/Sema7a/Tert | 11 |
| GO:0060993 | kidney morphogenesis | BP | 0.0777 | 1.5e-05 | 0.000477 | 3.32 | 0.000341 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | BP | 0.0777 | 1.5e-05 | 0.000477 | 3.32 | 0.000341 | Grik1/Begain/Drd4/Nlgn1/Abat/Celf4/Grin2b/Slc8a3 | 8 |
| GO:0010720 | positive regulation of cell development | BP | 0.0397 | 1.53e-05 | 0.000481 | 3.32 | 0.000344 | Ptprz1/Sema5a/Mdk/Nog/Hoxd11/Igf1/Ptprd/Plxnb1/Kdr/Hoxa11/Has2/Lrp2/Sema7a/Serpine2/Cxcl12 | 15 |
| GO:0048015 | phosphatidylinositol-mediated signaling | BP | 0.0588 | 1.54e-05 | 0.000481 | 3.32 | 0.000344 | Hgf/Myo16/Angpt1/Car8/Igf1/Plxnb1/Kdr/Serpine2/Npr3/Agap2 | 10 |
| GO:0048562 | embryonic organ morphogenesis | BP | 0.042 | 1.55e-05 | 0.000481 | 3.32 | 0.000344 | Gli1/Ihh/Stra6/Col11a1/Mmp16/Wnt11/Nog/Hoxd10/Tbx2/Hoxd11/Hoxa11/Foxf1/Osr2/Hoxd9 | 14 |
| GO:0003015 | heart process | BP | 0.048 | 1.73e-05 | 0.00053 | 3.28 | 0.000379 | Kcnd3/Sgcd/Adra1a/Scn4b/Scn1a/Ednrb/Stc1/Calca/Nos3/Hopx/Edn3/Cacna1e | 12 |
| GO:0048017 | inositol lipid-mediated signaling | BP | 0.0578 | 1.79e-05 | 0.000545 | 3.26 | 0.00039 | Hgf/Myo16/Angpt1/Car8/Igf1/Plxnb1/Kdr/Serpine2/Npr3/Agap2 | 10 |
| GO:0003002 | regionalization | BP | 0.039 | 1.89e-05 | 0.000569 | 3.25 | 0.000407 | Gli1/Sfrp2/Hhip/Wnt11/Lef1/Nog/Hoxd10/Hoxd11/Gpc3/Hoxa11/Foxf1/Hoxa10/Lrp2/Hoxd9/Dzip1l | 15 |
| GO:0061448 | connective tissue development | BP | 0.0441 | 1.91e-05 | 0.000569 | 3.25 | 0.000407 | Ihh/Col11a1/Sfrp2/Mdk/Nog/Pkdcc/Hoxd11/Igf1/Hoxa11/Stc1/Cbs/Osr2/Gdf7 | 13 |
| GO:0055123 | digestive system development | BP | 0.0647 | 1.92e-05 | 0.000569 | 3.25 | 0.000407 | Ihh/Stra6/Sfrp2/Pkdcc/Tbx2/Igf1/Ednrb/Foxf1/Clmp | 9 |
| GO:0051216 | cartilage development | BP | 0.0516 | 1.97e-05 | 0.000569 | 3.25 | 0.000407 | Ihh/Col11a1/Sfrp2/Mdk/Nog/Pkdcc/Hoxd11/Hoxa11/Stc1/Cbs/Osr2 | 11 |
| GO:0001657 | ureteric bud development | BP | 0.0748 | 1.98e-05 | 0.000569 | 3.25 | 0.000407 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0072163 | mesonephric epithelium development | BP | 0.0748 | 1.98e-05 | 0.000569 | 3.25 | 0.000407 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0072164 | mesonephric tubule development | BP | 0.0748 | 1.98e-05 | 0.000569 | 3.25 | 0.000407 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0001505 | regulation of neurotransmitter levels | BP | 0.0472 | 2.02e-05 | 0.000574 | 3.24 | 0.000411 | Syn2/Maob/Slc6a2/Rims3/Adra1a/Drd4/Nlgn1/Abat/Edn3/P2rx2/Ppfia2/Cadps | 12 |
| GO:0010811 | positive regulation of cell-substrate adhesion | BP | 0.0643 | 2.04e-05 | 0.000574 | 3.24 | 0.000411 | Vit/Col26a1/Mdk/Npnt/Kdr/Spock2/Foxf1/Has2/Nid1 | 9 |
| GO:1904036 | negative regulation of epithelial cell apoptotic process | BP | 0.113 | 2.1e-05 | 0.000588 | 3.23 | 0.00042 | Angpt1/Sema5a/Mdk/Igf1/Kdr/Tert | 6 |
| GO:0061326 | renal tubule development | BP | 0.0741 | 2.12e-05 | 0.000589 | 3.23 | 0.000421 | Wnt6/Nog/Hoxd11/Adamts16/Npnt/Gpc3/Hoxa11/Cd44 | 8 |
| GO:0031345 | negative regulation of cell projection organization | BP | 0.0512 | 2.15e-05 | 0.000593 | 3.23 | 0.000424 | Ptprz1/Efnb3/Map2/Sema5a/Sema5b/Nlgn1/Dab1/Ntm/Evl/Grin2b/Sema7a | 11 |
| GO:0048704 | embryonic skeletal system morphogenesis | BP | 0.0734 | 2.27e-05 | 0.00062 | 3.21 | 0.000444 | Col11a1/Mmp16/Nog/Hoxd10/Hoxd11/Hoxa11/Osr2/Hoxd9 | 8 |
| GO:0006816 | calcium ion transport | BP | 0.0364 | 2.33e-05 | 0.00063 | 3.2 | 0.000451 | Nalcn/Spink1/Ryr1/Adra1a/Drd4/Igf1/Jph4/Slc24a4/Ednrb/Stc1/Calca/Nos3/Cacna1e/Grin2b/Slc8a3/Cxcl12 | 16 |
| GO:0072132 | mesenchyme morphogenesis | BP | 0.111 | 2.34e-05 | 0.00063 | 3.2 | 0.000451 | Wnt11/Lef1/Nog/Tbx2/Foxf1/Nos3 | 6 |
| GO:0034329 | cell junction assembly | BP | 0.0363 | 2.39e-05 | 0.00064 | 3.19 | 0.000458 | Gap43/Gpc6/Sdk2/Wnt11/St8sia2/Ptpro/Nlgn1/Ptprd/Plxnb1/Nptx1/Kdr/Epb41l3/Lrrc4b/Cdh6/Cdh2/Cdh10 | 16 |
| GO:0070482 | response to oxygen levels | BP | 0.043 | 2.44e-05 | 0.000641 | 3.19 | 0.000458 | Kcnk3/Kcnd2/Ryr1/Ddah1/Abat/Kdr/Pygm/Cbs/P2rx2/Grin2b/Tert/Slc8a3/Mmp2 | 13 |
| GO:0010001 | glial cell differentiation | BP | 0.0463 | 2.45e-05 | 0.000641 | 3.19 | 0.000458 | Gap43/Ptprz1/Lef1/Mdk/Nog/Igf1/Lamc3/Cdh2/Dab1/Serpine2/Wasf3/Slc8a3 | 12 |
| GO:0060740 | prostate gland epithelium morphogenesis | BP | 0.152 | 2.47e-05 | 0.000641 | 3.19 | 0.000458 | Nog/Igf1/Cyp7b1/Cd44/Mmp2 | 5 |
| GO:0062149 | detection of stimulus involved in sensory perception of pain | BP | 0.152 | 2.47e-05 | 0.000641 | 3.19 | 0.000458 | Scn1a/Calca/Grin2b/Ano1/Cxcl12 | 5 |
| GO:0031346 | positive regulation of cell projection organization | BP | 0.0361 | 2.53e-05 | 0.000651 | 3.19 | 0.000466 | Ptprz1/Hgf/Fez1/Sema5a/Dcc/Acsl6/Mdk/Lrrc7/Nlgn1/Ptprd/Plxnb1/Tenm3/Sema7a/Serpine2/Cxcl12/Ccdc88a | 16 |
| GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | BP | 0.0714 | 2.76e-05 | 0.000706 | 3.15 | 0.000505 | Prtg/Hmcn1/Sdk2/Pcdh18/Cdh6/Cdh2/Tenm3/Cdh10 | 8 |
| GO:0048706 | embryonic skeletal system development | BP | 0.0616 | 2.84e-05 | 0.000718 | 3.14 | 0.000514 | Ihh/Col11a1/Mmp16/Nog/Hoxd10/Hoxd11/Hoxa11/Osr2/Hoxd9 | 9 |
| GO:0060512 | prostate gland morphogenesis | BP | 0.147 | 2.87e-05 | 0.000718 | 3.14 | 0.000514 | Nog/Igf1/Cyp7b1/Cd44/Mmp2 | 5 |
| GO:0099560 | synaptic membrane adhesion | BP | 0.147 | 2.87e-05 | 0.000718 | 3.14 | 0.000514 | Gpc6/Lrrc4c/Nlgn1/Ptprd/Lrrc4b | 5 |
| GO:0019932 | second-messenger-mediated signaling | BP | 0.0421 | 3.1e-05 | 0.000765 | 3.12 | 0.000547 | Spink1/Sgcd/Drd4/Nr5a2/Igf1/Slc24a4/Kdr/Ednrb/Cbs/Nos3/Gnai1/Edn3/Ptgfr | 13 |
| GO:0050770 | regulation of axonogenesis | BP | 0.0541 | 3.19e-05 | 0.000779 | 3.11 | 0.000557 | Efnb3/Map2/Sema5a/Lrrc4c/Sema5b/Plxnb1/Cdh2/Dab1/Sema7a/Cxcl12 | 10 |
| GO:0010463 | mesenchymal cell proliferation | BP | 0.105 | 3.2e-05 | 0.000779 | 3.11 | 0.000557 | Ihh/Wnt11/Tbx2/Kdr/Gpc3/Foxf1 | 6 |
| GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | BP | 0.143 | 3.32e-05 | 0.000802 | 3.1 | 0.000574 | Pcdh18/Nlgn1/Cdh6/Cdh2/Cdh10 | 5 |
| GO:0002062 | chondrocyte differentiation | BP | 0.0696 | 3.34e-05 | 0.000802 | 3.1 | 0.000574 | Ihh/Col11a1/Sfrp2/Mdk/Pkdcc/Hoxd11/Hoxa11/Osr2 | 8 |
| GO:0099150 | regulation of postsynaptic specialization assembly | BP | 0.222 | 3.44e-05 | 0.000819 | 3.09 | 0.000586 | Gap43/Ptprd/Nptx1/Lrrc4b | 4 |
| GO:0035725 | sodium ion transmembrane transport | BP | 0.06 | 3.51e-05 | 0.000832 | 3.08 | 0.000595 | Nalcn/Slc6a2/Drd4/Scn4b/Slc24a4/Scn1a/Hecw2/Slc9a7/Slc8a3 | 9 |
| GO:0006939 | smooth muscle contraction | BP | 0.0684 | 3.78e-05 | 0.00089 | 3.05 | 0.000636 | Adra1a/Tbx2/Npnt/Abat/Ednrb/Calca/Edn3/P2rx2 | 8 |
| GO:0030111 | regulation of Wnt signaling pathway | BP | 0.0409 | 4.16e-05 | 0.000973 | 3.01 | 0.000696 | Gli1/Sfrp2/Wnt11/Lef1/Sema5a/Apcdd1/Mdk/Nog/Ptpro/Gpc3/Cdh2/Nkd1/Tert | 13 |
| GO:1905874 | regulation of postsynaptic density organization | BP | 0.211 | 4.32e-05 | 0.001 | 3 | 0.000717 | Ptprd/Nptx1/Lrrc4b/Cdh2 | 4 |
| GO:2000177 | regulation of neural precursor cell proliferation | BP | 0.0667 | 4.54e-05 | 0.00105 | 2.98 | 0.000748 | Gli1/Ptprz1/Mdk/Nog/Igf1/Cdh2/Lrp2/Smarca1 | 8 |
| GO:0007416 | synapse assembly | BP | 0.0518 | 4.57e-05 | 0.00105 | 2.98 | 0.000749 | Gap43/Gpc6/Sdk2/St8sia2/Nlgn1/Ptprd/Plxnb1/Nptx1/Lrrc4b/Cdh2 | 10 |
| GO:0048588 | developmental cell growth | BP | 0.0433 | 4.71e-05 | 0.00106 | 2.97 | 0.000761 | Map2/Sema5a/St8sia2/Col14a1/Adra1a/Sema5b/Igf1/Cpne5/Itga4/Edn3/Sema7a/Cxcl12 | 12 |
| GO:0060324 | face development | BP | 0.0984 | 4.73e-05 | 0.00106 | 2.97 | 0.000761 | Stra6/Lef1/Nog/Crispld1/Itga4/Mmp2 | 6 |
| GO:1904019 | epithelial cell apoptotic process | BP | 0.0661 | 4.81e-05 | 0.00107 | 2.97 | 0.000769 | Angpt1/Sema5a/Mdk/Igf1/Itga4/Kdr/Tert/Bok | 8 |
| GO:0003018 | vascular process in circulatory system | BP | 0.0468 | 4.85e-05 | 0.00108 | 2.97 | 0.00077 | Angpt1/Adra1a/Ddah1/Kdr/Ednrb/Calca/Cbs/Nos3/Edn3/Mmp2/Npr3 | 11 |
| GO:0097106 | postsynaptic density organization | BP | 0.132 | 5e-05 | 0.0011 | 2.96 | 0.000788 | Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2 | 5 |
| GO:0050982 | detection of mechanical stimulus | BP | 0.0968 | 5.19e-05 | 0.00114 | 2.94 | 0.000813 | Col11a1/Igf1/Scn1a/Grin2b/Serpine2/Cxcl12 | 6 |
| GO:0031589 | cell-substrate adhesion | BP | 0.0375 | 5.36e-05 | 0.00117 | 2.93 | 0.000836 | Ptprz1/Vit/Col26a1/Angpt1/Mdk/Lamc3/Itga4/Npnt/Kdr/Spock2/Foxf1/Has2/Frem1/Nid1 | 14 |
| GO:0008016 | regulation of heart contraction | BP | 0.0508 | 5.43e-05 | 0.00118 | 2.93 | 0.000841 | Kcnd3/Adra1a/Scn4b/Ednrb/Stc1/Calca/Nos3/Hopx/Edn3/Cacna1e | 10 |
| GO:0043010 | camera-type eye development | BP | 0.0374 | 5.52e-05 | 0.00119 | 2.93 | 0.000849 | Ndp/Cyp1b1/Ihh/Stra6/Sdk2/Nhs/Tbx2/Tub/Lamc3/Kdr/Celf4/Rcn1/Tenm3/Osr2 | 14 |
| GO:0045879 | negative regulation of smoothened signaling pathway | BP | 0.128 | 5.69e-05 | 0.00122 | 2.92 | 0.000869 | Gli1/Hhip/Gpc3/Serpine2/Kif7 | 5 |
| GO:0060079 | excitatory postsynaptic potential | BP | 0.0761 | 5.88e-05 | 0.00125 | 2.9 | 0.000894 | Grik1/Begain/Drd4/Nlgn1/Celf4/Grin2b/Slc8a3 | 7 |
| GO:0003007 | heart morphogenesis | BP | 0.0423 | 5.98e-05 | 0.00126 | 2.9 | 0.000904 | Ihh/Col11a1/Sfrp2/Wnt11/Angpt1/Ryr1/Nog/Tbx2/Foxf1/Nos3/Has2/Lrp2 | 12 |
| GO:0003205 | cardiac chamber development | BP | 0.05 | 6.16e-05 | 0.00129 | 2.89 | 0.000926 | Stra6/Col11a1/Sfrp2/Wnt11/Nog/Tbx2/Foxf1/Nos3/Lrp2/Kif7 | 10 |
| GO:0048880 | sensory system development | BP | 0.035 | 6.34e-05 | 0.00131 | 2.88 | 0.000936 | Ndp/Tmem132e/Cyp1b1/Ihh/Stra6/Sdk2/Nhs/Tbx2/Tub/Lamc3/Kdr/Celf4/Rcn1/Tenm3/Osr2 | 15 |
| GO:0010977 | negative regulation of neuron projection development | BP | 0.0556 | 6.4e-05 | 0.00131 | 2.88 | 0.000936 | Ptprz1/Efnb3/Map2/Sema5a/Sema5b/Nlgn1/Dab1/Ntm/Sema7a | 9 |
| GO:0048565 | digestive tract development | BP | 0.0635 | 6.42e-05 | 0.00131 | 2.88 | 0.000936 | Ihh/Stra6/Sfrp2/Pkdcc/Tbx2/Ednrb/Foxf1/Clmp | 8 |
| GO:0060325 | face morphogenesis | BP | 0.125 | 6.44e-05 | 0.00131 | 2.88 | 0.000936 | Stra6/Lef1/Nog/Crispld1/Mmp2 | 5 |
| GO:0060351 | cartilage development involved in endochondral bone morphogenesis | BP | 0.125 | 6.44e-05 | 0.00131 | 2.88 | 0.000936 | Ihh/Hoxd11/Hoxa11/Stc1/Cbs | 5 |
| GO:0009100 | glycoprotein metabolic process | BP | 0.0392 | 6.46e-05 | 0.00131 | 2.88 | 0.000936 | Ihh/Col11a1/St6gal2/St8sia2/Igf1/Itm2a/Spock2/Tet1/St8sia4/Chst10/Gxylt2/Tmtc1/Soat1 | 13 |
| GO:0045165 | cell fate commitment | BP | 0.0418 | 6.61e-05 | 0.00133 | 2.88 | 0.000953 | Gap43/Ihh/Wnt6/Sfrp2/Wnt11/AW551984/Ptch2/Hoxd10/Tbx2/Kdr/Hoxa11/Gdf7 | 12 |
| GO:0050768 | negative regulation of neurogenesis | BP | 0.0552 | 6.71e-05 | 0.00135 | 2.87 | 0.000962 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Dab1/Sema7a/Tert | 9 |
| GO:0008589 | regulation of smoothened signaling pathway | BP | 0.0745 | 6.75e-05 | 0.00135 | 2.87 | 0.000962 | Gli1/Ihh/Hhip/Evc/Gpc3/Serpine2/Kif7 | 7 |
| GO:0014706 | striated muscle tissue development | BP | 0.0414 | 7.3e-05 | 0.00145 | 2.84 | 0.00103 | Gli1/Col11a1/Lef1/AW551984/Angpt1/Col14a1/Sgcd/Nog/Adra1a/Tbx2/Igf1/Lrp2 | 12 |
| GO:0048286 | lung alveolus development | BP | 0.0909 | 7.4e-05 | 0.00146 | 2.84 | 0.00104 | Stra6/Pkdcc/Igf1/Kdr/Foxf1/Hopx | 6 |
| GO:0014013 | regulation of gliogenesis | BP | 0.062 | 7.58e-05 | 0.00148 | 2.83 | 0.00106 | Ptprz1/Mdk/Nog/Igf1/Dab1/Lrp2/Tert/Serpine2 | 8 |
| GO:0060438 | trachea development | BP | 0.182 | 7.94e-05 | 0.00155 | 2.81 | 0.00111 | Lef1/Edaradd/Foxf1/Ano1 | 4 |
| GO:0001707 | mesoderm formation | BP | 0.0896 | 8.05e-05 | 0.00155 | 2.81 | 0.00111 | Sfrp2/Wnt11/Lef1/Nog/Hoxa11/Foxf1 | 6 |
| GO:0060350 | endochondral bone morphogenesis | BP | 0.0896 | 8.05e-05 | 0.00155 | 2.81 | 0.00111 | Ihh/Mmp16/Hoxd11/Hoxa11/Stc1/Cbs | 6 |
| GO:0051961 | negative regulation of nervous system development | BP | 0.0539 | 8.09e-05 | 0.00155 | 2.81 | 0.00111 | Prtg/Efnb3/Map2/Sema5a/Nog/Sema5b/Dab1/Sema7a/Tert | 9 |
