Extreme pH Profiles
Pasqualina Vonlanthen, David Stappard & Jens Daniel Müller
12 April, 2024
Last updated: 2024-04-12
Checks: 7 0
Knit directory:
bgc_argo_r_argodata/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9ff080a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/MHWs_categorisation.Rmd
Modified: analysis/_site.yml
Modified: analysis/load_broullon_DIC_TA_clim.Rmd
Modified: code/Workflowr_project_managment.R
Modified: code/start_background_job.R
Modified: code/start_background_job_load.R
Modified: code/start_background_job_partial.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/extreme_pH.Rmd) and HTML
(docs/extreme_pH.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | c076fba | mlarriere | 2024-04-12 | Build site. |
| html | 91f08a6 | mlarriere | 2024-04-07 | Build site. |
| html | db21f55 | mlarriere | 2024-04-06 | Build site. |
| html | c851476 | mlarriere | 2024-04-01 | Build site. |
| Rmd | 3ee6c97 | mlarriere | 2024-04-01 | extreme_pH building test |
| html | f9de50e | ds2n19 | 2024-01-01 | Build site. |
| html | 07d4eb8 | ds2n19 | 2023-12-20 | Build site. |
| html | fa6cf38 | ds2n19 | 2023-12-14 | Build site. |
| Rmd | 64fd104 | ds2n19 | 2023-12-14 | revised coverage analysis and SO focused cluster analysis. |
| html | f110b74 | ds2n19 | 2023-12-13 | Build site. |
| Rmd | fa9795c | ds2n19 | 2023-12-12 | dependencies listed are start of markdown files. |
| Rmd | a434982 | ds2n19 | 2023-12-11 | test run of coverage maps |
| html | e60ebd2 | ds2n19 | 2023-12-07 | Build site. |
| html | 23a24d1 | ds2n19 | 2023-12-05 | Revised extreme identification. Adjusted profiles for cluster analysis based on max anomaly. |
| Rmd | 3cb4b17 | ds2n19 | 2023-12-04 | Cluster under surface extreme. |
| Rmd | fa1083d | ds2n19 | 2023-12-01 | Additional analysis to cluster process. |
| html | cec2a2a | ds2n19 | 2023-11-24 | Build site. |
| Rmd | 3dc557d | ds2n19 | 2023-11-24 | Switched to new profile details. |
| Rmd | 59f5cc4 | ds2n19 | 2023-11-23 | Moved spatiotemporal analysis to use aligned profiles. |
| html | ff31fef | ds2n19 | 2023-11-20 | Build site. |
| Rmd | 44ce113 | ds2n19 | 2023-11-20 | Switched to new profile details. |
| html | 80c16c2 | ds2n19 | 2023-11-15 | Build site. |
| html | 56c8f49 | ds2n19 | 2023-10-20 | Build site. |
| html | 1cd9ec1 | ds2n19 | 2023-10-19 | Build site. |
| Rmd | 6dfb0be | ds2n19 | 2023-10-19 | moved from month by month regression to annual with monthly |
| html | 879821d | ds2n19 | 2023-10-18 | Build site. |
| Rmd | dba28d5 | ds2n19 | 2023-10-18 | Clean up BGC load and re-run coverage and extreme packages. |
| html | 93b4545 | ds2n19 | 2023-10-18 | Build site. |
| html | 2efb8f2 | ds2n19 | 2023-10-17 | Build site. |
| Rmd | a8f4bb0 | ds2n19 | 2023-10-17 | standard range v climatology, season order resolved and count labels to |
| html | a97d9c1 | ds2n19 | 2023-10-17 | Build site. |
| Rmd | 4293339 | ds2n19 | 2023-10-17 | standard range v climatology, season order resolved and count labels to |
| html | 4b55c43 | ds2n19 | 2023-10-12 | Build site. |
| html | 1ae81b3 | ds2n19 | 2023-10-11 | reworked core load process to work initially by year and then finally create consolidated all years files. |
| html | 44f5720 | ds2n19 | 2023-10-09 | manual commit |
| Rmd | ce19a66 | ds2n19 | 2023-10-04 | Revised version of OceanSODA product -v2023 |
| html | 7b3d8c5 | pasqualina-vonlanthendinenna | 2022-08-29 | Build site. |
| html | bdd516d | pasqualina-vonlanthendinenna | 2022-05-23 | Build site. |
| Rmd | b41e65f | pasqualina-vonlanthendinenna | 2022-05-23 | recreate data in bgc_argo_preprocessed_data |
| html | ae0f995 | jens-daniel-mueller | 2022-05-12 | Build site. |
| Rmd | 018f4b4 | jens-daniel-mueller | 2022-05-12 | scaled DIC to 2019 (rather than 2015) |
| html | 71e58d6 | jens-daniel-mueller | 2022-05-12 | Build site. |
| Rmd | 1bdcd6e | jens-daniel-mueller | 2022-05-12 | revised color scale for argo location map |
| html | 4173c20 | jens-daniel-mueller | 2022-05-12 | Build site. |
| Rmd | 78acca9 | jens-daniel-mueller | 2022-05-12 | run with DIC clim scaled to 2016 |
| html | dfe89d7 | jens-daniel-mueller | 2022-05-12 | Build site. |
| html | 710edd4 | jens-daniel-mueller | 2022-05-11 | Build site. |
| Rmd | 2f20a76 | jens-daniel-mueller | 2022-05-11 | rebuild all after subsetting AB profiles and code cleaning |
| html | b917bd0 | jens-daniel-mueller | 2022-05-11 | Build site. |
| Rmd | 86144c6 | jens-daniel-mueller | 2022-05-11 | rerun with flag A and B subset |
| html | ca30beb | pasqualina-vonlanthendinenna | 2022-05-05 | Build site. |
| Rmd | bb146f4 | pasqualina-vonlanthendinenna | 2022-05-05 | updated map colors and plotting |
| html | 4cf88e4 | pasqualina-vonlanthendinenna | 2022-05-05 | Build site. |
| Rmd | 3bde57b | pasqualina-vonlanthendinenna | 2022-05-05 | added argo profile locations |
| html | f46b9da | pasqualina-vonlanthendinenna | 2022-05-05 | Build site. |
| Rmd | ebcc576 | pasqualina-vonlanthendinenna | 2022-05-05 | added argo profile locations |
| html | 708f923 | pasqualina-vonlanthendinenna | 2022-05-04 | Build site. |
| Rmd | d569024 | pasqualina-vonlanthendinenna | 2022-05-04 | added number of profiles to plot |
| html | b923426 | pasqualina-vonlanthendinenna | 2022-05-03 | Build site. |
| Rmd | ab0c45f | pasqualina-vonlanthendinenna | 2022-05-03 | corrected pH profile depth axis |
| html | 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 | Build site. |
| Rmd | a0139c2 | pasqualina-vonlanthendinenna | 2022-05-03 | updated figure aspect for pH profiles |
| html | a107add | pasqualina-vonlanthendinenna | 2022-05-03 | Build site. |
| Rmd | 6c57ac4 | pasqualina-vonlanthendinenna | 2022-05-03 | added pH anomaly profiles |
| html | 6a6e874 | pasqualina-vonlanthendinenna | 2022-04-29 | Build site. |
| html | 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 | Build site. |
| Rmd | 8b582f0 | pasqualina-vonlanthendinenna | 2022-04-29 | added broullon climatology page, argo locations |
| html | e61c08e | pasqualina-vonlanthendinenna | 2022-04-27 | Build site. |
| Rmd | 9664e0e | pasqualina-vonlanthendinenna | 2022-04-27 | added temp data page, changed double extremes |
| html | 10036ed | pasqualina-vonlanthendinenna | 2022-04-26 | Build site. |
| html | c03dd24 | pasqualina-vonlanthendinenna | 2022-04-20 | Build site. |
| html | 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 | Build site. |
| Rmd | d21c526 | pasqualina-vonlanthendinenna | 2022-04-11 | cleaned up code |
| Rmd | f3ca885 | pasqualina-vonlanthendinenna | 2022-04-07 | added OceanSODA-Argo SST comparison |
| html | 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 | Build site. |
| Rmd | 72a65a7 | pasqualina-vonlanthendinenna | 2022-04-05 | added new biomes to extreme pH |
| html | eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 | Build site. |
| Rmd | c4d4031 | pasqualina-vonlanthendinenna | 2022-03-31 | extended OceanSODA to 1995 for extreme detection |
| html | a2271df | pasqualina-vonlanthendinenna | 2022-03-30 | Build site. |
| Rmd | 25d5eed | pasqualina-vonlanthendinenna | 2022-03-30 | updated figure aspects |
| html | 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 | Build site. |
| Rmd | 5432acf | pasqualina-vonlanthendinenna | 2022-03-29 | added january pH climatology |
| html | dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 | Build site. |
| Rmd | b9a42f9 | pasqualina-vonlanthendinenna | 2022-03-29 | added january plots and changed pH anomaly detection to mean |
| html | 6dd0945 | pasqualina-vonlanthendinenna | 2022-03-25 | Build site. |
| Rmd | 5b93849 | pasqualina-vonlanthendinenna | 2022-03-25 | added climatology pages |
| html | e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 | Build site. |
| Rmd | e4d1d1e | pasqualina-vonlanthendinenna | 2022-03-15 | updated to new only flag A data |
| html | c8451b9 | pasqualina-vonlanthendinenna | 2022-03-14 | Build site. |
| html | 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 | Build site. |
| Rmd | f0fde29 | pasqualina-vonlanthendinenna | 2022-03-11 | changed anomaly detection to 1x1 grid with old data |
| html | 520dafe | pasqualina-vonlanthendinenna | 2022-03-08 | Build site. |
| Rmd | b1bb0ec | pasqualina-vonlanthendinenna | 2022-03-08 | subsetted profiles with flag A only for extremes |
| html | 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 | Build site. |
| Rmd | 18dff1b | pasqualina-vonlanthendinenna | 2022-03-08 | subsetted profiles with flag A only for extremes |
| html | 33238fe | pasqualina-vonlanthendinenna | 2022-03-02 | Build site. |
| Rmd | 17af9b5 | pasqualina-vonlanthendinenna | 2022-03-02 | added January 2018 profile |
| html | 97a098b | pasqualina-vonlanthendinenna | 2022-03-02 | Build site. |
| Rmd | 5a073cb | pasqualina-vonlanthendinenna | 2022-03-02 | removed facet wrap |
| html | 9d97f25 | pasqualina-vonlanthendinenna | 2022-03-02 | Build site. |
| Rmd | 9ccabc6 | pasqualina-vonlanthendinenna | 2022-03-02 | removed facet wrap |
| html | e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 | Build site. |
| Rmd | 6ca535c | pasqualina-vonlanthendinenna | 2022-03-01 | updated profiles |
| html | da665ab | pasqualina-vonlanthendinenna | 2022-03-01 | Build site. |
| Rmd | 57ada58 | pasqualina-vonlanthendinenna | 2022-03-01 | updated figure aspects |
| html | 5ef4df2 | pasqualina-vonlanthendinenna | 2022-03-01 | Build site. |
| Rmd | 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 | plotted Atlantic mean seasonal profiles |
| html | 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 | plotted Atlantic mean seasonal profiles |
| Rmd | 73463cc | pasqualina-vonlanthendinenna | 2022-03-01 | changed line thickness for H and L raw profiles |
| html | c4362e5 | pasqualina-vonlanthendinenna | 2022-02-28 | Build site. |
| Rmd | 5b0901d | pasqualina-vonlanthendinenna | 2022-02-28 | corrected dates and titles |
| html | ab29b31 | pasqualina-vonlanthendinenna | 2022-02-28 | Build site. |
| Rmd | 6e27fb1 | pasqualina-vonlanthendinenna | 2022-02-28 | update with eval = false for single profile line thickness |
| html | d299359 | pasqualina-vonlanthendinenna | 2022-02-28 | Build site. |
| Rmd | aad1df4 | pasqualina-vonlanthendinenna | 2022-02-28 | plotted specific profiles |
| html | 21582c5 | pasqualina-vonlanthendinenna | 2022-02-25 | Build site. |
| Rmd | fe0b970 | pasqualina-vonlanthendinenna | 2022-02-25 | plotted line profiles and changed HNL colors |
| html | fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 | Build site. |
| Rmd | 64c2c71 | pasqualina-vonlanthendinenna | 2022-02-25 | plotted line profiles and changed HNL colors |
| html | daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 | Build site. |
| Rmd | 4557a6e | pasqualina-vonlanthendinenna | 2022-02-24 | added st dev for pH profiles |
| html | 08c8d4b | pasqualina-vonlanthendinenna | 2022-02-23 | Build site. |
| Rmd | 7517b78 | pasqualina-vonlanthendinenna | 2022-02-23 | updated regression and merging for extreme_pH |
| html | 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 | Build site. |
| Rmd | 48803ef | pasqualina-vonlanthendinenna | 2022-02-23 | updated regression and merging for extreme_pH |
| html | 905d82f | pasqualina-vonlanthendinenna | 2022-02-15 | Build site. |
| html | 54ea512 | pasqualina-vonlanthendinenna | 2022-02-10 | Build site. |
| html | f2fa56a | pasqualina-vonlanthendinenna | 2022-02-10 | Build site. |
| Rmd | eda8ca8 | pasqualina-vonlanthendinenna | 2022-02-10 | code review |
| html | 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 | Build site. |
| Rmd | ecf2f74 | pasqualina-vonlanthendinenna | 2022-02-03 | corrected surface mean pH |
| html | 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 | Build site. |
| Rmd | dcc269c | pasqualina-vonlanthendinenna | 2022-02-03 | changed figure aspect |
| html | 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 | Build site. |
| Rmd | 3f38f15 | pasqualina-vonlanthendinenna | 2022-02-03 | corrected mean argo surface pH |
| html | d7debab | pasqualina-vonlanthendinenna | 2022-02-02 | Build site. |
| Rmd | a73c7cf | pasqualina-vonlanthendinenna | 2022-02-02 | changed to log scale and mean surface argo ph |
| html | d14b7f1 | pasqualina-vonlanthendinenna | 2022-02-02 | Build site. |
| Rmd | bb15149 | pasqualina-vonlanthendinenna | 2022-02-02 | changed map figure aspect |
| html | 31e4d42 | pasqualina-vonlanthendinenna | 2022-02-02 | Build site. |
| Rmd | 09ab7e9 | pasqualina-vonlanthendinenna | 2022-02-02 | changed map figure aspect |
| html | 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 | Build site. |
| Rmd | ce1bbab | pasqualina-vonlanthendinenna | 2022-02-02 | updated bar charts and argo vs oceansoda ph |
| html | de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 | Build site. |
| Rmd | db007b5 | pasqualina-vonlanthendinenna | 2022-02-01 | updated figure aspect |
| html | 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 | Build site. |
| Rmd | f62e851 | pasqualina-vonlanthendinenna | 2022-02-01 | added flat maps, bar charts and OceanSODA vs argo pH |
| html | 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 | Build site. |
| Rmd | b45a03e | pasqualina-vonlanthendinenna | 2022-02-01 | added sigma maps and log transform depth |
| html | 28c8d17 | jens-daniel-mueller | 2022-01-29 | Build site. |
| Rmd | c0c12d0 | jens-daniel-mueller | 2022-01-28 | code cleaning: basinmask and regression |
| html | cfd734c | jens-daniel-mueller | 2022-01-28 | Build site. |
| Rmd | 5024768 | jens-daniel-mueller | 2022-01-28 | code review: basinmask and regression |
