Standardized Precipitation Index (SPI)
Last updated: 2021-03-24
Checks: 5 1
Knit directory:
thesis/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210321) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness,
e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the results is
critical for reproducibility. To start using Git, open the Terminal and type
git init in your project directory.
This project is not being versioned with Git. To obtain the full
reproducibility benefits of using workflowr, please see
?wflow_start.
1 Loading the data
files = list.files("../data/vector/extraction/", pattern = "_SPI", full.names = T)
data = lapply(files, function(x){
filename = str_split(basename(x), "_")[[1]]
if(length(filename) == 3){
unit = filename[1]
buffer = as.numeric(filename[2])
var = str_remove(filename[3], ".gpkg")
} else {
unit = filename[1]
buffer = 0
var = str_remove(filename[2], ".gpkg")
}
layers = ogrListLayers(x)
layers = layers[grep("attr_", layers)]
data = do.call(cbind, lapply(layers, function(l){
tmp = st_read(x, layer = l, quiet = TRUE)
lnames = c("SPI1", "SPI3", "SPI6", "SPI12")
names(tmp) = paste(l, lnames, sep = "-")
tmp
}))
data$id = 1:nrow(data)
data %>%
as_tibble () %>%
gather("tmp", "value", -id) %>%
separate(tmp, into = c("time","SPI"), sep = "-SPI") %>%
mutate(SPI = as.factor(SPI),
time = str_remove(time, "attr_"),
unit = unit, buffer = buffer, var = var)
})
data = do.call(rbind, data)
str(data)tibble [7,142,400 × 7] (S3: tbl_df/tbl/data.frame)
$ id : int [1:7142400] 1 2 3 4 5 6 7 8 9 10 ...