| GO:0042692 | muscle cell differentiation | BP | 0.0342 | 8.2e-05 | 0.00156 | 2.81 | 0.00112 | AW551984/Ryr1/Col14a1/Sgcd/Adra1a/Tbx2/Igf1/Npnt/Ednrb/Tmod2/Cdh2/Foxf1/Hopx/P2rx2/Cxcl12 | 15 |
| GO:0045880 | positive regulation of smoothened signaling pathway | BP | 0.116 | 9.18e-05 | 0.00174 | 2.76 | 0.00125 | Gli1/Ihh/Evc/Gpc3/Kif7 | 5 |
| GO:0050769 | positive regulation of neurogenesis | BP | 0.0403 | 9.45e-05 | 0.00178 | 2.75 | 0.00127 | Ptprz1/Sema5a/Mdk/Nog/Igf1/Ptprd/Plxnb1/Kdr/Lrp2/Sema7a/Serpine2/Cxcl12 | 12 |
| GO:0006820 | anion transport | BP | 0.0321 | 0.000101 | 0.00188 | 2.73 | 0.00134 | Car4/Slc1a3/Grik1/Pla2g10/Slc13a5/Slc2a3/Drd4/Abat/Slc12a8/Ano2/Stc1/Lrp2/Grin2b/Ano1/Slc7a2/Gabra4 | 16 |
| GO:0071695 | anatomical structure maturation | BP | 0.04 | 0.000101 | 0.00188 | 2.73 | 0.00134 | Epha8/Ihh/Pla2g10/Spink1/Ryr1/Igf1/Slc24a4/Plxnb1/Kdr/Ednrb/Calca/Mmp2 | 12 |
| GO:0050771 | negative regulation of axonogenesis | BP | 0.0857 | 0.000103 | 0.00191 | 2.72 | 0.00137 | Efnb3/Map2/Sema5a/Sema5b/Dab1/Sema7a | 6 |
| GO:0006874 | cellular calcium ion homeostasis | BP | 0.032 | 0.000105 | 0.00194 | 2.71 | 0.00139 | Kcnk3/Ryr1/Adra1a/Slc24a4/Kdr/Ednrb/Pygm/Stc1/Calca/Edn3/P2rx2/Ptgfr/Cacna1e/Grin2b/Bok/Slc8a3 | 16 |
| GO:0035296 | regulation of tube diameter | BP | 0.052 | 0.000106 | 0.00194 | 2.71 | 0.00139 | Adra1a/Kdr/Ednrb/Calca/Cbs/Nos3/Edn3/Mmp2/Npr3 | 9 |
| GO:0097746 | blood vessel diameter maintenance | BP | 0.052 | 0.000106 | 0.00194 | 2.71 | 0.00139 | Adra1a/Kdr/Ednrb/Calca/Cbs/Nos3/Edn3/Mmp2/Npr3 | 9 |
| GO:0014065 | phosphatidylinositol 3-kinase signaling | BP | 0.0588 | 0.00011 | 0.002 | 2.7 | 0.00143 | Hgf/Myo16/Angpt1/Igf1/Plxnb1/Kdr/Serpine2/Agap2 | 8 |
| GO:0035150 | regulation of tube size | BP | 0.0517 | 0.000111 | 0.002 | 2.7 | 0.00143 | Adra1a/Kdr/Ednrb/Calca/Cbs/Nos3/Edn3/Mmp2/Npr3 | 9 |
| GO:0048332 | mesoderm morphogenesis | BP | 0.0845 | 0.000112 | 0.00201 | 2.7 | 0.00144 | Sfrp2/Wnt11/Lef1/Nog/Hoxa11/Foxf1 | 6 |
| GO:0001708 | cell fate specification | BP | 0.0686 | 0.000113 | 0.00203 | 2.69 | 0.00145 | Ihh/Sfrp2/Wnt11/Ptch2/Hoxd10/Tbx2/Hoxa11 | 7 |
| GO:0050918 | positive chemotaxis | BP | 0.111 | 0.000115 | 0.00204 | 2.69 | 0.00146 | Angpt1/Sema5a/Kdr/Lrp2/Cxcl12 | 5 |
| GO:0033002 | muscle cell proliferation | BP | 0.0425 | 0.000116 | 0.00205 | 2.69 | 0.00146 | Gli1/Angpt1/Vipr2/Nog/Drd4/Tbx2/Igf1/Nos3/Tert/Mmp2/Npr3 | 11 |
| GO:0048638 | regulation of developmental growth | BP | 0.0348 | 0.000118 | 0.00209 | 2.68 | 0.00149 | Gli1/Map2/Sema5a/H19/Col14a1/Nog/Sema5b/Tbx2/Igf1/Cpne5/Nkd1/Hopx/Sema7a/Cxcl12 | 14 |
| GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | BP | 0.068 | 0.00012 | 0.00211 | 2.68 | 0.00151 | Hgf/Angpt1/Igf1/Plxnb1/Kdr/Serpine2/Agap2 | 7 |
| GO:0030509 | BMP signaling pathway | BP | 0.0511 | 0.000121 | 0.00211 | 2.68 | 0.00151 | Sfrp2/Lef1/Nog/Fst/Rnf165/Kdr/Gpc3/Lrp2/Gdf7 | 9 |
| GO:0032102 | negative regulation of response to external stimulus | BP | 0.0347 | 0.000122 | 0.00211 | 2.68 | 0.00151 | Acod1/Hgf/Pla2g10/Sema5a/Cd109/Mdk/Sema5b/Igf1/Abat/Tfpi/Foxf1/Cd44/Sema7a/Serpine2 | 14 |
| GO:0043271 | negative regulation of ion transport | BP | 0.0508 | 0.000126 | 0.00218 | 2.66 | 0.00156 | Maob/Spink1/Drd4/Abat/Hecw2/Stc1/Calca/Nos3/Serpine2 | 9 |
| GO:0030510 | regulation of BMP signaling pathway | BP | 0.0673 | 0.000128 | 0.0022 | 2.66 | 0.00157 | Sfrp2/Nog/Fst/Rnf165/Kdr/Gpc3/Lrp2 | 7 |
| GO:0050807 | regulation of synapse organization | BP | 0.0418 | 0.000132 | 0.00226 | 2.65 | 0.00162 | Gpc6/St8sia2/Rims3/Ptpro/Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2/Ppfia2/Grin2b | 11 |
| GO:0007620 | copulation | BP | 0.16 | 0.000134 | 0.00227 | 2.64 | 0.00163 | Drd4/Abat/Ednrb/Serpine2 | 4 |
| GO:0035988 | chondrocyte proliferation | BP | 0.16 | 0.000134 | 0.00227 | 2.64 | 0.00163 | Ihh/Mmp16/Lef1/Stc1 | 4 |
| GO:0050890 | cognition | BP | 0.0362 | 0.000141 | 0.00237 | 2.62 | 0.0017 | Stra6/Ptprz1/Mdk/Nog/Drd4/Igf1/Jph4/Cyp7b1/Tmod2/Chst10/Cacna1e/Grin2b/Slc8a3 | 13 |
| GO:0001558 | regulation of cell growth | BP | 0.0325 | 0.000144 | 0.00241 | 2.62 | 0.00172 | Sfrp2/Map2/Wnt11/Tro/Sema5a/Ptch2/Col14a1/Sema5b/Igf1/Cpne5/Epb41l3/Cd44/Sema7a/Serpine2/Cxcl12 | 15 |
| GO:1903522 | regulation of blood circulation | BP | 0.0414 | 0.000146 | 0.00243 | 2.61 | 0.00174 | Kcnd3/Adra1a/Scn4b/Ednrb/Stc1/Calca/Nos3/Hopx/Edn3/Cacna1e/Mmp2 | 11 |
| GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | BP | 0.08 | 0.000151 | 0.00251 | 2.6 | 0.00179 | Hgf/Angpt1/Igf1/Plxnb1/Kdr/Agap2 | 6 |
| GO:0060326 | cell chemotaxis | BP | 0.0382 | 0.000154 | 0.00254 | 2.6 | 0.00182 | Cxcl15/Hgf/Lef1/Sema5a/Mdk/Ptpro/Cyp7b1/Kdr/Ednrb/Calca/Edn3/Cxcl12 | 12 |
| GO:0035137 | hindlimb morphogenesis | BP | 0.104 | 0.000156 | 0.00257 | 2.59 | 0.00183 | Aff3/Hoxd10/Gpc3/Osr2/Hoxd9 | 5 |
| GO:0045766 | positive regulation of angiogenesis | BP | 0.0492 | 0.000162 | 0.00264 | 2.58 | 0.00189 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Ddah1/Kdr/Nos3/Tert | 9 |
| GO:1904018 | positive regulation of vasculature development | BP | 0.0492 | 0.000162 | 0.00264 | 2.58 | 0.00189 | Cyp1b1/Hgf/Sfrp2/Sema5a/Mdk/Ddah1/Kdr/Nos3/Tert | 9 |
| GO:0048738 | cardiac muscle tissue development | BP | 0.0407 | 0.000166 | 0.00269 | 2.57 | 0.00192 | Gli1/Col11a1/AW551984/Angpt1/Col14a1/Sgcd/Nog/Adra1a/Tbx2/Igf1/Lrp2 | 11 |
| GO:0050803 | regulation of synapse structure or activity | BP | 0.0406 | 0.000172 | 0.00276 | 2.56 | 0.00198 | Gpc6/St8sia2/Rims3/Ptpro/Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2/Ppfia2/Grin2b | 11 |
| GO:0010810 | regulation of cell-substrate adhesion | BP | 0.0441 | 0.000175 | 0.00278 | 2.56 | 0.00199 | Ptprz1/Vit/Col26a1/Mdk/Npnt/Kdr/Spock2/Foxf1/Has2/Nid1 | 10 |
| GO:0007269 | neurotransmitter secretion | BP | 0.0486 | 0.000176 | 0.00278 | 2.56 | 0.00199 | Syn2/Rims3/Adra1a/Drd4/Nlgn1/Edn3/P2rx2/Ppfia2/Cadps | 9 |
| GO:0099643 | signal release from synapse | BP | 0.0486 | 0.000176 | 0.00278 | 2.56 | 0.00199 | Syn2/Rims3/Adra1a/Drd4/Nlgn1/Edn3/P2rx2/Ppfia2/Cadps | 9 |
| GO:1903305 | regulation of regulated secretory pathway | BP | 0.0486 | 0.000176 | 0.00278 | 2.56 | 0.00199 | Rims3/Adra1a/Drd4/Syt13/Nlgn1/Foxf1/P2rx2/Ppfia2/Cadps | 9 |
| GO:0014821 | phasic smooth muscle contraction | BP | 0.148 | 0.000183 | 0.00287 | 2.54 | 0.00205 | Tbx2/Ednrb/Edn3/P2rx2 | 4 |
| GO:0099545 | trans-synaptic signaling by trans-synaptic complex | BP | 0.273 | 0.000185 | 0.0029 | 2.54 | 0.00207 | Efnb3/Nlgn1/Ptprd | 3 |
| GO:0001654 | eye development | BP | 0.0333 | 0.000191 | 0.00296 | 2.53 | 0.00212 | Ndp/Cyp1b1/Ihh/Stra6/Sdk2/Nhs/Tbx2/Tub/Lamc3/Kdr/Celf4/Rcn1/Tenm3/Osr2 | 14 |
| GO:0008344 | adult locomotory behavior | BP | 0.0631 | 0.000192 | 0.00296 | 2.53 | 0.00212 | Efnb3/Hoxd10/Drd4/Scn1a/Dab1/Hoxd9/Cxcl12 | 7 |
| GO:0099175 | regulation of postsynapse organization | BP | 0.0631 | 0.000192 | 0.00296 | 2.53 | 0.00212 | Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2/Ppfia2/Grin2b | 7 |
| GO:0002027 | regulation of heart rate | BP | 0.0625 | 0.000203 | 0.00311 | 2.51 | 0.00223 | Kcnd3/Adra1a/Scn4b/Ednrb/Calca/Edn3/Cacna1e | 7 |
| GO:0150063 | visual system development | BP | 0.033 | 0.000205 | 0.00313 | 2.5 | 0.00224 | Ndp/Cyp1b1/Ihh/Stra6/Sdk2/Nhs/Tbx2/Tub/Lamc3/Kdr/Celf4/Rcn1/Tenm3/Osr2 | 14 |
| GO:0019233 | sensory perception of pain | BP | 0.0537 | 0.000206 | 0.00313 | 2.5 | 0.00224 | Kcnd2/Scn1a/Ednrb/Calca/Cacna1e/Grin2b/Ano1/Cxcl12 | 8 |
| GO:0008038 | neuron recognition | BP | 0.098 | 0.000209 | 0.00315 | 2.5 | 0.00225 | Gap43/Ptprz1/Prtg/Efnb3/Sema5a | 5 |
| GO:0003231 | cardiac ventricle development | BP | 0.053 | 0.000226 | 0.00339 | 2.47 | 0.00242 | Stra6/Col11a1/Sfrp2/Wnt11/Nog/Foxf1/Nos3/Lrp2 | 8 |
| GO:0061756 | leukocyte adhesion to vascular endothelial cell | BP | 0.0962 | 0.000229 | 0.00343 | 2.47 | 0.00245 | Mdk/Itga4/Chst2/Cxcl12/Podxl2 | 5 |
| GO:0009952 | anterior/posterior pattern specification | BP | 0.0426 | 0.000231 | 0.00344 | 2.46 | 0.00246 | Sfrp2/Lef1/Nog/Hoxd10/Hoxd11/Gpc3/Hoxa11/Foxf1/Hoxa10/Hoxd9 | 10 |
| GO:0003206 | cardiac chamber morphogenesis | BP | 0.0526 | 0.000236 | 0.0035 | 2.46 | 0.0025 | Col11a1/Sfrp2/Wnt11/Nog/Tbx2/Foxf1/Nos3/Lrp2 | 8 |
| GO:1904861 | excitatory synapse assembly | BP | 0.138 | 0.000243 | 0.00359 | 2.44 | 0.00257 | Nlgn1/Ptprd/Nptx1/Lrrc4b | 4 |
| GO:0014015 | positive regulation of gliogenesis | BP | 0.0732 | 0.000247 | 0.00364 | 2.44 | 0.0026 | Ptprz1/Mdk/Nog/Igf1/Lrp2/Serpine2 | 6 |
| GO:0030850 | prostate gland development | BP | 0.0943 | 0.000251 | 0.00364 | 2.44 | 0.00261 | Nog/Igf1/Cyp7b1/Cd44/Mmp2 | 5 |
| GO:2000351 | regulation of endothelial cell apoptotic process | BP | 0.0943 | 0.000251 | 0.00364 | 2.44 | 0.00261 | Angpt1/Sema5a/Itga4/Kdr/Tert | 5 |
| GO:0098659 | inorganic cation import across plasma membrane | BP | 0.0603 | 0.000252 | 0.00364 | 2.44 | 0.00261 | Kcnj3/Kcnd3/Slc24a4/Slc12a8/Cacna1e/Slc9a7/Slc8a3 | 7 |
| GO:0099587 | inorganic ion import across plasma membrane | BP | 0.0603 | 0.000252 | 0.00364 | 2.44 | 0.00261 | Kcnj3/Kcnd3/Slc24a4/Slc12a8/Cacna1e/Slc9a7/Slc8a3 | 7 |
| GO:0007159 | leukocyte cell-cell adhesion | BP | 0.0339 | 0.000264 | 0.0038 | 2.42 | 0.00272 | Ihh/Efnb3/Lef1/Cd83/Mdk/Igf1/Itga4/Chst2/Calca/Cd44/Has2/Cxcl12/Podxl2 | 13 |
| GO:0035249 | synaptic transmission, glutamatergic | BP | 0.0598 | 0.000266 | 0.00381 | 2.42 | 0.00272 | Grid1/Grik1/Gria4/Nlgn1/Cdh2/Grin2b/Serpine2 | 7 |
| GO:0032892 | positive regulation of organic acid transport | BP | 0.0926 | 0.000274 | 0.00392 | 2.41 | 0.0028 | Grik1/Pla2g10/Acsl6/Abat/Grin2b | 5 |
| GO:0031099 | regeneration | BP | 0.0513 | 0.000281 | 0.004 | 2.4 | 0.00286 | Gli1/Gap43/Hgf/Mdk/Igf1/Hopx/Mmp2/Cxcl12 | 8 |
| GO:2000300 | regulation of synaptic vesicle exocytosis | BP | 0.0714 | 0.000282 | 0.004 | 2.4 | 0.00286 | Rims3/Adra1a/Drd4/Nlgn1/P2rx2/Ppfia2 | 6 |
| GO:0006836 | neurotransmitter transport | BP | 0.0413 | 0.000292 | 0.00412 | 2.38 | 0.00295 | Syn2/Slc6a2/Rims3/Adra1a/Drd4/Nlgn1/Edn3/P2rx2/Ppfia2/Cadps | 10 |
| GO:0001662 | behavioral fear response | BP | 0.0909 | 0.000299 | 0.00419 | 2.38 | 0.00299 | Ankfn1/Mdk/Drd4/Cacna1e/Grin2b | 5 |
| GO:1902476 | chloride transmembrane transport | BP | 0.0909 | 0.000299 | 0.00419 | 2.38 | 0.00299 | Slc1a3/Slc12a8/Ano2/Ano1/Gabra4 | 5 |
| GO:0051966 | regulation of synaptic transmission, glutamatergic | BP | 0.0706 | 0.000301 | 0.00419 | 2.38 | 0.003 | Grik1/Gria4/Nlgn1/Cdh2/Grin2b/Serpine2 | 6 |
| GO:0048570 | notochord morphogenesis | BP | 0.231 | 0.000316 | 0.00432 | 2.36 | 0.00309 | Gli1/Wnt11/Nog | 3 |
| GO:0061036 | positive regulation of cartilage development | BP | 0.129 | 0.000317 | 0.00432 | 2.36 | 0.00309 | Mdk/Pkdcc/Hoxd11/Hoxa11 | 4 |
| GO:2000352 | negative regulation of endothelial cell apoptotic process | BP | 0.129 | 0.000317 | 0.00432 | 2.36 | 0.00309 | Angpt1/Sema5a/Kdr/Tert | 4 |
| GO:0072577 | endothelial cell apoptotic process | BP | 0.0893 | 0.000325 | 0.0044 | 2.36 | 0.00315 | Angpt1/Sema5a/Itga4/Kdr/Tert | 5 |
| GO:0046928 | regulation of neurotransmitter secretion | BP | 0.0574 | 0.000343 | 0.00462 | 2.34 | 0.0033 | Rims3/Adra1a/Drd4/Nlgn1/Edn3/P2rx2/Ppfia2 | 7 |
| GO:0007613 | memory | BP | 0.0497 | 0.000348 | 0.00467 | 2.33 | 0.00334 | Mdk/Nog/Drd4/Igf1/Cyp7b1/Chst10/Grin2b/Slc8a3 | 8 |
| GO:1903039 | positive regulation of leukocyte cell-cell adhesion | BP | 0.0403 | 0.000355 | 0.00474 | 2.32 | 0.00339 | Ihh/Efnb3/Lef1/Cd83/Mdk/Igf1/Itga4/Chst2/Cd44/Has2 | 10 |
| GO:1990138 | neuron projection extension | BP | 0.0441 | 0.000363 | 0.0048 | 2.32 | 0.00343 | Map2/Sema5a/St8sia2/Sema5b/Cpne5/Itga4/Edn3/Sema7a/Cxcl12 | 9 |
| GO:0003151 | outflow tract morphogenesis | BP | 0.0682 | 0.000363 | 0.0048 | 2.32 | 0.00343 | Sfrp2/Wnt11/Ryr1/Nog/Tbx2/Lrp2 | 6 |
| GO:0021675 | nerve development | BP | 0.0682 | 0.000363 | 0.0048 | 2.32 | 0.00343 | Slc1a3/Rnf165/Slc24a4/Itga4/Nptx1/Serpine2 | 6 |