| html | 5635ef2 | pasqualina-vonlanthendinenna | 2022-01-27 | Build site. |
| Rmd | 23dc282 | pasqualina-vonlanthendinenna | 2022-01-27 | failed attempt at updating basinmask and regression |
| html | c44ff0f | pasqualina-vonlanthendinenna | 2022-01-25 | Build site. |
| Rmd | 3851824 | pasqualina-vonlanthendinenna | 2022-01-25 | added basin-mean profiles |
| html | 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 | Build site. |
| Rmd | 825a50a | pasqualina-vonlanthendinenna | 2022-01-25 | added seasonal and biome profiles |
| html | 3ae43e4 | pasqualina-vonlanthendinenna | 2022-01-24 | Build site. |
| Rmd | 3f8e824 | pasqualina-vonlanthendinenna | 2022-01-24 | updated 24/01 |
| html | 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | e72d7ca | pasqualina-vonlanthendinenna | 2022-01-21 | updated linear regression to monthly |
| html | 587755e | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | 7a9209b | pasqualina-vonlanthendinenna | 2022-01-21 | updated threshold calculation 2 |
| html | c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 | Build site. |
| Rmd | 58b3b3b | pasqualina-vonlanthendinenna | 2022-01-21 | updated threshold calculation |
| html | ed3fef2 | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | 3d2f8fc | jens-daniel-mueller | 2022-01-07 | code review |
| html | 486c9c8 | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | e9ad067 | jens-daniel-mueller | 2022-01-07 | code review |
| html | 343689f | pasqualina-vonlanthendinenna | 2022-01-06 | Build site. |
| Rmd | f53cc2d | pasqualina-vonlanthendinenna | 2022-01-06 | updated profile page |
| html | b8a6482 | pasqualina-vonlanthendinenna | 2022-01-03 | Build site. |
| Rmd | 054f8a6 | pasqualina-vonlanthendinenna | 2022-01-03 | added Argo profiles |
Task
Compare depth profiles of normal pH and of extreme pH, as identified in the surface OceanSODA pH data product. BGC pH profiles have already been validate for coverage and aligned to climatology depths.
Dependencies
OceanSODA.rds - bgc preprocessed folder, created by OceanSODA_argo_pH.
pH_bgc_va.rds - bgc preprocessed folder, created by ph_align_climatology.
pH_anomaly_va.rds - bgc preprocessed folder, created by ph_align_climatology.
Outputs
OceanSODA_pH_anomaly_field_01.rds (or _02.rds)
OceanSODA_global_pH_anomaly_field_01.rds (or _02.rds)
theme_set(theme_bw())
HNL_colors <- c("H" = "#b2182b",
"N" = "#636363",
"L" = "#2166ac")
HNL_colors_map <- c('H' = 'red3',
'N' = 'transparent',
'L' = 'blue3')
# opt_min_profile_range
# profiles with profile_range >= opt_min_profile_range will be selected 1 = profiles of at least 614m, 2 = profiles of at least 1225m, 3 = profiles of at least 1600m
opt_min_profile_range = 3
# opt_extreme_determination
# 1 - based on the trend of de-seasonal data - we believe this results in more summer extremes where variation tend to be greater.
# 2 - based on the trend of de-seasonal data by month. grouping is by lat, lon and month.
opt_extreme_determination <- 2Load data
path_argo <- '/nfs/kryo/work/updata/bgc_argo_r_argodata'
path_argo_preprocessed <- paste0(path_argo, "/preprocessed_bgc_data")
path_emlr_utilities <- "/nfs/kryo/work/jenmueller/emlr_cant/utilities/files/"
path_argo <- '/nfs/kryo/work/datasets/ungridded/3d/ocean/floats/bgc_argo'
# /nfs/kryo/work/datasets/ungridded/3d/ocean/floats/bgc_argo/preprocessed_bgc_data
path_argo_preprocessed <- paste0(path_argo, "/preprocessed_bgc_data")# load in new Mayot biomes
nm_biomes <- read_rds(file = paste0(path_argo_preprocessed, "/nm_biomes.rds"))
# WOA 18 basin mask
basinmask <-
read_csv(
paste(path_emlr_utilities,
"basin_mask_WOA18.csv",
sep = ""),
col_types = cols("MLR_basins" = col_character())
)
basinmask <- basinmask %>%
filter(MLR_basins == unique(basinmask$MLR_basins)[1]) %>%
select(-c(MLR_basins, basin))
# OceanSODA
OceanSODA <- read_rds(file = paste0(path_argo_preprocessed, "/OceanSODA.rds"))
OceanSODA <- OceanSODA %>%
mutate(year = year(date),
month = month(date))
# load validated and vertically aligned pH profiles,
full_argo <-
read_rds(file = paste0(path_argo_preprocessed, "/pH_bgc_va.rds")) %>%
filter(profile_range >= opt_min_profile_range) %>%
mutate(date = ymd(format(date, "%Y-%m-15")))
# map data
map <-
read_rds(paste(path_emlr_utilities,
"map_landmask_WOA18.rds",
sep = ""))Regions
Biome grid reduction
map +
geom_tile(data = nm_biomes,
aes(x = lon,
y = lat,
fill = biome_name))+
lims(y = c(-85, -30))+
scale_fill_brewer(palette = 'Dark2')+
labs(title = 'Mayot biomes pre-grid reduction')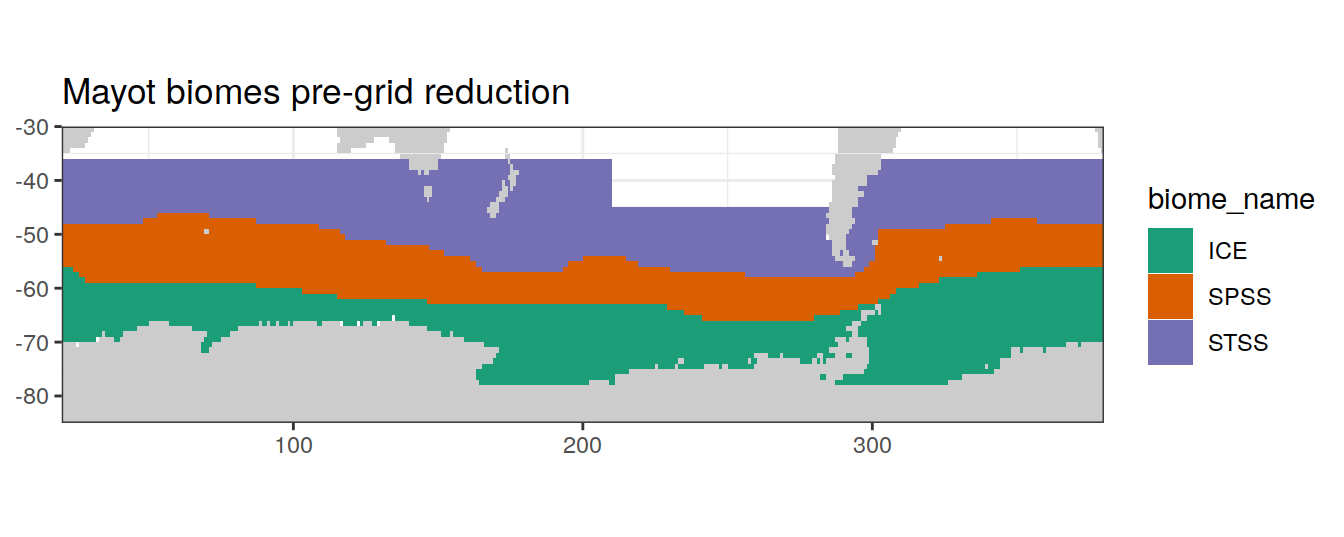
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| d14b7f1 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
# Commented
# nm_biomes_2x2 <- nm_biomes %>%
# mutate(lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
# lon = as.numeric(as.character(lon)),
# lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
# lat = as.numeric(as.character(lat)))
#
# nm_biomes_2x2 <- nm_biomes_2x2 %>%
# count(lon, lat, biome_name) %>%
# group_by(lon, lat) %>%
# slice_max(n, with_ties = FALSE) %>%
# ungroup()
# New
nm_biomes <- nm_biomes %>%
count(lon, lat, biome_name) %>%
group_by(lon, lat) %>%
slice_max(n, with_ties = FALSE) %>%
ungroup()
# Commented
#rm(nm_biomes)# map+
# geom_tile(data = nm_biomes_2x2,
# aes(x = lon,
# y = lat,
# fill = biome_name))+
# lims(y = c(-85, -30))+
# scale_fill_brewer(palette = 'Dark2')+
# labs(title = 'Mayot biomes post-grid reduction')Basin grid reduction
# basinmask <- basinmask %>%
# filter(lat < -30)map +
geom_tile(data = basinmask,
aes(x = lon,
y = lat,
fill = basin_AIP))+
scale_fill_brewer(palette = 'Dark2')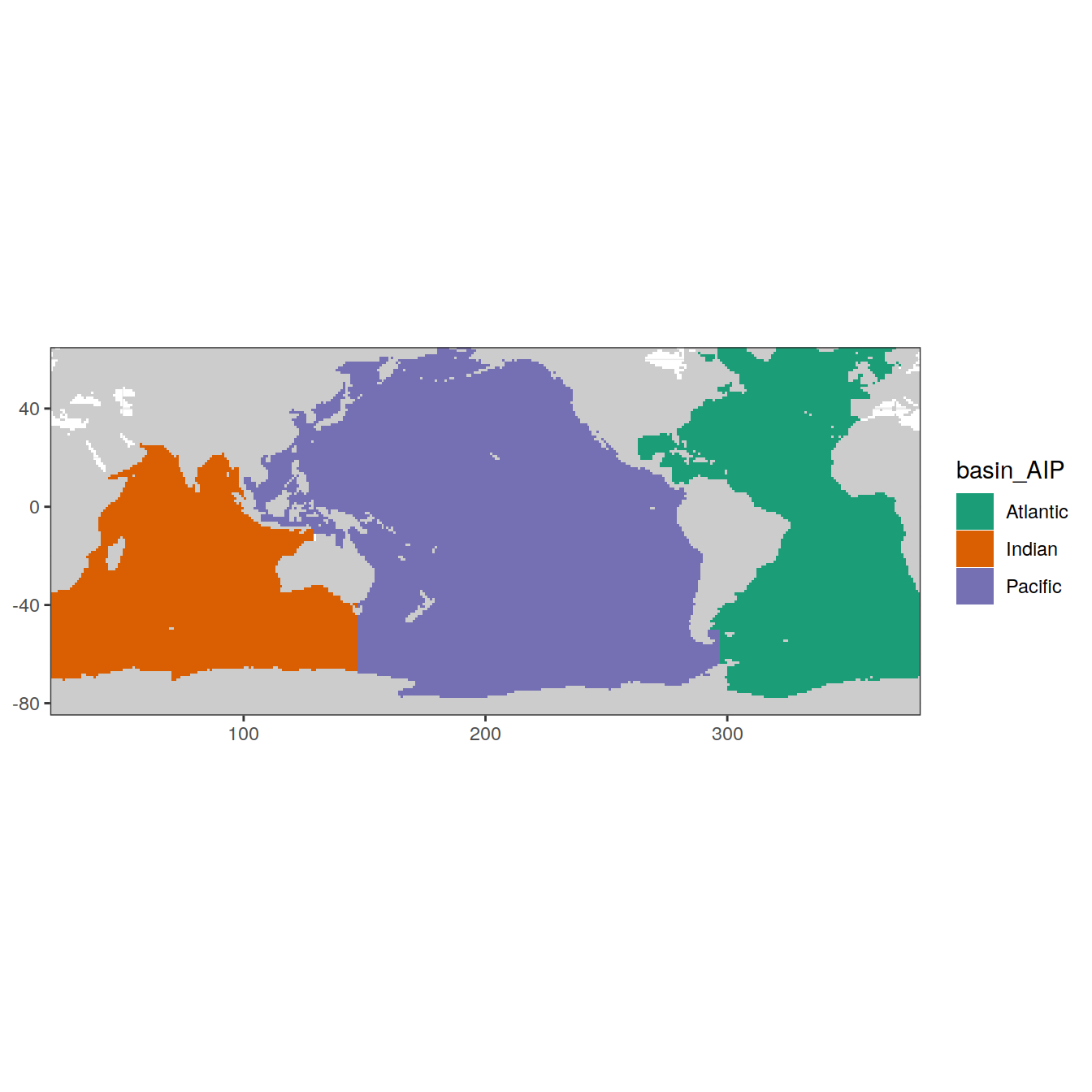
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f110b74 | ds2n19 | 2023-12-13 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| d14b7f1 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
# Commented
# basinmask_2x2 <- basinmask %>%
# mutate(
# lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
# lat = as.numeric(as.character(lat)),
# lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
# lon = as.numeric(as.character(lon))
# )
#
# # assign basins from each pixel to to each 2 Lon x Lat pixel, based on the majority of basins in each 2x2 grid
#
# basinmask_2x2 <- basinmask_2x2 %>%
# count(lon, lat, basin_AIP) %>%
# group_by(lon, lat) %>%
# slice_max(n, with_ties = FALSE) %>%
# ungroup() %>%
# select(-n)
# Added
basinmask <- basinmask %>%
count(lon, lat, basin_AIP) %>%
group_by(lon, lat) %>%
slice_max(n, with_ties = FALSE) %>%
ungroup() %>%
select(-n)
# commented
#rm(basinmask)# map+
# geom_tile(data = basinmask_2x2 %>% filter(lat < -30),
# aes(x = lon,
# y = lat,
# fill = basin_AIP))+
# lims(y = c(-85, -30))+
# scale_fill_brewer(palette = 'Dark2')OceanSODA
Monthly clim surface pH
OceanSODA <- OceanSODA %>%
group_by(lon, lat, month) %>%
mutate(clim_ph = mean(ph_total, na.rm = TRUE),
clim_diff = ph_total - clim_ph,
.after = ph_total) %>%
ungroup()Grid reduction
# Note: While reducing lon x lat grid,
# we keep the original number of observations
# OceanSODA_2x2 <- OceanSODA %>%
# mutate(
# lat_raw = lat,
# lon_raw = lon,
# lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
# lat = as.numeric(as.character(lat)),
# lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
# lon = as.numeric(as.character(lon))) # regrid into 2x2º grid
OceanSODA <- OceanSODA %>%
mutate(
lat_raw = lat,
lon_raw = lon)
# rm(OceanSODA)Apply region masks
# # keep only Southern Ocean data
# OceanSODA_2x2_SO <- inner_join(OceanSODA_2x2, nm_biomes_2x2)
#
# # add in basin separations
# OceanSODA_2x2_SO <- inner_join(OceanSODA_2x2_SO, basinmask_2x2)
# # expected number of rows from -30 to -70º latitude, 360º longitude, for 12 months, 8 years:
# # 40 lat x 360 lon x 12 months x 8 years = 1 382 400 rows
# # actual number of rows: 919 768
#
# OceanSODA_2x2_SO <- OceanSODA_2x2_SO %>%
# filter(!is.na(ph_total))
## keep only Southern Ocean data
# OceanSODA_SO <- inner_join(OceanSODA, nm_biomes %>%
# select(-n))
# add in basin separations
OceanSODA <- inner_join(OceanSODA, basinmask)
OceanSODA <- OceanSODA %>%
filter(!is.na(ph_total))Maps
OceanSODA %>%
filter(year == 2020) %>%
ggplot(aes(lon_raw, lat_raw, fill = clim_ph)) +
geom_tile() +
scale_fill_viridis_c() +
facet_wrap(~ month, ncol = 2) +
coord_quickmap(expand = 0)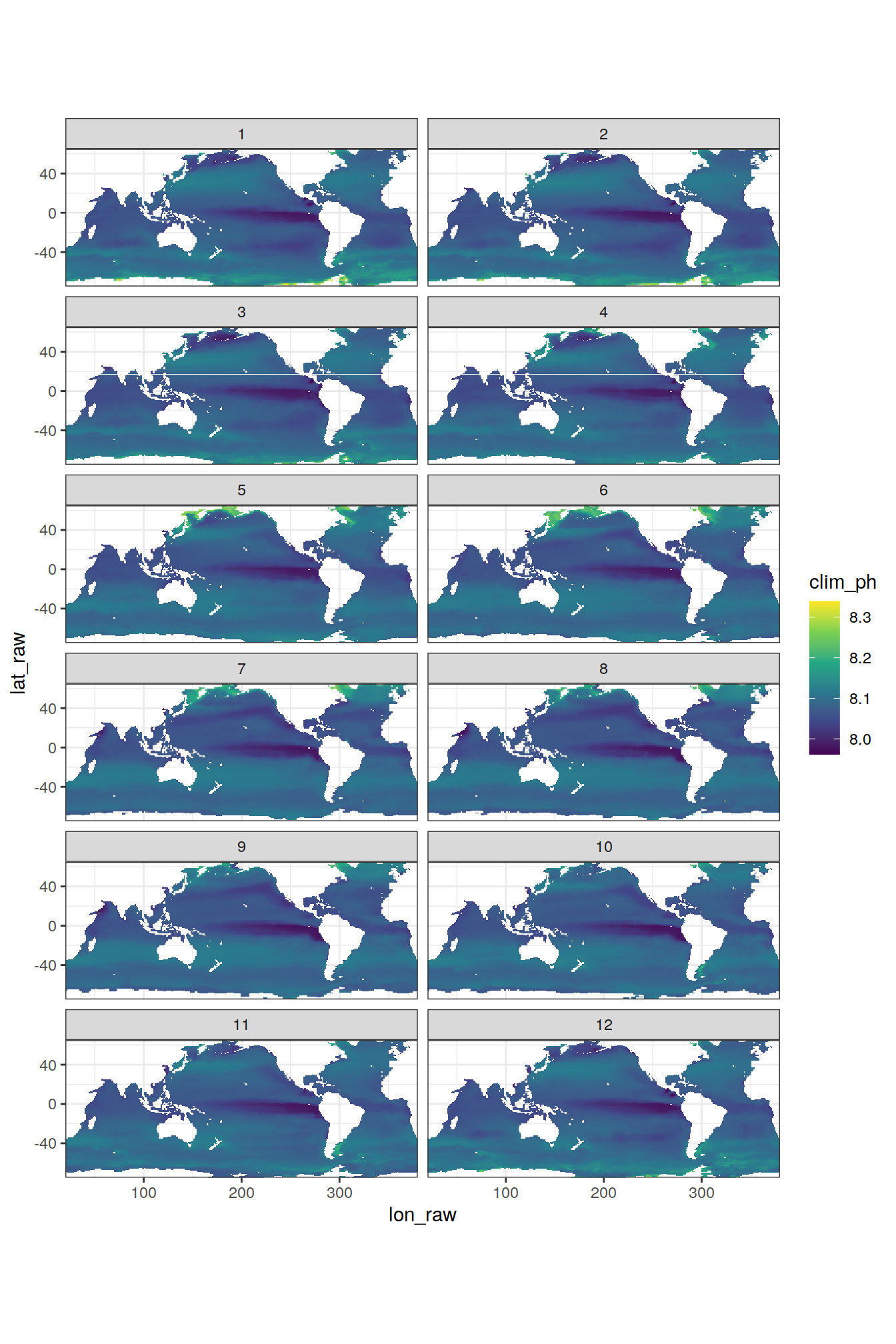
# map of climatological OceanSODA pH
OceanSODA %>%
group_split(month) %>%
#head(1) %>%
map(
~map+
geom_tile(data = .x,
aes(x = lon_raw,
y = lat_raw,
fill = clim_ph))+
scale_fill_viridis_c()+
labs(title = paste('climatological OceanSODA pH (1995-2020) month:', unique(.x$month)))
)[[1]]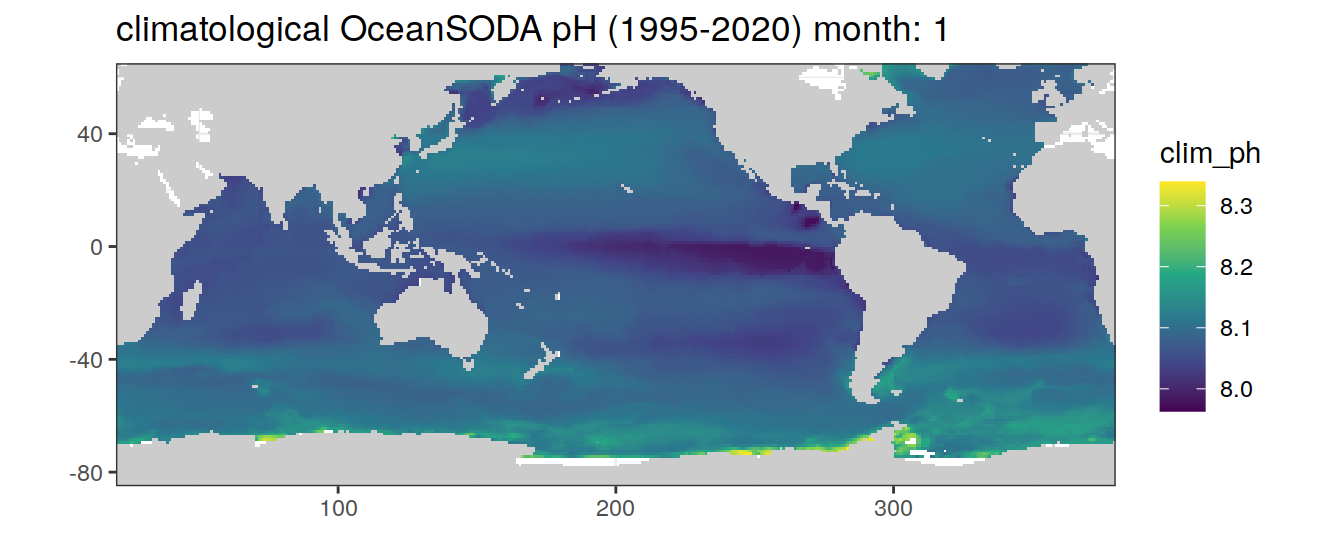
[[2]]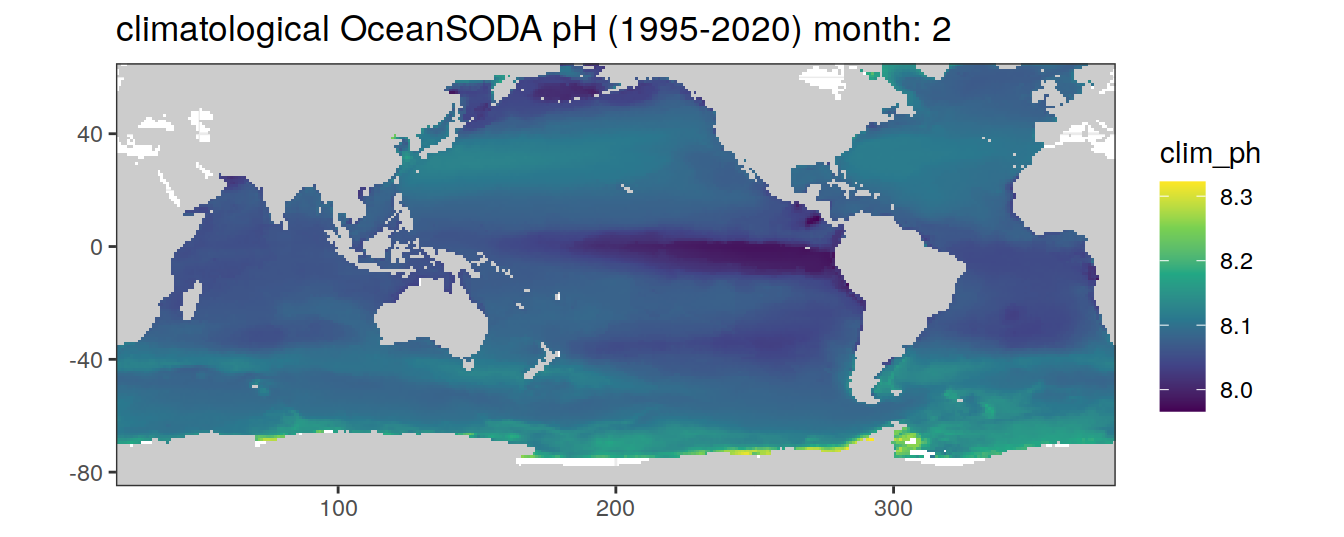
[[3]]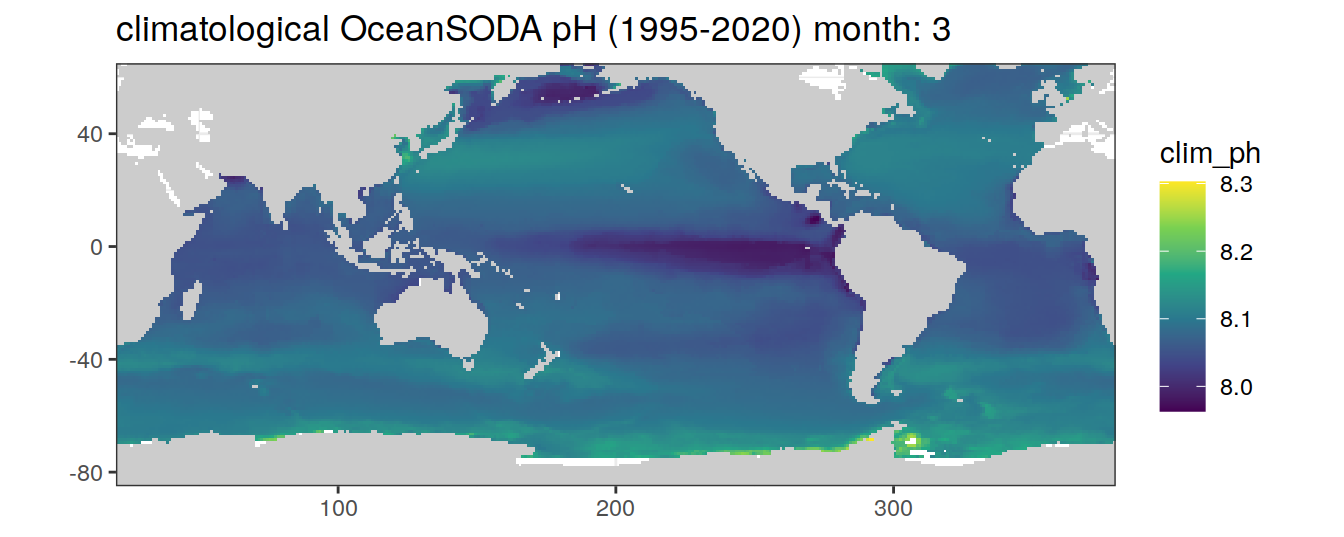
[[4]]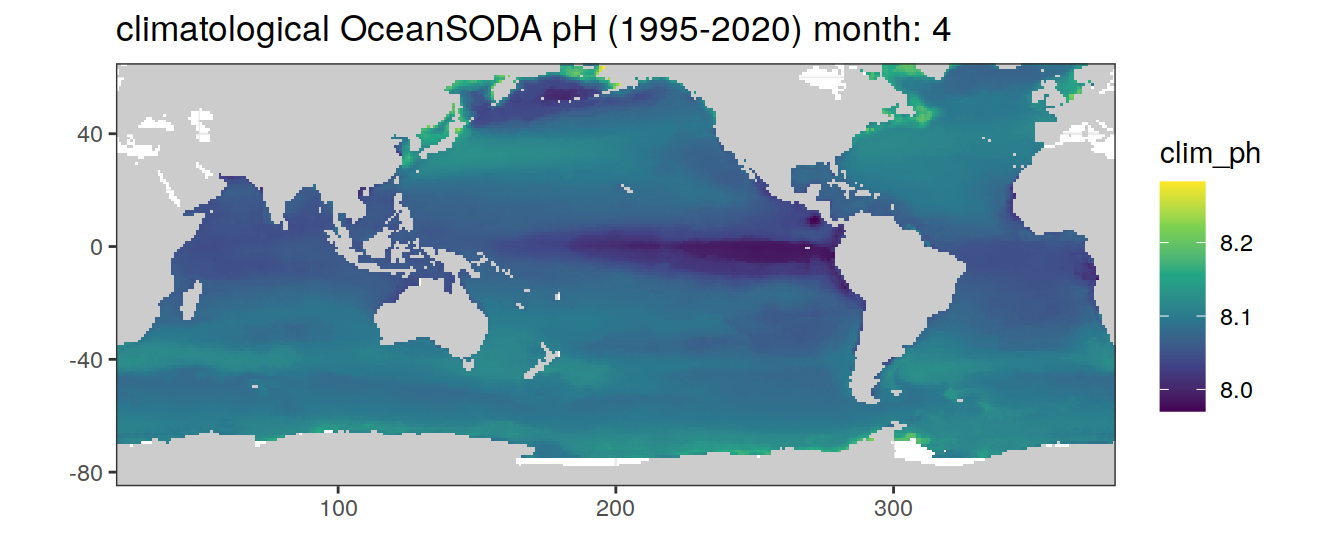
[[5]]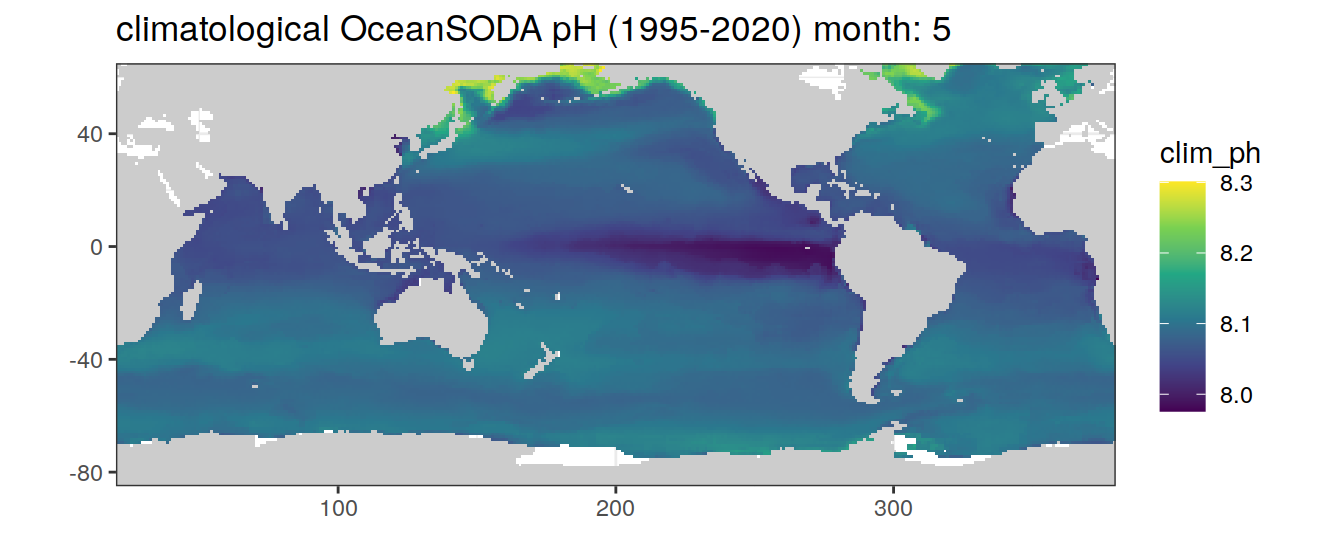
[[6]]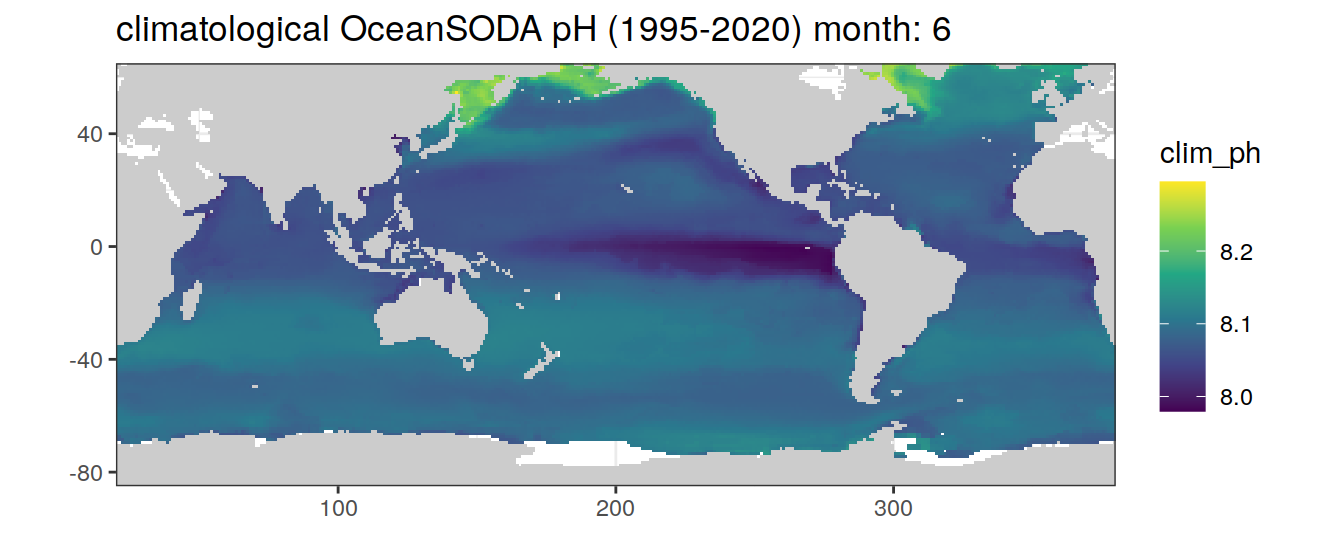
[[7]]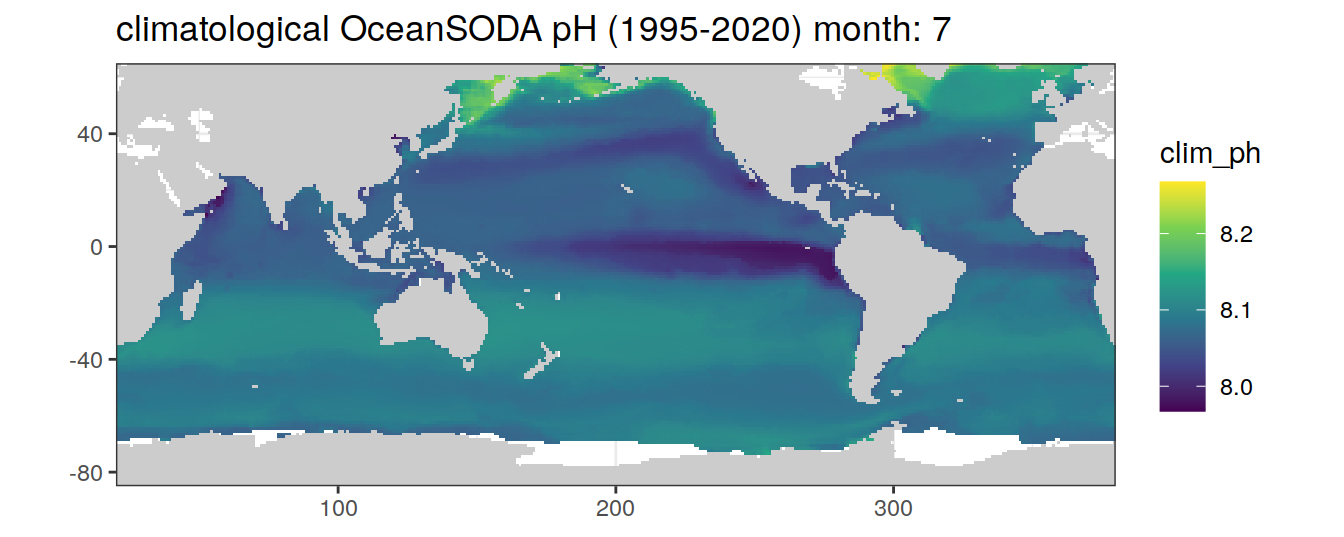
[[8]]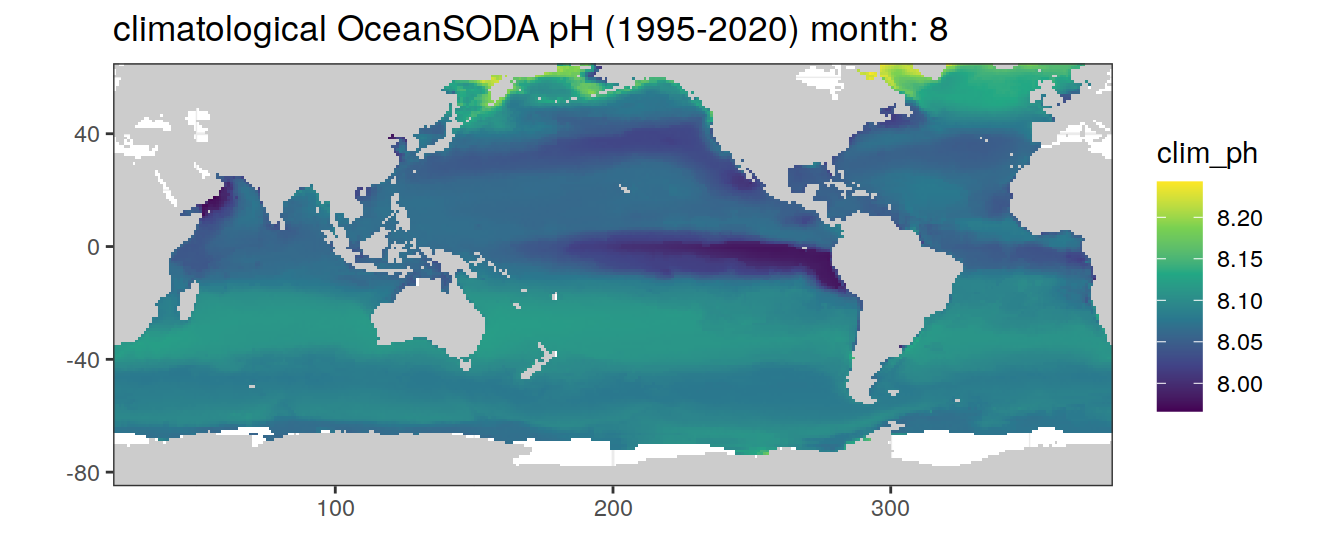
[[9]]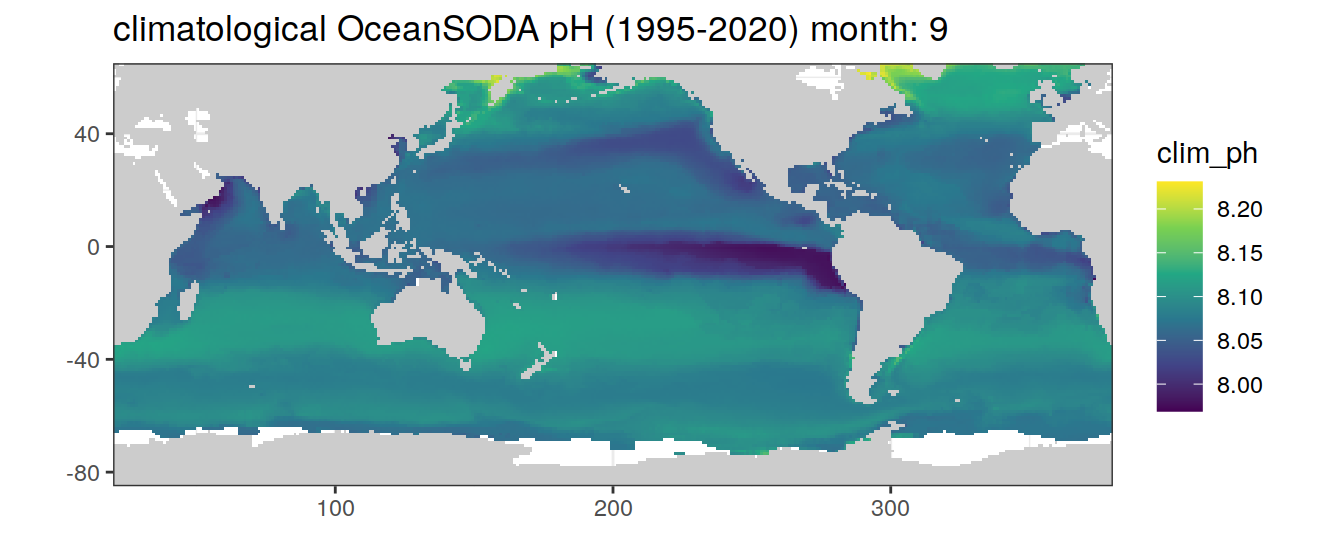
[[10]]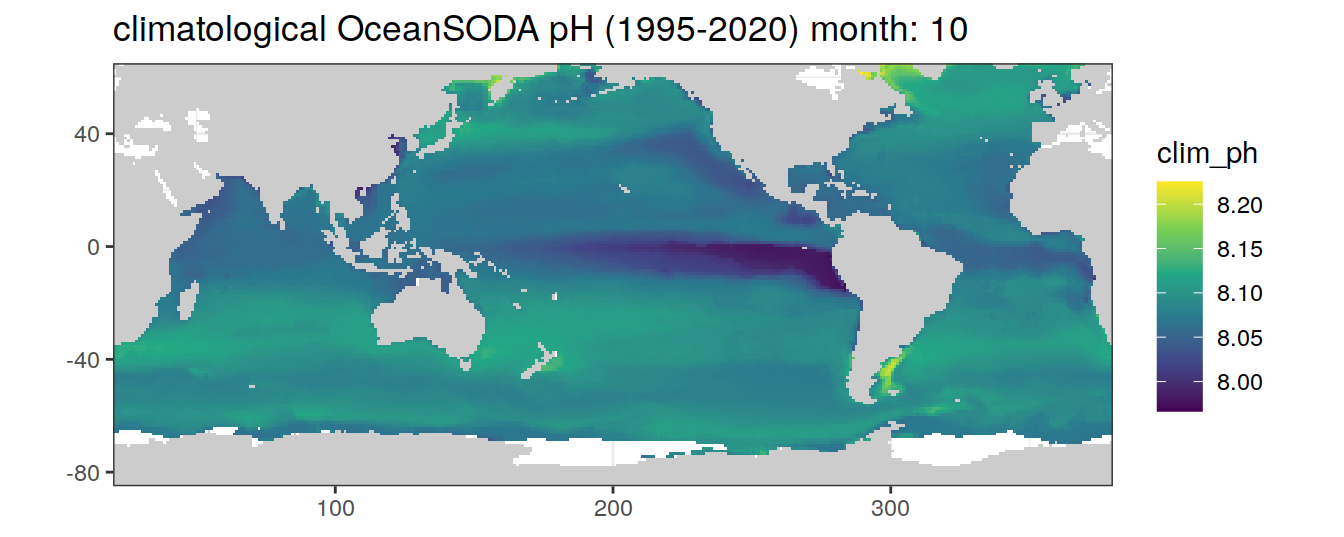
[[11]]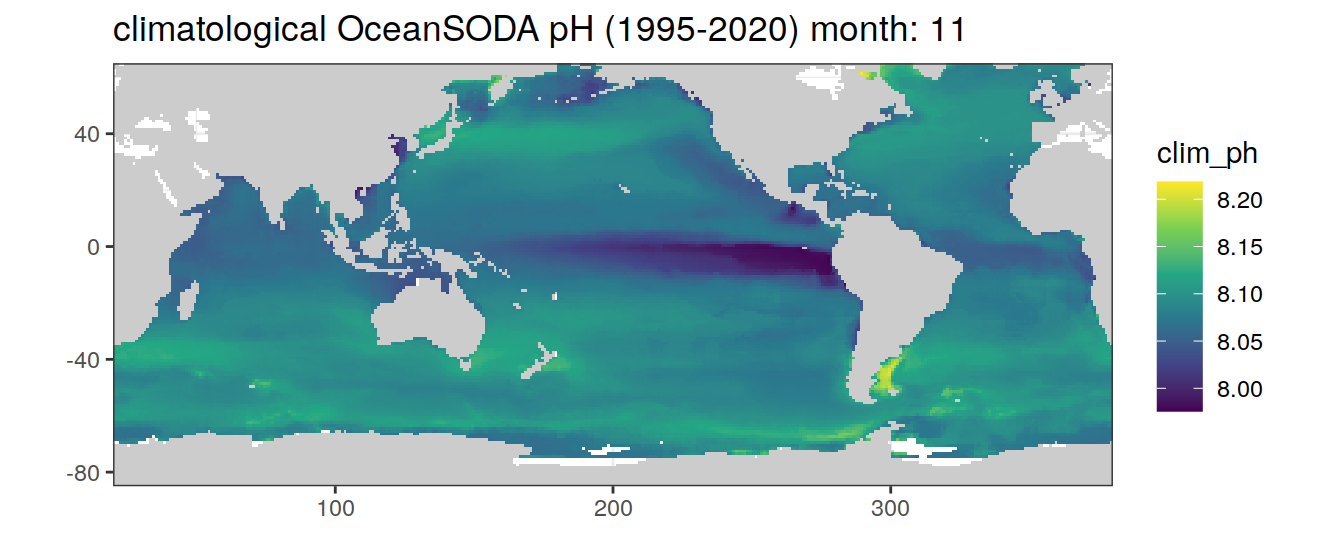
[[12]]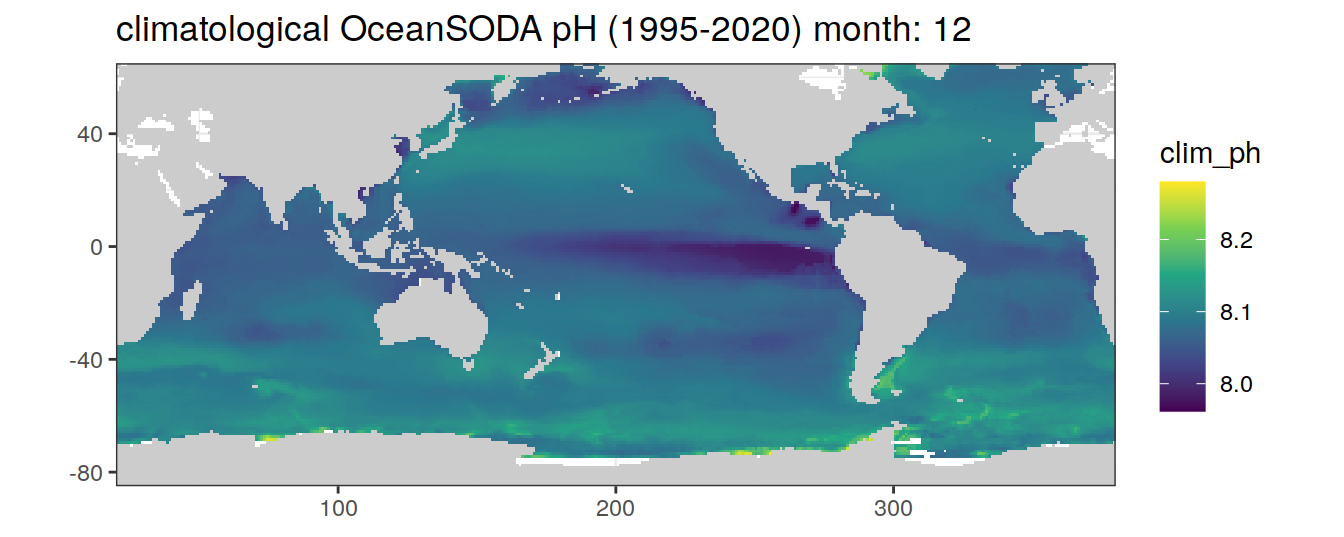
# map of monthly anomaly relative to climatology
OceanSODA %>%
group_split(month) %>%
#head(1) %>%
map(
~ map +
geom_tile(data = .x,
aes(
x = lon_raw,
y = lat_raw,
fill = clim_diff
)) +
scale_fill_divergent(mid = 'grey80') +
facet_wrap( ~ year, ncol = 2) +
labs(
title = paste(
'in-situ OceanSODA pH - clim OceanSODA pH, month:',
unique(.x$month)
)
) +
theme(legend.position = 'right')
)[[1]]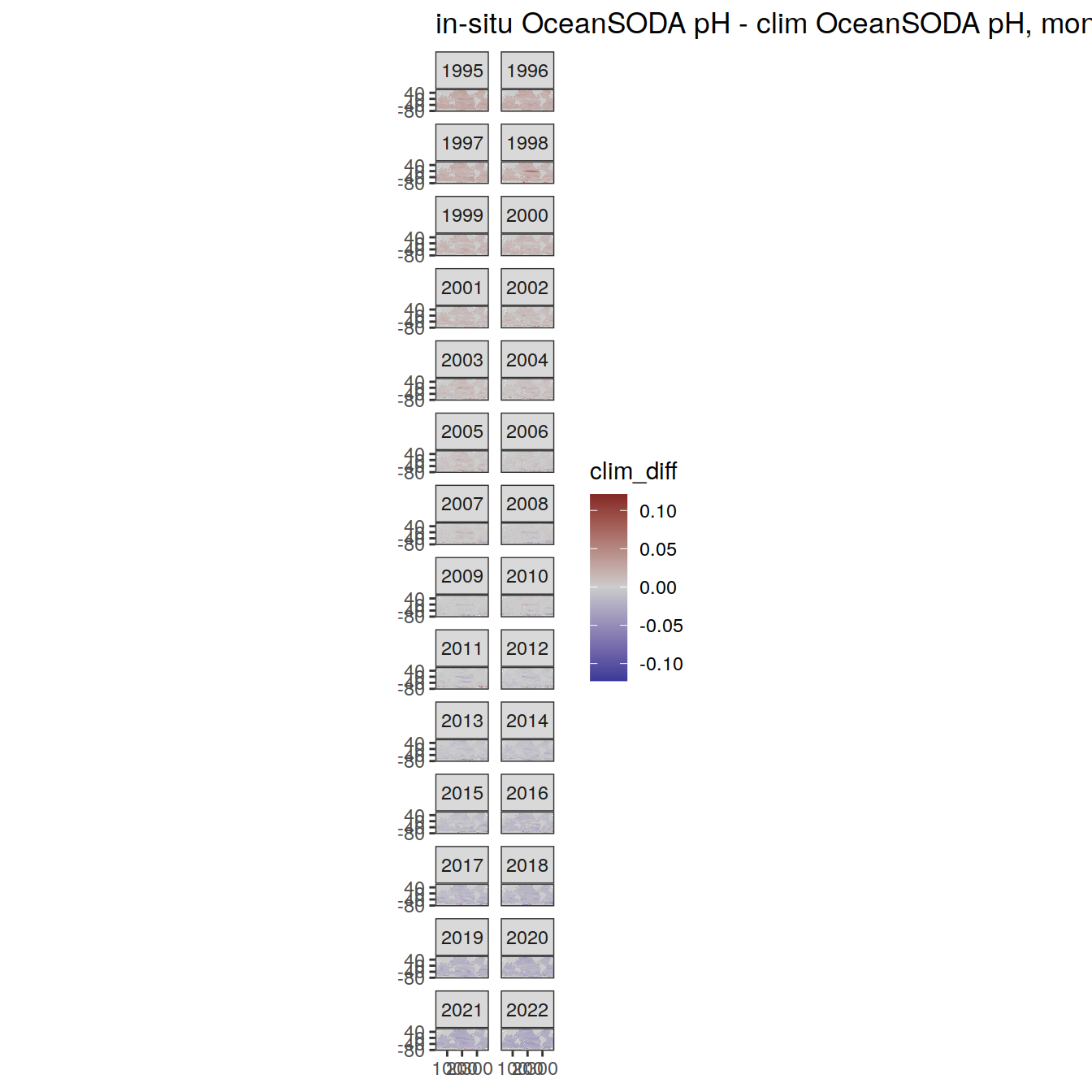
[[2]]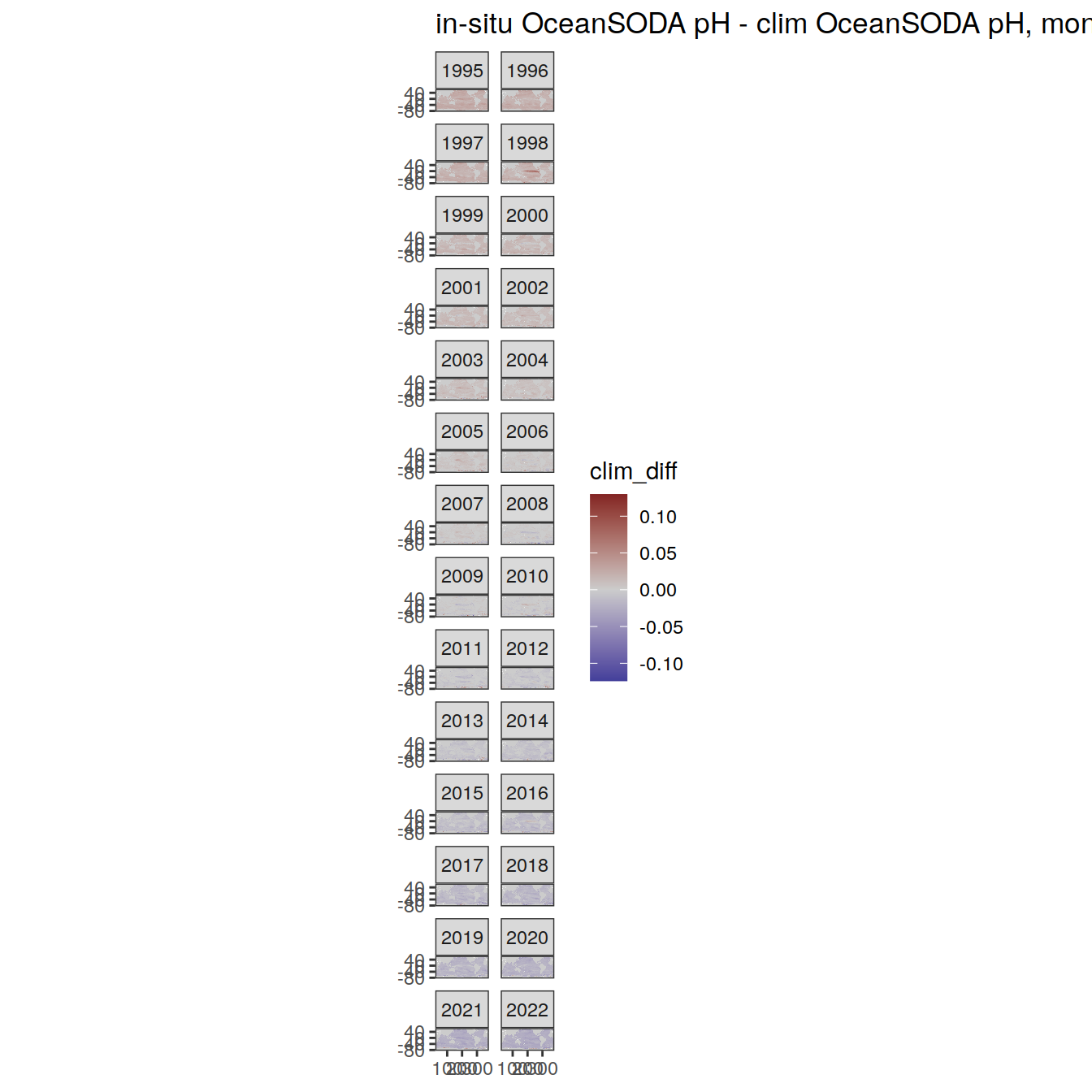
[[3]]
[[4]]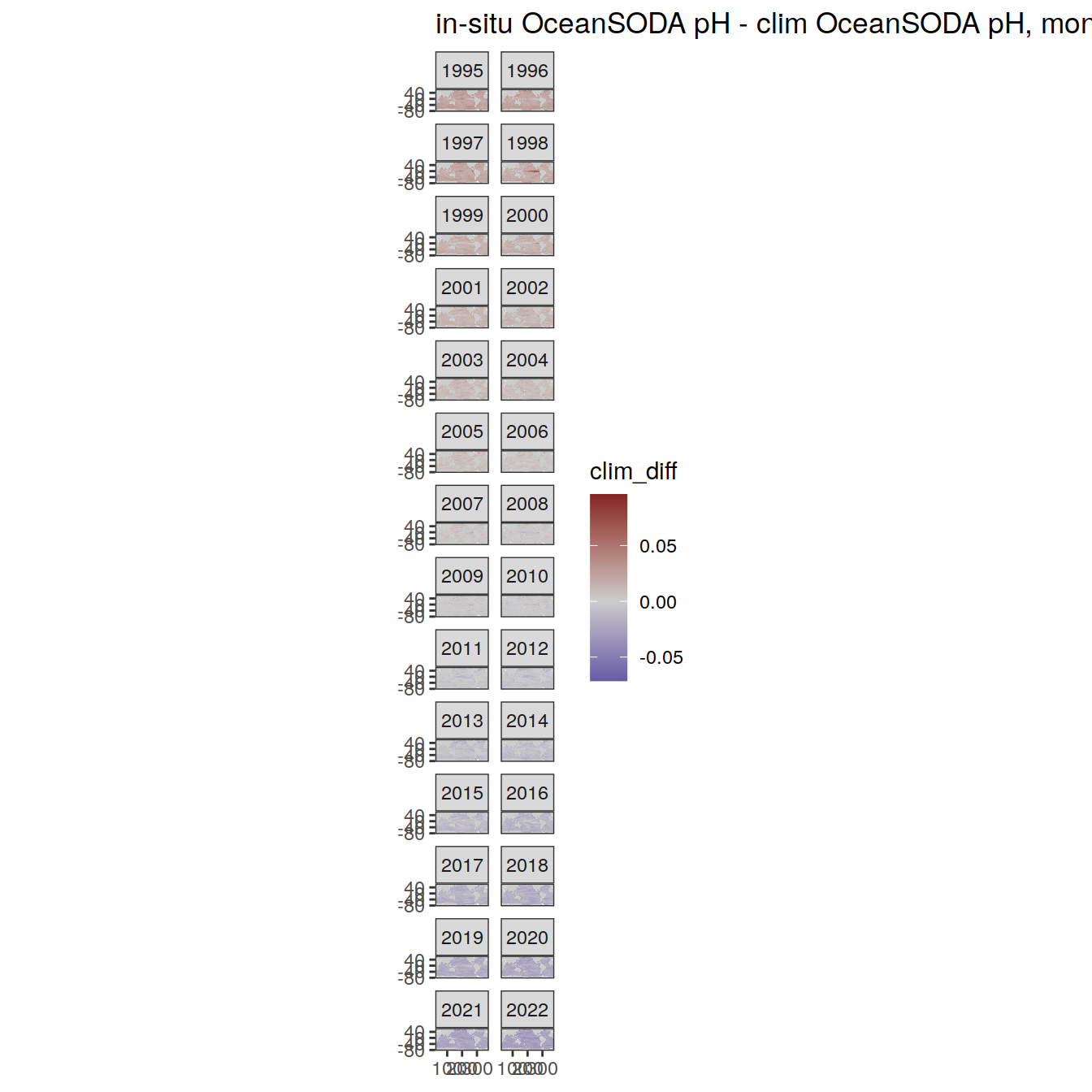
[[5]]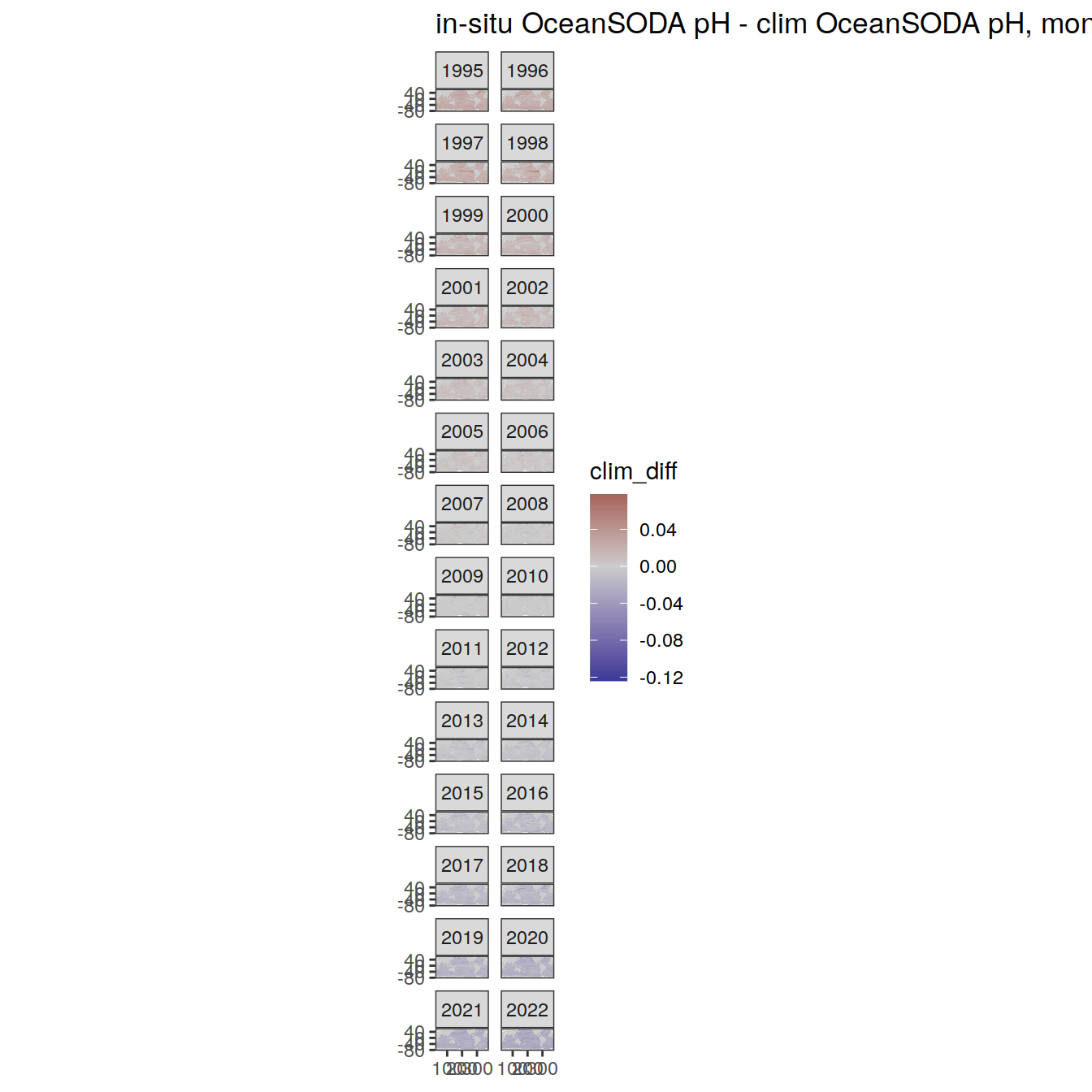
[[6]]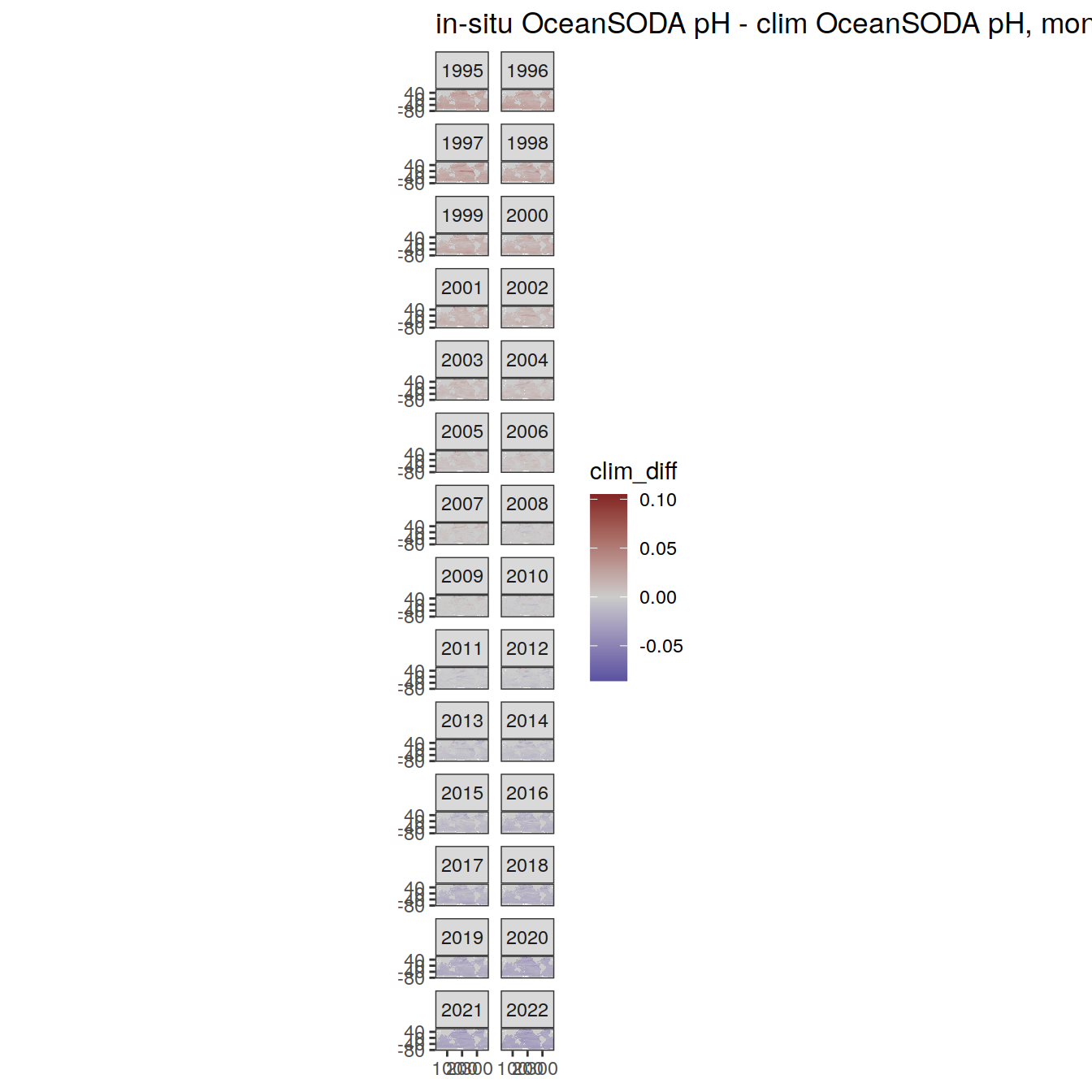
[[7]]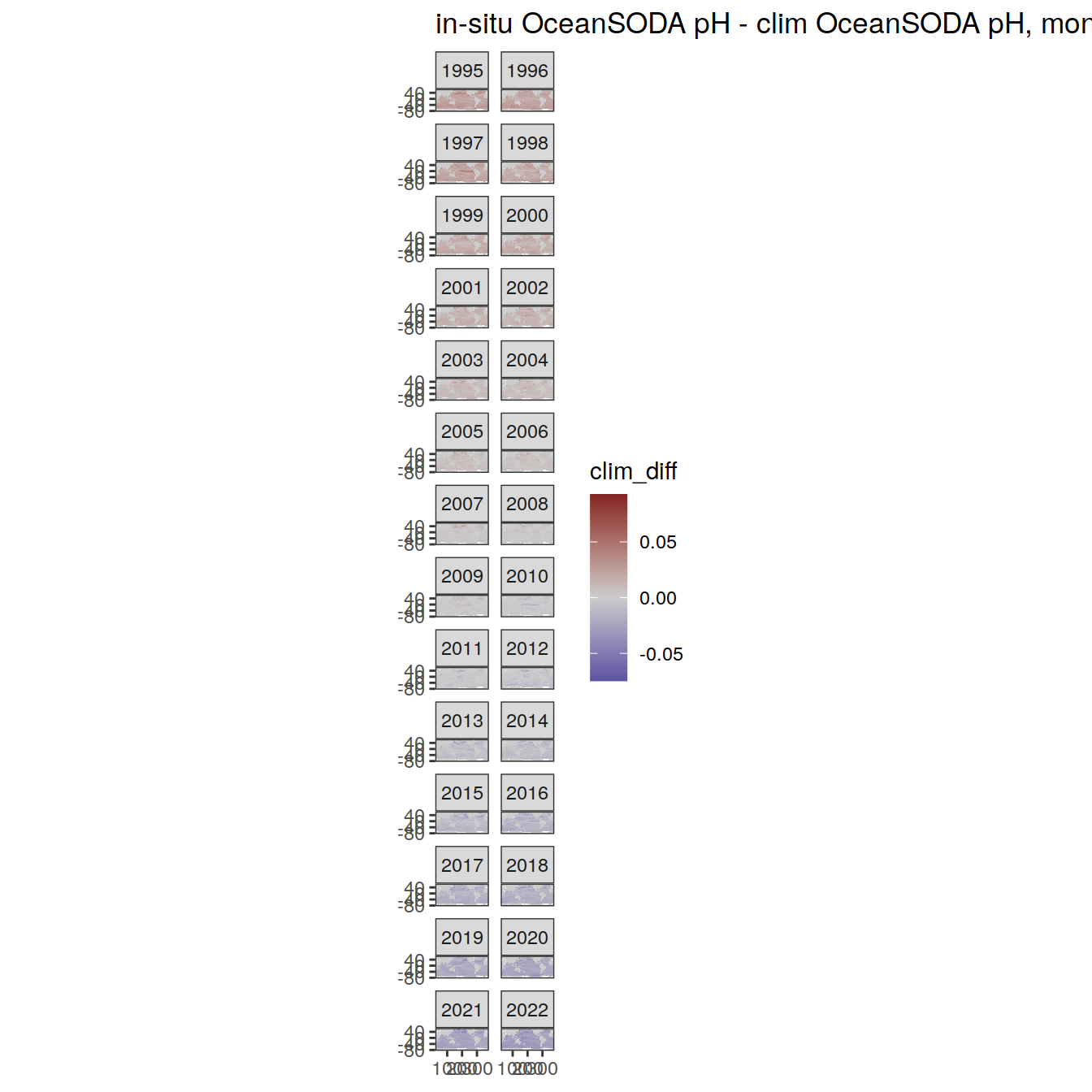
[[8]]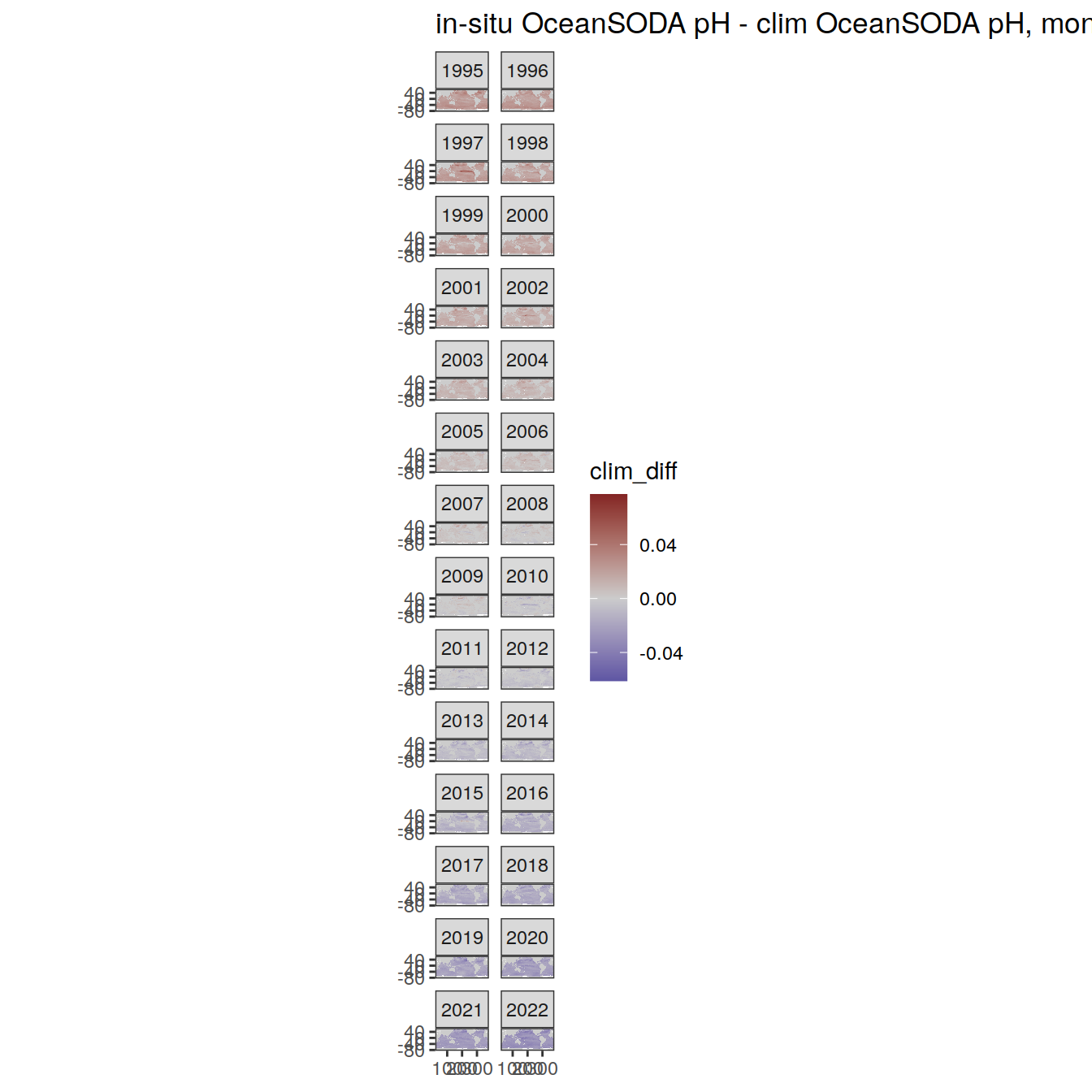
[[9]]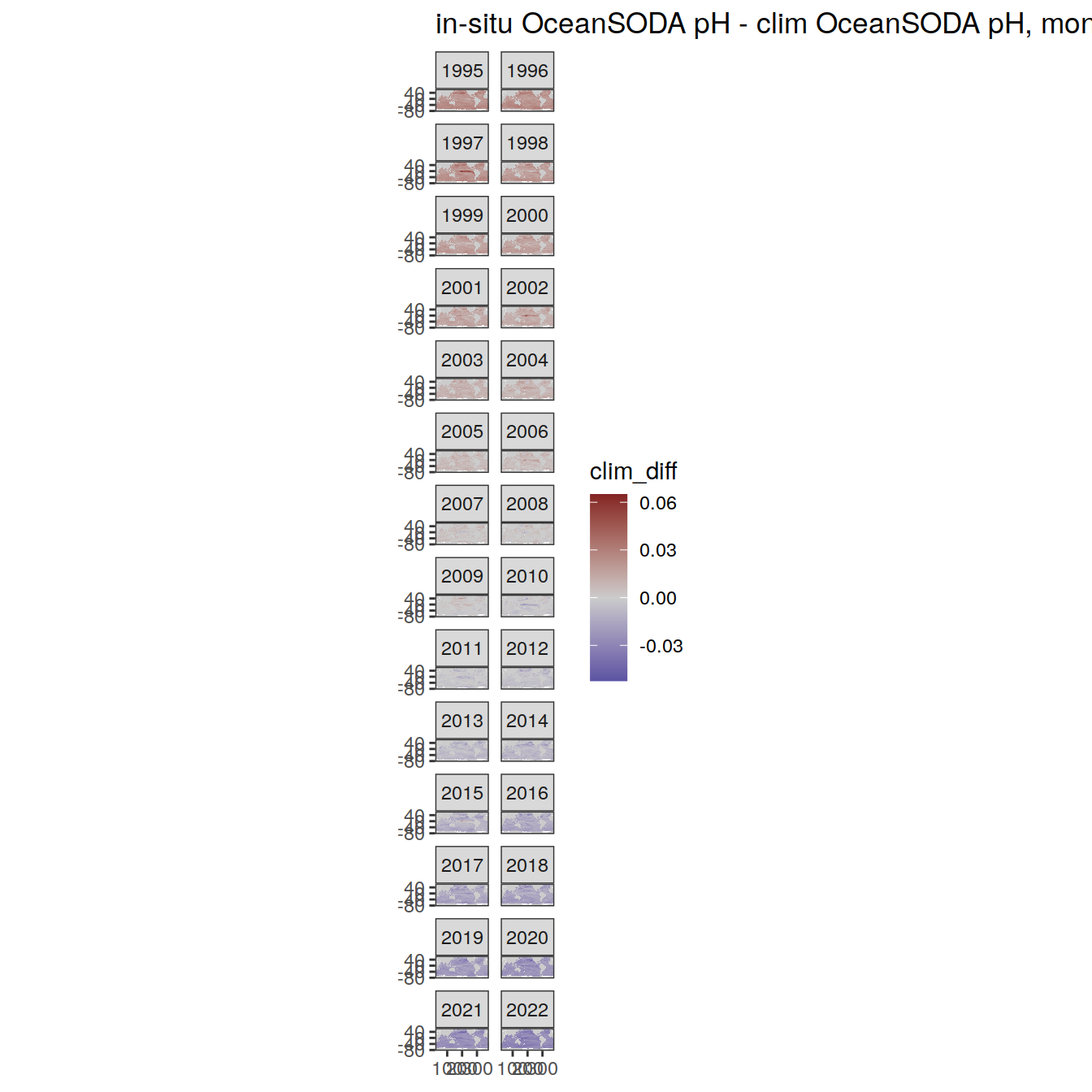
[[10]]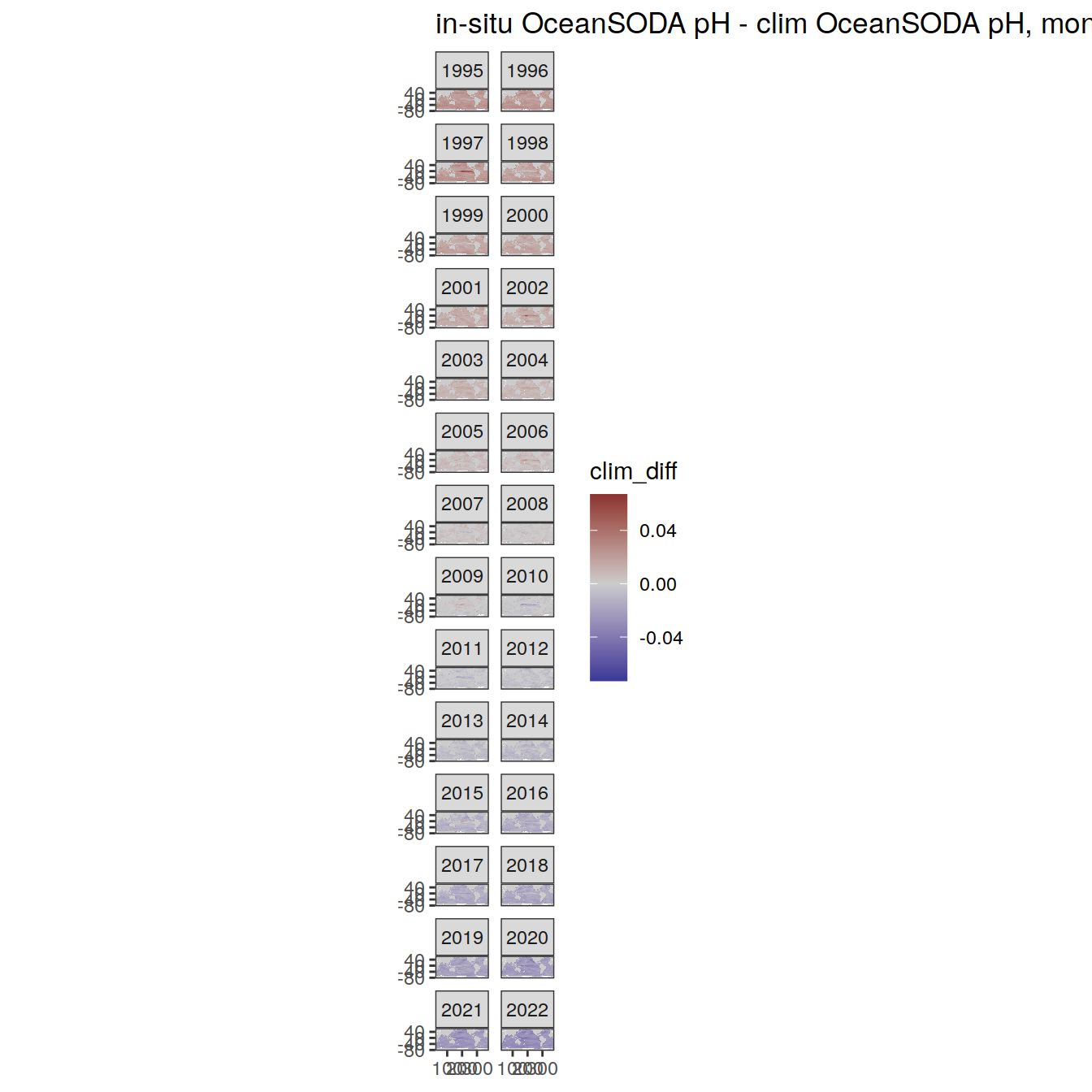
[[11]]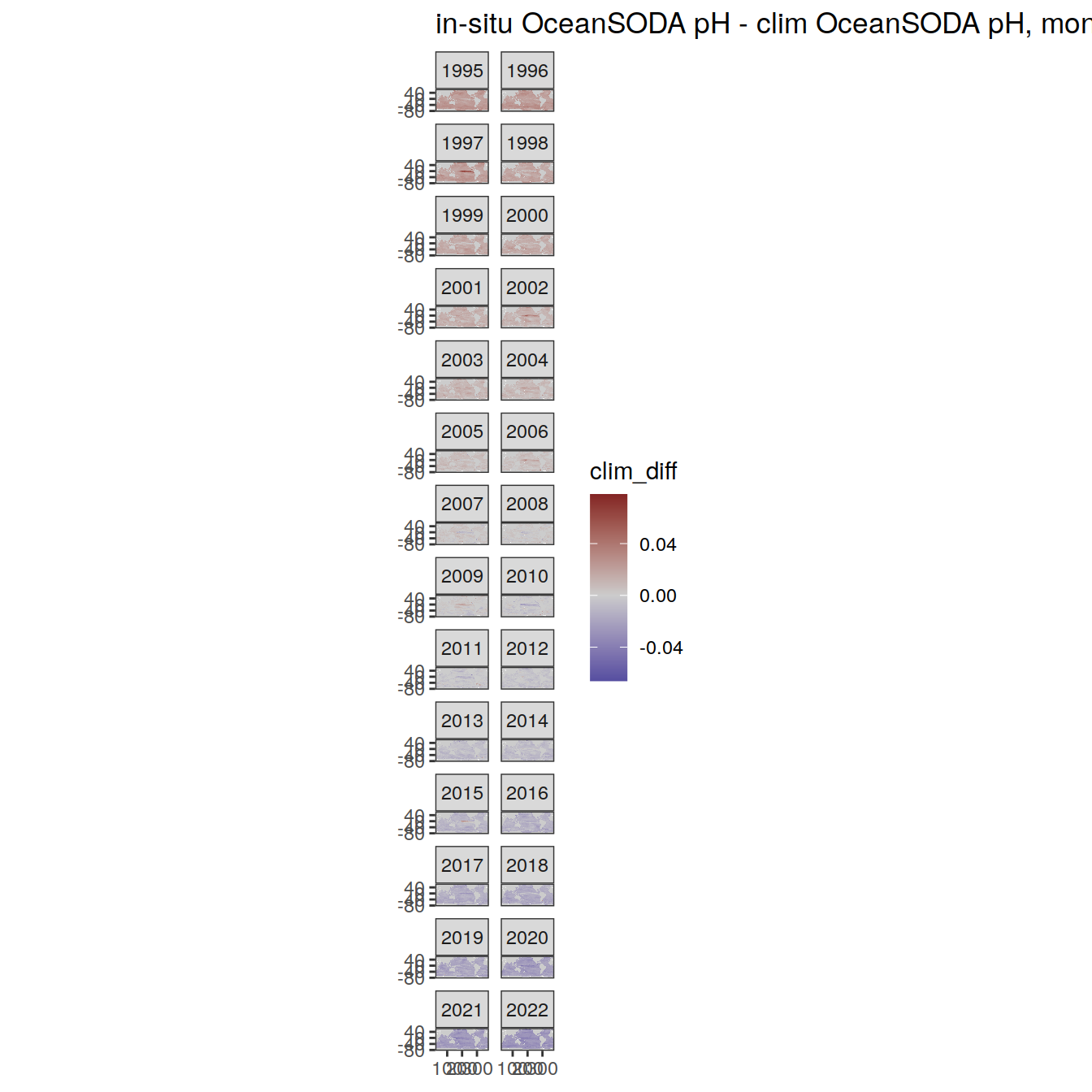
[[12]]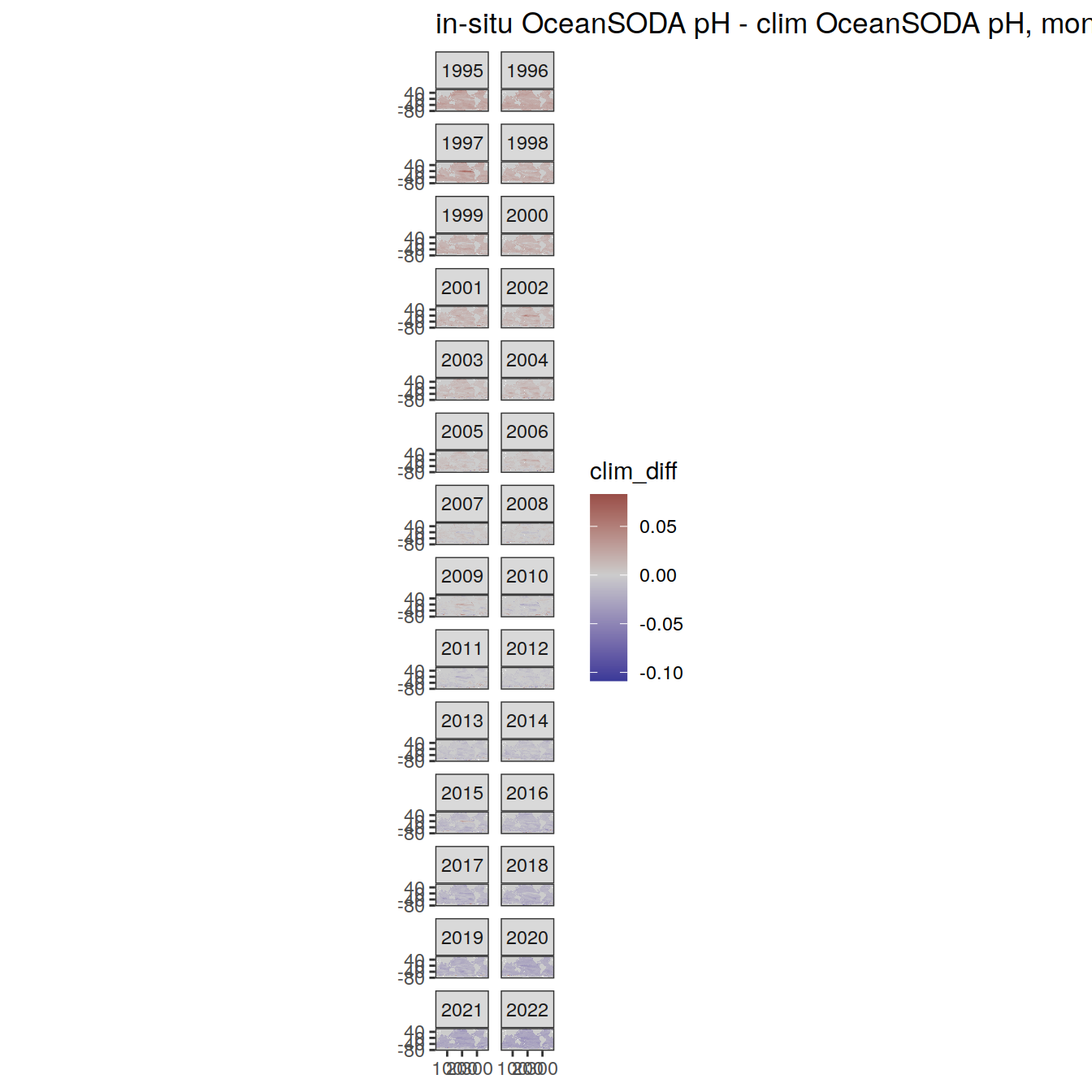
OceanSODA pH anomalies
Grid level
Climatological thresholds
Fit lm models
OceanSODA <- OceanSODA %>%
mutate(decimal_year = decimal_date(date), .after = year)
# fit a linear regression of OceanSODA pH against time (temporal trend)
# in each lat/lon/month grid
# in each lat/lon/month grid, month is used depending on opt_extreme_determination
if (opt_extreme_determination == 1){
OceanSODA_regression <- OceanSODA %>%
nest(data = -c(lon, lat)) %>%
mutate(
fit = map(
.x = data,
.f = ~ lm(clim_diff ~ decimal_year, data = .x)
),
tidied = map(.x = fit, .f = tidy),
glanced = map(.x = fit, .f = glance),
augmented = map(.x = fit, .f = augment)
)
} else if (opt_extreme_determination == 2){
OceanSODA_regression <- OceanSODA %>%
nest(data = -c(lon, lat, month)) %>%
mutate(
fit = map(
.x = data,
.f = ~ lm(clim_diff ~ decimal_year, data = .x)
),
tidied = map(.x = fit, .f = tidy),
glanced = map(.x = fit, .f = glance),
augmented = map(.x = fit, .f = augment)
)
}
OceanSODA_regression_tidied <- OceanSODA_regression %>%
select(-c(data, fit, augmented, glanced)) %>%
unnest(tidied)
OceanSODA_regression_tidied <- OceanSODA_regression_tidied %>%
select(lat:estimate) %>%
pivot_wider(names_from = term,
values_from = estimate) %>%
rename(intercept = `(Intercept)`,
slope = decimal_year)
OceanSODA_regression_data <- OceanSODA_regression %>%
select(-c(fit, tidied, glanced, augmented)) %>%
unnest(data)
OceanSODA_regression_augmented <- OceanSODA_regression %>%
select(-c(fit, tidied, glanced, data)) %>%
unnest(augmented) %>%
select(lat:decimal_year, .resid)
OceanSODA_regression_augmented <- bind_cols(
OceanSODA_regression_augmented,
OceanSODA_regression_data %>% select(
date, basin_AIP,
clim_ph, ph_total,
lon_raw, lat_raw))
OceanSODA_regression_glanced <- OceanSODA_regression %>%
select(-c(data, fit, tidied, augmented)) %>%
unnest(glanced)Fit mean
# identify the mean value
# in each lat/lon/month grid
# OceanSODA_regression_tidied <- OceanSODA_SO %>%
# # filter(basin_AIP == "Indian",
# # biome_name == "SPSS",
# # lon < 40) %>%
# group_by(lon, lat, month) %>%
# summarise(slope = 0,
# intercept = mean(clim_diff, na.rm = TRUE)) %>%
# ungroup()
#
# OceanSODA_regression_glanced <- OceanSODA_SO %>%
# # filter(basin_AIP == "Indian",
# # biome_name == "SPSS",
# # lon < 40) %>%
# group_by(lon, lat, month) %>%
# summarise(sigma = sd(clim_diff, na.rm = TRUE)) %>%
# ungroup()
#
# OceanSODA_regression_augmented <- OceanSODA_SO %>%
# # filter(basin_AIP == "Indian",
# # biome_name == "SPSS",
# # lon < 40) %>%
# mutate(.resid = clim_diff)Slope maps
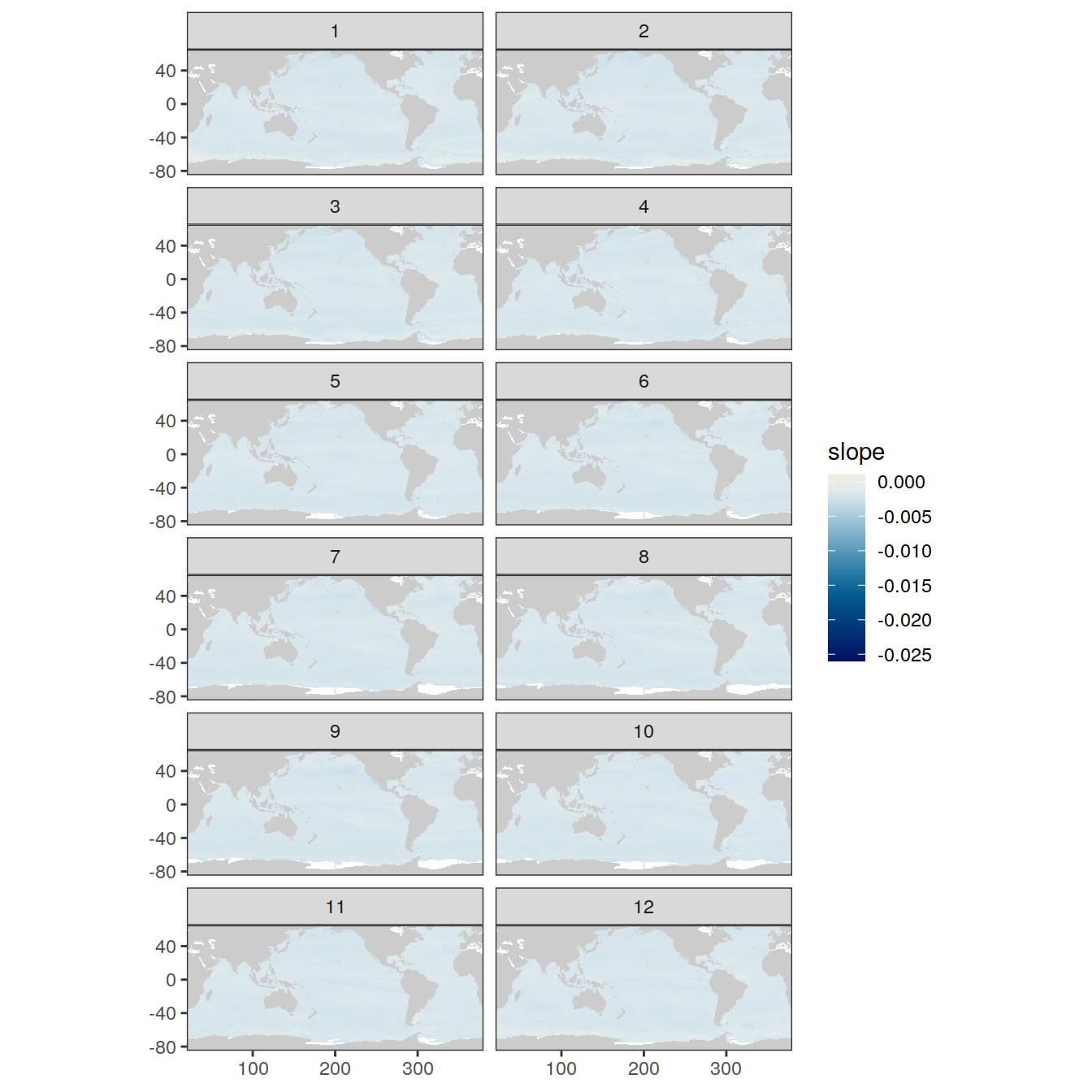
if (opt_extreme_determination == 1){
map +
geom_tile(data = OceanSODA_regression_tidied,
aes(x = lon,
y = lat,
fill = slope)) +
scale_fill_scico(palette = 'vik', midpoint = 0)
} else if (opt_extreme_determination == 2){
map +
geom_tile(data = OceanSODA_regression_tidied,
aes(x = lon,
y = lat,
fill = slope)) +
scale_fill_scico(palette = 'vik', midpoint = 0) +
facet_wrap( ~ month, ncol = 2)
}
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f110b74 | ds2n19 | 2023-12-13 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 31e4d42 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
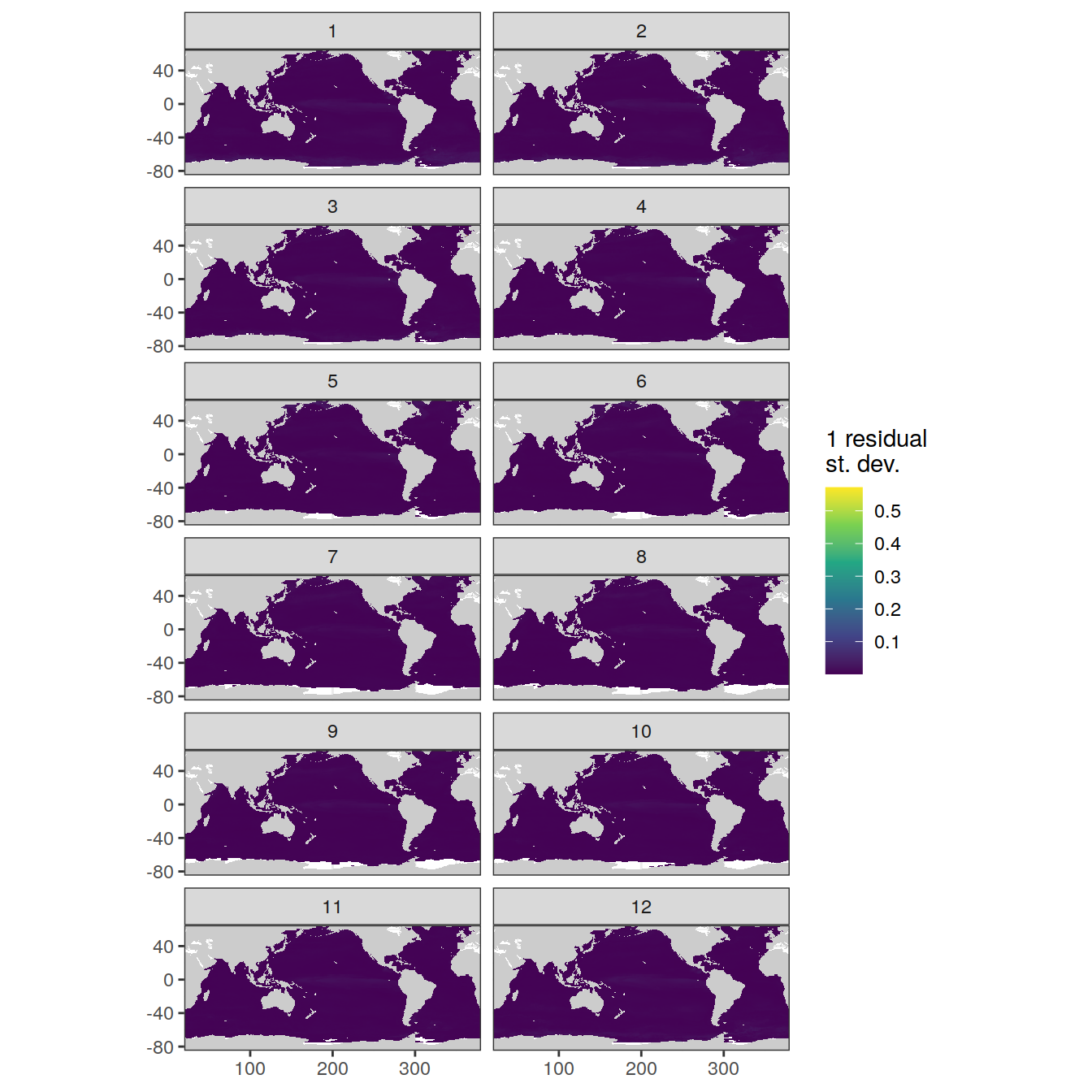
Sigma maps
if (opt_extreme_determination == 1){
map +
geom_tile(data = OceanSODA_regression_glanced,
aes(x = lon,
y = lat,
fill = sigma)) +
scale_fill_viridis_c() +
labs(fill = '1 residual \nst. dev.')