$ time : chr [1:7142400] "2000-01" "2000-01" "2000-01" "2000-01" ...
$ SPI : Factor w/ 4 levels "1","12","3","6": 1 1 1 1 1 1 1 1 1 1 ...
$ value : num [1:7142400] 0.907 0.946 0.968 0.237 0.323 ...
$ unit : chr [1:7142400] "basins" "basins" "basins" "basins" ...
$ buffer: num [1:7142400] 100 100 100 100 100 100 100 100 100 100 ...
$ var : chr [1:7142400] "SPI" "SPI" "SPI" "SPI" ...2 Missing Values
data %>%
group_by(unit, buffer) %>%
summarise(N = n(), isna = sum(is.na(value)), isnotna = sum(!is.na(value)), perc = sum(is.na(value)) / n() * 100)# A tibble: 8 x 6
# Groups: unit [2]
unit buffer N isna isnotna perc
<chr> <dbl> <int> <int> <int> <dbl>
1 basins 0 972480 23966 948514 2.46
2 basins 50 972480 18275 954205 1.88
3 basins 100 972480 18234 954246 1.88
4 basins 200 972480 18234 954246 1.88
5 states 0 813120 19167 793953 2.36
6 states 50 813120 15249 797871 1.88
7 states 100 813120 15247 797873 1.88
8 states 200 813120 15246 797874 1.883 Time Series
data %>%
filter(buffer==0, unit == "states") %>%
mutate(time = as.Date(paste0(time, "-01"))) %>%
group_by(time, unit, SPI) %>%
summarise(value = median(value, na.rm = T)) %>%
mutate(sign = if_else(value>0, "pos", "neg"),
cap = paste0("Scale of ", SPI, " months"),
scale = as.numeric(as.character(SPI))) %>%
mutate(cap = fct_reorder(cap, scale, min)) %>%
ggplot(aes(x=time, y=value, fill = sign)) +
geom_bar(stat = "identity", show.legend = FALSE) +
#scale_x_date(date_breaks = "year", date_labels = "%Y") +
#scale_y_continuous(breaks = seq(-2, 2, 1)) +
scale_fill_manual(values = c("#99000d", "#034e7b")) +
labs(y = "SPI", x = "") +