| GO:0060828 | regulation of canonical Wnt signaling pathway | BP | 0.0402 | 0.000366 | 0.00482 | 2.32 | 0.00345 | Gli1/Sfrp2/Wnt11/Sema5a/Mdk/Nog/Ptpro/Gpc3/Cdh2/Nkd1 | 10 |
| GO:0051480 | regulation of cytosolic calcium ion concentration | BP | 0.0327 | 0.000372 | 0.00488 | 2.31 | 0.00349 | Kcnk3/Ryr1/Adra1a/Slc24a4/Kdr/Ednrb/Calca/P2rx2/Ptgfr/Cacna1e/Grin2b/Bok/Slc8a3 | 13 |
| GO:0014032 | neural crest cell development | BP | 0.0674 | 0.000386 | 0.00504 | 2.3 | 0.0036 | Sema5a/Sema5b/Ednrb/Cdh2/Edn3/Sema7a | 6 |
| GO:0099151 | regulation of postsynaptic density assembly | BP | 0.214 | 4e-04 | 0.0052 | 2.28 | 0.00372 | Ptprd/Nptx1/Lrrc4b | 3 |
| GO:0033687 | osteoblast proliferation | BP | 0.121 | 0.000405 | 0.00522 | 2.28 | 0.00373 | Plxnb1/Osr2/Nell1/Npr3 | 4 |
| GO:0048566 | embryonic digestive tract development | BP | 0.121 | 0.000405 | 0.00522 | 2.28 | 0.00373 | Ihh/Stra6/Pkdcc/Foxf1 | 4 |
| GO:0048841 | regulation of axon extension involved in axon guidance | BP | 0.121 | 0.000405 | 0.00522 | 2.28 | 0.00373 | Sema5a/Sema5b/Sema7a/Cxcl12 | 4 |
| GO:0090287 | regulation of cellular response to growth factor stimulus | BP | 0.0367 | 0.000409 | 0.00524 | 2.28 | 0.00375 | Sfrp2/Hhip/Angpt1/Cd109/Nog/Fst/Rnf165/Npnt/Kdr/Gpc3/Lrp2 | 11 |
| GO:0098815 | modulation of excitatory postsynaptic potential | BP | 0.0833 | 0.000449 | 0.00574 | 2.24 | 0.00411 | Drd4/Nlgn1/Celf4/Grin2b/Slc8a3 | 5 |
| GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.0391 | 0.000455 | 0.00578 | 2.24 | 0.00413 | Sfrp2/Cd109/Nog/Fst/Rnf165/Npnt/Kdr/Gpc3/Lrp2/Gdf7 | 10 |
| GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.118 | 0.000455 | 0.00578 | 2.24 | 0.00413 | Mdk/Itga4/Chst2/Cxcl12 | 4 |
| GO:0048640 | negative regulation of developmental growth | BP | 0.0547 | 0.000458 | 0.0058 | 2.24 | 0.00415 | Map2/Sema5a/H19/Nog/Sema5b/Nkd1/Sema7a | 7 |
| GO:0042476 | odontogenesis | BP | 0.0543 | 0.00048 | 0.00603 | 2.22 | 0.00431 | Wnt6/Lef1/Apcdd1/Fst/Slc24a4/Edaradd/Osr2 | 7 |
| GO:0031641 | regulation of myelination | BP | 0.082 | 0.000485 | 0.00605 | 2.22 | 0.00433 | Ptprz1/Hgf/Igf1/Cdh2/Wasf3 | 5 |
| GO:0042596 | fear response | BP | 0.082 | 0.000485 | 0.00605 | 2.22 | 0.00433 | Ankfn1/Mdk/Drd4/Cacna1e/Grin2b | 5 |
| GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | BP | 0.2 | 0.000496 | 0.00616 | 2.21 | 0.0044 | Sfrp2/Gpc3/Nkd1 | 3 |
| GO:0003279 | cardiac septum development | BP | 0.0538 | 0.000503 | 0.00623 | 2.21 | 0.00445 | Stra6/Wnt11/Nog/Tbx2/Nos3/Lrp2/Kif7 | 7 |
| GO:0003401 | axis elongation | BP | 0.114 | 0.00051 | 0.00629 | 2.2 | 0.0045 | Sfrp2/Wnt11/Areg/Nkd1 | 4 |
| GO:0042475 | odontogenesis of dentin-containing tooth | BP | 0.0638 | 0.000517 | 0.00636 | 2.2 | 0.00455 | Wnt6/Lef1/Apcdd1/Fst/Slc24a4/Edaradd | 6 |
| GO:0014031 | mesenchymal cell development | BP | 0.0632 | 0.000547 | 0.00661 | 2.18 | 0.00473 | Sema5a/Sema5b/Ednrb/Cdh2/Edn3/Sema7a | 6 |
| GO:0014910 | regulation of smooth muscle cell migration | BP | 0.0632 | 0.000547 | 0.00661 | 2.18 | 0.00473 | Cyp1b1/Mdk/Drd4/Igf1/Has2/Tert | 6 |
| GO:0048864 | stem cell development | BP | 0.0632 | 0.000547 | 0.00661 | 2.18 | 0.00473 | Sema5a/Sema5b/Ednrb/Cdh2/Edn3/Sema7a | 6 |
| GO:0061053 | somite development | BP | 0.0632 | 0.000547 | 0.00661 | 2.18 | 0.00473 | Ihh/Sfrp2/Wnt11/Lef1/Nog/Foxf1 | 6 |
| GO:0022407 | regulation of cell-cell adhesion | BP | 0.0299 | 0.000551 | 0.00663 | 2.18 | 0.00474 | Ihh/Efnb3/Lef1/Cd83/Mdk/Igf1/Itga4/Abat/Chst2/Tenm3/Cd44/Has2/Serpine2/Cxcl12 | 14 |
| GO:0001755 | neural crest cell migration | BP | 0.0794 | 0.000564 | 0.00676 | 2.17 | 0.00484 | Sema5a/Sema5b/Ednrb/Edn3/Sema7a | 5 |
| GO:0035115 | embryonic forelimb morphogenesis | BP | 0.111 | 0.000569 | 0.00678 | 2.17 | 0.00485 | Hoxd11/Hoxa11/Osr2/Hoxd9 | 4 |
| GO:0070509 | calcium ion import | BP | 0.0625 | 0.000578 | 0.00687 | 2.16 | 0.00491 | Spink1/Slc24a4/Stc1/Cacna1e/Slc8a3/Cxcl12 | 6 |
| GO:0060026 | convergent extension | BP | 0.188 | 0.000605 | 0.00715 | 2.15 | 0.00511 | Sfrp2/Wnt11/Nkd1 | 3 |
| GO:0014911 | positive regulation of smooth muscle cell migration | BP | 0.0781 | 0.000606 | 0.00715 | 2.15 | 0.00511 | Cyp1b1/Mdk/Igf1/Has2/Tert | 5 |
| GO:0048846 | axon extension involved in axon guidance | BP | 0.108 | 0.000633 | 0.00734 | 2.13 | 0.00525 | Sema5a/Sema5b/Sema7a/Cxcl12 | 4 |
| GO:0050901 | leukocyte tethering or rolling | BP | 0.108 | 0.000633 | 0.00734 | 2.13 | 0.00525 | Itga4/Chst2/Cxcl12/Podxl2 | 4 |
| GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | BP | 0.108 | 0.000633 | 0.00734 | 2.13 | 0.00525 | Sfrp2/Wnt11/Gpc3/Nkd1 | 4 |
| GO:1902284 | neuron projection extension involved in neuron projection guidance | BP | 0.108 | 0.000633 | 0.00734 | 2.13 | 0.00525 | Sema5a/Sema5b/Sema7a/Cxcl12 | 4 |
| GO:0007548 | sex differentiation | BP | 0.0347 | 0.000646 | 0.00745 | 2.13 | 0.00533 | Tex15/Stra6/Sfrp2/Angpt1/Fst/Kdr/Hoxa11/Hoxa10/Nos3/Lrp2/Mmp2 | 11 |
| GO:0032835 | glomerulus development | BP | 0.0769 | 0.000651 | 0.00748 | 2.13 | 0.00535 | Angpt1/Nog/Ptpro/Ednrb/Nid1 | 5 |
| GO:0001649 | osteoblast differentiation | BP | 0.0405 | 0.000668 | 0.00765 | 2.12 | 0.00547 | Gli1/Ihh/Sfrp2/Wnt11/Nog/Igf1/Areg/Npnt/Nell1 | 9 |
| GO:0030902 | hindbrain development | BP | 0.0449 | 0.000677 | 0.00772 | 2.11 | 0.00552 | Gli1/Lef1/Mdk/Nog/Igf1/Dab1/Cbs/Serpine2 | 8 |
| GO:0001938 | positive regulation of endothelial cell proliferation | BP | 0.0606 | 0.000681 | 0.00772 | 2.11 | 0.00552 | Sema5a/Mdk/Igf1/Itga4/Kdr/Cxcl12 | 6 |
| GO:0014033 | neural crest cell differentiation | BP | 0.0606 | 0.000681 | 0.00772 | 2.11 | 0.00552 | Sema5a/Sema5b/Ednrb/Cdh2/Edn3/Sema7a | 6 |
| GO:0016079 | synaptic vesicle exocytosis | BP | 0.0511 | 0.000688 | 0.00773 | 2.11 | 0.00553 | Rims3/Adra1a/Drd4/Nlgn1/P2rx2/Ppfia2/Cadps | 7 |
| GO:0042633 | hair cycle | BP | 0.0511 | 0.000688 | 0.00773 | 2.11 | 0.00553 | Pla2g10/Apcdd1/Cd109/Ptch2/Fst/Edaradd/Tert | 7 |
| GO:0045776 | negative regulation of blood pressure | BP | 0.0758 | 0.000698 | 0.00782 | 2.11 | 0.00559 | Adra1a/Abat/Kdr/Calca/Nos3 | 5 |
| GO:1905606 | regulation of presynapse assembly | BP | 0.105 | 0.000701 | 0.00783 | 2.11 | 0.0056 | Gpc6/Nlgn1/Ptprd/Lrrc4b | 4 |
| GO:0015849 | organic acid transport | BP | 0.0343 | 0.000716 | 0.00797 | 2.1 | 0.0057 | Slc1a3/Grik1/Pla2g10/Slc13a5/Acsl6/Drd4/Abat/Rbp7/Lrp2/Grin2b/Slc7a2 | 11 |
| GO:0009101 | glycoprotein biosynthetic process | BP | 0.0368 | 0.000727 | 0.00804 | 2.09 | 0.00575 | St6gal2/St8sia2/Igf1/Itm2a/Tet1/St8sia4/Chst10/Gxylt2/Tmtc1/Soat1 | 10 |
| GO:1904889 | regulation of excitatory synapse assembly | BP | 0.176 | 0.000729 | 0.00804 | 2.09 | 0.00575 | Ptprd/Nptx1/Lrrc4b | 3 |
| GO:1905564 | positive regulation of vascular endothelial cell proliferation | BP | 0.176 | 0.000729 | 0.00804 | 2.09 | 0.00575 | Mdk/Igf1/Itga4 | 3 |
| GO:0010976 | positive regulation of neuron projection development | BP | 0.04 | 0.000735 | 0.00807 | 2.09 | 0.00577 | Ptprz1/Hgf/Fez1/Dcc/Acsl6/Mdk/Lrrc7/Tenm3/Serpine2 | 9 |
| GO:0051588 | regulation of neurotransmitter transport | BP | 0.0504 | 0.000749 | 0.00821 | 2.09 | 0.00587 | Rims3/Adra1a/Drd4/Nlgn1/Edn3/P2rx2/Ppfia2 | 7 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | BP | 0.034 | 0.000772 | 0.00844 | 2.07 | 0.00603 | Ptprz1/Hgf/Sfrp2/Angpt1/Spink1/Pkdcc/Adra1a/Igf1/Areg/Kdr/Cd44 | 11 |
| GO:0070374 | positive regulation of ERK1 and ERK2 cascade | BP | 0.0396 | 0.000782 | 0.00849 | 2.07 | 0.00607 | Angpt1/Dcc/Adra1a/Igf1/Npnt/Kdr/Cd44/Sema7a/Cxcl12 | 9 |
| GO:0099504 | synaptic vesicle cycle | BP | 0.0396 | 0.000782 | 0.00849 | 2.07 | 0.00607 | Syn2/Rims3/Adra1a/Drd4/Nlgn1/Cdh2/P2rx2/Ppfia2/Cadps | 9 |
| GO:0018212 | peptidyl-tyrosine modification | BP | 0.0336 | 0.000833 | 0.00901 | 2.05 | 0.00644 | Ptprz1/Hgf/Sfrp2/Angpt1/Spink1/Pkdcc/Adra1a/Igf1/Areg/Kdr/Cd44 | 11 |
| GO:0001704 | formation of primary germ layer | BP | 0.0583 | 0.000838 | 0.00902 | 2.04 | 0.00645 | Sfrp2/Wnt11/Lef1/Nog/Hoxa11/Foxf1 | 6 |
| GO:0021510 | spinal cord development | BP | 0.0583 | 0.000838 | 0.00902 | 2.04 | 0.00645 | Vit/Dcc/Nog/Hoxd10/Dab1/Gdf7 | 6 |
| GO:0051146 | striated muscle cell differentiation | BP | 0.0335 | 0.000854 | 0.00912 | 2.04 | 0.00652 | AW551984/Ryr1/Col14a1/Sgcd/Adra1a/Igf1/Tmod2/Cdh2/Hopx/P2rx2/Cxcl12 | 11 |
| GO:0051926 | negative regulation of calcium ion transport | BP | 0.0725 | 0.000856 | 0.00912 | 2.04 | 0.00652 | Spink1/Drd4/Stc1/Calca/Nos3 | 5 |
| GO:0060425 | lung morphogenesis | BP | 0.0725 | 0.000856 | 0.00912 | 2.04 | 0.00652 | Hhip/Nog/Tbx2/Igf1/Foxf1 | 5 |
| GO:0032230 | positive regulation of synaptic transmission, GABAergic | BP | 0.167 | 0.000868 | 0.00919 | 2.04 | 0.00657 | Grik1/Adra1a/Nlgn1 | 3 |
| GO:0097091 | synaptic vesicle clustering | BP | 0.167 | 0.000868 | 0.00919 | 2.04 | 0.00657 | Syn2/Nlgn1/Cdh2 | 3 |
| GO:0086003 | cardiac muscle cell contraction | BP | 0.0714 | 0.000913 | 0.00962 | 2.02 | 0.00688 | Kcnd3/Sgcd/Scn4b/Scn1a/Stc1 | 5 |
| GO:0050728 | negative regulation of inflammatory response | BP | 0.0486 | 0.000923 | 0.00969 | 2.01 | 0.00693 | Acod1/Hgf/Pla2g10/Mdk/Igf1/Foxf1/Cd44 | 7 |
| GO:0007612 | learning | BP | 0.0428 | 0.000933 | 0.00976 | 2.01 | 0.00698 | Stra6/Nog/Drd4/Jph4/Chst10/Cacna1e/Grin2b/Slc8a3 | 8 |
| GO:0014909 | smooth muscle cell migration | BP | 0.0566 | 0.000974 | 0.0101 | 1.99 | 0.00725 | Cyp1b1/Mdk/Drd4/Igf1/Has2/Tert | 6 |
| GO:0015844 | monoamine transport | BP | 0.0566 | 0.000974 | 0.0101 | 1.99 | 0.00725 | Maob/Slc6a2/Syt13/Abat/Cxcl12/Cadps | 6 |
| GO:0034765 | regulation of ion transmembrane transport | BP | 0.0281 | 0.001 | 0.0104 | 1.98 | 0.00744 | Kcnb2/Nalcn/Kcnj3/Kcnd2/Kcna6/Kcnd3/Drd4/Scn4b/Nlgn1/Scn1a/Hecw2/Edn3/Kcnq5/Cacna1e | 14 |
| GO:0070989 | oxidative demethylation | BP | 0.158 | 0.00102 | 0.0105 | 1.98 | 0.00752 | Cyp3a59/Cyp3a25/Tet1 | 3 |
| GO:0071456 | cellular response to hypoxia | BP | 0.0561 | 0.00102 | 0.0105 | 1.98 | 0.00752 | Kcnk3/Kcnd2/Ddah1/Cbs/Tert/Slc8a3 | 6 |
| GO:0010092 | specification of animal organ identity | BP | 0.0952 | 0.00103 | 0.0105 | 1.98 | 0.00752 | Wnt11/Hoxd11/Hoxa11/Lrp2 | 4 |
| GO:0090175 | regulation of establishment of planar polarity | BP | 0.0952 | 0.00103 | 0.0105 | 1.98 | 0.00752 | Sfrp2/Wnt11/Gpc3/Nkd1 | 4 |
| GO:0099174 | regulation of presynapse organization | BP | 0.0952 | 0.00103 | 0.0105 | 1.98 | 0.00752 | Gpc6/Nlgn1/Ptprd/Lrrc4b | 4 |
| GO:0061035 | regulation of cartilage development | BP | 0.0694 | 0.00104 | 0.0106 | 1.98 | 0.00755 | Mdk/Nog/Pkdcc/Hoxd11/Hoxa11 | 5 |
| GO:2000179 | positive regulation of neural precursor cell proliferation | BP | 0.0694 | 0.00104 | 0.0106 | 1.98 | 0.00755 | Gli1/Mdk/Nog/Igf1/Lrp2 | 5 |
| GO:0001508 | action potential | BP | 0.0476 | 0.00104 | 0.0106 | 1.98 | 0.00755 | Kcnd2/Kcnd3/Adra1a/Scn4b/Scn1a/P2rx2/Grin2b | 7 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | BP | 0.0307 | 0.00109 | 0.011 | 1.96 | 0.00784 | Sfrp2/Lef1/Cd109/Nog/Fst/Rnf165/Npnt/Kdr/Gpc3/Lrp2/Btbd11/Gdf7 | 12 |
| GO:0008202 | steroid metabolic process | BP | 0.0325 | 0.00109 | 0.011 | 1.96 | 0.00784 | Apob/Cyp3a59/Cyp1b1/Cyp3a25/Hmgcs2/Nr5a2/Igf1/Cyp7b1/Ednrb/Chst10/Soat1 | 11 |
| GO:0045055 | regulated exocytosis | BP | 0.0348 | 0.00109 | 0.011 | 1.96 | 0.00785 | Syn2/Rims3/Adra1a/Drd4/Syt13/Nlgn1/Foxf1/P2rx2/Ppfia2/Cadps | 10 |
| GO:1905330 | regulation of morphogenesis of an epithelium | BP | 0.0685 | 0.0011 | 0.0111 | 1.96 | 0.00792 | Hgf/Mdk/Nog/Tbx2/Nkd1 | 5 |
| GO:0008406 | gonad development | BP | 0.0375 | 0.00116 | 0.0115 | 1.94 | 0.00824 | Sfrp2/Angpt1/Fst/Kdr/Hoxa11/Hoxa10/Nos3/Lrp2/Mmp2 | 9 |
| GO:1903510 | mucopolysaccharide metabolic process | BP | 0.0676 | 0.00117 | 0.0117 | 1.93 | 0.00835 | Angpt1/Spock2/Ednrb/Cd44/Has2 | 5 |
| GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | BP | 0.15 | 0.00119 | 0.0118 | 1.93 | 0.00841 | Calca/Ano1/Cxcl12 | 3 |
| GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | BP | 0.15 | 0.00119 | 0.0118 | 1.93 | 0.00841 | Scn1a/Grin2b/Cxcl12 | 3 |