} else if (opt_extreme_determination == 2){
map +
geom_tile(data = OceanSODA_regression_glanced,
aes(x = lon,
y = lat,
fill = sigma)) +
scale_fill_viridis_c() +
labs(fill = '1 residual \nst. dev.') +
facet_wrap(~month, ncol = 2)
}
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f110b74 | ds2n19 | 2023-12-13 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 31e4d42 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
Anomaly identification
Calculate OceanSODA pH anomalies: L for abnormally low, H for abnormally high, N for normal pH
# when the in-situ OceanSODA pH is lower than the 5th percentile (predicted - 2*residual.st.dev), assign 'L' for low extreme
# when the in-situ OceanSODA pH exceeds the 95th percentile (predicted + 2*residual.st.dev), assign 'H' for high extreme
# when the in-situ OceanSODA pH is within 95% of the range, then assign 'N' for normal pH
# combine observations and regression statistics
if (opt_extreme_determination == 1){
OceanSODA_extreme_grid <-
full_join(
OceanSODA_regression_augmented,
OceanSODA_regression_glanced %>%
select(lat:lon, sigma)
)
} else if (opt_extreme_determination == 2){
OceanSODA_extreme_grid <-
full_join(
OceanSODA_regression_augmented,
OceanSODA_regression_glanced %>%
select(lat:month, sigma)
)
}
# identify observations in anomaly classes
OceanSODA_extreme_grid <- OceanSODA_extreme_grid %>%
mutate(
ph_extreme = case_when(
.resid < -sigma*2 ~ 'L',
.resid > sigma*2 ~ 'H',
TRUE ~ 'N'
)
)
OceanSODA_extreme_grid <- OceanSODA_extreme_grid %>%
mutate(ph_extreme = fct_relevel(ph_extreme, "H", "N", "L"))
# combine with regression coefficients
OceanSODA_extreme_grid <-
full_join(OceanSODA_extreme_grid,
OceanSODA_regression_tidied)
OceanSODA_extreme_grid <- OceanSODA_extreme_grid %>%
mutate(year = year(date),
month = month(date),
.after = decimal_year)Write anomalies to file
# Restrict to SO by inner join to nm_biomes
OceanSODA_SO_extreme_grid <- inner_join(OceanSODA_extreme_grid, nm_biomes %>%
select(-n))
# if (opt_extreme_determination == 1){
# OceanSODA_SO_extreme_grid %>%
# write_rds(file = paste0(path = path_argo_preprocessed, "/OceanSODA_pH_anomaly_field_01.rds"))
# OceanSODA_extreme_grid %>%
# write_rds(file = paste0(path = path_argo_preprocessed, "/OceanSODA_global_pH_anomaly_field_01.rds"))
# } else if (opt_extreme_determination == 2){
# OceanSODA_SO_extreme_grid %>%
# write_rds(file = paste0(path = path_argo_preprocessed, "/OceanSODA_pH_anomaly_field_02.rds"))
# OceanSODA_extreme_grid %>%
# write_rds(file = paste0(path = path_argo_preprocessed, "/OceanSODA_global_pH_anomaly_field_02.rds"))
# }if (opt_extreme_determination == 1){
OceanSODA_SO_extreme_grid %>%
group_split(lon, lat) %>%
head(8) %>%
map(
~ ggplot(data = .x) +
geom_point(aes(
x = year,
y = clim_diff,
col = ph_extreme
)) +
geom_abline(data = .x, aes(slope = slope,
intercept = intercept)) +
geom_abline(
data = .x,
aes(slope = slope,
intercept = intercept + 2 * sigma),
linetype = 2
) +
geom_abline(
data = .x,
aes(slope = slope,
intercept = intercept - 2 * sigma),
linetype = 2
) +
labs(title = paste(
fititle = paste("lon:", unique(.x$lon),
"| lat:", unique(.x$lat))
)) +
scale_color_manual(values = HNL_colors)
)
} else if (opt_extreme_determination == 2){
OceanSODA_SO_extreme_grid %>%
group_split(lon, lat, month) %>%
head(8) %>%
map(
~ ggplot(data = .x) +
geom_point(aes(
x = year,
y = clim_diff,
col = ph_extreme
)) +
geom_abline(data = .x, aes(slope = slope,
intercept = intercept)) +
geom_abline(
data = .x,
aes(slope = slope,
intercept = intercept + 2 * sigma),
linetype = 2
) +
geom_abline(
data = .x,
aes(slope = slope,
intercept = intercept - 2 * sigma),
linetype = 2
) +
labs(title = paste(fititle = paste(
"lon:", unique(.x$lon),
"| lat:", unique(.x$lat),
"| month:", unique(.x$month)
))) +
scale_color_manual(values = HNL_colors)
)
}[[1]]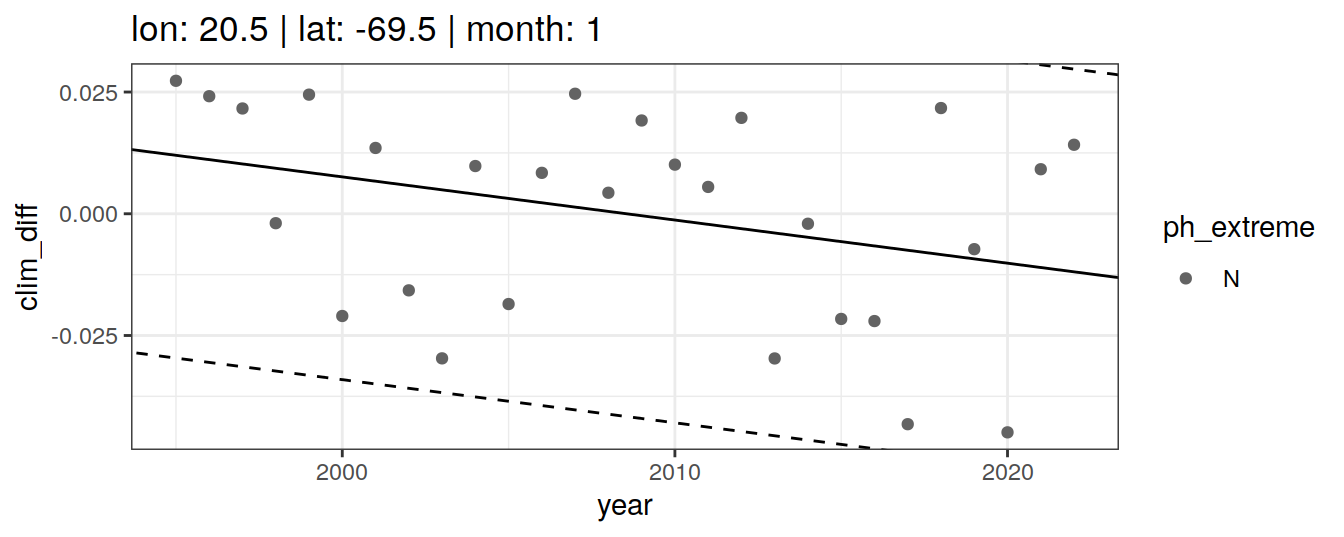
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[2]]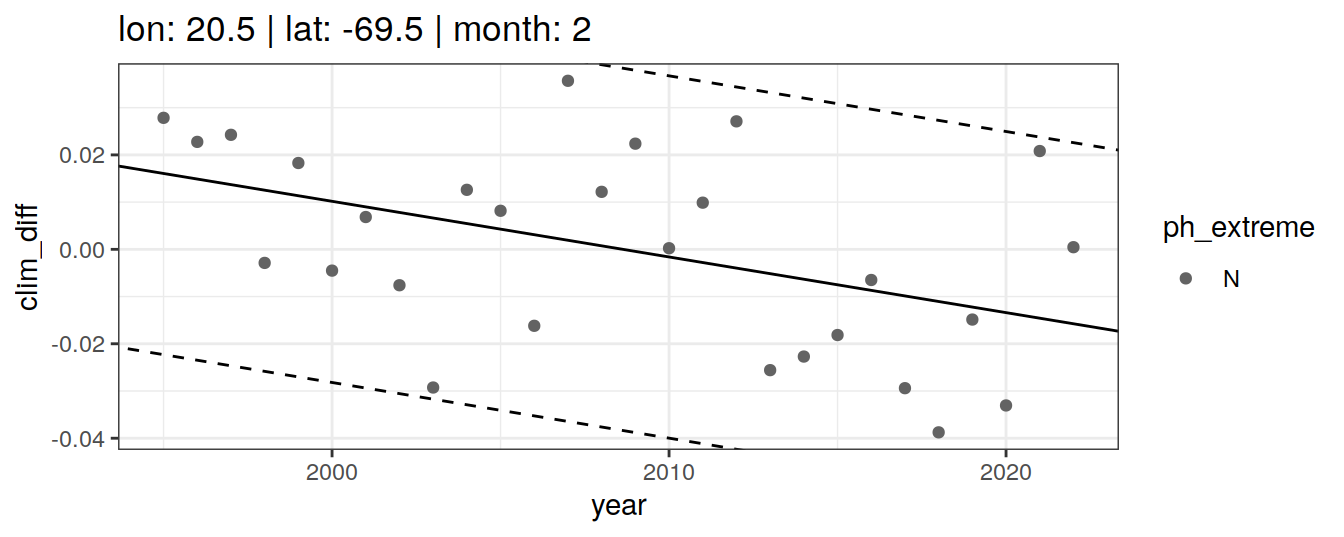
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[3]]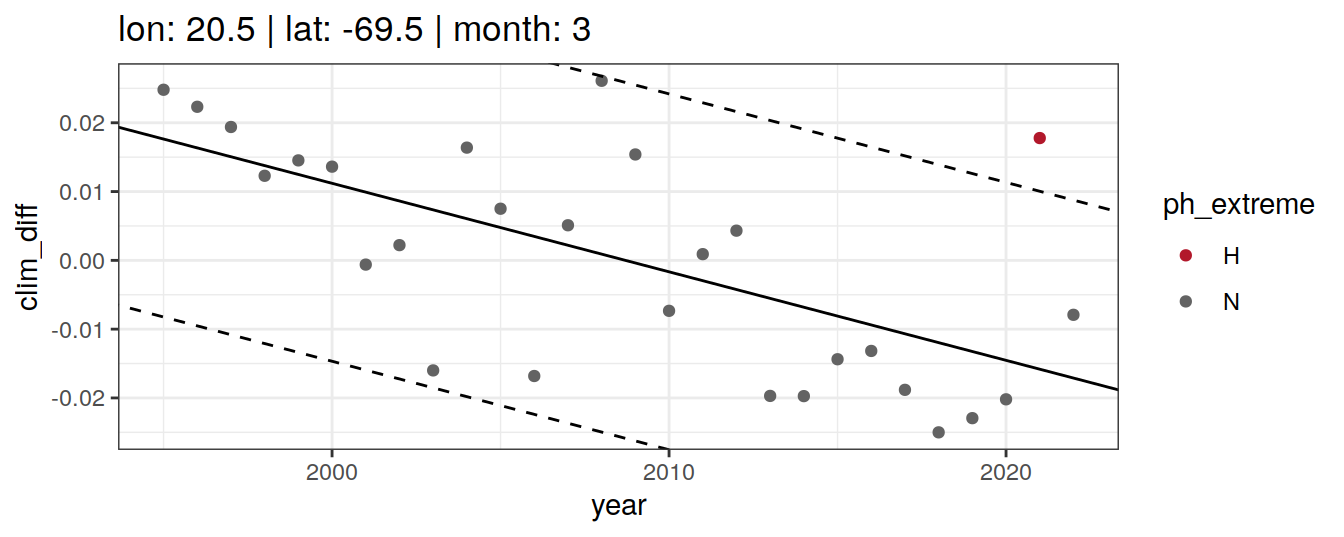
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[4]]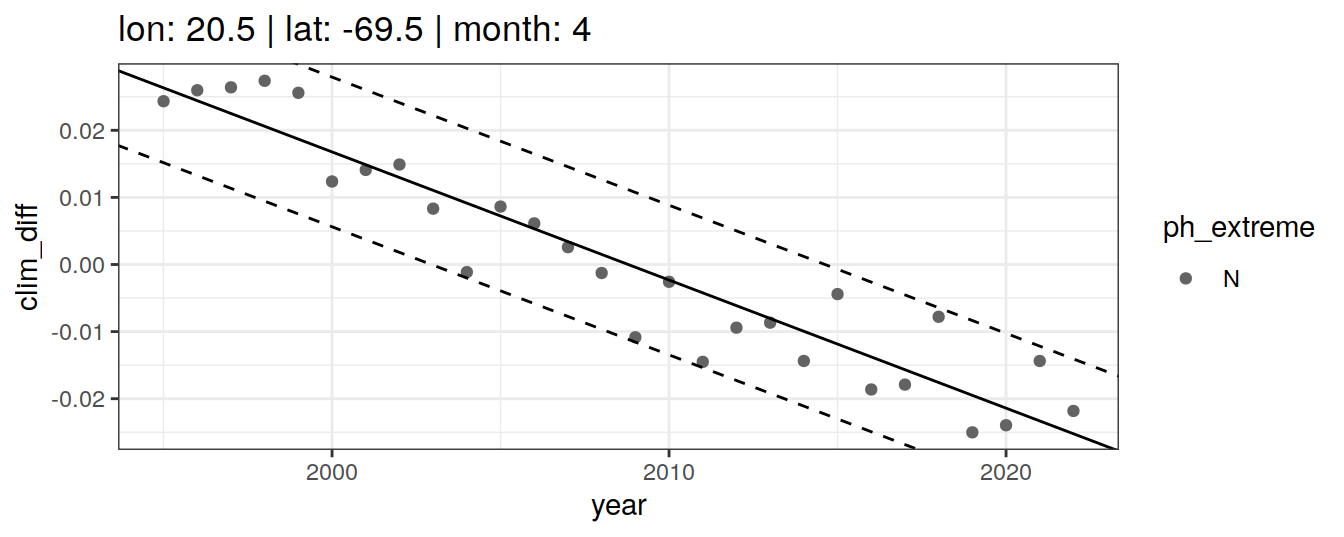
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[5]]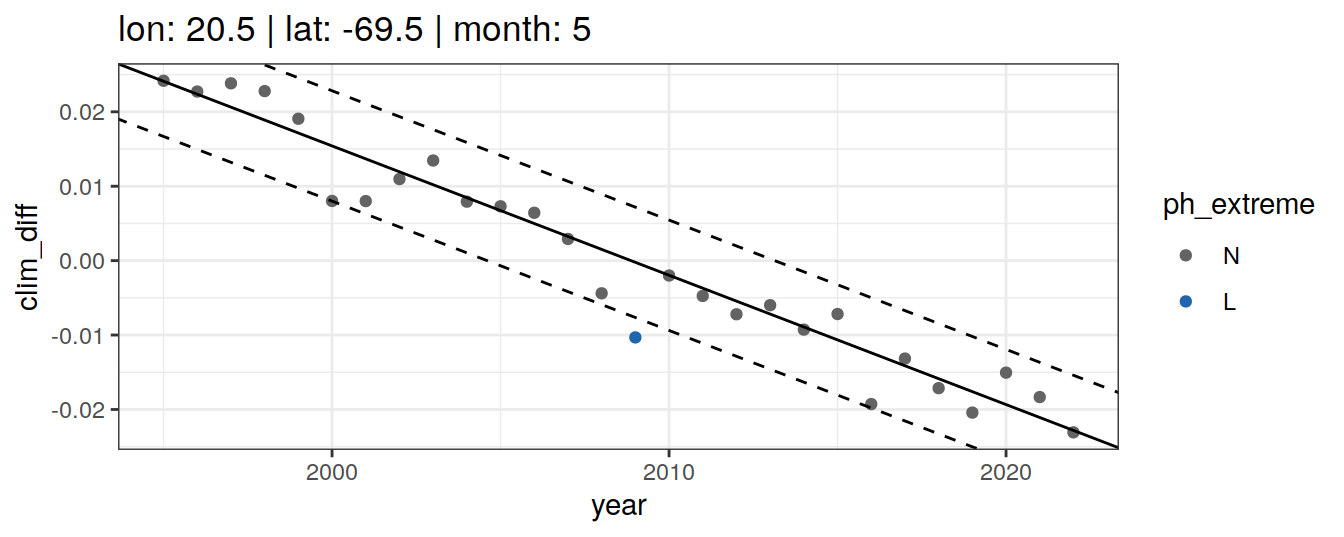
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[6]]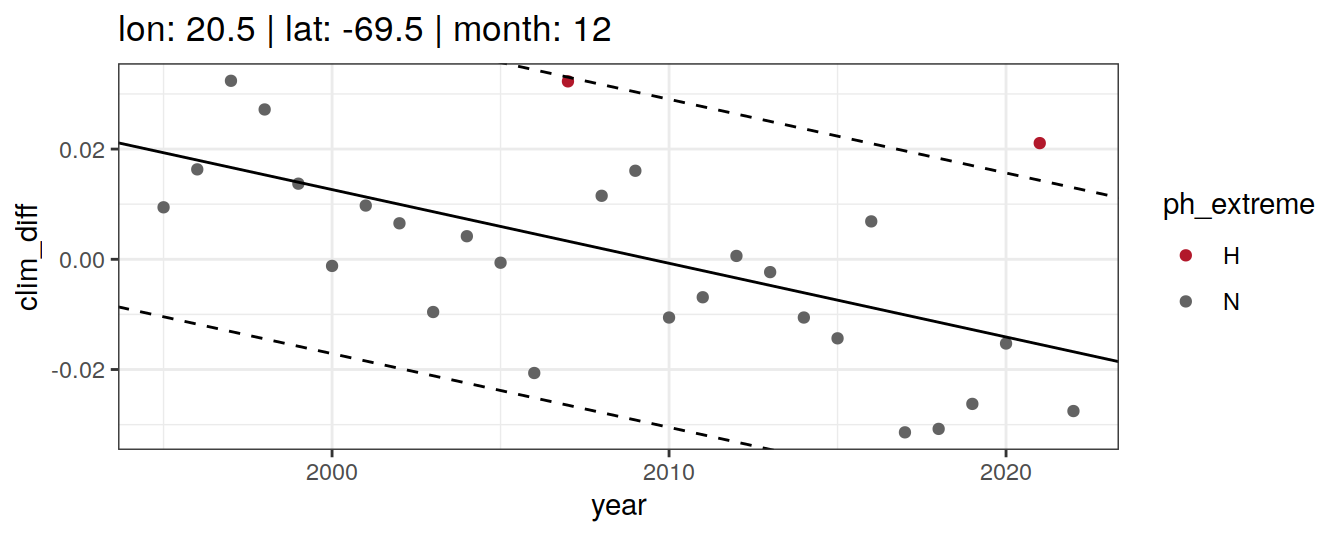
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
[[7]]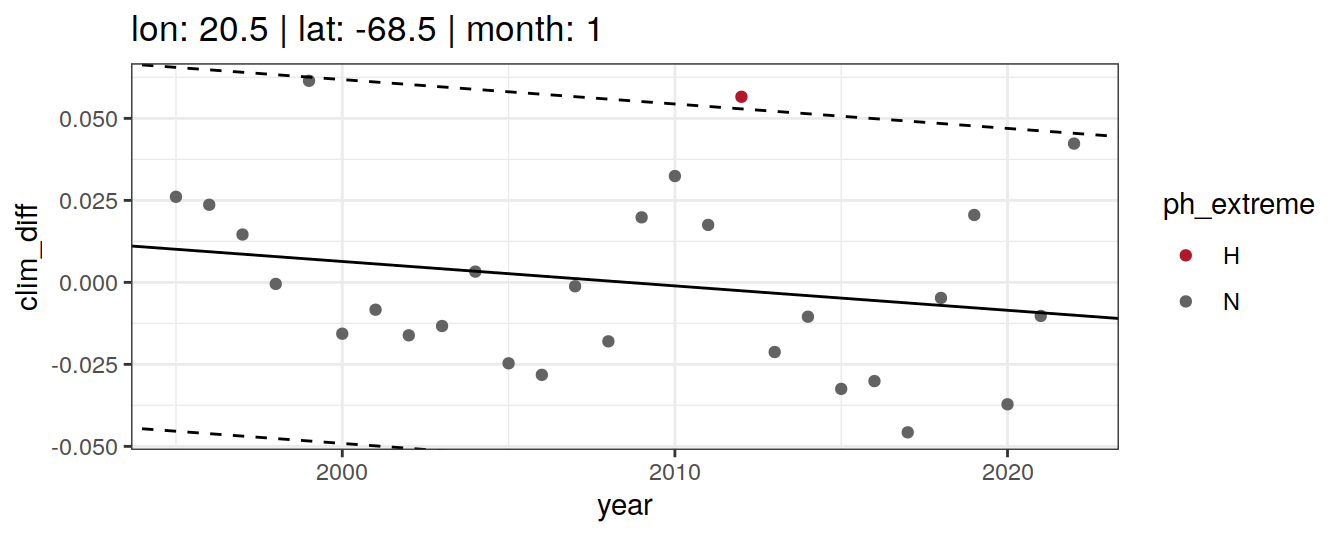
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
[[8]]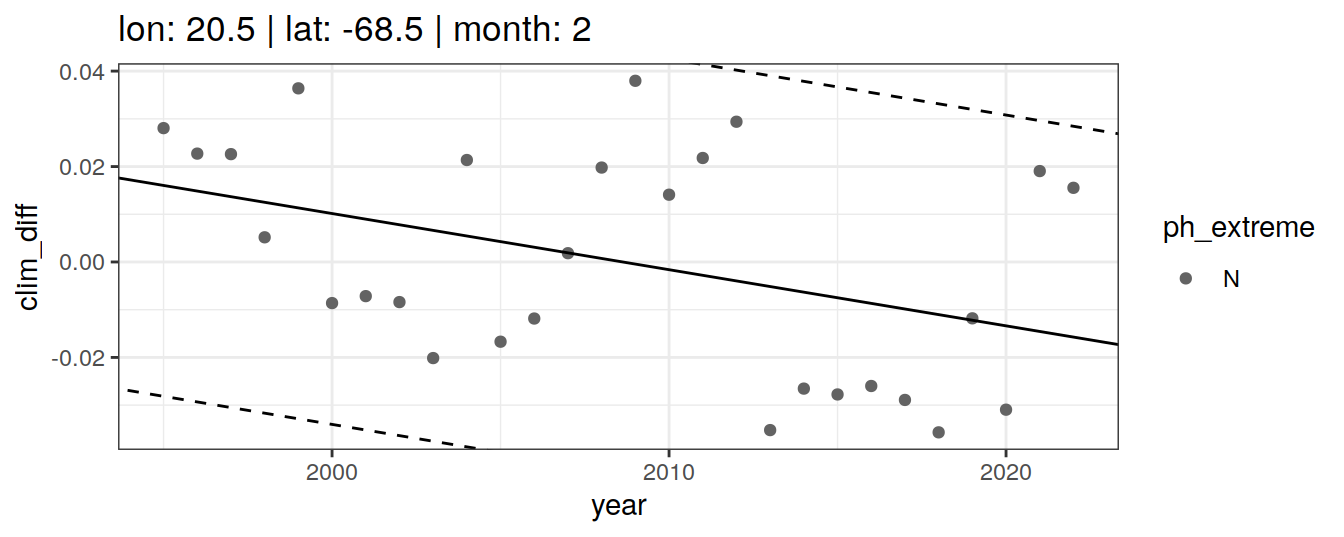
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
Anomaly maps
#use the original lat/lon grid to plot the extreme on a 1x1
#(anomalies detected with 1995-2020 data but mapped only from 2013 to 2020)
OceanSODA_SO_extreme_grid %>%
filter(year >= 2013) %>%
group_split(month) %>%
#head(1) %>%
map(
~map +
geom_tile(data = .x,
aes(x = lon_raw,
y = lat_raw,
fill = ph_extreme))+
scale_fill_manual(values = HNL_colors_map)+
facet_wrap(~year, ncol = 2)+
lims(y = c(-85, -30))+
labs(title = paste('month:', unique(.x$month)),
fill = 'pH')
)[[1]]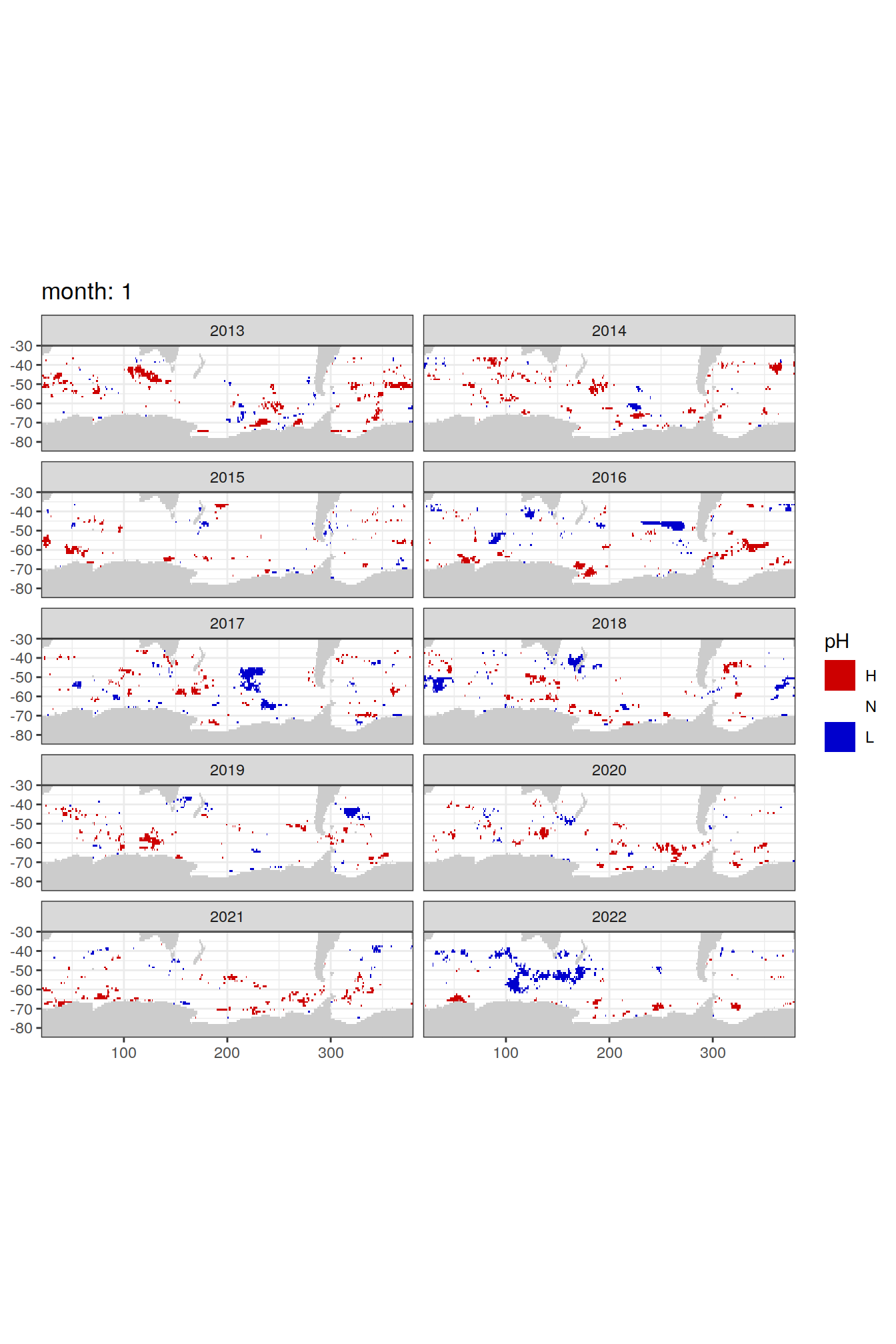
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| da665ab | pasqualina-vonlanthendinenna | 2022-03-01 |
[[2]]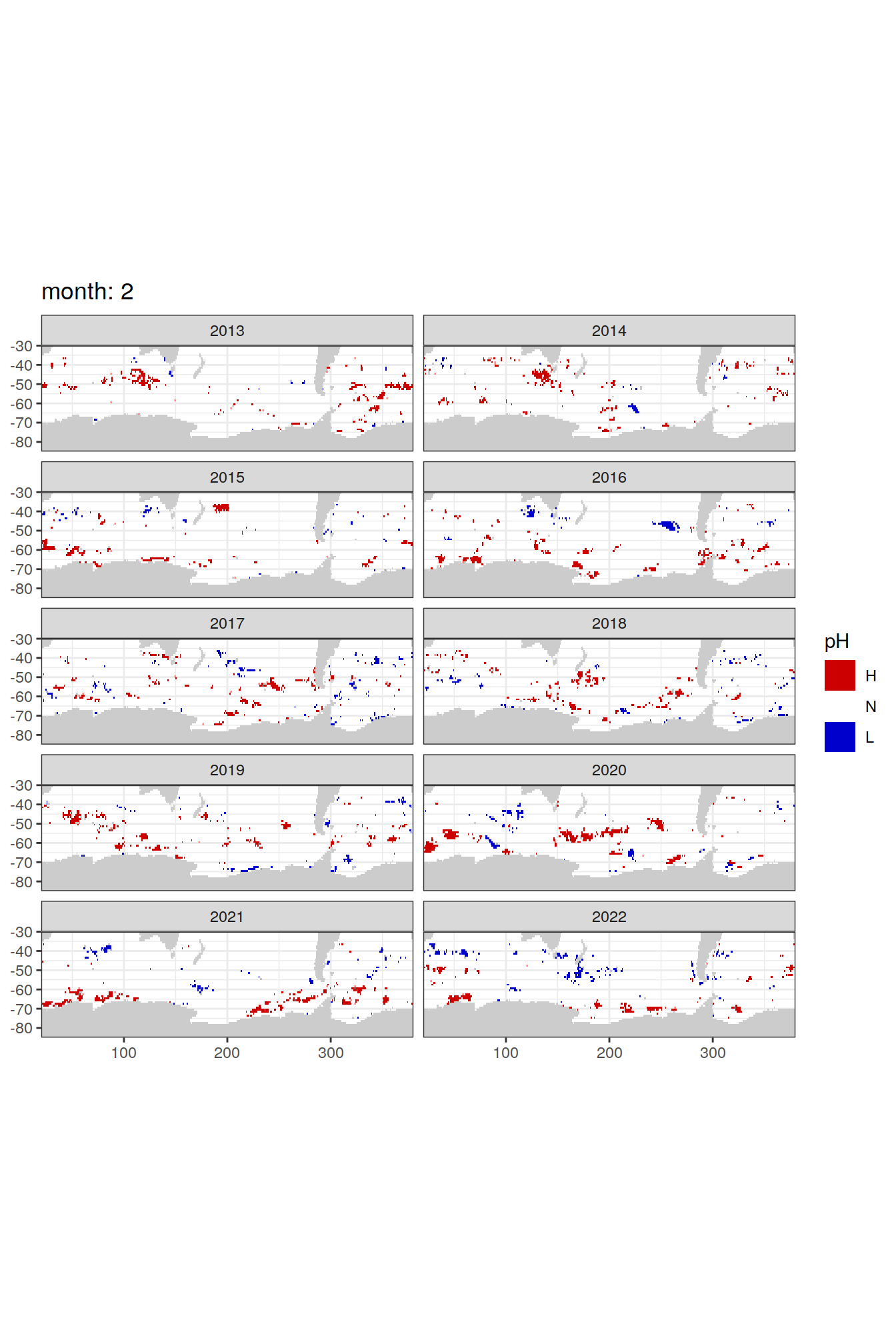
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| da665ab | pasqualina-vonlanthendinenna | 2022-03-01 |
[[3]]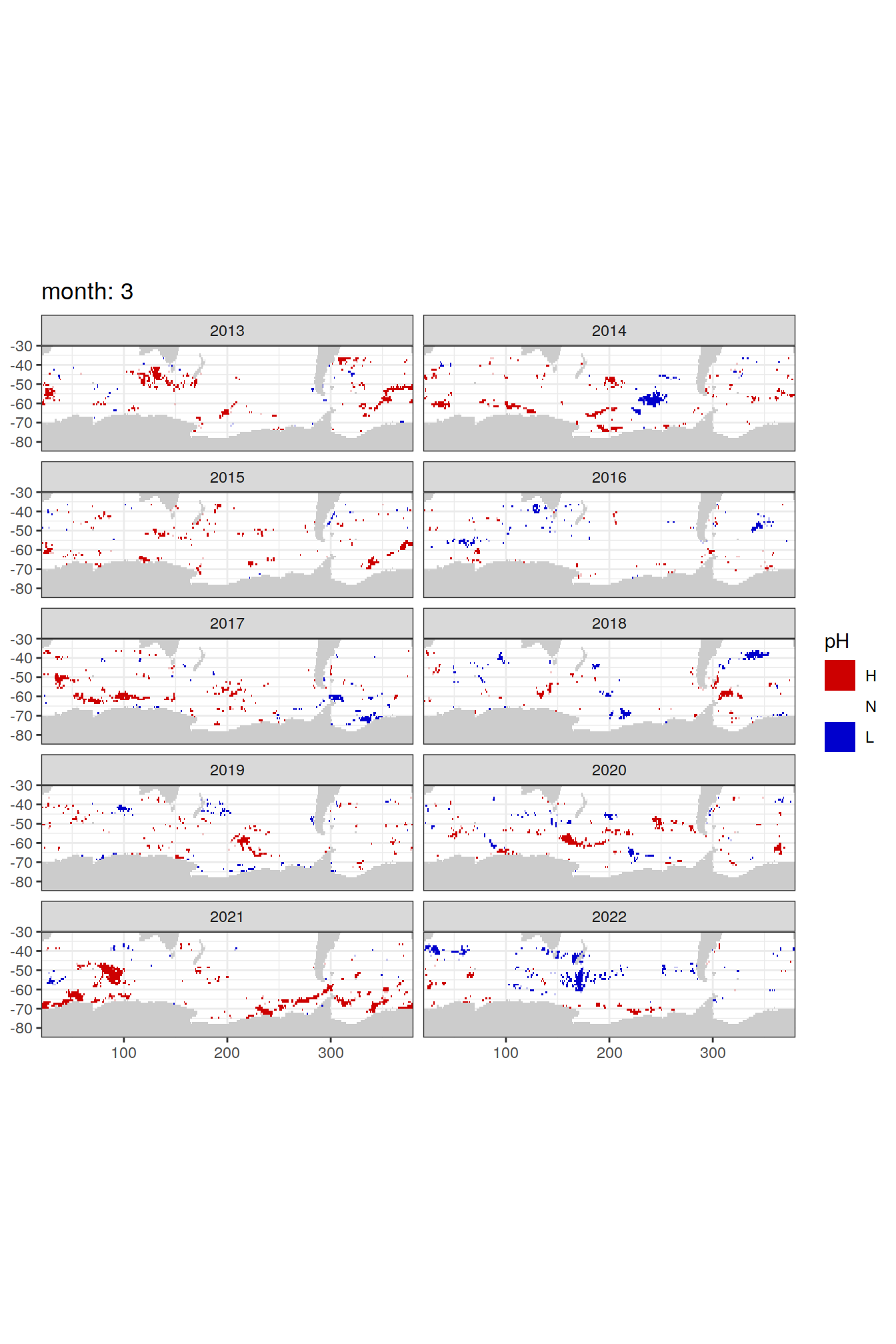
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| da665ab | pasqualina-vonlanthendinenna | 2022-03-01 |
[[4]]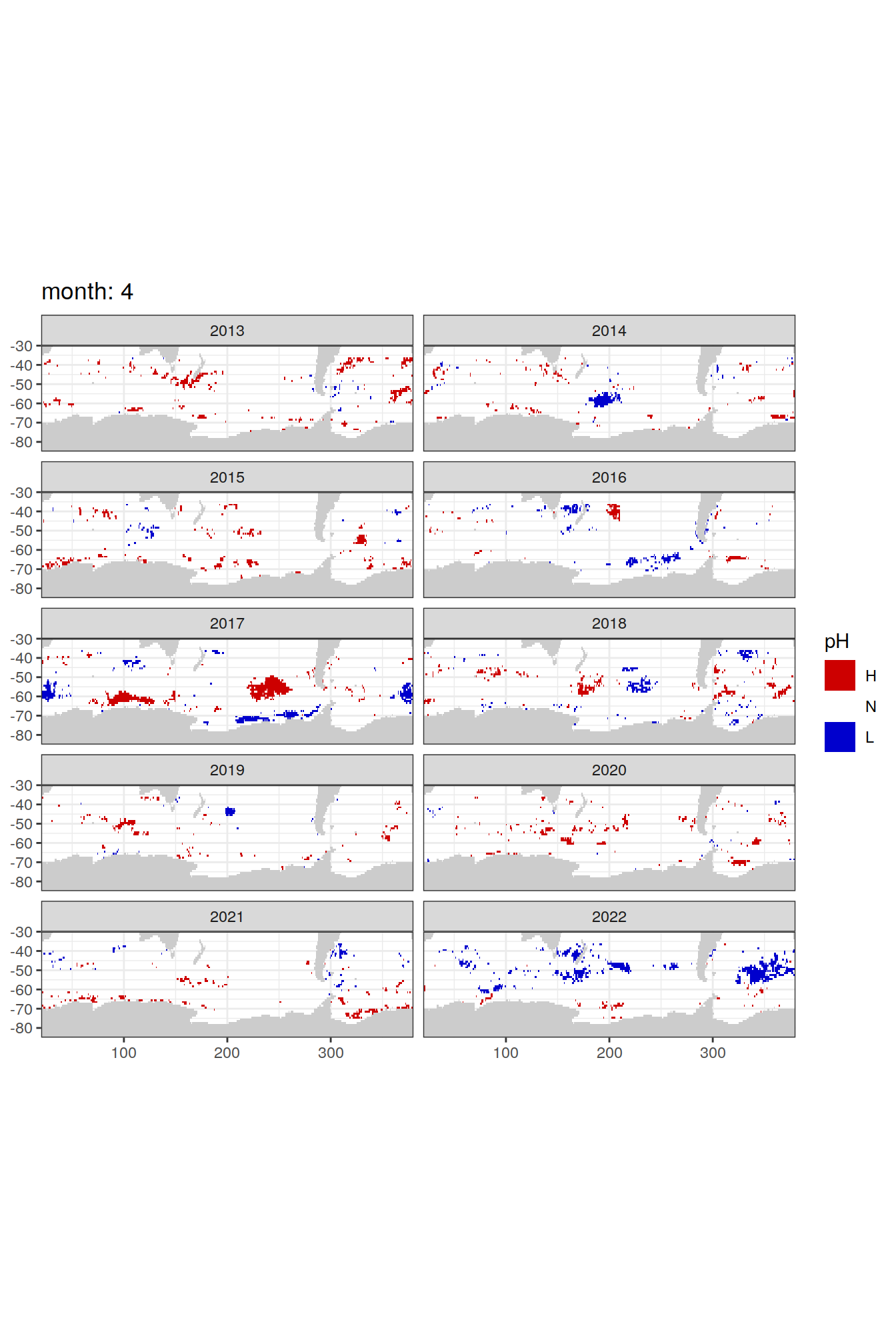
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
[[5]]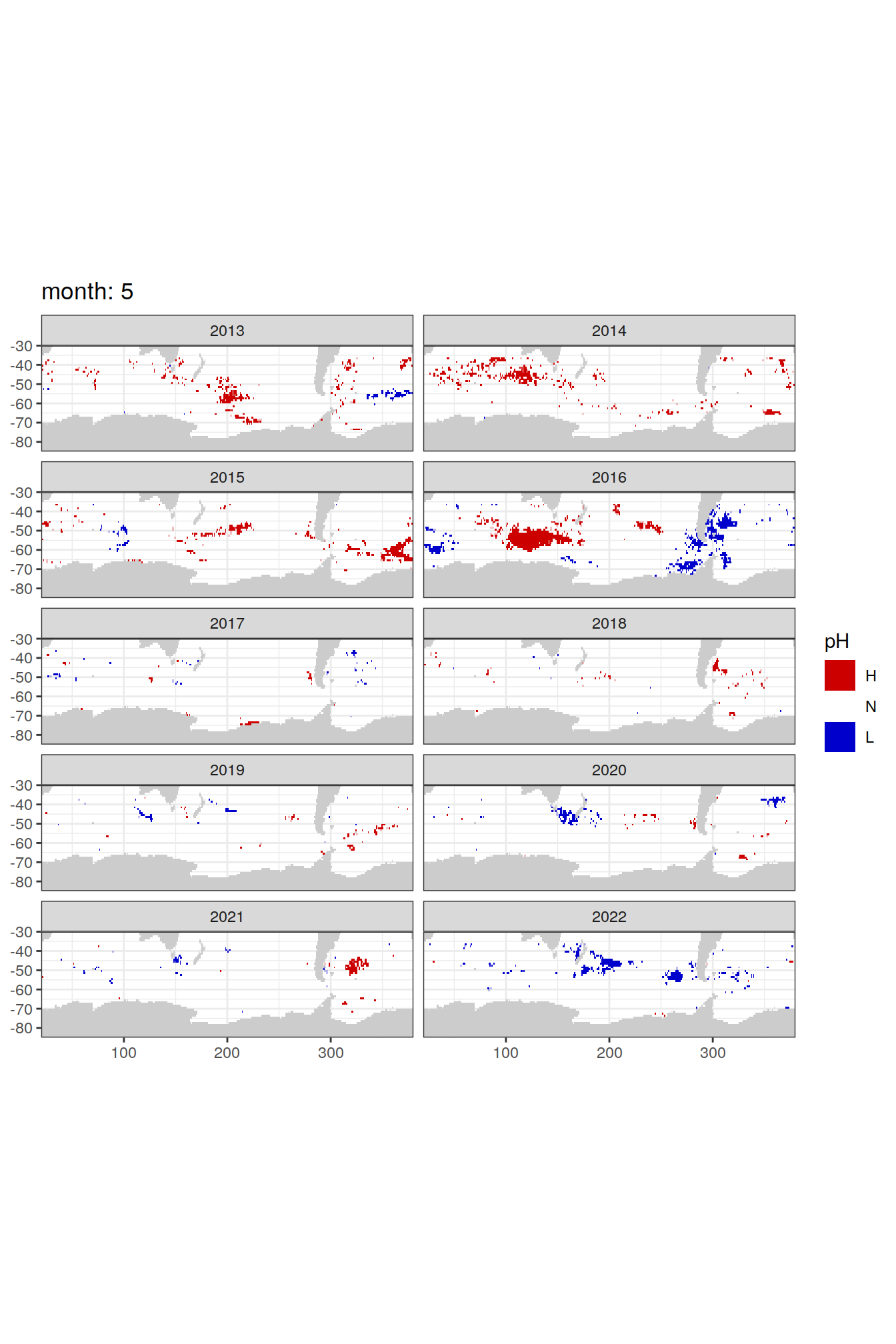
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
[[6]]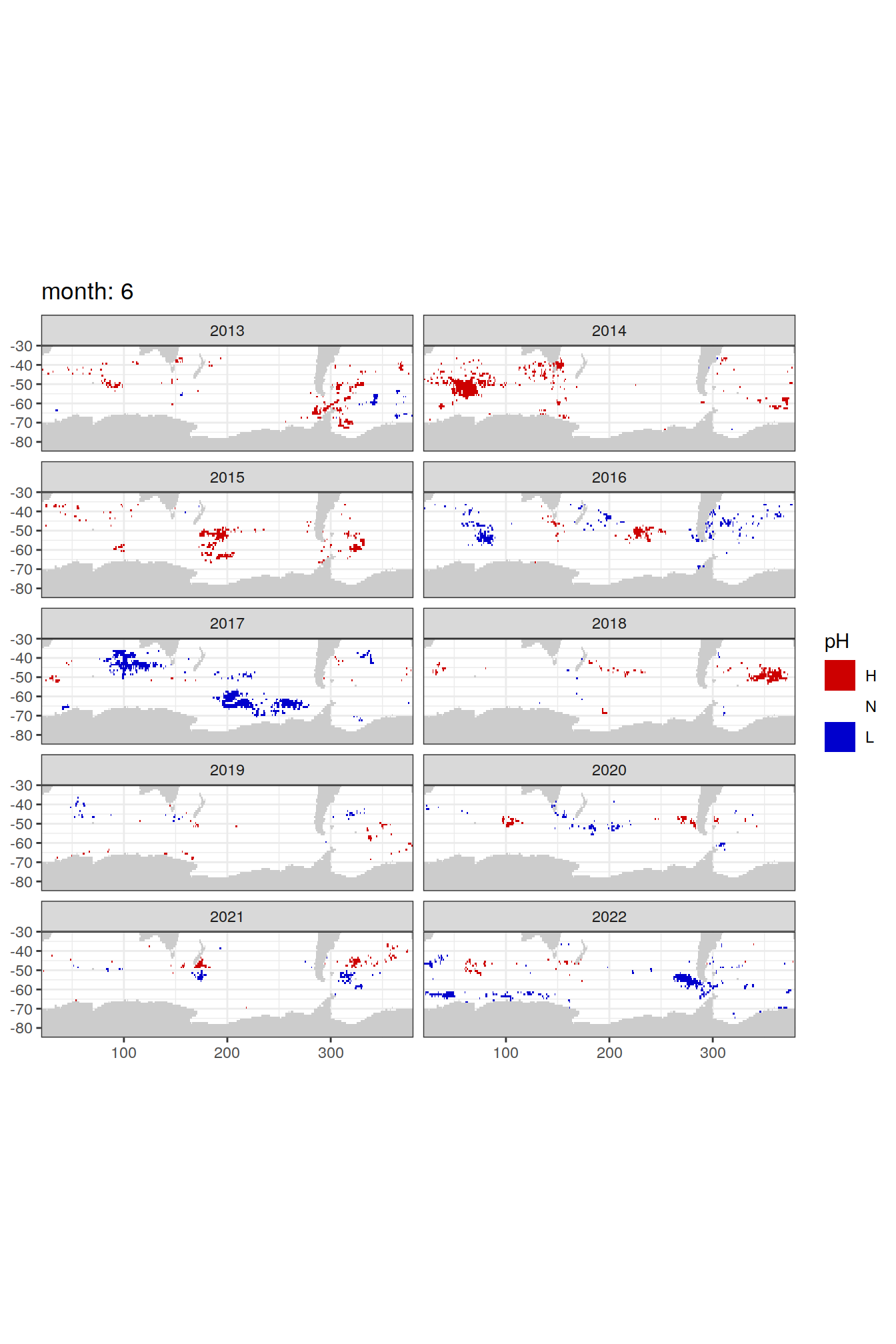
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
[[7]]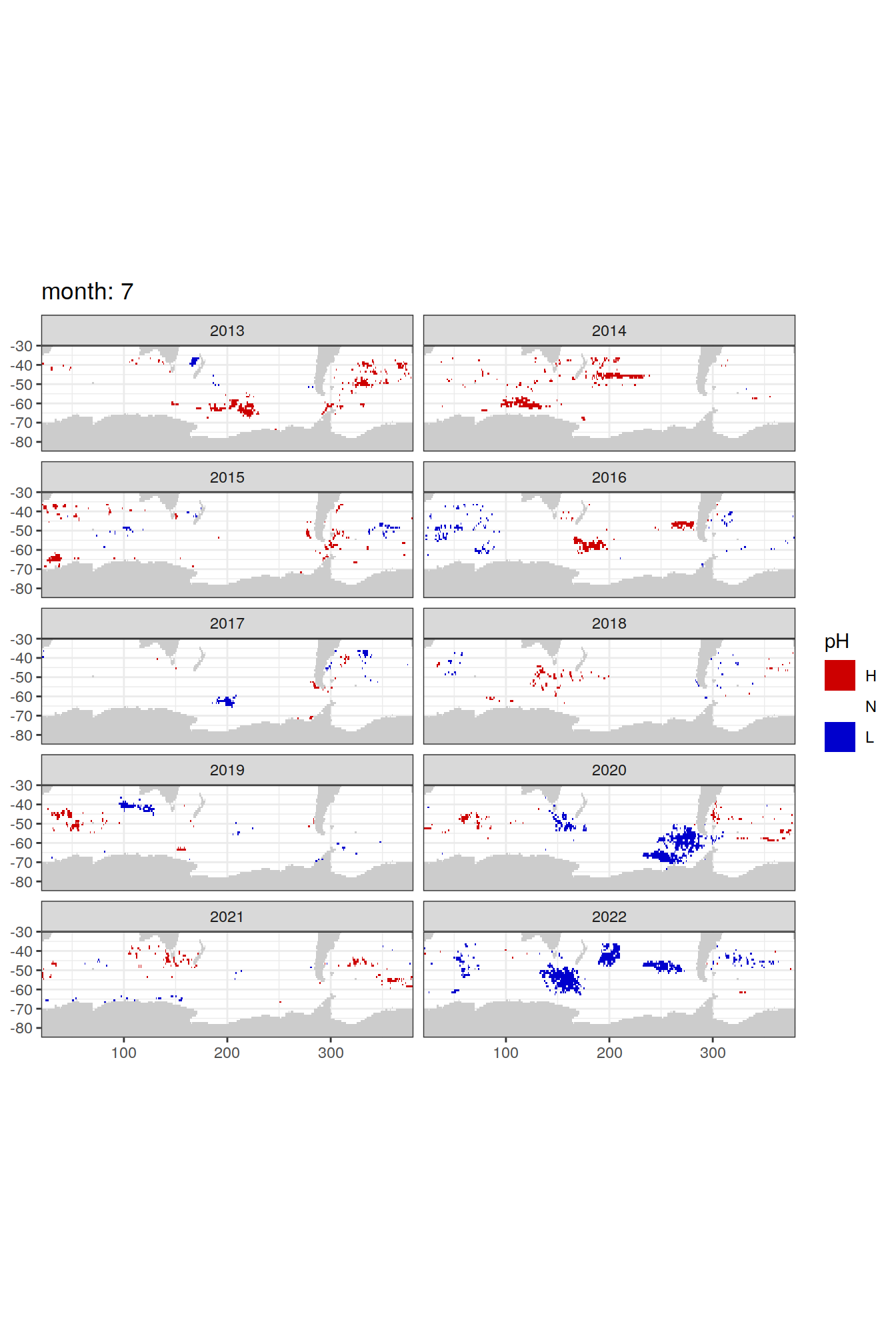
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
[[8]]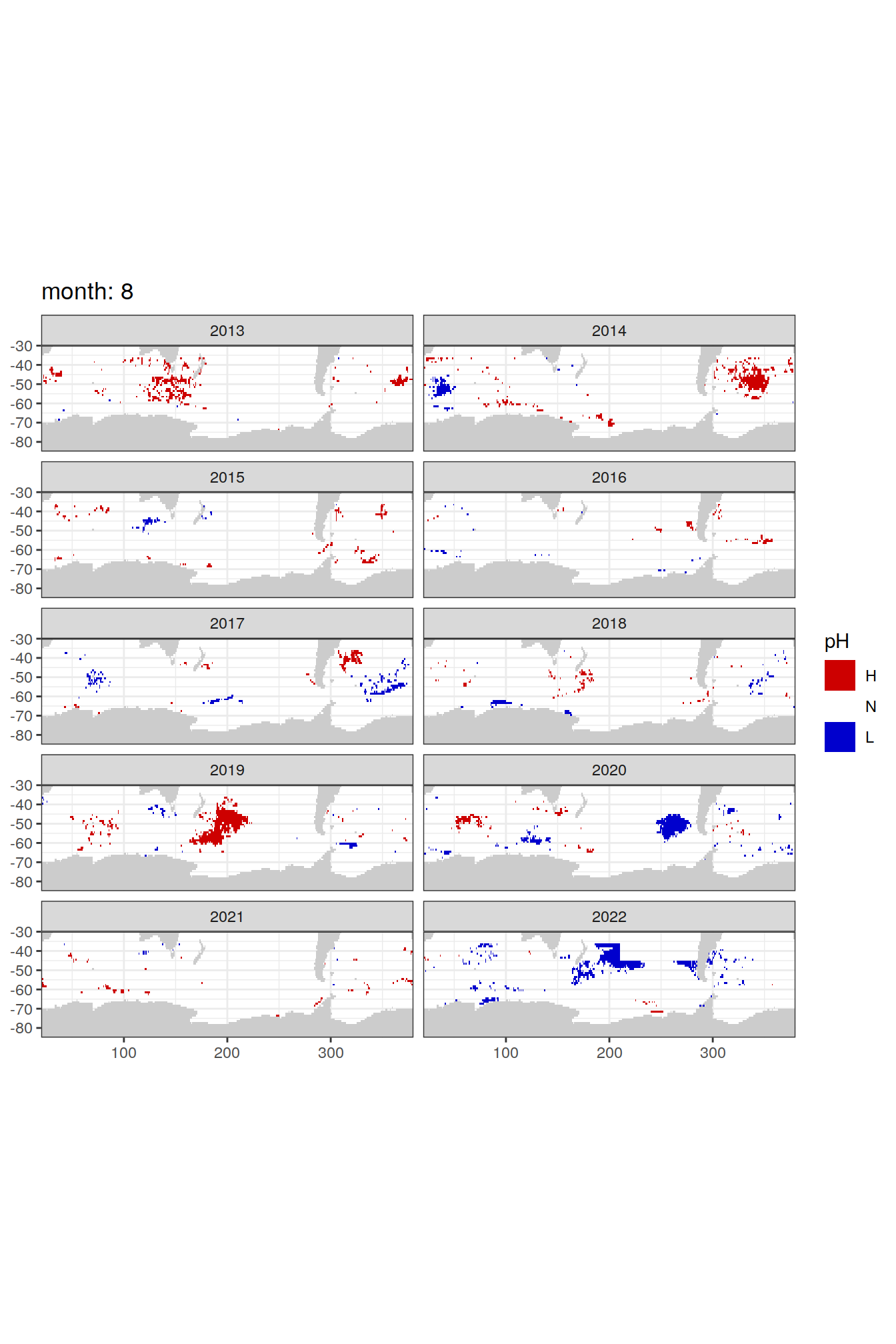
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
[[9]]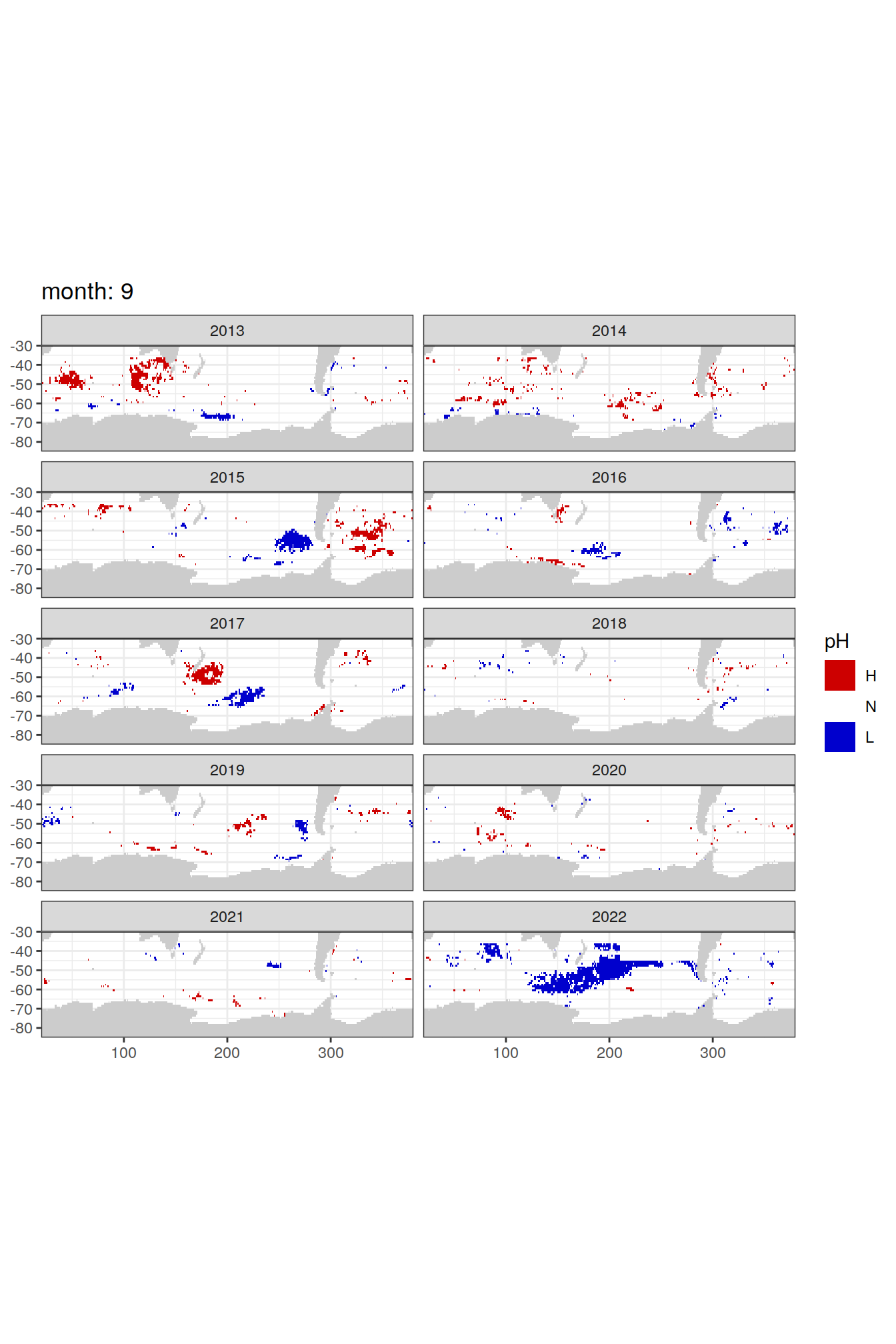
[[10]]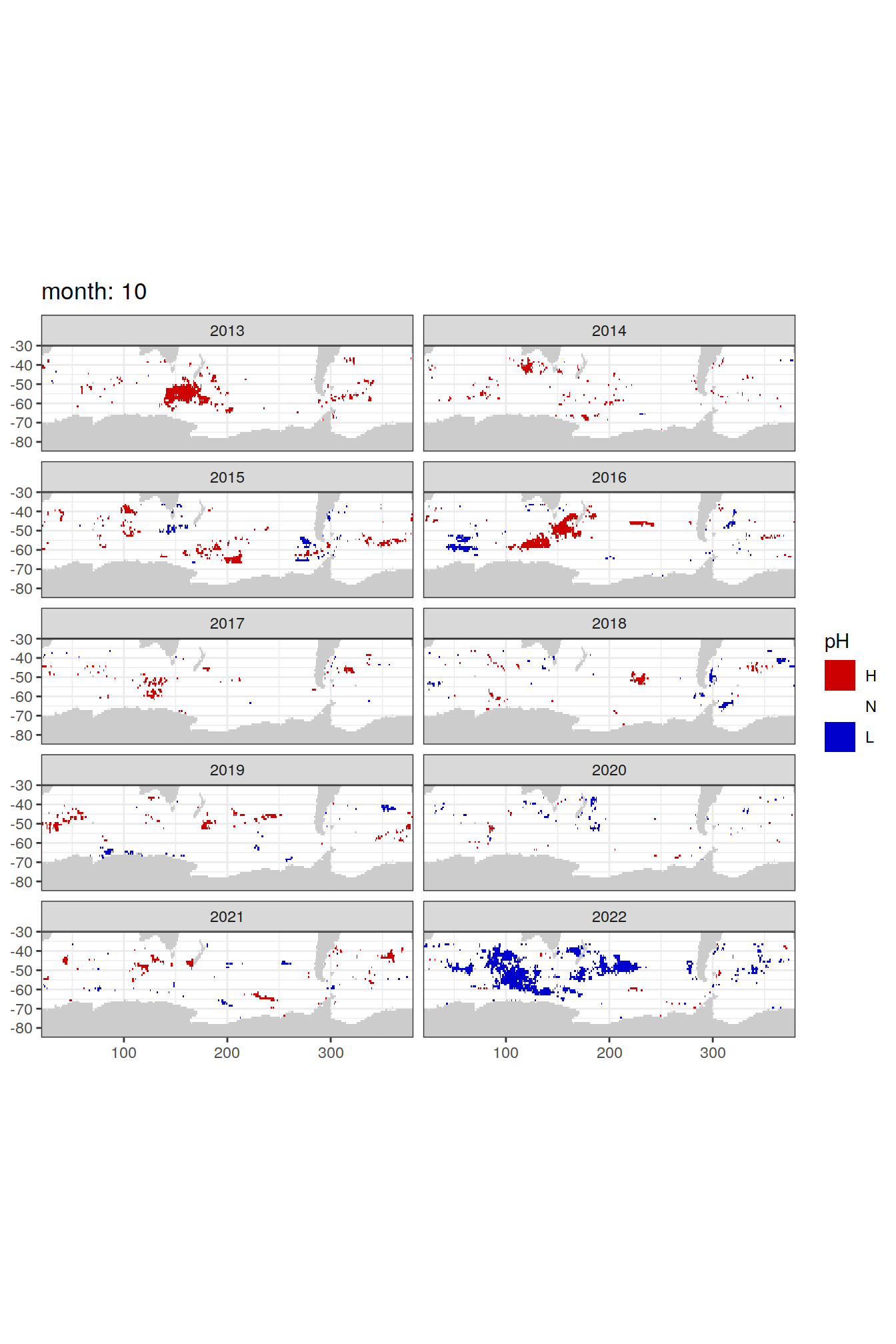
[[11]]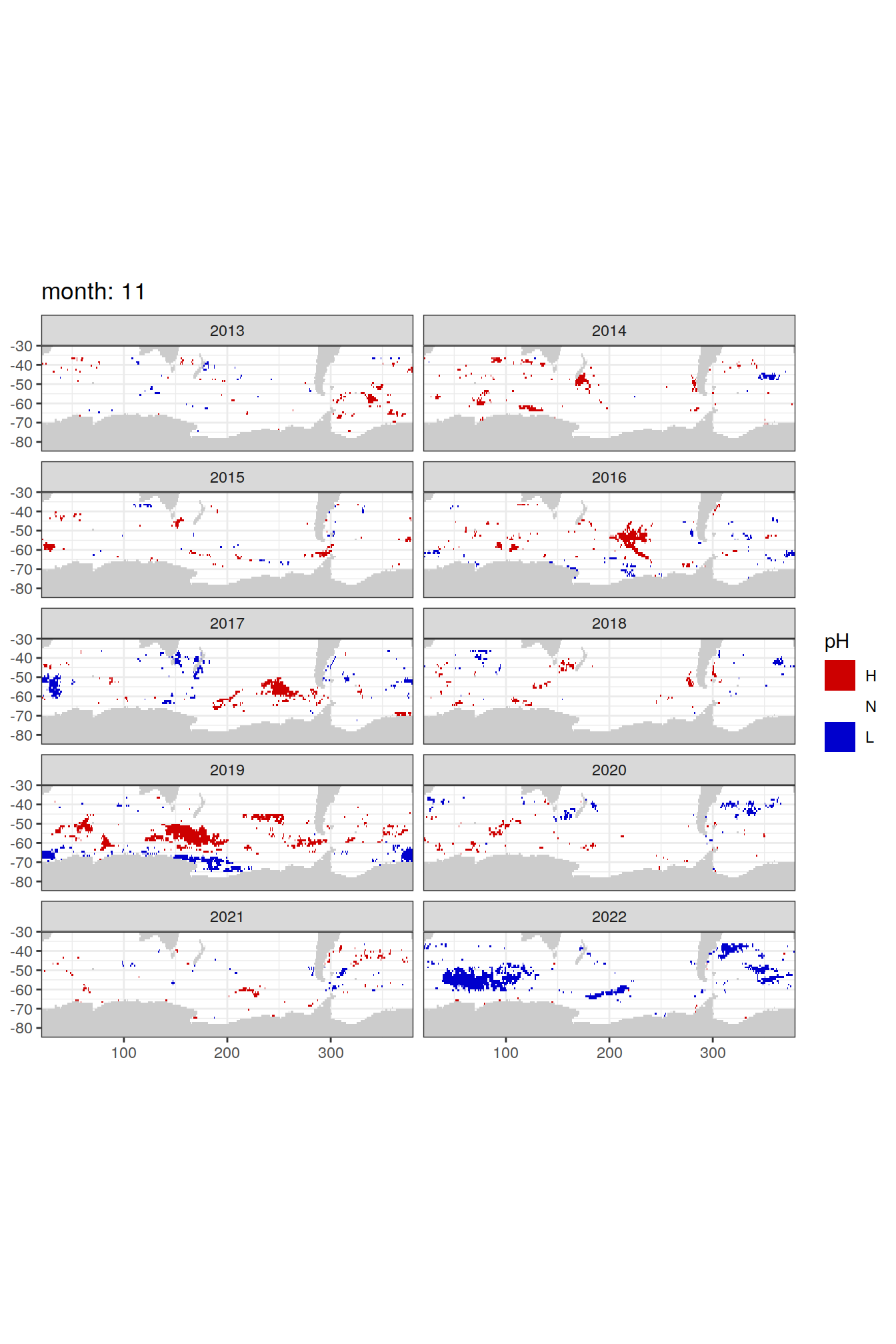
[[12]]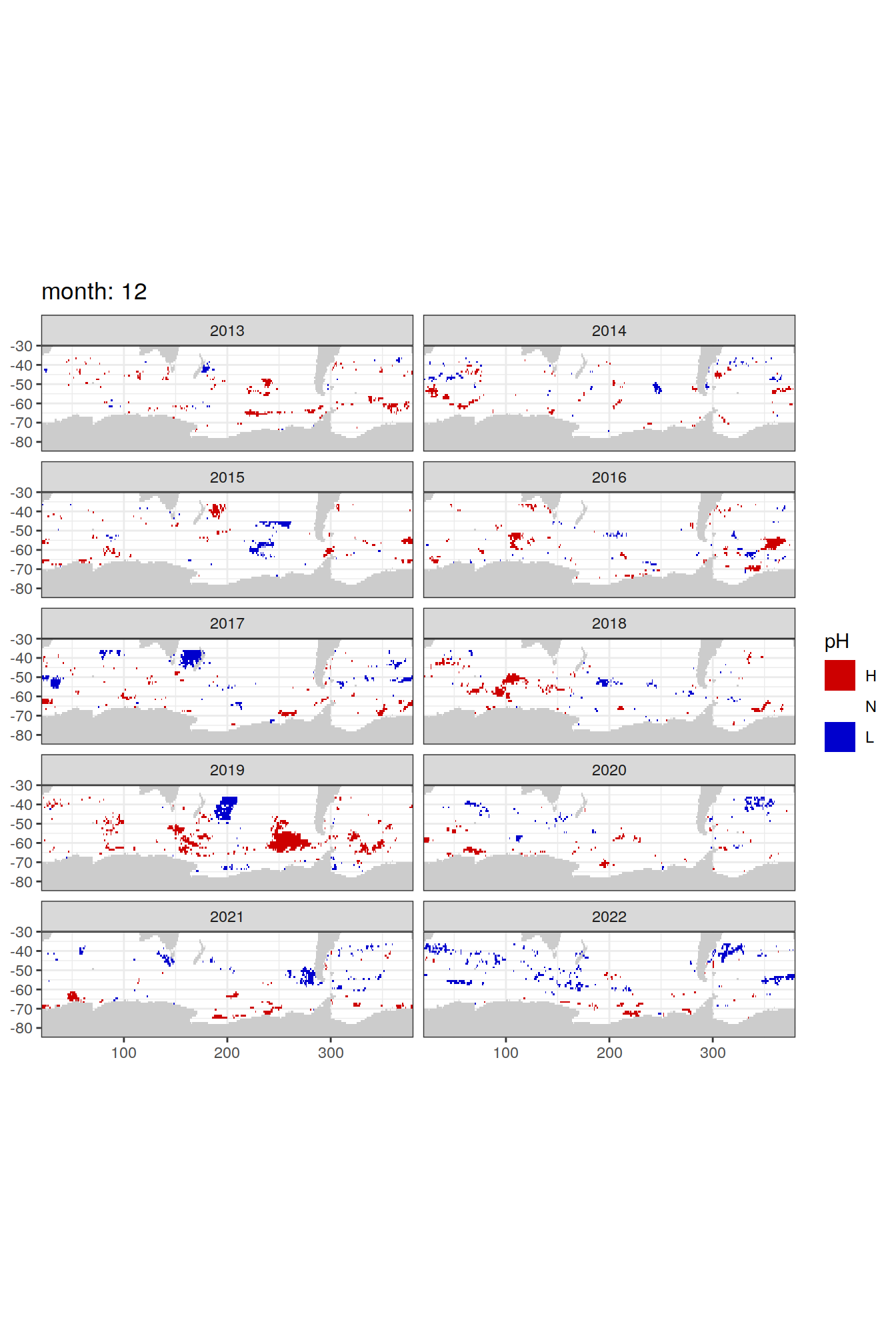
Anomaly time series
# calculate a regional mean pH for each biome, basin, and ph extreme (H/L/N) and plot a timeseries
OceanSODA_SO_extreme_grid %>%
group_by(year, biome_name, basin_AIP, ph_extreme) %>%
summarise(ph_regional = mean(ph_total, na.rm = TRUE)) %>%
ungroup() %>%
ggplot(aes(x = year, y = ph_regional, col = ph_extreme))+
geom_point(size = 0.3)+
geom_line()+
scale_color_manual(values = HNL_colors) +
facet_grid(basin_AIP~biome_name)+
theme(legend.position = 'bottom')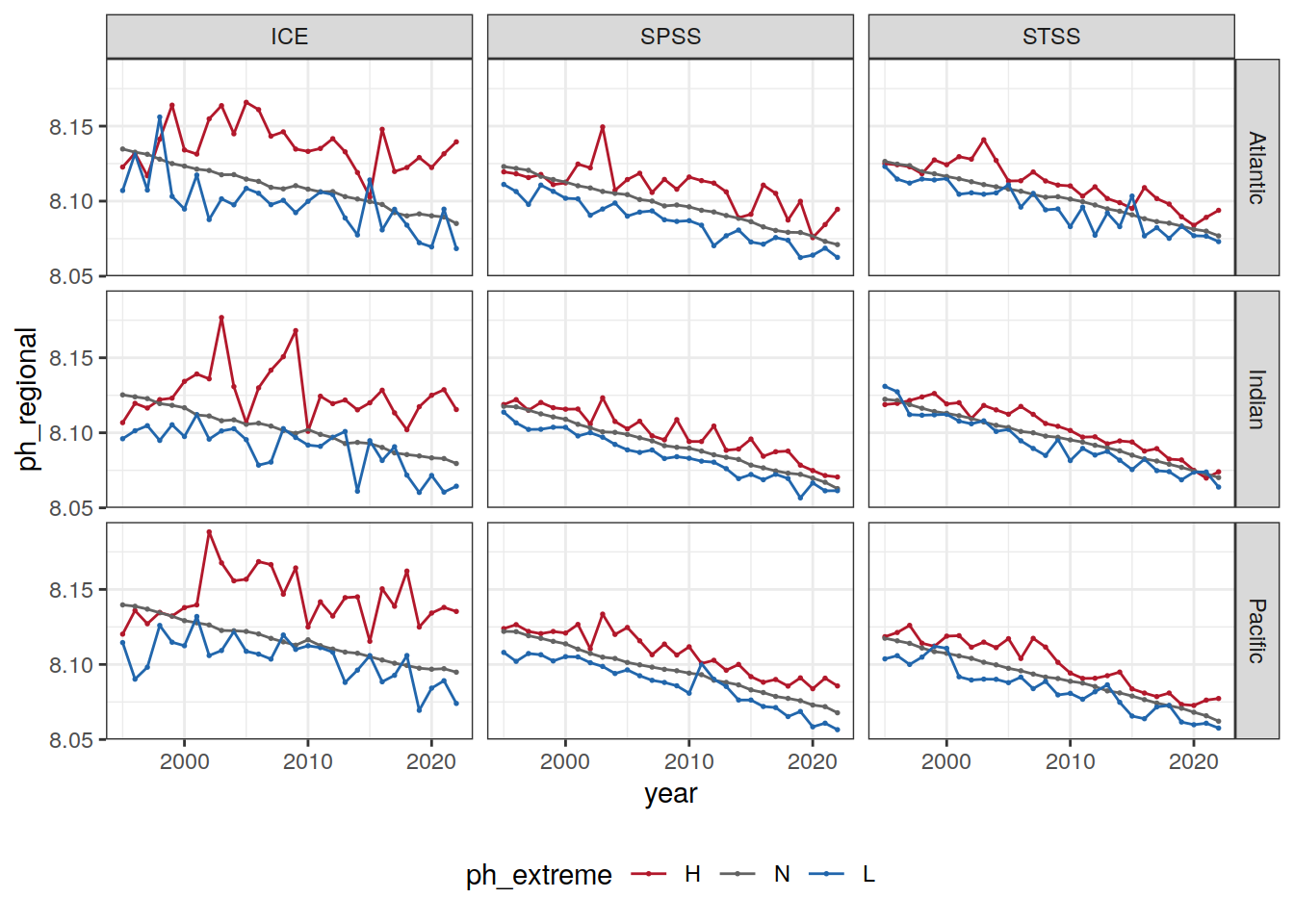
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| 587755e | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
Anomaly histogram
OceanSODA_SO_extreme_grid %>%
ggplot(aes(ph_total, col = ph_extreme)) +
geom_density() +
scale_color_manual(values = HNL_colors) +
facet_grid(basin_AIP ~ biome_name) +
coord_cartesian(xlim = c(8, 8.2)) +
labs(x = 'value',
y = 'density',
col = 'pH anomaly') +
theme(legend.position = 'bottom')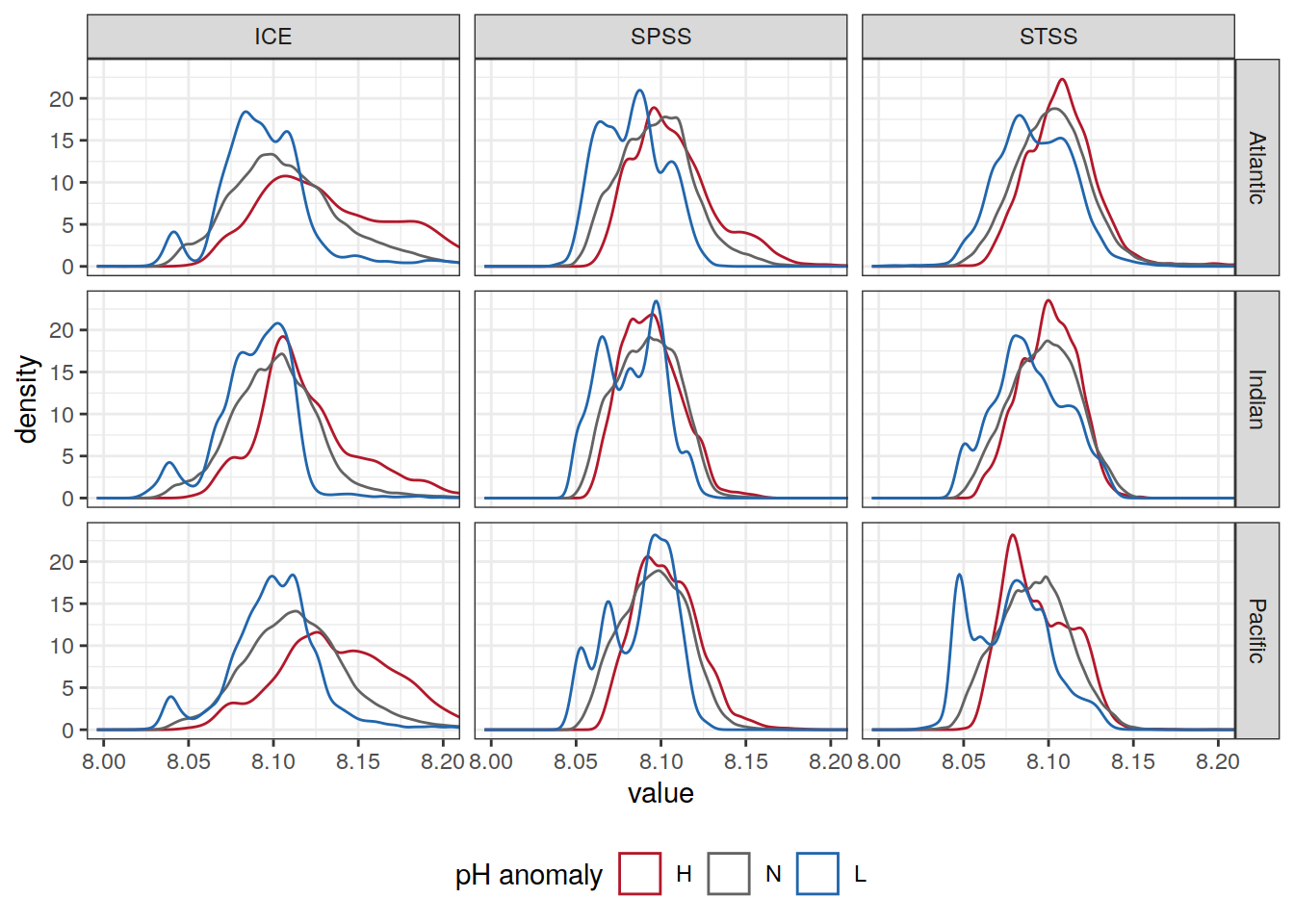
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 8805f99 | pasqualina-vonlanthendinenna | 2022-04-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| 962cdb9 | pasqualina-vonlanthendinenna | 2022-01-25 |
| 6b22341 | pasqualina-vonlanthendinenna | 2022-01-21 |
| 587755e | pasqualina-vonlanthendinenna | 2022-01-21 |
| c96ad5e | pasqualina-vonlanthendinenna | 2022-01-21 |
| 486c9c8 | jens-daniel-mueller | 2022-01-07 |
Argo
Grid reduction
# Note: While reducing lon x lat grid,
# we keep the original number of observations
# full_argo_2x2 <- full_argo %>%
# mutate(
# lat_raw = lat,
# lon_raw = lon,
# lat = cut(lat, seq(-90, 90, 2), seq(-89, 89, 2)),
# lat = as.numeric(as.character(lat)),
# lon = cut(lon, seq(20, 380, 2), seq(21, 379, 2)),
# lon = as.numeric(as.character(lon))) # re-grid to 2x2
full_argo <- full_argo %>%
mutate(
lat_raw = lat,