theme_classic() +
facet_wrap(~cap, nrow = 4)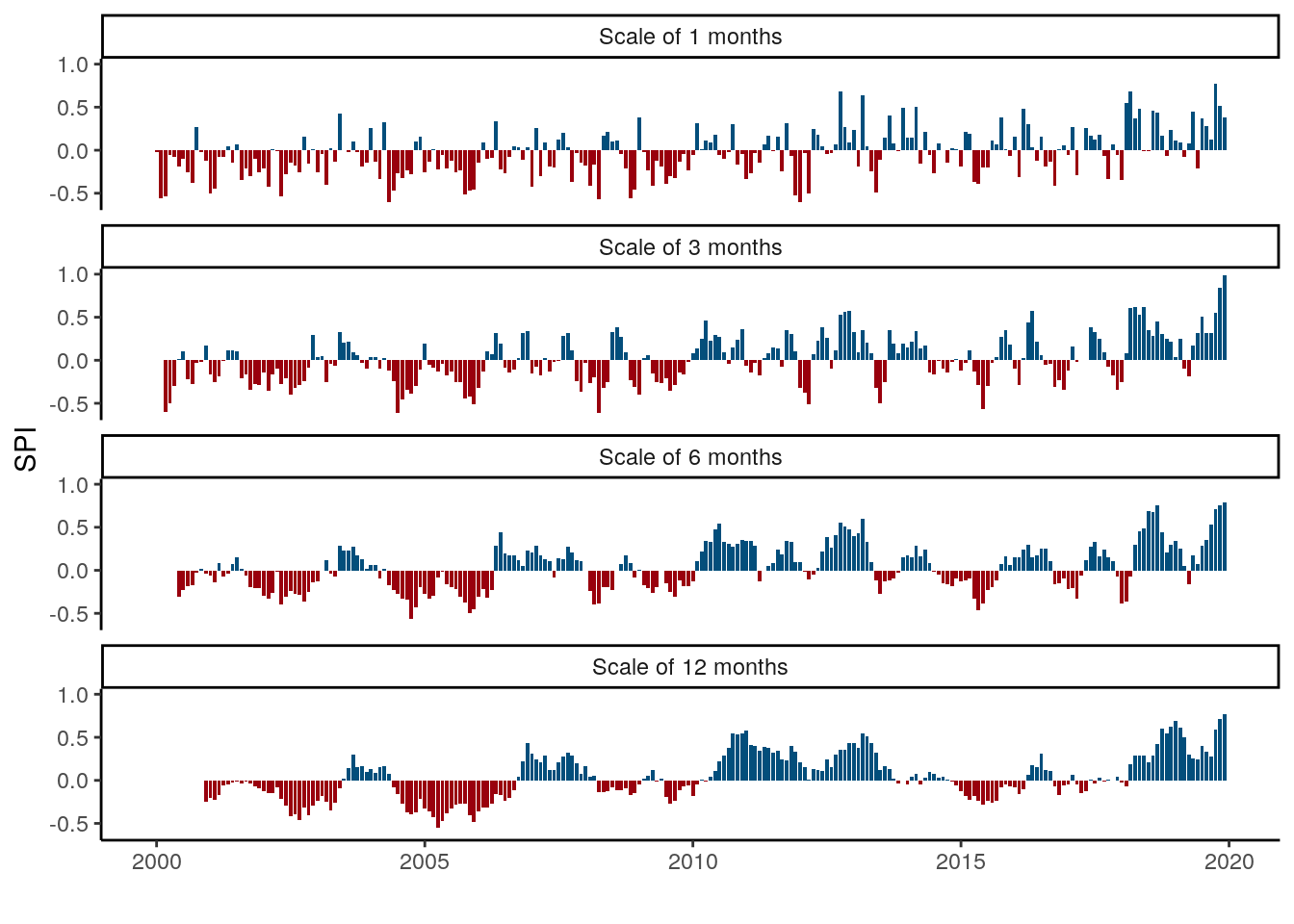
data %>%
filter(buffer==0, unit == "basins") %>%
mutate(time = as.Date(paste0(time, "-01"))) %>%
group_by(time, unit, SPI) %>%
summarise(value = median(value, na.rm = T)) %>%
mutate(sign = if_else(value>0, "pos", "neg"),
cap = paste0("Scale of ", SPI, " months"),
scale = as.numeric(as.character(SPI))) %>%
mutate(cap = fct_reorder(cap, scale, min)) %>%
ggplot(aes(x=time, y=value, fill = sign)) +
geom_bar(stat = "identity", show.legend = FALSE) +
#scale_x_date(date_breaks = "year", date_labels = "%Y") +
#scale_y_continuous(breaks = seq(-2, 2, 1)) +
scale_fill_manual(values = c("#99000d", "#034e7b")) +
labs(y = "SPI", x = "") +
theme_classic() +
facet_wrap(~cap, nrow = 4)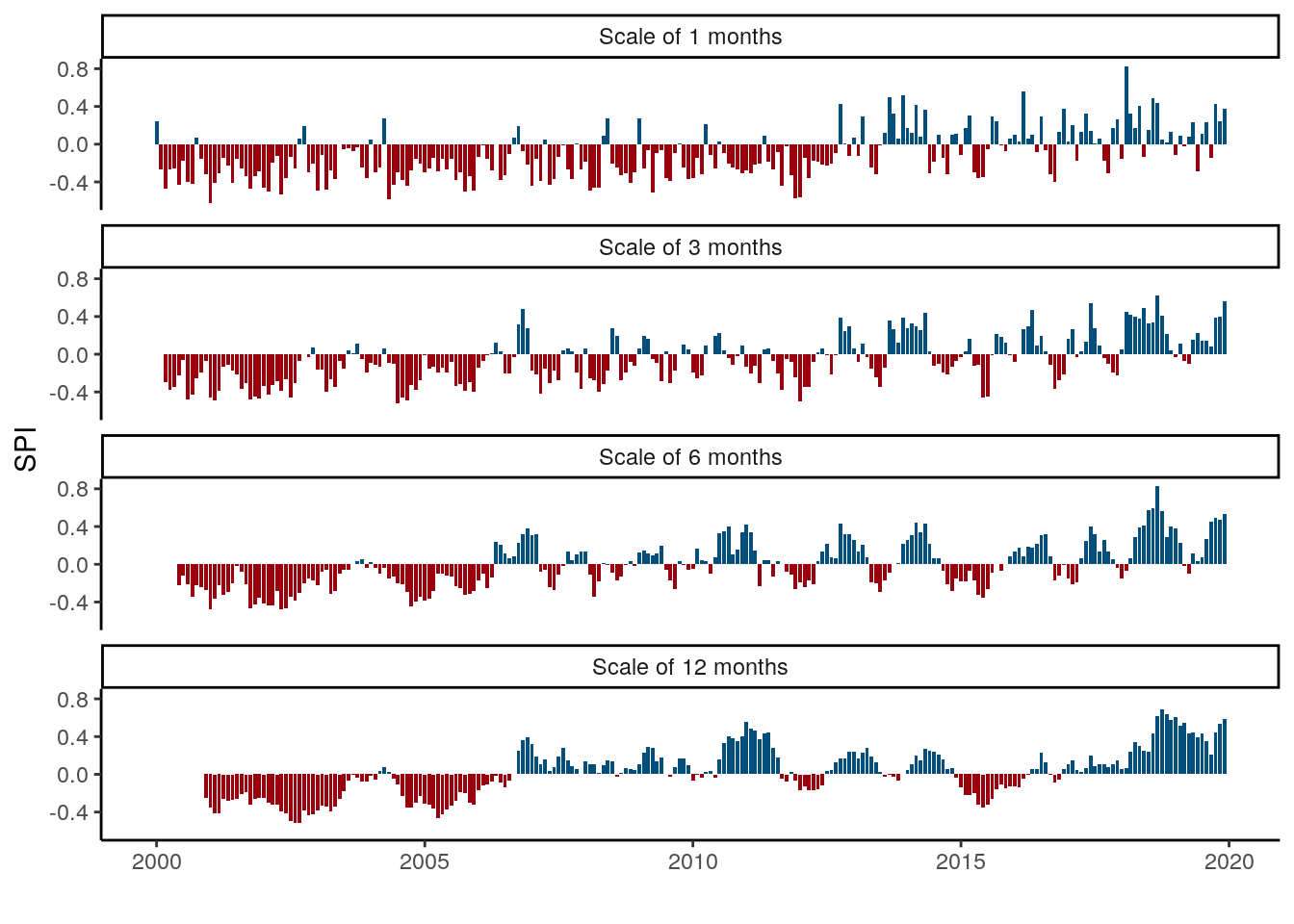
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Debian GNU/Linux 10 (buster)
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.3.5.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.7.9.2 rgdal_1.5-18 countrycode_1.2.0 welchADF_0.3.2
[5] rstatix_0.6.0 ggpubr_0.4.0 scales_1.1.1 RColorBrewer_1.1-2
[9] latex2exp_0.4.0 cubelyr_1.0.0 gridExtra_2.3 ggtext_0.1.1
[13] magrittr_2.0.1 tmap_3.2 sf_0.9-7 raster_3.4-5
[17] sp_1.4-4 forcats_0.5.0 stringr_1.4.0 purrr_0.3.4
[21] readr_1.4.0 tidyr_1.1.2 tibble_3.0.6 tidyverse_1.3.0
[25] huwiwidown_0.0.1 kableExtra_1.3.1 knitr_1.31 rmarkdown_2.7.3
[29] bookdown_0.21 ggplot2_3.3.3 dplyr_1.0.2 devtools_2.3.2
[33] usethis_2.0.0
loaded via a namespace (and not attached):
[1] readxl_1.3.1 backports_1.2.0 workflowr_1.6.2