| GO:1903236 | regulation of leukocyte tethering or rolling | BP | 0.15 | 0.00119 | 0.0118 | 1.93 | 0.00841 | Itga4/Chst2/Cxcl12 | 3 |
| GO:1903037 | regulation of leukocyte cell-cell adhesion | BP | 0.0321 | 0.00122 | 0.012 | 1.92 | 0.0086 | Ihh/Efnb3/Lef1/Cd83/Mdk/Igf1/Itga4/Chst2/Cd44/Has2/Cxcl12 | 11 |
| GO:0010464 | regulation of mesenchymal cell proliferation | BP | 0.0909 | 0.00123 | 0.012 | 1.92 | 0.0086 | Ihh/Wnt11/Kdr/Foxf1 | 4 |
| GO:0048645 | animal organ formation | BP | 0.0667 | 0.00125 | 0.0122 | 1.91 | 0.00872 | Wnt11/Nog/Hoxd11/Hoxa11/Lrp2 | 5 |
| GO:0019722 | calcium-mediated signaling | BP | 0.0408 | 0.00126 | 0.0122 | 1.91 | 0.00876 | Sgcd/Drd4/Nr5a2/Igf1/Slc24a4/Kdr/Ednrb/Ptgfr | 8 |
| GO:0030900 | forebrain development | BP | 0.0302 | 0.00126 | 0.0122 | 1.91 | 0.00876 | Gli1/Lef1/Sema5a/Mdk/Nog/Cdh2/Dnah5/Dab1/Lrp2/Sema7a/Cxcl12/Gdf7 | 12 |
| GO:0046660 | female sex differentiation | BP | 0.0461 | 0.00126 | 0.0122 | 1.91 | 0.00876 | Stra6/Angpt1/Fst/Kdr/Nos3/Lrp2/Mmp2 | 7 |
| GO:0055017 | cardiac muscle tissue growth | BP | 0.0536 | 0.0013 | 0.0125 | 1.9 | 0.00894 | Gli1/Col14a1/Nog/Adra1a/Tbx2/Igf1 | 6 |
| GO:0045137 | development of primary sexual characteristics | BP | 0.0369 | 0.0013 | 0.0125 | 1.9 | 0.00894 | Sfrp2/Angpt1/Fst/Kdr/Hoxa11/Hoxa10/Nos3/Lrp2/Mmp2 | 9 |
| GO:0050905 | neuromuscular process | BP | 0.0404 | 0.00134 | 0.0129 | 1.89 | 0.00924 | Ankfn1/Stra6/Slc1a3/Hoxd10/Jph4/Scn1a/Grin2b/Slc8a3 | 8 |
| GO:2001234 | negative regulation of apoptotic signaling pathway | BP | 0.0366 | 0.00137 | 0.0131 | 1.88 | 0.0094 | Hgf/Sfrp2/Nog/Igf1/Cd44/Tert/Bok/Cxcl12/Agap2 | 9 |
| GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | BP | 0.143 | 0.00138 | 0.0131 | 1.88 | 0.0094 | Adra1a/Calca/P2rx2 | 3 |
| GO:0032332 | positive regulation of chondrocyte differentiation | BP | 0.143 | 0.00138 | 0.0131 | 1.88 | 0.0094 | Pkdcc/Hoxd11/Hoxa11 | 3 |
| GO:0097107 | postsynaptic density assembly | BP | 0.143 | 0.00138 | 0.0131 | 1.88 | 0.0094 | Ptprd/Nptx1/Lrrc4b | 3 |
| GO:0006029 | proteoglycan metabolic process | BP | 0.0649 | 0.0014 | 0.0132 | 1.88 | 0.00944 | Ihh/Col11a1/Igf1/Spock2/Chst10 | 5 |
| GO:0043113 | receptor clustering | BP | 0.0649 | 0.0014 | 0.0132 | 1.88 | 0.00944 | Lrrc7/Nlgn1/Itga4/Cdh2/Grin2b | 5 |
| GO:0060976 | coronary vasculature development | BP | 0.0649 | 0.0014 | 0.0132 | 1.88 | 0.00944 | Angpt1/Sgcd/Gpc3/Lrp2/Kif7 | 5 |
| GO:0098661 | inorganic anion transmembrane transport | BP | 0.0649 | 0.0014 | 0.0132 | 1.88 | 0.00944 | Slc1a3/Slc12a8/Ano2/Ano1/Gabra4 | 5 |
| GO:0017157 | regulation of exocytosis | BP | 0.0364 | 0.00141 | 0.0132 | 1.88 | 0.00946 | Rims3/Adra1a/Drd4/Syt13/Nlgn1/Foxf1/P2rx2/Ppfia2/Cadps | 9 |
| GO:0022409 | positive regulation of cell-cell adhesion | BP | 0.0336 | 0.00145 | 0.0135 | 1.87 | 0.00967 | Ihh/Efnb3/Lef1/Cd83/Mdk/Igf1/Itga4/Chst2/Cd44/Has2 | 10 |
| GO:0002691 | regulation of cellular extravasation | BP | 0.087 | 0.00145 | 0.0135 | 1.87 | 0.00968 | Mdk/Itga4/Chst2/Cxcl12 | 4 |
| GO:0098656 | anion transmembrane transport | BP | 0.0396 | 0.00153 | 0.0142 | 1.85 | 0.0102 | Slc1a3/Slc13a5/Slc12a8/Ano2/Lrp2/Ano1/Slc7a2/Gabra4 | 8 |
| GO:0015711 | organic anion transport | BP | 0.0312 | 0.00154 | 0.0143 | 1.85 | 0.0102 | Car4/Slc1a3/Grik1/Pla2g10/Slc13a5/Slc2a3/Drd4/Abat/Lrp2/Grin2b/Slc7a2 | 11 |
| GO:0043616 | keratinocyte proliferation | BP | 0.0851 | 0.00157 | 0.0145 | 1.84 | 0.0104 | Cd109/Mdk/Fst/Has2 | 4 |
| GO:0045667 | regulation of osteoblast differentiation | BP | 0.0443 | 0.00158 | 0.0145 | 1.84 | 0.0104 | Gli1/Sfrp2/Nog/Igf1/Areg/Npnt/Nell1 | 7 |
| GO:0060080 | inhibitory postsynaptic potential | BP | 0.136 | 0.00159 | 0.0145 | 1.84 | 0.0104 | Grik1/Drd4/Abat | 3 |
| GO:0060749 | mammary gland alveolus development | BP | 0.136 | 0.00159 | 0.0145 | 1.84 | 0.0104 | Areg/Foxf1/Agap2 | 3 |
| GO:0061377 | mammary gland lobule development | BP | 0.136 | 0.00159 | 0.0145 | 1.84 | 0.0104 | Areg/Foxf1/Agap2 | 3 |
| GO:0030308 | negative regulation of cell growth | BP | 0.0392 | 0.00162 | 0.0148 | 1.83 | 0.0106 | Sfrp2/Map2/Wnt11/Tro/Sema5a/Sema5b/Sema7a/Serpine2 | 8 |
| GO:0030517 | negative regulation of axon extension | BP | 0.0833 | 0.0017 | 0.0155 | 1.81 | 0.0111 | Map2/Sema5a/Sema5b/Sema7a | 4 |
| GO:0099003 | vesicle-mediated transport in synapse | BP | 0.0354 | 0.00171 | 0.0155 | 1.81 | 0.0111 | Syn2/Rims3/Adra1a/Drd4/Nlgn1/Cdh2/P2rx2/Ppfia2/Cadps | 9 |
| GO:0003208 | cardiac ventricle morphogenesis | BP | 0.0617 | 0.00176 | 0.0159 | 1.8 | 0.0114 | Col11a1/Sfrp2/Nog/Foxf1/Lrp2 | 5 |
| GO:0099173 | postsynapse organization | BP | 0.0386 | 0.00178 | 0.0161 | 1.79 | 0.0115 | Gap43/Nlgn1/Ptprd/Nptx1/Lrrc4b/Cdh2/Ppfia2/Grin2b | 8 |
| GO:0045765 | regulation of angiogenesis | BP | 0.0326 | 0.0018 | 0.0162 | 1.79 | 0.0116 | Cyp1b1/Hgf/Sfrp2/Hhip/Sema5a/Mdk/Ddah1/Kdr/Nos3/Tert | 10 |
| GO:0090257 | regulation of muscle system process | BP | 0.0352 | 0.0018 | 0.0162 | 1.79 | 0.0116 | Ryr1/Adra1a/Igf1/Npnt/Abat/Stc1/Calca/Nos3/Slc8a3 | 9 |
| GO:0042053 | regulation of dopamine metabolic process | BP | 0.13 | 0.00181 | 0.0162 | 1.79 | 0.0116 | Maob/Drd4/Abat | 3 |
| GO:0050961 | detection of temperature stimulus involved in sensory perception | BP | 0.13 | 0.00181 | 0.0162 | 1.79 | 0.0116 | Calca/Ano1/Cxcl12 | 3 |
| GO:0071526 | semaphorin-plexin signaling pathway | BP | 0.0816 | 0.00184 | 0.0163 | 1.79 | 0.0117 | Sema5a/Sema5b/Plxnb1/Sema7a | 4 |
| GO:0007498 | mesoderm development | BP | 0.05 | 0.00184 | 0.0163 | 1.79 | 0.0117 | Sfrp2/Wnt11/Lef1/Nog/Hoxa11/Foxf1 | 6 |
| GO:0014812 | muscle cell migration | BP | 0.05 | 0.00184 | 0.0163 | 1.79 | 0.0117 | Cyp1b1/Mdk/Drd4/Igf1/Has2/Tert | 6 |
| GO:0045123 | cellular extravasation | BP | 0.061 | 0.00186 | 0.0164 | 1.78 | 0.0117 | Mdk/Itga4/Chst2/Cxcl12/Podxl2 | 5 |
| GO:0051963 | regulation of synapse assembly | BP | 0.0496 | 0.00192 | 0.017 | 1.77 | 0.0121 | Gpc6/St8sia2/Nlgn1/Ptprd/Nptx1/Lrrc4b | 6 |
| GO:0050954 | sensory perception of mechanical stimulus | BP | 0.0323 | 0.00193 | 0.017 | 1.77 | 0.0122 | Col11a1/Slc1a3/Igf1/Tub/Scn1a/P2rx2/Lrp2/Grin2b/Serpine2/Cxcl12 | 10 |
| GO:0006937 | regulation of muscle contraction | BP | 0.0427 | 0.00195 | 0.0171 | 1.77 | 0.0122 | Ryr1/Adra1a/Npnt/Abat/Stc1/Calca/Slc8a3 | 7 |
| GO:0007338 | single fertilization | BP | 0.0427 | 0.00195 | 0.0171 | 1.77 | 0.0122 | Pla2g10/Spink1/Hoxd10/Hoxd11/Hoxa11/Hoxa10/Hoxd9 | 7 |
| GO:0055021 | regulation of cardiac muscle tissue growth | BP | 0.0602 | 0.00196 | 0.0171 | 1.77 | 0.0122 | Gli1/Col14a1/Nog/Tbx2/Igf1 | 5 |
| GO:1901342 | regulation of vasculature development | BP | 0.0322 | 0.00198 | 0.0172 | 1.76 | 0.0123 | Cyp1b1/Hgf/Sfrp2/Hhip/Sema5a/Mdk/Ddah1/Kdr/Nos3/Tert | 10 |
| GO:0003197 | endocardial cushion development | BP | 0.08 | 0.00198 | 0.0172 | 1.76 | 0.0123 | Nog/Tbx2/Foxf1/Nos3 | 4 |
| GO:0060419 | heart growth | BP | 0.0492 | 0.002 | 0.0174 | 1.76 | 0.0124 | Gli1/Col14a1/Nog/Adra1a/Tbx2/Igf1 | 6 |
| GO:0030879 | mammary gland development | BP | 0.0424 | 0.00202 | 0.0175 | 1.76 | 0.0125 | Lef1/Tbx2/Igf1/Areg/Foxf1/Hoxd9/Agap2 | 7 |
| GO:0030903 | notochord development | BP | 0.125 | 0.00205 | 0.0176 | 1.75 | 0.0126 | Gli1/Wnt11/Nog | 3 |
| GO:0031643 | positive regulation of myelination | BP | 0.125 | 0.00205 | 0.0176 | 1.75 | 0.0126 | Hgf/Igf1/Wasf3 | 3 |
| GO:0048333 | mesodermal cell differentiation | BP | 0.125 | 0.00205 | 0.0176 | 1.75 | 0.0126 | Sfrp2/Hoxa11/Foxf1 | 3 |
| GO:0006809 | nitric oxide biosynthetic process | BP | 0.0595 | 0.00206 | 0.0177 | 1.75 | 0.0126 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2 | 5 |
| GO:0010631 | epithelial cell migration | BP | 0.0319 | 0.00207 | 0.0177 | 1.75 | 0.0127 | Cyp1b1/Angpt1/Sema5a/Igf1/Kdr/Stc1/Calca/Nos3/Has2/Evl | 10 |
| GO:0010466 | negative regulation of peptidase activity | BP | 0.0344 | 0.00211 | 0.018 | 1.75 | 0.0128 | Hgf/Sfrp2/Spink1/Cd109/Igf1/Gpc3/Tfpi/Cd44/Serpine2 | 9 |
| GO:0090132 | epithelium migration | BP | 0.0317 | 0.00217 | 0.0184 | 1.73 | 0.0132 | Cyp1b1/Angpt1/Sema5a/Igf1/Kdr/Stc1/Calca/Nos3/Has2/Evl | 10 |
| GO:0051954 | positive regulation of amine transport | BP | 0.0769 | 0.00229 | 0.0193 | 1.71 | 0.0138 | Grik1/Abat/Grin2b/Cxcl12 | 4 |
| GO:0007263 | nitric oxide mediated signal transduction | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Spink1/Cbs/Nos3 | 3 |
| GO:0010884 | positive regulation of lipid storage | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Apob/Pla2g10/Vstm2a | 3 |
| GO:0010996 | response to auditory stimulus | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Stra6/Slc1a3/Mdk | 3 |
| GO:0042069 | regulation of catecholamine metabolic process | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Maob/Drd4/Abat | 3 |
| GO:0101023 | vascular endothelial cell proliferation | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Mdk/Igf1/Itga4 | 3 |
| GO:1905562 | regulation of vascular endothelial cell proliferation | BP | 0.12 | 0.00232 | 0.0193 | 1.71 | 0.0138 | Mdk/Igf1/Itga4 | 3 |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | BP | 0.0338 | 0.00233 | 0.0194 | 1.71 | 0.0139 | Ptprz1/Hgf/Sfrp2/Angpt1/Spink1/Adra1a/Igf1/Areg/Cd44 | 9 |
| GO:0032890 | regulation of organic acid transport | BP | 0.0575 | 0.00241 | 0.0199 | 1.7 | 0.0143 | Grik1/Pla2g10/Acsl6/Abat/Grin2b | 5 |
| GO:0060411 | cardiac septum morphogenesis | BP | 0.0575 | 0.00241 | 0.0199 | 1.7 | 0.0143 | Wnt11/Nog/Tbx2/Nos3/Lrp2 | 5 |
| GO:0032355 | response to estradiol | BP | 0.0755 | 0.00245 | 0.0202 | 1.69 | 0.0145 | Areg/Nos3/Ptgfr/Mmp2 | 4 |
| GO:0070169 | positive regulation of biomineral tissue development | BP | 0.0755 | 0.00245 | 0.0202 | 1.69 | 0.0145 | Wnt6/Pkdcc/Osr2/Nell1 | 4 |
| GO:0001738 | morphogenesis of a polarized epithelium | BP | 0.0568 | 0.00253 | 0.0207 | 1.68 | 0.0148 | Sfrp2/Wnt11/Gpc3/Nkd1/Foxf1 | 5 |
| GO:0003281 | ventricular septum development | BP | 0.0568 | 0.00253 | 0.0207 | 1.68 | 0.0148 | Stra6/Wnt11/Nog/Nos3/Lrp2 | 5 |
| GO:0051937 | catecholamine transport | BP | 0.0568 | 0.00253 | 0.0207 | 1.68 | 0.0148 | Slc6a2/Syt13/Abat/Cxcl12/Cadps | 5 |
| GO:0030282 | bone mineralization | BP | 0.0469 | 0.00255 | 0.0208 | 1.68 | 0.0149 | Wnt11/Pkdcc/Igf1/Gpc3/Osr2/Nell1 | 6 |
| GO:0048639 | positive regulation of developmental growth | BP | 0.0364 | 0.0026 | 0.021 | 1.68 | 0.015 | Gli1/Sema5a/Tbx2/Igf1/Cpne5/Hopx/Sema7a/Cxcl12 | 8 |
| GO:0060065 | uterus development | BP | 0.115 | 0.0026 | 0.021 | 1.68 | 0.015 | Stra6/Hoxa11/Hoxa10 | 3 |
| GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | BP | 0.115 | 0.0026 | 0.021 | 1.68 | 0.015 | Mdk/Itga4/Chst2 | 3 |
| GO:2000050 | regulation of non-canonical Wnt signaling pathway | BP | 0.115 | 0.0026 | 0.021 | 1.68 | 0.015 | Sfrp2/Gpc3/Nkd1 | 3 |
| GO:0007617 | mating behavior | BP | 0.0741 | 0.00263 | 0.021 | 1.68 | 0.015 | Drd4/Abat/Ednrb/Serpine2 | 4 |
| GO:0010883 | regulation of lipid storage | BP | 0.0741 | 0.00263 | 0.021 | 1.68 | 0.015 | Apob/Pla2g10/Mest/Vstm2a | 4 |
| GO:0032330 | regulation of chondrocyte differentiation | BP | 0.0741 | 0.00263 | 0.021 | 1.68 | 0.015 | Mdk/Pkdcc/Hoxd11/Hoxa11 | 4 |
| GO:0099054 | presynapse assembly | BP | 0.0741 | 0.00263 | 0.021 | 1.68 | 0.015 | Gpc6/Nlgn1/Ptprd/Lrrc4b | 4 |
| GO:0033674 | positive regulation of kinase activity | BP | 0.0276 | 0.00263 | 0.021 | 1.68 | 0.015 | Epha8/Angpt1/Adra1a/Drd4/Igf1/Areg/Kdr/Dab1/Calca/Edn3/Ccdc88a/Agap2 | 12 |
| GO:0030048 | actin filament-based movement | BP | 0.0465 | 0.00265 | 0.0211 | 1.68 | 0.0151 | Kcnd3/Sgcd/Scn4b/Scn1a/Stc1/Evl | 6 |
| GO:0050805 | negative regulation of synaptic transmission | BP | 0.0562 | 0.00266 | 0.0211 | 1.68 | 0.0151 | Grik1/Grid2ip/Drd4/Celf4/Gnai1 | 5 |
| GO:0006941 | striated muscle contraction | BP | 0.0402 | 0.00272 | 0.0215 | 1.67 | 0.0154 | Kcnd3/Sgcd/Adra1a/Scn4b/Scn1a/Stc1/Slc8a3 | 7 |
| GO:0070997 | neuron death | BP | 0.0274 | 0.00278 | 0.0218 | 1.66 | 0.0156 | Ptprz1/Angpt1/Dcc/St8sia2/Mdk/Igf1/Kdr/Dpysl4/Grin2b/Tert/Bok/Agap2 | 12 |