lon_raw = lon)Apply region masks
# keep only Southern Ocean argo data
full_argo_SO <- inner_join(full_argo, nm_biomes)
# add in basin separations
full_argo_SO <- inner_join(full_argo_SO, basinmask)Join OceanSODA
# rename OceanSODA columns
OceanSODA_SO_extreme_grid <- OceanSODA_SO_extreme_grid %>%
select(-c(lon, lat)) %>%
rename(OceanSODA_ph = ph_total,
lon = lon_raw,
lat = lat_raw) %>%
filter(year >= 2013)
# combine the argo profile data to the surface extreme data
profile_extreme <- inner_join(
full_argo %>%
select(
file_id,
year,
month,
date,
lon,
lat,
depth,
pH
),
OceanSODA_SO_extreme_grid %>%
select(
year,
month,
date,
lon,
lat,
OceanSODA_ph,
ph_extreme,
clim_ph,
clim_diff,
biome_name,
basin_AIP
)
)
# profile_extreme <- profile_extreme %>%
# unite('platform_cycle', platform_number:cycle_number, sep = '_', remove = FALSE)Location of Argo profiles
OceanSODA_SO_extreme_grid %>%
group_split(month) %>%
# head(1) %>%
map(
~ map +
geom_tile(
data = .x,
aes(x = lon,
y = lat,
fill = ph_extreme),
alpha = 0.5
) +
scale_fill_manual(values = HNL_colors_map) +
new_scale_fill() +
geom_tile(
data = profile_extreme %>%
distinct(lon, lat, file_id, year, month),
aes(
x = lon,
y = lat,
fill = 'argo\nprofiles',
height = 1,
width = 1
),
alpha = 0.5
) +
scale_fill_manual(values = "springgreen4",
name = "") +
facet_wrap( ~ year, ncol = 1) +
lims(y = c(-85, -30)) +
labs(title = paste('month:', unique(.x$month)))
)[[1]]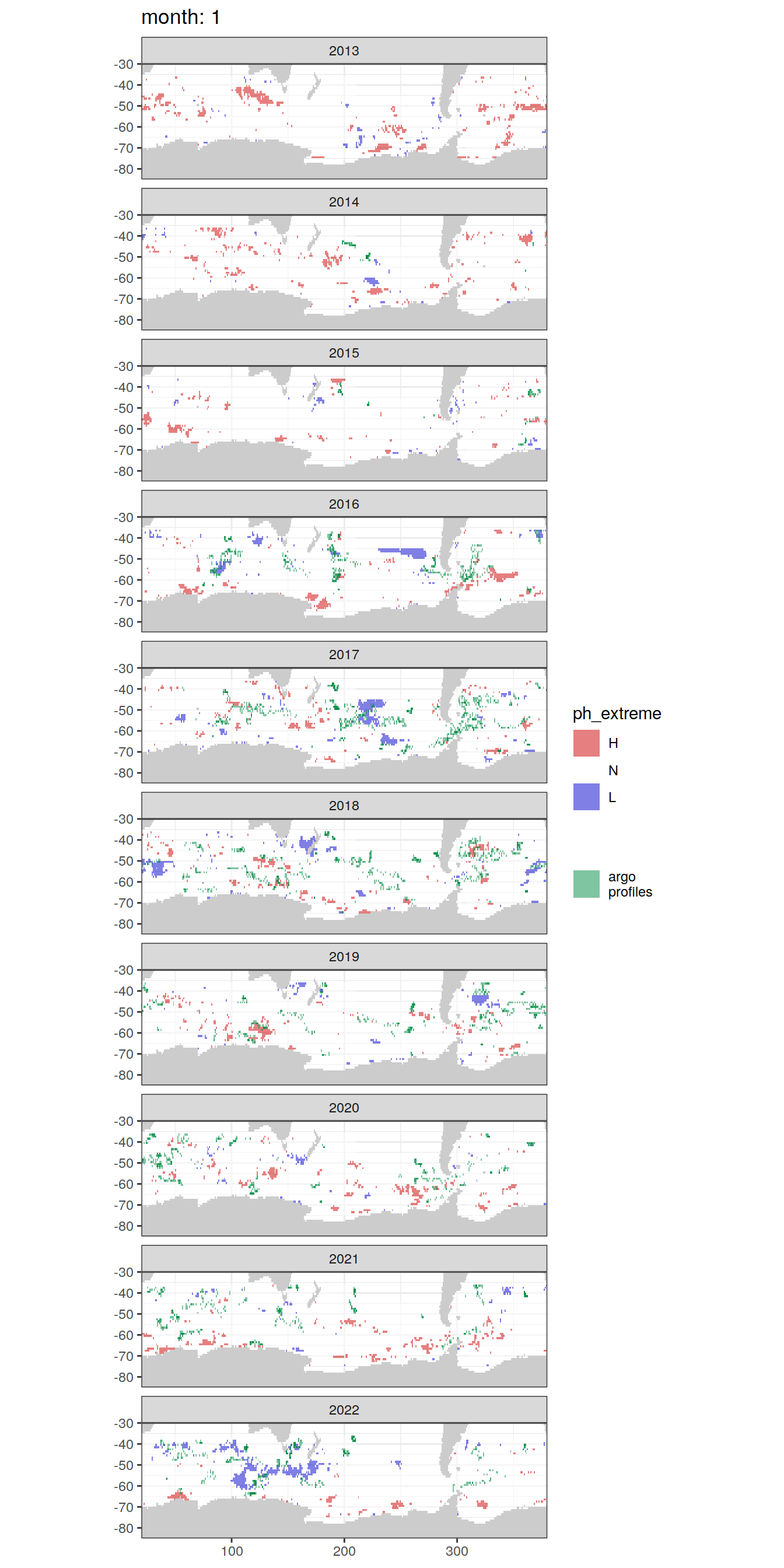
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| ca30beb | pasqualina-vonlanthendinenna | 2022-05-05 |
| 4cf88e4 | pasqualina-vonlanthendinenna | 2022-05-05 |
| f46b9da | pasqualina-vonlanthendinenna | 2022-05-05 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[2]]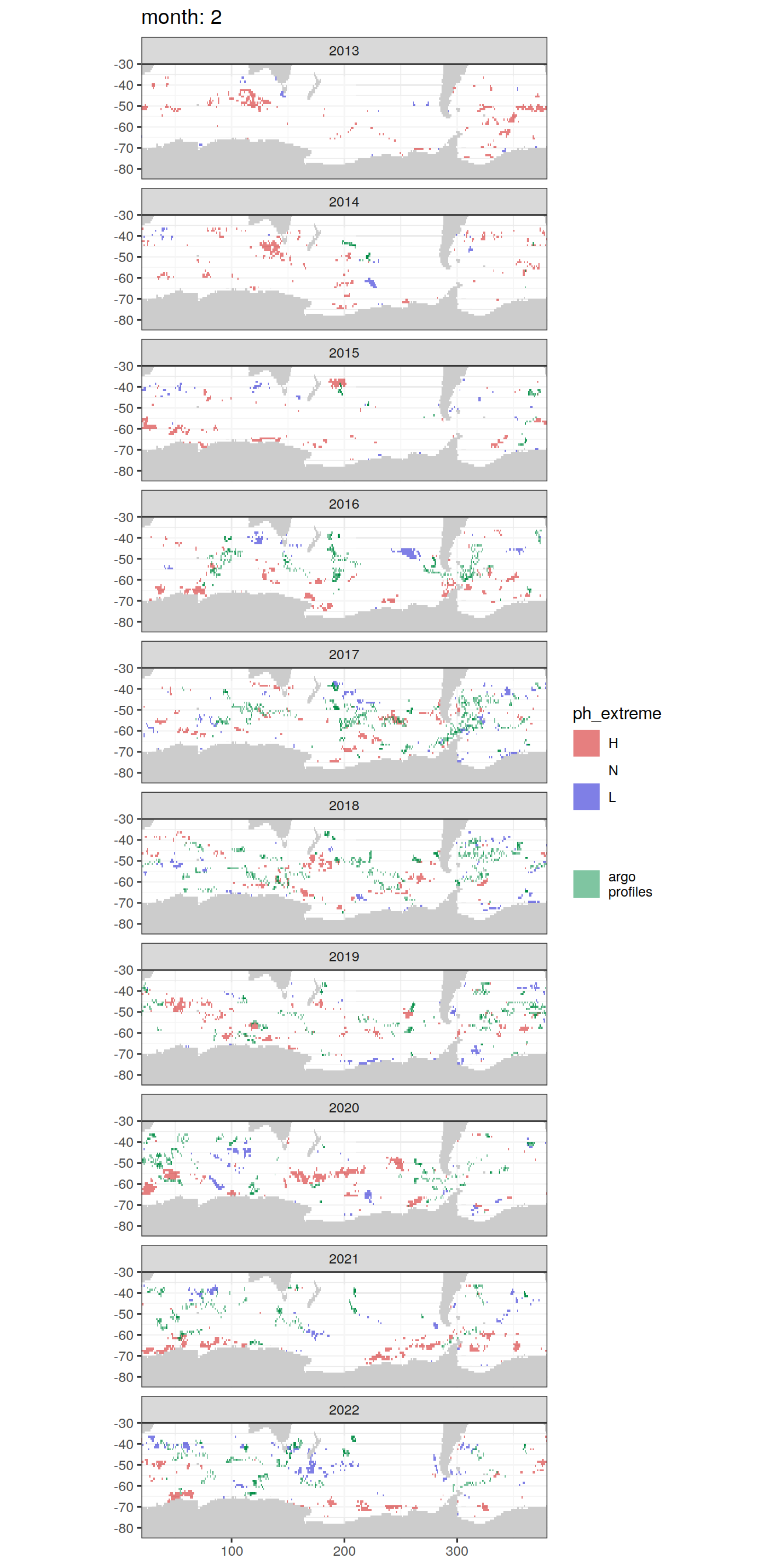
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[3]]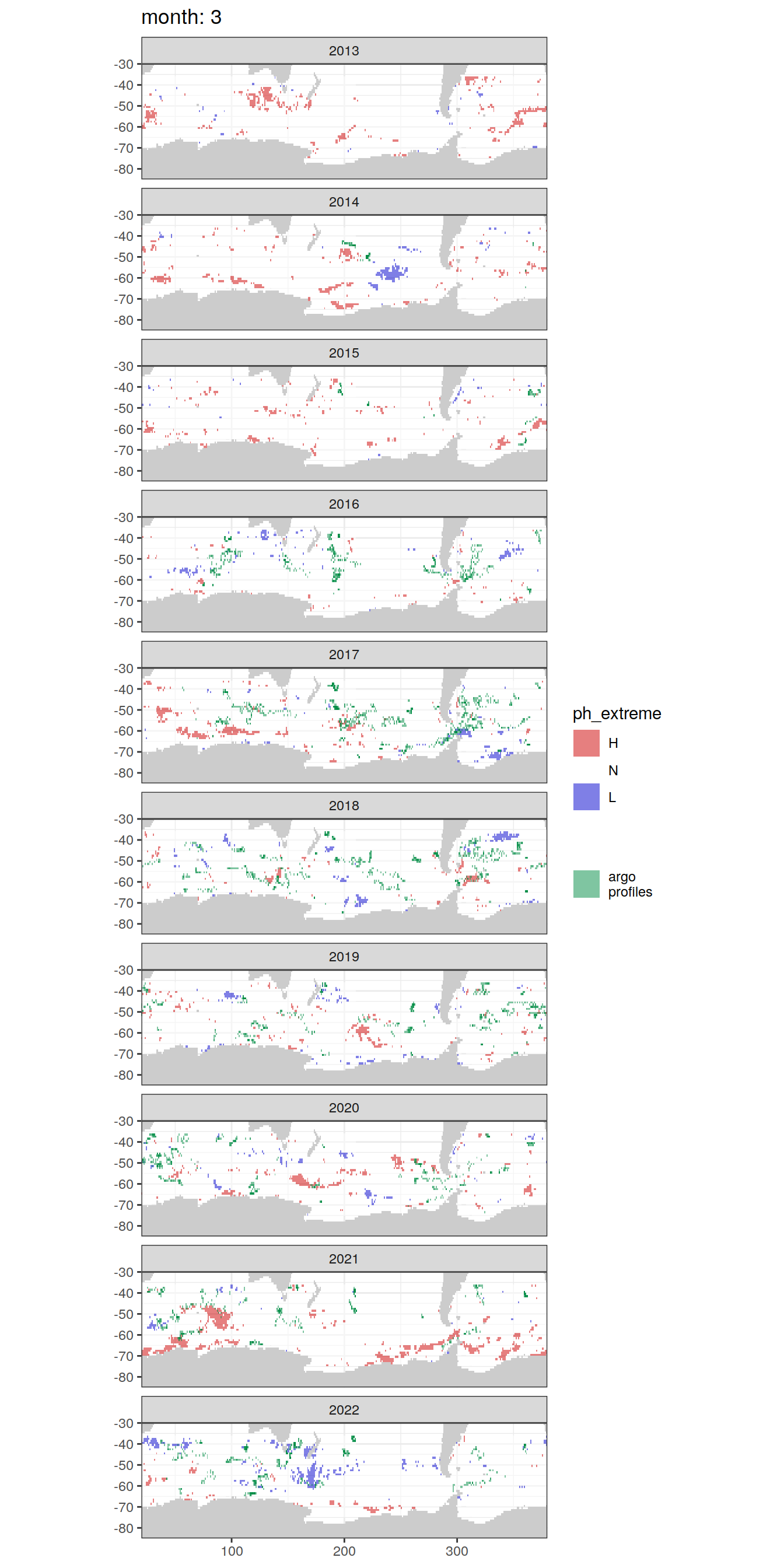
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[4]]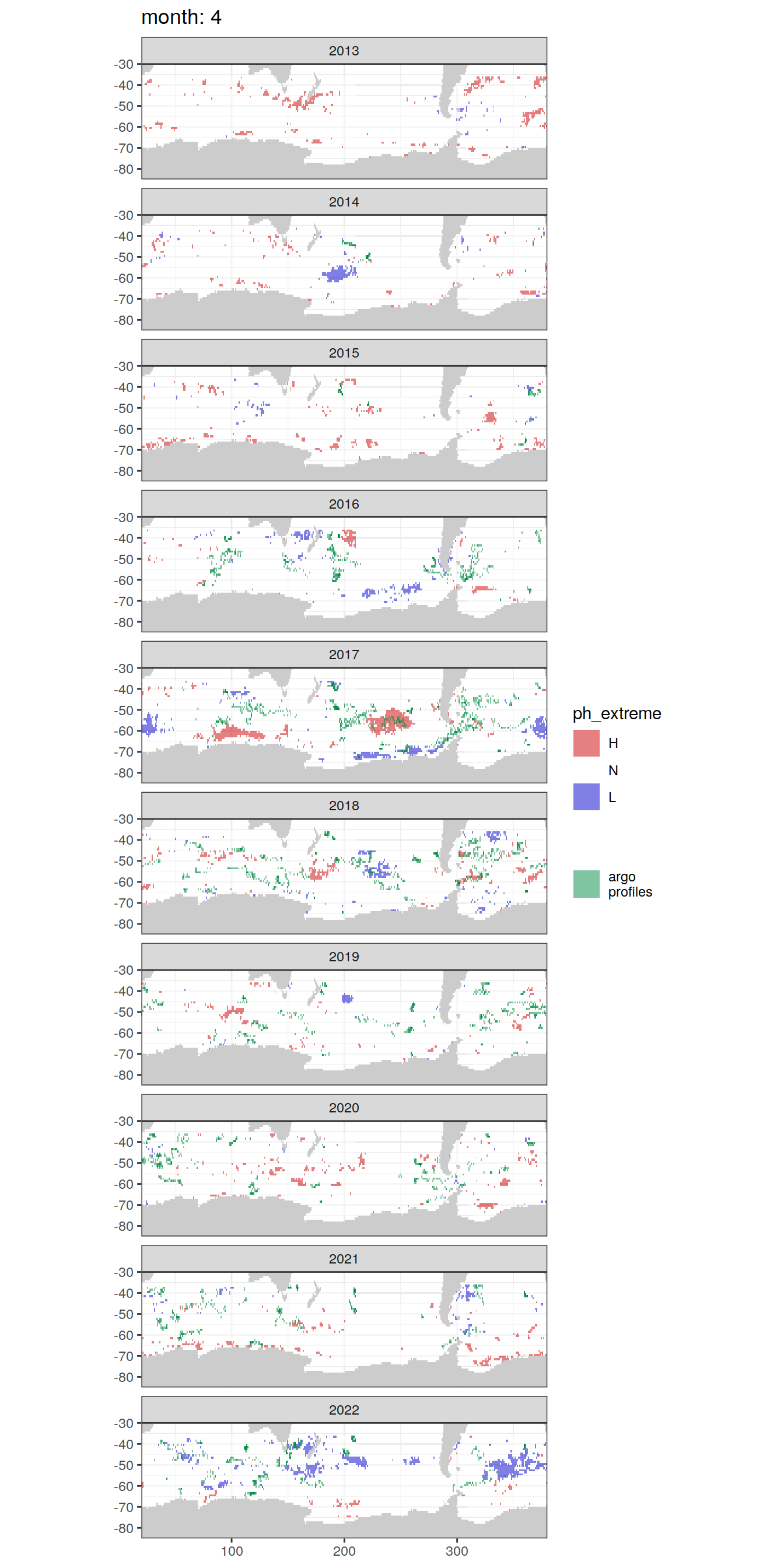
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[5]]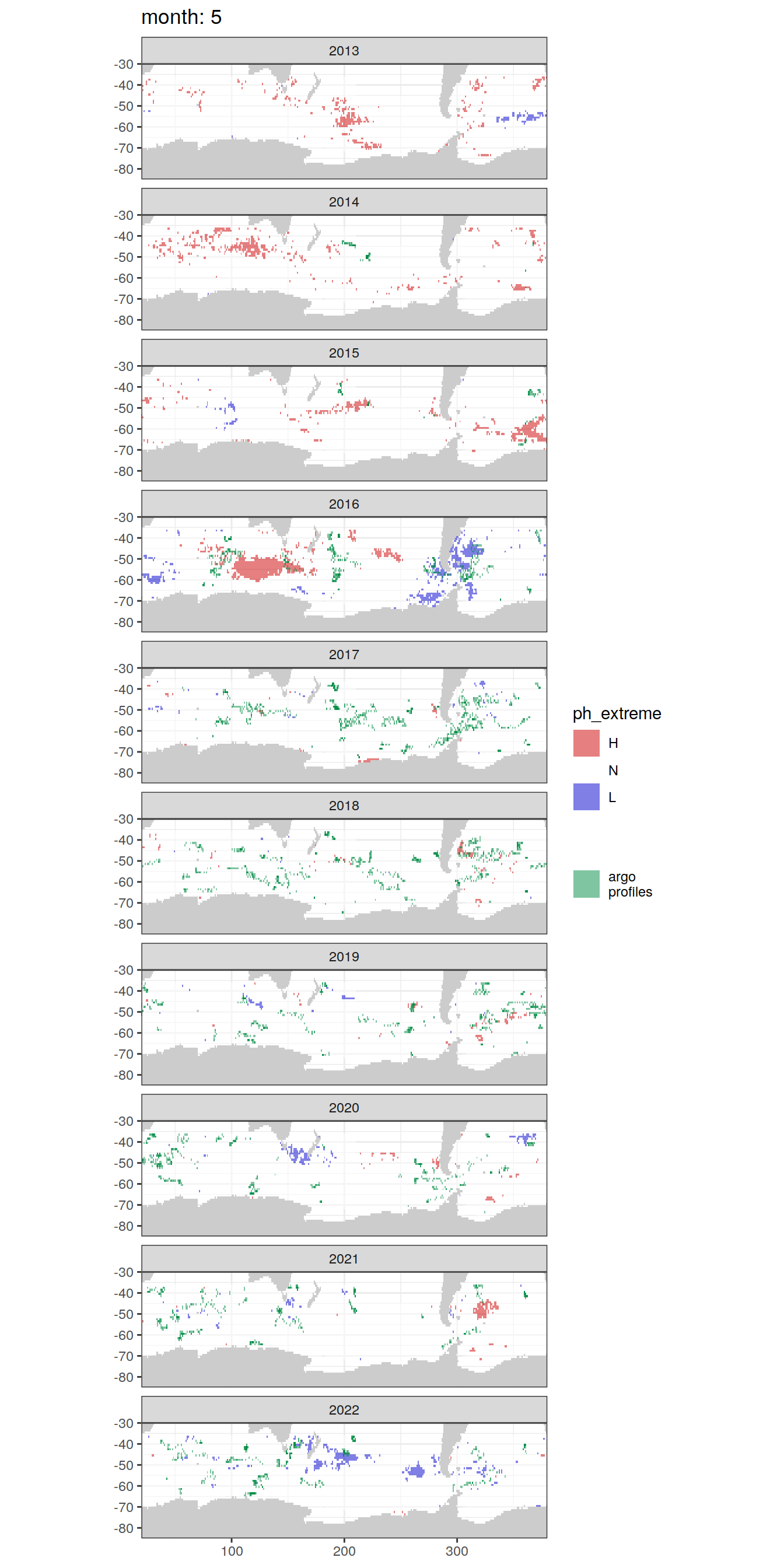
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[6]]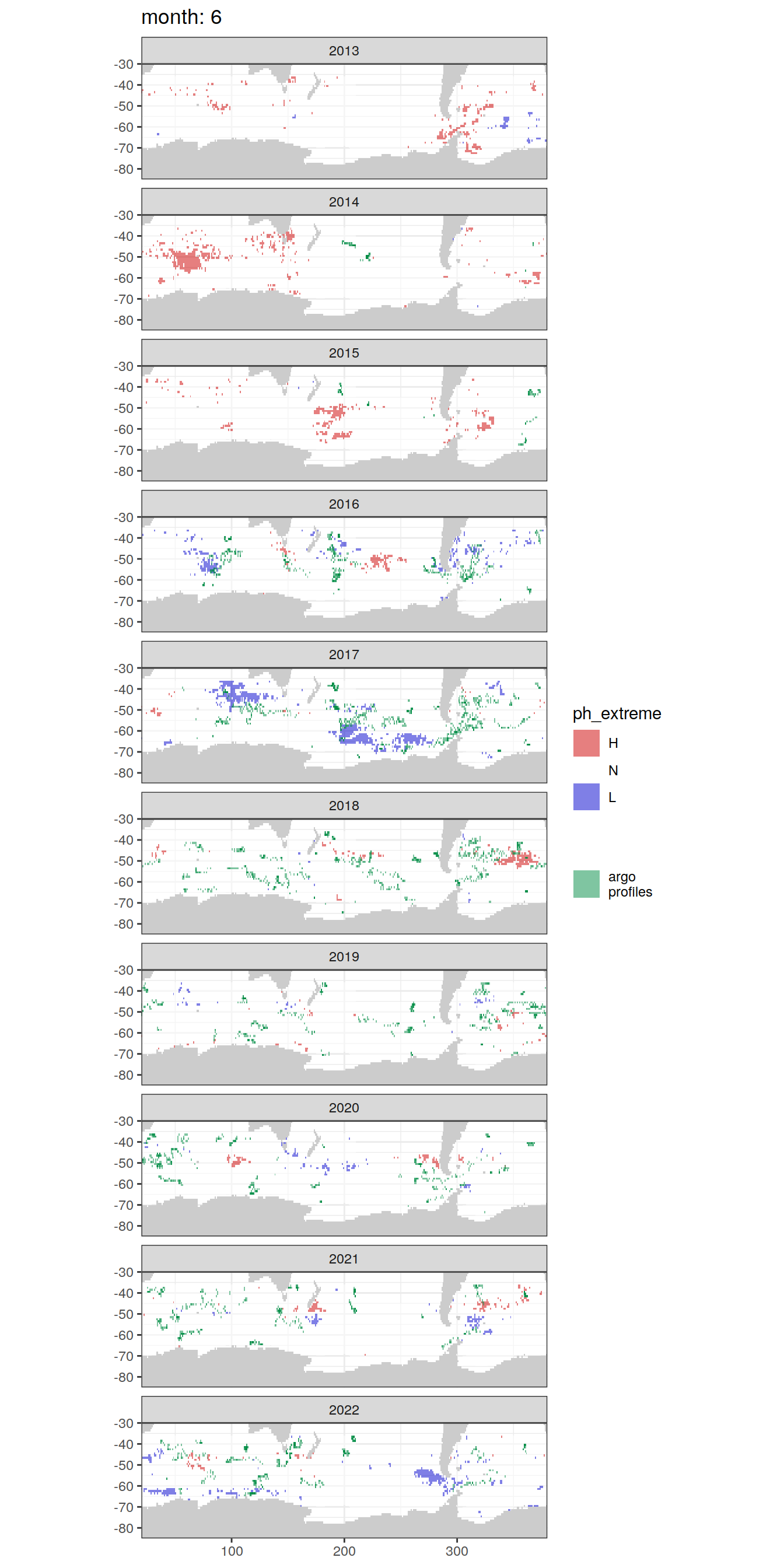
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[7]]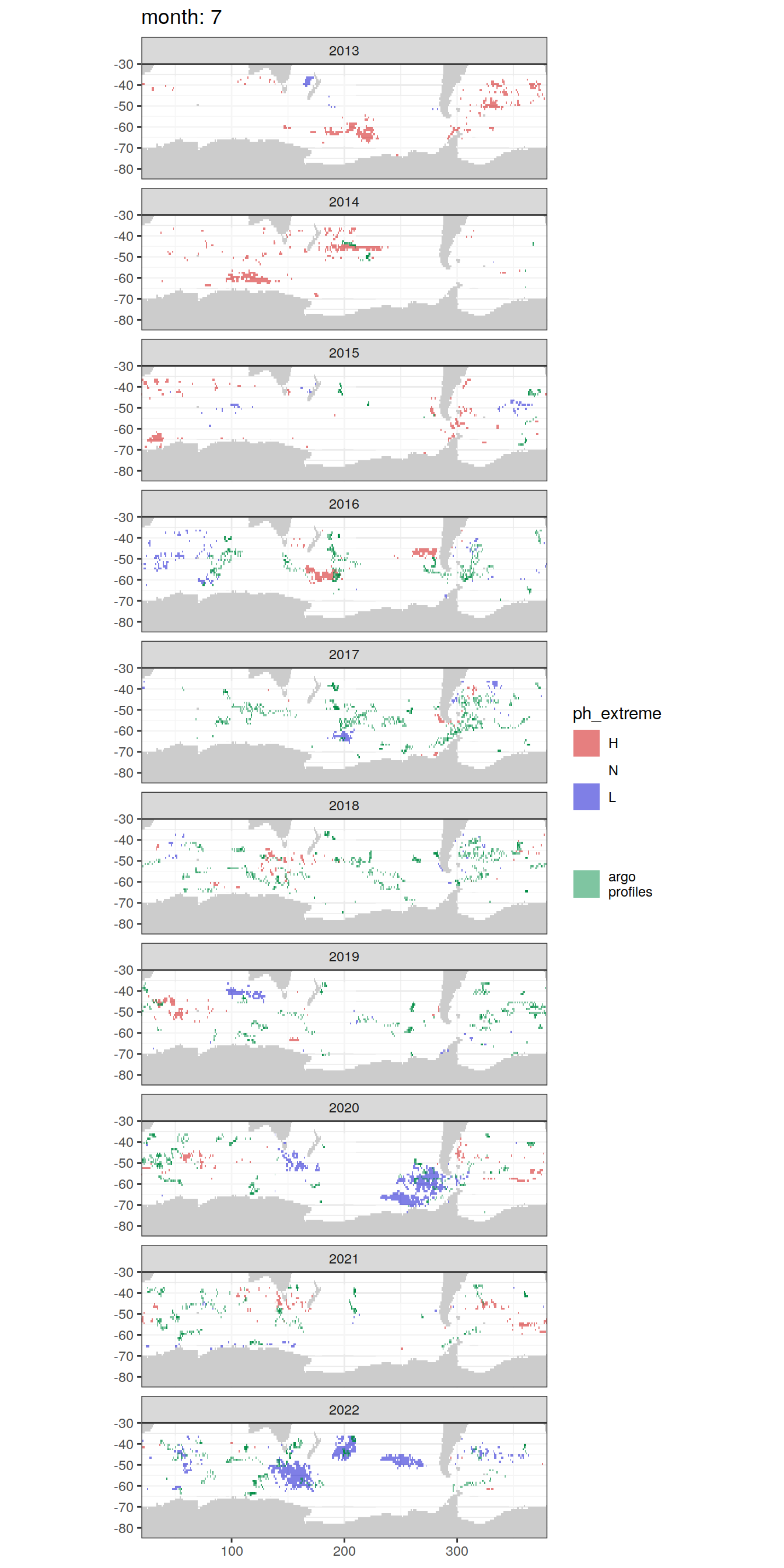
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[8]]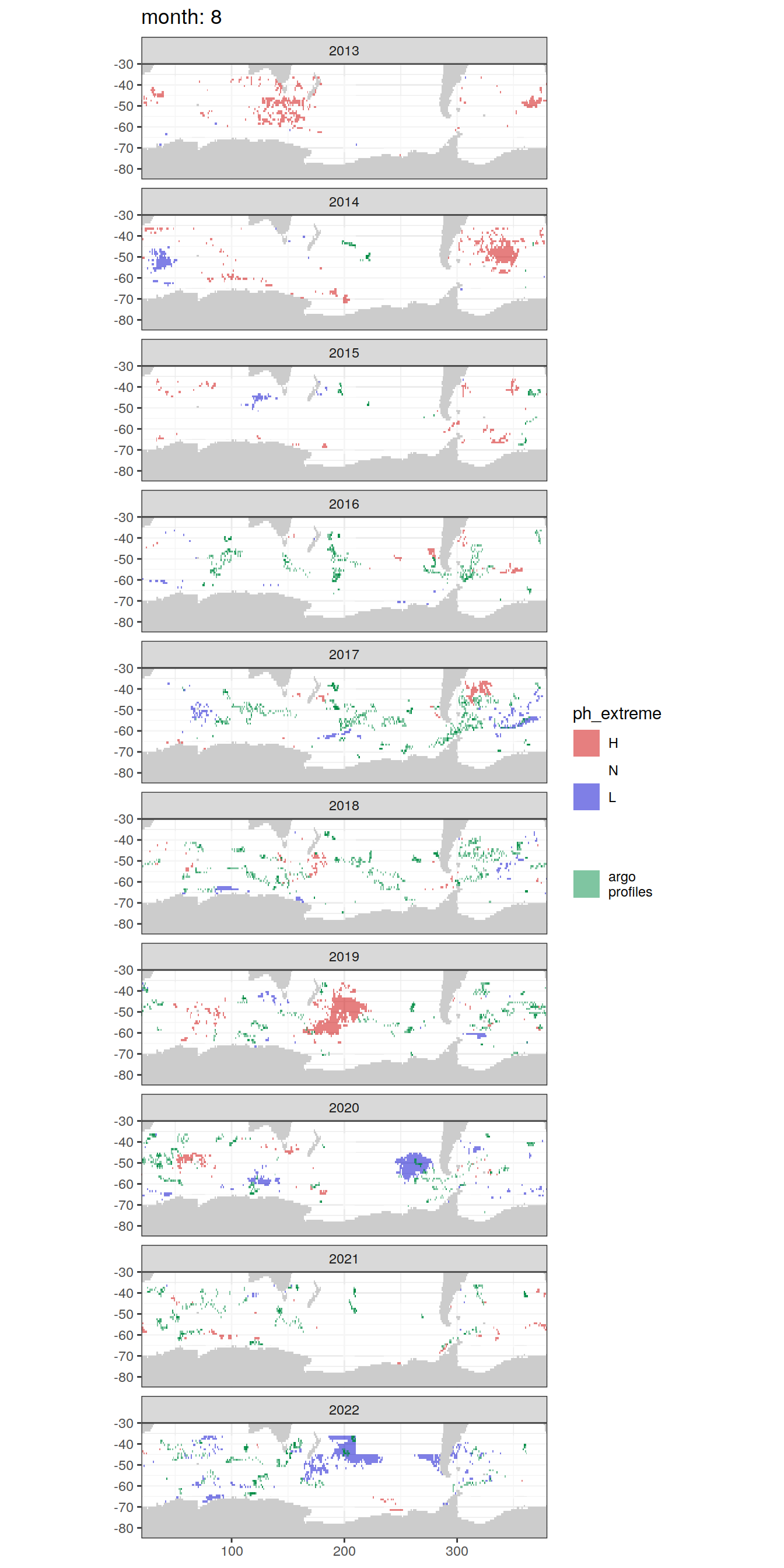
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[9]]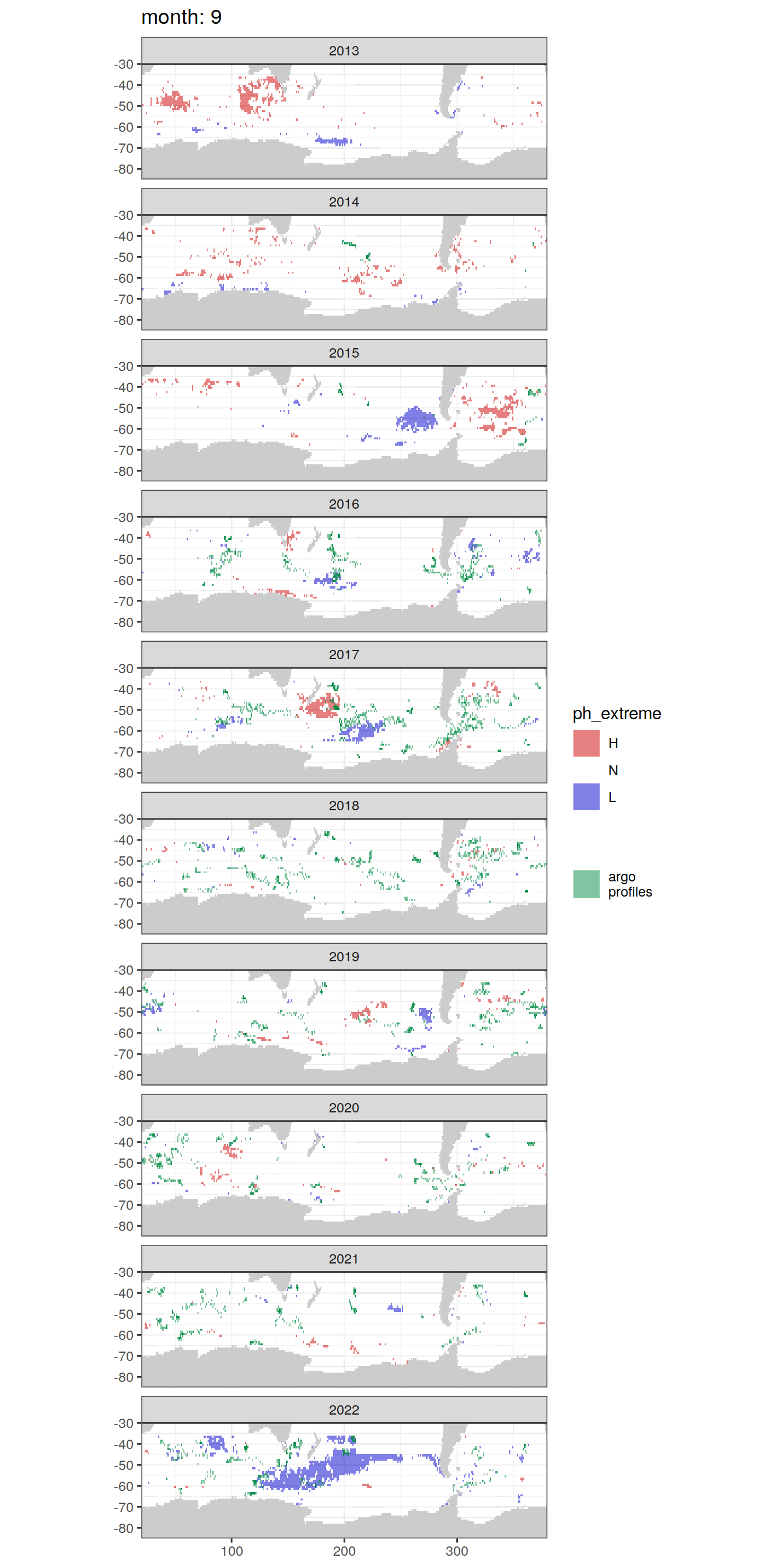
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[10]]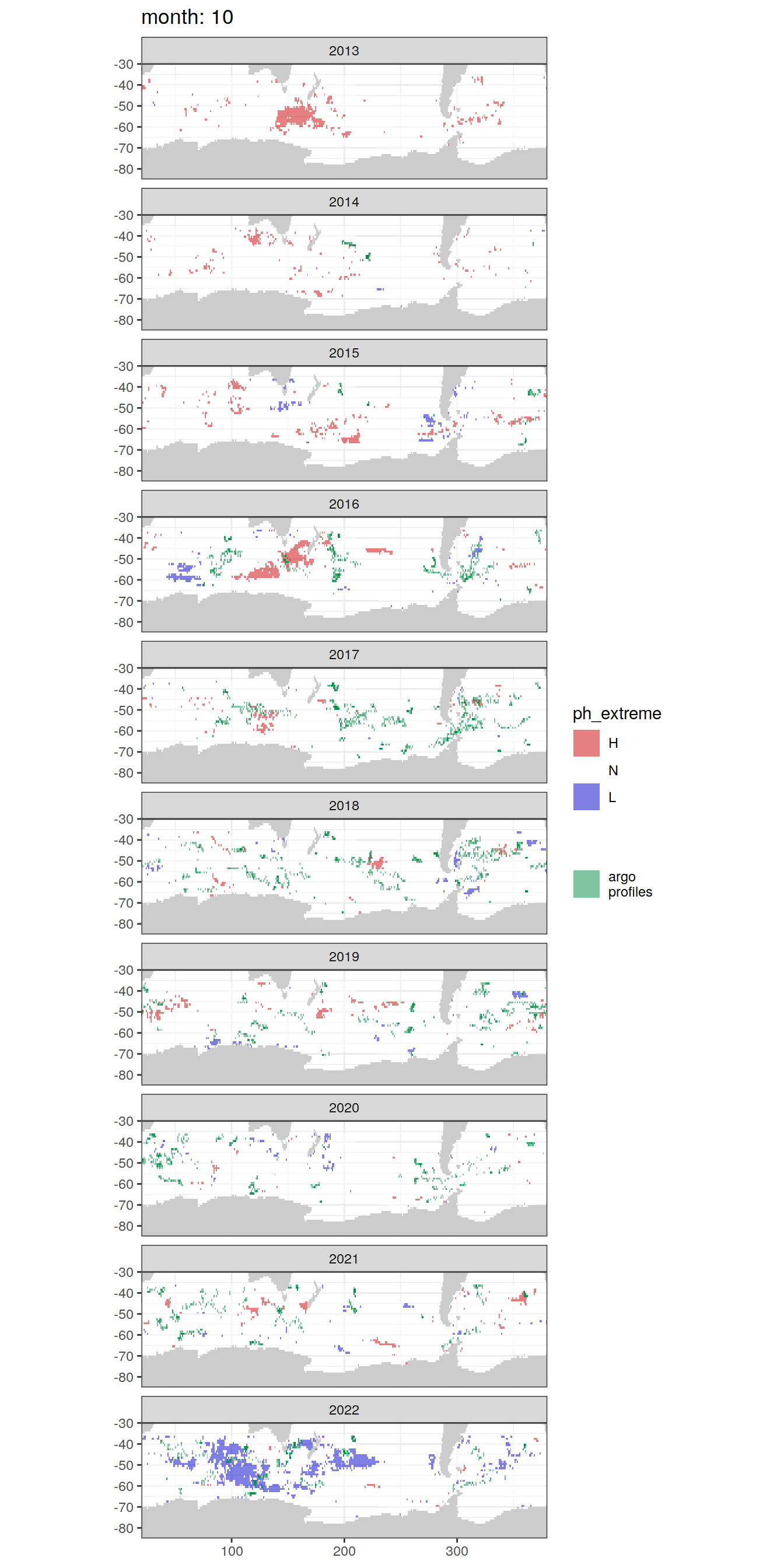
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[11]]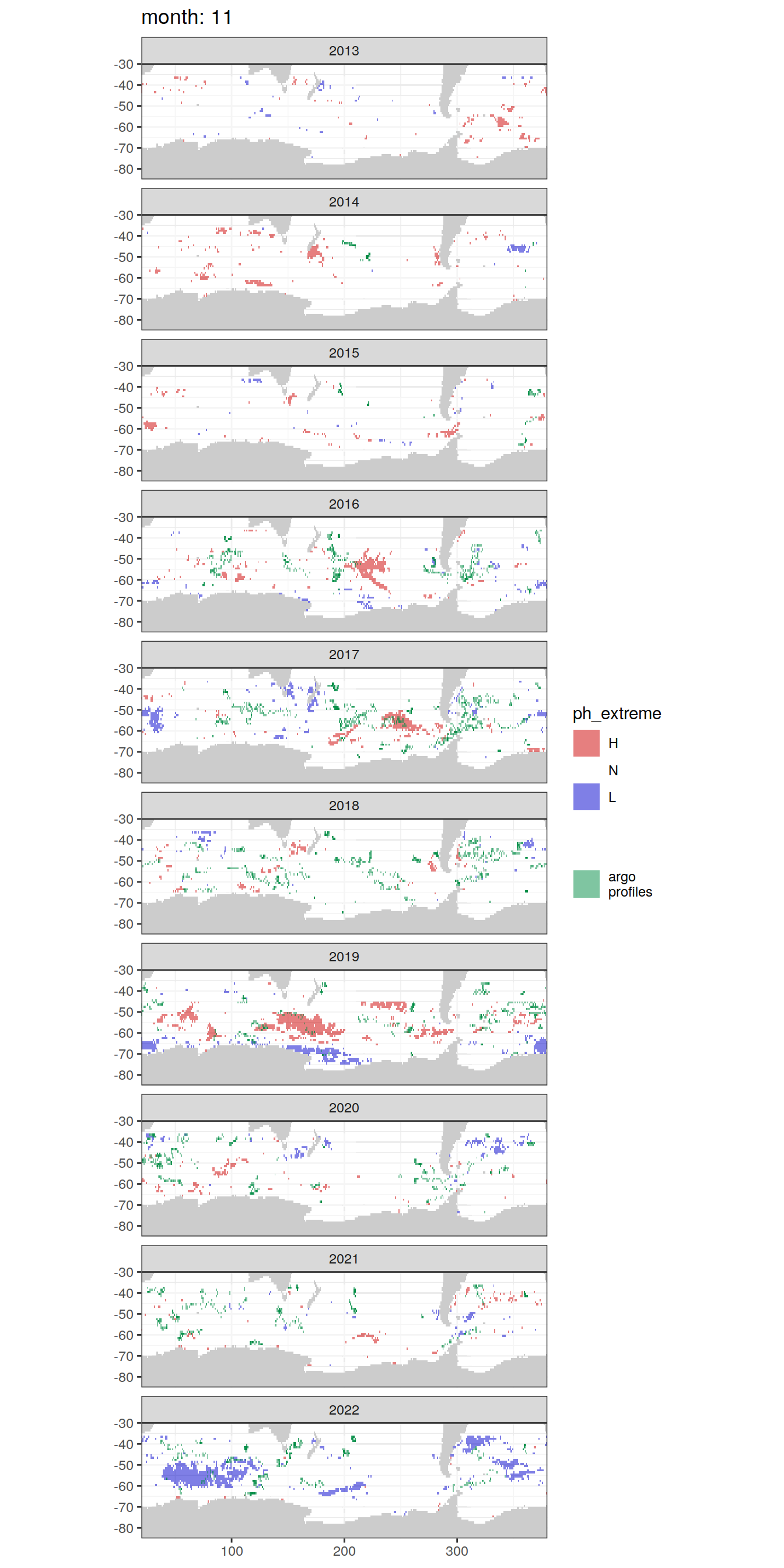
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
[[12]]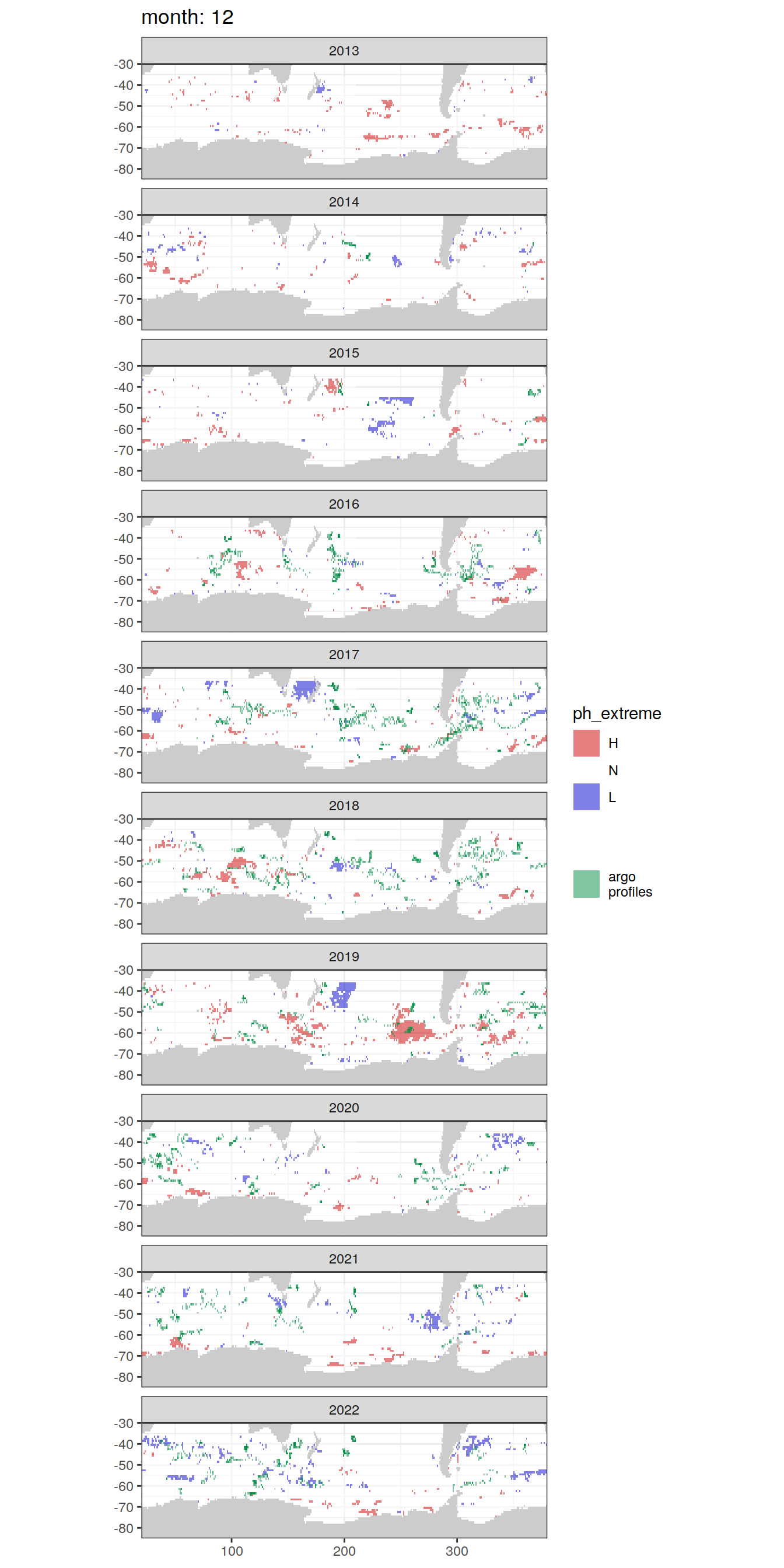
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 71e58d6 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| 710edd4 | jens-daniel-mueller | 2022-05-11 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| 2d44f8a | pasqualina-vonlanthendinenna | 2022-04-29 |
Plot profiles
Argo profiles plotted according to the surface OceanSODA pH
L profiles correspond to a surface acidification event (low pH), as recorded in OceanSODA
H profiles correspond to an event of high surface pH, as recorded in OceanSODA
N profiles correspond to normal surface OceanSODA pH
Raw
profile_extreme %>%
group_split(biome_name, basin_AIP, year) %>%
head(6) %>%
map(
~ ggplot(
data = .x,
aes(
x = pH,
y = depth,
group = file_id,
col = ph_extreme
)
) +
geom_path(data = .x %>% filter(ph_extreme == 'N'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.3) +
geom_path(data = .x %>% filter(ph_extreme == 'H' | ph_extreme == 'L'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.5)+
scale_y_reverse() +
scale_color_manual(values = HNL_colors) +
facet_wrap(~ month, ncol = 6) +
labs(
x = 'Argo pH (total scale)',
y = 'depth (m)',
title = paste(
unique(.x$basin_AIP),
"|",
unique(.x$year),
"| biome:",
unique(.x$biome_name)
),
col = 'OceanSODA pH \nanomaly'
)
)[[1]]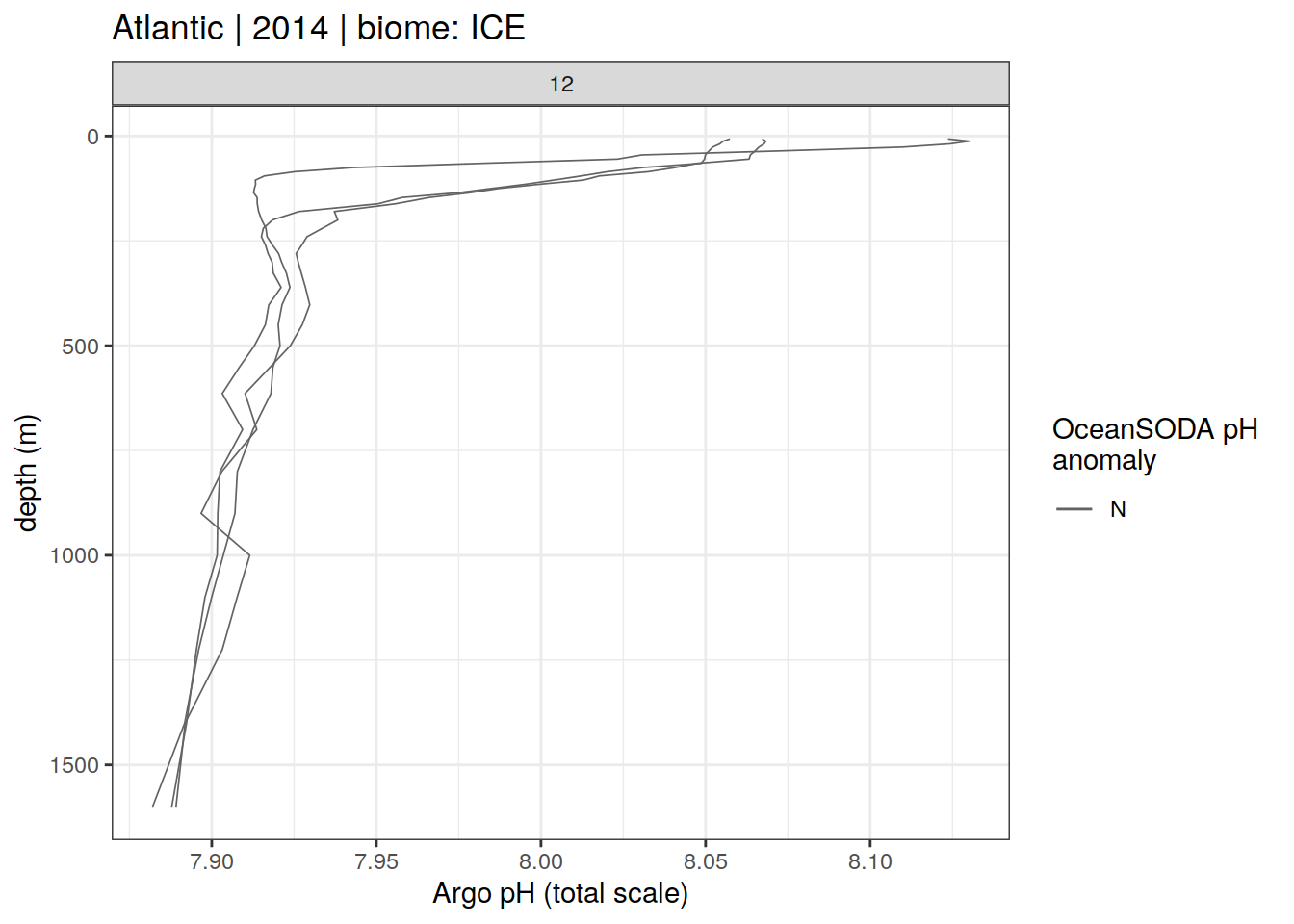
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| ff31fef | ds2n19 | 2023-11-20 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[2]]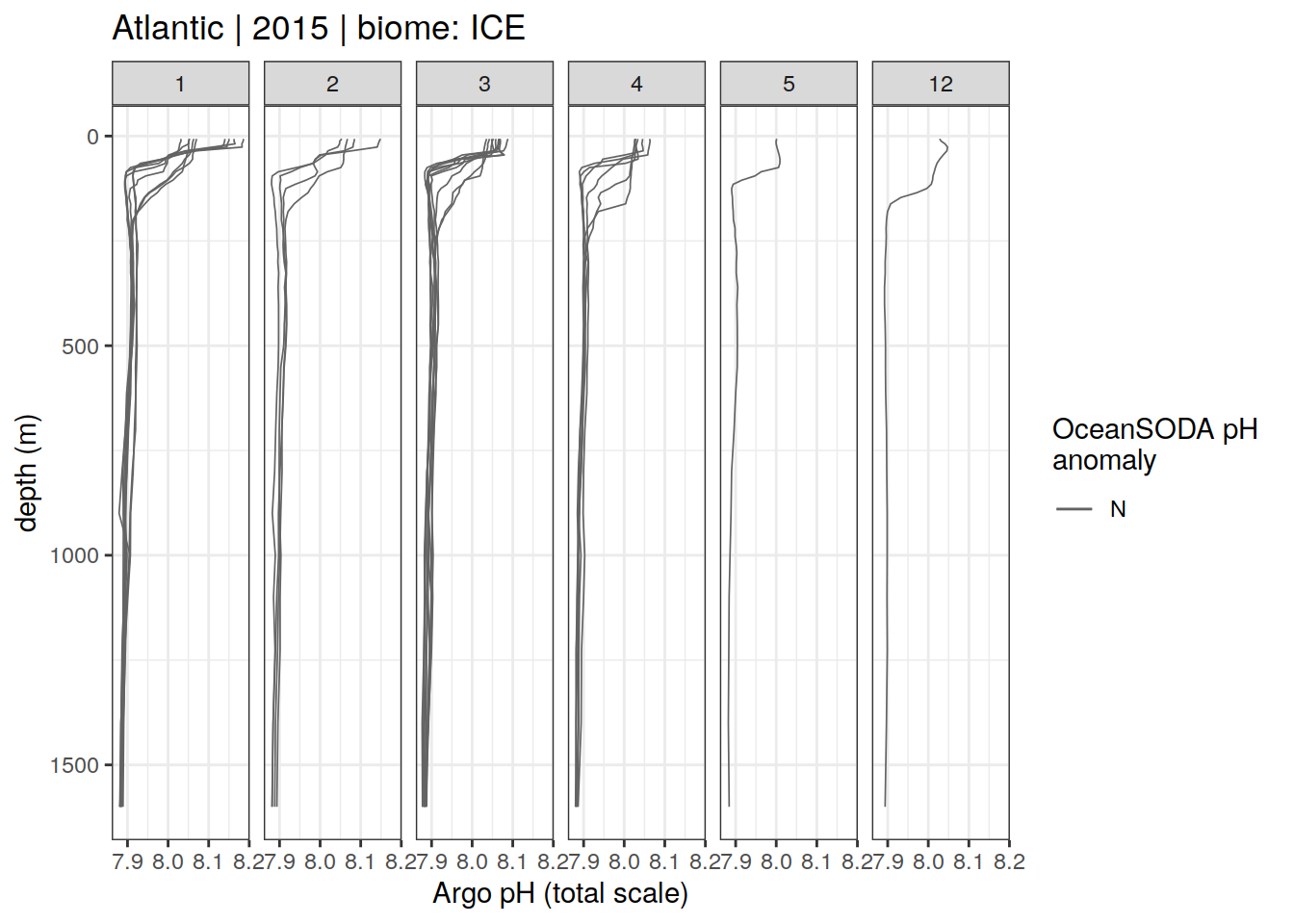
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[3]]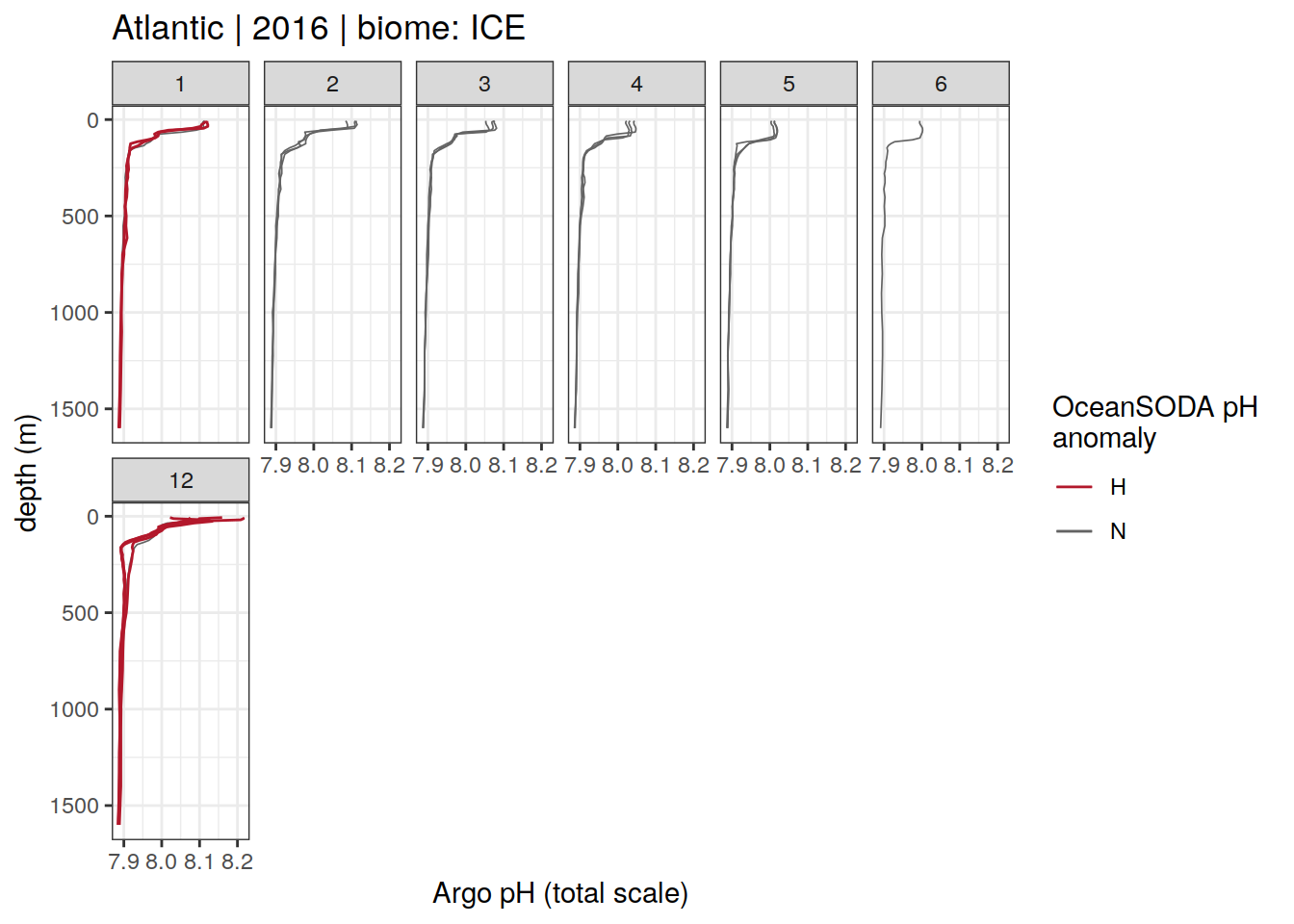
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[4]]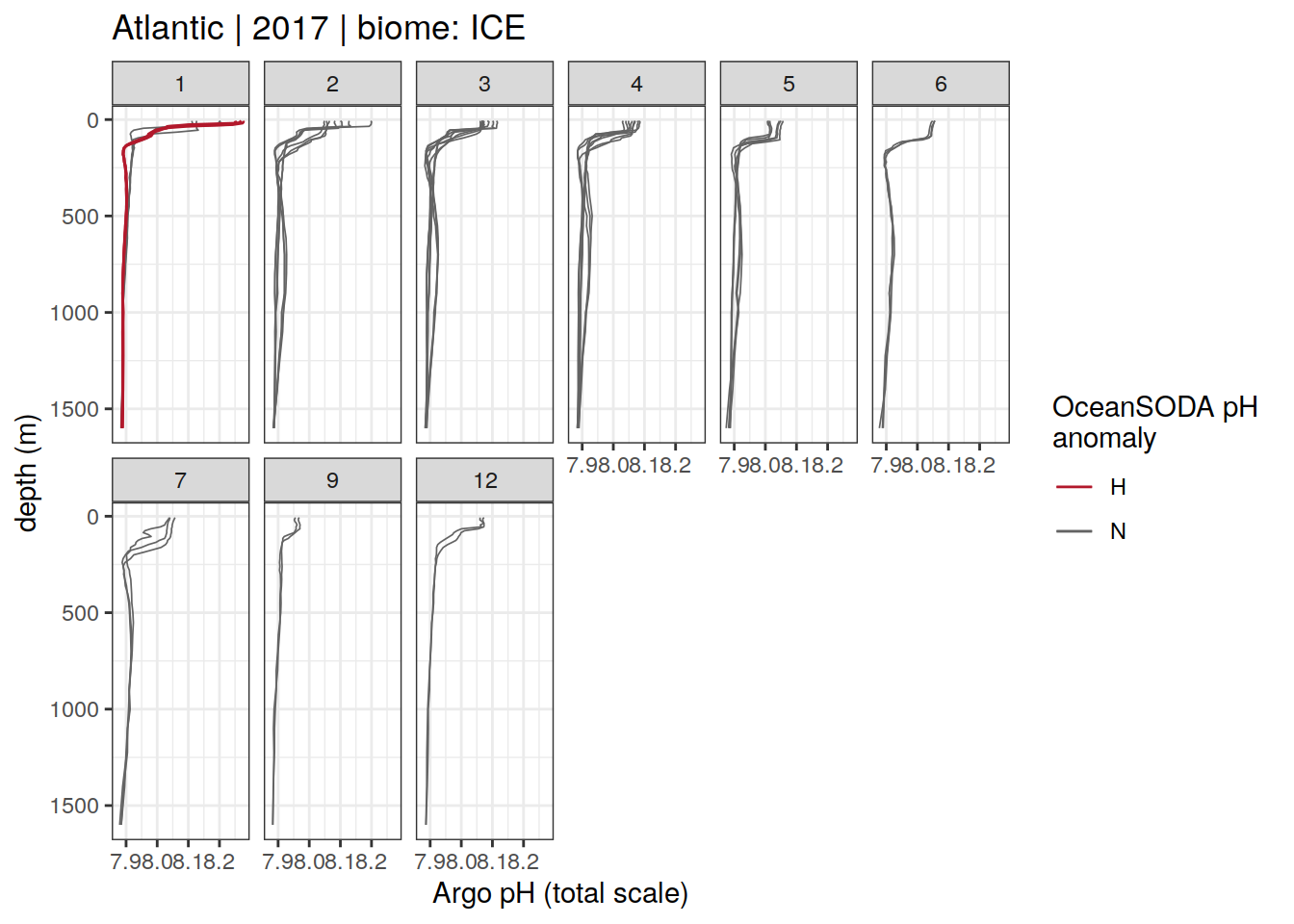
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[5]]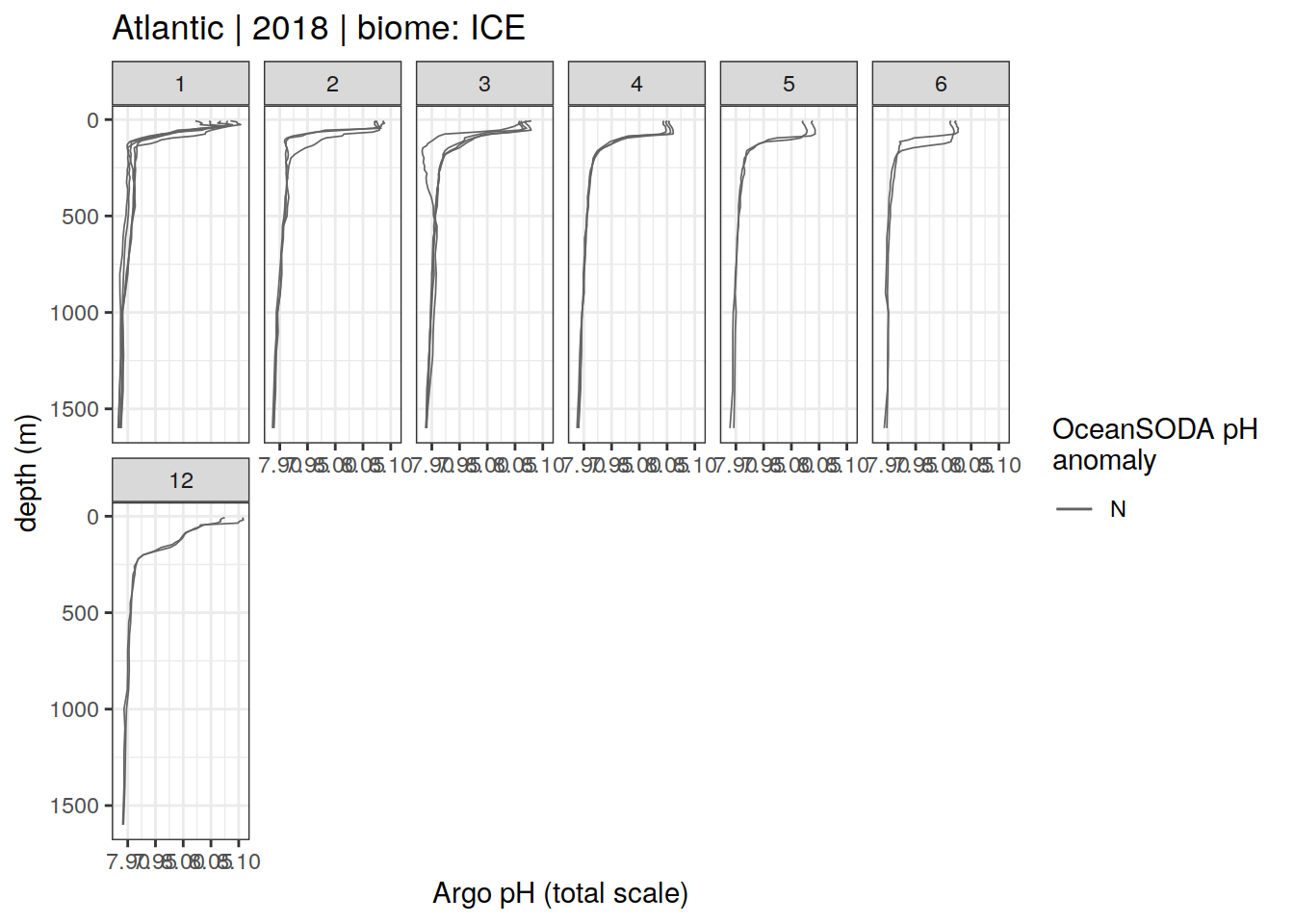
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[6]]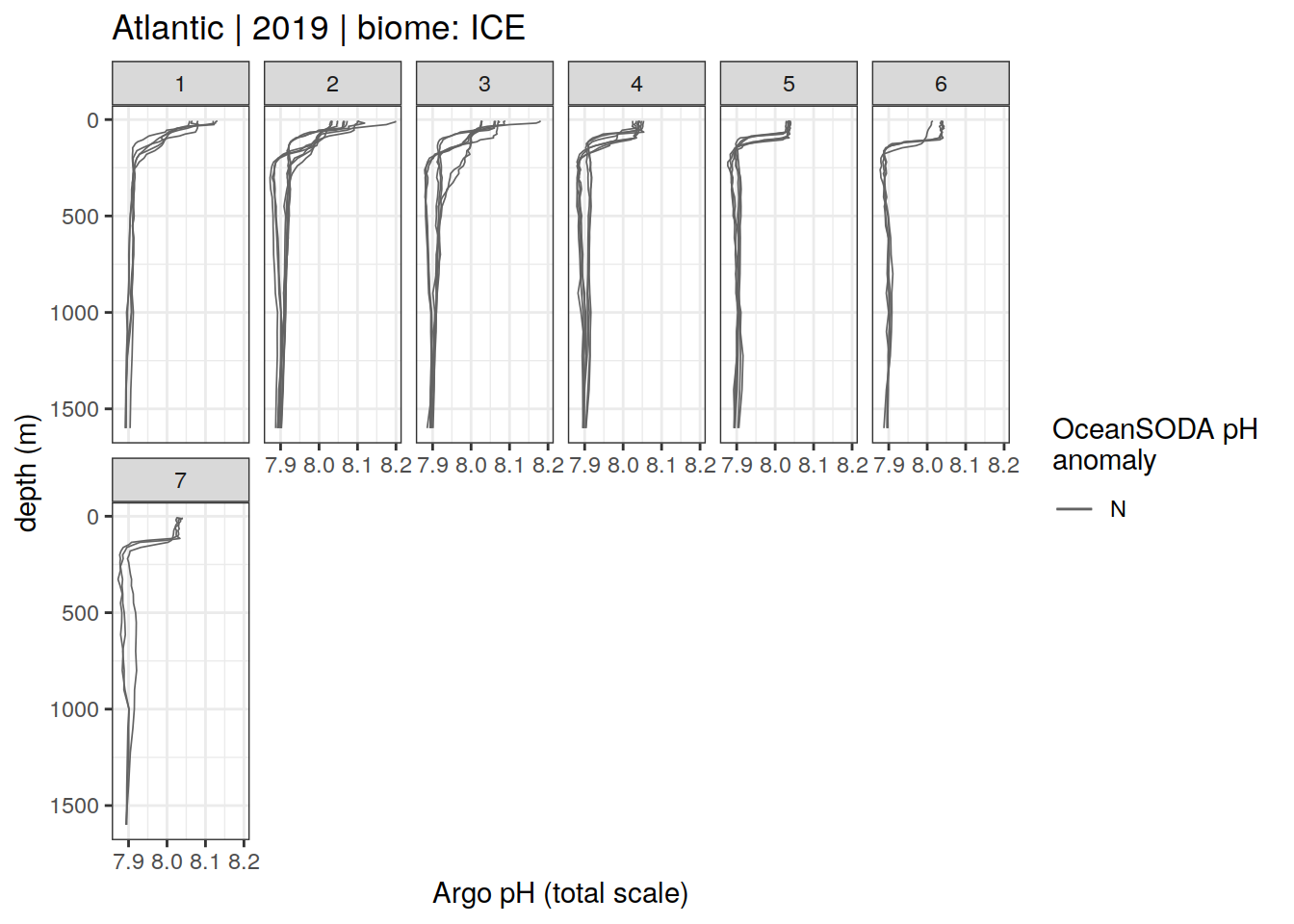
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 8ef1277 | pasqualina-vonlanthendinenna | 2022-03-01 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
Atl, STSS biome, Aug 2017
# plot profiles for the Atlantic basin, biome 1, month 08, 2017
OceanSODA_SO_extreme_grid_2017 <-
OceanSODA_SO_extreme_grid %>%
filter(date == '2017-08-15')
map+
geom_tile(data = OceanSODA_SO_extreme_grid_2017,
aes(x = lon,
y = lat,
fill = ph_extreme))+
scale_fill_manual(values = HNL_colors_map)+
lims(y = c(-85, -30))+
labs(title = 'August 2017',
fill = 'OceanSODA pH \nextreme')
profile_extreme_Atl_2017 <- profile_extreme %>%
filter(date == '2017-08-15',
basin_AIP == 'Atlantic',
biome_name == 'STSS')
profile_extreme_Atl_2017 %>%
ggplot(aes(y = depth,
x = pH,
group = file_id,
col = ph_extreme))+
geom_path(data = profile_extreme_Atl_2017 %>% filter(ph_extreme == 'N'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.3)+
geom_path(data = profile_extreme_Atl_2017 %>% filter(ph_extreme == 'H'| ph_extreme == 'L'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.5)+
scale_y_reverse()+
scale_color_manual(values = HNL_colors)+
labs(title = 'Atlantic basin, STSS biome, August 2017',
col = 'OceanSODA\npH anomaly',
x = 'Argo pH')
rm(OceanSODA_SO_extreme_grid_2017, profile_extreme_Atl_2017)Atl, STSS biome, Dec 2017
# Plot profiles for the Pacific basin, biome 3, months 12, 2017
OceanSODA_SO_extreme_grid_2017 <-
OceanSODA_SO_extreme_grid %>%
filter(date == '2017-12-15')
map+
geom_tile(data = OceanSODA_SO_extreme_grid_2017,
aes(x = lon,
y = lat,
fill = ph_extreme))+
scale_fill_manual(values = HNL_colors_map)+
lims(y = c(-85, -30))+
labs(title = 'December 2017',
fill = 'OceanSODA pH \nextreme')
profile_extreme_Atl_2017 <- profile_extreme %>%
filter(date == '2017-12-15',
basin_AIP == 'Atlantic',
biome_name == 'STSS')
profile_extreme_Atl_2017 %>%
ggplot(aes(y = depth,
x = pH,
group = file_id,
col = ph_extreme))+
geom_path(data = profile_extreme_Atl_2017 %>% filter(ph_extreme == 'N'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.3)+
geom_path(data = profile_extreme_Atl_2017 %>% filter(ph_extreme == 'H' | ph_extreme == 'L'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.5)+
scale_y_reverse()+
scale_color_manual(values = HNL_colors)+
labs(title = 'Atlantic basin, STSS biome, December 2017',
col = 'OceanSODA\npH anomaly',
x = 'Argo pH')
rm(OceanSODA_SO_extreme_grid_2017, profile_extreme_Atl_2017)Atl, STSS biome, Jan 2018
OceanSODA_SO_extreme_grid_2018 <-
OceanSODA_SO_extreme_grid %>%
filter(date == '2018-01-15')
map+
geom_tile(data = OceanSODA_SO_extreme_grid_2018,
aes(x = lon,
y = lat,
fill = ph_extreme))+
scale_fill_manual(values = HNL_colors_map)+
lims(y = c(-85, -30))+
labs(title = 'January 2018',
fill = 'OceanSODA pH \nextreme')
profile_extreme_Atl_2018 <- profile_extreme %>%
filter(date == '2018-01-15',
basin_AIP == 'Atlantic',
biome_name == 'STSS')
profile_extreme_Atl_2018 %>%
ggplot(aes(y = depth,
x = pH,
group = file_id,
col = ph_extreme))+
geom_path(data = profile_extreme_Atl_2018 %>% filter(ph_extreme == 'N'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.3)+
geom_path(data = profile_extreme_Atl_2018 %>% filter(ph_extreme == 'H' | ph_extreme == 'L'),
aes(x = pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.5)+
scale_y_reverse()+
scale_color_manual(values = HNL_colors)+
labs(title = 'Atlantic basin, STSS biome, January 2018',
col = 'OceanSODA\npH anomaly',
x = 'Argo pH')