[4] lwgeom_0.2-5 splines_3.6.3 crosstalk_1.1.0.1
[7] leaflet_2.0.3 digest_0.6.27 htmltools_0.5.1.1
[10] fansi_0.4.2 memoise_1.1.0 openxlsx_4.2.3
[13] remotes_2.2.0 modelr_0.1.8 prettyunits_1.1.1
[16] colorspace_2.0-0 rvest_0.3.6 haven_2.3.1
[19] xfun_0.21 leafem_0.1.3 callr_3.5.1
[22] crayon_1.4.0 jsonlite_1.7.2 lme4_1.1-26
[25] glue_1.4.2 stars_0.4-3 gtable_0.3.0
[28] webshot_0.5.2 car_3.0-10 pkgbuild_1.2.0
[31] abind_1.4-5 DBI_1.1.0 Rcpp_1.0.5
[34] viridisLite_0.3.0 gridtext_0.1.4 units_0.6-7
[37] foreign_0.8-71 htmlwidgets_1.5.3 httr_1.4.2
[40] ellipsis_0.3.1 farver_2.0.3 pkgconfig_2.0.3
[43] XML_3.99-0.3 dbplyr_2.0.0 utf8_1.1.4
[46] labeling_0.4.2 tidyselect_1.1.0 rlang_0.4.10
[49] later_1.1.0.1 tmaptools_3.1 munsell_0.5.0
[52] cellranger_1.1.0 tools_3.6.3 cli_2.3.0
[55] generics_0.1.0 broom_0.7.2 evaluate_0.14
[58] yaml_2.2.1 processx_3.4.5 leafsync_0.1.0
[61] fs_1.5.0 zip_2.1.1 nlme_3.1-150
[64] xml2_1.3.2 compiler_3.6.3 rstudioapi_0.13
[67] curl_4.3 png_0.1-7 e1071_1.7-4
[70] testthat_3.0.1 ggsignif_0.6.0 reprex_0.3.0
[73] statmod_1.4.35 stringi_1.5.3 highr_0.8
[76] ps_1.5.0 desc_1.2.0 lattice_0.20-41
[79] Matrix_1.2-18 nloptr_1.2.2.2 classInt_0.4-3
[82] vctrs_0.3.6 pillar_1.4.7 lifecycle_0.2.0
[85] data.table_1.13.2 httpuv_1.5.5 R6_2.5.0
[88] promises_1.1.1 KernSmooth_2.23-18 rio_0.5.16
[91] sessioninfo_1.1.1 codetools_0.2-16 dichromat_2.0-0
[94] boot_1.3-25 MASS_7.3-53 assertthat_0.2.1
[97] pkgload_1.1.0 rprojroot_2.0.2 withr_2.4.1
[100] parallel_3.6.3 hms_1.0.0 grid_3.6.3
[103] minqa_1.2.4 class_7.3-17 carData_3.0-4
[106] git2r_0.27.1 base64enc_0.1-3