| GO:0006835 | dicarboxylic acid transport | BP | 0.0556 | 0.00279 | 0.0218 | 1.66 | 0.0156 | Slc1a3/Slc13a5/Abat/Lrp2/Grin2b | 5 |
| GO:0019915 | lipid storage | BP | 0.0556 | 0.00279 | 0.0218 | 1.66 | 0.0156 | Apob/Pla2g10/Mest/Soat1/Vstm2a | 5 |
| GO:0031016 | pancreas development | BP | 0.0556 | 0.00279 | 0.0218 | 1.66 | 0.0156 | Ihh/Nr5a2/Igf1/Cdh2/Foxf1 | 5 |
| GO:0046209 | nitric oxide metabolic process | BP | 0.0556 | 0.00279 | 0.0218 | 1.66 | 0.0156 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2 | 5 |
| GO:0042552 | myelination | BP | 0.04 | 0.00281 | 0.0218 | 1.66 | 0.0156 | Ptprz1/Hgf/Igf1/Epb41l3/Cdh2/Wasf3/Slc8a3 | 7 |
| GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | BP | 0.0727 | 0.00281 | 0.0218 | 1.66 | 0.0156 | Adra1a/Ednrb/Nos3/Edn3 | 4 |
| GO:0048546 | digestive tract morphogenesis | BP | 0.0727 | 0.00281 | 0.0218 | 1.66 | 0.0156 | Ihh/Stra6/Sfrp2/Foxf1 | 4 |
| GO:0015740 | C4-dicarboxylate transport | BP | 0.111 | 0.0029 | 0.0224 | 1.65 | 0.016 | Slc1a3/Slc13a5/Abat | 3 |
| GO:0016048 | detection of temperature stimulus | BP | 0.111 | 0.0029 | 0.0224 | 1.65 | 0.016 | Calca/Ano1/Cxcl12 | 3 |
| GO:0048843 | negative regulation of axon extension involved in axon guidance | BP | 0.111 | 0.0029 | 0.0224 | 1.65 | 0.016 | Sema5a/Sema5b/Sema7a | 3 |
| GO:0060420 | regulation of heart growth | BP | 0.0549 | 0.00293 | 0.0226 | 1.65 | 0.0161 | Gli1/Col14a1/Nog/Tbx2/Igf1 | 5 |
| GO:0036294 | cellular response to decreased oxygen levels | BP | 0.0455 | 0.00297 | 0.0228 | 1.64 | 0.0163 | Kcnk3/Kcnd2/Ddah1/Cbs/Tert/Slc8a3 | 6 |
| GO:0060048 | cardiac muscle contraction | BP | 0.0455 | 0.00297 | 0.0228 | 1.64 | 0.0163 | Kcnd3/Sgcd/Adra1a/Scn4b/Scn1a/Stc1 | 6 |
| GO:0001974 | blood vessel remodeling | BP | 0.0714 | 0.003 | 0.0229 | 1.64 | 0.0164 | Igf1/Itga4/Cbs/Nos3 | 4 |
| GO:0007588 | excretion | BP | 0.0714 | 0.003 | 0.0229 | 1.64 | 0.0164 | Mdk/Adra1a/Ednrb/Stc1 | 4 |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | BP | 0.0714 | 0.003 | 0.0229 | 1.64 | 0.0164 | Angpt1/Igf1/Kdr/Nos3 | 4 |
| GO:1903035 | negative regulation of response to wounding | BP | 0.0543 | 0.00307 | 0.0233 | 1.63 | 0.0167 | Cd109/Mdk/Abat/Tfpi/Serpine2 | 5 |
| GO:2001057 | reactive nitrogen species metabolic process | BP | 0.0543 | 0.00307 | 0.0233 | 1.63 | 0.0167 | Cyp1b1/Igf1/Ddah1/Nos3/Slc7a2 | 5 |
| GO:0008366 | axon ensheathment | BP | 0.0393 | 0.00309 | 0.0233 | 1.63 | 0.0167 | Ptprz1/Hgf/Igf1/Epb41l3/Cdh2/Wasf3/Slc8a3 | 7 |
| GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | BP | 0.0352 | 0.00314 | 0.0237 | 1.63 | 0.0169 | Vipr2/Adra1a/Drd4/Calca/Gnai1/Ptgfr/Adgrl4/Npr3 | 8 |
| GO:0007369 | gastrulation | BP | 0.0391 | 0.00318 | 0.0238 | 1.62 | 0.017 | Sfrp2/Wnt11/Lef1/Nog/Hoxa11/Foxf1/Osr2 | 7 |
| GO:0060402 | calcium ion transport into cytosol | BP | 0.0391 | 0.00318 | 0.0238 | 1.62 | 0.017 | Ryr1/Adra1a/Slc24a4/Calca/Cacna1e/Grin2b/Slc8a3 | 7 |
| GO:0031646 | positive regulation of nervous system process | BP | 0.0702 | 0.0032 | 0.0238 | 1.62 | 0.017 | Hgf/Igf1/Abat/Wasf3 | 4 |
| GO:0035567 | non-canonical Wnt signaling pathway | BP | 0.0702 | 0.0032 | 0.0238 | 1.62 | 0.017 | Sfrp2/Wnt11/Gpc3/Nkd1 | 4 |
| GO:0086001 | cardiac muscle cell action potential | BP | 0.0702 | 0.0032 | 0.0238 | 1.62 | 0.017 | Kcnd2/Kcnd3/Scn4b/Scn1a | 4 |
| GO:0010959 | regulation of metal ion transport | BP | 0.0269 | 0.00322 | 0.0238 | 1.62 | 0.017 | Spink1/Drd4/Scn4b/Igf1/Jph4/Hecw2/Stc1/Calca/Nos3/Edn3/Serpine2/Cxcl12 | 12 |
| GO:0033688 | regulation of osteoblast proliferation | BP | 0.107 | 0.00322 | 0.0238 | 1.62 | 0.017 | Plxnb1/Nell1/Npr3 | 3 |
| GO:0043931 | ossification involved in bone maturation | BP | 0.107 | 0.00322 | 0.0238 | 1.62 | 0.017 | Ryr1/Igf1/Plxnb1 | 3 |
| GO:0061484 | hematopoietic stem cell homeostasis | BP | 0.107 | 0.00322 | 0.0238 | 1.62 | 0.017 | Crispld1/Gprasp2/Emcn | 3 |
| GO:0030595 | leukocyte chemotaxis | BP | 0.0351 | 0.00323 | 0.0238 | 1.62 | 0.017 | Cxcl15/Mdk/Ptpro/Cyp7b1/Ednrb/Calca/Edn3/Cxcl12 | 8 |
| GO:0043542 | endothelial cell migration | BP | 0.0351 | 0.00323 | 0.0238 | 1.62 | 0.017 | Cyp1b1/Angpt1/Sema5a/Igf1/Kdr/Stc1/Calca/Nos3 | 8 |
| GO:0048660 | regulation of smooth muscle cell proliferation | BP | 0.0389 | 0.00328 | 0.0241 | 1.62 | 0.0173 | Vipr2/Drd4/Igf1/Nos3/Tert/Mmp2/Npr3 | 7 |
| GO:0050806 | positive regulation of synaptic transmission | BP | 0.0299 | 0.00337 | 0.0246 | 1.61 | 0.0176 | Slc1a3/Grik1/Gria4/Nog/Rims3/Adra1a/Nlgn1/Grin2b/Serpine2/Slc8a3 | 10 |
| GO:0030203 | glycosaminoglycan metabolic process | BP | 0.0532 | 0.00337 | 0.0246 | 1.61 | 0.0176 | Angpt1/Spock2/Ednrb/Cd44/Has2 | 5 |
| GO:0051899 | membrane depolarization | BP | 0.0532 | 0.00337 | 0.0246 | 1.61 | 0.0176 | Grik1/Scn4b/Scn1a/Kdr/Bok | 5 |
| GO:0060041 | retina development in camera-type eye | BP | 0.0387 | 0.00339 | 0.0246 | 1.61 | 0.0176 | Ndp/Cyp1b1/Ihh/Sdk2/Tub/Lamc3/Celf4 | 7 |
| GO:0015872 | dopamine transport | BP | 0.069 | 0.00341 | 0.0247 | 1.61 | 0.0176 | Slc6a2/Syt13/Abat/Cxcl12 | 4 |
| GO:0021587 | cerebellum morphogenesis | BP | 0.069 | 0.00341 | 0.0247 | 1.61 | 0.0176 | Gli1/Dab1/Cbs/Serpine2 | 4 |
| GO:0042311 | vasodilation | BP | 0.069 | 0.00341 | 0.0247 | 1.61 | 0.0176 | Kdr/Ednrb/Calca/Npr3 | 4 |
| GO:1901214 | regulation of neuron death | BP | 0.0281 | 0.00344 | 0.0248 | 1.6 | 0.0178 | Ptprz1/Angpt1/Dcc/St8sia2/Mdk/Igf1/Kdr/Grin2b/Tert/Bok/Agap2 | 11 |
| GO:0045685 | regulation of glial cell differentiation | BP | 0.0526 | 0.00353 | 0.0254 | 1.6 | 0.0182 | Ptprz1/Mdk/Nog/Dab1/Serpine2 | 5 |
| GO:0001759 | organ induction | BP | 0.103 | 0.00356 | 0.0256 | 1.59 | 0.0183 | Wnt11/Hoxd11/Hoxa11 | 3 |
| GO:0030318 | melanocyte differentiation | BP | 0.103 | 0.00356 | 0.0256 | 1.59 | 0.0183 | Adamts20/Ednrb/Edn3 | 3 |
| GO:0001935 | endothelial cell proliferation | BP | 0.0383 | 0.0036 | 0.0257 | 1.59 | 0.0184 | Sema5a/Mdk/Igf1/Itga4/Kdr/Calca/Cxcl12 | 7 |
| GO:0030199 | collagen fibril organization | BP | 0.0678 | 0.00363 | 0.0258 | 1.59 | 0.0184 | Cyp1b1/Col11a1/Sfrp2/Col14a1 | 4 |
| GO:0031279 | regulation of cyclase activity | BP | 0.0678 | 0.00363 | 0.0258 | 1.59 | 0.0184 | Vipr2/Calca/Nos3/Npr3 | 4 |
| GO:0045778 | positive regulation of ossification | BP | 0.0678 | 0.00363 | 0.0258 | 1.59 | 0.0184 | Pkdcc/Calca/Osr2/Nell1 | 4 |
| GO:1903793 | positive regulation of anion transport | BP | 0.0678 | 0.00363 | 0.0258 | 1.59 | 0.0184 | Grik1/Pla2g10/Abat/Grin2b | 4 |
| GO:0030177 | positive regulation of Wnt signaling pathway | BP | 0.0432 | 0.00383 | 0.0272 | 1.57 | 0.0194 | Sfrp2/Lef1/Sema5a/Gpc3/Nkd1/Tert | 6 |
| GO:0002028 | regulation of sodium ion transport | BP | 0.0515 | 0.00386 | 0.0272 | 1.57 | 0.0195 | Drd4/Scn4b/Hecw2/Nos3/Serpine2 | 5 |
| GO:0097479 | synaptic vesicle localization | BP | 0.0667 | 0.00386 | 0.0272 | 1.57 | 0.0195 | Syn2/Map2/Nlgn1/Cdh2 | 4 |
| GO:0048596 | embryonic camera-type eye morphogenesis | BP | 0.1 | 0.00393 | 0.0275 | 1.56 | 0.0197 | Ihh/Stra6/Tbx2 | 3 |
| GO:0051957 | positive regulation of amino acid transport | BP | 0.1 | 0.00393 | 0.0275 | 1.56 | 0.0197 | Grik1/Abat/Grin2b | 3 |
| GO:0060563 | neuroepithelial cell differentiation | BP | 0.1 | 0.00393 | 0.0275 | 1.56 | 0.0197 | Wnt11/Lef1/Cdh2 | 3 |
| GO:0070977 | bone maturation | BP | 0.1 | 0.00393 | 0.0275 | 1.56 | 0.0197 | Ryr1/Igf1/Plxnb1 | 3 |
| GO:0042632 | cholesterol homeostasis | BP | 0.051 | 0.00403 | 0.0281 | 1.55 | 0.0201 | Apob/Pla2g10/Nr5a2/Cyp7b1/Soat1 | 5 |
| GO:0070252 | actin-mediated cell contraction | BP | 0.051 | 0.00403 | 0.0281 | 1.55 | 0.0201 | Kcnd3/Sgcd/Scn4b/Scn1a/Stc1 | 5 |
| GO:0046942 | carboxylic acid transport | BP | 0.0311 | 0.00403 | 0.0281 | 1.55 | 0.0201 | Slc1a3/Grik1/Pla2g10/Slc13a5/Drd4/Abat/Lrp2/Grin2b/Slc7a2 | 9 |
| GO:0010656 | negative regulation of muscle cell apoptotic process | BP | 0.0656 | 0.00409 | 0.0284 | 1.55 | 0.0203 | Sfrp2/Gria4/Mdk/Igf1 | 4 |
| GO:0099172 | presynapse organization | BP | 0.0656 | 0.00409 | 0.0284 | 1.55 | 0.0203 | Gpc6/Nlgn1/Ptprd/Lrrc4b | 4 |
| GO:0048659 | smooth muscle cell proliferation | BP | 0.0372 | 0.00417 | 0.0288 | 1.54 | 0.0206 | Vipr2/Drd4/Igf1/Nos3/Tert/Mmp2/Npr3 | 7 |
| GO:0055092 | sterol homeostasis | BP | 0.0505 | 0.00421 | 0.029 | 1.54 | 0.0208 | Apob/Pla2g10/Nr5a2/Cyp7b1/Soat1 | 5 |
| GO:0048675 | axon extension | BP | 0.0423 | 0.00426 | 0.0293 | 1.53 | 0.021 | Map2/Sema5a/Sema5b/Edn3/Sema7a/Cxcl12 | 6 |
| GO:0003085 | negative regulation of systemic arterial blood pressure | BP | 0.0968 | 0.00432 | 0.0296 | 1.53 | 0.0211 | Adra1a/Kdr/Calca | 3 |
| GO:0050951 | sensory perception of temperature stimulus | BP | 0.0968 | 0.00432 | 0.0296 | 1.53 | 0.0211 | Calca/Ano1/Cxcl12 | 3 |
| GO:0001736 | establishment of planar polarity | BP | 0.0645 | 0.00434 | 0.0296 | 1.53 | 0.0211 | Sfrp2/Wnt11/Gpc3/Nkd1 | 4 |
| GO:0021575 | hindbrain morphogenesis | BP | 0.0645 | 0.00434 | 0.0296 | 1.53 | 0.0211 | Gli1/Dab1/Cbs/Serpine2 | 4 |
| GO:0051339 | regulation of lyase activity | BP | 0.0645 | 0.00434 | 0.0296 | 1.53 | 0.0211 | Vipr2/Calca/Nos3/Npr3 | 4 |
| GO:0051932 | synaptic transmission, GABAergic | BP | 0.0645 | 0.00434 | 0.0296 | 1.53 | 0.0211 | Grik1/Adra1a/Nlgn1/Gabra4 | 4 |
| GO:0043524 | negative regulation of neuron apoptotic process | BP | 0.0368 | 0.00441 | 0.03 | 1.52 | 0.0215 | Ptprz1/Angpt1/Mdk/Kdr/Tert/Bok/Agap2 | 7 |
| GO:0010632 | regulation of epithelial cell migration | BP | 0.0332 | 0.0045 | 0.0305 | 1.52 | 0.0218 | Angpt1/Sema5a/Igf1/Kdr/Stc1/Nos3/Has2/Evl | 8 |
| GO:0045664 | regulation of neuron differentiation | BP | 0.0332 | 0.0045 | 0.0305 | 1.52 | 0.0218 | Sfrp2/Fez1/Nlgn1/Rgs6/Ednrb/Dab1/Cxcl12/Gdf7 | 8 |
| GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | BP | 0.0495 | 0.00458 | 0.0308 | 1.51 | 0.022 | Adra1a/Ednrb/Calca/Ano1/Npr3 | 5 |
| GO:0060021 | roof of mouth development | BP | 0.0495 | 0.00458 | 0.0308 | 1.51 | 0.022 | Wnt11/Lef1/Pkdcc/Tbx2/Osr2 | 5 |
| GO:0007164 | establishment of tissue polarity | BP | 0.0635 | 0.0046 | 0.0309 | 1.51 | 0.0221 | Sfrp2/Wnt11/Gpc3/Nkd1 | 4 |
| GO:0050727 | regulation of inflammatory response | BP | 0.0285 | 0.00467 | 0.0313 | 1.51 | 0.0224 | Acod1/Hgf/Pla2g10/Mdk/Igf1/Ednrb/Foxf1/Cd44/Sema7a/Slc7a2 | 10 |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | BP | 0.0365 | 0.00467 | 0.0313 | 1.51 | 0.0224 | Ptprz1/Hgf/Angpt1/Adra1a/Igf1/Areg/Cd44 | 7 |
| GO:0003416 | endochondral bone growth | BP | 0.0938 | 0.00473 | 0.0315 | 1.5 | 0.0225 | Kdr/Evc/Stc1 | 3 |
| GO:0060977 | coronary vasculature morphogenesis | BP | 0.0938 | 0.00473 | 0.0315 | 1.5 | 0.0225 | Angpt1/Sgcd/Lrp2 | 3 |
| GO:0098703 | calcium ion import across plasma membrane | BP | 0.0938 | 0.00473 | 0.0315 | 1.5 | 0.0225 | Slc24a4/Cacna1e/Slc8a3 | 3 |
| GO:0006054 | N-acetylneuraminate metabolic process | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | St6gal2/Npl | 2 |
| GO:0006527 | arginine catabolic process | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | Ddah1/Nos3 | 2 |
| GO:0060385 | axonogenesis involved in innervation | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | Itga4/Nptx1 | 2 |
| GO:0097105 | presynaptic membrane assembly | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | Nlgn1/Ptprd | 2 |
| GO:0097119 | postsynaptic density protein 95 clustering | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | Nlgn1/Cdh2 | 2 |
| GO:2000807 | regulation of synaptic vesicle clustering | BP | 0.2 | 0.00481 | 0.0316 | 1.5 | 0.0226 | Nlgn1/Cdh2 | 2 |
| GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | BP | 0.0625 | 0.00486 | 0.0319 | 1.5 | 0.0228 | Drd4/Igf1/Tert/Mmp2 | 4 |
| GO:0007204 | positive regulation of cytosolic calcium ion concentration | BP | 0.0282 | 0.00495 | 0.0324 | 1.49 | 0.0232 | Ryr1/Adra1a/Slc24a4/Kdr/Ednrb/Calca/Ptgfr/Cacna1e/Grin2b/Slc8a3 | 10 |
| GO:0010469 | regulation of signaling receptor activity | BP | 0.0408 | 0.00503 | 0.0329 | 1.48 | 0.0235 | Begain/Nog/Nlgn1/Slc24a4/Areg/Nptx1 | 6 |