rm(OceanSODA_SO_extreme_grid_2018, profile_extreme_Atl_2018)Averaged profiles
# calculate mean profiles in each basin and biome, for each month between 2014 and 2021
# cut depth levels at 10, 20, .... etc m
# add seasons
# Dec, Jan, Feb <- summer
# Mar, Apr, May <- autumn
# Jun, Jul, Aug <- winter
# Sep, Oct, Nov <- spring
profile_extreme <- profile_extreme %>%
mutate(
season = case_when(
between(month, 3, 5) ~ 'autumn',
between(month, 6, 8) ~ 'winter',
between(month, 9, 11) ~ 'spring',
month == 12 | 1 | 2 ~ 'summer'
),
season_order = case_when(
between(month, 3, 5) ~ 2,
between(month, 6, 8) ~ 3,
between(month, 9, 11) ~ 4,
month == 12 | 1 | 2 ~ 1
),
.after = date
) Overall mean
profile_extreme_mean <- profile_extreme %>%
group_by(ph_extreme, depth) %>%
summarise(ph_mean = mean(pH, na.rm = TRUE),
ph_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
profile_extreme_mean %>%
arrange(depth) %>%
ggplot(aes(
x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme
)) +
geom_ribbon(aes(xmin = ph_mean - ph_std,
xmax = ph_mean + ph_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = "Overall mean",
col = 'OceanSODA\npH anomaly \n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly \n(mean ± st dev)',
y = 'depth (m)',
x = 'Argo mean pH') +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))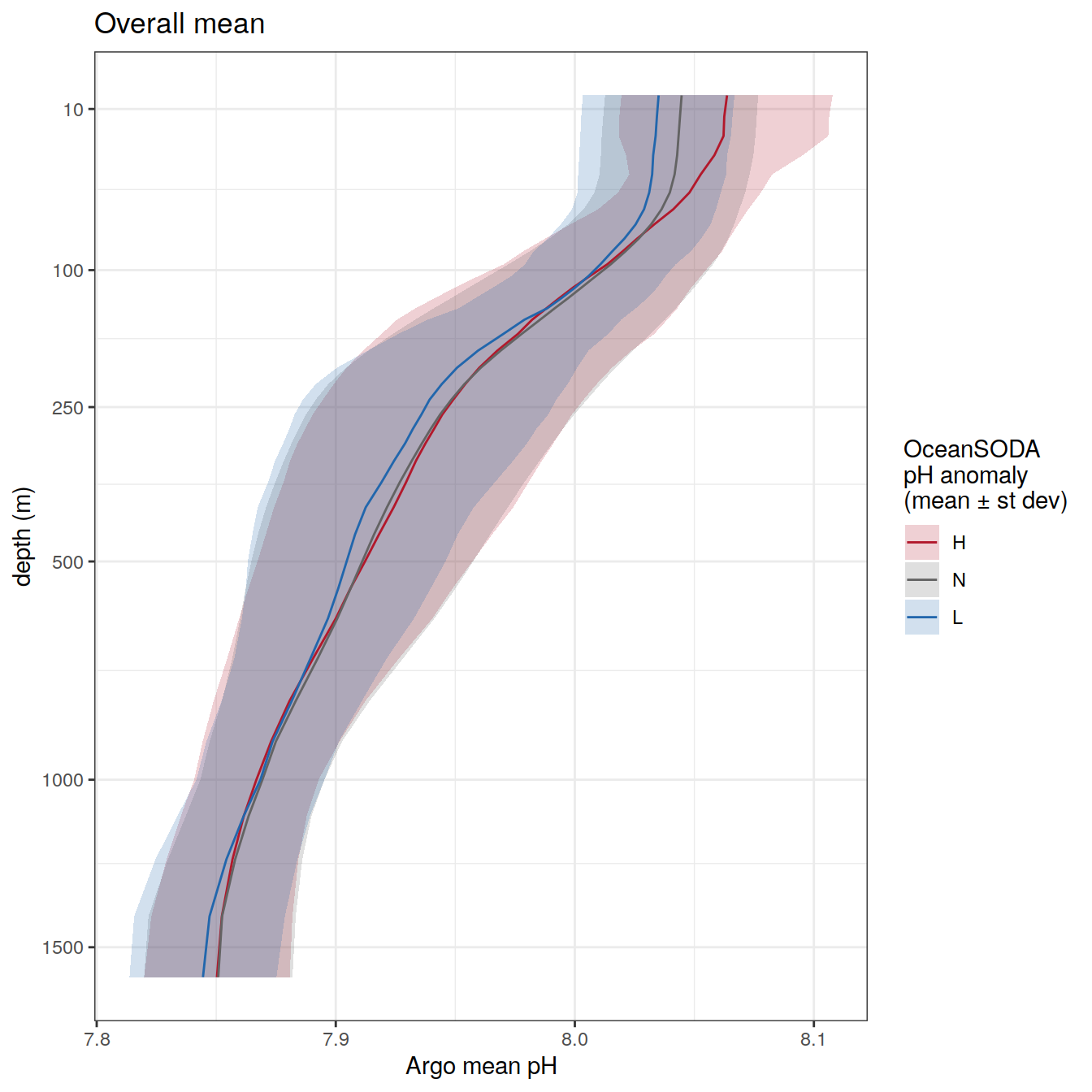
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
rm(profile_extreme_mean)Number of profiles
profile_count_mean <- profile_extreme %>%
distinct(ph_extreme, file_id) %>%
count(ph_extreme)
profile_count_mean %>%
ggplot(aes(x = ph_extreme, y = n, fill = ph_extreme))+
geom_col(width = 0.5)+
scale_y_continuous(trans = 'log10')+
labs(y = 'log(number of profiles)',
title = 'Number of profiles')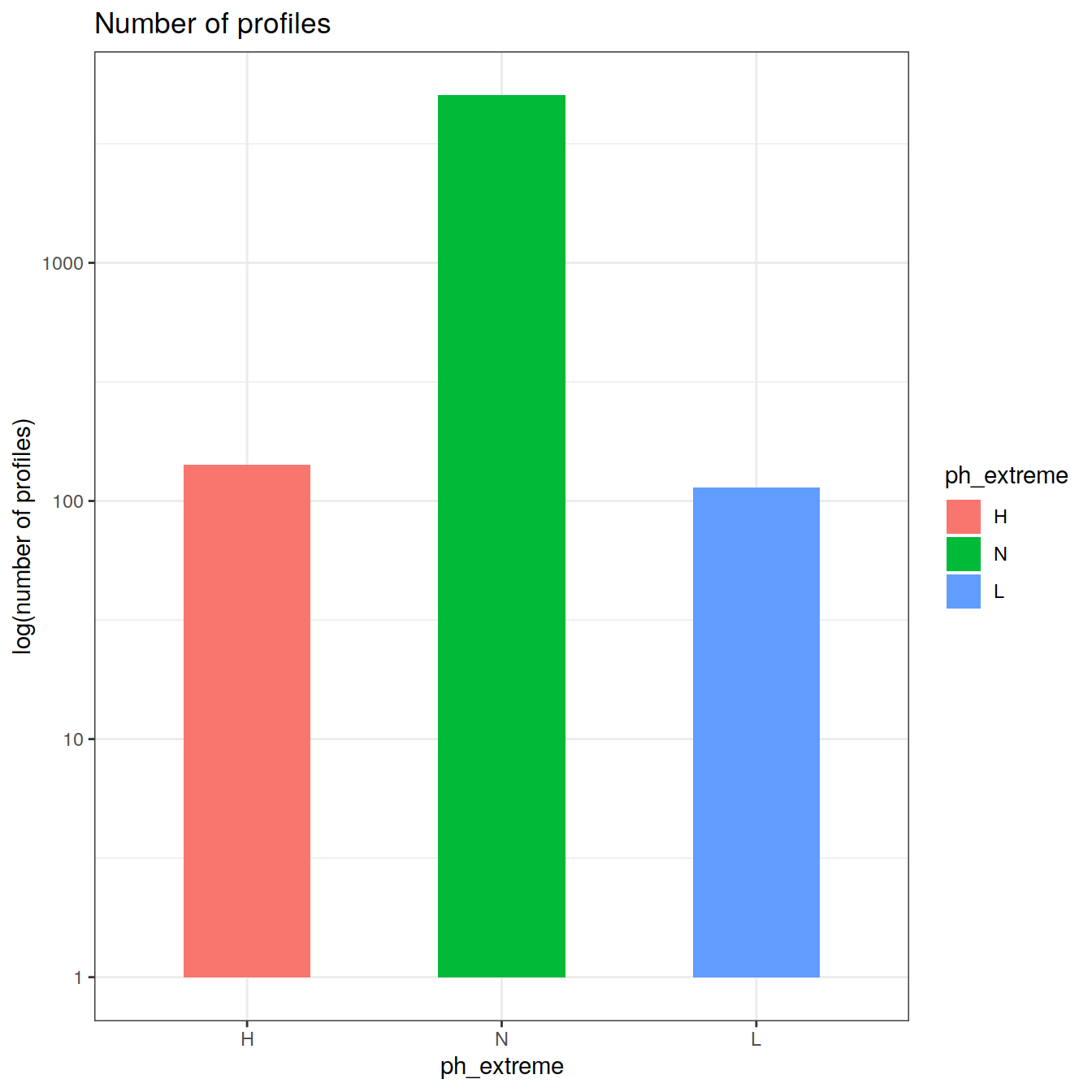
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
# rm(profile_count_mean)Surface Argo pH vs surface OceanSODA pH (20 m)
# calculate surface-mean argo pH, for each profile
surface_ph_mean <- profile_extreme %>%
filter(depth <= 20) %>%
group_by(ph_extreme, file_id) %>%
summarise(argo_surf_ph = mean(pH, na.rm = TRUE),
OceanSODA_surf_ph = mean(OceanSODA_ph, na.rm = TRUE)) %>%
ungroup()
surface_ph_mean %>%
group_by(ph_extreme) %>%
group_split(ph_extreme) %>%
map(
~ggplot(data = .x, aes(x = OceanSODA_surf_ph,
y = argo_surf_ph))+
geom_bin2d(data = .x, aes(x = OceanSODA_surf_ph,
y = argo_surf_ph)) +
scale_fill_viridis_c()+
geom_abline(slope = 1, intercept = 0)+
coord_fixed(ratio = 1,
xlim = c(7.9, 8.21),
ylim = c(7.9, 8.21))+
# facet_grid(basin_AIP ~ biome) +
labs(title = paste('pH extreme:', unique(.x$ph_extreme)),
x = 'OceanSODA pH',
y = 'Argo pH')
)[[1]]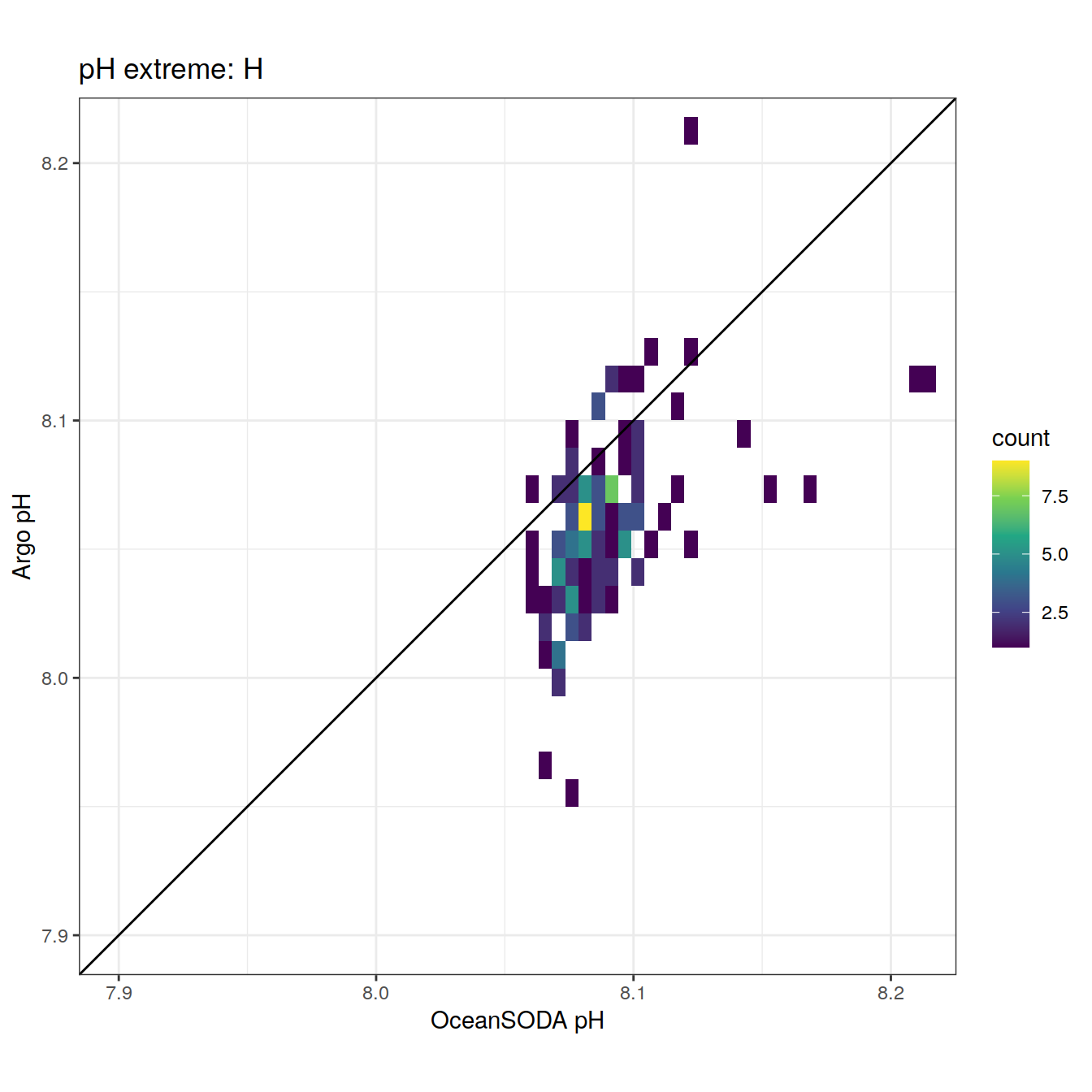
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[2]]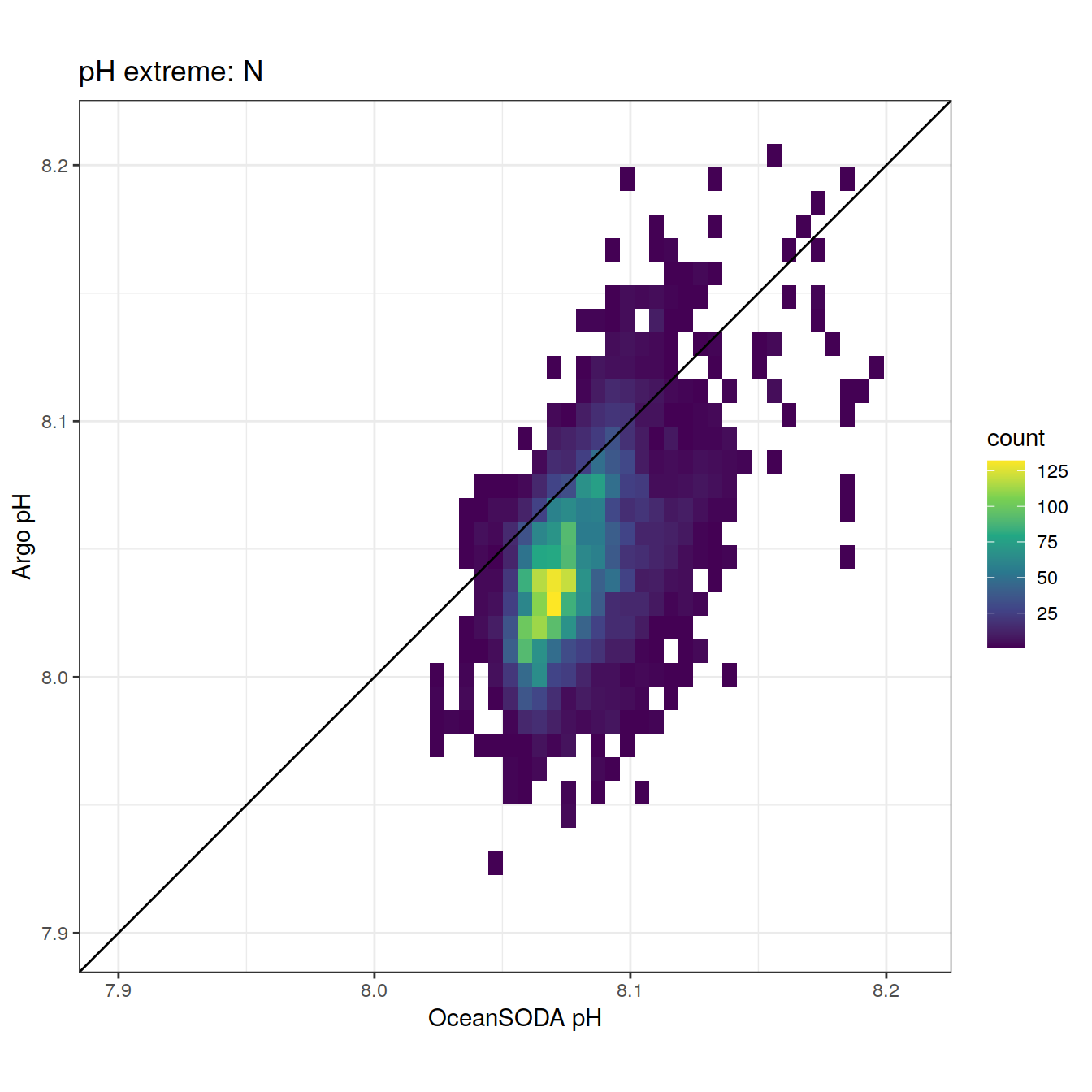
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[3]]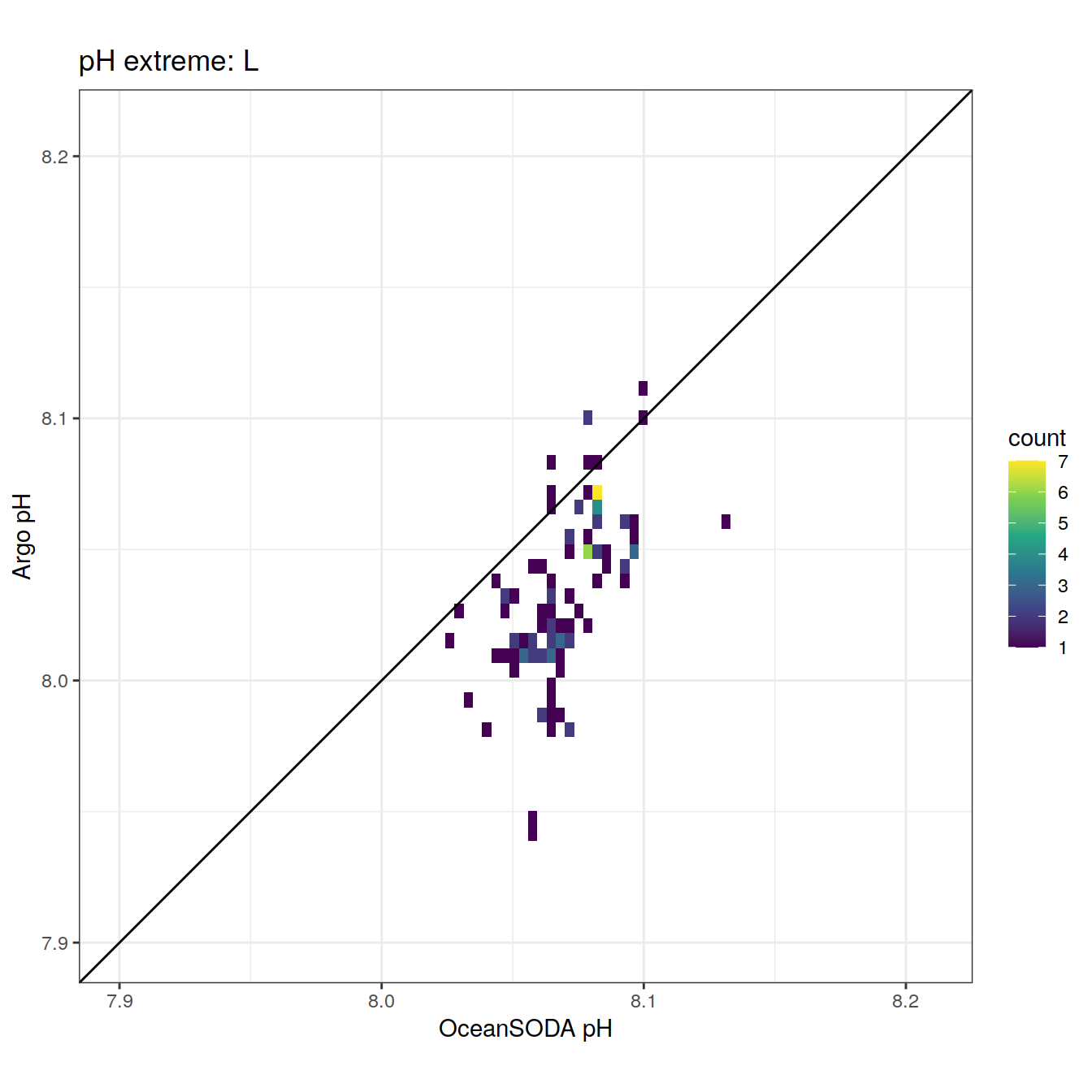
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
rm(surface_ph_mean)January Overall Mean Profiles
profile_extreme_mean_jan <- profile_extreme %>%
filter(month == 1) %>%
group_by(ph_extreme, depth) %>%
summarise(ph_mean = mean(pH, na.rm = TRUE),
ph_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
profile_extreme_mean_jan %>%
arrange(depth) %>%
ggplot(aes(
x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme
)) +
geom_ribbon(aes(xmin = ph_mean - ph_std,
xmax = ph_mean + ph_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = "Overall mean January profiles",
col = 'OceanSODA\npH anomaly \n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly \n(mean ± st dev)',
y = 'depth (m)',
x = 'Argo mean pH') +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))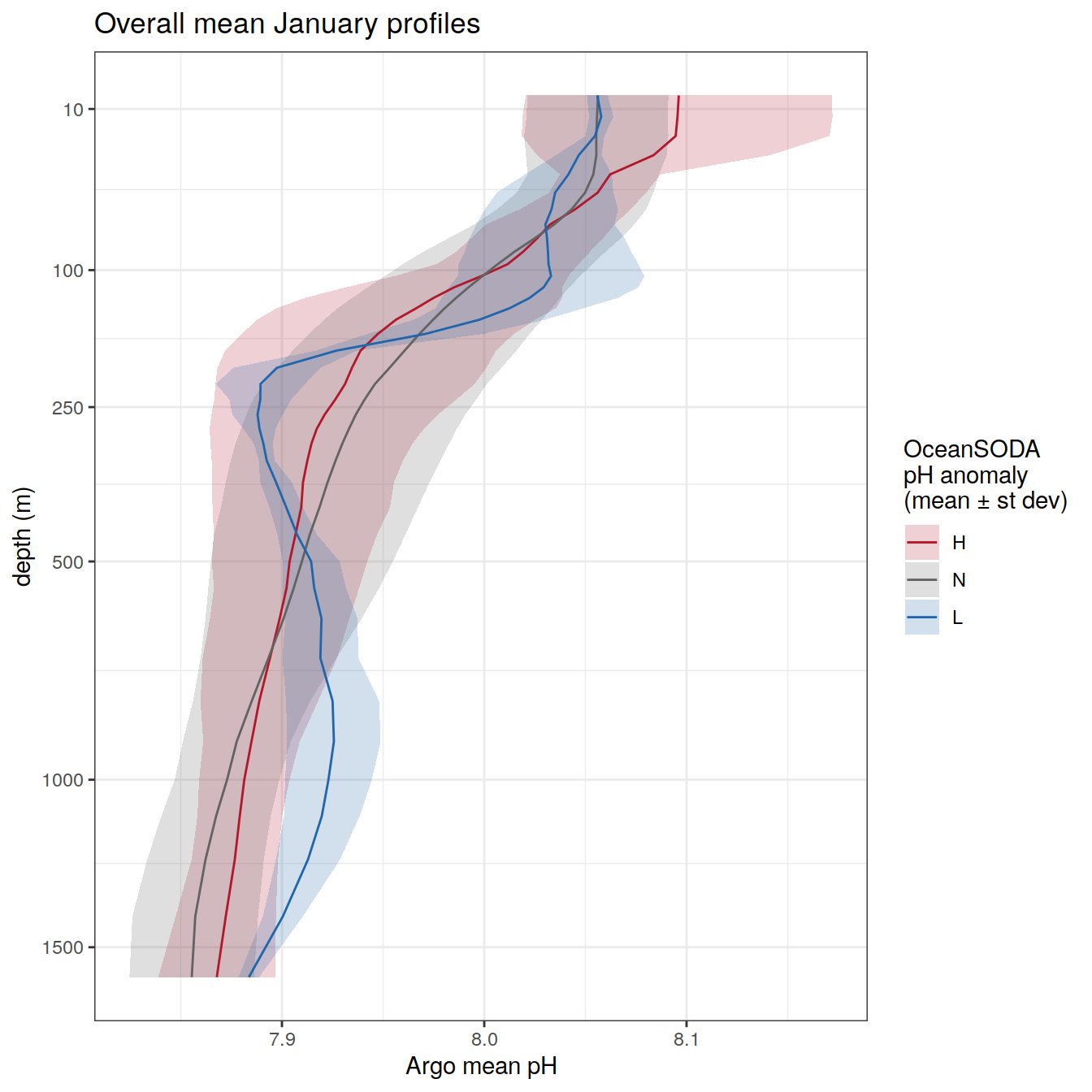
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
rm(profile_extreme_mean_jan)Season x Mayot biome
profile_extreme_biome <- profile_extreme %>%
group_by(season_order, season, biome_name, ph_extreme, depth) %>%
summarise(ph_biome = mean(pH, na.rm = TRUE),
ph_biome_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
facet_label <- as_labeller(c("1"="summer",
"2"="autumn",
"3"="winter",
"4"="spring",
"ICE" = "ICE",
"SPSS" = "SPSS",
"STSS" = "STSS",
"Atlantic" = "Atlantic",
"Indian" = "Indian",
"Pacific" = "Pacific"
))
profile_extreme_biome %>%
ggplot(aes(
x = ph_biome,
y = depth,
group = ph_extreme,
col = ph_extreme
)) +
geom_ribbon(aes(xmin = ph_biome - ph_biome_std,
xmax = ph_biome + ph_biome_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(col = 'OceanSODA\npH anomaly \n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly \n(mean ± st dev)',
y = 'depth (m)',
x = 'Biome mean Argo pH',
title = 'Biome-mean Argo profiles') +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500))) +
facet_grid(season_order ~ biome_name, labeller = facet_label)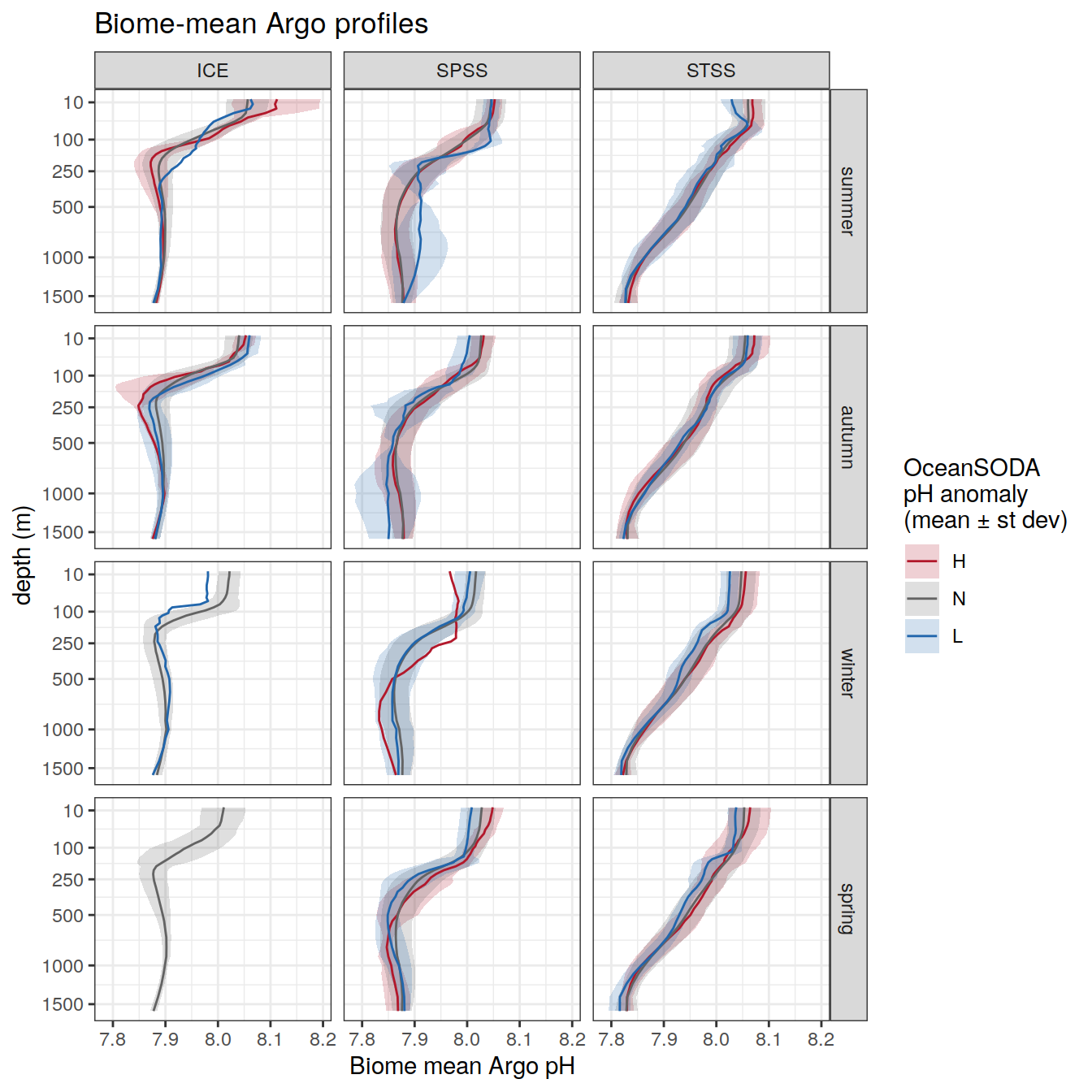
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
rm(profile_extreme_biome)Number of profiles season x biome
profile_count_biome <- profile_extreme %>%
distinct(season_order, season, biome_name, ph_extreme, file_id) %>%
group_by(season_order, season, biome_name, ph_extreme) %>%
count(ph_extreme)
profile_count_biome %>%
ggplot(aes(x = ph_extreme, y = n, fill = ph_extreme))+
geom_col(width = 0.5)+
facet_grid(season_order ~ biome_name, labeller = facet_label)+
scale_y_continuous(trans = 'log10')+
labs(y = 'log(number of profiles)',
title = 'Number of profiles season x biome')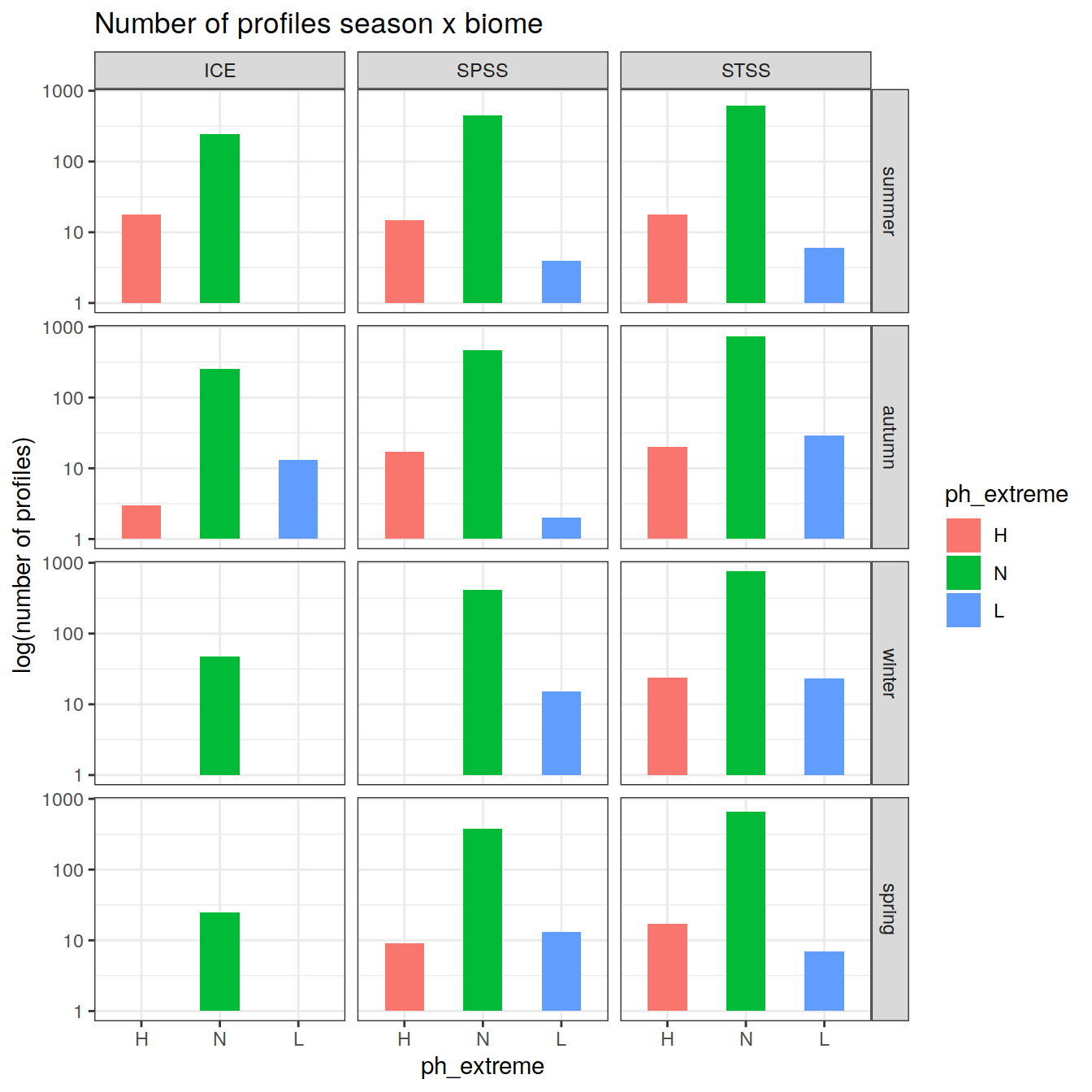
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
# rm(profile_count_biome)Surface Argo vs surface OceanSODA pH (20 m) season x biome
surface_ph_biome <- profile_extreme %>%
filter(depth <= 20) %>%
group_by(season_order, season, biome_name, ph_extreme, file_id) %>%
summarise(argo_surf_ph = mean(pH, na.rm=TRUE),
OceanSODA_surf_ph = mean(OceanSODA_ph, na.rm = TRUE))
surface_ph_biome %>%
group_by(ph_extreme) %>%
group_split(ph_extreme) %>%
map(
~ggplot(data = .x, aes(x = OceanSODA_surf_ph,
y = argo_surf_ph))+
geom_bin2d(data = .x, aes(x = OceanSODA_surf_ph,
y = argo_surf_ph)) +
scale_fill_viridis_c()+
geom_abline(slope = 1, intercept = 0)+
coord_fixed(ratio = 1,
xlim = c(7.94, 8.21),
ylim = c(7.94, 8.21))+
facet_grid(season_order~biome_name, labeller = facet_label) +
labs(title = paste( 'pH extreme:', unique(.x$ph_extreme)),
x = 'OceanSODA pH',
y = 'Argo pH')
)[[1]]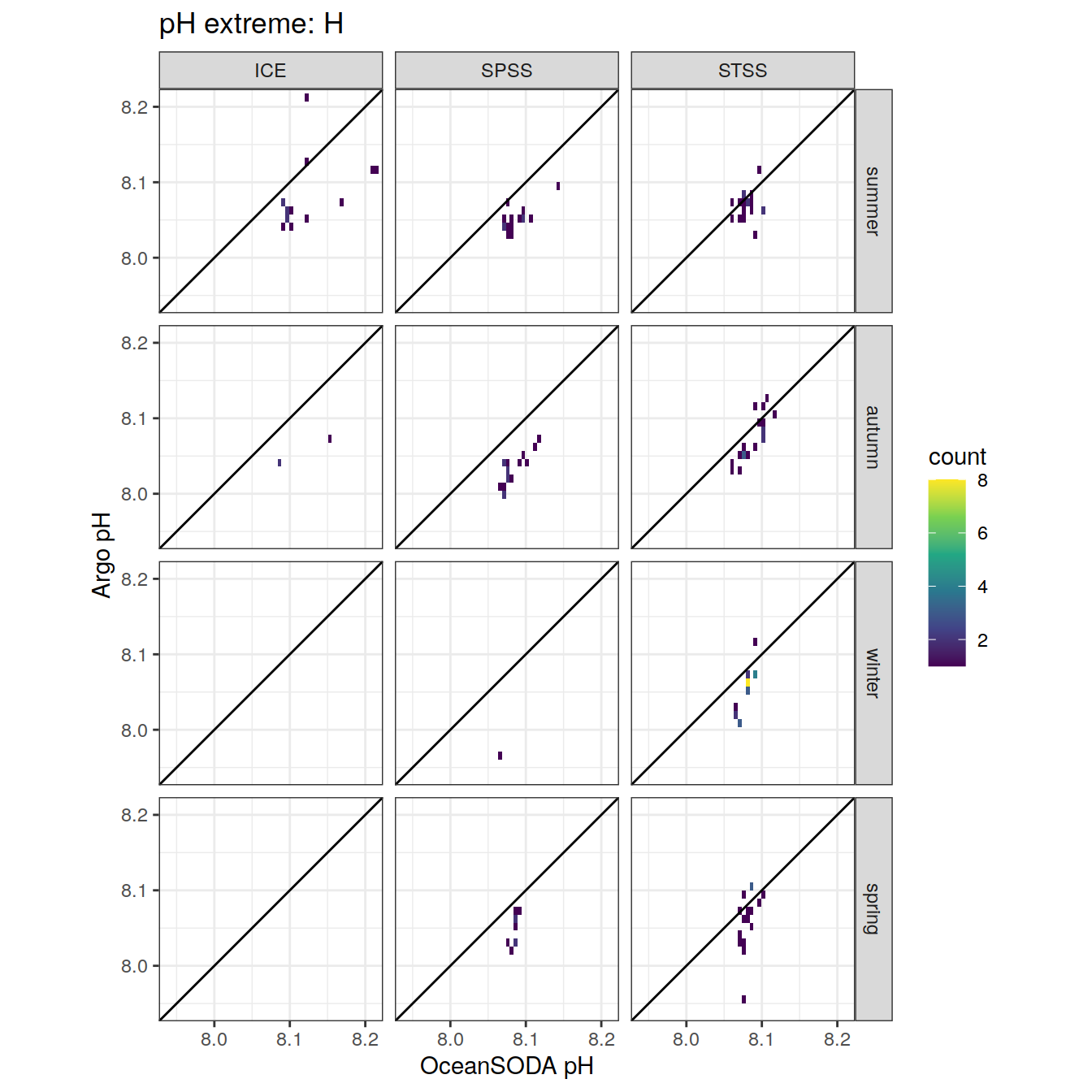
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[2]]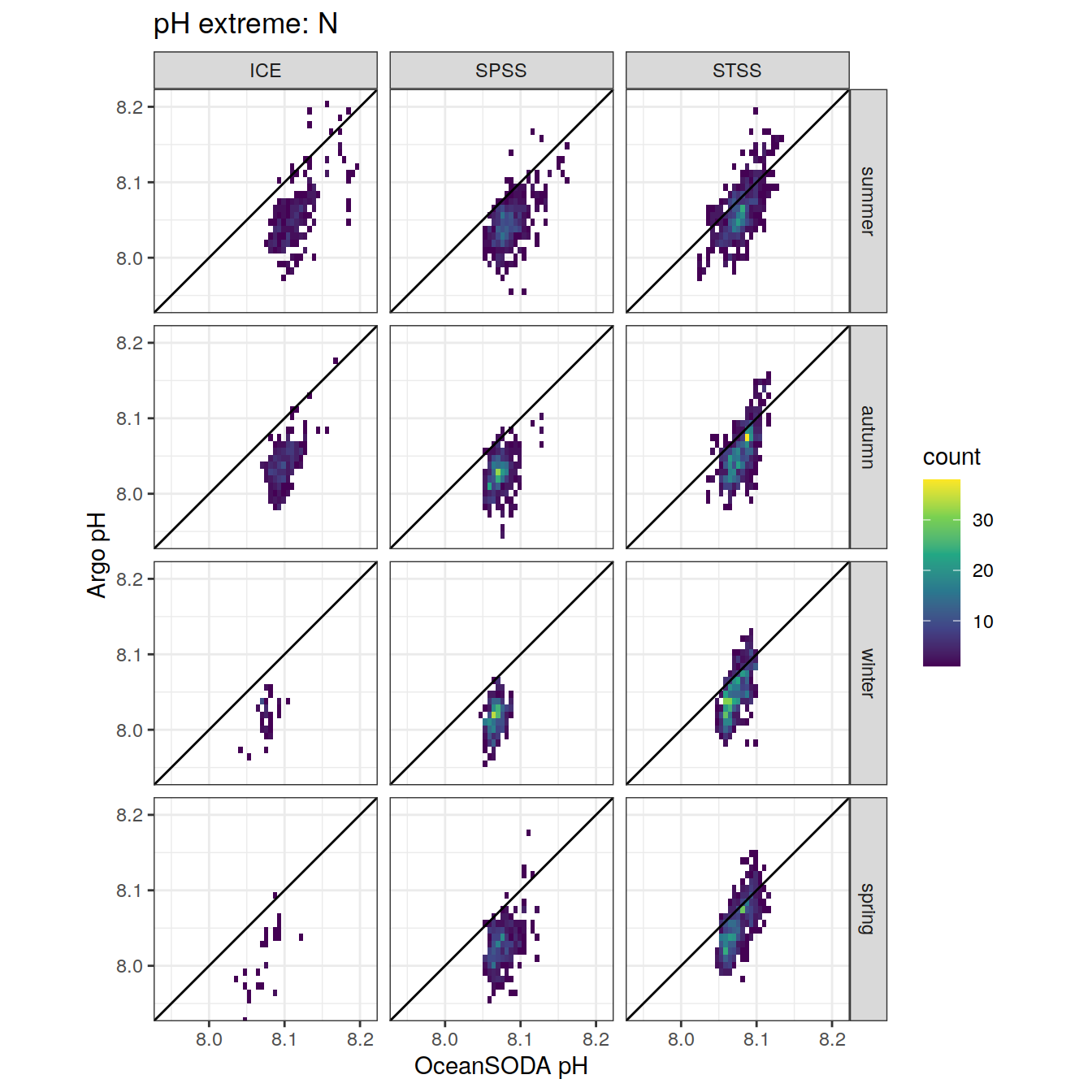
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[3]]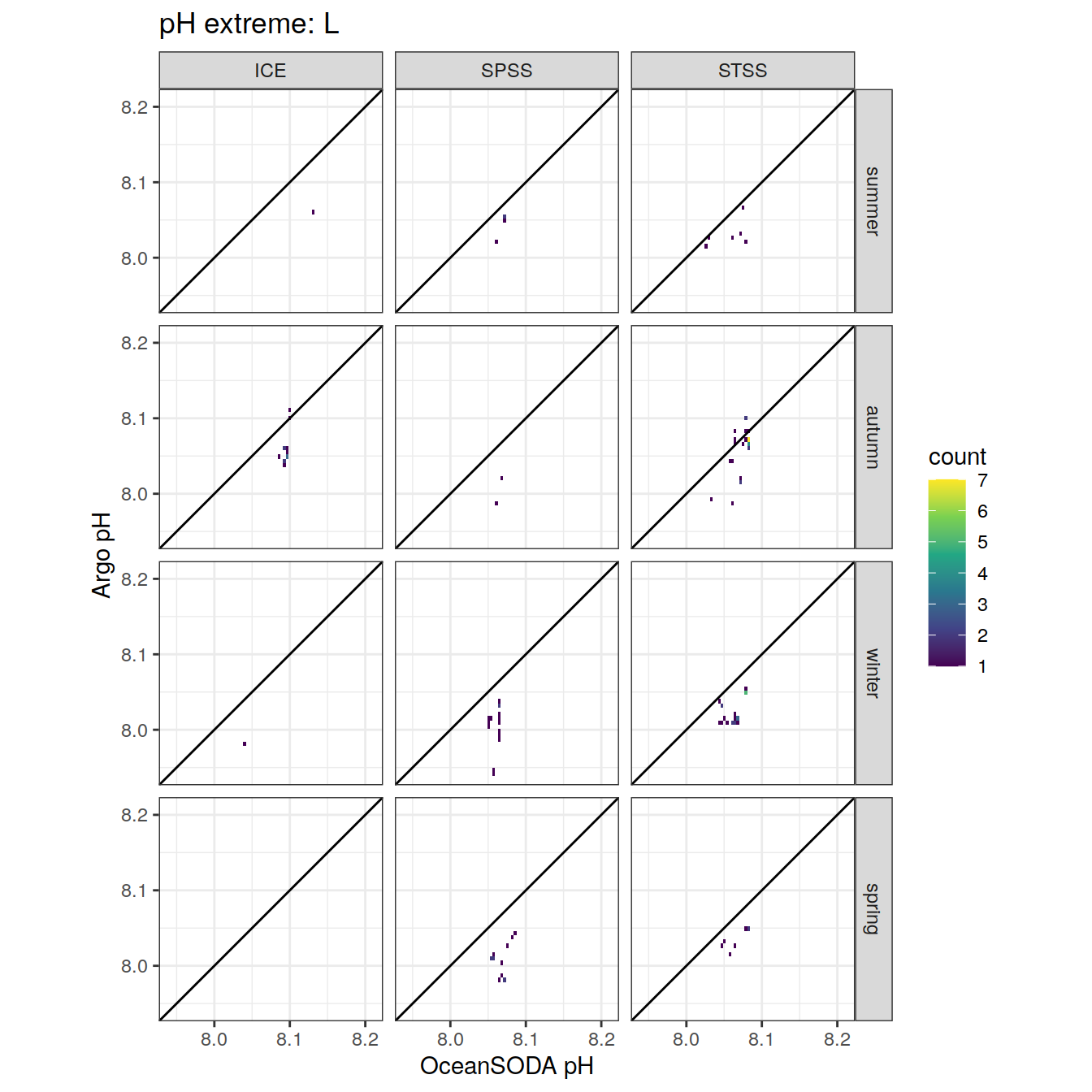
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
rm(surface_ph_biome)Season x basin
profile_extreme_basin <- profile_extreme %>%
group_by(season_order, season, basin_AIP, ph_extreme, depth) %>%
summarise(ph_basin = mean(pH, na.rm = TRUE),
ph_basin_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
profile_extreme_basin %>%
ggplot(aes(x = ph_basin,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_ribbon(aes(xmax = ph_basin + ph_basin_std,
xmin = ph_basin - ph_basin_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.3)+
geom_path()+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
labs(col = 'OceanSODA\npH anomaly\n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly\n(mean ± st dev)',
y = 'depth (m)',
x = 'Basin mean Argo pH',
title = 'Basin-mean Argo profiles')+
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500))) +
facet_grid(season_order~basin_AIP, labeller = facet_label)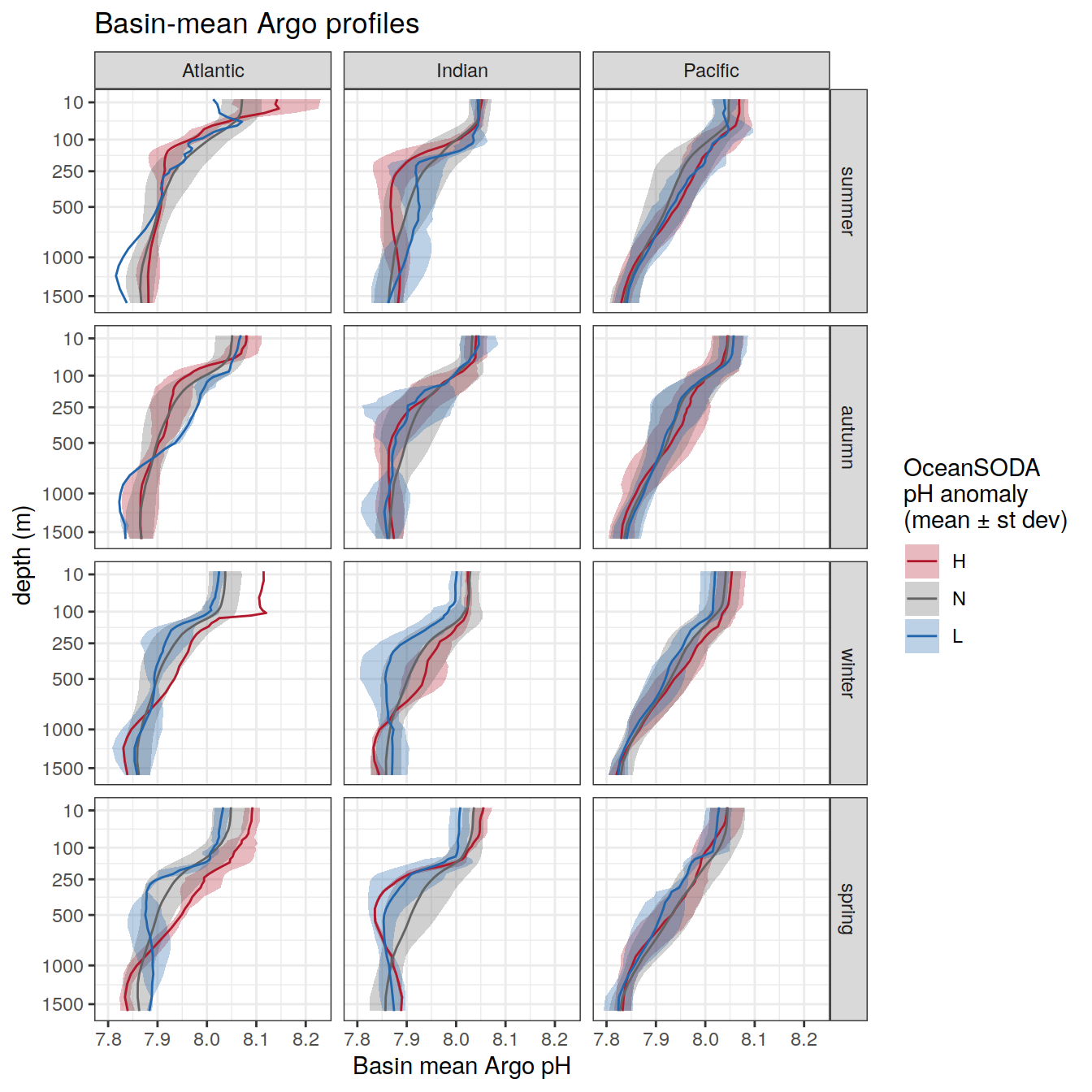
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
| cfd734c | jens-daniel-mueller | 2022-01-28 |
| c44ff0f | pasqualina-vonlanthendinenna | 2022-01-25 |
rm(profile_extreme_basin)Number of profiles season x basin
profile_count_basin <- profile_extreme %>%
distinct(season_order, season, basin_AIP, ph_extreme, file_id) %>%
group_by(season_order, season, basin_AIP, ph_extreme) %>%
count(ph_extreme)
profile_count_basin %>%
ggplot(aes(x = ph_extreme, y = n, fill = ph_extreme))+
geom_col(width = 0.5)+
facet_grid(season_order~basin_AIP, labeller = facet_label)+
scale_y_continuous(trans = 'log10')+
labs(y = 'log(number of profiles)',
title = 'Number of profiles season x basin')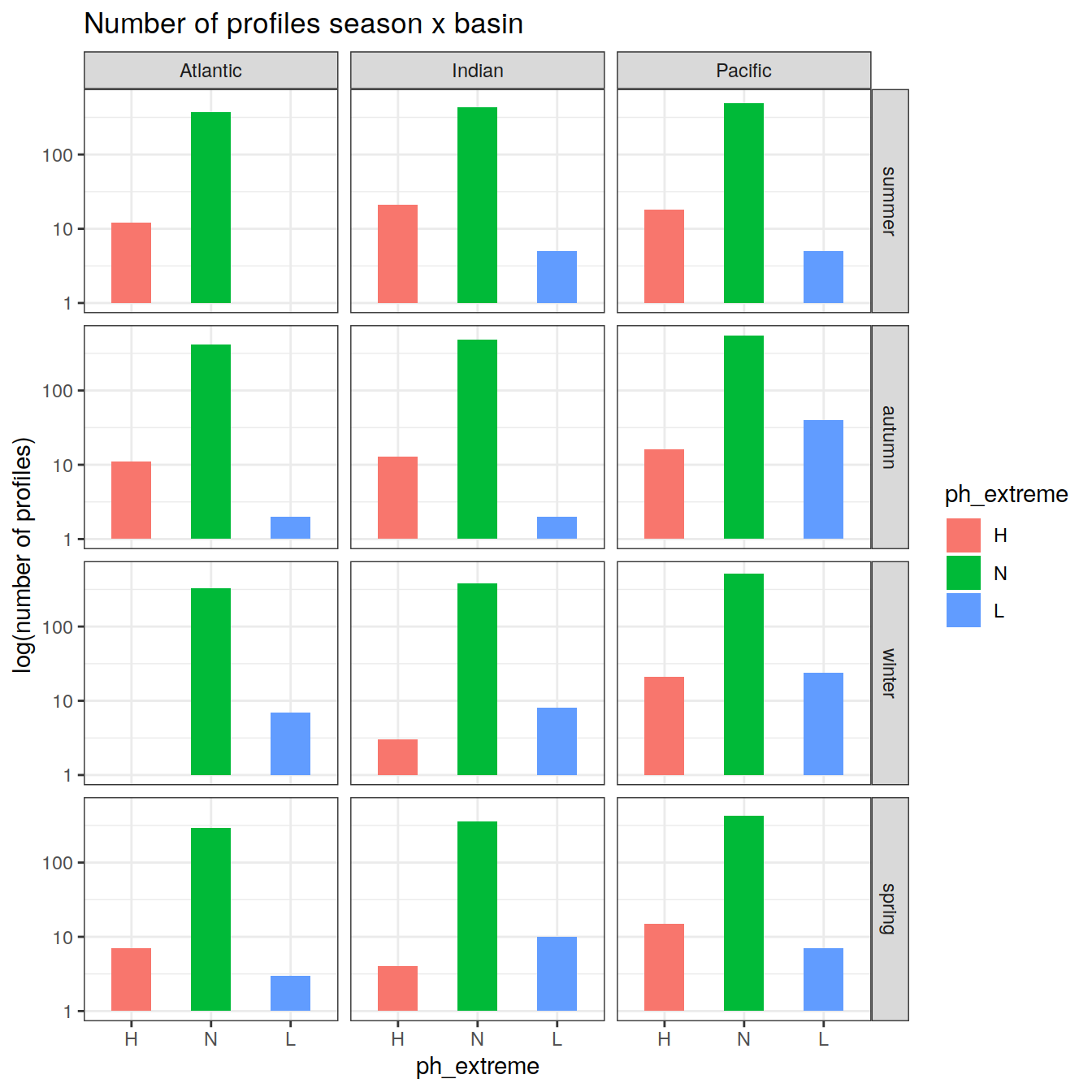
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
# rm(profile_count_basin)Surface Argo vs surface OceanSODA pH (20 m) season x basin
# calculate surface-mean argo pH to compare against OceanSODA surface pH (one value)
surface_ph_basin <- profile_extreme %>%
filter(depth <= 20) %>%
group_by(season_order, season, basin_AIP, ph_extreme, file_id) %>%
summarise(surf_argo_ph = mean(pH, na.rm=TRUE),
surf_OceanSODA_ph = mean(OceanSODA_ph, na.rm = TRUE))
surface_ph_basin %>%
group_by(ph_extreme) %>%
group_split(ph_extreme) %>%
map(
~ggplot(data = .x, aes(x = surf_OceanSODA_ph,
y = surf_argo_ph))+
geom_bin2d(data = .x, aes(x = surf_OceanSODA_ph,
y = surf_argo_ph)) +
scale_fill_viridis_c()+
geom_abline(slope = 1, intercept = 0)+
coord_fixed(ratio = 1,
xlim = c(7.94, 8.21),
ylim = c(7.94, 8.21))+
facet_grid(season_order~basin_AIP, labeller = facet_label) +
labs(title = paste('pH extreme:', unique(.x$ph_extreme)),
x = 'OceanSODA pH',
y = 'Argo pH')
)[[1]]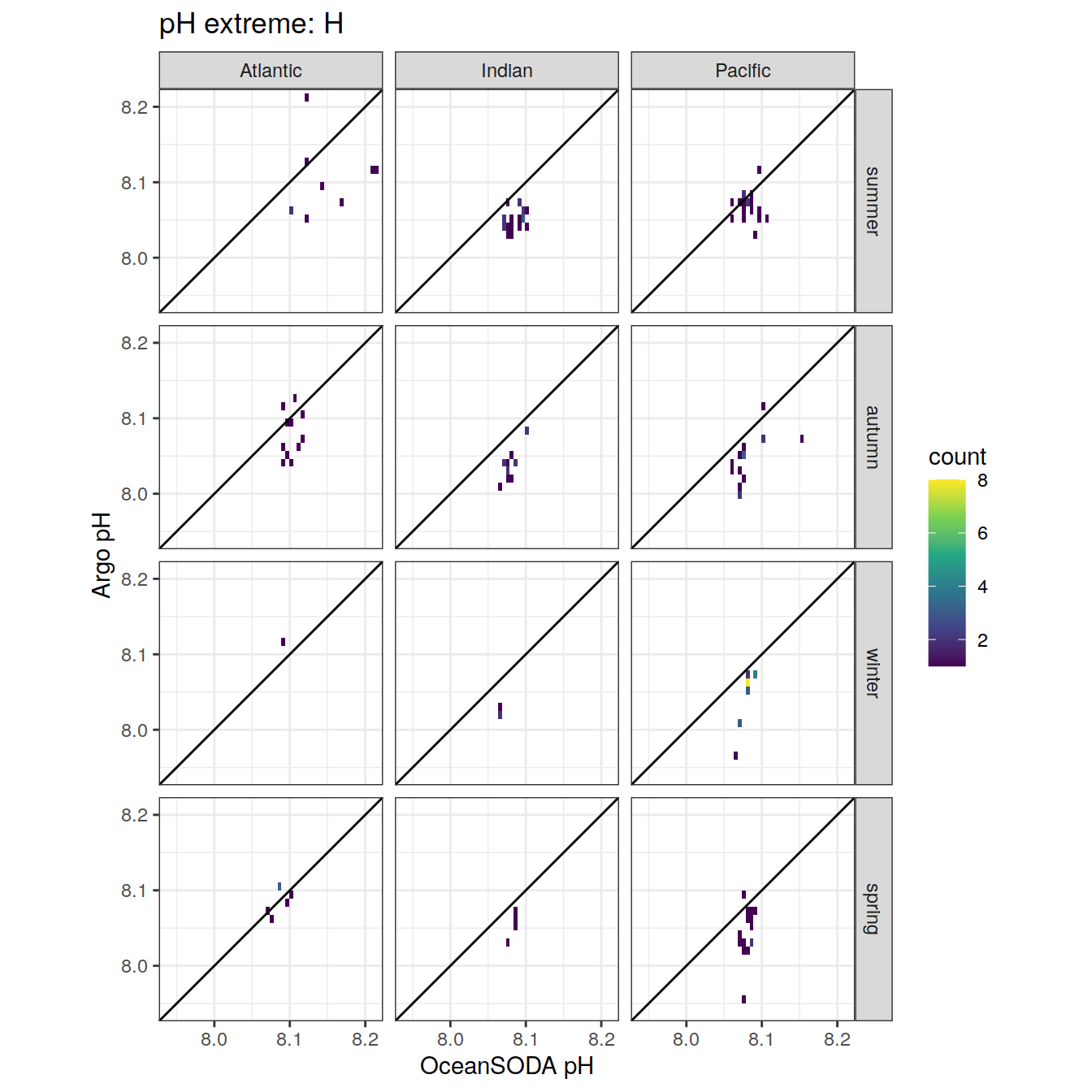
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[2]]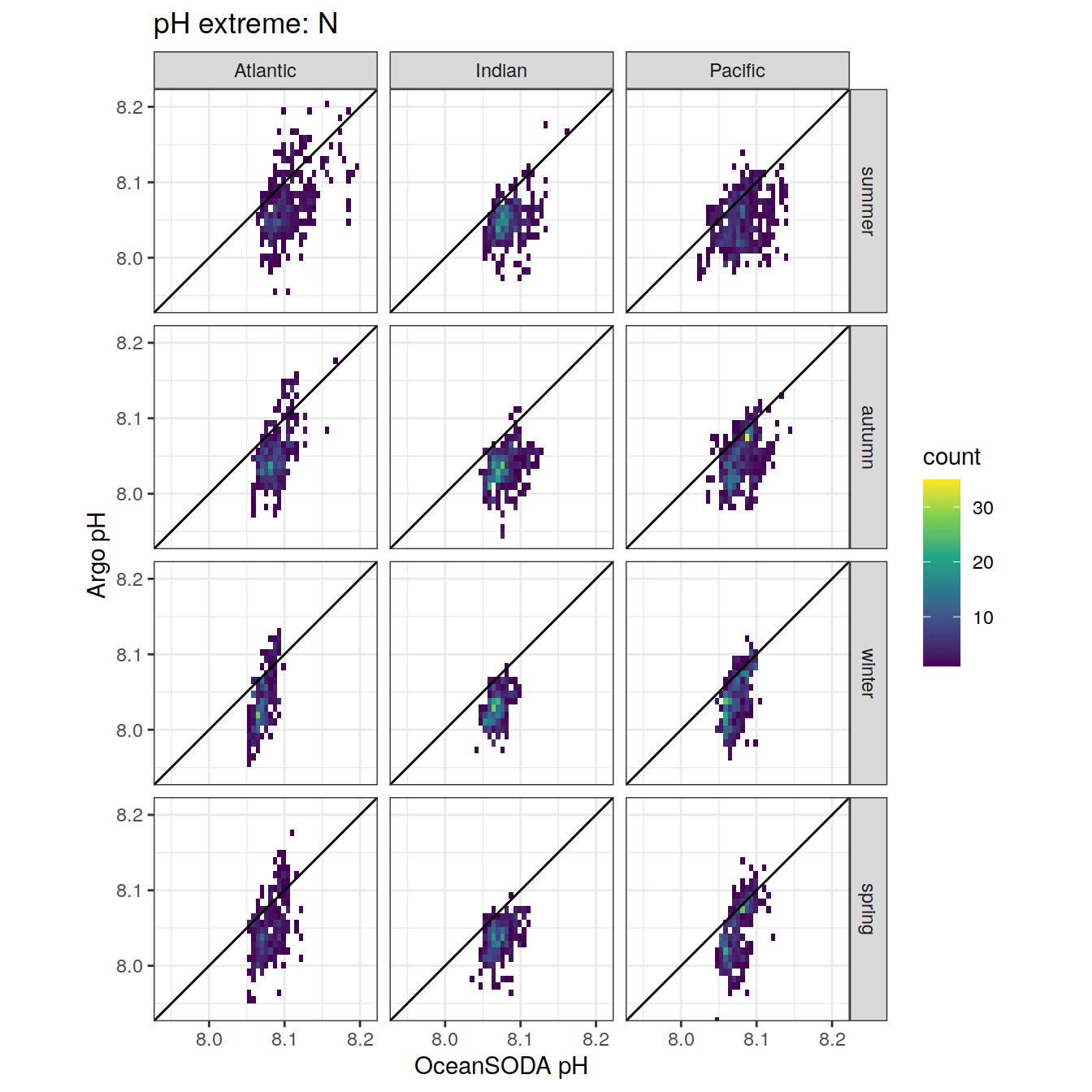
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[3]]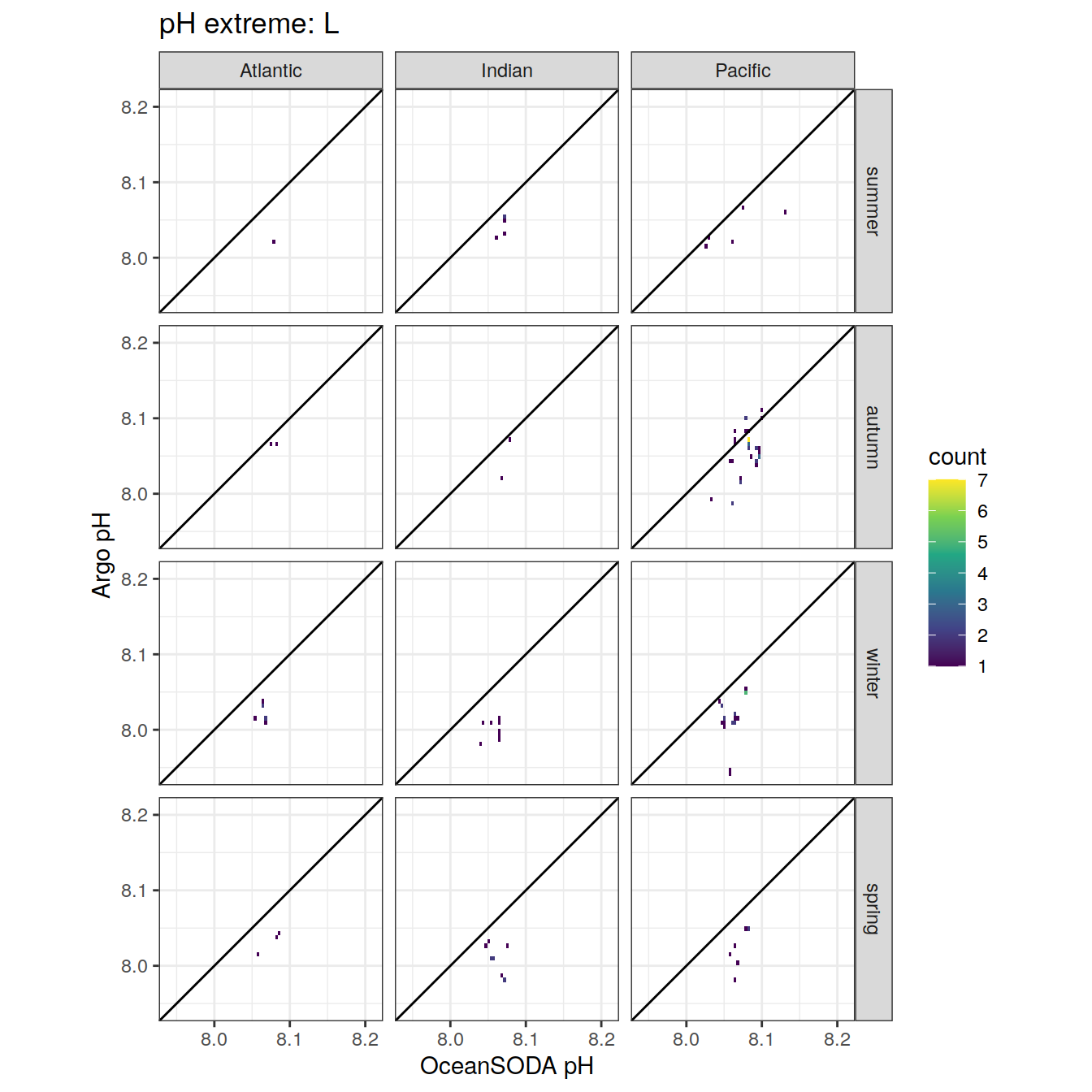
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
rm(surface_ph_basin)Biome-basin January mean
profile_extreme_biome_basin_jan <- profile_extreme %>%
filter(month == 1) %>%
group_by(biome_name, basin_AIP, ph_extreme, depth) %>%
summarise(ph_mean = mean(pH, na.rm = TRUE),
ph_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
profile_extreme_biome_basin_jan %>%
arrange(depth) %>%
ggplot(aes(x = ph_mean,
y = depth)) +
geom_ribbon(aes(xmin = ph_mean - ph_std,
xmax = ph_mean + ph_std,
fill = ph_extreme),
alpha = 0.2)+
geom_path(aes(x = ph_mean,
col = ph_extreme))+
facet_grid(basin_AIP~biome_name)+
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = "Basin-biome-mean January profiles",
col = 'OceanSODA\nph anomaly \n(mean ± st dev)',
fill = 'OceanSODA\nph anomaly \n(mean ± st dev)',
y = 'sqrt(depth)',
x = 'mean Argo pH)') +
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))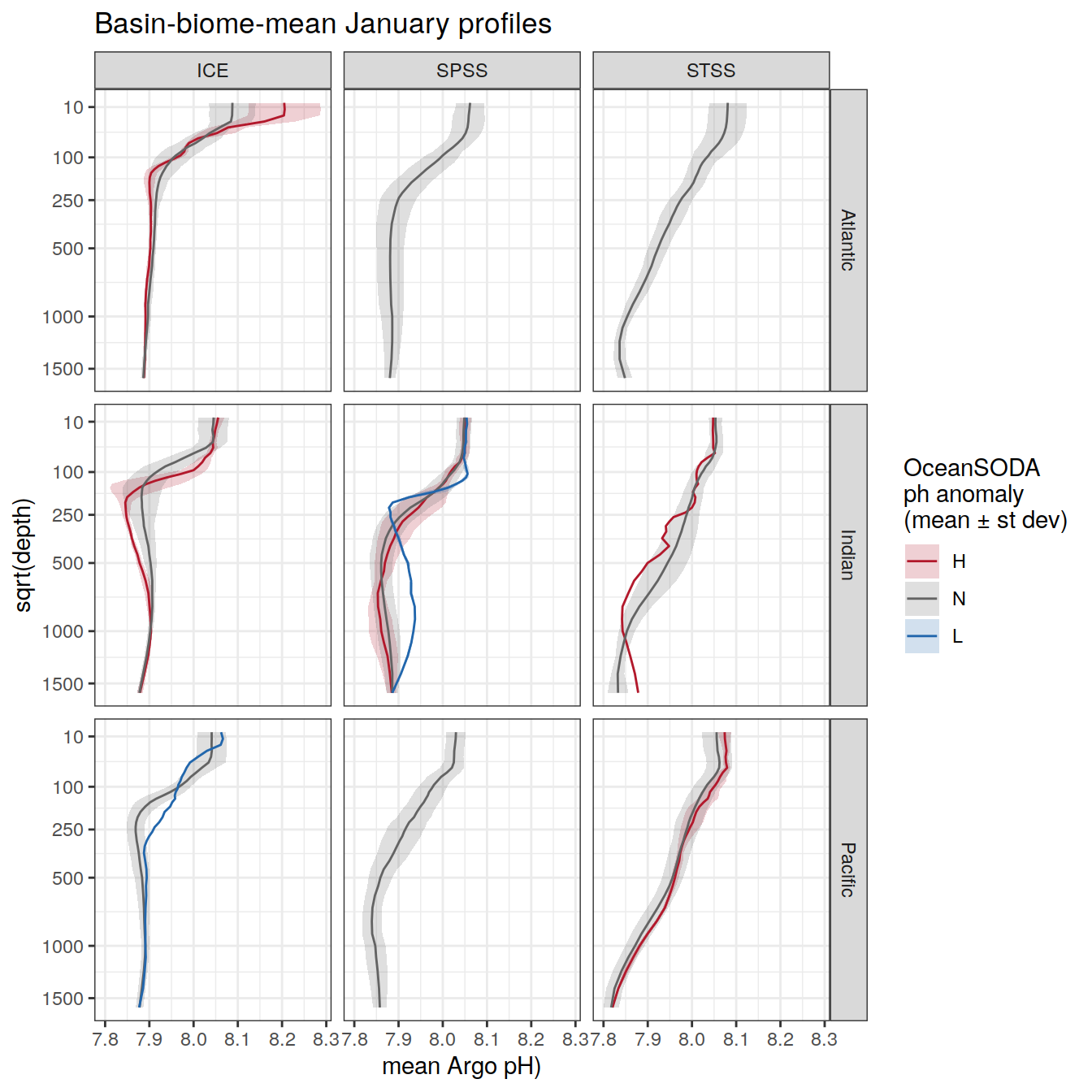
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| a2271df | pasqualina-vonlanthendinenna | 2022-03-30 |
rm(profile_extreme_biome_basin_jan)Season x biome x basin
profile_extreme_season <- profile_extreme %>%
group_by(season_order, season, biome_name, basin_AIP, ph_extreme, depth) %>%
summarise(ph_mean = mean(pH, na.rm = TRUE),
ph_std = sd(pH, na.rm = TRUE)) %>%
ungroup()
profile_extreme_season %>%
arrange(depth) %>%
group_split(season_order) %>%
# head(1) %>%
map(
~ ggplot(
data = .x,
aes(
x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme
)
) +
geom_ribbon(aes(xmax = ph_mean + ph_std,
xmin = ph_mean - ph_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = paste("season:", unique(.x$season)),
col = 'OceanSODA\npH anomaly\n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly\n(mean ± st dev)',
y = 'sqrt(depth)',
x = 'Argo pH') +
scale_y_continuous(
trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500))
) +
facet_grid(basin_AIP ~ biome_name)
)[[1]]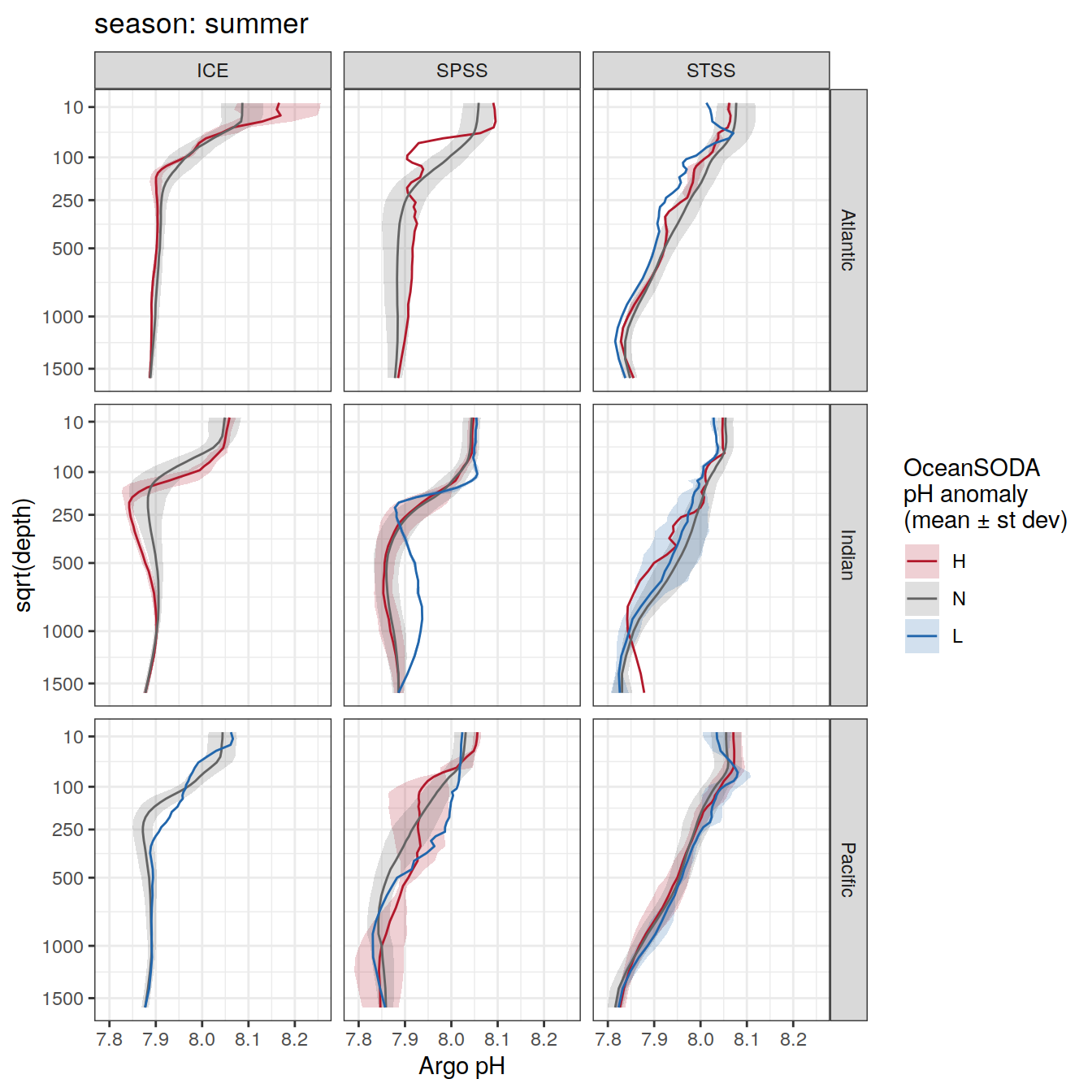
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[2]]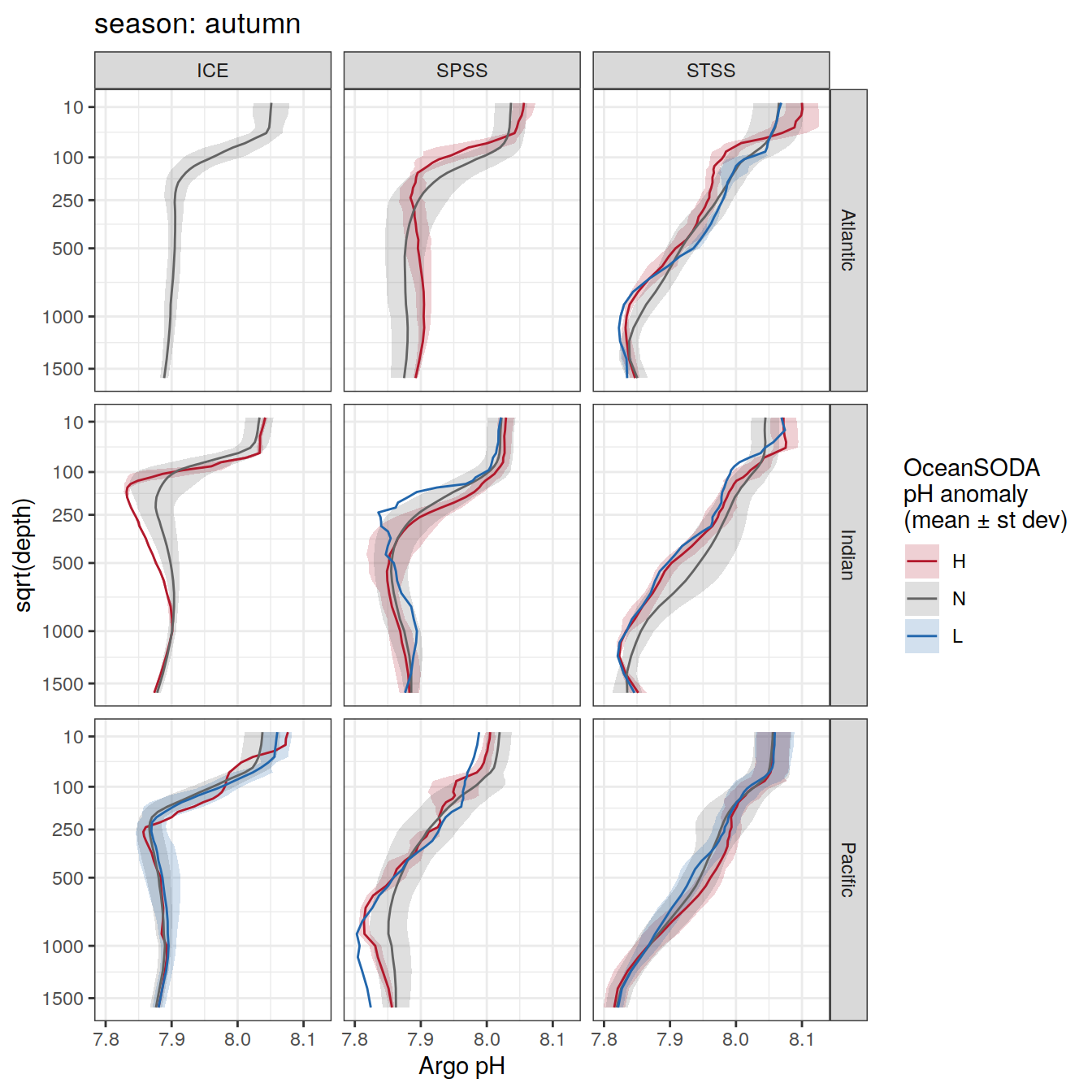
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[3]]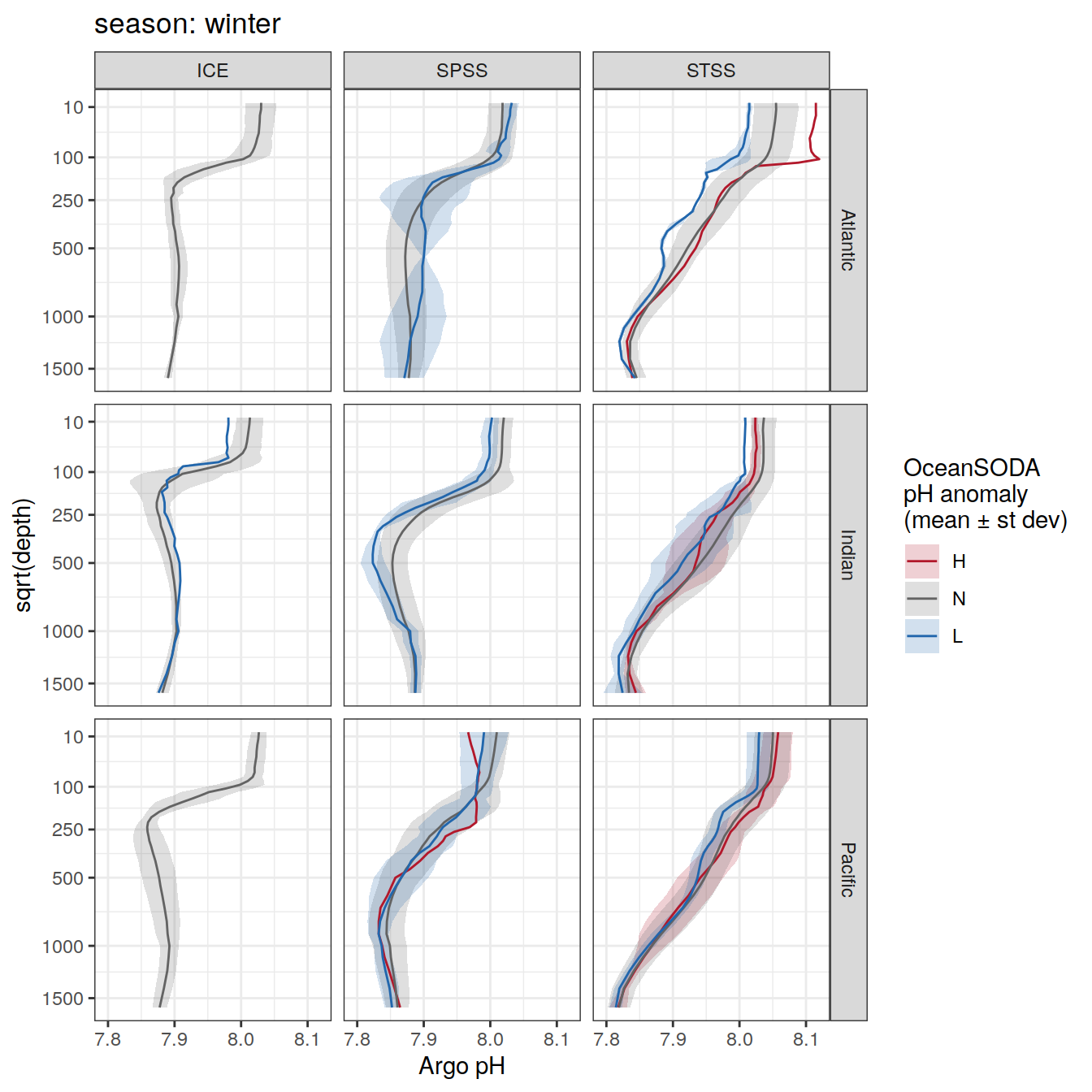
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
[[4]]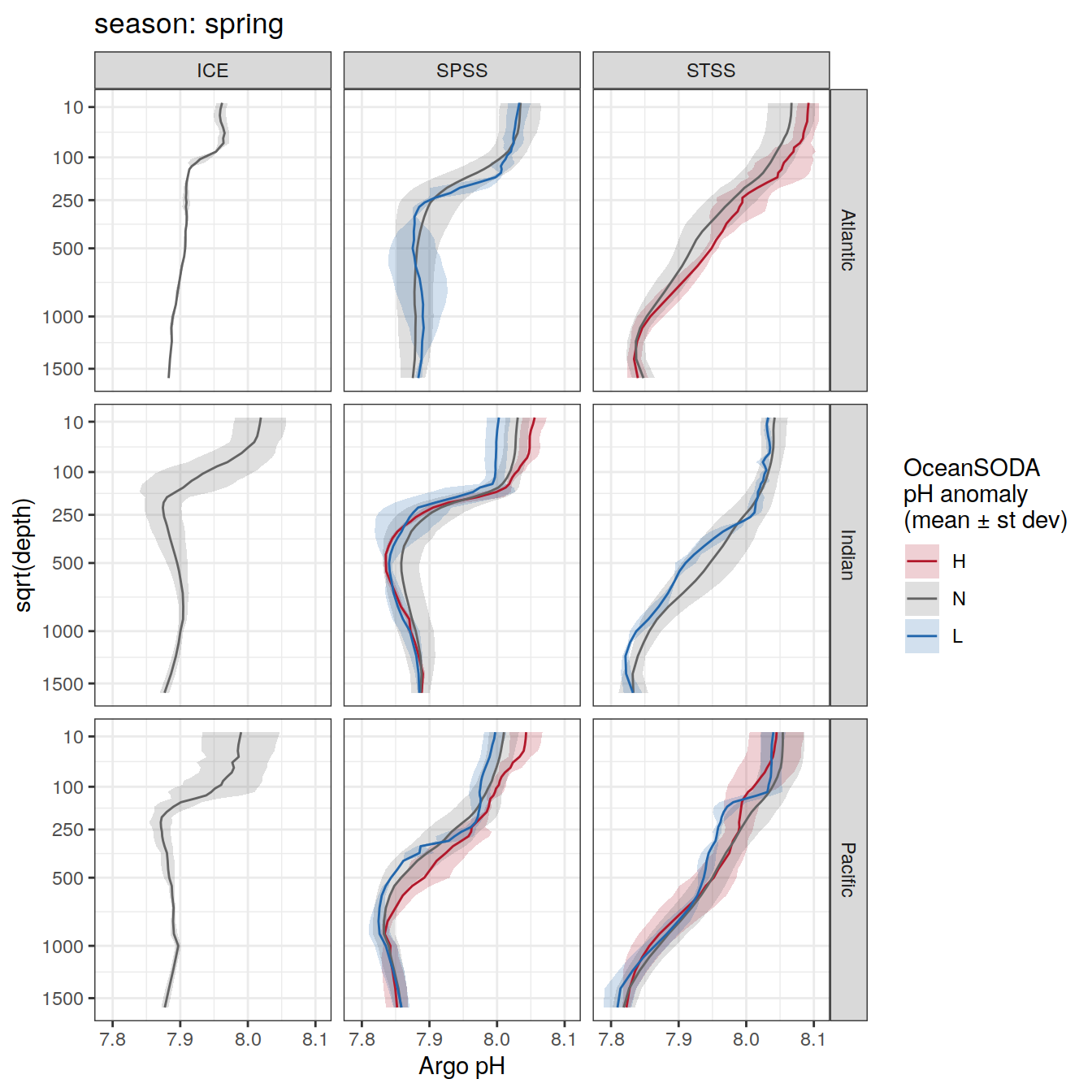
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| daa0a8f | pasqualina-vonlanthendinenna | 2022-02-24 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44fcfb6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 28c8d17 | jens-daniel-mueller | 2022-01-29 |
Number of profiles per season x biome x basin x pH extreme
profile_count_season <- profile_extreme %>%
distinct(season_order, season, biome_name, basin_AIP,
ph_extreme, file_id) %>%
group_by(season_order, season, biome_name, basin_AIP, ph_extreme) %>%
count(ph_extreme)
profile_count_season %>%
group_by(season_order) %>%
group_split(season_order) %>%
map(
~ggplot()+
geom_col(data =.x,
aes(x = ph_extreme,
y = n,
fill = ph_extreme),
width = 0.5)+
facet_grid(basin_AIP ~ biome_name)+
scale_y_continuous(trans = 'log10')+
labs(y = 'log(number of profiles)',
title = paste('season:', unique(.x$season)))
)[[1]]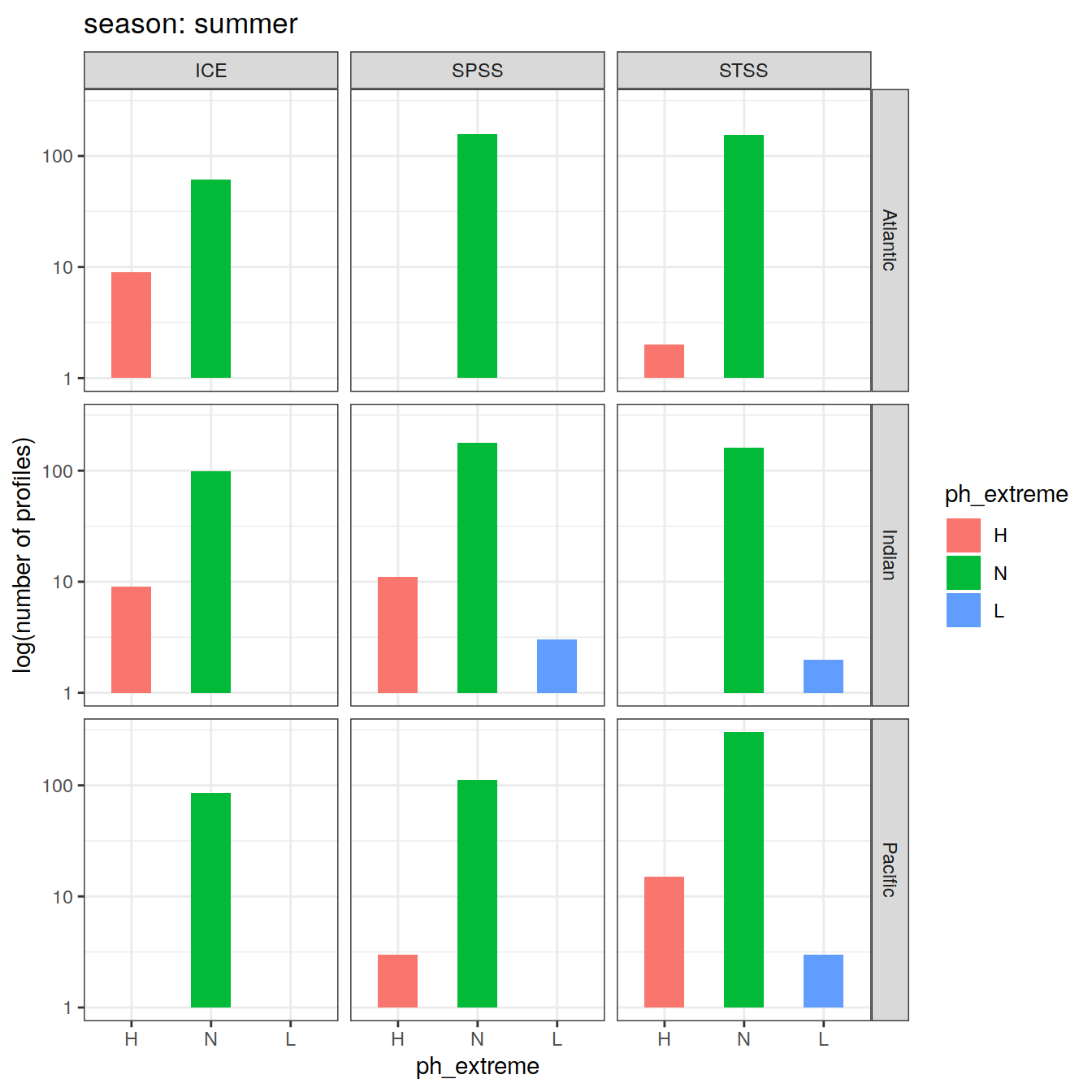
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[2]]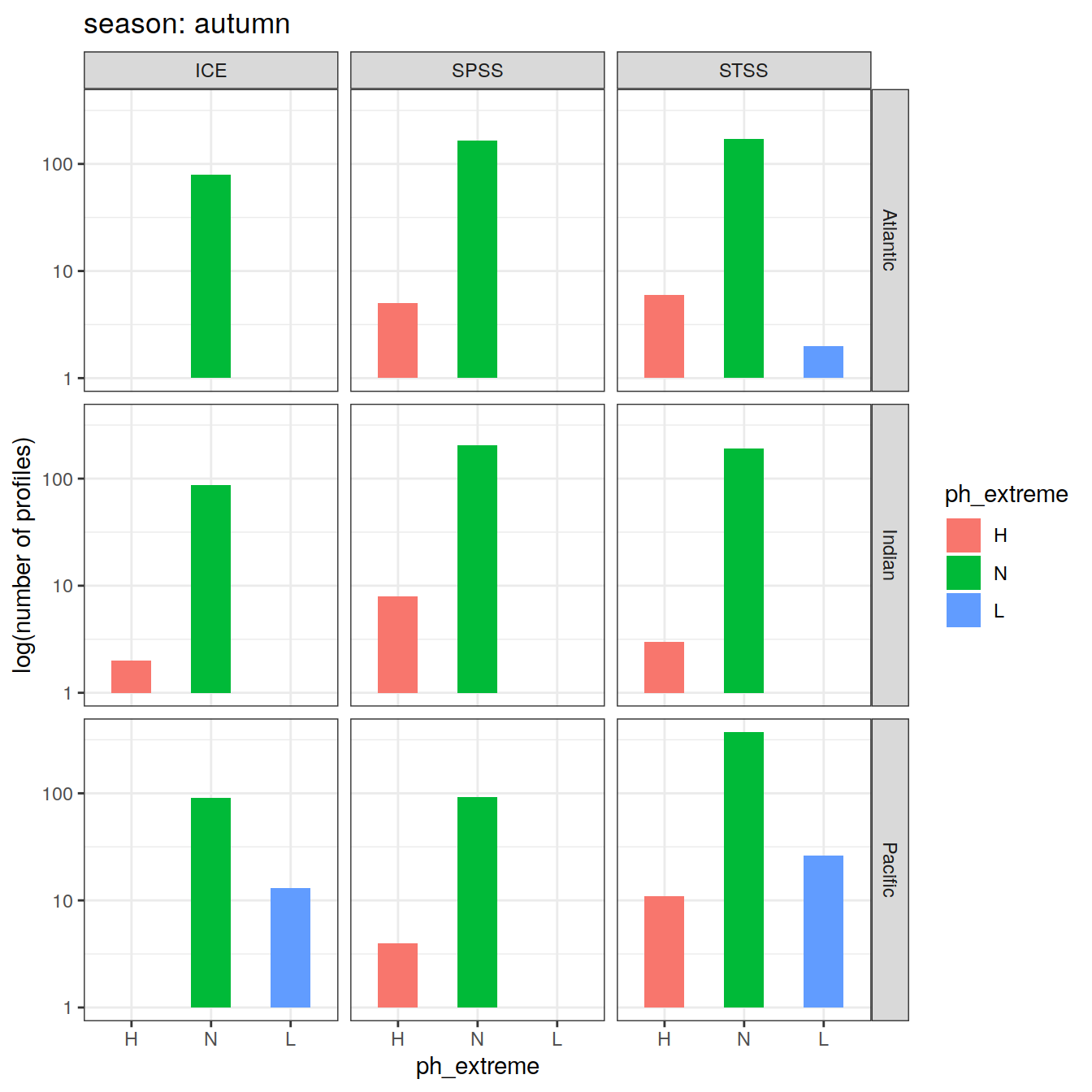
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[3]]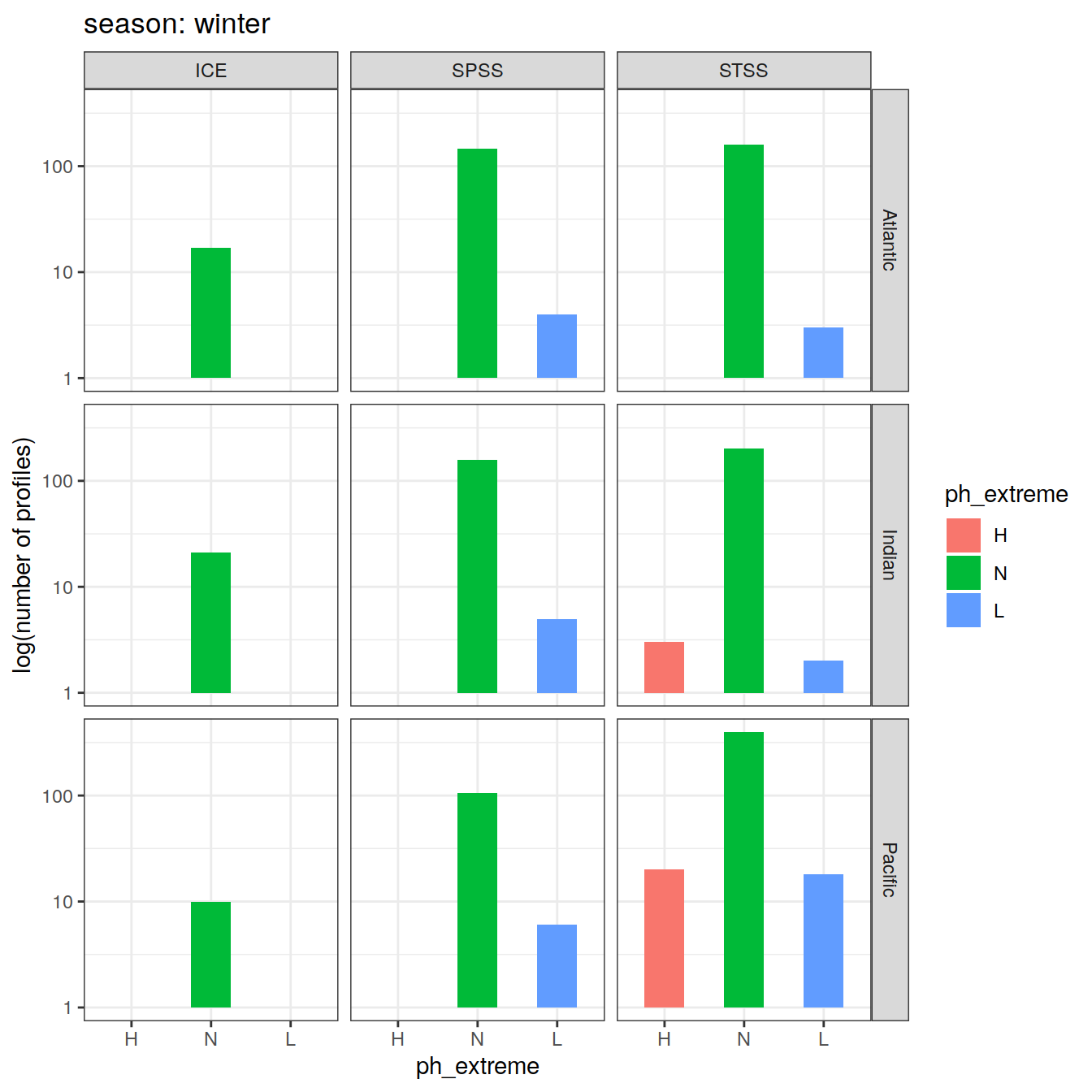
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[4]]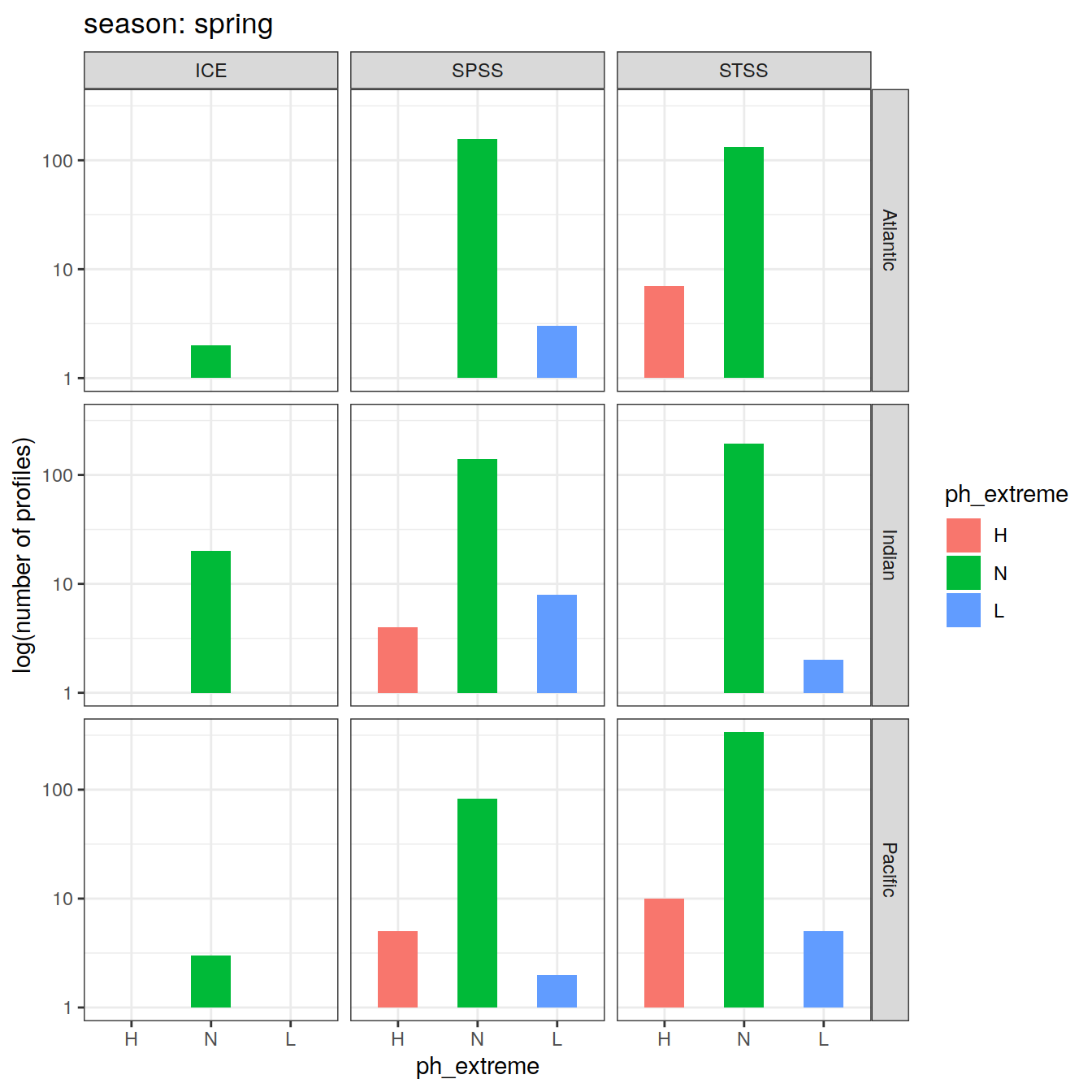
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 2d1bdae | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
# rm(profile_count_season)Surface OceanSODA pH vs surface Argo pH (20 m)
# calculate surface-mean argo pH, for each season x biome x basin x ph extreme
surface_ph_season <- profile_extreme %>%
filter(depth <= 20) %>%
group_by(season_order,
season,
basin_AIP,
biome_name,
ph_extreme,
file_id) %>%
summarise(surf_argo_ph = mean(pH, na.rm=TRUE),
surf_OceanSODA_ph = mean(OceanSODA_ph, na.rm = TRUE))
surface_ph_season %>%
group_by(season_order, ph_extreme) %>%
group_split(season_order, ph_extreme) %>%
map(
~ggplot(data = .x, aes(x = surf_OceanSODA_ph,
y = surf_argo_ph))+
geom_bin2d(data = .x, aes(x = surf_OceanSODA_ph,
y = surf_argo_ph)) +
scale_fill_viridis_c()+
geom_abline(slope = 1, intercept = 0)+
coord_fixed(ratio = 1,
xlim = c(7.94, 8.21),
ylim = c(7.94, 8.21))+
facet_grid(basin_AIP ~ biome_name) +
labs(title = paste('season:', unique(.x$season),
'| pH extreme:', unique(.x$ph_extreme)),
x = 'OceanSODA pH',
y = 'Argo pH')
)[[1]]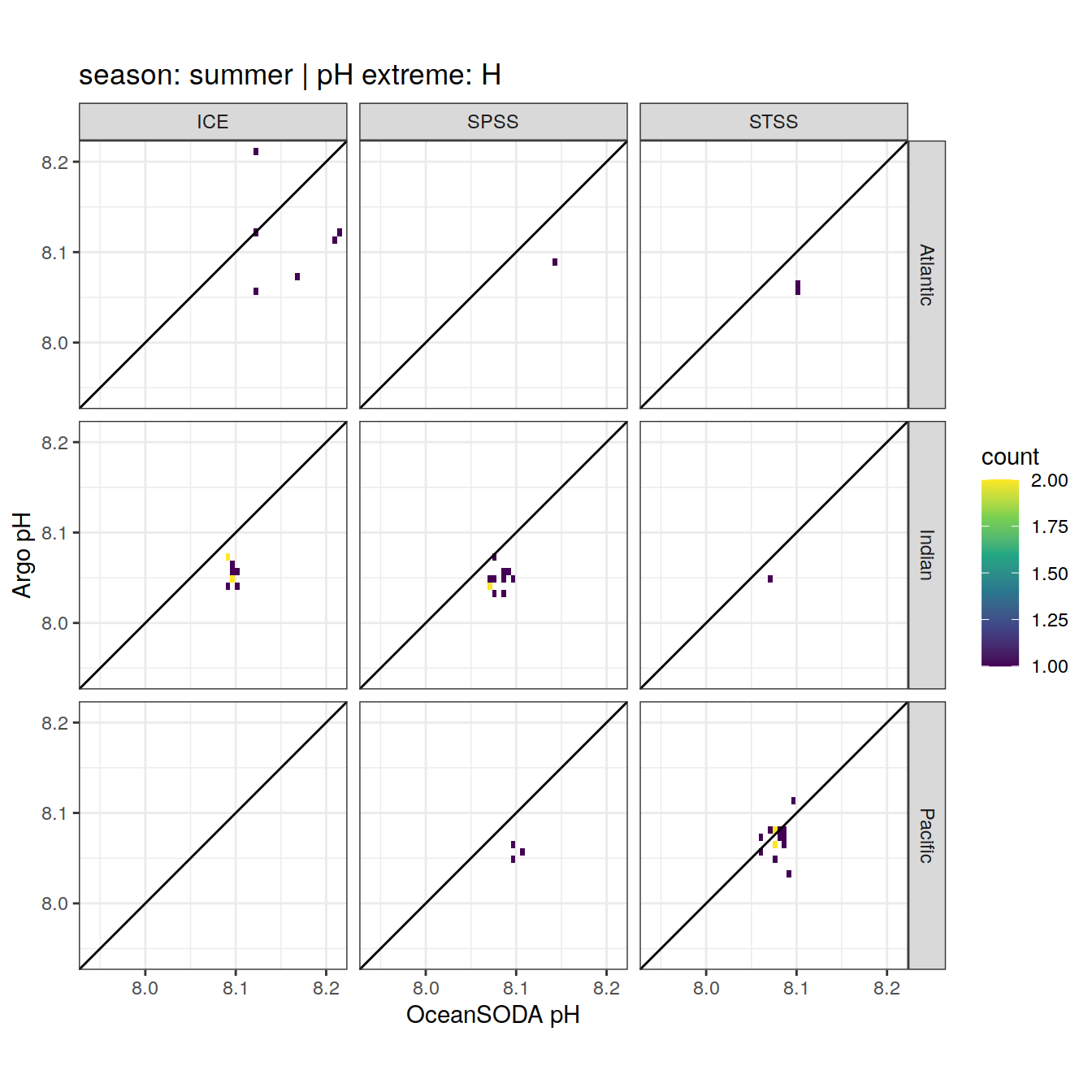
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[2]]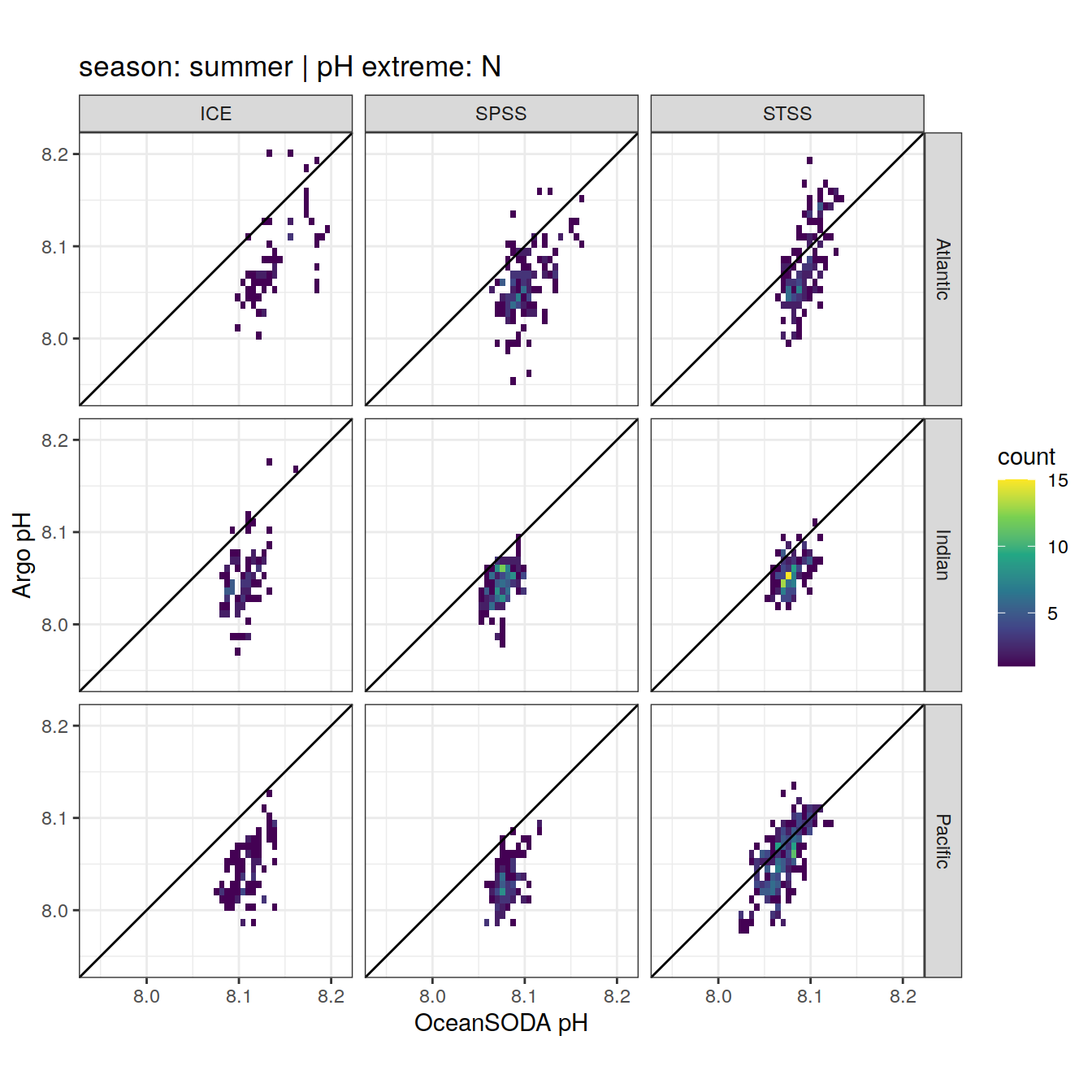
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[3]]
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[4]]
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
| de183c6 | pasqualina-vonlanthendinenna | 2022-02-01 |
| 44a2ec3 | pasqualina-vonlanthendinenna | 2022-02-01 |
[[5]]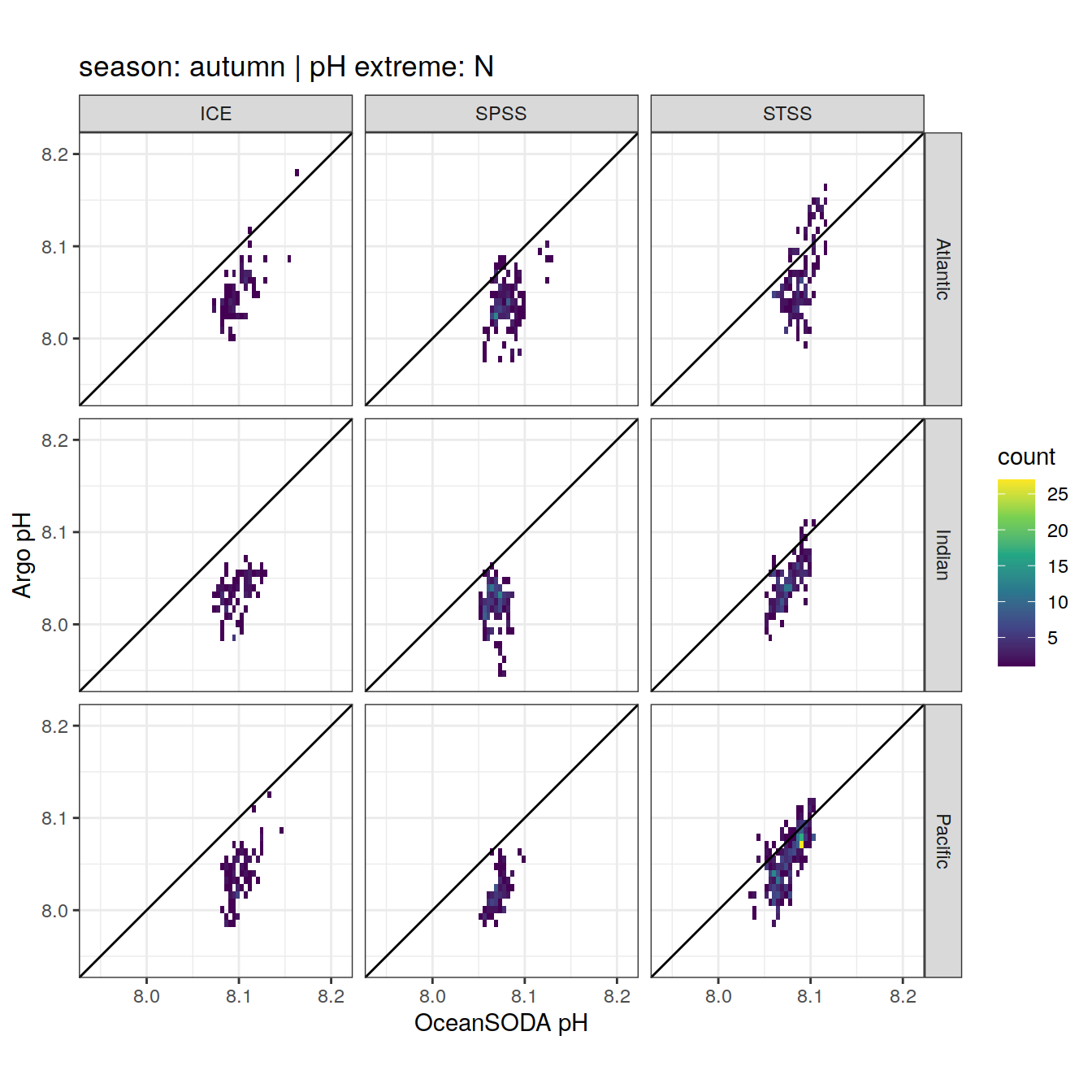
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[6]]
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[7]]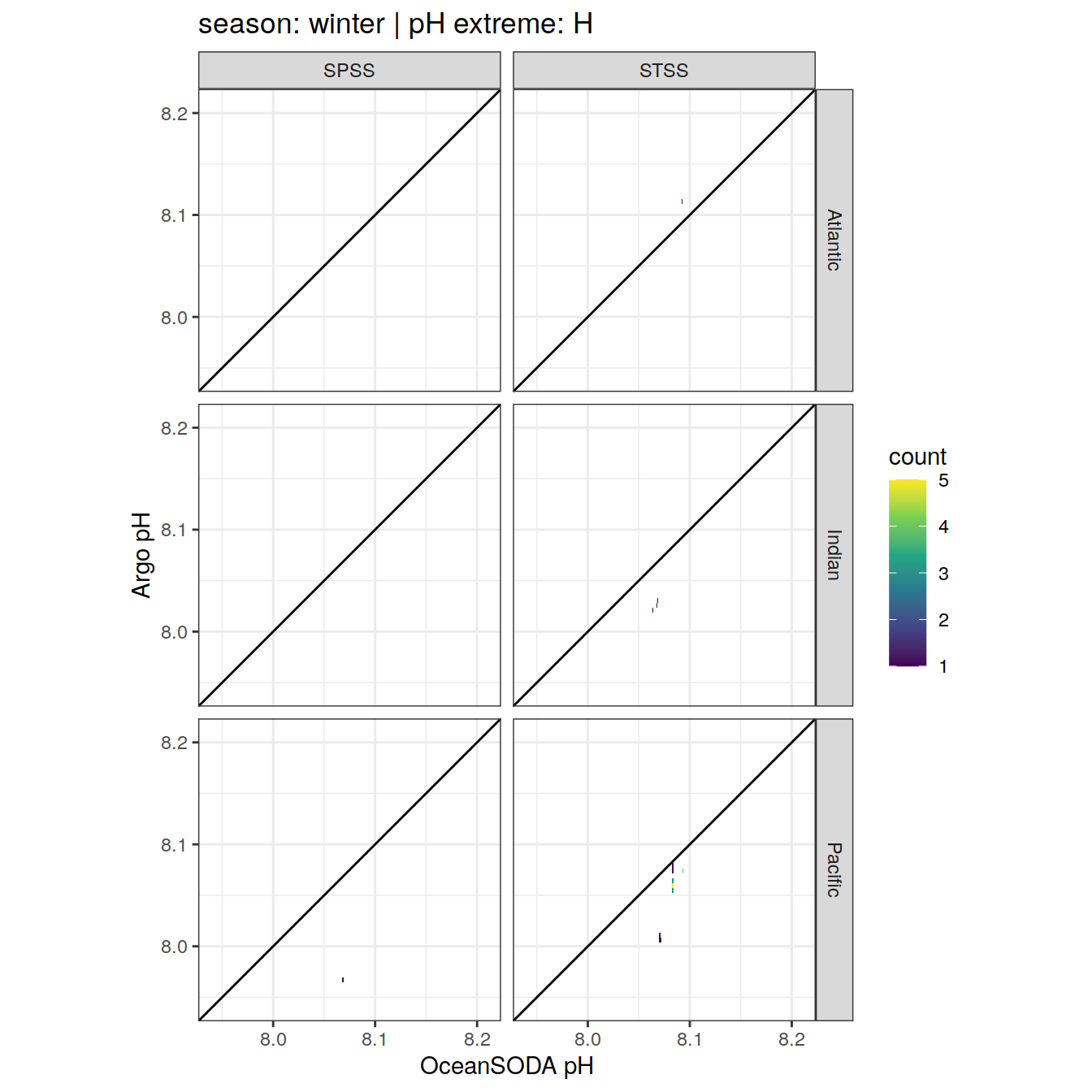
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[8]]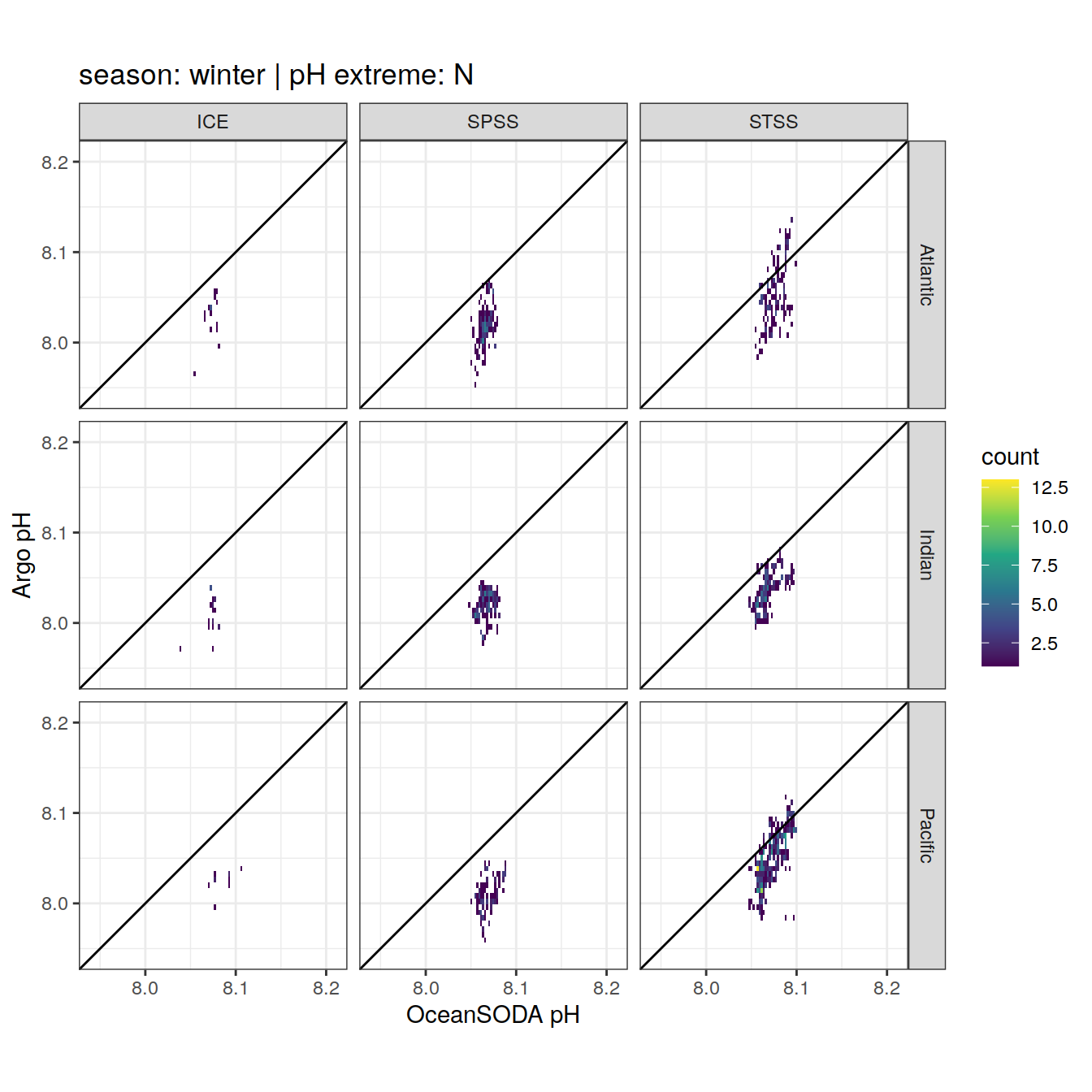
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[9]]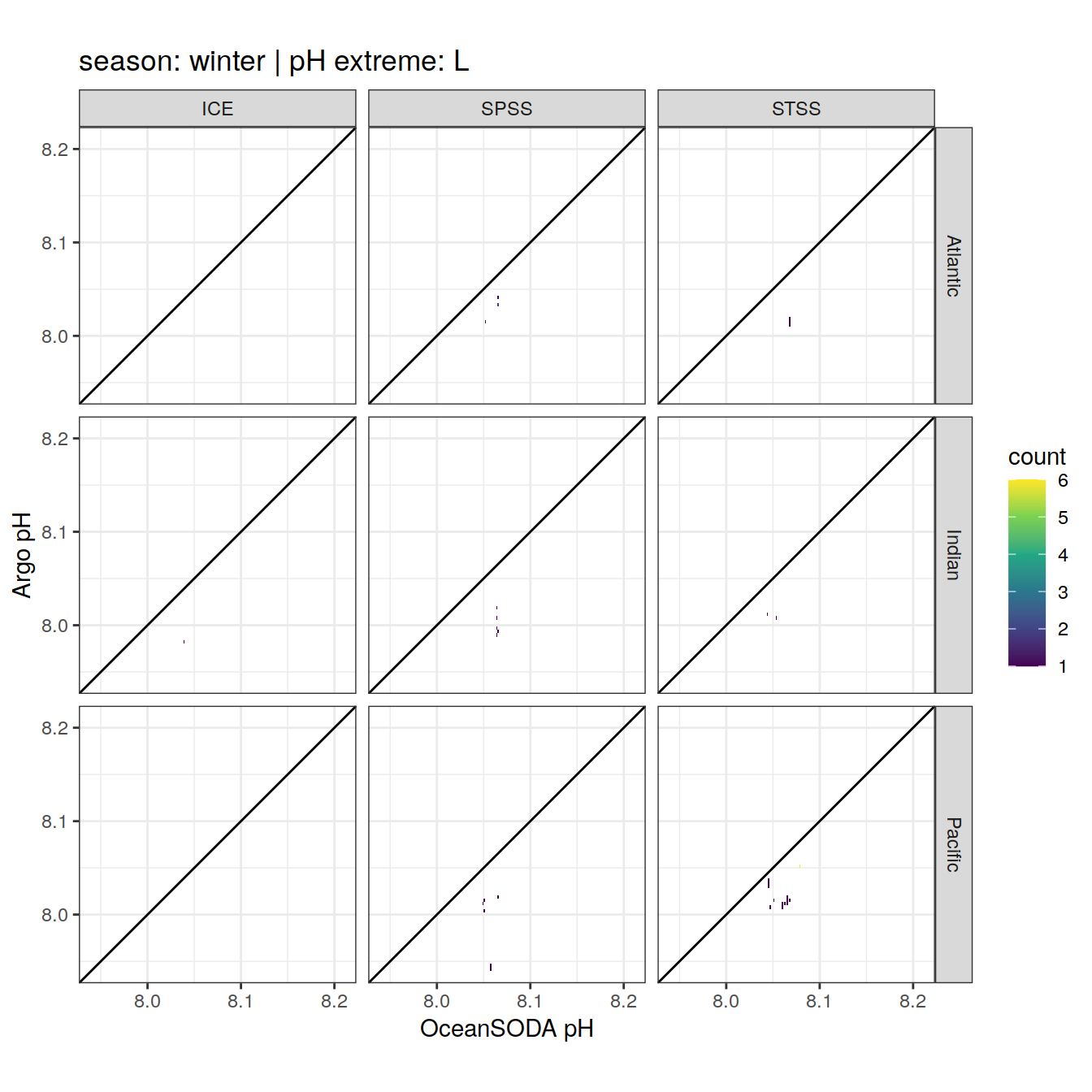
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[10]]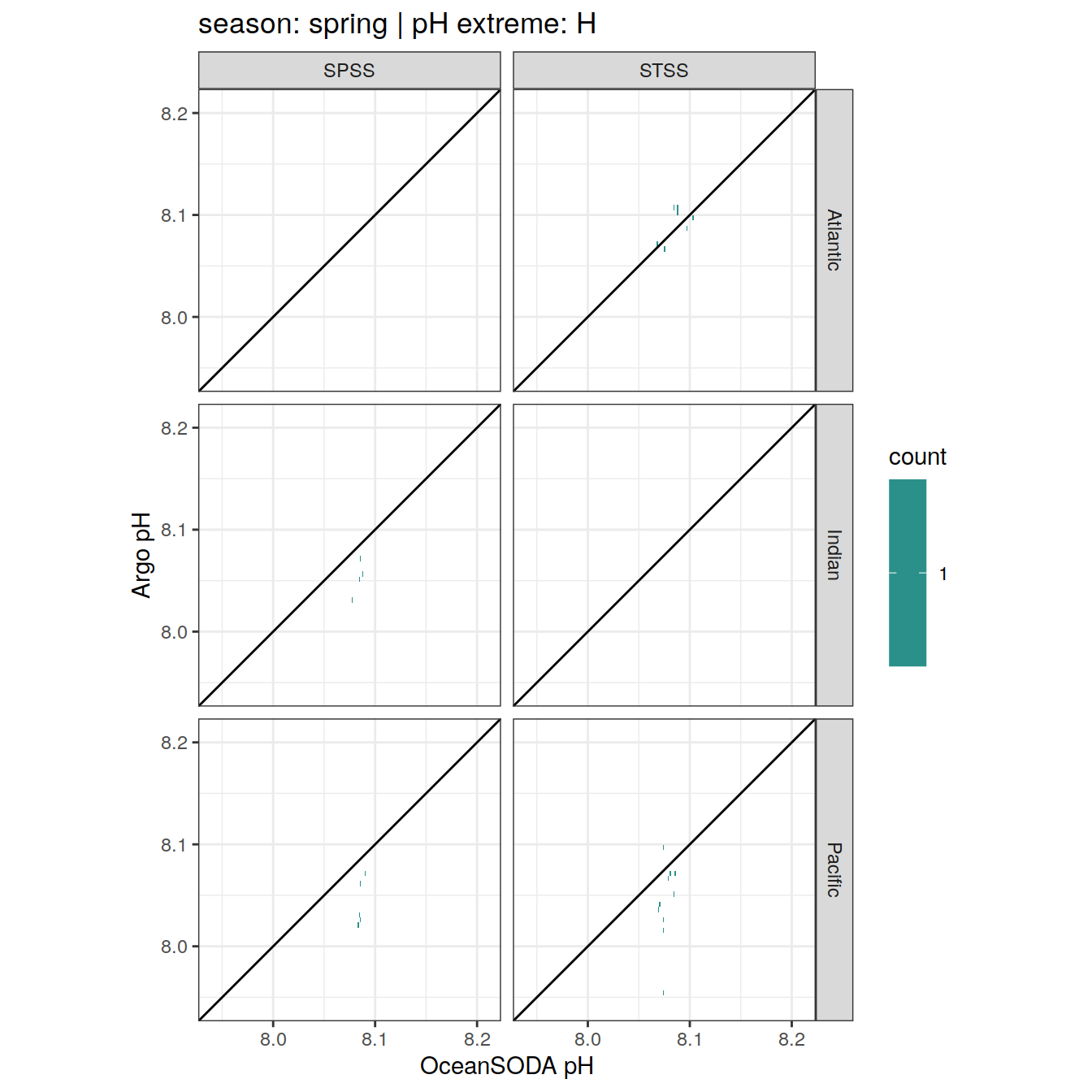
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[11]]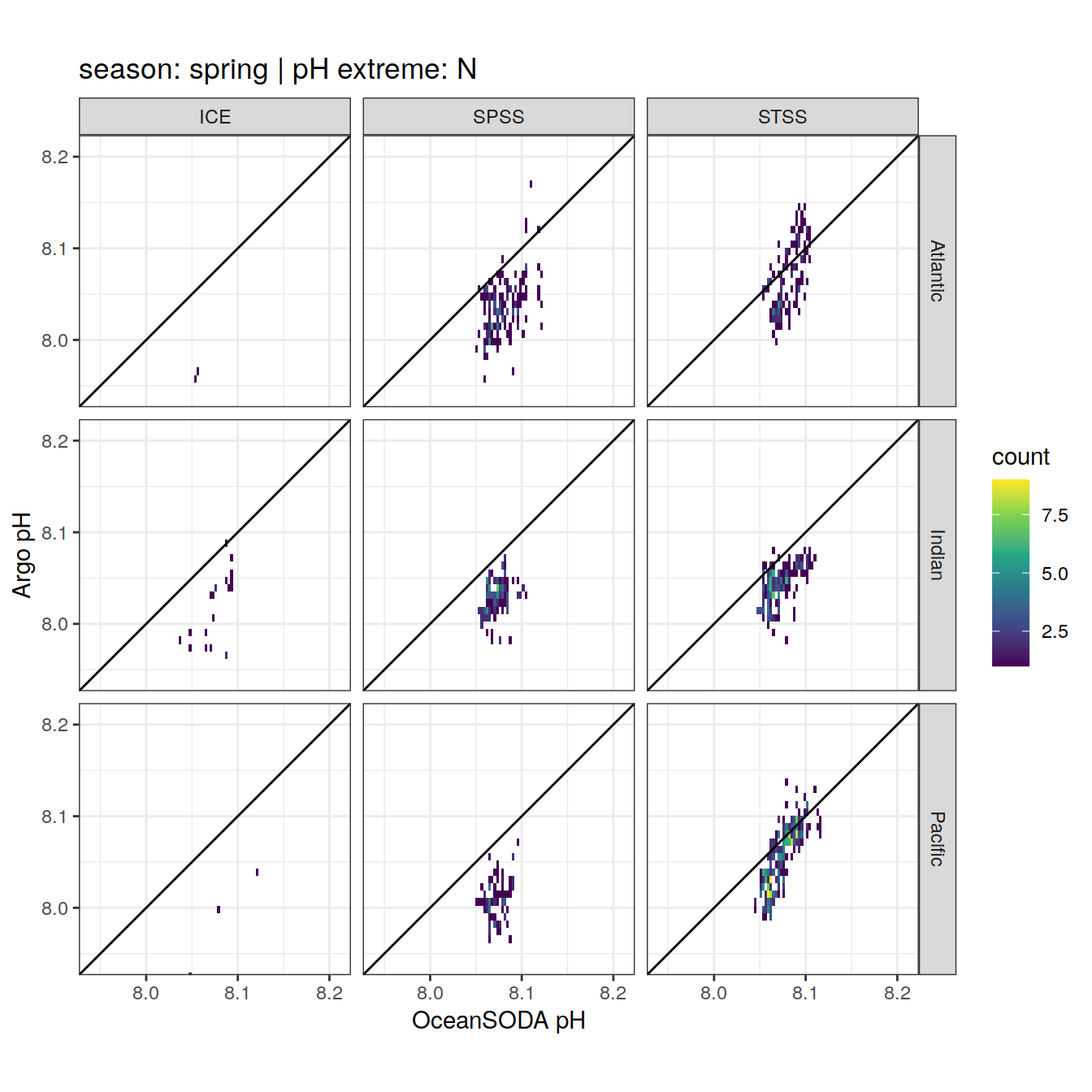
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
[[12]]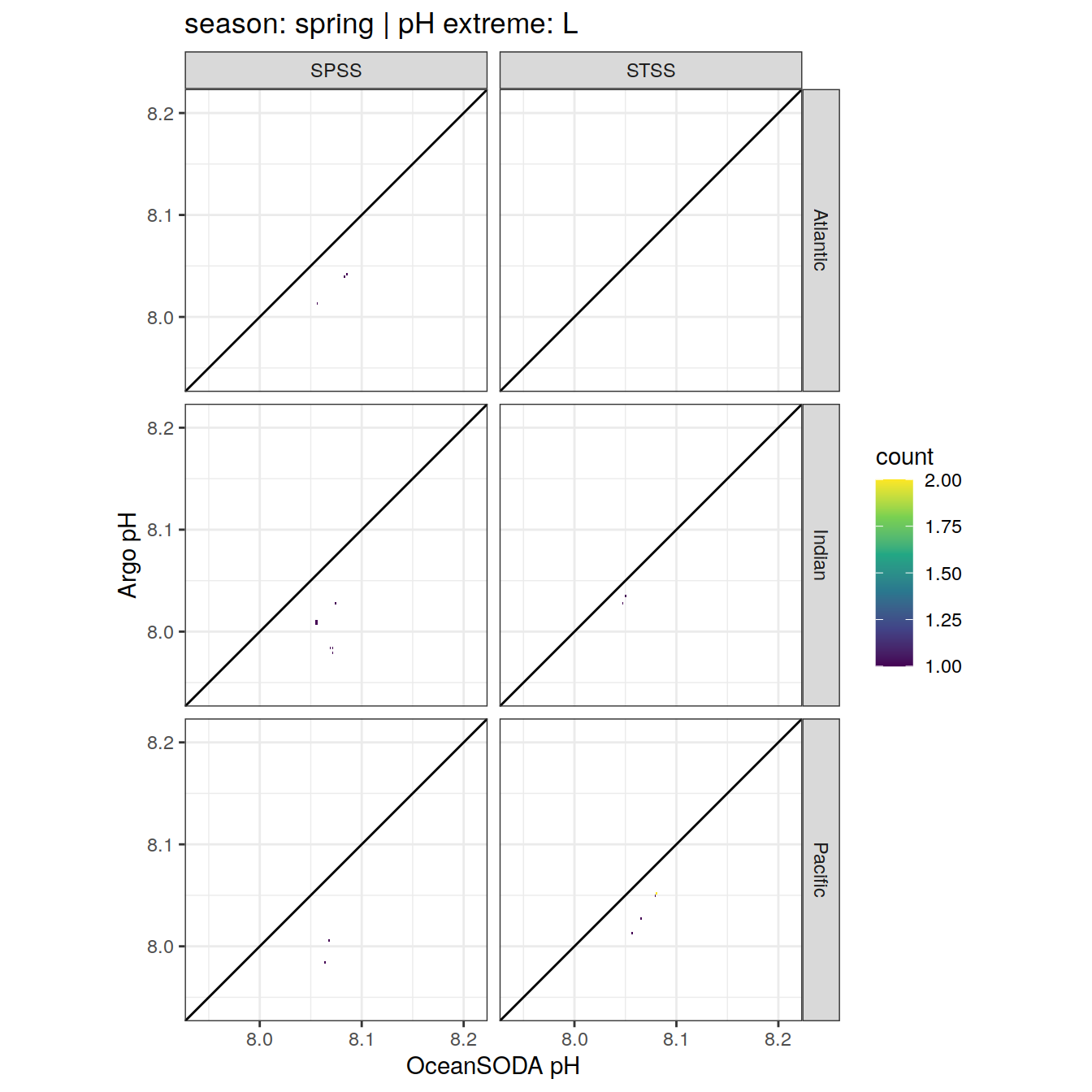
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| fd521d1 | pasqualina-vonlanthendinenna | 2022-02-25 |
| 71ced5f | pasqualina-vonlanthendinenna | 2022-02-23 |
| 25c9e6b | pasqualina-vonlanthendinenna | 2022-02-03 |
| 71958c4 | pasqualina-vonlanthendinenna | 2022-02-03 |
| d7debab | pasqualina-vonlanthendinenna | 2022-02-02 |
| 7376be6 | pasqualina-vonlanthendinenna | 2022-02-02 |
rm(surface_ph_season)Atl, SPSS biome, winter
profile_extreme_season %>%
filter(basin_AIP == 'Atlantic',
season == 'winter',
biome_name == 'SPSS') %>%
arrange(depth) %>%
ggplot(aes(x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme)) +
geom_ribbon(aes(xmax = ph_mean + ph_std,
xmin = ph_mean - ph_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = 'Atlantic basin, SPSS biome, winter',
col = 'OceanSODA\npH anomaly\n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly\n(mean ± st dev)',
y = 'sqrt(depth)',
x = 'Argo pH') +
scale_y_continuous(
trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))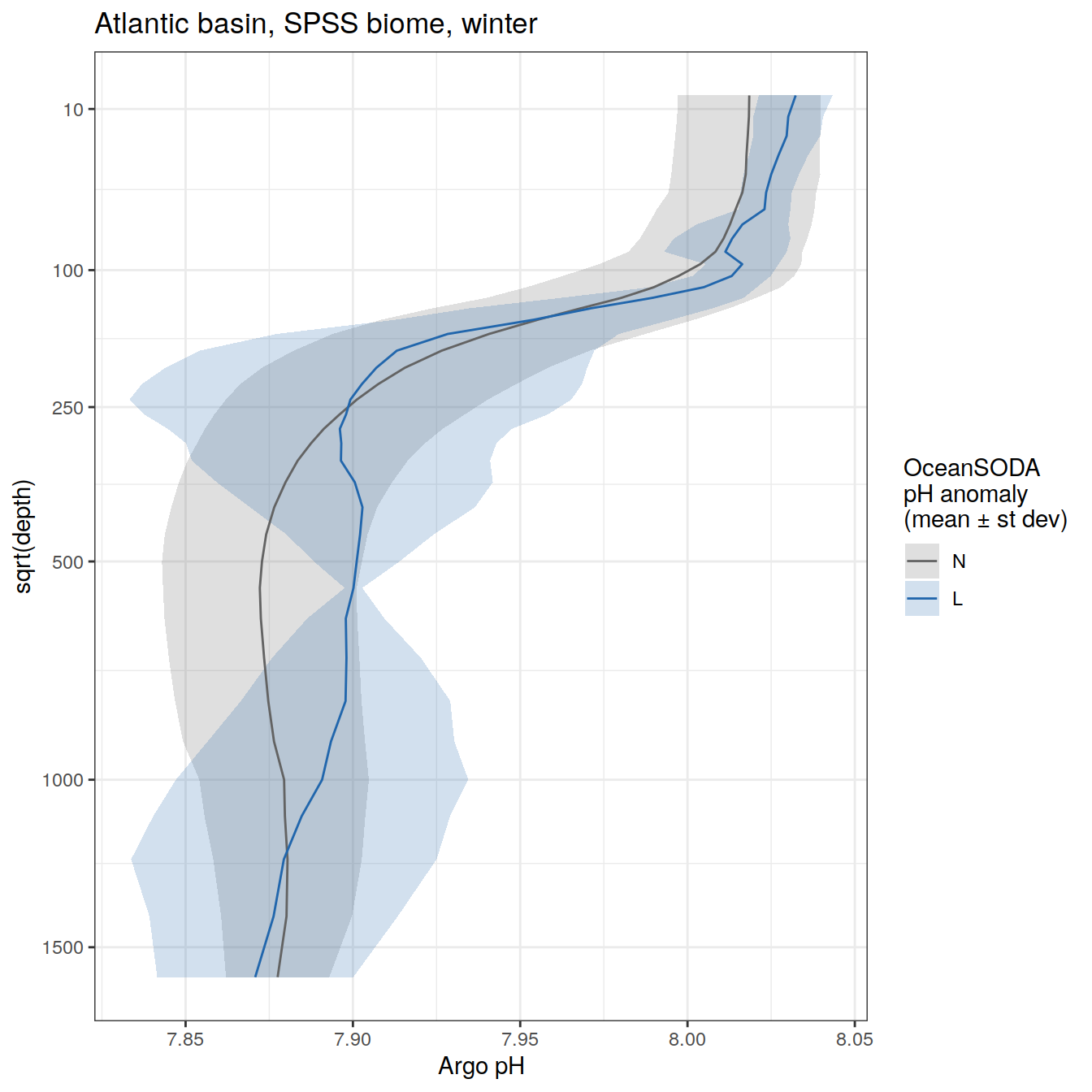