| GO:0003229 | ventricular cardiac muscle tissue development | BP | 0.0615 | 0.00514 | 0.0335 | 1.47 | 0.024 | Col11a1/Col14a1/Nog/Lrp2 | 4 |
| GO:1902656 | calcium ion import into cytosol | BP | 0.0909 | 0.00516 | 0.0336 | 1.47 | 0.024 | Slc24a4/Cacna1e/Slc8a3 | 3 |
| GO:0070167 | regulation of biomineral tissue development | BP | 0.0481 | 0.00518 | 0.0337 | 1.47 | 0.0241 | Wnt6/Pkdcc/Nos3/Osr2/Nell1 | 5 |
| GO:0071453 | cellular response to oxygen levels | BP | 0.0405 | 0.0052 | 0.0337 | 1.47 | 0.0241 | Kcnk3/Kcnd2/Ddah1/Cbs/Tert/Slc8a3 | 6 |
| GO:0045860 | positive regulation of protein kinase activity | BP | 0.0279 | 0.00534 | 0.0346 | 1.46 | 0.0247 | Angpt1/Adra1a/Drd4/Igf1/Areg/Dab1/Calca/Edn3/Ccdc88a/Agap2 | 10 |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | BP | 0.0476 | 0.00539 | 0.0349 | 1.46 | 0.0249 | Hgf/Sfrp2/Igf1/Tert/Agap2 | 5 |
| GO:0051047 | positive regulation of secretion | BP | 0.0264 | 0.00544 | 0.0351 | 1.46 | 0.0251 | Grik1/Pla2g10/Igf1/Nlgn1/Abat/Ednrb/Edn3/Grin2b/Ano1/Cxcl12/Cadps | 11 |
| GO:0009953 | dorsal/ventral pattern formation | BP | 0.0472 | 0.00561 | 0.0359 | 1.44 | 0.0257 | Gli1/Hhip/Nog/Hoxd11/Hoxa11 | 5 |
| GO:0003156 | regulation of animal organ formation | BP | 0.0882 | 0.00561 | 0.0359 | 1.44 | 0.0257 | Wnt11/Hoxd11/Hoxa11 | 3 |
| GO:0048799 | animal organ maturation | BP | 0.0882 | 0.00561 | 0.0359 | 1.44 | 0.0257 | Ryr1/Igf1/Plxnb1 | 3 |
| GO:1990874 | vascular associated smooth muscle cell proliferation | BP | 0.0597 | 0.00572 | 0.0365 | 1.44 | 0.0261 | Drd4/Igf1/Tert/Mmp2 | 4 |
| GO:0006821 | chloride transport | BP | 0.0467 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Slc1a3/Slc12a8/Ano2/Ano1/Gabra4 | 5 |
| GO:0003139 | secondary heart field specification | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Wnt11/Lrp2 | 2 |
| GO:0010749 | regulation of nitric oxide mediated signal transduction | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Spink1/Cbs | 2 |
| GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Gli1/Igf1 | 2 |
| GO:0030202 | heparin metabolic process | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Angpt1/Ednrb | 2 |
| GO:0033689 | negative regulation of osteoblast proliferation | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Plxnb1/Nell1 | 2 |
| GO:0036302 | atrioventricular canal development | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Tbx2/Has2 | 2 |
| GO:0060405 | regulation of penile erection | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Drd4/Ednrb | 2 |
| GO:0061299 | retina vasculature morphogenesis in camera-type eye | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Ndp/Cyp1b1 | 2 |
| GO:0090037 | positive regulation of protein kinase C signaling | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Wnt11/Adra1a | 2 |
| GO:0097104 | postsynaptic membrane assembly | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Nlgn1/Cdh2 | 2 |
| GO:1902513 | regulation of organelle transport along microtubule | BP | 0.182 | 0.00584 | 0.0365 | 1.44 | 0.0261 | Map2/Fez1 | 2 |
| GO:0060401 | cytosolic calcium ion transport | BP | 0.0348 | 0.00597 | 0.0372 | 1.43 | 0.0266 | Ryr1/Adra1a/Slc24a4/Calca/Cacna1e/Grin2b/Slc8a3 | 7 |
| GO:0010837 | regulation of keratinocyte proliferation | BP | 0.0857 | 0.00609 | 0.0376 | 1.42 | 0.0269 | Cd109/Mdk/Has2 | 3 |
| GO:0035116 | embryonic hindlimb morphogenesis | BP | 0.0857 | 0.00609 | 0.0376 | 1.42 | 0.0269 | Aff3/Gpc3/Osr2 | 3 |
| GO:0045987 | positive regulation of smooth muscle contraction | BP | 0.0857 | 0.00609 | 0.0376 | 1.42 | 0.0269 | Adra1a/Npnt/Abat | 3 |
| GO:0048710 | regulation of astrocyte differentiation | BP | 0.0857 | 0.00609 | 0.0376 | 1.42 | 0.0269 | Nog/Dab1/Serpine2 | 3 |
| GO:0007517 | muscle organ development | BP | 0.0274 | 0.0061 | 0.0376 | 1.42 | 0.0269 | Stra6/Col11a1/Lef1/Angpt1/Ryr1/Nog/Hoxd10/P2rx2/Lrp2/Hoxd9 | 10 |
| GO:0051346 | negative regulation of hydrolase activity | BP | 0.0273 | 0.00621 | 0.0382 | 1.42 | 0.0273 | Hgf/Sfrp2/Spink1/Cd109/Igf1/Gpc3/Tfpi/Nos3/Cd44/Serpine2 | 10 |
| GO:0015850 | organic hydroxy compound transport | BP | 0.0314 | 0.00628 | 0.0386 | 1.41 | 0.0276 | Apob/Stra6/Maob/Slc6a2/Syt13/Abat/Cxcl12/Cadps | 8 |
| GO:0010595 | positive regulation of endothelial cell migration | BP | 0.0459 | 0.00631 | 0.0387 | 1.41 | 0.0277 | Angpt1/Sema5a/Igf1/Kdr/Nos3 | 5 |
| GO:0006940 | regulation of smooth muscle contraction | BP | 0.058 | 0.00635 | 0.0389 | 1.41 | 0.0278 | Adra1a/Npnt/Abat/Calca | 4 |
| GO:0031503 | protein-containing complex localization | BP | 0.0343 | 0.00646 | 0.0394 | 1.4 | 0.0282 | Gpc6/Sgcd/Lrrc7/Drd4/Nlgn1/Tub/Nptx1 | 7 |
| GO:0001837 | epithelial to mesenchymal transition | BP | 0.0387 | 0.00648 | 0.0394 | 1.4 | 0.0282 | Sfrp2/Wnt11/Lef1/Mdk/Nog/Has2 | 6 |
| GO:0007043 | cell-cell junction assembly | BP | 0.0387 | 0.00648 | 0.0394 | 1.4 | 0.0282 | Wnt11/Ptpro/Epb41l3/Cdh6/Cdh2/Cdh10 | 6 |
| GO:0048593 | camera-type eye morphogenesis | BP | 0.0387 | 0.00648 | 0.0394 | 1.4 | 0.0282 | Ihh/Stra6/Sdk2/Tbx2/Kdr/Tenm3 | 6 |
| GO:0006022 | aminoglycan metabolic process | BP | 0.0455 | 0.00655 | 0.0397 | 1.4 | 0.0284 | Angpt1/Spock2/Ednrb/Cd44/Has2 | 5 |
| GO:0002053 | positive regulation of mesenchymal cell proliferation | BP | 0.0833 | 0.00659 | 0.0397 | 1.4 | 0.0284 | Ihh/Kdr/Foxf1 | 3 |
| GO:0003203 | endocardial cushion morphogenesis | BP | 0.0833 | 0.00659 | 0.0397 | 1.4 | 0.0284 | Nog/Tbx2/Nos3 | 3 |
| GO:0008045 | motor neuron axon guidance | BP | 0.0833 | 0.00659 | 0.0397 | 1.4 | 0.0284 | Nog/Rnf165/Cxcl12 | 3 |
| GO:0048048 | embryonic eye morphogenesis | BP | 0.0833 | 0.00659 | 0.0397 | 1.4 | 0.0284 | Ihh/Stra6/Tbx2 | 3 |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | BP | 0.0833 | 0.00659 | 0.0397 | 1.4 | 0.0284 | Grik1/Kdr/Grin2b | 3 |
| GO:0042246 | tissue regeneration | BP | 0.0571 | 0.00668 | 0.04 | 1.4 | 0.0286 | Gap43/Mdk/Igf1/Hopx | 4 |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | BP | 0.0571 | 0.00668 | 0.04 | 1.4 | 0.0286 | Vipr2/Drd4/Nos3/Npr3 | 4 |
| GO:0010634 | positive regulation of epithelial cell migration | BP | 0.0385 | 0.00668 | 0.04 | 1.4 | 0.0286 | Angpt1/Sema5a/Igf1/Kdr/Nos3/Has2 | 6 |
| GO:0021549 | cerebellum development | BP | 0.045 | 0.0068 | 0.0407 | 1.39 | 0.0291 | Gli1/Mdk/Dab1/Cbs/Serpine2 | 5 |
| GO:0048872 | homeostasis of number of cells | BP | 0.0269 | 0.00693 | 0.0411 | 1.39 | 0.0294 | Tex15/Pla2g10/Col14a1/Crispld1/Gprasp2/Emcn/P4htm/Cdh2/Nos3/Cd44 | 10 |
| GO:0007501 | mesodermal cell fate specification | BP | 0.167 | 0.00696 | 0.0411 | 1.39 | 0.0294 | Sfrp2/Hoxa11 | 2 |
| GO:0042135 | neurotransmitter catabolic process | BP | 0.167 | 0.00696 | 0.0411 | 1.39 | 0.0294 | Maob/Abat | 2 |
| GO:0048934 | peripheral nervous system neuron differentiation | BP | 0.167 | 0.00696 | 0.0411 | 1.39 | 0.0294 | Hoxd10/Hoxd9 | 2 |
| GO:0048935 | peripheral nervous system neuron development | BP | 0.167 | 0.00696 | 0.0411 | 1.39 | 0.0294 | Hoxd10/Hoxd9 | 2 |
| GO:0099519 | dense core granule cytoskeletal transport | BP | 0.167 | 0.00696 | 0.0411 | 1.39 | 0.0294 | Map2/Ppfia2 | 2 |
| GO:0017156 | calcium-ion regulated exocytosis | BP | 0.0563 | 0.00702 | 0.0413 | 1.38 | 0.0296 | Syn2/Rims3/Syt13/Cadps | 4 |
| GO:0050432 | catecholamine secretion | BP | 0.0563 | 0.00702 | 0.0413 | 1.38 | 0.0296 | Syt13/Abat/Cxcl12/Cadps | 4 |
| GO:0060441 | epithelial tube branching involved in lung morphogenesis | BP | 0.0811 | 0.00712 | 0.0418 | 1.38 | 0.0299 | Hhip/Tbx2/Foxf1 | 3 |
| GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | BP | 0.0811 | 0.00712 | 0.0418 | 1.38 | 0.0299 | Igf1/Tert/Mmp2 | 3 |
| GO:0022408 | negative regulation of cell-cell adhesion | BP | 0.0337 | 0.00715 | 0.0419 | 1.38 | 0.03 | Ihh/Lef1/Mdk/Abat/Cd44/Serpine2/Cxcl12 | 7 |
| GO:1901215 | negative regulation of neuron death | BP | 0.0307 | 0.00718 | 0.042 | 1.38 | 0.03 | Ptprz1/Angpt1/Mdk/Igf1/Kdr/Tert/Bok/Agap2 | 8 |
| GO:0030516 | regulation of axon extension | BP | 0.0442 | 0.00732 | 0.0428 | 1.37 | 0.0306 | Map2/Sema5a/Sema5b/Sema7a/Cxcl12 | 5 |
| GO:0048168 | regulation of neuronal synaptic plasticity | BP | 0.0556 | 0.00737 | 0.0429 | 1.37 | 0.0307 | Grik1/Nog/Kdr/Grin2b | 4 |
| GO:0055008 | cardiac muscle tissue morphogenesis | BP | 0.0556 | 0.00737 | 0.0429 | 1.37 | 0.0307 | Col11a1/Angpt1/Nog/Lrp2 | 4 |
| GO:0090596 | sensory organ morphogenesis | BP | 0.0282 | 0.00755 | 0.0439 | 1.36 | 0.0314 | Ihh/Stra6/Col11a1/Sdk2/Nog/Tbx2/Kdr/Tenm3/Osr2 | 9 |
| GO:0098868 | bone growth | BP | 0.0789 | 0.00767 | 0.0445 | 1.35 | 0.0318 | Kdr/Evc/Stc1 | 3 |
| GO:0048167 | regulation of synaptic plasticity | BP | 0.0265 | 0.00771 | 0.0447 | 1.35 | 0.032 | Grik1/Nog/Grid2ip/Rims3/Jph4/Nlgn1/Kdr/Grin2b/Serpine2/Slc8a3 | 10 |
| GO:0061045 | negative regulation of wound healing | BP | 0.0548 | 0.00773 | 0.0447 | 1.35 | 0.032 | Cd109/Abat/Tfpi/Serpine2 | 4 |
| GO:0031032 | actomyosin structure organization | BP | 0.0329 | 0.0081 | 0.046 | 1.34 | 0.0329 | Kank4/Wnt11/Frmd3/Epb41l3/Tmod2/Evl/Ccdc88a | 7 |
| GO:1902115 | regulation of organelle assembly | BP | 0.0329 | 0.0081 | 0.046 | 1.34 | 0.0329 | Gap43/Fez1/Adamts16/Ptprd/Nptx1/Lrrc4b/Ccdc88a | 7 |
| GO:0001942 | hair follicle development | BP | 0.0431 | 0.00816 | 0.046 | 1.34 | 0.0329 | Pla2g10/Apcdd1/Cd109/Fst/Edaradd | 5 |
| GO:0051952 | regulation of amine transport | BP | 0.0431 | 0.00816 | 0.046 | 1.34 | 0.0329 | Grik1/Syt13/Abat/Grin2b/Cxcl12 | 5 |
| GO:0003157 | endocardium development | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Angpt1/Kdr | 2 |
| GO:0010838 | positive regulation of keratinocyte proliferation | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Mdk/Has2 | 2 |
| GO:0014052 | regulation of gamma-aminobutyric acid secretion | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Grik1/Abat | 2 |
| GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Angpt1/Kdr | 2 |
| GO:0031223 | auditory behavior | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Stra6/Slc1a3 | 2 |
| GO:0033240 | positive regulation of cellular amine metabolic process | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Maob/Abat | 2 |
| GO:0034374 | low-density lipoprotein particle remodeling | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Apob/Pla2g10 | 2 |
| GO:0034392 | negative regulation of smooth muscle cell apoptotic process | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Gria4/Igf1 | 2 |
| GO:0035810 | positive regulation of urine volume | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Ednrb/Npr3 | 2 |
| GO:0048340 | paraxial mesoderm morphogenesis | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Wnt11/Lef1 | 2 |
| GO:0097090 | presynaptic membrane organization | BP | 0.154 | 0.00817 | 0.046 | 1.34 | 0.0329 | Nlgn1/Ptprd | 2 |
| GO:0031281 | positive regulation of cyclase activity | BP | 0.0769 | 0.00825 | 0.0461 | 1.34 | 0.033 | Vipr2/Calca/Nos3 | 3 |
| GO:0033238 | regulation of cellular amine metabolic process | BP | 0.0769 | 0.00825 | 0.0461 | 1.34 | 0.033 | Maob/Drd4/Abat | 3 |
| GO:0050931 | pigment cell differentiation | BP | 0.0769 | 0.00825 | 0.0461 | 1.34 | 0.033 | Adamts20/Ednrb/Edn3 | 3 |
| GO:0051180 | vitamin transport | BP | 0.0769 | 0.00825 | 0.0461 | 1.34 | 0.033 | Stra6/Slc2a3/Lrp2 | 3 |
| GO:1904645 | response to amyloid-beta | BP | 0.0769 | 0.00825 | 0.0461 | 1.34 | 0.033 | Igf1/Itga4/Mmp2 | 3 |
| GO:0052547 | regulation of peptidase activity | BP | 0.0248 | 0.00839 | 0.0468 | 1.33 | 0.0335 | Hgf/Sfrp2/Spink1/Cd109/Igf1/Gpc3/Tfpi/Cd44/Grin2b/Serpine2/Bok | 11 |
| GO:0044070 | regulation of anion transport | BP | 0.0427 | 0.00845 | 0.0471 | 1.33 | 0.0337 | Grik1/Pla2g10/Abat/Stc1/Grin2b | 5 |
| GO:0042982 | amyloid precursor protein metabolic process | BP | 0.0533 | 0.00849 | 0.0471 | 1.33 | 0.0337 | Rtn1/Igf1/Itm2a/Soat1 | 4 |
| GO:0045669 | positive regulation of osteoblast differentiation | BP | 0.0533 | 0.00849 | 0.0471 | 1.33 | 0.0337 | Sfrp2/Igf1/Npnt/Nell1 | 4 |
| GO:0097120 | receptor localization to synapse | BP | 0.0533 | 0.00849 | 0.0471 | 1.33 | 0.0337 | Gpc6/Lrrc7/Nlgn1/Nptx1 | 4 |
| GO:0070372 | regulation of ERK1 and ERK2 cascade | BP | 0.0276 | 0.00864 | 0.0479 | 1.32 | 0.0342 | Angpt1/Dcc/Adra1a/Igf1/Npnt/Kdr/Cd44/Sema7a/Cxcl12 | 9 |
| GO:0008585 | female gonad development | BP | 0.0424 | 0.00875 | 0.0483 | 1.32 | 0.0345 | Angpt1/Fst/Kdr/Nos3/Mmp2 | 5 |
| GO:0051897 | positive regulation of protein kinase B signaling | BP | 0.0424 | 0.00875 | 0.0483 | 1.32 | 0.0345 | Angpt1/Sema5a/Igf1/Lrp2/Cxcl12 | 5 |
| GO:0009311 | oligosaccharide metabolic process | BP | 0.075 | 0.00884 | 0.0487 | 1.31 | 0.0348 | St6gal2/St8sia2/St8sia4 | 3 |
| GO:0051349 | positive regulation of lyase activity | BP | 0.075 | 0.00884 | 0.0487 | 1.31 | 0.0348 | Vipr2/Calca/Nos3 | 3 |
| GO:0060249 | anatomical structure homeostasis | BP | 0.0274 | 0.00897 | 0.0493 | 1.31 | 0.0353 | Tex15/Ihh/Angpt1/Col14a1/Tub/Kdr/Calca/Nos3/P2rx2 | 9 |
| GO:0022405 | hair cycle process | BP | 0.042 | 0.00905 | 0.0495 | 1.31 | 0.0354 | Pla2g10/Apcdd1/Cd109/Fst/Edaradd | 5 |
| GO:0045666 | positive regulation of neuron differentiation | BP | 0.042 | 0.00905 | 0.0495 | 1.31 | 0.0354 | Fez1/Rgs6/Dab1/Cxcl12/Gdf7 | 5 |
| GO:0043270 | positive regulation of ion transport | BP | 0.0274 | 0.00914 | 0.0499 | 1.3 | 0.0357 | Grik1/Pla2g10/Drd4/Scn4b/Abat/Stc1/Edn3/Grin2b/Cxcl12 | 9 |
| GO:0099699 | integral component of synaptic membrane | CC | 0.124 | 2.27e-11 | 6.89e-09 | 8.16 | 5.06e-09 | Kcnj3/Efnb3/Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Ptprd/Lrrc4b/Cdh2/Cacna1e/Grin2b | 14 |
| GO:0099240 | intrinsic component of synaptic membrane | CC | 0.114 | 7.24e-11 | 1.1e-08 | 7.96 | 8.08e-09 | Kcnj3/Efnb3/Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Ptprd/Lrrc4b/Cdh2/Cacna1e/Grin2b | 14 |
| GO:0097060 | synaptic membrane | CC | 0.0564 | 3.48e-10 | 3.09e-08 | 7.51 | 2.27e-08 | Ptprz1/Kcnj3/Efnb3/Grid1/Grik1/Kcnd2/Gria4/Slc6a2/Dcc/Kcnd3/Lrrc4c/Grid2ip/Rims3/Ptpro/Nlgn1/Ptprd/Lrrc4b/Cdh2/P2rx2/Cacna1e/Grin2b/Gabra4 | 22 |
| GO:0062023 | collagen-containing extracellular matrix | CC | 0.0556 | 4.64e-10 | 3.09e-08 | 7.51 | 2.27e-08 | Ndp/Gpc6/Col11a1/Ptprz1/Hmcn1/Vit/Angpt1/Col14a1/Col4a6/Colec12/Igf1/Lamc3/Npnt/Spock2/Gpc3/Emid1/Tgfbi/Col9a2/Serpine2/Frem1/Lama4/Nid1 | 22 |
| GO:0098984 | neuron to neuron synapse | CC | 0.0582 | 5.09e-10 | 3.09e-08 | 7.51 | 2.27e-08 | Gap43/Syn2/Efnb3/Grid1/Map2/Grik1/Kcnd2/Gria4/Dcc/Kcnd3/Lrrc4c/Lrrc7/Ptpro/Ptprd/Epb41l3/Lrrc4b/Cdh2/Dab1/P2rx2/Grin2b/Slc8a3 | 21 |
| GO:0099060 | integral component of postsynaptic specialization membrane | CC | 0.164 | 1.02e-09 | 5.18e-08 | 7.29 | 3.81e-08 | Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Lrrc4b/Cdh2/Grin2b | 10 |
| GO:0098948 | intrinsic component of postsynaptic specialization membrane | CC | 0.159 | 1.42e-09 | 6.18e-08 | 7.21 | 4.54e-08 | Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Lrrc4b/Cdh2/Grin2b | 10 |
| GO:0099572 | postsynaptic specialization | CC | 0.0568 | 1.99e-09 | 6.84e-08 | 7.17 | 5.02e-08 | Gap43/Syn2/Grid1/Map2/Grik1/Kcnd2/Gria4/Dcc/Kcnd3/Lrrc4c/Lrrc7/Ptpro/Nlgn1/Epb41l3/Lrrc4b/Cdh2/Dab1/P2rx2/Grin2b/Slc8a3 | 20 |
| GO:0099055 | integral component of postsynaptic membrane | CC | 0.129 | 2.02e-09 | 6.84e-08 | 7.17 | 5.02e-08 | Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Lrrc4b/Cdh2/Cacna1e/Grin2b | 11 |
| GO:0098936 | intrinsic component of postsynaptic membrane | CC | 0.124 | 3.33e-09 | 1.01e-07 | 6.99 | 7.44e-08 | Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Lrrc4b/Cdh2/Cacna1e/Grin2b | 11 |
| GO:0014069 | postsynaptic density | CC | 0.0576 | 4.1e-09 | 1.13e-07 | 6.95 | 8.32e-08 | Gap43/Syn2/Grid1/Map2/Grik1/Kcnd2/Gria4/Dcc/Kcnd3/Lrrc4c/Lrrc7/Ptpro/Epb41l3/Lrrc4b/Cdh2/Dab1/P2rx2/Grin2b/Slc8a3 | 19 |
| GO:0032279 | asymmetric synapse | CC | 0.056 | 6.35e-09 | 1.61e-07 | 6.79 | 1.18e-07 | Gap43/Syn2/Grid1/Map2/Grik1/Kcnd2/Gria4/Dcc/Kcnd3/Lrrc4c/Lrrc7/Ptpro/Epb41l3/Lrrc4b/Cdh2/Dab1/P2rx2/Grin2b/Slc8a3 | 19 |
| GO:0034702 | ion channel complex | CC | 0.0578 | 2.56e-08 | 5.99e-07 | 6.22 | 4.4e-07 | Kcnb2/Kcnj3/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Ano2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1/Gabra4 | 17 |
| GO:0099634 | postsynaptic specialization membrane | CC | 0.118 | 2.83e-08 | 6.14e-07 | 6.21 | 4.51e-07 | Grid1/Kcnd2/Dcc/Kcnd3/Lrrc4c/Ptpro/Nlgn1/Lrrc4b/Cdh2/Grin2b | 10 |
| GO:0043197 | dendritic spine | CC | 0.0707 | 3.71e-08 | 7.53e-07 | 6.12 | 5.52e-07 | Ptprz1/Slc1a3/Kcnd2/Gria4/Kcnd3/Grid2ip/Lrrc7/Drd4/Ptpro/Nlgn1/P2rx2/Ppfia2/Grin2b/Slc8a3 | 14 |
| GO:0044309 | neuron spine | CC | 0.069 | 5.08e-08 | 9.65e-07 | 6.02 | 7.08e-07 | Ptprz1/Slc1a3/Kcnd2/Gria4/Kcnd3/Grid2ip/Lrrc7/Drd4/Ptpro/Nlgn1/P2rx2/Ppfia2/Grin2b/Slc8a3 | 14 |
| GO:0045211 | postsynaptic membrane | CC | 0.0586 | 5.5e-08 | 9.84e-07 | 6.01 | 7.22e-07 | Ptprz1/Grid1/Grik1/Kcnd2/Gria4/Dcc/Kcnd3/Lrrc4c/Grid2ip/Ptpro/Nlgn1/Lrrc4b/Cdh2/Cacna1e/Grin2b/Gabra4 | 16 |
| GO:0098978 | glutamatergic synapse | CC | 0.0526 | 1e-07 | 1.69e-06 | 5.77 | 1.24e-06 | Syn2/Efnb3/Grid1/Kcnd2/Gria4/Begain/Lrrc4c/Drd4/Ptpro/Nlgn1/Ptprd/Nptx1/Lrrc4b/Ppfia2/Grin2b/Wasf3/Cadps | 17 |
| GO:0034703 | cation channel complex | CC | 0.0625 | 1.72e-07 | 2.75e-06 | 5.56 | 2.02e-06 | Kcnb2/Kcnj3/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Kcnq5/Cacna1e/Unc80/Grin2b | 14 |
| GO:1902495 | transmembrane transporter complex | CC | 0.0457 | 7.28e-07 | 1.11e-05 | 4.96 | 8.12e-06 | Kcnb2/Kcnj3/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Ano2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1/Gabra4 | 17 |
| GO:0042734 | presynaptic membrane | CC | 0.0758 | 1.8e-06 | 2.49e-05 | 4.6 | 1.83e-05 | Kcnj3/Efnb3/Grik1/Slc6a2/Rims3/Ptprd/Cdh2/P2rx2/Cacna1e/Grin2b | 10 |
| GO:0099061 | integral component of postsynaptic density membrane | CC | 0.154 | 3.63e-06 | 4.6e-05 | 4.34 | 3.37e-05 | Grid1/Dcc/Lrrc4c/Ptpro/Lrrc4b/Grin2b | 6 |
| GO:0099146 | intrinsic component of postsynaptic density membrane | CC | 0.146 | 4.91e-06 | 5.97e-05 | 4.22 | 4.38e-05 | Grid1/Dcc/Lrrc4c/Ptpro/Lrrc4b/Grin2b | 6 |
| GO:0034705 | potassium channel complex | CC | 0.0899 | 5.55e-06 | 6.48e-05 | 4.19 | 4.76e-05 | Kcnb2/Kcnj3/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Kcnq5 | 8 |
| GO:0042383 | sarcolemma | CC | 0.0606 | 1.32e-05 | 0.000143 | 3.84 | 0.000105 | Car4/Kcnj3/Kcnd2/Ryr1/Sgcd/Adra1a/Scn1a/Cdh2/Nos3/Slc8a3 | 10 |
| GO:0098839 | postsynaptic density membrane | CC | 0.1 | 4.61e-05 | 0.000457 | 3.34 | 0.000335 | Grid1/Dcc/Lrrc4c/Ptpro/Lrrc4b/Grin2b | 6 |
| GO:0005604 | basement membrane | CC | 0.0672 | 4.66e-05 | 0.000457 | 3.34 | 0.000335 | Hmcn1/Col4a6/Lamc3/Npnt/Tgfbi/Frem1/Lama4/Nid1 | 8 |
| GO:0045121 | membrane raft | CC | 0.0356 | 0.000106 | 0.000948 | 3.02 | 0.000696 | Angpt1/Kcnd2/Dcc/Kcnd3/Rgs16/Kdr/Ednrb/Cdh2/Tfpi/Nos3/Gnai1/Has2/Lrp2/Ano1 | 14 |
| GO:0099056 | integral component of presynaptic membrane | CC | 0.114 | 0.000109 | 0.000948 | 3.02 | 0.000696 | Kcnj3/Efnb3/Ptprd/Cdh2/Cacna1e | 5 |
| GO:0098857 | membrane microdomain | CC | 0.0355 | 0.000109 | 0.000948 | 3.02 | 0.000696 | Angpt1/Kcnd2/Dcc/Kcnd3/Rgs16/Kdr/Ednrb/Cdh2/Tfpi/Nos3/Gnai1/Has2/Lrp2/Ano1 | 14 |
| GO:0008076 | voltage-gated potassium channel complex | CC | 0.08 | 0.000162 | 0.00137 | 2.86 | 0.001 | Kcnb2/Kcnj3/Kcnd2/Kcna6/Kcnd3/Kcnq5 | 6 |
| GO:0034706 | sodium channel complex | CC | 0.148 | 0.000192 | 0.00157 | 2.8 | 0.00116 | Grik1/Gria4/Scn4b/Scn1a | 4 |
| GO:0098889 | intrinsic component of presynaptic membrane | CC | 0.098 | 0.000221 | 0.00177 | 2.75 | 0.0013 | Kcnj3/Efnb3/Ptprd/Cdh2/Cacna1e | 5 |
| GO:0043195 | terminal bouton | CC | 0.0667 | 0.000437 | 0.00341 | 2.47 | 0.0025 | Grik1/Gria4/Drd4/Calca/P2rx2/Grin2b | 6 |
| GO:0005790 | smooth endoplasmic reticulum | CC | 0.114 | 0.000534 | 0.00406 | 2.39 | 0.00298 | Map2/Ryr1/Rtn1/Jph4 | 4 |
| GO:0005614 | interstitial matrix | CC | 0.176 | 0.000756 | 0.00547 | 2.26 | 0.00401 | Vit/Col14a1/Igf1 | 3 |
| GO:0043679 | axon terminus | CC | 0.0421 | 0.00112 | 0.0079 | 2.1 | 0.0058 | Grik1/Gria4/Kcna6/Drd4/Calca/P2rx2/Grin2b/Slc8a3 | 8 |
| GO:0016528 | sarcoplasm | CC | 0.0588 | 0.00229 | 0.0155 | 1.81 | 0.0114 | Car4/Ryr1/Sgcd/Jph4/Pygm | 5 |
| GO:0031253 | cell projection membrane | CC | 0.0312 | 0.00266 | 0.0174 | 1.76 | 0.0128 | Gap43/Ptprz1/Car4/Map2/Evc2/Evc/Epb41l3/Cd44/Lrp2/Gabra4 | 10 |
| GO:0031225 | anchored component of membrane | CC | 0.0387 | 0.00362 | 0.022 | 1.66 | 0.0161 | Gpc6/Cntn3/Car4/Cd109/Gpc3/Ntm/Sema7a | 7 |
| GO:0044853 | plasma membrane raft | CC | 0.0441 | 0.00366 | 0.022 | 1.66 | 0.0161 | Kcnd2/Kcnd3/Cdh2/Tfpi/Nos3/Has2 | 6 |
| GO:0097440 | apical dendrite | CC | 0.103 | 0.00369 | 0.022 | 1.66 | 0.0161 | Map2/Grin2b/Osbp2 | 3 |
| GO:0043292 | contractile fiber | CC | 0.0338 | 0.00439 | 0.0252 | 1.6 | 0.0185 | Ryr1/Adra1a/Scn1a/Npnt/Tmod2/Pygm/Grin2b/Mmp2 | 8 |
| GO:0030175 | filopodium | CC | 0.0505 | 0.00443 | 0.0252 | 1.6 | 0.0185 | Gap43/Ptprz1/Map2/Lrrc7/Nlgn1 | 5 |
| GO:0016342 | catenin complex | CC | 0.0968 | 0.00447 | 0.0252 | 1.6 | 0.0185 | Cdh6/Cdh2/Cdh10 | 3 |
| GO:0030426 | growth cone | CC | 0.0345 | 0.00671 | 0.0364 | 1.44 | 0.0267 | Ptprz1/Map2/Fez1/Dcc/Itga4/Tmod2/Lrp2 | 7 |
| GO:0032809 | neuronal cell body membrane | CC | 0.0789 | 0.00793 | 0.042 | 1.38 | 0.0309 | Kcnb2/Kcnd2/Slc6a2 | 3 |
| GO:0016529 | sarcoplasmic reticulum | CC | 0.0526 | 0.00927 | 0.0478 | 1.32 | 0.0351 | Car4/Ryr1/Sgcd/Pygm | 4 |
| GO:0022836 | gated channel activity | MF | 0.0619 | 3.54e-10 | 1.05e-07 | 6.98 | 7.91e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Ano2/P2rx2/Kcnq5/Cacna1e/Grin2b/Ano1/Gabra4 | 20 |
| GO:0005216 | ion channel activity | MF | 0.0525 | 4.24e-10 | 1.05e-07 | 6.98 | 7.91e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Slc24a4/Scn1a/Ano2/Trpm3/P2rx2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1/Gabra4 | 23 |
| GO:0005261 | cation channel activity | MF | 0.0601 | 6.03e-10 | 1.05e-07 | 6.98 | 7.91e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Slc24a4/Scn1a/Trpm3/P2rx2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1 | 20 |
| GO:0050839 | cell adhesion molecule binding | MF | 0.0619 | 9.95e-10 | 1.17e-07 | 6.93 | 8.87e-08 | Ptprz1/Prtg/Lrrc4c/Igf1/Ptpro/Nlgn1/Ptprd/Itga4/Npnt/Kdr/Cdh6/Cdh2/Tgfbi/Nos3/Tenm3/Grin2b/Sema7a/Cxcl12/Cdh10 | 19 |
| GO:0046873 | metal ion transmembrane transporter activity | MF | 0.0523 | 1.13e-09 | 1.17e-07 | 6.93 | 8.87e-08 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Slc1a3/Grik1/Slc5a11/Slc13a5/Kcnd2/Slc6a2/Kcna6/Kcnd3/Ryr1/Scn4b/Slc24a4/Scn1a/Slc12a8/Kcnq5/Cacna1e/Grin2b/Slc9a7/Slc8a3 | 22 |
| GO:0015267 | channel activity | MF | 0.0477 | 2.66e-09 | 1.98e-07 | 6.7 | 1.5e-07 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Slc24a4/Scn1a/Ano2/Trpm3/P2rx2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1/Gabra4 | 23 |
| GO:0022803 | passive transmembrane transporter activity | MF | 0.0477 | 2.66e-09 | 1.98e-07 | 6.7 | 1.5e-07 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Grid1/Grik1/Kcnd2/Gria4/Kcna6/Kcnd3/Ryr1/Scn4b/Slc24a4/Scn1a/Ano2/Trpm3/P2rx2/Kcnq5/Cacna1e/Unc80/Grin2b/Ano1/Gabra4 | 23 |
| GO:0005244 | voltage-gated ion channel activity | MF | 0.0729 | 2.13e-08 | 1.32e-06 | 5.88 | 9.94e-07 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Kcnd2/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Kcnq5/Cacna1e/Grin2b/Ano1 | 14 |
| GO:0022832 | voltage-gated channel activity | MF | 0.0725 | 2.27e-08 | 1.32e-06 | 5.88 | 9.94e-07 | Kcnk3/Kcnb2/Nalcn/Kcnj3/Kcnd2/Kcna6/Kcnd3/Ryr1/Scn4b/Scn1a/Kcnq5/Cacna1e/Grin2b/Ano1 | 14 |
| GO:0015081 | sodium ion transmembrane transporter activity | MF | 0.0759 | 4.74e-07 | 2.25e-05 | 4.65 | 1.7e-05 | Nalcn/Slc1a3/Grik1/Slc5a11/Slc13a5/Slc6a2/Scn4b/Slc24a4/Scn1a/Slc9a7/Slc8a3 | 11 |