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| a2271df | pasqualina-vonlanthendinenna | 2022-03-30 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
Atl, STSS biome, summer
profile_extreme_season %>%
filter(basin_AIP == 'Atlantic',
season == 'summer',
biome_name == 'STSS') %>%
arrange(depth) %>%
ggplot(aes(x = ph_mean,
y = depth,
group = ph_extreme,
col = ph_extreme)) +
geom_ribbon(aes(xmax = ph_mean + ph_std,
xmin = ph_mean - ph_std,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_path() +
scale_color_manual(values = HNL_colors) +
scale_fill_manual(values = HNL_colors)+
labs(title = 'Atlantic basin, STSS biome, summer',
col = 'OceanSODA\npH anomaly\n(mean ± st dev)',
fill = 'OceanSODA\npH anomaly\n(mean ± st dev)',
y = 'sqrt(depth)',
x = 'Argo pH') +
scale_y_continuous(
trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))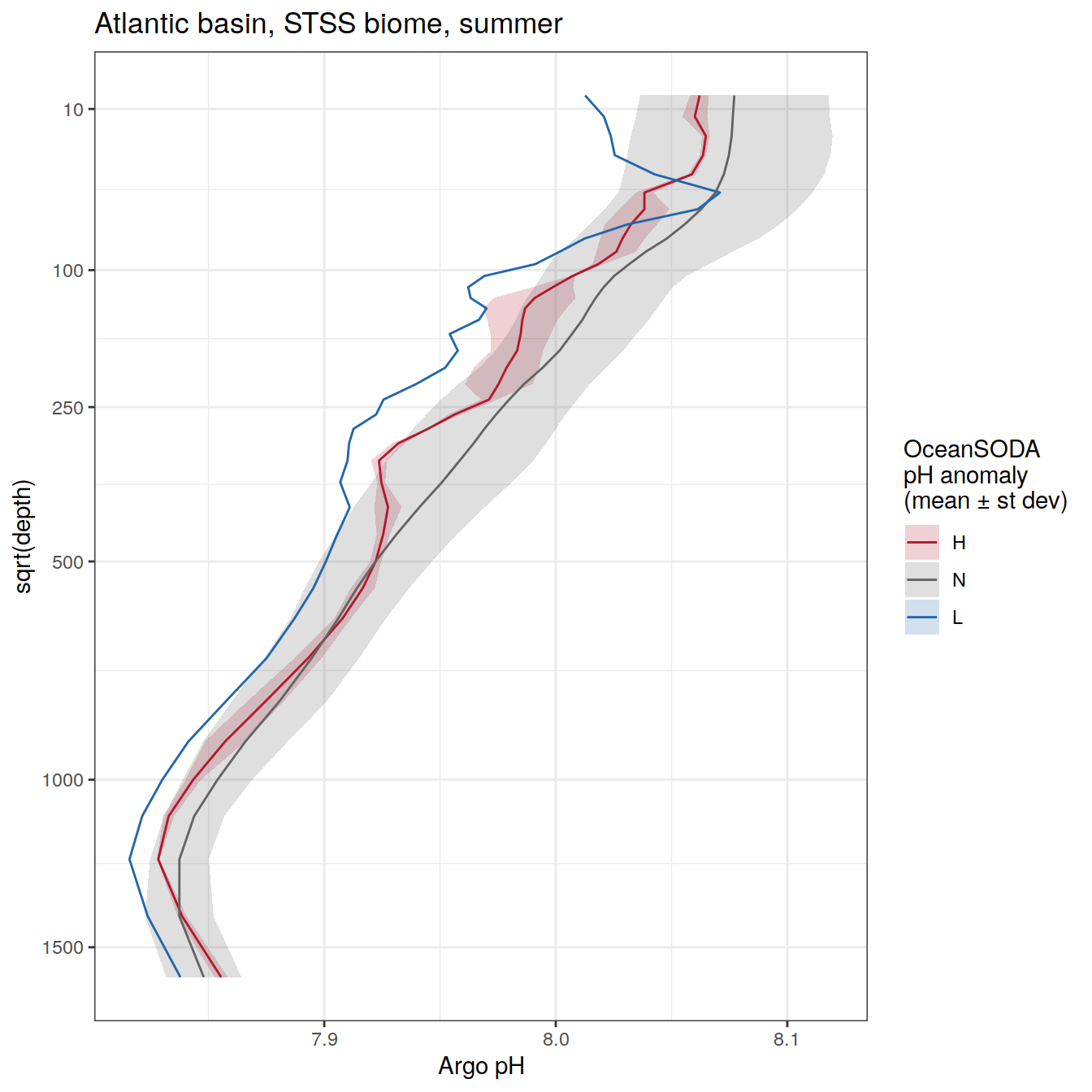
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 9875dd0 | pasqualina-vonlanthendinenna | 2022-04-05 |
| eb8e3be | pasqualina-vonlanthendinenna | 2022-03-31 |
| a2271df | pasqualina-vonlanthendinenna | 2022-03-30 |
| 968ad85 | pasqualina-vonlanthendinenna | 2022-03-29 |
| dfd75e9 | pasqualina-vonlanthendinenna | 2022-03-29 |
| e12a216 | pasqualina-vonlanthendinenna | 2022-03-15 |
| 1ffe07f | pasqualina-vonlanthendinenna | 2022-03-11 |
| 7540ae4 | pasqualina-vonlanthendinenna | 2022-03-08 |
| e4188d2 | pasqualina-vonlanthendinenna | 2022-03-01 |
Anomaly profiles
Read anomalies
# profile_extreme_binned <- profile_extreme %>%
# group_by(lon, lat, year, month, file_id,
# biome_name, basin_AIP, ph_extreme,
# depth, season, season_order) %>%
# summarise(ph_adjusted_binned = mean(pH, na.rm = TRUE)) %>%
# ungroup()# broullon_clim <- read_rds(file = paste0(path_argo_preprocessed,
# '/broullon_TA_DIC_clim_SO_pH.rds'))
#
# # compatibility with profile_extreme
# broullon_clim <- broullon_clim %>%
# mutate(depth_broullon = depth)
#
# # grid average climatological temp into the argo depth bins
# broullon_clim <- broullon_clim %>%
# mutate(
# depth = cut(
# depth_broullon,
# breaks = c(0, 10, 20, 30, 50, 70, 100, 300, 500, 800, 1000, 1500, 2000),
# include.lowest = TRUE,
# labels = as.factor(unique(profile_extreme$depth))[1:12]
# ),
# depth = as.numeric(as.character(depth))
# )
#
#
# # calculate mean climatological pH per depth bin
# broullon_clim_binned <- broullon_clim %>%
# group_by(lon, lat, depth, month, basin_AIP, biome_name) %>%
# summarise(clim_ph_binned = mean(pH, na.rm = TRUE)) %>%
# ungroup()
#
#
# # join climatology and ARGO profiles
# remove_clim <- inner_join(profile_extreme_binned,
# broullon_clim_binned)
remove_clim <-
read_rds(file = paste0(path_argo_preprocessed, "/pH_anomaly_va.rds")) %>%
filter(profile_range >= opt_min_profile_range) %>%
mutate(date = ymd(format(date, "%Y-%m-15")))
remove_clim <- inner_join(
remove_clim %>%
select(
file_id,
year,
month,
date,
lon,
lat,
depth,
pH,
clim_pH,
anomaly_pH
),
OceanSODA_SO_extreme_grid %>%
select(
year,
month,
date,
lon,
lat,
OceanSODA_ph,
ph_extreme,
biome_name,
basin_AIP
)
)
remove_clim <- remove_clim %>%
mutate(
season = case_when(
between(month, 3, 5) ~ 'autumn',
between(month, 6, 8) ~ 'winter',
between(month, 9, 11) ~ 'spring',
month == 12 | 1 | 2 ~ 'summer'
),
season_order = case_when(
between(month, 3, 5) ~ 2,
between(month, 6, 8) ~ 3,
between(month, 9, 11) ~ 4,
month == 12 | 1 | 2 ~ 1
),
.after = date
) Profiles - Absolute
remove_clim %>%
group_split(biome_name, basin_AIP, year) %>%
head(6) %>%
map(
~ ggplot() +
geom_path(
data = .x %>%
filter(ph_extreme == 'N'),
aes(
x = pH,
y = depth,
group = file_id,
col = ph_extreme
),
size = 0.3
) +
geom_path(
data = .x %>%
filter(ph_extreme == 'H' | ph_extreme == 'L'),
aes(
x = pH,
y = depth,
group = file_id,
col = ph_extreme
),
size = 0.5
) +
geom_point(
data = .x,
aes(x = clim_pH,
y = depth,
col = ph_extreme),
size = 0.5
) +
scale_y_reverse() +
scale_color_manual(values = HNL_colors) +
labs(
x = 'Argo pH',
y = 'depth (m)',
title = paste(
"Biome:",
unique(.x$biome_name),
"| basin:",
unique(.x$basin_AIP),
" | ",
unique(.x$year)
),
col = 'OceanSODA pH \nanomaly'
)
)[[1]]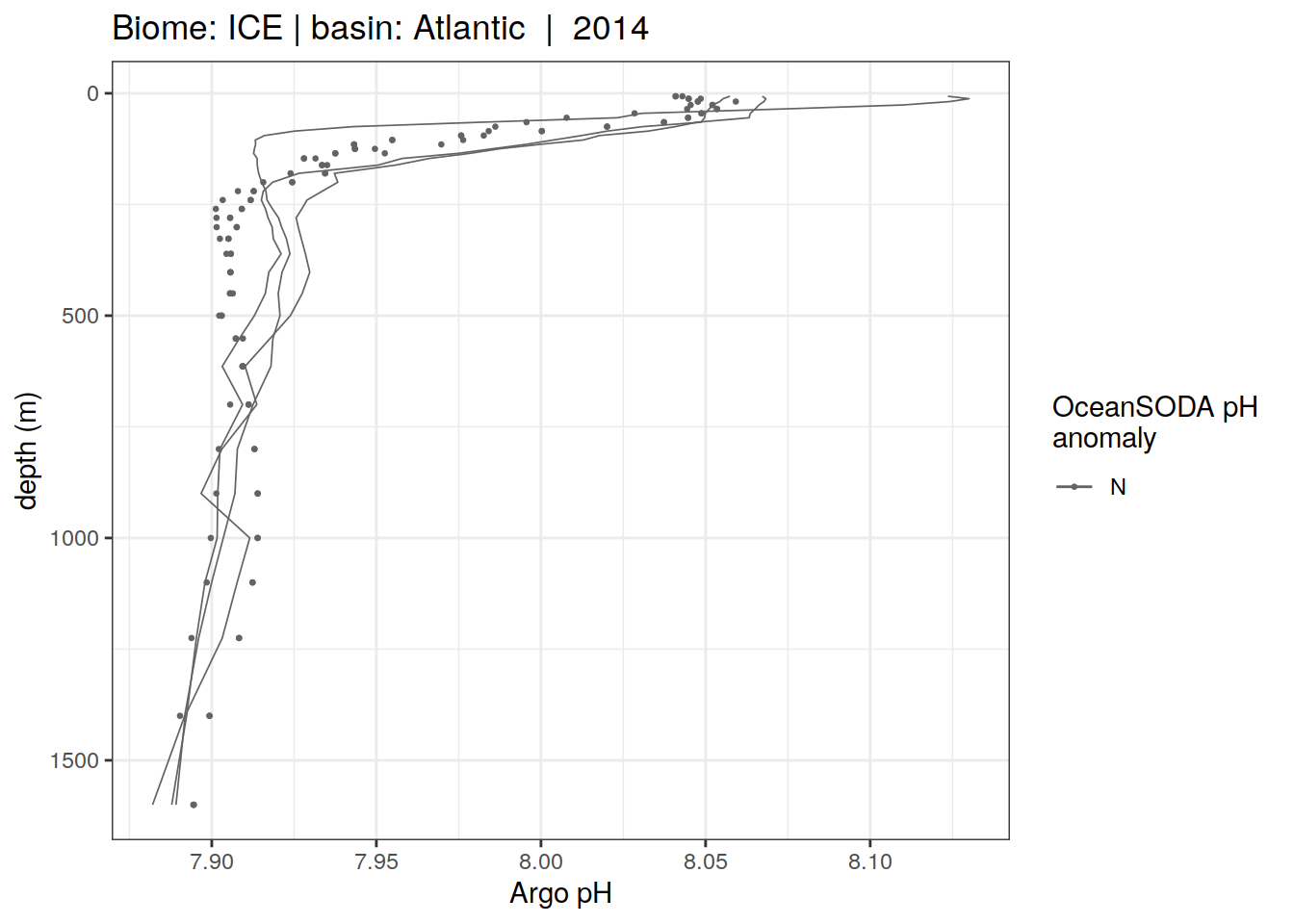
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[2]]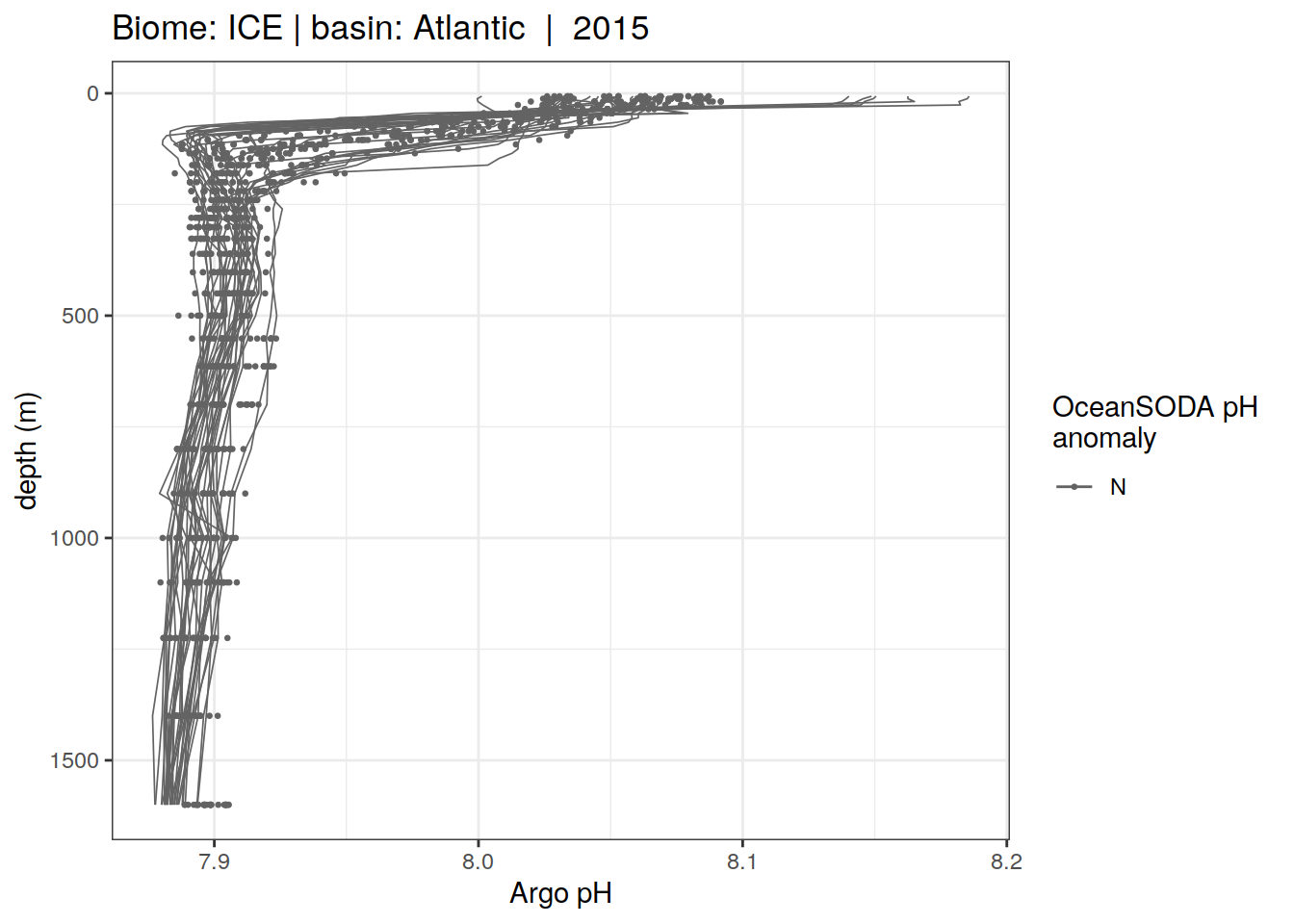
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[3]]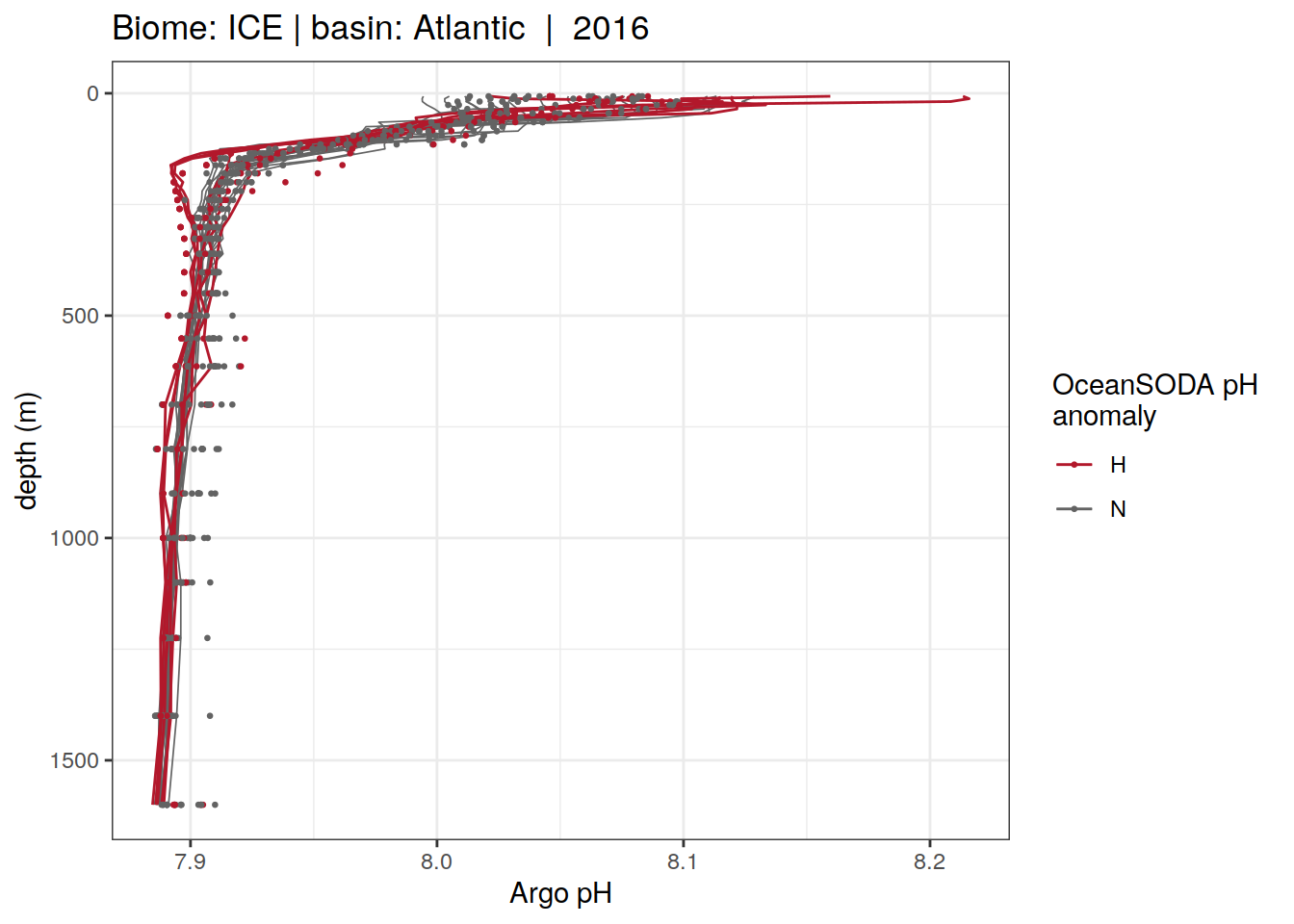
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[4]]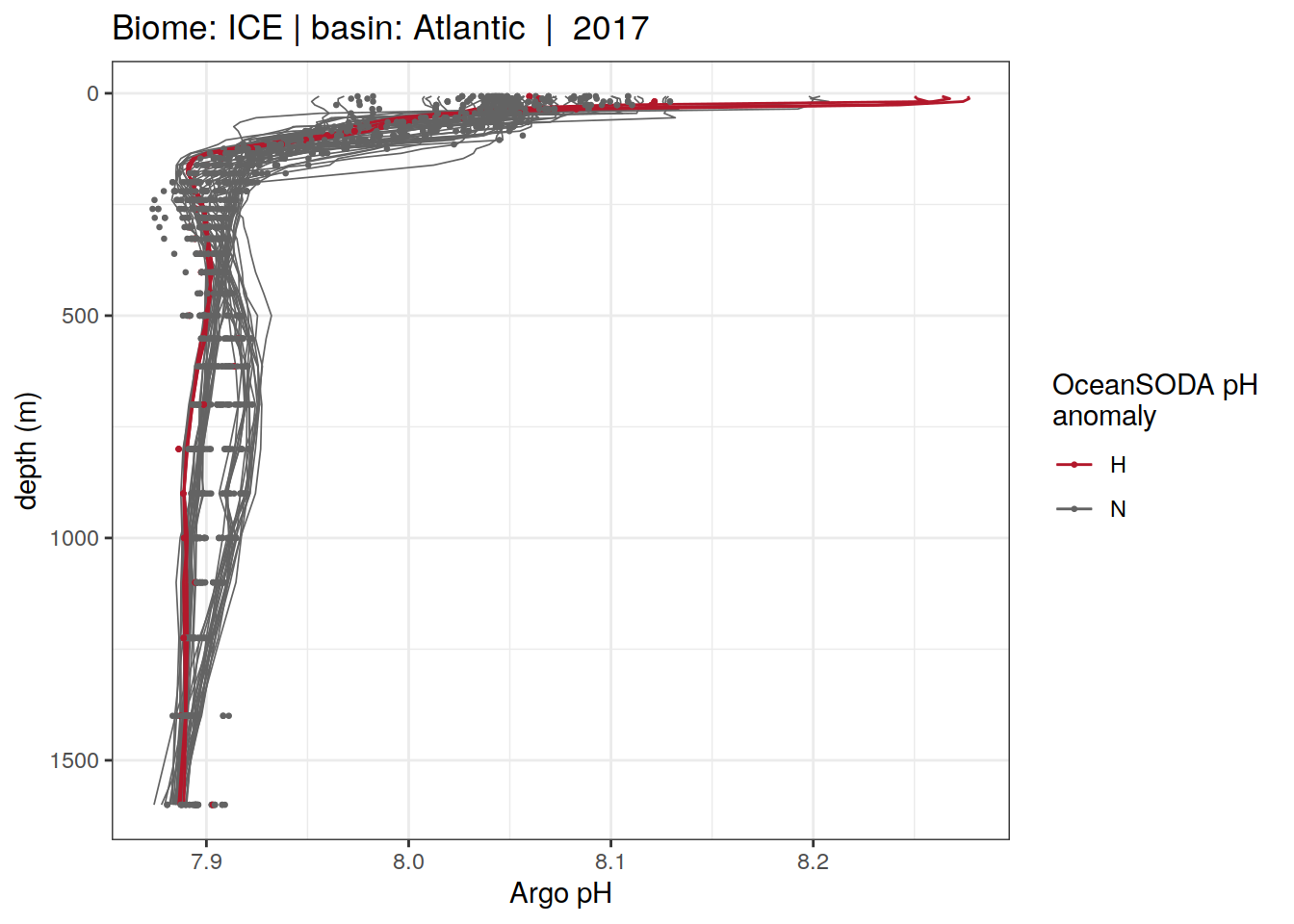
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[5]]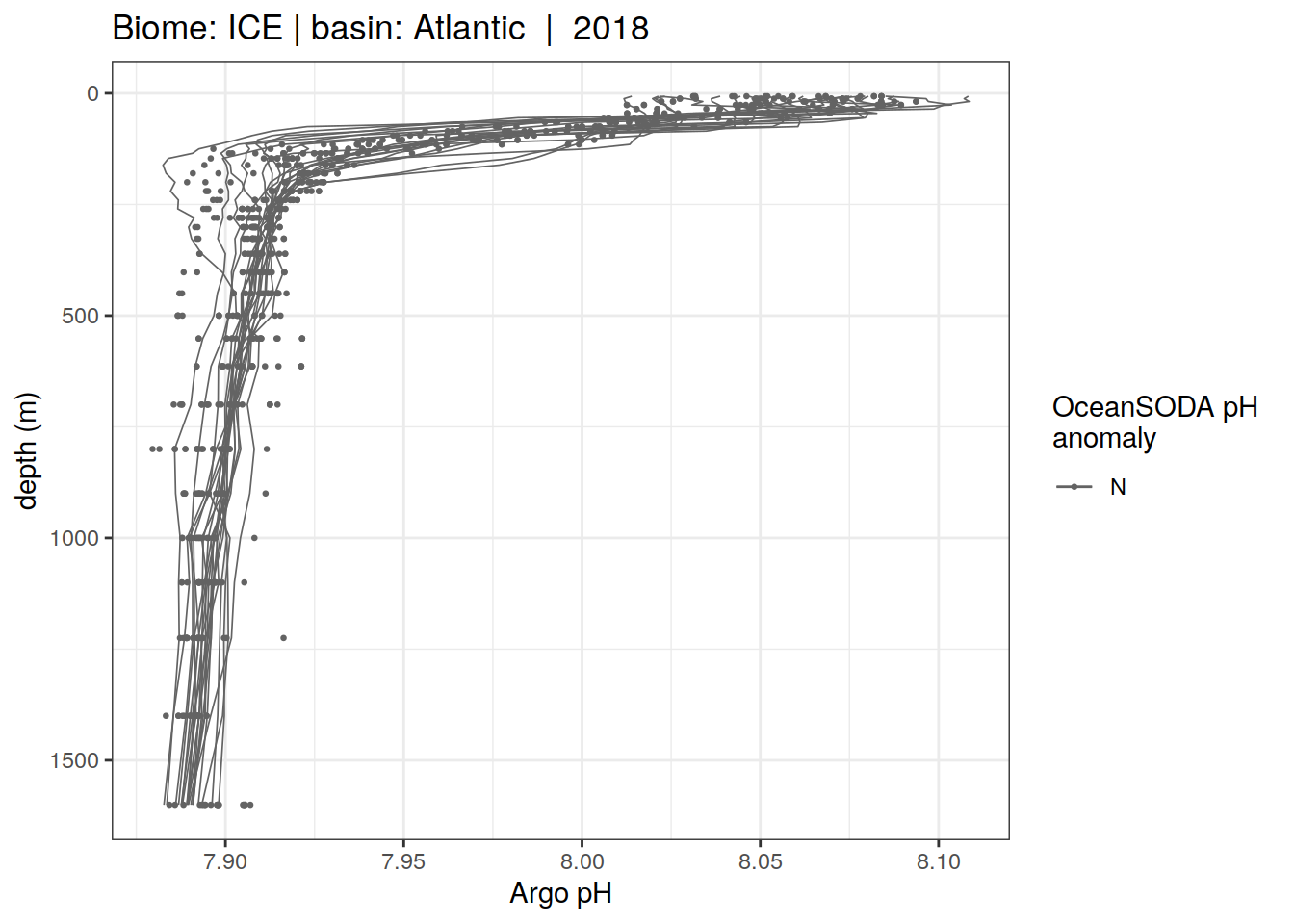
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[6]]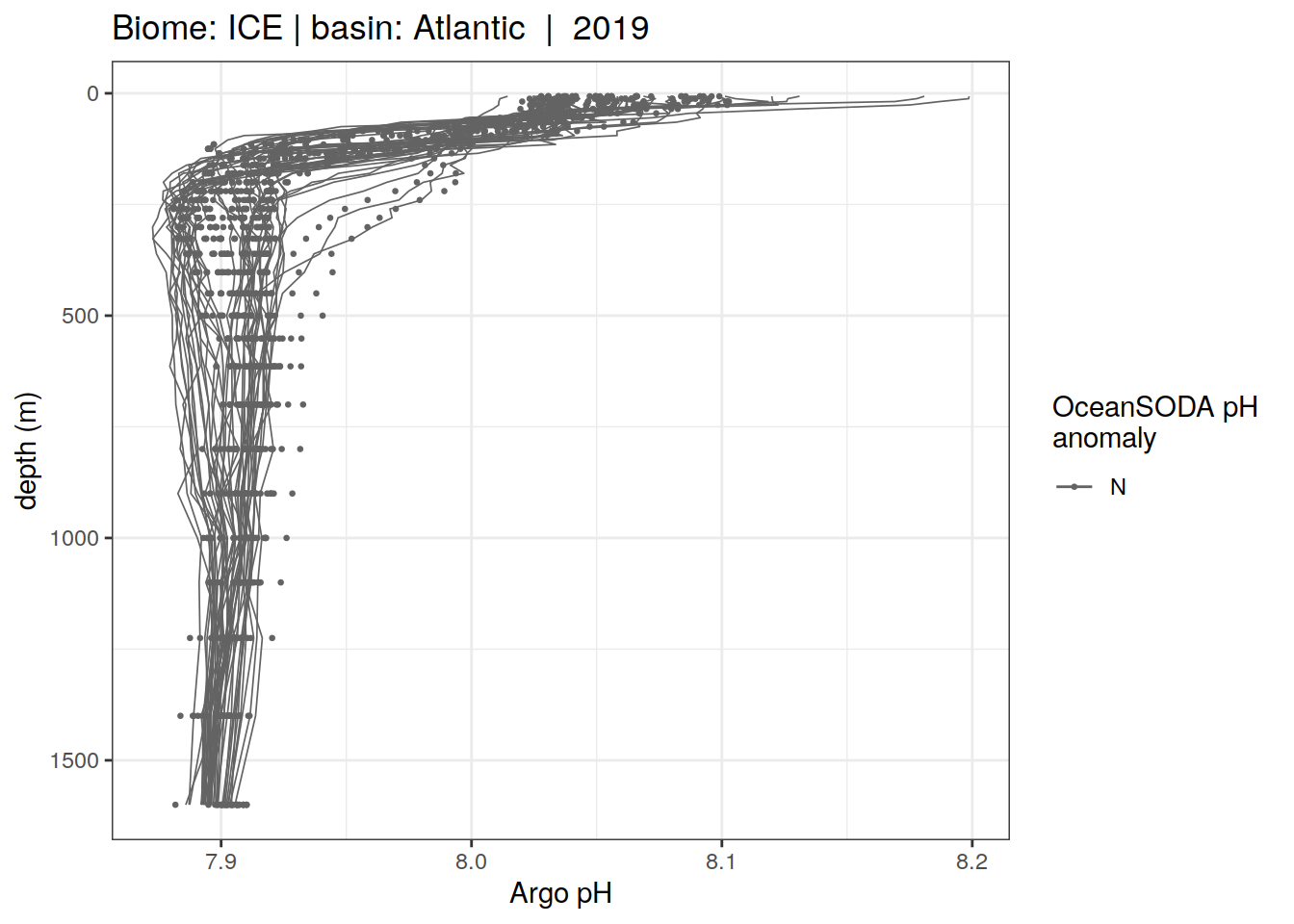
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
Profiles - Anomaly
remove_clim %>%
group_split(month) %>%
#head(1) %>%
map(
~ggplot()+
geom_path(data = .x %>% filter(ph_extreme == 'N'),
aes(x = anomaly_pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.2)+
geom_path(data = .x %>% filter(ph_extreme == 'H'| ph_extreme == 'L'),
aes(x = anomaly_pH,
y = depth,
group = file_id,
col = ph_extreme),
size = 0.3)+
geom_vline(xintercept = 0)+
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
facet_grid(basin_AIP~biome_name)+
labs(title = paste('month:', unique(.x$month)))
)[[1]]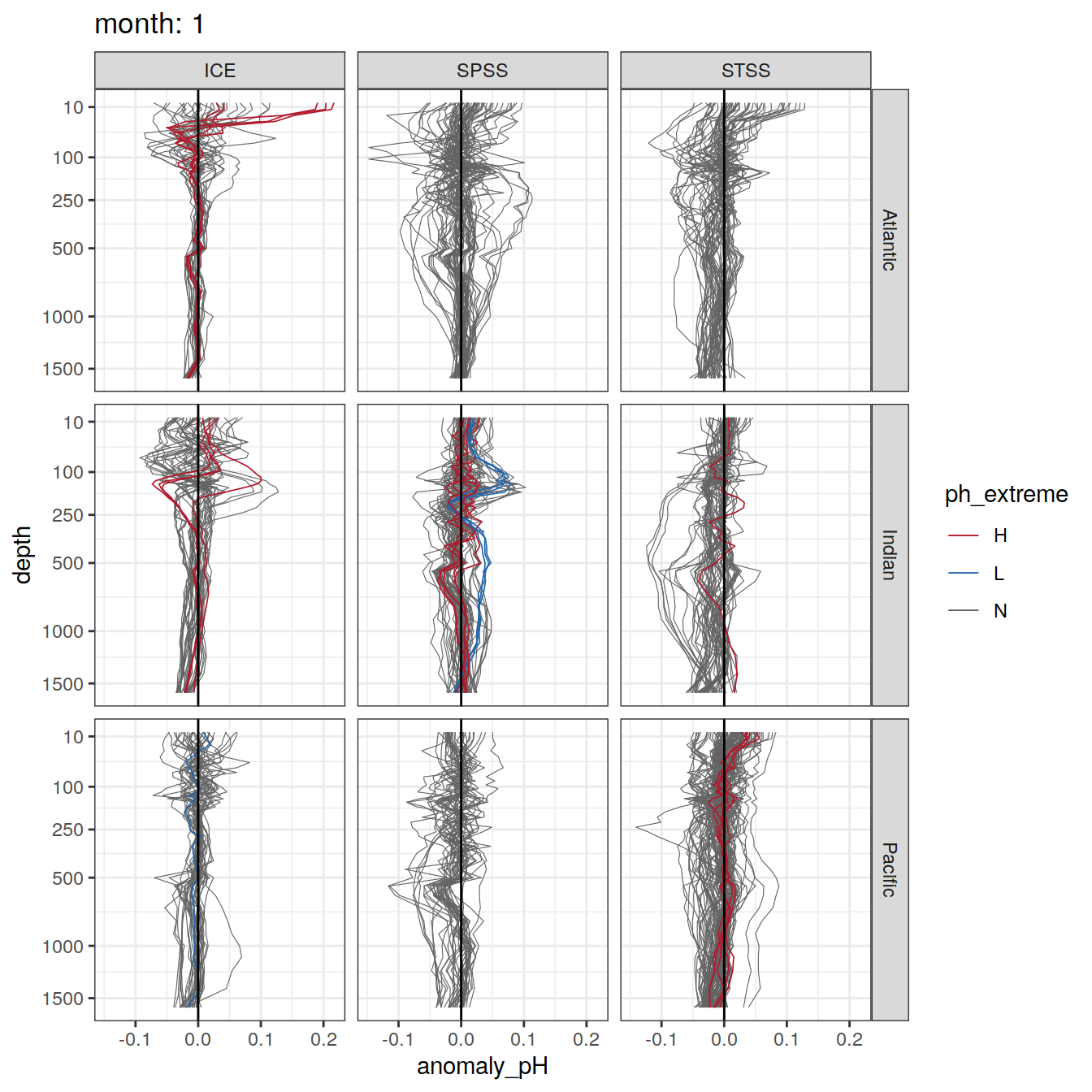
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[2]]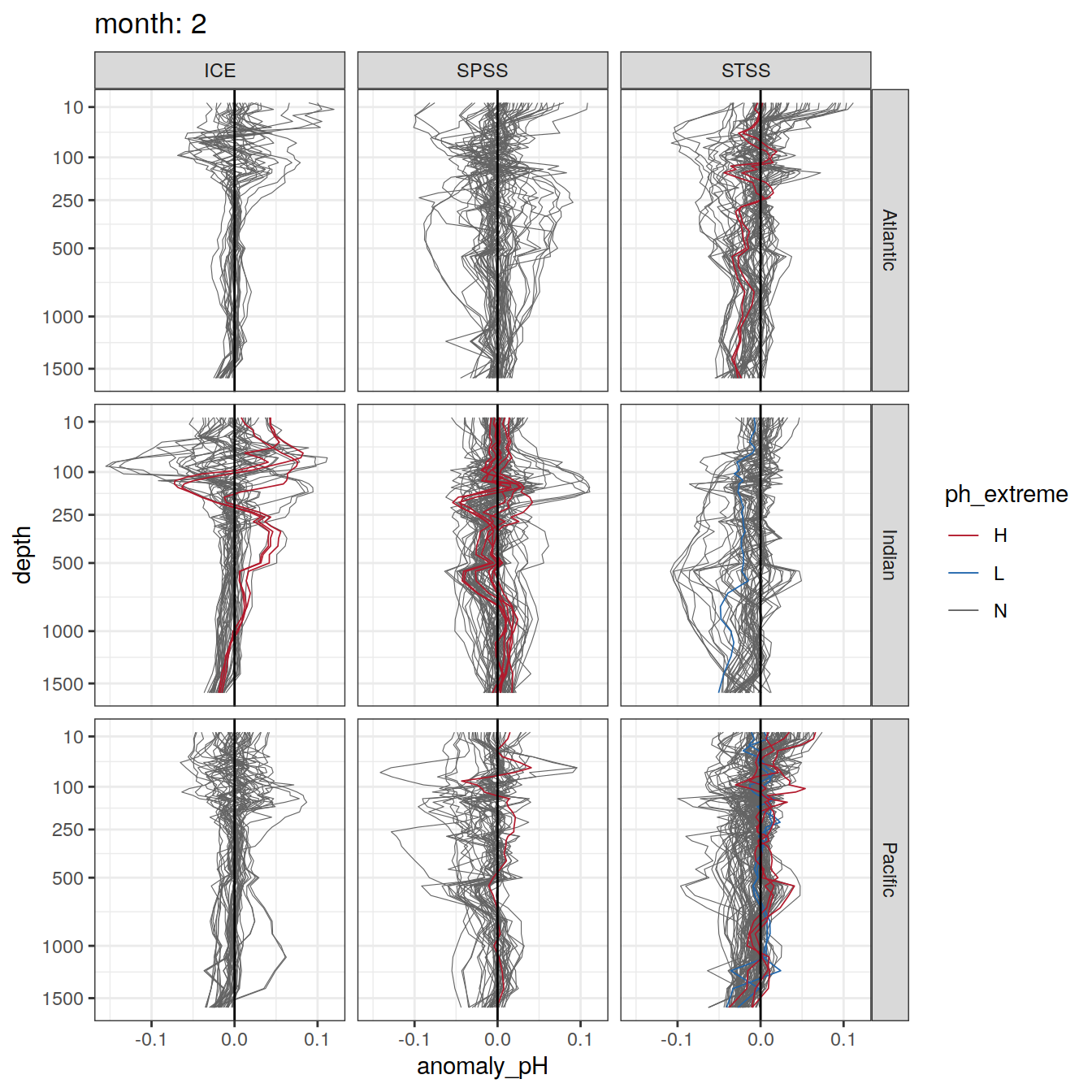
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[3]]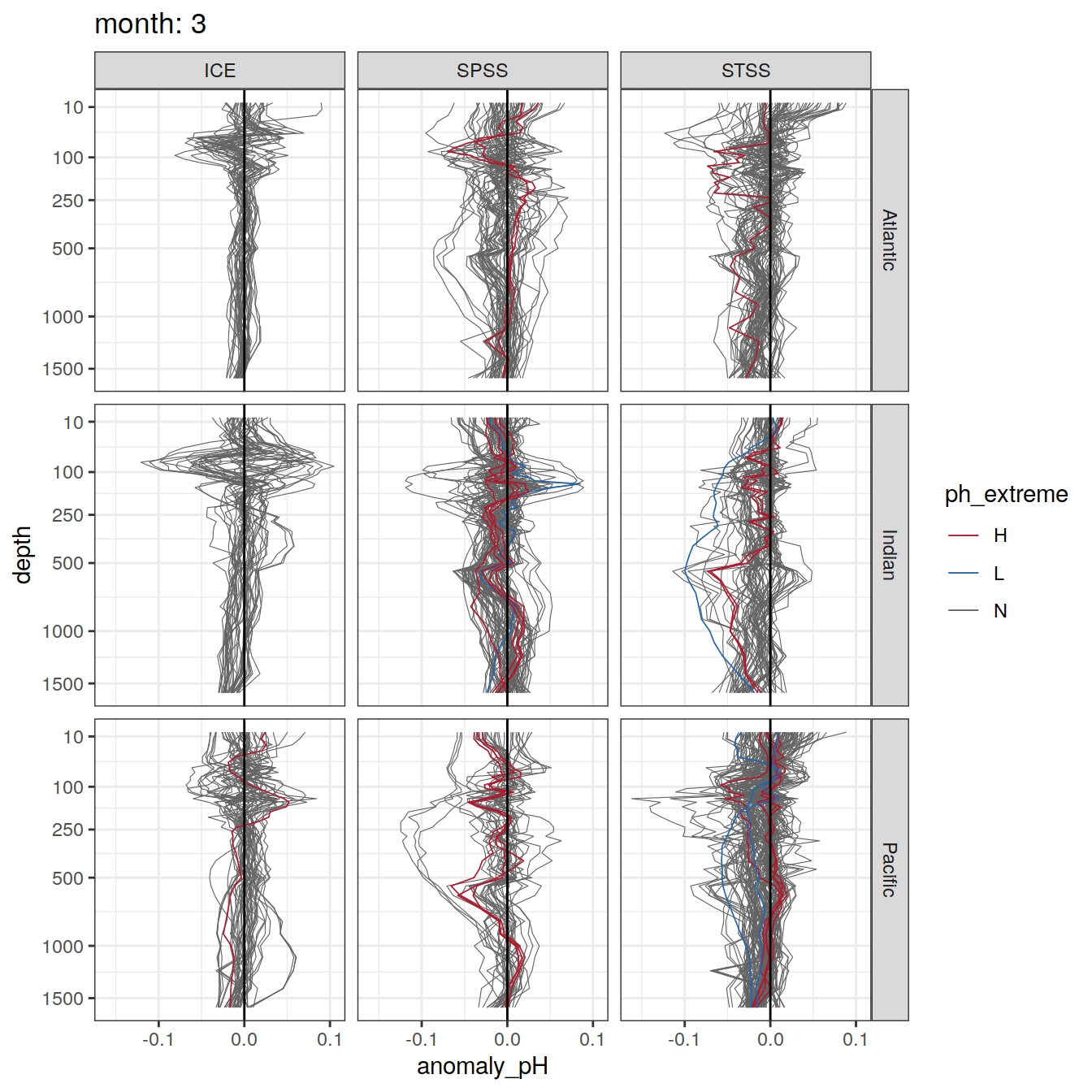
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[4]]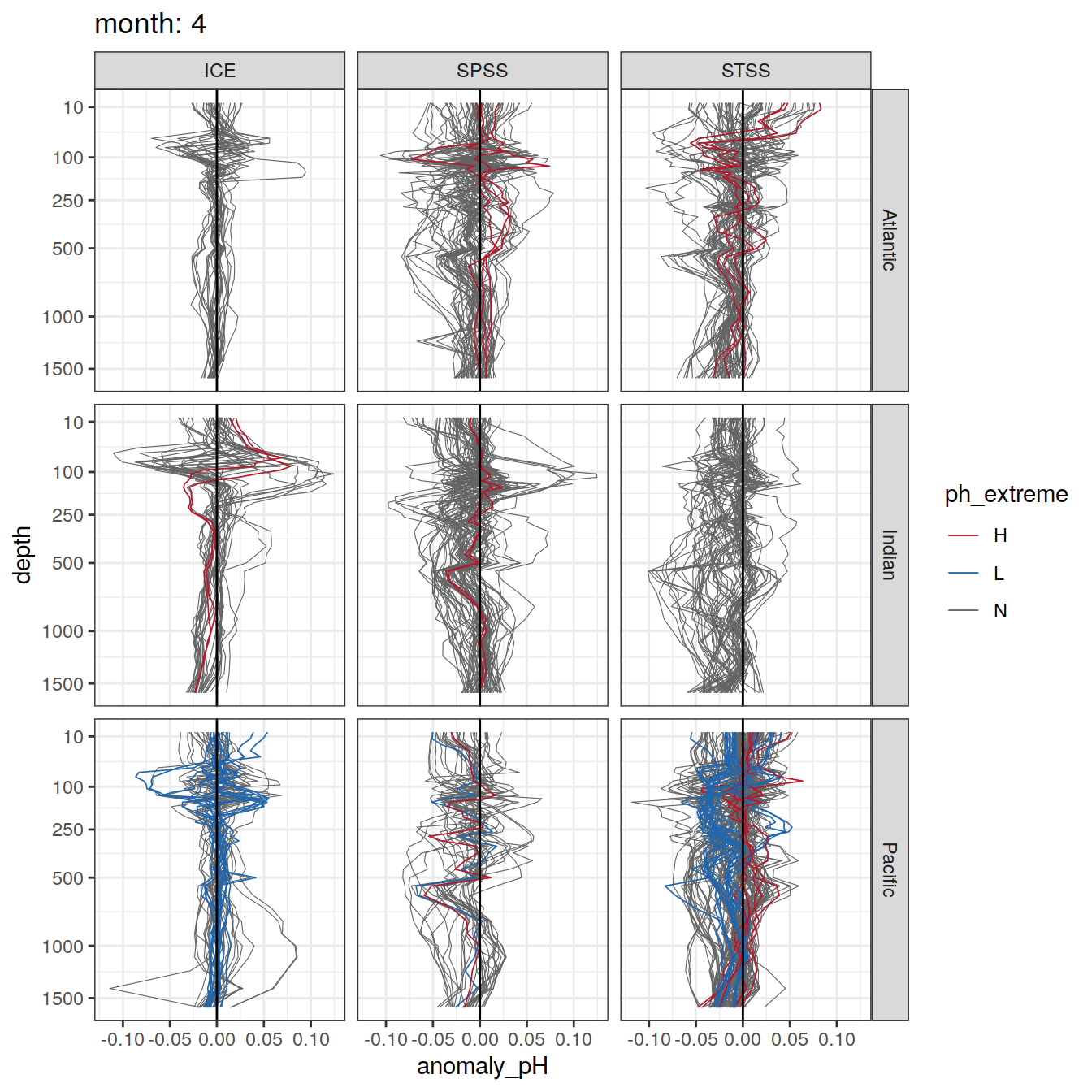
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[5]]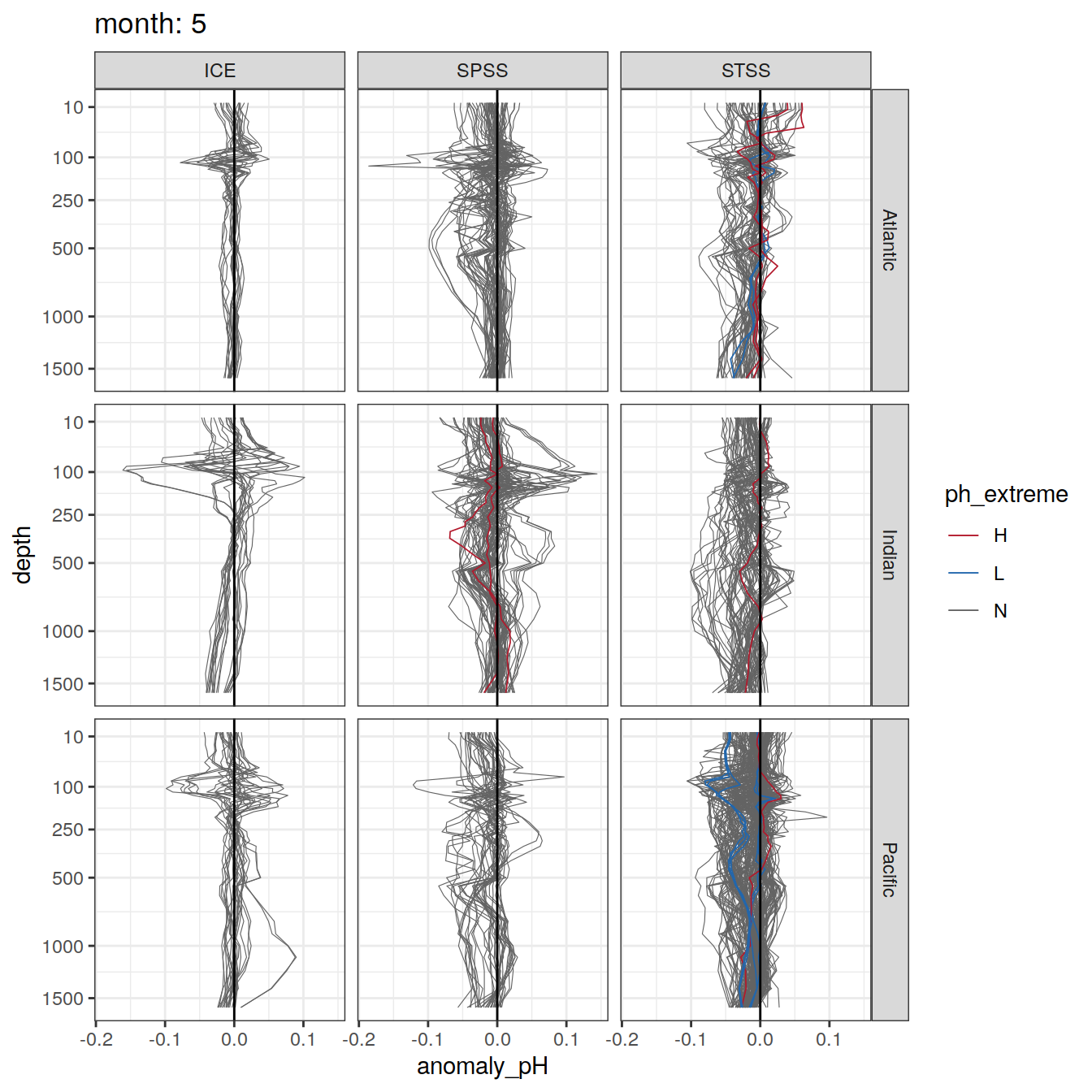
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[6]]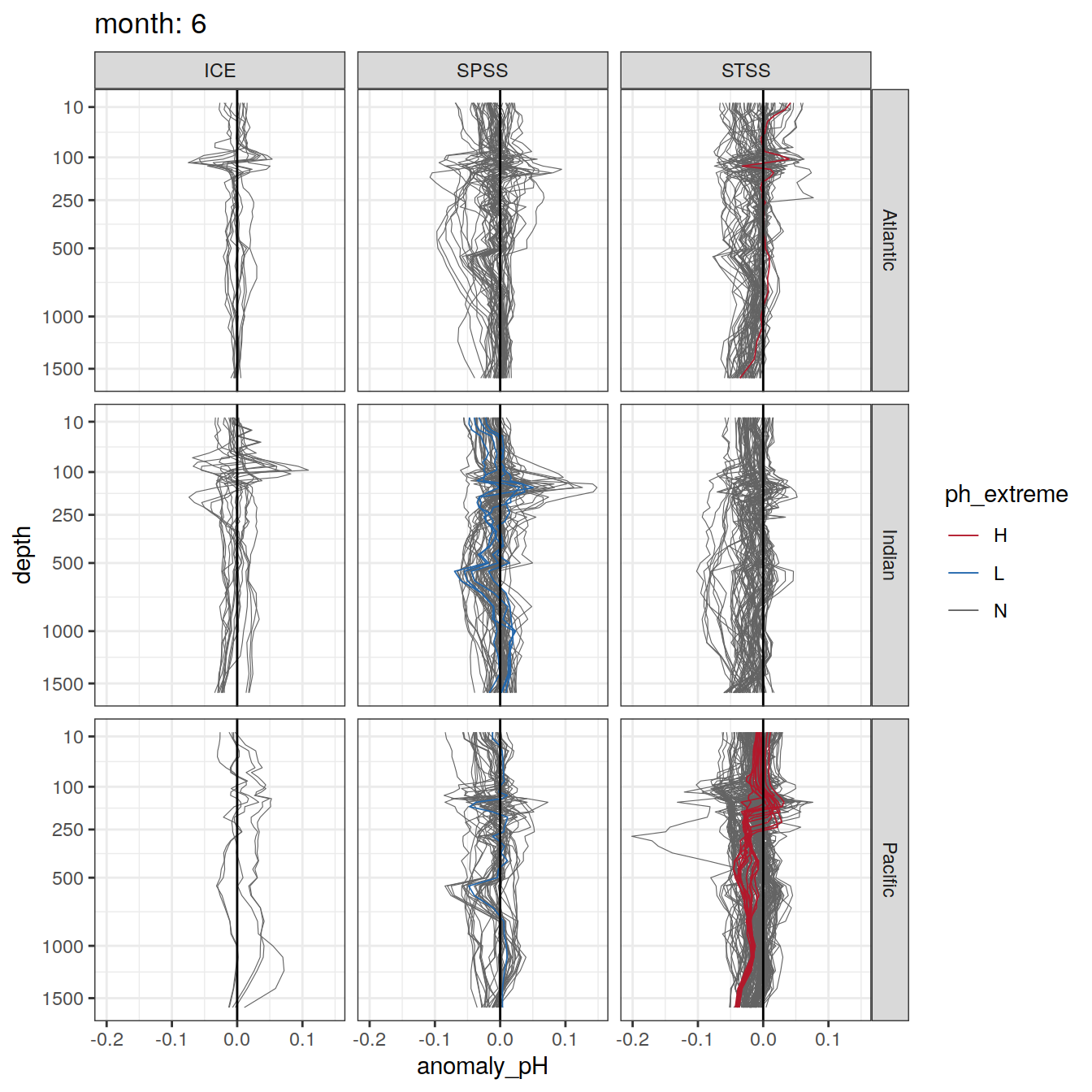
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[7]]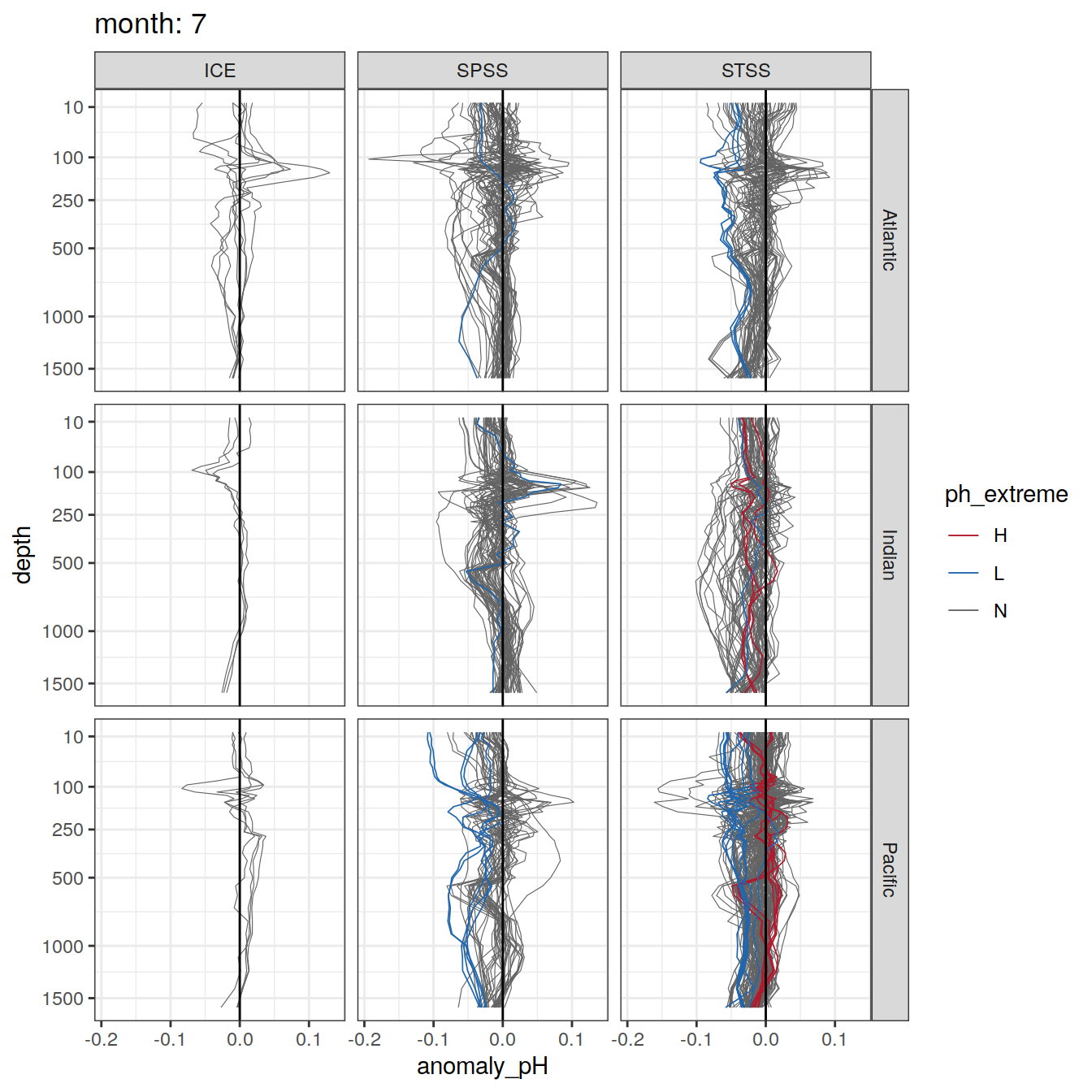
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[8]]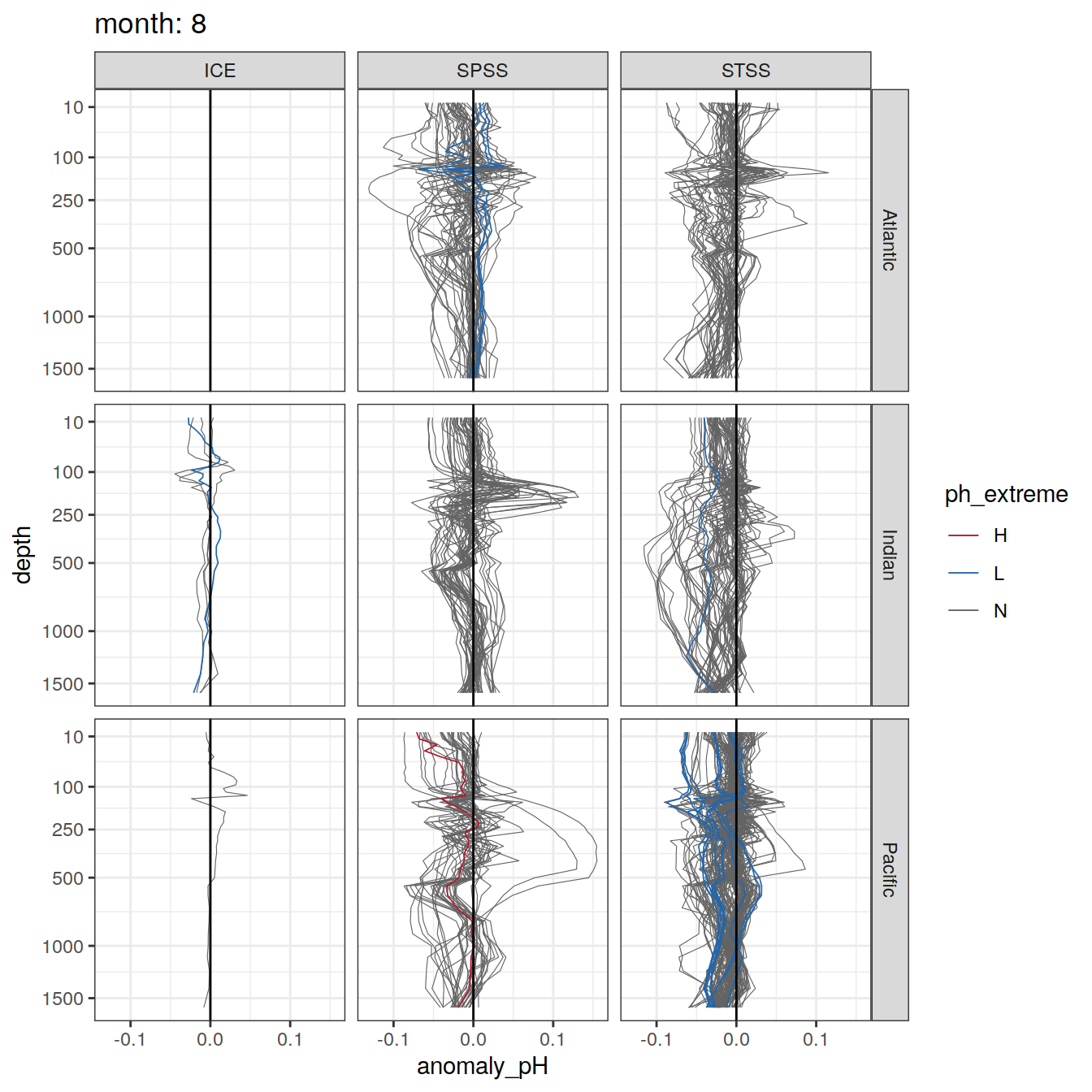
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[9]]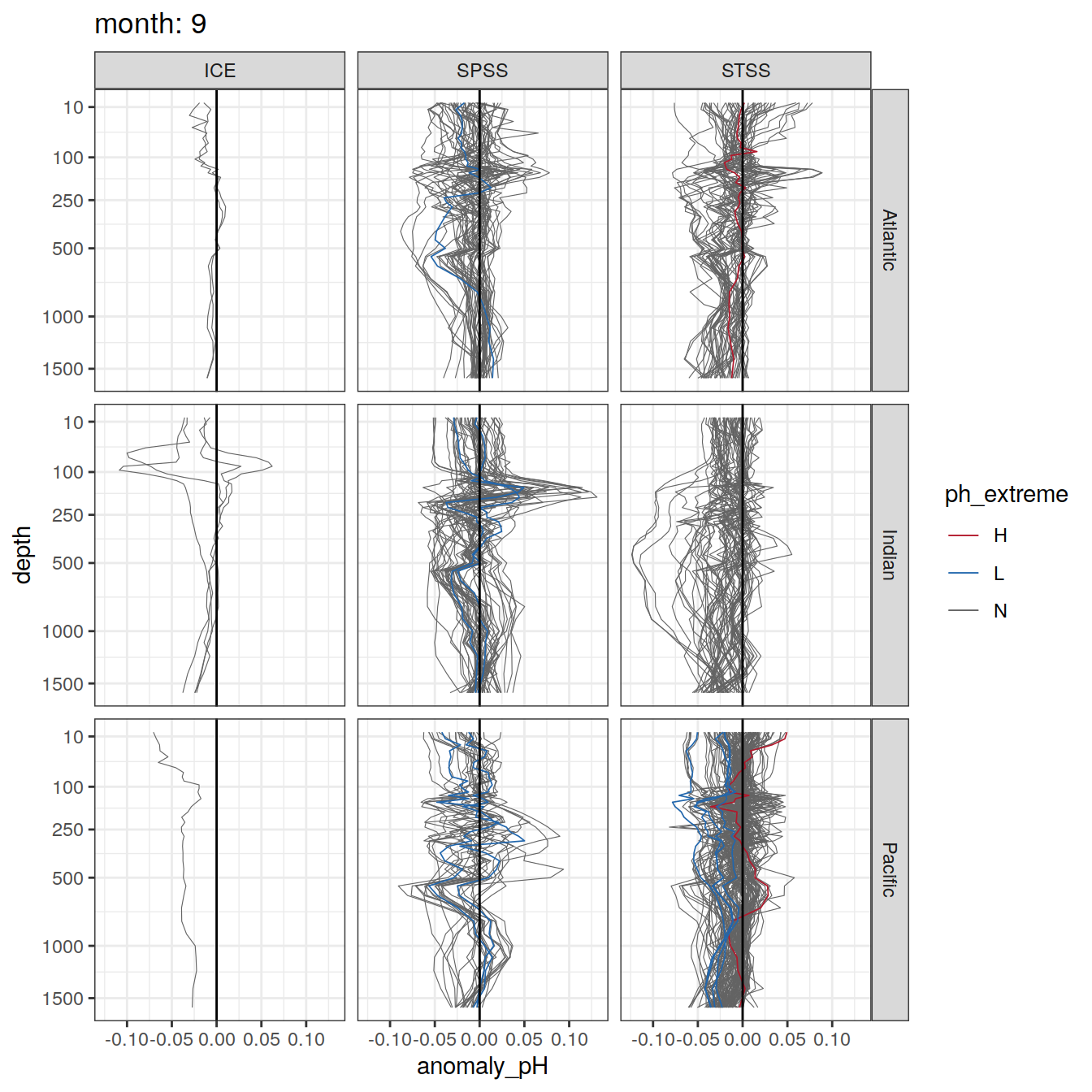
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[10]]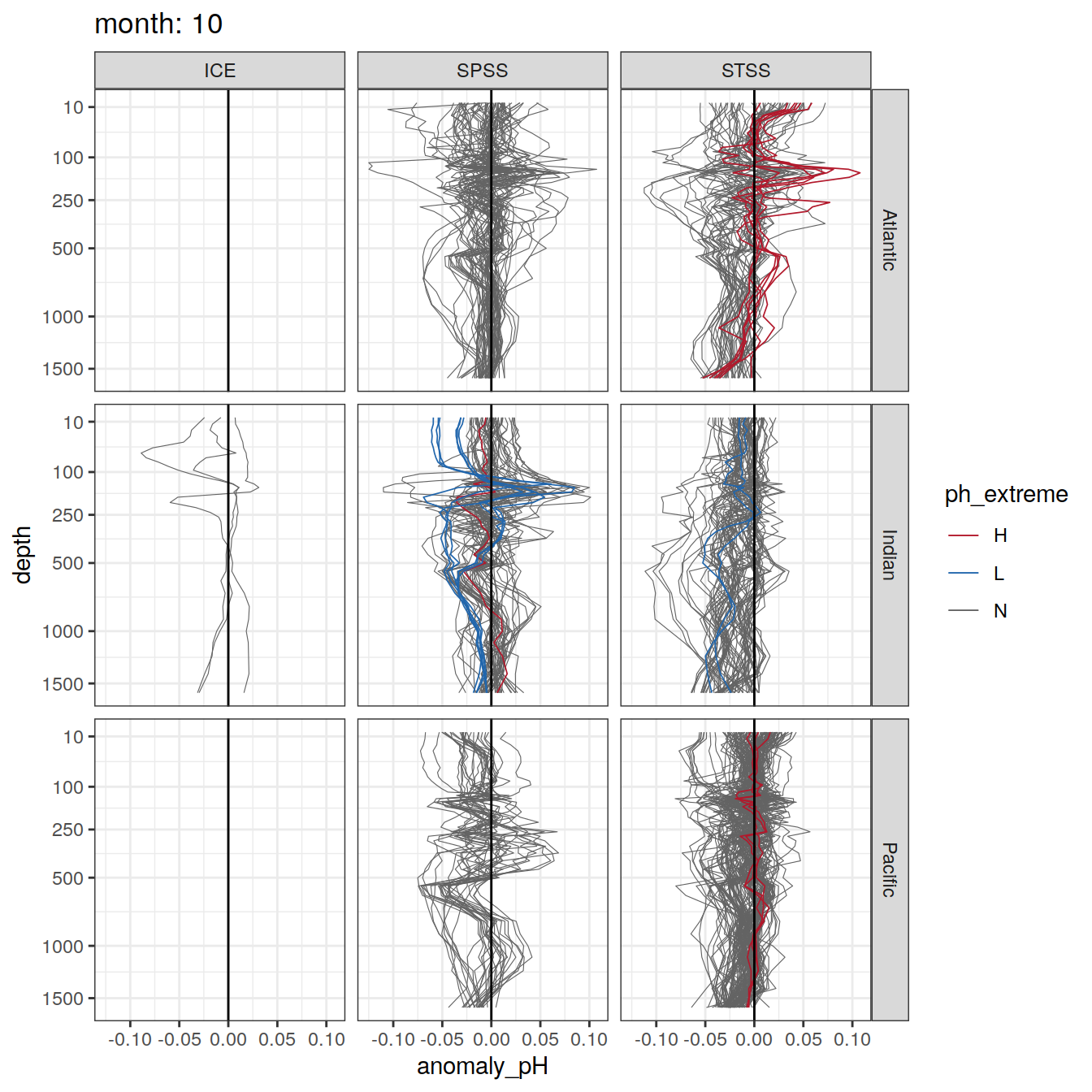
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| 91f08a6 | mlarriere | 2024-04-07 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[11]]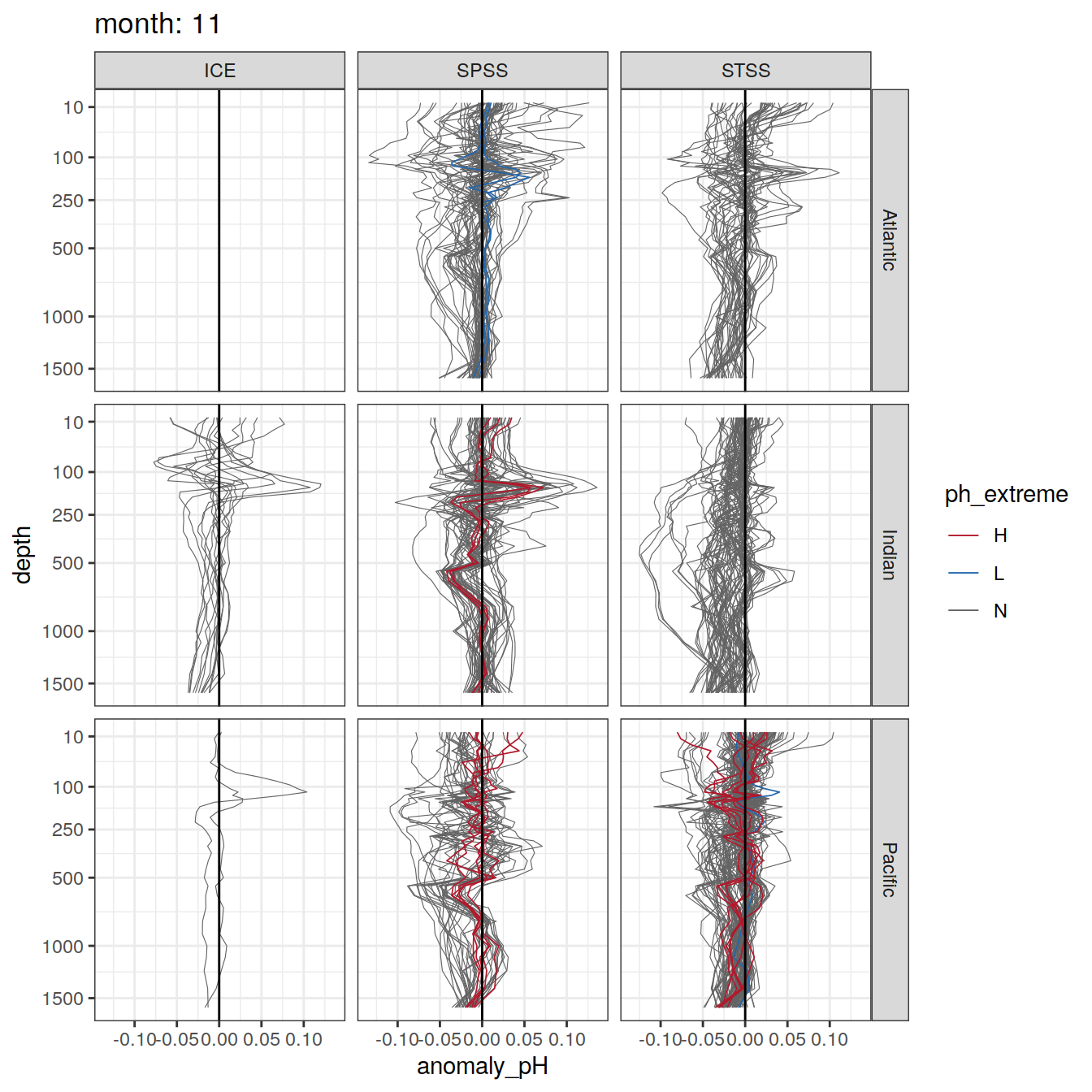
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[12]]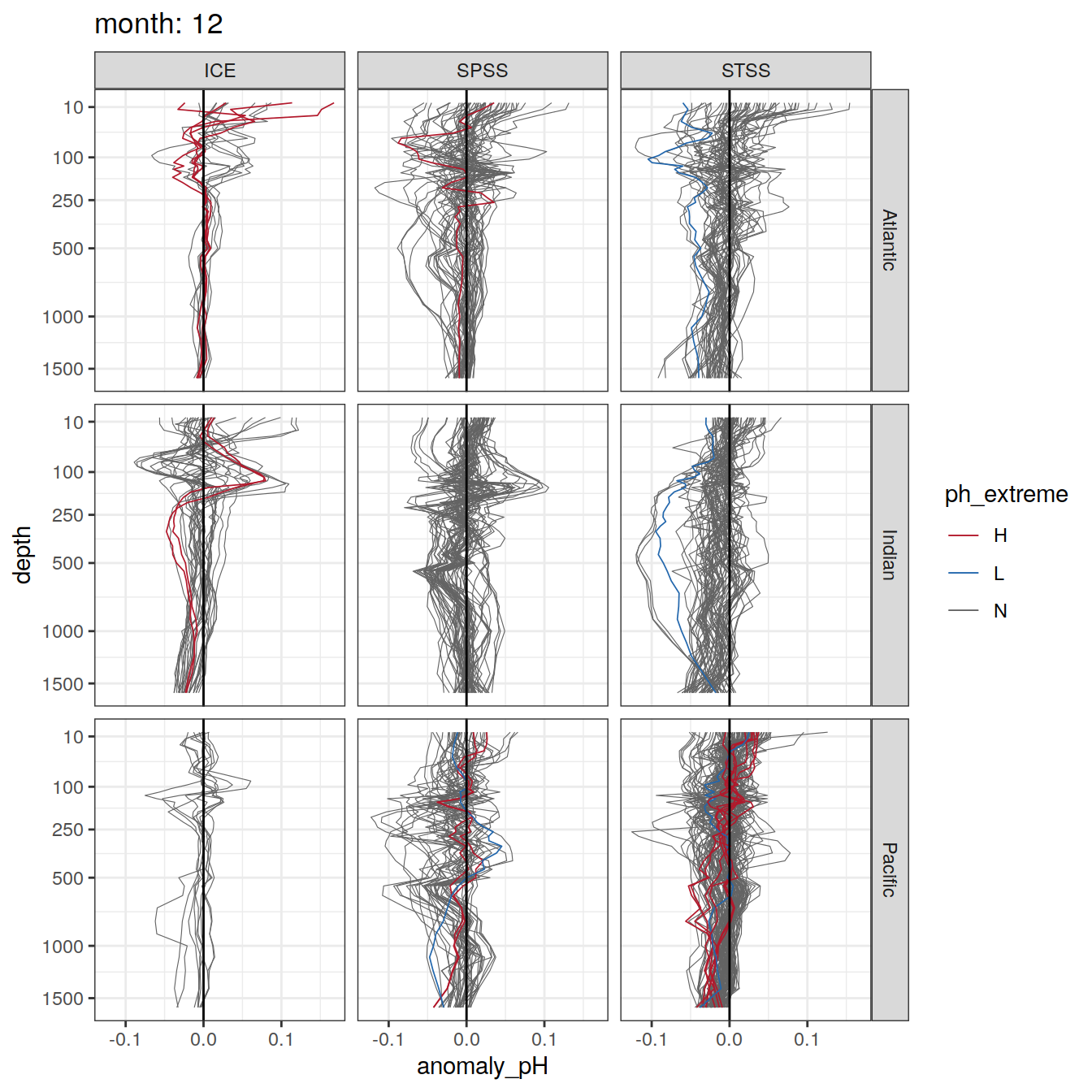
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
Overall mean anomaly profiles
remove_clim_overall_mean <- remove_clim %>%
group_by(ph_extreme, depth) %>%
summarise(ph_anomaly_mean = mean(anomaly_pH, na.rm = TRUE),
ph_anomaly_sd = sd(anomaly_pH, na.rm = TRUE)) %>%
ungroup()
remove_clim_overall_mean %>%
ggplot()+
geom_path(aes(x = ph_anomaly_mean,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_ribbon(aes(xmax = ph_anomaly_mean + ph_anomaly_sd,
xmin = ph_anomaly_mean - ph_anomaly_sd,
y = depth,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_continuous(trans = trans_reverser("sqrt"),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
# geom_text_repel(data = profile_count_mean,
# aes(x = -0.075,
# y = 1500,
# label = paste0(n),
# col = ph_extreme),
# size = 7,
# segment.color = 'transparent')+
geom_text(data = profile_count_mean[2,],
aes(x = -0.07,
y = 1200,
label = paste0(n),
col = ph_extreme),
size = 6)+
geom_text(data = profile_count_mean[1,],
aes(x = -0.07,
y = 1400,
label = paste0(n),
col = ph_extreme),
size = 6)+
geom_text(data = profile_count_mean[3,],
aes(x = -0.07,
y = 1600,
label = paste0(n),
col = ph_extreme),
size = 6)+
coord_cartesian(xlim = c(-0.08, 0.08))+
scale_x_continuous(breaks = c(-0.08, -0.04, 0, 0.04, 0.08))+
labs(title = 'Overall Mean Anomaly Profiles')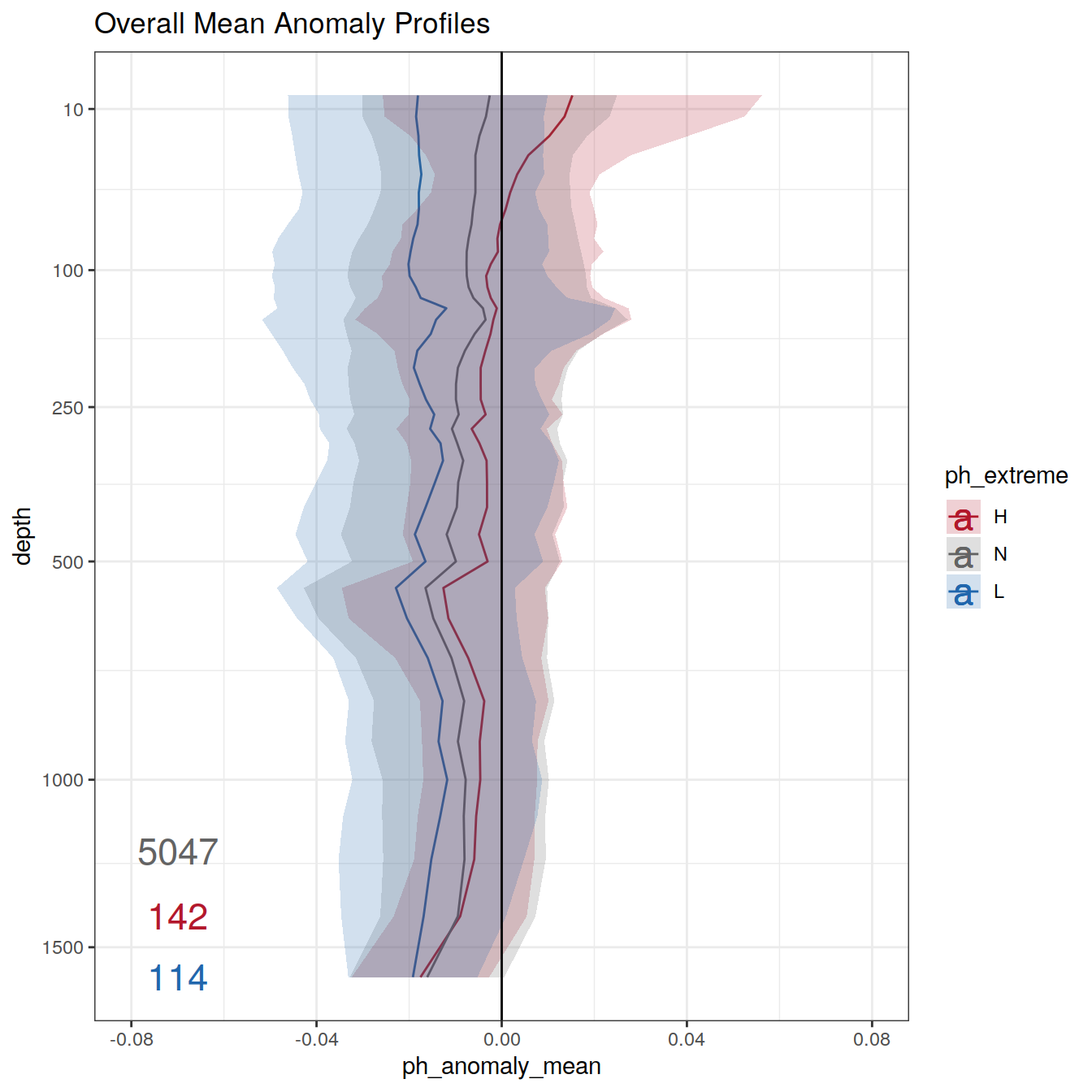
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 708f923 | pasqualina-vonlanthendinenna | 2022-05-04 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
rm(remove_clim_overall_mean, profile_count_mean)Biome-mean anomaly profiles
remove_clim_biome <- remove_clim %>%
group_by(depth, ph_extreme, biome_name, season_order, season) %>%