| GO:0015079 | potassium ion transmembrane transporter activity | MF | 0.0701 | 1.05e-06 | 4.55e-05 | 4.34 | 3.43e-05 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcnd2/Kcna6/Kcnd3/Slc24a4/Slc12a8/Kcnq5/Slc9a7 | 11 |
| GO:0022843 | voltage-gated cation channel activity | MF | 0.0704 | 3.1e-06 | 0.000124 | 3.91 | 9.4e-05 | Kcnk3/Kcnb2/Kcnj3/Kcnd2/Kcna6/Kcnd3/Ryr1/Kcnq5/Cacna1e/Grin2b | 10 |
| GO:0004970 | ionotropic glutamate receptor activity | MF | 0.222 | 3.42e-05 | 0.00127 | 2.89 | 0.000963 | Grid1/Grik1/Gria4/Grin2b | 4 |
| GO:0001664 | G protein-coupled receptor binding | MF | 0.0406 | 4.39e-05 | 0.00143 | 2.84 | 0.00108 | Cxcl15/Ndp/Wnt6/Wnt11/Ptch2/Gprasp2/Tub/Ednrb/Calca/Gnai1/Edn3/Grin2b/Cxcl12 | 13 |
| GO:0005249 | voltage-gated potassium channel activity | MF | 0.0795 | 4.39e-05 | 0.00143 | 2.84 | 0.00108 | Kcnk3/Kcnb2/Kcnj3/Kcnd2/Kcna6/Kcnd3/Kcnq5 | 7 |
| GO:0005267 | potassium channel activity | MF | 0.0661 | 4.78e-05 | 0.00146 | 2.83 | 0.00111 | Kcnk3/Kcnb2/Kcnj3/Grik1/Kcnd2/Kcna6/Kcnd3/Kcnq5 | 8 |
| GO:0005230 | extracellular ligand-gated ion channel activity | MF | 0.087 | 9.45e-05 | 0.00246 | 2.61 | 0.00186 | Grid1/Grik1/Gria4/P2rx2/Grin2b/Gabra4 | 6 |
| GO:0045296 | cadherin binding | MF | 0.087 | 9.45e-05 | 0.00246 | 2.61 | 0.00186 | Ptpro/Kdr/Cdh6/Cdh2/Nos3/Cdh10 | 6 |
| GO:0098960 | postsynaptic neurotransmitter receptor activity | MF | 0.087 | 9.45e-05 | 0.00246 | 2.61 | 0.00186 | Grid1/Grik1/Gria4/Drd4/Grin2b/Gabra4 | 6 |
| GO:0015276 | ligand-gated ion channel activity | MF | 0.0593 | 0.000103 | 0.00257 | 2.59 | 0.00194 | Kcnj3/Grid1/Grik1/Gria4/Ryr1/P2rx2/Grin2b/Gabra4 | 8 |
| GO:0019842 | vitamin binding | MF | 0.0588 | 0.000109 | 0.00258 | 2.59 | 0.00195 | Stra6/Abat/P3h3/Pygm/P4htm/Rbp7/Cbs/Sptlc3 | 8 |
| GO:0022834 | ligand-gated channel activity | MF | 0.0584 | 0.000115 | 0.0026 | 2.59 | 0.00196 | Kcnj3/Grid1/Grik1/Gria4/Ryr1/P2rx2/Grin2b/Gabra4 | 8 |
| GO:0008509 | anion transmembrane transporter activity | MF | 0.0383 | 0.000148 | 0.00321 | 2.49 | 0.00243 | Slc1a3/Grik1/Slc13a5/Slc6a2/Slc2a3/Slc24a4/Slc12a8/Ano2/Slc9a7/Ano1/Slc7a2/Gabra4 | 12 |
| GO:0008066 | glutamate receptor activity | MF | 0.154 | 0.000156 | 0.00326 | 2.49 | 0.00246 | Grid1/Grik1/Gria4/Grin2b | 4 |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | MF | 0.0441 | 0.000173 | 0.00347 | 2.46 | 0.00263 | Cyp3a59/Cyp1b1/Cyp3a25/Fads2/Cyp7b1/P3h3/Tet1/P4htm/Nos3/Fads1 | 10 |
| GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential | MF | 0.0962 | 0.000228 | 0.0044 | 2.36 | 0.00333 | Grid1/Grik1/Gria4/Grin2b/Gabra4 | 5 |
| GO:0005178 | integrin binding | MF | 0.0516 | 0.000268 | 0.00498 | 2.3 | 0.00376 | Ptprz1/Igf1/Itga4/Npnt/Kdr/Tgfbi/Sema7a/Cxcl12 | 8 |
| GO:0043177 | organic acid binding | MF | 0.0513 | 0.00028 | 0.00502 | 2.3 | 0.00379 | Slc1a3/Grik1/Ddah1/P3h3/P4htm/Nos3/P2rx2/Grin2b | 8 |
| GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential | MF | 0.0909 | 0.000298 | 0.00517 | 2.29 | 0.00391 | Grid1/Grik1/Gria4/Grin2b/Gabra4 | 5 |
| GO:0101020 | estrogen 16-alpha-hydroxylase activity | MF | 0.231 | 0.000316 | 0.0053 | 2.28 | 0.00401 | Cyp3a59/Cyp1b1/Cyp3a25 | 3 |
| GO:0022824 | transmitter-gated ion channel activity | MF | 0.0877 | 0.000352 | 0.00556 | 2.26 | 0.0042 | Grid1/Grik1/Gria4/Grin2b/Gabra4 | 5 |
| GO:0022835 | transmitter-gated channel activity | MF | 0.0877 | 0.000352 | 0.00556 | 2.26 | 0.0042 | Grid1/Grik1/Gria4/Grin2b/Gabra4 | 5 |
| GO:0016595 | glutamate binding | MF | 0.214 | 0.000399 | 0.00611 | 2.21 | 0.00461 | Slc1a3/Grik1/Grin2b | 3 |
| GO:0030020 | extracellular matrix structural constituent conferring tensile strength | MF | 0.105 | 0.000699 | 0.0104 | 1.98 | 0.00786 | Col11a1/Col14a1/Col4a6/Col9a2 | 4 |
| GO:0099094 | ligand-gated cation channel activity | MF | 0.0583 | 0.000834 | 0.0121 | 1.92 | 0.00912 | Kcnj3/Grik1/Gria4/Ryr1/P2rx2/Grin2b | 6 |
| GO:0016597 | amino acid binding | MF | 0.0714 | 0.00091 | 0.0128 | 1.89 | 0.00967 | Slc1a3/Grik1/Ddah1/Nos3/Grin2b | 5 |
| GO:0005539 | glycosaminoglycan binding | MF | 0.0386 | 0.000933 | 0.0128 | 1.89 | 0.00967 | Apob/Col11a1/Vit/Sema5a/Mdk/Spock2/Cd44/Nell1/Serpine2 | 9 |
| GO:0004222 | metalloendopeptidase activity | MF | 0.0566 | 0.000969 | 0.0129 | 1.89 | 0.00978 | Mmp16/Adamts20/Adamts16/Adam33/Adamts19/Mmp2 | 6 |
| GO:0030594 | neurotransmitter receptor activity | MF | 0.0545 | 0.00117 | 0.0149 | 1.83 | 0.0113 | Grid1/Grik1/Gria4/Drd4/Grin2b/Gabra4 | 6 |
| GO:0005272 | sodium channel activity | MF | 0.0909 | 0.00122 | 0.0152 | 1.82 | 0.0115 | Nalcn/Grik1/Scn4b/Scn1a | 4 |
| GO:0008373 | sialyltransferase activity | MF | 0.143 | 0.00138 | 0.0167 | 1.78 | 0.0126 | St6gal2/St8sia2/St8sia4 | 3 |
| GO:0030215 | semaphorin receptor binding | MF | 0.136 | 0.00158 | 0.0187 | 1.73 | 0.0142 | Sema5a/Sema5b/Sema7a | 3 |
| GO:0015491 | cation:cation antiporter activity | MF | 0.125 | 0.00205 | 0.0237 | 1.62 | 0.0179 | Slc24a4/Slc9a7/Slc8a3 | 3 |
| GO:0030170 | pyridoxal phosphate binding | MF | 0.0769 | 0.00228 | 0.0255 | 1.59 | 0.0193 | Abat/Pygm/Cbs/Sptlc3 | 4 |
| GO:0005248 | voltage-gated sodium channel activity | MF | 0.12 | 0.00231 | 0.0255 | 1.59 | 0.0193 | Nalcn/Scn4b/Scn1a | 3 |
| GO:0005506 | iron ion binding | MF | 0.0412 | 0.00238 | 0.0255 | 1.59 | 0.0193 | Cyp3a59/Cyp1b1/Cyp3a25/Cyp7b1/P3h3/Tet1/P4htm | 7 |
| GO:0070279 | vitamin B6 binding | MF | 0.0755 | 0.00245 | 0.0255 | 1.59 | 0.0193 | Abat/Pygm/Cbs/Sptlc3 | 4 |
| GO:0015294 | solute:cation symporter activity | MF | 0.0526 | 0.00351 | 0.0331 | 1.48 | 0.025 | Slc1a3/Slc5a11/Slc13a5/Slc6a2/Slc12a8 | 5 |
| GO:0015298 | solute:cation antiporter activity | MF | 0.103 | 0.00356 | 0.0331 | 1.48 | 0.025 | Slc24a4/Slc9a7/Slc8a3 | 3 |
| GO:0017147 | Wnt-protein binding | MF | 0.103 | 0.00356 | 0.0331 | 1.48 | 0.025 | Sfrp2/Apcdd1/Ptpro | 3 |
| GO:0045499 | chemorepellent activity | MF | 0.103 | 0.00356 | 0.0331 | 1.48 | 0.025 | Sema5a/Sema5b/Sema7a | 3 |
| GO:0050750 | low-density lipoprotein particle receptor binding | MF | 0.103 | 0.00356 | 0.0331 | 1.48 | 0.025 | Apob/Sacs/Lrp2 | 3 |
| GO:0031406 | carboxylic acid binding | MF | 0.038 | 0.00368 | 0.0337 | 1.47 | 0.0255 | Slc1a3/Grik1/P3h3/P4htm/Rbp7/Nos3/Grin2b | 7 |
| GO:0008237 | metallopeptidase activity | MF | 0.0378 | 0.0038 | 0.0341 | 1.47 | 0.0258 | Cpxm2/Mmp16/Adamts20/Adamts16/Adam33/Adamts19/Mmp2 | 7 |
| GO:0005251 | delayed rectifier potassium channel activity | MF | 0.1 | 0.00392 | 0.0346 | 1.46 | 0.0262 | Kcnb2/Kcna6/Kcnq5 | 3 |
| GO:0022853 | active ion transmembrane transporter activity | MF | 0.0336 | 0.00415 | 0.0361 | 1.44 | 0.0273 | Slc1a3/Slc5a11/Slc13a5/Slc6a2/Slc24a4/Slc12a8/Slc9a7/Slc8a3 | 8 |
| GO:0015293 | symporter activity | MF | 0.042 | 0.00438 | 0.0374 | 1.43 | 0.0283 | Slc1a3/Slc5a11/Slc13a5/Slc6a2/Slc24a4/Slc12a8 | 6 |
| GO:0016836 | hydro-lyase activity | MF | 0.0635 | 0.00458 | 0.0378 | 1.42 | 0.0286 | Cyp1b1/Car4/Car8/Cbs | 4 |
| GO:0015291 | secondary active transmembrane transporter activity | MF | 0.0331 | 0.00459 | 0.0378 | 1.42 | 0.0286 | Slc1a3/Slc5a11/Slc13a5/Slc6a2/Slc24a4/Slc12a8/Slc9a7/Slc8a3 | 8 |
| GO:0005516 | calmodulin binding | MF | 0.0365 | 0.00464 | 0.0378 | 1.42 | 0.0286 | Gap43/Map2/Ryr1/Slc24a4/Nos3/Kcnq5/Slc8a3 | 7 |
| GO:0015368 | calcium:cation antiporter activity | MF | 0.2 | 0.0048 | 0.0379 | 1.42 | 0.0287 | Slc24a4/Slc8a3 | 2 |
| GO:0070643 | vitamin D 25-hydroxylase activity | MF | 0.2 | 0.0048 | 0.0379 | 1.42 | 0.0287 | Cyp3a59/Cyp3a25 | 2 |
| GO:0070325 | lipoprotein particle receptor binding | MF | 0.0909 | 0.00515 | 0.0396 | 1.4 | 0.03 | Apob/Sacs/Lrp2 | 3 |
| GO:0008083 | growth factor activity | MF | 0.0405 | 0.00517 | 0.0396 | 1.4 | 0.03 | Hgf/Mdk/Igf1/Areg/Cxcl12/Gdf7 | 6 |
| GO:0015370 | solute:sodium symporter activity | MF | 0.058 | 0.00633 | 0.0478 | 1.32 | 0.0361 | Slc1a3/Slc5a11/Slc13a5/Slc6a2 | 4 |
| GO:0005231 | excitatory extracellular ligand-gated ion channel activity | MF | 0.0833 | 0.00658 | 0.049 | 1.31 | 0.037 | Grik1/P2rx2/Grin2b | 3 |
| GO:0015108 | chloride transmembrane transporter activity | MF | 0.045 | 0.00677 | 0.0497 | 1.3 | 0.0376 | Slc6a2/Slc12a8/Ano2/Ano1/Gabra4 | 5 |
Visualisation
Biological Process
bp_dot[[3]]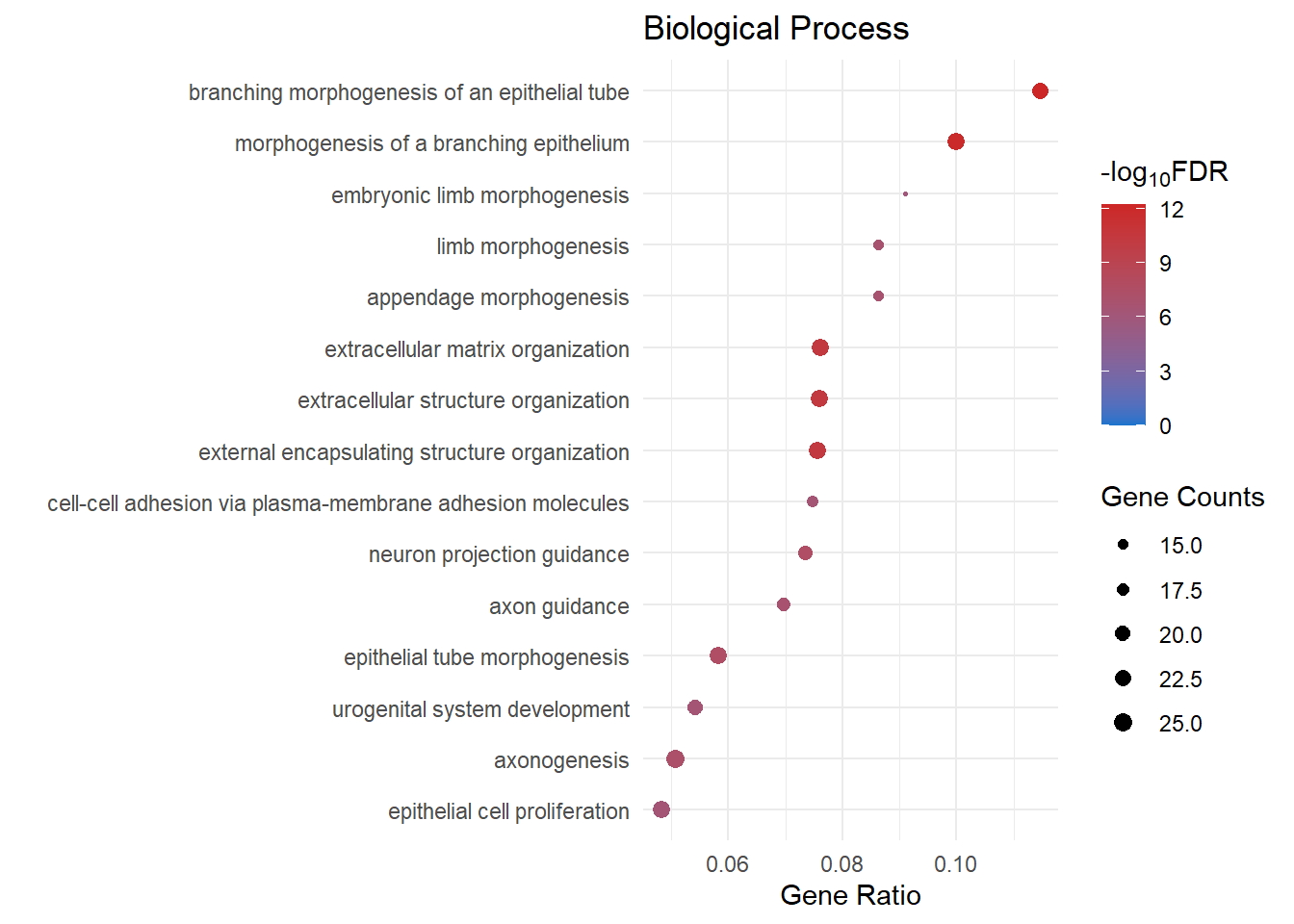
Molecular Function
mf_dot[[3]]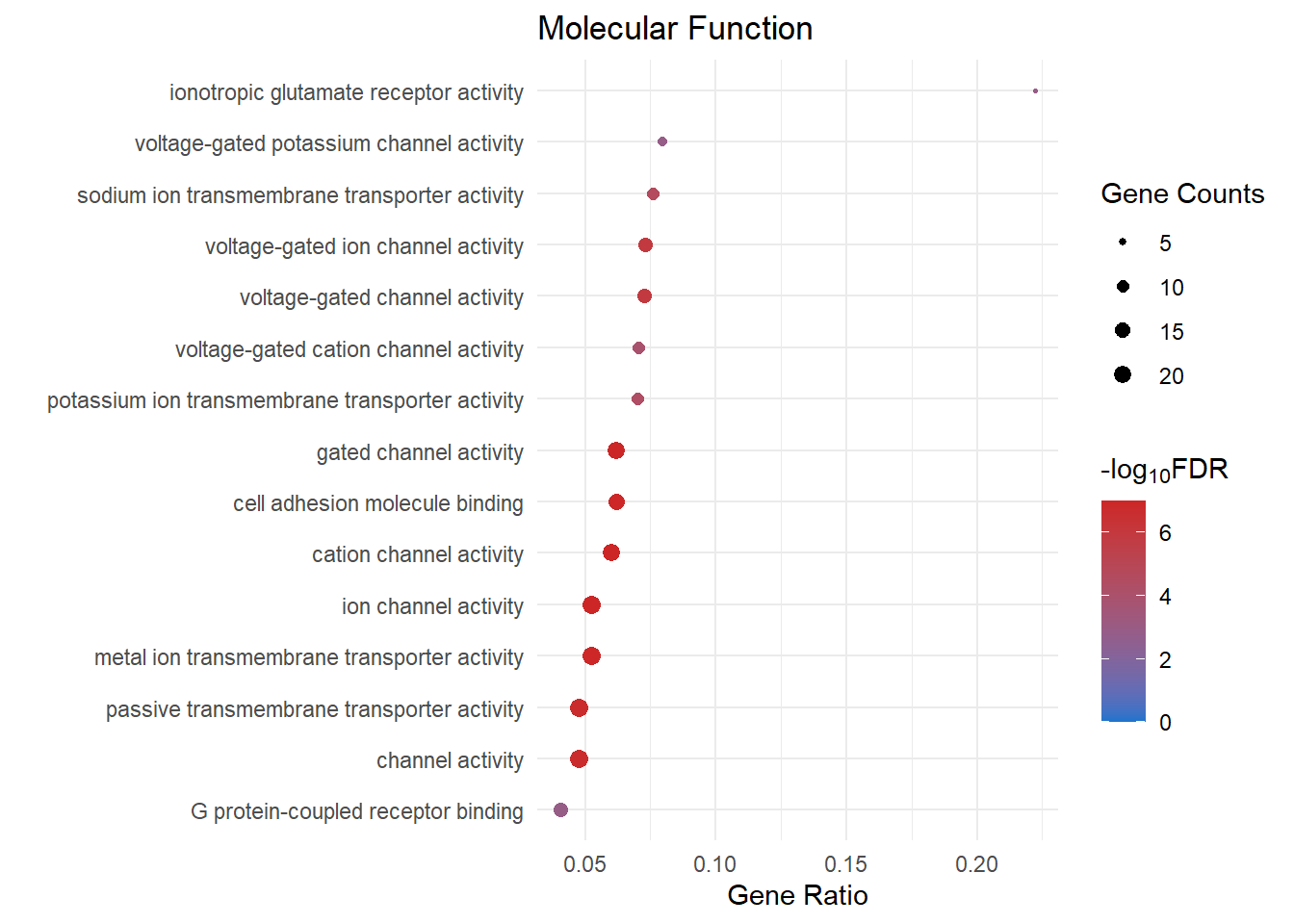
Cellular Components
cc_dot[[3]]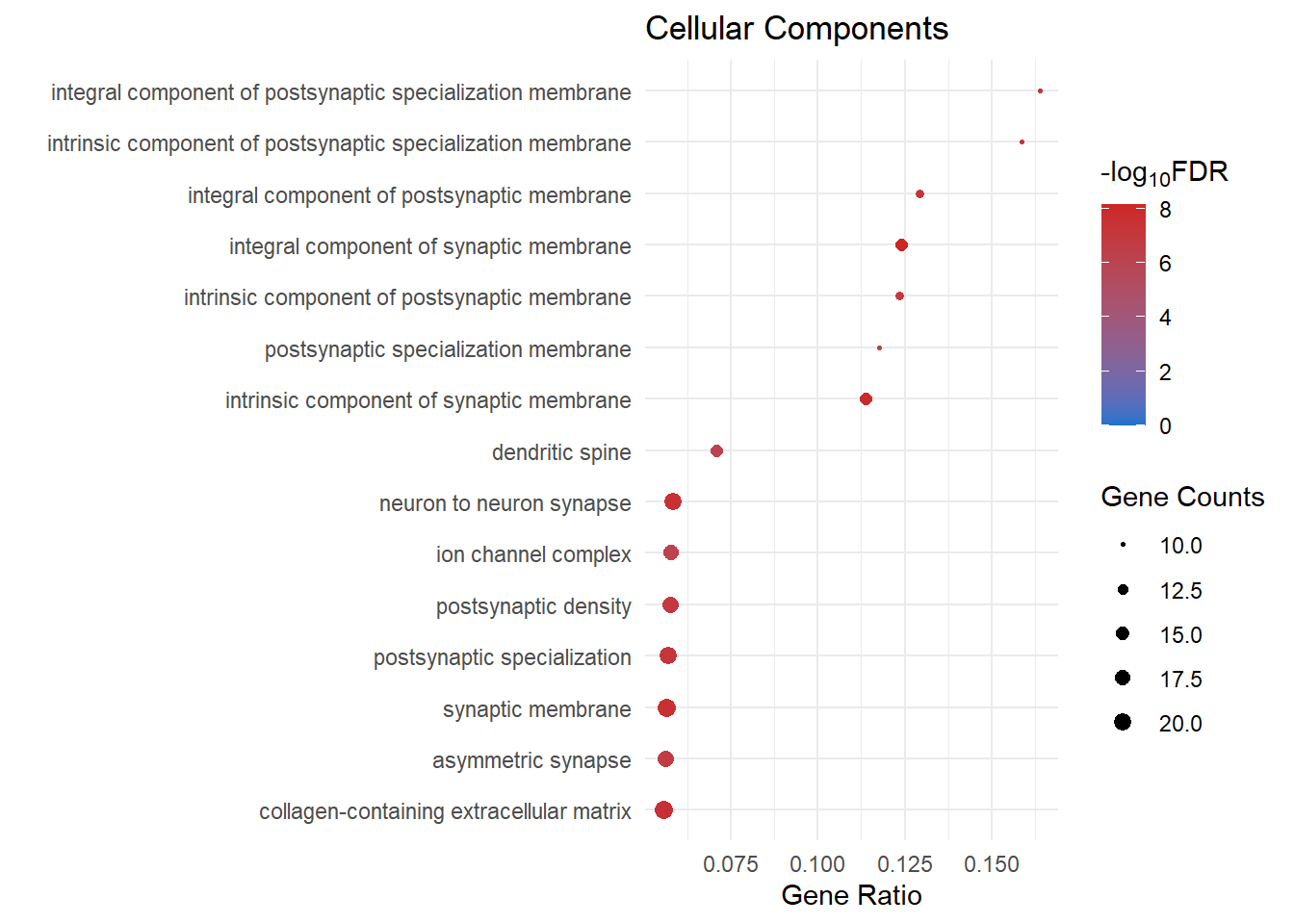
Upset
upset[[3]]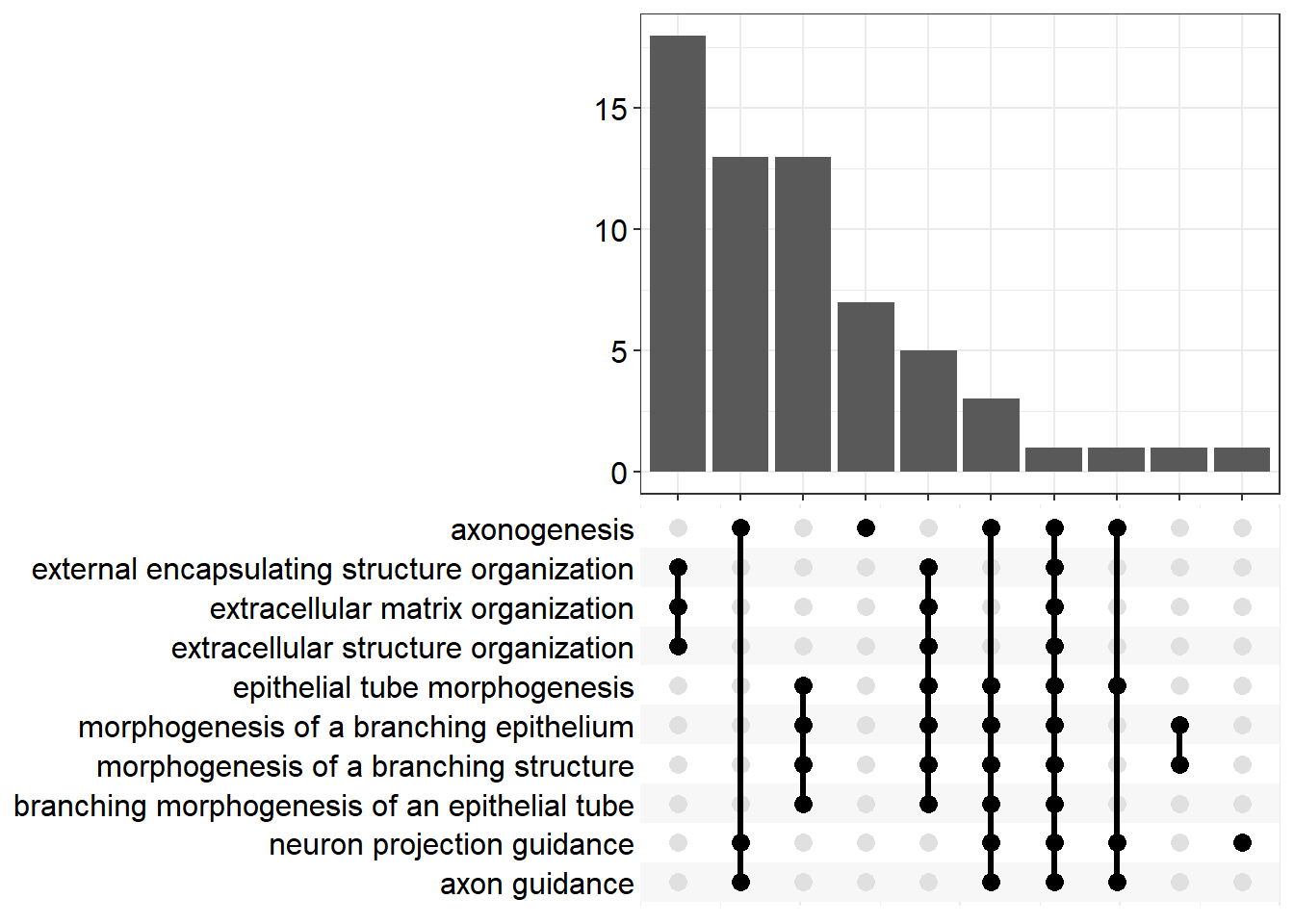
Export Data
# save to excel
writexl::write_xlsx(x = enrichGO_sig, here::here("3_output/enrichGO_sig.xlsx"))
saveRDS(object = enrichGO_sig,file = here::here("0_data/RDS_objects/enrichGO_sig.rds"))
saveRDS(object = enrichGO,file = here::here("0_data/RDS_objects/enrichGO.rds"))
sessionInfo()R version 4.2.1 (2022-06-23 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_Australia.utf8 LC_CTYPE=English_Australia.utf8
[3] LC_MONETARY=English_Australia.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Australia.utf8
attached base packages:
[1] stats4 grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] enrichplot_1.16.2 org.Mm.eg.db_3.15.0 AnnotationDbi_1.58.0
[4] IRanges_2.30.1 S4Vectors_0.34.0 Biobase_2.56.0
[7] BiocGenerics_0.42.0 clusterProfiler_4.4.4 Glimma_2.6.0
[10] edgeR_3.38.4 limma_3.52.4 ggrepel_0.9.1
[13] ggbiplot_0.55 scales_1.2.1 plyr_1.8.7
[16] gridExtra_2.3 kableExtra_1.3.4 forcats_0.5.2
[19] stringr_1.4.1 purrr_0.3.5 tidyr_1.2.1
[22] ggplot2_3.3.6 tidyverse_1.3.2 reshape2_1.4.4
[25] tibble_3.1.8 readr_2.1.3 magrittr_2.0.3
[28] dplyr_1.0.10
loaded via a namespace (and not attached):
[1] utf8_1.2.2 tidyselect_1.2.0
[3] RSQLite_2.2.18 htmlwidgets_1.5.4
[5] BiocParallel_1.30.3 scatterpie_0.1.8
[7] munsell_0.5.0 ragg_1.2.3
[9] codetools_0.2-18 withr_2.5.0
[11] colorspace_2.0-3 GOSemSim_2.22.0
[13] highr_0.9 knitr_1.40
[15] rstudioapi_0.14 DOSE_3.22.1
[17] labeling_0.4.2 MatrixGenerics_1.8.1
[19] git2r_0.30.1 GenomeInfoDbData_1.2.8
[21] polyclip_1.10-0 bit64_4.0.5
[23] farver_2.1.1 rprojroot_2.0.3
[25] downloader_0.4 vctrs_0.4.2
[27] treeio_1.20.2 generics_0.1.3
[29] xfun_0.33 R6_2.5.1
[31] GenomeInfoDb_1.32.4 graphlayouts_0.8.2
[33] locfit_1.5-9.6 bitops_1.0-7
[35] cachem_1.0.6 fgsea_1.22.0
[37] gridGraphics_0.5-1 DelayedArray_0.22.0
[39] assertthat_0.2.1 promises_1.2.0.1
[41] ggraph_2.1.0 googlesheets4_1.0.1
[43] gtable_0.3.1 tidygraph_1.2.2
[45] workflowr_1.7.0 rlang_1.0.6
[47] genefilter_1.78.0 systemfonts_1.0.4
[49] splines_4.2.1 lazyeval_0.2.2
[51] gargle_1.2.1 broom_1.0.1
[53] yaml_2.3.5 modelr_0.1.9
[55] backports_1.4.1 httpuv_1.6.6
[57] qvalue_2.28.0 tools_4.2.1
[59] ggplotify_0.1.0 ellipsis_0.3.2
[61] jquerylib_0.1.4 RColorBrewer_1.1-3
[63] Rcpp_1.0.9 zlibbioc_1.42.0
[65] RCurl_1.98-1.9 viridis_0.6.2
[67] SummarizedExperiment_1.26.1 haven_2.5.1
[69] fs_1.5.2 here_1.0.1
[71] data.table_1.14.2 DO.db_2.9
[73] reprex_2.0.2 googledrive_2.0.0
[75] whisker_0.4 matrixStats_0.62.0
[77] hms_1.1.2 patchwork_1.1.2
[79] evaluate_0.17 xtable_1.8-4
[81] XML_3.99-0.11 readxl_1.4.1
[83] ggupset_0.3.0 compiler_4.2.1
[85] writexl_1.4.0 crayon_1.5.2
[87] shadowtext_0.1.2 htmltools_0.5.3
[89] ggfun_0.0.7 later_1.3.0
[91] tzdb_0.3.0 geneplotter_1.74.0
[93] aplot_0.1.8 lubridate_1.8.0
[95] DBI_1.1.3 tweenr_2.0.2
[97] dbplyr_2.2.1 MASS_7.3-57
[99] Matrix_1.5-1 cli_3.4.1
[101] parallel_4.2.1 igraph_1.3.5
[103] GenomicRanges_1.48.0 pkgconfig_2.0.3
[105] xml2_1.3.3 ggtree_3.4.4
[107] svglite_2.1.0 annotate_1.74.0
[109] bslib_0.4.0 webshot_0.5.4
[111] XVector_0.36.0 rvest_1.0.3
[113] yulab.utils_0.0.5 digest_0.6.29
[115] Biostrings_2.64.1 rmarkdown_2.17
[117] cellranger_1.1.0 fastmatch_1.1-3
[119] tidytree_0.4.1 lifecycle_1.0.3
[121] nlme_3.1-157 jsonlite_1.8.2
[123] viridisLite_0.4.1 fansi_1.0.3
[125] pillar_1.8.1 lattice_0.20-45
[127] KEGGREST_1.36.3 fastmap_1.1.0
[129] httr_1.4.4 survival_3.3-1
[131] GO.db_3.15.0 glue_1.6.2
[133] png_0.1-7 bit_4.0.4
[135] ggforce_0.4.1 stringi_1.7.8
[137] sass_0.4.2 blob_1.2.3
[139] textshaping_0.3.6 DESeq2_1.36.0
[141] memoise_2.0.1 ape_5.6-2