summarise(ph_anomaly_mean = mean(anomaly_pH, na.rm = TRUE),
ph_anomaly_sd = sd(anomaly_pH, na.rm = TRUE))
remove_clim_biome %>%
ggplot()+
geom_path(aes(x = ph_anomaly_mean,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_ribbon(aes(xmax = ph_anomaly_mean + ph_anomaly_sd,
xmin = ph_anomaly_mean - ph_anomaly_sd,
y = depth,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_continuous(trans = trans_reverser('sqrt'),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))+
facet_grid(season_order~biome_name, labeller = facet_label)+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
# geom_text_repel(data = profile_count_biome,
# aes(x = -0.085,
# y = 1500,
# label = paste0(n),
# col = ph_extreme),
# size = 4,
# segment.color = 'transparent')+
geom_text(data = profile_count_biome %>% filter (ph_extreme == 'N'),
aes(x = -0.07,
y = 800,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_biome %>% filter (ph_extreme == 'H'),
aes(x = -0.07,
y = 1200,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_biome %>% filter (ph_extreme == 'L'),
aes(x = -0.07,
y = 1600,
label = paste0(n),
col = ph_extreme),
size = 4)+
coord_cartesian(xlim = c(-0.08, 0.08))+
scale_x_continuous(breaks = c(-0.08, -0.04, 0, 0.04, 0.08))+
labs(title = 'Biome-mean anomaly profiles')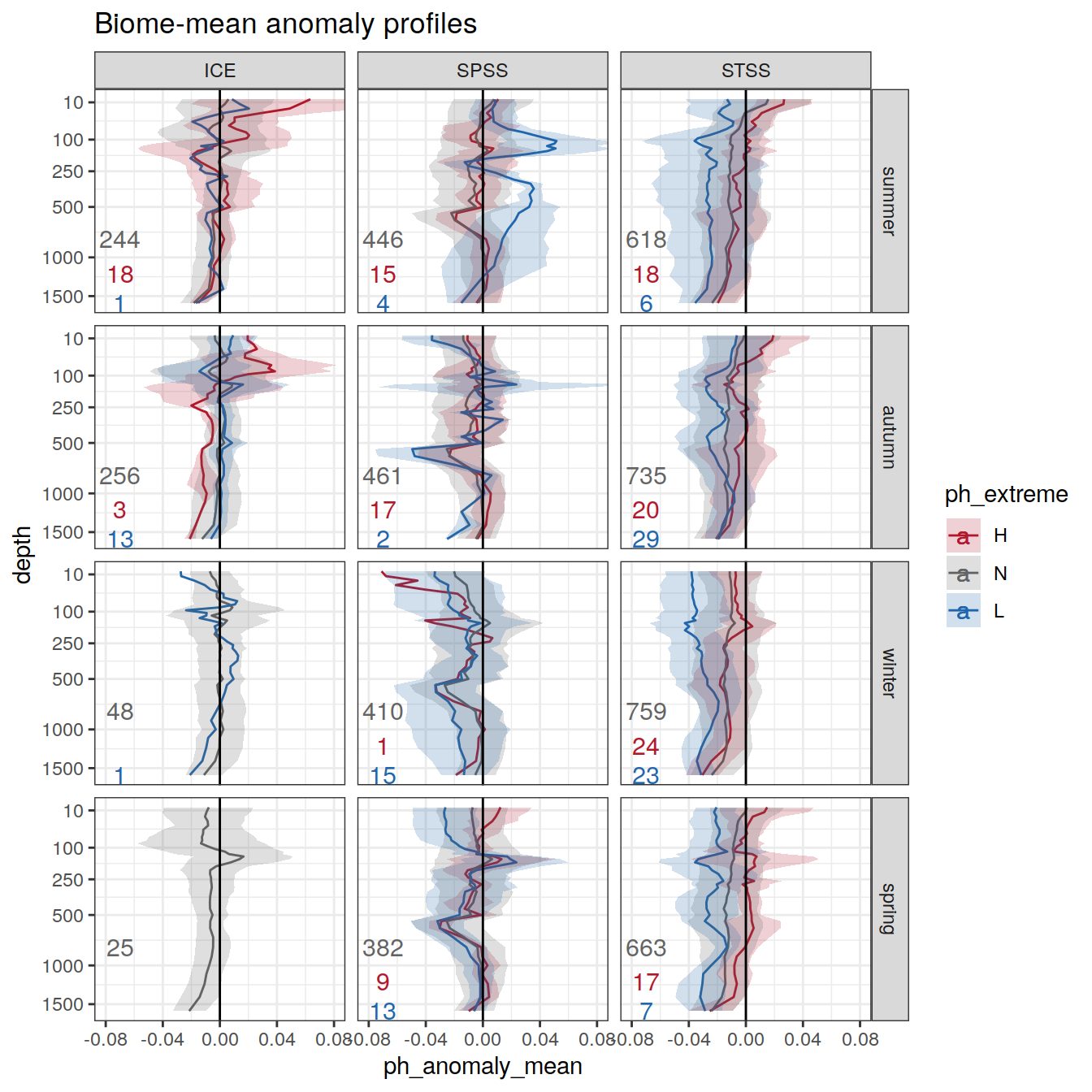
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 708f923 | pasqualina-vonlanthendinenna | 2022-05-04 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
rm(remove_clim_biome, profile_count_biome)Basin-mean anomaly profiles
remove_clim_basin <- remove_clim %>%
group_by(depth, ph_extreme, basin_AIP, season_order, season) %>%
summarise(ph_anomaly_mean = mean(anomaly_pH, na.rm = TRUE),
ph_anomaly_sd = sd(anomaly_pH, na.rm = TRUE))
remove_clim_basin %>%
ggplot()+
geom_path(aes(x = ph_anomaly_mean,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_ribbon(aes(xmax = ph_anomaly_mean + ph_anomaly_sd,
xmin = ph_anomaly_mean - ph_anomaly_sd,
y = depth,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
scale_y_continuous(trans = trans_reverser('sqrt'),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))+
geom_vline(xintercept = 0)+
facet_grid(season_order~basin_AIP, labeller = facet_label)+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
# geom_text_repel(data = profile_count_basin,
# aes(x = -0.1,
# y = 1500,
# label = paste0(n),
# col = ph_extreme),
# size = 4,
# segment.color = 'transparent')+
geom_text(data = profile_count_basin %>% filter (ph_extreme == 'N'),
aes(x = -0.07,
y = 800,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_basin %>% filter (ph_extreme == 'H'),
aes(x = -0.07,
y = 1200,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_basin %>% filter (ph_extreme == 'L'),
aes(x = -0.07,
y = 1600,
label = paste0(n),
col = ph_extreme),
size = 4)+
coord_cartesian(xlim = c(-0.08, 0.08))+
scale_x_continuous(breaks = c(-0.08, -0.04, 0, 0.04, 0.08))+
labs(title = 'Basin-mean anomaly profiles')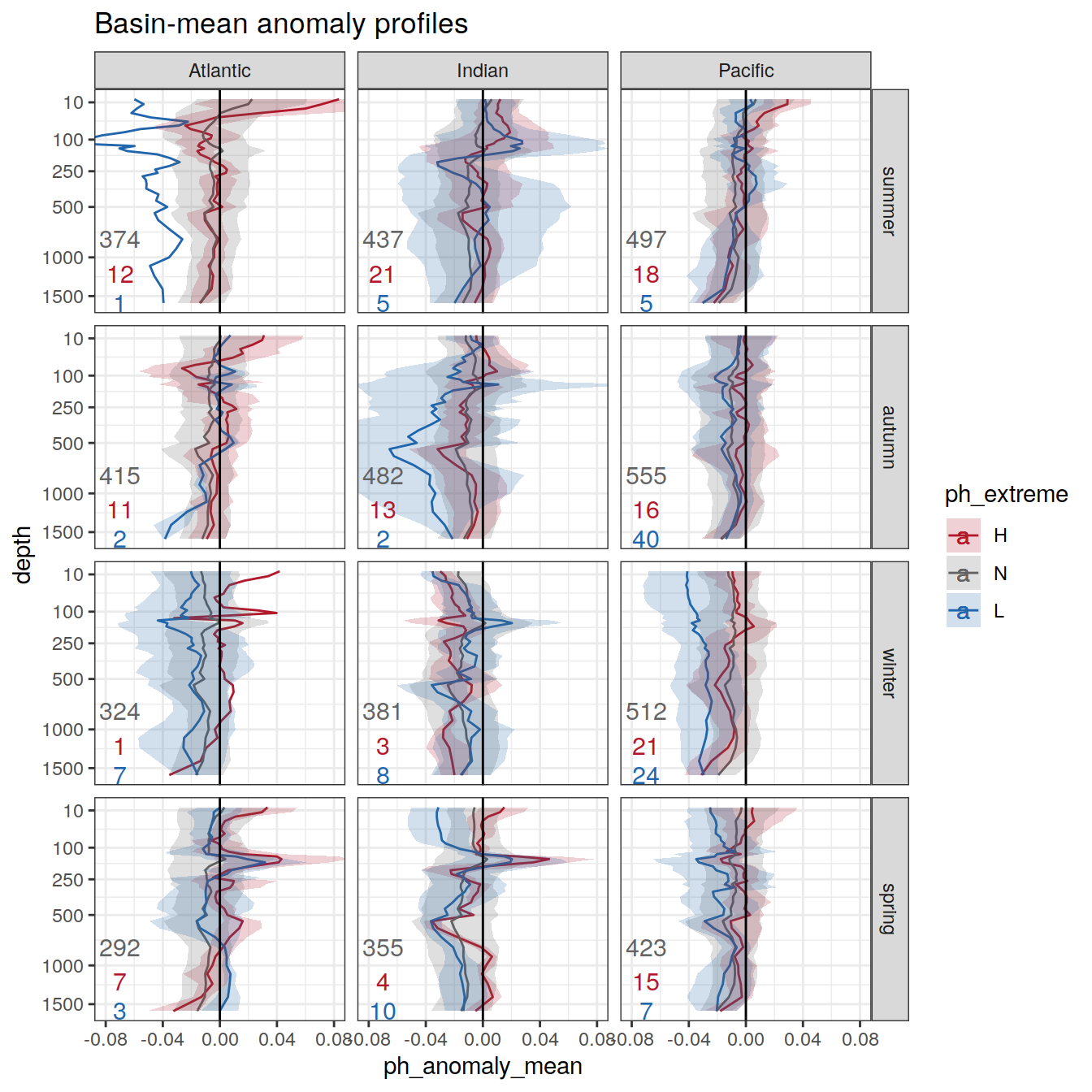
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| 708f923 | pasqualina-vonlanthendinenna | 2022-05-04 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
rm(remove_clim_basin, profile_count_basin)Basin-biome-mean anomaly profiles
remove_clim_basin_biome <- remove_clim %>%
group_by(depth, ph_extreme, season_order, season, basin_AIP, biome_name) %>%
summarise(ph_anomaly_mean = mean(anomaly_pH, na.rm=TRUE),
ph_anomaly_sd = sd(anomaly_pH, na.rm = TRUE))
remove_clim_basin_biome %>%
group_by(season_order) %>%
group_split(season_order) %>%
map(
~ggplot()+
geom_path(data = .x,
aes(x = ph_anomaly_mean,
y = depth,
group = ph_extreme,
col = ph_extreme))+
geom_ribbon(data = .x,
aes(xmax = ph_anomaly_mean+ph_anomaly_sd,
xmin = ph_anomaly_mean-ph_anomaly_sd,
y = depth,
group = ph_extreme,
fill = ph_extreme),
col = NA,
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_continuous(trans = trans_reverser('sqrt'),
breaks = c(10, 100, 250, 500, seq(1000, 5000, 500)))+
facet_grid(basin_AIP~biome_name)+
scale_color_manual(values = HNL_colors)+
scale_fill_manual(values = HNL_colors)+
geom_text(data = profile_count_season %>% filter (ph_extreme == 'N' & season == unique(.x$season)),
aes(x = -0.07,
y = 800,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_season %>% filter (ph_extreme == 'H' & season == unique(.x$season)),
aes(x = -0.07,
y = 1200,
label = paste0(n),
col = ph_extreme),
size = 4)+
geom_text(data = profile_count_season %>% filter (ph_extreme == 'L' & season == unique(.x$season)),
aes(x = -0.07,
y = 1600,
label = paste0(n),
col = ph_extreme),
size = 4)+
coord_cartesian(xlim = c(-0.08, 0.08))+
scale_x_continuous(breaks = c(-0.08, -0.04, 0, 0.04, 0.08))+
labs(title = paste0('basin-biome-mean profiles ', unique(.x$season)))
)[[1]]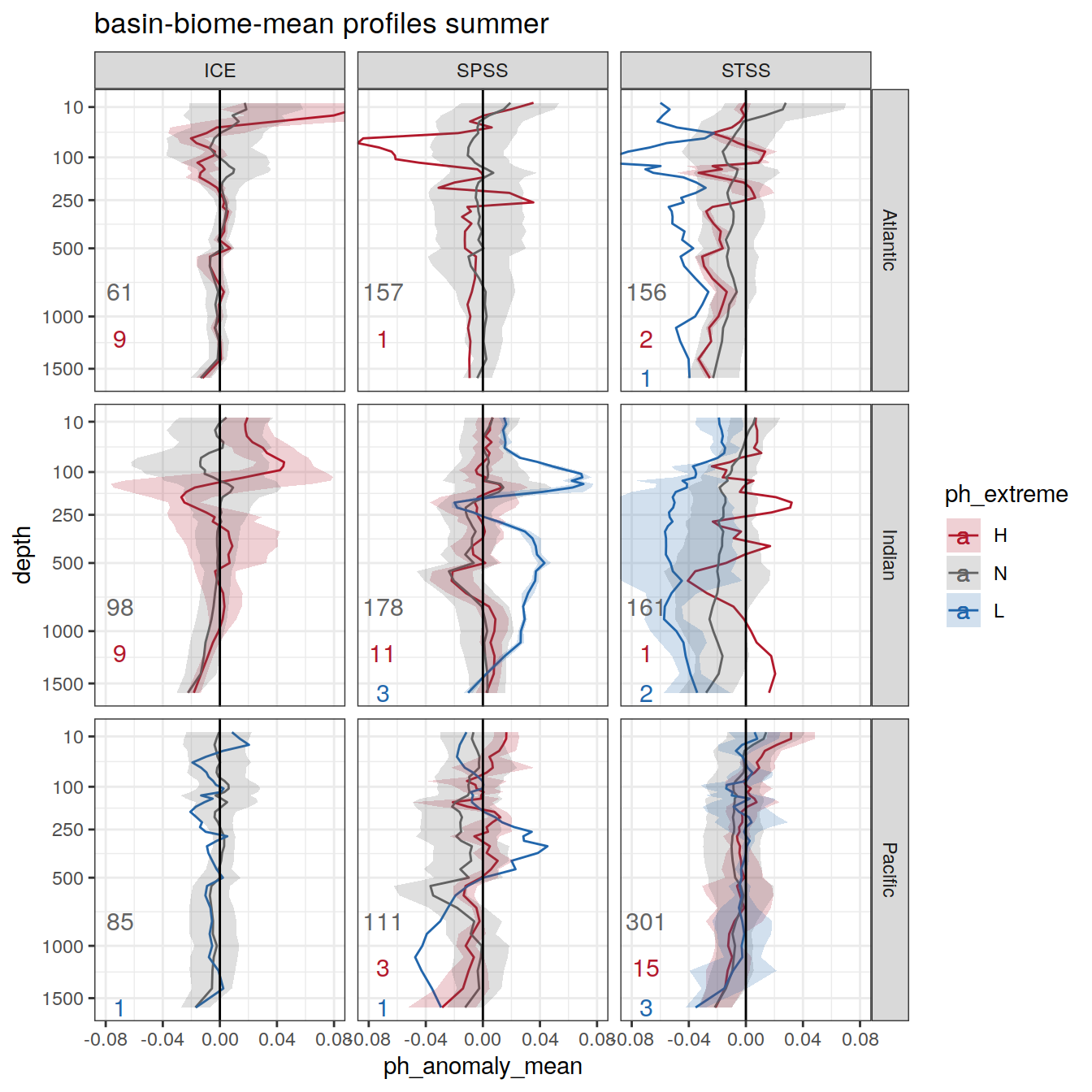
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 2efb8f2 | ds2n19 | 2023-10-17 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[2]]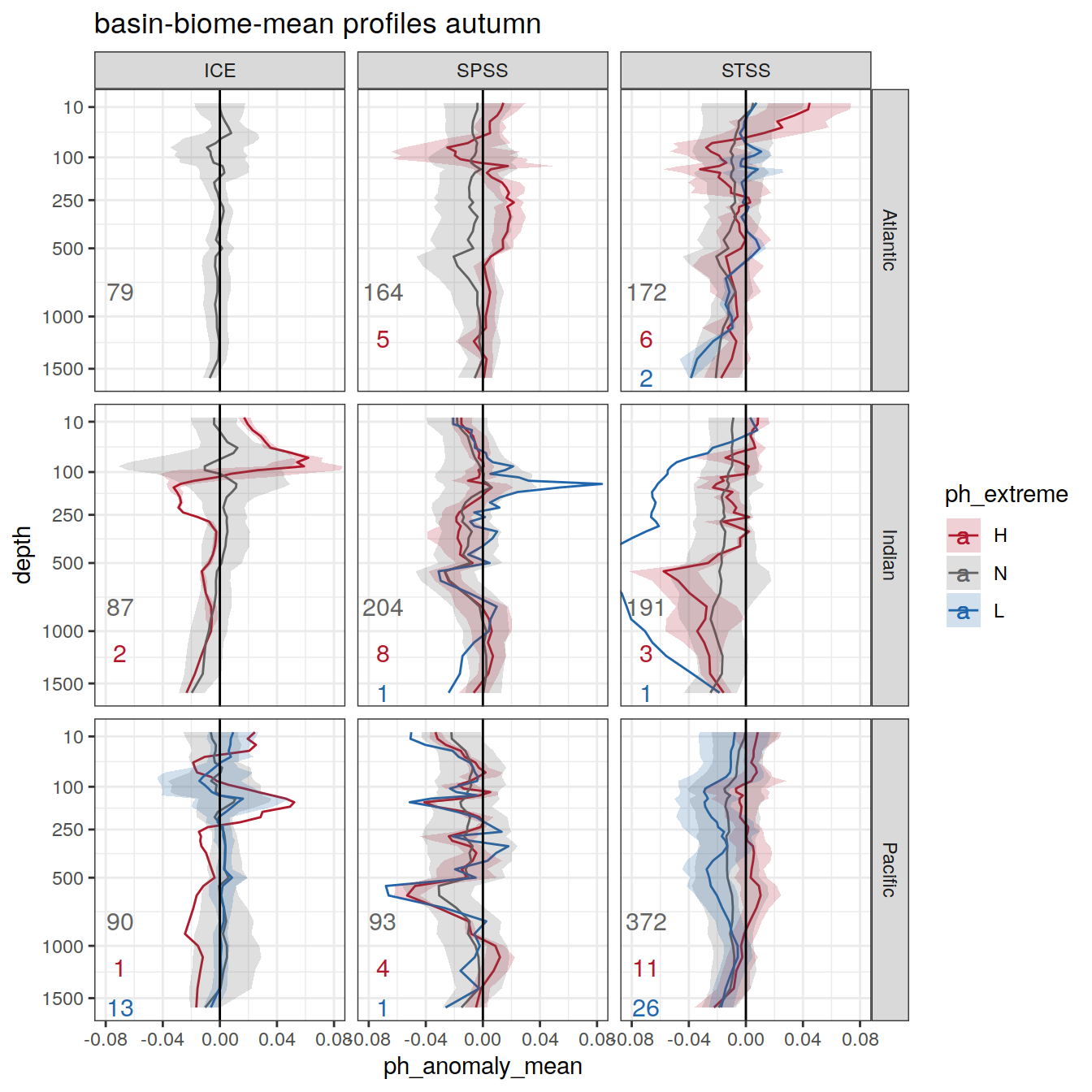
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 2efb8f2 | ds2n19 | 2023-10-17 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[3]]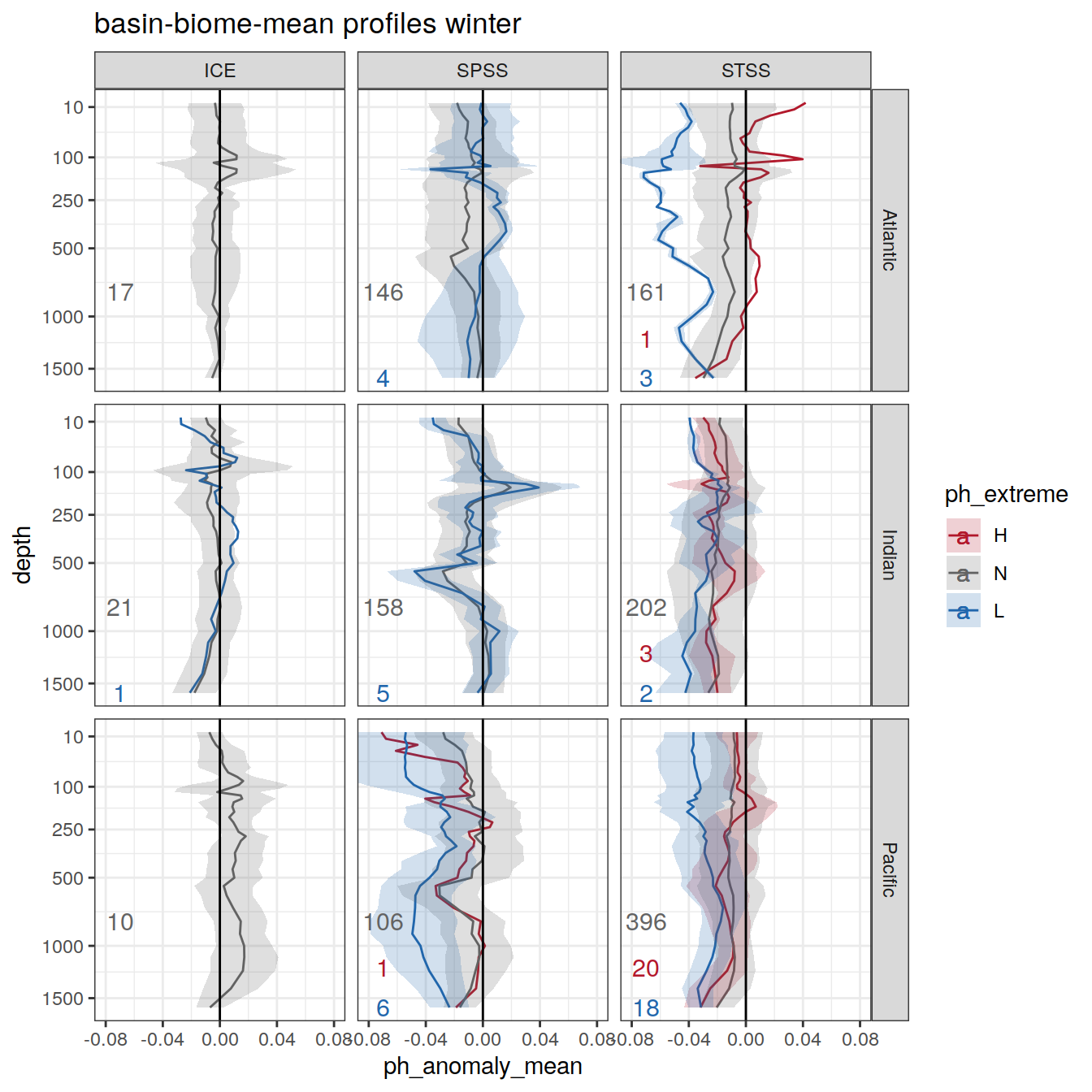
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 2efb8f2 | ds2n19 | 2023-10-17 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
[[4]]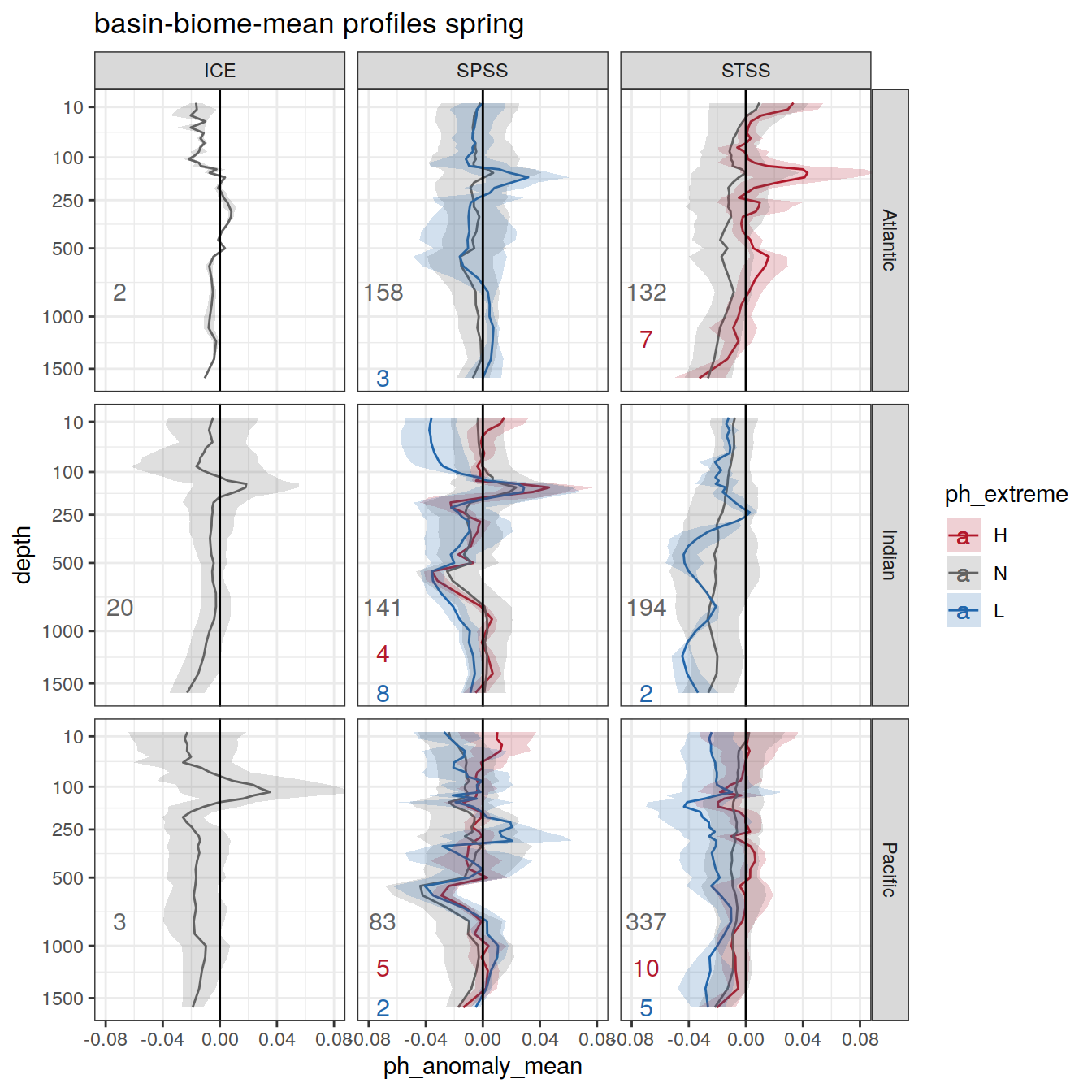
| Version | Author | Date |
|---|---|---|
| c076fba | mlarriere | 2024-04-12 |
| f9de50e | ds2n19 | 2024-01-01 |
| 23a24d1 | ds2n19 | 2023-12-05 |
| cec2a2a | ds2n19 | 2023-11-24 |
| ff31fef | ds2n19 | 2023-11-20 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| 1cd9ec1 | ds2n19 | 2023-10-19 |
| 879821d | ds2n19 | 2023-10-18 |
| 2efb8f2 | ds2n19 | 2023-10-17 |
| a97d9c1 | ds2n19 | 2023-10-17 |
| 1ae81b3 | ds2n19 | 2023-10-11 |
| 44f5720 | ds2n19 | 2023-10-09 |
| ae0f995 | jens-daniel-mueller | 2022-05-12 |
| 4173c20 | jens-daniel-mueller | 2022-05-12 |
| b917bd0 | jens-daniel-mueller | 2022-05-11 |
| b923426 | pasqualina-vonlanthendinenna | 2022-05-03 |
| 4e3571d | pasqualina-vonlanthendinenna | 2022-05-03 |
| a107add | pasqualina-vonlanthendinenna | 2022-05-03 |
rm(remove_clim_basin_biome)
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.5
Matrix products: default
BLAS: /usr/local/R-4.2.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.2.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggnewscale_0.4.8 ggrepel_0.9.2 ggforce_0.4.1 metR_0.13.0
[5] scico_1.3.1 ggOceanMaps_1.3.4 ggspatial_1.1.7 broom_1.0.5
[9] lubridate_1.9.0 timechange_0.1.1 forcats_0.5.2 stringr_1.5.0
[13] dplyr_1.1.3 purrr_1.0.2 readr_2.1.3 tidyr_1.3.0
[17] tibble_3.2.1 ggplot2_3.4.4 tidyverse_1.3.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 sf_1.0-9 bit64_4.0.5
[4] RColorBrewer_1.1-3 httr_1.4.4 rprojroot_2.0.3
[7] tools_4.2.2 backports_1.4.1 bslib_0.4.1
[10] utf8_1.2.2 R6_2.5.1 KernSmooth_2.23-20
[13] rgeos_0.5-9 DBI_1.2.2 colorspace_2.0-3
[16] raster_3.6-11 withr_2.5.0 sp_1.5-1
[19] tidyselect_1.2.0 bit_4.0.5 compiler_4.2.2
[22] git2r_0.30.1 cli_3.6.1 rvest_1.0.3
[25] xml2_1.3.3 labeling_0.4.2 sass_0.4.4
[28] checkmate_2.1.0 scales_1.2.1 classInt_0.4-8
[31] proxy_0.4-27 digest_0.6.30 rmarkdown_2.18
[34] pkgconfig_2.0.3 htmltools_0.5.8.1 highr_0.9
[37] dbplyr_2.2.1 fastmap_1.1.0 rlang_1.1.1
[40] readxl_1.4.1 rstudioapi_0.15.0 farver_2.1.1
[43] jquerylib_0.1.4 generics_0.1.3 jsonlite_1.8.3
[46] vroom_1.6.0 googlesheets4_1.0.1 magrittr_2.0.3
[49] Rcpp_1.0.10 munsell_0.5.0 fansi_1.0.3
[52] lifecycle_1.0.3 terra_1.7-65 stringi_1.7.8
[55] whisker_0.4 yaml_2.3.6 MASS_7.3-58.1
[58] grid_4.2.2 parallel_4.2.2 promises_1.2.0.1
[61] crayon_1.5.2 lattice_0.20-45 haven_2.5.1
[64] hms_1.1.2 knitr_1.41 pillar_1.9.0
[67] codetools_0.2-18 reprex_2.0.2 glue_1.6.2
[70] evaluate_0.18 data.table_1.14.6 modelr_0.1.10
[73] tweenr_2.0.2 vctrs_0.6.4 tzdb_0.3.0
[76] httpuv_1.6.6 cellranger_1.1.0 polyclip_1.10-4
[79] gtable_0.3.1 assertthat_0.2.1 cachem_1.0.6
[82] xfun_0.35 e1071_1.7-12 later_1.3.0
[85] viridisLite_0.4.1 class_7.3-20 googledrive_2.0.0
[88] gargle_1.2.1 memoise_2.0.1 workflowr_1.7.0
[91] units_0.8-0 ellipsis_0.3.2